
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Sustain. Food Syst. , 23 May 2024
Sec. Agroecology and Ecosystem Services
Volume 8 - 2024 | https://doi.org/10.3389/fsufs.2024.1329422
This article is part of the Research Topic Optimizing Natural Features and BMPs in Agroecosystems Through a One-Health Approach View all 5 articles
Introduction: Agricultural drainage ditches are essential for maintaining flow efficiency and are often managed by brushing (i.e., vegetation clearing along ditch banks) and dredging. These maintenance practices not only impact flow but also the ditches’ capacity to process and assimilate agricultural chemicals. However, the influence of ditch management on microbial activities, essential in processing and transforming these contaminants, remains underexplored.
Methods: We conducted a four-year surface water sampling campaign in two long-established (over 40 years) agricultural drainage ditches situated in eastern Ontario, Canada. One drainage ditch underwent intensive bank brushing and channel dredging, while the other remained unmanaged. We assessed the impact of these contrasting ditch management practices on the diversity and compositional structure of surface water microbiome communities, with a particular focus on the distribution of genes associated with nitrogen (N) cycling, using shotgun metagenomics sequencing and an assembly-based metagenome approach.
Results and discussion: From 117 surface water samples, we reconstructed 157 metagenome-assembled genomes (MAGs), predominantly from the Patescibacteria superphylum. Brushing alone, conducted about 10 months prior to dredging, had an insignificant impact on the aquatic microbial community. However, dredging led to a significant reduction in community diversity and abundance of genes affiliated with denitrification, dissimilatory nitrate reduction to ammonia, and complete nitrification through comammox. Ditch management resulted in noticeable shifts in the microbial community, evidenced by the enrichment of taxa in Polaromonas, Emticicia, Flectobacillus, and Patescibacteria in the water of the managed ditch sites. The metagenomes of these taxa harbor genes involved in various nitrogen biotransformation pathways. Interestingly, following dredging, the levels of ammonia and ammonium, nitrite, and total Kjeldahl N in the managed ditch did not increase, but significantly decreased; likely due to ditch substrate removal and potentially increased flow efficiency and dilution. Being one of the few studies conducted to date, this study provided unique insights into the consequences of drainage ditch management on freshwater microbiomes involved in N cycling.
A sustainable agro-ecosystem relies on the ecological services provided by natural and semi-natural landscape features (Bennett et al., 2021; Damphousse et al., 2024). Artificial subsurface drainage, commonly known as tile drainage, is designed to drain sub-surface water from fields to facilitate crop productivity (Schwab et al., 1966). Agricultural drainage ditches receive tile and field runoff water that can be laden with agro-chemicals, fecal pollution from livestock waste, and waterborne zoonotic pathogens from livestock waste (Kladivko et al., 2001; Wilkes et al., 2014; Sunohara et al., 2015; Hruby et al., 2016; Hunting et al., 2017; Räty et al., 2020). In humid temperate agro-ecosystems, agricultural drainage ditches are common, for example, covering >1 km for every square kilometer of watershed (Rideout et al., 2022). The management of these ditches is essential to maintain efficient water flow (Needelman et al., 2007; Kavanah et al., 2017). Common management practices include sediment dredging, removal of in-stream vegetation, and varying degrees of vegetation clearance along the banks—a process known as brushing. Brushing not only facilitates access to dredging equipment but also reduces shading effects by woody vegetation on adjacent crop fields (Dollinger et al., 2015). The frequency and extent of brushing and dredging are determined by factors such as sedimentation levels, vegetation’s impact on water flow, aesthetic considerations, field shading, and the size of the catchment area (Levavasseur et al., 2014; Kavanah et al., 2017). The impact of ditch management practices on the ecosystem services provided by drainage ditch ecosystems warrants greater attention (Rideout et al., 2022). This is particularly crucial given that these ditches are among the few semi-naturalized features in otherwise depauperate agricultural landscapes (Herzon and Helenius, 2008; Dollinger et al., 2015; Dyck et al., 2021; Cital et al., 2022; Rideout et al., 2022).
A significant gap in knowledge exists concerning how ditch management affects microbial communities, which critically govern the cycling, dissipation, and transformation of agro-chemicals and pathogens in the aquatic and semi-aquatic ditch environments (Shen et al., 2016; Soana et al., 2017; Kumwimba et al., 2018; Xu et al., 2022). Nitrogen (N), originating from both organic and inorganic fertilizers, can leach into tile drains, or be carried in runoff (often as nitrate) into drainage ditch networks (Randall et al., 1997; Dinnes et al., 2002; Sunohara et al., 2014). Excessive N in agricultural waterways can critically contribute to eutrophication, as evidenced in large scale in hypoxic zones in the Gulf of Mexico (Rabalais and Turner, 2019). Romanelli et al. (2020) demonstrated that nitrified ammonium from fertilizers accounted for 50–75% of nitrate in streams. Wu et al. (2023) found that nutrient input was the primary driver of significant greenhouse gas (GHG) production and emissions in agricultural ditch systems. Shi et al. (2023) found that a subset of the stream microbial community in agricultural drainage ditches, specifically the conditionally rare taxa, were predominantly composed of N-cycling bacteria. This bacterial group was influenced by factors such as nutrient loading and flow dynamics in drainage ditches. The processes including denitrification, dissimilatory nitrate reduction to ammonium (DNRA), and anaerobic ammonium oxidation, play key roles in nitrate transformation in waterways (Domangue and Mortazavi, 2018). In particular, anaerobic ammonium oxidation can lead to sediment N loss via the production of dinitrogen (N2) gas in agricultural drainage ditches (Shen et al., 2016). However, examining how the effects of contrasting drainage ditch management practices influence microbial communities involved in N cycling remains largely unexplored. Such information is crucial for understanding how to optimize the management of these features to minimize GHG and reduce impacts on ecological and public health endpoints.
Nitrite and nitrate can be removed from the aquatic system by denitrifying bacteria, which can result in in-stream or near-stream N2O fluxes (Roland et al., 2018; Groffman et al., 2021; Wu et al., 2023); N2O being a potent GHG. Moreover, ammonium-oxidizing bacteria can transform ammonium to N2, which is not a greenhouse gas (Schubert et al., 2006; Roland et al., 2018; Almaraz et al., 2023). DNRA retains nitrate in the environment by converting it into ammonium, thereby making N bioavailable to other organisms (Murphy et al., 2016). Anammox bacteria, identified in agricultural ditches, have been linked to the removal of ammonium and the production of N2 gas (Shen et al., 2016). Furthermore, Chen et al. (2018) found that nitrifiers were more abundant in agricultural ditch waters compared with larger water courses influenced by a wider range of land use activities and dilution influences. This pattern was particularly notable in spring when the flow of surface water was relatively higher in the region of study. As mentioned previously, maintenance practices such as brushing of ditch bank vegetation and channel dredging can significantly affect vegetation, temperature, substrate, aeration, and hydrological regime of these systems. Cui et al. (2020) reported that the removal of vegetation in agricultural ditches significantly lowers sediment bacterial community diversity and its ability to remove agricultural N compared to undisturbed sediment. Consequently, these practices have the potential to substantially alter the environmental conditions that govern N-metabolizing bacteria and denitrifying and nitrifying the genes they contain (Rich and Myrold, 2004; Dandie et al., 2011; Waymouth et al., 2021).
This study examined the diversity and abundance of bacteria and genes affiliated with N cycling in water from experimental agriculture drainage ditches with contrasting ditch management practices. One ditch recently underwent brushing of bank vegetation and dredging, while the other remained relatively undisturbed since its inception. We hypothesized that there would be a notable decrease in the diversity of the microbial community associated with N cycling in the managed ditch due to the physical removal of organic substrate and vegetation that provide conditions suitable for their growth and proliferation (Veraart et al., 2017; Cui et al., 2020). To evaluate the diversity and functional characteristics of the aquatic microbial community, we employed a metagenome-assembled genome (MAG) approach (Zhou et al., 2022). The MAG community consists of reconstructed genomes derived from shotgun metagenomic sequencing data of in-stream water samples. These genomes typically represent microorganisms that were sufficiently abundant and had adequate sequencing coverage for assembly. By analyzing the dynamics of the MAG community, this study aimed to provide insights into the functional capabilities and metabolic pathways associated with N cycling under contrasting, yet common, agricultural drainage ditch management conditions.
This study was conducted in the South Nation River basin in eastern Ontario, Canada (Figure 1). The river basin is characterized by mixed but primarily agricultural land use. Water samples were collected from two municipal drains, which serve as agricultural drainage ditches, from 2017 to 2021. These drains are part of the Environmental Change One Health Observatory (ECO2) study, which has been operational under various names since the early 2000s (Lyautey et al., 2007; Schmidt et al., 2013; Wilkes et al., 2014). Before this study, both drainage ditches had not undergone any significant ditch management activities (outside of things such as periodic dismantling of beaver dams and removal of dead tree falls), such as brushing and dredging, for over 40 years. Before the 2018 water sampling season, one drainage ditch (hereafter referred to as the “managed ditch”) underwent vegetation brushing followed by dredging to help restore full flow capacity in the ditch. The managed drainage ditch covers an effective surface contributing area of 225 ha (Wilkes et al., 2014). The paired drainage ditch, which remained unmanaged, has an effective surface contributing area of 467 ha (Wilkes et al., 2014). Two sampling sites, SN18 and SN19, are situated on the unmanaged drainage ditch, with SN18 positioned 1.45 km downstream of SN19. These sites serve as redundant monitoring sites and are not considered replicates in a strict sense. Similarly, in the managed drainage ditch, there were two sites, SN20 and SN21, with SN20 being located approximately 1 km downstream of SN21. These two sites also functioned as redundant monitoring points.
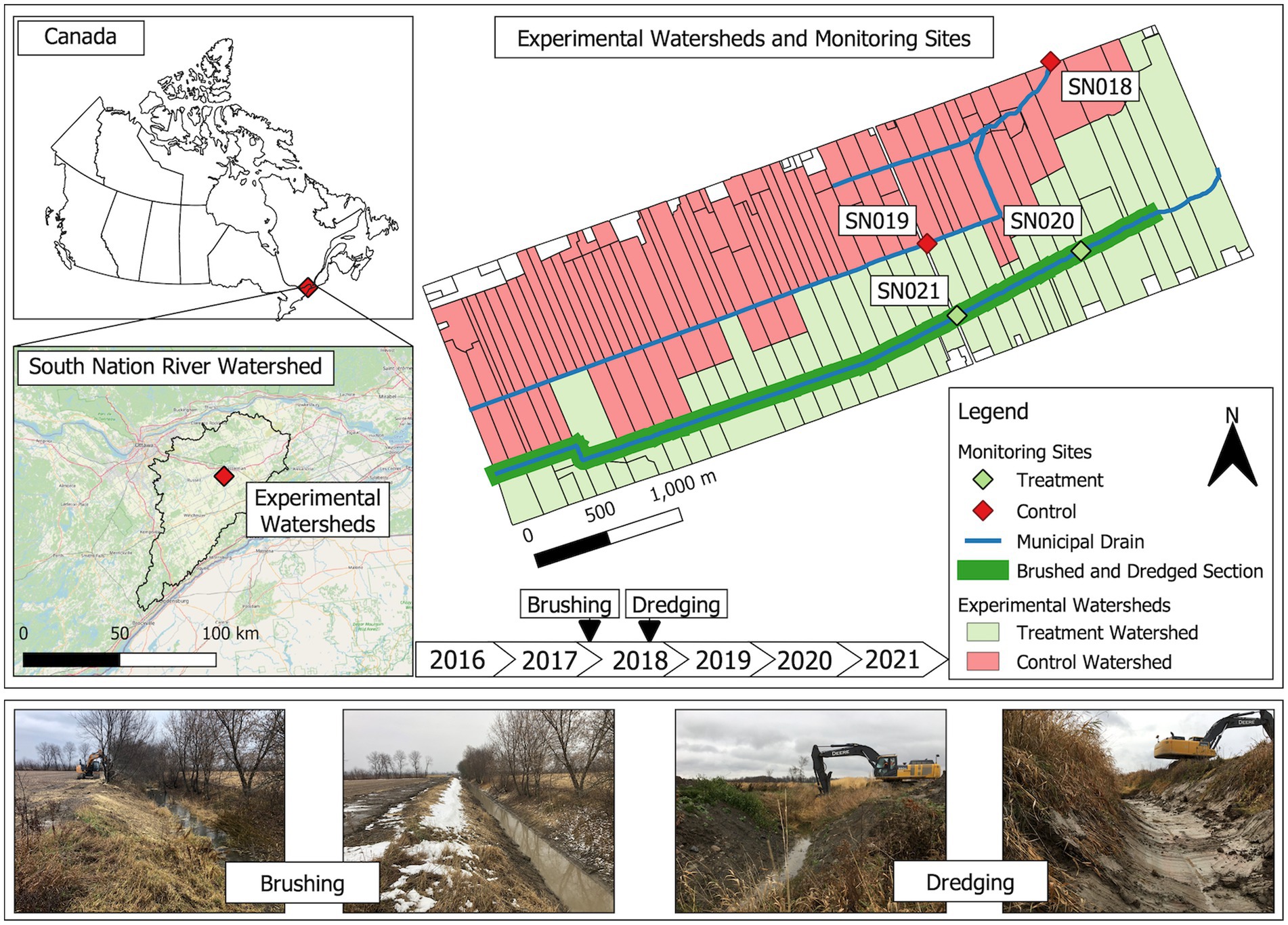
Figure 1. Sampling sites. This map shows the South Nation River basin in eastern Ottawa, Ontario, Canada, highlighting two distinct watersheds. Sites SN18 and SN19 are in the unmanaged ditch and function as control sites. Sites SN20 and SN21 are in the managed ditch that underwent ditch interventions, with brushing in the winter of 2018 before the sampling season, and dredging in the fall of 2018 after the sampling season.
The brushing began on 28 February 2018, prior to the 2018 water sampling season. Vegetation brushing was conducted to remove woody vegetation from the managed drainage using an excavator equipped with a brush mower attachment. This process cut and sheared the woody vegetation while leaving roots intact, thus maintaining slope integrity and allowing for regrowth over time, as described by Kavanah et al. (2017) (Figure 1).
Subsequently, dredging was carried out along the managed ditch using an excavator starting on 29 October 2018, beginning at the east end of the ditch. This occurred after crops had been harvested, lasting for approximately a week. The dredging process involved removing channel vegetation, sediments, and debris from the channel bottom to the lower mid-slope of the ditch. The excavation depth reached approximately 30 cm below the pre-excavation sediment level in the middle of the channel, with gradually shallower depths from mid-channel to the mid-slopes. The excavation of the organic rich sediments and organic matter in the managed ditch exposed varved clay substrates (Cummings et al., 2011; Blume et al., 2022). This approach was purposefully aggressive with respect to how ditches are generally managed in the region, though not uncommon in many areas of the world where agricultural drainage ditches predominate. The aggressive approach was employed to provide highly contrasting ditch management conditions for experimental purposes (Figure 1). The dredged material was either spread across adjacent fields or used in other field locations, depending on the landowner’s preference. Annual agricultural activities, such as crop types, pesticide application, and nutrient and soil management, were effectively similar among the paired experimental watersheds in this study (Sunohara et al., 2015).
A total of 117 surface water samples were collected from the top 0–5 cm of the water column from all sites every 2 weeks from April to November, spanning the years from 2017 to 2021. However, in 2020, the second-year post-dredging, we were unable to access these sampling sites due to Canadian federal legislation related to the COVID-19 pandemic. The water samples were collected using 2 L sterile Polyethylene Terephthalate (PET) bottles (Systems Plus, Baden, ON, Canada) and were transported to coolers in the laboratories in Agriculture and Agri-Food Canada (AAFC)‘s Ottawa Research and Development Centre (ORDC). Upon arrival, the samples were stored at 4°C until they were ready for processing.
Water physicochemical parameters, including temperature, pH, dissolved oxygen, electrical conductivity, turbidity, and oxidation–reduction potential, were measured in the field using a YSI PRO DSS sonde (YSI Incorporated, Yellow Springs, OH, United States), according to the methods given in the study by Wilkes et al. (2011). To ensure consistency with historical data in water nutrient characterization, water samples were analyzed at the City of Ottawa, Robert O. Pickard Environmental Center (ROPEC) Laboratory. Ammonia (NH3) and ammonium (NH4) contents were analyzed using the automated phenate method, following the Standard Method 4,500-NH3. Nitrite (NO2−) and nitrate (NO3−) levels were determined by ion chromatography following the Standard Method 4110B. Total Kjeldahl nitrogen (TKN) was analyzed using the automated phenate method preceded by sulfuric acid digestion, following the Standard Method 4,500-Norg. Total phosphorus was measured using the automated ascorbic acid method preceded by persulfate digestion, following the Standard Method 4,500-P F. Total suspended solids were quantified through ignite filter and gravimetry following the Standard Method E3188. Dissolved organic carbon (DOC) and total organic carbon (TOC) were determined using ultraviolet persulfate oxidation following the Standard Method 5310C [see Zolfaghari et al. (2019)]. For this study, concentrations were focused on as they were deemed more formative to microbial responses than loads.
At sampling sites, SN18 and SN20, stream discharge (flow rate) measurements were made using an ISCO 4150 area velocity flow module (Teledyne Isco, Inc.) and low-profile area velocity sensors within circular road culverts. Open channel area velocity calculations (flow = velocity x cross-sectional area) were used to determine stream discharge (Grant and Dawson, 2001), and stage-discharge rating curves were developed to predict discharge during periods when velocity data were unavailable, according to Sunohara et al. (2015). Stream discharge (m3 s−1) was converted to watershed unit area drainage values (m3 ha−1 15-min−1) by scaling measurements to 15-min interval volumes and dividing by watershed contributing areas. The watershed unit area drainage volumes from SN18 and SN20 were temporally collocated and compared using linear regression to assess slope changes associated with drainage ditch interventions (Sunohara et al., 2015).
Water samples were initially filtered through a 0.7 μm borosilicate glass filter (Thermo Fisher, Ottawa, ON, Canada). The filtrates were then passed through a 0.22 μm nitrocellulose filter (Millipore, Billerica, MA, United States) to prevent clogging. Total environmental DNA was extracted from 0.22 μm filters using the PowerSoil DNA Extraction Kit (MD, United States), following the manufacturer’s instructions. Sequencing libraries were prepared using the Illumina Nextera XT Kit, following the manufacturer’s protocol. For the samples from 2017 to 2019, paired-end sequencing data at 2×150 bp were obtained using an Illumina HiSeq 2,500 sequencer at the National Research Council of Canada (Montreal, Quebec, Canada). Samples from 2021 were sequenced on the Illumina NovaSeq 6,000 at Nucleic Acid Solutions from the National Research Council of Canada (Saskatoon, Saskatchewan, Canada).
To ensure reliability and comparability of shotgun metagenomics data across different years and address batch effect issues, we implemented specific steps as follows: (1) quality filtering of reads to reduce technical variability across batches and (2) read mapping and normalization of coverage calculations using Anvi’o platform (Eren et al., 2021). In brief, raw sequencing data of all DNA samples were populated and processed simultaneously using the default settings of the respective software unless stated otherwise. Raw reads were trimmed with Trimmomatic v.0.39 (Bolger et al., 2014) with the following parameters: LEADING:3, TRAILING:3, SLIDINGWINDOW:4:15, MINLEN:36, and HEADCROP:6. Adapters were removed using BBDuk.1 Due to the large volume of sequencing data obtained from all samples, we opted to perform co-assembly of the sequencing data for each year. This was done using MEGAHIT v.1.2.9, setting a minimum contig length of 1 kbp (Li et al., 2015).
Gene prediction for contigs was conducted using Prodigal v.2.6.3 (Hyatt et al., 2010). We then mapped quality-trimmed reads to the assembled contigs using Bowtie2 v.7.5.0 (Langmead and Salzberg, 2012). The recruited reads were stored as BAM files using Samtools v.1.7 (Li et al., 2009). Sample-specific contig binning was independently conducted using CONCOCT v.1.1.0 (Alneberg et al., 2014) and MetaBat2 v.2.12.1 (Kang et al., 2019). Consensus bins were created using Das_Tool v.1.1.3 (Sieber et al., 2018). Further manual refinement of these bins was performed using the anvi-refine function within the Anvi’o platform (Eren et al., 2021). Bins that were greater than 50% complete and had less than 10% redundancy (or contamination) were retained, in accordance with the Minimum Information about a Metagenome-Assembled Genome (MIMAG) standards established by the Genomic Standards Consortium (Bowers et al., 2017).
To consolidate metagenome-assembled genomes (MAGs) from different years, bins were dereplicated using the anvi-dereplicate-genomes function in Anvi’o. This process employs fastANI v.1.1 with a similarity threshold to resolve bins that are similar across different years (Jain et al., 2018). This pipeline produced a total of 157 genomic bins. Taxonomical predictions were performed using GTDB-tk v.2.1.0 (Chaumeil et al., 2020) with reference to the Genome Taxonomy Database (GTDB) Release 207 v.2 (Parks et al., 2021). For predicting gene functions and reconstructing metabolic pathways, the anvi-estimate-metabolism function from Anvi’o (Eren et al., 2021) was utilized, referencing the Kyoto Encyclopedia of Genes and Genomes (KEGG) database (Kanehisa and Goto, 2000).
The normalization process within the Anvi’o pipeline (Eren et al., 2021) involves deriving both taxonomic and gene abundance data from the number of reads that aligned to each contig. Specifically, for each contig, the process counts the reads from a given sample that map to it. These counts are then normalized by dividing them by the total number of reads from all samples that map to that contig. This step is crucial for adjusting for differences in sample size and sequencing depth, thereby enabling a more accurate and comparable analysis across different samples.
We generated the maximum likelihood tree of the MAGs, incorporating GTDB’s representative genomes (Parks et al., 2021), using GToTree v1.7.00 (Lee, 2019). This was done utilizing the prepackaged single-copy gene sets for bacteria (172 target genes) and archaea (76 target genes). Gene prediction on the input genomes, provided as fasta files, was performed with Prodigal v2.6.3 (Hyatt et al., 2010). HMMER3 v3.3.2 (Eddy, 2011) identified the target genes, which were then individually aligned using Muscle v5.1 (Edgar, 2022). These alignments were trimmed with trimAl v1.4.rev15 (Capella-Gutiérrez et al., 2009) and concatenated for phylogenetic tree reconstruction using FastTree2 v2.1.11 (Price et al., 2010). TaxonKit (Shen and Ren, 2021) linked full lineages to their respective taxonomic IDs. Finally, the phylogenetic tree was annotated, pruned, and visualized using iTOL v5 (Letunic and Bork, 2021).
All statistical analyses were performed in R (v4.2.1) (R Core Team, 2022), except where noted otherwise. The two drainage ditches, although adjacent to each other, are situated in separate surface and subsurface (tile) drainage catchment areas (Sunohara et al., 2015). For analysis purposes, each pair of sites, SN18 + SN19 in the unmanaged ditch and SN20 + SN21 in the managed ditch, was treated as watershed blocks, functioning as redundant monitoring sites. There may be some level of spatial autocorrelation, given that SN18 is downstream of SN19 and SN20 is downstream of SN21. To account for this factor, we utilized linear mixed-effects (LME) models to assess the impact of ditch management practices, season, and year on water physiochemical properties, abundance and diversity of MAGs, and genes and metabolic pathways associated with N cycling. The analyses were performed using the lme function in the nlme package (Pinheiro et al., 2018), with a significance threshold set at p ≤ 0.05.
To develop the LME models, ditch management practices and sampling season were set as fixed effects to evaluate their influence on specific response variables. The models aimed to assess the impact of brushing and dredging practices across different seasons, including both their individual and combined effects. To account for within-group correlations and potential non-independence of observations, sampling year and week were incorporated as random effects in our models. Watershed blocks were also considered as a random effect to address spatial variability. This modeling approach allowed us to effectively control for temporal and spatial variations while focusing on the interactions and main effects of management practices and seasonal changes. Post hoc analyses were conducted using the Bonferroni–Holm correction for multiple comparisons across different conditions (e.g., ditch management, sampling seasons, and/or years), utilizing the glht function from the multcomp package (Bretz et al., 2010).
The assumptions of the LME models were evaluated and visualized using several diagnostic tools. Homoscedasticity and linearity were assessed with residuals versus fitted plot, while normality was checked using a normal Q–Q plot. Autocorrelation in the residuals was examined using the acf R function. Additionally, Cook’s distance plots were employed to identify influential data points (Supplementary Figure S2). Notably, these plots revealed that the influential data points, which significantly deviated from the mean, were primarily found in summer samples from 2021. These points, suggestive of potential treatment effects, were deliberately retained in the analysis to ensure that any significant biological changes were not overlooked.
All alpha-diversity indices, such as the Shannon–Weiner and Simpson indices, were obtained using the diversity function from the vegan package. The Shannon–Weiner-based True Diversity (ShanTD) (Jost, 2006) was derived by exponentiating the Shannon–Weiner index. A permutational multivariate analysis of variance (PERMANOVA) analysis was conducted to compare the aquatic microbial community compositional structure between sites, utilizing the Adonis function from the vegan package (Oksanen et al., 2019). The community compositional heterogeneity was visualized through double principal coordinate analysis (dPCoA), which was performed using the DPCoA command in the phyloseq package (McMurdie and Holmes, 2013). For both the PERMANOVA and dPCoA analyses, the abundance matrices (taxa or genes) were clr-transformed prior to calculating the Aitchison distance. Additionally, we used the envfit function in the vegan package to project the water physicochemical vectors onto the dPCoA ordination plots, to assess their impact on microbial community compositional changes. The correlation plot (Supplementary Figure S1) for water physicochemical properties was generated using the levelplot function in the lattice R package (Sarkar, 2008).
Interestingly, linear regression analysis of measured flow from site SN18 (unmanaged) and SN20 (managed) for pre-management years (2016–17), and post-management (2019–21) years, showed there was no significant difference in regression slopes between the two time periods at the 0.05 level.
The initial objective was to examine the impact of brushing and dredging on the physicochemical properties of ditch water and their potential influences on the aquatic microbiome. LME analyses revealed no significant differences in most water physicochemical properties between the managed and unmanaged ditch sites in the year 2017, the year before ditch management intervention began (Figure 2). However, nitrate (Figure 2, F = 4.70, Padj = 0.04) and nitrite (Figure 2, F = 5.70, Padj = 0.03) concentrations were significantly higher in the unmanaged ditches compared with the managed ditch in 2017, prior to maintenance activities. The managed sites showed no significant changes in water physicochemical properties during the 2018 water sampling period following brushing at the beginning of the year (vs. 2017), although Dyck et al. (2021) reported brushing-induced changes in turbidity, conductivity, and water temperatures of ditch streams.
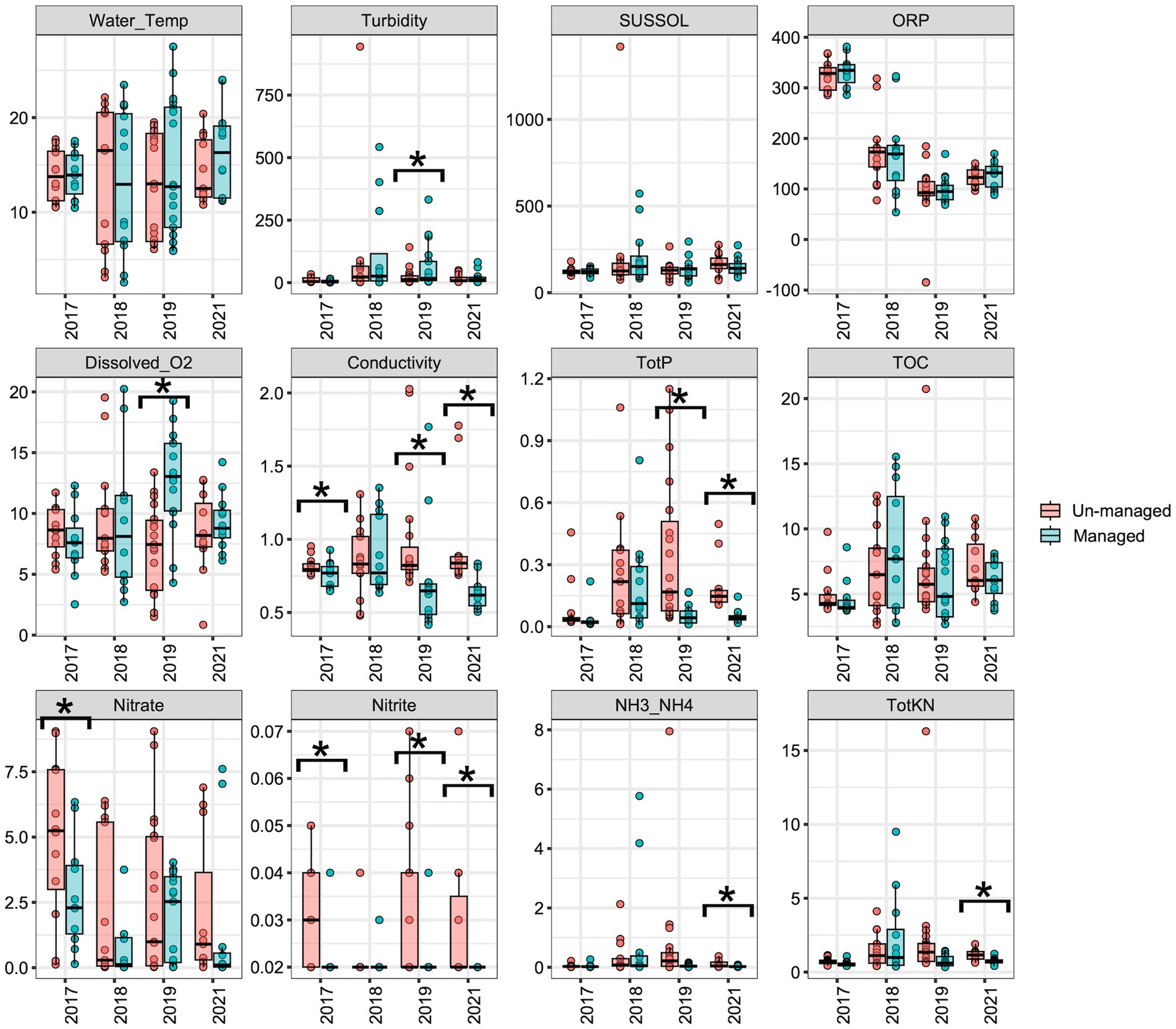
Figure 2. Boxplots displaying the variation of physicochemical properties in managed versus unmanaged ditches over the sampling years. An asterisk (*) indicates significant differences (p ≤ 0.05) between managed and unmanaged ditches, as determined by linear mixed-effects analyses.
For the 2019 water sampling period, following dredging, the previous fall, turbidity significantly increased (Figure 2; F = 4.60, Padj = 0.04), while total phosphorus (Figure 2; F = 15.0, Padj = 0.001) and nitrite concentrations (Figure 2; F = 6.40, Padj = 0.02) were significantly lower in the managed ditch than in the unmanaged ditch. After 3 years of dredging, in 2021, conductivity (Figure 2; F = 10.00, Padj = 0.004), total phosphorus (Figure 2; F = 15.00, Padj = 0.001), nitrite (Figure 2; F = 5.10, Padj = 0.04), NH3/NH4 (Figure 2; F = 6.50, Padj = 0.02), and total Kjeldahl nitrogen (TotKN) (Figure 2; F = 19.00, Padj < 0.001) were significantly lower in the managed ditch than in the unmanaged ditch. The decrease in phosphorus and nitrogen species concentrations in the managed ditch compared to the unmanaged ditch is consistent with the load-based findings by Smith and Huang (2010). Reductions may have been due to the removal of sediments and soils that had accumulated these nutrient species prior to the ditch management activities, increased flow efficiency in the managed system augmenting dilution and more rapid off-site mass export of nutrients, and potentially oxidation of reduced sediments in the channel (Smith and Huang, 2010).
Furthermore, the analysis of water physicochemical properties through LME models over 4 years reveals a complex interplay between ditch management practices and seasonal changes in water quality. In 2017 (before brushing) and 2018 (following brushing in winter), the models that included interaction term (ditch management x season) did not generally enhance the interpretation of water property changes—conductivity in 2017 being the exception—as shown in Supplementary Table S2. In contrast, an LME analysis showed that the impact of ditch management on total phosphorous (TotP, p = 0.0051) and water temperature (Water_Temp, p = 0.0341) was notably season-dependent in 2019, the first year after dredging. Similarly, in 2021 (third-year post-dredging), conductivity (p = 0.0005) and ammonia/ammonium (NH3_NH4, p = 0.0001) also displayed differences between the seasons (Supplementary Table S2). These observations may indicate that the managed ditch exhibits heightened responsiveness to seasonal changes in weather patterns following the removal of vegetation.
Shotgun metagenomics sequencing of 130 stream samples yielded approximately 7.96 billion nucleotides across four sampling years. The assembly of the sequencing data resulted in 3.4 million contigs, organized into 354 bins. After dereplication, we retained 157 MAGs that met the criteria of being >0.5 Mbp in size, with >50% completion and <10% redundancy (Supplementary Table S1). Of the 157 MAGs, the majority were assigned to the bacterial superphylum Patescibacteria (77 MAGs, 47.9% relative abundance), followed by Proteobacteria (30 MAGs, 31.4% relative abundance) and Bacteroidota (22 MAGs, 11.2% relative abundance). Of the 77 Patescibacteria MAGs, 59 were assigned to the UBA9983_A order. Furthermore, 48 Patescibacteria MAGs clustered within a clade alongside the representative genome from species CAIJXO01 (in the class Paceibacteria), which can be found ubiquitously in freshwater and marine environments (Figure 3). All Proteobacteria MAGs were assigned to the Burkholderiaceae family, including 10 Burkholderiaceae MAGs distributed among NBD-18 (3 MAGs), Limnohabitans (2), Polaromonas (1), UBA954 (1), Malikia (1), Sphaerotilus hippei (1), and Rhizobacter (1). Within the Bacteroidota group (7 MAGs), four were assigned to the Crocinitomicaceae family [UBA952 (2), CAIUKX01 (1), and an unidentified genus] and three were assigned to the Chitinophagaceae family [Sediminibacterium (2) and Lacibacter (1)]. Additionally, two MAGs were classified under the archaeal phylum Nanoarchaeota, with one assigned to the GW2011-AR6 family and the other one assigned to the UBA73 family.
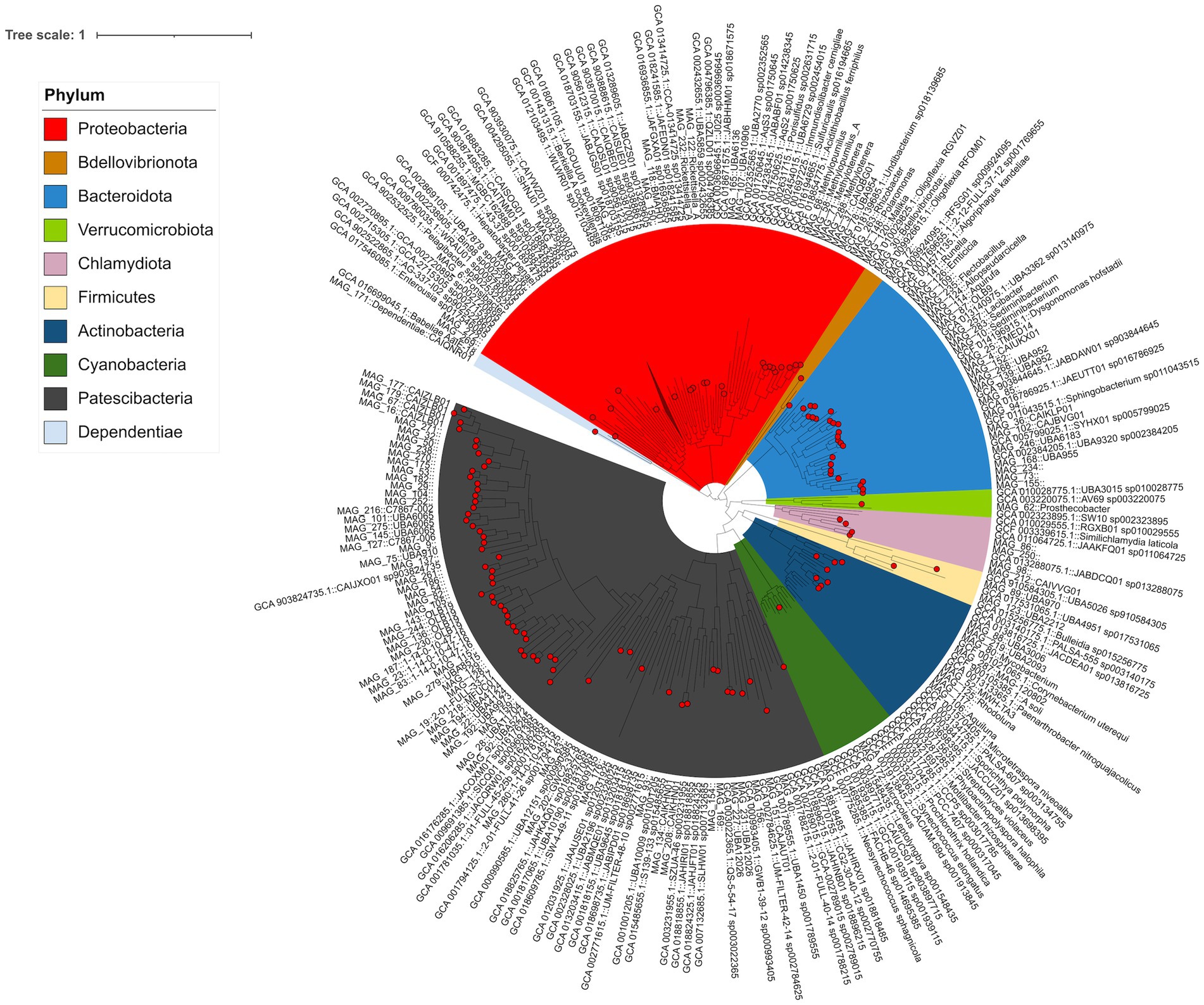
Figure 3. Bacterial phylogenetic tree reconstructed using a set of 172 single-copy genes, with MAG IDs including taxonomic assignments using GTDB-tk.
The LME models were used to assess how ditch maintenance influenced the diversity and compositional structure of the MAG community. The models found no significant difference in the Shannon–Weiner-based True Diversity (ShanTD) index between managed and unmanaged ditch systems in 2017 (pre-intervention), 2018 (post-brushing), and 2019 (post-dredging) (Figure 4A; Padj > 0.05 for each comparison). However, by 2021 (third-year post-dredging), the ShanTD index significantly decreased in managed ditch sites compared to their unmanaged counterparts (Figure 4A; Padj = 0.01), indicating a management-related decline in microbial diversity. Additionally, we observed a progressive decline in the ShanTD index within managed ditch sites from 2017 to 2019, suggesting a cumulative impact of maintenance activities on microbial diversity. In contrast, the ShanTD index for the unmanaged ditch sites expectedly remained relatively unchanged during the same period. When exploring the interactive effects of ditch management practices and seasonal variations, our LME models suggested that seasonality did not significantly influence the effects of the management practices regarding the MAG community (Supplementary Table S3).
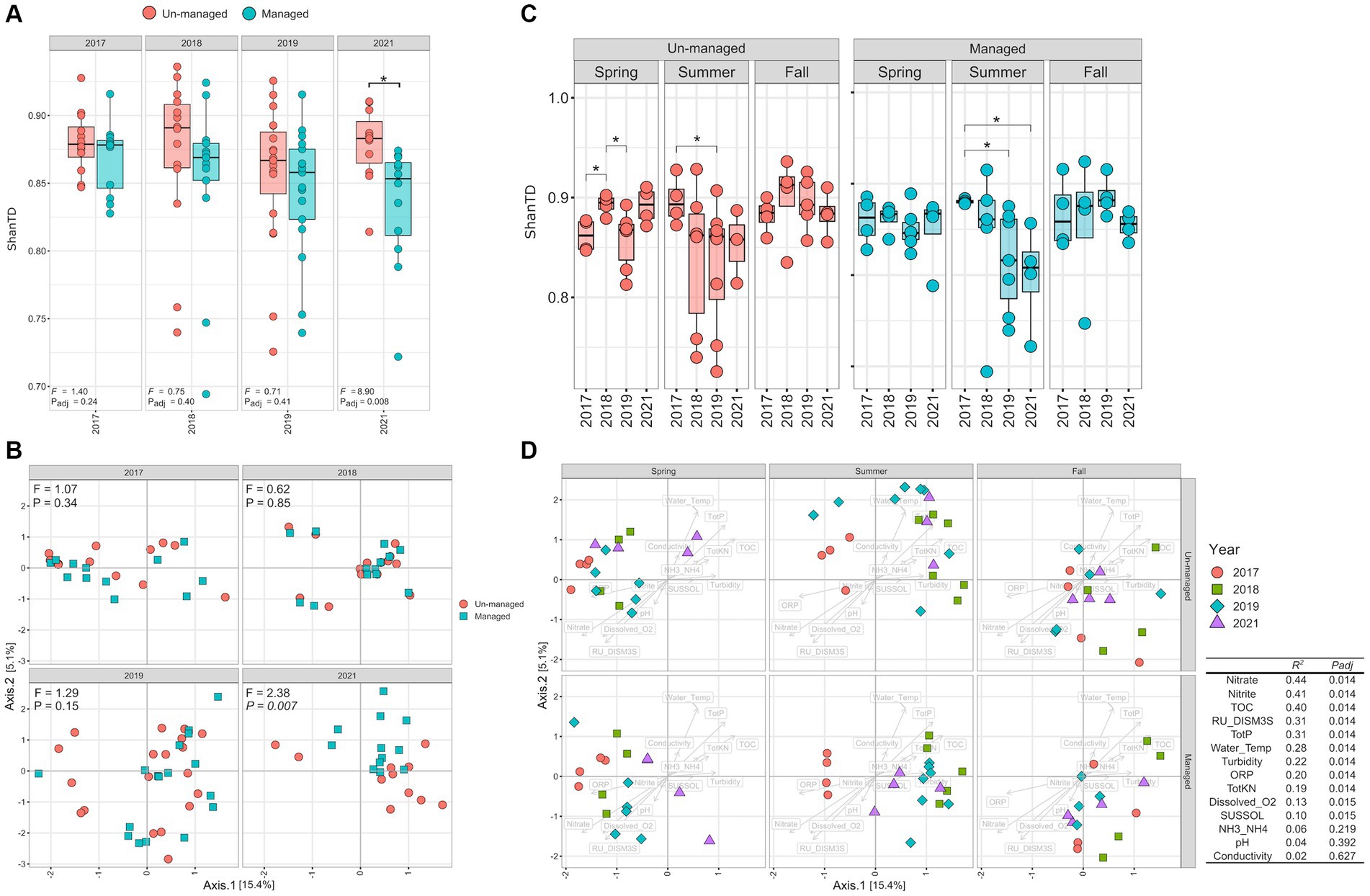
Figure 4. MAG community diversity. (A) Shannon–Weiner true diversity (ShanTD) comparison between managed and unmanaged sites over sampling years. (B) Community compositional shifts depicted by dPCoA plots. (C) Seasonal ShanTD comparison within managed and unmanaged ditches. (D) Seasonal shifts in community compositional structure within managed and unmanaged ditches. The table displays how water physicochemical properties influence the microbial community’s compositional structure, as determined by the envfit analysis provided in the vegan R package.
Double Principal Coordinate Analysis (dPCoA) demonstrated that ditch management accounted for over 60% of the variance in the compositional structure of the MAG community between managed and unmanaged sites (Figure 4B). PERMANOVA analysis revealed significant shifts in the MAG community composition within managed ditches at different stages: pre-intervention, post-brushing, and following dredging (R2 = 0.21, Padj < 0.001). Additionally, a marked divergence in the compositional structure of MAG communities between managed and unmanaged ditches was observed after dredging in 2019, with the differences becoming significantly pronounced by 2021 (Figure 4B; F = 2.38, Padj = 0.007). These findings imply that the influence of management practices on MAG community structure evolved gradually over time, rather than occurring as an immediate shift.
We further investigated the impact of seasonal variations on the alpha-diversity of the MAG community following ditch management interventions (Figure 4C). Alpha-diversity, which is a measure of species diversity within a community, is indicative of the potential functional versatility of the ecosystem. Overall, alpha diversity, quantified by the ShanTD, showed significant seasonal fluctuations, with the summer season exhibiting the most considerable diversity changes compared to spring and fall across both managed and unmanaged ditch sites (Figure 4C). Notably, there was a consistent decrease in the ShanTD during the summer months from 2018 to 2021 at all sites. This downward trend was more marked in the managed watershed, especially after dredging (2019 and 2021, Padj ≤ 0.05), than during the pre-intervention (2017) and post-brushing (2018) phases (Figure 4C). Although brushing appeared to marginally decrease MAG community diversity at managed sites, this reduction was not statistically significant. These observations suggest that dredging had a more substantial impact on the diversity of the ditch microbiome than brushing, particularly under drier conditions prevalent in the summer where the contrasting ditch substrates would be more exposed to flow-rain event-based interactions. This suggests that the diversity of the microbiome, and thereby its functional capacity, may be more sensitive to ditch management practices during periods of lower water availability, such as the summer season.
Similarly, PERMANOVA analysis revealed more pronounced changes in the MAG community’s compositional structure were observed during the summer season (Padj < 0.001) at both managed and unmanaged ditch sites. However, it is important to note that at the unmanaged site, summer samples from 2018, 2019, and 2021 did not form distinct clustering based on dPCoA analysis. In contrast, samples from the managed watershed from summers of the following intervention years—2018 (post-brushing; R2 = 0.17, Padj = 0.054), 2019 (post-dredging; R2 = 0.18, Padj = 0.006), and 2021 (3 years post-maintenance; R2 = 0.26, Padj = 0.006)—showed distinct clustering, setting them apart from the pre-maintenance summers of 2017. The differences in MAG community compositional structure during summer between the unmanaged and managed ditches may reflect the impact of reduced connectivity between agricultural inputs, different substrates, and in-stream processes under drier conditions. Although speculative, the absence of notable seasonal variations in microbiome community structure during spring and fall, observed at both managed and unmanaged sites, may be attributed to the overriding influence of tile drainage flow and inputs from adjacent agricultural areas (Figure 4D).
We further explored the influence of water physicochemical properties on the MAG community’s heterogeneity as represented by dPCoA plots (Figure 4D). Our analysis revealed that several water physicochemical parameters had a moderate influence (0 < R2 < 0.5) on community compositional structure. These parameters included total organic carbon (TOC, R2 = 0.37, Padj < 0.001), nitrate (R2 = 0.32, Padj < 0.001), dissolved oxygen (R2 = 0.29, Padj < 0.001), water temperature (R2 = 0.24, Padj < 0.001), total phosphorus (TotP: R2 = 0.21, Padj < 0.001), total Kjeldahl nitrogen (TotKN: R2 = 0.10, Padj = 0.008), ORP (R2 = 0.11, Padj = 0.02), and conductivity (R2 = 0.072, Padj = 0.04). Seasonal patterns also emerged, linking specific water physicochemical properties to the sampling seasons across both unmanaged and managed ditches. Specifically, higher relative levels of nitrate, dissolved oxygen, and ORP were associated with spring sampling conditions. In contrast, water temperature, total organic carbon, total phosphorus, and total Kjeldahl nitrogen were relatively higher during the summer sampling periods. Additionally, higher conductivity was predominantly observed during fall sampling periods.
Overall, the abundance of genes affiliated with N cycling, based on KEGG annotations, exhibited an upward trend in their abundance within the unmanaged ditch from 2017 to 2019, stabilizing at elevated levels in 2021. Conversely, at the managed ditch, the abundance of N-cycling genes decreased following the dredging (from 2019 onward) (Supplementary Figure S3; Padj < =0.05). Similarly, the abundance of genes related to other functional categories, such as ATP synthesis, as well as carbon, methane, and sulfur metabolism pathways, also declined following dredging. This reduction persisted from 2019 to 2021 compared with the baseline levels observed in 2017 and with the parallel data from unmanaged ditches in the same timeframe. However, these downward trends did not reach statistical significance (p ≥ 0.05) (Supplementary Figure S3). In contrast, genes involved in photosynthesis demonstrated a significant increase in the abundance at managed ditch after dredging in 2019 and 2021, relative to the pre-intervention data from 2017 (Supplementary Figure S3; Padj = 0.01) but noted a decrease by 2021.
We employed Linear Discriminant Analysis Effect Size (LEfSe) analysis to identify MAGs and N-cycling genes that were overrepresented (LDA score > 2.0) in the managed ditch following brushing and/or dredging. Notably, no MAGs were overrepresented at the unmanaged ditch at significant levels across the years. However, LEfSe analysis revealed distinct patterns in the managed ditch. In 2017, before any ditch interventions, Saccharimonadia (a genus within the Candidatus Saccharibacteria phylum, previously known as TM7) was significantly overrepresented (Figure 5A). The water sampling season in 2018, which followed brushing and preceded dredging, observed a predominance of Actinomycetia. For the subsequent water sampling year in 2019, marked by both post-brushing and post-dredging, we observed a significant overrepresentation of Bacteroidota spp., Emticicia spp., Polaromonas spp., and OLB19 (affiliated with Candidatus Paceibacteria). By 2021, members of the Simkaniaceae family within the Chlamydiota phylum were notably more prevalent in managed ditch sites.
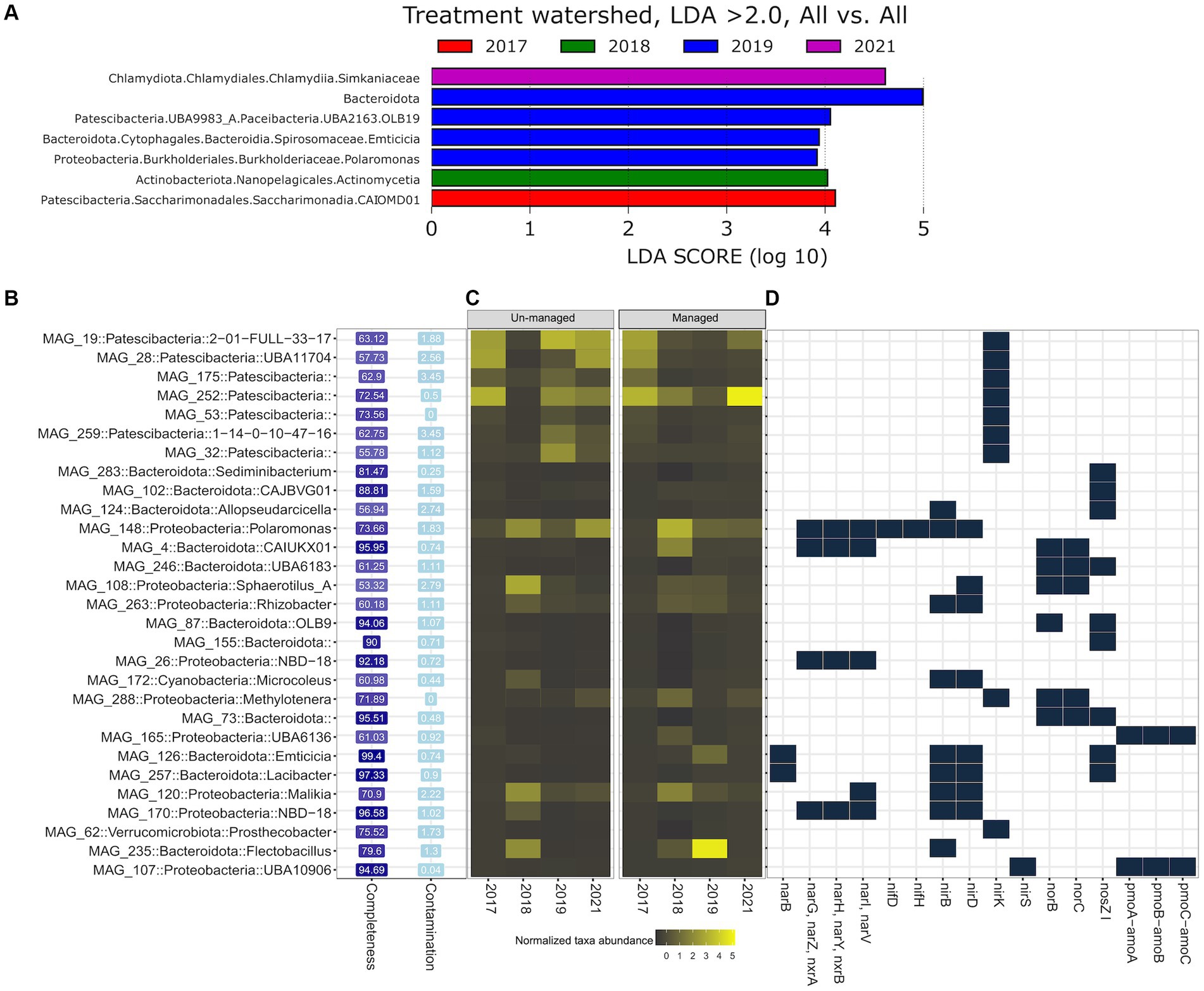
Figure 5. Differential analysis of MAGs and N-cycling genes between managed and unmanaged ditch sites. (A) Taxa overrepresented in managed sites according to LEfSe analysis, with no taxa overrepresented in unmanaged sites. (B) MAGs containing at least one N metabolic gene. (C) Relative abundance of MAGs in managed and unmanaged ditch sites across sampling years. (D) Presence and absence of N-cycling genes within each MAG.
From the 157 MAGs functionally annotated against the KEGG orthology to predict genes associated with N metabolism pathways (KEGG pathway: map00910), we identified 29 MAGs harboring one or more N-cycling genes. These 29 MAGs spanned five bacterial phyla, namely, Bacteroidota (n = 11, relative abundance = 11%), Patescibacteria (n = 7, 23%), Proteobacteria (n = 9, 53%), Cyanobacteria (n = 1, 10%), and Verrucomicrobiota (n = 1, 2%) (Figure 5B). LEfSe analysis indicated that some MAGs belonging to Flectobacillus (in Bacteroidota), Emticicia, Polaromonas, and Patescibacteria, which contained N-cycling genes, were significantly more abundant in the managed ditch in 2019, following dredging activities. For example, MAG 235 (completeness = 79.60%, contamination = 1.30%) was classified within the Flectobacillus genus and found to carry a singular N-cycling gene, nirB, which codes for the large subunit of nitrite reductase (NADH) involved in the assimilatory nitrate reduction pathway. In the genomes of all MAGs classified under Patescibacteria, only one N functional gene, nirK, associated with denitrification, was identified. In contrast, MAG 126 (completeness = 99.40%, contamination = 0.74%), belonging to the Emticicia genus, exhibited a diverse array of denitrification and assimilatory nitrate reductase genes, including narB, nirB/nirD, and nosZI (Figure 5D). Additionally, MAG 148 (completeness = 73.66%, contamination = 1.83%), identified as a member of the Polaromonas genus, harbored genes associated with various N-cycling processes, such as nitrogen fixation (nifD/nifH), denitrification (nirB/nirD/narG/narH/narI), and DNRA (nirB).
To assess the impact of ditch management on the distribution of the N-cycling pathway and genes, we normalized the abundance of each N-cycling gene and corresponding KEGG modules over time for comparative analysis. Across both the managed and unmanaged ditches, there was an upward trend in the abundance of N-cycling pathways and genes from 2017 to 2019, which then diminished in 2021 (Figures 6, 7). This trend may reflect the general influences of annual-seasonal variations in agricultural practices and environmental conditions, and not just ditch management interventions, as evidenced by the parallel patterns in both watersheds. To further investigate these observations, we carried out LME modeling, using sampling year and watershed block as random effects and ditch management as a fixed effect. The LME model revealed that in 2017, prior to ditch management interventions, the abundance of most N-cycling pathways was comparable between the two drainage ditches. However, there was a notable exception for the denitrification gene nosZI, which was significantly more abundant in unmanaged sites compared to the managed sites (Figure 7; F = 4.00, Padj = 0.05). In 2018, following the brushing intervention, no notable changes in N-cycling pathways or gene abundance were detected. However, in 2019, after dredging activities, a significant decrease in the denitrification pathway was observed within the managed watershed (Figure 6; F = 4.90, Padj = 0.04). While not statistically significant, a general decline in the abundance of most denitrification genes was also observed in the managed ditch relative to the unmanaged reference ditch. By 2021, after 3 years of the ditch management interventions, the denitrification pathway (Figure 6; F = 7.10, Padj = 0.02) and associated genes narG/narH/narI (Figure 7; F = 6.80, Padj = 0.02), as well as norB and norC (Figure 7; F = 8.00, Padj = 0.01), were found to be more abundant in the unmanaged ditch. Other pathways, such as complete ammonia oxidation (commamox) (Figure 6; F = 9.20, Padj = 0.01) and dissimilatory nitrate reduction to ammonium (Figure 6; F = 12.00, Padj = 0.003), also exhibited reduced abundance in the managed ditch compared to the unmanaged ditch. Furthermore, N fixation genes, nifD/nifH, were significantly less abundant in the managed ditch than in the unmanaged ditch in 2021, 3 years after dredging activities (Figure 7; F = 10.00, Padj = 0.01).
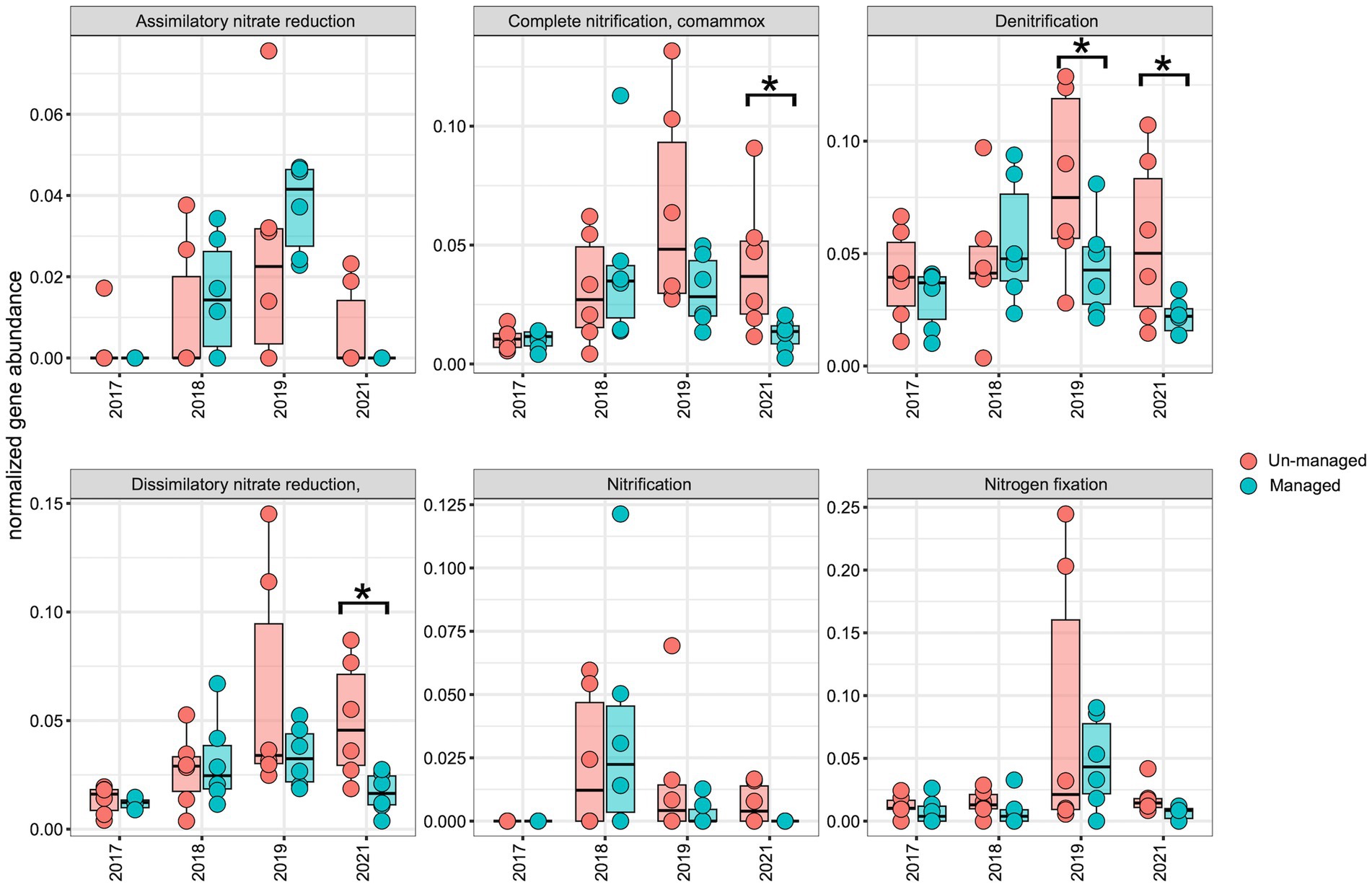
Figure 6. Boxplots representing the abundance of N-cycling pathways in managed and unmanaged ditch sites over the sampling years. An asterisk (*) marks significant differences (p ≤ 0.05) as determined by linear mixed-effects analysis.
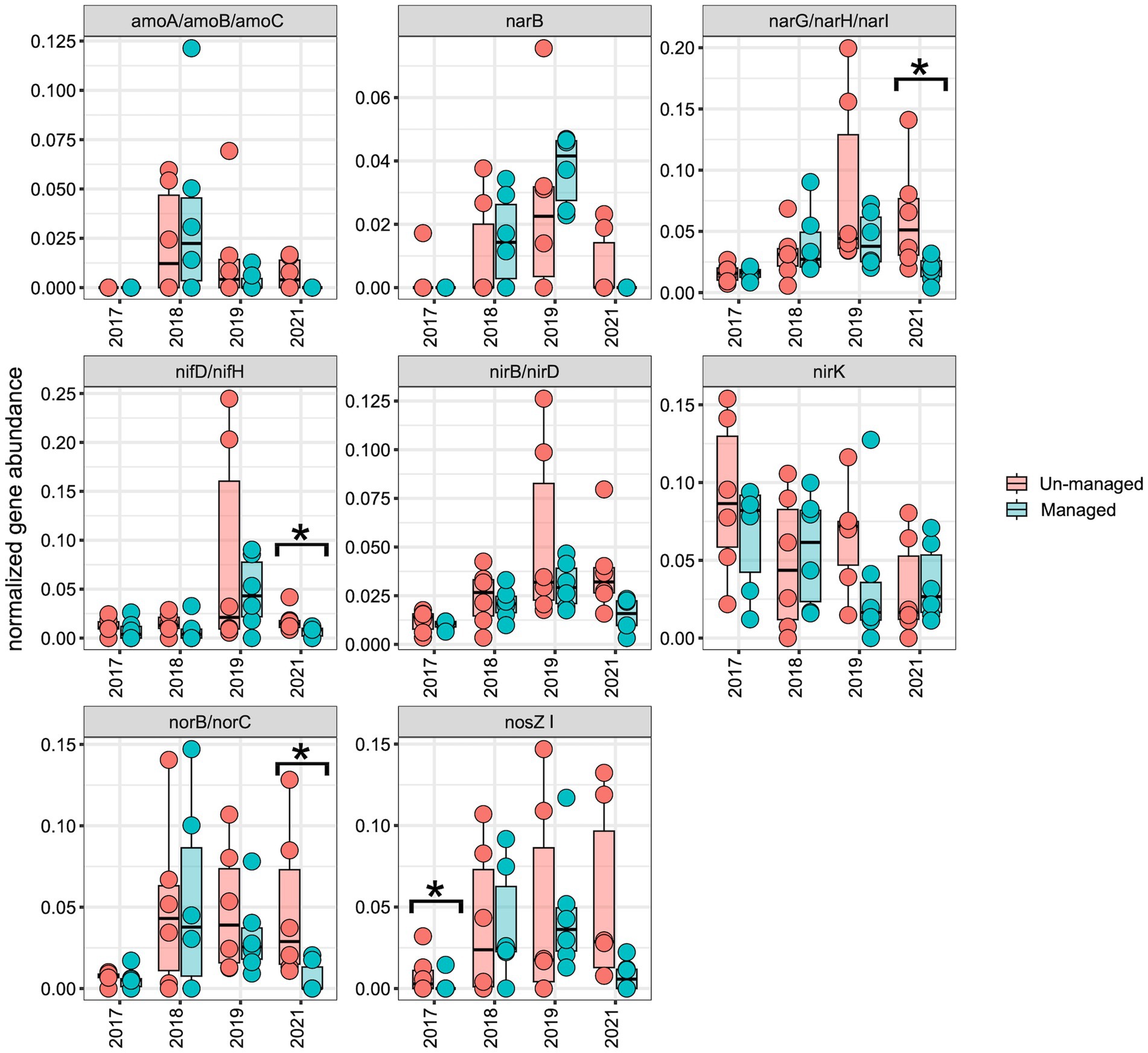
Figure 7. Boxplots representing the abundance of N-cycling genes. An asterisk (*) highlights significant differences (p ≤ 0.05) between managed and unmanaged ditches, as determined by linear mixed-effects analysis. Notably, denitrification genes (narG, narH, narI, norB, and norC) and nitrogen fixation genes (nifD, nifH) were significantly less abundant in managed ditch sites than in unmanaged ditch sites 3 years after interventions.
Agricultural drainage ditches receive drainage and runoff from nearby farmlands, which act as primary receptors of agrochemicals and organic fertilizer pollutants from agricultural activities (Wilkes et al., 2014; Sunohara et al., 2015; Damphousse et al., 2024). These ditches harbor and receive microorganisms essential for the assimilation, dissipation, and transformation of such chemical and organic pollutants (Dollinger et al., 2015). Drainage ditch management practices, such as dredging and vegetation clearing, can significantly influence these processes by altering factors such as sunlight exposure, sediment/soil substrate type and exposure, temperatures of water, and water flow dynamics. Given their indispensable role in facilitating agricultural drainage in the areas they serve, it is essential to optimize ditch management strategies to help preserve and enhance the ecosystem services these ditches offer (Collins et al., 2019; Rideout et al., 2022; Damphousse et al., 2024). The primary objective of this study is to assess the impact of agricultural drainage ditch management practices, specifically brushing and dredging, on the freshwater microbial community’s function in N cycling, which plays a critical role in processes such as eutrophication and production of greenhouse gasses (Schindler, 2006; Thompson et al., 2019).
We employed a shotgun metagenomics approach to obtain a comprehensive genomic snapshot of the stream’s microbial community under different states of ditch management (Carraro et al., 2018). This method enables the assessment of both the taxonomic composition and the potential functional capabilities of the stream microbiome, encompassing dead, inactive, or live microorganisms. It is important to note that while shotgun metagenomics provides a broad overview of the biotic disposition of the system, metatranscriptomics (high-throughput sequencing of RNAs of mixed specimens) can offer higher fidelity insights into the active functional processes of the microbiome at the time of sampling, which was not the initial intent of this watershed-scale study. Moreover, RNA levels can change rapidly (Xu and Asakawa, 2021), indicating that metatranscriptomic profiles might represent a highly transient state that was not fully representative of the more broader generalized condition of the watershed system. Although using both shotgun metagenomics and metatranscriptomics approaches would provide a more comprehensive understanding of a microbiome, the combined cost can be prohibitive. Therefore, the shotgun metagenomics approach was chosen for the objectives of our study.
Brushing and dredging interventions in this study were performed in a purposefully aggressive manner to represent a “worse-case management scenario,” allowing comparison of the aquatic microbiomes associated with distinctly contrasting management conditions. Our findings suggest that within the first few years following drainage ditch brushing and dredging, when the managed and unmanaged ditch conditions were at their most contrasting biophysical state in terms of vegetation and ditch channel morphology and condition, the freshwater microbial communities in the managed ditch experienced significant changes. The process of sediment dredging not only directly removes channel vegetation and organically enriched sediments but also exposes other sediment substrates in the dredged area, in this case, tight varved clays. Cui et al. (2020) showed that vegetated agricultural drainage ditches foster habitats more conducive to wildlife, thus enhancing bacterial abundance and diversity. Additionally, root exudates, including those from hydrophilic plants in the ditch proper, can promote the growth of heterotrophic denitrifying bacteria in constructed wetland (Wu et al., 2017). Consequently, the removal of vegetation and sediment through dredging can trigger a shift in the local microbial ecosystem, at least within the first few years following the interventions.
Consistent with previous studies (Wu et al., 2017; Cui et al., 2020), this study documented a notable decrease in the MAG community’s overall diversity after the intervention in the managed ditch sites (Figure 4), especially during the summer growing seasons (Figures 4C,D). Shi et al. (2023) made similar observations, noting greater variability in the microbiomes of agricultural drainage ditches during summer, in contrast to the aquatic microbiome derived from the samples of larger water courses. Notably, the re-establishment of vegetation in the agricultural ditches after interventions may take years, and our study indicates that 3 years post-intervention, the microbial community did not recover to pre-intervention diversity levels, despite the introduction of microorganisms through the runoff and tile drainage processes such as post-ditch management (Milsom et al., 2004; Teurlincx et al., 2018). This suggests that the microbial ecosystems in such environments are complex processes that might require significant time to (re)stabilize and/or reach pre-management conditions.
Ditch vegetation can foster a diverse microbiome capable of converting N derivatives into forms that are more readily absorbed by plants in ditch ecosystems (Hunting et al., 2017; Kumwimba et al., 2018; Vymazal and Březinová, 2018; Cui et al., 2020). Specifically, Zhang et al. (2020) investigated three pilot-scale simulated drainage ditches planted with Myriophyllum aquaticum. The authors found that these vegetated ditches achieved 76–89% ammonium removal efficiency at low temperatures, with microbial processes accounting for 39–55% of this removal, according to mass balance calculations. They further identified nitrification and denitrification as the primary nitrogen removal mechanisms under these conditions. It is reasonable to expect the removal of sediment and the rhizosphere of aquatic plants, leading to a decline in microorganisms involved in N cycling. Indeed, in this study, we noted a marked decrease in the abundance of N-cycling genes following dredging activities. Specifically, 3 years post-dredging, the detected levels of genes associated with complete ammonia oxidation (comammox), dissimilatory nitrate reduction to ammonium, and denitrification were significantly reduced at our study sites (Figures 6, 7). This decrease in N-cycling bacteria and genes could signify a diminished ability to process nitrate and ammonia, possibly leading to increased nitrogen loading downstream. The duration of this reduction in N-cycling capability remains uncertain. However, the extreme conditions of ditch management investigated in this study provide a unique perspective on microbial dynamics. As the ditches gradually recover, in terms of both substrate and vegetation, we anticipate a resurgence of N-cycling microorganisms, which will enhance the capacity of the ecosystem for nitrogen processing.
Following the dredging intervention in the managed sites, Emticicia, Flectobacillus, Polaromonas, and Simkaniaceae significantly increased in abundance compared to that of pre-interventions (Figure 5A). Emticicia and Flectobacillus, both from the Spirosomaceae family, have distinct ecological niches and functions. Emticicia has been isolated from freshwater environments, facilitating microalgal growth and possessing denitrifying genes, suggesting its role in N-cycling (Xie et al., 2016; Ngo et al., 2017; Toyama et al., 2019). Flectobacillus, though primarily associated with Antarctic marine settings, is also present in freshwater ecosystems, indicating its adaptability (McGuire et al., 1987; Šimek et al., 2007). It is not clear why Simkaniaceae sp. increased in abundance 3 years post-dredging (Figure 5). The members of Simkaniaceae can be intracellular parasites of whiteflies, fish, and unicellular eukaryotes (Everett et al., 1999, 2005; Nylund et al., 2015; Burnard et al., 2017; Lienard et al., 2017). Polaromonas, which is prevalent in cold environments such as Arctic waters, was found to increase in abundance in the managed ditch post-dredging. The MAGs associated with Polaromonas were predicted to contain genes crucial for nitrogen processing, including nitrate reductase and nitrite oxidoreductase (Figure 5D), highlighting their potential role in mitigating nitrogen pollution.
Furthermore, we noted a significant shift in the microbial population, which was highlighted by an increased abundance of Patescibacteria (Figure 5D). After finding in diverse aquatic settings, either as free-living organisms or episymbionts alongside Nitrospirota, this phylum plays a versatile role in aquatic ecosystems (Beam et al., 2020; Tian et al., 2020; Chaudhari et al., 2021). Interestingly, all MAGs attributed to Patescibacteria exclusively carried the nirK gene, encoding the nitrite reductase that transforms nitrite into nitrous oxide, suggesting a simplified lifestyle adapted to their environmental niche (Herrmann et al., 2019; Tian et al., 2020). In this study, the abundant recovery of Patescibacteria MAGs indicates their potentially significant role in the ecosystem. Further insights come from Williams et al. (2021) and Seymour et al. (2023), who identified that novel Patescibacteria members, including those from the Cloacimonadota and Omnitrophota clades, are characterized by obligate fermentative heterotrophs and specialized metabolic pathways. These findings indicate complex metabolic interactions within the microbial community, particularly with hydrogen-consuming bacteria, that facilitate their survival. In our context, the Patescibacteria phylum emerges as a foundational element for the microbial community’s succession toward stability or another state condition, potentially providing key metabolites to partners such as Nitrospirota (Chaudhari et al., 2021). The observed proliferation of Patescibacteria, alongside genera such as Emticicia, Flectobacillus, and Polaromonas, suggests the capacity of the ecosystem to process nitrogen compounds, including nitrate, nitrite, and ammonia/ammonium (Figure 5A). This synergistic effect might explain the changes we documented, positioning these microbial communities as pivotal in the influence of N loading in water and gaseous N emissions. However, these speculations would demand further exploration, including nitrogen dynamics studies and culture-based experiments.
In this study, we investigated the influences of agricultural drainage ditch management practices, bank brushing, and channel dredging on the microbiome and N-cycling genes within two paired, small, agricultural watersheds, employing a metagenome-assembled genome approach.
Our study found that brushing alone had a minimal effect on the microbial community structure and function within the ditch water. However, dredging, particularly when considered in conjunction with prior brushing, had a significant impact, altering both the microbial community composition and the abundance of N-cycling genes. Notably, post-dredging, there was an increased presence of specific bacterial taxa, such as Polaromonas, Emticicia, Flectobacillus, and Patescibacteria, in the managed ditch sites. These changes also extended to the genes involved in N cycling within the microbiome. The implications of these shifts with respect to N loading or concentrations in the water and gaseous N effluxes are currently uncertain.
In summary, while in some circumstances, the management of agricultural drainage ditches is essential for optimizing crop productivity by improving field drainage efficiency, our study revealed that such interventions can significantly alter ditch microbial communities that are responsible for processing and assimilation of N within these receptors of field agricultural drainage and runoff. Nevertheless, the long-term consequences of these changes could not be fully elucidated in this heuristic study; however, this study provided some indications of transitional microbiome shifts that could variably influence the fate and transport of N species agro-ecosystems where drainage ditches are managed.
The datasets presented in this study can be found in online repositories. The raw paired-end shotgun metagenomics sequencing data have been deposited in the Sequence Read Archive (SRA), under the Bioproject accession PRJNA1029945, Biosample accessions SAMN38026186 – SAMN38026302.
GG: Data curation, Formal analysis, Investigation, Software, Visualization, Writing – original draft, Writing – review & editing. DL: Conceptualization, Funding acquisition, Investigation, Methodology, Resources, Writing – review & editing. IK: Conceptualization, Funding acquisition, Investigation, Methodology, Project administration, Resources, Supervision, Writing – review & editing. MS: Investigation, Methodology, Writing – review & editing. EC: Investigation, Methodology, Writing – review & editing. WC: Conceptualization, Data curation, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Writing – review & editing.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This study was financially supported by the Agriculture and Agri-Food Canada (AAFC)-funded A-base project J-002305 (Environmental Change Onehealth Observatory, ECO2) and the Government of Canada’s Genomics Research and Development Initiative (GRDI) Shared Priority Project J-001263 titled “Metagenomics Based Ecosystem Biomonitoring (Ecobiomics).”
We thank Michel Cloutier (retired), Rafik Assabgui, and co-op students from Agriculture and Agri-Food Canada for collecting and processing environmental samples. We also thank Charles Greer, Sylvie Sanschagrin, and David Konkin from the National Research Council for their help in preparing samples for high-throughput sequencing.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fsufs.2024.1329422/full#supplementary-material
Almaraz, M., Groffman, P. M., Silver, W. L., Hall, S. J., Lin, Y., O’Connell, C., et al. (2023). Dinitrogen emissions dominate nitrogen gas emissions from soils with low oxygen availability in a moist tropical Forest. J. Geophys. Res. Biogeo. 128:e2022JG007210. doi: 10.1029/2022JG007210
Alneberg, J., Bjarnason, B. S., de Bruijn, I., Schirmer, M., Quick, J., Ijaz, U. Z., et al. (2014). Binning metagenomic contigs by coverage and composition. Nat. Methods 11, 1144–1146. doi: 10.1038/nmeth.3103
Beam, J. P., Becraft, E. D., Brown, J. M., Schulz, F., Jarett, J. K., Bezuidt, O., et al. (2020). Ancestral absence of Electron transport chains in Patescibacteria and DPANN. Front. Microbiol. 11:1848. doi: 10.3389/fmicb.2020.01848
Bennett, E. M., Baird, J., Baulch, H., Chaplin-Kramer, R., Fraser, E., Loring, P., et al. (2021). “Chapter one - ecosystem services and the resilience of agricultural landscapes” in Advances in ecological research. eds. D. A. Bohan and A. J. Vanbergen (Academic Press), 1–43.
Blume, O., Guitard, E., Crann, C., Orekhov, M., Amos, R., Clark, I., et al. (2022). Relationships between carbon age and CO2 efflux in agricultural and drainage ditch soils using the thermonuclear bomb pulse. Vadose Zone J. 21:e20208. doi: 10.1002/vzj2.20208
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Bowers, R. M., Kyrpides, N. C., Stepanauskas, R., Harmon-Smith, M., Doud, D., Reddy, T. B. K., et al. (2017). Minimum information about a single amplified genome (MISAG) and a metagenome-assembled genome (MIMAG) of bacteria and archaea. Nat. Biotechnol. 35, 725–731. doi: 10.1038/nbt.3893
Bretz, F., Hothorn, T., and Westfall, P., (2010). Multiple comparisons using R. New York: Chapman and Hall/CRC.
Burnard, D., Weaver, H., Gillett, A., Loader, J., Flanagan, C., and Polkinghorne, A. (2017). Novel Chlamydiales genotypes identified in ticks from Australian wildlife. Parasit. Vectors 10:46. doi: 10.1186/s13071-017-1994-y
Capella-Gutiérrez, S., Silla-Martínez, J. M., and Gabaldón, T. (2009). trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25, 1972–1973. doi: 10.1093/bioinformatics/btp348
Carraro, L., Hartikainen, H., Jokela, J., Bertuzzo, E., and Rinaldo, A. (2018). Estimating species distribution and abundance in river networks using environmental DNA. Proc. Natl. Acad. Sci. 115, 11724–11729. doi: 10.1073/pnas.1813843115
Chaudhari, N. M., Overholt, W. A., Figueroa-Gonzalez, P. A., Taubert, M., Bornemann, T. L. V., Probst, A. J., et al. (2021). The economical lifestyle of CPR bacteria in groundwater allows little preference for environmental drivers. Environ. Microbiome 16:24. doi: 10.1186/s40793-021-00395-w
Chaumeil, P.-A., Mussig, A. J., Hugenholtz, P., and Parks, D. H. (2020). GTDB-Tk: a toolkit to classify genomes with the genome taxonomy database. Bioinformatics 36, 1925–1927. doi: 10.1093/bioinformatics/btz848
Chen, W., Wilkes, G., Khan, I. U., Pintar, K. D., Thomas, J. L., Lévesque, C. A., et al. (2018). Aquatic bacterial communities associated with land use and environmental factors in agricultural landscapes using a metabarcoding approach. Front. Microbiol. 9:2301. doi: 10.3389/fmicb.2018.02301
Cital, F., Ramírez-Hernández, J., García-Hernández, J., García-Leyva, G., Rodríguez-Burgueño, J. E., and Ramírez-Barreto, M. E. (2022). Ecosystem services (ES) provided by ditches in a desert agricultural valley. Ecol. Eng. 174:106462. doi: 10.1016/j.ecoleng.2021.106462
Collins, S. J., Bellingham, L., Mitchell, G. W., and Fahrig, L. (2019). Life in the slow drain: landscape structure affects farm ditch water quality. Sci. Total Environ. 656, 1157–1167. doi: 10.1016/j.scitotenv.2018.11.400
Cui, N., Zhang, X., Cai, M., Zhou, L., Chen, G., and Zou, G. (2020). Roles of vegetation in nutrient removal and structuring microbial communities in different types of agricultural drainage ditches for treating farmland runoff. Ecol. Eng. 155:105941. doi: 10.1016/j.ecoleng.2020.105941
Cummings, D. I., Gorrell, G., Guilbault, J.-P., Hunter, J. A., Logan, C., Ponomarenko, D., et al. (2011). Sequence stratigraphy of a glaciated basin fill, with a focus on esker sedimentation. GSA Bull. 123, 1478–1496. doi: 10.1130/B30273.1
Damphousse, L., Van Goethem, K., Carroll, E., Stammler, K., and Febria, C. (2024). Ecological impacts of management practices in agricultural drain networks: a literature synthesis. Can. Water Resour. J. 0, 1–26. doi: 10.1080/07011784.2023.2295330
Dandie, C. E., Wertz, S., Leclair, C. L., Goyer, C., Burton, D. L., Patten, C. L., et al. (2011). Abundance, diversity and functional gene expression of denitrifier communities in adjacent riparian and agricultural zones. FEMS Microbiol. Ecol. 77, 69–82. doi: 10.1111/j.1574-6941.2011.01084.x
Dinnes, D. L., Karlen, D. L., Jaynes, D. B., Kaspar, T. C., Hatfield, J. L., Colvin, T. S., et al. (2002). Nitrogen management strategies to reduce nitrate leaching in tile-drained Midwestern soils. Agron. J. 94, 153–171. doi: 10.2134/agronj2002.1530
Dollinger, J., Dagès, C., Bailly, J.-S., Lagacherie, P., and Voltz, M. (2015). Managing ditches for agroecological engineering of landscape. A review. Agron. Sustain. Dev. 35, 999–1020. doi: 10.1007/s13593-015-0301-6
Domangue, R. J., and Mortazavi, B. (2018). Nitrate reduction pathways in the presence of excess nitrogen in a shallow eutrophic estuary. Environ. Pollut. 238, 599–606. doi: 10.1016/j.envpol.2018.03.033
Dyck, A., Robinson, S. A., Young, S. D., Renaud, J. B., Sabourin, L., Lapen, D. R., et al. (2021). The effects of ditch Management in Agroecosystems on embryonic and tadpole survival, growth, and development of northern leopard frogs (Lithobates pipiens). Arch. Environ. Contam. Toxicol. 81, 107–122. doi: 10.1007/s00244-021-00836-0
Eddy, S. R. (2011). Accelerated profile HMM searches. PLoS Comput. Biol. 7:e1002195. doi: 10.1371/journal.pcbi.1002195
Edgar, R. C. (2022). Muscle5: High-accuracy alignment ensembles enable unbiased assessments of sequence homology and phylogeny. Nat Commun. 13, 6968. doi: 10.1038/s41467-022-34630-w
Eren, A. M., Kiefl, E., Shaiber, A., Veseli, I., Miller, S. E., Schechter, M. S., et al. (2021). Community-led, integrated, reproducible multi-omics with anvi’o. Nat. Microbiol. 6, 3–6. doi: 10.1038/s41564-020-00834-3
Everett, K. D., Bush, R. M., and Andersen, A. A. (1999). Emended description of the order Chlamydiales, proposal of Parachlamydiaceae fam. Nov. and Simkaniaceae fam. Nov., each containing one monotypic genus, revised taxonomy of the family Chlamydiaceae, including a new genus and five new species, and standards for the identification of organisms. Int. J. Syst. Bacteriol. 49, 415–440. doi: 10.1099/00207713-49-2-415
Everett, K. D. E., Thao, M., Horn, M., Dyszynski, G. E., and Baumann, P. (2005). Novel chlamydiae in whiteflies and scale insects: endosymbionts “Candidatus Fritschea bemisiae” strain Falk and “Candidatus Fritschea eriococci” strain elm. Int. J. Syst. Evol. Microbiol. 55, 1581–1587. doi: 10.1099/ijs.0.63454-0
Grant, D. M., and Dawson, B. D. (2001). ISCO open channel flow measurement handbook. 5th. ISCO Inc., Lincoln, NE.
Groffman, P. M., Rosi, E. J., and Fulweiler, R. W. (2021). “Chapter 8 - the nitrogen cycle” in Fundamentals of ecosystem science (second edition). eds. K. C. Weathers, D. L. Strayer, and G. E. Likens (Academic Press), 161–188.
Herrmann, M., Wegner, C.-E., Taubert, M., Geesink, P., Lehmann, K., Yan, L., et al. (2019). Predominance of Cand. Patescibacteria in groundwater is caused by their preferential mobilization from soils and flourishing under oligotrophic conditions. Front. Microbiol. 10:1407. doi: 10.3389/fmicb.2019.01407
Herzon, I., and Helenius, J. (2008). Agricultural drainage ditches, their biological importance and functioning. Biol. Conserv. 141, 1171–1183. doi: 10.1016/j.biocon.2008.03.005
Hruby, C. E., Soupir, M. L., Moorman, T. B., Shelley, M., and Kanwar, R. S. (2016). Effects of tillage and poultry manure application rates on Salmonella and fecal indicator bacteria concentrations in tiles draining Des Moines lobe soils. J. Environ. Manag. 171, 60–69. doi: 10.1016/j.jenvman.2016.01.040
Hunting, E. R., Barmentlo, S. H., Schrama, M., van Bodegom, P. M., Zhai, Y., and Vijver, M. G. (2017). Agricultural constraints on microbial resource use and niche breadth in drainage ditches. PeerJ 5:e4175. doi: 10.7717/peerj.4175
Hyatt, D., Chen, G.-L., LoCascio, P. F., Land, M. L., Larimer, F. W., and Hauser, L. J. (2010). Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11:119. doi: 10.1186/1471-2105-11-119
Jain, C., Rodriguez-R, L. M., Phillippy, A. M., Konstantinidis, K. T., and Aluru, S. (2018). High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat. Commun. 9:5114. doi: 10.1038/s41467-018-07641-9
Kanehisa, M., and Goto, S. (2000). KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30. doi: 10.1093/nar/28.1.27
Kang, D. D., Li, F., Kirton, E., Thomas, A., Egan, R., An, H., et al. (2019). MetaBAT 2: an adaptive binning algorithm for robust and efficient genome reconstruction from metagenome assemblies. PeerJ 7:e7359. doi: 10.7717/peerj.7359
Kavanah, R. J., Wren, L., and Hoggarth, C. T. (2017). Guidance for maintaining and repairing municipal drains in Ontario. ON, Canada: Central and Artic Region Fisheries and Oceans Canada.
Kladivko, E. J., Brown, L. C., and Baker, J. L. (2001). Pesticide transport to subsurface tile drains in humid regions of North America. Crit. Rev. Environ. Sci. Technol. 31, 1–62. doi: 10.1080/20016491089163
Kumwimba, M. N., Meng, F., Iseyemi, O., Moore, M. T., Zhu, B., Tao, W., et al. (2018). Removal of non-point source pollutants from domestic sewage and agricultural runoff by vegetated drainage ditches (VDDs): design, mechanism, management strategies, and future directions. Sci. Total Environ. 639, 742–759. doi: 10.1016/j.scitotenv.2018.05.184
Langmead, B., and Salzberg, S. L. (2012). Fast gapped-read alignment with bowtie 2. Nat. Methods 9, 357–359. doi: 10.1038/nmeth.1923
Lee, M. D. (2019). GToTree: a user-friendly workflow for phylogenomics. Bioinformatics 35, 4162–4164. doi: 10.1093/bioinformatics/btz188
Letunic, I., and Bork, P. (2021). Interactive tree of life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 49, W293–W296. doi: 10.1093/nar/gkab301
Levavasseur, F., Biarnès, A., Bailly, J. S., and Lagacherie, P. (2014). Time-varying impacts of different management regimes on vegetation cover in agricultural ditches. Agric. Water Manag. 140, 14–19. doi: 10.1016/j.agwat.2014.03.012
Li, H., Handsaker, B., Wysoker, A., Fennell, T., Ruan, J., Homer, N., et al. (2009). The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079. doi: 10.1093/bioinformatics/btp352
Li, D., Liu, C.-M., Luo, R., Sadakane, K., and Lam, T.-W. (2015). MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics 31, 1674–1676. doi: 10.1093/bioinformatics/btv033
Lienard, J., Croxatto, A., Gervaix, A., Lévi, Y., Loret, J.-F., Posfay-Barbe, K. M., et al. (2017). Prevalence and diversity of Chlamydiales and other amoeba-resisting bacteria in domestic drinking water systems. New Microbes and New Infect. 15, 107–116. doi: 10.1016/j.nmni.2016.10.003
Lyautey, E., Lapen, D. R., Wilkes, G., McCleary, K., Pagotto, F., Tyler, K., et al. (2007). Distribution and characteristics of Listeria monocytogenes isolates from surface waters of the south Nation River watershed, Ontario, Canada. Appl. Environ. Microbiol. 73, 5401–5410. doi: 10.1128/AEM.00354-07
McGuire, A. J., Franzmann, P. D., and McMeekin, T. A. (1987). Flectobacillus glomeratus sp. nov., a curved, non-motile, pigmented bacterium isolated from Antarctic marine environments. Syst. Appl. Microbiol. 9, 265–272. doi: 10.1016/S0723-2020(87)80032-2
McMurdie, P. J., and Holmes, S. (2013). Phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 8:e61217. doi: 10.1371/journal.pone.0061217
Milsom, T. P., Sherwood, A. J., Rose, S. C., Town, S. J., and Runham, S. R. (2004). Dynamics and management of plant communities in ditches bordering arable fenland in eastern England. Agric. Ecosyst. Environ. 103, 85–99. doi: 10.1016/j.agee.2003.10.012
Murphy, A. E., Anderson, I. C., Smyth, A. R., Song, B., and Luckenbach, M. W. (2016). Microbial nitrogen processing in hard clam (Mercenaria mercenaria) aquaculture sediments: the relative importance of denitrification and dissimilatory nitrate reduction to ammonium (DNRA). Limnol. Oceanogr. 61, 1589–1604. doi: 10.1002/lno.10305
Needelman, B. A., Kleinman, P. J. A., Strock, J. S., and Allen, A. L. (2007). Drainage ditches: improved management of agricultural drainage ditches for water quality protection: An overview. J. Soil Water Conserv. 62, 171–178.
Ngo, H. T. T., Trinh, H., Yang, J.-E., Won, K.-H., Chu, D.-H., Kook, M., et al. (2017). Emticicia aquatilis sp. nov., isolated from a freshwater sample. Int. J. Syst. Evol. Microbiol. 67, 1703–1708. doi: 10.1099/ijsem.0.001840
Nylund, S., Steigen, A., Karlsbakk, E., Plarre, H., Andersen, L., Karlsen, M., et al. (2015). Characterization of ‘Candidatus Syngnamydia salmonis’ (Chlamydiales, Simkaniaceae), a bacterium associated with epitheliocystis in Atlantic salmon (Salmo salar L.). Arch. Microbiol. 197, 17–25. doi: 10.1007/s00203-014-1038-3
Oksanen, J., Blanchet, F. G., Friendly, M., Kindt, R., Legendre, P., McGlinn, D., et al. (2019). “Vegan: community ecology package (version 2.5-6)” in The comprehensive R archive network
Parks, D. H., Chuvochina, M., Rinke, C., Mussig, A. J., Chaumeil, P.-A., and Hugenholtz, P. (2021). GTDB: an ongoing census of bacterial and archaeal diversity through a phylogenetically consistent, rank normalized and complete genome-based taxonomy. Nucleic Acids Res. 50, D785–D794. doi: 10.1093/nar/gkab776
Pinheiro, J., Bates, D., DebRoy, S., and Sarkar, D.R Core Team (2018). {nlme}: Linear and Nonlinear Mixed Effects Models. Available at: https://CRAN.R-project.org/package=nlme
Price, M. N., Dehal, P. S., and Arkin, A. P. (2010). FastTree 2--approximately maximum-likelihood trees for large alignments. PLoS One 5:e9490. doi: 10.1371/journal.pone.0009490
R Core Team (2022). R: A language and environment for statistical computing. Available at: https://www.R-project.org/
Rabalais, N. N., and Turner, R. E. (2019). Gulf of Mexico hypoxia: past, present, and future. Limnol. Oceanogr. Bull. 28, 117–124. doi: 10.1002/lob.10351
Randall, G. W., Huggins, D. R., Russelle, M. P., Fuchs, D. J., Nelson, W. W., and Anderson, J. L. (1997). Nitrate losses through subsurface tile drainage in conservation reserve program, alfalfa, and row crop systems. J. Environ. Qual. 26, 1240–1247. doi: 10.2134/jeq1997.00472425002600050007x
Räty, M., Järvenranta, K., Saarijärvi, E., Koskiaho, J., and Virkajärvi, P. (2020). Losses of phosphorus, nitrogen, dissolved organic carbon and soil from a small agricultural and forested catchment in east-Central Finland. Agric. Ecosyst. Environ. 302:107075. doi: 10.1016/j.agee.2020.107075
Rich, J. J., and Myrold, D. D. (2004). Community composition and activities of denitrifying bacteria from adjacent agricultural soil, riparian soil, and creek sediment in Oregon, USA. Soil Biol. Biochem. 36, 1431–1441. doi: 10.1016/j.soilbio.2004.03.008
Rideout, N. K., Lapen, D. R., Peters, D. L., and Baird, D. J. (2022). Ditch the low flow: agricultural impacts on flow regimes and consequences for aquatic ecosystem functions. Ecohydrology 15:e2364. doi: 10.1002/eco.2364
Roland, F. A. E., Darchambeau, F., Borges, A. V., Morana, C., De Brabandere, L., Thamdrup, B., et al. (2018). Denitrification, anaerobic ammonium oxidation, and dissimilatory nitrate reduction to ammonium in an east African great Lake (Lake Kivu). Limnol. Oceanogr. 63, 687–701. doi: 10.1002/lno.10660
Romanelli, A., Soto, D. X., Matiatos, I., Martínez, D. E., and Esquius, S. (2020). A biological and nitrate isotopic assessment framework to understand eutrophication in aquatic ecosystems. Sci. Total Environ. 715:136909. doi: 10.1016/j.scitotenv.2020.136909
Schindler, D. W. (2006). Recent advances in the understanding and management of eutrophication. Limnol. Oceanogr. 51, 356–363. doi: 10.4319/lo.2006.51.1_part_2.0356
Schmidt, P. J., Pintar, K. D. M., Fazil, A. M., Flemming, C. A., Lanthier, M., Laprade, N., et al. (2013). Using Campylobacter spp. and Escherichia coli data and Bayesian microbial risk assessment to examine public health risks in agricultural watersheds under tile drainage management. Water Res. 47, 3255–3272. doi: 10.1016/j.watres.2013.02.002
Schubert, C. J., Durisch-Kaiser, E., Wehrli, B., Thamdrup, B., Lam, P., and Kuypers, M. M. M. (2006). Anaerobic ammonium oxidation in a tropical freshwater system (Lake Tanganyika). Environ. Microbiol. 8, 1857–1863. doi: 10.1111/j.1462-2920.2006.01074.x
Schwab, G. O., Taylor, G. S., Fouss, J. L., and Stibbe, E. (1966). Crop Response From Tile and Surface Drainage. Soil Science Society of America Journal. 30, 634–637. doi: 10.2136/sssaj1966.03615995003000050028x
Seymour, C. O., Palmer, M., Becraft, E. D., Stepanauskas, R., Friel, A. D., Schulz, F., et al. (2023). Hyperactive nanobacteria with host-dependent traits pervade Omnitrophota. Nat. Microbiol. 8, 727–744. doi: 10.1038/s41564-022-01319-1
Shen, W., and Ren, H. (2021). TaxonKit: a practical and efficient NCBI taxonomy toolkit. J. Genet. Genomics 48, 844–850. doi: 10.1016/j.jgg.2021.03.006
Shen, L., Zheng, P., and Ma, S. (2016). Nitrogen loss through anaerobic ammonium oxidation in agricultural drainage ditches. Biol. Fertil. Soils 52, 127–136. doi: 10.1007/s00374-015-1058-4
Shi, Y., Khan, I. U. H., Radford, D., Guo, G., Sunohara, M., Craiovan, E., et al. (2023). Core and conditionally rare taxa as indicators of agricultural drainage ditch and stream health and function. BMC Microbiol. 23:62. doi: 10.1186/s12866-023-02755-7
Sieber, C. M. K., Probst, A. J., Sharrar, A., Thomas, B. C., Hess, M., Tringe, S. G., et al. (2018). Recovery of genomes from metagenomes via a dereplication, aggregation and scoring strategy. Nat. Microbiol. 3, 836–843. doi: 10.1038/s41564-018-0171-1
Šimek, K., Weinbauer, M. G., Hornák, K., Jezbera, J., Nedoma, J., and Dolan, J. R. (2007). Grazer and virus-induced mortality of bacterioplankton accelerates development of Flectobacillus populations in a freshwater community. Environ. Microbiol. 9, 789–800. doi: 10.1111/j.1462-2920.2006.01201.x
Smith, R. D., and Huang, C. (2010). Assessing nutrient transport following dredging of agricultural drainage ditches. Trans. ASABE 53, 429–436. doi: 10.13031/2013.29583
Soana, E., Balestrini, R., Vincenzi, F., Bartoli, M., and Castaldelli, G. (2017). Mitigation of nitrogen pollution in vegetated ditches fed by nitrate-rich spring waters. Agric. Ecosyst. Environ. 243, 74–82. doi: 10.1016/j.agee.2017.04.004
Sunohara, M. D., Craiovan, E., Topp, E., Gottschall, N., Drury, C. F., and Lapen, D. R. (2014). Comprehensive nitrogen budgets for controlled tile drainage Fields in eastern Ontario, Canada. J. Environ. Qual. 43, 617–630. doi: 10.2134/jeq2013.04.0117
Sunohara, M., Gottschall, N., Wilkes, G., Craiovan, E., Topp, E., Que, Z., et al. (2015). Long-term observations of nitrogen and phosphorus export in paired-agricultural watersheds under controlled and conventional tile drainage. J. Environ. Qual. 44, 1589–1604. doi: 10.2134/jeq2015.01.0008
Teurlincx, S., Verhofstad, M. J. J. M., Bakker, E. S., and Declerck, S. A. J. (2018). Managing successional stage heterogeneity to maximize landscape-wide biodiversity of aquatic vegetation in ditch networks. Front. Plant Sci. 9:01013. doi: 10.3389/fpls.2018.01013
Thompson, R. L., Lassaletta, L., Patra, P. K., Wilson, C., Wells, K. C., Gressent, A., et al. (2019). Acceleration of global N2O emissions seen from two decades of atmospheric inversion. Nat. Clim. Chang. 9, 993–998. doi: 10.1038/s41558-019-0613-7
Tian, R., Ning, D., He, Z., Zhang, P., Spencer, S. J., Gao, S., et al. (2020). Small and mighty: adaptation of superphylum Patescibacteria to groundwater environment drives their genome simplicity. Microbiome 8:51. doi: 10.1186/s40168-020-00825-w
Toyama, T., Hanaoka, T., Yamada, K., Suzuki, K., Tanaka, Y., Morikawa, M., et al. (2019). Enhanced production of biomass and lipids by Euglena gracilis via co-culturing with a microalga growth-promoting bacterium, Emticicia sp. EG3. Biotechnol. Biofuels 12:205. doi: 10.1186/s13068-019-1544-2
Veraart, A. J., Dimitrov, M. R., Schrier-Uijl, A. P., Smidt, H., and de Klein, J. J. M. (2017). Abundance, activity and community structure of Denitrifiers in drainage ditches in relation to sediment characteristics, vegetation and Land-use. Ecosystems 20, 928–943. doi: 10.1007/s10021-016-0083-y
Vymazal, J., and Březinová, T. D. (2018). Removal of nutrients, organics and suspended solids in vegetated agricultural drainage ditch. Ecol. Eng. 118, 97–103. doi: 10.1016/j.ecoleng.2018.04.013
Waymouth, V., Miller, R. E., Kasel, S., Ede, F., Bissett, A., and Aponte, C. (2021). Soil bacterial community responds to Land-use change in riparian ecosystems. Forests 12:157. doi: 10.3390/f12020157
Wilkes, G., Brassard, J., Edge, T. A., Gannon, V., Gottschall, N., Jokinen, C. C., et al. (2014). Long-term monitoring of waterborne pathogens and microbial source tracking markers in paired agricultural watersheds under controlled and conventional tile drainage management. Appl. Environ. Microbiol. 80, 3708–3720. doi: 10.1128/AEM.00254-14
Wilkes, G., Edge, T., Gannon, V., Jokinen, C., Lyautey, E., Neumann, N., et al. (2011). Associations among pathogenic bacteria, parasites, and environmental and land use factors in multiple mixed-use watersheds. Water Res. 45, 5807–5825. doi: 10.1016/j.watres.2011.06.021
Williams, T. J., Allen, M. A., Berengut, J. F., and Cavicchioli, R. (2021). Shedding light on microbial “dark matter”: insights into novel Cloacimonadota and Omnitrophota from an Antarctic Lake. Front. Microbiol. 12:741077. doi: 10.3389/fmicb.2021.741077
Wu, W., Niu, X., Yan, Z., Li, S., Comer-Warner, S. A., Tian, H., et al. (2023). Agricultural ditches are hotspots of greenhouse gas emissions controlled by nutrient input. Water Res. 242:120271. doi: 10.1016/j.watres.2023.120271
Wu, H., Wang, X., He, X., Zhang, S., Liang, R., and Shen, J. (2017). Effects of root exudates on denitrifier gene abundance, community structure and activity in a micro-polluted constructed wetland. Sci. Total Environ. 598, 697–703. doi: 10.1016/j.scitotenv.2017.04.150
Xie, B., Liu, B., Yi, Y., Yang, L., Liang, D., Zhu, Y., et al. (2016). Microbiological mechanism of the improved nitrogen and phosphorus removal by embedding microbial fuel cell in anaerobic–anoxic–Oxic wastewater treatment process. Bioresour. Technol. 207, 109–117. doi: 10.1016/j.biortech.2016.01.090
Xu, Z., and Asakawa, S. (2021). A model explaining mRNA level fluctuations based on activity demands and RNA age. PLoS Comput. Biol. 17:e1009188. doi: 10.1371/journal.pcbi.1009188
Xu, M., Xu, R., Shen, X., Gao, P., Xue, Z., Huang, D., et al. (2022). The response of sediment microbial communities to temporal and site-specific variations of pollution in interconnected aquaculture pond and ditch systems. Sci. Total Environ. 806:150498. doi: 10.1016/j.scitotenv.2021.150498
Zhang, S., Liu, F., Huang, Z., Xiao, R., Zhu, H., and Wu, J. (2020). Are vegetated drainage ditches effective for nitrogen removal under cold temperatures? Bioresour. Technol. 301:122744. doi: 10.1016/j.biortech.2020.122744
Zhou, Y., Liu, M., and Yang, J. (2022). Recovering metagenome-assembled genomes from shotgun metagenomic sequencing data: methods, applications, challenges, and opportunities. Microbiol. Res. 260:127023. doi: 10.1016/j.micres.2022.127023
Zolfaghari, K., Wilkes, G., Bird, S., Ellis, D., Pintar, K. D. M., Gottschall, N., et al. (2019). Chlorophyll-a, dissolved organic carbon, turbidity and other variables of ecological importance in river basins in southern Ontario and British Columbia. Canada. Environ. Monit. Assess. 192:67. doi: 10.1007/s10661-019-7800-x
Keywords: agricultural drainage ditch, aquatic microbiome, brushing, dredging, ditch management, metagenome assembled genome, nitrogen cycling, shotgun metagenomics
Citation: Guo G, Lapen DR, Khan IUH, Sunohara M, Craiovan E and Chen W (2024) Examining the impact of agricultural drainage ditch management on in-stream bacterial communities involved in nitrogen cycling: insights from the Environmental Change One Health Observatory (ECO2). Front. Sustain. Food Syst. 8:1329422. doi: 10.3389/fsufs.2024.1329422
Received: 28 October 2023; Accepted: 25 March 2024;
Published: 23 May 2024.
Edited by:
Vassilios D. Litskas, Independent Researcher, Lefkosia, CyprusReviewed by:
Michalis D. Omirou, Agricultural Research Institute, Cyprus© 2024 His Majesty the King in Right of Canada, as represented by the Minister of Agriculture and Agri-Food Canada for the contribution of Galen Guo, David R. Lapen, Izhar U.H. Khan, Mark Sunohara, Emilia Craiovan, and Wen Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wen Chen, d2VuLmNoZW5AYWdyLmdjLmNh
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.