- 1Department of Plant and Soil Sciences, University of Pretoria, Hatfield, Pretoria, South Africa
- 2Department of Science and Innovation, National Research Foundation Centre of Excellence in Food Security, Pretoria, South Africa
- 3Food and Markets Department, Natural Resources Institute, University of Greenwich, London, United Kingdom
Background: Multidrug-resistant extended-spectrum β-lactamase (ESBL)-producing Enterobacterales is regarded as a critical health issue, yet, surveillance in the water-plant-food interface remains low, especially in Africa.
Objectives: The objective of the study was to elucidate the distribution and prevalence of antimicrobial resistance in clinically significant members of the Enterobacterales order isolated from the water-plant-food interface in Africa.
Methods: A literature search was conducted using six online databases according to the PRISMA guidelines. All available published studies involving phenotypic and genotypic characterization of ESBL-producing Enterobacterales from water, fresh produce or soil in Africa were considered eligible. Identification and characterization methods used as well as a network analysis according to the isolation source and publication year were summarized. Analysis of Escherichia coli, Salmonella spp. and Klebsiella pneumoniae included the calculation of the multiple antibiotic resistance (MAR) index according to isolation sources and statistical analysis was performed using RStudio.
Results: Overall, 51 studies were included for further investigation. Twelve African countries were represented, with environmental AMR surveillance studies predominantly conducted in South Africa. In 76.47% of the studies, occurrence of antimicrobial resistant bacteria was investigated in irrigation water samples, while 50.98% of the studies included fresh produce samples. Analysis of bacterial phenotypic antimicrobial resistance profiles were reported in 94.12% of the studies, with the disk diffusion method predominantly used. When investigating the MAR indexes of the characterized Escherichia coli, Klebsiella pneumoniae and Salmonella spp., from different sources (water, fresh produce or soil), no significant differences were seen across the countries. The only genetic determinant identified using PCR detection in all the studies was the blaCTX − M resistance gene. Only four studies used whole genome sequence analysis for molecular isolate characterization.
Discussion: Globally, AMR surveillance programmes recognize ESBL- and carbapenemase-producing Enterobacterales as vectors of great importance in AMR gene dissemination. However, in low- and middle-income countries, such as those in Africa, challenges to implementing effective and sustainable AMR surveillance programmes remain. This review emphasizes the need for improved surveillance, standardized methods and documentation of resistance gene dissemination across the farm-to-fork continuum in Africa.
1. Introduction
Antimicrobial resistance (AMR) is regarded as one of the top ten threats to global health (WHO, 2020). This follows as the emergence and spread of drug-resistant pathogens that have acquired new resistance mechanisms continue to threaten the effectiveness of clinically important antibiotics to treat common infections (Koutsoumanis et al., 2021; Rahman et al., 2022). Globally, major organizations including the World Health Organization (WHO), Food and Agriculture Organization of the United Nations (FAO), World Organization of Animal Health (OIE) and the European Commission (EC) have recognized the need for further investigation and a multidisciplinary approach to combatting AMR (Koutsoumanis et al., 2021). However, in low and middle income countries (LMICs), such as those in Africa, challenges to implementing effective and sustainable AMR surveillance programmes remain (Elton et al., 2020). This follows as LMICs often lack the necessary infrastructural and institutional capacities and effective reporting systems to roll out sustainable surveillance programmes (Elton et al., 2020).
Many different sources and routes for human acquisition of antimicrobial resistant bacteria are recognized, including human-to-human transmission, direct contact with food-producing animals and pets, foodborne transmission as well as the environment (Koutsoumanis et al., 2021). In LMICs, the main driver of AMR is reported to be transmission and not antimicrobial use (Koutsoumanis et al., 2021). Non-human sources of pathogens such as extended-spectrum β-lactamase (ESBL)-producing Escherichia coli and plasmid mediated AmpC (pAmpC)-producing E. coli has been reported, with the need for longitudinal studies and continuous monitoring (Mughini-gras et al., 2019). Furthermore, the Centers for Disease Control and Prevention (CDC) recently reported that urgent AMR threats in the United States (US) include carbapenem-resistant Enterobacterales, while ESBL-producing Enterobacterales, drug-resistant nontyphoidal Salmonella, and drug-resistant Salmonella serotype Typhi are regarded as serious threats, among others (CDC, 2019). The Enterobacterales form part of the normal epiphytic microflora of fruit and vegetables, and include members ubiquitous in terrestrial and aquatic environments, as well as human foodborne pathogens including pathogenic E. coli and Salmonella spp. (Rajwar et al., 2015). Moreover, Enterobacterales are adapted to sharing genetic material and often clinically significant resistance genes through carriage on mobile genetic elements (Partridge, 2015). Recently, a comprehensive assessment of the global burden of AMR stated that the six leading pathogens for deaths associated with resistance included E. coli, followed by Staphylococcus aureus, Klebsiella pneumoniae, Streptococcus pneumoniae, Acinetobacter baumannii, and Pseudomonas aeruginosa (Murray et al., 2022).
In the last decade, an increased emphasis on the role of the environment in dissemination of AMR has been reported (WHO, 2015; Koutsoumanis et al., 2021). Furthermore, authors have reported on the importance of an integrated One Health approach for developing and implementing mitigation strategies in combatting AMR (White and Hughes, 2019; Ikhimiukor et al., 2022). The One Health concept in AMR mitigation strategies recognizes that humans, animals (including wildlife), environments, and ecosystems are key priorities (White and Hughes, 2019). To date, most AMR surveillance studies, especially in LMICs in Africa, have focussed on humans and animals. As an example, from 901 studies in LMIC-based studies in 2000-2018, the rapid increasing trends of AMR in the food-animal sector for common indicator pathogens such as E. coli, Campylobacter spp., Salmonella spp., and S. aureus have been reported (Ikhimiukor et al., 2022). The main objective of the current study was to elucidate the distribution and prevalence of AMR in clinically significant members of the Enterobacterales family isolated from the water-plant-food nexus in Africa.
2. Materials and methods
The review and meta-analysis of occurrence included published articles from January 2010 – December 2022 and was compiled according to the Preferred Reporting Items for Systematic Reviews and Meta-Analysis (PRISMA) guidelines (Page et al., 2021).
2.1. Search strategy
Information on published articles from all countries on the African continent as defined by the African Union (African Union Commission, 2022) that reported on the occurrence of multidrug resistant Enterobacterales in the water-plant-food interface were included. Two authors independently performed comprehensive literature searches using five online databases: Google Scholar, PubMed Database, EBSCOhost Online Research Databases, Science Direct and Semantic Scholar. Boolean operators (“AND”, “OR”) were applied to search the articles and only English publications were included. The predefined search terms “(extended-spectrum beta-lactamase or extended-spectrum or beta-lactamase or ESBL or ESBL-producing) AND (Enterobacteriaceae or Enterobacterales) AND Africa AND (water or irrigation water or vegetables or fruit or fresh produce or produce or soil)” were used to retrieve relevant articles published within the chosen timeframe. The WHO Global Action Plan to tackle AMR (GAP-AMR) as well as the Global Antimicrobial Resistance and Use Surveillance System (GLASS) was established in 2015 and national action plans developed in individual countries subsequently followed (WHO, 2020; Willemsen et al., 2022). For the current review, the authors chose a timeframe that included environmental AMR surveillance studies 5 years prior to the launch of GLASS up to the most recent articles available in 2022, to provide an overview of current analysis of environmental AMR profiles in African countries over at least a decade.
2.2. Study inclusion and exclusion criteria
Publications were independently reviewed by two authors (LR and ED) to determine eligibility and duplicate entries were identified by considering the title, authors and the year of publication. Inclusion criteria comprised of all available full text articles involving phenotypic and genotypic characterization of ESBL-producing Enterobacterales from water, fresh produce or soil in Africa. More specifically, publications that described the occurrence of antimicrobial resistant bacteria in fresh produce production, including soil, harvested produce and associated irrigation water, as well as fresh produce at retail were considered eligible. Given the diversity of plant-associated bacteria, only members within the Enterobacterales order were included in the current review. This follows as a dramatic escalation in AMR among bacteria, especially members of the Enterobacteriaceae have been noted globally, resulting in ESBL-producing Enerobacteriaceae forming part of the WHO list of critical priority pathogens that pose the greatest threat to human health (WHO, 2017a; Lynch and Clark, 2021). Studies that focused on wastewater, human- or animal health and studies that did not identify the bacterial organisms to at least genus level were excluded (Figure 1). Furthermore, studies that focused on transmission of AMR or multidrug resistant bacteria through non-plant food sources (e.g. dairy, aquaculture or meat products) were not considered eligible for the current review. All published articles available on the selected databases at the time of data extraction (n = 922) were individually reviewed and those not meeting the pre-defined inclusion criteria were excluded from the final articles for analysis.
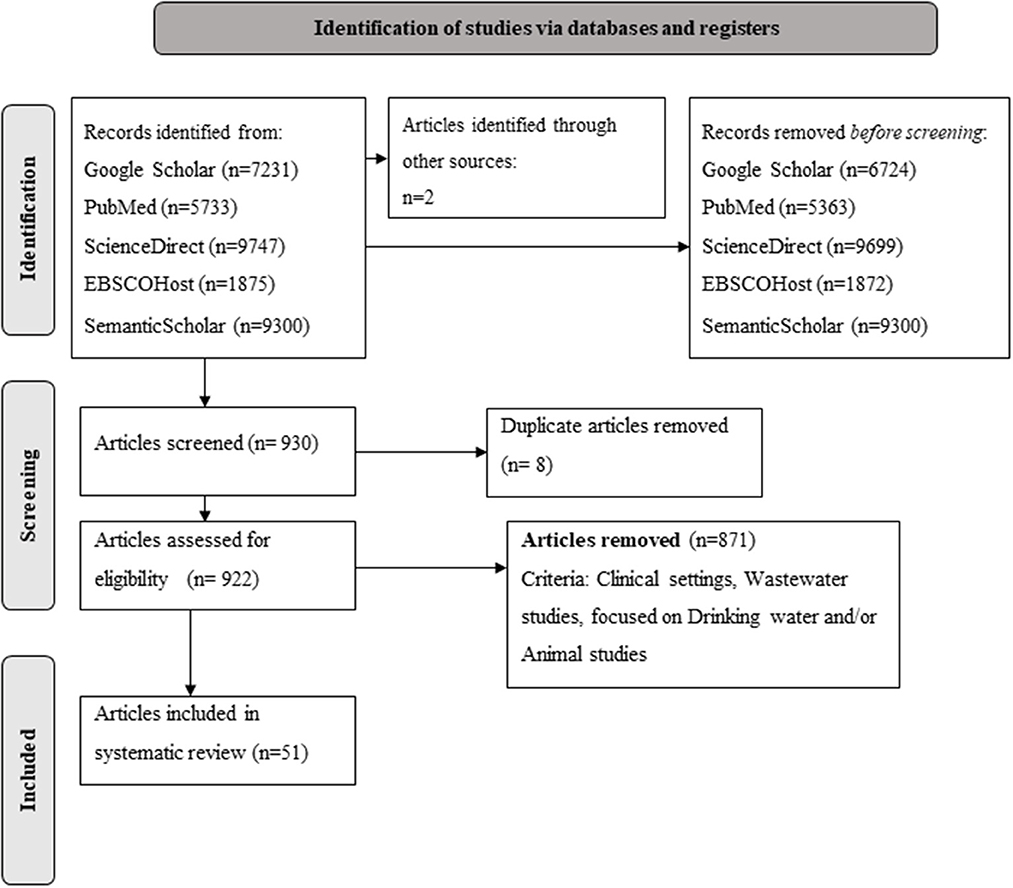
Figure 1. PRISMA flow diagram (Page et al., 2021) summarizing the process and selection of literature on occurrence and characterization of extended-spectrum beta-lactamase producing Enterobacterales in the water-plant-food nexus in Africa.
2.3. Data synthesis, analysis and reporting
Overall, 51 studies were included for further investigation. The publication year, isolation source (water, soil or fresh produce), percentage bacterial occurrence, and identification and characterization methods used were summarized for all publications (Supplementary Table 1). Network analysis was carried out using UCINET® 6 for Windows (Borgatti et al., 2002), with matrices created of the articles based on the isolation source and publication year and country. NetDraw2.175 was used to draw the network.
For selected bacterial species (E. coli, K. pneumoniae and Salmonella spp.) isolated from the different sources, where the information was not already included in the published results and possible to calculate, the multiple antibiotic resistance (MAR) indexes were calculated for analysis of the potential health risk (data not shown). The MAR index for each bacterial species in the respective studies was calculated as x/(y.z) where x represents the aggregate resistance of antibiotics to all isolates, while y represents the total number of antibiotics and z the number of isolates from the isolation source (Riaz et al., 2011). The β-lactamase genes detected in E. coli, K. pneumoniae and Salmonella spp. across the different countries were represented using DataWrapper (Lorenz et al., 2012) and Microsoft Excel and PowerPoint.
2.4. Statistical analysis
Data were analyzed using RStudio (RStudio Team, 2020). The Shapiro-Wilk test was performed on the standardized residuals to test for deviations from normality (Shapiro and Wilk, 1965). ANOVA was used to test for significant differences between the MAR indexes of the characterized E. coli, K. pneumoniae and Salmonella spp. per country and publication year, respectively. Student's protected t-LSD (Least significant difference) were calculated at a 5% significance level to compare significant source effects of the MAR indexes for the characterized isolates (Snedecor and Cochran, 1980).
3. Results
3.1. General overview
Based on the eligibility criteria (Figure 1), a total of 51 articles were included in the systematic review. The included studies represented 12 African countries (Figure 2). The majority of the studies were conducted in South Africa (n = 20), followed by Nigeria (n = 10), Tunisia (n = 6), Algeria (n = 4), Benin (n = 2), Ghana (n = 2), Morocco (n = 2), Egypt (=1), Tanzania (n = 1), Sudan (n = 1), Kenya (n = 1), and Democratic Republic of the Congo (n = 1).
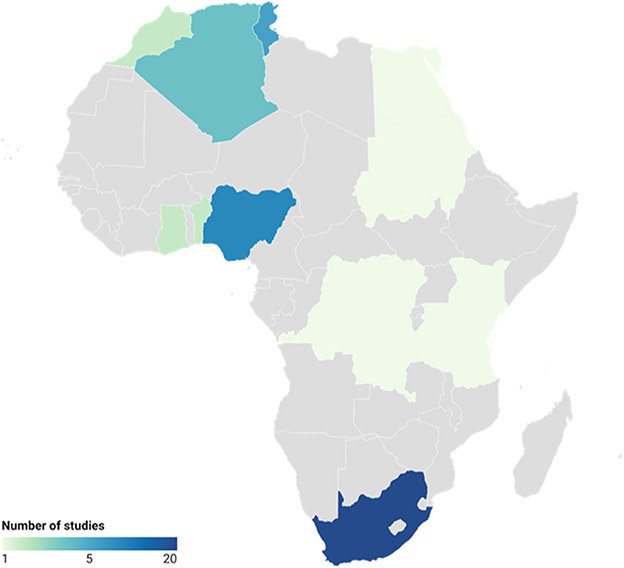
Figure 2. African countries (n = 12) represented by the data analyzed across studies (n = 51) related to antimicrobial resistance.
The occurrence of antimicrobial resistant bacteria in irrigation water samples were evaluated in the majority (76.47%) of the studies (Figure 3). Where indicated, the type of irrigation water included predominantly surface water (river) sources, followed by borehole, ponds, wells, streams and/or canals. In nine studies, irrigation water in conjunction with associated irrigated fresh produce were analyzed, while another eight studies focussed on bacterial isolation and characterization from water and soil in the agricultural environments (Figure 3). Six studies included analysis of irrigation water, soil and associated fresh produce, while eleven studies focussed on isolation and characterization of bacteria from fresh produce only and one focussed on soil analysis only (Figure 3). Overall, the studies that included water, soil and/or fresh produce samples were predominantly conducted in South Africa (n = 13), while studies that focussed on fresh produce or water samples only, were conducted mostly in Nigeria (n = 8) (Figure 3). The majority of the studies were published in 2020 (n = 13), followed by 2015 and 2022 (n = 7, respectively) and 2021 (n = 5).
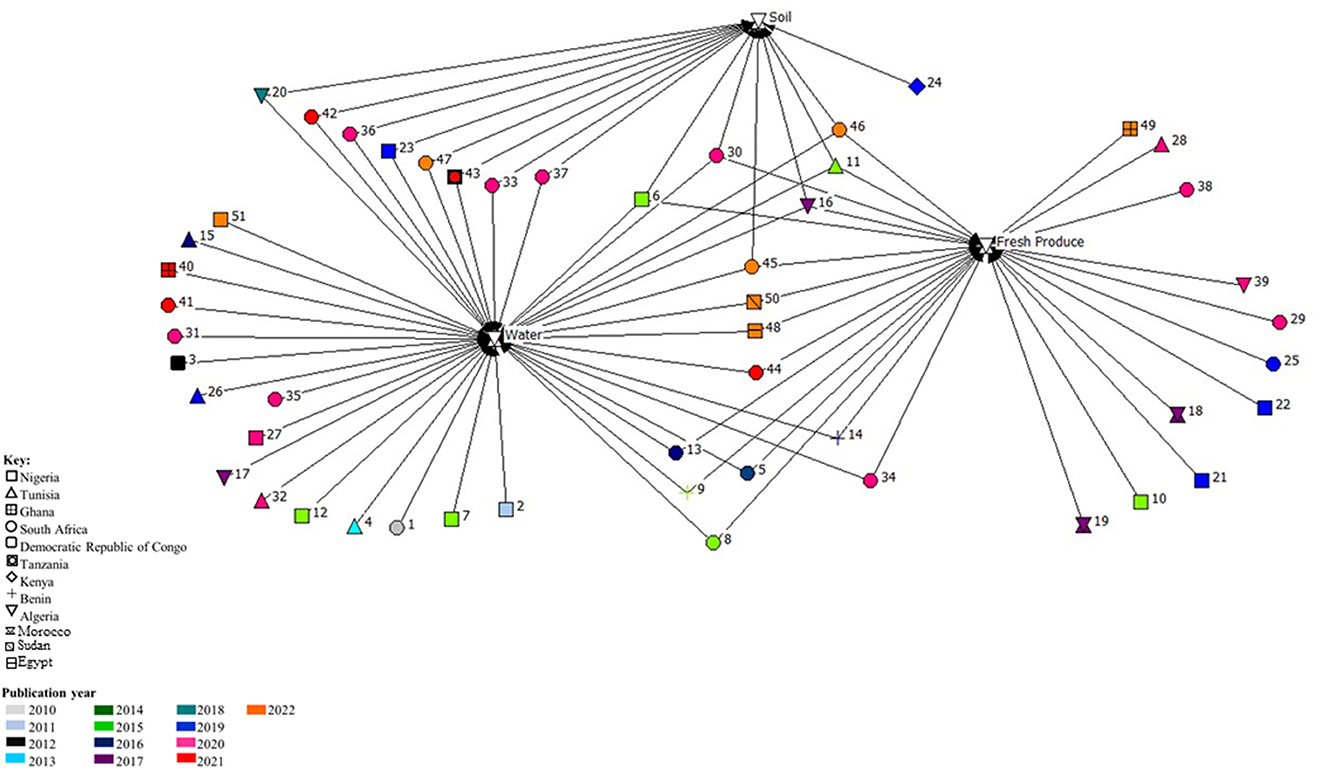
Figure 3. Network analysis of studies done in Africa on the occurrence and characterization of antimicrobial resistant Enterobacterales isolated from the water-soil-plant environment from 2010 to 2022. The numbers represent the articles (n = 51) included in the systematic review, while the colors represent the publication year and shapes the different countries.
3.2. Identification and characterization of bacterial isolates
Enterobacterales from 15 different genera were isolated and characterized for antimicrobial resistance, either phenotypically, genotypically, or both, in 12 African countries. Bacterial isolate identification was performed using principally three methods, alone or in combination, that included biochemical tests, PCR and/or mass spectrometry (Supplementary Table 1). Matrix assisted laser desorption ionization time of flight (MALDI-ToF) mass spectrometry identification appeared to be the gold standard for isolate identification in South African studies, used in 60% (n = 12), followed by 16S rDNA PCR identification (n = 5), biochemical tests (n = 2) and one study that used the OmniLog system for identification. In the other African countries, biochemical tests [analytical profile index (API) or indole testing] were predominantly used for isolate identification (n = 18), followed by MALDI-ToF (n = 7) and 16S rDNA PCR identification (n = 4). One study in the Democratic Republic of the Congo used only selective media for isolate identification, while the Phoenix 100 phenotyping system and a combination of biochemical tests and PCR was used in two studies in Tunisia, respectively.
The 51 studies included in the current review reported on isolation and characterization of 15 different Enterobacterales genera from irrigation water sources, soil and fresh produce (Supplementary Table 1). In total, 20 (39.22%) articles focussed on E. coli only, three (5.88%) on Klebsiella pneumoniae, two (3.92%) on Salmonella spp., and one each on Citrobacter spp., and Enterobacter spp. In the remaining 24 articles that focused on the Enterobacterales family, the most frequently reported bacteria were Klebsiella spp. (43.14%), followed by Citrobacter spp. (35.29%), Enterobacter spp. and E. coli (33.33% each), Serratia spp. (15.69%) and Salmonella spp. and Proteus spp. (11.76% each).
Analysis of phenotypic antimicrobial resistance profiles of the isolates were reported in 94.12% of the studies, with the disk diffusion method predominantly used (Supplementary Table 1). Phenotypic results analysis mainly relied on the interpretive criteria of the Clinical and Laboratory Standards Institute (CLSI, including NCCLS, n = 31), followed by the European Committee of Antimicrobial Susceptibility Testing (EUCAST, n = 3) and the Antibiogram Committee of the French Society of Microbiology (n = 4). Furthermore, in seven studies, both the CLSI and EUCAST criteria were used for results interpretation. Additionally, 72.55% of the studies (n = 37) included PCR detection of the resistance genes, with DNA sequencing as a complementary test to the PCR included in 31.37% of the studies. Of the 31.37%, only 7.84% (n = 4) used the whole genome sequencing (WGS) technique for further characterization.
3.3. Shared resistance genes in potential human pathogens within the water-plant-food interface
The 37 studies that included PCR analysis of resistance genes were predominantly in South Africa (n = 13), followed by Tunisia and Nigeria (n = 6 each), Benin, Morocco and Algeria (n = 2 each) and one study each in Kenya, Sudan, Ghana, Egypt and the Democratic Republic of Congo. Overall, the greatest number of resistance genes were identified in isolates from water samples, however, this comes with a caveat that more studies focussed on water sample analysis (n = 39) alone or in combination with fresh produce and/or soil (Figure 3). In South Africa, Nigeria, Algeria and Tunisia, the blaCTX − M ESBL resistance gene was identified in isolates from water, fresh produce and/or soil. Furthermore, the blaCTX − M resistance gene was the only genetic determinant identified in most of the studies that included PCR analysis, with the exception of studies in Ghana, Morocco and Egypt where the blaTEM gene was predominantly identified. Additionally, beta-lactamase genes including blaSHV, blaTEM, blaOXA, as well as AmpC resistance genetic determinants (blaFOX, blaMOX, blaCIT), sulfonamides and tetracyclines were identified in isolates from water, fresh produce and soil in the South African studies. Isolates from water samples in Nigeria predominantly harbored genes which contributed to resistance against tetracyclines, followed by aminoglycosides (aminoglycoside kinase, aph). In water sample isolates from both South Africa and Tunisia, the blaVIM and blaIMP carbapenem resistance genes were identified. Additionally, the blaKPC and blaNDM carbapenem resistance genes were identified in isolates from water samples, while the blaGES carbapenem resistance gene was present in isolates from water and fresh produce, and the mcr gene was detected from isolates in soil samples, all in studies conducted in South Africa. The NDM carbapenem resistance gene was also identified in isolates from soil samples in Nigeria and water samples from Egypt.
3.4. Further analysis of selected potential human pathogens from the water-soil-fresh produce environment
3.4.1. Escherichia coli
In studies from all nine countries where E. coli was isolated, water samples predominantly included river water used for fresh produce irrigation in urban areas. River water was reported to be impacted by anthropogenic activities (agricultural, industrial and/or domestic) in all the included studies that investigated the presence of multidrug resistant Enterobacterales. The fresh produce samples were purchased at open air markets or formal retailers and farm fresh produce and soil samples included soil from the field where fresh produce was harvested. At least nine classes of antibiotics were included for phenotypic antimicrobial resistance screening in most of the studies that focussed on characterization of E. coli. The dominant resistance patterns of the E. coli isolates included resistance to antibiotics within the tetracycline, penicillin and sulfonamide antibiotic classes, followed by aminoglycosides and quinolones. For the studies where calculations were possible, isolated E. coli had multiple antimicrobial resistance (MAR) indexes of ≥ 0.2, except for two studies in South Africa in 2014 and 2016 (Supplementary Table 2). These studies were conducted in Tunisia, Nigeria, Algeria, Morocco, Sudan, Ghana or South Africa, with significant differences in the MAR indexes between certain countries (p = 0.006) (Figure 4). With a one-way ANOVA, sufficient evidence was given that the MAR indexes of characterized E. coli per publication year did not differ significantly (p = 0.18). Furthermore, no significant differences were seen in the overall MAR indexes of the characterized E. coli from different sources being either water (p = 0.215), fresh produce (p = 0.435) or soil (p = 0.471) samples throughout the study period. Overall, 17 studies that included PCR analysis (either presence/absence detection or further sequencing), screened for resistance genes in E. coli isolated from water, soil or fresh produce samples. The greatest diversity of β-lactamase genes was found in E. coli isolated from samples analyzed in South Africa (Figure 5). In isolates from Tunisia, South Africa, Nigeria, Kenya, Algeria, Sudan, Morocco and Benin, both the blaTEM and blaCTX − M genetic determinants were found (Figure 5). Where sequencing was done, the blaTEM genetic determinants included TEM-1, TEM-2, TEM-3 and TEM-215 in E. coli isolates from South African studies and TEM-15 in isolates from Tunisia. The blaCTX − M genetic determinants included CTX-M-15 (Tunisia, South Africa, Sudan, Algeria and Nigeria), CTX-M-55 (Tunisia and South Africa), CTX-M-14 (Morocco) and CTX-M-1, CTX-M-3, CTX-M-2, CTX-M-14, CTX-M-8/25, CTX-M-27, CTX-M-9 (South Africa).
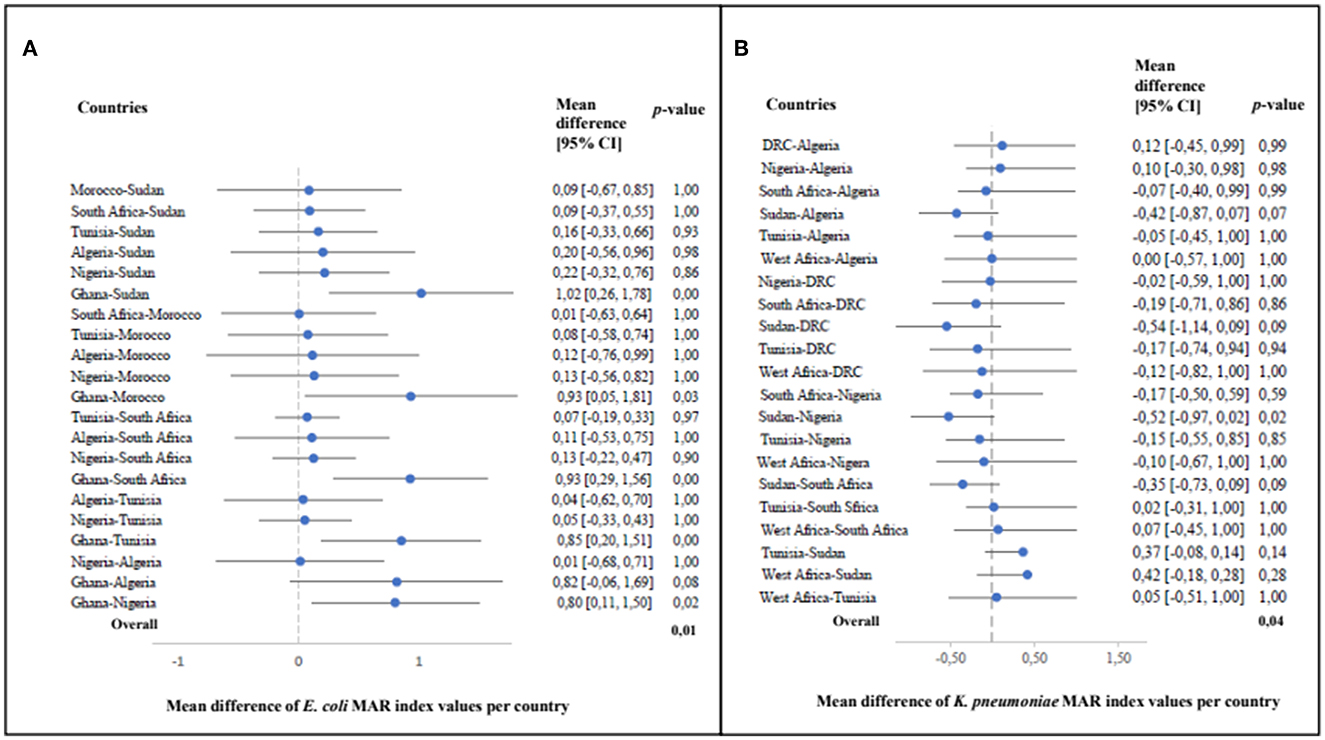
Figure 4. The mean differences in multiple antimicrobial resistance (MAR) index values of Escherichia coli (A) and Klebsiella pneumoniae (B) between different countries with corresponding 95% confidence intervals in the individual studies (E. coli: p = 0.01 and K. pneumoniae: p = 0.04) systematically reviewed across African countries (2010–2022).
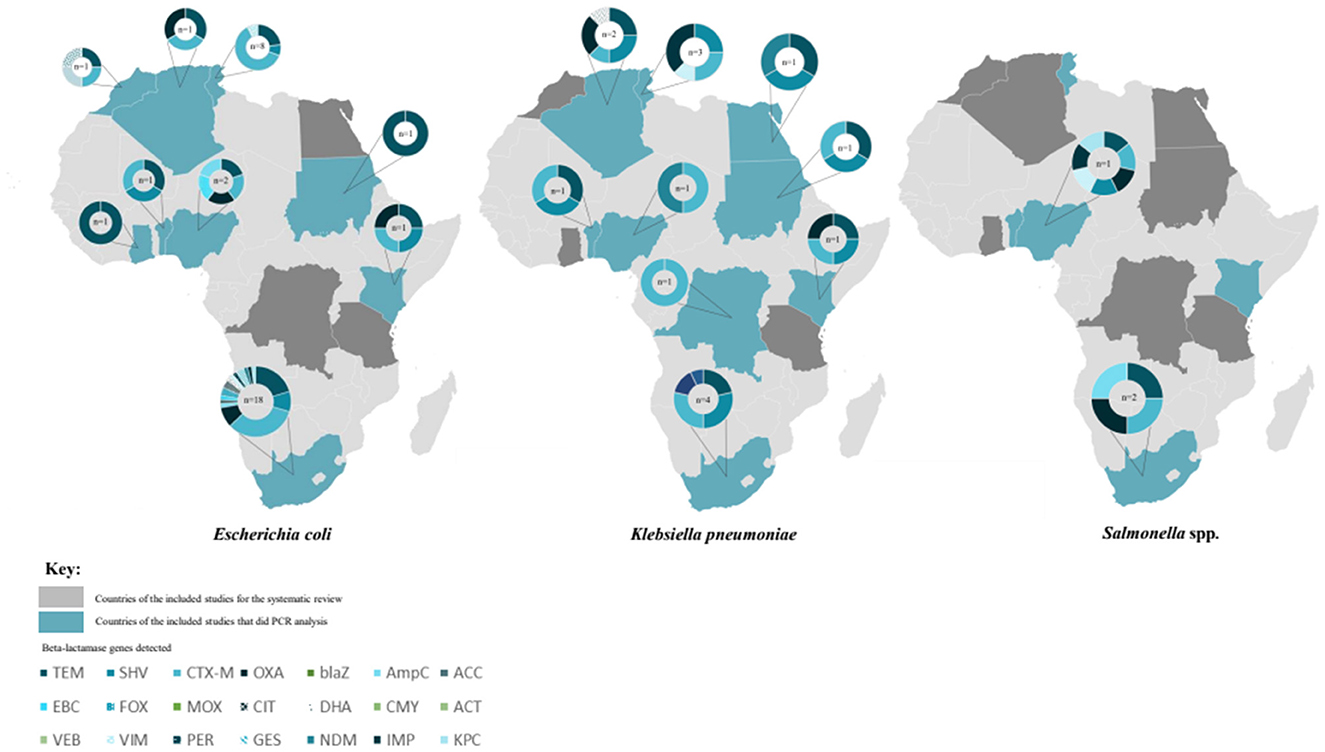
Figure 5. Beta-lactamase genes detected in Escherichia coli, Klebsiella pneumoniae and Salmonella spp. isolated from water, soil and/or fresh produce samples in different African countries between 2010 and 2022. The numbers in the middle of each circle indicates the number of studies where the genes were detected. In total, 21 different β-lactamase genes were detected across the different countries, indicated with different colors or patterns on the same circle.
3.4.2. Klebsiella pneumoniae
The samples where K. pneumoniae were isolated included mainly water, followed by fresh produce and soil. The soil sampled in the included studies were either close to food vending sites or where fresh produce were harvested in the field. The articles that focussed on characterization of K. pneumoniae only and included phenotypic characterization (Mouss et al., 2016; Zekar et al., 2020) tested seven classes of antibiotics for phenotypic resistance screening. This included penicillins, cephalosporins, carbapenems, aminoglycosides, quinolones, fluoroquinolones and sulfonamides. Similarly, where several members of the Enterobacterales family (including K. pneumoniae) were characterized, antibiotics which are usually used to treat infections by these pathogens, were included. Resistance to penicillin such as amoxycillin/clavulanic acid (Augmentin) were dominant, followed by resistance to aminoglycosides (including tobramycin) and cephalosporins such as ceftriaxone and cefotaxime. Overall, the MAR indexes ranged between 0.12 and 0.83 (Supplementary Table 2), which is an indicator that the K. pneumoniae isolates were from potentially high-risk environments where antibiotics are indiscriminately used (Krumperman, 1983; Fadare et al., 2020; Veloo et al., 2022). Significant differences were observed in the MAR indexes of the characterized K. pneumoniae per country (p = 0.04) as well as publication year (p = 0.07). However, the MAR indexes of the characterized K. pneumoniae from the different isolation sources i.e., water (p = 0.84), fresh produce (p = 0.63) and soil (p = 0.84) were not significantly different. In all nine countries where PCR analysis were included, K. pneumoniae strains harboring CTX-M genetic determinants were found (Figure 5). Where sequencing was done, the CTX-M variants included CTX-M-1 in isolates from the Democratic Republic of the Congo and CTX-M-15 in K. pneumoniae isolates from Tunisia, Nigeria and South Africa. Carbapenem resistance genes identified in K. pneumoniae included VIM and IMP variants in isolates from Tunisia, GES and OXA-48 genetic determinants in K. pneumoniae from South Africa and Algeria and the NDM-1 carbapenem resistance determinant in K. pneumoniae isolated from samples in Nigeria and Egypt.
3.4.3. Salmonella spp.
The studies where Salmonella spp. were detected were all conducted in Nigeria or South Africa. From the studies in Nigeria, samples that tested positive for Salmonella spp. predominantly included irrigation water, followed by fresh produce and soil. The Salmonella spp. positive samples from studies conducted in South Africa predominantly water samples, followed by soil samples from the fields in fresh produce production in selected studies. All samples came from urban areas where the river water used for irrigation. The three Salmonella spp. focussed articles were conducted in Nigeria and South Africa between 2011 and 2015 (Akinyemi et al., 2011; Abakpa et al., 2015; Raseala et al., 2020). Different antibiotics were tested in the respective studies, however, dominant resistance patterns in all three studies included resistance against antibiotics within the penicillin class (ampicillin or augmentin), sulfonomides, tetracyline and/or aminoglycosides. Similarly, multiple resistance to antibiotics from at least three different classes were seen in the isolates from the four studies that focussed on isolation of Enterobacterales and detected Salmonella spp. (Odigie et al., 2013; Richter et al., 2019; Iwu et al., 2020; Akinola et al., 2022). The MAR indexes were ≥0.2 in six of the studies (Supplementary Table 2), with no significant difference in MAR index values of characterized Salmonella spp. per country (p = 0.51) or publication year (p = 0.92). The MAR indexes of the characterized Salmonella spp. from water, fresh produce and soil were not significantly different (p = 0.64, p = 0.62, p = 0.87, respectively). Only three studies included PCR analysis of resistance genes in isolated Salmonella spp. (Figure 4). In these studies (Nigeria and South Africa), the β-lactamase genetic determinants included blaTEM and blaCTX − M, and blaOXA while Salmonella spp. isolated from samples in Nigeria additionally harbored VIM, IMP and KPC genetic determinants.
4. Discussion
The role of the environment in dissemination of AMR is increasingly being reported (Koutsoumanis et al., 2021). This meta-analysis summarized the published data available on the occurrence of clinically significant Enterobacterales harboring antibiotic resistance genes, including genes expressing broad-spectrum β-lactamases, ESBLs and/or AmpC β-lactamases, isolated from the water-plant-food nexus in Africa. Environmental antibiotic resistant bacteria are typically classified as (i) carriers, that have a role in dissemination of resistance genes but cannot colonize the human body, and (ii) vectors, comprising of ARB that can colonize and invade the human body (Manaia, 2017). Globally, AMR surveillance programmes recognize ESBL- and carbapenemase-producing Enterobacterales as vectors of great importance in AMR gene dissemination (WHO, 2014, 2020). Based on eligibility criteria within the systematic approach used, 51 studies were identified for the current meta-analysis.
It is well known that the environment contains a natural antimicrobial resistance gene pool as well as resistance genes resulting from anthropogenic activities (Manaia, 2017). Previous research have further demonstrated that water is a unifying transmission pathway for dissemination of AMR throughout different environments (Liguori et al., 2022). Moreover, irrigation water is regarded as one of the main routes of transmission of human pathogenic bacteria onto fresh produce. At least 13 of the studies in the current analysis, from six different countries, all demonstrated that multidrug resistant potential pathogens such as E. coli, K. pneumoniae, Salmonella spp., Enterobacter spp., Serratia fonticola and Citrobacter freundii were present in fresh produce and associated irrigation water (Supplementary Table 1), which corresponds to global studies linking irrigation water to fresh produce contamination (Blaak et al., 2014; Ye et al., 2017; Vital et al., 2018; Banach and Van Der Fels-Klerx, 2020).
The current review also elucidated that water was mainly investigated in environmental AMR studies in Africa between 2010 and 2022. These studies had heterogeneous study locations and methods used, with E. coli predominantly isolated and characterized. The WHO recently published an integrated global surveillance protocol on ESBL-producing E. coli as an indicator (WHO, 2021). The procedures described were specifically designed to be conducted in a harmonized manner to provide the opportunity to increase capacities and to build national integrated surveillance systems for AMR within a One Health approach (WHO, 2021). The environmental aspect of this protocol proposes to detect and quantify ESBL-producing E. coli in contamination hotspot sources including surface water such as rivers that receive wastewater (WHO, 2021). However, the current metadata analysis showed that the methodology, including the frequency and number of samples analyzed, isolation, identification and characterization methods used (even within countries) differed, which contrasts the WHO proposed protocol to conduct research in a harmonized manner.
The results from the current review showed that for studies in South Africa and Algeria, MALDI-ToF was predominantly used for identification of the potential pathogens, while countries including Nigeria, Tunisia, Ghana, Egypt, Morocco and Kenya among others, predominantly used biochemical tests such as API strips for isolate identification. Recently, the reproducibility and accuracy of MALDI-ToF mass spectrometry was evaluated through comprehensive comparison studies in the clinical field (Hou et al., 2019). The authors concluded that the high-throughput, cost effective method with high accuracy resulted in MALDI-ToF mass spectrometry superseding previous conventional molecular or biochemical identification systems (Hou et al., 2019). Moreover, globally, MALDI-ToF is increasingly being used in food safety following approval by the U.S. Food and Drug Administration (Cheng et al., 2016). This follows as fast and consistent detection of foodborne pathogens in a cost-effective manner is vital in food safety analyses where time sensitivity represents an important factor (Cheng et al., 2016; Elbehiry et al., 2017). In many LMICs however, researchers still rely on biochemical tests for isolate identification due to infrastructural or funding constraints. This was evident in the current review where selected studies in Nigeria, South Africa and Tunisia as well as the studies from Benin, Ghana, Egypt, Morocco, Kenya and Tanzania all utilized biochemical tests, predominantly the API20E panels, which are reported to give efficient economical identification (Popov et al., 2022). From the current study, ESBL-producing Enterobacterales predominantly included E. coli and K. pneumoniae isolates. Of note was that 39.22% of the studies focussed on isolation and characterization of E. coli only, therefore, firm conclusions on the prevalence of ESBL-producers in the water-plant-food environment from these African countries cannot be drawn from this data alone. The WHO has reported that E. coli and K. pneumoniae, amongst other third-generation cephalosporin-resistant Enterobacterales, are categorized as a critical priority for global antimicrobial resistance research and development (WHO, 2017a).
Across all countries from the current meta-analysis, the Kirby-Bauer disk diffusion method was mainly used for phenotypic AMR analysis, with the CLSI criteria predominantly followed. However, the antibiotics included in analysis differed across studies and countries, therefore, no conclusion regarding phenotypic resistance patterns of potential pathogens isolated from the different matrices could be reported. Of note is that multidrug resistance (MDR), which is defined as resistance to more than one antibiotic class (Magiorakos et al., 2012), was reported in the majority of the studies. In 2014, the WHO released the first report on AMR surveillance and highlighted the need for an improved and coordinated global effort, while the FAO reported in 2018 on the need for increased environmental antimicrobial resistance surveillance (WHO, 2014; FAO, 2018). Phenotypic resistance to antimicrobials considered as critically important in clinical treatment were detected in the isolates from the water-plant-food nexus in the current meta-analysis. As an example, E. coli isolated from fresh produce and water across all countries showed resistance against β-lactams including penicillins like amoxicillin/clavulanic acid, cephalosporins, carbapenems and aminoglycosides including gentamycin, among others. Furthermore, both Abakpa et al. (2015) and Richter et al. (2020) reported on the occurrence of MDR Salmonella spp. isolated from irrigation water and fresh produce. Antibiotics from classes including β-lactams (amoxicillin/clavulanic acid, cefoxitin, imipenem), cephalosporins (cefepime), tetracylines, and aminoglycosides (gentamycin) amongst other, formed part of the antibiogram profiles in Nigeria and South Africa, respectively. Previous studies have reported that penicillins (specifically amoxicillin/clavulanic acid) are the most commonly prescribed antibiotics in hospital settings in Nigeria as well as South Africa (Okoro et al., 2019; Alabi and Essack, 2022). Tetracycline resistance was also evident in a high number of environmental bacterial isolates from the current review. The use of antibiotics in food animals and overuse in clinical settings have been linked to transmission and rise in environmental multidrug resistant bacteria (Jones-Dias et al., 2016). Along with spread of multidrug-resistance bacteria across all One Health domains, outbreaks of ESBL- and carbapenemase-producing bacteria present a serious challenge to clinicians, with the increasing occurrence in environments posing a public health concern.
The blaCTX − M ESBL resistance gene was identified in all the studies that included PCR analysis in the current review. Similarly, Muthupandian et al. (2018) reported that class A and class D ESBLs are common in bacteria from clinical settings in Africa, with the CTX-M-15 gene being the most prevalent. Screening of resistance to carbapenem antibiotics were included in 25 (59.5%) of the studies from seven different countries in the current review. Imipenem, conferring resistance to Ambler Class B carbapenem-resistant metallo-β-lactamases (Sawa et al., 2020) was the dominant antibiotic included in phenotypic screening, followed by meropenem, which confers resistance to Ambler Class A carbapenemases (Sawa et al., 2020). Carbapenem antimicrobials are considered the last-resort antibiotics for treatment of infections caused by third- and fourth generation cephalosporin resistant and multidrug-resistant bacteria (Sheu et al., 2019). Similar to the results from the current review, Brunn et al. (2022) reported on the presence of carbapenem resistance in environmental reservoirs globally and suggested that wastewater treatment plant effluents were a primary environmental contaminating source. In the subset of studies that included genotypic analysis in the current review, only selected studies (n = 9) screened for presence of carbapenem resistance genes. The KPC genetic determinant was identified in isolates from water samples, while the GES carbapenem genetic determinant was detected in isolates from water and fresh produce samples.
Class A carbapenemases, which include KPC and GES, are plasmid-encoded and frequently detected in clinically significant Klebsiella spp. and Pseudomonas aeruginosa (Sawa et al., 2020). In the current review, these genes were present in E. coli and K. pneumoniae isolates in selected studies conducted in South Africa. Perovic et al. (2016) reported that the introduction of carbapenemase-producing Enterobacterales in South Africa was molecularly confirmed at the end 2011. Furthermore, following environmental introduction, that rapid gene dissemination occurs (Perovic et al., 2016). Recently, Ragheb et al. (2022) reported on the genetic environments of carbapenmases from a One Health perspective in Africa. The authors concluded that the most commonly found carbapenemases were variations of NDM, OXA-48 and VIM being reported from clinical, animal and environmental samples.
Class B carbapenemases are typically encoded on a plasmid, transposon, integron, or chromosome and include the IMP, VIM and NDM genetic determinants (Sawa et al., 2020), supporting rapid gene dissemination across the different one health domains. Interestingly, studies in South Africa from the current review only reported on presence of the VIM and IMP genetic determinants in E. coli isolates, while the same genetic determinants were reported in K. pneumoniae only, in similar studies in Tunisia. Additionally, the NDM carbapenem resistance gene was detected in soil K. pneumoniae isolates from Nigeria, water E. coli isolates from South Africa and fresh produce K. pneumoniae isolates from Egypt. The results from the current review reiterates that clinically significant antibiotic resistant bacteria are no longer restricted to hospital settings, supporting the WHO (2017c) findings that global research and development strategies should include antibiotics active against more common community bacteria.
Similar to Ragheb et al. (2022), the results from the current review showed that detection of ESBL- and carbapenemase-producing Enterobacterales in Africa were relatively low as only nine countries from the entire continent have done environmental AMR surveillance in the current review. Moreover, only studies from six countries have included PCR analysis of β-lactamase genes. However, it must be reiterated that the low prevalence of these multidrug-resistant critical priority pathogens does not represent low environmental occurrence, but rather limited research on the role of the environment in dissemination of AMR within Africa. In selected studies from the current review, multidrug resistant potential pathogens were detected in different samples along the fresh produce farm-to-fork continuum. The mobility of antimicrobial resistance genes in association with mobile genetic elements and the subsequent potential of these genes to move across species, highlights the vital importance of standardized methods, achievable for LMICs as well as, to establish internationally comparable baseline environmental occurrence data (Ikhimiukor et al., 2022; Liguori et al., 2022). From a food safety perspective, knowledge of AMR dissemination along the food supply chain, especially in food crops commonly consumed raw, is critically important for risk communication, intervention and public health (WHO, 2017b).
The inclusion of WGS has been reported as a promising tool for estimation of ARB in a one health context (Aslam et al., 2021). In the current review, only four studies included WGS analysis (Adelowo et al., 2020; Le Terrier et al., 2020; Zekar et al., 2020; Altayb et al., 2022), conducted in Nigeria, Algeria and Sudan, respectively. Additionally, a follow-up WGS analysis study for further characterization of isolates previously published was found in South Africa (Richter et al., 2021), however, this study was not included in the current meta-analysis, to avoid duplication of isolate occurrence information. Whole genome sequencing has successfully been incorporated in integrated food safety surveillance programs in high-income countries (Brown et al., 2019). Across borders, platforms such as GenomeTrakr and PulseNet International are used to share outbreak investigation data, however, there is still a long way to go in the ability to attribute sporadic illness to specific food categories (Brown et al., 2019). As sequencing costs reduce and methods for designing and interpreting metagenomic studies improve, the use of WGS in surveillance studies, especially in infrastructure and capacity-limited LMICs, are increasingly becoming a possibility (Ikhimiukor et al., 2022). The globalization of food supply chains necessitates an integrated surveillance system and platform to share molecular data for food safety purposes. The addition of WGS analysis to phenotypic AMR surveillance studies can aid in understanding the genetic basis of AMR mechanisms and distinguish isolates with phenotypically identical antibiograms, in addition to virulence genes and single-nucleotide polymorphism (SNP) analysis typically used in food safety surveillance and foodborne outbreak investigations (Brown et al., 2019; WHO, 2020). Subsequently, valuable information on the pathways of AMR dissemination across all sectors can be obtained.
5. Conclusion
The available data on occurrence of multidrug-resistant Enterobacterales in environmental settings in Africa emphasizes the need for improved surveillance and documentation of resistance gene dissemination across the farm-to-fork continuum globally. This follows as clinically significant bacterial isolates were found in various water sources and fresh produce. Furthermore, these human pathogenic microbes harbored resistance traits that corresponded to antibiotics often used in clinical settings as well as animal husbandry. The information obtained from the current review could however not be used to determine the extent of the human health risk in consumption of fresh produce where ESBL-producing potential pathogens were present. In addition to a need for harmonized methodology, the cost-effectiveness of One Health AMR surveillance systems, especially in LMICs, should also be considered and further investigated to inform the development and effective and efficient systems in Africa. The results further posed a challenge in comparative studies, as standardized methods were not utilized across the board. Although limited environmental AMR surveillance studies were found in comparison to published data on AMR surveillance in human and animal health, this review showed the vital importance of including information from the water-plant-food nexus in food safety surveillance programs related to AMR in a One Health context. It was further highlighted that comparable indicators to monitor AMR in food crop value chains is necessary. Establishing WGS as a surveillance tool in addition to phenotypic data in AMR surveillance studies will provide comprehensive information to inform comparable national and international actions plans against AMR.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.
Author contributions
LR, ED, SD, and LK contributed to the conceptualization of the study. LR and ED performed the data extraction. LR summarized the data, prepared all figures, and performed the statistical analysis. All authors contributed to the manuscript revision, read, and approved the submitted version.
Acknowledgments
The authors would like to acknowledge the support of Ms. Liesl Morey from the Agricultural Research Council of South Africa's Biometry Unit as well as Dr. J Gokul for support through the statistical analysis.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fsufs.2023.1106082/full#supplementary-material
References
Abakpa, G. O., Umoh, V. J., Ameh, J. B., Yakubu, S. E., Kwaga, J. K. P., Kamaruzaman, S., et al. (2015). Diversity and antimicrobial resistance of Salmonella enterica isolated from fresh produce and environmental samples. Environ. Nanotechnol. Monit. Manag. 3, 38–46. doi: 10.1016/j.enmm.2014.11.004
Adelowo, O. O., Ikhimiukor, O. O., Knecht, C., Vollmers, J., Bhatia, M., Kaster, A. K., et al. (2020). A survey of extended-spectrum betalactamase-producing Enterobacteriaceae in urban wetlands in southwestern Nigeria as a step towards generating prevalence maps of antimicrobial resistance. PLoS ONE 15, 1–19. doi: 10.1371/journal.pone.0229451
African Union Commission. African Union Handbook 2022: A Guide for Those Working With and Within the African Union. Ministry of Foreign Affairs and Trade (2022).
Akinola, T., Onyeaghasiri, O., Oluranti, F. U. O. O., and Oladapo Elutade, O. (2022). NDM Assessment of well water as a reservoir for extended-spectrum β-lactamases (ESBL) and carbapenem resistant Enterobacteriaceae from Iwo, Osun state, Nigeria. J. Microbiol. 14, 351–61. doi: 10.18502/ijm.v14i3.9772
Akinyemi, K. O., Iwalokun, B. A., Foli, F., Oshodi, K., and Coker, A. O. (2011). Prevalence of multiple drug resistance and screening of enterotoxin (stn) gene in Salmonella enterica serovars from water sources in Lagos, Nigeria. Public Health 125, 65–71. doi: 10.1016/j.puhe.2010.11.010
Alabi, M. E., and Essack, S. Y. (2022). Antimicrobial Resistance Antibiotic prescribing amongst South African general practitioners in private practice : an analysis of a health insurance database. JAC-Antimicrobial Resist. 4, 1–8. doi: 10.1093/jacamr/dlac101
Altayb, H. N., Elbadawi, H. S., Alzahrani, F. A., Baothman, O., Kazmi, I., Nadeem, M. S., et al. (2022). Co-Occurrence of β-Lactam and Aminoglycoside Resistance Determinants among Clinical and Environmental Isolates of Klebsiella pneumoniae and Escherichia coli: A Genomic Approach. Pharmaceuticals 15, 8. doi: 10.3390/ph15081011
Aslam, B., Khurshid, M., Arshad, M. I., Muzammil, S., Rasool, M. H., Shahid, A., et al. (2021). Antibiotic resistance: one health one world outlook. Front. Cell. Infect. Microbiol. 11, 1–20. doi: 10.3389/fcimb.2021.771510
Banach, J. L., and Van Der Fels-Klerx, H. J. (2020). Microbiological reduction strategies of irrigation water for fresh produce. J. Food Prot. 83, 1072–1087. doi: 10.4315/JFP-19-466
Blaak, H., van Hoek, A. H. A. M., Veenman, C., Docters van Leeuwen, A. E., and Lynch, G. (2014). Extended spectrum Beta-lactamase- and contitutively AmpC-producing Enterobacteriaceae on fresh procuce and in the agricultural environment. Int. J. Food Microbiol. 8, 168–169. doi: 10.1016/j.ijfoodmicro.2013.10.006
Borgatti, S. P., Everett, M. G., and Freeman, L. C. (2002). Ucinet 6 for Windows: Software for Social Network Analysis. Harvard, MA: Analytic Technologies.
Brown, E., Dessai, U., Mcgarry, S., and Gerner-Smidt, P. (2019). Use of whole-genome sequencing for food safety and public health in the United States. Foodborne Pathog. Dis. 16, 441–450. doi: 10.1089/fpd.2019.2662
Brunn, A., Kadri-Alabi, Z., Moodley, A., Guardabassi, L., Taylor, P., Mateus, A., et al. (2022). Characteristics and global occurrence of human pathogens harboring antimicrobial resistance in food crops: a scoping review. Front. Sustain. Food Syst. 6, 1–19. doi: 10.3389/fsufs.2022.824714
Cheng, K., Chui, H., Domish, L., Hernandez, D., and Wang, G. (2016). Recent development of mass spectrometry and proteomics applications in identification and typing of bacteria. Proteomics Clin. Appl. 10, 346–357. doi: 10.1002/prca.201500086
Elbehiry, A., Marzouk, E., Hamada, M., Al-Dubaib, M., Alyamani, E., Moussa, I. M., et al. (2017). Application of MALDI-TOF MS fingerprinting as a quick tool for identification and clustering of foodborne pathogens isolated from food products. New Microbiol. 40, 269–278.
Elton, L., Thomason, M. J., Tembo, J., Velavan, T. P., Pallerla, S. R., Arruda, L. B., et al. (2020). Antimicrobial resistance preparedness in sub-Saharan African countries. Antimicrob. Resis 9, 1–11. doi: 10.1186/s13756-020-00800-y
Fadare, F. T., Adefisoye, M. A., and Okoh, A. I. (2020). Occurrence, identification, and antibiogram signatures of selected Enterobacteriaceae from Tsomo and Tyhume rivers in the Eastern Cape Province, Republic of South Africa. PLoS ONE 15, 1–27. doi: 10.1371/journal.pone.0238084
FAO (2018). Antimicrobial Resistance and Foods of Plant Origin. Available online at: http://www.fao.org/3/BU657en/bu657en.pdf (accessed November 12, 2022).
Hou, T. Y., Chiang-Ni, C., and Teng, S. H. (2019). Current status of MALDI-TOF mass spectrometry in clinical microbiology. J. Food Drug Anal. 27, 404–414. doi: 10.1016/j.jfda.2019.01.001
Ikhimiukor, O. O., Odih, E. E., Donado-godoy, P., and Okeke, I. N. (2022). A bottom-up view of antimicrobial resistance transmission in developing countries. Nat. Microbiol. 7, 24. doi: 10.1038/s41564-022-01124-w
Iwu, C. D., Plessis, E. M., Korsten, L., Nontongana, N., and Okoh, A. I. (2020). Antibiogram signatures of some enterobacteria recovered from irrigation water and agricultural soil in two district municipalities of south africa. Microorganisms 8, 1–19. doi: 10.3390/microorganisms8081206
Jones-Dias, D., Manageiro, V., Ferreira, E., Barreiro, P., Vieira, L., Moura, I. B., et al. (2016). Architecture of class 1, 2. and 3 integrons from gram negative bacteria recovered among fruits and vegetables. Front. Microbiol. 7, 1–13. doi: 10.3389/fmicb.2016.01400
Koutsoumanis, K., Allende, A., Álvarez-Ordóñez, A., Bolton, D., Bover-Cid, S., Chemaly, M., et al. (2021). Role played by the environment in the emergence and spread of antimicrobial resistance (AMR) through the food chain. EFSA J. 19, 6651. doi: 10.2903/j.efsa.2021.6651
Krumperman, P. H. (1983). Multiple antibiotic resistance indexing of Escherichia coli to identify high-risk sources contamination of foods. Appl. Environ. Microbiol. 46, 165–170. doi: 10.1128/aem.46.1.165-170.1983
Le Terrier, C., Masseron, A., Uwaezuoke, N. S., Edwin, C. P., Ekuma, A. E., Olugbeminiyi, F., et al. (2020). Wide spread of carbapenemase-producing bacterial isolates in a Nigerian environment. J. Glob. Antimicrob. Resist. 21, 321–323. doi: 10.1016/j.jgar.2019.10.014
Liguori, K., Keenum, I., Davis, B. C., Calarco, J., Milligan, E., Harwood, V. J., et al. (2022). Antimicrobial resistance monitoring of water environments: a framework for standardized methods and quality control. Environ. Sci. Technol. 56, 9149–9160. doi: 10.1021/acs.est.1c08918
Lorenz, M., Aisch, G., and Kokkelink, D. (2012). Datawrapper: Create Charts and Maps. Available online at: https://www.datawrapper.de/.
Lynch, J. P., and Clark, N. M. (2021). Escalating antimicrobial resistance among Enterobacteriaceae: focus on carbapenemases. Expert Opinion Pharmacother. 22, 1455–1473. doi: 10.1080/14656566.2021.1904891
Magiorakos, A. P., Srinivasan, A., Carey, R. B., Carmeli, Y., Falagas, M. E., Giske, C. G., et al. (2012). Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 18, 268–281. doi: 10.1111/j.1469-0691.2011.03570.x
Manaia, C. M. (2017). Assessing the risk of antibiotic resistance transmission from the environment to humans: non-direct proportionality between abundance and risk. Trends Microbiol. 25, 173–181. doi: 10.1016/j.tim.2016.11.014
Moussé, W., Noumavo, P. A., Chabi, N. W., Sina, H., Tohoyessou, M. G., Ahoyo, T. A., et al. (2016). Phenotypic and genotypic characterization of extended spectrum β -lactamase klebsiella pneumoniae and phenotypic and genotypic characterization of extended spectrum β -lactamase klebsiella pneumoniae and fluorescent pseudomonas spp. Strains Mark. 7, 192–204. doi: 10.4236/fns.2016.73021
Mughini-gras, L., Dorado-garcía, A., Duijkeren, E., Van Bunt, G., Dierikx, C. M., and Bonten, M. J. M. (2019). Articles Attributable sources of community-acquired carriage of Escherichia coli containing β-lactam antibiotic resistance genes: a population-based modelling study. Lancet 3, 357–369. doi: 10.1016/S2542-5196(19)30130-5
Murray, C. J., Ikuta, K. S., Sharara, F., Swetschinski, L., Robles Aguilar, G., Gray, A., et al. (2022). Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet 399, 629–655. doi: 10.1016/S0140-6736(21)02724-0
Muthupandian, S., Ramachandran, B., and Barabadi, H. (2018). The prevalence and drug resistance pattern of extended spectrum β-lactamases (ESBLs) producing Enterobacteriaceae in Africa. Microb. Pathog. 114, 180–192. doi: 10.1016/j.micpath.2017.11.061
Odigie, I. E., Akinbo, B. D., Atere, A. D., Idemudia, N. L., and Asuquo, A. (2013). Antimicrobial susceptibility of some members of enterobacteriaceae isolated from salad vegetables in calabar. J. Chem. Inf. Model. 53, 1689–1699. Available online at: https://www.semanticscholar.org/paper/Antimicrobial-Susceptibility-Of-Some-Members-Of-In-Odigie-Akinbo/8b894fad3d89cc77e94bfe0f894677bb48667fb2
Okoro, R. N., Nmeka, C., and Erah, P. O. (2019). Antibiotics prescription pattern and determinants of utilization in the national health insurance scheme at a tertiary hospital in Nigeria. Afr. Health Sci. 19, 2356–2364. doi: 10.4314/ahs.v19i3.8
Page, M. J., McKenzie, J. E., Bossuyt, P. M., Boutron, I., Hoffmann, T. C., Mulrow, C. D., et al. (2021). The PRISMA 2020 statement: an updated guideline for reporting systematic reviews. Int. J. Surg. 88, 1–11. doi: 10.1016/j.ijsu.2021.105906
Partridge, S. R. (2015). Resistance mechanisms in Enterobacteriaceae. Pathology 47, 276–284. doi: 10.1097/PAT.0000000000000237
Perovic, O., Britz, E., Chetty, V., and Singh-Moodley, A. (2016). Molecular detection of carbapenemase-producing genes in referral enterobacteriaceae in South Africa: a short report. South African. Med. J. 106, 975–977. doi: 10.7196/SAMJ.2016.v106i10.11300
Popovć, N. T., Kepec, S., Kazazić, S. P., Strunjak-Perović, I., Bojanić, K., Rakovac, R., et al. (2022). Identification of environmental aquatic bacteria by mass spectrometry supported by biochemical differentiation. PLoS ONE 17, 1–13. doi: 10.1371/journal.pone.0269423
Ragheb, S. M., Govinden, U., and Sekyere, J. O. (2022). Genetic support of carbapenemases : a One Health systematic review and meta-analysis of current trends in Africa. Ann. N. Y. Acad. Sci. 1509, 50–73. doi: 10.1111/nyas.14703
Rahman, M., Alam, M. U., Luies, S. K., Kamal, A., Ferdous, S., Lin, A., et al. (2022). Contamination of fresh produce with antibiotic-resistant bacteria and associated risks to human health: a scoping review. Int. J. Environ. Res. Public Health 19, 1–15. doi: 10.3390/ijerph19010360
Rajwar, A., Srivastava, P., and Sahgal, M. (2015). Microbiology of fresh produce: route of contamination, detection methods and remedy. Crit. Rev. Food Sci. Nutr. 56, 2383–2390. doi: 10.1080/10408398.2013.841119
Raseala, C. M., Ekwanzala, M. D., and Momba, M. N. B. (2020). Shared extended-spectrum β-lactamase-producing Salmonella serovars between agricultural and aquatic environments revealed through inva amplicon sequencing. Microorganisms 8, 1–18. doi: 10.3390/microorganisms8121898
Riaz, S., Faisal, M., and Hasnain, S. (2011). Antibiotic susceptibility pattern and multiple antibiotic resistances (MAR) calculation of extended spectrum β- lactamase (ESBL) producing Escherichia coli and klebsiella species in Pakistan. Afr. J. Biotechnol. 10, 6325–6331. doi: 10.5897/AJB11.2449
Richter, L., du Plessis, E. M., Duvenage, S., Allam, M., Ismail, A., Korsten, L., et al. (2021). Whole genome sequencing of extended-spectrum- and AmpC- β-Lactamase-positive enterobacterales isolated from spinach production in Gauteng Province, South Africa. Front. Microbiol. 12, 734649. doi: 10.3389/fmicb.2021.734649
Richter, L., Du Plessis, E. M., Duvenage, S., and Korsten, L. (2019). Occurrence, identification, and antimicrobial resistance profiles of extended-spectrum and ampc β-lactamase-producing enterobacteriaceae from fresh vegetables retailed in Gauteng Province, South Africa. Foodborne Pathog. Dis. 16, 421–427. doi: 10.1089/fpd.2018.2558
Richter, L., Du Plessis, E. M., Duvenage, S., and Korsten, L. (2020). Occurrence, phenotypic and molecular characterization of extended-spectrum- and AmpC- β-lactamase producing enterobacteriaceae isolated from selected commercial spinach supply chains in South Africa. Front. Microbiol. 11, 1–10. doi: 10.3389/fmicb.2020.00638
RStudio Team (2020). RStudio: Integrated Development Environment for R. Available online at: http://www.rstudio.com/ (accessed February 21, 2023).
Sawa, T., Kooguchi, K., and Moriyama, K. (2020). Molecular diversity of extended-spectrum β-lactamases and carbapenemases, and antimicrobial resistance. J. Intensive Care 8, 1–13. doi: 10.1186/s40560-020-0429-6
Shapiro, S. S., and Wilk, M. B. (1965). An analysis of variance test for normality (complete samples). Biometrika 52, 591–611. doi: 10.1093/biomet/52.3-4.591
Sheu, C. C., Chang, Y. T., Lin, S. Y., Chen, Y. H., and Hsueh, P. R. (2019). Infections caused by carbapenem-resistant Enterobacteriaceae: an update on therapeutic options. Front. Microbiol. 10, 80. doi: 10.3389/fmicb.2019.00080
Snedecor, G. W., and Cochran, W. G. (1980). Statistical Methods. 7th Edn. Iowa: Iowa State University Press.
Veloo, Y., Thahir, S. S. A., Rajendiran, S., Hock, L. K., Ahmad, N., Muthu, V., et al. (2022). Multidrug-resistant gram-negative bacteria and extended-spectrum ß-lactamase-producing klebsiella pneumoniae from the poultry farm environment. Am. J. Vet. Res. 48, 186–242. doi: 10.1128/spectrum.02694-21
Vital, P. G., Zara, E. S., Paraoan, C. E. M., Dimasupil, M. A. Z., Abello, J. J. M., Santos, I. T. G., et al. (2018). Antibiotic resistance and extended-spectrum beta-lactamase production of Escherichia coli isolated from irrigation waters in selected urban farms in Metro Manila. Philippines. 20, 1–11. doi: 10.3390/w10050548
White, A., and Hughes, J. M. (2019). Critical importance of a one health approach to antimicrobial resistance. Ecohealth 16, 404–409. doi: 10.1007/s10393-019-01415-5
WHO (2017a). Global Priority List of Antibiotic-Resistant Bacteria to Guide Research, Discovery, and Development of New Antibiotics. Geneva, Switzerland.
WHO (2017b). Integrated Surveillance of Antimicrobial Resistance in Foodborne Bacteria: Application of a One Health Approach. Available online at: https://apps.who.int/iris/bitstream/handle/10665/255747/9789241512411-eng.pdf;jsessionid=1710DEAA0E355CBCB99450559C0DD0C8?sequence=1%0A http://apps.who.int/iris/bitstream/10665/255747/1/9789241512411-eng.pdf?ua=1%0Ahttp://apps.who.int/iris/bitstream/10665/ (accessed November 18, 2022).
WHO (2017c). Prioritization of Pathogens to Guide Discovery, Research and Development of New Antibiotics for Drug-Resistant Bacterial Infections, Including Tuberculosis. Geneva, Switzerland.
WHO (2020). GLASS Whole-Genome Sequencing for Surveillance of Antimicrobial Resistance: Global Antimicrobial Resistance and Use Surveillance System (GLASS). Available online at: https://www.who.int/health-topics/antimicrobial-resistance (accessed November 18, 2022).
WHO (2021). Global Tricycle Surveillance – ESBL E.coli - Integrated Global Surveillance on ESBL-producing E. coli Using a “One Health” Approach: Implementation and Opportunities. Available online at: http://apps.who.int/bookorders (accessed November 18, 2022).
Willemsen, A., Reid, S., and Assefa, Y. (2022). A review of national action plans on antimicrobial resistance: strengths and weaknesses. Antimicrob. Resist. Infect. Control. 11:1-13. doi: 10.1186/s13756-022-01130-x
Ye, Q., Wu, Q., Zhang, S., Zhang, J., Yang, G., Wang, J., et al. (2017). Characterization of extended-spectrum β-lactamase-producing enterobacteriaceae from retail food in China. Front. Microbiol. 9, 1709. doi: 10.3389/fmicb.2018.01709
Keywords: multidrug resistance (MDR), ESBLs, environmental AMR surveillance, foodborne pathogens, low and middle-income countries (LMICs), meta-analysis, Enterobacterales
Citation: Richter L, Du Plessis EM, Duvenage S and Korsten L (2023) Prevalence of extended-spectrum β-lactamase producing Enterobacterales in Africa's water-plant-food interface: A meta-analysis (2010–2022). Front. Sustain. Food Syst. 7:1106082. doi: 10.3389/fsufs.2023.1106082
Received: 23 November 2022; Accepted: 06 March 2023;
Published: 28 March 2023.
Edited by:
Barbara Häsler, Royal Veterinary College (RVC), United KingdomReviewed by:
Abdelaziz Ed-Dra, Sultan Moulay Slimane University, MoroccoKalmia Kniel, University of Delaware, United States
Copyright © 2023 Richter, Du Plessis, Duvenage and Korsten. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Erika M. Du Plessis, ZXJpa2EuZHVwbGVzc2lzQHVwLmFjLnph
 Loandi Richter
Loandi Richter Erika M. Du Plessis
Erika M. Du Plessis Stacey Duvenage
Stacey Duvenage Lise Korsten
Lise Korsten