- 1College of Life Sciences, Inner Mongolia Agriculture University, Hohhot, Inner Mongolia, China
- 2Plant Protection Research Institute, Inner Mongolia Academy of Agricultural and Animal Husbandry Sciences, Hohhot, Inner Mongolia, China
Plant roots shape the rhizosphere microbiome, recruiting microbes with beneficial functions. While genetically engineered crops offer yield advantages, their impacts on rhizosphere microbial communities remain understudied. This study evaluated the effects of transgenic maize, alongside a non-transgenic counterpart, on rhizosphere bacterial and fungal community composition using 16S rRNA and ITS amplicon sequencing. Additionally, glyphosate was used to evaluate its impact on microbial assembly and the magnitude of its effect at various maize growth stages. The results showed that transgenic maize D105 line significantly increased bacterial alpha diversity but not fungal diversity. Beta diversity analysis showed clear separation between bacterial and fungal communities at higher glyphosate treatment. Specific bacterial taxa such as Pseudomonas and Sphingomonas were enriched, while fungal taxa such as Ascomycota, Lasiosphaeriaceae, Verticillium were differentially abundant in glyphosate treatments. LEfSe analysis identified distinct enrichment patterns of bacterial (Proteobacteria and Actinobacteria) and fungal taxa (Verticillium and Guehomyces) associated with the transgenic line and glyphosate levels. KEGG functional analysis suggested potential impacts on bacterial metabolic pathways and shifts in fungal trophic modes (saprotrophs, pathogens) within the rhizosphere microbiome. This research provides insights into the classification, functional relationships, and underlying mechanisms shaping microbial communities carrying insect resistance and glyphosate resistance traits.
1 Introduction
Maize (Zea mays L.) is the most widely planted, potentially high-yield, and useful grain crop, and approximately one billion tons of maize are produced every year across the world (Warren et al., 2019). To increase the yield of maize, the farmland always be fertilized and watered which induces the overgrowth of weeds. Weeds are the major biotic barriers as they compete with maize for sunlight, water, and nutrients (Martin, 2020; Zhu et al., 2020). In China, approximately 27 million hectares of maize were seriously affected by weeds every year, leading to a 20–30% decrease in maize yield.
With the global cultivation of transgenic crops, the herbicide-tolerant transgenic maize, containing a glyphosate-insensitive 5-enolpyruvyl-shikimate-3-phosphatase synthase (EPSPS) gene, was widely planted for weed management (Staller and Staller, 2010; Vázquez et al., 2021). Glyphosate-tolerant transgenic maize offers benefits such as effective weed control through the use of glyphosate, a broad-spectrum herbicide, without damaging the crop. This can lead to higher yields, simplified weed management, and environmental benefits such as reduced soil erosion and carbon sequestration through conservation tillage practices enabled by glyphosate tolerance (Brookes and Dinh, 2021; Yu et al., 2023). However, there are concerns about the development of glyphosate-resistant weeds, reducing the technology’s effectiveness over time. The potential impact on non-target organisms such as beneficial insects, soil microorganisms, and aquatic life, especially at higher application rates, is a concern (Wang et al., 2022). While glyphosate residues are generally considered safe at established tolerance levels, there are concerns about potential health risks from long-term exposure and environmental accumulation. Issues of monopolization, higher seed costs, and public skepticism due to perceived risks and ethical concerns can also hinder adoption.
The soil rhizosphere harbors a diverse and intricate microbial community that plays a pivotal role in various ecological processes, including oxidation, nitrification, ammonification, nitrogen fixation, and sulfurization. These microorganisms are highly sensitive to the secretion of exogenous gene products by transgenic crops (Dilnashin et al., 2020). Assessing the changes in soil microbial community structure and diversity is of paramount significance for evaluating the environmental safety of transgenic crops. Previous studies have reported varying effects of transgenic plant releases on soil microbial communities (Zhang et al., 2019). While numerous studies have demonstrated no significant or only minor and transient changes in the soil microbial community, others have observed major consequences on soil microbial populations, soil enzyme activities, or the structure of a microbial community (Bunge et al., 2006). These divergent findings underscore the complexity of soil ecosystems, which represent one of the richest microbial communities on Earth. Consequently, further research on a case-by-case basis is necessary to evaluate the effects of new transgenic plant releases on soil microorganisms, with a particular emphasis on rhizosphere microbial communities (Eichmann et al., 2021).
The rhizosphere is a hotspot for complex plant–microbe interactions influencing plant growth and nutrient cycling (Hinsinger et al., 2009; Mommer et al., 2016). Within this zone, root exudates facilitate selective enrichment of microbial taxa, known as the rhizosphere effect. Given the inconsistent effects of transgenic crops on soil microbial communities, a study investigated a double-transgenic insect-resistant and glyphosate-tolerant maize variety. To combat this issue, a double-transgenic insect-resistant and glyphosate-tolerant maize variety containing the EPSPS and Cry1ab genes was selected (Mesnage et al., 2013). Using high-throughput sequencing, its effects on the composition and function of the soil microbiome of maize across the soil–plant continuum were determined (Jia et al., 2017). At the seedling stage and tasseling stage, a field experiment was conducted for a comprehensive analysis of microbial communities comprising bacterial and fungal biomasses. In addition, glyphosate has been reported to contaminate soil and potentially affect the structure and function of microbial communities (Peiffer et al., 2013; Trivedi et al., 2021). Therefore, glyphosate was used to evaluate its impact on microbial assembly and the magnitude of its effect at various maize growth stages.
2 Materials and methods
2.1 Plant materials
The transgenic maize line Yuanke 105D, containing an EPSPS gene and a Cry1ab gene, confers glyphosate-tolerant and lepidopteran insect resistance. In total, 41% glyphosate isopropylammonium salt (PD20160174), a broad-spectrum systemic herbicide and crop desiccant, was obtained from Beijing Zhongbao Green Agriculture Technology Group Company. The wild maize line Yuanke 105 was used as a control in this study. A field trial was conducted at the Biological Breeding Research and Experimental Base of the Inner Mongolia Academy of Agricultural and Animal Husbandry Sciences, which was situated in Tuoxian town, Hohhot City, China (43°30’N, 111°23′E). The experimental plots were established on a saline tidal soil, which were characterized by an approximate water content of 11%, an organic matter content of 6 g/kg, and a soil pH ranging from 8.2 to 8.3.
2.2 Experiment design and sampling
The field experiment was conducted using a structured shelter planting method, adhering to local conventional farming management practices. Each experimental plot measured 30 m2, with a spacing of 120 cm between adjacent plots. The maize was sown in rows spaced 50 cm apart, with a plant-to-plant distance of 25 cm within each row. The transgenic maize line Yuanke 105D, harboring the EPSPS gene for glyphosate tolerance, was subjected to varying glyphosate application rates at the seedling stage. An HD400 sprayer equipped with a fan nozzle was employed for the foliar application of glyphosate at rates of 0 g/ha (control), 922 g/ha, 1,230 g/ha, and 1,537 g/ha to the respective Yuanke 105D plots.
To investigate the potential effects of the transgenic maize line and glyphosate application on the rhizosphere soil microbial community, soil samples were collected at seedling and tasseling stage. There are five maize plots with different treatments, named 105, 105D-0, 105D-150, 105D-200, and 105D-250. Rhizosphere soil samples were obtained by carefully excavating the root systems and brushing off the soil tightly adhering to the root surface. Six biological replicates were collected from each plot, thoroughly mixed, and placed in plastic bags containing ice packs to maintain sample integrity during transportation.
2.3 Metagenomic DNA extraction from rhizosphere soil samples
Total metagenomic DNA was extracted from approximately 0.5 g of maize rhizosphere soil samples using the HiPure Universal DNA Kit, following the manufacturer’s protocol. This commercial kit employs a robust and standardized methodology for efficient recovery of high-quality DNA from environmental samples. The concentrations of the metagenomic DNA obtained from each biological replicate were quantified using a NanoDrop 2000 spectrophotometer (García-Alegría et al., 2020). The completeness of DNA was determined by 1% agarose gel electrophoresis. These high-quality metagenomic DNA samples with high integrity and concentrations exceeding 15 ng/μl were stored at −80°C to ensure long-term preservation and minimize potential degradation.
2.4 rDNA amplification and sequencing
To characterize the bacterial and fungal communities present in the rhizosphere soil samples, amplicon sequencing targeting the 16S rRNA gene and internal transcribed spacer (ITS) regions was performed (Quast et al., 2013). All DNA samples were dispatched to Nanjing Genepioneer Biotechnologies (Nanjing, China) for library preparation and sequencing on the Illumina MiSeq platform, utilizing the MiSeq Reagent Kit. For profiling the bacterial communities, the full-length 16S rRNA gene was amplified using the universal primer pair 341F (5’-CCTACGGGNGGCWGCAG-3′) and 806R (5’-GGACTACHVGGGTWTCTAAT-3′) (Yilmaz et al., 2014). To assess the fungal community composition, the internal transcribed spacer (ITS) region 1 was amplified using the primers ITS1F (5’-CTTGGTCATTTAGAGGAAGTAA-3′) and ITS1R (5’-GCTGCGTTCTTCATCGATGC-3′). The ITS region, located between the small and large subunit rRNA genes, is a widely used marker for fungal identification and taxonomic classification due to its high degree of sequence variation among different fungal species (Nilsson et al., 2019).
2.5 Data analyses
The 16S rRNA gene and ITS amplicon sequencing data were processed using a comprehensive bioinformatics pipeline to characterize the bacterial and fungal communities present in the rhizosphere soil samples. The bacterial 16S rRNA gene sequences were analyzed using the SILVA 138/16S rRNA database for taxonomic assignment (Schloss et al., 2009; Cole et al., 2014). Quality control measures were implemented to ensure data integrity, including the detection and removal of chimeric sequences using UCHIME (v1.9.1) software. Alpha diversity indices, such as Shannon and Chao1, were calculated using MOTHUR (v1.30.2) software to assess the richness and diversity of bacterial communities across samples. Differences in richness and diversity between experimental groups were statistically evaluated using two-way analysis of variance (ANOVA) with Tukey’s post-hoc test. For the analysis of fungal communities, the ITS amplicon sequences were taxonomically assigned using the Unite 8.0/ITS fungi database. To investigate the overall community composition and assess the similarities or dissimilarities between samples, beta diversity analysis was performed using principal coordinate analysis (PCoA) implemented in the QIIME (v1.9.1) software package (Chen et al., 2021). The resulting PCoA plots were generated using the R packages “vegan,” “ecodist,” and “ggplot2” in R (v4.1.3), enabling the visual representation and interpretation of beta diversity patterns (Adams et al., 2013).
Data were subjected to a one-way analysis of variance with factors of treatments and expressed as means + SD. Comparisons between any two groups were performed by unpaired Student’s t-tests. **Indicates a significant difference at p-values of 0.01 compared with the control. *Indicates a significant difference at p-values of 0.05 compared with the control.
3 Results
3.1 Overall analysis of 16S rDNA
A total of 601,358 amplicon sequence variants (ASVs) were generated using the 341F-806R primer pair. On average, each rhizosphere soil replicate sample yielded 60,135 ASV pairs, providing a robust dataset for downstream analysis of the bacterial community composition and diversity. For the characterization of fungal communities, the ITS1F-ITS1R primer pair, targeting the ITS region, was employed. This approach resulted in the generation of 1,087,847 ASV pairs, with an average of 108,784 ASV pairs per rhizosphere soil replicate sample (Supplementary Table S1).
3.2 Alpha diversity analysis of rhizosphere microbial communities
The assessment of alpha diversity, which reflects the richness and evenness of species within a community, was conducted for the rhizosphere soil microbial communities at the seedling and tasseling stages of maize growth. The rarefaction analysis, based on the normalized observed amplicon sequence variants (ASVs), Chao1, and Shannon indices, revealed that the rarefaction curves nearly reached saturation plateaus. The mean and standard deviation of these indices were calculated based on all biological replicates of the rhizosphere soil samples. This approach enabled the assessment of changes in microbial community richness and diversity associated with the different maize lines under investigation.
The non-transgenic control maize line (105) did not significantly influence bacterial richness or diversity, as evidenced by the Chao1 and Shannon indices, at both the seedling and tasseling stages (Figures 1A,B). However, the transgenic maize line (D105) exhibited a significant increase in bacterial Shannon diversity during the flowering/tasseling stage (Figure 1B), suggesting a potential impact on the evenness of bacterial taxa within the rhizosphere community. In contrast to the observed effects on bacterial communities, neither the non-transgenic control (105) nor the transgenic line (D105) had a significant impact on fungal alpha diversity, as measured by the Chao1 and Shannon indices, at both the seedling and tasseling stages (Figures 1C,D). This finding indicates that the richness and evenness of fungal taxa within the rhizosphere were not substantially altered by the presence of the transgenic maize line.
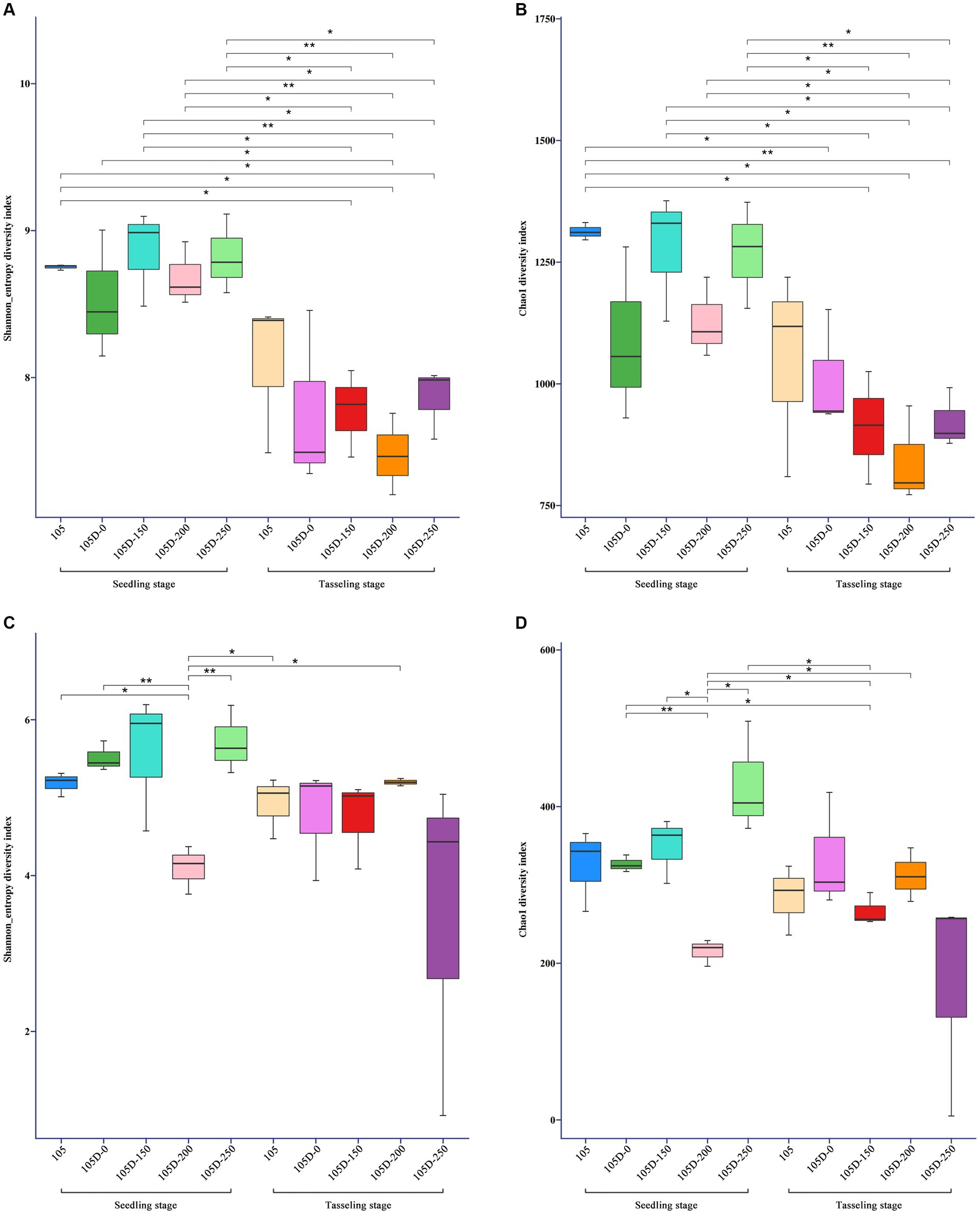
Figure 1. The Chao 1 (species richness) and Shannon (species diversity) indexes of bacterial (A,B) and fungal (C,D) communities within rhizosphere soil compartments of control 105 and glyphosate-treated transgenic 105D maize lines at seedling stage and tasseling stage. One hundred and five represents the wild maize line Yuanke 105. 105D-0, 105D-150, 105D-200, and 105D-250 represent the transgenic maize lines Yuanke 105D treated with 0, 922, 1,230, and 1,537 g of glyphosate per hectare. *Significant at the 0.05 probability level. **Significant at the 0.01 probability level.
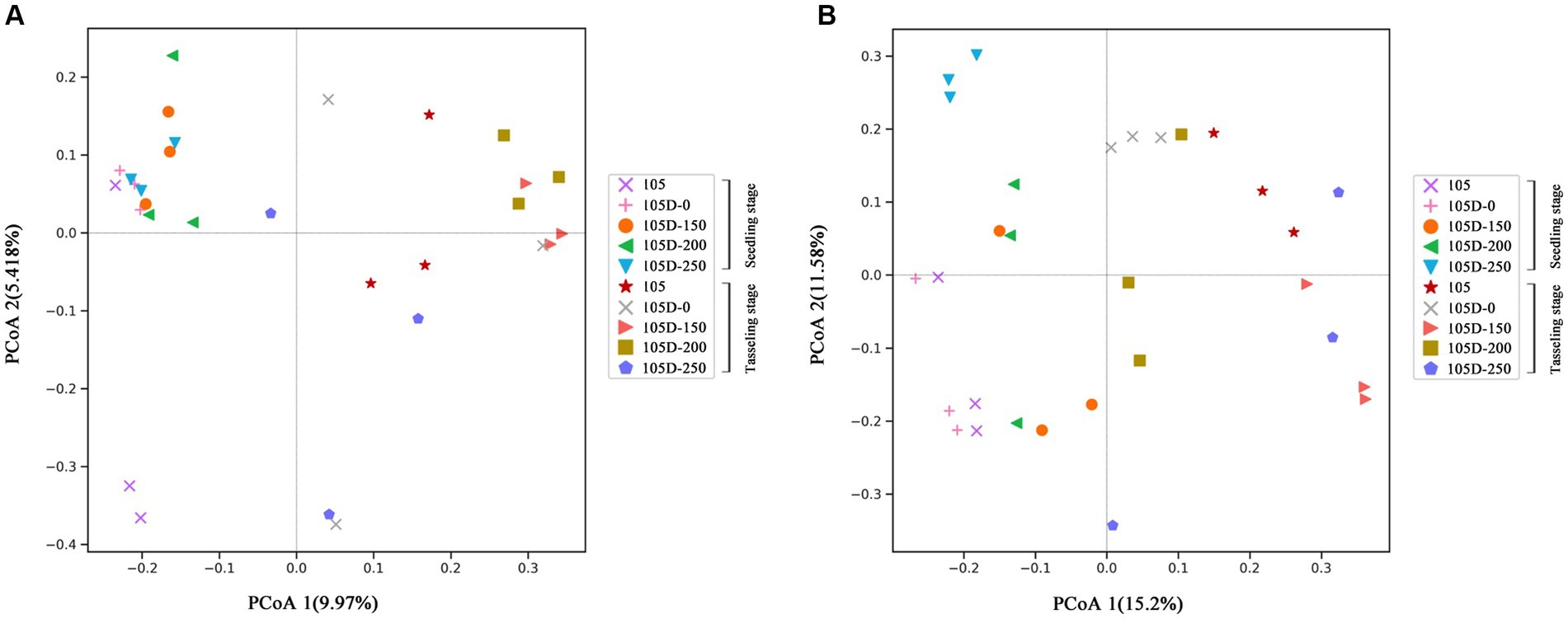
Figure 2. Principal component analysis based on the distance matrix calculated using the Bray–Curtis distance matrix for rhizosphere soil. The rhizosphere soil compartments cause the largest source of variance among bacterial (A) and fungal (B) communities of control 105 and glyphosate-treated transgenic 105D maize lines at seedling stage and tasseling stage.
3.3 Beta diversity analysis of rhizosphere microbial communities
To investigate the differences in microbial community composition among the rhizosphere soil samples, beta diversity analysis was performed using principal coordinate analysis (PCoA). The PCoA based on bacterial amplicon sequence variant (ASV) composition revealed a clear separation between the rhizosphere soil replicates of the non-transgenic control maize line (105) and the transgenic line (D105) with different glyphosate treatments, along the first principal coordinate axis (PCoA1) at both the seedling and tasseling stages (Figure 2A). The PCoA1 axis accounted for 15.38% of the total variance in bacterial community composition. However, within the transgenic D105 line, the bacterial ASV composition did not exhibit distinct clustering among the different glyphosate treatment levels.
The analysis of fungal community composition using PCoA demonstrated distinct patterns across the growth stages (Figure 2B). At the seedling stage, the rhizosphere soil replicates of the transgenic D105 line treated with higher glyphosate rates (105D-250, 105D-200, and 105D-150) were clearly separated from the non-transgenic control (105) along the PCoA axes. This separation became more pronounced at the tasseling stage, where the fungal communities of all transgenic D105 treatments, irrespective of glyphosate rate, were distinctly clustered apart from the non-transgenic control (105). The first principal coordinate axis (PCoA1) explained 26.78% of the total variance in fungal community composition.
3.4 Alterations in bacterial community composition
We assessed the taxonomic distributions of bacterial ASVs at different classification levels. At the phylum level, 20 dominant bacterial phyla were assigned, which comprised 98% of the entire bacterial community. The most prevalent bacterial phylum was Proteobacteria, which was then followed by Acidobacteriota, Actinobacteriota, Chloroflexi, Bacteroidota, Gemmatimonadota, Planctomycetota, and Patescibacteria (Figure 3A). The relative abundance of Proteobacteria, Actinobacteriota, and Bacteroidota increased obviously from the seedling stage to the tasseling stage, while Planctomycetota and Gemmatimonadota decreased. Altogether, the composition of bacterial communities at the phylum level has no significant differences among the rhizosphere soils of the transgenic maize 105D with different glyphosate treatments compared with the control maize 105.
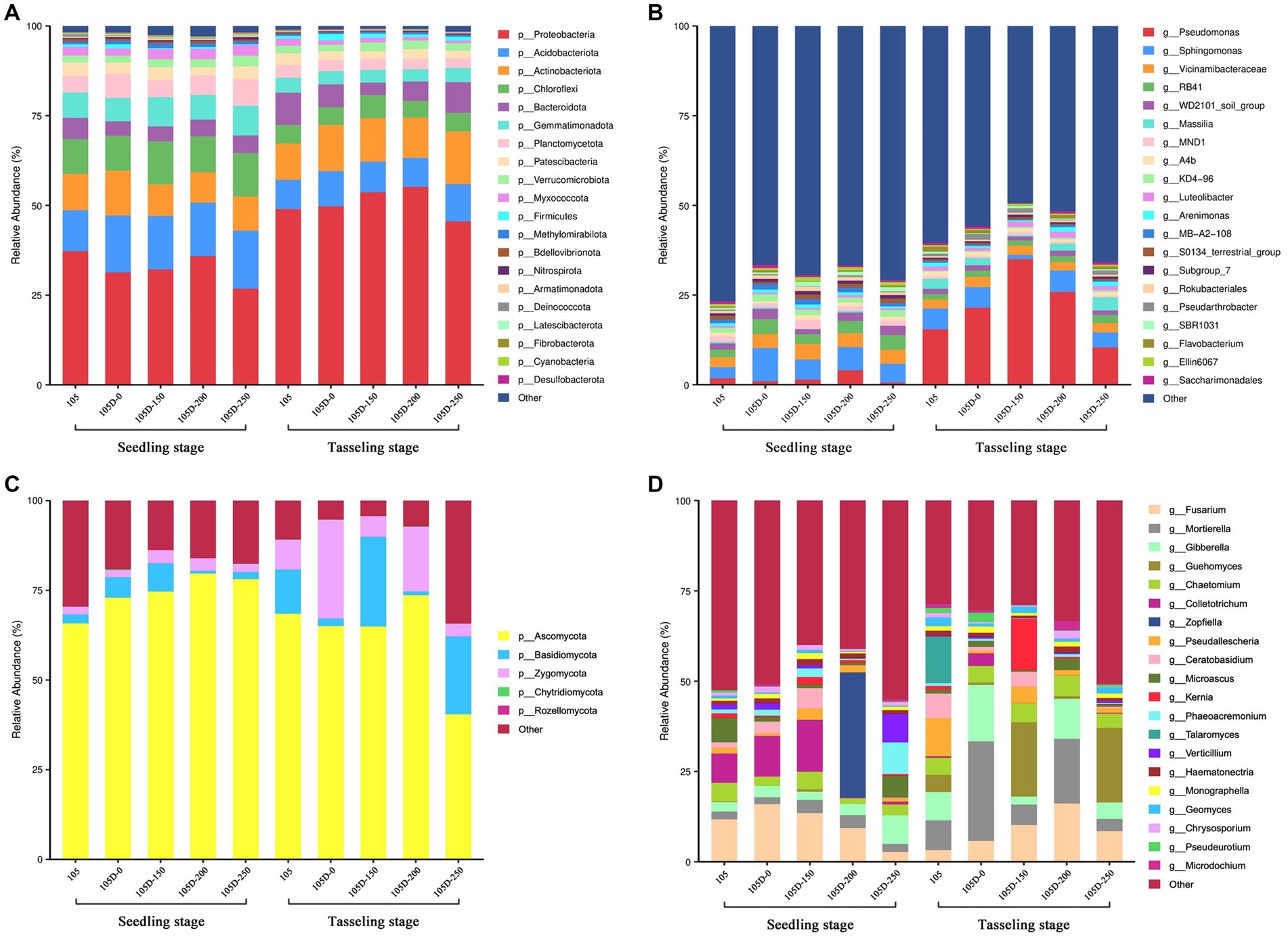
Figure 3. Composition and structure of bacterial and fungal communities of control 105 and glyphosate-treated transgenic 105D maize lines at seedling stage and tasseling stage. (A,B) Represent the phylum and genus levels of bacteria, respectively. (C,D) Represent the phylum and genus levels of fungi, respectively. The top 20 relative abundances are shown, and the remaining abundances are indicated as ‘Others’.
At the class level, 20 dominant bacterial classes were assigned, which comprised more than 50% of the entire bacterial community. The abundant classes were Sphingomonas, Vicinamibacteraceae, RB41, and WD2101 at the seedling stage, while Pseudomonas, Sphingomonas, and Vicinamibacteraceae were the most abundant bacteria at the tasseling stage (Figure 3B). The relative abundance of the Pseudomonas class increased sharply from the seedling stage to the tasseling stage.
Compared with the two growth stages, the abundance of Pseudomonas species significantly increased, and transgenic 105D showed a trend of increasing and decreasing with the concentration of glyphosate treatment. The proportion of other bacterial genera decreased, and the decrease was lower when spraying high-concentration glyphosate compared with other samples. The control 105 of Sphingomonas increased, while the transgenic 105D decreased. The decrease was slower under high-concentration glyphosate treatment. The abundance proportion of Vicinamido bacteriaceae genus and Acidobacterium RB41 slightly decreased, and there was no significant difference among the various varieties.
3.5 Alterations in fungal community composition
The taxonomic composition in the rhizosphere soil of 105D and its control 105 at the phylum level is shown in Figure 3C, and the most abundant phylum was Ascomycota at the seedling stage and tasseling stage, which was followed by Basidiomycota and Zygomycota. At the seedling stage, only the relative abundance of Ascomycota was higher in the rhizosphere soil of 105D compared with that of the control 105, which suggested that the relative abundances of Ascomycota were increased in the rhizosphere soil of 105D with glyphosate treatment (Figure 3C). At the tasseling stage, both the relative abundances of Basidiomycota and Zygomycota increased, whereas the relative abundance of Ascomycota decreased in the rhizosphere soils of 105D and control 105.
At the class level, 20 dominant fungal classes were assigned, which comprised approximately 50–75% of the entire fungal community at the seedling stage and tasseling stage. The abundant classes were Fusarium, Chaetomium, Mortierella, and Gibberella at the seedling stage, while Mortierella, Fusarium, Gibberella, and Chaetomium were the most abundant fungi at the tasseling stage (Figure 3D). The relative abundance of the Fusarium class was decreased in the rhizosphere soil of 105D with glyphosate treatment at the seedling stage, which suggested that glyphosate has a negative effect on Fusarium.
3.6 LEfSe of the bacterial and fungal communities in the rhizosphere
To gain deeper insights into the rhizosphere microbial communities associated with the transgenic maize line (D105) and different glyphosate treatment levels, we employed the linear discriminant analysis effect size (LEfSe) method. This approach enabled the identification of bacterial and fungal taxa that were differentially abundant among the non-transgenic control (105), transgenic untreated (105D-0), and transgenic lines treated with varying glyphosate concentrations (105D-150, 105D-200, and 105D-250) at both the seedling and tasseling stages.
At the seedling stage, LEfSe analysis revealed 88 distinctly abundant bacterial taxa among the 5 groups, with a linear discriminant analysis (LDA) score higher than 3.5 (Figure 4A). The non-transgenic control (105) was enriched in taxa belonging to the order Xanthomonadales and the family Xanthomonadaceae. In contrast, the transgenic untreated line (105D-0) exhibited enrichment of Acidimicrobia, Vicinamibacteria, and Vicinamibacterales. The transgenic line treated with a low glyphosate concentration (105D-150) was characterized by an abundance of Gemmatimonadales, Gemmatimonadaceae, and Gemmatimonadetes, while the highest glyphosate treatment (105D-250) favored the enrichment of Acidobacteriota, Chloroflexi, and Planctomycetes. At the tasseling stage, 46 bacterial taxa were identified as differentially abundant among the groups (Figure 4B). The non-transgenic control (105) exhibited enrichment of Bacteroidota and Bacteroidia. The transgenic line treated with a low glyphosate concentration (105D-150) was characterized by an abundance of Pseudomonas, Pseudomonadanceae, Pseudomonadales, and Gammaproteobacteria. The intermediate glyphosate treatment (105D-200) favored the enrichment of Proteobacteria, while the highest glyphosate concentration (105D-250) promoted the abundance of Alphaproteobacteria, Actinobacteria, Micrococcales, and Micrococcaceae.
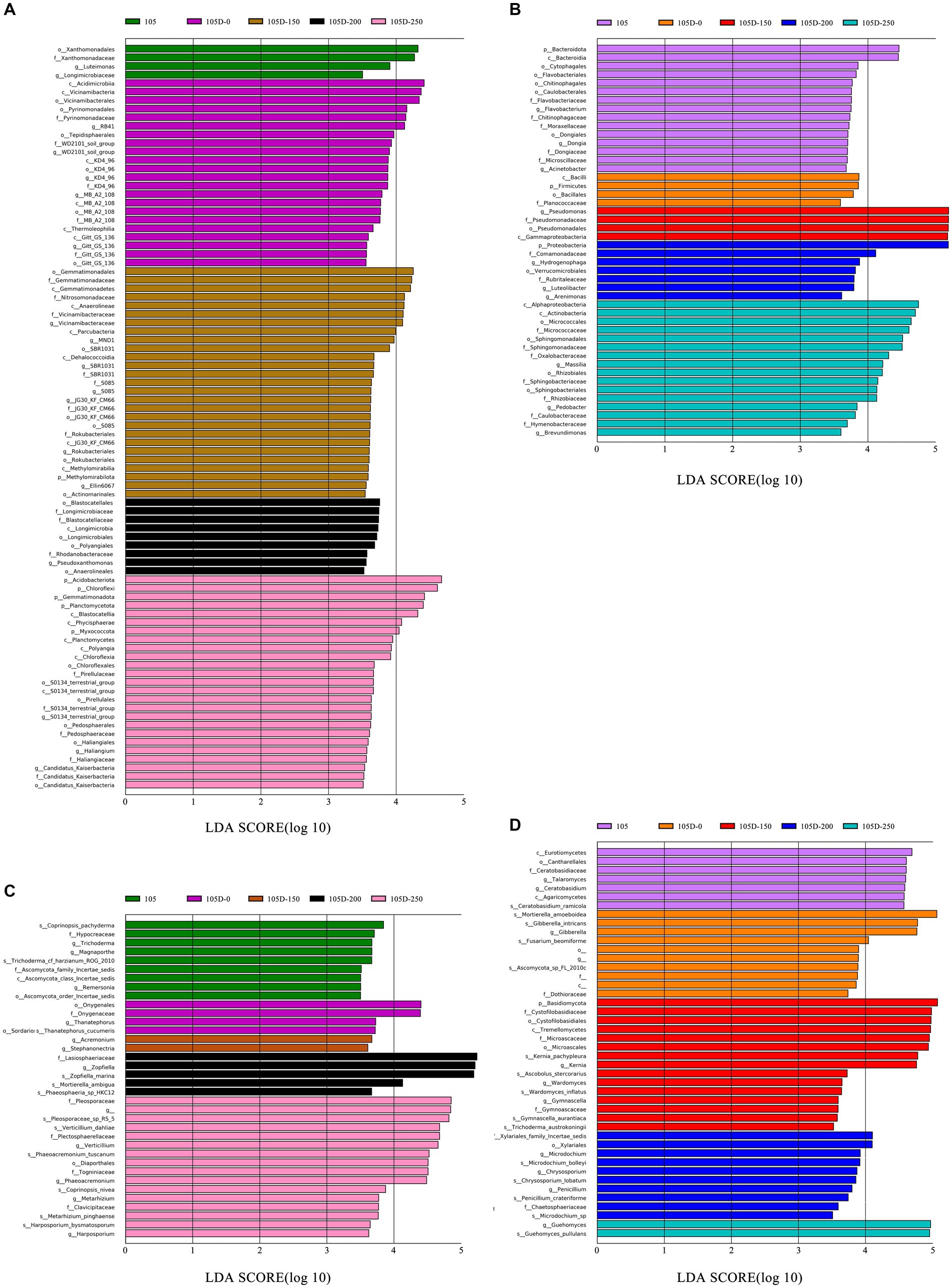
Figure 4. LEfSe analysis at multiple taxonomic levels comparing the rhizobiome soil of control 105 and glyphosate-treated transgenic 105D maize lines at seedling stage and tasseling stage. Histogram of the LDA scores greater than 3.5 computed for groups with differential abundance among the rhizobacterial communities at seedling stage (A) and tasseling stage (B). Histogram of the LDA scores greater than 3.5 computed for groups with differential abundance among the rhizofungal communities at seedling stage (C) and tasseling stage (D). The length of each bar represents the contribution of species with significant differences in abundance.
Regarding the fungal communities, 36 abundant taxa significantly differed across the groups at the seedling stage, with an LDA score higher than 3.5 (Figure 4C). The non-transgenic control (105) exhibited enrichment of nine fungal taxa. The transgenic untreated line (105D-0) was characterized by an abundance of Cnygenales and Onygenaceae. The transgenic line treated with an intermediate glyphosate concentration (105D-200) favored the enrichment of Lasiosphaeriaceae, Zopfiella, and Zopfiella_marina, while the highest glyphosate treatment (105D-250) promoted the abundance of Pleosporaceae, Plectosphaerellaceae, and Verticillium_dahliae.
At the tasseling stage, 44 fungal taxa were identified as differentially abundant among the groups (Figure 4D). The non-transgenic control (105) exhibited enrichment of Eurotiomycetes, Ceratobasidiaceae, and Cantharellales. The transgenic untreated line (105D-0) was characterized by an abundance of Mortierella_amoeboidea, Gibberella_intricans, and Gibberella. The transgenic line treated with a low glyphosate concentration (105D-150) favored the enrichment of Basidiomycota, Cystofilobasidiaceae, and Tremellomycetes, while the intermediate glyphosate treatment (105D-200) promoted the abundance of Xylariales. The highest glyphosate concentration (105D-250) favored the enrichment of Guehomyces and Guehomyces_pullulans.
3.7 KEGG functional annotation analysis of bacterial and fungal communities
To gain insights into the functional potential of the rhizosphere microbial communities associated with the transgenic maize line and different glyphosate treatments, we performed KEGG (Kyoto Encyclopedia of Genes and Genomes) functional annotation analysis. This approach enabled the assessment of the abundance and distribution of various metabolic pathways and trophic modes within the bacterial and fungal communities.
The abundance of KEGG primary pathway functions related to metabolism, environmental information processing, cellular processes, and genetic information processing in bacterial communities was analyzed across the non-transgenic control (105), transgenic untreated (105D-0), and transgenic lines treated with varying glyphosate concentrations (105D-150, 105D-200, and 105D-250) at both the seedling and tasseling stages (Figure 5A). Notably, the abundance of metabolic pathways was the highest across all groups, followed by genetic information processing, environmental information processing, and cellular processes. Functional systems associated with human diseases and organismal systems did not exhibit significant changes among the different groups. However, minor variations were observed in the abundance of environmental information processing pathways between the groups.
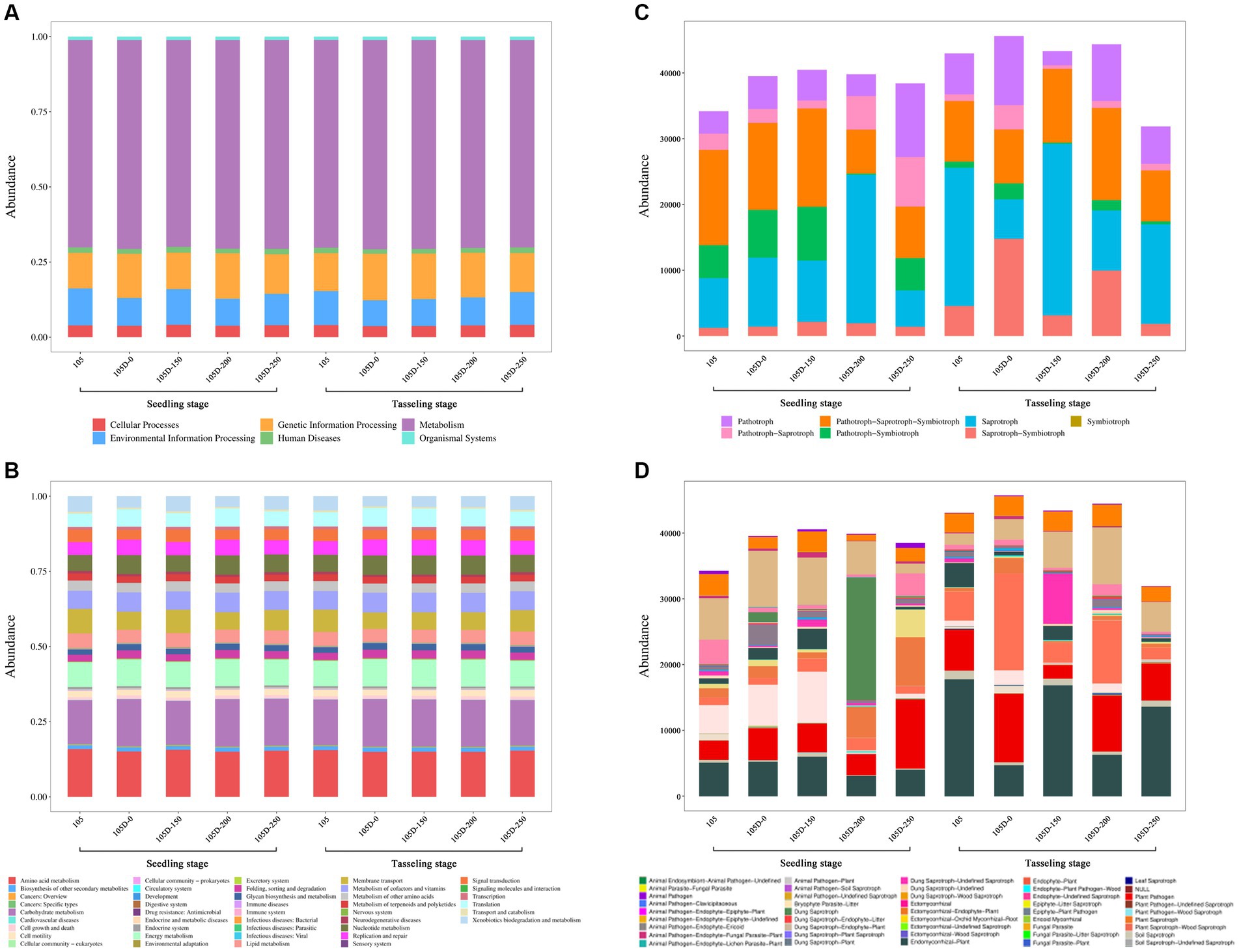
Figure 5. Functional analysis of different species in the rhizobiome soil of control 105 and glyphosate-treated transgenic 105D maize lines at the seedling stage and tasseling stage. (A) Bacterial KEGG primary functional abundance. (B) Bacterial KEGG secondary functional abundance. (C) Fungal KEGG primary functional abundance. (D) Fungal KEGG secondary functional abundance.
Furthermore, the abundance of PICRUSt2 secondary pathway functions was analyzed (Figure 5B). The pathways related to amino acid metabolism, carbohydrate metabolism, and energy metabolism were the most abundant. Notably, no obvious changes were observed among the 42 secondary pathway annotations at both the seedling and tasseling stages, suggesting a relatively stable functional profile within the bacterial communities.
The KEGG functional annotation analysis of fungal communities revealed seven trophic types: saprophytic, pathotroph, symbiotroph, pathotroph–saprotroph, pathotroph–symbiotroph, saprotroph–symbiotroph, and pathotroph–saprotroph–symbiotroph (Figure 5C). The saprophytic trophic type exhibited the highest proportion, while the pathotroph–symbiotroph trophic type had the lowest abundance. Subclass analysis of functional groups revealed that the dominant functional groups across the samples were animal pathogen–endophyte–fungal parasite–plant pathogen–wood saprotroph, animal pathogen–dung saprotroph–endophyte–epiphyte–plant saprotroph–wood saprotroph, undefined saprotroph, and plant pathogen. The observed patterns in metabolic pathways and trophic types may have implications for nutrient cycling, plant–microbe interactions, and ecosystem processes within the rhizosphere microbiome (Figure 5D).
4 Discussion
The soil microbiome plays a pivotal role in promoting plant growth and productivity, driving biogeochemical cycles, and contributing to global food security (Caporaso et al., 2010; Hu et al., 2022). As such, understanding the potential impacts of genetically modified crops on soil microbial communities is of paramount importance. This study provides a comprehensive characterization of the maize-associated microbiome, encompassing both bacterial and fungal communities, through high-throughput amplicon sequencing of field-grown samples.
Based on the sequencing of bacterial 16S rRNA and fungal ITS1 regions, principal coordinate analysis (PCoA) and Chao1 analyses revealed distinct clustering patterns among the rhizosphere microbial communities associated with the non-transgenic control (105), transgenic (D105), and different glyphosate treatment levels (105D-0, 105D-150, 105D-200, and 105D-250) at both the seedling and tasseling stages.
In this study, in addition to the non-transgenic control (105), the application of glyphosate had a substantial impact on soil characteristics and the dynamics of microbial communities. Following glyphosate application, the abundance of certain bacterial genera, such as Pseudomonas, exhibited significant changes, while the proportion of other genera, such as Sphingomonas, Vicinamidophilus, and Acidobacterium, was differentially affected depending on the glyphosate concentration and the presence of the transgenic trait. At the seedling stage, the transgenic maize line appeared to promote the abundance of Ascomycota and Basidiomycota phyla while inhibiting other fungal phyla. Conversely, at the tasseling stage, the transgenic line exhibited an inhibitory effect on Ascomycota and Basidiomycota while promoting the abundance of Fusobacteria. These findings suggest that different fungal phyla may respond differently to the presence of the transgenic trait and glyphosate stress across different growth stages.
LEfSe analysis of the rhizosphere microbial community revealed that the transgenic maize line had no significant effect on fungal communities during the seedling stage. However, low concentrations of glyphosate appeared to inhibit fungal abundance, while higher concentrations had a promoting effect. At the tasseling stage, the transgenic line promoted an increase in fungal abundance, with low glyphosate concentrations exhibiting a promoting effect and high concentrations of an inhibitory effect, demonstrating an opposite trend compared with the seedling stage (Figure 4).
Within the bacterial communities, KEGG functional annotation analysis indicated that the transgenic maize line had an inhibitory effect on the abundance of pathotrophs (plant pathogen) at both seedling and tasseling stages, with no clear pattern in response to glyphosate. The abundance and distribution of various metabolic pathways and trophic modes exhibited no difference between control and transgenic lines. However, within the fungal communities, the abundance of pathotroph–saprotroph–symbiotroph was inhibited at the seedling stage but unaffected at tasseling stage. The abundance of saprotroph (decomposer) was promoted at seedling stage but inhibited at tasseling stage by transgenic maize. Low glyphosate promoted, while high concentrations inhibited the abundance of animal pathogen–dung, saprotroph–endophyte-epiphyte–plant saprotroph–wood saprotroph, and animal pathogen–endophyte–plant undefined saprotroph. These shifts in microbial trophic modes suggest impacts on nutrient cycling, plant–microbe interactions, and rhizosphere processes, warranting further mechanistic investigations.
These findings highlight the complex interplay between genetically modified crops, herbicide application, and soil microbial communities, underscoring the importance of considering both bacterial and fungal components of the microbiome. While the introduction of transgenic traits and glyphosate application can modulate the abundance and composition of certain microbial taxa, the observed effects appear to be context-dependent, which are influenced by factors, such as plant developmental stage, soil conditions, and herbicide concentration.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Author contributions
GW: Conceptualization, Data curation, Investigation, Methodology, Project administration, Resources, Writing – review & editing. SY: Visualization, Writing – review & editing, Software, Data curation. SF: Data curation, Formal Analysis, Software, Validation, Visualization, Writing – review & editing. GZ: Data curation, Formal analysis, Investigation, Methodology, Resources, Supervision, Writing – review & editing. XHe: Data curation, Formal analysis, Investigation, Resources, Supervision, Writing – review & editing. XHa: Conceptualization, Data curation, Formal analysis, Methodology, Project administration, Resources, Supervision, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was supported by the “Germplasm Innovation and Molecular Breeding of Forage Crops and Useful Microbiology” team, project number: TD202103.
Acknowledgments
The authors thank the Plant Protection Research Institute, Inner Mongolia Academy of Agricultural and Animal Husbandry Sciences for providing a research site.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fsufs.2024.1421837/full#supplementary-material.
References
Adams, R. I., Miletto, M., Taylor, J. W., and Bruns, T. D. (2013). Dispersal in microbes: fungi in indoor air are dominated by outdoor air and show dispersal limitation at short distances. ISME J. 7, 1262–1273. doi: 10.1038/ismej.2013.28
Brookes, G., and Dinh, T. X. (2021). The impact of using genetically modified (gm) corn/maize in vietnam: results of the first farm-level survey. Gm Crops Food 12, 71–83. doi: 10.1080/21645698.2020.1816800
Bunge, J., Epstein, S. S., and Peterson, D. G. (2006). Comment on" computational improvements reveal great bacterial diversity and high metal toxicity in soil". Science 313:918. doi: 10.1126/science.1126593
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F. D., Costello, E. K., et al. (2010). Qiime allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336. doi: 10.1038/nmeth.f.303
Chen, K.-H., Longley, R., Bonito, G., and Liao, H. L. (2021). A two-step Pcr protocol enabling flexible primer choice and high sequencing yield for Illumina MiSeq meta-barcoding. Agronomy 11:1274. doi: 10.3390/agronomy11071274
Cole, J. R., Wang, Q., Fish, J. A., Chai, B., McGarrell, D. M., Sun, Y., et al. (2014). Ribosomal database project: data and tools for high throughput rRNA analysis. Nucleic Acids Res. 42, D633–D642. doi: 10.1093/nar/gkt1244
Dilnashin, H., Birla, H., Hoat, T. X., Singh, H. B., Singh, S. P., and Keswani, C. (2020). Applications of agriculturally important microorganisms for sustainable crop production. Mol. Aspects Plant Benef. Microbes Agric. 2020, 403–415. doi: 10.1016/B978-0-12-818469-1.00033-X
Eichmann, R., Richards, L., and Schäfer, P. (2021). Hormones as go-betweens in plant microbiome assembly. Plant J. 105, 518–541. doi: 10.1111/tpj.15135
García-Alegría, A. M., Anduro-Corona, I., Pérez-Martínez, C. J., Guadalupe Corella-Madueño, M. A., Rascón-Durán, M. L., and Astiazaran-Garcia, H. (2020). Quantification of Dna through the NanoDrop spectrophotometer: methodological validation using standard reference material and Sprague Dawley rat and human DNA. Int J Anal Chem 2020, 1–9. doi: 10.1155/2020/8896738
Hinsinger, P., Bengough, A. G., Vetterlein, D., and Young, I. M. (2009). Rhizosphere: biophysics, biogeochemistry and ecological relevance. Plant Soil 321, 117–152. doi: 10.1007/s11104-008-9885-9
Hu, H. W., Chen, Q. L., and He, J. Z. (2022). The end of hunger: fertilizers, microbes and plant productivity. Microb. Biotechnol. 15, 1050–1054. doi: 10.1111/1751-7915.13973
Jia, Z., Cai, Y., Yun, J., and Du, W. (2017). Comparison of soil microbiome by single cell technology, classical microscope methods and high-throughput MiSeq sequencing. Acta Microbiol Sin. 57, 899–919. doi: 10.13343/j.cnki.wsxb.20170067
Martin, R. J. (2020). Weed research issues, challenges, and opportunities in Cambodia. Crop Prot. 134:104288. doi: 10.1016/j.cropro.2017.06.019
Mesnage, R., Clair, E., Gress, S., Then, C., Székács, A., and Séralini, G. E. (2013). Cytotoxicity on human cells of Cry1Ab and Cry1Ac Bt insecticidal toxins alone or with a glyphosate-based herbicide. J. Appl. Toxicol. 33, 695–699. doi: 10.1002/jat.2712
Mommer, L., Hinsinger, P., Prigent-Combaret, C., and Visser, E. J. (2016). Advances in the rhizosphere: stretching the interface of life. Plant Soil 407, 1–8. doi: 10.1007/s11104-016-3040-9
Nilsson, R. H., Larsson, K. H., Taylor, A. F. S., Bengtsson-Palme, J., Jeppesen, T. S., Schigel, D., et al. (2019). The unite database for molecular identification of fungi: handling dark taxa and parallel taxonomic classifications. Nucleic Acids Res. 47, D259–D264. doi: 10.1093/nar/gky1022
Peiffer, J. A., Spor, A., Koren, O., Jin, Z., Tringe, S. G., Dangl, J. L., et al. (2013). Diversity and heritability of the maize rhizosphere microbiome under field conditions. Proc. Natl. Acad. Sci. 110, 6548–6553. doi: 10.1073/pnas.1302837110
Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., Schweer, T., Yarza, P., et al. (2013). The Silva ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596. doi: 10.1093/nar/gks1219
Schloss, P. D., Westcott, S. L., Ryabin, T., Hall, J. R., Hartmann, M., Hollister, E. B., et al. (2009). Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75, 7537–7541. doi: 10.1128/AEM.01541-09
Staller, J. E., and Staller, J. E. (2010). Scientific, Botanical, and Biological Research on Maize. Berlin, Heidelberg: Springer.
Trivedi, P., Leach, J. E., Tringe, S. G., Sa, T., and Singh, B. K. (2021). Author correction: plant–microbiome interactions: from community assembly to plant health. Nat. Rev. Microbiol. 19:72. doi: 10.1038/s41579-020-00490-8
Vázquez, M. B., Moreno, M. V., Amodeo, M. R., and Bianchinotti, M. V. (2021). Effects of glyphosate on soil fungal communities: a field study. Rev. Argent. Microbiol. 53, 349–358. doi: 10.1016/j.ram.2020.10.005
Wang, W., Nie, Y., Tian, H., Quan, X., Li, J., Shan, Q., et al. (2022). Microbial community, co-occurrence network relationship and fermentation lignocellulose characteristics of Broussonetia papyrifera ensiled with wheat bran. Microorganisms 10:2015. doi: 10.3390/microorganisms10102015
Warren, R. J., Candeias, M., Labatore, A., Olejniczak, M., and Yang, L. (2019). Multiple mechanisms in woodland plant species invasion. J. Plant Ecol. 12, 201–209. doi: 10.1093/jpe/rty010
Yilmaz, P., Parfrey, L. W., Yarza, P., Gerken, J., Pruesse, E., Quast, C., et al. (2014). The Silva and "all-species living tree project (LTP)" taxonomic frameworks. Nucleic Acids Res. 42, D643–D648. doi: 10.1093/nar/gkt1209
Yu, X., Sun, Y., Lin, C., Wang, P., Shen, Z., and Zhao, Y. (2023). Development of transgenic maize tolerant to both glyphosate and glufosinate. Agronomy 13:226. doi: 10.3390/agronomy13010226
Zhang, X., Zhang, L., Zhang, L., Ji, Z., Shao, Y., Zhou, H., et al. (2019). Comparison of rhizosphere bacterial communities of reed and Suaeda in Shuangtaizi River estuary, Northeast China [J]. Mar. Pollut. Bull. 140, 171–178. doi: 10.1016/j.marpolbul.2019.01.041
Keywords: glyphosate, rhizosphere microbial communities, transgenic maize line D105, high-throughput sequencing, agricultural production
Citation: Wang G, Yang S, Feng S, Zhao G, He X and Han X (2024) Impact of glyphosate on the rhizosphere microbial communities of a double-transgenic maize line D105. Front. Sustain. Food Syst. 8:1421837. doi: 10.3389/fsufs.2024.1421837
Edited by:
Moslem Bahmankar, Agricultural Research, Education and Extension Organization (AREEO), IranReviewed by:
Aliakbar Zare, Soil and Water Research Institute, IranSwarnalee Dutta, Jeonbuk National University, Republic of Korea
Saeed Shahbazi, Safiabad Agriculnd Education Center, Iran
Copyright © 2024 Wang, Yang, Feng, Zhao, He and Han. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaodong Han, aHhkb25AaW1hdS5lZHUuY24=
 Guihua Wang1
Guihua Wang1 Shuqian Yang
Shuqian Yang Shilin Feng
Shilin Feng Guofen Zhao
Guofen Zhao Xiaodong Han
Xiaodong Han