- 1U.S. Dairy Forage Research Center, USDA-ARS, Madison, WI, United States
- 2Department of Biochemistry, University of Wisconsin-Madison, Madison, WI, United States
Aboveground plant material is subject to stress-induced senescence after harvest. For flowers, vegetables and forages, this process can severely impact quality. Proteases, in particular, are responsible for post-harvest protein degradation and contribute to cellular breakdown. Alfalfa is a high-quality forage and a perennial source of protein for dairy cows. Its nutritional value and suitability for sustainable agricultural production are also driving interest in the possible use of alfalfa for human consumption, however, the issue of rapid protein degradation after harvest needs to be addressed before the protein can be extracted for human use on a large scale. We have optimized a cost-effective, medium-throughput, fluorescence intensity-based protease activity assay specifically for aboveground plant tissue that balances precision and throughput, the latter of which has been a barrier to further study. This method will allow higher-throughput phenotyping of plant protease activity than is currently feasible: one person can process approximately 120 samples per day, compared to approximately 25 with previous methods. We have used this method in alfalfa to assess protease activity during wilting at pH 7.4 and pH 6.3.
1 Introduction
Stress-induced senescence in plants occurs as a normal physiological response to tissue damage; it serves to liberate and transport nutrients to growing parts of the plant through the phloem if the tissue is still connected (Roberts et al., 2012; Maillard et al., 2015). This process causes major changes to the cellular architecture and content of the harvested plant material including a decrease in chlorophyll content, degradation and overall decrease in protein, and cell death. Proteases–enzymes that hydrolyze proteins—do much of the work in regard to senescence, beginning with degrading the chloroplast and hydrolyzing rubisco in order to mobilize nitrogen (Roberts et al., 2012; Buet et al., 2019). The quality of certain crops is particularly affected by this process after harvest, including vegetables, flowers, and forages (Muck et al., 2003; Brasil and Siddiqui, 2018; Liu et al., 2022).
Alfalfa is a high protein, nitrogen-fixing, perennial forage that has traditionally been used as a feed for dairy cows and other ruminants. High-quality alfalfa contains up to 30% protein in its leaves, most of which is in the form of rubisco. Compared to seed crops such as soybean and pea, alfalfa is able to make more protein per acre. Because of its abundance and sustainability, there is a renewed interest in alfalfa as a source of protein for human diets, for example, as an alternative for protein from soybean and pea (Edwards et al., 1975; Hadidi et al., 2023). However, extracting protein from the leaves is challenging. Unlike seed protein, which is predominantly storage protein recalcitrant to degradation, leaf protein begins to degrade rapidly upon harvest. Plant-derived proteases are particularly active in alfalfa and lead to enormous losses of true protein after harvesting and ensiling (up to 87%; Scalet et al., 1984; Muck, 1987). In order to use alfalfa leaf protein for human consumption, it will be imperative to develop strategies for mitigating post-harvest losses of intact protein through modifying the actions of endogenous proteases.
Alfalfa proteases and proteolysis have been studied in order to gain a better understanding of alfalfa post-harvest biology with the intent of mitigating protein losses and nitrogen inefficiency in the agroecosystem context. Studies have shown that alfalfa maintains a high rate of protein degradation after 24 h of wilting, and the process continues at a lower rate during ensiling and storage (Papadopoulos and McKersie, 1983; Scalet et al., 1984; Muck, 1987; Jones et al., 1995). It has been shown that protease activity in alfalfa has approximately 14% narrow sense heritability, suggesting that breeding for reduced protease activity post-harvest may be possible, and investigating management strategies to mitigate this proteolysis will be important (Bowley and McKersie, 1987).
Despite progress that has been made in understanding post-harvest protease activity in alfalfa, the breadth and scope of previous studies have been limited by the methods of measuring protease activity that are available. The most accurate method of measuring protease activity and proteolysis is by measuring a change in soluble non-protein nitrogen (McKersie, 1985). This method takes several steps and is carried out over a few days or an entire week. Other methods that measure protease activity in a sample such as one that uses casein as a substrate and Folin’s reagent to measure the byproducts of its hydrolysis, are cumbersome, limited in sensitivity, and not particularly scalable (Cupp-Enyard, 2008). Commercially available kits that use a fluorescently labeled substrate are promising because they are higher throughput and relatively sensitive, however, they were not developed to work with green plant material and have other drawbacks. Because of these limitations, it would be difficult to assess protease activity in a sufficiently large number of samples to profile its phenological and genetic bases, e.g., to screen hundreds or thousands of alfalfa varieties to begin breeding for reduced protease activity. We hypothesized that commercially available fluorescent protease activity kit protocols could be modified to effectively and efficiently screen green plant material.
Accordingly, we present a method for measuring protease activity in green plant samples that is precise, scalable, and suitable for analyzing many samples in a relatively short period of time (Figure 1).
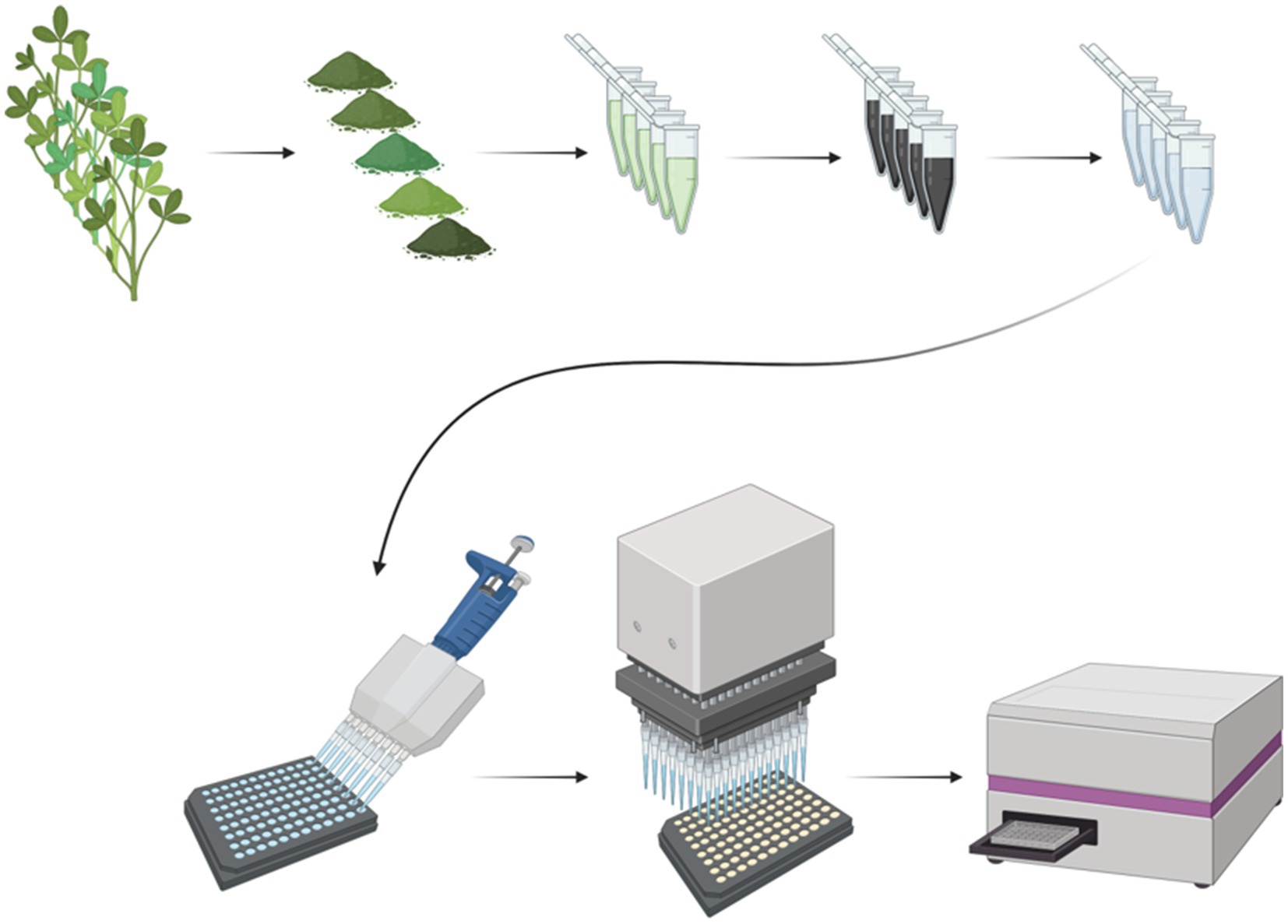
Figure 1. Overview of the protease assay for green plant material. The top row of images depicts aboveground plant sample processing for downstream use in the protease assay. Green plant samples are freeze-dried and ground, extracted in buffer, and finally treated with activated carbon to remove chlorophyll. The bottom row of the figure depicts how the protease assay is run. Standards, plant samples, and blanks are loaded onto a black 96-well plate. Then, the substrate is loaded into each well at the exact same time with a 96-well plate loader (pipette). Finally, the plate is incubated at room temperature, and the fluorescence intensity is read with a plate reader. Created with Biorender (biorender.com).
2 Methods
The objective of this method is to precisely measure protease activity of green plant samples in a medium-throughput, 96-well plate format. The overview of the workflow for this assay is depicted in Figure 1.
2.1 Plant material
Alfalfa clones from a commercial breeding population were grown in a greenhouse in containers containing potting soil. At approximately six weeks of growth, containers were randomized in the greenhouse and three biological replicates were harvested and allowed to wilt in the greenhouse at the following timepoints: 0 h, 2 h, 4 h, 5 h, and 24 h. After wilting, alfalfa samples were placed into plastic bags and immediately flash frozen in liquid nitrogen. They were stored at −80°C until they were freeze dried in a VirTis model 36DX66 freeze dryer (Gardiner, NY). After freeze drying for approximately 4 days, they were ground with an UDY mill equipped with a 1-mm screen (Fort Collins, CO) and stored in airtight plastic containers at room temperature. Care was taken to avoid any exposure to moisture.
2.2 Plant sample preparation and extraction
Microcentrifuge tubes (1.5 mL) were prepared by adding two glass beads (4 mm diameter) for later extraction of the samples. Using an analytical balance (Mettler Toledo; Columbus, OH), 25 mg (± <1 mg) of freeze dried, ground alfalfa samples were added to the centrifuge tubes containing the glass beads. The exact weights of the alfalfa samples were recorded for each sample. Dry samples can be stored in the tubes at room temperature if they are kept dry and are not exposed to moisture.
The protease assay was carried out in two different buffers. Phosphate buffer (PB) at pH 7.4 was chosen because it is in the range of the pH recommended in the kit protocol for maximizing fluorescence of the cleaved substrate. 4-(2-sulfonatoethyl) morpholin-4-ium (MES) buffer pH 6.3 was chosen to simulate plant physiological pH.
Phosphate and MES buffers were prepared (0.1 M and 5 mM, respectively) and the pH of the solutions were adjusted using a Mettler Toledo pH electrode and sterilized by autoclaving. The protease assay was performed on the same wilting timepoint samples in the two different buffers (one plate per buffer) on two different days. One milliliter of the appropriate buffer was added to the alfalfa samples (three biological replicates per time point, 15 samples total per buffer). Tubes were vortexed briefly to ensure full and homogeneous suspension of powdered plant material in buffer. The samples were processed as quickly as possible and kept on ice between steps.
The tubes were shaken at medium speed for 15 s in a Retsch Ball Mill MM 400 (Haan, Germany), taking care that each sample was processed in the same manner. After shaking, the tubes were allowed to rest at room temperature for 10 min.
The samples in buffer were centrifuged in an Eppendorf Centrifuge 5,418 R Microcentrifuge (Hamburg, Germany) for 1 min at 16,873× gravity (maximum speed). Seven hundred microliters of the supernatant were transferred into an empty 1.5 mL tube. The new tube was centrifuged for 1 min at maximum speed. Six hundred milliliters of the supernatant were transferred into a tube containing 10 mg (±2 mg, exact weight was not recorded) DARCO® 100 mesh particle size activated charcoal (Millipore Sigma; Burlington, MA) and vortexed briefly.
After vortexing, the samples were incubated for 5 min at room temperature, then centrifuged for 1 min at maximum speed. Five hundred microliters of the supernatant were transferred into the final tube. The samples were centrifuged again for 1 min at maximum speed to ensure that any leftover activated charcoal was at the bottom of the tube and the samples were visually clear and colorless. At this step, the samples may be frozen at −80°C and used in the assay at a later time, as long as all of the samples are treated in the same manner. However, they may only be frozen once before being used to minimize the effects of repeated freeze–thaw cycles. Ideally, the samples should be used in the assay the same day they are extracted and kept on ice before use.
2.3 Protease assay
The FITC-casein substrate and the trypsin standards used for this protease assay were from the Pierce Fluorescent Protease kit from Thermo Fisher (Watham, MA), based on the method by Twining (1984). The substrate and standards were prepared according to the manufacturer’s instructions. Briefly, the FITC-casein substrate was diluted to a working concentration of 10 μg/mL in sterile micropure water and mixed very gently (never vortexed). Fifteen milliliters were made per plate that was run. The substrate was diluted the same day that it was used and kept on ice in the dark. One aliquot of trypsin (included in the Pierce Fluorescent Protease kit) was diluted in the appropriate buffer (0.1 PB or 5 mM MES buffer). Three serial dilutions of the trypsin standards were made in the respective buffers. The resulting final dilution that was subsequently used as the most concentrated trypsin standard used for the assay was 1,000 ng/mL.
Two hundred microliter Corning black (sides and bottom), opaque 96-well plates (Corning, NY) were used for the assay. For the standard curve, two technical replicates of each of the following trypsin concentrations (one standard curve was included per buffer) were pipetted into the wells by performing 0.5 times serial dilutions on the plate with a Rainin multichannel pipette (Oakland, CA). The final trypsin concentrations included in the standard curve were as follows: 1,000 ng/mL, 500 ng/mL, 250 ng/mL 125 ng/mL, 62.5 ng/mL, 31.25 ng/mL, 15.625 ng/mL, 7.8125 ng/mL.
For the plant samples, 75 μL of the appropriate buffer (0.1 PB or 5 mM MES) and 25 μL of the extracted samples were pipetted into wells on the black 96-well plate using a multichannel pipette and a Rainin single-channel pipette (both Rainin). One hundred microliters of the substrate solution were added to all of the samples and standards at the exact same time using a Rainin MicroPro semi-automated 96-channel pipette and corresponding Rainin MPR 300 μL LS 4800/50 tips. The plate was incubated for 10 min at room temperature. The fluorescence intensity was measured on a Tecan Spark Multimode Microplate Reader (Zürich, Switzerland) with excitation/ emission of 485/538 nm.
The protease assay was run under different conditions in order to determine important parameters of the assay. For example, one plate was run with 0.5 times serial dilutions of a plant sample in PB and MES buffers. Readings were taken with the plate reader at multiple timepoints over a 120-min incubation period. A repeat of the protease assay of the wilting timepoint samples were also run in MES buffer pH 6.3.
2.4 Data analysis
The regression equations of the trypsin standard curves were calculated using R Statistical Software core functions (v4.4.1; R Core Team, 2021). The concentration of trypsin in ng/mL was designated as the independent variable and plotted on the x-axis, the total fluorescence of these trypsin standards (two technical replicates) was designated as the dependent variable and plotted on the y-axis. The type of curve, either polynomial or power, was determined by the R-squared value. The standard curve data were also tested for heteroscedasticity using the Breusch-Pagan test function in the lmtest package in R (Zeileis and Hothorn, 2002).
The trypsin standard curve of the wilting timepoints run in PB pH 7.4 was a power curve (y = 20905.45x0.119334) with an R-squared value of 0.995. The regression equation of the trypsin curve run with the wilting timepoints in MES buffer pH 6.3 was y = −0.00998x2 + 45.52x + 16,670 with an R-squared value of 0.9933. The mean fluorescence of three technical replicates of the plant samples were used as input into the regression equations and the corresponding relative concentration was calculated using custom functions in R. The relative protease activity calculated from the trypsin standard curves of all of the samples (at pH 7.4 and 6.3) were then normalized to the exact weight of the samples that were previously recorded and corrected for the 0.25 times dilution of the samples. The result was protease activity potential of the samples per gram dry matter. The mean of the three biological replicates were calculated and plotted along with the standard error.
The precision of the protease within and between plates was calculated by taking the mean absolute deviation of the biological replicates.
3 Results
The method presented here is based on a commercially available 96-well plate assay that uses casein bound to fluorescein as the substrate and trypsin as the standard. When the proteases present in a plant sample break down the substrate, the fluorescent molecule is liberated, and the fluorescence intensity is measured with a plate reader. Even though this substrate and the standard are sold for the purpose of measuring protease activity generally, chlorophyll interferes with the fluorescence of the cleaved substrate. Furthermore, it is not able to precisely measure more than one column (or row) of samples in a 96-well plate at a time. We have optimized this assay for use in green plant tissue and have used it to assess protease activity in alfalfa over 24 h of wilting (five timepoints) at two different pHs.
3.1 Activated charcoal treatment during extraction
The activated charcoal was important to reduce the amount of chlorophyll in the alfalfa samples because chlorophyll interferes with the fluorescence detection in the plate reader. For each sample type, it is important to determine the correct activated charcoal treatment required to reduce chlorophyll content. The minimum amount of activated charcoal should be used to effectively reduce chlorophyll, and more than one treatment may be necessary. After the samples were treated with activated charcoal, the samples were clear. If samples are black after charcoal treatment, it indicates that the samples contain activated charcoal, in this case, the supernatant should be transferred to another tube and centrifuged again. If the samples are green, they should be treated with activated charcoal again to eliminate the residual chlorophyll.
If the sample has very little chlorophyll, or the samples are going to be diluted before the reaction occurs, the activated charcoal may not be needed. In that case, one or two wells per sample in the 96-well plate should be blanks (plant sample in buffer without substrate added) and the fluorescence should be subtracted from the total fluorescence of the sample incubated with substrate. This may also be done in order to test the efficacy of the charcoal treatment—the fluorescence of the plant sample should be greatly reduced compared to the untreated plant sample.
3.2 Appropriate dilution of the plant samples
The exact dilution for the samples was determined by running several plates with a trypsin standard curve and a range of dilutions of the alfalfa samples. In this specific case, a dilution of 0.25 times was found to be within the fluorescence of the trypsin standard curve at both pH 7.5 and pH 6.3. The dilution factor of each sample type should be empirically determined in the appropriate buffer. The resulting fluorescence must be within the range of the standard curve for proper interpretation of the results.
3.3 Timing of substrate delivery
It is imperative that the FITC-casein substrate solution is delivered to all of the wells at the exact same time. Hydrolysis of the substrate begins immediately and there is a large effect of time on the fluorescence readings (Figure 2). Attempts at pipetting the substrate solution into each column with an 8-channel pipette caused issues with the fluorescence reading. A trypsin standard of 1,000 ng/mL was used in every well. The substrate was pipetted with an 8-channel pipette over 160 s and the plate was incubated at room temperature for an hour before being read with a plate reader. The fluorescence intensity of the trypsin samples of the same concentration decreased consistently until 105 s, then the fluorescence intensity decreased precipitously (Figure 2). To circumvent this issue, the substrate must be delivered to all of the wells containing the samples and standards at the exact same time by using either a 96-well plate loader or a 96-channel pipette.
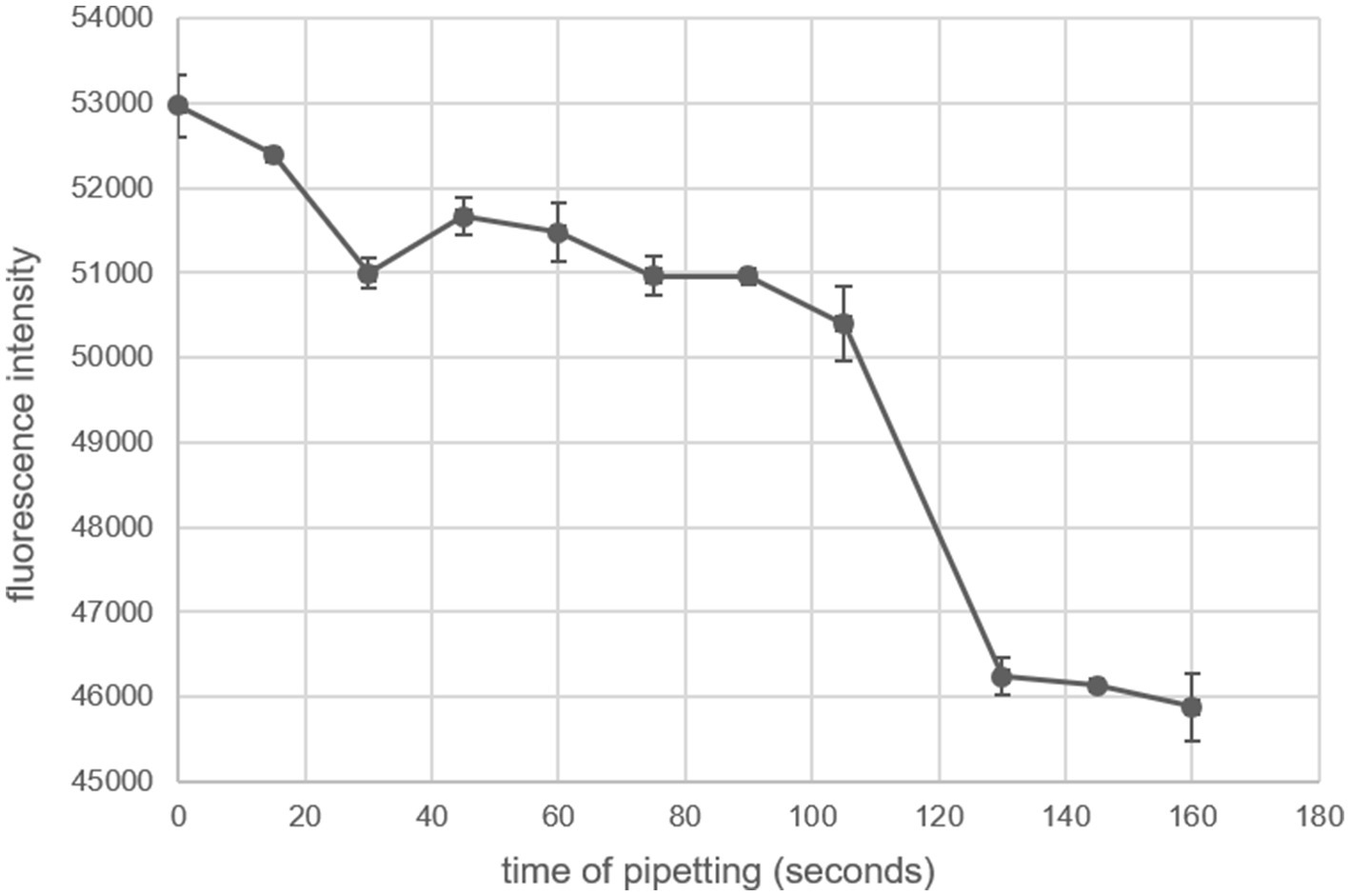
Figure 2. Effect of substrate delivery timing on fluorescence intensity. The FITC-casein substrate was pipetted with an 8-channel pipette at different times onto a 96-well plate containing 1,000 ng/mL trypsin in 0.1 M PB buffer pH 7.4. Time of pipetting is relative to the first delivery of substrate at time = 0. The plate was incubated for 1 h at room temperature. Error bars are standard error; each timepoint is the mean of eight technical replicates.
3.4 Data analysis
Heteroscedasticity of the trypsin standard curve was tested using the Breusch-Pagan test. The curve was found not to be heteroscedastic and therefore assumed to be homoscedastic (p-value was 0.3142; Supplementary Figure S1A). The regression equations were therefore used directly for calculating the protease activity of the plant samples because transformation of the data was not necessary to collapse the differences in variation.
It was also determined that including a blank (defined as 0 ng/mL trypsin incubated with the substrate) was not necessary for subsequent calculations. First, the standard curves are typically fit with either polynomial or power equations, which precludes inclusion of a zero concentration datapoint. It was also previously determined that the lowest concentration of trypsin that should be included in the standard curve is 7.8125 ng/mL (data not shown). Subtracting the blank fluorescence from the rest of the standard curve fluorescence data reveals a similar regression equation and does not impact the plant sample calculations. For example, when a protease assay of the trypsin standard curve and an alfalfa sample was performed in MES buffer pH 6.3, the regression equations were the same with and without the blank fluorescence subtracted, except for the intercept (Supplementary Figure S1B). Furthermore, the calculated protease activity of diluted plant samples was very similar when calculated using the standard curve/ plant sample without and with subtracting the blank (451.93 and 451.94, respectively). Therefore, including a blank seems dispensable and this could save room on the 96-well plate for more plant samples.
The time of incubation was also found to be an important factor to consider. It was found that the trypsin standard curve changes shape over time as the substrate gets converted to product. At very early timepoints, the curve is quadratic, opening upward (a > 0), then the curve is quadratic opening downward (a < 0). Finally, the curve becomes a power curve as it levels off as the reaction goes to completion at higher trypsin concentrations (Supplementary Figure S2). It was determined that using a reading when the trypsin standard curve is quadratic opening upward yields the most consistent results when the plant samples are calculated. The exact timing of this is variable and factors such as ambient temperature may affect it. Taking a few readings over time will help ensure that a reading is taken at the proper time (for example, five readings taken between 40 min and 90 min).
3.5 Protease activity of alfalfa samples over 24 h of wilting
Protease activity of greenhouse grown alfalfa samples harvested and wilted over five timepoints (0 h, 2 h, 4 h, 5 h, and 24 h) was tested at two different pHs. There were three biological replicates per time point. Each sample also had three technical replicates on the assay plate. The protease assay was performed in 0.1 M PB pH 7.4 and 5 mM MES buffer pH 6.3. The relative protease activity at pH 6.3 was approximately 13 to 40 times greater than the activity of the same samples at pH 7.4. At pH 7.4, the protease activity decreased steadily over the wilting time period. At pH 6.3, the protease activity stayed relatively high over the 24-h wilting period (Figure 3).
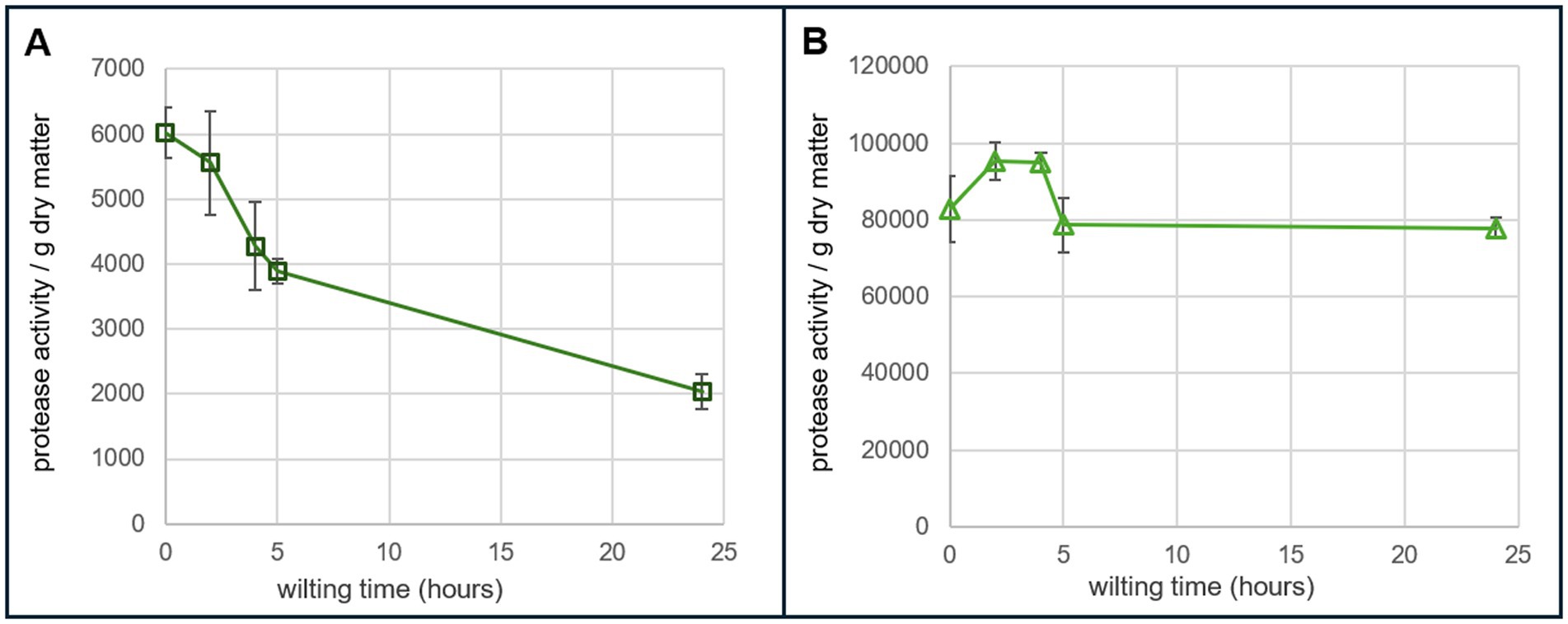
Figure 3. Protease activity in alfalfa over 24 h of wilting. (A) Protease activity per gram of dry matter of greenhouse grown alfalfa performed in 0.1 PB buffer at pH 7.4. The equation of trypsin standard curve that was used for calculations was y = 20905.45x0.119334 with an R-squared of 0.995. (B) Protease activity per gram of dry matter of greenhouse grown alfalfa performed in 5 mM MES buffer at pH 6.3. The equation of trypsin standard curve that was used for calculations was y = −0.00998x2 + 45.52x + 16,670 with an R-squared of 0.9933. Each timepoint consisted of three biological replicates and three technical replicates per biological replicate.
The same wilting samples were tested again in MES buffer pH 6.3 in order to determine repeatability of the assay. For the biological replicates within and the mean of the wilting timepoints between plates, the precisions were greater than 80%, based on the mean absolute deviation. The largest deviation value was between the two-hour wilting point samples (19.97%). This timepoint seems particularly variable and may make sense based on what we know about increases in protease activity at harvest, although more testing should be done at this timepoint. In contrast, the smallest calculated deviation value was 0.53%, which was the between-plate deviation of the zero-hour wilting timepoint; the flash frozen plant samples that were not allowed to wilt.
3.6 Limitations of the assay
This assay measures protease activity of a plant sample relative to a trypsin, a standard protease. The disadvantage of this assay is that it is not quantitative and therefore accuracy cannot be determined. When screening many plant samples across numerous plates, each plate must have a trypsin standard curve under the same conditions as the samples.
Trypsin is a well-studied, pure protease and the plant samples that are tested are a complex mixture of many different proteases found in a dynamic environment. The enzyme kinetics between the two samples are inherently different and therefore the comparison is not exact. This assay may benefit from additional changes that could mitigate some of the effects of these issues.
4 Discussion
This fluorescence-based protease assay has been developed for screening green plant samples, which is not feasible with current methods. This protocol increases the throughput from about 25 samples per day using casein and Folin’s reagent, to approximately 120 plant samples per day over three 96-well plates. Furthermore, this assay has increased sensitivity because the product of the substrate hydrolysis is fluorescent as opposed to colorimetric. The disadvantage of this increased sensitivity is the effect that time of pipetting has on fluorescence readings. This is overcome through delivering the substrate to the samples and standards on the plate at the same time with a 96-channel plate loader. Another challenge is that plant sample autofluorescence interferes with the fluorescence reading (Donaldson, 2020). This is addressed by removing interfering substances in the plant samples with activated charcoal.
Furthermore, the assay measures protease activity of a plant sample relative to a trypsin, a standard protease. A trypsin standard curve that is typically carried out over 16 wells on the 96-well plate, two replicates for each of eight concentrations, is required for each plate that is run. Depending on incubation time, ideally between 10 and 60 min, the type of regression applied to the standard curve may be different. Ideally, the plate should be measured when the shape of the curve is a parabola opening down (quadratic equation with a < 0). This increases consistency during the data analysis. The R-squared values of the standard curves are typically greater than 0.9.
Another important aspect to note is that this assay measures proteolysis potential of a sample. The protein that is broken down (FITC-casein) is added into the system exogenously and therefore this does not measure, directly or indirectly, the amount of endogenous protein that is broken down. There are different mechanisms that plants use to restrict protein breakdown via proteases (Fernández-Fernández et al., 2023). One such method is restricting substrate access through binding to specialized metabolites. One example of this is o-quinone in red clover (Sullivan and Hatfield, 2006). In a case such as red clover, the protease activity assay may suggest that protein breakdown in the plant is higher than actual plant protein degradation. This is because the added substrate would not be bound to the specialized metabolite and therefore would not be protected from protease activity. In these cases, it is important to verify results by directly measuring endogenous protein degradation, such as by measuring soluble nonprotein nitrogen generation in a sample or quantifying intact protein in a sample.
The developed assay was applied to alfalfa samples that were wilted over a period of 24 h. A previous study investigated protease activity of alfalfa over three wilting timepoints using a method that measures change in absorbance after the addition of azocasein as the substrate; it was carried out at a low pH of 4.5 to mimic silage conditions (Papadopoulos and McKersie, 1983). At pH 4.5, protease activity in alfalfa steadily decreases over a 24-h period of wilting but remains high compared to other forages. In the present study, the protease activity of alfalfa was measured at five time points over 24 h of wilting at pH 7.4 and pH 6.3 (Figure 3). pH 7.4 was chosen to test the fluorescence of the assay because overall FITC fluorescence is higher at neutral-to-alkaline pH. pH 6.3 was chosen in order to test the efficacy of the assay at plant physiological pH and also to gain an understanding of the action of plant proteases within the plant during wilting, which has not been investigated previously. Proteases are sensitive to pH and different classes function optimally at different pHs. For pH 7.4, the overall protease activity decreased over 24 h; similar to what was seen at pH 4.5. At pH 6.3, the protease activity was approximately 13 to 40 times higher compared to the activity at pH 7.4 and stays very high even at 24 h (Figure 3). This indicates that the proteases within the plant are more active at plant physiological pH (pH 6.3) than pH 7.4 after harvest and this is captured with the protease assay.
Importantly, in PB pH 7.4, trypsin hydrolyzes the FITC-casein substrate rapidly and even at 10 min of incubation the standard curve leveled off. Furthermore, the plant samples did not perform well at this pH, which can be seen by the low calculated protease activity in Figure 3. This highlights the importance of testing various buffers and pHs to ensure that the conditions are appropriate for the samples being tested.
The method presented here is a cost-effective, medium throughput fluorescence intensity-based protease activity assay optimized for green plant tissue. This method will allow one person to process approximately 5–6 times more samples per day (120 vs. 25) from green plant tissue without interference from chloroplast/chlorophyll autofluorescence. We have used this method to assess protease activity during wilting in alfalfa at pH 7.4 and pH 6.3 to alleviate the analysis bottleneck limiting further study of alfalfa as a source of human-edible protein.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.
Author contributions
CS-A: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Resources, Supervision, Writing – original draft, Writing – review & editing. AM: Investigation, Writing – original draft, Writing – review & editing. KP-B: Conceptualization, Resources, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was funded through congressional allocation to USDA ARS project 5090-21500-001-000-D. The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Acknowledgments
The authors would like to thank Melanie Katawczik for her invaluable troubleshooting help and overall guidance.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fsufs.2024.1496189/full#supplementary-material
References
Bowley, S. R., and McKersie, B. D. (1987). Genetic variance of proteolytic activity in alfalfa herbage. Can. J. Plant Sci. 67, 159–165. doi: 10.4141/cjps87-019
Brasil, I. M., and Siddiqui, M. W. (2018). “Chapter 1 – Postharvest quality of fruits and vegetables: An overview” in Preharvest modulation of postharvest fruit and vegetable quality. ed. M. W. Siddiqui (Cambridge, Massachusetts: Academic Press), 1–40.
Buet, A., Costa, M. L., Martínez, D. E., and Guiamet, J. J. (2019). Chloroplast protein degradation in senescing leaves: proteases and lytic compartments. Front. Plant Sci. 10:747. doi: 10.3389/fpls.2019.00747
Cupp-Enyard, C. (2008). Sigma’s non-specific protease activity assay – casein as a substrate. J. Vis. Exp. 19:e899. doi: 10.3791/899
Edwards, R. H., Miller, R. E., De Fremery, D., Knuckles, B. E., Bickoff, E. M., and Kohler, G. O. (1975). Pilot plant production of an edible white fraction leaf protein concentrate from alfalfa. J. Agric. Food Chem. 23, 620–626. doi: 10.1021/jf60200a046
Fernández-Fernández, Á. D., Stael, S., and Van Breusegem, F. (2023). Mechanisms controlling plant proteases and their substrates. Cell Death Differ. 30, 1047–1058. doi: 10.1038/s41418-023-01120-5
Hadidi, M., Orellana Palacios, J. C., McClements, D. J., Mahfouzi, M., and Moreno, A. (2023). Alfalfa as a sustainable source of plant-based food proteins. Trends Food Sci. Technol. 135, 202–214. doi: 10.1016/j.tifs.2023.03.023
Jones, B. A., Hatfield, R. D., and Muck, R. E. (1995). Characterization of proteolysis in alfalfa and red clover. Crop Sci. 35, 537–541. doi: 10.2135/cropsci1995.0011183X003500020043x
Liu, R., Zuo, X., Chen, Y., Qian, Z., Xu, C., Wang, L., et al. (2022). Transcriptional regulation in leaves of cut Chrysanthemum (Chrysanthemum morifolium) ‘FenDante’ in response to post-harvest ethylene treatment. Horticulturae 8:573. doi: 10.3390/horticulturae8070573
Maillard, A., Diquélou, S., Billard, V., Laîné, P., Garnica, M., Prudent, M., et al. (2015). Leaf mineral nutrient remobilization during leaf senescence and modulation by nutrient deficiency. Front. Plant Sci. 6:317. doi: 10.3389/fpls.2015.00317
McKersie, B. D. (1985). Effect of pH on proteolysis in ensiled legume forage. Agron. J. 77, 81–86. doi: 10.2134/agronj1985.00021962007700010019x
Muck, R. E. (1987). Dry matter level effects on alfalfa silage quality. Trans. ASAE 30:7-0014. doi: 10.13031/2013.30393
Muck, R. E., Moser, L. E., and Pitt, R. E. (2003). “Postharvest factors affecting ensiling” in Silage science and technology (John Wiley & Sons, Ltd), 251–304.
Papadopoulos, Y. A., and McKersie, B. D. (1983). A comparison of protein degradation during wilting and ensiling of six forage species. Can. J. Plant Sci. 63, 903–912. doi: 10.4141/cjps83-114
R Core Team (2021). R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing.
Roberts, I. N., Caputo, C., Criado, M. V., and Funk, C. (2012). Senescence-associated proteases in plants. Physiol. Plant. 145, 130–139. doi: 10.1111/j.1399-3054.2012.01574.x
Scalet, M., Alpi, A., and Picciarelli, P. (1984). Proteolytic activities in alfalfa (Medicago sativa L.) leaves. J. Plant Physiol. 116, 133–145. doi: 10.1016/S0176-1617(84)80070-X
Sullivan, M. L., and Hatfield, R. D. (2006). Polyphenol oxidase ando‐diphenols inhibit postharvest proteolysis in red clover and alfalfa. Crop Sci. 46, 662–670. doi: 10.2135/cropsci2005.06-0132
Twining, S. S. (1984). Fluorescein isothiocyanate-labeled casein assay for proteolytic enzymes. Anal. Biochem. 143, 30–34. doi: 10.1016/0003-2697(84)90553-0
Zeileis, A., and Hothorn, T. (2002). Diagnostic checking in regression relationships. R News. Available at: https://CRAN.R-project.org/doc/Rnews/ (accessed December 17, 2024).
Keywords: protease, proteolysis, alfalfa, post-harvest, perennial agriculture
Citation: Stonoha-Arther C, Molodchenko A and Panke-Buisse K (2025) A medium-throughput protease assay for screening green plant material. Front. Sustain. Food Syst. 8:1496189. doi: 10.3389/fsufs.2024.1496189
Edited by:
Mohamed Ait-El-Mokhtar, University of Hassan II Casablanca, MoroccoReviewed by:
Marta R. M. Lima, Virginia Tech, United StatesJuan Valente Hidalgo Contreras, Colegio de Postgraduados (COLPOS), Mexico
Copyright © 2025 Stonoha-Arther, Molodchenko and Panke-Buisse. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Christina Stonoha-Arther, Y2hyaXN0aW5hLmFydGhlckB1c2RhLmdvdg==
 Christina Stonoha-Arther
Christina Stonoha-Arther Andrew Molodchenko
Andrew Molodchenko Kevin Panke-Buisse
Kevin Panke-Buisse