
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Sustain. Food Syst., 10 February 2023
Sec. Agroecology and Ecosystem Services
Volume 7 - 2023 | https://doi.org/10.3389/fsufs.2023.1125538
This article is part of the Research TopicCrop Pest Control and Pollination, volume IIView all 14 articles
 Luyun Luo1†
Luyun Luo1† Pei Wang2†
Pei Wang2† Diandong Wang1
Diandong Wang1 Xiaobin Shi2
Xiaobin Shi2 Jingwen Zhang3
Jingwen Zhang3 Zhixiang Zhao4
Zhixiang Zhao4 Jun Zeng5
Jun Zeng5 Jingjing Liao1
Jingjing Liao1 Zhuo Zhang2*
Zhuo Zhang2* Yong Liu2*
Yong Liu2*The Rhodopseudomonas palustris (R. palustris) PSB06 can promote crop growth, as it maybe regulates microbial communities in plant root soil, soil physicochemical properties, thus creating a favorable habitat for the crop growth. However, there are few studies on the yields and rhizosphere microbial community of R. palustris PSB06 agent. In the study, the high-throughput sequencing was used to study the changes of rhizosphere soil bacterial community after PSB06 treatment. The results indicated R. palustris PSB06 agent increased the pepper yield by 33.45% when compared to control group, with better effect than other treatments. And it also significantly increased soil nitrogen concentration. R. palustris PSB06 agent had improved pepper rhizosphere bacterial α diversity and changed the community structure. Acidobacteria, Proteobacteria, Actinomycetes and Firmicutes were dominant phyla in all the pepper rhizosphere soil samples. The results showed that soil bacterial community were significantly positively correlated with pH (R = 0.8537, P = 0.001) and total nitrogen (R = 0.4347, P = 0.003). The nine significantly enriched OTU in R.palustris PSB06 treatment (PB) group belong to Nitrososphaera (OTU_109, OTU_14, OTU_18, OTU_8), Lysobacter (OTU_2115, OTU_13), Arenimonas (OTU_26), Luteimonas (OTU_49), and Ramlibacter (OTU_70) were significantly positively correlated with the total yield of pepper (R > 0.5, P < 0.05). Overall, our results provide a theoretical basis for studying the microbial regulation of R.palustris PSB06 on rhizosphere soil.
Microorganisms play an important role in the process of matter cycling and energy flow in the ecosystem and maintain the stability of soil ecosystem function (Guo et al., 2021). Soil microorganisms, phychemical properties and enzymes are important components of soil ecosystem and key factors in regulating soil microecological environment and function. Soil microorganisms can convert organic matter in soil into nutrients needed for plant growth (Yin et al., 2013). Root exudates play an important role in biogeochemical cycling, regulation of rhizosphere ecological processes, and plant growth and development. They can attract beneficial microorganisms and affect the assembly of rhizosphere microbiota, thus improving the ability of plants to adapt to the environment (Bulgarelli et al., 2012). Plant rhizosphere growth-promoting bacteria are a kind of beneficial microorganisms that can stably survive in plant rhizosphere and can promote growth and control soil-borne diseases. After entering the rhizosphere environment, they colonize the root surface by interacting with microorganisms in plants and soil to promote crop growth and control plant diseases.
Photosynthetic bacteria (PSB) are a group of prokaryotes that perform photosynthesis without oxygen production, widely distributed in soil, paddy fields, swamps, lakes, rivers, oceans and other places (Hohmann-Marriott and Blankenship, 2012). Photosynthetic bacteria have a mutualistic symbiosis with plants. They can significantly improve the available nutrients in soil by fixing nitrogen and solubilizing phosphate, and it also are excellent growth promoting bacteria in plant rhizosphere (Ndona et al., 2011; Wang et al., 2019a). R. palustris that belongs to PSB, which is considered to be the most metabolized bacteria. It can decompose various carbon and nitrogen sources, while it is also an important source of growth promoting bacteria in the rhizosphere of plants (Merugu et al., 2011; Wong et al., 2014). R. palustris is also a promising biofertilizer, which not only supplies plant nutrients through nitrogen fixation, but also increases the utilization rate of synthetic nitrogen fertilizer, thereby improving crop yield and soil fertility and promoting crop growth (Kornochalert et al., 2014; Nunkaew et al., 2014). R. palustris PS3 has a growth-promoting effect on a variety of crops (Wong et al., 2014; Lee et al., 2016). During leaf growth development, R. palustris PS3 inoculation can promote plant growth by enhancing nitrate absorption (Hsu et al., 2021). Arashida et al. (2019) co-cultured Bacillus subtilis and the purple non-sulfur bacterium R. palustris in nitrogen-free medium, and observed diazotrophic growth in the subculture. Foliar spraying of R. palustris can change the soil microbial community of stevia to promote plant growth (Xu et al., 2016). And R. palustris GJ-22 was proved that can also promote crop growth by producing IAA (Su et al., 2017). At the same time, studies on rice also showed that the inoculation of R. palustris could promote the plant growth, increase the crop yield and change the soil microbial community (Kantha et al., 2015; Luo et al., 2019). Therefore, R. palustris can effectively reduce the use of chemical fertilizers in agriculture, and has a broad application prospect in reducing the application of chemical fertilizers.
As an effective biological agent, R. palustris PSB06 has been registered as a pesticide. In this study, we compared the differences of rhizosphere bacterial communities of R. palustris PSB06 and other treatments in the field, analyzed the correlation between bacterial communities and the yield and physicochemical properties of pepper, and focused on the population differences of bacterial communities. Our studies will afford a theoretical basis for reveal the mechanism of R. palustris PSB06 affecting pepper yield and regulating rhizosphere environment.
Field experiment was conducted in Maidi Village (112°28′54″E, 29°29′52″N), Yueshi Town, Huarong County, Yueyang City, Hunan Province from March 15 to June 18 in 2018. The pepper variety selected in this experiment was “Xiangyan 15”, which was grown in large quantities in Hunan Province from Longping Seed Industry Company. The strain Rhodopseudomonas palustris (R. palustris) PSB06 (CCTCC No: M2012518) was used in the study with 107 cfu/g from Hunan Institute of Plant Protection. And the planting soil had a pH = 7.74, soil matter = 15.1 mg·g−1, available K = 122.70 mg·g−1, available P = 43.08 mg·g−1, total N = 1328.69 mg·g−1, total P = 864.50 mg·g−1, and total K = 1818.61 mg·g−1. The field experiment was carried out with five groups, one group is the soil samples before the experiment, and other four groups are the experimental treatment group. LB: soil samples before planting; PB: R. palustris PSB06 fermentation broth; MB: Farmhouse manure (2 ton·mu−1 of chicken manure); CB: Conventional fertilizer (51% Sanning compound fertilizer, N: P2O5:K2O: 25:10:16:2 ton·mu−1, Hubei Sanning Chemical Co., Ltd.) and CKB: Fresh water. Each experimental treatment was repeated for seven times, with a total of twenty-eight plots, and fifty pepper plants were planted in each plot. The plots were arranged in completely random groups. The pepper was treated with R. palustris PSB06 agent at 7-leaf stage, with 30 mL per plant, and the root was irrigated once every 7 days for 3 times in total. After 30 days of field application, five-point sampling method was used to randomly select sampling points to collect the rhizosphere and surrounding soil of pepper. Soil samples of four pepper plants were collected in each site and mixed as one repeated soil sample.
Pepper fruits were collected since the first fruiting time after treatment, and collected every ten days and three times in total. The peppers were collected and weighed in each time, while the yield was recorded.
The pepper root surrounding soil was collected by shaking root method and then air-dried. The physiochemical properties of the air-dried soil samples were determined. The pH of the manure samples was measured in the aqueous extract (soil: deionized water = 1:2.5) using a multi-parameter water quality-monitoring instrument. Total potassium (TK, measured according to flame atomic absorption spectrophotometric method, GB 9836-1998), total nitrogen (TN, measured according to the modified Kjeldahl method, HJ/T 707-2014), total phosphorus (TP, measured according to the sodium hydrogen carbonate solution-Mo-Sb anti spectrophotometric method, HJ/T 704-2014), and organic matter (OM, measured according to the method for determination of soil organic matter, GB9834-1988) were measured by the Institute of Soil Science, Chinese Academy of Sciences (Nanjing, China).
The pepper rhizosphere soil samples were collected at the 7 day after the third irrigation. The roots were removed by shaking off the root method, and the roots were placed in a conical flask containing 100 mL 0.05 mol/L phosphoric acid buffer (pH = 7.0). Place four plants per conical flask and shake the conical flask from side to side to wash thoroughly the roots of the plants so that the soil is fully mixed into the buffer. Remove the cleaned root blocks from the conical flask with sterile forceps, then pour the remaining mixture into a 50 mL sterile plastic sterile centrifuge tube at a high speed of 13,000 rpm for 3 min to allow the soil to fully settle in the centrifuge tube and discard the supernatant. Then 50 mL centrifuge tube containing the soil sample was placed in a freeze-drying apparatus for freeze-drying. After freeze-drying, the soil samples were thoroughly ground with a sterile mortar, filtered through an 80-mesh sieve, and stored in a refrigerator at −20°C. 0.5 g of each sample was accurately weighed for DNA extraction that using the Fast DNA Spin Kit for Soil (MP Biomedicals, USA) according to the Kit instructions.
The total DNA concentration of samples were determined by NanoDrop 2000, with the A260/A280 value required between 1.8 and 2.0. The genomic DNA concentration of all samples was quantified to 30 ng·μL −1 before amplification. Taking the total DNA of the sample as the template, Universal primers 515F (5 '-GTGCCAGCMGCCGCGGTAA-3') and 806R (5 '-GGACTACHVGGGTWTCTAAT-3') were used for PCR amplification of bacterial 16S rDNA fragments with 6 bp barcode (Wang et al., 2015). The PCR reaction system as follows: 5 μL 10×PCR buffer (containing 20 mmol·L−1 MgCl2), 4 μL dNTP (10 mmol·L−1), 1 U Taq DNA polymerase, 1 μL DNA template, and sterilized ddH2O supplemented to 50 μL. PCR reaction conditions: pre-denaturation at 95°C for 10 min, after denaturation at 95°C for 45 s, annealing at 55°C for 1 min, extension at 72°C for 45 s, cycling for 35 times. Finally, it was extended at 72°C for 10 min and stored at 4°C at constant temperature. The purified PCR products were sent to Nanjing Puvekon Biotechnology Co., Ltd. (Nanjing, China) for sequencing. The clean reads were deposited into the NCBI Sequence Read Archive (SRA) database (Accession Number: PRJNA869895).
Raw sequence data reads were processed with an in-house pipeline (http://mem.rcees.ac.cn:8080). In brief, a separate sample was generated according to different barcodes and primers, allowing for one mismatch. Paired-end reads with at least 30 bp overlap were combined by the FLASH program (Magoč and Salzberg, 2011), and filtered by Btrim program with Quality Score <20 (Kong, 2011). Then we discarded the sequences with either an ambiguous base or <200 bp. The UPARSE algorithms were used to detecte and remove chimera sequences (Edgar, 2013). Low abundance OTUs ( ≤ 1 count) were eliminated from the OTU table. The bacterial representative sequences for each OTU were assigned to different taxonomic groups using the RDP Classifier database (Silva database 132 version). The resampled OTU table, which was obtained by resampled randomly with the lowest sequence number, was used for the subsequent analysis. The α-diversity was assessed using the Chao1, Observed_richness, Shannon and Inv_simpson index. Weighted principal coordination analysis (Weighted_PCoA) based on unifrac matrix, multi-response permutation procedures (MRPP), analysis of similarities (ANOSIM), and Adonis were used to test the bacterial community differences among the groups. The mantel test and canonical correspondence analysis (CCA) were used to analyze the relationship between environmental factors and bacterial communities. The key bacterial taxa responsible for discrimination between two groups were identified using linear discriminant analysis (LEfSe) with linear discriminant analysis (LDA) = 3.
The difference of α-diversity index and yield of pepper among different treatments were evaluated by one-way ANOVA after multiple comparisons based on Duncan algorithm using IBM SPSS for Windows (v.22.0) software. The results were presented as mean ± standard error (SE). The Student t test was used to assessed the difference between two groups of soil physichemical properties and relative abundance of top 30 genera, and statistical significant level was set at P < 0.05 by the software Microsoft Excel 2019. The spearman correlation coefficient was used to analyze the correlations between the core genera of the four treatment groups and yield, and the results were visualized using Cytoscape 3.6.0 software.
The total yield and yield of each time points of pepper were calculated, and the results were shown in Figure 1. The yield of pepper in R. palustris PSB06 root-irrigation treatment (PB) and farm manure treatment (MB) groups was significantly higher than that in the control group when the pepper was collected at first and second sampling time (P < 0.05), and there was no significant difference between formal fertilizer treatment (CB) group and control group (CKB). But the yield of pepper in the experimental group was significantly higher than that in the control group at third sampling time (P < 0.05). The total yield of pepper in PB, MB and CB groups was significantly higher than that in control group (P < 0.05) and highest in PB group. Compared with the control group, the total yield of pepper in PB, MB and CB groups increased by 33.45, 28.44, and 11.03%, respectively.
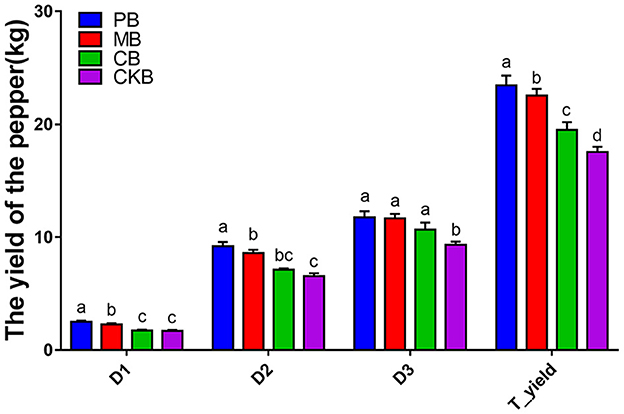
Figure 1. The total pepper yield and yield at different time points. a–d indicate the statistical diferences at P < 0.05 for one-way ANOVA.
Physiochemical properties of soil were measured before planting and 30 days after treatment, and the results were shown in Figure 2. Before planting pepper, the pH, total nitrogen (TN), available kalium (AK) values of pre-planting soil sample group (LB) were the highest and significantly higher than control group (CKB), but no significant difference at organic matter (OM), total phosphorus (TP), available phosphorus (AP) concentration. The pH of PB, MB and CB groups were significantly lower than those of CKB group (P < 0.05), and the pH of PB group was the lowest (pH = 6.9).
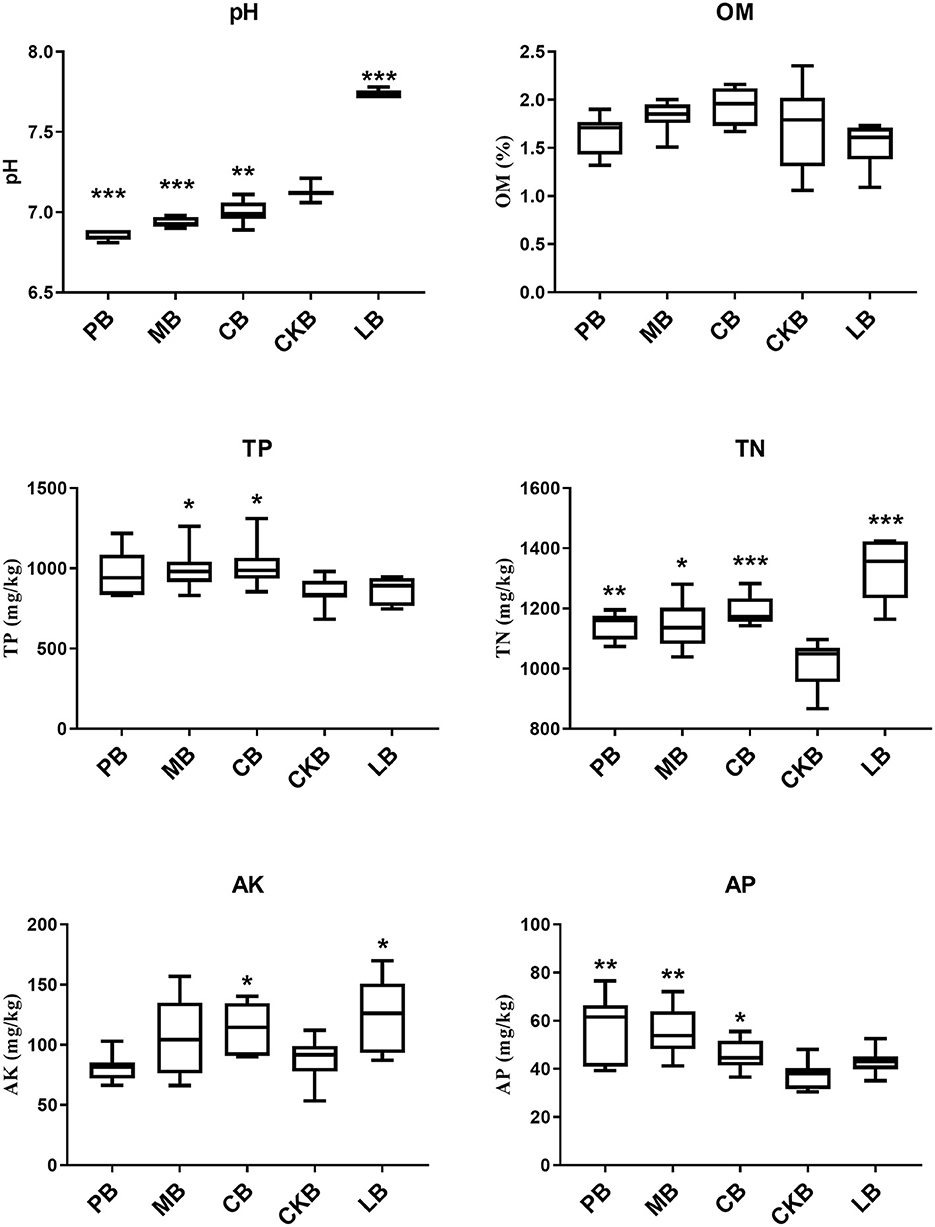
Figure 2. The physicochemical properties of different treatments. PB, R. palustris PSB06 root-irrigation treatment; MB, farm manure treatment; CB, formal fertilizer treatment; CKB, control treatment; LB, pre-planting soil sample group, n = 7. “*”, “**”, “***” indicate significant difference between treatments and control group, significance level at P < 0.05, P < 0.01, P < 0.001.
The contents of TN and AP in three experimental groups were significantly higher than those in CKB group (1011.86 mg/kg) (P < 0.05), the concentration of AP in PB group (56.78 mg/kg) was the highest, and the concentration of AP in PB group was 49.88% higher than control group. There was no significant difference in OM concentration between treatment and control group. The concentration of TP in CB and MB groups was significantly higher than that in control group (P < 0.05). The AP concentration of CB group was significantly higher than CKB, but there was no significant difference between PB, MB groups and CKB group.
A total of 1,648,432 high quality sequences were obtained from the raw data of 35 soil samples in the field experiment after a series of quality controls, with the sequence number ranging from 23,339 to 87,188. A total of 4861 OTU representative sequences were obtained from each group of samples by clustering at 97% similarity. The bacterial α diversity index of the pre-planting soil sample group (LB) and four treatment groups were shown in Table 1. The bacterial α-diversity index (Chao1, Observed_richness, Shannon, and Inv_Simpson index) of the PB group was significantly higher than other four treatment groups. The bacterial α-diversity index (Observed_richness, Shannon index) of the PB and CB group was significantly higher than control group (CKB) while no significant difference between MB and CKB group. The bacterial α-diversity index (Chao1 and Inv_Simpson index) of the PB and CB group was higher than CKB group but no significant difference among these groups.
A total of 4861 operational taxa (OTU) were identified from 35 soil samples in the field. The dominant phyla and class is shown in Supplementary Figure S1A. The dominant phylum in LB group were Proteobacteria (38.87%), Actinobacteria (11.68%), Thaumarchaeota (7.48%), and Bacteroidetes (7.82%), respectively. And Acidobacteria, Proteobacteria, Actinobacteria, and Firmicutes all were dominant phyla in the four groups of pepper rhizosphere soil samples. At the class level, the dominant populations were shown in Supplementary Figure S1B, and all OTUs are divided into 80 class. The dominant class in LB soil samples were α -proteobacteria, β -proteobacteria, γ -proteobacteria and δ -proteobacteria, Actinobacteria and Acidobacteria Gp6. The dominant class in the four groups pepper rhizosphere soil samples were α -proteobacteria, β -proteobacteria, γ -proteobacteria and Bacilli, respectively. At the phylum identification level, the dominant bacterial groups in the LB group (relative abundance greater than 4%) were Proteobacteria, Actinobacteria, Thaumarchaeota, Bacteroidetes and Acidobacteria. The dominant phyla in soil samples of PB group were Proteobacteria (58.47%), Actinobacteria (12.18%), and Firmicutes (9.46%), and Thaumarchaeota (Chigarchaea: 6.19%). The dominant phyla in the soil samples of the MB group were Proteobacteria (71.79%), Actinobacteria (11.56%), and Firmicutes (4.91%), while conventional fertilizer treatment (CB) group were Proteobacteria (54.57%), Actinobacteria (14.11%), Firmicutes (8.69%), and Bacteroidetes (14.11%). 7.22%), and Acidobacteria (5.48%). And in the control group (CKB), Proteobacteria (69.50%), Actinobacteria (12.60%), and Firmicutes (5.95%) were the dominant phyla.
At the genus level, the differences of the top 30 genera among the five groups were analyzed, and the results were shown in Supplementary Figure S2. The results showed that the relative abundance of Flavobacterium, Gaiella, Gp6, Nitrososphaera, Phycicoccus and Arenimonas in LB group was significantly higher than that in CKB group but significantly lower in Aeromicrobium, Bacillus, Ensifer, Enterobacter, Factibacillus, Lechevalieria, Lysobacter, Nocardioides, Paenibacillus, Pseudomonas, Pseudoxanthomonas, Rhizobium, Shinella, Sphingobium, Streptophyta, Streptomyces, Variovorax. The relative abundance of Gaiella, Gp6, Nitrososphaera, Ramlibacter, and Arenimonas in PB group was significantly higher than that in control group while lower in Enterobacter, Nocardioides, Pseudomonas, Rhizobium, and Streptophyta. The relative abundance of Gaiella, Gp6 in MB group was significantly higher than that in control group while lower in Marmoricola, Sphingomonas and Streptophyta. The relative abundance of Agromyces, Arthrobacter, Dyadobacter, Gaiella, Lysobacter, Nitrososphaera, Phycicoccus, Pseudoxanthomonas, Ramlibacter and Variovorax in the CB group was significantly higher than that in control group while lower in Ensifer, Enterobacter, Lechevalieria, Rhizobium and Sphingomonas.
The differences of the rhizosphere soil bacterial community structure among soil samples were compared, and the results were shown in Figure 3 and Supplementary Table S1. Principal coordinate analysis (PCoA) and dissimilarity analysis showed that there were significant differences in the bacterial community structure between the the pre-planting soil samples and the four pepper rhizosphere soil samples. The PCoA results indicated that the bacterial communities of the pre-planting soil samples and the four pepper rhizosphere soil samples were significantly separated while the bacterial communities of the four treatment groups were also significantly separated from each other. PCoA1 and pCoA2 accounted for 77.03% of the total variation. The results of dissimilarity analysis (MRPP, ANOSIM, and ADONIS) based on Bray-Curtis matrix showed that there were significant differences between LB and CKB group (P < 0.01), and the three experimental groups (PB, MB, CB) were also significantly different from CKB group (P < 0.05).
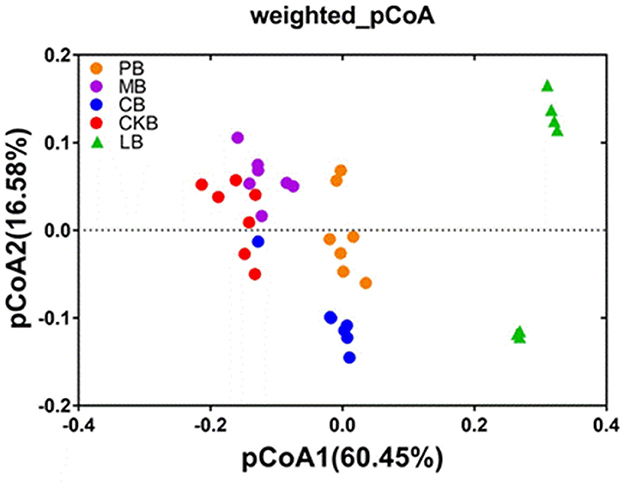
Figure 3. Principal coordinate analysis (PCoA) of bacterial communities among different treatments. PB, R. palustris PSB06 root-irrigation treatment; MB, farm manure treatment; CB, formal fertilizer treatment; CKB, control treatment; LB, soil samples before planting, n = 7.
The results showed that the total yield was correlated with pH (R = −0.651, P < 0.001) and TK (R = −0.4237, P < 0.05). In addition, TP was positively correlated with AP and TN, while pH was negatively correlated with TP, TN, and AP. Mantel test analysis was used to assess the relationship between bacterial community structure and environmental factors (Table 2). The results showed that soil bacterial community were significantly positively correlated with pH (R = 0.8537, P = 0.001) and TN (R = 0.4347, P = 0.003) (Supplementary Figure S3).
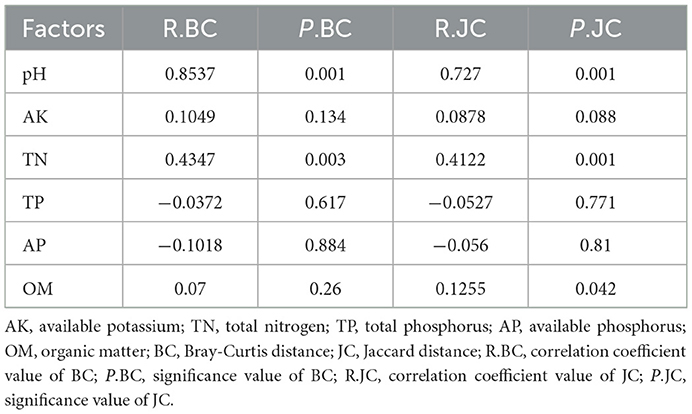
Table 2. The mantel test result between environmental factors and bacterial communities based on Bray-Curtis and Jaccard distances.
In addition, CCA was used to evaluate the soil bacterial community and physiochemical properties among all groups, as shown in Supplementary Figure S4. Soil OM, AP, pH, TN, and AP were significantly correlated with the bacterial community structure between the pre-planting soil samples group (LB) and four treatment group samples, and CCA1 and CCA2 accounted for 86.53% of the total variation. The Total (N, P), available (P, K), and pH were significantly correlated with the soil community structure, explaining 54.60% of the total variation, which CCA1 and CCA2 explained 31.96 and 22.64% of the variation, respectively.
The LefSe method was used to screen significantly enriched OTU in the three treatment groups and the control group, among which 12 OTU were enriched in the PB group, 19 OTU in the CB group and 4 OTU in the MB group (Figure 4). And 9, 5, 2 enriched OTU in the PB, CB, MB group were significantly positively correlated with the total pepper yields (R > 0.5, P < 0.05), respectively (Figure 4A). These OTU mainly belong to Thaumarchaeota, Proteobacteria, Actinobacteria, and Verrucomicrobia. The significantly enriched OTU in PB group belong to Nitrososphaera (OTU_109, OTU_14, OTU_18, OTU_8), Lysobacter (OTU_2115, OTU_13), Arenimonas (OTU_26), Luteimonas (OTU_49), and Ramlibacter (OTU_70). And the significantly enriched OTU in CB group belong to Nitrososphaera (OTU_18, OTU_109), Lysobacter (OTU_13), Phycicoccus (OTU_31) and Terrimicrobium (OTU_68) while Acinetobacter (OTU_55, OTU_74) in the MB group.
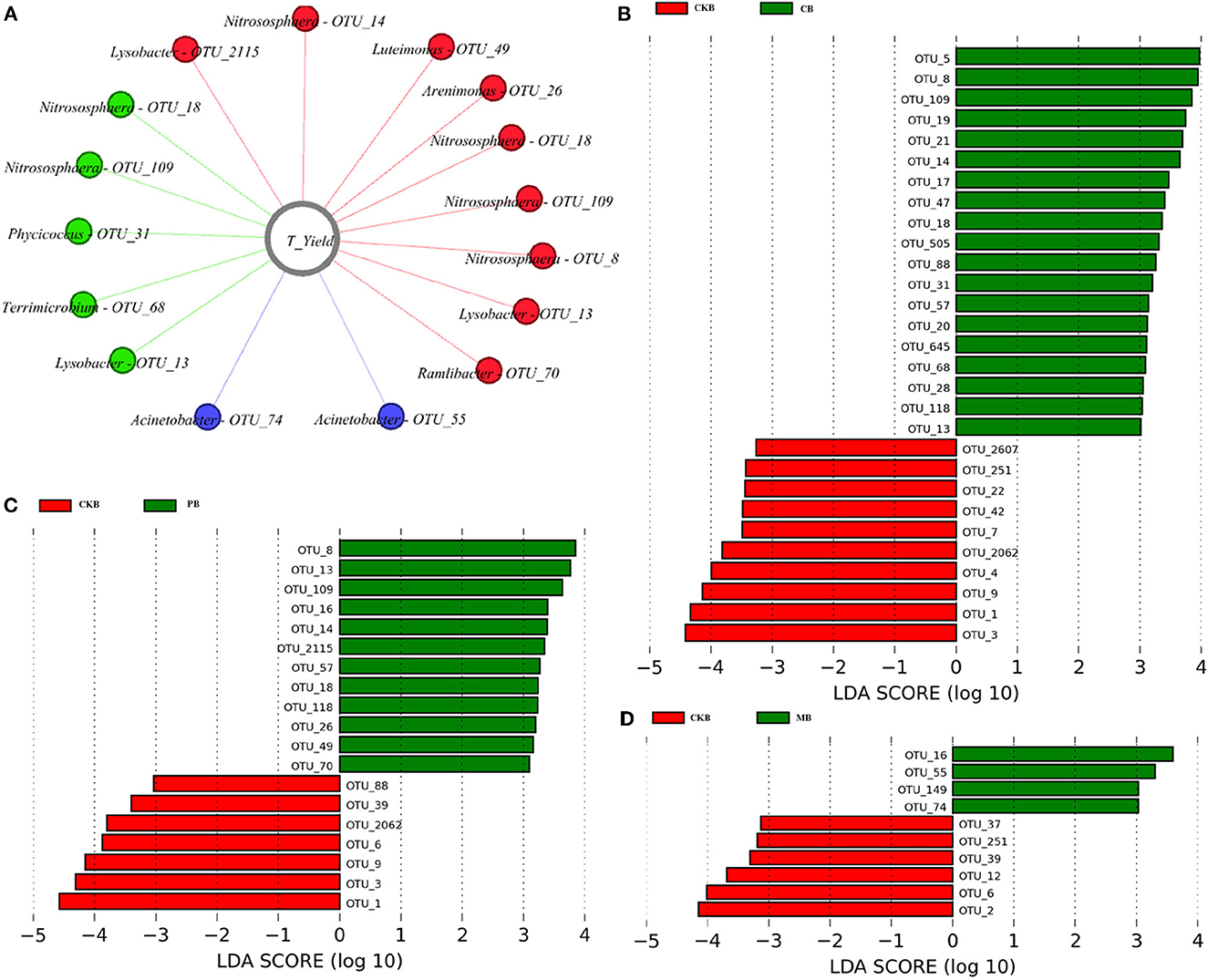
Figure 4. LefSe analysis between three treatment groups and the control group. (A) The enriched significantly OTU in the PB, CB, MB group were significantly positively correlated with the total yield of pepper (R > 0.5, P < 0.05), respectively. (B) The significantly enriched OTU between CB and CK group. (C) The significantly enriched OTU between PB and CK group. (D) The significantly enriched OTU between MB and CK group.
It is an environmentally friendly approach to improve the crop yield and soil environment by introducing beneficial microbes to the agriculture ecological system. Previous study also indicated that soil microbial inoculants can enhanced nutrient uptake and stimulated plant growth accumulation after whole-inoculation procedures (Wang et al., 2019a). In our study, the total yield of pepper treated with R. palustris PSB06, farm manure and conventional fertilizer was significantly higher than control group (P < 0.05), which increased by 33.45, 28.44, and 11.03%, respectively. And it indicated that R. palustris PSB06 inoculations significantly increased pepper yield, with better yield increase than other treatments. In previous studies, R. palustris is also a promising biofertilizer, which improving crop yield and soil fertility by supplies plant nutrients through nitrogen fixation and increases the utilization rate of synthetic nitrogen fertilizer (Kornochalert et al., 2014; Nunkaew et al., 2014). R. palustris PS3 has a growth-promoting effect on a variety of crops (Wong et al., 2014; Lee et al., 2016). In agricultural production, the application of R. palustris PSB06 as a biofertilizer to reduce or replace the use of manure and chemical fertilizers will effectively reduce the emission of pollutants in the environment and the environmental safety problems caused by the overuse of chemical fertilizers to a certain extent.
Soil microbes play an important role in agroecosystems by mediating biogeochemical and nutrient transformation (Banerjee et al., 2018; Zhu et al., 2018). The rhizosphere is the most active area in the plant-microbe interactions system. Some studies have evaluated the influences of different microbial inoculants on soil properties and plant nutrient uptake under controlled conditions (Qiao et al., 2019; Wang et al., 2019b, 2021a). The composition and assembly process of rhizosphere bacterial communities may be affected by different rhizosphere environments. In this study, the soil pH of R. palustris PSB06, farm manure and conventional fertilizer group was significantly lower than control group, and the total nitrogen concentration was increased by 13.22, 12.92, and 18.19%, respectively. Previous studies indicated that each environmental variables may have different driving effects on community assembly and composition (Lee et al., 2017; Wang et al., 2017; Zhao et al., 2017). Conventional fertilizers and farm fertilizers contain a large amount of nitrogen and phosphorus elements, which were directly absorbed and utilized by plants when applied to the environment. R. palustris PSB06, as a bacteria with nitrogen fixation function, it can indirectly promote the pepper growth and development by producing nutrients needed by plants through biological nitrogen fixation. In addition, we also analyzed the association between rhizosphere bacterial microbiota and the corresponding environmental factors. The results showed that soil bacterial community were significantly positively correlated with pH (R = 0.8537, P = 0.001) and TN (R = 0.4347, P = 0.003). It indicated that the soil TN and pH significantly affected rhizosphere bacterial community composition, and further demonstrated the function of nitrogen in microbial community changes and pepper growth and development. The changes in soil chemical factors due to R. palustris PSB06 inoculation, such as nitrogen and pH, were the dominant factors explaining the succession of the resident community. Kuramae et al. (2010) also reported that soil pH significantly altered the microbial secondary succession, the soil pH in inoculated treatments significantly differed from that in non-inoculated soil. Wang et al. (2019a) found that the phosphate-solubilizing bacteria possess the ability to produce organic acid and release nutrient contents, thus leading to a decrease of the soil pH and changes in the related nutrient contents. In the present study, the concentration of TN were significantly increased but slight difference among available nutrients compared to the control group after the PSB06 inoculation application. Previous study also showed that the addition of bacterial growth medium had a very limited effect on soil available nutrients (Wang et al., 2021b). As a efficient and ecofriendly soil management strategy, microbial inoculants are applied for improving crop productivity and soil properties by colonizing the rhizosphere and increasing nutrient availability to the host plant (Yilmaz and Sönmez, 2017; Berg et al., 2020; Pagnani et al., 2020).
Soil microorganisms are the main drivers of soil ecosystem functioning (Zhong et al., 2020). However, the native soil microbial community is sensitive to exogenous disturbances and natural climate change (Hartmann et al., 2015; Suleiman et al., 2016). Human disturbance of farmland soil can greatly affect crop rhizosphere processes, especially rhizosphere microbial communities (Mariotte et al., 2018). The importance of the root-associated microbial community for plant growth and development has been widely recognized (Wagner et al., 2014; Debenport et al., 2015). Previous study suggested that invasion by a single strain may change microbial community composition and function, and diversity determines the outcome of biotic invasions (Mallon et al., 2018). Mawarda et al. (2020) also indicated that the deliberate release of microbial inoculants may affect resident microbiome interactions. Dissimilarities among rhizosphere bacterial community compositions at different treatments indicate that changes in the rhizosphere environment occur after factitious interference. In our study, principal coordinates analysis (PCoA) demonstrated that bacterial community structure between the experimental group and control group after application, indicating that R. palustris PSB06 irrigation changed the bacterial community structure in rhizosphere soil. In addtion, the α-diversity of rhizosphere bacterial community at R. palustris PSB06 group was significantly higher than control group, reflecting the application on the evolution of rhizosphere bacterial community.
Soil-resident microbial communities are frequently subjected to biotic disturbances, including beneficial microbial inoculants and harmful pathogens, which can change microbial community succession, composition, and diversity (Xiong et al., 2017; Lourenco et al., 2018). In our study, we defined the taxonomic structure of the pepper root microbiota, which mainly comprising Acidobacteria, Proteobacteria, Actinomycetes, and Firmicutes. Acidobacteria and Acidobacteria are the dominant phyla in rhizosphere soil and widely distributed in soil around the world, indicating that pepper rhizosphere bacteria also follow the general rule of bacterial community establishment (Delgado-Baquerizo et al., 2018). Acidobacteria can offers efficient carbon and nitrogen cycling from soil organic matter, while Proteobacteria can release nutrients from complexes of organo-mineral that facilitate plant growth (Lugtenberg and Kamilova, 2009; Eilers et al., 2010). Actinobacteria are generally defined as copiotrophic bacteria, while Acidobacteria are oligotrophic bacteria (Dai et al., 2018). Compared with the control group, PSB06 treatment group decreased the relative abundance of Proteobacteria and increased the relative abundance of Firmicutes. Wang et al. (2021b) found that the relative abundances of families like Xanthomonadaceae significantly increased after PSB treatments application, suggesting that the introduction of PSB changed specific resident microbial populations. The microbial invasions frequently usually start with a dominating microbial population and have an impact on the native soil microbiome (Mallon et al., 2018). Previous studies have attempted to evaluate the impacts on the microbial community in the rhizosphere of the introduction of plant-growth-promoting rhizobacteria (PGPR), it indicated that microbial inoculants may alter the resident community composition by causing resource competition, synergistic effects, and antagonistic effects (Zhang et al., 2019; Zhuang et al., 2021). These changes in the abundances of some taxa after the initial disturbance due to microbial inoculation may be a result of competition for resources in the soil (Krause et al., 2014).
Understanding the interactions among microbial taxa and environmental factors can reveal the complex microbial community structure and detect potential keystone species (Wang et al., 2015; He et al., 2017). Therefore, we screened the enriched OTUs in different treatments and analyzed their correlation with yield. The results showed that the PB group have more positive OTUs than other treatments and control group. The nine significantly enriched OTU in PB group belong to Nitrososphaera (OTU_109, OTU_14, OTU_18, OTU_8), Lysobacter (OTU_2115, OTU_13), Arenimonas (OTU_26), Luteimonas (OTU_49), and Ramlibacter (OTU_70) were significantly positively correlated with the total pepper yield (R > 0.5, P < 0.05). The yield improvement may be the result of microbial-crop interaction, our results indicated that cooperative microbial interactions may play an critical role in soil microbial assembly and may benefit plant growth and development. The functional study of these bacteria is also the focus of our next research. So, isolation and functional verification of rhizosphere microbiota is necessary for future work, especially for potential keystone species. It will help further our understanding of this microecosystem in the crop rhizosphere.
R. palustris PSB06 treatment increased the pepper yield and improved the rhizosphere soil microbial environment by increasing the bacterial alpha diversity and changing the rhizosphere bacterial structure, which created a more healthy soil environment for pepper growth.
The data presented in the study are deposited in the SRA repository, accession number PRJNA869895. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA869895.
LL, PW, and ZZ performed the experiments and analyzed the data. ZXZ, JWZ, and XS prepared the figures and tables. LL, YL, and DW conceived and designed the experiments. JZ and JL analyzed the data, authored or reviewed drafts of the manuscript, and approved the final draft. All authors contributed to the article and approved the submitted version.
This work was supported by the National Key R&D Program of China (2022YFD1400700), National Science Foundation of China (32272517), Scientifc and Technological Research Program of Chongqing Municipal Education Commission (2022NSCQ-MSX1554), 4 Hunan Agricultural Science and Technology Innovation Fund Project (2022CX68), and commonweal project of Xinjiang agricultural sciences (XJGYX2019-20).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fsufs.2023.1125538/full#supplementary-material
Arashida, H., Kugenuma, T., Watanabe, M., and Maeda, I. (2019). Nitrogen fixation in Rhodopseudomonas palustris co-cultured with Bacillus subtilis in the presence of air. J. Biosci. Bioeng. 127, 589–593. doi: 10.1016/j.jbiosc.10010
Banerjee, S., Schlaeppi, K., and van der Heijden, M. G. A. (2018). Keystone taxa as drivers of microbiome structure and functioning. Nat. Rev. Microbiol. 16, 567–576. doi: 10.1038/s41579-018-0024-1
Berg, S., Dennis, P. G., Paungfoo-Lonhienne, C., Anderson, J., Robinson, N., Brackin, R., et al. (2020). Effects of commercial microbial biostimulants on soil and root microbial communities and sugarcane yield. Biol. Fertil. Soils. 56, 565–580. doi: 10.1007/s00374-019-01412-4
Bulgarelli, D., Schlaeppi, K., Spaepen, S., Themaat, E. V. L. V., and Schulze-Lefert, P. (2012). Structure and functions of the bacterial microbiota of plants. Annu. Rev. Plant Biol. 64, 807–838. doi: 10.1146/annurev-arplant-050312-120106
Dai, Z. M., Su, W. Q., Chen, H. H., Barberán, A., Zhao, H. C., Yu, M. J., et al. (2018). Long-term nitrogen fertilization decreases bacterial diversity and favors the growth of Actinobacteria and Proteobacteria in agro-ecosystems across the globe. Glob. Change Biol. 24, 3452–3461. doi: 10.1111/gcb.14163
Debenport, S. J., Assigbetse, K., Bayala, R., Chapuis-Lardy, L., Dick, R. P., Gardener, B. B. M., et al. (2015). Association of shifting populations in the root zone microbiome of millet with enhanced crop productivity in the Sahel region (Africa). Appl. Environ. Microbiol. 81, 2841–2851. doi: 10.1128/AEM.04122-14
Delgado-Baquerizo, M., Oliverio, A. M., Brewer, T. E., Benavent-González, A., Eldridge, D. J., Bardgett, R. D., et al. (2018). A global atlas of the dominant bacteria found in soil. Science. 359, 320–325. doi: 10.1126/science.aap9516
Edgar, R. C. (2013). UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. methods. 10, 996–998. doi: 10.1038/nmeth.2604
Eilers, K. G., Lauber, C. L., Knight, R., and Fierer, N. (2010). Shifts in bacterial community structure associated with inputs of low molecular weight carbon compounds to soil. Soil Biol. Biochem. 42, 896–903. doi: 10.1016/j.soilbio.02003
Guo, X. L., Wan, Y. Q., Shakeel, M., Wang, D. L., and Xiao, L. H. (2021). Effect of mycorrhizal fungi inoculation on bacterial diversity, community structure and fruit yield of blueberry. Rhizosphere 19, 100360. doi: 10.1016/j.rhisph.2021.100360
Hartmann, M., Frey, B., Mayer, J., Mäder, P., and Widmer, F. (2015). Distinct soil microbial diversity under long-term organic and conventional farming. ISME J. 9, 1177–1194. doi: 10.1038/ismej.2014.210
He, D., Shen, W. J., Eberwein, J., Zhao, Q., Ren, L. J., Wu, Q. L. L., et al. (2017). Diversity and co-occurrence network of soil fungi are more responsive than those of bacteria to shifts in precipitation seasonality in a subtropical forest. Soil Biol. Biochem. 115, 499–510. doi: 10.1016/j.soilbio.09023
Hohmann-Marriott, M. F., and Blankenship, R. E. (2012). The photosynthetic world. Springer Netherlands. 3, 32. doi: 10.1007./978-94-007-1579-0_1
Hsu, S. H., Shen, M. W., Chen, J. C., Lur, H. S., and Liu, C. T. (2021). The photosynthetic bacterium Rhodopseudomonas palustris strain PS3 exerts plant growth-promoting effects by stimulating nitrogen uptake and elevating auxin levels in expanding leaves. Front. Plant. Sci. 12, 573634. doi: 10.3389/fpls.2021.573634
Kantha, T., Kantachote, D., and Klongdee, N. (2015). Potential of biofertilizers from selected Rhodopseudomonas palustris strains to assist rice (Oryza sativa L. subsp.indica) growth under salt stress and to reduce greenhouse gas emissions. Ann. Microbiol. 65, 2109–2118. doi: 10.1007/s13213-015-1049-6
Kong, Y. (2011). Btrim: a fast, lightweight adapter and quality trimming program for next-generation sequencing technologies. Genomics. 98, 152–153. doi: 10.1016/j.ygeno.05009
Kornochalert, N., Kantachote, D., Chaiprapat, S., and Techkarnjanaruk, S. (2014). Use of Rhodopseudomonas palustris P1 stimulated growth by fermented pineapple extract to treat latex rubber sheet wastewater to obtain single cell protein. Ann. Microbiol. 64, 1021–1032. doi: 10.1007/s13213-013-0739-1
Krause, S., Le Roux, X., Niklaus, P. A., Van Bodegom, P. M., Lennon, J. T., Bertilsson, S., et al. (2014). Trait-based approaches for understanding microbial biodiversity and ecosystem functioning. Front. Microbiol. 5, 251. doi: 10.3389/fmicb.2014.00251
Kuramae, E. E., Gamper, H. A., Yergeau, E., Piceno, Y. M., Brodie, E. L., Desantis, T. Z., et al. (2010). Microbial secondary succession in a chronosequence of chalk grasslands. ISME J. 4, 711–715. doi: 10.1038/ismej.2010.11
Lee, S. H., Sorensen, J. W., Grady, K. L., Tobin, T. C., and Shade, A. (2017). Divergent extremes but convergent recovery of bacterial and archaeal soil communities to an ongoing subterranean coal mine fire. ISME J. 11, 1447–1459. doi: 10.1038/ismej.2017.1
Lee, S. K., Lur, H. S., Lo, K. J., Cheng, K. C., Chuang, C. C., Tang, S. J., et al. (2016). Evaluation of the effects of different liquid inoculant formulations on the survival and plant-growth-promoting efficiency of Rhodopseudomonas palustris strain PS3. Appl. Microbiol. Biot. 100, 7977–7987. doi: 10.1007/s00253-016-7582-9
Lourenco, K. S., Suleiman, A. K. A., Pijl, A., van Veen, J. A., Cantarella, H., Kuramae, E. E., et al. (2018). Resilience of the resident soil microbiome to organic and inorganic amendment disturbances and to temporary bacterial invasion. Microbiome. 6, 142. doi: 10.1186/s40168-018-0525-1
Lugtenberg, B., and Kamilova, F. (2009). Plant-growth-promoting rhizobacteria. Annu. Rev. Microbiol. 63, 541–556. doi: 10.1146/annurev.micro.62.081307.162918
Luo, L. Y., Wang, P., Zhai, Z. Y., Su, P., Tan, X. Q., Zhang, D. Y., et al. (2019). The effects of Rhodopseudomonas palustris PSB06 and CGA009 with different agricultural applications on rice growth and rhizosphere bacterial communities. AMB Express. 9, 173. doi: 10.1186/s13568-019-0897-z
Magoč, T., and Salzberg, S. L. (2011). FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27, 2957–2963. doi: 10.1093/bioinformatics/btr507
Mallon, C. A., Le Roux, X., van Doorn, G. S., Dini-Andreote, F., Poly, F., Salles, J. F., et al. (2018). The impact of failure: unsuccessful bacterial invasions steer the soil microbial community away from the invader's niche. ISME J. 12, 728–741. doi: 10.1038/s41396-017-0003-y
Mariotte, P., Mehrabi, Z., Bezemer, T. M., Deyn, De., Kulmatiski, G. B., Drigo, B., et al. (2018). Plant-soil feedback: bridging natural and agricultural sciences. Trends Ecol. Evol. 33, 129–142. doi: 10.1016/j.tree.11005
Mawarda, P. C., Le Roux, X., Van Elsas, J. D., and Salles, J. F. (2020). Deliberate introduction of invisible invaders: a critical appraisal of the impact of microbial inoculants on soil microbial communities. Soil Biol. Biochem. 148, 107874. doi: 10.1016/j.soilbio.2020.107874
Merugu, R., Rudra, M. P. P., Rao, A. S., Ramesh, D., Nageshwari, B., and Rajyalaxmi, K. Influence of different cultural conditions on photoproduction of hydrogen by Rhodopseudomonas palustris KU003. (2011). ISRN Renew. Energy. 2011. doi: 10.5402./2011/328984
Ndona, R. K., Friedel, J. K., Spornberger, A., Rinnofner, T., and Jezik, K. (2011). “Effective micro-organisms” (EM): an effective plant strengthening agent for tomatoes in protected cultivation. Biol. Agric. Hortic. 27, 189–204. doi: 10.1080/01448765.2011.9756647
Nunkaew, T., Kantachote, D., Kanzaki, H., Nitoda, T., and Ritchie, R. J. (2014). Effects of 5-aminolevulinic acid (ALA)-containing supernatants from selected Rhodopseudomonas palustris strains on rice growth under NaCl stress, with mediating effects on chlorophyll, photosynthetic electron transport and antioxidative enzymes. Electron. J. Biotechn. 17, 19–26. doi: 10.1016/j.ejbt.12004
Pagnani, G., Galieni, A., Stagnari, F., Pellegrini, M., Del Gallo, M., Pisante, M., et al. (2020). Open field inoculation with PGPR as a strategy to manage fertilization of ancient Triticum genotypes. Biol. Fertil Soils. 56, 111–124. doi: 10.1007/s00374-019-01407-1
Qiao, C. C., Penton, C. R., Xiong, W., Liu, C., Wang, R. F., Liu, Z. Y., et al. (2019). Reshaping the rhizosphere microbiome by bio-organic amendment to enhance crop yield in a maize-cabbage rotation system. Appl. Soil Ecol. 142, 136–146. doi: 10.1016/j.apsoil.2
Su, P., Tan, X. Q., Li, C. G., Zhang, D. Y., Cheng, J. E., Zhang, S. B., et al. (2017). Photosynthetic bacterium Rhodopseudomonas palustris GJ-22 induces systemic resistance against viruses. Micro. Biotechnol. 10, 612–624. doi: 10.1111/1751-7915.12704
Suleiman, A. K. A., Gonzatto, R., Aita, C., Lupatini, M., Jacques, R. J. S., Kuramae, E. E., et al. (2016). Temporal variability of soil microbial communities after application of dicyandiamide-treated swine slurry and mineral fertilizers. Soil Biol. Biochem. 97, 71–82. doi: 10.1016/j.soilbio.03002
Wagner, M. R., Lundberg, D. S., Coleman-Derr, D., Tringe, S. G., Dangl, J. L., Mitchell-Olds, T., et al. (2014). Natural soil microbes alter flowering phenology and the intensity of selection on flowering time in a wild arabidopsis relative. Ecol. Lett. 17, 717–726. doi: 10.1111/ele.12276
Wang, X. B., Van Nostrand, J. D., Deng, Y., Lü, X. T., Wang, C., Zhou, J. Z., et al. (2015). Scale-dependent effects of climate and geographic distance on bacterial diversity patterns across northern China's grasslands. FEMS Microbiol. Ecol. 91, fiv133. doi: 10.1093./femsec/fiv133
Wang, Y. S., Li, C. N., Kou, Y. P., Wang, J. J., Tu, B., Li, H., et al. (2017). Soil pH is a major driver of soil diazotrophic community assembly in Qinghai-Tibet alpine meadows. Soil Biol. Biochem. 115, 547–555. doi: 10.1016/j.soilbio.09024
Wang, Z. K., Chen, Z. Y., and Fu, X. X. (2019b). Integrated effects of co-inoculation with phosphate-solubilizing bacteria and N2-fixing bacteria on microbial population and soil amendment under C deficiency. Int. J. Environ. Res. Public Health. 16, 2442. doi: 10.3390/ijerph16132442
Wang, Z. K., Chen, Z. Y., Kowalchuk, G. A., Xu, Z. H., Fu, X. X., Kuramae, E. E., et al. (2021a). Succession of the resident soil microbial community in response to periodic inoculations. Appl. Environ. Microbiol. 87, e00046–21. doi: 10.1128/AEM.00046-21
Wang, Z. K., Chen, Z. Y., Xu, Z. H., and Fu, X. X. (2019a). Effects of phosphate-solubilizing bacteria and N2-fixing bacteria on nutrient uptake, plant growth, and bioactive compound accumulation in Cyclocarya paliurus (Batal.) Iljinskaja. Forests. 10, 772. doi: 10.3390/F10090772
Wang, Z. K., Xu, Z. H., Chen, Z. Y., Kowalchuk, G. A., Fu, X. X., Kuramae, E. E., et al. (2021b). Microbial inoculants modulate growth traits, nutrients acquisition and bioactive compounds accumulation of Cyclocarya paliurus (Batal.) Iljinskaja under degraded field condition. Forest Ecol. Manage. 482, 118897. doi: 10.1016/j.foreco.2020.118897
Wong, W. T., Tseng, C. H., Hsu, S. H., Lur, H. S., Mo, C. W., Huang, C. N., et al. (2014). Promoting effects of a single Rhodopseudomonas palustris inoculant on plant growth by Brassica rapa chinensis under low fertilizer input. Microb. Environ. 29, 303–313. doi: 10.1264/jsme2.ME14056
Xiong, W., Guo, S., Jousset, A., Zhao, Q. Y., Wu, H. S., Li, R., et al. (2017). Bio-fertilizer application induces soil suppressiveness against Fusarium wilt disease by reshaping the soil microbiome. Soil Biol. Biochem. 114, 238–247. doi: 10.1016/j.soilbio.07016
Xu, J. B., Feng, Y. Z., Wang, Y. L., Luo, X. S., Tang, J. W., Lin, X. G., et al. (2016). The foliar spray of Rhodopseudomonas palustris grown under Stevia residue extract promotes plant growth via changing soil microbial community. J. Soil Sediment. 16, 916–923. doi: 10.1007/s11368-015-1269-1
Yilmaz, E., and Sönmez, M. (2017). The role of organic/bio-fertilizer amendment on aggregate stability and organic carbon content in different aggregate scales. Soil Tillage Res. 168, 118–124. doi: 10.1016/j.still.01003
Yin, R., Deng, H., Wang, H. L., and Zhang, B. (2013). Vegetation type affects soil enzyme activities and microbial functional diversity following re-vegetation of a severely eroded red soil in sub-tropical China. Catena. 115, 96–103. doi: 10.1016/j.catena.11015
Zhang, Y., Gao, X., Shen, Z. Z., Zhu, C. Z., Jiao, Z. X., Li, R., et al. (2019). Pre-colonization of PGPR triggers rhizosphere microbiota succession associated with crop yield enhancement. Plant Soil. 439, 553–567. doi: 10.1007/s11104-019-04055-4
Zhao, D., Cao, X. Y., Huang, R., Zeng, J., Shen, F, Xu, H. M., et al. (2017). The heterogeneity of composition and assembly processes of the microbial community between different nutrient loading lake zones in Taihu Lake. Appl. Microbiol. Biotechnol. 101, 1–11. doi: 10.1007/s00253-017-8327-0
Zhong, Y. Q. W., Liu, J., Jia, X. Y., Shangguan, Z. P., Wang, R. W., Yan, W. M., et al. (2020). Microbial community assembly and metabolic function during wheat straw decomposition under different nitrogen fertilization treatments. Biol. Fertil. Soils. 56, 697–710. doi: 10.1007/s00374-020-01438-z
Zhu, G. B., Wang, S. Y., Li, Y. X., Zhuang, L. J., Zhao, S. Y., Wang, C., et al. (2018). Microbial pathways for nitrogen loss in an upland soil. Environ. Microbiol. 20, 1723–1738. doi: 10.1111/1462-2920.14098
Keywords: Rhodopseudomonas palustris PSB06, yield, diversity, bacterial community, rhizosphere
Citation: Luo L, Wang P, Wang D, Shi X, Zhang J, Zhao Z, Zeng J, Liao J, Zhang Z and Liu Y (2023) Rhodopseudomonas palustris PSB06 agent enhance pepper yield and regulating the rhizosphere microecological environment. Front. Sustain. Food Syst. 7:1125538. doi: 10.3389/fsufs.2023.1125538
Received: 16 December 2022; Accepted: 25 January 2023;
Published: 10 February 2023.
Edited by:
Xingyuan Men, Shandong Academy of Agricultural Sciences, ChinaReviewed by:
Yu Shi, Henan University, ChinaCopyright © 2023 Luo, Wang, Wang, Shi, Zhang, Zhao, Zeng, Liao, Zhang and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhuo Zhang,  lionkingno.1@163.com; Yong Liu,
lionkingno.1@163.com; Yong Liu,  haoasliu@163.com
haoasliu@163.com
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.