
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Oncol. , 07 June 2022
Sec. Gastrointestinal Cancers: Colorectal Cancer
Volume 12 - 2022 | https://doi.org/10.3389/fonc.2022.893591
This article is part of the Research Topic Precision Medicine in Gastrointestinal Cancers View all 24 articles
Numerous studies have shown that long noncoding RNAs (lncRNAs) play a critical role in the malignant progression of cancer. However, the potential involvement of lncRNAs in colon adenocarcinoma (COAD) remains unexplored. In this study, the expression of lncRNA SNHG7 in colon cancer tissues and its correlation with clinical characteristics were analyzed based on data from The Cancer Genome Atlas (TCGA) database. SNHG7 was found to be highly expressed in 17 types of cancer, including COAD. Next, TCGA data were further investigated to identify differentially expressed genes, and Gene Ontology and Kyoto Encyclopedia of Genes and Genomes analyses were performed. In addition, the relationship between SNHG7 expression and clinical features were analyzed. SNHG7 expression was found to be a potentially valuable indicator for COAD diagnosis and prognosis. Finally, gene set enrichment analysis showed that SNHG7 may affect lupus erythematosus and reactome cellular senescence, possibly influencing the prognosis of patients with COAD. Altogether, these results suggest that SNHG7 may be associated with the occurrence and development of COAD, having potential diagnostic and prognostic value.
Colon adenocarcinoma (COAD) is the second most lethal malignancy worldwide, which is currently treated surgically and/or using chemotherapy and radiotherapy (1). Although the overall survival (OS) rate has improved, invasion and metastasis remain the main death cause among patients with COAD (2). Extensive studies have shown that tumor biomarkers are highly sensitive and specific for diagnosing and monitoring tumors (3). Therefore, there is a critical need to identify new diagnostic or prognostic biomarkers and develop novel therapeutic strategies for COAD.
Long noncoding RNAs (lncRNAs) are a class of noncoding RNA transcripts that are over 200 nucleotides in length. The dysregulation of lncRNAs is closely related to various major diseases, including cancer (4). Many studies have shown that cancer- associated lncRNAs are involved in the regulation of tumor proliferation, invasion, and metastasis; thus, are considered to be a class of potential candidate biomarkers for cancer diagnosis and therapy (5). For example, the lncRNA HOTAIR is an oncogene that is upregulated in breast cancer tissues and is closely related to poor prognosis and tumor metastasis (6). MALAT1 (metastasis-associated lung adenocarcinoma transcript 1) is a lncRNA that was originally found to be abundantly expressed in metastatic carcinoma cells and to be significantly upregulated in various types of cancer, such as breast cancer (7) and non-small cell lung cancer (8), being suggested as a prognostic biomarker and potential therapeutic target for metastatic cancers (9). H19 is an estrogen-regulated lncRNA transcript whose aberrant expression is closely associated with cell proliferation and migration in a variety of cancers, such as gastric, gallbladder, and pancreatic cancers (10). Although lncRNAs have been broadly recognized to play important regulatory roles in human cancers, few have been demonstrated to function in COAD, and most of their mechanisms are largely unknown.
Small nucleolar RNA host genes (SNHGs) are newly recognized lncRNAs that have oncogenic roles in various cancers (11). Members of the SNHG family have been shown to regulate cellular proliferation, apoptosis, invasion, and migration in multiple cancers (12). LncRNA SNHG7 is closely related to the occurrence, development, and carcinogenesis potential of numerous cancers, including lung, gastric and cervical cancer, as well as renal cell carcinoma and hepatocellular carcinoma (13–15). Nevertheless, few reports have explored the impact of SNHG7 on COAD. This study aimed to investigate the relationship between the expression of SNHG7 and the prognosis of COAD using bioinformatics tools.
RNA sequencing data from 521 COAD samples and associated clinical information were obtained from The Cancer Genome Atlas (TCGA) database (https://portal.gdc.cancer.gov/). Another RNA sequencing data of 698 COAD samples and clinical information were also included for validation. RNA sequencing data were converted from fragments per kilobyte per million (FPKM) to the transcripts per million reads (TPM) format, and compared according to the corresponding clinicopathological information. As all data collected was publicly available, informed consent and ethical approval were not necessary to obtain.
To clarify the association between SNHG7 expression and clinical features of COAD, Wilcoxon signed-rank test and logistic regression were performed. The detailed clinicopathological characteristics of the patients with COAD are listed in Table 1.
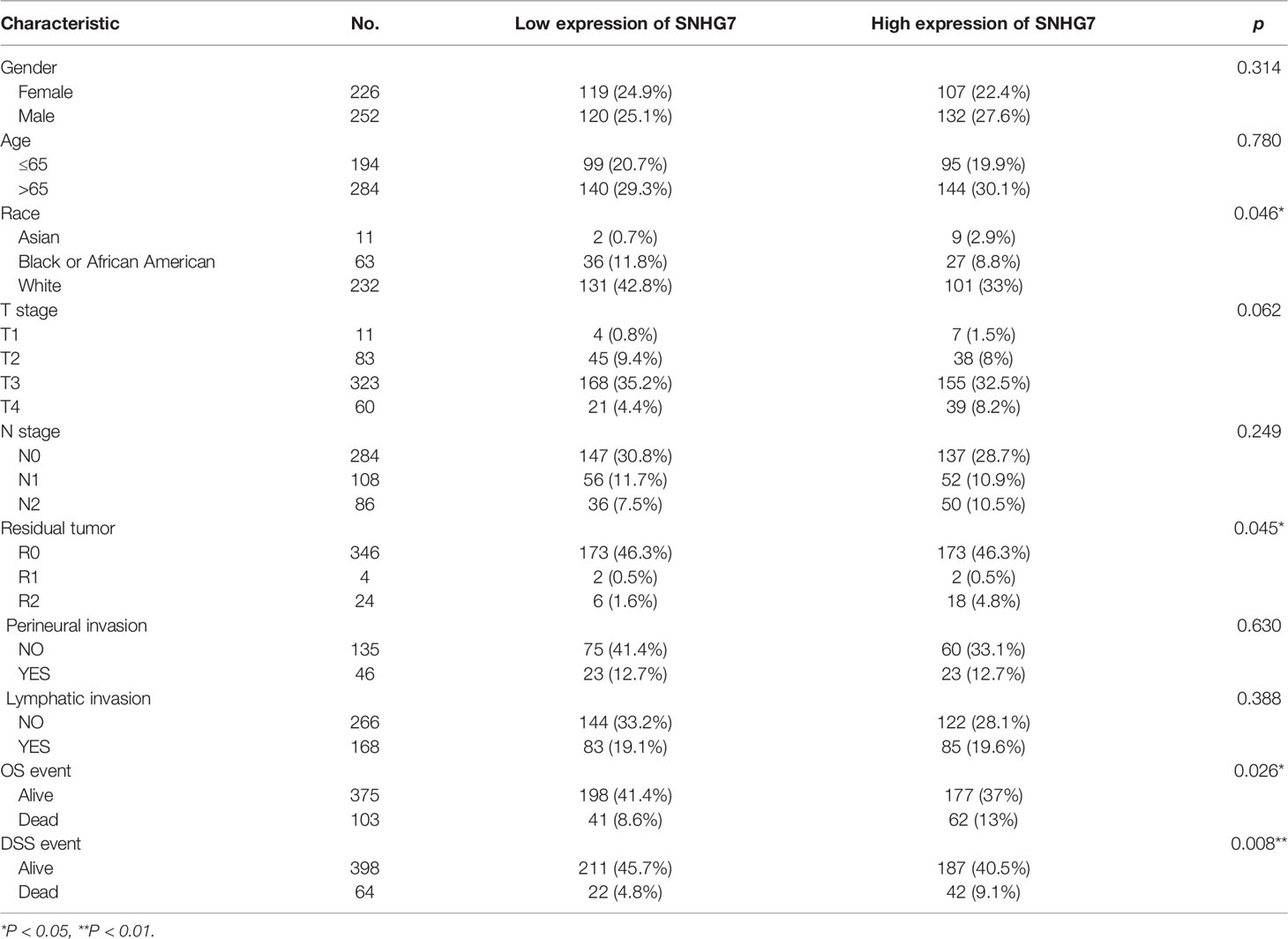
Table 1 The relationships between SNHG7 expression and clinicopathological features in COAD patients.
To assess the predictive potential of SNHG7 for COAD diagnosis, SNHG7 expression in COAD and normal tissues was compared using receiver operating characteristic (ROC) analysis. COAD and corresponding normal tissue data were obtained from the TCGA database. The analysis was performed using the R package “pROC” (version1.17.0.1), and the visualization was achieved using “ggplot2” (version 3.3.3).
Kaplan–Meier analysis, and univariate and multivariate Cox regression analyses were used for prognosis analysis. Nomograms were created using the R packages “rms” (version 6.2-0) and “survival” (version 3.2-10). R (v3.6.3; R Foundation for Statistical Computing, Vienna, Austria) was used to conduct all statistical studies, with p-values below 0.05 deemed significant.
COAD gene expression data in the HTSeq-TPM format were obtained from TCGA for analysis. SNHG7 coexpressed genes were screened using Pearson correlation coefficients (|r| > 0.4 and p < 0.001). To explore the possible biological functions and signaling pathways affected by SNHG7, the R package “cluster Profiler” was used to perform GO and KEGG analyses of coexpressed genes, with p < 0.05 deemed statistically significant. GO analysis included biological processes (BP), cell composition (CC), and molecular function (MF).
GSEA is a computational method used to determine whether an a priori defined gene set exhibits statistically significant and consistent differences between two biological states (16). In the present study, we elucidated the survival differences between groups with high and low SNHG7 expression using GSEA. Gene set permutations were performed 1,000 times for each analysis. The expression of SNHG7 was used as the phenotypic label. The nominal p-value and normalized enrichment score (NES) were used to identify the pathways enriched for each phenotype.
Based on TCGA data analysis, we found that SNHG7 was upregulated in 17 of the 33 cancer types investigated, including cholangiocarcinoma (CHOL), prostate adenocarcinoma (PRAD), and thyroid carcinoma (THCA) (Figure 1A). Further analysis showed that SNHG7 expression was much higher in patients with COAD than that in normal tissues (p < 0.001, Figure 1B). These findings indicate that SNHG7 may play a significant regulatory role in the progression of COAD.
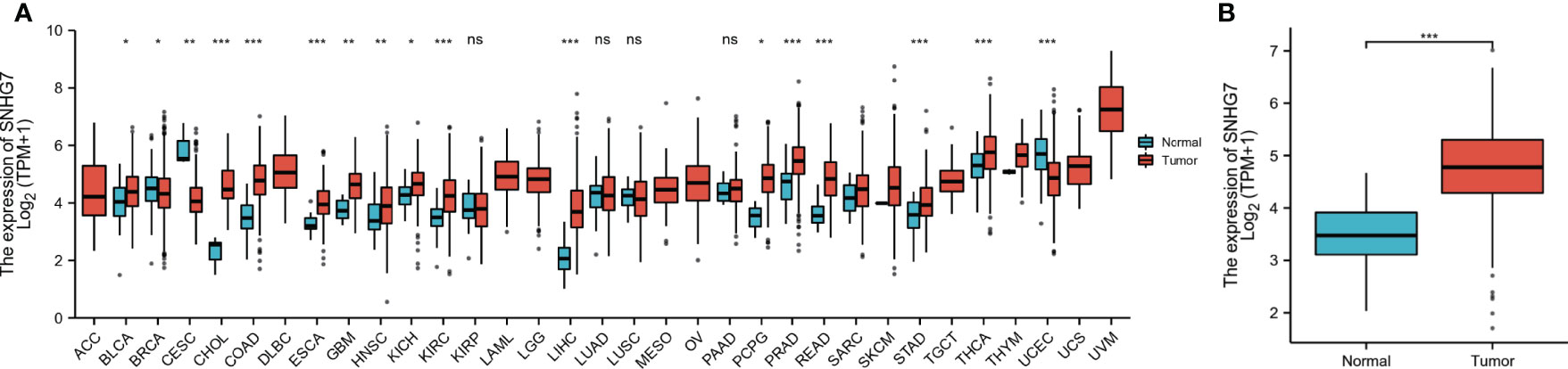
Figure 1 Expression level of SNHG7 in different tumors and in COAD. (A) The comparison of SNHG7 expression between normal and tumor tissue in different types of cancers based on TCGA database. (B) SNHG7 expression is significantly higher in COAD (n = 480) than normal tissue (n = 41). ns, P ≥ 0.05, *P < 0.05, **P < 0.01, ***P < 0.001.
Clinical information, including sex, age, race, T stage, N stage, residual tumor, perineural invasion, lymphatic invasion, OS, and disease-specific survival (DSS) (Table 1), for 521 COAD patients was obtained from TCGA database. SNHG7 expression was not only significantly correlated with race (p < 0.05) and residual tumor (p < 0.05), but was also closely correlated with OS (p < 0.05) and DSS (p < 0.01). No correlation was observed between SNHG7 expression and the other clinicopathological characteristics.
ROC curves were used to evaluate the potential of SNHG7 expression to identify patients with COAD. SNHG7 expression had high sensitivity and specificity for COAD diagnosis, with an area under the curve (AUC) of 0.912 (95% confidence interval [CI], 0.878–0.947) (Figure 2A). Further analysis showed that SNHG7 expression could diagnose T and N stages, with AUC values of 0.913 (95% CI: 0.878–0.947) and 0.921 (95% CI: 0.887–0.955), respectively (Figures 2B, C). Taken together, these results suggested that SNHG7 expression could represent a valuable tool to diagnosis COAD.
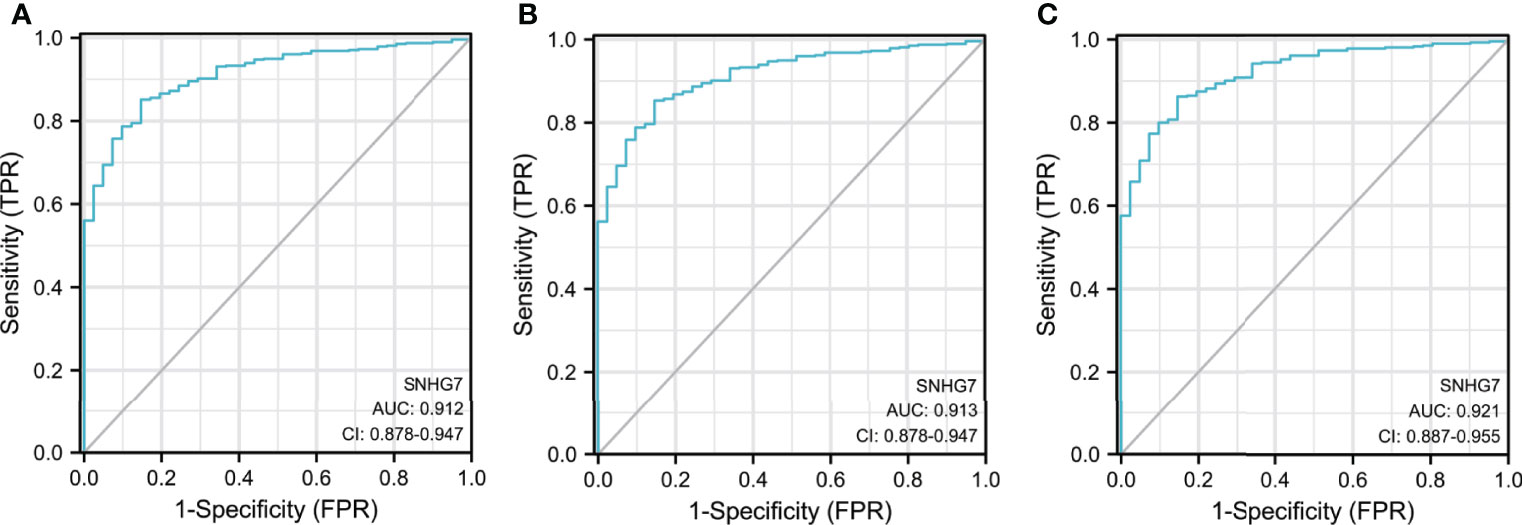
Figure 2 The diagnostic value of SNHG7 in COAD patients. (A) ROC curve of SNHG7 in diagnosing of COAD patients. (B) ROC curve of SNHG7 in T staging of COAD patients. (C) ROC curve of SNHG7 in M staging of COAD patients and 0.921 (95% CI: 0.887-0.955), respectively (Figure 2B&C).
Kaplan–Meier analysis was used to confirm whether SNHG7 expression could be used for the prediction of clinical outcomes among patient with COAD. Overall, high SNHG7 expression was associated to shorter OS (hazard ratio [HR]: 1.85, p = 0.002) and DSS (HR: 2.35, p = 0.001) in COAD patients (Figures 3A, B). To determine whether SNHG7 expression had a predictive value for clinical outcomes, we also performed univariate and multivariate Cox regression analyses. As shown in Table 2, SNHG7 expression (HR: 1.847, 95% CI: 1.244–2.741, p = 0.002) was an independent risk factor for OS. Age, T stage, pathological stage, lymphatic invasion, and carcinoembryonic antigen (CEA) level also showed prognostic advantages for clinical outcomes. In addition, the N stage showed prognostic advantages in both univariate (HR: 4.051, 95% CI: 2.593–6.329, p < 0.001) and multivariate (HR: 6.048, 95% CI: 1.006–36.361, p = 0.049) Cox regression analyses. Furthermore, SNHG7 expression, T stage, N stage, pathological stage, perineural invasion, lymphatic invasion, and CEA level were all independent risk factors for DSS (Table 3). N stage (HR: 2.933, 95% CI: 0.218–39.407, p = 0.021) was an independent risk factor for DSS.
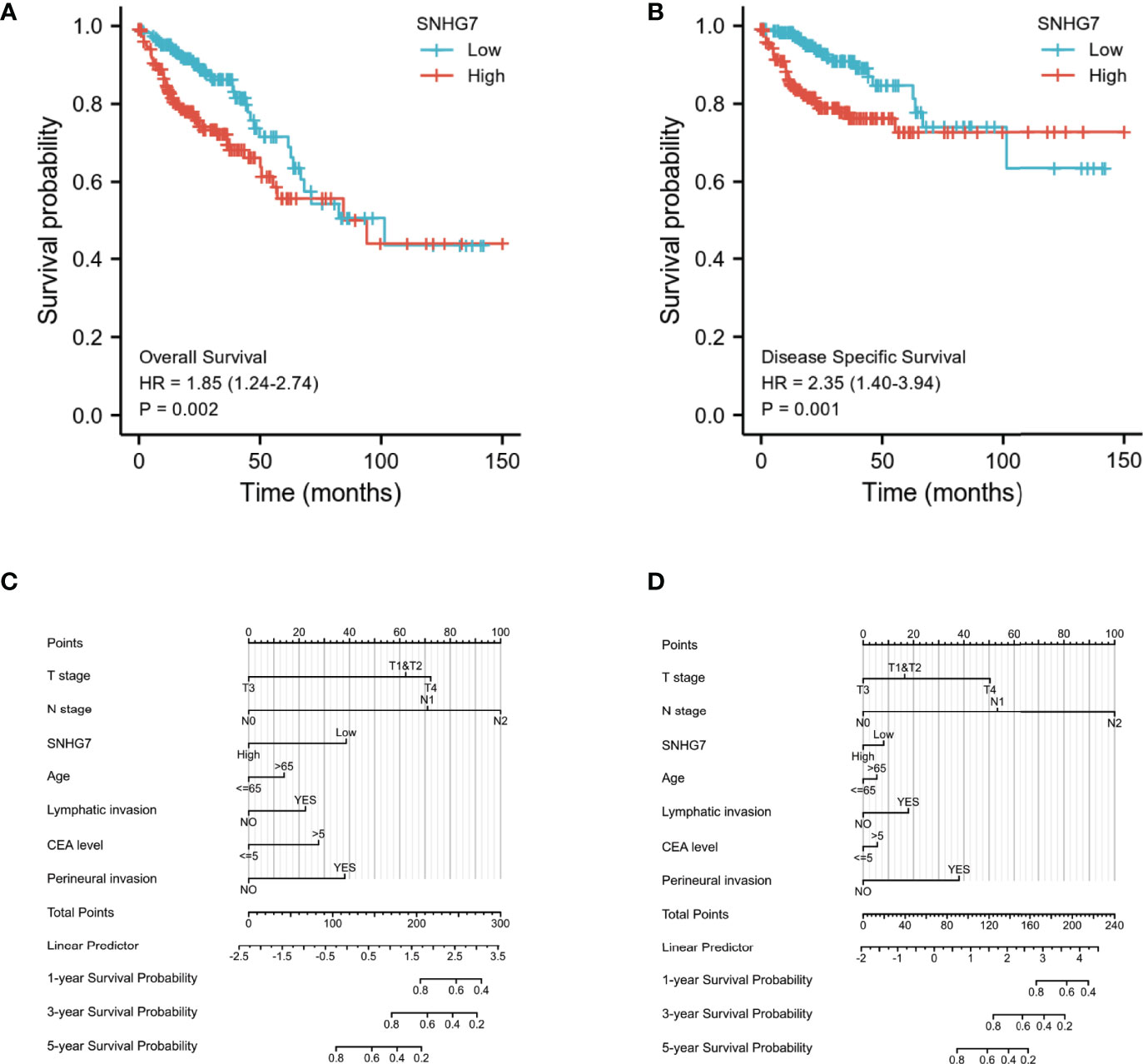
Figure 3 Relationship between SNHG7 expression and survival rate in COAD analyzed by Kaplan–Meier (A, B). Construction and validation of nomograms based on SNHG7 expression (C, D). (A) The relationship between overall survival and SNHG7 expression. (B) The relationship between disease-specific survival and SNHG7 expression. (C) Nomograms constructed to establish SNHG7 expression-based risk scoring models for 1-, 3-, and 5-year overall survival. (D) Nomograms constructed to establish SNHG7 expression-based risk scoring models for 1-, 3-, and 5-year disease-specific survival.
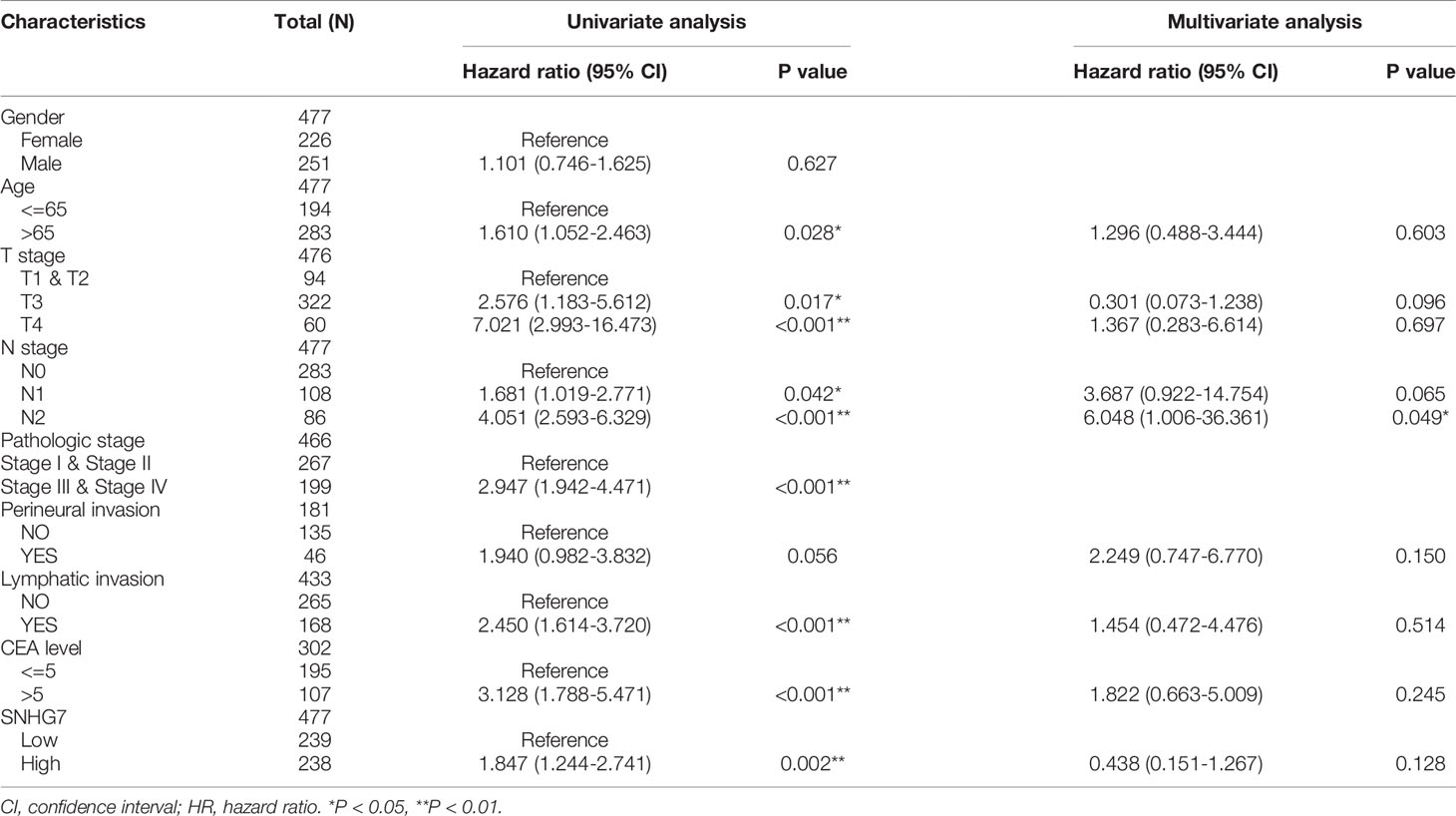
Table 2 Univariate and multivariate Cox proportional hazards analysis of SNHG7 expression and OS (overall survival) for patients with COAD in the validation cohort.
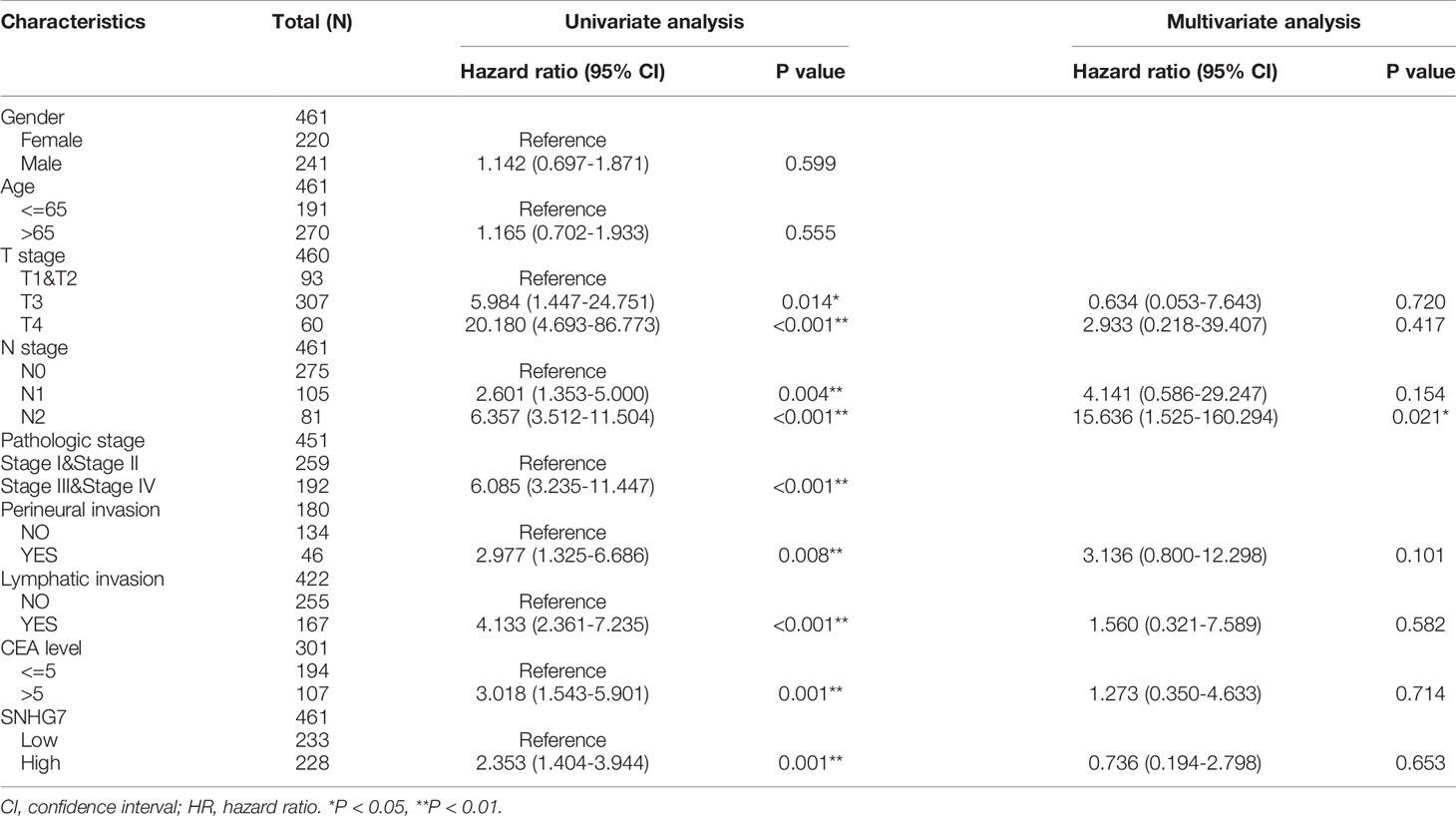
Table 3 Univariate and multivariate Cox proportional hazards analysis of SNHG7 expression and DSS (Disease Specific Survival) for patients with COAD in the validation cohort.
Based on the significant prognostic factors identified in the Cox regression analysis, prognostic nomograms were designed. Age, pathological stage, perineural or lymphatic invasion, and SNHG7 expression were included in the nomogram to predict OS (C-index = 0.836) (Figure 3C) and DSS (C-index = 0.875) (Figure 3D). These results indicated that SNHG7 expression was not only significantly upregulated in COAD but also had prognostic value, suggesting that SNHG7 has important regulatory functions in this type of cancer.
To screen for coexpressed genes of SNHG7, Pearson correlation coefficients were set as |r| > 0.4 and p < 0.001. The top 20 positively and negatively correlated coexpressed genes of SNHG7 are displayed in the form of a heatmap (Figure 4).
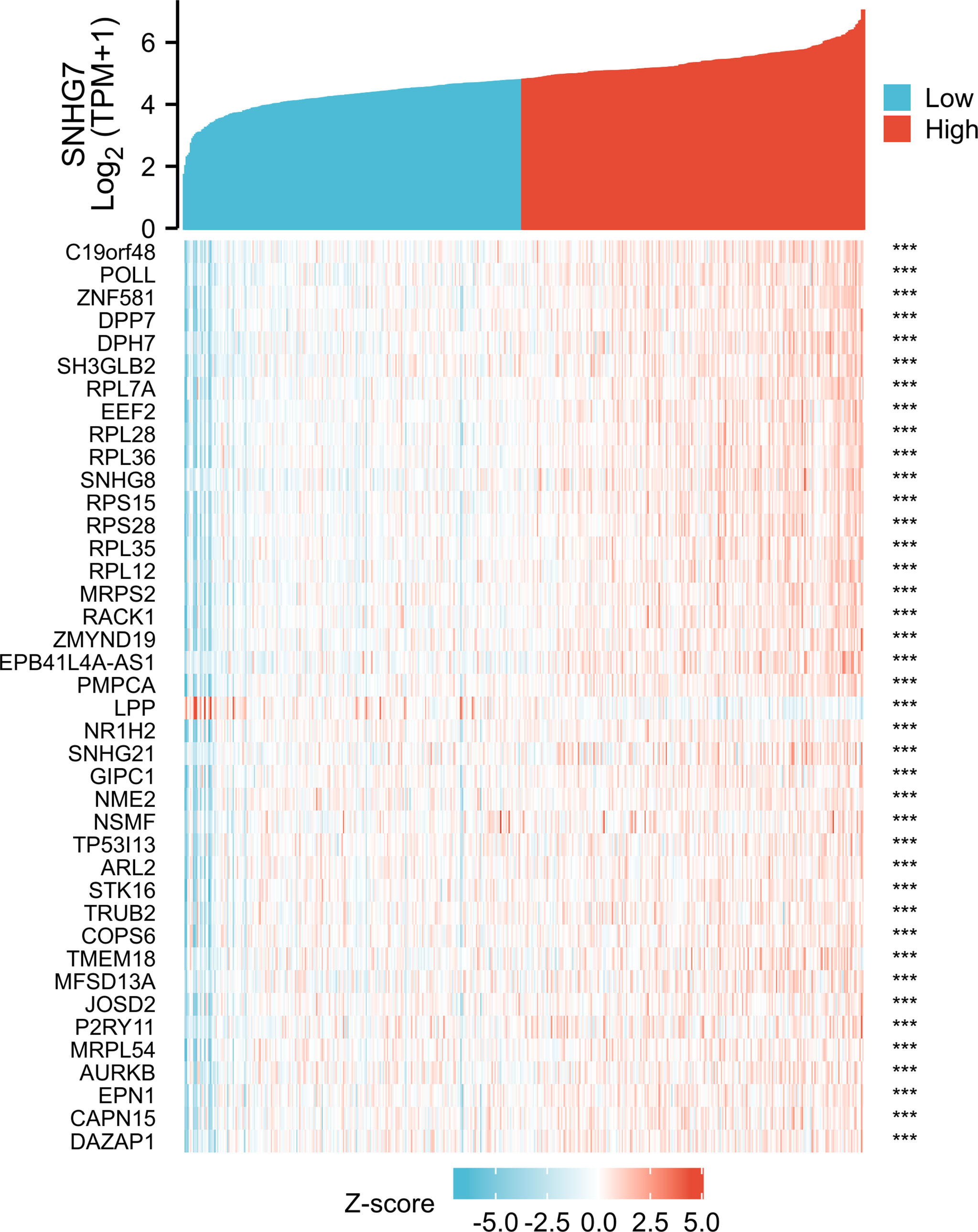
Figure 4 The top 20 genes with positive and negative co-expression of SNHG7 in TCGA database according to Heatmap in COAD. ***P<0.001.
Next, we performed GO and KEGG analysis of SNHG7- associated DEGs in COAD. GO analysis demonstrated that genes of GO-BP terms were significantly enriched in viral gene expression, viral transcription, establishment of protein localization to the endoplasmic reticulum (ER), and protein targeting to the ER. For GO-CC terms, the genes were mainly located in the cytosolic part, ribosome, ribosomal subunit, and the cytosolic ribosome. In GO-MF analysis, genes were enriched in the structural constituent of ribosomes, 5′–3′ RNA polymerase activity, DNA-directed 5′– 3′ RNA polymerase activity, and RNA polymerase II activity (Table 4; Figure 5A). As shown in Figure 5B; Table 5, KEGG pathway analysis indicated that the top pathways were mainly associated with Huntington’s disease, ribosome, amyotrophic lateral sclerosis, spliceosome, and RNA polymerases.
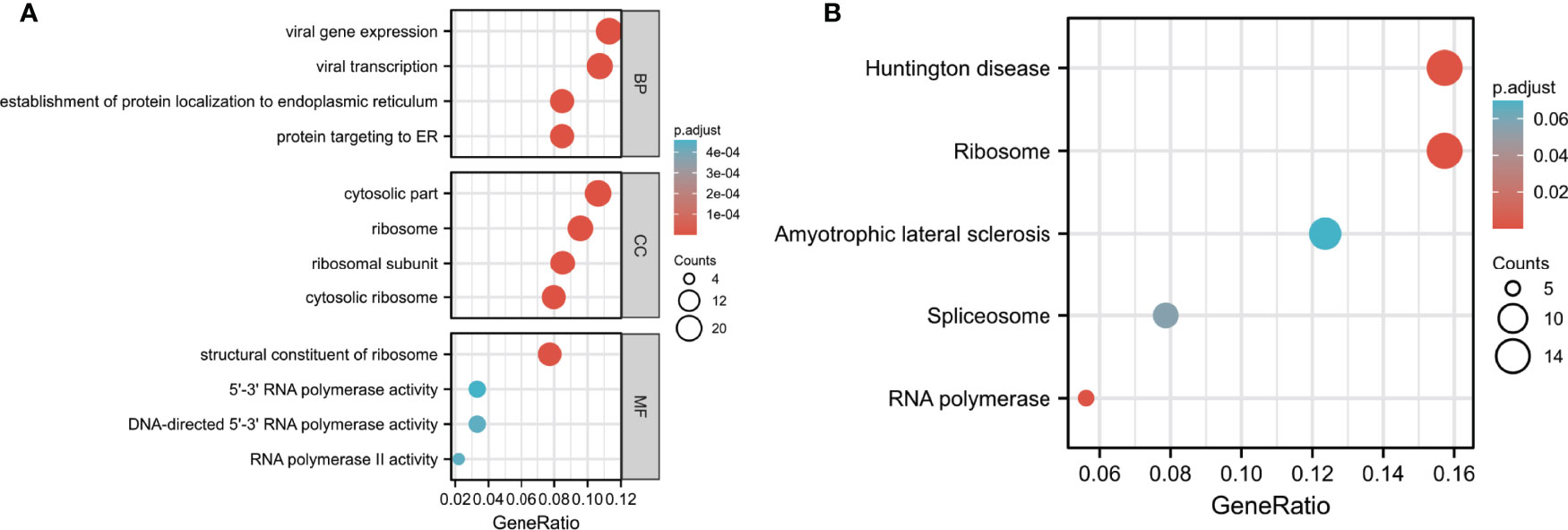
Figure 5 GO and KEGG pathway analyses of DEGs in COAD. (A) Biological process (BF), cell components (CC), and molecular function (MF) enrichment analyses of DEGs. (B) KEGG pathway analysis of DEGs.
To further clarify biological functions of SNHG7 in COAD, GSEA enrichment analysis was performed on the high and low expression datasets of SNHG7. Significant differences (false discovery rate < 0.25, adjusted p < 0.05) were observed in the enrichment of the MSigDB Collection (c2.cp.v7.2.symbols.gmt). The most markedly enriched signaling pathways were screened based on their NES (Figures 6A, B). The results illustrated that lupus erythematosus (NES = −1.628, adjusted p = 0.048) and reactome cellular senescence (NES = −1.413, adjusted p = 0.048) were mainly enriched in the highly expressed SNHG7 phenotype.
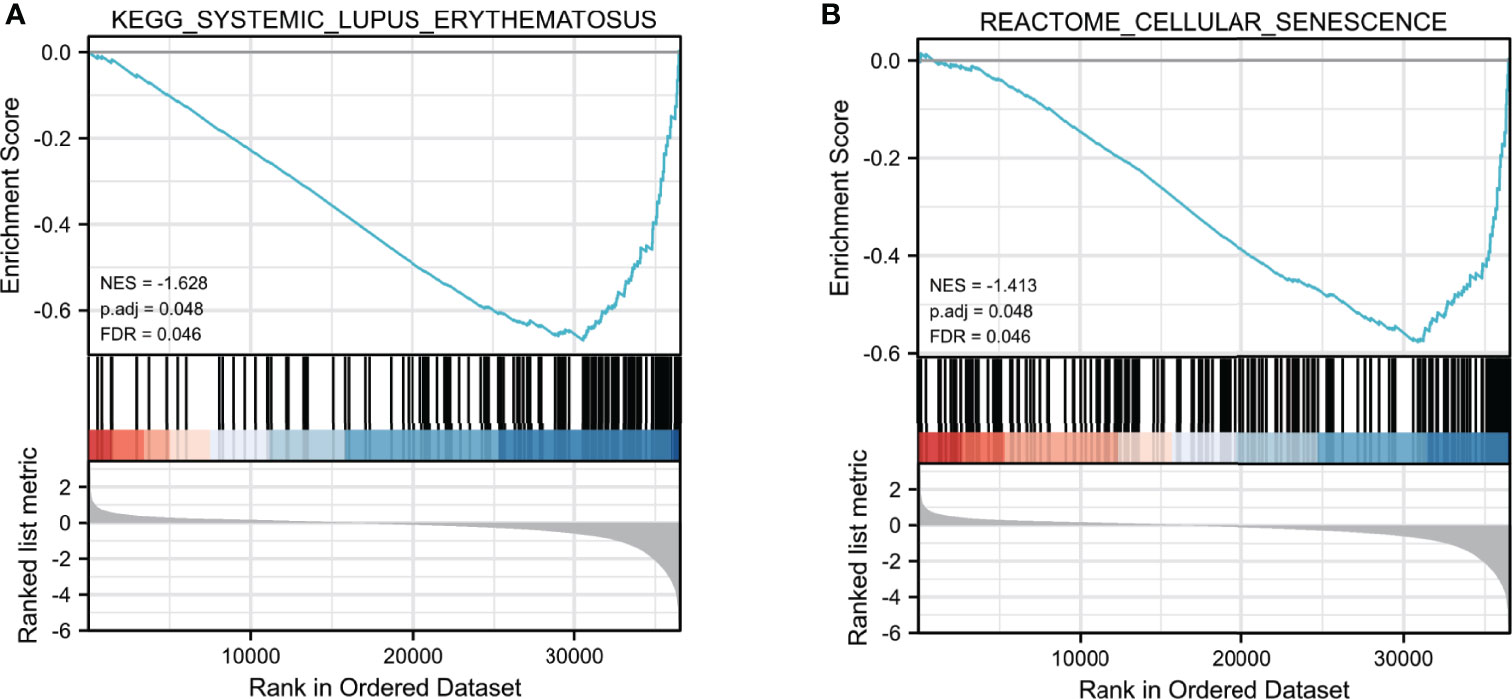
Figure 6 Enrichment plots from GSEA analysis. GSEA analysis showed that Lupus erythematosus (A) and Reactome cellular senescence (B) were differentially enriched between high- and low-SNHG7 expression groups.
To validate the prognostic robustness and clinical reproducibility of SNHG7, an independent cohort available at TCGA database, comprising 698 samples, was also analyzed. As shown in Figure 7A, SNHG7 expression was significantly upregulated in the tumor group as compared with normal tissues (p < 0.001). Similarly, ROC curve analysis also indicated that SNHG7 had a very high diagnostic value in COAD (AUC = 0.911, 95% CI: 0.879−0.943) (Figure 7B). Analysis of the OS (Figure 7C; HR: 1.55, p = 0.014) and DSS (Figure 7D; HR: 2.04, p = 0.002) of these patients further suggested that high SNHG7 expression was correlated with poor prognosis in COAD. These results were consistent with the conclusions of SNHG7 in cohort of 521 samples, indicating that the diagnostic and prognostic value of SNHG7 in COAD is credible and reproducible.
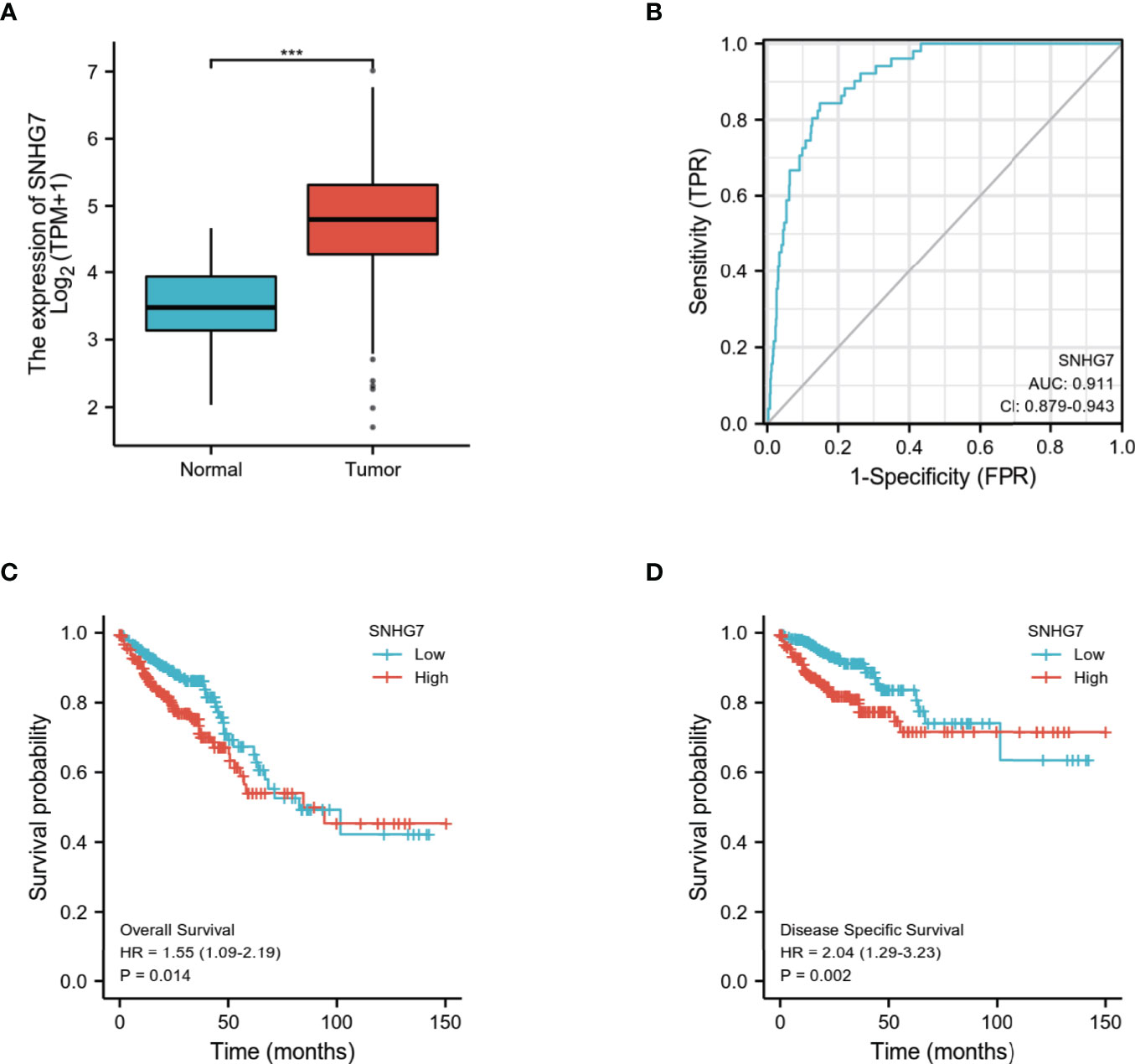
Figure 7 Validation of differential expression, prognostic and diagnostic value of SNHG7 in COAD (n=698) (A) SNHG7 expression is significantly higher in COAD (n = 647) than normal tissue (n = 51). ***P < 0.001. (B) ROC curve of SNHG7 in diagnosing of COAD patients. (C) The relationship between overall survival and SNHG7 expression. (D) The relationship between disease-specific survival and SNHG7 expression.
Currently, the 5-year survival rate of early COAD exceeds 70−90%; nonetheless, the curative effect for advanced COAD is still not ideal, which is mainly due to its high recurrence and metastasis rate (17, 18). Therefore, the development of biomarkers aiding early differential diagnosis and predicting COAD progression is of major importance both for research and therapeutic evolution (19). It has been established that lncRNAs may be potential diagnostic and/or prognostic markers for clinical applications. In particular, many lncRNA biomarkers were reported for colorectal cancer (20, 21).
SNHG7, which is a member of the SNHG family, is differentially expressed in various malignant tumors (13–15). Noteworthy, recent studies revealed that SNHG7 has a regulatory role in colorectal cancer. For example, SNHG7 is an oncogenic biomarker in COAD, and it interacts with miR-193b (22) and positively regulates GALNT1 levels through sponging miR-216b in colorectal cancer (23). Moreover, SNHG7 and FAIM2 are upregulated in colorectal cancer tissues compared with normal adjacent tissues (24). However, the possible clinical significance and prognostic/diagnostic value of SNHG7
in COAD remain unclear. Therefore, the development of new and effective biomarkers for the prognosis and early diagnosis of COAD would be beneficial to enhance the treatment and prognosis of patients.
To gain a comprehensive understanding of the role of SNHG7 in COAD, we first identified the differential expression of SNHG7 using publicly available pan-cancer data. We confirmed that SNHG7 is differentially expressed in multiple tumors; in particular SNHG7 expression was significantly upregulated in COAD compared with other tumors. These findings suggest that SNHG7 differential expression may be tissue- specific and it may have an important regulatory role in COAD.
To further test our hypothesis, we analyzed the clinical relationship of SNHG7 in COAD by univariate and multivariate Cox regression analyses. We discovered a strong associated between SNHG7 expression and race, residual tumor, OS, and DSS of COAD patients, with SNHG7 expression appearing to be higher in patients with certain characteristics, such as specific race and with residual tumor. Moreover, we demonstrated that high SNHG7 expression was associated with significantly shorter OS and DSS in COAD patients, but was also an independent risk factor for OS and DSS.
Histopathological characteristics have been implicated as prognostic predictors, such as tumor stages, perineural invasion, and lymphatic invasion (25). Our results also confirmed that these three prognostic predictors were closely related to poor prognosis for OS and DSS in COAD patient with high SNHG7 expression. Noteworthily, in line with our SNHG7-based predicted outcome, univariate Cox regression analysis showed that high CEA levels, which are an independent prognostic factor and can be used for
TNM staging of COAD, reflected poor prognosis in COAD (26). Hence, the remarkable predictive ability of SNHG7 expression suggests its potential as a prognostic biomarker of poor survival in COAD.
In addition, we explored the potential functions and underlying mechanisms of action of SNHG7 in COAD. GO and KEGG analyses revealed that both ribosome and RNA polymerase were closely related to SNHG7 based on the functional annotation of SNHG7-related DEGs. These results also indicated that SNHG7 expression is closely associated with COAD.
The accuracy of a diagnostic tool is based on the area under the ROC curve; the closer the area under the ROC curve is to 1, the better the diagnostic potential of the tool (27). Our results consistently revealed that high SNHG7 expression led to advanced COAD, indicating that SNHG7 expression had high sensitivity and specificity for COAD diagnosis. Assessment of an independent COAD cohort further confirmed the differential expression of SNHG7, and its diagnostic and prognostic value in COAD, indicating that SNHG7 is reliable and reproducible as a prognostic and diagnostic biomarker of COAD.
In conclusion, this study demonstrated that COAD is associated with high SNHG7 expression and that SNHG7 is a reliable biomarker for the diagnosis and prognosis of COAD. Hence, these findings may represent new foundations for the development of enhanced diagnostic and prognostic strategies for COAD.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
The studies involving human participants were reviewed and approved by The TCGA database. The patients/participants provided their written informed consent to participate in this study. Written informed consent was obtained from the individual(s) for the publication of any potentially identifiable images or data included in this article.
MH: design. CJ and SQ: acquisition of data. CJ, SQ, and TL: analysis and interpretation of data. CJ and MH: writing, review, and/or revision of the manuscript. SQ and MH: study supervision. All authors contributed to the article and approved the submitted version.
This research was supported by a grant from Department of Science and Technology of Jilin Province.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We would like to thank Editage (www.editage.cn) for English language editing.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2022.893591/full#supplementary-material
1. Chen EX, Jonker DJ, Loree JM, Kennecke HF, Berry SR, Couture F, et al. Effect of Combined Immune Checkpoint Inhibition vs Best Supportive Care Alone in Patients With Advanced Colorectal Cancer: The Canadian Cancer Trials Group CO.26 Study. JAMA Oncol (2020) 6(6):831–38. doi: 10.1001/jamaoncol
2. Zhang WJ, Hu CG, Luo HL, Zhu ZM. Activation of P2×7 Receptor Promotes the Invasion and Migration of Colon Cancer Cells via the STAT3 Signaling. Front Cell Dev Biol (2020) 8:586555. doi: 10.3389/fcell.2020.586555
3. Goodall J, Mateo J, Yuan W, Mossop H, Porta N, Miranda S, et al. Circulating Cell-Free DNA to Guide Prostate Cancer Treatment With PARP Inhibition. Cancer Discovery (2017) 7(9):1006–17. doi: 10.1158/2159-8290.CD-17-0261
4. Volders PJ, Anckaert J, Verrheggen K, Nuytens J, Martens L, Mestdagh P, et al. LNCipedia 5: Towards a Reference Set of Human Long non-Coding RNAs. Nucleic Acids Res (2019) 47(D1):D135–9. doi: 10.1093/nar/gky1031
5. Shih JW, Chiang WF, Wu AT, Wu MH, Wang LY, Yu YL, et al. Long Noncoding RNA LncHIFCAR/MIR31HG is a HIF-1α Co-Activator Driving Oral Cancer Progression. Nat Commun (2017) 8:15874. doi: 10.1038/ncomms15874
6. Grembergen OV, Bizet M, Bony EJ, Calonne E, Putmans P, Brohée S, et al. Portraying Breast Cancers With Long Noncoding RNAs. Sci Adv (2016) 2(9):e1600220. doi: 10.1126/sciadv.1600220
7. Kim J, Piao HL, Kim BJ, Yao F, Han Z, Wang Y, et al. Long Noncoding RNA MALAT1 Suppresses Breast Cancer Metastasis. Nat Genet (2018) 50(12):1705–15. doi: 10.1038/s41588-018-0252-3
8. Schmidt LH, Görlich D, Spieker T, Rohde C, Schuler M, Mohr M, et al. Prognostic Impact of Bcl-2 Depends on Tumor Histology and Expression of MALAT-1 lncRNA in non-Small-Cell Lung Cancer. J Thorac Oncol (2014) 9(9):1294–304. doi: 10.1097/JTO.0000000000000243
9. Qu D, Sun WW, Li L, Ma L, Sun L, Jin X, et al. Long Noncoding RNA MALAT1 Releases Epigenetic Silencing of HIV-1 Replication by Displacing the Polycomb Repressive Complex 2 From Binding to the LTR Promoter. Nucleic Acids Res (2019) 47(6):3013–27. doi: 10.1093/nar/gkz117
10. Li X, Liu R, Yang J, Sun L, Zhang L, Jiang Z, et al. The Role of Long Noncoding RNA H19 in Gender Disparity of Cholestatic Liver Injury in Multidrug Resistance 2 Gene Knockout Mice. Hepatology (2017) 66(3):869–84. doi: 10.1002/hep.29145
11. Shi J, Ding W, Lu H. Identification of Long Non-Coding RNA SNHG Family as Promising Prognostic Biomarkers in Acute Myeloid Leukemia. Onco Targets Ther (2020) 13:8441–50. doi: 10.2147/OTT.S265853
12. Qin Y, Sun W, Wang Z, Dong W, He L, Zhang T, et al. Long Non-Coding Small Nucleolar RNA Host Genes (SNHGs) in Endocrine- Related Cancers. Onco Targets Ther (2020) 13:7699–717. doi: 10.2147/OTT.S267140
13. She K, Yan H, Huang J, Zhou H, He J. miR-193b Availability is Antagonized by LncRNA-SNHG7 for FAIM2-Induced Tumour Progression in non-Small Cell Lung Cancer. Cell Prolif (2018) 51(1):e12406. doi: 10.1111/cpr.12406
14. Zhang Y, Yuan Y, Zhang Y, Cheng L, Zhou X, Chen K. SNHG7 Accelerates Cell Migration and Invasion Through Regulating miR-34a- Snail-EMT Axis in Gastric Cancer. Cell Cycle (2020) 19(1):142–52. doi: 10.1080/15384101
15. Wu F, Sui Y, Wang Y, Xu T, Fan L, Zhu H. Long Noncoding RNA SNHG7, a Molecular Sponge for microRNA-485, Promotes the Aggressive Behavior of Cervical Cancer by Regulating Pak4. Onco Targets Ther (2020) 13:685–99. doi: 10.2147/OTT.S232542
16. Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene Set Enrichment Analysis: A Knowledge-Based Approach for Interpreting Genome-Wide Expression Profiles. Proc Natl Acad Sci U S A. (2005) 102(43):15545– 50. doi: 10.1073/pnas.0506580102
17. Yuan Y, Chen J, Wang J, Xu M, Zhang Y, Sun P, et al. Development and Clinical Validation of a Novel 4-Gene Prognostic Signature Predicting Survival in Colorectal Cancer. Front Oncol (2020) 10:595. doi: 10.3389/fonc.2020.00595
18. Weng L, Ma J, Jia YP, Wu SQ, Liu BY, Cao Y, et al. MiR-4262 Promotes Cell Apoptosis and Inhibits Proliferation of Colon Cancer Cells: Involvement of GALNT4. Am J Transl Res (2018) 10(12):3969–77.
19. Bereczki E, Branca RM, Francis PT, Pereira JB, Baek JH, Hortobágyi T, et al. Synaptic Markers of Cognitive Decline in Neurodegenerative Diseases: A Proteomic Approach. Brain (2018) 141(2):582–95. doi: 10.1093/brain/awx352
20. Liu Z, Liu L, Weng S, Guo C, Dang Q, Xu H, et al. Machine Learning-Based Integration Develops an Immune-Derived lncRNA Signature for Improving Outcomes in Colorectal Cancer. Nat Commun (2022) 13(1):816. doi: 10.1038/s41467-022-28421-6
21. Liu Z, Guo C, Dang Q, Wang L, Liu L, Weng S, et al. Integrative Analysis From Multi-Center Studies Identities a Consensus Machine Learning-Derived lncRNA Signature for Stage II/III Colorectal Cancer. EBioMedicine (2022) 75:103750. doi: 10.1016/j.ebiom.2021.103750
22. Liu KL, Wu J, Li WK, Li NS, Li Q, Lao YQ. LncRNA SNHG7 is an Oncogenic Biomarker Interacting With MicroRNA- 193b in Colon Carcinogenesis. Clin Lab (2019) 65(11):1–10. doi: 10.7754/Clin.Lab.2019.190501
23. Shan Y, Ma J, Pan Y, Hu J, Liu B, Jia L. LncRNA SNHG7 Sponges miR-216b to Promote Proliferation and Liver Metastasis of Colorectal Cancer Through Upregulating GALNT1. Cell Death Dis (2018) 9(7):722. doi: 10.1038/s41419-018-0759-7
24. Ziaee F, Hajjari M, Kazeminezhad RS, Behmanesh M. SNHG7 and FAIM2 are Up-Regulated and Co-Expressed in Colorectal Adenocarcinoma Tissues. Klin Onkol (2020) 33(6):445–9. doi: 10.48095/ccko2020445
25. Martin LW, D’Cunha J, Wang X, Herzan D, Gu L, Abraham N, et al. Detection of Occult Micrometastases in Patients With Clinical Stage I Non- Small-Cell Lung Cancer: A Prospective Analysis of Mature Results of CALGB 9761 (Alliance). J Clin Oncol (2016) 34(13):1484–91. doi: 10.1200/JCO.2015.63.4543
26. Lu Y, Wu S, Cui C, Yu M, Wang S, Yue Y, et al. Gene Expression Along With Genomic Copy Number Variation and Mutational Analysis Were Used to Develop a 9-Gene Signature for Estimating Prognosis of COAD. Onco Targets Ther (2020) 13:10393–408. doi: 10.2147/OTT.S255590
Keywords: SNHG7, bioinformatics, diagnosis, prognosis, colon adenocarcinoma
Citation: Jiang C, Qu S, Liu T and Hao M (2022) Long Noncoding RNA SNHG7 Is a Diagnostic and Prognostic Marker for Colon Adenocarcinoma. Front. Oncol. 12:893591. doi: 10.3389/fonc.2022.893591
Received: 10 March 2022; Accepted: 09 May 2022;
Published: 07 June 2022.
Edited by:
Norfilza M. Mokhtar, National University of Malaysia, MalaysiaReviewed by:
Lifeng Li, First Affiliated Hospital of Zhengzhou University, ChinaCopyright © 2022 Jiang, Qu, Liu and Hao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Miao Hao, miaohao@jlu.edu.cn
†These authors share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.