
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci. , 24 February 2022
Sec. Marine Fisheries, Aquaculture and Living Resources
Volume 9 - 2022 | https://doi.org/10.3389/fmars.2022.843637
This article is part of the Research Topic Omics and Fish Nutrition View all 12 articles
Reducing dependency on dietary fish meal (FM) and fish oil (FO) is extremely important for the sustainable development of the aquaculture industry. However, the metabolic consequences and mechanisms underlying the replacement of dietary FM and FO by terrestrial proteins (TPs) and lipids remain unclear. To reveal the effects of replacing dietary FM and/or FO on the metabolic changes, the integrated analysis of metabolomics and transcriptomics were employed to evaluate the changes in metabolites and genes of rainbow trout (Oncorhynchus mykiss) feeding different experimental diets. Four diets were formulated for the 84-day duration of the experiment: control group (FMFO), FM and vegetable oil (FMVO), terrestrial protein and FO (TPFO), and terrestrial protein and vegetable oil (TPVO). Integrated metabolomic and transcriptomic analyses revealed the significant difference in the metabolic pathways of O. mykiss among the three replacement schemes, i.e., single replacement of dietary FM by TP, single replacement of dietary FO by VO, and combined replacement of FM by TP and FO by VO. The combined replacement of FM and FO by TP and VO, respectively, disturbed immune function, energy metabolism, cellular protein biosynthesis capacity, and lipid metabolism of O. mykiss. The reduction of antioxidant capacity was only observed in individuals feeding diets with replacement of FM by TP. Furthermore, as soon as the dietary FM and/or FO were reduced, cellular protein biosynthesis ability was suppressed and accompanied by higher energy consumption in response to fluctuations of dietary quality, resulting in reduced growth performance. Interestingly, adenylosuccinate and adenosine monophosphate involved in purine metabolism were induced by both individual and combined replacement of FM and FO by TPs and lipids, respectively. It suggested that these two metabolites might be potential biomarkers for O. mykiss fed diets with reduction of FM and/or FO. This study constitutes a new understanding of the molecular and metabolic mechanisms of O. mykiss in response to the replacement of dietary FM and/or FO by TP and/or VO, respectively, and built a theoretical basis for further improvement of aquafeed formulation and sustainable development of aquaculture.
Fish meal (FM) and fish oil (FO) are known as the most desirable ingredients used in the formulation of aquafeeds (Turchini et al., 2019). Firstly, FM is regarded as the most reliable protein source owing to its relatively high protein level, excellent amino acid (AA) composition, good palatability, and digestibility to fulfill the dietary requirement of aquatic species (Olsen and Hasan, 2012). Moreover, FO constitutes one of the major dietary components in aquafeed to provide energy and essential fatty acids (EFA), especially long-chain ω3 polyunsaturated fatty acids, which are short in vegetable oil (VO) (Geay et al., 2015). Such long-chain unsaturated FAs are crucial for the growth, health, and development of aquatic animals (Tocher, 2010). However, overfishing of wild fish to produce FM and FO for aquaculture use is unsustainable and as such reducing the reliance on aquaculture feeds, composed of FM and FO is essential for the sustainable development aquaculture industry (Klinger and Naylor, 2012).
The possibility of feeding farmed fish with diets based on various alternative protein sources, mainly terrestrial proteins (TPs), has been evaluated by numerous studies (Panserat et al., 2009; Glencross et al., 2011; Messina et al., 2013; Oliva-Teles et al., 2015; Rimoldi et al., 2018; Choi et al., 2019; Randazzo et al., 2021). Plant proteins are considered the desirable alternative source for FM because of their reasonable price, consistent quality, and stable supply (Oliva-Teles et al., 2015). However, the unbalanced AAs profiles and presence of antinutritional factors (ANFs) limit the application of plant-based proteins in aquafeed (Krogdahl et al., 2003; Espe et al., 2006). The depression of growth performance caused by the replacement of about 80% dietary FM with one protein source has been documented in carnivorous fish species (Dias et al., 2005). Animal proteins typically have adequate AA profiles, suitable digestible phosphorus, and the absence of ANFs compared to plant proteins. However, the relatively high ash content in some animal proteins can negatively affect the utilization of dietary macromolecules by aquatic species, which results in repressed growth performance (Bureau et al., 1999; Moreno-Arias et al., 2018). Therefore, one strategy to overcome such shortcomings is to combine multiple alternative dietary protein sources to achieve a complementary effect and to offset the undesirable ingredients present in the alternative sources, thus improving the growth performance of farmed fish (Li S. et al., 2021). In addition to the unfavorable components of alternative protein sources, the energy metabolism of fish needs to be considered. It is known that due to the limitation of carbohydrate utilization in fish, dietary proteins (especially several AAs) are typically utilized preferentially as fuel for energy metabolism in fish (Jia et al., 2017). Therefore, the development of alternative FM diets must ensure a sufficient supply of AAs for protein synthesis and energy metabolism.
As regards FO, VO is widely considered as the potential alternative ingredient that can effectively replace dietary FO at high levels without significant effect on the growth performance of fish (Hixson et al., 2014; Betancor et al., 2015; Oliva-Teles et al., 2015; Grayson and Dabrowski, 2020; Lu et al., 2020). Despite the desired positive results, the FA composition of VO can negatively affect the lipid metabolism of the farmed fish. Additionally, previous studies indicated that unbalanced dietary FA profiles can induce intestinal enteritis and oxidative stress, which is the response of the immune system to external physical or chemical stimuli (Petropoulos et al., 2009). Thus, both the growth performance of the fish and the quality of the final product needs to be taken into account when replacing dietary FO with alternative sources. For this consideration, several solutions have been developed to reduce the negative effects of replacing FO with VO on the metabolism and product quality of aquatic species. For example, the use of new oil sources (Hixson et al., 2014; Grayson and Dabrowski, 2020) and transgenic plants (Betancor et al., 2015) have been tested. As such, further understanding of lipid metabolism in fish feeding diets with replacement of FO by alternative sources is required.
Considering that partial replacement of dietary FM or FO has been successfully achieved, it is feasible to conduct investigations to evaluate the effects of combined replacement of dietary FM and FO. Previous studies have investigated the effects of combined replacement of FM and FO on fish. For instance, Torrecillas et al. (2017) successfully reduced dietary FM and FO levels of European sea bass (Dicentrarchus labrax) down to 10 and 3%, respectively. Glencross et al. (2016) achieved the total replacement of dietary FO by ricebran oil and 90% dietary FM by TP in Asian seabass (Lates calcarifer). Uncertainty and controversy exist regarding the effects of supplementary TP and lipid in feed on fish metabolism and growth. A high level of dietary FM and FO substitution, for example, resulted in reduced growth performance after 1 year due to the unbalanced AA profiles (Torstensen et al., 2008). Moreover, most current studies about the replacement of dietary FM and FO were focused on the growth performance of fish while reports regarding the effects of such replacement on fish metabolism are scarce so far.
Fish metabolism involves a series of complex biological processes with the regulations of numerous genes and the production of various metabolites. Recently, metabolomics and transcriptomics have been increasingly used to study the physiological and molecular mechanisms of organisms in response to external stimuli (Yan et al., 2018; Kong et al., 2020). Transcriptome analysis can help to determine important information about gene expression levels and is a valuable tool for understanding biological information. However, transcriptome sequencing cannot reflect the metabolic levels in an organism. Meanwhile, metabolomics has been successfully employed in nutrition research because it can reveal the metabolite profiles of an organism and further clarification on its metabolic response to various dietary ingredients (Wei et al., 2017; Alfaro and Young, 2018). As such, a simultaneous analysis combining transcriptomic and metabolomic information can compensate for the shortage of the application of a single method and improve the systematics and accuracy for understanding the physiological processes and the metabolic and molecular mechanisms of the target organism (Hao et al., 2019; Li Y. et al., 2021).
As the second-largest salmonid species in terms of total production worldwide, rainbow trout (Oncorhynchus mykiss) is one of the most commercially important species (FAO, 2020). O. mykiss aquaculture consumes considerable FM and FO as the main ingredients of fish diets. As such, it is economically and ecologically significant to minimize the usage of FM and FO by means of replacing such feed ingredients with alternative terrestrial sources. The present investigation is aimed to evaluate the effects of combined replacement of dietary FM and FO on the metabolism of O. mykiss and the molecular mechanism based on the integrated information from transcriptomic and metabolomic analyses to provide a scientific base for the development of feed techniques in O. mykiss farming.
Four experimental diets with isonitrogenous (461.9 g/kg) and isolipidic (160.4 g/kg) were formulated. The use of alternative sources of FM and FO in the present study was referred to previous studies on O. mykiss (Panserat et al., 2009; Rimoldi et al., 2018), and the feed formulation was appropriately modified according to the nutritional requirements of juvenile O. mykiss (Guillaume et al., 2001). The specific compositions of the four diets are as follows: (1) FMFO diet only containing FM and FO as the protein and lipid sources, respectively, (2) FMVO diet containing FM and 80% VO plus 20% FO, (3) TPFO diet containing FO and 70% TP plus 30% FM, (4) TPVO diet containing 70% TP plus 30% FM and 80% VO plus 20% FO. The feed formulation and proximate analysis are shown in Table 1. All feed ingredients were completely mixed and pelletized, dried in an oven at 55°C for 16 h, and frozen at −20°C for future use.
The feeding experiment was carried out in Haiyang Yellow Sea Aquatic Product Co., Ltd. (Shandong, China). All O. mykiss obtained from Wanzefeng Fishery Company (Rizhao, Shandong, China) were acclimated to the experimental conditions for 14 days. A total of 240 O. mykiss with an average weight of 38.09 ± 0.73 g were randomly assigned to 12 aquariums with a volume of 400 L, i.e., 20 fish in each tank. Three tanks of fish were grouped as one of the four feed treatments and fed with one of the four diets three times every day (at 8:00, 15:00, and 21:00) for 84 consecutive days. Feces and residual feeds were collected after one hour of feeding, and the weight of residual feed was recorded. The temperature was kept constant at 16 ± 0.5 °C with automatic regulators. The dissolved oxygen was kept at no less than 6.0 mg L–1. An artificially controlled photoperiod was established as 12 h light: 12 h dark during the feeding experiment. Sufficient aeration ensured homogenous environmental conditions for each aquarium.
All individuals in each replicate were weighed to determine the final body weight (FBW), specific growth rate (SGR), feed efficiency (FE), protein efficiency ratio (PER), and lipid efficiency ratio (LER) at the end of the experiment. All individuals were anesthetized before tissue sample collection. Condition factor (K) and hepatosomatic index (HSI) of O. mykiss were calculated using the body weight, length, and liver weight of four fish from each tank. And feces samples were collected for apparent digestibility coefficient (ADC) analyses. Three individuals were collected from each group for RNA-seq, and six fish from each treatment were collected for metabolomic analysis. And the liver of each individual was quickly removed and frozen at −80°C ultra-low temperature refrigerator until subsequent multi-omics analysis.
TRIzol® Reagent was applied to extract the total RNA from the liver and genomic DNA was cleared by DNase I (TaKara). RNA quality was detected by 2100 Bioanalyser (Agilent) and ND-2000 (NanoDrop Technologies) was applied for quantification. Isolation of mRNA by oligo(dT) beads following the polyA selection method, and sequencing libraries were constructed by the TruSeqTM RNA sample of preparation Kit from Illumina (San Diego, CA, United States). The Illumina HiSeq X Ten/NovaSeq 6000 sequencer was employed to sequence the Paired-end RNA-seq (2 × 150 bp read length). Finally, the clean reads were obtained from the raw reads being trimmed. All RNA-seq data have been uploaded to the NCBI Sequence Read Archive (SRA) database, accession NO: PRJNA788999. Gene abundances were quantified by RSEM1 and normalized based on transcripts per million reads value. Essentially, differential expression analysis was performed using the DESeq2 with P < 0.05, and |log2FC|> 1. In addition, the Kyoto Encyclopedia of Genes and Genomes (KEGG) database was applied to detect which differentially expressed genes (DEGs) were significantly enriched at P < 0.05. KOBAS2 was applied for KEGG pathway analysis (Xie et al., 2011).
Ten DEGs were randomly selected to validate the RNA-seq results by qRT-PCR. Primers for qRT-PCR were designed using Primer Premier 5.0 software, and primers’ details were listed in Supplementary Table 1. EF1α and β-actin were employed as the internal control for qRT-PCR. The temperature programming conditions were: denaturation at 95°C for 30 s, followed by 35 cycles of 95°C for 10 s, 60°C for 30 s, 95°C for 15 s, 60°C for 60 s, and 95°C for 1 s. Gene expression results were obtained using the 2–ΔΔCt method (Livak and Schmittgen, 2001).
For metabolomics analysis, the 50 mg solid sample from each group was separated with 400 μL of the solution containing methanol and water 4:1 (v/v). The samples were homogenized by a tissue crusher at 50 Hz for 6 min and then sonicated for 30 min at 40 kHz and 5°C. The samples were placed for 30 min at −20°C to achieve protein precipitation. The mixture was centrifuged at 4°C, 13,000g for 15 min, and the supernatant was separated for subsequent analysis.
The LC-MS analysis was conducted using an ExionLCTMAD system (AB Sciex, United States) equipped with an ACQUITY UPLC BEH C18 column (100 mm × 2.1 mm i.d., 1.7 μm; Waters, Milford, CT, United States). The UPLC system worked in tandem with a quadrupole time-of-flight mass spectrometer (Triple TOF™ 5600+, AB Sciex, United States) that has an electrospray ionization (ESI) source with both positive and negative ionization modes of operation.
The positive and negative data obtained from the LC-MS analysis were combined and loaded into SIMCA software (V14.1, Umea, Sweden) for orthogonal projections for latent structures-discriminant analysis (OPLS-DA). Afterward, sevenfold cross-validation was performed. Differential metabolites (DMs) were identified according to the variable importance in projection values (VIP > 1) obtained from the OPLS-DA model and P < 0.05 from the Student’s t-test. KEGG3 was employed for annotating metabolites and searching for metabolite pathways. The scipy stats4 was employed to confirm the statistically significantly enriched pathway by Fisher’s exact test.
Kyoto Encyclopedia of Genes and Genomes was applied to annotate and enrich the metabolic pathways mapped by DMs and DEGs of O. mykiss, and DMs and DEGs enriched to the shared KEGG pathways were filtered for subsequent analysis. Pearson correlation analysis was utilized to identify the correlation between DMs and DEGs O. mykiss using the Cytoscape software. A heatmap was used to show the correlation of the DMs and DEGs, respectively. P < 0.05 was considered a statistically significant level of difference.
The growth performance of fish was determined by the following equations:
where Wf and W0 represent final and initial body weight of O. mykiss, t represents the duration of experiment. L represents the length of O. mykiss.
All results are expressed as mean ± SD. The growth performance of O. mykiss was analyzed using two-way ANOVA with the percentage of dietary FM or FO as fixed factors. P < 0.05 indicated that the data has statistically significant differences. All analyses were performed on the SPSS 21.0 software.
After the 84-d experiment, no interaction term between dietary levels of FM and FO on growth performance or feed utilization were observed in O. mykiss (Table 2). Significant differences in the growth performance in terms of FBW, SGR, and K and the feed utilization in terms of ADC were observed between the two FM inclusion levels (P < 0.05) and HSI showed a significant difference in O. mykiss between the two FO levels (P < 0.05). In addition, there was no significant difference of FE, PER, and LER in O. mykiss fed diets with replacement of dietary FM or FO by TP or VO, respectively (P > 0.05).
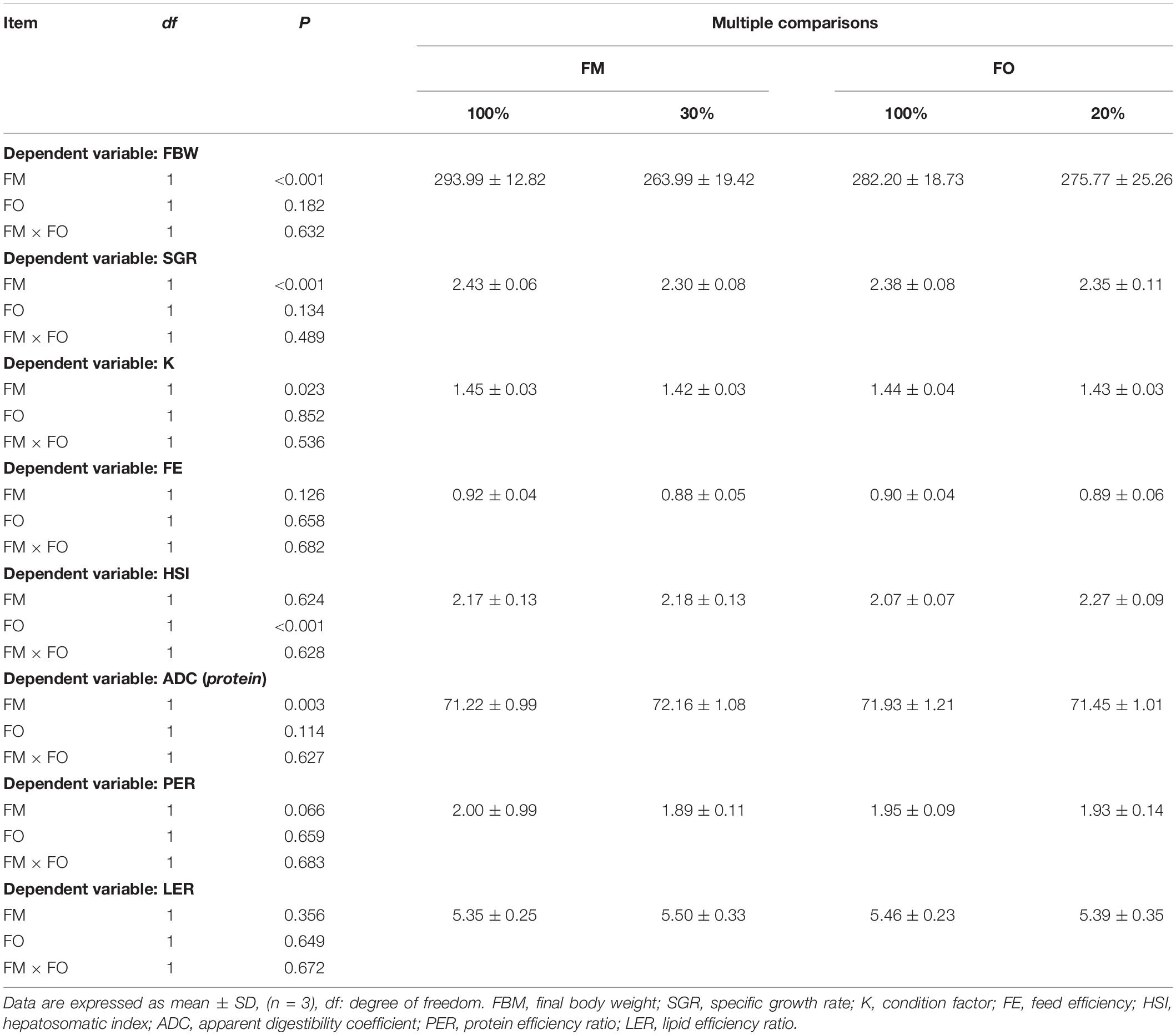
Table 2. Two-way factorial analysis of variance (ANOVA) for the growth performance and feed utilization of O. mykiss fed different diets.
A total of 518 and 93 DEGs were identified between FMFO vs. TPFO and FMVO vs. TPVO, respectively. There were 24 overlapping DEGs between FMFO vs. TPFO and FMVO vs. TPVO (Figure 1A). In the comparison of FMFO vs. TPFO, 378 and 140 genes were upregulated and downregulated, respectively. Meanwhile, there were 48 and 45 genes showing up-regulated and down-regulated expression in FMVO vs. TPVO, respectively (Figure 1B and Supplementary Figure 1).
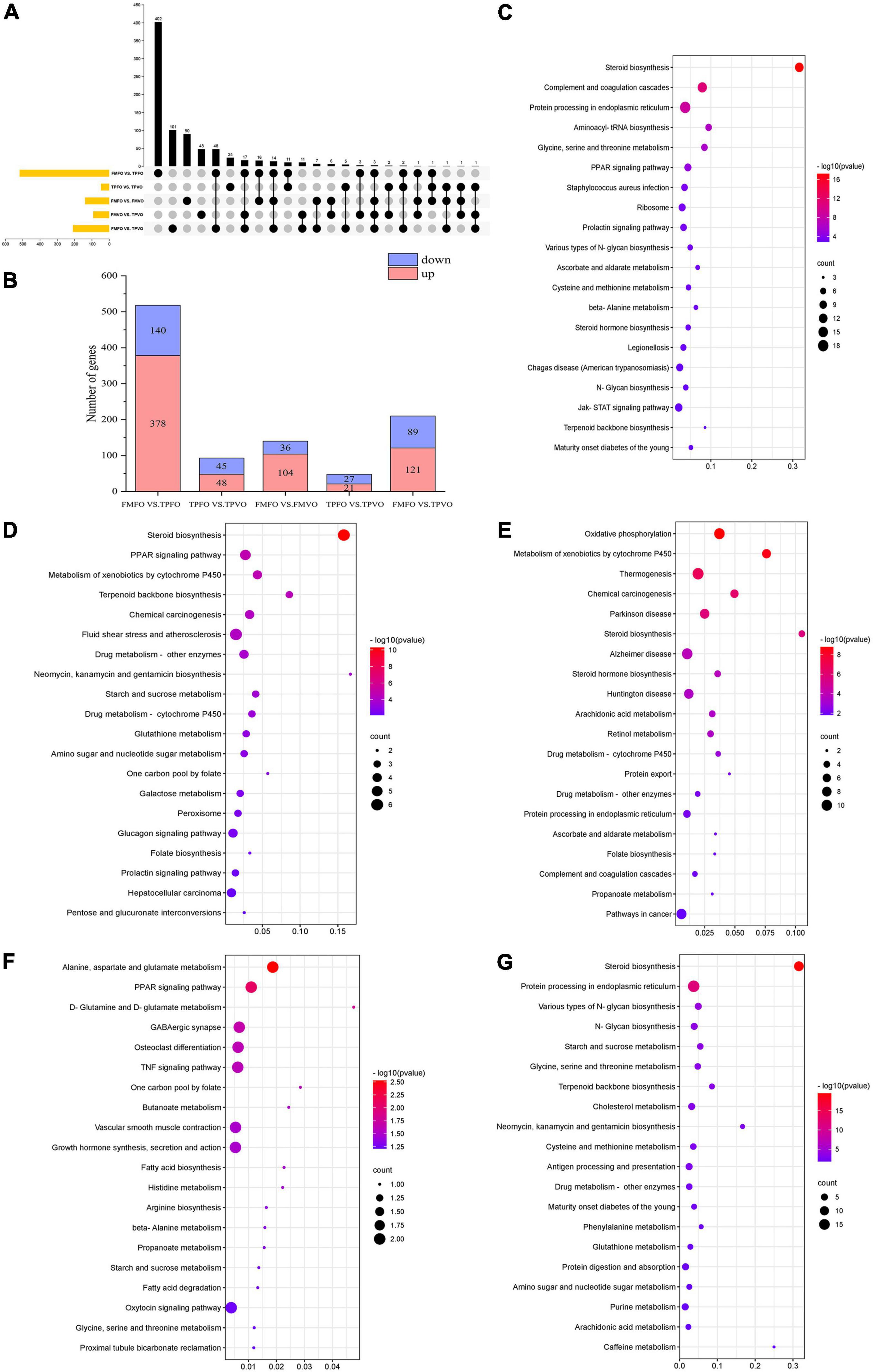
Figure 1. Transcriptomic analysis of O. mykiss fed diets with replacement of FM and/or FO. The number of up- and down-regulated DEGs were identified in the FM replacement and FO replacement (A,B). KEGG pathways analysis of DEGs in response to FM replacement (C,D), FO replacement (E,F), and combined replacement of FM and FO (G). Pathway enrichment analysis plots (top 20) of upregulated expressed metabolisms according to P < 0.05.
To further explore the metabolic pathways of the replacement of FM by TP, the DEGs in each group were annotated (Supplementary Table 2). Based on significant pathways (P < 0.05), KEGG was applied to correlate the DEGs with metabolic pathways. It was determined that most of DEGs in the comparison of FMFO vs. TPFO were enriched in “steroid biosynthesis,” “aminoacyl-tRNA biosynthesis,” “protein processing in endoplasmic reticulum,” “glycine, serine and threonine metabolism,” and “PPAR signaling pathway” (Figure 1C), while most of DEGs in the comparison of FMVO vs. TPVO were enriched in “steroid biosynthesis,” “PPAR signaling pathway,” “glutathione metabolism,” and “pentose and glucuronate interconversions” (Figure 1D).
There were 140 and 48 DEGs identified in FMFO vs. FMVO and TPFO vs. TPVO, respectively. A total of 3 overlapping DEGs between FMFO vs. TPFO and FMVO vs. TPVO. In the comparison of FMFO vs. FMVO, 104 genes were expressed up-regulated and 36 genes were expressed down-regulated. Meanwhile, 21 and 27 gene expressions were up- and down-regulated in TPFO vs. TPVO, respectively (Supplementary Figure 1).
To further explore the metabolic pathways of the replacement of FO by VO, the DEGs in each group were annotated (Supplementary Table 3). Based on significant pathways (P < 0.05), KEGG was employed to correlate the DEGs with metabolic pathways. It was determined that most of DEGs in FMFO vs. FMVO were enriched in “oxidative phosphorylation,” “steroid biosynthesis,” and “arachidonic acid metabolism” (Figure 1E), while most of DEGs in TPFO vs. TPVO were enriched in “alanine, aspartate and glutamate metabolism,” “PPAR signaling pathway,” “D-glutamine, and D-glutamate metabolism” and “fatty acid biosynthesis” (Figure 1F).
A total of 210 DEGs were identified in FMFO vs. TPVO. Compared with the control group, 121 and 89 genes showed up-regulated and down-regulated expression in the TPVO group, respectively (Supplementary Figure 1).
To further explore the metabolic pathways of the combined replacement of FM and FO by TP and VO, respectively, the DEGs in each group were annotated (Supplementary Table 4). Based on pathway P < 0.05, KEGG was employed to correlate the DEGs with metabolic pathways. It was determined that most of DEGs in FMFO vs. TPVO were enriched in “steroid biosynthesis,” “protein processing in endoplasmic reticulum,” “glycine, serine and threonine metabolism,” “purine metabolism,” “phenylalanine metabolism,” and “arachidonic acid metabolism” (Figure 1G).
In this LC-MS analysis, a total of 12,928 valid peaks were extracted. To maximize the discrimination between the replacement of dietary FM and/or FO by TP and/or VO and the control groups, and OPLS-DA was applied to determine the differences in metabolite levels between comparable groups. R2Y represents the percentage of all sample variables explained by the model. Q2 represents the percentage of all sample variables predicted by the model. OPLS-DA score plots of FMFO vs. TPFO, FMVO vs. TPVO, FMFO vs. FMVO, TPFO vs. TPVO, and FMFO vs. TPVO had the cumulative values of R2Y being 0.909, 0.927, 0.967, 0.976, and 0.975, and Q2 being 0.838, 0837, 0.767, 0.629, and 0.923, respectively, indicating that the model derived by OPLS-DA were highly plausible (Supplementary Figure 2) and can be employed in the further analysis.
A total of 204 and 199 DMs were identified between FMFO vs. TPFO and FMVO vs. TPVO, respectively. In the comparison of FMFO vs. TPFO, there were 70 and 134 DMs upregulated and downregulated, respectively. In the comparison of FMVO vs. TPVO, 42 and 157 DMs were upregulated and downregulated, respectively. There were 127 overlapping metabolites between FMFO vs. TPFO and FMVO vs. TPVO (Supplementary Figure 3).
To identify the potential metabolic pathways affected by the replacement of FM by TP, KEGG was applied to correlate the DMs with metabolic pathways. The results of pathway analysis described in detail the metabolic pathway changes related to the replacement of FM by TP(Figure 2). The most closely related metabolic pathways of FMFO vs. TPFO and FMVO vs. TPVO were shown in Supplementary Table 5.
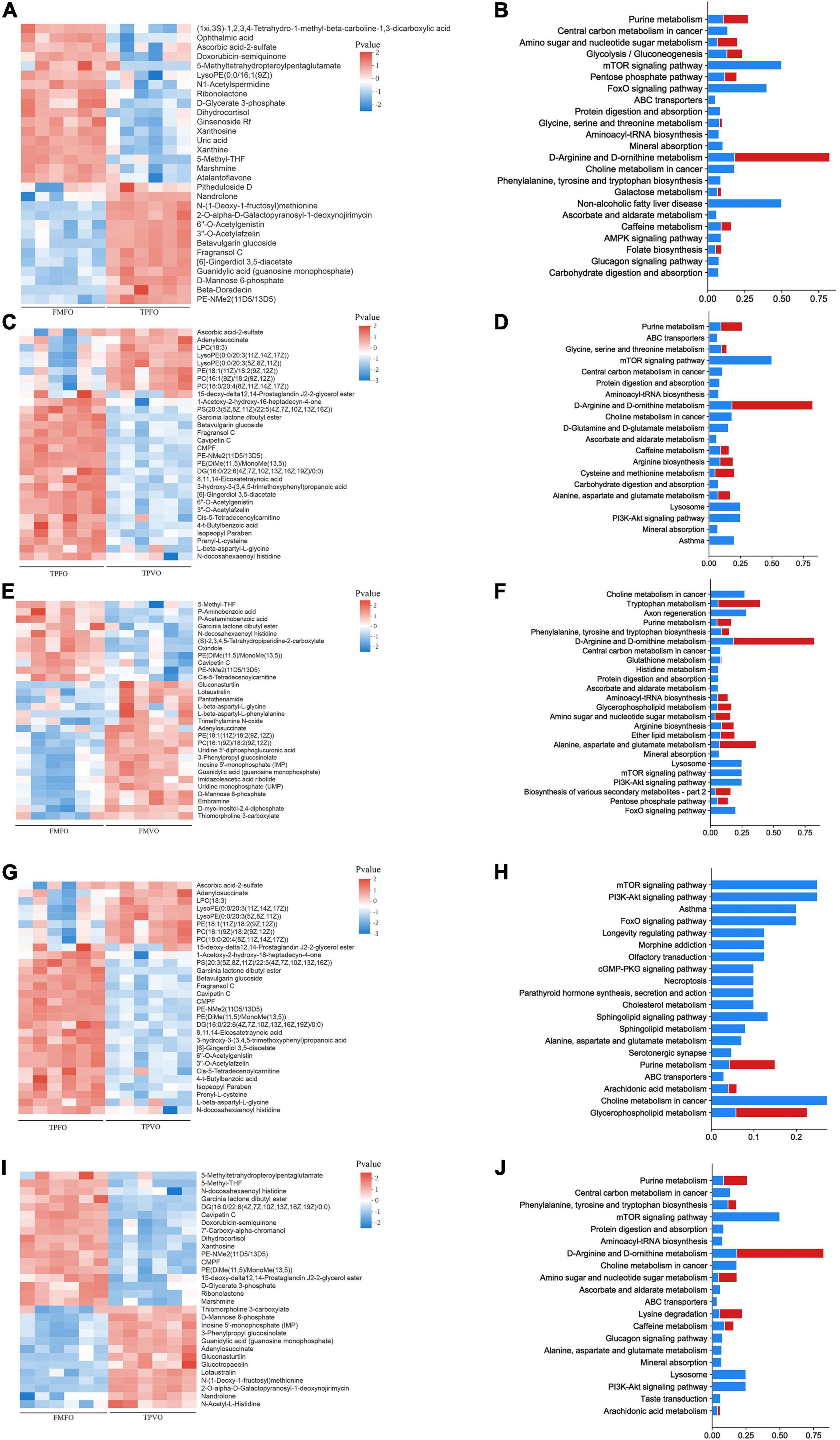
Figure 2. Metabolomic profiles of O. mykiss fed diets with replacement of FM and/or FO. Heatmaps and pathway analysis plots of DMs in FM replacement (A–D), FO replacement (E–H), and combined replacement of FM and FO (I,J). The blue bar in pathway analysis plots showed enrichment ratio and the red bar showed pathway impact value calculated from pathway topology analysis.
There were 167 and 122 DMs identified in FMFO vs. FMVO and TPFO vs. TPVO, respectively. In the comparison of FMFO vs. FMVO, 87 DMs were expressed up-regulated and 80 DMs were expressed down-regulated. In the comparison of TPFO vs. TPVO, 36 and 86 DMs were upregulated and downregulated, respectively. A total of 67 overlapping metabolites between FMFO vs. FMVO and TPFO vs. TPVO (Supplementary Figure 3).
To identify the potential metabolic pathways affected by the replacement of FO by VO, KEGG was applied to correlate the DMs with metabolic pathways. The results of pathway analysis described in detail the metabolic pathway changes related to the replacement of FO by VO (Figure 2). The most closely related metabolic pathways of FMFO vs. FMVO and TPFO vs. TPVO were shown in Supplementary Table 6.
A total of 220 DMs were identified between FMFO vs. TPVO. Compared to the control group, 63 and 157 DMs were upregulated and downregulated in the combined replacement of FM and FO by TP and VO, respectively (Supplementary Figure 3).
To identify the potential metabolic pathways affected by the combined replacement of dietary FM and FO, KEGG was applied to correlate the DMs with metabolic pathways. The results of pathway analysis described in detail the metabolic pathway changes related to the combined replacement of dietary FM and FO by TP and VO, respectively (Figure 2). The most closely related metabolic pathways of FMFO vs. TPVO were shown in Supplementary Table 7.
The integrative analysis of transcriptome and metabolome revealed the shared KEGG pathways of DEGs and DMs of O. mykiss fed with reduction of dietary FM and/or FO in the diets (Supplementary Table 8). The shared DEGs and DMs involved in these metabolic pathways were shown in Supplementary Table 9. For the replacement of FM by TP, 32 shared KEGG pathways were mapped from 63 and 42 shared KEGG pathways between FMFO vs. TPFO and FMVO vs. TPVO, respectively. Among these pathways, 10 significantly important metabolic pathways were identified via topological pathway analysis (“purine metabolism,” “cysteine and methionine metabolism,” “lysine degradation,” “arginine biosynthesis,” “glycolysis/gluconeogenesis,” “phenylalanine metabolism,” “amino sugar and nucleotide sugar metabolism,” “glycine, serine and threonine metabolism,” “pentose and glucuronate interconversions,” and “glutathione metabolism”) (Figure 3A). For the replacement of FO by VO, 6 shared KEGG pathways were mapped from 33 and 10 shared KEGG pathways in FMFO vs. FMVO and TPFO vs. TPVO, respectively. Among these pathways, six significantly important metabolic pathways were identified via topological pathway analysis (“purine metabolism,” “glycine, serine and threonine metabolism,” “aminoacyl-tRNA biosynthesis,” “cysteine and methionine metabolism,” “sphingolipid metabolism,” and “alanine, aspartate and glutamate metabolism”) (Figure 3B). For the combined replacement of FM and FO by TP and VO, respectively, 52 shared KEGG pathways were mapped from FMFO vs. TPVO. Among these pathways, 11 significantly important metabolic pathways were identified via topological pathway analysis (“tryptophan metabolism,” “purine metabolism,” “lysine degradation,” “amino sugar and nucleotide sugar metabolism,” “arginine biosynthesis,” “glycolysis/gluconeogenesis,” “phenylalanine metabolism,” “citrate cycle (TCA cycle),” “glycerophospholipid metabolism,” “pentose and glucuronate interconversions,” and “cysteine and methionine metabolism”) (Figure 3C). Correlation analysis calculated by Pearson showed tight correlations between DEGs and DMs, which further emphasized the correlation between each DEG and each DM (Figure 4).
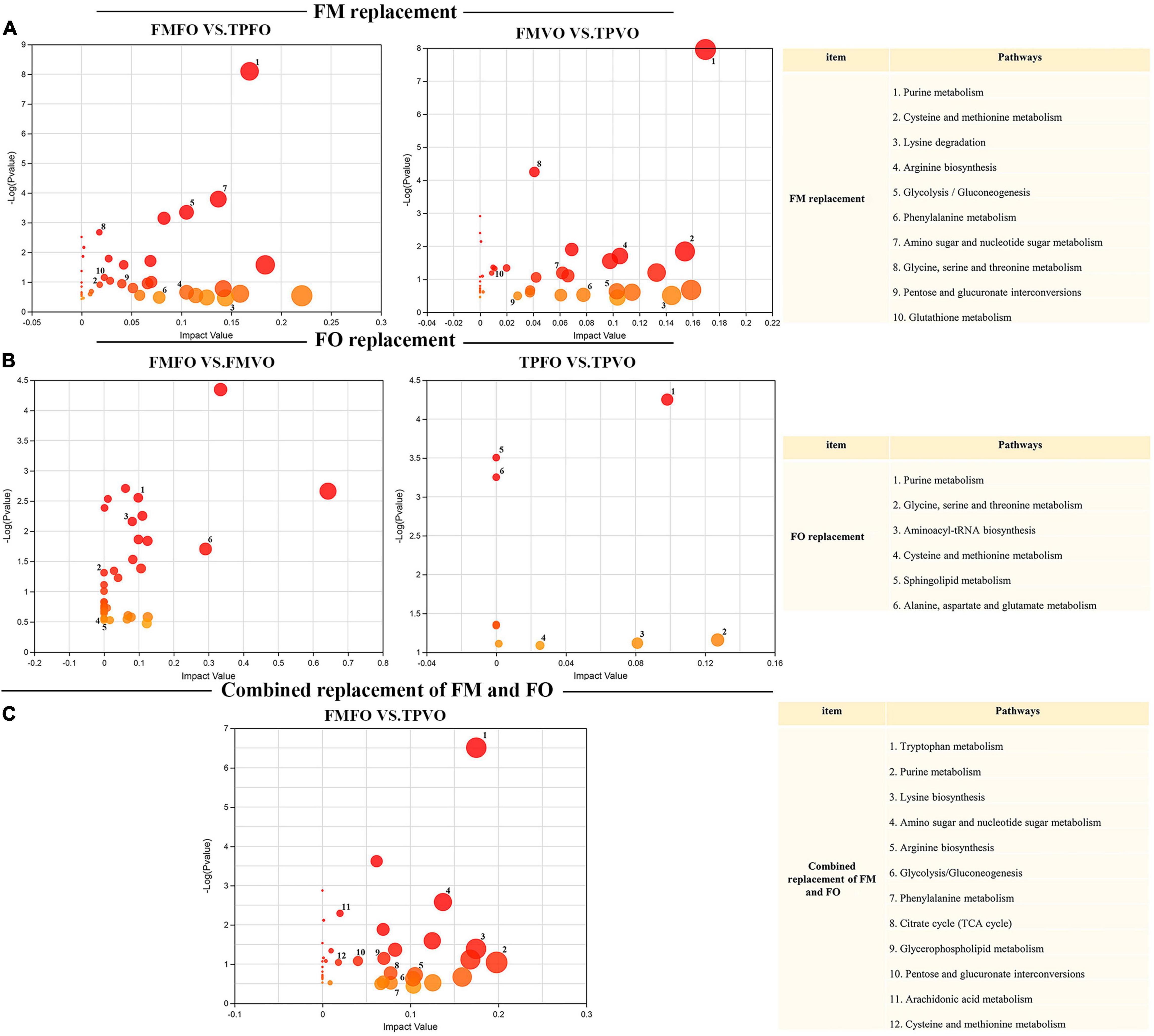
Figure 3. Significant specific pathway identified by multi-omics analysis between FM or FO replacement and combined replacement FM and FO. Pathway analysis of DMs from common pathways shared with DEGs was plotted at the replacement of FM (A), replacement of FO (B), and combined replacement of FM and FO (C). Map00230: purine metabolism; map00310: lysine degradation; map00220: arginine biosynthesis; map00010: glycolysis/gluconeogenesis; map00360: phenylalanine metabolism; map00520: amino sugar and nucleotide sugar metabolism; map00260: glycine, serine, and threonine metabolism; map00040: pentose and glucuronate interconversions; map00480: glutathione metabolism; map00380: tryptophan metabolism; map00020: citrate cycle (TCA cycle); map00564: glycerophospholipid metabolism.
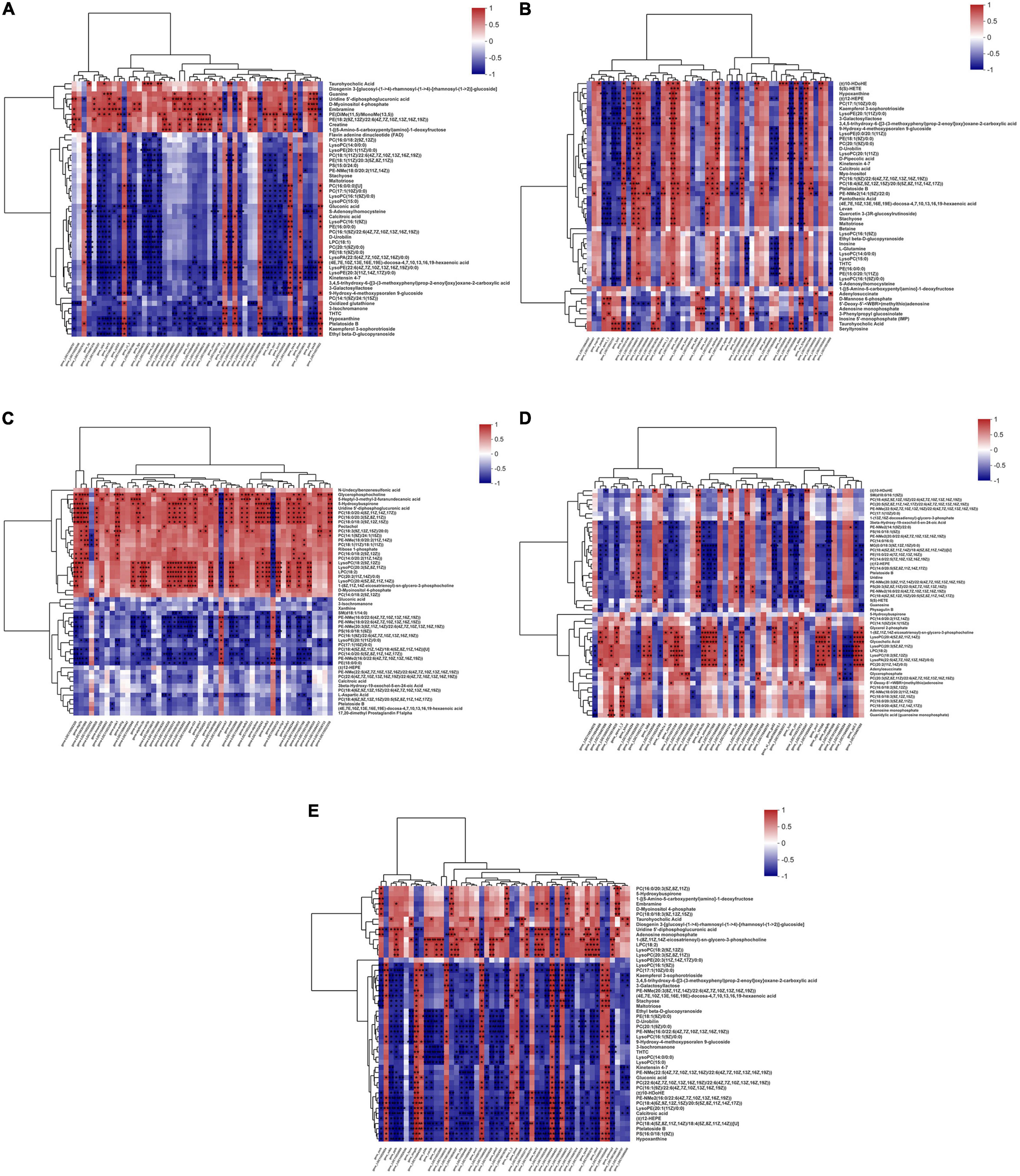
Figure 4. Correlation plots of the correlations between metabolome and transcriptome. The correlation plots with genes in columns and metabolites in rows to reveal the connection between genes and metabolites of O. mykiss fed diets with FM replacement (A,B), FO replacement (C,D), and combined replacement FM and FO (E). The red and blue showed the positive and negative correlation between transcriptomics and metabolomics, respectively. The symbol “*” indicates significant correlation between the gene and metabolites (P < 0.05).
Shared DMs and DEGs of individual and combined replacement of FM and FO by TP and VO, respectively, were further aggregated to explore the metabolic process that might occur in the liver of O. mykiss (Table 3). A comprehensive metabolic network of these shared metabolites and genes involving the replacement of dietary FM and/or FO by TP and/or VO was shown in Figure 5. Adenylosuccinate and adenosine monophosphate were all upregulated in replacement of dietary FM and/or FO by TP and/or VO and were involved in purine metabolism. The high concentration of 2-(formamido)-N1-(5′-phosphoribosyl) acetamidine, but low contents of xanthosine, xanthine, and urate which possibly due to the low expression of PNP and XDH were found in purine metabolism. The low concentrations of R-S-glutathione, 5-oxoproline, 5-L-glutamyl-L-alanine, and low expression levels of OPLAH, which are involved in the glutathione metabolism, were identified in fish feeding diets with replacement of FM. Moreover, the concentrations of L-tryptophan and anthranilate were both downregulated, but high expression levels of ACAT2 were identified in the combined replacement of dietary FM and FO by TP and VO, respectively. In glycerophospholipid metabolism, lysine metabolism, and tryptophan metabolism, 1-acyl-SN-glycero-3-phosphocholine, phosphatidylcholine (lecithin), PCYT1, L-lysine, and L-2-aminoadipate were downregulated in combined replacement of dietary FM and FO by TP and VO, respectively. In addition, the low expression levels of D-glycerate 3-phosphate and D-glucuronic acid, but the high expression levels of UDP-D-glucuronate may be mainly controlled by the up-regulation of upstream DEGs, which were found in glycolysis/gluconeogenesis and pentose and glucuronate interconversions. Furthermore, L-arginine, L-phenylalanine, and D-glucuronic acid, which are involved in arginine biosynthesis, phenylalanine metabolism, and amino sugar and nucleotide sugar metabolism were expressed at low levels in individual replacement of dietary FM by TP and combined replacement of dietary FM and FO by TP and VO, respectively.
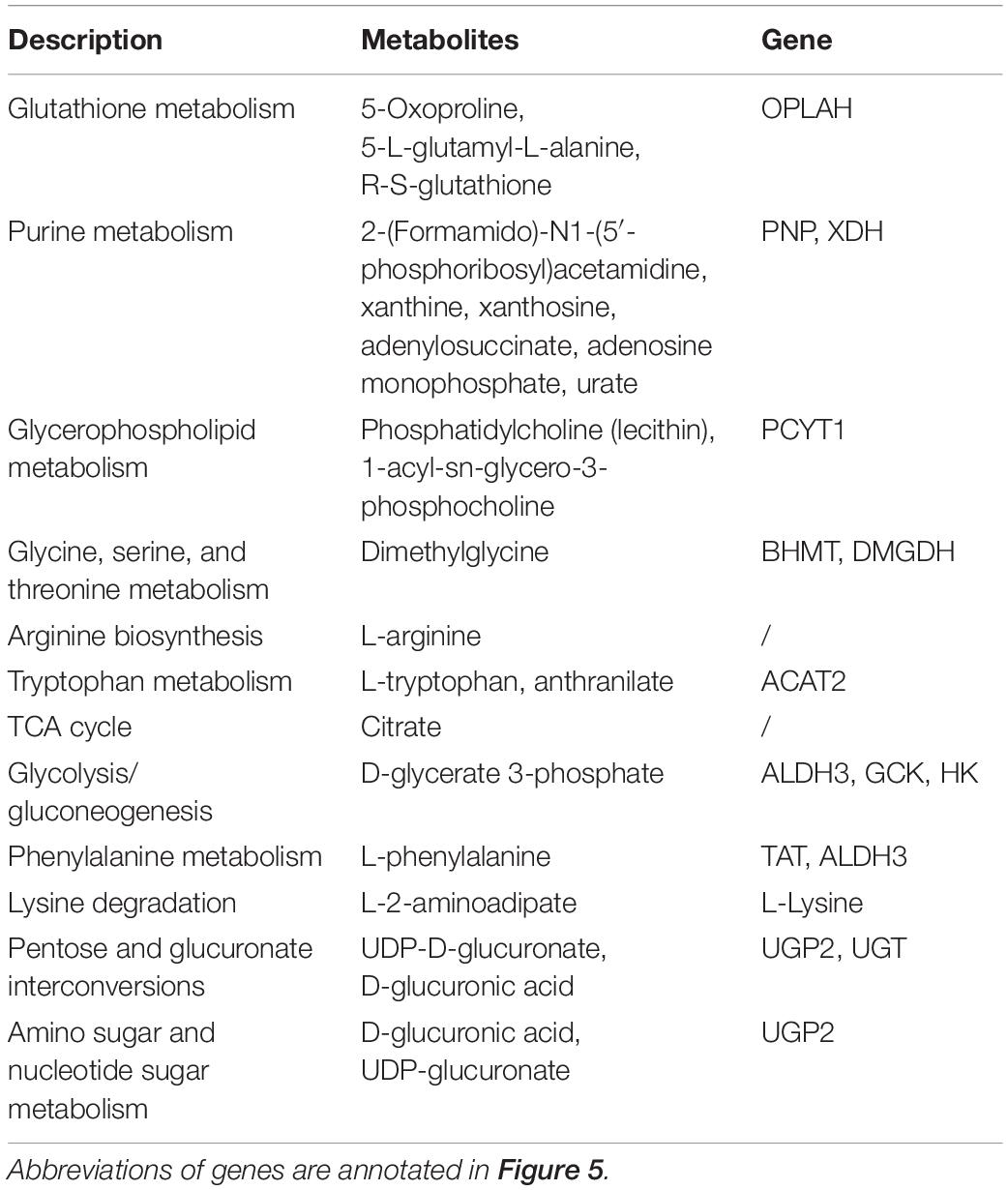
Table 3. Shared DMs and DEGs in the KEGG pathway of O. mykiss which fed diets with replacement of FM and/or FO.
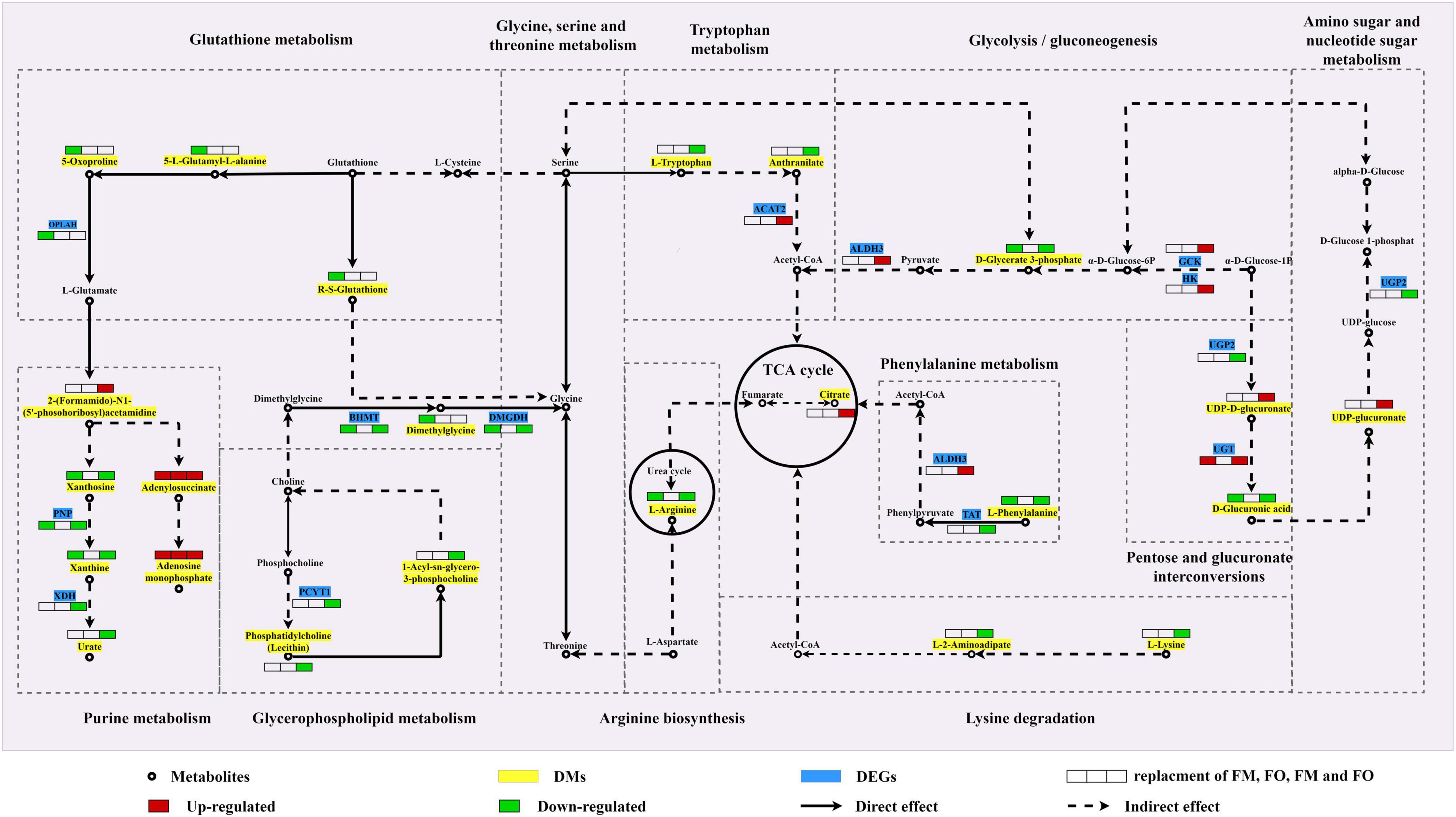
Figure 5. A hypothetical integrated metabolic network of key differential metabolites and genes in significant pathways identified by multi-omics analysis between FM and FO replacement and combined replacement FM and FO based on the KEGG pathway database. Map00230: purine metabolism; map00310: lysine degradation; map00220: arginine biosynthesis; map00010: glycolysis/gluconeogenesis; map00360: phenylalanine metabolism; map00520: amino sugar and nucleotide sugar metabolism; map00260: glycine, serine, and threonine metabolism; map00040: pentose and glucuronate interconversions; map00480: glutathione metabolism; map00380: tryptophan metabolism; map00020: citrate cycle (TCA cycle); map00564: glycerophospholipid metabolism. Metabolites were marked as circles, differential metabolites as green frames, and differential genes as blue frames. The red/green boxes (from left to right, replacement of dietary FM, FO, FM and FO, respectively) show high/low expression of alternative FM and/or FO. OPLAH: 5-oxoprolinase; PNP: purine-nucleoside phosphorylase; XDH: xanthine dehydrogenase/oxidase; PCYT1: choline-phosphate cytidylyltransferase; BHMT: betaine-homocysteine S-methyltransferase; DMGDH: dimethylglycine dehydrogenase; ACAT2: acetyl-CoA C-acetyltransferase; TAT: tyrosine aminotransferase; ALDH3: aldehyde dehydrogenase; GCK: glucokinase; HK: hexokinase; UGP2: UTP-glucose-1-phosphate uridylyltransferase; UGT: glucuronosyltransferase.
In our study, the averages of 44.5, 44.2, 44.8, and 43.3 million clean reads were localized to the O. mykiss genome from the FMFO, FMVO, TPFO, and TPVO groups, respectively (Supplementary Table 10). The GC contents of the four groups showed the average values with 49.9, 49.88, 49.09, and 49.3%, respectively. And Q30 contents were on the average of 95.24, 94.89, 95.06, and 95.10%, respectively. The average map rates of the four groups were 94.75, 94.82, 93.64, and 94.23%, respectively (Supplementary Table 10). These data suggested that the sequencing results were favorable and that the results of the subsequent transcriptome analysis were dependable. To further confirm the accuracy and reliability of transcriptomic data, qRT-PCR was conducted to validate the RNA-seq results of 10 DEGs, and the expression patterns were in accordance with the trend of RNA-seq results (Supplementary Figure 4).
The growth performance results obtained in the present study were consistent with those previously reported in African catfish (Clarias gariepinus) (Sourabié et al., 2018), and dietary protein sources were the main limiting factor affecting growth performance. In terms of FO, replacing dietary FO with VO did not compromise the growth performance of O. mykiss, which was consistent with the previous studies with partial replacement FO in farmed fish (Glencross et al., 2003; Hardy, 2010). In contrast, individuals feeding diets with reduced dietary FM presented lower feed utilization than fish feeding diets with higher dietary FM content, resulting in poor growth performance regardless of the dietary lipid sources. This result was in agreement with the previous investigations in European sea bass (D. labrax) (Torrecillas et al., 2017) and Atlantic cod (Gadus morhua L.) (Hansen et al., 2007) that accepted the reduction dietary FM diets. The response of O. mykiss to the replacement of FM by TP was mainly associated with the reduction in feed intake and FE (Torstensen et al., 2008). Particularly, some ingredients present in alternative protein sources, such as unbalanced AAs profiles and anti-nutritional factors repress the growth performance of fish (Li et al., 2014). Additionally, fish feeding diets with reduction of dietary FM and FO appeared to have similar protein and lipid utilization efficiencies to those of individuals in the FMFO group, as no significant differences in PER and LER were observed between individuals in the four experimental groups. Similar results have also been documented in Atlantic salmon (Torstensen et al., 2008) and Pacific yellowtail (Seriola lalandi) (Nuche-Pascual et al., 2018).
The immune system of fish is the first line of defense against external pathogens and a crucial mechanism in organisms (Saurabh and Sahoo, 2008). In this study, we identified pathways that might be involved in the immune response, such as arginine biosynthesis, amino and nucleotide sugars metabolism, and tryptophan metabolism. Arginine is known to be a multifunctional AA closely related to immune system regulation, mediating immunosuppressive mechanisms while maintaining hepatic urea cycle activity for ammonia detoxification (Wu et al., 2014; Azeredo et al., 2015). Thus, the low concentration of arginine revealed by multi-omics data indicated that the depletion of free arginine and low levels of availability, caused by the individual replacement of FM by TP and combined replacement of dietary FM and FO by TP and VO, respectively, resulted in impaired immunity in the liver of O. mykiss. Amino sugar and nucleotide sugars metabolism is also an important immune-related pathway that is associated with external environmental stress (Guo et al., 2014; Xu et al., 2016). In the present investigation, D-glucuronic acid and UGP2 were downregulated in response to the individual replacement of dietary FM by TP and the combined replacement of FM and FO by TP and VO, respectively. Meanwhile, anthranilate is an important intermediate in tryptophan metabolism and involves the synthesis of nonsteroidal anti-inflammatory drugs (NSAIDs) (Sorgdrager et al., 2019). NSAIDs can induce the synthesis of nonsteroidal anti-inflammatory molecules that signal the immune system response and prevent inflammation (Zummo et al., 2012). The low concentrations of L-tryptophan and anthranilate in the present study implied that NSAIDs associated with anthranilate might be inhibited, which might affect the immunity of O. mykiss fed diets with combined replacement of FM and FO by TP and VO, respectively. Furthermore, previous reports have demonstrated that numerous energy supplies are crucial for the immune defense processes (Zhang et al., 2019). ACAT2, related to the production of acetyl-CoA, was upregulated and promoted its participation in the TCA cycle, suggesting that fish require additional energy to maintain immune function. Therefore, these results indicated that individuals feeding diets with combined replacement of FM and FO might lead to increasingly higher energy requirements of fish. More attention should be paid to the design of diet formulation to prevent the growth performance of fish from being affected by excessive substitution leading to immune dysfunction and energy requirements that are difficult to meet.
Under physiological homeostasis state, organisms will produce reactive oxygen species and maintain the antioxidant defenses by providing a set of mechanisms to equilibrium. Although antioxidant defense in fish is influenced by nutritional factors, little is known about how the oxidative status in fish responds to the dietary ingredients (Rueda-Jasso et al., 2004). In the present study, the low concentrations of R-S-glutathione, 5-oxoproline, 5-L-glutamyl-L-alanine, and OPLAH in individuals fed a reduction FM diet, indicating that glutathione metabolism was repressed. Furthermore, these metabolites are commonly used as potential biomarkers for hepatic glutathione status assessment (Geenen et al., 2013). Particularly, R-S-glutathione, one of the transformation products of glutathione formation complexes, transports into the specific position, which is a common detoxification mechanism (Liang et al., 2016). Altering the level of dietary FM has been confirmed to affect the antioxidant capacity, thereby repressing the growth and development of Pacific white shrimp (Xie et al., 2020), Barramundi (L. calcarifer) (Chaklader et al., 2020), and European sea bass (Guerreiro et al., 2015). Previous study has reported that antioxidant defense is a metabolic process that requires massive energy expenditure in response to external stimuli (Xie et al., 2020). Meanwhile, individuals fed the replacement of FM by TP diet required more energy supply to deal with the oxidative stress caused by the reduction of dietary FM and support the subsequent growth requirements. Given this requirement, the TCA cycle, pentose and glucuronate interconversions, and glycolysis/gluconeogenesis were activated to provide sufficient energy for the growth and antioxidative requirements of O. mykiss. Therefore, our results suggested that the replacement of dietary FM by TP represses the antioxidant system of O. mykiss, which could result in lower antioxidant and detoxifying capabilities.
The TCA cycle is a central pathway for most metabolic pathways such as AA, lipid, and carbohydrate, and is an important metabolic pathway that provides energy to the organism (Akram, 2014). Comparative multi-omics data in fish between FMFO and TPVO showed several essential intermediates involved in the TCA cycle, including L-tryptophan, L-lysine, and L-phenylalanine (Mishra et al., 2017), all of which were downregulated in TPVO compared with FMFO. The low concentrations of these metabolites might be consumed for the energy metabolism of O. mykiss. Meanwhile, adenylosuccinate is the precursor of fumaric acid for further synthesis of citrate, which plays a crucial role in energy metabolism. Citrate is an important intermediate of the TCA cycle and is closely associated with ATP generation. The multi-omics data showed the high concentration of citrate possibly indicated that fish feeding diets with combined replacement of FM and FO by TP and VO, respectively, activated the TCA cycle to supply sufficient energy to accommodate the shift dietary quality. Huang et al. (2017) reported that the decreased AA may be associated with the increased energy requirements to maintain body homeostasis in response to external stimuli. In the present study, the low concentrations of L-tryptophan, L-lysine, and L-phenylalanine were observed in TPVO, indicating that the activated TCA cycle may be a strategy for O. mykiss to cope with the combined replacement of FM and FO by TP and VO, respectively. Furthermore, Li et al. (2009) reported that the decrease in phenylalanine could depress growth performance and even negatively affect the survival rate of fish. It was speculated that the poor feeding intake rates and growth performance of O. mykiss might result from the low concentration of L-phenylalanine in the liver of O. mykiss.
Glycolysis/gluconeogenesis is an important metabolic pathway to maintain glucose homeostasis (Metón et al., 2003). D-glycerate 3-phosphate, a precursor of tryptophan in organisms, is an important intermediate in glycolysis metabolism to produce pyruvate, the final product of glycolysis, which is required to activate the TCA cycle (Tikunov et al., 2014). In our work, the low concentration of D-glycerate 3-phosphate in fish feeding diets with individual replacement of dietary FM and combined replacement of dietary FM and FO by TP and VO, respectively, indicated the consumption of D-glycerate 3-phosphate. Furthermore, the UDP-D-glucuronate, GCK, HK, and ALDH3 are important metabolite and genes involved in the glycolysis/gluconeogenesis, and glucose and pentose interconversions pathways were increased in individuals fed diets with reduction of both FM and FO. The results suggested that the energy metabolism of O. mykiss was activated and provided sufficient energy in response to the fluctuation of dietary quality. Thus, once the metabolic energy is consumed in maintaining the adverse reactions caused by the diets, less energy is available for the growth of the fish.
Nucleotide metabolism plays an important physiological and biochemical role in organisms, mediating energy metabolism and signal transmission (Cosgrove, 1998). De novo synthesis of nucleotides can satisfy the normal growth of organisms, but when nucleotide metabolism is affected, the immune function of organs such as the liver may be impaired (Grimble and Westwood, 2000). The present study showed that nucleotide metabolism, especially purine metabolism in O. Mykiss, was significantly affected by the fluctuation of dietary quality. The low concentrations of several purines, including xanthosine, xanthine, and urate may be caused by the downregulated PNP and XDH that could promote the conversion of xanthosine to urate (Baccolini and Witte, 2019). Purines are the main components and metabolites of nucleotides, which can be absorbed by transporters and reused for protein synthesis (Mohlmann et al., 2010). However, the concentrations of purine in fish feeding diets with individual replacement of dietary FM by TP and combined replacement of FM and FO by TP and VO, respectively, were low, suggesting that purine synthesis might be repressed and thus affect the protein synthesis. Moreover, it is worth noting that as soon as the dietary FM and/or FO were reduced, both adenylosuccinate and adenosine monophosphate of O. mykiss were upregulated. Most purine nucleotides required for cell replication are derived from the purine biosynthesis pathway, and adenylosuccinate plays a considerable role in de novo biosynthesis of purine and replication (Yuan et al., 2011). However, the high concentrations of these metabolites employed for cell replication were detected in the liver, suggesting that the replacement of dietary FM and FO by TP and VO, respectively, repressed the cell protein synthesis ability of O. mykiss. Therefore, the results suggested that the impaired growth performance of O. mykiss might be associated with the replacement of dietary FM and/or FO by TP and/or VO, especially the inhibition of cellular protein biosynthesis induced by the combined replacement of FM and FO by TP and VO, respectively.
In terms of lipid metabolism, most DEGs and DMs were associated with glycerophospholipid metabolism. For instance, phosphatidylcholine (PC) is one of the main components of biofilms (Zong et al., 2018), and is crucial for the structure and function of these membranes (Gibellini and Smith, 2010). Previous studies reported that PC could bloke the damage of cell membrane that is caused by active free radicals through antioxidant effect, accelerate the lipolysis in the liver, and promote liver fibrosis in collagenase active link (Navder et al., 1997; Mi et al., 2000). In our study, PCYT1 was downregulated in the liver of O. mykiss fed reduction of both FM and FO diet, and resulted in low concentration of PC and 1-acyl-sn-glycero-3-phosphocholine, which might reflect significant changes in cell membrane composition. Additionally, previous investigations have demonstrated that the FA profiles of cell membranes were significantly affected by dietary FA profiles (Oxley et al., 2010) which is critical for fluidity and permeability of cell membranes, thereby affecting the function of immune receptors (Arts and Kohler, 2009). Therefore, in our study, the individual reduction of FM and combined replacement of FM and FO by TP and VO, respectively, may impair the immune function and cell membrane fluidity of O. mykiss to limit the normal physiological activities and growth performance of fish.
Integrated metabolomics and transcriptomics were used to investigate the effects of the replacement of dietary FM and/or FO by TP and/or VO, respectively, on changes of metabolites and genes in O. mykiss. The specific metabolic pathways of O. mykiss fed diet with combined replacement of FM and FO by TP and VO, respectively, were identified, mainly repressing immune function, cellular protein biosynthesis capacity, and lipid metabolism. The depression of antioxidant capacity was only observed in fish feeding diets with the replacement of dietary FM by TP. Moreover, as soon as the dietary FM and/or FO were reduced, the cellular protein biosynthesis ability of O. mykiss was repressed and accompanied by higher energy consumption in response to fluctuations of dietary quality, resulting in reduced growth performance. Furthermore, both adenylosuccinate and adenosine monophosphate related to cellular protein biosynthesis ability were significantly affected by both individual and combined replacement of FM and FO by TP and VO, respectively, suggesting that purine metabolism and its related two metabolites were potential biomarkers for O. mykiss fed diets with reduction of dietary FM and/or FO. Overall, our study provides new insights and theoretical basis into the metabolic and molecular mechanism behind the growth of O. mykiss response to the diet with reduction of dietary FM and/or FO.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA788999.
The animal study was reviewed and approved by the Regulations of the Administration of Affairs Concerning Experimental Animals of China and Regulations of the Administration of Affairs Concerning Experimental Animals of Shandong Province.
YC completed the data analysis and manuscript writing. QG and SD designed the study. QG was the major instructor. XL, YW, and ZD helped to collect the sample. YZ revised the manuscript. All authors contributed to the study and approved the submitted version.
This study was funded by the National Key R&D Program of China (Project No. 2019YFD0901000) and the National Natural Science Foundation of China (Grant No. 31872575).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We acknowledge those who donated the experimental animals and those who helped support this study at the Ocean University of China.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2022.843637/full#supplementary-material
Akram, M. (2014). Citric acid cycle and role of its intermediates in metabolism. Cell Biochem. Biophys. 68, 475–478. doi: 10.1007/s12013-013-9750-1
Alfaro, A. C., and Young, T. (2018). Showcasing metabolomic applications in aquaculture: a review. Rev. Aquac. 10, 135–152. doi: 10.1111/raq.12152
Arts, M. T., and Kohler, C. C. (2009). “Health and condition in fish: the influence of lipids on membrane competency and immune response,” in Lipids in Aquatic Ecosystems, ed. M. T. Arts (New York, NY: Springer Press), 237–256. doi: 10.1007/978-0-387-89366-2_10
Azeredo, R., Pérez-Sánchez, J., Sitjà-Bobadilla, A., Fouz, B., Tort, L., Aragão, C., et al. (2015). European sea bass (Dicentrarchus labrax) immune status and disease resistance are impaired by arginine dietary supplementation. PLoS One 10:e0139967. doi: 10.1371/journal.pone.0139967
Baccolini, C., and Witte, C. P. (2019). AMP and GMP catabolism in Arabidopsis converge on Xanthosine, which is degraded by a nucleoside hydrolase heterocomplex. Plant Cell 31, 734–751. doi: 10.1105/tpc.18.00899
Betancor, M. B., Sprague, M., Sayanova, O., Usher, S., Campbell, P. J., Napier, J. A., et al. (2015). Evaluation of a high-EPA oil from transgenic Camelina sativa in feeds for Atlantic salmon (Salmo salar L.): effects on tissue fatty acid composition, histology and gene expression. Aquaculture 444, 1–12. doi: 10.1016/j.aquaculture.2015.03.020
Bureau, D. P., Harris, A. M., and Cho, C. Y. (1999). Apparent digestibility of rendered animal protein ingredients for rainbow trout (Oncorhynchus mykiss). Aquaculture 180, 345–358. doi: 10.1016/s0044-8486(99)00210-0
Chaklader, M. R., Siddik, M. A. B., and Fotedar, R. (2020). Total replacement of fishmeal with poultry by-product meal affected the growth, muscle quality, histological structure, antioxidant capacity, and immune response of juvenile barramundi, Lates calcarifer. PLoS One 15:0242079. doi: 10.1371/journal.pone.0242079
Choi, D. G., He, M., Fang, H., Wang, X., Li, X., and Leng, X. (2019). Replacement of fish meal with two fermented soybean meals in diets for rainbow trout (Oncorhynchus mykiss). Aquac. Nutr. 26, 37–46. doi: 10.1111/anu.12965
Dias, J., Alvarez, M. J., Arzel, J., Corraze, G., Diez, A., Bautista, J. M., et al. (2005). Dietary protein source affects lipid metabolism in the European seabass (Dicentrarchus labrax). Comp. Biochem. Physiol. A 142, 19–31. doi: 10.1016/j.cbpb.2005.07.005
Espe, M., Lemme, A., Petri, A., and El-Mowafi, A. (2006). Can Atlantic salmon grow on diets devoid of fish meal? Aquaculture 255, 255–262. doi: 10.1016/j.aquaculture.2005.12.030
FAO (2020). The State of World Fisheries and Aquaculture, Meeting the Sustainable Development Goals. Rome: FAO.
Geay, F., Wenon, D., Mellery, J., Tinti, E., Mandiki, S. N. M., Tocher, D. R., et al. (2015). Dietary linseed oil reduces growth while differentially impacting LC-PUFA synthesis and accretion into tissues in Eurasian perch (Perca fluviatilis). Lipids 50, 1219–1232. doi: 10.1007/s11745-015-4079-8
Geenen, S., Yates, J. W. T., Kenna, J. G., Bois, F. Y., Wilson, I. D., and Westerhoff, H. V. (2013). Multiscale modelling approach combining a kinetic model of glutathione metabolism with PBPK models of paracetamol and the potential glutathione-depletion biomarkers ophthalmic acid and 5-oxoproline in humans and rats. Integr. Biol. 5, 877–888. doi: 10.1039/c3ib20245c
Gibellini, F., and Smith, T. K. (2010). The Kennedy pathway-de novo synthesis of phosphatidylethanolamine and phosphatidylcholine. IUBMB Life 62, 414–428. doi: 10.1002/iub.337
Glencross, B., Blyth, D., Irvin, S., Bourne, N., Campet, M., Boisot, P., et al. (2016). An evaluation of the complete replacement of both fishmeal and fish oil in diets for juvenile Asian seabass, Lates calcarifer. Aquaculture 451, 298–309. doi: 10.1016/j.aquaculture.2015.09.012
Glencross, B., Hawkins, W., and Curnow, J. (2003). Evaluation of canola oils as alternative lipid resources in diets for juvenile red seabream, Pagrus auratus. Aquac. Nutr. 9, 305–315. doi: 10.1046/j.1365-2095.2003.00257.x
Glencross, B., Rutherford, N., and Jones, B. (2011). Evaluating options for fishmeal replacement in diets for juvenile barramundi (Lates calcarifer). Aquac. Nutr. 17, 722–732. doi: 10.1111/j.1365-2095.2010.00834.x
Grayson, J., and Dabrowski, K. (2020). Partial and total replacement of fish oil with fatty acid ethyl esters in the starter diets of rainbow trout (Oncorhynchus mykiss). Aquaculture 522:735018. doi: 10.1016/j.aquaculture.2020.735018
Grimble, G. K., and Westwood, O. M. R. (2000). “Nucleotides,” in Nutrition and Immunology: Principles and Practice, eds J. B. German and C. L. Keen (Totowa, NJ: Humana Press), 135–144. doi: 10.1007/978-1-59259-709-3_11
Guerreiro, I., Couto, A., Perez-Jimenez, A., Oliva-Teles, A., and Enes, P. (2015). Gut morphology and hepatic oxidative status of European sea bass (Dicentrarchus labrax) juveniles fed plant feedstuffs or fishmeal-based diets supplemented with short-chain fructo-oligosaccharides and xylo-oligosaccharides. Br. J. Nutr. 114, 1975–1984. doi: 10.1017/s0007114515003773
Guillaume, J., Kaushik, S. J., Bergot, P., and Métailler, R. (2001). Nutrition and Feeding of Fish and Crustaceans. New York, NY: Springer Press. doi: 10.1016/s0044-8486(02)00261-2
Guo, C., Huang, X., Yang, M., Wang, S., Ren, S., Li, H., et al. (2014). GC/MS-based metabolomics approach to identify biomarkers differentiating survivals from death in crucian carps infected by Edwardsiella tarda. Fish Shellfish Immunol. 39, 215–222. doi: 10.1016/j.fsi.2014.04.017
Hansen, A., Rosenlund, G., Karlsen, Ø., Koppe, W., and Hemre, G. (2007). Total replacement of fish meal with plant proteins in diets for Atlantic cod (Gadus morhua L.) I–effects on growth and protein retention. Aquaculture 272, 599–611. doi: 10.1016/j.aquaculture.2007.08.034
Hao, R. J., Du, X. D., Yang, C. Y., Deng, Y. W., Zheng, Z., and Wang, Q. H. (2019). Integrated application of transcriptomics and metabolomics provides insights into unsynchronized growth in pearl oyster Pinctada fucata martensii. Sci. Total Environ. 666, 46–56. doi: 10.1016/j.scitotenv.2019.02.221
Hardy, R. W. (2010). Utilization of plant proteins in fish diets: effects of global demand and supplies of fishmeal. Aquac. Res. 41, 770–776. doi: 10.1111/j.1365-2109.2009.02349.x
Hixson, S. M., Parrish, C. C., and Anderson, D. M. (2014). Full substitution of fish oil with camelina (Camelina sativa) oil, with partial substitution of fish meal with camelina meal, in diets for farmed Atlantic salmon (Salmo salar) and its effect on tissue lipids and sensory quality. Food Chem. 157, 51–61. doi: 10.1016/j.foodchem.2014.02.026
Huang, S. S., Benskin, J. P., Veldhoen, N., Chandramouli, B., Butler, H., Helbing, C. C., et al. (2017). A multi-omic approach to elucidate low-dose effects of xenobiotics in zebrafish (Danio rerio) larvae. Aquat. Toxicol. 182, 102–112. doi: 10.1016/j.aquatox.2016.11.016
Jia, S., Li, X., Zheng, S., and Wu, G. (2017). Amino acids are major energy substrates for tissues of hybrid striped bass and zebrafish. Amino Acids 49, 2053–2063. doi: 10.1007/s00726-017-2481-7
Klinger, D., and Naylor, R. (2012). Searching for solutions in aquaculture: charting a sustainable course. Annu. Rev. Environ. Resour. 37, 247–276. doi: 10.1146/annurev-environ-021111-161531
Kong, T., Lin, S., Ren, X., Li, S., and Gong, Y. J. (2020). Transcriptome and metabolome integration analysis of mud crab Scylla paramamosain challenged to Vibrio parahaemolyticus infection. Fish Shellfish Immunol. 103, 430–437. doi: 10.1016/j.fsi.2020.05.069
Krogdahl, A., Bakke-McKellep, A. M., and Baeverfjord, G. (2003). Effects of graded levels of standard soybean meal on intestinal structure, mucosal enzyme activities, and pancreatic response in Atlantic salmon (Salmo salar L.). Aquac. Nutr. 9, 361–371. doi: 10.1046/j.1365-2095.2003.00264.x
Li, P., Mai, K. S., Trushenski, J., and Wu, G. Y. (2009). New developments in fish amino acid nutrition: towards functional and environmentally oriented aquafeeds. Amino Acids 37, 43–53.
Li, S., Dai, M., Qiu, H., and Chen, N. (2021). Effects of fishmeal replacement with composite mixture of shrimp hydrolysate and plant proteins on growth performance, feed utilization, and target of rapamycin pathway in largemouth bass, Micropterus salmoides. Aquaculture 533:736185. doi: 10.1016/j.aquaculture.2020.736185
Li, Y., Ai, Q. H., Mai, K. S., Xu, W., Deng, J. M., and Cheng, Z. Y. (2014). Comparison of high-protein soybean meal and commercial soybean meal partly replacing fish meal on the activities of digestive enzymes and aminotransferases in juvenile Japanese seabass, Lateolabrax japonicus (Cuvier, 1828). Aquac. Res. 45, 1051–1060. doi: 10.1111/are.12042
Li, Y., Niu, D. H., Wu, Y. H., Dong, Z. G., and Li, J. L. (2021). Integrated analysis of transcriptomic and metabolomic data to evaluate responses to hypersalinity stress in the gill of the razor clam (Sinonovacula constricta). Comp. Biochem. Phys. D 38:100793. doi: 10.1016/j.cbd.2021.100793
Liang, T., Ding, H., Wang, G., Kang, J., Pang, H., and Lv, J. (2016). Sulfur decreases cadmium translocation and enhances cadmium tolerance by promoting sulfur assimilation and glutathione metabolism in Brassica chinensis L. Ecotoxicol. Environ. Saf. 124, 129–137. doi: 10.1016/j.ecoenv.2015.10.011
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2 (-Delta Delta C(T)) Method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lu, J., Tibbetts, S. M., Lall, S. P., and Anderson, D. M. (2020). Use of dietary oil, solvent-extracted meal and protein concentrate from Camelina sativa for rainbow trout, Oncorhynchus mykiss, at the early fry stage. Aquaculture 524:735252. doi: 10.1016/j.aquaculture.2020.735252
Messina, M., Piccolo, G., Tulli, F., Messina, C., Cardinaletti, G., and Tibaldi, E. (2013). Lipid composition and metabolism of European sea bass (Dicentrarchus labrax L.) fed diets containing wheat gluten and legume meals as substitutes for fish meal. Aquaculture 33, 169–175. doi: 10.1016/j.aquaculture.2012.11.005
Metón, I., Fernandez, F., and Baanante, I. V. (2003). Short- and long-term effects of refeeding on key enzyme activities in glycolysis-gluconeogenesis in the liver of gilthead seabream (Sparus aurata). Aquaculture 225, 99–107. doi: 10.1016/s0044-8486(03)00281-3
Mi, L. J., Mak, K. M., and Lieber, C. S. (2000). Attenuation of alcohol-induced apoptosis of hepatocytes in rat livers by polyenylphosphatidylcholine (PPC). Alcohol. Clin. Exp. Res. 24, 207–212. doi: 10.1111/j.1530-0277.2000.tb04592.x
Mishra, P., Gong, Z. Y., and Kelly, B. C. (2017). Assessing biological effects of fluoxetine in developing zebrafish embryos using gas chromatography-mass spectrometry based metabolomics. Chemosphere 188, 157–167. doi: 10.1016/j.chemosphere.2017.08.149
Mohlmann, T., Bernard, C., Hach, S., and Neuhaus, H. E. (2010). Nucleoside transport and associated metabolism. Plant Biol. 12, 26–34. doi: 10.1111/j.1438-8677.2010.00351.x
Moreno-Arias, A., López-Elías, J. A., Martínez-Córdova, L. R., Ramírez-Suárez, J. C., Carvallo-Ruiz, M. G., García-Sánchez, G., et al. (2018). Effect of fishmeal replacement with a vegetable protein mixture on the amino acid and fatty acid profiles of diets, biofloc and shrimp cultured in BFT system. Aquaculture 483, 53–62. doi: 10.1016/j.aquaculture.2017.10.011
Navder, K. P., Baraona, E., and Lieber, C. S. (1997). Polyenylphosphatidylcholine attenuates alcohol-induced fatty liver and hyperlipemia in rats. J. Nutr. 127, 1800–1806. doi: 10.1093/jn/127.9.1800
Nuche-Pascual, M. T., Lazo, J. P., Ruiz-Cooley, R. I., and Herzka, S. Z. (2018). Amino acid-specific δ15N trophic enrichment factors in fish fed with formulated diets varying in protein quantity and quality. Ecol. Evol. 8, 9192–9217. doi: 10.1002/ece3.4295
Oliva-Teles, A., Enes, P., and Peres, H. (2015). “Replacing fishmeal and fish oil in industrial aquafeeds for carnivorous fish,” in Feed and Feeding Practices in Aquaculture, ed. D. A. Davis (Cambridge, MA: Elsevier Press), 203–233. doi: 10.1016/B978-0-08-100506-4.00008-8
Olsen, R. L., and Hasan, M. R. (2012). A limited supply of fishmeal: impact on future increases in global aquaculture production. Trends Food Sci. Technol. 27, 120–128. doi: 10.1016/j.tifs.2012.06.003
Oxley, A., Jolly, C., Eide, T., Jordal, A. E. O., Svardal, A., and Olsen, R. E. (2010). The combined impact of plant-derived dietary ingredients and acute stress on the intestinal arachidonic acid cascade in Atlantic salmon (Salmo salar). Br. J Nutr. 103, 851–861. doi: 10.1017/S0007114509992467
Panserat, S., Hortopan, G., Plagnes-Juan, E., Kolditz, C., Lansard, M., Skiba-Cassy, S., et al. (2009). Differential gene expression after total replacement of dietary fish meal and fish oil by plant products in rainbow trout (Oncorhynchus mykiss) liver. Aquaculture 294, 123–131. doi: 10.1016/j.aquaculture.2009.05.013
Petropoulos, I. K., Thompson, K. D., Morgan, A., Dick, J. R., Tocher, D. R., and Bell, J. G. (2009). Effects of substitution of dietary fish oil with a blend of vegetable oils on liver and peripheral blood leucocyte fatty acid composition, plasma prostaglandin E2 and immune parameters in three strains of Atlantic salmon (Salmo salar). Aquac. Nutr. 15, 596–607. doi: 10.1111/j.1365-2095.2008.00627.x
Randazzo, B., Zarantoniello, M., Gioacchini, G., Cardinaletti, G., Belloni, A., Giorgini, E., et al. (2021). Physiological response of rainbow trout (Oncorhynchus mykiss) to graded levels of Hermetia illucens or poultry by-product meals as single or combined substitute ingredients to deitary plant proteins. Aquaculture 538:73650. doi: 10.1016/j.aquaculture.2021.736550
Rimoldi, S., Terova, G., Ascione, C., Giannico, R., and Brambilla, F. (2018). Next generation sequencing for gut microbiome characterization in rainbow trout (Oncorhynchus mykiss) fed animal by-product meals as an alternative to fishmeal protein sources. PLoS One 13:e0193652. doi: 10.1371/journal.pone.0193652
Rueda-Jasso, R., Conceiçao, L. E., Dias, J., De Coen, W., Gomes, E., Rees, J. F., et al. (2004). Effect of dietary non-protein energy levels on condition and oxidative status of Senegalese sole (Solea senegalensis) juveniles. Aquaculture 231, 417–433. doi: 10.1016/s0044-8486(03)00537-4
Saurabh, S., and Sahoo, P. K. (2008). Lysozyme: an important defence molecule of fish innate immune system. Aquac. Res. 39, 223–239. doi: 10.1111/j.1365-2109.2007.01883.x
Sorgdrager, F. J., Naudé, P. J., Kema, I. P., Nollen, E. A., and Deyn, P. P. (2019). Tryptophan metabolism in inflammaging: from biomarker to therapeutic target. Front. Immunol. 10:2565. doi: 10.3389/fimmu.2019.02565
Sourabié, A., Mandiki, S., Geay, F., Sene, T., Toguyeni, A., and Kestemont, P. (2018). Fish proteins not lipids are the major nutrients limiting the use of vegetable ingredients in catfish nutrition. Aquac. Nutr. 24, 1393–1405. doi: 10.1111/anu.12676
Tikunov, A. P., Stoskopf, M. K., and Macdonald, J. M. (2014). Fluxomics of the eastern oyster for environmental stress studies. Metabolites 4, 53–70. doi: 10.3390/metabo4010053
Tocher, D. R. (2010). Fatty acid requirements in ontogeny of marine and freshwater fish. Aquac. Res. 41, 717–732. doi: 10.1111/j.1365-2109.2008.02150.x
Torrecillas, S., Robaina, L., Caballero, M. J., Montero, D., Calandra, G., Mompel, D., et al. (2017). Combined replacement of fishmeal and fish oil in European sea bass (Dicentrarchus labrax): production performance, tissue composition and liver morphology. Aquaculture 509, 246–248. doi: 10.1016/j.aquaculture.2017.03.031
Torstensen, B. E., Espe, M., Sanden, M., Stubhaug, I., Waagbo, R., Hemre, G. I., et al. (2008). Novel production of Atlantic salmon (Salmo salar) protein based on combined replacement of fish meal and fish oil with plant meal and vegetable oil blends. Aquaculture 285, 193–200. doi: 10.1016/j.aquaculture.2008.08.025
Turchini, G. M., Trushenski, J. T., and Glencross, B. D. (2019). Thoughts for the future of aquaculture nutrition: realigning perspectives to reflect contemporary issues related to judicious use of marine resources in aquafeeds. North Am. J. Aquac. 81, 13–39. doi: 10.1002/naaq.10067
Wei, Y. L., Liang, M. Q., Mai, K. S., Zheng, K. K., and Xu, H. G. (2017). The effect of ultrafiltered fish protein hydrolysate levels on the liver and muscle metabolic profile of juvenile turbot (Scophthalmus maximus L.) by 1H NMR-based metabolomics studies. Aquac. Res. 48, 3515–3527. doi: 10.1111/are.13178
Wu, G., Bazer, F. W., Dai, Z., Li, D., Wang, J., and Wu, Z. (2014). Amino acid nutrition in animals: protein synthesis and beyond. Annu. Rev. Anim. Biosci. 2, 387–417. doi: 10.1146/annurev-animal-022513-114113
Xie, C., Miao, X., Huang, J., Ding, Y., Wu, J., Dong, S., et al. (2011). KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 39, 316–322. doi: 10.1093/nar/gkr483
Xie, S. W., Wei, D., Chen, S. J., Zhuang, Z. X., Yin, P., Liu, Y. J., et al. (2020). Dietary fishmeal levels affect anti-oxidative ability and metabolomics profile of juvenile Pacific white shrimp, Litopenaeus vannamei. Aquac. Nutr. 26, 978–989. doi: 10.1111/anu.13055
Xu, Z., Li, T., Li, E., Chen, K., Ding, Z., Qin, J. G., et al. (2016). Comparative transcriptome analysis reveals molecular strategies of oriental river prawn Macrobrachium nipponense in response to acute and chronic nitrite stress. Fish Shellfish Immunol. 48, 254–265. doi: 10.1016/j.fsi.2015.12.005
Yan, J., Risacher, S. L., Shen, L., and Saykin, A. J. (2018). Network approaches to systems biology analysis of complex disease: integrative methods for multi-omics data. Brief. Bioinf. 19, 1370–1381. doi: 10.1093/bib/bbx066
Yuan, T., Gu, J. R., Gu, W. B., Wu, J., Ge, S. R., and Xu, H. (2011). Molecular cloning, characterization and expression analysis of adenylosuccinate lyase gene in grass carp (Ctenopharyngodon idella). Mol. Biol. Rep. 38, 2059–2065. doi: 10.1007/s11033-010-0331-8
Zhang, H., Wang, H., Chen, H., Wang, M. Q., Zhou, Z., Qiu, L. M., et al. (2019). The transcriptional response of the Pacific oyster Crassostrea gigas under simultaneous bacterial and heat stresses. Dev. Comp. Immunol. 94, 1–10. doi: 10.1016/j.dci.2019.01.006
Zong, L., Xing, J. P., Liu, S., Liu, Z. Q., and Song, F. R. (2018). Cell metabolomics reveals the neurotoxicity mechanism of cadmium in PC12 cells. Ecotoxicol. Environ. Saf. 147, 26–33. doi: 10.1016/j.ecoenv.2017.08.028
Zummo, F., Marineo, S., Pace, A., Civiletti, F., Giardina, A., and Puglia, A. M. (2012). Tryptophan catabolism via kynurenine production in Streptomyces coelicolor: identification of three genes coding for the enzymes of tryptophan to anthranilate pathway. Appl. Microbiol. Biotechnol. 94, 719–728. doi: 10.1007/s00253-011-3833-y
Keywords: fish meal, fish oil, replacement, Oncorhynchus mykiss, metabolomics, transcriptomics
Citation: Cao Y, Gao Q, Li X, Zhou Y, Dong S, Wang Y and Dai Z (2022) Integrated Analysis of Metabolomics and Transcriptomics for Assessing Effects of Fish Meal and Fish Oil Replacement on the Metabolism of Rainbow Trout (Oncorhynchus mykiss). Front. Mar. Sci. 9:843637. doi: 10.3389/fmars.2022.843637
Received: 26 December 2021; Accepted: 02 February 2022;
Published: 24 February 2022.
Edited by:
Kang-le Lu, Jimei University, ChinaReviewed by:
Linghong Miao, Freshwater Fisheries Research Center, Chinese Academy of Fishery Sciences, ChinaCopyright © 2022 Cao, Gao, Li, Zhou, Dong, Wang and Dai. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qinfeng Gao, cWZnYW9Ab3VjLmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.