
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Microbiol. , 11 December 2023
Sec. Food Microbiology
Volume 14 - 2023 | https://doi.org/10.3389/fmicb.2023.1289937
This article is part of the Research Topic The Weissella and Periweissella genera: Taxonomy, Detection, Safety and Their Application in Food and Health View all articles
 Vincenzina Fusco1
Vincenzina Fusco1 Daniele Chieffi1
Daniele Chieffi1 Francesca Fanelli1
Francesca Fanelli1 Marco Montemurro1
Marco Montemurro1 Carlo Giuseppe Rizzello2
Carlo Giuseppe Rizzello2 Charles M. A. P. Franz3*
Charles M. A. P. Franz3*Bacteria belonging to the genera Weissella and Periweissella are lactic acid bacteria, which emerged in the last decades for their probiotic and biotechnological potential. In 2015, an article reviewing the scientific literature till that date on the taxonomy, ecology, and biotechnological potential of the Weissella genus was published. Since then, the number of studies on this genus has increased enormously, several novel species have been discovered, the taxonomy of the genus underwent changes and new insights into the safety, and biotechnological and probiotic potential of weissellas and periweissellas could be gained. Here, we provide an updated overview (from 2015 until today) of the taxonomy, ecology, safety, biotechnological, and probiotic potential of these lactic acid bacteria.
The genus Weissella was first described by Collins et al. (1993), who isolated a group of Leuconostoc-like microorganisms during a survey of the lactic acid microbiota of dry naturally fermented Greek sausage. The novel genus was named Weissella M.L. dim. fem. after Norbert Weiss, acknowledged microbiologist which largely contributed to the lactic acid bacteria (LAB) taxonomy. In this first description, seven species were biochemically characterized and differentiated by 16S rRNA gene sequence analysis: W. confusa comb. nov. (described in Holzapfel and Kandler, 1969; Kandler and Weiss, 1986) W. halotolerans comb. nov. (described in Kandler et al., 1983), W. kandleri (described in Holzapfel and van Wyk, 1982), W. minor comb. nov. (described in Kandler et al., 1983), W. paramesenteroides (described by Garvie, 1967, 1986), W. viridescens (described by Niven and Evans, 1957; Kandler and Weiss, 1986), which is the type species of this genus, and W. hellenica sp. nov. (Collins et al., 1993). Sources of isolation and type strains are indicated in Table 1.
In 2000, Tanasupawat et al. (2000) isolated some LAB from fermented fish in Thailand whose DNA–DNA genetic relatedness toward the previously described Weissella species allowed the authors to assign them as W. thailandensis sp. nov. and identified the type strain as FS61-1T. In 2002, three additional species were described and included into the Weissella genus: W. cibaria, W. soli, and W. koreensis. W. cibaria was described by Björkroth et al. (2002): The authors selected 37 isolates, from humans and animal clinical samples as well as from foods sold in Malaysia and differentiated this species based on DNA–DNA reassociation experiments, which showed hybridization levels below 49% toward W. confusa. The type strain of this species is W. cibaria CCUG 41967T, which was isolated from the popular food ingredient Chili Bo. W. soli was isolated from garden soil by Magnusson et al. (2002) and showed relatedness to W. kandleri and W. confusa (95.5 and 95.3% 16S rRNA gene sequence identity, respectively). Lee et al. (2002) isolated from kimchi (a Korean fermented vegetable food) some strains with 97.2% 16S rRNA gene sequence identity to W. kandleri. The novel species was named W. koreensis, and the designated type strain was KCTC 3621T.
In 2010, Padonou et al. (2010) characterized the novel species W. beninensis sp. nov., isolated from submerged fermenting cassava in Ketou, Benin, and, since this species was demonstrated as being motile, they emended the description of Weissella genus that until then only comprised non-motile species. W. fabaria and W. fabalis were described in De Bruyne et al. (2010) and Snauwaert et al. (2013), respectively. W. fabaria LMG 24289T was isolated from traditional heap fermentations of Ghanaian cocoa beans and, although it showed 99.5% 16S rRNA gene sequence identity toward W. ghanensis LMG 24286T, DNA–DNA hybridization and metabolic characteristics recognize them as a separate species. W. fabalis LMG 26217T was isolated from a Brazilian cocoa bean fermentation and had the highest 16S rRNA gene sequence identity toward W. fabaria LMG 24289T (97.7 %). The confirmation that this strain could have been recognized as a novel species was achieved by pheS gene sequence analysis, DNA–DNA hybridization, the MALDI-TOF MS profile, and biochemical analysis.
W. diestrammenae was isolated from the gut of a camel cricket (Diestrammena coreana) in 2013 by Oh et al. (2013) and showed the highest 16S rRNA gene sequence identity to W. koreensis (97.7 %). The type strain of this species is W. diestrammenae DSM 27940T. In the same period, Tohno et al. (2013) conducted a taxonomic study on SG25T and SG23. These LAB strains, Gram-positive and catalase-negative, were isolated from grains of fermented Japanese rice (Oryza sativa L. subsp. japonica). Genetic analysis showed that W. soli was the closest relative to this novel W. oryzae species (96.9% of 16S rRNA gene sequence identity). W. uvarum was described in 2014 by Nisiotou et al., who isolated this species while studying the microbiota of wine grapes (Vitis vinifera L.) from the Nemea region in Greece. Although W. uvarum 16S rRNA gene sequence showed a high identity to that of W. minor NRIC 1625T (99.1 %), the isolated strains were assigned to a novel species for their capacity to grow at 42°C and to ferment specific carbohydrates, by using a genetic approach including randomly amplified polymorphic DNA (RAPD), restriction enzyme analysis-pulse field gel electrophoresis (REA-PFGE), and repetitive sequence-based PCR (rep-PCR analyses).
The name W. bombi was assigned by Praet et al. (2015) to a novel LAB isolated from the bumble bee gut in the region of Ghent, Belgium, which showed a 99.6% 16S sequence identity to that of W. hellenica LMG 15125T. The analysis of the pheS gene sequences, DNA G+C content analysis, (GTG)5-PCR fingerprinting, DNA–DNA hybridization experiments, and a biochemical characterization led to the assignment of this novel species, whose type strain is LMG 28290T. W. ceti was first isolated in 2011 by Vela et al. (2011) within a study aimed at assessing the microbiota of beaked whales (Mesoplodon bidens). The type strain of this species is CCUG 59653T. In 2019, Heo et al. (2019) isolated from the gut of an insect, Cryptocercus kyebangensis sampled from mountains in South Korea, a novel organism that in a 16S rRNA gene phylogenetic tree analysis clustered with W. ghanensis, W. beninensis, W. fabaria, and W. fabalis-type strains and showed a sequence identity of 95.9% with both W. beninensis 2L24P13T and W. ghanensis LMG 24286T. Genomic and phenotypic analyses allowed the description of the novel species W. cryptocerci.
W. muntiaci was characterized by Lin et al. (2020), who isolated this Gram-positive strain from feces of a barking deer (Muntiacus reevesi) in Taiwan. W. muntiaci 8H-2T showed a 16S rRNA gene sequence identity with the type strains of W. oryzae, W. confusa, W. cibaria, and W. soli of 99.2, 97.8, 97.6, and 97.3 %, respectively. In the same year, Li et al. (2020) characterized three Gram-positive bacterial strains, named as X0750T, X0401, and X0278. The type strain, isolated from a traditional yogurt of the Chines Saga County, showed a 16S rRNA gene sequence 94.4–100 % identical to that of the type strains of W. hellenica, W. bombi, W. paramesenteroides, W. jogaejeotgali, W. thailandensis, W. oryzae, W. cibaria, and W. confusa. The resulting novel species was designated as W. sagaensis.
In 2021, Hyun et al. (2021) isolated some weissellas from the intestine of the diving beetle Cybister lewisianus that showed 98.9% 16S rRNA gene sequence identity and 79.5% ANI to W. koreensis KCTC 3621T. Based on phylogenetic, chemotaxonomic, phenotypic, and genotypic analyses, named this novel species W. coleopterorum, whose type strain is HDW19T.
Until 2022, the genus counted 26 species, including W. jogaejeotgali which was described by Lee et al. (2015b), but then was identified as a later heterotypic synonym of W. thailandensis, described by Tanasupawat et al. (2000), as well as W. kimchi, which was first described by Choi et al. (2002), but later correctly identified as the later heterotypic synonym of W. cibaria (Ennahar and Cai, 2004), described by Björkroth et al. (2002).
In 2022, Bello et al. (2022) revised the taxonomy of the genera Leuconostoc, Convivina, Oenococcus, Fructobacillus, and Weissella by using the concatenated sequences of 498 core proteins and the 16S rRNA gene phylogeny. The authors then transferred five species that clustered in a separate clade from the genus Weissella into the proposed novel genus Periweissella (P.): P. cryptocerci (basonym W. cryptocerci; Heo et al., 2019), P. beninensis (basonym W. beninensis; Padonou et al., 2010), P. fabalis (basonym W. fabalis; Snauwaert et al., 2013), P. fabaria (basonym W. fabaria; De Bruyne et al., 2010), and P. ghanensis (basonym W. ghanensis; De Bruyne et al., 2010) (Table 1). The same assumption was achieved by Fanelli et al. (2022) who, in the same year, showed that Weissella species could be clustered into six different species groups by a genome-based phylogenomic analysis, with one including the five species that Bello proposed as belonging to a separate novel genus. Furthermore, the phylogenomic species group clustering, in many cases, overlapped with the carbohydrate metabolism pathways (Fanelli et al., 2022).
In 2023, a novel species, namely W. fangxianensis sp. nov., was described by Xiang et al. (2023), who isolated three LAB strains from rice wine starter used in Fangxian County (China). This novel species was described using a polyphasic approach, based on genomic and metabolic analyses. Both 16S rRNA gene sequence and genomic phylogeny placed this species close to W. thailandensis and W. paramesenteroides. The type strain of this species is HBUAS51963T (Table 1).
Both the Weissella and Periweissella genera are members of the phylum Bacillota, class Bacilli, order Lactobacillales, and family Lactobacillaceae, recently revised by Zheng et al. (2020), who merged it with the Leuconostocaceae family.
All Weissella, such as other LAB, are Gram-positive and catalase-negative. They occur in short rods with tapered rounded ends or ovoid cells (Collins et al., 1993; Björkroth et al., 2014), and they have a tendency toward pleomorphism. Weissellas do not produce spores, and they are facultatively anaerobic chemoorganotrophs fermenting glucose heterofermentatively by way of the hexose-monophosphate and phosphoketolase pathways that deliver lactic acid as end products [DL or D(-), depending on the species], carbon dioxide, and ethanol and/or acetic acid. For their growth, weissellas have an obligate need for vitamins, peptides, amino acids, fatty acids, nucleic acids, and fermentable carbohydrates. Not all Weissella species hydrolyze arginine. Growth at 15°C has been detected for all weissellas, while only some species are able to grow at 42–45°C (Björkroth et al., 2014). All Weissella strains are able to utilize maltotriose, D-fructose, N-acetyl-D-glucosamine, turanose, D-mannose, and α-D glucose palatinose. The peptidoglycan of weissellas typically contains lysine as diamino acid and apart from W. kandleri which contains also glycine, and all weissellas contain alanine or alanine and serine in the interpeptide bridge (Holzapfel and van Wyk, 1982).
There are currently 20 Weissella species which, based on phylogenomic comparison by Fanelli et al. (2022) and the genomic data available for the W. fangxianensis species (Xiang et al., 2023), can be grouped into five species groups, i.e., the W. kandleri species group (W. kandleri, W. soli, W. diestrammenae, W. coleopterorum, W. koreensis), the W. oryzae species group (W. oryzae, W. muntiaci), the W. halotolerans group (W. halotolerans, W. ceti, W. uvarum, W. minor, W. viridescens), the W. confusa species group (W. confusa, W. cibaria), and the W. paramesenteroides species group (W. thailandensis, W. fangxianensis, W. paramesenteroides, W. bombi, W. hellenica, and W. sagaensis). The description of the type species and species in alphabetical order follows below.
Description of the type species:
Synonyms: Lactobacillus viridescens Niven and Evans 1957, p. 758; Lactobacillus corynoides subsp. corynoides Kandler and Abo-Elnaga, 1966, p. 573. Note that in the Approved List of Bacterial Names L. viridescens is incorrectly cited as Lactobacillus viridescens Kandler and Abo-Elnaga 1966, p. 573. For this strain, the utilization of sucrose concomitant with a possible production of dextran has not been studied.
vi.ri.des'cens. M.L. pres. part. viridescens, growing green, greening.
W. viridescens cells, which are non-motile, appear as irregular rods, occurring singly or in pairs, with rounded tapered ends. The interpeptide bridge of the peptidoglycan contains lysine, alanine, and serine. W. viridescens produces DL lactic acid from glucose, whereas it does not utilize arginine. The type strain (ATCC 12706T = DSM 20410T = LMG 3507T) was isolated from a discolored meat product. The mol% GC of the DNA is 41–44, and the GenBank accession nos. for the 16S rRNA gene sequence are ABO23236, M23040, and X52568.
Description of other Weissella species:
bom'bi. L. n. bombus a boom, a deep hollow noise, buzzing, also the zoological genus name of the bumble bee: N.L. gen. n. bombi of Bombus, of a bumble bee.
Cells, which are non-motile, occur as elongated cocci, in pairs or chains. W. bombi does not grow at 45°C and produces D(-) lactic acid from glucose. The type strain, isolated from the gut of a B. terrestris bumble bee, is LMG 28290T (=DSM 28794T). The mol% GC content of the DNA is 37.2, and the GenBank 16S rRNA gene sequence accession number is LK054487.
ce.ti. L. gen. n. ceti of a whale.
W. ceti are short rod-shaped or coccoid cells, which are non-motile and occur singly or in pairs. It can grow at 22°C and 37°C but not at 15°C or 42°C. W. ceti produces DL lactic acid from glucose and does not produce dextran from sucrose. The type strain hydrolyzes arginine, while this feature is variable among the other strains of this species. The type strain, isolated from the spleen of a beaked whale (Mesoplodon bidens), is CECT 7719T = LMG 30639T. The DNA G+C content of the type strain is 39.2 mol%, and the GenBank accession no. of the 16S rRNA nucleotide sequence is FN813251.
ci.ba'ri.a. L. adj. cibaria, pertaining to food.
W. cibaria cells are non-motile and occur as short rods in pairs. The interpeptide bridge structure is Lys-Ala-(Ser)-Ala. W. cibaria produces DL lactic acid from glucose and grows at 45°C and 15°C but not at 4°C. It produces ammonia from arginine and dextran from sucrose. The type strain, isolated from Malaysian chili bo, is DSM 15878T = LMG 17699T. It has a mol% GC content of the DNA of 44, and the 16S rRNA gene sequence GenBank accession n. is AJ295989.
co.le.o.pte.ro'rum. N.L. gen. pl. n. coleopterorum of Coleoptera, the name of the order of the beetles from which the strain was first isolated.
W. coleopterorum cells are typically non-motile and rod-shaped. It grows at 4–37°C. The amino acids in the peptidoglycan structure are alanine and lysine. Its DNA has a mol% GC content of 37.2%. The type strain isolated from the intestine of the diving beetle Cybister lewisianus is JCM 33684T = KCTC 43114T. The GenBank/EMBL/DDBJ accession number of the 16S rRNA gene sequence of the type strain is MN099422.
Synonyms: Lactobacillus confusus Garvie and Tilbury 1972, p. 396; Lactobacillus coprophilus subsp. confusus Holzapfel and Kandler 1969, p. 665.
con.fu'sus. L. v. confundere to confuse: L. past. part. confusus confused.
W. confusa has non-motile cells occurring as short rods, singly or (rarely) in short chains, with a tendency to be thicker at one of the ends. Lys-Ala is the amino acid that occurs in the interpeptide bridge of the peptidoglycan structure. The lactic acid enantiomers produced from glucose metabolism are DL. Growth at 45°C is variable with some strains capable of good growth at this temperature. Dextran is produced from sucrose and ammonia from arginine. The type strain, isolated from sugar cane, is DSM 20196T = ATCC 10881T = LMG 9497T. The DNA has a mol% GC content of 45–47, and the 16S rRNA gene sequence GenBank accession nos. are AB023241 and M23036.
di.es.tram.me'nae. N.L. gen. n. diestrammenae of Diestrammina, referring to Diestrammena coreana, a camel cricket from the gut of which this bacterium was isolated.
W. diestrammenae does not exhibit motility, and the cell's morphologies are coccoid or rod-shaped. It is capable of growth at 4–37°C in 0 to 4% NaCl. The bacteria can hydrolyze arginine to ammonia and produce D(-) lactic acid from glucose. The amino acids in the cell wall are Lys-Ala-Ser. The DNA has mol% GC content of 45, and the type strain, isolated from the gut of a camel cricket (Diestrammenae coreana) in South Korea, is JCM 18559T = DSM 27940T = LMG 30643T. The GenBank/EMBL/DDBJ accession number for the 16S rRNA gene of the type strain is JQ646523.
fang. xian. en'is. N.L. fem. adj. fangxianensis pertaining to Fangxian county, a city located in Hubei Province, PR China, where the type strain was first isolated.
Cells are non-motile, non-spore-forming and spherical. Growth occurs at 15–37°C but not at 10°C or 40°C, with optimum between 25 and 35°C. The genomic DNA mol% GC content is 38.6 mol%. The type strain, isolated from rice wine starter in Fangxian county Hubei Province, PR China, in 2021, is GDMCC 1.3506T = JCM 35803T.
Synonym: Lactobacillus halotolerans Kandler, Schillinger and Weiss, 1983, 672. Effective publication: kandler, Schillinger and Weisss, 1983, p. 283.
ha.lo.to'le.rans. Gr. n. hals, halos salt; L. part. adj. tolerans, tolerating, enduring; N.L. part.adj. halotolerans, salt-tolerating.
W. halotolerans does not exhibit motility and cells appear as irregular, short or even coccoid rods, with rounded to tapered ends with a tendency to form coiling chains. Cells were observed also to clump together. Lys-Ala-Ser is the amino acid composition of the interpeptide bridge of the peptidoglycan structure. W. halotolerans produces DL lactic acid from glucose and does not grow at 45°C, whereas it grows in 12% NaCl, with very weak growth occurring at 14% NaCl. Arginine is not metabolized, and dextran production from sucrose has not been investigated.
hel.le'ni.ca. Gr. masc. adj. hellênikos, Greek; N.L. fem. adj. hellenica, Greece, from where the bacterium was first isolated.
The non-motile cells of this species are spherical but sometimes also show a lenticular morphology and generally occur in pairs or short chains. A tendency to associate in clusters was observed. W. hellenica grow at 10°C and 4°C (delayed) but not at 37°C. All strains produce D(-) lactic acid from glucose. W. hellenica does not hydrolyze arginine and does not produce slime from sucrose. The cell wall murein is type Lys-L-Ala-L-Ser(L-Ala). The DNA base compositions of strains ranged between 39.4 and 40.0 mol% GC, respectively, and the type strain, isolated from fermented sausages, is NCFB 2973F = DSM 7378T = LMG 15125T. The 16S ribosomal gene GenBank accession number is NR_118771.1.
Synonym: Lactobacillus kandleri Holzapfel and van Wyk 1983, 439. Effective publication: Holzapfel and van Wyk 1982, 501.
kand'le.ri. M.L. gen. n. kandleri, of Kandler; named for O. Kandler, a German microbiologist.
W. kandleri cells were observed to be non-motile of partly irregular rod shape, and they were found to occur singly as well as in in pairs but seldom in short chains. Lys-Ala-Gly-Ala2 was the amino acid determined to occur in the interpeptide bridge of the peptidoglycan structure. W. kandleri produces DL lactic acid from glucose, ammonia from arginine, and dextran from sucrose. It does not grow at 45°C. The type strain, isolated from a desert spring, is DSM 20595T = LMG 18979T which has a 39 mol% GC content in the DNA. The GenBank accession numbers of the 16S rRNA gene are AB022922 and M23038.
ko.re.en'sis. N.L. adj. koreensis of Korea, where the novel organisms were isolated.
Cells are irregular, short, and rod-shaped or coccoid. No growth occurs at 42°C, but it grows at 10 and 37°C. It is capable of arginine hydrolysis and production of dextran from sucrose and D(-) lactic acid from glucose metabolism. The DNA has a G-C content of 37 mol%, and the cell wall was shown to contain Lys-Ala-Ser. The type strain, isolated from the traditional Korean fermented vegetable kimchee, is DSM 15830T = KCCM 41516T = JCM 11263T. The 16S rDNA sequences of the type strain have the GenBank/EMBL/DDBJ accession number AY035891.
Synonyms: Lactobacillus minor (Kandler, Schillinger and Weiss 1983, 672. Effective publication: Kandler, Schillinger and Weiss, 1983, 284. (Lactobacillus corynoides subsp. minor Abo-Elnaga and Kandler 1965, 128; Lactobacillus viridescens subsp. minor Kandler and Abo-Elnaga, 1966, 754).
mi'nor. L. comp. adj. minor smaller.
W. minor shows non-motile cells appearing as irregular short rods, occurring in pairs or short chains, with rounded to tapered ends often bent with unilateral swellings. The amino acid composition of the peptidoglycan structure is Lys-Ser-Ala2. It produces DL lactate from glucose and ammonia from arginine but does not produce dextran from sucrose. It does not grow at 45°C. The mol% GC content of the DNA is 44, and the type strain, which stems from the sludge of milking machines, is DSM 20014T = LMG 9847T. The GenBank accession no. of the 16S rRNA gene is M23039.
mun.ti′a.ci. N.L. gen. n. muntiaci of Muntiacus, a genus of the muntjacs, barking deer of Taiwan, from which the type strain was isolated.
W. muntiaci shows non-motile cells appearing as short rods occurring singly and rarely in pairs. It grows at 10 to 37°C but not at 4, 45, and 50°C. It produces ammonia from arginine and D(-) lactic acid from glucose. The amino acid composition of the peptidoglycan structure is A3α (L-Lys–L-Ala–L-Ser) with the presence of Glu, Ser, Ala, and Lys in a molar ratio of 1: 1: 3:1. The type strain was obtained from the feces of the Formosan barking deer. This feces were collected in the Fushan Botanical Garden, Yilan County, Taiwan, in 2017. The type strain is BCRC 81133T = NBRC 113537T. The genomic mol% GC content is 40.5. The 16S rRNA gene sequence of the type strain has the GenBank/EMBL/DDBJ accession number MK774696.
o.ry'za.e. L. gen. n. oryzae of rice, from which the type strain was isolated.
W. oryzae shows non-motile cells that are irregular, short rod-shaped, or coccoid and occur singly or in pairs and/or short chains. The peptidoglycan structure consists of glutamic acid, lysine, serine, and alanine. W. oryzae grows at 10–42°C but not at 4 or 50°C. It is a facultatively anaerobic lactic acid bacterium that does not produce dextran from sucrose and produces D(-) from glucose. The type strain DSM 25784T = LMG 30913T originates from fermented rice grain that was obtained in Tochigi, Japan. This strain has a DNA with mol%GC content of 40.6 mol%, while its 16S rRNA gene sequence has the GenBank/EMBL/DDBJ accession number AB690345.
pa.ra.me.sen.ter.oi'des. Gr. prep. para resembling; M.L. mesenteroides a specific epithet; M.L. adj. paramesenteroides, resembling Leuconostoc mesenteroides.
W. paramesenteroides has non-motile cells that are spherical but often also lenticular, occurring in pairs and chains. It grows at 30°C but optimally at 18–24°C. Lys-Ala2 or Lys-Ser-Ala2 is the amino acid occurring in the peptidoglycan structure. W. paramesenteroides produces D(-) lactate from glucose but not ammonium from arginine and dextran from sucrose. The DNA of the species has a mol% GC content ranging from 37 to 38, and the type strain, that originated from a dairy source, is DSM 20288T = LMG 9852T. The GenBank accession nos. for the 16S rRNA gene are AB023238, M23033, and X95982.
sa. ga. en′sis. N.L. fem. adj. sagaensis, pertaining to Saga County, a county located in Tibet Autonomous Region, PR China, where the bacterium was isolated.
W. sagaensis are non-motile cells which appear as rods that can occur singly, in pairs or in short chains. This bacterium can grow at 10–37°C but not at 5 or 45°C. It produces D(-) lactate from glucose and does not hydrolyze arginine. The interpeptide bridge contains serine and alanine. The DNA of the type strain has a mol% GC of 36.7. The culture collection numbers for the type strain are NCIMB 15192T = LMG 31184T, and the 16S rRNA gene sequence is stored in GenBank/ENA/DDBJ under the accession number LC438526.
so'li. L. n. solum soil; L. gen. n. soli, of the soil.
W. soli shows non-motile cells, occurring singly or in pairs, that are rod-shaped and often thickened at one end. The composition of the interpeptide bridge of the peptidoglycan structure is not known. W. soli produces D(-) lactate from glucose, dextran from sucrose, and not ammonia from arginine. It grows at 4–40°C but not at 45°C. The type strain that stems from soil is DSM 14420T = LMG 20113T, and the DNA of this strain has a mol% GC content of 43. The GenBank accession no. of the 16S rRNA gene is AY028260.
thai.lan'den.sis M.L. fem. adj. thailandensis pertaining to Thailand, where the strains were first isolated.
W. thailandensis occurs as non-motile coccoid cells arranged either in pairs or chains. D(-) is the major lactic acid enantiomer produced from glucose. It does not hydrolyze arginine and does not produce slime from sucrose. It grows at 25 to 37°C but not at 42°C, and the peptidoglycan structure contains L-Lys-L-Ala. The strains of this species have mol% GC contents of their DNA ranging from 38 ± 0 to 41.2. The type strain stems from pla-ra which is a Thai fermented fish and received the culture collection numbers DSM 15832T = LMG 19821T = JCM 10695T. The DDBJ accession number for the 16S rRNA gene sequence of the type strain is AB023838.
u.va'rum. L. fem. gen. pl. n. uvarum of grapes, where the type strain was isolated.
W. uvarum has non-motile cells appearing as cocci or short rods that occur singly, in pairs or short chains. It can grow at both 15 and 42°C but not at 4 or 45°C. It produces D(-) lactate from glucose, ammonia from arginine, and not dextran from sucrose. The type strain of this species is the only strain among weissellas and periweissellas to utilize D-arabitol and D-sorbitol (Fanelli et al., 2022). The mol% GC of the DNA is 39.1, and the type strain, isolated from grapes from the region of Nemea located in Greece, is DSM 28060T = LMG 30647T. The GenBank/EMBL/DDBJ accession number for the 16S rRNA gene sequence of the type strain is KF999666.
Bello et al. (2022) showed Weissella species to occur in two distinct clades in a core protein tree derived from the genomes, and comparative analyses, furthermore, identified various conserved signature indels in signature specific for the members of the two clades. The Weissellas, therefore, could be shown not to constitute a monophyletic group but instead comprise two distinct and unrelated clades, namely, a “main clade” and “clade 2.” The clade 2 Weissellas shared the presence of five conserved signature indels in the proteins amidophosphoribosyltransferase protein, DEAD/DEAH box helicase, ArgR family transcriptional regulator, Flp pilus assembly complex ATPase component (TadA), and hydroxyethylthiazole kinase. Species of clade 2 were thus proposed to belong to a novel genus, i.e., Periweissella gen. nov. (Bello et al., 2022). Periweissellas are Gram-positive and obligately heterofermentative bacteria that appear as non-spore-forming short rods or cocci. They grow at temperatures 15–37°C (optimum 28–30°C), and their DNA has a mol% GC content ranging from 35.4 to 41.1. Several species of this genus may hydrolyze arginine (Bello et al., 2022). The P. beninensis, P. fabalis-, P. fabaria-, and P. ghanensis-type strains are capable of utilizing α-ketobutyric acid, glycyl-L-methionine, and pyruvic acids, while only P. fabaria and P. fabalis are able to utilize D-malic acid, i-erythritol, and D-trehalose. The P. fabaria-type strain is the only strain among these which is able to utilize formic acid, while the P. fabalis-type strain metabolizes fumaric acid, glycyl-L-glutamine, and α-cyclodextrin (Fanelli et al., 2022).
Recent studies demonstrate that with the exception of P. cryptocerci, all Periweissella species possess genetic loci coding for flagellar-related proteins (Fanelli et al., 2023a; Qiao et al., 2023), and flagellar structures have been detected in P. beninensis-, P. ghanensis-, P. fabalis-, and P. fabaria-type strains (Qiao et al., 2023).
Pe. ri. weiss. el'la. Gr. prep. peri, about, around or nearby; N.L. fem. dim. n. Weissella, a bacterial genus named after Norbert Weiss, a German microbiologist; N.L. fem. dim. n. Periweissella, a genus about or nearby Weissella.
Description of the type species:
Synonym Weissella ghanensis De Bruyne, Camu, Lefebvre, de Vuyst, and Vandamme 2008, 2723vp
gha.nen'sis. N.L. fem. adj. ghanensis, pertaining to Ghana.
Cells are small rods appearing singly, in pairs or short chains. The type strain produces both the DL lactic acid enantiomers (DL 90:10) from glucose. W. ghanensis produces ammonium from arginine and slime from glucose. The mol% GC content of the DNA is 40.0, and the type strain (DSM 19935T = LMG 24286T) was isolated from Ghanaian cocoa heaps undergoing fermentation. The GenBank accession no. of the 16S rRNA gene is AM882997.
Description of other species:
Synonym Weissella beninensis Padonou, Schillinger, Nielsen, Franz, Hansen, Hounhouigan, Nago, and Jakobsen, 2010, 2196VP
ben.in.en'sis. N.L. fem. adj. beninensis, pertaining to Benin.
P. beninensis exhibits motility, and the cells were shown to possess with peritrichous flagella. Cells were determined to be short and rod-shaped or coccoid. Cells were observed to occur singly, in pairs or short chains. The bacterium is capable of growth at 15°C but not at 45°C. It hydrolyzes arginine and produces DL lactate from glucose. Most strains were observed to produce dextran from sucrose. Among weissellas and periweissellas, P. beninensis utilizes the widest range of carbohydrates tested (Fanelli et al., 2022). Indeed, the type strain of this species metabolizes α-D-lactose, D-melibiose, D-galactose, β-methyl-D-galactoside, pyruvic acid methyl ester, lactulose, sucrose, uridine-5'-monophosphate, and D-raffinose (Fanelli et al., 2022). The type strain was isolated from cassava fermentations in Ketou, Benin, and the mol% GC content is 37. The type strain is DSM 22752T (=LMG 25373T). The GenBank accession no. for the 16S rRNA gene sequence is EU439435.
Synonym Weissella cryptocerci Heo, Hamada, Cho, Weon, Kim, Hong, Kim, and Kwon 2019, 2805VP
cryp.to.cer'ci. N.L. gen. n. cryptocerci, of Cryptocercus, a genus of insect from which the species was isolated.
P. cryptocerci dos not exhibit motility, and cells appear to be rod-shaped. The bacterium grows at 4–35°C and can produce DL lactate from glucose. It does not hydrolyze arginine and does not produce slime from sucrose. The cell wall peptidoglycan is type A4α, characterized by an interpeptide bridge of Gly-D-Glu. The mol% GC content is 41.1, and the type strain, isolated from the gut of the insect Cryptocercus kyebangensis, was obtained in the mountainous area of Seoraksan, Yangyang-gun, Republic of Korea. The type strain was deposited as KACC 18423T = NBRC 113066T. Its GenBank accession number of the 16S rRNA gene is MK395366.
Synonym Weissella fabalis Snauwaert, Papalexandratou, De Vuyst, and Vandamme 2013, 1714VP
fa.ba'lis. L. fem. adj. fabalis of or belonging to beans.
P. fabalis did not show motility, and the cells were observed to be of coccoid morphology, occurring singly, in pairs or in short chains. The bacterium can grow at temperatures ranging from 15 to 37°C and in the presence of 5–6% NaCl but not in the presence of 7–8% NaCl. It produces ammonia from arginine and D-lactic acid from glucose. The DNA of the type strain has a mol% GC content of 37. The type strain, isolated from a Brazilian cocoa bean box fermentation carried out in Ilhéus, Bahia, Brazil, in 2007, is LMG 26217T = DSM 28407T. The 16S rRNA gene nucleotide sequence has the GenBank/EMBL/DDBJ accession number HE576795.
Synonym Weissella fabaria De Bruyne, Camu, De Vuyst, and Vandamme 2010, 2002VP
fa.ba'ri.a. L. fem. adj. fabaria of or belonging to beans.
P. fabaria was described to be non-motile, and the cells were determined to have a coccoid morphology, occurring singly, in pairs or short chains. Bacteria of this species produce both the D and L lactic acid enantiomers in a ratio 9:1. They grow at 15–37°C and produce ammonia from arginine and slime from glucose. L-Lys–L-Ala–L-Ser is the amino acid present in the peptidoglycan structure. The DNA of the type strain has a mol% GC content of 38.2 mol%. The type strain, isolated from a Ghanaian cocoa fermentation in 2004, is LMG 24289T = DSM 21416T. The 16S rRNA gene of the type strain has the GenBank/EMBL/DDBJ accession number FM179678.
From 2015 (Fusco et al., 2015) up to date, no advancements have been achieved in the isolation of weissellas: modified CHALMERS (Pepe et al., 2001) and de Man, Rogosa, and Sharpe (MRS) (DeMan et al., 1960) broth enrichment combined with plating on MRS agar added with 2,3,5-triphenyltetrazolium chloride (TTC) (Zamudio-Maya et al., 2008), remain the only media that, among other LAB, allow the differentiation and isolation of weissellas, while Leuconostoc selective medium (LUSM) (Benkerroum et al., 1993), sourdough bacteria (SDB) medium (Kline and Sugihara, 1971), and MRS, allow the isolation of weissellas apart from presumptive lactobacilli and Leuconostoc. As for the identification of weissellas, biochemical methods such as those based on the comparison of total soluble cell protein patterns (Dicks, 1995; Tsakalidou et al., 1997) and profiles of cellular fatty acids (Samelis et al., 1998), as well as commercial identification kits such as the Phoenix Automated Microbiology System (Becton Dickinson Diagnostic Systems, Sparks, MD), theVitek2 system (Bio Merieux, Marcy l'Etoile, France), the API50 CHL kit (BioMérieux, Lyon, France) (Lee K. W. et al., 2012), and the RapID™ STR System (Thermo Scientific, Hudson, NH, USA), do not allow an accurate and reliable identification (Fusco et al., 2015; Sturino, 2018). The culture-based identification of weissellas has been improved by matrix-assisted laser desorption–ionization time of flight mass spectrometry (MALDI-TOF MS) (Albesharat et al., 2011; Fairfax et al., 2014; Lee M. R. et al., 2015; Kim et al., 2017, 2021a,b; Nacef et al., 2017; Wang et al., 2020b; Joglekar et al., 2023).
As for the DNA-based taxonomical methods, to overcome the low reliability of 16S rRNA gene sequencing for discriminating highly phylogenetically related weissellas (Kulwichit et al., 2007; Fairfax et al., 2014; Medford et al., 2014; Joglekar et al., 2023), several methods have been developed such as ribotyping (Björkroth et al., 2002), amplified ribosomal DNA restriction analysis (ARDRA) (Jang et al., 2021), denaturing gradient gel electrophoresis (DGGE) of PCR amplified fragments of the 16S rRNA gene (Walter et al., 2001), and sequence typing of pheS, gyrB, and dnaA genes, with the pheS gene providing the better taxonomic resolution (Joglekar et al., 2023). A genus-specific PCR assay, targeting the 16Sr RNA gene, was developed by Schillinger et al. (2008) for the differentiation of Weissella and Leuconostoc. Fusco et al. (2011) developed a species-specific PCR for Weissella confusa from an AFLP (amplified fragment length polymorphism)-derived marker, whereas a conventional PCR and a real-time PCR were developed by Snyder et al. (2015) for the identification and quantification of W. ceti NC36. A real-time PCR assay was developed by Gómez-Rojo et al. (2015) to quantitatively detect W. viridescens in blood sausages, whereas following a pan-genome analysis, Kim et al. (2022b) designed species-specific pairs of primers for the real-time PCR detection of 11 Weissella species. Finally, Ma et al. (2022) developed an aptasensor based on fluorescence polarization for the detection of W. viridescens.
Culture-independent approaches including PCR-DGGE and next-generation sequencing approaches such as metagenetics and metagenomics have allowed the detection of weissellas in various ecological niches (Table 2). Whole-genome sequencing is another approach that is being widely used to identify and characterize weissellas (Benomar et al., 2011; Kim et al., 2011; Amari et al., 2012; Lee J. H. et al., 2012; Figueiredo et al., 2014a,b, 2015; Tanizawa et al., 2014; Malik et al., 2016; Heng et al., 2017; Ku et al., 2017; Li et al., 2017; Du et al., 2018; Garcia-Cancino et al., 2019; Kwak et al., 2019; Panthee et al., 2019; Lin et al., 2020; Månberger et al., 2020; Baugh et al., 2021; Contente et al., 2021; Jang et al., 2021; Patrone et al., 2021; Yuan et al., 2021; Apostolakos et al., 2022; Fanelli et al., 2022; Fukuda and Nolasco-Hipolito, 2022; Surachat et al., 2022; Teixeira et al., 2022).
Weissella and Periweissella species may play a role in the fermentation process of products both intended for human and animal consumption. They have been described as components of the fermentative microbiota of crop silages intended as animal feed (Otoni et al., 2018; Dong et al., 2020; Wang et al., 2020a; Wen Fang Wu Wu et al., 2021), where Weissella spp. may be generally involved in early stages of fermentation (0–7 days) or even remain relatively stable during the subsequent stages of the process (Wang et al., 2020a). Apart from such products that are not intended for human nutrition, Fusco et al. (2015) reported that Weissella and Periweissella species are also found in various fermented foods including wheat sourdough, cheeses, fermented meat-, milk-, fish-, and plant-based products (Fusco et al., 2015). However, in the last decade, a growing number of studies are aiming to characterize the microbiota of various fermented foods, helping to increase the awareness that Weissella and Periweissella may play a role in a wide variety of traditional and novel fermented foods (Table 3).
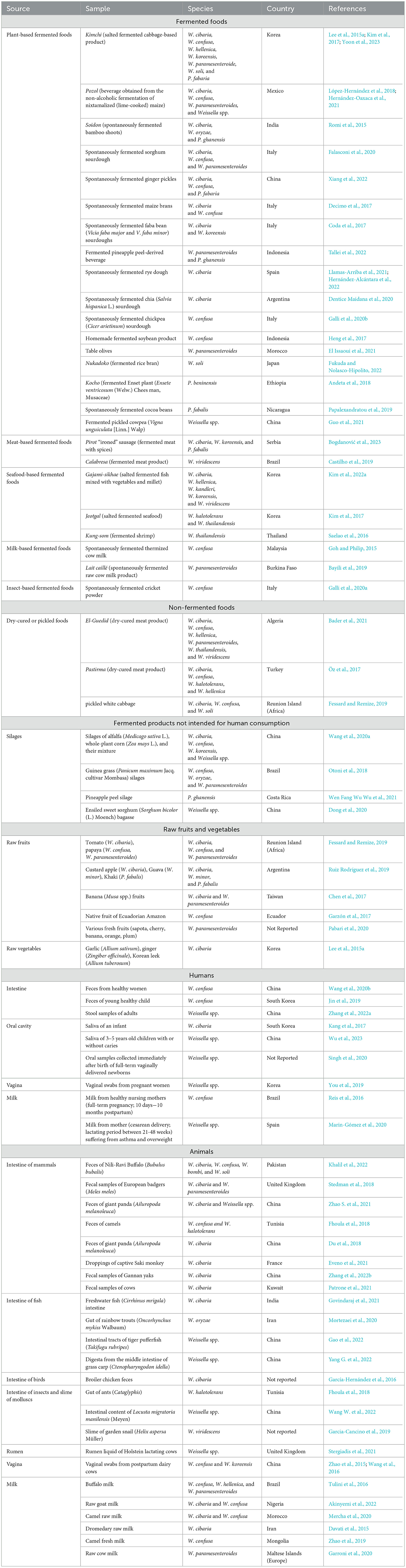
Table 3. Various isolation sources of Weissella and Periweissella species reported from 2015 up to date.
The increasingly popular and worldwide consumed kimchi, a salted fermented cabbage-based Korean food (Lee et al., 2022), is a well-known source of W. cibaria, W. confusa, W. koreensis, W. hellenica, W. paramesenteroides, W. soli, and P. fabaria (Lee et al., 2015a; Kim et al., 2017; Yoon et al., 2023, Table 3). Moreover, Weissella and Periweissella spp. are also being reported as part of the inhabiting microbiota also in other lesser-known traditional fermented products (Table 3). For instance, W. paramesenteroides was detected during the production of lait caillé, a spontaneously fermented traditional raw milk product made in Burkina Faso (Bayili et al., 2019). In particular, Bayili et al. (2019) found that W. paramesenteroides was more abundant during the early stages of fermentations (0–7 h), while it could no longer be recovered later, until the end of fermentation (59 h) (Bayili et al., 2019). The presence of Weissella is also reported in novel fermented products that are being developed to meet the growing human dietary needs. Galli et al. (2020a) recently detected W. confusa (along with other LAB belonging to Latilactobacillus, Lactiplantibacillus, Lactococcus, and Enterococcus genera) during the spontaneous fermentation of a cricket powder, which was propagated through a backslopping procedure. Coda et al. (2017) reported the presence of Weissella during the spontaneous backslop-propagated fermentation of two faba bean (Vicia faba minor and Vicia faba major) flours that could be used as substitutes of animal-derived protein sources. It was found that, among LAB, W. koreensis showed one of the highest incidences of occurrence in the tested faba bean sourdoughs, although also the presence of W. cibaria was reported during the fermentation of these sourdough samples (Coda et al., 2017). Furthermore, Decimo et al. (2017) found W. cibaria and/or W. confusa during the initial stages of spontaneous fermentation of two types of commercial native maize brans, which could potentially be exploited as functional food in human nutrition. Apart from legume- and cereal-derived fermented products, Weissella species may also be found during the fermentation of oilseed- and other naturally gluten-free cereal-derived products, representing alternative foods for people with celiac disease (Falasconi et al., 2020). In particular, W. cibaria was detected during the spontaneous fermentation of chia (Salvia hispanica L.) sourdough, being found as one of the dominant species at the final stages (8–10 days) of a backslopping-propagated fermentation (Dentice Maidana et al., 2020), while W. paramesenteroides, W. confusa, and W. cibaria were detected at the early stages (0–1 days) of the spontaneous fermentation of a sorghum sourdough, similarly obtained using a backslopping procedure (Falasconi et al., 2020).
Therefore, as other LAB, Weissella and Periweissella species often participate during the fermentation process of various products owing to their enzymatic capabilities that are particularly adapted toward carbohydrate metabolism, as was recently reported (Hernández-Oaxaca et al., 2021; Fanelli et al., 2022). Nevertheless, the dynamics and microbial successions during the fermentation process are likely to be influenced by (i) the initial microbial composition of the raw materials, (ii) the physico-chemical composition of the products to be fermented, (iii) the fermentation conditions (e.g., temperature and oxygen availability) and procedures applied, and (iv) the metabolic interactions with other microorganisms constituting the microbiota. Additionally, Weissella species have been also detected in some pickle- and dry-cured products (Table 3), necessitating more targeted studies to better understand and describe the role of Weissella and Periweissella within the fermentative and curing processes that to date remains not yet fully clarified.
Weissella and Periweissella spp. can also be associated with raw foods (Table 3), including animal and human milk.
In particular, apart from human-derived milk (Oikonomou et al., 2020; Mantziari and Rautava, 2021), and beyond the detection of Weissella spp. in milk of commonly raised dairy animals such as cows, ewes, and goats, as well as milk of companion animals, such as dogs (Fusco et al., 2015), Weissella is being also reported in milk of less common domesticated animals. In particular, although enterococci were the most frequently isolated LAB, W. cibaria was found in the raw milk from dromedary in Iran (Davati et al., 2015), while W. confusa, either alone or together with W. cibaria, was isolated from raw camel milk in Mongolia and Morocco (Zhao et al., 2019; Mercha et al., 2020). Although, to the best of our knowledge, the ecology and the relevant sources of Weissella species in milk have not been clearly elucidated, various mechanisms are hypothesized to shape the milk microbiota both in humans and animals. Apart from the mere contamination of milk immediately after excretion, due to the presence of microorganisms that inhabit the skin or originate from fecal or environment contamination, three other mechanisms may represent possible sources of the milk microbiota, as was suggested for milk from both humans and cows (Oikonomou et al., 2020; Mantziari and Rautava, 2021). These include the (i) enteromammary pathway through which dendritic cells or macrophages transfer bacteria from the maternal gut to the mammary gland and then release the bacteria in the milk, (ii) the retrograde backflow of bacteria from the skin, the environment, or the offspring oral cavity during suckling or milking, and (iii) the presence of a resident microbiota in the mammary tissue (Oikonomou et al., 2020; Mantziari and Rautava, 2021). Weissella species, that are known to occur in different areas of the human and animal body including rumen and vagina (Table 3), are also found in the oral cavity of young children, as well as in human and animal intestine (detected mainly by fecal sampling) (Table 3); therefore, these may represent possible starting sites for Weissella to reach the human and animal milk, needing further investigations.
Notably, Weissella spp. can inhabit the intestinal tract of both vertebrates and invertebrates, the latter including insects and molluscs (Table 3), where they may be associated with the healthy status of the gut. In this regard, a recent study conducted on the feces of giant pandas found in healthy sub-adult animals a higher abundance of Weissella spp., including W. cibaria, when compared to sub-adult animals suffering from anorexia (Zhao S. et al., 2021). Interestingly, a symbiotic relationship among Weissella and Leuconostoc, Bacillus, and Streptococcus genera appeared to occur in the gut of the analyzed subjects, and, conversely, a decrease in Weissella and Streptococcus and increase in Clostridium could be the cause of the reported anorexia symptoms (Zhao S. et al., 2021).
Weissella and Periweissella may thus inhabit various ecological niches, with W. cibaria, W. confusa, and W. paramesenteroides being frequently reported in different sources (Table 3). Less knowledge is available for other species, especially those lastly described (W. coleopterorum, P. cryptocerci, W. muntiaci, W. fangxianensis, and W. sagaensis) for which, apart from the studies that recently reported their primary source of isolation (Table 1), no further knowledge is substantially available. This, therefore, deserves future investigations to clearly identify their relevant niches and better understand the ecological role also of the novel Weissella and Periweissella species.
Due to a long history of safety and the optimal pro-technological characteristics, LAB can be considered as the most important microbial group acting as starters in traditional and novel fermented foods and many of their functions have long been investigated and understood. Among these, Weissella spp. strains own numerous technological and functional properties and frequently play a significant part in food preservation and health benefits; thus, they have been evaluated as innovative starter cultures with an industrially significant interest (Fessard and Remize, 2017). However, their use as starters for food and beverage fermentation such as their inclusion in commercial products is still limited due to the lack of safety evaluation by two major food safety authorities, namely, the Food and Drug Administration (FDA) and the European Food Safety Authority (EFSA), which still do not consider any strain belonging to this genus as GRAS or QPS, respectively. A paucity of scientific data evaluating the safety aspects, antibiotic resistance pattern, potential biogenic amine synthesis, and infection risk partly explains such neglecting (Fessard and Remize, 2017; Ahmed et al., 2022). In the last years, weissellas characterization including technological and functionality assessments was widely performed to demonstrate also the health effects of single strains in accordance to the definition of probiotics by the International Scientific Association for Probiotics and Prebiotic (ISAPP). Therefore, the weissellas isolates from different fermented products have been characterized as potential starters to be used in food processing (Fusco et al., 2015), increasing their possible applications (Figure 1).
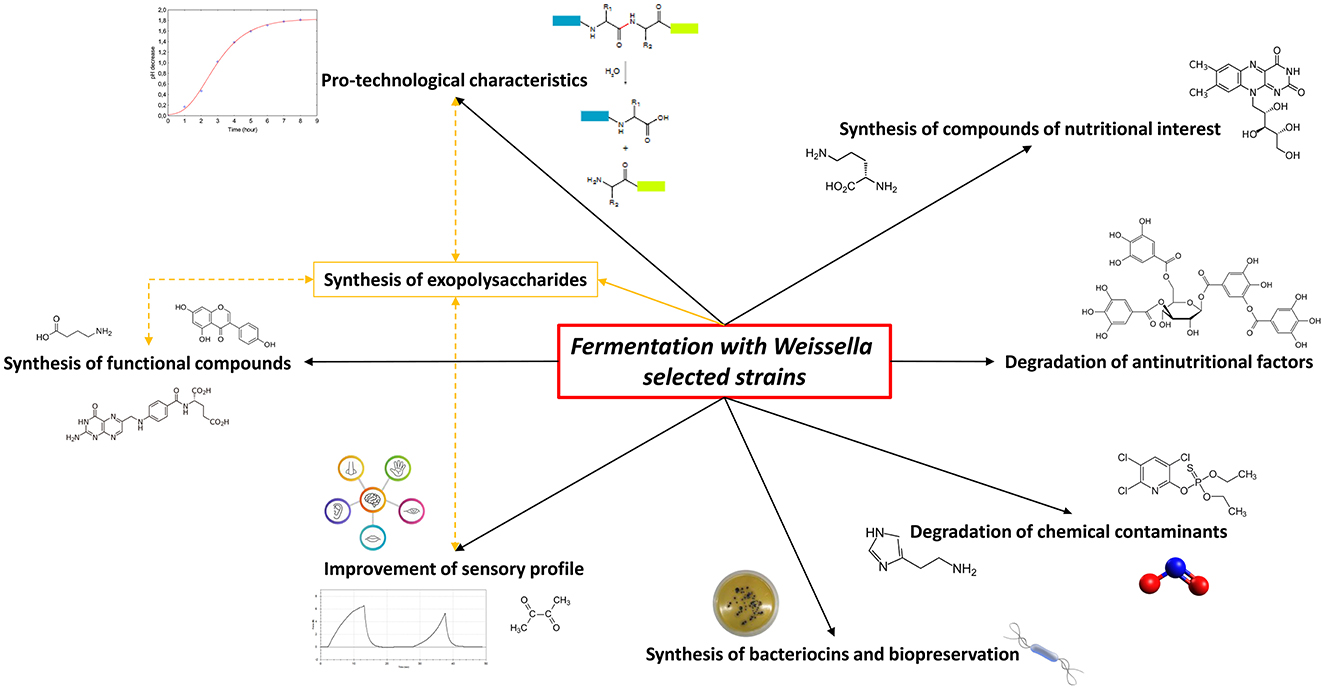
Figure 1. Potential of Weissella spp. selected strains as starters in food fermentation applications. The yellow lines indicate the correlation between the exopolysaccharides production by Weissella spp. and the improvement of food characteristics.
The suitability of W. cibaria and W. confusa to be used as starters for bread making was analyzed following an integrated approach including phenotypic, genotypic, and metabolomic characteristics. W. cibaria-com2 was identified (Lopez et al., 2022) as a strain able to perform an intense proteolysis in wheat flour doughs leading to release huge amounts of numerous amino acids and peptides, some of which belong to the branched-chain amino acid-derived compounds (BCAA). Thus, the differential metabolite profile of that strain was partially explained by a genome investigation for putative differences in peptidases, proteases, and amino acid/peptide transporters (Lopez et al., 2022). In particular, a higher presence of amino acid permeases, peptidases (C40 family), and oligopeptide ABC transporters was found in this strain as compared to other Weissella spp. strains (Lopez et al., 2022). Proteolysis is indeed considered as a key process in food fermentation leading toward a higher impact on the technological, nutritional, organoleptic, and functional features of the fermented foods.
W. confusa SD8 was used for making a sorghum sourdough in the study of Olojede et al. (2022) and was found to confer an optimal specific volume to the bread and moreover also contributed to tannin degradation (Olojede et al., 2022). Tannins are very abundant in sorghum flour (such as in other minor cereals, pseudocereals, and legumes) and considered as antinutritional factor since they are able to bind proteins making them refractory to digestion and are responsible for the bitter taste of different plant-based ingredients.
A W. koreensis strain (DB1) isolated from kimchi producing high levels of ornithine was proposed as a functional starter culture for rice bran fermentation (Yeong et al., 2020). Ornithine is produced in microbes from arginine throughout the intracellular arginine deiminase pathway (Yeong et al., 2020). Different Weissella strains were also proposed as starters for kimchi production due to their technological characteristics, even though these were also found to exhibit a weak hemolytic activity (Jeong and Lee, 2015). W. cibaria M3 was used (as mixed starter, in association with Lactococcus lactis) to produce a typical Chinese fermented fish product (Chouguiyu), showing optimal organoleptic characteristics (Bao et al., 2018). A folate-producing strain of W. cibaria (PL17-3) was also selected for the production of fermented fish (Deatraksa et al., 2018). Sixteen Weissella isolates from artisanal Brazilian cheeses were identified as potential starter candidates for the dairy industry owed to their high acidification ability, diacetyl production, and proteolytic activity (Teixeira et al., 2021). A selected W. cibaria strain was demonstrated to improve the organoleptic profile of Sichuan pickle when used in association with Lactiplantibacillus plantarum (Xiang et al., 2020).
W. cibaria 30 and W. cibaria 64, isolated from tropical fruits, were included in a list of starters for fruits and vegetables fermentation, whereas W. soli 58 showed the characteristics of a potential preservative culture for fruits and vegetables (Fessard and Remize, 2019). W. cibaria FB069 was tested as a starter to produce functional fermented soymilk. The addition of xylooligosaccharides before the fermentation process led to considerable increase of the acidification rate, viscosity, and W. cibaria FB069 growth. Moreover, the synbiotic fermented product obtained was characterized by increased levels of dextran, folate, GABA, genistein, and daidzein, and it was able to decrease the proliferation of Caco-2 and HCT116 cell lines (Le et al., 2020).
A spontaneous mutant strain, W. cibaria BAL3C-5 B2, was selected among different parental and mutant strains of W. cibaria to produce a content riboflavin bread, characterized by concentrations over 0.1 mg of riboflavin in 100 g of bread (Hernández-Alcántara et al., 2022). Moreover, the use of Weissella strains was also demonstrated in the production of baker's yeast-free bread (Lopez et al., 2022).
LAB able to produce EPS play a pivotal role in industry for the development of functional food and are also used as coadjutants or starter cultures for the development of yogurt and other traditional fermented foods (Zhu et al., 2018). In these products, the EPS production by LAB starter cultures may occur in situ. As a consequence, since LAB EPS improve the texture and rheology of fermented foods by acting as natural biothickeners, the use of food additives, such as pectin and starch, may be avoided (Zhu et al., 2018). Furthermore, LAB EPS may act as probiotic and prebiotic due to their immunoregulatory, antitumoral, and antioxidant activities, as well as cholesterol-lowering ability, and prebiotic effects (Zhu et al., 2018) (for the probiotic and prebiotic role of the weissellas' EPS, see the subheader “Exopolysaccharides produced by weissellas” in the paragraph on the probiotic potential below). Among LAB, weissellas can generate high content of EPS without excessive amount of acetate which represents undesired compound in some products (e.g., beverages) also in the presence of added sugar usually used to maximize the EPS production. Indeed, the addition of sucrose during weissellas fermentation leads to dextran production and the use of fructose as a carbon source and not as electron acceptor, thus generating minimal quantities of acetate instead of mannitol (Rolim et al., 2019).
Weissella confusa A16, a strain characterized by a high level synthesis of dextran, was efficiently used for the EPS enrichment of a brewer's spent grain added with 4% of sucrose, aimed at improving its technological properties and contributing to its recycle as food ingredient (Koirala et al., 2021). As natural structure-forming agent, dextran has already been used as a food additive. Its supplementation to food formulations efficiently improved the technological properties of different raw materials such as fiber- and protein-rich matrices (such as cereal by-products), allowing their use as ingredients in food production chains (Koirala et al., 2021). Viscosity increase in fermented brewers' spent grain supplemented with sucrose occurred first after 10 h of fermentation and increased until 24 h concomitantly with dextran accumulation. The dextran content after 24 h was ~1% on the total weight of the brewers' spent grain (Koirala et al., 2021). A dextransucrase gene was identified in W. confusa A16 that showed a typical inducible characteristic, with an intense upregulation occurring at 10 h. EPS produced by a strain isolated from distiller grains of Chinese Baijiu, namely, Weissella cibaria NC516.11, was proven to be able to promote the cross-linking of starch molecules, thus increasing the water-holding capacity (Li et al., 2022). Dynamic rheology indicated that the aqueous solutions of EPS are pseudoplastic fluids, and their addition to gluten-free ingredients increases the viscoelastic features of the dough (Li et al., 2022). The use of EPS producing strain in bread making was extensively exploited in both gluten-free and gluten-containing bakery products. Weissella cibaria P9 was used to produce gluten-free bread also including sucrose-containing flour instead of sugar as a precursor (Montemurro et al., 2021, 2023). Moreover, the in situ production of EPS by weissellas for improving the steamed bread quality, usually associated with increase of the specific volume, enhance of the texture, and decrease of the staling rate, was recently confirmed (Xu et al., 2020; Sha et al., 2023).
A wholemeal quinoa beverage was also fermented with W. cibaria MG1 with the aim to produce a yogurt-like product that showed high water-holding capacity, viscosity, and EPS concentration as a consequence of the fermentation (Zannini et al., 2018). It was hypothesized that the high EPS (dextran) concentration was responsible for the optimal structural properties of the fermented matrix (Zannini et al., 2018).
Other plant-derived substrates were also efficiently in situ enriched with EPS using selected strains of Weissella spp., such as faba bean flour (Xu et al., 2017; Rizzello et al., 2019), chickpea (Galli et al., 2020a), and rye (Kajala et al., 2016). A W. cibaria strain (SJ14) isolated from Sichuan paocai (a type of Chinese pickles), characterized by strong salt tolerance, acidification, and nitrite depletion capacities, was identified as a heteropolysaccharide producer. Moreover, a strong antioxidant activity of its EPS was demonstrated (Zhu et al., 2018).
The potential probiotic and exopolysaccharide-producing strain W. confusa VP30 was isolated from young children's feces, and its EPS was characterized and quantified. Moreover, the safety was assessed with the aim of applying the strain in food production (Jin et al., 2019). Lastly, a wild W. minor (W4451) strain was demonstrated to be able to significantly increase milk viscosity and was therefore proposed as starter for the dairy sector (Bancalari et al., 2020).
For the antimicrobial activity of weissellas and periweissellas in detail, see the subheader “Antimicrobial activity of weissellas” in the paragraph on the probiotic potential below.
The bacteriocin weissellicin D was produced by the strain W. hellenica D1501 in fermented pork and showed antimicrobial activity against Staphylococcus aureus, Listeria monocytogenes, and E. coli (Chen et al., 2014a). W. hellenica D1501 was therefore also tested as starter to produce a tofu with long shelf life (Chen et al., 2014b). W. cibaria D30 was used in cottage cheese after whey separation from the curd and not only demonstrated inhibitory activity against L. monocytogenes ATCC 15313 but also increased the antioxidant properties of the product (Kariyawasam et al., 2019).
One of a possible solution for overcoming the still not authorized use of weissellas in food production is the use of cell-free suspension (CFS), as reviewed by Aggarwal et al. (2022) and Ahmed et al. (2022). CFS is considered postbiotics according to the definition of ISAPP, describing them as a “preparation of inanimate microorganisms and/or their components that confer a health benefit on the host.” Weissella cibaria CMU, an oral care probiotic, was discovered to produce and release secreted proteins, organic acid, and hydrogen peroxide with antibacterial activity against periodontal pathogens (Lim et al., 2018). The CFS from W. viridescens WV20-15 was tested, excluding the effect of organic acids and hydrogen peroxide, to control Listeria monocytogenes 10403S. Inhibitory compounds of proteinaceous nature, probably bacteriocins, decreased the production of microbial biofilm and eradicated preformed biofilms on different materials. Moreover, a significant reduction of L. monocytogenes 10403S growth was found on chilled pork (Yang C. et al., 2022). The partially purified bacteriocin 7293 obtained from W. hellenica BCC 7293 CFS was effectively used to produce an antimicrobial biodegradable food packaging applied in PLA/SP film. In vitro assays demonstrated the inhibition of both Gram-positive (Staphylococcus aureus and Listeria monocytogenes) and Gram-negative bacteria (Escherichia coli, Salmonella enterica serovar Typhimurium, Pseudomonas aeruginosa, and Aeromonas hydrophila). Moreover, the innovative packaging was used to avoid the proliferation of the pathogenic microorganisms in a challenge test of inhibition of chilled pangasius filet (Woraprayote et al., 2018).
Hamoud and Sifour (2021) demonstrated that the potentially probiotic strain Weissella confusa Lb. Con was able to survive in MRS broth at a concentration of 200 μg/ml of chlorpyrifos, being able also to degrade about 25% of this pesticide. Considering the wide use of this pesticide to control foliar insects in different vegetables, these results suggested the potential use of this strain in the decontamination of food matrices or in probiotic formulations, aiming at the in vivo reduction of pesticide toxicity. Liu et al. (2020) tested W. cibaria X31 and W. confusa L2 as low nitrite dry-fermented sausages starters. The final product was characterized by high growth rate of both inoculated microorganisms, high degree of redness, high proteolysis rate, and decreased residual nitrites and S. enterica growth. Fermented meat can contain not only nitrite but also high amounts of biogenic amines. The use of both Lactiplantibacillus plantarum His6 and Weissella viridescens F2 as starters for Roucha production led to the decrease of histamine and tyramine of ~50%, due to their amine oxidase activity and the conversion into aldehyde, hydrogen peroxide, and ammonia (Han J. et al., 2022).
In 2015, Fusco et al. (2015) reviewed all the cases of clinical infections with weissellas that occurred until that year. From 2015 to date, further cases have occurred, all involving W. confusa strains (Table 4). As for Weissella infections in animals from 2015 to date, only five cases have been reported, with W. ceti as the etiological agent of weissellosis in rainbow trouts (Castrejón-Nájera et al., 2018; Mitomi et al., 2018; Medina et al., 2020; Vásquez-Machado et al., 2020).
No studies have been published so far about the pathogenic potential of Periweissella species.
Since 2015, when Fusco et al. (2015) provided an overview of studies published until that year on the probiotic potential of weissellas, many further articles on the same topic have been published up to date. As reported in Tables 5, 6, most of the studies focused on W. confusa and W. cibaria strains isolated from various ecological niches. For the majority of the potentially probiotic strains, a safety assessment consisting of investigations into the antibiotic susceptibility and the hemolytic activity was performed. However, Sturino (2018) carried out a literature-based safety assessment of W. confusa, concluding that many strains of this species can be safely used for poultry in direct-fed microbial products. Cupi and Elvig-Jørgensen (2019) assessed the toxicological safety of W. confusa by in vivo, in vitro, and ex vivo studies. In the tested conditions, no toxic effects were shown by W. confusa allowing to conclude that this species could be used as a safe direct-fed microbial product (Cupi and Elvig-Jørgensen, 2019). However, for their studies, they used “an off powder of freeze-dried bacteria composed of almost entirely W. confusa,” but no specification on the composition in strain/strains of this powder was made. By contrast, Bourdichon et al. (2021) used 46 strains of W. confusa (17 of clinical and 26 of food origin) to provide a safety assessment based on their hemolytic activity and antibiotic susceptibility, as well as on the search of antibiotic resistance genes, virulence determinants, and genes coding for deleterious metabolites (such as biogenic amines) within their genomes. Moreover, a literature search was conducted to find reports of infection caused by strains of W. confusa (Bourdichon et al., 2021). This study allowed Bourdichon et al. (2021) to consider W. confusa as “safe for use in the food chain, food culture for fermentation, or as probiotic strain candidate”.
Jang et al. (2021) performed a safety assessment of W. cibaria JW15 by phenotypic (antibiotic susceptibility, production of toxic metabolites, and hemolytic activity) and genotypic analyses (whole-genome sequencing and search of virulence and antibiotic resistance genes), confirming the safety of this strain. The probiotic potential and the safety of four Periweissella and two Weissella type strains were assessed by Fanelli et al. (2023b) using phenotypic and genotypic methods. The potential probiotic P. beninensis-type strain results the only safe candidate. Furthermore, these authors confirmed the necessity of assessing the probiotic potential and safety of weissellas and periweissellas on a strain-specific basis (Fanelli et al., 2023b). Among Lactobacillaceae, Weissella species are the second only to Lacticaseibacillus rhamnosus causing opportunistic (hospital-acquired) infections, but even in the case of Lc. rhamnosus, EFSA has expressed its opinion on the safety of certain strains of this species to be used as technological additive for all animal species (EFSA Panel on Additives Products or Substances used in Animal Feed et al., 2017, 2021).
While until 2014 few studies investigated the probiotic potential of weissellas by way of in vitro and in vivo studies (Fusco et al., 2015), from 2014 up to date, the number of such studies has increased greatly (Tables 5, 6), but, as reported in Table 5, those in vivo studies mainly involved W. cibaria strains, apart from some W. confusa and W. paramesenteroides strains and one W. viridescens strain. However, in addition to the study of Park et al. (2022a), who demonstrated the functional laxative effects of milk fermented with a probiotic W. confusa strain in loperamide-induced constipation in rats, none of the in vivo studies administered food or feed containing weissellas strains to animals or humans.
According to Fusco et al. (2015), up to the year 2014, 12 articles were published on the screening of Weissella strains for their antimicrobial activity. From 2014 up to date, further investigations have investigated weissellas for their antagonistic activity against pathogens (Jang et al., 2016; Shah et al., 2016; Das Purkhayastha et al., 2017; Ye K. et al., 2018; Yu et al., 2018; Dey et al., 2019; Tenea and Israel Lara, 2019; Dinoto et al., 2021; Yeu et al., 2021; Styková et al., 2022; Yang C. et al., 2022; Yao D. et al., 2022; El-Mekkawy et al., 2023; Fanelli et al., 2023b; Table 5). In addition, Kang and Park (2022) demonstrated the in vitro inactivation of respiratory viruses, namely, human respiratory syncytial virus (RSC) and the influenza A virus (H1N1) and rotavirus by the oral probiotic strain W. cibaria CMS1, isolated from the saliva of Korean children with healthy oral cavity.
As reported by Fusco et al. (2015), up to 2014, six bacteriocins were detected in five Weissella strains. From 2014 up to date, further bacteriocins have been discovered in these bacteria. In particular, weissellicin 110 produced by a strain isolated from the yan-dong-gua (fermented wax gourd), namely, W. cibaria 860106, was characterized by Wu et al. (2015). The bacteriocin A3 produced by W. confusa A3, a strain of dairy origin, was isolated and characterized by Goh and Philip (2015). This bacteriocin inhibited the growth of strains of P. aeruginosa, Bacillus cereus, Enterococcus faecium, Micrococcus luteus, E. coli, and Lactococcus lactis (Goh and Philip, 2015). Two putatively novel bacteriocins, namely, bacteriocin 7193A and bacteriocin 7293B, produced by W. hellenica BCC 7293, isolated from Nham (Thai fermented pork sausage) were isolated by Woraprayote et al. (2015). Apart from some Gram-positive strains including Staphylococcus aureus ATCC 23235 and S. aureus ATCC 25923, both bacteriocins were found active against Gram-negative foodborne pathogens such as Salmonella enterica serovar Typhimurium, Aeromonas hydrophila, Escherichia coli, and Pseudomonas aeruginosa.
W. confusa MBF8-1, isolated from a Indonesian home-made soya product, revealed bacteriocin-like inhibitory substance (BLIS) activity against some Gram-positive bacteria including six W. confusa strains, two W. cibaria strains, a Leuconostoc mesenteroides strain, a Macrococcus luteus, and a Lactococcus lactis strain (Malik et al., 2016). Malik et al. (2016) demonstrated that the so called weissellicin MBF was encoded by a large plasmid, pWcMBF8-1. Subsequently, Sartono et al. (2019) demonstrated spermicidal and antibacterial activity against the indicator bacterium Leuconostoc mesenteroides of the bacteriocin-like peptides of W. confusa MBF8-1. BLIS that is active against non-specified Gram-positive and Gram-negative bacteria was found also in W. confusa LM85 by Kaur and Tiwari (2016).
Dubey and Jeevaratnam (2018) demonstrated the antimicrobial activity of W. confusa AJ79, isolated from fermented butter, against numerous pathogenic bacteria such as Citrobacter freundii, Aeromonas hydrophila, Bacillus cereus, B. subtilis, Mycobacterium smegmatis, E. coli, B. licheniformis, L. monocytogenes, Clostridium sporogenes, Klebsiella pneumoniae, C. perfringens, Micrococcus luteus, Pseudomonas aeruginosa, Vibrio parahaemolyticus, S. aureus, S. epidermidis, and Proteus vulgaris. These authors also isolated and characterized the relevant class II bacteriocin named BAC79 from this strain (Dubey and Jeevaratnam, 2018). Kariyawasam et al. (2019) successfully investigated the antilisterial effect of W. cibaria D30, isolated from Korean kimchi, when used as protective culture in the production of cottage cheese, but they did not ascertain to what kind of antimicrobial compound this effect was due.
Teixeira et al. (2022) sequenced the genome of W. cibaria W25, isolated from a Brazilian pasture samples of a Brazilian dairy farm, and found that this strain had the possibility of producing two different bacteriocins. Thereafter, the same authors (Teixeira et al., 2023) demonstrated a putative bacteriocinogenic activity of W. cibaria W25 against Salmonella enterica Newport, Kocuria rhizophila, Listeria innocua, and E. coli strains. The same antimicrobial spectrum was observed also in the neutralized supernatant of W. cibaria W42, isolated from the soil of a Brazilian dairy farm (Teixeira et al., 2023). Moreover, Teixeira et al. (2023) sequenced also the genomes of W. cibaria W42 and W. cibaria W21 isolated from pasture samples of a Brazilian dairy farm. In these strains, they also found the putative gene for a bacteriocin identified as bacteriocin_IIc (Teixeira et al., 2023), but the strain W21 lacks the transport-related gene and its neutralized supernatant lost antimicrobial activity, whereas the supernatant of W25 and W42 lost the antimicrobial activity only when treated with proteinase K (Teixeira et al., 2023).
Apart from antimicrobial activity of weissellas, the study of Li et al. (2021) also demonstrated the cadmium biosorption of W. viridescens ZY-6, isolated from fermented pickles, whereas Kinoshita et al. (2016) demonstrated the biosorption properties of W. viridescens MY 205 isolated from bovine intestine for the periodic group 12 metals cadmium (Cd), mercury (Hg), and zinc (Zn). Considering the detrimental effects of heavy metal pollution of food and beverages on human and animal health, the role of weissellas as heavy metal sorbents may be of high importance.
Weissellas may also play an important role in binding of aflatoxins. Kavitake et al. (2020), for example, demonstrated that a strain of W. confusa, isolated from an Indian traditional fermented food (Idli batter), produced a galactan exopolysaccharide with aflatoxin B1-binding activity. As in the case of heavy metals discussed above, this may prevent absorption of the aflatoxin in the human body once the food with the bacteria is ingested. However, more research in this would be required.
Among the metabolites produced by weissellas, exopolysaccharides (EPSs) play an important role in several beneficial and technological functions. Since 2015, when Fusco et al. (2015) reviewed the research published on weissellas' EPS up to that date, a plethora of papers were published dealing with the characterization of weissellas producing EPS (Table 7). Mainly homo- but also heteropolysaccharides have been found in W. cibaria and W. confusa strains of various origin, with technological and functional properties (Table 7). The latter mainly include antioxidant, antibacterial, antifungal, anti-inflammatory, and prebiotic functions, which have been demonstrated in vitro (Table 7), while few in vivo studies have demonstrated the amelioration of functional constipation in rats and humans (Table 7).
In the last decades the number of articles published on weissellas is increased enormously. Novel species have been discovered, the taxonomy of the genus has changed so that a new genus, namely Periweissella, has been derived from the previous genus Weissella, and new insights into the safety, biotechnological, and probiotic potential of weissellas and periweissellas have been provided. Regarding their technological characteristics, the weissellas and periweissellas have excellent potential with reference to their proteolytic and saccharolytic, as well as antimicrobial and EPS production activities, thus contributing greatly to the safety and functionality of the products. As for the safety, biotechnological, and probiotic potential, most studies focused on weissellas, mainly W. confusa and W. cibaria, but the most recent studies are focusing also on periweissellas and other species of the Weissella genus. Apart from one safety study carried out on 46 W. confusa strain allowing to define this species as safe, for the other Weissella and Periweissella species the safety assessment was carried out on few strains some of which resulted to be unsafe. These groups of LAB have a high potential for biotechnological application, and the probiotic potential of numerous strains is being demonstrated. However, for their application in food, a strain-based assessment of their safety still remains mandatory.
VF: Conceptualization, Methodology, Project administration, Writing – original draft, Writing – review & editing. DC: Writing – original draft, Writing – review & editing. FF: Writing – original draft, Writing – review & editing. MM: Writing – original draft, Writing – review & editing. CR: Writing – original draft, Writing – review & editing. CF: Funding acquisition, Writing – original draft, Writing – review & editing.
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Adebayo-Tayo, B., Ishola, R., and Oyewunmi, T. (2018). Characterization, antioxidant and immunomodulatory potential on exopolysaccharide produced by wild type and mutant Weissella confusa strains. Biotechno. Rep. 19, e00271. doi: 10.1016/j.btre.2018.e00271
Adesulu-Dahunsi, A. T., Sanni, A. I., and Jeyaram, K. (2018b). Production, characterization and in vitro antioxidant activities of exopolysaccharide from Weissella cibaria GA44. LWT Food Sci. Technol. 87, 432–442. doi: 10.1016/j.lwt.2017.09.013
Adesulu-Dahunsi, A. T., Sanni, A. I., Jeyaram, K., Ojediran, J. O., Ogunsakin, A. O., Banwo, K., et al. (2018a). Extracellular polysaccharide from Weissella confusa OF126: production, optimization and characterization. Int. J. Macromol. 111, 514–525. doi: 10.1016/j.ijbiomac.2018.01.060
Aggarwal, S., Sabharwal, V., Kaushik, P., Joshi, A., Aayushi, A., Suri, M., et al. (2022). Postbiotics: from emerging concept to application. Front. Sust. Food Syst. 6, 887642. doi: 10.3389/fsufs.2022.887642
Ahmed, S., Singh, S., Singh, V., Roberts, K. D., Zaidi, A., Rodriguez-Palacios, A., et al. (2022). The Weissella genus: clinically treatable bacteria with antimicrobial/probiotic effects on inflammation and cancer. Microorganisms 10, 2427. doi: 10.3390/microorganisms10122427
Akinyemi, M. O., Ogunremi, O. R., Adeleke, R. A., and Ezekiel, C. N. (2022). Probiotic potentials of lactic acid bacteria and yeasts from raw goat milk in Nigeria. Prob. Antimicrob. Prot. 15, 1–8. doi: 10.1007/s12602-022-10022-w
Albesharat, R., Ehrmann, M. A., Korakli, M., Yazaji, S., and Vogel, R. F. (2011). Phenotypic and genotypic analyses of lactic acid bacteria in local fermented food, breast milk and faeces of mothers and their babies. Syst. Appl. Microbiol. 34, 148–155. doi: 10.1016/j.syapm.2010.12.001
Amari, M., Laguerre, S., Vuillemin, M., Robert, H., Loux, V., Klopp, C., et al. (2012). Genome sequence of Weissella confusa LBAE C39-2, isolated from a wheat sourdough. J. Bacteriol. 194, 1608–1609. doi: 10.1128/JB.06788-11
Amer, M. N., Atwa, N. A., Eldiwany, A. I., Elgammal, E. W., Dawoud, I. E., Rashad, F. M., et al. (2022). Anticancer and antioxidant activities of L-asparaginase produced by local Weissella paramesenteroides MN2C2 strain. Egypt. J. Chem. 65, 409–419. doi: 10.21608/EJCHEM.2021.102465.4832
Ampe, F., ben Omar, N., Moizan, C., Wacher, C., and Guyot, J. P. (1999). Polyphasic study of the spatial distribution of microorganisms in Mexican pozol, a fermented maize dough, demonstrates the need for cultivation-independent methods to investigate traditional fermentations. Appl. Environ. Microbiol. 65, 5464–5473. doi: 10.1128/AEM.65.12.5464-5473.1999
Anandharaj, M., Sivasankari, B., Santhanakaruppu, R., Manimaran, M., Parveen Rani, R., Sivakumar, S., et al. (2015). Determining the probiotic potential of cholesterol-reducing Lactobacillus and Weissella strains isolated from gherkins (fermented cucumber) and south Indian fermented koozh. Res. Microbiol. 166, 428–439. doi: 10.1016/j.resmic.2015.03.002
Andeta, A. F., Vandeweyer, D., Woldesenbet, F., Eshetu, F., Hailemicael, A., Woldeyes, F., et al. (2018). Fermentation of enset (Ensete ventricosum) in the Gamo highlands of Ethiopia: physicochemical and microbial community dynamics. Food Microbiol. 73, 342–350. doi: 10.1016/j.fm.2018.02.011
Apostolakos, I., Paramithiotis, S., and Mataragas, M. (2022). Functional and safety characterization of Weissella paramesenteroides strains isolated from dairy products through whole-genome sequencing and comparative genomics. Dairy 3, 799–813. doi: 10.3390/dairy3040055
Azim, A., Singh, N., Venkatesh, V., Verma, S., and Agarwal, A. (2023). Weissella confusa causing vancomycin-resistant septicemia infection in a pediatric patient: a case report from a University Teaching Hospital in North India. Cureus 15, e38292. doi: 10.7759/cureus.38292
Bader, R., Becila, S., Ruiz, P., Djeghim, F., Sanah, I., Boudjellal, A., et al. (2021). Physicochemical and microbiological characteristics of El-Guedid from meat of different animal species. Meat Sci. 171, 108277. doi: 10.1016/j.meatsci.2020.108277
Bancalari, E., Alinovi, M., Bottari, B., Caligiani, A., Mucchetti, G., Gatti, M., et al. (2020). Ability of a wild Weissella strain to modify viscosity of fermented milk. Front. Microbiol. 10, 3086. doi: 10.3389/fmicb.2019.03086
Bao, R., Liu, S., Ji, C., Liang, H., Yang, S., Yan, X., et al. (2018). Shortening fermentation period and quality improvement of fermented fish, Chouguiyu, by co-inoculation of Lactococcus lactis M10 and Weissella cibaria M3. Front. Microbiol. 9, 3003. doi: 10.3389/fmicb.2018.03003
Baruah, R., and Goyal, A. (2015). Hyper glucansucrase, glucan and oligosaccharide producing novel Weissella cibaria RBA12 isolated from Pummelo (Citrus maxima). Ann. Microbiol. 65, 2301–2310. doi: 10.1007/s13213-015-1072-7
Baruah, R., Maina, N. H., Katina, K., Juvonen, R., and Goyal, A. (2017). Functional food application of dextran from Weissella cibaria RBA12 from pummelo (Citrus maxima). Int. J. Food Microbiol. 242, 124–131. doi: 10.1016/j.ijfoodmicro.2016.11.012
Baugh, A. V., Howarth, T. M., West, K. L., Kerr, L. E. J., Love, J., Parker, D. A., et al. (2021). Draft genome sequence of Weissella paramesenteroides STCH-BD1, isolated from ensiled sorghum bicolor. Microbiol. Res. Announce. 10, e01328–e01320. doi: 10.1128/MRA.01328-20
Bayili, G. R., Johansen, P., Nielsen, D. S., Sawadogo-Lingani, H., Ouedraogo, G. A., Diawara, B., et al. (2019). Identification of the predominant microbiota during production of lait caillé, a spontaneously fermented milk product made in Burkina Faso. World J. Microbiol. Biotechnol. 35, 100. doi: 10.1007/s11274-019-2672-3
Bello, S., Rudra, B., and Gupta, R. S. (2022). Phylogenomic and comparative genomic analyses of Leuconostocaceae species: identification of molecular signatures specific for the genera Leuconostoc, Fructobacillus and Oenococcus and proposal for a novel genus Periweissella gen. nov. Int. J. Syst. Evol. Microbiol. 72, 005284. doi: 10.1099/ijsem.0.005284
Benhouna, I. S., Heumann, A., Rieu, A., Guzzo, J., Kihal, M., Bettache, G., et al. (2019). Exopolysaccharide produced by Weissella confusa: chemical characterization, rheology and bioactivity. Int. Dairy J. 90, 88–94. doi: 10.1016/j.idairyj.2018.11.006
Benkerroum, N., Misbah, M., Sandine, W. E., and Elaraki, A. T. (1993). Development and use of a selective medium for isolation of Leuconostoc spp. from vegetables and dairy products. Appl. Environ. Microbiol. 59, 607–609. doi: 10.1128/aem.59.2.607-609.1993
Benomar, N., Abriouel, H., Lee, H., Cho, G. S., Huch, M., Pulido, R. P., et al. (2011). Genome sequence of Weissella thailandensis fsh4-2. J. Bacteriol. 193, 5868. doi: 10.1128/JB.05883-11
Berhe, T., Ipsen, R., Seifu, E., Kurtu, M. Y., Fugl, A., Hansen, E. B., et al. (2019). Metagenomic analysis of bacterial community composition in Dhanaan: Ethiopian traditional fermented camel milk. FEMS Microbiol. Lett. 366. fnz128. doi: 10.1093/femsle/fnz128
Björkroth, J., Dicks, L. M. T., and Endo, A. (2014). The Genus Weissella. Lactic Acid Bacteria. New York, NY: Wiley.
Björkroth, K. J., Schillinger, U., Geisen, R., Weiss, N., Hoste, B., Holzapfel, W. H., et al. (2002). Taxonomic study of Weissella confusa and description of Weissella cibaria sp. nov., detected in food and clinical samples. Int. J. Syst. Evol. Microbiol. 52, 141–148. doi: 10.1099/00207713-52-1-141
Bogdanović, S., Stanković, S., Berić, T., Tomasevic, I., Heinz, V., Terjung, N., et al. (2023). Bacteriobiota and chemical changes during the ripening of traditional fermented “pirot 'ironed' sausage”. Foods 12, 664. doi: 10.3390/foods12030664
Bourdichon, F., Patrone, V., Fontana, A., Milani, G., and Morelli, L. (2021). Safety demonstration of a microbial species for use in the food chain: Weissella confusa. Int. J. Food Microbiol. 339, 109028. doi: 10.1016/j.ijfoodmicro.2020.109028
Cao, J., Wu, C., Wang, K., Hu, H., Duan, J., Zhao, B., et al. (2021). Metagenomic profiling reveals dominance of gram-positive bacteria in the gut microbiome shifts associated with immunoglobulin A vasculitis (Henoch-Schönlein Purpura). Clin. Transl. Immunol. 10, e1342. doi: 10.1002/cti2.1342
Castilho, N. P. A., Colombo, M., Oliveira, L. L., Todorov, S. D., and Nero, L. A. (2019). Lactobacillus curvatus UFV-NPAC1 and other lactic acid bacteria isolated from calabresa, a fermented meat product, present high bacteriocinogenic activity against Listeria monocytogenes. BMC Microbiol. 19, 63. doi: 10.1186/s12866-019-1436-4
Castrejón-Nájera, J., Ortega, C., Fajardo, R., Irgang, R., Tapia-Cammas, D., Poblete-Morales, M., et al. (2018). Isolation characterization, virulence potential of Weissella ceti responsible for weissellosis outbreak in rainbow trout (Oncorhynchus mykiss) cultured in Mexico. Transbound. Emerg. Dis. 65, 1401–1407. doi: 10.1111/tbed.12978
Cheaito, R. A., Awar, G., Alkozah, M., Cheaito, M. A., and El Majzoub, I. (2020). Meningitis due to Weissella confusa. The Am. J. Emerg. Med. 38, 1298e1–1298e3. doi: 10.1016/j.ajem.2020.02.004
Chen, C., Chen, X., Jiang, M., Rui, X., Li, W., Dong, M., et al. (2014a). A newly discovered bacteriocin from Weissella hellenica D1501 associated with Chinese Dong fermented meat (Nanx Wudl). Food Control 42, 116–124. doi: 10.1016/j.foodcont.2014.01.031
Chen, C., Liu, Y., Tian, H., Ai, L., and Yu, H. (2023). (2020). Metagenomic analysis reveals the impact of JIUYAO microbial diversity on fermentation and the volatile profile of Shaoxing-jiu. Erratum Food Microbiol. 111, 104205. doi: 10.1016/j.fm.2022.104205
Chen, C., Rui, X., Lu, Z., Li, W., and Dong, M. (2014b). Enhanced shelf-life of tofu by using bacteriocinogenic Weissella hellenica D1501 as bioprotective cultures. Food Control 46, 203–209. doi: 10.1016/j.foodcont.2014.05.004
Chen, Y. S., Liao, Y. J., Lan, Y. S., Wu, H. C., and Yanagida, F. (2017). Diversity of lactic acid bacteria associated with banana fruits in Taiwan. Curr. Microbiol. 74, 484–490. doi: 10.1007/s00284-017-1213-2
Cheng, W., Chen, X., Zeng, H., and Xue, X. (2023). Association between microbial community composition and quality indicators of strong-flavor Daqu of different producing regions in China. CyTA – J. Food 211, 82–92. doi: 10.1080/19476337.2022.2162974
Choi, H. J., Cheigh, C. I., Kim, S. B., Lee, J. C., Lee, D. W., Choi, S. W., et al. (2002). Weissella kimchii sp. nov., a novel lactic acid bacterium from kimchi. Int. J. Syst. Evol. Microbiol. 52, 507–511. doi: 10.1099/00207713-52-2-507
Choi, S. I., You, S., Kim, S., Won, G. Y., and Kang, C. H. (2021). Weissella cibaria MG5285 and Lactobacillus reuteri MG5I49 attenuated fat acculumation in adipose and hepatic steatosis in high-fat diet-induced C57BL/6J obese mice. Food Nutr. Res. 65, 8087. doi: 10.29219/fnr.v65.8087
Coda, R., Kianjam, M., Pontonio, E., Verni, M., Cagno, D., Katina, R., et al. (2017). Sourdough-type propagation of faba bean flour: dynamics of microbial consortia and biochemical implications. Int. J. Food Microbiol. 248, 10–21. doi: 10.1016/j.ijfoodmicro.2017.02.009
Collins, M. D., Samelis, J., Metaxopoulos, J., and Wallbanks, S. (1993). Taxonomic studies on some leuconostoc-like organisms from fermented sausages: description of a new genus Weissella for the Leuconostoc paramesenteroides group of species. J. Appl. Bacteriol. 75, 595–603. doi: 10.1111/j.1365-2672.1993.tb01600.x
Contente, D., Feito, J., Díaz-Formoso, L., Gómez-Sala, B., Borrero, J., Peña, N., et al. (2021). Draft Genome sequence of Weissella cibaria P71, a promising aquaculture probiotic strain isolated from Common Octopus (Octopus vulgaris). Microbiol. Res. Announc. 10, e0079221. doi: 10.1128/MRA.00792-21
Cupi, D., and Elvig-Jørgensen, S. G. (2019). Safety assessment of Weissella confusa- a direct-fed microbial candidate. Regulat. Toxicol. Pharmacol. 107, 104414. doi: 10.1016/j.yrtph.2019.104414
Das Purkhayastha, S., Bhattacgarya, M. K., Prasad, H. K., Upadhyaya, H., Das Laala, S., Pal, K., et al. (2017). Antibacterial activity of Weissella confusa isolated from vaginal swab of Indian women. Int. J. Adv. Chem. Eng. Biol. Sci. 4, 98–101. doi: 10.15242/IJACEBS.A0217021
Davati, N., Tabatabaee Yazdi, F., Zibaee, S., Shahidi, F., and Edalatian, M. R. (2015). Study of lactic acid bacteria community from raw milk of Iranian one humped camel and evaluation of their probiotic properties. Jundishapur J. Microbiol. 8, e16750. doi: 10.5812/jjm.8(5)2015.16750
De Bruyne, D., Camu, K., De Vuyst, N. L., and Vandamme, P. (2010). Weissella fabaria sp. nov., from a Ghanaian cocoa fermentation. Int. J. Syst. Evol. Microbiol. 60, 1999–2005. doi: 10.1099/ijs.0.019323-0
De Bruyne, K., Camu, N., Lefebvre, K., De Vuyst, L., and Vandamme, P. (2008). Weissella ghanensis Sp. nov., isolated from a Ghanaian cocoa fermentation. Int. J. Syst. Evol. Microbiol. 58, 2721–2725. doi: 10.1099/ijs.0.65853-0
Deatraksa, J., Sunthornthummas, S., Rangsiruji, A., Sarawaneeyaruk, S., Suwannasai, N., Pringsulaka, O., et al. (2018). Isolation of folate-producing Weissella spp. from Thai fermented fish (Plaa Som Fug). LWT Food Sci. Technol. 89, 388–391. doi: 10.1016/j.lwt.2017.11.016
Decimo, M., Quattrini, M., Ricci, G., Fortina, M. G., Brasca, M., Silvetti, T., et al. (2017). Evaluation of microbial consortia and chemical changes in spontaneous maize bran fermentation. AMB Express 7, 205. doi: 10.1186/s13568-017-0506-y
DeMan, J. C., Rogosa, M., and Sharpe, M. E. (1960). A medium for the cultivation of lactobacilli. J. Appl. Bacteriol. 23, 130–135. doi: 10.1111/j.1365-2672.1960.tb00188.x
Dentice Maidana, S., Aristimuño Ficoseco, C., Bassi, D., Cocconcelli, P. S., Puglisi, E., Savoy, G., et al. (2020). Biodiversity and technological-functional potential of lactic acid bacteria isolated from spontaneously fermented chia sourdough. Int. J. Food Microbiol. 316, 108425. doi: 10.1016/j.ijfoodmicro.2019.108425
Devi, P. B., Kavitake, D., Jayamanohar, J., and Shetty, P. H. (2021). Preferential growth stimulation of probiotic bacteria by galactan exopolysaccharide from Weissella confusa KR780676. Food Res. Int. 143, 110333. doi: 10.1016/j.foodres.2021.110333
Dey, D. K., and Kang, S. C. (2020). Weissella confusa DD_A7 pre-treatment to zebrafish larvae ameliorates the inflammation response against Escherichia coli O157:H7. Microbiol. Res. 237, 126489. doi: 10.1016/j.micres.2020.126489
Dey, D. K., Khan, I., and Kang, S. C. (2019). Anti-bacterial susceptibility profiling of Weissella confusa DD_A7 against the multidrug-resistant ESBL-positive E. coli. Microb. Pathog. 128, 119–130. doi: 10.1016/j.micpath.2018.12.048
Dicks, L. M. T. (1995). Relatedness of Leuconostoc species of the Leuconostoc sensu stricto line of descent, Leuconostoc oenos and Weissella paramesenteroides revealed by numerical analysis of total soluble cell protein patterns. Syst. Appl. Microbiol. 18, 99–102. doi: 10.1016/S0723-2020(11)80455-8
Dinoto, A., Suswati, S., Handayani, R., and Julistiono, H. (2021). “Weissella paramesenteroides from intestine of indonesian eel (Anguilla bicolor McClelland) and their potential antimicrobial property,” in AIP Conference Proceedings. Melvile, NY: AIP Publishing.
Do, K. H., Park, H. E., Kang, M. S., Kim, J. T., Yeu, J. E., Lee, W. K., et al. (2019). Effects of Weissella cibaria CMU on halitosis and calculus, plaque, and gingivitis indices in beagles. J. Vet. Dent. 36, 135–142. doi: 10.1177/0898756419872562
Do, K. H., Seo, K., Kim, S., Kim, S., Park, G. Y., Kang, M. S., et al. (2022). Therapeutic efficacy of Weissella cibaria CMU and CMS1 on allergic inflammation exacerbated by diesel exhaust particulate matter in a murine asthma model. Medicina 58, 1310. doi: 10.3390/medicina58091310
Dong, M., Li, Q., Xu, F., Wang, S., Chen, J., Li, W., et al. (2020). Effects of microbial inoculants on the fermentation characteristics and microbial communities of sweet sorghum bagasse silage. Sci. Rep. 10, 837. doi: 10.1038/s41598-020-57628-0
Du, X., Dai, F., Yao, F., Tan, M., and Pan, Q. (2018). Genome Sequence of Weissella cibaria M2, a potential probiotic strain isolated from the feces of a giant panda. Microbiol. Res. Announc. 7, e01121–e01118. doi: 10.1128/MRA.01121-18
Dubey, A. K., and Jeevaratnam, K. (2015). Structural characterization and functional evaluation of an exopolysaccharide produced by Weissella confusa AJ53, an isolate from fermented Uttapam Batter supplemented with Piper betle L. leaves. Food Sci. Biotechnol. 24, 2117–2124. doi: 10.1007/s10068-015-0281-y
Dubey, A. K., and Jeevaratnam, K. (2018). Purification and characterization of an antibacterial peptide (bacteriocin) produced by Weissella confusa AJ79. IOSR J. Pharm. Biol. Sci. 13, 17–24. doi: 10.9790/3008-1302031724
EFSA Panel on Additives and Products or Substances used in Animal Feed, Bampidis, V. Azimonti G. Bastos M. D. L. Christensen H. Dusemund B. (2021). Safety and efficacy of a feed additive consisting of Lacticaseibacillus rhamnosus (formerly Lactobacillus rhamnosus) IMI 507023 for all animal species (ALL-TECHNOLOGY (IRELAND) LIMITED [Alltech Ireland]). EFSA J. 19, e06700. doi: 10.2903/j.efsa.2021.6700
EFSA Panel on Additives and Products or Substances used in Animal Feed, Rychen, G. Aquilina G. Azimonti G. Bampidis V. Bastos M. D. L. (2017). Safety and efficacy of Lactobacillus rhamnosus DSM 29226 as a silage additive for all animal species. EFSA J. 15, e04673. doi: 10.2903/j.efsa.2017.4673
El Issaoui, K., Khay, E. O., Abrini, J., Zinebi, S., Amajoud, N., Senhaji, N. S., et al. (2021). Molecular identification and antibiotic resistance of bacteriocinogenic lactic acid bacteria isolated from table olives. Arch. Microbiol. 203, 597–607. doi: 10.1007/s00203-020-02053-0
Elizaquível, P., Pérez-Cataluña, A., Yépez, A., Aristimuño, C., Jiménez, E., Cocconcelli, P. S., et al. (2015). Pyrosequencing vs. culture-dependent approaches to analyze lactic acid bacteria associated to chicha, a traditional maize-based fermented beverage from Northwestern Argentina. Int. J. Food Microbiol. 198, 9–18. doi: 10.1016/j.ijfoodmicro.2014.12.027
El-Mekkawy, R. M., Hamour, N. E., Hassanein, W. A., and Allam, A. A. (2023). Evaluation of the antibacterial activity of Weissella confusa K3 cell-free supernatant against extended-spectrum βeta lactamase (ESBL) producing uropathogenic Escherichia coli U60. Saudi J. Biol. Sci. 30, 103595. doi: 10.1016/j.sjbs.2023.103595
Emamjomeh, M., Mohd Hashim, A., Abdul-Mutalib, N. A., Mokhtar, N. F. K., Mustapha, N. A., Maeda, T., et al. (2023). Profiling bacterial communities and foodborne pathogens on food-associated surface following contact with raw beef, chicken and pork using 16S amplicon metagenomics. Food Control 149, 109698. doi: 10.1016/j.foodcont.2023.109698
Ennahar, S., and Cai, Y. (2004). Genetic evidence that Weissella kimchii Choi et al. 2002 is a later heterotypic synonym of Weissella cibaria Björkroth et al. 2002. Int. J. Syst. Evol. Microbiol. 54, 463–465. doi: 10.1099/ijs.0.02783-0
Escobar-Zepeda, A., Sanchez-Flores, A., and Quirasco Baruch, M. (2016). Metagenomic analysis of a Mexican ripened cheese reveals a unique complex microbiota. Food Microbiol. 57, 116–127. doi: 10.1016/j.fm.2016.02.004
Espinoza-Monje, M., Campos, J., Alvarez Villamil, E., Jerez, A., Dnetice Maidana, S., Elean, M., et al. (2021). Characterization of Weissella viridescens UCO-SMC3 as a potential probiotic for the skin: its beneficial role in the pathogenesis of acne vulgaris. Microorganisms 9, 1486. doi: 10.3390/microorganisms9071486
Eveno, M., Salouhi, A., Belguesmia, Y., Bazinet, L., Gancel, F., Fliss, I., et al. (2021). Biodiversity and phylogenetic relationships of novel bacteriocinogenic strains isolated from animal's droppings at the zoological garden of Lille, France. Probiotics Antimicrob. Proteins 13, 218–228. doi: 10.1007/s12602-020-09657-4
Fairfax, M. R., Lephart, P. R., and Salimnia, H. (2014). Weissella confusa: problems with identification of an opportunistic pathogen that has been found in fermented foods and proposed as a probiotic. Front. Microbiol. 5, 254. doi: 10.3389/fmicb.2014.00254
Falasconi, I., Fontana, A., Patrone, V., Rebecchi, A., Duserm Garrido, G., Principato, L., et al. (2020). Genome-assisted characterization of Lactobacillus fermentum, Weissella cibaria, and Weissella confuse strains isolated from sorghum as starters for sourdough fermentation. Microorganisms 8, 1388. doi: 10.3390/microorganisms8091388
Fanelli, F., Montemurro, M., Chieffi, D., Cho, G. S., Franz, C. M. A. P., Dell'Aquila, A., Rizzello, C. G., and Fusco, V. (2022). Novel insights into the phylogeny and biotechnological potential of Weissella species. Front. Microbiol. 13, 914036. doi: 10.3389/fmicb.2022.914036
Fanelli, F., Montemurro, M., Chieffi, D., Cho, G.-S., Low, H.-Z., Hille, F., et al. (2023a). Motility in Periweissella species: genomic and phenotypic characterization and update on motility in Lactobacillaceae. Microorganisms 11, 2923. doi: 10.3390/microorganisms11122923
Fanelli, F., Montemurro, M., Verni, M., Garbetta, A., Bavaro, A. R., Chieffi, D., et al. (2023b). Probiotic potential and safety assessment of type strains of Weissella and Periweissella species. Microbiol. Spectr. 11, e0304722. doi: 10.1128/spectrum.03047-22
Fang, R. S., Dong, Y. C., Chen, F., and Chen, Q. H. (2015). Bacterial diversity analysis during the fermentation processing of traditional Chinese yellow rice wine revealed by 16S rDNA 454 pyrosequencing. J. Food Sci. 80, M2265–M2271. doi: 10.1111/1750-3841.13018
Fatmawati, N. N. D., Gotoh, K., Mayura, I. P. B., Nocianitri, K. A., Suwardana, G. N. R., Komalasari, N. L. G. Y., et al. (2020). Enhancement of intestinal epithelial barrier function by Weissella confusa F213 and Lactobacillus rhamnosus FBB81 probiotic candidates in an in vitro model of hydrogen peroxide-induced inflammatory bowel disease. BMC Res. Notes 13, 489. doi: 10.1186/s13104-020-05338-1
Fernández-Niño, M., Rodríguez-Cubillos, M. J., Herrera-Rocha, F., Anzola, J. M., Cepeda-Hernández, M. L., Aguirre Mejía, J. L., et al. (2021). Dissecting industrial fermentations of fine flavour cocoa through metagenomic analysis. Sci. Rep. 11, 8638. doi: 10.1038/s41598-021-88048-3
Fessard, A., and Remize, F. (2017). Why are Weissella spp. not used as commercial starter cultures for food fermentation? Fermentation 3, 38. doi: 10.3390/fermentation3030038
Fessard, A., and Remize, F. (2019). Genetic and technological characterization of lactic acid bacteria isolated from tropically grown fruits and vegetables. Int. J. Food Microbiol. 301, 61–72. doi: 10.1016/j.ijfoodmicro.2019.05.003
Fhoula, I., Rehaiem, A., Najjari, A., Usai, D., Boudabous, A., Sechi, L. A., et al. (2018). Functional probiotic assessment and in vivo cholesterol-lowering efficacy of Weissella sp. associated with arid lands living-hosts. Biomed Res. Int. 2018, 1654151. doi: 10.1155/2018/1654151
Figueiredo, H. C., Leal, C. A., Dorella, F. A., Carvalho, A. F., Soares, S. C., Pereira, F. L., et al. (2014a). Complete genome sequences of fish pathogenic Weissella ceti strains WS74 and WS105. Genome Announc. 2, e01014–e01014. doi: 10.1128/genomeA.01014-14
Figueiredo, H. C., Leal, G., Pereira, F. L., Soares, S. C., Dorella, F. A., Carvalho, A. F., et al. (2014b). Whole-genome sequence of Weissella ceti strain WS08, isolated from diseased rainbow trout in Brazil. Genome Announc. 2, e00851–e00814. doi: 10.1128/genomeA.00851-14
Figueiredo, H. C., Soares, S. C., Pereira, F. L., Dorella, F. A., Carvalho, A. F., Teixeira, J. P., et al. (2015). Comparative genome analysis of Weissella ceti, an emerging pathogen of farm-raised rainbow trout. BMC Genomics 16, 1095. doi: 10.1186/s12864-015-2324-4
Fonseca, J. F., Alvim, L. B., Nunes, Á. C., Oliveira, F. M. S., Amaral, R. S., Caliari, M. V., et al. (2019). Probiotic effect of Bifidobacterium longum 51A and Weissella paramesenteroides WpK4 on gerbils infected with Giardia lamblia. J. Appl. Microbiol. 127, 1184–1191. doi: 10.1111/jam.14338
Fuka, M. M., Wallisch, S., Engel, M., Welzl, G., Havranek, J., Schloter, M., et al. (2013). Dynamics of bacterial communities during the ripening process of different Croatian cheese types derived from raw ewe's milk cheeses. PLoS ONE 8, e80734. doi: 10.1371/journal.pone.0080734
Fukuda, D., and Nolasco-Hipolito, C. (2022). Draft genome sequence of Weissella soli strain DB-2, isolated from Nukadoko. Microbiol. Res. Announc. 11, e0116021. doi: 10.1128/MRA.01160-21
Fusco, V., Quero, G. M., Cho, G.-.S., Kabisch, J., Meske, D., Neve, H., et al. (2015). The genus Weissella: taxonomy, ecology and biotechnological potential. Front. Microbiol. 6, 155. doi: 10.3389/fmicb.2015.00155
Fusco, V., Quero, G. M., Stea, G., Morea, M., and Visconti, A. (2011). Novel PCR-based identification of Weissella confusa using an AFLP-derived marker. Int. J. Food Microbiol. 145, 437–443. doi: 10.1016/j.ijfoodmicro.2011.01.015
Galli, V., Venturi, M., Coda, R., Maina, N. H., and Granchi, L. (2020b). Isolation and characterization of indigenous Weissella confusa for in situ bacterial exopolysaccharides (EPS) production in chickpea sourdough. Food Res. Int. 138, 109785. doi: 10.1016/j.foodres.2020.109785
Galli, V., Venturi, M., Pini, N., and Granchi, L. (2020a). Technological feature assessment of lactic acid bacteria isolated from cricket powder's spontaneous fermentation as potential starters for cricket-wheat bread production. Foods 9, 1322. doi: 10.3390/foods9091322
Gao, L., Zhang, Z., Xing, Z., Li, Q., Kong, N., Wang, L., et al. (2022). The variation of intestinal autochthonous bacteria in cultured tiger pufferfish Takifugu rubripes. Front. Cell. Infect. Microbiol. 12, 1062512. doi: 10.3389/fcimb.2022.1062512
Garcia-Cancino, A., Albarracin, L., Espinoza-Monje, M., Campos-Martin, J., Garcia-Castillo, V., Nakano, Y., et al. (2019). Draft genome sequence of Weissella viridescens UCO-SMC3, isolated from the slime of Helix aspersa Müller snails. Microbiol. Res. Announc. 8, e01654–e01618. doi: 10.1128/MRA.01654-18
García-Hernández, Y., Pérez-Sánchez, T., Boucourt, R., Balcázar, J. L., Nicoli, J. R., Moreira-Silva, J., et al. (2016). Isolation, characterization and evaluation of probiotic lactic acid bacteria for potential use in animal production. Res. Vet. Sci. 108, 125–132. doi: 10.1016/j.rvsc.2016.08.009
Garroni, E., Doulgeraki, A. I., Pavli, F., Spiteri, D., and Valdramidis, V. P. (2020). Characterization of indigenous lactic acid bacteria in cow milk of the Maltese islands: a geographical and seasonal assessment. Microorganisms 8, 812. doi: 10.3390/microorganisms8060812
Garvie, E. I. (1967). The growth factor and amino acid requirements of species of the genus Leuconostoc, including Leuconostoc paramesenteroides (sp.nov.) and Leuconostoc oenos. J. Gen. Microbiol. 48, 439–447. doi: 10.1099/00221287-48-3-439
Garvie, E. I. (1986). “Genus Leuconostoc,” in Bergey's Manual of Systematic Bacteriology, Vol. 2, eds P. H. A. Sneath, N. S. Mair, M. E. Sharpe, and J. G. Holt (Baltimore, MD: Williams and Wilkins), 1071–1075.
Garzón, K., Ortega, C., and Tenea, G. N. (2017). Characterization of bacteriocin-producing lactic acid bacteria isolated from native fruits of Ecuadorian Amazon. Polish J. Microbiology 66, 473–481. doi: 10.5604/01.3001.0010.7037
Goh, H. F., and Philip, K. (2015). Purification and characterization of bacteriocin produced by Weissella confusa A3 of dairy origin. PLoS ONE 10, e0140434. doi: 10.1371/journal.pone.0140434
Gómez-Rojo, E. M., Romero-Santacreu, L., Jaime, I., and Rovira, J. (2015). A novel real-time PCR assay for the specific identification and quantification of Weissella viridescens in blood sausages. Int. J. Food Microbiol. 215, 16–24. doi: 10.1016/j.ijfoodmicro.2015.08.002
Govindaraj, K., Samayanpaulraj, V., Narayanadoss, V., and Uthandakalaipandian, R. (2021). Isolation of lactic acid bacteria from intestine of freshwater fishes and elucidation of probiotic potential for aquaculture application. Probiotics Antimicrob. Proteins 13, 1598–1610. doi: 10.1007/s12602-021-09811-6
Guo, Z., Wang, Y., Xiang, F., Hou, Q., and Zhang, Z. (2021). Bacterial diversity in pickled cowpea (Vigna unguiculata [Linn.] Walp) as determined by Illumina MiSeq sequencing and culture-dependent methods. Curr. Microbiol. 78, 1286–1297. doi: 10.1007/s00284-021-02382-3
Hamoud, N. E. H., and Sifour, M. (2021). Biodegradation of chlorpyrifos by a Weissella confusa strain and evaluation of some probiotic traits. Arch. Microbiol. 203, 3615–3621. doi: 10.1007/s00203-021-02353-z
Han, H. S., Yum, H., Cho, Y. D., and Kim, S. (2022). Improvement of halitosis by probiotic bacterium Weissella cibaria CMU: a randomized controlled trial. Front. Microbiol. 14, 1108762. doi: 10.3389/fmicb.2023.1108762
Han, J., Lin, X., Liang, H., Zhang, S., Zhu, B., Ji, C., et al. (2022). Improving the safety and quality of Roucha using amine-degrading lactic acid bacteria starters. Food Res. Int. 161, 111918. doi: 10.1016/j.foodres.2022.111918
Heng, N. C., Yeh, C. W., and Malik, A. (2017). Draft genome sequence of Weissella confusa MBF8-1, a glucansucrase- and bacteriocin-producing strain isolated from a homemade soy product. Genome Announc. 5, e01497–e01416. doi: 10.1128/genomeA.01497-16
Heo, J., Hamada, M., Cho, H., Weon, H. Y., Kim, J. S., Hong, S. B., et al. (2019). Weissella cryptocerci sp. nov., isolated from gut of the insect Cryptocercus kyebangensis. Int. J. Syst. Evol. Microbiol.gy 69, 2801–2806. doi: 10.1099/ijsem.0.003564
Hernández-Alcántara, A. M., Chiva, R., Mohedano Bonillo, M. L., Russo, P., Ruiz-Masó, J. A., Solar, G. D., et al. (2022). Weissella cibaria riboflavin-overproducing and dextran-producing strains useful for the development of functional bread. Front. Nutr. 9, 978831. doi: 10.3389/fnut.2022.978831
Hernández-Oaxaca, D., López-Sánchez, R., Lozano, L., Wacher-Rodarte, C., Segovia, L., López Munguía, A., et al. (2021). Diversity of Weissella confuse in Pozol and its carbohydrate metabolism. Front. Microbiol. 12, 629449. doi: 10.3389/fmicb.2021.629449
Hisham, M. B., Hashim, A. M., Mohd Hanafi, N., Abdul Rahman, N., Abdul Mutalib, N. E., Tan, C. K., et al. (2022). Bacterial communities associated with silage of different forage crops in Malaysian climate analysed using 16S amplicon metagenomics. Sci. Rep. 12, 7107. doi: 10.1038/s41598-022-08819-4
Holzapfel, W. H., and Kandler, O. (1969). Zur Taxonomie der Gattung Lac~obacillus Beijerinck. VI Lactobaciflus coprophilus subsp. confusus nov. subsp., eine neue unterart der untergattung betabacterium. Zentralblatt f i r Bakteriol. Mikrobiol. Hyg. 123, 657–666.
Holzapfel, W. H., and van Wyk, E. P. (1982). Lactobacillus kandleri sp. nov., a new species of the subgenus Betabacterium with glycinein the peptidoglycan. Zentralbl. Bakteriol. Parasitenkd. Infektionskr. Hyg. C3, 495–502. doi: 10.1016/S0721-9571(82)80007-0
Hong, Y. F., Lee, Y. D., Park, J. Y., Kim, S., Lee, Y. W., Jeon, B., et al. (2016). Lipoteichoic acid isolated from Weissella cibaria increases cytokine production in human monocyte-like THP-1 cells and mouse splenocytes. J. Microbiol. Biotechnol. 26, 1198–1205. doi: 10.4014/jmb.1601.01047
Huang, L., Cui, K., Mao, W., Du, Y., Yao, N., Li, Z., et al. (2020). Weissella cibaria attenuated LPS-induced dysfunction of intestinal epithelial barrier in a Caco-2 cell monolayer model. Front. Microbiol. 11, 2039. doi: 10.3389/fmicb.2020.02039
Hurt, W., Savarimuthu, S., Mughal, N., and Moor, L. S. P. (2021). A rare case of Weissella confusa endocarditis. Clin. Inf. Prac. 12, 100078. doi: 10.1016/j.clinpr.2021.100078
Hyun, D. W., Lee, J. Y., Sung, H., Kim, P. S., Jeong, Y. S., Lee, J. Y., et al. (2021). Brevilactibacter coleopterorum sp. nov., isolated from the intestine of the dark diving beetle Hydrophilus acuminatus, and Weissella coleopterorum sp. nov., isolated from the intestine of the diving beetle Cybister lewisianus. Int. J. Syst. Evol. Microbiol. 71, 779. doi: 10.1099/ijsem.0.004779
Jang, H. J., Kang, M. S., Yi, S. H., Hong, J. Y., and Hong, S. P. (2016). Comparative study on the characteristics of Weissella cibaria CMU and probiotic strains for oral care. Molecules 21, 1752. doi: 10.3390/molecules21121752
Jang, Y. J., Gwon, H. M., Jeong, W. S., Yeo, S. H., and Kim, S. Y. (2021). Safety evaluation of Weissella cibaria JW15 by phenotypic and genotypic property analysis. Microorganisms 9, 2450. doi: 10.3390/microorganisms9122450
Jeong, D. W., and Lee, J. H. (2015). Antibiotic resistance, hemolysis and biogenic amine production assessments of Leuconostoc and Weissella isolates for kimchi starter development. LWT Food Sci. Technol. 64, 1078–1084. doi: 10.1016/j.lwt.2015.07.031
Jeong, S. H., Jung, J. Y., Lee, S. H., Jin, H. M., and Jeon, C. O. (2013). Microbial succession and metabolite changes during fermentation of dongchimi, traditional Korean watery kimchi. Int. J. Food Microbiol. 164, 46–53. doi: 10.1016/j.ijfoodmicro.2013.03.016
Jin, H., Jeong, Y., Yoo, S. H., Johnston, T. V., Ku, S., Ji, G. E., et al. (2019). Isolation and characterization of high exopolysaccharide-producing Weissella confusa VP30 from young children's feces. Microb. Cell Fact. 18, 1–13. doi: 10.1186/s12934-019-1158-1
Jin, H., Park, J., Li, L., Ji, G. E., Johnston, T. V., Choe, D., et al. (2023). A randomized, double-blind, controlled human study: the efficacy of exopolysaccharides in milk fermented by Weissella confusa VP30 (VP30-EPS) to ameliorate functional constipation. J. Funct. Foods 104, 105491. doi: 10.1016/j.jff.2023.105491
Joglekar, A., Nimonkar, Y., Bajaj, A., and Prakash, O. (2023). Resolution of inter/intraspecies variation in Weissella group requires multigene analysis and functional characterization. J. Basic Microbiol. 63, 140–155. doi: 10.1002/jobm.202200357
Jung, J. Y., Lee, S. H., Kim, J. M., Park, M. S., Bae, J. W., Hahn, Y., et al. (2011). Metagenomic analysis of kimchi, a traditional Korean fermented food. Appl. Environ. Microbiol. 77, 2264–2274. doi: 10.1128/AEM.02157-10
Justé, A., Malfliet, S., Waud, M., Crauwels, S., Cooman, D., Aerts, L., et al. (2014). Bacterial community dynamics during industrial malting, with an emphasis on lactic acid bacteria. Food Microbiol. 39, 39–46. doi: 10.1016/j.fm.2013.10.010
Kahyani, F., Pirali-Kheirabadi, E., Shafiei, S., and Masouleh, A. S. (2021). Effect of dietary supplementation of potential probiotic Weissella confusa on innate immunity, immune-related genes expression, intestinal microbiota and growth performance of rainbow trout (Oncorhynchus mykiss). Aquacult. Nutr.tion 27, 1411–1420. doi: 10.1111/anu.13279
Kajala, I., Mäkel,ä, J., Coda, R., Shukla, S., Shi, Q., Maina, N. H., et al. (2016). Rye bran as fermentation matrix boosts in situ dextran production by Weissella confusa compared to wheat bran. Appl. Microbiol. Biotechnol. 100, 3499–3510. doi: 10.1007/s00253-015-7189-6
Kandler, O., Schillinger, U., and Weiss, N. (1983). Lactobacillus halotolerans sp. nov., nom. rev. and Lactobacillus minor sp. nov., nom. rev. Syst. Appl. Microbiol. 4, 284–285. doi: 10.1016/S0723-2020(83)80056-3
Kandler, O., and Weiss, N. (1986). “Genus Lactobacillus,” in Bergey's Manual of Systematic Bacteriology, Vol. 2, ed. P. H. A. Sneath, N. S. Mair, and M. E. Sharpepp (Baltimore, MD: Williams and Wilkins), 1209–1234.
Kang, M. S., Lee, D. S., Kim, M., Lee, S. A., and Nam, S. H. (2021). A randomized, double-blind placebo-controlled trial to assess the acidogenic potential of dental biofilms through a tablet containing Weissella cibaria CMU. Int. J. Environ. Res. Public Health 18, 4674. doi: 10.3390/ijerph18094674
Kang, M. S., Lee, D. S., Lee, S. A., Kim, M. S., and Nam, S. H. (2020). Effects of probiotic bacterium Weissella cibaria CMU on periodontal health and microbiota: a randomised, double-blind, placebo-controlled trial. BMC Oral Health 20, 243. doi: 10.1186/s12903-020-01231-2
Kang, M. S., and Park, G. Y. (2022). In vitro inactivation of respiratory viruses and rotavirus by the oral probiotic strain Weissella cibaria CMS1. Probiotics Antimicrob. Proteins 14, 760–766. doi: 10.1007/s12602-022-09947-z
Kang, M. S., Park, G. Y., Kim, J. H., Lee, A. R., and Pik, H.-. D. (2023b). Probiotic Weissella cibaria displays antibacterial and anti-biofilm effect against cavity-causing Streptococcus mutans. Microb. Pathog. 180, 106151. doi: 10.1016/j.micpath.2023.106151
Kang, M. S., Park, G. Y., and Lee, A. R. (2023a). In vitro preventive effect and mechanism of action of Weissella cibaria CMU against Streptococcus mutans biofilm formation and periodontal pathogens. Microorganisms 11, 962. doi: 10.3390/microorganisms11040962
Kang, M. S., Yeu, J. E., Oh, J. S., Shin, B. A., and Kim, J. H. (2017). Complete genome sequences of Weissella cibaria Strains CMU, CMS1, CMS2, and CMS3 isolated from infant saliva in South Korea. Genome Announc. 5, e01103–e01117. doi: 10.1128/genomeA.01103-17
Kanjan, P., Kimtun, A., Chaimongkol, S., and Sakpetch, P. (2022). Probiotic Weissella cibaria KY10 derived from digestive tract of healthy shrimp exhibits strong antibacterial effects against Vibrio parahaemolyticus causing AHPND in shrimp. Aquac. Res. 53, 2597–2607. doi: 10.1111/are.15777
Kariyawasam, K. M. G. M. M., Jeewanthi, R. K. C., Lee, N. K., and Paik, H. D. (2019). Characterization of cottage cheese using Weissella cibaria D30: physicochemical, antioxidant, and antilisterial properties. J. Dairy Sci. 102, 3887–3893. doi: 10.3168/jds.2018-15360
Kaur, R., and Tiwari, S. K. (2016). Isolation, identification and characterization of Pediococcus pentosaceus LB44 and Weissella confusa LM85 for the presence of bacteriocin-like inhibitory substances (BLIS). Microbiology 85, 540–547. doi: 10.1134/S0026261716050088
Kavitake, D., Balyan, S., Devi, P. B., and Shetty, P. H. (2019). Interface between food grade flavor and water soluble galactan biopolymer to form a stable water-in-oil-in-water emulsion. Int. J. Biol. Macromol. 135, 445–452. doi: 10.1016/j.ijbiomac.2019.05.199
Kavitake, D., Singh, S. P., Kandasamy, S., Devi, P. B., and Shetty, P. H. (2020). Report on aflatoxin-binding activity of galactan exopolysaccharide produced by Weissella confusa KR780676, 3. Biotechnology 10, 181. doi: 10.1007/s13205-020-02173-w
Kelkar, A., Mailman, J., and Chung, S. (2021). (2021). Caught by confusa: a rare case of Weisella confusa in a liver transplant patient. Chest 160, A842–3. doi: 10.1016/j.chest.2021.07.790
Khalil, T., Okla, M. K., Al-Qahtani, W. H., Ali, F., Zahra, M., Shakeela, Q., et al. (2022). Tracing probiotic producing bacterial species from gut of buffalo (Bubalus bubalis), South-East-Asia. Braz. J. Biol. 84, e259094. doi: 10.1590/1519-6984.259094
Khanlari, Z., Moayedi, A., Ebrahimi, P., Khomeiri, M., and Sadeghi, A. (2021). Enhancement of γ-aminobutyric acid (GABA) content in fermented milk by using Enterococcus faecium and Weissella confusa isolated from sourdough. Food Proc. Preserv. 45, e15869. doi: 10.1111/jfpp.15869
Kim, B., Seo, W. T., Kim, M. G., et al. (2012). Metagenomic lactic acid bacterial diversity during Mulkimchi fermentation based on 16S rRNA sequence. J. Korean Soc. Appl. Biol. Chem. 55, 787–792. doi: 10.1007/s13765-012-2185-3
Kim, D. H., Kang, M. S., Yeu, J. E., Lee, M. G., and Cho, J. W. (2020a). Inhibitory effect of the probiotic bacteria, Weissella cibaria CMU on halitosis: a randomized placebo-controlled study. J. Korean Acad. Oral Health 44, 246–252. doi: 10.11149/jkaoh.2020.44.4.246
Kim, D. S., Choi, S. H., Kim, D. W., Nam, S. H., Kim, R. N., Kang, A., et al. (2011). Genome sequence of Weissella cibaria KACC 11862. J. Bacteriol. 193, 797–798. doi: 10.1128/JB.01342-10
Kim, E., Cho, E. J., Yang, S. M., Kim, M. J., and Kim, H. Y. (2021a). Novel approaches for the identification of microbial communities in kimchi: MALDI-TOF MS analysis and high-throughput sequencing. Food Microbiol. 94, 103641. doi: 10.1016/j.fm.2020.103641
Kim, E., Cho, Y., Lee, Y., Han, S. K., Kim, C. G., Choo, D. W., et al. (2017). A proteomic approach for rapid identification of Weissella species isolated from Korean fermented foods on MALDI-TOF MS supplemented with an in-house database. Int. J. Food Microbiol. 243, 9–15. doi: 10.1016/j.ijfoodmicro.2016.11.027
Kim, E., Won, J. E., Yang, S. M., Kim, H. J., and Kim, H. Y. (2022a). Diversity of a lactic acid bacterial community during fermentation of gajami-sikhae, a traditional Korean fermented fish, as determined by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Foods 11, 909. doi: 10.3390/foods11070909
Kim, E., Yang, S. M., and Kim, H. Y. (2021b). Analysis of cultivable microbial community during kimchi fermentation using MALDI-TOF MS. Foods 10, 1068. doi: 10.3390/foods10051068
Kim, E., Yang, S. M., Kim, I. S., and Kim, H. Y. (2022b). Identification of novel molecular targets for Weissella species-specific real time PCR based on pangenome analysis. Appl. Microbiol. Biotechnol. 106, 4157–4168. doi: 10.1007/s00253-022-12003-z
Kim, H. J., Yeu, J. E., Lee, D. S., and Kang, M. S. (2019). Weissella cibaria CMU suppresses mgl gene expression and enzyme activity associated with bad breath. Int. J. Oral Biol. 44, 152–159. doi: 10.11620/IJOB.2019.44.4.152
Kim, H. Y., Bong, Y. J., Jeong, J. K., Lee, S., Kim, B. Y., Park, K. Y., et al. (2016). Heterofermentative lactic acid bacteria dominate in Korean commercial kimchi. Food Sci. Biotechnol. 25, 541–545. doi: 10.1007/s10068-016-0075-x
Kim, J. W., Jung, B. H., Lee, J. H., Yoo, K. Y., Lee, H., Kang, M. S., et al. (2020b). Effect of Weissella cibaria on the reduction of periodontal tissue destruction in mice. J. Periodontol. 91, 1367–1374. doi: 10.1002/JPER.19-0288
Kim, T. W., Lee, J. H., Kim, S. E., Park, M. H., Chang, H. C., Kim, H. Y., et al. (2009). Analysis of microbial communities in doenjang, a Korean fermented soybean paste, using nested PCR-denaturing gradient gel electrophoresis. Int. J. Food Microbiol. 131, 265–271. doi: 10.1016/j.ijfoodmicro.2009.03.001
Kinoshita, H., Ohtake, F., Ariga, Y., and Kimura, K. (2016). Comparision and characterization of biosorption by Weissella viridescens MY 205 of periodic group 12 metal ions. Anim. Sci. J. 87, 271–276. doi: 10.1111/asj.12425
Kline, L., and Sugihara, T. F. (1971). Microorganisms of the San Francisco sourdough process. II. Isolation and characterization of undescribed bacterial species responsible for souring activity. Appl. Microbiol. 21, 459–465. doi: 10.1128/am.21.3.459-465.1971
Koirala, P., Maina, N. H., Nihtil,ä, H., Katina, K., and Coda, R. (2021). Brewers' spent grain as substrate for dextran biosynthesis by Leuconostoc pseudomesenteroides DSM20193 and Weissella confusa A16. Microb. Cell Fact. 20, 1–13. doi: 10.1186/s12934-021-01515-4
Ku, H. J., Kim, Y. T., and Lee, J. H. (2017). Genomic insights of Weissella jogaejeotgali FOL01T reveals its food fermentation ability and human gut adaptive potential for probiotic applications in food industries. J. Microbiol. Biotechnol. 27, 943–946. doi: 10.4014/jmb.1702.02054
Kulwichit, W., Nilgate, S., Chatsuwan, T., Krajiw, S., Unhasuta, C., and Chongthaleong, A. (2007). Accuracies of Leuconostoc phenotypic identification: a comparison of API systems and conventional phenotypic assays. BMC Infect. Dis. 7, 69. doi: 10.1186/1471-2334-7-69
Kwak, M. J., Choi, S. B., Kim, B. Y., and Chun, J. (2019). Genome-based reclassification of Weissella jogaejeotgali as a later heterotypic synonym of Weissella thailandensis. Int. J. Syst. Evol. Microbiol. 69, 3672–3675. doi: 10.1099/ijsem.0.003315
Kwon, A., and Park, Y. S. (2021). Immunostimulatory activity of synbiotics using Lactococcus lactis SG-030 and glucooligosaccharides from Weissella cibaria YRK005. Microorganisms 9, 2437. doi: 10.3390/microorganisms9122437
Lakra, A. K., Domdi, L., Hanjon, G., Tilwani, M. Y., and Arul, V. (2020a). Some probiotic potential of Weissella confusa MD1 and Weissella cibaria MD2 isolated from fermented batter. LWT Food Sci. Technol. 125, 109261. doi: 10.1016/j.lwt.2020.109261
Lakra, A. K., Domdi, L., Tilwani, Y. M., and Arul, V. (2020b). Physicochemical and functional characterization of mannan exopolysaccharide from Weissella confusa MD1 with bioactivities. Int. J. Biol. Macromol. 143, 797–805. doi: 10.1016/j.ijbiomac.2019.09.139
Lakra, A. K., Ramatchandirane, M., Kumar, S., Suchiang, K., and Arul, V. (2021). Physico-chemical characterization and aging effects of fructan exopolysaccharide produced by Weissella cibaria MD2 on Caenorhabditis elegans. LWT Food Sci. Technol. 143, 111100. doi: 10.1016/j.lwt.2021.111100
Le, B., Ngoc, A. P. T., and Yang, S. H. (2020). Synbiotic fermented soymilk with Weissella cibaria FB069 and xylooligosaccharides prevents proliferation in human colon cancer cells. J. Appl. Microbiol. 128, 1486–1496. doi: 10.1111/jam.14551
Le, B., and Yang, S. H. (2018). Isolation of Weissella strains as potent probiotics to improve antioxidant activity of salted squid by fermentation. J. Appl. Biol. Chem. 61, 93–100. doi: 10.3839/jabc.2018.014
Lee, D. S., Lee, S. A., Kim, M., Nam, S. H., and Kang, M. S. (2020). Reduction of halitosis by a tablet containing Weissella cibaria CMU: a randomized, double-blind, placebo-controlled study. J. Med. Food 23, 649–657. doi: 10.1089/jmf.2019.4603
Lee, H. A., Song, B. R., Kim, H. R., Kim, J. E., Yun, W. B., Park, J. J., et al. (2017). Butanol extracts of Asparagus cochinchinensis fermented with Weissella cibaria inhibit iNOS-mediated COX-2 induction pathway and inflammatory cytokines in LPS-stimulated RAW264.7 macrophage cells. Exp. Ther. Med. 14, 4986–4994. doi: 10.3892/etm.2017.5200
Lee, H. W., Yoon, S. R., Dang, Y. M., Yun, J. H., Jeong, H., Kim, K. N., et al. (2022). Metatranscriptomic and metataxonomic insights into the ultra-small microbiome of the Korean fermented vegetable, kimchi. Front. Microbiol. 13, 1026513. doi: 10.3389/fmicb.2022.1026513
Lee, J. H., Bae, J. W., and Chun, J. (2012). Draft genome sequence of Weissella koreensis KCTC 3621T. J. Bacteriol. 194, 5711–5712. doi: 10.1128/JB.01356-12
Lee, J. S., Heo, G. Y., Lee, J. W., Oh, Y. J., Park, J. A., Park, Y. H., et al. (2005). Analysis of kimchi microflora using denaturing gradient gel electrophoresis. Int. J. Food Microbiol. 102, 143–150. doi: 10.1016/j.ijfoodmicro.2004.12.010
Lee, J. S., Lee, K. C., Ahn, J. S., Mheen, T. I., Pyun, Y. R., and Park, Y. H. (2002). Weissella koreensis sp. nov., isolated from kimchi. Int. J. Syst. Evol. Microbiol. 52, 1257–1261. doi: 10.1099/00207713-52-4-1257
Lee, K. W., Park, J. Y., Jeong, H. R., Heo, H. J., Han, N. S., Kim, J. H., et al. (2012). Probiotic properties of Weissella strains isolated from human faeces. Anaerobe 18, 96–102. doi: 10.1016/j.anaerobe.2011.12.015
Lee, M., Song, J. H., Jung, M. Y., Lee, S. H., and Chang, J. Y. (2017). Large-scale targeted metagenomics analysis of bacterial ecological changes in 88 kimchi samples during fermentation. Food Microbiol. 66, 173–183. doi: 10.1016/j.fm.2017.05.002
Lee, M. R., Tsai, C. J., Teng, S. H., and Hsueh, P. R. (2015). Identification of Weissella species by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Front. Microbiol. 6, 1246. doi: 10.3389/fmicb.2015.01246
Lee, S. H., Jung, J. Y., and Jeon, C. O. (2015a). Source tracking and succession of kimchi lactic acid bacteria during fermentation. J. Food Sci. 80, M1871–M1877. doi: 10.1111/1750-3841.12948
Lee, S. H., Ku, H. J., Ahn, M. J., Hong, J. S., Lee, S. H., Shin, H., et al. (2015b). Weissella jogaejeotgali sp. nov., isolated from jogae jeotgal, a traditional Korean fermented seafood. Int. J. Syst. Evol. Microbiol. 65, 4674–4681. doi: 10.1099/ijsem.0.000631
Lee, Y. J., Lee, A., Yoo, H. J., Kim, M., and Noh, G. M. (2018). Supplementation with the probiotic strain Weissella cibaria JW15 enhances natural killer cell activity in nondiabetic subjects. J. Funct. Foods 48, 153–158. doi: 10.1016/j.jff.2018.07.009
Li, J., Ai, L., Xu, F., Hu, X., Yao, Y., Wang, L., et al. (2022). Structural characterization of exopolysaccharides from Weissella cibaria NC516, 11. in distiller grains and its improvement in gluten-free dough. Int. J. Biol. Macromol. 199, 17–23. doi: 10.1016/j.ijbiomac.2021.12.089
Li, S. W., Chen, Y. S., Lee, Y. S., Yang, C. H., Srionnual, S., Wu, H. C., et al. (2017). Comparative genomic analysis of bacteriocin-producing Weissella cibaria 110. Appl. Microbiol. Biotechnol. 101, 1227–1237. doi: 10.1007/s00253-016-8073-8
Li, W., Chen, Y., and Wang, T. (2021). Cadmium biosorption by lactic acid bacteria Weissella viridescens ZY-6. Food Control 123, 107747. doi: 10.1016/j.foodcont.2020.107747
Li, Y. Q., Tian, W. L., and Gu, C. T. (2020). Weissella sagaensis sp. nov., isolated from traditional Chinese yogurt. Int. J. Syst. Evol. Microbiol. 70, 2485–2492. doi: 10.1099/ijsem.0.004062
Liang, H., Chen, H., Zhang, W., Yu, C., Ji, C., Lin, X., et al. (2018). Investigation on microbial diversity of industrial Zhacai paocai during fermentation using high-throughput sequencing and their functional characterization. LWT Food Sci. Technol. 91, 460–466. doi: 10.1016/j.lwt.2018.01.088
Lim, H. S., Yeu, J. E., Hong, S. P., and Kang, M. S. (2018). Characterization of antibacterial cell-free supernatant from oral care probiotic Weissella cibaria, CMU. Molecules 23, 1984. doi: 10.3390/molecules23081984
Lim, S. K., Kwon, M. S., Lee, J., Oh, Y. J., Jang, J. Y., Lee, J. H., et al. (2017). Weissella cibaria WIKIM28 ameliorates atopic dermatitis-like skin lesions by inducing tolerogenic dendritic cells and regulatory T cells in BALB/c mice. Sci. Rep. 7, 40040. doi: 10.1038/srep40040
Lin, S. T., Wang, L. T., Wu, Y. C., Guu, J. J., Tamura, T., Mori, K., et al. (2020). Weissella muntiaci sp. nov., isolated from faeces of Formosan barking deer (Muntiacus reevesi). Int. J. Syst. Evol. Microbiol. 70, 1578–1584. doi: 10.1099/ijsem.0.003937
Liu, D., Zhang, C., Zhang, J., Xin, X., and Liao, X. (2021). Metagenomics reveals the formation mechanism of flavor metabolites during the spontaneous fermentation of potherb mustard (Brassica juncea var. multiceps). Food Res. Int. 148, 110622. doi: 10.1016/j.foodres.2021.110622
Liu, X., Qu, H., Gou, M., Guo, H., Wang, L., Yan, X., et al. (2020). Application of Weissella cibaria X31 or Weissella confusa L2 as a starter in low nitrite dry-fermented sausages. Int. J. Food Eng. 16, 344. doi: 10.1515/ijfe-2019-0344
Llamas-Arriba, M. G., Hernández-Alcántara, A. M., Mohedano, M. L., Chiva, R., Celador-Lera, L., Velázquez, E., et al. (2021). Lactic acid bacteria isolated from fermented doughs in Spain produce dextrans and riboflavin. Foods 10, 2004. doi: 10.3390/foods10092004
Lobo, R. E., Orrillo, P. A., Ribotta, S. B., De Valdez, J., Garcia, G. F., Cabello, M. S., et al. (2022). Structural characterization of a homopolysaccharide produced by Weissella cibaria FMy 2-21-1 and its potential application as a green corrion inhibiting film. Int. J. Biol. Macromol. 212, 193–201. doi: 10.1016/j.ijbiomac.2022.05.105
Lopez, C. M., Rocchetti, G., Fontana, A., Lucini, L., and Rebecchi, A. (2022). Metabolomics and gene-metabolite networks reveal the potential of Leuconostoc and Weissella strains as starter cultures in the manufacturing of bread without baker's yeast. Food Res. Int. 162, 112023. doi: 10.1016/j.foodres.2022.112023
López-Hernández, M., Rodríguez-Alegría, M. E., López-Munguía, A., and Wacher, C. (2018). Evaluation of xylan as carbon source for Weissella spp., a predominant strain in pozol fermentation. LWT Food Sci. Technol. 89, 192–197. doi: 10.1016/j.lwt.2017.10.030
Månberger, A., Verbrugghe, P., Guð*mundsdóttir, E. E., Santesson, S., Nilsson, A., Hreggvið*sson, G. Ó., et al. (2020). Taxogenomic assessment and genomic characterisation of Weissella cibaria strain 92 able to metabolise oligosaccharides derived from dietary fibres. Sci. Rep. 10, 5853. doi: 10.1038/s41598-020-62610-x
Ma, P., Duan, N., Ye, H., Xia, Y., Ding, Z., Wang, Z., et al. (2022). Selection, truncation and fluorescence polarization based aptasensor for Weissella viridescens detection. Talanta 246, 123499. doi: 10.1016/j.talanta.2022.123499
Magnusson, J., Jonsson, H., Schnürer, J., and Roos, S. (2002). Weissella soli sp. nov., a lactic acid bacterium isolated from soil. Int. J. Syst. Evol. Microbiol. 52, 831–834. doi: 10.1099/00207713-52-3-831
Malang, S. K., Maina, N. H., Schwab, C., Tenkanen, M., and Lacroix, C. (2015). Characterization of exopolysaccharide and ropy capsular polysaccharide formation by Weissella. Food Microbiol. 46, 418–427. doi: 10.1016/j.fm.2014.08.022
Malik, A., Sheilla, S., Firdausi, W., Handayani, T., and Saepudin, E. (2015). Sucrase activity and exopolysaccharide partial characterization from three Weissella confusa strains. HYATI J. Biosci. 22, 130–135. doi: 10.1016/j.hjb.2015.10.003
Malik, A., Sumayyah, S., Yeh, C. W., and Heng, N. C. (2016). Identification and sequence analysis of pWcMBF8-1, a bacteriocin-encoding plasmid from the lactic acid bacterium Weissella confusa. FEMS Microbiol. Lett. 363, fnw059. doi: 10.1093/femsle/fnw059
Mantziari, A., and Rautava, S. (2021). Factors influencing the microbial composition of human milk. Semin. Perinatol. 45, 151507. doi: 10.1016/j.semperi.2021.151507
Marin-Gómez, W., Grande, M. J., Pérez-Pulido, R., Galvez, A., and Lucas, R. (2020). Changes in the bacterial diversity of human milk during late lactation period (weeks 21 to 48). Foods 9, 1184. doi: 10.3390/foods9091184
Marino, M., De Dubsky, G. D., Wittenau, G., Saccà, E., Cattonaro, F., and Spadotto, A. (2019). Metagenomic profiles of different types of Italian high-moisture Mozzarella cheese. Food Microbiol. 79, 123–131. doi: 10.1016/j.fm.2018.12.007
Martín, R., Heilig, H. G., Zoetendal, E. G., Jiménez, E., Fernández, L., Smidt, H., et al. (2007). Cultivation-independent assessment of the bacterial diversity of breast milk among healthy women. Res. Microbiol. 158, 31–37. doi: 10.1016/j.resmic.2006.11.004
Masoud, W., Vogensen, F. K., Lillevang, S., Abu Al-Soud, W., Sørensen, S. J., Jakobsen, M., et al. (2012). The fate of indigenous microbiota, starter cultures, Escherichia coli, Listeria innocua and Staphylococcus aureus in Danish raw milk and cheeses determined by pyrosequencing and quantitative real time (qRT)-PCR. Int. J. Food Microbiol. 153, 192–202. doi: 10.1016/j.ijfoodmicro.2011.11.014
Massasati, S. A., and Waseem, S. (2023). A novel case of Weissella confusa infective endocarditis of a bio-prosthetic valve: management and treatment. Cureus 15, e37514. doi: 10.7759/cureus.37514
Medford, R., Patel, S. N., and Evans, G. A. (2014). A confusing case-Weissella confusa prosthetic joint infection: a case report and review of the literature. Can. J. Infect. Dis. Med. Microbiol. 25, 173–175. doi: 10.1155/2014/745856
Medina, M., Fernandez-Espinel, C., Sotil, G., Yunis, A. J., and Flores-Dominick, V. (2020). First description of Weissella ceti associated with mortalities in farmed rainbow trout (Oncorhynchus mykiss) in Peru. Aquaculture 529, 735608. doi: 10.1016/j.aquaculture.2020.735608
Mercha, I., Lakram, N., Kabbour, M. R., Bouksaim, M., Zkhiri, F., El Maadoudi, E. H., et al. (2020). Probiotic and technological features of Enterococcus and Weissella isolates from camel milk characterised by an Argane feeding regimen. Arch. Microbiol. 202, 2207–2219. doi: 10.1007/s00203-020-01944-6
Mitomi, K., Hoai, T. D., Nishiki, I., and Yoshida, T. (2018). First isolation of Weissella ceti responsible for outbreaks of weissellosis in farmed rainbow trout in Japan. J. Fish Dis. 41, 847–850. doi: 10.1111/jfd.12787
Montemurro, M., Beccaccioli, M., Perri, G., Rizzello, C. G., Reverberi, M., Pontonio, E., et al. (2023). A chestnut-hemp type-II sourdough to improve technological, nutritional, and sensory properties of gluten-free bread. Int. J. Food Microbiol. 404, 110322. doi: 10.1016/j.ijfoodmicro.2023.110322
Montemurro, M., Pontonio, E., and Rizzello, C. G. (2021). Design of a “clean-label” gluten-free bread to meet consumers demand. Foods 10, 462. doi: 10.3390/foods10020462
Mortezaei, F., Royan, M., Allaf Noveirian, H., Babakhani, A., Alaie Kordghashlaghi, H., Balcázar, J. L., et al. (2020). In vitro assessment of potential probiotic characteristics of indigenous Lactococcus lactis and Weissella oryzae isolates from rainbow trout (Oncorhynchus mykiss Walbaum). J. Appl. Microbiol. 129, 1004–1019. doi: 10.1111/jam.14652
Mutungwazi, A., Ijoma, G. N., Ogola, H. J. O., and Matambo, T. S. (2022). Physico-chemical and metagenomic profile analyses of animal manures routinely used as inocula in anaerobic digestion for biogas production. Microorganisms 10, 671. doi: 10.3390/microorganisms10040671
Nacef, M., Chevalier, M., and Chollet, S. (2017). MALDI-TOF mass spectrometry for the identification of lactic acid bacteria isolated from a French cheese: the Maroilles. Int. J. Food Microbiol. 247, 2–8. doi: 10.1016/j.ijfoodmicro.2016.07.005
Nath, S., Roy, M., Sikidar, J., Deb, B., Sharma, I., Guha, A., et al. (2021). Characterization and in-vitro screening of probiotic potential of novel Weissella confusa strain GCC_19R1 isolated from fermented sour rice. Curr. Res. Biotechnol. 3, 99–108. doi: 10.1016/j.crbiot.2021.04.001
Nie, Z., Zheng, Y., Du, H., Xie, S., and Wang, M. (2015). Dynamics and diversity of microbial community succession in traditional fermentation of Shanxi aged vinegar. Food Microbiol. 47, 62–68. doi: 10.1016/j.fm.2014.11.006
Nisiotou, A., Dourou, D., Filippousi, M. E., Banilas, G., and Tassou, C. (2014). Weissella uvarum sp. nov., isolated from wine grapes. Int. J. Syst. Evol. Microbiol. 64, 3885–3890. doi: 10.1099/ijs.0.066209-0
Niven, C. F., and Evans, J. B. (1957). Lactobacillus viridescens nov. spec., a heterofermentative species that produces a green dis- coloration of cured meat pigments. J. Bacteriology 73, 758–759. doi: 10.1128/jb.73.6.758-759.1957
Oh, S. J., Shin, N. R., Hyun, D. W., Kim, P. S., Kim, J. Y., Kim, M. S., et al. (2013). Weissella diestrammenae Sp. nov., isolated from the gut of a camel cricket (Diestrammena coreana). Int. J. Syst. Evol. Microbiol. 63(Pt 8), 2951–2956. doi: 10.1099/ijs.0.047548-0
Oikonomou, G., Addis, M. F., Chassard, C., Nader-Macias, M. E. F., Grant, I., Delbès, C., et al. (2020). Milk microbiota: what are we exactly talking about? Front. Microbiol. 11, 60. doi: 10.3389/fmicb.2020.00060
Ojekunle, O., Banwo, K., and Sanni, A. I. (2017). In vitro and in vivo evaluation of Weissella cibaria and Lactobacillus plantarum for their protective effect against cadmium and lead toxicities. Lett. Appl. Microbiol. 64, 379–385. doi: 10.1111/lam.12731
Oladajo, B. O., and Oluwasola, H. M. (2021). Oral administration of Weissella and Pediococcus sp alleviates formalin induced inflammation in rats by cytokine modulation. IFE J. Sci. 23, 95–104. doi: 10.4314/ijs.v23i2.10
Oladejo, B. O., and Oluwasola, J. M. (2022). Hematological modulatory effects of Weissella and Pediococcus sp. on formalin-induced inflammation in wistar rats. Asian J. Biotechnol. 19, 28–34. doi: 10.13057/biotek/c190105
Olojede, A. O., Sanni, A. I., Banwo, K., and Michael, T. (2022). Improvement of texture, nutritional qualities, and consumers' perceptions of sorghum-based sourdough bread made with Pediococcus pentosaceus and Weissella confusa strains. Fermentation 8, 32. doi: 10.3390/fermentation8010032
Ono, H., Nishio, S., Tsurii, J., Kawamoto, T., Sonomoto, K., Nakayama, J., et al. (2014). Monitoring of the microbiota profile in nukadoko, a naturally fermented rice bran bed for pickling vegetables. J. Biosci. Bioeng. 118, 520–525. doi: 10.1016/j.jbiosc.2014.04.017
Otoni, I., Paula, R., Alves, J., Roseira, J., Pimentel, F., Ribeiro, K., et al. (2018). PSVIII-35 Isolation and identification of lactic acid bacteria in guinea grass silages. J. Anim. Sci. 96, 219. doi: 10.1093/jas/sky404.476
Öz, E., Kaban, G., Bariş, Ö., and Kaya, M. (2017). Isolation and identification of lactic acid bacteria from pastirma. Food Control 77, 158–162. doi: 10.1016/j.foodcont.2017.02.017
Pabari, K., Pithva, S., Kothari, C., Purama, R. K., Kondepudi, K. K., Vyas, B. R. M., et al. (2020). Evaluation of probiotic properties and prebiotic utilization potential of Weissella paramesenteroides isolated from fruits. Probiotics Antimicrob. Proteins 12, 1126–1138. doi: 10.1007/s12602-019-09630-w
Padonou, S. W., Schillinger, U., Nielsen, D. S., Franz, C. M. A. P., Hansen, M., Hounhouigan, J. D., et al. (2010). Weissella beninensis sp. nov., a motile lactic acid bacterium from submerged cassava fermentations, and emended description of the genus Weissella. Int. J. Syst. Evol. Microbiol. 60, 2193–2198. doi: 10.1099/ijs.0.014332-0
Panthee, S., Paudel, A., Blom, J., Hamamoto, H., and Sekimizu, K. (2019). Complete genome sequence of Weissella hellenica 0916-4-2 and its comparative genomic analysis. Front. Microbiol. 10, 1619. doi: 10.3389/fmicb.2019.01619
Papalexandratou, Z., Kaasik, K., Kauffmann, L. V., Skorstengaard, A., Bouillon, G., Espensen, J. L., et al. (2019). Linking cocoa varietals and microbial diversity of Nicaraguan fine cocoa bean fermentations and their impact on final cocoa quality appreciation. Int. J. Food Microbiol. 304, 106–118. doi: 10.1016/j.ijfoodmicro.2019.05.012
Park, H. E., Do, K. H., and Lee, W. K. (2019). The immune-modulating effects of viable Weissella cibaria JW15 on RAW 264.7 macrophage cells. J. Biomed. Res. 34, 36–43. doi: 10.7555/JBR.33.20190095
Park, H. E., Kang, K. W., Kim, B. S., Lee, S. M., and Lee, W. K. (2017). Immunomodulatory potential of Weissella cibaria in aged C57BL/6J mice. J. Microbiol. Biotechnol. 27, 2094–2103. doi: 10.4014/jmb.1708.08016
Park, H. E., and Lee, W. K. (2018). Immune enhancing effects of Weissella cibaria JW15 on BALB/c mice immunosuppressed by cyclophosphamide. J. Funct. Foods 49, 518–525. doi: 10.1016/j.jff.2018.09.003
Park, S., Saravanakumar, K., Sathiyaseelan, A., Park, S. J., Hu, X., Wang, M.-.H., et al. (2022a). Cellular antioxidant properties of nontoxic exopolysaccharide extracted from Lactobacillales (Weissella cibaria) isolated from Korean kimchi. LWT – Food Sci. Technol. 154, 112727. doi: 10.1016/j.lwt.2021.112727
Park, S. H., Lee, M. R., Yang, S. Y., Lee, J. Y., Lee, H. H., Seong, Y. J., et al. (2022b). In vivo functional effects of Weissella confusa VP30 exopolysacchariders on looperamide-induced constipation in rats. Food Sci. Biotechnol. 31, 1703–1715. doi: 10.1007/s10068-022-01159-z
Patrone, V., Al-Surrayai, T., Romaniello, F., Fontana, A., Milani, G., Sagheddu, V., et al. (2021). Integrated phenotypic-genotypic analysis of candidate probiotic Weissella cibaria strains isolated from dairy cows in Kuwait. Probiotics Antimicrob. Proteins 13, 809–823. doi: 10.1007/s12602-020-09715-x
Pepe, O., Villani, F., and Coppola, S. (2001). Differential viable count of mixed starter cultures of lactic acid bacteria in doughs by using modified Chalmers medium. Microbiol. Res. 155, 351–354. doi: 10.1016/S0944-5013(01)80015-X
Petrovici, A. R., Rosca, I., Stoica, I., Silion, M., Nicolescu, A., Dodi, G., et al. (2018). Biosynthesis of exopolysaccharides by Weissella confusa in a new culture medium. Romanian Biotechnol. Lett. 23, 13637.
Prado, G. K. S., Torrinha, K. C., Cruz, R. E., Gonçalves, A. B. B., Silva, C. A. V., Oliveira, F. M. S., et al. (2020). Weissella paramesenteroides WpK4 ameliorate the experimental amoebic colitis by increasing the expression of MUC-2 and the intestinal epithelial regeneration. J. Appl. Microbiol. 129, 1706–1719. doi: 10.1111/jam.14671
Praet, J., Meeus, I., Cnockaert, M., Houf, K., Smagghe, G., Vandamme, P., et al. (2015). Novel lactic acid bacteria isolated from the bumble bee gut: Convivina intestini gen. nov., sp. nov., Lactobacillus bombicola sp. nov., and Weissella bombi sp. nov. Antonie Van Leeuwenhoek 107, 1337–1349. doi: 10.1007/s10482-015-0429-z
Qiao, N., Bechtner, J., Cnockaert, M., Depoorter, E., Díaz-Muñoz, C., Vandamme, P., et al. (2023). Comparative genomic analysis of periweissella and the characterization of novel motile species. Appl. Environ. Microbiol. 89, e0103423. doi: 10.1128/aem.01034-23
Reis, N. A., Saraiva, M. A., Duarte, E. A., de Carvalho, H., Vieira, E. A. B. B., and Evangelista-Barreto, N. S. (2016). Probiotic properties of lactic acid bacteria isolated from human milk. J. Appl. Microbiol. 121, 811–820. doi: 10.1111/jam.13173
Rizzello, C. G., Coda, R., Wang, Y., Verni, M., Kajala, I., Katina, K., et al. (2019). Characterization of indigenous Pediococcus pentosaceus, Leuconostoc kimchii, Weissella cibaria and Weissella confusa for faba bean bioprocessing. Int. J. Food Microbiol. 302, 24–34. doi: 10.1016/j.ijfoodmicro.2018.08.014
Roh, S. W., Kim, K. H., Nam, Y. D., Chang, H. W., Park, E. J., Bae, J. W., et al. (2010). Investigation of archaeal and bacterial diversity in fermented seafood using barcoded pyrosequencing. ISME Journal 4, 1–16. doi: 10.1038/ismej.2009.83
Rolim, P. M., Hu, Y., and Gänzle, M. G. (2019). Sensory analysis of juice blend containing isomalto-oligosaccharides produced by fermentation with Weissella cibaria. Food Res. Int. 124, 86–92. doi: 10.1016/j.foodres.2018.08.089
Romi, W., Ahmed, G., and Jeyaram, K. (2015). Three-phase succession of autochthonous lactic acid bacteria to reach a stable ecosystem within 7 days of natural bamboo shoot fermentation as revealed by different molecular approaches. Mol. Ecol. 24, 3372–3389. doi: 10.1111/mec.13237
Ruiz Rodríguez, L. G., Mohamed, F., Bleckwedel, J., Medina, R., De Vuyst, D., Hebert, L., et al. (2019). Diversity and functional properties of lactic acid bacteria isolated from wild fruits and flowers present in Northern Argentina. Front. Microbiol. 10, 1091. doi: 10.3389/fmicb.2019.01091
Rüstemoglu, M., Erkan, M. E., Cengiz, G., and Hajyzadeh, M. (2023). Bacterial metagenome profiling of hand-made herby cheese samples utilizing high-throughput sequencing to detect geographical indication and marketing potential. Heliyon 9, e13334. doi: 10.1016/j.heliyon.2023.e13334
Saelao, S., Keasuwan, S., Benjakul, S., and Maneerat, S. (2016). Safety evaluation and bacterial community of kung-som using PCR-DGGE technique. Songklanakarin J. Sci. Technol. 38, 413–419.
Salazar, J. K., Carstens, C. K., Ramachandran, P., Shazer, A. G., Narula, S. S., Reed, E., et al. (2018). Metagenomics of pasteurized and unpasteurized gouda cheese using targeted 16S rDNA sequencing. BMC Microbiol. 18, 189. doi: 10.1186/s12866-018-1323-4
Samelis, J., Rementzis, J., Tsakalidou, E., and Metaxopoulos, J. (1998). Usefulness of rapid GC analysis of cellular fatty acids for distinguishing Weissella viridescens, Weissella paramesenteroides, Weissella hellenica and some non-identifiable, arginine-negative Weissella strains of meat origin. Syst. Appl. Microbiol. 21, 60–65. doi: 10.1016/S0723-2020(98)80031-3
Sandes, S., Figueiredo, N., Pedroso, S., Sant'Anna, F., Acurcio, L., and Abatemarco Junior, M. (2020). Weissella paramesenteroides WpK4 plays an immunobiotic role in gut-brain axis, reducing gut permeability, anxiety-like and depressive-like behaviors in murine models of colitis and chronic stress. Food Res. Int. 137, 109741. doi: 10.1016/j.foodres.2020.109741
Sartono, G., Rizqiyah, I., Asmarinah Heng, N. C. K., and Malik, A. (2019). Three bacteriocin peptides from a lactic acid bacterium Weissella confusa MBF8-1 with spermicidal activity. Curr. Pharmacol. Biotechnol. 20, 766–771. doi: 10.2174/1389201020666190617163310
Schillinger, U., Boehringer, B., Wallbaum, S., Caroline, L., Gonfa, A., Huch Née Kostinek, M., et al. (2008). A genus-specific PCR method for differentiation between Leuconostoc and Weissella and its application in identification of heterofermentative lactic acid bacteria from coffee fermentation. FEMS Microbiol. Lett. 286, 222–226. doi: 10.1111/j.1574-6968.2008.01286.x
Sha, H. Y., Wang, Q. Q., and Li, Z. J. (2023). Comparison of the effect of exopolysaccharide-producing lactic acid bacteria from sourdough on dough characteristics and steamed bread quality. Int. J. Food Sci. Technol. 58, 378–386. doi: 10.1111/ijfs.15881
Shah, N., Patel, A., Ambalam, P., et al. (2016). Determination of an antimicrobial activity of Weissella confusa, Lactobacillus fermentum, and Lactobacillus plantarum against clinical pathogenic strains of Escherichia coli and Staphylococcus aureus in co-culture. Ann. Microbiol. 66, 1137–1143. doi: 10.1007/s13213-016-1201-y
Sharma, S., Kandasamy, S., Kavitake, D., and Shetty, P. H. (2018). Probiotic characterization and antioxidant properties of Weissella confusa KR780676, isolated from an Indian fermented food. LWT Food Sci. Technol. 97, 53–60. doi: 10.1016/j.lwt.2018.06.033
Siavoshi, F., Ebrahimi, H., and Sarrafnejad, A. (2021). Weissella confusa with thermostable β-hemolytic exopolysaccharide. Toxicon 202, 67–74. doi: 10.1016/j.toxicon.2021.09.015
Silva, M. S., Ramos, C. L., González-Avila, M., Gschaedler, A., Arrizon, J., Schwan, F. R., et al. (2017). Probiotic properties of Weissella cibaria and Leuconostoc citreum isolated from tejuino – a typical Mexican beverage. LWT Food Sci. Technol. 86, 227–232. doi: 10.1016/j.lwt.2017.08.009
Singh, P., Rajora, P., Parihar, A. S., Kaur, P., Gandhi, P., Gandhi, V., et al. (2020). Evaluation of effect of gestational diabetes mellitus on composition of the initial oral microbiota of neonates. Adv. Biomed. Res. 9, 78. doi: 10.4103/abr.abr_179_20
Singh, S., Bhatia, R., Singh, A., Singh, P., Kaur, R., Khare, P., et al. (2018). Probiotic attributes and prevention of LPS-induced pro-inflammatory stress in RAW264.7 macrophages and human intestinal epithelial cell line (Caco-2) by newly isolated Weissella cibaria strains. Food Function 9, 1254–1264. doi: 10.1039/C7FO00469A
Snauwaert, I., Papalexandratou, Z., Vuyst, D. e, and Vandamme, L. (2013). Characterization of strains of Weissella fabalis sp. nov. and Fructobacillus tropaeoli from spontaneous cocoa bean fermentations. Int. J. Syst. Evol. Microbiol. 63, 1709–1716. doi: 10.1099/ijs.0.040311-0
Snyder, A. K., Hinshaw, J. M., and Welch, T. J. (2015). Diagnostic tools for rapid detection and quantification of Weissella ceti NC36 infections in rainbow trout. Lett. Appl. Microbiol. 60, 103–110. doi: 10.1111/lam.12365
Song, Q., Wang, B., Han, Y., and Zhou, Z. (2022). Metagenomics reveals the diversity and taxonomy of carbohydrate-active enzymes and antibiotic resistance genes in suancai bacterial communities. Genes 13, 773. doi: 10.3390/genes13050773
Spiegelhauer, M. R., Yusibova, M., Rasmussen, I. K. B., Fuglsang, K. A., Thomsen, K., Andersen, L., et al. (2020). A case report of polymicrobial bacteremia with Weissella confusa and comparison of previous treatment for successful recovery with a review of the literature. Access Microbiol. 2, acmi000119. doi: 10.1099/acmi.0.000119
Stedman, A., De Maluquer, D., Motes, C., Lesellier, S., Dalley, D., and Chambers, M. (2018). Lactic acid bacteria isolated from European badgers (Meles meles) reduce the viability and survival of Bacillus Calmette-Guerin (BCG) vaccine and influence the immune response to BCG in a human macrophage model. BMC Microbiol. 18, 74. doi: 10.1186/s12866-018-1210-z
Stergiadis, S., Cabeza-Luna, I., Mora-Ortiz, M., Stewart, R. D., Dewhurst, R. J., Humphries, D. J., et al. (2021). Unravelling the role of rumen microbial communities, genes, and activities on milk fatty acid profile using a combination of omics approaches. Front. Microbiol. 11, 590441. doi: 10.3389/fmicb.2020.590441
Sturino, J. M. (2018). Literature-based safety assessment of an agriculture- and animal-associated microorganism: Weissella confusa. Reg. Toxicol. Pharmacol. 95, 142–152. doi: 10.1016/j.yrtph.2018.03.013
Styková, E., Nemcová, R., Madar, M., Bujnáková, D., Mucha, R., Gancarčíková, S., et al. (2022). Antibiofilm Activity of Weissella spp. and Bacillus coagulans isolated from equine skin against Staphylococcus aureus. Life 12, 2135. doi: 10.3390/life12122135
Sun, H. Y., Kim, K. P., Bae, C. H., Choi, A. J., Paik, H. D., Kim, I. H., et al. (2019). Evaluation of Weissella cibaria JW15 probiotic derived from fermented korean vegetable product supplementation in diet on performance characteristics in adult beagle dog. Animals 9, 581. doi: 10.3390/ani9080581
Surachat, K., Kantachote, D., Wonglapsuwan, M., Chukamnerd, A., Deachamag, P., Mittraparp-Arthorn, P., et al. (2022). Complete genome sequence of Weissella cibaria NH9449 and comprehensive comparative-genomic analysis: genomic diversity and versatility trait revealed. Front. Microbiol. 13, 826683. doi: 10.3389/fmicb.2022.826683
Tallei, T. E., Fatimawali, Y. A., Kusumawaty, D., Effendi, Y., Park, M. N., and Alhumaydhi, F. A. (2022). Predictive microbial community and functional gene expression profiles in pineapple peel fermentation using 16S rRNA gene sequences. Fermentation 8, 194. doi: 10.3390/fermentation8050194
Tanasupawat, S., Shida, O., Okada, S., and Komagata, K. (2000). Lactobacillus acidipiscis sp. nov. and Weissella thailandensis sp. nov., isolated from fermented fish in Thailand. Int. J. Syst. Evol. Microbiol. 50, 1479–1485. doi: 10.1099/00207713-50-4-1479
Tang, X., Liu, N., Huang, W., Cheng, X., Wang, F., Zhang, B., et al. (2018). Syneresis rate, water distribution, and microstructure of wheat starch gel during freeze-thaw process: role of a high molecular weight dextran produced by Weissella confusa WS813 from traditional sourdough. Cereal Chem. 95, 117–129. doi: 10.1094/CCHEM-08-17-0174-R
Tang, X., Zhang, B., Huang, W., Ma, Z., Zhang, F., Wang, F., et al. (2019). Hydration, water distribution and microstructure of gluten during freeze thaw process: role of a high molecular weight dextran produced by Weissella confusa WS813. Food Hydrocoll. 90, 377–384. doi: 10.1016/j.foodhyd.2018.10.025
Tanizawa, Y., Fujisawa, T., Mochizuki, T., Kaminuma, E., Suzuki, Y., Nakamura, Y., et al. (2014). Draft genome sequence of Weissella oryzae SG25T, isolated from fermented rice grains. Genome Announc. 2, e00667–e00614. doi: 10.1128/genomeA.00667-14
Teixeira, C. G., Fusieger, A., Martins, E., De Freitas, D., Vakarelova, R., Nero, M., et al. (2021). Biodiversity and technological features of Weissella isolates obtained from Brazilian artisanal cheese-producing regions. LWT Food Sci. Technol. 147, 111474. doi: 10.1016/j.lwt.2021.111474
Teixeira, C. G., Rodrigues, R. D. S., Yamatogi, R. S., Lucau-Danila, A., Drider, D., Nero, L. A., et al. (2022). Genomic analyses of Weissella cibaria W25, a potential bacteriocin-producing strain isolated from pasture in Campos das Vertentes, Minas Gerais, Brazil. Microorganisms 10, 314. doi: 10.3390/microorganisms10020314
Teixeira, C. G., Rodrigues, R. D. S., Yamatogi, R. S., Lucau-Danila, A., Nero, L. A., Carvalho, A. F., et al. (2023). Genome analyses of Weissella strains isolated from Campos das Vertentes Minas Gerais, Brazil revealed new bacteriocins with a large spectrum. Food Bioscience 52, 102421. doi: 10.1016/j.fbio.2023.102421
Tenea, G. N., and Israel Lara, M. (2019). Antimicrobial compounds produced by Weissella confusa Cys2-2 strain inhibit Gram-negative bacteria growth. CYTA J. Food 17, 105–111. doi: 10.1080/19476337.2018.1561520
Tohno, M., Kitahara, M., Inoue, H., Uegaki, R., Irisawa, T., Ohkuma, M., et al. (2013). Weissella oryzae sp. nov., isolated from fermented rice grains. Int. J. Syst. Evol. Microbiol. 63, 1417–1420. doi: 10.1099/ijs.0.043612-0
Tsakalidou, E., Samelis, J., Metaxopoulos, J., and Kalantzopoulos, G. (1997). A typical Leuconostoc-like Weissella strains isolated from meat, sharing low phenotypic relatedness with the so far recognized arginine-negative Weissella spp. as revealed by SDS-Page of whole cell proteins. Syst. Appl. Microbiol. 20, 659–664. doi: 10.1016/S0723-2020(97)80039-2
Tulini, F. L., Hymery, N., Haertlé, T., Le Blay, G., and Martinis, D. e. E. C. (2016). Screening for antimicrobial and proteolytic activities of lactic acid bacteria isolated from cow, buffalo and goat milk and cheeses marketed in the southeast region of Brazil. J. Dairy Res. 83, 115–124. doi: 10.1017/S0022029915000606
Vasanthakumari, D. S., Harikumar, S., Beena, D. J., Pandey, A., and Nampoothiri, K. M. (2015). Physicochemical characterization of an exopolysaccharide produced by a newly isolated Weissella cibaria. Appl. Biochem. Biotechnol. 176, 140–153. doi: 10.1007/s12010-015-1586-2
Vasquez, A., Pancholi, P., and Balada-Llasat, J. M. (2015). Photo quiz: confusing bacteremia in a Crohn's disease patient. J. Clin. Microbiol. 53, 759. doi: 10.1128/JCM.00941-13
Vásquez-Machado, G., Rubiano-Garzón, M., Yepes Blandón, J., Gordillo, D., and Avila-Coy, J. (2020). Weissellosis in rainbow trout in Colombia. Braz. J. Vet. Pathol. 13, 575–80. doi: 10.24070/bjvp.1983-0246.v13i3p575-580
Vela, A. I., Fernández, A., De Bernaldo, H., Quirós, Y., Herráez, P., Domínguez, L., et al. (2011). Weissella ceti sp. nov., isolated from beaked whales (Mesoplodon bidens). Int. J. Syst. Evol. Microbiol. 61, 2758–2762. doi: 10.1099/ijs.0.028522-0
Walter, J., Hertel, C., Tannock, G. W., Lis, C. M., Munro, K., and Hammes, W. P. (2001). Detection of Lactobacillus, Pediococcus, Leuconostoc, and Weissella species in human feces by using group specific PCR primers and denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 67, 2578–2585. doi: 10.1128/AEM.67.6.2578-2585.2001
Wang, H., Wei, Q., Gui, S., Feng, Y., Zhang, Y., Liu, Y., et al. (2017). Metagenomic profiling of the bacterial community changes from koji to mash stage in the brewing of soy sauce. Polish J. Microbiology 66, 537–541. doi: 10.5604/01.3001.0010.7097
Wang, J., Sun, C., Liu, C., Yang, Y., and Lu, W. (2016). Comparison of vaginal microbial community structure in healthy and endometritis dairy cows by PCR-DGGE and real-time PCR. Anaerobe 38, 1–6. doi: 10.1016/j.anaerobe.2015.11.004
Wang, L., Si, W., Xue, H., and Zhao, X. (2017). A fibronectin-binding protein (FbpA) of Weissella cibaria inhibits colonization and infection of Staphylococcus aureus in mammary glands. Cell Microbiol. 19, 12731. doi: 10.1111/cmi.12731
Wang, M., Franco, M., Cai, Y., and Yu, Z. (2020a). Dynamics of fermentation profile and bacterial community of silage prepared with alfalfa, whole-plant corn and their mixture. Anim. Feed Sci. Technol. 270, 114702. doi: 10.1016/j.anifeedsci.2020.114702
Wang, Q., Liu, Y., and Yin, X. (2022). Comparison of gut bacterial communities of Locusta migratoria manilensis (Meyen) reared on different food plants. Biology 11, 1347. doi: 10.3390/biology11091347
Wang, W., Li, S., Heng, X., and Chu, W. (2022). Weissella confusa CGMCC 19,308 strain protects against oxidative stress, increases lifespan and bacterial disease resistance in Caenorhabditis elegans. Probiotics Antimicrob. Proteins 14, 121–129. doi: 10.1007/s12602-021-09799-z
Wang, W., Liu, W., and Chu, W. (2020b). Isolation and preliminary screening of potentially probiotic Weissella confusa strains from healthy human feces by culturomics. Microb. Pathog. 147, 104356. doi: 10.1016/j.micpath.2020.104356
Wang, X., Zhang, F., Wang, L., and Pan, L. (2023). Structure and effect on Bifidobacterium longum of exopolysaccharide produced by Weissella cibaria using cost-effective substrates. Food Quality Safety 7, 1–7. doi: 10.1093/fqsafe/fyad012
Wen Fang Wu Wu, J., Redondo-Solano, M., Uribe, L., WingChing-Jones, R., Usaga, J., and Barboza, N. (2021). First characterization of the probiotic potential of lactic acid bacteria isolated from Costa Rican pineapple silages. PeerJ 9, e12437. doi: 10.7717/peerj.12437
Wijarnpreecha, K., and Fontana, R. J. (2022). Watch out for the saurkraut: Weissella confusa endocarditis in a liver transplant candidate. Clinical Liver Dis. 20, 188–193. doi: 10.1002/cld.1256
Woraprayote, W., Pumpuang, L., Tosukhowong, A., Roytrakul, S., Perez, R. H., Zendo, T., et al. (2015). Two putatively novel bacteriocins active against gram-negative foodborne pathogens produced by Weissella hellenica BCC 7293. Food Control 55, 176–184. doi: 10.1016/j.foodcont.2015.02.036
Woraprayote, W., Pumpuang, L., Tosukhowong, A., Zendo, T., Sonomoto, K., Benjakul, S., et al. (2018). Antimicrobial biodegradable food packaging impregnated with bacteriocin 7293 for control of pathogenic bacteria in pangasius fish fillets. LWT Food Sci. Technol. 89, 427–433. doi: 10.1016/j.lwt.2017.10.026
Wu, H. C., Srionnual, S., Yanagida, F., and Chen, Y. S. (2015). Detection and characterization of Weissellicin 110, a bacteriocin produced by Weissella cibaria. Iran J. Biotechnol. 13, e1159. doi: 10.15171/ijb.1159
Wu, L., Ma, B., Yu, F., Ma, Z., Meng, Q., Li, Z., et al. (2023). Salivary microbiome diversity in Chinese children with various caries states. Clin. Oral Investig. 27, 773–785. doi: 10.1007/s00784-022-04825-y
Xia, Y., Qin, S., and Shen, Y. (2019). Probiotic potential of Weissella strains isolated from horse feces. Microb. Pathog. 132, 117–123. doi: 10.1016/j.micpath.2019.04.032
Xia, Y., Zhou, W., Du, Y., Wang, Y., Zhu, M., Zhao, Y., et al. (2023). Difference of microbial community and gene composition with saccharification function between Chinese nongxiangxing daqu and jiangxiangxing daqu. J. Sci. Food Agric. 103, 637–647. doi: 10.1002/jsfa.12175
Xiang, F., Dong, Y., Cai, W., Zhao, H., Liu, H., Shan, C., et al. (2023). Comparative genomic analysis of the genus Weissella and taxonomic study of Weissella fangxianensis sp. nov., isolated from Chinese rice wine starter. Int. J. Syst. Evol. Microbiol. 73, 5870. doi: 10.1099/ijsem.0.005870
Xiang, W. L., Zhang, N. D., Lu, Y., Zhao, Q. H., Xu, Q., Rao, Y., et al. (2020). Effect of Weissella cibaria co-inoculation on the quality of Sichuan Pickle fermented by Lactobacillus plantarum. LWT Food Sci. Technol. 121, 108975. doi: 10.1016/j.lwt.2019.108975
Xiang, W. L., Zhao, Q. H., Lu, Y., Tang, J., Cai, T., Rao, Y., et al. (2022). Tetracycline residue alters profile of lactic acid bacterial communities and metabolites of ginger pickle during spontaneous fermentation. Food Res. Int. 155, 111109. doi: 10.1016/j.foodres.2022.111109
Xu, D., Hu, Y., Wu, F., Jin, Y., Xu, X., Gänzle, M. G., et al. (2020). Comparison of the functionality of exopolysaccharides produced by sourdough lactic acid bacteria in bread and steamed bread. J. Agric. Food Chem. 68, 8907–8914. doi: 10.1021/acs.jafc.0c02703
Xu, Y., Wang, Y., Coda, R., Säde, E., Tuomainen, P., Tenkanen, M., et al. (2017). In situ synthesis of exopolysaccharides by Leuconostoc spp. and Weissella spp. and their rheological impacts in fava bean flour. Int. J. Food Microbiol. 248, 63–71. doi: 10.1016/j.ijfoodmicro.2017.02.012
Yadav, M., and Sunita Shukla, P. (2022). Probiotic potential of Weissella paramesenteroides MYPS5.1 isolated from customary dairy products and its therapeutic application. Biotechnology 12, 19. doi: 10.1007/s13205-021-03074-2
Yalmanci, D., Dertli, E., Tekin-Caknak, Z. H., and Karasu, S. (2023). The stabilization of low-fat mayonnaise by whey protein isolate-microbial exopolysaccharides (Weissella confusa W-16 strain) complex. Int. J. Food Sci. Technol. 58, 1304–1316. doi: 10.1111/ijfs.16287
Yalmanci, D., Ispirli, H., and Dertli, E. (2022). Identification of lactic acid bacteria (LAB) from pre-fermented liquids of selected cereals and legumes and characterization of their exopolysaccharides (EPS). Food Biosci. 50, 102014. doi: 10.1016/j.fbio.2022.102014
Yang, C., Zhang, Y., and Yu, T. (2022). Application of the cell-free supernatant from Weissella viridescens to control Listeria monocytogenes. Czech J. Food Sci. 40, 290–297. doi: 10.17221/6/2022-CJFS
Yang, G., Xiang, Y., Wang, S., Tao, Y., Xie, L., Bao, L., et al. (2022). Response of intestinal microbiota to the variation in diets in grass carp (Ctenopharyngodon idella). Metabolites 12, 1115. doi: 10.3390/metabo12111115
Yang, Y., Wang, S. T., Lu, Z. M., Zhang, X. J., Chai, L. J., Shen, C. H., et al. (2021). Metagenomics unveils microbial roles involved in metabolic network of flavor development in medium-temperature daqu starter. Food Res. Int. 140, 110037. doi: 10.1016/j.foodres.2020.110037
Yao, D., Wang, X., Ma, L., Wu, M., Xu, L., Yu, Q., et al. (2022). Impact of Weissella cibaria BYL4.2 and its supernatants on Penicillium chrysogenum metabolism. Front. Microbiol. 13, 983613. doi: 10.3389/fmicb.2022.983613
Yasir, M., Al-Zahrani, I. A., Bibi, F., Abd El Ghany, M., and Azhar, E. I. (2022). New insights of bacterial communities in fermented vegetables from shotgun metagenomics and identification of antibiotic resistance genes and probiotic bacteria. Food Res. Int. 157, 111190. doi: 10.1016/j.foodres.2022.111190
Ye, G., Chen, Y., Wang, C., Yang, R., and Bin, X. (2018). Purification and characterization of exopolysaccharide produced by Weissella cibaria YB-1 from pickle Chinese cabbage. Int. J. Biol. Macromol. 120, 1315–1321. doi: 10.1016/j.ijbiomac.2018.09.019
Ye, K., Liu, J., Hunag, Y., Wang, K., and Zhou, G. (2018). Effects of two Weissella viridescens strains on Listeria monocytogenes growth at different initial inoculum proportions. CYTA – J. Food 16, 299–305. doi: 10.1080/19476337.2017.1401667
Yeong, M. S., Hee, M. S., and Choon, C. H. (2020). Characterization of high-ornithine-producing Weissella koreensis DB1 isolated from kimchi and its application in rice bran fermentation as a starter culture. Foods 9, 1545. doi: 10.3390/foods9111545
Yeu, J. E., Lee, H. G., Park, G. Y., Lee, J., and Kang, M. S. (2021). Antimicrobial and antibiofilm activities of Weissella cibaria against pathogens of upper respiratory tract infections. Microorganisms 9, 1181. doi: 10.3390/microorganisms9061181
Yin, L., Zgang, Y., Azi, F., Tejkye, M., Zhou, J., Li, X., et al. (2022). Soybean whey bio-processed using Weissella hellenica D1501 protects neuronal PC12 cells against oxidative damage. Front. Nutr. 9, 833555. doi: 10.3389/fnut.2022.833555
Yoon, J. A., Kwun, S. Y., Park, E. H., and Kim, M. D. (2023). Complete genome sequence data of Leuconostoc mesenteroides KNU-2 and Weissella hellenica MBEL1842 isolated from kimchi. Data Brief 47, 108919. doi: 10.1016/j.dib.2023.108919
You, Y. A., Kwon, E. J., Choi, S. J., Hwang, H. S., Choi, S. K., Lee, S. M., et al. (2019). Vaginal microbiome profiles of pregnant women in Korea using a 16S metagenomics approach. American J. Rep. Immunol. 82, e13124. doi: 10.1111/aji.13124
Yu, H. S., Jang, J., Lee, N. K., and Paik, H. D. (2019). Evaluation of the probiotic characteristics and prophylactic potential of Weissella cibaria strains isolated from kimchi. LWT Food Sci. Technol. 112, 108229. doi: 10.1016/j.lwt.2019.05.127
Yu, H. S., Lee, N. K., Choi, A. J., Choe, J. S., Bae, C. H., Paik, H. D., et al. (2018). Antagonistic and antioxidant effect of probiotic Weissella cibaria JW15. Food Sci. Biotechnol. 28, 851–855. doi: 10.1007/s10068-018-0519-6
Yu, Y., Li, L., Xu, Y., Li, H., Yu, Y., Xu, Z., et al. (2022). Metagenomics reveals the microbial community responsible for producing biogenic amines during mustard [Brassica juncea (L.)] fermentation. Front. Microbiol. 13, 824644. doi: 10.3389/fmicb.2022.824644
Yuan, S., Wang, Y., Zhao, F., and Kang, L. (2021). Complete genome sequence of Weissella confusa LM1 and comparative genomic analysis. Front. Microbiol. 12, 749218. doi: 10.3389/fmicb.2021.749218
Zamudio-Maya, M., Narváez-Zapata, J., and Rojas-Herrera, R. (2008). Isolation and identification of lactic acid bacteria from sediments of a coastal marsh using a differential selective medium. Lett. Appl. Microbiol. 46, 402–407. doi: 10.1111/j.1472-765X.2008.02329.x
Zannini, E., Jeske, S., Lynch, K. M., and Arendt, E. K. (2018). Development of novel quinoa-based yoghurt fermented with dextran producer Weissella cibaria MG1. Int. J. Food Microbiol. 268, 19–26. doi: 10.1016/j.ijfoodmicro.2018.01.001
Zhang, J., Liu, S., Sun, H., Jiang, Z., Xu, Y., Mao, J., et al. (2022). (2022). Metagenomics-based insights into the microbial community profiling and flavor development potentiality of baijiu Daqu and huangjiu wheat Qu. Erratum Food Res Int. 157, 111448. doi: 10.1016/j.foodres.2022.111448
Zhang, J., Wang, X., Huo, D., Li, W., Hu, Q., Xu, C., et al. (2016). Metagenomic approach reveals microbial diversity and predictive microbial metabolic pathways in Yucha, a traditional Li fermented food. Sci. Rep. 6, 32524. doi: 10.1038/srep32524
Zhang, Q., Wang, M., Ma, X., Li, Z., Jiang, C., Pan, Y., et al. (2022b). In vitro investigation on lactic acid bacteria isolatedfrom Yak faeces for potential probiotics. Front. Cell. Infect. Microbiol. 12, 984537. doi: 10.3389/fcimb.2022.984537
Zhang, Y., Chen, H., Lu, M., Cai, J., Lu, B., Luo, C., et al. (2022a). Habitual diet pattern associations with gut microbiome diversity and composition: results from a chinese adult cohort. Nutrients 14, 2639. doi: 10.3390/nu14132639
Zhao, C., Su, W., Mu, Y., Mu, Y., and Jiang, L. (2021). Integrative metagenomics-metabolomics for analyzing the relationship between microorganisms and non-volatile profiles of traditional xiaoqu. Front. Microbiol. 11, 617030. doi: 10.3389/fmicb.2020.617030
Zhao, D., Jiang, J., Liu, L., Wang, S., Ping, W., Ge, J., et al. (2021). Characterization of exopolysaccharides produced by Weissella confusa XG-3 and their potential biotechnological applications. Int. J. Biol. Macromol. 178, 306–315. doi: 10.1016/j.ijbiomac.2021.02.182
Zhao, J., Fan, H., Kwok, L. Y., Guo, F., Ji, R., Ya, M., et al. (2019). Analyses of physicochemical properties, bacterial microbiota, and lactic acid bacteria of fresh camel milk collected in Inner Mongolia. J. Dairy Sci. 103, 106–116. doi: 10.3168/jds.2019-17023
Zhao, J., Wang, J., Yang, Y., Li, X., and Sun, C. (2015). In vitro assessment of probiotic properties of lactic acid bacteria isolated from vaginas of healthy cows. Indian J. Anim. Res. 49, 355–359. doi: 10.5958/0976-0555.2015.00049.7
Zhao, S., Li, C., Zhu, T., Jin, L., Deng, W., Zhao, K., et al. (2021). Diversity and composition of gut bacterial community in giant panda with anorexia. Curr. Microbiol. 78, 1358–1366. doi: 10.1007/s00284-021-02424-w
Zheng, J., Wittouck, S., Salvetti, E., Franz, C. M. A. P., Harris, H. M. B., Mattarelli, P., et al. (2020). A taxonomic note on the genus Lactobacillus: description of 23 novel genera, emended description of the genus Lactobacillus beijerinck 1901, and union of Lactobacillaceae and Leuconostocaceae. Int. J. Syst. Evol. Microbiol. 70, 2782–2758. doi: 10.1099/ijsem.0.004107
Zheng, W., Zhang, Z., Liu, C., Qiao, Y., Zhou, D., Qu, J., et al. (2015). Metagenomic sequencing reveals altered metabolic pathways in the oral microbiota of sailors during a long sea voyage. Sci. Rep. 5, 9131. doi: 10.1038/srep09131
Zhu, K. K., Yang, B. T., Hao, Z. P., Li, H. Z., Cong, W., Kang, Y. H., et al. (2022). Dietary supplementation with Weissella cibaria C-10 and Bacillus amyloliquefaciens T-5 ehnance immunity against Aeromonas veronii infection in crucian carp (Carassiu auratus). Microb. Pathog. 167, 105559. doi: 10.1016/j.micpath.2022.105559
Keywords: Weissella, Periweissella, lactic acid bacteria, probiotic, bacteriocin, prebiotic, exopolysaccharides, fermented food
Citation: Fusco V, Chieffi D, Fanelli F, Montemurro M, Rizzello CG and Franz CMAP (2023) The Weissella and Periweissella genera: up-to-date taxonomy, ecology, safety, biotechnological, and probiotic potential. Front. Microbiol. 14:1289937. doi: 10.3389/fmicb.2023.1289937
Received: 06 September 2023; Accepted: 14 November 2023;
Published: 11 December 2023.
Edited by:
Michael Gänzle, University of Alberta, CanadaReviewed by:
Ömer Şimşek, Yildiz Technical University, TürkiyeCopyright © 2023 Fusco, Chieffi, Fanelli, Montemurro, Rizzello and Franz. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Charles M. A. P. Franz, charles.franz@mri.bund.de
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.