
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Sustain. Food Syst. , 16 November 2023
Sec. Crop Biology and Sustainability
Volume 7 - 2023 | https://doi.org/10.3389/fsufs.2023.1253735
This article is part of the Research Topic Biostimulants for Climate-Smart and Sustainable Agriculture View all 12 articles
Plants face numerous challenges in novel and harsh environments, including altered precipitation regimes, salinity, extreme temperatures, increased atmospheric CO2, nutrient deficiency, heavy metals, and oxygen. Drought remains a major constraint to crop productivity and meeting food demand, with the frequency, intensity, and duration of drought expected to raise in the coming century. The “cry for help” hypothesis proposes that timely recruiting of the microbiome by plants may confer benefits in stress alleviation, plant growth, fitness, and health. The root-associated microbiome harbors 10–100 times more functional genes than the host, which can significantly stimulate the metabolic and genetic potential of plant–microbiome assembly. However, cross-talk among drought and the root-associated microbes, and among the root-associated microbiome and the host-plant, is less well understood. Understanding the molecular aspect of multiple mechanisms by which microbes associate with plants during drought stress is of fundamental importance in plant biology and agriculture. In this review, we examine the progress in research on the response of plant and its microbiome assemblages and interactions to drought stress, including the impact of drought and root exudates on host resilience. We delve into the potential of ‘omics’ technologies to unravel the signaling networks underlying these interactions and the multiway interactions that occur among the host and its associated microbiome. We then discuss the shortfalls, challenges, and future research directions in this field. Overall, we argue that harnessing/manipulating the crop microbiome presents a promising strategy for improving agricultural systems in the face of global climate change.
Plants are continuously exposed to harsh environmental conditions, including water scarcity, salinity, heat waves, elevated CO2 levels, heavy metals accumulation, and soil poverty, which challenge their adaptability and resilience (Shen et al., 2020). Drought remains a serious impediment to crop productivity and food security, especially with the increase in its frequency, duration, and intensity under global climate change (Canarini et al., 2021; Anli et al., 2022). It disrupts plant metabolism, reduces photosynthetic activity, and induces electrolyte disturbances and reactive oxygen species (ROS) accumulation, causing cell apoptosis and plant death (Boutasknit et al., 2021; Benaffari et al., 2022; Toubali et al., 2022).
Drought stress affects plant functioning, which in turn alters plant metabolism and root exudates, influencing the plant rhizosphere microbiome. Plant exudates, such as sugars, vitamins, organic acids, amino acids, fatty acids, flavonoids, carboxylic acids, benzoxazinoids, and ethylene (ET), play a crucial role in selectively recruiting rhizospheric microbial communities (Vives-Peris et al., 2020). Phytohormones, such as jasmonic acid (JA) and salicylic acid (SA), and their signaling pathways significantly impact plant microbiome structure (French et al., 2019). Drought may also impact plant microbiome association, putting selective pressure on its components to endure stressful conditions. Inducing glycerol-3-phosphate (G3P) synthesis during drought, for example, favors Actinobacteria in the rhizosphere, which promotes plant fitness and health under water deficit (Xu et al., 2018b). Drought also induces a decline in SA production, which significantly impacts the formation of both exo- and endogen microbiome (Lebeis et al., 2015). Many investigations have revealed that during drought stress, plants promote monoderm bacteria (or gram-positive) over diderm bacteria (or gram-negative) in the rhizosphere (Naylor et al., 2017; Naylor and Coleman-Derr, 2018). Drought reduces iron and phytosiderophore availability in the rhizosphere, favoring Actinobacteria that may thrive in such environment and promote plant performances (Xu et al., 2021). A recent study by Santos-Medellín et al. (2021) revealed that drought can have long-lasting impact on the rhizospheric microbiome. They discovered that the rice root-associated microbiome structure was severely altered during a short period of water deficit but retrieved to its predrought state after recovery. However, extended water stress had serious and long-term impact on the endosphere community, which were not completely recovered even after rewatering. According to the same study, the abundance of Actinobacteria recorded a significant increase after prolonged drought, accounting for >80% of the bacterial community after the drought period. In the same vein, water shortage intervals the establishment of sorghum root microbial communities during the early development and plays a role in restructuring the root microbiome by increasing enrichment in monoderm bacteria and their activity (Xu et al., 2018b). Various studies have demonstrated that drought has a substantial impact on the activity and composition of plant root-associated bacteria in a manner that is remarkably preserved regardless of host species and sites (Karlowsky et al., 2018; De Vries et al., 2019; Williams and De Vries, 2020). The analysis of co-cited references in Figure 1 identified emerging trends and research hotspots in the field. Nine clusters were identified, with each cluster corresponding to a specific line of research. All clusters, except for “Cluster #1: Antibiotic resistance genes,” were closely related to the topic of plant-microbe interactions under drought. “Cluster #0: Water deficit” contained the majority of the nodes and has been widely reported. “Cluster #1: Antibiotic resistance genes” and “Cluster #2: Abiotic stress tolerance” were the most active areas of research. Recent research has focused on plant tolerance to water stress and their associated microorganisms, as seen in “Cluster #0: Water deficit,” “Cluster #2: Abiotic stress tolerance,” “Cluster #4: Microbial communities,” “Cluster #5: Plant responses,” and “Cluster #7: Crop resiliency”. These results emphasize the significance of understanding the role of plant-microbe interactions for mitigating the effects of drought and enhancing plant resilience. The evolving trends and research priorities in this field underscore the important role performed by plant-associated microorganisms.
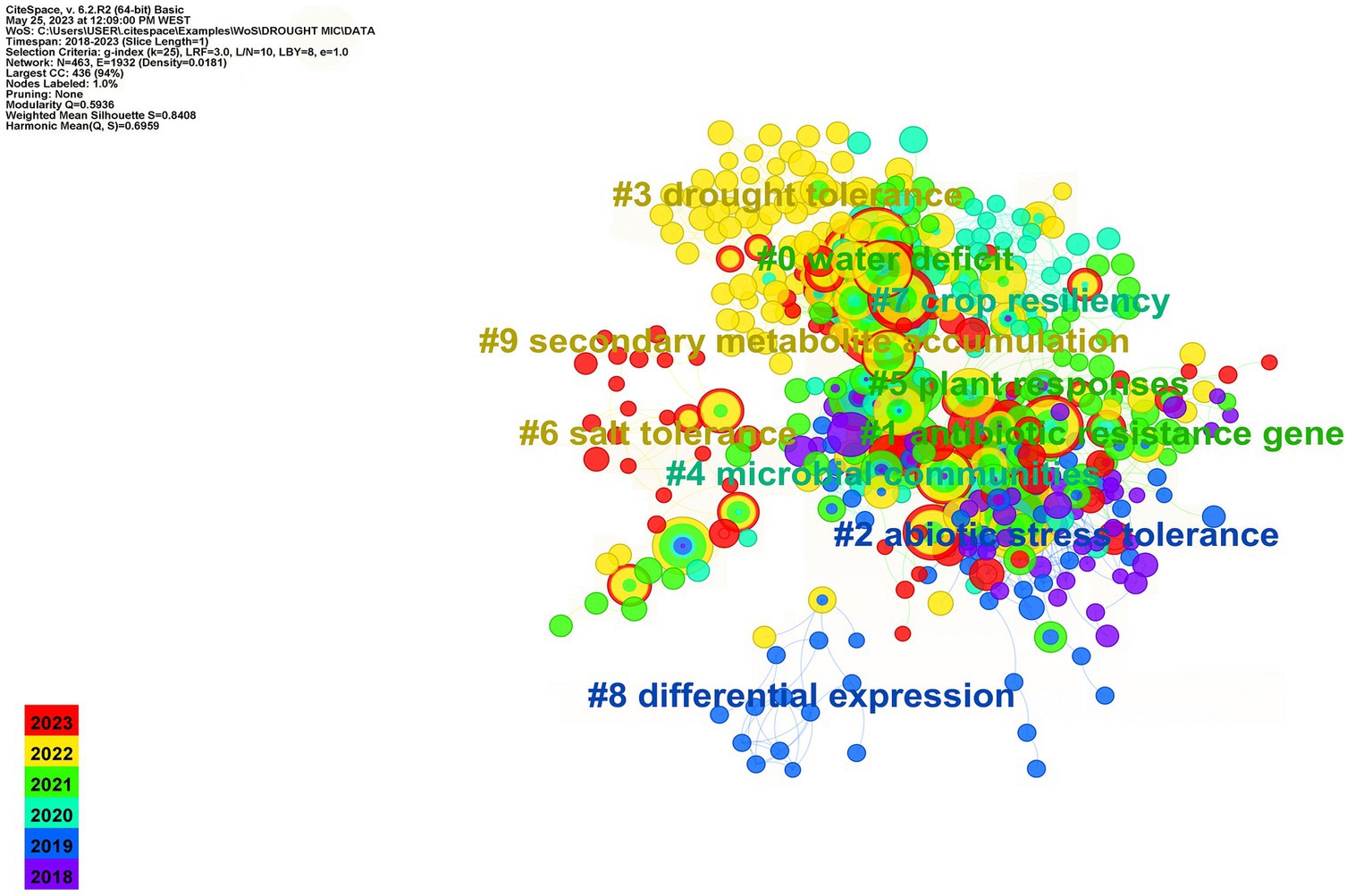
Figure 1. Co-cited references of keyword co-occurrence clustering analysis in the study of plant–microbiome interactions under drought from 2018 to 2023.
The root-associated microbiome, including plant growth-promoting fungi (PGPF) and plant growth-promoting bacteria (PGPB), may promote plants’ resistance and adaptation to water deficiency (Xu and Coleman-Derr, 2019; Table 1). Plant tolerance to drought is increased either directly by members of the root microbial community or indirectly via microbe-microbe interactions (Hartman and Tringe, 2019; Abbott et al., 2021). Microbiome may help host resist water deficit by activating transcriptional reprogramming of multiple genes and transcription factors (TFs) regulating plant defense. Microbes, for instance, have recently been shown to induce various genes expression under drought stress.
Although the composition of the plant-associated microbial communities has been studied in a great number of plant species, less interest has been paid to the effect of water shortage on the root-associated microbiota. There is a lack of understanding of the ‘cross-talk’ among drought, the root-associated microbiome, as well as between the root-associated microbial communities and the host-plant. Understanding the molecular patterns that orchestrate the assemblage of microbial communities with plants under drought stress is critical for the plant survival and fitness. Thus, knowledge of plant-microbe-drought regulation will be necessary for the design of environmentally-friendly crop management strategies and sustainable agriculture under changing environments. This review summarizes the research advances on the shaping of plant microbiome under water deficit conditions and the mechanisms by which microbiome could alleviate the adverse impact of drought on plants. Finally, we highlight the gaps, challenges, and perspectives of future research in this ‘ménage à trois’ that could help harnessing the microbiota to improve drought outcomes.
Plants continuously release exudates into their surroundings, in the form of liquid, solid, or gaseous compounds, through their leaves, shoots, or roots. Plants exuded c. 11% of the net fixed C or 27% of the C assigned to roots to the rhizosphere. The amount of these compounds varies depending on multiple factors, such as plant age, species, and nutritional performance (Bais et al., 2006; Jones et al., 2009; Nakayama and Tateno, 2018). Root exudates contain various substances, including primary and secondary metabolites and phytohormones, such as ET, JA, SA, indole 3-acetic acid (IAA), abscisic acid (ABA), gibberellic acid (GA), and cytokinins (CKs). Primary metabolites consist of sugars, amino acids, and organic acids, while secondary metabolites include flavonoids, glucosinolates, phytoalexins, triterpenes, and benzoxainoids (Badri et al., 2008; Pang et al., 2021). For instance, tomato plants exude a mixture of metabolites, including organic acids, steroidal glycoalkaloids, acylsugars, and hydroxycinnamic acid derivatives, into the rhizosphere (Korenblum et al., 2020).
Microbes diverge in their functioning and metabolism, and their sensitivity to water availability varies. Thus, drought can directly impact the assembly of plant microbiome. Plant surface microbiome, i.e., phyllosphere, is likely to be significantly altered by drought since environmental conditions vary more rapidly than those inside plant tissue (i.e., endosphere), which is more stable (Trivedi et al., 2020). Most of the microbes from the bulk soil (a source of potential microbes to colonize plant roots) are directly influenced by external climatic factors, including drought. On the other hand, rhizosphere microbiome are directly altered by these factors and indirectly by plant responses, such as changes in host morphology, physiology, immune system, and root exudation (Figure 2). Previous studies suggest a consistent response of the host-associated microbiome to water deficiency (Vescio et al., 2021; Wipf et al., 2021; Aslam et al., 2022; Tebele et al., 2023). Under water limitation, several plant species recruit monoderm bacteria that are resistant to dryness owing to their thicker cell walls and reduce diderm bacteria in the roots and rhizosphere (Naylor et al., 2017; Naylor and Coleman-Derr, 2018).
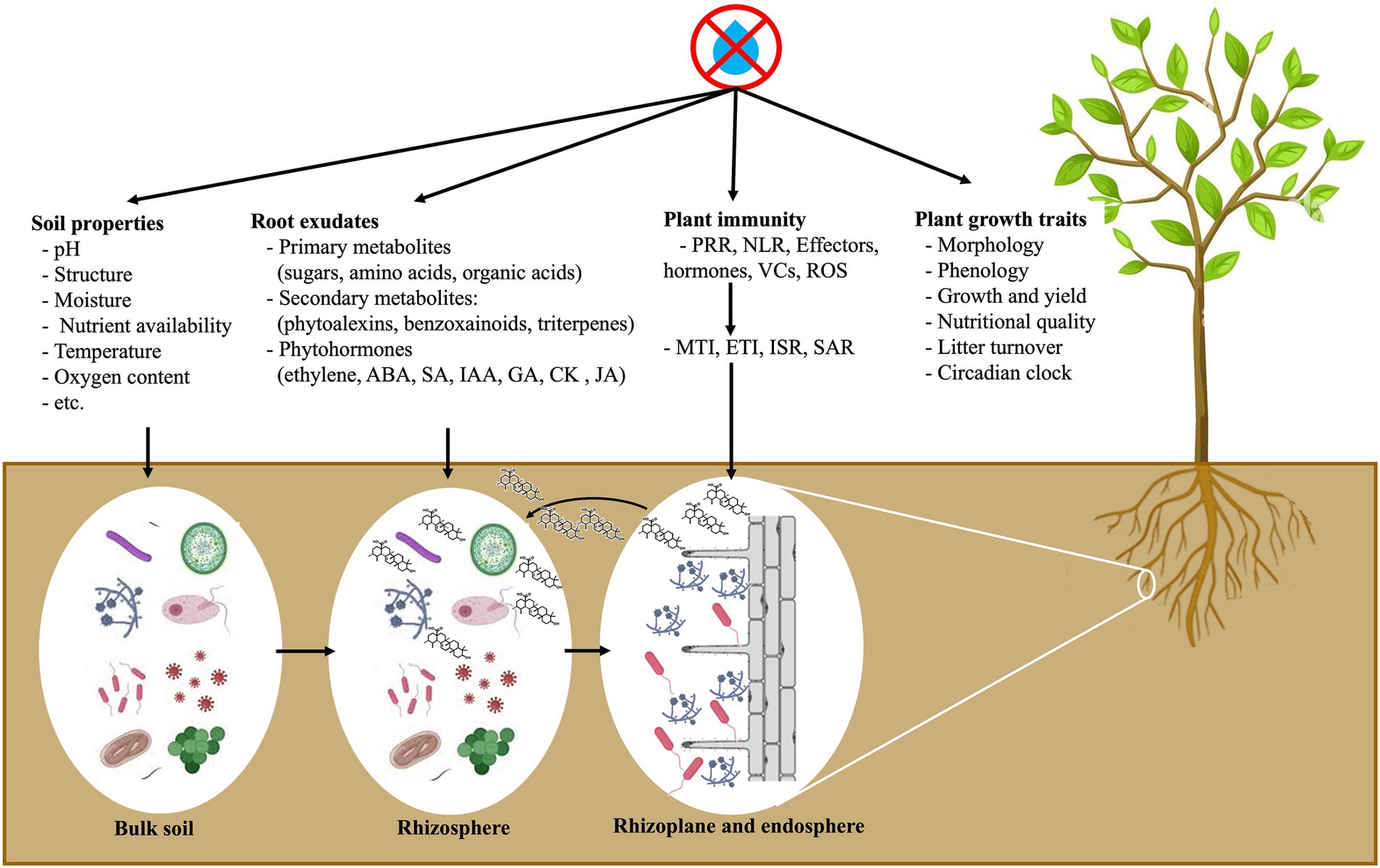
Figure 2. Drought-mediated rhizosphere microbiota recruitment: roots actively regulate rhizosphere microbial structure by secreting specific chemicals. ABA, abscissic acid; CK, cytokinin; ETI, effector-triggered immunity; GA, gibberllic acid; IAA, indole-3-acetic acid; ISR, induced systemic resistance; JA, jasmonic acid; MTI, microbe-associated molecular patterns (MAMP)-triggered immunity; NLR, leucine-rich repeats; PRR, plant pattern-recognition receptors; ROS, reactive oxygen species; SA, salicylic acid; SAR, systemic acquired resistance; VCs, volatile compounds.
Understanding how drought stress affects plant–microbiome assemblage is still a challenging task owing to the multilayers and complex interconnections that orchestrate this ‘triangle’ interaction. Chemical signals exchange heavily influences plant-microbiome communication under water scarcity (Figure 2). For example, under stressful environments, plants have evolved a ‘cry for help’ mediated exudation, resulting in the recruitment of stress-mitigating microbes (Liu et al., 2020). Plants have a complex microbial recruiting system to select the most beneficial microorganisms to incorporate into plant tissues, discriminating between friendly and beneficial or hostile and harmful interactions (Hacquard et al., 2017; Teixeira et al., 2019; Zhang et al., 2023). The host plant possesses protective mechanisms to perform this selection: (i) pattern recognition receptors (PRRs), the initial mechanism of defense to identify microbe-associated molecular patterns (MAMPs), such as fungal chitin or bacterial flagellin, leading to MAMP-triggered immunity (MTI) and (ii) nucleotide-binding leucine-rich repeat (NLR) proteins as a second defense mechanism to identify pathogen effectors, inducing effector-triggered immunity (ETI). Changes in plant immunity caused by drought could structure the plant-microbiome assembly, especially inside host plant tissues. Suppression of ETI may interfere with host-mediated management of microbe colonization and may result in microbial dysbiosis inside plant tissues. The suppression of ETI can also be a new pattern used by plants to lessen their defense mechanisms, allowing beneficial microbes to colonize roots and promote stress mitigation-related gene transcripts.
The mutual communication involving the plant defense system and the microbiome structures the plant-microbiome assemblage. Plants control immune response in rapidly changing environmental conditions using active but tightly controlled modifications in different hormone pathways (i.e., ET, SA, ABA, and JA; Li et al., 2021; Ait-El-Mokhtar et al., 2022). Lebeis et al. (2015) reported that drought reduces SA production, which is involved in the assembly of both endophytic and epiphytic microbiome. SA can promote or inhibit microbial community growth directly on microbiome members or through established signaling pathways based on hormonal cross-talk. For instance, ABA-induced production under water scarcity interferes with the SA-mediated immune pathway. Drought-mediated alterations in plant hormones may differ depending on the plant developmental stage and tissue type. For example, maize grown under water limitation boosts benzoxazinoid defense system in plant aboveground part while stimulating terpenoid phytoalexins in the belowground tissues (Vaughan et al., 2018). Alterations in the distribution or allocation of diverse signaling molecules or defense metabolites as a result of drought may have an additional impact on microbiome assembly.
It is worth noting that in host-microbiome research, core-and-hub microbiota concepts are gaining traction (Singh B. K. et al., 2020). These refer to the microbiota that exist in a specific species regardless of environmental conditions, growing season, or management practices and perform critical host functions (Trivedi et al., 2020). Given their significance, it is crucial to understand how drought affects the ‘core-and-hub’ microbiota that may organize community-scale functions in plant-microbiome communications. An enlarged understanding of the ecological vectors that orchestrate microbiome response to water scarcity will promote our knowledge of microbiome traits that boost plant performances under changing environments.
Plant and microbiota have a two-way communication, both underground and aboveground, that enables them to sense and respond to stress conditions. In response to stress, plants release a range of metabolites that attract specific microorganisms capable of enhancing their tolerance to stress (Bai et al., 2022). According to the ‘cry for help’ hypothesis, plants recruit particular microbiome communities that aid them in managing stress (Liu et al., 2020). This concept was first observed when plants grown in soils deficient in phosphorus and nutrient-supplying arbuscular mycorrhizal fungi (AMF), and nitrogen recruited nitrogen-fixing rhizobia (Carbonnel and Gutjahr, 2014; Nishida and Suzaki, 2018). The ‘cry for help’ assumption is also applicable to plants experiencing drought stress, as the microbiome composition in roots significantly changes by promoting actinobacteria and other gram-positive bacteria over gram-negative ones (Timm et al., 2018). Plants may selectively recruit drought-tolerant microbes that have evolved from repetitive drought periods, resulting in beneficial and efficient plant-microbiome interactions that enhance the performance of both host and microbes (Naylor and Coleman-Derr, 2018). Terhorst et al. (2014) demonstrated that drought-stressed Brassica rapa plants exhibit higher and more diverse bacterial richness around their roots in comparison to the controls.
Santos-Medellín et al. (2021) revealed that the above- and below-ground microbiome undergo a plant-driven change in response to water limitation, resulting in an increase in drought-tolerant endophytic monoderm bacteria that may help mitigate drought. The same authors demonstrated how persistent drought can permanently impede plant endophytic microbiome growth. This effect persists even after the drought constraint is alleviated. Their research revealed how long-lasting drought stress may shift microbial community composition, which may affect plant fitness. They also identified promising candidates for microbiome engineering to create performant microbial assemblages against water deficit, including drought-resistant endophytic microorganisms that increased in abundance in the endosphere after drought stress. Active microorganism recruitment under stressful conditions appears to be a common evolutionary mechanism to promote plant performances. Nonetheless, the strategies that allow plant host to incorporate external signals during symbiotic microbes’ recruitment and the host genetic characters controlling this recruitment are still under investigations. These processes are orchestrated by multilayer components involving plants, microorganisms, soil, and environmental traits that shape the final result. The accumulation of stress-related factors in plant roots during drought, including G3P and pipecolic acid, has been linked to actinobacteria enrichment in the rhizosphere (Knight et al., 2018; Caddell et al., 2020; Table 2). Understanding the underground signals communication is still in its early stages, particularly in drought-stressed hosts that shape their tolerant microbes.
A dynamic understanding of the role of metabolites and their genomic features is essential to comprehend the multilayered complexity of the ‘cry for help’ theory in host microbe assembly before and after drought conditions. This will provide new insight into creating drought-tolerant microbial consortia for sustainable agriculture. Recently, Bai et al. (2022) proposed a model based on the ‘cry for help’ hypothesis for plant microbiome recruitment under water scarcity (Figure 2). Under drought conditions, plants undergo molecular and metabolic readjustments and produce specific root metabolites. The root exudates may promote the reprogramming and restructuring of the microbiome by recruiting selective drought-tolerant microorganisms with a vast arsenal of functional enzymes. Subsequently, drought-resistant microbiota can mitigate stress impact and deliver nutrients to host plants via multiple direct and indirect mechanisms.
The plant and its associated microbiome form a harmonized and functional entity known as a holobiont (Bordenstein and Theis, 2015), in which evolutionary selection occurs not only between the host and the associated microbes but also among the microbes (Ait-El-Mokhtar and Baslam, 2023). To maintain the harmony of this association, systems of coordination among the microbial communities and with the host plant are necessary. Our understanding of the acquisition of microbes in the environment and the rules governing their association in the plant holobiont is very limited. It is widely believed that during root penetration into bulk soil, the soil microbiome progressively differentiates into the rhizosphere microbiome via contact with rhizodeposits, which have a significant effect on the microbiota composition (Tian et al., 2020; Figure 2). After this initial community shift, the microbiota composition is finely adjusted in specific compartments (rhizosphere, rhizoplane, or endosphere; Bulgarelli et al., 2013). Plant genetic factors, including root exudate quality and quantity and root morphology, have been known to shape the rhizospheric microbial communities (Sasse et al., 2018). Other factors, including the plant developmental stage, plant immune system, and season, have also been shown to play a significant role in shaping the rhizosphere microbiome (Hassani et al., 2018). The presence of a core microbiota in some crops, irrespective of fertilization management or soil origin, suggests that these communities are partly assembled and selected by plants. In general, it is believed that the plant selects its microbial associates via the action of its root exudates. Yet, this unidirectional recruitment is being questioned (Uroz et al., 2019) because the co-evolution of the plant-microbe holobiont suggests bidirectional interactions, particularly in terms of rhizosphere microbiome shaping (Figure 3).
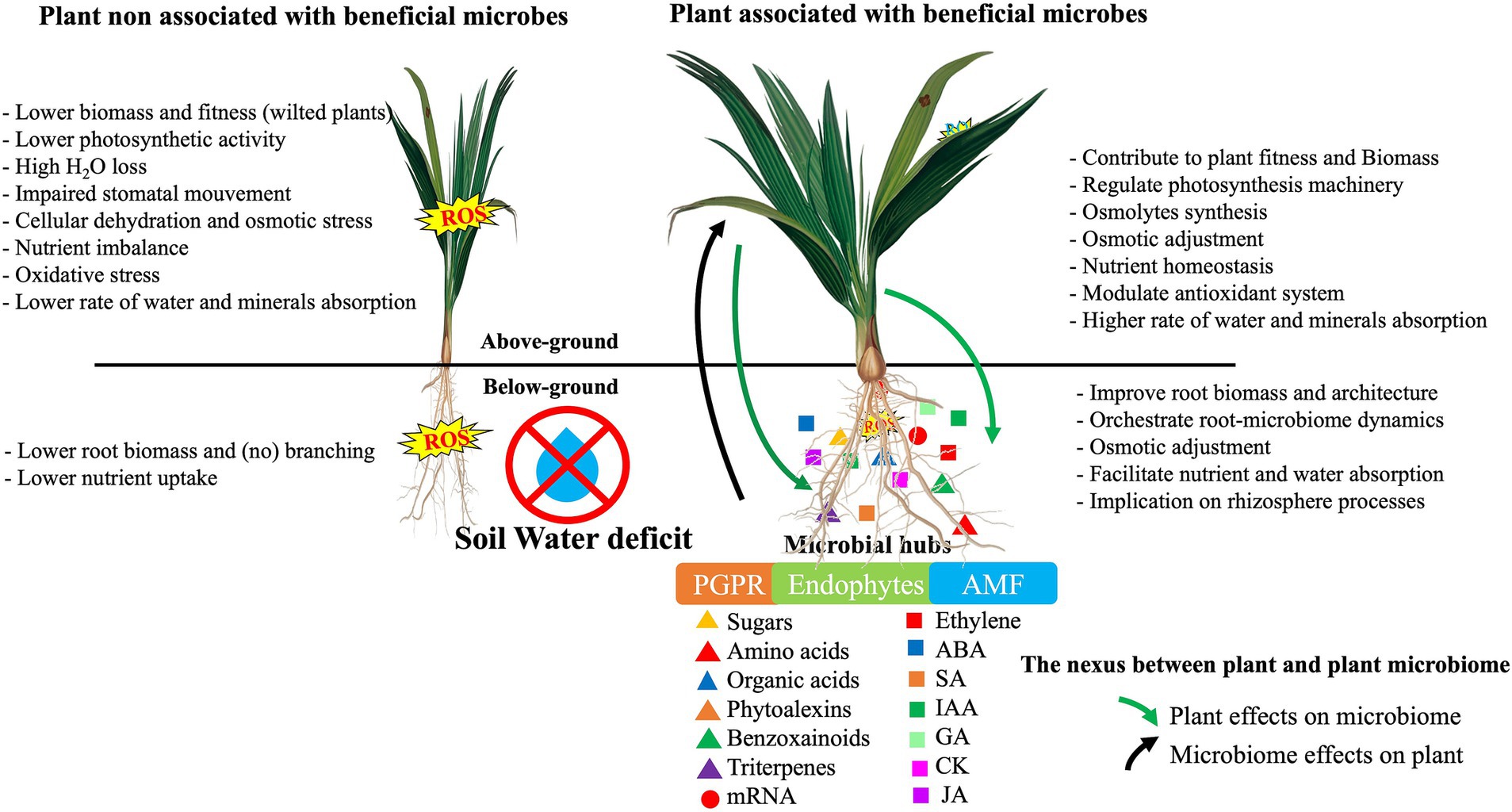
Figure 3. Overview of the mechanisms by which plant mediates rhizosphere microbiota to mitigate drought through signalling integrators and metabolic changes. ABA, Abscissic acid; CK, Cytokinin; GA, Gibberllic acid; IAA, Indole-3- acetic acid; JA, Jasmonic acid; mRNA, microRNA.
Conventionally, root exudate secretion was thought to be a passive process facilitated by various pathways, such as diffusion across the root cell membrane, ionic channels, and vesicle transport (Baetz and Martinoia, 2014). The chemical characteristics of the exuded molecules determine the type of the secretion pathway. Diffusion is involved in the exudation of metabolites with low molecular weight, such as amino acids, sugars, carboxylic acids, and phenolics. This process is caused by the difference in the concentrations of compounds between root cell cytoplasm and rhizosphere, and it may be influenced by root membrane permeability, root cell integrity, and compound polarity (Badri and Vivanco, 2009). Ionic channel pathways are involved in the exudation of carbohydrates and particular carboxylates, including oxalate and malate (released in large amounts), which are transported through membranes via a transport mechanism performed by proteins rather than diffusion. Two types of anionic channels are involved in this process: SLow Anion Channels (SLACs), which are activated in many seconds, and QUick Anion Channels (QUACs), which take a few milliseconds to be activated (Dreyer et al., 2012). The third form of passive transport pathways is vesicle transport (exocytosis) responsible for the exudation of high molecular weight metabolites kept inside vesicles (Badri and Vivanco, 2009). Exuded compounds are produced by the endoplasmic reticulum or the Golgi apparatus and aid in pathogen defense (Weston et al., 2012). In contrast, the active transport pathway of root-secreted metabolites is mediated by plasmatic membrane proteins (Baetz and Martinoia, 2014): ATP-Binding Cassette (ABC) and Multidrug and Toxic Compound Extrusion (MATE) (Kang et al., 2011). Protein-mediated root exudation may take three forms and depends on their specificity: transporters exuding multiple compounds, compounds secreted via various membrane transporters in the rhizosphere, and metabolites secreted by a single transporter. ABC transporters are classified as primary transporters owing to the use of ATP hydrolysis for the necessary energy to transport varied solutes (Jones and George, 2002; Orelle et al., 2018). The gene names encoding these transporters have evolved over time. Nevertheless, another classification has been established based on the organization of TMD and NBD domains, assembling the various members into nine families termed with letters ranging from A to I, despite the fact that family H does not exist in plants (Verrier et al., 2008). Some studies have looked into the involvement of ABC transporters in root exudation and highlighted their significance in this process (Badri et al., 2008, 2009; Olanrewaju et al., 2019).
Recent studies have shown that shifting from naturally occurring microbiome to adapted microbial communities to the stressful condition enables the plant holobiont to quickly adapt to changing environments (Pantigoso et al., 2022; Faist et al., 2023; Li et al., 2023). Rice provides an example of plant-mediated microbial abundance modulation, where under water deficiency, the microbiome composition shifts to a greater abundance of drought-resistant plant growth promoting rhizobacteria (PGPR) strains (Santos-Medellín et al., 2017). Changes in root exudation caused by water shortage may be the source of the enhancement of several types of microorganisms. Stressful soil conditions, along with microbial composition changes, can influence rhizosphere microbial activity. The modulation of the rhizosphere microbiome is influenced not only by plants but also by microbes. By exuding phytohormones, antimicrobials, volatile compounds (VCs), and quorum-sensing, plants may regulate the plant environment and even benefit their hosts (Venturi and Keel, 2016). Under water limitation conditions, the rhizosphere microbiome is shaped by microbial interactions among members and their preferences for specific metabolites (Zhalnina et al., 2018). Microbial interactions may alter gene expression within communicating microbes (Sasse et al., 2018), indicating that microbial interactions affect both the function and shape of the microbiome. According to Uroz et al. (2019), both partners are ecological engineers of the holobiont because they regulate plant-associated microbes. Therefore, microbial communications (and plant-modulating microbiome) in the rhizosphere play a critical role in shaping plant-associated microbiome.
Using metatranscriptomic analysis to study the wheat holobiont under water deficiency, Pande et al. (2023) revealed that the microbial associates were the most responsive to water deficit. The majority of the differentially abundant (DA) genes were associated with bacteria in the rhizosphere and fungi in the roots when comparing drought-stressed wheat and control treatments. In the rhizosphere, Actinobacteria were overrepresented in positively regulated DA transcripts, while Acidobacteria and Proteobacteria were overrepresented in negatively regulated ones. These authors demonstrated that certain transcripts were more abundant in the roots and rhizosphere under severe water stress, including heat shock proteins (HSPs), as well as carbohydrate and amino acid transport and metabolism-related transcripts. Another metatranscriptomic study reported that drought stress promoted the transcripts of Proteobacteria and Bacteroidetes, while reducing those of Actinobacteria and Acidobacteria (Tartaglia et al., 2023). This study also indicated an over-expression of universal stress proteins by Proteobacteria and Bacteroidetes in water-stressed treatment compared to the control. Furthermore, Xu et al. (2018a,b) revealed greater enrichment in various Actinobacteria and Chloroflexi taxa and a decline in the abundance of different Acidobacteria and Deltaproteobacteria taxa under drought conditions. This change was coupled with an increase in G3P-related transcripts in Actinobacteria.
In this cross-kingdom communication, there has been a recent surge of interest in small RNAs, specifically microRNAs, as active ‘hormone-like’ mediators in cell-to-cell communication (Leitão et al., 2020). Arabidopsis and Botrytis cinerea, a fungal pathogen, were the first to demonstrate the bi-directional cross-kingdom RNAi via sRNA trafficking (Wang et al., 2016). Still, multiple studies are emphasizing the function of microRNA exchanges in non-pathogenic associations with a focus on the host-microbiota communication in the gut (Liu et al., 2016; Bi et al., 2020; Casado-Bedmar and Viennois, 2022). Intestinal epithelial microRNAs have been reported to influence the composition of the gut microbiome by penetrating specific bacteria and controlling the transcription of a large number of genes involved in sugar degradation and housekeeping, thereby influencing the bacterial fitness (Gao et al., 2019). After their ingestion, plants secrete extracellular vesicles (EVs) in the gut, where specific bacteria can uptake them with their microRNA, causing an alteration in their gene expression (Mu et al., 2014; Teng et al., 2018). As a result, their metabolite production and secretion could be modified, resulting in differential growth of bacterial strains that interact with these specific bacteria. Following these advances, Middleton et al. (2021) propose that plants and their associated rhizosphere microbes interact through microRNAs, that modulate the composition and activity of rhizospheric microbiota. Strikingly, hundreds of microRNAs have been found in host root tissues (Breakfield et al., 2012). It is therefore assumed that some microRNAs, selected through co-evolution with nearby microorganisms, could be secreted through EVs structure in the rhizosphere.
Taking a step further, Escudero-Martinez et al. (2022) applied metagenomics data to map the plant genetic factors regulating microbiota in the rhizosphere of domesticated and wild barley genotypes. The authors identified a small number of loci that have a significant impact on the rhizospheric microbiome composition. One of those loci, called QRMC-3HS, emerged as a major determining factor of microbiota composition. A comparative root RNA-seq profiling of soil-grown sibling lines with contrasting QRMC-3HS alleles and presenting distinct microbiotas enabled the identification of three primary candidate genes: Nucleotide-Binding-Leucine-Rich-Repeat (NLR) gene and two other genes from QRMC-3HS. The NLR gene encodes an NLR protein, one of two types of immune system receptors involved in the microbe proliferation recognition through effector recognition (Jones and Dangl, 2006). The first gene of QRMC-3HS is differently regulated in sibling lines with different microbiotas, and it is unclear how it mechanistically contributes to rhizospheric plant-microbiota interactions because it encodes an unknown protein. The second gene encodes a xyloglucan endotransglucosylase/hydrolase (XTH) enzyme involved in the cleavage and/or rearrangement of xyloglucans (Yang et al., 2019), which are the most abundant hemicellulosic compounds in plant primary cell walls (Ezquer et al., 2020). The XTH gene may still be involved in microbiota shaping through cell wall polysaccharide modification closely related to plant-microbe interactions (Vorwerk et al., 2004). In Arabidopsis, cell wall traits serve as recruitment cues for almost 50% of the endogenous root microbial communities (Bulgarelli et al., 2012). In addition, cell wall alterations underpin some of the gene ontology classes identified in genome-wide association mapping experiments carried out using this plant (Horton et al., 2014). Improved adaptation to soil physicochemical conditions may represent further involvement of XTH genes in plant-microbiome interactions. Han et al. (2017) previously reported that XTH genes are involved in drought stress tolerance.
As previously discussed, soil microbes provide various benefits to plants, with root-associated microbiome playing a key role in determining host fitness and performance under various environments, including drought. Root microbiome composition is influenced by both plant and environmental factors. Plant-associated microbiome may boost host development under drought stress by stimulating different layers of plant tolerance mechanisms (Table 1). Among these microbes, PGPRs are abundant in the rhizospheric area and have been shown to be one of the most successful strategies for mitigating the adverse effect of water shortage on the host plant (Etesami and Jeong, 2018). PGPRs enhance crop plant resistance by increasing the production of osmolytes (including glycine betaine and proline), accumulating secondary metabolites, and modulating the expression of a myriad of host drought-related genes.
Vurukonda et al. (2016) revealed that PGPRs associated with stressed plants are crucial for inducing systemic tolerance (IST). A metabolome study of drought-stressed Sorghum bicolor primed with Bacillus and Pseudomonas isolates showed an induction of signature metabolic profiles and biomarkers related to IST in the host (Carlson et al., 2020). The findings revealed substantial treatment-related differential metabolic reprogramming among rhizobacteria-treated and control plants. This was correlated to the ability of the selected isolates to preserve host plants against drought stress by up-regulating IAA, CK, SA, GA, JA, brassinosteroïds (BRs), sphingosine, psychosine, osmolytes and antioxidants, and down-regulating ET production. Raheem et al. (2018) found that IAA and other auxins produced by PGPR bacteria enhance plant performance under drought stress, as demonstrated by Bacillus amyloliquefaciens, an auxin-producing bacteria isolated from Acacia arabica rhizospheric areas in arid climate. The production of exopolysaccharide by PGPRs is another cue inducing plant drought tolerance. Using a Bacillus amyloliquefaciens exopolysaccharide-deficient mutant (epsC), Lu et al. (2018) demonstrated that the epsC gene is a key gene involved in drought tolerance in Arabidopsis (Table 3). The authors highlighted the effect of ET and JA in inducing IST and up-regulating many drought-resilient genes (including ERD1, LEA14, RD17, and RD29A) in leaves. Staudinger et al. (2016) used proteomics to show that Sinorhizobium sp -inoculated Medicago truncatula grown under drought induced hormonal crosstalk and increased JA translational regulation, playing a role in increased leaf maintenance in nodulated plants during drought. PGPRs have been shown to accelerate the antioxidant enzymes and osmoprotectants biosynthesis in drought-stressed plants. By using various formulations of PGPRs isolated from Megathyrsus maximus rhizosphere in dry areas, Moreno-Galván et al. (2020) showed that the isolates improved Megathyrsus drought tolerance by up-regulating proline biosynthesis and down-regulating malondialdehyde (MDA) concentration and glutathione reductase activity. Recently, Singh D. P. et al. (2020) revealed that the overexpression of genes encoding enzymes responsible for phenylpropanoid has been linked to ROS scavenging in rice plants inoculated with Pseudomonas and Trichoderma and grown under water scarcity conditions. Similarly, upregulation of genes encoding PiP, DHN, and DREB contributes to the improvement of drought resistance in plants treated with PGPRs. Drought-stressed tomato plants treated with PGPRs yielded enhanced growth traits, water status, proline biosynthesis, antioxidant enzyme defense (i.e., ascorbate peroxidase, catalase, and glutathione peroxidase), and H2O2 accumulation compared to the controls (Abbasi et al., 2020). The authors demonstrated that PGPR-treated tomato plants showed altered stress signaling and regulatory networks controlling the expression of target genes related to plant response to drought, such as ERF1 and WRKY70. Woo et al. (2020) reported that Bacillus subtilis boosted drought resilience in Arabidopsis and Brassica by up-regulating the expression of drought-sensor genes, including NCED3, RAB18, RD29B, and RD20 in Arabidopsis and WRKY7, CSD3, and DREB1D in Brassica.
Several studies have shown that specialized microbiome may alleviate plant drought stress (Mathur and Roy, 2021; Aslam et al., 2022; Singh et al., 2023). Endophytes, microorganisms living in the endosphere without inducing disease symptoms, confer stress resilience to host species and perform a significant function in the survival of some plants in high-stress environments. Various bacterial and fungal endophytes have been reported to increase plant drought resilience. Bacterial endophytes isolated from drought-stressed maize roots improved plant fitness due to biosynthesis of IAA, GA, and CK, 1-aminocyclopropane-1-carboxylate (ACC) deaminase activity and siderophore biosynthesis (Sandhya et al., 2017). Drought-stressed sorghum plants treated with a consortium of root bacterial endophytes (Enterobacter cloacae, Enterobacter sp., Ochrobactrum sp., and Microbacterium sp.) resulted in higher growth and osmotic adjustment (Govindasamy et al., 2020). Drought tolerance mediated through microbial inoculation has been associated with enhanced proline accumulation and proline-related biosynthesis genes, SbP5CS 1 and SbP5CS 2. Interestingly, Vigani et al. (2019) found that root endophytic bacteria conferred drought resistance in Capsicum annuum by modifying vacuolar H+ pyrophosphatases (VPP) expression, which aided in preserving osmotic balance. Strains of Bacillus endophytes found in Lepidium perfoliatum roots were responsible for the formation of biofilm on roots, resulting in drought resistance and seedling germination (Li et al., 2017). Piriformospora indica, a well-known root fungal endophyte, colonizes the roots of many plant species and provides multifaceted amenities, including drought alleviation (Gill et al., 2016). Metabolome and proteome profiling analysis of barley plants inoculated with P. indica- grown under drought stress showed that inoculation redistributes resources, maintains the aquaporins (AQPs) presence, and promotes energy modulation, photorespiration protective proteins and transporters production, primary metabolism, and autophagy in water-stressed plants (Ghaffari et al., 2019). Inoculation of rice roots with P. indica increased growth, biomass accumulation, mineral nutrition (Zn and P), and upregulated P5CS genes in drought-stressed plants (Saddique et al., 2018). González-Teuber et al. (2018) reported that inoculating the roots of Chenopodium quinoa with the fungal endophyte Penicillium minioluteum primarily conferred drought tolerance through significant adjustments in below-ground biomass, photosynthesis, water-use efficiency (WUE), and photochemical efficiency.
AM symbiosis is one of the most complex and mutualistic interactions that plants have evolved to cope with droughts. Following a molecular dialog between the two partners involving “branching factors,” “mycorrhizal factors,” and common symbiotic signaling pathways, the AMF enters the plant roots and establish arbuscule formations ensuring nutrient exchange. Plant hormones act as central regulators of the development of the plant-AMF interaction (Santner et al., 2009; Charpentier et al., 2014; Etemadi et al., 2014), and auxins have potential key role in this interaction (Takeda et al., 2015; Jin et al., 2016). The modulation of AMF-mediated host drought tolerance is an extremely intricate process implicating multiple metabolites and pathways. Particularly, AMF can directly improve nutrients and water absorption and transport, boost host osmotic regulation, and increase plant gas exchange capacity, WUE, and antioxidant defense (Osakabe et al., 2014; Ruiz-Lozano et al., 2016). Zhang et al. (2019) found that a combination of Funneliformis mosseae, Rhizoglomus intraradices, and Diversispora versiformis improved the survival of Zenia insignis subjected to water scarcity through regulating osmolytes accumulation, antioxidant enzyme activity, and plant N and P uptake. Drought-stressed trifoliate oranges inoculated with F. mosseae have less oxidative stress via the increase of H2O2 efflux from the root system (Santner et al., 2009; Jin et al., 2016). In addition, the regulation of polyamine metabolism-associated gene transcripts by AMF has been identified to play a pivotal role in drought resilience (Zhang et al., 2020). Taking a step further downstream, several AM-specific host genes and proteins have been reported to play a key role in promoting plant water stress resilience. The different mechanisms involved in conferring drought resilience to mycorrhizal plants have been previously reviewed (Cheng S. et al., 2021; Wang et al., 2023). Drought-induced genes and compounds can be divided into: functional genes playing a direct role in stress [i.e., late embryogenesis abundant (LEA) proteins, AQPs, sugar, and proline] and regulatory genes implicated in the regulation of gene expression and the transduction signals (i.e., stress-related TFs) and signal molecules, such as calmodulin-binding protein. Many aspect of the molecular regulatory network linking AM symbiosis and drought stress have been elucidated, including the identification of several gene/protein functions (Bahadur et al., 2019; Ho-Plágaro and García-Garrido, 2022). Various TFs, such as AP2/ERF family, GRAS family, and MYB family have been found to be specifically induced by AMF under drought conditions, thereby contributing to the modulation of stress by phytohormones and other molecular signaling pathways (Wang et al., 2023). Jia-Dong et al. (2019) revealed that AM symbiosis significantly activated the expression of root tip AQPs PtTIP1;2, PtTIP1;3, and PtTIP4;1 in drought-stressed trifoliate orange. In the same vein, it has been reported that drought stress induced the up-regulation of eight AQP-associated genes in AMF-treated plants (Recchia et al., 2018). An interactive impact of AMF and drought was recorded on the over-expression of MAPK pathway gene that triggers an improvement in photosynthetic efficiency, osmolyte production, and antioxidant defense (Huang et al., 2020). Testerink and Munnik (2005) showed the role of ABA in up-regulating resistance genes expression to mitigate drought-induced damages. Under water limitation, lettuce inoculated with G. intraradices exhibited high expression of the Lsnced gene, which encodes a pivotal enzyme in the production of ABA. In contrast, the expression of this gene in roots was not affected by the exogenous application of ABA. This suggests that mycorrhizal plants adjust the endogenous ABA levels more efficiently and quickly than non-AM plants, leading to a more appropriate equilibrium between water acquisition and leaf transpiration under water deficit (Aroca et al., 2008). In a proteomic study on wheat, differential expression proteins involved in cell wall integrity and carbohydrate production were observed, while most stress-associated molecules, including enzymes involved in ET biosynthesis, were downregulated (Bernardo et al., 2017).
Recently, Poveda (2020) found an increase in the secretion of the fungal enzyme chorismate mutase by using mutant strains of Trichoderma parareese. This increase conferred tolerance to rapeseed plants grown under drought conditions by increasing gene expression (i.e., NCED3 and PYL4) related to the hormonal pathways of ABA. Notably, colonization of Trichoderma sp. on rapeseed roots increased under stressed condition, initiating a myriad of host metabolic pathways. Comparative qRT-PCR analyzes with the chorismate mutase-silenced strain connected this enzyme to drought-tolerant mechanisms owing to its involvement in ACCO1 (1-Aminocyclopropane-1-carboxylic acid oxidase) and ERF1 downregulation, and ABA pathway genes NCED3 upregulation. In addition, Bashyal et al. (2021) used transcriptome profiling of droughted- Trichoderma harzianum-inoculated rice and revealed the upregulation of 1,053 genes and the downregulation of 733 genes in stressed T. harzianum-inoculated plants. Most photosynthetic and antioxidative genes, including plastocyanin, PSI subunit Q, PSII subunit PSBY, small chain of Rubisco, proline-rich protein, osmoproteins, stress-induced proteins, AQPs, and chaperonins, were exclusively expressed in stressed T. harzianum-inoculated rice. Using the enrichment analysis, the same authors showed that the metabolic (38%) and pathways involved in the synthesis of secondary metabolites (25%), phenyl propanoid (7%), carbon metabolism (6%), and glutathione metabolism (3%) were the most enriched pathways.
The co-inoculation with different members of plant microbiome was also reported to enhance host drought tolerance. Eshaghi Gorgi et al. (2022) studied the effect of the dual application of PGPR and AMF on biomass accumulation, water status, photosynthetic pigments, and proline content of drought-stressed Melissa officinalis. They demonstrated that the combined microbial treatment increased all these parameters under drought stress. The same authors also reported that leaves chemical composition of secondary metabolites was altered by PGPR+AMF inoculation. Another study showed that the co-inoculation of Common myrtle with Funneliformis mosseae and Rhizophagus irregularis AMF strains and Pseudomonas fluorescens and P. putida PGPR strains boosted seedlings survival, growth fitness, and (non-)enzymatic antioxidant accumulation while reducing electrolyte leakage, and MDA and proline concentrations under drought stress (Azizi et al., 2021). In the same vein, the combined application of Glomus versiforme and Bacillus methylotrophicus recorded significant drought resistance through improving growth and photosynthetic performances, nutrition status, phenols and flavonoids accumulation, antioxidant enzymatic system and ABA and IAA concentrations in tobacco plants under drought stress (Begum et al., 2022). Singh D. P. et al. (2020) revealed that Trichoderma and Pseudomonas primed rice seeds induced significant increase in the transcript levels of multiple genes involved in the antioxidant enzymes and phenylpropanoid biosynthesis pathway in seedlings grown under drought stress.
Several studies have revealed the significant beneficial effects of combining microbes and others biostimulants in mitigating drought stress. The application of AMF and organic amendment improved the tolerance of pistachio seedlings to drought stress through enhancing soil physicochemical and biological traits, as well as plant nutrient uptake (Paymaneh et al., 2023). Soussani et al. (2023) demonstrated that the application of AMF and/or compost promoted tomato growth, yield and fruit bioactive compounds, while reducing oxidative stress and enhancing the efficiency of the antioxidant enzyme system under water stress. The combined effect of AMF, PGPR, and compost boosted tomato growth fitness, fruit yield and quality, while increasing drought stress tolerance (Tahiri et al., 2022). A two-year field experiment showed that the application of different biostimulants such AMF, PGPR, and seaweed extract increased wheat root volume, membrane stability index, leaf relative water content (RWC), and photosynthetic pigment content, which promoted plant resilience under water shortage conditions (Najafi Vafa et al., 2022).
The rhizosphere microbiome induces a profound influence on host physiology, and both monocrop and rotational crops exhibit significant variations in the regulation of different genes related to phytohormones and plant defense system (Li et al., 2021). Managing aboveground biodiversity can increase the diversity of plant-associated microbiome and plant immunity, resulting in substantial economic and ecological benefits. Substantial progress has been made in understanding how drought affects the molecular machinery that drive the connection between aboveground and belowground diversity. In fact, drought can promote plants to support root biomass production or form novel and stronger associations with AMF to provide distant water from the rhizospheric area to the plants. These phenotypic and biotic alterations have the potential to significantly affect soil physical properties. Elucidating how combined changes in abiotic components affect plant adaptation allows for anticipating resilience and productivity. However, it will be necessary to understand the spatial and temporal dynamics of ecosystems and to consider multiple ecosystem dimensions, especially physical properties.
Plants possess cognitive capacities and may obtain, process, and memorize information that may adjust their behavior to (upcoming) natural signals (Michmizos and Hilioti, 2019). The cognitive abilities of plants can be extended to their surrounding environment either through root functions or beneficial symbiotic microorganisms (Parise et al., 2020). However, the operation of a plant cognitive system is still largely unknown given the lack of a nervous communication system, as is the case with animals. Well-identified learning and memory mechanisms at cellular and molecular levels may provide a plausible pathway toward adaptation built on prior stimuli such as prolonged water stress. However, determining the role of plant gnosophysiology–a newly emerging scientific field challenges conventional perspectives and delves into the concept that plants possess the ability to respond to stimuli, and learn from them, and make decisions ensuring their survival–and its relative contribution to drought resilience will require substantial theoretical and experimental evidence.
Due to the importance of plant and soil-rhizosphere microbiome diversity in boosting host defense and resilience to environmental constraints, a drought-related decrease in biodiversity has the potential to impact the abiotic resilience of plant communities with serious implications for adaptation. Enhanced plant diversity offers various PRR reservoirs capable of recognizing a variety of MAMPs, thereby boosting immunological defense mechanisms. Systemic signals can then be transmitted from one plant species to another via VCs (produced by microbes and plants) or root exudates, which impact primary productivity and host tolerance (Weisskopf et al., 2021).
By inducing the transcriptional reprogramming of numerous genes and TFs involved in a range of plant defense systems, microbial communities can assist host-plants endure water deficiency (Figure 4). A recent study revealed that under stressful conditions, microorganisms may produce ABA or ABA analogs and trigger essential genes that produce ABA, such ABA2, ABA3, and NCED3 (Gowtham et al., 2021). ABA plays a variety of roles, from detecting environmental signals to activating transcription as an adaptive mechanism to regulating a range of developmental, physio-biochemical, and cellular, properties. According to Woo et al. (2020), B. subtilis strain GOT9 boosted water stress tolerance in A. thaliana by inducing the expression of several NCED genes, which control crucial ABA production-related enzymes. Additionally, a large number of genes implicated in the control of ABA drought signaling pathways (Takahashi et al., 2018), including DREB1D, RAB18, RD20, RD29B, CSD3, WRKY7, and ERF1 are induced by plant-associated microorganisms (Abbasi et al., 2020; Woo et al., 2020).
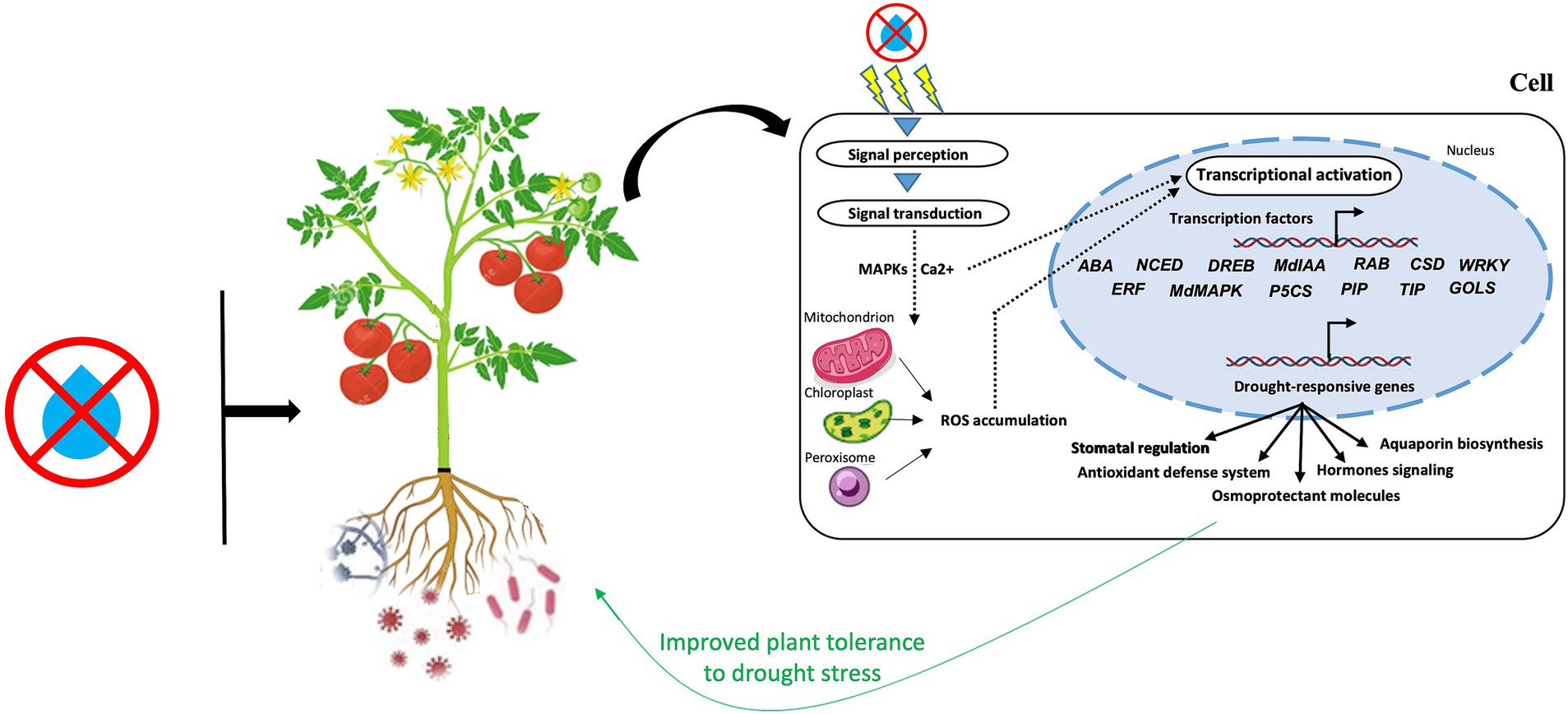
Figure 4. A schematic diagram highlighting the major signaling players involved in the microbiome-plant-drought axis. MAPKs, mitogen-activated protein kinases; ROS, reactive oxygen species.
Furthermore, the transcriptional activity of MAPKs genes, including MdMAPK7-1, MdMAPK16-2, MdMAPK17, and MdMAPK20-1, which are crucial for signal transduction during stress, have been reported to increase in AMF-treated apple seedlings (Huang et al., 2020). Ca2+-dependent protein kinases (CDPKs) and MAPKs perform a significant function in the signals transduction to the nucleus during drought stress through multiple TFs (i.e., DREB, ABRE, MYB/MYC, WRKY, and NAC) involved in drought-tolerant genes regulation (Ali et al., 2022). In the same vein, a recent study in maize reported that pyrroline-5-carboxylate synthase (P5CS) genes, responsible of proline production, including SbP5CS 1 and SbP5CS 2, were upregulated under water stress (Govindasamy et al., 2020). Similar findings were made by Saddique et al. (2018), who found that P. indica-treated rice showed P5CS genes upregulation and improved phosphate and zinc uptake in order to alleviate the negative impact of water deficiency. In addition, Pseudomonas simiae-inoculated soybean enhanced fitness and water stress resistance through improving the transcription of several important genes, including water transporters (TIP and PIP) and osmoprotectants (P5CS, DREB/EREB, GOLS) (Vaishnav and Choudhary, 2019). The aforementioned alterations in the expression of proline-related genes were strongly linked with plant morphological and physiological adaptation to water deficit effects.
AQPs are considered as a main actor in the cell transport system. Jia-Dong et al. (2019) have suggested that these proteins are frequently linked to the process of nutrient exchange during mycorrhizal symbiosis. Zou et al. (2019) proposed an AMF-boosted mechanism that enhances the tolerance of trifoliate orange to water deficiency by up-and-down regulating the expression of specific AQPs genes. This study confirms the essential and direct involvement of these fungi in host water stress resistance. In fact, MdIAA24 gene overexpression, which controls apples mycorrhizal symbiosis by regulating strigolactones production, was associated with general plant drought tolerance, including improvements in RWC, stomatal conductance, and osmotic adjustment (Huang et al., 2021). Metabolomic and proteomic investigations have revealed the upregulation of multiple essential genes controlling transporters, signaling proteins, major metabolic enzymes, and oxidative stress-related proteins following the application of P. indica in barley under drought condition (Ghaffari et al., 2019). However, further research in other species is necessary to determine whether these gene expression patterns are universal.
In addition to plants inoculated with beneficial microorganisms, pathogen-infected plants have also shown improved tolerance to drought stress. For instance, rice infected with brome mosaic virus (BMV) and beet plants infected with cucumber mosaic virus (CMV) have shown enhanced production of various antioxidants and osmoprotectants, which promote their tolerance to drought stress (Xu et al., 2008). Another study demonstrated that the viral protein 2b, which impacts host ABA signaling and RNA silencing pathways, provided drought tolerance to Arabidopsis plants in the case of CMV (Westwood et al., 2013). When Nicotiana benthamiana was treated with Yellowtail flower mild mottle virus, its ability to withstand water stress was also enhanced (Dastogeer et al., 2018). The effects of two ascomycete fungal endophytes isolated from wild Australian Nicotiana growing in an arid area that were examined in conjunction with this virus on plant gene expression and osmolytes and antioxidants production under drought stress were comparable to the virus effect. These outcomes indicated that the responses of plants infected with viruses and fungi to water deficiency are somewhat similar.
The ubiquitous presence of the soil microbiome and its impact on various aspects of plant functioning under drought stress have made it increasingly challenging to isolate specific aspects of the microbiome-plant-drought axis. This highlights the need for multi-disciplinary interventions to uncover opportunities and strategies for enhancing crop drought resistance (Hartman and Tringe, 2019; Ali et al., 2022). Multidomain research approaches that combine plant resilience, microbiome recruitment, and the interactions among the different (a)biotic components of the ecosystem have proven to be valuable strategies. They establish connections that modulate microbiome composition and activity, leading to improved drought tolerance (Trivedi et al., 2022). Yet, the full potential of the host microbiome in sustainable agriculture is not yet fully realized due to multiple factors, including soil type, plant genotype, microbial interactions, agricultural practices, and the complex interplay among these components (Busby et al., 2017; Soman et al., 2017; Schmidt et al., 2019). Moreover, microbiome engineering faces challenges in achieving accurate and long-lasting positive effects on plants. Multiple interconnected factors, such as the richness and complexity of microbial communities and alterations in microbiota functioning during host development stages, contribute to the complexities that limit the effectiveness of microbiome engineering.
To maximize the benefits of plant microbiome assembly under drought stress, it is crucial to investigate the genetic complexity of both plants and their associated microbiome, the heterogeneity of their environment, and the metabolic patterns that influence the host microbiome. This requires the application of inclusive and systemic biological methods coupled with advanced multiomics techniques. While existing tools and methodologies have made significant advancement in understanding the effect of the microbiome on host resistance to water stress, there are still many gaps to be filled in order to make substantial progress toward microbiome-targeted approaches for enhancing crop production and resilience under drought stress.
Many approaches such as culture-dependent, culture-independent, and reductionist synthetic communities (SynCom) have been primarily used to study the root microbiome. However, reproducing a suitable natural conditions required for the development of different microbes in the laboratory is challenging, and a large number of them cannot be cultured, leading to a loss of microbiome information (Hill et al., 2000). However culture-dependent methods, including the reductionist SynCom method remain highly efficient for microbiome studies (Bai et al., 2015; Zhang et al., 2021). DNA fingerprinting and phospholipid fatty acid approaches, as culture-independent methods, focus on studying the composition and diversity of the entire microbiome community. However, these approaches provide less data in comparison to the current advances in metaomics, sequencing, and computational techniques (Jo et al., 2020). By utilizing advanced sequencing methods, studies have achieved unprecedented precision and comprehensiveness in understanding the composition of microbial communities under drought condition (Lundberg et al., 2012; Liu et al., 2019). Furthermore, the development of metagenomics, metatranscriptomics, and metaproteomics approaches enable a thorough understanding of microbiome function under drought stress (Liu et al., 2021).
The census method, which includes metagenomic and amplicons statistics, offers a comprehensive understanding of plant associated-microbes, which is essential for studying natural microbiome facts. On the other hand, the reductionist SynCom method links plant molecular biology to microbial ecology (Guttman et al., 2014; Mendes et al., 2014; Bulgarelli et al., 2015; Edwards et al., 2015; Liu et al., 2019). The census approach provides data for the reductionist investigation of plant-microbiome interactions. By combining this information with data on isolated strains, an extensive range of representative SynCom can be created, providing accurate genetic information (Bulgarelli et al., 2012; Vorholt et al., 2017). These SynComs are then used to simulate plant-microbiome interactions, allowing the study of the components that orchestrate microbial communities and validate their monitoring functions and molecular strategies throughout plant growth and development, even under stressful conditions, such as drought (Bai et al., 2015; Castrillo et al., 2017; Chen et al., 2020; Finkel et al., 2020).
Multiomics approaches offer a valuable tool for addressing the challenging task of translating plant alterations at the genetic, proteomic, or metabolomic levels. The integration of multiomics data-driven science has significantly advanced our understanding of microbiome composition and functional responses in intricate environments, such as the rhizosphere, where the complex network of microbial connections orchestrates plant behavior under stressful conditions. The combination of these approaches has led to remarkable progress in plant-microbiome-related research.
Each technique employed in multiomics approaches brings unique advantages, but no single approach can provide a comprehensive understanding of the mechanisms governing the assembly of plant-associated microbiome under drought stress. Therefore, it is crucial to integrate various research approaches to pave the way toward a comprehensive understanding and utilization of the microbiome in future studies, with the aim of developing targeted strategies to boost plant resilience to water deficit stress.
Unlocking the intricate interactions between plant and their microbiome under water limitation and their implications for plant fitness and productivity are crucial steps toward harnessing the potential of the microbiome to promote host adaptation to environmental perturbation. Exploring the extent of these interactions at evolutionary, ecological, biochemical, and molecular levels can provide valuable insights for advancing our system-level understanding and inform microbial approaches to improve host resistance and fitness. Several key research directions should be prioritized in the future:
Gain a complete comprehension of how drought impacts the assembly and functions of the plant microbiome across different temporal and spatial scales.
Investigate the effect of drought on plant fitness and defense system, elucidating the specific alterations in photosynthates, root exudates, and defense mechanisms, and their influence on the assembly and functions of the plant microbiome.
Determine the changes in major signaling cascades and metabolite profiles and their interactions, elucidating their roles in shaping host plant and microbial functions and fitness.
Increase our understanding of the biosynthetic pathways, genetics, and mechanisms of action of drought-responsive phytohormones, as well as their effects on plant-microbiome and microbe-microbe interactions.
Identify the frequency and duration of drought necessary for the eco-evolutionary adaptation of plant microbiome and the establishment of drought resilience in host plant.
Acquire cutting-edge knowledge about the molecular interactions that orchestrate plant-microbe interactions under drought conditions.
Develop methods for in situ manipulation of the plant and associated-microbiome to mitigate the detrimental effects of drought on crop productivity.
Microorganisms exhibit remarkable resilience to harsh environments, triggering various signaling cascades and metabolic processes that enable them to swiftly adapt to changing conditions. The soil microbiome, with its manifold benefits for plants, has the ability to mitigate the adverse impact of drought on crops (Anli et al., 2020; Boutasknit et al., 2020; Ben-Laouane et al., 2021; Meddich et al., 2021). For instance, certain ectomycorrhizal fungal (EMF) species in the rhizosphere have been found to colonize the roots of specific pinyon pine genotypes, enhancing their drought resistance (Lau and Lennon, 2012). The varying drought tolerance among different genotypes can be attributed to the presence of these EMF communities, which represent an extended genetic repertoire of the host plant and may serve as a key strategy for drought resilience. The association between host plant and diverse bacterial/fungal communities, possessing potent metabolic and biogeochemical functions, can facilitate plant adaptation to water scarcity. Through long-term field and greenhouse studies coupled with microbial community sequencing, researchers have uncovered links between host-determined EMF communities and disparities in plant performance. Moreover, they have discovered that drought-resistant genotypes exhibit more intense colonization by Geospora EMF species. Adaptation has been a subject of extensive discussion in evolutionary biology and ecology. Gehring et al. (2017) revealed that the adaptation does not necessarily involve drastic changes in phenotype but rather subtle variations that affect how the host forms associations with microscopic rhizospheric fungi. Many questions remain unanswered; (1) What are the host traits that control specific interactions with microbial communities? Are they morphological, chemical, or even phenological? Speculation can be made regarding the involvement of genes and chemical signaling molecules that underlie host-mycorrhizal interactions, (2) How significant are plant-associated soil microbial communities in drought adaptation? The fact that genotypic difference in drought resistance were observed only in pinyon pine associated with microbes suggests that the plant’s ability to form specific microbial associations is the primary driver of drought resistance.
Given that host genotypes shape distinct soil microbial communities that perform various biogeochemical and metabolic functions (Edwards et al., 2015; Wagner et al., 2016), the potential adaptation of plant traits controlling plant-microbe interactions becomes significant. A comprehensive understanding of host-microbiome interactions at biochemical and genomic levels will enhance our knowledge of drought adaptation and contribute to improved models of host responses to water deficiency. Recognizing that genotypes exhibit differential interactions with aboveground and belowground microbes may revolutionize the strategies employed to enhance plant agronomic traits and sustain global food security in the face of increasing drought trends.
During periods of water deficiency, plants heavily rely on microbiome to perform essential functions, including nutrient uptake and stress adaptation. In this review, we have synthesized the current understanding of plant-microbiome interactions under drought stress, which ultimately shape the composition of root-associated microbes. However, there is a lack of research investigating the root-associated microbiota specifically under drought conditions. The limited knowledge about the intricate connections among microbes and their modulation under drought, as well as their complex communication with the host plant, highlight the need for further investigation. Therefore, it is imperative to conduct more research on the plant–microbiome-drought axis, with a particular emphasis on omics-based techniques that integrate genomics, metagenomics, proteomics and metabolomics. Such approaches will unravel the mechanisms orchestrating the adaptation of plant microbiome assembly to drought trends and provide insights to identify microbial communities that confer drought resilience and enhance plant performance under water deficient conditions.
MA: Data curation, Formal analysis, Investigation, Methodology, Resources, Software, Validation, Writing – original draft. AM: Investigation, Supervision, Validation, Writing – review & editing. MB: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing – review & editing.
This work was supported as part of FOSC project (Sus-Agri-CC) from the European Union’s Horizon 2020 research and innovation program under grant agreement N°862555 and as a part of PRIMA project (SALAM-MED) from the European Union’s Horizon 2020 research and innovation program under grant agreement N°2131.
MB acknowledges the support of the Faculty of Sciences Semlalia (Marrakesh, Morocco). We apologize to colleagues whose work could not be referenced due to length constraints.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abbasi, S., Sadeghi, A., and Safaie, N. (2020). Streptomyces alleviate drought stress in tomato plants and modulate the expression of transcription factors ERF1 and WRKY70 genes. Sci. Hortic. (Amsterdam). 265:109206. doi: 10.1016/j.scienta.2020.109206
Abbott, K. C., Eppinga, M. B., Umbanhowar, J., Baudena, M., and Bever, J. D. (2021). Microbiome influence on host community dynamics: conceptual integration of microbiome feedback with classical host–microbe theory. Ecol. Lett. 24, 2796–2811. doi: 10.1111/ele.13891
Ait-El-Mokhtar, M., and Baslam, M. (2023). Holo-omic applications to unveil microbiome shaping sustainable horticultural production. Front. Sustain. Food Syst. 7, 1–18. doi: 10.3389/fsufs.2023.1151367
Ait-El-Mokhtar, M., El Amerany, F., Fakhech, A., Akensous, F.-Z., Ait-Rahou, Y., Ben-Laouane, R., et al. (2022). “Cereals and Phytohormones under drought stress” in Sustainable Remedies for Abiotic Stress in Cereals (Berlin: Springer), 313–350.
Ali, S., Tyagi, A., Park, S., Mir, R. A., Mushtaq, M., Bhat, B., et al. (2022). Deciphering the plant microbiome to improve drought tolerance: mechanisms and perspectives. Environ. Exp. Bot. 201:104933. doi: 10.1016/j.envexpbot.2022.104933
Anli, M., Baslam, M., Tahiri, A.-I., Raklami, A., Boutasknit, A., Symanczik, S., et al. (2020). Biofertilizers as strategies to improve photosynthetic apparatus, growth, and drought stress tolerance in the date palm. Front. Plant Sci. 11:818. doi: 10.3389/fpls.2020.516818
Anli, M., Boutasknit, A., Ben-Laoaune, R., Ait-El-Mokhtar, M., Fakhech, A., El Modafar, C., et al. (2022). “Use of biostimulants to improve drought tolerance in cereals” in Sustainable Remedies for Abiotic Stress in Cereals (Singapore: Springer Nature), 519–555.
Aroca, R., Vernieri, P., and Ruiz-Lozano, J. M. (2008). Mycorrhizal and non-mycorrhizal Lactuca sativa plants exhibit contrasting responses to exogenous ABA during drought stress and recovery. J. Exp. Bot. 59, 2029–2041. doi: 10.1093/jxb/ern057
Asaf, S., Khan, A. L., Khan, M. A., Imran, Q. M., Yun, B. W., and Lee, I. J. (2017). Osmoprotective functions conferred to soybean plants via inoculation with Sphingomonas sp. LK11 and exogenous trehalose. Microbiol. Res. 205, 135–145. doi: 10.1016/j.micres.2017.08.009
Aslam, M. M., Okal, E. J., Idris, A. L., Qian, Z., Xu, W., Karanja, J. K., et al. (2022). Rhizosphere microbiomes can regulate plant drought tolerance. Pedosphere 32, 61–74. doi: 10.1016/S1002-0160(21)60061-9
Azizi, S., Kouchaksaraei, M. T., Hadian, J., Nosrat Abad, A. R. F., Modarres Sanavi, S. A. M., Ammer, C., et al. (2021). Dual inoculations of arbuscular mycorrhizal fungi and plant growth-promoting rhizobacteria boost drought resistance and essential oil yield of common myrtle. For. Ecol. Manag. 497:119478. doi: 10.1016/j.foreco.2021.119478
Badri, D. V., Loyola-Vargas, V. M., Broeckling, C. D., De-la-Peña, C., Jasinski, M., Santelia, D., et al. (2008). Altered profile of secondary metabolites in the root exudates of arabidopsis ATP-binding cassette transporter mutants. Plant Physiol. 146, 762–771. doi: 10.1104/pp.107.109587
Badri, D. V., Quintana, N., El Kassis, E. G., Kim, H. K., Choi, Y. H., Sugiyama, A., et al. (2009). An ABC transporter mutation alters root exudation of phytochemicals that provoke an overhaul of natural soil microbiota. Plant Physiol. 151, 2006–2017. doi: 10.1104/pp.109.147462
Badri, D. V., and Vivanco, J. M. (2009). Regulation and function of root exudates. Plant Cell Environ. 32, 666–681. doi: 10.1111/j.1365-3040.2009.01926.x
Baetz, U., and Martinoia, E. (2014). Root exudates: the hidden part of plant defense. Trends Plant Sci. 19, 90–98. doi: 10.1016/j.tplants.2013.11.006
Bahadur, A., Batool, A., Nasir, F., Jiang, S., Mingsen, Q., Zhang, Q., et al. (2019). Mechanistic insights into arbuscular mycorrhizal Fungi-mediated drought stress tolerance in plants. Int. J. Mol. Sci. 20:4199. doi: 10.3390/ijms20174199
Bai, B., Liu, W., Qiu, X., Zhang, J., Zhang, J., and Bai, Y. (2022). The root microbiome: community assembly and its contributions to plant fitness. J. Integr. Plant Biol. 64, 230–243. doi: 10.1111/jipb.13226
Bai, Y., Müller, D. B., Srinivas, G., Garrido-Oter, R., Potthoff, E., Rott, M., et al. (2015). Functional overlap of the Arabidopsis leaf and root microbiota. Nature 528, 364–369. doi: 10.1038/nature16192
Bais, H. P., Weir, T. L., Perry, L. G., Gilroy, S., and Vivanco, J. M. (2006). The role of root exudates in rhizosphere interactions with plants and other organisms. Annu. Rev. Plant Biol. 57, 233–266. doi: 10.1146/annurev.arplant.57.032905.105159
Bashyal, B. M., Parmar, P., Zaidi, N. W., and Aggarwal, R. (2021). Molecular programming of drought-challenged Trichoderma harzianum-bioprimed Rice (Oryza sativa L.). Front. Microbiol. 12:165. doi: 10.3389/fmicb.2021.655165
Begum, N., Wang, L., Ahmad, H., Akhtar, K., Roy, R., Khan, M. I., et al. (2022). Co-inoculation of arbuscular mycorrhizal Fungi and the plant growth-promoting Rhizobacteria improve growth and photosynthesis in tobacco under drought stress by up-regulating antioxidant and mineral nutrition metabolism. Microb. Ecol. 83, 971–988. doi: 10.1007/s00248-021-01815-7
Benaffari, W., Boutasknit, A., Anli, M., Ait-El-mokhtar, M., Ait-Rahou, Y., Ben-Laouane, R., et al. (2022). The native arbuscular mycorrhizal Fungi and Vermicompost-based organic amendments enhance soil fertility, growth performance, and the drought stress tolerance of quinoa. Plan. Theory 11, 1–29. doi: 10.3390/plants11030393
Ben-Laouane, R., Ait-El-Mokhtar, M., Anli, M., Boutasknit, A., Ait Rahou, Y., Raklami, A., et al. (2021). Green compost combined with mycorrhizae and rhizobia: a strategy for improving alfalfa growth and yield under field conditions. Gesunde Pflanz 73, 193–207. doi: 10.1007/s10343-020-00537-z
Bernardo, L., Morcia, C., Carletti, P., Ghizzoni, R., Badeck, F. W., Rizza, F., et al. (2017). Proteomic insight into the mitigation of wheat root drought stress by arbuscular mycorrhizae. J. Proteome 169, 21–32. doi: 10.1016/j.jprot.2017.03.024
Bi, K., Zhang, X., Chen, W., and Diao, H. (2020). MicroRNAs regulate intestinal immunity and gut microbiota for gastrointestinal health: a comprehensive review. Genes (Basel) 11:716. doi: 10.3390/genes11091075
Bordenstein, S. R., and Theis, K. R. (2015). Host biology in light of the microbiome: ten principles of holobionts and hologenomes. PLoS Biol. 13, 1–23. doi: 10.1371/journal.pbio.1002226
Boutasknit, A., Baslam, M., Ait-El-Mokhtar, M., Anli, M., Ben-Laouane, R., Ait-Rahou, Y., et al. (2021). Assemblage of indigenous arbuscular mycorrhizal fungi and green waste compost enhance drought stress tolerance in carob (Ceratonia siliqua L.) trees. Sci. Rep. 11, 1–23. doi: 10.1038/s41598-021-02018-3
Boutasknit, A., Baslam, M., Ait-El-mokhtar, M., Anli, M., Ben-Laouane, R., Douira, A., et al. (2020). Arbuscular mycorrhizal fungi mediate drought tolerance and recovery in two contrasting carob (Ceratonia siliqua L.) ecotypes by regulating stomatal, water relations, and (in)organic adjustments. Plan. Theory 9:80. doi: 10.3390/plants9010080
Breakfield, N. W., Corcoran, D. L., Petricka, J. J., Shen, J., Sae-Seaw, J., Rubio-Somoza, I., et al. (2012). High-resolution experimental and computational profiling of tissue-specific known and novel miRNAs in Arabidopsis. Genome Res. 22, 163–176. doi: 10.1101/gr.123547.111
Breuillin-Sessoms, F., Floss, D. S., Karen Gomez, S., Pumplin, N., Ding, Y., Levesque-Tremblay, V., et al. (2015). Suppression of arbuscule degeneration in Medicago truncatula phosphate transporter4 mutants is dependent on the ammonium transporter 2 family protein AMT2;3. Plant Cell 27, 1352–1366. doi: 10.1105/tpc.114.131144
Bulgarelli, D., Garrido-Oter, R., Münch, P. C., Weiman, A., Dröge, J., Pan, Y., et al. (2015). Structure and function of the bacterial root microbiota in wild and domesticated barley. Cell Host Microbe 17, 392–403. doi: 10.1016/j.chom.2015.01.011
Bulgarelli, D., Rott, M., Schlaeppi, K., Loren, V., van Themaat, E., Ahmadinejad, N., et al. (2012). Revealing structure and assembly cues for Arabidopsis root-inhabiting bacterial microbiota. Nature 488, 91–95. doi: 10.1038/nature11336
Bulgarelli, D., Schlaeppi, K., Spaepen, S., Van Themaat, E. V. L., and Schulze-Lefert, P. (2013). Structure and functions of the bacterial microbiota of plants. Annu. Rev. Plant Biol. 64, 807–838. doi: 10.1146/annurev-arplant-050312-120106
Busby, P. E., Soman, C., Wagner, M. R., Friesen, M. L., Kremer, J., Bennett, A., et al. (2017). Research priorities for harnessing plant microbiomes in sustainable agriculture. PLoS Biol. 15, 1–14. doi: 10.1371/journal.pbio.2001793
Caddell, D. F., Louie, K., Bowen, B., Sievert, J. A., Hollingsworth, J., Dahlberg, J., et al. (2020). Drought shifts sorghum root metabolite and microbiome profiles and enriches the stress response factor pipecolic acid. bioRxiv 47, 248–250. doi: 10.1101/2020.11.08.373399
Canarini, A., Schmidt, H., Fuchslueger, L., Martin, V., Herbold, C. W., Zezula, D., et al. (2021). Ecological memory of recurrent drought modifies soil processes via changes in soil microbial community. Nat. Commun. 12, 1–14. doi: 10.1038/s41467-021-25675-4
Carbonnel, S., and Gutjahr, C. (2014). Control of arbuscular mycorrhiza development by nutrient signals. Front. Plant Sci. 5, 1–6. doi: 10.3389/fpls.2014.00462
Carlson, R., Tugizimana, F., Steenkamp, P. A., Dubery, I. A., Hassen, A. I., and Labuschagne, N. (2020). Rhizobacteria-induced systemic tolerance against drought stress in Sorghum bicolor (L.) Moench. Microbiol. Res. 232:126388. doi: 10.1016/j.micres.2019.126388
Casado-Bedmar, M., and Viennois, E. (2022). MicroRNA and gut microbiota: tiny but mighty—novel insights into their cross-talk in inflammatory bowel disease pathogenesis and therapeutics. J. Crohns Colitis 16, 992–1005. doi: 10.1093/ecco-jcc/jjab223
Castrillo, G., Teixeira, P. J. P., Paredes, S. H., Law, T. F., De Lorenzo, L., Feltcher, M. E., et al. (2017). Root microbiota drive direct integration of phosphate stress and immunity. Nature 543, 513–518. doi: 10.1038/nature21417
Chareesri, A., De Deyn, G. B., Sergeeva, L., Polthanee, A., and Kuyper, T. W. (2020). Increased arbuscular mycorrhizal fungal colonization reduces yield loss of rice (Oryza sativa L.) under drought. Mycorrhiza 30, 315–328. doi: 10.1007/s00572-020-00953-z
Charpentier, M., Sun, J., Wen, J., Mysore, K. S., and Oldroyd, G. E. D. (2014). Abscisic acid promotion of arbuscular mycorrhizal colonization requires a component of the PROTEIN PHOSPHATASE 2A complex. Plant Physiol. 166, 2077–2090. doi: 10.1104/pp.114.246371
Chen, T., Nomura, K., Wang, X., Sohrabi, R., Xu, J., Yao, L., et al. (2020). A plant genetic network for preventing dysbiosis in the phyllosphere. Nature 580, 653–657. doi: 10.1038/s41586-020-2185-0
Chen, C., Xin, K., Liu, H., Cheng, J., Shen, X., Wang, Y., et al. (2017). Pantoea alhagi, a novel endophytic bacterium with ability to improve growth and drought tolerance in wheat. Sci. Rep. 7, 1–14. doi: 10.1038/srep41564
Cheng, S., Zou, Y. N., Kuča, K., Hashem, A., Abd-Allah, E. F., and Wu, Q. S. (2021). Elucidating the mechanisms underlying enhanced drought tolerance in plants mediated by arbuscular mycorrhizal Fungi. Front. Microbiol. 12:473. doi: 10.3389/fmicb.2021.809473
Cheng, H. Q., Zou, Y. N., Wu, Q. S., and Kuča, K. (2021). Arbuscular mycorrhizal Fungi alleviate drought stress in trifoliate Orange by regulating H+-ATPase activity and gene expression. Front. Plant Sci. 12, 1–9. doi: 10.3389/fpls.2021.659694
Dastogeer, K. M. G., Li, H., Sivasithamparam, K., Jones, M. G. K., and Wylie, S. J. (2018). Fungal endophytes and a virus confer drought tolerance to Nicotiana benthamiana plants through modulating osmolytes, antioxidant enzymes and expression of host drought responsive genes. Environ. Exp. Bot. 149, 95–108. doi: 10.1016/j.envexpbot.2018.02.009
De Vries, F. T., Williams, A., Stringer, F., Willcocks, R., McEwing, R., Langridge, H., et al. (2019). Changes in root-exudate-induced respiration reveal a novel mechanism through which drought affects ecosystem carbon cycling. New Phytol. 224, 132–145. doi: 10.1111/nph.16001
Dreyer, I., Gomez-Porras, J. L., Riaño-Pachón, D. M., Hedrich, R., and Geiger, D. (2012). Molecular evolution of slow and quick anion channels (SLACs and QUACs/ALMTs). Front. Plant Sci. 3, 1–12. doi: 10.3389/fpls.2012.00263
Edwards, J., Johnson, C., Santos-Medellín, C., Lurie, E., Podishetty, N. K., Bhatnagar, S., et al. (2015). Structure, variation, and assembly of the root-associated microbiomes of rice. Proc. Natl. Acad. Sci. U. S. A. 112, E911–E920. doi: 10.1073/pnas.1414592112
Escudero-Martinez, C., Coulter, M., Alegria Terrazas, R., Foito, A., Kapadia, R., Pietrangelo, L., et al. (2022). Identifying plant genes shaping microbiota composition in the barley rhizosphere. Nat. Commun. 13, 1–14. doi: 10.1038/s41467-022-31022-y
Eshaghi Gorgi, O., Fallah, H., Niknejad, Y., and Barari Tari, D. (2022). Effect of plant growth promoting rhizobacteria (PGPR) and mycorrhizal fungi inoculations on essential oil in Melissa officinalis L. under drought stress. Biologia (Bratisl) 77, 11–20. doi: 10.1007/s11756-021-00919-2
Etemadi, M., Gutjahr, C., Couzigou, J. M., Zouine, M., Lauressergues, D., Timmers, A., et al. (2014). Auxin perception is required for arbuscule development in arbuscular mycorrhizal symbiosis. Plant Physiol. 166, 281–292. doi: 10.1104/pp.114.246595
Etesami, H., and Jeong, B. R. (2018). Silicon (Si): review and future prospects on the action mechanisms in alleviating biotic and abiotic stresses in plants. Ecotoxicol. Environ. Saf. 147, 881–896. doi: 10.1016/j.ecoenv.2017.09.063
Ezquer, I., Salameh, I., Colombo, L., and Kalaitzis, P. (2020). Plant cell walls tackling climate change: biotechnological strategies to improve crop adaptions and photosynthesis in response to global warming. Plan. Theory 9:212. doi: 10.3390/plants9020212
Faist, H., Trognitz, F., Antonielli, L., Symanczik, S., White, P. J., and Sessitsch, A. (2023). Potato root-associated microbiomes adapt to combined water and nutrient limitation and have a plant genotype-specific role for plant stress mitigation. Environ. Microbiome 18, 1–19. doi: 10.1186/s40793-023-00469-x
Finkel, O. M., Salas-González, I., Castrillo, G., Conway, J. M., Law, T. F., Teixeira, P. J. P. L., et al. (2020). A single bacterial genus maintains root growth in a complex microbiome. Nature 587, 103–108. doi: 10.1038/s41586-020-2778-7
French, E., Ghaste, M., Widhalm, J. R., and Iyer-Pascuzzi, A. S. (2019). Defense hormones modulate root microbiome diversity and composition in tomato. bioRxiv 2019:656769. doi: 10.1101/656769
Gao, Z., Han, M., Hu, Y., Li, Z., Liu, C., Wang, X., et al. (2019). Effects of continuous cropping of sweet potato on the fungal community structure in Rhizospheric soil. Front. Microbiol. 10:269. doi: 10.3389/fmicb.2019.02269
Gehring, C. A., Sthultz, C. M., Flores-Rentería, L., Whipple, A. V., and Whitham, T. G. (2017). Tree genetics defines fungal partner communities that may confer drought tolerance. Proc. Natl. Acad. Sci. U. S. A. 114, 11169–11174. doi: 10.1073/pnas.1704022114
Ghaffari, M. R., Mirzaei, M., Ghabooli, M., Khatabi, B., Wu, Y., Zabet-Moghaddam, M., et al. (2019). Root endophytic fungus Piriformospora indica improves drought stress adaptation in barley by metabolic and proteomic reprogramming. Environ. Exp. Bot. 157, 197–210. doi: 10.1016/j.envexpbot.2018.10.002
Gill, S. S., Gill, R., Trivedi, D. K., Anjum, N. A., Sharma, K. K., Ansari, M. W., et al. (2016). Piriformospora indica: potential and significance in plant stress tolerance. Front. Microbiol. 7, 1–20. doi: 10.3389/fmicb.2016.00332
González-Teuber, M., Urzúa, A., Plaza, P., and Bascuñán-Godoy, L. (2018). Effects of root endophytic fungi on response of Chenopodium quinoa to drought stress. Plant Ecol. 219, 231–240. doi: 10.1007/s11258-017-0791-1
Govindasamy, V., George, P., Kumar, M., Aher, L., Raina, S. K., Rane, J., et al. (2020). Multi-trait PGP rhizobacterial endophytes alleviate drought stress in a senescent genotype of Sorghum Sorghum bicolor L Moench. 3 Biotech 10, 1–14. doi: 10.1007/s13205-019-2001-4
Gowtham, H. G., Duraivadivel, P., Ayusman, S., Sayani, D., Gholap, S. L., Niranjana, S. R., et al. (2021). ABA analogue produced by Bacillus marisflavi modulates the physiological response of Brassica juncea L. under drought stress. Appl. Soil Ecol. 159:103845. doi: 10.1016/j.apsoil.2020.103845
Guttman, D. S., McHardy, A. C., and Schulze-Lefert, P. (2014). Microbial genome-enabled insights into plant-microorganism interactions. Nat. Rev. Genet. 15, 797–813. doi: 10.1038/nrg3748
Hacquard, S., Spaepen, S., Garrido-Oter, R., and Schulze-Lefert, P. (2017). Annual review of phytopathology interplay between innate immunity and the plant microbiota. Annu. Rev. Phytopathol. 2017:516. doi: 10.1146/annurev-phyto-080516-
Han, Y., Han, S., Ban, Q., He, Y., Jin, M., and Rao, J. (2017). Overexpression of persimmon DkXTH1 enhanced tolerance to abiotic stress and delayed fruit softening in transgenic plants. Plant Cell Rep. 36, 583–596. doi: 10.1007/s00299-017-2105-4
Hartman, K., and Tringe, S. G. (2019). Interactions between plants and soil shaping the root microbiome under abiotic stress. Biochem. J. 476, 2705–2724. doi: 10.1042/BCJ20180615
Hassani, M. A., Durán, P., and Hacquard, S. (2018). Microbial interactions within the plant holobiont. Microbiome 6:58. doi: 10.1186/s40168-018-0445-0
He, F., Sheng, M., and Tang, M. (2017). Effects of rhizophagus irregularis on photosynthesis and antioxidative enzymatic system in Robinia pseudoacacia L. Under drought stress. Front. Plant Sci. 8, 1–14. doi: 10.3389/fpls.2017.00183
He, F., Zhang, H., and Tang, M. (2016). Aquaporin gene expression and physiological responses of Robinia pseudoacacia L. to the mycorrhizal fungus Rhizophagus irregularis and drought stress. Mycorrhiza 26, 311–323. doi: 10.1007/s00572-015-0670-3
Hill, G. T., Mitkowski, N. A., Aldrich-Wolfe, L., Emele, L. R., Jurkonie, D. D., Ficke, A., et al. (2000). Methods for assessing the composition and diversity of soil microbial communities. Appl. Soil Ecol. 15, 25–36. doi: 10.1016/S0929-1393(00)00069-X
Ho-Plágaro, T., and García-Garrido, J. M. (2022). Molecular regulation of arbuscular mycorrhizal Symbiosis. Int. J. Mol. Sci. 23:5960. doi: 10.3390/ijms23115960
Horton, M. W., Bodenhausen, N., Beilsmith, K., Meng, D., Muegge, B. D., Subramanian, S., et al. (2014). Genome-wide association study of Arabidopsis thaliana leaf microbial community. Nat. Commun. 5, 1–7. doi: 10.1038/ncomms6320
Huang, D., Ma, M., Wang, Q., Zhang, M., Jing, G., Li, C., et al. (2020). Arbuscular mycorrhizal fungi enhanced drought resistance in apple by regulating genes in the MAPK pathway. Plant Physiol. Biochem. 149, 245–255. doi: 10.1016/j.plaphy.2020.02.020
Huang, D., Wang, Q., Jing, G., Ma, M., Li, C., and Ma, F. (2021). Overexpression of MdIAA24 improves apple drought resistance by positively regulating strigolactone biosynthesis and mycorrhization. Tree Physiol. 41, 134–146. doi: 10.1093/treephys/tpaa109
Jia-Dong, H., Tao, D., Hui-Hui, W., Zou, Y. N., Wu, Q. S., and Kamil, K. (2019). Mycorrhizas induce diverse responses of root TIP aquaporin gene expression to drought stress in trifoliate orange. Sci. Hortic. (Amsterdam) 243, 64–69. doi: 10.1016/j.scienta.2018.08.010
Jiao, J., Ma, Y., Chen, S., Liu, C., Song, Y., Qin, Y., et al. (2016). Melatonin-producing endophytic bacteria from grapevine roots promote the abiotic stress-induced production of endogenous melatonin in their hosts. Front. Plant Sci. 7, 1–13. doi: 10.3389/fpls.2016.01387
Jin, Y., Liu, H., Luo, D., Yu, N., Dong, W., Wang, C., et al. (2016). DELLA proteins are common components of symbiotic rhizobial and mycorrhizal signalling pathways. Nat. Commun. 7:12433. doi: 10.1038/ncomms12433
Jo, J., Oh, J., and Park, C. (2020). Microbial community analysis using high-throughput sequencing technology: a beginner’s guide for microbiologists. J. Microbiol. 58, 176–192. doi: 10.1007/s12275-020-9525-5
Jones, J. D. G., and Dangl, J. L. (2006). The plant immune system. Nature 444, 323–329. doi: 10.1038/nature05286
Jones, P. M., and George, A. M. (2002). Mechanism of ABC transporters: a molecular dynamics simulation of a well characterized nucleotide-binding subunit. Proc. Natl. Acad. Sci. U. S. A. 99, 12639–12644. doi: 10.1073/pnas.152439599
Jones, D. L., Nguyen, C., and Finlay, R. D. (2009). Carbon flow in the rhizosphere: carbon trading at the soil-root interface. Plant Soil 321, 5–33. doi: 10.1007/s11104-009-9925-0
Kang, J., Park, J., Choi, H., Burla, B., Kretzschmar, T., Lee, Y., et al. (2011). Plant ABC Transporters. Arabidopsis Book 9:e0153. doi: 10.1199/tab.0153
Karlowsky, S., Augusti, A., Ingrisch, J., Akanda, M. K. U., Bahn, M., and Gleixner, G. (2018). Drought-induced accumulation of root exudates supports post-drought recovery of microbes in mountain grassland. Front. Plant Sci. 871, 1–16. doi: 10.3389/fpls.2018.01593
Khan, A. L., Waqas, M., Kang, S. M., Al-Harrasi, A., Hussain, J., Al-Rawahi, A., et al. (2014). Bacterial endophyte Sphingomonas sp. LK11 produces gibberellins and IAA and promotes tomato plant growth. J. Microbiol. 52, 689–695. doi: 10.1007/s12275-014-4002-7
Knight, R., Vrbanac, A., Taylor, B. C., Aksenov, A., Callewaert, C., Debelius, J., et al. (2018). Best practices for analysing microbiomes. Nat. Rev. Microbiol. 16, 410–422. doi: 10.1038/s41579-018-0029-9
Korenblum, E., Dong, Y., Szymanski, J., Panda, S., Jozwiak, A., Massalha, H., et al. (2020). Rhizosphere microbiome mediates systemic root metabolite exudation by root-to-root signaling. Proc. Natl. Acad. Sci. U. S. A. 117, 3874–3883. doi: 10.1073/pnas.1912130117
Lau, J. A., and Lennon, J. T. (2012). Rapid responses of soil microorganisms improve plant fitness in novel environments. Proc. Natl. Acad. Sci. U. S. A. 109, 14058–14062. doi: 10.1073/pnas.1202319109
Lebeis, S. L., Paredes, S. H., Lundberg, D. S., Breakfield, N., Gehring, J., McDonald, M., et al. (2015). Salicylic acid modulates colonization of the root microbiome by specific bacterial taxa. Science 80, 860–864. doi: 10.1126/science.aaa8764
Leitão, A. L., Costa, M. C., Gabriel, A. F., and Enguita, F. J. (2020). Interspecies communication in holobionts by non-coding RNA exchange. Int. J. Mol. Sci. 21:333. doi: 10.3390/ijms21072333
Li, T., and Chen, B.-D. (2013). Arbuscular mycorrhizal fungi improving drought tolerance of maize plants by up-regulation of aquaporin gene expressions in roots and the fungi themselves. Chinese J. Plant Ecol. 36, 973–981. doi: 10.3724/sp.j.1258.2012.00973
Li, Y., Cheng, C., and An, D. (2017). Characterisation of endophytic bacteria from a desert plant Lepidium perfoliatum L. Plant Prot. Sci. 53, 32–43. doi: 10.17221/14/2016-PPS
Li, N., Euring, D., Cha, J. Y., Lin, Z., Lu, M., Huang, L. J., et al. (2021). Plant hormone-mediated regulation of heat tolerance in response to global climate change. Front. Plant Sci. 11, 1–11. doi: 10.3389/fpls.2020.627969
Li, G., Wang, K., Qin, Q., Li, Q., Mo, F., Nangia, V., et al. (2023). Integrated microbiome and Metabolomic analysis reveal responses of rhizosphere bacterial communities and root exudate composition to drought and genotype in Rice (Oryza sativa L.). Rice 16, 1–17. doi: 10.1186/s12284-023-00636-1
Lim, J. H., and Kim, S. D. (2013). Induction of drought stress resistance by multi-functional PGPR Bacillus licheniformis K11 in pepper. Plant Pathol. J. 29, 201–208. doi: 10.5423/PPJ.SI.02.2013.0021
Liu, H., Brettell, L. E., Qiu, Z., and Singh, B. K. (2020). Microbiome-mediated stress resistance in plants. Trends Plant Sci. 25, 733–743. doi: 10.1016/j.tplants.2020.03.014
Liu, S., Da Cunha, A. P., Rezende, R. M., Cialic, R., Wei, Z., Bry, L., et al. (2016). The host shapes the gut microbiota via fecal MicroRNA. Cell Host Microbe 19, 32–43. doi: 10.1016/j.chom.2015.12.005
Liu, Y. X., Qin, Y., and Bai, Y. (2019). Reductionist synthetic community approaches in root microbiome research. Curr. Opin. Microbiol. 49, 97–102. doi: 10.1016/j.mib.2019.10.010
Liu, Y. X., Qin, Y., Chen, T., Lu, M., Qian, X., Guo, X., et al. (2021). A practical guide to amplicon and metagenomic analysis of microbiome data. Protein Cell 12, 315–330. doi: 10.1007/s13238-020-00724-8
Liu, C. Y., Zhang, F., Zhang, D. J., Srivastava, A., Wu, Q. S., and Zou, Y. N. (2018). Mycorrhiza stimulates root-hair growth and IAA synthesis and transport in trifoliate orange under drought stress. Sci. Rep. 8, 1–9. doi: 10.1038/s41598-018-20456-4
Lu, X., Liu, S. F., Yue, L., Zhao, X., Zhang, Y. B., Xie, Z. K., et al. (2018). Epsc involved in the encoding of exopolysaccharides produced by Bacillus amyloliquefaciens FZB42 act to boost the drought tolerance of Arabidopsis thaliana. Int. J. Mol. Sci. 19:795. doi: 10.3390/ijms19123795
Lundberg, D. S., Lebeis, S. L., Paredes, S. H., Yourstone, S., Gehring, J., Malfatti, S., et al. (2012). Defining the core Arabidopsis thaliana root microbiome. Nature 488, 86–90. doi: 10.1038/nature11237
Mathur, P., and Roy, S. (2021). Insights into the plant responses to drought and decoding the potential of root associated microbiome for inducing drought tolerance. Physiol. Plant. 172, 1016–1029. doi: 10.1111/ppl.13338
Meddich, A., Ait Rahou, Y., Boutasknit, A., Ait-El-Mokhtar, M., Fakhech, A., Lahbouki, S., et al. (2021). Role of mycorrhizal fungi in improving the tolerance of melon (Cucumus melo) under two water deficit partial root drying and regulated deficit irrigation. Plant Biosyst. 2021, 1–22. doi: 10.1080/11263504.2021.1881644
Mendes, L. W., Kuramae, E. E., Navarrete, A. A., Van Veen, J. A., and Tsai, S. M. (2014). Taxonomical and functional microbial community selection in soybean rhizosphere. ISME J. 8, 1577–1587. doi: 10.1038/ismej.2014.17
Michmizos, D., and Hilioti, Z. (2019). A roadmap towards a functional paradigm for learning & memory in plants. J. Plant Physiol. 232, 209–215. doi: 10.1016/j.jplph.2018.11.002
Middleton, H., Yergeau, É., Monard, C., Combier, J.-P., and El Amrani, A. (2021). Rhizospheric plant–microbe interactions: miRNAs as a key mediator. Trends Plant Sci. 26, 132–141. doi: 10.1016/j.tplants.2020.09.005
Moreno-Galván, A. E., Cortés-Patiño, S., Romero-Perdomo, F., Uribe-Vélez, D., Bashan, Y., and Bonilla, R. R. (2020). Proline accumulation and glutathione reductase activity induced by drought-tolerant rhizobacteria as potential mechanisms to alleviate drought stress in Guinea grass. Appl. Soil Ecol. 147:103367. doi: 10.1016/j.apsoil.2019.103367
Mu, J., Zhuang, X., Wang, Q., Jiang, H., Deng, Z. B., Wang, B., et al. (2014). Interspecies communication between plant and mouse gut host cells through edible plant derived exosome-like nanoparticles. Mol. Nutr. Food Res. 58, 1561–1573. doi: 10.1002/mnfr.201300729
Najafi Vafa, Z., Sohrabi, Y., Mirzaghaderi, G., and Heidari, G. (2022). Soil microorganisms and seaweed application with supplementary irrigation improved physiological traits and yield of two dryland wheat cultivars. Front. Plant Sci. 13:5090. doi: 10.3389/fpls.2022.855090
Nakayama, M., and Tateno, R. (2018). Solar radiation strongly influences the quantity of forest tree root exudates. Trees Struct. Funct. 32, 871–879. doi: 10.1007/s00468-018-1685-0
Naylor, D., and Coleman-Derr, D. (2018). Drought stress and root-associated bacterial communities. Front. Plant Sci. 8, 1–16. doi: 10.3389/fpls.2017.02223
Naylor, D., Degraaf, S., Purdom, E., and Coleman-Derr, D. (2017). Drought and host selection influence bacterial community dynamics in the grass root microbiome. ISME J. 11, 2691–2704. doi: 10.1038/ismej.2017.118
Nishida, H., and Suzaki, T. (2018). Nitrate-mediated control of root nodule symbiosis. Curr. Opin. Plant Biol. 44, 129–136. doi: 10.1016/j.pbi.2018.04.006
Olanrewaju, O. S., Ayangbenro, A. S., Glick, B. R., and Babalola, O. O. (2019). Plant health: feedback effect of root exudates-rhizobiome interactions. Appl. Microbiol. Biotechnol. 103, 1155–1166. doi: 10.1007/s00253-018-9556-6
Orelle, C., Durmort, C., Mathieu, K., Duchêne, B., Aros, S., Fenaille, F., et al. (2018). A multidrug ABC transporter with a taste for GTP. Sci. Rep. 8, 1–14. doi: 10.1038/s41598-018-20558-z
Osakabe, Y., Osakabe, K., Shinozaki, K., and Tran, L. S. P. (2014). Response of plants to water stress. Front. Plant Sci. 5, 1–8. doi: 10.3389/fpls.2014.00086
Pande, P. M., Azarbad, H., Tremblay, J., St-Arnaud, M., and Yergeau, E. (2023). Metatranscriptomic response of the wheat holobiont to decreasing soil water content. ISME Commun. 3, 1–13. doi: 10.1038/s43705-023-00235-7
Pang, Z., Chen, J., Wang, T., Gao, C., Li, Z., Guo, L., et al. (2021). Linking plant secondary metabolites and plant microbiomes: a review. Front. Plant Sci. 12:276. doi: 10.3389/fpls.2021.621276
Pantigoso, H. A., Newberger, D., and Vivanco, J. M. (2022). The rhizosphere microbiome: plant–microbial interactions for resource acquisition. J. Appl. Microbiol. 133, 2864–2876. doi: 10.1111/jam.15686
Parise, A. G., Gagliano, M., and Souza, G. M. (2020). Extended cognition in plants: is it possible? Plant Signal. Behav. 15:1661. doi: 10.1080/15592324.2019.1710661
Paymaneh, Z., Sarcheshmehpour, M., Mohammadi, H., and Askari Hesni, M. (2023). Vermicompost and/or compost and arbuscular mycorrhizal fungi are conducive to improving the growth of pistachio seedlings to drought stress. Appl. Soil Ecol. 182:104717. doi: 10.1016/j.apsoil.2022.104717
Poveda, J. (2020). Trichoderma parareesei favors the tolerance of rapeseed (Brassica napus L.) to salinity and drought due to a Chorismate mutase. Agronomy 10:118. doi: 10.3390/agronomy10010118
Raheem, A., Shaposhnikov, A., Belimov, A. A., Dodd, I. C., and Ali, B. (2018). Auxin production by rhizobacteria was associated with improved yield of wheat (Triticum aestivum L.) under drought stress. Arch. Agron. Soil Sci. 64, 574–587. doi: 10.1080/03650340.2017.1362105
Recchia, G. H., Konzen, E. R., Cassieri, F., Caldas, D. G. G., and Tsai, S. M. (2018). Arbuscular mycorrhizal symbiosis leads to differential regulation of drought-responsive genes in tissue-specific root cells of common bean. Front. Microbiol. 9, 1–24. doi: 10.3389/fmicb.2018.01339
Ruiz-Lozano, J. M., Aroca, R., Zamarreño, Á. M., Molina, S., Andreo-Jiménez, B., Porcel, R., et al. (2016). Arbuscular mycorrhizal symbiosis induces strigolactone biosynthesis under drought and improves drought tolerance in lettuce and tomato. Plant Cell Environ. 39, 441–452. doi: 10.1111/pce.12631
Saddique, M. A. B., Ali, Z., Khan, A. S., Rana, I. A., and Shamsi, I. H. (2018). Inoculation with the endophyte Piriformospora indica significantly affects mechanisms involved in osmotic stress in rice. Rice 11, 1–12. doi: 10.1186/s12284-018-0226-1
Sandhya, V., Shrivastava, M., Ali, S. Z., and Prasad, S. S. K. (2017). Endophytes from maize with plant growth promotion and biocontrol activity under drought stress. Russ. Agric. Sci. 43, 22–34. doi: 10.3103/S1068367417010165
Santner, A., Calderon-Villalobos, L. I. A., and Estelle, M. (2009). Plant hormones are versatile chemical regulators of plant growth. Nat. Chem. Biol. 5, 301–307. doi: 10.1038/nchembio.165
Santos-Medellín, C., Edwards, J., Liechty, Z., Nguyen, B., and Sundaresan, V. (2017). Drought stress results in a compartment-specific restructuring of the rice root-associated microbiomes. MBio 8, 1–15. doi: 10.1128/mBio.00764-17
Santos-Medellín, C., Liechty, Z., Edwards, J., Nguyen, B., Huang, B., Weimer, B. C., et al. (2021). Prolonged drought imparts lasting compositional changes to the rice root microbiome. Nat. Plants 7, 1065–1077. doi: 10.1038/s41477-021-00967-1
Sasse, J., Martinoia, E., and Northen, T. (2018). Feed your friends: do Plant exudates shape the root microbiome? Trends Plant Sci. 23, 25–41. doi: 10.1016/j.tplants.2017.09.003
Schmidt, J. E., Kent, A. D., Brisson, V. L., and Gaudin, A. C. M. (2019). Agricultural management and plant selection interactively affect rhizosphere microbial community structure and nitrogen cycling. Microbiome 7, 1–18. doi: 10.1186/s40168-019-0756-9
Shen, M., Huang, W., Chen, M., Song, B., Zeng, G., and Zhang, Y. (2020). (Micro)plastic crisis: un-ignorable contribution to global greenhouse gas emissions and climate change. J. Clean. Prod. 254:120138. doi: 10.1016/j.jclepro.2020.120138
Shu, B., Jue, D., Zhang, F., Zhang, D., Liu, C., Wu, Q., et al. (2020). Genome-wide identification and expression analysis of the citrus calcium-dependent protein kinase (CDPK) genes in response to arbuscular mycorrhizal fungi colonization and drought. Biotechnol. Biotechnol. Equip. 34, 1304–1314. doi: 10.1080/13102818.2020.1837011
Singh, B. K., Liu, H., and Trivedi, P. (2020). Eco-holobiont: a new concept to identify drivers of host-associated microorganisms. Environ. Microbiol. 22, 564–567. doi: 10.1111/1462-2920.14900
Singh, A., Mazahar, S., Chapadgaonkar, S. S., Giri, P., and Shourie, A. (2023). Phyto-microbiome to mitigate abiotic stress in crop plants. Front. Microbiol. 14, 1–19. doi: 10.3389/fmicb.2023.1210890
Singh, D. P., Singh, V., Gupta, V. K., Shukla, R., Prabha, R., Sarma, B. K., et al. (2020). Microbial inoculation in rice regulates antioxidative reactions and defense related genes to mitigate drought stress. Sci. Rep. 10, 1–17. doi: 10.1038/s41598-020-61140-w
Soman, C., Li, D., Wander, M. M., and Kent, A. D. (2017). Long-term fertilizer and crop-rotation treatments differentially affect soil bacterial community structure. Plant Soil 413, 145–159. doi: 10.1007/s11104-016-3083-y
Soussani, F. E., Boutasknit, A., Ben-Laouane, R., Benkirane, R., Baslam, M., and Meddich, A. (2023). Arbuscular mycorrhizal Fungi and compost-based biostimulants enhance fitness, physiological responses, yield, and quality traits of drought-stressed tomato plants. Plants 12:856. doi: 10.3390/plants12091856
Staudinger, C., Mehmeti-Tershani, V., Gil-Quintana, E., Gonzalez, E. M., Hofhansl, F., Bachmann, G., et al. (2016). Evidence for a rhizobia-induced drought stress response strategy in Medicago truncatula. J. Proteome 136, 202–213. doi: 10.1016/j.jprot.2016.01.006
Sun, Z., Song, J., Xin, X., Xie, X., and Zhao, B. (2018). Arbuscular mycorrhizal fungal 14-3-3 proteins are involved in arbuscule formation and responses to abiotic stresses during AM symbiosis. Front. Microbiol. 9, 1–17. doi: 10.3389/fmicb.2018.00091
Tahiri, A., Meddich, A., Raklami, A., Alahmad, A., Bechtaoui, N., Anli, M., et al. (2022). Assessing the potential role of compost, PGPR, and AMF in improving tomato plant growth, yield, fruit quality, and water stress tolerance. J. Soil Sci. Plant Nutr. 22, 743–764. doi: 10.1007/s42729-021-00684-w
Takahashi, F., Suzuki, T., Osakabe, Y., Betsuyaku, S., Kondo, Y., Dohmae, N., et al. (2018). A small peptide modulates stomatal control via abscisic acid in long-distance signalling. Nature 556, 235–238. doi: 10.1038/s41586-018-0009-2
Takeda, N., Handa, Y., Tsuzuki, S., Kojima, M., Sakakibara, H., and Kawaguchi, M. (2015). Gibberellins interfere with symbiosis signaling and gene expression and alter colonization by arbuscular mycorrhizal fungi in Lotus japonicus. Plant Physiol. 167, 545–557. doi: 10.1104/pp.114.247700
Tartaglia, M., Ranauda, M. A., Falzarano, A., Maisto, M., Postiglione, A., Prigioniero, A., et al. (2023). Metatranscriptomics of pastures under drought stress show a rhizospheric meta-organism reshape. Rhizosphere 26:100687. doi: 10.1016/j.rhisph.2023.100687
Tebele, S. M., Marks, R. A., and Farrant, J. M. (2023). Exploring the root-associated microbiome of the resurrection plant Myrothamnus flabellifolia. Plant Soil 2023:6019. doi: 10.1007/s11104-023-06019-1
Teixeira, P. J. P., Colaianni, N. R., Fitzpatrick, C. R., and Dangl, J. L. (2019). Beyond pathogens: microbiota interactions with the plant immune system. Curr. Opin. Microbiol. 49, 7–17. doi: 10.1016/j.mib.2019.08.003
Teng, Y., Ren, Y., Sayed, M., Hu, X., Lei, C., Kumar, A., et al. (2018). Plant-derived Exosomal MicroRNAs shape the gut microbiota. Cell Host Microbe 24, 637–652.e8. doi: 10.1016/j.chom.2018.10.001
Terhorst, C. P., Lennon, J. T., and Lau, J. A. (2014). The relative importance of rapid evolution for plant-microbe interactions depends on ecological context. Proc. R. Soc. B Biol. Sci. 281:20140028. doi: 10.1098/rspb.2014.0028
Testerink, C., and Munnik, T. (2005). Phosphatidic acid: a multifunctional stress signaling lipid in plants. Trends Plant Sci. 10, 368–375. doi: 10.1016/j.tplants.2005.06.002
Tian, T., Reverdy, A., She, Q., Sun, B., and Chai, Y. (2020). The role of rhizodeposits in shaping rhizomicrobiome. Environ. Microbiol. Rep. 12, 160–172. doi: 10.1111/1758-2229.12816
Timm, C. M., Carter, K. R., Carrell, A. A., Jun, S., Jawdy, S. S., Vélez, J. M., et al. (2018). Abiotic Stresses Shift Belowground. MSystems 3, 1–17. doi: 10.1128/mSystems.00070-17
Timmusk, S., Abd El-Daim, I. A., Copolovici, L., Tanilas, T., Kännaste, A., Behers, L., et al. (2014). Drought-tolerance of wheat improved by rhizosphere bacteria from harsh environments: enhanced biomass production and reduced emissions of stress volatiles. PLoS One 9:86. doi: 10.1371/journal.pone.0096086
Toubali, S., Ait-El-Mokhtar, M., Boutasknit, A., Anli, M., Ait-Rahou, Y., Benaffari, W., et al. (2022). Root reinforcement improved performance, productivity, and grain bioactive quality of field-Droughted quinoa (Chenopodium quinoa). Front. Plant Sci. 13:484. doi: 10.3389/fpls.2022.860484
Trivedi, P., Batista, B. D., Bazany, K. E., and Singh, B. K. (2022). Plant–microbiome interactions under a changing world: responses, consequences and perspectives. New Phytol. 234, 1951–1959. doi: 10.1111/nph.18016
Trivedi, P., Leach, J. E., Tringe, S. G., Sa, T., and Singh, B. K. (2020). Plant–microbiome interactions: from community assembly to plant health. Nat. Rev. Microbiol. 18, 607–621. doi: 10.1038/s41579-020-0412-1
Uroz, S., Courty, P. E., and Oger, P. (2019). Plant symbionts are engineers of the plant-associated microbiome. Trends Plant Sci. 24, 905–916. doi: 10.1016/j.tplants.2019.06.008
Vaishnav, A., and Choudhary, D. K. (2019). Regulation of drought-responsive gene expression in Glycine max L. Merrill is mediated through Pseudomonas simiae strain AU. J. Plant Growth Regul. 38, 333–342. doi: 10.1007/s00344-018-9846-3
Vaughan, M. M., Block, A., Christensen, S. A., Allen, L. H., and Schmelz, E. A. (2018). The effects of climate change associated abiotic stresses on maize phytochemical defenses. Phytochem. Rev. 17, 37–49. doi: 10.1007/s11101-017-9508-2
Venturi, V., and Keel, C. (2016). Signaling in the rhizosphere. Trends Plant Sci. 21, 187–198. doi: 10.1016/j.tplants.2016.01.005
Verrier, P. J., Bird, D., Burla, B., Dassa, E., Forestier, C., Geisler, M., et al. (2008). Plant ABC proteins–a unified nomenclature and updated inventory. Trends Plant Sci. 13, 151–159. doi: 10.1016/j.tplants.2008.02.001
Vescio, R., Malacrinò, A., Bennett, A. E., and Sorgonà, A. (2021). Single and combined abiotic stressors affect maize rhizosphere bacterial microbiota. Rhizosphere 17, 1–6. doi: 10.1016/j.rhisph.2021.100318
Vigani, G., Rolli, E., Marasco, R., Dell’Orto, M., Michoud, G., Soussi, A., et al. (2019). Root bacterial endophytes confer drought resistance and enhance expression and activity of a vacuolar H+-pumping pyrophosphatase in pepper plants. Environ. Microbiol. 21, 3212–3228. doi: 10.1111/1462-2920.14272
Vives-Peris, V., De Ollas, C., Gómez-Cadenas, A., and Pérez-Clemente, R. M. (2020). Root exudates: from plant to rhizosphere and beyond. Plant Cell Rep. 39, 3–17. doi: 10.1007/s00299-019-02447-5
Vorholt, J. A., Vogel, C., Carlström, C. I., and Müller, D. B. (2017). Establishing causality: opportunities of synthetic communities for plant microbiome research. Cell Host Microbe 22, 142–155. doi: 10.1016/j.chom.2017.07.004
Vorwerk, S., Somerville, S., and Somerville, C. (2004). The role of plant cell wall polysaccharide composition in disease resistance. Trends Plant Sci. 9, 203–209. doi: 10.1016/j.tplants.2004.02.005
Vurukonda, S. S. K. P., Vardharajula, S., Shrivastava, M., and SkZ, A. (2016). Enhancement of drought stress tolerance in crops by plant growth promoting rhizobacteria. Microbiol. Res. 184, 13–24. doi: 10.1016/j.micres.2015.12.003
Wagner, M. R., Lundberg, D. S., Del Rio, T. G., Tringe, S. G., Dangl, J. L., and Mitchell-Olds, T. (2016). Host genotype and age shape the leaf and root microbiomes of a wild perennial plant. Nat. Commun. 7, 1–15. doi: 10.1038/ncomms12151
Wang, M., Weiberg, A., Lin, F. M., Thomma, B. P. H., Da Huang, H., and Jin, H. (2016). Bidirectional cross-kingdom RNAi and fungal uptake of external RNAs confer plant protection. Nat. Plants 2, 1–10. doi: 10.1038/nplants.2016.151
Wang, C. J., Yang, W., Wang, C., Gu, C., Niu, D. D., Liu, H. X., et al. (2012). Induction of drought tolerance in cucumber plants by a consortium of three plant growth-promoting Rhizobacterium strains. PLoS One 7, 1–10. doi: 10.1371/journal.pone.0052565
Wang, Y., Zou, Y. N., Shu, B., and Wu, Q. S. (2023). Deciphering molecular mechanisms regarding enhanced drought tolerance in plants by arbuscular mycorrhizal fungi. Sci. Hortic. (Amsterdam) 308:111591. doi: 10.1016/j.scienta.2022.111591
Weisskopf, L., Schulz, S., and Garbeva, P. (2021). Microbial volatile organic compounds in intra-kingdom and inter-kingdom interactions. Nat. Rev. Microbiol. 19, 391–404. doi: 10.1038/s41579-020-00508-1
Weston, L. A., Ryan, P. R., and Watt, M. (2012). Mechanisms for cellular transport and release of allelochemicals from plant roots into the rhizosphere. J. Exp. Bot. 63, 3445–3454. doi: 10.1093/jxb/ers054
Westwood, J. H., Mccann, L., Naish, M., Dixon, H., Murphy, A. M., Stancombe, M. A., et al. (2013). A viral RNA silencing suppressor interferes with abscisic acid-mediated signalling and induces drought tolerance in Arabidopsis thaliana. Mol. Plant Pathol. 14, 158–170. doi: 10.1111/j.1364-3703.2012.00840.x
Williams, A., and De Vries, F. T. (2020). Plant root exudation under drought: implications for ecosystem functioning. New Phytol. 225, 1899–1905. doi: 10.1111/nph.16223
Wipf, H. M. L., Bùi, T. N., and Coleman-Derr, D. (2021). Distinguishing between the impacts of heat and drought stress on the root microbiome of Sorghum bicolor. Phytobiomes J. 5, 166–176. doi: 10.1094/PBIOMES-07-20-0052-R
Woo, O. G., Kim, H., Kim, J. S., Keum, H. L., Lee, K. C., Sul, W. J., et al. (2020). Bacillus subtilis strain GOT9 confers enhanced tolerance to drought and salt stresses in Arabidopsis thaliana and Brassica campestris. Plant Physiol. Biochem. 148, 359–367. doi: 10.1016/j.plaphy.2020.01.032
Xie, X., Huang, W., Liu, F., Tang, N., Liu, Y., Lin, H., et al. (2013). Functional analysis of the novel mycorrhiza-specific phosphate transporter PT1 and PHT1 family from Astragalus sinicus during the arbuscular mycorrhizal symbiosis. New Phytol. 198, 836–852. doi: 10.1111/nph.12188
Xu, P., Chen, F., Mannas, J. P., Feldman, T., Sumner, L. W., and Roossinck, M. J. (2008). Virus infection improves drought tolerance. New Phytol. 180, 911–921. doi: 10.1111/j.1469-8137.2008.02627.x
Xu, L., and Coleman-Derr, D. (2019). Causes and consequences of a conserved bacterial root microbiome response to drought stress. Curr. Opin. Microbiol. 49, 1–6. doi: 10.1016/j.mib.2019.07.003
Xu, L., Dong, Z., Chiniquy, D., Pierroz, G., Deng, S., Gao, C., et al. (2021). Genome-resolved metagenomics reveals role of iron metabolism in drought-induced rhizosphere microbiome dynamics. Nat. Commun. 12:553. doi: 10.1038/s41467-021-23553-7
Xu, L., Li, T., Wu, Z., Feng, H., Yu, M., Zhang, X., et al. (2018a). Arbuscular mycorrhiza enhances drought tolerance of tomato plants by regulating the 14-3-3 genes in the ABA signaling pathway. Appl. Soil Ecol. 125, 213–221. doi: 10.1016/j.apsoil.2018.01.012
Xu, L., Naylor, D., Dong, Z., Simmons, T., Pierroz, G., Hixson, K. K., et al. (2018b). Drought delays development of the sorghum root microbiome and enriches for monoderm bacteria. Proc. Natl. Acad. Sci. U. S. A. 115, E4284–E4293. doi: 10.1073/pnas.1717308115
Yang, Y., Zhang, X., Wang, H., Fu, X., Wen, X., Zhang, C., et al. (2019). How understory vegetation affects the catalytic properties of soil extracellular hydrolases in a Chinese fir (Cunninghamia lanceolata) forest. Eur. J. Soil Biol. 90, 15–21. doi: 10.1016/j.ejsobi.2018.11.004
Zhalnina, K., Louie, K. B., Hao, Z., Mansoori, N., Da Rocha, U. N., Shi, S., et al. (2018). Dynamic root exudate chemistry and microbial substrate preferences drive patterns in rhizosphere microbial community assembly. Nat. Microbiol. 3, 470–480. doi: 10.1038/s41564-018-0129-3
Zhang, D., He, J., Cheng, P., Zhang, Y., Khan, A., Wang, S., et al. (2023). 4-methylumbelliferone (4-MU) enhances drought tolerance of apple by regulating rhizosphere microbial diversity and root architecture. Hortic. Res. 10:99. doi: 10.1093/hr/uhad099
Zhang, J., Liu, Y. X., Guo, X., Qin, Y., Garrido-Oter, R., Schulze-Lefert, P., et al. (2021). High-throughput cultivation and identification of bacteria from the plant root microbiota. Nat. Protoc. 16, 988–1012. doi: 10.1038/s41596-020-00444-7
Zhang, Z., Zhang, J., Xu, G., Zhou, L., and Li, Y. (2019). Arbuscular mycorrhizal fungi improve the growth and drought tolerance of Zenia insignis seedlings under drought stress. New For. 50, 593–604. doi: 10.1007/s11056-018-9681-1
Zhang, F., Zou, Y. N., Wu, Q. S., and Kuča, K. (2020). Arbuscular mycorrhizas modulate root polyamine metabolism to enhance drought tolerance of trifoliate orange. Environ. Exp. Bot. 171:3926. doi: 10.1016/j.envexpbot.2019.103926
Keywords: plant-associated microbiome, biostimulants, plant resilience, ‘omics’, signaling networks, water deficit, root exudates, multiway interactions
Citation: Ait-El-Mokhtar M, Meddich A and Baslam M (2023) Plant-microbiome interactions under drought—insights from the molecular machinist’s toolbox. Front. Sustain. Food Syst. 7:1253735. doi: 10.3389/fsufs.2023.1253735
Received: 06 July 2023; Accepted: 02 November 2023;
Published: 16 November 2023.
Edited by:
Raffaella Balestrini, National Research Council (CNR), ItalyReviewed by:
Raj Majumdar, Agricultural Research Service (USDA), United StatesCopyright © 2023 Ait-El-Mokhtar, Meddich and Baslam. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Marouane Baslam, bW1tLmJhc2xhbUBnbWFpbC5jb20=
†Present address: Marouane Baslam, GrowSmart, Seoul, Republic of Korea
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.