- 1Department of Emergency Medicine, Camden Clark Medical Center, West Virginia University, Parkersburg, WV, United States
- 2Department of Pathology, Feinberg School of Medicine, Northwestern University, Chicago, IL, United States
The Acari Hypothesis posits that acarians, i.e., mites and ticks, are operative agents of allergy. It derived from observations that allergens are molecular elements of acarians or acarian foodstuffs. A corollary of The Hypothesis provides how acarian dietary elements are selected as allergens; namely, a pattern recognition receptor native to the acarian digestive tract complexes with dietary molecules problematic to the acarian. By virtue of its interspecies operability, the receptor then enables not only removal of the dietary elements by the acarian immune system, but also—should such a complex be inoculated into a human—production of an element-specific IgE. Because pattern recognition receptors bind to molecules problematic to the organism from which the receptors originate, it follows that molecules targeted by adaptive IgE, i.e., allergens, must be problematic to acarians. This claim is supported by evidence that host organisms, when infested by acarians, upregulate representative members of allergenic molecular families. Appreciation of the relationship between allergens and acarians provides insight well beyond allergy, shedding light also on the anti-acarian defenses of many living things, especially humans.
1 Introduction
In hope of identifying a unifying principle on the nature of allergenicity, existing allergy research has focused primarily on structural features of allergenic molecules. Indeed, most allergens derive from about 30–40 molecular families (1). Although within a given family significant sequential and structural homologies exist, allergenic molecules from different families vary greatly. Consequently, no truly unifying principle has come from structural analyses alone.
A teleological rationalization of allergenicity was proposed by Profet in 1991 (2). It postulates that allergens are molecules generally toxic to humans, with mast cell-mediated immune activation constituting a last line of defense against allergens as toxins. Indeed, such rationalization is supported by allergens attributable to venomous hymenopterans, e.g., Apis mellifera, the Western honeybee. With specific regard to A. mellifera, at least 12 allergens have been identified, all of which are expressed in the bee venom sac, venom duct and/or hypopharyngeal gland (3). Thus, at least in the case of Western honeybees, IgE does appear to target materials that might adversely affect human physiology.
Although the proposal of Profet accounts reasonably well for the allergenicity of molecules expressed by honeybees, it does not account for the inadequacy of the response directed against those molecules, i.e., the gross mechanical reflexes elicited by IgE do not effectively deter the sting of a bee. Furthermore, if honeybee venom is threatening to all humans, then why do so few humans elicit an IgE-mediated response following envenomation? In the context of otherwise benign allergens, the proposal is even more problematic. Pollens, as examples, cause debilitating IgE-mediated symptoms in many humans (4).
The Acari Hypothesis was conceived only very recently (5–8). Just as does the proposal of Profet, it foregoes structural considerations in the assignment of allergenicity using, instead, teleological considerations for that purpose. The Hypothesis proposes most, if not all, allergens derive from acarians or their foodstuffs. One of its corollaries offers mechanistic understanding relevant to the elicitation of IgE. It proposes an acarian pattern recognition receptor, perhaps a fibrinogen-related protein (FReP) operating within the acarian digestive tract, elicits IgE in humans following inoculation of the receptor and its associated materials (6).
Acarian FRePs, like many FRePs of other species, function as innate immune effectors (9). They bind and agglutinate materials that display molecular patterns dangerous to the acarian, earmarking those materials for phagocytosis and degradation (10). The fibrinogen-like domain of some FRePs contains multiple binding sites that can operate independently or synergistically (11). This enables them to recognize a vast array of endogenous and exogenous ligands. As envisaged per The Hypothesis, acarian FRePs may complex with 2 categories of allergens: (1) acarian immunoregulators, e.g., Der p 2, a major allergen of the human dust mite and a homolog of human myeloid differentiation factor 2 (12), and/or (2) molecular elements of acarian foodstuffs, Figure 1. Following inoculation into a human, complexes incorporating category 1 allergens prime the recipient for felicitous IgE directed against the acarian. In contrast, complexes incorporating category 2 allergens prime the recipient for specious IgE directed against the acarian dietary element. There is yet a third category of allergens: molecules that cross-react with IgE directed against category 1 or 2 allergens. Allergens from category 3 are always problematic. They often derive from invertebrates, e.g., insects, mollusks or helminths, because the allergenic molecules of mites, e.g., tropomyosin or paramyosin, share strong homology with the molecules of other invertebrates (13–17).
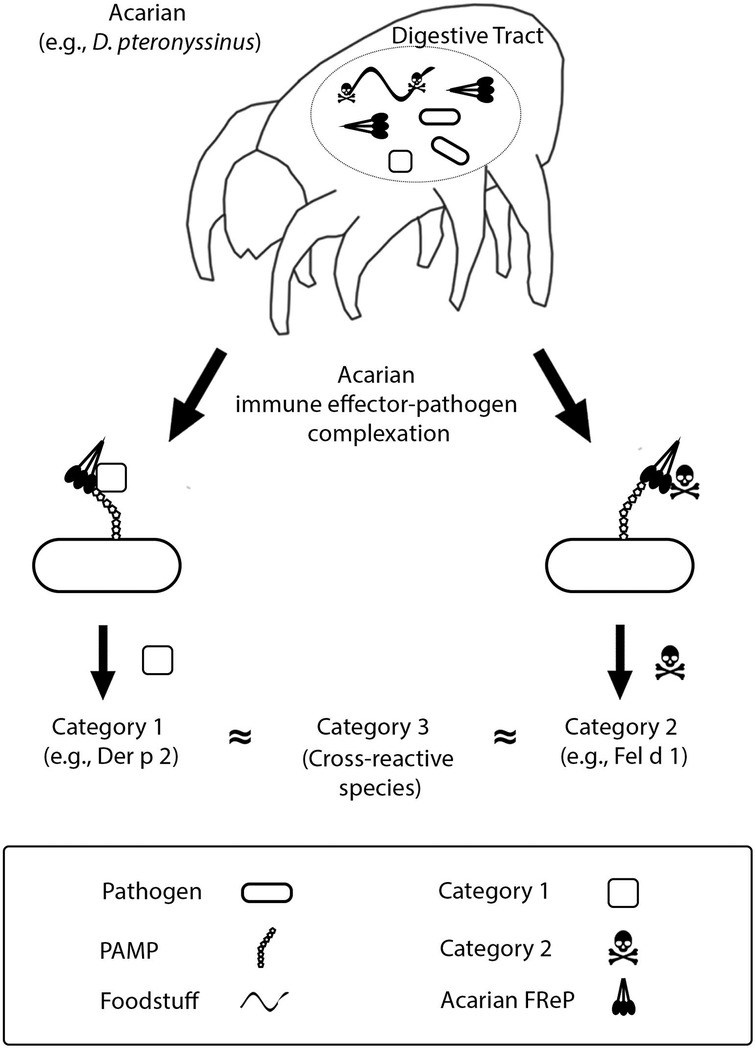
Figure 1. Deciphering allergenicity. See text and previous installments of this series for details (5–8).
Indeed, the tropomyosin family exemplifies the complicated nature of IgE reactivity. Cockroach tropomyosins, which are homologous with acarian tropomyosins, are strongly allergenic (18–20). Thus, they may be category 3 allergens in persons sensitized to acarian tropomyosin. Cockroaches can also serve as phoretic hosts or foodstuffs for some acarian species (21, 22), rendering them a source of category 2 allergen. In short, depending upon the situation, cockroach materials are sometimes sensitizing and other times cross-reacting. This situational dependence undoubtedly also applies to other important allergens, even ones that derive from acarians.
In all instances, the human immune system construes the allergen as acarian in nature. Following subsequent exposure, the system activates reflexes intended to clear acarians from epithelial surfaces, e.g., cough, spit, itch, vomit, defecate, etc.
The Acari Hypothesis explains the observations that gave rise to the Profet proposal. It also accounts for the shortcomings of that proposal. Category 2 allergens are materials uniformly toxic to acarians, not to humans. Although some category 2 allergens are toxic to humans as well, e.g., melittin, most are problematic only to the acarian species from which the sensitizing complex originated.
Researchers studying plant immunity have classified 19 groups of molecules participating in plant defense against pathogens and phytophagous arthropods, i.e., pathogenesis-related proteins (PR 1–19) (23, 24). PR proteins are well-represented among human allergens (25), entirely consistent with the idea that IgE-targeted materials are toxic to acarians. From among the groups of PR proteins, the ones best exemplifying this unifying allergenic attribute are the defensins, cystatins, peroxidases, chitinases, and PR-10 proteins. That being the case, the remainder of this report focuses on them and three other families of common category 2 allergens: lipocalins, secretoglobins and palate lung nasal clone (PLUNC) proteins.
2 Defensins
The defensins are cysteine-rich, cationic immuno-polypeptides expressed by many plants and animals (26, 27). The group includes at least two protein superfamilies, each of which took a convergent evolutionary path predicated on the utility of a structural motif consisting of a cysteine-dense core and exteriorly displayed loops (28, 29). Defensins are well-represented within the human allergen database and include molecules from ragweed, peanut, celery and soybean (30).
Functionally, defensins have antimicrobial activity against a diverse microbial repertoire (31, 32). Their antimicrobial activity is believed to be due to direct interaction with cell membranes, causing lytic defects (33). Some defensins also inhibit α-amylase (34, 35), an enzyme commonly expressed within the digestive tract of arthropods, including acarians (36). Inhibition of α-amylase limits the ability of arthropods to digest foodstuffs. Consistent with an anti-acarian role, organisms upregulate defensins in response to acarian infestation. As an example, A. mellifera is subject to parasitism by the mite, Varroa destructor (37). Upon infestation by the mite, defensins are upregulated in A. mellifera, and increasing mite infestation increases defensin expression (38, 39). Because defensins inhibit acarian digestion, it is reasonable to assume acarian immune effectors operating within the acarian digestive tract complex with, and neutralize, defensins.
Plant defensins are categorized in the PR-12 grouping, and their role in defense against pathogens and phytophagous acarians is very well-established. As an example, Arabidopsis upregulates defensins in response to herbivory by the spider mite, Tetranychus urticae (40).
Humans also produce defensins, and there is evidence human defensins are involved in IgE-mediated disease. Human β-defensin-1 (HBD-1) and human β-defensin-2 (HBD-2) are innate immune effectors constitutively secreted by epithelial cells and leukocytes (41, 42). Consistent with the ideas that IgE-mediated disease is caused by acarian species and HBDs participate in human anti-acarian defense, levels of HBD-1 and -2 are altered in allergic disease (43, 44) and functional mutations in HBD-1 and -2 predispose children to both atopic dermatitis and asthma (45, 46).
3 Cystatins
Cystatins are a family of small proteins that are also well-represented in the database of the allergens of humans. They include molecules from plants, e.g., ragweed and kiwi (47–49), and mammals, e.g., canines and felines (50, 51). Plant cystatins are commonly referred to as phytocystatins and are assigned to the PR-6 grouping. The primary function of phytocystatins is inhibition of cysteine proteases, including those used by acarians to digest foodstuffs (52). Such inhibition deprives acarians of essential nutrients.
There are many examples of plant species that upregulate phytocystatins in direct response to acarian herbivory (53). As one example, phytocystatins expressed by barley protect against acarian parasitism (54). Indeed, transgenic expression of barley phytocystatins by wheat and maize confers anti-acarian protection (55). Maize, too, upregulates phytocystatins, especially in response to parasitism by mites T. urticae and Oligonychus pratensis (56). As another example, a cystatin expressed in the seeds of chestnuts inactivates Der f 1, the dominant allergen and well-described cysteine protease of the dust mite, Dermatophagoides farinae (57, 58).
Dermatophagoides is a genus of polyphagous synanthropic acarian proposed by The Acari Hypothesis to be a primary etiological agent of allergic disease. To digest human materials, the mites use cysteine proteases, like Der p 1 and Der f 1 (59). As do plants, humans express their own array of cystatins. Cystatin A is a cysteine protease inhibitor expressed by human keratinocytes (60). It neutralizes Der f 1 (61). Thus, just as phytocystatins defend plants against herbivory by phytophagous acarians, human cystatins defend humans against carnivory by anthropophagous acarians. Consistent with the proposal that mites are the etiological agents of allergy, a mutation of cystatin A is associated with development of atopic dermatitis (62). Another human cystatin, the salivary cystatin SN, has also been shown to inhibit mite proteases, and its expression is upregulated in asthmatic patients (63).
4 Peroxidases
Plant peroxidases are another family of molecules used by plants to defend against pathogens and phytophagous acarians. Peroxidases are assigned to the PR-9 grouping. They generate and detoxify peroxide, thereby regulating reactive oxygen species, which are directly toxic to numerous pathogens (23, 64, 65).
Many plant species use peroxidase in defense against acarian infestation. As examples, Solanum dulcamara upregulates peroxidase following its infestation by the gall mite, Aceria cladophthirus (66), and peroxidase activity confers resistance to T. urticae infestation by both the hops plant, Humulus lupulus, and the cassava plant, Manihot esculenta (67, 68). As in the cases of other PR proteins, peroxidases are well-represented among the allergens of humans. They include enzymes expressed by wheat, bananas and tomatoes (69–71).
Just as humans express analogs of plant defensins and phytocystatins, so, too, do humans express a peroxidase, and that peroxidase is involved in allergic inflammation. In humans, eosinophils mediate the late-stage inflammation characteristic of allergic disease (72). The cells drive inflammation via the release of chemical mediators, including eosinophilic peroxidase (73, 74). Obvious parallels between the secretory milieu of plants following acarian infestation and that of humans during bouts of allergic disease lend further credence to the proposal IgE is intended to target acarian species.
5 Chitinases
Chitin [β- (1–4)-poly-N-acetyl-D-glucosamine] is the primary structural component of acarian exoskeletons (75, 76). Because it is essential to acarian physiology, many organisms have evolved enzymes to degrade chitin, i.e., chitinases, to defend against acarian threats. As examples, maize and barley upregulate chitinase expression in response to infestation by phytophagous mites (56).
Using current nomenclature, and based on structural characteristics, chitinases are categorized into PR groupings 3, 4, 8 and 11 (77). Chitinases are well-represented in the database of known allergens of humans, and they include molecules from bananas, avocados, tomatoes and maize (78–81).
The human proteome includes two chitinases, chitotriosidase 1 (CHIT1) and acid mammalian chitinase (AMCase), and evidence indicates both enzymes play a role in IgE-mediated disease. CHIT1 is expressed by epithelial cells and monocytes (82). AMCase is expressed by monocytes/macrophages, lung epithelial cells and natural killer cells (83). Levels of CHIT1 and AMCase are elevated in persons with allergic disease (84–86). Additionally, genetic variations in AMCase predispose to bronchial asthma (87). These observations highlight yet other instances in which diverse organisms use a shared means to defend against acarian species.
6 PR-10 proteins and lipocalins
PR-10 proteins are another group of phytoproteins that participate in plant defense against pathogens and phytophagous acarians. Although their exact mechanism-of-action is unknown, it likely relates to a large hydrophobic cavity that spans the length of the molecules (88). The cavity, which binds a wide variety of ligands, is suspected to sequester and/or deliver hydrophobic molecules (88), perhaps ones vital to acarian metabolism/physiology.
The PR-10 proteins are well-represented in the database of known allergens of humans and include molecules from—among many other sources—birch pollen (Bet v 1), cherries, peanuts, tomatoes, celery and walnuts (89–93). They, too, are upregulated in response to acarian infestation. As an example, the subterranean clover, Trifolium subterraneum, upregulates a PR-10 protein in response to infestation by the red-legged earth mite, Halotydeus destructor (94).
Although PR-10 proteins do not have obvious mammalian analogues, they share structural homology with lipocalins (95), a family of proteins expressed by bacteria, plants and animals, including mammals (96, 97). Just as the structure of PR-10 proteins features a large central cavity for transporting and/or sequestering hydrophobic molecules, so does the structure of lipocalins (98). Given this feature, it is not unreasonable to think lipocalins share the anti-acarian activity of PR-10 proteins. Importantly, the secretory lipocalins of domesticated mammals are a major source of the allergens of humans (99). As examples, the lipocalins, Can f 1, Can f 2, Can f 4 and Can f 6 are allergens derived from canines (100).
Relatedly, humans secrete apolipoprotein D (apo D), a lipocalin, onto their epithelial surfaces (101, 102). Apo D, produced by eccrine glands, is the third most abundant protein in the sweat of healthy humans (102). In the context of The Hypothesis, it was recently theorized that the expansion of eccrine glands across the epidermal surface of catarrhine primates was a consequence of an acarian-related evolutionary pressure (8). As a consequence of that pressure, eccrine gland secretions evolved to become the primary anti-acarian defense of humans. Thus, the inclusion of apo D in those secretions is entirely consistent with an anti-acarian role for lipocalins.
Catarrhine primates are not the only mammals subject to acarian ectoparasitism. Just as catarrhines developed an expanded eccrine glandular system to deal with acarian threats, other mammals evolved other means to protect from acarians. And just as IgE informs on the anti-acarian activities of plant proteins, so, too, does it inform on the anti-acarian activities of mammalian proteins. Although the library of the allergens of humans continues to expand, mammalian allergens from two molecular families, secretoglobins and PLUNC proteins, are already well-described. These last two will be elaborated next, in the context of The Hypothesis.
7 Secretoglobins
Secretoglobins are another family of proteins against which humans produce IgE (103). They are expressed by mammals exclusively, and representative molecules are typically secreted onto epithelial surfaces (104). They are found in abundance within the surface liquids of the uterus, prostate, lungs, and lacrimal and salivary glands (104). Secretoglobins are released as disulfide-linked dimers that, like PR-10 proteins and lipocalins, have an internal hydrophobic cavity that binds and transports small hydrophobic molecules (104). Two secretoglobins are well-described allergens. Fel d 1, a major allergen from felines, is expressed in cat saliva. It is also secreted by cat sebaceous glands (105) and, therefore, present in exfoliated feline scale and related detritus. Ory c 3, a major allergen from rabbits, is present on rabbit fur (106). Although the functions of these allergenic molecules are not yet well-defined, their localization to epidermis ideally positions them to defend against acarian ectoparasitism.
The best studied human secretoglobin, secretoglobin family 1A member 1 (SCGB1A1), is expressed by clara cells, which are non-ciliated secretory cells of the upper airways (107). In comparison to healthy controls, asthmatics have lower levels of SCGB1A1 in both serum and bronchoalveolar fluid (108), and mutations in SCGB1A1 are associated with increased asthmatic risk (109). If, as proposed, asthma is an IgE-associated bronchospastic disease attributable to acarian species, and SCGB1A1 has an anti-acarian role, then one would expect both low levels and mutations of SCGB1A1 to influence the development of the disease.
8 PLUNC proteins
PLUNC proteins are a molecular family unique to air-breathing vertebrates (110). Like secretoglobins, they are secreted by mammals onto epithelial surfaces. PLUNC proteins are expressed abundantly in liquids that bathe oral, nasal and upper respiratory airways, where they act as surfactants and participate in mucosal immunity (111). Examples of major allergens include Fel d 8, which is secreted from the salivary glands of cats (112), and Ecu c 4, i.e., latherin, the dominant protein of equine sweat (113, 114).
Equine sweat is fundamentally different from human sweat, having developed via a distinct evolutionary pathway. Whereas human sweat is made by eccrine glands, equine sweat is made by apocrine glands and is much more protein-rich than its human counterpart (115). Like human sweat, equine sweat contains a lipocalin, in this case, the allergen, Ecu c 1 (114). Given the targeting of molecules in equine sweat by IgE, it is tempting to speculate that equine sweat, like human sweat, developed in response to acarian-related evolutionary pressure.
The most studied of the human PLUNC proteins is BPI fold-containing family A, member 1 (BPIFA1), a protein expressed in upper airways and involved in innate immunity (116, 117). Two experimental findings support the notion human PLUNC proteins participate in anti-acarian defense. Firstly, levels of BPIFA1 are markedly diminished in the airways of asthmatics (118). Secondly, BPIFA1 is cleaved and inactivated by Der p 1, an allergenic cysteine protease of the dust mite, Dermatophagoides pteronyssinus (119). Given the similarities of the epithelial secretions of diverse lineages of mammalian species and the proposed role of human epithelial secretions in anti-acarian defense, it appears the Acari significantly influenced mammalian evolution, particularly that having to do with epithelial surfaces.
9 Closing
By means of direct recognition and subsequent opsonization, some acarian immune effectors that operate in the acarian digestive tract neutralize pathogenic/toxic materials. According to The Acari Hypothesis, those effectors have interspecies operability and are involved in initiation of the adaptive IgE response of humans. If this is true, then it stands to reason non-acarian materials targeted by IgE must be pathogenic/toxic to acarians. Such reasoning is certainly valid for the allergens reviewed in this report: defensins, cystatins, peroxidases, chitinases, and PR-10 proteins.
As proposed and elaborated elsewhere, following a near extinction event suffered by our ancestral catarrhine primates, sweating evolved and displaced IgE as dominant anti-acarian defense (8). Because sweating has, in recent times, been abrogated by human hygiene, IgE has returned to the forefront as an operational anti-acarian defense. However, having evolved long ago to engage the parasitic predecessors of contemporary synanthropic mites, the IgE response is now largely anachronistic. As an example, Dermatophagoides, a genera of contemporary dust mites whose diet is exceptionally diverse and includes bacteria and fungi (120, 121), is a member of the family, Pyroglyphidae (122). Pyroglyphidae descended from Psoroptidia, a parvorder of acarians that parasitizes birds and mammals (123). Pyroglyphidae followed an exceedingly unusual evolutionary pathway, having evolved from parasites to free-living scavengers (124).
Because: (1) allergenicity depends upon the dietary choices of parasitizing acarians, and (2) the ancestors of synanthropic acarians that now cause allergy were carnivorous, our mammalian ancestors might have been at risk for IgE-mediated red meat allergy had they not themselves expressed galactose-α-1,3-galactose (α-gal) (125). It was only after catarrhine primates stopped expressing α-gal that the possibility of allergy to the disaccharide could exist. Then, only after parasitic mites became polyphagous did allergies beyond red meat become possible. In short, allergies suffered by humans today are consequences of modern-day hygiene and modern-day diversification of the diets of domestic Pyroglyphidae.
The Acari Hypothesis is yet unproven. It assumes a role for FRePs in the development of mammalian Th2 immunity. Further, it attributes to mammals the ability to utilize acarian FRePs to direct the IgE arm of the Th2 response. Extensive testing is required to determine whether either of these is valid. If both are valid, however, then elucidation of how IgE is elicited yields information relevant to more than just allergy. Perhaps most importantly, it facilitates the identification of all manner of molecules and mechanisms used by lifeforms in defense against acarians. Although this report focuses on endogenous defensive molecules of plants and animals, the defenses of plants and animals likely also include materials expressed by constituents of their respective microbiomes. As demonstrated by the pathophysiology of atopic dermatitis, IgE targets an extensive array of molecules expressed by Malassezia spp., indicating this genus must be problematic to acarians (126, 127).
Amongst fungal species, Malassezia is unusual in that members of the genus lack a gene for fatty acid synthase (128). Absent this enzyme, Malassezia cannot generate fatty acids essential for survival. Consequently, the fungus has an absolute requirement for exogenous fatty acids. Human sebum is composed primarily of triglycerides and free fatty acids (129). Given the nutrient requirements of Malassezia, it comes as no surprise secretion of sebum onto mammalian epidermis yields an environment especially conducive to malassezial growth. Indeed, Malassezia is the dominant eukaryote of the epidermal microbiome of adult humans (130).
The apparent anti-acarian activity of Malassezia as well as the eminent suitability of mammalian skin for malassezial growth makes it likely the relationship between mammals and Malassezia is one of mutualism, wherein mammals supply Malassezia with essential lipids and the fungus defends humans from acarians. A role for sebaceous glands in human anti-acarian defense is supported by the epidemiology of allergy, the incidence of which is highest during childhood (131–133) and lowest following puberty, when sebaceous gland output and Malassezia colonization rise precipitously (134–137). The impact of the mutualism of humans and Malassezia on the epidemiology of allergy is elaborated in the next installment of this series (ACR, submitted).
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author contributions
AR: Conceptualization, Investigation, Writing – original draft, Writing – review & editing. GR: Conceptualization, Resources, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was supported in part by funding to GR from the Department of Pathology, Feinberg School of Medicine, Northwestern University.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Dramburg S, Hilger C, Santos AF, de las Vecillas L, Aalberse RC, Acevedo N, et al. EAACI molecular allergology user’s guide 2.0. Pediatr Allergy Immunol. (2023) 34(Suppl 28):e13854. doi: 10.1111/pai.13854
2. Profet M. The function of allergy: immunological defense against toxins. Q Rev Biol. (1991) 66(1):23–62. doi: 10.1086/417049
3. Burzyńska M, Piasecka-Kwiatkowska D. A review of honeybee venom allergens and allergenicity. Int J Mol Sci. (2021) 22(16):8371. doi: 10.3390/ijms22168371
4. D'Amato G, Murrieta-Aguttes M, D'Amato M, Ansotegui IJ. Pollen respiratory allergy: is it really seasonal? World Allergy Organ J. (2023) 16(7):100799. doi: 10.1016/j.waojou.2023.100799
5. Retzinger AC, Retzinger GS. Mites, ticks, anaphylaxis and allergy: the acari hypothesis. Med Hypotheses. (2020) 144:110257. doi: 10.1016/j.mehy.2020.110257
6. Retzinger AC, Retzinger GS. The acari hypothesis, II: interspecies operability of pattern recognition receptors. Pathogens. (2021) 10(9):1220. doi: 10.3390/pathogens10091220
7. Retzinger AC, Retzinger GS. The acari hypothesis, III: atopic dermatitis. Pathogens. (2022) 11(10):1083. doi: 10.3390/pathogens11101083
8. Retzinger AC, Retzinger GS. The acari hypothesis, IV: revisiting the role of hygiene in allergy. Front Allergy. (2024) 5:1415124. doi: 10.3389/falgy.2024.1415124
9. Hanington PC, Zhang SM. The primary role of fibrinogen-related proteins in invertebrates is defense, not coagulation. J Innate Immun. (2010) 3(1):17–27. doi: 10.1159/000321882
10. Honig Mondekova H, Sima R, Urbanova V, Kovar V, Rego ROM, Grubhoffer L, et al. Characterization of ixodes ricinus fibrinogen-related proteins (ixoderins) discloses their function in the tick innate immunity. Front Cell Infect Microbiol. (2017) 7:509. doi: 10.3389/fcimb.2017.00509
11. Bidula S, Sexton DW, Schelenz S. Ficolins and the recognition of pathogenic microorganisms: an overview of the innate immune response and contribution of single nucleotide polymorphisms. J Immunol Res. (2019) 2019:1. doi: 10.1155/2019/3205072
12. Trompette A, Divanovic S, Visintin A, Blanchard C, Hegde RS, Madan R, et al. Allergenicity resulting from functional mimicry of a toll-like receptor complex protein. Nature. (2009) 457(7229):585–8. doi: 10.1038/nature07548
13. Shafique RH, Inam M, Ismail M, Chaudhary FR. Group 10 allergens (tropomyosins) from house-dust mites may cause covariation of sensitization to allergens from other invertebrates. Allergy Rhinol (Providence). (2012) 3(2):e74–90. doi: 10.2500/ar.2012.3.0036
14. Bernardini R, Mistrello G, Novembre E, Roncarolo D, Zanotta S, Lombardi E, et al. Cross-Reactivity between IgE-binding proteins from Anisakis simplex and Dermatophagoides pteronyssinus. Int J Immunopathol Pharmacol. (2005) 18(4):671–5. doi: 10.1177/039463200501800408
15. Wong L, Huang CH, Lee BW. Shellfish and house dust mite allergies: is the link tropomyosin? Allergy Asthma Immunol Res. (2016) 8(2):101–6. doi: 10.4168/aair.2016.8.2.101
16. De Marchi L, Wangorsch A, Zoccatelli G. Allergens from edible insects: cross-reactivity and effects of processing. Curr Allergy Asthma Rep. (2021) 21(5):35. doi: 10.1007/s11882-021-01012-z
17. Yu C, Ding X, Gao X, Lin H, Ullah Khan M, Lin H, et al. Immunological cross-reactivity involving mollusc species and mite-mollusc and cross-reactive allergen PM are risk factors of mollusc allergy. J Agric Food Chem. (2022) 70(1):360–72. doi: 10.1021/acs.jafc.1c05421
18. Ayuso R, Reese G, Leong-Kee S, Plante M, Lehrer SB. Molecular basis of arthropod cross-reactivity: igE-binding cross-reactive epitopes of shrimp, house dust mite and cockroach tropomyosins. Int Arch Allergy Immunol. (2002) 129(1):38–48. doi: 10.1159/000065172
19. Patel S, Meher BR. A review on emerging frontiers of house dust mite and cockroach allergy research. Allergol Immunopathol (Madr). (2016) 44(6):580–93. doi: 10.1016/j.aller.2015.11.001
20. Uzel A, Capan N, Canbakan S, Yurdakul AS, Dursun B. Evaluation of the relationship between cockroach sensitivity and house-dust-mite sensitivity in Turkish asthmatic patients. Respir Med. (2005) 99(8):1032–7. doi: 10.1016/j.rmed.2004.12.013
21. Mills RR. Biotic associations and medical importance. In: Bell WJ, Adiyodi KG, editors. The American Cockroach. 1st ed Cambridge, England: The University Press (1981). p. 11–2.
22. Yusof AM. Cockroaches as potential mechanical vectors for mites infestation the first report in kuantan. Int J Zoo Res. (2018) 14:71–6. doi: 10.3923/ijzr.2018.71.76
23. Dos Santos C, Franco OL. Pathogenesis-related proteins (PRs) with enzyme activity activating plant defense responses. Plants (Basel). (2023) 12(11):2226. doi: 10.3390/plants12112226
24. Stintzi A, Heitz T, Prasad V, Wiedemann-Merdinoglu S, Kauffmann S, Geoffroy P, et al. Plant ‘pathogenesis-related’ proteins and their role in defense against pathogens. Biochimie. (1993) 75(8):687–706. doi: 10.1016/0300-9084(93)90100-7
25. Sinha M, Singh RP, Kushwaha GS, Iqbal N, Singh A, Kaushik S, et al. Current overview of allergens of plant pathogenesis related protein families. ScientificWorldJournal. (2014) 2014:1. doi: 10.1155/2014/543195
26. Parisi K, Shafee TMA, Quimbar P, van der Weerden NL, Bleackley MR, Anderson MA. The evolution, function and mechanisms of action for plant defensins. Semin Cell Dev Biol. (2019) 88:107–18. doi: 10.1016/j.semcdb.2018.02.004
27. Ganz T. Defensins: antimicrobial peptides of vertebrates. C R Biol. (2004) 327(6):539–49. doi: 10.1016/j.crvi.2003.12.007
28. Shafee TM, Lay FT, Hulett MD, Anderson MA. The defensins consist of two independent, convergent protein superfamilies. Mol Biol Evol. (2016) 33(9):2345–56. doi: 10.1093/molbev/msw106
29. Shafee TM, Lay FT, Phan TK, Anderson MA, Hulett MD. Convergent evolution of defensin sequence, structure and function. Cell Mol Life Sci. (2017) 74(4):663–82. doi: 10.1007/s00018-016-2344-5
30. Cosi V, Gadermaier G. The role of defensins as pollen and food allergens. Curr Allergy Asthma Rep. (2023) 23(6):277–85. doi: 10.1007/s11882-023-01080-3
31. Ganz T. Defensins: antimicrobial peptides of innate immunity. Nat Rev Immunol. (2003) 3(9):710–20. doi: 10.1038/nri1180
32. Kudryashova E, Seveau SM, Kudryashov DS. Targeting and inactivation of bacterial toxins by human defensins. Biol Chem. (2017) 398(10):1069–85. doi: 10.1515/hsz-2017-0106
33. Hein MJA, Kvansakul M, Lay FT, Phan TK, Hulett MD. Defensin-lipid interactions in membrane targeting: mechanisms of action and opportunities for the development of antimicrobial and anticancer therapeutics. Biochem Soc Trans. (2022) 50(1):423–37. doi: 10.1042/BST20200884
34. Pelegrini PB, Lay FT, Murad AM, Anderson MA, Franco OL. Novel insights on the mechanism of action of alpha-amylase inhibitors from the plant defensin family. Proteins. (2008) 73(3):719–29. doi: 10.1002/prot.22086
35. Bukhteeva I, Hrunyk NI, Yusypovych YM, Shalovylo YI, Kovaleva V, Nesmelova IV. Structure, dynamics, and function of PsDef2 defensin from Pinus sylvestris. Structure. (2022) 30(5):753–762.e5. doi: 10.1016/j.str.2022.03.001
36. Sánchez-Monge R, García-Casado G, Barber D, Salcedo G. Interaction of allergens from house-dust mite and from cereal flours: Dermatophagoides pteronyssinus α-amylase (Der p 4) and wheat and rye α-amylase inhibitors. Allergy. (1996) 51(3):176–80. doi: 10.1111/j.1398-9995.1996.tb04583.x
37. Noël A, Le Conte Y, Mondet F. Varroa destructor: how does it harm Apis mellifera honey bees and what can be done about it? Emerg Top Life Sci. (2020) 4(1):45–57. doi: 10.1042/ETLS20190125
38. Zaobidna EA, Żółtowska K, Łopieńska-Biernat E. Varroa destructor induces changes in the expression of immunity-related genes during the development of Apis mellifera worker and drone broods. Acta Parasitol. (2017) 62(4):779–89. doi: 10.1515/ap-2017-0094
39. Khongphinitbunjong K, de Guzman LI, Tarver MR, Rinderer TE, Chen Y, Chantawannakul P. Differential viral levels and immune gene expression in three stocks of Apis mellifera induced by different numbers of varroa destructor. J Insect Physiol. (2015) 72:28–34. doi: 10.1016/j.jinsphys.2014.11.005
40. Santamaría ME, Arnaiz A, Velasco-Arroyo B, Grbic V, Diaz I, Martinez M. Arabidopsis response to the spider mite tetranychus urticae depends on the regulation of reactive oxygen species homeostasis. Sci Rep. (2018) 8(1):9432. doi: 10.1038/s41598-018-27904-1
41. Semple F, Dorin JR. β-defensins: multifunctional modulators of infection, inflammation and more? J Innate Immun. (2012) 4(4):337–48. doi: 10.1159/000336619
42. Seo SJ, Ahn SW, Hong CK, Ro BI. Expressions of β-defensins in human keratinocyte cell lines. J Dermatol Sci. (2001) 27(3):183–91. doi: 10.1016/S0923-1811(01)00135-9
43. Bogefors J, Kvarnhammar AM, Höckerfelt U, Cardell LO. Reduced tonsillar expression of human β-defensin 1, 2 and 3 in allergic rhinitis. FEMS Immunol Med Microbiol. (2012) 65(3):431–8. doi: 10.1111/j.1574-695X.2012.00959.x
44. Niyonsaba F, Kiatsurayanon C, Ogawa H. The role of human β-defensins in allergic diseases. Clin Exp Allergy. (2016) 46(12):1522–30. doi: 10.1111/cea.12843
45. Borchers NS, Santos-Valente E, Toncheva AA, Wehkamp J, Franke A, Gaertner VD, et al. Human β-defensin 2 mutations are associated with asthma and atopy in children and its application prevents atopic asthma in a mouse model. Front Immunol. (2021) 12:636061. doi: 10.3389/fimmu.2021.636061
46. Prado-Montes de Oca E, García-Vargas A, Lozano-Inocencio R, Gallegos-Arreola MP, Sandoval-Ramírez L, Dávalos-Rodríguez NO, et al. Association of β-defensin 1 single nucleotide polymorphisms with atopic dermatitis. Int Arch Allergy Immunol. (2007) 142(3):211–8. doi: 10.1159/000097023
47. Rogers BL, Pollock J, Klapper DG, Griffith IJ. Sequence of the proteinase-inhibitor cystatin homologue from the pollen of Ambrosia artemisiifolia (short ragweed). Gene. (1993) 133(2):219–21. doi: 10.1016/0378-1119(93)90641-F
48. Bublin M, Mari A, Ebner C, Knulst A, Scheiner O, Hoffmann-Sommergruber K, et al. Ige sensitization profiles toward green and gold kiwifruits differ among patients allergic to kiwifruit from 3 European countries. J Allergy Clin Immunol. (2004) 114(5):1169–75. doi: 10.1016/j.jaci.2004.07.016
49. Rassam M, Laing WA. Purification and characterization of phytocystatins from kiwifruit cortex and seeds. Phytochemistry. (2004) 65(1):19–30. doi: 10.1016/j.phytochem.2003.09.019
50. van Hage M, Käck U, Asarnoj A, Konradsen JR. An update on the prevalence and diagnosis of cat and dog allergy—emphasizing the role of molecular allergy diagnostics. Mol Immunol. (2023) 157:1–7. doi: 10.1016/j.molimm.2023.03.003
51. Ichikawa K, Vailes LD, Pomes A, Chapman MD. Identification of a novel cat allergen–cystatin. Int Arch Allergy Immunol. (2001) 124:55–6. doi: 10.1159/000053667
52. Tremblay J, Goulet MC, Vorster J, Goulet C, Michaud D. Harnessing the functional diversity of plant cystatins to design inhibitor variants highly active against herbivorous arthropod digestive proteases. FEBS J. (2022) 289(7):1827–41. doi: 10.1111/febs.16288
53. Martinez M, Santamaria ME, Diaz-Mendoza M, Arnaiz A, Carrillo L, Ortego F, et al. Phytocystatins: defense proteins against phytophagous insects and acari. Int J Mol Sci. (2016) 17(10):1747. doi: 10.3390/ijms17101747
54. Santamaria ME, Diaz-Mendoza M, Perez-Herguedas D, Hensel G, Kumlehn J, Diaz I, et al. Overexpression of HvIcy6 in barley enhances resistance against tetranychus urticae and entails partial transcriptomic reprogramming. Int J Mol Sci. (2018) 19(3):697. doi: 10.3390/ijms19030697
55. Carrillo L, Martinez M, Ramessar K, Cambra I, Castañera P, Ortego F, et al. Expression of a barley cystatin gene in maize enhances resistance against phytophagous mites by altering their cysteine-proteases. Plant Cell Rep. (2011) 30(1):101–12. doi: 10.1007/s00299-010-0948-z
56. Bui H, Greenhalgh R, Ruckert A, Gill GS, Lee S, Ramirez RA, et al. Generalist and specialist Mite herbivores induce similar defense responses in maize and barley but differ in susceptibility to benzoxazinoids. Front Plant Sci. (2018) 9:1222. doi: 10.3389/fpls.2018.01222
57. Pernas M, Sánchez-Monge R, Gómez L, Salcedo G. A chestnut seed cystatin differentially effective against cysteine pro-teinases from closely related pests. Plant Mol Biol. (1998) 38(6):1235–42. doi: 10.1023/A:1006154829118
58. Pernas M, Sanchez-Ramos I, Sanchez-Monge R, Lombardero M, Arteaga C, Castañera P, et al. Der p 1 and der f 1, the highly related and major allergens from house dust mites, are differentially affected by a plant cystatin. Clin Exp Allergy. (2000) 30(7):972–8. doi: 10.1046/j.1365-2222.2000.00845.x
59. Vidal-Quist JC, Ortego F, Hernández-Crespo P. Contribution of cysteine and serine proteases to proteolytic digestion in an allergy-eliciting house dust mite. J Insect Physiol. (2021) 133:104285. doi: 10.1016/j.jinsphys.2021.104285
60. Takahashi H, Honma M, Ishida-Yamamoto A, Namikawa K, Kiyama H, Iizuka H. Expression of human cystatin A by keratinocytes is positively regulated via the ras/MEKK1/MKK7/JNK signal transduction pathway but negatively regulated via the ras/raf-1/MEK1/ERK pathway. J Biol Chem. (2001) 276(39):36632–8. doi: 10.1074/jbc.M102021200
61. Kato T, Takai T, Mitsuishi K, Okumura K, Ogawa H. Cystatin A inhibits IL-8 production by keratinocytes stimulated with der p 1 and der f 1: biochemical skin barrier against mite cysteine proteases. J Allergy Clin Immunol. (2005) 116(1):169–76. doi: 10.1016/j.jaci.2005.03.044
62. Vasilopoulos Y, Cork MJ, Teare D, Marinou I, Ward SJ, Duff GW, et al. A nonsynonymous substitution of cystatin A, a cysteine protease inhibitor of house dust mite protease, leads to decreased mRNA stability and shows a significant association with atopic dermatitis. Allergy. (2007) 62(5):514–9. doi: 10.1111/j.1398-9995.2007.01350.x
63. Yao L, Yuan X, Fu H, Guo Q, Wu Y, Xuan S, et al. Epithelium-derived cystatin SN inhibits house dust mite protease activity in allergic asthma. Allergy. (2023) 78(6):1507–23. doi: 10.1111/all.15739
64. Kidwai M, Ahmad IZ, Chakrabarty D. Class III peroxidase: an indispensable enzyme for biotic/abiotic stress tolerance and a potent candidate for crop improvement. Plant Cell Rep. (2020) 39(11):1381–93. doi: 10.1007/s00299-020-02588-y
65. Almagro L, Gómez Ros LV, Belchi-Navarro S, Bru R, Ros Barceló A, Pedreño MA. Class III peroxidases in plant defence reactions. J Exp Bot. (2009) 60(2):377–90. doi: 10.1093/jxb/ern277
66. Bronner R, Westphal E, Dreger F. Enhanced peroxidase activity associated with the hypersensitive response of Solanum dulcamara to the gall mite Aceria cladophthirus (Acari: Eriophyoidea). Can J Bot. (1991) 69(10):2192–6. doi: 10.1139/b91-275
67. Trevisan MTS, Scheffer JJC, Verpoorte R. Peroxidase activity in hop plants after infestation by red spider mites. Crop Prot. (2003) 22(2):423–4. doi: 10.1016/S0261-2194(02)00151-5
68. Liang X, Chen Q, Lu H, Wu C, Lu F, Tang J. Increased activities of peroxidase and polyphenol oxidase enhance cassava resistance to tetranychus urticae. Exp Appl Acarol. (2017) 71(3):195–209. doi: 10.1007/s10493-017-0125-y
69. Sánchez-Monge R, García-Casado G, López-Otín C, Armentia A, Salcedo G. Wheat flour peroxidase is a prominent al-lergen associated with baker’s asthma. Clin Exp Allergy. (1997) 27(10):1130–7. doi: 10.1111/j.1365-2222.1997.tb01149.x
70. Suriyamoorthy P, Madhuri A, Tangirala S, Michael KR, Sivanandham V, Rawson A, et al. Comprehensive review on banana fruit allergy: pathogenesis, diagnosis, management, and potential modification of allergens through food processing. Plant Foods Hum Nutr. (2022) 77(2):159–71. doi: 10.1007/s11130-022-00976-1
71. Weangsripanaval T, Nomura N, Moriyama T, Ohta N, Ogawa T. Identification of suberization-associated anionic pe-roxidase as a possible allergenic protein from tomato. Biosci Biotechnol Biochem. (2003) 67(6):1299–304. doi: 10.1271/bbb.67.1299
72. Minai-Fleminger Y, Levi-Schaffer F. Mast cells and eosinophils: the two key effector cells in allergic inflammation. Inflamm Res. (2009) 58(10):631–8. doi: 10.1007/s00011-009-0042-6
73. Choi Y, Jeon H, Yang EA, Yoon JS, Kim HH. Nasal eosinophilia and eosinophil peroxidase in children and adolescents with rhinitis. Korean J Pediatr. (2019) 62(9):353–9. doi: 10.3345/kjp.2019.00318
74. Wang Z, DiDonato JA, Buffa J, Comhair SA, Aronica MA, Dweik RA, et al. Eosinophil peroxidase catalyzed protein carbamylation participates in asthma. J Biol Chem. (2016) 291(42):22118–35. doi: 10.1074/jbc.M116.750034
75. Cote J, Ada E, Hochberg R. Elemental enrichment of the exoskeleton in three species of tick (Arachnida: Ixodidae). J Parasitol. (2020) 106(6):742–54. doi: 10.1645/20-95
76. Zhao YE, Wang ZH, Xu Y, Xu JR, Liu WY, Wei M, et al.; cloning and sequence analysis of chitin synthase gene fragments of demodex mites. J Zhejiang Univ Sci B. (2012) 13(10):763–8. doi: 10.1631/jzus.B1200155
77. Khan RS, Iqbal A, Bibi A, Khalil I, Islam ZU, Jan F, et al. Plant chitinases: types, structural classification, antifungal potential and transgenic expression in plants for enhanced disease resistance. Plant Cell Tiss Organ Cult. (2024) 156(75). doi: 10.1007/s11240-024-02696-7
78. Sanchez-Monge R, Blanco C, Díaz-Perales A, Collada C, Carrillo T, Aragoncillo C, et al. Isolation and charac-terization of major banana allergens: identification as fruit class I chitinases. Clin Exp Allergy. (1999) 29(5):673–80. doi: 10.1046/j.1365-2222.1999.00526.x
79. Diaz-Perales A, Collada C, Blanco C, Sánchez-Monge R, Carrillo T, Aragoncillo C, et al. Class I chitinases with hevein-like domain, but not class II enzymes, are relevant chestnut and avocado allergens. J Allergy Clin Immunol. (1998) 102(1):127–33. doi: 10.1016/S0091-6749(98)70063-6
80. Volpicella M, Leoni C, Fanizza I, Placido A, Pastorello EA, Ceci LR. Overview of plant chitinases identified as food allergens. J Agric Food Chem. (2014) 62(25):5734–42. doi: 10.1021/jf5007962
81. Volpicella M, Leoni C, Fanizza I, Distaso M, Leoni G, Farioli L, et al. Characterization of maize chitinase-A, a tough allergenic molecule. Allergy. (2017) 72(9):1423–9. doi: 10.1111/all.13164
82. Kanneganti M, Kamba A, Mizoguchi E. Role of chitotriosidase (chitinase 1) under normal and disease conditions. J Epithel Biol Pharmacol. (2012) 5:1–9. doi: 10.2174/1875044301205010001
83. Zhu Z, Zheng T, Homer RJ, Kim YK, Chen NY, Cohn L, et al. Acidic mammalian chitinase in asthmatic Th2 inflammation and IL-13 pathway activation. Science. (2004) 304(5677):1678–82. doi: 10.1126/science.1095336
84. Hong JY, Kim M, Sol IS, Kim KW, Lee CM, Elias JA, et al. Chitotriosidase inhibits allergic asthmatic airways via regulation of TGF-β expression and Foxp3+ treg cells. Allergy. (2018) 73(8):1686–99. doi: 10.1111/all.13426
85. Ramanathan M Jr, Lee WK, Lane AP. Increased expression of acidic mammalian chitinase in chronic rhinosinusitis with nasal polyps. Am J Rhinol. (2006) 20(3):330–5. doi: 10.2500/ajr.2006.20.2869
86. Sadeghi HR, Pirayesh A, Shahsavan S, Amani S, Amini SA, Samani KG, et al. Correlation of acidic mammalian chitinase expression with disease severity in patients with moderate/severe persistent allergic rhinitis. Cent Eur J Immunol. (2020) 45(3):294–300. doi: 10.5114/ceji.2020.101251
87. Bierbaum S, Nickel R, Koch A, Lau S, Deichmann KA, Wahn U, et al. Polymorphisms and haplotypes of acid mammalian chitinase are associated with bronchial asthma. Am J Respir Crit Care Med. (2005) 172(12):1505–9. doi: 10.1164/rccm.200506-890OC
88. Fernandes H, Michalska K, Sikorski M, Jaskolski M. Structural and functional aspects of PR-10 proteins. FEBS J. (2013) 280(5):1169–99. doi: 10.1111/febs.12114
89. Neudecker P, Schweimer K, Nerkamp J, Scheurer S, Vieths S, Sticht H, et al. Allergic cross-reactivity made visible: solution structure of the major cherry allergen pru av 1. J Biol Chem. (2001) 276(25):22756–63. doi: 10.1074/jbc.M101657200
90. Aglas L, Soh WT, Kraiem A, Wenger M, Brandstetter H, Ferreira F. Ligand binding of PR-10 proteins with a particular focus on the bet v 1 allergen family. Curr Allergy Asthma Rep. (2020) 20(7):25. doi: 10.1007/s11882-020-00918-4
91. Włodarczyk K, Smolińska B, Majak I. Tomato allergy: the characterization of the selected allergens and antioxidants of tomato (Solanum lycopersicum)-A review. Antioxidants. (2022) 11(4):644. doi: 10.3390/antiox11040644
92. Führer S, Unterhauser J, Zeindl R, Eidelpes R, Fernández-Quintero ML, Liedl KR, et al. The structural flexi-bility of PR-10 food allergens. Int J Mol Sci. (2022) 23(15):8252. doi: 10.3390/ijms23158252
93. Wangorsch A, Jamin A, Lidholm J, Gräni N, Lang C, Ballmer-Weber B, et al. Identification and implication of an allergenic PR-10 protein from walnut in birch pollen associated walnut allergy. Mol Nutr Food Res. (2017) 61(4). doi: 10.1002/mnfr.201600902
94. Broderick K, Pittock C, Arioli T, Creaser EH, Weinman JJ, Rolfe BG. Pathogenesis-related proteins in Trifolium subterraneum: a general survey and subsequent characterisation of a protein inducible by ethephon and redlegged earth mite attack. Funct Plant Biol. (1997) 24:819–29. doi: 10.1071/PP97022
95. Roth-Walter F, Gomez-Casado C, Pacios LF, Mothes-Luksch N, Roth GA, Singer J, et al. Bet v 1 from birch pollen is a lipocalin-like protein acting as allergen only when devoid of iron by promoting Th2 lymphocytes. J Biol Chem. (2014) 289(25):17416–21. doi: 10.1074/jbc.M114.567875
96. Diez-Hermano S, Ganfornina MD, Skerra A, Gutiérrez G, Sanchez D. An evolutionary perspective of the lipocalin protein family. Front Physiol. (2021) 12:718983. doi: 10.3389/fphys.2021.718983
97. Ganfornina MD, Gutiérrez G, Bastiani M, Sánchez D. A phylogenetic analysis of the lipocalin protein family. Mol Biol Evol. (2000) 17(1):114–26. doi: 10.1093/oxfordjournals.molbev.a026224
98. Flower DR. The lipocalin protein family: structure and function. Biochem J. (1996) 318:1–14. doi: 10.1042/bj3180001
99. Virtanen T, Kinnunen T, Rytkönen-Nissinen M. Mammalian lipocalin allergens–insights into their enigmatic allergenicity. Clin Exp Allergy. (2012) 42(4):494–504. doi: 10.1111/j.1365-2222.2011.03903.x
100. Yamamoto K, Ishibashi O, Sugiura K, Ubatani M, Sakaguchi M, Nakatsuji M, et al. Crystal structure of the dog allergen can f 6 and structure-based implications of its cross-reactivity with the cat allergen fel d 4. Sci Rep. (2019) 9(1):1503. doi: 10.1038/s41598-018-38134-w
101. Rassart E, Desmarais F, Najyb O, Bergeron KF, Mounier C. Apolipoprotein D. Gene. (2020) 756:144874. doi: 10.1016/j.gene.2020.144874
102. Csősz É, Emri G, Kalló G, Tsaprailis G, Tőzsér J. Highly abundant defense proteins in human sweat as revealed by targeted proteomics and label-free quantification mass spectrometry. J Eur Acad Dermatol Venereol. (2015) 29(10):2024–31. doi: 10.1111/jdv.13221
103. Zahradnik E, Raulf M. Respiratory allergens from furred mammals: environmental and occupational exposure. Vet Sci. (2017) 4(3):38. doi: 10.3390/vetsci4030038
104. Jackson BC, Thompson DC, Wright MW, McAndrews M, Bernard A, Nebert DW, et al. Update of the human secretoglobin (SCGB) gene superfamily and an example of 'evolutionary bloom' of androgen-binding protein genes within the mouse Scgb gene superfamily. Hum Genomics. (2011) 5(6):691–702. doi: 10.1186/1479-7364-5-6-691
105. Kaiser L, Grönlund H, Sandalova T, Ljunggren HG, van Hage-Hamsten M, Achour A, et al. The crystal structure of the major cat allergen fel d 1, a member of the secretoglobin family. J Biol Chem. (2003) 278(39):37730–5. doi: 10.1074/jbc.M304740200
106. Hilger C, Kler S, Arumugam K, Revets D, Muller CP, Charpentier C, et al. Identification and isolation of a fel d 1-like molecule as a major rabbit allergen. J Allergy Clin Immunol. (2014) 133(3):759–66. doi: 10.1016/j.jaci.2013.04.034
107. Singh G, Katyal SL. Clara cell proteins. Ann N Y Acad Sci. (2000) 923:43–58. doi: 10.1111/j.1749-6632.2000.tb05518.x
108. Guerra S, Vasquez MM, Spangenberg A, Halonen M, Martin RJ. Club cell secretory protein in serum and bron-choalveolar lavage of patients with asthma. J Allergy Clin Immunol. (2016) 138(3):932–934.e1. doi: 10.1016/j.jaci.2016.03.047
109. Nie W, Xue C, Chen J, Xiu Q. Secretoglobin 1A member 1 (SCGB1A1) +38A/G polymorphism is associated with asthma risk: a meta-analysis. Gene. (2013) 528(2):304–8. doi: 10.1016/j.gene.2013.06.049
110. Bingle CD, LeClair EE, Havard S, Bingle L, Gillingham P, Craven CJ. Phylogenetic and evolutionary analysis of the PLUNC gene family. Protein Sci. (2004) 13(2):422–30. doi: 10.1110/ps.03332704
111. Gakhar L, Bartlett JA, Penterman J, Mizrachi D, Singh PK, Mallampalli RK, et al. PLUNC is a novel airway surfactant protein with anti-biofilm activity. PLoS One. (2010) 5(2):e9098. doi: 10.1371/journal.pone.0009098
112. Popescu FD, Ganea CS, Panaitescu C, Vieru M. Molecular diagnosis in cat allergy. World J Methodol. (2021) 11(3):46–60. doi: 10.5662/wjm.v11.i3.46
113. Victor S, Binnmyr J, Lampa E, Rask-Andersen A, Elfman L. Levels of horse allergen equ c 4 in dander and saliva from ten horse breeds. Clin Exp Allergy. (2019) 49(5):701–11. doi: 10.1111/cea.13362
114. McDonald RE, Fleming RI, Beeley JG, Bovell DL, Lu JR, Zhao X, et al. Latherin: a surfactant protein of horse sweat and saliva. PLoS One. (2009) 4(5):e5726. doi: 10.1371/journal.pone.0005726
115. Eckersall PD, Kerr MG, Snow DH. An investigation into the proteins of horse sweat (Equus caballus). Comp Biochem Physiol Part B. (1982) 73(2):375–8. doi: 10.1016/0305-0491(82)90300-5
116. Campos MA, Abreu AR, Nlend MC, Cobas MA, Conner GE, Whitney PL. Purification and characterization of PLUNC from human tracheobronchial secretions. Am J Respir Cell Mol Biol. (2004) 30(2):184–92. doi: 10.1165/rcmb.2003-0142OC
117. Liu Y, Bartlett JA, Di ME, Bomberger JM, Chan YR, Gakhar L, et al. SPLUNC1/BPIFA1 contributes to pulmonary host defense against Klebsiella pneumoniae respiratory infection. Am J Pathol. (2013) 182(5):1519–31. doi: 10.1016/j.ajpath.2013.01.050
118. Wu T, Huang J, Moore PJ, Little MS, Walton WG, Fellner RC, et al. Identification of BPIFA1/SPLUNC1 as an epithelium-derived smooth muscle relaxing factor. Nat Commun. (2017) 8:14118. doi: 10.1038/ncomms14118
119. Zhang R, Trower J, Wu T. Degradation of bacterial permeability family member A1 (BPIFA1) by house dust mite (HDM) cysteine protease der p 1 abrogates immune modulator function. Int J Biol Macromol. (2020) 164:4022–31. doi: 10.1016/j.ijbiomac.2020.08.214
120. Naegele A, Reboux G, Scherer E, Roussel S, Millon L. Fungal food choices of Dermatophagoides farinae affect indoor fungi selection and dispersal. Int J Environ Health Res. (2013) 23(2):91–5. doi: 10.1080/09603123.2012.699029
121. Molva V, Nesvorna M, Hubert J. Feeding interactions between microorganisms and the house dust mites dermatopha-goides pteronyssinus and dermatophagoides farinae (Astigmata: pyroglyphidae). J Med Entomol. (2019) 56(6):1669–77. doi: 10.1093/jme/tjz089
122. Klimov PB, Bochkov AV, OConnor BM. Phylogenetic position of the house dust mite subfamily guatemalichinae (acariformes: pyroglyphidae) based on integrated molecular and morphological analyses and different measures of support. Cladistics. (2016) 32(3):261–75. doi: 10.1111/cla.12126
123. Klimov PB, OConnor BM. Origin and higher-level relationships of psoroptidian mites (Acari: Astigmata: Psoroptidia): evidence from three nuclear genes. Mol Phylogenet Evol. (2008) 47(3):1135–56. doi: 10.1016/j.ympev.2007.12.025
124. Klimov PB, OConnor B. Is permanent parasitism reversible?–critical evidence from early evolution of house dust mites. Syst Biol. (2013) 62(3):411–23. doi: 10.1093/sysbio/syt008
125. Galili U. Paleo-immunology of human anti-carbohydrate antibodies preventing primate extinctions. Immunology. (2023) 168(1):18–29. doi: 10.1111/imm.13582
126. Zhang E, Tanaka T, Tajima M, Tsuboi R, Kato H, Nishikawa A, et al. Anti-Malassezia-specific IgE antibodies production in japanese patients with head and neck atopic dermatitis: relationship between the level of specific IgE antibody and the colonization frequency of cutaneous Malassezia species and clinical severity. J Allergy. (2011) 2011:1. doi: 10.1155/2011/645670
127. Mayser P, Gross A. Ige antibodies to malassezia furfur, M. sympodialis and pityrosporum orbiculare in patients with atopic dermatitis, seborrheic eczema or pityriasis versicolor, and identification of respective allergens. Acta Derm Venereol. (2000) 80(5):357–61. doi: 10.1080/000155500459303
128. Triana S, de Cock H, Ohm RA, Danies G, Wösten HAB, Restrepo S, et al. Lipid metabolic versatility in malassezia spp. Yeasts studied through metabolic modeling. Front Microbiol. (2017) 8:1772. doi: 10.3389/fmicb.2017.01772
129. Picardo M, Ottaviani M, Camera E, Mastrofrancesco A. Sebaceous gland lipids. Dermatoendocrinol. (2009) 1(2):68–71. doi: 10.4161/derm.1.2.8472
130. Belvoncikova P, Splichalova P, Videnska P, Gardlik R. The human mycobiome: colonization, composition and the role in health and disease. J Fungi. (2022) 8(10):1046. doi: 10.3390/jof8101046
131. Pakkasela J, Ilmarinen P, Honkamäki J, Tuomisto LE, Andersén H, Piirilä P, et al. Age-specific incidence of allergic and non-allergic asthma. BMC Pulm Med. (2020) 20(1):9. doi: 10.1186/s12890-019-1040-2
132. Bylund S, Kobyletzki LB, Svalstedt M, Svensson Å. Prevalence and incidence of atopic dermatitis: a systematic review. Acta Derm Venereol. (2020) 100(12):adv00160. doi: 10.2340/00015555-3510
133. Willits EK, Park MA, Hartz MF, Schleck CD, Weaver AL, Joshi AY. Food allergy: a comprehensive popula-tion-based cohort study. Mayo Clin Proc. (2018) 93(10):1423–30. doi: 10.1016/j.mayocp.2018.05.031
134. Pochi PE, Strauss JS, Downing DT. Age-related changes in sebaceous gland activity. J Invest Dermatol. (1979) 73(1):108–11. doi: 10.1111/1523-1747.ep12532792
135. Cotterill JA, Cunliffe WJ, Williamson B, Bulusu L. Age and sex variation in skin surface lipid composition and sebum excretion rate. Br. J. Dermatol. (1972) 87:333–40. doi: 10.1111/j.1365-2133.1972.tb07419.x
136. Jo JH, Deming C, Kennedy EA, Conlan S, Polley EC, Ng WI, et al. Diverse human skin fungal communities in children converge in adulthood. J. Investig. Dermatol. (2016) 136:2356–63. doi: 10.1016/j.jid.2016.05.130
Keywords: the acari hypothesis, allergenicity, FReP, defensins, cystatins, peroxidases, chitinases, PR-10 proteins
Citation: Retzinger AC and Retzinger GS (2024) The Acari Hypothesis, V: deciphering allergenicity. Front. Allergy 5:1454292. doi: 10.3389/falgy.2024.1454292
Received: 24 June 2024; Accepted: 18 October 2024;
Published: 1 November 2024.
Edited by:
Wai Tuck Soh, Max Planck Institute for Multidisciplinary Sciences, GermanyReviewed by:
Alexis Labrada, National Center of Bioproducts (BIOCEN), CubaAlain Jacquet, Chulalongkorn University, Thailand
Copyright: © 2024 Retzinger and Retzinger. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Andrew C. Retzinger, YW5kcmV3LnJldHppbmdlckBnbWFpbC5jb20=
 Andrew C. Retzinger
Andrew C. Retzinger Gregory S. Retzinger
Gregory S. Retzinger