- 1Department of Medicine and Surgery, Pediatric Unit, University of Perugia, Perugia, Italy
- 2European Biomedical Research Institute of Salerno (EBRIS), Salerno, Italy
- 3Department of Neurosciences, Mental Health and Sensory Organs (NESMOS), Faculty of Medicine and Psychology, Sapienza University of Rome, Pediatric Unit, Sant'Andrea University Hospital, Rome, Italy
The gut barrier encompasses several interactive, physical, and functional components, such as the gut microbiota, the mucus layer, the epithelial layer and the gut mucosal immunity. All these contribute to homeostasis in a well-regulated manner. Nevertheless, this frail balance might be disrupted for instance by westernized dietary habits, infections, pollution or exposure to antibiotics, thus diminishing protective immunity and leading to the onset of chronic diseases. Several gaps of knowledge still exist as regards this multi-level interaction. In this review we aim to summarize current evidence linking food antigens, microbiota and gut permeability interference in diverse disease conditions such as celiac disease (CeD), non-celiac wheat sensitivity (NCWS), food allergies (FA), eosinophilic gastrointestinal disorder (EOGID) and irritable bowel syndrome (IBS). Specific food elimination diets are recommended for CeD, NCWS, FA and in some cases for EOGID. Undoubtfully, each of these conditions is very different and quite unique, albeit food antigens/compounds, intestinal permeability and specific microbiota signatures orchestrate immune response and decide clinical outcomes for all of them.
Introduction
The gut barrier is a selective filter for the gastrointestinal (GI) tract, which is exposed to a wide range of external antigens, including food, microbes, and toxins (1). This barrier encompasses several interactive, physical, and functional components: (1) the gut microbiota; (2) the mucus layer; (3) the epithelial layer; (4) the mucosal immunity. Each unit is key in shielding the host from harmful ingested antigens. The microbial barrier, or gut microbiota, consists of trillions of microorganisms and acts as a significant protective shield against pathogens. It contributes to host health, facilitating nutrient absorption, metabolism, and immune regulation. This multitude of microorganisms can induce immune cell maturation and appropriate response for host defense (2). The trajectories of microbiota composition vary during life. Neonatal gut microbiota is composed predominantly by anaerobic bacteria, with a greater abundance of Bifidobacteria in those who are breastfed (3).
After the first few years of life, the microbiota becomes more stable and “adult-like” in its composition (4), but it could be still perturbated by dietary choices, hormones, antibiotics and pollution (5–8). The resistance and resilience of the gut microbiota depends on various reasons: - diversity; - functional redundancy;—adaptive capacity;—intermicrobial interactions;—host immunity;—diet;—environmental consistency, which means the stability of nutrients, pH and oxygen level;—gut habitat complexity, i.e., having different anatomical and functional units such as folds and crypts allowing more stable bacterial reservoirs (9).
The mucus layer consists of an extracellular coat located outside the intestinal epithelial cells (IECs). Mucus is composed of mucins, principally Muc17 in the small intestine and Muc2 in the colon, which are glycoproteins overlapping each other in a net-like fashion (10).
Epithelial cells lining the intestine are vital for maintaining a tight interplay between the luminal environment and the host. This epithelium comprises several cell types derived from stem cells that constantly replenish differentiated intestinal epithelial cells (IECs), eventually shedding into the lumen (11). IEC subsets are broadly categorized into absorptive and secretory epithelial lineages. Most epithelial cells are absorptive enterocytes, albeit their active involvement in mucosal immunology needs to be further elucidated. Secretory IECs include: -enteroendocrine cells; -Paneth cells, which sustain stem cells and secrete antimicrobial peptides; -goblet cells, which deploy mucus production; -tuft cells, which detect luminal signals and promote type 2 immunity (12, 13). Differently from absorptive and secretory cells, Microfold (M) cells constitute a unique IEC subset that allow the trans-cytoplasmatic transportation of luminal antigens to immune cells into the lamina propria (14).
Altogether, epithelial cells throughout the intestinal tract orchestrate nutrient absorption, pathogen defense, and immune regulation. Antigens that reach the intestinal epithelium can be transported by different routes depending on their size and solubility. Soluble antigens are absorbed by enterocytes and transported mainly via the transcellular route. Under homeostatic conditions tight junctions between enterocytes prevent the paracellular passing of antigens, preserving the cell polarity necessary for directional functions. Several proteins, such as angulins, occludins, and claudins constitute these junctions (15).
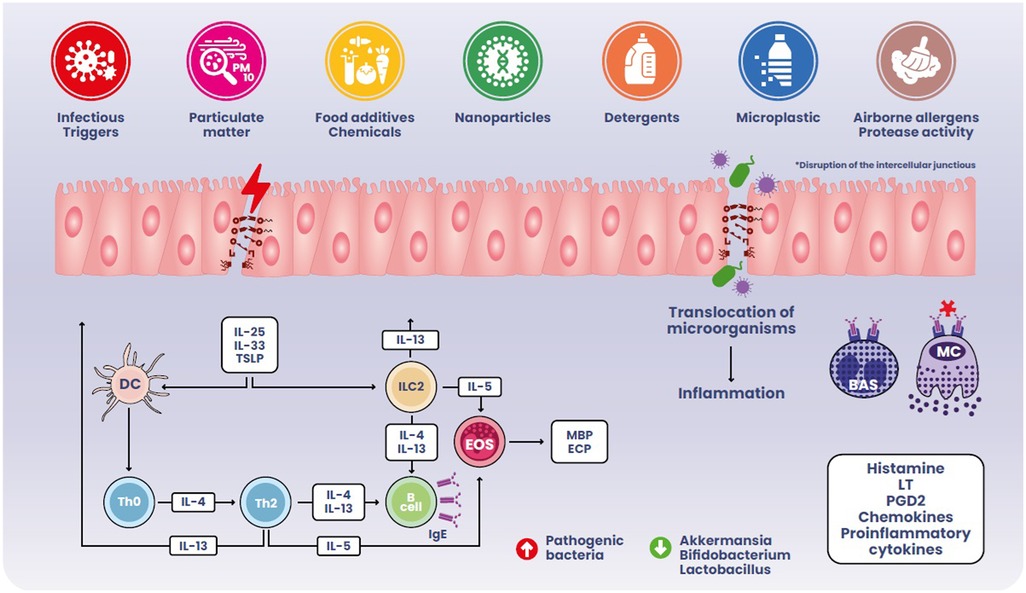
The presence of numerous foreign antigens in the intestinal lumen implies the accumulation of immune cell populations within the GI tract. Nonetheless, immune cell subsets in the intestine coexist to uphold homeostasis towards various stimuli in a well-regulated manner. Disrupting the mechanisms underlying this balance diminishes protective immunity and leads to the onset of chronic inflammatory diseases as exemplified in Figure 1.
In this review we aim to summarize current evidence linking food antigens, intestinal microbiota and gut permeability interference in diverse disease conditions such as celiac disease (CeD), non-celiac wheat sensitivity (NCWS), food allergies (FA), eosinophilic gastrointestinal disorder (EOGID) and irritable bowel syndrome (IBS).
Food antigens clashing into the barrier
Gluten, celiac disease and non-celiac wheat sensitivity
Celiac disease (CeD) is a systemic immunological disorder triggered by the intake of gluten and related prolamins in genetically at-risk individuals, i.e., those bearing HLA-DQ2 or HLA-DQ8 haplotypes (16). It presents with a wide range of clinical symptoms, the presence of CeD-specific antibodies and immune-mediated intestinal damage. While CeD can occur at any age, most at-risk individuals are diagnosed during their early childhood, typically within the first five years, as shown also by several prospective studies (17–19). Due to its diverse systemic and malabsorptive symptoms, CeD is often described as a “clinical chameleon,” which can pose challenges for primary care practitioners (20). Currently, many individuals with CeD experience few symptoms or predominantly exhibit extraintestinal issues, while only a minority manifests the “classical” signs of malabsorption characterized by weight loss, failure to thrive, and persistent diarrhea (21). The only effective treatment for CeD is a strict gluten-free diet (GFD), albeit several investigational drugs are in phase 2 clinical trials for non-dietary/complementary management strategies (22).
In CeD patients, gluten-dependent increased intestinal permeability is a paradigm of the disease, priming and perpetrating intestinal damage on a gluten-containing diet. Zonulin is a gastrointestinal paracrine hormone that, together with other growth factors, negatively affects intercellular integrity by disassembling tight junctions (23).
Among the various potential stimuli in the intestinal lumen that can induce zonulin release, small intestinal exposure to microorganisms and gluten have been identified as the most potent triggers (24, 25). From an evolutionary perspective, the zonulin-induced opening of the paracellular route might act as an ultimate defensive process that washes out microorganisms, working alongside other similar mechanisms, such as the IL-22-mediated modifications of intestinal permeability against bacteria (26).
Gliadins are gluten-complex proteins usually rich in prolines and glutamines, which are not digested by intestinal enzymes. Few peptides that residue from partial digestion of gliadins impact intestinal barrier function by triggering the release of zonulin upon interaction with the chemokine receptor CXCR3 (27). Gliadin thus rapidly and temporarily enhances zonulin-dependent paracellular permeability of the gut, regardless of disease status (28). Notably, once the GFD is commenced, the intestinal paracellular permeability is switched off (29).
Genetic studies have supported the role of the paracellular pathway in gluten transport within the lamina propria, which have found a connection between specific tight junction-related genes and CeD (30, 31). In CeD, the loss of barrier function is the first step towards the loss of tolerance to gluten and the consequent damage to the intestinal mucosa. This is witnessed by the fact that healthy first-degree relatives of patients with CeD might present with increased intestinal permeability (32). A preliminary study from the CD-GEMM (Celiac Disease Genomic Environmental Microbiome and Metabolomic Study) prospective cohort suggests that in infancy the onset of CeD is preceded by an increase of zonulin levels months before the disease manifests. This rise is associated with a medical history of multiple courses of antibiotics, thus linking microbiota disturbances to the subsequent loss of immune tolerance towards gluten (33). In addition, infections by enteroviruses and rotaviruses have been accounted as putative triggers for CeD onset in longitudinal studies (34, 35). Barrier breach and antigen molecular mimicry could be hypothesized as underling mechanisms preceding disease inception (36, 37).
Within the spectrum of gluten-dependent disorders, another condition is the so called non-coeliac wheat sensitivity (NCWS), currently defined as a wide set of intestinal and extraintestinal symptoms after gluten consumption in subjects for whom CeD and wheat allergy have been thoroughly excluded (38).
However, with except of symptoms such as brain fog, fatigue, and joint pain, several features of NCWS overlap with functional GI disorders, particularly IBS, thus posing a challenging task for physicians who deal with these patients. To discriminate the two conditions, Barbaro et al. have demonstrated that increased serum levels of zonulin are more frequently present in NCWS rather than in IBS, therefore highlighting within a clinical framework the role of intestinal permeability in this condition (39). In addition, Ahmed et al. very recently reported that the presence of conventional anti-gliadin antibodies (AGA) is strongly predictive of NCWS among IBS patients (40). Since AGA have been previously associated with increased intestinal permeability (41), this evidence reinforces the role of “leaky” gut in NCWS.
Another in vivo study highlighted intestinal epithelial defects at fluorescein-based confocal microscopy a few minutes after wheat exposure in several patients with IBS-like symptoms. This was linked to a heightened expression of the pore-forming protein claudin-2, thus suggesting that a leaky gut seems to coexist with NCWS misdiagnosed as IBS (42). Furthermore, another group has reported elevated serum levels of lipopolysaccharide-binding protein in patients with NCWS, advocating an increased passage of microbial components from the lumen to the systemic circulation. Notably, this phenomenon seems to revert on a GFD (43).
Notwithstanding these data from pre-clinical and clinical studies, it should be considered that other translational studies on this topic have shown contrasting results.
For example, Sapone et al. showed similar permeability parameters in the small bowel of NCWS individuals and patients with dyspepsia using the lactulose/mannitol ratio (44). Another study measured the increase in permeability assessed as modification of transepithelial electrical resistance from ex vivo intestinal biopsies after exposure to gliadin: no increase was found in those from NCWS subjects compared with those from healthy controls (45). Nevertheless, it should be considered that both these studies were affected by small sample size and that both methods used to determine intestinal permeability are pretty variable.
Exposure to wheat components other than gluten are currently under scrutiny as possible etiology drivers for NCWS. Amylase-trypsin inhibitors (ATIs) are water-soluble globulins from wheat that play a role in grain maturation, carbohydrate storage and defense from parasites. They are major stimulators of innate immune cells but initially they were not considered to directly affect intestinal permeability (46).
However, in a model of specific pathogen-free mice lacking Toll-like receptors, a Canadian group has reported that ATIs can increase intestinal permeability assessed by Ussing chambers (47). Of note, tissue conductance was ameliorated when specific strains of Lactobacilli expressing enzymes for digesting ATIs were provided in the medium. This experiment has shown an exciting attempt to counteract barrier dysfunction in gluten-related disorders. Nevertheless, for the time being evidence has been scant regarding the possibility of modulating intestinal permeability through other probiotic interventions.
There is extensive literature on numerous strains of Lactobacilli and Bifidobacteria possessing proteolytic enzyme machinery for gluten. Still, these have been tested mainly for improving clinical outcomes, and they were not assessed as regards peptide residual interference on intestinal permeability (48). Fewer studies focused on probiotics and intestinal permeability in the setting of experimental models for CeD. An interesting study from Lamas et al. highlighted in NOD/DQ8 mice the indirect action of L. reuteri that reduced intestinal permeability by enhancing the tryptophan-derivates pathways (49). Moreover, on Caco-2 cells, a specific strain of Bifidobacterium (B. lactis) could inhibit the gliadin-induced increase in epithelial permeability, and this effect of the strain was clearly dose-dependent (50). Another preclinical study on L. rhamnosus GG showed that this latter strain sustained the expression of the intercellular junction proteins, thus contrasting the increase in intestinal permeability in the context of murine enteropathy induced by pepsin-trypsin-digested gliadins (51).
Food allergens: food allergy and eosinophilic GI disease
Food allergy (FA) is defined as “an adverse health effect arising from a specific immune response that occurs reproducibly on exposure to a given food”. FA is a growing public health concern, with increasing prevalence in Western countries. It currently affects 8% of children and 3% of adults, with prevalence rising every year and significantly varying depending on the method of assessment (self-reported FA vs. gold standard oral food challenge) (52–54). According to the pathophysiologic immune mechanisms, it can be classified into IgE-mediated, non-IgE-mediated (cell-mediated) or mixed (IgE and cell-mediated) (55, 56).
Post-translational modifications of antigens, including processes such as glycosylation, phosphorylation, acetylation and hydroxylation, may alter the allergenic potential of food proteins (57). These modifications may enhance protein stability, modify epitopes that immune cells recognize or affect how proteins are processed and presented to the immune system. As a result, the same protein can elicit different immune responses depending on its post-translationally modified forms, contributing to the variability observed in allergic reactions among individuals. For example, glycosylation of casein hinders its full digestion, therefore its allergenicity is enhanced (58). As regards peanut allergy, boiled vs. roasted peanut proteins show a marked difference in their ability to trigger IgE response. Boiling tends to denature some of the proteins responsible for allergic reactions and this can lead to a less severe allergic response in sensitive individuals (59). In contrast, roasting provokes the Maillard reaction, which in its early phases is a glycation of proteins; glycated peanut proteins are more likely to trigger allergic reactions due to a preferential recognition by dendritic cells (60).
Although genetic factors play a role, FAs are increasing in prevalence at a higher rate that cannot be explained only by genetics (61).
It has been known for years that many environmental factors are associated with the development of FAs, such as caesarean section, formula feeding, early antibiotic exposure, not having siblings or pets; nevertheless, the connection between epidemiological data and the underlying immune mechanism remains elusive. This gap is being filled by increasing evidence about gut microbiota as one of the main actors in the complex mechanism of sensitization. Indeed, the previous mentioned epidemiological factors have been shown as culprits in the development of dysbiosis (62–67).
The gut microbiota, which consists of trillions of microorganisms including bacteria, viruses, fungi, and archaea, plays a vital role in maintaining immune homeostasis and protecting the epithelial barrier.
Human milk microbiota and the birth canal in vaginal delivery are the two main sources through which infants can acquire microbes colonizing gut lumen. Moreover, children ingest every day with breast milk an amount of bacteria ranging between 1 × 105 and 1 × 107, which is almost 30% of whole infant bacteria pool (68).
Human milk could play a pivotal role in preventing or promoting allergy development by modulating gut microbiota biodiversity and gut barrier function due to the activity of molecules such as milk oligosaccharides, glycomacropeptides, lactoferrin, defensins, and other metabolites (69). Human milk oligosaccharides (HMO) are non-digestible oligosaccharides having a very important probiotic role: they inhibit bacterial adhesion to intestinal mucosa surface and prevent colonization of certain bacteria strains (70). Casein glycomacropeptides, defensins and tryptophan metabolites have also shown specific antimicrobial properties (71). Finally, lactoferrin, a protein that binds and separates iron from bacterial pathogens, has both antiviral and antibacterial activities (72).
The composition of human milk microbiota may be influenced by many factors such as stage of lactation, maternal body mass index (BMI), age and diet, geographical location, socioeconomic status, use of antibiotics and probiotics during pregnancy (73–77).
As regards specific microbiota signatures and atopy, Lachnospiraceae have shown to be well represented in children allergic to cow's milk (78); moreover, Canadian Health Infant longitudinal development (CHILD) study has shown that a reduction of genera such as Lachnospira, Veilonella, Faecalibacterium and Rothia, and an abundance of Clostridium difficile and Staphylococcus aureus are linked with asthma development later in life (79).
An early gut colonization by Bacteroides fragilis expressing polysaccharide A could inhibit Treg proliferation, therefore predisposing towards allergy (80).
It has been also highlighted that early colonization with Clostridium genera is related to an increased IL-22 secretion by group-3 innate lymphoid cell and T-helper 17 and specific IgE production in children with FA (81).
The importance of microbiota-host relationship in allergy is witnessed by murine models of germ-free mice displaying elevated blood levels of IgE. Instead, after microbial colonization which starts between birth and the first week, IgE production is completely inhibited (82).
Among the important functions that intestinal microbiota plays in the host, its contribution to immune tolerance for orally administered antigens is key. Oral tolerance is a state of systemic unresponsiveness that should be the default response to food antigens in the GI tract. This is an active process that begins with the uptake of potential food antigens by immune cells of the gut-associated lymphoid tissue (GALT) in the small intestine (83). Antigens are then degraded into small peptides inside vesicles, loaded into MHC-II in endosomes and released to be taken up by CD103 + dendritic cells (84, 85). Antigens with resistance to proteolysis can reach the basolateral membrane in an intact form (86). M cells can endocytose antigens and myeloid cells such as dendritic cells and/or macrophages may directly capture potential food antigens from the gut lumen by extending a process through a tight junction (periscoping behaviour) or by extending a process through a transcellular pore in an M cell (87, 88). These dendritic cells can then migrate from the lamina propria to the draining lymph nodes where the dendritic cells express transforming growth factor-β (TGFβ) and retinoic acid, inducing naive T cells to differentiate into regulatory T (Treg) cells; therefore, the cell-mediated trafficking of antigen to the secondary lymphoid tissue promotes the establishment of tolerance (89, 90). Tregs regulate the immune response by producing inhibitory mediators, such as IL-10 and TGF-β (91).
Dendritic cells also imprint GI homing capacity, allowing the recently primed Tregs to home back to the lamina propria where they interact with macrophages that produce IL-10 and expand. In addition, anergy and T cell depletion have also been shown as mechanisms of oral tolerance, with anergy referring to T-cell unresponsiveness to the antigen and depletion to the apoptosis of antigen-specific T cells. These two mechanisms cooperate depending on the dose of antigen exposure: high dose antigen exposure induces anergy or depletion while low dose antigen leads to induction of Tregs (92).
The microbiota plays a key role in the development of oral tolerance by multiple ways. Intestinal bacteria stimulate the production of mucus glycoproteins, sealing the intestinal barrier and protecting the epithelium from the growth of pathogenetic bacteria. In addition, they have a role in the modulation of Th1/Th2 response in favor of a tolerogenic immune response and in the activation of Tregs: these events are mediated by metabolites such as butyrate, a short-chain fatty acid (SCFA) produced by the fermentation of dietary fiber in the colon, which has a strong immunoregulatory effect (93, 94).
On the other hand, several studies in mice have demonstrated an association between compositional and functional changes of the gut microbiota, also known as dysbiosis, and development and progression of FA (95, 96). Early studies showed that germ-free mice were unable to achieve oral tolerance to food allergens (97). Recent studies have further shown that transferring gut microbiota from patients with FA to germ-free mice can transmit susceptibility to FA (98). Conversely, germ-free mice colonized with bacteria from healthy infants were protected against anaphylactic responses to a cow's milk allergen (99). Certain microbial orders, including Clostridiales and Lactobacillales, have been related to suppression of FA in mouse studies (100). Similarly, Bacteroidales and Enterobacteriales have been described to have both beneficial and detrimental effects with regards to FA (101, 102).
The hypothesis on gut dysbiosis preceding FA relies on several basic science and clinical studies showing proinflammatory microbiota changes nurturing a chronic, low-grade, inflammation (103–120). Moreover, intestinal dysbiosis cooperates with a leaky gut barrier in FA pathogenesis (121–123). External factors (particularly the consumption of ultra-processed foods, but also other dietary factors, infections, inflammatory bowel diseases etc.) can influence both the gut microbiota and epithelial barriers leading to a pro-inflammatory milieu: when the epithelial barrier is damaged, antigens may cross freely, causing the release of pro-inflammatory epithelial-derived molecules like IL-25, IL-33 and thymic stromal lymphopoietin (TSLP), which promote T-naïve cell differentiation into Th2 cells, IgE class-switching, and tissue accumulation of mast cells and eosinophils (124). After sensitization to a food allergen, allergen-specific IgE antibodies bind to basophils and mast cells surface IgE receptors. A subsequent exposure to the allergen can cause these cells to immediately release histamine and other proinflammatory mediators, such as leukotrienes and prostaglandins, that lead to tissue inflammation and recruitment of inflammatory cells, worsening gut permeability and amplifying type-2 inflammation (125). Several studies have identified a mutual link between increased intestinal permeability and FA: not only the establishment of FA is favored by a leaky intestinal barrier, but also the allergic response further impacts intestinal permeability allowing the translocation of allergenic molecules (126–128).
Complementary food introduction is known to influence gut microbiota composition and future health outcomes. The first 1,000 days of life indeed seem to represent the critical window of opportunity for microbiota modulation. A delayed introduction of solid food could cause a lag in microbial maturation increasing susceptibility to allergies and obesity (129). On the other hand, an earlier introduction could expose infants to potential pathogens and allergens. Therefore, the timing of solid food introduction should be balanced between these risks and benefits (130, 131). Particularly, a meta-analysis on the timing of introduction of egg and peanut in infant diet showed indication to introduce egg at age 4–6 months and peanut at age 4–11 months (in an age-appropriate form to avoid risk of inhalation), as this behavior was associated with reduced egg and peanut allergy (132). Notably, this is strikingly conflicting with the previous recommendation to delay the introduction of allergenic foods to the infant diet to prevent sensitization (133).
An earlier introduction of multiple allergenic foods was associated with reduced IgE-mediated allergy, consistent with the findings of the Preventing Atopic Dermatitis and Allergies in Children (PreventADALL) trial. Even though safety data were generally reassuring, their findings were limited by high rates of withdrawal from the intervention (134).
Breast-feeding and diet diversity promote a healthy microbiota not only during the weaning period, but also over the whole critical time of the two first years of life. After this period, the gut microbiome tends to acquire an adult-like configuration with distinct microbial community composition and functions. A varied and healthy diet appears to confer a positive modulation of gut microbiota: a significant association between higher diet diversity and a lower prevalence of parent-reported, doctor diagnosed food allergy has been demonstrated (135).
The immunologic basis explaining how diet diversity potentially affects allergy outcomes may be mediated by the induction of tolerance mechanisms including T and B regulatory cells, immune regulatory cytokines and suppressed IgE antibodies.
A more diverse diet may indirectly affect tolerance development via an effect on the microbiota as increased diet diversity leads to increased microbial diversity in infants at weaning. Moreover, a more diverse diet may also lead to exposure to different food antigens that impact on the development of immune tolerance, though this may be “low dose” exposure, supporting recent randomized controlled trials regarding early allergen introduction (136).
Though there is a need to harmonize study methods and define diet diversity for studying adequate dietary intake and allergy outcomes, diet seems to be a modifiable factor that can be used to prevent or manage allergic disease (137).
Restrictive dietary regimens of children with specific diseases (FA, inborn errors of metabolism, CeD, etc.) may alter the gut microbiota triggering the overgrowth of intestinal inflammation. Elimination diets that decrease SCFA-producing bacteria, such as the gluten-free diet, phenylketonuria diet, and ketogenic diets and those with overall low consumption of a plant-based products may have negative effects on the microbiota (138–141).
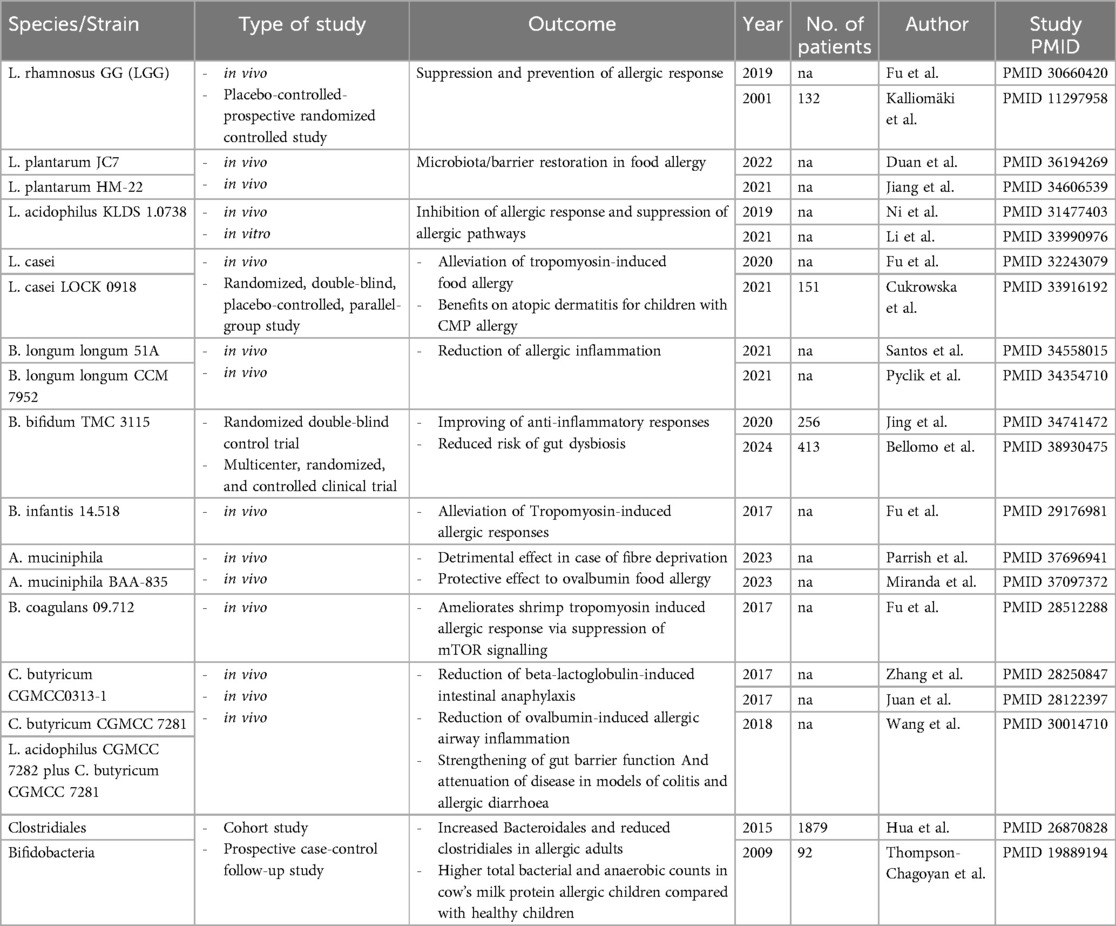
Despite a significant impact on some pre-clinical and clinical aspects, current evidence does still not support a clear recommendation on the use of probiotics in the prevention or treatment of FA. Clear information on the specific strain, dosage, and adequate duration of therapy is lacking. Moreover, future strain-specific studies should ideally take into account that gut microbiota also reflects the integrity of the gut barrier and any supplementation approach should contextually aim for its restoration. A summary of specific bacteria/probiotics and their impact on food allergies is presented in Table 1.
Eosinophilic gastrointestinal diseases (EGIDs) are a group of chronic inflammatory disorders characterized by eosinophilic infiltration into various segments of the GI tract, including eosinophilic esophagitis (EoE), eosinophilic gastritis (EoG), and eosinophilic colitis (EoC) (142). The pathophysiology of these conditions is multifactorial, involving complex interactions between environmental triggers, such as food antigens, immune responses, and alterations in the epithelial barrier and the gut microbiota (143, 144). Recent studies have provided both pre-clinical and clinical evidence that these diseases are associated with significant changes in gut permeability, membrane integrity, and microbita composition (145). These alterations appear to play a pivotal role in disease progression and symptom manifestation.
One of the critical components of EGIDs is the dysfunction of the intestinal epithelial barrier, which is often referred to the aforementioned leaky gut. This term describes the increased permeability of the gut lining. In EGIDs, intestinal barrier is compromised, leading to increased intestinal permeability (146). Pre-clinical models have demonstrated that this increased permeability may facilitate the translocation of food antigens and bacterial products into the lamina propria, thereby triggering or exacerbating local immune responses. Specifically, it has been shown that eosinophils, key effector cells in these diseases, are recruited to the gut in response to antigen exposure, where they release pro-inflammatory mediators such as cytokines, chemokines, and granule proteins (including major basic protein and eosinophil-derived neurotoxin), which further disrupt epithelial integrity and amplify the inflammatory response.
Clinically, patients with EGIDs exhibit a range of symptoms that are closely linked to this compromised epithelial barrier. Small bowel permeability is overall increased in patients with active EoE, and is normal in patients with EoE in remission when compared to healthy controls (147). In eosinophilic gastritis, patients may present with abdominal pain, nausea, vomiting, and malnutrition, symptoms that are also likely exacerbated by a dysfunctional epithelial barrier that allows for abnormal exposure to luminal antigens and microbial products. Endoscopic evaluations in these patients often reveal mucosal erythema, edema, and furrowing, further evidence of the inflammation and tissue damage caused by increased permeability (148).
A growing body of literature also highlights the significant role of the microbiome in the pathogenesis of EGIDs. Alterations in the composition and diversity of the gut microbiota, known as dysbiosis, have been also implicated in EGIDs. In the context of EoE, for instance, several studies have reported significant differences in the esophageal microbiota of patients with the disease compared to healthy controls. Specifically, there is evidence of a relative abundance of Streptococcus species in the esophagus of EoE patients, which may contribute to disease pathogenesis by promoting a pro-inflammatory environment that favors eosinophil recruitment and activation (149).
The increase in permeability is believed to result from a disruption in the regulation of tight junction proteins, specifically claudin 1 (CLDN1) and zonula occludens 1 (ZO-1) (150). However, it is possible that the compromised mucosal integrity observed in EoE may be secondary to byproducts of the inflammatory infiltrate, suggesting that it could be a consequence rather than a cause of the inflammation. Previous research has indicated that the inflammatory cytokines associated with EoE, including IL-13 and IL-5, have the ability to downregulate key barrier proteins such as desmoglein (DSG1) and filaggrin (FLG) (151, 152). Interestingly, in the context of atopic dermatitis, similar barrier dysfunctions have been observed not only in the skin but also in the small intestine (153). This raises an important question: is the esophagus the only site of allergen penetration in EoE, or could allergens also enter through the small intestine, potentially triggering or exacerbating inflammation in the esophagus? The current evidence on whether the duodenum plays a role in EoE is conflicting. One study in adults found increased small intestinal permeability (147), while another study in children did not show such findings (154). Regardless, the integrity of the mucosa in both the esophagus and possibly the small intestine may be crucial to understanding the disease's pathology, and restoring this integrity may be key to achieving complete histological remission. Elemental diet studies, which exclude food allergens, have demonstrated significant clinical and pathological improvements in EoE (155, 156). However, the impact of eliminating food allergens on mucosal integrity has not yet been explored in these patients. In addition to changes in microbial composition, recent research has suggested that the function of the microbiota may also be altered in EGIDs (157). The gut microbiome is known to produce a variety of metabolites that influence both local and systemic immune responses. For example, SCFA have been shown to have anti-inflammatory effects and to promote the integrity of the epithelial barrier (158). However, in patients with EGIDs, there may be a reduction in the production of these beneficial metabolites, leading to a dysregulated immune response and further compromising barrier function. This hypothesis is supported by a study demonstrating that the dominant taxa in patients with EGIDs was increased (Streptococcus in esophagus; Prevotella in stomach) (149). Specific taxa were associated with active disease for both EoE (Streptococcus, Gemella) and EoG (Leptotrichia), although highly individualized communities likely impacted statistical testing. Stool analyses did not correlate with bacterial communities found in mucosal biopsy samples and was similar in patients and controls. Therefore, further study is needed to determine if therapeutic interventions contribute to the observed community differences.
Nevertheless, pre-clinical models of EGIDs have provided valuable insights into the mechanisms by which microbiota alterations contribute to disease pathogenesis (159). In mouse models of EoE, for example, it has been shown that antibiotic treatment, which disrupts the gut microbiota, can exacerbate disease symptoms and lead to increased eosinophilic infiltration of the esophagus. Conversely, the administration of probiotics or microbiota-targeted therapies has been found to reduce eosinophil levels and improve epithelial barrier function, suggesting that modulating the gut microbiota may be a promising therapeutic strategy for patients with EGIDs (160, 161).
The clinical implications of these findings are striking. Current treatment options for EGIDs primarily focus on suppressing the immune response and reducing eosinophilic inflammation through the use of corticosteroids, biologic agents (such as anti-IL-5 and anti-IL-4Rα therapies), and dietary interventions (such as elimination diets that remove suspected food triggers). However, emerging evidence suggests that targeting the gut microbiota may represent a novel therapeutic approach for these diseases (162). For example, clinical trials are currently underway to evaluate the efficacy of fecal microbiota transplantation (FMT) and other microbiota-targeted therapies in patients with EoE and other EGIDs. These therapies aim to restore a healthy microbial balance in the gut, thereby improving epithelial barrier function and reducing inflammation.
The need for further research is evident, particularly in exploring how dietary interventions and microbiota-modulating therapies affect barrier integrity and inflammation. Understanding the mechanisms by which allergens and microbial products interact with the immune system in these diseases will be crucial for developing more effective, targeted treatments aimed at both preventing eosinophil infiltration and restoring mucosal health.
Other food components as barrier disruptors: emulsifiers, plastic nanoparticles and advanced glycation end products
A higher consumption of processed foods in Western and Westernized countries has unfortunately been driven by the increase in productivity in modern societies. These foods are rich in food additives such as emulsifiers, plastic nanoparticles and AGEs (advanced glycation end products) which can all pose risks for human health (163).
Emulsifiers play a very important role in industrial food production, helping to combine immiscible ingredients such as oil and water, and can be commonly found in many processed foods. They are widely used in industrial processed food as they optimize food properties and help to exalt appearance, texture and flavor. Main food emulsifiers include guar gum (E412) found in cheese and other dairy products, lecithin (E32) commonly used in chocolate, xanthan gum (E415) usually found in mayonnaise, carrageenan (E407) found in ice creams/desserts and polysorbates (E432-436) found in ice cream, cakes and oils (164).
Polysorbate—60, polysorbate—80, carboxymethylcellulose (CMC), glyceryl monolaurate and carrageenan are mainly studied and, as shown in vitro and in animal models, they are known for their detrimental effect on human health and intestinal inflammation due to their impact on gut microbiota and gut permeability (165–177).
Emulsifiers may determine gut dysbiosis and reduce biodiversity of gut microbiota: many studies have proven an emulsifier-driven reduction of Lactobacillales and Clostridiales such as Faecalibacterium genera and specifically Faecalibacterium prausnitzii in inflammatory bowel disease (IBD); on the contrary, an increase of other specific commensals such as Escherichia, Roseburia, Bradyrhizobium and Turicibacter genera have been also described on a emulsifier-rich diet (178).
As shown in murine and human models, mucus layer thickness was reduced upon exposure to CMC and polysorbate 80, common emulsifiers used in foods, and there were also an increased bacterial penetration of the mucus and a shift to proinflammatory microbiota composition (179, 180).
It has been demonstrated that polysorbate 80 could increase the viscosity of mucus, due to smaller pores in mucus layer and those changes could accelerate movement of some bacteria as Escherichia Coli and modify interaction with other bacteria (181).
These changes in intestinal barrier could determine an increased permeability and a greater bacterial translocation. In fact, it has also been shown that exposure to carrageenan changed location of zonula occludens (Z0-1) from peripheral to more central position in cell membrane and altered actine filaments (182). Carrageenan also causes non-reversible modification in microbiota composition, bacterial density and enhancement of pro-inflammatory pathways (168, 183). These mechanisms could be crucial for determining intestinal inflammation at the onset or perpetrating IBDs.
In addition, CMC and polysorbate 80 also have a pro-inflammatory effects, as they induce an increased intestinal production of tumor necrosis factor (TNF)-α, IL-1 β, IL-6, IL-8 and activated B cells unleashed by Toll-like Receptor-4 (TLR4) (184).
As regards the possible involvement of emulsifiers in Th2-type adaptive immunity, in murine models a dietary exposure to CMC and polysorbate 80 increased IL-4 and IL-5 gene expression; specifically, the latter upregulated gene involved in histamine synthesis, IL-4, IL-5, IL-13, IL-33 and other genes involved in mast cell activation (185).
Another important role in barrier disfunction could be played by plastic nanoparticles (NP), which are commonly found in human food, due to an increased release of plastic waste in the environment. Among plastic particles, those with less than 100 nm are more easily internalized by enterocytes through endocytosis, avoiding tight junction defense system. Their role, especially for long term exposure, is still to be completely defined as a health risk factor, both alone or as a cofactor when epithelial barrier presents a dysfunction. While several inorganic plant-derived NP may reduce diversity of microbiota (186), other NP might interfere with immune system and gut-brain axis, through an exacerbation of oxidative stress or an increase in gut permeability (187, 188).
A summary of studies on specific food additives and their impact on gut permeability/inflammation/microbiota is shown in Table 2.
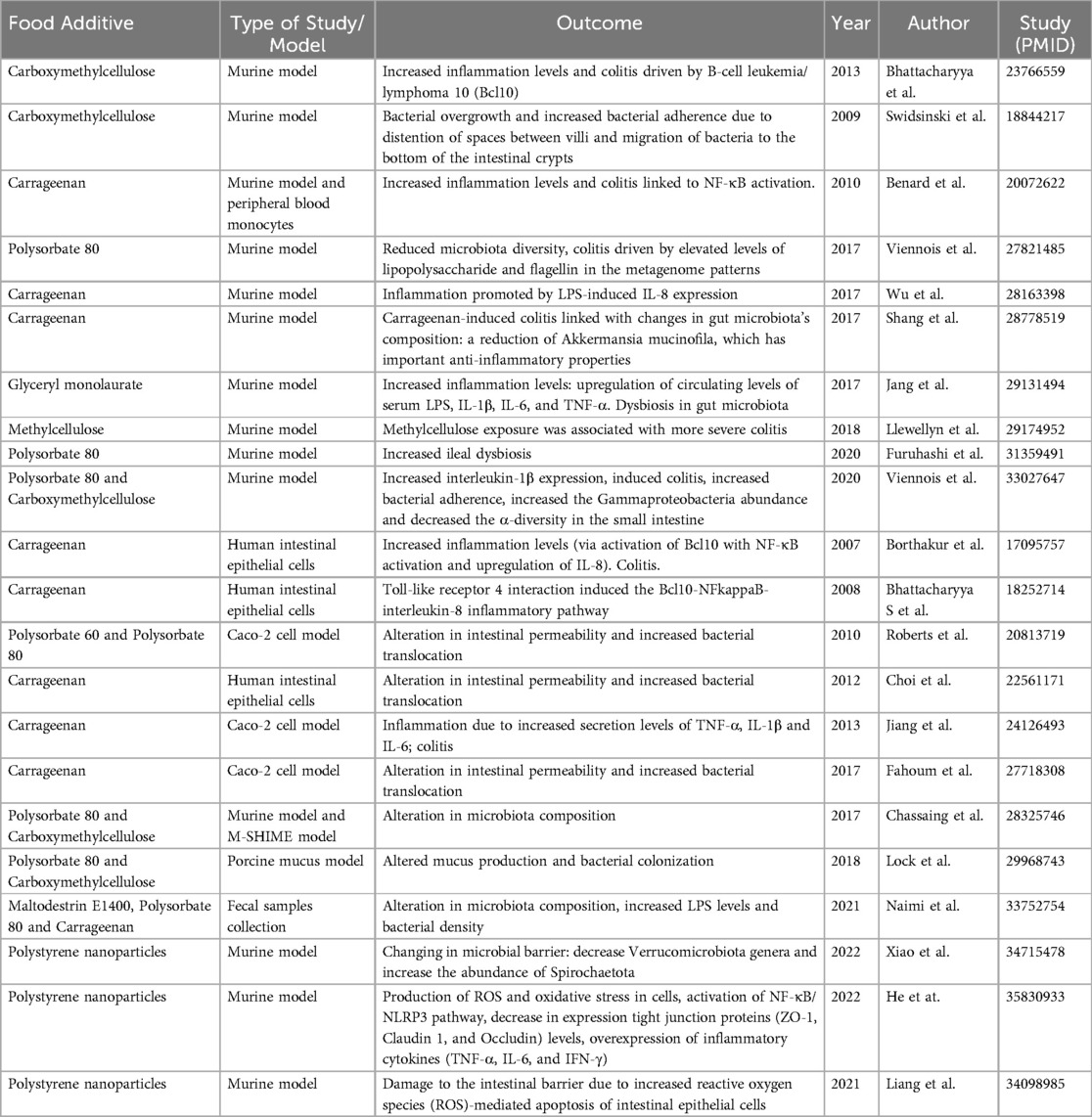
Table 2. Literature summary of specific food additives/nanoplastic compounds and their impact on gut permeability/inflammation/microbiome.
AGEs are a heterogenous group of compounds which are formed during the Maillard reaction, a non-enzymatic reaction or glycation that occurs between reducing-sugars and free aminoacidic group of proteins, peptides or free amino acids. This process is very common in the food industry because it helps to improve taste, consistency, color and aroma (189). Many human and animal studies point out that AGEs could be partially absorbed in the intestinal lumen and could distribute across various tissues and organs. About 60% of absorbed AGEs were found after 3 days especially in kidney and liver, but they were also found in heart, lung and spleen (190, 191). AGEs could also be endogenously produced. An accumulation of endogenous and exogenous AGEs could increase the production of ROS and determine cellular dysfunction and apoptosis. In fact, these compounds can interact with surface receptors of the antigen presenting cells and with RAGE, the receptor for advanced glycation end products (192–194). An exposure to AGEs could determine in human enterocytes a sizable increase in the production of IL-25 e IL-33, thus shaping immune response to Th2-pathways and allergic inflammation (195). Furthermore, it has been demonstrated that AGEs also cause a downregulation of tight junction protein expression, with a higher mucosal permeability and higher susceptibility to environmental triggers, amplifying the inflammatory response and potentially induce also epigenetic modifications (196). This alteration can theoretically facilitate the development of FA due to an increased exposure to food antigens. In addition, AGEs can also increase oxidative stress, as we know that the binding of their cellular receptor (RAGE) on enterocyte's surface leads to the activation of an extracellular kinase pathway (ERK 1 and 2) and NF-kb pathway, which bring to ROS production (197).
AGEs could also have an extra impact on the development of food allergy. Heilmann et al. have shown in a murine study that the specific glycation of food antigens could have allergenic potential by influencing T-cell immunogenicity. Their study showed an increased production of several cytokines (IL-2, IL-17, IFN-y) with glycated-ovalbumin compared to native ovalbumin (198).
The brain-gut-microbiome axis and its barrier
Most functions of GI physiology are influenced by neural control, both enteric nervous system (ENS) and autonomic nervous system (ANS). The influence on the gut is bidirectional, as the gut also sends information to the systems through complex pathways to achieve homeostasis, and alterations in this communication have been proven to be associated with diseases. Cross-communication between the brain and gut occurs through multiple biological axes involving the above-mentioned neural network, the neuroendocrine and immune system, and metabolic pathways, enabling bidirectional communication (199). The intestine physically communicates with the brain through 2 neuroanatomical pathways: the ANS and the vagus nerve (VN). The afferent VN innervates the mucosal and muscle layers of the gut, senses stimuli as luminal volume through mechanoreceptors or chemical stimuli (hormones, neurotransmitters, and metabolites) through chemoreceptors, and transmits these signals to the brain. On the other hand, the efferent VN transfers information from the central nervous system (CNS) to the gut. The other bidirectional exchange is guaranteed via the enteric nervous system (ENS) in the intestine, which is connected to the ANS and VN in the spinal cord and transmits information to the brainstem nuclei (200). Recently, the microbiome has emerged as an integral player in gut-brain communication, and the concept of a microbiome-gut-brain axis has substituted the previous gut-brain axis entity. Gut microbiota interplays with the human host in a mutually beneficial relationship, influencing the development and maturation of the immune, endocrine, and nervous systems. Microbiota composition depends on various factors including the physiological changes in the gastrointestinal tract such as motility and secretion, which are tightly regulated by the central nervous and enteric nervous systems (201). Changes in small intestine or large intestine motility, such as diarrhea or constipation, can cause dysbiosis (202). At the same time, several studies in germ-free animals have shown an abnormal gut motility and altered perception of inflammatory pain, stressing out the importance of microbiota in the regulation of gut motility (203, 204).
Moreover, the central nervous system can control microorganisms by affecting the secretion of serotonin, cytokines, catecholamines, and dynorphin from enteroendocrine cells, immune cells, and nerves, responsible for mucus production, secretory functions, mucosal immune responses, and direct changes in intestinal mucosal permeability (205). Despite the physical separation constituted by the intestinal epithelium, microbiota can also affect the brain-gut axis: the VN can sense microbial signals (i.e., bacterial metabolites) or can be influenced via microbiota-mediated modulation of enteroendocrine cells in the gut epithelium (206). Microbial metabolites include metabolites generated by the host and modified by the gut microbes, such as secondary bile acids, dietary product-derived molecules (compound K), or de novo synthesized compounds such as SCFAs. SCFAs may deeply affect gastrointestinal physiology, influencing peristalsis, visceral pain, epithelial proliferation, barrier function, host immunity, but also bacterial pathogenesis itself (207). SCFAs can epigenetically modulate epithelial cells; for example, butyrate enhances mucus production, activating the MUC2 promoter and enhancing histone acetylation in cell cultures. Moreover, SCFAs can bind to specific receptors like G- protein-coupled receptors (GPR), inducing the production of chemokines and cytokines. For example, butyrate induces the IL-18 excretion in epithelial cells by binding to GPR109a, thus protecting the colon against inflammation and carcinogenesis (208).
On the contrary, gut microbiota can also directly transmit signals to submucosal afferent nerve receptors which acts as microbe-associated molecular patterns (MAMPs) such as peptidoglycan or lipopolysaccharide (LPS) in the cell wall. MAMPs can activate various immune cells, especially innate immune cells such as macrophages, neutrophils, and dendritic cells. Inflammatory cytokines such as IL-1α, IL-1β, TNF-α, and IL-6 produced by these cells can pass through the blood-brain barrier and affect brain function by acting on receptors expressed on neurons and glial cells, especially microglia (209, 210). LPS can also directly affect the brain through the blood-brain barrier (211).
Gut microbiota can also influence the host's metabolic state and neuroendocrine system. For example, SCFAs produced by microbes regulate the expression and secretion of glucagon-like peptide-1 (GLP-1) through the free fatty acid receptor of L cells, controlling insulin release and appetite (212). In addition, secondary bile acids metabolized by microbes activate the G protein-coupled bile acid receptor (TGR5) of L cells to secrete peptide YY (PYY) and GLP-1 (213). PYY inhibits GI motility, slows food intake, decreases appetite, and increases energy consumption.
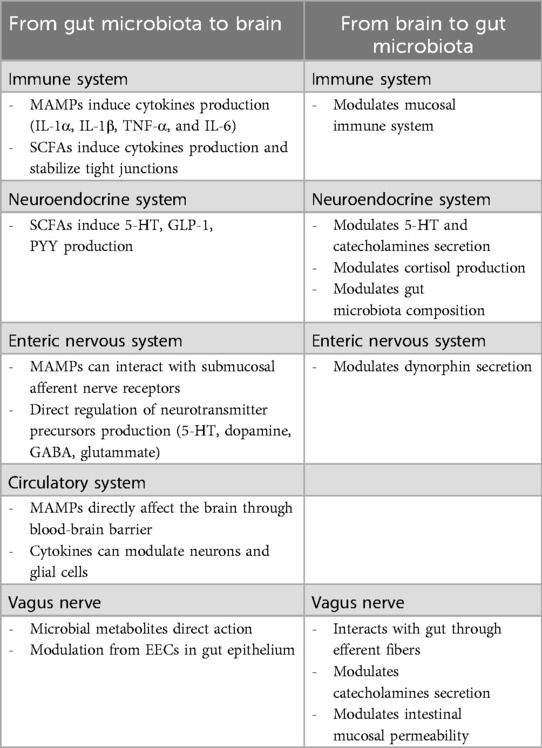
The intestinal microbiota can also affect the secretion of serotonin (5-HT), a neurotransmitter fundamental for different gastrointestinal functions, such as sensory-motor function and gut homeostasis. Specific spore-forming bacteria from both humans and mice can increase colonic and serum 5-HT levels in germ-free (GF) mice and ameliorate GF-associated gut dysmotility by producing SCFAs (214), which increase 5-HT production by enteroendocrine cells (EECs) (215, 216). Moreover, SCFAs can increase serotonin production by increasing the expression of tryptophan hydroxylase-1 in ECCs (217). In addition, indole produced by microorganisms metabolizing tryptophan can stimulate ECC receptors to promote serotonin secretion (218). Interestingly, the gut microbiota can directly produce neurotransmitters such as serotonin, dopamine, epinephrine, norepinephrine, γ-aminobutyric acid, and acetylcholine (219, 220). However, neurotransmitters have large molecular weights and cannot pass through the blood-brain barrier, making it difficult for neurotransmitters in the periphery to affect the central nervous system directly. It is possible that the gut microbiota can regulate and influence the metabolism or production of precursors that can pass through the blood-brain barrier (221). Some crosstalk pathways between brain and gut-microbiota are summarized in Table 3.
Microbial activity is fundamental in regulating epithelial permeability: indole derivatives, bile acid metabolites, conjugated fatty acids, polyamines, and polyphenolic derivatives are other microbial-derived compounds that actively regulate gut barrier function (222). In addition, gut microbiota as well as bacterial and viral infections can also influence intestinal barrier function through the modulation of tight junction expression and assembly (223, 224). In a Canadian study, germ-free mice proved to have a higher colonic expression of claudin-1 and occludin and a lower paracellular uptake of probes as compared with conventional mice, suggesting that commensal microbiota may control colonic tight junction proteins and paracellular permeability (225). The same study demonstrated that transplantation of fecal microbiota from healthy humans can restore the barrier features (paracellular permeability, colonic barrier structure) of conventional mice within a week. Altogether, these data suggest that gut microbiota is crucial in preserving the integrity of the intestinal barrier and preventing the systemic spread of potentially harmful antigens (226).
In particular, specific gut microbiota components may modulate the intestinal permeability differently. For example, colonization of germ-free mice with Bacteroides or Escherichia coli Nissle 1917 (EcN) led to the up-regulation of genes encoding for proteins such as small proline-rich protein-2 (sprr2a) and ZO-1 responsible for cellular adhesion (227, 228). Moreover, it has been demonstrated that the increase in paracellular permeability in patients with IBS could be ameliorated by EcN, thus reducing abdominal pain and bloating (229).
The gut microbiota might also overstimulate the immune system by facilitating a leaky gut, leading to the release of immune mediators, such as histamine, tryptase, serotonin, polyunsaturated fatty acids (12-hydroperoxyeicosatetraenoic acid, 15-hydroxyeicosatetraenoic acid, 5-hydroxyeicosatetraenoic acid, 5-oxoeicosatetraenoic acid and leukotriene B4), known to evoke sensory afferent over-stimulation and pain (230–232). Moreover, some recent studies confirmed that IBS is associated with reduced stability and biodiversity of the gut microbiota (233). Also, an Italian cross-sectional study revealed significant differences among IBS subtypes in the distribution of Clostridiales. Relative Clostridiales abundance was correlated with significant differences in the level of fecal SCFAs, which together were associated with altered fecal cytokine levels (234).
Similarly, in clear-cut inflammatory conditions such as IBD, microbiota dysbiosis has been well described in association with an impairment of intestinal permeability. The intestinal barrier may represent the target of mediators released by inflammatory cells in the lamina propria, and disruption of the physiologic barrier would then allow the passage of antigen, leading to further inflammation in a self-maintaining pathological inflammatory process and antagonizing tissue repair (235). In patients with IBD, there has been a loss of biodiversity (with a decreased concentration of Firmicutes species and Akkermansia muciniphila) and microbial stability, while an increased concentration of Proteobacteria such as Enterobacteriaceae, Bilophila, and Bacteroidetes (236, 237). Furthermore, in patients with IBD, a reduction in SCFA-producing bacteria such as Faecalibacterium prausnitzii has been observed (238). F. prausnitzii is well-known to have anti-inflammatory properties through its ability to produce butyrate, thus regulating T regulatory cell and T helper 17 (239). Doubtlessly, these changes may alter intestinal barrier integrity, potentially resulting in increased immune responses and the diffusion of pathogens into the intestinal tissues. The intestinal inflammatory process depends on the TLR pathway, responsible for the downstream production of cytokines such as TNF-α and IFN-γ, further fueling increased permeability both in IBD and IBS (240, 241).
Conclusion
From the gathered evidence, it is clear that food antigens might interfere with gut permeability in diverse ways and in several diseases. This interference, however, does not rely on one-to-one relationship, given the fact that genetic background, barrier disruptors, microbiota perturbances and inflammation interact in a multidimensional fashion on a path towards disease onset. Unfavorable dietary choices and consequential changes in gut microbiota can promote intestinal barrier dysfunction and, subsequently, loss of tolerance towards food antigen and mucosal inflammation, with effects not only restricted to the gastrointestinal tract. On the other hand, inflammatory mediators can increase mucosal permeability and influence gut microbiota, leading to a vicious and self-maintaining circle.
Several gaps of knowledge still exist as regards this multi-level interaction. For example, it remains elusive why only one twin from monozygotic twins, with almost overlapping genetics, similar microbiota and diet, might develop a multifactorial disease, while the other twin might not. System biology approaches could hopefully sort out many unanswered questions if properly applied to a large set of metadata and biological samples from prospective cohorts.
By advancing our understanding of these intricate and interactive processes, future personalized therapies could more effectively alleviate the burden of food-triggered diseases and improve quality of life for affected individuals.
Author contributions
FV: Conceptualization, Data curation, Project administration, Resources, Software, Validation, Writing – original draft, Writing – review & editing, Methodology. MM: Conceptualization, Validation, Writing – original draft, Writing – review & editing, Data curation, Methodology. GM: Data curation, Writing – original draft. GF: Data curation, Writing – original draft. MP: Data curation, Writing – original draft. GDN: Data curation, Methodology, Project administration, Resources, Writing – original draft, Writing – review & editing. GDC: Data curation, Methodology, Resources, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
Acknowledgments
All the authors acknowledge Prof Alberto Verrotti di Pianella for his continuous encouragement towards this project.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Valitutti F, Fasano A. Breaking down barriers: how understanding celiac disease pathogenesis informed the development of novel treatments. Dig Dis Sci. (2019) 64(7):1748–58. doi: 10.1007/s10620-019-05646-y
2. Kedia S, Ahuja V. Human gut microbiome: a primer for the clinician. JGH Open. (2023) 7(5):337–50. doi: 10.1002/jgh3.12902
3. Ogawa K, Ben RA, Pons S, de Paolo MI, Bustos Fernández L. Volatile fatty acids, lactic acid, and pH in the stools of breast-fed and bottle-fed infants. J Pediatr Gastroenterol Nutr. (1992) 15(3):248–52. doi: 10.1097/00005176-199210000-00004
4. Ku HJ, Kim YT, Lee JH. Microbiome study of initial gut microbiota from newborn infants to children reveals that diet determines its compositional development. J Microbiol Biotechnol. (2020) 30(7):1067–71. doi: 10.4014/jmb.2002.02042
5. Ramaboli MC, Ocvirk S, Khan Mirzaei M, Eberhart BL, Valdivia-Garcia M, Metwaly A, et al. Diet changes due to urbanization in South Africa are linked to microbiome and metabolome signatures of Westernization and colorectal cancer. Nat Commun. (2024) 15(1):3379. doi: 10.1038/s41467-024-46265-0
6. Liwinski T, Auer MK, Schröder J, Pieknik I, Casar C, Schwinge D, et al. Gender-affirming hormonal therapy induces a gender-concordant fecal metagenome transition in transgender individuals. BMC Med. (2024) 22(1):346. doi: 10.1186/s12916-024-03548-z
7. Panzer JJ, Maples C, Meyer MP, Tillotson G, Theis KR, Chopra T. Gut microbiome alpha diversity decreases in relation to body weight, antibiotic exposure, and infection with multidrug-resistant organisms. Am J Infect Control. (2024) 52(6):707–11. doi: 10.1016/j.ajic.2023.12.017
8. Cheng SL, Hedges M, Keski-Rahkonen P, Chatziioannou AC, Scalbert A, Chung KF, et al. Multiomic signatures of traffic-related air pollution in London reveal potential short-term perturbations in gut microbiome-related pathways. Environ Sci Technol. (2024) 58(20):8771–82. doi: 10.1021/acs.est.3c09148
9. Vliex LMM, Penders J, Nauta A, Zoetendal EG, Blaak EE. The individual response to antibiotics and diet—insights into gut microbial resilience and host metabolism. Nat Rev Endocrinol. (2024) 20(7):387–98. doi: 10.1038/s41574-024-00966-0
10. Pelaseyed T, Hansson GC. Membrane mucins of the intestine at a glance. J Cell Sci. (2020) 133(5):jcs240929. doi: 10.1242/jcs.240929
11. Beumer J, Clevers H. Cell fate specification and differentiation in the adult mammalian intestine. Nat Rev Mol Cell Biol. (2021) 22(1):39–53. doi: 10.1038/s41580-020-0278-0
12. Philpott JD, Rodriguez Hovnanian KM, Stefater-Richards M, Mehta NM, Martinez EE. The enteroendocrine axis and its effect on gastrointestinal function, nutrition, and inflammation. Curr Opin Crit Care. (2024) 30(4):290–7. doi: 10.1097/MCC.0000000000001175
13. Silverman JB, Vega PN, Tyska MJ, Lau KS. Intestinal tuft cells: morphology, function, and implications for human health. Annu Rev Physiol. (2024) 86:479–504. doi: 10.1146/annurev-physiol-042022-030310
14. Nakamura Y, Kimura S, Hase K. M cell-dependent antigen uptake on follicle-associated epithelium for mucosal immune surveillance. Inflamm Regen. (2018) 38:15. doi: 10.1186/s41232-018-0072-y
15. Horowitz A, Chanez-Paredes SD, Haest X, Turner JR. Paracellular permeability and tight junction regulation in gut health and disease. Nat Rev Gastroenterol Hepatol. (2023) 20(7):417–32. doi: 10.1038/s41575-023-00766-3
16. Husby S, Koletzko S, Korponay-Szabó I, Kurppa K, Mearin ML, Ribes-Koninckx C, et al. European society paediatric gastroenterology, hepatology and nutrition guidelines for diagnosing coeliac disease 2020. J Pediatr Gastroenterol Nutr. (2020) 70(1):141–56. doi: 10.1097/MPG.0000000000002497
17. Valitutti F, Leonard MM, Kenyon V, Montuori M, Piemontese P, Francavilla R, et al. Early antibody dynamics in a prospective cohort of children at risk of celiac disease. Am J Gastroenterol. (2023) 118(3):574–7. doi: 10.14309/ajg.0000000000002192
18. Lionetti E, Castellaneta S, Francavilla R, Pulvirenti A, Tonutti E, Amarri S, et al. Introduction of gluten, HLA status, and the risk of celiac disease in children. N Engl J Med. (2014) 371(14):1295–303. doi: 10.1056/NEJMoa1400697
19. Vriezinga SL, Auricchio R, Bravi E, Castillejo G, Chmielewska A, Crespo Escobar P, et al. Randomized feeding intervention in infants at high risk for celiac disease. N Engl J Med. (2014) 371(14):1304–15. doi: 10.1056/NEJMoa1404172
20. Fasano A. Celiac disease–how to handle a clinical chameleon. N Engl J Med. (2003) 348(25):2568–70. doi: 10.1056/NEJMe030050
21. Stroud C, Almilaji O, Nicholas D, Kirkham S, Surgenor SL, Williams I, et al. Evolving patterns in the presentation of coeliac disease over the last 25 years. Frontline Gastroenterol. (2020) 11(2):98–103. doi: 10.1136/flgastro-2018-101170
22. Discepolo V, Kelly CP, Koning F, Schuppan D. How future pharmacologic therapies for celiac disease will complement the gluten-free diet. Gastroenterology. (2024) 167(1):90–103. doi: 10.1053/j.gastro.2024.02.050
23. Tripathi A, Lammers KM, Goldblum S, Shea-Donohue T, Netzel-Arnett S, Buzza MS, et al. Identification of human zonulin, a physiological modulator of tight junctions, as prehaptoglobin-2. Proc Natl Acad Sci. (2009) 106(39):16799–804. doi: 10.1073/pnas.0906773106
24. Asmar R E, Panigrahi P, Bamford P, Berti I, Not T, Coppa GV, et al. Host-dependent zonulin secretion causes the impairment of the small intestine barrier function after bacterial exposure. Gastroenterology. (2002) 123(5):1607–15. doi: 10.1053/gast.2002.36578
25. Lammers KM, Khandelwal S, Chaudhry F, Kryszak D, Puppa EL, Casolaro V, et al. Identification of a novel immunomodulatory gliadin peptide that causes interleukin-8 release in a chemokine receptor CXCR3-dependent manner only in patients with coeliac disease. Immunology. (2011) 132(3):432–40. doi: 10.1111/j.1365-2567.2010.03378.x
26. Tsai PY, Zhang B, He WQ, Zha JM, Odenwald MA, Singh G, et al. IL-22 upregulates epithelial claudin-2 to drive diarrhea and enteric pathogen clearance. Cell Host Microbe. (2017) 21(6):671–81.e4. doi: 10.1016/j.chom.2017.05.009
27. Lauxmann MA, Vazquez DS, Schilbert HM, Neubauer PR, Lammers KM, Dodero VI. From celiac disease to coccidia infection and vice-versa: the polyQ peptide CXCR3-interaction axis. Bioessays. (2021) 43(12):e2100101. doi: 10.1002/bies.202100101
28. Kelly CP, Green PH, Murray JA, Dimarino A, Colatrella A, Leffler DA, et al. Larazotide acetate in patients with coeliac disease undergoing a gluten challenge: a randomised placebo-controlled study. Aliment Pharmacol Ther. (2013) 37(2):252–62. doi: 10.1111/apt.12147
29. Pizzuti D, Bortolami M, Mazzon E, Buda A, Guariso G, D'Odorico A, et al. Transcriptional downregulation of tight junction protein ZO-1 in active coeliac disease is reversed after a gluten-free diet. Dig Liver Dis. (2004) 36(5):337–41. doi: 10.1016/j.dld.2004.01.013
30. Wapenaar MC, Monsuur AJ, van Bodegraven AA, Weersma RK, Bevova MR, Linskens RK, et al. Associations with tight junction genes PARD3 and MAGI2 in Dutch patients point to a common barrier defect for coeliac disease and ulcerative colitis. Gut. (2007) 57(4):463–7. doi: 10.1136/gut.2007.133132
31. Jauregi-Miguel A, Santin I, Garcia-Etxebarria K, Olazagoitia-Garmendia A, Romero-Garmendia I, Sebastian-delaCruz M, et al. MAGI2 gene region and celiac disease. Front Nutr. (2019) 6:187. doi: 10.3389/fnut.2019.00187
32. Mishra A, Prakash S, Sreenivas V, Das TK, Ahuja V, Gupta SD, et al. Structural and functional changes in the tight junctions of asymptomatic and serology-negative first-degree relatives of patients with celiac disease. J Clin Gastroenterol. (2016) 50(7):551–60. doi: 10.1097/MCG.0000000000000436
33. DaFonte TM, Valitutti F, Kenyon V, Locascio JJ, Montuori M, Francavilla R, et al. Zonulin as a biomarker for the development of celiac disease. Pediatrics. (2024) 153(1):e2023063050. doi: 10.1542/peds.2023-063050
34. Kahrs CR, Chuda K, Tapia G, Stene LC, Mårild K, Rasmussen T, et al. Enterovirus as trigger of coeliac disease: nested case-control study within prospective birth cohort. Br Med J. (2019) 364:l231. doi: 10.1136/bmj.l231
35. Kemppainen KM, Lynch KF, Liu E, Lönnrot M, Simell V, Briese T, et al. Factors that increase risk of celiac disease autoimmunity after a gastrointestinal infection in early life. Clin Gastroenterol Hepatol. (2017) 15(5):694–702.e5. doi: 10.1016/j.cgh.2016.10.033
36. Guttman JA, Finlay BB. Tight junctions as targets of infectious agents. Biochim Biophys Acta. (2009) 1788(4):832–41. doi: 10.1016/j.bbamem.2008.10.028
37. Dolcino M, Zanoni G, Bason C, Tinazzi E, Boccola E, Valletta E, et al. A subset of anti-rotavirus antibodies directed against the viral protein VP7 predicts the onset of celiac disease and induces typical features of the disease in the intestinal epithelial cell line T84. Immunol Res. (2013) 56(2-3):465–76. doi: 10.1007/s12026-013-8420-0
38. Fasano A, Sapone A, Zevallos V, Schuppan D. Nonceliac gluten sensitivity. Gastroenterology. (2015) 148(6):1195–204. doi: 10.1053/j.gastro.2014.12.049
39. Barbaro MR, Cremon C, Morselli-Labate AM, Di Sabatino A, Giuffrida P, Corazza GR, et al. Serum zonulin and its diagnostic performance in non-coeliac gluten sensitivity. Gut. (2020) 69(11):1966–74. doi: 10.1136/gutjnl-2019-319281
40. Ahmed A, Dixit K, Singh A, Agarwal A, Mehtab W, Prasad S, et al. Sieving out non-celiac gluten sensitivity amongst patients with irritable bowel syndrome. Dig Liver Dis. (2024) 56(3):451–7. doi: 10.1016/j.dld.2023.10.014
41. Barbato M, Maiella G, Di Camillo C, Guida S, Valitutti F, Lastrucci G, et al. The anti-deamidated gliadin peptide antibodies unmask celiac disease in small children with chronic diarrhoea. Dig Liver Dis. (2011) 43(6):465–9. doi: 10.1016/j.dld.2010.12.006
42. Fritscher-Ravens A, Pflaum T, Mösinger M, Ruchay Z, Röcken C, Milla PJ, et al. Many patients with irritable bowel syndrome have atypical food allergies not associated with immunoglobulin E. Gastroenterology. (2019) 157(1):109–18.e5. doi: 10.1053/j.gastro.2019.03.046
43. Uhde M, Ajamian M, Caio G, De Giorgio R, Indart A, Green PH, et al. Intestinal cell damage and systemic immune activation in individuals reporting sensitivity to wheat in the absence of coeliac disease. Gut. (2016) 65(12):1930–7. doi: 10.1136/gutjnl-2016-311964
44. Sapone A, Lammers KM, Casolaro V, Cammarota M, Giuliano MT, De Rosa M, et al. Divergence of gut permeability and mucosal immune gene expression in two gluten-associated conditions: celiac disease and gluten sensitivity. BMC Med. (2011) 9:23. doi: 10.1186/1741-7015-9-23
45. Hollon J, Puppa EL, Greenwald B, Goldberg E, Guerrerio A, Fasano A. Effect of gliadin on permeability of intestinal biopsy explants from celiac disease patients and patients with non-celiac gluten sensitivity. Nutrients. (2015) 7(3):1565–76. doi: 10.3390/nu7031565
46. Schuppan D, Zevallos V. Wheat amylase trypsin inhibitors as nutritional activators of innate immunity. Dig Dis. (2015) 33(2):260–3. doi: 10.1159/000371476
47. Caminero A, McCarville JL, Zevallos VF, Pigrau M, Yu XB, Jury J, et al. Lactobacilli degrade wheat amylase trypsin inhibitors to reduce intestinal dysfunction induced by immunogenic wheat proteins. Gastroenterology. (2019) 156(8):2266–80. doi: 10.1053/j.gastro.2019.02.028
48. Ramedani N, Sharifan A, Gholam-Mostafaei FS, Rostami-Nejad M, Yadegar A, Ehsani-Ardakani MJ. The potentials of probiotics on gluten hydrolysis; a review study. Gastroenterol Hepatol Bed Bench. (2020) 13(Suppl1):S1–7.33584998
49. Lamas B, Hernandez-Galan L, Galipeau HJ, Constante M, Clarizio A, Jury J, et al. Aryl hydrocarbon receptor ligand production by the gut microbiota is decreased in celiac disease leading to intestinal inflammation. Sci Transl Med. (2020) 12(566):eaba0624. doi: 10.1126/scitranslmed.aba0624
50. Lindfors K, Blomqvist T, Juuti-Uusitalo K, Stenman S, Venäläinen J, Mäki M, et al. Live probiotic Bifidobacterium lactis bacteria inhibit the toxic effects induced by wheat gliadin in epithelial cell culture. Clin Exp Immunol. (2008) 152(3):552–8. doi: 10.1111/j.1365-2249.2008.03635.x
51. Orlando A, Linsalata M, Bianco G, Notarnicola M, D'Attoma B, Scavo MP, et al. Lactobacillus rhamnosus GG protects the epithelial barrier of wistar rats from the pepsin-trypsin-digested gliadin (PTG)-induced enteropathy. Nutrients. (2018) 10(11):1698. doi: 10.3390/nu10111698
52. Gupta RS, Warren CM, Smith BM, Blumenstock JA, Jiang J, Davis MM, et al. The public health impact of parent-reported childhood food allergies in the United States. Pediatrics. (2018) 142(6):e20181235. doi: 10.1542/peds.2018-1235
53. Di Costanzo M, Vella A, Infantino C, Morini R, Bruni S, Esposito S, et al. Probiotics in infancy and childhood for food allergy prevention and treatment. Nutrients. (2024) 16(2):297. doi: 10.3390/nu16020297
54. Peters RL, Mavoa S, Koplin JJ. An overview of environmental risk factors for food allergy. Int J Environ Res Public Health. (2022) 19(2):722. doi: 10.3390/ijerph19020722
55. Boyce JA, Assa'ad A, Burks AW, Jones SM, Sampson HA, Wood RA, et al. Guidelines for the diagnosis and management of food allergy in the United States: report of the NIAID-sponsored expert panel. J Allergy Clin Immunol. (2010) 126(6 Suppl):S1–58. doi: 10.1016/j.nut.2010.12.001
56. Cosme-Blanco W, Arroyo-Flores E, Ale H. Food allergies. Pediatr Rev. (2020) 41(8):403–15. doi: 10.1542/pir.2019-0037
57. Costa J, Villa C, Verhoeckx K, Cirkovic-Velickovic T, Schrama D, Roncada P, et al. Are physicochemical properties shaping the allergenic potency of animal allergens? Clin Rev Allergy Immunol. (2022) 62(1):1–36. doi: 10.1007/s12016-020-08826-1
58. Boutrou R, Jardin J, Blais A, Tomé D, Léonil J. Glycosylations of kappa-casein-derived caseinomacropeptide reduce its accessibility to endo- but not exointestinal brush border membrane peptidases. J Agric Food Chem. (2008) 56(17):8166–73. doi: 10.1021/jf801140d
59. Vissers YM, Blanc F, Skov PS, Johnson PE, Rigby NM, Przybylski-Nicaise L, et al. Effect of heating and glycation on the allergenicity of 2S albumins (Ara h 2/6) from peanut. PLoS One. (2011) 6(8):e23998. doi: 10.1371/journal.pone.0023998
60. Mueller GA, Maleki SJ, Johnson K, Hurlburt BK, Cheng H, Ruan S, et al. Identification of Maillard reaction products on peanut allergens that influence binding to the receptor for advanced glycation end products. Allergy. (2013) 68(12):1546–54. doi: 10.1111/all.12261
61. Agache I, Miller R, Gern JE, Hellings PW, Jutel M, Muraro A, et al. Emerging concepts and challenges in implementing the exposome paradigm in allergic diseases and asthma: a Practall document. Allergy. (2019) 74(3):449–63. doi: 10.1111/all.13690
62. du Toit G, Tsakok T, Lack S, Lack G. Prevention of food allergy. J Allergy Clin Immunol. (2016) 137(4):998–1010. doi: 10.1016/j.jaci.2016.02.005
63. Niewiem M, Grzybowska-Chlebowczyk U. Intestinal barrier permeability in allergic diseases. Nutrients. (2022) 14(9):1893. doi: 10.3390/nu14091893
64. De Paepe E, Plekhova V, Vangeenderhuysen P, Baeck N, Bullens D, Claeys T, et al. Integrated gut metabolome and microbiome fingerprinting reveals that dysbiosis precedes allergic inflammation in IgE-mediated pediatric cow’s milk allergy. Allergy. (2024) 79(4):949–63. doi: 10.1111/all.16005
65. Poto R, Fusco W, Rinninella E, Cintoni M, Kaitsas F, Raoul P, et al. The role of gut microbiota and leaky gut in the pathogenesis of food allergy. Nutrients. (2024) 16(1):92. doi: 10.3390/nu16010092
66. Molloy J, Allen K, Collier F, Tang ML, Ward AC, Vuillermin P. The potential link between gut microbiota and IgE-mediated food allergy in early life. Int J Environ Res Public Health. (2013) 10(12):7235–56. doi: 10.3390/ijerph10127235
67. Peroni DG, Nuzzi G, Trambusti I, Di Cicco ME, Comberiati P. Microbiome composition and its impact on the development of allergic diseases. Front Immunol. (2020) 11:700. doi: 10.3389/fimmu.2020.00700
68. Cabrera-Rubio R, Collado MC, Laitinen K, Salminen S, Isolauri E, Mira A. The human milk microbiome changes over lactation and is shaped by maternal weight and mode of delivery. Am J Clin Nutr. (2012) 96(3):544–51. doi: 10.3945/ajcn.112.037382
69. van den Elsen LWJ, Garssen J, Burcelin R, Verhasselt V. Shaping the gut microbiota by breastfeeding: the gateway to allergy prevention? Front Pediatr. (2019) 7:47. doi: 10.3389/fped.2019.00047
70. Zivkovic AM, German JB, Lebrilla CB, Mills DA. Human milk glycobiome and its impact on the infant gastrointestinal microbiota. Proc Natl Acad Sci. (2011) 108(Suppl 1):4653–8. doi: 10.1073/pnas.1000083107
71. van den Elsen LWJ, Kollmann TR, Verhasselt V. Microbial antigen in human milk: a natural vaccine? Mucosal Immunol. (2022) 15(6):1058–9. doi: 10.1038/s41385-022-00561-4
72. Ongena R, Dierick M, Vanrompay D, Cox E, Devriendt B. Lactoferrin impairs pathogen virulence through its proteolytic activity. Front Vet Sci. (2024) 11:1428156. doi: 10.3389/fvets.2024.1428156
73. Soto A, Martín V, Jiménez E, Mader I, Rodríguez JM, Fernández L. Lactobacilli and bifidobacteria in human breast milk: influence of antibiotherapy and other host and clinical factors. J Pediatr Gastroenterol Nutr. (2014) 59(1):78–88. doi: 10.1097/MPG.0000000000000347
74. Hermansson H, Kumar H, Collado MC, Salminen S, Isolauri E, Rautava S. Breast milk microbiota is shaped by mode of delivery and intrapartum antibiotic exposure. Front Nutr. (2019) 6:4. doi: 10.3389/fnut.2019.00004
75. Khodayar-Pardo P, Mira-Pascual L, Collado MC, Martínez-Costa C. Impact of lactation stage, gestational age and mode of delivery on breast milk microbiota. J Perinatol. (2014) 34(8):599–605. doi: 10.1038/jp.2014.47
76. Li SW, Watanabe K, Hsu CC, Chao SH, Yang ZH, Lin YJ, et al. Bacterial composition and diversity in breast milk samples from mothers living in Taiwan and Mainland China. Front Microbiol. (2017) 8:965. doi: 10.3389/fmicb.2017.00965
77. Kumar H, du Toit E, Kulkarni A, Aakko J, Linderborg KM, Zhang Y, et al. Distinct patterns in human milk microbiota and fatty acid profiles across specific geographic locations. Front Microbiol. (2016) 7:1619. doi: 10.3389/fmicb.2016.01619
78. Fazlollahi M, Chun Y, Grishin A, Wood RA, Burks AW, Dawson P, et al. Early-life gut microbiome and egg allergy. Allergy. (2018) 73(7):1515–24. doi: 10.1111/all.13389
79. Azad MB, Konya T, Guttman DS, Field CJ, Sears MR, HayGlass KT, et al. Infant gut microbiota and food sensitization: associations in the first year of life. Clin Exp Allergy. (2015) 45(3):632–43. doi: 10.1111/cea.12487
80. An D, Oh SF, Olszak T, Neves JF, Avci FY, Erturk-Hasdemir D, et al. Sphingolipids from a symbiotic microbe regulate homeostasis of host intestinal natural killer T cells. Cell. (2014) 156(1-2):123–33. doi: 10.1016/j.cell.2013.11.042
81. Stefka AT, Feehley T, Tripathi P, Qiu J, McCoy K, Mazmanian SK, et al. Commensal bacteria protect against food allergen sensitization. Proc Natl Acad Sci. (2014) 111(36):13145–50. doi: 10.1073/pnas.1412008111
82. Cahenzli J, Köller Y, Wyss M, Geuking MB, McCoy KD. Intestinal microbial diversity during early-life colonization shapes long-term IgE levels. Cell Host Microbe. (2013) 14(5):559–70. doi: 10.1016/j.chom.2013.10.004
83. Tordesillas L, Berin MC. Mechanisms of oral tolerance. Clin Rev Allergy Immunol. (2018) 55(2):107–17. doi: 10.1007/s12016-018-8680-5
84. Mallegol J, Van Niel G, Lebreton C, Lepelletier Y, Candalh C, Dugave C, et al. T84-intestinal epithelial exosomes bear MHC class II/peptide complexes potentiating antigen presentation by dendritic cells. Gastroenterology. (2007) 132(5):1866–76. doi: 10.1053/j.gastro.2007.02.043
85. Farache J, Koren I, Milo I, Gurevich I, Kim KW, Zigmond E, et al. Luminal bacteria recruit CD103+dendritic cells into the intestinal epithelium to sample bacterial antigens for presentation. Immunity. (2013) 38(3):581–95. doi: 10.1016/j.immuni.2013.01.009
86. Ménard S, Cerf-Bensussan N, Heyman M. Multiple facets of intestinal permeability and epithelial handling of dietary antigens. Mucosal Immunol. (2010) 3(3):247–59. doi: 10.1038/mi.2010.5
87. Chieppa M, Rescigno M, Huang AY, Germain RN. Dynamic imaging of dendritic cell extension into the small bowel lumen in response to epithelial cell TLR engagement. J Exp Med. (2006) 203(13):2841–52. doi: 10.1084/jem.20061884
88. Lelouard H, Fallet M, de Bovis B, Méresse S, Gorvel JP. Peyer’s patch dendritic cells sample antigens by extending dendrites through M cell-specific transcellular pores. Gastroenterology. (2012) 142(3):592–601.e3. doi: 10.1053/j.gastro.2011.11.039
89. Bakdash G, Vogelpoel LT, van Capel TM, Kapsenberg ML, de Jong EC. Retinoic acid primes human dendritic cells to induce gut-homing, IL-10-producing regulatory T cells. Mucosal Immunol. (2015) 8(2):265–78. doi: 10.1038/mi.2014.64
90. Esterházy D, Loschko J, London M, Jove V, Oliveira TY, Mucida D. Classical dendritic cells are required for dietary antigen-mediated induction of peripheral T(reg) cells and tolerance. Nat Immunol. (2016) 17(5):545–55. doi: 10.1038/ni.3408
91. Akdis CA, Akdis M. Mechanisms of immune tolerance to allergens: role of IL-10 and Tregs. J Clin Invest. (2014) 124(11):4678–80. doi: 10.1172/JCI78891
92. Friedman A. Induction of anergy in Th1 lymphocytes by oral tolerance. Importance of antigen dosage and frequency of feeding. Ann N Y Acad Sci. (1996) 778:103–10. doi: 10.1111/j.1749-6632.1996.tb21119.x
93. Kozakova H, Schwarzer M, Tuckova L, Srutkova D, Czarnowska E, Rosiak I, et al. Colonization of germ-free mice with a mixture of three lactobacillus strains enhances the integrity of gut mucosa and ameliorates allergic sensitization. Cell Mol Immunol. (2016) 13(2):251–62. doi: 10.1038/cmi.2015.09
94. Cait A, Cardenas E, Dimitriu PA, Amenyogbe N, Dai D, Cait J, et al. Reduced genetic potential for butyrate fermentation in the gut microbiome of infants who develop allergic sensitization. J Allergy Clin Immunol. (2019) 144(6):1638–47.e3. doi: 10.1016/j.jaci.2019.06.029
95. Lin R, Zhou L, Zhang J, Wang B. Abnormal intestinal permeability and microbiota in patients with autoimmune hepatitis. Int J Clin Exp Pathol. (2015) 8(5):5153–60.26191211
96. Leonard MM, Valitutti F, Karathia H, Pujolassos M, Kenyon V, Fanelli B, et al. Microbiome signatures of progression toward celiac disease onset in at-risk children in a longitudinal prospective cohort study. Proc Natl Acad Sci. (2021) 118(29):e2020322118. doi: 10.1073/pnas.2020322118
97. Parrish A, Boudaud M, Grant ET, Willieme S, Neumann M, Wolter M, et al. Akkermansia muciniphila exacerbates food allergy in fibre-deprived mice. Nat Microbiol. (2023) 8(10):1863–79. doi: 10.1038/s41564-023-01464-1
98. Abdel-Gadir A, Stephen-Victor E, Gerber GK, Noval Rivas M, Wang S, Harb H, et al. Microbiota therapy acts via a regulatory T cell MyD88/RORγt pathway to suppress food allergy. Nat Med. (2019) 25(7):1164–74. doi: 10.1038/s41591-019-0461-z
99. Feehley T, Plunkett CH, Bao R, Choi Hong SM, Culleen E, Belda-Ferre P, et al. Healthy infants harbor intestinal bacteria that protect against food allergy. Nat Med. (2019) 25(3):448–53. doi: 10.1038/s41591-018-0324-z
100. Huang YJ, Marsland BJ, Bunyavanich S, O'Mahony L, Leung DY, Muraro A, et al. The microbiome in allergic disease: current understanding and future opportunities-2017 PRACTALL document of the American academy of allergy, asthma & immunology and the European academy of allergy and clinical immunology. J Allergy Clin Immunol. (2017) 139(4):1099–110. doi: 10.1016/j.jaci.2017.02.007
101. Atarashi K, Tanoue T, Shima T, Imaoka A, Kuwahara T, Momose Y, et al. Induction of colonic regulatory T cells by indigenous clostridium species. Science. (2011) 331(6015):337–41. doi: 10.1126/science.1198469
102. Hua X, Goedert JJ, Pu A, Yu G, Shi J. Allergy associations with the adult fecal microbiota: analysis of the American gut project. EBioMedicine. (2016) 3:172–9. doi: 10.1016/j.ebiom.2015.11.038
103. Zelante T, Iannitti RG, Cunha C, De Luca A, Giovannini G, Pieraccini G, et al. Tryptophan catabolites from microbiota engage aryl hydrocarbon receptor and balance mucosal reactivity via interleukin-22. Immunity. (2013) 39(2):372–85. doi: 10.1016/j.immuni.2013.08.003
104. Agus A, Planchais J, Sokol H. Gut microbiota regulation of tryptophan metabolism in health and disease. Cell Host Microbe. (2018) 23(6):716–24. doi: 10.1016/j.chom.2018.05.003
105. Fu G, Zhao K, Chen H, Wang Y, Nie L, Wei H, et al. Effect of 3 lactobacilli on immunoregulation and intestinal microbiota in a β-lactoglobulin-induced allergic mouse model. J Dairy Sci. (2019) 102(3):1943–58. doi: 10.3168/jds.2018-15683
106. Kalliomäki M, Salminen S, Arvilommi H, Kero P, Koskinen P, Isolauri E. Probiotics in primary prevention of atopic disease: a randomised placebo-controlled trial. Lancet. (2001) 357(9262):1076–9. doi: 10.1016/S0140-6736(00)04259-8
107. Ni WW, Zhang QM, Zhang X, Li Y, Yu SS, Wu HY, et al. Modulation effect of lactobacillus acidophilus KLDS 1.0738 on gut microbiota and TLR4 expression in β-lactoglobulin-induced allergic mice model. Allergol Immunopathol (Madr). (2020) 48(2):149–57. doi: 10.1016/j.aller.2019.06.002
108. Li A, Yang J, Zhang C, Chi H, Li T, Zhang J, et al. Lactobacillus acidophilus KLDS 1.0738 inhibits TLR4/NF-κB inflammatory pathway in β-lactoglobulin-induced macrophages via modulating miR-146a. J Food Biochem. (2021) 45(10):e13662. doi: 10.1111/jfbc.13662
109. Fu L, Xie M, Wang C, Qian Y, Huang J, Sun Z, et al. Lactobacillus Casei Zhang alleviates shrimp tropomyosin-induced food allergy by switching antibody isotypes through the NF-κB-dependent immune tolerance. Mol Nutr Food Res. (2020) 64(10):e1900496. doi: 10.1002/mnfr.201900496
110. Cukrowska B, Ceregra A, Maciorkowska E, Surowska B, Zegadło-Mylik MA, Konopka E, et al. The effectiveness of probiotic lactobacillus rhamnosus and lactobacillus casei strains in children with atopic dermatitis and cow’s milk protein allergy: a multicenter, randomized, double blind, placebo controlled study. Nutrients. (2021) 13(4):1169. doi: 10.3390/nu13041169
111. Santos SS, Miranda VC, Trindade LM, Cardoso VN, Reis DC, Cassali GD, et al. Bifidobacterium longum subsp. longum 51A attenuates signs of inflammation in a murine model of food allergy. Probiotics Antimicrob Proteins. (2023) 15(1):63–73. doi: 10.1007/s12602-021-09846-9
112. Pyclik MJ, Srutkova D, Razim A, Hermanova P, Svabova T, Pacyga K, et al. Viability status-dependent effect of bifidobacterium longum ssp. longum CCM 7952 on prevention of allergic inflammation in mouse model. Front Immunol. (2021) 12:707728. doi: 10.3389/fimmu.2021.707728
113. Jing W, Liu Q, Wang W. Bifidobacterium bifidum TMC3115 ameliorates milk protein allergy in by affecting gut microbiota: a randomized double-blind control trial. J Food Biochem. (2021) 45(11):e13591. doi: 10.1111/jfbc.13591
114. Bellomo AR, Rotondi G, Rago P, Bloise S, Di Ruzza L, Zingoni A, et al. Effect of bifidobacterium bifidum supplementation in newborns born from cesarean section on atopy, respiratory tract infections, and dyspeptic syndromes: a multicenter, randomized, and controlled clinical trial. Microorganisms. (2024) 12(6):1093. doi: 10.3390/microorganisms12061093
115. Fu L, Song J, Wang C, Fu S, Wang Y. Potentially alleviates shrimp tropomyosin-induced allergy by tolerogenic dendritic cell-dependent induction of regulatory T cells and alterations in gut microbiota. Front Immunol. (2017) 8:1536. doi: 10.3389/fimmu.2017.01536
116. Miranda VC, Souza RO, Quintanilha MF, Gallotti B, Assis HC, Faria AMC, et al. A next-generation bacteria (Akkermansia muciniphila BAA-835) presents probiotic potential against ovalbumin-induced food allergy in mice. Probiotics Antimicrob Proteins. (2024) 16(3):737–51. doi: 10.1007/s12602-023-10076-4
117. Fu L, Peng J, Zhao S, Zhang Y, Su X, Wang Y. Lactic acid bacteria-specific induction of CD4+ Foxp3+ T cells ameliorates shrimp tropomyosin-induced allergic response in mice via suppression of mTOR signaling. Sci Rep. (2017) 7(1):1987. doi: 10.1038/s41598-017-02260-8
118. Zhang J, Su H, Li Q, Wu H, Liu M, Huang J, et al. Oral administration of Clostridium butyricum CGMCC0313-1 inhibits β-lactoglobulin-induced intestinal anaphylaxis in a mouse model of food allergy. Gut Pathog. (2017) 9:11. doi: 10.1186/s13099-017-0160-6
119. Juan Z, Zhao-Ling S, Ming-Hua Z, Chun W, Hai-Xia W, Meng-Yun L, et al. Oral administration of Clostridium butyricum CGMCC0313-1 reduces ovalbumin-induced allergic airway inflammation in mice. Respirology. (2017) 22(5):898–904. doi: 10.1111/resp.12985
120. Thompson-Chagoyan OC, Vieites JM, Maldonado J, Edwards C, Gil A. Changes in faecal microbiota of infants with cow’s milk protein allergy–a Spanish prospective case-control 6-month follow-up study. Pediatr Allergy Immunol. (2010) 21(2):e394–400. doi: 10.1111/j.1399-3038.2009.00961.x
121. Duan C, Ma L, Yu J, Sun Y, Liu L, Ma F, et al. Oral administration of Lactobacillus plantarum JC7 alleviates OVA-induced murine food allergy through immunoregulation and restoring disordered intestinal microbiota. Eur J Nutr. (2023) 62(2):685–98. doi: 10.1007/s00394-022-03016-5
122. Jiang S, Hou Y, Meng L, Pu X, Zhu X, Tuo Y, et al. Effect of Lactiplantibacillus plantarum HM-22 on immunoregulation and intestinal microbiota in α-lactalbumin-induced allergic mice. Food Funct. (2021) 12(19):8887–98. doi: 10.1039/D1FO01703A
123. Wang Y, Gu Y, Fang K, Mao K, Dou J, Fan H, et al. Lactobacillus acidophilus and clostridium butyricum ameliorate colitis in murine by strengthening the gut barrier function and decreasing inflammatory factors. Benef Microbes. (2018) 9(5):775–87. doi: 10.3920/BM2017.0035
124. Divekar R, Kita H. Recent advances in epithelium-derived cytokines (IL-33, IL-25, and thymic stromal lymphopoietin) and allergic inflammation. Curr Opin Allergy Clin Immunol. (2015) 15(1):98–103. doi: 10.1097/ACI.0000000000000133
125. Celebi Sözener Z, Cevhertas L, Nadeau K, Akdis M, Akdis CA. Environmental factors in epithelial barrier dysfunction. J Allergy Clin Immunol. (2020) 145(6):1517–28. doi: 10.1016/j.jaci.2020.04.024
126. Chen T, Liu X, Ma L, He W, Li W, Cao Y, et al. Food allergens affect the intestinal tight junction permeability in inducing intestinal food allergy in rats. Asian Pac J Allergy Immunol. (2014) 32(4):345–53. doi: 10.12932/AP0443.32.4.2014
127. Troncone R, Caputo N, Florio G, Finelli E. Increased intestinal sugar permeability after challenge in children with cow’s milk allergy or intolerance. Allergy. (1994) 49(3):142–6. doi: 10.1111/j.1398-9995.1994.tb00816.x
128. Ballegaard AR, Bøgh KL. Intestinal protein uptake and IgE-mediated food allergy. Food Res Int. (2023) 163:112150. doi: 10.1016/j.foodres.2022.112150
129. Bäckhed F, Roswall J, Peng Y, Feng Q, Jia H, Kovatcheva-Datchary P, et al. Dynamics and stabilization of the human gut microbiome during the first year of life. Cell Host Microbe. (2015) 17(5):690–703. doi: 10.1016/j.chom.2015.04.004
130. Caffarelli C, Di Mauro D, Mastrorilli C, Bottau P, Cipriani F, Ricci G. Solid food introduction and the development of food allergies. Nutrients. (2018) 10(11):1790. doi: 10.3390/nu10111790
131. Koukou Z, Papadopoulou E, Panteris E, Papadopoulou S, Skordou A, Karamaliki M, et al. The effect of breastfeeding on food allergies in newborns and infants. Children (Basel). (2023) 10(6):1046. doi: 10.3390/children10061046
132. Scarpone R, Kimkool P, Ierodiakonou D, Leonardi-Bee J, Garcia-Larsen V, Perkin MR, et al. Timing of allergenic food introduction and risk of immunoglobulin E-mediated food allergy: a systematic review and meta-analysis. JAMA Pediatr. (2023) 177(5):489–97. doi: 10.1001/jamapediatrics.2023.0142
133. Halken S, Muraro A, de Silva D, Khaleva E, Angier E, Arasi S, et al. EAACI guideline: preventing the development of food allergy in infants and young children (2020 update). Pediatr Allergy Immunol. (2021) 32(5):843–58. doi: 10.1111/pai.13496
134. Skjerven HO, Lie A, Vettukattil R, Rehbinder EM, LeBlanc M, Asarnoj A, et al. Early food intervention and skin emollients to prevent food allergy in young children (PreventADALL): a factorial, multicentre, cluster-randomised trial. Lancet. (2022) 399(10344):2398–411. doi: 10.1016/S0140-6736(22)00687-0
135. Roduit C, Frei R, Depner M, Schaub B, Loss G, Genuneit J, et al. Increased food diversity in the first year of life is inversely associated with allergic diseases. J Allergy Clin Immunol. (2014) 133(4):1056–64. doi: 10.1016/j.jaci.2013.12.1044
136. Du Toit G, Foong RX, Lack G. Prevention of food allergy—early dietary interventions. Allergol Int. (2016) 65(4):370–7. doi: 10.1016/j.alit.2016.08.001
137. Zhong C, Guo J, Tan T, Wang H, Lin L, Gao D, et al. Increased food diversity in the first year of life is inversely associated with allergic outcomes in the second year. Pediatr Allergy Immunol. (2022) 33(1):e13707. doi: 10.1111/pai.13707
138. Golfetto L, de Senna FD, Hermes J, Beserra BT, França F, Martinello F. Lower bifidobacteria counts in adult patients with celiac disease on a gluten-free diet. Arq Gastroenterol. (2014) 51(2):139–43. doi: 10.1590/S0004-28032014000200013
139. Bassanini G, Ceccarani C, Borgo F, Severgnini M, Rovelli V, Morace G, et al. Phenylketonuria diet promotes shifts in firmicutes populations. Front Cell Infect Microbiol. (2019) 9:101. doi: 10.3389/fcimb.2019.00101
140. Xie G, Zhou Q, Qiu CZ, Dai WK, Wang HP, Li YH, et al. Ketogenic diet poses a significant effect on imbalanced gut microbiota in infants with refractory epilepsy. World J Gastroenterol. (2017) 23(33):6164–71. doi: 10.3748/wjg.v23.i33.6164
141. Zhao L, Zhang F, Ding X, Wu G, Lam YY, Wang X, et al. Gut bacteria selectively promoted by dietary fibers alleviate type 2 diabetes. Science. (2018) 359(6380):1151–6. doi: 10.1126/science.aao5774
142. Guajardo JR, Plotnick LM, Fende JM, Collins MH, Putnam PE, Rothenberg ME. Eosinophil-associated gastrointestinal disorders: a world-wide-web based registry. J Pediatr. (2002) 141(4):576–81. doi: 10.1067/mpd.2002.127663
143. Menard-Katcher C, Aceves S. Pathophysiology and clinical impact of esophageal remodeling and fibrosis in eosinophilic esophagitis. Immunol Allergy Clin North Am. (2024) 44(2):129–43. doi: 10.1016/j.iac.2023.12.002
144. Dunn JLM, Spencer LA. Pathophysiology of non-esophageal eosinophilic gastrointestinal disorders. Immunol Allergy Clin North Am. (2024) 44(2):299–309. doi: 10.1016/j.iac.2024.01.003
145. McGowan EC, Singh R, Katzka DA. Barrier dysfunction in eosinophilic esophagitis. Curr Gastroenterol Rep. (2023) 25(12):380–9. doi: 10.1007/s11894-023-00904-6
146. Talley NJ, Burns GL, Hoedt EC, Duncanson K, Keely S. Beyond eosinophilic esophagitis: eosinophils in gastrointestinal disease-new insights, “new” diseases. J Can Assoc Gastroenterol. (2023) 6(6):199–211. doi: 10.1093/jcag/gwad046
147. Katzka DA, Geno DM, Blair HE, Lamsam JL, Alexander JA, Camilleri M. Small intestinal permeability in patients with eosinophilic oesophagitis during active phase and remission. Gut. (2015) 64(4):538–43. doi: 10.1136/gutjnl-2013-305882
148. Greuter T, Katzka D. Endoscopic features of eosinophilic gastrointestinal diseases. Immunol Allergy Clin North Am. (2024) 44(2):357–68. doi: 10.1016/j.iac.2024.01.007
149. Furuta GT, Fillon SA, Williamson KM, Robertson CE, Stevens MJ, Aceves SS, et al. Mucosal microbiota associated with eosinophilic esophagitis and eosinophilic gastritis. J Pediatr Gastroenterol Nutr. (2023) 76(3):347–54. doi: 10.1097/MPG.0000000000003685
150. Yuki T, Tobiishi M, Kusaka-Kikushima A, Ota Y, Tokura Y. Impaired tight junctions in atopic dermatitis skin and in a skin-equivalent model treated with interleukin-17. PLoS One. (2016) 11(9):e0161759. doi: 10.1371/journal.pone.0161759
151. Blanchard C, Stucke EM, Burwinkel K, Caldwell JM, Collins MH, Ahrens A, et al. Coordinate interaction between IL-13 and epithelial differentiation cluster genes in eosinophilic esophagitis. J Immunol. (2010) 184(7):4033–41. doi: 10.4049/jimmunol.0903069
152. Sherrill JD, Kc K, Wu D, Djukic Z, Caldwell JM, Stucke EM, et al. Desmoglein-1 regulates esophageal epithelial barrier function and immune responses in eosinophilic esophagitis. Mucosal Immunol. (2014) 7(3):718–29. doi: 10.1038/mi.2013.90
153. Bjarnason I, Goolamali SK, Levi AJ, Peters TJ. Intestinal permeability in patients with atopic eczema. Br J Dermatol. (1985) 112(3):291–7. doi: 10.1111/j.1365-2133.1985.tb04856.x
154. Leung AJ, Persad S, Slae M, Abdelradi A, Kluthe C, Shirton L, et al. Intestinal and gastric permeability in children with eosinophilic esophagitis and reflux esophagitis. J Pediatr Gastroenterol Nutr. (2015) 60(2):236–9. doi: 10.1097/MPG.0000000000000590
155. Peterson KA, Byrne KR, Vinson LA, Ying J, Boynton KK, Fang JC, et al. Elemental diet induces histologic response in adult eosinophilic esophagitis. Am J Gastroenterol. (2013) 108(5):759–66. doi: 10.1038/ajg.2012.468
156. Kelly KJ, Lazenby AJ, Rowe PC, Yardley JH, Perman JA, Sampson HA. Eosinophilic esophagitis attributed to gastroesophageal reflux: improvement with an amino acid-based formula. Gastroenterology. (1995) 109(5):1503–12. doi: 10.1016/0016-5085(95)90637-1
157. Mennini M, Tambucci R, Riccardi C, Rea F, De Angelis P, Fiocchi A, et al. Eosinophilic esophagitis and microbiota: state of the art. Front Immunol. (2021) 12:595762. doi: 10.3389/fimmu.2021.595762
158. Du Y, He C, An Y, Huang Y, Zhang H, Fu W, et al. The role of short chain fatty acids in inflammation and body health. Int J Mol Sci. (2024) 25(13):7379. doi: 10.3390/ijms25137379
159. Massimino L, Barchi A, Mandarino FV, Spanò S, Lamparelli LA, Vespa E, et al. A multi-omic analysis reveals the esophageal dysbiosis as the predominant trait of eosinophilic esophagitis. J Transl Med. (2023) 21(1):46. doi: 10.1186/s12967-023-03898-x
160. Brusilovsky M, Bao R, Rochman M, Kemter AM, Nagler CR, Rothenberg ME. Host-microbiota interactions in the esophagus during homeostasis and allergic inflammation. Gastroenterology. (2022) 162(2):521–34.e8. doi: 10.1053/j.gastro.2021.10.002
161. Holvoet S, Doucet-Ladevèze R, Perrot M, Barretto C, Nutten S, Blanchard C. Beneficial effect of lactococcus lactis NCC 2287 in a murine model of eosinophilic esophagitis. Allergy. (2016) 71(12):1753–61. doi: 10.1111/all.12951
162. Dai YX, Shi CB, Cui BT, Wang M, Ji GZ, Zhang FM. Fecal microbiota transplantation and prednisone for severe eosinophilic gastroenteritis. World J Gastroenterol. (2014) 20(43):16368–71. doi: 10.3748/wjg.v20.i43.16368
163. Lawrence MA, Baker PI. Ultra-processed food and adverse health outcomes. Br Med J. (2019) 365:l2289. doi: 10.1136/bmj.l2289
164. De Siena M, Raoul P, Costantini L, Scarpellini E, Cintoni M, Gasbarrini A, et al. Food emulsifiers and metabolic syndrome: the role of the gut microbiota. Foods. (2022) 11(15):2205. doi: 10.3390/foods11152205
165. Bhattacharyya S, Xue L, Devkota S, Chang E, Morris S, Tobacman JK. Carrageenan-induced colonic inflammation is reduced in Bcl10 null mice and increased in IL-10-deficient mice. Mediators Inflamm. (2013) 2013:397642. doi: 10.1155/2013/397642
166. Swidsinski A, Ung V, Sydora BC, Loening-Baucke V, Doerffel Y, Verstraelen H, et al. Bacterial overgrowth and inflammation of small intestine after carboxymethylcellulose ingestion in genetically susceptible mice. Inflamm Bowel Dis. (2009) 15(3):359–64. doi: 10.1002/ibd.20763
167. Benard C, Cultrone A, Michel C, Rosales C, Segain JP, Lahaye M, et al. Degraded carrageenan causing colitis in rats induces TNF secretion and ICAM-1 upregulation in monocytes through NF-kappaB activation. PLoS One. (2010) 5(1):e8666. doi: 10.1371/journal.pone.0008666
168. Wu W, Zhen Z, Niu T, Zhu X, Gao Y, Yan J, et al. Carrageenan enhances lipopolysaccharide-induced interleukin-8 secretion by stimulating the Bcl10-NF-. Mediators Inflamm. (2017) 2017:8634865. doi: 10.1155/2017/8634865
169. Shang Q, Sun W, Shan X, Jiang H, Cai C, Hao J, et al. Carrageenan-induced colitis is associated with decreased population of anti-inflammatory bacterium, akkermansia muciniphila, in the gut microbiota of C57BL/6J mice. Toxicol Lett. (2017) 279:87–95. doi: 10.1016/j.toxlet.2017.07.904
170. Jiang Z, Zhao M, Zhang H, Li Y, Liu M, Feng F. Antimicrobial emulsifier-glycerol monolaurate induces metabolic syndrome, gut microbiota dysbiosis, and systemic low-grade inflammation in low-fat diet fed mice. Mol Nutr Food Res. (2018) 62(3):1700547. doi: 10.1002/mnfr.201700547
171. Llewellyn SR, Britton GJ, Contijoch EJ, Vennaro OH, Mortha A, Colombel JF, et al. Interactions between diet and the intestinal microbiota alter intestinal permeability and colitis severity in mice. Gastroenterology. (2018) 154(4):1037–46.e2. doi: 10.1053/j.gastro.2017.11.030
172. Furuhashi H, Higashiyama M, Okada Y, Kurihara C, Wada A, Horiuchi K, et al. Dietary emulsifier polysorbate-80-induced small-intestinal vulnerability to indomethacin-induced lesions via dysbiosis. J Gastroenterol Hepatol. (2020) 35(1):110–7. doi: 10.1111/jgh.14808
173. Bhattacharyya S, Gill R, Chen ML, Zhang F, Linhardt RJ, Dudeja PK, et al. Toll-like receptor 4 mediates induction of the Bcl10-NFkappaB-interleukin-8 inflammatory pathway by carrageenan in human intestinal epithelial cells. J Biol Chem. (2008) 283(16):10550–8. doi: 10.1074/jbc.M708833200
174. Roberts CL, Keita AV, Duncan SH, O'Kennedy N, Söderholm JD, Rhodes JM, et al. Translocation of Crohns disease Escherichia coli across M-cells: contrasting effects of soluble plant fibres and emulsifiers. Gut. (2010) 59(10):1331–9. doi: 10.1136/gut.2009.195370
175. Choi HJ, Kim J, Park SH, Do KH, Yang H, Moon Y. Pro-inflammatory NF-κB and early growth response gene 1 regulate epithelial barrier disruption by food additive carrageenan in human intestinal epithelial cells. Toxicol Lett. (2012) 211(3):289–95. doi: 10.1016/j.toxlet.2012.04.012
176. Jiang HY, Wang F, Chen HM, Yan XJ. κ-carrageenan induces the disruption of intestinal epithelial Caco-2 monolayers by promoting the interaction between intestinal epithelial cells and immune cells. Mol Med Rep. (2013) 8(6):1635–42. doi: 10.3892/mmr.2013.1726
177. Lock JY, Carlson TL, Wang CM, Chen A, Carrier RL. Acute exposure to commonly ingested emulsifiers alters intestinal mucus structure and transport properties. Sci Rep. (2018) 8(1):10008. doi: 10.1038/s41598-018-27957-2
178. Naimi S, Viennois E, Gewirtz AT, Chassaing B. Direct impact of commonly used dietary emulsifiers on human gut microbiota. Microbiome. (2021) 9(1):66. doi: 10.1186/s40168-020-00996-6
179. Chassaing B, Koren O, Goodrich JK, Poole AC, Srinivasan S, Ley RE, et al. Dietary emulsifiers impact the mouse gut microbiota promoting colitis and metabolic syndrome. Nature. (2015) 519(7541):92–6. doi: 10.1038/nature14232
180. Chassaing B, Van de Wiele T, De Bodt J, Marzorati M, Gewirtz AT. Dietary emulsifiers directly alter human microbiota composition and gene expression ex vivo potentiating intestinal inflammation. Gut. (2017) 66(8):1414–27. doi: 10.1136/gutjnl-2016-313099
181. Viennois E, Bretin A, Dubé PE, Maue AC, Dauriat CJG, Barnich N, et al. Dietary emulsifiers directly impact adherent-invasive E. coli gene expression to drive chronic intestinal inflammation. Cell Rep. (2020) 33(1):108229. doi: 10.1016/j.celrep.2020.108229
182. Fahoum L, Moscovici A, David S, Shaoul R, Rozen G, Meyron-Holtz EG, et al. Digestive fate of dietary carrageenan: evidence of interference with digestive proteolysis and disruption of gut epithelial function. Mol Nutr Food Res. (2017) 61(3):1600545. doi: 10.1002/mnfr.201600545
183. Viennois E, Merlin D, Gewirtz AT, Chassaing B. Dietary emulsifier-induced low-grade inflammation promotes colon carcinogenesis. Cancer Res. (2017) 77(1):27–40. doi: 10.1158/0008-5472.CAN-16-1359
184. Borthakur A, Bhattacharyya S, Dudeja PK, Tobacman JK. Carrageenan induces interleukin-8 production through distinct Bcl10 pathway in normal human colonic epithelial cells. Am J Physiol Gastrointest Liver Physiol. (2007) 292(3):G829–38. doi: 10.1152/ajpgi.00380.2006
185. Harusato A, Chassaing B, Dauriat CJG, Ushiroda C, Seo W, Itoh Y. Dietary emulsifiers exacerbate food allergy and colonic type 2 immune response through microbiota modulation. Nutrients. (2022) 14(23):4983. doi: 10.3390/nu14234983
186. Xiao J, Jiang X, Zhou Y, Sumayyah G, Zhou L, Tu B, et al. Results of a 30-day safety assessment in young mice orally exposed to polystyrene nanoparticles. Environ Pollut. (2022) 292(Pt B):118184. doi: 10.1016/j.envpol.2021.118184
187. Liang B, Zhong Y, Huang Y, Lin X, Liu J, Lin L, et al. Underestimated health risks: polystyrene micro- and nanoplastics jointly induce intestinal barrier dysfunction by ROS-mediated epithelial cell apoptosis. Part Fibre Toxicol. (2021) 18(1):20. doi: 10.1186/s12989-021-00414-1
188. He Y, Li Z, Xu T, Luo D, Chi Q, Zhang Y, et al. Polystyrene nanoplastics deteriorate LPS-modulated duodenal permeability and inflammation in mice via ROS drived-NF-κB/NLRP3 pathway. Chemosphere. (2022) 307(Pt 1):135662. doi: 10.1016/j.chemosphere.2022.135662
189. Briceno Noriega D, Zenker HE, Croes CA, Ewaz A, Ruinemans-Koerts J, Savelkoul HFJ, et al. Receptor mediated effects of advanced glycation end products (AGEs) on innate and adaptative immunity: relevance for food allergy. Nutrients. (2022) 14(2):371. doi: 10.3390/nu14020371
190. Koschinsky T, He CJ, Mitsuhashi T, Bucala R, Liu C, Buenting C, et al. Orally absorbed reactive glycation products (glycotoxins): an environmental risk factor in diabetic nephropathy. Proc Natl Acad Sci U S A. (1997) 94(12):6474–9. doi: 10.1073/pnas.94.12.6474
191. Förster A, Kühne Y, Henle T. Studies on absorption and elimination of dietary maillard reaction products. Ann N Y Acad Sci. (2005) 1043:474–81. doi: 10.1196/annals.1333.054
192. Liu Z, Ma Y, Cui Q, Xu J, Tang Z, Wang Y, et al. Toll-like receptor 4 plays a key role in advanced glycation end products-induced M1 macrophage polarization. Biochem Biophys Res Commun. (2020) 531(4):602–8. doi: 10.1016/j.bbrc.2020.08.014
193. He S, Hu Q, Xu X, Niu Y, Chen Y, Lu Y, et al. Advanced glycation end products enhance M1 macrophage polarization by activating the MAPK pathway. Biochem Biophys Res Commun. (2020) 525(2):334–40. doi: 10.1016/j.bbrc.2020.02.053
194. Han X, Ma W, Zhu Y, Sun X, Liu N. Advanced glycation end products enhance macrophage polarization to the M1 phenotype via the HIF-1α/PDK4 pathway. Mol Cell Endocrinol. (2020) 514:110878. doi: 10.1016/j.mce.2020.110878
195. Paparo L, Coppola S, Nocerino R, Pisapia L, Picariello G, Cortese M, et al. How dietary advanced glycation end products could facilitate the occurrence of food allergy. J Allergy Clin Immunol. (2024) 153(3):742–58. doi: 10.1016/j.jaci.2023.11.023
196. Qiang X, Liang S, Lv Y, Wang X, Zhang H, Zhan J. Advanced glycation end products (AGEs) impair the intestinal epithelial barrier via STAT3 activation mediated by macrophages. Food Chem Toxicol. (2024) 192:114966. doi: 10.1016/j.fct.2024.114966
197. Samak G, Narayanan D, Jaggar JH, Rao R. Cav1.3 channels and intracellular calcium mediate osmotic stress-induced N-terminal c-Jun kinase activation and disruption of tight junctions in Caco-2 CELL MONOLAYERS. J Biol Chem. (2011) 286(34):30232–43. doi: 10.1074/jbc.M111.240358
198. Heilmann M, Wellner A, Gadermaier G, Ilchmann A, Briza P, Krause M, et al. Ovalbumin modified with pyrraline, a Maillard reaction product, shows enhanced T-cell immunogenicity. J Biol Chem. (2014) 289(11):7919–28. doi: 10.1074/jbc.M113.523621
199. Gwak MG, Chang SY. Gut-brain connection: microbiome, gut barrier, and environmental sensors. Immune Netw. (2021) 21(3):e20. doi: 10.4110/in.2021.21.e20
200. Carabotti M, Scirocco A, Maselli MA, Severi C. The gut-brain axis: interactions between enteric microbiota, central and enteric nervous systems. Ann Gastroenterol. (2015) 28(2):203–9.25830558
201. Martin CR, Osadchiy V, Kalani A, Mayer EA. The brain-gut-microbiome axis. Cell Mol Gastroenterol Hepatol. (2018) 6(2):133–48. doi: 10.1016/j.jcmgh.2018.04.003
202. Vandeputte D, Falony G, Vieira-Silva S, Tito RY, Joossens M, Raes J. Stool consistency is strongly associated with gut microbiota richness and composition, enterotypes and bacterial growth rates. Gut. (2016) 65(1):57–62. doi: 10.1136/gutjnl-2015-309618
203. Gustafsson BE, Midtvedt T, Strandberg K. Effects of microbial contamination on the cecum enlargement of germfree rats. Scand J Gastroenterol. (1970) 5(4):309–14. doi: 10.1080/00365521.1970.12096595
204. Amaral FA, Sachs D, Costa VV, Fagundes CT, Cisalpino D, Cunha TM, et al. Commensal microbiota is fundamental for the development of inflammatory pain. Proc Natl Acad Sci. (2008) 105(6):2193–7. doi: 10.1073/pnas.0711891105
205. Quigley EMM. Gut microbiome as a clinical tool in gastrointestinal disease management: are we there yet? Nat Rev Gastroenterol Hepatol. (2017) 14(5):315–20. doi: 10.1038/nrgastro.2017.29
206. Bonaz B, Bazin T, Pellissier S. The vagus nerve at the interface of the microbiota-gut-brain axis. Front Neurosci. (2018) 12:49. doi: 10.3389/fnins.2018.00049
207. Barbara G, Barbaro MR, Fuschi D, Palombo M, Falangone F, Cremon C, et al. Inflammatory and microbiota-related regulation of the intestinal epithelial barrier. Front Nutr. (2021) 8:718356. doi: 10.3389/fnut.2021.718356
208. Singh N, Gurav A, Sivaprakasam S, Brady E, Padia R, Shi H, et al. Activation of Gpr109a, receptor for niacin and the commensal metabolite butyrate, suppresses colonic inflammation and carcinogenesis. Immunity. (2014) 40(1):128–39. doi: 10.1016/j.immuni.2013.12.007
209. Ma Q, Xing C, Long W, Wang HY, Liu Q, Wang RF. Impact of microbiota on central nervous system and neurological diseases: the gut-brain axis. J Neuroinflammation. (2019) 16(1):53. doi: 10.1186/s12974-019-1434-3
210. Sampson TR, Mazmanian SK. Control of brain development, function, and behavior by the microbiome. Cell Host Microbe. (2015) 17(5):565–76. doi: 10.1016/j.chom.2015.04.011
211. Lee A, Lee JY, Jung SW, Shin SY, Ryu HS, Jang SH, et al. Brain-gut-microbiota axis. Korean J Gastroenterol. (2023) 81(4):145–53. doi: 10.4166/kjg.2023.028
212. Tolhurst G, Heffron H, Lam YS, Parker HE, Habib AM, Diakogiannaki E, et al. Short-chain fatty acids stimulate glucagon-like peptide-1 secretion via the G-protein-coupled receptor FFAR2. Diabetes. (2012) 61(2):364–71. doi: 10.2337/db11-1019
213. Ridlon JM, Kang DJ, Hylemon PB. Bile salt biotransformations by human intestinal bacteria. J Lipid Res. (2006) 47(2):241–59. doi: 10.1194/jlr.R500013-JLR200
214. Furusawa Y, Obata Y, Fukuda S, Endo TA, Nakato G, Takahashi D, et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature. (2013) 504(7480):446–50. doi: 10.1038/nature12721
215. Reigstad CS, Salmonson CE, Rainey JF, Szurszewski JH, Linden DR, Sonnenburg JL, et al. Gut microbes promote colonic serotonin production through an effect of short-chain fatty acids on enterochromaffin cells. FASEB J. (2015) 29(4):1395–403. doi: 10.1096/fj.14-259598
216. Yano JM, Yu K, Donaldson GP, Shastri GG, Ann P, Ma L, et al. Indigenous bacteria from the gut microbiota regulate host serotonin biosynthesis. Cell. (2015) 161(2):264–76. doi: 10.1016/j.cell.2015.02.047
217. Goldstein AM, Hofstra RM, Burns AJ. Building a brain in the gut: development of the enteric nervous system. Clin Genet. (2013) 83(4):307–16. doi: 10.1111/cge.12054
218. Ye L, Bae M, Cassilly CD, Jabba SV, Thorpe DW, Martin AM, et al. Enteroendocrine cells sense bacterial tryptophan catabolites to activate enteric and vagal neuronal pathways. Cell Host Microbe. (2021) 29(2):179–96.e9. doi: 10.1016/j.chom.2020.11.011
219. Valles-Colomer M, Falony G, Darzi Y, Tigchelaar EF, Wang J, Tito RY, et al. The neuroactive potential of the human gut microbiota in quality of life and depression. Nat Microbiol. (2019) 4(4):623–32. doi: 10.1038/s41564-018-0337-x
220. Miri S, Yeo J, Abubaker S, Hammami R. Neuromicrobiology, an emerging neurometabolic facet of the gut microbiome? Front Microbiol. (2023) 14:1098412. doi: 10.3389/fmicb.2023.1098412
221. Carecho R, Carregosa D, Dos Santos CN. Low molecular weight (poly)phenol metabolites across the blood-brain barrier: the underexplored journey. Brain Plast. (2021) 6(2):193–214. doi: 10.3233/BPL-200099
222. Ghosh S, Whitley CS, Haribabu B, Jala VR. Regulation of intestinal barrier function by microbial metabolites. Cell Mol Gastroenterol Hepatol. (2021) 11(5):1463–82. doi: 10.1016/j.jcmgh.2021.02.007
223. Allam-Ndoul B, Castonguay-Paradis S, Veilleux A. Gut microbiota and intestinal trans-epithelial permeability. Int J Mol Sci. (2020) 21(17):6402. doi: 10.3390/ijms21176402
224. Lee YY, Annamalai C, Rao SSC. Post-infectious irritable bowel syndrome. Curr Gastroenterol Rep. (2017) 19(11):56. doi: 10.1007/s11894-017-0595-4
225. Hayes CL, Dong J, Galipeau HJ, Jury J, McCarville J, Huang X, et al. Commensal microbiota induces colonic barrier structure and functions that contribute to homeostasis. Sci Rep. (2018) 8(1):14184. doi: 10.1038/s41598-018-32366-6
226. De Palma G, Collins SM, Bercik P. The microbiota-gut-brain axis in functional gastrointestinal disorders. Gut Microbes. (2014) 5(3):419–29. doi: 10.4161/gmic.29417
227. Hooper LV, Wong MH, Thelin A, Hansson L, Falk PG, Gordon JI. Molecular analysis of commensal host-microbial relationships in the intestine. Science. (2001) 291(5505):881–4. doi: 10.1126/science.291.5505.881
228. Ukena SN, Singh A, Dringenberg U, Engelhardt R, Seidler U, Hansen W, et al. Probiotic Escherichia coli Nissle 1917 inhibits leaky gut by enhancing mucosal integrity. PLoS One. (2007) 2(12):e1308. doi: 10.1371/journal.pone.0001308
229. Barbaro MR, Fuschi D, Cremon C, Carapelle M, Dino P, Marcellini MM, et al. Escherichia coli Nissle 1917 restores epithelial permeability alterations induced by irritable bowel syndrome mediators. Neurogastroenterol Motil. (2018) 30(8):e13388. doi: 10.1111/nmo.13388
230. Cenac N, Bautzova T, Le Faouder P, Veldhuis NA, Poole DP, Rolland C, et al. Quantification and potential functions of endogenous agonists of transient receptor potential channels in patients with irritable bowel syndrome. Gastroenterology. (2015) 149(2):433–44.e7. doi: 10.1053/j.gastro.2015.04.011
231. Bautzova T, Hockley JRF, Perez-Berezo T, Pujo J, Tranter MM, Desormeaux C, et al. 5-oxoETE Triggers nociception in constipation-predominant irritable bowel syndrome through MAS-related G protein-coupled receptor D. Sci Signal. (2018) 11(561):eaal2171. doi: 10.1126/scisignal.aal2171
232. Barbara G, Wang B, Stanghellini V, de Giorgio R, Cremon C, Di Nardo G, et al. Mast cell-dependent excitation of visceral-nociceptive sensory neurons in irritable bowel syndrome. Gastroenterology. (2007) 132(1):26–37. doi: 10.1053/j.gastro.2006.11.039
233. Durbán A, Abellán JJ, Jiménez-Hernández N, Artacho A, Garrigues V, Ortiz V, et al. Instability of the faecal microbiota in diarrhoea-predominant irritable bowel syndrome. FEMS Microbiol Ecol. (2013) 86(3):581–9. doi: 10.1111/1574-6941.12184
234. Gargari G, Taverniti V, Gardana C, Cremon C, Canducci F, Pagano I, et al. Fecal Clostridiales distribution and short-chain fatty acids reflect bowel habits in irritable bowel syndrome. Environ Microbiol. (2018) 20(9):3201–13. doi: 10.1111/1462-2920.14271
235. Kuo WT, Zuo L, Odenwald MA, Madha S, Singh G, Gurniak CB, et al. The tight junction protein ZO-1 is dispensable for barrier function but critical for effective mucosal repair. Gastroenterology. (2021) 161(6):1924–39. doi: 10.1053/j.gastro.2021.08.047
236. Alipour M, Zaidi D, Valcheva R, Jovel J, Martínez I, Sergi C, et al. Mucosal barrier depletion and loss of bacterial diversity are primary abnormalities in paediatric ulcerative colitis. J Crohns Colitis. (2016) 10(4):462–71. doi: 10.1093/ecco-jcc/jjv223
237. Png CW, Lindén SK, Gilshenan KS, Zoetendal EG, McSweeney CS, Sly LI, et al. Mucolytic bacteria with increased prevalence in IBD mucosa augment in vitro utilization of mucin by other bacteria. Am J Gastroenterol. (2010) 105(11):2420–8. doi: 10.1038/ajg.2010.281
238. Machiels K, Joossens M, Sabino J, De Preter V, Arijs I, Eeckhaut V, et al. A decrease of the butyrate-producing species Roseburia hominis and Faecalibacterium prausnitzii defines dysbiosis in patients with ulcerative colitis. Gut. (2014) 63(8):1275–83. doi: 10.1136/gutjnl-2013-304833
239. Zhou L, Zhang M, Wang Y, Dorfman RG, Liu H, Yu T, et al. Faecalibacterium prausnitzii produces butyrate to maintain Th17/Treg balance and to ameliorate colorectal colitis by inhibiting histone deacetylase 1. Inflamm Bowel Dis. (2018) 24(9):1926–40. doi: 10.1093/ibd/izy182
240. Woznicki JA, Saini N, Flood P, Rajaram S, Lee CM, Stamou P, et al. TNF-α synergises with IFN-γ to induce caspase-8-JAK1/2-STAT1-dependent death of intestinal epithelial cells. Cell Death Dis. (2021) 12(10):864. doi: 10.1038/s41419-021-04151-3
Keywords: intestinal permeability, gut barrier, celiac disease, food allergy, eosinophilic gastrointestinal disorders, irritable bowel syndrome
Citation: Valitutti F, Mennini M, Monacelli G, Fagiolari G, Piccirillo M, Di Nardo G and Di Cara G (2025) Intestinal permeability, food antigens and the microbiome: a multifaceted perspective. Front. Allergy 5:1505834. doi: 10.3389/falgy.2024.1505834
Received: 3 October 2024; Accepted: 16 December 2024;
Published: 9 January 2025.
Edited by:
Eishin Morita, Shimane University, JapanReviewed by:
Alex Dent, Indiana University, Purdue University Indianapolis, United StatesMarat Khodoun, University of Cincinnati, United States
Copyright: © 2025 Valitutti, Mennini, Monacelli, Fagiolari, Piccirillo, Di Nardo and Di Cara. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Francesco Valitutti, ZnJhbmNlc2NvLnZhbGl0dXR0aUB1bmlwZy5pdA==
 Francesco Valitutti
Francesco Valitutti Maurizio Mennini
Maurizio Mennini Gianluca Monacelli1
Gianluca Monacelli1 Marisa Piccirillo
Marisa Piccirillo Giovanni Di Nardo
Giovanni Di Nardo Giuseppe Di Cara
Giuseppe Di Cara