
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Sustain. Food Syst., 15 September 2023
Sec. Sustainable Food Processing
Volume 7 - 2023 | https://doi.org/10.3389/fsufs.2023.1208970
This article is part of the Research TopicOmics Technology in Food and Agricultural NutritionView all 4 articles
Introduction: Kefir grains with efficient proteolytic system is an excellent starter culture for the production of bioactive peptides and milk products. This study explores the casein peptides derived from fermented bovine milk by kefir grains using the peptidomics approaches. The angiotensin converting enzyme (ACE) inhibitory activity of these peptides were also investigated.
Methods: After fermentation, peptidomics based on the LC-MS/MS was used to investigate the dynamic profile and the structure specificity of generated peptides. The ACE inhibitory activity of peptides was determined by measuring the amount of hippuric acid (HA) by a spectrophotometer at 228 nm.
Results: The results indicated that the cell envelope proteinases (CEPs) were the PI-/PIII-type. A total of 122 peptides were identified. The β-casein was preferentially hydrolyzed by kefir grains, and the main hydrolysis regions were f57-93, f132-160 and f192-209. The αs1-, and κ-casein were also hydrolyzed by a weaker degree. In the process of fermentation, the accumulated peptides increased with the fermentation time. The fermentation products exhibited ACE inhibitory activity, and this bioactivity remained 63% after simulated gastrointestinal (GI) digestion in vitro. Additionally, 14 Pro-containing peptides with ACE inhibitory activity were also identified.
Conclusion: These results provide new insights and evidence to investigate the bioactive milk peptides generated by kefir grains fermentation, as well as a reference for the development of functional foods.
Kefir grains are small, gelatinous, cauliflower-shaped clusters of bacteria and fungus that are used to ferment milk or other liquids (Purutoğlu et al., 2020; González-Orozco et al., 2022). To date, a wide range of microbiota have been identified from kefir grains and the Lacotacillus is the predominant microbial species. Other bacterial genus, such as Leuconostoc, Lactococcus, Streptococcus, Acetobacter, etc. are also identified (Chang-Liao et al., 2020). As for the fungus in kefir grains, yeasts are the most abundant species. Low abundance of molds including Alternaria, Aspergillus, Malassezia have been also found (Dertli and Con, 2017). Kefir grains are usually white or yellow in color and have a slightly rubbery texture. However, the characteristics including size, shape, and color will change depending on the type of milk or liquid they are fermented in and the specific strains of bacteria and yeast present in the grains (Leite et al., 2013; Shahabi-Ghahfarrokhi et al., 2015; Guzel-Seydim et al., 2021). Kefir grains are highly valued for their ability to produce a probiotic-rich products that is packed with beneficial bacteria and yeast. One of the most common use for kefir grains is to ferment milk. In the past several years, various health benefits exerted by milk products fermented by kefir grains have been documented (Reid, 2015; Champagne et al., 2018). In most cases, some beneficial effects of these fermented milk products are due to the generation of secondary metabolites, especially the bioactive peptides (Ebner et al., 2015).
In fermented milk, the bioactive peptides are released from caseins through the enzyme system of kefir grains. Potential advantages associated with these peptides include enhancing immune function, antioxidant, antibacterial, mineral binding, and angiotensin converting enzyme (ACE) inhibitory activity (Ebner et al., 2015). In addition, the differences of microorganism in kefir grains will be crucial to the quantity and category of bioactive peptides. Cell envelope proteinases (CEPs) of microorganism (especially the Lacotacillus) catalyze the first step of hydrolysis of milk proteins into peptides. Up to now, six different types of CEPs including PrtH, PrtB, PrtP, PrtL, PrtR, and PrtS have been identified (Solieri et al., 2018). Previous study indicated that microorganisms in kefir grains could significantly influence immunoregulatory properties of rats which may be related to release of different bioactive peptides (Davras et al., 2018). However, the substrate specificities of CEPs are variable at the level of inter-species and intra-species which will influence peptide composition in the final fermentation product. Therefore, large-scale and high-resolution mass spectrometry (MS) are necessary to determine the peptide profiles in fermented food with kefir grains.
In recent years, peptidomics approaches are gaining ever-growing attention in the field of food science. Food peptidomics is defined as the collection of peptides existing in food matrix or generated during food processing, storage or digestion (Martini et al., 2021). The recent development in high-throughput peptidomics techniques using high-resolution MS makes it possible to investigate the peptide profiles and the peptide production mechanisms in food. Peptidomics has therefore proved to be a suitable approach to monitor food protein hydrolysis and peptide generation (De Cicco et al., 2019). For example, Raveschot et al. (2020) recently exploited multiparametric analysis to identify the best bioactive peptides-producers among 120 LAB strains isolated from different Mongolian dairy fermented food.
In the present study, we investigated the peptide profiles in fermented casein by kefir grains using the peptidomics approach. Meanwhile, the ACE inhibitory effects of fermented products were also determined before and after the simulated in vitro gastrointestinal (GI) digestion. This study is expected to give researchers some new insights into how proteins may be degraded by the kefir grains.
Kefir grains were purchased from Beinuo Biological Corporation (Shanghai, China). Skim milk powder (3.4 g pro/100 g) were purchased from brightdairy Dairy Co., Ltd. (Shanghai, China). Alcalase (P4860, >2.4 U/g), pepsin (P7000, >250 units/mg powder), pancreatin (P1750, 4 × USP specifications), ACE (from rabbit lung), Hippuryl-histidiyl-leucine (HHL; substrate for ACE) were purchased from the Sigma-Aldrich Chemical Co. (Shanghai, China). Acetonitrile (HPLC grade) and trifluoroacetic acid (TFA, HPLC grade) were obtained from Fisher Scientific Inc. (Hudson, NH, USA). All the other reagents were of analytical grade.
The purchased kefir grains were sub-cultured in skim milk according to the method of Garofalo et al. (2015) with slight modifications. Kefir grains were first inoculated into the sterile skim milk (95°C, 30 min) in the concentration of 12.5% (w/v), and then incubated for 3 months at 28°C. Grains were transferred two times a week.
The diversity of bacterial and fungal in kefir grains were determined. The DNA of bacterial and fungal were extracted by Ezup Column Bacteria Genomic DNA Purification Kit (Sangon Biotech, Shanghai). Full-length of bacterial 16S rRNA genes were amplified with the universal primers 27F (AGAGTTTGATCCTGGCTCAG) and 1492R (TACGGCTACCT- TGTTACGACTT). 5.8S rDNA genes for fungal was amplified with the primers ITS 1 (CTTGGTCA TTTAGAGGAAGTAA) and ITS 4 (TCCTCCGCTTATTGATATGC). A set of 10-base barcodes for every DNA sample was added to the forward and reverse PCR primers. The PCR was run on a mastercycler gradient (Bio-radT100, USA). The total reaction volume was 25 μL, containing 12.5 μL 2 × Taq PCR MasterMix, 1 μL Primer (5 μM), 2 μL template DNA, and 9.5 μL ddH2O. The amplification program was as follows: 95°C for 5 min; 35 cycles of 95°C for 1 min, 55°C for 1 min, 72°C for 90 s, and with a final extension at 72°C for another10 min. The PCR products were purified using a GeneJET Gel Extraction Kit (Thermo Scientific, USA). 16S rRNA, ITS 1 and ITS 4 genes were sequenced by PacBio Sequel and Illumina Miseq platform, respectively. After that, the SMRT Link (version 7.0) and Illumina Analysis Pipeline (version 2.6) were used to conduct the image analysis, base calling and error estimation of bacterial and fungal raw data.
The milk fermentation process was conducted according to the previous study (Solieri et al., 2015). Briefly, kefir grains were first inoculated in MRS broth for activation at 37°C. Then the microorganisms were rinsed three times using 50 mmol Trise-HCl buffer (pH 6.5) and re-suspended in 11% (w/w) skimmed bovine milk. The number of microorganisms was adjusted to 1 × 108 ~ 6 × 108 CFU/mL. After that, 2% (v/v) of suspension containing kefir grains was inoculated to skimmed bovine milk prepared by ultra-high temperature sterilization technology (UHT). Fermentation was carried out for 24 h at 37°C at 10 rpm. Samples at 6, 12, 18 and 24 h were gathered for the following analysis.
The consumption of casein during the fermentation was determined using a RP-HPLC method with gradient elution (Bonfatti et al., 2008). The samples were performed on a Shimadzu LC-15C HPLC system equipped with a column (ZORBAX SB-C8, 4.6 mm i.d. × 250 mm, 5 μm, Agilent Technologies, USA). Solvent A was 0.1% (v/v) TFA in water, and solvent B was 0.1% (v/v) TFA in acetonitrile. Separations were carried out with the following program: 33–35% solvent B from 0 to 5 min, 35–37% solvent B from 5 to 9 min, 37–40% solvent B from 9 to 18 min, 40–41% solvent B from 18 to 22 min, 41% solvent B from 22 to 27 min, 41–43% solvent B from 27 to 28 min, 43–45% solvent B from 28 to 36 min, 45–33% solvent B from 36 to 37 min. The flow rate was 0.5 mL/min, column temperature was 45°C, and the peaks were detected at 214 nm.
The molecular weight distribution of peptides in fermented samples in the present study was determined using size exclusion chromatography according to our previous study (Wang and Li, 2017). Briefly, the pH of samples was first adjusted to 4.6 with 10% TCA, centrifuged (12,000 × g) at 4°C for 10 min and the supernatant was collected. Peptide molecular distribution of each sample was determined by HPLC equipped with a TSK gel G2000 SWXL column. Five reference substances including aprotinin (6,512 Da), bacitracin (1,423 Da), WPWW (674 Da), NCS (322 Da) and Gly-Sar (146 Da) were used to fit the calibration curve. The peptide contents in different samples were determined by using Pierce™ Quantitative Colorimetric Peptide Assay (Catalog number 23275).
The fermented samples were applied to sequential GI digestion according to our previous study (Wang and Li, 2017). Briefly, fermented samples were incubated with pepsin (1:50, w/w, enzyme/sample, pH 2.0) for 2 h at 37°C, and then the pH was adjusted to 7.5 with 1 M NaOH. The gastric digest was further hydrolyzed by pancreatin (1:50, enzyme/gastric digest, w/w) at 37°C for another 4 h. The simulated pancreatic digestion was terminated by heating solution in boiling water for 10 min.
The ACE inhibitory activity was determined by measuring the amount of hippuric acid (HA) generated from HHL according to previous studies with a few modifications (Abubakar et al., 1998; Lan et al., 2015). ACE was dissolved in 0.2 M borate buffer (pH 8.3) containing 1.0 M NaCl at the concentration of 0.5 U/mL, and the HHL were also dissolved in the same buffer at the concentration of 12.5 mM. Then 75 μL ACE solution and 50 μL HHL solution were mixed and incubated for 60 min at 37°C. The reaction was terminated by adding 150 μL 1 M HCl. The absorbance of HA was monitored at 228 nm with a spectrophotometer after ethyl acetate extraction.
The peptidomics analysis of the fermented milk was performed on a qTOF mass spectrometer Q-TOF-2 (Waters/Micromass, Manchester, UK) equipped with a nanoESI source. The TOF analyzer was calibrated using Glu-fib (Sigma-Aldrich, Shanghai, China). Before MS analysis, peptide mixtures were desalted using C18 Zip-Tip pre-packed micro-columns (Millipore, Bedford, MA, USA). The mobile phase A was 3% acetonitrile solution with 0.1% formic acid and B was 95% acetonitrile solution with 0.1% formic acid. The gradient elution was performed as follows: 0% B for 1 min, linearly 90% B in 60 min, and 90% B for 10 min.
For peptide sequencing, the MS/MS data for each peptide were processed by the MassLynx Maximum Entropy 3 (MaxEnt 3), and converted into PKL files, which were suitable for further analysis. The nano-ESI fragment ion peak lists, generated as PKL files, were analyzed by searching sequence databases with Mascot (Matrix Science, London, UK). Additionally, automated de novo sequencing combined with database searching was performed on the qTOF-MS/MS data using the MassLynx (version 4.1, Waters). Peptides identified were further investigated in related to the BIOPEP database1 for the ACE inhibitory activity.
The cleavage specificity of CEPs toward the milk protein was determined according to the previous study (Schechter, 2012). The P1 and P1’ subsites were designated as the amino acid residues in the N-terminal direction and C-terminal direction, respectively. P1 subsite will interact with the S1 subsite in CEPs and P1’ will interact with the S1’ subsite in CEPs. Therefore, the P1-P1’ peptide bond was the hydrolyzed bond. The quantitatively analysis for each specific amino acid was calculated, and the calculation process was as following (Solieri et al., 2018):
For amino acid A in position n (P1 or P1’ subsite), the cleavage probability of P1-P1’ peptide bond will be:
Where the TotalAn is the total amino acid A cleaved in position n, and the TotalApro is the total amino acid A in protein. Therefore, the mean cleavage probability of the protein is:
The coefficient Kn was defined as the ratio of Pn and , which showed the positive or negative influence of the amino acid A in the P1-P1’ bond:
When Kn>0 indicated the positive influence of the amino acid A in the P1-P1’ bond, and Kn<0 indicated a negative influence in the bond.
All data are presented as mean ± standard deviation (SD) for three replicates. One way ANOVA and Tukey analyses were performed to determine differences between samples, using the SPSS 19.0 (SPSS Inc., Chicago, IL, USA).
From the taxonomic point of view, 99.96% of the identified bacteria was firmicutes, and Lactobacillus was the predominant genus (99.95%). This result was in accordance with that of a previous study which reported that the most abundant bacterial population in kefir grains was Lactobacillus (Garofalo et al., 2015). The other identified bacterial genera in kefir grains included Pseudomonas, Acetobacter, Delftia, Bacillus, Lactococcus, Acinetobacter, and their abundances were lower than 0.5% (Figure 1A). At the species level, Lactobacillus kefir and Lactobacillus casei were the predominant species which the total abundances were more than 99%. It is reported that the strong biofilm formation capacity of Lactobacillus kefir is a key factor to maintaining the shape and size of kefir grains (Garofalo et al., 2015; Wang et al., 2021). Besides, another 5 species with low abundance were also detected, and the Lactobacillus helveticus was the most abundant one. Lactobacillus helveticus is important for the flavor of fermented milk product because it will produce the nutty flavors and prevent the bitter taste (Griffiths and Tellez, 2013). Previous study also reported that the fermented bovine milk by Lactobacillus helveticus contained ACE inhibitory peptides which could help to decrease the blood pressure (Aihara et al., 2005). As for the fungal diversity, Ascomycota was the most abundant phyla (over 92%). Several previous studies about the microflora of kefir grains from different regions also found that Ascomycota was the predominant phyla (Dertli and Con, 2017; Wang et al., 2021). At the genus level, Saccharomyces and Kluyveromyces were the most abundant fungal (54.76 and 41.55%, respectively), followed the Kazachstania which accounted for 2.33% (Figure 1B). This result was in agreement with previous studies about the fungal composition of kefir grains (Marsh et al., 2013; Gut et al., 2019). Additionally, Trichoderma, Penicillium, Epicocum, Sarcinomyces, Chaetomium, and Myceliophthora were also detected with the abundance lower than 1%.
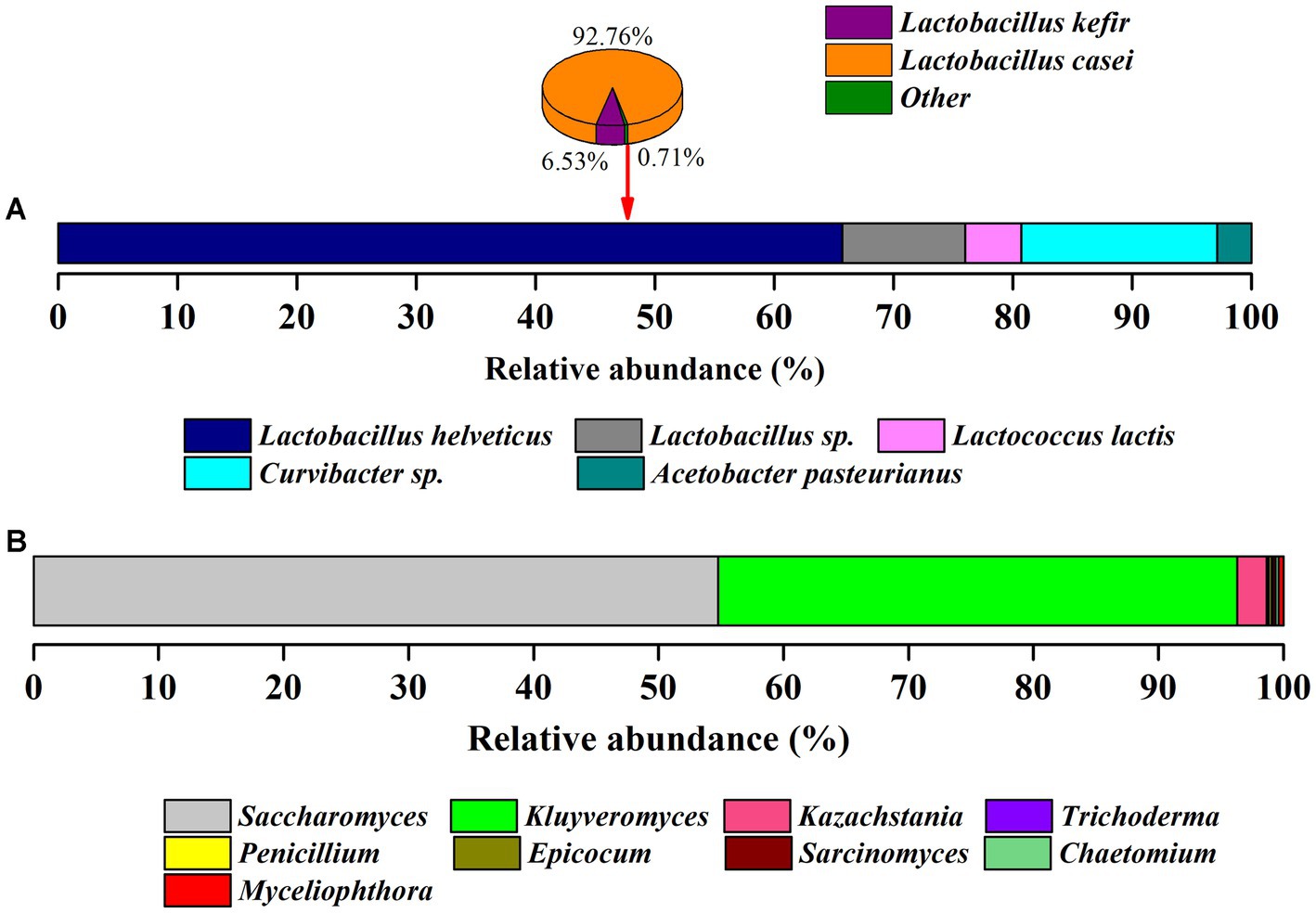
Figure 1. The relative abundance of microorganisms in kefir grains. (A) Bacterial species community. (B) Fungal species community.
About 80% of the bovine milk is casein which can be subdivided into β-casein, αs1-casein, αs2-casein and κ-casein. In the fermented process, the consumption of these four casein fractions by kefir grains was determined. As shown in Figure 2, all the hydrolyzed casein fractions increased with the extension of fermentation time. The hydrolyzed contents of β- and αs1-casein were always higher than that of αs2 and κ-casein (p < 0.05). This may be related to the abundance of β- and αs1-casein. It is reported that the β-, αs1-, αs2- and κ-casein occurs in the approximate proportions 4:4:1:1 in milk (Visser et al., 1991). At 6 h, the hydrolysis degrees of four casein fractions were very low, 1.13 mg/mL for β-casein, 1.05 mg/mL for αs1-casein, 0.33 mg/mL for αs2-casein and 0.17 mg/mL for κ-casein. However, at the end of fermentation of 24 h, a 6.8–12.6 fold increase were observed. The difference in hydrolysis degree may be caused by the preference of CEPs from the kefir grains, which would be discussed in the following sections.
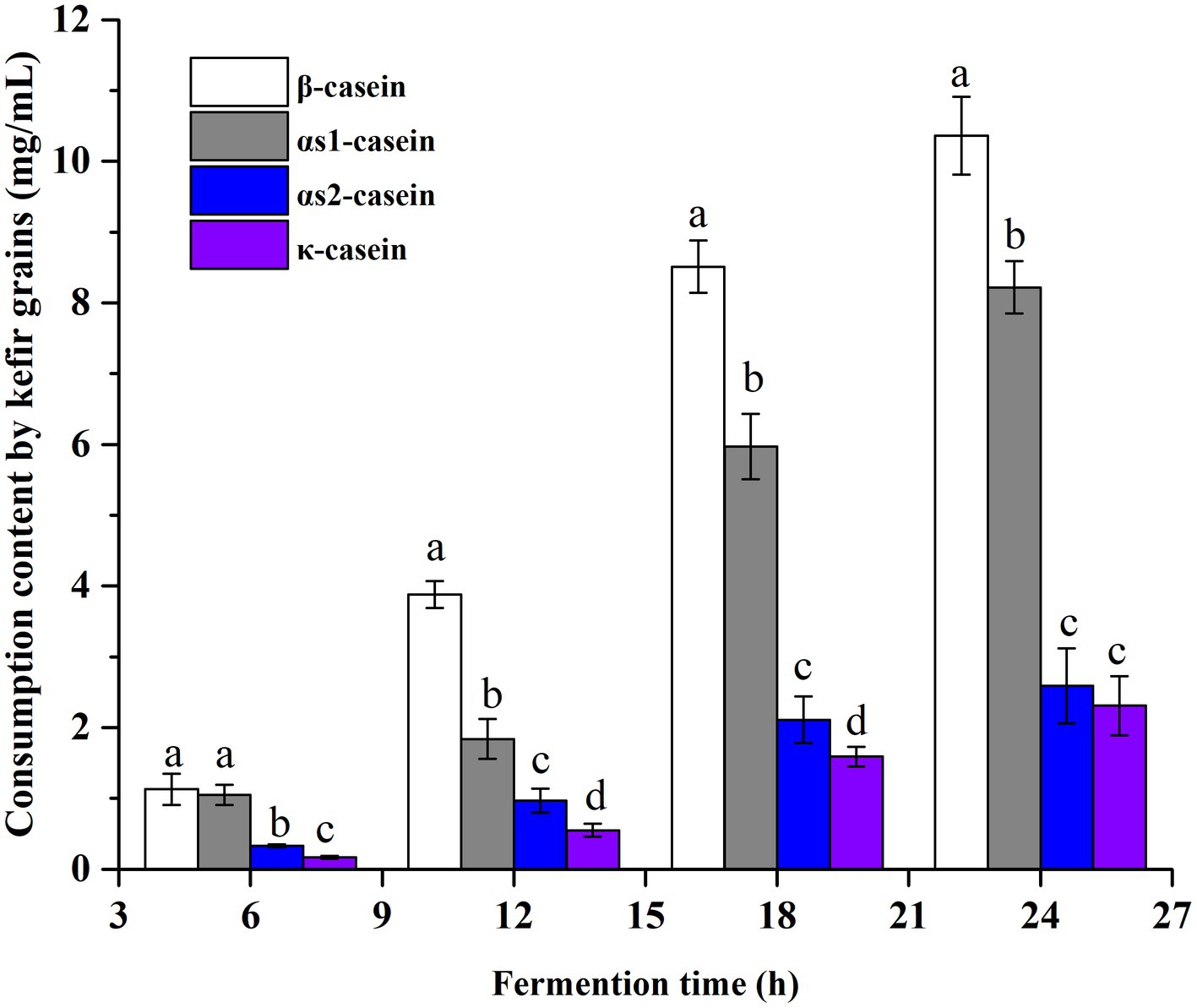
Figure 2. The consumption contents of four different casein fractions (β-, αs1-, αs2- and κ-casein) by kefir grains at different fermentation time points. Four casein fractions were determined by RP-HPLC, and the quantitative calculations of their content were performed by the peak area.
In the present study, the molecular weight of generated peptides was divided into four fractions by size exclusion chromatography, i.e., > 5,000 Da, 3,000–5,000 Da, 1,000–3,000 Da, and < 1,000 Da. As shown in Figure 3 the total content of peptides increased at first and then decreased. The maximum concentration of peptides (9.29 mg/mL) was obtained at 18 h, and then decreased to 8.49 mg/mL at 24 h. In addition, the peptides with low molecular weights increased with the extension of fermentation time. In prior period of fermentation (6 h and 12 h), the content of high molecular weight peptides (> 3,000 Da) was more than 70%, and this proportion decreased to 56.3% for 18 h and 40.8% for 24 h, respectively.
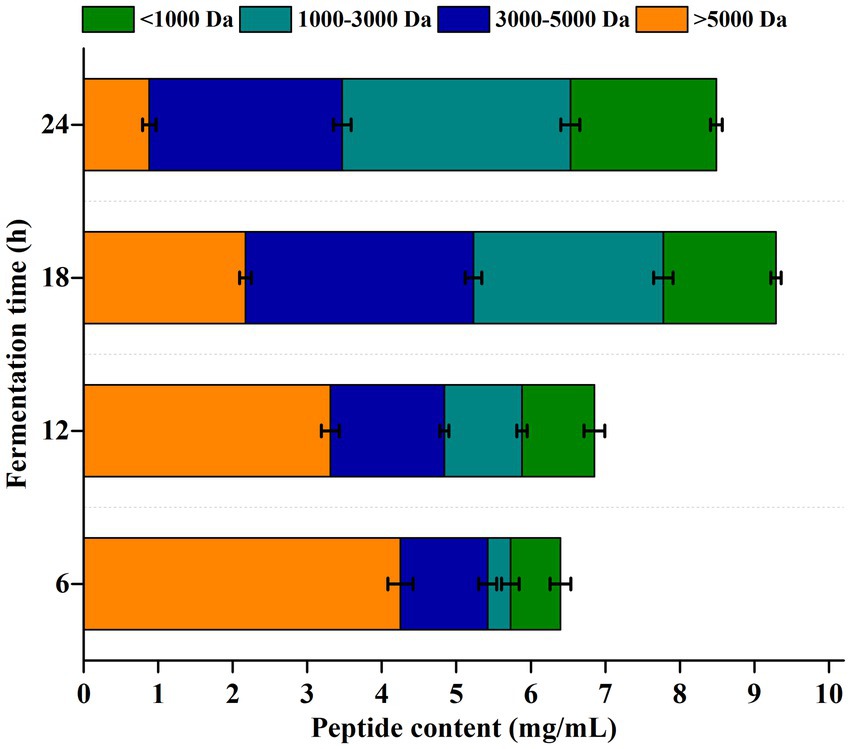
Figure 3. Molecular weight distribution of peptides generated in the fermentation process. Peptides were divided into four fractions by size exclusion chromatography, i.e., > 5,000 Da, 3,000–5,000 Da, 1,000–3,000 Da and < 1,000 Da.
The full set of peptides generated in the fermentation process was analyzed by mass spectrometry. A total of 122 milk peptides were released by the CEPs of kefir grains at the end of fermentation (Supplementary Tables S1–S4). According to the protein origin of these identified peptides, β-casein was the most preferred substrate of kefir grains. In particular, 81 peptides were derived from β-casein, which accounted for 66.4% of the total identified peptides. Followed by the κ-casein-derived peptides and αs1-casein-derived peptides, which were 19 peptides (15.6%) and 17 peptides (13.9%), respectively. For the αs2-casein, only 5 peptides (4.1%) were detected in the fermentation system indicating the poor capacity of CEPs to hydrolyze αs2-casein. The significant difference between peptides derived from different casein fractions may be related to the CEPs of microorganism in kefir grains, especially the Lactococcus lactis. CEPs can be classified according to the degradation patterns toward β-, αs1-, αs2- and κ-casein (Kunji et al., 1996). Generally, two kinds of CEPs have been described, PI-type and PIII-type. The primary substrate of PI-type is β-casein, and it can also hydrolyze the κ-casein to a lesser extent. For PIII-type CEPs, they can degrade β-, αs1-, and κ-casein equally (Visser et al., 1986; Pritchard and Coolbear, 1993). In addition, an intermediate proteases named PI-/PIII-type has also been identified, which can cleave β-casein as with PI-type and α-, and κ-casein to a lesser extent (Exterkate et al., 1993; Sadat-Mekmene et al., 2011a; Villegas et al., 2015). In the present study, the kefir grains exhibited a predominant hydrolyzation activity toward β-casein, and a lower proteolytic activity toward αs1-casein. These results indicated that the CEPs activity of kefir grains in this study may be the PI-/PIII-type.
The cleavage specificity of kefir grains toward β-casein was shown in Supplementary Figure S1. Sixty five different cleavage sites were detected, which was 31.3% of the total peptide bonds present in β-casein. This result indicated that the kefir grains have a broad cleavage specificity. According to the amino acid sequence of the identified peptides (Supplementary Table S1), we found that the cleavage sites were distributed throughout the β-casein sequence (mainly in f57-93, f132-160 and f192-209), rather than located only at C- or N-terminus. This finding was in accordance with cleavage of proteinase isolated from Lb. rhamnosus (CGMCC11055), which the cleavage sites were also distributed along the entire β-casein sequence (Guo et al., 2016). However, previous study reported that the proteinases from lactobacilli would preferentially degrade the C-terminal of β-casein (Lozo et al., 2011).
The cleavage probability (%Pn) at P1 and P1’ positions by kefir grains were also calculated. As shown in Table 1, when the P1 positions were the amino acids of Met, Trp, Gln and Asn, the %Pn ≥ 50%, which indicated the preference of CEPs toward these amino acid residues. As for the P1’ position, preferentially cleavage of CEPs were the amino acid residues of Val, Trp, Tyr, Ser, Asp., Arg and His. Furthermore, coefficients Kn were calculated to quantify the influence of different amino acid residues on the P1-P1’cleavage probabilities. As shown in Figure 4A, the Met, Trp, Gln, Asn in P1 position and Met, Ser, His in P1’ position showed the strongest positive effects on the cleavage of CEPs. The amino acids residues of Leu and His in P1 position and Val, Tyr, Asp, Arg in P1’ position also exhibited weaker positive effects on the cleavage. On the contrary, amino acid residues of Val, Gly, Tyr, and Asp in the P1 position and Ile, Pro in the P1’ position showed strong inhibition effects on the cleavage occurrence. Similarly, the amino acid residues of Ile, Pro in P1 position and Val, Phe, Gln, Glu in the P1’ position exhibited weaker inhibition effects toward the cleavage of CEPs. Usually, the CEPs preferentially degrade hydrophobic and negatively charged amino acid residues (Monnet et al., 1992; Hebert et al., 2008; Lozo et al., 2011). However, the cleavage preference for amino acids of CEPs was varied depend on the strains. For instance, CEP from Lb. rhamnosus strain CGMCC11055 the Pro was preferred in both P1 and P1’ positions (Guo et al., 2016), whereas the Pro at both P1 and P1’ positions strongly inhibited the CEP cleavage activity in strain NCDO763 (Monnet et al., 1992). The kefir grains used in the present study consist of various organisms, the cleavage specificity of CEPs may be different from that of CEP from a single strain.
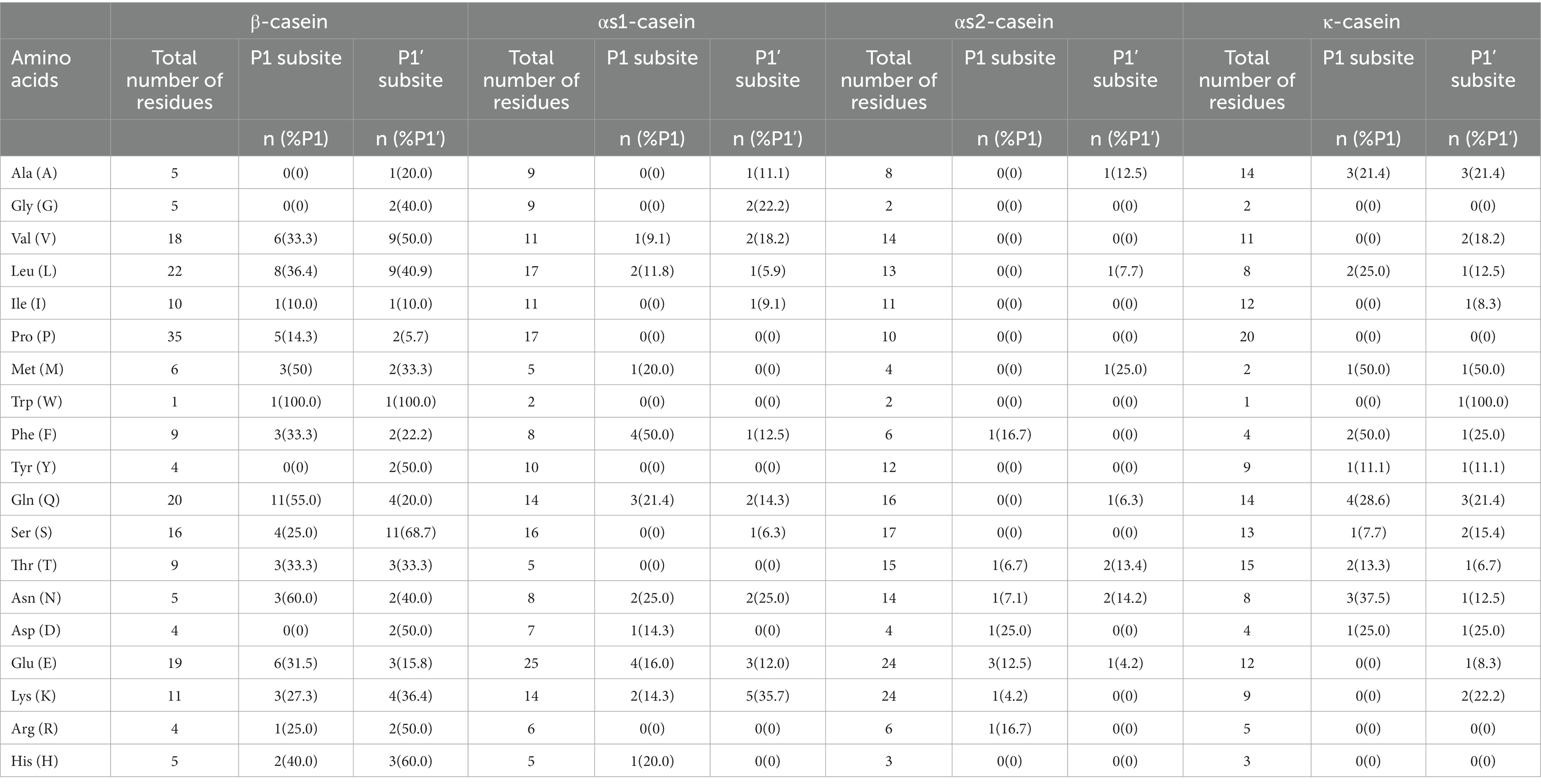
Table 1. The amino acid occurrence and cleavage probability (%Pn) caused by cell envelope proteinases (CEPs) on four casein fractions in the P1 and P1’ subsites.
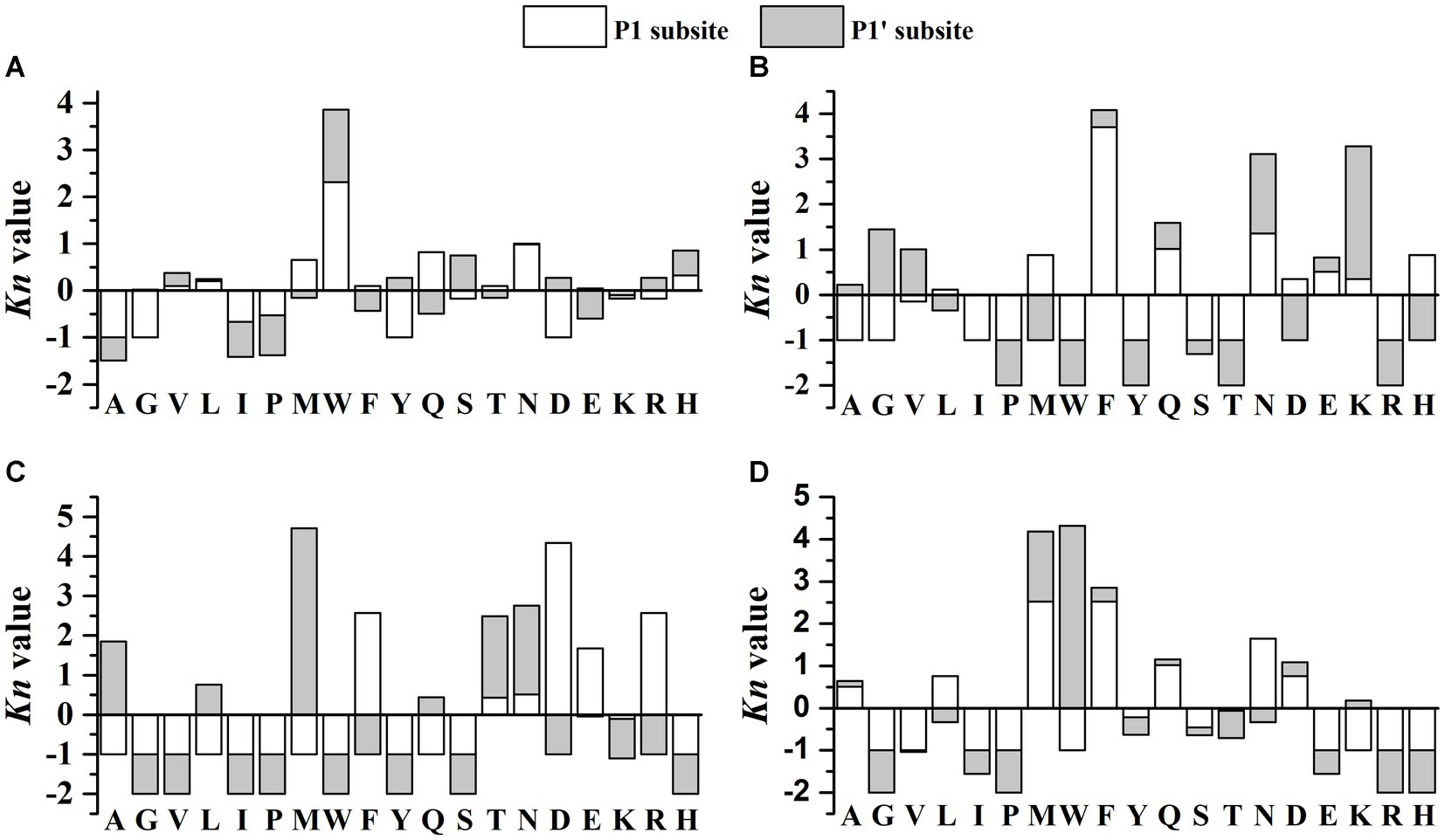
Figure 4. The cleavage preference (expressed as Kn) toward different amino acid residues at P1 and P1’ positions. (A) β-casein (B) αs1-casein (C) αs2-casein (D) κ-casein. Positive and negative values of Kn represent positive or negative effects exerted by each amino acid residue on the cleavage of the P1-P1’ bond, respectively.
In total, 21 different cleavage sites were detected and constituted 10.6% of all the peptide bonds in αs1-casein (Supplementary Figure S2). Although the amount of αs1-casein is comparable with that of β-casein, the number of cleavage sites was only one-third of that of β-casein. Additionally, most of the cleavages sites (61.9%) was distributed at the N-terminal. These cleavage occurrence may be related to the CEPs in Lactobacillus which can possess two CEPs named PrtH1 and PrtH2. It is reported that when only PrtH2 was present, 22–30% of peptides were released from αs1-casein, and the percentage increased to 41–49% when both CEPs were expressed (Sadat-Mekmene et al., 2011a). In the perspective of peptide percentage, it is likely that only PrtH2 was expressed in the kefir grains. Compare with the hydrolyzation of purified casein, the number of αs1-casein-derived peptides will drastic decrease when the strains were grown in milk (Sadat-Mekmene et al., 2011b).
For the cleavage probability analysis, only the amino acid residue Phe in the P1 position possessed %Pn = 50% (Table 1), suggesting the low preference toward αs1-casein. The calculated coefficients Kn showed that Phe, Asn in P1 position and Asn, Lys in P1’ position exerted strong positive effects on the cleavage of CEPs (Figure 4B). In contrast, almost 9 amino acid residues (Ala, Gly, Ile, Pro, Trp, Tyr, Ser, Thr, Arg) in P1 position and 8 amino acid residues (Pro, Met, Trp, Tyr, Thr, Asp, Arg, His) in P1’ position showed strong inhibition effects on the cleavage probability. The αs1-casein fragment (f1-23) is usually used to classify the CEPs according to their cleavage specificities (Exterkate et al., 1993). In the present study, 5 cleavage sites including H8-Q9, Q9-G10, L16-N17, N17-E18, E18-N19 were detected (Supplementary Figure S2). Most of these cleavage sites are typical of PI-/PIII-type protease (Fernandez-Espla et al., 2000; Hebert et al., 2008). This result was agree with the previous findings in section of “peptidomics analysis” in the present study.
According to the peptidomics analysis, only 5 peptides and 9 different cleavage sites (4.4% of the total peptide bonds) were detected from the αs2-casein (Supplementary Table S3 and Supplementary Figure S3). This is probably related to the structure of αs2-casein, which consists of more α-helix in the interior regions limiting the accessibility of CEPs (Farrell et al., 2009; Sadat-Mekmene et al., 2011a). As for the cleavage probability analysis, the %Pn of all amino acid residues was below 50% suggesting low cleavage probability of CEPs toward αs2-casein (Table 1). As shown in Figure 4C, the amino acid residues of Phe, Asp., Glu, Arg in P1 position and Ala, Met, Thr, Asn in P1’ position showed strong positive effects on cleavage. However, there were 12 amino acid residues both in P1 position and P1’ position exerted inhibition effects on the cleavage. It is reported that the cleavage sites of PrtH2 toward αs2-casein are mainly located at fragment of f97-162 (Sadat-Mekmene et al., 2011b). The detected 5 peptides of this study were mainly located in fragment of f115-207. Since the fragment of f126-207 are hydrophilic (Miclo et al., 2012), the PrtH2 preferred to degrade the hydrophilic regions.
A total of 21 different cleavage sites were observed, which consisted of 12.5% of the total peptide bonds present in κ-casein (Supplementary Figure S4). And these cleavage sites were distributed along with the sequence of κ-casein. For the cleavage probability, the Met, Phe in P1 position and Met, Trp in P1’ position were the preferred amino acid residues for cleavage (Table 1). The coefficients Kn showed that Met, Phe, Asn in P1 position and Met, Trp in P1’ position exhibited strong positive influence on the cleavage of CEPs. Leu, Gln and Asp in P1 position showed weaker positive effects. By in contrast, 9 amino acid residues (Gly, Val, Ile, Pro, Trp, Glu, Lys, Arg, His) in P1 position and 7 amino acid residues (Gly, Ile, Pro, Thr, Glu, Arg, His) in P1’ position showed strong inhibition effects on the cleavage probability (Figure 4D).
The identified peptides in the fermented products were searched against the milk bioactive peptide database and BIOPEP database (Minkiewicz et al., 2008; Nielsen et al., 2017) for finding peptides matched with the known ACE inhibitory peptides. As shown in Table 2, 14 peptides (11.5% of the total identified peptides) were designated with the ACE inhibitory activity. Among these peptides, 12 peptides were derived from β-casein mainly distributed in the region of f47-90 and f132-208, and the other two peptides were from αs1-casein. Four peptides of YPFPGPIPN, LHLPLP, KVLPVPQ and RPKHPIKHQ have been proven to reduce the blood pressure in spontaneously hypertensive rats (Saito et al., 2000; Fuglsang et al., 2003; Quirós et al., 2007; Garcia-Tejedor et al., 2015; Martini et al., 2020). The other 10 peptides have been proven the in vitro antihypertensive activity.
Usually, the resistance to the GI protease is a prerequisite for bioactive peptides to exert their physiological effects (Wang and Li, 2017). In the present study, the ACE inhibitory activities of the fermented milk at different time points were determined before and after the simulated GI digestion (Figure 5). The inhibitory activity increased with the extension of fermentation time, suggesting the continuous generation of ACE inhibitory peptides. At 24 h of fermentation, the inhibitory activities for before and after GI digestion were 73.5 and 46.3%, respectively. The activity retention rate was about 63%, which indicated that these ACE inhibitory peptides possessed well digestion stability. It is notable that, the ACE inhibitory activity at 6 h increased after simulated GI digestion. The degree of hydrolysis and peptide content were low at the beginning of fermentation, and the protease in GI tract might exert positive effect on the peptide releasing. In previous studies, peptides containing Pro in their sequences were proven to be resistant to the digestive proteases (Tagliazucchi et al., 2016; Ma et al., 2021). As shown in Table 2, all the 14 identified ACE inhibitory peptides were Pro-containing peptides and the number of Pro ranged from 1 to 4.
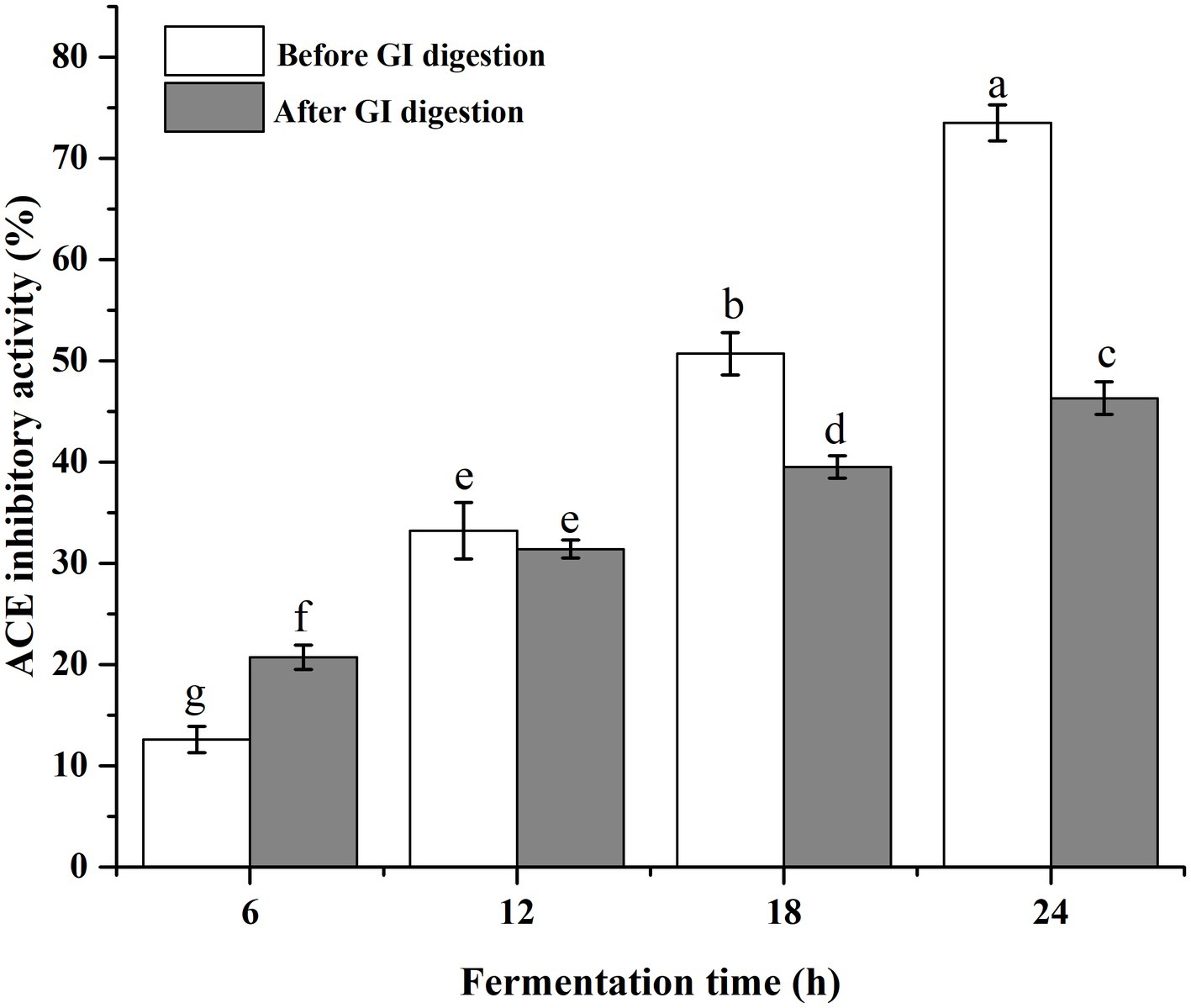
Figure 5. The ACE inhibitory activity of fermented milk products before and after the simulated gastrointestinal digestion.
Kefir grains are excellent starters for fermented milk products with diverse physiological functions. Usually, the bioactive peptides generated during the fermentation are responsible for these functions. The analysis and exploration of generated bioactive peptides in fermented milk are therefore important. The developments of peptidomics and high-resolution MS make it possible for this problem. In the present study, peptidomics approaches were used for exploring the peptide profiles in fermented skim milk by kefir grains. A total of 122 peptides were detected, and 66.4% were released from β-casein by CEPs which were mainly the PI-/PIII-type. Meanwhile, the fermented milk showed ACE inhibitory effect, which 63% of the activity were retained after the simulated in vitro GI digestion. Fourteen ACE inhibitory peptides containing Pro were identified according to the online database. However, more in vitro and in vivo experiments are needed to further verify the ACE inhibitory activity of these peptides.
The data presented in the study are publicly available. This data can be found here: (link: https://pan.baidu.com/s/1_cxJUee-q_3JWKWLaCYOKA accession number: a2b3).
BW, SX, and YC performed the project, methodology, and validation. JW provided the supervision. BW wrote the manuscript. SX and XC analyzed the data. All authors have read and approved the manuscript for publication.
This study was supported by Guangdong Basic and Applied Basic Research Foundation (2020A1515110211), Project of Educational Commission of Guangdong Province of China (2021KTSCX132), and Research start-up funds of DGUT (211135027).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fsufs.2023.1208970/full#supplementary-material
Abubakar, A., Saito, T., Kitazawa, H., Kawai, Y., and Itoh, T. (1998). Structural analysis of new antihypertensive peptides derived from cheese whey protein by proteinase K digestion. J. Dairy Sci. 81, 3131–3138. doi: 10.3168/jds.S0022-0302(98)75878-3
Aihara, K., Kajimoto, O., Hirata, H., Takahashi, R., and Nakamura, Y. (2005). Effect of powdered fermented milk with Lactobacillus helveticus on subjects with high-normal blood pressure or mild hypertension. J. Am. Coll. Nutr. 24, 257–265. doi: 10.1080/07315724.2005.10719473
Bonfatti, V., Grigoletto, L., Cecchinato, A., Gallo, L., and Carnier, P. (2008). Validation of a new reversed-phase high-performance liquid chromatography method for separation and quantification of bovine milk protein genetic variants. J. Chromatogra. A 1195, 101–106. doi: 10.1016/j.chroma.2008.04.075
Champagne, C. P., da Cruz, A. G., and Daga, M. (2018). Strategies to improve the functionality of probiotics in supplements and foods. Curr. Opin. Food Sci. 22, 160–166. doi: 10.1016/j.cofs.2018.04.008
Chang-Liao, W. P., Lee, A., Chiu, Y. H., Chang, H. W., and Liu, J. R. (2020). Isolation of a Leuconostoc mesenteroides strain with anti-porcine epidemic diarrhea virus activities from kefir grains. Front. Microbiol. 11:1578. doi: 10.3389/fmicb.2020.01578
Davras, F., Tas, T. K., and Guzel-Seydim, Z. B. (2018). Immunological effects of kefir produced from kefir grains versus starter cultures when fed to mice. Funct. Foods Health Dis. 8, 367–423. doi: 10.31989/ffhd.v8i8.533
De Cicco, M., Mamone, G., Di Stasio, L., Ferranti, P., Addeo, F., and Picariello, G. (2019). Hidden “digestome”: current analytical approaches provide incomplete peptide inventories of food digests. J. Agric. Food Chem. 67, 7775–7782. doi: 10.1021/acs.jafc.9b02342
Dertli, E., and Con, A. H. (2017). Microbial diversity of traditional kefir grains and their role on kefir aroma. LWT-Food Sci. Technol. 85, 151–157. doi: 10.1016/j.lwt.2017.07.017
Ebner, J., Arslan, A. A., Fedorova, M., Hoffmann, R., Küçükçetin, A., and Pischetsrieder, M. (2015). Peptide profiling of bovine kefir reveals 236 unique peptides released from caseins during its production by starter culture or kefir grains. J. Proteome 117, 41–57. doi: 10.1016/j.jprot.2015.01.005
Exterkate, F. A., Alting, A. C., and Bruinenberg, P. G. (1993). Diversity of cell envelope proteinase specificity among strains of Lactococcus lactis and its relationship to charge characteristics of the substrate-binding region. App. Environ. Microbiol. 59, 3640–3647. doi: 10.1128/aem.59.11.3640-3647.1993
Farrell, H. M. Jr., Malin, E. L., Brown, E. M., and Mora-Gutierrez, A. (2009). Review of the chemistry of αS2-casein and the generation of a homologous molecular model to explain its properties. J. Dairy Sci. 92, 1338–1353. doi: 10.3168/jds.2008-1711
Fernandez-Espla, M. D., Garault, P., Monnet, V., and Rul, F. (2000). Streptococcus thermophilus cell-wall anchored proteinase: release, purification, and biochemical and genetic characterization. App. Environ. Microbiol. 66, 4772–4778. doi: 10.1128/AEM.66.11.4772-4778.2000
Fuglsang, A., Rattray, F. P., Nilsson, D., and Nyborg, N. C. (2003). Lactic acid bacteria: inhibition of angiotensin converting enzyme in vitro and in vivo. Anton Leeuw. Int. J. G. 83, 27–34. doi: 10.1023/A:1022993905778
Garcia-Tejedor, A., Sanchez-Rivera, L., Recio, I., Salom, J. B., and Manzanares, P. (2015). Dairy Debaryomyces hansenii strains produce the antihypertensive casein-derived peptides LHLPLP and HLPLP. LWT Food Sci. Technol. 61, 550–556. doi: 10.1016/j.lwt.2014.12.019
Garofalo, C., Osimani, A., Milanovic, V., Aquilanti, L., De Filippis, F., Stellato, G., et al. (2015). Bacteria and yeast microbiota in milk kefir grains from different Italian regions. Food Microbiol. 49, 123–133. doi: 10.1016/j.fm.2015.01.017
González-Orozco, B. D., García-Cano, I., Jiménez-Flores, R., and Alvárez, V. B. (2022). Invited review: milk kefir microbiota-direct and indirect antimicrobial effects. J. Dairy Sci. 105, 3703–3715. doi: 10.3168/jds.2021-21382
Griffiths, M. W., and Tellez, A. M. (2013). Lactobacillus helveticus: the proteolytic system. Front. Microbiol. 4:30. doi: 10.3389/fmicb.2013.00030
Guo, T., Ouyang, X., Xin, Y., Wang, Y., Zhang, S., and Kong, J. (2016). Characterization of a new cell envelope proteinase PrtP from Lactobacillus rhamnosus CGMCC11055. J. Agric. Food Chem. 64, 6985–6992. doi: 10.1021/acs.jafc.6b03379
Gut, A. M., Vasiljevic, T., Yeager, T., and Donkor, O. N. (2019). Characterization of yeasts isolated from traditional kefir grains for potential probiotic properties. J. Funct. Foods 58, 56–66. doi: 10.1016/j.jff.2019.04.046
Guzel-Seydim, Z. B., Gökırmaklı, Ç., and Greene, A. K. (2021). A comparison of milk kefir and water kefir: physical, chemical, microbiological and functional properties. Trends Food Sci. Tech. 113, 42–53. doi: 10.1016/j.tifs.2021.04.041
Hebert, E. M., Mamone, G., Picariello, G., Raya, R. R., Savoy, G., Ferranti, P., et al. (2008). Characterization of the pattern of as1- and β-casein breakdown and release of bioactive peptide by a cell envelope proteinase from Lactobacillus delbrueckii subsp. lactis CRL 581. App. Environ. Microbiol. 74, 3682–3689. doi: 10.1128/AEM.00247-08
Kunji, E. R. S., Mierau, I., Hagting, A., Poolman, B., and Konings, W. N. (1996). The proteolytic systems of lactic acid bacteria. Anton Leeuw. Int. J. G. 70, 187–221. doi: 10.1007/BF00395933
Lan, X. D., Liao, D. K., Wu, S. G., Wang, F., Sun, J. H., and Tong, Z. F. (2015). Rapid purification and characterization of angiotensin converting enzyme inhibitory peptides from lizard fish protein hydrolysates with magnetic affinity separation. Food Chem. 182, 136–142. doi: 10.1016/j.foodchem.2015.02.004
Leite, A. M. D. O., Miguel, M. A. L., Peixoto, R. S., Rosado, A. S., Silva, J. T., and Paschoalin, V. M. F. (2013). Microbiological, technological and therapeutic properties of kefir: a natural probiotic beverage. Braz. J. Microbiol. 44, 341–349. doi: 10.1590/S1517-83822013000200001
Lozo, J., Strahinic, I., Dalgalarrondo, M., Chobert, J. M., Haertlé, T., and Topisirovic, C. (2011). Comparative analysis of β-casein proteolysis by PrtP proteinase from Lactobacillus paracasei subsp. paracasei BGHN14, PrtR proteinase from lacto- bacillus rhamnosus BGT10 and PrtH proteinase from Lactobacillus helveticus BGRA43. Int. Dairy J. 21, 863–868. doi: 10.1016/j.idairyj.2011.05.002
Ma, Y., Hou, Y., Han, B., Xie, K., Zhang, L., and Zhou, P. (2021). Peptidome comparison following gastrointestinal digesta of bovine versus caprine milk serum. J. Dairy Sci. 104, 47–60. doi: 10.3168/jds.2020-18471
Marsh, A. J., O’Sullivan, O., Hill, C., Ross, R. P., and Cotter, P. D. (2013). Sequencing-based analysis of the bacterial and fungal composition of kefir grains and milks from multiple sources. PLoS One 8:e69371. doi: 10.1371/journal.pone.0069371
Martini, S., Conte, A., and Tagliazucchi, D. (2020). Effect of ripening and in vitro digestion on the evolution and fate of bioactive peptides in Parmigiano-Reggiano cheese. Int. Dairy J. 105:104668. doi: 10.1016/j.idairyj.2020.104668
Martini, S., Solieri, L., and Tagliazucchi, D. (2021). Peptidomics: new trends in food science. Curr. Opin. Food Sci. 39, 51–59. doi: 10.1016/j.cofs.2020.12.016
Miclo, L., Roux, E., Genay, M., Brusseaux, E., Poirson, C., Jameh, N., et al. (2012). Variability of hydrolysis of β-, αs1-, and αs2-caseins by 10 strains of Streptococcus thermophilus and resulting bioactive peptides. J. Agric. Food Chem. 60, 554–565. doi: 10.1021/jf202176d
Minkiewicz, P., Dziuba, J., Iwaniak, A., Dziuba, M., and Darewicz, M. (2008). BIOPEP database and other programs for processing bioactive peptide sequences. J. AOAC Int. 91, 965–980. doi: 10.1093/jaoac/91.4.965
Monnet, V., Ley, J. P., and Gonzalez, S. (1992). Substrate specificity of the cell envelope-located proteinase of Lactococcus lactis subsp. lactis NCDO 763. Int. J. Biochem. 24, 707–718. doi: 10.1016/0020-711x(92)90004-k
Nielsen, S. D., Beverly, R. L., Qu, Y., and Dallas, D. C. (2017). Milk bioactive peptide database: a comprehensive database of milk protein-derived bioactive peptides and novel visualization. Food Chem. 232, 673–682. doi: 10.1016/j.foodchem.2017.04.056
Pritchard, G. G., and Coolbear, T. (1993). The physiology and biochemistry of the proteolytic system in lactic acid bacteria. FEMS Microbiol. Rev. 12, 179–206. doi: 10.1111/j.1574-6976.1993.tb00018.x
Purutoğlu, K., İspirli, H., Yüzer, M. O., Serencam, H., and Dertli, E. (2020). Diversity and functional characteristics of lactic acid bacteria from traditional kefir grains. Int. J. Dairy Technol. 73, 57–66. doi: 10.1111/1471-0307.12633
Quirós, A., Ramos, M., Muguerza, B., Delgado, M. A., Miguel, M., Aleixandre, A., et al. (2007). Identification of novel antihypertensive peptides in milk fermented with Enterococcus faecalis. Int. Dairy J. 17, 33–41. doi: 10.1016/j.idairyj.2005.12.011
Raveschot, C., Cudennec, B., Deracinois, B., Frémont, M., Vaeremans, M., Dugersuren, J., et al. (2020). Proteolytic activity of Lactobacillus strains isolated from Mongolian traditional dairy products: a multiparametric analysis. Food Chem. 304, 125–415. doi: 10.1016/j.foodchem.2019.125415
Reid, G. (2015). The growth potential for dairy probiotics. Int. Dairy J. 49, 16–22. doi: 10.1016/j.idairyj.2015.04.004
Schechter, I. (2012). Reprint of “On the size of the active site in proteases. I. Papain.” Biochem. Bioph. Res. Co. 425, 497–502.
Sadat-Mekmene, L., Genay, M., Atlan, D., Lortal, S., and Gagnaire, V. (2011a). Original features of cell-envelope proteinases of Lactobacillus helveticus. A review. Int. J. Food Microbiol. 146, 1–13. doi: 10.1016/j.ijfoodmicro.2011.01.039
Sadat-Mekmene, L., Jardin, J., Corre, C., Mollé, D., Richoux, R., Delage, M. M., et al. (2011b). Simultaneous presence of PrtH and PrtH2 proteinases in Lactobacillus helveticus strains improves breakdown of the pure αs1-casein. App. Environ. Microbiol. 77, 179–186. doi: 10.1128/AEM.01466-10
Saito, T., Nakamura, T., Kitazawa, H., Kawai, Y., and Itoh, T. (2000). Isolation and structural analysis of antihypertensive peptides that exist naturally in gouda cheese. J. Dairy Sci. 83, 1434–1440. doi: 10.3168/jds.S0022-0302(00)75013-2
Shahabi-Ghahfarrokhi, I., Khodaiyan, F., Mousavi, M., and Yousefi, H. (2015). Effect of γ-irradiation on the physical and mechanical properties of kefiran biopolymer film. Int. J. Biol. Macromol. 74, 343–350. doi: 10.1016/j.ijbiomac.2014.11.038
Solieri, L., Rutella, G. S., and Tagliazucchi, D. (2015). Impact of non-starter lactobacilli on release of peptides with angiotensin-converting enzyme inhibitory and antioxidant activities during bovine milk fermentation. Food Microbiol. 51, 108–116. doi: 10.1016/j.fm.2015.05.012
Solieri, L., De Vero, L., and Tagliazucchi, D. (2018). Peptidomic study of casein proteolysis in bovine milk by Lactobacillus casei PRA205 and Lactobacillus rhamnosus PRA331. Int. Dairy J. 85, 237–246. doi: 10.1016/j.idairyj.2018.06.010
Tagliazucchi, D., Helal, A., Verzelloni, E., Bellesia, A., and Conte, A. (2016). Composition and properties of peptides that survive standardised in vitro gastro-pancreatic digestion of bovine milk. Int. Dairy J. 61, 196–204. doi: 10.1016/j.idairyj.2016.06.002
Villegas, J. M., Brown, L., Savoy de Giori, G., and Hebert, E. M. (2015). Characterization of the mature cell surface proteinase of Lactobacillus delbrueckii subsp. lactis CRL581. Appl. Microbiol. Biotechnol. 99, 4277–4286. doi: 10.1007/s00253-014-6258-6
Visser, S., Exterkate, F. A., Slangen, C. J., and de Veer, J. C. M. (1986). Comparative study of action of cell wall proteinases from various strains of Streptococcus cremoris on bovine as1-, β-, and κ-casein. App. Environ. Microbiol. 52, 1162–1166. doi: 10.1128/aem.52.5.1162-1166.1986
Visser, S., Slangen, C. J., and Rollema, H. S. (1991). Phenotyping of bovine milk proteins by reversed-phase high-performance liquid chromatography. J. Chromatogr. A 548, 361–370. doi: 10.1016/S0021-9673(01)88619-2
Wang, B., and Li, B. (2017). Effect of molecular weight on the transepithelial transport and peptidase degradation of casein-derived peptides by using Caco-2 cell model. Food Chem. 218, 1–8. doi: 10.1016/j.foodchem.2016.08.106
Keywords: peptidomics, casein degradation, bioactive peptides, kefir fermentation, ACE inhibitory activity
Citation: Wang B, Xiao S, Cai Y, Chen X and Wang J (2023) Peptidomics approaches to the discovery and ACE inhibitory effect of casein peptides derived from fermented bovine milk by kefir grains. Front. Sustain. Food Syst. 7:1208970. doi: 10.3389/fsufs.2023.1208970
Received: 20 April 2023; Accepted: 25 August 2023;
Published: 15 September 2023.
Edited by:
Yangchao Luo, University of Connecticut, United StatesCopyright © 2023 Wang, Xiao, Cai, Chen and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jihui Wang, d2FuZ2ppaHVpQGRndXQuZWR1LmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.