- Centre Borelli, ENS Paris-Saclay, Université de Paris, CNRS, INSERM, SSA, Gif-sur-Yvette, France
Introduction: The recognized credibility crisis in scientific research has led to an increasing focus on reproducibility studies, particularly in computer science. Existing studies predominantly examine specific technological aspects of reproducibility but neglect the critical interplay between authors and publishers in enabling reproducible computational scientific research.
Methods: A systematic review was conducted following the PRISMA Literature Review methodology, complemented by a Journals Survey. This approach enabled a comprehensive analysis of reproducibility policies and software/data management practices in scientific computing journals.
Results: The survey revealed significant variability in reproducibility policies and practices across computer science journals. Many gaps and challenges were identified, including inconsistencies in policy enforcement, lack of standardized tools, and insufficient recognition of software as a research artifact. The analysis highlighted the potential of Reproducibility as a Service (RaaS) as an innovative solution to address these challenges.
Discussion: This study underscores the need for improved standardization and implementation of reproducibility policies. Strategies to enhance reproducibility include fostering collaboration among authors, publishers, and technology providers, as well as recognizing software as a critical research output. The findings aim to guide stakeholders in bridging the current gaps and advancing the reproducibility of computational scientific articles.
1 Introduction
Reproducibility is a broad and complex topic strongly related to the history of science and knowledge (Ivie and Thain, 2018) reflected in the cumulative technological and scientific development of humanity (Hughes, 2001). Such development has been based on the evolutionary capacity of human beings to build new knowledge from previous discoveries and achievements, passing this knowledge to new generations through a continuous cycle of refinement. The evolution of science through the reproducibility of knowledge could be metaphorically compared to the natural mechanisms of DNA replication (Hu et al., 2020) transmitted from generation to generation in a continuous refinement cycle. Within these reproducible mechanisms, scientific journals play a significant role in the communication, divulgation, corroboration, validation, and acceptance of reliable and trustworthy knowledge.
The reproducibility of knowledge has recently become relevant to the scientific community given that there is a growing concern for ethics and transparency in the research results in scientific publications in the so-called reproducibility crisis (Plavén-Sigray et al., 2017; Gundersen, 2020). In addition, with the boom in artificial intelligence/machine learning (ML/AI), publications have evolved toward data-centric and model-centric developments that have forced journals to adapt their publishing models to new dynamics accelerated by technological changes (Hutson, 2018).
In response to these developments, several recent articles have hypothesized what the future of academic publishing will be like (Dodds, 2019; Baillieul et al., 2018; Ahmed et al., 2023), analyzing important changes, proposing technological tools (Kitchenham et al., 2020; Anchundia and Fonseca, 2020), and identifying significant gaps in publishing policies (Stoddart, 2016; Kapoor and Narayanan, 2022; Lucic et al., 2022; Ahmed et al., 2022).
This article accounts for policies implemented by publishers and their evolution, which are crucial for understanding their evaluation processes oriented to the reproducibility of knowledge, and facilitate participation, understanding, and dissemination of research to general public readers and the scientific community.
This article analyzes the journal policies concerning the reproducibility of knowledge addressed to trustworthiness and transparency through a survey of computer science journals indexed in SCOPUS and WoS and makes a systematic PRISMA-based literature review. Our main purpose is to answer the following questions: What is the reproducibility gap resulting from the credibility crisis, and what are the mutualized efforts by authors and publishers to bridge the reproducibility gap in AI/ML computer science research?
Figure 1 summarizes three perspectives/facets that have been addressed and discussed in this paper: efforts required by authors (Section 6.1) and by publishers (Section 6.2), and the technological evolution required to reproduce results (Section 3). The combined efforts of these three actors are required to close the reproducibility gap.
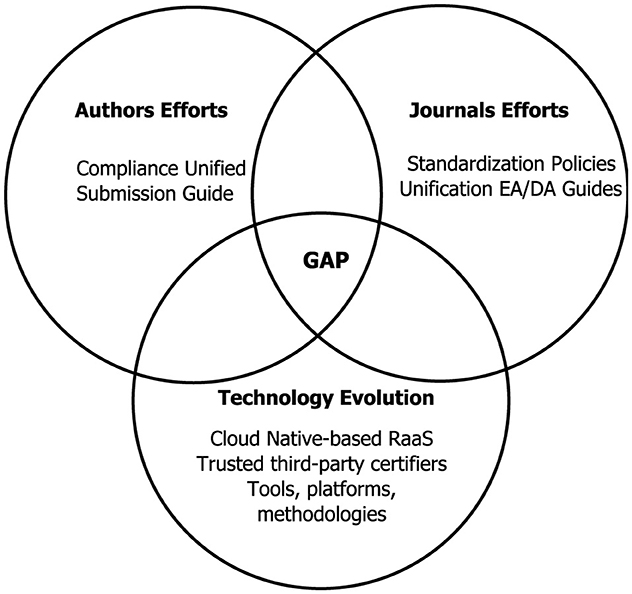
Figure 1. This diagram summarizes three perspectives/facets that have been analyzed and discussed in this paper: efforts required by authors (Section 6.1) and by publishers (Section 6.2), and the technological evolution required to reproduce results (Section 3). The combined efforts of these three actors are required to close the reproducibility gap.
This article is structured as follows: Section 2 provides the fundamentals and definitions of reproducibility, including key terms (Section 2), types of reproducibility (Section 2.1), and how reproducibility is measured and evaluated (Section 2.2). We establish the consensus in terminology and definitions used throughout the article, and we discuss the corresponding difficulties, definitions, and measures related to reproducibility, which allows us to define the technological evolution (Baillieul et al., 2018) as necessary to reproduce results and the fundamental strategies of reproducibility, as outlined in Section 3.
Section 3 explores strategies and technological evolution necessary for reproducibility, addressing topics such as open-source software and repositories (Section 3.1), open data formats (Section 3.2), system architecture (Section 3.3), and tools and platforms (Section 3.4). We also highlight the role of stakeholders (Diaba-Nuhoho and Amponsah-Offeh, 2021) in ensuring the reproducibility of computational scientific articles. Section 3.4 presents a landscape of existing tools, data management platforms, and techniques that are helpful in reproducible research. Best practices in data management are covered in Section 3.6, and Section 3.7 discusses the role of scientific publishers in reproducible research, including new types of publications with code and the challenges of evaluating research artifacts.
In Section 4, a survey of 16 computer science journals provides insights into experiences implementing data-code sharing policies based on the reviewed reproducibility platforms and technologies. Section 5 includes our technological discussion of the previous topics, with a focus on dilemmas like virtualization solutions versus dependency management (Section 5.1) and the shared responsibility between authors and publishers supported by technological evolution (Section 5.2).
Section 6 analyzes the combined efforts required by authors (Section 6.1) and publishers (Section 6.2) to close the reproducibility gap. Emerging dilemmas regarding reproducibility sharing policies are discussed in Sections 5.1 and 6.3, which explore the possibility of regarding reproducibility as a service provided by a trusted third party, the consideration of software as valuable research artifacts, and how to appropriately reward authors. Finally, the paper concludes in Section 7.
2 Fundamentals and definitions of reproducibility
Several studies (Sculley et al., 2015; Kitzes et al., 2018; Baker et al., 2019; Parashar et al., 2022; Thompson and Burnett, 2012; Raghupathi et al., 2022; Cacho and Taghva, 2020; Hummel and Manner, 2024) have addressed reproducibility from different points of view, as Gundersen (2021) reproducibility is considered a fundamental part of the scientific method. However, to our knowledge, no studies have holistically reviewed the different dimensions and strategies of reproducibility in computer science, i.e., to consider their essential participation within an end-to-end data science project/experiment life cycle. This life cycle begins from scientific research and ends in mass industrial production for final customers. The life cycle also incorporates the responsibilities of the main stakeholders (Diaba-Nuhoho and Amponsah-Offeh, 2021; Feger and Woźniak, 2022; Macleod and the University of Edinburgh Research Strategy Group, 2022) in this process (e.g., journals, authors, industry, and the scientific community).
The report from the National Academies of Sciences, Engineering, and Medicine (NASEM) (Committee on Reproducibility and Replicability in Science et al., 2019) is a reference reproducibility study that gathers contributions from relevant specialized researchers. It focuses on strategies for obtaining consistent computational results using the same input data, computational steps, methods, code, analysis conditions, and replicability to obtain consistent results across studies. In NASEM's definitions, reproducibility involves the original data and code, whereas replicability is related to the collection of new data and similar methods used in previous studies.
The simplest definition of reproducibility extended and used in the different studies is the one proposed by ACM in version 1.1 of their Artifact Review and Badging report,1 as shown in Figure 2.
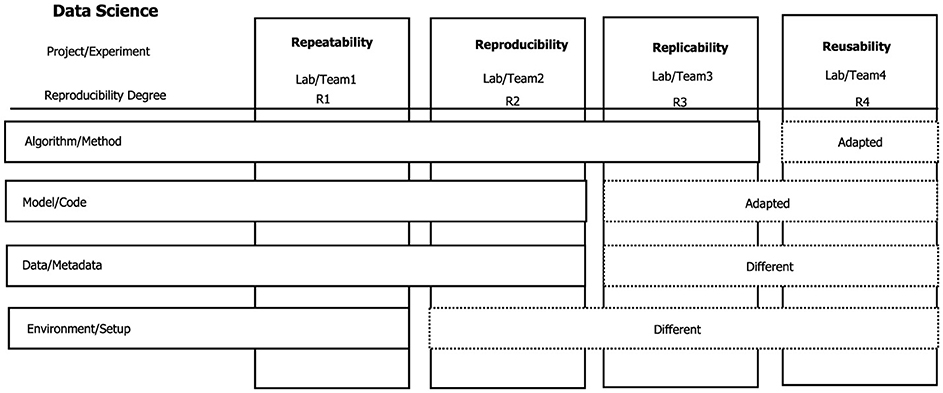
Figure 2. Definition of reproducibility, replicability, repeatability, and reusability (4R) (Benureau and Rougier, 2018). Different degrees of reproducibility can be considered according to the characteristics of the particular experiment or project.
Reproducibility (different team, same experimental setup): the experiment is done with different equipment, different environment, and same code/algorithm. Repeatability (same team, same experimental setup): the experiment is done by the same team, same environment (software/hardware), and same code/algorithm. Replicability (different team, different experimental setup): the experiment is done with different equipment, different environment, different code, and same algorithm. Reusability (different equipment, different, and partial experimental configuration): the experiment is carried out with different equipment, different environments, different codes, and the algorithm partially implemented.
There is still some discussion, in some cases even confusion, about the definitions (Plesser, 2018) even from a taxonomic point of view (Essawy et al., 2020; Heroux et al., 2018). A very different interpretation of reproducibility is presented in the reproducibility article (Lin and Zhang, 2020), where it is a continuous improvement process, rather than an achievable objective. However, following the discussions with the National Information Standards Organization (NISO), ACM accepted the recommendation to harmonize its terminology and definitions with those widely used in the community of scientific research. In this way, it interchanged the terms reproducibility and replicability with the existing definitions proposed by ACM to ensure consistency.
Figure 3A shows, according to these definitions, the minimal set of components one eventually should consider to make software reproducible. Figure 3B shows a generalization of an architecture for reproducible projects or experiments. It is made of basic blocks interconnected to build complex systems, applications, and workflows.
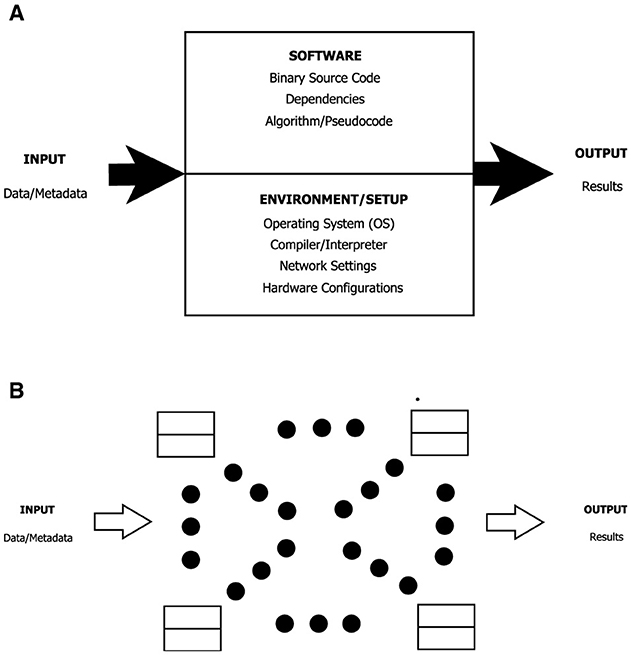
Figure 3. Generalization of architecture allowing for reproducible projects or experiments. (A) Basic block software/data and hardware/environment reproducibility. (B) System/flow reproducibility.
In this chapter, we specifically discuss the reproducibility of complex AI/ML data science projects. A scientific publication in AI/ML can range from effectively a model developed in an experiment by a single researcher for a tiny device to large implementations of Distributed Big Data supercomputing developed by large consortiums of universities, governmental, or research institutions.
2.1 Types of reproducibility
Defining reproducibility is as important as determining the types of reproducibility, considering the nuances that conceptually appear when studying the various cases and possibilities. The term reproducibility is acceptable in the case where the same input can lead to statically equivalent same results. It is also important to note that reproducibility in data science does not necessarily imply obtaining the same numerical result from the same numerical input.
Previous studies (Gundersen and Kjensmo, 2018; Raghupathi et al., 2022) have defined three degrees of reproducibility: R1 (Experiment, Data, Method), R2 (Data, Method), and R3 (Method). It is only sometimes possible to obtain the same numerical result from different realizations of an experiment. In that case, we can consider the following definitions (Impagliazzo et al., 2022):
• Experimental reproducibility: similar input (data) + similar experimental protocol → similar results.
• Statistical reproducibility: similar input (data) + same analysis → same conclusions [independently from (random) sampling variability].
• Computational reproducibility: same input (data) + same code/software + same software environment → exact same bitwise results.
2.2 Measure and evaluate reproducibility
One difficulty is to measure (Rosenblatt et al., 2023) the degrees of reproducibility depending on the complexity, type of reproducibility, type of data (Ahmed et al., 2022), and field of the research study (Raff, 2020) given that despite what is generally expected from computer systems and as has been shown in multiple positions (Gundersen and Kjensmo, 2018; Raghupathi et al., 2022; Bailey, 2020; Jalal Apostal et al., 2020), executing the same code on a different machine does not generate the same numerical result, but it can be established that a result is statistically similar (Raff, 2019). Another approach is to calculate the probability that a particular experiment gives comparable results (Nordling and Peralta, 2022).
The survival analysis proposed in Raff (2020) permits the extraction of new insights that better explain past longitudinal data and extend a recent data set with reproduction times, taking into account the number of days it took to reproduce an article (Collberg and Proebsting, 2016). Additionally, to measure support for reproducibility in several data management platforms, Gundersen et al. (2022) proposed a quantitative method.
This point is crucial because it is imperative to measure reproducibility to evaluate the degree and percentage of reproducibility of an article. As will be seen in the artifact evaluation Section 3.9, there is a wide disparity among journals/conferences in the criteria and policies for describing and evaluating artifacts, partly due to the difficulty in measuring reproducibility.
3 Reproducibility strategies and technological evolution
Motivated by the great reproducibility challenges (Schelter et al., 2015; Freire et al., 2012), the extensive literature on data science projects include current approaches for executing big data science projects (Saltz and Krasteva, 2022) and the best coding practices to ensure reproducibility (Gonzalo Rivero, 2020). Different strategies have been proposed (LeVeque et al., 2012) to tackle the problem of reproducibility of scientific works, specifically in AI/ML.
The size and scope of data science projects can range from small projects of the Internet of Things (IoT) (Ray, 2022) to very large high-end distributed HPC (Pouchard et al., 2019). Complex infrastructure is required for the latter, e.g., for the recently popular large language models (LLM). As such, different strategies are required to address the complexity of each specific project and experiment.
This section surveys the most relevant characteristics that can be considered as general reproducibility strategies (Gundersen, 2021), such as the use of open source software, open repositories, open data formats, the use of well-established methodologies, the following of good practices, and the use of system architectures which are typically used in systems dedicated to run AI/ML applications.
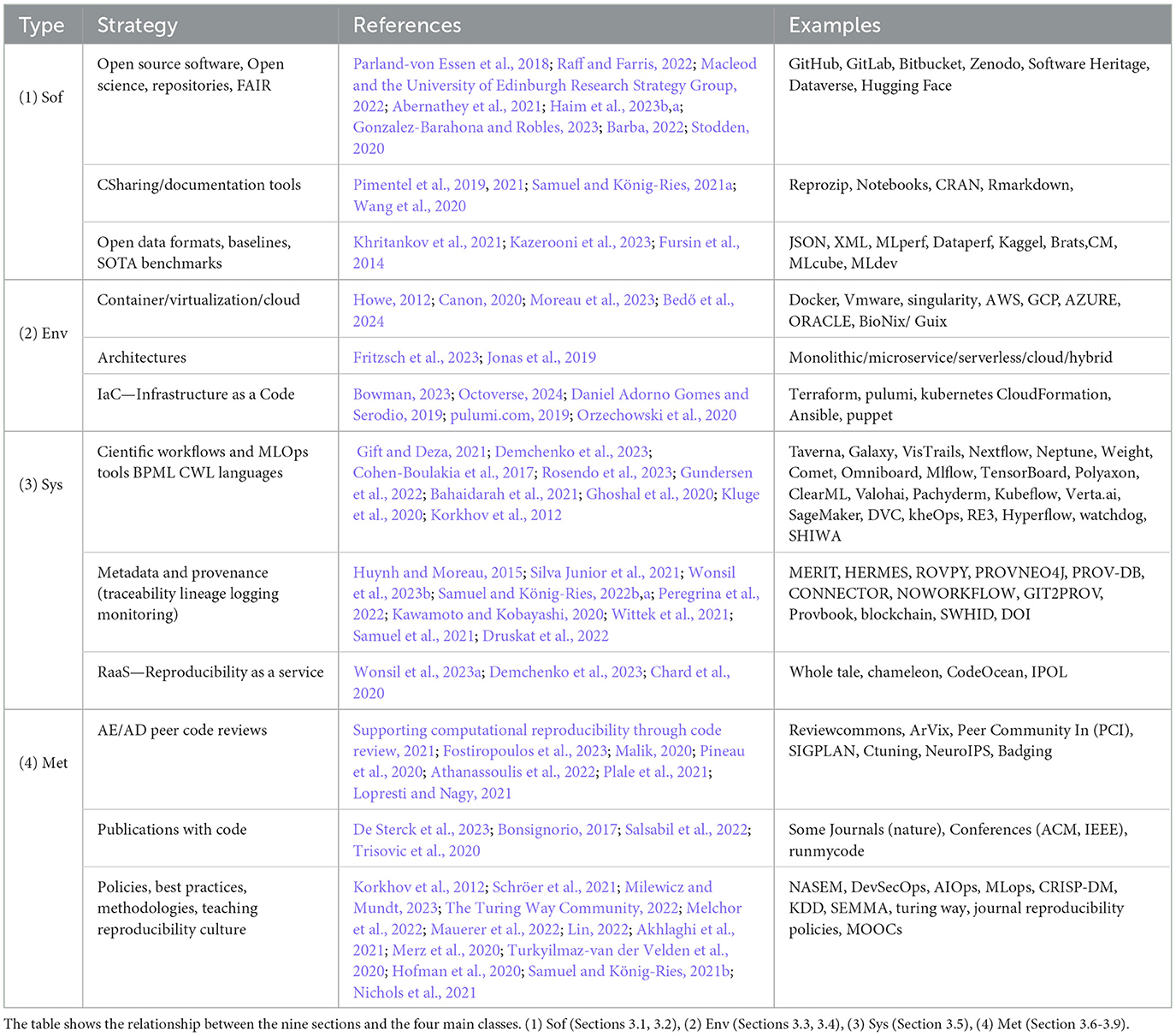
We divide the strategies identified in literature into four main classes: software and data, environment, system data management and workflows, and methods. Each strategy can be part of a more complex one. For example, workflows can be made of containers, and code publications strategies require the open code/data repositories strategy.
1. Software and data reproducibility
• Adoption of free and open source software.
• Tools with the potential to be used as reproducibility tools, for example, notebooks.
• Standardized automation benchmarks, open dataset formats, state-of-the-art model baselines.
2. Environment reproducibility
• Software reproducibility: containers and virtualization.
• System architecture: monolithic, microservices, serverless functions.
• Hardware reproducibility, including ambient configuration. For example, Infrastructure as Code.
3. System and workflow reproducibility
• Metadata and provenance (lineage and traceability).
• Reproducibility as a Service. This includes third-party specialized and trusted entities that certify reproducibility. They typically also offer services for the execution of algorithms on the infrastructure they provide.
4. Methodological reproducibility
• Adoption of good practices and methodologies.
• Teaching and reproducibility culture.
• Performing evaluation specifically for research code and data artifacts.
• Publications with code (journals and conferences).
3.1 Open source software and open repositories
Uploading code and data to a public repository and labeling it as open-source software might be considered a sufficient guarantee of reproducibility and transparency in research (Macleod and the University of Edinburgh Research Strategy Group, 2022; Barba, 2022). However, there have been objections to this approach (Abernathey et al., 2021) as well as proposals for evaluating the reproducibility level (Gonzalez-Barahona and Robles, 2023). It cannot be ensured that code will not be modified after publication2 or that the code is executed in the same environment, dependencies, and parameters. In many cases, the full reproduction of a work cannot be achieved and often requires contacting the authors to obtain detailed information. The authors themselves may even be unable to replicate the experiment due to changes in their research infrastructure, lack of documentation, or the code being outdated as the project evolves (Stodden et al., 2018).
Significant examples of these repositories, reproducibility initiatives, technological infrastructures, and open source communities include, GitHub, Bitbucket, GitLab, Zenodo, the Open Science Framework,3 OpenAIRE (Open Access Infrastructure for Research in Europe), COAR (Confederation of Open Access Repositories), the French open document repository HAL, EOSC (European Open Science Cloud), HuggingFace, the Harvard DLhub,4 and Dataverse,5 among many others. Certain studies have proposed concrete solutions to the problem of sufficient guarantee of reproducibility, such as reproducibility and scientific software transparency initiatives (Haim et al., 2023a,b; Stodden, 2020).
The above examples show how the data generally seek to comply with FAIR principles for Research Software (FAIR4RS) (Barker et al., 2022),6 which is briefly defined as findable, accessible, interoperable, and reusable. An open-source code may end up being non-reproducible without proper access to the data. Findable: Metadata are assigned a globally unique and persistent identifier, for example, the minimal viable identifiers (minids) or software Heritage SWHIDs. Accessible: The metadata are retrievable by its identifier using a standardized communication protocol; Interoperable: Metadata use a formal, accessible, shared, and widely applicable specification for knowledge representation; Reusable: Metadata are described in detail with a plurality of precise and relevant attributes.
Applying the FAIR principles to data, specifically their R (Reuse) component, allows for the promotion of the reproducibility of scientific publications. These principles aim to categorize the data more extensively and systematically (Parland-von Essen et al., 2018), as a means to improve research data services. They also promote a convenient tripartite categorization of research data artifacts. Although many data science projects and research laboratories have adopted the FAIR principles, each research study represents a particular case. Therefore, the challenge of complying with the FAIR principles is usually only partially achieved (Albertoni et al., 2023). Furthermore, there are discrepancies in how to implement them, from considering how to handle big data and using cloud-native repositories (Abernathey et al., 2021) to smaller scale data science projects requiring affordable sharing (Vanschoren et al., 2014). Because each team and laboratory establish their own means of complying with the guiding principles, determining, and to a certain extent, auditing the degree of FAIR compliance is particularly challenging.
3.2 Open data formats and benchmarking
There are cases where it is not viable to publish datasets and codes because they contain sensitive data (e.g., medical data corresponding to individuals) or industrial secrets (Rosenblatt et al., 2023) (e.g., patents). In these cases, the assessment of the reproducibility of the methods is compromised. Therefore, the concept of federated learning (Karargyris et al., 2023; Baracaldo et al., 2022) is being developed as a novel paradigm, which is based on decentralized and private data for the shared training of models.
However, these cases are exceptional and scarce, and, in general, it is possible to approach open science by using open datasets, standardized formats, baselines, and benchmarks (Vitek and Kalibera, 2011), allowing the scientific community to check the results of published methods reliably.
Even when data cannot be made available for confidentiality reasons, benchmarking and comparing results without accessing the data can be relied on. Several tools have been recently developed for this purpose (Vitek and Kalibera, 2011), including DataPerf, Mlperf, Collective Mind (Fursin et al., 2014), ReQuEST (Fursin, 2018), or MLCommons7 with MLcube8 among others. Such tools attempt to determine the state of the art in specific disciplines by comparing the performances of various scores (e.g., precision and recall). Competitions such as Kaggle9 or BRATS (brain tumor segmentation) (Kazerooni et al., 2023) challenge participants to make predictions using published open datasets and have become a reference for the industry to evaluate and compare models.
3.3 System architecture
Two major trends can be identified in architecture for AI/ML systems: the deployment of microservices and the use of serverless functions.
Microservices allow the building of scalable and flexible software systems for which each component works independently and can be reused in different contexts. Because many applications of AI/ML require large resources for computations and storage, they are usually deployed as a distributed system. Compared to monolithic architectures (Fritzsch et al., 2023), microservices allow different modules to work autonomously and subsequently contribute to the reproducibility and understanding of the system.
In particular, microservices can help reproduce scientific experiments and improve the portability and reusability of the software. By being divided into isolated components of an experiment into microservices, the flexibility and modularity of the software can be increased, making it easier to adapt the code for new tests or experiments and lessening the dependency on software specific to a development environment.
Serverless computing is a popular cloud-based computing model (Jonas et al., 2019) that is similarly related to functionality and dependency isolation. Here, the cloud provider manages the server infrastructure and platform resources, allowing developers to focus on application logic. Depending on the provider, the functionalities can also be referred to as lambda-functions.10 Using these serverless functions is beneficial for reproducibility in computer science, as it reduces the complexity and variability of the underlying infrastructure and enables greater modularity and automation when developing applications and services.
3.4 Tools and platforms
This section surveys tools and platforms commonly used in AI/ML applications and how they contribute to reproducibility. Specifically, we focus on containers, cloud computing, and the Infrastructure as Code (IaC) technique.
3.4.1 Notebooks
In data science, notebooks have been popularized because they allow the incorporation of executable code, rich visualization, and documentation in the same document. It has recently become a common practice to publish and share work with notebooks, providing a step forward for reproducibility. However, it has been shown (Pimentel et al., 2019) that this approach has some deficiencies, such as needing more version control. Very recent studies (Samuel and Mietchen, 2023) have also studied the low degree of reproducibility of Jupyter notebooks in biomedical publications.
Several solutions have been proposed to address these challenges (Pimentel et al., 2021), including the use of Python scripts and the adoption of best practices for documentation, version control, and additional packages. For instance, the ReproduceMeGit tool analyzes the reproducibility of ML pipelines in Notebooks (Samuel and König-Ries, 2021a) and Osiris (Wang et al., 2020).
3.4.2 Containers and cloud computing
Advances in cloud computing and containerization have undoubtedly contributed to the reproducibility of large distributed systems.
These systems are complex and have several interacting components (Wolke et al., 2016; Congo, 2015) along a pipeline. Control over the execution environment is required to reproduce the experiments and even trust them. Given a code, the associated data, and the execution pipeline, we should be able to obtain the same results repeatedly. To achieve the same results, the pipelines, dependencies of the software, and the environment need to be perfectly defined. Virtual machines and lightweight containers such as Docker help define and fix the execution environment (Howe, 2012). We could summarize these two concepts as follows:
• Virtualization = data + code + environment.
• Cloudcomputing = data + code + environment + resources + services.
We address the topics on lightweight containers such as Docker, the MLOps methodology, the management of scientific workflows, and techniques such as IaC below.
3.4.3 Docker containers
Since their appearance in 2007, Docker containers have quickly become popular in computer systems as a fundamental reproducibility tool. Its lightweight nature allows several containers to be dedicated to small microservices on the same machine, with limited consumption and sharing of resources. This feature is a significant advantage concerning complete virtual machines such as VMWare or Hyper-V. The light containers eventually allow for better reuse, and many infrastructures are migrating to containers, e.g., RE3 (Bahaidarah et al., 2021).
Docker is one of the most efficient and widely used tools today, with applications for reproducibility. However, it is still limited (Canon, 2020) in certain aspects of reproducibility compared to container alternatives such as singularity containers for HPC.
The emergence of containerization technologies such as Docker and orchestrators such as Kubernetes (Orzechowski et al., 2020) has allowed the rapid development and automation (Bahaidarah et al., 2021; Vasyukov and Petrov, 2018) of experiment pipelines, thus making the reproduction of complex computationally intensive experiments possible. Therefore, these experiments can be divided into different functional blocks that can be easily integrated, as shown in Figure 3B.
For example, Repo2Docker (Forde et al., 2018) can, with Binder, fetch a notebook for a given repository, create a proper execution environment, and run it inside a container. This action makes the experiment publicly available for anyone to reproduce the results. Specifically for HPC, there are initiatives such as The Extreme-Scale Scientific Software Stack (E4S), a community effort to provide open-source software packages for developing, deploying, and running scientific applications on high-performance computing (HPC) platforms. As an essential contribution to the reproducibility of such a complex, E4S builds from source code and provides containers of a comprehensive collection of HPC software packages.
3.5 Reproducibility of workflows/pipelines and scientific experiments
Many scientific experiments comprise pipelines that concatenate several processes (Sugimura and Hartl, 2018). In terms of reproducibility over time, highly specialized platforms have been developed to manage these complex workflow management systems (Steidl et al., 2023) [e.g., watchdog (Kluge et al., 2020)], tools (Samuel et al., 2021), roadmaps (Da Silva et al., 2021), and general frameworks are proposed (Melchor et al., 2022). They allow researchers to focus on solving their specific scientific problems, rather than the underlying infrastructure, networking, or other technical characteristics (Françoise et al., 2021). Despite the significant step forward, many interoperability and reproducibility difficulties persist (Prabhu and Fox, 2020; Ghoshal et al., 2020). Because there are myriad possibilities of languages, open and private infrastructures that are currently available or under development in the ecosystem of AI/ML data science technologies need to be considered.
3.5.1 Workflow management systems
Scientific workflow management systems (Meng and Thain, 2017) help manage complex, cloud-distributed workflows (Rosendo et al., 2023), and automate repetitive processes (Cohen-Boulakia et al., 2017). They also enable detailed documentation and workflow sharing with other researchers, thus helping improve the reproducibility of results and speeding up scientific research (Plale et al., 2021).
Direct acyclic graphs (DAG) represent workflows in software computer design (Santana-Perez and Pérez-Hernández, 2015). In these graphs, a task starts in a particular node to be processed and then transferred to the next one in the chain until the final result is available in the last node. As pointed out in Section 2 and described in Figure 3B, the pipeline of the workflows and the node themselves need to follow well-established reproducibility principles to obtain reliable results, including access to the source code running the computations, as well as an accurate description of the environment, the use of FAIR data, and the use of open data formats for interoperability, among others.
Each scientific community has developed its own workflow managers. Some well-known workflow managers include Taverna (Hull et al., 2006) (bioinformatics, cheminformatics, and ever social sciences), the Galaxy project (The Galaxy Community et al., 2024) (bioinformatics), OpenAlea (Pradal et al., 2019) (Botanics), Chimera11 (cheminformatics), or Pegasus (Deelman et al., 2004) (physics and bioinformatics), Knime12 (semantic workflow), Makeflow (Albrecht et al., 2012) (data-intensive workflow). Pegasus was the workflow management system used by LIGO for the first detection of gravitational waves and became very popular in the physics community.
The criteria to establish the reproducibility of a given pipeline can vary significantly between different communities. Although the basic principles between different workflows remain the same (see Section 3), their specificities depend on the field. Cohen-Boulakia et al. (2017) address this point in their study, which analyzed three cases of the use of in silico experiments in the domain of biological sciences with Taverna, Galaxy, OpenAlea, VisTrails, and Nextflow, proposing different criteria and discussing these reproducible environments based on Docker, Vagrant, Conda, and ReproZip.
3.5.2 MLOps and reproducibility
The significant increase in articles on AI/ML inevitably forces workflows to adapt toward novel requirements in the management of both data and software(code) because both are a source and contribution to knowledge in these articles. Therefore, analyzing tools, infrastructures, and technologies must evolve to support constraints linked to these management requirements. In this sense, SciOps (Johnson, 2024), AIOps/MLops have evolved from the DevOps/DevSecOps (Development—Operations) concept to cover several reproducibility management infrastructures for computer-based scientific articles.
Transferring knowledge and prototypes from academic to industrial environments is often challenging (Breck et al., 2017). There are very specific methodologies for software development in the industry, such as DevOps, which includes continuous integration/continuous delivery (CI/CD). However, in the academic environment, these practices are only sometimes followed, explained in part by authors' need for more career rewards (as discussed in Section 6.2.2).
MLOps (Gift and Deza, 2021) can be considered the natural evolution of the DevOps best practices components adapted to the particular needs of ML-based software development (Amershi et al., 2019). In general terms, within data science projects, MLOps tries to harmonize the practices of two environments with very different characteristics, such as academic/research environments with ML production environments for a final client, where reproducibility plays a crucial role. MLops harmonization is an end-to-end process from the research model to the last model, exploited by the end customer or reproducibility reviewer. Few studies deal with MLops from the point of view of reproducibility. Among these, Gundersen et al. (2022) does an excellent analysis of the reproducibility of various MLops tools.
Other studies have proposed benchmarks for different MLops features (Schlegel and Sattler, 2022), as both open and proprietary software (Preprint, 2021). These tools are equally important when a journal requires data and software management. The most relevant tools from our review of the literature include the following: Neptune,13 Comet,14 Weights&Biases,15 Sacred + Omniboard,16 Polyaxon,17 ClearML,18 Pachyderm,19 MLflow, Tensorboard,20 and Collective Knowledge (Fursin, 2020a). Table 7 in the Appendix benchmark different MLops tools for more detail on its features.
With the emerging Internet of Things (IoT) technology and the advances in smaller devices with significant computing power, simplified ML models at the edge are possible with TinyMLOps (Ray, 2022). Significant reproducibility challenges appear considering the substantial energy consumption restrictions, limited computing capacity, and heterogeneity between different devices and technologies. In addition, it is no longer possible to containerize and virtualize with Docker.
3.5.3 Workflow languages
Despite the efforts to unify existing workflows, each community has kept its own particularities, including the language to define the pipelines (Cohen-Boulakia et al., 2017). This fragmentation (Adams et al., 2020) makes it harder for integration and interoperability between different academic groups. Indeed, some of the groups use a very particular language for their workflows.
There are initiatives such as SHIWA (SHaring Interoperable Workflows for Large-Scale Scientific Simulations on Available DCIs) (Korkhov et al., 2012) that try to provide a solution to this problem of interoperability. Multiple organizations and providers of workflow systems have also jointly worked to propose the Common Workflow Language (CWL) (Crusoe et al., 2022; Demchenko et al., 2023) to standardize the pipelines around a common language.
Those specifications propose a conceptual workflow language to describe high-level scientific tasks, aiming to promote workflow specification portability and reusability and address the heterogeneity of workflow languages.
3.5.4 Infrastructure as code (IaC)
Much attention is focused on source code and containerization to address reproducibility, but unfortunately, only a little attention is paid to hardware (Bowman, 2023). With the rise of cloud computing technologies, the possibility of replicating the exact execution environment for an experiment is viable. Indeed, for reproducibility purposes, it is required to define the characteristics of hardware, such as the type of CPU, TPU, GPU, memory amount, or network architecture. This is especially important for an extensive distributed system such as HPC applications.
In this respect, IaC provides several advantages toward reproducibility in computer science. One of the main benefits is that IaC allows researchers to define and control their infrastructure accurately in a format that can be easily stored, versioned, and shared, making it easy to reproduce experiments and obtain the same results at each execution. Defining infrastructure as code discharges from manually configuring infrastructure resources allows researchers to easily version and share the infrastructure configuration with colleagues. According to the Octave 2022 report,21 the Hashicorp Configuration Language (HCL) programming Terraform languages were widely used by developers in 2022, indicating that IaC practices are becoming quite popular for GitHub projects.
Additionally, IaC can improve consistency and accuracy by ensuring that all infrastructure instances are created and configured identically. This helps ensure that the test conditions are the same each time an experiment is performed. IaC in the academic environment can significantly help in many aspects, such as the quality of software developed, and is a step forward in the reproducibility of scientific research. As a recent example, Daniel Adorno Gomes and Serodio (2019) managed to define a complete experiment with IaC from a unique high-level code with Pulumi (pulumi.com, 2019).
3.5.5 Provenance and metadata traceability of artifacts
Provenance refers to how the origin (Silva Junior et al., 2021) of the artifacts of an experiment is documented in metadata. Provenance documentation is a commonly used technique to improve the reproducibility of scientific workflows and research artifacts. There are numerous articles proposing tools such as ProvStore (Huynh and Moreau, 2015), ReproZip (Chirigati et al., 2013), MERIT (Wonsil et al., 2023b), CAESAR (Samuel and König-Ries, 2022a), HERMES (Druskat et al., 2022), Provbook (Samuel et al., 2021) in several different disciplines and research areas (Samuel and König-Ries, 2022b), demonstrating how it can help improve traceability, linage and transparency of results.
The PROV standards allow the task to be carried out (see Openprovenance,22 for example. However, it needs to be completed and can be generalized to multiple cases and languages. The preceding requires the use of permanent, Unique Identifiers and tools that manage this aspect to have correct traceability of data sources and artifacts, even using new technologies such as blockchain (Wittek et al., 2021) InterPlanetary File System (IPFS) (Kawamoto and Kobayashi, 2020) to achieve traceability and lineage of software or code snippets.
3.5.6 Reproducibility as a Service (RaaS)
The Reproducibility as a Service (RaaS) concept was proposed in 2021 by Wolsin (Wonsil et al., 2023a). A strategy based on RaaS takes advantage of the availability of cloud computing technology to offer reproducibility services. This strategy includes the reproduction and research artifacts after the execution of software in the controlled environment and its evaluation, validation, and certification (related to this, see Section 3.9 about code review). In addition, granting reproducibility badges, tracking software provenance, or assigning persistent identifiers to software at different granularity levels. Another responsibility of RaaS is to manage the underlying architecture in a way that makes it easier for authors to share and execute their code depending on the chosen complexity, from bare-metal infrastructure to fully managed services. Figure 4 shows how a RaaS architecture is organized in a complex system.
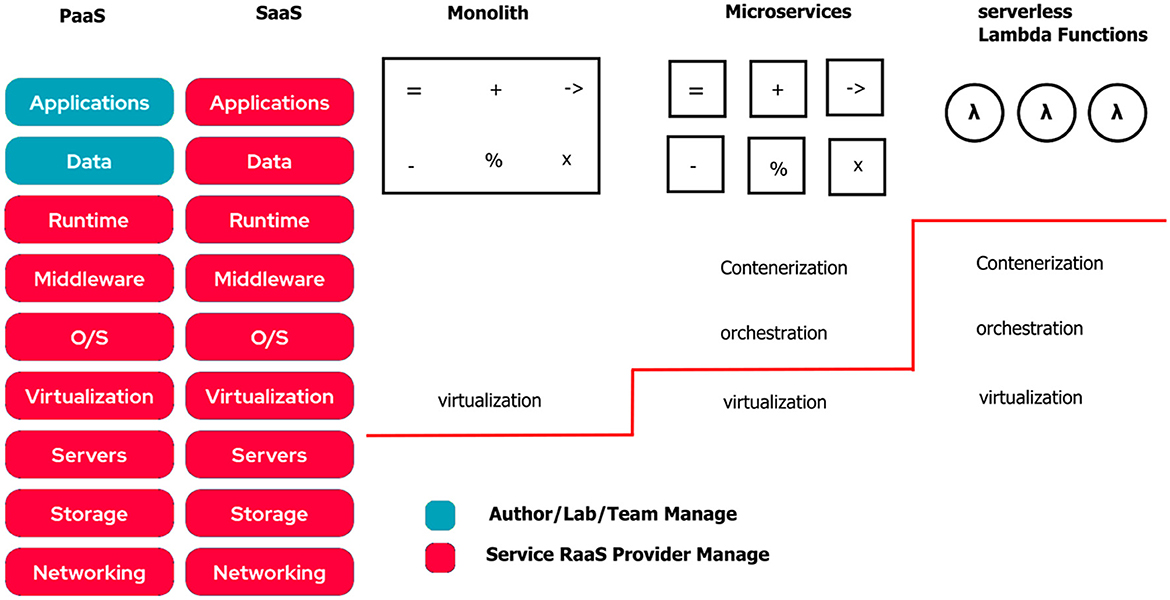
Figure 4. Representation of a RaaS-managed cloud infrastructure. The description layers, microservices, the serverless approach, and taking care of the granularity of software help the reproducibility of a complex system.
Crick et al. (2015) to make a first approach to offering reproducibility services for journals/conferences from an empirical and quantitative point of view. They presented a cyber-infrastructure and the associated workflow for a reproducibility service as a high-level technical specification without delving into technical details. On the other hand, the study by Demchenko et al. (2023) addresses the topic of provisioning on demand in research environments. It introduces the concept of Platform Research Infrastructure as a Service (PRIaaS) to ensure data quality and support effective data sharing.
For example, the IPOL journal (Colom et al., 2015) also partially meets the attributes of what can be considered as a RaaS tool, together with the article, makes available a technological platform for the creation and execution of online demos (simplified demonstrations of algorithms).
Equally, among other existing reference platforms, we could mention CodeOcean, Chameleon, Replicate,23 TXYZ.AI,24 and Whole Tale. They allow code to be executed in a wide range of languages but are still maintained at the demo level with certain technical restrictions to offer the mentioned features. They start to be actively taken into account by publishers.
3.6 Best practices and data management methodologies
Agility and security are among the many quality attributes of software (Milewicz and Mundt, 2023), even though most of them are not specifically designed for reproducibility in computer science. However, data project management methodologies and well-known best practice guides are applied widely across the AI/ML industry to improve reproducibility.
Several studies (Schelter et al., 2015) and best practices guides (Stodden and Miguez, 2014) have proposed different tools for the management of data science project artifacts (Schlegel and Sattler, 2022) as well as methodologies. For example, Goodman et al. propose 10 simple rules to achieve reproducibility. The Turing Way handbook also provides a relevant compilation of good practices (The Turing Way Community, 2022) for reproducible, ethical, and collaborative data science projects.
There is a consensus that one of the main factors limiting the success of data science projects is the need for reproducibility in the management platforms (Baillieul et al., 2018; AlNoamany and Borghi, 2018; Martinez et al., 2021). Of the many methodologies available, the most popular are CRISP-DM (Schröer et al., 2021), KDD, SEMMA, Microsoft TDSP, Agile DS Lifecycle, Domino, DS Lifecycle, IBM FMDS, RAMSYS, and MIDST, among others. These are widely used in the industry, especially CRISP-DM.
Indeed, as observed in the scarce literature, there needs to be a standard or unified methodology focused on reproducibility for data management. So far, only good practices, recommendations (Turkyilmaz-van der Velden et al., 2020; Merz et al., 2020; Samuel and König-Ries, 2021b; Nichols et al., 2021), and guides from different fields of computer science for different needs are available.
3.7 Scientific publishers and reproducible research
Historically, one of the primary forms of communication, recognition, socialization, and validation before the scientific community are the articles published in journals and conferences (Stodden et al., 2018). Publishers de facto become auditors of the scientific activity, and indeed, the metrics (impact factor and others); they have established are the typical indicators used to evaluate researchers in their career and their advancement. Publishers have, therefore, a responsibility to assure the scientific integrity of the work they make public, along with their own interest in maintaining their own reputation. This responsibility includes not only avoiding fraud but also establishing clear quality criteria. In scientific publications, reproducibility is fundamental as it allows others to verify if the same or equivalent results are obtained when repeating the experiment, thus allowing them to refuse a paper containing wrong or inaccurate claims. As pointed out by Heesen (2017), the work that is not widely shared is not really scientific work.
3.8 Publications with code
Associating source code with a particular publication is gaining popularity in the scientific and technical community (Bonsignorio, 2017). It allows for greater transparency and reproducibility, which is essential to guarantee the quality and reliability of the results (De Sterck et al., 2023). However, the reproducibility aspects of this practice are evaluated in the Dataverse repository (Trisovic et al., 2020).
Many conferences have started to request that the source code be given and made public. Others go one step further and perform an exhaustive evaluation of the artifacts. For example, Checklist NeuroIPS (Pineau et al., 2020) is a widely recognized checklist for the reproducibility assessment of conference papers.
From many examples, we might include Code Ocean, used by IEEE's publishers after the integration of the CodeOcean's platform as a computational research platform, Whole Tale (Chard et al., 2020) allowing researchers to create and share scientific narratives that include data, code, and runtime environments (Brinckman et al., 2019; Chard et al., 2019), Binder as a platform that allows users to create and share code execution environments online, making it easy to reproduce and distribute results, PapersWithCode with open resources on ML, ReproducedPapers with open teaching and structuring machine learning reproducibility (Yildiz et al., 2021), or the ReScience Journal (Rougier and Hinsen, 2019) which replicates computations from independent open-source implementations of original research and the advanced Chameleon25 large-scale edge to cloud tool (Keahey et al., 2019), CatalyzeX,26 ScienceCast,27 DagsHub,28 and CentML.29
Unfortunately, in many cases, this is limited to providing a non-persistent link (Salsabil et al., 2022; Idrissou et al., 2022) to the source code repository in public platforms (see Section 3.1). Moreover, each journal sets its own strict criteria, formats, and procedures for authors. Aspects such as consistency, reproducibility, and reusability cannot be appropriately tracked or audited by other teams and research over time, thus limiting their impact (Raff and Farris, 2022).
3.9 Review of research artifacts
First, it must be understood that for different reasons (Gomes et al., 2022) an article is not 100% reproducible, but relatively certain elements (e.g., computational artifacts, pseudocode, algorithms, and demos) that the author decides to share and considers sufficient grounds to legitimize their results.
The evaluation criteria for accepting articles for publication are traditionally well-defined for scientific journals. They are typically based on originality, novelty, or overall scientific interest. However, when considering a publication as the article and all major research artifacts, including source code, the criteria are relaxed, if considered at all. When the evaluation considers the associated source code, it is required to establish the proper evaluation criteria for peer review (Supporting computational reproducibility through code review, 2021).
Conferences have started to publish guides containing checklists for evaluating artifacts and to grant the so-called reproducibility badges (Frery et al., 2020; Athanassoulis et al., 2022) if the conditions are met. Among the most important conferences, we can cite the checklist of NeurIPS 2019,30 the ACM reproducibility badges,31 and other initiatives such as the Unified Artifact Appendix and the Reproducibility Checklist,32 the CTuning artifact evaluate,33 or the Empirical Evaluation Guidelines SIGPLAN NISO RP-31-2021,34 among others.
Following several of the published guides, recently, the SC23 supercomputing conference (one of the most important conferences in HPC) (Plale et al., 2021) adopted the Reproducibility Initiative, where accepted papers with available artifacts were acknowledged with the corresponding ACM badges. The use of blockchain technology for artifact traceability has also been proposed (Radha et al., 2021; Kawamoto and Kobayashi, 2020). CTuning has participated in the artifact evaluation task for different ACM conferences (Fursin, 2020c) and has defined a more detailed Unified Artifact Appendix and the Reproducibility Checklist based on the previous evaluation experience in ACM ASPLOS, MLSys, MICRO, and SCC'23 conferences.
Other specialized scientific journals have already implemented specific criteria to a greater or lesser degree. For example, checklist for Artifacts Description/Artifacts Evaluation (AD/AE) reproducibility (Fostiropoulos et al., 2023; Malik, 2020) for data science experiments and projects of different publishers.
Eventually reproducibility-certifying agencies have started to offer their evaluation as a service in different disciplines working with sensitive or confidential data, outsourcing this function as a trusted third party. Recently, Cascad (Pérignon et al., 2019) has been proposed in the field of economics and management (Radha et al., 2021).
From our review of the data above, we observe that the existing criteria are still quite varied, not standardized, complex for the authors to fulfill, and time-consuming on the reviewer's side. Table 3 extensively summarizes the reproducibility strategies and technologies reviewed in this work. However, how they are implemented according to the reproducibility policies of the different scientific journals needs to be analyzed. From an empirical point of view, our survey provides insights on applying these strategies directly from participating journals, and Table 4 summarizes different evaluation/description (AD/AE) guides.
4 Survey on policies of computer science journals
4.1 Related work
The existing literature concludes that there is still incipient and timid progress toward implementing sharing and open science policies in scientific works (Stodden et al., 2013, 2018). The traditional peer review scheme is maintained, with slight variations, and it is, in general, limited to encouraging the publication of the source code and data in software repositories (Lewis, 2023; Stodden et al., 2012).
For example, The Diamond OA Journals Study (Bosman et al., 2021) makes a general survey; in our case, the results of question 41 are highlighted. To the question “Do you have any policy or practice to stimulate open sharing of research data?” 42% of the respondents declared to have a policy or practice to stimulate open sharing of research data. In the same survey, Question 54 asked, “Does the journal require linking to data, code, and other research results?” and although there is not much information available from journals about requiring links to data, code, and other research outputs in DOAJ, from the survey data, the study found that nearly half of respondents reported not requiring this, against 24.8% who do. For more than 25%, the answer was “No” or “Unknown.”
The above questions are undoubtedly limited to code-sharing policies in journals but do not delve into actual reproducibility policies through article automation, evaluation, and preservation of reproducibility technologies. This inconsistency represents a dilemma that is discussed in Section 6.3.
In the article Vasilevsky et al. (2017), the authors reviewed 318 biomedical journals manually to analyze the journals' data-sharing requirements and characteristics. 11.9% of journals analyzed explicitly stated that data sharing was required as a condition of publication. 9.1% of journals required data sharing but did not state that it would affect publication decisions. 23.3% of journals had a statement encouraging authors to share their data but did not require it.
Another contribution by Konkol et al. (2020) from the point of view of the analysis of reproducibility technologies for publishing computational research concludes that still, publishing reproducible articles is a demanding task and not achieved simply by providing access to code scripts and data files. Several platforms were analyzed, including Whole Tale, ReproZip, REANA, o2r, Manuscripts, Gigantum, Galaxy, eLife RDS, Code Ocean, Binder, its limitations, and the facilities it offers for authors. The previous article is complemented by the study by Willis and Stodden (2020). In the study by Malik (2020), the technical difficulties are discussed, and the benefits of implementing Artifact Description and evaluation policies for presenting scientific articles to journals and conferences.
We observe that the percentage of implementation of concrete reproducibility policies is still low at this moment.
4.2 Questions and answers
In our case, this study analyzes the problem from the point of view of practical implementation of policies by publishers, based on your opinion and experiences with the following research questions: best way to make reproducibility policy mandatory, or instead an incentive policy for authors and reviewers, to allow publishers improve the quality and impact of their publications? What type of technological infrastructure best supports these types of reproducibility policies?
Question 1. Do you want to be mentioned in the acknowledgment section as a Survey participant? Despite having a policy of sharing and publishing code and data implemented at some level, some publishers 26.7% refrained from being mentioned, probably due to not being able to match several items. Indeed, the survey asked very specific questions about the implementation of infrastructures and technical details. Some publishers requested to be considered anonymous in this question.
Question 2. Respondent's role in the scientific journal The answers came from a variety of different roles, with a predominance of eight editors-in-chief.
Question 3. Do you have a Reproducibility policy or similar in your guidelines for authors? A large majority (75 %) of the respondents indicate that they do have a reproducibility policy. we can observe that there is a significant percentage (41.7%) of journals which request reproducibility as an essential condition for publication and thus make it mandatory. This decision has important consequences and, in general, it is counterproductive for almost all journals to add extra requirements for the publication because it reduced the publication rate.35 On the other hand, it improves the overall quality of the publications.
Question 4. If you wish, you can indicate the link to the policy of the scientific journal or guides for authors. Nine journals provided a link to their reproducibility policy.
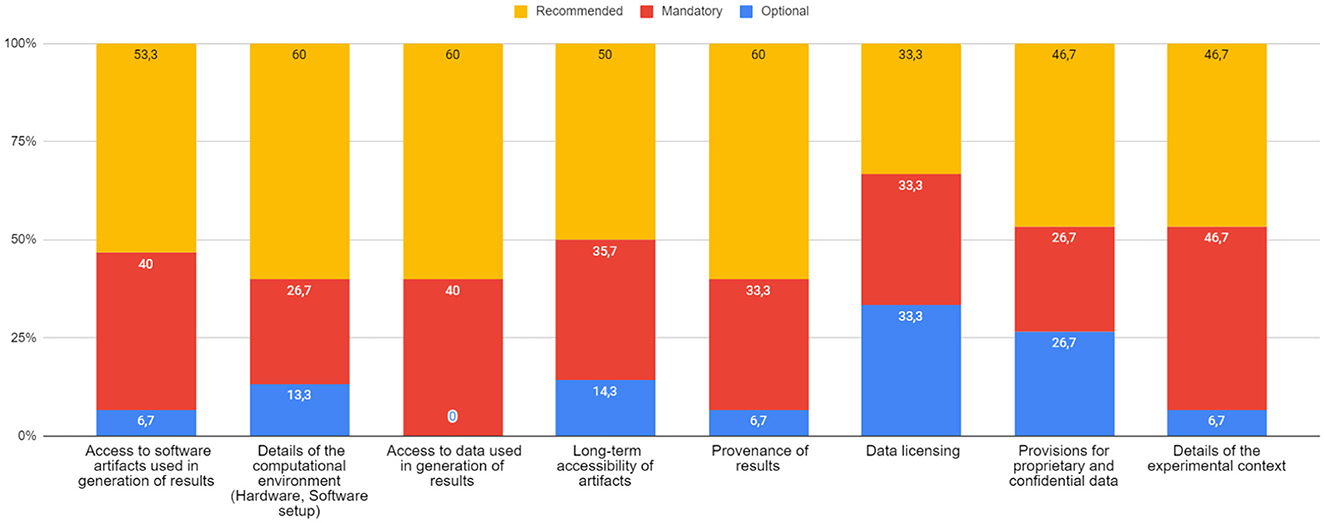
Question 5. How do you think the reproducibility policy requirements should be? In this question we asked about what should be the most significant requirements for a reproducibility policy, regardless whether the journal actually implemented them or not. The results are given in Figure 5, with a variety of different preferences and showing, in any case, gradual interest toward making them mandatory.
Question 6. Do you follow any guide or checklist for the evaluation of research artifacts? If so, which one? The responses were very varied, which shows the lack of standardization in this matter, or don't have 36.8%. The problem of the evaluation of the research artifacts has been extensively studied, yet without much agreement or formalization.
Question 7. Journal access modality Most of the journals answered that their publication modality was open-access 56.3%.
Question 8. What is the range of your APC (Article Publication Charges)? The APC are very relevant for the discussion about how the reproducibility costs are shared between authors, publishers, and technology providers. Free publication costs predominate in the responses 50%. Between $50 and $500, 12.5%; between $500 and $1,000, 18.8%; between $2,000 and $3,000, 18.8%. In addition to question #7, it is an indicator that the business model of these journals is based on open platforms and repositories.
Question 9. Preferred sharing method This question confirms that free open platforms are used to share the code, and the number of journals that owe third parties or have their own technological storage infrastructure is very low: 18.8%, public online repository, 25%, journal hosted own infrastructure, and 31.3% multiple methods equally recommended.
Question 10. How compliant is your publication of software and data policy with FAIR-TLC? The answers indicate the increasing level of implementation of the reproducibility policies, considering that most of the articles are accessible and reusable but still low in the other attributes. This could be explained because the use of open repositories limits the journals to offer the other attributes satisfactorily.
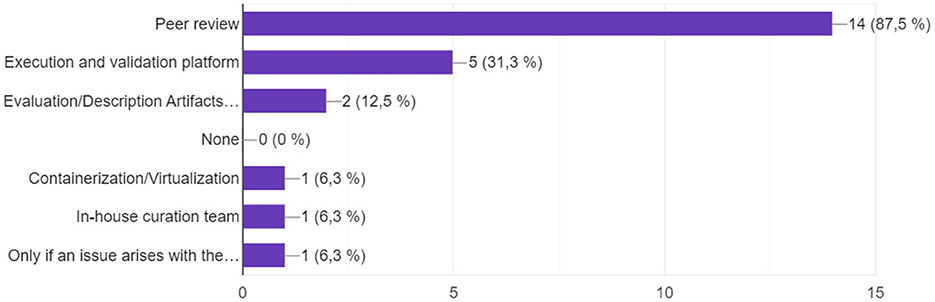
Question 11. Reproducibility validation method
The results (Figure 6) show that the traditional peer review model for article validation and acceptance is maintained, compared to other more automated forms of reproducibility validation. Therefore, validating the legitimacy of an article rests on one or two experts as well as their own available testing resources. Two responses to Evaluation/Description Artifacts checklist and one open response, “Only if an issue arises with the paper.”
Question 12. If you request to share the source code. What platforms or repositories do you recommend for sharing code? If others, you can write those you recommend
The results (Figure 7) describe show that GitHub if the preferred specialized platform, although more for developers than for publishing research results. Zenodo, on the other hand, allows the citation of code and data through its identifiers but remains a simple non peer-reviewed repository. There is therefore still a significant lack of automation in the policies of code and data for reproducibility purposes, to validate the legitimacy and quality of the articles.
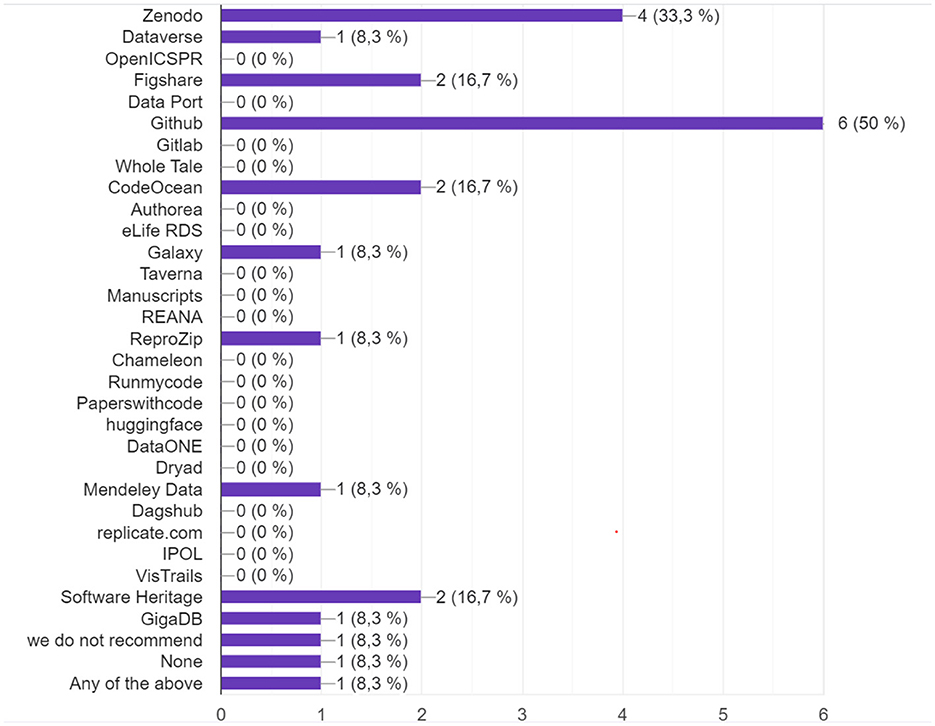
Figure 7. If you request to share source code, what platforms or repositories do you recommend for sharing code?
5 Reproducibility technological discussion
As pointed out by several of the review studies, reproducibility is greatly beneficial for both authors and journals. Such benefits include greater credibility and recognition (Ghimpau, 2019), research results are accurate and reliable, increased visibility and impact (Boulbes et al., 2018), facilitate collaboration and reuse (AlNoamany and Borghi, 2018), and increase credibility and confidence in the results (Samuel and König-Ries, 2021b; Gupta et al., 2022).
In this section, we discuss reproducibility insights from the point of view of technological evolution. These insights are based on the strategies, which are presented as a reproducibility fundamental lever and support. We analyzed how these interrelate with different challenges, problems, and solutions that have been proposed in other studies, as well as how they relate to the above benefits. In particular, we discuss the issue of the responsibility of authors and publishers, including their efforts toward reproducibility, the possibility of understanding reproducibility as a service, and the impact of considering software as an essential research artifact and the reward to researchers.
5.1 Dilemma: virtualization solution or dependency
As described in the technological evolution Section 3.4, many of the reproducibility tools and platforms (e.g., Workflows) proposed so far are entirely based on container technology.
Docker, or more generally lightweight virtualization, is considered the holy grail of reproducibility. As seen in Section 3.4, many solutions to computer reproducibility are based on this technology. Following the popularization of agile methodologies demonstrated by the landscape and the containerization strategy, many reproducibility problems have been partially solved with docker (Moreau et al., 2023; Canon, 2020). However, many signs suggest that the practice of lightweight virtualization is being abused. As noted earlier, Docker is practical, light, and facilitates many processes previously tedious; however, it is not a tool specifically designed for reproducibility and, therefore, cannot be used indiscriminately to hide bad practices.
The possibility of packaging, freezing, and porting a code to any infrastructure and maintaining stable functionality over time make it attractive in the scientific world; however, as stated in Fursin (2020b), this indiscriminate use brings great inconveniences.
At this point, it is necessary to analyze the problem of reproducibility, repeatability, containers, and development in depth. The problem is that two characteristics are desirable in systems, but actually, they are antagonists. On the one hand, we want robust systems that will not break after an update. The classic example is a Python program that uses PyPI packages that, even if the user sets the versions in a virtual environment, the libraries may not be available in a particular version of Python. In that case, many system designers opt for virtualization.
The containers ensure reproducibility, given that the complete environment is fixed. However, proper attention is not paid to the maintenance of the container. In that case, it might face security problems because if the environment is fixed and not updated, the libraries will stop receiving bug fixes and security updates.
Docker is undoubtedly a useful tool that allows fixing the execution environment, but maintenance is still required. Regular automatic testing is recommended.
5.2 The shared responsibility between authors and publishers
As discussed in Sections 3.4 and 3.7, complying with the criteria for reproducibility implies some costs (e.g., working time, economic infrastructure resources). It also implies a mutualized shared responsibility between authors and scientific journals. Share responsibility also needs a commitment to transparency and reproducibility (Haibe-Kains et al., 2020). Despite the analysis of the reproducibility stakeholders in Diaba-Nuhoho and Amponsah-Offeh (2021), Feger and Woźniak (2022), and Macleod and the University of Edinburgh Research Strategy Group (2022), the roles and relationship between authors and publishers still need to be clarified: indeed, much of the reproducibility burden relies on the authors.
Authors are responsible for providing detailed information about the methods and techniques used in their research and making public the data and codes used to generate the results. They must also ensure that their results are replicable, and thus, they can be verified by peers. On the other hand, publishers are responsible for establishing clear policies and guidelines (Stodden et al., 2018) for processing the submitted scientific articles adequately and verifying and guaranteeing the results' transparency and reproducibility.
Guaranteeing and legitimizing the reproducibility of scientific work in AI/ML implies assuming significant economic and time costs (Poldrack, 2019) depending on the size and complexity of the research project. These cannot be assumed only by the researcher.
5.2.1 Reproducibility cost
Estimating the cost of reproducibility is not easy because the cost can be assessed from the execution of a simple container on a personal laptop as well as a distributed execution of software in the cloud, with the market costs per hour of CPU, GPUs, and storage depending on each provider and their business model (e.g., GCP, Amazon, Azure, Oracle, and others).
Existing virtualization, containerization techniques, and cloud computing infrastructure are crucial elements of this problem (Howe, 2012). Therefore, the costs related to cloud computing are relevant to reproducibility.
It is essential to highlight these associated costs (Armbrust, 2009) and the implications for the scientific parties that have a role in the reproducibility of the scientific work. One can describe the main technological costs for the reproducibility of computational projects and experiments as follows:
where CR is the total reproducibility cost, CHD the cost of hosting data, CHC the cost of hosting code, and CRC the cost of running code.
The problem of increasingly complex research projects is not specific to computer science but common to other disciplines, especially when they combine different fields. Take, for example, the case of bioinformatics.
Researchers in this field need to have not only knowledge of biology but also the skills to operate software, data formats of research artifacts, and running environments. Specialized expertise is typically required in Python, R (Gandrud, 2020), diverse operating systems, database management, and complex platforms such as Galaxy (Afgan et al., 2018).
IaC, virtualization and containerization, and cloud computing approaches help address this divisional responsibility in a simplified manner. They allow the author's steps to be tracked so that other researchers can repeat and reproduce the experiment in the same environment. However, these approaches still require a high level of computing skills, which should not necessarily be assumed only by the authors. Section 6.2.1 discusses the Reproducibility as a Service (RaaS) strategy and how it can be applied to manage this shared responsibility.
6 Reproducibility efforts of authors and publishers
6.1 Effort of authors
In the case of authors, from the technical and methodological limitations and for several of the reasons discussed, we can deduce that, in most cases, a scientific article cannot be reproduced in its entirety (100%). Authors generally choose to reproduce only parts of the algorithms, demos, or data essential to support the conclusions.
Therefore, authors work to achieve 100% reproducibility in their study or to fully clarify the reasons that prevented reaching this objective, complying with the policies and requirements of journals and conferences.
The improvement of the writing quality in scientific articles and the associated documentation of the software has already been studied (Mack, 2018) and represents an additional effort for the author. In practice, however, these aspects are sometimes overlooked. We can easily identify articles with confusing writing unnecessarily overloaded with complex academic jargon. Such writing is challenging to interpret and, consequently, very hard or impossible to reproduce.
6.1.1 Reproducibility guide for authors when submitting their research
From our analysis of the shared responsibility between authors and journals and the most recent technological advances in computing, we shall discuss the efforts required by each of these two actors.
It is still challenging for journals and authors to close the gap in a mutual effort, and it is even more complex when the authors must comply with article submission guidelines between different journals. An article comprises theoretical and computational parts that can only be reproduced in a certain percentage and specific components that only the author is responsible for defining and specifying with the greatest of details and following a standard guide that avoids reprocessing between publishers.
MICRO2023 is a recent experience toward unified EA (artifact evaluation) guides and procedures,36 which allows speed up the AE process. In a conference where artifacts can be complex and time-consuming to evaluate, 25% of the submitted artifacts were awarded the artifact reusable badge. In this context, practices such as Reviewers performing an initial “smoke test” (for example, installing the artifact or resolving access/environment/setup issues) were developed. In addition, they reviewed the key claims of the paper and the artifact. Similarly, two surveys were conducted consulting authors and evaluators to seek feedback on the AE process. Essential insights are derived from this survey, especially in enabling authors and reviewers to quickly iterate on artifacts efficiently and seamlessly in a reasonable time. For example, Reviewers suggested that requesting authors to prepare a subset of simulations (and/or representative checkpoints) would be a good practice. Results “will appear in the ACM/IEEE MICRO 2023 conference front-matter”37 and support a trend toward improvements to the process and clearer and standardized instructions preferable to most subjective assessment of other experiences.
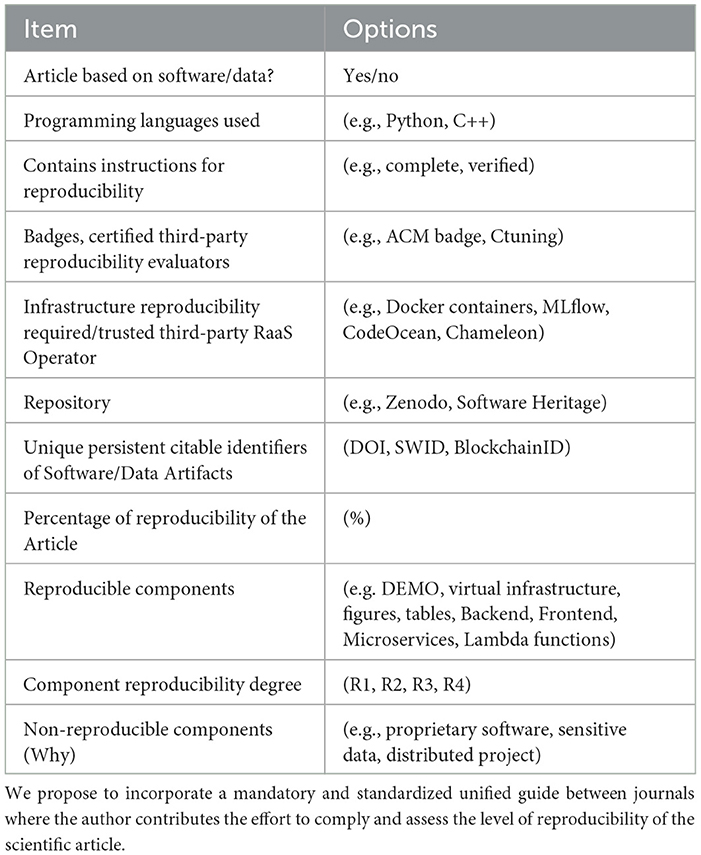
Therefore, in addition to standardizing the different evaluation and description guides of artifacts, we propose to incorporate a mandatory and standardized unified guide between journals where the author contributes the effort to comply and assess the level of reproducibility of their scientific article (see Table 1).
6.2 Efforts of publishers
In the case of publishers, given the comprehensive typology of articles submitted to journals and their reproducibility costs, it is economically unfeasible that they maintain their own reproducibility infrastructure. Because of such costs, journals today tend to rely on third trusted parties. Here, we discuss how the Reproducibility as a Service (RaaS) methodology could help discharge authors from the burden of running code and maintaining a complex reproducibility infrastructure. We also discuss how publishers consider software as more valued research artifacts and thus properly reward authors. Finally, we provide a brief gap analysis from our survey results in Section 4.2.
6.2.1 Reproducibility as a Service
As pointed out in Section 5.2, reaching reproducibility might require a significant technological investment for some projects, which should not be assumed only by the authors but shared with publishers and offered by specialized third parties. One particular strategy, Reproducibility as a Service (RaaS) (Wonsil et al., 2023a), might be helpful for this purpose.
As introduced in Section 3, RaaS is an approach to address non-reproducibility in scientific research by providing access to tools and resources that researchers and industrial actors can use to replicate experiments and data science projects. RaaS also facilitates, manages, and overcomes many of the limitations and barriers that we have identified in our literature review.
According to Brundage et al. (2020), RaaS could be labeled as any third-party service made of tools that allow the reproducibility of scientific work. They propose using the existing cloud computing tools to offer a service that fills the gap between two major requirements to achieve reproducibility. First, actions are taken by researchers who want to facilitate reproducibility. They provide a detailed procedure that allows the ability to obtain the result artifacts and the exact execution environment. Second, actions taken by publishers or the industry validate reproducibility.
Additionally, there are trusted third parties that deal with big data projects and confidentiality issues of sensitive datasets. They aim to reduce the need for the strong computing skills typically required to study in complex AI/ML data science projects. Cloud Native-based RaaS (Wonsil et al., 2023a) adds an additional standardized layer with simplified interfaces for IaC, virtualization, and cloud computing (see Figure 4). Examples of these tools include Invenio,38 Eprints,39 DSpace,40 among others.
Although not particularly adapted to complex workflow systems and user interactivity, the partnership between IEEE and Code Ocean41 is an excellent example of the relationship between a journal and a third party that offers reproducibility services in the cloud. The code from IEEE articles can be browsed, discovered (assigned a DOI), run, modified, and eventually built the researcher's study on the cloud without any complex setup.
6.2.2 Culture of software as a valuable research artifact and reward to authors
Traditionally, the published computer science article has been considered the most important and rewarded (Parsons et al., 2019) research artifact, leaving aside software production. Universities, research centers, and evaluation committees often consider the number of articles published in high-impact factor journals and the number of citations as the major criterion for hiring a researcher, increasing the salary, and career evolution, among other incentives. Consequently, researchers typically do not invest considerable resources in the reproducibility of the results, the quality of the produced software, or even the possibility of publishing the software itself. For example, the study by Gomes et al. (2022) and Baker (2016) focus on the barriers (Anzt et al., 2020) concerning why authors might be reluctant to share code and data in their publications.
Many research projects are based on software contributed by others, including libraries, applications, or complete frameworks, and in many cases, there is no explicit recognition of the authors of the third-party software. The recent incident about the vulnerability in the log4j library (Hiesgen et al., 2022) is an excellent example of a widely spread software used by hundreds of companies, not necessarily acknowledging the library's authors.
Moreover, some of the current incentives produce perverse behaviors in a hyper-competitive environment (Edwards and Roy, 2017), which certainly goes against ethics and scientific transparency. They have also promoted the rise of predatory journals (Cukier et al., 2020); some authors are discouraged from improving the quality of their papers in the so-called publish or perish race.
The lack of incentives for researchers and software developers to produce quality and reproducible software has a clear negative impact (Ke et al., 2023) on the development of Open Science. Fortunately, the criteria for evaluating researchers are evolving in parallel and going in the right direction (Technopolis, 2020).
Relatedly, the reproducibility culture has also been analyzed in previous studies (Karathanasis et al., 2022; Mauerer et al., 2022; Hofman et al., 2020; Fund, 2023; Lin, 2022), as has teaching reproducibility in academic environments to young students and Massive Open Online Courses (MOOC).42,43,44
Different studies (Parsons et al., 2019; Smith et al., 2016) have focused on analyzing the citation of scientific software and data publishers (Cousijn et al., 2018) as a natural need to implement FAIR and paper-with-code strategies. To this purpose, FORCE11 (The Future of Research Communications and e-Scholarship) (Smith et al., 2016) provides guidelines for citation software and data.
Software needs to be properly cited and preserved. Given their dynamic and changing nature, these two requirements are certainly not easy to fulfill. Indeed, millions of software repositories are constantly being updated at every instant in GitHub and other repositories.
The Software Heritage project, supported by UNESCO, is one important step forward in both citation and perpetual preservation of software via proper identifiers, such as the SWHID (Di Cosmo et al., 2022). Zenodo also provides a Digital Object Identifier (DOI) and Chameleon a QR-code to reference the code. It is also a great source of information for determining the provenance of software contributions.
Regarding using badges as an incentive for author reproducibility, it must be observed that they really impact the researcher's reputation in the same way as the popularized and mature badge system awarded in e-learning by important companies, academies, to certify technical skills (Stefaniak and Carey, 2019) published on reputable platforms such as Credly45 and easily shared on Linkedin,46 which allow the candidate to reinforce their CV and demonstrate to the employers in a competitive labor market. Currently, at least 138 computer science journals award reproducibility badges.47
As pointed out by Dozmorov (2018), GitHub is currently the most complete database to measure the impact of software. Interestingly, they concluded that the number of forks as a measure of software impact is not correlated with the number of citations associated with a scientific paper. This finding, at first counter-intuitive, shows that citation indices (such as the h-index and others) must fully explain the true impact of the scientific work and the associated software. The consequence is, therefore, that producing quality software is neither properly promoted nor taken into account for researchers' career advancement.
There is, therefore, a need for metrics that are specific to software, beyond indirect measures such as the number of forks or stars in public repositories. Strategies such as RaaS and more adapted metrics such as the scientific impact factor (SIF) (Lippi and Mattiuzzi, 2017) could be of great help rather than the H-index or impact factor of the journal (FIJ), Clarivative,48 or Altmetric49 indicators.
Our recommendation is depicted in Figure 8. It includes incentives for all the actors, as well as for third parties that implement permanent and long-term reproducibility infrastructures supporting the editorial business.
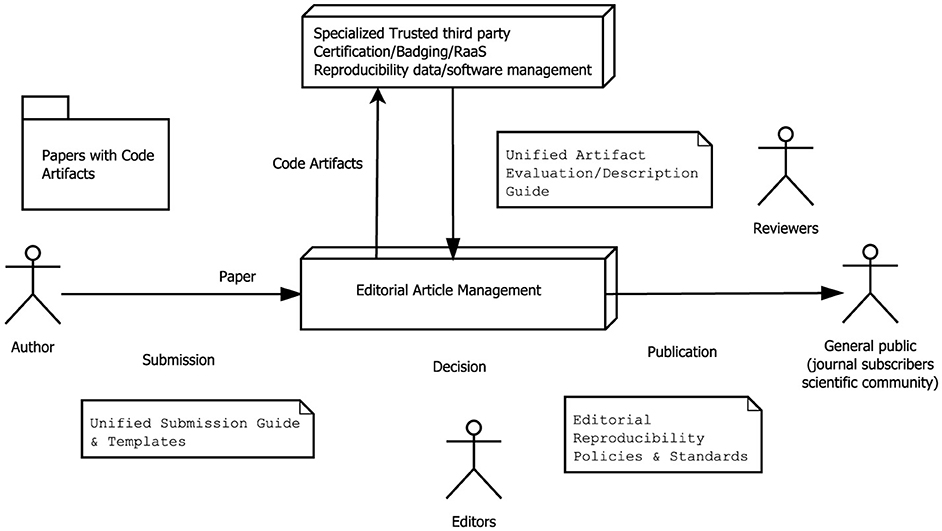
Figure 8. Proposal for a future editorial process on which the article and code are published as a whole, and third parties certify reproducibility.
6.3 Dilemma: reproducibility sharing policies
At this point, an important clarification must be made, journal reproducibility policies should not be confused with traditional open access and open science initiatives. It could even be considered an open topic that requires standardization. In Stodden et al. article (Stodden et al., 2018), the authors made a first approximation to the analysis of data and code of the publications in policy adoption by Journals. Consequently, this work analyzes (Stodden et al., 2013, 2012) the journal policy implementation and effectiveness for computational reproducibility, however a clear concept of “reproducibility policies” is not consolidated. This leads us to consider that journals face an important dilemma in defining their internal policy of just limiting themselves to a code and data sharing policy or going further in defining veritable and strict automation tools and reproducibility evaluation article reproducibility policy. Maintaining a sustainable cost and business model at the same time.
6.4 Brief gap analysis
We provided a small gap analysis of the level of implementation of reproducibility policies that we observed in our survey. We intended to bring together all the elements of analysis. We include technological aspects and the efforts required by both authors and publishers to help close or, at least, reduce the reproducibility gap. Aspects such as the standardization and implementation of reproducibility policies, adaptation of business models, and association with specialized third parties are considered. These recommendations come from analyzing the answers in our survey results (Section 4.2).
Table 2 shows the journal policy evaluation gap in identified key aspects, indicating the survey question that helps evaluate the percentage of implementation of the reproducibility policies. With this table, each journal is evaluated regarding its reproducibility policies and the effort it must make in the key aspects identified.
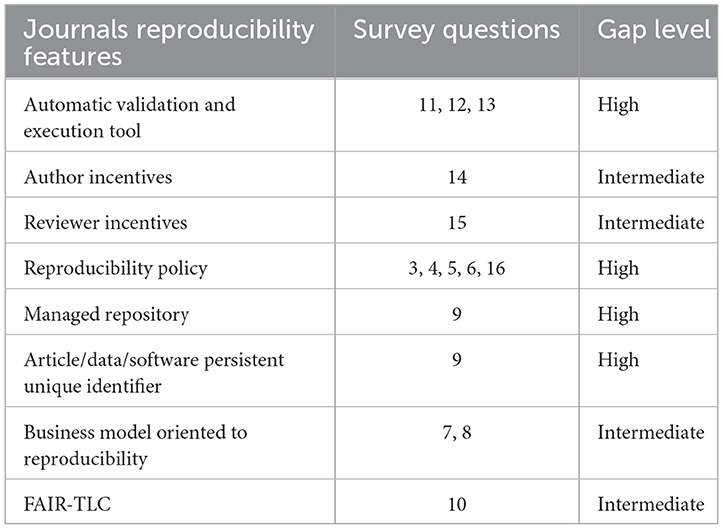
Table 2. The gap in the implementation of the journal policies, along with the related survey's questions.
It can observed from the answers that there is a low percentage of implementation of reproducibility policies, as well as the low use of technological tools for automation, validation, and sustainability of reproducibility in the long term (longevity of reproducibility).
This is explained by the fact that there is still no consensus and standardization on what should be a good reproducibility policy for journals, as well as the lack of a developed and mature market of trusted specialized RaaS services.
To determine the gap, we used the following qualitative ranking:
• High: when there is a complete lack of accomplishment or implementation of the criterion.
• Intermediate: when there is the presence of an initiative with immature development of the criterion.
• Low: when there is a complete and functional implementation of the criterion.
Unfortunately, there is a significant gap in implementing some aspects, mainly those related to automation, establishing reproducibility policies, managing repositories, and using persistent identifiers for the research artifacts, including software. Other aspects, such as orienting the business model toward reproducibility itself or the use of FAIR data seem to be more developed.
Despite the observed gap, there is an opportunity to reduce the reproducibility gap with the common efforts of authors, publishers, and technological providers. See Tables 1–3 for more details.
6.5 Classification of reproducibility criteria
Table 4 shows the classification of reproducibility criteria for artifacts description (AD) and artifacts evaluation (AE) used in the review of publications.
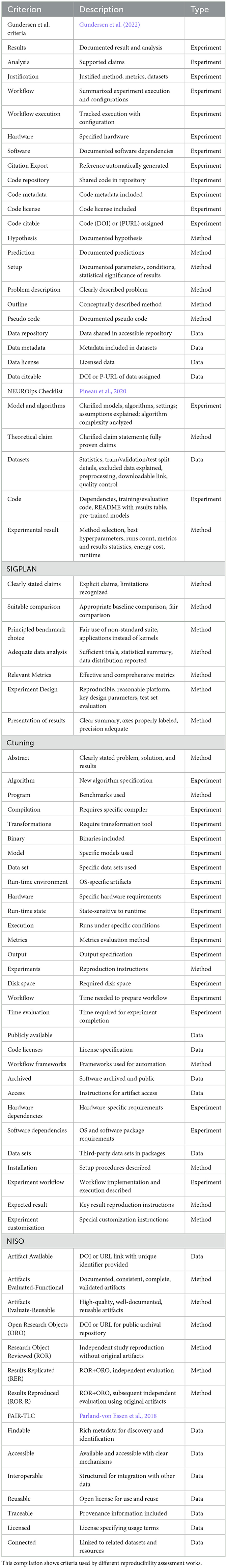
Table 4. Classification of reproducibility criteria for artifact description (AD) and artifact evaluation (AE).
7 Conclusion
As a consequence of the aforementioned credibility crisis, in this study, we have addressed the problem of reproducibility, specifically in computer science, including ML/AI projects, from diverse reproducibility stakeholder points of view. We establish insights from the best practices, frameworks, methodologies, and technologies available at the moment.
In computer science, the variety of languages, new developments, platforms, frameworks, hardware, and architectures on which the code of scientific articles can be run are vast.
We conclude that the high cost of guaranteeing the reproducibility of a software project is not adequately rewarded at this moment to the reproducibility stakeholders. Considering the costs in own infrastructure that would be required, we still need to consider business models that encourage investment by third parties in infrastructure and thus guarantee longevity and perennity in the reproducibility of scientific publications.
It is convenient to define a new metric equivalent of the Impact Factor, which could be used specifically for software. This could help properly reward the effort of software developers by acknowledging them clearly as co-authors of the scientific study and measuring the real impact of their contributions in reproducible computer science projects.
There is a low level of implementation of reproducibility policies in journals. For the moment, the traditional peer review evaluation methodologies are preferred. The responsibility of the validation is mainly on the expertise of the reviewers chosen by the editors and the few functional tests of the artifacts that they can do with their limited testing infrastructure.
We conclude that mutually beneficial relationship should be established between authors, reviewers, and publishers to balance the benefits and costs. Therefore it is imperative to bring together coordinated efforts to agree on standardized guides for authors to submit articles, unified reproducibility policies and artifact evaluation criteria from editors, supported by the reproducibility strategies and technological evolution discussed in this article. Consequently, there is a promising future with opportunities and potential to reduce the reproducibility gap identified with the joint effort of all actors involved to ensure reliability and trustworthiness in the knowledge conveyed by computer science-based publications.
In the current circumstances, readers and the general public could be considered passive actors. However, it is interesting to suggest that in the future, strategies and policies will be created that promote the more active participation of readers (e.g., likes and followers) in reviewing inconsistencies or errors in research claims as an important support to the scientific community.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://doi.org/10.5281/zenodo.14561905.
Author contributions
JH: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Resources, Software, Validation, Visualization, Writing – original draft, Writing – review & editing. MC: Conceptualization, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Supervision, Validation, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. SESAME's OVD-SaaS project from Région Île de France and BPI France for its financial support, also to the Ministry of Science, Technology and Innovation of Colombia (Minciencias), within the framework of Call 885 of 2020, to finance the Doctorates of Excellence Program.
Acknowledgments
The authors would like to thank the computer science journals that participated in the survey for their time spent carefully answering the questions.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplemental data
Survey Reproducibility in Computer Science Scientific Journals - Questions Form
Computer Science Journals List, Journals that issue Open Science Badges
PRISMA WoS Articles list
PRISMA Scopus Articles list
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcomp.2024.1491823/full#supplementary-material
Footnotes
1. ^https://www.acm.org/publications/policies/artifact-review-and-badging-current
2. ^Indeed, the history of a repository in GitHub can be altered with a hard push command or using the corresponding tools provided by GitHub.
6. ^https://www.rd-alliance.org/groups/fair-research-software-fair4rs-wg/outputs/?output=94498
8. ^https://mlcommons.org/en/mlcube/
10. ^Note that, despite their name, they are totally unrelated to lambda calculus!
11. ^https://www.rbvi.ucsf.edu/chimera/
16. ^https://github.com/IDSIA/sacred
19. ^https://www.pachyderm.com/
20. ^https://www.tensorflow.org/tensorboard
21. ^https://octoverse.github.com/
22. ^https://openprovenance.org/store/)
25. ^https://www.chameleoncloud.org/
26. ^https://www.catalyzex.com/
29. ^https://centml.ai/centml-platform-launch/
30. ^https://nips.cc/Conferences/2019/CallForPapers
31. ^https://www.acm.org/publications/policies/artifact-review-badging
32. ^https://ctuning.org/ae/checklist.html
33. ^https://ctuning.org/ae/reviewing.html
34. ^https://www.sigplan.org/Resources/EmpiricalEvaluation/
35. ^See https://scholarlykitchen.sspnet.org/2018/09/25/does-adopting-a-strict-data-sharing-policy-affect-submissions/.
36. ^https://ctuning.org/ae/micro2023.html
37. ^https://www.linkedin.com/pulse/micro-2023-artifact-evaluation-report-56th-ieeeacm-symposium-fursin-bsgwe/
38. ^http://invenio-software.org
41. ^https://codeocean.com/signup/ieee
42. ^https://www.fun-mooc.fr/en/courses/reproducible-research-methodological-principles-transparent-scie/
43. ^https://www.coursera.org/learn/reproducible-research
44. ^https://learning.edx.org/course/course-v1:HarvardX+PH527x+1T2020
46. ^https://www.linkedin.com/
References
(2021). Supporting computational reproducibility through code review. Nat. Hum. Behav. 5, 965–966. doi: 10.1038/s41562-021-01190-w
Abernathey, R. P., Augspurger, T., Banihirwe, A., Blackmon-Luca, C. C., Crone, T. J., Gentemann, C. L., et al. (2021). Cloud-native repositories for big scientific data. Comput. Sci. Eng. 23, 26–35. doi: 10.1109/MCSE.2021.3059437
Adams, M., Hense, A. V., and ter Hofstede, A. H. (2020). YAWL: an open source business process management system from science for science. SoftwareX 12:100576. doi: 10.1016/j.softx.2020.100576
Afgan, E., Baker, D., Batut, B., Van Den Beek, M., Bouvier, D., Čech, M., et al. (2018). The galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Res. 46, W537–W544. doi: 10.1093/nar/gky379
Ahmed, A., Al-Khatib, A., Boum, Y., Debat, H., Gurmendi Dunkelberg, A., Hinchliffe, L. J., et al. (2023). The future of academic publishing. Nat. Hum. Behav. 7, 1021–1026. doi: 10.1038/s41562-023-01637-2
Ahmed, H., Tchoua, R., and Lofstead, J. (2022). “Measuring reproduciblity of machine learning methods for medical diagnosis,” in 2022 Fourth International Conference on Transdisciplinary AI (TransAI) (Laguna Hills, CA: IEEE), 9–16. doi: 10.1109/TransAI54797.2022.00008
Akhlaghi, M., Infante-Sainz, R., Roukema, B. F., Khellat, M., Valls-Gabaud, D., Baena-Galle, R., et al. (2021). Toward long-term and archivable reproducibility. Comput. Sci. Eng. 23, 82–91. doi: 10.1109/MCSE.2021.3072860
Albertoni, R., Colantonio, S., Skrzypczyński, P., and Stefanowski, J. (2023). Reproducibility of machine learning: terminology, recommendations and open issues. arXiv [Preprint]. arXiv:2302.12691. doi: 10.48550/arXiv.2302.12691
Albrecht, M., Donnelly, P., Bui, P., and Thain, D. (2012). “Makeflow: a portable abstraction for data intensive computing on clusters, clouds, and grids,” in Proceedings of the 1st ACM SIGMOD Workshop on Scalable Workflow Execution Engines and Technologies (Scottsdale, AZ: ACM), 1–13. doi: 10.1145/2443416.2443417
AlNoamany, Y., and Borghi, J. A. (2018). Towards computational reproducibility: researcher perspectives on the use and sharing of software. PeerJ Comput. Sci. 4:e163. doi: 10.7717/peerj-cs.163
Amershi, S., Begel, A., Bird, C., DeLine, R., Gall, H., Kamar, E., et al. (2019). “Software engineering for machine learning: a case study,” in 2019 IEEE/ACM 41st International Conference on Software Engineering: Software Engineering in Practice (ICSE-SEIP) (Montreal, QC: IEEE), 291–300. doi: 10.1109/ICSE-SEIP.2019.00042
Anchundia, C. E., and Fonseca, C. E. R. (2020). Resources for reproducibility of experiments in empirical software engineering: topics derived from a secondary study. IEEE Access 8, 8992–9004. doi: 10.1109/ACCESS.2020.2964587
Anzt, H., Bach, F., Druskat, S., Löffler, F., Loewe, A., Renard, B. Y., et al. (2020). An environment for sustainable research software in Germany and beyond: current state, open challenges, and call for action. F1000Res. 9:295. doi: 10.12688/f1000research.23224.1
Athanassoulis, M., Triantafillou, P., Appuswamy, R., Bordawekar, R., Chandramouli, B., Cheng, X., et al. (2022). Artifacts availability & reproducibility (VLDB 2021 round table). ACM SIGMOD Rec. 51, 74–77. doi: 10.1145/3552490.3552511
Bahaidarah, L., Hung, E., Oliveira, A. F. D. M., Penumaka, J., Rosario, L., Trisovic, A., et al. (2021). Toward reusable science with readable code and reproducibility. arXiv [Preprint]. arXiv:2109.10387. doi: 10.48550/arXiv.2109.10387
Bailey, D. H. (2020). Reproducibility and variable precision computing. Int. J. High Perform. Comput. Appl. 34, 483–490. doi: 10.1177/1094342020938424
Baillieul, J., Grenier, G., and Setti, G. (2018). Reflections on the future of research curation and research reproducibility [point of view]. Proc. IEEE 106, 779–783. doi: 10.1109/JPROC.2018.2816618
Baker, M. (2016). Why scientists must share their research code. Nature. doi: 10.1038/nature.2016.20504
Baker, N., Alexander, F., Bremer, T., Hagberg, A., Kevrekidis, Y., Najm, H., et al. (2019). Workshop report on basic research needs for scientific machine learning: core technologies for artificial intelligence. Tech. Rep. 1478744. doi: 10.2172/1478744
Baracaldo, N., Anwar, A., Purcell, M., Rawat, A., Sinn, M., Altakrouri, B., et al. (2022). Towards an accountable and reproducible federated learning: a factsheets approach. arXiv [Preprint]. arXiv:2202.12443. doi: 10.48550/ARXIV.2202.12443
Barba, L. A. (2022). Defining the role of open source software in research reproducibility. Computer 55, 40–48. doi: 10.1109/MC.2022.3177133
Barker, M., Chue Hong, N. P., Katz, D. S., Lamprecht, A.-L., Martinez-Ortiz, C., Psomopoulos, F., et al. (2022). Introducing the FAIR principles for research software. Sci. Data 9:622. doi: 10.1038/s41597-022-01710-x
Bedő, J., Di Stefano, L., and Papenfuss, A. T. (2024). Unifying package managers, workflow engines, and containers: computational reproducibility with BioNix. GigaScience 9:giaa121. doi: 10.1093/gigascience/giaa121
Benureau, F. C. Y., and Rougier, N. P. (2018). Re-run, repeat, reproduce, reuse, replicate: transforming code into scientific contributions. Front. Neuroinform. 11:69. doi: 10.3389/fninf.2017.00069
Bonsignorio, F. (2017). A new kind of article for reproducible research in intelligent robotics [from the field]. IEEE Robot. Autom. Mag. 24, 178–182. doi: 10.1109/MRA.2017.2722918
Bosman, J., Frantsvåg, J. E., Kramer, B., Langlais, P.-C., and Proudman, V. (2021). OA Diamond Journals Study. Part 1: Findings. Technical report, Zenodo. doi: 10.5281/zenodo.4558704
Boulbes, D. R., Costello, T., Baggerly, K., Fan, F., Wang, R., Bhattacharya, R., et al. (2018). A survey on data reproducibility and the effect of publication process on the ethical reporting of laboratory research. Clin. Cancer Res. 24, 3447–3455. doi: 10.1158/1078-0432.CCR-18-0227
Bowman, R. W. (2023). Improving instrument reproducibility with open source hardware. Nat. Rev. Methods Primers 3:27. doi: 10.1038/s43586-023-00218-x
Breck, E., Cai, S., Nielsen, E., Salib, M., and Sculley, D. (2017). “The ML test score: a rubric for ml production readiness and technical debt reduction,” in 2017 IEEE International Conference on Big Data (Big Data) (Boston, MA: IEEE), 1123–1132. doi: 10.1109/BigData.2017.8258038
Brinckman, A., Chard, K., Gaffney, N., Hategan, M., Jones, M. B., Kowalik, K., et al. (2019). Computing environments for reproducibility: capturing the “whole tale". Future Gener. Comput. Syst. 94, 854–867. doi: 10.1016/j.future.2017.12.029
Brundage, M., Avin, S., Wang, J., Belfield, H., Krueger, G., Hadfield, G., et al. (2020). Toward trustworthy AI development: mechanisms for supporting verifiable claims. arXiv [Preprint]. arXiv:2004.07213. doi: 10.48550/ARXIV.2004.07213
Cacho, J. R. F., and Taghva, K. (2020). “The state of reproducible research in computer science,” in 17th International Conference on Information Technology-New Generations (ITNG 2020), ed. S. Latifi (Cham: Springer International Publishing), 519–524. doi: 10.1007/978-3-030-43020-7_68
Canon, R. S. (2020). “The role of containers in reproducibility,” in 2020 2nd International Workshop on Containers and New Orchestration Paradigms for Isolated Environments in HPC (CANOPIE-HPC) (Atlanta, GA: IEEE), 19–25. doi: 10.1109/CANOPIEHPC51917.2020.00008
Chard, K., Gaffney, N., Hategan, M., Kowalik, K., Ludäscher, B., McPhillips, T., et al. (2020). “Toward enabling reproducibility for data-intensive research using the whole tale platform,” in Advances in Parallel Computing, eds. I. Foster, G. R. Joubert, L. Kuc?era, W. E. Nagel, and F. Peters (IOS Press). doi: 10.3233/APC200107
Chard, K., Gaffney, N., Jones, M. B., Kowalik, K., Ludäscher, B., Nabrzyski, J., et al. (2019). “Implementing computational reproducibility in the whole tale environment,” in Proceedings of the 2nd International Workshop on Practical Reproducible Evaluation of Computer Systems (Phoenix, AZ: ACM), 17–22. doi: 10.1145/3322790.3330594
Chirigati, F., Shasha, D., and Freire, J. (2013). “Reprozip: using provenance to support computational reproducibility,” in 5th USENIX Conference on Theory and Practice of Provenance (TaPP'13). Available at: https://fchirigati.com/files/papers/chirigati-tapp2013.pdf
Cohen-Boulakia, S., Belhajjame, K., Collin, O., Chopard, J., Froidevaux, C., Gaignard, A., et al. (2017). Scientific workflows for computational reproducibility in the life sciences: status, challenges and opportunities. Future Gener. Comput. Syst. 75, 284–298. doi: 10.1016/j.future.2017.01.012
Collberg, C., and Proebsting, T. (2016). Repeatability in computer systems research. Commun. ACM 59, 62–69. doi: 10.1145/2812803
Colom, M., Kerautret, B., Limare, N., Monasse, P., and Morel, J.-M. (2015). “IPOL: a new journal for fully reproducible research; analysis of four years development,” in 2015 7th International Conference on New Technologies, Mobility and Security (NTMS) (Paris: IEEE), 1–5. doi: 10.1109/NTMS.2015.10287266500
Committee on Reproducibility and Replicability in Science Board on Behavioral, Cognitive, and Sensory Sciences, Committee on National Statistics, Division of Behavioral and Social Sciences and Education, Nuclear and Radiation Studies Board, Division on Earth and Life Studies. (2019). Reproducibility and Replicability in Science. Washington, DC: National Academies Press.
Congo, F. (2015). Building a Cloud Service for Reproducible Simulation Management. Austin, TX, 187–193. doi: 10.25080/Majora-7b98e3ed-01d
Cousijn, H., Kenall, A., Ganley, E., Harrison, M., Kernohan, D., Lemberger, T., et al. (2018). A data citation roadmap for scientific publishers. Sci. Data 5:180259. doi: 10.1038/sdata.2018.259
Crick, T., Hall, B. A., and Ishtiaq, S. (2015). Reproducibility as a technical specification. arXiv [Preprint]. arXiv:1504.01310. doi: 10.48550/ARXIV.1504.01310
Crusoe, M. R., Abeln, S., Iosup, A., Amstutz, P., Chilton, J., Tijanić, N., et al. (2022). Methods included: standardizing computational reuse and portability with the common workflow language. Commun. ACM 65, 54–63. doi: 10.1145/3486897
Cukier, S., Helal, L., Rice, D. B., Pupkaite, J., Ahmadzai, N., Wilson, M., et al. (2020). Checklists to detect potential predatory biomedical journals: a systematic review. BMC Med. 18:104. doi: 10.1186/s12916-020-01566-1
Da Silva, R. F., Casanova, H., Chard, K., Altintas, I., Badia, R. M., Balis, B., et al. (2021). “A community roadmap for scientific workflows research and development,” in 2021 IEEE Workshop on Workflows in Support of Large-Scale Science (WORKS) (St. Louis, MO: IEEE), 81–90. doi: 10.1109/WORKS54523.2021.00016
Daniel Adorno Gomes, P. M., and Serodio, C. (2019). “Infrastructure-as-code for scientific computing environments,” in CENTRIC 2019: The Twelfth International Conference on Advances in Human-Oriented and Personalized Mechanisms.
De Sterck, H., Shu, C.-W., and Abgrall, R. (2023). Enhancing reproducibility of research papers in SISC, JSC and JCP. J. Sci. Comput. 95:77. doi: 10.1007/s10915-023-02193-7
Deelman, E., Blythe, J., Gil, Y., Kesselman, C., Mehta, G., Patil, S., et al. (2004). “Pegasus: mapping scientific workflows onto the grid,” inGrid Computing, ed. M. D. Dikaiakos (Berlin: Springer), 11–20. doi: 10.1007/978-3-540-28642-4_2
Demchenko, Y., Gallenmuller, S., Fdida, S., Andreou, P., Crettaz, C., Kirkeng, M., et al. (2023). “Experimental research reproducibility and experiment workflow management,” in 2023 15th International Conference on COMmunication Systems & NETworkS (COMSNETS) (Bangalore: IEEE), 835–840. doi: 10.1109/COMSNETS56262.2023.10041378
Di Cosmo, R., Gruenpeter, M., and Zacchiroli, S. (2022). 204.4 identifiers for digital objects: the case of software source code preservation. OSF. doi: 10.17605/OSF.IO/KDE56
Diaba-Nuhoho, P., and Amponsah-Offeh, M. (2021). Reproducibility and research integrity: the role of scientists and institutions. BMC Res. Notes 14:451. doi: 10.1186/s13104-021-05875-3
Dodds, F. (2019). The future of academic publishing: revolution or evolution revisited. Learn. Publ. 32, 345–354. doi: 10.1002/leap.1258
Dozmorov, M. G. (2018). GitHub statistics as a measure of the impact of open-source bioinformatics software. Front. Bioeng. Biotechnol. 6:198. doi: 10.3389/fbioe.2018.00198
Druskat, S., Bertuch, O., Juckeland, G., Knodel, O., and Schlauch, T. (2022). Software publications with rich metadata: state of the art, automated workflows and HERMES concept. arXiv [Preprint]. arXiv:2201.09015. doi: 10.48550/arXiv.2201.09015
Edwards, M. A., and Roy, S. (2017). Academic research in the 21st century: maintaining scientific integrity in a climate of perverse incentives and hypercompetition. Environ. Eng. Sci. 34, 51–61. doi: 10.1089/ees.2016.0223
Essawy, B. T., Goodall, J. L., Voce, D., Morsy, M. M., Sadler, J. M., Choi, Y. D., et al. (2020). A taxonomy for reproducible and replicable research in environmental modelling. Environ. Modell. Softw. 134:104753. doi: 10.1016/j.envsoft.2020.104753
Feger, S. S., and Woźniak, P. W. (2022). Reproducibility: a researcher-centered definition. Multimodal Technol. Interact. 6:17. doi: 10.3390/mti6020017
Forde, J., Head, T., Holdgraf, C., Panda, Y., Nalvarete, G., Ragan-Kelley, B., et al. (2018). Reproducible Research Environments With Repo2Docker. Openreview. Available at: https://openreview.net/pdf?id=B1lYOwuoxm
Fostiropoulos, I., Brown, B., and Itti, L. (2023). “Reproducibility requires consolidated artifacts,” in 2023 IEEE/ACM 2nd International Conference on AI Engineering – Software Engineering for AI (CAIN) (Melbourne, VIC: IEEE), 100–101. doi: 10.1109/CAIN58948.2023.00025
Françoise, J., Caramiaux, B., and Sanchez, T. (2021). “Marcelle: composing interactive machine learning workflows and interfaces,” in The 34th Annual ACM Symposium on User Interface Software and Technology (New York, NY: ACM), 9–53. doi: 10.1145/3472749.3474734
Freire, J., Bonnet, P., and Shasha, D. (2012). “Computational reproducibility: state-of-the-art, challenges, and database research opportunities,” in Proceedings of the 2012 ACM SIGMOD International Conference on Management of Data (Scottsdale, AZ: ACM), 593–596. doi: 10.1145/2213836.2213908
Frery, A. C., Gomez, L., and Medeiros, A. C. (2020). A badging system for reproducibility and replicability in remote sensing research. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 13, 4988–4995. doi: 10.1109/JSTARS.2020.3019418
Fritzsch, J., Bogner, J., Haug, M., da Silva, A. C. F., Rubner, C., and Saft, S. (2023). Adopting microservices and DevOps in the cyber-physical systems domain: a rapid review and case study. software: practice and experience. Softw. Pract. Exp. 53, 790–810. doi: 10.1002/spe.3169
Fund, F. (2023). “We need more reproducibility content across the computer science curriculum,” in Proceedings of the 2023 ACM Conference on Reproducibility and Replicability (Santa Cruz, CA: ACM), 97–101. doi: 10.1145/3589806.3600033
Fursin, G. (2018). “Invited talk abstract: introducing ReQuEST: an open platform for reproducible and quality-efficient systems-ML tournaments,” in 2018 1st Workshop on Energy Efficient Machine Learning and Cognitive Computing for Embedded Applications (EMC2) (Williamsburg, VA: IEEE), 3. doi: 10.1109/EMC2.2018.00008
Fursin, G. (2020a). Collective knowledge: organizing research projects as a database of reusable components and portable workflows with common APIs. arXiv [Preprint]. arXiv:2011.01149. doi: 10.48550/ARXIV.2011.01149
Fursin, G. (2020b). The collective knowledge project: making ML models more portable and reproducible with open APIs, reusable best practices and MLOps. arXiv [Preprint]. arXiv:2006.07161. doi: 10.48550/ARXIV.2006.07161
Fursin, G. (2020c). Enabling reproducible ML and Systems research: the good, the bad, and the ugly. Zenodo. doi: 10.5281/ZENODO.4005773
Fursin, G., Miceli, R., Lokhmotov, A., Gerndt, M., Baboulin, M., Malony, A. D., et al. (2014). Collective mind: towards practical and collaborative auto-tuning. Sci. Program. 22, 309–329. doi: 10.1155/2014/797348
Gandrud, C. (2020). Reproducible research with R and RStudio. The R series, 3rd Edn. Boca Raton, FL: CRC Press. doi: 10.1201/9780429031854
Ghimpau, V. (2019). “Incentives, rewards, and recognition - what really motivates a researcher?,” in Judging Research (MDPI). doi: 10.3390/books978-3-03928-315-6-11
Ghoshal, D., Paine, D., Pastorello, G., Elbashandy, A., Gunter, D., Amusat, O., et al. (2020). “Experiences with reproducibility: case studies from scientific workflows,” in Proceedings of the 4th International Workshop on Practical Reproducible Evaluation of Computer Systems (Stockholm: ACM), 3–8. doi: 10.1145/3456287.3465478
Gift, N., and Deza, A. (2021). Practical MLOps: operationalizing machine learning models, 1st Edn. Sebastopol, CA: O'Reilly Media Inc. OCLC: on1249501065.
Gomes, D. G. E., Pottier, P., Crystal-Ornelas, R., Hudgins, E. J., Foroughirad, V., Sánchez-Reyes, L. L., et al. (2022). Why don't we share data and code? Perceived barriers and benefits to public archiving practices. Proc. R. Soc. B Biol. Sci. 289:20221113. doi: 10.1098/rspb.2022.1113
Gonzalez-Barahona, J. M., and Robles, G. (2023). Revisiting the reproducibility of empirical software engineering studies based on data retrieved from development repositories. Inf. Softw. Technol. 164:107318. doi: 10.1016/j.infsof.2023.107318
Gundersen, O. E. (2020). The reproducibility crisis is real. AI Mag. 41, 103–106. doi: 10.1609/aimag.v41i3.5318
Gundersen, O. E. (2021). The fundamental principles of reproducibility. Philos. Trans. R. Soc. A Math. Phys. Eng. Sci. 379:20200210. arXiv:2011.10098 [cs]. doi: 10.1098/rsta.2020.0210
Gundersen, O. E., and Kjensmo, S. (2018). State of the art: reproducibility in artificial intelligence. Proc. AAAI Conf. Artif. Intell. 32:11503. doi: 10.1609/aaai.v32i1.11503
Gundersen, O. E., Shamsaliei, S., and Isdahl, R. J. (2022). Do machine learning platforms provide out-of-the-box reproducibility? Future Gener. Comput. Syst. 126, 34–47. doi: 10.1016/j.future.2021.06.014
Gupta, A., Wright, C., Ganapini, M. B., Sweidan, M., and Butalid, R. (2022). State of AI ethics report. arXiv [Preprint]. arXiv:2202.07435. doi: 10.48550/arXiv.2202.07435
Haibe-Kains, B., Adam, G. A., Hosny, A., Khodakarami, F., Massive Analysis Quality Control (MAQC) Society Board of Directors, and Waldron, L. (2020). Transparency and reproducibility in artificial intelligence. Nature 586, E14-E16. doi: 10.1038/s41586-020-2766-y
Haim, A., Shaw, S., and Heffernan, N. (2023a). “How to open science: a principle and reproducibility review of the learning analytics and knowledge conference,” in LAK23: 13th International Learning Analytics and Knowledge Conference (Arlington, TX: ACM), 156–164. doi: 10.1145/3576050.3576071
Haim, A., Shaw, S. T., and Heffernan, N. T. (2023b). “How to open science: promoting principles and reproducibility practices within the artificial intelligence in education community,” in Artificial Intelligence in Education. Posters and Late Breaking Results, Workshops and Tutorials, Industry and Innovation Tracks, Practitioners, Doctoral Consortium and Blue Sky, Vol. 1831, eds. N. Wang, G. Rebolledo-Mendez, V. Dimitrova, N. Matsuda, and O. C. Santos (Cham: Springer Nature Switzerland), 74–78. doi: 10.1007/978-3-031-36336-8_11
Heesen, R. (2017). Communism and the incentive to share in science. Philos. Sci. 84, 698–716. doi: 10.1086/693875
Heroux, M. A., Barba, L., Parashar, M., Stodden, V., and Taufer, M. (2018). Toward a Compatible Reproducibility Taxonomy for Computational and Computing Sciences. Tech. Rep. SAND2018-11186. doi: 10.2172/1481626
Hiesgen, R., Nawrocki, M., Schmidt, T. C., and Wählisch, M. (2022). The race to the vulnerable: measuring the log4j shell incident. arXiv [Preprint]. arXiv:2205.02544. doi: 10.48550/arXiv.2205.02544
Hofman, J. M., Goldstein, D. G., Sen, S., and Poursabzi-Sandegh, F. (2020). “Expanding the scope of reproducibility research through data analysis replications,” in Companion Proceedings of the Web Conference 2020 (Taipei: ACM), 567–571. doi: 10.1145/3366424.3383417
Howe, B. (2012). Virtual appliances, cloud computing, and reproducible research. Comput. Sci. Eng. 14, 36–41. doi: 10.1109/MCSE.2012.62
Hu, Y., Tareen, A., Sheu, Y.-J., Ireland, W. T., Speck, C., Li, H., et al. (2020). Evolution of DNA replication origin specification and gene silencing mechanisms. Nat. Commun. 11:5175. doi: 10.1038/s41467-020-18964-x
Hughes, T. (2001). “History of technology,” in International Encyclopedia of the Social & Behavioral Sciences (Amsterdam: Elsevier), 6852–6857. doi: 10.1016/B0-08-043076-7/02648-6
Hull, D., Wolstencroft, K., Stevens, R., Goble, C., Pocock, M. R., Li, P., et al. (2006). Taverna: a tool for building and running workflows of services. Nucleic Acids Res. 34, W729-W732. doi: 10.1093/nar/gkl320
Hummel, T., and Manner, J. (2024). “A literature review on reproducibility studies in computer science,” in Proceedings of the 16th ZEUS Workshop on Services and Their Composition (ZEUS 2024) (CEUR). Available at: https://ceur-ws.org/Vol-3673/paper9.pdf
Hutson, M. (2018). Artificial intelligence faces reproducibility crisis. Science 359, 725–726. doi: 10.1126/science.359.6377.725
Huynh, T. D., and Moreau, L. (2015). “ProvStore: a public provenance repository,” in Provenance and Annotation of Data and Processes, Vol. 8628, eds. B. Ludäscher, and B. Plale (Cham: Springer International Publishing), 275–277. doi: 10.1007/978-3-319-16462-5_32
Idrissou, A., Zamborlini, V., and Kuhn, T. (2022). “Documenting the creation, manipulation and evaluation of links for reuse and reproducibility,” in Knowledge Engineering and Knowledge Management, Vol. 13514, eds. O. Corcho, L. Hollink, O. Kutz, N. Troquard, and F. J. Ekaputra (Cham: Springer International Publishing), 81–96. doi: 10.1007/978-3-031-17105-5_6
Impagliazzo, R., Lei, R., Pitassi, T., and Sorrell, J. (2022). “Reproducibility in learning,” in Proceedings of the 54th Annual ACM SIGACT Symposium on Theory of Computing (Rome: ACM), 818–831. doi: 10.1145/3519935.3519973
Ivie, P., and Thain, D. (2018). Reproducibility in scientific computing. ACM Comput. Surv. 51, 63:1–63:36. doi: 10.1145/3186266
Jalal Apostal, S. F., Apostal, D., and Marsh, R. (2020). “Improving numerical reproducibility of scientific software in parallel systems,” in 2020 IEEE International Conference on Electro Information Technology (EIT) (Chicago, IL: IEEE), 066–074. doi: 10.1109/EIT48999.2020.9208338
Johnson, E. C. (2024). SciOps: achieving productivity and reliability in data-intensive research. arXiv [Preprint]. arXiv:2401.00077. doi: 10.48550/arXiv.2401.00077
Jonas, E., Schleier-Smith, J., Sreekanti, V., Tsai, C.-C., Khandelwal, A., Pu, Q., et al. (2019). Cloud programming simplified: a Berkeley view on serverless computing. arXiv [Preprint]. arXiv:1902.03383. doi: 10.48550/ARXIV.1902.03383
Kapoor, S., and Narayanan, A. (2022). Leakage and the reproducibility crisis in ML-based science. arXiv [Preprint]. arXiv:2207.07048. doi: 10.48550/ARXIV.2207.07048
Karargyris, A., Umeton, R., Sheller, M. J., Aristizabal, A., George, J., Wuest, A., et al. (2023). Federated benchmarking of medical artificial intelligence with MedPerf. Nat. Mach. Intell. 5, 799–810. doi: 10.1038/s42256-023-00652-2
Karathanasis, N., Hwang, D., Heng, V., Abhimannyu, R., Slogoff-Sevilla, P., Buchel, G., et al. (2022). Reproducibility efforts as a teaching tool: a pilot study. PLoS Comput. Biol. 18:e1010615. doi: 10.1371/journal.pcbi.1010615
Kawamoto, Y., and Kobayashi, A. (2020). “AI pedigree verification platform using blockchain,” in 2020 2nd Conference on Blockchain Research & Applications for Innovative Networks and Services (BRAINS) (Paris: IEEE), 204–205. doi: 10.1109/BRAINS49436.2020.9223307
Kazerooni, A. F., Khalili, N., Liu, X., Haldar, D., Jiang, Z., Anwar, S. M., et al. (2023). The brain tumor segmentation (BraTS) challenge 2023: focus on pediatrics (CBTN-CONNECT-DIPGR-ASNR1229 MICCAI BraTS-PEDs). arXiv [Preprint]. arXiv:2305.17033v7. doi: 10.48550/ARXIV.2305.17033
Ke, Q., Gates, A. J., and Barabási, A.-L. (2023). A network-based normalized impact measure reveals successful periods of scientific discovery across disciplines. Proc. Nat. Acad. Sci. 120:e2309378120. doi: 10.1073/pnas.2309378120
Keahey, K., Riteau, P., Stanzione, D., Cockerill, T., Mambretti, J., Rad, P., et al. (2019). “Chameleon: a scalable production testbed for computer science research,” in Contemporary High Performance Computing, eds. K. Keahey, P. Riteau, D. Stanzione, T. Cockerill, J. Mambretti, P. Rad, et al. (Boca Raton, FL: CRC Press), 123–148. doi: 10.1201/9781351036863-5
Khritankov, A., Pershin, N., Ukhov, N., and Ukhov, A. (2021). MLDev: data science experiment automation and reproducibility software. arXiv [Preprint]. arXiv:2107.12322. doi: 10.48550/arXiv.2107.12322
Kitchenham, B., Madeyski, L., and Brereton, P. (2020). Meta-analysis for families of experiments in software engineering: a systematic review and reproducibility and validity assessment. Empirical. Softw. Eng. 25, 353–401. doi: 10.1007/s10664-019-09747-0
Kitzes, J., Turek, D., and Deniz, F. editors (2018). The practice of reproducible research: case studies and lessons from the data-intensive sciences. Oakland, CA: University of California Press.
Kluge, M., Friedl, M.-S., Menzel, A. L., and Friedel, C. C. (2020). Watchdog 2.0: new developments for reusability, reproducibility, and workflow execution. GigaScience 9:giaa068. doi: 10.1093/gigascience/giaa068
Konkol, M., Nüst, D., and Goulier, L. (2020). Publishing computational research - a review of infrastructures for reproducible and transparent scholarly communication. Res. Integr. Peer Rev. 5:10. doi: 10.1186/s41073-020-00095-y
Korkhov, V., Krefting, D., Montagnat, J., Truong-Huu, T., Kukla, T., Terstyanszky, G., et al. (2012). Shiwa workflow interoperability solutions for neuroimaging data analysis. Stud. Health Technol. Inform. 175, 109–10.
LeVeque, R. J., Mitchell, I. M., and Stodden, V. (2012). Reproducible research for scientific computing: tools and strategies for changing the culture. Comput. Sci. Eng. 14, 13–17. doi: 10.1109/MCSE.2012.38
Lewis, T.-M. (2023). From policy to practice: How journal-based data policies encourage scientists' adoption of reproducible research practices (PhD thesis). Chapel Hill, NC: The University of North Carolina at Chapel Hill University Libraries.
Lin, J. (2022). Building a culture of reproducibility in academic research. arXiv [Preprint]. arXiv:2212.13534. doi: 10.48550/ARXIV.2212.13534
Lin, J., and Zhang, Q. (2020). “Reproducibility is a process, not an achievement: the replicability of IR reproducibility experiments,” in Advances in Information Retrieval, Volume 12036, eds. J. M. Jose, E. Yilmaz, J. Magalhães, P. Castells, N. Ferro, M. J. Silva, and F. Martins (Cham: Springer International Publishing), 43–49. doi: 10.1007/978-3-030-45442-5_6
Lippi, G., and Mattiuzzi, C. (2017). Scientist impact factor (SIF): a new metric for improving scientists' evaluation? Ann. Transl. Med. 5:24. doi: 10.21037/atm.2017.06.24
Lopresti, D., and Nagy, G. (2021). “Reproducibility: evaluating the evaluations,” in Reproducible Research in Pattern Recognition, Vol. 12636, eds. B. Kerautret, M. Colom, A. Krähenbühl, D. Lopresti, P. Monasse, and H. Talbot (Cham: Springer International Publishing), 12–23. doi: 10.1007/978-3-030-76423-4_2
Lucic, A., Bleeker, M., Jullien, S., Bhargav, S., and de Rijke, M. (2022). Reproducibility as a mechanism for teaching fairness, accountability, confidentiality, and transparency in artificial intelligence. Proc. AAAI Conf. Artif. Intell. 36, 12792–12800.doi: 10.1609/aaai.v36i11.21558
Mack, C. A. (2018). How to Write a Good Scientific Paper. Bellingham: SPIE Press. OCLC: on1019885580. doi: 10.1117/3.2317707
Macleod, M., and the University of Edinburgh Research Strategy Group (2022). Improving the reproducibility and integrity of research: what can different stakeholders contribute? BMC Res. Notes 15:146. doi: 10.1186/s13104-022-06030-2
Malik, T. (2020). “Artifact description/artifact evaluation: a reproducibility bane or a boon,” in Proceedings of the 4th International Workshop on Practical Reproducible Evaluation of Computer Systems (Stockholm: ACM), 1. doi: 10.1145/3456287.3465479
Martinez, I., Viles, E., and Olaizola, I. G. (2021). “A survey study of success factors in data science projects,” in 2021 IEEE International Conference on Big Data (Big Data) (Orlando, FL: IEEE), 2313–2318. doi: 10.1109/BigData52589.2021.9671588
Mauerer, W., Klessinger, S., and Scherzinger, S. (2022). “Beyond the badge: reproducibility engineering as a lifetime skill,” in Proceedings of the 4th International Workshop on Software Engineering Education for the Next Generation (Pittsburgh, PA: ACM), 1–4. doi: 10.1145/3528231.3528359
Melchor, F., Rodriguez-Echeverria, R., Conejero, J. M., Prieto, A. E., and Gutiérrez, J. D. (2022). “A model-driven approach for systematic reproducibility and replicability of data science projects,” in Advanced Information Systems Engineering, Vol. 13295, eds. X. Franch, G. Poels, F. Gailly, and M. Snoeck (Cham: Springer International Publishing), 147–163. doi: 10.1007/978-3-031-07472-1_9
Meng, H., and Thain, D. (2017). Facilitating the reproducibility of scientific workflows with execution environment specifications. Procedia Comput. Sci. 108, 705–714. doi: 10.1016/j.procs.2017.05.116
Merz, K. M., Amaro, R., Cournia, Z., Rarey, M., Soares, T., Tropsha, A., et al. (2020). Editorial: Method and data sharing and reproducibility of scientific results. J. Chem. Inf. Model. 60, 5868–5869. doi: 10.1021/acs.jcim.0c01389
Milewicz, R., and Mundt, M. (2023). “Towards evidence-based software quality practices for reproducibility: preliminary results and research directions,” in Proceedings of the 2023 ACM Conference on Reproducibility and Replicability (Santa Cruz, CA: ACM), 85–88. doi: 10.1145/3589806.3600040
Moreau, D., Wiebels, K., and Boettiger, C. (2023). Containers for computational reproducibility. Nat. Rev. Methods Primers 3, 1–16. doi: 10.1038/s43586-023-00236-9
Nichols, J. D., Oli, M. K., Kendall, W. L., and Boomer, G. S. (2021). A better approach for dealing with reproducibility and replicability in science. Proc. Natl. Acad. Sci. U S A 118:e2100769118. doi: 10.1073/pnas.2100769118
Nordling, T., and Peralta, T. M. (2022). A literature review of methods for assessment of reproducibility in science. Res. Sq. doi: 10.21203/rs.3.rs-2267847/v1
Octoverse (2024). State of the Octoverse 2024. Available at: https://octoverse.github.com/ (accessed December 24, 2024).
Orzechowski, M., Baliś, B., Słota, R. G., and Kitowski, J. (2020). “Reproducibility of computational experiments on kubernetes-managed container clouds with hyperflow,” in Computational Science - ICCS 2020, Vol. 12137, eds. V. V. Krzhizhanovskaya, G. Závodszky, M. H. Lees, J. J. Dongarra, P. M. A. Sloot, S. Brissos, and J. Teixeira (Cham: Springer International Publishing), 220–233. doi: 10.1007/978-3-030-50371-0_16
Parashar, M., Heroux, M. A., and Stodden, V. (2022). Research reproducibility. Computer 55, 16–18. doi: 10.1109/MC.2022.3176988
Parland-von Essen, J., Fält, K., Maalick, Z., Alonen, M., and Gonzalez, E. (2018). Supporting FAIR data: categorization of research data as a tool in data management. Informaatiotutkimus 37:76084. doi: 10.23978/inf.76084
Parsons, M. A., Duerr, R. E., and Jones, M. B. (2019). The history and future of data citation in practice. Data Sci. J. 18:52. doi: 10.5334/dsj-2019-052
Peregrina, J. A., Ortiz, G., and Zirpins, C. (2022). “Towards a metadata management system for provenance, reproducibility and accountability in federated machine learning,” in Advances in Service-Oriented and Cloud Computing, Vol. 1617, eds. C. Zirpins, G. Ortiz, Z. Nochta, O. Waldhorst, J. Soldani, M. Villari, and D. Tamburri (Cham: Springer Nature Switzerland), 5–18. doi: 10.1007/978-3-031-23298-5_1
Pérignon, C., Gadouche, K., Hurlin, C., Silberman, R., and Debonnel, E. (2019). Certify reproducibility with confidential data. Science 365, 127–128. doi: 10.1126/science.aaw2825
Pimentel, J. F., Murta, L., Braganholo, V., and Freire, J. (2019). “A large-scale study about quality and reproducibility of jupyter notebooks,” in 2019 IEEE/ACM 16th International Conference on Mining Software Repositories (MSR) (New York, NY: ACM), 507–517. ISSN: 2574-3864. doi: 10.1109/MSR.2019.00077
Pimentel, J. F., Murta, L., Braganholo, V., and Freire, J. (2021). Understanding and improving the quality and reproducibility of Jupyter notebooks. Empir. Softw. Eng. 26:65. doi: 10.1007/s10664-021-09961-9
Pineau, J., Vincent-Lamarre, P., Sinha, K., Larivière, V., Beygelzimer, A., d'Alché Buc, F., et al. (2020). Improving reproducibility in machine learning research (a report from the neurips 2019 reproducibility program). arXiv [Preprint]. arXiv:2003.12206. doi: 10.48550/arXiv.2003.12206
Plale, B. A., Malik, T., and Pouchard, L. C. (2021). Reproducibility practice in high-performance computing: community survey results. Comput. Sci. Eng. 23, 55–60. doi: 10.1109/MCSE.2021.3096678
Plavén-Sigray, P., Matheson, G. J., Schiffler, B. C., and Thompson, W. H. (2017). The readability of scientific texts is decreasing over time. Elife 6:e27725. doi: 10.7554/eLife.27725
Plesser, H. E. (2018). Reproducibility vs. replicability: a brief history of a confused terminology. Front. Neuroinform. 11:76. doi: 10.3389/fninf.2017.00076
Poldrack, R. A. (2019). The costs of reproducibility. Neuron 101, 11–14. doi: 10.1016/j.neuron.2018.11.030
Pouchard, L., Baldwin, S., Elsethagen, T., Jha, S., Raju, B., Stephan, E., et al. (2019). Computational reproducibility of scientific workflows at extreme scales. Int. J. High Perform. Comput. Appl. 33, 763–776. doi: 10.1177/1094342019839124
Pradal, C., Fournier, C., Boudon, F., Valduriez, P., Pacitti, E., Guédon, Y. Y., et al. (2019). OpenAlea.
Radha, S. K., Taylor, I., Nabrzyski, J., and Barclay, I. (2021). Verifiable badging system for scientific data reproducibility. Blockchain Res. Appl. 2:100015. doi: 10.1016/j.bcra.2021.100015
Raff, E. (2019). A step toward quantifying independently reproducible machine learning research. arXiv [Preprint]. arXiv:1909.06674. doi: 10.48550/ARXIV.1909.06674
Raff, E. (2020). Research reproducibility as a survival analysis. arXiv [Preprint]. arXiv:2012.09932. doi: 10.48550/arXiv.2012.09932
Raff, E., and Farris, A. L. (2022). A siren song of open source reproducibility. arXiv [Preprint]. arXiv:2204.04372. doi: 10.48550/arXiv.2204.04372
Raghupathi, W., Raghupathi, V., and Ren, J. (2022). Reproducibility in computing research: an empirical study. IEEE Access 10, 29207–29223. doi: 10.1109/ACCESS.2022.3158675
Ray, P. P. (2022). A review on TinyML: state-of-the-art and prospects. J. King Saud Univ. Comput. Inf. Sci. 34, 1595–1623. doi: 10.1016/j.jksuci.2021.11.019
Rosenblatt, L., Herman, B., Holovenko, A., Lee, W., Loftus, J., McKinnie, E., et al. (2023). Epistemic parity: reproducibility as an evaluation metric for differential privacy. Proc. VLDB Endowment 16, 3178–3191. doi: 10.14778/3611479.3611517
Rosendo, D., Keahey, K., Costan, A., Simonin, M., Valduriez, P., Antoniu, G., et al. (2023). “KheOps: cost-effective repeatability, reproducibility, and replicability of edge-to-cloud experiments,” in Proceedings of the 2023 ACM Conference on Reproducibility and Replicability (Santa Cruz, CA: ACM), 62–73. doi: 10.1145/3589806.3600032
Rougier, N. P., and Hinsen, K. (2019). “ReScience C: a journal for reproducible replications in computational science,” in Reproducible Research in Pattern Recognition, Vol. 11455, eds. B. Kerautret, M. Colom, D. Lopresti, P. Monasse, and H. Talbot (Cham: Springer International Publishing), 150–156. doi: 10.1007/978-3-030-23987-9_14
Salsabil, L., Wu, J., Choudhury, M. H., Ingram, W. A., Fox, E. A., Rajtmajer, S. M., et al. (2022). A study of computational reproducibility using URLs linking to open access datasets and software,” in Companion Proceedings of the Web Conference 2022 (Lyon: ACM), 784–788. doi: 10.1145/3487553.3524658
Saltz, J. S., and Krasteva, I. (2022). Current approaches for executing big data science projects-a systematic literature review. PeerJ Comput. Sci. 8:e862. doi: 10.7717/peerj-cs.862
Samuel, S., and König-Ries, B. (2021a). “ReproduceMeGit: a visualization tool for analyzing reproducibility of jupyter notebooks,” in Provenance and Annotation of Data and Processes, Vol. 12839, eds. B. Glavic, V. Braganholo, and D. Koop (Cham: Springer International Publishing), 201–206. doi: 10.1007/978-3-030-80960-7_12
Samuel, S., and König-Ries, B. (2021b). Understanding experiments and research practices for reproducibility: an exploratory study. PeerJ 9:e11140. doi: 10.7717/peerj.11140
Samuel, S., and König-Ries, B. (2022a). A collaborative semantic-based provenance management platform for reproducibility. PeerJ Comput. Sci. 8:e921. doi: 10.7717/peerj-cs.921
Samuel, S., and König-Ries, B. (2022b). End-to-End provenance representation for the understandability and reproducibility of scientific experiments using a semantic approach. J. Biomed. Semantics 13:1. doi: 10.1186/s13326-021-00253-1
Samuel, S., Löffler, F., and König-Ries, B. (2021). “Machine learning pipelines: provenance, reproducibility and FAIR data principles,” in Provenance and Annotation of Data and Processes, Volume 12839, eds. B. Glavic, V. Braganholo, and D. Koop (Cham: Springer International Publishing), 226–230. doi: 10.1007/978-3-030-80960-7_17
Samuel, S., and Mietchen, D. (2023). Computational reproducibility of jupyter notebooks from biomedical publications. GigaScience 13:giad113. doi: 10.1093/gigascience/giad113
Santana-Perez, I., and Pérez-Hernández, M. S. (2015). Towards reproducibility in scientific workflows: an infrastructure-based approach. Sci. Program. 2015:e243180. doi: 10.1155/2015/243180
Schelter, S., Biessmann, F., Januschowski, T., Salinas, D., Seufert, S., and Szarvas, G. (2015). On challenges in machine learning model management. IEEE Data Eng. Bull.
Schlegel, M., and Sattler, K.-U. (2022). Management of machine learning lifecycle artifacts: a survey. arXiv [Preprint]. arXiv:2210.11831. doi: 10.48550/ARXIV.2210.11831
Schröer, C., Kruse, F., and Marx Gómez, J. (2021). A systematic literature review on applying crisp-dm process model. Procedia Comput. Sci. 181, 526–534. doi: 10.1016/j.procs.2021.01.199
Sculley, D., Holt, G., Golovin, D., Davydov, E., Phillips, T., Ebner, D., et al. (2015). “Hidden technical debt in machine learning systems,” in Advances in Neural Information Processing Systems, Vol. 28, eds. C. Cortes, N. Lawrence, D. Lee, M. Sugiyama, and R. Garnett (Newry: Curran Associates, Inc).
Silva Junior, D., Pacitti, E., Paes, A., and De Oliveira, D. (2021). Provenance-and machine learning-based recommendation of parameter values in scientific workflows. PeerJ Comput. Sci. 7:e606. doi: 10.7717/peerj-cs.606
Smith, A. M., Katz, D. S., Niemeyer, K. E., and FORCE11 Software Citation Working Group (2016). Software citation principles. PeerJ Comput. Sci. 2:e86. doi: 10.7717/peerj-cs.86
Stefaniak, J., and Carey, K. (2019). Instilling purpose and value in the implementation of digital badges in higher education. Int. J. Educ. Technol. High. Educ. 16:44. doi: 10.1186/s41239-019-0175-9
Steidl, M., Felderer, M., and Ramler, R. (2023). The pipeline for the continuous development of artificial intelligence models-current state of research and practice. J. Syst. Softw. 199:111615. doi: 10.1016/j.jss.2023.111615
Stoddart, C. (2016). Is there a reproducibility crisis in science? Nature d41586-019-00067-3. doi: 10.1038/d41586-019-00067-3
Stodden, V. (2020). “Beyond open data: a model for linking digital artifacts to enable reproducibility of scientific claims,” in Proceedings of the 3rd International Workshop on Practical Reproducible Evaluation of Computer Systems (Stockholm: ACM), 9–14. doi: 10.1145/3391800.3398172
Stodden, V., Guo, P., and Ma, Z. (2012). How Journals Are Adopting Open Data and Code Policies. Digital Library of the Commons Indiana University Libraries.
Stodden, V., Guo, P., and Ma, Z. (2013). Toward reproducible computational research: an empirical analysis of data and code policy adoption by journals. PLoS ONE 8:e67111. doi: 10.1371/journal.pone.0067111
Stodden, V., and Miguez, S. (2014). Best practices for computational science: Software infrastructure and environments for reproducible and extensible research. J. Open Res. Softw. 2:e21. doi: 10.5334/jors.ay
Stodden, V., Seiler, J., and Ma, Z. (2018). An empirical analysis of journal policy effectiveness for computational reproducibility. Proc. Nat. Acad. Sci. 115, 2584–2589. doi: 10.1073/pnas.1708290115
Sugimura, P., and Hartl, F. (2018). Building a reproducible machine learning pipeline. arXiv [Preprint]. arXiv:1810.04570. doi: 10.48550/arXiv.1810.04570
Technopolis (2020). Science europe study on research assessment practices. Zenodo. doi: 10.5281/ZENODO.4915998
The Galaxy Community Abueg, L. A. L. Afgan E. Allart O. Awan A. H. Bacon W. A. . (2024). The Galaxy platform for accessible, reproducible, and collaborative data analyses: 2024 update. Nucleic Acids Res. 52, W83–W94. doi: 10.1093/nar/gkae410
The Turing Way Community (2022). The Turing Way: A Handbook for Reproducible, Ethical and Collaborative Research. doi: 10.5281/ZENODO.3233853
Trisovic, A., Durbin, P., Schlatter, T., Durand, G., Barbosa, S., Brooke, D., et al. (2020). “Advancing computational reproducibility in the dataverse data repository platform,” in Proceedings of the 3rd International Workshop on Practical Reproducible Evaluation of Computer Systems (Stockholm: ACM), 15–20. doi: 10.1145/3391800.3398173
Turkyilmaz-van der Velden, Y., Dintzner, N., and Teperek, M. (2020). Reproducibility starts from you today. Patterns 1:100099. doi: 10.1016/j.patter.2020.100099
Vanschoren, J., Braun, M., and Ong, C. S. (2014). Open Science in Machine Learning. Implementing Reproducible Research.
Vasilevsky, N. A., Minnier, J., Haendel, M. A., and Champieux, R. E. (2017). Reproducible and reusable research: are journal data sharing policies meeting the mark? PeerJ 5:e3208. doi: 10.7717/peerj.3208
Vasyukov, A., and Petrov, I. (2018). Using computing containers and continuous integration to improve numerical research reproducibility. Int. J. Comput. 30, 27–33. Available at: https://ijcjournal.org/index.php/InternationalJournalOfComputer/article/view/1249
Vitek, J., and Kalibera, T. (2011). “Repeatability, reproducibility, and rigor in systems research,” in Proceedings of the ninth ACM international conference on Embedded software (Taipei: ACM), 33–38. doi: 10.1145/2038642.2038650
Wang, J. Kuo, T.-y., Li, L., and Zeller, A. (2020). “Restoring reproducibility of Jupyter notebooks,” in Proceedings of the ACM/IEEE 42nd International Conference on Software Engineering: Companion Proceedings (Seoul: ACM), 288–289. doi: 10.1145/3377812.3390803
Willis, C., and Stodden, V. (2020). Trust but verify: how to leverage policies, workflows, and infrastructure to ensure computational reproducibility in publication. Harvard Data Sci. Rev. 2. doi: 10.1162/99608f92.25982dcf
Wittek, K., Wittek, N., Lawton, J., Dohndorf, I., Weinert, A., Ionita, A., et al. (2021). “A blockchain-based approach to provenance and reproducibility in research workflows,” in 2021 IEEE International Conference on Blockchain and Cryptocurrency (ICBC) (Sydney, NSW: IEEE), 1–6. doi: 10.1109/ICBC51069.2021.9461139
Wolke, A., Bichler, M., Chirigati, F., and Steeves, V. (2016). Reproducible experiments on dynamic resource allocation in cloud data centers. Inf. Syst. 59, 98–101. doi: 10.1016/j.is.2015.12.004
Wonsil, J., Boufford, N., Agrawal, P., Chen, C., Cui, T., Sivaram, A., et al. (2023a). Reproducibility as a service. Softw. Pract. Exp. 53, 1543–1571. doi: 10.1002/spe.3202
Wonsil, J., Sullivan, J., Seltzer, M., and Pocock, A. (2023b). “Integrated reproducibility with self-describing machine learning models,” in Proceedings of the 2023 ACM Conference on Reproducibility and Replicability (Santa Cruz, CA: ACM), 1–14. doi: 10.1145/3589806.3600039
Keywords: repeatability, reproducibility, replicability, reusability, data science ML/AI, RaaS, scientific journal, trustworthy research
Citation: Hernandez JA and Colom M (2025) Reproducible research policies and software/data management in scientific computing journals: a survey, discussion, and perspectives. Front. Comput. Sci. 6:1491823. doi: 10.3389/fcomp.2024.1491823
Received: 13 September 2024; Accepted: 06 December 2024;
Published: 15 January 2025.
Edited by:
Guiming Luo, Tsinghua University, ChinaReviewed by:
Shadi Abudalfa, King Fahd University of Petroleum and Minerals, Saudi ArabiaKazem Taghva, University of Nevada, Las Vegas, United States
Leonardo Candela, National Research Council (CNR), Italy
Copyright © 2025 Hernandez and Colom. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jose Armando Hernandez, am9zZS5oZXJuYW5kZXpfZ29uemFsZXpAZW5zLXBhcmlzLXNhY2xheS5mcg==
 Jose Armando Hernandez
Jose Armando Hernandez Miguel Colom
Miguel Colom