- 1Department of Molecular Biology and Biotechnology, Pan African University Institute for Basic Sciences, Technology and Innovation (PAUSTI), Nairobi, Kenya
- 2Department of Horticulture and Food Security, Jomo Kenyatta University of Agriculture and Technology (JKUAT), Nairobi, Kenya
- 3Department of Botany, Jomo Kenyatta University of Agriculture and Technology (JKUAT), Nairobi, Kenya
Bambara groundnut (Vigna subterranean) is a resilient leguminous crop that can withstand drought conditions and is commonly grown in arid regions where soils are degraded and of low fertility status. Although the crop can fix nitrogen, its yields frequently fall short of their maximum potential which can be attributed to association with ineffective rhizobia strains. In the present study, we isolated, screened, and in-vitro characterized Bradyrhizobium isolates from the root nodules of Bambara groundnuts with plant growth promoting properties for potential use as bioinoculant. Root nodules were sampled from JKUAT farm where healthy Bambara groundnut plants were growing. Ten isolates with slow growth rate were isolated. The 10 isolates were screened using morphological, biochemical and molecular (16S rRNA gene sequencing) techniques. The sequence analysis established that all isolates were congeneric with Bradyrhizobium. In addition, all the isolates showed Nitrogen-fixing potentials and also had significant (p < 0.005) ability to solubilize phosphate in the range of 0.77 ± 0.771–3.22 ± 0.368 phosphate solubilizing index. Further, isolates P4A17, P4A18, P4A16, P4A6 and C2 produced IAA concentrations of 54.97 ± 3.21–108 ± 12.10 μg/mL. However, none of the isolates could produce HCN. The isolates were assessed further on their ability to grow in varied physiological conditions. Among them, P4A6 and P4A18 demonstrated higher growth potential under pH 3, pH 5, pH 9, pH 11, 1% NaCl, 3% NaCl, 5% NaCl and high temperature range of 40°C–50°C than the rest of the isolates. Given the in-vitro results, these isolates are promising bioinoculant (biofertilizer) candidates and, ought to be further tested in greenhouse and field conditions for Bambara groundnut production.
1 Introduction
Bambara groundnut (BGN) (Vigna subterranean) is a legume in the same genera as cowpea (Vigna unguiculata) (Swanevelder, 1998). According to multiple reports, the crop is thought to have originated from West Africa, mainly the northeastern region of Nigeria (Mayes et al., 2019), but has been cultivated across Africa and certain areas of Asia (Singh and Basu, 2016). Subsistence farmers primarily grow the plant for its pods, which offer a nutritionally balanced meal containing carbohydrates, protein, and minerals, and more recently as a cash crop in western Kenya. In addition, Bambara groundnut is a hardy crop which withstands drought and flourishes in hot and arid regions, thereby earning the status of a climate-adapted crop which can play a role in diversifying global food systems. Unfortunately, the crop yields in small holder farms frequently fall below the expected yields (Khan et al., 2020). The low yields can be partly linked to a decline in soil fertility, high cost of commercial inputs, intensification of agriculture, and crucially, lack of improved varieties.
Legumes provide almost 20 million tons of nitrogen (N) to agriculture each year through nitrogen fixation (Herridge et al., 2008). Nitrogen fixation may only occur when effective rhizobia strains are present, whether naturally occurring in the soil, or imported as commercial inoculants.
Both rapid and slow-growing rhizobia have successfully established associations with Bambara groundnut resulting in symbiotic nitrogen fixation (Jaiswal and Dakora, 2019). Bradyrhizobium manausense, Bradyrhizobium japonicum, Bradyrhizobium elkanii, Bradyrhizobium subterraneum, and Bradyrhizobium vignae are among the Bradyrhizobium strains that nodulate Bambara groundnut (Grönemeyer et al., 2016). The mechanism of symbiotic N-fixation can be utilized by man in the development of biofertilizers which are cost effective and environmentally friendly compared to commercially available N-fertilizers. In this regard, the significant impact of cost-effective and environmentally friendly biofertilizers on promoting plant growth, and boosting crop productivity cannot be underestimated especially in the face of threats to global food security (Fasusi et al., 2021). Therefore, efficient rhizobia as an inoculant to legumes can lower the cost of production and enhance grain production (Matiru and Dakora, 2004). However, the widespread practice of inoculation remains limited, and its efficacy in eliciting favorable outcomes remain uncertain. The use of rhizobia inoculants has however been successful for various legumes, including Bambara groundnut (Adjei et al., 2022). These have shown the potential of this option to increase Bambara groundnut production in Kenya and other growing areas.
To enhance the inoculation success of tropical pulses, Mathu et al. (2012) proposed utilizing native rhizobia as inoculants. Moreover, efficient rhizobia are abundantly available in native soils and may provide a source of superior strains for local inoculant production (Mathu et al., 2012). Given the diverse rhizobia species that form nodules on legumes, it is crucial to consistently investigate new growing regions to determine native Bradyrhizobium with efficiency in forming nodules and enhancing the growth of underutilized crop species like Bambara groundnut. Therefore, this work aims to identify, screen, and in-vitro characterize Bradyrhizobium isolates from the root nodules of Bambara groundnut to identify those with high plant growth promoting properties for potential use as bioinoculants.
2 Materials and methods
2.1 Samples source
Root nodules were sampled from Jomo Kenyatta University of Agriculture and Technology (JKUAT) farm on which Bambara groundnuts (BGN) were grown. This farm has deep clay soils (vertisols) and had no history of commercial rhizobia inoculation. The coordinates of the field are, −1.090806, 37.012611; −1.090757, 37.012697; −1.090857, 37.012654 and −1.090803, 37.012721.
2.2 Sampling
Root samples were collected from different plots when the Bambara groundnut crop (variety: IITA-1472; Black seeds with white spotted eyes) was at the flowering stage of growth. Root samples were collected at five (5) sampling points in each plot following a procedure reported by Barillot et al. (2013). Using a sterile hand shovel and a cutting tool, BGN roots were dug out while ensuring that the root system remained whole, and thereafter the roots were shaken to remove the bulk soil (non-rhizosphere soil). The sampling was done at a depth of 5–40 cm to ensure that all the roots were collected. Most sampled roots had a length of 22–35 cm. Samples from the field were stored in sterile zip lock bags in an ice box and transported to the laboratory where they were put in storage at 4°C prior to isolation of the endophytic bacteria. Samples were labeled according to the plot number from which the samples were taken. Thus P1, P2, P3, P4 refers to Plot 1, Plot 2, Plot 3 and Plot 4, respectively.
2.3 Isolation of bacteria from root nodules
Isolation of rhizobacteria was conducted according to the protocol of Somasegaran and Hoben (1994), whereby each root was carefully washed under tap water to get rid of soil particles and the root nodules captured using a 5 mm sieve. Intact nodules were thereafter gently rinsed and then air dried under the laminar flow hood (LabTech DAIHAN). Detached root nodules appeared pinkish, and these were dipped in 70% (v/v) ethanol for 10 s to surface sterilize and then immersed in 10% (v/v) sodium hypochlorite (JIK) for four (4) minutes. They were subsequently rinsed with sterile distilled water (six times) to remove traces of sodium hypochlorite. Thereafter, a droplet of sterile distilled water was applied to the surface of sterile glass slide and then, the nodules (one or more depending on the size) were ground with the aid of a blunt sterile glass rod. A loopful of exudates from the nodules were streaked directly onto plates containing yeast extract mannitol agar (YEMA) supplemented with Congo red (Vincent, 1970). After inoculation, the plates were well labeled with the sample codes and wrapped with parafilm and inverted. The plates were enclosed in aluminum foil to create darkness which is required for growth, thereafter, the plates were incubated at 28°C ± 2°C for five (5) to fifteen (15) days with daily observations. Growth attributes were recorded during daily observations. After growth was observed over the period, the isolated colonies (≤2 mm) from each plate were repeatedly sub-cultured to get pure colonies. Isolates were labeled based on the plot numbers as well as the plant from which the isolate was obtained. Thus, P4A6 indicates Plot 4 plant A isolate 6.
2.4 Confirmation and preservation of Bradyrhizobium isolates
Bromothymol Blue (BTB) as an indicator was used to distinguish bacteria by assessing their capacity to metabolize carbohydrates and produce acidic byproducts (Al-Hossainy et al., 2021). Isolates were streaked on BTB supplemented YEMA plates and incubated at 28°C ± 2°C for 2–10 days while monitoring the color changes of the media. The isolates that were positive were expected to retain a blue color and others to remain unchanged (green) on the BTB agar medium, whereas the negative isolates were expected to turn yellow as a result of acid production by the fast-growing Rhizobium sp. (Dinkwar et al., 2020). These isolates were subsequently inoculated into YEM broth and allowed to incubate 28°C ± 2°C for 5–10 days. Thereafter, growth of isolates was confirmed by observing the turbidity of the broth in comparison to an uninoculated broth. The purified isolates were preserved in 20% glycerol (v/v) and YEM broth in a ratio of 3:7, respectively. To avoid freeze shocking the bacteria, they were put in 4°C for 3 days to acclimatize to the cold conditions before being transferred to −20°C for long term preservation.
2.5 Morphological and biochemical characterization
The colony morphologies were observed based on form, elevation, margin, size, texture, surface, and opacity, with reference to standard microbiological techniques (Fayaz et al., 2022). These characteristics were used to aid in characterizing the isolates and narrowing down the number of isolates in the process. Further, Gram staining was performed according to Smith and Hussey (2005). The slides were observed under a light microscope with a magnification of ×100 (OPTIKA light microscope). Red or pink colored isolates were recorded as Gram negative.
2.5.1 Biochemical characterization
Biochemical tests (oxidase, catalase and citrate test) were performed following the procedures described by Abiola and Oyetayo (2016). Thus, for the oxidase test, a sterile loop was used to select a colony of bacteria and evenly spread over the oxidase disc. The reactions were observed within 5–10 s at 28°C. A deep purple coloration was seen from oxidase positive bacteria while no color change indicated negative. With the catalase test, a loopful of isolates from well-isolated colonies were put on microscope slides. A droplet of 3% H2O2 solution was put on the colonies on the slide and then covered with a lid. Gas bubbles were observed as white foam which indicated the presence of catalase enzyme and hence positive. To assesses the capacity of bacteria to utilize citrate, Simon’s citrate agar (HiMEDIA-M099) was prepared based on the guidelines of the manufacturer. Thereafter, they were poured into glass tubes and autoclaved at 121°C for 15 min, then tilted and allowed to cool and solidify. The glass tubes were inoculated by single surface streaking, and incubated at 28°C for seven (7) days; observations were made on color changes, where green to blue indicated use of the citrate, hence positive.
2.6 Molecular characterization based on 16S rRNA sequencing
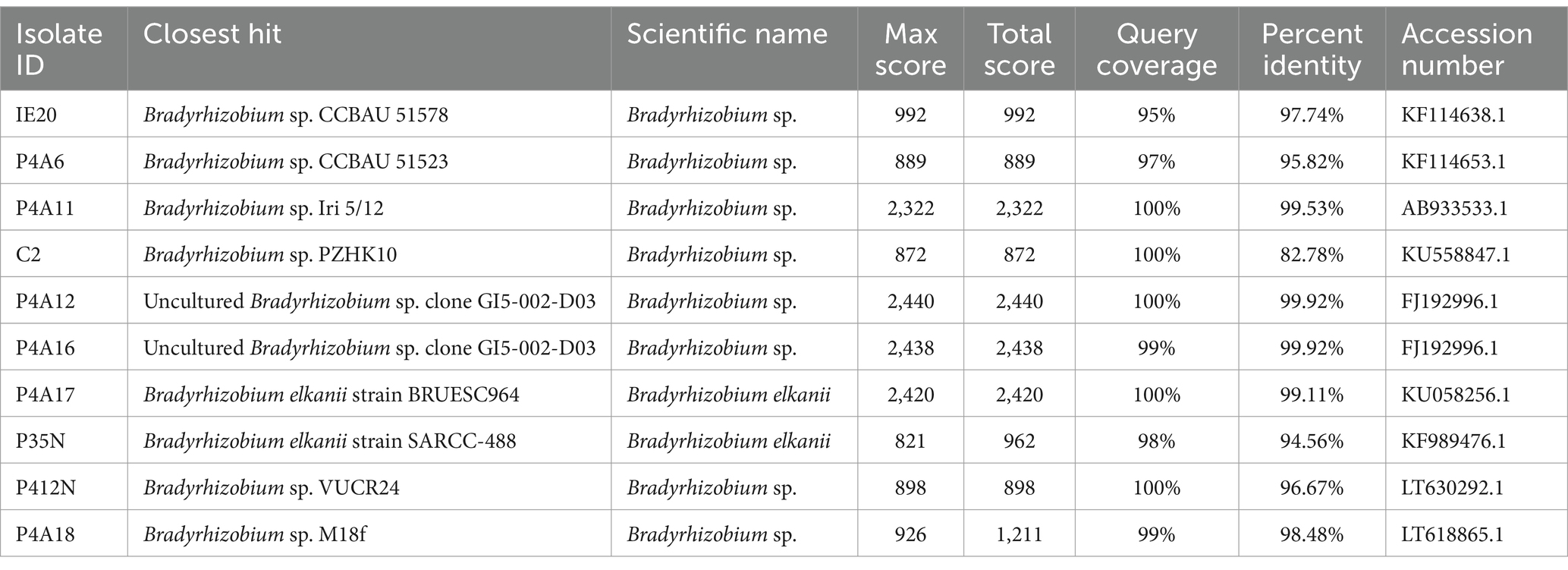
DNA was extracted from pure colonies of suspected Bradyrhizobium isolates using the Quick-DNA Fungal/Bacterial Miniprep kit (D6005) as per ZYMO Research’s protocol. A nanodrop spectrophotometer (JENWAY Genova Nano) was used to check the concentration (17.086–111.53 ng/μL), purity, and contamination of the genomic DNA. The 16S rRNA gene was targeted and amplified using Polymerase Chain Reaction (PCR) in a thermocycler (ProFlex Base PCR system) with universal primers; fD1 (27F) (AGAGTTTGA TCMTGGCTCAG) and rP3 (1492R) (TACGGYTACCTTGTTA CGACTT) (Issifu et al., 2022). The volume of the PCR reaction mixture for amplifying the 16S rRNA was 50 μL. It composed of 2 μL of the purified bacterial DNA (17.086–111.53 ng/μL) as the template, One Taq 2x Master mix 25 μL, forward primer 1 μL (1 μM), reverse primer 1 μL (1 μM), and nuclease free water 21 μL. The master mix and the primers were used as negative control, while genomic DNA from Lactoccocus. spp. was the positive control. The cycling reaction was performed using the ProFlex Base PCR system set up as; initial denaturation at 95°C for 5 min, then 35 cycles of denaturation at 95°C for 3 min, annealing at 57°C for 30 s and extension 72°C at 1 and half minutes, lastly a final extension of 72°C for 5 min (Marinkovic et al., 2017). After the reaction was complete, the PCR products were loaded and run on a 1% (w/v) agarose gel in 1X TAE buffer after staining with SafeView (ABM G108) alongside a one (1) kilo base pair ladder (N3232S). The PCR amplicons were checked by gel electrophoresis, purified, labeled accordingly, and shipped to Macrogen Asia Pacific Pte. Ltd. for sequencing. The DNA sequence data from Macrogen Asia Pacific Pte. Ltd. were cleaned to remove background “noise” and edited using Snapgene and BioEdit. These sequences were uploaded unto Basic Local Alignment Search Tool (BLASTn) on the National Centre for Biotechnology Information (NCBI) website1 to identify them. Sequences with the highest percentage identity, good query coverage and score from the BLASTn results were then selected and retrieved for multiple sequence alignment using MUSCLE alignment in the MEGA 11. The aligned sequences were used to perform phylogenetic analysis, using MEGA11 to construct a cladogram based on the neighbor-joining method with a bootstrap of 1,000 to ensure a good number of replication (Tamura et al., 2021). Consensus sequences of 540–1,355 bp of the 16S-rRNA partial sequences were used to construct a Neighbor-Joining (NJ) phylogenetic tree together with top hits from the BLASTn output (Table 1). The sequence groups were categorized into two (2) key clades based on their clustering with strains from the NCBI database. The evolutionary changes over time were calculated based on the p-distance method, expressed in terms of the number of base changes per site.
2.7 In-vitro evaluation of Bradyrhizobium species for N-fixing and plant-beneficial characteristics
The Bradyrhizobium isolates were evaluated for N-fixing potential using NFb medium (Baldani et al., 2014). An inoculation loop was used to pick a single colony from plates containing purified colonies. This was spot inoculated in the media and incubated at 28°C for 5–10 days. Successful N-fixation was centered on growth of the bacteria isolates on the NFb media and a visible color change of the media from green to blue. The growth of bacterial isolates on the agar plates containing NFb medium was described as stated by Issifu et al. (2022). A non-inoculated medium was used as the negative control.
The solubilization ability of phosphate (P) by the Bradyrhizobium isolates was tested using Pikovskaya’s (PVK) agar medium in replicates (Ajilogba et al., 2022). P-solubilization (PS) was detected as a formation of clear zones surrounding colonies with the ability. The size of the halos and the colonies were measured (cm), and used to infer the P-solubilizing ability of isolates based on the phosphate solubilization index (PSI). This was given as the ratio of the total diameter (cm) (zone + colony) to the colony diameter (cm) (Chaudhary et al., 2021). Observations were also recorded as Positive (+) if a halo was formed and Negative (−) if there was no halo. A non-inoculated medium was the control.
Where: PSI = phosphate solubilization index, Dc = Colony diameter, Dhz = Halo zone diameter
To determine HCN production by the isolates, the method of Issifu et al. (2022) was used with a slight modification (Yeast Extract Mannitol Agar was used in place of Nutrient agar).
To evaluate the capacity of the isolates to synthesize indole-3-acetic acid (IAA) both qualitatively and quantitatively, Tryptone broth (HiMEDIA-M463) was used as the growth media. A loopful of colonies were inoculated into each tube containing broth. The tubes were incubated at 28°C for 5–10 days. To detect production of IAA, Kovac’s reagent was added directly into each tube to cover the top of the Tryptone broth and these tubes were incubated in the dark for 1–2 h at 28°C. The formation of a pink layer on the top of the Tryptone broth after incubation indicated positive (+) for IAA, but Negative (−) if there was no detected IAA. An un-inoculated medium served as the negative control. To measure the quantity of IAA produced, 1 mL of broth was taken into sterile 1.5 mL micro-centrifuge tubes and then centrifuged at 10,000 rpm for ten (10) minutes, afterwards the supernatants were pipetted. Standard concentrations of 1–100 μg/mL was made using standard IAA (LOBA CHEMIE), 200 μL of the standard concentrations as well as the supernatant from the centrifugation were pipetted into a sterile 96 well plate in triplicates. A volume of 100 μL of Kovac’s reagent (HiMEDIA) was pipetted into each well, then the plates were placed in the dark to incubate for 1–2 h at 28°C before the optical densities (ODs) (0.102, 0.107, 0.102, 0.111, 0.105, 0.122, 0.137, 0.123, 0.155 and 0.202) were read using a microplate reader (BioTek ELX 800) at an absorbance of 630 nm. The ODs and the known concentrations were used to plot a standard curve which was used to compute the concentrations produced by the various isolates.
2.8 Growth response of Bradyrhizobium species under various physiological conditions (salinity, temperature and pH)
The capability of Bradyrhizobium cultures to propagate and be viable in different physiological conditions (Salinity, temperature and pH) was evaluated by inoculating bacterial isolates into YEMA media containing 1, 3, 5 and 10% of NaCl, with 0.001% NaCl as the control, then also at incubation temperatures of 25°C–30°C and 40°C–50°C, and finally pH conditions of 3.0, 5.0, 9.0 and 11.0 using 0.1 M HCl for acid and 0.1 M NaOH for adjustments. The petri dishes were incubated at 28°C for 5–10 days. Growth of bacterial isolates under the different conditions were qualitatively documented as Maximum growth (+++), Moderate growth (++), Minimum/low growth (+), and No growth (−).
2.9 Evaluating the effectiveness of Bradyrhizobium strains (P4A6 and P4A18) for nodules formation
Two isolates (P4A6 and P4A18) were selected for authentication based on their N-fixing and PGP abilities, as well as their growth potential under harsh physiological conditions. Based on the data, all potential isolates were ranked and the topmost 2 were selected. Importantly, the most effective strains have a better potential to improve crop productivity, hence selection of the two strains. The isolates were revived in Yeast Extract Mannitol (YEM) broth for 10 days. Selected Bambara groundnut seeds (IITA-1472) were washed with sterile water and surfaced sterilized with 3.5% sodium hypochlorite, and later coated with the respective inoculum using Arabic gum as an adhesive (0.5 g of gum: 5 mL broth). Thereafter, the treated seeds were allowed to air dry under before planting. To ascertain the effect of the treatments on the crops, a nutrient limiting soil type (red soil) was used. This was further divided into sterile (autoclaved continuously for 6 h to get rid of living organisms) and non-sterile soil; this aided in understanding the competiveness of the isolates in the presence of other soil microbes. Three healthy growing plants from each treatment were gently uprooted after 60 days of planting, and the nodules data was recorded. The treatments were as follows: No inoculation, Commercial inoculant (formulated in powdered form; thus the carrier material was a coarse matter mixed with the bacteria), P4A18 and P4A6 (both isolates were in broth but were mixed with Arabic cum [0.5 g of gum in 5 mL broth] for adhesion to the seed). All treatments were in replicates.
2.10 Data analysis
GenStat version 18 was used to compute the means of the amount of IAA produced as well as the PSI. One-way ANOVA was performed and the means separation was done using Fisher protected LSD. Heatmaps were generated using R statistical software version 4.4.1.
2.11 Accession numbers
Bradyrhizobium isolates identified from this study have been deposited in NCBI GenBank under the following accession numbers: PQ269135, PQ269136, PQ269137, PQ269138, PQ412527, PQ269139, PQ269140, PQ269141, PQ269142 and PQ269143.
3 Results
3.1 Bacteria isolated from root nodules
A total of one hundred and ninety-nine (199) bacteria isolates were obtained from the root samples. A large proportion (95%) of the isolates were fast-growing and turned the YEMA-BTB media yellow within 3–5 days of incubation. These were noted as possible Rhizobium and other bacteria, and hence were excluded from the selection. Ten isolates were slow-growing on YEMA/BTB media and were presumptively identified as Bradyrhizobium. These isolates include: P4A6, P4A16, P4A18, C2, IE20, P4A11, P4A12, P4A17, P412N and P35N. These were subsequently characterized and evaluated in-vitro for their plant growth-promoting (PGP) capabilities.
3.2 Morphological characteristics and differences among isolates based on acid/alkaline producing abilities
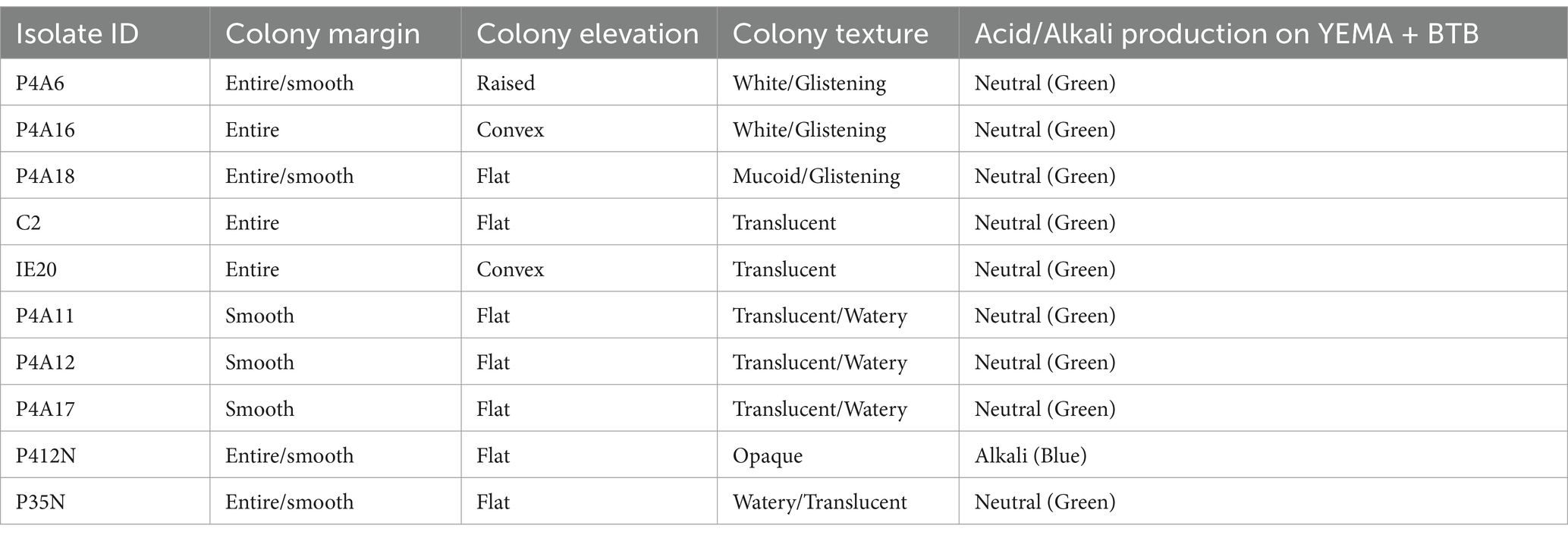
The bacteria isolates showed differences in their morphological characteristics. Specifically, the colony elevation and the colony texture varied between isolates. Three (3) isolates were raised and formed a convex shape while seven (7) remained flat on the growth media. Regarding the colony texture, six (6) were translucent with only one (1) being mucoid (Table 2). However, all isolates were small with smooth, entire and circular colonies. The cellular characterization based on gram staining revealed that the isolates were gram negative: they appeared pink when viewed under the microscope. None of the isolates absorbed Congo red dye upon incubation in darkness for 5–15 days. The slow growing isolates (5–15 days to grow) maintained the green color of the media, while some increased the pH and turned the YEMA-BTB media blue, hence the 10 isolates were noted as Bradyrhizobium.
3.3 Biochemical characteristics
The results obtained from the biochemical tests are presented on Figure 1. All isolates were positive for both the oxidase and catalase tests. Two isolates (P35N and P412N) proved to use citrate as a carbon source, while the remaining eight (8) were negative for citrate utilization.
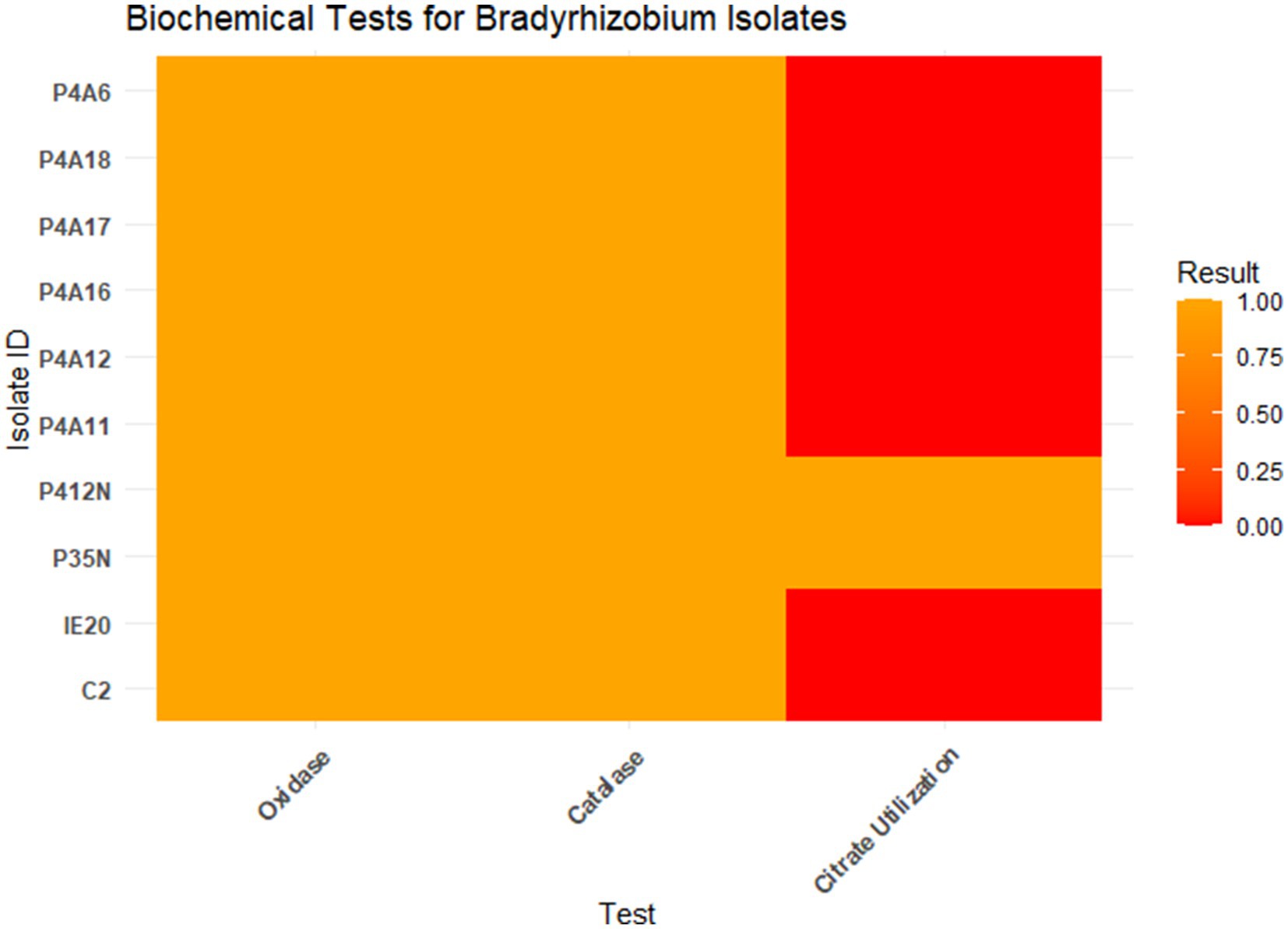
Figure 1. Biochemical reactions of the various Bradyrhizobium isolates. Orange = Positive test and Red = Negative test.
3.4 Molecular characteristics of isolates determined using 16S rRNA sequencing
The DNA sequence based on 16S-rRNA matched the morphological characterization. The 16S rRNA genes of each of the 10 Bambara groundnut bacteria was amplified by the PCR step, this yielded band sizes of about 1,465 base pairs (bp) before sequencing. Based on homology search against existing sequences in NCBI database it was revealed that, all the isolates were confirmed as Bradyrhizobium species (Table 1). Specifically, the percentage of identity to the closest species ranged from 82.78 to 99.92% (Table 1).
3.4.1 Phylogenetics of Bradyrhizobium isolates
All the isolates from this study clustered together in clade 1. From this, P412N stood out as a single subclade (Figure 2). However, this isolate had a similarity of 96.67% with Bradyrhizobium reference strain (Table 1). The evolutionary relations of the isolates as established by means of the Neighbor-Joining method. The lengths of the branches share units (0.05) with the established evolutionary distances.
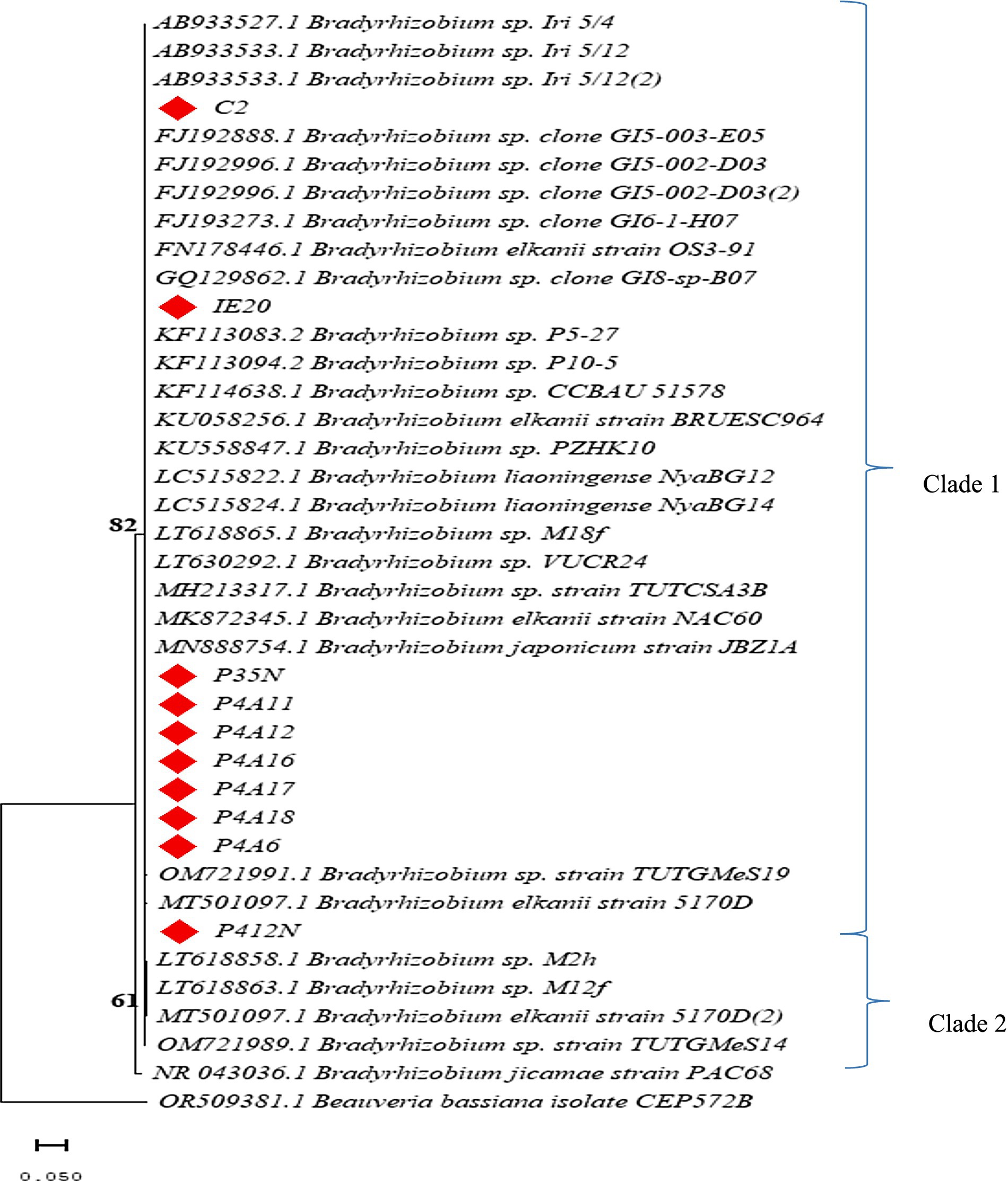
Figure 2. Neighbor-joining phylogenetic tree illustrating the relatedness of the ten (10) confirmed isolates and top selected Bradyrhizobium species. Filled red diamond shapes are isolates from this study. Beauvaria bassiana was used as the outgroup.
3.5 N-fixation and in-vitro potential of plant beneficial traits
The N-fixing and plant growth promotion (PGP) tests revealed that, all ten (10) Bradyrhizobium isolates had nitrogen fixing and phosphate solubilizing abilities. Specifically, isolate P4A18 demonstrated a greater ability to fix nitrogen by exhibiting maximum growth. Five (5) isolates had IAA production ability only. All isolates did not show HCN production ability. Isolates that tested positive for at least one of these assays were classified as elite isolates.
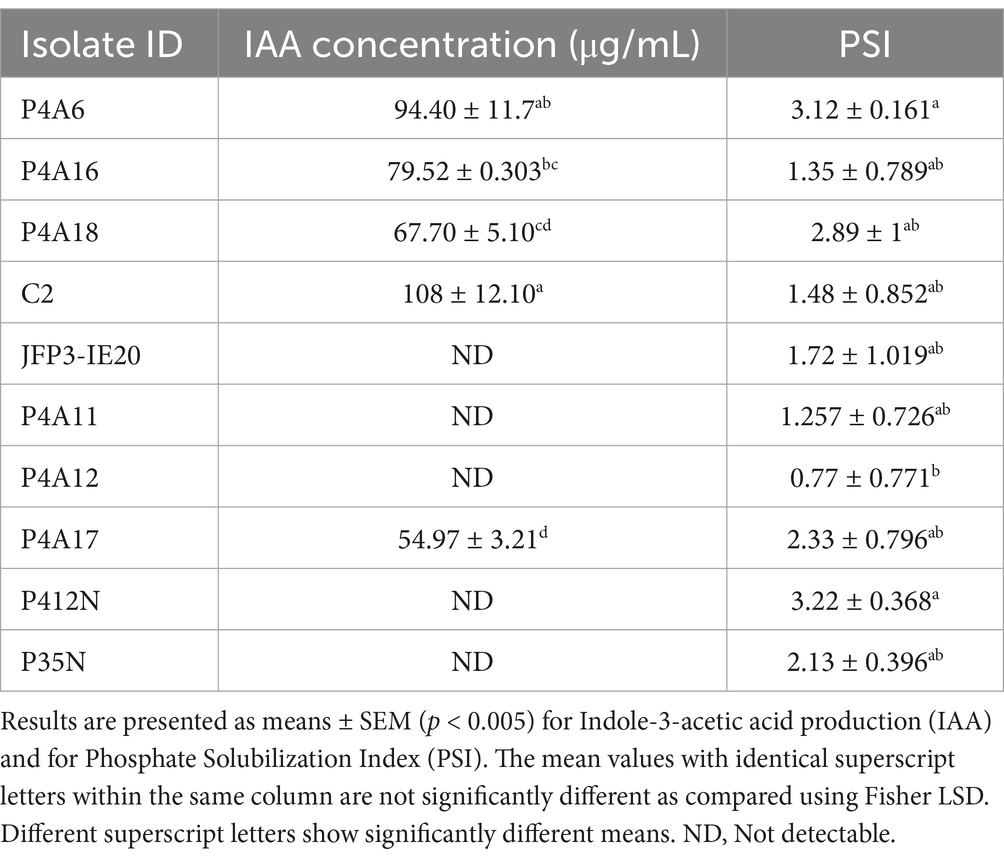
Phosphate-solubilizing isolates were distinctively recognized by the appearance of a transparent halo surrounding their colonies as a result of phosphate solubilization. The results showed that all isolates solubilized the inorganic tricalcium phosphate (Figure 3). However, the solubilizing activities varied in relation to the diameter of the halo zones formed. (Table 3). The diameters ranged from 0.77 to 3.22 cm, with isolates P4A6 and P412N demonstrating statistically (p < 0.005) significant p-solubilizing ability (3.12 ± 0.161and 3.223.22 ± 0.368, df = 2 respectively), but these were significantly different (p > 0.005) from the other isolates (Table 3). In contrast, isolate P4A12 recorded the least p solubilization (0.77 ± 0.771) (Table 3).
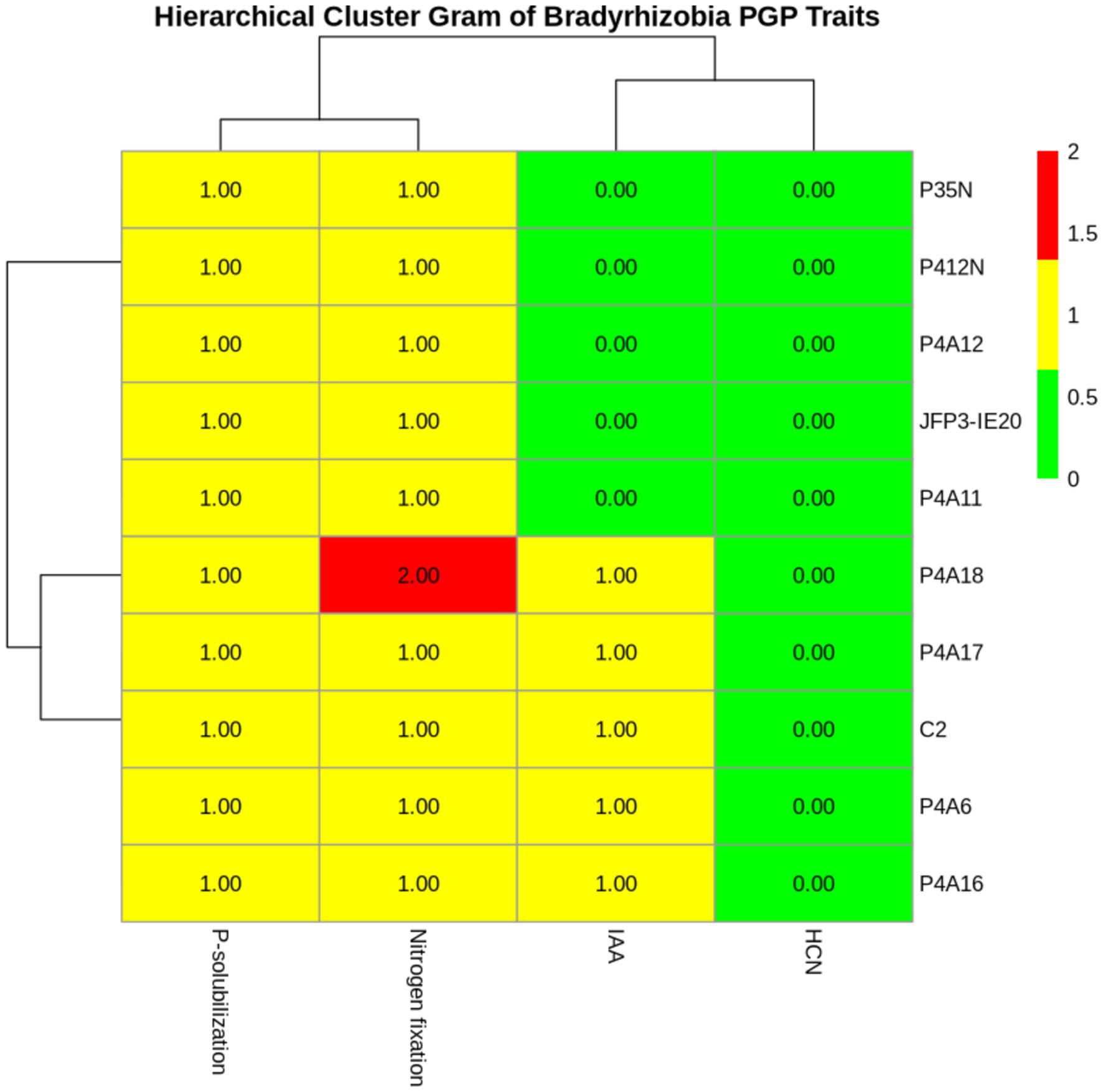
Figure 3. Cluster-gram of multiple PGP traits of Bradyrhizobium isolates. 0.00 (Green) = No activity; 1.00 (Yellow) = Moderate activity; and 2.00 (Red) = Maximum activity.
From the data in Table 3, it is apparent that the isolates showed clear differences (p < 0.005 and F-value = 62.88) in synthesizing IAA. Of the 10 isolates, 50% produced a measurable quantity of IAA that varied from 54.97 ± 3.21 μg/mL to 108 ± 12.10 μg/mL with statistically significant (p < 0.005) differences between the levels, while the remaining isolates were non-detectable (Table 3). Isolate C2 produced the highest amount of IAA (108 ± 12.10 μg/mL), then isolate P4A6 (94.40 ± 11.7 μg/mL). The lowest level of IAA was recorded from isolate P4A17 (54.97 ± 3.21 μg/mL) (Table 3).
3.6 Growth response under varying physiological stresses (pH, salinity and temperature)
Growth of all the Bradyrhizobium isolates was highest at all control conditions (0.001% NaCl, pH 6.6–7 and temperature of 25°C–30°C). The results obtained from these tests are presented on Figure 4. All Bradyrhizobium isolates showed varying growth potential across all conditions (Figure 4). The Bradyrhizobium isolates exhibited variability in their responses to sodium chloride (NaCl) concentrations. All isolates thrived in the media with 0.001% NaCl (control). An increase in NaCl concentration led to a decrease in the NaCl tolerance by the isolates. Of the 10 Bradyrhizobium isolates tested, all grew well on media containing 1% NaCl, 6 grew in 3% NaCl, same six isolates extended their growth tolerance in 5% NaCl, and none grew in 10% NaCl. With pH, all the isolates showed minimum to moderate growth in pH 5 and pH 9. However, with the extremes such as pH 3 and pH 11, only five (5) and eight (8) isolates were able to grow, respectively. Isolate P4A18 (Bradyrhizobium sp.) showed maximum growth even at pH 11, more than the control pH range of 6.6–7.0. Regarding the ability to tolerate varying thermal conditions, four (4) isolates showed growth potentials at 40°C–50°C. Among these, only P4A6 (Bradyrhizobium sp.) showed maximum growth at this high temperature.
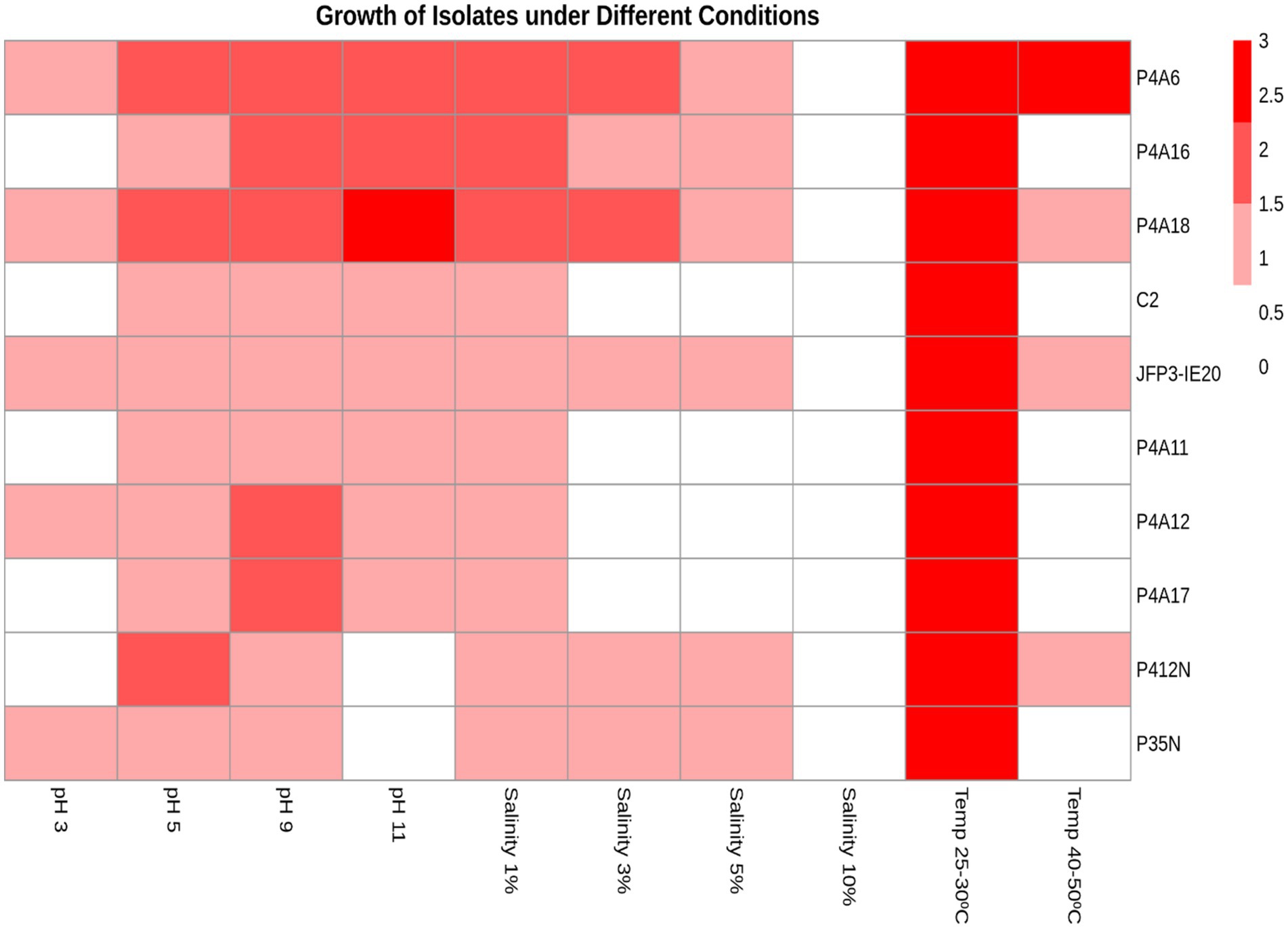
Figure 4. Heatmap showing growth responses of isolates under variable conditions of pH, salinity and temperature. 0 = No growth (White), 1 = Minimum growth (Pink), 2 = Moderate growth (Red), and 3 = Maximum growth (Deep red).
3.7 Authentication of nodule formation abilities of selected isolates (P4A6 and P4A18)
In authenticating the ability of the strains to nodulate Bambara groundnuts, it was observed that, P4A6 recorded the highest number of nodules in sterile (35) and non-sterile (41) soils, respectively. P4A18 recorded 26 and 29 nodules per plant in sterile and non-sterile soils. The lowest number of nodules was 14.67 recorded by the commercial inoculant in sterile soils (Figure 5).
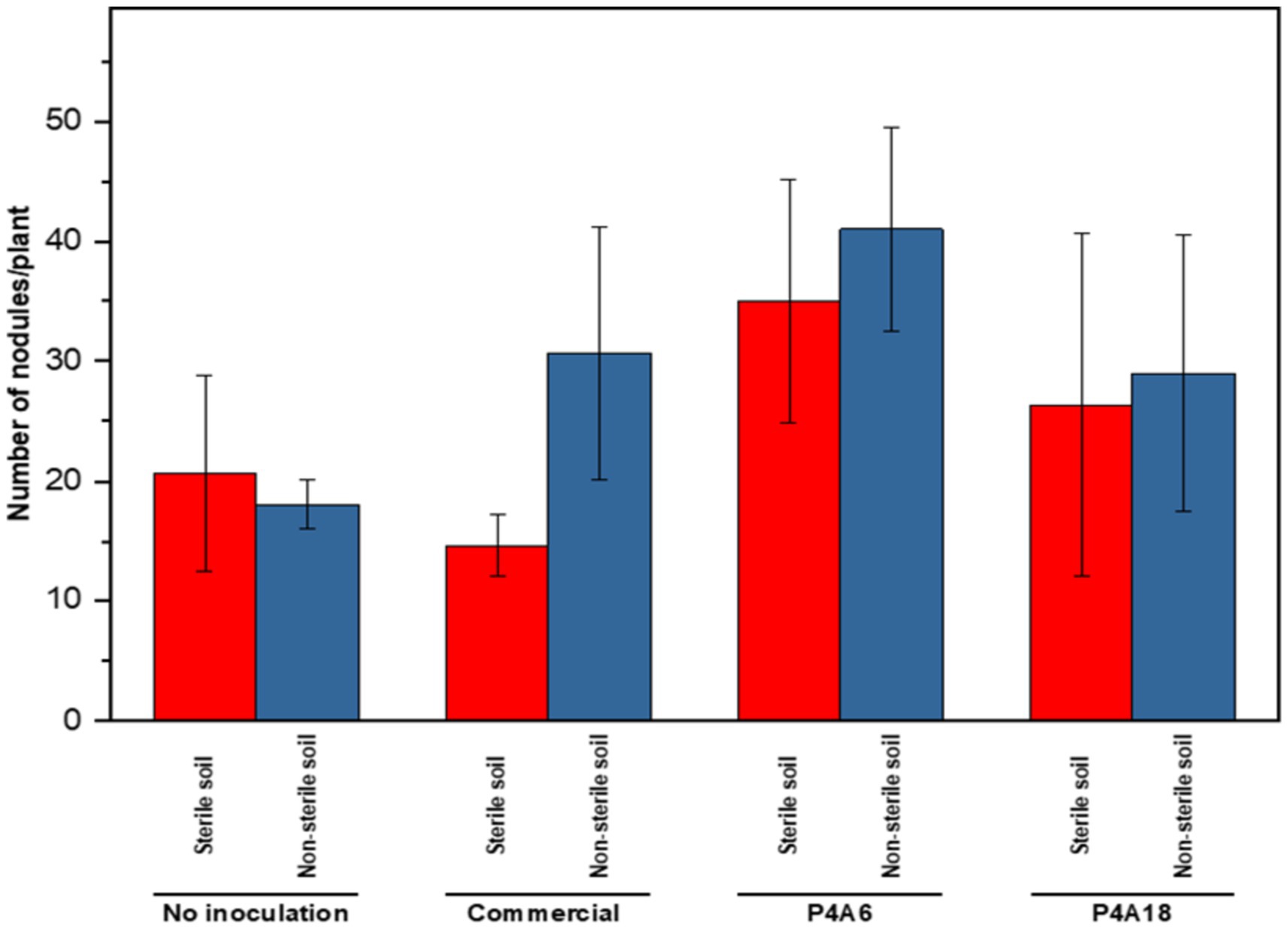
Figure 5. Number of nodules formed per plant based on the activities of each treatment. Values are means (n = 3) ± standard error bars with significance at p < 0.05.
4 Discussion
Utilizing rhizobial inoculants is a highly efficient approach for advancing sustainable agriculture and addressing the interconnected challenges with global food security and environmental stability (Kebede, 2021). The incorporation of plant growth-promoting (PGP) bacteria is critical for encouraging the sustained production of climate-adapted crops such as Bambara groundnut (BGN). This study isolated elite Bradyrhizobium isolates with PGP abilities from BGN root nodules which agrees with Puozaa et al. (2017), who reported that, Bradyrhizobium species is the predominant bacteria responsible for nodulation in BGN. Primarily, the isolates displayed a range of phenotypic features on the appearance, size (diameter), color, and texture of their colonies. Kaur et al. (2012) documented the presence of diverse physical characteristics and cultural attributes of isolates in their study on soybean, and Muindi et al. (2021) published the isolation of beneficial bacteria from common bean with varying growth characteristics. Morphological studies of the colonies and cells revealed little to no absorption of the Congo red (CR) dye by the bacteria. This characteristic was unique to bacteria in the rhizobia family. Out of the 199 isolates, ten (10) showed the capacity to produce alkaline; causing a color change of the media from green to blue, while some retained the green color of the media when grown on YEMA-BTB. Our present study supports the results of Fossou et al. (2020), who indicated that Bradyrhizobium does not always maintain green coloration in the presence of BTB. This suggests that the indicator may lack the ability to accurately differentiate between Rhizobium, Agrobacterium, and Bradyrhizobium. Ultimately, the 10 isolates were considered as potential Bradyrhizobium species (Somasegaran and Hoben, 1994), despite the wide variety of strains within this genus.
Biochemically, all isolates were positive for oxidase and catalase tests. As reported, Bradyrhizobium species possess both the oxidase and the catalase enzymes, except rare cases like Bradyrhizobium ivorense (Fossou et al., 2020). The ability of isolates P35N and P412N to utilize citrate contradicts the findings of Kaur et al. (2012), and Somasegaran and Hoben (1994) who reported that Bradyrhizobium isolates do not utilize citrate. Nevertheless, the results of this investigation are in accordance with the results of Deshmukh et al. (2013), who documented the capacity of Bradyrhizobium to metabolize citrate. This finding is particularly intriguing because Bradyrhizobium isolates mostly exhibit specificity toward carbon sources, predominantly six-carbon sugars such as mannitol, sucrose, and sorbitol. Despite the specificity in carbon utilization, these isolates associate symbiotically with the Bambara groundnut crop and derive organic compounds from the root exudates, hence there could be a higher amount of citrate being exuded by the plant roots which led to expression of citrate transport and metabolism genes in isolates P35N and p412N. The potential for these isolates to represent distinct strains or species due to variations in their citrate metabolism genes should not be disregarded.
We conducted molecular studies using 16S-rRNA sequencing to validate the morphological identification of the isolates. This approach allowed for the identification and assessment of the genetic variability among the isolates due to its universal presence in all bacteria. The analysis in this study revealed that all isolates derived from the root nodules of Bambara groundnut belong to the Bradyrhizobium genus. Our findings are consistent with prior studies undertaken by Grönemeyer et al. (2016) and Grönemeyer and Reinhold-Hurek (2018), who reported the identification of diverse plant growth-promoting (PGP) Bradyrhizobium from Bambara groundnut nodules. Isolate C2 identified as Bradyrhizobium sp. exhibited a considerably lower percent identity (82.78%) to sequences on NCBI database. This suggests that the isolate may belong to a very distinct species that may be different from the other isolates (Idris Hassen et al., 2014). Within the clustering analysis, isolates P412N identified as Bradyrhizobium sp. exhibited a degree of variation by forming its own subclade, suggesting that it may also be of distinct Bradyrhizobium sp. This therefore implies that, conducting a more in-depth study of constitutive and symbiotic genes may uncover the precise species within the Bradyrhizobium lineage that nodulate Bambara groundnut. This identification is essential in creating a catalog of native Bambara groundnut symbionts, which can be utilized to develop inoculants capable of enhancing yield.
The growth and production of crops are heavily reliant on the presence of necessary nutrients. PGP bacteria provide benefits to plants by producing certain chemicals, aiding in the absorption of soil nutrients, and protecting plants from pathogens (Hayat et al., 2010). The stimulation of legume growth and development may be attributed to the advantageous activities of bacteria, such as generation of plant growth hormones, nitrogen fixation, hydrogen cyanide (HCN) production, and phosphate (P) solubilization. Bambara groundnut, similar to other legumes, establishes symbiotic relationships with nitrogen-fixing soil microorganisms, commonly Bradyrhizobium (Grönemeyer et al., 2014). Evidently, this study demonstrates that all ten (10) isolates exhibited growth on NFB basal medium. Notably, isolate P4A18; identified as Bradyrhizobium sp. showed a superior ability to fix nitrogen due to its maximum growth on N-free medium. All isolates thereafter exhibited a substantial increase in size and had a media color change from green to blue. Our investigation corroborates the findings of Grönemeyer et al. (2016) regarding the nitrogen-fixing capabilities of Bradyrhizobium isolates from BGN. As nitrogen fixation is crucial in agricultural systems, especially in smallholder farming systems where the availability of chemically formulated nitrogen fertilizer is limited due to high cost and scarcity (Houlton et al., 2019), Bradyrhizobium isolates obtained in this study could effectively enhance Bambara groundnut cultivation when used as bioinoculants.
PGP Bradyrhizobium can solubilize insoluble phosphorus (P) (a limiting factor for crop production), and convert it into plant-available forms. This trait is particularly significant in environments with limited phosphorus availability, particularly African soils. It was revealed in this study that all isolates exhibited the capacity to solubilize phosphate. In addition, our study found that isolates P4A12 (0.77 ± 0.771b), P4A18 (2.89 ± 1ab), P4A6 (3.12 ± 0.161a), and P412N (3.22 ± 0.368a) exhibited significantly distinct phosphate solubilizing indices (PSI). P4A12 exhibited the least ability to solubilize phosphorus, whereas P412N demonstrated the highest solubilization index. The discrepancies in solubilization levels can be related to the variations in Bradyrhizobium species. A prior study has shown that the phosphate solubilization index (PSI) of plant growth-promoting (PGP) isolates varied from 1.14 to 3.61 indices (Jain et al., 2021), this range aligns with the indices recorded from this study. The isolates from this study have shown the ability to dissolve inorganic phosphates and enhance the accessibility of phosphorus. Bradyrhizobium isolates that solubilize phosphates can be used as biofertilizers to increase the production of Bambara groundnuts especially at their early growth stages.
One common way that PGPR work, is through synthesizing phytohormones, such as IAA (Rico-Jiménez et al., 2023). IAA plays a role in root initiation, cells development, and increasing the surface area of roots. Through in-vitro screening, it was revealed that isolates C2 (Bradyrhizobium sp.) and P4A6 (Bradyrhizobium sp.) produced the highest levels of IAA, with concentrations of 108 μg/mL and 94.40 μg/mL, respectively. The values reported in our study were significantly greater than those reported by Chukwuneme et al. (2020). The ability of bacterial isolates to synthesize indole-3-acetic acid (IAA) demonstrates their capability to stimulate plant growth hormones or growth regulators at different levels, as observed in this investigation. The findings generated from this present research align with previous studies that investigated PGP bacteria in the rhizosphere, which produced IAA concentrations ranging from 72.93 ± 0.2 μg/mL to 82.48 ± 0.9 μg/mL (Maya et al., 2022). It is crucial to note that some isolates in this research were unable to synthesize indole-3-acetic acid (IAA). The variance in the production of IAA by PGP bacteria aligns with the findings of Maya et al. (2020), who noted different production capacities across different bacterial isolates. The variance observed may be attributed to the different biochemical routes of the Bradyrhizobium isolates, the expression of growth development-related genes, and enzymes that play a role in conversion of active free IAA into conjugated forms (Patten and Glick, 1996).
Due to its potent toxicity against phytopathogens, agriculture has widely employed hydrogen cyanide (HCN) as a bio-control agent. However, none of the Bradyrhizobium isolates found in this study were capable of producing HCN. The present study contradicts the earlier research conducted by Tamiru and Muleta (2018), which demonstrated the efficacy of Bradyrhizobium in managing black root rot in Faba beans based on their HCN production abilities. While our findings differ from the results from Deshwal et al. (2003) and Tamiru and Muleta (2018), they however align with Ajilogba et al. (2022), which indicated no synthesis of HCN by PGP bacteria isolated from Bambara groundnut. Although the isolates do not possess the potential to produce HCN, it cannot be definitively inferred that crops will be very susceptible to phytopathogen infections. Perhaps the inability to produce HCN in these isolates could be attributed to their genetic composition, where the ability to produce HCN may be repressed due to the absence of pathogen threats in the local soils.
The key parameters that mostly limit symbiotic N-fixation in legume-rhizobia interaction are changes in temperatures, pH, and soil salinity. Therefore, only tolerant strains would be able to survive under harsh physiological conditions (Berrada et al., 2012). Salinity is a significant global constraint that impedes plant growth by influencing several physiological processes. Thus, it is crucial to evaluate native Bradyrhizobium species that exhibit enhanced efficiency in facilitating atmospheric nitrogen fixation and enhancing plant growth even under various salt conditions. From our study, all isolates grew in the presence of 1% NaCl, with varying levels of growth at 3% NaCl and 5% NaCl. This finding aligns with Elsheikh and Wood (1989) who indicated that different species of Bradyrhizobium can vary significantly in their tolerance levels. However, all of the isolates exhibited an inability to thrive in the presence of 10% NaCl, indicating their susceptibility to elevated salt concentrations. Deshwal and Chaubey (2014) also observed similar findings. Higher salt concentrations can harm bacteria population due to both direct toxicity and indirect osmotic stress. In general, isolates P4A6 (Bradyrhizobium sp.) and P4A18 (Bradyrhizobium sp.) exhibited superior growth capabilities under different salt conditions, which relates to the report of Ahmad et al. (2013), who indicated that PGP bacteria were tolerant to salt and showed improved growth promotion of Vigna radiata. This innate ability of salt tolerance can be related to the capacity of isolates to buildup protective organic osmolytes, such as amino acids or carbohydrates. Importantly, the potential of these isolates to thrive under high saline conditions gives them a high competitive advantage in the rhizosphere to survive and nodulate host plants particularly when soils have high salt levels.
The Bradyrhizobium-legume symbiosis is affected by high temperatures because it makes it harder for bacteria to attach to root hair, which stops root hair from growing and infection threads from being formed. In tropical regions, the temperature can reach as high as 42°C–47°C, especially given the recent change in climate. Under such conditions, temperature-tolerant Bradyrhizobium might be beneficial to farmers, even in unfavorable field conditions. Therefore, in our investigation, we noticed that P4A6 (Bradyrhizobium sp.) showed maximum growth at temperatures of 40°C–50°C, whereas P4A18 (Bradyrhizobium sp.), IE20 (Bradyrhizobium sp.), and P412N (Bradyrhizobium sp.) showed minimum growth (Figure 4). The results are consistent with an earlier research on strains obtained from Egypt, which demonstrated tolerance to temperatures of 40°C (Moawad and Beck, 1991). The inherent thermal tolerance of the four isolates may have significantly enabled their growth at these temperatures (40°C–50°C), rather than solely attributing the growth potential to environmental factors. When the temperature was lowered to the ideal 25°C–30°C, all the samples exhibited the highest level of growth on the YEMA media. This agrees with the results of Kaur et al. (2012), who observed that Bradyrhizobium exhibited the highest rate of growth at the optimum temperature of 28°C.
The pH tolerance test conducted for this study demonstrated that P4A6 (Bradyrhizobium sp.), P4A18 (Bradyrhizobium sp.), IE20 (Bradyrhizobium sp.), P4A12 (Bradyrhizobium sp.), and P35N (Bradyrhizobium elkanii) can withstand pH levels of 3. This aligns with the research of Chhetri et al. (2019), which showed that Bradyrhizobium sp. can withstand acidic conditions. Nevertheless, the remaining isolates in our investigation were unable to withstand the pH of 3, resulting in no observable growth. According to Richardson and Simpson (1989), even small changes in pH can have a substantial impact on the growth of nodule bacteria. Thus, all the Bradyrhizobium isolates exhibited minimal to moderate growth at pH 5 and pH 9. However, P4A18 (Bradyrhizobium sp.) exhibited optimal development even at a higher pH of 11, but P412N (Bradyrhizobium sp.) and P35N (Bradyrhizobium elkanii) were unable to tolerate alkaline conditions, and hence could not grow. According to Msaddak et al. (2017), Bradyrhizobium sp. obtained from L. luteus species in Tunisia could not survive under pH conditions of 12. Reasonably, intracellular pH of bacteria is minimally influenced by external acidity, but their metabolic pathways may become inefficient or even toxic when exposed to alkaline conditions (Graham et al., 1994). Bradyrhizobium strains exhibit varied capacity in tolerating different pH levels that impact their overall propagation under various environments.
The effect of the Bradyrhizobium strains on nodules formation was evident on the varied number of nodules formed by the plants as shown on Figure 5. Nodule formulation is impacted by the genetic traits of the strains. Different strains may generate a higher number of nodules than others, which may produce fewer or less efficient nodules as alluded to by Allito et al. (2020). Additionally, even though all the strains were from the same Bambara groundnut variety, the host plant could have exhibited strain specific association and this may have influenced the number of nodules formed. Aside these factors, the availability of soil (red soil) nutrients such as phosphorus, and environmental conditions may have also impacted the strains responses in promoting nodule formation. Plants associate symbiotically with several microorganisms in the rhizosphere. Due to this diversity in population and need to survive, there is competition for resources (especially carbon) among the soil microbes such that the fittest usually outcompete the rest consequently posing a challenge in terms of inoculant viability (Tabassum et al., 2017). Our results showed no significant differences in nodule number between the two soil treatments for most of the strains except for the commercial one. This was probably because a relatively ineffective bacterial strain was used in the production of the commercial inoculant or storage conditions affected the effectiveness of the strain. In addition, legumes often form ineffective, white nodules which do not fix any nitrogen, thus the total nodule number per plant may not necessarily be positively correlated with the fixation process (Horváth et al., 2011; Kalo et al., 2005; Smit et al., 2005). Nonetheless, in some cases, a positive relationship between nodule number and fixation was reported (Oladzad et al., 2020). An optimal number of nodules is necessary to ensure a balanced energy usage by plants as this would tend to benefit plant growth and even yield due to the reduced wastage of energy resources. P4A6 clearly demonstrates a better potential for nodule formation as observed in both soil treatments, while P4A18 recorded a smaller number. To validate the potential of these isolates, further open field evaluation will be recommended.
5 Conclusion
This study set out to identify elite plant growth promoting Bradyrhizobium from the nodules of Bambara groundnuts. The Bradyrhizobium isolates from our study demonstrated varying morphological, biochemical and molecular characteristics, this suggest the presence of diversity among the indigenous Bradyrhizobium isolates which could be exploited in bioinoculant processing. Despite all 10 isolates having promising N-fixing potentials, two isolates: P4A6 and P4A18 were better than the rest and were selected as potential sources of bioinoculants for Bambara groundnuts. Hence, these isolates require extensive study to evaluate their symbiotic efficiency in greenhouse and field environments in order to utilize them in commercial production of effective bioinoculants.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
Author contributions
OS: Conceptualization, Formal analysis, Investigation, Methodology, Writing – original draft, Writing – review & editing. SG: Conceptualization, Supervision, Validation, Writing – review & editing. JW: Conceptualization, Supervision, Validation, Writing – review & editing. VN: Supervision, Validation, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. The study was funded by the African Union through the Pan African University scholarship which included research and publication of this article.
Acknowledgments
The authors greatly appreciate the African Union’s (AU) provision of scholarship and research funds to support this study through the Pan African University Institute for Basic Sciences, Technology and Innovation (PAUSTI). We further thank Dr. Muazu Issifu and Portia Osei-Obeng for their suggestions.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that no Gen AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Footnotes
References
Abiola, C., and Oyetayo, V. O. (2016). Isolation and biochemical characterization of microorganisms associated with the fermentation of kersting’s groundnut (Macrotyloma geocarpum). Res. J. Microbiol. 11, 47–55. doi: 10.3923/jm.2016.47.55
Adjei, J. A., Aserse, A. A., Yli-Halla, M., Ahiabor, B. D. K., Abaidoo, R. C., and Lindstrom, K. (2022). Phylogenetically diverse Bradyrhizobium genospecies nodulate Bambara groundnut (Vigna subterranea L. Verdc) and soybean (Glycine max L. Merril) in the northern savanna zones of Ghana. FEMS Microbiol. Ecol. 98, 1–17. doi: 10.1093/femsec/fiac043
Ahmad, I., Akhtar, M. J., Asghar, H. N., and Khalid, M. (2013). Influence of Rhizobium applied in combination with micronutrients on mungbean. Pak. J. Life Soc. Sci. 11, 53–59.
Ajilogba, F. C., Babalola, O. O., Adebola, P., and Adeleke, R. (2022). Bambara groundnut rhizobacteria antimicrobial and biofertilization potential. Front. Plant Sci. 13, 1–16. doi: 10.3389/fpls.2022.854937
Al-Hossainy, A. A., Ibrahim, A., Mogharbel, R. T., and Ibrahim, S. M. (2021). Synthesis of novel keto-bromothymol blue in different media using oxidation–reduction reactions: combined experimental and DFT-TDDFT computational studies. Chem. Pap. 75, 3103–3118. doi: 10.1007/s11696-021-01540-y
Allito, B. B., Ewusi-mensah, N., and Logah, V. (2020). Legume-Rhizobium strain specificity enhances nutrition and nitrogen fixation in Faba bean (Vicia faba L.). Agronomy 10:826. doi: 10.3390/agronomy10060826
Baldani, J. I., Reis, V. M., Videira, S. S., Boddey, L. H., and Baldani, V. L. D. (2014). The art of isolating nitrogen-fixing bacteria from non-leguminous plants using N-free semi-solid media: a practical guide for microbiologists. Plant Soil 384, 413–431. doi: 10.1007/s11104-014-2186-6
Barillot, C. D. C., Sarde, C. O., Bert, V., Tarnaud, E., and Cochet, N. (2013). A standardized method for the sampling of rhizosphere and rhizoplan soil bacteria associated to a herbaceous root system. Ann. Microbiol. 63, 471–476. doi: 10.1007/s13213-012-0491-y
Berrada, H., Nouioui, I., Houssaini, M. I., Naïma El Ghachtouli, M. G., and Benbrahim, K. F. (2012). Phenotypic and genotypic characterizations of rhizobia isolated from root nodules of multiple legume species native of fez, Morocco. Afric. J. Microbiol. Res. 6, 5314–5324. doi: 10.5897/ajmr11.1505
Chaudhary, T., Gera, R., and Shukla, P. (2021). Deciphering the potential of rhizobium pusense mb-17a, a plant growth-promoting root endophyte, and functional annotation of the genes involved in the metabolic pathway. Front. Bioeng. Biotechnol. 8, 1–12. doi: 10.3389/fbioe.2020.617034
Chhetri, T. K., Subedee, B. R., and Pant, B. (2019). Isolation, identification and production of encapsulated Bradyrhizobium japonicum and study on their viability. Nepal J. Biotechnol. 7, 39–49. doi: 10.3126/njb.v7i1.26950
Chukwuneme, C. F., Babalola, O. O., Kutu, F. R., and Ojuederie, O. B. (2020). Characterization of actinomycetes isolates for plant growth promoting traits and their effects on drought tolerance in maize. J. Plant Interact. 15, 93–105. doi: 10.1080/17429145.2020.1752833
Deshmukh, V. V., Mane, S. S., Gade, R. M., Ingle, R. W., and Joshi, M. S. (2013). Biochemical studies of Bradyrhizobium japonicum isolates. Am. Int. J. Nat. Sci. 4, 53–57.
Deshwal, V. K., and Chaubey, A. (2014). Isolation and characterization of Rhizobium leguminosarum from root nodule of Pisum sativum L. J. Acad. Ind. Res. 2:464.
Deshwal, V. K., Dubey, R. C., and Maheshwari, D. K. (2003). Isolation of plant growth-promoting strains of Bradyrhizobium (Arachis) sp. with biocontrol potential against Macrophomina phaseolina causing charcoal rot of peanut. Curr. Sci. 84, 443–448.
Dinkwar, G., Thakur, K., Bramhankar, S., Pillai, T., Isokar, S., Ravali, T., et al. (2020). Biochemical and physiological characterization of Bradyrhizobium japonicum. Int. J. Chem. Stud. 8, 1589–1594. doi: 10.22271/chemi.2020.v8.i2x.8989
Elsheikh, E. A. E., and Wood, M. (1989). Response of chickpea and soybean rhizobia to salt: osmotic and specific ion effects of salts. Soil Biol. Biochem. 21, 889–895. doi: 10.1016/0038-0717(89)90077-1
Fasusi, O. A., Cruz, C., and Babalola, O. O. (2021). Agricultural sustainability: microbial biofertilizers in rhizosphere management. Agriculture (Switzerland) 11, 1–19. doi: 10.3390/agriculture11020163
Fayaz, U., Banday, S., Bhat, A. H., Khan, N. A., and Shahnaz, E. (2022). Morphological characterization and biological management of Gloeosporium ampelophagum (pass.) Sacc causing anthracnose of grapes in India. International. J. Phytopathol. 11, 181–194. doi: 10.33687/phytopath.011.02.4314
Fossou, R. K., Pothier, J. F., Zézé, A., and Perret, X. (2020). Bradyrhizobium ivorense sp. nov. as a potential local bioinoculant for Cajanus cajan cultures in Côte d’ivoire. Int. J. Syst. Evol. Microbiol. 70, 1421–1430. doi: 10.1099/ijsem.0.003931
Graham, P. H., Draeger, K. J., Ferrey, M. L., Conroy, M. J., Hammer, B. E., Martinez, E., et al. (1994). Acid pH tolerance in strains of Rhizobium and Bradyrhizobium, and initial studies on the basis for acid tolerance of Rhizobium trpici UMR1899. Can. J. Microbiol. 40, 198–207. doi: 10.1139/m94-033
Grönemeyer, J. L., Hurek, T., Bünger, W., and Reinhold-Hurek, B. (2016). Bradyrhizobium vignae sp. nov., a nitrogen-fixing symbiont isolated from effective nodules of Vigna and Arachis. Int. J. Syst. Evol. Microbiol. 66, 62–69. doi: 10.1099/ijsem.0.000674
Grönemeyer, J. L., Kulkarni, A., Berkelmann, D., Hurek, T., and Reinhold-Hurek, B. (2014). Rhizobia indigenous to the Okavango region in sub-saharan Africa: diversity, adaptations, and host specificity. Appl. Environ. Microbiol. 80, 7244–7257. doi: 10.1128/AEM.02417-14
Grönemeyer, J. L., and Reinhold-Hurek, B. (2018). Diversity of Bradyrhizobium in subsahara Africa: a rich resource. Front. Microbiol. 9:2194. doi: 10.3389/fmicb.2018.02194
Hayat, R., Ali, S., Amara, U., Khalid, R., and Ahmed, I. (2010). Soil beneficial bacteria and their role in plant growth promotion: a review. Ann. Microbiol. 60, 579–598. doi: 10.1007/s13213-010-0117-1
Herridge, D., Peoples, M., and Boddey, R. (2008). Global inputs of biological nitrogen fixation in agricultural systems. Plant Soil 311, 1–18. doi: 10.1007/s11104-008-9668-3
Horváth, B., Yeun, L. H., Domonkos, Á., Halász, G., Gobbato, E., Ayaydin, F., et al. (2011). Medicago truncatula IPD3 is a member of the common symbiotic signaling pathway required for rhizobial and mycorrhizal symbioses. Mol. Plant-Microbe Interact. 24, 1345–1358. doi: 10.1094/MPMI-01-11-0015
Houlton, B. Z., Almaraz, M., Aneja, V., Austin, A. T., Bai, E., Cassman, K. G., et al. (2019). A world of cobenefits: solving the global nitrogen challenge. Earth’s Future 7, 865–872. doi: 10.1029/2019EF001222
Idris Hassen, A., Lebogang Bopape, F., and Trytsman, M. (2014). Nodulation study and characterization of rhizobial microsymbionts of forage and pasture legumes in South Africa. World J. Agric. Res. 2, 93–100. doi: 10.12691/wjar-2-3-2
Issifu, M., Songoro, E. K., Niyomukiza, S., Ateka, E. M., Onguso, J., and Ngumi, V. W. (2022). Identification and in vitro characterization of plant growth-promoting Pseudomonas spp. isolated from the rhizosphere of tomato (Lycopersicum esculentum) plants in Kenya. Univ. J. Agric. Res. 10, 667–681. doi: 10.13189/ujar.2022.100608
Jain, D., Sharma, J., Kaur, G., Bhojiya, A. A., Chauhan, S., Sharma, V., et al. (2021). Phenetic and molecular diversity of nitrogen fixating plant growth promoting Azotobacter isolated from semiarid regions of India. Biomed. Res. Int. 2021:6686283. doi: 10.1155/2021/6686283
Jaiswal, S. K., and Dakora, F. D. (2019). Widespread distribution of highly adapted Bradyrhizobium species nodulating diverse legumes in Africa. Front. Microbiol. 10, 1–16. doi: 10.3389/fmicb.2019.00310
Kalo, P., Gleason, C., Edwards, A., Marsh, J., Mitra, R. M., Hirsch, S., et al. (2005). Nodulation signaling in legumes requires NSP2, a member of the GRAS family of transcriptional regulators. Science 308, 1786–1789. doi: 10.1126/science.1110951
Kaur, H., Sharma, P., Kaur, N., and Gill, B. S. (2012). Phenotypic and biochemical characterization of Bradyrhizobium and Ensifer spp. isolated from soybean rhizosphere. Biosci. Discov. 3, 40–46.
Kebede, E. (2021). Competency of rhizobial inoculation in sustainable agricultural production and biocontrol of plant diseases. Front. Sustain. Food Syst. 5, 1–22. doi: 10.3389/fsufs.2021.728014
Khan, M. M. H., Rafii, M. Y., Ramlee, S. I., Jusoh, M., and Mamun, A. (2020). Genetic variability, heritability, and clustering pattern exploration of Bambara groundnut (Vigna subterranea L. Verdc) accessions for the perfection of yield and yield-related traits. Biomed. Res. Int. 2020:2195797. doi: 10.1155/2020/2195797
Marinkovic, J., Bjelic, D., Tintor, B., Ignjatov, M., Nikolic, Z., Djukic, V., et al. (2017). Molecular identification of Bradyrhizobium japonicum strains isolated from root nodules of soybean (Glycine max L.). Zbornik Matice Srpske Za Prirodne Nauke 132, 49–56. doi: 10.2298/zmspn1732049m
Mathu, S., Herrmann, L., Pypers, P., Matiru, V., Mwirichia, R., and Lesueur, D. (2012). Potential of indigenous Bradyrhizobium versus commercial inoculants to improve cowpea (Vigna unguiculata L. walp.) and green gram (Vigna radiata L. wilczek.) yields in Kenya. Soil Sci. Plant Nutr. 58, 750–763. doi: 10.1080/00380768.2012.741041
Matiru, V. N., and Dakora, F. D. (2004). Potential use of rhizobial bacteria as promoters of plant growth for increased yield in landraces of African cereal crops. Afr. J. Biotechnol. 3, 1–7. doi: 10.5897/ajb2004.000-2002
Maya, B. K. C., Gauchan, D. P., Khana, S. N., Chimouriya, S., and Lamichhane, J. (2020). Extraction of indole-3-acetic acid from plant growth promoting rhizobacteria of bamboo rhizosphere and its effect on biosynthesis of chlorophyll in bamboo seedlings. Indian J. Agric. Res. 54, 781–786. doi: 10.18805/IJARe.A-5578
Maya, K. C. B., Prasad Gauchan, D., Khanal, S. N., and Lamichhane, J. (2022). Isolation and characterization of plant growth promoting rhizobacteria from bamboo rhizosphere and their role in plant growth promotion. Nepal J. Sci. Technol. 21, 1–12. doi: 10.3126/njst.v21i1.49908
Mayes, S., Ho, W. K., Chai, H. H., Gao, X., Kundy, A. C., Mateva, K. I., et al. (2019). “Bambara groundnut: an exemplar underutilised legume for resilience under climate change” in Planta, vol. 250 (Berlin Heidelberg: Springer), 803–820.
Moawad, H., and Beck, D. P. (1991). Some characteristics of Rhizobium leguminosarum isolates from uninoculated field-grown lentil. Soil Biol. Biochem. 23, 933–937. doi: 10.1016/0038-0717(91)90173-H
Msaddak, A., Rejili, M., Durán Wendt, D., Rey, L., Imperial, J., Palacios, J., et al. (2017). Members of microvirga and Bradyrhizobium genera are native endosymbiotic bacteria nodulating Lupinus luteus in northern Tunisian soils. FEMS Microbiol. Ecol. 93:fix068. doi: 10.1093/femsec/fix068
Muindi, M. M., Muthini, M., Njeru, E. M., and Maingi, J. (2021). Symbiotic efficiency and genetic characterization of rhizobia and non rhizobial endophytes associated with cowpea grown in semi-arid tropics of Kenya. Heliyon 7:e06867. doi: 10.1016/j.heliyon.2021.e06867
Oladzad, A., González, A., Macchiavelli, R., de Jensen, C. E., Beaver, J., Porch, T., et al. (2020). Genetic factors associated with nodulation and nitrogen derived from atmosphere in a middle american common bean panel. Front. Plant Sci. 11:576078. doi: 10.3389/fpls.2020.576078
Patten, C. L., and Glick, B. R. (1996). Bacterial biosynthesis of indole-3-acetic acid. Can. J. Microbiol. 42, 207–220. doi: 10.1139/m96-032
Puozaa, D. K., Jaiswal, S. K., and Dakora, F. D. (2017). African origin of Bradyrhizobium populations nodulating Bambara groundnut (Vigna subterranea L. Verdc) in Ghanaian and south African soils. PLoS One 12, e0184943–e0184923. doi: 10.1371/journal.pone.0184943
Richardson, A. E., and Simpson, R. J. (1989). Acid-tolerance of rhizobium growing in an acid soil. Soil Biol. Biochem. 21, 87–96. doi: 10.1016/0038-0717(89)90016-3
Rico-Jiménez, M., Muñoz-Mira, S., Lomas-Martínez, C., Krell, T., and Matilla, M. A. (2023). Regulation of indole-3-acetic acid biosynthesis and consequences of auxin production deficiency in Serratia plymuthica. Microb. Biotechnol. 16, 1671–1689. doi: 10.1111/1751-7915.14296
Singh, A. L., and Basu, M. S. (2016). Bambara groundnut: its physiology and introduction in India. Adv. Plant Physiol. 235–249.
Smith, A., and Hussey, M. (2005). Gram tain protocols. Washington, DC: American Society for Microbiology, 1–9.
Smit, P., Raedts, J., Portyanko, V., Debelle, F., Gough, C., Bisseling, T., et al. (2005). NSP1 of the GRAS protein family is essential for rhizobial nod factor-induced transcription. Science 308, 1789–1791. doi: 10.1126/science.1111025
Somasegaran, P., and Hoben, H. J. (1994). Handbook for rhizobia. In Handbook for Rhizobia. New York, NY, USA: Springer-Verlag New York, Inc.
Swanevelder, C. J. (1998). Bambara food for Africa. Pretoria-South Africa: National Department of Agriculture ARC - Grain Crops Institute 5–16.
Tabassum, B., Khan, A., Tariq, M., Ramzan, M., Iqbal Khan, M. S., Shahid, N., et al. (2017). Bottlenecks in commercialisation and future prospects of PGPR. Appl. Soil Ecol. 121, 102–117. doi: 10.1016/j.apsoil.2017.09.030
Tamiru, G., and Muleta, D. (2018). The effect of rhizobia isolates against black root rot disease of Faba bean (Vicia faba L) caused by fusarium solani. Open Agric. J. 12, 131–147. doi: 10.2174/1874331501812010131
Tamura, K., Stecher, G., and Kumar, S. (2021). MEGA11: molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 38, 3022–3027. doi: 10.1093/molbev/msab120
Keywords: Bradyrhizobium, plant growth promotion, Bambara groundnuts, 16S rRNA, biofertilizer
Citation: Seidu OA, Githiri SM, Wesonga JM and Ngumi VW (2025) Isolation, screening and in-vitro characterization of plant growth-promoting Bradyrhizobium isolates from the nodules of Bambara groundnut (Vigna subterranean) for potential use as bioinoculants. Front. Sustain. Food Syst. 8:1506346. doi: 10.3389/fsufs.2024.1506346
Edited by:
Maryke T. Labuschagne, University of the Free State, South AfricaReviewed by:
Eastonce Tendayi Gwata, University of Venda, South AfricaTheophilus Kwabla Tengey, CSIR-Savanna Agricultural Research Institute, Ghana
Copyright © 2025 Seidu, Githiri, Wesonga and Ngumi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Osman Abdulai Seidu, YWJkdWxhaS5vc21hbkBzdHVkZW50cy5qa3VhdC5hYy5rZQ==
 Osman Abdulai Seidu
Osman Abdulai Seidu Stephen Mwangi Githiri
Stephen Mwangi Githiri John Mwibanda Wesonga
John Mwibanda Wesonga Victoria Wambui Ngumi3
Victoria Wambui Ngumi3