- 1Yellow Sea Fisheries Research Institute, Chinese Academy of Fishery Sciences, Qingdao, China
- 2Laboratory for Marine Fisheries Science and Food Production Processes, Qingdao National Laboratory for Marine Science and Technology, Qingdao, China
Omics approaches provide more metabolic information to explain the relationship between dietary nutrition and fish growth. This study aimed to explore the metabolome and proteome response of turbot (Scophthalmus maximus) fed diets containing lysine and leucine in free and dipeptide forms by the approaches of integrated liquid chromatography-tandem mass spectrometry (LC-MS/MS)-based metabolomics and isobaric tags for relative and absolute quantification (iTRAQ)-based proteomics. Plant protein-based diets were formulated to contain the equivalent of lysine and leucine in free amino acid [crystalline amino acid (CAA)] and synthetic Lys-Leu (Lys-Leu) forms. The metabolome and proteome profiles of the liver were screened in fish fed either the CAA diet or the Lys-Leu diet after an 8-week feeding trial. Fish fed the Lys-Leu diet showed a significantly higher final body weight and a specific growth rate compared with fish fed the CAA diet. Protein- and amino acid-related metabolic processes in the liver were identified between the Lys-Leu and CAA groups based on differential metabolites and proteins. The proteolytic enzymes and amino acid transporters from differential proteins of the liver showed that the process of protein digestion and absorption may be affected by the different forms of lysine and leucine in the feed. A mechanistic target of rapamycin complex 1 and ubiquitin proteasome pathways were identified by differential proteins, which were involved in the processes of protein synthesis and degradation in the liver. Lysine degradation, tryptophan metabolism, alanine, aspartate, and glutamate metabolism, arginine biosynthesis, arginine and proline metabolism, and glycine, serine, and threonine metabolism were identified based on differential metabolites and proteins, which showed that the metabolism of various amino acids, including lysine, had been affected by both the CAA and Lys-Leu groups. In conclusion, the data of integrated metabonomics and proteomics suggested that different forms of lysine and leucine in the feed may affect liver metabolic processes including protein digestion and absorption, protein synthesis and degradation, and amino acid metabolism. In addition, a good correlation between differential metabolites and proteins was observed in amino acid metabolism by using the approaches of integrated LC-MS/MS-based metabolomics and iTRAQ-based proteomics.
Introduction
With the rising demand and price of fish meal, expanding the amount of plant protein in aquafeeds was an effective strategy to replace fish meal (Daniel, 2017). When more plant ingredients were formulated in diets, it was also necessary to supplement essential amino acids based on the requirement of fish (Furuya et al., 2004; El-Husseiny et al., 2017). Nutritionists proposed that essential amino acids in feed could be supplemented with synthetic peptides or crystalline forms (Dabrowski et al., 2003; Ostaszewska et al., 2013). Those studies in fish species mainly focused on a comparison of dietary amino acid sources in peptides or free amino acids forms using classical nutritional methods, such as the evaluation of growth and feed utilization and the analysis of digestive enzyme activity and amino acid composition of fish whole body (Mamauag et al., 2012; Kim and Lee, 2013; Kamaszewski et al., 2019, 2020). Regarding turbot (Scophthalmus maximus), our previous studies showed that the utilization efficiency of dietary Lys-Leu dipeptide was not only higher than that of dietary crystalline lysine and leucine but also higher than that of dietary Lys-Leu-Lys/Leu-Lys-Leu, Lys-Gly, or Gly-Leu (Wei et al., 2020a,b). However, the effect of dietary amino acid in free and peptide forms on fish growth was still controversial in different fish species, such as carp (Cyprinus carpio) (Kamaszewski et al., 2014), red sea bream (Pagrus major) (Mamauag et al., 2012), and olive flounder (Paralichthys olivaceus) (Rahimnejad and Lee, 2014). It implied that the application of classical nutritional methods may not comprehensively understand how the forms of dietary amino acids regulated the metabolic process of fish, which, in turn, affected their growth. Hence, in order to address this issue, omics approaches, including metabolomics and proteomics, should be introduced in fish nutrition (Sales et al., 2014).
Metabolomics and proteomics are the functional continuation of transcriptomics and can be used to further elucidate the role of nutritional changes in diets on fish growth by understanding the metabolic pathways (de Vareilles et al., 2012; Rodrigues et al., 2012; Ceciliani et al., 2018). Metabolomics can help to gain insight into the alterations of metabolism by identifying small molecule metabolites (Deborde et al., 2021). Proteomics focuses on diverse processes, such as cytoskeletal dynamics, energy and carbohydrate metabolism, lipoprotein metabolism, amino acid metabolism, and protein metabolism, by identifying proteins in the cell and tissue (de Vareilles et al., 2012). However, to date, only a few studies had explored the response of fish to dietary amino acid imbalance or protein source changes by metabolomics or proteomics (Gómez-Requeni et al., 2011; de Vareilles et al., 2012; Wei et al., 2017a,b; Deborde et al., 2021). For instance, Gómez-Requeni et al. (2011) and de Vareilles et al. (2012) reported that a proteomics approach was used to explore the effects of dietary lysine imbalances on the protein expression of whole body or muscle in zebrafish (Danio rerio). Our previous studies evaluated the metabolome profile of liver and muscle in turbot fed diets containing fish meal and fish protein hydrolysate (low molecular weight peptides and free amino acids) (Wei et al., 2017a,b). Omics approaches in those studies were often used separately–proteomics and metabolomics–and an integrated analysis using these approaches was rarely conducted in fish nutrition. Thus, in order to provide more metabolic information to explain the relationship between dietary nutrition and fish growth, it was necessary to combine metabolomics with proteomics in nutritional studies with fish (Guo et al., 2019; Min et al., 2020; Yu et al., 2020).
The aim of this study was to explore the liver metabolome and proteome response of turbot fed diets containing lysine and leucine in free and dipeptide forms. The growth and the feed utilization of turbot are investigated after an 8-week feeding trial (Wei et al., 2020a), and the metabolome and proteome profiles of the liver were screened by the approaches of integrated liquid chromatography-tandem mass spectrometry (LC-MS/MS)-based metabolomics and isobaric tags for relative and absolute quantification (iTRAQ)-based proteomics.
Materials and Methods
Experimental Diets and Feeding Trail
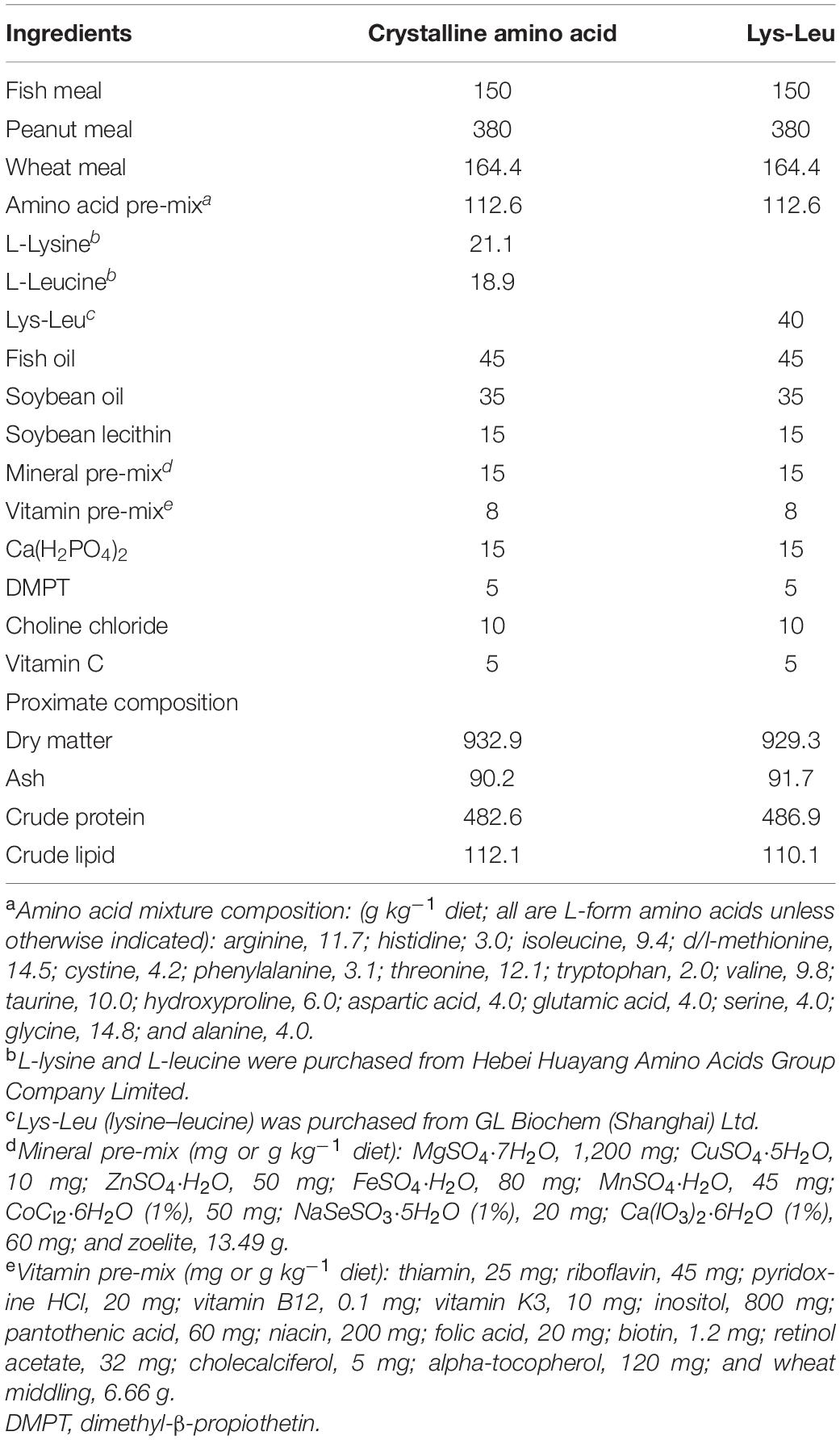
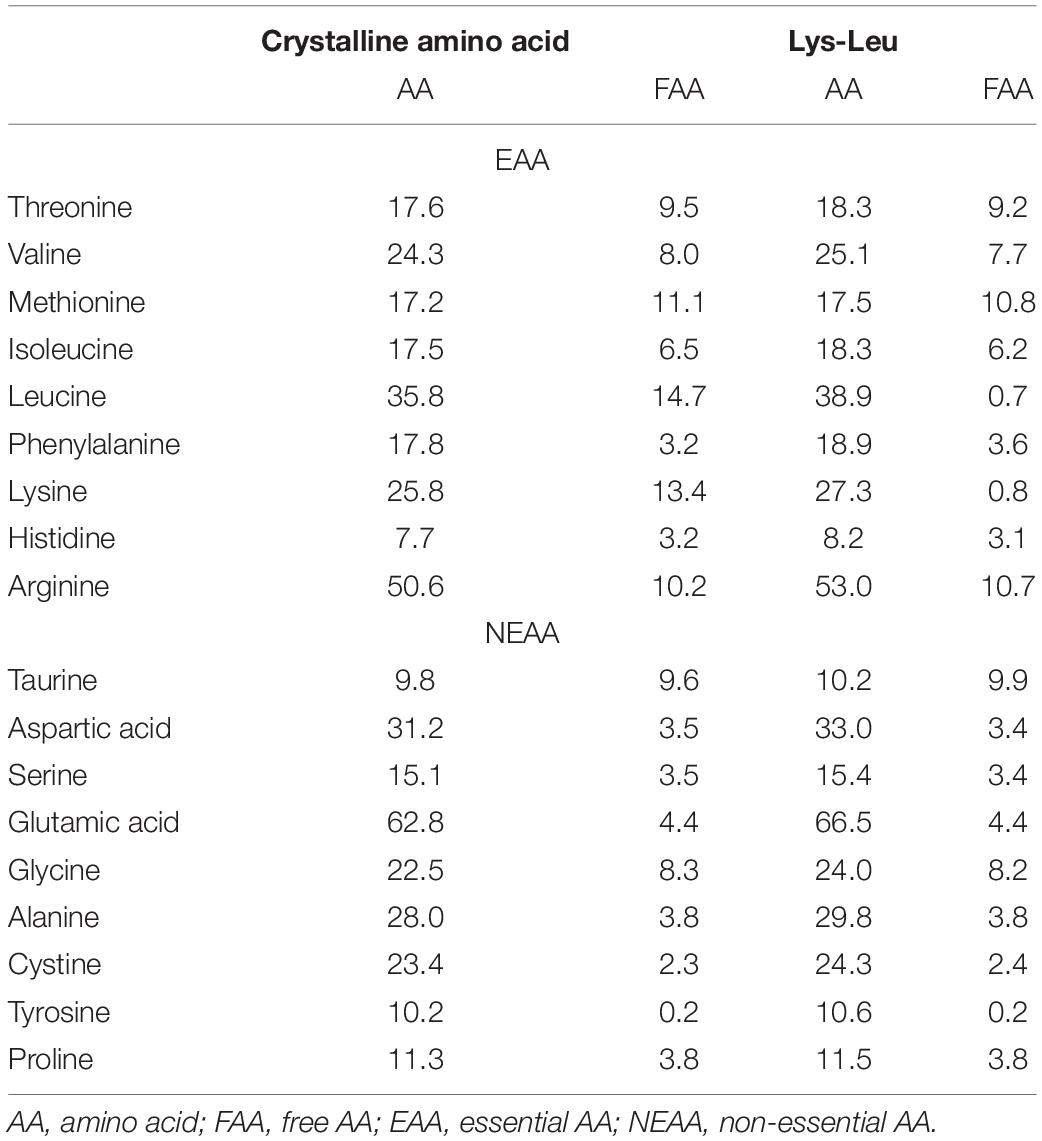
Our previous study provided evidence that the inclusion of dietary dipeptide formed by lysine and leucine was effective for growth and feed utilization in turbot (Wei et al., 2020a). Thus, the equivalent of lysine and leucine, but in different forms (free or dipeptide), was used in plant protein-based diets (Table 1). Briefly, two isoproteic (480 g kg–1 crude protein) diets were formulated to contain 150 g kg–1 fish meal and 380 g kg–1 peanut meal. Then, one of them was supplemented with crystalline lysine (21.1 g kg–1) and leucine (18.9 g kg–1) [the Crystalline amino acid (CAA) diet]; the other was supplemented with synthetic Lys-Leu (40.0 g kg–1) (the Lys-Leu diet). CAA pre-mix without lysine and leucine was added to the diets according to Peres and Oliva-Teles (2005). The analysis of amino acid composition in diets revealed that the contents of total lysine and leucine were similar in the two diets, while the contents of free lysine and leucine in the CAA diet were higher than those in the Lys-Leu diet (Table 2).
Turbot was purchased and cultured at Yantai Tianyuan Aquatic Product Co., Ltd. (Shandong, China). The fish were transported from a cemented tank (5.0 m × 5.0 m × 1.5 m) to a large cylindrical fiberglass tank (water volume, 300 L) and fed a commercial feed for 2 weeks to acclimate to new culture conditions. Then, 150 fish with initial weight (11.98 ± 0.03 g) were stocked in six tanks (water volume, 120 L; 25 fish per tank). Each diet was assigned to three replicate tanks. The fish were hand-fed to apparent satiation twice daily (06:30 and 16:30) in a flow-through system with continuous aeration and water flow rate of 3 L min–1. The uneaten feed was siphoned from the tank bottom and weighed to accurately estimate the feed consumed. The feeding trail lasted for 8 weeks from August to October. During this period, natural photoperiod was used, and water temperature, pH, salinity, and dissolved oxygen were 15 ± 1°C, 7.7 ± 0.2, 32 ppt, and 7 mg L–1, respectively. At the termination of the feeding trail, the fish were starved for 24 h, and then the number and weight of the fish from each tank were recorded. Three fish from each tank were collected at 2 h after feeding and then anesthetized with eugenol (1:10,000). After blood sampling, the liver sample of each fish was flash-frozen in liquid nitrogen and then stored at −80°C before use. The experimental protocols followed the guidelines approved by the Institutional Animal Care and Use Committee of the Yellow Sea Fisheries Research Institute.
Non-targeted Metabolomic and Analysis
Extraction of Metabolites
Metabolites of the liver in turbot were extracted using a modification of the organic protein precipitation method (Chen et al., 2018; Dong et al., 2020). The liver sample (25 mg) was transferred into a tube with 800 μl frozen solution of methanol/acetonitrile/water (2:2:1) and two small steel balls.
The sample was homogenized using a bead mill homogenizer (TissueLyser LT; Qiagen Hilden, Germany) at 60 Hz for 4 min, the steel balls were taken out, and then the sample was sonicated (80 Hz) in an ice bath for 10 min. The mixture was placed at −20°C for 120 min and then centrifuged at 25,000 × g for 15 min at 4°C. The supernatant was collected and transferred to a new tube. Subsequently, the centrifugation was repeated once, and 600 μl of supernatant was collected and freeze-dried. Finally, the dried supernatant sample was dissolved in 50% methanol solution, sonicated (80 Hz) in an ice bath for 10 min, and then centrifuged at 25,000 × g for 15 min at 4°C. The supernatant with metabolites was collected again for a subsequent analysis. In addition, 50 μl from each supernatant sample was pooled as the quality control (QC) sample.
LC-MS/MS Analysis
The supernatant with metabolites were determined by a liquid chromatography (LC)-mass spectrometer (MS) system. The injection volume for each supernatant sample was 5 μl. An ultra-performance liquid chromatography (UPLC) (Waters Corp., United Kingdom) was performed using an ACQUITY UPLC HSS T3 column (internal diameter, 2.1 mm; column length, 100 mm; and particle size, 1.8 μm; Waters Corp., United Kingdom). After the metabolites from the supernatant sample were eluted, a high-resolution tandem mass spectrometer (Xevo G2 XS Q-TOF, Waters Corp., United Kingdom) was used in both the positive and the negative ion modes. Mass spectrometry data were achieved by Centroid MSE mode (where E represents the collision energy). The centroid mode range was 50–1,200 Da, and the scan time was 0.2 s. During MS/MS acquisition, all precursors were fragmented with 20–40 eV, and the scan time was 0.2 s. In the process of data acquisition, mass accuracy was calibrated by acquiring the leucine–encephalin signal every 3 s. Furthermore, in order to evaluate the stability of the instrument system, a QC sample was collected for every 10 samples.
Data Processing and Statistical Data Analysis
The raw data from the mass spectrometer were imported into Progenesis QI (version 2.2) for a data matrix to obtain metabolite-related mass-to-charge ratio (m/z), retention time, and ion intensity. The normalized peak data were pre-processed using metaX software, including imputing the remaining peaks with missing values using the k-nearest neighbor algorithm, removing low-quality ions (50% of QC samples or 80% of biological samples), and then filtering out ions with a relative standard deviation >30% in QC samples (Wen et al., 2017). Differential metabolites in the liver of different treatments were identified using univariate analysis and multivariate statistics. Univariate analysis was performed by the methods of t-test and fold change (FC) analysis (P < 0.05; FC ≥ 1.2 or ≤0.8333). Multivariate statistics were performed by partial least squares-discriminate analysis (PLS-DA) with variable importance for the projection (VIP) values (≥1.0). Heat map was used to visualize differential metabolites, and Kyoto Encyclopedia of Genes and Genomes (KEGG)1 was used to help in understanding the functions of metabolites.
Proteomics Analysis
Protein Extraction and Digestion
Proteins of the liver in turbot were extracted using the method reported by Wu et al. (2019). The liver tissue from each fish was homogenized in lysis buffer 3 [8 mol/L urea, 40 mmol/L Tris–HCl, or tetraethylammonium bicarbonate (TEAB) with 1 mmol/L phenylmethanesulfonyl fluoride, 2 mmol/L ethylenediaminetetraacetic acid, and 10 mmol/L dithiothreitol (DTT), pH 8.5] by TissueLyser (Qiagen, Hilden, Germany). After centrifugation at 25,000 × g for 20 min at 4°C, 10 mmol/L DTT was added to the supernatant at 56°C for 1 h, and then the mixture was incubated with 55 mmol/L iodoacetamide in the dark at room temperature for 45 min. The precipitated protein was washed three times with acetone. The quality and the quantity of the protein sample were measured by sodium dodecyl sulfate polyacrylamide gel electrophoresis and Bradford method, respectively.
The protein solutions (100 μg) were digested with 2.5 μg Trypsin gold (Promega, Madison, WI, United States) at 37°C for 4 h. Trypsin gold was added once more with the same ratio, and trypsin digestion was continued at 37°C for 8 h. After trypsin digestion, the peptides were desalted with a Strata X C18 column (Phenomenex, Torrance, CA, United States) and vacuum-dried.
iTRAQ Labeling and Peptide Fractionation
The vacuum-dried peptides were dissolved in 30 μl of 0.5 mol/L TEAB with vortexing. After the iTRAQ labeling reagents were recovered to ambient temperature, peptide labeling was performed by iTRAQ Reagent 8-plex Kit (Applied Biosystems, Foster City, CA, United States). The labeled samples were desalted with a Strata X C18 column and then vacuum-dried.
The peptides were separated using a Shimadzu LC-20AB HPLC Pump system (Kyoto, Japan) equipped with Gemini C18 column (internal diameter, 4.6 mm; column length, 250 mm; and particle size, 5 μm; Phenomenex, Torrance, CA, United States). The peptides were dissolved with buffer A [5% acetonitrile (ACN), 95% H2O, pH 9.8] to 2 ml. Elution is monitored by measuring absorbance at 214 nm, and fractions are collected every 1 min. The eluted peptides are pooled as 20 fractions and vacuum-dried.
LC-MS/MS Analysis
Each fraction was reconstituted in buffer A (2% ACN and 0.1% formic acid) and centrifuged at 20,000 × g for 10 min. The supernatant was loaded on an Ultimate 3000 UHPLC system (Thermo Scientific, Waltham, MA, United States), equipped with a trap and an analytical column. The samples were desalted in a trap column and then eluted into a nanocapillary C18 column (inner diameter, 75 μm; column diameter, 3 μm; and column length, 25 μm). The peptides separated from nano-high-performance liquid chromatography were subjected to tandem mass spectrometry Q Exactive HF X (Thermo Fisher Scientific, San Jose, CA, United States) for data-dependent acquisition detection by nano-electrospray ionization.
Protein Identification and Functional Annotation
The raw data was firstly transformed into mascot generic format (MGF) for bioinformatics analysis. Then, the exported MGF files were searched using the Mascot search engine (version 2.3.02; Matrix Science, London, United Kingdom). For protein quantitation, an automated software (IQuant software) was used to analyze the labeled peptides with isobaric tags (Wen et al., 2014). The peptide-spectrum matches were pre-filtered at a false discovery rate (FDR) of 1% to assess the confidence of the peptides. Then, identified peptide sequences were assembled into proteins. In order to reduce the rate of false-positive at the protein level, the proteins were identified at a FDR of 1% (Savitski et al., 2015). Proteins with 1.2-fold change (mean value) and P-value (t-test) less than 0.05 were defined as differential proteins. Cluster analysis was used to visualize differential proteins. For the pathway analysis, KEGG was used to predict the main metabolic pathways of the differential proteins.
Results
Growth Performance
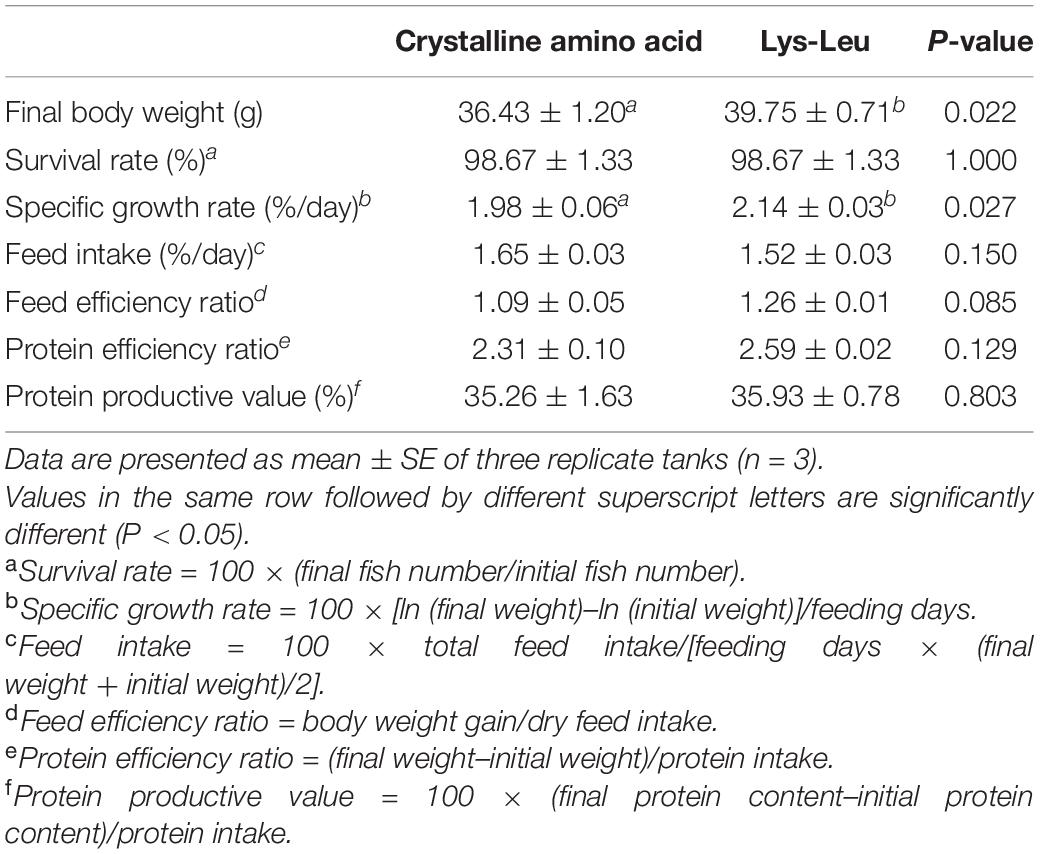
The analysis of t-test revealed that the final body weight (P = 0.022) and the specific growth rate (P = 0.027) offish fed the Lys-Leu diet were significantly higher than those of fish fed the CAA diet (Table 3). However, no significant differences among survival rate, feed intake, feed efficiency ratio, protein efficiency ratio, and protein productive value were observed between dietary treatments (P > 0.05).
Differential Metabolites of the Liver Between Dietary Treatments
Differential metabolites were screened from all identified metabolites based on P < 0.05, FC ≥ 1.2, or FC ≤ 0.8333 in the t-test and VIP ≥ 1 in the PLS-DA model (Figure 1). A total of 1,140 and 661 identified metabolites were obtained between the CAA and Lys-Leu treatments in the POS and the NEG modes, respectively (Supplementary Tables 1, 2 and the annotation information of metabolites is provided in Supplementary Table 3 based on compound.ID). Then, the KEGG database was used to annotate and enrich the pathway analysis of these differential metabolites from the POS and the NEG modes.
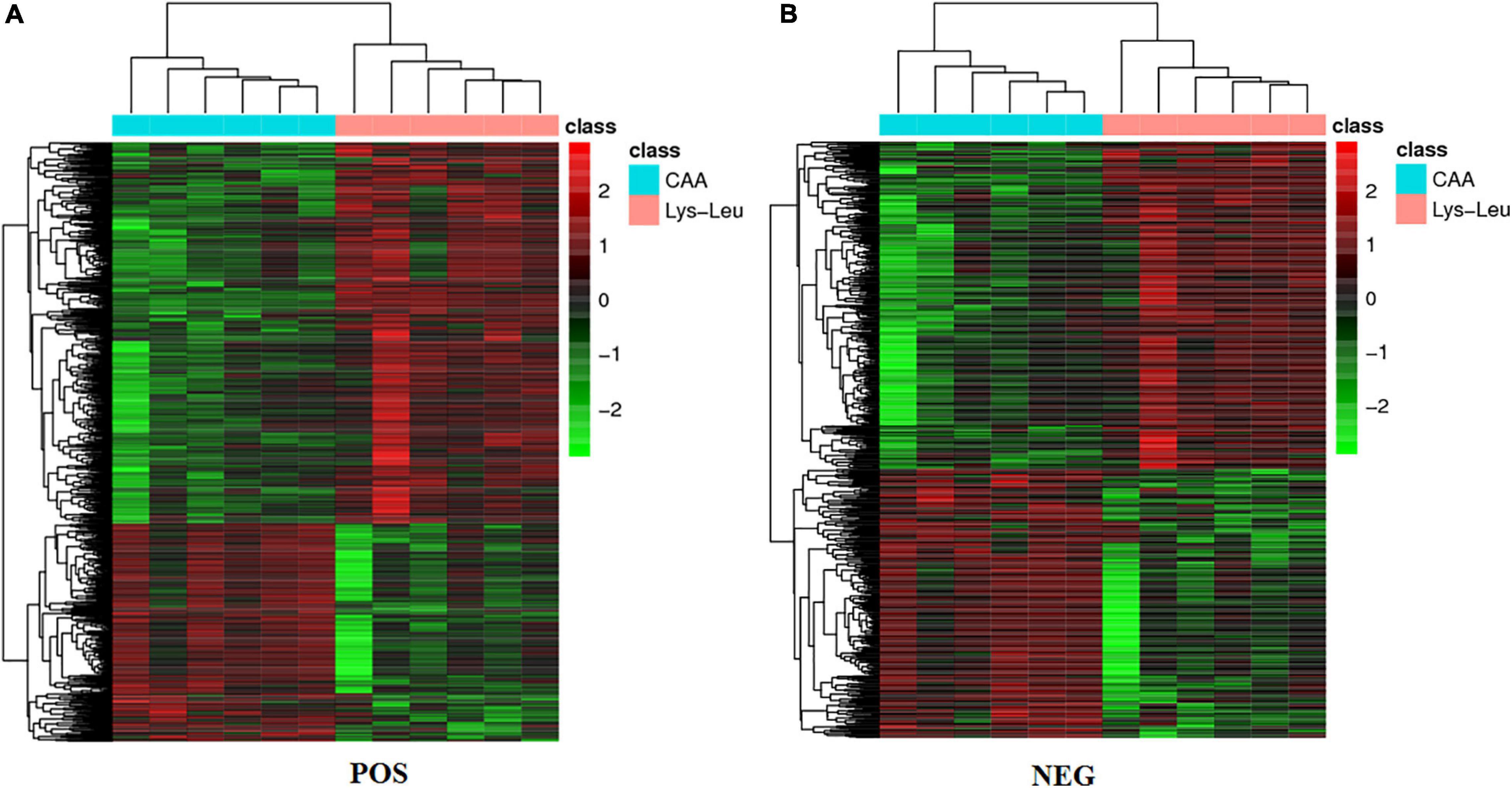
Figure 1. Heat maps of differential metabolites between the CAA and Lys-Leu treatments. (A) The positive ion mode. (B) The negative ion mode. Red color represents the value that is higher than the mean value. Green color represents the value that is lower than the mean value. Black color represents the value that is close to the mean value.
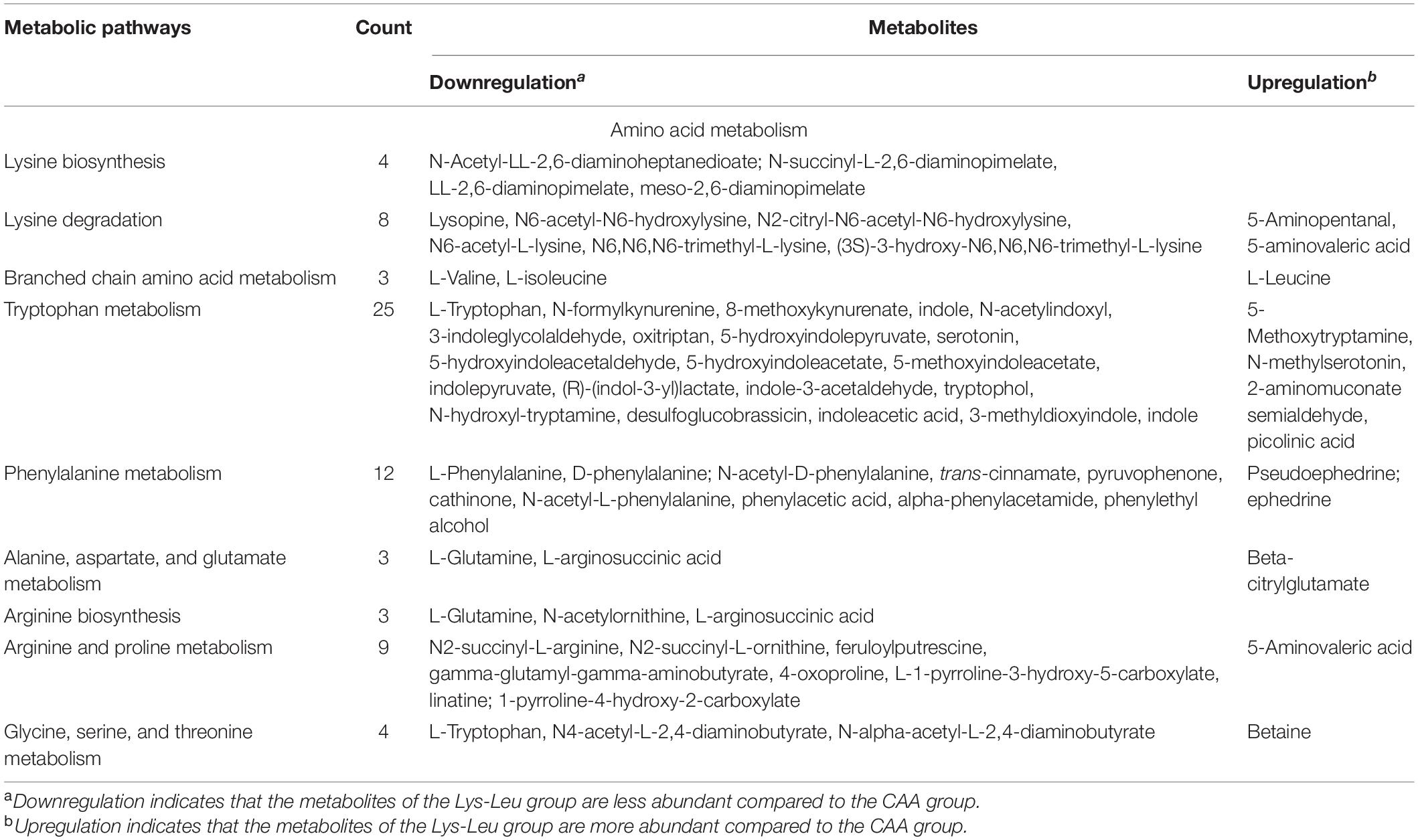
Key metabolic pathways of protein- and amino acid-related metabolic processes were selected from 148 metabolic pathways in positive ion mode and 89 metabolic pathways in negative ion mode (Table 4). Lysine biosynthesis and degradation as well as valine, leucine, and isoleucine metabolism in the liver were affected when lysine and leucine were added to diets in the form of dipeptide (Lys-Leu) or free amino acids. In addition to lysine and leucine metabolism, differential metabolites between the Lys-Leu and CAA groups were involved in other amino acid metabolisms, such as tryptophan metabolism, phenylalanine metabolism, alanine, aspartate, and glutamate metabolism, arginine biosynthesis, arginine and proline metabolism, and glycine, serine, and threonine metabolism.
Differential Proteins of the Liver Between Dietary Treatments
According to iTRAQ quantification, 935,928 spectrums were generated and 32,261 peptides and 5,708 proteins were identified with 1% FDR. A total of 618 differential proteins were identified between the Lys-Leu and CAA treatments. Among them, 431 (Supplementary Table 4) significantly downregulated and 187 (Supplementary Table 5) significantly upregulated differential proteins were screened using IQuant software based on FC > 1.2 and P < 0.05 (Figure 2). Then, the database of KEGG was used for the functional annotation of differential proteins.
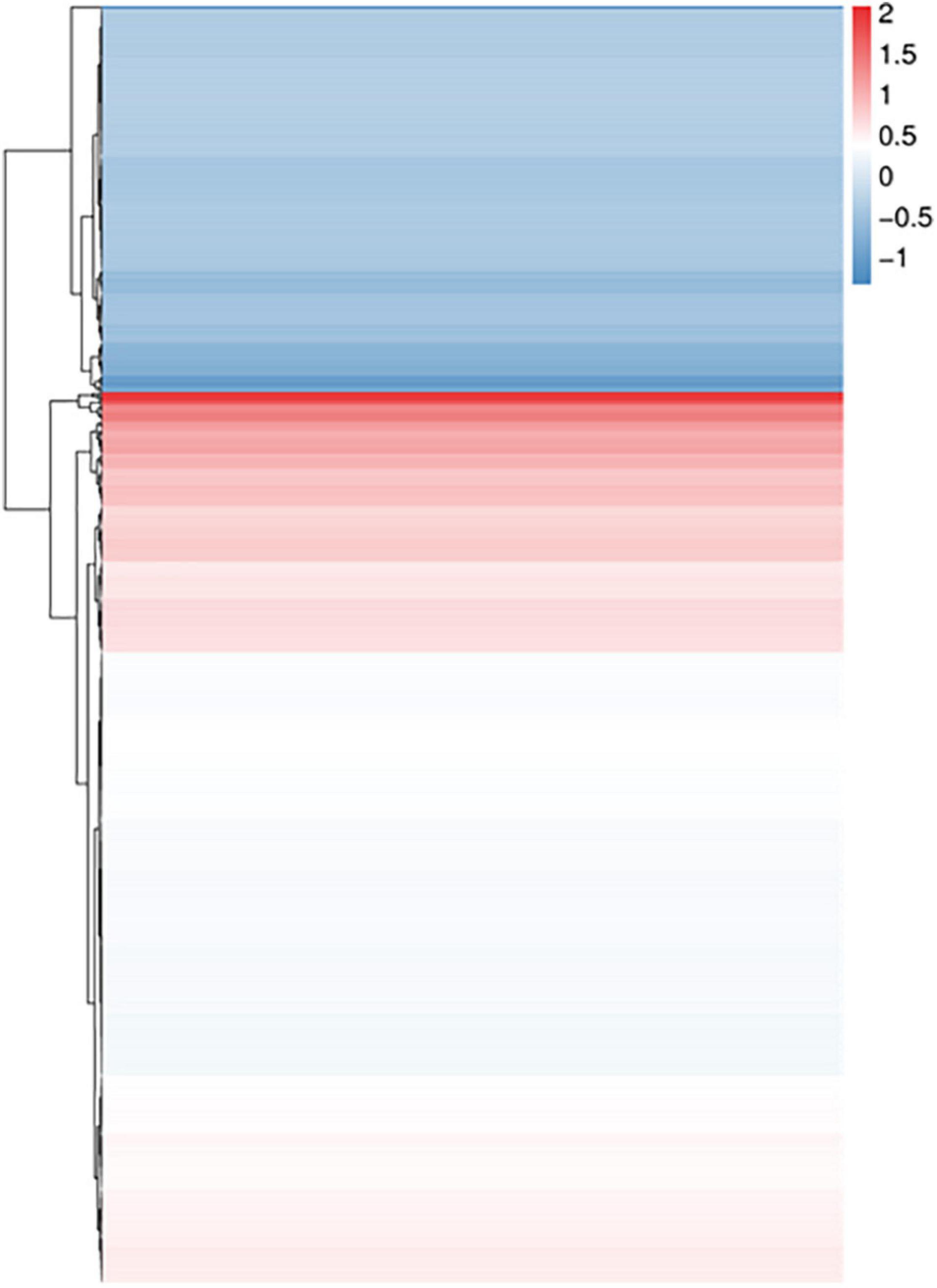
Figure 2. Cluster analysis of differential proteins between the CAA and Lys-Leu treatments. Blue color means downregulation of differential proteins, and red color means upregulation of differential proteins.
Key metabolic pathways of protein- and amino acid-related metabolic processes were selected from 305 metabolic pathways using the KEGG database (Table 5). For protein digestion and absorption, digestive enzymes including trypsin, pancreatic elastase II, carboxypeptidase A1, carboxypeptidase A2, and carboxypeptidase B as well as amino acid transporter including CD98 heavy chain (SLC3A2) were downregulated in the Lys-Leu group compared with the CAA group. For protein synthesis and degradation, the differential proteins between the Lys-Leu and CAA groups were enriched in mammalian target of rapamycin (mTOR) signaling pathway and ubiquitin-mediated proteolysis. For amino acid metabolism, the differential proteins between the Lys-Leu and CAA groups were involved in lysine degradation as well as other amino acid metabolisms (tryptophan metabolism, alanine, aspartate, and glutamate metabolism, arginine biosynthesis, arginine and proline metabolism, and glycine, serine, and threonine metabolism).
Discussion
Studies of the comparison of dietary amino acid sources in dipeptides or free amino acid forms showed that olive flounder (P. olivaceus) could utilize lysine or leucine of dipeptide form more efficiently than that of free amino acid form (Kim and Lee, 2013; Rahimnejad and Lee, 2014). The data of growth performance in this study had been provided in a separate study (Wei et al., 2020a), which suggested that similar results to those of olive flounder were observed in turbot regarding the utilization of lysine and leucine in dipeptide and free amino acid forms. However, in addition to the analysis of growth performance, our previous study only investigated the effects of the different forms of dietary amino acids on the concentrations of serum-free amino acids and the expressions of intestinal amino acids and peptide transporters (Wei et al., 2020a). This study attempted to use the approaches of integrated metabolomics and proteomics to comprehensively elucidate the effects of dietary lysine and leucine in the forms of dipeptides and free amino acids on protein- and amino acid-related metabolic processes.
Protein Digestion and Absorption
Trypsin, pancreatic elastase II, carboxypeptidase A1, carboxypeptidase A2, and carboxypeptidase B are proteolytic enzymes, and their main physiological function is to digest the protein (Cohen et al., 1981; Neurath, 1984; Zambonino Infante and Cahu, 2007). In this study, the expressions of trypsin, pancreatic elastase II, carboxypeptidase A1, carboxypeptidase A2, and carboxypeptidase B were downregulated in fish fed the Lys-Leu diet compared with fish fed the CAA diet (Table 5), which showed that dietary synthetic Lys-Leu may decrease the activities of proteolytic enzymes compared to the dietary free form of lysine and leucine. However, the growth performance in the Lys-Leu group was higher than that in the CAA group. Obviously, the activities of proteolytic enzymes and the growth were contradictory between the two treatments. A possible reason was that the proteolytic enzyme activities were related to the sampling time. At the sampling time (2 h after feeding) of this study, it happened that proteolytic enzyme activities decreased in the Lys-Leu group compared to the CAA group, but regardless of whether proteolytic enzyme activities increased or decreased between the two treatments, different forms of lysine and leucine in the feed may firstly cause changes in the activities of proteolytic enzymes and then affect the digestion of dietary protein. For amino acid absorption, the proteome profile of the liver also showed a lower expression of the heavy chain of the 4F2 antigen (4F2hc) in the Lys-Leu treatment. The 4F2hc, named CD98 heavy chain, is a type II membrane N-glycoprotein that forms heterodimers with associated transporters. These glycoprotein-associated amino acid transporters include L-type amino acid transporters (4F2hc/LAT1 and 4F2hc/LAT2), y+ L-type amino acid transporters (4F2hc/y+LAT1 and 4F2hc/y+LAT2), the alanine, serine, and cysteine transporter 1 (4F2hc/Asc-1), and the cystine and glutamate exchange transporter (4F2hc/xCT), which play a key role in the transport of a variety of amino acids including lysine and leucine (Fotiadis et al., 2013). Thus, the proteome profile of the liver in the process of protein digestion and absorption showed that the different forms of dietary lysine and leucine not only affect the digestion of dietary protein but also affect the absorption of a variety of amino acids.
Protein Synthesis and Degradation
Protein synthesis can be promoted by activating the mechanistic target of rapamycin complex 1 (mTORC1) (Laplante and Sabatini, 2012). As a member of the branched-chain amino acid family, leucine is considered as the potent activator of mTORC1 (Kim and Guan, 2011). Therefore, we hypothesized that free or dipeptide leucine in this study may impact protein synthesis by regulating the mTORC1 signaling pathway. Obviously, differential proteins in the liver supported this hypothesis (Table 5). Based on the proteome profile of the liver, the effects of dietary lysine and leucine in free and dipeptide forms on the mTORC1 signaling pathway were achieved by two upstream signals. One of the upstream signals may be phosphatidylinositol 3-kinase (PI3K)-protein kinase B (Akt) signals. Compared with fish fed the CAA diet, the mTORC1 signaling pathway was affected by downregulating the expression of insulin and protein kinase B (Akt2) and upregulating the expression of tuberous sclerosis 2 in fish fed the Lys-Leu diet (Ma and Blenis, 2009). The other upstream signal may be the cellular entry of leucine because 4F2hc/LAT1 and 4F2hc/LAT2 are considered as the primary routes (Kim and Guan, 2011). In the present study, the downregulated expression of 4F2hc in fish fed the Lys-Leu indicated that it may directly affect the mTORC1 signaling pathway through the transport of leucine.
Ubiquitin-mediated proteolysis is the primary pathway for the degradation of intracellular proteins in all tissues (Lecker et al., 2006). At the transcription level, our previous studies found that dietary dipeptides, tripeptides, and free forms of lysine and leucine may affect protein degradation by regulating the expression of muscle RING finger 1 (ubiquitin protein ligase-related gene) (Wei et al., 2020b). Similarly, in this study, differential proteins related to ubiquitin-mediated proteolysis were identified between the Lys-Leu and CAA groups (Table 5). In the ubiquitin proteasome pathway (UPP), there are distinct enzymatic components, among which E1 (ubiquitin-activating enzyme) is responsible for activating the C-terminal glycine of ubiquitin, E2s (ubiquitin carrier or conjugating proteins) are responsible for transporting the activated ubiquitin, and E3s (ubiquitin protein ligase) is responsible for catalyzing the covalent binding of ubiquitin to the substrate (Ciechanover et al., 2000; Lecker et al., 2006). Based on the differential proteins in the liver, different forms of lysine and leucine in the feed may regulate protein degradation via the E2s and E3s of the UPP.
Amino Acid Metabolism
Since this study focused on the different forms of lysine and leucine in feed, the metabolism of lysine and leucine in the liver was analyzed firstly. Lysine biosynthesis and degradation as well as branched-chain amino acid (valine, leucine, and isoleucine) metabolism were annotated using differential metabolites between dietary treatments (Table 4). The de novo biosynthesis of lysine contains two main pathways: the diaminopimelate and the α-aminoadipate pathways (Hall and da Costa, 2018). The differential metabolites showed that different forms of dietary lysine and leucine may affect the diaminopimelate pathway, which belonged to the aspartate-derived biosynthetic family. For lysine degradation, the pipecolate and saccharopine pathways were considered as the main pathways in the liver (Pena et al., 2016). However, by combining the data of metabolomics and proteomics, it was shown that dietary lysine in dipeptide and free amino acid forms may not mainly affect the two degradation pathways of lysine but also affected the synthesis of carnitine using lysine as a precursor (Vaz and Wanders, 2002). For branched-chain amino acid metabolism, since these three amino acids (leucine, isoleucine, and valine) may share common transport systems and the same enzymes of degradation (Harper et al., 1984; Peganova and Eder, 2003), antagonistic effects had been found in some fish species (Yamamoto et al., 2004). In this study, a higher level of leucine and lower levels of valine and isoleucine in liver metabolites were observed in fish fed the Lys-Leu diet compared to fish fed the CAA diet, which indicated that different forms of dietary lysine and leucine may affect the antagonism between branched-chain amino acids.
In addition to the metabolism related to lysine and leucine, the data of metabolomics and proteomics showed that tryptophan metabolism, alanine, aspartate, and glutamate metabolism, arginine biosynthesis, arginine and proline metabolism, and glycine, serine, and threonine metabolism were identified between dietary treatments (Tables 4, 5). For tryptophan metabolism, tryptophan, and leucine share 4F2hc/LAT1, which may lead to the concentrations of these two amino acids showing opposite trends in the liver (Table 4). A lower level of tryptophan in the Lys-Leu group may further affect tryptophan degradation in the downstream pathways of serotonin, indoleacetic acid, and kynuric acid (Hoseini et al., 2019). For alanine, aspartate, and glutamate metabolism, arginine biosynthesis, and arginine and proline metabolism, different forms of dietary lysine and leucine may mainly affect glutamine and glutaminase, which may be a central mediator metabolite and enzyme. Glutamine may not only be used as the carbon for arginine and proline synthesis but may also be converted into glutamate by glutaminase (Curthoys and Watford, 1995; Morris, 2016). In glycine, serine, and threonine metabolism, the choline–glycine pathway may be responsible for the methyl donor (Wang et al., 2013). According to metabolomics and proteomics data, dietary Lys-Leu upregulated the level of betaine and downregulated the enzyme activity of betaine-homocysteine S-methyltransferase. Thus, it implied that different forms of amino acids in the feed may affect the supply of methyl via the choline–glycine pathway.
Conclusion
In this study, a total of 1,140 (POS mode) and 661 (NEG mode) differential metabolites as well as a total of 618 differential proteins were identified between the Lys-Leu and CAA treatments. Based on integrated data of differential metabolites and proteins, the protein- and amino acid-related metabolic processes of the liver included protein digestion and absorption, protein synthesis and degradation, and amino acid metabolism. In the process of protein metabolism, the differential proteins of the liver were related to proteolytic enzymes and amino acid transporters as well as mTORC1 and UPPs, respectively. In the process of amino acid metabolism, differential metabolites and proteins of the liver were related to lysine degradation, tryptophan metabolism, alanine, aspartate, and glutamate metabolism, arginine biosynthesis, arginine and proline metabolism, and glycine, serine, and threonine metabolism. Thus, the mechanism of the effects of dietary lysine and leucine on the growth of turbot can be understood from three aspects: protein digestion and absorption, protein synthesis and degradation, and amino acid metabolism. In addition, amino acid metabolism, in this study, revealed a good correlation between differential metabolites and protein using the approaches of integrated LC–MS/MS-based metabolomics and iTRAQ-based proteomics.
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD025276.
Ethics Statement
The animal study was reviewed and approved by Institutional Animal Care and Use Committee of the Yellow Sea Fisheries Research Institute.
Author Contributions
ML, YW, and HX conceived and designed this experiment, drafted and reviewed the manuscript. BL accomplished turbot rearing, the feeding trial, and the sampling process. All authors reviewed and commented on the manuscript.
Funding
This research was supported by the National Natural Science Foundation of China (31902387, 31972803, and 31672663), the Central Public-Interest Scientific Institution Basal Research Fund, CAFS (2020TD48), and China Agriculture Research System of MOF and MARA (CARS-47).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2021.691404/full#supplementary-material
Footnotes
References
Ceciliani, F., Lecchi, C., Urh, C., and Sauerwein, H. (2018). Proteomics and metabolomics characterizing the pathophysiology of adaptive reactions to the metabolic challenges during the transition from late pregnancy to early lactation in dairy cows. J. Proteomics 178, 92–106. doi: 10.1016/j.jprot.2017.10.010
Chen, X. Q., Elsheikha, H. M., Hu, R. S., Hu, G. X., Guo, S. L., Zhou, C. X., et al. (2018). Hepatic metabolomics investigation in acute and chronic murine toxoplasmosis. Front. Cell. Infect. Microbiol. 8:189. doi: 10.3389/fcimb.2018.00189
Ciechanover, A., Orian, A., and Schwartz, A. L. (2000). Ubiquitin-mediated proteolysis: biological regulation via destruction. Bioessays 22, 442–451. doi: 10.1002/(SICI)1521-1878(200005)22:5<442::AID-BIES6<3.0.CO;2-Q
Cohen, T., Gertler, A., and Birk, Y. (1981). Pancreatic proteolytic enzymes from carp (Cyprinus carpio)-II. Kinetic properties and inhibition studies of trypsin, chymotrypsin and elastase. Comp. Biochem. Physiol. B 69, 647–653. doi: 10.1016/0305-0491(81)90365-5
Curthoys, N. P., and Watford, M. (1995). Regulation of glutaminase activity and glutamine metabolism. Annu. Rev. Nutr. 15, 133–159. doi: 10.1146/annurev.nu.15.070195.001025
Dabrowski, K., Lee, K. J., and Rinchard, J. (2003). The smallest vertebrate, teleost fish, can utilize synthetic dipeptide-based diets. J. Nutr. 133, 4225–4229. doi: 10.1093/jn/133.12.4225
Daniel, N. (2017). Status of aquaculture with respect to nutrition and feed. Int. J. Fish. Aquat. Stud. 5, 333–345.
de Vareilles, M., Conceição, L. E., Gómez-Requeni, P., Kousoulaki, K., Richard, N., Rodrigues, P. M., et al. (2012). Dietary lysine imbalance affects muscle proteome in zebrafish (Danio rerio): a comparative 2D-DIGE study. Mar. Biotechnol. 14, 643–654. doi: 10.1007/s10126-012-9462-3
Deborde, C., Hounoum, B. M., Moing, A., Maucourt, M., Jacob, D., Corraze, G., et al. (2021). Putative imbalanced amino acid metabolism in rainbow trout long term fed a plant-based diet as revealed by 1H-NMR metabolomics. J. Nutr. Sci. 10:e13. doi: 10.1017/jns.2021.3
Dong, X., Liu, Q., Kan, D., Zhao, W., Guo, H., and Lv, L. (2020). Effects of ammonia-N exposure on the growth, metabolizing enzymes, and metabolome of Macrobrachium rosenbergii. Ecotoxicol. Environ. Safe. 189:110046. doi: 10.1016/j.ecoenv.2019.110046
El-Husseiny, O., El-Haroun, E. R., Goda, A., Hassan, I., Woodward, B., and Suloma, A. (2017). Effects of lysine and tryptophan supplementations in plant protein-based diets on the performance of Nile tilapia (Oreochromis niloticus). J. Appl. Aquac. 29, 266–276. doi: 10.1080/10454438.2017.1364684
Fotiadis, D., Kanai, Y., and Palacín, M. (2013). The SLC3 and SLC7 families of amino acid transporters. Mol. Aspects Med. 34, 139–158. doi: 10.1016/j.mam.2012.10.007
Furuya, W. M., Pezzato, L. E., Barros, M. M., Pezzato, A. C., Furuya, V. R., and Miranda, E. C. (2004). Use of ideal protein concept for precision formulation of amino acid levels in fish-meal-free diets for juvenile Nile tilapia (Oreochromis niloticus L.). Aquac. Res. 35, 1110–1116. doi: 10.1111/j.1365-2109.2004.01133.x
Gómez-Requeni, P., de Vareilles, M., Kousoulaki, K., Jordal, A. E. O., Conceição, L. E., and Rønnestad, I. (2011). Whole body proteome response to a dietary lysine imbalance in zebrafish Danio rerio. Comp. Biochem. Physiol. Part D Genomics Proteomics 6, 178–186. doi: 10.1016/j.cbd.2011.02.002
Guo, C., Sun, D., Wang, X., and Mao, S. (2019). A combined metabolomic and proteomic study revealed the difference in metabolite and protein expression profiles in ruminal tissue from goats fed hay or high-grain diets. Front. Physiol. 10:66. doi: 10.3389/fphys.2019.00066
Hall, C. J., and da Costa, T. P. S. (2018). Lysine: biosynthesis, catabolism and roles. Wiki J. Sci. 1:4. doi: 10.15347/wjs/004
Harper, A. E., Miller, R., and Block, K. P. (1984). Branched-chain amino acid metabolism. Ann. Rev. Nutr. 4, 409–454. doi: 10.1146/annurev.nu.04.070184.002205
Hoseini, S. M., Pérez-Jiménez, A., Costas, B., Azeredo, R., and Gesto, M. (2019). Physiological roles of tryptophan in teleosts: current knowledge and perspectives for future studies. Rev. Aquacult. 11, 3–24. doi: 10.1111/raq.12223
Kamaszewski, M., Ostaszewska, T., Napora-Rutkowski, Ł, Wójcik, M., and Dabrowski, K. (2019). The role of dipeptide on fish growth and digestive enzyme activity modulation in common carp (Cyprinus carpio L.). Anim. Sci. Pap. Rep. 37, 75–86.
Kamaszewski, M., Prasek, M., Ostaszewska, T., and Dabrowski, K. (2014). The influence of feeding diets containing wheat gluten supplemented with dipeptides or free amino acids on structure and development of the skeletal muscle of carp (Cyprinus carpio). Aquac. Int. 22, 259–271. doi: 10.1007/s10499-013-9683-0
Kamaszewski, M., Wójcik, M., Krawczyńska, A., and Ostaszewska, T. (2020). The influence of diet containing wheat gluten supplemented with dipeptides or amino acids on the morphology of white muscle of yellow perch (Perca flavescens). Animals 10:388. doi: 10.3390/ani10030388
Kim, J., and Guan, K. L. (2011). Amino acid signaling in TOR activation. Annu. Rev. Biochem. 80, 1001–1032. doi: 10.1146/annurev-biochem-062209-094414
Kim, S. S., and Lee, K. J. (2013). Comparison of leucine requirement in olive flounder (Paralichthys olivaceus) by free or synthetic dipeptide forms of leucine. Anim. Feed Sci. Tech. 183, 195–201. doi: 10.1016/j.anifeedsci.2013.05.008
Laplante, M., and Sabatini, D. M. (2012). mTOR signaling in growth control and disease. Cell 149, 274–293. doi: 10.1016/j.cell.2012.03.017
Lecker, S. H., Goldberg, A. L., and Mitch, W. E. (2006). Protein degradation by the ubiquitin–proteasome pathway in normal and disease states. J. Am. Soc. Nephrol. 17, 1807–1819. doi: 10.1681/ASN.2006010083
Ma, X. M., and Blenis, J. (2009). Molecular mechanisms of mTOR-mediated translational control. Nat. Rev. Mol. Cell Bio. 10, 307–318. doi: 10.1038/nrm2672
Mamauag, R. E. P., Gao, J., Thanh Nguyen, B., Ragaza, J. A., Koshio, S., Ishikawa, M., et al. (2012). Supplementations of dl-methionine and methionine dipeptide in diets are effective for the development and growth of larvae and juvenile red sea bream, Pagrus major. J. World Aquac. Soc. 43, 362–374. doi: 10.1111/j.1749-7345.2012.00563.x
Min, C. W., Hyeon, H., Gupta, R., Park, J., Cheon, Y. E., Lee, G. H., et al. (2020). Integrated proteomics and metabolomics analysis highlights correlative metabolite-protein networks in soybean seeds subjected to warm-water soaking. J. Agric. Food Chem. 68, 8057–8067. doi: 10.1021/acs.jafc.0c00986
Morris, S. M. Jr. (2016). Arginine metabolism revisited. J. Nutr. 146, 2579S–2586S. doi: 10.3945/jn.115.226621
Neurath, H. (1984). Evolution of proteolytic enzymes. Science 224, 350–357. doi: 10.1126/science.6369538
Ostaszewska, T., Dabrowski, K., Kamaszewski, M., Kwasek, K., Grodzik, M., and Bierla, J. (2013). The effect of dipeptide, Lys-Gly, supplemented diets on digestive tract histology in juvenile yellow perch (Perca flavescens). Aquac. Nutr. 19, 100–109. doi: 10.1111/j.1365-2095.2012.00948.x
Peganova, S., and Eder, K. (2003). Interactions of various supplies of isoleucine, valine, leucine and tryptophan on the performance of laying hens. Poultry Sci. 82, 100–105. doi: 10.1093/ps/82.1.100
Pena, I. A., Marques, L. A., Laranjeira, A. B., Yunes, J. A., Eberlin, M. N., and Arruda, P. (2016). Simultaneous detection of lysine metabolites by a single LC–MS/MS method: monitoring lysine degradation in mouse plasma. Springerplus 5:172. doi: 10.1186/s40064-016-1809-1
Peres, H., and Oliva-Teles, A. (2005). The effect of dietary protein replacement by crystalline amino acid on growth and nitrogen utilization of turbot Scophthalmus maximus juveniles. Aquaculture 250, 755–764. doi: 10.1016/j.aquaculture.2005.04.046
Rahimnejad, S., and Lee, K. J. (2014). Comparison of free and dipeptide lysine utilization in diets for juvenile olive flounder Paralichthys olivaceus. Fish. Aquat. Sci. 17, 433–439. doi: 10.5657/FAS.2014.0433
Rodrigues, P. M., Silva, T. S., Dias, J., and Jessen, F. (2012). Proteomics in aquaculture: applications and trends. J. Proteomics 75, 4325–4345. doi: 10.1016/j.jprot.2012.03.042
Sales, N. M. R., Pelegrini, P. B., and Goersch, M. C. (2014). Nutrigenomics: definitions and advances of this new science. J. Nutr. Metabol. 2014:202759. doi: 10.1155/2014/202759
Savitski, M. M., Wilhelm, M., Hahne, H., Kuster, B., and Bantscheff, M. (2015). A scalable approach for protein false discovery rate estimation in large proteomic data sets. Mol. Cell. Proteomics 14, 2394–2404. doi: 10.1074/mcp.M114.046995
Vaz, F. M., and Wanders, R. J. (2002). Carnitine biosynthesis in mammals. Biochem. J. 361, 417–429. doi: 10.1042/bj3610417
Wang, W., Wu, Z., Dai, Z., Yang, Y., Wang, J., and Wu, G. (2013). Glycine metabolism in animals and humans: implications for nutrition and health. Amino Acids 45, 463–477. doi: 10.1007/s00726-013-1493-1
Wei, Y., Li, B., Xu, H., and Liang, M. (2020a). Effects of lysine and leucine in free and different dipeptide forms on the growth, amino acid profile and transcription of intestinal peptide, and amino acid transporters in turbot (Scophthalmus maximus). Fish Physiol. Biochem. 46, 1795–1807. doi: 10.1007/s10695-020-00828-2
Wei, Y., Liang, M., Mai, K., Zheng, K., and Xu, H. (2017a). 1H NMR−based metabolomics studies on the effect of size-fractionated fish protein hydrolysate, fish meal and plant protein in diet for juvenile turbot (Scophthalmus maximus L.). Aquac. Nutr. 23, 523–536. doi: 10.1111/anu.12420
Wei, Y., Liang, M., Mai, K., Zheng, K., and Xu, H. (2017b). The effect of ultrafiltered fish protein hydrolysate levels on the liver and muscle metabolic profile of juvenile turbot (Scophthalmus maximus L.) by 1H NMR-based metabolomics studies. Aquac. Res. 48, 3515–3527. doi: 10.1111/are.13178
Wei, Y., Xu, H., and Liang, M. (2020b). Amino acid absorption and protein synthesis responses of turbot Scophthalmus maximus to lysine and leucine in free, dipeptide and tripeptide forms. Aquac. Nutr. 26, 358–367. doi: 10.1111/anu.12998
Wen, B., Mei, Z., Zeng, C., and Liu, S. (2017). metaX: a flexible and comprehensive software for processing metabolomics data. BMC Bioinformatics 18:183. doi: 10.1186/s12859-017-1579-y
Wen, B. O., Zhou, R., Feng, Q., Wang, Q., Wang, J., and Liu, S. (2014). IQuant: an automated pipeline for quantitative proteomics based upon isobaric tags. Proteomics 14, 2280–2285. doi: 10.1002/pmic.201300361
Wu, X., Pan, X., Cao, S., Xu, F., Lan, L., Zhang, Y., et al. (2019). iTRAQ-based quantitative proteomic analysis provides insights into strong broodiness in Muscovy duck (Cairina moschata) combined with metabolomics analysis. J. Proteomics 204, 103401. doi: 10.1016/j.jprot.2019.103401
Yamamoto, T., Shima, T., and Furuita, H. (2004). Antagonistic effects of branched-chain amino acids induced by excess protein-bound leucine in diets for rainbow trout (Oncorhynchus mykiss). Aquaculture 232, 539–550. doi: 10.1016/S0044-8486(03)00543-X
Yu, K., Matzapetakis, M., Horvatić, A., Terré, M., Bach, A., Kuleš, J., et al. (2020). Metabolome and proteome changes in skeletal muscle and blood of pre-weaning calves fed leucine and threonine supplemented diets. J. Proteomics 216, 103677. doi: 10.1016/j.jprot.2020.103677
Keywords: dipeptide, free amino acid, liver, metabolomics, proteomics, turbot
Citation: Wei Y, Li B, Xu H and Liang M (2021) Liver Metabolome and Proteome Response of Turbot (Scophthalmus maximus) to Lysine and Leucine in Free and Dipeptide Forms. Front. Mar. Sci. 8:691404. doi: 10.3389/fmars.2021.691404
Received: 06 April 2021; Accepted: 26 May 2021;
Published: 23 June 2021.
Edited by:
Songlin Li, Shanghai Ocean University, ChinaReviewed by:
Fei Song, South China Normal University, ChinaMansour Torfi Mozanzadeh, South Iran Aquaculture Research Center, Iran
Dizhi Xie, South China Agricultural University, China
Copyright © 2021 Wei, Li, Xu and Liang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mengqing Liang, liangmq@ysfri.ac.cn
 Yuliang Wei
Yuliang Wei