- 1Liaoning Key Laboratory of Marine Animal Immunology and Disease Control, Dalian Ocean University, Dalian, China
- 2Liaoning Key Laboratory of Marine Animal Immunology, Dalian Ocean University, Dalian, China
- 3Dalian Key Laboratory of Aquatic Animal Disease Prevention and Control, Dalian Ocean University, Dalian, China
- 4Laboratory of Marine Fisheries Science and Food Production Process, Qingdao National Laboratory for Marine Science and Technology, Qingdao, China
Intestinal autochthonous bacteria play important roles in the maintenance of the physiological homeostasis of animals, especially contributing to the host immune system. In the present study, the variation of autochthonous bacterial community in the intestinal tract of 2-7 months-old tiger pufferfish Takifugu rubripes and bacterial communities in the seawater of recirculating aquaculture system (RAS) and the following offshore sea cage aquaculture system (OSCS) were analyzed during the aquaculture period from May to October 2021. Proteobacteria was found to be the most dominant phyla in both intestinal and seawater bacterial communities, which accounted for 68.82% and 65.65% of the total bacterial abundance, respectively. Arcobacter was the most core bacterial taxon in the intestinal bacterial community, with the most dominant abundance (42.89%) at the genus level and dominant positions in co-occurrence relationships with other bacterial taxa (node-betweenness value of 150). Enterococcaceae was specifically enriched in the intestinal bacterial community of pufferfishes from RAS, while Vibrionaceae was enriched in the intestinal bacterial community from OSCS. The F-values of beta diversity analysis between intestinal and seawater bacterial communities generally increased from May (6.69) to October (32.32), indicating the increasing differences between the intestinal and seawater bacterial communities along with the aquaculture process. Four bacterial taxa of Weissella sp., Akkermansia muciniphila, Dietzia sp. and Psychrobacter pacificensis had significant correlations with immune response parameters, and they were suggested to be the indicators for immune status and pathological process of pufferfish. The knowledge about the specific core bacteria, potentially pathogenic bacteria and the change of bacterial community in the intestinal tract of cultured pufferfish is of great scientific significance and will contribute to the understanding of intestinal bacterial homeostasis and biosecurity practice in pufferfish aquaculture.
Introduction
Tiger pufferfish (Takifugu rubripes) is one of model organisms, as well as a highly promising finfish species for aquaculture (Tao et al., 2012; Fishery Bureau of China Agriculture Department, 2021; Gao et al., 2022b), while the diseases of cultured pufferfish caused by intestinal microbiota disturbances and intestinal pathogens have been reported in recent years (Kodama et al., 2014; Sun et al., 2018; Li et al., 2019). The fish intestinal bacteria, especially autochthonous bacteria, contributes greatly to the immune response, development and growth of the hosts (Burns et al., 2016). The fish intestinal tract is therefore regarded as a critical immune organ and play important roles in the maintenance of physiological homeostasis and health of the hosts (Duan et al., 2020). The diversities, functions and symbiotic features of the intestinal bacteria have been investigated in some fish species, such as rainbow trout (Oncorhynchus mykiss), common carp (Cyprinus carpio) and turbot (Scophthalmus maximus) (Desai et al., 2012; Liu et al., 2019; Meng et al., 2021). The knowledge about the change of intestinal bacterial community during the aquaculture period of pufferfish will be crucial for the disease prevention and control.
Intestinal bacteria is constrained by many factors, such as environmental abiotic and biotic factors, host immune responses and nutritional status, among which environmental factors have a considerable impact on the structure and composition of the intestinal bacteria of fish through constant contact (Hopkins et al., 2002; Dethlefsen et al., 2006; Tanaka et al., 2009; Turnbaugh et al., 2009; Dehler et al., 2017; Kokou et al., 2019). There are two different aquaculture systems for pufferfish, named offshore sea cage aquaculture system (OSCS) and recirculating aquaculture system (RAS). The fingerlings are reared in RAS until around 10 g of body weight in late June and then transferred to OSCS. Fish are switched back to RAS in late October to overwinter and then transferred to OSCS again next May until harvest. The two aquaculture systems have different environmental factors, including environmental bacterial community, water temperature, dissolved oxygen and inorganic substance concentrations. The environmental factors in RAS are more restricted and stable than those in OSCS due to artificial manipulation. The pufferfishes in different aquaculture systems always display a wide variation in health status, survival rate and flesh quality (Jia et al., 2019), and it is partly due to the effect of environmental factors of different aquaculture systems on intestinal bacteria and physiological metabolism of pufferfish.
The intestinal bacteria is always viewed as an “extra organ” due to its important role in intestinal protection and homeostasis (Dehler et al., 2017). Intestinal bacteria not only hinders pathogens to colonize in the intestinal tract through antibiosis or niche exclusion, but also provides protection for host health by stimulating the immune response of mammals (Webb et al., 2016). Recently, the fish intestinal bacteria was also reported to form a complex and dynamic microbial ecosystem, which contributed to immune system priming, disease prevention and host nutrient acquisition (Gómez and Balcázar, 2008; Banerjee and Ray, 2017). Besides the variations of bacterial communities in healthy fish (de Bruijn et al., 2018), the effect of environmental factors on intestinal bacteria (Wang et al., 2018) and interactions between intestinal bacteria and immune response (Gómez and Balcázar, 2008; Nie et al., 2017) were also drew much interest recently. The immune system impacts the ecology of commensal bacterial communities, and the bacteria were also found to be able to elicit a mild inflammatory reaction and activate the innate immune response after they were colonized in zebrafish larvae (Montalban-Arques et al., 2015), and induce oxidative stress (Yin et al., 2015; Bandeira Junior and Baldisserotto, 2021). Some intestinal bacterial taxa were also reported as health indicators in marine invertebrate species (Xiong et al., 2015; Huang et al., 2018). However, the variation of the bacterial community of intestinal tract in pufferfish under different culture systems and the relationship between the intestinal bacteria and immune response are still not clear.
In this study, the characteristics of both intestinal and environmental bacterial communities were analyzed using high-throughput sequencing technologies with the main objectives to (1) understand the change of autochthonous bacterial community in the intestinal tract of cultured pufferfish, (2) compare the characteristics of the autochthonous bacterial communities in the in intestinal tract of pufferfish from RAS and OSCS systems, (3) explore the relationship between the intestinal bacterial community and the parameters involved in immune response including oxidative stress parameters as well as cytokines, which are secreted by the cells of immune system in immune responses and act as the means of communication (Stow et al., 2009). It is hopefully to identify the indicators of bacterial taxa for immune status and pathological process of pufferfish.
Materials and methods
Sample collection
The samples were collected in Dalian, China, from RAS system in May and June 2021, and OSCS system from July to October 2021. The geographic coordinates of RAS and OSCS are N39°36’20”–E122°51’12” and N39°28’12”–E123°4’59”, respectively. The samples were collected twice a month from the two systems at the same time interval, and nine fish were collected each time. The fish were anesthetized and dissected. The digestive tracts were excised, and the intestinal contents were squeezed out. The intestinal tract were rinsed three times with peptone water (1g/L bacterial peptone and 8.5 g/L NaCl) to remove the non-adherent bacteria and capture adherent bacteria (Ringø, 1993). Then, 0.1 g of the treated intestinal tract was taken from one individual, and the intestinal tracts from three individuals were pooled as one sample. For spleen samples, 0.1 g spleen was collected from one individual, and the spleens from three individuals were pooled as one sample. There were three replicates of spleen and intestinal tract samples for each sampling time, and they were stored at -80°C for further processing. Three-liter seawater (1 L for one sample) was collected at a depth of 1 m and then stored at 4°C for the analyses of water quality. Other 4 L seawater (1 L for one sample) was filtered using 0.22 μm pore size membranes (Sagon, Shanghai, China) to enrich the bacteria cells and stored at -80°C for DNA extraction.
High−throughput sequencing and data processing
The bacteria genomic DNA from pufferfish intestinal tract and seawater were extracted by the Soil DNA Kit (Omega, Norcross, GA, USA) and Water DNA Kit (Omega, Norcross, GA, USA) respectively. DNA quality and quantity were analyzed with 1% agarose gel electrophoresis and NanoDrop spectrophotometer (Thermo Fisher Scientific, USA), respectively. The V3–V4 region of 16S rDNA genes was amplified using the primers of 515F (GTGBCAGCMGCCGCGGTAA) and 806R (GGACTACHVGGGTWTCTAAT). The high-throughput sequencing of V3–V4 region of 16S rDNA gene was performed on the Illumina HiSeq platform by Novogene (Beijing, China). The raw reads were trimmed and denoised by Quantitative Insights into Microbial Ecology 2 (QIIME 2) using “dada2 denoise-single” with default parameters and “–p-trim-left 0” and “–p-trunc-len 250”, to remove singletons and chimeric sequences and obtain amplicon sequence variants (ASVs) (Bolyen et al., 2019). Handled by QIIME 2 pipeline with the clean reads, taxonomy was assigned at a single nucleotide level with a feature classifier against the SILVA 138 SSU database, and alpha and beta diversity were analyzed by Chao1 index and unweighted UniFrac distances, respectively.
Bioinformatics analysis
MicrobiomeAnalyst (http://www.microbiomeanalyst.ca) (Chong et al., 2020) was used to analyze the abundance and diversity of the bacterial community and generate visual exploration, and to conduct linear discriminant analysis effect size (LEfSe) analysis and identify the significantly differentially abundant taxa between intestinal and seawater bacterial communities, with P-value cutoff of 0.1 and Log LDA score of 2.0. The functional profiles and metabolic pathways were analyzed and predicted with Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt) 2 software with the KEGG database for annotation (Douglas et al., 2020). STAMP software (version 2.1.3) was used for functional profile analysis (Parks et al., 2014). The co-occurrence ecological network was built using the online Molecular Ecological Network Analysis (MENA) pipeline (http://ieg2.ou.edu/MENA) (Deng et al., 2012), and the network construction was conducted using Cytoscape software (version 3.7.2) (Shannon et al., 2003).
Determination of water quality indexes
Water temperature, dissolved oxygen (DO), pH and salinity were monitored by a YSI Professional Plus meter (YSI, Yellow Springs, Ohio, USA) in situ. The concentrations of TAN, NO2–N, NO3–N and PO4-P of the seawater samples were analyzed using the indophenol blue spectrophotometric method, N-(1-naphthyl)-ethylenediamine dihydrochloride spectrophotometric method, Zn-Cd reduction method and Phosphomolybdenum blue spectrophotometric method, respectively, according to the procedures of the National Specification for Marine Monitoring (SOA of China, 2007).
Immune response parameter analysis
The activities of superoxide dismutase (SOD) in spleens were analyzed by water-soluble tetrazolium salt (WST-1) assay with assay kit (Jiancheng, Nanjing, China; Product code: A001-3-2). The contents of malondialdehyde (MDA) in spleens were analyzed by 2-thiobarbituric acid (TBA) method with assay kit (Jiancheng, Nanjing, China; Product code: A003-1-2). The experiments were carried out according to the manufacturer’s introductions. The SOD activity unit was defined as the amount of enzyme that can convert 1 μmol substrate. The total RNA was extracted from the spleen samples using TRIzol™ Reagent. RNA concentration was measured using a NanoDrop spectrophotometer, and the purity and integrity were examined through electrophoresis analysis. The first strand of cDNA was synthesized using the total mRNA as a template and oligo (dT)17-adaptor as a primer by PrimeScript™ RT Reagent Kit with gDNA Eraser (TaKaRa, Dalian, China) according to the manufacturer’s instruction. The synthesis reaction was conducted at 42°C for 5 min and terminated by heating at 85°C for 5 s. Temporal expression of IL-1β, TNF-α, IL10 and IL17-AF mRNA were measured by quantitative real-time PCR (qRT-PCR) using SYBR Green Master Mix (TaKaRa, Japan). The primer information was listed in Table 1. The expression of β-actin mRNA was used as the internal control. All reactions were performed in an ABI PRISM 7500 Detection System (Applied Biosystems, USA). The mRNA expressions were analyzed by the 2-ΔΔCT method (Livak and Schmittgen, 2001).
Statistical analysis
The statistical analysis was conducted using one-way ANOVA by Statistical Package for Social Sciences (SPSS, version 26.0) (Field, 2013). The ASVs that were available in more than half of samples were screened for the correlation analysis with immune response parameters. The correlations between the abundance of bacterial taxa and immune response parameters were investigated through Spearman’s and Pearson’s correlation analysis using SPSS 26. The data were presented as means ± standard deviations with three parallel replicates. P-value < 0.05 was considered statistically significant.
Results
Composition and co-occurrence of intestinal bacterial community
A total of 2,865,508 reads were obtained after the high−throughput sequencing of intestinal tract samples. The mean read depth per sample was 79,597.4 sequences. The average length of the sequences was 253 bp. Across all samples, a total of 2,610 ASVs were detected in initial identification, with 1,979,446 appearance frequency in all samples. In total, 16 phyla were observed, and eight phyla were identified with abundance > 1%. Proteobacteria (68.82%), Firmicutes (16.09%), Cyanobacteria (4.35%), Spirochaetes (4.12%) and Tenericutes (2.01%) were dominant phyla in the intestinal bacterial community, accounting for 95.39% of the total abundance (Figure 1A). At the genus level, the five top dominant taxa were Arcobacter (42.89%), Photobacterium (5.33%), Vagococcus (5.16%), Weissella (3.21%) and Vibrio (2.7%), accounting for around 60% of the total abundance, and 31.27% of ASVs were failed to be annotated at the genus level. Arcobacter was found to be the most core bacterial taxon in the intestinal bacterial community of pufferfish (Figure 1B). When the detection threshold was set at 0.01%, the prevalence of Arcobacter was 97.2%; when the detection threshold was set at 0.321%, the prevalence of Arcobacter was still above 50% (Figure 1C). The co-occurrence analysis was conducted to investigate the interaction relationships among bacterial taxa and construct the microbial co-occurrence networks (Figure 1D). Among all genera identified in this analysis, Ruminococcus processed the dominant position in the co-occurrence relationships with the node-degree value of 51. Ruminococcus and Arcobacter were the top two betweenness hubs with the node-betweenness values of 190 and 150, respectively.
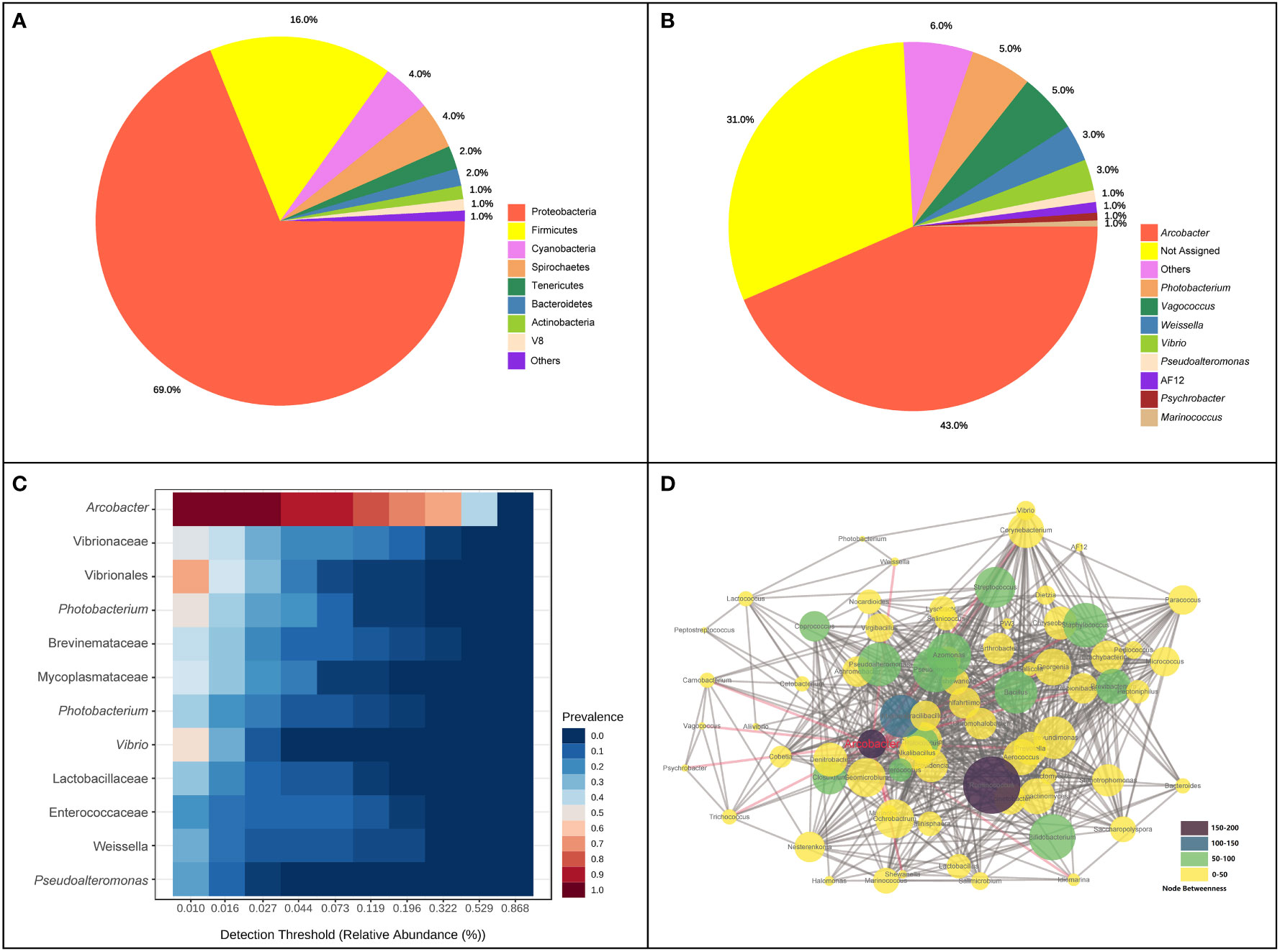
Figure 1 Composition and co-occurrence of intestinal bacterial community of pufferfish using all samples in this study. (A) The proportion of intestinal bacterial abundance at phylum level; (B) The proportion of intestinal bacterial abundance at genus level; (C) Core bacterial taxa in the intestinal bacterial community; (D) Microbial co-occurrence networks at the genus level.
The difference of intestinal bacterial communities in pufferfishes from RAS and OSCS systems
The abundance of the intestinal bacterial community in OSCS was about two-fold higher than that in RAS. At the phylum level, the relative ASV abundance of Spirochaetes and Bacteroidetes in OSCS (6.026% and 2.400%, respectively) were obviously higher than that in RAS (0.096% and 0.226%, respectively) (Figure 2A). The alpha diversity range of the intestinal bacterial community in OSCS was more variable than that in RAS. The result of beta diversity analysis showed that there was a distinct separation of intestinal bacterial communities between RAS and OSCS samples (Figure 2B). In LEfSe analysis, significant enrichments of ASVs were observed. The top-4 enriched ASVs in OSCS were Vibrionaceae, Brevinemataceae, Photobacterium and Photobacterium, with the LDA score of -3.19, -3.0, -2.9 and -2.8, respectively. The top-4 enriched ASVs in RAS were Vagococcus, Enterococcaceae, Vagococcus and Proteobacteria, with the LDA score of 3.13, 2.95, 2.9 and 2.77, respectively (Figure 2C). The functional profiles and metabolic pathways of intestinal bacterial communities were analyzed and predicted, and the functional pathways of Vibrio cholerae infection and Vibrio cholerae pathogenic cycle were found to be significantly more enriched in the intestinal bacteria community in OSCS than that in RAS (Figure 2D).
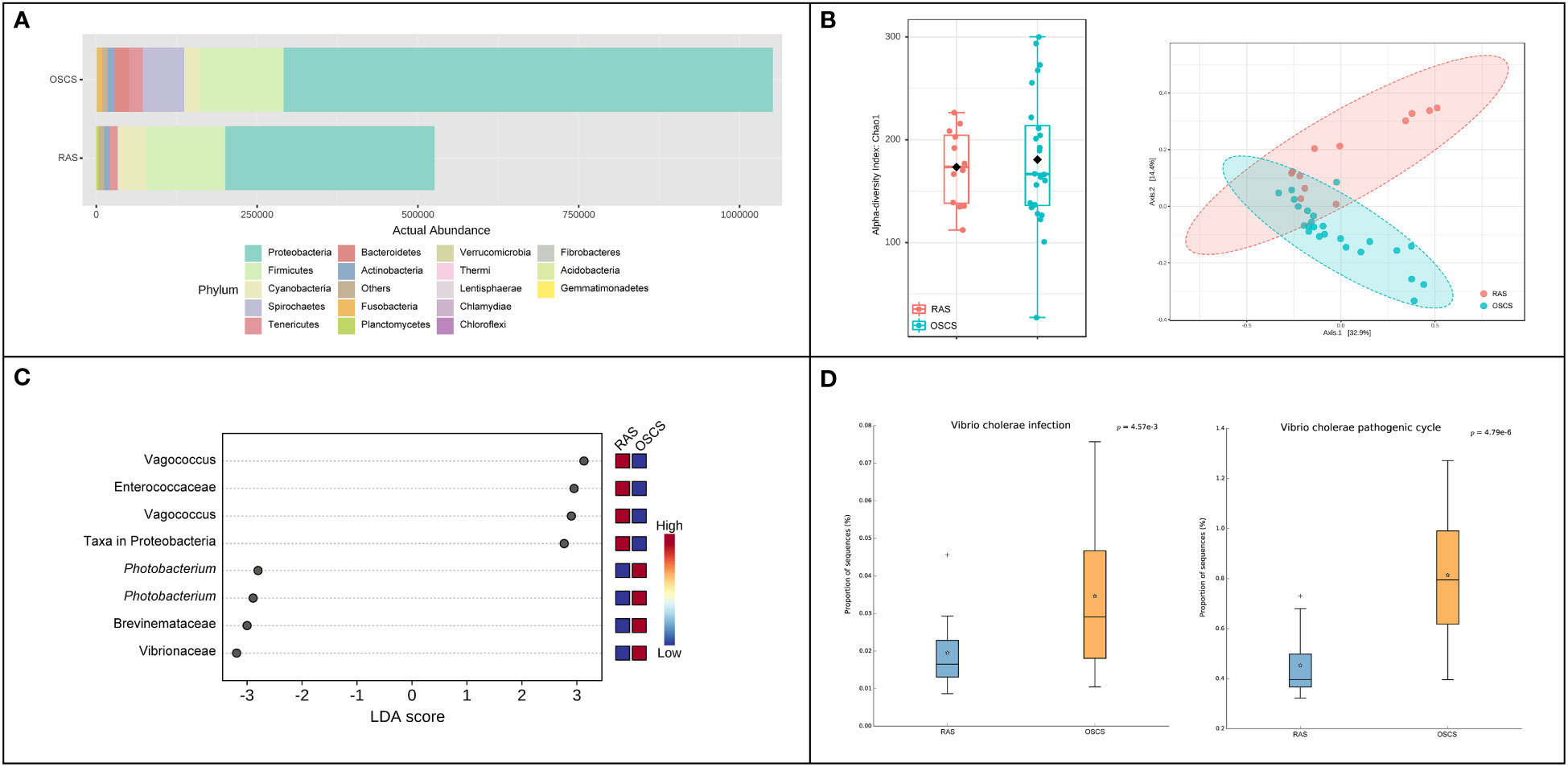
Figure 2 Intestinal bacterial communities in RAS and OSCS. (A) Bacterial abundance at phylum level; (B) LEfSe analysis of ASVs in intestinal bacterial communities between RAS and OSCS samples; (C) Alpha and beta diversity analyses of intestinal bacterial communities between RAS and OSCS samples; (D) Differences of functional pathways of intestinal bacterial communities between RAS and OSCS samples.
Changes of water quality indexes in RAS and OSCS systems
The water temperature in RAS ranged from 17.2°C to 22.7°C, with the average value of 19.9°C, and the water temperature in OSCS ranged from 16.6°C to 24.7°C, with the average value of 22.0°C (Figure 3). The water temperature decreased to 20.6°C in early July due to rainfalls. pH in RAS ranged from 7.53 to 7.89, with the average value of 7.73, and pH in OSCS ranged from 7.60 to 8.09, with the average value of 7.74. The DO concentration varied and remained above 5 mg/L from May to October. The salinity ranged from 25.94 to 30.27 and reached the lowest value in late July due to rainfalls. The concentrations of TAN, NO2-N, NO3-N and PO4-P were generally higher in RAS than those in OSCS, with the highest values of 0.911, 0.105, 1.148 and 0.592 mg/L in RAS, respectively (Figure 3). The concentrations of TAN, NO2-N, NO3-N and PO4-P were found to have significant correlations with the aquaculture system (RAS/OSCS) (P < 0.05), with the correlation values of -0.81, -0.63, -0.83 and -0.81, respectively (Table S1). The concentration of PO4-P had significant correlations with the concentrations of TAN, NO2-N and NO3-N (P < 0.05), with the correlation values of 0.78, 0.67 and 0.91, respectively. The concentration of TAN had significant correlations with the concentrations of NO2-N and NO3-N (P < 0.05), with the correlation values of 0.70 and 0.61, respectively. The correlation analysis between water quality index and the beta diversity of intestinal bacterial community was conducted, and no significant correlation (P < 0.05) in both Spearman’s correlation analysis and Pearson’s correlation analysis was found (Table S2).
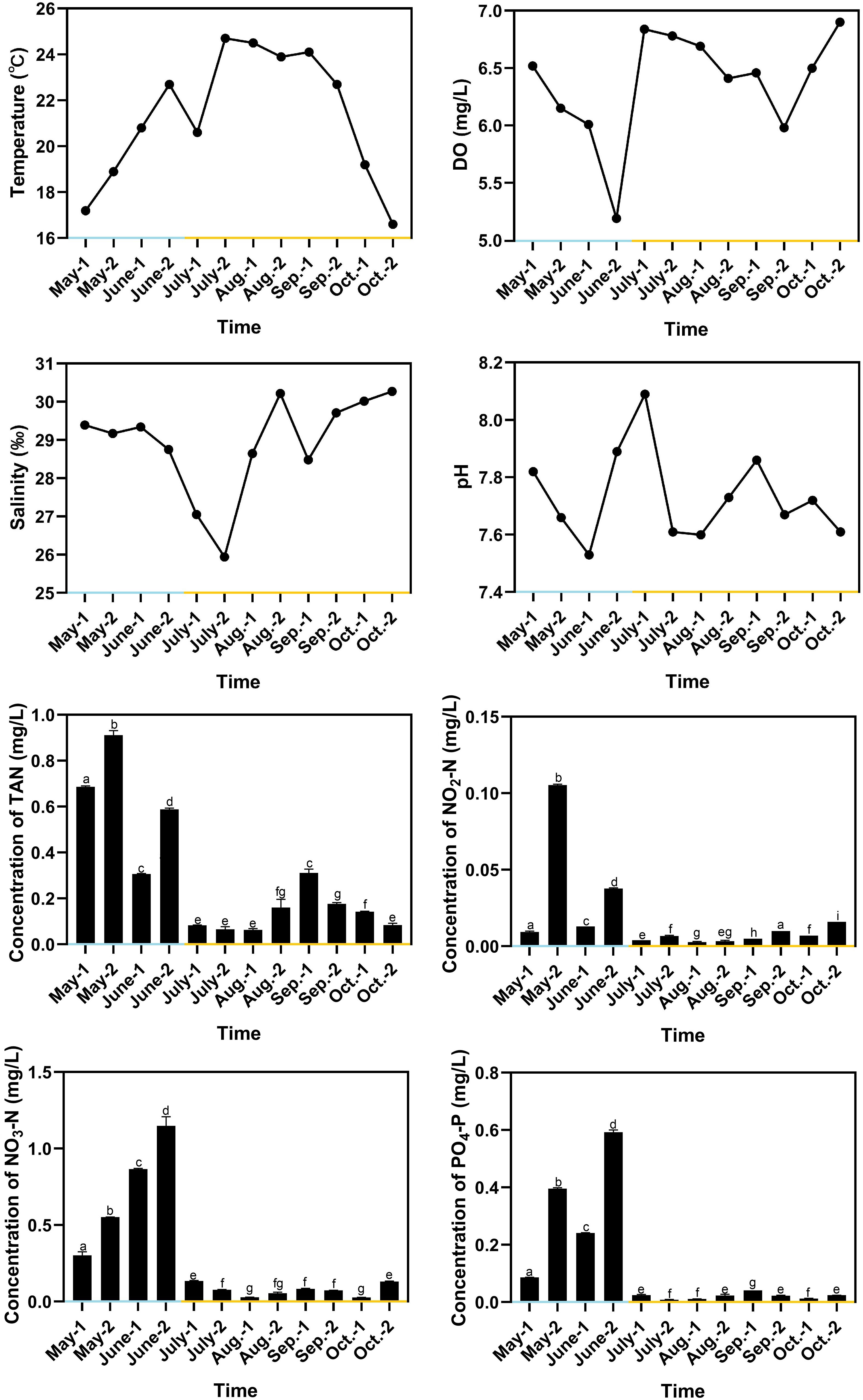
Figure 3 Variation of water quality indexes during aquaculture period. Blue x-axis indicates RAS aquaculture period, and yellow x-axis indicates OSCS aquaculture period. The letters above the bars indicate the significance of differences between groups; the groups with different letters have significant differences (P < 0.05).
Comparison between intestinal and seawater bacterial communities during the aquaculture period
In seawater, a total of 27 identified phyla were observed, and six phyla were identified with abundance > 1%. Proteobacteria (65.65%), Bacteroidetes (19.18%), Actinobacteria (6.02%), Cyanobacteria (3.85%) and Verrucomicrobia (2.46%) were dominant phyla in seawater bacterial community, accounting for 95.39% of the total abundance (Figure 4A). The correlation analysis of the abundance of the bacterial phyla that shared by intestinal tract and seawater samples was conducted, and it was found that the correlation values ranged from -0.323 to 0.188, and no significant correlation was observed (Table S3). There were noticeable differences in some phyla between the intestinal tract and seawater bacterial communities, including Bacteroidetes, Firmicutes, Actinobacteria, Spirochaetes, etc. According to LEfSe analysis, several bacterial taxa were identified to have significant differences between the intestinal tract and seawater bacterial communities, including Firmicutes, Bacteroidetes, Actinobacteria, SAR406 and Proteobacteria (Figure 4B). The PCA analysis of bacterial function showed clearly separate clustering of microbial function for intestinal and seawater bacterial communities (Figure 4C). There were significant differences in a lot of microbial functions, with the most prominent functions of synthesis and degradation of ketone bodies, secondary bile acid biosynthesis, bacterial chemotaxis, biosynthesis of ansamycins and valine, leucine and isoleucine degradation, etc. (Figure 4D). The F-values of beta diversity analyses generally increased with the sampling months, with the lowest value of 6.69 in May and the highest value of 32.32 in October (Figures 5A-G). The alpha diversity of the seawater bacterial community was significantly higher than that of the intestinal bacterial community (Figure 5H).
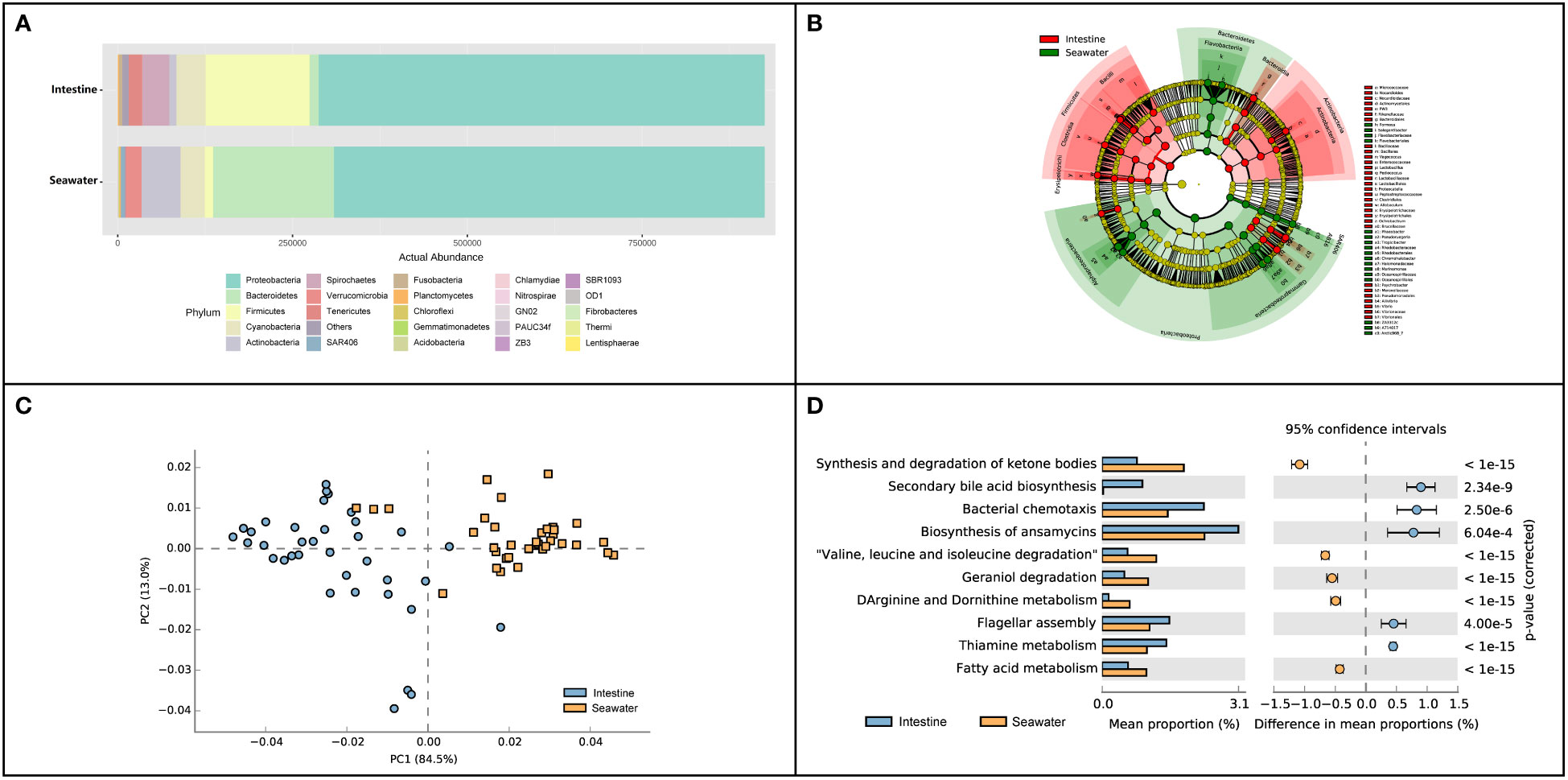
Figure 4 Composition and function differences between intestinal and seawater bacterial communities. (A) Bacterial abundance at the phylum level; (B) LEfSe analysis of ASVs between intestinal and seawater bacterial communities; (C) Principal components analysis (PCA) of bacterial functional characterization; (D) The differences of functional pathways between intestinal and seawater bacterial communities.
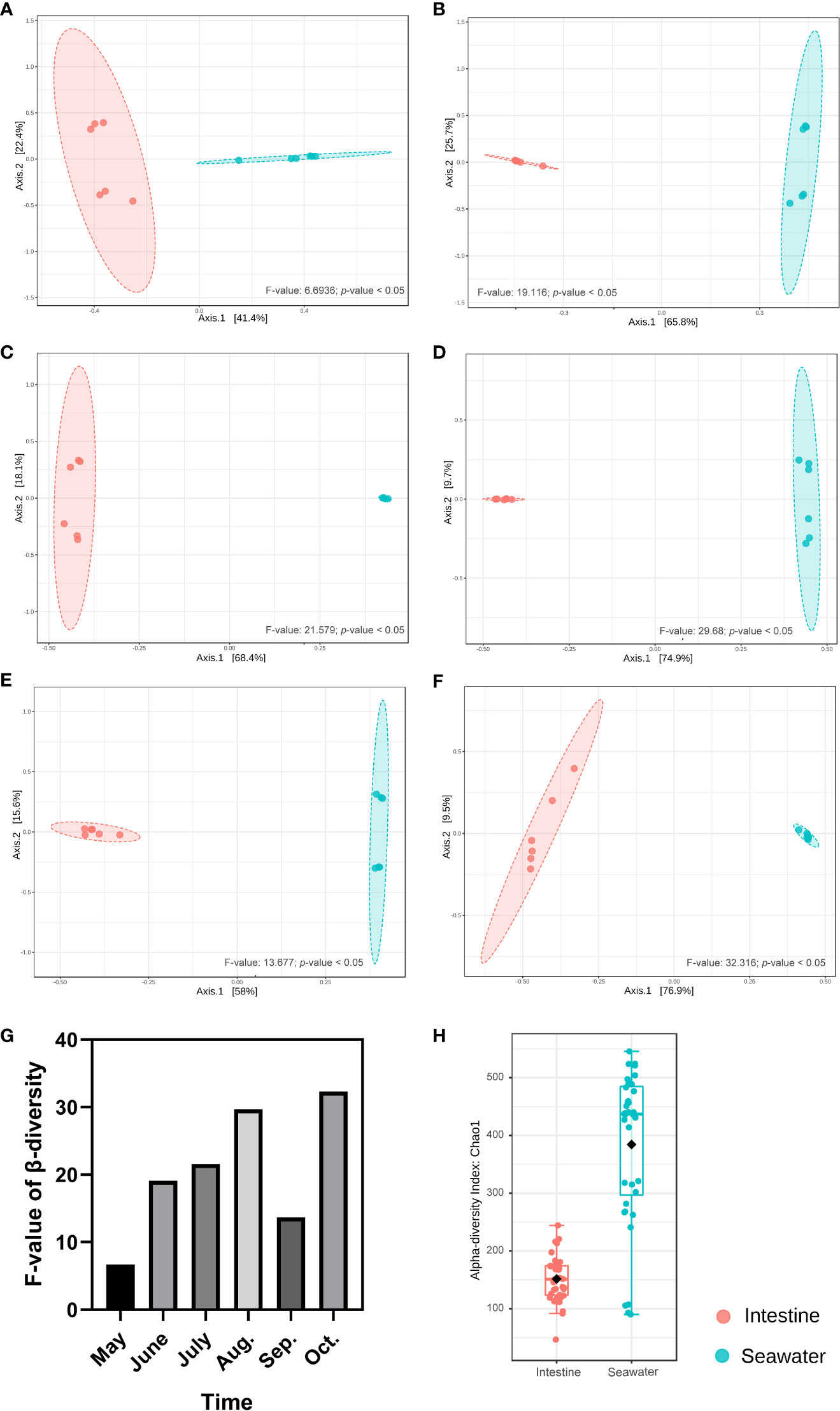
Figure 5 Beta and alpha diversity analyses between intestinal and seawater bacterial communities. (A-F) Beta diversity analyses of samples from May to October; (G) The F-values of beta diversity analyses in different months; (H) Alpha diversity analysis between intestinal and seawater bacterial communities.
Changes of immune response parameters
The MDA contents in spleen reached a peak of 11.55 nmol/mgprot in late July, which was significantly higher than those in the other sampling time points (Figure 6). The SOD activities decreased in early August to 48.98 U/mgprot, which was lower than those in most of the other sampling time points. The minimum and maximum values of the relative expression levels of IL-1β were observed in early September and early October, which were 0.81-fold and 4.47-fold of that in June, respectively. The relative expression levels of IL-1β significantly decreased in early September to 0.25-fold (P < 0.05) and 0.34-fold of that in late August and late September respectively. The minimum and maximum values of the relative expression levels of TNF-α were observed in late October and early July, which were 0.097-fold and 1.28-fold of that in June, respectively. There was a drastic drop in the relative expression levels of TNF-α in late July, and the low levels were maintained until the end of the experiment. The minimum and maximum values of the relative expression levels of IL10 were observed in June and late August, respectively, and the value in late August was 12.30-fold of that in June and was significantly higher than those in the other sampling time points. The minimum and maximum values of the relative expression levels of IL17-AF were observed in late October and early July, which were 0.13-fold and 2.78-fold of that in June, respectively. The relative expression levels of IL17-AF decreased gradually during July and August, with an increase from September to early October and a decrease in late October (Figure 6). The correlation analysis between water quality index and immune response parameter was conducted, and no significant correlation (P < 0.05) in both Spearman’s correlation analysis and Pearson’s correlation analysis was found (Table S4).
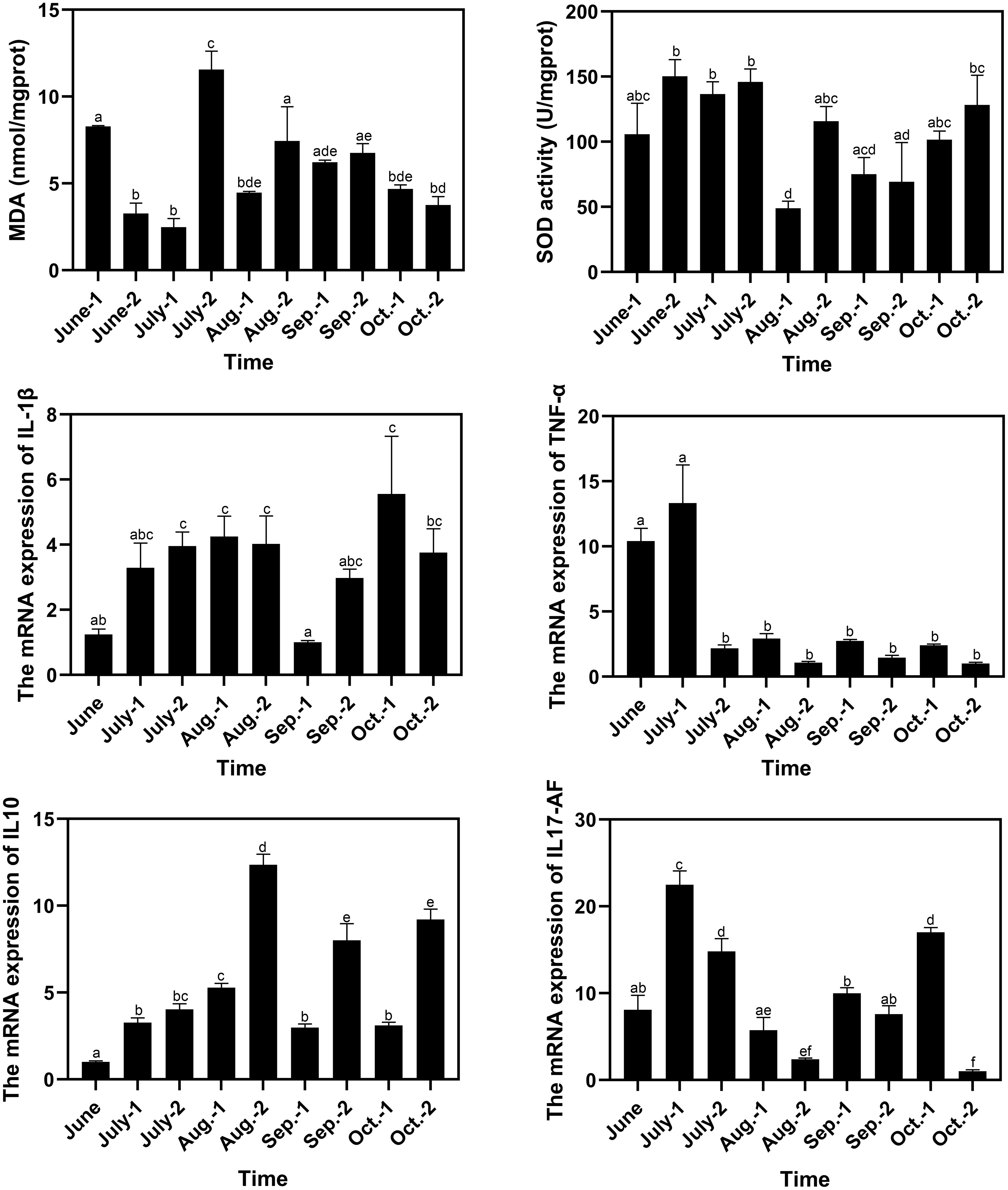
Figure 6 Variation of immune response parameters of pufferfish during the aquaculture period. The letters above the bars indicate the significance of differences between groups; the groups with different letters have significant differences (P < 0.05).
Correlation analysis of intestinal bacterial taxa and immune response parameters
All ASVs of the intestinal bacterial community were examined in all the 36 samples, and 114 ASVs were identified for the correlation analysis with immune response parameters. Among the 114 ASVs, 27 ASVs had significant correlations with one or more immune response parameters through Spearman’s correlation analysis or Pearson’s correlation analysis. The correlation values between the 27 ASVs and the immune response parameters in Spearman’s correlation analysis and Pearson’s correlation analysis ranged in 0.667-0.832 and 0.666-0.898 respectively (Figure 7). Arcobacter was found to have significant correlation with MDA content with the correlation value of -0.72 (P < 0.05) in Pearson’s correlation analysis, while without significant correlation with MDA content in Spearman’s correlation analysis. A total of seven ASVs were found to have significant correlations with one or more immune response parameters through both Spearman’s correlation analysis and Pearson’s correlation analysis (Table S5). They were annotated as Weissella sp., Akkermansia muciniphila, Dietzia sp., Psychrobacter pacificensis, Actinomycetales sp., Betaproteobacteria sp. and Actinomycetales sp.
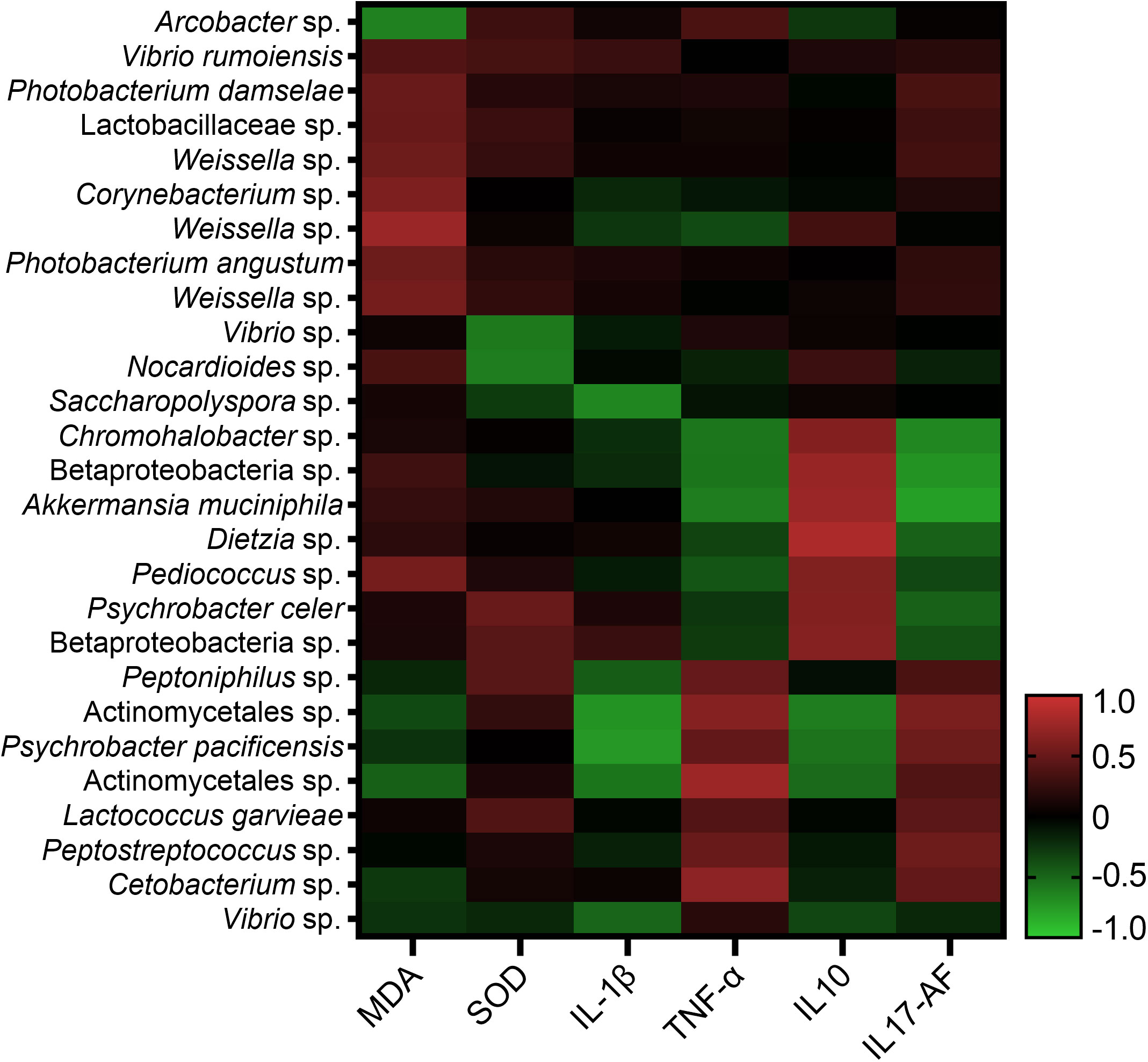
Figure 7 Correlation analysis of 27 ASVs of intestinal bacterial community and immune response parameters.
Discussion
The intestinal bacteria of fish can be broadly divided into autochthonous species that colonized the intestinal mucosa and the allochthonous species that free live and could not attach to the intestinal mucosa (Dehler et al., 2017). The autochthonous bacteria on the intestinal mucosa was obviously different from the allochthonous bacteria in digesta (Feng et al., 2011). The autochthonous bacteria always plays more important roles than allochthonous bacteria, as they could block the attachment sites of pathogens and provide disease prevention support to hosts (Banerjee and Ray, 2017). The allochthonous bacteria in the intestinal tract of pufferfish has been investigated in different development stages (Tanasomwang, 1989) and feeding with specific commercial feed formulations (Su et al., 2017). The present study was focused on the autochthonous bacterial community on pufferfish intestinal mucosa in order to illuminate its characteristics during aquaculture period and the possible relationship with immune response. The predominant phyla in the intestinal bacterial community were Proteobacteria, Firmicutes, Cyanobacteria, Spirochaetes and Tenericutes. Proteobacteria was found to be the most dominant phyla, which accounted for 68.82% of the total bacterial abundance. The result was consistent with previous studies on relative abundance of Proteobacteria in fish intestinal tract with 45.5 ± 18.3% in rainbow trout (Ingerslev et al., 2014) and 59-87% in common carp (Li et al., 2013).
The predominant genera in the intestinal bacterial community were Arcobacter, Photobacterium, Vagococcus, Weissella and Vibrio, which was different from the previous studies of pufferfish (Talwar et al., 2018; Ou et al., 2019). Arcobacter, Photobacterium and Vibrio contains some common pathogenic species for aquatic species. For example, Photobacterium damselae is a neuraminidases producer and infectious agent and is associated with the skin ulcers of fish (Egerton et al., 2018). Vagococcus fluvialis was reported to be a good candidate of probiotic as it showed good protection against Vibrio challenge (Sorroza et al., 2012). Some species in Weissella are proposed as probiotics, whereas other species might be opportunistic pathogens (Mortezaei et al., 2020). The previous studies were focused on the allochthonous bacteria, and Vibrio, Enterobacter, Bacillus, Bacteroides and Escherichia were reported to be the predominant genera (Li et al., 2015; Su et al., 2017). Vibrio was found be the dominant genus in the autochthonous intestinal bacteria in this study and also in the allochthonous intestinal bacteria in other literatures (Li et al., 2015; Yu et al., 2019; Kong et al., 2021). Vibrio is one of the most important bacterial genera in aquaculture and occupies the species as opportunistic pathogens causing infections such as V. anguillarum, V. salmonicida and V. vulnificus (Austin et al., 2007), and also has probiotic such as V. alginolyticus (Austin et al., 1995). It was reported that the intestinal tracts of marine fish are predominantly colonized by the members of the genus Vibrio (Shiina et al., 2006). In addition, there were some differences in intestinal bacterial communities between pufferfish and other fish species (e.g., Actinobacteria and Bacteroides usually dominant in other fish species) (Li et al., 2016; Nie et al., 2017), indicating that host phylogeny played important roles in the determination of the composition of fish intestinal bacteria (Xiong et al., 2019).
In the present study, Arcobacter was found to be the most core bacterial taxon in the intestinal bacterial community of pufferfish, with the most dominant abundance at the genus level and dominant positions in co-occurrence relationships with other bacterial taxa. Arcobacter has been reported to be dominant in the intestinal bacterial community of pufferfish (not specific for autochthonous bacteria) under some conditions, such as excess dietary astaxanthin (ASTX, 500 mg/kg) and arachidonic acid (ARA, 23.41 g/kg) (Ou et al., 2019; Yu et al., 2019). In the present study, Arcobacter was first found to be consistently highly enriched during the aquaculture period. Arcobacter is always regarded as emergent enteric pathogens and potential zoonotic agents, and it is frequently isolated from ill animals and humans with enteritis (Phillips, 2001; Ou et al., 2019). Arcobacter was also reported to be dominant in ulcer mucus surfaces of fish and moribund marine invertebrates (Lokmer and Mathias Wegner, 2015; Karlsen et al., 2017). However, pufferfishes did not show obvious disease symptoms under the high abundance of Arcobacter in the present study, indicating that Arcobacter might play other roles instead of pathogens in pufferfish. Immune system of mucosal tissues influences pathobionts decisions to be pathogenic or nonpathogenic. Arcobacter was also reported to be the dominant genus in the intestinal tract of deep-sea hagfishes (Lian et al., 2021). Phage genome was found to be inserted and integrated into the genome of Arcobacter with RM system, multiple TA systems and two CRISPR-Cas systems. It was speculated that Arcobacter might have the ability to resist viruses and could help relieve viral pressure for the hosts of hagfishes. Arcobacter was thus suggested to play important roles in the intestinal autochthonous microbiota of healthy pufferfish, and further study is needed to clarify the mechanism.
The mean levels of alpha diversity of intestinal bacterial communities in RAS and OSCS systems were very close, but the alpha diversity range in OSCS was more variable than that in RAS. The separation of beta diversity unweighted UniFrac distances between RAS and OSCS samples was probably caused by the differences in aquaculture systems between RAS and OSCS systems, including water environment, developmental stages of pufferfish and dietary composition (Kokou et al., 2019). The significant correlations among the concentrations of nutritive salts and between the concentrations of nutritive salts and aquaculture systems in this study indicated that different aquaculture systems occupied specific water environment qualities, which may affect the intestinal bacterial community. Intestinal bacterial diversity could be used as a biomarker of fish health because lower bacterial diversity was frequently detected in diseased fish (Xiong et al., 2019). The information of bacterial diversity variation in fish intestinal bacterial communities in RAS and OSCS systems is helpful for early warning of intestinal bacteria disorder and disease occurrence.
In regards to the differences of intestinal bacterial communities between RAS and OSCS, the abundances of some common phyla were found to be relatively higher in OSCS than that in RAS such as Spirochaetes and Bacteroidetes, which is consistent with previous studies (Shiina et al., 2006; Dehler et al., 2017; Su et al., 2017). Spirochaetes forms a monophyletic phylum of bacteria and has several genera that contain pathogenic species, which could lead to intestinal spirochetosis (Hampson and Ahmed, 2009). Spirochaetes was commonly detected in the digestive tracts of arthropods and several species of mammals and regularly found deep in marine sediments and soils, but at low concentrations in the intestinal microbiota of fish (Yoshida et al., 2022). The Spirochaetes abundance in the seawater in OSCS system was extremely lower than that in the intestine samples in this study, suggesting that the high abundance of Spirochaetes in the intestine samples in OSCS system was not caused by the passive transfer from seawater but possibly due to the active selection by fish. Bacteroidetes is one of the largest phyla of Gram-negative bacteria inhabiting mammal gastrointestinal tract and is considered a cornerstone of sophisticated homeostasis in fish (Gibiino et al., 2018; Talwar et al., 2018; Wang et al., 2018). The low Bacteroidetes/Firmicutes ratio could lead to obesity in mammals and fast growth of fish (Li et al., 2013). The lower Bacteroidetes/Firmicutes ratio in the intestine samples in RAS compared with that in OSCS in this study may suggest the intestinal bacteria in RAS have more favorable roles in growth promotion than that in OSCS. According to LEfSe analysis, three of the top-4 enriched ASVs of intestinal bacterial communities in RAS belonged to Enterococcaceae, and three of the top-4 enriched ASVs in OSCS belonged to Vibrionaceae. Enterococcaceae performs wide diversity of functions in intestinal microbiota. Enterococcaceae spp. was reported to have an active role in maintaining immunologic homeostasis within the pouch mucosa (Smith et al., 2005; Scarpa et al., 2011), and Enterococcaceae in intestine also functioned as a master regulator of host defense against radiation, protecting both hematopoietic and gastrointestinal systems (Guo et al., 2020). Vibrionaceae occupies some pathogenic species especially in the genus Vibrio, and the functional pathways of intestinal bacteria related to Vibrio were also found to be more enriched in OSCS than that in RAS in this study. The enriched intestinal bacterial taxa in OSCS, such as Spirochaetes and Vibrionaceae, contain some bacterial species that are pathogenic to aquatic animals, while in most cases they do not cause disease. Pathogenic bacteria could be more prevalent and cause infection and disease only when the balance of intestinal commensal microbiota is disturbed (de Bruijn et al., 2018). It is suggested that the intestinal bacteria in OSCS are more enriched in potentially pathogenic species, and more attention should be paid to that during the aquaculture of pufferfish.
The alpha diversity of the seawater bacterial community was significantly higher than that of the intestinal bacterial community, which was consistent with previous reports (Duan et al., 2020; Kuang et al., 2020). It is reasonable that the water environment provides ideal habitat for various bacterial communities to have high diversity, and intestinal bacteria are constrained by abiotic and biotic factors, such as nutrition, salinity and host immune system (Kuang et al., 2020). However, there were some similarities of bacterial communities between intestinal and seawater samples. For instance, Proteobacteria and Cyanobacteria were found to be dominant phyla in both intestinal and seawater bacterial communities. Nevertheless, no significant correlation was found in the abundance of the bacterial phyla that shared by intestinal tract and seawater samples, and more differences were found in bacterial communities between intestinal and seawater samples, in terms of bacterial community taxa and functions. It is suggested that the changes of bacterial communities in intestinal tract and seawater are independent during the aquaculture period. Compared with terrestrial animals, environmental bacteria has a considerable impact on bacterial compositions of the fish intestinal tract due to constant contact with water (Kuang et al., 2020). Fish select and enrich the intestinal bacteria from the surrounding water during early development (Vadstein et al., 2018), and the environmental bacteria can be transferred from the water environment to the intestinal tract at any time through water ingestion (Kuang et al., 2020). At the same time, a series of exogenous and endogenous factors could affect the colonization process of bacteria in the fish intestinal tract, among which food sources, developmental stage and physiological status are of the most significance. Therefore, intestinal bacteria are not a passive collection of the environmental free-living bacterial communities. Intestinal bacterial community of aquatic species always exhibit stability, and the bacterial community in the hemolymph of Pacific oyster (Crassostrea gigas) was stable from June to August based on the survey in Germany and Netherlands (Lokmer et al., 2016). In the present study, the increasing F-values of the beta diversity with the sampling months indicated that the seawater bacteria play important roles in intestinal bacterial colonization during early development, and the differences between the intestinal bacteria and seawater bacteria tended to increase along with the aquaculture process, when the transfer from RAS to OSCS will further increase the differences.
The intestinal bacteria can participate in the host inflammatory response by cytokine-microbiota or microbiota-cytokine modulation interactions and influence the immune homeostasis and stress response (Mendes et al., 2019). From July to August, the pufferfish had been transferred from RAS to OSCS, and MDA contents, SOD activities and the relative expression levels of TNF-α, IL10 and IL17-AF changed significantly, indicating the immune response of pufferfish. On one hand, the pufferfishes had to acclimate to the new environment of the offshore sea cage. The water quality and seawater bacterial composition in OSCS were much more complex than that in RAS, which could induce stress on pufferfish. On the other hand, biotic and abiotic stresses happened more frequently in summer than that in the other seasons. For instance, the abundance of some marine pathogenic bacteria generally increased with the elevated water temperature (Oberbeckmann et al., 2012; Froelich et al., 2015), and the abundance of Vibrio was found to reached a peak in summer in the North Yellow Sea, where the present study was conducted (Gao et al., 2022a). The increasing pathogen abundance might raise the stress of immune response and induce acute inflammatory processes and cytokine variations in July and August. In addition, the elevated water temperature and salinity variation caused by rainfall were other factors causing abiotic stress on pufferfish (Liu et al., 2014). Therefore, the immune response of pufferfish should be monitored more strictly during the aquaculture period from July to August, to ensure the safety of aquaculture.
In the present study, four bacterial taxa at the genus level, Weissella sp., Akkermansia muciniphila, Dietzia sp. and Psychrobacter pacificensis, were found to have significant correlations with one or more immune response parameters through both Spearman’s correlation analysis and Pearson’s correlation analysis. The species in Weissella and Dietzia include both probiotics and opportunistic pathogens (Click, 2011; Mortezaei et al., 2020; Brown et al., 2022). Akkermansia spp. are widely present in the intestinal tract of human and animals and are negatively associated with metabolic disorder in clinical studies (Derrien et al., 2017; Parolini, 2019). The knowledge of the function of Psychrobacter pacificensis, which was first isolated from deep seawater, in intestinal microbiota is very limited, while it was speculated to be evolved from a pathobiont (Welter et al., 2021). The maintenance of health status of fish always relies on the balance between the immune system and endogenous bacteria, especially the autochthonous bacteria (Gómez and Balcázar, 2008). The intestinal bacteria is able to modulate the host immune system by regulating expressions of immunity-related genes in the hosts and producing various substances, such as antimicrobial compounds and sebastenoic acid (Rawls et al., 2007). In addition, the colonizing of autochthonous bacteria provides colonization resistance to other bacteria introduced later and prevents the invasion of potentially pathogenic bacteria. Recently, the specific bacteria taxa in the shrimp intestinal tract were reported as indicators of shrimp health and disease (Xiong et al., 2015; Huang et al., 2018). The correlations between intestinal autochthonous bacteria taxa and immune response parameters in spleen were analyzed in this study to identify the bacteria taxa that may be related to immune response. However, there are some other important immune organs that were not included in the immune response analysis of the present study, such as head kidney, as well as intestinal tract, which is the first barrier between host and external environment. It should be noted that further analysis on the immune response parameters in other immune organs will be necessary to fully understand the mechanism of how intestinal autochthonous bacteria modulate immune response. Overall, in this study, the above four bacterial taxa were suggested to be the indicators for immune status and pathological process of pufferfish.
Conclusion
The characteristics of both intestinal autochthonous bacteria of pufferfish and seawater bacteria were analyzed during the aquaculture period in RAS and OSCS. Arcobacter was found to be the core bacterial taxon in the intestinal autochthonous bacteria of pufferfish during the aquaculture period. Increasing differences were found between the intestinal and seawater bacterial communities along with the aquaculture process. Four bacterial taxa had significant correlations with immune response parameters, and they were suggested to be the indicators for immune status and pathological process of pufferfish. The results will contribute to the understanding of intestinal bacterial homeostasis and biosecurity practice in pufferfish aquaculture.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/, PRJNA884649.
Ethics statement
The animal study was reviewed and approved by ethics committee of Dalian Ocean University.
Author contributions
LG, investigation, sample collection, methodology, formal analysis, and writing original draft. ZZ, ZX, and QL, investigation and sample collection. NK, sample collection. LW, writing review and editing, supervision, and funding acquisition. LS, writing review and editing, supervision, and funding acquisition. All authors contributed to the article and approved the submitted version.
Funding
This research was supported by Liaoning Science and Technology Major Special Project (2020JH1/10200002), National key R&D Program of China (2018YFD0900501), the Fund for Outstanding Talents and Innovative Team of Agricultural Scientific Research in MARA, the Research Foundation for Climbing Scholar in Liaoning, the Scientific Research Project of Liaoning Province Department of Education (No. QL201902), the Research Foundation for Talented Scholars in Dalian Ocean University (to LW), Liaoning Applied Basic Research Program (2022JH2/101300140) and the Science and Technology Innovation Fund of Dalian (2021JJ13SN73).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2022.1062512/full#supplementary-material
References
Austin, B., Austin, D. A., Munn, C. (2007). Bacterial fish pathogens: Disease of farmed and wild fish (Chichester: Springer).
Austin, B., Stuckey, L., Robertson, P., Effendi, I., Griffith, D. (1995). A probiotic strain of vibrio alginolyticus effective in reducing diseases caused by aeromonas salmonicida, vibrio anguillarum and vibrio ordalii. J. Fish Dis. 18 (1), 93–96. doi: 10.1111/j.1365-2761.1995.tb01271.x
Bandeira Junior, G., Baldisserotto, B. (2021). Fish infections associated with the genus aeromonas: A review of the effects on oxidative status. J. Appl. Microbiol. 131 (3), 1083–1101. doi: 10.1111/jam.14986
Banerjee, G., Ray, A. K. (2017). Bacterial symbiosis in the fish gut and its role in health and metabolism. Symbiosis 72 (1), 1–11. doi: 10.1007/s13199-016-0441-8
Bolyen, E., Rideout, J. R., Dillon, M. R., Bokulich, N. A., Abnet, C. C., Al Ghalith, G. A., et al. (2019). Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 37 (8), 852–857. doi: 10.1038/s41587-019-0209-9
Brown, W. D., Feinberg, N., Stedman, E., Dejace, J., Hale, A. J. (2022). Dietzia cinnamea: An increasingly recognized human pathogen. IDCases 29, e01539. doi: 10.1016/j.idcr.2022.e01539
Burns, A. R., Stephens, W. Z., Stagaman, K., Wong, S., Rawls, J. F., Guillemin, K., et al. (2016). Contribution of neutral processes to the assembly of gut microbial communities in the zebrafish over host development. ISME J. 10 (3), 655–664. doi: 10.1038/ismej.2015.142
Chong, J., Liu, P., Zhou, G., Xia, J. (2020). Using MicrobiomeAnalyst for comprehensive statistical, functional, and meta-analysis of microbiome data. Nat. Protoc. 15 (3), 799–821. doi: 10.1038/s41596-019-0264-1
Click, R. E. (2011). Successful treatment of asymptomatic or clinically terminal bovine mycobacterium avium subspecies paratuberculosis infection (Johne's disease) with the bacterium dietzia used as a probiotic alone or in combination with dexamethasone: Adaption to chronic human diarrheal diseases. Virulence 2 (2), 131–143. doi: 10.4161/viru.2.2.15647
de Bruijn, I., Liu, Y., Wiegertjes, G. F., Raaijmakers, J. M. (2018). Exploring fish microbial communities to mitigate emerging diseases in aquaculture. FEMS Microbiol. Ecol. 94 (1), 161. doi: 10.1093/femsec/fix161
Dehler, C. E., Secombes, C. J., Martin, S. A. (2017). Environmental and physiological factors shape the gut microbiota of Atlantic salmon parr (Salmo salar l.). Aquaculture 467, 149–157. doi: 10.1016/j.aquaculture.2016.07.017
Deng, Y., Jiang, Y. H., Yang, Y., He, Z., Luo, F., Zhou, J. (2012). Molecular ecological network analyses. BMC Bioinform. 13 (1), 1–20. doi: 10.1186/1471-2105-13-113
Derrien, M., Belzer, C., de Vos, W. M. (2017). Akkermansia muciniphila and its role in regulating host functions. Microb. Pathogen. 106, 171–181. doi: 10.1016/j.micpath.2016.02.005
Desai, A. R., Links, M. G., Collins, S. A., Mansfield, G. S., Drew, M. D., Van Kessel, A. G., et al. (2012). Effects of plant-based diets on the distal gut microbiome of rainbow trout (Oncorhynchus mykiss). Aquaculture 350, 134–142. doi: 10.1016/j.aquaculture.2012.04.005
Dethlefsen, L., Eckburg, P. B., Bik, E. M., Relman, D. A. (2006). Assembly of the human intestinal microbiota. Trends Ecol. Evol. 21 (9), 517–523. doi: 10.1016/j.tree.2006.06.013
Douglas, G. M., Maffei, V. J., Zaneveld, J. R., Yurgel, S. N., Brown, J. R., Taylor, C. M., et al. (2020). PICRUSt2 for prediction of metagenome functions. Nat. Biotechnol. 38 (6), 685–688. doi: 10.1038/s41587-020-0548-6
Duan, Y., Huang, J., Wang, Y., Zhang, J. (2020). Characterization of bacterial community in intestinal and rearing water of penaeus monodon differing growth performances in outdoor and indoor ponds. Aquac. Res. 51 (10), 4279–4289. doi: 10.1111/are.14770
Egerton, S., Culloty, S., Whooley, J., Stanton, C., Ross, R. P. (2018). The gut microbiota of marine fish. Front. Microbiol. 9, 873. doi: 10.3389/fmicb.2018.00873
Feng, J. B., Luo, P., Dong, J. D., Hu, C. Q. (2011). Intestinal microbiota of mangrove red snapper (Lutjanus argentimaculatus forsskål 1775) reared in sea cages. Aquac. Res. 42 (11), 1703–1713. doi: 10.1111/j.1365-2109.2010.02768.x
Field, A. (2013). Discovering statistics using IBM SPSS statistics, SAGE (Los Angeles: SAGE Publications Ltd.).
Fishery Bureau of China Agriculture Department (2021). China Fishery statistical yearboo (Beijing: China Agriculture Press).
Froelich, B., Ayrapetyan, M., Fowler, P., Oliver, J., Noble, R. (2015). Development of a matrix tool for the prediction of vibrio species in oysters harvested from north Carolina. Appl. Environ. Microb. 81 (3), 1111–1119. doi: 10.1128/AEM.03206-14
Gao, X., Wang, X., Wang, X., Li, H., Xu, L., Fang, Y., et al. (2022b). Effect of winter feeding frequency on growth performance, biochemical blood parameters, oxidative stress, and appetite-related genes in takifugu rubripes. fish. Physiol. Biochem. 48, 1167–1181. doi: 10.1007/s10695-022-01107-y
Gao, L., Yu, Z., Liu, C., Kong, N., Wang, L., Song, L. (2022a). Characteristics and particularities of bacterial community variation in the offshore shellfish farming waters of the north yellow Sea. Front. Mar. Sci. 9, 997817. doi: 10.3389/fmars.2022.997817
Gibiino, G., Lopetuso, L. R., Scaldaferri, F., Rizzatti, G., Binda, C., Gasbarrini, A. (2018). Exploring bacteroidetes: Metabolic key points and immunological tricks of our gut commensals. Digest. Liver. Dis. 50 (7), 635–639. doi: 10.1016/j.dld.2018.03.016
Gómez, G. D., Balcázar, J. L. (2008). A review on the interactions between gut microbiota and innate immunity of fish. FEMS Microbiol. Immunol. 52 (2), 145–154. doi: 10.1111/j.1574-695X.2007.00343.x
Guo, H., Chou, W., Lai, Y., Liang, K., Tam, J. W., Brickey, W. J., et al. (2020). Multi-omics analyses of radiation survivors identify radioprotective microbes and metabolites. Science 370 (6516), eaay9097. doi: 10.1126/science.aay9097
Hampson, D. J., Ahmed, N. (2009). Spirochaetes as intestinal pathogens: Lessons from a brachyspira genome. Gut Pathog. 1 (1), 1–3. doi: 10.1186/1757-4749-1-10
Hopkins, M., Sharp, R., Macfarlane, G. (2002). Variation in human intestinal microbiota with age. Digest. Liver. Dis. 34, S12–S18. doi: 10.1016/S1590-8658(02)80157-8
Huang, F., Pan, L., Song, M., Tian, C., Gao, S. (2018). Microbiota assemblages of water, sediment, and intestine and their associations with environmental factors and shrimp physiological health. Appl. Microbiol. Biot. 102 (19), 8585–8598. doi: 10.1007/s00253-018-9229-5
Ingerslev, H., von Gersdorff Jørgensen, L., Strube, M. L., Larsen, N., Dalsgaard, I., Boye, M., et al. (2014). The development of the gut microbiota in rainbow trout (Oncorhynchus mykiss) is affected by first feeding and diet type. Aquaculture 424, 24–34. doi: 10.1016/j.aquaculture.2013.12.032
Jia, Y., Jing, Q., Zhai, J., Guan, C., Huang, B. (2019). Alternations in oxidative stress, apoptosis, and innate-immune gene expression at mRNA levels in subadult tiger puffer (Takifugu rubripes) under two different rearing systems. Fish Shellfish Immunol. 92, 756–764. doi: 10.1016/j.fsi.2019.07.016
Karlsen, C., Ottem, K., Brevik, Ø.J., Davey, M., Sørum, H., Winther-Larsen, H. (2017). The environmental and host-associated bacterial microbiota of Arctic seawater-farmed Atlantic salmon with ulcerative disorders. J. Fish Dis. 40 (11), 1645–1663. doi: 10.1111/jfd.12632
Kodama, H., Otani, K., Iwasaki, T., Takenaka, S., Horitani, Y., Togase, H. (2014). Metabolomic investigation of pathogenesis of myxosporean emaciation disease of tiger puffer fish takifugu rubripes. J. Fish Dis. 37 (7), 619–627. doi: 10.1111/jfd.12154
Kokou, F., Sasson, G., Friedman, J., Eyal, S., Ovadia, O., Harpaz, S., et al. (2019). Core gut microbial communities are maintained by beneficial interactions and strain variability in fish. Nat. Microbiol. 4 (12), 2456–2465. doi: 10.1038/s41564-019-0560-0
Kong, Y., Liao, Z., Ma, X., Liang, M., Xu, H., Mai, K., et al. (2021). Effects of different dietary lipid levels on intestinal mucosal barrier and microbial community of juvenile tiger puffer takifugu rubripes. Aquacult. Nutr. 27 (5), 1626–1639. doi: 10.1111/anu.13302
Kuang, T., He, A., Lin, Y., Huang, X., Liu, L., Zhou, L. (2020). Comparative analysis of microbial communities associated with the gill, gut, and habitat of two filter-feeding fish. Aquac. Rep. 18, 100501. doi: 10.1016/j.aqrep.2020.100501
Lian, C., Zhu, F., Wei, Z., He, L. (2021). Composition and potential functions of the dominant microbiota in deep-sea hagfish gut from the south China Sea. Deep-Sea Res. I: Oceanogr. Res. Pap. 169, 103488. doi: 10.1016/j.dsr.2021.103488
Li, T., Long, M., Ji, C., Shen, Z., Gatesoupe, F., Zhang, X., et al. (2016). Alterations of the gut microbiome of largemouth bronze gudgeon (Coreius guichenoti) suffering from furunculosis. Sci. Rep. 6 (1), 1–9. doi: 10.1038/srep30606
Li, Q., Qiao, G., Wang, L., Zhang, J., Li, R., Ni, P., et al. (2019). Studies on bacterial diversity and vibrio harveyi distribution associated with diseased fugu (Takifugu rubripes) in northeastern China. Pak. J. Zool. 51 (1), 67–77. doi: 10.17582/journal.pjz/2019.51.1.67.77
Liu, Y., Chen, Z., Dai, J., Yang, P., Xu, W., Ai, Q., et al. (2019). Sodium butyrate supplementation in high-soybean meal diets for turbot (Scophthalmus maximus l.): Effects on inflammatory status, mucosal barriers and microbiota in the intestine. Fish Shellfish Immunol. 88, 65–75. doi: 10.1016/j.fsi.2019.02.064
Liu, Y., Zhou, Q., Zhang, H., Jiang, C., Zhang, F. (2014). Effect of temperature on growth and sex differentiation at early developmental stage of redfin puffer (Takifugu rubripes). South China Fish. Sci. 10 (5), 24–29.
Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCT method. Methods 25 (4), 402–408. doi: 10.1006/meth.2001.1262
Li, X., Yan, Q., Xie, S., Hu, W., Yu, Y., Hu, Z. (2013). Gut microbiota contributes to the growth of fast-growing transgenic common carp (Cyprinus carpio l.). PloS One 8 (5), e64577. doi: 10.1371/journal.pone.0064577
Li, Y., Zhang, T., Zhang, C., Zhu, Y., Ding, J., Ma, Y. (2015). Bacterial diversity in the intestine of young farmed puffer fish takifugu rubripes. Chin. J. Oceanol. Limnol. 33 (4), 913–918. doi: 10.1007/s00343-015-4219-2
Lokmer, A., Goedknegt, M. A., Thieltges, D. W., Fiorentino, D., Kuenzel, S., Baines, J. F., et al. (2016). Spatial and temporal dynamics of pacific oyster hemolymph microbiota across multiple scales. Front. Microbiol. 7, 1367. doi: 10.3389/fmicb.2016.01367
Lokmer, A., Mathias Wegner, K. (2015). Hemolymph microbiome of pacific oysters in response to temperature, temperature stress and infection. ISME J. 9 (3), 670–682. doi: 10.1038/ismej.2014.160
Mendes, V., Galvao, I., Vieira, A. T. (2019). Mechanisms by which the gut microbiota influences cytokine production and modulates host inflammatory responses. J. Interf. Cytok. Res. 39 (7), 393–409. doi: 10.1089/jir.2019.0011
Meng, X., Wu, S., Hu, W., Zhu, Z., Yang, G., Zhang, Y., et al. (2021). Clostridium butyricum improves immune responses and remodels the intestinal microbiota of common carp (Cyprinus carpio l.). Aquaculture 530, 735753. doi: 10.1016/j.aquaculture.2020.735753
Montalban-Arques, A., De Schryver, P., Bossier, P., Gorkiewicz, G., Mulero, V., Gatlin, D. M., III, et al. (2015). Selective manipulation of the gut microbiota improves immune status in vertebrates. Front. Immunol. 6, 512. doi: 10.3389/fimmu.2015.00512
Mortezaei, F., Royan, M., Allaf Noveirian, H., Babakhani, A., Alaie Kordghashlaghi, H., Balcázar, J. (2020). In vitro assessment of potential probiotic characteristics of indigenous lactococcus lactis and weissella oryzae isolates from rainbow trout (Oncorhynchus mykiss walbaum). J. Appl. Microbiol. 129 (4), 1004–1019. doi: 10.1111/jam.14652
Nie, L., Zhou, Q., Qiao, Y., Chen, J. (2017). Interplay between the gut microbiota and immune responses of ayu (Plecoglossus altivelis) during vibrio anguillarum infection. Fish Shellfish Immunol. 68, 479–487. doi: 10.1016/j.fsi.2017.07.054
Oberbeckmann, S., Fuchs, B. M., Meiners, M., Wichels, A., Wiltshire, K. H., Gerdts, G. (2012). Seasonal dynamics and modeling of a vibrio community in coastal waters of the north Sea. Microb. Ecol. 63 (3), 543–551. doi: 10.1007/s00248-011-9990-9
Ou, W., Liao, Z., Yu, G., Xu, H., Liang, M., Mai, K., et al. (2019). The effects of dietary astaxanthin on intestinal health of juvenile tiger puffer takifugu rubripes in terms of antioxidative status, inflammatory response and microbiota. Aquacult. Nutr. 25 (2), 466–476. doi: 10.1111/anu.12872
Parks, D. H., Tyson, G. W., Hugenholtz, P., Beiko, R. G. (2014). STAMP: statistical analysis of taxonomic and functional profiles. Bioinform 30 (21), 3123–3124. doi: 10.1093/bioinformatics/btu494
Parolini, C. (2019). Effects of fish n-3 PUFAs on intestinal microbiota and immune system. Mar. Drugs 17 (6), 374. doi: 10.3390/md17060374
Phillips, C. (2001). Arcobacters as emerging human foodborne pathogens. Food. Control 12 (1), 1–6. doi: 10.1016/S0956-7135(00)00024-4
Rawls, J. F., Mahowald, M. A., Goodman, A. L., Trent, C. M., Gordon, J. I. (2007). In vivo imaging and genetic analysis link bacterial motility and symbiosis in the zebrafish gut. Proc. Natl. Acad. Sci. U.S.A 104 (18), 7622–7627. doi: 10.1073/pnas.0702386104
Ringø, E. (1993). The effect of chromic oxide (Cr2O3) on aerobic bacterial populations associated with the intestinal epithelial mucosa of Arctic charr, salvelinus alpinus (L.). Can. J. Microbiol. 39 (12), 1169–1173. doi: 10.1139/m93-177
Scarpa, M., Grillo, A., Faggian, D., Ruffolo, C., Bonello, E., D’Incà, R., et al. (2011). Relationship between mucosa-associated microbiota and inflammatory parameters in the ileal pouch after restorative proctocolectomy for ulcerative colitis. Surgery 150 (1), 56–67. doi: 10.1016/j.surg.2011.02.009
Shannon, P., Markiel, A., Ozier, O., Baliga, N. S., Wang, J. T., Ramage, D., et al. (2003). Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13 (11), 2498–2504. doi: 10.1101/gr.1239303
Shiina, A., Itoi, S., Washio, S., Sugita, H. (2006). Molecular identification of intestinal microflora in takifugu niphobles. Comp. Biochem. Physiol. Part D Genomics Proteomics 1 (1), 128–132. doi: 10.1016/j.cbd.2005.10.001
Smith, F., Coffey, J., Kell, M., O'Sullivan, M., Redmond, H., Kirwan, W. (2005). A characterization of anaerobic colonization and associated mucosal adaptations in the undiseased ileal pouch. Colorectal. Dis. 7 (6), 563–570. doi: 10.1111/j.1463-1318.2005.00833.x
SOA of China (2007). Specification for marine monitoring (Beijing, China: China Quality and Standards Publishing & Media Co., Ltd.).
Sorroza, L., Padilla, D., Acosta, F., Román, L., Grasso, V., Vega, J., et al. (2012). Characterization of the probiotic strain vagococcus fluvialis in the protection of European sea bass (Dicentrarchus labrax) against vibriosis by vibrio anguillarum. Vet. Microbiol. 155 (2-4), 369–373. doi: 10.1016/j.vetmic.2011.09.013
Stow, J. L., Low, P. C., Offenhäuser, C., Sangermani, D. (2009). Cytokine secretion in macrophages and other cells: pathways and mediators. Immunobiology 214 (7), 601–612. doi: 10.1016/j.imbio.2008.11.005
Su, P., Han, Y., Jiang, C., Ma, Y., Pan, J., Liu, S., et al. (2017). Effects of chitosan-oligosaccharides on growth performance, digestive enzyme and intestinal bacterial flora of tiger puffer (Takifugu rubripes temminck et schlegel 1850). J. Appl. Ichthyol. 33 (3), 458–467. doi: 10.1111/jai.13282
Sun, H., Luo, Z., Zhang, Z., Hao, S., Ding, S. (2018). Recovery of listonella anguillarum from diseased pufferfish (Takifugu rubripes) in China. Isr. J. Aquac. 70, 1500. doi: 10.46989/001c.20935
Talwar, C., Nagar, S., Lal, R., Negi, R. K. (2018). Fish gut microbiome: current approaches and future perspectives. Indian J. Microbiol. 58 (4), 397–414. doi: 10.1007/s12088-018-0760-y
Tanaka, S., Kobayashi, T., Songjinda, P., Tateyama, A., Tsubouchi, M., Kiyohara, C., et al. (2009). Influence of antibiotic exposure in the early postnatal period on the development of intestinal microbiota. FEMS Microbiol. Immunol. 56 (1), 80–87. doi: 10.1111/j.1574-695X.2009.00553.x
Tanasomwang, V. (1989). Intestinal microflora of rockfish sebastes schlegeli, tiger puffer takifugu rubripes and red grouper epinephelus akaara at their larval and juvenile stages. Bull. Jpn. Soc Sci. Fish. 55 (8), 1371–1377. doi: 10.2331/suisan.55.1371
Tao, N., Wang, L., Gong, X., Liu, Y. (2012). Comparison of nutritional composition of farmed pufferfish muscles among fugu obscurus, fugu flavidus and fugu rubripes. J. Food Compost. Anal. 28 (1), 40–45. doi: 10.1016/j.jfca.2012.06.004
Turnbaugh, P. J., Hamady, M., Yatsunenko, T., Cantarel, B. L., Duncan, A., Ley, R. E., et al. (2009). A core gut microbiome in obese and lean twins. Nature 457 (7228), 480–484. doi: 10.1038/nature07540
Vadstein, O., Attramadal, K. J., Bakke, I., Forberg, T., Olsen, Y., Verdegem, M., et al. (2018). Managing the microbial community of marine fish larvae: a holistic perspective for larviculture. Front. Microbiol. 9, 1820. doi: 10.3389/fmicb.2018.01820
Wang, A. R., Ran, C., Ringø, E., Zhou, Z. G. (2018). Progress in fish gastrointestinal microbiota research. Rev. Aquac. 10 (3), 626–640. doi: 10.1111/raq.12191
Webb, C. R., Koboziev, I., Furr, K. L., Grisham, M. B. (2016). Protective and pro-inflammatory roles of intestinal bacteria. Pathophysiology 23 (2), 67–80. doi: 10.1016/j.pathophys.2016.02.002
Welter, D. K., Ruaud, A., Henseler, Z. M., De Jong, H. N., van Coeverden de Groot, P., Michaux, J., et al. (2021). Free-living, psychrotrophic bacteria of the genus psychrobacter are descendants of pathobionts. Msystems 6 (2), e00258–e00221. doi: 10.1128/mSystems.00258-21
Xiong, J., Nie, L., Chen, J. (2019). Current understanding on the roles of gut microbiota in fish disease and immunity. Zool. Res. 40 (2), 70. doi: 10.24272/j.issn.2095-8137.2018.069
Xiong, J., Wang, K., Wu, J., Qiuqian, L., Yang, K., Qian, Y., et al. (2015). Changes in intestinal bacterial communities are closely associated with shrimp disease severity. Appl. Microbiol. Biot. 99 (16), 6911–6919. doi: 10.1007/s00253-015-6632-z
Yin, F., Gong, H., Ke, Q., Li, A. (2015). Stress, antioxidant defence and mucosal immune responses of the large yellow croaker pseudosciaena crocea challenged with cryptocaryon irritans. Fish Shellfish Immunol. 47 (1), 344–351. doi: 10.1016/j.fsi.2015.09.013
Yoshida, M.-a., Tanabe, T., Akiyoshi, H., Kawamukai, M. (2022). Gut microbiota analysis of blenniidae fishes including an algae-eating fish and clear boundary formation among isolated vibrio strains. Sci. Rep. 12 (1), 1–13. doi: 10.1038/s41598-022-08511-7
Keywords: pufferfish Takifugu rubripes, intestinal autochthonous bacteria, seawater bacteria, immune response, bacterial indicator
Citation: Gao L, Zhang Z, Xing Z, Li Q, Kong N, Wang L and Song L (2022) The variation of intestinal autochthonous bacteria in cultured tiger pufferfish Takifugu rubripes. Front. Cell. Infect. Microbiol. 12:1062512. doi: 10.3389/fcimb.2022.1062512
Received: 06 October 2022; Accepted: 29 November 2022;
Published: 13 December 2022.
Edited by:
Assunta Liberti, Stazione Zoologica Anton Dohrn Napoli, ItalyReviewed by:
Patricia Diaz-Rosales, Centro de Investigación en Sanidad Animal, CISA (INIA-CSIC), SpainMaria M. Costa, Instituto de Investigaciones Marinas, Spain
Claudia Ibacache, Universidad de Valparaiso, Chile
Copyright © 2022 Gao, Zhang, Xing, Li, Kong, Wang and Song. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Linsheng Song, bHNoc29uZ0BkbG91LmVkdS5jbg==
 Lei Gao
Lei Gao Ziyang Zhang1,2,3
Ziyang Zhang1,2,3 Lingling Wang
Lingling Wang Linsheng Song
Linsheng Song