- 1Research Unit for Comparative Microbiome Analysis, Helmholtz Munich, German Research Center for Environmental Health, Neuherberg, Germany
- 2Department of Environmental Medicine, Faculty of Medicine, University of Augsburg, Augsburg, Germany
- 3Insitute of Environmental Medicine, Helmholtz Munich, German Research Center for Environmental Health, Neuherberg, Germany
- 4Christine Kühne Center for Allergy Research and Education, Davos, Switzerland
- 5Chair of Environmental Microbiology, TUM School of Life Sciences Weihenstephan, Technical University of Munich, Freising, Germany
Atopic dermatitis (AD) is a prevalent inflammatory skin disorder characterized by chronic inflammation, skin barrier dysfunction, and microbial dysbiosis, with Staphylococcus aureus playing a significant role in its pathogenesis. This paper explores the strain diversity and microevolution of S. aureus within AD patients, emphasizing how specific strains adapt to the altered skin environment, exacerbating the condition. The review emphasizes the significance of variation in specific functional genes among S. aureus strains, which enhances their ability to adapt to different microenvironments and shapes their pathogenic potential. It also discusses how mobile genetic elements, particularly prophages, contribute to genetic diversity and drive the virulence and antibiotic resistance of S. aureus in AD, highlighting the clinical challenges posed by these strain-specific factors in managing the disease. The paper advocates for the integration of advanced genomic tools such as whole-genome sequencing and machine learning to develop targeted therapies. By focusing on the genetic adaptability of S. aureus and its impact on AD, this review underscores the need for strain-specific diagnostics and personalized treatment strategies to improve patient outcomes.
1 Introduction
S. aureus is a highly adaptable and versatile bacterium known for its ability to inhabit a wide range of hosts, including humans (1), livestock, and domestic pets (2, 3), suggesting a potential for cross-species transmission. It has also been detected in various environmental settings (4). This bacterium has a long evolutionary history as both a commensal and an opportunistic pathogen, reflecting its remarkable ability to thrive in diverse ecological niches. In humans, S. aureus commonly colonizes the nose, ear, and skin as a commensal bacterium in about 20–30% of the population (1). However, its presence is not purely neutral; S. aureus is also associated with several atopic diseases, such as AD (5) and hay fever in the nose (6), demonstrating its dual role in human health as both a harmless inhabitant and a potential trigger for allergic reactions.
The ability of S. aureus to exist in such a wide range of environments and hosts raises significant questions about its genetic adaptability: does the core genome of S. aureus, which includes genes common to all strains, provide the necessary flexibility for adaptation and phenotypic changes across different ecological niches, or is it possible that the pan-genome, which encompasses the entire set of genes present across various strains of S. aureus, allows specific strains to colonize distinct niches? For example, in the context of AD, the affected skin regions can be considered as a distinctive environment with an increased selection pressure, which might promote the evolution of S. aureus (7, 8). Therefore, understanding the drivers for microevolution of S. aureus will improve our understanding of the commensal or pathogenic nature of strains which will allow for a more precise identification of pathogenic strains, which might facilitate in the future the development of new forms of therapy for AD patients.
In this paper, we summarize the current stage of knowledge on strain diversification of S. aureus in AD patients and vote for a strain-specific identification of S. aureus also for diagnostic purposes.
2 Discussion
2.1 Staphylococcus aureus prevalence in AD patients
AD stands as a significant clinical and public health concern worldwide, characterized by chronic inflammation, intense itching, and a compromised skin barrier (9). The global prevalence of AD has been increasing over the past decades, affecting over 20% of children and 10% of adults worldwide (10). The 2019 Global Burden of Diseases data shows 171 million cases of AD worldwide, with a 28.6% increase in prevalence since 1990 (133 million) and a strong association with higher socioeconomic development (11). This rise not only highlights the pressing need for effective management strategies but also underscores the multifactorial nature of AD, influenced by genetic, immunological, psychological, and environmental factors like the skin microbiome (12).
Central to this discussion on AD pathology is the role of S. aureus. While S. aureus often coexists harmlessly with its host, its colonization is not just prevalent in individuals with AD but also plays a pivotal role in exacerbating the condition. Numerous studies have revealed a significantly higher rate of S. aureus colonization in AD patients compared to healthy individuals (13–15). The primary source of S. aureus contamination in AD patients is self-colonization from the nasal passages (11), spreading to the skin through direct contact, especially in areas with a compromised skin barrier. Research indicates that up to 70–90% of AD patients are colonized by S. aureus on lesional skin, a strong contrast to 20–30% in healthy individuals colonized by S. aureus in the nose (16, 17). This heightened prevalence is not limited to lesional skin; non-lesional skin of AD patients also exhibits higher colonization rates (17, 18). AD is more prevalent and severe in children, particularly infants and young children aged 5–9 years, while adults experience a lower prevalence and more localized symptoms as they age. In pediatric AD patients, the prevalence of S. aureus nasal colonization ranges from 46.4 to 82.2%, compared to 19.4 to 34.1% in the control group, while in adults with AD, nasal colonization rates are between 62.0 and 69.8% (19). For skin colonization, the rate of S. aureus increases with age in AD patients, with colonization rates of 50% in infants, 80% in children, and 87.5% in adults for acute lesions, and 18.5% in infants, 41.8% in children, and 48.9% in adults for chronic lesions (20). This increased burden of S. aureus correlates with both skin microbiome dysbiosis and heightened disease severity during flares (13, 14).
2.2 Pathogenic mechanisms of Staphylococcus aureus in AD patients
The impact of S. aureus colonization on AD symptoms is profound and multifaceted, involving the direct effects of bacterial colonization, the impact of toxins produced by S. aureus, and the induction of inflammatory responses that worsen AD symptoms (Figure 1). The abnormal proliferation of S. aureus in AD leads to dysbiosis of the skin microbiome, significantly reducing overall microbial diversity and weakening the skin barrier. This increases susceptibility to irritants, allergens, and pathogens (21), disrupts the skin immune response, and exacerbates inflammation (22). Recent studies reveal that early-onset barrier dysfunction triggers immune responses to commensal organisms, with S. aureus forming antimicrobial-resistant biofilms that influence keratinocyte biology and contribute to AD-associated inflammation (23). In addition, IL-9 is differentially produced by skin-tropic and extracutaneous memory T cells in response to various allergens like house dust mites and staphylococcal enterotoxin B, enabling patient stratification based on allergen sensitization and highlighting the key role of S. aureus in immune polarization in AD patients (24). Genetic mutations, such as those in the filaggrin gene, impair the skin barrier and create a favorable environment for S. aureus colonization (25). Psychological factors like stress impose unneglectable influences on the skin’s immune response, as stress-induced neuropeptide mediators affect immune and skin cells, increasing mast cell activity and nerve interactions (26, 27). Yet, direct studies specifically linking stress to particular strains of S. aureus are limited. The altered skin microenvironment in AD, including reduced antimicrobial peptide production (28) and increased skin pH, further supports S. aureus proliferation in a pH-dependent manner (29).
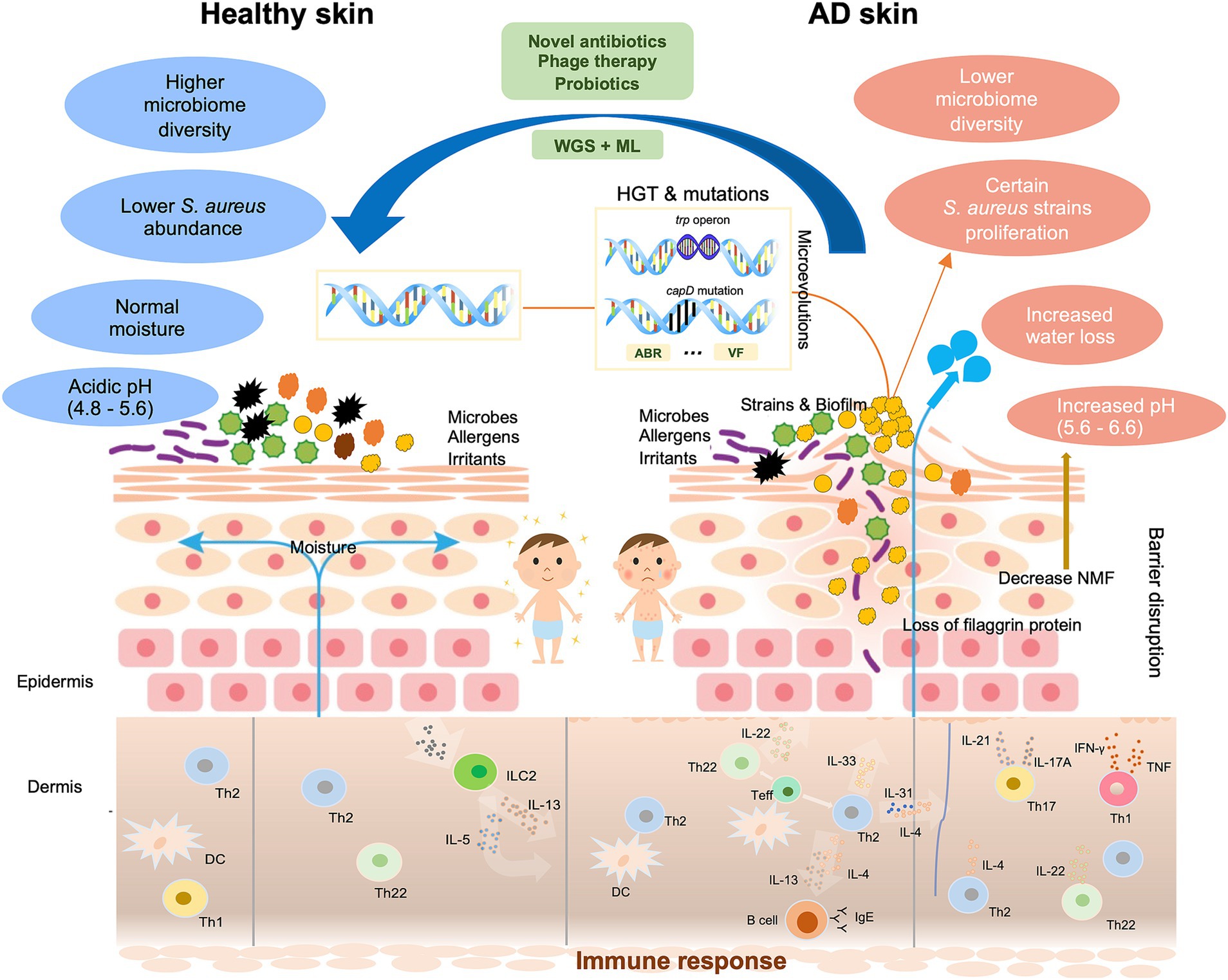
Figure 1. Schematic diagram of the multifaceted impact of microevolution on the pathogenicity of Staphylococcus aureus in AD. Normal skin is clinically intact with balanced diverse bacteria and physiological traits. AD skin exhibits the disrupted epidermal barrier like the flaggrin gene mutation, reduced skin microbiome diversity, increased water loss, and elevated pH. In affected skin areas where the altered microenvironment exerts significant selection pressure on certain Staphylococcus aureus strains, these strains proliferate, and heightened immune responses are triggered: Langerhans cells and inflammatory epidermal dendritic cells, which carry specific IgE attached to the high-affinity IgE receptor, as well as dermal dendritic cells, facilitate the introduction of allergens and antigens. Cytokines IL-4, IL-13, and IL-31 of the type-2 class directly stimulate sensory nerves, leading to itchiness. These changes make AD skin more susceptive to allergens, irritants, and pathogens. Mutation and HGT may occur in certain AD-associated strains, which result in increased production of VFs, and acquisition of ABR genes, and genetic variations that allow better adhesion (capD gene mutation), enhanced metabolic capabilities (trp operon), biofilm formation. Proposed therapeutic approaches include phage therapy, probiotics, and the integration of WGS and ML to develop targeted treatments. WGS, whole genome sequencing. ML, machine learning. ABR, antibiotic resistance. VF, virulence factor. HGT, horizontal gene transfer. DC, dendritic cell. IFN, interferon. IL, interleukin. ILC, innate lymphoid cell. Teff, effector T cell. Th, T-helper cell. TNF, tumor necrosis factor.
S. aureus binds more efficiently to AD skin due to specific receptors and binding proteins like fibronectin-and fibrinogen-binding proteins, which are upregulated in AD’s inflammatory milieu (30, 31). Once colonized, S. aureus exacerbates AD through the production of virulence factors (VF) that act as superantigens, inducing a disproportionate immune response and increased inflammation (30, 32). S. aureus also secretes enzymes like proteases and lipases that degrade crucial skin barrier proteins, increasing permeability and facilitating the entry of other allergens and pathogens (33, 34). Furthermore, the ability of S. aureus to form biofilms complicates eradication, as biofilms resist antimicrobial agents and host immune responses, making colonization persistent (35). These interactions highlight the need for targeted therapies addressing both the pathogen and immune dysregulation to effectively manage AD.
2.3 Strain diversity, microevolution, and geographical scale for Staphylococcus aureus diversification
The clinical implications of S. aureus in AD are significantly influenced by its strain diversity. Previous research has shown significant differences between S. aureus strains from AD patients and healthy individuals (14, 36, 37), indicating that AD-associated strains have a selective advantage in inflamed skin. Additionally, studies further revealed a high degree of strain diversity of S. aureus within AD (7, 8), with specific strains or clonal complexes (CC) of S. aureus like CC1 being more frequently associated with AD (36, 38), highlighting the significance of strain diversity.
The diversity of S. aureus strains complicates the management of AD in several ways.
1. Certain strains might be more pathogenic than others due to their secretion of strain-specific VFs. Understanding these strain-specific factors is crucial for developing targeted interventions. For instance, some strains exhibit heightened virulence through the production of specific toxins or enzymes that support cell wall anchoring and thus further disrupt the skin barrier or modulate the immune response more aggressively, leading to more severe AD flare-ups (25, 39). The variation in superantigens activates distinct subsets of T cell receptors, giving S. aureus strains unique T cell-avoidance capabilities, resulting in persistent infections and chronic inflammation (40).
2. S. aureus strains might possess distinct unique antibiotic resistance (ABR) profiles, which compounds the challenge of targeting S. aureus, especially for severe inflammatory AD conditions. In the UK, researchers found a higher prevalence of specific S. aureus CCs in AD patients, with over 80% of these strains carrying a plasmid with the ß-lactamase gene, conferring penicillin resistance—an attribute not found in the control group (36). Similarly, S. aureus isolates from AD showed higher fusidic acid resistance (41), making infections harder to treat.
3. S. aureus may possess strain-specific functions or pathways that confer a selective advantage in the AD microenvironment. A cohort study from Japan discovered that dysfunctional mutations in the accessory gene regulatory (Agr) quorum-sensing system were more common in the HE group, linking a functional Agr system to AD (42). An Japanese study discovered that only ST97A, a subclade of S. aureus detected in lesional AD skin, possessed the complete tryptophan (trp) operon, enabling bacterial survival without exogenous trp on AD skin, where the trp level was significantly reduced (43). A recent global study revealed that adaptive capsule loss via capD mutations, essential for S. aureus capsular polysaccharide synthesis, was more common in AD, enhancing adherence to AD skin (7). Studies also showed that S. aureus strains exhibit heterogeneous siderophore production within the host (44). In the context of AD, S. aureus strains with robust siderophore production can dominate the skin microbiome by outgrowing other bacteria with less efficient iron scavenging mechanisms. This competitive edge can lead to a higher relative abundance of S. aureus in AD patients, exacerbating the condition due to its pathogenic potential.
These results underscore the critical importance of strain-level variation in S. aureus, highlighting the need for precise and individualized approaches in managing AD. Furthermore, these findings emphasize the necessity of considering geographical scale when examining the diversification mechanisms of S. aureus strains. Most observed differences have been region-specific, suggesting these adaptations are spontaneous rather than indicative of a general distinction between AD and HE strains. The broader question of whether these genomic differences and adaptations occur locally or globally remains unresolved. Additionally, a recent study revealed that patient co-factors such as age, sex, and race also significantly impact the variation (5). This raises relevant questions: to what extent do health status, geographical location/scale, and patient co-factors influence S. aureus diversification? Which factors are more critical? Furthermore, given the frequent interactions between humans, livestock, and pets, could these animals serve as hotspots for microevolutionary changes? These questions underscore the complex interplay of factors affecting S. aureus diversification.
2.4 Role of prophages in Staphylococcus aureus adaptability and pathogenicity
Overall, around 15–20% of S. aureus genome is characterized by the presence of numerous mobile genetic elements such as plasmids, transposons, and bacteriophages (45). This genomic plasticity could be a key driver of its pathogenic potential. These MGEs facilitate genetic exchanges, conferring new metabolic capabilities and enabling the rapid acquisition of genes encoding ABRs and VFs. Such genomic adaptability allows S. aureus to swiftly respond to environmental selective pressures, making it a formidable pathogen.
Bacteriophages, known for their host specificity and diverse functional gene repertoire (46), significantly influence bacterial evolution. Embedded within the bacterial genome, prophages are bacteriophage DNA that can be lytic or lysogenic. In particular, lysogenic prophages, dormant within the host genome, could introduce new genes, enhance bacterial metabolic capabilities, and potentially increase pathogenicity, leading to important clinical implications (47). Research indicates that the primary mechanism of horizontal gene transfer in S. aureus is bacteriophage-mediated transduction (48). Therefore, prophages play a pivotal role in the diversification and pathogenicity of S. aureus, significantly influencing its genetic diversity, virulence, and ABR (57). For example, prophages in S. aureus sequence type 398 carry a set of genes encoding VFs, including a tyrosine recombinase linked to biofilm-associated staphylococcal infections, an ATP-dependent Clp protease, and an autolysin Atl involved in mediating adhesion to host tissue (49). Despite advances in understanding S. aureus genetic diversity, the extent to which prophages contribute to this genetic diversity and their link to AD severity remains underexplored. Therefore, profiling the prophages in S. aureus strains is crucial for understanding the relationship between genomic composition, phage-driven adaptation, and resultant phenotypic outcomes of S. aureus strains.
2.5 Future directions in research and treatment
AD is a prevalent and complex condition posing a significant global public health challenge. The diverse strains of S. aureus in AD, varying in virulence, ABR, and metabolic potential, call for advanced research methodologies and innovative treatment strategies.
Traditional treatments for AD, such as systemic glucocorticosteroids and ciclosporin, remain effective for more severe cases. In addition, therapies like topical or systemic antibiotics, topical corticosteroids, calcineurin inhibitors, and supportive options like bleach baths or antiseptics washes are often recommended. However, frequent use of antiseptics can disrupt the balance of the skin’s microbial community, thus should be limited to specific cases and combined with strategies aimed at restoring and maintaining a healthy skin microbiome. Meanwhile, new therapies have been introduced, including biologics like dupilumab and tralokinumab, along with Janus kinase inhibitors such as abrocitinib, baricitinib, and upadacitinib, which offer targeted immunomodulatory effects (50). Addressing psychological stress through behavioral therapy and psychopharmacologic agents (e.g., anxiolytics) could be an additional measure.
Whole-genome sequencing (WGS) and machine learning (ML) technologies offer unprecedented opportunities to decipher the subtle differences between S. aureus strains from AD and HE (51, 52). WGS has revolutionized our understanding of the genetic diversity of S. aureus, allowing for the detailed characterization of strain-specific markers and the identification of VF and ABR genes (53). When coupled with ML algorithms, WGS data can be analyzed to predict strain pathogenicity, ABR profiles, and potential VFs with high accuracy. This integrative approach facilitates the identification of novel biomarkers for S. aureus strains that are specifically associated with AD severity, enabling the development of more advanced diagnostic tools and personalized treatment approaches. Additionally, high-throughput isolation techniques are needed, as current molecular data may lack the necessary resolution for strain-level analyses. Moreover, creating and studying mutants can offer valuable insights into gene functions by allowing researchers to observe the effects of specific gene deletions or modifications, which can uncover potential targets for therapeutic intervention. To gain a systematic perspective, a global approach to characterizing strain variation and S. aureus’s role in AD across different populations and regions can provide valuable insights. Furthermore, alternative therapies like phage therapy and probiotics show promise in restoring skin microbiome balance (54–56) and reducing the risk of dysbiosis associated with broad-spectrum antibiotics. By employing these advanced methods, we can potentially uncover new therapeutic targets or diagnostic markers, facilitating personalized treatment approaches and the development of targeted therapies.
3 Conclusion
In conclusion, this review underscores the critical role of S. aureus strain diversity and microevolution in the pathogenesis of AD. The variation in specific functional genes among S. aureus strains enables these bacteria to adapt to the unique microenvironment of AD-affected skin, contributing to their increased pathogenic potential. Additionally, the presence of MGEs, especially prophages, further drives the genetic variability and virulence of these strains, complicating the clinical management of AD. The review highlights the necessity of incorporating advanced genomic tools, such as WGS and ML, to better understand the complex interactions between S. aureus and the host, ultimately leading to more precise diagnostics and personalized treatment strategies. Addressing these strain-specific factors is essential for developing effective therapies that target the root causes of S. aureus pathogenicity in AD, potentially improving patient outcomes and reducing disease burden.
Author contributions
ZW: Conceptualization, Formal analysis, Investigation, Methodology, Resources, Software, Validation, Visualization, Writing – original draft, Writing – review & editing. CH: Conceptualization, Project administration, Resources, Supervision, Writing – review & editing. CT-H: Conceptualization, Project administration, Resources, Supervision, Writing – review & editing. MR: Conceptualization, Project administration, Resources, Supervision, Writing – review & editing. MS: Conceptualization, Funding acquisition, Project administration, Resources, Supervision, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This study was supported by internal funding from the Helmholtz Munich.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Wertheim, HF, Melles, DC, Vos, MC, van Leeuwen, W, van Belkum, A, Verbrugh, HA, et al. The role of nasal carriage in Staphylococcus aureus infections. Lancet Infect Dis. (2005) 5:751–62. doi: 10.1016/S1473-3099(05)70295-4
2. Howden, BP, Giulieri, SG, Wong Fok Lung, T, Baines, SL, Sharkey, LK, Lee, JYH, et al. Staphylococcus aureus host interactions and adaptation. Nat Rev Microbiol. (2023) 21:380–95. doi: 10.1038/s41579-023-00852-y
3. Haag, AF, Fitzgerald, JR, and Penadés, JR. Staphylococcus aureus in animals. Microbiol Spectr. (2019) 7:19. doi: 10.1128/microbiolspec.gpp3-0060-2019
4. Kozajda, A, Jeżak, K, and Kapsa, A. Airborne Staphylococcus aureus in different environments—a review. Environ Sci Pollut Res Int. (2019) 26:34741–53. doi: 10.1007/S11356-019-06557-1
5. Rauer, L, Reiger, M, Bhattacharyya, M, Brunner, PM, Krueger, JG, Guttman-Yassky, E, et al. Skin microbiome and its association with host cofactors in determining atopic dermatitis severity. J Eur Acad Dermatology Venereol. (2023) 37:772–82. doi: 10.1111/JDV.18776
6. Janeczek, K, Emeryk, A, Zimmer, Ł, Poleszak, E, and Ordak, M. Nasal carriage of Staphylococcus aureus in children with grass pollen-induced allergic rhinitis and the effect of polyvalent mechanical bacterial lysate immunostimulation on carriage status: a randomized controlled trial. Immunity, Inflamm Dis. (2022) 10:e584. doi: 10.1002/IID3.584
7. Key, FM, Khadka, VD, Romo-González, C, Blake, KJ, Deng, L, Lynn, TC, et al. On-person adaptive evolution of Staphylococcus aureus during treatment for atopic dermatitis. Cell Host Microbe. (2023) 31:593–603.e7. doi: 10.1016/j.chom.2023.03.009
8. Conte, AL, Brunetti, F, Marazzato, M, Longhi, C, Maurizi, L, Raponi, G, et al. Atopic dermatitis-derived Staphylococcus aureus strains: what makes them special in the interplay with the host. Front Cell Infect Microbiol. (2023) 13:1194254. doi: 10.3389/fcimb.2023.1194254
9. Langan, SM, Irvine, AD, and Weidinger, S. Atopic dermatitis. Lancet. (2020) 396:345–60. doi: 10.1016/S0140-6736(20)31286-1
10. Weidinger, S, and Novak, N. Atopic dermatitis. Lancet. (2016) 387:1109–22. doi: 10.1016/S0140-6736(15)00149-X
11. Shin, YH, Hwang, J, Kwon, R, Lee, SW, Kim, MS, Il, SJ, et al. Global, regional, and national burden of allergic disorders and their risk factors in 204 countries and territories, from 1990 to 2019: a systematic analysis for the global burden of disease study 2019. Allergy. 78:2232–54. doi: 10.1111/ALL.15807
12. Mu, Z, and Zhang, J. The role of genetics, the environment, and epigenetics in atopic dermatitis. Adv Exp Med Biol. (2020) 1253:107–40. doi: 10.1007/978-981-15-3449-2_4
13. Kong, HH, Oh, J, Deming, C, Conlan, S, Grice, EA, Beatson, MA, et al. Temporal shifts in the skin microbiome associated with disease flares and treatment in children with atopic dermatitis. Genome Res. (2012) 22:850–9. doi: 10.1101/gr.131029.111
14. Byrd, AL, Deming, C, Cassidy, SKB, Harrison, OJ, Ng, WI, Conlan, S, et al. Staphylococcus aureus and Staphylococcus epidermidis strain diversity underlying pediatric atopic dermatitis. Sci Transl Med. (2017) 9:eaal465. doi: 10.1126/scitranslmed.aal4651
15. De Tomassi, A, Reiter, A, Reiger, M, Rauer, L, Rohayem, R, Traidl-Hoffmann, C, et al. Combining 16S sequencing and qPCR quantification reveals Staphylococcus aureus driven bacterial overgrowth in the skin of severe atopic dermatitis patients. Biomol Ther. (2023) 13:1030. doi: 10.3390/biom13071030
16. Tauber, M, Balica, S, Hsu, CY, Jean-Decoster, C, Lauze, C, Redoules, D, et al. Staphylococcus aureus density on lesional and nonlesional skin is strongly associated with disease severity in atopic dermatitis. J Allergy Clin Immunol. (2016) 137:1272–1274.e3. doi: 10.1016/j.jaci.2015.07.052
17. Totté, JEE, van der Feltz, WT, Hennekam, M, van Belkum, A, van Zuuren, EJ, and Pasmans, SGMA. Prevalence and odds of Staphylococcus aureus carriage in atopic dermatitis: a systematic review and meta-analysis. Br J Dermatol. (2016) 175:687–95. doi: 10.1111/bjd.14566
18. Alsterholm, M, Strömbeck, L, Ljung, A, Karami, N, Widjestam, J, Gillstedt, M, et al. Variation in Staphylococcus aureus colonization in relation to disease severity in adults with atopic dermatitis during a fivemonth follow-up. Acta Derm Venereol. (2017) 97:802–7. doi: 10.2340/00015555-2667
19. Sangaphunchai, P, Kritsanaviparkporn, C, and Treesirichod, A. Association between Staphylococcus Aureus colonization and pediatric atopic dermatitis: a systematic review and Meta-analysis. Indian J Dermatol. (2023) 68:619–27. doi: 10.4103/ijd.ijd_453_22
20. Park, HY, Kim, CR, Huh, IS, Jung, MY, Seo, EY, Park, JH, et al. Staphylococcus aureus colonization in acute and chronic skin lesions of patients with atopic dermatitis. Ann Dermatol. (2013) 25:410–6. doi: 10.5021/ad.2013.25.4.410
21. Lynde, CW, Andriessen, A, Bertucci, V, McCuaig, C, Skotnicki, S, Weinstein, M, et al. The skin microbiome in atopic dermatitis and its relationship to emollients. J Cutan Med Surg. (2016) 20:21–8. doi: 10.1177/1203475415605498
22. Naz, S, Ali, Z, Minhas, A, Fatima, A, and Waseem, S. Generation of dysbiotic microbiota in cutaneous leishmaniasis and enhancement of skin inflammation. Microb Pathog. (2023) 181:106202. doi: 10.1016/j.micpath.2023.106202
23. Hammond, M, Gamal, A, Mukherjee, PK, Damiani, G, McCormick, TS, Ghannoum, MA, et al. Cutaneous dysbiosis may amplify barrier dysfunction in patients with atopic dermatitis. Front Microbiol. (2022) 13:944365. doi: 10.3389/fmicb.2022.944365
24. García-Jiménez, I, Sans-de San Nicolás, L, Curto-Barredo, L, Bertolín-Colilla, M, Sensada-López, E, Figueras-Nart, I, et al. Heterogeneous IL-9 production by circulating skin-tropic and Extracutaneous memory T cells in atopic dermatitis patients. Int J Mol Sci. (2024) 25:8569. doi: 10.3390/ijms25168569
25. Riethmuller, C, McAleer, MA, Koppes, SA, Abdayem, R, Franz, J, Haftek, M, et al. Filaggrin breakdown products determine corneocyte conformation in patients with atopic dermatitis. J Allergy Clin Immunol. (2015) 136:1573–1580.e2. doi: 10.1016/j.jaci.2015.04.042
26. Courtney, A, and Su, JC. The psychology of atopic dermatitis. J Clin Med. (2024) 13:1602. doi: 10.3390/jcm13061602
27. Buske-Kirschbaum, A, Gierens, A, Höllig, H, and Hellhammer, DH. Stress-induced immunomodulation is altered in patients with atopic dermatitis. J Neuroimmunol. (2002) 129:161–7. doi: 10.1016/S0165-5728(02)00168-6
28. Kim, J, Kim, BE, Ahn, K, and Leung, DYM. Interactions between atopic dermatitis and Staphylococcus aureus infection: clinical implications. Allergy, Asthma Immunol Res. (2019) 11:593–603. doi: 10.4168/aair.2019.11.5.593
29. Hülpüsch, C, Tremmel, K, Hammel, G, Bhattacharyya, M, de Tomassi, A, Nussbaumer, T, et al. Skin pH–dependent Staphylococcus aureus abundance as predictor for increasing atopic dermatitis severity. Allergy Eur J Allergy Clin Immunol. (2020) 75:2888–98. doi: 10.1111/all.14461
30. Foster, TJ, Geoghegan, JA, Ganesh, VK, and Höök, M. Adhesion, invasion and evasion: the many functions of the surface proteins of Staphylococcus aureus. Nat Rev Microbiol. (2014) 12:49–62. doi: 10.1038/nrmicro3161
31. Fleury, OM, McAleer, MA, Feuillie, C, Formosa-Dague, C, Sansevere, E, Bennett, DE, et al. Clumping factor B promotes adherence of Staphylococcus aureus to corneocytes in atopic dermatitis. Infect Immun. (2017) 85:12. doi: 10.1128/IAI.00994-16
32. Berube, BJ, and Wardenburg, JB. Staphylococcus aureus α-toxin: nearly a century of intrigue. Toxins (Basel). (2013) 5:1140–66. doi: 10.3390/toxins5061140
33. Nakatsuji, T, Chen, TH, Two, AM, Chun, KA, Narala, S, Geha, RS, et al. Staphylococcus aureus exploits epidermal barrier defects in atopic dermatitis to trigger cytokine expression. J Invest Dermatol. (2016) 136:2192–200. doi: 10.1016/j.jid.2016.05.127
34. Sonesson, A, Przybyszewska, K, Eriksson, S, Mörgelin, M, Kjellström, S, Davies, J, et al. Identification of bacterial biofilm and the Staphylococcus aureus derived protease, staphopain, on the skin surface of patients with atopic dermatitis. Sci Rep. (2017) 7:8689. doi: 10.1038/s41598-017-08046-2
35. Di, DEG, Cavallo, I, Capitanio, B, Ascenzioni, F, Pimpinelli, F, Morrone, A, et al. Staphylococcus aureus and the cutaneous microbiota biofilms in the pathogenesis of atopic dermatitis. Microorganisms. (2019) 7:301. doi: 10.3390/microorganisms7090301
36. Harkins, CP, Pettigrew, KA, Oravcová, K, Gardner, J, Hearn, RMR, Rice, D, et al. The microevolution and epidemiology of Staphylococcus aureus colonization during atopic eczema disease flare. J Invest Dermatol. (2018) 138:336–43. doi: 10.1016/j.jid.2017.09.023
37. Wang, Z, Hülpüsch, C, Foesel, B, Traidl-Hoffmann, C, Reiger, M, and Schloter, M. Genomic and functional divergence of Staphylococcus aureus strains from atopic dermatitis patients and healthy individuals: insights from global and local scales. Microbiol Spectr. (2024) 12:19. doi: 10.1128/spectrum.00571-24
38. Clausen, ML, Edslev, SM, Andersen, PS, Clemmensen, K, Krogfelt, KA, and Agner, T. Staphylococcus aureus colonization in atopic eczema and its association with filaggrin gene mutations. Br J Dermatol. (2017) 177:1394–400. doi: 10.1111/bjd.15470
39. Spaulding, AR, Salgado-Pabón, W, Kohler, PL, Horswill, AR, Leung, DYM, and Schlievert, PM. Staphylococcal and streptococcal superantigen exotoxins. Clin Microbiol Rev. (2013) 26:422–47. doi: 10.1128/CMR.00104-12
40. Thammavongsa, V, Kim, HK, Missiakas, D, and Schneewind, O. Staphylococcal manipulation of host immune responses. Nat Rev Microbiol. (2015) 139:529–43. doi: 10.1038/nrmicro3521
41. Harkins, CP, McAleer, MA, Bennett, D, McHugh, M, Fleury, OM, Pettigrew, KA, et al. The widespread use of topical antimicrobials enriches for resistance in Staphylococcus aureus isolated from patients with atopic dermatitis. Br J Dermatol. (2018) 179:951–8. doi: 10.1111/bjd.16722
42. Nakamura, Y, Takahashi, H, Takaya, A, Inoue, Y, Katayama, Y, Kusuya, Y, et al. Staphylococcus Agr virulence is critical for epidermal colonization and associates with atopic dermatitis development. Sci Transl Med. (2020) 12:4068. doi: 10.1126/SCITRANSLMED.AAY4068
43. Obata, S, Hisatsune, J, Kawasaki, H, Fukushima-Nomura, A, Ebihara, T, Arai, C, et al. Comprehensive genomic characterization of Staphylococcus aureus isolated from atopic dermatitis patients in Japan: correlations with disease severity, eruption type, and anatomical site. Microbiol Spectr. (2023) 11:e0523922. doi: 10.1128/spectrum.05239-22
44. Perry, WJ, Spraggins, JM, Sheldon, JR, Grunenwald, CM, Heinrichs, DE, Cassat, JE, et al. Staphylococcus aureus exhibits heterogeneous siderophore production within the vertebrate host. Proc Natl Acad Sci USA. (2019) 116:21980–2. doi: 10.1073/pnas.1913991116
45. Malachowa, N, and Deleo, FR. Mobile genetic elements of Staphylococcus aureus. Cell Mol Life Sci. (2010) 67:3057–71. doi: 10.1007/S00018-010-0389-4
46. Koskella, B, and Meaden, S. Understanding bacteriophage specificity in natural microbial communities. Viruses. (2013) 5:806–23. doi: 10.3390/v5030806
47. Gummalla, VS, Zhang, Y, Te, LY, and Wu, VCH. The role of temperate phages in bacterial pathogenicity. Microorganisms. (2023) 11:541. doi: 10.3390/microorganisms11030541
48. McCarthy, AJ, Loeffler, A, Witney, AA, Gould, KA, Lloyd, DH, and Lindsay, JA. Extensive horizontal gene transfer during staphylococcus aureus co-colonization in vivo. Genome Biol Evol. (2014) 6:2697–708. doi: 10.1093/gbe/evu214
49. Diene, SM, Corvaglia, AR, François, P, and van der Mee-Marquet, N. Regional infection control Group of the Centre Region. Prophages and adaptation of Staphylococcus aureus ST398 to the human clinic. BMC Genomics. (2017) 18:133. doi: 10.1186/s12864-017-3516-x
50. Werfel, T, Heratizadeh, A, Aberer, W, Augustin, M, Biedermann, T, Bauer, A, et al. S3 guideline atopic dermatitis: part 2 - systemic treatment. JDDG J der Dtsch Dermatologischen Gesellschaft. (2024) 22:307–20. doi: 10.1111/ddg.15229
51. Hernández Medina, R, Kutuzova, S, Nielsen, KN, Johansen, J, Hansen, LH, Nielsen, M, et al. Machine learning and deep learning applications in microbiome research. ISME Commun. (2022) 2:98–7. doi: 10.1038/s43705-022-00182-9
52. Kwong, JC, Mccallum, N, Sintchenko, V, and Howden, BP. Whole genome sequencing in clinical and public health microbiology. Pathology. (2015) 47:199–210. doi: 10.1097/PAT.0000000000000235
53. Lan, R, and Reeves, PR. Intraspecies variation in bacterial genomes: the need for a species genome concept. Trends Microbiol. (2000) 8:396–401. doi: 10.1016/S0966-842X(00)01791-1
54. Hülpüsch, C, Rohayem, R, Reiger, M, and Traidl-Hoffmann, C. Exploring the skin microbiome in atopic dermatitis pathogenesis and disease modification. J Allergy Clin Immunol. (2024) 154:31–41. doi: 10.1016/j.jaci.2024.04.029
55. Nakatsuji, T, Chen, TH, Butcher, AM, Trzoss, LL, Nam, SJ, Shirakawa, KT, et al. A commensal strain of Staphylococcus epidermidis protects against skin neoplasia. Sci Adv. (2018) 4:eaao4502. doi: 10.1126/sciadv.aao4502
56. Nakatsuji, T, Hata, TR, Tong, Y, Cheng, JY, Shafiq, F, Butcher, AM, et al. Development of a human skin commensal microbe for bacteriotherapy of atopic dermatitis and use in a phase 1 randomized clinical trial. Nat Med. (2021) 27:700–9. doi: 10.1038/s41591-021-01256-2
Keywords: atopic dermatitis, Staphylococcus aureus , strain diversity, microevolution, prophage, antibiotic resistance, horizontal gene tranfer
Citation: Wang Z, Hülpüsch C, Traidl-Hoffmann C, Reiger M and Schloter M (2024) Understanding the role of Staphylococcus aureus in atopic dermatitis: strain diversity, microevolution, and prophage influences. Front. Med. 11:1480257. doi: 10.3389/fmed.2024.1480257
Edited by:
Luis F. Santamaria-Babí, University of Barcelona, SpainReviewed by:
Giovanni Damiani, University of Milan, ItalyRegina Fölster-Holst, University Medical Center Schleswig-Holstein, Germany
Copyright © 2024 Wang, Hülpüsch, Traidl-Hoffmann, Reiger and Schloter. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhongjie Wang, emhvbmdqaWUud2FuZ0BoZWxtaG9sdHotbXVuaWNoLmRl
†These authors have contributed equally to this work and share first authorship
 Zhongjie Wang
Zhongjie Wang Claudia Hülpüsch2,3,4†
Claudia Hülpüsch2,3,4† Michael Schloter
Michael Schloter