
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Sustain. Food Syst. , 18 March 2024
Sec. Nutrition and Sustainable Diets
Volume 8 - 2024 | https://doi.org/10.3389/fsufs.2024.1364403
The intestinal microbiota plays a crucial role in human health, yet the impact of high-resistant starch and low-glutelin rice in the gut of obese individuals remains unexplored. In this study, different types of rice, namely japonica rice (control), low-glutelin content 1 rice (Lgc1), high-resistant starch and low-glutelin content 1 rice (HR + Lgc1), and commercially available low-glycemic index rice (LowGI), were utilized as samples to examine their in vitro digestion and fermentation, as well as the impact of HR + Lgc1 on the intestinal microbiota in obese individuals. The findings revealed that HR + Lgc1 rice exhibited lower in vitro digestion rates for starch and protein compared to the other three types. Following 24 h of in vitro fermentation, the total short-chain fatty acid content in HR + Lgc1 was 108.67 mmol/L, which was 60.33, 51.26, and 17.29% that in control, Lgc1, and LowGI, respectively. Moreover, HR + Lgc1 rice displayed an inhibitory effect on the production of harmful branched-chain fatty acid metabolites. Analysis through 16S rDNA sequencing indicated that, after fermentation, HR + Lgc1 significantly enhanced the abundance of beneficial bacteria such as Bifidobacterium, Parabacteroides, and Faecalibacterium in the gut, whereas it reduced the abundance of harmful bacteria such as Escherichia-shigella. Our findings may contribute to the development of new foods rich in dietary fiber to improve the digestive function of patients with multiple comorbidities.
• HR + Lgc1 has high-resistant starch and low-glutelin content.
• HR+Lgc1 rice exhibited lower in vitro digestion rates for starch and protein.
• HR+Lgc1 was utilized by gut microbiota to promote the production of short chain fatty acids.
• HR + Lgc1 can regulate the intestinal flora of obese patients and increase the abundance of beneficial bacteria.
The intestinal microbiota is composed of “1014” microorganisms, categorized into four bacterial phyla: Firmicutes, Bacteroidetes, Actinobacteria, and Proteobacteria. The major phyla, Firmicutes and Bacteroidetes, constitute 60–75% and 30–40% of the microbiota, respectively, and exert significant effects on human health (Wang Y. et al., 2019). The intestinal microbiota plays a crucial role in various aspects of human health, for example, influencing the fermentation of dietary fiber and prebiotics in the gut, and is closely related to immune function. Additionally, it can generate beneficial substances, including short-chain fatty acids (SCFAs) such as acetic acid, propionic acid, butyric acid, and lactic acid (Stewart et al., 2004).
SCFAs exert positive effects on the human body. For example, acetic acid contributes to the development of peripheral tissues, propionic acid facilitates the growth of liver cells, and butyric acid stimulates the growth of intestinal epithelial cells (Blaut and Clavel, 2007). Furthermore, SCFAs aid in reducing the pH of intestinal contents, enhancing mineral absorption by increasing mineral solubility, and regulating the body’s glucose and lipid metabolism (Lin et al., 2012). These findings underscore the crucial role of the intestinal microbiota in physiological processes in the host, including metabolic events, blood-tissue barrier integrity, and immune system function.
A recent study has reported a correlation between the production of abnormal metabolites by the gut flora and the onset and progression of obesity in patients (Gurung et al., 2020). The gut microbiota actively participates in shaping the immune system’s maturation and essential metabolic pathways, such as the fermentation of sugars and proteins, as well as the metabolism of bile acids and exogenous substances (Gomes et al., 2018). The dysregulation of microbial structure can affect human health, leading to various digestive system and metabolic diseases such as obesity (Fang et al., 2021). Obesity, recognized as a pressing global health challenge, has multifaceted causes. There are not only unhealthy habits such as high intake of fat and monosaccharides, but also environmental conditions, stressors and genetic factors, as well as the influence of intestinal flora and short-chain fatty acid (Li et al., 2015). Excessive fat accumulation can disrupt normal bodily functions, leading to a spectrum of diseases and conditions such as cardiovascular, respiratory, endocrinological, and social-psychological problems (Barczyńska et al., 2018). Therefore, the seriousness of obesity should not be underestimated. Taken together, these findings underscore the potential therapeutic benefits of methods that may enhance the microbial structure in individuals struggling with obesity.
Dietary fiber in food can reduce the risk of chronic diseases such as diabetes, obesity, cardiovascular diseases, and certain cancers (Landman and Quévrain, 2016). Dietary fiber, as an active substance, plays a very important role in the regulation of the human body (Wang et al., 2023). Resistant starch (RS), a type of dietary fiber in grains, cannot be digested in the small intestine. It is fermented by colonic bacteria to produce beneficial substances such as short-chain fatty acids, lactic acid, and small amounts of gas, while also reducing the pH of the large intestine and affecting the metabolism of glucose in the human body (Hamaker and Tuncil, 2014). In recent years, RS has been reported to promote human health and reverse certain diseases (Keenan et al., 2015). For instance, RS supplements can improve levels of fasting blood glucose, insulin, and low-density lipoprotein cholesterol, especially in obese and diabetic patients (DeMartino and Cockburn, 2020). Low-amylose rice can meet the special needs of patients with kidney disease, and it can be used as a dietary supplement for these patients because it can reduce protein intake to alleviate disease symptoms (Yang et al., 2022). Additionally, low-amylose rice can promote the metabolism of short-chain fatty acids and lactic acid, as well as the growth of probiotics while inhibiting the production of branched-chain fatty acids and ammonia, and the growth of harmful bacteria (Li et al., 2023). Currently, research is underway to understand the effects of high-resistant starch and low-amylose rice on the intestinal flora. However, there is no study on the impact of high-resistant starch and low-amylose rice on the intestinal microbiota in humans. Therefore, investigating the replacement of meals with high-resistant starch and low-amylose rice is of great importance for improving the microbial structure of obese individuals or those with multiple comorbidities.
Conducting gastrointestinal studies in humans requires a considerable investment of time and cost. As a result, there has been a gradual shift towards in vitro digestion simulation experiments that replicate the conditions of the human gastrointestinal tract. In vitro dynamic simulation, in contrast to static simulation, not only addresses the limitations of the latter but also replicates the physiological processes of the gastrointestinal tract. This approach streamlines the sampling process, facilitating the detection of changes at various stages of digestion (Bellmann et al., 2019).
In this study, an in vitro simulated model of digestion that used four types of rice, namely japonica rice (control), low-glutelin content 1 rice (Lgc1), high-resistant starch and low-glutelin content 1 (HR + Lgc1), and commercially available low-glycemic index rice (LowGI), was employed. Fecal samples were collected from obese patients, and the investigation utilized an independently developed biomimetic-colon-model dynamic simulation device for in vitro fermentation (Zhang et al., 2022). Therefore, this study aimed to examine the impact of different types of rice on the intestinal microbiota and elucidate the mechanism by which high-resistant starch and low-amylose rice affect the microbial structure of obese individuals.
Japonica rice (control) was cultivated in the fields of Nanjing, Jiangsu, whereas low-glutelin content 1 (Lgc1) rice was cultivated in the laboratory. The glutelin content of the dominant mutant Low glutelin content 1 (Lgc1) is lower than that of the wild-type (Iida et al., 1993), a characteristic attributed to the silencing of two highly homologous glutelin genes that form a double-stranded RNA molecule caused by a 3.5-kb deletion between GluB4 and GluB5. High-resistant starch and low-glutelin content 1 (HR + Lgc1) rice was obtained by disrupting the sbeIIb gene (LOC_Os02g32660) in Lgc1 (Li Z. et al., 2021; Li Z. T. et al., 2021). Low glycemic index (LowGI) rice was purchased from a local supermarket. All reagents used in this study were purchased from Shanghai Guoyao Chemical Reagent Co., Ltd., (Shanghai, China).
The Goni method (GonI et al., 1997) was employed to determine the resistant starch content. The procedure involved weighing 100 mg of each type of rice, adding 10 mL of KCl-HCl buffer and 3 mg of pepsin, and shaking at 40°C for 1 h. Subsequently, 9 mL of 0.1 mol/L Tris-maleate buffer was added, the pH was adjusted to 6.9 with HCl or NaOH, and α-amylase was added. The mixture was shaken at 37°C for 16 h and centrifuged (3,000 g, 15 min). The supernatant was collected, and the precipitate was washed with distilled water and centrifuged. The supernatants were combined, and the above steps were repeated. To the sample, 3 mL of distilled water was added, followed by 3 mL of 4 mol/L KOH solution, and the mixture was oscillated at room temperature for 30 min. Subsequently, 5.5 mL of 2 mol/L HCl and 3 mL of 0.4 mol/L sodium acetate buffer were added, the pH was adjusted to 4.75, and glucoamylase was added. The mixture was shaken at 60°C for 45 min and centrifuged. The supernatant was collected, and the precipitate was washed with distilled water and centrifuged. The supernatants were combined, and a stock volume solution based on the resistant starch content was prepared.
The GOPOD kit was used to measure the reducing sugar content. To each test tube, 0.1 mL of the supernatant and 3.0 mL of the GOPOD reagent were added, followed by incubation at 50°C for 20 min. For the blank, 0.1 mL of acetic acid–sodium acetate buffer (pH 4.5) and 3.0 mL of GOPOD were used. For the glucose standard solution, 0.1 mL of D-glucose (1 mg/mL) and 3.0 mL of GOPOD were used. The absorbance of the solution was measured at 510 nm relative to the blank.
Resistant starch (g/100 g sample) (sample containing >10% RS) = ΔE × F/W × volume of the test solution × 0.9.
Where ΔE is the absorbance value relative to the blank; F = 100 (μg of D-glucose)/absorbance value of 100 μg of D-glucose.
To determine the protein content in rice, 100 mg of rice flour was added to 800 μL of protein extraction buffer, followed by incubation in a water bath at 28°C for 30 min and centrifuged (11,180 g, 15 min). Subsequently, 100 μL of the solution was combined with 40 μL of 5× loading buffer, again followed by incubation in a water bath at 100°C for 10 min and centrifugation to obtain the supernatant (Chen et al., 2022). Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was performed according to the method of Laemmli (1970), with a 12% separation gel and a 5% concentration gel. After electrophoresis, the gel was stained with 0.1% Coomassie Brilliant Blue R-250 for 2 h and destained until the background was clear.
Simulated gastric fluid digestion was performed as previously described (Tian et al., 2023). An electrolyte solution mimicking the fluid of the gastrointestinal tract was prepared by dissolving 1.10 g/L KCl, 3.10 g/L NaCl, 1.50 g/L CaCl2, and 6.0 g/L NaHCO3 in deionized water and adjusting the pH to 2.0 with 0.1 M HCl. Subsequently, 15.0 mL of electrolyte solution, 3.6 mg of porcine pepsin (final concentration, 2000 U/mL), 3.8 mg of porcine lipase (final concentration, 20 U/mL), and 0.3 mL of 1.0 M CH3COONa solution (pH 5.0) were combined, and the pH was adjusted to 2.0 with 0.1 M HCl to obtain the simulated gastric fluid. To 5 mL of simulated gastric fluid, 1 g of freeze-dried rice flour was added, followed by incubation at 37°C for 2 h while stirring at 300 rpm. The reaction was terminated by adjusting the pH to 7.0 with 1 M NaHCO3. To prepare the intestinal electrolyte solution, 5.40 g/L NaCl, 6.50 g/L KCl, and 3.30 g/L CaCl2 were dissolved in deionized water, and the pH was adjusted to 7.0 with 1 M NaHCO3. To 10.0 mL of intestinal electrolyte solution, 1.3 mg of pancreatic protease, 40.0 mL of bile salt solution (4%, w/w), 10.0 mL of pancreatic enzyme solution (7%, w/w), and 10 mL of sodium acetate buffer containing 0.24 mL of amylase were added, and the pH was adjusted to 7.0 with 0.1 M NaOH. Subsequently, 5 mL of gastric digestion sample (sample digested in simulated gastric fluid for 2 h) was combined with 5.0 mL of simulated small intestine fluid, mixed, and incubated at 37°C for 4 h while stirring at 300 rpm. Starch digestion mainly occurs in the small intestine, while protein digestion occurs in gastric and intestinal fluids. The protein digestion rate was assayed at 0, 60, 120, 180, 240, 300, and 360 min according to the method of Gwala et al. (2020), and these time points spanned gastric and intestinal digestion. To precipitate proteins, 1 mL of the sample was combined with an equal volume of 10% trichloroacetic acid, followed by incubation for 1 h, centrifuged (11,180 g, 10 min), collection of the precipitate, determination of the nitrogen content by the Kjeldahl method (Lynch and Barbano, 1999), and calculation of the protein digestion rate.
The starch digestion rate was determined as previously described (Zheng et al., 2018), with minor modifications. The starch digestion rate was assayed at 0, 5, 10, 15, 20, 30, 45, 60, 90, 120, 180, and 240 min. To deactivate the digestive enzymes, 200 μL of sample was combined with 1.8 mL of anhydrous ethanol at the indicated time points. Subsequently, the samples were centrifuged at 3000 g for 10 min, combined with 50 μL of the supernatant in 1.5 mL of glucose oxidase reagent, and incubated at 50°C for 20 min. The absorbance of each sample was measured at 540 nm using a spectrophotometer (Unico, Shanghai, China). The glucose concentration was calculated, and the value was converted to the starch digestion ratio.
According to a previously described method (Li Z. et al., 2021; Li Z. T. et al., 2021; Solaesa et al., 2022; Sun et al., 2022), in vitro fermentation was conducted using undigested rice flour. On the day of the fermentation experiment, fresh fecal samples were collected from three obese volunteers aged between 24 and 32 years, with a body mass index of 25–35 kg/m2. These volunteers had not experienced any digestive system diseases in the 3 months leading up to the fermentation test, had not taken any antibiotics or probiotic products, and voluntarily provided written informed consent. Three portions of each fecal sample were added to 0.1 M phosphate buffer (pH 7.2), diluted to make a 10% fecal slurry (w/v), homogenized, and filtered through four layers of sterile gauze. The in vitro digestion model was one part of Bionic Gastrointestinal Reactor (BGR) developed by Li Z. et al. (2021) and Li Z. T. et al. (2021). For the BGR, before initiating the experiment, the system was purged with nitrogen three times to eliminate oxygen from the system. Subsequently, 230 mL of Brain Heart Infusion medium was added and autoclaved at 115°C for 20 min. After sterilization, when the reaction vessel cooled to around 37°C, 5 mL of filtered fecal slurry and 5 mL of phosphate buffer containing 5 g of freeze-dried rice flour were injected into the sampling port of the biomimetic gastrointestinal reactor for fermentation at 37°C. Samples were collected at 0, 6, 12, and 24 h and stored at −80°C for subsequent analysis. Each fermentation consisted of three independent replicates.
The bacterial cell density (OD600) of each type of rice flour after in vitro fermentation as measured at 600 nm using the Spark Multimode Plate Reader (Tecan, Männedorf, Switzerland). The content of SCFAs was determined by gas chromatography as previously described (Chen et al., 2017; Sun et al., 2022). In brief, 1.0 mL of sample was combined with 50 μL of phosphoric acid (34%), and the SCFA content was analyzed using the Shimadzu GC-2014 system (Kyoto, Japan) equipped with a flame ionization detector (FID) and a DB-FFAP column (30 m × 0.25 mm × 0.25 μm). The operating conditions for gas chromatography were as follows: nitrogen was used as the carrier gas with a flow rate of 60 mL/min; the injector temperature was set to 250°C; the FID detector was set to 300°C; the oven temperature was initially set to 100°C, maintained for 5 min, increased to 250°C at a rate of 10°C/min, and then maintained for 12 min.
DNA was extracted from the fermentation samples using the TIANamp Stool DNA kit (Tiangen Biotech, Beijing, China), and DNA samples were sent to Novo Gene Bioinformatics Technology Co., Ltd., (Beijing, China) for microbial DNA analysis. For each sample, the V4 region of bacterial 16S rDNA was sequenced using the Illumina MiSeq platform (San Diego, CA, United States). Sequence reads and operational taxonomic units formed the basis for all analyses (Sun et al., 2022).
Each experiment was conducted three independent times. Data were analyzed using analysis of variance and Tukey’s test, with p < 0.05 indicating a statistically significant difference and statistically analyzed using SPSS software. Graphs were plotted using Origin 2019 software (Irvine, CA, United States).
Starch stands out as the predominant nutritional element in rice. Conventional rice having a high content of branched-chain starch that is almost devoid of resistant starch, which is rapidly digested by the human body, resulting in a high glycemic index. Conversely, rice with elevated levels of resistant starch digests at a slower pace than conventional rice, offering benefits to individuals with diabetes and kidney disease. In our study, HR + Lgc1 rice displayed a high-resistant starch content of 10.3%, surpassing the resistant starch content of commercially available LowGI rice, which stood at 8.6% and was lower than HR + Lgc1 (Figure 1A). Both the control and Lgc1 rice showed negligible resistant starch content, aligning with typical rice varieties. Besides starch-related characteristics, another advantageous trait of HR + Lgc1 rice is its low protein content. Glutelin, which is abundant in lysine, makes up approximately 80% of the total protein dry weight of rice and is easily digestible by the human body (Wang et al., 2005). However, excessive absorption of soluble proteins may disrupt protein metabolism, particularly in patients with kidney disease (Iida et al., 2004). To assess changes in rice flour protein composition, total protein was extracted with protein extract solution and SDS-PAGE was used to separate total proteins in rice flour. The results showed that both control and LowGI varieties exhibited prominent bands corresponding to the acidic subunit of glutelin at approximately 33 kD and the basic subunit at approximately 22–23 kD, consistent with typical rice varieties. Conversely, the band corresponding to the 13 kD alcohol-soluble protein was relatively weak (Figure 1B). In HR + Lgc1 and Lgc1 rice, there was a significant reduction in the intensity of bands corresponding to the two glutelin subunits, while the band of the alcohol-soluble protein showed a noticeable enhancement. Therefore, our experimental material exhibits two key characteristics: a high content of resistant starch and a low content of glutelin.
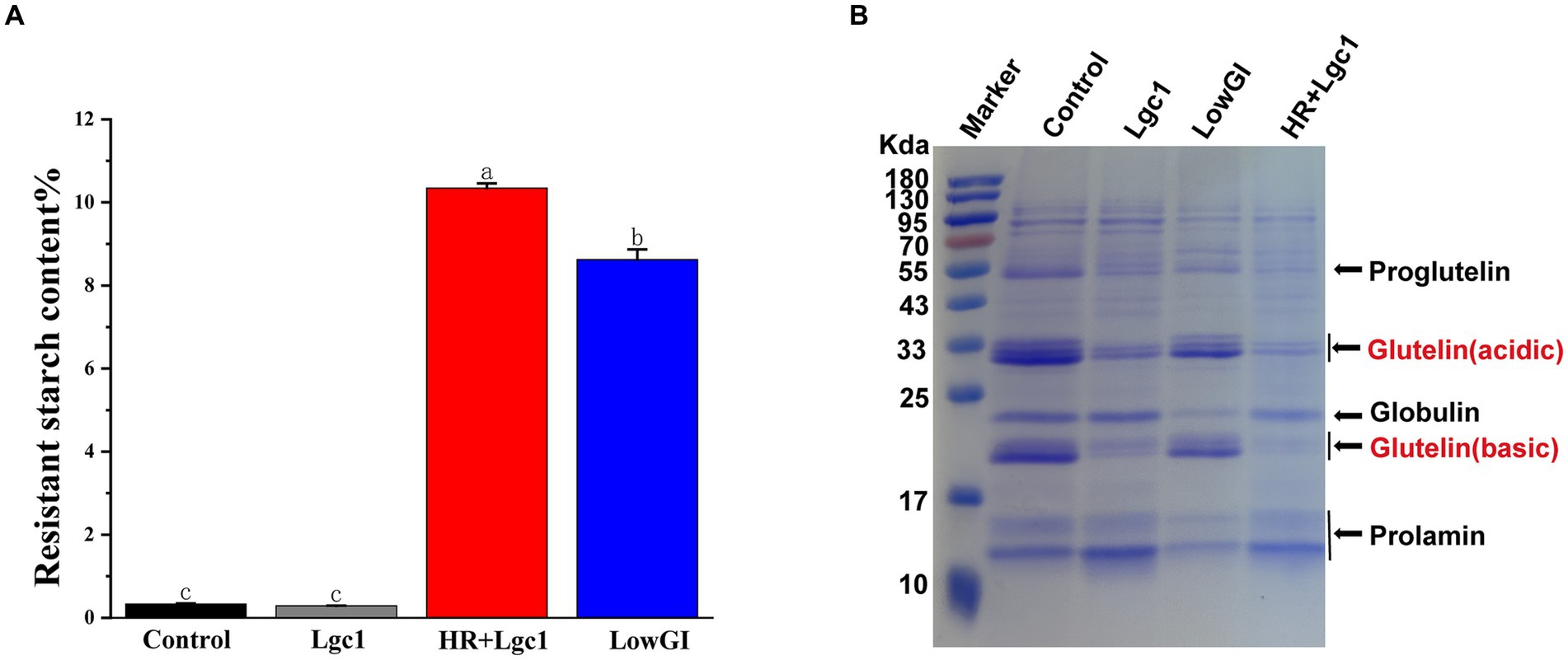
Figure 1. Resistant starch and protein content. (A) Resistant starch content of four types of rice, namely control, Lgc1, HR + Lgc1, and LowGI. (B) SDS-PAGE of proteins extracted from rice grains of control, Lgc1, HR + Lgc1, and LowGI. The experiments were repeated three independent times. Differences marked with the same lowercase letter in the graph indicate non-significant differences (p > 0.05), and different lowercase letters indicate significant differences (p < 0.05).
Due to the higher content of resistant starch and the cross-linking of insoluble proteins and starch, HR + Lgc1 exhibited significantly lower digestibility of both starch and protein compared to the other rice varieties (Lu et al., 2022; Li et al., 2023). A previous study has reported that rice protein components can reduce the digestibility of rice starch, indicating the potential of controlling rice starch digestibility by modifying rice protein composition (Solaesa et al., 2022). The results showed that, during the gastrointestinal digestion period, the starch digestibility of the four rice varieties was initially fast and then slow, releasing a substantial amount of reducing sugar under the influence of digestive enzymes. However, as digestion progressed, starch hydrolysis slowed, resulting in a decrease in the release of reducing sugar. Notably, the control demonstrated the highest starch digestibility, whereas the starch digestion rates of LowGI and HR + Lgc1, characterized by high-resistant starch content, were significantly lower than those of the other rice varieties. Lgc1, influenced by protein components, had a lower starch digestibility than control (Figure 2A), consistent with a previous study (Li et al., 2023). Conventional rice is characterized by a high glutelin content, which is a water-soluble protein that is easily hydrolyzed. Conversely, HR + Lgc1 rice has a lower glutelin content and a higher alcohol-soluble protein content, making it less susceptible to hydrolysis (He et al., 2021). In addition, HR + Lgc1 rice has a lower protein digestibility (Figure 2B). The starch and protein digestibility of HR + Lgc1 rice was lower than that of the control, Lgc1, and LowGI rice (Figure 2). The starch hydrolysis rates of HR + Lgc1 at various digestion stages were significantly lower than those of the other rice varieties, indicating that HR + Lgc1 converts starch into sugar more slowly in the human gastrointestinal tract. This suggests that HR + Lgc1 rice is particularly well-suited for individuals with diabetes, kidney disease, and those intolerant to sugar.
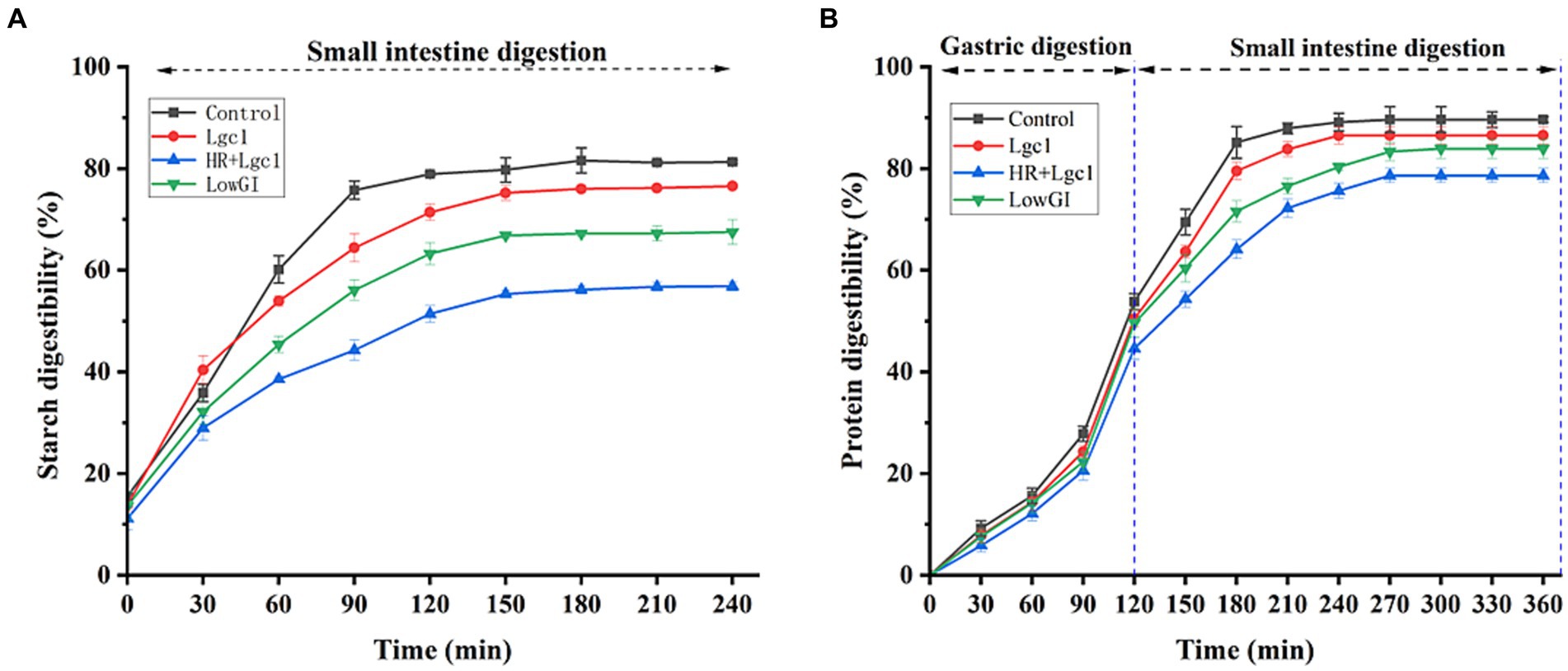
Figure 2. Starch and protein digestibility of four types of rice in dynamic digestion simulation. Starch digestibility (A) and protein digestibility (B) of four types of rice, namely control, Lgc1, HR + Lgc1, and LowGI in dynamic digestion simulation.
The intestinal microbiota absorbs and utilizes carbohydrates to produce various metabolites, primarily SCFAs such as acetate, propionate, butyrate, and valerate. SCFAs play diverse roles and serve as the main energy source for intestinal cells, with an increase in dietary fiber intake resulting in higher SCFA production. After 24 h of fermentation, the total SCFA content for the blank, control, Lgc1, HR + Lgc1, and LowGI were 45.21, 67.78, 71.84, 108.67, and 92.65 mmol/L, respectively (Figure 3A). Notably, the concentrations of acetate, propionate, butyrate, and valerate in HR + Lgc1 rice were 37.91, 35.03, 18.65, and 17.1 mmol/L, respectively, all surpassing those of the other rice varieties (Figure 3A). This was attributed to the higher resistant starch content of HR + Lgc1, which provided more carbohydrates to the intestinal microbiota for SCFA production. The total SCFAs content of LowGI was higher than that of the blank, control, and Lgc1 but lower than that of HR + Lgc1, indicating the crucial role of resistant starch in promoting SCFAs production by intestinal microbiota. The total SCFA content of Lgc1 was 71.84 mmol, slightly higher than that of conventional rice, consistent with a previous study (Li et al., 2023). These results indicate that HR + Lgc1 rice not only promotes the growth of intestinal microbiota but also provides more carbon sources to stimulate the production of beneficial SCFAs. As HR + Lgc1 provides more carbon sources for microbial utilization following gastrointestinal digestion. The total branched-chain fatty acid (BCFA) content of HR + Lgc1 was 0.73 mmol/L, LowGI was 0.91 mmol/L, Lgc1 was1.12 mmoL/L, control was 1.17 mmoL/L, blank was 1.34 mmol/L. HR + Lgc1 markedly lower than that of the blank, control, Lgc1, and LowGI (Figure 3B), with isobutyrate and isovalerate deemed detrimental to intestinal cells, metabolic processes, and human health (Maathuis et al., 2009).
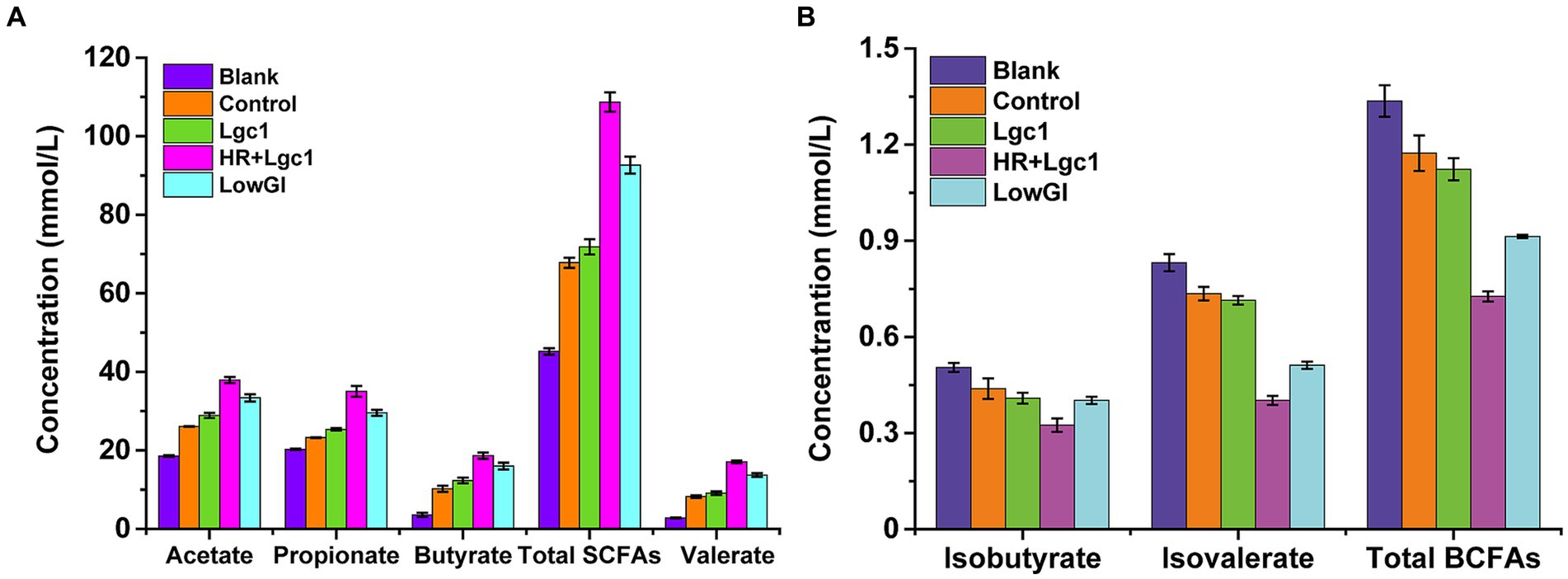
Figure 3. Effect of four types of rice on the production of metabolites by the gut microbiota in dynamic fermentation simulation. (A) SCFA and lactic acid content of intestinal flora fermented with different carbon sources for 24 h. (B) BCFA and ammonia content of intestinal flora fermented with different carbon sources for 24 h. The data are shown as the mean ± SD (n = 3) and analyzed using one-way ANOVA, followed by Tukey’s test.
To evaluate the impact of different types of rice on the microbial structure in obese patients, the sequences of 16S rDNA were analyzed after fermentation. A total of 1,166,025 valid reads were obtained from five groups, namely blank, control, Lgc1, HR + Lgc1, and LowGI (each with three replicates), with an average of 77,735 valid reads per sample. After fermentation, the total number of species in the gut microbiota for the four rice varieties was significantly higher than that of the control, with the highest species richness observed in HR + Lgc1 (Figure 4A). Compared to the other three types of rice, HR + Lgc1 more effectively increased the richness and evenness of the intestinal microbiota (Figure 4B). The Chao1 plot estimated the total number of species in the intestinal microbiota (Figure 4C), whereas the Shannon box plot showed the total number and proportion of classifications in the intestinal flora, with higher microbial diversity and more even species distribution (Figure 4D). Following fermentation, HR + Lgc1 exhibited the highest total number of species and the greatest microbial diversity compared to the control, Lgc1, HR + Lgc1, and LowGI. Therefore, it can be concluded that HR + Lgc1 more effectively enhances the diversity of the gut microbiota and regulates the composition of the intestinal flora compared to the control, Lgc1, and LowGI.
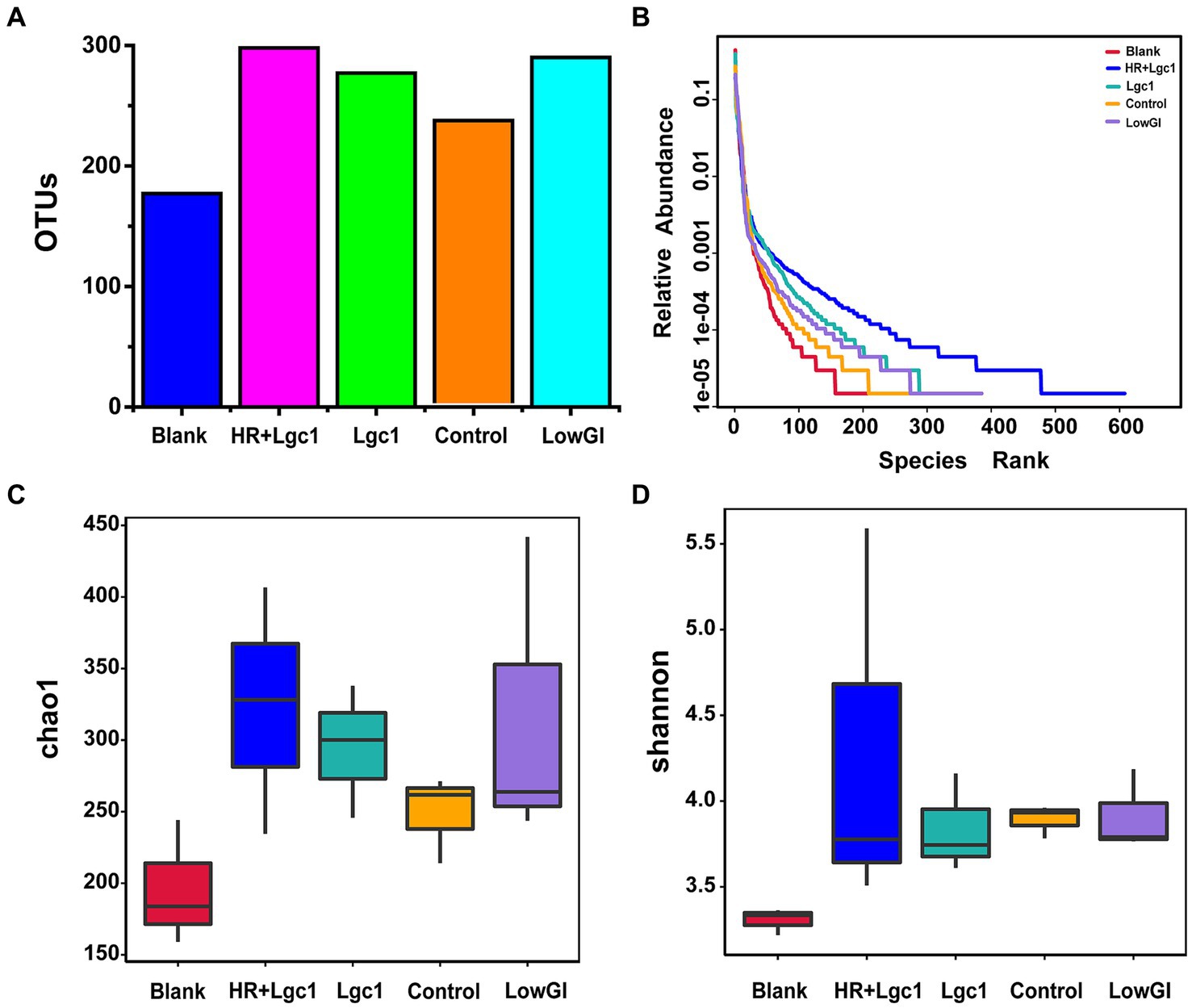
Figure 4. Effects of four types of rice on intestinal flora. (A) Operational taxonomic units (OTUs): abundance of intestinal flora. (B) Rank abundance of intestinal flora. (C) Chao1: total number of bacterial species. (D) Shannon: total number and proportion of intestinal flora.
To further investigate the impact of HR + Lgc1 rice on the microbial structure, we conducted an analysis to observe the changes in microbiota composition during in vitro fermentation of HR + Lgc1. After analyzing the changes after fermentation of the four rice varieties, it was revealed that the dominant phyla were Proteobacteria, Firmicutes, and Bacteroidetes, constituting over 95% of the total number of microbes (Figure 5A). After in vitro fermentation, compared to the blank, there was a significant increase in the abundance of Bacteroidetes and Firmicutes, while the abundance of Proteobacteria decreased, with the most pronounced changes observed in HR + Lgc1, followed by LowGI. This phenomenon may be attributed to HR + Lgc1 rice providing more carbon sources but fewer proteins, peptides, and free amino acids. Bacteroidetes and Firmicutes are known to be beneficial bacteria in the gastrointestinal environment of humans (Walsh et al., 2014). After in vitro fermentation, compared to the blank, the abundance of Proteobacteria in HR + Lgc1 decreased from 54.85 to 32.22%. Proteobacteria is a recognized marker of microbial dysbiosis, which is detrimental because it can influence the microbial structure through changes in population abundance (Chen et al., 2016). The results indicate that HR + Lgc1 rice exerts a regulatory effect on the gut microbiota of obese patients.
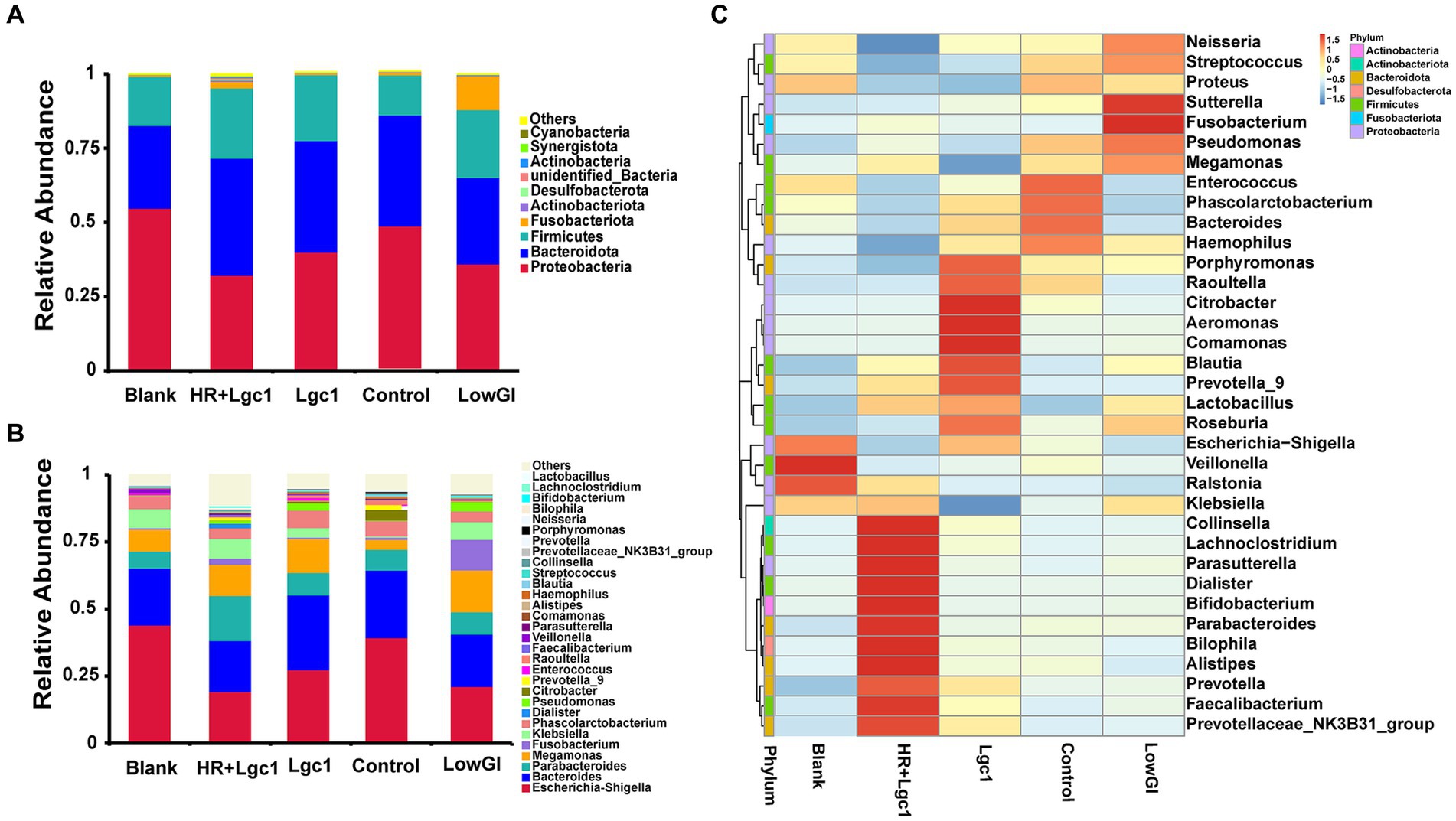
Figure 5. Effect of four types of rice on the microbial structure in obese patients. (A) Gut microbial composition at the phylum level. (B) Gut microbial composition at the genus level. (C) Heatmap of gut microbial composition at the genus level.
At the genus level, there were notable differences in the microbial structure among the four rice varieties after in vitro fermentation of samples (Figures 5B,C). The results revealed that the relative abundance of Lachnoclostridium, Bifidobacterium, Parabacteroides, and Faecalibacterium significantly increased in HR + Lgc1 compared to the other rice varieties. Lachnoclostridium, which produces acetate from monosaccharides and disaccharides, effectively stabilizes the gastrointestinal environment through anti-inflammatory and immune-suppressive effects (Kang et al., 2021). Bifidobacterium improves digestion, exhibits antimicrobial and antiviral properties, possesses anti-inflammatory effects, improves blood sugar levels, lowers lipid levels, enhances immunity, possesses antioxidant activity, and prevents eczema, relieves stress, and alleviates allergies (Henrick et al., 2021). Parabacteroides has been negatively correlated with conditions such as obesity, non-alcoholic fatty liver disease, and diabetes, suggesting a potentially positive regulatory role in glucose and lipid metabolism (Wang K. et al., 2019). Faecalibacterium, recognized as a typical butyrate-producing bacteria, is known for its anti-inflammatory effects, protective impact on digestive health, promotion of intestinal barrier function, and enhancement of metabolism and immune function (Effendi et al., 2022). Furthermore, compared to the other groups, the abundance of Escherichia-shigella significantly decreased in HR + Lgc1 and LowGI. Escherichia-shigella is a Gram-negative bacterium responsible for bacterial dysentery (shigellosis), which is characterized by invasion and inflammation of the intestinal epithelium. It is considered an indicator of microbial imbalance in the gut (Belotserkovsky and Sansonetti, 2018). Therefore, HR + Lgc1 may enhance the intestinal microbiota in obese individuals by elevating the ecological balance of the gut microbiota and serving as a potential prebiotic (Henrick et al., 2021).
This study employed japonica rice (control), low-glutelin content 1 (Lgc1) rice, high-resistant starch and low-glutelin content 1 (HR + Lgc1) rice, and low-glycemic index (LowGI) rice as carbon sources in a simulated colon reactor to ferment the fecal microbiota of obese individuals. The research aimed to investigate the impact of HR + Lgc1 rice on the microbial structure in the gut as well as the metabolic by-products. After measuring the SCFA content in the simulated colon reactor, it was observed that the total SCFA content of Lgc1 was 6% higher than that of the control, the SCFA content of LowGI rice was 28.97% higher than that of Lgc1, and the SCFA content of HR + Lgc1 was 17.29% higher than that of LowGI. Currently, rice marketed for populations dealing with obesity or kidney disease is primarily high-resistant starch and low-protein rice. Compared to the other types of rice, HR + Lgc1 demonstrated a more significant effect in promoting the production of beneficial SCFAs in the intestine and inhibiting the generation of undesirable metabolites such as isobutyrate acid and isovaleric acid. The 16S rDNA results indicated that HR + Lgc1 could enhance the diversity and richness of the intestinal microbiota. Through 16S rDNA gene sequencing, it was observed that HR + Lgc1 could enhance the abundance of beneficial bacteria in the gut, including Lachnoclostridium, Bifidobacterium, Parabacteroides, and Faecalibacterium, while reducing the abundance of harmful bacteria such as Escherichia-shigella. In summary, high-resistant starch and low-protein rice has a positive impact on the gut microbial structure and metabolism in obese individuals. The results of the study indicate that HR + lgc1 rice has the potential to be processed into different varieties of food in the future market, or health products such as intestinal prebiotics, which brings more good news for obese people. However, in this study, we did not know the specific mechanism by which HR + Lgc1 rice can enrich intestinal beneficial flora; Whether the combination of resistant starch and changes in other protein components of rice produce different effects on intestinal flora. This provides a direction for us to further study the diet structure of obese people in the future.
The datasets presented in this study can be found in online repositories. This data can be found here: NCBI database http://www.ncbi.nlm.nih.gov/bioproject/1079074 accession number: PRJNA1079074.
TZ: Writing – original draft, Writing – review & editing. ZT: Writing – original draft, Writing – review & editing. RL: Data curation, Methodology, Supervision, Writing – review & editing. LG: Formal analysis, Project administration, Validation, Writing – review & editing. WL: Investigation, Software, Writing – review & editing. YY: Funding acquisition, Resources, Visualization, Writing – review & editing. BZ: Funding acquisition, Resources, Visualization, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was supported by Jiangsu Agricultural Science and Technology Innovation Fund (cx (21)1002).
We would like to thank A&L Scientific Editing (www.alpublish.com) for its linguistic assistance during the preparation of this manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fsufs.2024.1364403/full#supplementary-material
Barczyńska, R., Litwin, M., Sliżewska, K., Szalecki, M., Berdowska, A., Bandurska, K., et al. (2018). Bacterial microbiota and fatty acids in the Faeces of overweight and obese children. Pol. J. Microbiol. 67, 339–345. doi: 10.21307/pjm-2018-041
Bellmann, S., Krishnan, S., de Graaf, A., de Ligt, R. A., Pasman, W. J., Minekus, M., et al. (2019). Appetite ratings of foods are predictable with an in vitro advanced gastrointestinal model in combination with an in silico artificial neural network. Food Res. Int. 122, 77–86. doi: 10.1016/j.foodres.2019.03.051
Belotserkovsky, I., and Sansonetti, P. J. (2018). Shigella and enteroinvasive Escherichia coli. Curr. Top. Microbiol. Immunol. 416, 1–26. doi: 10.1007/82_2018_104
Blaut, M., and Clavel, T. (2007). Metabolic diversity of the intestinal microbiota: implications for health and disease. J. Nutr. 10, 208–212. doi: 10.1089/jmf.2005.067
Chen, D., Chen, G., Wan, P., Hu, B., Chen, L., Ou, S., et al. (2017). Digestion under saliva, simulated gastric and small intestinal conditions and fermentation in vitro of polysaccharides from the flowers of Camellia sinensis induced by human gut microbiota. Food Funct. 8, 4619–4629. doi: 10.1039/c7fo01024a
Chen, Z., Du, H., Tao, Y., Xu, Y., Wang, F., Li, B., et al. (2022). Efficient breeding of low glutelin content rice germplasm by simultaneous editing multiple glutelin genes via CRISPR/Cas9. Plant Sci. 324:111449. doi: 10.1016/j.plantsci.2022.111449
Chen, C., Huang, Q., Fu, X., and Liu, R. H. (2016). In vitro fermentation of mulberry fruit polysaccharides by human fecal inocula and impact on microbiota. Food Funct. 7, 4637–4643. doi: 10.1039/c6fo01248e
DeMartino, P., and Cockburn, D. W. (2020). Resistant starch: impact on the gut microbiome and health. Curr. Opin. Biotechnol. 61, 66–71. doi: 10.1016/j.copbio.2019.10.008
Effendi, R., Anshory, M., Kalim, H., Dwiyana, R. F., Suwarsa, O., Pardo, L. M., et al. (2022). Akkermansia muciniphila and Faecalibacterium prausnitzii in immune-related diseases. Microorganisms. 10:2382. doi: 10.3390/microorganisms10122382
Fang, Q., Zheng, B., Liu, N., Liu, J., Liu, W., Huang, X., et al. (2021). Trimethylamine N-oxide exacerbates renal inflammation and fibrosis in rats with diabetic kidney disease. Front. Physiol. 12:682482. doi: 10.3389/fphys.2021.682482
Gomes, A. C., Hoffmann, C., and Mota, J. F. (2018). The human gut microbiota: metabolism and perspective in obesity. Gut Microbes 9, 1–18. doi: 10.1080/19490976.2018.1465157
GonI, I., Garcia-Alonso, A., and Saura-Calixto, F. (1997). A starch hydrolysis procedure to estimate glycemic index. Nutr. Res. 17, 427–437. doi: 10.1016/S0271-5317(97)00010-9
Gurung, M., Li, Z., You, H., Rodrigues, R., Jump, D. B., Morgun, A., et al. (2020). Role of gut microbiota in type 2 diabetes pathophysiology. EBioMedicine 51:102590. doi: 10.1016/j.ebiom.2019.11.051
Gwala, S., Pallares Pallares, A., Pälchen, K., Hendrickx, M., and Grauwet, T. (2020). In vitro starch and protein digestion kinetics of cooked Bambara groundnuts depend on processing intensity and hardness sorting. Food Res. Int. 137:109512. doi: 10.1016/j.foodres.2020.109512
Hamaker, B. R., and Tuncil, Y. E. (2014). A perspective on the complexity of dietary fiber structures and their potential effect on the gut microbiota. J. Mol. Biol. 426, 3838–3850. doi: 10.1016/j.jmb.2014.07.028
He, W., Wang, L., Lin, Q., and Yu, F. (2021). Rice seed storage proteins: biosynthetic pathways and the effects of environmental factors. J. Integr. Plant Biol. 63, 1999–2019. doi: 10.1111/jipb.13176
Henrick, B. M., Rodriguez, L., Lakshmikanth, T., Pou, C., Henckel, E., Arzoomand, A., et al. (2021). Bifidobacteria-mediated immune system imprinting early in life. Cell 184, 3884–3898.e11. doi: 10.1101/2020.10.24.353250
Iida, S., Amano, E., and Nishio, T. (1993). A rice (Oryza sativa L.) mutant having a low content of glutelin and a high content of prolamine. Theor Appl Genet. 87, 374–378. doi: 10.1007/bf01184926
Iida, S., Sunohara, Y., Maeda, H., Matsushita, K., Nemoto, H., Ishii, T., et al. (2004). A new rice cultivar with good eating quality (low amylose) and low glutelin protein, “LGC Soft”. Bull. Natl. Agric. Res. Center Western Region.
Kang, L., Li, P., Wang, D., Wang, T., Hao, D., and Qu, X. (2021). Alterations in intestinal microbiota diversity, composition, and function in patients with sarcopenia. Sci. Rep. 11:4628. doi: 10.1038/s41598-021-84031-0
Keenan, M. J., Zhou, J., Hegsted, M., Pelkman, C., Durham, H. A., Coulon, D. B., et al. (2015). Role of resistant starch in improving gut health, adiposity, and insulin resistance. Adv. Nutr. 6, 198–205.
Laemmli, U. K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685. doi: 10.1038/227680a0
Landman, C., and Quévrain, E. (2016). Gut microbiota: description, role and pathophysiologic implications. Rev. Med. Interne 37, 418–423. doi: 10.1016/j.revmed.2015.12.012
Li, Q., Blume, S. W., Huang, J. C., Hammer, M., and Ganz, M. L. (2015). Prevalence and healthcare costs of obesity-related comorbidities: evidence from an electronic medical records system in the United States. J. Med. Econ. 18, 1020–1028. doi: 10.3111/13696998.2015.1067623
Li, Z., Guo, D., Li, X., Tang, Z., Ling, X., Zhou, T., et al. (2021). Heat-moisture treatment further reduces in vitro digestibility and enhances resistant starch content of a high-resistant starch and low-Glutelin Rice. Food Secur. 10:2562. doi: 10.3390/foods10112562
Li, Z. T., Han, S. X., Pu, J. Y., Wang, Y. Y., Jiang, Y., Gao, M. J., et al. (2023). In vitro digestion and fecal fermentation of low-gluten Rice and its effect on the gut microbiota. Foods 12:855. doi: 10.3390/foods12040855
Li, Z. T., Hu, G. A., Zhu, L., Zhao, Z. C., Jiang, Y., Gao, M. J., et al. (2021). In vitro digestion and fecal fermentation of highly resistant starch rice and its effect on the gut microbiota. Food Chem. 361:130095. doi: 10.1016/j.foodchem.2021.130095
Lin, H. V., Frassetto, A., Kowalik Jr, E. J., Nawrocki, A. R., Lu, M. M., Kosinski, J. R., et al. (2012). Butyrate and propionate protect against diet-induced obesity and regulate gut hormones via free fatty acid receptor 3-independent mechanisms. PLoS One 7:e35240. doi: 10.1371/journal.pone.0035240
Lu, X., Ma, R., Zhan, J., Wang, F., and Tian, Y. (2022). The role of protein and its hydrolysates in regulating the digestive properties of starch: a review. Trends Food Sci. Technol. 125, 54–65. doi: 10.1016/j.tifs.2022.04.027
Lynch, J. M., and Barbano, D. M. (1999). Kjeldahl nitrogen analysis as a reference method for protein determination in dairy products. J. AOAC Int. 82, 1389–1398. doi: 10.1021/jf990185x
Maathuis, A., Hoffman, A., Evans, A., Sanders, L., and Venema, K. (2009). The effect of the undigested fraction of maize products on the activity and composition of the microbiota determined in a dynamic in vitro model of the human proximal large intestine. J. Am. Coll. Nutr. 28, 657–666. doi: 10.1080/0731-5724.2009.10719798
Solaesa, Á. G., Villanueva, M., Vela, A. J., and Ronda, F. (2022). Impact of microwave radiation on in vitro starch digestibility, structural and thermal properties of rice flour. From dry to wet treatments. Int. J. Biol. Macromol. 222, 1768–1777. doi: 10.1016/j.ijbiomac.2022.09.262
Stewart, C. S., Duncan, S. H., and Cave, D. R. (2004). Oxalobacter formigenes and its role in oxalate metabolism in the human gut. FEMS Microbiol. Lett. 230, 1–7. doi: 10.1016/s03781097-(03)00864-4
Sun, Y., Tang, Z., Hao, T., Qiu, Z., and Zhang, B. (2022). Simulated digestion and fermentation in vitro by obese human gut microbiota of Sulforaphane from broccoli seeds. Food Secur. 11:4016. doi: 10.3390/foods11244016
Tian, J., Wang, X., Zhang, X., Chen, X., Dong, M., Rui, X., et al. (2023). Artificial simulated saliva, gastric and intestinal digestion and fermentation in vitro by human gut microbiota of intrapolysaccharide from paecilomyces cicadae TJJ1213. Food Sci Hum. 12, 622–633. doi: 10.1016/j.fshw.2022.07.065
Walsh, C. J., Guinane, C. M., O'Toole, P. W., and Cotter, P. D. (2014). Beneficial modulation of the gut microbiota. FEBS Lett. 588, 4120–4130. doi: 10.1016/j.febslet.2014.03.035
Wang, Y., Chen, J., Song, Y. H., Zhao, R., Xia, L., Chen, Y., et al. (2019). Effects of the resistant starch on glucose, insulin, insulin resistance, and lipid parameters in overweight or obese adults: a systematic review and meta-analysis. Nutr. Diabetes 9:19. doi: 10.1038/s41387-019-0086-9
Wang, A., Li, L., Zhang, S., Wang, J., Lu, Z., Lu, F., et al. (2023). Screening and identification of a lignocellulose-degrading strain and its application in highland barley fermentation. Food Mater Res. 3:6. doi: 10.48130/FMR-2023-0006
Wang, K., Liao, M., Zhou, N., Bao, L., Ma, K., Zheng, Z., et al. (2019). Parabacteroides distasonis alleviates obesity and metabolic dysfunctions via production of succinate and secondary bile acids. Cell Rep. 26, 222–235.e5. doi: 10.1016/j.celrep.2018.12.028
Wang, Y. H., Liu, S. J., Ji, S. L., Zhang, W. W., Wang, C. M., Jiang, L., et al. (2005). Fine mapping and marker-assisted selection (MAS) of a low glutelin content gene in rice. Cell Res. 15, 622–630. doi: 10.1038/sj.cr.7290332
Yang, Y., Shen, Z., Li, Y., Xu, C., Xia, H., Zhuang, H., et al. (2022). Rapid improvement of rice eating and cooking quality through gene editing toward glutelin as target. J. Integr. Plant Biol. 64, 1860–1865. doi: 10.1111/jipb.13334
Zhang, W., Hu, B., Liu, C., Hua, H., Guo, Y., Cheng, Y., et al. (2022). Comprehensive analysis of Sparassis crispa polysaccharide characteristics during the in vitro digestion and fermentation model. Food Res Int. 154:111005. doi: 10.1016/j.foodres.2022.111005
Keywords: high-resistant starch and low-glutelin rice, obesity, in vitro fermentation, gut microbiota, short-chain fatty acids
Citation: Zhou T, Tang Z, Liu R, Gui L, Luo W, Yang Y and Zhang B (2024) High-resistant starch and low-glutelin content 1 rice benefits gut function in obese patients. Front. Sustain. Food Syst. 8:1364403. doi: 10.3389/fsufs.2024.1364403
Received: 02 January 2024; Accepted: 20 February 2024;
Published: 18 March 2024.
Edited by:
Kathleen L. Hefferon, Cornell University, United StatesReviewed by:
Digambar Kavitake, National Institute of Nutrition (ICMR), IndiaCopyright © 2024 Zhou, Tang, Liu, Gui, Luo, Yang and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Baolong Zhang, emhibDIyNDhAaG90bWFpbC5jb20=
†These authors share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.