- 1Science Center for Future Foods, Jiangnan University, Wuxi, Jiangsu, China
- 2Engineering Research Center of Ministry of Education on Food Synthetic Biotechnology, Jiangnan University, Wuxi, Jiangsu, China
Fibroblasts are important components of animal tissues such as muscle and skin, as they are the major producers of various matrix proteins. Matrix proteins such as collagen play an important role in meat products by providing unique nutrition, texture, and flavor. Cultured meat is an innovative meat alternative produced by culturing animal cells, but currently, relatively few studies have been conducted using fibroblasts as seed cells for cultured meat manufacturing. In this work, we first developed an innovative digestion-friction method for isolating fibroblasts from porcine skin efficiently and cost-effectively. After optimizing the enzymatic digestion and physical friction conditions, 2.39 ± 0.28 × 105 fibroblasts were obtained from 1 cm2 of porcine skin tissue, which was about 9 times higher than the conventional tissue explant method. In addition, we identified an edible bio-ink composed of gelatin and chitosan that has good printing properties and supports fibroblast adhesion and growth. Furthermore, we fabricated fibroblast-based cultured meat by 3D printing with an initial cell density of 1.0 × 107 mL−1 and evaluated its texture and nutritional properties. This work provides valuable insights and references for introducing fibroblasts into the production of cultured meat that is more comparable to structured animal meat.
1 Introduction
Cultured meat, also known as cultivated meat or cell-based meat, is a novel food produced using cell culture, tissue engineering, and food processing technologies (Guan et al., 2021; Liu et al., 2023). Cultured meat technology is a promising alternative to conventional farming, as it is expected to address resource shortages, environmental pollution, and animal welfare issues (Zhang et al., 2020). For both technological development and industrial production of cultured meat, seed cells are undoubtedly the most important. In general, muscle satellite cells are the most preferred seed cells because of their ability to proliferate and differentiate into skeletal muscle fibers, endowing cultured meat with the nutritional and processing properties of native muscle tissue (Choi et al., 2021; Guan et al., 2022, 2023; Yan et al., 2022; Zhu et al., 2022). Other cell types such as fibro-adipogenic progenitor (FAP) and adipose-derived stem cells (ADSCs) are of interest because they can generate fat cells, providing meat flavor and juicy mouthfeel to cultured meat (Dohmen et al., 2022; Liu et al., 2023).
In addition to muscle fibers and fat tissues, natural meat products contain other components such as connective tissue and skin, which provide different textures and nutrition from muscle and fat, but are neglected in most cultured meat studies. In both connective tissue and skin, the main components are fibroblasts and their secreted extracellular matrix (ECM) proteins such as collagen, elastin, and fibronectin (Plikus et al., 2021). The ECM surrounds and supports muscle fibers and skin layers, constituting a complex and compact tissue structure that provides strong mechanical strength to animal bodies as well as different physiological functions (Ahmad et al., 2021). At the same time, these ECM proteins not only absorb seasonings and enhance the overall flavor of the dish but are also beneficial to the skin, bones, and joints after consumption. Therefore, it is important to introduce fibroblasts in the production of cultured meat to increase the content of ECM ingredients, thereby enhancing the structural integrity and improving the nutrition and mouthfeel of cultured meat. Currently in the field of cultured meat, although there are studies on fibroblasts, the aims are mainly to generate muscle or fat cells. Jeong et al. and Ma et al. employed cellular reprogramming techniques to induce trans-differentiation of fibroblasts into skeletal muscle cells, and thereby used for the fabrication of muscle tissues (Jeong et al., 2022; Ma et al., 2023). Alternatively, fibroblasts have been induced to differentiate into adipocytes, thus enhancing the production of adipose tissue (Pasitka et al., 2023). However, there is still a lack of studies that directly address the application of fibroblasts in the preparation of meat-related connective tissue and skin.
Isolating and culturing seed cells is the first step in cultured meat production. The skin is easily accessible and rich in fibroblasts, making it the primary tissue site for the isolation and extraction of fibroblasts. In previous studies, several methods have been applied to obtain fibroblasts from mouse and human skins. As a simple one, fibroblasts can be isolated by cultivating adherent skin tissue pieces (Borges et al., 2020), but this method is time-consuming with a limited number of cells being obtained. The enzymatic digestion method first degrades the ECM in the skin tissue using proteases such as collagenase, thus usually achieving relatively high isolation efficiency and cell number of fibroblast (Wang et al., 2004). However, compared to humans and mice, the skin of livestock such as pigs and cows is thicker and more compact, so conventional enzymatic digestion processes may not be effective in isolating fibroblasts from livestock (Boroda et al., 2020). In addition to tissue explanting and enzymatic digestion, physical friction is another attractive method to promote cell release during the tissue dissociation process. In previous research, keratinocytes were isolated by gently rubbing the mouse epidermis after incubating with a small amount of enzyme (Li et al., 2017). Therefore, the combined use of enzymatic digestion and physical friction methods would be a promising method for the efficient and cost-effective acquisition of fibroblasts from domestic animals. There is still a lack of systematic studies on the procedures and condition parameters for the isolation of fibroblasts by the combination of enzymatic digestion and physical friction.
The application of fibroblasts offers the possibility of manufacturing structured cultured meat containing connective tissue or skin, which involves three-dimensional (3D) culture of cells. In previous studies, textured soy protein (TSP) was used as the scaffold to support the 3D growth of seed cells, and structured cultured meat samples were obtained after seeding and culture of bovine satellite cells (Ben-Arye et al., 2020). Besides, some research teams employed hydrogels as the substrate for 3D cell culture and achieved the desired shapes with silicone molds (Zhu et al., 2022). However, the cultured meat samples manufactured by the above methods were still far from real meat regarding the external shape and internal structure, and their manufacturing processes were also limited in terms of automation and standardization. Alternatively, 3D printing technology offers a promising solution for fabricating structured cultured meat. 3D bio-printing is commonly employed to manufacture 3D biological structures by accurately positioning bio-ink layer-by-layer, which allows flexibility in shaping cell cultures into any desired forms and adjusting the proportions of different components in cultured meat (Kang et al., 2021; Guo et al., 2023). Suitable bio-inks are a prerequisite for 3D bioprinting, which must support the adhesive growth of the target cells and possess printable properties. In addition, printing parameters are crucial factors in 3D printing that affect the quality and appearance of the sample and the efficiency of cell seeding. Therefore, it is necessary to determine the optimal bio-ink and associated printing parameters for fibroblasts, which lay the foundation for the fabrication of structured culture meat containing connective tissue or skin.
In this study, we first developed an optimized method to efficiently isolate fibroblasts from porcine skin tissue by combining enzymatic digestion and physical friction. Then, we identified the optimal food hydrocolloid that supported the adhesion and growth of fibroblasts and prepared bio-inks with printable properties. Finally, we determined the optimal initial cell number for 3D bio-printing and fabricated fibroblast-based cultured meat samples with improved texture and nutrition (Figure 1).
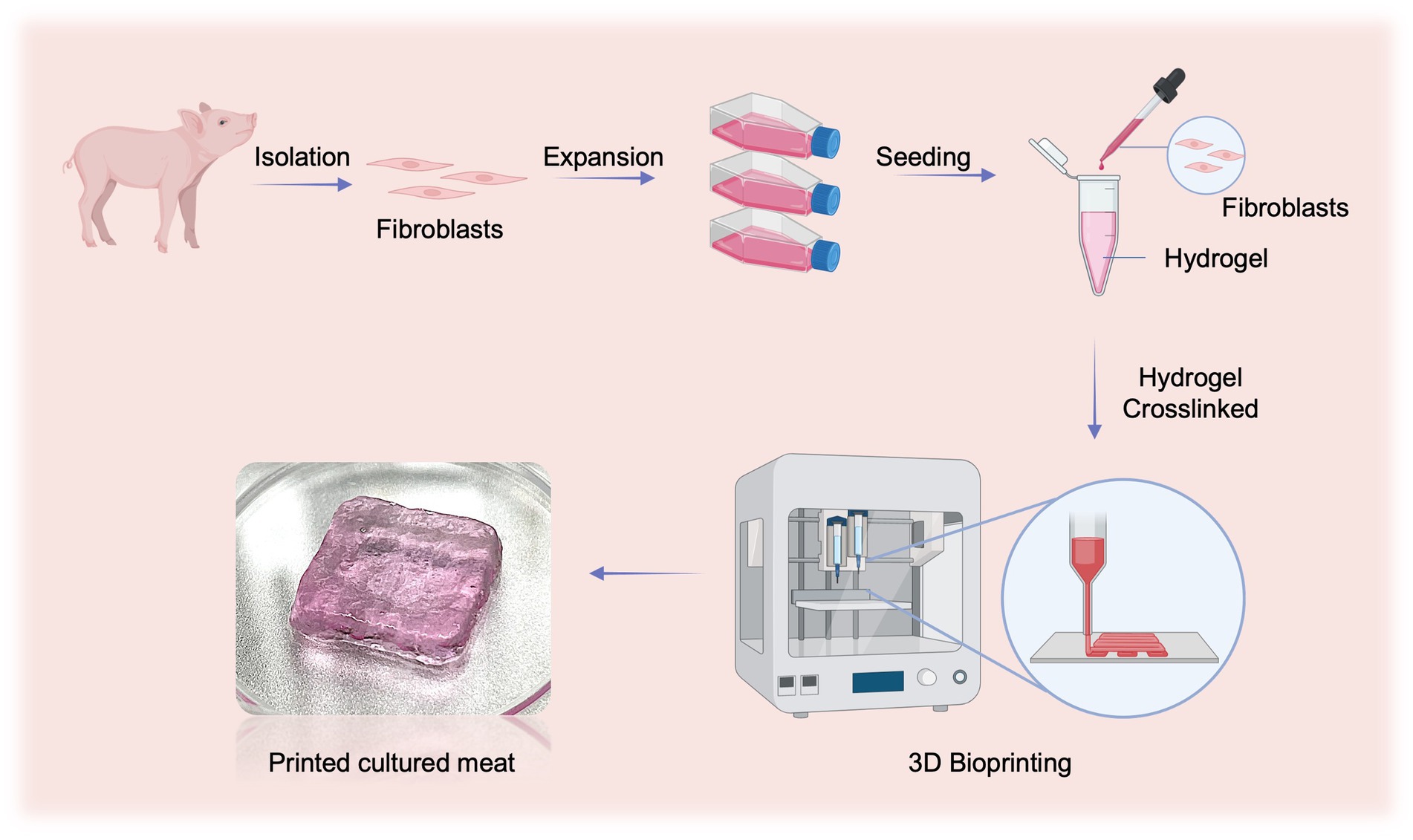
Figure 1. Schematic illustration of preparing fibroblasts-based cultured meat. This figure is created with BioRender.com.
2 Materials and methods
2.1 Tissue collection
Skin samples were collected from piglets aged 3–7 days and sterilized with 75% ethanol solution. After removing hair and subcutaneous tissues carefully, the skin samples were washed three times with sterile phosphate-buffered saline (PBS) and cut into 1 × 1 cm2 pieces. The animal experimental protocols were approved by the Institutional Animal Care and Use Committee of Jiangnan University (JN. No. 20211030p0020301[431]).
2.1.1 Isolation of fibroblasts by the enzyme digestion with friction (DF) method
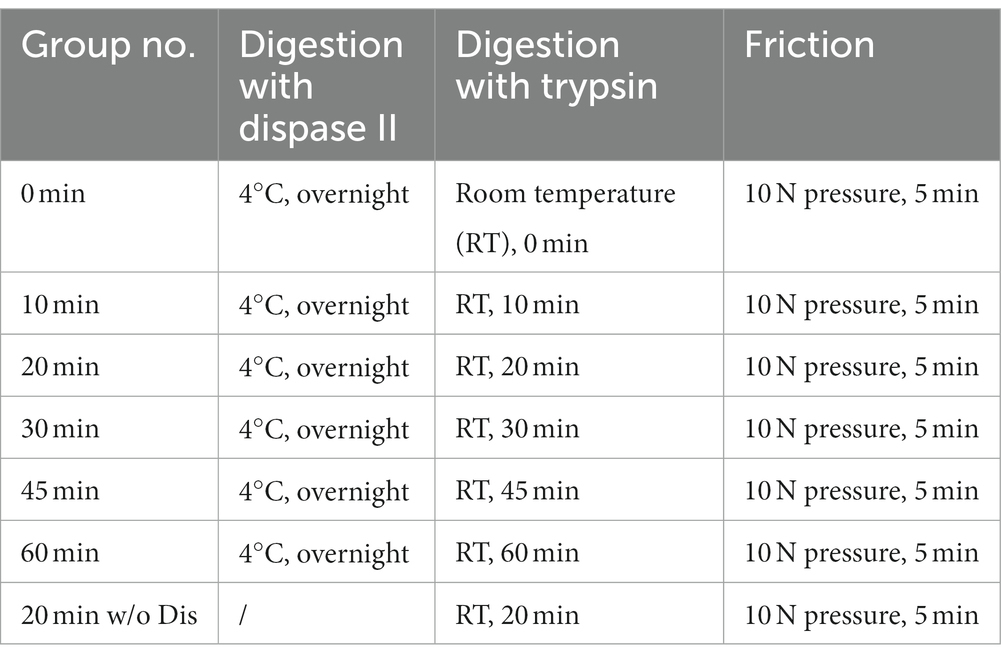
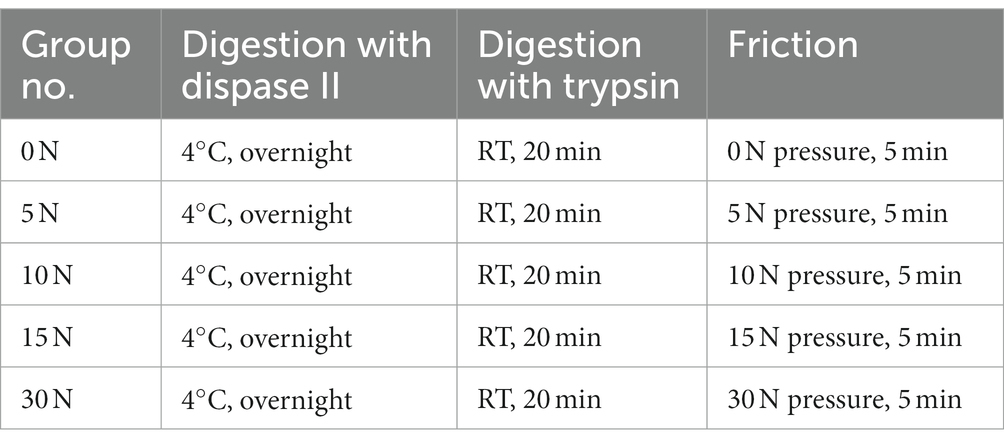
First, porcine skin pieces were incubated in Dispase II solution (4 mg mL−1, Roche, Switzerland) at 4°C overnight and the epidermis was separated from the dermis. Then, the dermal fragments were digested with 0.25% trypsin (v/v; Gibco, USA) for 0, 10, 20, 30, 45, or 60 min at room temperature. For samples that were not subjected to Dispase II digestion, they were placed in PBS at 4°C overnight followed by digestion with 0.25% trypsin. To terminate the digestion process, 1 mL of complete medium was added to the dermal fragment, which was DMEM (Gibco, USA) supplemented with 10% Fetal Bovine Serum (v/v; Gibco, USA) and 1% Penicillin–Streptomycin (v/v; Gibco, USA). Subsequently, the dermal fragments were pressed against the petri dish with a thrust meter (Nscing Es, China) and rubbed back and forth for 5 min at a consistent pressure of 0, 5, 10, 15, or 30 N to release individual cells. Then, the medium containing released cells was aspirated and filtered out with a 40 μm cell strainer (Biosharp, China). The obtained cells were counted after centrifugation and cultured in the complete medium at 37°C, 5% CO2. The cell survival rate at 36 h was calculated by the following formula:
where CN36h is the live cell number after 36 h of culture and CN is the number of initially isolated fibroblasts.
2.1.2 Isolation of fibroblasts by the tissue explant method
The skin samples were sectioned into 1 × 1 mm2 small pieces and placed in 12-well plates with a drop of complete medium on each tissue piece. After incubation at 37°C for 4–6 h to ensure attachment of the tissue to the bottom of the plate, 1 mL of complete medium was carefully added to each well. After several days of culture in the incubator at 37°C and 5% CO2, the tissue pieces were removed when cells were obviously migrated from the edges of tissue explants (TE). The cells were then counted and used for subsequent studies.
2.2 Immunofluorescent staining
The cell sample was first subjected to fixation using 4% paraformaldehyde (Sinopharm, China) for 15 min at 4°C and then washed with PBS. After permeabilizing the cell membrane with 0.5% Triton X-100 (Thermo, USA), the cell sample was incubated with the blocking buffer at room temperature for 30 min, which was PBS containing 1% BSA (m/v; Solarbio, China), 2.25% glycine (m/v; Solarbio, China), and 0.1% Tween 20 (v/v; Solarbio, China). Next, the cell sample was incubated with the primary antibody at 4°C overnight, followed by washed three times with PBS. Then, the secondary antibody was added to the cell sample and incubated at room temperature for 2 h. Finally, the nuclei were stained with DAPI solution (1:1000; Thermo, USA) and the cell sample was observed under a fluorescence microscope (Mshot Co., Ltd., Guangzhou, China). The antibodies used in this work were anti-Collagen Type I antibody (1:600; Proteintech, China), anti-Vimentin antibody (1:400; Cell Signal Technology, USA), and Goat anti-Mouse/Rabbit-488 Secondary antibody (1,500; Proteintech, China).
2.3 Gelatin-based hydrogel preparation
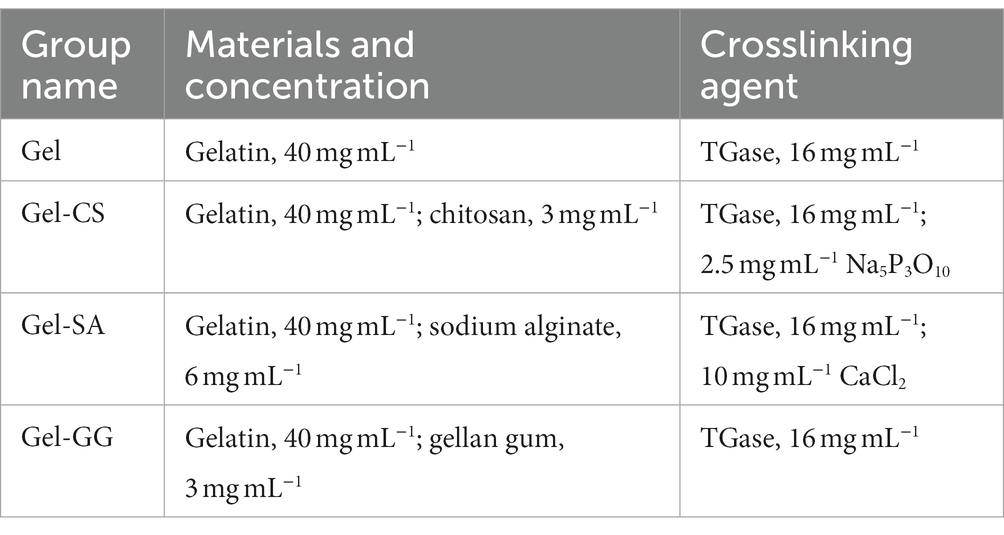
Gelatin (Gel), sodium alginate (SA), and gellan gum (GG) powder were purchased from Sigma Aldrich Life Science (St. Louis, MO). Chitosan (CS, deacetylation degree ≥95%) and sodium tripolyphosphate (Na5P3O10) powder were provided by Aladdin Industrial Corporation (Shanghai, China). Calcium chloride (CaCl2) powder was supplied by Sinopharm Chemical Reagent (Beijing, China), and transglutaminase (TGase) powder was procured from Dongsheng Biotechnology (Taixing, China). These powders were configured into working solutions according to the concentrations in Table 1. The Gel, Gel-SA, Gel-CS, and Gel-GG hydrogel precursors were prepared by mixing Gel with TGase, and then with SA, CS, and GG solutions, respectively. These hydrogel precursors were incubated at 37°C for 3 h to crosslink Gel in the presence of TGase. To prepare completely cross-linked hydrogels, the Gel-SA and Gel-CS hydrogels were then incubated with CaCl2 and Na5P3O10 solutions, respectively, for 15 min at room temperature to cross-link SA and CS. After cross-linking, the hydrogels were washed three times with PBS.
2.4 Rheological measurements
The storage modulus (G’) and loss modulus (G’) of the various hydrogels were determined by a rotary rheometer (DHR3, TA, USA) in the oscillation frequency mode. Each group of hydrogels was placed between parallel plates with a diameter of 40 mm with a gap of 1,000 μm at 20°C, under the condition of strain γ = 0.1% and angular frequency ω = 0.1–125 rad·s−1. The viscosity of different hydrogel groups was measured by steady rate sweeps at 20°C, varying the shear rate from 0.1 to 10 s−1.
2.5 3D bioprinting
In this study, an extrusion 3D bioprinter (Regenovo, China) was employed to fabricate structures by 3D bioprinting. For testing the printing performance of different hydrogels, four hydrogel precursors were first prepared according to the method in Section 2.3 and loaded into printing syringes (Regenovo, China), respectively. Subsequently, the syringes were kept at 4°C (referred to as “No crosslinking”) or 37°C (referred to as “Gel crosslinking”) for 3 h, and then a single-layer structure was printed at 20°C using a 0.1 mm diameter needle (Regenovo, China).
To print hydrogels embedded with fibroblasts, cells were first mixed with the Gel-CS hydrogel precursor at various concentrations and then loaded into printing syringes. Then, the hydrogels were printed using a 0.1 mm diameter needle at 20°C after incubating at 37°C for 3 h and then cross-linked using the Na5P3O10 solution.
2.6 Cell viability assay
2.6.1 Assessment of fibroblast adhesion and proliferation rates on the surface of hydrogels (2D culture)
The adhesion and proliferation rates of fibroblasts were analyzed by the Alamar Blue assay. First, a standard curve was established using a gradient of fibroblast concentrations. Specifically, fibroblasts were seeded in a 48-well plate at concentrations of 0, 2.5 × 104, 3.75 × 104, 5 × 104, 6.25 × 104, 7.5 × 104, and 8.75 × 104 cells mL−1. After 6 h of incubation, the medium was aspirated and cells were washed with PBS. The 10% Alamar Blue solution (v/v; Keygen, China) was then added and incubated at 37°C for 5 h. The fluorescence intensity was measured at an excitation wavelength of 530 nm and an emission wavelength of 590 nm using a microplate reader (BioTek, Winooski, VT, USA) and the standard curve was obtained.
The Gel, Gel-SA, Gel-CS, and Gel-GG hydrogels were prepared in a 48-well plate as described in Section 2.3. Porcine fibroblasts at a concentration of 2.5 × 104 cells mL−1 were seeded on the surface of different hydrogels and cultured for 6 h and 102 h, respectively. After washing with PBS 3 times to remove unattached cells, the 10% Alamar Blue was added to each well and incubated for 5 h at 37°C. Then, the fluorescence intensity of each well was measured. The cell number was calculated from the standard curve and the cell adhesion and proliferation rates were calculated using the following formulas:
where CN6h is the number of attached fibroblasts on the surface of hydrogels at 6 h and CN is the number of attached fibroblasts on the bottom of wells without hydrogels at 6 h.
where CN102h is the number of attached fibroblasts on the surface of hydrogels at 102 h and CN6h is the number of attached fibroblasts on the hydrogels at 6 h.
2.6.2 Assessment of the proliferation rate of fibroblasts embedded in hydrogels (3D culture)
First, 100 μL of various hydrogel precursors were mixed with 1.0 × 105 porcine fibroblasts and then dispensed into individual wells of a 48-well plate. After complete cross-linking of hydrogels as according to Section 2.3, they were cultured with the complete medium at 37°C, 5% CO2. The cell proliferation rate was analyzed by the Alamar Blue assay at 0 h and 96 h as described in 2.6.1. The fibroblast proliferation rate was calculated as follows:
where CN96h is the number of fibroblasts at 96 h and CN0h is the number of fibroblasts at the beginning of the culture (0 h).
2.7 Degradation rate test of hydrogels
The hydrogels embedded with fibroblasts were prepared as described above and the initial weight M0 was recorded. Then, the hydrogel was immersed in the complete medium for 7 days with medium change every other day. The weight of the hydrogel after draining the surface water droplets was recorded daily as Mn (n = 1, 2, 3, …, 7). The degradation rate was calculated as follows:
where M0 represents the initial weight of the hydrogel, while Mn denotes the weight of the hydrogel on the n-th day.
2.8 Cell live/dead staining
The Calcein-AM/PI cell viability assay kit (Beyotime, China) was used for live/dead staining as described by the instruction. Briefly, the cell samples were thoroughly washed with PBS and then incubated with Calcein-AM/PI solutions for 10 min at 37°C. After washing the samples with PBS three times, the samples were observed and imaged using a confocal laser microscope (Nikon, Japan).
2.9 Amino acid analysis
The hydrogels embedded with fibroblasts were prepared as described above and cultured with the complete medium for 7 days. Then, amino acids were detected using a reversed-phase high-performance liquid chromatography (RP-HPLC) system (Agilent Technologies, USA) as described previously (Guan et al., 2023).
2.10 Statistical analysis
Data were visualized using GraphPad Prism 9 and presented as mean ± SD. Comparison between two groups was analyzed by an unpaired two-tailed Student’s t-test and comparison among ≥3 groups was determined by one-way ANOVA (followed by Duncan’s test) using SPSS. Statistical significance was defined as p < 0.05. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
3 Results
3.1 Efficient isolation of porcine fibroblasts using the DF method
The skin tissue is primarily composed of three layers, including the epidermal layer, the dermal layer, and the subcutaneous tissue, among which fibroblasts are mainly distributed in the dermal layer (Vig et al., 2017). In this study, we used dispase II to separate the dermal and epidermal, then used trypsin to digest and gently rubbed the skin to isolate fibroblasts inside (Figure 2). To achieve efficient isolation of fibroblasts from porcine skin tissues using enzyme digestion combined with physical friction, there is a need to investigate the optimal time of enzyme digestion and the pressure of friction, both of which are important factors affecting the number and viability of the released cells. Therefore, we first investigated the effect of trypsin digestion time on the efficiency of fibroblast isolation under the same dispase II digestion and friction conditions (Table 2). The results showed that extending the trypsin incubation time from 0 min to 20 min resulted in a doubling of the number of cells released, but further extension of the digestion time did not increase the cell number (Figures 3A,B). Moreover, after 36 h of culture, the 20 min group showed the highest number of live cells and cell survival rate (Figures 3C,D). Meanwhile, we tested the necessity of using dispase II to separate epidermal and dermal before trypsin digestion. Compared to the 20 min group that underwent dispase II digestion, the 20 min w/o Dis group exhibited lower cell number after isolation, live cell number, and cell survival rate (Figures 3B–D), indicating the important role of dispase II digestion. The results above showed that a combination of overnight dispase II digestion and 20-min trypsin digestion was the optimal digestion approach for the isolation of porcine skin fibroblasts.
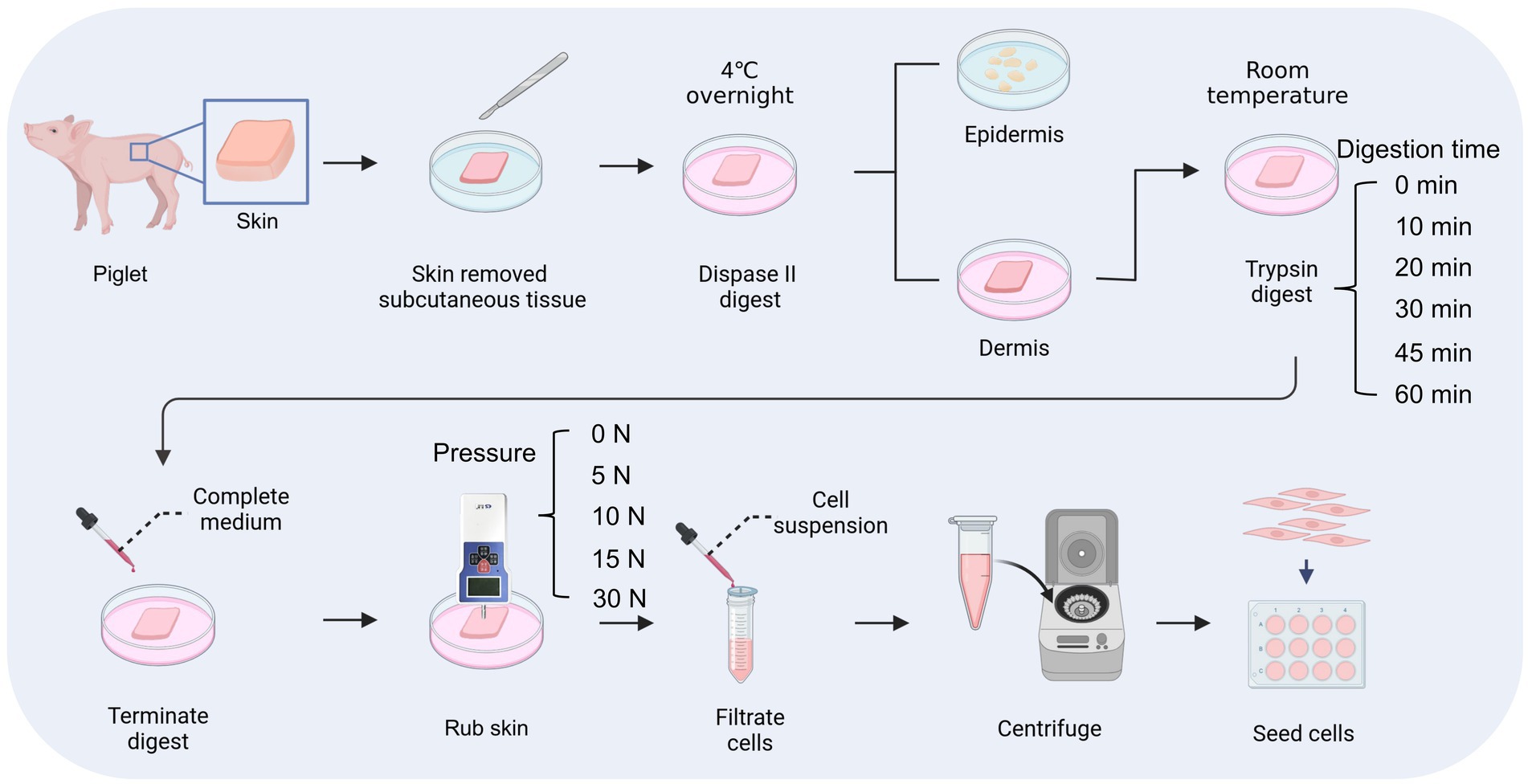
Figure 2. Process of digestion and friction method. This figure is created with BioRender.com.
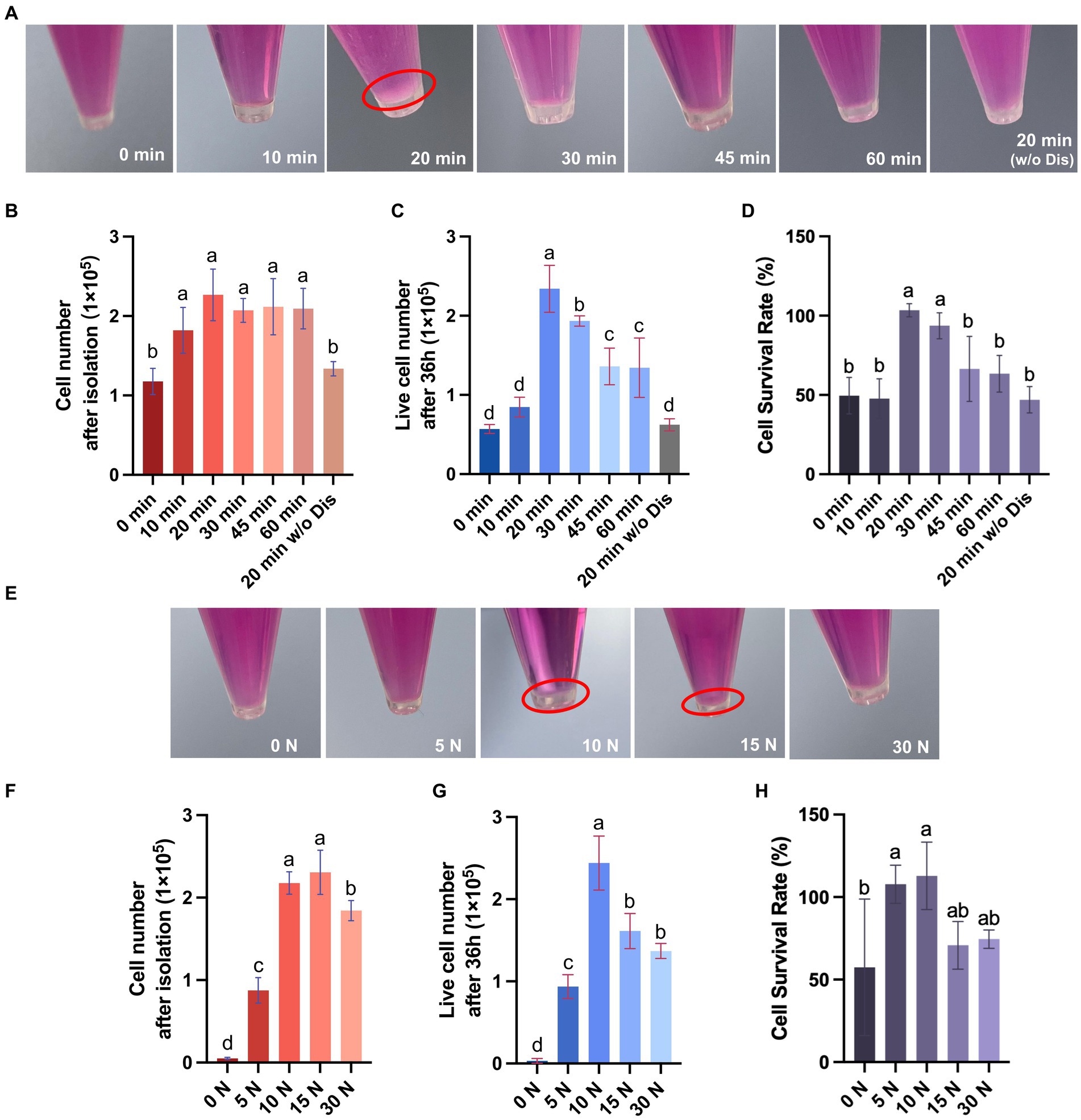
Figure 3. Porcine dermal fibroblasts can be efficiently obtained by the digestive-friction method. (A) Representative pictures of cell pellets obtained by isolation under various enzyme digestion conditions. (B) The number of cells obtained by isolation under various enzyme digestion conditions. (C) The number of live cells after 36 h of culture for different enzyme digestion groups. (D) The cell survival rate at 36 h for different enzyme digestion groups. (E) Representative pictures of cell pellets obtained by isolation under various friction pressures. (F) The number of cells obtained by isolation under various friction pressures. (G) The number of live cells after 36 h of culture for different friction groups. (H) The cell survival rate at 36 h for different friction groups. The values with different superscripts are significantly different, p < 0.05.
To further explore the impact of varying friction pressures on the efficiency of fibroblast isolation, we conducted experiments under the previously established optimal enzyme digestion conditions (Table 3). The results indicate that extending the friction pressure from 0 to 15 N increased the number of cells released two-fold, but a further increase of the friction pressure to 30 N resulted in a decrease in the cell number, probably due to cell damage caused by high friction pressure (Figures 3E,F). Additionally, upon 36 h of culture, the group subjected to a 10 N friction pressure exhibited the most favorable outcomes, recording the highest number of live cells and the best cell survival rate (Figures 3G,H). In summary, the most efficient isolation process of fibroblast from porcine skin tissue involves three major steps: (1) incubating skin pieces with cold Dispase II overnight to separate the epidermal and dermal layers, (2) digesting dermal parts with trypsin at room temperature for 20 min to degrade ECM components, (3) rubbing the dermal parts with a 10 N pressure for 5 min to release cells. By calculating the live cell number after 36 h of culture, we obtained 2.39 ± 0.28 × 105 cells from 1 cm2 of porcine skin.
3.2 Comparison of the DF and TE methods for isolation of porcine skin fibroblasts
The TE method is a conventional method for fibroblast isolation, which involves obtaining skin from pigs, culturing the skin pieces in petri dishes under suitable conditions, and obtaining migrating fibroblasts from the periphery of tissue pieces (Figure 4A; Diar-Bakirly and El-Bialy, 2021). Thus, we also performed the fibroblast isolation with the TE method and compared the cell isolation efficiency and cell purity between the DF and TE methods. In the TE method, it took approximately 6 days for the cells to migrate outward from the tissue pieces whereas the DF method obtained cell suspensions for culture on the 1st day (Figures 4B,C). Besides, after 9 days of culture from isolation, the number of cells obtained from 1 cm2 of porcine skin with the DF method was 9 times higher than that with the TE method (Figure 4D). Furthermore, immunofluorescence analysis of the isolated cells for vimentin and collagen type I was performed (Kuwabara and Tallquist, 2017; Plikus et al., 2021). The results showed that fibroblasts isolated by both DF and TE methods expressed vimentin and collagen at high levels, confirming the high purity of fibroblasts isolated by both methods (Figures 4E–H). Collectively, the DF method enabled more efficient isolation of fibroblasts from porcine skin without loss of cell purity.
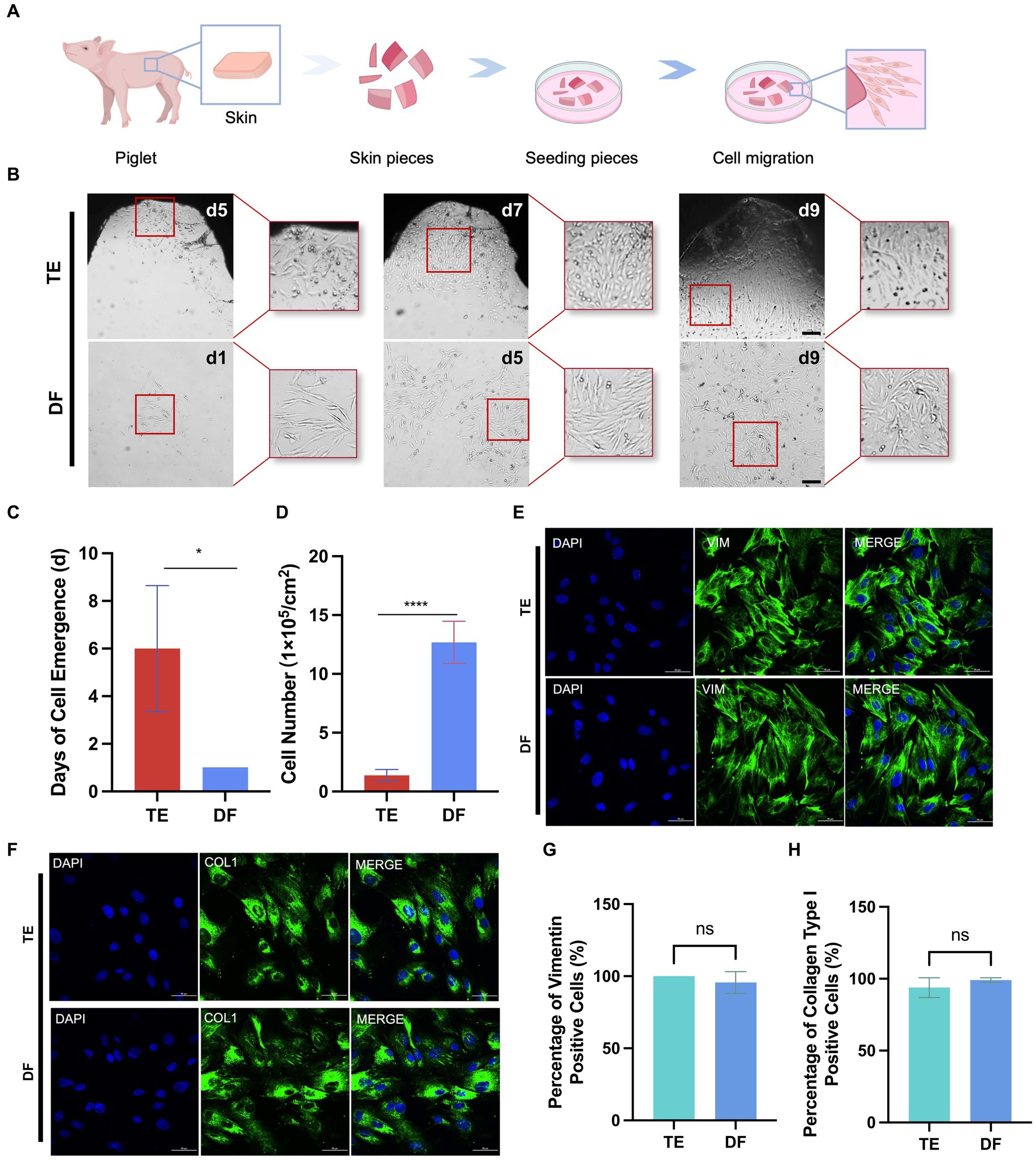
Figure 4. Comparison of DF and TE methods for isolation of porcine fibroblasts. (A) Schematic diagram of the workflow for isolation of fibroblasts using the TE method. (B) Representative bright field images of cells at different times after isolation using DF and TE methods. Scale bars, 200 μm. (C) The days of cell emergence using TE and DF methods. (D) The number of cells at day 9 after cell isolation using DF and TE methods. (E) Representative vimentin immunofluorescence images of cells obtained with DF and TE methods. Scale bars, 50 μm. (F) Representative collagen type I immunofluorescence images of cells obtained with DF and ADH methods. Scale bars, 50 μm. (G) The percentage of vimentin-positive cells isolated by TE and DF methods. (H) The percentage of collagen type I positive cells isolated by TE and DF methods. *p < 0.05, **p < 0.01.
3.3 Identification of candidate bio-inks for 3D bioprinting of fibroblasts
Next, we aimed to identify suitable bio-inks formulations that could be applied to 3D bio-printing of fibroblasts. Gelatin (Gel) is a cost-effective, edible, and cell-compatible hydrogel material, but its mechanical properties are relatively poor. Food hydrocolloids such as SA, CS, and GG possess good mechanical properties but poor compatibility with cells (Chen X. B. et al., 2023). Therefore, we combined Gel with different food hydrocolloids and investigated their physical properties and cell compatibility, to determine the optimal bio-ink formulation for 3D bio-printing. Transglutaminase (TGase) was used for Gel crosslinking to ensure the edibility of the bio-inks. To minimize the introduction of exogenous materials, we tested the gel-forming properties of different concentrations of Gel, so as to determine the lowest Gel working concentration. As shown in Figures 5A, 40 mg mL−1 was the minimum Gel concentration that maintained stable gelation at 37°C after TGase cross-linking. Therefore, we prepared four hydrogels with 40 mg mL−1 of Gel, including pure Gel, Gel-SA, Gel-CS, and Gel-GG, and evaluated their suitability for 3D printing applications.
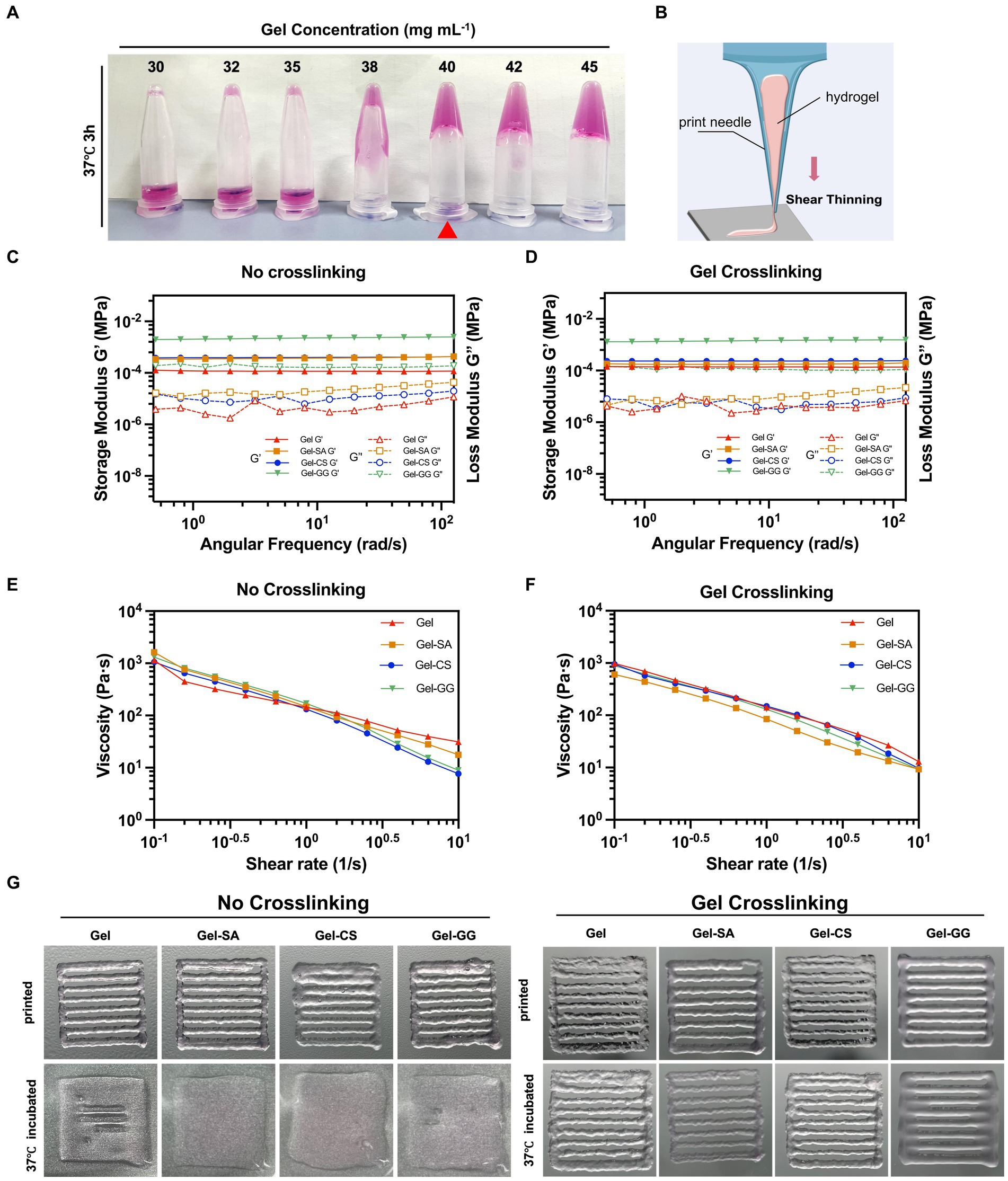
Figure 5. Printable properties tests of Gel-derived hydrogels. (A) Photos of varying concentrations of Gel hydrogels cross-liked with TGase after incubating at 37°C for 3 h and then inverted. (B) Illustration of shear thinning. (C) Storage/loss modulus of pure Gel, Gel-SA, Gel-CS, and Gel-GG hydrogels without Gel cross-linking. (D) Storage/loss modulus of pure Gel, Gel-SA, Gel-CS, and Gel-GG hydrogels subjected to TGase-mediated Gel crosslinking. (E) Viscosity of pure Gel, Gel-SA, Gel-CS, and Gel-GG hydrogels without Gel cross-linking. (F) Viscosity of pure Gel, Gel-SA, Gel-CS, and Gel-GG hydrogels with Gel crosslinking. (G) Photos of pure Gel, Gel-SA, Gel-CS, and Gel-GG hydrogels after printing under no cross-linking and Gel crosslinking conditions.
A solid extrusion 3D printer was employed in this work, which requires the bio-ink to remain solid during the printing process. Therefore, we assessed the modulus and viscosity of four hydrogels before and after Gel crosslinking with TGase, denoted as the ‘No crosslinking’ group and ‘Gel crosslinking’ group, respectively, according to 2.5. The energy storage modulus (denoted as G’) and the loss modulus (denoted as G”) are key parameters characterizing the elasticity and viscosity of substances, respectively (Oroian et al., 2018; Wei et al., 2023). Regardless of whether or not the Gel was crosslinked in the hydrogels, the G’ and G” curves of pure Gel, Gel-SA, Gel-CS, and Gel-GG hydrogels demonstrated no intersection points, and the G’ values consistently exceeded the G” values, indicating that these hydrogels were able to form stable elastic structures over a broad spectrum of shear frequencies (Figures 5C,D). Notably, among the four hydrogels, the pure Gel hydrogel exhibited the lowest G’ and G” values, suggesting that the composition of Gel with SA, CS, and GG improved the overall stiffness and elasticity of hydrogels.
The rheological property of “shear-thinning” in bio-inks plays an important role in determining whether they can be continuously extruded during printing (Highley et al., 2015). As shown in Figures 5E,F, the viscosity of all hydrogels decreased rapidly with increasing shear rates both in the no crosslinking group and Gel crosslinking group, suggesting the good printability of these hydrogels. Subsequent printing experiments demonstrated that all hydrogels could be consistently extruded through the print needle to draw the desired geometry (Figure 5G). However, the printed samples that were not cross-linked did not retain their original shape when placed in the 37°C incubator. In contrast, pre-crosslinking of Gel before printing ensured that the samples maintained their structural integrity at 37°C. Therefore, prior to 3D printing, we pre-crosslinked the Gel using TGase by incubation at 37°C for 3 h.
3.4 Effect of hydrogels on the adhesion and proliferation of fibroblasts in 2D and 3D cultures
Next, we investigated the effect of different hydrogels on the adhesion and proliferation of fibroblasts in 2D and 3D culture conditions. 2D culture refers to seeding cells onto the surface of crosslinked solid hydrogels and then culturing. 3D culture involves mixing cells with liquid hydrogel precursors and then completely crosslinking to solid so that cells are encapsulated inside the hydrogels when culturing (Figure 6A). In 2D cultures, fibroblasts exhibited satisfactory adhesion and proliferation rates on the Gel-CS surface (Figures 6B–D). Consistently, in 3D cultures, fibroblasts in Gel-CS hydrogels also exhibited superior proliferation capability and highest cell viability after 96 h of culturing (Figures 6F–H). In addition, among different hydrogels embedded with fibroblasts, the degradation rate of Gel-CS was relatively slower than Gel (Figure 6E). Taken together, we selected Gel-CS hydrogel as the preferred bio-ink for 3D printing of fibroblasts.
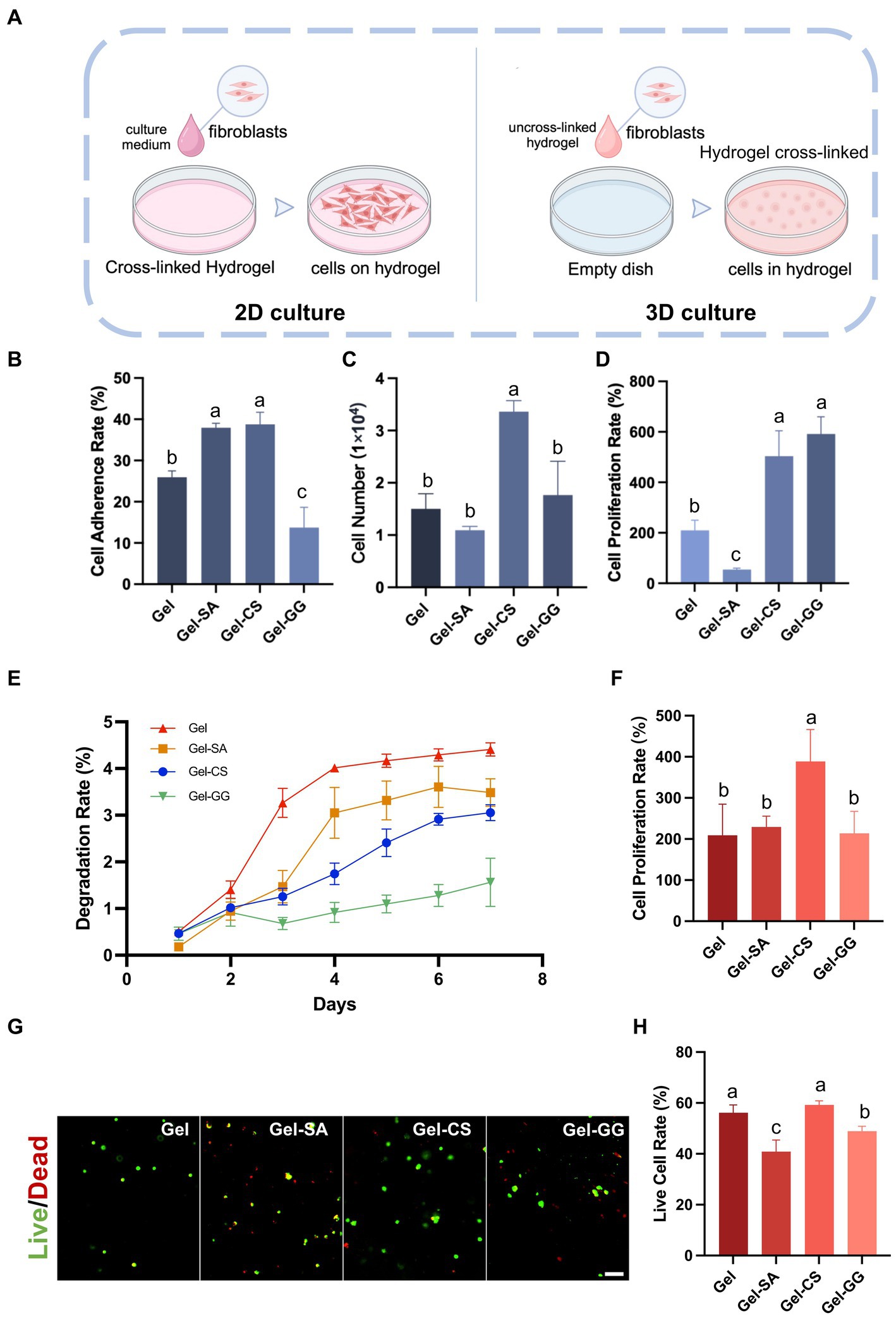
Figure 6. Suitability of fibroblasts on different hydrogels. (A) Schematic of 2D culture and 3D culture of fibroblasts on hydrogels. (B) Cell adherence rate of fibroblasts on various hydrogel surfaces after 6 h of culture (2D culture). (C) Cell number of fibroblasts on various hydrogel surfaces after 96 h of culture (2D culture). (D) Cell proliferation rate of fibroblasts on various hydrogel surfaces after 96 h of culture (2D culture). (E) The degradation rate of hydrogels containing fibroblasts inside (3D culture). (F) Cell proliferation rate of fibroblasts within hydrogels after 96 h of culture (3D culture). (G) Live and dead cell staining of fibroblasts within hydrogels (3D culture) after 96 h of culture. Scale bars, 100 μm. (H) The live cell rate is calculated according to (G). The values with different superscripts are significantly different, p < 0.05.
3.5 Comparison of cell seeding efficiency between 3D printing and dripping cell concentrates
In previous 3D cell culture studies, in addition to seeding cells by 3D printing cell-hydrogel mixtures (Print), dropping cell concentrates into printed scaffolds (Drip) was also a common method of cell seeding (Figure 7A; Yan et al., 2022). Here, we performed both methods to seed fibroblasts and compared their seeding efficiency and cell survival. After fibroblasts were seeded and incubated for 6 h, the number of cells on the scaffolds in the Print group was significantly higher than that in the Drip group (Figure 7B). Consistently, the cell retention rate was close to 100% in the Mix group but less than 50% in the Drip group (Figure 7C), suggesting that the Drip method might result in a large number of cells leaking out of the scaffold. Subsequently, we examined the printed samples after DAPI staining. The printed samples presented a cross-shaped structure, so we subdivided each cross-section into four regions and counted the number of cell nuclei in each region (Figure 7D). The cell nuclei number in each region shown in the boxplot was more aggregated in the Print group, indicating that the cells in the Print group were more evenly distributed than those in the Drip group (Figures 7D,E). Additionally, the live cell rates of both groups were approximately 100%, indicating that cells were in good condition after seeding (Figures 7F,G). Collectively, Print was selected as the preferred method for seeding fibroblasts in the Gel-CS scaffolds.
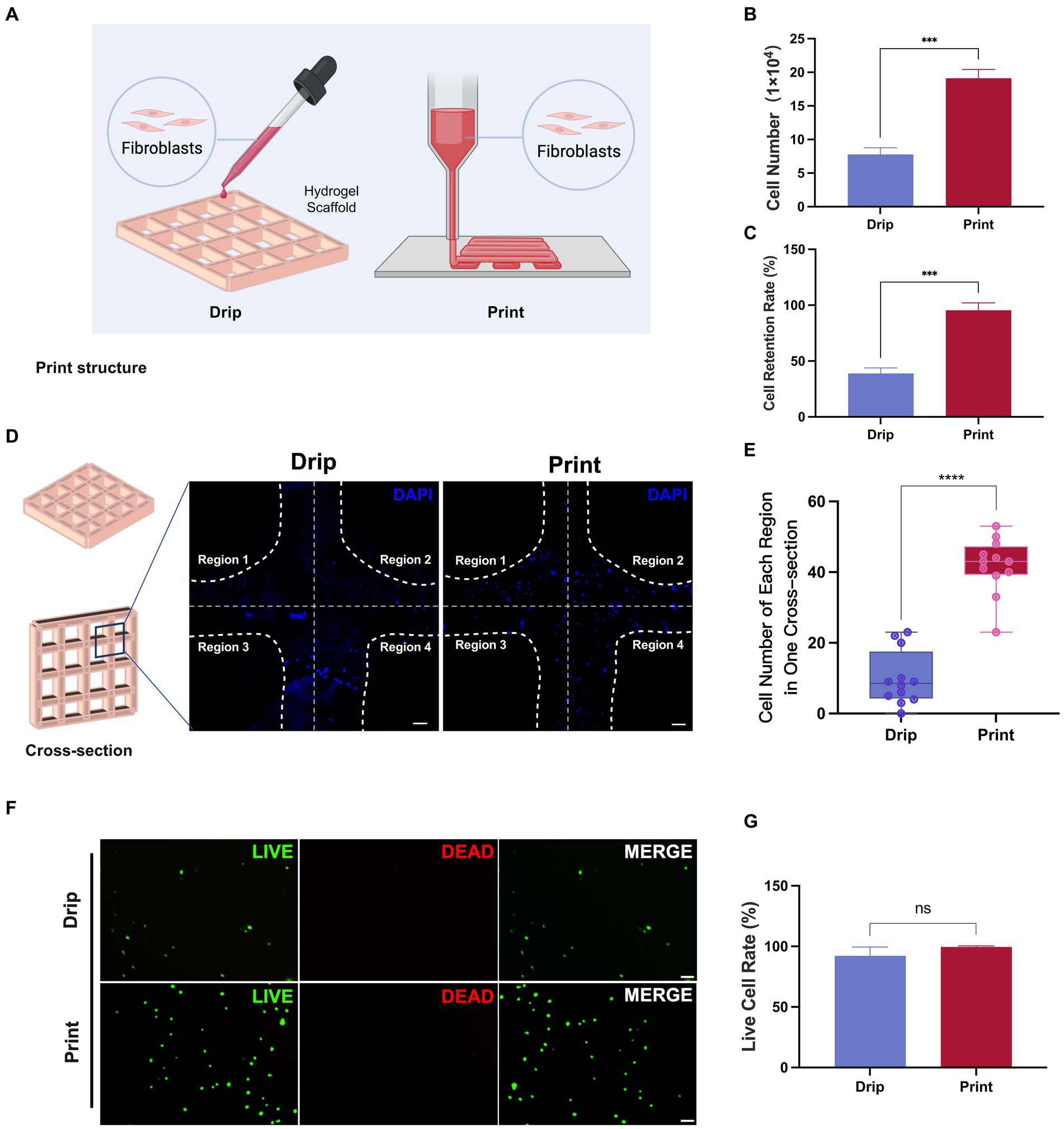
Figure 7. Comparison of cell number, distribution, and retention between Drip and Print methods. (A) Illustration diagram of cell seeding with Drip and Print methods. (B,C) The number of cells on the scaffold and cell retention rate were calculated at 6 h after cell seeding with Drip and Print methods. (D) DAPI staining images of Drip and Print samples (divided into four regions by white broken lines). Scale bars, 100 μm. (E) Boxplot of cell number of each region in one cross-section according to (D). (F) Live and dead cell staining after 24 h of culture. Scale bars, 100 μm. (G) Living cell rate according to (F). ***p < 0.001, ****p < 0.0001.
3.6 Fabrication and evaluation of porcine fibroblast-based cultured meat
Next, we investigated what initial cell concentration was most suitable for 3D printing and cell growth. We fabricated different samples at cell concentrations of 0 × 106, 1.0 × 106, 5.0 × 106, 1.0 × 107, and 1.5 × 107 mL−1, respectively. We found that as the cell density increased, the number of cells overflowing out of the hydrogel scaffolds during the printing process gradually increased. When the cell density reached 1.5 × 107 mL−1, the number of cells overflowing into the printing gaps increased significantly (Figures 8A,B). Therefore, we fabricated cultured meat samples by 3D printing with porcine fibroblasts at cell concentrations of 1.0 × 107 mL−1 and below. After printing, these samples were cultured in vitro for 8 days. It could be observed that the structure of the cell-containing hydrogel samples was able to remain intact, whereas the empty hydrogel scaffolds showed collapse and dissolution (Figures 8C,D). In addition, the number of cells in the hydrogel samples increased with time. After 8 days of culture, the sample printed at the cell concentration of 1.0 × 107 mL−1 achieved the highest total cell number (Figure 8E).
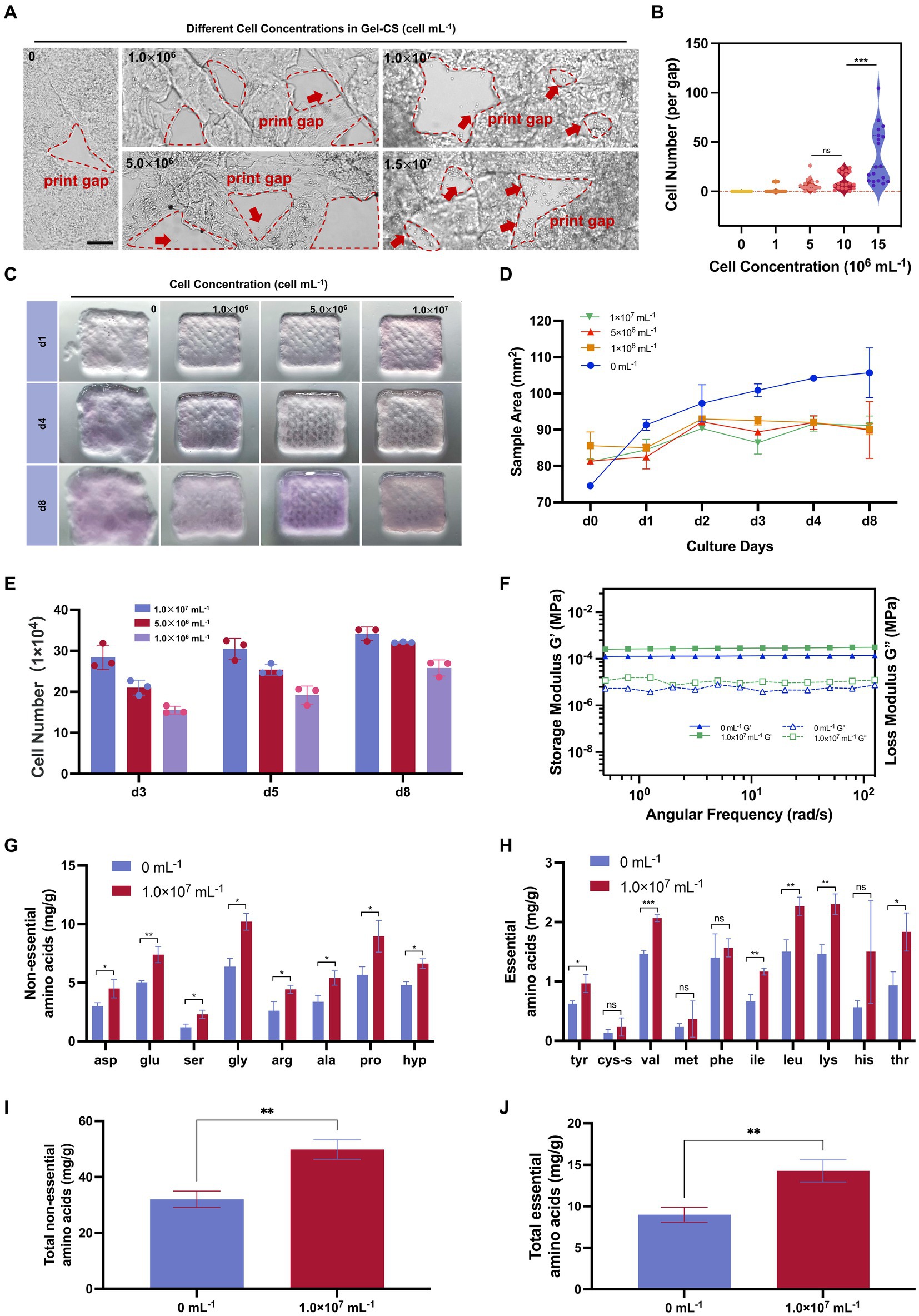
Figure 8. Fibroblast can make skin-derived cultured meat providing abundant amino acids. (A) Illustration of printed samples with various cell concentrations. Scale bars, 200 μm. (B) Overflowed cell number per grid of printed samples according to (A). (C) Illustration of printed samples with various cell concentrations with continuous culturing. (D) Dispersal area of samples according to (C). (E) Cell number after several days cultured of various printed samples. (F) Storage/loss modulus of 0 mL−1 and 1.0 × 107 mL−1 groups after 8 days of culturing. (G) Non-essential amino acids of 0 mL−1 and 1.0 × 107 mL−1 groups after 8 days of culturing. (H) Essential amino acids of 0 mL−1 and 1.0 × 107 mL−1 groups after 8 days of culturing. (I) Total non-essential amino acids of 0 mL−1 and 1.0 × 107 mL−1 groups after 8 days of culturing. (J) Total essential amino acids of 0 mL−1 and 1.0 × 107 mL−1 groups after 8 days of culturing. *p < 0.05, **p < 0.01, ***p < 0.001.
Furthermore, we carried out the rheological analysis and amino acid analysis on day 8. The rheological analysis showed that both G’ and G” were higher in the 1.0 × 107 mL−1 group than in the hydrogel scaffold without cells, suggesting that the incorporation of cells improved the mechanical strength of the whole sample (Figure 8F). Furthermore, the total amount of essential and non-essential amino acids in the fibroblast-containing samples was significantly higher than in the pure scaffold samples (Figures 8G–J). In particular, the three amino acids that make up collagen, glycine, proline, and hydroxyproline, were significantly increased in samples containing fibroblasts. Collectively, the addition of fibroblasts improved the stiffness of hydrogel scaffolds and provided the nutritional composition for manufacturing cultured meat products.
4 Discussion
In previous studies, combinations of different enzymes have been employed to separate primary cells such as satellite cells and fibroblasts from animal tissues efficiently (Edelman and Redente, 2018; Widbiller et al., 2019; Li et al., 2022). The use of enzymes in combination can expand the advantages and compensate for the deficiencies of a single enzyme, thus obtaining a greater number of cells. In this study, we used a combination of Dispase II and Trypsin, two commonly used and inexpensive proteases, for the digestion of porcine skin. Dispase II specializes in disrupting intercellular half-desmosomes while retaining intact desmosomes, making it suitable for separating the dermis and epidermis (Kitano and Okada, 1983). Trypsin interrupts both half-desmosomes and desmosomes, so it is more potent at digesting tissues but may cause more damage to cells (Takahashi et al., 1985). In our study, the data indicates that an extension in trypsin digestion time initially leads to an increase followed by a decrease in cell release. This suggests that prolonged digestion may compromise cell structure, thereby diminishing release efficiency. Concurrently, with extended digestion, the post-36-h viability of the harvested cells shows a similar trend, peaking and then declining to approximately 50%. This pattern likely reflects cell mortality due to overextended digestion, resulting in the liberation of cells inclusive of a significant proportion of non-viable cells, which adversely affects the efficiency of viable cell release.
Porcine skin is more than five times thicker than mouse and human skin and has a much higher ECM content, making it difficult to achieve effective cell release by enzymatic digestion alone (Boroda et al., 2020). Therefore, we carried out physical friction after enzyme digestion to further dissociate the intercellular matrix structure that was not enzymatically digested, which could increase the efficiency of cell release and save costs (Li et al., 2017; Hosseini et al., 2020). Meanwhile, our findings indicate that an increase in pressure applied during the friction of the dermis leads to a reduction in the number of cells released and a decrease in the post-36-h viability of the harvested cells. This phenomenon might be attributed to the disruption of the intact cellular structure caused by excessive mechanical forces. Consequently, we optimized the pressure applied during friction to minimize undesired cell damage. Finally, we developed an efficient and cost-effective process consisting of overnight Dispase II incubation, 20-min Trypsin digestion, and 10 N friction (referred to as the DF method) for the isolation of fibroblasts from porcine skin, which resulted in a 9-fold increase in the number of harvested cells as compared to the traditional tissue adhesion method. In addition, immunofluorescence demonstrated the high purity of fibroblasts isolated using the DF method.
Gelatin has been widely produced by hydrolyzing tissues such as animal skin and bone or by recombinant expression of microorganisms, and it can form stable hydrogels after cross-linking using TGase (Wang et al., 2017; Sonaye et al., 2022; Andreazza et al., 2023). TGase is a safe food additive that has been widely used to improve the flavor, texture, and appearance of meat products (Li et al., 2021; Chen Y. et al., 2023). In this study, we combined gelatin and three food-grade hydrocolloidal materials SA, CS, and GG to obtain bio-inks. Rheological testing revealed that the storage modulus (G’) of gelatin-hydrocolloid mixtures was higher than that of the pure gelatin group. Additionally, the degradation rate of the various materials containing cells was significantly lower than that of the pure gelatin group, indicating that the composite materials enhanced mechanical properties and increased resistance to degradation. These results suggest that gelatin and other food hydrocolloids form a double cross-linked network, enhancing the degree of polymer cross-linking. The formation of such a dual network is a common strategy to bolster the mechanical properties of food hydrogels (Khalesi et al., 2020). Additionally, the experimental results showed that the combination of Gel with each of these three hydrocolloids produced different effects on the adhesion and proliferation of porcine fibroblasts. Among them, the Gel-CS was the most effective in providing adhesion and growth conditions for porcine fibroblasts. This enhanced effect may be attributed to the fact that chitosan is a cationic polysaccharide that has a strong affinity for mammalian cells with negatively charged surfaces (Mazia et al., 1975; Ribas Fonseca et al., 2020). Additionally, the amino acid groups on CS contribute to enhanced cell adhesion and expansion, consequently promoting cell proliferation on Gel-CS (Wei et al., 2023).
In the realm of 3D bioprinting, the impact of shear forces generated by nozzle extrusion pressure on cellular viability is significant. Studies have shown that as the extrusion pressure increases, the viability of fibroblasts in the hydrogel gradually decreases (Fakhruddin et al., 2018). Contrarily, the extrusion pressure used in our study was moderate and did not affect cell viability. Additionally, our results show a direct relationship between the cell concentration in the hydrogel ink and the number of cells overflowing during printing, highlighting the importance of selecting the right cell concentration to prevent the wastage of cell resources. We also found that adding cells to the hydrogel stabilizes the structure of the printed sample. Rheological analysis revealed that the storage modulus (G’) of samples with cells is higher than that of hydrogel alone, indicating greater mechanical strength. We speculate that this may be due to the action of cell adhesion molecules, which enhance the overall structural strength of the hydrogel.
Nutrition is one of the most essential elements in evaluating cultured meat. Real meat products contain various cell types from tissues such as skeletal muscle, fat, connective tissue, and skin. Each cell type has unique nutritional properties, and their combined effect enables the meat to have complete amino acid profiles and favorable compositional ratios, thus representing a major source of high-quality protein. Previously, Kang et al. (2021) created cultured meat samples containing muscle stem cells, fat cells, and vessel cells to manufacture multicomponent cultured meat. More studies are still needed to determine whether cultured meat can have a similar nutritional profile as real meat. Guan et al. (2023) manufactured cultured meat samples by seeding porcine muscle stem cells on soy protein scaffolds and found that the amino acid content and quality of the cultured meat samples increased with the efficiency of myogenic differentiation and myotube formation. In this study, we focused on the nutritional profile of fibroblasts and found that different from previous studies, the addition of fibroblasts significantly elevated the content of collagen-specific amino acid hydroxyproline, and collagen-related amino acids proline and glycine (Shoulders and Raines, 2009; Yuswan et al., 2021; Melo et al., 2023), suggesting the importance of fibroblasts in the manufacture of ECM-rich tissues such as connective tissue and skin.
This study is focused on developing cultured meat that mimics the skin part of pork belly, using pig fibroblasts. While the final cultured product shows an increase in amino acids associated with collagen, its mechanical strength significantly differs from real meat. Further investigation into the growth mechanisms of fibroblasts in hydrogel scaffolds is necessary. This understanding will aid in promoting the accumulation of extracellular matrix and rearrangement of molecular structures within the hydrogel, ultimately enhancing the mechanical strength of the cultured meat.
5 Conclusion
In summary, we developed a DF method combining Dispase II and trypsin digestion and constant pressure friction and successfully isolated fibroblasts from porcine skin efficiently and cost-effectively. Furthermore, we determined a Gel-CS hydrogel material that well supports fibroblast proliferation and 3D printing and then fabricated fibroblast-based cultured meat samples. This study provides a highly efficient strategy for isolating fibroblasts from livestock skin and also offers a new idea for the cultured meat industry to manufacture more simulated cultured meat products.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics statement
The animal study was approved by Institutional Animal Care and Use Committee of Jiangnan University. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
RY: Formal analysis, Investigation, Methodology, Writing – original draft. ZF: Investigation, Methodology, Writing – review & editing. LW: Writing – review & editing, Data curation, Investigation. HT: Data curation, Investigation, Writing – review & editing. WS: Writing – review & editing, Data curation, Investigation. ML: Writing – review & editing, Data curation, Investigation. QL: Data curation, Investigation, Writing – review & editing. JC: Conceptualization, Funding acquisition, Resources, Writing – review & editing. XG: Funding acquisition, Methodology, Validation, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was funded by the Strategic Research and Consulting Project of Chinese Academy of Engineering (2023-XZ-79), the National Natural Science Foundation of China (32102132), the Starry Night Science Fund of Zhejiang University Shanghai Institute for Advanced Study (SN-ZJU-SIAS-0013), and the Natural Science Foundation of Jiangsu Province (BK20210462).
Acknowledgments
The authors extend their appreciation to the State Key Laboratory of Food Science and Resources, Jiangnan University for supporting the experiments on amino acid detection and analysis.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Ahmad, K., Lim, J. H., Lee, E. J., Chun, H. J., Ali, S., Ahmad, S. S., et al. (2021). Extracellular matrix and the production of cultured meat. Food Secur. 10, 1–14. doi: 10.3390/foods10123116
Andreazza, R., Morales, A., Pieniz, S., and Labidi, J. (2023). Gelatin-based hydrogels: potential biomaterials for remediation. Polymers 15, 1–12. doi: 10.3390/polym15041026
Ben-Arye, T., Shandalov, Y., Ben-Shaul, S., Landau, S., Zagury, Y., Ianovici, I., et al. (2020). Textured soy protein scaffolds enable the generation of three-dimensional bovine skeletal muscle tissue for cell-based meat. Nature Food 1, 210–220. doi: 10.1038/s43016-020-0046-5
Borges, A. A., Lira, G. P. O., Nascimento, L. E., Santos, M. V. O., Oliveira, M. F., Silva, A. R., et al. (2020). Isolation, characterization, and cryopreservation of collared peccary skin-derived fibroblast cell lines. PeerJ 8:e9136. doi: 10.7717/peerj.9136
Boroda, A. V., Kipryushina, Y. O., Golochvastova, R. V., Shevchenko, O. G., Shulgina, M. A., Efimova, K. V., et al. (2020). Isolation, characterization, and ecotoxicological application of marine mammal skin fibroblast cultures. In Vitro Cell. Develop. Biol. Anim. 56, 744–759. doi: 10.1007/s11626-020-00506-w
Chen, X. B., Fazel Anvari-Yazdi, A., Duan, X., Zimmerling, A., Gharraei, R., Sharma, N. K., et al. (2023). Biomaterials / bioinks and extrusion bioprinting. Bioact. Mater. 28, 511–536. doi: 10.1016/j.bioactmat.2023.06.006
Chen, Y., Lan, D., Wang, W., Zhang, W., and Wang, Y. (2023). Effect of transglutaminase-catalyzed crosslinking behavior on the quality characteristics of plant-based burger patties: A comparative study with methylcellulose. Food Chem. 428:136754. doi: 10.1016/j.foodchem.2023.136754
Choi, K. H., Yoon, J. W., Kim, M., Lee, H. J., Jeong, J., Ryu, M., et al. (2021). Muscle stem cell isolation and in vitro culture for meat production: A methodological review. Compr. Rev. Food Sci. Food Saf. 20, 429–457. doi: 10.1111/1541-4337.12661
Diar-Bakirly, S., and El-Bialy, T. (2021). Human gingival fibroblasts: isolation, characterization, and evaluation of CD146 expression. Saudi J. Biol. Sci. 28, 2518–2526. doi: 10.1016/j.sjbs.2021.01.053
Dohmen, R. G. J., Hubalek, S., Melke, J., Messmer, T., Cantoni, F., Mei, A., et al. (2022). Muscle-derived fibro-adipogenic progenitor cells for production of cultured bovine adipose tissue. NPJ Sci. Food 6:6. doi: 10.1038/s41538-021-00122-2
Edelman, B. L., and Redente, E. F. (2018). Isolation and characterization of mouse fibroblasts. Methods Mol. Biol. 1809, 59–67. doi: 10.1007/978-1-4939-8570-8_5
Fakhruddin, K., Hamzah, M. S. A., and Razak, S. I. A. (2018). “Effects of extrusion pressure and printing speed of 3D bioprinted construct on the fibroblast cells viability” in IOP conference series: Materials science and engineering, vol. 440 (IOP Publishing), 012042.
Guan, X., Lei, Q., Yan, Q., Li, X., Zhou, J., Du, G., et al. (2021). Trends and ideas in technology, regulation and public acceptance of cultured meat. Future Foods 3:100032. doi: 10.1016/j.fufo.2021.100032
Guan, X., Yan, Q., Ma, Z., and Zhou, J. (2023). Production of mature myotubes in vitro improves the texture and protein quality of cultured pork. Food Funct. 14, 3576–3587. doi: 10.1039/d3fo00445g
Guan, X., Zhou, J., Du, G., and Chen, J. (2022). Bioprocessing technology of muscle stem cells: implications for cultured meat. Trends Biotechnol. 40, 721–734. doi: 10.1016/j.tibtech.2021.11.004
Guo, X., Wang, D., He, B., Hu, L., and Jiang, G. (2023). 3D bioprinting of cultured meat: A promising avenue of meat production. Food Bioprocess Technol. 1–22. doi: 10.1007/s11947-023-03195-x
Highley, C. B., Rodell, C. B., and Burdick, J. A. (2015). Direct 3D printing of shear-thinning hydrogels into self-healing hydrogels. Adv. Mater. 27, 5075–5079. doi: 10.1002/adma.201501234
Hosseini, V., Kalantary-Charvadeh, A., Hasegawa, K., Nazari Soltan Ahmad, S., Rahbarghazi, R., Mahdizadeh, A., et al. (2020). A mechanical non-enzymatic method for isolation of mouse embryonic fibroblasts. Mol. Biol. Rep. 47, 8881–8890. doi: 10.1007/s11033-020-05940-3
Jeong, D., Seo, J. W., Lee, H. G., Jung, W. K., Park, Y. H., and Bae, H. (2022). Efficient myogenic/Adipogenic Transdifferentiation of bovine fibroblasts in a 3D bioprinting system for steak-type cultured meat production. Adv. Sci. 9:e2202877. doi: 10.1002/advs.202202877
Kang, D. H., Louis, F., Liu, H., Shimoda, H., Nishiyama, Y., Nozawa, H., et al. (2021). Engineered whole cut meat-like tissue by the assembly of cell fibers using tendon-gel integrated bioprinting. Nat. Commun. 12:5059. doi: 10.1038/s41467-021-25236-9
Khalesi, H., Lu, W., Nishinari, K., and Fang, Y. (2020). New insights into food hydrogels with reinforced mechanical properties: A review on innovative strategies. Adv. Colloid Interf. Sci. 285:102278. doi: 10.1016/j.cis.2020.102278
Kitano, Y., and Okada, N. (1983). Separation of the epidermal sheet by dispase. Br. J. Dermatol. 108, 555–560. doi: 10.1111/j.1365-2133.1983.tb01056.x
Kuwabara, J. T., and Tallquist, M. D. (2017). Tracking adventitial fibroblast contribution to disease: A review of current methods to identify resident fibroblasts. Arterioscler. Thromb. Vasc. Biol. 37, 1598–1607. doi: 10.1161/ATVBAHA.117.308199
Li, F., Adase, C. A., and Zhang, L. J. (2017). Isolation and culture of primary mouse keratinocytes from neonatal and adult mouse skin. J. Vis. Exp. 125, 1–6. doi: 10.3791/56027
Li, M., Wang, D., Fang, J., Lei, Q., Yan, Q., Zhou, J., et al. (2022). An efficient and economical way to obtain porcine muscle stem cells for cultured meat production. Food Res. Int. 162:112206. doi: 10.1016/j.foodres.2022.112206
Li, Q., Yi, S., Wang, W., Xu, Y., Mi, H., Li, X., et al. (2021). Different thermal treatment methods and TGase addition affect gel quality and flavour characteristics of Decapterus maruadsi surimi products. Food Secur. 11, 1–21. doi: 10.3390/foods11010066
Liu, P., Song, W., Bassey, A. P., Tang, C., Li, H., Ding, S., et al. (2023). Preparation and quality evaluation of cultured fat. J. Agric. Food Chem. 71, 4113–4122. doi: 10.1021/acs.jafc.2c08004
Ma, T., Ren, R., Yang, R., Zheng, X., Hu, Y., Zhu, G., et al. (2023). Transdifferentiation of fibroblasts into muscle cells to constitute cultured meat with tunable intramuscular fat deposition. bioRxiv 2010:2026.564179.
Mazia, D., Schatten, G., and Sale, W. (1975). Adhesion of cells to surfaces coated with polylysine. Applications to electron microscopy. J. Cell Biol. 66, 198–200. doi: 10.1083/jcb.66.1.198
Melo, M. M. P., Mesquita, R. B. R., Coscueta, E. R., Pintado, M. E., and Rangel, A. (2023). Assessment of collagen content in fish skin - development of a flow analysis method for hydroxyproline determination. Anal. Methods 15, 5901–5908. doi: 10.1039/d3ay01589k
Oroian, M., Ropciuc, S., and Paduret, S. (2018). Honey authentication using rheological and physicochemical properties. J. Food Sci. Technol. 55, 4711–4718. doi: 10.1007/s13197-018-3415-4
Pasitka, L., Cohen, M., Ehrlich, A., Gildor, B., Reuveni, E., Ayyash, M., et al. (2023). Spontaneous immortalization of chicken fibroblasts generates stable, high-yield cell lines for serum-free production of cultured meat. Nat. Food 4, 35–50. doi: 10.1038/s43016-022-00658-w
Plikus, M. V., Wang, X., Sinha, S., Forte, E., Thompson, S. M., Herzog, E. L., et al. (2021). Fibroblasts: origins, definitions, and functions in health and disease. Cell 184, 3852–3872. doi: 10.1016/j.cell.2021.06.024
Ribas Fonseca, L., Porto Santos, T., Czaikoski, A., and Lopes Cunha, R. (2020). Modulating properties of polysaccharides nanocomplexes from enzymatic hydrolysis of chitosan. Food Res. Int. 137:109642. doi: 10.1016/j.foodres.2020.109642
Shoulders, M. D., and Raines, R. T. (2009). Collagen structure and stability. Annu. Rev. Biochem. 78, 929–958. doi: 10.1146/annurev.biochem.77.032207.120833
Sonaye, S. Y., Ertugral, E. G., Kothapalli, C. R., and Sikder, P. (2022). Extrusion 3D (bio)printing of alginate-gelatin-based composite scaffolds for skeletal muscle tissue engineering. Materials 15, 1–24. doi: 10.3390/ma15227945
Takahashi, Y., Mutasim, D. F., Patel, H. P., Anhalt, G. J., Labib, R. S., and Diaz, L. A. (1985). The use of human pemphigoid autoantibodies to study the fate of epidermal basal cell hemidesmosomes after trypsin dissociation. J. Invest. Dermatol. 85, 309–313. doi: 10.1111/1523-1747.ep12276893
Vig, K., Chaudhari, A., Tripathi, S., Dixit, S., Sahu, R., Pillai, S., et al. (2017). Advances in skin regeneration using tissue engineering. Int. J. Mol. Sci. 18, 1–19. doi: 10.3390/ijms18040789
Wang, X., Ao, Q., Tian, X., Fan, J., Tong, H., Hou, W., et al. (2017). Gelatin-based hydrogels for organ 3D bioprinting. Polymers 9, 1–24. doi: 10.3390/polym9090401
Wang, H., Van Blitterswijk, C. A., Bertrand-De Haas, M., Schuurman, A. H., and Lamme, E. N. (2004). Improved enzymatic isolation of fibroblasts for the creation of autologous skin substitutes. In Vitro Cell. Dev. Biol. Anim. 40, 268–277. doi: 10.1290/0408055.1
Wei, L., Wang, X., Fu, J., Yin, J., and Hu, J. (2023). A physically cross-linked double network polysaccharides/Ca2+ hydrogel scaffold for skeletal muscle tissue engineering. Colloids Surf. A Physicochem. Eng. Asp. 668:131410. doi: 10.1016/j.colsurfa.2023.131410
Widbiller, M., Bucchi, C., Rosendahl, A., Spanier, G., Buchalla, W., and Galler, K. M. (2019). Isolation of primary odontoblasts: expectations and limitations. Aust. Endod. J. 45, 378–387. doi: 10.1111/aej.12335
Yan, Q., Fei, Z., Li, M., Zhou, J., Du, G., and Guan, X. (2022). Naringenin promotes Myotube formation and maturation for cultured meat production. Food Secur. 11, 1–16. doi: 10.3390/foods11233755
Yuswan, M. H., Jalil, N. H., Mohamad, H., Keso, S., Mohamad, N. A., Yusoff, T. S., et al. (2021). Hydroxyproline determination for initial detection of halal-critical food ingredients (gelatin and collagen). Food Chem. 337:127762. doi: 10.1016/j.foodchem.2020.127762
Zhang, G., Zhao, X., Li, X., Du, G., Zhou, J., and Chen, J. (2020). Challenges and possibilities for bio-manufacturing cultured meat. Trends Food Sci. Technol. 97, 443–450. doi: 10.1016/j.tifs.2020.01.026
Keywords: cultured meat, 3D bioprinting, fibroblasts, cell isolation, gelatin, chitosan, food hydrocolloid
Citation: Yang R, Fei Z, Wang L, Tang H, Sun W, Li M, Lei Q, Chen J and Guan X (2024) Highly efficient isolation and 3D printing of fibroblasts for cultured meat production. Front. Sustain. Food Syst. 8:1358862. doi: 10.3389/fsufs.2024.1358862
Edited by:
Na Zhang, Harbin University of Commerce, ChinaReviewed by:
Mecit Oztop, Middle East Technical University, TürkiyeJanpriy Sharma, Dr. B. R. Ambedkar National Institute of Technology, Jalandhar, India
Copyright © 2024 Yang, Fei, Wang, Tang, Sun, Li, Lei, Chen and Guan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jian Chen, amNoZW5AamlhbmduYW4uZWR1LmNu; Xin Guan, Z3VhbnhpbkBqaWFuZ25hbi5lZHUuY24=
 Ruoqing Yang
Ruoqing Yang Zhuocheng Fei1,2
Zhuocheng Fei1,2 Jian Chen
Jian Chen Xin Guan
Xin Guan