
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Sustain. Food Syst. , 12 February 2024
Sec. Agro-Food Safety
Volume 8 - 2024 | https://doi.org/10.3389/fsufs.2024.1362265
This article is part of the Research Topic The Hazards and Nutritional Benefits of Metal(loid)s in Food and Environment View all 7 articles
Introduction: Cadmium (Cd) is a highly toxic heavy metal which contaminates agricultural soils and is easily absorbed by plants. Brassica rapa L. is one of the most popular vegetables in China and is known to accumulate Cd in its roots and aerial tissues.
Methods: A highly Cd-resistant bacterium (‘CD2’) was isolated and identified. Its ability to immobilize Cd(II) in medium was studied. Strain CD2 were added into Cd-polluted soil to ameliorate Cd accumulation in B. rapa. The underlying mechanisms of ‘CD2’ to reduce Cd accumulation in B. rapa. were analyzed by transcriptomics.
Results and discussion: Strain CD2 was classified as belonging to the genus Stenotrophomonas. Strain CD2 was found to be able to remove 0.1 mmol/L Cd(II) after 36 h by intracellular sequestration and by producing biofilm, exopolysaccharide, and H2S. When applied to Cd-contaminated soil, ‘CD2’ significantly increased the content of nonbioavailable Cd by 212.70%. Furthermore, ‘CD2’-inoculated B. rapa exhibited a 51.16% decrease in the Cd content of roots and a 55.56% decrease in the Cd content of aerial tissues. Transcriptome analysis identified 424 differentially expressed genes (DEGs) in the roots and 501 DEGs in the aerial tissues of uninoculated Cd-exposed plants. By comparison, 1107 DEGs were identified in the roots and 1721 DEGs were identified in the aerial tissues of ‘CD2’-inoculated Cd-exposed plants. In both treatment groups, genes related to vacuolar sequestration were upregulated, resulting in inhibited Cd transport. In addition, both catalase and glutathione transferase were induced in uninoculated plants, while the oxidative stress-related genes CPK and RBOH belonged to ‘plant-pathogen interactions’ were upregulated in ‘CD2’-inoculated plants. Moreover, inoculation with ‘CD2’ resulted in the enrichment of phenylpropane metabolism; cutin, suberine, and wax biosynthesis; and the AP2, Dof, WOX, Trihelix, B3, EIL, and M-type_MADS transcription factors; as well as the downregulation of zinc transporters and blue copper proteins. All of these changes likely contributed to the reduced Cd accumulation in ‘CD2’-inoculated B. rapa. The results of this study suggest that Stenotrophomonas sp. CD2 may prove to be a useful inoculant to prevent Cd accumulation in B. rapa.
The heavy metal cadmium (Cd) is highly toxic to plants, animals, and humans. Worryingly, Cd can accumulate in vegetables, thereby threatening human health through the food chain (Kubier et al., 2019; Xia et al., 2021). Cd is categorized as a Class I human carcinogen by the International Agency for Research on Cancer (IARC), and has been linked to lung cancer, breast cancer, nephropathy, and osteoporosis, among other conditions (Waalkes, 2003; Akesson et al., 2008; Larsson and Wolk, 2016). Cd is widely used in the production of a variety of industrial products, including batteries, dyes, coatings, electroplating, cadmium quantum dots, alloys, and nuclear fission infrastructure (Turner, 2019). One-sixth of the world’s total Cd production comes from China, where Cd pollution has impacted 2.8 × 109 m2 of farmland (Liu et al., 2015). According to the Chinese Ministry of Environmental Protection, Cd has been the most common pollutant exceeding the Ministry’s limits, accounting for 7% of all samples (Zhao et al., 2015). In fact, Cd-polluted soil is beginning to seriously restrict crop production in China, posing a severe threat to the safety and quality of agricultural products. Thus, effective soil remediation techniques must be developed to mitigate or prevent the absorption of Cd by crops.
Cd is a transition metal with low reduction potential, making biological reduction difficult (Nancharaiah et al., 2015). Cd is primarily found in the divalent [Cd(II)] form, which is characterized by high solubility and mobility and which easily accumulates in the food chain (Kubier et al., 2019). Cd(II) can enter plant cells by way of various transporters. Cd(II) uptake occurs mainly via ZIP family (Guerinot, 2000), natural resistance-associated macrophage protein (NRAMP) family, and yellow stripe-like (YSL) family transporters (Thomine et al., 2000; Feng et al., 2017). However, the plants can fix metal ions absorbed into plant tissues. They can chelate Cd, store Cd in their vacuoles, or activate the antioxidant defense system to response to Cd toxicity (Moons, 2003; Kuramata et al., 2009; Zhang et al., 2010). For example, some low molecular weight chelators, such as glutathione (GSH), glutathione synthetase (GS), phosphatidylcholine (PC), and metallothioneins (MT), can bind to Cd and then mediate vacuoles through several transporters (Clemens et al., 2002; Clemens, 2006; Verbruggen et al., 2009). In addition, the plant can reduce the Cd absorption from soil. They can secrete organic acids, such as citric acid, malic acid, oxaloacetic acid, malonic acid, and tartaric acid by the roots (Anjum et al., 2015). Furthermore, microbes in soli can adsorb Cd on the cell surface using electronegative functional groups and exopolysaccharides (EPS), sequestering Cd inside cells using metallothionein and phytochelatin, or producing hydrogen sulfide (H2S) to coprecipitate with Cd (Xia et al., 2021). Both secreted acids and microbes can lead to the passivation of soluble metal ions in the soil and effectively prevents their absorption by plants (Anjum et al., 2015; Xia et al., 2021).
Soil microbes are important members of all terrestrial ecosystems. Studies suggest that certain microbes can not only passivate Cd in soil but also reduce the absorption of Cd by plants. For example, Cd(II)-resistant Cupriavidus taiwanensis KKU2500-3 can colonize rice tissues and reduce Cd absorption in Cd-polluted soil (Punjee et al., 2018). Both Methylobacterium oryzae CBMB20 and Burkholderia spp. CBMB40 can promote growth and reduce Ni and Cd absorption in tomato (Madhaiyan et al., 2007). In addition, Pseudomonas fluorescens UW4 can reduce Cd stress in lettuce and Enterobacter asburiae NC16 can reduce Cd absorption in wheat (Albano and Macfie, 2016; Zhou et al., 2019). Given these results, the use of microbes is increasingly seen as a potentially low-cost and environmentally friendly method to remediate Cd-polluted soils and to prevent the absorption of Cd by crop plants.
Chinese cabbage (Brassica rapa L.) is one of the most popular and widely cultivated vegetables in China. Unfortunately, B. rapa is known to accumulate Cd in its roots and edible tissues (He et al., 2013; Wu et al., 2022). Recently, the B. rapa Cd stress response has been studied using transcriptomic and proteomic approaches (Sun et al., 2023; Yu et al., 2023). Notably, several Cd-resistant microbes have been found to reduce Cd absorption by B. rapa, including Pseudomonas sp. B7 (Wu et al., 2022), Enterobacter sp. A11, and Comamonas sp. A23 (Wang et al., 2020). However, the mechanism by which these microbes alter Cd dynamics and toxicity in B. rapa is unclear. In this work, we isolated a highly Cd(II)-resistant bacterium (Stenotrophomonas sp. CD2) from Cd-contaminated soil and studied its ability to immobilize Cd(II) in medium and in soil. In addition, we evaluated the ability of strain CD2 to ameliorate Cd accumulation in B. rapa. Finally, we utilized transcriptomics to discover the mechanisms underlying the ability of “CD2” to reduce Cd accumulation in B. rapa.
The strain CD2 was isolated from Cd-contaminated soil in Daye City, Hubei, China (N 29°59′41″, E114°56′56″). Soil samples were plated on LB (Luria-Bertani) plates containing 2 mM CdCl2, which were cultivated at 28°C for 7 d to obtained strain CD2. After cultivation on LB plates for 2 days at 28°C, the colonies of strain CD2 were observed. Strain CD2 were cultivated in LB medium until reaching an OD600 of 1.0. Subsequently, cells were collected by centrifugation at 8000 g for 5 min at 4°C to observe cells morphology using scanning electron microscopy (SEM) by Wuhan Detection of Technical Sousepad Ltd., Wuhan, China. Additionally, the genome of strain CD2 was sequenced by Wuhan Bio-Broad Co., Ltd., Wuhan, China, and then then annotated using the NCBI Prokaryotic Genome Annotation Pipeline in combination with GeneMarkS+ (Tatusova et al., 2016; Haft et al., 2018; Li et al., 2021). The extracted 16S rRNA gene sequences from its genome were used to construct a neighbor-joining (NJ) phylogenetic tree of strain CD2 using MEGA version11.0 software (Tamura et al., 2021).
The strain CD2 was cultured in LB medium at 28°C with shaking at 150 rpm to until reaching an OD600 of 1.0. Then, 1% (v/v) fresh culture was inoculated into LB medium with different heavy metal(loid)s and incubated at 28°C with shaking at 150 rpm for further analysis. The OD600 of the cultures was measured after 48 h. The different metals and their corresponding concentrations were as follows: CdCl2[Cd(II)]: 0.5, 1.0, 1.5, 2.0, 2.5 mM; ZnCl2[Zn(II)]: 1.0, 2.0, 4.0, 6.0, 7.0 mM; CuCl2[Cu(II)]: 1.0, 2.0, 4.0, 5.0, 6.0 mM; K2CrO4 [Cr(VI)]: 1.0, 3.0, 6.0, 8.0, 10.0 mM; NaAsO2[As(III)]: 1.0, 2.0, 4.0, 8.0, 10.0 mM; C8H4K2O12Sb2S3(H2O)[Sb(III)]: 1.0, 3.0, 6.0, 8.0, 10.0 mM.
1% (v/v) fresh culture was inoculated into 100 mL of LB medium with or without the addition of 0.1 mM Cd(II) to assess the ability of strain CD2 to remove Cd(II). The LB medium without inoculating strain CD2 was as a control. The cultures were incubated at 28°C with shaking at 150 rpm. Culture samples were collected at the indicated times to measure the OD600, followed by centrifugation at 12,000 g for 5 min to separate supernatant and pellets. The supernatant were used to measure the Cd(II) concentration by atomic absorption spectrometry (TAS-990\u00B0F, Persee). The pellets were used to measure the Cd(II) concentration in different cell components including extracellular immobilization and intracellular sequestration (Wu et al., 2022).
The biofilm content and exopolysaccharide (EPS) were detected using the crystal violet staining method and LB-aniline blue plates, respectively (O’Toole and Kolter, 1998; Ashraf et al., 2004). The H2S were detected by the lead acetate test paper (Wang et al., 2020). Strain CD2 were cultured in LB medium with or without 0.1 mmol/L Cd(II) at 28°C with shaking at 150 rpm. Cells were harvested at both 24 h and 36 h, followed by freeze-drying under vacuum conditions using a vacuum (Labconco) for FTIR determination conducted by Wuhan Detection of Technical Sousepad Ltd., Wuhan, China.
We carried out pot experiments to explore the ability of strain CD2 to remediate Cd(II)-polluted soil and reduce Cd(II) accumulation in B. rapa. The Cd-free potting soil was collected from farmland in Ezhou, Hubei, China. The pH of pot soil was 6.32 ± 0.05. To this soil was added 2 mg/kg Cd(II), and then the soil was mixed thoroughly and equilibrated for 14 d. The Cd concentration in soil at 0 d and 14 d after 2 mg/kg added to the soil were 1.95 ± 0.13 mg/Kg and 2.03 ± 0.22 mg/Kg, respectively. The experiment consisted of three treatment groups: Control (no Cd(II) and no “CD2”), Cd(II) (2 mg/kg Cd(II)), and CD2 + Cd(II) (2 mg/kg Cd(II) and “CD2” inoculation). Each treatment consisted of 5 replicates. Three seedlings of similar size were transplanted into each pot containing 2.5 kg of soil. Stenotrophomonas sp. CD2 was cultured in lysogeny broth (LB) to OD600 = 1.0, at which point the cells were harvested, washed with 0.9% NaCl, and then resuspended in water. Five-hundred milliliters of this bacterial suspension (107 cfu/g) was added to each “CD2 + Cd(II)” experimental pot. Potted plants were grown in a greenhouse for 32 d, with an average temperature of 25.0°C and average of 12 h of light per day. Following harvest, wet weight, dry weight, and Cd content in roots and aerial tissues (shoots and leaves) were quantified (Li et al., 2014). Rhizosphere soil was collected to evaluate changes in Cd speciation using a modified European Community Bureau of Reference (BCR) method (Kartal et al., 2006). Specifically, the exchangeable and carbonate-bound, and reducible fractions were classified as “bioavailable Cd,” while the oxidizable and residual fractions were classified as “nonbioavailable Cd.”
After 32 d of growth under greenhouse conditions, the plants were harvested and their roots and aerial tissues (shoots and leaves) were separated. Total RNA was extracted from each set of tissues (roots and aerial parts) using TRIzol Reagent (Invitrogen, MA, United States). RNA concentration was quantified using a NanoDrop 2000 (ThermoFisher Scientific, MA, United States) spectrometer. An RNA-Seq library was constructed using 3 μg of RNA, and the library was paired-end (PE) sequenced using an Illumina sequencing platform. Fastp (v0.22.0) (Chen et al., 2018) was used to filter the raw data and remove connectors and low-quality reads in order to obtain clean, high-quality reads. HTSeq (v0.9.1) (Anders et al., 2015) was used to compare the Read Count value of each gene to quantify gene expression. HISeq was used to compare the Read Count value of each gene to quantify gene expression, which was standardized as Fragments Per Kilo bases per Million fragments (FPKM). The raw data were stored in NCBI (PRJNA1064471). DESeq (v1.38.3) was used to identify differentially expressed genes (DEGs) according to the following criteria: log2FoldChange > 1 and p-value <0.05. The DEGs were then functionally annotated using the Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) databases. Transcription factors (TFs) were identified using both the Animal Transcription Factor Database (AnimalTFDB) (Hu et al., 2019) and Plant Transcription Factor Database (PlantTFDB) (Tian et al., 2020), and the number of DEGs predicted to be TFs was counted.
Strain CD2 was isolated from Cd-contaminated soil in Daye, Hubei, China. Colonies of strain CD2 were light yellow, smooth, and circular, with a diameter of approximately 1.0 mm (Supplementary Figure S1A). Scanning electron microscopy revealed that individual “CD2” cells were rod-shaped and between 0.3 and 0.5 μm in diameter and 0.7–1.5 μm in length (Supplementary Figure S1B).
Genome-wide sequences for strain CD2 have been deposited in DDBJ/EMBL/GenBank under accession number CP102248. The strain CD2 genome was 4.93 Mb in size, with a GC content of 66.2%. In addition, its genome contained 4854 genes, including 3273 protein-coding genes. The 16S rRNA gene sequences (1,545 bp) of strain CD2 shared the highest similarity with Stenotrophomonas geniculata ATCC 19374 (99.64%) and Stenotrophomonas maltophilia NCTC10257 (99.43%), according to EzBioCloud and NCBI analysis. A neighbor-joining (NJ) phylogenetic analysis indicated that strain CD2 was related to Stenotrophomonas geniculata ATCC 19374 (Supplementary Figure S2). Together, these results indicated that strain CD2 belongs to genus Stenotrophomonas.
Supplementary Figure S3 shows the resistance of strain CD2 to different heavy metal(oid)s. The minimum inhibitory concentration (MIC) values of strain CD2 to Cd(II), Zn(II), Cu(II), and Cr(VI) were found to be 2.5, 7.0, 5.0, and 8.0 mM, respectively. Furthermore, the MICs of As(III) and Sb(III) were > 10 mM. These results suggest that strain CD2 is resistant to multiple heavy metal(oid)s.
In order to evaluate the ability of Cd(II)-resistant “CD2” to immobilize Cd(II), we generated growth curves, Cd(II) immobilization curves, and Cd(II) distribution curves (Figure 1). Exposure to 0.1 mM Cd(II) had little effect on the growth of “CD2” between 0 h and 48 h, but inhibited growth somewhat after 48 h (Figure 1A). The concentration of Cd(II) in the supernatant was decreased from 100 to 83.36 μM within 24 h, and decreased sharply between 24–36 h to 0 μM (Figure 1B). Meanwhile, by 36 h, the intracellular Cd(II) concentration increased to 34.98 μM and 60.39 μM of Cd(II) was immobilized by extracellular adsorption. These results suggest that strain CD2 adsorbs more Cd(II) than it sequesters (Figure 1B).
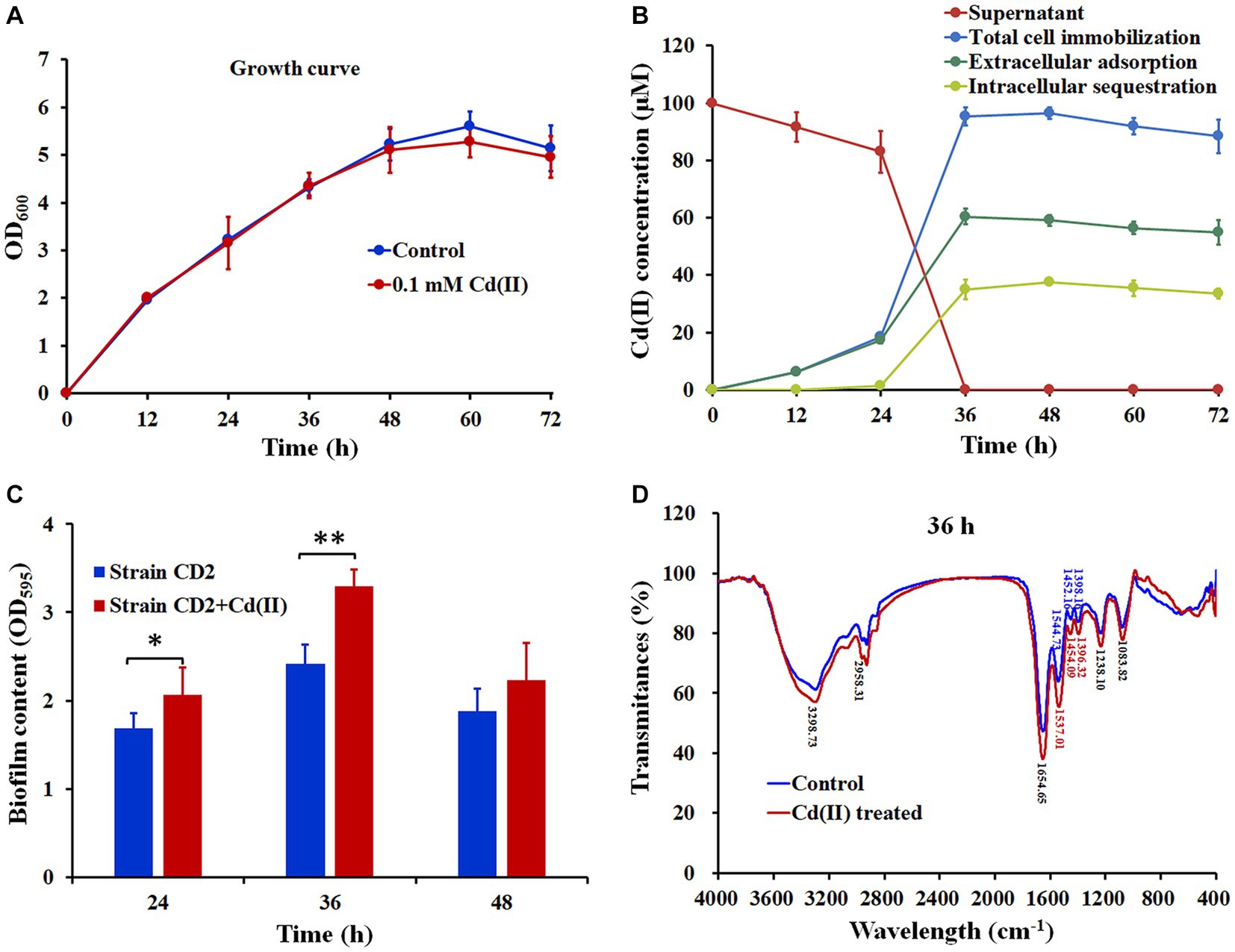
Figure 1. Growth curves, Cd(II) immobilization, biofilm production and FTIR spectrum of Stenotrophomonas sp. CD2. (A) Growth curves of strain CD2 in LB broth with or without 0.1 mmol/L Cd(II). (B) Cd(II) immobilization curves of strain CD2 and Cd(II) distribution curves in different cellular components. (C) Biofilm production of strain CD2 with or without 0.1 mmol/L Cd(II). (D) FTIR spectra of strain CD2 with or without 0.1 mmol/L Cd(II) at 36 h. The data were expressed as the mean ± standard deviation (n = 3).
To explore the mechanisms by which strain CD2 immobilizes Cd(II), we studied the ability of strain CD2 to produce EPS, H2S, and biofilm, as well as performed Fourier Transform Infrared (FTIR) spectroscopy. “CD2” colonies appeared blue when cultured on LB-aniline blue plates, indicating EPS production (Supplementary Figure S4A). Lead acetate test paper turned black when exposed to Cd(II)-containing and CD(II)-free medium, indicating that “CD2” produces H2S and that Cd(II) promotes H2S production (Supplementary Figure S4B). In addition, “CD2” was able to produce biofilm under control and Cd(II)-exposed conditions. Notably, “CD2” produced significantly more biofilm when exposed to Cd(II) than under control conditions at both 24 h and 36 h (Figure 1C). Analysis of the FTIR spectrum indicated that the transmittance of hydroxyl (3298.73 nm) was reduced from 61.30 to 57.02%, methyl C-H (2958.31 nm) was reduced from 95.84 to 94.66%, and ene hydroxyl (1654.65 nm) was reduced from 47.29 to 38.18% in the presence of Cd(II) (Figure 1D). The aliphatic nitro compound peak changed from 1544.73 nm to 1537.01 nm and the carboxyl peak changed from 1398.16 nm to 1396.32 nm, while the transmittance of aliphatic nitro compounds was reduced from 64.04 to 55.42% and of carboxyl groups from 83.77 to 79.75% (Figure 1D). Together, it appears that a combination of hydroxyl, methyl, ene hydroxyl, and carboxyl groups, as well as aliphatic nitro compounds, may be involved in the extracellular adsorption of Cd(II).
In order to evaluate the ability of strain CD2 to alter Cd(II) accumulation in B. rapa and immobilize Cd(II) in soil, we performed pot experiments. As shown in Figure 2A, the Cd concentrations in roots and aerial tissues were 1.29 mg/Kg and 0.36 mg/Kg in Cd-exposed plants, respectively. In contrast, the Cd concentrations in roots and aerial tissues were 0.63 mg/Kg and 0.16 mg/Kg in Cd-exposed plants inoculated with “CD2”, respectively (Figure 2A). Compared with uninoculated Cd-contaminated soil, the bioavailable Cd content decreased by 53.79% and the nonbioavailable Cd content significantly increased by 212.70% in “CD2”-inoculated soil (Figure 2B). These results suggest that strain CD2 can not only reduce the bioaccumulation of Cd(II) in plants but also immobilize Cd(II) in soil.
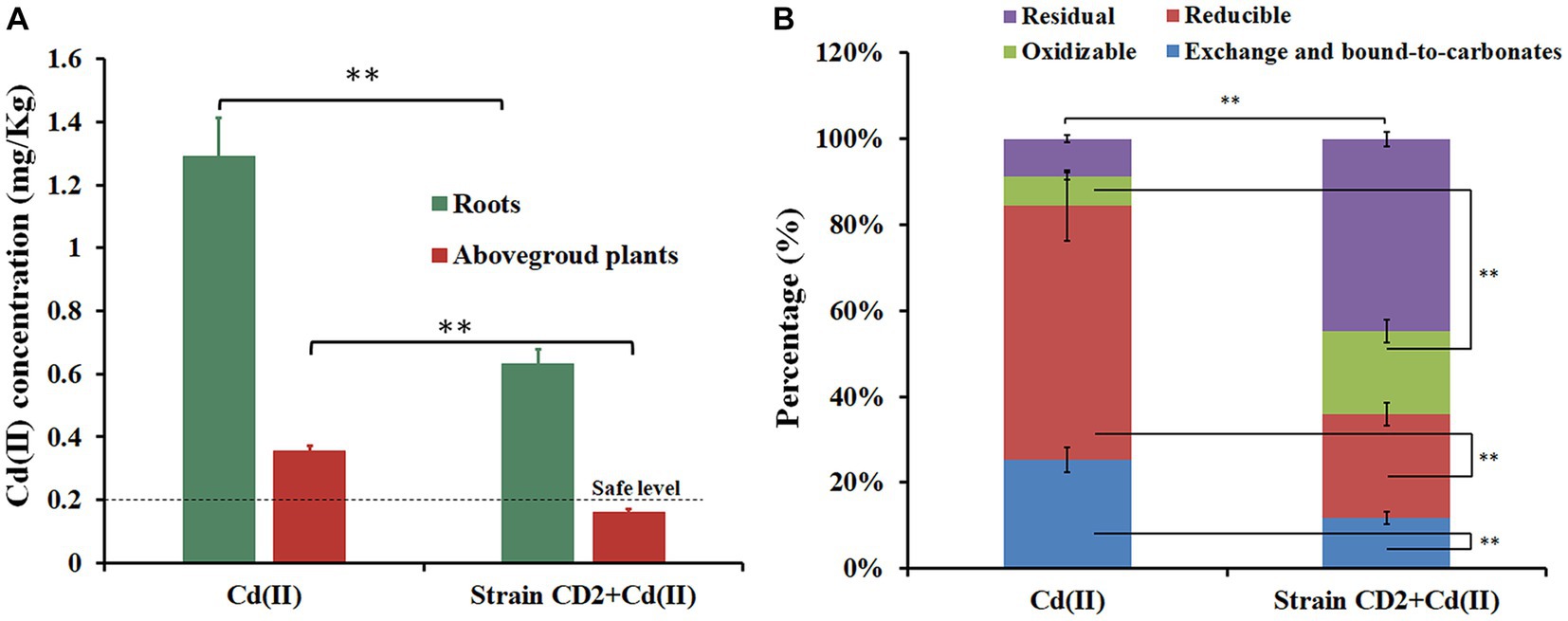
Figure 2. The effect of adding strain CD2 on Cd content in Brassica rapa L. (A) and Cd speciation in rhizosphere soil (B). **p-value <0.01.
We performed a transcriptomic analysis of the roots and aerial tissues of B. rapa inoculated with “CD2” in order to better understand the interactions between “CD2”, B. rapa, and Cd(II) exposure. Six treatment groups were analyzed: R-control (roots, no “CD2” or Cd(II) exposure), R-Cd (roots, with Cd(II) exposure), R-CD2 + Cd (roots, with “CD2” and Cd(II) exposure), L-control (aerial tissues, no “CD2” or Cd(II) exposure), L-Cd (aerial tissues, with Cd(II) exposure), and L-CD2 + Cd (aerial tissues, with “CD2” and Cd(II) exposure). Compared with R-control, R-Cd and R-CD2 + Cd contained 424 and 1,107 DEGs, respectively. Among these, R-Cd contained 258 upregulated DEGs and 166 downregulated DEGs, while R-CD2 + Cd contained 608 upregulated DEGs and 499 downregulated DEGs (Figures 3A,B). Compared with L-control, L-Cd and L-CD2 + Cd contained 501 and 1721 DEGs, respectively. Among these, L-Cd contained 223 upregulated DEGs and 278 downregulated DEGs, while L-CD2 + Cd contained 832 upregulated DEGs and 889 downregulated DEGs (Figures 3C,D). Comparisons between R-Cd vs. R-control, R-CD2 + Cd vs. R-control, L-Cd vs. L-control, and L-CD2 + Cd vs. L-control revealed the presence of 258, 911, 285, and 1,457 unique DEGs, respectively (Figure 3E). These results suggest that inoculation with strain CD2 results in greater differential gene expression in Cd(II)-exposed B. rapa.
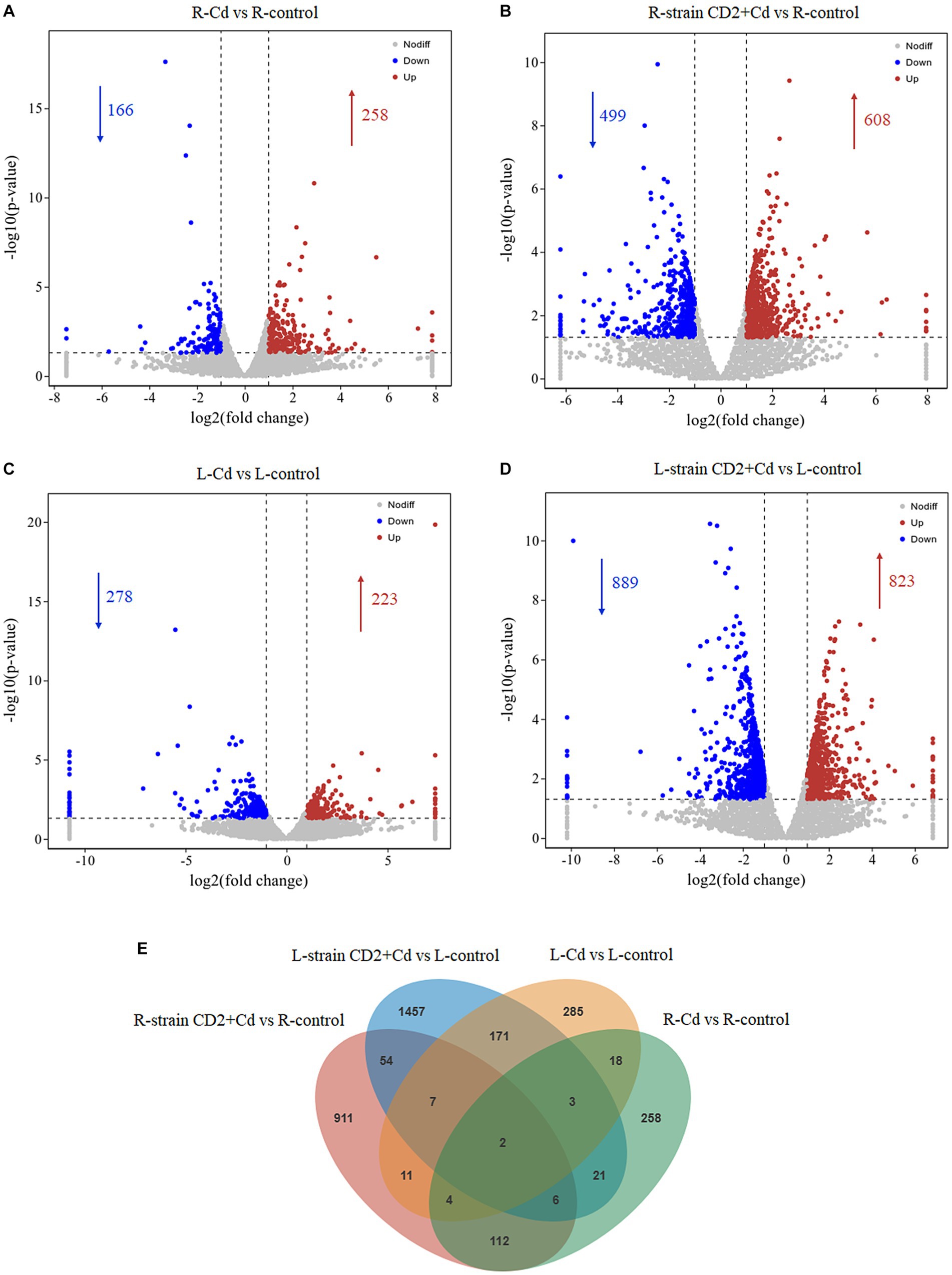
Figure 3. Volcano plots and Venn diagrams of differentially expressed genes (DEGs) in different comparative groups. (A) Volcano plot of significantly up- or down-regulated genes in roots of Cd vs. control group. (B) Volcano plot of significantly up- or down-regulated genes in roots of strain CD2 + Cd vs. control group. (C) Volcano plot of significantly up- or down-regulated genes in aerial tissues (shoots and leaves) of Cd vs. control group. (D) Volcano plot of significantly up- or down-regulated genes in aerial tissues of strain CD2 + Cd vs. control group. (E) Venn diagrams of DEGs in different comparative groups.
In order to clarify the primary biological functions of these DEGs, we performed GO and KEGG enrichment analyses. GO enrichment analysis divides functions into three categories: biological processes (BP), molecular functions (MF), and cellular components (CC). Compared to control roots, uninoculated Cd-exposed roots were enriched in 239 GO terms, including response to stimulus, response to chemical, oxidation–reduction process, and oxidoreductase activity, among other functions. Meanwhile, “CD2”-inoculated Cd-exposed roots were enriched in 442 GO terms, including membrane, cell periphery, plasma membrane, oxidation–reduction process, and oxidoreductase activity, among other functions (Figures 4A,B). Compared with control aerial tissues, the aerial tissues of uninoculated Cd-exposed plants were enriched in 342 GO terms, including organic acid metabolic process, oxoacid metabolic process, secondary metabolic process, and monocarboxylic acid metabolic process, among other functions. Finally, the aerial tissues of “CD2”-inoculated Cd-exposed plants were enriched in 441 GO terms, including nucleolus, thylakoid, ribosome biogenesis, and photosynthetic membrane, among other functions (Figures 4C,D). These results suggest that the Cd-response mechanisms differ between strain CD2-inoculated and uninoculated plants.
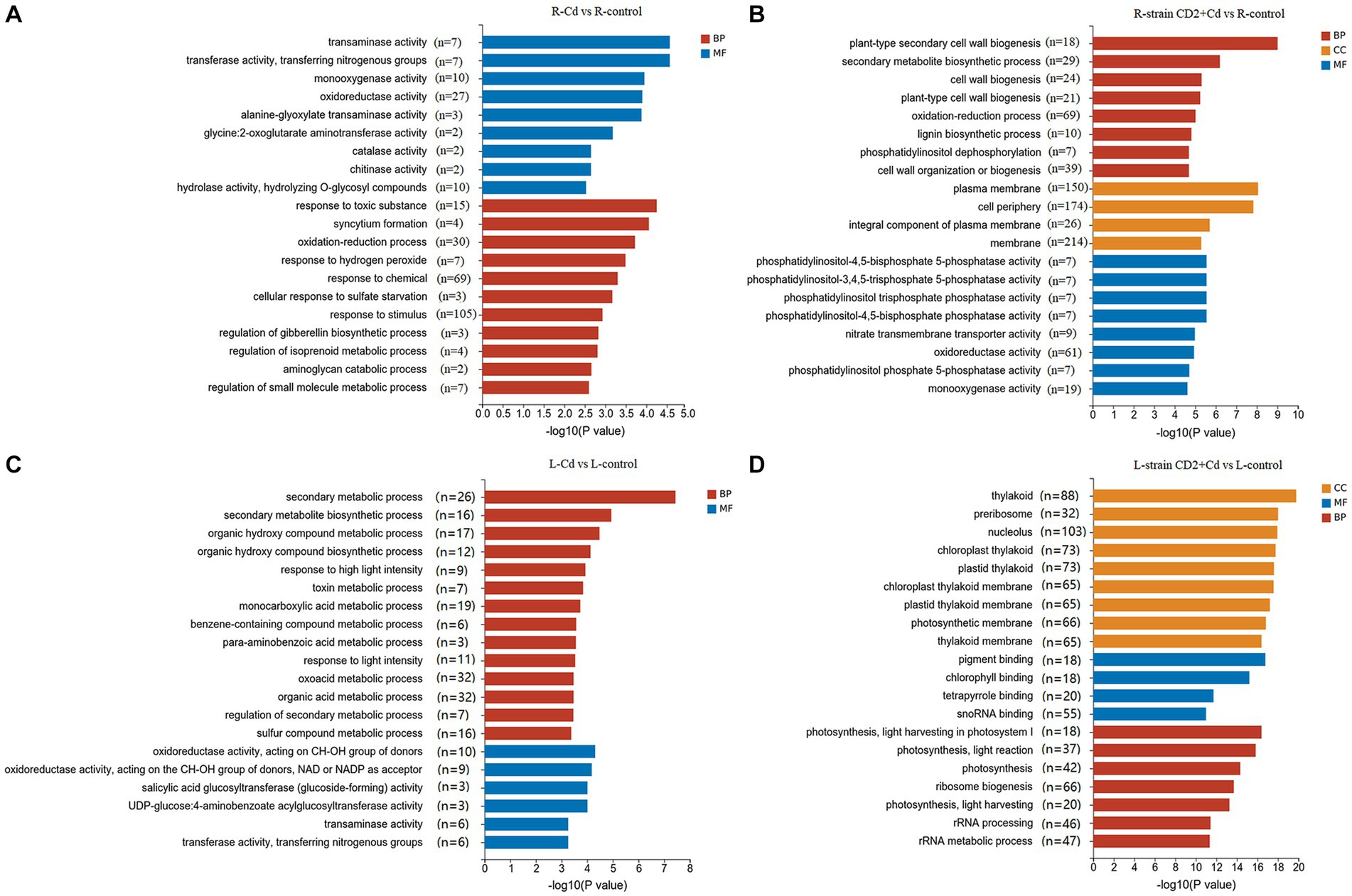
Figure 4. GO term enrichments based on DEGs under Cd stress. (A) Roots added Cd alone. (B) Roots added strain CD2 and Cd. (C) Aboveground parts added Cd alone. (D) Aboveground parts added strain CD2 and Cd.
Figure 5 shows the results of the KEGG pathway enrichment analysis of DEGs. Compared with control roots, uninoculated Cd-exposed roots were mainly enriched in glycine, serine and threonine metabolism; beta-alanine metabolism; other glycan degradation; glyoxylate and dicarboxylate metabolism; and glutathione metabolism. Meanwhile, “CD2”-inoculated Cd-exposed roots were mainly enriched in glucosinolate biosynthesis; phenylpropanoid biosynthesis; plant-pathogen interaction; cysteine and methionine metabolism; and cutin, suberine, and wax biosynthesis (Figures 5A,B). Compared with control aerial tissues, the aerial tissues of uninoculated Cd-exposed plants were mainly enriched in glycosphingolipid biosynthesis – ganglio series, fatty acid elongation, glycosaminoglycan degradation, pentose phosphate pathway, and glutamate metabolism. Finally, the aerial tissues of “CD2”-inoculated Cd-exposed plants were mainly enriched in photosynthesis – antenna proteins, ribosome biogenesis in eukaryotes, pentose phosphate pathway, fatty acid elongation, and glutathione metabolism (Figures 5C,D). These metabolic pathways have been reported as being responsive to Cd toxicity in other plants (Ma et al., 2022). These results suggest that the Cd(II)-response mechanisms of roots are more varied than those of aerial tissues.
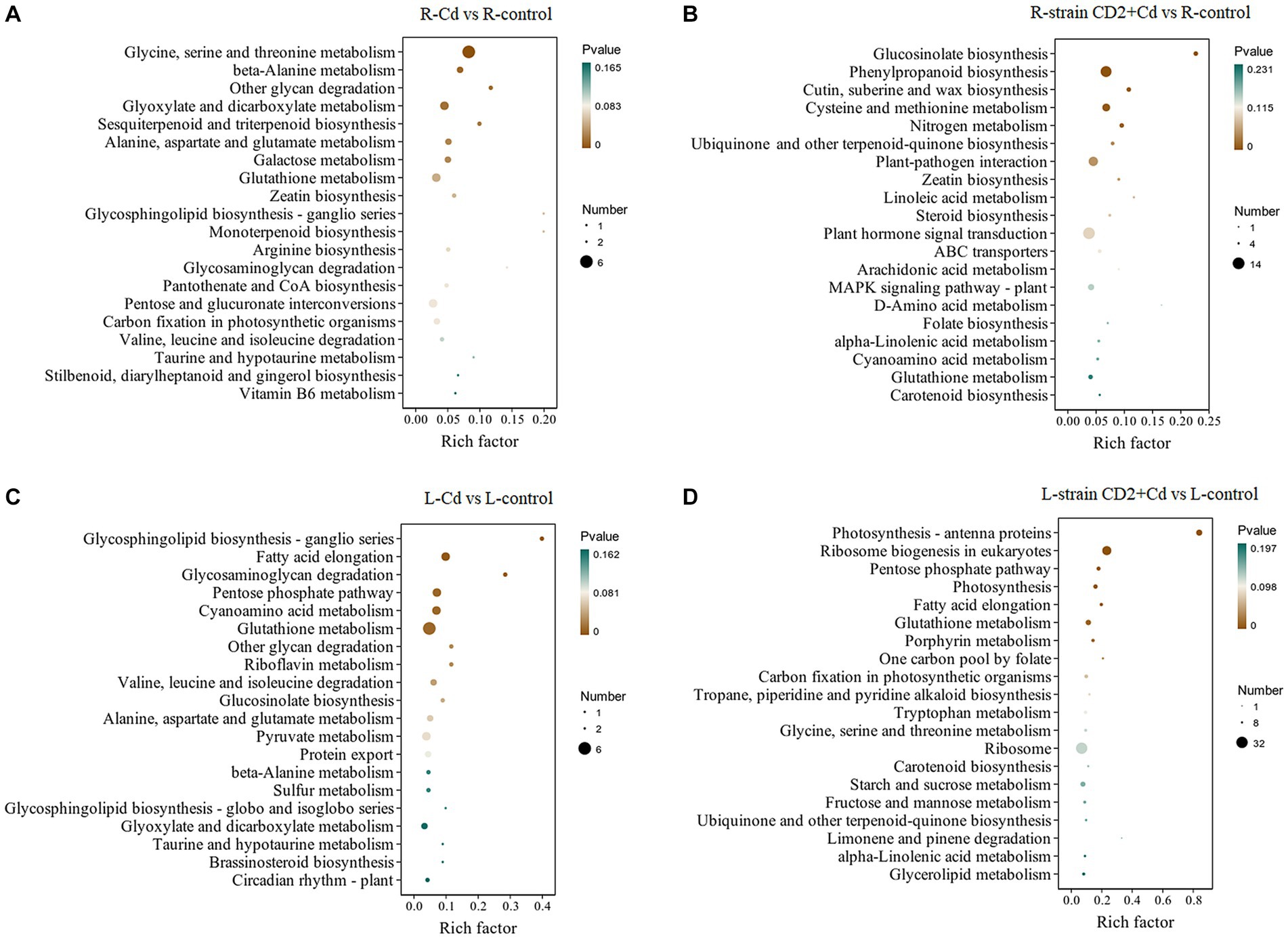
Figure 5. KEGG pathway enrichments based on DEGs under Cd stress. (A) Roots added Cd salone. (B) Roots added strain CD2 and Cd. (C) Aboveground parts added Cd alone. (D) Aboveground parts added strain CD2 and Cd.
TFs are well known to mediate plant responses to diverse stressors. In R-Cd vs. R-control, 36 DEGs were identified to belong to 16 families of TFs, including ERF, G2-like, MYB, NAC, and bHLH (Figure 6A). In R-CD2 + Cd vs. R-control, 107 DEGs were identified to belong to 29 families of TFs, including ERF, bHLH, MYB, NAC, MIKC_MADS, and B3 (Figure 6B). In L-Cd vs. L-control, 38 DEGs were identified to belong to 16 families of TFs, including ERF, MYB, WRKY, LBD, and bHLH (Figure 6C). Finally, in L-CD2 + Cd vs. L-control, 117 DEGs were identified to belong to 32 families of TFs, including bHLH, ERF, MYB, NAC, WRKY, and bZIP (Figure 6D). Among these comparisons, the most common Cd-responsive TF families were ERF, bHLH, and MYB. Notably, inoculation with “CD2” resulted in an increased number and diversity of Cd-responsive TFs.
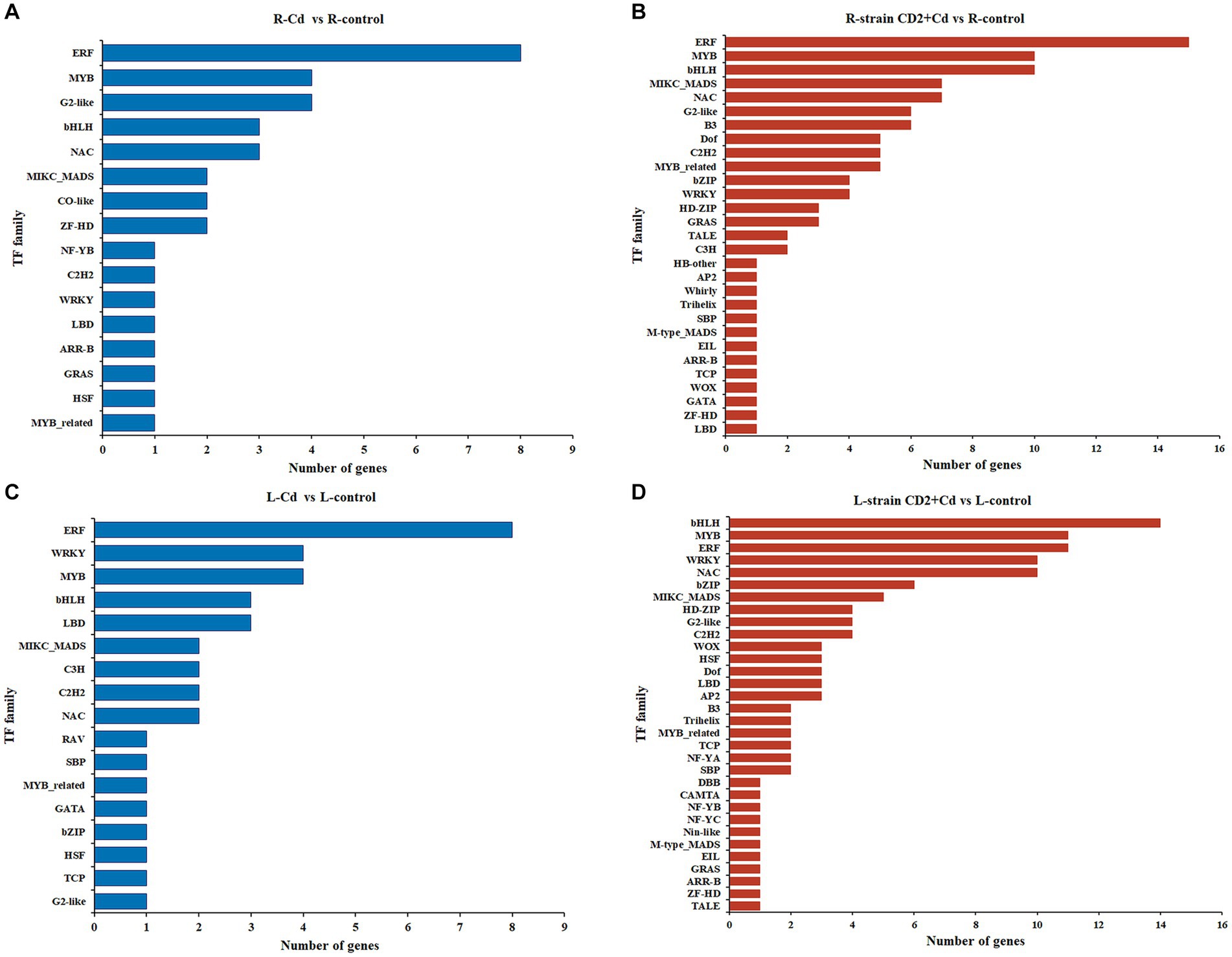
Figure 6. The number and distribution of transcription factors families under Cd stress. (A) Roots added Cd alone. (B) Roots added strain CD2 and Cd. (C) Aboveground parts added Cd alone. (D) Aboveground parts added strain CD2 and Cd.
Chinese cabbage (B. rapa.) is one of the most popular and widely cultivated vegetables in China. However, the risk of Cd contamination of this popular crop is increasing as more farmland becomes polluted with this toxic heavy metal (Zhao et al., 2015). Here, we tested the ability of the Cd-resistant bacterial strain CD2 to reduce Cd(II) accumulation in B. rapa (Figure 2). Strain CD2 was identified as belonging to the genus Stenotrophomonas (Supplementary Figure S1) and was found to be able to immobilize 0.1 mmol/L Cd(II) within 36 h (Figure 1). Several bacterial species and strains have been reported to immobilize Cd(II). For example, Pseudomonas sp. B7 can remove 0.1 mmol/L Cd(II) within 60 h (Wu et al., 2022). Enterobacter sp. A11, Comamonas sp. A23, and a co-culture of the two, can remove 0.1 mmol/L Cd(II) in around 48 h (Wang et al., 2020). Notably, the immobilization ability of strain CD2 was stronger than that of these bacteria. Microbes resist and immobilize Cd(II) through several mechanisms (Ammendola et al., 2014; Ziller and Fraissinet-Tachet, 2018; Gallardo-Benavente et al., 2019; Shi et al., 2020; Xia et al., 2021). Our research suggests that strain CD2 relies on a combination of intracellular sequestration and extracellular adsorption mediated by biofilm and EPS. In addition, strain CD2 was found to produce H2S, which could lead to the production of CdS precipitates. The genome of strain CD2 contained a multitude of efflux proteins such as CusA, CzcD, and MntH; Cd(II)-binding proteins such as CadW; and genes involved in the production of biofilms, EPS, and H2S (Supplementary Table S1). Therefore, our phenotypic observations were in good agreement with our genotypic analysis.
Inoculation with strain CD2 was found to effectively reduce the Cd content in the roots and aerial tissues of B. rapa. Specifically, “CD2”-inoculation reduced the Cd content of roots by 51.16% and of aerial tissues by 55.56%, compared to uninoculated Cd-exposed plants. Moreover, the residual Cd content in the aerial tissues (edible parts) met food security standards (0.2 mg/kg) in ‘CD2-inoculated plants (Codex Alimentarius Commission, 2010; European Union, 2011; Ministry of Health (MOH), 2012). In addition, adding strain CD2 to the soil significantly increased the nonbioavailable Cd content and reduced the bioavailable Cd content. This suggests that strain CD2 may prevent Cd accumulation in plants by reducing the bioavailable Cd(II) content of the soil itself. Notably, this mechanism has been reported in other microbe-plant systems (Shi et al., 2020; Wang et al., 2020; Wu et al., 2022; Lou et al., 2024). The addition of strain CD2 to the soil effectively reduces the Cd accumulation in B. rapa during pot experiments. The future research will primarily focus on evaluating the efficacy of strain CD2 in mitigating Cd accumulation in plants when applied to real-world soil contaminated with Cd.
Transcriptomic analysis can identify key genes and expression patterns, as well as clarify the mechanisms, associated with the plant response to heavy metal stress. Several metabolic pathways have been implicated in the plant response to Cd(II) stress (Thomine et al., 2000; Anjum et al., 2015). Surprisingly, we found that different functions and pathways were activated in uninoculated and “CD2”-inoculated Cd(II)-exposed plants, according to GO and KEGG enrichment analyses. According to the GO enrichment analysis, inoculation with “CD2” affected the expression of genes related to cell wall biosynthesis in roots and membranes (plasma, thylakoid, chloroplast, and photosynthetic) in aerial tissues. Similar results have been reported in Cd-stressed maize and mustard (Brassica juncea) (Peng et al., 2015; Li et al., 2023). Studies suggest that the biosynthesis of cutin, suberine, wax, and phenylpropanoid confers abiotic stress resistance in plants. This is because these lipophilic cell wall barriers mediate the flux of gasses, water, and solutes (Pollard et al., 2008). The phenylpropane metabolic pathway precedes the biosynthesis of flavonoids, which participate in the abiotic stress response through a wide range of chemical reactions (Gan et al., 2022). We found that “CD2”-inoculation resulted in an increase in DEGs related to cutin, suberine, wax, and phenylpropanoid biosynthesis (Figure 5B). These results suggest that inoculation with “CD2” increases the Cd-stress resistance of B. rapa by regulating secondary metabolism.
In plants, excessive Cd accumulation results in reactive oxygen species (ROS) metabolism disturbance. In response, plants have evolved antioxidant mechanisms to maintain physiological homeostasis (Li et al., 2022). We identified several Cd-responsive DEGs related to oxidative stress resistance in both the GO and KEGG enrichment analyses. In uninoculated Cd-exposed roots, these DEGs were enriched in the GO terms catalase (2 upregulated), antioxidant (4 upregulated and 1 downregulated), glutathione transferase activity (2 upregulated and 1 downregulated), and response to hydrogen peroxide (3 upregulated and 4 downregulated). Meanwhile, in uninoculated Cd-exposed aerial tissues, these DEGs were only enriched in glutathione transferase (2 upregulated and 2 downregulated) (Supplementary Table S2). Following inoculation with “CD2”, Cd-exposed roots were found to be enriched in hydrogen peroxide (4 upregulated and 5 downregulated) while Cd-exposed aerial tissues were found to be enriched in glutathione transferase (10 downregulated). KEGG enrichment analysis indicated that glutathione metabolism was enriched significantly in both the roots and aerial tissues of uninoculated Cd-exposed plants, but only in the aerial tissues of “CD2”-inoculated Cd-exposed plants. In addition, “plant-pathogen interaction” was enriched significantly in the roots of “CD2”-inoculated Cd-exposed plants (Figure 5B). The ‘plant-pathogen interaction’ KEGG category includes important signaling molecules which mediate the interaction between microbes and plants. Two genes in this category (CPK and RBOH) were upregulated in the roots of Cd-stress plants (Supplementary Table S3). Both of these genes confer oxidative stress resistance through regulating Ca2+ dynamics. Together, these results indicate that Cd-induced oxidative stress was more severe in roots than in aerial tissues. This may be related to the relatively lower content of Cd in aerial tissues than in roots. Moreover, inoculation with “CD2” appears to reduce oxidative stress in B. rapa through a ‘plant-pathogen interaction’-related mechanism.
Transporter proteins are responsible for sequestering Cd into vacuoles, thereby preventing the transport of Cd from roots to shoots (Zhang et al., 2010). Several upregulated proteins related to vacuolar sequestration were identified in the roots of Cd-exposed B. rapa, including three in uninoculated plants and two in “CD2”-inoculated plants. These results suggest that vacuolar sequestration may inhibit Cd transport in both inoculated and uninoculated plants. In plants, Cd(II) uptake and accumulation are also affected by ion channel proteins and transporter proteins such as ZIPs, ABC transporters, and ATPases. We observed that Cd exposure changed the expression of ABC transporters, ATPases, sodium/calcium exchanger proteins, and potassium channel proteins in uninoculated or “CD2”-inoculated plants (Supplementary Table S3). Additionally, inoculation with “CD2” resulted in the downregulation of a ZIP family Zn-transporter and three blue copper proteins (BCPs) in roots (Supplementary Table S3). BCPs can bind Cd(II) or participate in Cd(II) efflux (Wang et al., 2023). Thus, it appears that inoculation with “CD2” may negatively impact the expression of Zn transporters and BCPs, resulting in reduced Cd accumulation in B. rapa.
A growing body of research indicates that TFs are important regulators of the plant response to abiotic stress. In particular, the WRKY, MYB, bHLH, ERF, ZIP, and C2H2 TFs have been implicated in the Cd stress response (Tokumoto et al., 2019; He et al., 2021). We found that the ERF, MYB, C2H2, NAC, WRKY, and G2-like TFs were highly responsive to Cd in the roots and aerial tissues of both uninoculated and “CD2”-inoculated plants (Figure 6), suggesting that these TFs represent an innate Cd stress-resistance mechanism in B. rapa. In addition, inoculation with “CD2” resulted in the upregulation of AP2, Dof, WOX, Trihelix, B3, EIL, and M-type MADS TFs in both roots and aerial tissues. Dof TFs are specific to plants, and play important roles in growth, development, seed germination, photoresponse, storage protein accumulation, biological stress, carbon and nitrogen metabolism, secondary metabolite biosynthesis, and stress response (Zou and Sun, 2023). In Arabidopsis thaliana, Dof4.2 regulates the accumulation of flavonoids under stressful conditions (Skirycz et al., 2007). According to KEGG analysis, inoculation with “CD2” resulted in induction of the phenylpropane metabolic pathway, which precedes stress-reducing flavonoid biosynthesis (Gan et al., 2022). Therefore, inoculation with strain CD2 may help defend plants against Cd stress through a mechanism mediated by Dof and flavonoid biosynthesis. The more experiments would be carried out in the future to detect the function of Dof and flavonoid biosynthesis pathway in resisting Cd.
In this study, the highly Cd(II)-resistant Stenotrophomonas sp. CD2 was isolated from Cd-contaminated soil. This strain was found to be able to immobilize Cd(II) through both intracellular sequestration and extracellular adsorption related to the production of biofilm, EPS, and H2S. Application of “CD2” to Cd-polluted soil resulted in reduced bioavailability of Cd(II) and reduced Cd(II) accumulation in B. rapa. Moreover, the Cd content of the aerial tissues (edible parts) of “CD2”-inoculated plants was found to meet food security standards. Transcriptomic analysis revealed that uninoculated B. rapa resisted Cd-induced ROS toxicity mainly through the expression of catalase and glutathione transferase, while “CD2”-inoculated plants exhibited an increase in ‘plant-pathogen interaction’ pathways. Both uninoculated and inoculated plants were able to prevent the transportation of Cd from roots to shoots by vacuolar sequestration. In addition, inoculation with “CD2” was found to inhibit the expression of Zn transporters and BCPs; alter the expression of various TFs (in particular, Dof) and the biosynthesis of phenylpropane, cutin, suberine, and wax. The results of this study suggest that Stenotrophomonas sp. CD2 may prove to be a useful inoculant to prevent Cd accumulation in B. rapa.
The bacterial strain Stenotrophomonas sp. CD2 has been deposited as a patent strain in China Center for Type Culture Collection (http://www.cctcc.org/) under the accession number of CCTCC M 20231348.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ncbi.nlm.nih.gov/genbank/, CP102248 https://www.ncbi.nlm.nih.gov/, PRJNA1064471.
XF: Conceptualization, Writing – review & editing, Project administration, Data curation. KY: Data curation, Investigation, Methodology, Writing – original draft. QP: Data curation, Formal analysis, Writing – original draft. RL: Data curation, Formal analysis, Writing – original draft. YZ: Conceptualization, Project administration, Writing – original draft.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was funded by the National Natural Science Foundation of China (32100102).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fsufs.2024.1362265/full#supplementary-material
Akesson, A., Julin, B., and Wolk, A. (2008). Long-term dietary cadmium intake and postmenopausal endometrial cancer incidence: a population-based prospective cohort study. Cancer Res. 68, 6435–6441. doi: 10.1158/0008-5472.CAN-08-0329
Albano, L. J., and Macfie, S. M. (2016). Investigating the ability of Pseudomonas fluorescens UW4 to reduce cadmium stress in Lactuca sativa via an intervention in the ethylene biosynthetic pathway. Can. J. Microbiol. 62, 1057–1062. doi: 10.1139/cjm-2016-0315
Ammendola, S., Cerasi, M., and Battistoni, A. (2014). Deregulation of transition metals homeostasis is a key feature of cadmium toxicity in Salmonella. Biometals 27, 703–714. doi: 10.1007/s10534-014-9763-2
Anders, S., Pyl, P. T., and Huber, W. (2015). HTSeq-a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169. doi: 10.1093/bioinformatics/btu638
Anjum, N. A., Hasanuzzaman, M., Hossain, M. A., Thangavel, P., Roychoudhury, A., Gill, S. S., et al. (2015). Jacks of metal/metalloid chelation trade in plants — an overview. Front. Plant Sci. 6:192. doi: 10.3389/fpls.2015.00192
Ashraf, M., Hasnain, S., Berge, O., and Mahmood, T. (2004). Inoculating wheat seedlings with exopolysaccharide-producing bacteria restricts sodium uptake and stimulates plant growth under salt stress. Biol. Fert. Soils 40, 157–162. doi: 10.1007/s00374-004-0766-y
Chen, S., Zhou, Y., Chen, Y., and Gu, J. (2018). Fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890. doi: 10.1093/bioinformatics/bty560
Clemens, S. (2006). Toxic metal accumulation, responses to exposure and mechanisms of tolerance in plants. Biochimie 88, 1707–1719. doi: 10.1016/j.biochi.2006.07.003
Clemens, S., Palmgren, M. G., and Krämer, U. (2002). A long way ahead: understanding and engineering plant metal accumulation. Trends Plant Sci. 7, 309–315. doi: 10.1016/S1360-1385(02)02295-1
Codex Alimentarius Commission . (2010). Codex general standard for contaminants and toxins in food and feed, CODEX STAN 193–1995: Codex Alimentarius commission.
European Union (2011). Commission regulation (EC) no 420/2011 of 29 April 2011 amending regulation (EC) no 1881/2006 setting maximum levels for certain contaminants in foodstuffs. Off. J. Eur. Commun. 111, 5–6.
Feng, S., Tan, J., Zhang, Y., Liang, S., Xiang, S., Wang, H., et al. (2017). Isolation and characterization of a novel cadmium-regulated yellow stripe-like transporter (SnYSL3) in Solanum nigrum. Plant Cell Rep. 36, 281–296. doi: 10.1007/s00299-016-2079-7
Gallardo-Benavente, C., Carrión, O., Todd, J. D., Pieretti, J. C., Seabra, A. B., Durán, N., et al. (2019). Biosynthesis of CdS quantum dots mediated by volatile sulfur compounds released by antarctic Pseudomonas fragi. Front. Microbiol. 10, 1866–1880. doi: 10.3389/fmicb.2019.01866
Gan, C., Liu, Z., Pang, B., Zuo, D., Hou, Y., Zhou, L., et al. (2022). Integrative physiological and transcriptome analyses provide insights into the cadmium (cd) tolerance of a cd accumulator: Erigeron canadensis. BMC Genomics 23:778. doi: 10.1186/s12864-022-09022-5
Guerinot, M. L. (2000). The ZIP family of metal transporters. BBA-Biomembranes 1465, 190–198. doi: 10.1016/S0005-2736(00)00138-3
Haft, D. H., DiCuccio, M., Badretdin, A., Brover, V., Chetvernin, V., O'Neill, K., et al. (2018). RefSeq: an update on prokaryotic genome annotation and curation. Nucleic Acids Res. 46, D851–D860. doi: 10.1093/nar/gkx1068
He, H., Ye, Z., Yang, D., Yan, J., Xiao, L., Zhong, T., et al. (2013). Characterization of endophytic Rahnella sp. JN6 from Polygonum pubescens and its potential in promoting growth and cd, Pb, Zn uptake by Brassica napus. Chemosphere 90, 1960–1965. doi: 10.1016/j.chemosphere.2012.10.057
He, Q. H., Zhou, T., Sun, J. K., Wang, P., Yang, C. P., Bai, L., et al. (2021). Transcriptome profiles of leaves and roots of goldenrain tree (Koelreuteria paniculata Laxm.) in response to cadmium stress. Int. J. Environ. Res. Public Health 18:12046. doi: 10.3390/ijerph182212046
Hu, H., Miao, Y. R., Jia, L. H., Yu, Q. Y., Zhang, Q., and Guo, A. Y. (2019). AnimalTFDB 3.0: a comprehensive resource for annotation and prediction of animal transcription factors. Nucleic Acids Res. 47, D33–D38. doi: 10.1093/nar/gky822
Kartal, S., Aydin, Z., and Tokalioglu, S. (2006). Fractionation of metals in street sediment samples by using the BCR sequential extraction procedure and multivariate statistical elucidation of the data. J. Hazard. Mater. 132, 80–89. doi: 10.1016/j.jhazmat.2005.11.091
Kubier, A., Wilkin, R. T., and Pichler, T. (2019). Cadmium in soils and groundwater: a review. Appl. Geochem. 108, 104388–104316. doi: 10.1016/j.apgeochem.2019.104388
Kuramata, M., Masuya, S., Takahashi, Y., Kitagawa, E., Inoue, C., Ishikawa, S., et al. (2009). Novel cysteine-rich peptides from Digitaria ciliaris and Oryza sativa enhance tolerance to cadmium by limiting its cellular accumulation. Plant Cell Physiol. 50, 106–117. doi: 10.1093/pcp/pcn175
Larsson, S. C., and Wolk, A. (2016). Urinary cadmium and mortality from all causes, cancer and cardiovascular disease in the general population: systematic review and meta-analysis of cohort studies. Int. J. Epidemiol. 45, 782–791. doi: 10.1093/ije/dyv086
Li, C., Cao, Y., Li, T., Guo, M., Ma, X., Zhu, Y., et al. (2022). Changes in antioxidant system and sucrose metabolism in maize varieties exposed to cd. Environ. Sci. Pollut. Res. Int. 29, 64999–65011. doi: 10.1007/s11356-022-20422-8
Li, H., Liu, Y., Zeng, G., Zhou, L., Wang, X., Wang, Y., et al. (2014). Enhanced efficiency of cadmium removal by Boehmeria nivea (L.) gaud. In the presence of exogenous citric and oxalic acids. J. Environ. Sci. 26, 2508–2516. doi: 10.1016/j.jes.2014.05.031
Li, W., O’Neill, K. R., Haft, D. H., DiCuccio, M., Chetvernin, V., Badretdin, A., et al. (2021). RefSeq: expanding the prokaryotic genome annotation pipeline reach with protein family model curation. Nucleic Acids Res. 49, D1020–D1028. doi: 10.1093/nar/gkaa1105
Li, L., Wang, S., Wu, S., Rao, S., Li, L., Cheng, S., et al. (2023). Morphological and physiological indicators and transcriptome analyses reveal the mechanism of selenium multilevel mitigation of cadmium damage in Brassica juncea. Plants (Basel). 12:1583. doi: 10.3390/plants12081583
Liu, F., Liu, X. N., Ding, C., and Wu, L. (2015). The dynamic simulation of rice growth parameters under cadmium stress with the assimilation of multi-period spectral indices and crop model. Field Crop Res. 183, 225–234. doi: 10.1016/j.fcr.2015.08.004
Lou, Y. X., Liao, M., Lu, X. X., Xu, N., Xie, X. M., and Gao, W. M. (2024). Unveiling the performance of a novel alkalizing bacterium Enterobacter sp. LYX-2 in immobilization of available cd. J. Environ. Sci. 137, 245–257. doi: 10.1016/j.jes.2023.02.010
Ma, X., Zhao, X., Zhang, Q., Zhou, Z. H., Dou, Y. B., Ji, W., et al. (2022). Comparative transcriptome analysis of broccoli seedlings under different cd exposure levels revealed possible pathways involved in hormesis. Sci. Hortic. 304:111330. doi: 10.1016/j.scienta.2022.111330
Madhaiyan, M., Poonguzhali, S., and Sa, T. (2007). Metal tolerating methylotrophic bacteria reduces nickel and cadmium toxicity and promotes plant growth of tomato (Lycopersicones culentum L.). Chemosphere 69, 220–228. doi: 10.1016/j.chemosphere.2007.04.017
Ministry of Health (MOH) . (2012). National Food Safety Standard-Contaminant Limit in food, GB 2762–2012. Beijing, China: Ministry of Health of P. R. China.
Moons, A. (2003). OsPDR9, which encodes a PDR-type ABC transporter, is induced by heavy metals, hypoxic stress and redox perturbations in rice roots. FEBS Lett. 553, 370–376. doi: 10.1016/S0014-5793(03)01060-3
Nancharaiah, Y. V., Venkata, M. S., and Lens, P. N. (2015). Metals removal and recovery in bioelectrochemical systems: a review. Bioresour. Technol. 195, 102–114. doi: 10.1016/j.biortech.2015.06.058
O’Toole, G., and Kolter, R. (1998). Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol. Microbiol. 30, 295–304. doi: 10.1046/j.1365-2958.1998.01062.x
Peng, H., He, X., Gao, J., Ma, H., Zhang, Z., Shen, Y., et al. (2015). Transcriptomic changes during maize roots development responsive to cadmium (cd) pollution using comparative RNAseq-based approach. Biochem. Biophys. Res. Commun. 464, 1040–1047. doi: 10.1016/j.bbrc.2015.07.064
Pollard, M., Beisson, F., Li, Y., and Ohlrogge, J. B. (2008). Building lipid barriers: biosynthesis of cutin and suberin. Trends Plant Sci. 13, 236–246. doi: 10.1016/j.tplants.2008.03.003
Punjee, P., Siripornadulsil, W., and Siripornadulsil, S. (2018). Reduction of cadmium uptake in rice endophytically colonized with the cadmium-tolerant bacterium Cupriavidus taiwanensis KKU2500-3. Can. J. Microbiol. 64, 131–145. doi: 10.1139/cjm-2017-0198
Shi, Z., Zhang, Z., Yuan, M., Wang, S., Yang, M., Yao, Q., et al. (2020). Characterization of a high cadmium accumulating soil bacterium, Cupriavidus sp. WS2. Chemosphere 247:125834. doi: 10.1016/j.chemosphere.2020.125834
Skirycz, A., Jozefczuk, S., Stobiecki, M., Muth, D., Zanor, M. I., Witt, I., et al. (2007). Transcription factor AtDOF4;2 affects phenylpropanoid metabolism in Arabidopsis thaliana. New Phytol. 175, 425–438. doi: 10.1111/j.1469-8137.2007.02129.x
Sun, Y., Liu, X., Li, W., Wang, X., Zhong, X., Gao, Y., et al. (2023). The regulatory metabolic networks of the Brassica campestris L. hairy roots in response to cadmium stress revealed from proteome studies combined with a transcriptome analysis. Ecotoxicol. Environ. Saf. 263:115214. doi: 10.1016/j.ecoenv.2023.115214
Tamura, K., Stecher, G., and Kumar, S. (2021). MEGA11: molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 38, 3022–3027. doi: 10.1093/molbev/msab120
Tatusova, T., DiCuccio, M., Badretdin, A., Chetvernin, V., Nawrocki, E. P., Zaslavsky, L., et al. (2016). NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 44, 6614–6624. doi: 10.1093/nar/gkw569
Thomine, S., Wang, R., Ward, J. M., Crawford, N. M., and Schroeder, J. I. (2000). Cadmium and iron transport by members of a plant metal transporter family in Arabidopsis with homology to Nramp genes. Proc. Nat. Acad. Sci. U. S. A. 97, 4991–4996. doi: 10.1073/pnas.97.9.4991
Tian, F., Yang, D. C., Meng, Y. Q., Jin, J., and Gao, G. (2020). PlantRegMap: charting functional regulatory maps in plants. Nucleic Acids Res. 48, D1104–D1113. doi: 10.1093/nar/gkz1020
Tokumoto, M., Lee, J. Y., and Satoh, M. (2019). Transcription factors and downstream genes in cadmium toxicity. Biol. Pharm. Bull. 42, 1083–1088. doi: 10.1248/bpb.b19-00204
Turner, A. (2019). Cadmium pigments in consumer products and their health risks. Sci. Total Environ. 657, 1409–1418. doi: 10.1016/j.scitotenv.2018.12.096
Verbruggen, N., Hermans, C., and Schat, H. (2009). Mechanisms to cope with arsenic or cadmium excess in plants. Curr. Opin. Plant Biol. 12, 364–372. doi: 10.1016/j.pbi.2009.05.001
Waalkes, M. P. (2003). Cadmium carcinogenesis. Mutat. Res. 533, 107–120. doi: 10.1016/j.mrfmmm.2003.07.011
Wang, X., Hu, K., Xu, Q., Lu, L., Liao, S., and Wang, G. (2020). Immobilization of cd using mixed Enterobacter and Comamonas bacterial reagents in pot experiments with Brassica rapa L. Environ. Sci. Technol. 54, 15731–15741. doi: 10.1021/acs.est.0c03114
Wang, X., Xu, Q., Hu, K., Wang, G., and Shi, K. (2023). A coculture of Enterobacter and Comamonas species reduces cadmium accumulation in rice. Mol. Plant-Microbe Interact. 36, 95–108. doi: 10.1094/MPMI-09-22-0186-R
Wu, S., Zhou, Z., Zhu, L., Zhong, L., Dong, Y., Wang, G., et al. (2022). Cd immobilization mechanisms in a Pseudomonas strain and its application in soil cd remediation. J. Hazard. Mater. 425:127919. doi: 10.1016/j.jhazmat.2021.127919
Xia, X., Wu, S., Zhou, Z., and Wang, G. (2021). Microbial cd(II) and Cr(VI) resistance mechanisms and application in bioremediation. J. Hazard. Mater. 401:123685. doi: 10.1016/j.jhazmat.2020.123685
Yu, Y., Wang, Q., Wan, Y., Huang, Q., and Li, H. (2023). Transcriptome analysis reveals different mechanisms of selenite and selenate regulation of cadmium translocation in Brassica rapa. J. Hazard. Mater. 452:131218. doi: 10.1016/j.jhazmat.2023.131218
Zhang, Z. C., Chen, B. X., and Qiu, B. S. (2010). Phytochelatin synthesis plays a similar role in shoots of the cadmium hyperaccumulator Sedum alfredii as in non-resistant plants. Plant Cell Environ. 33, 1248–1255. doi: 10.1111/j.1365-3040.2010.02144.x
Zhao, F. J., Ma, Y., Zhu, Y. G., Tang, Z., and McGrath, S. P. (2015). Soil contamination in China: current status and mitigation strategies. Environ. Sci. Technol. 49, 750–759. doi: 10.1021/es5047099
Zhou, C., Ge, N., Guo, J., Zhu, L., Ma, Z., Cheng, S., et al. (2019). Enterobacter asburiae reduces cadmium toxicity in maize plants by repressing iron uptake-associated pathways. J. Agric. Food Chem. 67, 10126–10136. doi: 10.1021/acs.jafc.9b03293
Ziller, A., and Fraissinet-Tachet, L. (2018). Metallothionein diversity and distribution in the tree of life: a multifunctional protein. Metallomics 10, 1549–1559. doi: 10.1039/C8MT00165K
Keywords: Brassica rapa L., cadmium, Stenotrophomonas , Cd accumulation, transcriptome
Citation: Fan X, Yuan K, Peng Q, Lv R and Zheng Y (2024) Stenotrophomonas strain CD2 reduces cadmium accumulation in Brassica rapa L. Front. Sustain. Food Syst. 8:1362265. doi: 10.3389/fsufs.2024.1362265
Received: 28 December 2023; Accepted: 26 January 2024;
Published: 12 February 2024.
Edited by:
Juan Han, Chinese Academy of Agricultural Sciences, ChinaReviewed by:
Xiangjun Zhou, Hubei Normal University, ChinaCopyright © 2024 Fan, Yuan, Peng, Lv and Zheng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xia Fan, ZmFueGlhQGhnbnUuZWR1LmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.