- 1Department of Translational Medical Science, University of Naples Federico II, Naples, Italy
- 2ImmunoNutritionLab at the CEINGE Advanced Biotechnologies Research Center, University of Naples Federico II, Naples, Italy
- 3Department of Biomedicine and Prevention, University of Rome “Tor Vergata”, Rome, Italy
- 4Task Force on Microbiome Studies, University of Naples Federico II, Naples, Italy
- 5European Laboratory for the Investigation of Food-Induced Diseases, University of Naples Federico II, Naples, Italy
Increasing evidence suggests that alterations in the gut microbiome (GM) play a pivotal role in the pathogenesis of pediatric food allergy (FA). This scoping review analyzes the current evidence on GM features associated with pediatric FAs and highlights the importance of the GM as a potential target of intervention for preventing and treating this common condition in the pediatric age. Following the Preferred Reporting Items for Systematic Reviews and Meta-Analysis guidelines, we searched PubMed and Embase using the keywords (gut microbiome OR dysbiosis OR gut microbiota OR microbiome signatures) AND (food allergy OR IgE-mediated food allergy OR food protein-induced allergic proctocolitis OR food protein-induced enterocolitis OR non-IgE food allergy OR cow milk allergy OR hen egg allergy OR peanut allergy OR fish allergy OR shellfish allergy OR tree nut allergy OR soy allergy OR wheat allergy OR rice allergy OR food sensitization). We included 34 studies reporting alterations in the GM in children affected by FA compared with healthy controls. The GM in pediatric FAs is characterized by a higher abundance of harmful microorganisms (e.g., Enterobacteriaceae, Clostridium sensu stricto, Ruminococcus gnavus, and Blautia spp.) and lower abundance of beneficial bacteria (e.g., Bifidobacteriaceae, Lactobacillaceae, some Bacteroides species). Moreover, we provide an overview of the mechanisms of action elicited by these bacterial species in regulating immune tolerance and of the main environmental factors that can modulate the composition and function of the GM in early life. Altogether, these data improve our knowledge of the pathogenesis of FA and can open the way to innovative diagnostic, preventive, and therapeutic strategies for managing these conditions.
1 Introduction
Food allergy (FA) is one of the most common chronic conditions in the pediatric age, and its prevalence has increased in recent decades (1). Increased FA prevalence is paralleled with a worsening of the clinical picture, leading to a significant increase in hospital admissions for food-induced anaphylaxis (2, 3). This dramatic scenario has a direct impact on the psychological burden of patients and their parents as well as on the costs for the families and healthcare systems (4, 5).
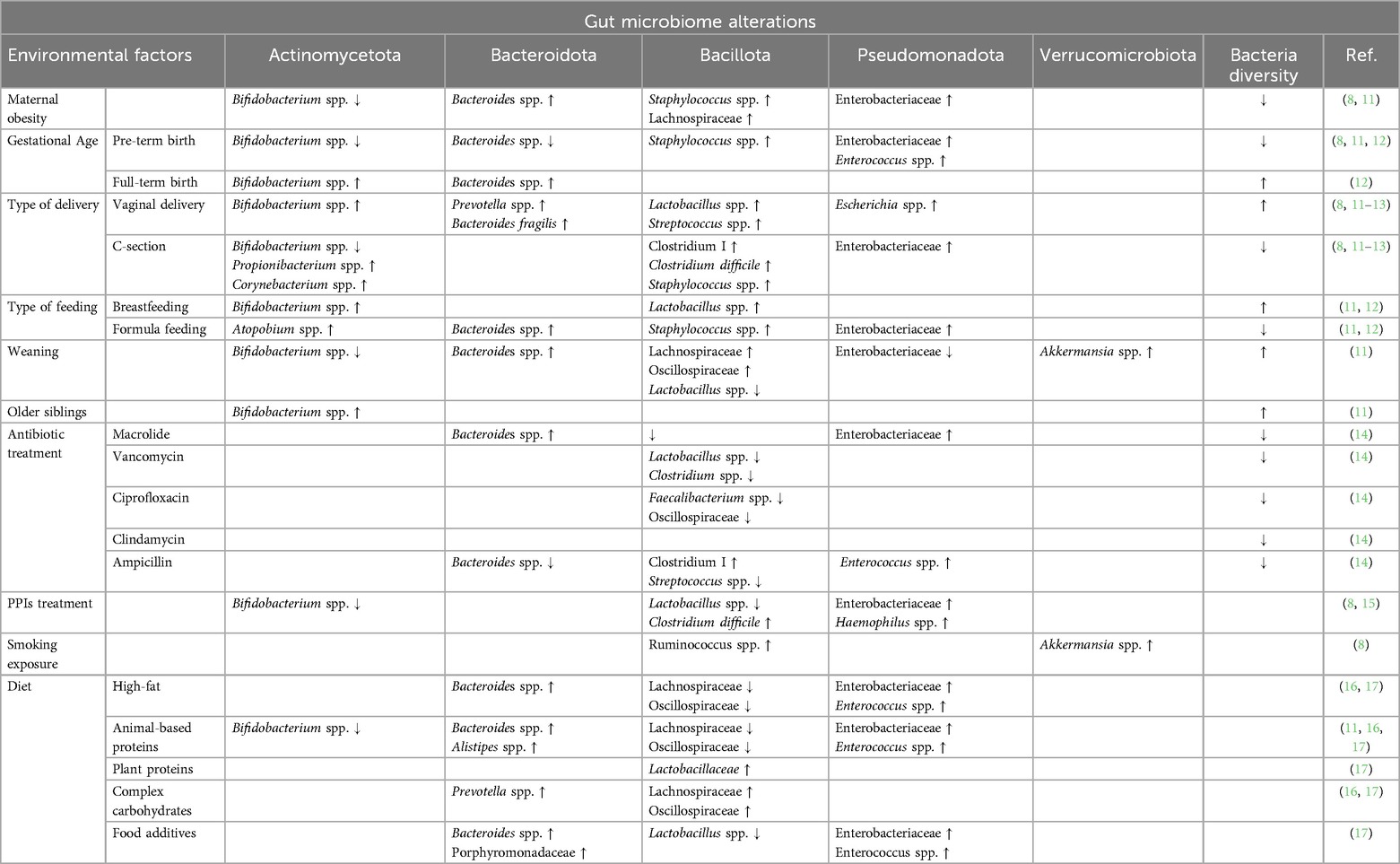
Food allergy results from a breakdown of the immune tolerance mechanisms (6). The induction and maintenance of immune tolerance is ensured by a dynamic and bidirectional link between the gut microbiome (GM) and immune system, defining the concept of the GM-immune system axis. Several environmental factors have been indicated as modulators of this axis, especially in the first 1,000 days of life (Table 1) (7–9). During this critical period, the development of the structure and function of the GM drives the development of the immune system (10). Thus, environmental factors, shaping the composition of the GM, influence immune system development and establish a child's current and future health status. Recent evidence shows that early exposure to drugs, such as antibiotics and gastric acidity inhibitors, antiseptic agents, ultra-processed foods (UPFs), and pollutants negatively influences the GM and increases the risk of developing chronic non-communicable diseases, including FA (10).
Gut microbiome is physically and functionally linked to the immune system; indeed, some beneficial bacteria that produce immunomodulatory compounds, such as the short-chain fatty acid (SCFA) butyrate, increase the rate of regulatory T cells (Tregs) (i.e., crucial cells involved in immune tolerance) (18). By contrast, the loss of beneficial bacterial species has been associated not only with a reduction in these beneficial compounds but also the production of pro-inflammatory mediators, such as lipopolysaccharides (LPSs), that can contribute to the development of FA (19).
All these data suggest that the balance of beneficial and detrimental bacteria in the GM could be involved in the occurrence of FA, which paves the way to innovative preventive and therapeutic strategies for one of the most prevalent and expensive diseases of the pediatric age (20). This scoping review analyzes the current evidence on GM features associated with pediatric FA and highlights the importance of the GM as a potential target of intervention for preventing and treating this common condition in the pediatric age.
2 Methods
2.1 Search strategy
The Embase and PubMed databases were searched with medical subject heading (MeSH) keywords to identify studies of interest. The search was last performed on 1 March 2024. The following MeSH terms were used: (gut microbiome OR dysbiosis OR gut microbiota OR microbiome signatures) AND (food allergy OR IgE-mediated food allergy OR food protein-induced allergic proctocolitis OR food protein-induced enterocolitis OR non-IgE food allergy OR cow milk allergy OR hen egg allergy OR peanut allergy OR fish allergy OR shellfish OR tree nuts OR soy allergy OR wheat allergy OR rice allergy OR food sensitization).
The filters used for all databases were language (English only) and time (articles retrieved between 1 January 2011 and 29 February 2024). In addition, the filters applied for PubMed were studies on humans and age (child, birth–18 years; newborn, birth–1 month; infant, birth–23 months; infant, 1–23 months; child, 6–12 years; adolescent, 13–18 years; and preschool child, 2–5 years). Additional filters used for Embase were studies on humans and age (newborn, infant, child, preschool child, school child, and adolescent).
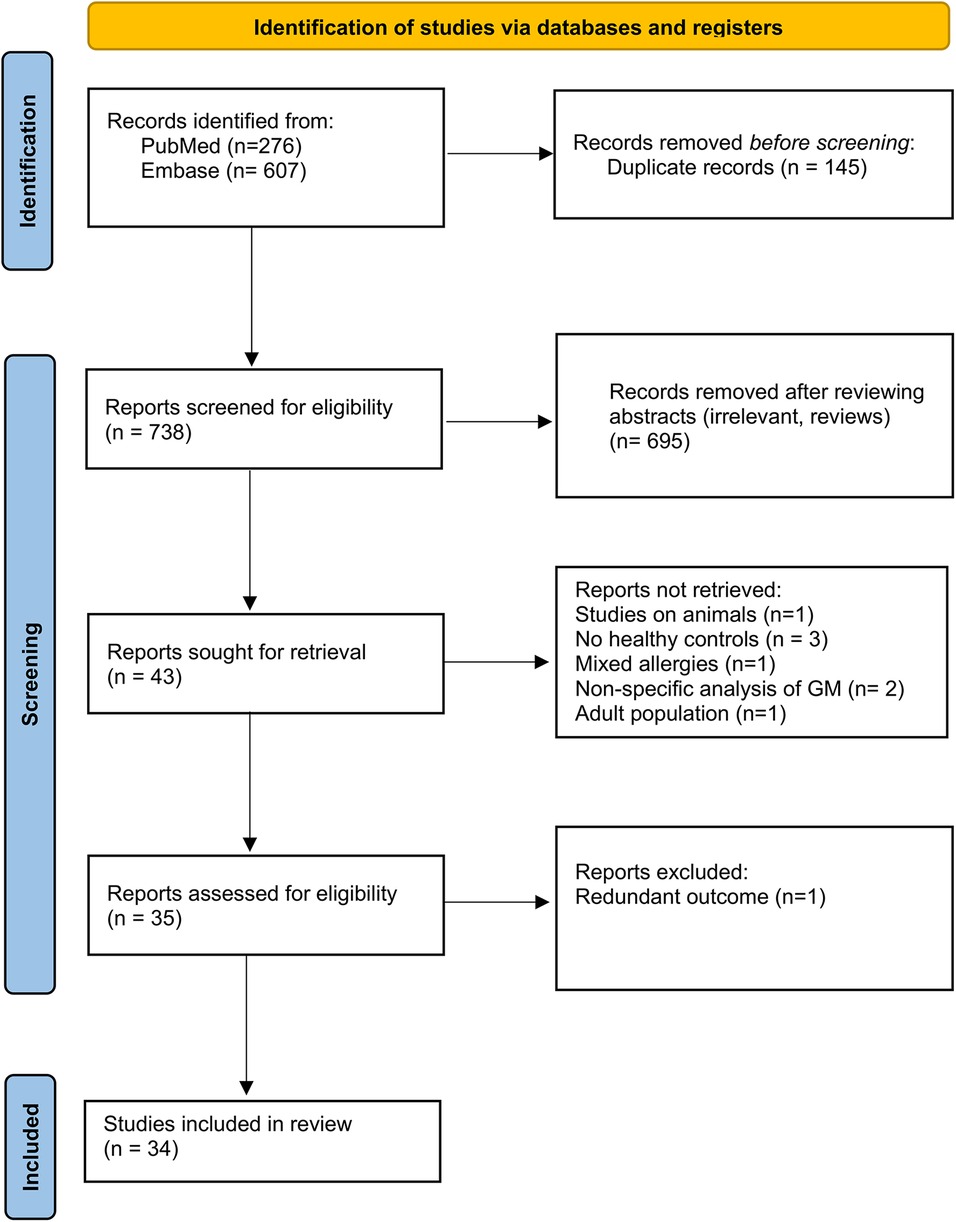
This study was conducted following the recommendations of the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidelines (21). The search strategy and PRISMA flow chart are detailed in Figure 1.
2.2 Selection criteria and data extraction
Two investigators (MF and LC) independently reviewed the titles and abstracts of the original articles extracted in the initial search and selected the observational studies, clinical trials, cross-sectional studies, and case–control studies regarding GM composition and its alterations in pediatric FA and food sensitization in comparison with age- and sex-matched healthy controls that were available as full-text original peer-reviewed articles. Reviews were excluded. In the included studies, the analysis was performed on the stool microbiome. Only pediatric subjects (aged 0–18 years), irrespective of gender, race, geographic location, mode of delivery, and feeding, were chosen for this review. All types of pediatric FA were included regardless of the allergen causing the disease and were as follows: IgE- (including food sensitization) and non-IgE-mediated FA. The main technique used for analyzing GM composition in the included studies was 16S rRNA sequencing or shotgun sequencing due to the comprehensive profiling of the microbiome that these methods allow. The relevant data from the included studies were extracted independently by MF and LC. Any disagreements were solved by discussion, and if not resolved, by consulting a third member of the team (RC).
The bacteria were classified into phylum, family, genus, and species. If the study only offered one of the taxa classifications (i.e., the genus), the National Center for Biotechnology Information (NCBI) taxonomy browser for bacteria was used to determine the rest of the categories upstream of the one specified. NCBI's up-to-date taxonomic classification was used, and this resulted in some phyla and genera being renamed and reclassified.
2.3 Data items
The data extracted included the number of participants in the FA and healthy groups, age of subjects, geographical location, methodology of microbiome analysis used, study design, FA features, and GM signature.
3 Results
3.1 Studies record
As reported in Figure 1, the search yielded a cumulative total of 883 records. After duplicate removal, in the screening phase, 34 papers were included. Studies on animals, studies that did not include a healthy control population, and studies describing participants as “allergic” without specifying the type of allergy were excluded. However, studies involving animals with the primary objective of transferring human stools into gnotobiotic mice were included if microbiome analysis was conducted before the transfer. Studies using supplemented feeding formulas to assess their effect on microbiome composition were excluded unless baseline microbiome analysis was also performed prior to treatment. The characteristics of the studies identified as eligible evidence for this scoping review are presented in Table 2.
3.2 Study characteristics
Of the 34 studies included in the review, most (n = 24) were case–control studies, eight were prospective cohort studies and two were randomized controlled trials. The year of publication ranged from 2011 to 2023. In terms of geographical region, eight were conducted in the USA, seven in China, four in Italy, four in Spain, four in Japan, and the others were conducted in Poland, Norway, the United Kingdom, Canada, Israel, Taiwan, and Australia. The sample size of the studies ranged from 6 to 447 subjects, representing a total population of 2,822 study participants.
Seven studies enrolled patients with both IgE-mediated and non-IgE-mediated FA without distinguishing between these two categories. Nineteen studies focused on patients with only IgE-mediated FA or food sensitization, and eight studies enrolled patients with non-IgE-mediated FA.
4 Gut microbiome structure
All studies included in our review reported alterations in the structure of the GM in pediatric FA patients and in subjects sensitized to food allergens compared with healthy controls. No differences were reported according to the sex of the subjects and the food antigens responsible for the disease. Most of the available data referred to patients already affected by FA; only five studies prospectively investigated the structure of the GM and the subsequent occurrence of FA (25–50). These studies showed that GM dysbiosis could precede the occurrence of FA by 3–6 months and persist during the disease course (25–50). Among these studies, Martin et al. (48) analyzed a population of children affected by food protein-induced allergic proctocolitis (FPIAP) and compared GM features before (pre-symptomatic), during (symptomatic), and after the resolution of the disease. Interestingly, they reported a progressive increase in the abundance of the Enterobacteriaceae family from the pre-symptomatic group to the symptomatic stage (48). Moreover, in the same study, an increased abundance of the Lactobacillus genus was observed when comparing children with active disease with children who acquired immune tolerance, supporting the hypothesis that these bacteria could be linked to the development of immune tolerance (48). Joseph et al. prospectively evaluated the structure of the GM in children who developed IgE-mediated FA or who did not develop FA (healthy controls), collecting stool samples at 1 and 6 months of life. The results of the study highlighted an overall delayed maturity of the structure of the GM as a typical feature of the intestinal microbial community in FA children (44).
Four studies investigated the structure of the GM in children with food antigen sensitization (29, 36, 38, 40). In food-sensitized children, there is a lower overall GM richness at 3 months of life and an increased Enterobacteriaceae/Bacteroidaceae ratio (E/B ratio), before the development of sensitization at 12 months (29). Accordingly, Chen et al. found that food-sensitized subjects exhibited a lower total microbiota diversity (p = 0.01) and a lower diversity of the bacterial phylum Bacteroidetes (p = 0.02) compared with healthy controls (36). On the contrary, no differences in microbial diversity measures were detected between food sensitization and food allergy cases and controls. However, at the genus level, Haemophilus, Dialister, Dorea, and Clostridium were significantly underrepresented among subjects with food sensitization (40). Finally, in a prospective study comparing cow's milk-sensitized subjects vs. cow's milk allergy patients vs. healthy controls, sensitized subjects had the highest number of total and rare operational taxonomic units (OTUs) (i.e., observed and Chao I index, respectively), whereas cow’s milk allergy was characterized by the lowest OTU richness (38).
Considering the relevant role elicited by the age of the subject on the maturation of the GM (11, 56), we divided the included studies according to the age of the enrolled subjects. We classified the available data into the following three categories according to the participant's age: (1) from 0 to 6 months; (2) from 6 to 12 months; and (3) >12 months of age. All data obtained in our study are summarized in Tables 3–8, which show the bacterial taxa that were found to be increased or reduced in at least two studies.
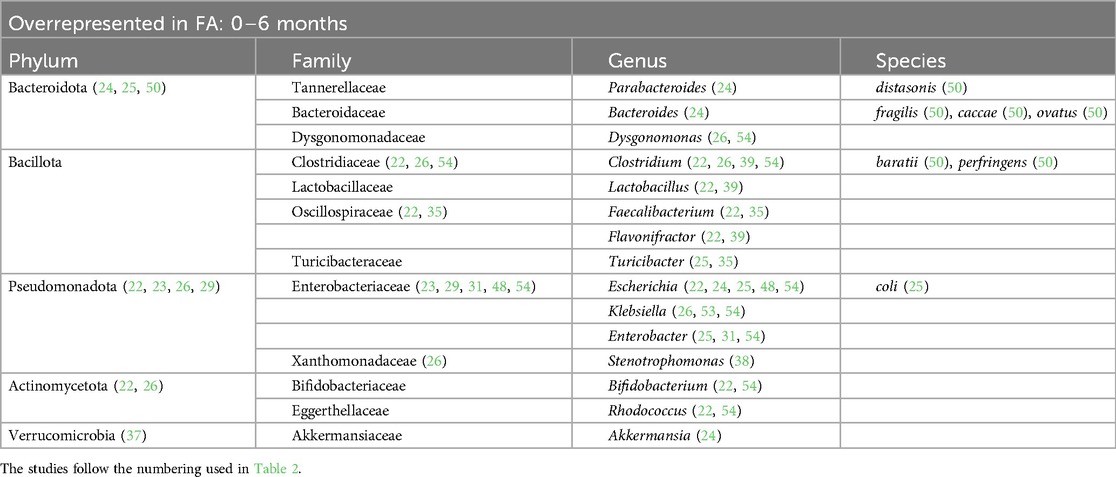
Table 3. Bacterial species overrepresented in the GM of children affected by FA in the first 6 months of life.
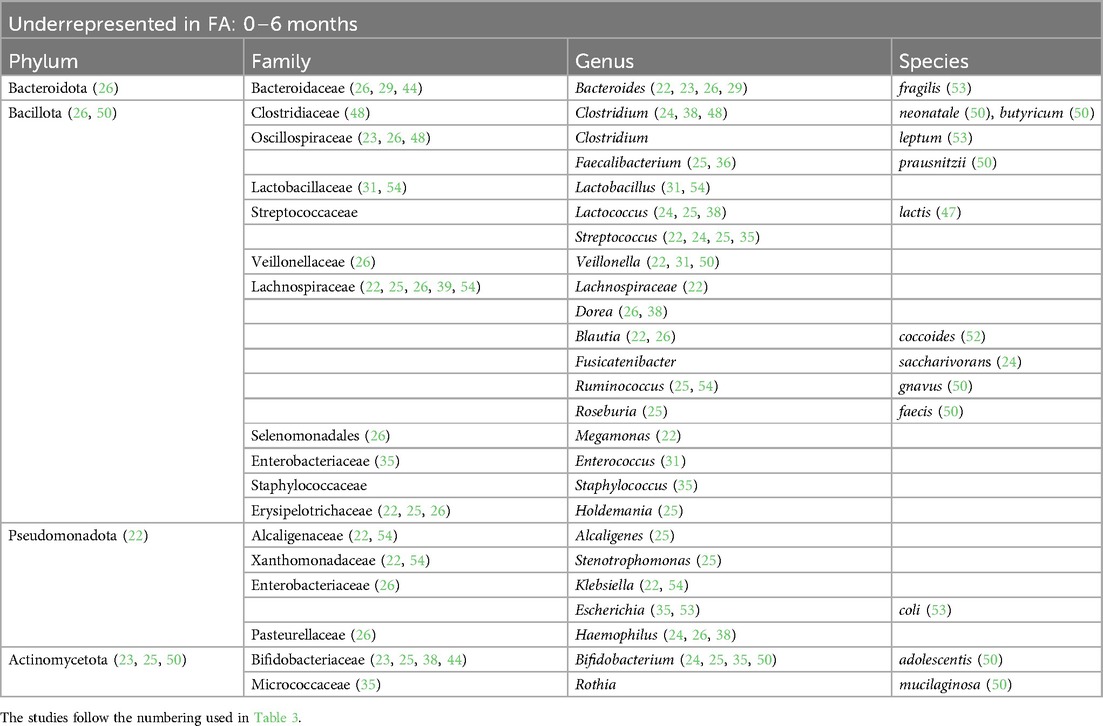
Table 4. Bacterial species that are underrepresented in the GM of children affected by FA in the first 6 months of life.
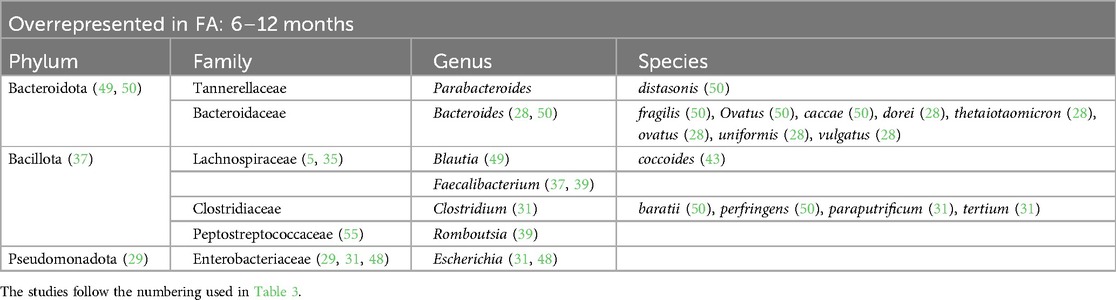
Table 5. Bacterial species overrepresented in the GM of children affected by FA from 6 to 12 months of life.
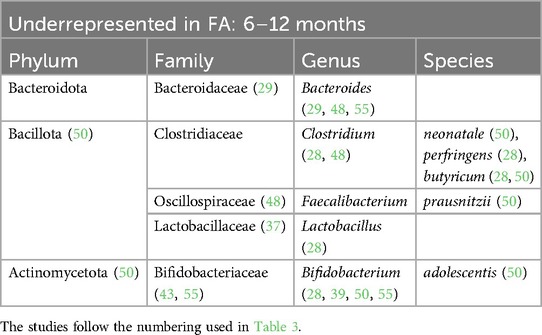
Table 6. Bacterial species that are underrepresented in the GM of children affected by FA from 6 to 12 months of life.
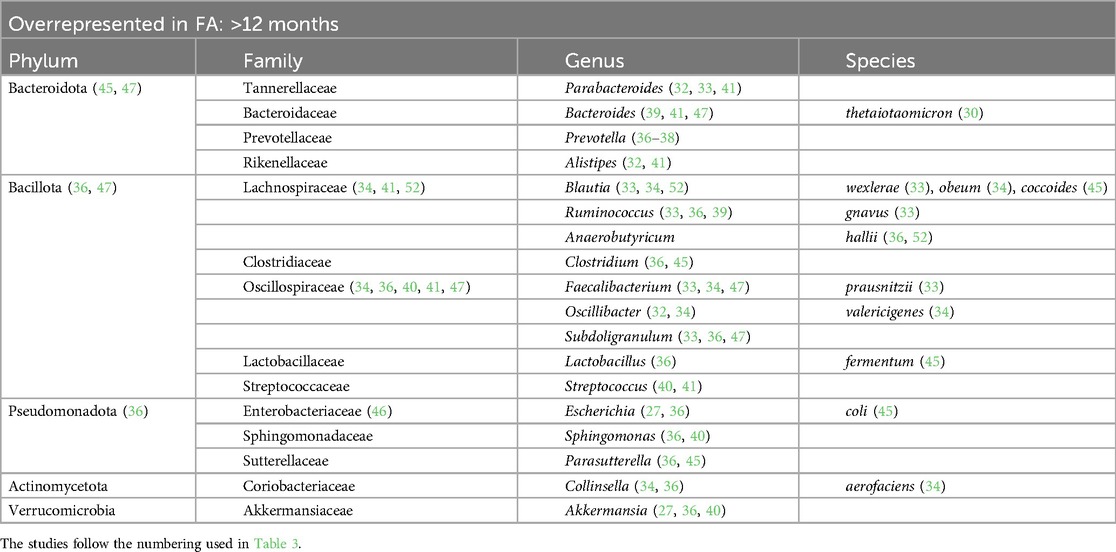
Table 7. Bacterial species overrepresented in the GM of children affected by FA with more than 12 months of life.
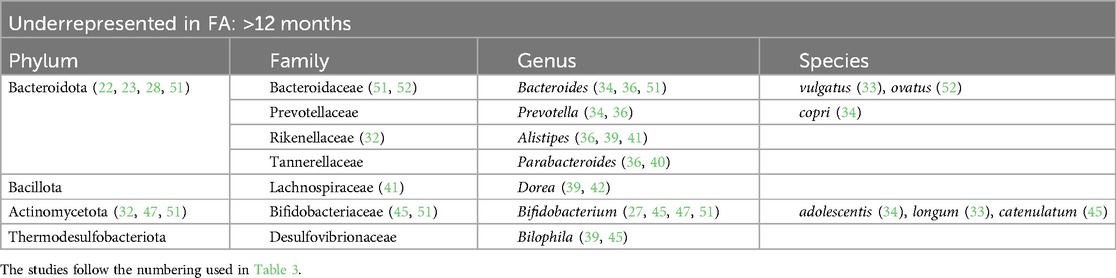
Table 8. Bacterial species that are underrepresented in the GM of children affected be FA with more than 12 months of life.
At the phylum level, we found that in children affected by FA aged 0–6 months there was an increase in Pseudomonadota (22, 23, 26, 29) and a decrease in Actinomycetota (23, 25, 50). In children aged >12 months there was a decrease in the phylum Bacteroidota (22, 23, 40, 51).
At the family level, we found an increase in Enterobacteriaceae within the GM of FA children (22–25, 29, 30, 36, 45, 48, 49, 53, 54). In particular, the species Escherichia coli was overrepresented. The Enterobacteriaceae family was one of the most represented taxa in the GM of the healthy subjects in the first 6 months of life and its abundance gradually decreased with growth. By contrast, this shift was not reported in FA children, resulting in an increase in the Enterobacteriaceae/Bacteroides ratio (23, 29), resembling the features of an immature microbial ecosystem (57).
During the first 6 months of life, we also noticed an increase in the Clostridiaceae family with some species that were overrepresented, such as Clostridium baratii, Clostridium perfringens, Clostridium paraputrificum, and Clostridium tertium (22, 31, 39, 54), and other species that were decreased, such as Clostridium neonatale and Clostridium butyricum, in FA children compared with healthy controls (28, 31, 50). A decrease in Streptococcaceae (the genera Lactococcus and Streptococcus) (22, 24, 25, 31, 35, 36, 38, 55) and Erysipelotrichaceae (22, 25, 26) was also found in FA children aged 0–6 months.
Another common FA feature was the reduction in the beneficial Bifidobacteriaceae family reported in all ages (23, 25, 28, 33–35, 38, 39, 44, 45, 47, 50, 51, 55). In particular, the Bifidobacterium genus, which belongs to this family, is an important source of beneficial immunomodulatory compounds, such as SCFAs, and constitutes one of the prevalent taxa in the first months of life in healthy breastfed infants (11).
Moreover, we found a clear reduction in the Lactobacillaceae family (the genera Lactobacillus and Weissella) in the GM of FA children, especially in the first 6 months of life (24, 28, 31, 54). The decrease in this family could precede the onset of FA. In addition, in children affected by cow milk protein allergy (CMPA), it has been demonstrated that the total abundance of the Lactobacillaceae family increases with the development of immune tolerance (25).
The Bacteroidaceae family was decreased in the GM of FA children (26, 29, 44, 51, 52) but there are conflicting data on the genus Bacteroides. This could be related to the large number of species belonging to this genus. According to some studies, the species Bacteroides thetaiotaomicron, Bacteroides uniformis, Bacteroides ovatus, Bacteroides caccae, and Bacteroides fragilis are overrepresented in the GM of FA children (22, 24, 28, 30, 39, 41, 49, 50). In other studies, other species belonging to the Bacteriodes genus were underrepresented in FA children (23, 29, 33, 37, 44, 48, 51, 53–55), e.g., Bacteroides vulgatus, Bacteroides dorei, and B. fragilis. In addition in this case, some species (e.g., B. fragilis) were overrepresented (50) or underrepresented (53) in FA children, complicating the interpretation of this aspect. These apparently conflicting data could be explained by the strain-specificity that was not considered in these studies (58).
Lachnospiraceae is another pivotal family that can distinguish FA vs. healthy children. In particular, the genera Lachnospira (33, 42), Anaerostipes (22, 33, 42), Lachnobacterium (50), and Ruminococcus (33, 36, 39) were increased in the microbial ecosystem of the FA children aged more than 6 months (28, 34, 41, 52, 55), whereas this family was decreased in the first 6 months of life (22, 25, 26, 39, 54). Dorea and Blautia, two bacterial genera belonging to this family, were decreased in FA children compared with healthy controls in the first 6 months of life (22, 26, 38, 52). According to some studies, the increase in some members of the Lachnospiraceae family could precede the development of immune tolerance, as also demonstrated in children with CMPA (35). Conversely, in another study, the relative abundance at baseline of Lachnospiraceae was significantly higher in infants who did not outgrow CMPA than in infants who outgrew CMPA (59).
Regarding the Oscillospiraceae family, there is still contrasting evidence and no agreement in the selected studies. In FA pediatric patients, this family was underrepresented in the first 6 months of life (23, 26, 48) and overrepresented in children aged >6 months (22, 35, 36, 40).
One of the main species involved in SCFA metabolism, Faecalibacterium prausnitzii, has been found to be both increased (33, 34, 37, 39, 47) and decreased (26, 38, 50) in FA children in relation to age. The metagenomic approach once again clarified that the same bacterial species can exert different actions. In a previous study by our group, we observed the abundance of this bacterium in the GM of FA children, and we also described the presence of different clades of F. prausnitzii in healthy and allergic children (clade A being characteristic of allergic children), characterized by distinct metabolic activities (33). Conversely, according to the two studies included in the review that analyzed the GM composition in children affected by food protein-induced enterocolitis syndrome (FPIES), the abundance of F. prausnitzii was decreased (50, 52). In pediatric FA patients aged >12 months, two other bacterial genera, Akkermansia (32, 34, 42) and Bilophila (39, 45), were decreased.
5 Discussion
In this scoping review, we provided updated information on GM features associated with the occurrence of pediatric FA. In addition, we provided data on the structure and function of the GM at different ages and information regarding the most relevant environmental factors impacting them. The final aim was to make data accessible and engaging for physicians and researchers to help the development of innovative, preventive, and therapeutic strategies for managing FA.
Our results confirmed the presence of alterations in the structure and function of the GM in pediatric patients affected by FA. Impaired GM maturation, marked by a reduction in age-related GM maturation, is a hallmark of FA occurrence (60, 61). FA children have a lower GM diversity, and their microbial ecosystem is unbalanced, with a loss of beneficial species and an increase in species associated with alterations in the gut barrier, inflammation, and the Th2 response (61). Data regarding selected bacteria species seem to be conflicting but these discrepancies should be interpreted considering that bacteria belonging to the same species can exert both beneficial and detrimental effects on the gut barrier and immune system (33, 58, 62).
The term “dysbiosis” refers to the disruption of the normal homeostasis of the microbial ecosystem inhabiting the human intestinal tract, involving quantitative and qualitative alterations in the predominant bacterial species (63). Dysbiosis, however, does not merely mean alterations in the structure but also in GM function, which stimulates the immune system in a pro-inflammatory direction (64). This condition precedes the occurrence of FA (48) and is driven by the influence of several environmental factors (see Table 1).
Several mechanisms of action have been proposed to explain how GM dysbiosis and dysfunction can facilitate the occurrence of FA (see Figure 2). First, the loss of beneficial species, such as Bifidobacterium, Lactobacillus, and certain species within the Lachnospiraceae family, facilitate the alteration in gut barrier homeostasis. These bacterial strains are responsible for producing SCFAs as butyrate, through the fermentation of non-digestible dietary fibers, which increase gut barrier function by acting on enterocytes and immune cells (65). Metabolomic analyses conducted on stool samples from allergic children demonstrated a reduction in butyrate production (43, 49). The SCFAs exert their effects on the host primarily through G protein-coupled receptors (GPR41, GPR45, and GPR109A) promoting immune tolerance (66). Butyrate serves as an energy substrate for colonocytes, modulating their metabolism and promoting the expression of mucins. By inhibiting histone deacetylases (HDAC), it exerts an epigenetic effect on immune cells such as dendritic cells (DCs), stimulating the production of IL-18, which has a key role in maintaining barrier function, and IL-10 and IL-23. The SCFAs stimulate the proliferation and activation of Treg cells through the same epigenetic mechanism. Among other effects, they facilitate class switching and promote IgA production through B lymphocytes, improving gut barrier function (66). In this way, the reduction of butyrate-producing bacteria has been associated with an imbalance in the Th1/Th2 ratio toward Th2 responses and a decrease in Treg cells, resulting in the loss of oral tolerance and development of FA (67–70).
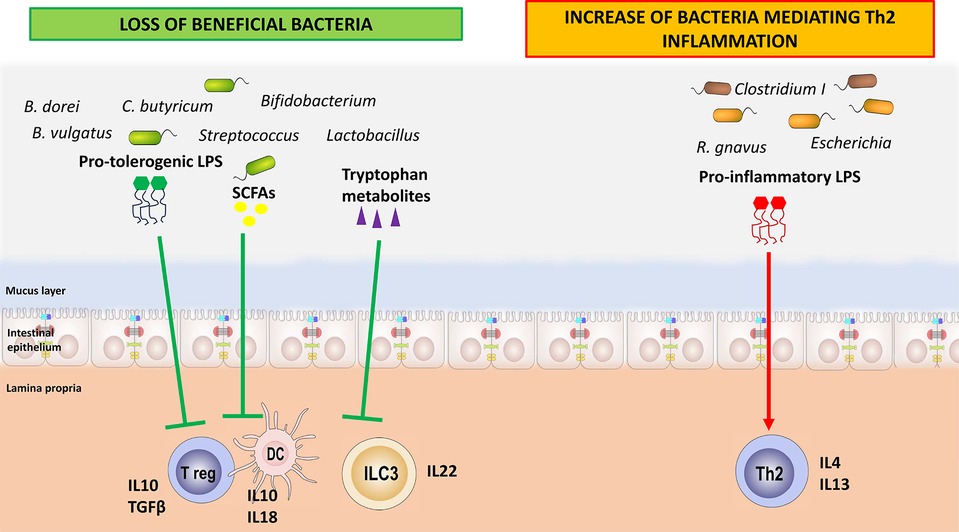
Figure 2. How the loss of beneficial bacteria and the increase in harmful bacteria may play a role in food allergy pathogenesis. The loss of beneficial bacteria and of their immune-regulatory metabolites leads to a breakdown in immune tolerance mechanisms. Concomitantly, the higher rate of harmful bacteria is associated with a huge release of pro-inflammatory compounds, such as specific lipopolysaccharides, resulting in the activation of T helper 2 cells.
In our previous study, we described an increase in Lactobacillus and SCFA-producing bacteria belonging to the Lachnospiraceae family, such as Roseburia, Coprococcus, and Blautia, in CMPA infants who acquired immune tolerance after a 6-month dietary treatment with extensively hydrolyzed casein formula (EHCF) supplemented with the probiotic Lactobacillus rhamnosus GG (35). Martin et al. also described an increased abundance of Lactobacillus in children affected by FPIAP who developed immune tolerance (48). In another study, we also found that a higher abundance of Lachnospiraceae (specifically Lachnospira pectinoschiza and Anaerostipes hadrus) and Bifidobacterium in the stools of allergic children at diagnosis correlated with the development of immune tolerance after 36 months of follow-up (33).
Another important class of compounds produced by a healthy GM, identified through metabolomics, are tryptophan metabolites such as indole, indole-3-acetic acid, skatole, indole-3-aldehyde, and indole-lactic acid (67). Several bacterial strains, such as Lactobacillus spp., Bifidobacterium spp., and B. fragilis that are underrepresented in the GM of FA children share this metabolic pathway, which contributes to the maintenance of intestinal homeostasis by preventing colonization by potentially pathogenic organisms and reducing inflammation (66).
Through the activation of the aryl hydrocarbon receptor (AhR), tryptophan derivatives promote the production of IL-22 by type 3 innate lymphoid cells (ILC3s) and RORγt+ T cells (71). IL-22 belongs to the family of IL-10, the main anti-inflammatory cytokine, and allows the production of antimicrobial peptides and the expression of tight junctions proteins (72). On the other hand, 3-indole-lactic acid inhibits the polarization of CD4+ Th lymphocytes toward the Th17 lineage and instead promotes the development of Treg cells (67).
In addition, the microbiome is responsible for another metabolic activity, less studied in the context of FA, which is the production of secondary bile acids (66). Primary bile acids are produced via host cholesterol catabolism in the liver and are further secreted by the gall bladder into the gut lumen after conjugation (66). Bile acids that escape from reabsorption in the terminal ileum can be metabolized by the GM into secondary bile acids by certain bacterial genera, such as Lactobacillus, Bifidobacterium, Bacteroides fragilis, and some species of Clostridium (66). Campbell et al. found that one of these compounds, 3β-hydroxydeoxycholic acid (isoDCA), increased Foxp3 expression in T lymphocytes by acting on DCs in an immunoregulatory way (73). Metabolomic studies on FA and other allergic conditions, such as asthma, have revealed distinct profiles of secondary bile acids, indicating an alteration in this metabolic pathway that is specific for the type of allergic disorder (74).
Gut microbiome interacts with the immune system also through bacterial components such as LPSs and flagellins. Changes to the structure of these bacterial components can have significant impacts on signaling so that they can exert anti-inflammatory or pro-inflammatory activities (75). In particular, LPS is a glycoconjugate part of the outer cell membrane of Gram-negative bacteria (76). Fragments of LPS are released into the environment by the bacteria during the continuous process of outer bacterial membrane renewal (77). Lipid A is the main immunostimulatory portion of LPS (78). It is recognized by the toll-like receptor 4 (TLR4)/myeloid differentiation factor 2 (MD-2) receptor complex, leading to the production of pro-inflammatory mediators through the NF-κB pathway, thus initiating and modulating the immune response. The activation of the TLR4/MD-2-dependent response is largely determined by the chemical structure of lipid A (78). The Enterobacteriaceae family produces an LPS with pronounced pro-inflammatory activity, which can disrupt the gut barrier and drive the production of pro-inflammatory cytokines such as IL-4, IL-13, and TNFα (75). LPS derived from Bacteroides sp., which is diminished in stool samples from children affected by FA, does not seem to stimulate this pro-inflammatory pathway. It elicits a tolerogenic response with the increase in IL-10 and TGFβ (79). Capsular polysaccharide A (PSA) of commensal B. fragilis can be identified via TLR2 and TLR1 heterodimers together with Dectin, resulting in glycogen synthase kinase 3b (GSK3b) inactivation, which in turn promotes the cAMP response element binding protein (CREB)–associated activation of anti-inflammatory genes and Treg cell differentiation (80).
In a recent study from our group, we purified fecal LPSs and lipid A from healthy children and children affected by IgE-mediated FA (76). We provided evidence for a different immunoregulatory action elicited by LPS on peripheral blood mononuclear cells collected from healthy donors, suggesting that LPSs from healthy individuals can protect against the occurrence of FA, and LPSs from children affected by FA can promote an allergic response (76).
The significance of dysbiosis in the pathogenesis of FA is further reinforced by studies on fecal microbiota transplantation (FMT) (39, 54, 81, 82). FMT of feces from allergic children into germ-free (GF) mice results in a deficit in the total number of Treg lymphocytes in the lamina propria, which are known to play a crucial role in immune tolerance. Specifically, the quantity of Tregs exhibits a positive correlation with the abundance of bacteria from the genera Bacteroides and Lactobacillus, as well as with the family Lachnospiraceae. Conversely, there is a negative correlation with Raoultella and Clostridium sensu stricto (54).
Furthermore, the bacterial colonization of GF mice with fecal samples from healthy children shields mice from an anaphylactic response to beta-lactoglobulin challenge, and this response does not occur when fecal samples from CMPA infants are transferred to mice (82). According to this study, the protective action of the healthy microbiome is primarily linked to the abundance of the Lachnospiraceae family (82).
Bacteriotherapy with certain consortia of Clostridiales and Bacteroidales also protects sensitized mice from developing FA symptoms, increasing the number of Tregs in the lamina propria (LP) after FMT (39), thereby supporting the possible protective role of these bacterial strains against FA. Atarashi et al. also demonstrate the ability of some bacterial families belonging to Clostridiales, such as Lachnospiraceae and Oscillospiraceae, to induce Treg cells in a murine model (81).
Finally, to translate the GM concept in clinical practice, it is crucial to define the environmental factors inducing GM dysbiosis and dysfunction, and then the occurrence of FA. In Table 1, we summarize the actual available evidence on environmental factors influencing the alteration in the GM and the occurrence of FA. Since intrauterine life, many environmental factors actively modulate the structure of the microbiota from both the maternal and the fetal side (83). The maternal GM during pregnancy influences the newborn's GM composition, impacting their risk of developing allergic conditions (61). In particular, maternal obesity (8, 11), an unbalanced diet during pregnancy (11, 16, 84), the use of antibiotics during pregnancy (14, 16, 61, 85, 86), and exposure to smoke (8) act from the maternal side, reducing GM biodiversity and increasing detrimental bacteria linked to the occurrence of FA.
Birth is a crucial moment for the colonization of the newborn's gut. Gestational age and the mode of delivery have a huge impact on GM composition (8, 11–13). Pre-term birth and cesarean section (C-section) are linked to the development of allergic conditions (8, 11–13). Vaginally delivered infants meet the maternal vaginal and fecal microbiota, which results in neonatal gut colonization by vagina-associated microbes such as Lactobacillus (87). In contrast, C-section-delivered infants are not directly exposed to maternal microbes and are thus more likely to become colonized by environmental microorganisms from the maternal skin, hospital staff, or hospital environment (87). Then, the type of infant feeding (breastfeeding vs. formula feeding) and the diet after weaning play a crucial role in modulating the structure and function of the GM (11, 17, 88).
The housing setting, increased hygiene, and the presence of siblings and pets can also modulate GM composition (11). Broad-spectrum antibiotics can affect the abundances of 30% of the bacteria in the gut community, causing rapid and significant drops in taxonomic richness, diversity, and evenness (89, 90). Antibiotic-induced GM alterations can remain for long periods of time, spanning months and even years (15, 91, 92).
Some therapeutic strategies have been proposed to counteract GM dysbiosis, such as probiotics, prebiotics, postbiotics, and synbiotics (93–96). Several clinical studies and a systematic review and meta-analysis suggest that active diet therapy with EHCF supplemented with the probiotic L. rhamnosus GG (LGG) may reduce disease duration in children with CMPA (95–99), the occurrence of functional gastrointestinal disorders (100), and other allergic manifestations (eczema, urticaria, asthma, and rhinoconjunctivitis) later in the life compared with other formulas (95, 96, 98, 101). In addition, substitutive infant formulas have emerged as a major cost driver in the management of pediatric patients with CMPA, as it is estimated to be up to six times more expensive to feed a child with this condition (102, 103). Therefore, these data highlight the role of EHCF + LGG not only in improving clinical outcomes but also in reducing the cost of patient management, providing a cost-effective nutritional strategy for healthcare systems in the management of infants with CMPA (104–106). It is important to underline that the beneficial results observed in FA children treated with LGG could not necessarily be extended to other probiotic strains. Data on other probiotic strains with regard to the modulation of the structure of the GM in FA children are conflicting (107). More evidence is needed to demonstrate the role of these strategies in preventing the onset of FA (108) or alternatively modifying its natural history.
6 Conclusion
Our knowledge on the link between GM dysbiosis and FA (i.e., the GM-immune system axis) is continuosly enriched with new insights (109, 110). New investigations are advocated to better define the role of other components of the GM, such as viruses, fungi, and archaea (111, 112), in the regulation of the GM-immune system axis.
It is now clear that GM dysbiosis is strongly associated with immune system dysfunction. Thus, the loss of some beneficial bacteria species in favor of harmful ones is not merely an alteration of the structure of the GM but implies a GM dysfunction that impairs immune system development and activity in the pediatric age. Concomitantly, the finding that several environmental factors actively modulate the GM-immune system axis opens the way for the development of new strategies aimed at preventing and treating FA with a possible positive impact also on the development and treatment of other non-communicable diseases.
Author contributions
MF: Conceptualization, Data curation, Formal Analysis, Investigation, Methodology, Resources, Visualization, Writing – original draft, Writing – review & editing. LC: Data curation, Formal Analysis, Methodology, Supervision, Validation, Writing – original draft, Writing – review & editing. SC: Writing – review & editing. FO: Writing – review & editing. AM: Writing – review & editing. MC: Writing – original draft, Writing – review & editing. RN: Supervision, Writing – review & editing. RBC: Conceptualization, Funding acquisition, Methodology, Project administration, Supervision, Validation, Visualization, Writing – review & editing.
Funding
The authors declare financial support was received for the research, authorship, and/or publication of this article. The study was supported by the Department of Translational Medical Science of the University of Naples Federico II (Naples, Italy), which received funding from the National Recovery and Resilience Plan (NRRP), Mission 4 Component 2 Investment 1.3—Call for tender No. 341 of 15 March 2022 of Italian Ministry of University and Research funded by the European Union—NextGenerationEU. Project code PE00000003, Concession Decree No. 1550 of 11 October 2022, adopted by the Italian Ministry of University and Research, CUP E63C22002030007, Project title “ON Foods—Research and innovation network on food and nutrition Sustainability, Safety and Security—Working ON Foods” and from the Italian Ministry of Health-Health Operational Plan Trajectory 5-Line of action “Creation of an action program for the fight against malnutrition in all its forms and for the dissemination of the principles of the Mediterranean diet” (Mediterranean Diet for Human Health Lab, “MeDiHealthLab”, code T5-AN-07, CUP E63C22002570006).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Nocerino R, Carucci L, Coppola S, Cecere G, Micillo M, Castaldo T, et al. Epidemiology of paediatric Italian food allergy: results of the EPIFA study. J Allergy Clin Immunol Glob. (2024) 3(3):100246. doi: 10.1016/j.jacig.2024.100246
2. Nocerino R, Leone L, Cosenza L, Berni Canani R. Increasing rate of hospitalizations for food-induced anaphylaxis in Italian children: an analysis of the Italian ministry of health database. J Allergy Clin Immunol. (2015) 135(3):833–5.e3. doi: 10.1016/j.jaci.2014.12.1912
3. Warren CM, Jiang J, Gupta RS. Epidemiology and burden of food allergy. Curr Allergy Asthma Rep. (2020) 20(2):6. doi: 10.1007/s11882-020-0898-7
4. Fong AT, Ahlstedt S, Golding MA, Protudjer JLP. The economic burden of food allergy: what we know and what we need to learn. Curr Treat Options Allergy. (2022) 9(3):169–86. doi: 10.1007/s40521-022-00306-5
5. Golding MA, Batac ALR, Gunnarsson NV, Ahlstedt S, Middelveld R, Protudjer JLP. The burden of food allergy on children and teens: a systematic review. Pediatr Allergy Immunol. (2022) 33(3):e13743. doi: 10.1111/pai.13743
6. Lopes JP, Sicherer S. Food allergy: epidemiology, pathogenesis, diagnosis, prevention, and treatment. Curr Opin Immunol. (2020) 66:57–64. doi: 10.1016/j.coi.2020.03.014
7. Nance CL, Deniskin R, Diaz VC, Paul M, Anvari S, Anagnostou A. The role of the microbiome in food allergy: a review. Children (Basel). (2020) 7(6):50. doi: 10.3390/children7060050
8. Jeong S. Factors influencing development of the infant microbiota: from prenatal period to early infancy. Clin Exp Pediatr. (2021) 65(9):439–47. doi: 10.3345/cep.2021.00955
9. Davis EC, Jackson CM, Ting T, Harizaj A, Järvinen KM. Predictors and biomarkers of food allergy and sensitization in early childhood. Ann Allergy Asthma Immunol. (2022) 129(3):292–300. doi: 10.1016/j.anai.2022.04.025
10. Akagawa S, Kaneko K. Gut microbiota and allergic diseases in children. Allergol Int. (2022) 71(3):301–9. doi: 10.1016/j.alit.2022.02.004
11. Milani C, Duranti S, Bottacini F, Casey E, Turroni F, Mahony J, et al. The first microbial colonizers of the human gut: composition, activities, and health implications of the infant gut microbiota. Microbiol Mol Biol Rev. (2017) 81(4):e00036–17. doi: 10.1128/MMBR.00036-17
12. Rinninella E, Raoul P, Cintoni M, Franceschi F, Miggiano GAD, Gasbarrini A, et al. What is the healthy gut microbiota composition? A changing ecosystem across age, environment, diet, and diseases. Microorganisms. (2019) 7(1):14. doi: 10.3390/microorganisms7010014
13. Salminen S, Gibson GR, McCartney AL, Isolauri E. Influence of mode of delivery on gut microbiota composition in seven year old children. Gut. (2004) 53(9):1388–9. doi: 10.1136/gut.2004.041640
14. Diesner SC, Bergmayr C, Pfitzner B, Assmann V, Krishnamurthy D, Starkl P, et al. A distinct microbiota composition is associated with protection from food allergy in an oral mouse immunization model. Clin Immunol. (2016) 173:10–8. doi: 10.1016/j.clim.2016.10.009
15. Weersma RK, Zhernakova A, Fu J. Interaction between drugs and the gut microbiome. Gut. (2020) 69(8):1510–9. doi: 10.1136/gutjnl-2019-320204
16. Zmora N, Suez J, Elinav E. You are what you eat: diet, health and the gut microbiota. Nat Rev Gastroenterol Hepatol. (2019) 16(1):35–56. doi: 10.1038/s41575-018-0061-2
17. Adolph TE, Zhang J. Diet fuelling inflammatory bowel diseases: preclinical and clinical concepts. Gut. (2022) 71(12):2574–86. doi: 10.1136/gutjnl-2021-326575
18. Yang W, Yu T, Huang X, Bilotta AJ, Xu L, Lu Y, et al. Intestinal microbiota-derived short-chain fatty acids regulation of immune cell IL-22 production and gut immunity. Nat Commun. (2020) 11(1):4457. doi: 10.1038/s41467-020-18262-6
19. Campbell E, Hesser LA, Berni Canani R, Carucci L, Paparo L, Patry RT, et al. A lipopolysaccharide-enriched cow’s milk allergy microbiome promotes a TLR4-dependent proinflammatory intestinal immune response. J Immunol. (2024) 212(4):702–14. doi: 10.4049/jimmunol.2300518
20. Paparo L, Picariello G, Bruno C, Pisapia L, Canale V, Sarracino A, et al. Tolerogenic effect elicited by protein fraction derived from different formulas for dietary treatment of cow’s milk allergy in human cells. Front Immunol. (2021) 11:604075. doi: 10.3389/fimmu.2020.604075
21. Tricco AC, et al. PRISMA extension for scoping reviews (PRISMA-ScR): checklist and explanation. Ann Intern Med. (2018) 169(7):467–73. doi: 10.7326/M18-0850
22. Ling Z, Li Z, Liu X, Cheng Y, Luo Y, Tong X, et al. Altered fecal microbiota composition associated with food allergy in infants. Appl Environ Microbiol. (2014) 80(8):2546–54. doi: 10.1128/AEM.00003-14
23. Dong P, Feng JJ, Yan DY, Lyu YJ, Xu X. Early-life gut microbiome and cow’s milk allergy—a prospective case—control 6-month follow-up study. Saudi J Biol Sci. (2018) 25(5):875–80. doi: 10.1016/j.sjbs.2017.11.05
24. Łoś-Rycharska E, Gołębiewski M, Sikora M, Grzybowski T, Gorzkiewicz M, Popielarz M, et al. A combined analysis of gut and skin microbiota in infants with food allergy and atopic dermatitis: a pilot study. Nutrients. (2021) 13(5):1682. doi: 10.3390/nu13051682
25. Wang S, Zhang R, Li X, Gao Y, Dai N, Wei Y, et al. Relationship between maternal-infant gut microbiota and infant food allergy. Front Microbiol. (2022) 13:933152. doi: 10.3389/fmicb.2022.933152
26. Yu Z, Yue L, Yang Z, Wang Y, Wang Z, Zhou F, et al. Impairment of intestinal barrier associated with the alternation of intestinal flora and its metabolites in cow’s milk protein allergy. Microb Pathog. (2023) 183:106329. doi: 10.1016/j.micpath.2023.106329
27. Yan X, Yan J, Xiang Q, Dai H, Wang Y, Fang L, et al. Early-life gut microbiota in food allergic children and its impact on the development of allergic disease. Ital J Pediatr. (2023) 49(1):148. doi: 10.1186/s13052-023-01557-x
28. Mauras A, Wopereis H, Yeop I, Esber N, Delannoy J, Labellie C, et al. Gut microbiota from infant with cow’s milk allergy promotes clinical and immune features of atopy in a murine model. Allergy. (2019) 74(9):1790–3. doi: 10.1111/all.13787
29. Azad MB, Konya T, Guttman DS, Field CJ, Sears MR, HayGlass KT, et al. Infant gut microbiota and food sensitization: associations in the first year of life. Clin Exp Allergy. (2015) 45(3):632–43. doi: 10.1111/cea.12487
30. Zhang L, Chun Y, Ho HE, Arditi Z, Lo T, Sajja S, et al. Multiscale study of the oral and gut environments in children with high- and low-threshold peanut allergy. J Allergy Clin Immunol. (2022) 150(3):714–20.e2. doi: 10.1016/j.jaci.2022.04.026
31. Tanaka M, Korenori Y, Washio M, Kobayashi T, Momoda R, Kiyohara C, et al. Signatures in the gut microbiota of Japanese infants who developed food allergies in early childhood. FEMS Microbiol Ecol. (2017) 93(8):fix099. doi: 10.1093/femsec/fix099
32. Kourosh A, Luna RA, Balderas M, Nance C, Anagnostou A, Devaraj S, et al. Fecal microbiome signatures are different in food-allergic children compared to siblings and healthy children. Pediatr Allergy Immunol. (2018) 29(5):545–54. doi: 10.1111/pai.12904
33. De Filippis F, Paparo L, Nocerino R, Della Gatta G, Carucci L, Russo R, et al. Specific gut microbiome signatures and the associated pro-inflammatory functions are linked to pediatric allergy and acquisition of immune tolerance. Nat Commun. (2021) 12(1):5958. doi: 10.1038/s41467-021-26266-z
34. Goldberg MR, Mor H, Magid Neriya D, Magzal F, Muller E, Appel MY, et al. Microbial signature in IgE-mediated food allergies. Genome Med. (2020) 12(1):92. doi: 10.1186/s13073-020-00789-4
35. Berni Canani R, Sangwan N, Stefka AT, Nocerino R, Paparo L, Aitoro R, et al. Lactobacillus rhamnosus GG-supplemented formula expands butyrate-producing bacterial strains in food allergic infants. ISME J. (2016) 10(3):742–50. doi: 10.1038/ismej.2015.151
36. Chen CC, Chen KJ, Kong MS, Chang HJ, Huang JL. Alterations in the gut microbiotas of children with food sensitization in early life. Pediatr Allergy Immunol. (2016) 27(3):254–62. doi: 10.1111/pai.12522
37. Fazlollahi M, Chun Y, Grishin A, Wood RA, Burks AW, Dawson P, et al. Early-life gut microbiome and egg allergy. Allergy. (2018) 73(7):1515–24. doi: 10.1111/all.13389
38. Savage JH, Lee-Sarwar KA, Sordillo J, Bunyavanich S, Zhou Y, O'Connor G, et al. A prospective microbiome-wide association study of food sensitization and food allergy in early childhood. Allergy. (2018) 73(1):145–52. doi: 10.1111/all.13232
39. Abdel-Gadir A, Stephen-Victor E, Gerber GK, Noval Rivas M, Wang S, Harb H, et al. Microbiota therapy acts via a regulatory T cell MyD88/RORγt pathway to suppress food allergy. Nat Med. (2019) 25(7):1164–74. doi: 10.1038/s41591-019-0461-z
40. Mennini M, Reddel S, Del Chierico F, Gardini S, Quagliariello A, Vernocchi P, et al. Gut microbiota profile in children with IgE-mediated cow’s milk allergy and cow’s milk sensitization and probiotic intestinal persistence evaluation. Int J Mol Sci. (2021) 22(4):1649. doi: 10.3390/ijms22041649
41. Lee KH, Guo J, Song Y, Ariff A, O’Sullivan M, Hales B, et al. Dysfunctional gut microbiome networks in childhood IgE-mediated food allergy. Int J Mol Sci. (2021) 22(4):2079. doi: 10.3390/ijms22042079
42. Inoue R, Sawai T, Sawai C, Nakatani M, Romero-Pérez GA, Ozeki M, et al. A preliminary study of gut dysbiosis in children with food allergy. Biosci Biotechnol Biochem. (2017) 81(12):2396–9. doi: 10.1080/09168451.2017.1383849
43. Chagoyan OC, Fallani M, Maldonado J, Vieites JM, Khanna S, Edwards C, et al. Faecal microbiota and short-chain fatty acid levels in faeces from infants with cow’s milk protein allergy. Int Arch Allergy Immunol. (2011) 156(3):325–32. doi: 10.1159/000323893
44. Joseph CL, Sitarik AR, Kim H, Huffnagle G, Fujimura K, Yong GJM, et al. Infant gut bacterial community composition and food-related manifestation of atopy in early childhood. Pediatr Allergy Immunol. (2022) 33(1):e13704. doi: 10.1111/pai.13704
45. Guo L, Bai H, Dong Y, Huang DX, Zhang X, Gong S, et al. Comparative analysis of fecal microbiota in 5-8-year-old children with and without cow milk protein allergy. Iran J Pediatr. (2016) 26(6):e6397. doi: 10.5812/jjp.6397
46. Yamagishi M, Akagawa S, Akagawa Y, Nakai Y, Yamanouchi S, Kimata T, et al. Decreased butyric acid-producing bacteria in gut microbiota of children with egg allergy. Allergy. (2021) 76(7):2279–82. doi: 10.1111/all.14795
47. Dong Y, Fei P, Han Y, Guo L. Characterization of fecal microbiota, short-chain fatty acids and lactic acid concentrations in 5-8-year-old children with cow milk protein allergy. Iran J Pediatr. (2018) 28(4):e64638. doi: 10.5812/ijp.64638
48. Martin VM, Virkud YV, Dahan E, Seay HL, Itzkovits D, Vlamakis H, et al. Longitudinal disease-associated gut microbiome differences in infants with food protein-induced allergic proctocolitis. Microbiome. (2022). doi: 10.1186/s40168-022-01322-y
49. Berni Canani R, De Filippis F, Nocerino R, Paparo L, Di Scala C, Cosenza L, et al. Gut microbiota composition and butyrate production in children affected by non-IgE-mediated cow’s milk allergy. Sci Rep. (2018) 8(1):12500. doi: 10.1038/s41598-018-30428-3
50. Su KW, Cetinbas M, Martin VM, Virkud YV, Seay H, Ndahayo R, et al. Early infancy dysbiosis in food protein-induced enterocolitis syndrome: a prospective cohort study. Allergy. (2023) 78(6):1595–604. doi: 10.1111/all.15644
51. Díaz M, Guadamuro L, Espinosa-Martos I, Mancabelli L, Jiménez S, Molinos-Norniella C, et al. Microbiota and derived parameters in fecal samples of infants with non-IgE cow’s milk protein allergy under a restricted diet. Nutrients. (2018) 10(10):1481. doi: 10.3390/nu10101481
52. Caparrós E, Cenit MC, Muriel J, Benítez-Páez A, Moreno MV, González-Delgado P, et al. Intestinal microbiota is modified in pediatric food protein-induced enterocolitis syndrome. J Allergy Clin Immunol Glob. (2022) 1(4):217–24. doi: 10.1016/j.jacig.2022.07.005
53. Kumagai H, Maisawa S, Tanaka M, Takahashi M, Takasago Y, Nishijima A, et al. Intestinal microbiota and secretory immunoglobulin A in feces of exclusively breast-fed infants with blood-streaked stools. Microbiol Immunol. (2012) 56(10):657–63. doi: 10.1111/j.1348-0421.2012.00487.x
54. Wang J, Zheng S, Yang X, Huazeng B, Cheng Q. Influences of non-IgE-mediated cow’s milk protein allergy-associated gut microbial dysbiosis on regulatory T cell-mediated intestinal immune tolerance and homeostasis. Microb Pathog. (2021) 158:105020. doi: 10.1016/j.micpath.2021.105020
55. Aparicio M, Cam Public Health Area PSGO, Alba C, Rodríguez JM, Fernández L. Microbiological and immunological markers in milk and infant feces for common gastrointestinal disorders: a pilot study. Nutrients. (2020) 12(3):634. doi: 10.3390/nu12030634
56. Ihekweazu FD, Versalovic J. Development of the pediatric gut microbiome: impact on health and disease. Am J Med Sci. (2018) 356(5):413–23. doi: 10.1016/j.amjms.2018.08.005
57. Shu SA, Yuen AWT, Woo E, Chu KH, Kwan HS, Yang GX, et al. Microbiota and food allergy. Clin Rev Allergy Immunol. (2019) 57(1):83–97. doi: 10.1007/s12016-018-8723-y
58. Wexler HM. Bacteroides: the good, the bad, and the nitty-gritty. Clin Microbiol Rev. (2007) 20(4):593–621. doi: 10.1128/CMR.00008-07
59. Hendrickx DM, An R, Boeren S, Mutte SK, Lambert JM, Belzer C. Assessment of infant outgrowth of cow’s milk allergy in relation to the faecal microbiome and metaproteome. Sci Rep. (2023) 13(1):12029. doi: 10.1038/s41598-023-39260-w
60. Hoskinson C, Dai DLY, Del Bel KL, Becker AB, Moraes TJ, Mandhane PJ, et al. Delayed gut microbiota maturation in the first year of life is a hallmark of pediatric allergic disease. Nat Commun. (2023) 14(1):4785. doi: 10.1038/s41467-023-40336-4
61. Gao Y, Stokholm J, O'Hely M, Ponsonby AL, Tang MLK, Ranganathan S, et al. Gut microbiota maturity mediates the protective effect of siblings on food allergy. J Allergy Clin Immunol. (2023) 152(3):667–75. doi: 10.1016/j.jaci.2023.02.034
62. Vacca M, Celano G, Calabrese FM, Portincasa P, Gobbetti M, De Angelis M. The controversial role of human gut Lachnospiraceae. Microorganisms. (2020) 8(4):573. doi: 10.3390/microorganisms8040573
63. Weiss GA, Hennet T. Mechanisms and consequences of intestinal dysbiosis. Cell Mol Life Sci. (2017) 74(16):2959–77. doi: 10.1007/s00018-017-2509-x
64. Levy M, Kolodziejczyk AA, Thaiss CA, Elinav E. Dysbiosis and the immune system. Nat Rev Immunol. (2017) 17(4):219–32. doi: 10.1038/nri.2017.7
65. Morrison DJ, Preston T. Formation of short chain fatty acids by the gut microbiota and their impact on human metabolism. Gut Microbes. (2016) 7(3):189–200. doi: 10.1080/19490976.2015.1134082
66. Wang G, Huang S, Wang Y, Cai S, Yu H, Liu H, et al. Bridging intestinal immunity and gut microbiota by metabolites. Cell Mol Life Sci. (2019) 76(20):3917–37. doi: 10.1007/s00018-019-03190-6
67. Mazmanian SK, Liu CH, Tzianabos AO, Kasper DL. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell. (2005) 122(1):107–18. doi: 10.1016/j.cell.2005.05.007
68. Ohnmacht C, Park JH, Cording S, Wing JB, Atarashi K, Obata Y, et al. The microbiota regulates type 2 immunity through RORγt+ T cells. Science. (2015) 349(6251):989–93. doi: 10.1126/science.aac4263
69. Chiu CY, Cheng ML, Chiang MH, Kuo YL, Tsai MH, Chiu CC, et al. Gut microbial-derived butyrate is inversely associated with IgE responses to allergens in childhood asthma. Pediatr Allergy Immunol. (2019) 30(7):689–97. doi: 10.1111/pai.13096
70. Macia L, Nanan R, Hosseini-Beheshti E, Grau GE. Host- and microbiota-derived extracellular vesicles, immune function, and disease development. Int J Mol Sci. (2019) 21(1):107. doi: 10.3390/ijms21010107
71. Kemter AM, Patry RT, Arnold J, Hesser LA, Campbell E, Ionescu E, et al. Commensal bacteria signal through TLR5 and AhR to improve barrier integrity and prevent allergic responses to food. Cell Rep. (2023) 42(10):113153. doi: 10.1016/j.celrep.2023.113153
72. Keir M, Yi Y, Lu T, Ghilardi N. The role of IL-22 in intestinal health and disease. J Exp Med. (2020) 217(3):e20192195. doi: 10.1084/jem.20192195
73. Campbell C, McKenney PT, Konstantinovsky D, Isaeva OI, Schizas M, Verter J, et al. Bacterial metabolism of bile acids promotes generation of peripheral regulatory T cells. Nature. (2020) 581(7809):475–9. doi: 10.1038/s41586-020-2193-0
74. Crestani E, Harb H, Charbonnier LM, Leirer J, Motsinger-Reif A, Rachid R, et al. Untargeted metabolomic profiling identifies disease-specific signatures in food allergy and asthma. J Allergy Clin Immunol. (2020) 145(3):897–906. doi: 10.1016/j.jaci.2019.10.014
75. Mohr AE, Crawford M, Jasbi P, Fessler S, Sweazea KL. Lipopolysaccharide and the gut microbiota: considering structural variation. FEBS Lett. (2022) 596(7):849–75. doi: 10.1002/1873-3468.14328
76. Di Lorenzo F, Paparo L, Pisapia L, Oglio F, Pither MD, Cirella R, et al. The chemistry of gut microbiome-derived lipopolysaccharides impacts on the occurrence of food allergy in the pediatric age. Front Mol Biosci. (2023) 13(10):1266293. doi: 10.3389/fmolb.2023.1266293
77. Di Lorenzo F, De Castro C, Silipo A, Molinaro A. Lipopolysaccharide structures of gram-negative populations in the gut microbiota and effects on host interactions. FEMS Microbiol Rev. (2019) 43(3):257–72. doi: 10.1093/femsre/fuz002
78. Garcia-Vello P, Di Lorenzo F, Zucchetta D, Zamyatina A, De Castro C, Molinaro A. Lipopolysaccharide lipid A: a promising molecule for new immunity-based therapies and antibiotics. Pharmacol Ther. (2022) 230:107970. doi: 10.1016/j.pharmthera.2021.107970
79. Di Lorenzo F, Pither MD, Martufi M, Scarinci I, Guzmán-Caldentey J, Łakomiec E, et al. Pairing Bacteroides vulgatus LPS structure with its immunomodulatory effects on human cellular models. ACS Cent Sci. (2020) 6(9):1602–16. doi: 10.1021/acscentsci.0c00791
80. Erturk-Hasdemir D, Oh SF, Okan NA, Stefanetti G, Gazzaniga FS, Seeberger PH, et al. Symbionts exploit complex signaling to educate the immune system. Proc Natl Acad Sci U S A. (2019) 116(52):26157–66. doi: 10.1073/pnas.1915978116
81. Atarashi K, Tanoue T, Oshima K, Suda W, Nagano Y, Nishikawa H, et al. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature. (2013) 500(7461):232–6. doi: 10.1038/nature12331
82. Feehley T, Plunkett CH, Bao R, Choi Hong SM, Culleen E, Belda-Ferre P, et al. Healthy infants harbor intestinal bacteria that protect against food allergy. Nat Med. (2019) 25(3):448–53. doi: 10.1038/s41591-018-0324-z
83. Celebi Sozener Z, Ozdel Ozturk B, Cerci P, Turk M, Gorgulu Akin B, Akdis M, et al. Epithelial barrier hypothesis: effect of the external exposome on the microbiome and epithelial barriers in allergic disease. Allergy. (2022) 77(5):1418–49. doi: 10.1111/all.15240
84. Venter C. Immunonutrition: diet diversity, gut microbiome and prevention of allergic diseases. Allergy Asthma Immunol Res. (2023) 15(5):545–61. doi: 10.4168/aair.2023.15.5.545
85. Vich Vila A, Collij V, Sanna S, Sinha T, Imhann F, Bourgonje AR, et al. Impact of commonly used drugs on the composition and metabolic function of the gut microbiota. Nat Commun. (2020) 11(1):362. doi: 10.1038/s41467-019-14177-z
86. Jordakieva G, Kundi M, Untersmayr E, Pali-Schöll I, Reichardt B, Jensen-Jarolim E. Country-wide medical records infer increased allergy risk of gastric acid inhibition. Nat Commun. (2019) 10(1):3298. doi: 10.1038/s41467-019-10914-6
87. Dominguez-Bello MG, Costello EK, Contreras M, Magris M, Hidalgo G, Fierer N, et al. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci U S A. (2010) 107(26):11971–5. doi: 10.1073/pnas.1002601107
88. Rodríguez JM, Murphy K, Stanton C, Ross RP, Kober OI, Juge N, et al. The composition of the gut microbiota throughout life, with an emphasis on early life. Microb Ecol Health Dis. (2015) 26:26050. doi: 10.3402/mehd.v26.26050
89. Dethlefsen L, Relman DA. Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Proc Natl Acad Sci U S A. (2011) 108(Suppl 1):4554–61. doi: 10.1073/pnas.1000087107
90. De La Cochetière MF, Durand T, Lepage P, Bourreille A, Galmiche JP, Doré J. Resilience of the dominant human fecal microbiota upon short-course antibiotic challenge. J Clin Microbiol. (2005) 43(11):5588–92. doi: 10.1128/JCM.43.11.5588-5592.2005
91. Dethlefsen L, Huse S, Sogin ML, Relman DA. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol. (2008) 6(11):e280. doi: 10.1371/journal.pbio.0060280
92. Jernberg C, Löfmark S, Edlund C, Jansson JK. Long-term ecological impacts of antibiotic administration on the human intestinal microbiota. ISME J. (2007) 1(1):56–66. doi: 10.1038/ismej.2007.3
93. Bunyavanich S, Berin MC. Food allergy and the microbiome: current understandings and future directions. J Allergy Clin Immunol. (2019) 144(6):1468–77. doi: 10.1016/j.jaci.2019.10.019
94. Di Costanzo M, Carucci L, Berni Canani R, Biasucci G. Gut microbiome modulation for preventing and treating pediatric food allergies. Int J Mol Sci. (2020) 21(15):5275. doi: 10.3390/ijms21155275
95. Berni Canani R, Nocerino R, Terrin G, Frediani T, Lucarelli S, Cosenza L, et al. Formula selection for management of children with cow’s milk allergy influences the rate of acquisition of tolerance: a prospective multicenter study. J Pediatr. (2013) 163(3):771–7.e1. doi: 10.1016/j.jpeds.2013.03.008
96. Berni Canani R, Di Costanzo M, Bedogni G, Amoroso A, Cosenza L, Di Scala C, et al. Extensively hydrolyzed casein formula containing Lactobacillus rhamnosus GG reduces the occurrence of other allergic manifestations in children with cow’s milk allergy: 3-year randomized controlled trial. J Allergy Clin Immunol. (2017) 139(6):1906–13.e4. doi: 10.1016/j.jaci.2016.10.050
97. Blázquez AB, Berin MC. Microbiome and food allergy. Transl Res. (2017) 179:199–203. doi: 10.1016/j.trsl.2016.09.003
98. Tan W, Zhou Z, Li W, Lu H, Qiu Z. Lactobacillus rhamnosus GG for cow’s milk allergy in children: a systematic review and meta-analysis. Front Pediatr. (2021) 22(9):727127. doi: 10.3389/fped.2021.727127
99. Berni Canani R, Nocerino R, Terrin G, Coruzzo A, Cosenza L, Leone L, et al. Effect of Lactobacillus GG on tolerance acquisition in infants with cow’s milk allergy: a randomized trial. J Allergy Clin Immunol. (2012) 129:580–2.e5. doi: 10.1016/j.jaci.2011.10.004
100. Nocerino R, Di Costanzo M, Bedogni G, Cosenza L, Maddalena Y, Di Scala C, et al. Dietary treatment with extensively hydrolyzed casein formula containing the probiotic Lactobacillus rhamnosus GG prevents the occurrence of functional gastrointestinal disorders in children with cow’s milk allergy. J Pediatr. (2019) 213:137–42. doi: 10.1016/j.jpeds.2019.06.004
101. Nocerino R, Bedogni G, Carucci L, Cosenza L, Cozzolino T, Paparo L, et al. The impact of formula choice for the management of pediatric cow’s milk allergy on the occurrence of other allergic manifestations: the atopic march cohort study. J Pediatr. (2021) 232:183–91.e3. doi: 10.1016/j.jpeds.2021.01.059
102. Fiocchi A, Schunemann H, Ansotegui I, Assa’ad A, Bahna S, Canani RB, et al. The global impact of the DRACMA guidelines cow’s milk allergy clinical practice. World Allergy Organ J. (2018) 11:2. doi: 10.1186/s40413-017-0179-7
103. Bilaver LA, Chadha AS, Doshi P, O’Dwyer L, Gupta RS. Economic burden of food allergy: a systematic review. Ann Allergy Asthma Immunol. (2019) 122:373–80.e1. doi: 10.1016/j.anai.2019.01.014
104. Ovcinnikova O, Panca M, Guest JF. Cost-effectiveness of using an extensively hydrolyzed casein formula plus the probiotic Lactobacillus rhamnosus GG compared to an extensively hydrolyzed formula alone or an amino acid formula as first-line dietary management for cow’s milk allergy in the US. Clinicoecon Outcomes Res. (2015) 7:145–52. doi: 10.2147/CEOR.S75071
105. Guest JF, Singh H. Cost-effectiveness of using an extensively hydrolyzed casein formula supplemented with Lactobacillus rhamnosus GG in managing IgE-mediated cow’s milk protein allergy in the UK. Curr Med Res Opin. (2019) 35:1677–85. doi: 10.1080/03007995.2019.1612339
106. Guest JF, Weidlich D, Mascuñan Díaz JI, Díaz JJ, Ojeda PM, Ferrer-González JP, et al. Relative cost effectiveness of using an extensively hydrolyzed casein formula containing the probiotic Lactobacillus rhamnosus GG in managing infants with cow’s milk allergy in Spain. Clinicoecon Outcomes Res. (2015) 7:583–91. doi: 10.2147/CEOR.S89347
107. Santos SCD, Konstantyner T, Cocco RR. Effects of probiotics in the treatment of food hypersensitivity in children: a systematic review. Allergol Immunopathol (Madr). (2020) 48(1):95–104. doi: 10.1016/j.aller.2019.04.009
108. Osborn DA, Sinn JK. Probiotics in infants for prevention of allergic disease and food hypersensitivity. Cochrane Database Syst Rev. (2007) 17(4):CD006475. doi: 10.1002/14651858
109. Berni Canani R, Gilbert JA, Nagler CR. The role of the commensal microbiota in the regulation of tolerance to dietary allergens. Curr Opin Allergy Clin Immunol. (2015) 15(3):243–9. doi: 10.1097/ACI.0000000000000157
110. Augustine T, Kumar M, Al Khodor S, van Panhuys N. Microbial dysbiosis tunes the immune response towards allergic disease outcomes. Clin Rev Allergy Immunol. (2023) 65(1):43–71. doi: 10.1007/s12016-022-08939-9
111. Seed PC. The human mycobiome. Cold Spring Harb Perspect Med. (2014) 5(5):a019810. doi: 10.1101/cshperspect.a019810
Keywords: microbiota, short-chain fatty acids, probiotics, allergy, dysbiosis, immune tolerance, cow milk protein allergy, children
Citation: Farnetano M, Carucci L, Coppola S, Oglio F, Masino A, Cozzolino M, Nocerino R and Berni Canani R (2024) Gut microbiome features in pediatric food allergy: a scoping review. Front. Allergy 5:1438252. doi: 10.3389/falgy.2024.1438252
Received: 25 May 2024; Accepted: 13 August 2024;
Published: 25 September 2024.
Edited by:
Ronald van Ree, Amsterdam University Medical Center, NetherlandsReviewed by:
Maurizio Mennini, Sapienza University of Rome, ItalyIsabella Pali-Schöll, University of Veterinary Medicine Vienna, Austria
Copyright: © 2024 Farnetano, Carucci, Coppola, Oglio, Masino, Cozzolino, Nocerino and Berni Canani. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Roberto Berni Canani, YmVybmlAdW5pbmEuaXQ=
†These authors have contributed equally to this work
 Margherita Farnetano
Margherita Farnetano Laura Carucci
Laura Carucci Serena Coppola1,2
Serena Coppola1,2 Franca Oglio
Franca Oglio Antonio Masino
Antonio Masino Rita Nocerino
Rita Nocerino Roberto Berni Canani
Roberto Berni Canani