- 1Zhongzhi International Institute of Agricultural Biosciences, Shunde Graduate School, Research Center of Biology and Agriculture, University of Science and Technology Beijing (USTB), Beijing, China
- 2Beijing Beike Institute of Precision Medicine and Health Technology, Beijing, China
- 3Beijing Engineering Laboratory of Main Crop Bio-Tech Breeding, Beijing International Science and Technology Cooperation Base of Bio-Tech Breeding, Beijing Solidwill Sci-Tech Co., Ltd., Beijing, China
Probiotics are known to contribute to the anti-oxidation, immunoregulation, and aging delay. Here, we investigated the extension of lifespan by fermented pickles-origin Pediococcus acidilactici (PA) in Caenorhabditis elegans (C. elegans), and found that PA promoted a significantly extended longevity of wild-type C. elegans. The further results revealed that PA regulated the longevity via promoting the insulin/IGF-1 signaling, JNK/MAPK signaling but not TOR signaling in C. elegans, and that PA reduced the reactive oxygen species (ROS) levels and modulated expression of genes involved in fatty acids uptake and lipolysis, thus reducing the fat accumulation in C. elegans. Moreover, this study identified the nrfl-1 as the key regulator of the PA-mediated longevity, and the nrfl-1/daf-18 signaling might be activated. Further, we highlighted the roles of one chloride ion exchanger gene sulp-6 in the survival of C. elegans and other two chloride ion channel genes clh-1 and clh-4 in the prolonged lifespan by PA-feeding through the modulating expression of genes involved in inflammation. Therefore, these findings reveal the detailed and novel molecular mechanisms on the longevity of C. elegans promoted by PA.
Introduction
Population aging has been a global problem, leading to a growing of longer healthy life expectancy (1). There will be huge numbers of elderly people (65 years old and above), which is over 20% of the total population across world at 2050 (2). Some countries are facing a challenge of rapid population aging, such as South Korea, Japan, and China. Aging and longevity are multifactorial complex processes, involving in both genetic and environmental factors (3).
Gut microbiota is the largest internal microenvironment in human body to communicate with the external world and its composition is shaped predominantly by environmental factors (4). In addition, microbiota acts as a mediator between host genes and environmental factors (5). Gut microbiota, with a 10-fold cell number and a 100-fold gene number greater than those of human, has been accepted as a potential marker in aging (6). Gut microbes that influence the immune system and metabolic process may shape the energy harvest, endow defense to pathogens, reduce oxidative stress, confer abilities to alleviate stress, and improve anti-inflammatory effect in anti-aging (7, 8). Probiotics, mainly Lactobacillus and Bifidobacterium from lactic acid bacteria (LAB) which are widely recognized beneficial bacteria for a long time and have been proved to increase the longevity of Caenorhabditis elegans (C. elegans) (9–12).
C. elegans is an ideal model for the longevity research of host-bacteria interaction due to a short life span, fast reproducible life, and using bacteria as food (13). Many longevity-associated signaling pathways, such as the insulin/insulin-like growth factor-1 (IGF-1) signaling, NSY-1/PMK-1 MAP kinase signaling, and Target of Rapamycin (TOR) signaling were found in C. elegans (14, 15). In recent years, new signaling pathways have been found, such as ATFS-1/unfolded protein response (UPRmt) and HLH-30/TFEB-autophagy (16, 17). However, the mechanism of gut microbes regulating longevity is not totally understood and there are other longevity associated signaling pathways yet to be investigated.
Pediococcus acidilactici (PA), also a member of LAB, was reported to regulate gut microbiota, reduce inflammation, release bacteriocin, and inhibit pathogenic bacteria in animals (18–20). PA is a strain isolated from naturally fermented pickles in Chongqing, China. In this study, we examined whether PA can extend the lifespan of C. elegans and explored the mechanism of its anti-aging effect.
Materials and Methods
Bacterial Strains and Caenorhabditis elegans Strains
Escherichia coli strain OP50 was provided by the Institute of Biophysics of Chinese Academy of Sciences and grown in Luria-Bertani (LB) broth (Oxoid, United Kingdom) at 37°C for 18 h. PA was isolated from fermented pickles and grown in de Man, Rogosa, and Sharpe (MRS) broth (Oxoid, United Kingdom) at 37°C for 18 h.
Wild-type N2, CB1370 [daf-2 (e1370) II], VC1167 [sulp-6(ok1586)], RB1139 [clh-4(ok1162)], XA900 [clh-1(qa900)], VC1795 [nrfl-1(ok2292)], VC222 [raga-1 (ok386) II], GR1307 [daf-16 (mgDf50) I], QQ202 [daf-2(cv20[daf-2:gfp])III], TJ356 [zIs356 (daf-16:GFP) IV], and VC8 [jnk-1(gk7)IV] were purchased from Caenorhabditis Genetics Center (CGC, University of Minnesota, Twin Cities, United States). All C. elegans strains were cultured at 20°C on the Nematode Growth Medium (NGM) plates using living E. coli strain OP50 as food.
Longevity Assays in Caenorhabditis elegans
To measure the lifespan of C. elegans accurately, age-synchronized worms of the wild-type strain (N2) were grown on NGM agar plates containing live E. coli OP50 at 20°C, until they reached the young adult stage. Approximately 80–100 age-synchronized worms were then transferred to fresh NGM plates covered with lawns of E. coli OP50 or PA (21). The plates were incubated at 20°C, the worms were transferred every other day thereafter. The worms that showed no reaction to gentle stimulation were scored as dead, whereas the animals that crawled off the plate, exploded, or bagged were censored, the surviving nematodes were transferred to a fresh plate, and the numbers of live and dead worms were scored. Two independent lifespan assays were performed.
Measurement of Body Size
For the body length measurement, approximately 20 wild-type L4 larvae were randomly selected and inoculated to bacterial NGM plates covered with the lawns of E. coli OP50 or PA. From the first day that C. elegans were transferred to fresh bacteria spreading NGM plates, the sizes of live worms were examined every other day until reaching 5 days of age, according to the previous research (22). The worms were visualized with a microscope (Olympus SZ61) using a 10× objective lens of a bright-field microscope. The area of worm’s projection was analyzed as the body size using the Olympus view software.
Brood Size Measurement
The L4-stage worms were placed on bacterial NGM plates covered with the lawns of E. coli OP50 or PA and incubated at 20°C. These worms were transferred to fresh bacterial NGM plates every 24 h until the end of ovulation. The progeny was counted each day. This assay was performed for six consecutive days, which was the reproductive period of the worms. The sum of the plate algebra was taken as the total sub algebra of the worm. Brood size evaluation of each bacterial species contained twenty worms, and one worm was detected on one plate.
Assessment of Pharyngeal Pumping Rate
The N2 worms were used to evaluate the age-related decline in muscle function by pharyngeal pumping rate measurement at room temperature (10). L4-stage worms were transferred to NGM plates covered with the lawns of E. coli OP50 or PA and incubated at 20°C. The pharyngeal pumping rates were analyzed on days 3, 6, and 9 by counting the pumping frequency of the terminal pharyngeal bulb of each worm within 60 s. Three independent trials were performed with ten worms for each bacterial species.
Measurement of Intracellular Reactive Oxygen Species Level
Dichlorofluorescin diacetate (H2DCF-DA, Sigma) was used to determine the intracellular reactive oxygen species (ROS) concentration in N2 nematodes. L4-stage worms were treated with E. coli OP50 or PA for 4 days, and collected with M9 buffer. Bacteria were removed by washing three times with M9 buffer. Approximately 35 nematodes were transferred to a transparent-bottom black 96-well plate (Corning, United States). The plate containing 100 μl of H2DCF-DA at 100 mM diluted with phosphate buffer (pH 7.2). The fluorescence intensity was measured by a fluorescence microplate reader (Molecular Devices SpectraMax250, United States) at 485/535 nm, and samples were measured every 20 min for a total of 160 min (23).
DAF-16 Localization and DAF-2 Fluorescence Assay
The strain TJ356 (with DAF-16:GFP) was used to detect the intracellular DAF-16 localization. Then, 3-day-old adult worms (L4 stage) were transferred to NGM plates covered with the lawns of PA or E. coli OP50. The worms were placed for 5 days at 20°C, collected and washed three times by M9 buffer. The worms were fixed on the slide containing 2% agarose, anaesthetized with 10 mM imidazole (Sigma), and covered with coverslips. The worms were photographed by using a green excitation channel of confocal scanning laser microscope. Images were analyzed using the Image J 8.4 software (24). The percentage of daf-16 localization (cytosolic, nuclear, and both) was calculated according to a recent report (25). The same method was used for the detection of DAF-2 fluorescence from QQ202 strain.
Oil Red O Staining
The L4-stage worms were placed on NGM plates covered with the lawns of PA or E. coli OP50. The plates were incubated at 25°C for 4 days. The worms were collected with phosphate buffer saline (PBS, pH 7.2) plus with 1% Triton to avoid the adhesion of the worms and reduce the loss of the worms during operation. Paraformaldehyde solution (4%, w/v) was used for fixation (4°C, 15 min). Then, the worms were subjected to freeze-thaw cycles two times between a −80°C freezing for 20 min and 25°C water thawing for 5 min (26). In this way, the cuticle of the worms could be broken and the worms were fully stained. Discarding the supernatant after centrifuging, the worms were washed once by M9 buffer. Then, the worms were dehydrated in 60% isopropanol for 15 min in the shaker at the maximum speed. Subsequently, the worms were stained by incubation with 1 ml Oil Red O stain (Beyotime, China) for 4–6 h. Stained worms were washed to remove the dyestuff, fixed on a slide containing 2% agarose, and visualized with Leica IIC50W microscope. The image J 8.4 software was used to determine the lipid content of staining worms. At least three independent determinations were repeated.
BODIPY C12 Uptake Assay
C1-BODIPY-C12 (Molecular Probes, Invitrogen) was dissolved in dimethyl sulfoxide (DMSO, Sigma) to obtain 5 mM stock solution. Then, the stock solution was diluted to 1 mM with PBS. Furthermore, 0.5 ml diluted solution was loaded to the surface of empty NGM plates. The worms were quickly transferred to the plate and placed at 20°C for 6 h (27). Finally, after washing three times with M9 buffer, the worms were visualized using the green excitation channel of confocal scanning laser microscope TCS SP8 (Leica, Wetzlar, Germany). Fluorescence intensity was measured using the image J 8.4 software.
RNA Isolation and Quantitative Real-Time PCR (RT-qPCR) Analysis
Following an 8-day treatment with PA or E. coli OP50, the worms were washed with M9 buffer for three times, n = 200. Total RNA was extracted using TRIzol reagent (TIANGEN Biotech Co., Ltd., Beijing, China), and quantified with a NanoDrop 2000 spectrophotometer (Thermo Scientific, United States). The cDNA was synthesized by reverse transcription using the 5× All-In-One RT MasterMix (Applied Biological Materials Inc., G490, BC, Canada) according to the manufacturer’s instructions. Software Primer premier 6.0 was used to design oligonucleotides for quantitative real-time PCR (RT-qPCR) (Supplementary Table 1). The RT-qPCR was performed with TB Green® Premix Ex Taq™ (TAKARA Inc., Dalian, China) and QuantStudio 5 Real-Time PCR Systems (Applied Biosystems) (28). Three independent experiments were performed, the relative gene expression was determined using the 2–ΔΔCT method.
Statistical Analysis
Statistical analysis was presented using unpaired Student’s t-test and one-way ANOVA was compared in multiple groups using GraphPad Prism version 8.0 software (GraphPad Software, San Diego, CA, United States). Data were expressed as means ± SD. The log rank test was used to test the significance of life span in different experimental treatment groups. Compared with the control group, statistical differences at the p < 0.05 or p < 0.01 level were defined to be significant.
Results
Evaluation of the Lifespan Prolongation of Caenorhabditis elegans Mediated by Pediococcus acidilactici
Recent advances in gut microbiota are rapidly propagating into the aging targets discovery and anti-aging probiotics screening practices (29). As the common probiotics, Lactobacillus and Bifidobacterium are well established for life extension research in C. elegans (30). However, PA has been mainly focused on its antimicrobial effects and bacteriocin production, while the anti-aging effects of PA are largely unknown. As shown in Figure 1A, the prolongation of C. elegans lifespan in the probiotic PA group was significantly higher than that in the control group (p < 0.001) fed by E. coli OP50. Moreover, we observed that PA-feeding had no significant influence on the basic physiology traits of the worms, such as body size, pumping rates, reproductive performance (progeny number), and brood size (Figures 1B–E).
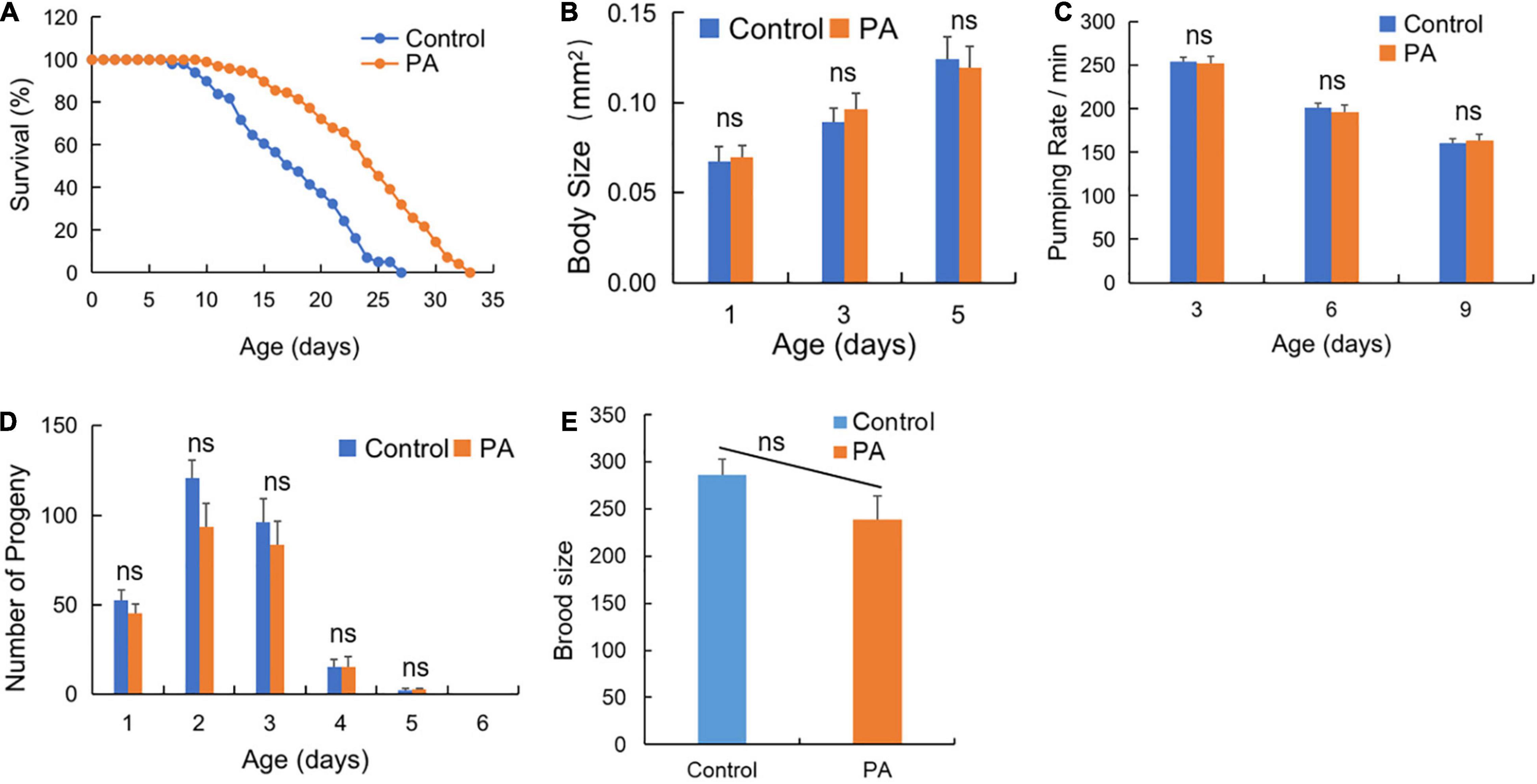
Figure 1. Pediococcus acidilactici (PA) extends lifespan but not affects basic physiology of wild-type Caenorhabditis elegans (C. elegans). (A) Comparison of PA and Escherichia coli (E. coli) OP50 on the survival of C. elegans, n = 100. (B) The effects of PA on body size of C. elegans, n = 20. (C) The effects of PA on the pumping rate of C. elegans. (D) The effects of PA on the reproductive capacity of C. elegans, n = 20. (E) The effects of PA on brood size of C. elegans. ns, non-significant.
daf-16-Dependent Signaling Pathway Contributes to the Pediococcus acidilactici-Mediated Longevity of Caenorhabditis elegans
Common conserved signaling pathways have a profound impact on host prolongevity, such as daf-2/daf-16 in insulin/IGF-1 signaling and jnk-1/daf-16 in JNK/MAPK signaling (31). To elucidate the mechanism on lifespan extension mediated by PA, we investigated the fluorescent tracer of daf-2 and daf-16 in C. elegans and the effects of PA on the lifespan prolongation of C. elegans under four mutant backgrounds, such as daf-2 (CB1370), daf-16 (GR1307), raga-1 (VC222), and jnk-1 (gk7). As a result, deficiency in daf-2, daf-16, or jnk-1 cannot extend the longevity of C. elegans by PA (Figures 2A–C), while PA extends the longevity of C. elegans with TOR deficiency (Figure 2D), suggesting that the PA-mediated longevity was dependent on insulin/IGF-1 signaling (daf-2/daf-16) and JNK/MAPK signaling (jnk-1/daf-16) but not TOR signaling (raga-1). In addition, the relocation of daf-16 to cell nucleus affects the phosphorylation of daf-16 which is required for life span extension (32). In daf-16:GFP worms, PA increased the nuclear accumulation of daf-16 at 2 or 5 days probiotic intake (Figure 2E). PA can induce an increase of nuclear daf-16:GFP translocation (E. coli OP50: 6.67%; PA: 40%) and a reduction of daf-16:GFP in the cytosolic (E. coli OP50: 46.67%; PA: 26.67%) at 5 days (Figure 2F). Additionally, we observed that PA declined the fluorescence intensity of daf-2 in daf-2:GFP worms (Figure 2G). These results indicated that PA prolonged the longevity via daf-2/daf-16 mediated insulin/IGF-1 signaling, jnk-1/daf-16 mediated JNK/MAPK signaling but not raga-1 dependent TOR signaling.
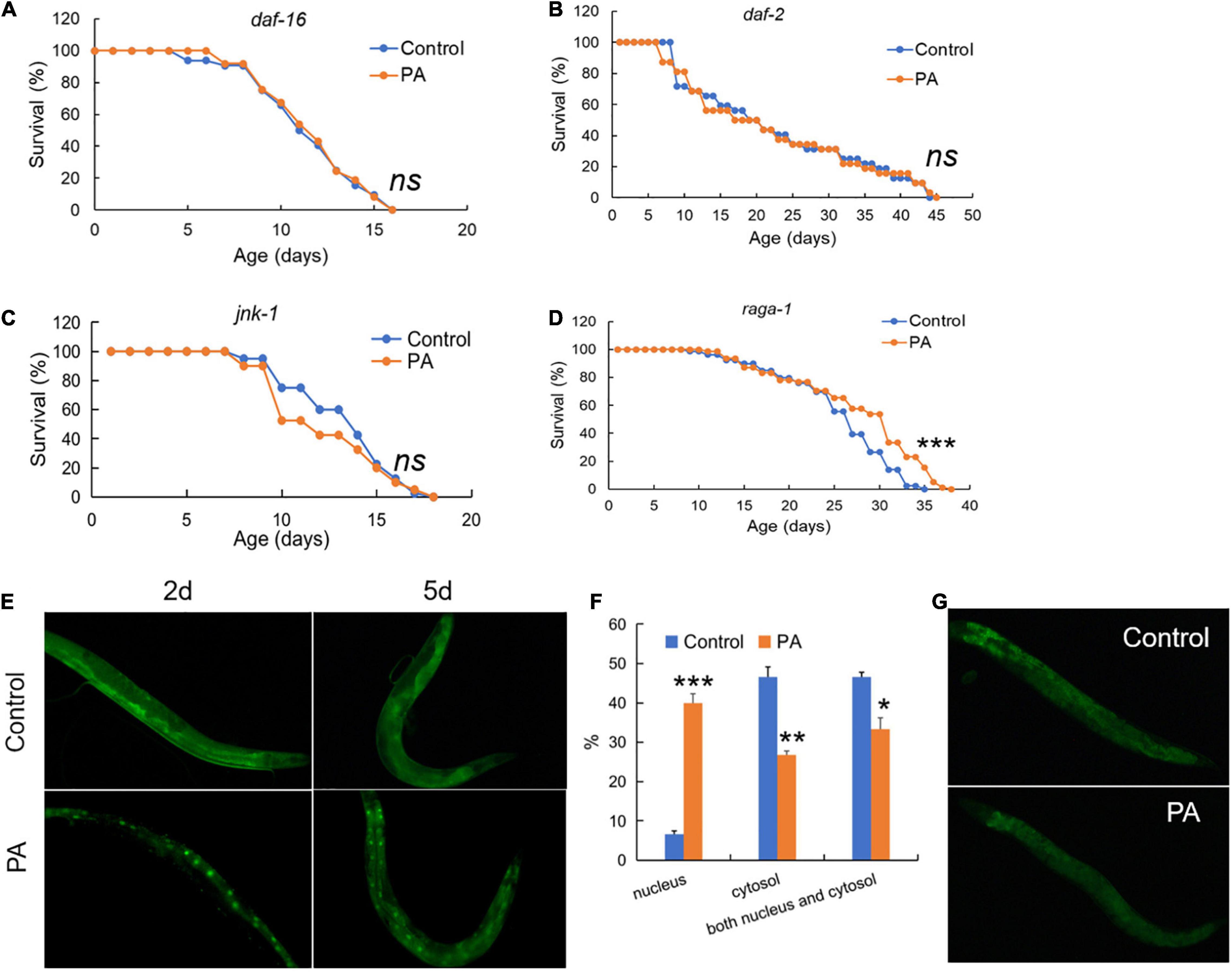
Figure 2. Pediococcus acidilactici extends lifespan of C. elegans by regulating both daf-2 and jnk-1/daf-16 signaling. (A) The survival rate of daf-16 (mgDf50) worms by PA. The control group is E. coli OP50 feeding. (B) The survival rate of daf-2 (CB1370) worms, n = 80. (C) The survival rate of jnk-1 (gk7) worms by PA. (D) The survival rate of raga-1 (VC222) worms by PA, n = 80. ns, non-significant; *p < 0.05; **p < 0.01; ***p < 0.001. (E) Feeding daf-16:GFP worms with PA for 2–5 days activates daf-16 expression with the increased nuclear accumulation of daf-16 protein. (F) Comparing percentage of daf-16 localization data (cytosolic, nuclear, and both) between E. coli control and PA-treated C. elegans. (G) Feeding daf-2:GFP worms with PA for 5 days represses daf-2 expression.
Pediococcus acidilactici Reduces Fat Accumulation in Caenorhabditis elegans
Generally, dye-labeled lipid assays can provide the robust mapping of lipid accumulation especially the intestinal lipid storage in C. elegans since the fat is mainly stored in intestinal and epidermal cell (33). Oil Red O is a lysochrome and highly soluble dye in lipids, and can stain triglycerides and lipoproteins in C. elegans (34). Compared with the transparent body of C. elegans, Oil Red O shows obvious red lipid droplets to allow the qualitative evaluation of lipid distribution (35). Our results revealed a significant decrease in Oil Red O intensity in the worms treated with the probiotic PA compared with control (Figures 3A,B). Recent studies have revealed that regulating lipid metabolism is crucial for longevity in C. elegans and the content of triglycerides is a marker reflecting the energy levels (36). PA-treated C. elegans reflect a lower fat mass and lower energy, thus contributed to lifespan extension (Figures 3A,B). To explore the cause of lower fat storage, the dietary fat source was detected. As a result, the fluorescence intensity of the BODIPY-labeled fatty acids was significantly decreased in the worms fed with PA compared with the OP50-fed control, suggesting a decrease in fatty acids uptake (Figures 3C,D). In addition, we examined the expression of three key genes involved in the lipocatabolic pathway. As expected, PA-feeding significantly increased the expression of sod-3, fat-4, and lipl-4 genes encoding superoxide dismutase, fatty acid desaturase, and triglyceride lipase, respectively (Figure 3E). Excess ROS are genotoxic and can cause oxidative damage within the cell. Further accumulated oxidative stress can induce lipid peroxidation and protein homeostasis interference (37). Here, we found that the fluorescence intensity reflecting ROS levels was significantly reduced by PA-feeding compared with the OP50-fed control (Figures 3F,G), indicating that PA intake can protect host from ROS-induced oxidative damage and lipid peroxidation product accumulation.
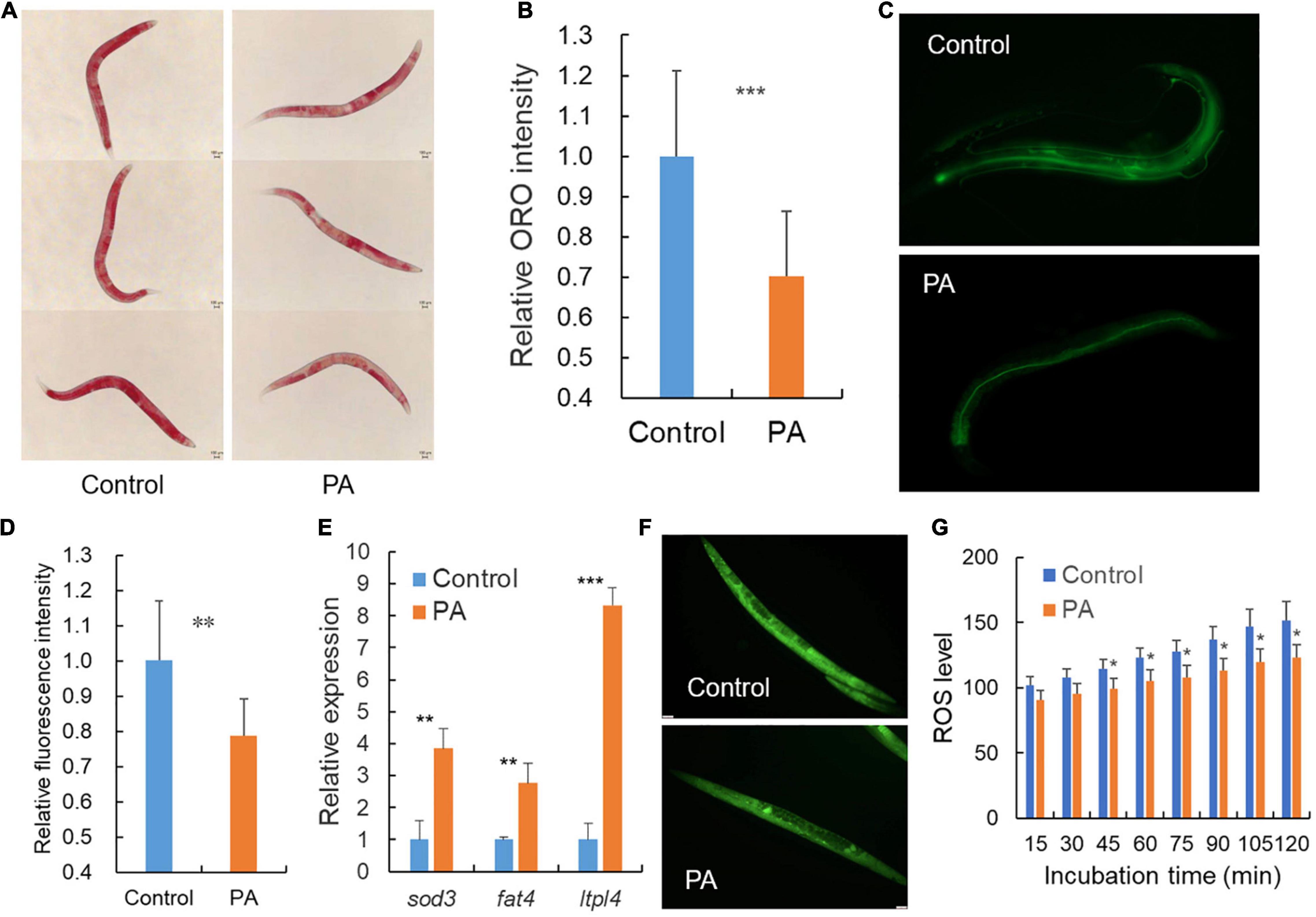
Figure 3. Pediococcus acidilactici affects the lipid accumulation of wild-type C. elegans. (A) Representative images of Oil Red O staining after treatment with E. coli OP50 or PA in C. elegans, n = 45. (B) Relative quantification of the Oil Red O staining intensity in (A). (C) Representative fluorescence images of fatty acids staining after treatment with E. coli OP50 or PA in C. elegans, n = 20. (D) Relative quantification of the green fluorescence intensity in (C). (E) Transcript expression of lipid related genes in PA-fed C. elegans compared with control E. coli OP50. (F) Representative images of fluorescence labeled reactive oxygen species (ROS) in C. elegans, n = 35. (G) Relative quantification of the green fluorescence labeled ROS, *p < 0.05; **p < 0.01; ***p < 0.001.
Pediococcus acidilactici Promotes the Survival Rate and Age of Caenorhabditis elegans by Regulating the Expression of Genes Related to Chloride Ion and Immune Response
To address the novel targets of PA-mediated longevity, we examined the expression of chloride ion related genes in PA- or OP50-fed wild-type C. elegans. We found that PA-fed C. elegans showed 11-fold increase in sulp-6, 13-fold increase in clh-1, eight-fold increase in clh-4, and six-fold increase in nrfl-1 transcripts compared with the OP50-fed control (Figure 4A). Most importantly, PA significantly promoted the survival rate and age of clh-1, clh-4, and sulp-6 mutant worms (Figures 4B1–B3). In addition, PA had no effect on the longevity of nrfl-1 mutant worms (Figure 4B4).
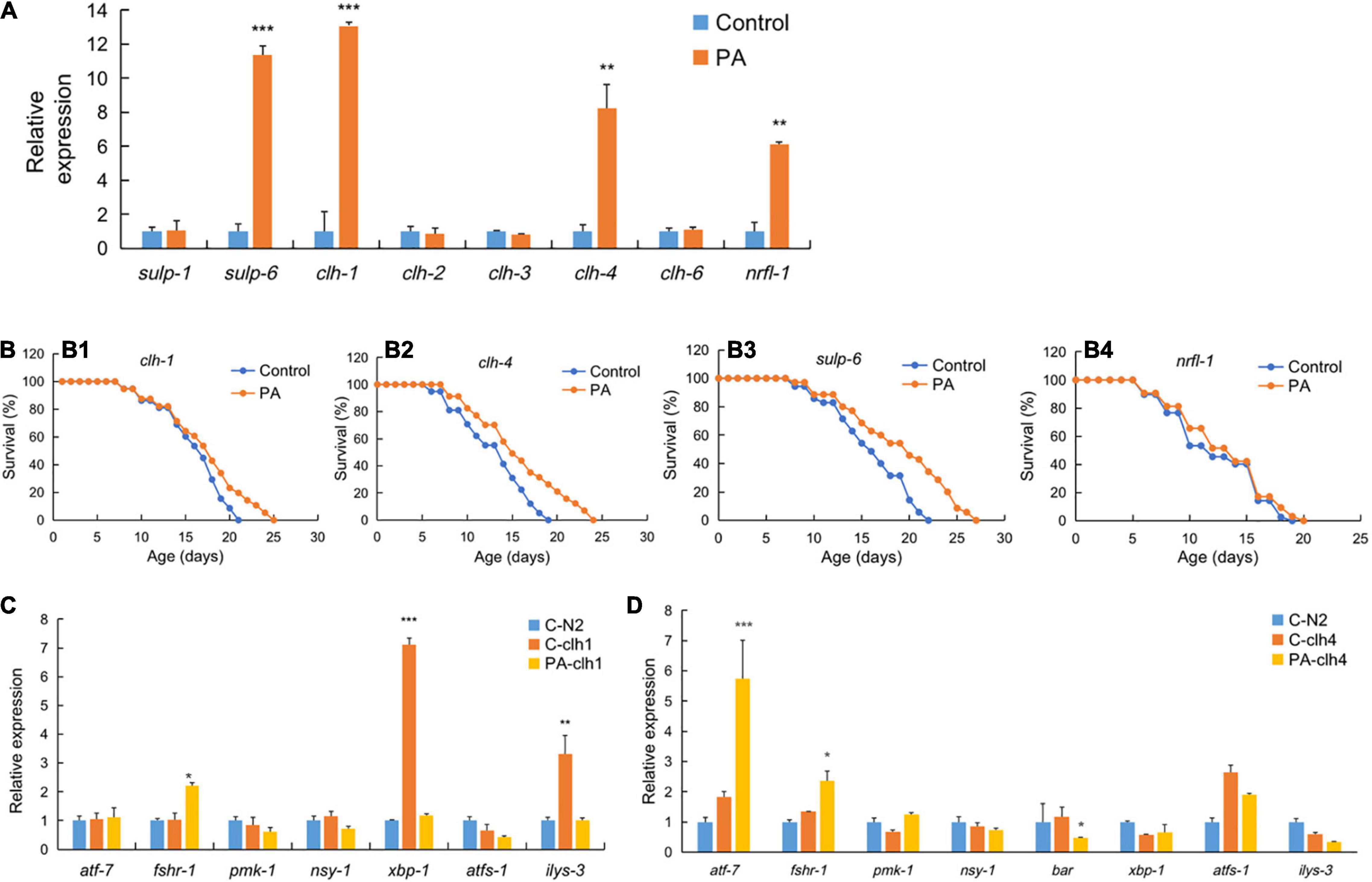
Figure 4. The regulation of genes expression and survival of different mutant worms by PA feeding. (A) Expression of chloride ion related genes in PA-fed C. elegans compared with control E. coli OP50. (B) The survival rates of clh-1 (XA900, B1), clh-4 (RB1139, B2), and sulp-6 (VC1167, B3), and nrfl-1 (VC1795, B4) worms by PA feeding, n = 80. (C,D) The expression of inflammatory or anti-inflammatory genes in PA-fed clh1 (C) or clh4 (D) gene mutant C. elegans compared with the mutant control and wild-type N2 feeding E. coli OP50, *p < 0.05; **p < 0.01; ***p < 0.001.
The immunoregulatory effect is one of the most important functions of probiotics. Modulating immune response is crucial for the survival of C. elegans (38). We examined whether PA regulates the expression of eight genes (atf-7, fshr-1, pmk-1, nsy-1, bar, xbp-1, atfs-1, and ilys-3) involved in the inflammation of clh-1 and clh-4 mutant worms (Figures 4C,D). C. elegans is the absence of most part of mammalian innate immune response, e.g., no mobile immune cells and not producing inflammatory cytokines. However, it contains a conserved principal NSY-1/SEK-1/PMK-1 mitogen-activated protein kinase (MAPK) signaling pathway (39). For example, atf-7 can directly regulate the genes of host defense and the anti-inflammatory PMK-1/ATF-7 signaling can contribute to innate immune response to infection (40). The G protein coupled with receptor fshr-1, in parallel to the p38 MAPK pathway, is another important effector triggering innate immune response to ingested pathogens in C. elegans (41). Xbox binding protein (xbp-1), encoded by an inflammatory gene, can increase the endoplasmic reticulum (ER) folding capacity in the PMK-1-mediated defenses (42). Similarly, the beta-catenin homolog bar-1 is important for the inflammation of S. aureus infection (43). Ilys-3, an invertebrate-type lysozyme gene in C. elegans, can be upregulated in the intestine with an inflammatory ERK-dependent manner when challenged with pathogens (44). In this study, it is observed that the gene expression of both xbp-1 and ilys-3 was relatively lower in PA-fed clh-1 and OP50-fed wild-type control when compared with the OP50-fed clh-1 mutant worms, while fshr-1 expression was significantly increased in PA-fed clh-1 mutant worms (Figure 4C). Meanwhile, we observed a significant increase in atf-7 and fshr-1 expression, but a significant reduction in bar expression in PA-fed clh-4 mutant worms compared with the OP50-fed clh-4 mutant worms (Figure 4D), implying that PA has immunoregulatory effects on the survival of C. elegans.
Schematic Representation of Pediococcus acidilactici-Mediated Lifespan Extension in Caenorhabditis elegans
The probiotic mechanism of PA-feeding effects on C. elegans was summarized in Figure 5. In this study, the probiotic PA significantly prolonged the lifespan of C. elegans without an influence on reproductive performance. Then, the PA failed to modify the lifespan of daf-2, daf-16 and jnk-1 mutant worms, implying that the classic Insulin/IGF-1 and JNK/MAPK signaling pathways were implicated in the PA regulated lifespan extension. But this lifespan extension was not dependent on TOR signaling since PA can extend the longevity in the raga-1 mutant worms. As a downstream of daf-16, sod-3 was required for the antioxidative process to reduce ROS levels. The increased expression levels of sod-3, fat-4, and ltpl-4 are contributed to the reducing lipid disposition, and thus promoting longevity. Our findings also pointed to a novel requirement for the sulp-6, nrfl-1, clh-1, and clh-4 expression regulating chloride ion related process, emphasizing the possibility that chloride ion is tightly linked to the promoted longevity. The longevity initiated by PA intake was dependent on nrfl-1 and the nrfl-1/daf-18 signaling, upstream of Insulin/IGF-1 signaling, might be activated. The clh-1 or clh-4 mutations affected the inflammatory gene expression, and PA regulated some anti-inflammatory gene expression and improved longevity. Thus, these data suggest that the PA prolonged lifespan of C. elegans by regulating the Insulin/IGF-1 signaling and JNK/MAPK signaling, reducing lipid accumulation, and regulating chloride ion dependent genes, but not dependent on TOR signaling.
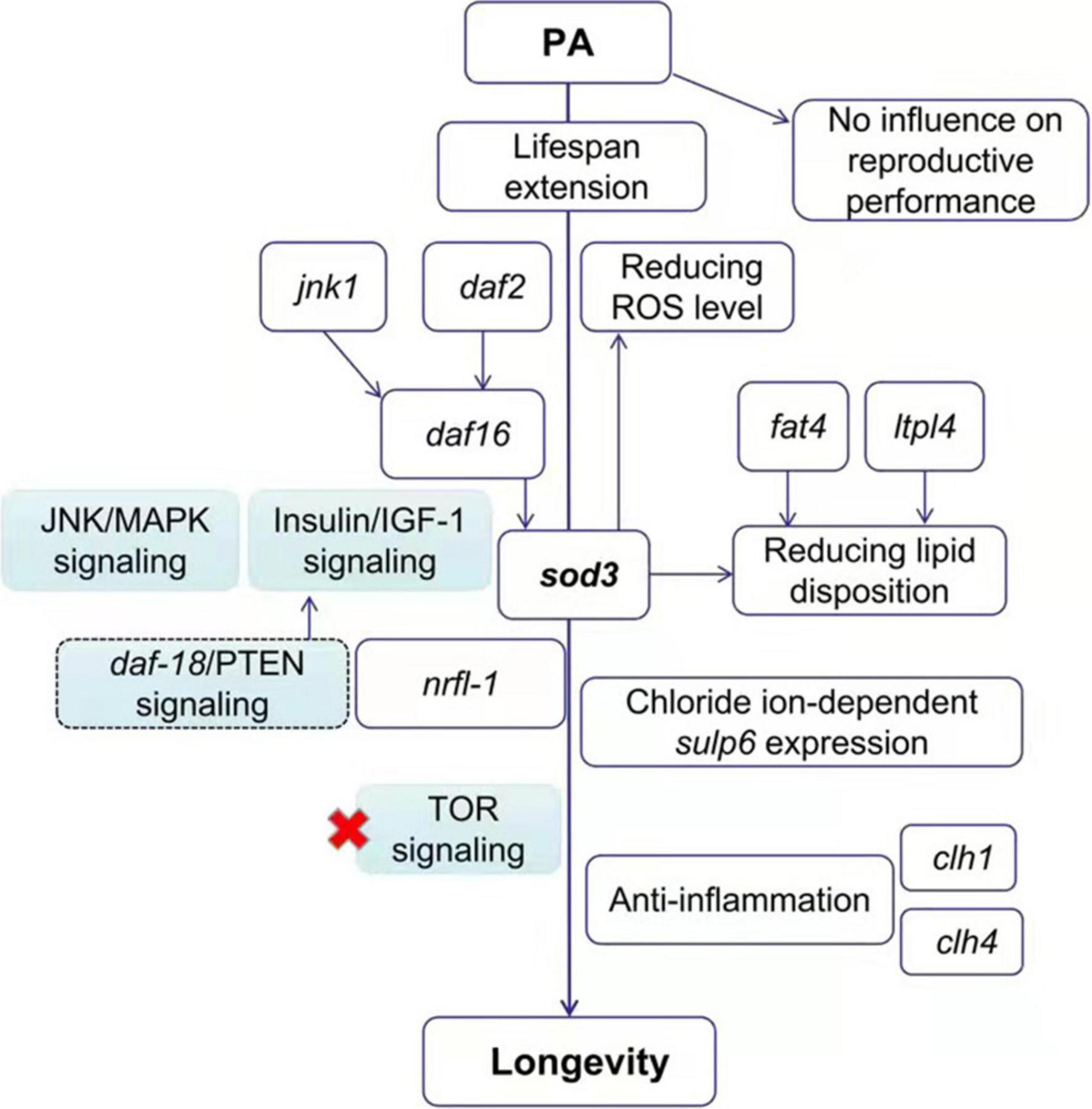
Figure 5. The proposed mechanism of PA-feeding effects on the longevity of C. elegans. The PA increases the lifespan of nematodes via the Insulin/IGF-1 signaling (daf-2/daf-16) and JNK/MAPK signaling (jnk1/daf-16) pathway but not Target of Rapamycin (TOR) pathway, inhibiting the fat accumulation, and regulating some of the chloride ion dependent genes.
Discussion
Gut beneficial microbes play an important role in the production of age-related metabolites, oxidative stress, and immunoregulation in aging (45). Here, we performed an in vivo study to investigate whether probiotics can exhibit anti-aging effect and lead to longevity in the C. elegans, and found the extension of lifespan by fermented pickles-origin PA in C. elegans (Figures 1–4). Up to date, a lot of probiotics, most of which are LAB strains, have been shown to provide lifespan benefits in C. elegans (30). A large-scale strain screening experiment reveals that some LAB strains isolated from Kimchi can prolong lifespan in C. elegans (46). Concerning the probiotic safety, feeding the worms with microbes might not affect the reproduction development of the worms. As shown by the results, PA feeding displayed a non-obvious effect on body size, brood size, pharyngeal pumping rate, and reproduction of these worms (Figure 1), though the previous study revealed that body-size reducing was an effect of dietary restriction (47). Thus, the prolonged lifespan effect of PA is independent on dietary restriction. Another TOR pathway evidence further indicates that PA extends lifespan independent on dietary restriction since the reduced TOR signaling is a primary mechanism involved in dietary restriction-mediated extending longevity (48). Some probiotic candidates, such as Weissella species and L. salivarius can affect the reproduction and locomotor activity which might need some further safety evaluation before application (49, 50).
Several lifespan extension signaling pathways have been identified in C. elegans. Among them, the Insulin/IGF-1 signaling plays the critical role in longevity of the C. elegans (51). In this study, we demonstrated that the Insulin/IGF-1 pathway, such as the receptor daf-2 and the downstream transcription factor daf-16, was involved in the PA-mediated prolongevity effects. Other studies prove that some probiotic strains can influence aging and longevity via the daf-2/daf-16 insulin signaling pathway (52–54). However, the prolongevity regulated by L. gasseri SBT2055 in C. elegans is independent on the daf-2/daf-16 pathway, and Bifidobacterium longum BB68 regulates the lifespan via daf-16 independent on the daf-2 pathway, suggesting that the probiotics are strain specific (10, 55). Moreover, the JNK/MAPK signaling is involved in the PA-mediated longevity of C. elegans. As an upstream regulator, jnk-1 interacts with daf-16 and is considered to be prominently responsible for the immune response and stress (51). B. longum BB68 is confirmed to activate the TIR-1/JNK-1/daf-16 signaling and positively regulates longevity (55). Besides, the pro-longevity effects of PA are independent on the raga-1 pathway.
Reactive oxygen species are known to cause damage to DNA and cumulative genomic damage accelerates aging and influences the health span (56). In this study, the PA showed significant higher ROS-scavenging ability compared with control, and promoted the anti-oxidative sod-3 gene expression (Figure 3). B. longum BB68 is also demonstrated to increase sod-3 expression which is downstream of the daf-16 signaling and might contribute to extending the lifespan of C. elegans (55). Meanwhile, PA could reduce the absorption of fluorescence-labeled total fatty acids in the intestine, enhance the fatty acid conversion via fat-4 activation, and increase the lipid hydrolysis via lipl-4 upregulation, thus reducing fat storage in body as proved by the observation of oil red staining (Figure 3). Consistently, an earlier report finds that the lipid hydrolysis and disposition are modulated by aspirin through fat-4 and lipl-4 activation (27). A probiotic Lactobacillus rhamnosus CNCM I-3690 is found to inhibit the total fat deposit in C. elegans via another fatty acid desaturase gene fat-7 (54).
To investigate whether PA-feeding activates the chloride channels, we detected the mRNA expression of chloride related genes and performed survival assays on two highly activated gene mutant worms by PA. clh-1, clh-2, clh-3, and clh-4 are orthologs of human CLC2 gene which is associated with retina degeneration, constipation, intestinal mucosa repair, epilepsy, and so on (57). Another sulp-6 is an ortholog of human SLC26A3, SLC26A4, and SLC26A5 genes. SLC26A3 is a key chloride-bicarbonate exchanger protein contributed to infectious diarrhea and inflammatory bowel disease (IBD) (58). SLC26A4 and SLC26A5 genes are risk loci for human asthma and hearing loss, respectively (59, 60). Taken together, chloride ion-dependent genes, such as sulp-6, clh-1, and clh-4 are associated with the integrity of intestinal mucosa barrier. Consistently, sulp-6, clh-1, or clh-4 mutant worms had a decreased lifespan compared with the N2 worms (Figure 4). The significant effect on lifespan of the sulp-6, clh-1, or clh-4 mutant worms by PA might be beneficial to intestinal mucosa barrier. More importantly, we identified the nrfl-1 as the key regulator of longevity initiated by PA intake. nrfl-1, which encodes a human ortholog of Na+/H+ exchanger regulatory factor-1 (NHERF1, also called SLC9A3R1), can interact with chloride CFTR channel and regulate the concentration of Cl–/HCO3– during the capacitation of spermatozoa (61). There exists a SLC9A3R1/PTEN signaling that NHERF can recruit PTEN to bind PDGFR on membrane, leading to an inhibition of PTEN signaling pathway (62). Another study confirmed that PTEN (a worm ortholog of daf-18) signaling was upstream of insulin/IGF-1 signaling (63). Thus, we propose a mechanism that the nrfl-1/daf-18 signaling was stimulated by PA feeding, which subsequently activated insulin/IGF-1 signaling, and might play a role in PA-mediated extending lifespan (Figure 5).
The chloride ion plays an important role in the cell-volume regulation, mucus secretion, neuroexcitation development, and so on (64). The increasing evidence reveals that chloride channels and chloride ion dependent genes are involved in various diseases, such as cystic fibrosis, epilepsy, osteopetrosis, myotonia, and hyperekplexia (65). Its beneficial effect regulated by probiotic Lactobacillus acidophilus on host stimulation of Cl–/OH– exchange activity was first reported in animals (66). Besides, Bifidobacterium breve C50 has also been confirmed to promote intestinal homeostasis by controlling chloride ion secretion (67). More importantly, a body tissue chloride ion influx in high fat-fed rats is found in Lactobacillus casei intervention group and subsequently this probiotic can prevent colitis by activating chloride ion dependent channel CFTR in mice (68, 69). In this study, the mRNA increase of chloride channel clh-1 and clh-4 as well as chloride exchanger sulp6 which promote the lifespan of clh-1 or clh-4 mutant worms might contribute to enhance the intestinal electrolyte absorption and underlie the potential effects of PA (Figure 4). Further, the mRNA increase of chloride-related nrfl-1 but not prolonging the lifespan of nrfl-1 mutant worm by PA demonstrated that the PA-mediated lifespan extending was dependent on nrfl-1. Besides, we found that PA could increase anti-inflammatory atf-7 and fshr-1 mRNA levels in clh-1 or clh-4 mutant worms whereas downregulate inflammatory xbp-1 and ilys-3 mRNA levels in clh-1 mutant and bar mRNA level in clh-4 mutant (Figure 4) (70). This might be contributed to the PA-mediated extending lifespan of clh-1 or clh-4 mutant worms.
In conclusion, the PA with extending the longevity of C. elegans were widely investigated in this study (Figure 5). On one hand, the PA could modulate the daf-2 expression and nucleus location of daf-16 via the Insulin/IGF-1 pathway, and the increased sod-3 expression was the downstream of daf-16. Meanwhile, the PA not only decreased the intestinal fatty acids absorption, but also regulated the lipid hydrolysis and disposition via fat-4 and lipl-4 expression, thus reducing ROS accumulation. On the other hand, PA could significantly alter the chloride ion-associated genes mediated inflammatory state and extend the longevity of C. elegans. Therefore, it is speculated that the PA might play an important role in the anti-aging activities via the modulation of chloride ion-related genes, providing a new perspective to explore the lifespan extending effect. Besides, the study has a vital referential significance for other probiotics screening and provides the theoretical basis for its future application.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author Contributions
YZ, XWa, and XWe designed experiments. RH and YZ carried out the experiments, analyzed the experimental results, prepared the original draft, and performed the statistical analysis. WQ, YLe, YLo, and XL finished the validation. YZ, XWa, and XWe reviewed and edited the manuscript. XWa and JL acquired resources. All authors contributed to the article and approved the submitted version.
Funding
This research was supported by the National Key Research and Development Program of China (2018YFC2000504); the Fundamental Research Funds for the Central Universities (No. 06500060); the Beijing Talents Foundation (Nos. 2017000021223ZK27 and 2017000021223ZK29); and the Beijing Municipal Science and Technology Project (No. Z181100009318004).
Conflict of Interest
YZ, YLe, YLo, XL, JL, XWa, and XWe were employed by Beijing Solidwill Sci-Tech Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fnut.2022.821685/full#supplementary-material
References
1. Cheng XJ, Yang Y, Schwebel DC, Liu ZY, Li L, Cheng PX, et al. Population ageing and mortality during 1990-2017: a global decomposition analysis. PLoS Med. (2020) 17:17. doi: 10.1371/journal.pmed.1003138
2. Li JM, Han XL, Zhang XX, Wang SX. Spatiotemporal evolution of global population ageing from 1960 to 2017. BMC Public Health. (2019) 19:15. doi: 10.1186/s12889-019-6465-2
3. Zhavoronkov A, Li R, Ma C, Mamoshina P. Deep biomarkers of aging and longevity: from research to applications. Aging (Albany NY). (2019) 11:10771–80. doi: 10.18632/aging.102475
4. Woodhams DC, Bletz MC, Becker CG, Bender HA, Buitrago-Rosas D, Diebboll H, et al. Host-associated microbiomes are predicted by immune system complexity and climate. Genome Biol. (2020) 21:20. doi: 10.1186/s13059-019-1908-8
5. Zheng DP, Liwinski T, Elinav E. Interaction between microbiota and immunity in health and disease. Cell Res. (2020) 30:492–506. doi: 10.1038/s41422-020-0332-7
6. Huttenhower C, Gevers D, Knight R, Abubucker S, Badger JH, Chinwalla AT, et al. Structure, function and diversity of the healthy human microbiome. Nature. (2012) 486:207–14. doi: 10.1038/nature11234
7. DeJong EN, Surette MG, Bowdish DME. The gut microbiota and unhealthy aging: disentangling cause from consequence. Cell Host Microbe. (2020) 28:180–9. doi: 10.1016/j.chom.2020.07.013
8. Ctoi AF, Corina A, Katsiki N, Vodnar DC, Andreicut AD, Stoian AP, et al. Gut microbiota and aging-A focus on centenarians. Biochim Biophys Acta Mol Basis Dis. (2020) 1866:165765. doi: 10.1016/j.bbadis.2020.165765
9. Sharma K, Pooranachithra M, Balamurugan K, Goel G. Multivariate analysis of increase in life span of Caenorhabditis elegans through intestinal colonization by indigenous probiotic strains. Probiotics Antimicrob Proteins. (2019) 11:865–73. doi: 10.1007/s12602-018-9420-0
10. Nakagawa H, Shiozaki T, Kobatake E, Hosoya T, Moriya T, Sakai F, et al. Effects and mechanisms of prolongevity induced by Lactobacillus gasseri SBT2055 in Caenorhabditis elegans. Aging Cell. (2016) 15:227–36. doi: 10.1111/acel.12431
11. Barros PP, Scorzoni L, Ribeiro FC, Fugisaki LRO, Fuchs BB, Mylonakis E, et al. Lactobacillus paracasei 28.4 reduces in vitro hyphae formation of Candida albicans and prevents the filamentation in an experimental model of Caenorhabditis elegans. Microb Pathog. (2018) 117:80–7. doi: 10.1016/j.micpath.2018.02.019
12. Komura T, Ikeda T, Yasui C, Saeki S, Nishikawa Y. Mechanism underlying prolongevity induced by bifidobacteria in Caenorhabditis elegans. Biogerontology. (2013) 14:73–87. doi: 10.1007/s10522-012-9411-6
13. Kumar A, Baruah A, Tomioka M, Iino Y, Kalita MC, Khan M. Caenorhabditis elegans: a model to understand host-microbe interactions. Cell Mol Life Sci. (2020) 77:1229–49. doi: 10.1007/s00018-019-03319-7
14. Li WJ, Wang CW, Tao L, Yan YH, Zhang MJ, Liu ZX, et al. Insulin signaling regulates longevity through protein phosphorylation in Caenorhabditis elegans. Nat Commun. (2021) 12:4568. doi: 10.1038/s41467-021-24816-z
15. Irazoqui JE, Urbach JM, Ausubel FM. Evolution of host innate defence: insights from Caenorhabditis elegans and primitive invertebrates. Nat Rev Immunol. (2010) 10:47–58. doi: 10.1038/nri2689
16. Han B, Sivaramakrishnan P, Lin CJ, Neve IAA, He J, Tay LWR, et al. Microbial genetic composition tunes host longevity. Cell. (2017) 169:1249–62. doi: 10.1016/j.cell.2017.05.036
17. Dinic M, Herholz M, Kacarevic U, Radojevic D, Novovic K, Dokic J, et al. Host-commensal interaction promotes health and lifespan in Caenorhabditis elegans through the activation of HLH-30/TFEB-mediated autophagy. Aging (Albany NY). (2021) 13:8040–54. doi: 10.18632/aging.202885
18. Wang S, Yao B, Gao H, Zang J, Tao S, Zhang S, et al. Combined supplementation of Lactobacillus fermentum and Pediococcus acidilactici promoted growth performance, alleviated inflammation, and modulated intestinal microbiota in weaned pigs. BMC Vet Res. (2019) 15:239. doi: 10.1186/s12917-019-1991-9
19. Tan K, Deng D, Ma X, Cui Y, Tian Z. Pediococcus acidilactici P25 protected Caenorhabditis elegans against enterotoxigenic Escherichia coli K88 infection and transcriptomic analysis of its potential mechanisms. Biomed Res Int. (2020) 2020:7340312. doi: 10.1155/2020/7340312
20. Ashouri G, Mahboobi Soofiani N, Hoseinifar SH, Jalali SAH, Morshedi V, Valinassab T, et al. Influence of dietary sodium alginate and Pediococcus acidilactici on liver antioxidant status, intestinal lysozyme gene expression, histomorphology, microbiota, and digestive enzymes activity, in Asian sea bass (Lates calcarifer) juveniles. Aquaculture. (2020) 518:734638. doi: 10.1016/j.aquaculture.2019.734638
21. Sanzo-Machuca A, Moreno JMM, Casado-Navarro R, Karakuzu O, Guerrero-Gomez D, Fierro-Gonzalez JC, et al. Redox-dependent and redox-independent functions of Caenorhabditis elegans thioredoxin 1. Redox Biol. (2019) 24:8. doi: 10.1016/j.redox.2019.101178
22. Li HM, Roxo M, Cheng XL, Zhang SX, Cheng HR, Wink M. Pro-oxidant and lifespan extension effects of caffeine and related methylxanthines in Caenorhabditis elegans. Food Chem X. (2019) 1:9. doi: 10.1016/j.fochx.2019.100005
23. Zhang XY, Li W, Tang YZ, Lin CX, Cao Y, Chen YJ. Mechanism of pentagalloyl glucose in alleviating fat accumulation in Caenorhabditis elegans. J Agric Food Chem. (2019) 67:14110–20. doi: 10.1021/acs.jafc.9b06167
24. Mouchiroud L, Houtkooper RH, Moullan N, Katsyuba E, Ryu D, Canto C, et al. The NAD(+)/sirtuin pathway modulates longevity through activation of mitochondrial UPR and FOXO signaling. Cell. (2013) 154:430–41. doi: 10.1016/j.cell.2013.06.016
25. Phulara SC, Pandey S, Jha A, Chauhan PS, Gupta P, Shukla V. Hemiterpene compound, 3,3-dimethylallyl alcohol promotes longevity and neuroprotection in Caenorhabditis elegans. Geroscience. (2021) 43:791–807. doi: 10.1007/s11357-020-00241-w
26. O’Rourke EJ, Soukas AA, Carr CE, Ruvkun G. C. elegans major fats are stored in vesicles distinct from lysosome-related organelles. Cell Metab. (2009) 10:430–5. doi: 10.1016/j.cmet.2009.10.002
27. Huang XB, Mu XH, Wan QL, He XM, Wu GS, Luo HR, et al. Aspirin increases metabolism through germline signalling to extend the lifespan of Caenorhabditis elegans. PLoS One. (2017) 12:14. doi: 10.1371/journal.pone.0184027
28. Zhang Q, Wu XY, Chen P, Liu LM, Xin N, Tian Y, et al. The mitochondrial unfolded protein response is mediated cell-non-autonomously by retromer-dependent Wnt signaling. Cell. (2018) 174:870–83.e17. doi: 10.1016/j.cell.2018.06.029
29. Vaiserman AM, Koliada AK, Marotta F. Gut microbiota: a player in aging and a target for anti-aging intervention. Ageing Res Rev. (2017) 35:36–45. doi: 10.1016/j.arr.2017.01.001
30. Roselli M, Schifano E, Guantario B, Zinno P, Uccelletti D, Devirgiliis C. Caenorhabditis elegans and probiotics interactions from a prolongevity perspective. Int J Mol Sci. (2019) 20:14. doi: 10.3390/ijms20205020
31. Lapierre LR, Hansen M. Lessons from C. elegans: signaling pathways for longevity. Trends Endocrinol Metab. (2012) 23:637–44. doi: 10.1016/j.tem.2012.07.007
32. Wolff S, Ma H, Burch D, Maciel GA, Hunter T, Dillin A. SMK-1, an essential regulator of DAF-16-mediated longevity. Cell. (2006) 124:1039–53. doi: 10.1016/j.cell.2005.12.042
33. Mullaney BC, Ashrafi K. C-elegans fat storage and metabolic regulation. Biochim Biophys Acta Mol Cell Biol Lipids. (2009) 1791:474–8. doi: 10.1016/j.bbalip.2008.12.013
34. Koopman R, Schaart G, Hesselink MKC. Optimisation of oil red O staining permits combination with immunofluorescence and automated quantification of lipids. Histochem Cell Biol. (2001) 116:63–8.
35. Escorcia W, Ruter DL, Nhan J, Curran SP. Quantification of lipid abundance and evaluation of lipid distribution in Caenorhabditis elegans by Nile red and oil red O staining. J Vis Exp. (2018) 6:57352. doi: 10.3791/57352
36. Mutlu AS, Duffy J, Wang MC. Lipid metabolism and lipid signals in aging and longevity. Dev Cell. (2021) 56:1394–407. doi: 10.1016/j.devcel.2021.03.034
37. Miranda-Vizuete A, Veal EA. Caenorhabditis elegans as a model for understanding ROS function in physiology and disease. Redox Biol. (2017) 11:708–14. doi: 10.1016/j.redox.2016.12.020
38. Kumar S, Egan BM, Kocsisova Z, Schneider DL, Murphy JT, Diwan A, et al. Lifespan extension in C-elegans caused by bacterial colonization of the intestine and subsequent activation of an innate immune response. Dev Cell. (2019) 49:100. doi: 10.1016/j.devcel.2019.03.010
39. Pukkila-Worley R, Ausubel FM. Immune defense mechanisms in the Caenorhabditis elegans intestinal epithelium. Curr Opin Immunol. (2012) 24:3–9. doi: 10.1016/j.coi.2011.10.004
40. Fletcher M, Tillman EJ, Butty VL, Levine SS, Kim DH. Global transcriptional regulation of innate immunity by ATF-7 in C. elegans. PLoS Genet. (2019) 15:14. doi: 10.1371/journal.pgen.1007830
41. Powell JR, Kim DH, Ausubel FM. The G protein-coupled receptor FSHR-1 is required for the Caenorhabditis elegans innate immune response. Proc Natl Acad Sci U S A. (2009) 106:2782–7. doi: 10.1073/pnas.0813048106
42. Richardson CE, Kinkel S, Kim DH. Physiological IRE-1-XBP-1 and PEK-1 signaling in Caenorhabditis elegans larval development and immunity. PLoS Genet. (2011) 7:10. doi: 10.1371/journal.pgen.1002391
43. Shi YJ, Li Q, Shao ZY. Wnts promote synaptic assembly through T-cell specific transcription factors in Caenorhabditis elegans. Front Molec Neurosci. (2018) 11:14. doi: 10.3389/fnmol.2018.00194
44. Gravato-Nobre MJ, Vaz F, Filipe S, Chalmers R, Hodgkin J. The invertebrate lysozyme effector ILYS-3 is systemically activated in response to danger signals and confers antimicrobial protection in C. elegans. PLoS Pathog. (2016) 12:42. doi: 10.1371/journal.ppat.1005826
45. Lu MY, Wang Z. Microbiota and aging. Adv Exp Med Biol. (2018) 1086:141–56. doi: 10.1007/978-981-13-1117-8-9
46. Lee J, Yun HS, Cho KW, Oh S, Kim SH, Chun T, et al. Evaluation of probiotic characteristics of newly isolated Lactobacillus spp.: immune modulation and longevity. Int J Food Microbiol. (2011) 148:80–6. doi: 10.1016/j.ijfoodmicro.2011.05.003
47. Walker G, Houthoofd K, Vanfleteren JR, Gems D. Dietary restriction in C. elegans: from rate-of-living effects to nutrient sensing pathways. Mech Ageing Dev. (2005) 126:929–37. doi: 10.1016/j.mad.2005.03.014
48. Stanfel MN, Shamieh LS, Kaeberlein M, Kennedy BK. The TOR pathway comes of age. Biochim Biophys Acta Gen Subj. (2009) 1790:1067–74. doi: 10.1016/j.bbagen.2009.06.007
49. Zhao Y, Zhao L, Zheng XN, Fu TJ, Guo HY, Zhang H, et al. Lactobacillus salivarius strain FDB89 induced longevity in Caenorhabditis elegans by dietary restriction. J Microbiol. (2013) 51:183–8. doi: 10.1007/s12275-013-2076-2
50. Lee J, Kwon G, Lim YH. Elucidating the mechanism of Weissella-dependent lifespan extension in Caenorhabditis elegans. Sci Rep. (2015) 5:14. doi: 10.1038/srep17128
51. Marudhupandiyan S, Balamurugan K. Intrinsic JNK-MAPK pathway involvement requires daf-16-mediated immune response during Shigella flexneri infection in C-elegans. Immunol Res. (2017) 65:609–21. doi: 10.1007/s12026-016-8879-6
52. Martorell P, Llopis S, Gonzalez N, Chenoll E, Lopez-Carreras N, Aleixandre A, et al. Probiotic strain Bifidobacterium animalis subsp lactis CECT 8145 reduces fat content and modulates lipid metabolism and antioxidant response in Caenorhabditis elegans. J Agric Food Chem. (2016) 64:3462–72. doi: 10.1021/acs.jafc.5b05934
53. Kato M, Hamazaki Y, Sun S, Nishikawa Y, Kage-Nakadai E. Clostridium butyricum MIYAIRI 588 increases the lifespan and multiple-stress resistance of Caenorhabditis elegans. Nutrients. (2018) 10:13. doi: 10.3390/nu10121921
54. Grompone G, Martorell P, Llopis S, Gonzalez N, Genoves S, Mulet AP, et al. Anti-Inflammatory Lactobacillus rhamnosus CNCM I-3690 strain protects against oxidative stress and increases lifespan in Caenorhabditis elegans. PLoS One. (2012) 7:13. doi: 10.1371/journal.pone.0052493
55. Zhao L, Zhao Y, Liu RH, Zheng XN, Zhang M, Guo HY, et al. The transcription factor DAF-16 is essential for increased longevity in C. elegans exposed to Bifidobacterium longum BB68. Sci Rep. (2017) 7:7. doi: 10.1038/s41598-017-07974-3
56. Khan SS, Singer BD, Vaughan DE. Molecular and physiological manifestations and measurement of aging in humans. Aging Cell. (2017) 16:624–33. doi: 10.1111/acel.12601
57. Wang HW, Xu MH, Kong QJ, Sun P, Yan FY, Tian WY, et al. Research and progress on ClC-2 (Review). Mol Med Rep. (2017) 16:11–22. doi: 10.3892/mmr.2017.6600
58. Yu Q. Slc26a3 (DRA) in the gut: expression, function, regulation, role in infectious diarrhea and inflammatory bowel disease. Inflamm Bowel Dis. (2021) 27:575–84. doi: 10.1093/ibd/izaa256
59. Yick CY, Zwinderman AH, Kunst PW, Grunberg K, Mauad T, Dijkhuis A, et al. Transcriptome sequencing (RNA-Seq) of human endobronchial biopsies: asthma versus controls. Eur Respir J. (2013) 42:662–70. doi: 10.1183/09031936.00115412
60. Hosoya M, Fujioka M, Kobayashi R, Okano H, Ogawa K. Overlapping expression of anion exchangers in the cochlea of a non-human primate suggests functional compensation. Neurosci Res. (2016) 110:1–10. doi: 10.1016/j.neures.2016.04.002
61. Lobo M, Amaral MD, Zaccolo M, Farinha CM. EPAC1 activation by cAMP stabilizes CFTR at the membrane by promoting its interaction with NHERF1. J Cell Sci. (2016) 129:2599–612. doi: 10.1242/jcs.185629
62. Natalia S, Jan TG, Miguel V, Wiljan JA, Rafael P. PTEN-PDZ domain interactions: binding of PTEN to PDZ domains of PTPN13. Methods. (2015) 77–78:147–56. doi: 10.1016/j.ymeth.2014.10.017
63. Park HE, Hwang W, Ham S, Kim E, Altintas O, Park S, et al. A PTEN variant uncouples longevity from impaired fitness in Caenorhabditis elegans with reduced insulin/IGF-1 signaling. Nat Commun. (2021) 12:5631. doi: 10.1038/S41467-021-25920-W
64. Duran C, Thompson CH, Xiao QH, Hartzell HC. Chloride channels: often enigmatic, rarely predictable. Annu Rev Physiol. (2010) 72:95–121. doi: 10.1146/annurev-physiol-021909-135811
65. Verkman AS, Galietta LJV. Chloride channels as drug targets. Nat Rev Drug Discov. (2009) 8:153–71. doi: 10.1038/nrd2780
66. Raheja G, Singh V, Ma K, Boumendjel R, Borthakur A, Gill RK, et al. Lactobacillus acidophilus stimulates the expression of SLC26A3 via a transcriptional mechanism. Am J Physiol Gastroint Liver Physiol. (2010) 298:G395–401. doi: 10.1152/ajpgi.00465.2009
67. Heuvelin E, Lebreton C, Bichara M, Cerf-Bensussan N, Heyman MA. Bifidobacterium probiotic strain and its soluble factors alleviate chloride secretion by human intestinal epithelial cells. J Nutr. (2010) 140:7–11. doi: 10.3945/jn.109.114553
68. Zhang Y, Guo X, Guo JL, He QW, Li H, Song YQ, et al. Lactobacillus casei reduces susceptibility to type 2 diabetes via microbiota-mediated body chloride ion influx. Sci Rep. (2014) 4:10. doi: 10.1038/srep05654
69. Cohen LB, Troemel ER. Microbial pathogenesis and host defense in the nematode C-elegans. Curr Opin Microbiol. (2015) 23:94–101. doi: 10.1016/j.mib.2014.11.009
Keywords: longevity, C. elegans, probiotic, Pediococcus acidilactici, chloride ion related genes
Citation: Hu R, Zhang Y, Qian W, Leng Y, Long Y, Liu X, Li J, Wan X and Wei X (2022) Pediococcus acidilactici Promotes the Longevity of C. elegans by Regulating the Insulin/IGF-1 and JNK/MAPK Signaling, Fat Accumulation and Chloride Ion. Front. Nutr. 9:821685. doi: 10.3389/fnut.2022.821685
Received: 14 January 2022; Accepted: 21 February 2022;
Published: 01 April 2022.
Edited by:
Quancai Sun, Jiangsu University, ChinaReviewed by:
Virendra Shukla, Hebrew University of Jerusalem, IsraelWei Zhao, Jiangnan University, China
Jing Wang, Beijing Technology and Business University, China
Copyright © 2022 Hu, Zhang, Qian, Leng, Long, Liu, Li, Wan and Wei. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiangyuan Wan, d2FueGlhbmd5dWFuQHVzdGIuZWR1LmNu; Xun Wei, d2VpeHVuQHVzdGIuZWR1LmNu
†These authors have contributed equally to this work
 Rui Hu1,2†
Rui Hu1,2† Xiangyuan Wan
Xiangyuan Wan