
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Sustain. Food Syst., 11 January 2024
Sec. Agro-Food Safety
Volume 7 - 2023 | https://doi.org/10.3389/fsufs.2023.1305914
This article is part of the Research TopicMycotoxins and Their Management in Sustainable Food SystemsView all 5 articles
 Herbert Michlmayr1*
Herbert Michlmayr1* Gerlinde Wiesenberger1
Gerlinde Wiesenberger1 Krisztian Twaruschek1
Krisztian Twaruschek1 Florian Kastner1
Florian Kastner1 Marta M. Sopel2†
Marta M. Sopel2† Christian Hametner3
Christian Hametner3 Franz Berthiller2
Franz Berthiller2 Gerhard Adam1
Gerhard Adam1The use of lactic acid bacteria as a low-cost sustainable management tool to prevent further build-up of Fusarium mycotoxins during grain storage is increasingly propagated. It has been reported that even deoxynivalenol contamination already formed in the field can be reduced by such treatment in unknown ways. An enigmatic deoxynivalenol derivative, 3-lactyl-deoxynivalenol, has been reported already in 1982 as the toxic principle of Fusarium-infected barley from China, but very little is known about this metabolite. Here, we show that the enzymatic machinery of Fusarium graminearum is sufficient for its biosynthesis. Similarly, when challenged with propionic acid, F. graminearum can form a novel modified mycotoxin, 3-propionyl-deoxynivalenol. Lactic acid and propionic acid are first converted into lactyl-CoA and propionyl-CoA, respectively. These acyl-CoA derivatives can subsequently be used by the 3-O-acyltransferase encoded by TRI101. We expressed the respective genes in E. coli and utilized the affinity-purified proteins for enzymatic synthesis of the reference substances 3-lactyl- and 3-propionyl-deoxynivalenol. The structures of the purified compounds were confirmed by nuclear magnetic resonance spectroscopy. Preliminary toxicological assessment using in vitro translation assays indicated residual toxicity, most likely due to reactivation of deoxynivalenol by de-acylation. In conclusion, this study reports a method to synthesize 3-lactyl- and 3-propionyl-deoxynivalenol reference substances, which will be highly useful to determine occurrences of these acylated deoxynivalenol-derivatives in cereal samples and to perform more detailed studies to evaluate their toxicological relevance.
The Fusarium mycotoxin deoxynivalenol (DON) is a major contaminant of small grain cereals and maize worldwide. In many countries, maximum tolerated levels have been enacted for food commodities (Van Egmond et al., 2007; European Commission, 2023). Contaminated grain unfit for human consumption is utilized primarily as animal feed, and also guideline levels for different feed commodities and animal species exist in the EU to protect animal health and to avoid reduced animal performance (European Commission, 2016). Deoxynivalenol is produced primarily in the field by Fusarium graminearum and related species, which infect small grain cereals and maize. Yet, the mycotoxin contamination can further increase post-harvest under suboptimal storage conditions (Garcia-Cela et al., 2018). Reducing the grain humidity by drying and cool storage conditions are energy demanding. With increasing energy costs, controlling temperature and humidity for contaminated low-value feed is economically questionable. Therefore, treatment with lactic acid or with lactic acid bacteria is increasingly propagated to prevent further fungal growth and mycotoxin production, numerous reviews exist on this topic (Dalié et al., 2010; Pawlowska et al., 2012; Oliveira et al., 2014; Hassan et al., 2016; Sadiq et al., 2019; Maidana et al., 2022; Smaoui et al., 2022).
Interestingly, DON already present in the harvested material can also be reduced by lactic acid bacteria. Various in vitro studies pointed to the binding of DON to cell wall components as the main mechanism causing this effect (Franco et al., 2011). However, the use of solvent extraction protocols that also capture DON bound to cell wall components indicated that the total DON content can in fact be reduced by lactic acid bacteria treatment (Juodeikiene et al., 2023).
Deoxynivalenol and other trichothecenes inhibit protein biosynthesis by eukaryotic ribosomes. For self-protection in DON-producing Fusarium, after the first toxic intermediate isotrichodermol is formed, the C3-hydroxy group is acetylated in all later intermediates of DON biosynthesis (McCormick et al., 2011). Acetylation of the C3-OH has been reported as a detoxification reaction (Kimura et al., 1998; Payros et al., 2016) and molecular docking analysis indicates that 3-acetyl-DON (3-ADON) is sterically hindered to interact with the ribosomal target site (Dellafiora et al., 2017). During DON biosynthesis 3,15-diacetyl-DON is initially formed, exported by the TRI12-encoded transporter and extracellularly converted to either 3-ADON or 15-acetyl-DON (15-ADON), depending on the allele of the esterase encoded by TRI8 (Alexander et al., 2011). In planta, there is further rapid deacetylation of both forms to DON (Schmeitzl et al., 2015a), so that typically less than 10% of DON still occurs as 3-ADON or 15-ADON in grain. In the plant, DON (and also 15-ADON) can be glycosylated and thus detoxified to DON-3-O-glucoside (D3G) and the (unstable) compound 15-acetyl-DON-3-O-glucoside (Schmeitzl et al., 2015b). Depending on the plant species and cultivar, typically about 20% of DON can be present as glucoside. Lactic acid bacteria possess highly active β-glucosidases capable of hydrolyzing D3G back to toxic DON (Berthiller et al., 2011; Michlmayr et al., 2015). Treatment with lactic acid bacteria should therefore reduce the DON-glucoside content, and consequently increase DON concentrations. However, in a recent study analyzing such derivatives (except 15-acetyl-DON-3-O-glucoside, for which no standard is available), a strong reduction of the DON-derivatives was observed after treatment with various bacterial strains (Juodeikiene et al., 2023). A barley whole-meal sample with initially 6.5 mg/kg DON after 48-h treatment with a Lactobacillus casei strain at 30°C contained only 52% of the initial value of DON, and 11% of the initial D3G concentration. Also, the concentrations of acetylated forms were reduced: of the predominant 15-ADON, only 65% remained of the initial 929 μg/kg. Apparently, the treatment with lactic acid bacteria indeed reduced the detectable DON concentrations, potentially by conversion into to date unknown DON-metabolites.
A candidate for such a metabolite is the enigmatic 3-lactyl-DON. This compound had already been reported in 1982 as the toxic principle of Fusarium-infected barley from Shanghai causing vomiting in humans and swine (Xu et al., 1982). A sample of that material was later transferred to the US, and the structure of the trimethyl-silyl-derivative was confirmed by nuclear magnetic resonance spectroscopy (NMR), as reported in an influential book chapter (Mirocha et al., 2003). The gas chromatography–mass spectrometry (GC–MS) spectrum of this derivative was then reported (Rodrigues-Fo et al., 2002), without providing information on the origin of this material.
The aim of the present study was to elucidate the biosynthetic origin of 3-lactyl-DON by investigating different possible biochemical routes, including lactic acid bacteria, wheat cells and Fusarium graminearum. Using proteins heterologously expressed in E. coli, we could enzymatically synthesize 3-lactyl-DON and the novel metabolite 3-propionyl-DON. We further purified and characterized these compounds in order to obtain analytical standards and to generate sufficient amounts for a preliminary toxicological assessment.
Two commercial preparations of lactic acid bacteria (Ombe immune, Ombe intim, STADA Arzneimittel GmbH, Vienna) were obtained from a local pharmacy and used for the pilot experiment. One capsule containing 106 lyophilized bacteria, either a mix of two Lactobacillus plantarum strains (CECT 7315 and CECT 7316), or a mix of Lactobacillus rhamnosus (GR-1) and Lactobacillus reuteri (RC-14) were resuspended in 1 mL of MRS medium (Sigma 1106610500) containing 200 mg/L deoxynivalenol. Portions of 300 μL were transferred to 2 mL screw-cap micro tubes (Sarstedt) and incubated for three days at three different temperatures (20°C, 30°C, 37°C) without shaking. Then 300 μL ethanol was added to each tube, and after vortexing the content was transferred to Eppendorf tubes and centrifuged for 10 min at 14,650 rpm. The supernatant was diluted (50 μL with 450 μL 50% ethanol), and centrifuged again. Another 1:10 dilution step was prepared before transfer to vials for high performance liquid chromatography (HPLC).
A suspension culture (PC-998) of Triticum aestivum (derived from cv. Heines Koga II) was obtained from DSMZ (Braunschweig, Germany) and cultured in B5 medium (Duchefa Biochemie G0209) at 20°C. Cells were distributed to 2.2 mL Eppendorf tubes with cut (wide bore) tips, so that after a gentle centrifugation (5 min at 1000 rpm) and removal of the supernatant about 100 mg wet weight of cells were present in each tube. Fresh medium with DON (final concentration 75 mg/L) was added to a final volume of 500 μL, and stock solutions (10%) of the acids were added to reach final concentrations of 0.1% or 0.5% of either racemic lactic acid or propionic acid, respectively. After 22 h of incubation (20°C, shaking at 100 rpm) one volume of ethanol was added (to avoid adsorption), and the mix centrifuged gently for 5 min. Then 400 μL supernatant were removed and centrifuged for 10 min at 14,680 rpm. After another 1:10 dilution step and centrifugation, the supernatant was transferred to HPLC vials.
The strain F. graminearum PH-1 (NRRL 31084) and a tri5::loxP mutant thereof [strain dTRI5#11–19, (Seidl et al., 2022)] was used for the acid and DON treatment. For testing production of metabolites on media with added acids, first 5 mL precultures in 2SM medium (Lowe et al., 2012) in 50 mL flasks were inoculated with 105 conidiospores in 100 μL. After 2 days another 5 mL of medium with DON and acids was added, resulting in a final concentration of 200 mg/L DON and 1% of either acetic acid, DL-lactic acid or propionic acid (in triplicate). The cultures were incubated for 1 week (30°C, 150 rpm). With a wide bore 1 mL tip, 700 μL medium (and some mycelium) was transferred to an Eppendorf tube and an equal volume of ethanol was added. After centrifugation, 100 μL supernatant was removed and diluted with 900 μL 50% aqueous ethanol. After another 1:5 dilution, the supernatant was transferred to HPLC vials for measurements.
The expression vector for the FgTri101 (Garvey et al., 2008) was kindly provided by Prof. Ivan Rayment (Univ. Wisconsin, Madison). The prpE gene of E. coli from strain K894 (Yale Stock Center # 7785, https://cgsc.biology.yale.edu/Strain.php?ID=90741) was amplified with primers 4,469 and 4,470 (see Table 1 for sequences), the product cleaved with NdeI and XhoI and cloned into the respective sites of pET21a. The resulting sequenced vector (pHM34) was used for expression of a prpE protein with C-terminal 6xHis tag. The three exons of the putative Fusarium propionyl-CoA Ligase FGSG_10126 were amplified with three primer pairs with overlaps to the pET21a vector or the next exon, respectively (see Table 2). The resulting sequence confirmed plasmid is pHM60. For improved protein expression, the FGSG_10126 insert was cut out of pHM60 with NdeI and XhoI and ligated into the pCA02b (Michlmayr et al., 2018) backbone previously digested with these enzymes. The resulting vector (pHM61) was used to express a fusion protein with N-terminal maltose binding protein and C-terminal 6xHis tag.
Protein production was carried out with E. coli T7 Express (New England Biolabs, Ipswich, MA, USA) in terrific broth with 1 mM isopropyl β-D-thiogalactopyranoside (IPTG) added at the exponential phase (OD ≈ 0.5), followed by incubation for 20 h at 20°C. Cells were harvested at 6000 x g for 15 min and resuspended in 50 mM phosphate buffer (pH 7.4), 500 mM NaCl, 20 mM imidazole. The cells were sonicated on ice 3–4 × 1 min with intervals to cool on a Bandelin sonopuls HD 4100 at 60% amplitude, the cell lysate was cleared at 30,000 x g, 30 min. Protein purifications were carried out using 1 mL HisTrap crude FF columns (Cytiva, Marlborough, MA). Unbound protein was washed out with the buffer specified above and bound protein was eluted with 500 mM imidazole in the same buffer. Afterwards, buffer change to 50 mM phosphate (pH 7.0), 50 mM KCl, 10% glycerol was carried out with Amicon® Ultra-15 Centrifugal Filter Unit 10 kDa (Millipore, Burlington, MA, USA), the proteins were stored at −80°C.
Enzymatic reactions with the one-step purified proteins were initially performed in a 200 μL scale from which 20 μL aliquots were removed and stopped by addition of 180 μL methanol at different time points. The reaction mix contained the respective acid salt and other cosubstrates (cofactors) in excess over DON (34 μM). prpE and Tri101 were added to concentrations of 1 mg/mL and 0.8 mg/mL, as judged from the Bradford Assay. Subsequent conversion experiments with prpE and Tri101 were conducted several times independently under different conditions to achieve sufficient conversion yields at DON concentrations up to 10 mM. The final preparative setup for the synthesis of acyl-DON derivatives contained 0.1 M Tris/Cl pH 7, 20 mM acid salt (propionic acid, L-lactic acid, D-lactic acid; titrated to pH 7), 0.5 mM CoA, 20 mM ATP, 5 mM MgCl2 and 10 mM DON. After 24 h, samples were taken for HPLC analysis and the reactions were stored at −20°C until the metabolites were purified by preparative HPLC. Assays with the FGSG_10126 gene product were performed under the same conditions as above, except for using 0.75 mg/mL purified FGSG_10126 and 34 μM DON.
Determination of DON, 3-ADON, 3-lactyl-DON and 3-propionyl-DON was performed by HPLC coupled to tandem mass spectrometry (HPLC–MS/MS). An Agilent 1100 HPLC system (Waldbronn, Germany) was coupled to a Sciex 3500 QqQ tandem mass spectrometer (Framingham, MA, US). A Phenomenex Gemini C18 column (Torrance, CA, US), 4.6 × 150 mm, 5 μm particle size, was used for separation. Mobile phase A was water, mobile phase B was 98% aqueous methanol, and both phases contained 5 mM ammonium acetate. Injections of 25 μL into a flow rate of 800 μL/min were used at a column temperature of 25°C. 10% mobile phase B were kept constant in the first minute after injection, then a linear gradient to 100% followed till minute 6.0. After 3 min washout and 3 min re-equilibration, the run ended at 12.0 min. The Turbo V electrospray source (Sciex) was operated at 550°C with gases 1 and 2 both at 50 psi each. Negative ionization was performed at −4,000 V spray voltage. The curtain gas was set to 35 psi. The following selected reaction monitoring (SRM) transitions were used with a dwell time of 25 ms each (analyte, Q1 mass > Q3 mass, declustering potential, collision energy): DON (355.1 Da > 59.1 Da, −65 V, −44 eV; 355.1 Da > 265.1 Da, −65 V, −20 eV); 3-ADON (397.1 Da > 59.1 Da, −60 V, −52 eV; 397.1 Da > 307.1 Da, −60 V, −20 eV); 3-lactyl-DON (427.1 Da > 59.1 Da, −60 V, −52 eV; 427.1 Da > 337.1 Da, −60 V, −20 eV); 3-propionyl-DON (411.1 Da > 59.1 Da, −60 V, −52 eV; 411.1 Da > 321.1 Da, −60 V, −20 eV). Analyst software version 1.6.2 (Sciex) was used for acquisition and data evaluation. Purification of both compounds was achieved using an Agilent 1,100 series preparative HPLC system. A Waters (Vienna, Austria) Sunfire C18 OBD preparative column (19 × 100 mm, 5 μm particle size) was used for separation with pure water and pure methanol as mobile phases. A flow rate of 16 mL/min was employed at 25°C. Various injection volumes up to 900 μL were used. The gradient was kept constant at 20% B for 1 min, followed by a linear gradient to 100% B at 8 min. Afterwards, the column was washed (3 min) and re-equilibrated (3 min). UV at 220 nm was used to detect all trichothecenes. A peak-triggered collection was performed into 30 mL glass vials. Under the selected conditions 3-lactyl-DON eluted at 5.33 min, while 3-propionyl-DON eluted at 6.42 min. The fractions were pooled, methanol was evaporated and the remaining aqueous solution was frozen. After lyophilization, white powders were obtained which were used for further structural elucidation by NMR.
MS/MS characterization was performed on a 6500+ QTrap mass spectrometer (Sciex) in enhanced product ion (EPI) mode at a collision energy of 30 eV.
NMR spectra were obtained on a Bruker Avance IIIHD 600 FT-NMR spectrometer (600 MHz for 1H, 150 MHz for 13C). The samples were dissolved in MeOH-d4 and measured at ambient temperature using a Cryoprobe Prodigy™ probehead. Chemical shifts were established on the basis of residual solvent resonances (3.31 ppm for 1H, 49.15 ppm for 13C) and expressed relative to tetramethylsilane.
A bottom agar of 15 mL YPD (1% yeast extract, 2% peptone, 2% glucose, 1% agarose) was prepared in a 9 cm Petri dish. A preculture of yeast strain YZGA515 (relevant genotype pdr5 pdr10 pdr15 ayt1) was diluted back to an OD600 of about 0.3. Three mL of YPD top agar (with 0.6% agarose) were equilibrated at 42°C. After mixing with 100 μL yeast suspension this overlay was poured on the bottom agar. After solidification at room temperature, UV treated antibiotic discs were placed on the agar surface with a sterile forceps, and 5 μL of test solutions (10 mg/mL DON, acyl-DONs) were added.
For testing inhibition of protein synthesis, the coupled in vitro transcription/translation systems with animal ribosomes (TnT® T7 coupled Rabbit Reticulocyte Lysate System) and plant ribosomes (TnT® T7 Coupled Wheat Germ Extract System) by Promega (Madison, WI, USA) were used as previously described (Varga et al., 2015). After stopping the reactions with cycloheximide, 5-μL aliquots were removed and added to 45 μL of acetonitrile (ACN). For the time course experiment translation assays were set up as before and 10-μL samples were removed immediately after mixing and subsequently every 10 min (up to one hour) and added to 5 volumes of ACN. The samples were centrifuged at room temperature for 10 min at maximum speed (app. 20,000 x g) and 45 μL of the supernatant were subjected to HPLC analysis.
In the original publication (Xu et al., 1982), no details were given about the circumstances leading to the formation of the reported 3-lactyl-DON. Our working hypothesis was that Fusarium-infected grain with a high DON content was stored moist at high temperatures, which could have favored growth of lactic acid bacteria naturally occurring in grain. Under such sourdough-like conditions, autoinhibitory levels of lactic acid might accumulate within days, so that both precursors of 3-lactyl-DON (lactic acid and DON) are present. Alternatively, highly Fusarium-contaminated grain might have been treated with lactic acid. In principle, each of the three organisms in this system could potentially be responsible for production of 3-lactyl-DON: (a) either the lactic acid bacteria exposed to DON and lactic acid, (b) the plant cells, or (c) Fusarium graminearum.
To test this, we first incubated different lactic acid bacteria in MRS medium containing 200 mg/L DON. After incubation for three days at three different temperatures, no compound with the predicted mass of 3-lactyl-DON was detected by high performance liquid chromatography coupled to mass spectrometry (HPLC–MS). Later, after discovery of 3-propionyl-DON (see below), also six strains of propionic acid bacteria were tested, again with negative result.
Plants contain multiple members of the acyl-activating enzyme superfamily, some of which could potentially also activate short chain carboxylic acids to the respective acyl-CoA derivatives (Shockey et al., 2003). Since we did not have a barley cell line at hand, we have tested whether cells of a wheat suspension culture maintained in the lab could form 3-lactyl-DON when challenged with DON (75 mg/L) and with either lactic acid or propionic acid (0.1 and 0.5%, respectively). No conversion was detectable by HPLC–MS in either case.
We next tested F. graminearum, and a trichothecene biosynthesis deficient tri5 mutant to avoid possible interference of de novo synthesis with the metabolization of externally added DON. F. graminearum is sensitive to its own toxin, but can adapt and become resistant to high concentrations by inducing genes encoding drug efflux transporters (O’Mara et al., 2023). We therefore added a DON concentration (200 mg/L) that should be sufficient to reach the cytosol and a medium (2SM) in which DON production normally occurs (Lowe et al., 2012). DON was added to this medium (containing 1% glycerol and 4% sucrose as carbon sources) together with one of the following acids at a final concentration of 1% (w/v): acetic acid, DL-lactic acid and propionic acid. After one week of incubation, 3-acetyl-DON was found in all samples: in the acetic acid treated sample, on average 166 μg/L was found, while the DL-lactic acid treated samples contained higher levels (472 μg/L), and in the propionic acid treated sample, only 52 μg/L on average were present. 3-lactyl-DON was only found in traces (below limit of quantification). Yet, in the propionic acid-treated sample, clear evidence for 3-propionyl-DON was found, later quantified to be about 135 μg/L.
This result provides strong evidence that the Fusarium gene products are sufficient for production of at least propionyl-DON. We hypothesized that a likely candidate for the 3-O-acetyltransferase is Tri101, which might have relaxed substrate specificity and in addition to acetyl-CoA might also be able to utilize propionyl-CoA and lactyl-CoA. In propionate-grown E. coli or Salmonella typhimurium and other bacteria, an operon is induced, which includes the gene prpE (Tsang et al., 1998; Armando et al., 2012), coding for propionyl-CoA synthetase, an enzyme that also accepts lactate as substrate (Megraw et al., 1965). To identify similar gene products in F. graminearum PH-1, we performed a BLASTp search with prpE as query, revealing two “probable acetyl-CoA synthetase” genes (FGSG_00330, FGSG_01743) and also an additional gene annotated as “related to acetyl coenzyme A synthetase” (FGSG_10126). Interestingly, this protein is more closely related to prpE than to the other two Fusarium acetyl-CoA synthetases.
To test the hypothesis that the products of either prpE or FGSG_10126, and the protein encoded by TRI101 (FGSG_07896) catalyze the synthesis of lactyl- and propionyl-DON, we set out to reconstitute these reactions in vitro. We obtained a previously described F. graminearum Tri101 expression vector (Garvey et al., 2008). Plasmids for the overexpression of the prpE protein and F. graminearum FGSG_10126 in E. coli were constructed (see Material and Methods) and the respective enzymes were purified by affinity chromatography.
As shown schematically in Figure 1, prpE uses ATP and coenzyme A to activate propionic acid (propanoate + ATP + CoA < => AMP + diphosphate + propanoyl-CoA). The formed propanoyl-CoA (and in analogy, lactyl-CoA from lactate) can subsequently be used by Tri101 to produce 3-propionyl- or 3-lactyl-DON. This reaction also regenerates free CoA. We first tested these reactions in a small scale pilot experiment with 10 mg/L (34 μM) DON and other substrates/cofactors in molar excess. This showed that the combination of prpE and Tri101 was indeed able to form the respective acyl-DON derivatives. As shown in Figure 2, propionyl-, lactyl and acetyl-DON could be clearly separated from DON in LC–MS. Due to the unavailability of analytical standards, exact quantification of reaction products was not possible at the time these experiments were conducted. Judged from DON-consumption, we estimated complete conversion of DON to propionyl-DON within 24 h. The prpE protein was also able to activate acetic acid, since 3-acetyl-DON formation was observed when acetic acid was added. The combination of prpE and Tri101 could further convert DON to lactyl-DON with both lactic acid isomers, although conversion was incomplete in both cases. The reaction with L-lactic acid was driven to near completion (approximately 90% conversion estimated from DON consumption) within 24 h. With D-lactic acid as substrate, less than 50% of DON was converted to D-lactyl-DON within 24 h.
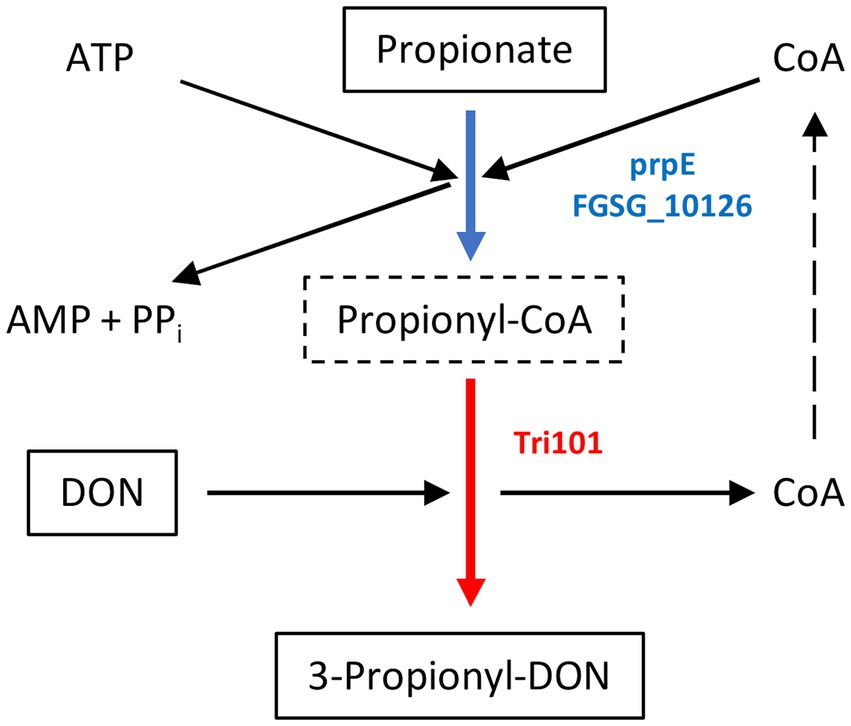
Figure 1. Mechanism of 3-propionyl-DON formation. Propionate (or lactate in case of 3-lactyl-DON formation) is activated by a broad specificity acyl-CoA synthetase (E. coli prpE or F. graminearum FGSG_10126 gene products). The 3-O-acetyltransferase encoded by TRI101 can use lactyl-CoA or propionyl-CoA instead of acetyl-CoA to generate 3-lactyl-DON or 3-propionyl-DON (instead of the common 3-acetyl-DON).
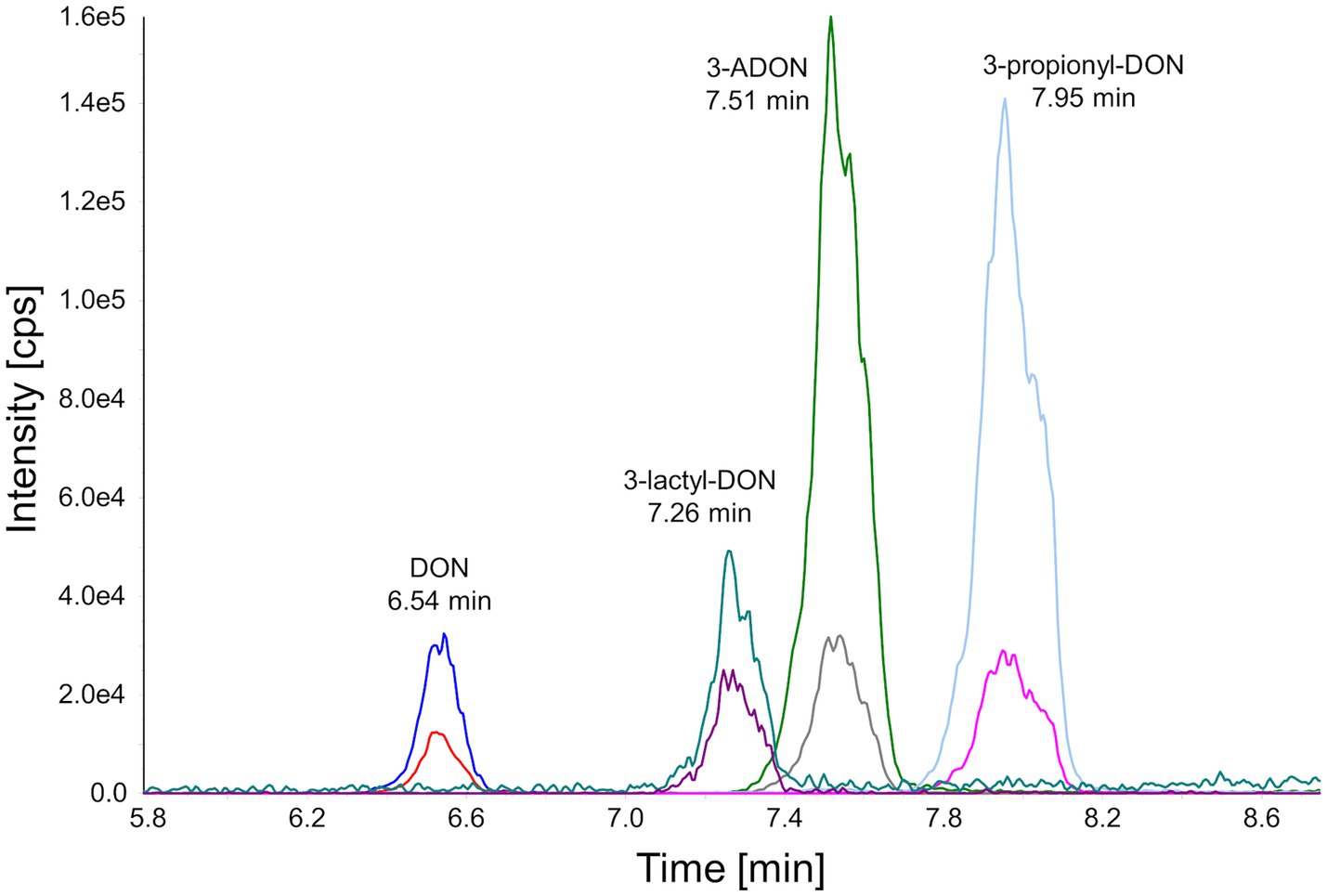
Figure 2. Chromatographical separation of DON and its acyl-derivatives. The selected reaction monitoring chromatogram was obtained after injection of an 1.00 mg/L standard into the HPLC-MS/MS system.
The putative acetyl coenzyme A synthetase related FGSG_10126 gene product was able to activate propanoate and both lactate isomers under similar conditions (i.e., 34 μM DON). As with prpE, in combination with Tri101, we observed almost complete conversion of DON to its propionyl- and acetyl- derivatives (100 and 91% conversion, respectively; means of two independent replicates). As observed with prpE, the conversion of DON to L-lactyl-DON and D-lactyl-DON was incomplete and amounted to about 80 and 70%, respectively within 24 h.
Using prpE and Tri101, we subsequently scaled up the reactions to total amount 10 mg of DON (about 10 mM DON in reactions) in order to synthesize sufficient amounts of acyl-DON derivatives for purification and further characterization. This confirmed the observations in the above-mentioned small-scale experiments, being complete conversion to propionyl-DON, but not to lactyl-DON (both isomers). Increasing enzyme concentrations (prpE, Tri101) could not improve the yields. The products were purified by preparative HPLC and subjected to structural characterization by mass spectrometry and NMR (Table 2). The LC–MS/MS fragmentation patterns of 3-L-lactyl-DON and 3-propionyl-DON are shown in Figure 3, indicating the typical pattern of loss of the C15-OH (Figure 3).
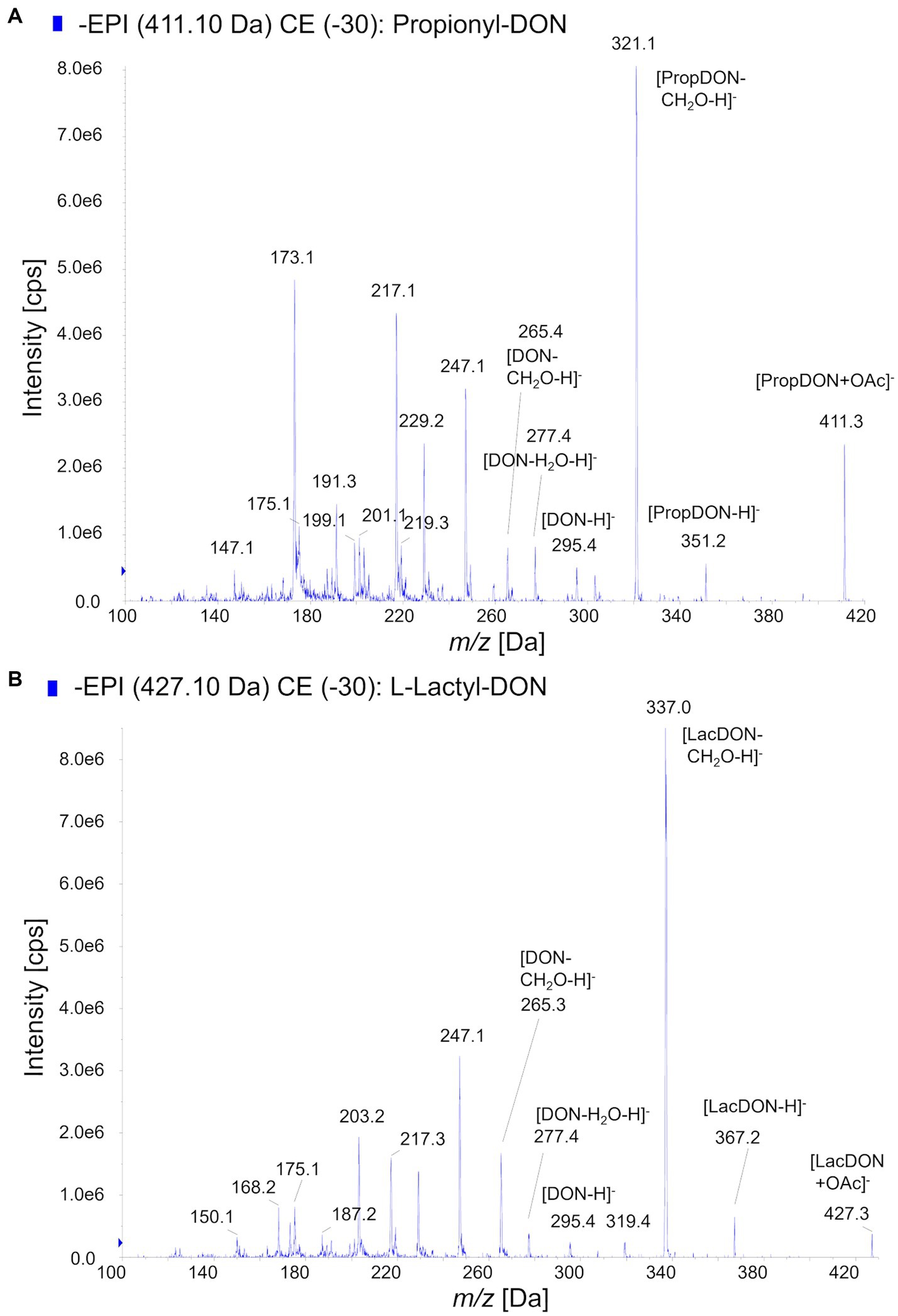
Figure 3. MS/MS-Fragmentation patterns of acyl-DON derivatives. (A) Propionyl-DON, (B) L-lactyl-DON.
3-acetyl-DON is sterically hindered to bind to the A-site of ribosomes (to block entry of aminoacyl-tRNAs) and therefore does not inhibit translation as DON does. Therefore, our expectation was that the derivatives with the even bulkier acyl-groups should likewise be non-toxic. First we performed a simple agar diffusion test using the toxin-sensitive yeast strain YZGA515 (Poppenberger et al., 2003). While application of 50 μg of DON to the discs produced a clear halo with about 24 mm diameter, this was clearly not the case with equal amounts of 3-acetyl-DON, 3-L-lactyl-DON and propionyl DON. Yet, a tiny halo (comparable to about 1 mg/mL DON) appeared with propionyl-DON (Figure 4).
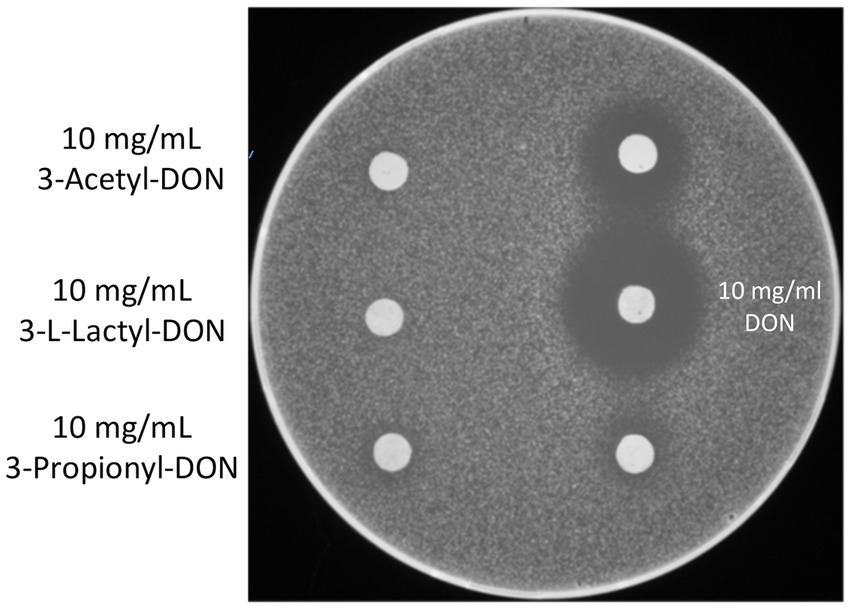
Figure 4. Agar diffusion test - inhibition zones. The toxin sensitive strain YZGA515 was used as indicator organism. On the right, 5 μL of DON was spotted as control (upper disc 5 mg/mL, on the lower disc 2.5 mg/mL).
As this assay is quite insensitive, only semi-quantitative, and could furthermore be confounded by different uptake of the compounds, we also tested if the two metabolites were able to inhibit plant and mammalian ribosomes similar to DON. We performed coupled in vitro transcription and translation assays using rabbit reticulocyte lysates and wheat germ extracts in presence of DON (as a control), lactyl-DON and propionyl-DON. When compared to DON, the two substances exhibit much lower, but still surprisingly high inhibition in both translation reactions. Interestingly, the inhibitory effect of the two substances was quite different: while propionyl-DON was less inhibitory than lactyl-DON for rabbit ribosomes, the order was reversed in case of the reaction with wheat ribosomes (Figure 5).
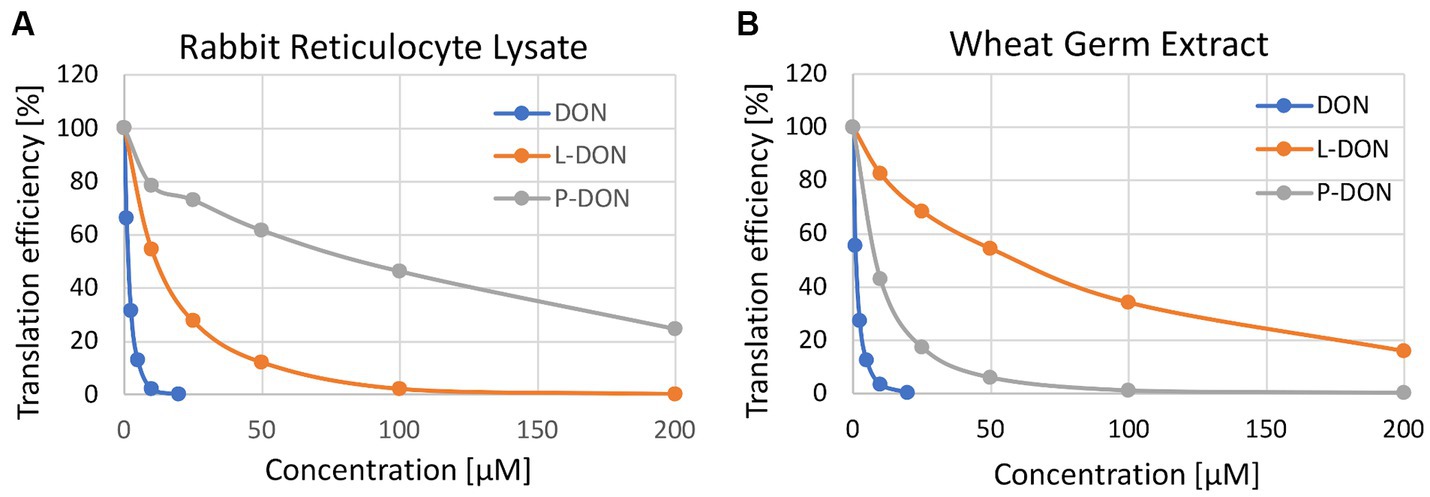
Figure 5. In vitro toxicity of deoxynivalenol (DON), lactyl-DON (L-DON) and propionyl-DON (P-DON) in (A) rabbit reticulocyte lysate and (B) wheat germ extract-based translation assays.
As stated above, one would not expect any interference of lactyl- and propionyl-DON with ribosomes, and thus no inhibitory effect at all. One explanation for the inhibitory effect on both mammalian and plant ribosomes might be de-acylation of the compounds in the course of the translation reactions, thus leading to release of DON, which in turn inhibits translation. To test if this is the case, we set up two different experiments: (1) After stopping the translation assays with cycloheximide, samples were removed and subjected to HPLC analysis. At the end point of the translation assays (20 min) we found considerable amounts of DON and reduced concentrations of the respective metabolites, e.g., in the reaction where 200 μM lactyl-DON was incubated with rabbit reticulocyte lysate, we found 2.7 μM DON, a concentration sufficient to completely inhibit translation (see Figure 5) and an even higher DON concentration was detected in the 200 μM sample of propionyl-DON in the wheat germ extract (app. 4.4 μM). (2) For a time-course experiment, we incubated each substance with either the rabbit reticulocyte lysate or with wheat germ extract and determined the fate of the metabolites over time. The concentrations of both substances decreased over a time course of 60 min with concomitant increase of DON, albeit to a different extent. While propionyl-DON was quite stable in the rabbit reticulocyte lysate (with about 12% reduction after 60 min incubation), about 31% were converted into DON in the wheat germ extract. On the other hand, almost all of the lactyl-DON (83%) disappeared after 60 min in presence of the rabbit reticulocyte lysate, but only 22% in the wheat germ extract (not shown). These observations align perfectly well with the results of the translation assays, which in this case do not reflect toxicological differences between the compounds, but in the presence of contaminating carboxylesterases in the ribosome preparations from wheat germ and rabbit reticulocytes.
Formation of 3-lactyl-DON has two prerequisites: an enzyme capable of activating lactate to lactyl-CoA and a transferase with substrate specificity for DON that can utilize this co-substrate. Lactyl-CoA is not a usual metabolite of lactate, which is normally converted into pyruvate by lactate dehydrogenase (lactate + NAD+ < => pyruvate + NADH + H+) and subsequently by the pyruvate dehydrogenase complex to acetyl-CoA (pyruvate + CoA + NAD+ < => acetyl-CoA + CO2 + NADH + H+). While genes with similarity to prpE exist in many bacteria, this does not seem to be the case for lactic acid bacteria based on BLASTp searches. On the other hand, ubiquitous acetyl-CoA synthetases may have a side activity with lactate and propionate (at least in vitro without competing acetate). We therefore could not exclude that the lactic acid bacteria are responsible for 3-lactyl-DON synthesis. Little is known about bacterial acyltransferases that might use DON as substrate. Acetylation activity for the C4-OH of nivalenol has been described for a Pseudomonas strain (Tanaka et al., 2016), but no gene is known. Recently an Acinetobacter pittii acyltransferase involved in transformation of deoxynivalenol to 3-acetyl-deoxynivalenol was identified (Liu et al., 2023). In general, de-acetylation of acetyl-DON by bacteria is more frequently encountered (Young et al., 2007).
Plants could theoretically activate lactate to lactyl-CoA, as they have a rich gene inventory of acyl-transferases. The ability to acetylate T-2 toxin at the C3-OH has been demonstrated for wheat (Nathanail et al., 2015), barley (Meng-Reiterer et al., 2015) and oat (Meng-Reiterer et al., 2016). The obtained negative result in our study does not completely rule out the possibility that plants have the ability to synthesize 3-lactyl-DON. The high DON concentration employed will block transcription, so if de novo gene expression would be necessary, such an induced enzyme would not be expressed and overlooked. Also, the concentration of the acids (0.1 and 0.5%) may have been too low, but was chosen so out of concern not to immediately inactivate the sensitive suspension cells with higher levels.
Our results clearly show that the gene products of Fusarium are sufficient to generate 3-lactyl-DON. The FGSG_10126 propionyl-CoA-synthetase is not only similar in sequence to E. coli prpE, but could fully substitute this enzyme in the in vitro assay. A BLASTp search indicates that this gene is highly conserved in other Fusarium species and other fungi. Likewise, genes encoding products with lower similarity to Tri101, with activity against DON exist in other fungi, also in non-trichothecene producers (Khatibi et al., 2011). Even baker’s yeast has a DON-inactivating 3-O-acetyltransferase, Ayt1 (Alexander et al., 2002), so it remains an open question for further research whether 3-lactyl- or 3-propionyl-DON formation capability could be more widespread. Another topic for future research is also to explore under which conditions lactyl-DON (and propionyl-DON) are naturally formed. In our experiment with Fusarium treated with 1% lactic acid, we did not see measurable formation of 3-lactyl-DON, but a 2.8-fold increase of 3-acetyl-DON. Lactate under mild stress conditions may still have been converted to pyruvate and acetate, while in case of propionic acid, clear evidence for formation of propionyl-CoA was obtained. Assuming that Tri101 has a preference for acetyl-CoA, high intracellular concentrations of lactic acid or propionic acid are a likely prerequisite for formation of the respective CoA-derivatives in concentrations that these can compete with acetyl-CoA in the transferase reaction. The toxicity of the acids for fungi depends strongly on the dissociation grade of the acid, or pH of the medium. Removing weak acids with an ATP-driven efflux carrier (as in yeast by the product of PDR12 (Piper, 1998), is rather costly for cells, and if also DON is diffusing back into Fusarium cells, it might be beneficial to switch to the short chain acids present in high concentrations in the medium and to activate these for self-protection. Future experiments are needed to test whether under higher lactate concentrations, 3-lactyl-DON formation indeed occurs. Interestingly, it has been reported (Humer et al., 2016) that treatment of Fusarium-contaminated grain with 5% lactic acid already after 5 h led to a reduction of DON (58% remaining of the initial 6 mg/kg), while D3G remained unchanged.
Concerning the toxicity of lactyl-DON, already the initial publication reported that 3-lactyl-DON is still toxic for animals (rabbit skin test) and also showed phytotoxicity (inhibition of germination of pea seeds). In principle, both 3-lactyl-DON and 3-propionyl-DON should be unable to inhibit eukaryotic ribosomes, which is the primary mode of toxicity of trichothecenes. Yet, the situation is most likely similar to the case of 3-acetyl-DON, which is also rapidly de-acetylated and converted into toxic DON. While it has long been known that human carboxylesterases can deacetylate trichothecenes (Johnsen et al., 1986), little information exists about the contribution of individual gene products. The gene family of human and animal carboxylesterases is of limited complexity (Hosokawa et al., 2007). In contrast, plants possess a large gene family, and only few members are biochemically characterized. It will be interesting to test whether a Brachypodium carboxylesterase shown to deacetylate 3-acetyl-DON (Schmeitzl et al., 2015a) can also hydrolyze 3-lactyl- and/or 3-propionyl-DON. The results from the in vitro translation assays are suggestive of a different preference of plant and animal carboxylesterases contaminating the ribosomal preparations obtained from wheat germ and rabbit reticulocytes. Such an effect is known for the conversion of 3-ADON into DON and has previously also been shown in case of the Type-A trichothecenes NX-2/NX-3 (Varga et al., 2015). Another issue is the Fusarium chemotype. In the dominant 15-acetyl-DON producing strains, 3-acetyl-DON is rapidly hydrolyzed to DON by the respective Tri8 allele. Potentially, the difference in formation of propionyl-DON versus lactyl-DON in the tri5 mutant of PH-1, which is a 15-acetyl-DON producer, could also be due to different hydrolysis after extended incubation.
In summary, our research has shed light on the enzymatic machinery possibly responsible for formation of the enigmatic lactyl-DON and the novel propionyl-DON. More detailed biochemical analyses of prpE/FGSG_10126 and homologous acyl-CoA synthetases as well as Tri101 homologs regarding substrate specificities and reaction kinetics will be highly interesting. Synthesis and purification of the acylated derivatives further allowed us to perform preliminary toxicological analyses, implying residual toxicity most likely due to rapid de-acylation by carboxylesterases. The presented method to generate and purify these compounds will now permit further studies to evaluate occurrences and more detailed toxicological analyses. Many questions are open regarding the relevance of these compounds in case lactic or propionic acid are used as chemicals to stabilize contaminated feed (or are used in bread making). It should also be clarified whether the use of lactic acid bacteria starter cultures to stabilize grain in storage, or to prevent toxin buildup in barley during malting for beer production (Liske et al., 2000; Oliveira et al., 2015) could lead to formation of these modified toxins. Furthermore, lactic acid bacteria have also been suggested as biocontrol agents in the field to reduce the DON content (Byrne et al., 2022). In case relevant concentrations of the novel modified toxins indeed occur under such conditions, it seems warranted to investigate the toxicological effects in human and animal cells.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was funded by the Austrian Science Fund FWF (SFB Fusarium, F37 & P 33011).
We thank Ivan Rayment (University of Wisconsin, Madison, WI, USA) for providing the Tri101 expression vector and Konrad Domig and Johanna Brändle (BOKU, Food Microbiology and Hygiene Laboratory) for providing coded commercial strains of propionic acid bacteria. We also thank Li Guofen for translating the Chinese article first describing 3-lactyl-DON for us.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Alexander, N. J., McCormick, S. P., and Hohn, T. M. (2002). The identification of the Saccharomyces cerevisiae gene AYT1 (ORF-YLL063c) encoding an acetyltransferase. Yeast 19, 1425–1430. doi: 10.1002/yea.924
Alexander, N. J., McCormick, S. P., Waalwijk, C., Van Der Lee, T., and Proctor, R. H. (2011). The genetic basis for 3-ADON and 15-ADON trichothecene chemotypes in fusarium. Fungal Genet. Biol. 48, 485–495. doi: 10.1016/j.fgb.2011.01.003
Armando, J. W., Boghigian, B. A., and Pfeifer, B. A. (2012). LC-MS/MS quantification of short-chain acyl-CoA’s in Escherichia coli demonstrates versatile propionyl-CoA synthetase substrate specificity: comparison of PrpE enzymes in E. coli via LC-MS/MS. Lett. Appl. Microbiol. 54, 140–148. doi: 10.1111/j.1472-765X.2011.03184.x
Berthiller, F., Krska, R., Domig, K. J., Kneifel, W., Juge, N., Schuhmacher, R., et al. (2011). Hydrolytic fate of deoxynivalenol-3-glucoside during digestion. Toxicol. Lett. 206, 264–267. doi: 10.1016/j.toxlet.2011.08.006
Byrne, M. B., Thapa, G., Doohan, F., and Burke, J. I. (2022). Lactic acid Bacteria as potential biocontrol agents for Fusarium head blight disease of spring barley. Front. Microbiol. 13:912632. doi: 10.3389/fmicb.2022.912632
Dalié, D. K. D., Deschamps, A. M., and Richard-Forget, F. (2010). Lactic acid bacteria – potential for control of mould growth and mycotoxins: a review. Food Control 21, 370–380. doi: 10.1016/j.foodcont.2009.07.011
Dellafiora, L., Galaverna, G., and Dall’Asta, C. (2017). In silico analysis sheds light on the structural basis underlying the ribotoxicity of trichothecenes—a tool for supporting the hazard identification process. Toxicol. Lett. 270, 80–87. doi: 10.1016/j.toxlet.2017.02.015
European Commission (2016). Commission recommendation (EU) 2016/1319 of 29 July 2016 amending recommendation 2006/576/EC as regards deoxynivalenol, zearalenone and ochratoxin a in pet food (text with EEA relevance). Available at: http://data.europa.eu/eli/reco/2016/1319/oj/eng [Accessed September 27, 2023].
European Commission (2023). Commission regulation (EU) 2023/915 of 25 April 2023 on maximum levels for certain contaminants in food and repealing regulation (EC) no 1881/2006 (text with EEA relevance). Available at: https://eur-lex.europa.eu/legal-content/EN/TXT/?uri=CELEX%3A32023R0915.
Franco, T. S., Garcia, S., Hirooka, E. Y., Ono, Y. S., and Dos Santos, J. S. (2011). Lactic acid bacteria in the inhibition of Fusarium graminearum and deoxynivalenol detoxification: bacterial removal of deoxynivalenol. J. Appl. Microbiol. 111, 739–748. doi: 10.1111/j.1365-2672.2011.05074.x
Garcia-Cela, E., Kiaitsi, E., Medina, A., Sulyok, M., Krska, R., and Magan, N. (2018). Interacting environmental stress factors affects targeted metabolomic profiles in stored natural wheat and that inoculated with F. graminearum. Toxins 10:56. doi: 10.3390/toxins10020056
Garvey, G. S., McCormick, S. P., and Rayment, I. (2008). Structural and functional characterization of the TRI101 Trichothecene 3-O-acetyltransferase from Fusarium sporotrichioides and Fusarium graminearum. J. Biol. Chem. 283, 1660–1669. doi: 10.1074/jbc.M705752200
Hassan, Y. I., Zhou, T., and Bullerman, L. B. (2016). Sourdough lactic acid bacteria as antifungal and mycotoxin-controlling agents. Food Sci. Technol. Int. 22, 79–90. doi: 10.1177/1082013214565722
Hosokawa, M., Furihata, T., Yaginuma, Y., Yamamoto, N., Koyano, N., Fujii, A., et al. (2007). Genomic structure and transcriptional regulation of the rat, mouse, and human carboxylesterase genes. Drug Metab. Rev. 39, 1–15. doi: 10.1080/03602530600952164
Humer, E., Lucke, A., Harder, H., Metzler-Zebeli, B., Böhm, J., and Zebeli, Q. (2016). Effects of citric and lactic acid on the reduction of deoxynivalenol and its derivatives in feeds. Toxins 8:285. doi: 10.3390/toxins8100285
Johnsen, H., Odden, E., Lie, Ø., Johnsen, B. A., and Fonnum, F. (1986). Metabolism of T-2 toxin by rat liver carboxylesterase. Biochem. Pharmacol. 35, 1469–1473. doi: 10.1016/0006-2952(86)90111-5
Juodeikiene, G., Trakselyte-Rupsiene, K., Reikertaite, K., Janic Hajnal, E., Bartkevics, V., Pugajeva, I., et al. (2023). Influence of biotreatment on Hordeum vulgare L. cereal wholemeal contamination and enzymatic activities. Foods 12:1050. doi: 10.3390/foods12051050
Khatibi, P. A., Newmister, S. A., Rayment, I., McCormick, S. P., Alexander, N. J., and Schmale, D. G. (2011). Bioprospecting for trichothecene 3- O -acetyltransferases in the fungal genus Fusarium yields functional enzymes with different abilities to modify the mycotoxin deoxynivalenol. Appl. Environ. Microbiol. 77, 1162–1170. doi: 10.1128/AEM.01738-10
Kimura, M., Kaneko, I., Komiyama, M., Takatsuki, A., Koshino, H., Yoneyama, K., et al. (1998). Trichothecene 3-O-acetyltransferase protects both the producing organism and transformed yeast from related mycotoxins. J. Biol. Chem. 273, 1654–1661. doi: 10.1074/jbc.273.3.1654
Liske, R., Niessen, L., and Vogel, R. (2000). Potential of lactic acid bacteria to reduce the growth of Fusarium culmorum in the malting process. Mycotoxin Res. 16, 62–65. doi: 10.1007/BF02942983
Liu, Y., Xu, L., Shi, Z., Wang, R., Liu, Y., Gong, Y., et al. (2023). Identification of an Acinetobacter pittii acyltransferase involved in transformation of deoxynivalenol to 3-acetyl-deoxynivalenol by transcriptomic analysis. Ecotoxicol. Environ. Saf. 263:115395. doi: 10.1016/j.ecoenv.2023.115395
Lowe, R., Jubault, M., Canning, G., Urban, M., and Hammond-Kosack, K. E. (2012). “The induction of mycotoxins by trichothecene producing fusarium species” in Plant fungal pathogens methods in molecular biology. eds. M. D. Bolton and B. P. H. J. Thomma (Totowa, NJ: Humana Press), 439–455.
Maidana, L., De Souza, M., and Bracarense, A. P. F. R. L. (2022). Lactobacillus plantarum and deoxynivalenol detoxification: a concise review. J. Food Prot. 85, 1815–1823. doi: 10.4315/JFP-22-077
McCormick, S. P., Stanley, A. M., Stover, N. A., and Alexander, N. J. (2011). Trichothecenes: from simple to complex mycotoxins. Toxins 3, 802–814. doi: 10.3390/toxins3070802
Megraw, R. E., Reeves, H. C., and Ajl, S. J. (1965). Formation of lactyl-coenzyme a and pyruvyl-coenzyme a from lactic acid by Escherichia coli. J. Bacteriol. 90, 984–988. doi: 10.1128/jb.90.4.984-988.1965
Meng-Reiterer, J., Bueschl, C., Rechthaler, J., Berthiller, F., Lemmens, M., and Schuhmacher, R. (2016). Metabolism of HT-2 toxin and T-2 toxin in oats. Toxins 8:364. doi: 10.3390/toxins8120364
Meng-Reiterer, J., Varga, E., Nathanail, A. V., Bueschl, C., Rechthaler, J., McCormick, S. P., et al. (2015). Tracing the metabolism of HT-2 toxin and T-2 toxin in barley by isotope-assisted untargeted screening and quantitative LC-HRMS analysis. Anal. Bioanal. Chem. 407, 8019–8033. doi: 10.1007/s00216-015-8975-9
Michlmayr, H., Varga, E., Malachová, A., Fruhmann, P., Piątkowska, M., Hametner, C., et al. (2018). UDP-glucosyltransferases from rice, Brachypodium, and barley: substrate specificities and synthesis of type a and B Trichothecene-3-O-β-d-glucosides. Toxins 10:111. doi: 10.3390/toxins10030111
Michlmayr, H., Varga, E., Malachova, A., Nguyen, N. T., Lorenz, C., Haltrich, D., et al. (2015). A versatile family 3 glycoside hydrolase from Bifidobacterium adolescentis hydrolyzes β-glucosides of the fusarium mycotoxins deoxynivalenol, nivalenol, and HT-2 toxin in cereal matrices. Appl. Environ. Microbiol. 81, 4885–4893. doi: 10.1128/AEM.01061-15
Mirocha, C. J., Xie, W. P., and Filho, E. R. (2003). “Chemistry and detection of Fusarium mycotoxins” in Fusarium head blight of wheat and barley. eds. K. J. Leonard and W. R. Bushnell (St. Paul: American Phytopathological Society (APS Press)), 144–164.
Nathanail, A. V., Varga, E., Meng-Reiterer, J., Bueschl, C., Michlmayr, H., Malachova, A., et al. (2015). Metabolism of the Fusarium mycotoxins T-2 toxin and HT-2 toxin in wheat. J. Agric. Food Chem. 63, 7862–7872. doi: 10.1021/acs.jafc.5b02697
O’Mara, S. P., Broz, K., Schwister, E., Singh, L., Dong, Y., Elmore, J. M., et al. (2023). The Fusarium graminearum transporters Abc1 and Abc6 are important for xenobiotic resistance, trichothecene accumulation, and virulence to wheat. Phytopathology 113, 1916–1923. doi: 10.1094/PHYTO-09-22-0345-R
Oliveira, P., Brosnan, B., Jacob, F., Furey, A., Coffey, A., Zannini, E., et al. (2015). Lactic acid bacteria bioprotection applied to the malting process. Part II: substrate impact and mycotoxin reduction. Food Control 51, 444–452. doi: 10.1016/j.foodcont.2014.11.011
Oliveira, P. M., Zannini, E., and Arendt, E. K. (2014). Cereal fungal infection, mycotoxins, and lactic acid bacteria mediated bioprotection: from crop farming to cereal products. Food Microbiol. 37, 78–95. doi: 10.1016/j.fm.2013.06.003
Pawlowska, A. M., Zannini, E., Coffey, A., and Arendt, E. K. (2012). ‘Green preservatives’: combating Fungi in the food and feed industry by applying antifungal lactic acid Bacteria. Advances in Food and Nutrition Research 66, 217–238. doi: 10.1016/B978-0-12-394597-6.00005-7
Payros, D., Alassane-Kpembi, I., Pierron, A., Loiseau, N., Pinton, P., and Oswald, I. P. (2016). Toxicology of deoxynivalenol and its acetylated and modified forms. Arch. Toxicol. 90, 2931–2957. doi: 10.1007/s00204-016-1826-4
Piper, P. (1998). The Pdr12 ABC transporter is required for the development of weak organic acid resistance in yeast. EMBO J. 17, 4257–4265. doi: 10.1093/emboj/17.15.4257
Poppenberger, B., Berthiller, F., Lucyshyn, D., Sieberer, T., Schuhmacher, R., Krska, R., et al. (2003). Detoxification of the Fusarium mycotoxin deoxynivalenol by a UDP-glucosyltransferase from Arabidopsis thaliana. J. Biol. Chem. 278, 47905–47914. doi: 10.1074/jbc.M307552200
Rodrigues-Fo, E., Mirocha, C. J., Xie, W., Krick, T. P., and Martinelli, J. A. (2002). Electron ionization mass spectral fragmentation of deoxynivalenol and related tricothecenes. Rapid Commun. Mass Spectrom. 16, 1827–1835. doi: 10.1002/rcm.796
Sadiq, F. A., Yan, B., Tian, F., Zhao, J., Zhang, H., and Chen, W. (2019). Lactic acid bacteria as antifungal and anti-mycotoxigenic agents: a comprehensive review. Comp. Rev. Food Sci. Food Safe 18, 1403–1436. doi: 10.1111/1541-4337.12481
Schmeitzl, C., Varga, E., Warth, B., Kugler, K., Malachová, A., Michlmayr, H., et al. (2015a). Identification and characterization of carboxylesterases from Brachypodium distachyon deacetylating trichothecene mycotoxins. Toxins 8:6. doi: 10.3390/toxins8010006
Schmeitzl, C., Warth, B., Fruhmann, P., Michlmayr, H., Malachová, A., Berthiller, F., et al. (2015b). The metabolic fate of Deoxynivalenol and its acetylated derivatives in a wheat suspension culture: identification and detection of DON-15-O-glucoside, 15-acetyl-DON-3-O-glucoside and 15-acetyl-DON-3-sulfate. Toxins 7, 3112–3126. doi: 10.3390/toxins7083112
Seidl, B., Rehak, K., Bueschl, C., Parich, A., Buathong, R., Wolf, B., et al. (2022). Gramiketides, novel polyketide derivatives of Fusarium graminearum, are produced during the infection of wheat. J. Fungi 8:1030. doi: 10.3390/jof8101030
Shockey, J. M., Fulda, M. S., and Browse, J. (2003). Arabidopsis contains a large superfamily of acyl-activating enzymes. Phylogenetic and biochemical analysis reveals a new class of acyl-coenzyme a Synthetases. Plant Physiol. 132, 1065–1076. doi: 10.1104/pp.103.020552
Smaoui, S., Agriopoulou, S., D’Amore, T., Tavares, L., and Mousavi Khaneghah, A. (2022). The control of Fusarium growth and decontamination of produced mycotoxins by lactic acid bacteria. Crit. Rev. Food Sci. Nutr. 63, 11125–11152. doi: 10.1080/10408398.2022.2087594
Tanaka, A., Saikawa, S., Suzuki, T., Echigo, A., Maeda, K., Sato, M., et al. (2016). Acetyltransferase activity in Pseudomonas sp. capable of acetylating the C-4 hydroxyl group of nivalenol-type trichothecenes. J. Gen. Appl. Microbiol. 62, 326–329. doi: 10.2323/jgam.2016.05.002
Tsang, A. W., Horswill, A. R., and Escalante-Semerena, J. C. (1998). Studies of regulation of expression of the propionate (prpBCDE) operon provide insights into how Salmonella typhimurium LT2 integrates its 1,2-Propanediol and propionate catabolic pathways. J. Bacteriol. 180, 6511–6518. doi: 10.1128/JB.180.24.6511-6518.1998
Van Egmond, H. P., Schothorst, R. C., and Jonker, M. A. (2007). Regulations relating to mycotoxins in food: perspectives in a global and European context. Anal. Bioanal. Chem. 389, 147–157. doi: 10.1007/s00216-007-1317-9
Varga, E., Wiesenberger, G., Hametner, C., Ward, T. J., Dong, Y., Schöfbeck, D., et al. (2015). New tricks of an old enemy: isolates of Fusarium graminearum produce a type a trichothecene mycotoxin. Environ. Microbiol. 17, 2588–2600. doi: 10.1111/1462-2920.12718
Xu, Y.-C., Huang, X., and Cai, Y. (1982). Isolation and structural study of a new mycotoxin - CBD2 (translated title). Acta Microbiol Sin. 22, 35–39.
Keywords: fusarium, mycotoxin, deoxynivalenol, lactic acid bacteria, grain storage, modified mycotoxin
Citation: Michlmayr H, Wiesenberger G, Twaruschek K, Kastner F, Sopel MM, Hametner C, Berthiller F and Adam G (2024) Enzymatic synthesis of the modified mycotoxins 3-lactyl- and 3-propionyl-deoxynivalenol. Front. Sustain. Food Syst. 7:1305914. doi: 10.3389/fsufs.2023.1305914
Received: 02 October 2023; Accepted: 20 December 2023;
Published: 11 January 2024.
Edited by:
John Franklin Leslie, Kansas State University, United StatesReviewed by:
Susan McCormick, United States Department of Agriculture (USDA), United StatesCopyright © 2024 Michlmayr, Wiesenberger, Twaruschek, Kastner, Sopel, Hametner, Berthiller and Adam. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Herbert Michlmayr, SGVyYmVydC5taWNobG1heXJAYm9rdS5hYy5hdA==
†Present address
Marta M. SopelWageningen Food Safety Research (WFSR), Wageningen University and Research (WUR), Wageningen, Netherlands
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.