- 1Department of Environmental Science, American University, Washington, DC, United States
- 2U.S. Geological Survey, Nebraska Cooperative Fish and Wildlife Research Unit, School of Natural Resources, University of Nebraska-Lincoln, Lincoln, NE, United States
- 3Chesapeake Biological Laboratory, University of Maryland Center for Environmental Science, Solomons, MD, United States
- 4U.S. Geological Survey, Alaska Science Center, Anchorage, AK, United States
- 5U.S. Geological Survey, Fort Collins Science Center, Fort Collins, CO, United States
Understanding changes at the base of the marine food web in the rapidly transforming Arctic is essential for predicting and evaluating ecosystem dynamics. The northern Bering Sea experienced record low sea ice in 2018, followed by the second lowest in 2019, highlighting the urgency of the issue for this region. In this study, we investigated the diet of the clam Macoma calcarea in the Pacific Arctic using DNA metabarcoding, employing 18S and rbcL markers to identify dietary components. Our findings revealed a strong dependence on pelagic diatoms, particularly Chaetoceros sp., with a near absence of ice algae in the clam diet. This pattern reflects the lack of lipid-rich ice algal production during these low sea ice events. Additionally, our analysis detected algae capable of producing harmful toxins, notably Alexandrium dinoflagellates, in the clam diet, underscoring the need for increased monitoring due to potential ecosystem and human health risks. This study demonstrates the utility of DNA metabarcoding in unraveling the complex dynamics of Arctic marine food webs and pelagic-benthic coupling, providing a glimpse of future conditions in a rapidly changing environment.
1 Introduction
The Pacific Arctic region is seasonally one of the most productive marine ecosystems in the world (Grebmeier et al., 2006). In the northern Bering and Chukchi seas, ice-associated blooms occur attached to the ice, as under ice blooms, and/or as marginal ice-edge blooms as seasonal sea ice retreats across the shallow, continental shelf (Arrigo et al., 2014; Waga et al., 2021; Nielsen et al., 2024). Sympagic (ice-associated) production, is largely dominated by diatoms and is an important resource for consumers because it provides an early pulse of lipid-rich food each spring, with some species aggregating and sinking rapidly to the seafloor, while pelagic algal blooms remain suspended in the water column for longer periods (Szymanski and Gradinger, 2016; Siddon et al., 2020). This flux of carbon results in a tightly coupled sympagic-pelagic-benthic system (Grebmeier et al., 1988, 2018). The seasonal cycle of sea ice retreat and formation over the shelf has led to persistent high benthic biomass at biological hotspots throughout the region, including bivalves, polychaete worms, amphipods and other invertebrates that are important prey items for larger benthic feeding marine mammals (Fay, 1982; Stewart et al., 2023). Despite the importance of diatoms to primary production and the marine food web in the Arctic, gaps remain in our knowledge regarding the response of diatom community composition to changes in sea ice dynamics, particularly in the northern Bering Sea and Bering Strait (Fukai et al., 2020). Moreover, the Pacific Arctic ecosystem is undergoing rapid changes driven by warming ocean temperatures and declining sea ice (Grebmeier, 2012; Huntington et al., 2020; Siddon et al., 2020), introducing additional uncertainty of future ecosystem response.
In 2018, the northern Bering Sea experienced record low winter sea ice, followed by the second lowest winter in 2019 (Stabeno et al., 2019; Thoman et al., 2020). This unprecedented low winter ice event was a result of persistent warm, southerly winds, late winter freeze up of sea ice in the southern Chukchi Sea in 2017 and 2018, and warmer than average ocean temperatures in the Bering Sea (Stabeno et al., 2019). Profound ecological implications were documented throughout the marine food web in response to this event (Siddon et al., 2020). Ecosystem responses were partly attributed to changes in the timing and quality of the spring bloom (Frey et al., 2018), leading to lipid-poor zooplankton and cascading impacts throughout the food web (Duffy-Anderson et al., 2019). The lack of bottom cold pool formation led to the northward expansion of groundfish, seabird die-offs due to starvation, and unusual mortality events of ice seals and gray whales (Duffy-Anderson et al., 2019; Siddon et al., 2020; Stewart et al., 2023). Increases in harmful algal blooms (HABs) were also reported (Anderson et al., 2022). As species that are adapted to the seasonal pulses of food from ice associated blooms are pushed to physiological limits or are outcompeted by boreal species, restructuring of regional ecosystems appears likely (Grebmeier, 2012; Moore et al., 2014). One potential outcome for the Bering and Chukchi Seas could become a system that is no longer dominated by the benthic food web, but rather a pelagic system (Moore and Stabeno, 2015; Kędra et al., 2019). The extreme conditions in 2018 and 2019 presented a glimpse into the future of projected climate scenarios and allows us to understand better how the system may respond.
One ecologically important bivalve in this region, the tellinid clam Macoma calcarea (Gmelin, 1791), commonly occurs throughout the Pacific Arctic (Grebmeier et al., 2018; Goethel et al., 2019; Lisitsyna et al., 2024; Gerasimova et al., 2019). This species dominates macrofaunal biomass and abundance, as documented through the coordinated Distributed Biological Observatory (DBO) in the northern Bering and Chukchi Seas. Specific hotspots of Macoma biomass and abundance include the site of an annually recurrent winter polynya south of St. Lawrence Island (“SLIP”) and north into the northeast Chukchi Sea near Barrow Canyon (Moore and Grebmeier, 2018). These bivalves are a frequent prey item for the benthic-feeding Pacific walrus (Odobenus rosmarus divergens), bearded seals (Erignathus barbatus), and spectacled eider (Somateria fischeri), and are strongly associated with foraging site selection (Jay et al., 2014; Sheffield and Grebmeier, 2009; Beatty et al., 2016; Lovvorn et al., 2003). As facultative deposit and suspension feeders, M. calcarea utilize both organic matter deposited on the seafloor and from the water column via siphon (Naumov, 2006; Lisitsyna et al., 2024). Previous research on M. calcarea suggests they consume mixtures of these sources in the Bering Sea (Oxtoby et al., 2016). However, it remains uncertain if bivalves selectively consume ice algae (Oxtoby et al., 2016; McMahon et al., 2006; Sun et al., 2009) and whether the redistribution and (or) reduction of sympagic production will be detrimental for this species in the future. Long-term observations of this ecosystem already indicate that northward shifts in M. calcarea distribution have occurred in the northern Bering Sea (Goethel et al., 2019), perhaps because of shifting food sources (Grebmeier, 2012). Additionally, the increasing occurrences of harmful algal blooms (HABs) in the region (Lefebvre et al., 2022; Anderson et al., 2022), underscores the importance of understanding the diets of M. calcarea and other bivalves because they serve as an entry point for phycotoxins into the marine food web and may affect subsistence resources for coastal communities, which use walrus and bearded seal for subsistence diets (Pućko et al., 2023). The specific ecological responses by M. calcarea to these extreme sea ice reduction events of 2018 and 2019 have not yet been fully documented.
DNA metabarcoding techniques are emerging as a useful approach in diverse feeding studies, including Arctic food web ecology. For example, the analysis of Pacific walrus fecal matter collected from ice floes revealed the utility of genetic diet analysis in a rapidly changing Arctic ecosystem (Sonsthagen et al., 2020). That approach confirmed the importance of tellinid bivalves in walrus diet as well as the biodiversity and spatial heterogeneity of benthic prey available to foraging walruses from any given ice floe (Sonsthagen et al., 2020). Analysis of the gut contents of Calanus copepods revealed ice algae consumption (Cleary et al., 2017). Beyond the Arctic, this technique has provided insights on other bivalve diets, such as Macoma balthica, which expanded our understanding of tellinid feeding plasticity and utilization of freshly deposited organic matter (Garrison et al., 2022). Therefore, we posit that DNA metabarcoding techniques can improve understanding of this common bivalve’s feeding ecology and broader ecosystem responses to sea ice dynamics in the Pacific Arctic region.
For this study, we applied the DNA metabarcoding technique to analyze the diet of M. calcarea during the summers of 2018 and 2019, with the objective of gaining an improved understanding of potential shifts at the base of the food web, specifically focusing on ice-associated and pelagic diatoms. With the unprecedented sea ice conditions during these years in the Bering Sea, we were able to examine M. calcarea diet and, to some extent, changes in the algal community composition during record low sea ice events. Noting that changes in sea ice have shown to influence the composition of algal communities (Szymanski and Gradinger, 2016; Ardyna and Arrigo, 2020), we hypothesized that M. calcarea diets would contain limited but detectable amounts of ice-associated diatoms relative to pelagic diatom species.
2 Methods
2.1 Sample collection
Benthic 0.1m2 Van Veen grab samples were collected in July from the Canadian Coast Guard ship Sir Wilfrid Laurier during 2018 and 2019 cruises in the Bering Sea and Chukchi Sea (Figure 1). M. calcarea were obtained from 11 sampling stations with 7–10 clams selected per station per year (n = 190) for dissection (Table 1). Clams were stored in collection bags by station and frozen at -20°C prior to dissection. Dissections occurred in 2020 between the months of January and October. The whole wet weight of collected clams (including shell) and length, height, and width were recorded. Clams were placed on an individual weighing boat that was changed for every sample. Stomach and digestive gland (hereafter “stomach”) tissues were excised from adductor muscles, mantle, foot, and gills (outer surface was not rinsed prior to excision). Dissection tools were cleaned with 10% bleach solution and rinsed with water between samples. Precautions were taken throughout to prevent cross-contamination of PCR products. Only stomach tissues were used to ascertain diatom prey of Macoma clams. DNA was obtained from clam stomachs (and their contents) using a DNeasy PowerSoil Pro Kit (Qiagen) and eluted in 50µL of Solution C6. The DNeasy PowerSoil Pro Kit protocol contains a vortex step with beads that homogenize the stomach contents prior to DNA extraction. A single extraction protocol was performed on each stomach. Most clams (n = 122 of 190) had stomach samples that were < 250 mg of tissue, the maximum amount suggested in the DNeasy PowerSoil Pro Kit protocol. Of the remaining 68 clams, most of the stomach tissue and associated content was subsampled and included in the DNA extraction protocol. The portion of the stomach that was not included in the DNA extraction protocol was largely extraneous tissue. Extraction blanks were included every 23 samples.
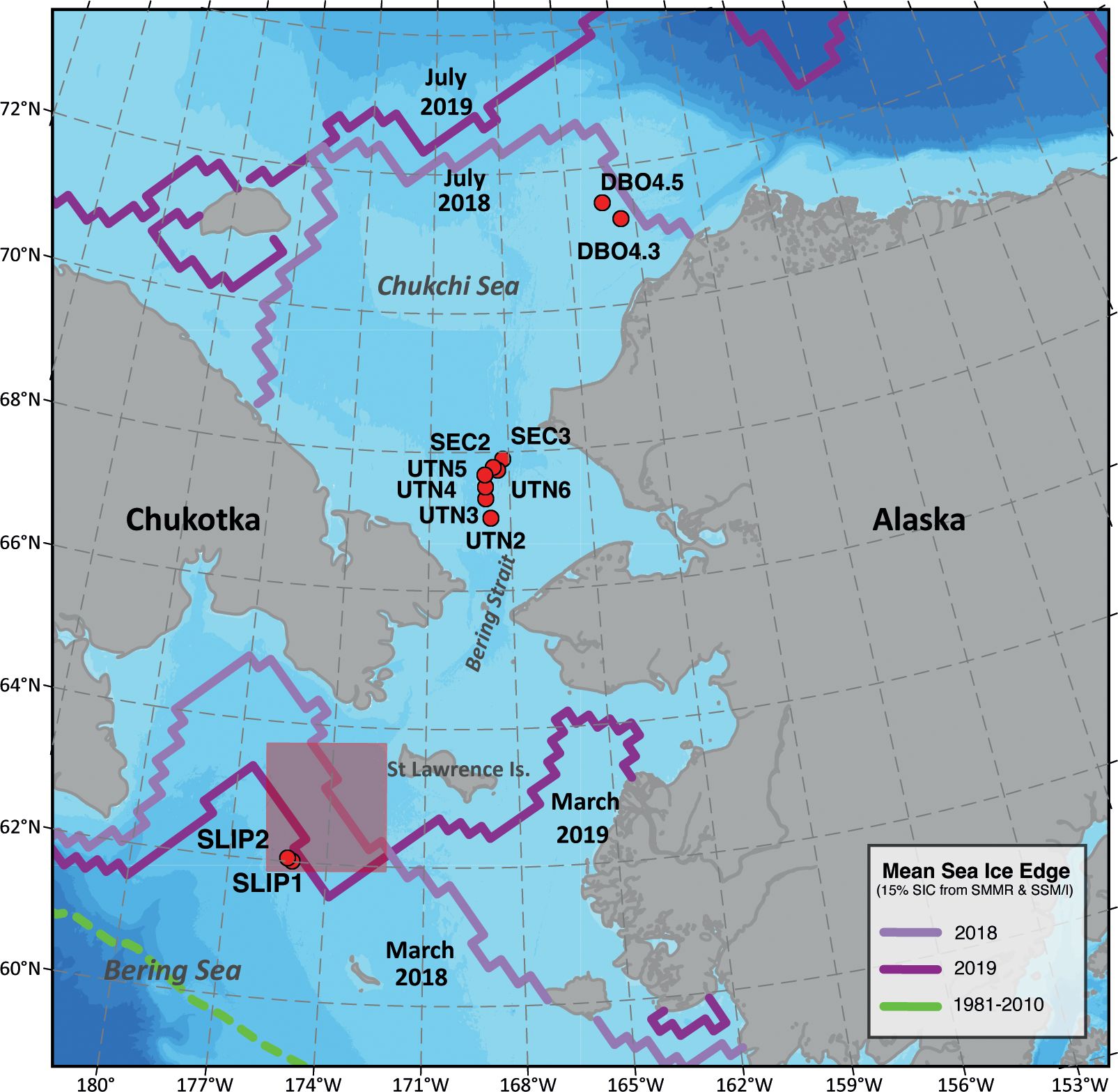
Figure 1. Map of sample collection locations on the 2018 and 2019 CCGC Sir Wilfred Laurier cruises as part of the Distributed Biological Observatory (DBO). Stations in the northern Bering Sea are near the location of the annually recurrent St. Lawrence Island polynya (SLIP, shaded box). UTN and SEC stations are located in the southeast Chukchi Sea (DBO 3 region) and DBO 4.3 and 4.5 are located in the northeast Chukchi Sea (DBO 4 region). Monthly mean sea ice extent data are derived from the Scanning Multichannel Microwave Radiometer (SMMR) instrument on the Nimbus-7 satellite and the Special Sensor Microwave/Imager (SSM/I), obtained from the National Snow and Ice Data Center (Fetterer et al., 2017).
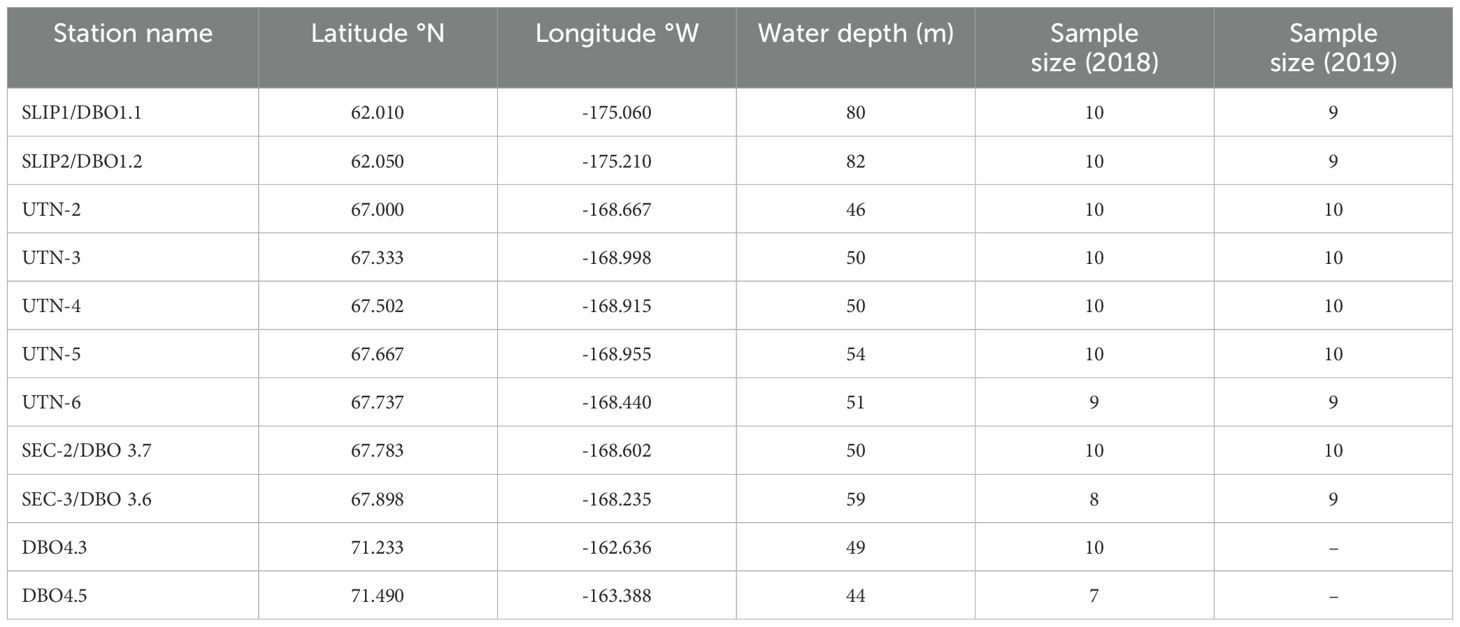
Table 1. Summary of sample collection Location and sample size (n) of tellinid clams collected in 2018 and 2019 on the Canadian Coast Guard Cutter Sir Wilfred Laurier as part of the Distributed Biological Observatory program (https://dbo.cbl.umces.edu/).
2.2 Library preparation
We performed DNA metabarcoding on two genes, the 18S V9 SSU ribosomal RNA gene (18S rDNA) and the large subunit of ribulose bisphosphate carboxylase (rbcL) gene located in the chloroplast genome. We selected the 18S gene because it is broadly amplified in eukaryotes and its wide use in the literature allows for cross-study comparisons as well as a richer reference database from public repositories. We also amplified the rbcL gene to focus on organisms with chlorophyll, which are the presumed primary diet of Macoma clams (Lovvorn et al., 2005).
We followed the 18S Illumina Amplicon Protocol adapted from the Earth Microbiome Project (Amaral-Zettler et al., 2009) that amplifies ~260 base pairs (bp; +/- 50 bp). A Macoma clam blocking primer was designed and included in reactions to limit amplification of host DNA during PCR (Vestheim and Jarman, 2008). Macoma balthica, M. nasuta, and M. secta 18S sequences were obtained from the National Center for Biotechnology Information (NCBI) GenBank and the blocking primer was constructed at a region with conserved sites among species with a C3 spacer at the 3’ end (ClamBlock_1391F: GCCCGTCGCTACGACCGATTGT[I][I][I][I][I]TTAATGAGCGAT[SpC3]). No sequence data was available for M. calcarea. The Earth Microbiome Project protocol amplifies 18S in a single PCR step. Eur1391F and EurBr primers contain the Illumina adapter sequence, 18S locus specific primer, with the EurBr_R primer fitted with a unique 12 bp Golay barcode (Thompson et al., 2017). PCR amplifications were performed in 25µL volumes containing 4µL of genomic DNA, 1X Phusion High-Fidelity Master Mix with HF buffer (Thermo Scientific), 4% bovine serum albumin (BSA), 0.4µM Euk1391F primer and 0.6µM ClamBlock_1391F with 0.4µM EurBr primer added separately to each well. A negative control was included in each PCR amplification. Thermocycler conditions were 98°C for 30 s, 35 cycles of 98°C for 10 s, 65°C for 15 s, 57°C for 30 s, 72°C for 40 s, and concluded with 72°C for 5 min. PCR amplifications were performed in triplicate. Products were visually inspected on an 1.5% agarose gel to verify successful amplification and pooled by sample.
We adapted the protocol by Vasselon et al. (2017) for amplification of 312 bp of rbcL. Specifically, we adapted the primers to conform to Illumina adapters for library preparation on an Illumina platform using a two-step PCR protocol. Briefly, locus-specific primers Diat_rbcL_708F_1, Diat_rbcL_708F_2, Diat_rbcL_708F_3, rbcL_R3_1 and rbcL_R3_2 were fitted with Illumina Nextera overhangs on the 5’ end (F: TCGTCGGCAGCGTCAGATGTGTATAAGAGACAG; R: GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAG). Initial PCR reactions were carried out in 25µL volumes containing 4µL of genomic DNA from excised stomachs, 1X Phusion High-Fidelity Master Mix with HF buffer (Thermo Scientific), 4% Bovine serum albumin (BSA), 0.4µM primer pool Diat_rbcL_708F and 0.4µM primer pool Diat_rbcL_R3. Thermocycler conditions were 98°C for 30 s, 30 cycles of 98°C for 10 s, 55°C for 30 s, 72°C for 40 s, and concluded with 72°C for 5 min. PCR amplifications were performed in triplicate. Products were visually inspected on an 1.5% agarose gel to verify successful amplification. PCR products were pooled by sample, and excess dNTPs and primers removed with ExoSAP-IT (USB Corporation, Cleveland, OH). The second PCR reactions incorporated Nextera indices and adaptors and were carried out in 15µL volumes containing 1µL pooled PCR product, 1X Phusion High-Fidelity Master Mix with HF buffer, and 1.0µM each of P5 and P7 adaptors. Thermocycler conditions were the same as above for rbcL except the number of cycles was reduced to 20 and the annealing temperature was increased to 60°C.
PCR products for 18S and rbcL marker were quantified using a Broad Range Quant-iT dsDNA Assay Kit (Invitrogen, Carlsbad, California, USA). 18S and rbcL sample libraries with unique barcodes or index pairs were pooled in equimolar concentration (sets of 48 samples for 18S and 96 samples for rbcL) by locus, purified with a 1.8x concentration of AMPure XP beads (Beckman Coulter, Inc.) to remove excess dNTPs and primer, then quantified using a High Sensitivity Quant-iT dsDNA Assay Kit (Invitrogen, Carlsbad, CA). Pooled libraries (8–10pM library and 30% PhiX) for each locus were sequenced on an Illumina MiSeq (250 bp pair-end reads) at the U.S. Geological Survey Alaska Science Center. Extraction blanks and PCR negatives failed to generate libraries that could be sequenced. Sequences and metadata are deposited in the NCBI database under BioProject (PRJNA1190063; BioSample accessions SAMN45024226-45024419). Detailed sample information is available in Sonsthagen et al. (2024).
2.3 Data assembly
Reads were trimmed with BBDuk (Bushnell, 2025) using a modulus trim of 5 and a Phred-scaled minimum quality of 10. Read pairs were then merged with vsearch v. 2.14.1 (Rognes et al., 2016), requiring an overlap of 100 (rbcL) or 50 (18S) and a maximum difference in the overlap region set to 6%. For the 18S locus, the “fastq_allowmergestagger” option was set since many amplicons are expected to be shorter than the cycle number. For 18S, both merged read pairs and the first read of unmerged read pairs were analyzed, whereas unmerged rbcL read pairs were discarded. Primers were not trimmed because the primers were not degenerate and thus did not affect the distribution of alignment scores. Each locus was then clustered within each sample with Swarm v. 2 (Mahé et al., 2015) with the “fastidious” option invoked to link adjacent low-abundance swarms. Small clusters of fewer than ten reads in a given sample were set to zero to reduce potential contamination or tag jumping errors.
2.4 Taxonomic assignment of operational taxonomic units
The resulting operational taxonomic units (OTUs) were assigned a taxonomy using the least common ancestor (LCA) approach. OTUs were first aligned to the nucleotide database of the NCBI with BLAST+ v. 2.10.0 (Camacho et al., 2009) with low complexity masking disabled and a maximum of 25 matches reported. The bit score was used as the scoring metric, and the assigned taxon was the lowest common ancestor of all taxa within 3% of the highest bit score, provided that the bit score was at least 400 for rbcL or 150 for 18S. Alignments to accessions with no established taxonomy (e.g., uncultured, environmental, or metagenomic sequences) were excluded. The LCA algorithm was then evaluated for each set of matched accessions at the standard ranks of species, genus, family, order, and class, as listed in the NCBI taxonomy database (accessed April 1, 2021). The cluster size was then considered to be the abundance of the OTU in the sample, and the taxon abundance was summed across all OTUs with the same assignment.
The LCA method is useful when well-scoped, discrete reference databases are not available, as is typically the case for these markers in marine environments (see Table 1 in Weigand et al., 2019, for example). LCA uses relative alignment-score distributions independently for each OTU, rather than probabilistic training on a discrete reference database that allows estimation of assignment confidence at species and genus level; otherwise these OTUs were demoted to the next taxonomic rank for re-evaluation. Specifically, for rbcL, species-level assignments required the average percent identity of matches to be 97% or greater and the maximum bit score of matches to be 500 or greater, whereas genus-level assignments required an average percent identity of 93% and a maximum bit score of at least 450. For 18S, species-level assignments required an average percent identity of 97% and genus-level assignments required an average percent identity of 93% (no minimum bit score was required due to the variation in amplicon size). Given that we are not aware of studies quantifying intraspecific and intergeneric variation at barcode loci in these taxa, these thresholds are necessarily somewhat arbitrary but were informed by the overall distribution of best alignment scores for OTUs at each locus (See Supplementary Figure S1).
2.5 Diet analysis
OTUs that were unlikely to be considered prey items (e.g., mammals) and known parasitic dinoflagellates and apicomplexans were filtered from the data set. Read counts were normalized to counts per million (cpm). We applied a 1% threshold to taxa within samples to account for inherent errors associated with amplification sequencing and alignment methods that make it difficult to distinguish among low-abundance prey, environmental background, amplification artifacts, and assignment errors (Deagle et al., 2019).
To assess the relative frequency of OTUs with the same taxonomic assignments across all sampling stations, we utilized heatmaps as a visualization tool. For the 18S gene data, we filtered the dataset to include only the top 15 taxa based on normalized read counts, due to the many low-abundance taxa assigned, the detection of which is expected to be highly stochastic, and which contribute minimal dietary information. This filtering allowed us to focus on the most abundant taxa, providing a clearer picture of the dominant prey items. The heatmaps were generated using normalized read counts, with color gradients representing the relative abundance of each OTU with similar taxonomic assignments across each station. This approach facilitated the identification of spatial patterns in the distribution of taxa and highlighted key differences in community composition among the sampling sites and years.
3 Results
3.1 Eukaryotic community composition based on 18S and rbcL markers
Initial trimming and filtering of nontarget taxa (see Methods) for the 18S marker resulted in a final read count of 235,064,859 (mean 1,237,184 per sample) prior to normalizing read counts and applying read count thresholds to samples. After normalizing samples, the lowest taxonomic rank that could be assigned to each OTU was as follows: 11% to class, 15% to order, 30% to family, 25% to genus, and 19% to species. The rbcL marker analysis resulted in a final read count of 221,898,210. The lowest taxonomic rank that could be assigned to each OTU was as follows: 3% to class, 3% to order, 21% to family, 27% to genus, and 46% to species. The read counts were grouped by station and means calculated for each OTU. A list of OTU and read count assignments are available at the Arctic Data Center (Koch et al., 2025).
To allow for the analysis of all available taxonomic ranks across the 18S gene data, read counts were then assigned to primary diet items by eukaryotic groups (Figure 2). These diet categories included cercozoans, prasinophytes, bacillariophytes (henceforth, referred to as diatoms), dinophytes (henceforth referred to as dinoflagellates), silicoflagellates, and other, which included raphidophytes, prymnesiophytes and ciliates. The relative 18S read count of each eukaryotic group by year and location indicated a dominance of diatoms in both years across nearly all stations. In 2018, SLIP1 (85%), DBO4.3 (89%) and DBO4.5 (86%) had the highest relative read count of diatoms. In 2019, UTN3 had ~92% relative read count of diatoms. In 2018, the proportional 18S read abundances of dinoflagellates in the clams’ diet at UTN3 and UTN4 increased by 43% and 41% respectively; these stations are located immediately north of the Bering Strait (Figure 1). In 2019, relative dinoflagellate 18S read counts increased moving north of the Bering Strait into the Chukchi Sea. In the southeast Chukchi Sea (SEC3), proportional read counts of dinoflagellates in the clam diets exceeded those for diatoms (67% versus 25%; Figure 2).
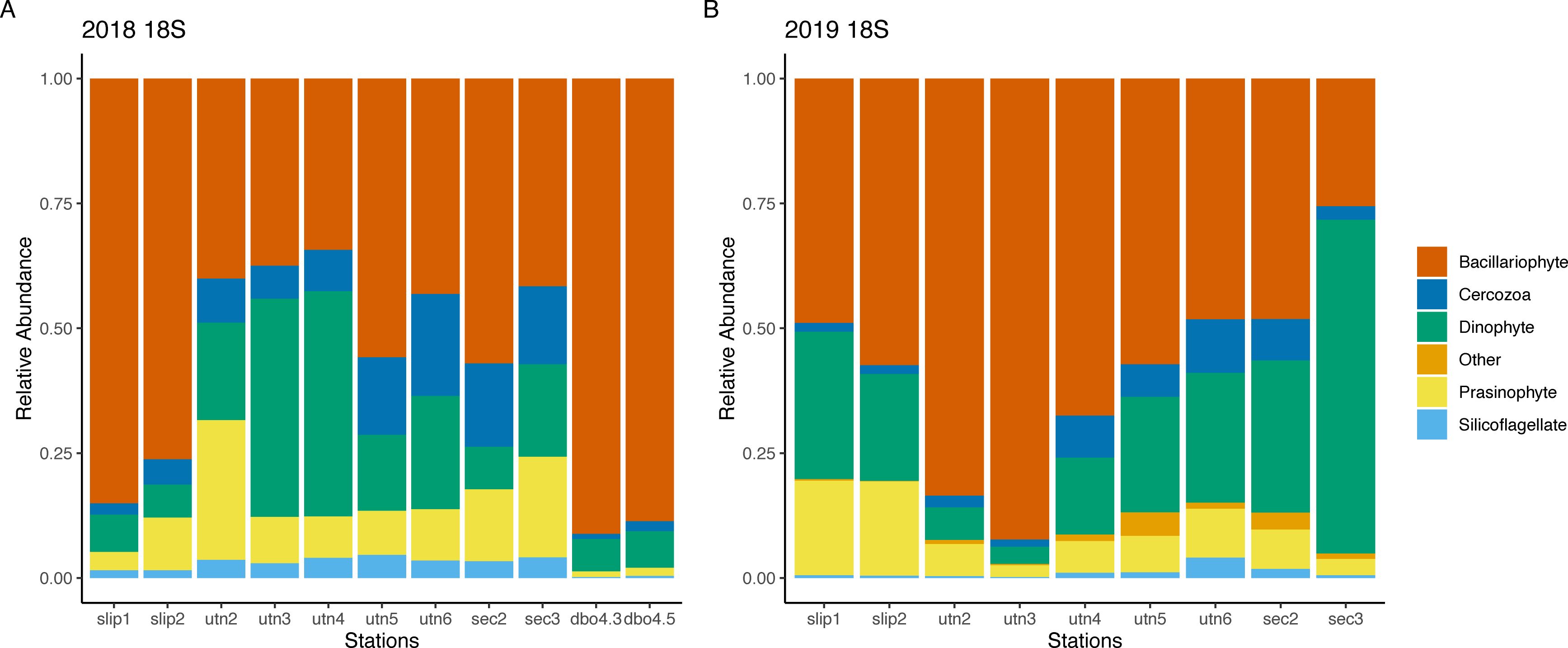
Figure 2. Proportions of normalized read abundance of eukaryotic groups within the summer diets of Macoma calcarea for the 18S rDNA dataset in (A) 2018 and (B) 2019. Stations are arranged geographically from south to north. DBO4.3 and DBO 4.5 were not sampled in 2019.
The proportions of prasinophytes, cercozoans, and silicoflagellates decreased within the 18S read counts from 2018 to 2019 (Figure 2). Of these groups, prasinophytes comprised a substantial proportion of all normalized 18S reads, which included the species Micromonas polaris and Pyramimonas parkea (Figures 3A, B). In 2018, the station UTN2 had the highest relative proportion of prasinophytes at 27% (Figure 2A). In 2019, this group was highest at SLIP1 and SLIP2 (~19% proportion of normalized read counts at both stations; Figure 2B).
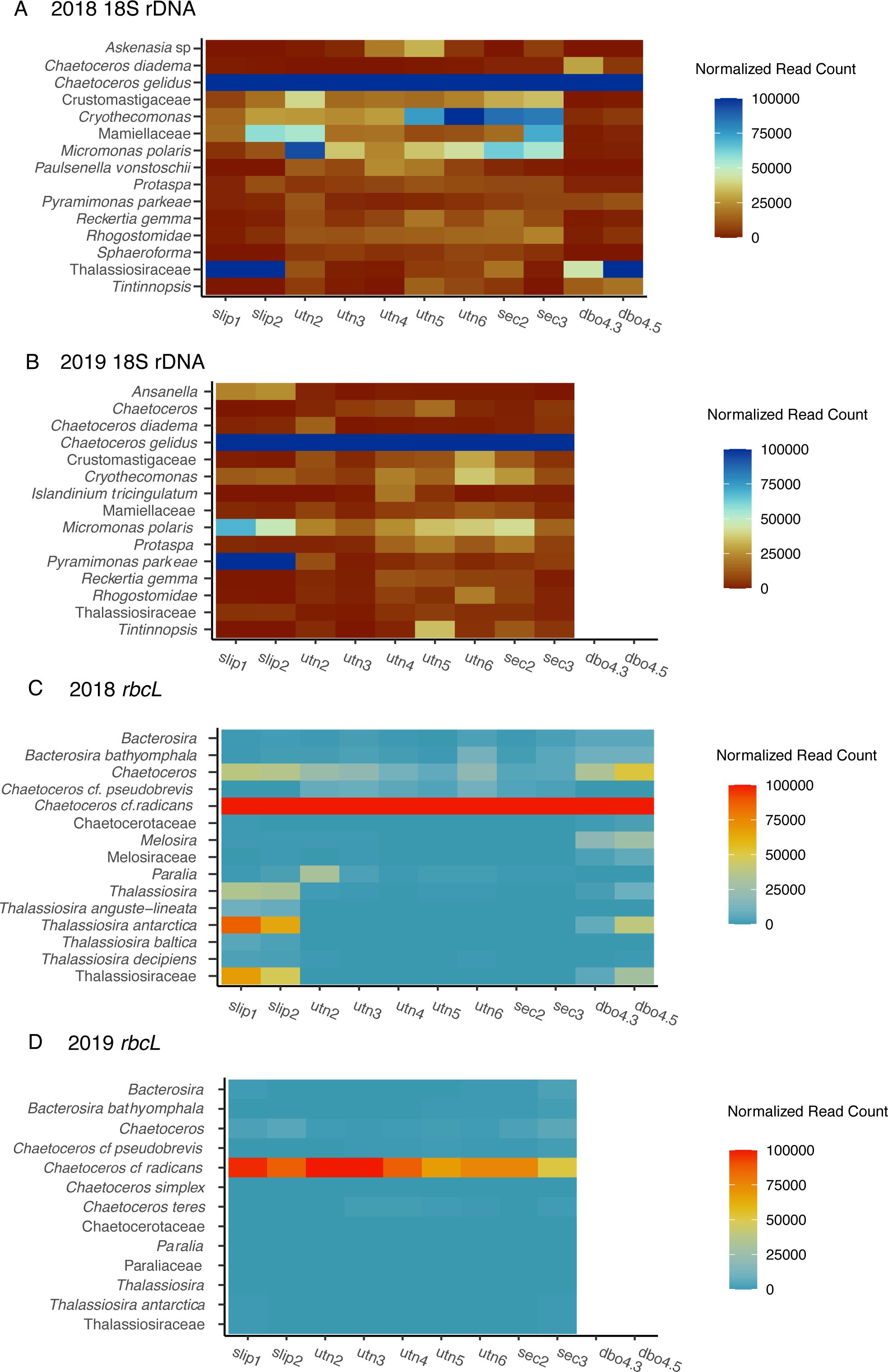
Figure 3. Heatmaps illustrating normalized read counts of operational taxonomic units (OTUs) across sampling stations for metabarcoding gene markers for top 15 taxa. Panels represent: (A) 2018 18S rDNA, (B) 2019 18S rDNA, (C) 2018 rbcL, and (D) 2019 rbcL (only 13 taxa had normalized read counts greater than zero). The y-axis lists OTUs grouped by shared taxonomic assignments, while the color gradient reflects normalized read counts by taxon at each station. Sampling stations are arranged geographically from south to north.
Dinoflagellates comprised a substantial proportion of the normalized 18S mean read counts in both years. In 2018, dinoflagellates ranged from 6% at DBO4.3 to 43% at UTN3 (Figure 2). In 2019, the range of proportional normalized read counts ranged from 3% at UTN3 to 67% at SEC3. We also detected frequent normalized read counts of the dinoflagellate Alexandrium in the 18S results throughout both years and stations (Figure 4). In 2018, the highest normalized read counts for Alexandrium occurred at station UTN2, which is north of Bering Strait (6803 counts) and these dinoflagellates were not detected in clams collected from the St. Lawrence Island polynya region, in one station immediately north of the Bering Strait (UTN3), or in the southeast Chukchi Sea (SEC stations). Normalized read counts ranged from 1046 to 3486 at the other UTN stations in 2018. In 2019, normalized read counts of Alexandrium were identified across all stations (noting that DBO4 was not sampled in 2019). Normalized read counts were greatest at UTN3 (3139 counts) and UTN4 (3849 counts). Detection of Alexandrium reads in the southeastern Chukchi Sea notably increased in 2019 compared to 2018.
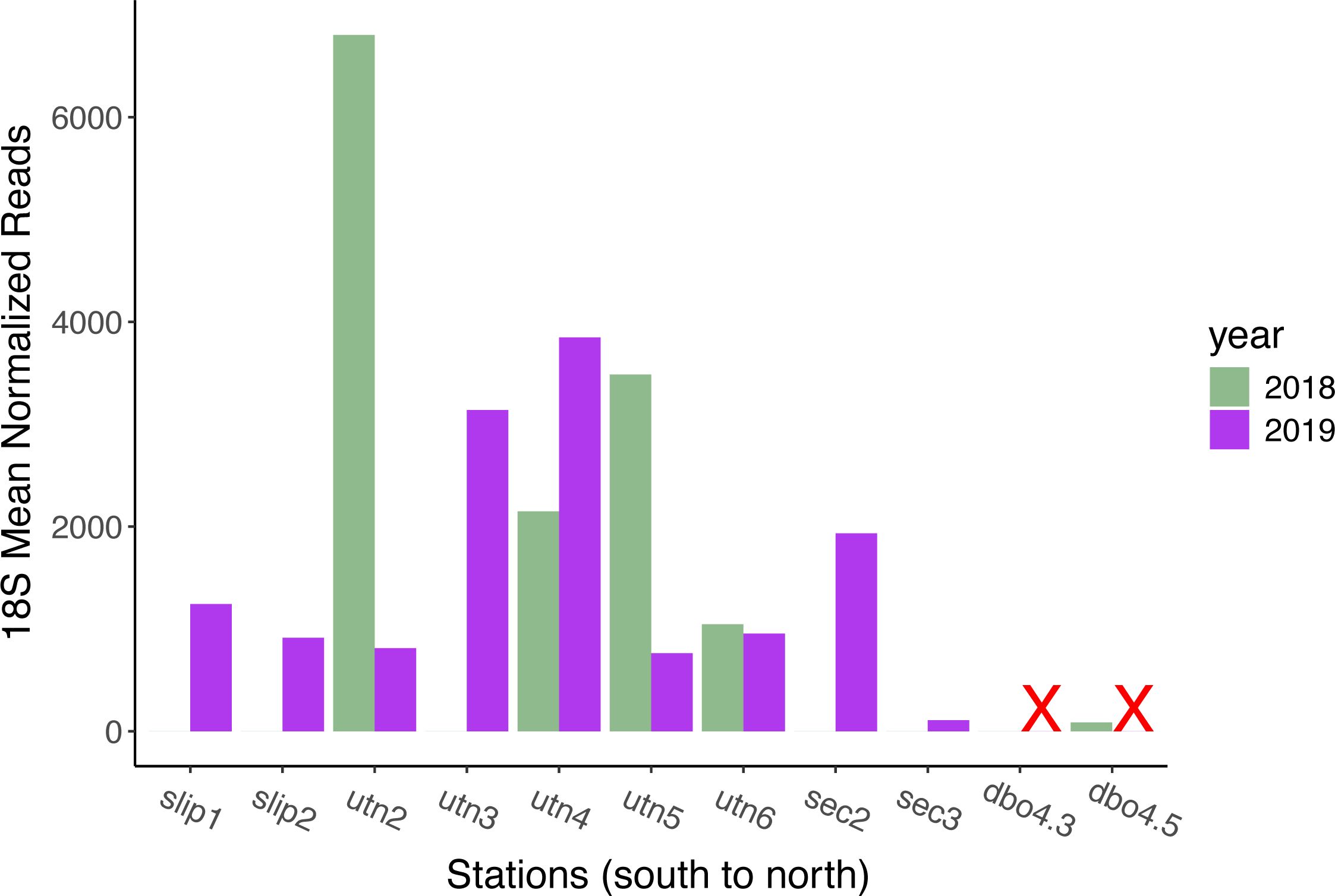
Figure 4. Comparison of mean normalized read counts for the harmful algae, Alexandrium, for 2018 (green) and 2019 (purple) based on 18S rDNA metabarcoding data. Sampling stations are arranged geographically from south to north. DBO4.3 and DBO4.5 were not sampled in 2019, indicated by an ‘X’.
To address our initial objective of identifying ice-associated and pelagic diatoms, and to explore the full extent of our dataset, we categorized the 18S and rbcL read counts for diatoms by family including: Chaetocerotaceae, Thalassiosiraceae, Melosiraceae, Paraliaceae, and two general categories, Other (centrics), and Other (pennates; Figure 5). These families were largely represented by the following genera: Chaetoceros, Thalassiosira, Melosira, and Paralia. The ‘other–centric’ diatom category included Biddulphia, Leptocylindrus, Minutocellus, Rhizosolenia, and Stephanopyxis. The ‘other–pennate’ diatom category included the genera Fragilariopsis and Navicula. Approximately 89% of the OTUs within the Chaetocerotaceae family were assigned to species level, and Chaetoceros is the probable genus represented. Similarly, ~80% of the Thalassiosiraceae OTU assignments were to genus or species-level for Thalassiosira and are the probable genus represented.
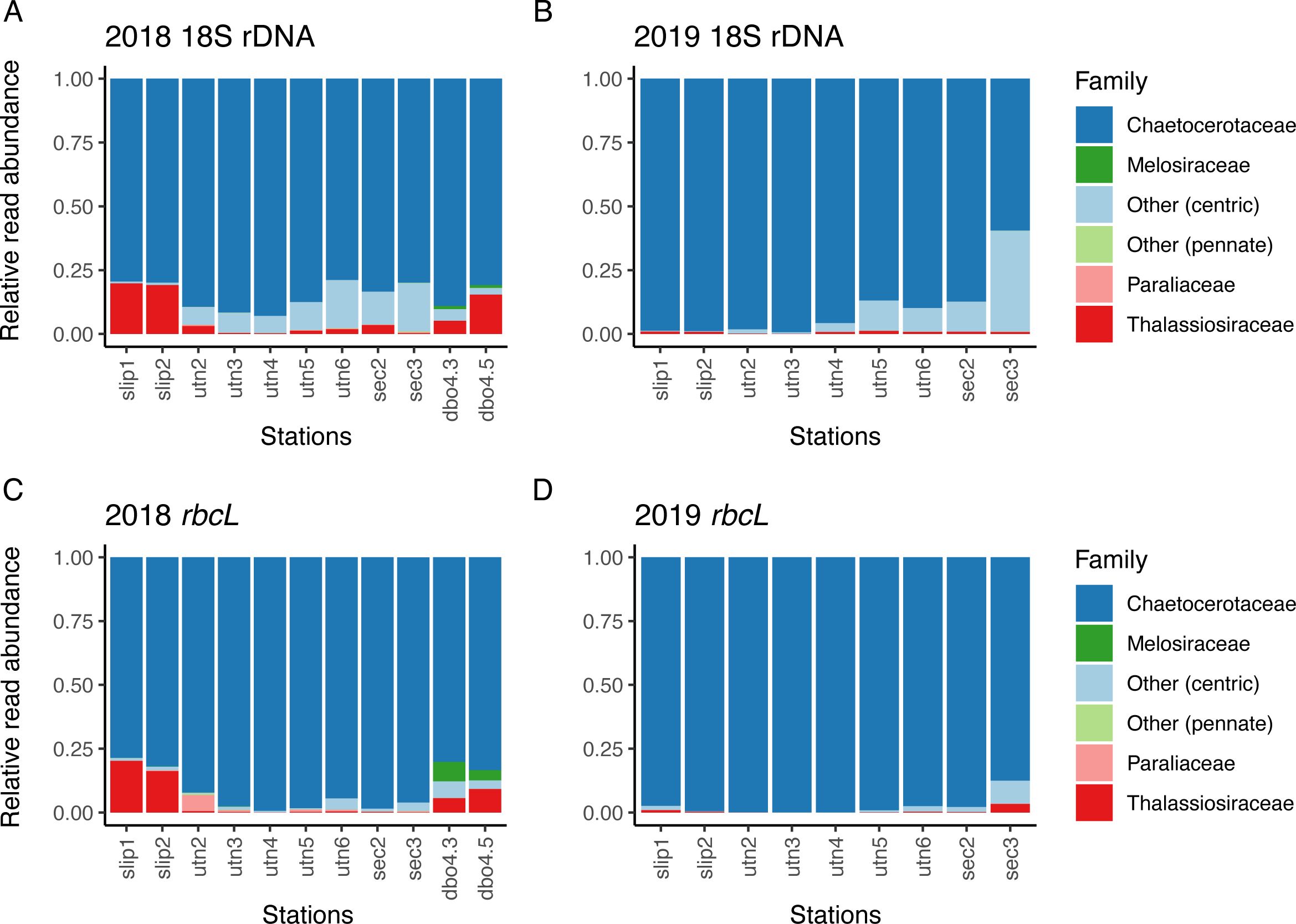
Figure 5. Proportions of normalized read abundance assigned to diatom families within the summer diets of Macoma calcarea for the 18S rDNA dataset in (A) 2018 and (B) 2019 and the rbcL marker datasets in (C) 2018 and (D) 2019. Sampling stations are arranged geographically from south to north. DBO4.3 and DBO 4.5 were not sampled in 2019.
In 2018 and 2019 across both 18S and rbcL markers, Chaetoceros sp. dominated clam diets, with dietary proportions greater than 75% in all stations in 2018 and greater than 80% at all stations in 2019, with the exception of SEC3 (Figure 3). Normalized read counts at SEC3 were composed of ~30% other (centrics), which could only be assigned to the order Thalassiosirales (Figure 5). Otherwise, the genera Thalassiosira was the second-most frequently occurring in read count abundance at most other stations. For the normalized read counts assigned to species, the 18S results indicated a dominance of Chaetoceros gelidus across all stations, while Chaetoceros radicans was the most abundant species assignment for the rbcL marker (Figure 3). Melosira sp., a centric, ice-associated species, increased in relative frequency (>1%) at the northernmost stations, DBO4.3 and DBO4.5 in 2018 (Figures 5A, C). The pennate ice-associated diatom, Nitszchia frigida, was not detected and nor was the pelagic diatom, Cylindrotheca closterium, major components of the diatom flux in 2018 and 2019 (Lalande et al., 2021; Fukai et al., 2020).
4 Discussion
4.1 Primary food sources
In this study, we explored the primary food sources for the clam, M. calcarea, in the Pacific Arctic during extreme low sea ice years, using DNA metabarcoding techniques to provide increased understanding of bivalve diets at a higher taxonomic resolution than possible through gut content and microscopy counts (Reid and Reid, 1969; North et al., 2014). Our findings are consistent with a diet consisting primarily of pelagic diatoms, specifically Chaetoceros (C. gelidus or C. radicans, Figure 3) as the dominant food source for these clams in July of 2018 and 2019. C. gelidus occasionally dominates the Arctic spring bloom (von Quillfeldt, 2000; Sergeeva et al., 2010). In the summer of 2018, C. gelidus was reported to be a major constituent of the diatom community in the Alaska Coastal Water (ACW) mass south of St. Lawrence Island (Fukai et al., 2020), which corroborates the high proportional reads of this species in our samples. While these two markers suggest different species, it is more likely that these markers represent the same species and exemplify how these two gene markers can produce differing results (Dermastia et al., 2023).
Linkages between the pelagic and benthic components of the ecosystem can further be interpreted using diatom fluxes from sediment traps located within the DBO regions 2 (closest to UTN2) and 3 (nearest SEC2), as reported by Lalande et al. (2021). The flux data from these sediment traps are not fully consistent with our data reported here but generally align with the proportion of normalized read counts for Chaetoceros and Thalassiosira that we observed. For example, in Lalande et al. (2021) the particle flux in 2018 following the spring bloom at DBO2 had a large proportion of Chaetoceros diatoms followed by Thalassiosira. In 2019, the spring bloom had a large proportion of Thalassiosira and Fragilariopsis. At DBO 3, large fluxes of Thalassiosira and Fragilariopsis also occurred from April through June in 2018 and 2019, but less so for Chaetoceros sp. Notably, Fragilariopsis, a pennate diatom, appeared minimally in the clam diets in both years, perhaps indicative of a preference for centric diatoms (Ward et al., 2004). A higher proportion of centric diatoms had also been observed for Macoma sp. in the northern Bering Sea based on gut content and microscopy analysis (North et al., 2014). The lack of Cylindrotheca closterium in our samples was notable, given the large fluxes of this pennate diatom in nearby sediment traps at DBO 2 and 3 (Lalande et al., 2021). Comparisons of 18S, rbcL, and microscopy methods in other Arctic studies also reported a lack of expected reads for C. closterium based on microscopy results (Dermastia et al., 2023). They concluded that these diatoms were previously misidentified by microscopy and were probably Pseudo-nitzschia sp., which look similar and were well-represented in their read assignments (Dermastia et al., 2023). Despite these specific inconsistencies, linkages between the diet of the clams and the sediment trap data underscores the importance of pelagic, centric diatoms as a significant resource for the benthic food web in this region and demonstrates the role of benthic-pelagic coupling.
Diet studies using biogeochemical approaches such as fatty acid biomarkers, stable isotope analysis, and (or) direct examination of gut contents of bivalves in the Pacific Arctic region previously indicated dietary plasticity and a strong reliance on pelagic phytoplankton (e.g., Budge et al., 2007; Iken et al., 2010; Oxtoby et al., 2016; Kędra et al., 2019; North et al., 2014). DNA metabarcoding is a promising addition to complement these methodologies, because it potentially facilitates study of a greater diversity and number of species that are not practical for other analytical techniques. Shifts in biodiversity are poorly understood in the Arctic (Feng et al., 2021), which underscores the complexity of Arctic marine food webs and the importance of adopting a multifaceted approach to understand the ecological dynamics of the changing Arctic environment. DNA metabarcoding therefore offers a comprehensive and nuanced perspective on the complex interactions within marine food webs, enhancing our understanding of ecosystem dynamics and biodiversity. This approach not only enables tracing of the intricate trophic links that sustain marine life but also provides the knowledge to predict and respond to the ecological shifts anticipated in a rapidly changing Arctic (Geraldi et al., 2024).
DNA metabarcoding is not without its limitations (Coissac et al., 2012). One significant constraint for our study was the differentiation between DNA derived from ingested food and DNA from environmental contamination or from the digestion of prey, which can complicate interpretations of diet (Garrison et al., 2022). More specifically, samples could potentially be contaminated with phytoplankton assemblages not actually consumed, but rather introduced into the interior clam tissues when the siphons retract into the shells and/or during digging with its foot (Reid and Reid, 1969). Bacterial breakdown of large diatoms may be necessary for incorporation into bivalve diets in this region (North et al., 2014). Another limitation in characterizing diet is the incomplete representation of potential prey items in reference databases, which can lead to the underestimation or misidentification of components within a diet (Dermastia et al., 2023). This gap in reference data particularly affects rare or less-studied species, whose DNA sequences might not yet be cataloged. Furthermore, DNA metabarcoding is sensitive to the relative abundance of DNA, meaning that it may overemphasize the presence of species that shed more DNA or whose DNA degrades more slowly, potentially skewing the perceived dietary composition (Murray et al., 2011; Garrison et al., 2023).
4.2 Phytoplankton and ice algal assemblages in extreme sea ice years
The winter of 2018 was remarkable for the lack of sea ice formation in the northern Bering Sea, which corresponded to more typical winter conditions of the southeastern Bering Sea (Stabeno and Bell, 2019; Thoman et al., 2020). As a result of a marine heat wave, sea ice was thinner and formed later in the Chukchi Sea in 2017, leading to the late arrival of sea ice in the Bering Sea in November (Stabeno and Bell, 2019). Warm, southerly winds in February blew the sea ice present in the Bering Strait region out by mid-February 2018 (Kikuchi et al., 2020). There was no sea ice present south of St. Lawrence Island in the spring of 2018, an event not identified in observed or reconstructed conditions since 1850 (Walsh et al., 2017). As a result, no ice-associated spring bloom occurred, but a phytoplankton bloom occurred ~1 month later than usual and only once the surface waters thermally stratified, in contrast to the salinity stratified surface waters typically resulting from ice melt (Duffy-Anderson et al., 2019; Kikcuhi et al., 2020). Accordingly, the summer phytoplankton community in the northern Bering Sea had transitioned from cold-water species in 2017 to more cosmopolitan species in 2018, including Chaetoceros sp (Fukai et al., 2020). The marine heat wave continued into 2019, the second lowest winter sea ice in the northern Bering Sea on record (Stabeno and Bell, 2019). In these years, the sea ice retreat in mid-winter provided unfavorable conditions for a spring ice-associated bloom due to lack of sunlight and atypical winter conditions, leading to another unusual spring phytoplankton bloom (Kikuchi et al., 2020). Therefore, the clams in our study (with the exception of DBO4.3 and DBO4.5 in 2018) were sampled approximately 5 months after sea ice retreat, from nutrient depleted water in the Bering Sea, with more cosmopolitan species of diatoms (Fukai et al., 2020).
The appearance of Melosira sp. from the DBO 4 region in 2018 suggests a nuanced aspect of food availability relative to sea ice persistence and timing. While we were not able to assign this OTU to species, Melosira arctica is the most probable species to be present based on taxonomic studies (Poulin et al., 2011, 2014). M. arctica are a centric, ice-associated colonial diatom typically associated with sub-ice habitat that are released as the ice retreats in July in high latitude locations (Booth and Horner, 1997; Lalande et al., 2019). In 2018, the sea ice had retreated from the Chukchi Sea (DBO4 region) in mid-June, approximately 3 weeks prior to sampling as opposed to the 5+ months in the northern Bering Sea (Perovich et al., 2018). Therefore, the presence of Melosira suggests that there was a recent ice algae bloom in the Chukchi Sea. While not represented in the 18S or rbcL marker reads, the presence of ice-associated species such as Nitzschia frigida have been observed in greater abundances within particle fluxes in the Chukchi Sea, highlighting the spatial variability in food sources (Koch et al., 2020b; Lalande et al., 2021). It is probable that ice algae and ice-associated blooms are an important component in the diets of Macoma clams under specific sea ice conditions and at certain times of the year.
Our findings were also consistent with lipid biomarkers analyzed from Macoma clams collected from the same grab samples as the 2018 sampling effort reported here (Koch et al., 2020a). The biomarker results also indicated a minimal or absent ice algal signature in the SLIP region and in the Bering Strait/southern Chukchi Sea, which was attributed in that study to clams suspension feeding rather than surface deposit feeding in the summer. Sub-surface deposit feeders in the northern Bering Sea have access to benthic reserves of ice-associated organic matter (North et al., 2014; Pirtle-Levy et al., 2009). With the early pulse of food from ice-associated blooms in the spring, benthic activity is stimulated, and bioturbation increases, leading to rapid utilization and burial of ice-associated organic matter (Grebmeier et al., 2006; Clough et al., 1997; McMahon et al., 2006). With this species capable of engaging in both suspension and surface deposit feeding, those results were likely reflective of the prevailing conditions in the water column following the most recent bloom, whereas subsurface deposit feeding bivalves in the sediments can access ice-associated organic carbon buried in the sediments long after the ice algal bloom has concluded (Koch et al., 2020a). Given the timing of sampling in 2018 relative to sea ice retreat and the overall lack of ice-associated production, ice-associated diatoms are unlikely to be a significant component of the summer diet of Macoma clams in the northern Bering Sea.
The contributions of the prasinophytes, Micromonas and Pyramimonas, to the diet of these clams were also potentially indicators of nutrient availability and bloom progression. Micromonas polaris had abundant read counts in both 2018 and 2019 throughout the region and Pyramimonas parkaeae had elevated normalized read counts in the St. Lawrence Island polynya region in 2019 (Figure 3). These species are indicative of shifting phytoplankton assemblage structures in a changing Arctic, where prasinophytes thrive under very low nitrogen (N) conditions while diatoms require high N and Si (Ardyna and Arrigo, 2020). Accordingly, in 2018, the spring bloom was delayed by ~1 month (Stabeno and Bell, 2019) and nutrients were depleted in the upper mixed layer resulting in non-diatom phytoplankton blooms (Fukai et al., 2020).
The temporal mismatch between our DNA metabarcoding of benthic consumers and the window of ice-associated production highlights the potential for additional work. Conducting metabarcoding studies throughout various seasonal transitions—from ice breakup through to resuspension and the autumn blooms—could provide a more comprehensive understanding of the variation in clam diets and resource utilization across different bloom phases. A similar approach examining the diet of the arctic copepod, Calanus glacialis, revealed similar complexities where higher proportions of ice-associated diatoms were observed at certain times of the year but also found higher abundances of the pelagic diatom, Thalassiosira, when these organisms were observed foraging directly under the sea ice (Cleary et al., 2017). There are gaps remaining in our understanding of ice-associated diatom community composition, species that span these niches (sympagic diatoms seeding phytoplankton blooms, and vice versa), and increasingly the role of under ice algal blooms (Ardyna et al., 2020).
4.3 Harmful algae in clam diets
Over the last few years, the Pacific Arctic has had increasing attention focused on harmful algal blooms (HABs), which may pose an escalating threat to coastal communities and marine ecosystems (Anderson et al., 2022; Lefebvre et al., 2022). Alexandrium catenella stands out for its toxicity and potential to disrupt marine food webs, particularly within bivalves, seabirds, and marine mammals, as well as impact human health (Lefebvre et al., 2022). Our study revealed the presence of Alexandrium sp. across all stations sampled during our study period, with the highest detection observed in 2019 at station UTN2. Like other organisms of interest in this study, we were only able to identify this OTU to genus, but A. catenella is widely distributed and abundant in this region as confirmed by other studies (Anderson et al., 2022; Brosnahan et al., 2020; Lefebvre et al., 2022). Additionally, over half of the 30 known species of Alexandrium produce paralytic shellfish toxins (Anderson et al., 2012). The widespread detection of Alexandrium in clams aligns with recent findings from Anderson et al. (2022) and Lefebvre et al. (2022), who reported large dinoflagellate cyst beds in the sediments at the same stations we sampled. Furthermore, these studies noted the presence of saxitoxins—a potent neurotoxin produced by Alexandrium—in clams and walruses in 2019 within the Bering Strait and southeast Chukchi Sea regions, areas that correspond to where we detected signatures of Alexandrium.
In addition to Alexandrium, we also detected the presence of other potentially harmful algal species, such as the raphidophyte Chattonella subsalsa (Hallegraeff and Hara, 2003), albeit less frequently than A. catenella. While these taxonomic assignments were rarer, they nonetheless signify the potential for early detection and monitoring of future HAB events in Alaska waters. The presence of these species, along with A. catenella, highlights the potential threat that HABs present to marine ecosystems and human health in the Arctic (Anderson et al., 2022; Pućko et al., 2023). Our findings contribute to the growing body of evidence indicating that monitoring and research into emerging HAB dynamics in a changing Pacific Arctic should be prioritized alongside efforts to manage their impacts on marine ecosystems and the health of coastal communities in Alaska (Anderson et al., 2022). The ability to use environmental DNA (eDNA) metabarcoding to rapidly identify harmful algal species has profound implications for public health, particularly in the context of monitoring and managing risks associated with paralytic shellfish toxins (Jacobs-Palmer et al., 2021). The detection of harmful species through this method not only highlights the direct links between marine biodiversity and human health but also the potential of eDNA metabarcoding as an accessible tool for supporting community-based monitoring programs (Larson et al., 2020).
5 Conclusion
Our study employed DNA metabarcoding techniques on Macoma calcarea, a common bivalve in the Pacific Arctic and important prey item for benthic-feeding marine mammals and waterfowl, to identify summer diet during two record low sea ice years. We conclude that both 18S and rbcL markers offered valuable and unique insights into the diet (e.g., the presence of harmful algae versus the specificity of diatom species assignments). Despite some of the current limitations, DNA metabarcoding remains a promising approach to advance ecological research in the Arctic, offering insights into complex dietary networks that were previously difficult to achieve. However, it is particularly important to carefully consider these limitations and address them in the study design and interpretation of results (de Sousa et al., 2019).
Based on recent projections, and as atmospheric warming conditions continue, the Arctic region may experience its first ice-free summer before 2030 (Heuzé and Jahn, 2024). The urgency to document changes with appropriate and emerging techniques is vital. These results specifically emphasize the importance of diatoms in the Arctic marine food web and prompt critical questions about the future of benthic-dominated ecosystems. While smaller diatom species and flagellates are predicted to become more prevalent in response to changing conditions in the Arctic (Ardyna and Arrigo, 2020), the implications of this community shift to the benthos remain uncertain. Additional research using DNA metabarcoding with multiple gene markers across various marine species and seasons holds promise as an informative approach to track the anticipated restructuring of the Pacific Arctic ecosystem.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Ethics statement
Ethical approval was not required for the study involving animals in accordance with the local legislation and institutional requirements because the study focused on invertebrates.
Author contributions
CK: Data curation, Formal analysis, Investigation, Methodology, Project administration, Software, Visualization, Writing – original draft, Writing – review & editing. SS: Conceptualization, Data curation, Funding acquisition, Investigation, Methodology, Project administration, Validation, Writing – original draft, Writing – review & editing. LC: Conceptualization, Funding acquisition, Investigation, Methodology, Resources, Supervision, Writing – original draft, Writing – review & editing. JG: Conceptualization, Funding acquisition, Investigation, Methodology, Resources, Supervision, Writing – original draft, Writing – review & editing. AR-B: Data curation, Formal analysis, Methodology, Validation, Writing – review & editing. RC: Data curation, Formal Analysis, Methodology, Validation, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. Funding was provided by the U.S. Geological Survey Changing Arctic Ecosystems initiative through the Wildlife Program of the Ecosystems Mission Area (SS) to support genetic analysis. Sample collection was supported through the Distributed Biological Observatory program funded by the United States National Science Foundation award 1917469 (JG and LC).
Acknowledgments
We thank the two reviewers for comments that helped improved earlier versions of the manuscript. The authors thank the captain and crew aboard the Canadian Coast Guard Cutter Sir Wilfrid Laurier for providing the means to collect the samples. We also thank Chad Jay for his participation and guidance on the design and implementation of this study and for graphics support from Alynne Bayard. Any use of trade, firm, or product names is for descriptive purposes only and does not imply endorsement by the U.S. Government.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2025.1480327/full#supplementary-material
Supplementary Figure 1 | Scatterplots indicating the distribution of alignment metrics for the top high-scoring pair (HSP) of each operational taxonomic unit (OTU) of each locus, aligned to the nucleotide database as described in the text. Bit score is a normalized measure of alignment score that is independent of the database searched (see BLAST+ software documentation, https://www.ncbi.nlm.nih.gov/books/NBK279690/).
References
Amaral-Zettler L. A., McCliment E. A., Ducklow H. W., Huse S. M. (2009). A method for studying protistan diversity using massively parallel sequencing of V9 hypervariable regions of small-subunit ribosomal RNA genes. PLOS ONE 4 (7), e6372. doi: 10.1371/journal.pone.0006372
Anderson D. M., Alpermann T. J., Cembella A. D., Collos Y., Masseret E., Montresor M. (2012). The globally distributed genus Alexandrium: Multifaceted roles in marine ecosystems and impacts on human health. Harmful Algae 14, 10–35. doi: 10.1016/j.hal.2011.10.012
Anderson D. M., Fachon E., Hubbard K., Lefebvre K. A., Lin P., Pickart R., et al. (2022). Harmful algal blooms in the Alaskan Arctic: An emerging threat as the ocean warms. Oceanography 35, 130–139. doi: 10.5670/oceanog.2022.121
Ardyna M., Arrigo K. R. (2020). Phytoplankton dynamics in a changing Arctic Ocean. Nat. Climate Change 10, 892–903. doi: 10.1038/s41558-020-0905-y
Ardyna M., Mundy C. J., Mayot N., Matthes L. C., Oziel L., Horvat C., et al. (2020). Under-ice phytoplankton blooms: Shedding light on the “invisible” part of Arctic primary production. Front. Mar. Sci. 7. doi: 10.3389/fmars.2020.608032
Arrigo K. R., Perovich D. K., Pickart R. S., Brown Z. W., van Dijken G. L., et al. (2014). Phytoplankton blooms beneath the sea ice in the Chukchi sea. Deep Sea Research Part II: Topical Studies in Oceanography 105, 1–16. doi: 10.1016/j.dsr2.2014.03.018
Beatty W. S., Jay C. V., Fischbach A. S., Grebmeier J. M., Taylor R. L., Blanchard A. L., et al. (2016). Space use of a dominant Arctic vertebrate: Effects of prey, sea ice, and land on Pacific walrus resource selection. Biol. Conserv. 203, 25–32. doi: 10.1016/j.biocon.2016.08.035
Booth B. C., Horner R. A. (1997). Microalgae on the arctic ocean section 1994: species abundance and biomass. Deep Sea Res. Part II: Topical Stud. Oceanography. 44, 1607–1622. doi: 10.1016/S0967-0645(97)00057-X
Brosnahan M. L., Fischer A. D., Lopez C. B., Moore S. K., Anderson D. M. (2020). Cyst-forming dinoflagellates in a warming climate. Harmful Algae 91, 101728. doi: 10.1016/j.hal.2019.101728
Budge S. M., Springer A. M., Iverson S. J., Sheffield G. (2007). Fatty acid biomarkers reveal niche separation in an Arctic benthic food web. Mar. Ecol. Prog. Ser. 336, 305–309. doi: 10.3354/meps336305
Bushnell B. (2025). BBMap - BBtools. Available online at: http://sourceforge.net/projects/bbmap/ (Accessed 06 Jan 2025).
Camacho C., Coulouris G., Avagyan V., Ma N., Papadopoulos J., Bealer K., et al. (2009). BLAST+: architecture and applications. BMC Bioinf. 10, 421. doi: 10.1186/1471-2105-10-421
Cleary A. C., Søreide J. E., Freese D., Niehoff B., Gabrielsen T. M. (2017). Feeding by Calanus glacialis in a high Arctic fjord: Potential seasonal importance of alternative prey. ICES J. Mar. Sci. 74, 1937–1946. doi: 10.1093/icesjms/fsx106
Clough L. M., Ambrose W. G., Kirk Cochran J., Barnes C., Renaud P. E., Aller R. C. (1997). Infaunal density, biomass and bioturbation in the sediments of the Arctic Ocean. Deep Sea Res. Part II: Topical Stud. Oceanography. 44, 1683–1704. doi: 10.1016/S0967-0645(97)00052-0
Coissac E., Riaz T., Puillandre N. (2012). Bioinformatic challenges for DNA metabarcoding of plants and animals. Mol. Ecol. 21, 1834–1847. doi: 10.1111/j.1365-294X.2012.05550.x
Deagle B. E., Thomas A. C., McInnes J. C., Clarke L. J., Vesterinen E. J., Clare E. L., et al. (2019). Counting with DNA in metabarcoding studies: How should we convert sequence reads to dietary data? Mol. Ecol. 28, 391–406. doi: 10.1111/mec.14734
Dermastia T., Vascotto I., Francé J., Stanković D., Mozetič P. (2023). Evaluation of the rbcL marker for metabarcoding of marine diatoms and inference of population structure of selected genera. Front. Microbiol. 14. doi: 10.3389/fmicb.2023.1071379
de Sousa L. L., Silva S. M., Xavier R. (2019). DNA metabarcoding in diet studies: Unveiling ecological aspects in aquatic and terrestrial ecosystems. Environ. DNA. 1, 199–214. doi: 10.1002/edn3.27
Duffy-Anderson J. T., Stabeno P., Andrews A. G. III, Cieciel K., Deary A., Farley E., et al. (2019). Responses of the northern Bering Sea and southeastern Bering Sea pelagic ecosystems following record-breaking low winter sea ice. Geophysical Res. Letters. 46, 9833–9842. doi: 10.1029/2019GL083396
Fay F. H. (1982). Ecology and biology of the Pacific walrus. Odobenus rosmarus divergens Illiger North Am. Fauna 74, 1–279. doi: 10.3996/nafa.74.0001
Feng Z., Ji R., Ashjian C., Zhang J., Campbell R., Grebmeier J. M. (2021). Benthic hotspots on the northern Bering and Chukchi continental shelf: Spatial variability in production regimes and environmental drivers. Prog. Oceanography 191, 102497. doi: 10.1016/j.pocean.2020.102497
Fetterer F., Knowles K., Meier W. N., Savoie M., Windnagel A. K. (2017). “Sea ice index (G02135, version 3),” in Monthly sea ice extent (National Snow and Ice Data Center, Boulder, Colorado, USA). Available at: https://noaadata.apps.nsidc.org/NOAA/G02135/north/monthly/shapefiles/shp_extent/ (Accessed December 12, 2024).
Frey K. E., Comiso J. C., Cooper L. W., Grebmeier J. M., Stock L. V. (2018). Arctic Ocean primary productivity: The response of marine algae to climate warming and sea ice decline [in Arctic Report Card 2018]. Available online at: https://www.arctic.noaa.gov/Report-Card (Accessed December 10, 2024).
Fukai Y., Abe Y., Matsuno K., Yamaguchi A. (2020). Spatial changes in the summer diatom community of the northern Bering Sea in 2017 and 2018. Deep Sea Res. Part II: Topical Stud. Oceanography 181–182, 104903. doi: 10.1016/j.dsr2.2020.104903
Garrison J. A., Motwani N. H., Broman E., Nascimento F. J. A. (2022). Molecular diet analysis enables detection of diatom and cyanobacteria DNA in the gut of Macoma balthica. PloS One 17, e0278070. doi: 10.1371/journal.pone.0278070
Geraldi N. R., Krause-Jensen D., Ørberg S. B., Frühe L., Sejr M. K., Hansen J. L. S., et al. (2024). Environmental drivers of Arctic communities based on metabarcoding of marine sediment eDNA. Proc. R. Soc. B: Biol. Sci. 291, 20231614. doi: 10.1098/rspb.2023.1614
Gerasimova A. V., Filippova N. A., Lisitsyna K. N., Filippov A. A., Nikishina D. V., Maximovich N. V. (2019). Distribution and growth of bivalve molluscs Serripes groenlandicus (Mohr) and Macoma calcarea (Gmelin) in the Pechora Sea. Polar Biol. 42, 1685–1702. doi: 10.1007/s00300-019-02550-z
Gmelin J. F. (1791). Systema naturae per regna tria naturae. 13 Vol. 1. Ed. Beer G. E. (Lipsiae [Leipzig]: Tome), 3021–3910.
Goethel C. L., Grebmeier J. M., Cooper L. W. (2019). Changes in abundance and biomass of the bivalve Macoma calcarea in the northern Bering Sea and the southeastern Chukchi Sea from 1998 to 2014, tracked through dynamic factor analysis models. Deep Sea Res. Part II: Topical Stud. Oceanography 162, 127–136. doi: 10.1016/j.dsr2.2018.10.007
Grebmeier J. M. (2012). Shifting patterns of life in the Pacific Arctic and Sub-Arctic seas. Annu. Rev. Mar. Sci. 4, 63–78. doi: 10.1146/annurev-marine-120710-100926
Grebmeier J. M., Frey K. E., Cooper L. W., Kędra M. (2018). Trends in benthic macrofaunal populations, seasonal sea ice persistence, and bottom water temperatures in the Bering Strait region. Oceanography 31, 136–151. doi: 10.5670/oceanog.2018.224
Grebmeier J. M., McRoy C. P., Feder H. M. (1988). Pelagic-benthic coupling on the shelf of the northern Bering and Chukchi seas. I. Food supply source and benthic biomass. Mar. Ecol. Prog. Ser. 48, 57–67. Available at: https://www.int-res.com/articles/meps/51/m051p253.pdf (Accessed June 1, 2024).
Grebmeier J. M., Overland J. E., Moore S. E., Farley E. V., Carmack E. C., Cooper L. W., et al. (2006). A major ecosystem shift in the northern Bering Sea. Science 311, 1461–1464. doi: 10.1126/science.1121365
Hallegraeff G., Hara Y. (2003). Taxonomy of harmful marine raphidophytes. Available online at: https://hdl.handle.net/102.100.100/532781 (Accessed 7 January 2025).
Heuzé C., Jahn A. (2024). The first ice-free day in the Arctic Ocean could occur before 2030. Nat. Commun. 15, 10101. doi: 10.1038/s41467-024-54508-3
Huntington H. P., Danielson S. L., Wiese F. K., Baker M., Boveng P., Citta J. J., et al. (2020). Evidence suggests potential transformation of the Pacific Arctic ecosystem is underway. Nat. Climate Change 10, 342–348. doi: 10.1038/s41558-020-0695-2
Iken K., Bluhm B., Dunton K. (2010). Benthic food-web structure under differing water mass properties in the southern Chukchi Sea. Deep Sea Res. Part II: Topical Stud. Oceanography 57, 71–85. doi: 10.1016/j.dsr2.2009.08.007
Jacobs-Palmer E., Gallego R., Cribari K., Keller A. G., Kelly R. P. (2021). Environmental DNA metabarcoding for simultaneous monitoring and ecological assessment of many harmful algae. Front. Ecol. Evol. 9. doi: 10.3389/fevo.2021.612107
Jay C. V., Grebmeier J. M., Fischbach A. S., McDonald T. L., Cooper L. W., Hornsby F. (2014). Pacific Walrus (Odobenus rosmarus divergens) Resource selection in the Northern Bering Sea. PLOS ONE 9, (4), e93035. doi: 10.1371/journal.pone.0093035
Kędra M., Cooper L. W., Zhang M., Biasatti D., Grebmeier J. M. (2019). Benthic trophic sensitivity to on-going changes in Pacific Arctic seasonal sea ice cover – Insights from the nitrogen isotopic composition of amino acids. Deep Sea Res. Part II: Topical Stud. Oceanography 162, 137–151. doi: 10.1016/j.dsr2.2019.01.002
Kikuchi G., Abe H., Hirawake T., Sampei M. (2020). Distinctive spring phytoplankton bloom in the Bering Strait in 2018: A year of historically minimum sea ice extent. Deep Sea Res. Part II: Topical Stud. Oceanography 181, 104905. doi: 10.1016/j.dsr2.2020.104905
Koch C. W., Cooper L. W., Grebmeier J. M., Frey K., Brown T. A. (2020a). Ice algae resource utilization by benthic macro- and megafaunal communities on the Pacific Arctic shelf determined through lipid biomarker analysis. Mar. Ecol. Prog. Ser. 651, 23–43. doi: 10.3354/meps13476
Koch C. W., Cooper L. W., Lalande C., Brown T. A., Frey K. E., Grebmeier J. M. (2020b). Seasonal and latitudinal variations in sea ice algae deposition in the northern Bering and Chukchi seas determined by algal biomarkers. PloS One 15, e0231178. doi: 10.1371/journal.pone.0231178
Koch C. W., Sonsthagen S., Cooper L. W., Cornman R. S., Grebmeier J. M., Riddle-Berntsen A. (2025). Operational Taxonomic Units (OTUs) for 18S ribosomal RNA (rDNA) and ribulose bisphosphate carboxylase (rbcL) gene markers in Macoma calcarea diets in the Bering and Chukchi Seas, (2018-2019). Arctic Data Center. doi. doi: 10.18739/A2R20RZ48
Lalande C., Grebmeier J. M., McDonnell A. M. P., Hopcroft R. R., ODaly S., Danielson S. L. (2021). Impact of a warm anomaly in the Pacific Arctic region derived from time-series export fluxes. PloS One 16, e0255837. doi: 10.1371/journal.pone.0255837
Lalande C., Nöthig E.-M., Fortier L. (2019). Algal export in the Arctic Ocean in times of global warming. Geophysical Res. Lett. 46, 5959–5967. doi: 10.1029/2019GL083167
Larson E. R., Graham B. M., Achury R., Coon J. J., Daniels M. K., Gambrell D. K., et al. (2020). From eDNA to citizen science: emerging tools for the early detection of invasive species. Front. Ecol. Environ. 18, 194–202. doi: 10.1002/fee.2162
Lefebvre K. A., Fachon E., Bowers E. K., Kimmel D. G., Snyder J. A., Stimmelmayr R., et al. (2022). Paralytic shellfish toxins in Alaskan Arctic food webs during the anomalously warm ocean conditions of 2019 and estimated toxin doses to Pacific walruses and bowhead whales. Harmful Algae 114, 102205. doi: 10.1016/j.hal.2022.102205
Lisitsyna K. N., Gerasimova A. V., Filippova N. A. (2024). Macoma calcarea (Gmelin 1791), a poorly studied bivalve, in the Kara Sea: Distribution and growth variability. Mar. Ecol. 45, e12798. doi: 10.1111/maec.12798
Lovvorn J. R., Cooper L. W., Brooks M. L., De Ruyck C. C., Bump J. K., Grebmeier J. M. (2005). Organic matter pathways to zooplankton and benthos under pack ice in late winter and open water in late summer in the north-central Bering Sea. Mar. Ecol. Prog. Ser. 291, 135–150. doi: 10.3354/meps291135
Lovvorn J. R., Richman S. E., Grebmeier J. M., Cooper L. W. (2003). Diet and body condition of spectacled eiders wintering in pack ice of the Bering Sea. Polar Biol. 26, 259–267. doi: 10.1007/s00300-003-0477-0
Mahé F., Rognes T., Quince C., de Vargas C., Dunthorn M. (2015). Swarm v2: Highly-scalable and high-resolution amplicon clustering. PeerJ 3, e1420. doi: 10.7717/peerj.1420
McMahon K. W., Ambrose W. G. Jr., Johnson B. J., Sun M.-Y., Lopez G. R., Clough L. M., et al. (2006). Benthic community response to ice algae and phytoplankton in Ny Ålesund, Svalbard. Mar. Ecol. Prog. Ser. 310, 1–14. doi: 10.3354/meps310001
Moore S. E., Grebmeier J. M. (2018). The Distributed Biological Observatory: Linking physics to biology in the Pacific Arctic Region. Arctic 71, 1–7. doi: 10.14430/arctic4606
Moore S. E., Logerwell E., Eisner L., Farley E. V., Harwood L. A., Kuletz K., et al. (2014). "Marine fishes, birds and mammals as sentinels of ecosystem variability and reorganization in the Pacific Arctic Region." in The Pacific Arctic Region: Ecosystem Status and Trends in a Rapidly Changing Environment, eds. J. M. Grebmeier and W. Maslowski. (Springer Netherlands: Dordrecht), 337–392. doi: 10.1007/978-94-017-8863-2_11
Moore S. E., Stabeno P. J. (2015). Synthesis of Arctic research (SOAR) in marine ecosystems of the Pacific Arctic. Prog. Oceanography 136, 1–11. doi: 10.1016/j.pocean.2015.05.017
Murray D. C., Bunce M., Cannell B. L., Oliver R., Houston J., White N. E., et al. (2011). DNA-based faecal dietary analysis: A comparison of qPCR and high throughput sequencing approaches. PloS One 6, e25776. doi: 10.1371/journal.pone.0025776
Naumov A. D. (2006). “Clams of the White Sea: ecological and faunistic analysis,” in Explorations of fauna of the seas, vol. 59. (Russian Academy of Science, Saint-Petersburg (in Russian), ISBN: 5–98092-010-2.351.
Nielsen J. M., Sigler M. F., Eisner L. B., Watson J. T., Rogers L. A., Bell S. W., et al. (2024). Spring phytoplankton bloom phenology during recent climate warming on the Bering Sea shelf. Progress in Oceanography 220, 103176. doi: 10.1016/j.pocean.2023.103176
North C. A., Lovvorn J. R., Kolts J. M., Brooks M. L., Cooper L. W., Grebmeier J. M. (2014). Deposit-feeder diets in the Bering Sea: Potential effects of climatic loss of sea ice-related microalgal blooms. Ecol. Appl. 24, 1525–1542. doi: 10.1890/13-0486.1
Oxtoby L. E., Budge S. M., Iken K., OBrien D. M., Wooller M. J. (2016). Feeding ecologies of key bivalve and polychaete species in the Bering Sea as elucidated by fatty acid and compound-specific stable isotope analyses. Mar. Ecol. Prog. Ser. 557, 161–175. doi: 10.3354/meps11863
Perovich D., Meier W., Tschudi M., Farrell S., Hendricks S., Gerland S., et al. (2018). Sea ice [in Arctic Report Card 2018]. Available online at: https://www.arctic.noaa.gov/Report-Card (Accessed December 10, 2024).
Pirtle-Levy R., Grebmeier J. M., Cooper L. W., Larsen I. L. (2009). Chlorophyll a in Arctic sediments implies long persistence of algal pigments. Deep Sea Res. Part II: Topical Stud. Oceanography 56, 1326–1338. doi: 10.1016/j.dsr2.2008.10.022
Poulin M., Daugbjerg N., Gradinger R., Ilyash L., Ratkova T., von Quillfeldt C. (2011). The pan-Arctic biodiversity of marine pelagic and sea-ice unicellular eukaryotes: A first-attempt assessment. Mar. Biodiversity 41, 13–28. doi: 10.1007/s12526-010-0058-8
Poulin M., Underwood G. J. C., Michel C. (2014). Sub-ice colonial. Melosira arctica Arctic first-year ice Diatom Res. 29, 213–221. doi: 10.1080/0269249X.2013.877085
Pućko M., Rourke W., Hussherr R., Archambault P., Eert J., Majewski A. R., et al. (2023). Phycotoxins in bivalves from the western Canadian Arctic: The first evidence of toxigenicity. Harmful Algae 127, 102474. doi: 10.1016/j.hal.2023.102474
Reid R. G. B., Reid A. (1969). Feeding processes of members of the genus Macoma (Mollusca : Bivalvia). Can. J. Zoology 47, 649–657. doi: 10.1139/z69-111
Rognes T., Flouri T., Nichols B., Quince C., Mahé F. (2016). VSEARCH: A versatile open-source tool for metagenomics. PeerJ 4, e2584. doi: 10.7717/peerj.2584
Sergeeva V. M., Sukhanova I. N., Flint M. V., Pautova L. A., Grebmeier J. M., Cooper L. W. (2010). Phytoplankton community in the western arctic in july–august 2003. Oceanology 50, 184–197. doi: 10.1134/S0001437010020049
Sheffield G., Grebmeier J. M. (2009). Pacific walrus (Odobenus rosmarus divergens): Differential prey digestion and diet. Mar. Mammal Sci. 25, 761–777. doi: 10.1111/j.1748-7692.2009.00316.x
Siddon E. C., Zador S. G., Hunt G. L. Jr. (2020). Ecological responses to climate perturbations and minimal sea ice in the northern Bering Sea. Deep Sea Res. Part II: Topical Stud. Oceanography 181, 104914. doi: 10.1016/j.dsr2.2020.104914
Sonsthagen S. A., Jay C. V., Cornman R. S., Fischbach A. S., Grebmeier J. M., Talbot S. L. (2020). DNA metabarcoding of feces to infer summer diet of Pacific walruses. Mar. Mammal Sci. 36, 1196–1211. doi: 10.1111/mms.12717
Sonsthagen S. A., Riddle-Berntsen A. E., Cornman R. S., Koch C. W., Cooper L. W., Grebmeier J. M. (2024). Macoma clam prey identified by DNA metabarcoding 2028 and 2019, Alaska, U.S. Geological Survey data release. doi: 10.5066/P138OPD2
Stabeno P. J., Bell S. W. (2019). Extreme conditions in the Bering Sea, (2017–2018): Record-breaking low sea-ice extent. Geophysical Res. Lett. 46, 8952–8959. doi: 10.1029/2019GL083816
Stabeno P. J., Bell S. W., Bond N. A., Kimmel D. G., Mordy C. W., Sullivan M. E. (2019). Distributed Biological Observatory Region 1: Physics, chemistry and plankton in the northern Bering Sea. Distributed Biol. Observatory: A Change Detection Array Pacific Arctic Region 162, 8–21. doi: 10.1016/j.dsr2.2018.11.006
Stewart J. D., Joyce T. W., Durban J. W., Calambokidis J., Fauquier D., Fearnbach H., et al. (2023). Boom-bust cycles in gray whales associated with dynamic and changing Arctic conditions. Science 382, 207–211. doi: 10.1126/science.adi1847
Sun M.-Y., Clough L. M., Carroll M. L., Dai J., Ambrose W. G., Lopez G. R. (2009). Different responses of two common Arctic macrobenthic species (Macoma balthica and Monoporeia affinis) to phytoplankton and ice algae: Will climate change impacts be species specific? J. Exp. Mar. Biol. Ecol. 376, 110–121. doi: 10.1016/j.jembe.2009.06.018
Szymanski A., Gradinger R. (2016). The diversity, abundance, and fate of ice algae and phytoplankton in the Bering Sea. Polar Biol. 39, 309–325. doi: 10.1007/s00300-015-1783-z
Thoman R. L., Bhatt U. S., Bieniek P. A., Brettschneider B. R., Brubaker M., Danielson S. L., et al. (2020). The record low Bering Sea ice extent in 2018. Bull. Am. Meteorological Soc. 101, S53–S58. Available at: https://www.jstor.org/stable/27032744 (Accessed December 10, 2024).
Thompson L. R., Sanders J. G., McDonald D., Amir A., Ladau J., Locey K. J., et al. (2017). A communal catalogue reveals Earths multiscale microbial diversity. Nature 551, 457–463. doi: 10.1038/nature24621
Vasselon V., Rimet F., Tapolczai K., Bouchez A. (2017). Assessing ecological status with diatoms DNA metabarcoding: Scaling-up on a WFD monitoring network (Mayotte Island, France). Ecol. Indic. 82, 1–12. doi: 10.1016/j.ecolind.2017.06.024
Vestheim H., Jarman S. N. (2008). Blocking primers to enhance PCR amplification of rare sequences in mixed samples – A case study on prey DNA in Antarctic krill stomachs. Front. Zoology 5, 12. doi: 10.1186/1742-9994-5-12
von Quillfeldt C. H. (2000). Common diatom species in Arctic spring blooms: Their distribution and abundance. Botanica marina 43, 499–516. doi: 10.1515/BOT.2000.050
Waga H., Eicken H., Hirawake T., Fukamachi Y. (2021). Variability in spring phytoplankton blooms associated with ice retreat timing in the Pacific Arctic from 2003–2019. PLOS ONE 16 (12), e0261418. doi: 10.1371/journal.pone.0261418
Walsh J. E., Fetterer F., Scott J. S., Chapman W. L. (2017). A database for depicting Arctic sea ice variations back to 1850. Geographical Rev. 107, 89–107. doi: 10.1111/j.1931-0846.2016.12195.x
Ward J., Evan J., Shumway S. E. (2004). Separating the grain from the chaff: particle selection in suspension- and deposit-feeding bivalves. J. Exp. Mar. Biol. Ecol. 300, 83–130. doi: 10.1016/j.jembe.2004.03.002
Keywords: Arctic, bivalves, diatoms, DNA metabarcoding, harmful algal blooms
Citation: Koch CW, Sonsthagen SA, Cooper LW, Grebmeier JM, Riddle-Berntsen AE and Cornman RS (2025) Prevalence of pelagic diatoms and harmful algae in tellinid bivalve diets during record low sea ice in the Pacific Arctic determined by DNA metabarcoding. Front. Mar. Sci. 12:1480327. doi: 10.3389/fmars.2025.1480327
Received: 13 August 2024; Accepted: 06 February 2025;
Published: 25 February 2025.
Edited by:
Paul Snelgrove, Memorial University of Newfoundland, CanadaReviewed by:
Terri Sutherland, Fisheries and Oceans Canada (DFO), CanadaRaphaelle Descoteaux, The University Centre in Svalbard, Norway
Copyright © 2025 Koch, Sonsthagen, Cooper, Grebmeier, Riddle-Berntsen and Cornman. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chelsea W. Koch, ckoch@american.edu
 Chelsea W. Koch
Chelsea W. Koch