
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci., 04 March 2022
Sec. Marine Conservation and Sustainability
Volume 9 - 2022 | https://doi.org/10.3389/fmars.2022.836941
 Ian French1
Ian French1 Benjamin J. Wainwright1,2*
Benjamin J. Wainwright1,2*Shark populations have declined by more than 70% over the past 50 years. These declines have largely been attributed to increases in fishing efforts. Despite increased public awareness surrounding the conservation of sharks, three-quarters of all oceanic shark species are currently considered at risk of extinction. Here, we use DNA barcoding to identify shark DNA found in pet food purchased within Singapore. We identified a number of sharks that have some degree of control over their trade exerted under the auspices of the Convention on International Trade in Endangered Species of Wild Fauna and Flora (CITES), or through their classification as threatened by the International Union for Conservation of Nature (IUCN). The most commonly identified shark was the blue shark, Prionace glauca, a species that is not listed in CITES or classified as threatened by the IUCN, but one which scientific evidence suggests is overexploited and should have its catch regulated. The second most commonly encountered species was the CITES Appendix II listed silky shark, Carcharhinus falciformis. None of the products specifically listed shark as an ingredient, listing only generic terms, such as “ocean fish,” “white fish,” and “white bait.” The vague terminology used to describe pet food ingredients, and in some cases, the mislabeling of contents, prevents consumers – in this case, pet owners – from making informed and environmentally conscious decisions; consequently, pet owners and animal lovers may unwittingly be contributing to the overfishing of endangered sharks.
The global demand for shark products, such as fins, liver oil, and meat, has caused precipitous declines in shark populations (MacNeil et al., 2020; Shiffman et al., 2020; Pacoureau et al., 2021). It is estimated that the abundance of oceanic sharks and rays has declined by 71% since 1970, with these declines attributed to an 18-fold increase in fishing efforts. Correspondingly, three-quarters of all oceanic shark species are now considered at risk of global extinction (Pacoureau et al., 2021). Further demonstrating these declines, reef sharks were absent in nearly a fifth of over 15,000 baited underwater video surveys performed throughout the world’s coastal zones and were completely absent from all surveys in several coastal nations (MacNeil et al., 2020).
Another contributor to these declines is the mislabeling of seafood products, either deliberate or accidental. The practice of mislabeling generally involves the substitution of a low-value product for one of a higher value to increase the economic gain (Chang et al., 2021; Neo et al., 2022). However, the use of generic catch-all terms, such as “fish,” “ocean fish,” and “white fish,” are becoming increasingly common (Cardeñosa, 2019). These all-encompassing terms eliminate the consumer’s ability to make an informed decision about which products they wish to purchase and use. For example, pet foods and cosmetics frequently contain shark products even when not shown on the ingredients list. Pet food contains shark meat, while squalene is frequently found in cosmetics and other beauty products; in both of these cases, DNA barcoding has highlighted the presence of endangered shark species (Cardeñosa, 2019). Squalene can originate from non-shark sources (e.g., plant products); however, DNA barcoding has confirmed that the squalene in some pet food products and cosmetics is derived from sharks, such as the hammerhead shark (Sphyrna lewini), that are listed as endangered by the International Union for the Conservation of Nature (IUCN; Cardeñosa, 2019). In both pet food and cosmetics where shark DNA has been detected, the shark was not listed as an ingredient; instead, catch-all generic terms were used to describe the contents. Consequently, the consumer would have no way of knowing that the purchased product contains sharks without using DNA-based testing methods.
The official DNA barcode for animal species identification is a fragment of the mitochondrial gene coding for cytochrome c oxidase 1 (COI), and this region with approximately 650 bp has been widely adopted (Folmer et al., 1994; Hebert et al., 2003; Ward et al., 2005, 2008). However, tinned products that are sold for consumption (e.g., pet food), or those used by humans (e.g., beauty products), are usually highly processed to avoid health risks. Pet food is frequently thermally treated to improve safety and shelf life through the destruction of fungal spores and bacteria (van Rooijen et al., 2013), and similar treatments are used to avoid microbial contamination in beauty products (Kim et al., 2020). These processes destroy or degrade DNA rendering the full 650 bp fragment, which is impossible to amplify and, therefore, unusable in the molecular identification of species. To circumvent these challenges, a number of mini-barcoding techniques have been developed which use only a small fragment (100–200 bp) of the original gene (Hajibabaei et al., 2006; Meusnier et al., 2008; Shokralla et al., 2015), and a shark-specific mini-barcode has been developed to allow DNA amplification from challenging or degraded samples (Cardeñosa et al., 2017). These shorter fragments can be used to successfully make shark species’ identifications in highly processed products and samples, such as dried fins, shark liver oil, and environmental DNA (eDNA; Wainwright et al., 2018; Ip et al., 2021; Liu et al., 2021).
There is a rapidly growing public interest and awareness in conservation, particularly for charismatic species, such as sharks (Shiffman et al., 2020). It is likely that many pet owners who are broadly interested in conservation, or more specifically in the protection of sharks, are unaware that they may be inadvertently feeding endangered species of sharks to their pets, or that through the use of cosmetics containing shark-derived squalene, they are unknowingly helping to fuel the global overfishing of sharks.
Previous work has applied the same techniques to examine the occurrence of sharks in pet food samples collected within the United States (Cardeñosa, 2019). Here, we use a mini-DNA barcoding approach to investigate the occurrence of shark meat in a variety of pet food products purchased in Singapore. The use of consistent techniques allows comparisons between regions and products.
In total, 45 different pet food products were purchased from 16 different brands in Singapore. Most products used the generic terms “fish,” “ocean fish,” “white bait,” or “white fish” in the ingredients list to describe their contents. Some specifically listing “tuna” or “salmon,” but others did not indicate fish of any sort as an ingredient. None of the pet food products collected in this work specifically listed sharks on the ingredient list.
DNA was extracted via Chelex 100 resin (Walsh et al., 2013); a 10% solution of Chelex (Bio-Rad) with nuclease-free water was prepared. Taking care to ensure complete mixing to prevent settling, 200 μl of this mix was then transferred to a PCR tube. A small, approximately 1–2 mm2 pet food sample, was added, briefly vortexed (5–10 s), and then centrifuged. When multiple samples were collected from the can or packet – corresponding to different types of meat observed – a separate extraction was performed for each. For DNA extraction, samples were initially heated at 60°C for 20 min, then, 99°C for 25 min, and stored at 4°C until PCR was performed.
Using the following primer pair, we attempted to amplify approximately 150 bp fragments of the COI gene. Forward primer is VF2 (5′- TCA ACC AAC CAC AAA GAC ATT GGC AC –3′) and reverse primer is Shark150R (5′ -AAG ATT ACA AAA GCG TGG GC-3′) (Cardeñosa et al., 2017). All reactions were performed in 25 μl volumes; each reaction contained 12.5 μl GoTaq Master Mix Green (Promega), 1 μl forward primer (10 μM), 1 μl reverse primer (10 μM), 9.5 μl water, and 1 μl of undiluted DNA template. The PCR thermal cycling conditions followed an initial denaturation period of 2 min at 94°C, followed by 35 cycles at 94°C for 1 min, 52°C for 1 min, 72°C for 1 min, and a final extension period of 10 min at 72°C.
Successful PCR amplification was verified on a 1% TBE agarose gel, and products were sent to Macrogen Korea for cleaning and Sanger sequencing. Geneious v2020.2.4 (Kearse et al., 2012) was used to view sequence data. Species identifications were made using The Barcode of Life Data System (BOLD)1, and the Nucleotide BLAST (BLASTn) function in GenBank2. Species identifications were considered positive if BOLD and BLAST identified the same species with a 100% match. If BOLD and BLAST indicated that the sequence matched more than one species with 100% similarity (i.e., a species-level match could not be made) we made a genus-level identification.
Of the 144 samples sequenced, 31% (n = 45) contained shark DNA and returned sequences that we were able to use to identify samples to either the genus or species level. Because we took more than one sample from each of the collected products (maximum five samples, minimum two samples) (Supplementary Table 1), if the same species were identified more than once in the same tin or packet, we considered it the same individual and counted it only once (Supplementary Table 1). The most commonly identified genus was Carcharhinus. The top three most frequently identified species were: (1) Prionace glauca (blue shark) was identified seven times, (2) Carcharhinus falciformis (silky shark) was seen five times, and, Triaenodon obesus (whitetip reef shark) was found four times. We documented five occurrences of C. falciformis, the silky shark which is listed on the Convention on International Trade in Endangered Species of Wild Fauna and Flora (CITES) appendix II, and several species were classified as threatened by the International Union for the Conservation of Nature (IUCN; Table 1 and Figure 1). The most commonly identified non-shark identifications were chicken (Gallus gallus), salmon, tuna, and the decapod, Litopenaeus vannamei, commonly known as the whiteleg shrimp.
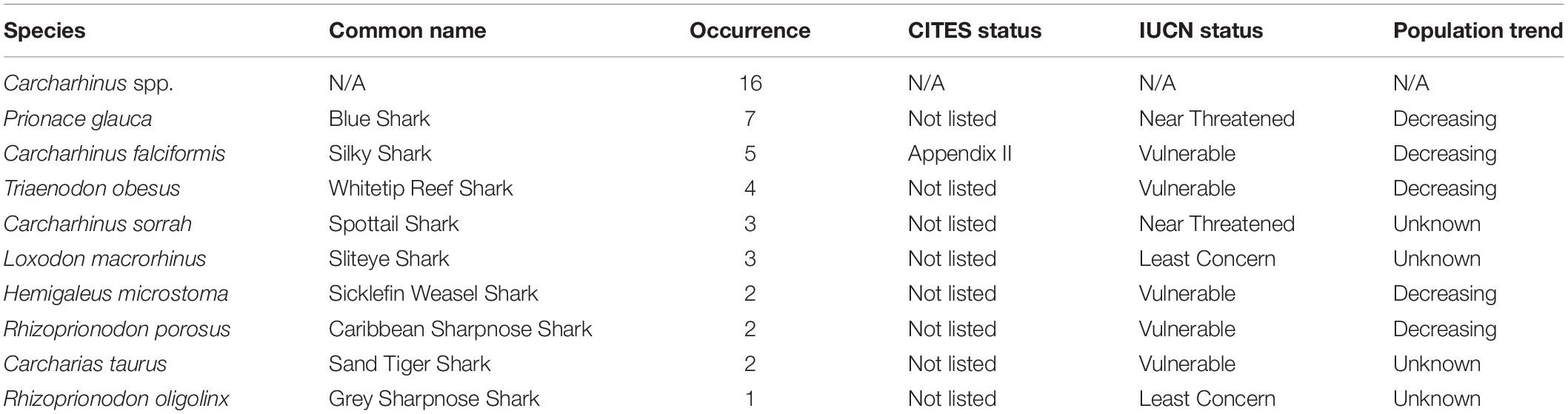
Table 1. Details of species identifications, including occurrence, CITES an IUCN status and the current population tend according to the IUCN Red List.
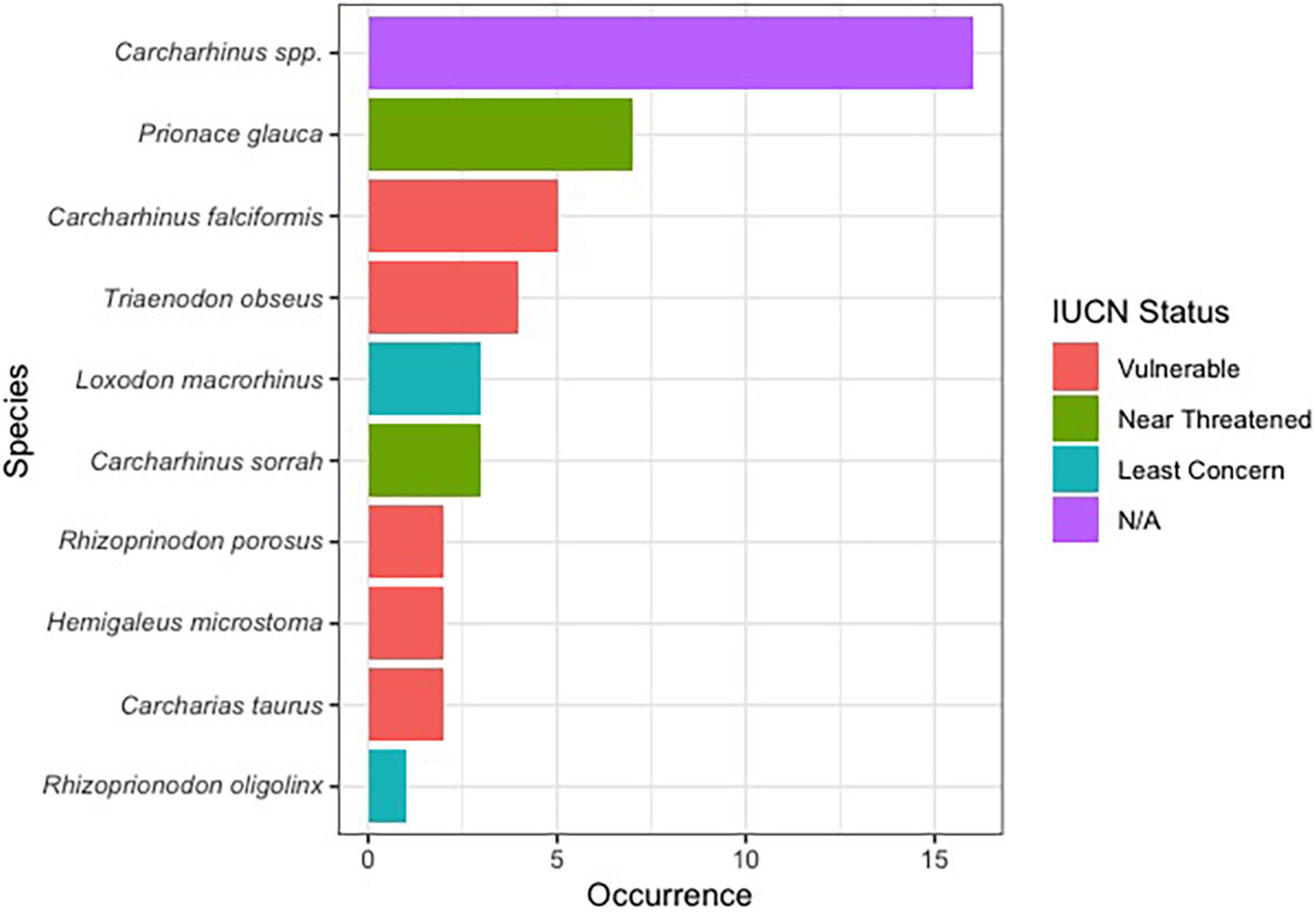
Figure 1. Bar plot showing the proportion of shark species identified in this work along with their IUCN red list status.
We detected shark DNA belonging to a number of species that are listed on CITES Appendix II or classified as endangered on the IUCN Red list. A listing on CITES Appendix II means that a species is not necessarily threatened with extinction, but its trade must be controlled to avoid overconsumption that is incompatible with its survival. Sharks were not listed as ingredients on any of the sampled products tested; while this is not illegal or a required procedure, we argue that many pet owners and lovers would be alarmed to find out that they are likely contributing to the unsustainable fishing practices that have caused massive declines in global shark populations. The overfishing of sharks can have negative impacts on the ecosystem’s stability and function, and their removal has even been linked to the amplification of detrimental climate change effects (Nowicki et al., 2021).
Despite taking numerous samples from each of the collected products, it is likely that we did not discover the full diversity of sharks from those samples. It is now possible to leverage the high throughput of a second (Illumina) and third (PacBio and Oxford Nanopore) generation DNA sequencing platforms to comprehensively sequence and identify the entire contents of a can, or packet of pet food (Ho et al., 2020). It is also highly probable that this approach will reveal other species that are not listed as ingredients; given the scale and extent of seafood mislabeling, some of these may also be endangered species (Shokralla et al., 2015).
An earlier study examining the contents of pet food in the United States similarly showed that sharks were frequently present (Cardeñosa, 2019). The most commonly encountered shark species in samples from the United States was the shortfin mako (Isurus oxyrinchus); this species was completely absent in all of our samples. In total, Cardeñosa (2019) was able to identify eighteen pet food-associated sequences to the species level, representing two different species (Isurus oxyrinchus n = 17, and Prionace glauca n = 1). It is unknown where the pet food in Cardeñosa (2019) was processed, but the samples purchased in our work were all produced, canned, or packaged in Thailand. Our samples constitute a more diverse collection of nine species of shark, possibly indicating that the pet food industry in the United States and Asia is supported by different supply chains. Like Cardeñosa (2019), we found the blue shark, Prionace glauca, in our samples, and other barcoding work performed in Southeast Asia has shown a high occurrence of the blue shark in the fin trade (Wainwright et al., 2018; Liu et al., 2021). Despite being one of the most frequently traded species in Hong Kong (Fields et al., 2018, 2020), the blue shark is not listed under CITES Appendices I or II and is only listed as near threatened on the IUCN red list. Blue sharks are recognized as a highly migratory species that are frequently encountered in pelagic longline fisheries as bycatch (Campana et al., 2002). Accidental catch of this fish could account for its high prevalence in the pet food industry; since a shark carcass is a low-value product once the fins have been removed, it is possible that the pet food industry processes this carcass instead of wasting it. Blue sharks do appear to be a victim of increasing fishing effort and are widely identified in the fin trade throughout Southeast Asia, and scientific evidence already suggests that blue shark fishing should be regulated (Simpfendorfer and Dulvy, 2017). This apparent high fishing pressure may warrant a greater degree of protection and justify listing blue sharks on the IUCN Red List and inclusion on the CITES Appendix II (Liu et al., 2021). The second most encountered shark in our pet food was the silky shark (C. falciformis). This species is recognized as the second most commonly encountered species in the Hong Kong shark fin trade, one of the world’s largest fin trading hubs (Cardeñosa et al., 2021). The frequent occurrence of silky sharks has been attributed to this species’ high bycatch rate where it is the most commonly encountered bycatch species in pelagic tuna fisheries (Hutchinson et al., 2015). As in the case of blue sharks, it is possible that once the silky sharks’ high-valued fins have been removed, their carcasses make their way into the pet food industry, thus accounting for its high prevalence in our samples.
We were unable to identify sixteen individuals past the level of the genus. This is consistent with other work that uses mini-DNA barcodes. These short fragments, while allowing us to amplify DNA from highly processed samples, lack the resolving power of a longer or full-length COI fragment to make species-level identifications. However, these smaller fragments are frequently the only ones that produce usable barcodes in processed samples where DNA is extremely likely to be degraded. Using these smaller fragments, we were able to detect shark DNA in a number of products that listed generic catch-all terms in their ingredient lists, such as “fish,” “white fish,” and “ocean fish.” Despite being specifically designed to amplify shark DNA, the primers used here still amplified and produced sequences that allowed the identification of a number of non-shark species, most frequently chicken (Gallus gallus). This is consistent with previous work showing that these primers amplify DNA from chicken and a number of teleost species (Cardeñosa, 2019).
Seafood fraud and the deliberate mislabeling or substitution of products is an increasingly recognized global problem that can both lead to and conceal the unsustainable use of marine resources (Luque and Donlan, 2019; Chang et al., 2021; Giagkazoglou et al., 2022; Neo et al., 2022).
Here, we show mislabeling is prevalent in the pet food trade and identify a number of shark species in pet food products, some of which are listed as endangered or have some degree of control imposed on their trade. We suggest that better labeling with globally implemented standards would help to avoid the overexploitation of endangered species such as sharks; doing so would also allow pet owners to have greater control over what they feed their pets.
We do not know whether the high incidence of sharks in pet food is an attempt to avoid the wastefulness of the shark fin trade where high-valued fins are retained and the low-valued carcasses are discarded; if this is the case, that may be commendable. However, we are skeptical that this is the sole reason that sharks end up in pet food. More likely, their presence demonstrates the high fishing pressure to which sharks are increasingly subjected. When viewed in the light of the facts that oceanic shark and ray populations have declined by 71% since 1970, and that three-quarters of all shark species are now considered at risk of global extinction, pet food is not a worthy use of these charismatic and ecologically important apex predators. Better labeling that avoids the currently used vague catch-all terminology would allow consumers to make more informed choices. This in turn would benefit shark populations by helping to mitigate unsustainable fishing and resource use incompatible with their continued survival.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethical review and approval was not required for the animal study because samples were collected from processed pet food purchased in public retail markets.
IF and BW contributed to conception and design of the study. IF performed lab work, analysis, and wrote the first draft of the manuscript. Both authors contributed to the manuscript, read, and approved the submitted version.
This work was supported by a Yale-NUS College Start-Up Fund.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2022.836941/full#supplementary-material
Campana, S., Gonzalez, P., and Joyce, W. (2002). Catch, Bycatch and Landings of Blue Shark (Prionace glauca) in the Canadian Atlantic Research Document 2002/101. Ottawa, ON: Fisheries and Oceans Canada, 42.
Cardeñosa, D. (2019). Genetic identification of threatened shark species in pet food and beauty care products. Conserv. Genet. 20, 1383–1387. doi: 10.1007/s10592-019-01221-0
Cardeñosa, D., Fields, A. T., Babcock, E., Shea, S. K. H., Feldheim, K. A., Kraft, D. W., et al. (2021). Indo-Pacific origins of silky shark fins in major shark fin markets highlight supply chains and management bodies key for conservation. Conserv. Lett. 14:e12780. doi: 10.1111/conl.12780
Cardeñosa, D., Fields, A., Abercrombie, D., Feldheim, K., Shea, S. K. H., and Chapman, D. D. (2017). A multiplex PCR mini-barcode assay to identify processed shark products in the global trade. PLoS One 12:e0185368. doi: 10.1371/journal.pone.0185368
Chang, C.-H., Tsai, M.-L., Huang, T.-T., and Wang, Y.-C. (2021). Authentication of fish species served in conveyor-belt sushi restaurants in Taiwan using DNA barcoding. Food Control 130:108264. doi: 10.1016/j.foodcont.2021.108264
Fields, A. T., Fischer, G. A., Shea, S. K. H., Zhang, H., Abercrombie, D. L., Feldheim, K. A., et al. (2018). Species composition of the international shark fin trade assessed through a retail-market survey in Hong Kong: shark fin trade. Conserv. Biol. 32, 376–389. doi: 10.1111/cobi.13043
Fields, A. T., Fischer, G. A., Shea, S. K. H., Zhang, H., Feldheim, K. A., and Chapman, D. D. (2020). DNA Zip-coding: identifying the source populations supplying the international trade of a critically endangered coastal shark. Anim. Conserv. 23:acv.12585. doi: 10.1111/acv.12585
Folmer, O., Black, M., Hoeh, W., Lutz, R., and Vrijenhoek, R. (1994). DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol. Mar. Biol. Biotechnol. 3, 294–299.
Giagkazoglou, Z., Griffiths, A. M., Imsiridou, A., Chatzispyrou, A., Touloumis, K., Hebb, J. L., et al. (2022). Flying under the radar: DNA barcoding ray wings in Greece detects protected species and umbrella labelling terms. Food Control 132:108517. doi: 10.1016/j.foodcont.2021.108517
Hajibabaei, M., Smith, M. A., Janzen, D. H., Rodriguez, J. J., Whitfield, J. B., and Hebert, P. D. N. (2006). A minimalist barcode can identify a specimen whose DNA is degraded. Mol. Ecol. Notes 6, 959–964. doi: 10.1111/j.1471-8286.2006.01470.x
Hebert, P. D. N., Cywinska, A., Ball, S. L., and deWaard, J. R. (2003). Biological identifications through DNA barcodes. Proc. Biol. Sci. 270, 313–321. doi: 10.1098/rspb.2002.2218
Ho, J. K. I., Puniamoorthy, J., Srivathsan, A., and Meier, R. (2020). MinION sequencing of seafood in Singapore reveals creatively l flatfishes, confused roe, pig DNA in squid balls, and phantom crustaceans. Food Control 112:107144. doi: 10.1016/j.foodcont.2020.107144
Hutchinson, M. R., Itano, D. G., Muir, J. A., and Holland, K. N. (2015). Post-release survival of juvenile silky sharks captured in a tropical tuna purse seine fishery. Mar. Ecol. Progr. Ser. 521, 143–154. doi: 10.3354/meps11073
Ip, Y. C. A., Chang, J. J. M., Lim, K. K. P., Jaafar, Z., Wainwright, B. J., and Huang, D. (2021). Seeing through sedimented waters: environmental DNA reduces the phantom diversity of sharks and rays in turbid marine habitats. BMC Ecol. Evol. 21:166. doi: 10.1186/s12862-021-01895-6
Kearse, M., Moir, R., Wilson, A., Stones-Havas, S., Cheung, M., Sturrock, S., et al. (2012). Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28, 1647–1649. doi: 10.1093/bioinformatics/bts199
Kim, H. W., Seok, Y. S., Cho, T. J., and Rhee, M. S. (2020). Risk factors influencing contamination of customized cosmetics made on-the-spot: evidence from the national pilot project for public health. Sci. Rep. 10:1561. doi: 10.1038/s41598-020-57978-9
Liu, C. J. N., Neo, S., Rengifo, N. M., French, I., Chiang, S., Ooi, M., et al. (2021). Sharks in hot soup: DNA barcoding of shark species traded in Singapore. Fisheries Res. 241:105994. doi: 10.1016/j.fishres.2021.105994
Luque, G. M., and Donlan, C. J. (2019). The characterization of seafood mislabeling: a global meta-analysis. Biol. Conserv. 236, 556–570. doi: 10.1016/j.biocon.2019.04.006
MacNeil, M. A., Chapman, D. D., Heupel, M., Simpfendorfer, C. A., Heithaus, M., Meekan, M., et al. (2020). Global status and conservation potential of reef sharks. Nature 583, 801–806. doi: 10.1038/s41586-020-2519-y
Meusnier, I., Singer, G. A., Landry, J.-F., Hickey, D. A., Hebert, P. D., and Hajibabaei, M. (2008). A universal DNA mini-barcode for biodiversity analysis. BMC Genomics 9:214. doi: 10.1186/1471-2164-9-214
Neo, S., Kibat, C., and Wainwright, B. J. (2022). Seafood mislabelling in Singapore. Food Control 135:108821. doi: 10.1016/j.foodcont.2022.108821
Nowicki, R. J., Thomson, J. A., Fourqurean, J. W., Wirsing, A. J., and Heithaus, M. R. (2021). Loss of predation risk from apex predators can exacerbate marine tropicalization caused by extreme climatic events. J. Anim. Ecol. 90, 2041–2052. doi: 10.1111/1365-2656.13424
Pacoureau, N., Rigby, C. L., Kyne, P. M., Sherley, R. B., Winker, H., Carlson, J. K., et al. (2021). Half a century of the global decline in oceanic sharks and rays. Nature 589, 567–571. doi: 10.1038/s41586-020-03173-9
Shiffman, D. S., Bittick, S. J., Cashion, M. S., Colla, S. R., Coristine, L. E., Derrick, D. H., et al. (2020). Inaccurate and biased global media coverage underlies public misunderstanding of shark conservation threats and solutions. iScience 23:101205. doi: 10.1016/j.isci.2020.101205
Shokralla, S., Hellberg, R. S., Handy, S. M., King, I., and Hajibabaei, M. (2015). A DNA mini-barcoding system for authentication of processed fish products. Sci. Rep. 5:15894. doi: 10.1038/srep15894
Simpfendorfer, C. A., and Dulvy, N. K. (2017). Bright spots of sustainable shark fishing. Curr. Biol. 27, R97–R98. doi: 10.1016/j.cub.2016.12.017
van Rooijen, C., Bosch, G., van der Poel, A. F. B., Wierenga, P. A., Alexander, L., and Hendriks, W. H. (2013). The Maillard reaction and pet food processing: effects on nutritive value and pet health. Nutr. Res. Rev. 26, 130–148. doi: 10.1017/S0954422413000103
Wainwright, B. J., Ip, Y. C. A., Neo, M. L., Chang, J. J. M., Gan, C. Z., Clark-Shen, N., et al. (2018). DNA barcoding of traded shark fins, meat and mobulid gill plates in Singapore uncovers numerous threatened species. Conserv. Genet. 19, 1393–1399. doi: 10.1007/s10592-018-1108-1
Walsh, P. S., Metzger, D. A., and Higuchi, R. (2013). Chelex 100 as a medium for simple extraction of DNA for PCR-based typing from forensic material. Biotechniques 54, 134–139. doi: 10.2144/000114018
Ward, R. D., Holmes, B. H., White, W. T., and Last, P. R. (2008). DNA barcoding Australasian chondrichthyans: results and potential uses in conservation. Mar. Freshw. Res. 59:57. doi: 10.1071/MF07148
Keywords: DNA barcoding, pet food, seafood fraud, seafood labeling, seafood traceability, shark conservation
Citation: French I and Wainwright BJ (2022) DNA Barcoding Identifies Endangered Sharks in Pet Food Sold in Singapore. Front. Mar. Sci. 9:836941. doi: 10.3389/fmars.2022.836941
Received: 16 December 2021; Accepted: 07 February 2022;
Published: 04 March 2022.
Edited by:
Annette Breckwoldt, Leibniz Centre for Tropical Marine Research (LG), GermanyReviewed by:
Véronique Merten, GEOMAR Helmholtz Centre for Ocean Research Kiel, Helmholtz Association of German Research Centres (HZ), GermanyCopyright © 2022 French and Wainwright. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Benjamin J. Wainwright, Ben.Wainwright@Yale-NUS.edu.sg
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.