- Department of Computer Science and Engineering, Amrita School of Computing, Amrita Vishwa Vidyapeetham, Chennai, India
Quantum Natural Language Processing (QNLP) is a relatively new subfield of research that extends the application of principles of natural language processing and quantum computing that has enabled the processing of complex biological information to unprecedented levels. The present comprehensive review analyses the potential of QNLP in influencing many branches of bioinformatics such as genomic sequence analysis, protein structure prediction, and drug discovery and design. To establish a correct background of QNLP techniques, this article is going to explore the basics of quantum computing including qubits, quantum entanglement, and quantum algorithms. The next section is devoted to the application of QNLP in the extraction of material and valuable information and knowledge related to drug discovery and development, prediction and assessment of drug-target interactions. In addition, the paper also explains the application of QNLP in protein structural prediction by quantum embedding, quantum simulation, and quantum optimization for exploring the sequence-structure relationship. However, this study also acknowledges the future of QNLP in bioinformatics in the discussion of the challenges and weaknesses of quantum hardware, data representation, encoding, and the construction and enhancement of the algorithms. This looks into real-life problems solved from industry applications, benchmarking and assessment criteria, and a comparison with other traditional NLP methods. Therefore, the review enunciates the research and application perspectives, as well as the developmental and implementation blueprint for QNLP in bioinformatics. The plan is as follows: its function is to achieve the objectives of precision medicine, new protein design, multi-omics, and green chemistry.
1 Introduction
Natural language processing (NLP) is a field of computer science and a subfield of artificial intelligence that aims to make computers understand human language. NLP uses computational linguistics, which is the study of how language works, and various models based on statistics, machine learning, and deep learning. Quantum Natural Language Processing (QNLP) is an integrative approach that encompasses natural language processing and physical theories taken from quantum mechanics to speed up the process of analyzing human language (Karamlou et al., 2022). By doing so, it can transform how humans engage with language-based data by proposing new and unique enhancements for a range of purposes such as Text Processing and Preprocessing; semantic analysis; text classification, sentiment analysis; information retrieval; and language translation. Because of principles as superposition and entanglement, QNLP can process enormous amounts of linguistic information simultaneously, which in turn entails a more efficient and comprehensive analysis of language than in classical NLP (Widdows et al., 2024). Besides, it is believed that QNLP should be more efficient and precise than conventional NLP due to the peculiarities of quantum computing as the solution to the problems associated with context-dependent and linguistic information.
Quantum systems, on which QNLP is based, are divided into two types: closed and open. Isolated quantum systems evolve under unitary time evolution, that is, by the Schrödinger equation. These systems are best used in theoretical work and form the basis of QNLP. On the other hand, the open quantum system has contact with other surroundings, the evolution is non-unitary due to dissipation and decoherence(Weidner et al., 2024). As a start for the exploration of QNLP techniques, this paper initially concentrates on closed quantum systems, although recognizing that open systems affected by Markovian and non-Markovian processes are essential in quantum computation. This work could be extended in future research by incorporating open quantum system models to expand the areas to which QNLP can be applied.
1.1 Background and motivation
In the past, ever since the development of modern computing, the application of algorithms and mathematical models in biological studies has expanded tremendously (Ofer et al., 2021). Natural language processing is now an indispensable method of data mining in bioinformatics to enable fast and efficient extraction of knowledge from bulk data. However, classical NLP techniques have problems with the organization of complexity, heterogeneity, and scale, typical for biological data, including scientific articles, databases, and experimental outcomes (Khurana et al., 2023). These limitations call for a change in the approach to computational strategies.
QNLP alleviates this problem by using quantum computing’s strengths of handling big data most efficiently. Quantum circuits and compositional vector-based semantics used in language tasks improve the simulation of biological processes such as interactions between molecules and genomics data analysis. For example, QNLP methods could bring a drastic improvement in such tasks as protein folding prediction, ligand binding constant estimation, and genome-wide sequence comparison. Since there is no currently implementable quantum hardware, the current quantum algorithms, simulators, and prototype quantum systems allow for research into QNLP in bioinformatics (Havlíček et al., 2019).
1.2 Biological challenges, scope, and contribution
Bioinformatics has a significant scope and potential contribution to Quantum Natural Language Processing (QNLP). The knowledge that can be utilized to enhance QNLP models is extracted from biological data using bioinformatics approaches, including text mining and natural language processing. An example of the advanced analytical skills that bioinformatics can contribute to QNLP (Huang et al., 2015) is the utilization of NLP approaches for the detection of noncoding RNA and the prediction of protein structure and function. Particularly in light of the quantum advantage in processing massive amounts of data (Kumar et al., 2024), the computational models and algorithms utilized in bioinformatics to manage huge datasets may be advantageous for QNLP. Furthermore, novel approaches to language processing in QNLP may be presented by the incorporation of bio-inspired models into computing, as elaborated in reference (Jiménez López, 2022).
At the time of biological data processing, however, conventional NLP methodologies confront several obstacles: The dualistic nature and uncertainty in achieving accurate reading and decoding of sentences in biology using typical natural language processing (NLP) models is sometimes difficult due to the terminology’s complexity, which often includes acronyms and multiple meanings (Locke et al., 2021). Integration and analysis of biological data are complicated by the heterogeneity of the data (Hilton et al., 2020). On the contrary, biological data sources include scientific articles, databases, experimental results, and scientific papers; each possesses its distinct organization, format, and nomenclature. Given the rapid expansion of biological data, traditional natural language processing (NLP) methods may face challenges in efficient processing and analyzing enormous datasets, leading to limitations in scalability and performance (Liu et al., 2024). In contrast to classical computers, quantum computers can revolutionize algorithm efficiency through the execution of operations that classical machines are incapable of. This can result in significant accelerations through the avoidance of superfluous computations. The quantum computers can execute intricate computations within days, which would require classical supercomputers an eternity to finish. Despite the lack of fully operational quantum hardware, efforts have continued to create and investigate quantum algorithms for natural language processing (NLP). Recent advancements in prototype construction, coupled with mathematical analysis and the introduction of high-performance quantum computer simulators, have facilitated the investigation of quantum algorithms for a wide range of biological applications (Ohno-Machado et al., 2013).
Although these difficulties highlight the need for novel computational techniques, QNLP presents encouraging paths to fill these gaps. This paper outlines a comprehensive approach to explore these possibilities, bridging theoretical and practical aspects of QNLP in bioinformatics.
1.3 Contribution and organization of the paper
The theoretical underpinnings and practical applications of quantum computing and QNLP in the domain of bioinformatics are encompassed within the scope of this article. In addition to outlining forthcoming opportunities and problems, it offers a complete assessment of the existing status of research in this location. The structure of the paper is as follows:
This research employs a systematic research methodology in Section 2 which outlines, detailing the search strategy, inclusion and exclusion criteria, and quality assessment process used for the review of QNLP in bioinformatics. The foundational principles of quantum computing and an assortment of QNLP approaches are presented in Section 3. Potential QNLP applications in various bioinformatics disciplines, including drug discovery and design, protein structure prediction, genomic sequence analysis, and biomedical literature mining, are examined in Section 4. The problems and limitations of QNLP in bioinformatics are examined in Section 5. These encompass constraints imposed by quantum hardware, concerns related to data representation and encoding, as well as the development and optimization of algorithms. Performance evaluation and comparative analysis are the subjects of Section 6. Evaluation metrics and a comparison of traditional NLP approaches versus QNLP methods are all covered. Section 7 provides a critical examination of the findings in relation to the research questions. In conclusion, Section 8 delves into prospective research avenues Future Research Directions, and Roadmap.
2 Research methodology
We examine Quantum Natural Language Processing (QNLP) and its bioinformatics applications in this exhaustive review. Our primary objective is to define the fundamental concepts of quantum computing and QNLP methodologies, with an emphasis on their potential advantages over conventional NLP approaches. Then, we examine the myriad bioinformatics applications of QNLP, which include biomedical literature mining, drug discovery and design, protein structure prediction, and genomic sequence analysis. Furthermore, we endeavor to recognize and investigate the barriers and restrictions that plague quantum natural language processing in the field of bioinformatics. These include limitations imposed by quantum hardware, complications related to data representation and encoding, as well as difficulties in developing and optimizing algorithms. To assess the practical implications and efficacy of QNLP, a comparative study is undertaken with traditional NLP methodologies. This analysis is substantiated by benchmarking outcomes and empirical case studies.
2.1 Research questions
Research Question 1: Investigate the application of quantum computing concepts and methodologies to natural language processing (QNLP).
Research Question 2: In what ways could QNLP be utilized to automate and enhance the drug discovery and design process, namely in literature mining, drug-target interaction prediction, and virtual screening?
Research Question 3: What is the performance of QNLP algorithms about traditional NLP techniques, as evaluated using benchmarking and criteria metrics?
Research Question 4: What are the current limitations and future opportunities for QNLP in bioinformatics?
Our objective is to furnish a comprehensive synopsis of the present state of QNLP in bioinformatics, expose areas where further research is warranted, and establish a framework for the advancement and adoption of QNLP in this field.
2.2 Search strategy
A comprehensive search was performed for this study, covering the period from 2013 to 2024. The search encompassed several reputable databases, such as PubMed, Scopus, IEEE Xplore, ACM Digital Library, and Web of Science. The extensive inquiry was motivated by the particular emphasis on the utilization of quantum-based techniques in addressing bioinformatics obstacles, such as drug development, Protein structure prediction, and genetic analysis, among others. With deliberate intention, we expanded our search beyond medical databases such as PubMed and Medline, which predominantly cover health informatics and biomedical subjects. Conversely, we investigated numerous databases about the domain of computer science (CS). The papers were identified by the utilization of several screening tasks and quantum computing-related keywords ‘Quantum Search Strategy,’ ‘Quantum Embeddings’, ‘Quantum Mapping’, ‘Quantum Superposition and Quantum Entanglement’, ‘Prediction’ and incorporating NLP keywords like ‘Relation Extraction’, ‘Name entity Recognition’, ‘Semantic Analysis, Sentiment Analysis’, ‘Knowledge discovery’, ‘Machine learning in NLP’.
2.3 Selection criteria
The criteria for including articles in this selection were as follows:
a. Articles must be written in English;
b. Publication date must be 2013 or later;
c. Articles must make initial and significant contributions to the field.
d. Articles must be published as original journal articles or conference proceedings.
This review did not include if:
a. The research was published in the form of a summary, research report, conference abstract, news article, internet-based material, or workshop report, or as a research protocol.
b. The study was identified as duplicates using a systematic approach which includes automatic detection through Zotero followed by manual verification.
c. Articles focused on research involving animals or non-human samples.
d. Articles did not address any of the research questions.
Quality Assessment Criteria (QAC) were created to guarantee the dependability and methodological soundness of the included studies. These standards assess the research based on its contributions to the area, methodological transparency, and relevancy. In addition to ensuring consistency in evaluating the caliber of research, the QACs aid in standardizing the inclusion process.
Each study has been evaluated using the QAC scoring system:
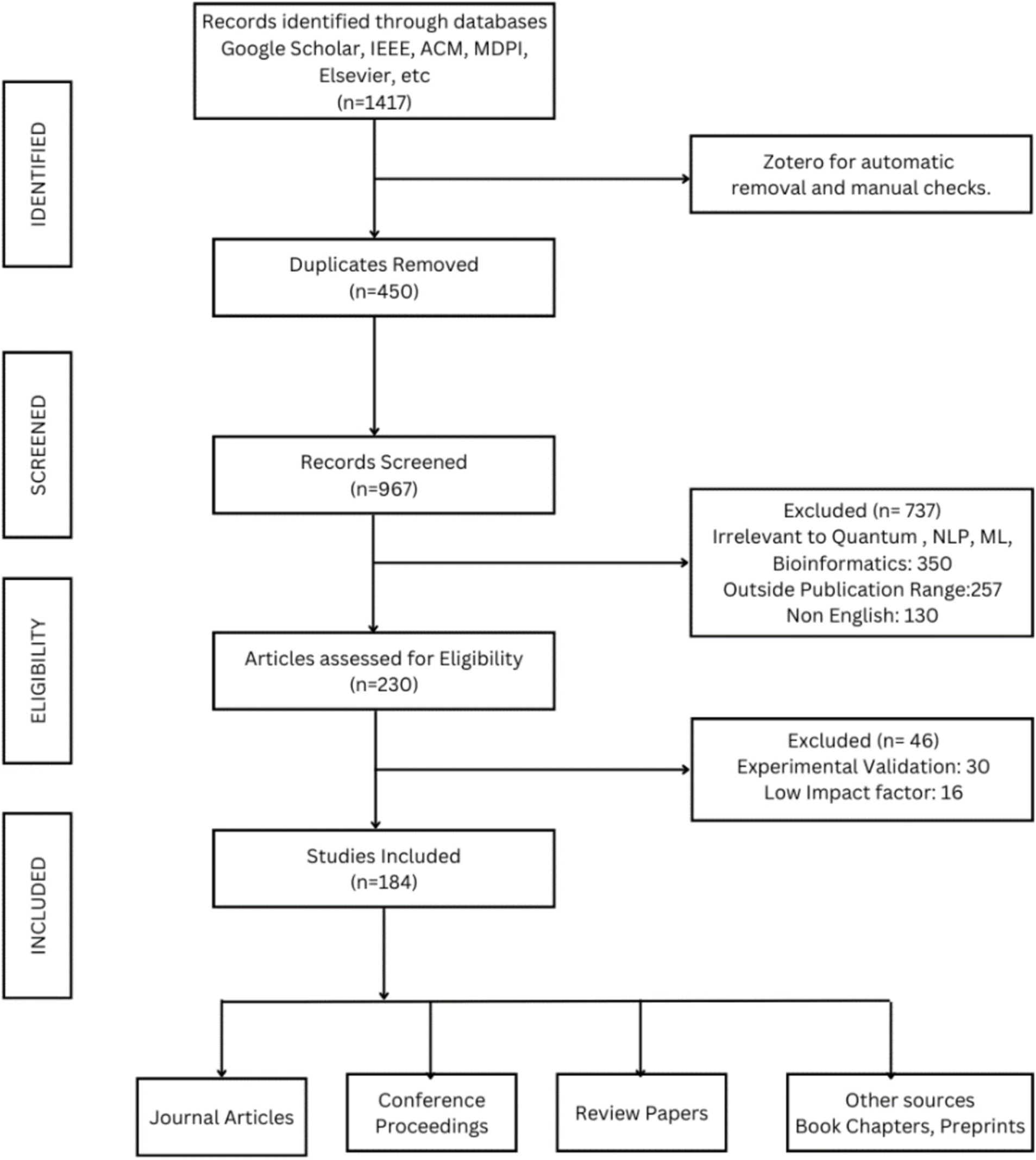
As shown in Figure 1, the initial search yielded a total of 1,417 records from the specified databases. After eliminating 450 duplicate articles, 967 records remained for screening. Subsequently, based on the pre-established inclusion criteria, 737 articles were excluded, leaving us with 230 articles for consideration in the second round of the selection process. Following a thorough examination of the full-text articles, a total of 184 papers were included in this systematic review. The next section begins with the review of the foundational principles of quantum computing and an assortment of QNLP approaches.
3 Quantum computing concepts and QNLP techniques
Quantum Natural Language Processing (QNLP) is an academic discipline that is founded upon the ideas and concepts of quantum computing, which are inherently distinct from the paradigms of classical computing. To comprehend the potential of QNLP and its bioinformatics applications, it is essential first to grasp quantum computing’s fundamentals. This section presents a comprehensive outline of the foundational principles, establishing the preparatory stage for the following discourse on QNLP methodologies and their ramifications within the realm of bioinformatics.
3.1 Fundamental quantum computing concepts
In contrast, quantum computers unlock an entirely new realm of potentialities. The initial obstacle in describing quantum computing is elucidating its information management system. Data is usually maintained in quantum bits, or qubits, which is a quantum version of classical computer bits present in a quantum processor. Integrating quantum computers in artificial intelligence (AI) has implications (Portugal, 2022). AI algorithms, which can be based on traditional computation techniques, could give by quantum processors the ability to gain vast processing capabilities. Defined by specific features such as superposition, interference, entanglement, DE coherence, gates, and circuits. For altering the way how AI deals with data, quantum computing opens the grounds for progressively complex and swift AI operations and the emergence of new quantum algorithms given these phenomena (Piattini et al., 2020).
3.1.1 Qubit
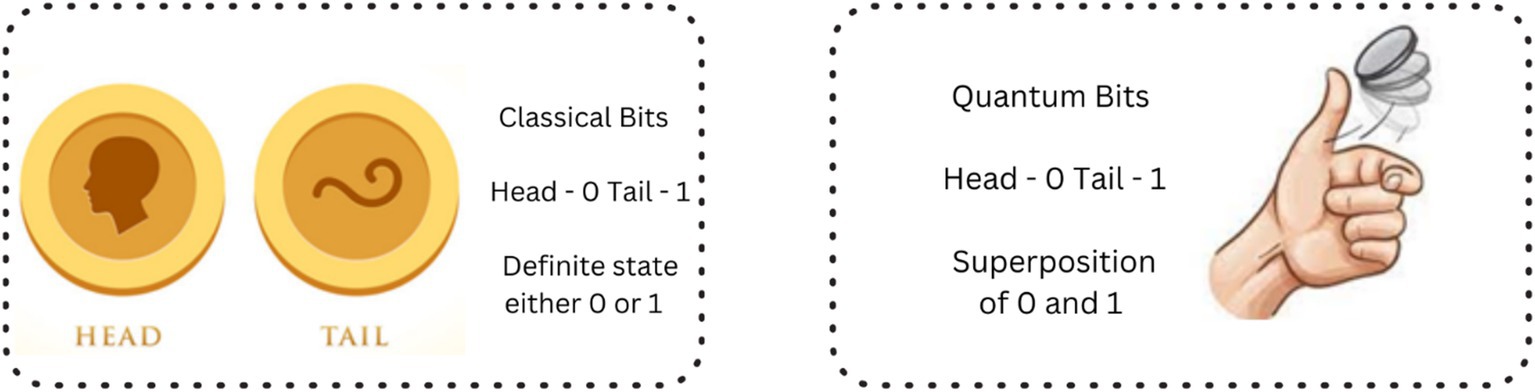
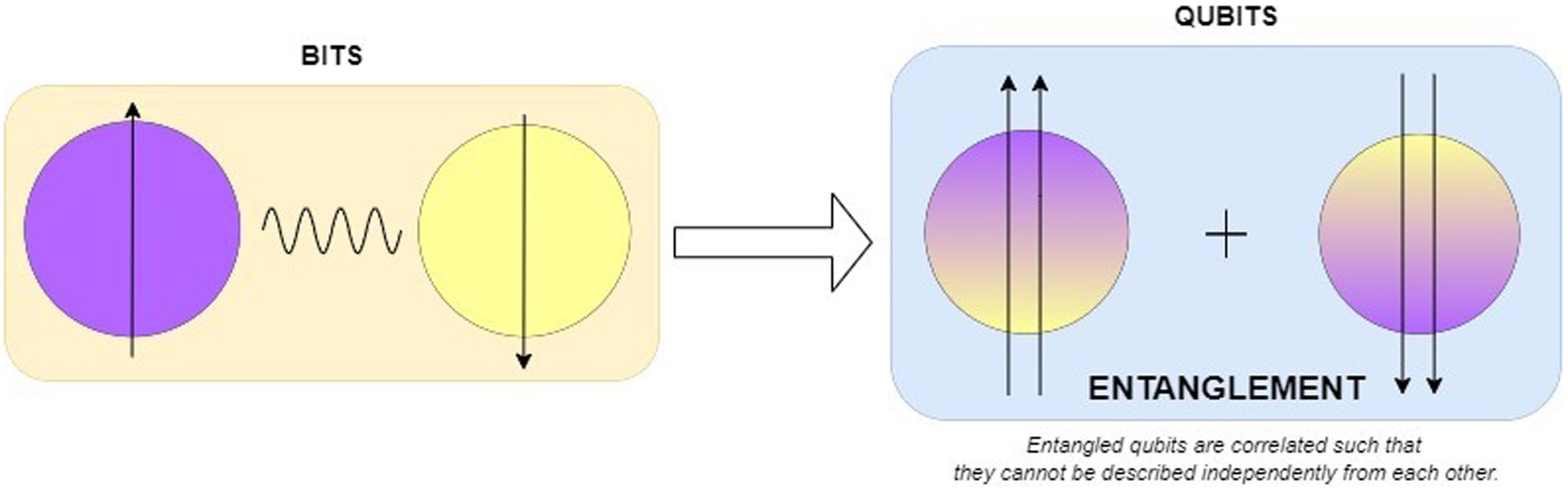
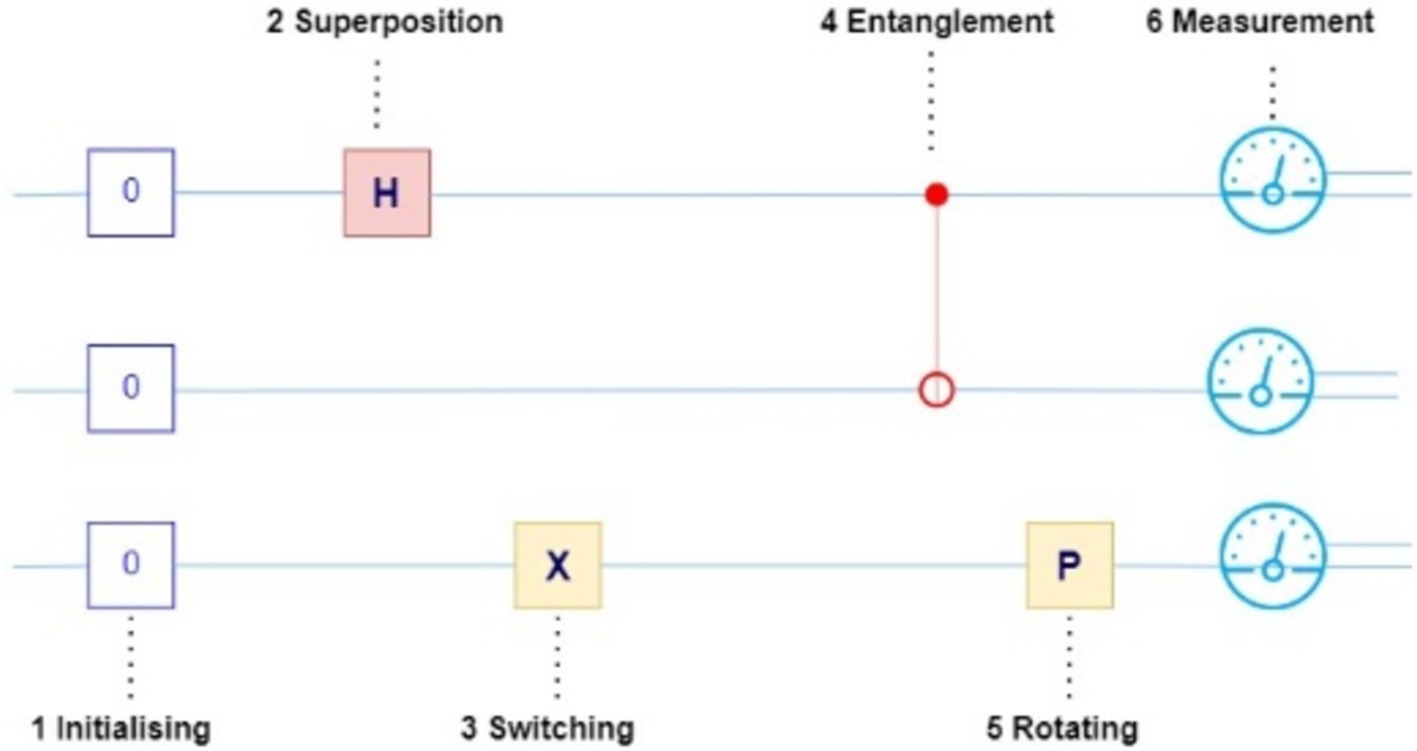
Quantum bits are actual physical systems of a photon with a specific polarization or an ion trapped in a magnetic field. a qubit is described as the basic information unit of a quantum computer. A qubit is different from a classical computer bit which can be either 0 or 1 at any one time but a qubit can be 0 and 1 simultaneously. Observe Figure 2 where the behavior of the coin shows the classical and quantum physics stating the key difference between the deterministic and probabilistic systems which introduces the quantum superposition and entanglement. In classical the coin has two possible states Head or Tail when it is spinning it lands on either head or tail so it has a chance of 50% for both head and tail. In quantum, the coin blends in both head and tail calculating the probability of the states and giving the state that has a high probability value. This is called superposition which can make quantum computers solve many problems altogether hence making them so efficient for specific tasks.
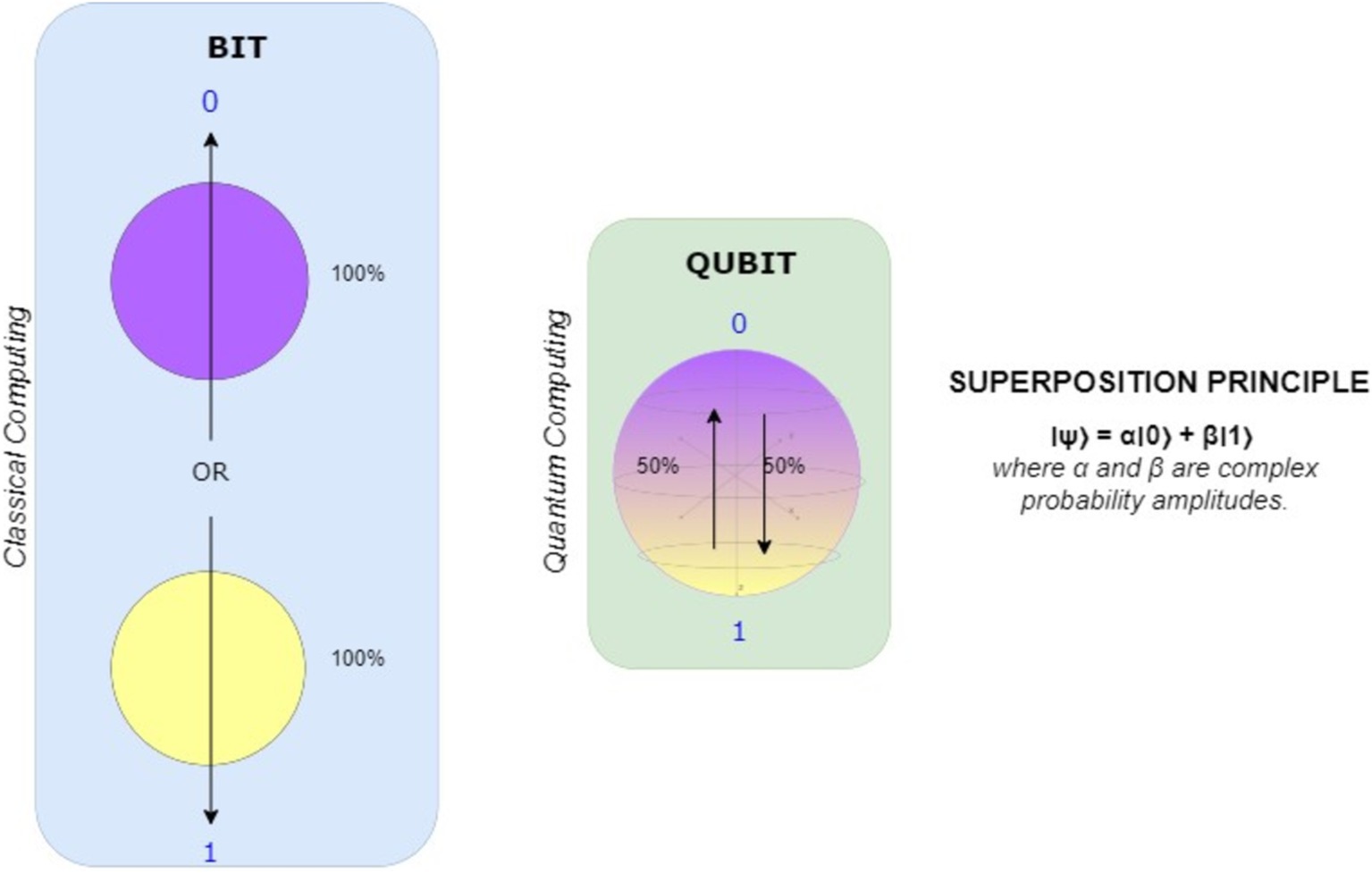
Qubits are basics units of quantum computer systems and are physically implemented through certain entities such as ions subjected to magnetic fields. A qubit can be in any state other than 0 and 1 as Schrodinger’s cat is alive and dead state at the same time (Black et al., 2002). A qubit exhibits one of the following collapse states when observed? Interactions with a single qubit can affect the entire state, as groups of qubits can correlate. Qubits, which are denoted by the complex coefficients α and β, exist in a superposition of the states 0 and 1, whereas bits are the fundamental units of traditional computing.
In Figure 3 watching a qubit changing its state, these coefficients pertain to physical measurements. In the domain of quantum information, the qubit, represented by the Dirac notation |0⟩ and consisting of two states, serves as the comparable entity. |1⟩, where |•⟩ denotes a quantum state. The main difference between quantum and classical information is that, as Equation 1 illustrates, a qubit can exist in any superposition of the states |0⟩ and |1⟩.
The frequencies of the distinct states in quantum computing are denoted by the complex coefficients α and β. As a fundamental principle of quantum physics, these amplitudes are highly susceptible to the impact of physical measurement. A qubit’s state will be altered during the measurement process, by the principles of quantum mechanics (Dejpasand and Sasani Ghamsari, 2023), if it is in a superposition of potential measurement outcomes. As a qubit collapses into its measured state, its amplitudes lose all information. Complex language patterns and relationships in biological data can be represented by qubits, which are capable of existing in superposition states. Simultaneously including numerous linguistic aspects or representations, QNLP models provide a more comprehensive analysis of biological texts, analogous to how a qubit can exist in a superposition of states.
3.1.2 Quantum entanglement
In addition, entanglement is a quantum phenomenon that qubits are capable of manifesting, in which the states of two qubits become coupled irrespective of their separation. It is easier to imagine two qubits as two magic dice. In classical the flipping of two dice results is independent of each other. As shown in Figure 4, in quantum they are “entangled,” then flipping one die immediately reveals the outcome of the other no matter where the two are located across the room, across the country, across the universe! Their results are perfectly correlated, even though the outcomes are random and unpredictable. This magical connection does not work the way we observe other connections that are usual to us but is a natural component of quantum mechanics.
The experimental data show that any quantum calculation that does not require entanglement can be carried out at a slightly slower pace on a classical computer. When computing is associated with systems, which involve many qubits that are susceptible to quantum entanglement, one fully understands the meaning of computing. Entanglement is defined as any process that takes place on a single qubit influences the total state of the whole set of qubits.
Figure 5 represents system consists of two qubits, with each qubit capable of existing in a superposition of the states |0⟩, |1⟩, the combined system can also exist in any superposition of the states |00⟩, |01⟩, |10⟩, |11⟩, and so forth (or any of the 2^N binary strings from |0…0.0⟩ to |1…0.1⟩ in the case of an N-qubit system). The so-called Bell states, which are significant in the context of quantum entanglement (Wong, 2019), are among these superpositions. This is illustrated by Equation 2.
The underlying assumption is based on the potential of the quantum computer to work at data-intensive large volumes. In order to understand this, let us consider a quantum system of N qubits at our disposal. If the state of the system is not entangled, the number of amplitudes in a state of this system is equal to 2^N, where amplitudes of states of each qubit in the system are summed. When the system becomes entangled, however, these amplitudes all become independent and the qubit register as a whole transform into a 2 N-dimensional vector (Tao, 2024). By modeling and analyzing interrelated links between biological things or concepts represented in textual data, the concept of quantum entanglement is utilized. Similar to how quantum entanglement enables the correlation of qubits, QNLP represents and comprehends the intricate interrelationships among proteins, illnesses, genes, and other biological components by employing this idea.
3.1.3 Quantum interference
Quantum interference is employed in the computation of quantum computers with the help of Equation 3, which is helpful. However, acknowledging the fact that quantum computers are inherently noisy and do not always provide accurate results and decisions, they always utilize probability to provide the best guess or most likely occurrence anticipated. Besides locations, quantum computers use the energy level of qubits or spin to do computations (Simmons, 2024). This can be expressed using the Born rule, which states that the probability of measuring a particular outcome x given a quantum state represented by a density matrix ρ is given by:
The weight of each state in the superposition is determined by the complex probability amplitudes, denoted as c1 and c2, which are represented by Equation 4. The interference of various states may result in interference effects in the final state ∣ψtotal⟩, which can manifest as observable phenomena like interference patterns in experiments involving interference, such as the double-slit experiment.
The interference element (c1c2*) in the superposition formula has the potential to induce either constructive or destructive interference, contingent upon the relative phases of 𝑐1 and 𝑐2. The interference behavior described here is an essential component of quantum physics and has far-reaching implications for quantum communication, computation, and other technologies.
The processing and analysis of textual data may be improved with the application of quantum interference (Torlai and Melko, 2020). Utilizing interference effects, QNLP models are capable of conducting more complex and context-aware analyses by modeling language elements and relationships as quantum states (Pseiner et al., 2024). Language modeling, sentiment analysis, and information extraction from biological texts are a few examples of the tasks in which this may result in enhanced precision.
3.1.4 Quantum gates
Quantum information can be manipulated by performing quantum gates wherein physical operations are utilized by using laser pulse for the ion qubits and optical elements for the photonic qubits. Unlike their more often than not conceptual definitions, quantum gates have to be described as unitary matrices by strict quantum mechanical demands. When a quantum gate is performed on at least two entangled qubits, then a 2 N × 2 N matrix is multiplied by a 2 N entity. The fact that quantum computers can register and manipulate roughly 2 N quantities of information using a number of operations equal to N forms the basis of a possible exponential quantum edge over classical computers.
For quantum gates to be used in the normalizing of quantum states, they have to be unit and linear, or act on superposition (Klimov et al., 2024). But, unlike classical computing that involves only one nontrivial gate, namely the NOT gate per bit, quantum computing can perform an infinite number of one-qubit quantum gates. It has been established that any quantum gate can be approximated from a basis set of gates which only includes the single qubit gate and the two qubit controlled NOT gate. The exponential number of gates may hinder the application of good approximations should the need arise in the future. Language properties and relationships can be transformed by quantum gates and therefore, more complex tasks can be performed (Van Vu et al., 2024).
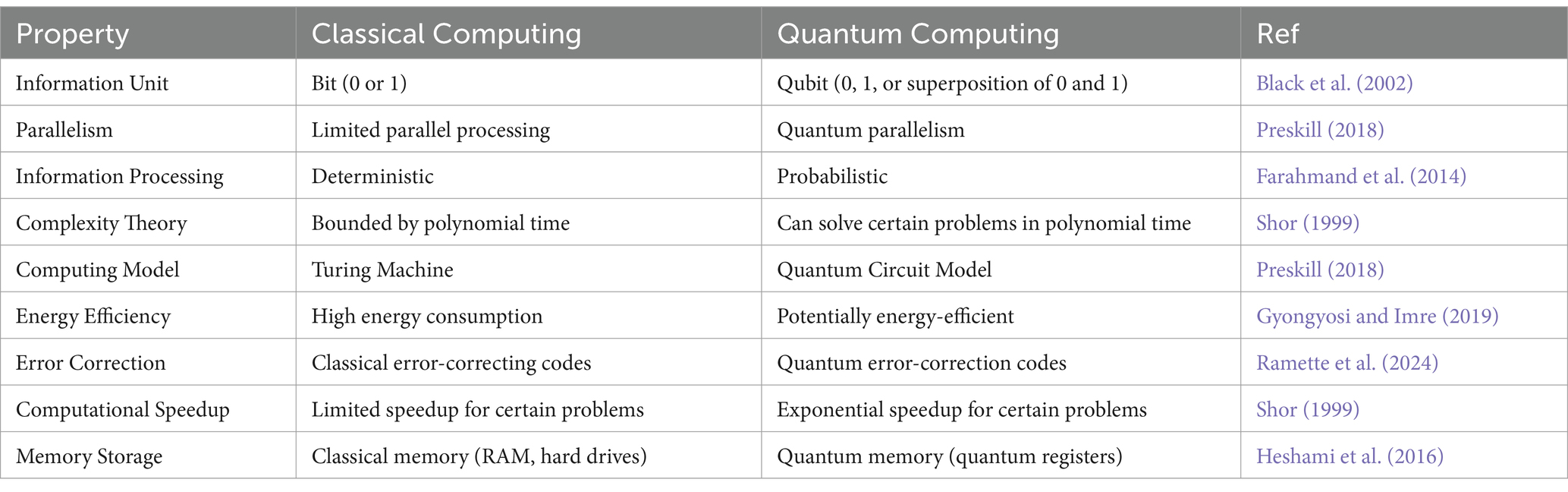
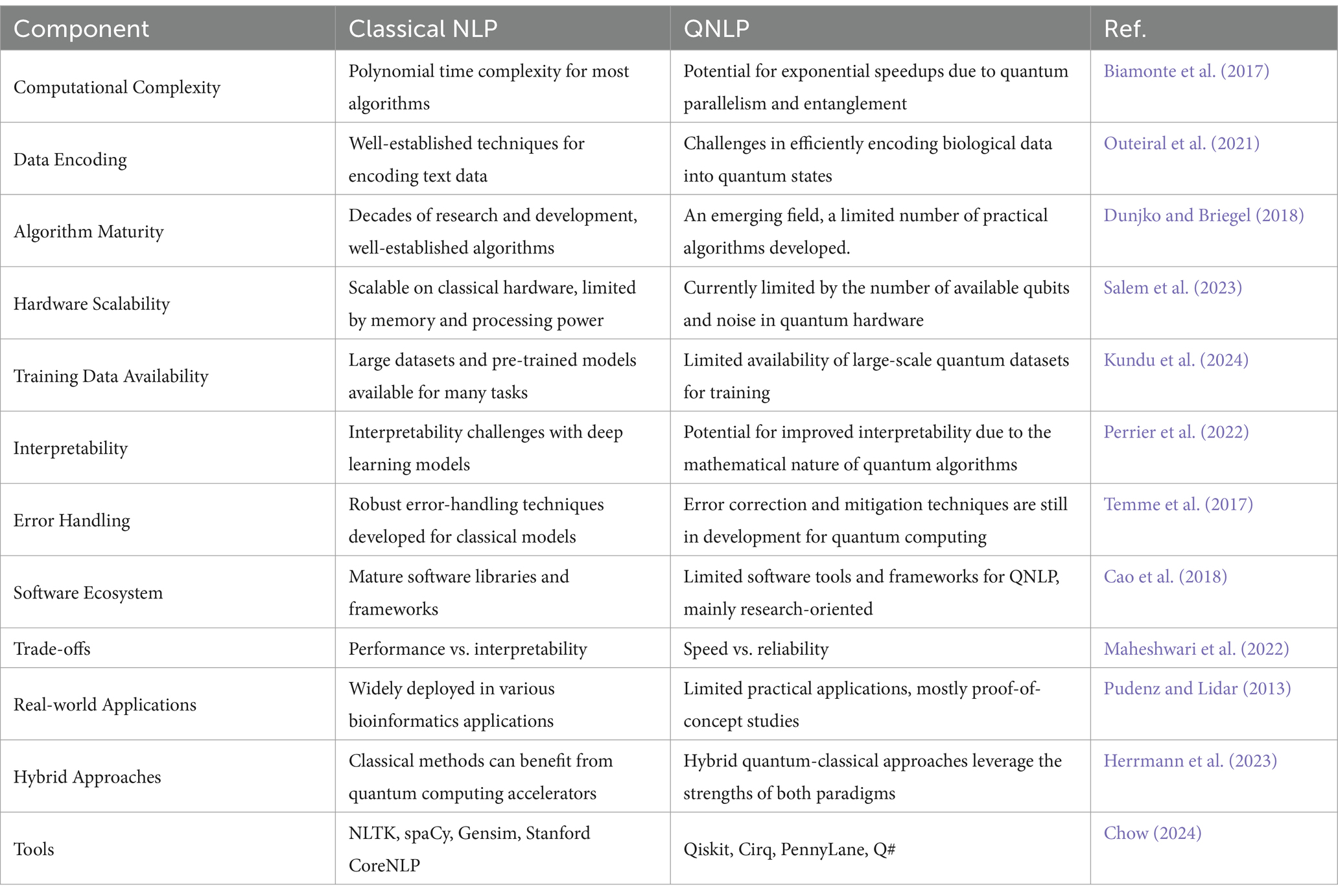
The development of quantum algorithms that are well suited to AI tasks such as data analysis, optimization, and machine learning has elicited a lot of attention from academic and industrial pioneers (Preskill, 2018). These algorithms exploit the specific features of quantum computers and, thus, perform calculations, which makes artificial intelligence operations more complex and faster compared to their counterparts based on classical computers (Patel et al., 2023). The properties of classical and quantum computing that distinguish their respective methods of operation are compared in Table 1.
Having discussed the fundamental principles of quantum computing, the next section focuses on how these principles are adapted and implemented in QNLP methodologies, particularly in bioinformatics applications.
3.2 Quantum natural language processing techniques
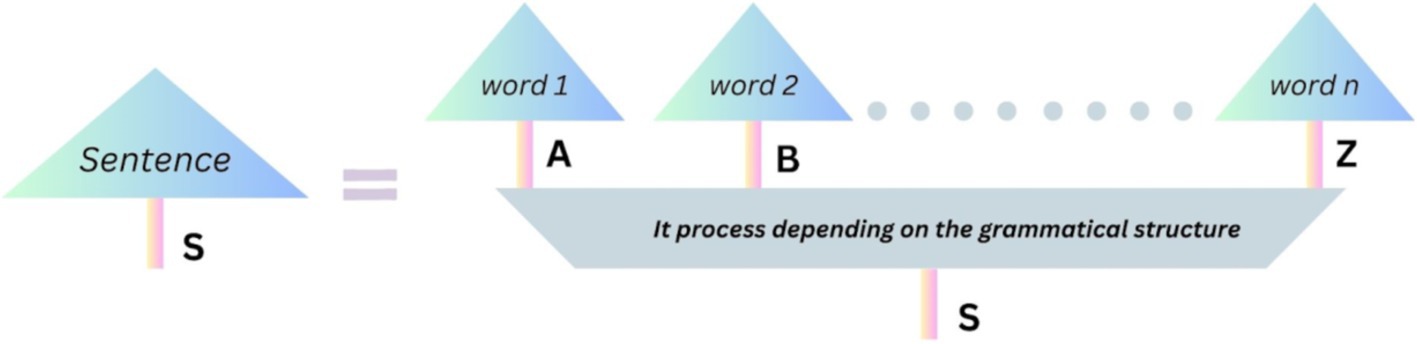
Quantum natural language processing (QNLP) improves natural language processing tasks through the utilization of quantum computing and parameterized quantum circuits to compute word embeddings. Drawing inspiration from categorical quantum mechanics, the DisCoCat framework, this approach transitions from grammatical structure to quantum processes via string diagrams. By employing the DisCoCat framework and Grover’s algorithm, the initial QNLP algorithm showcased a quadratic quantum acceleration in the domain of text categorization and the quantum language models which generates the text using the quantum algorithms by improving the model efficiency.
3.2.1 Quantum embedding
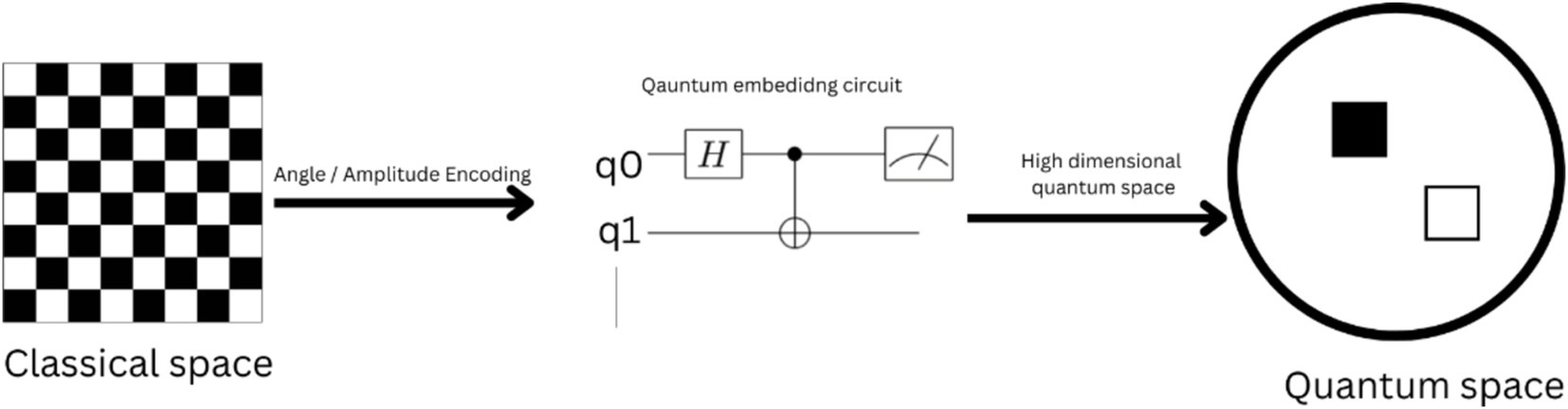
While we have the classical embeddings like Word2Vec or Glove, which map words in the high-dimensional vector space where quantum embedding maps words or phrases as quantum states as observed in Figure 6. This could lead to improved levels of comparison between text analysis and semantic similarity assessments of texts. In few of the research, the advantages of quantum embedding in NLP tasks were highlighted. One of the works suggested a quantum embedding model based on quantum circuits. By mapping words or phrases into quantum states through quantum gates, our paradigm allows more accurate and effective control and description of linguistic features compared to traditional methods (Nam and Nguyen., 2024). A unique approach inspired by quantum mechanics is presented in this study, which utilizes embeddings to facilitate biomedical text-mining tasks including entity detection and relation extraction. By exploiting quantum computing principles, this approach transforms high-dimensional quantum states into biological concepts and relationships (Samanta et al., 2016; Baiardi et al., 2023).
One approach is to use amplitude encoding, where each word is encoded as a quantum state is represented in Equation 5:
where, |i⟩ represents the basis states, and α_i are the complex amplitudes corresponding to the word w. An innovative methodology utilizes the principles of quantum computing to encode relationships and concepts in biomedicine into high-dimensional quantum states.
3.2.2 Discocat framework
Quantum Natural Language Processing (QNLP) is a recent and fascinating application of quantum computing that seeks to represent the meaning of sentences as vectors encoded into quantum computers (Abbaszade et al., 2021). It achieves this by extending the distributional meaning of words to encompass the compositional meaning of sentences, a concept known as the DisCoCat model (Martinez and Leroy-Meline, 2022). This model employs an algorithm based on tensor products to compose the vectors representing the meanings of words through the syntactic structure of the sentence. One striking aspect of this approach is that while the algorithm is inefficient on classical computers, it exhibits promising scalability when executed using quantum circuits.
One of the fundamental ideas underlying the convergence of quantum theory and natural language processing is the establishment of a direct link between linguistic features, such as syntactic structures and semantic meanings, and quantum states (Surov et al., 2021). As illustrated in Figure 5 (Yeung and Kartsaklis, 2021), the DisCoCat framework serves as a network-like language for accomplishing this relationship through the use of string diagrams as given in Figure 7. This approach is an integral part of the longstanding tradition of computational linguistics, which has sought efficient methods for describing language structures and meanings in machine-accessible formats (Tsujii, 2021). The distributional approach relies on statistical analysis of word contexts based on the distributional hypothesis. In contrast, the symbolic approach focuses on individual word meanings and the compositionality of sentences. The symbolic approach, rooted in theoretical linguistics, posits that the meaning of a sentence depends on the meanings of its constituent words and the grammar used to arrange them (Ganguly et al., 2022). One of paper shows how DisCoCat allows QNLP to classify biomedical abstracts by building sentence embeddings that retain structural and content-based characteristics. By mapping the syntactic dependencies of sentences to quantum states, the framework improves the identification of functional/contextual similarities in biology articles (Steedman and Baldridge, 2011). However, this approach has seen limited success in natural language processing applications, where the distributional paradigm, based on statistical analysis, currently dominates (Liu et al., 2024).
3.2.3 Quantum language models
Quantum language models (QLM) is a kind of quantum-inspired neural network model that defines language units, such as words and phrases, as quantum states in Hilbert space and create text using quantum algorithms, which may result in exponential speedups compared to classical models. Complex patterns in language data can be efficiently learned by the Quantum Boltzmann Machine (QBM), allowing for more precise language production and modeling (Wiebe et al., 2019). The present study introduces a novel quantum circuit-based QLM architecture and showcases its efficacy in various domains, including text classification, sentiment analysis, and language modeling (Shuyue et al., 2023). The existing body of research primarily represents word sequences as a classical mixture of word states, which fails to adequately utilize the capabilities of a probabilistic quantum description (Campbell et al., 2024). As of yet, a comprehensive quantum model that explicitly captures the non-classical correlations inside word sequences has not been created (Yu et al., 2020). A neural network architecture has been suggested, using an innovative Entanglement Embedding (EE) component, to convert word sequences into entangled pure states of many-body quantum systems. The word sequences exhibit robust quantum entanglement, a fundamental principle of quantum information and a sign of parallelized correlations among the words (Chen et al., 2023; Figure 8).
Equation 6 represents the quantum circuit to estimate the probability distribution over words, given a context as follows:
Where, |ψ(c)⟩ is the quantum state representing the context, U_w is a unitary operator corresponding to the word w, and ⟨ψ(c)|U_w|ψ(c)⟩ is the probability amplitude of observing w given c.
In the field of bioinformatics, QLMs predict the probability of a word in a context so that functions such as sequence alignment have been accomplished (Liang et al., 2023). Linguistic features are processed in quantum circuits which enhances the accuracy of sentiment to be derived from research abstracts or clinical data (D'Aloisio et al., 2024). Potential advantages of this quantum approach to estimating the probability distribution across words over classical methods include a more effective capacity to capture complex dependencies and context information (Jayanth et al., 2023).
These techniques not only capture complex linguistic relationships but also lay the foundation for addressing bioinformatics where semantic precision is critical. In the following section, we explore how Quantum techniques are used in the QNLP tasks.
3.3 Quantum techniques for QNLP
Quantum natural language processing extends the recent advances of classical machine learning and quantum machine learning to process language. Traditional deep learning methods like embeddings, neural networks, and transformers have paved the way for NLP progress by allowing functions such as text categorization, sentiment analysis, and translation. The above approaches are expanded on by QNLP but with the use of QML for scalability and efficiency. Two methods, Quantum Circuit Learning (QCL) and Quantum Kernel Learning (QKL), introduced in the paper, are designed to contribute to QNLP tasks as QNLP tasks involve encoding linguistic data into quantum states and pattern matching. Hybrid quantum-classical methods even extend the capacity of QNLP due to integration of quantum advantage with classical flexibility. For the first time, QNLP applies quantum optimization and quantum embeddings, thus connecting traditional NLP with quantum calculations and offering effective approaches to context-sensitive language processing, entity identification, and semantic search. It is in this regard that this relationship demonstrates how QNLP applies ML and QML frameworks to reinvent NLP in bioinformatics and more broadly.
3.3.1 Quantum machine learning in bioinformatics
Machine learning (ML) is a branch of Artificial intelligence that enables systems to learn patterns from data and make predictions. NLP focuses on equipping computers to comprehend, interpret, and generate human language and ML uses that data to generate predictions, decisions, and classifications. This ML integration in NLP is observed in Figure 9.
Quantum Machine Learning (QML) combines quantum computing with machine learning to enhance data processing by leveraging quantum properties like superposition and entanglement, offering exponential speedups and richer data representations (Das Sarma et al., 2019). Within QML, Quantum Natural Language Processing (QNLP) specializes in applying quantum principles to linguistic tasks, encoding text as quantum states and enabling efficient processing of language structures. Like sentiment analysis, semantic parsing, and relation extraction to scale efficiently while uncovering deeper patterns in language using quantum-enhanced embeddings and kernels. As Bioinformatics involves analyzing and interpreting large volumes of biological data, such as genomic sequences, protein structures, and gene expression data. QNLP benefits from QML’s advanced computation, many problems in this field can be formulated as machine learning tasks, such as classification, clustering, and pattern recognition (Repetto et al., 2024; Ghoabdi and Afsaneh, 2023).
Quantum machine learning (QML) has emerged as a promising approach to tackle these challenges, leveraging the principles of quantum mechanics to potentially enhance the computational capabilities of classical machine learning algorithms. The following concepts are provided for further enhancement of QML techniques.
3.3.1.1 Quantum circuit learning for bioinformatics
QML in bioinformatics is quantum circuit learning, which is the process of training parameterized quantum circuits to perform certain kinds of machine learning. These circuits can be represented as unitary operators U(θ), where θ is the set of trainable parameters. It is to find the best parameter θ* that may minimize a cost function C(θ) suitable for the bioinformatics problem in question. Mathematically, the optimization problem can be formulated as Equation 7:
The cost function C(θ) is then calculated through using the quantum circuit U(θ) on the input states that are equivalent to the biological data and measuring the output states. The derivatives of the cost function with respect to the parameters can be estimated by methods such as the parameter shift rules or analytical derivatives, and thus optimisation can be performed by gradient-based methods. Some of the works in the field of quantum circuit learning include protein structure prediction (Madsen et al., 2023), gene expression analysis (Navneet and Pokhrel, 2024), and genomic sequence classification (Zarei and Elaheh, 2024). Robert et al. (2021) developed a new method for the prediction of the secondary structure of proteins by using Quantum Circuit Learning (QCL). Their model employs a parameterized quantum circuit that takes an amino acid sequence of a protein as the input where QNLP could extract the secondary structure information from text-based annotations where further when coupled with quantum circuit learning to predict components like alpha helices or beta sheets, gives better precision and lesser time than other traditional methods. By adjusting the parameters of the quantum circuit with gradient information of the cost function, the model reduces the error between the predicted and actual secondary structures.
3.3.1.2 Quantum kernel methods for bioinformatics
Another subfield of QML in bioinformatics is the so-called quantum kernel methods, which apply quantum features to improve the efficiency of kernel-based machine learning techniques, including SVMs and Kernel methods. Quantum kernel methods are a very suitable addition to QNLP techniques since they provide a method of computing quantum similarity between quantum-encoded linguistic features. This approach may be applied to bioinformatics tasks for semantic classification, such as analyzing connections between drugs and diseases in biological texts.
In quantum kernel methods, the traditional kernel is substituted by a quantum kernel which is determined by the inner product of the quantum states corresponding to biological data. Mathematically, the quantum kernel between two data points x and y can be expressed as:
Where, in Equation 8 |ψ(x)⟩ and |ψ(y)⟩ are the quantum states of the data points x and y, respectively. Indeed, quantum kernel methods have been applied in other bioinformatics applications, including quantum machine learning for genomics data (Abbas, 2024), quantum kernel clustering for protein sequences (Sarkar, 2018), and quantum support vector machines for gene expression analysis (Ghosh et al., 2024). Ng et al. (2023) considered Quantum Kernel Support Vector Machines (QK-SVM) for classifying gene expression data derived from microarray experiments. Instead of using classical kernels such as linear or radial basis function (RBF), they suggested a quantum kernel that measures the similarity between the gene expression patterns with the help of their quantum state representations using QNLP. The developed QK-SVM algorithm uses this quantum kernel to classify gene expression datasets (Kang et al., 2019). The results of this study indicated that the proposed QK-SVM had a better performance of the classification than the classical SVM with traditional kernels on gene expression data sets.
3.3.1.3 Hybrid quantum-classical approaches
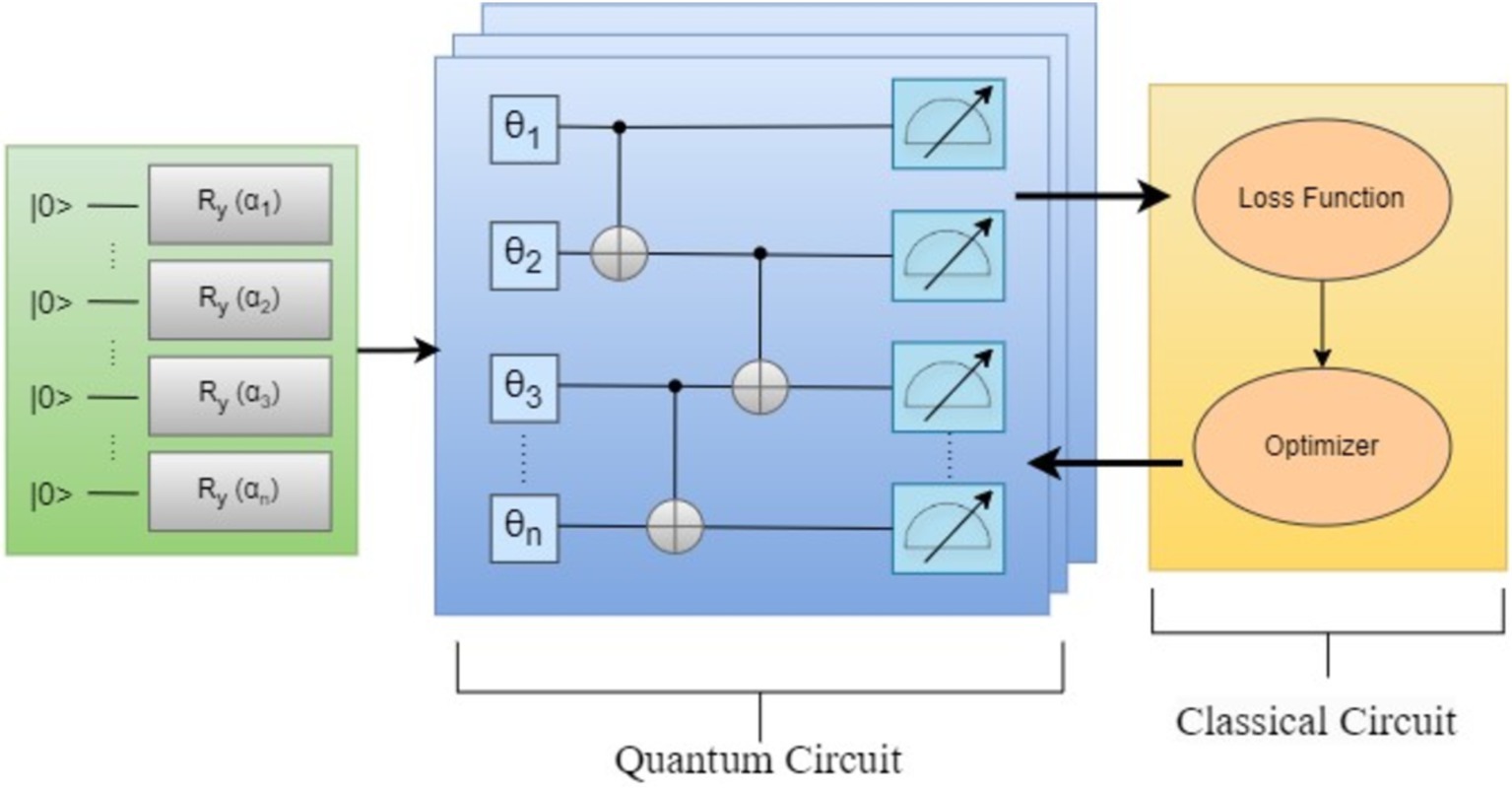
In addition to purely quantum approaches, hybrid quantum-classical algorithms have also been explored in bioinformatics. By utilizing classical models for extensive biomedical text analysis, hybrid approaches allow quantum circuits to encode context-sensitive language properties. Figure 10 shows both classical and quantum computing resources to solve complex problems more efficiently.
Figure 11 shows the high-level structure of the Hybrid quantum classical approach, there is a quantum circuit with quantum gates θ1, θ2, θ3, and θ4 acting on the input gates. The quantum circuit yields an output which becomes the input for the classical circuit. This approach combines both quantum and classical computing to determine parameters that minimize the loss function which can be any cost function or objective function depending on the use.
Multiple sequence Alignment (MSA) is a basic problem in computational biology that involves comparing biological sequences such as DNA, RNA, or protein sequences; Madsen et al. (2023) proposed a new hybrid quantum-classical algorithm for solving this problem. This algorithm utilizes quantum and classical hardware to solve the MSA problem more effectively. The quantum part uses quantum parallelism and superposition to investigate the massive solution space of potential alignments. The application of QML may be vital for solving challenging bioinformatics issues and improving the study of biological processes. Other research works by Cincio et al. (2020) and Hatakeyama-Sato et al. (2022) have investigated ways of reducing noise and errors in quantum machine learning to guide more efficient and useful applications in bioinformatics.
3.3.2 Quantum simulation
Quantum simulation involves using quantum computers to solve and model quantum systems which cannot even be approximated by classical computers due to the exponential resource requirement. It opens the chance to model complicated dependencies in the structures of language which is in concordance with the outlook of QNLP. Conducting new studies in this field has been made to explore many approaches to quantum simulation that can offer explanations of numerous quantum phenomena in various fields. A quantum circuit model which encodes the quantum system into a sequence of quantum gates acting on an initial state. As proposed in a recent study (Durán et al., 2023), the dynamics of a quantum system can be simulated using Equation 9:
Where |Ψ(0)⟩ is the initial state, U(t) is the time-evolution operator implemented as a quantum circuit, and |Ψ(t)⟩ is the final simulated state after time t. The study demonstrated an efficient simulation of quantum problems using this approach. Another study (Miranda et al., 2022) explored quantum annealing for simulating quantum systems, encoding the system into an energy landscape described by a quantum Hamiltonian. The quantum annealing process finds the ground state of the Hamiltonian, corresponding to the simulated system state.
Optimising new quantum algorithms and approximations of quantum circuits for large scale problems. In a recent work by Edward et al. (2024), the authors presented a quantum simulation strategy to simulate biomolecular events including protein folding changes using near-term quantum computers. They explained the basic ideas of the simulation of protein model and showed that quantum computers could be used for investigations of biomolecular systems. For this, they created a quantum algorithm that would be able to efficiently simulate quantum dynamics of the biomolecular system, using quantum characteristics such as superposition and entanglement. Magann et al. (2021) studied the application of quantum simulation in protein–ligand interactions because these interactions play a vital role in the discovery of drugs and engineering of proteins. They proposed a quantum algorithm for the quantum dynamics of a protein-ligand system and pondered over the benefits of applying quantum computing over the classical approach to this sphere. Cao et al. (2018) discussed the use of quantum simulation to study the Protein-DNA binding, which is critical in gene control and drug development. They designed a quantum algorithm for seeking an optimal binding of a protein to a DNA sequence. They debated the significance of such findings for future studies of gene regulation and the development of treatments that modify protein-DNA interactions.
3.3.3 Quantum optimization
The major issues in bioinformatics can be addressed using mathematical programming, which involves identifying the solution that provides the minimum or maximum value to a specified objective function. Such problems occur when the search space is large; therefore, it is computationally expensive when solved using conventional algorithms (Reali et al., 2017). The objective function is in turn mapped to a Quantum Hamiltonian which describes a wave operator for a quantum system. The goal is to find an optimal solution of the optimization problem which is the ground state energy value of the Hamiltonian.
Let us consider a general optimization problem:
Minimize f(x) Subject to x ∈ S where f(x) is the objective function, and S is the set of feasible solutions. In quantum optimization, this problem can be mapped to a quantum Hamiltonian, H, such that the ground state of H corresponds to the optimal solution(s) of the problem. Equation 10 expresses the quantum Hamiltonian mathematically by:
Where: 𝐻o represents the initial Hamiltonian, 𝐻𝑝 is the problem Hamiltonian encoding the objective function and constraints, and 𝛾 is a parameter controlling the weight of 𝐻𝑝 in the overall Hamiltonian.
The goal is to find the ground state of the Hamiltonian H, which can be achieved using quantum algorithms such as quantum annealing or the quantum approximate optimization algorithm (QAOA; Perdomo-Ortiz et al., 2012). Quantum annealing is a process where the quantum system is initialized in the ground state of H_0 and then slowly evolves towards the ground state of H by gradually increasing the value of γ. This process exploits quantum phenomena like superposition and tunneling to explore the vast solution space more efficiently than classical algorithms.
The QAOA is an iterative algorithm that alternates between applying a mixer operator and a phase operator to an initial quantum state. The mixer operator introduces quantum entanglement, while the phase operator encodes the objective function and constraints. The parameters of these operators are optimized to find an approximate solution to the optimization problem.
Mathematically, Equation 11 represents the QAOA as follows:
Where ∣ψ(γ,β) is the quantum state prepared by the circuit, parameterized by vectors 𝛾 and 𝛽,𝐻𝐵 and HC are the mixing and cost Hamiltonians respectively, representing the classical and quantum parts of the optimization problem,𝛽𝑝,𝛾𝑝 are the parameters controlling the evolution of the quantum state,∣𝜓0⟩ is an initial state, The circuit prepares a state that is expected to encode the optimal solution to the optimization problem and the parameters 𝛾 and 𝛽 are chosen to optimize the objective function.
These techniques optimize parameters in quantum linguistic embeddings, enabling tasks like accurate biomedical question-answering and semantic search. Quantum optimization has shown promise in multiple sequence alignment (MSA), which is a fundamental problem in bioinformatics. The objective of MSA is to discern areas of similarity across numerous biological sequences, hence offering valuable insights into functional conservation and evolutionary connections. Utilizing the quantum features of superposition and tunneling, a recent work (Benedetti et al., 2017) developed a quantum annealing methodology for MSA that more effectively explores the huge solution space than classical methods. Protein structure prediction is yet another bioinformatics use of quantum optimization. The optimization issue of deducing the three-dimensional structure of a protein from its amino acid sequence is extremely difficult and involves a large number of local minima. Scholars have investigated the application of quantum annealing and other techniques for quantum optimization to resolve this issue (Patti et al., 2022). Additionally, quantum optimization has been implemented to address the issue of inferring gene regulatory networks. The aforementioned networks symbolize the intricate interplay between genes and their regulatory factors. Deducing these networks from experimental data necessitates the resolution of extensive optimization challenges. A recent study (Mokhtari et al., 2024) introduced a method for deducing gene regulatory networks using quantum annealing, capitalizing on the quantum computer’s enhanced efficiency in traversing the extensive solution space. In addition, additional bioinformatics issues, including drug discovery (Onodera et al., 2023), phylogenetic tree reconstruction (Bach et al., 2024), and genomic sequence assembly (Boev et al., 2021), have demonstrated the potential of quantum optimization.
While these theoretical advances demonstrate the potential of QNLP, their true impact becomes evident in addressing practical challenges in bioinformatics. The following sections explore how these QNLP techniques are applied to bioinformatics problems, such as drug discovery, protein structure prediction, and genomic sequence analysis.
4 QNLP and its applications in bioinformatics
Exploring the application of quantum mechanics principles to the analysis of biological systems, quantum bioinformatics is an emerging field. The nature of computing operations, the platform type, and the type of biological data are the three key determinants upon which a comprehensive categorization system for quantum bioinformatics can be constructed, according to Marchetti et al. (2022). Calculations and tools for data mining activities, and the design, modeling, or creation of computational tools or optimization, are the two primary categories into which computational operations can be classified, according to this study. Furthermore, it is possible to categorize the platform under consideration as either classical or quantum computers and to differentiate between quantum biological data and classical biological data within the realm of biological information. The transformative potential of quantum computing algorithms in the field of computational biology has been recognized by numerous studies. These algorithms possess the capacity to solve, expedite, or improve the examination of a wide range of computational obstacles. The capacity to efficiently map multi-scale biological systems and genetic analysis onto quantum architectures is one example of the potential advantages that quantum computing algorithms may provide over their traditional counterparts, according to a paper by Sathan et al. (Sathan and Baichoo, 2024).
4.1 Applications of QNLP in bioinformatics
QNLP works with biological information of sequences such as DNA using quantum computing methods. Quantum scientists have developed large-scale QNLP models that can classify sequences: DNA included. By employing tensor networks, these models are ‘syntax aware’—they are concerned with structure and syntax from the outset. Thus, the models are more comprehensible and it takes fewer gate operations to work with them in order to gain an understanding of them. Incorporation of tensor networks and quantum theory in QNLP models allows the scientists to study the synergy of AI and quantum informatics in the sphere of bioinformatics. Due to such optimal functionalities like mid-circuit measurement and qubit reuse, quantum processors allow for the execution of circuits larger than what can be done on the quantum hardware (Nałęcz-Charkiewicz et al., 2024). This feature makes available QNLP models for the down and dirty bioinformatics applications featured in the subsequent sections.
4.2 Literature mining and knowledge extraction
The most common application of the Quantum Natural Language Processing methodologies in the pharmaceutical study is the knowledge mining and extraction from the biomedical literature. This is due to the exponential increase of data in the biomedical field and unstructured scientific literature that remains a challenge in the application of typical NLP methods as depicted in Figure 12.
In this regard, scholars have analyzed the potential of QML models and algorithms to search for information in biological text data as fast and accurately as possible. Other applications of QNLP include integration of QNLP with other quantum computing paradigms including quantum simulation and quantum machine learning techniques for Biomedical literature mining. They gathered knowledge graphs from biomedical literature using QNLP and used quantum graph neural networks to identify patterns, associations, and related entities in those knowledge graphs. The approach revealed relatively good effectiveness in the search for complex relations, such as higher-order patterns and nested relationships, which are critical for understanding the pathogenesis of diseases and identifying potential drug targets.
4.3 Drug discovery and design
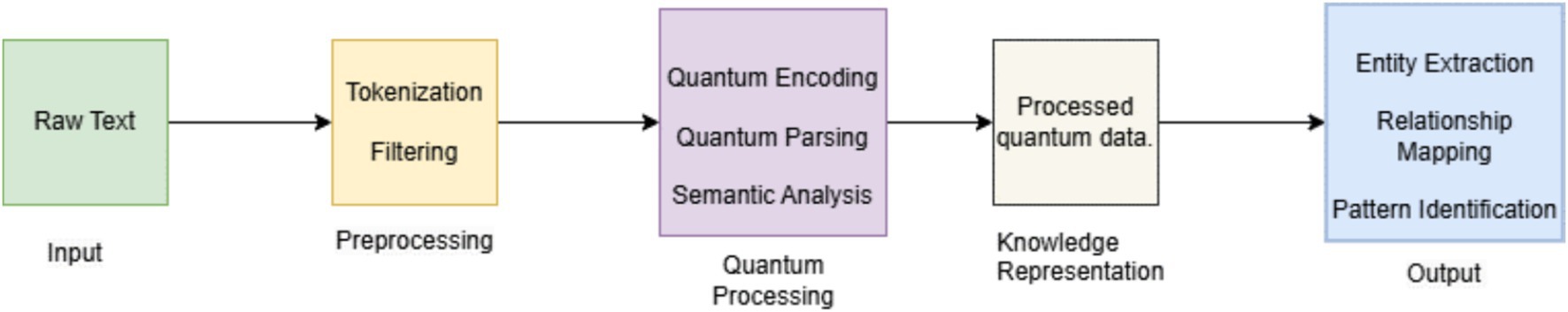
Analyzing huge quantities of biomedical literature and data is crucial to the drug discovery and design process to identify prospective therapeutic targets, develop lead compounds, and comprehend drug-target interactions. Figure 13 shows the procedure how Quantum Natural Language Processing (QNLP) methods, enable the extraction of knowledge from unstructured text input in a more precise and efficient manner.
4.3.1 Virtual screening and Lead compound identification
The process of drug discovery means virtual screening and identification of lead compounds as the major stages. These procedures estimate the affinities and selectivity of potential drugs to the target biomolecules. In this context, studies have been conducted to enhance the accuracy and efficiency of the procedures of QNLP, alongside machine learning and quantum computing models. In this approach, QNLP tools are used to search biomedical literature for information about potential drug candidates and their structures and targets (Gorgulla et al., 2022). This information is then employed and incorporated to commence quantum simulations of these drug candidates and the target biomolecules at the quantum level. QNLP can extract and incorporate useful knowledge from large volumes of text data more effectively than conventional approaches; quantum simulations allow for realistic modelling of the interaction of molecules and their properties.
4.3.2 Drug-target interaction prediction and analysis
Previous studies have revealed that QNLP methods are very useful in predicting and analyzing the drug-target interactions that are so important in the drug discovery phase. Some studies have investigated how quantum-based machine learning algorithms, using big databases of known drug–target associations and molecular conformations, can be used to predict new interactions and the underlying processes (Ginex et al., 2024). This approach shows the possibility of using QNLP in conjunction with other quantum computing methods for a comprehensive analysis of the interactions between drugs and targets for their use in drug discovery and development.
These techniques as a group demonstrate the usefulness of QNLP in understanding the drug discovery process. As we shall see, by utilizing properties of quantum computing that are intrinsically superior to classical computing, such as quantum parallelism and entanglement, one can hope for QNLP techniques to surpass classical models in terms of their ability to identify novel interactions and intricate patterns and structures within the data.
4.4 Protein structure prediction
Protein Structure Prediction (PSP) is an essential sub-discipline of computational biology, which involves predicting a protein’s tertiary structure and its secondary structures, such as helices and sheets from its amino acid sequence. Primary structure prediction mainly concerns with the local structures while the secondary structure predicts the local conformation and the tertiary structure concerns with over all three dimensional conformations. New opportunities for further PSP have opened due to the advances in quantum computing and quantum natural language processing, a process of identifying the three-dimensional conformation of the protein based on the amino acid sequence.
Accurate identification of PSP is essential in elucidating protein function and the mode of interaction in structural bioinformatics and for designing therapeutic strategies. Although, recent progress in computational techniques such as Alphafold and the availability of experimental structures, the protein folding problem remains challenging. This is a problem that has recently attracted the interest of the scientific community to be solved using a novel approach called Quantum Natural Language Processing (QNLP) that incorporates quantum computing. It uses quantum mechanism principles that facilitate better feature extraction and optimization of feature search space concerning sequence structure relationship.
Figure 14 shows the workflow for the prediction of protein structure using QNLP techniques. The process includes data pre-processing where protein sequences are retrieved from databases such as PDB and converted into quantum states and includes QNLP techniques such as quantum language models for sequence analysis, quantum kernel methods for structural similarities, model training using experimental datasets such as cryo-EM and X-ray crystallography results in predicted protein structure as the output. Despite the current limited development of QNLP for protein structure prediction, some investigations have been made to examine its advantages. These quantum models showed that by leveraging quantum phenomena such as superposition and entanglement, signal features could be represented with higher complexity and long range dependency could also be captured much better than with classical models. AlphaFold 2, a groundbreaking tool, combines evolutionary coupling with deep learning techniques to predict secondary structures alongside tertiary configurations. Predicting the tertiary structure of a protein, where the protein’s entire three-dimensional conformation is predicted, is still a computationally expensive task (Doga et al., 2024). QNLP compared to classical methods quantum embeddings can capture intricate sequence dependencies, knowledge integration where the unified representation of sequences, structure and experimental data. QNLP presents a promising avenue for enhancing protein structure prediction by enabling more efficient feature engineering, knowledge integration, and the development of quantum algorithms and simulations tailored for this challenging problem. The potential of QNLP to push the boundaries of computational protein structure prediction is evident in the growing body of literature in this field.
4.5 Genomic sequence analysis
Sequence comparison is one of the most basic tasks in bioinformatics and comprises sequence alignment, search for conserved motifs and patterns. They are important in characterizing biological systems, diagnosing diseases, and promoting development of individualized medication. These tasks have been traditionally solved by using well-known computational tools, namely the Smith-Waterman and Needleman-Wunsch ones. However, they are usually constrained in terms of the computational costs and time required when analyzing large scale genomic data. The development of quantum computing over the past few years including Quantum Natural Language Processing (QNLP) has brought new solutions to these problems.
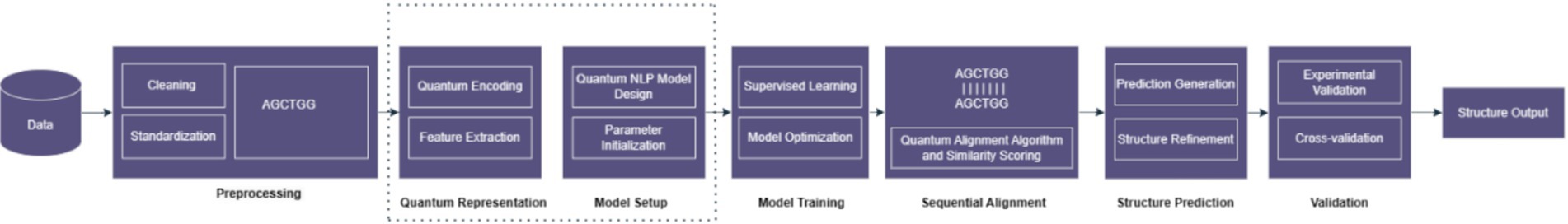
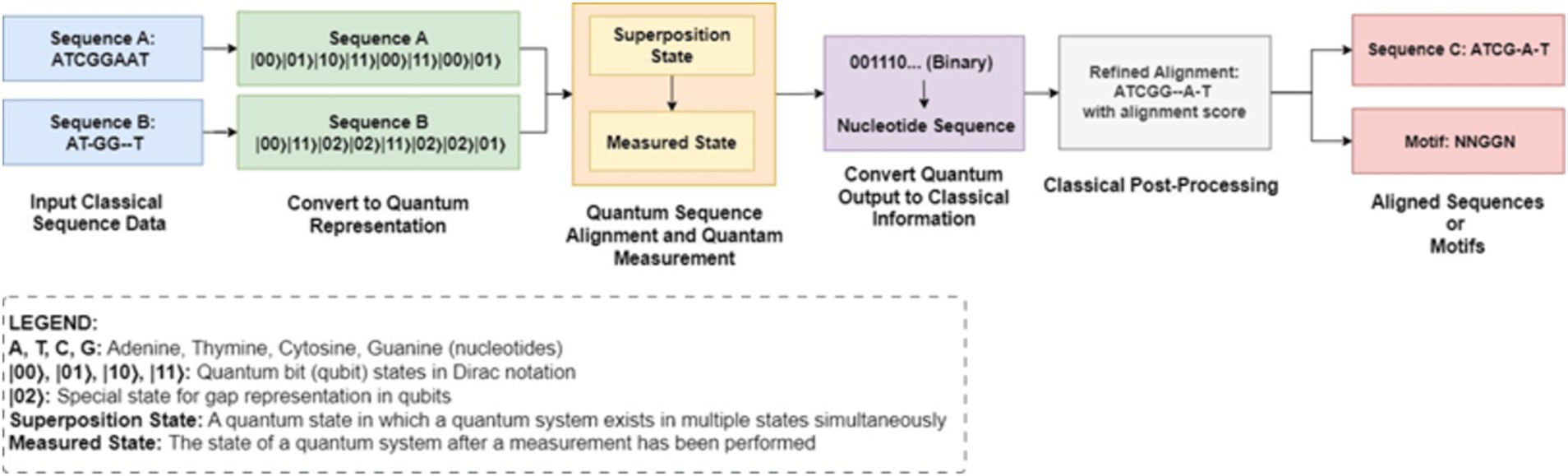
Figure 15 shows the hybrid approach combines classical NLP methods with quantum computing capabilities to potentially improve natural language processing tasks by leveraging quantum parallelism, quantum embedding spaces, or quantum algorithms for sequence alignment, a critical task in bioinformatics. It involves finding the optimal alignment between two or more biological sequences, such as DNA, RNA, or protein sequences. Homology search methods, such as Smith-Waterman and Needleman-Wunsch are based on dynamic programming of classical sequence alignment. Although they are quite efficient, their application degrades as the size of the data or the high dimensionality of the genomic data increases. Current complexity theory type item difficulties are addressable by quantum algorithms which depend on quantum superposition and entanglement.
Motif identification, the process of identifying recurring patterns or sequence motifs in genomic data, is another area where quantum algorithms have shown promise. Pattern recognition in genomic data is critical for identifying biological relationships and predicting disease markers. Classical methods rely heavily on statistical modeling and machine learning, but QNLP offers a transformative approach. By encoding genomic sequences into quantum states, QNLP leverages quantum embeddings and tensor-based models to capture complex relationships between data points.
Some of these works have been done in other genomic sequence analysis tasks that include gene prediction (Sun et al., 2012), phylogenetic tree reconstruction (Abdellah et al., 2023) and genome assembly (Kösoglu-Kind et al., 2023). Quantum simulation techniques are also being used for the study of gene interactions and for determining the impact of mutations at scales that have not been previously possible. These innovations illustrate how QNLP can be used to confront issues in genomic analysis, including multi-dimensional integration and noise immunity. The potential of QNLP in genomic sequence analysis is in the inherent optimization of the tools with high-throughput sequencing and other applications. More development in the QNLP field and in the next-generation quantum hardware, as well as in quantum algorithms, is essential to achieve the full potential of QNLP for genomics.
The discussed applications of QNLP in bioinformatics strengthen the efficiency in handling complex biological data using quantum approaches. The next section provides the case studies and how QNLP methodologies have been applied to solve complex challenges in bioinformatics, such as drug discovery, protein structure prediction, and genomic sequence analysis.
5 Case studies and real-world applications
5.1 Biomedical literature mining
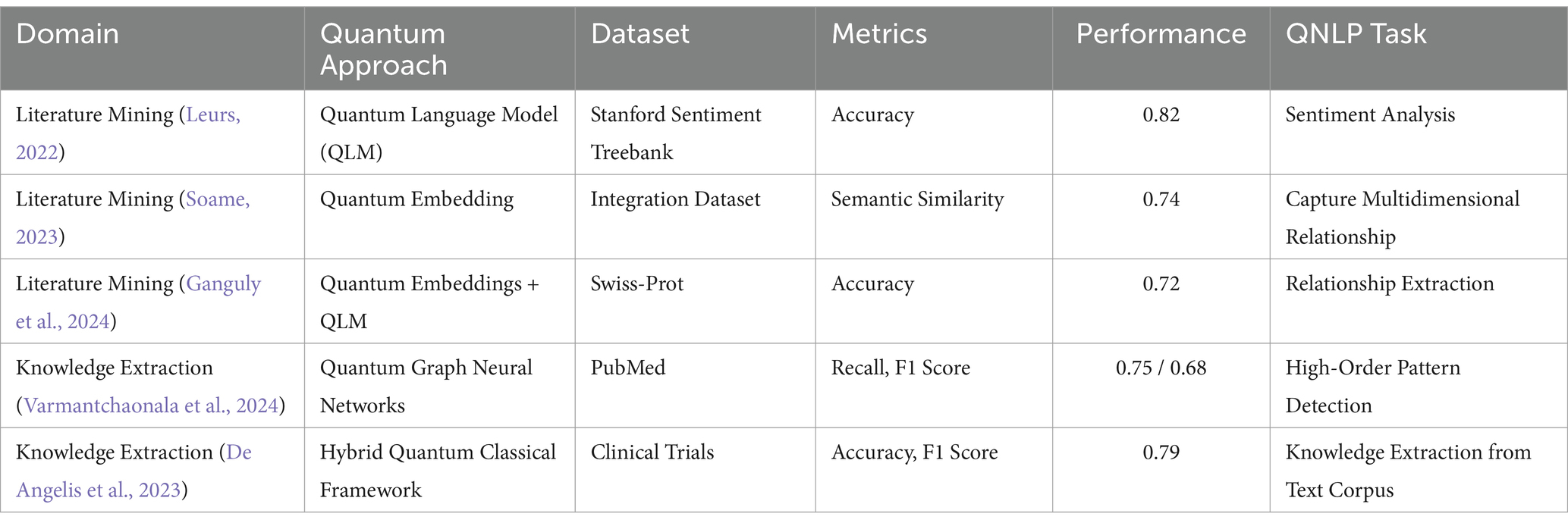
The biomedical field produces a large amount of articles every year, and therefore, it is difficult to find necessary information quickly. Prior NLP techniques are unable to process the biomedical text well because it is unstructured and the relationships between the terms are complex. This case study is concerned with the application of Quantum Natural Language Processing (QNLP) in biomedical text mining especially for named entity recognition (NER) and relation extraction. Another remarkable work has been done by Leurs (2022), which described how QLM can be used for mining biomedical literature. By employing quantum parallelism and entanglement, the authors employed a big number of biomedical articles to train a quantum language model. This model was then used to perform complex text processing tasks such as event extraction, named entity recognition and relation extraction. In the speed and accuracy of their approach to identify relevant information concerning new targets for therapy, existing drugs, and their interactions, the authors noted that the performance of their method outcompeted traditional approaches to NLP. Soame (2023) proposed an extension of the aforementioned works, which is a hybrid quantum-classical model for Knowledge Extraction and Biomedical literature mining. Thus, they were able to incorporate informative been using a combination of classical machine learning models and QNLP techniques from domain knowledge bases, experimental data, and scholarly literature. The following table summarizes the performance and applications of quantum approaches in these domains, showcasing the enhancements over classical methods.
In Table 2, quantum approaches show a general improvement in performance over literature mining and knowledge extraction problems in comparison to classical approaches. Quantum Language Models (QLMs) and embeddings improve the performance of sentiment analysis and the identification of semantic relations and Quantum Graph Neural Networks (QGNNs) help identify high-order patterns for knowledge graphs. The hybrid quantum-classical framework continues the integration process by adopting quantum optimization for knowledge extraction tasks. These results further emphasize that quantum computing offers an increasing role in revamping the traditional NLP process.
5.2 Quantum drug discovery
Drug development is one of the most significant real-world uses of QNLP in bioinformatics. Biogen and IonQ, a leading provider of quantum computing, partnered in 2021 to find prospective therapeutic targets and analyze biological data (IonQ, 2021). The alliance endeavors to speed the drug development process by utilizing the computing prowess of quantum computers, however, the precise particulars of their strategy remain undisclosed. Additionally, the biotechnology business Resilience has been actively investigating drug development solutions based on quantum computing. In a recent case study, they illustrated the utilization of quantum techniques, such as Grover’s algorithm and quantum annealing, for virtual screening and lead optimization (Salloum et al., 2024).
5.2.1 Quantum virtual screening and lead compound identification
Quantum machine learning models and quantum simulations have been shown in several recent works to be capable of predicting binding affinities and interactions between potential drugs and a target biomolecule, often a protein or nucleic acid. Mensa et al. presented a quantum machine learning model for virtual screening that utilized quantum features of the problem to incorporate quantum-mechanical effects inherent in molecular interactions (Mensa et al., 2022). Their approach demonstrated enhanced precision in the prediction of binding affinities from the traditional computational techniques. Arguing the same idea, Mohammed et al. (2017) have established a hybrid quantum mechanical/molecular mechanics model of virtual screening and identification of lead compounds. To this end, their model used both quantum simulations and machine learning algorithms to predict the binding affinities and interactions of the potential drugs with the target biomolecules. The authors showed that due to the application of quantum effects, the precision of predictions made with the help of the proposed model exceeded classical approaches, which indicates the applicability of quantum computing in this field. Thus, it has been suggested that for virtual screening and identification of lead compounds, it is possible to use hybrid quantum–classical methods based on the advantages of QNLP and quantum simulations. Ting and Caflisch (2010) described a pipeline that combines QNLP for mining chemical/biological information from text sources with quantum calculations for estimating the interactions of potential drugs with target biomolecules.
5.2.2 Drug-target interaction prediction and analysis
In a study by Sathan and Baichoo (2024) the authors proposed a quantum machine learning model for drug-target interaction prediction, demonstrating its potential to outperform classical models. Their approach involved training a quantum neural network on a large dataset of known drug-target interactions and molecular structures, leveraging the unique capabilities of quantum computing to capture the complex patterns and relationships within the data. Similarly, Veleiro et al. (2023) Combined Transformer and Graph Neural Networks, these architectures, models capture both global and local structural information of drugs and sequence features of targets, improving precision and recall in DTI predictions (Khurana et al., 2023).
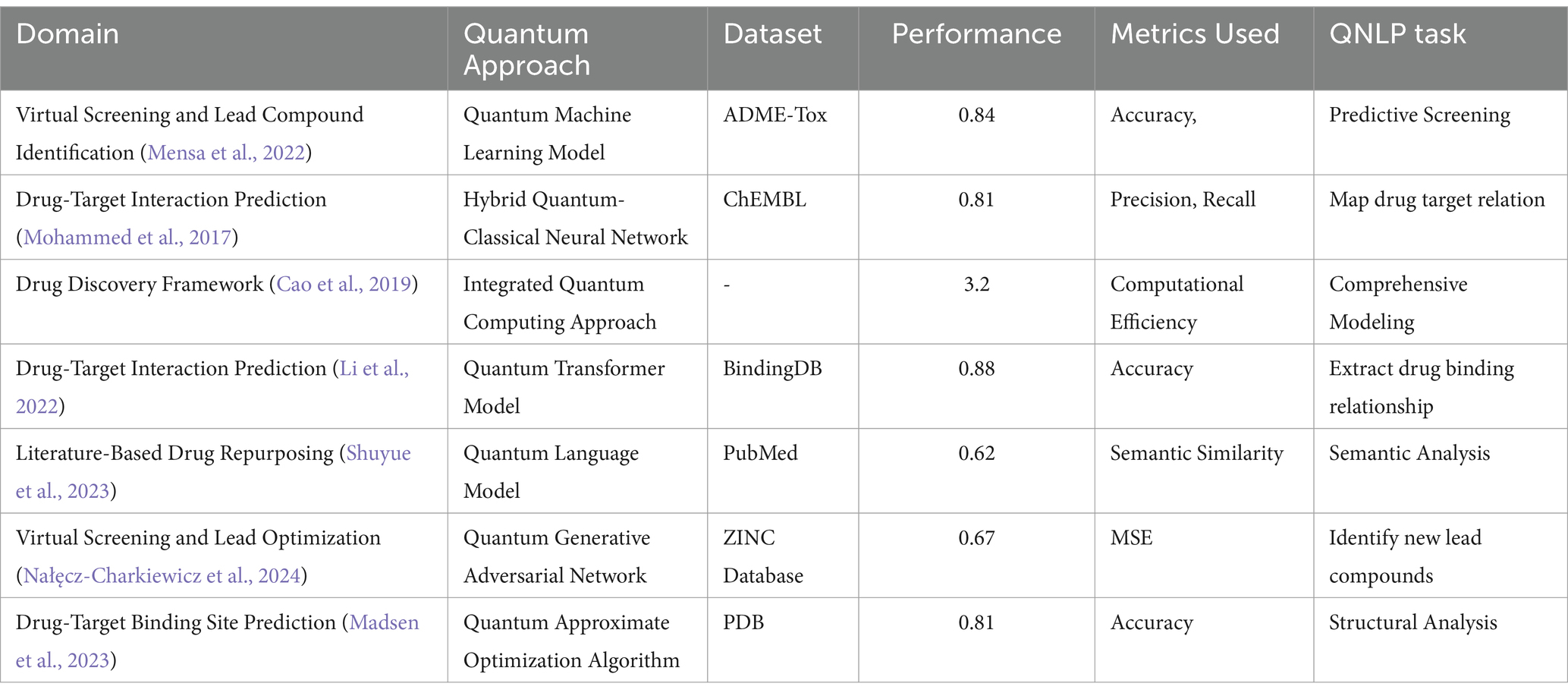
A notable study by Mohammed et al. (2017) further highlighted the potential of quantum machine learning models in this domain. The authors trained a hybrid quantum-classical neural network on a large dataset of drug-target interactions and molecular structures, demonstrating its ability to outperform state-of-the-art classical models in predicting novel interactions. Their work also explored the interpretability of the quantum model, providing insights into the underlying mechanisms of drug-target interactions. Xiong et al. (2023) proposed a framework called “Q-Drug” that aims to bring drug design into the quantum space using deep learning techniques. This framework incorporates QNLP for literature mining, quantum simulations for modeling molecular interactions, and quantum graph neural networks for predicting drug-target interactions. The integration of these components aims to provide a comprehensive and efficient computational framework for drug discovery and design. Similarly, a recent study by Sathan and Baichoo (2024) proposed a quantum graph neural network for drug-target interaction prediction, which utilizes QNLP for extracting relevant information from biomedical literature and integrates it with protein structure data and quantum simulations of molecular interactions. The Table 3 below provides details of quantum approaches applied to various drug discovery tasks, highlighting their datasets, performance metrics, and specific QNLP tasks.
The Table 3 presents different QNLP approaches, including quantum language models, quantum machine learning models (e.g., quantum neural networks, quantum transformers, quantum graph neural networks), quantum kernel methods, quantum generative adversarial networks, and quantum attention models. Various computational models in these tasks showcase their comparative analysis over classical NLP and QNLP methods. The research emphasized the capability of QNLP methods to efficiently investigate the extensive chemical space and detect prospective medication candidates. Nevertheless, these practical implementations also unveiled many obstacles. Prominent challenges that need to be addressed include the encoding and representation of data, the optimization of algorithms, and the restricted scalability of existing quantum technology (Selig et al., 2021). Furthermore, the implementation of QNLP methodologies in conjunction with pre-existing drug discovery pipelines and the analysis of quantum-derived outcomes presented pragmatic obstacles.
5.3 Protein structure prediction (PSP)
More recently, a work from researchers (Cherrat et al., 2024) proposed the Harvey et al., present complex-valued tensor network models where PT employs parameterised quantum circuits, thereby employing Hilbert space as the feature space for the sequence processing task. The models are connected to archive data in a tree like structure maximizing data correlation and compositional structure enhancing interpretability and the permanence of resource compression. The experimental results have established the use of the models in binary classification tasks using realistic datasets, proving the long-range correlation the models can tackle. This work can be considered as a major advancement in utilizing quantum machine learning for protein structure predictions that could further improve human health and welfare.
In a recent study, Drori et al. (2019) have investigated the possibility of using QNLP for secondary structure predictability classical methods like PSIPRED and SPIDER3, make use of the machine learning algorithms, such as the quantum embeddings and the quantum neural networks to model the sequence-structure map for proteins. Compared with the classical models, these quantum models showed that the utilization of quantum phenomena superposition and entanglement yielded better feature representation and long-range dependency capture (Boulebnane et al., 2022). In quantum computing, there are promising models known as Quantum Circuit Learning (QCL). Hatakeyama-Sato et al. (2022) has put forward a QCL model for protein sequences in which quantum embedding is used to improve the accuracy of the model by including secondary structure information in the quantum representation. Likewise, the quantum kernel methods like Quantum Kernel Support Vector Machines (QK-SVM) have shown better classification efficiency in identifying the secondary structures elements. Quantum computing takes it a step further Quantum computing takes it a step further. In this area, AlphaFold 2 has come a long way and gets to the experimental accuracy of many proteins using spatial graph networks. For example, in the work by Webber et al. (2022), the authors wanted to know if the quantum annealing technique can allow for the effective sampling of the conformational space of protein folding, which is a problem for standard molecular dynamics simulations. Edward et al. (2024) used quantum circuits to simulate the Protein folding dynamics to show the ability of quantum computers in processing larger and complex protein structures with better precision. Later suggested that QNLP should be combined with quantum annealing to probe the conformational landscape of the protein folding phenomenon. Through their work, they were able to show that folding dynamics could be approximated with better energy efficiency than those of classical molecular dynamics simulation. A recently published paper by Harvey et al. (2023) pointed out that the existing quantum hardware have several problems, such as noise, qubit error, and scalability, which would affect the application of quantum algorithms and simulations to large-scale protein structure prediction.
Table 4 specifically focuses on protein-related tasks, that detail advancements in protein structure prediction, folding, or interaction analysis and also requiring further innovations and hybrid approaches. This integrated representation can potentially capture complex patterns and relationships that are difficult to extract using classical methods, leading to improved structure prediction accuracy.
5.4 Genomic data analysis)
This research has shown that QNLP has a lot of promise for the analysis of genetic information. A few researchers from the University of Chicago have recently studied the possibility of using QAM to search and process genetic sequences (Wang et al., 2024). In their case study, the researchers came up with a QRAM framework that proved most effective in storing and accessing genetic data. In large scale genomic databases, the QRAM enabled fast search and pattern matching through conversion of genomic sequences into quantum states, thus outperforming other methods. While the QRAM was found to show promising results in controlled trials, the creators of the method encountered difficulties in trying to extend the method to handle the massive amounts of real genetic data that the world produces. In order to make the solutions practically usable, certain critical challenges need to be addressed including data encoding, the number of qubits needed and error mitigation (Guarasci et al., 2022).
One of the study, Daskin et al. (2014) applied Grover’s algorithm to sequence alignment, where quadratic time savings compared with the classical procedures are needed. Similarly, Khan et al. (2023) proposed a quantum algorithm for the pairwise sequence alignment of biological macromolecules and showed that the performance of the quantum algorithm is much higher than that of Smith-Waterman algorithm.
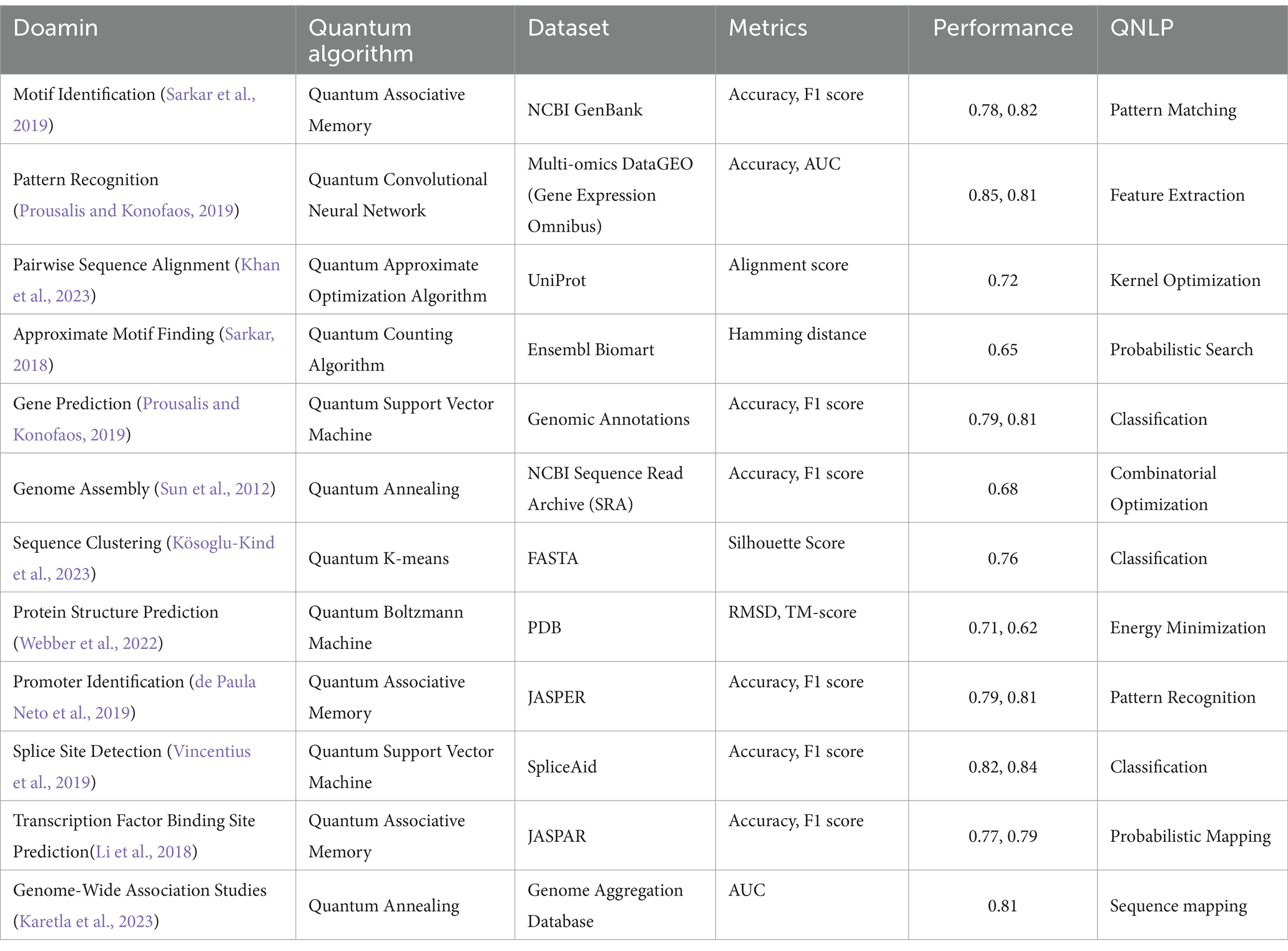
There are other domains as well, where quantum algorithms are useful are Motif identification is the process of finding sequence motifs that appear in genomic data. Go, First, Plans (2023) envisaged a quantum algorithm for motif identification that is superior to classical algorithms for certain circumstances. Sarkar et al. (2019) followed this up by providing a quantum algorithm for approximate motif discovery in DNA sequences, which can explore a solution space exponentially larger in polynomial time thanks to quantum parallelism. All of these show significant possibilities for thinking about regulatory components in DNA and RNA sequences. The Table 5 below provide key details, highlighting datasets, performance metrics,and specific QNLP tasks.
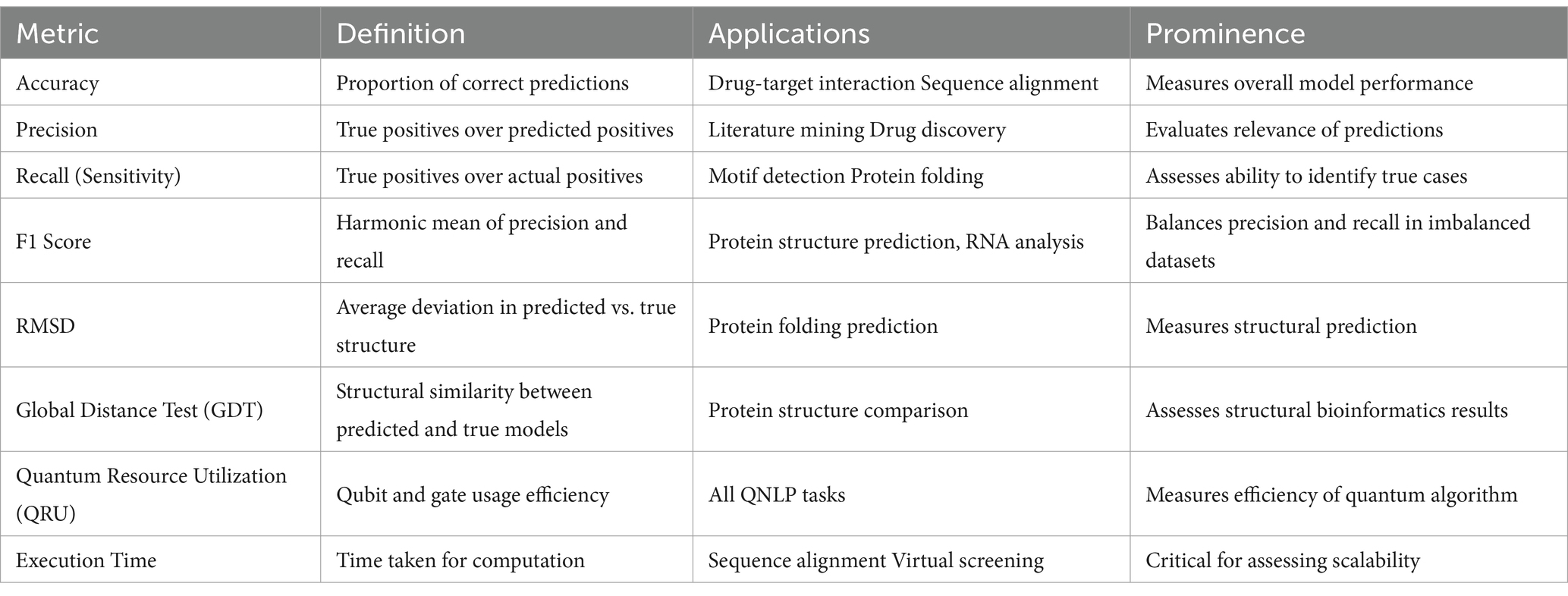
Data pattern recognition is especially essential in determining the existing or expected biological association and disease predictors in genomic data. While classical methods are statistical modeling and machine learning, QNLP brings a revolutionizing perspective. QNLP uses quantum embeddings and tensor based models to represent genomic sequences in terms of quantum states and to analyze relationships between the data points. Sarkar (2018) used quantum machine learning models for pattern recognition, and showed that these models perform better in terms of scalability and computational complexity. More development in the QNLP field and in the next-generation quantum hardware, as well as in quantum algorithms, is essential to achieve the full potential of QNLP for genomics. In light of the classical computational methods for genomic sequence analysis, the application of QNLP techniques presents an opportunity to tackle the limitations of these classical approaches, particularly in terms of computational complexity and scalability. Supplementary Figure 1 presents a comparison between classical NLP and QNLP for various applications where the X-axis represents the different NLP tasks and the y-axis represents performance metric scores ranging from 0 to 1.
For each task, it has two bars blue represents the performance score of classical NLP and orange represents the performance score of QNLP methods. As quantum models have given improvement in several bioinformatics domains such as drug discovery, protein folding, and genomic sequence analysis.
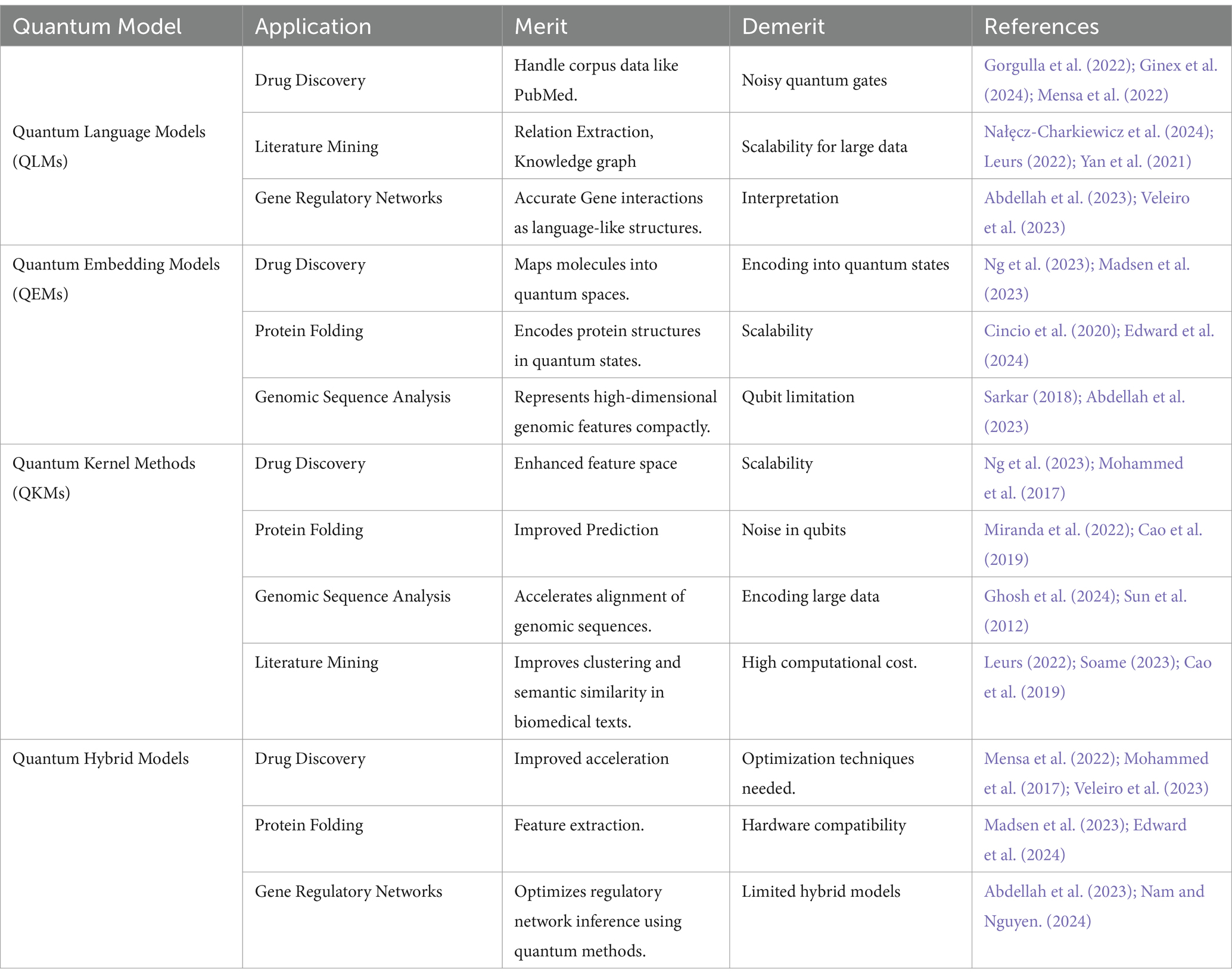
The following Table 6 synthesizes the main strengths and weaknesses of the main quantum approaches, as well as their applicability to certain tasks.
Despite the fact that QNLP has been identified as a relatively young subfield of bioinformatics, several research institutes and companies have started exploring its potential applications and realistic use cases. Such activities provide a vast amount of understanding about the potential and the limitations of the QNLP approaches as well as the challenges that surround their implementation and deployment.
The use of QNLP in bioinformatics and its application in Table 6 provide an understanding of the prospects and challenges in the field. Among the most important lessons acquired are:
a. The two constraints of encoding and representation of data are critical in determining the potential of quantum computing in handling of large data particularly in biological systems.
b. Algorithm optimization and efficient mapping on quantum processors both constitute core aspects of realizing quantifiable improvements compared to classical approaches.
c. Shortly, it might be crucial to utilize hybrid quantum-classical approaches to overcome the limitations of current quantum hardware and exploit the unique benefits of quantum computing and classical computing, respectively.
d. The collaboration of hardware engineers, domain specialists, and quantum algorithm researchers is of the utmost importance in the development of effective and functional QNLP solutions for the field of bioinformatics.
e. Interpretability and integration with current bioinformatics workflows are crucial considerations when putting QNLP techniques into practice.
With the continuous advancement of quantum computing technology and the accessibility of more potent quantum hardware, it is anticipated that the potential of QNLP in the field of bioinformatics will expand. To unlock the full potential of this growing industry, however, it will be crucial to address the obstacles mentioned in these studies.
The Case studies and applications of QNLP in bioinformatics highlight its transformative potential, but they also reveal key challenges, particularly with quantum hardware and algorithm scalability, which are explored in the following section on hardware limitations and future directions.
6 Challenges and limitations
6.1 Quantum hardware constraints
Noise and Qubit Decoherence: The practical implementation of QNLP algorithms in bioinformatics is currently constrained by the limitations of existing quantum hardware. One of the major challenges is the presence of noise and qubit errors, which can significantly impact the reliability and accuracy of quantum computations (Preskill, 2018; Daimon and Matsushita, 2024). Quantum systems are highly susceptible to environmental disturbances, such as electromagnetic fields, temperature fluctuations, and cosmic radiation, which can cause decoherence and errors in the qubit states. This shortens the time available for computations, particularly in tasks like large-scale genomic sequence analysis or protein folding prediction.
Quantum Error Correction: Robust quantum error correction is still under development. Mitigating these errors is crucial for the successful execution of quantum algorithms. Several error correction techniques have been proposed, including quantum error-correcting codes (Gowda et al., 2024) and fault-tolerant quantum computing (Wang and Liu, 2024). Error-prone calculations limit the scalability of QNLP for bioinformatics tasks where precision is critical, such as drug discovery or structural bioinformatics.
Limited Qubit Connectivity: Scalability is another significant challenge for quantum hardware. Current quantum computers have a limited number of qubits, typically in the range of tens or hundreds, which restricts the size and complexity of problems that can be addressed (Grover, 1996). Many bioinformatics applications, such as genome assembly, protein structure prediction, and large-scale sequence analysis, require processing vast amounts of data, necessitating quantum computers with thousands or millions of qubits that exceed the current hardware capabilities. Low gate fidelities and restricted qubit connectivity in current quantum systems affect the accuracy and efficiency of QNLP models.
Researchers are actively exploring various approaches to address these hardware constraints. One promising solution is the development of topological quantum computers, which leverage the principles of topological quantum field theory to achieve fault tolerance and scalability (Aïmeur et al., 2007). Additionally, quantum error mitigation techniques, such as zero-noise extrapolation (Cross et al., 2019) and probabilistic error cancellation (Zhou et al., 2020), aim to reduce the impact of hardware errors without the need for full-scale quantum error correction.
6.2 Data representation and encoding
Efficient data representation and encoding are critical for leveraging the potential of QNLP in bioinformatics applications. Biological data, such as genomic sequences, protein structures, and molecular interactions, often exhibit complex patterns and high-dimensional structures, which can be challenging to represent and process on quantum computers. One common approach for encoding biological data on quantum systems is the use of qubit representations, where each qubit or a set of qubits encodes specific aspects of the data (Quetschlich et al., 2022). In the case of DNA sequences, each nucleotide (A, T, C, G) can be mapped to a specific qubit state or a combination of qubit states. However, as the size of the biological data increases, the number of qubits required for encoding grows exponentially, leading to scalability issues. To address this challenge, researchers have explored various quantum data embedding techniques, such as amplitude encoding (Ibtehaz and Kihara, 2023) and quantum feature mapping (Kim et al., 2021). These methods aim to represent high-dimensional data in lower-dimensional quantum states, potentially reducing the number of qubits required and improving the efficiency of QNLP algorithms.
Another approach is the use of quantum machine learning models, which can learn efficient representations of biological data directly from quantum states (Nathans and Sterling, 2016). These models can leverage the principles of quantum mechanics, such as superposition and entanglement, to capture complex patterns and relationships in the data.
6.3 Algorithm development and optimization
In the following section we outline the challenges that have to be met for the development and optimization of quantum algorithms for QNLP particularly in bioinformatics. One of the significant issues is the lack of numerous realistic quantum algorithms for bioinformatics applications that provide effective solutions to particular real-world problems. As for quantum algorithms, there are some theoretical ones, like Grover’s algorithm (Grealey et al., 2022) and Shor’s algorithm (Shor, 1999), that provide more efficient solutions for some problems than classical ones, however, there can be found rather fewer works on how to use them for bioinformatics tasks with practical quantum computers.
As a result of this challenge, researchers have sought to employ integrated quantum-classical algorithms and data pre-processing, with quantum kernels. This approach will seek to take advantage of the two types of computing with a view of avoiding the current drawbacks of quantum computing. For data preparation and cleaning in genomic sequence analysis, classical methods can be applied, and for some particular computation, that requires heavy calculation, quantum method can be used, for example, pattern matching or sequence alignment. Another challenge experienced in algorithm development and optimization is the lack of benchmark and performance metrics. This is particularly important as the development of quantum algorithms and their use in bioinformatics continues particularly as it applies to the assessment of the efficiency of quantum computers as well as the comparison of quantum algorithms to their classical counterparts. This involves assessing aspects like runtime, precision, extensibility, and hardware consumption like the number of qubits, circuit depth.
Additionally, quantum algorithms themselves are not comprehensively optimized because their creation is based on the compromise between several parameters, including computation time, memory, and error. Thus, in the case of genomic sequence analysis, faster performing algorithms may have lower accuracy or may need more qubits which in turn causes more hardware limitations. The trade-offs between precision and speed are especially important to Algorithm designers and developers in the context of bioinformatics applications and they have to choose between these two factors based on their particular case.
Thirdly, the improvement of quantum algorithms is also important for practical applications at the same time. This may include circuit optimization (Zhou et al., 2020), quantum compiler optimization (Quetschlich et al., 2022), and quantum error mitigation techniques (Kim et al., 2021; Nathans and Sterling, 2016). These optimizations may enhance the general performance, decrease the demands on resources, and enhance the precision and stability of quantum computation for bioinformatics purposes.
6.4 Environmental sustainability and carbon footprint
Bioinformatics studies are more and more based on high-performance computing and big data processing, which generate high amounts of energy consumption and CO2 emissions. Solving this problem is necessary to achieve global sustainability objectives and decrease the impact of scientific computing on the environment. Some papers have compared the costs of different bioinformatics approaches to the environment and have given suggestions on how to reduce these costs.
A recent study by Nathans and Sterling (2016) compared the carbon footprint of typical bioinformatics tools and analyses based on RNA sequencing, GWAS, genome assembly, phylogenetic trees, metagenome, and molecular dynamics. The researchers used the Green Algorithms calculator to come up with the carbon emissions in kilograms of CO2 equivalent units (kgCO2e). They also identified the carbon cost of GWAS at the scale of a biobank, pointing out that the application of efficient codes and the use of low-carbon data centers are critical to the reduction of carbon footprint. In addition, (Grealey et al., 2022) also explored the effects of parallelization, the use of Central Processing Units and Graphics Processing Units, cloud and local computational resources, and geographical location on carbon footprint. The outcomes showed that applying more efficient parallelization strategies along with simple software updates could cut the carbon footprint of bioinformatics computations by half.
To address these challenges, a new field called environmentally sustainable computational science (ESCS) has developed and offers significant potential for enhancement. To support continued and sustainable growth in computational science, a more planned approach to awareness raising, the improvement of transparency, the better estimation of environmental costs, and the broader reporting of these costs are required (Stodden and Miguez, 2014). In support of this effort, the “GREENER” set of principles and best practices guidance has been developed to guide sustainable software development and deployment (Lannelongue et al., 2023). These environmental sustainability issues in bioinformatics are not only important for the sustainability context to meet the global sustainability objectives but also for the bioinformatics sustainability to be feasible (Selladurai et al., 2024) and sustainable in the future and large-scale uses. Efficiency improvement of the resource usage, energy-saving computing solutions, and cooperation between domain specialists, developers, and sustainability scholars are the key actions on the way to decreasing the carbon impact of the bioinformatics analyses. Furthermore, there has been suggestions that utilization of quantum computing in bioinformatics could help solve the problems by decreasing the carbon footprint of computationally rigorous tasks (Shaun et al., 2021). Specifically, quantum algorithms and simulations can lead to more efficient analysis in protein folding, genomic sequence alignments, molecular modeling, and others, thus decreasing resource usage and emissions (Wong and Chang, 2022).
Supplementary Figure 2 shows various challenges and limitations faced in the field of Quantum Computing and QNLP, ranging from hardware constraints to data representation, algorithm development, and environmental sustainability concerns.
Table 7 provides differences between computational characteristics, software ecosystems, and real-world applications. of Classical NLP and QNLP.
7 Performance evaluation and component analysis
7.1 Evaluation metrics
When it comes to the appropriate performance evaluation of QNLP algorithms in bioinformatics, a proper metric needs to be set up. This not only allows for making accurate and strict comparisons with classical NLP methods but also defines the direction of constructing and fine-tuning QNLP algorithms for particular tasks. The selection of performance indicators varies depending on the bioinformatics application area and its goals. When assessing the quality of protein modeling for example in protein structure prediction, the predicted models can be rated for accuracy by the root-mean-square deviation (RMSD) from the native structure or the Global Distance Test (GDT) scores (Kryshtafovych and Fidelis, 2009). Depending on task, in genomic sequence analysis one can use measures such as sensitivity, specificity, F1-score for the pattern recognition or alignment quality measures like Q-score (Yang et al., 2013). Besides, problem-specific objective functions, which are calculated based on the results of the algorithm, there are more universal criteria based on the evaluation of the number of computational operations, necessary memory space, and quantum resource usage, such as numbers of qubits, and the depth of the circuits.
7.1.1 Root-mean-square deviation
It is a fundamental metric in structural bioinformatics and computational chemistry for evaluating the accuracy of predicted protein structures. RMSD measures the average distance between the atoms of a predicted protein structure and the corresponding atoms in the experimentally determined native structure, providing a quantitative assessment of the structural similarity between the two structures (Olechnovič et al., 2019). The formula for calculating RMSD between two sets of coordinates, each containing n atoms, is given in Equation 12:
Here,and represent the coordinates of the 𝑖th atom in the predicted and native structures, respectively. The term denotes the Euclidean distance between the corresponding atoms in the two structures. In structural bioinformatics, RMSD is a key metric for assessing the quality of protein structure prediction algorithms and molecular docking simulations (Neveu et al., 2018). Low RMSD values indicate a high degree of similarity between the predicted and native structures, suggesting that the model accurately captures the protein’s folding pattern (Jumper et al., 2021). Researchers often use RMSD in conjunction with other metrics, such as Ramachandran plots and GDT (Global Distance Test), to provide a comprehensive evaluation of protein structure predictions and refine computational models for drug discovery and molecular biology applications.
7.1.2 Global distance test
It is another important metric used in structural bioinformatics to assess the quality of predicted protein structures. GDT measures the similarity between a predicted protein structure and the experimentally determined native structure by considering the distance between equivalent residues in the two structures (Poleksic, 2015). The GDT score is calculated as the percentage of residues in the predicted structure that are within a specified distance threshold of the corresponding residues in the native structure. The GDT score is typically calculated at different distance thresholds like 1 Å, 2 Å, 4 Å, and 8 Å to provide a comprehensive assessment of the structural similarity.
Mathematically, in Equation 13 the GDT score at a given distance threshold is calculated as follows:
D is the total number of residues (atoms) in the protein. GDTi is the fraction of residues for atom 𝑖 that fall within the distance thresholds.
The GDT score is expressed as a percentage, with higher scores indicating a greater degree of structural similarity between the predicted and native structures. A GDT score of 100% indicates perfect structural similarity, meaning that all residues in the predicted structure are within the specified distance threshold of the corresponding residues in the native structure. GDT provides a quantitative measure of the structural similarity between predicted and native structures. It complements other metrics such as RMSD and Ramachandran plots, offering researchers a comprehensive assessment of the accuracy of computational models in protein structure prediction.
7.1.3 F1-score
Equations 14–16 is important to assess the overall performance of a binary classification model. It is particularly useful when the class distribution is imbalanced (Nunn et al., 2021).
For calculating F1-score:
Where:
The F1 score ranges from 0 to 1, where a higher score indicates better model performance. It provides a balance between precision and recall, making it a useful metric for evaluating models, especially when there is an imbalance between the two classes. It is important to assess the overall performance of a binary classification model.
7.1.4 Quantum resource utilization
It is a metric that evaluates how efficiently a quantum algorithm utilizes quantum resources such as qubits, gates, and circuit depth. It is often used to compare the efficiency of different quantum algorithms in terms of resource consumption. The exact formula for calculating quantum resource utilization can vary depending on the specific context and the factors considered (Hansen et al., 2023). However, a general formula could be constructed as follows:
Where, the total number of physical qubits required for quantum computation, the depth of the quantum circuit represents the number of sequential gates that need to be applied, and the number of logical qubits required to represent the problem being implemented. Equation 17 provides a measure of how efficiently quantum resources (such as qubits and gates) are being utilized to solve a particular problem or implement an algorithm. A lower QRU indicates more efficient resource utilization, while a higher QRU indicates that more resources are required for the computation (Lubinski et al., 2023). Table 8 provide the overview of evaluation metrics and its significance.
QNLP is a captivating domain within the field of bioinformatics, presenting prospective benefits in terms of computational economy, precision, and performance when compared to traditional approaches. Although benchmarking studies have shown competitive outcomes for tasks such as genomic sequence analysis (Shiny Duela et al., 2023) and protein structure prediction (Chow, 2024), the current limitations in error correction overhead and quantum hardware scalability prevent the practical implementation of the theoretical speedups offered by QNLP (Saggi et al., 2024). To address these obstacles, hybrid quantum-classical methodologies have surfaced, which involve the strategic delegation of computationally demanding duties to quantum processors while making use of classical resources to preprocess and post-process data (Zhou et al., 2024).
Nevertheless, QNLP techniques encounter intrinsic drawbacks, such as the complexity of encoding data, challenges in optimizing algorithms, and the relative youth of quantum hardware in comparison to the firmly established classical NLP environment. Adoption of QNLP in bioinformatics will ultimately depend on continuing algorithm development, progress in quantum computing, and a prudent comparison of the capabilities of classical and hybrid techniques to application-specific performance, accuracy, and scalability needs. However, addressing these obstacles through collaborative research and technological advancements will not only overcome existing limitations but also set the stage for groundbreaking developments in bioinformatics. As we transition into discussing future directions, we focus on the transformative possibilities that lie ahead for QNLPFuture Research Directions and Roadmap.
7.2 Potential avenues and future prospects
With the ongoing progress in quantum computer technology, the domain of Quantum Natural Language Processing (QNLP) possesses tremendous potential to revolutionize bioinformatics and expand the frontiers of biological investigation and revelation. A multitude of burgeoning trends and prospects are positioned to influence the forthcoming implementations of QNLP within this field (Supplementary Figure 3).
7.2.1 Personalized medicine and precision therapeutics
A highly auspicious domain for QNLP to be implemented is customized medicine and precision treatments. QNLP techniques could facilitate a more precise and effective examination of individual genomic data by harnessing the computational capabilities of quantum computing. This, in turn, could pave the way for the creation of customized treatments and personalized pharmacological regimens (Chow, 2024). Equipped with extensive multi-omics and clinical data, quantum machine learning models can discern intricate patterns and correlations, hence enabling the identification of previously undiscovered biomarkers and therapeutic targets.
7.2.2 De novo protein design
De novo protein design and protein engineering are indispensable for the creation of novel biomaterials, enzymes, and medicines. When combined with quantum simulations and molecular modeling, QNLP methods have the potential to greatly revolutionize this field by enabling more accurate predictions of protein folding kinetics, structures, and functions (Shiny Duela et al., 2023). The application of quantum algorithms for the fast conformational sampling of proteins might help to develop new enzymes, designer proteins, and engineered biomaterials by defining sequences and structures that provide specific functions.
7.2.3 Multi-omics data integration
Using multiple omics data, including genomes, transcriptome, proteome, and metabolome, is crucial to develop systems-level models that capture complex cellular functions. Technique of QNLP can have a tremendous effect on this field as they can support the integration of data, patterns’ recognition, and knowledge search across different and diverse data (Saggi et al., 2024). It could be hypothesized that the existing relationships and interactions between multi-omics data sets could be learned by quantum machine learning and algorithms, providing new and previously unknown information about gene regulation networks, metabolic pathways, and disease etiology.
7.2.4 Quantum molecular simulations and drug design
The combination of QNLP methods with molecular quantum simulations and quantum chemistry calculations may open a new era in drug discovery. Thanks to quantum computer computing capacities, the scientists were able to create detailed models of what happens with proteins and ligands, biochemical reactions, and other complex biomolecular interactions with superior accuracy and efficiency (Zhou et al., 2024). The use of these simulations may result in profound understanding of the molecular interactions of drugs, which in turn may help in the rational design of novel therapeutic agents and optimizing their selectivity, pharmacokinetic profile and affinity.
7.2.5 Sustainable Bio-economies
QNLP methods can be beneficial for the development of new sustainable bioeconomies and the conservation of biological diversity. By applying QNLP, it is possible to analyze large biological data sets, which include ecological data, environmental monitoring data, and genomics data to improve the efficiency of sustainable practices in agriculture, forestry and ecosystem management (Pal et al., 2024). If, for instance, unstructured data from environmental reports and scientific research was subjected to QNLP methods, then there would be knowledge and information that is relevant to the development of sustainable bioeconomies and conservation of biotic diversity. QNLP is expected to have further potential in bioinformatics and related fields and the development of quantum computing technology will extend basic biological research and development. To achieve these prospective uses, however, will require collective endeavours to overcome the challenges that arise from the scale of quantum technology, the algorithms, the data encoding and the application.
7.2.6 Biodiversity conservation
There is a potential for QNLP techniques to contribute to sustainable bioeconomies and support of initiatives related to the conservation of biological diversity. QNLP could help promote the development of sustainable practices in different industries including agriculture and forestry (Kirubakaran and Midhunchakkaravarthy, 2024) because QNLP can process big biological data which include genomic data, ecological data and environmental data. From genomic data of crop species QNLP algorithms can identify the right crop cultivars for resistance and climatic adaptability by feature extraction (Prasad et al., 2024). For the enhancement of the conservation approaches and ecosystem management, it could also allow the integration of the data on biological diversity from different sources such as field surveys, eDNA, and remote sensing (Di Sipio et al., 2021). In addition, the knowledge acquisition and insight generation techniques applicable to sustainable economies and biodiverse conservation (Sood and Chauhan, 2024) could be used with QNLP techniques to extract information from such unstructured text data as environmental reports and research articles. It is also possible that some of the sustainable practices in industries such as agriculture, forestry, fishery, and biotechnology might be supported by QNLP through the improvement of efficient knowledge search and decision-making. The opportunity to change the approach to the management of natural systems, as well as the development of bio-economies, and the protection of the biological wealth of the planet for future generations, is in the use of QNLP for environmental sustainability and the conservation of biological diversity.
The potential applications of QNLP provides its capacity to improve in various domains, from personalized medicine to sustainable bio-economies and biodiversity conservation. These advancements underscore the transformative potential of QNLP in addressing some of the most challenges in bioinformatics and beyond. However, it helps in realizing these opportunities will require progress in quantum hardware, algorithm development, and data integration techniques. The following section delves into the roadmap for overcoming these challenges, outlining actionable strategies and collaborative efforts needed to fully harness the power of QNLP in future research and applications.
7.3 Roadmap and recommendations
To fully harness the capabilities of QNLP in the field of bioinformatics and effectively tackle the obstacles associated with environmental sustainability, it is imperative to establish a thorough roadmap and a set of suggestions is mentioned in Supplementary Figure 4. Collaboration among diverse stakeholders including university researchers, makers of hardware and software for quantum computing, experts in bioinformatics and sustainability, and industry partners is important for the formulation of this plan.
7.3.1 Quantum hardware development
The direction for the development of energy-saving quantum hardware technologies must be granted high importance. These include the analysis of new forms of qubit, such as quantum dots. Semiconductor nanostructures used in these qubits may allow them to operate at higher temperatures, thus rendering large-scale cooling unnecessary (Nayak et al., 2008). However, topological qubits, due to their grounding in the principles of topological matter, these qubits have an intrinsic immunity to external noise and dephasing. As a consequence, they might require less amount of energy to correct the errors and other expenses associated with the process. To achieve efficient quantum hardware, material scientists, specialists in energy efficiency, and engineers in quantum hardware will need to work together. To obtain reliable QNLP applications, it will be necessary to design quantum systems that are immune to errors. It is strongly believed that the performance of QNLP algorithms can be enhanced by the advancements in fault-tolerant architectures and quantum error-correcting codes. Further study of the more extensive quantum processors with more qubit numbers and longer coherence times is necessary. This would make it possible to apply more complicated QNLP models for tasks such as genome-wide association analysis and protein-ligand interaction analysis. Introducing louder architectures for quantum devices might reduce the impact of environmental decoherence and expand the usability of QNLP in actual problems of bioinformatics.
7.3.2 Algorithm research
In tandem with developments in hardware, the development and optimization of quantum algorithms for sustainable bioinformatics applications should be another top priority. This implies the examination of quantum algorithms that have been developed to tackle specific problems, and these are some of the problems that may be solved by quantum computing; sustainable agriculture, protection of biodiversity, and development of environmentally friendly drugs (Andersson et al., 2022). It is anticipated that QNLP together with future generations of quantum computers will revolutionize predictive bioinformatics. They may make the realistic quantum computer modelling of different biomolecular processes, for instance protein folding and gene regulation, feasible on FTQC. With better qubit coherence and coherence times and with the deployment of scalable architectures in the future quantum systems, the solution to the computational challenges of the emulation of sophisticated biological structures will be realized and quantum advancements in biological fields will be made. For example, the incorporation of hybrid quantum-classical systems to preprocess the data classically before going to quantum circuits for sequence alignment task could optimise it. Subsequent studies should focus on improving data encoding, quantum embeddings, and quantum-classical combinations in order to minimize the computational load and energy costs associated with them. (Nammouchi et al., 2023).
7.3.3 Sustainable software engineering
Prescribing and following the best practicable procedures for eco-friendly computational research and sustainable software engineering is vital to reducing the carbon impact of QNLP in bioinformatics. Applying ideas like the “GREENER” approach which aims at developing principles and recommendations for the sustainable software development, maximization of resource utilization efficiency such as memory, storage space, data transfer, as well as the incorporation of energy-efficient computing solutions are parts of this process (Lannelongue et al., 2023).
7.3.4 Environmental impact assessment
To provide impartial and consistent assessments of the ecological consequences of QNLP algorithms and approaches, it is imperative to establish standardized benchmarking frameworks and processes for environmental impact evaluation (Strubell et al., 2020). Measuring the energy usage of quantum and classical computational resources Assessing the carbon footprint associated with the deployment and operation of QNLP solutions Evaluating the efficient use of hardware resources, such as qubits, memory, and storage should be incorporated into these frameworks to enable comparisons with traditional methodologies and to direct the optimization of sustainable QNLP solutions (Liu et al., 2021).
7.3.5 Interdisciplinary collaboration
Establishing strong partnerships among quantum computing researchers, bioinformaticians, sustainability scientists, and industry stakeholders is imperative to effectively apply QNLP findings practically and sustainably. The establishment of interdisciplinary research institutes, collaborative initiatives, and platforms for knowledge exchange can expedite the development of sustainable QNLP solutions for bioinformatics and encourage the cross-pollination of ideas (Awschalom et al., 2021).
By executing this strategic blueprint and attending to these critical domains, the bioinformatics community can effectively utilize the paradigm-shifting capabilities of QNLP in a manner that is consistent with worldwide sustainability objectives and reduces the ecological repercussions of computational procedures. Creating libraries and tools for QNLP applications tailored to bioinformatics needs could accelerate the development and testing of novel algorithms on emerging quantum devices. Nevertheless, it is imperative to recognize that the achievement of sustainable QNLP in bioinformatics necessitates significant financial expenditures, enduring dedication, and interdisciplinary cooperation among many stakeholders (Quantum Technology and Application Consortium – QUTAC et al., 2021).
While the limitations of current quantum hardware present significant challenges, the future directions outlined suggest a promising trajectory for QNLP in bioinformatics. The discussion and conclusion section synthesizes the insights and their broader implications for the field.
8 Discussion
Research questions of this study were answered with theoretical and practical applications of quantum natural language processing in advancing bioinformatics. In the analysis of Research Question 1, we showed how QNLP applies basic tenets of quantum computing such as superposition, entanglement, and parallelism to analyze linguistic data. Such methods as quantum embeddings and the DisCoCat framework extend the capabilities of text analysis and allow for representation and recognizing patterns beyond the scope of classical NLP. To answer Research Question 2, we found out that QNLP algorithms outperform traditional methods of NLP especially when it comes to data scale. Grover’s algorithm speeds up the search by keywords, while quantum embeddings improve the language modeling tasks. Of these, the areas that best illustrate the growth of these principles are Bioinformatics, where scaling factors and precision are paramount. To answer Research Question 3, we presented the primary areas of application of QNLP in bioinformatics, such as drug discovery, protein folding, and genomic sequence analysis. In drug discovery, QNLP enhances the rate of literature review and virtual screening, as well as enhancing the identification of the interaction between drugs and targets. Likewise, in protein structure prediction, QNLP helps in understanding of large data sets and with that gives a ground for quantum computational simulations. These cases illustrate how QNLP is likely to bring about workflow optimization in bioinformatics and enhance the feasibility of several important processes. Last for Research Question 4, we looked at the current challenge and future prospect of QNLP in bioinformatics. The main issues are the limitations of quantum hardware like noise, qubit coherence, quantum hardware scaling and the problem of mapping biological data into quantum. Future opportunities lies in hybrid quantum-classical frameworks, noise-resilient algorithms, and advancements in quantum processors. As these limitations are addressed, QNLP is poised to become a powerful tool in personalized medicine, multi-omics integration, and environmental bioinformatics. This discussion ties the findings to the research questions, showing how QNLP can address pressing challenges in bioinformatics while outlining pathways for future exploration. By focusing on interdisciplinary collaboration and technological innovation, QNLP offers a promising avenue for advancing bioinformatics and related fields.
9 Conclusion
Quantum natural language processing (QNLP) is a new concept that represents a radical departure from the standard approach to bioinformatics through the application of quantum computing to transform the way biological information is analyzed and understood. The current comprehensive review has also discussed the theory and application, challenges and opportunities of QNLP in various areas such as genomics sequence analysis, protein structures prediction, and drug discovery. From the existing body of work and current industrial applications, it is obvious that QNLP has the potential to be more computationally efficient, accurate, and scalable than conventional NLP techniques despite being a relatively young field of study. By incorporating quantum characteristics such as entanglement, superposition, and parallelism, QNLP algorithms have shown promising results in several applications including sequence alignment, literature analysis, virtual library search, and protein folding. However, there are still some challenges that slow down the implementation of QNLP in the bioinformatics domain. These include limitations on the quantum hardware, and the challenge posed by data encoding, and the need for algorithm design and validation. To tackle these obstacles, it will be necessary for bioinformaticians, industry stakeholders, and quantum computing researchers to collaborate. Additionally, substantial investments will be needed in the development of sustainable software engineering practices, interdisciplinary education, and workforce development. QNLP can revolutionize fields like medicine and genomics. It could speed up the discovery of life-saving drugs, help doctors create personalized treatments based on a patient’s genetic code, and provide insights into diseases at a molecular level.
With great promise for revolutionary applications in personalized medicine, de novo protein design, multi-omics data integration, sustainable bioeconomics, and environmental sustainability, QNLP in bioinformatics has a bright future. Through the utilization of quantum computer computational capabilities and the integration of QNLP methodologies with molecular modeling and quantum simulations, scholars have the potential to unveil hitherto unexplored understandings of biological mechanisms, expedite the process of discovering new drugs, and establish environmentally sound approaches to ecosystem management and biodiversity preservation. With the ongoing advancements in quantum computing technology, the feasibility of incorporating QNLP into bioinformatics will grow substantially. This will facilitate the exploration of novel insights, the efficient analysis of data, and the creation of inventive resolutions for worldwide issues on healthcare, biotechnology, and environmental sustainability. By wholeheartedly adopting this burgeoning domain and following the strategic path delineated in this evaluation, the bioinformatics community can effectively utilize QNLP to its complete capacity, thereby propelling scientific advancement and making a positive and sustainable contribution to the future.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.
Author contributions
GP: Writing – original draft. RK: Writing – review & editing.
Funding
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcomp.2025.1464122/full#supplementary-material
SUPPLEMENTARY FIGURE 1 | Comparative performance.
SUPPLEMENTARY FIGURE 2 | Potential applications and future prospects.
SUPPLEMENTARY FIGURE 3 | Challenges and limitations.
SUPPLEMENTARY FIGURE 4 | Roadmap and recommendations.
References
Abbas, H. (2024). Quantum machine learning-models and algorithms: studying quantum machine learning models and algorithms for leveraging quantum computing advantages in data analysis, pattern recognition, and optimization. Aust. J. Mach. Learn. Res. Appl. 4, 221–232.
Abbaszade, M., Salari, V., Mousavi, S. S., Zomorodi, M., and Zhou, X. (2021). Application of quantum natural language processing for language translation. IEEE Access 9, 130434–130448. doi: 10.1109/ACCESS.2021.3108768
Abdellah, T., Merazka, F., and Zahra, H. F. (2023). “Quantum computing for DNA analysis and AI in genomics.” In 2023 20th ACS/IEEE International Conference on Computer Systems and Applications (AICCSA). pp. 1–6. IEEE.
Aïmeur, E., Brassard, G., and Gambs, S. (2007). “Quantum clustering algorithms.” In Proceedings of the 24th International Conference on Machine Learning. pp. 1–8. Association for Computing Machinery.
Andersson, M. P., Jones, M. N., Mikkelsen, K. V., You, F., and Mansouri, S. S. (2022). Quantum computing for chemical and biomolecular product design. Curr. Opin. Chem. Eng. 36:100754. doi: 10.1016/j.coche.2021.100754
Awschalom, D., Berggren, K. K., Bernien, H., Bhave, S., Carr, L. D., Davids, P., et al. (2021). Development of quantum interconnects (QuICs) for next-generation information technologies. PRX Quantum 2:017002. doi: 10.1103/PRXQuantum.2.017002
Bach, H. H., Nguyen, D. K., and Dung, N. N. V. (2024). “Quantum approach for constructing phylogenetic maximum parsimony tree” in Future data and security engineering. Big data, security and privacy, Smart City and industry 4.0 applications: 11th international conference on future data and security engineering, FDSE 2024, Binh Duong, Vietnam, November 27–29, 2024, proceedings, part II (Singapore: Springer Nature Singapore), 158–170.
Baiardi, A., Christandl, M., and Reiher, M. (2023). Quantum computing for molecular biology. Chembiochem 24:e202300120. doi: 10.1002/cbic.202300120
Benedetti, M., Realpe-Gómez, J., Biswas, R., and Perdomo-Ortiz, A. (2017). Quantum-assisted learning of hardware-embedded probabilistic graphical models. Phys. Rev. X 7:041052. doi: 10.1103/PhysRevX.7.041052
Bhuvaneswari, S., Deepakraj, R., Urooj, S., Sharma, N., and Pathak, N. (2023). Computational analysis: unveiling the quantum algorithms for protein analysis and predictions. IEEE Access 11, 94023–94033. doi: 10.1109/ACCESS.2023.3310812
Biamonte, J., Wittek, P., Pancotti, N., Rebentrost, P., Wiebe, N., and Lloyd, S. (2017). Quantum machine learning. Nature 549, 195–202. doi: 10.1038/nature23474
Black, P., Kuhn, D., and Williams, C. (2002). Quantum computing and communication. Adv. Comput. 56, 189–244. doi: 10.1016/S0065-2458(02)80007-9
Boev, A. S., Rakitko, A. S., Usmanov, S. R., Kobzeva, A. N., Popov, I. V., Ilinsky, V. V., et al. (2021). Genome assembly using quantum and quantum-inspired annealing. Sci. Rep. 11:13183. doi: 10.1038/s41598-021-88321-5
Boulebnane, S., Lucas, X., Meyder, A., Adaszewski, S., and Montanaro, A. (2022). Peptide conformational sampling using the quantum approximate optimization algorithm. NPJ Quantum Inform. 9, 1–12. doi: 10.1038/s41534-022-00685-9
Campbell, E., et al. (2024). A series of fast-paced advances in quantum error correction. Nat. Rev. Phys. 6, 160–161. doi: 10.1038/s42254-024-00706-3
Cao, Y., Romero, J., and Aspuru-Guzik, A. (2018). Potential of quantum computing for drug discovery. IBM J. Res. Dev. 62, 6:1–6:20. doi: 10.1147/JRD.2018.2888987
Cao, Y., Romero, J., Olson, J. P., Degroote, M., Johnson, P. D., Kieferová, M., et al. (2019). Quantum chemistry in the age of quantum computing. Chem. Rev. 119, 10856–10915. doi: 10.1021/acs.chemrev.8b00803
Chandarana, P., Hegade, N. N., Montalban, I., Solano, E., and Chen, X. (2023). Digitized counter diabatic quantum algorithm for protein folding. Phys. Rev. Appl. 20:014024. doi: 10.1103/PhysRevApplied.20.014024
Chapman, S., Adami, C., Wilke, C. O., and Kc, D. B. (2017). The evolution of logic circuits for the purpose of protein contact map prediction. PeerJ 5:e3139. doi: 10.7717/PEERJ.3139
Chen, Y., Pan, Y., and Dong, D. (2023). Quantum language model with entanglement embedding for question answering. IEEE Trans. Cybernet. 53, 3467–3478. doi: 10.1109/TCYB.2021.3131252
Cherrat, E. A., Kerenidis, I., Mathur, N., Landman, J., Strahm, M., and Li, Y. Y. (2024). Quantum vision transformers. Quantum 8:1265. doi: 10.22331/q-2024-02-22-1265
Cincio, L., Rudinger, K. M., Sarovar, M., and Coles, P. J. (2020). Machine learning of noise-resilient quantum circuits. Maryland, U.S: PRX Quantum.
Cross, A. W., Bishop, L. S., Sheldon, S., Nation, P. D., and Gambetta, J. M. (2019). Validating quantum computers using randomized model circuits. Phys. Rev. A 100:032328. doi: 10.1103/PhysRevA.100.032328
Daimon, S., and Matsushita, Y. (2024). Quantum circuit generation for amplitude encoding using a transformer decoder. Phys. Rev. Appl. 22:L041001. doi: 10.1103/PhysRevApplied.22.L041001
D'Aloisio, G., Fortz, S., Hanna, C., Fortunato, D., Bensoussan, A., Mendiluze Usandizaga, E., et al. (2024). “Exploring LLM-driven explanations for quantum algorithms.” in Proceedings of the 18th ACM/IEEE international symposium on empirical software engineering and measurement. pp. 475–481. Association for Computing Machinery.
Das Sarma, S., Deng, D.-L., and Duan, L.-M. (2019). Machine learning meets quantum physics. Phys. Today 72, 48–54. doi: 10.1063/PT.3.4164
Daskin, A., Grama, A., and Kais, S. (2014). Multiple network alignment on quantum computers. Quantum Inf. Process 13, 2653–2666. doi: 10.1007/s11128-014-0818-7
De Angelis, L., Baglivo, F., Arzilli, G., Privitera, G. P., Ferragina, P., Tozzi, A. E., et al. (2023). ChatGPT and the rise of large language models: the new AI-driven infodemic threat in public health. Front. Public Health 11:1166120. doi: 10.3389/fpubh.2023.1166120
de Paula Neto, F. M., da Silva, A. J., de Oliveira, W. R., and Ludermir, T. B. (2019). Quantum probabilistic associative memory architecture. Neurocomputing 351, 101–110. doi: 10.1016/j.neucom.2019.03.078
Dejpasand, M. T., and Sasani Ghamsari, M. (2023). Research trends in quantum computers by focusing on qubits as their building blocks. Quantum Rep. 5, 597–608. doi: 10.3390/quantum5030039
Di Sipio, R., Huang, J.-H., Chen, S. Y.-C., Mangini, S., and Worring, M. (2021). The dawn of quantum natural language processing. arXiv: Computation and language. Available at: https://arxiv.org/abs/2105.04058 (Accessed May 23, 2022).
Doga, H., Raubenolt, B., Cumbo, F., Joshi, J., DiFilippo, F. P., Qin, J., et al. (2024). A perspective on protein structure prediction using quantum computers. J. Chem. Theory Comput. 20, 3359–3378. doi: 10.1021/acs.jctc.4c00067
Drori, I., Thaker, D., Srivatsa, A., Jeong, D., Wang, Y., Nan, L., et al. (2019). Accurate protein structure prediction by embeddings and deep learning representations. arXiv preprint arXiv:1911.05531.
Dunjko, V., and Briegel, H. J. (2018). Machine learning & artificial intelligence in the quantum domain: A review of recent progress. Rep. Prog. Phys. 81:074001. doi: 10.1088/1361-6633/aab406
Durán, C., Carrasco, R., Soto, I., Galeas, I., Azócar, J., Peña, V., et al. (2023). Quantum algorithms: applications, criteria and metrics. Complex Intell. Syst. 9, 6373–6392. doi: 10.1007/s40747-023-01073-9
Edward, O., Pyzer-Knapp, A., and Curioni, A. (2024). Advancing biomolecular simulation through exascale HPC, AI, and quantum computing. Curr. Opin. Struct. Biol. 87:102826. doi: 10.1016/j.sbi.2024.102826
Farahmand, Y., Heidarnezhad, Z., Heidarnezhad, F., Muminov, K., and Heydari, F. (2014). A study of quantum information and quantum computers. Orient. J. Chem. 30, 601–606. doi: 10.13005/ojc/300227
Ganguly, S., Chandilkar, V., Jain, P., and Bertel, L. G. A. (2024). “Quantum graph neural networks based protein-ligand classification” in Artificial intelligence and knowledge processing. eds. R. V. Rodriguez, M. Rege, V. Piuri, G. Xu, and K. L. Ong, vol. 2127 (Springer), 139–149.
Ganguly, S., Morapakula, S. N., and Coronado, L. M. P. (2022). “Quantum natural language processing based sentiment analysis using Lambeq toolkit” in Proceedings of the 2022 second international conference on power, control and computing technologies (Los Alamitos, California: ICPC2T), 1–6. IEEE
Ghoabdi, M. Z., and Afsaneh, E. (2023). Quantum machine learning for untangling the real-world problem of cancers classification based on gene expressions. bioRxiv, 2023–08.
Ghosh, A., Fuad, M. D., and Bhattacharjee, S. (2024). Empirical quantum advantage analysis of quantum kernel in gene expression data. arXiv preprint arXiv:2411.07276.
Ginex, T., Vázquez, J., Estarellas, C., and Luque, F. J. (2024). Quantum mechanical-based strategies in drug discovery: finding the pace to new challenges in drug design. Curr. Opin. Struct. Biol. 87:102870. doi: 10.1016/j.sbi.2024.102870
Gorgulla, C., Jayaraj, A., Fackeldey, K., and Arthanari, H. (2022). Emerging frontiers in virtual drug discovery: from quantum mechanical methods to deep learning approaches. Curr. Opin. Chem. Biol. 69:102156. doi: 10.1016/j.cbpa.2022.102156
Gowda, D., Patil, H. Y., Abidin, S., Panda, R. A., and Suneetha, S. (2024). “Quantum machine learning for biomedical data analysis” in Quantum innovations at the Nexus of biomedical intelligence (Hershey, PA, USA: IGI Global), 180–205.
Grealey, J., Lannelongue, L., Saw, W.-Y., Marten, J., Méric, G., Ruiz-Carmona, S., et al. (2022). The carbon footprint of bioinformatics. Mol. Biol. Evol. 39:msac034. doi: 10.1093/molbev/msac034
Grover, L. K. (1996). “A fast quantum mechanical algorithm for database search.” in Proceedings of the Twenty-Eighth Annual ACM Symposium on Theory of Computing. pp. 212–219. ACM.
Guarasci, R., De Pietro, G., and Esposito, M. (2022). Quantum natural language processing: challenges and opportunities. Appl. Sci. 12:5651. doi: 10.3390/app12115651
Gyongyosi, L., and Imre, S. (2019). Quantum circuit design for objective function maximization in gate-model quantum computers. Quantum Inf. Process 18:225. doi: 10.1007/s11128-019-2326-2
Hansen, E., Joshi, S., and Rarick, H. (2023). “Resource estimation of quantum multiplication algorithms.” In 2023 IEEE international conference on quantum computing and engineering (QCE). pp. 199–202. IEEE.
Harvey, C., Yeung, R., and Meichanetzidis, K. (2023). Sequence processing with quantum tensor networks. arXiv preprint arXiv:2308.0786.
Hatakeyama-Sato, K., Igarashi, Y., Kashikawa, T., Kimura, K., and Oyaizu, K. (2022). Quantum circuit learning as a potential algorithm to predict experimental chemical properties. Dig. Dis. 2, 165–176. doi: 10.1039/D2DD00090C
Havlíček, V., Córcoles, A. D., Temme, K., Harrow, A. W., Kandala, A., Chow, J. M., et al. (2019). Supervised learning with quantum-enhanced feature spaces. Nature 567, 209–212. doi: 10.1038/s41586-019-0980-2
Herrmann, N., Arya, D., Doherty, M. W., Mingare, A., Pillay, J. C., Preis, F., et al. (2023). “Quantum utility – definition and assessment of a practical quantum advantage.” in 2023 IEEE international conference on quantum software (QSW). pp. 162–174.
Heshami, K., England, D. G., Humphreys, P. C., Bustard, P. J., Acosta, V. M., Nunn, J., et al. (2016). Quantum memories: emerging applications and recent advances. J. Mod. Opt. 63, 2005–2028. doi: 10.1080/09500340.2016.1148212
Hilton, C. B., Milinovich, A., and Felix, C. (2020). Personalized predictions of patient outcomes during and after hospitalization using artificial intelligence. NPJ Digit. Med. 3:51. doi: 10.1038/s41746-020-0249-z
Hong, Z., Wang, J., Qu, X., Zhu, X., Liu, J., and Xiao, J. (2021). “Quantum convolutional neural network on protein distance prediction.” in 2021 International Joint Conference on Neural Networks (IJCNN). pp. 1–8. IEEE.
Huang, T., Zeng, Z., Shi, H., Wu, Y., and Hong, Z. (2015). Survey of natural language processing techniques in bioinformatics. Comput. Math. Methods Med. 2015:674296. doi: 10.1155/2015/674296
Ibtehaz, N., and Kihara, D. (2023). “Application of sequence embedding in protein sequence-based predictions.” in Machine learning in bioinformatics of protein sequences: Algorithms, databases and resources for modern protein bioinformatics. pp. 31–55.
IonQ (2021). Biogen and IonQ partner to apply quantum computing to accelerate drug discovery. IonQ. Available at: https://ionq.com/posts/2021-11-16-biogen-and-ionq-partner (Accessed June 24, 2022).
Jayanth, K. K., Mohan, G. B., and Kumar, R. P. (2023). “Indian language analysis with XLM-RoBERTa: enhancing parts of speech tagging for effective natural language preprocessing.” in 2023 seventh international conference on image information processing (ICIIP). pp. 850–854. IEEE.
Jiménez López, M. D. (2022). “Processing natural language with biomolecules: Where linguistics, biology and computation meet” in Revolutions and revelations in computability. CiE 2022. Lecture notes in computer science. eds. U. Berger, J. N. Y. Franklin, F. Manea, and A. Pauly, vol. 13359 (Cham: Springer).
Jumper, J., Evans, R., Pritzel, A., Green, T., Figurnov, M., Ronneberger, O., et al. (2021). Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589. doi: 10.1038/s41586-021-03819-2
Kang, C., Huo, Y., Xin, L., Tian, B., and Yu, B. (2019). Feature selection and tumor classification for microarray data using relaxed lasso and generalized multi-class support vector machine. J. Theor. Biol. 463, 77–91. doi: 10.1016/j.jtbi.2018.12.010
Karamlou, A., Pfaffhauser, M., and Wootton, J. (2022). Quantum natural language generation on near-term devices. arXiv arXiv:2211.00727 [Preprint].
Karetla, G. R., Catchpoole, D., and Nguyen, Q. V. (2023). “Hybrid framework for genomic data classification using deep learning: QDeep_SVM” in Machine intelligence and data science applications: Proceedings of MIDAS 2022. eds. A. Ramdane-Cherif, T. P. Singh, R. Tomar, T. Choudhury, and J. S. Um (Singapore: Springer), 451–463.
Khan, M. R. I., Shahriar, S., and Rafid, S. F. (2023). A linear time quantum algorithm for pairwise sequence alignment. arXiv. arXiv:2307.04479 [Preprint].
Khurana, D., Koli, A., Khatter, K., and Singh, S. (2023). Natural language processing: state of the art, current trends and challenges. Multimed. Tools Appl. 82, 3713–3744. doi: 10.1007/s11042-022-13428-4
Kim, Y., Wood, C. J., Yoder, T. J., Merkel, S. T., Gambetta, J. M., Temme, K., et al. (2021). Scalable error mitigation for noisy quantum circuits produces competitive expectation values. Nat. Phys. 19, 752–759. doi: 10.1038/s41567-023-02225-9
Kirubakaran, A. P., and Midhunchakkaravarthy, J. (2024). “A hybrid application of quantum computing methodologies to AI techniques for paddy crop leaf disease identification” in Integrating blockchain and artificial intelligence for industry 4.0 innovations. eds. S. Goundar and R. Anandan (New York: Springer), 1–12.
Klimov, P. V., Bengtsson, A., Quintana, C., Bourassa, A., Hong, S., Dunsworth, A., et al. (2024). Optimizing quantum gates towards the scale of logical qubits. Nat. Commun. 15:2442. doi: 10.1038/s41467-024-46623-y
Kösoglu-Kind, B., Loredo, R., Grossi, M., Bernecker, C., Burks, J. M., and Buchkremer, R. (2023). A biological sequence comparison algorithm using quantum computers. Sci. Rep. 13:14552. doi: 10.1038/s41598-023-41086-5
Kryshtafovych, A., and Fidelis, K. (2009). Protein structure prediction and model quality assessment. Drug Discov. Today 14, 386–393. doi: 10.1016/j.drudis.2008.11.010
Kumar, P., Paul, R. K., Roy, H. S., Yeasin, M., Ajit,, and Paul, A. K. (2024). Big data analysis in computational biology and bioinformatics. Methods Mol. Biol. 2719, 181–197. doi: 10.1007/978-1-0716-3461-5_11
Kundu, D., Ghosh, A., Ekambaram, S., Wang, J., Dokholyan, N., and Ghosh, S. (2024). Application of quantum tensor networks for protein classification. bioRxiv, 2024–2003. doi: 10.1101/2024.03
Lannelongue, L., Bateman, A., Birney, E., Juckes, M., McEntyre, J., Reilly, G., et al. (2023). GREENER principles for environmentally sustainable computational science. Nat. Comp. Sci. 3, 514–521. doi: 10.1038/s43588-023-00461-y
Leurs, K. (2022). Design and implementation of a quantum kernel for natural language processing. arXiv.
Li, R. Y., Di Felice, R., Rohs, R., and Lidar, D. A. (2018). Quantum annealing versus classical machine learning applied to a simplified computational biology problem. NPJ Quantum Inform. 4:14. doi: 10.1038/s41534-018-0060-8
Li, F., Zhang, Z., Guan, J., and Zhou, S. (2022). Effective drug–target interaction prediction with mutual interaction neural network. Bioinformatics 38, 3582–3589. doi: 10.1093/bioinformatics/btac377
Liang, Z., Cheng, J., Yang, R., Ren, H., Song, Z., Wu, D., et al. (2023). Unleashing the potential of LLMs for quantum computing: A study in quantum architecture design. arXiv :arXiv:2307.08191 [Preprint].
Liu, J. G., and Wang, L. (2018). Differentiable learning of quantum circuit born machines. Phys. Rev. A 98:062324. doi: 10.1103/PhysRevA.98.062324
Liu, T., Wei, Y., and Wang, J. (2024). “Research on distributional compositional categorical model in both classical and quantum natural language processing.” in Proceedings of the SNPD 2024. pp. 1–6.
Liu, J., Yang, M., Yu, Y., Xu, H., Li, K., and Zhou, X. (2024). Large language models in bioinformatics: Applications and perspectives. ArXiv.
Liu, W., Zhao, J., Du, L., Padwal, H. H., and Vadivel, T. (2021). Intelligent comprehensive evaluation system using artificial intelligence for environmental evaluation. Environ. Impact Assess. Rev. 86:106495. doi: 10.1016/j.eiar.2020.106495
Locke, S., Bashall, A., Al-Adely, S., Moore, J., Wilson, A., and Kitchen, G. B. (2021). Natural language processing in medicine: A review. Trends Anaesthesia Crit. Care 38, 4–9. doi: 10.1016/j.tacc.2021.02.007
Lubinski, T., Johri, S., Varosy, P., Coleman, J., Zhao, L., Necaise, J., et al. (2023). Application-oriented performance benchmarks for quantum computing. IEEE Trans. Quant. Eng. 4, 1–32. doi: 10.1109/TQE.2023.3253761
Madsen, S., Marqversen, F., Rasmussen, S., and Zinner, N. (2023). Multi-sequence alignment using the quantum approximate optimization algorithm. arXiv preprint arXiv:2308.12103.
Magann, A. B., Grace, M. D., Rabitz, H. A., and Sarovar, M. (2021). Digital quantum simulation of molecular dynamics and control. Phys. Rev. Res. 3:023165. doi: 10.1103/PhysRevResearch.3.023165
Maheshwari, D., Garcia-Zapirain, B., and Sierra-Sosa, D. (2022). Quantum machine learning applications in the biomedical domain: A systematic review. IEEE Access 10, 80463–80484. doi: 10.1109/ACCESS.2022.3195044
Marchetti, L., Nifosì, R., Martelli, P. L., Da Pozzo, E., Cappello, V., Banterle, F., et al. (2022). Quantum computing algorithms: getting closer to critical problems in computational biology. Brief. Bioinform. 23:bbac437. doi: 10.1093/bib/bbac437
Martinez, V., and Leroy-Meline, G. (2022). A multiclass Q-NLP sentiment analysis experiment using DisCoCat. arXiv :arXiv:2209.03152. [Preprint].
Mensa, S., Şahin, M. E., Tacchino, F., Barkoutsos, P., and Tavernelli, I. (2022). Quantum machine learning framework for virtual screening in drug discovery: A prospective quantum advantage. arXiv.
Miranda, E. R., Yeung, R., Pearson, A., Meichanetzidis, K., and Coecke, B. (2022). “A quantum natural language processing approach to musical intelligence” in Quantum computer music: Foundations, methods and advanced concepts (Cham: Springer International Publishing), 313–356.
Mohammed, M., Al-Dabbagh, N., Salim, N., Mubarak, H., Ali, A., and Saeed, F. (2017). Quantum probability ranking principle for ligand-based virtual screening. J. Comput. Aided Mol. Des. 31, 365–378. doi: 10.1007/s10822-016-0003-4
Mokhtari, M., Khoshbakht, S., Ziyaei, K., Akbari, M. E., and Moravveji, S. S. (2024). New classifications for quantum bioinformatics: Q-bioinformatics, QCt-bioinformatics, QCg-bioinformatics, and QCr-bioinformatics. Brief. Bioinform. 25:bbae074. doi: 10.1093/bib/bbae074
Nałęcz-Charkiewicz, K., Charkiewicz, K., and Nowak, R. M. (2024). Quantum computing in bioinformatics: A systematic review mapping. Brief. Bioinform. 25:bbae391. doi: 10.1093/bib/bbae391
Nam, V., and Nguyen,. (2024). Quantum word embedding for machine learning. Phys. Scr. 99:086004. doi: 10.1088/1402-4896/ad6299
Nammouchi, A., Kassler, A., and Theorachis, A. (2023). Quantum machine learning in climate change and sustainability: A review. arXiv arXiv:2310.09162 [Preprint].
Nathans, J., and Sterling, P. (2016). How scientists can reduce their carbon footprint. eLife 5:e15928. doi: 10.7554/eLife.15928
Navneet, S., and Pokhrel, S. R. (2024). An independent implementation of quantum machine learning algorithms in Qiskit for genomic data. arXiv preprint arXiv:2405.09781.
Nayak, C., Simon, S. H., Stern, A., Freedman, M., and Das Sarma, S. (2008). Non-abelian anyons and topological quantum computation. Rev. Mod. Phys. 80, 1083–1159. doi: 10.1103/RevModPhys.80.1083
Neveu, E., Popov, P., Hoffmann, A., Migliosi, A., Besseron, X., Danoy, G., et al. (2018). RapidRMSD: rapid determination of RMSDs corresponding to motions of flexible molecules. Bioinformatics 34, 2757–2765. doi: 10.1093/bioinformatics/bty160
Ng, G. Y. L., Tan, S. C., and Ong, C. S. (2023). On the use of QDE-SVM for gene feature selection and cell type classification from scRNA-seq data. PLoS One 18:e0292961. doi: 10.1371/journal.pone.0292961
Nunn, A., Otto, C., Stadler, P., and Langenberger, D. (2021). Comprehensive benchmarking of software for mapping whole genome bisulfite data: from read alignment to DNA methylation analysis. Brief. Bioinform. 22:21. doi: 10.1093/bib/bbab021
Ofer, D., Brandes, N., and Linial, M. (2021). The language of proteins: NLP, machine learning & protein sequences. Comput. Struct. Biotechnol. J. 19, 1750–1758. doi: 10.1016/j.csbj.2021.03.022
Ohno-Machado, L., Nadkarni, P., and Johnson, K. (2013). Natural language processing: algorithms and tools to extract computable information from EHRs and from the biomedical literature. J. Am. Med. Inform. Assoc. 20:805. doi: 10.1136/amiajnl-2013-002214
Olechnovič, K., Monastyrskyy, B., Kryshtafovych, A., and Venclovas, Č. (2019). Comparative analysis of methods for evaluation of protein models against native structures. Bioinformatics 35, 937–944. doi: 10.1093/bioinformatics/bty760
Onodera, W., Hara, N., Aoki, S., Asahi, T., and Sawamura, N. (2023). Phylogenetic tree reconstruction via graph cut presented using a quantum-inspired computer. Mol. Phylogenet. Evol. 178:107636. doi: 10.1016/j.ympev.2022.107636
Outeiral, C., Strahm, M., Shi, J., Morris, G. M., Benjamin, S. C., and Deane, C. M. (2021). The prospects of quantum computing in computational molecular biology. Wiley Interdiscip. Rev. Comp. Molecul. Sci. 11:e1481. doi: 10.1002/wcms.1481
Pal, S., Bhattacharya, M., Lee, S. S., and Chakraborty, C. (2024). Quantum computing in the next-generation computational biology landscape: from protein folding to molecular dynamics. Mol. Biotechnol. 66, 163–178. doi: 10.1007/s12033-023-00765-4
Patel, H. B., Mishra, S., Jain, R., and Kansara, N. (2023). The future of quantum computing and its potential applications. J. Basic Sci. 23, 513–519.
Patti, T. L., Kossaifi, J., Anandkumar, A., and Yelin, S. F. (2022). Variational quantum optimization with multibasis encodings. Phys. Rev. Res. 4:033142. doi: 10.1103/PhysRevResearch.4.033142
Pecina, A., Eyrilmez, S. M., Köprülüoğlu, C., Miriyala, V. M., Lepšík, M., Fanfrlík, J., et al. (2020). SQM/COSMO scoring function: reliable quantum-mechanical tool for sampling and ranking in structure-based drug design. ChemPlusChem 85, 2362–2371. doi: 10.1002/cplu.202000120
Perdomo-Ortiz, A., Dickson, N., Drew-Brook, M., Rose, G., and Aspuru-Guzik, A. (2012). Finding low-energy conformations of lattice protein models by quantum annealing. Sci. Rep. 2:571. doi: 10.1038/srep00571
Perrier, E., Youssry, A., and Ferrie, C. (2022). QDataSet, quantum datasets for machine learning. Sci. Data 9:582. doi: 10.1038/s41597-022-01639-1
Piattini, M., Peterssen Nodarse, G., and Pérez-Castillo, R. (2020). Quantum computing: A new software engineering golden age. ACM SIGSOFT Softw. Eng. Notes 45, 12–14. doi: 10.1145/3402127.3402131
Poleksic, A. (2015). A polynomial time algorithm for computing the area under a GDT curve. Algor. Molecul. Biol. 10:27. doi: 10.1186/s13015-015-0058-0
Prasad, S. J. S., Suji Usharani, R., Sheryal, Oliver A., Sharma, D., Sureshkumar, M., and Bansod, P. (2024). Quantum computing-powered agricultural transformation: optimizing performance in farming.
Preskill, J. (2018). Quantum computing in the NISQ era and beyond. Quantum 2:79. doi: 10.22331/q-2018-08-06-79
Prousalis, K., and Konofaos, N. (2019). A quantum pattern recognition method for improving pairwise sequence alignment. Sci. Rep. 9:7226. doi: 10.1038/s41598-019-43697-3
Pseiner, J., Erhard, M., and Krenn, M. (2024). Quantum interference between distant creation processes. Phys. Rev. Res. 6:013294. doi: 10.1103/PhysRevResearch.6.013294
Pudenz, K. L., and Lidar, D. A. (2013). Quantum adiabatic machine learning. Quantum Inf. Process 12, 2027–2070. doi: 10.1007/s11128-012-0506-4
Quantum Technology and Application Consortium – QUTACAndreas, B., Guillaume, B., Julia, B., Thierry, A., Hans, E., Thomas, E., et al. (2021). Industry quantum computing applications. EPJ Quantum Technol. 8. doi: 10.1140/epjqt/s40507-021-00114-x
Quetschlich, N., Burgholzer, L., and Wille, R. (2022). Compiler optimization for quantum computing using reinforcement learning. arXiv. doi: 10.48550/arXiv.2212.04508
Ramette, J., Sinclair, J., Breuckmann, N. P., and Vuletić, V. (2024). Fault-tolerant connection of error-corrected qubits with noisy links. NPJ Quant. Inform. 10, 1–6. doi: 10.1038/s41534-024-00855-4
Reali, F., Priami, C., and Marchetti, L. (2017). Optimization algorithms for computational systems biology. Front. Appl. Math. Stat. 3:6. doi: 10.3389/fams.2017.00006
Repetto, V., Ceroni, E. G., Buonaiuto, G., and D'Aurizio, R. (2024). Quantum enhanced stratification of breast Cancer: Exploring quantum expressivity for real omics data. arXiv preprint arXiv:2409.14089.
Robert, A., Barkoutsos, P. K., Woerner, S., and Tavernelli, I. (2021). Resource-efficient quantum algorithm for protein folding. NPJ Quantum Inf. 7:38. doi: 10.1038/s41534-021-00368-4
Saggi, M. K., Bhatia, A. S., Isaiah, M., Gowher, H., and Kais, S. (2024). Multi-omic and quantum machine learning integration for lung subtypes classification. arXiv arXiv:2410.02085 [Preprint].
Salem, M., Merelo, J., Siarry, P., Bachir, R., Debakla, M., and Debbat, F. (2023). “Quantum natural language processing: A new and promising way to solve NLP problems” in Artificial intelligence: Theories and applications: First international conference, ICAITA 2022, Mascara, Algeria, November 7–8, 2022, revised selected papers. eds. M. Salem, J. J. Merelo, P. Siarry, R. B. Bouiadjra, M. Debakla, and F. Debbat, vol. 1769 (New York: Springer), 215–227.
Salloum, H., Lukin, R., and Mazzara, M. (2024). Quantum computing in drug discovery: A review of quantum annealing and gate-based approaches. doi: 10.13140/RG.2.2.32727.48805
Samanta, S., Choudhury, A., Dey, N., Ashour, A. S., and Balas, V. (2016). Quantum inspired evolutionary algorithm for scaling factors optimization during manifold medical information embedding. In Quantum Inspired Evol. Comp. (pp. 145–167). doi: 10.1016/B978-0-12-804409-4.00009-7
Sarkar, A., Al-Ars, Z., Almudever, C. G., and Bertels, K. (2019). An algorithm for DNA read alignment on quantum accelerators. arXiv preprint arXiv:1909.05563.
Sathan, D., and Baichoo, S. (2024). “Drug target interaction prediction using variational quantum classifier.” In Proceedings of the 2024 international conference on next generation computing applications (NextComp), Mauritius, pp. 1–7.
Selig, P., Murphy, N.R A. S., Redmond, D., and Caton, S. (2021). “A case for noisy shallow gate-based circuits in quantum machine learning.” In 2021 International Conference on Rebooting Computing (ICRC). pp. 24–34. IEEE.
Selladurai, P., Dahiya, R., Kandasamy, B., and Radhakrishnan, V. (2024). “Integrating quantum computing in bioinformatics and biomedical research” in Intelligent data analytics for bioinformatics and biomedical systems (Chapter 15). eds. N. Sharma, K. Cengiz, and P. Chatterjee.
Shaun, A., Abel, C. P., Verena, R., Sumir, P., Katherine, J., and Nicola, M. (2021). The development of a sustainable bioinformatics training environment within the H3Africa bioinformatics network (H3ABioNet). Front. Educ. 6:725702. doi: 10.3389/feduc.2021.725702
Shiny Duela, J., Umamageswari, A., Prabavathi, R., Umapathy, P., and Raja, K. (2023). “Quantum assisted genetic algorithm for sequencing compatible amino acids in drug design” in 2023 third international conference on advances in electrical, computing, communication and sustainable technologies (ICAECT), 1–7. doi: 10.1109/ICAECT57570.2023.10117673
Shor, P. W. (1999). Polynomial-time algorithms for prime factorization and discrete logarithms. Quantum Inf. Process 2, 303–332.
Shuyue, L., Shu, Z., Hongchao, S., Ruixing, L., Hexin, L., and Paola, G. (2023). “PQLM - Multilingual decentralized portable quantum language model.” IEEE International Conference on acoustics, Speech, and Signal Processing (ICASSP).
Simmons, S. (2024). Scalable fault-tolerant quantum technologies with silicon color centers. PRX Quantum 5:010102. doi: 10.1103/PRXQuantum.5.010102
Soame, R. (2023). “Quantum natural language processing: A new and promising way to solve NLP problems.” In Proceedings of the 2023 conference. pp. 215–227.
Soni, K. K., and Rasool, A. (2021). Quantum-based exact pattern matching algorithms for biological sequences. ETRI J. 43, 483–510. doi: 10.4218/etrij.2019-0589
Sood, V., and Chauhan, R. P. (2024). Quantum computing: impact on energy efficiency and sustainability. Expert Syst. Appl. 255:124401. doi: 10.1016/j.eswa.2024.124401
Steedman, M., and Baldridge, J. (2011). “Non-transformational syntax: Formal and explicit models of grammar” in Wiley-Blackwell companion to syntax. Eds. Robert D. B., and Kersti, B., Wiley-Blackwell, 1–27.
Stodden, V., and Miguez, S. (2014). Best practices for computational science: software infrastructure and environments for reproducible and extensible research. J. Open Res. Softw. 2:e21. doi: 10.5334/jors.ay
Strubell, E., Ganesh, A., and McCallum, A. (2020). “Energy and policy considerations for modern deep learning research.” in Proceedings of the AAAI Conference on Artificial Intelligence Vol. 34, No. 9. pp. 13693–13696.
Sun, J., Chen, W., Fang, W., Wun, X., and Xu, W. (2012). Gene expression data analysis with the clustering method based on an improved quantum-behaved particle swarm optimization. Eng. Appl. Artif. Intell. 25, 376–391. doi: 10.1016/j.engappai.2011.09.017
Surov, I. A., Semenenko, E., Platonov, A. V., Bessmertny, I. A., Galofaro, F., Toffano, Z., et al. (2021). Quantum semantics of text perception. Sci. Rep. 11:4193. doi: 10.1038/s41598-021-83490-9
Tao, Y. (2024). Quantum entanglement: principles and research progress in quantum information processing. Theoret. Nat. Sci. 30, 263–274. doi: 10.54254/2753-8818/30/20241130
Temme, K., Bravyi, S., and Gambetta, J. M. (2017). Error mitigation for short-depth quantum circuits. Phys. Rev. Lett. 119:180509. doi: 10.1103/PhysRevLett.119.180509
Ting, Z., and Caflisch, A. (2010). High-throughput virtual screening using quantum mechanical probes: discovery of selective kinase inhibitors. ChemMedChem 5, 1007–1014. doi: 10.1002/cmdc.201000085
Torlai, G., and Melko, R. G. (2020). Machine-learning quantum states in the NISQ era. Ann. Rev. Cond. Matter Physics 11, 325–344. doi: 10.1146/annurev-conmatphys-031119-050651
Tsujii, J. (2021). Natural language processing and computational linguistics. Comput. Linguist. 47, 1–21. doi: 10.1162/coli_a_00420
Van Vu, T., Kuwahara, T., and Saito, K. (2024). Fidelity-dissipation relations in quantum gates. Phys. Rev. Res. 6:033225. doi: 10.1103/PhysRevResearch.6.033225
Varmantchaonala, C. M., Fendji, J. L. K. E., Schöning, J., and Atemkeng, M. (2024). Quantum natural language processing: A comprehensive survey. IEEE Access 12, 99578–99598. doi: 10.1109/ACCESS.2024.3420707
Veleiro, U., de la Fuente, J., Pizurica, M., Pineda-Lucena, A., Ochoa, I., Gevaert, O., et al. (2023). GeNNius: An ultrafast drug-target interaction inference method based on graph neural networks. bioRxiv.
Vincentius, M., Zhao, J., Afek, A., Mielko, Z., and Gordân, R. (2019). QBiC-Pred: quantitative predictions of transcription factor binding changes due to sequence variants. Nucleic Acids Res. 47, W127–W135. doi: 10.1093/nar/gkz363
Wang, Y., Alexeev, Y., Jiang, L., Chong, F. T., and Liu, J. (2024). Fundamental causal bounds of quantum random access memories. NPJ Quant. Inform. 10, 1–7. doi: 10.1038/s41534-024-00848-3
Wang, Y., and Liu, J. (2024). Quantum machine learning: from NISQ to fault tolerance. ArXiv, abs/2401.11351. doi: 10.48550/arXiv.2401.11351
Webber, M., Elfving, V., Weidt, S., and Hensinger, W. K. (2022). The impact of hardware specifications on reaching quantum advantage in the fault tolerant regime. AVS Quantum Sci. 4:013801. doi: 10.1116/5.0073075
Weidner, C. A., Reed, E. A., Monroe, J., Sheller, B., O’Neil, S., Maas, E., et al. (2024). Robust quantum control in closed and open systems: theory and practice. Automatica 172:111987. doi: 10.48550/arXiv.2401.00294
Widdows, D., Alexander, A., Zhu, D., Zimmerman, C., and Majumder, A. (2024). Near-term advances in quantum natural language processing. Ann. Math. Artif. Intell. 92, 1249–1272. doi: 10.1007/s10472-024-09940-y
Wiebe, N., Bocharov, A., Smolensky, P., Troyer, M., and Svore, K. M. (2019). Quantum language processing. arXiv arXiv:1902.05162 [Preprint].
Wong, R., and Chang, W.-L. (2021). Quantum speedup for protein structure prediction. IEEE Trans. Nanobiosci. 20, 323–330. doi: 10.1109/TNB.2021.3065051
Wong, R., and Chang, W.-L. (2022). Fast quantum algorithm for protein structure prediction in hydrophobic-hydrophilic model. J. Parallel Distrib. Comp. 164, 178–190. doi: 10.1016/j.jpdc.2022.03.011
Xiong, Z., Cui, X., Lin, X., Ren, F., Liu, B., Li, Y., et al. (2023). Q-drug: A framework to bring drug design into quantum space using deep learning. arXiv preprint arXiv:2308.13171.
Yan, P., Li, L., Jin, M., and Zeng, D. (2021). Quantum probability-inspired graph neural network for document representation and classification. Neurocomputing 445, 276–286. doi: 10.1016/j.neucom.2021.02.060
Yang, L., Zhang, X., Wang, T., and Zhu, H. (2013). Large local analysis of the unaligned genome and its application. J. Comput. Biol. 20, 19–29. doi: 10.1089/cmb.2011.0052
Yeung, R., and Kartsaklis, D. (2021). A CCG-based version of the DisCoCat framework. arXiv :arXiv:2105.07720 [Preprint].
Yu, Y., Qiu, D., and Yan, R. (2020). “Quantum entanglement based sentence similarity computation.” in 2020 IEEE international conference on Progress in informatics and computing (PIC), Shanghai, China. pp. 250–257.
Zarei, M., and Elaheh, A. (2024). Potential of quantum machine learning for solving the real-world problem of cancer classification. Dis. Appl. Sci. 6:513. doi: 10.1007/s42452-024-06220-6
Zhou, Y., Chen, J., Li, W., Cheng, J., Karemore, G., Zitnik, M., et al. (2024). Quantum-machine-assisted drug discovery: Survey and perspective. arXiv. doi: 10.48550/arXiv.2408.13479
Keywords: quantum natural language processing, bioinformatics, sustainability, drug discovery, knowledge extraction, protein prediction, genome analysis
Citation: Pallavi G and Prasanna Kumar R (2025) Quantum natural language processing and its applications in bioinformatics: a comprehensive review of methodologies, concepts, and future directions. Front. Comput. Sci. 7:1464122. doi: 10.3389/fcomp.2025.1464122
Edited by:
Fasih Haider, University of Edinburgh, United KingdomReviewed by:
H. Z. Shen, Northeast Normal University, ChinaImane Guellil, University of Edinburgh, United Kingdom
Copyright © 2025 Pallavi and Prasanna Kumar. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rangarajan Prasanna Kumar, cl9wcmFzYW5uYWt1bWFyQGNoLmFtcml0YS5lZHU=
 Gundala Pallavi
Gundala Pallavi Rangarajan Prasanna Kumar*
Rangarajan Prasanna Kumar*