- Department of Agricultural, Food, Environmental, and Animal Science, University of Udine, Udine, Italy
Lacticaseibacillus casei N87 was used as starter culture for the production of fermented sausages. The strain was cultivated in anaerobic (A) and respiratory (growth in presence of oxygen and supplementation with haeme and menaquinone in the growth medium; R) conditions. Control without the starter culture inoculation and with the addition of 150 mg/kg of nitrate was also included. The effect on physico-chemical parameters (pH, Aw, weight loss, and color), microbial population, volatilome, proteolysis as well as the survival of the strain was evaluated during 90 days of ripening. Q-PCR and DGGE-PCR analyses demonstrated the ability of the strain used in this study to adapt to this environment and carry out the sausage's fermentation process. The inoculation of the strain did not have any effect on the Aw values, which decreased similarly in the different samples whereas the pH was lower in A samples (5.2) and the weight loss in R samples (2.5% less than the others). The color parameters of the samples inoculated with the starter cultures were comparable to those of the control added with nitrate. The concentration of aldehydes that usually are identified as marker of oxidation processes was similar in the samples inoculated with the starter cultures adapted under respiratory conditions and in the control. On the contrary, a higher level was detected in the samples inoculated with the starter cultivated under anaerobic conditions. The proteolysis that occurred during the ripening indicates the differentiation of the A samples from the others. Nonetheless, the volatile profiles of the inoculated fermented sausages were similar. The study demonstrated that aerobic adaptation of Lcb. casei N87 starter culture gave similar color parameters and amounts of aldehydes in sausages fermentations without nitrate compared to conventional fermentations with nitrate.
1. Introduction
Many efforts of the meat industry and researchers are focused on the development of new products with improved nutritional properties. The main strategies include the reduction and replacement of ingredients such as sodium chloride, nitrate/nitrite, and saturated fat, associated with increased risk of developing certain diseases, such as cardiovascular diseases, obesity, and some type of cancer, and the addition of functional components such as probiotics and prebiotics (Ursachi et al., 2020). Dry-fermented sausages are nowadays manufactured using nitrate and nitrite, added as potassium or sodium salts; their maximum permitted level of addition, established by the European Regulation (EC) No 1333/2008 (Berardi et al., 2021), is 150 mg/kg. These curing agents are used as antimicrobial for the control and prevention of Clostridium botulinum growth. In addition, they contribute to color formation, and they act as antioxidants inhibiting lipid oxidation and the development of rancid off-flavors (Honikel, 2008; Majou and Christieans, 2018). Although they are needed to ensure the quality and safety of food, concerns have been raised about their regular consumption. In fact, they can have an adverse effect on human health, causing allergic problems and reducing blood oxygen-carrying capacity. Nitrites can also form N-nitrosamines through reaction with secondary amines, which are implicated in the induction of certain cancers (Sindelar and Milkowski, 2012; Martínez et al., 2019). Currently, consumers' awareness about synthetic additives has increased, demanding for cleaner labels. Natural compounds such as plant extract have been proposed as alternatives to obtain nitrite-free meat products, assessing their impact on sensory and technological properties (Ferysiuk and Wójciak, 2020). Some emerging strategies to produce innovative healthier and functional food products also involve the use of microorganisms and their metabolites thereof. Lactic acid bacteria (LAB) have a long history in the production of several fermented foods. Their growth and metabolic activity can increase the shelf-life and the microbiological safety of certain food products and improve their technological and sensory qualities as well (Comi et al., 2016; Iacumin et al., 2021, 2022). In fermented sausages, they are represented mainly by lactobacilli, and they constitute a significant part of the natural microflora along with gram-positive coagulase-negative cocci (CNC) (Rantsiou et al., 2005; Morot-Bizot et al., 2006). The main function of LAB in meat fermentation is to obtain a rapid decrease of pH, limit the growth of pathogens and spoilage microorganisms and contribute to flavor formation (Lücke, 2000; Comi et al., 2020). Although fermentative metabolism is the most frequent pathway for energy production in LAB and is widely exploited in food production, respiratory capacity has begun to receive attention in recent years. Some LAB strains were found to be able to shift from fermentative to respiratory metabolism, in the presence of oxygen and supplementation with heme or heme and menaquinone (Lechardeur et al., 2011). Respiratory growth can increase biomass yield and improve resistance to oxidative stress, long-term survival, and freeze-drying stresses (Duwat et al., 2001; Zotta et al., 2017). Moreover, it has been observed that respiratory metabolism may influence flavor development and could be associated with increased antioxidant activity. The effect of the growth of Lacticaseibacillus casei N87 under respiratory conditions has been advantageously studied in Cheddar-type cheese production and wheat sourdough fermentation (Reale et al., 2016a,b). In these studies, the respirative culture increased the production of free amino acids, peptides, and aroma compounds such as diacetyl and acetoin, and reduced the oxidation of lipid and protein, compared to the anaerobic cultures. These results open the door for the development of starter cultures with additional functionality that can further optimize the sausages fermentation process. In light of the above, respiratory metabolism shows promising potential for the production and the use of competitive and functional starter cultures. For this reason, the aim of this study was to investigate the effect of the anaerobic and respiratory growth of Lcb. casei N87 during the production of fermented sausages. In particular, were evaluated the evolution of the microflora, the physico-chemical parameters (pH, Aw, weight loss, and color), the production of volatile compounds, the proteolysis, and the survival of the strain.
2. Materials and methods
2.1. Strain and culture conditions
Lacticaseibacillus casei N87 strain was used to promote the fermentation process in this work. The strain was maintained at the temperature of−80°C in cryovials containing DeMan, Rogosa and Sharp broth (MRS, Oxoid, Milan, Italy) supplemented with 20% of glycerol in the Culture Collection of the Laboratory of Food Microbiology, Università degli Studi di Udine, and routinely propagated in Weissella Medium Broth, pH 6.8 (WMB; Zotta et al., 2012), for 16 h at 37°C. At the time of use, the strain was revitalized in MRS broth (Oxoid, Milan, Italy) and then streaked on MRS Agar plates and incubated at 30°C for 48 h, to check the purity and prepare the inoculum for the following stages of the experiment. Lcb. casei N87 was then cultured in modified WMB (10 g/L glucose, without sodium acetate and sodium citrate; mWMB), in batch cultivations were carried out under anaerobic (nitrogen flow at 0.1 vol/vol/min; assay A) and respiratory (60% dissolved oxygen concentration, DO; supplementation of mWMB with 2.5 μg/mL hemin [Sigma-Aldrich, Milan, Italy] and 1 μg/mL menaquinone [Sigma-Aldrich, Milan, Italy]; assay R) conditions, as described by Ianniello et al. (2016). Bioreactors (3 l working volume; Applikon, Schiedam, the Netherlands) were inoculated (1% v/v) with an overnight (16 h, 37°C) WMB anaerobic pre-culture, washed twice in 20 mM potassium phosphate buffer pH 7 (PB7). DO was measured using a polarographic electrode (Applisens, Applikon) and was automatically controlled (ezControl controller, Applikon; set point 60%) by varying the stirrer speed (impeller speed from 200 to 800 rpm; two Rushton turbines, 45 mm diameter) and the opening (from 0 to 100%) of air flow valve (1 vol/vol/min maximum air flow). pH was controlled (pH 6.5) by automatic addition of sterile 4 eq/L NaOH, while foaming was controlled by automatic addition of a sterile 5% (v/v) Antifoam A solution. Adaptation was performed by culturing the cells three times. At the end of the adaptation period, the optical density (OD) at 600 nm (Abs 600; NanoDropTM 2000c, Thermo Fisher Scientific, Milan, Italy) was measured and a standard curve relating Abs 600 and cell number (CFU/mL) was calculated. Anaerobic (A) and respirative (R) grown cultures were recovered by centrifugation at 12,000 xg for 5 min, at 4°C, then washed twice in 20 mmol/L potassium phosphate buffer pH 7.0 (PB7) and re-suspended (to a final level of 108 CFU/mL) in reconstituted skimmed milk (RSM). Cultures were frozen at −80°C for 24 h and then freeze-dried with a Heto Drywinner 3 Benchtop freeze-dryer (Thermo Fisher Scientific, Milan, Italy), at −50°C, 0.1 hPa for 24 h. The freeze-dried cultures were stored for 30 days at −20°C and viable cells were enumerated (plate count on WMB agar, 37°C, 48 h, anaerobiosis) before use.
2.2. Fermented sausages production
Fermented sausages were prepared in a local meat factory using traditional techniques. The production involved the following ingredients: pork meat 64.8%, pork fat 25.2%, sodium chloride 2.6%, black pepper 0.07%, sugar 0.2%, and red wine mixed with garlic (360 mL in 45 kg). Sodium nitrate (E252) was added only in assay C (150 mg/kg), used as negative control, without the inoculation of N87 starter. The meat batter was minced and divided into three aliquots of 15 kg each, to produce three different lots of fermented sausages: (i) control (C, fermented sausages without the addition of starter culture); (ii) fermented sausages with the addition of Lcb. casei N87 adapted under anaerobic conditions (A); (iii) fermented sausages with the addition of Lcb. casei N87 adapted under respiratory conditions (R). The starter culture was inoculated with a final concentration of 6 Log CFU/g of meat batter. After the addition of the starter, each meat batters were minced and stuffed into natural casings, resulting in 25 cm long and 5 cm diameter fresh sausages. No smoking process was applied at any stage. Sausages were ripened for 90 days at the following conditions: the first 24 h at 4°C, then, 23°C and 99% RH (Relative Humidity) for 24 h, 19°C and 65% RH for 36 h, 17°C and 70% RH for 48 h, 15°C and 75% RH for 24 h, 14°C and 80% RH for 36 h, 14°C and 75% RH for 7 days, and 13°C and 75% RH for the last 75 days. Samplings were performed at 0, 2, 5, 7, 15, 30, 60, and 90 days. Analyses were performed in triplicate on three biological replicates per assay.
2.3. Microbiological analysis
Sausages were subjected to microbiological analysis to monitor the dynamic changes in the populations responsible for their ripening and their hygienic quality. Twenty-five grams of each sample were homogenized in 225 mL of saline-peptone water (8 g/L NaCl, 1 g/L bacteriological peptone, Oxoid, Milan, Italy) for 3 min in a Stomacher machine (PBI, Milan, Italy). Decimal dilutions were then prepared and the following analyses were carried out: (i) total aerobic count on Gelatin Sugar Free Agar, at 30°C for 48 h; (ii) lactobacilli on MRS with the addition of 0.01 mg/mL of the antimycotic agent containing natamycin (Delvocid®Instant, DSM Food Specialities, Delft, The Netherland), at 30°C for 48 h; (iii) lactococci on M17 Agar, at 30°C for 48 h (iv) coagulase-negative catalase-positive cocci (CNCPC) on Mannitol Salt Agar, at 30°C for 48 h; (v) total coliforms and Escherichia coli on ChromID coli® (BioMérieux, Florence, Italy), at 37°C for 48 h; (vi) Staphylococcus aureus on Baird-Parker Agar, at 37°C for 48 h; (vii) molds and yeasts on Malt Extract Agar with the addition of 0.025 mg/mL of tetracycline (Sigma-Aldrich, Milan, Italy), at 30°C for 48 h; (viii) enterococci on Kanamycin Aesculine Azide Agar, at 37°C for 48 h. MRS and M17 plates were incubated under restricted oxygen conditions by using an AnaeroGen kit (Oxoid, Milan, Italy). 25 g of sample were also diluted in 225 mL of Buffered-Peptone Water (BPW) and in Fraser Half broth, to investigate the presence of Salmonella spp. and Listeria monocytogenes according to the ISO methods 6579-1 and 11290/1, respectively. Unless otherwise specified, all media and ingredients were obtained from Oxoid (Milan, Italy). Colonies were counted, and the results were calculated as the means and standard deviation of three determinations per biological replicate and expressed as Log CFU/g (3).
2.4. Quantification of total and viable Lcb. casei/paracasei/rhamnosus
Total and viable cells of the species Lcb. casei, Lcb. paracasei and Lcb. rhamnosus were also quantified during fermentation using a culture-independent approach by quantitative PCR (qPCR). This approach was used to have an estimation of the concentration of the inoculated Lcb. casei species, comparing the results obtained from the DNA of the total cells (live and dead, qPCR) and the viable cells (PMA-qPCR). In this case, using a culture-independent approach also stressed cells, that possibly entered in viable but not culturable (VBNC) state were quantified. Two aliquots of 1 mL from the first dilution of each sample were collected. One aliquot was treated with propidium monoazide (PMA, Biotium, Inc., Hayward, CA, USA) and the other was not treated. PMA is able to bind the DNA of dead cells, preventing DNA amplification. Before the process with PMA, samples were centrifuged at 14,000 xg for 10 min, the pellets were washed twice and resuspended in 300 μL of sterile peptone water (maximum recovery diluent, Oxoid, Milan, Italy). After that, 1.4 μl of PMA 20 mM in 20% dimethyl sulfoxide (Sigma-Aldrich, Oakville, Ontario, Canada) was added. Each tube was incubated on ice in the dark for 5 min. For the photo-activation process, the tubes were placed in the PhAST Blue instrument (GenIUL, Barcelona, Spain) for 15 min at room temperature. The suspensions treated with PMA (PMA-qPCR) and the untreated (qPCR) were centrifuged at 14,000 xg for 7 min to pellet the cells. DNA was extracted from the pellets using the MasterPure™ Complete DNA and RNA Purification Kit (Lucigen, Middleton, WI, USA). DNA was resuspended in 40 μL of sterile distilled water. After extraction, the DNA concentration and purity were evaluated using a Nanodrop 2000c spectrophotometer (Thermo Fisher Scientific, Milan, Italy) and standardized to 25 ng/μL. Primers LCgprpoA-F2 (5′-CACTCAARATGAAYACYGATGA-3′) and LCgprpoA-R2 (5′-CGTGGTGAGATTGAGCCAT−3′) (Huang et al., 2011), targeting the rpoA gene of the formerly Lb. casei group, were chosen to perform qPCR. The qPCR mixture contained 10 μL of 2X SsoFast EvaGreen Supermix (Bio-Rad, Milan, Italy), 300 nM of each primer, and 2.5 ng/μL of DNA, and the volume was adjusted to 20 μl with sterile DNA-free Milli-Q water. RotorGene Q system (Qiagen, Milan, Italy) was used to perform qPCR with one cycle of initial denaturation of template DNA and activation of Taq polymerase at 95°C for 5 min, followed by 45 cycles of denaturation at 95°C for 20 s, primer annealing at 50°C for 20 s and extension at 72°C for 20 s. Fluorescence signal acquisition was performed during the extension step. To determine whether non-specific products or primer dimers had formed, the dissociation curves of the final products of each PCR were analyzed from 55 to 95°C (1°C interval). Standard curves were constructed as follows: Lcb. casei N87 grown in MRS broth (Oxoid, Milano, Italy) was enumerated at the exponential phase on MRS Agar (Oxoid, Milano, Italy) to determine the exact Log CFU/mL. DNA was extracted and amplified for each serial dilution. The signals produced in the qPCR reactions (threshold cycle, Ct) by each serial dilution were plotted against the CFU/mL, and standard curves were constructed. The construction of standard curves was performed using three replicates per each standard point, and each reaction was carried out in triplicate.
2.5. Lcb. casei presence confirmation during ripening by PCR-DGGE
Considering that the qPCR approach is group specific, and not only Lcb. casei could be detected, but also Lcb. paracasei and Lcb. rhamnosus, a confirmation step was performed. After counting, all MRS plates were used for bulk formation as previously described (Ercolini et al., 2001). In brief, the bulk formation was performed using all plates from the serial dilutions (from 10−2 to the last). For each dilution, all colonies present on the plate surface were suspended in an adequate volume of quarter strength Ringer's solution, harvested with a sterile pipette. To minimize the effects of different concentrations, all the suspensions were adjusted to give an optical density (OD) of 1 unit at 600 nm. Then, 1 mL of the bulk suspension was used for DNA extraction and subjected to molecular analysis. For DNA extraction, 1 (one) mL of each bulk suspension was centrifuged at 14,000xg for 10 min at 4°C to pellet the cells, and the pellet was subjected to DNA by using MasterPure™ Complete DNA and RNA Purification Kit (Lucigen, Middleton, WI, USA). Amplification was performed with a C1000 Touch™ Thermal Cycler (Bio-Rad, Milan, Italy) in a final volume of 50 μL containing: 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 3 mM MgCl2, 0.2 mM of each deoxynucleoside triphosphate dNTPs, 1.25 U Taq polymerase (Applied Biosystems™ AmpliTaq™, Thermo Fisher Scientific, Milan, Italy), 10 μmol of each primer (338f, with GC clamp underlined, 5'-CGC CCG CCG CGC GCG GCG GGC GGG GCG GGG GCA CGG GGG GCC TAC GGG AGG CAG CAG−3'; 518r, 5'-ATT ACC GCG GCT GCT GG-3') (Muyzer et al., 1993; Ampe et al., 1999) and the DNA template (DNA extracted from standard strains and MRS plates samples). The thermal cycler parameters were as follows: initial denaturation at 95°C for 5 min; 30 cycles of denaturation at 94°C for 1 min, annealing at 42°C for 1 min, and extension at 68°C for 1 min; and final extension at 68°C for 7 min. Five microliters of each PCR mixture were analyzed by electrophoresis in a 0.5x Tris-borate-EDTA (TBE) agarose gel added with EtBr 0.025% (v/v) (120 V for 30 min) before denaturing gradient gel electrophoresis (DGGE) analysis. The Dcode Universal Mutation Detection System™ (Bio-Rad, Milan, Italy) was used for DGGE analysis. Electrophoresis was performed in a polyacrylamide gel (8% [wt/v] acrylamide-bisacrylamide at 37.5:1, Bio-Rad, Milan, Italy) containing a 40–60% urea-formamide (Sigma-Aldrich, Milan, Italy) denaturing gradient. The gels were subjected to a constant voltage of 120 V for 5 h at 60°C. After electrophoresis, they were stained for 20 min in 1.25x Tris-acetate-EDTA (TAE) containing 1x SYBR green (final concentration; Molecular Probes, Eugene, USA) and visualized under UV light. Images of the gels were digitally captured with the BioImaging System GeneGenius (SynGene, Cambridge, United Kingdom). A reference pattern was established consisting of amplicons from 5 different bacterial species: Lcb. casei N87, Latilactobacillus curvatus DSMZ 20019, Levilactobacillus brevis DSMZ 20054, Lactiplantibacillus plantarum DSMZ 20174 and Latilactobacillus sakei DSMZ 6333 from the Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH, (Braunschweig, Germany). By including this standard reference pattern three times on each DGGE gel, resulting DGGE fingerprint band profiles from the different sausages were digitally normalized using Gel Compare 4.1 software (Applied Maths, Kortrijk, Belgium). Additionally, this reference pattern was used to obtain a preliminary identification of Lcb. casei strains. DGGE analyses were performed at least twice. Blocks of polyacrylamide gels containing selected DGGE bands corresponding to the reference position of Lcb. casei were excised with a gel cutting tips. Blocks were then transferred to 50 μL sterile water, and the DNA in the bands was left to diffuse overnight at 4°C. Two microliters of the eluted DNA were used for re-amplification, and PCR products generated with a GC-clamped primer were verified by DGGE. Only products migrating as a single band and at the same position with respect to the control were amplified, as described above, with the primer without the GC clamp. Products were then cloned in pGEM-T Easy vector (Promega, Milan, Italy) following the manufacturer's instructions. Clones were checked as described above (co-migration with control), and the inserts in appropriate clones were sequenced by a commercial facility (Eurofins Genomics GmbH, Ebersberg, Germany). Sequences were aligned in GeneBank using the Blast program (Altschul et al., 1997) to determine the closest known relatives of the partial 16S rRNA sequence obtained.
2.6. pH and Aw measurements
The pH potentiometric measurements were carried out with a pin electrode pH meter (Crison Basic 20, Barcelona, Spain) that was inserted directly into the sample. The water activity (Aw) was determined with an Aqua Lab 4 TE (Decagon Devices, Pullman, WA, USA). For both analyses, three independent measurements per biological replicate (3) were performed.
2.7. Colorimetric analysis and weight loss
The color changing of the samples were investigated using a Chromameter CR-200 colorimeter (Minolta, Osaka, Japan), for every sample three independent measurement were performed, in three different parts of the sausages. Chroma (C*) and Hue angle (hab) were calculated according to Salueña et al. (2019). For the weight loss evaluation, 10 samples per lot of production were weighed at each time of sampling.
2.8. Antioxidant abilities and volatiles profiles
Antioxidant abilities of the strain and volatile profiles of the samples were monitored at the end of ripening by solid-phase microextraction and gas chromatography (SPME-GC-MS) analysis. Five grams of each sample were placed in 10 mL vials sealed by PTFE/silicon septa. The samples were equilibrated for 10 min at 45°C and volatiles adsorbed for 40 min at 45°C on a fused silica fiber SPME-DVB-Carboxen/PDMS (85 μm, Supelco, Bellefonte, PA, USA). Adsorbed molecules were desorbed in GC for 10 min. For peak detection, an Agilent Hewlett – Packard 7890 GC equipped with MS detector 5977A and Autocamp PAL RSI 85 (Hewlett – Packard, Geneva, Switzerland) was used. A 30 meter for 0.25 mm of internal diameter Chrompack VF-Wax MS (Chrompack, Middelburg, Netherlands) was used. The conditions were the same as described by Serrazanetti et al. (2011). Volatile peak identification was carried out by computer matching of mass spectral data with those of the compounds contained in the Agilent Hewlett -gas Packard NIST 2014 mass spectral database and literature (Meynier et al., 1999; Procida et al., 1999; Bianchi et al., 2007; Di Cagno et al., 2008; Spaziani et al., 2009; Serrazanetti et al., 2011; Tabanelli et al., 2012, 2013; Sidira et al., 2015; Montanari et al., 2016). The quantitative data were normalized with respect to an internal standard (4-Methyl-2-pentanol, which was used at a final concentration of 50 mg/kg).
2.9. Proteolysis assessment
All samples were investigated to evaluate the proteolysis through sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) analysis, using Mini Protean II (Bio-Rad, Milan, Italy). A 13.5% acrylamide/bis-acrylamide running gel and 4% acrylamide/bis-acrylamide stacking gel were used. Total proteins were extracted as described by Còrdoba et al. (1994) with some modification: 2 g of samples were homogenized for 3 min with 40 mL of 1.1 M potassium iodide and 0.1 M sodium phosphate buffer (pH 7.4). Samples were centrifuged at 5000 xg for 15 min at 4°C. The obtained supernatants were filtered through a 0.45 μm and 0.22 μm filter and frozen until the time of use. Before the run, Bradford assay was performed in order to standardize the protein concentration to be loaded into the gel (30 μl). Fifteen (15) μL of sample were mixed with 15 μl of Laemmli sample buffer 2X (Bio-Rad, Milan, Italy) containing β-mercaptoethanol. The mixture was incubated at 100°C for 5 min. The standards used were Precision Plus Protein Standard (Bio-Rad, Milan, Italy). The wells of the gels were loaded with 25 μL of the denatured protein samples. The run was performed at 50 V for 15 min and 120 V for 90 min. Gels were stained for 1 h in 0.1% Coomassie Blue R250 dissolved in 50% (v/v) methanol and 7% (v/v) acetic acid solution and destained overnight in 5% (v/v) methanol and 7% (v/v) acetic acid solution. After destaining, gels were processed using the specialized software GelAnalyzer 2010 (Lazar and Lazar, 2010). The molecular weights of the bands were used to build up a single matrix, which was submitted to a two-way hierarchical analysis, obtaining a heatmap. In the heatmap, values are represented by cells colored according to the Z-values, which represent the observed values/standard deviation.
2.10. Statistical analysis
ANOVA analysis with Tukey's test, Principal Component Analysis (PCA), and the two-way hierarchical analysis were performed using R v 3.3.2 (R Development Core Team, 2011). Two PCA were performed to evaluate the association between the volatile compounds detected and the samples at the beginning and at the end of ripening. The first PCA was done using all the volatile compounds detected, the second was done considering the total amount of each chemical class.
3. Results and discussion
3.1. Evolution of the microflora
A wide variety of microbial species can grow in fermented sausages. Samples were subjected to microbiological analysis to investigate the effect of the inoculum performed on different microbial populations (Supplementary Table S1). Yeast population developed regardless of the inoculum (Figure 1A). Several authors have observed different values concerning the yeast population in salami samples, starting from 1 Log CFU/g in the raw material to 7.7 Log CFU/g at the end of the casing (Coppola et al., 2000; Gardini et al., 2001; Moretti et al., 2004; Comi et al., 2005; Cocolin et al., 2006, 2009; Aquilanti et al., 2007; Di Cagno et al., 2008). In our assays, yeasts were found at levels of 3.78 ± 0.23 Log CFU/g at day 0. They remained stable during the first 2 days of fermentation and then increased in all the assays of about 2 Log. Then slightly decreased of 1 Log till the end of ripening period. At 90 days significant differences were observed for the assay A in respect to the control (C) and R. Total aerobic counts (Figure 1B) increased during the fermentation period, reaching the maximum concentration (about 6.5 Log CFU/g) in the first 5 days of ripening and then remained stable, and no significant differences were observed among the three different theses. These results were similar to what other authors found (Comi et al., 2005; Cenci-Goga et al., 2008; Pisacane et al., 2015). Concerning enterococci (Figure 1C), they showed a non-linear behavior during the period monitored, and this has been observed also in other studies (Comi et al., 2005; Di Cagno et al., 2008; Pisacane et al., 2015). At the beginning of fermentation, the counts of the samples inoculated with Lcb. casei N87 grown under anaerobic conditions (A) were significantly higher compared to the others. However, with the progress of the fermentation, starting from day 15 and till the end, the starter culture grown under anaerobic conditions had the lowest counts, resulting in a reduction of 2-3 Log in respect to the other assay. Regarding CNCPC (Figure 1D), a significant difference was observed on day 0 and day 2 in assay C, with respect to assay A and R. This difference could be dependent on the sodium nitrate added in the batter of assay C. It is well known that bacteria, such as staphylococci, micrococci and also streptococci, grow and reduce nitrate to nitrite (Iacumin et al., 2019). Therefore, they grew more rapidly in assay C than in the other theses precisely due to the presence of this substrate. Starting from day 5, in all the fermentations CNCPC showed similar counts, and then they remained stable through the ripening. This trend has been observed also in other studies, in which (Coppola et al., 2000; Iacumin et al., 2006; Aquilanti et al., 2007; Silvestri et al., 2007; Di Cagno et al., 2008; Cocolin et al., 2009; Francesca et al., 2013) the growth of this microbial population fluctuates between 3 and 6 Log CFU/g (Coppola et al., 2000; Iacumin et al., 2006; Aquilanti et al., 2007; Silvestri et al., 2007; Di Cagno et al., 2008; Cocolin et al., 2009; Francesca et al., 2013). As observed, the addition of sodium nitrate allowed a more rapid increment in CNCP growth, but the difference was flattened rapidly (5 days) and no significant differences among the assay were then observed. LAB population (Figure 1E), at day 0 in control samples (spontaneous fermentation), was 4.08 Log CFU/g, reaching 6 Log CFU/g after 5 days and remaining stable until the end. Similar behavior was observed also by other authors (Coppola et al., 2000; Urso et al., 2006; Aquilanti et al., 2007; Cenci-Goga et al., 2008; Pisacane et al., 2015; Polka et al., 2015). However, Comi et al. (2005) reported higher LAB counts in naturally fermented sausages (8 Log CFU/g). As expected, the counts of samples A and R at day 0 were significantly higher compared to the control (p < 0.05), due to the inoculation performed. The lactococci counts (Figure 1F) showed no differences among the samples during the fermentation and, after an initial increase, they remained stable during the ripening. Similar counts have been found also in other studies (Cenci-Goga et al., 2008; Ranucci et al., 2013). Total coliforms (Figure 1G) showed a high variability during ripening. They resulted under the detection limit (< 1 Log CFU/g) in the raw materials of sample R, whereas the control samples and A samples had a count of 2.5 and 1.49 Log CFU/g, respectively. Throughout the period monitored, coliforms increased during the first 15 days of ripening in all the samples (4 Log CFU/g), before decreasing to counts similar to the raw material. These results agree with the data observed by Silvestri et al. (2007) and Cocolin et al. (2009). Although different authors have found coliforms in their studies, they were not detected at the end of the fermentation process (Moretti et al., 2004; Aquilanti et al., 2007; Pisacane et al., 2015). S. aureus and E. coli counts remained below the detection limit during ripening (2 and 1 Log CFU/g, respectively). Salmonella spp. and Listeria monocytogenes, investigated using ISO methods, were absent in 25 g of product at each time of sampling. These results, despite the presence of coliforms, underlined the good hygienic quality and safety of the raw materials. It can be concluded that the different adaptation methods didn't affect the microbial fermentation. Regardless of the adaptation method, in fact, fermentations in assays A and R proceeded correctly, following the same trend as in assay C. This was also confirmed by the absence of the development of pathogens.
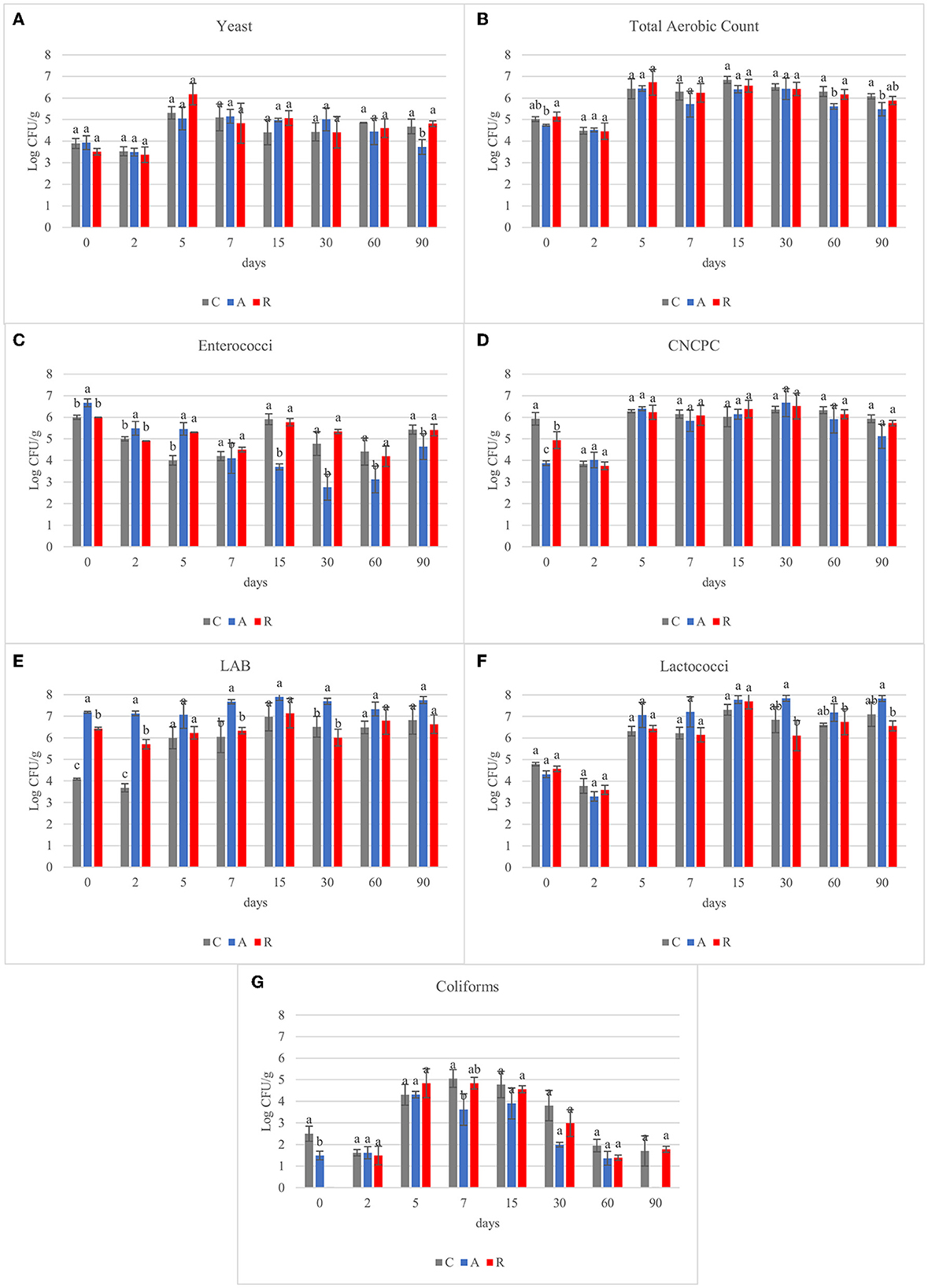
Figure 1. Results of microbiological analysis of carried out on the fermented sausages (non-inoculated, C; inoculated with Lcb. casei N87 cultivated under respiratory conditions, A; inoculated with Lcb. casei N87 cultivated under anaerobiosis, R) during ripening (Log CFU/g). (A) yeast; (B) total aerobic count; (C) enterococci; (D) coagulase-negative catalase-positive cocci, CNCPC; (E) lactic acid bacteria, LAB; (F) lactococci; (G) coliforms. Error bars refer to the standard deviations obtained from triplicate sample analysis. ANOVA analysis with Tukey's test was performed in samples, clustered according to microbial analysis and time of sampling. The results are showed as top letters on each bar.
3.2. Total and viable Lcb. casei quantification
The aim was to evaluate if Lcb. casei was able to survive to the environment and to the competition to the natural LAB flora, also considering the number of cells that could be entered in a state of VBNC for any reason. These cells, in fact, despite cannot be counted by cultural methods, can still have metabolic activity and influence the final characteristics of the product. So, to have a clear picture of the state of these inoculated cultures, and also to understand if the method of adaptation (R or A) could influence their survival during fermentation, this approach was chosen. In Figure 2 the results of qPCR analysis were reported. In control samples, the viable cells remained 2.5 Log CFU/g during ripening. These results are in accordance with the results obtained using DGGE analysis. In fact, no bands were highlighted in C samples from dilution 10−6 to 10−2, confirming that the species Lcb. casei was not present at concentration > 102 CFU/g (see “Section 3.3”). An exception can be observed at day 5, where a concentration of 2.38 Log CFU/g was detected (Figure 1A). It is worth to point out that using this technique all the three species of the formerly Lcb. casei group can be detected. For this reason, the concentration disclosed could be attributed also to Lcb. rhamnosus and Lcb. paracasei, since Lcb. casei was never detected using DGGE analysis. In assays R (Figure 2B) and A (Figure 2C), the total counts by qPCR and PMA-qPCR confirmed the Lcb. casei concentration of the inoculum being around 106 CFU/g. However, in R assays the concentration of viable cells were comparable to the total cells and remained substantially constant over time. Vice versa, in A assay, with anaerobic adaptation, the inoculated cells seemed to undergo a greater stress, in fact the viable cells (detected by PMA-qPCR) were 4.38 Log CFU/g at time 0, but this concentration then increased in the following days until it reached values higher than 7 Logs, exceeding the concentration found in theses R by at least 1 Log. In this specific context, is necessary to know if the Lcb. casei group was the main LAB in the samples. Comparing the viable and total counts to the MRS plate counts, it can be observed that Lcb. casei group was not the principal LAB group in control samples (Figure 2A). Conversely, the differences between the cells detected by PMA-qPCR and the number of colonies counted on MRS plates in samples R and A were noticeable only in some cases (Figures 2B, C, respectively). Several authors have affirmed that the growth of bacteria under sublethal stressing conditions (as it is the case of the cells used in A trials), which also include oxidative stress, results in an increment of the stress resistance (Ianniello et al., 2015, 2016; Ricciardi et al., 2019; Gao et al., 2022). This is in accordance with our results. In fact, the concentration of viable cells of samples R corresponds to the inoculum and did not change during ripening, remaining comparable with the total cells' concentration. Conversely, the strain adapted under anaerobic conditions decreased by almost 2 Log on day 0. However, during fermentation, the viable cells reached concentrations higher than those of samples R, showing that the initial stressing condition, that caused a decrement in the counts, was immediately overcome, thanks to the environmental conditions that were optimal for the growth characteristics of the inoculated strains.
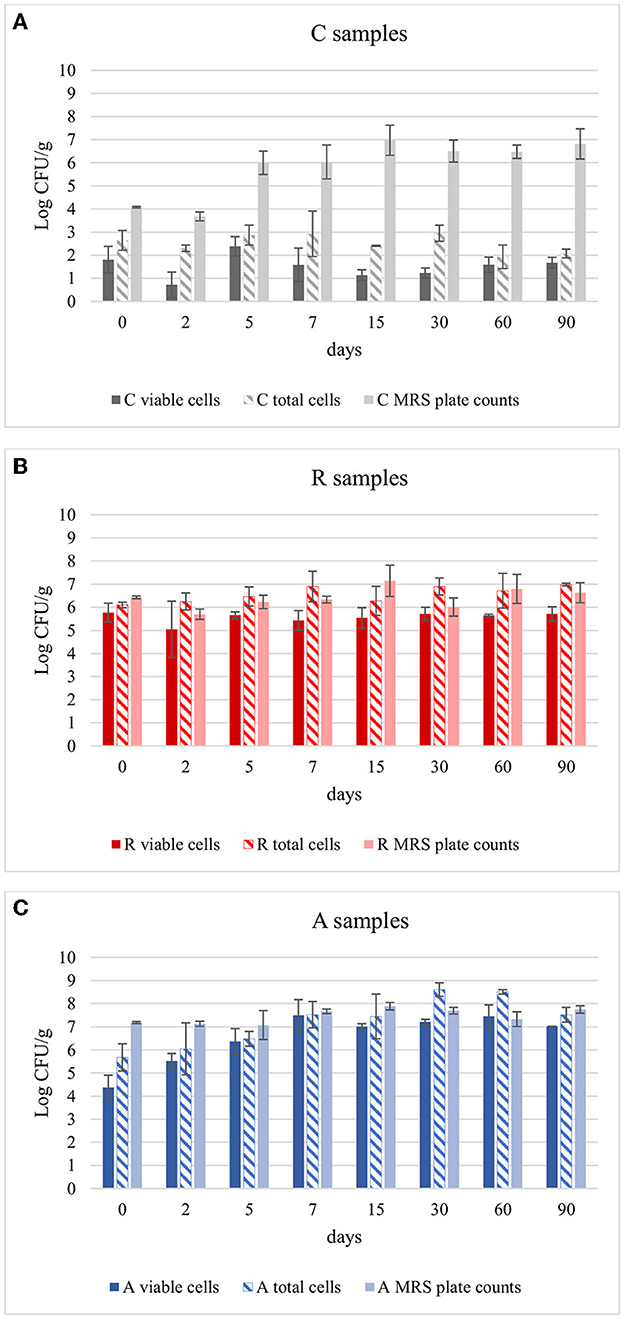
Figure 2. Comparison of Lcb. casei group viable cells (PMA-qPCR), total cells (live and dead, qPCR) and MRS plate count during the ripening of: (A) non-inoculated sausages, C; (B) sausages inoculated with Lcb. casei N87 cultivated under respiratory conditions, R; (C) sausages inoculated with Lcb. casei N87 cultivated under anaerobiosis, A. Error bars refer to the standard deviations obtained from triplicate sample analysis.
3.3. Presence of Lcb. casei N87 in samples
DGGE was performed to assess the presence of the inoculated starter cultures during the ripening. The band of Lcb. casei N87 was identified by comparing its relative position of migration in the gels with the DGGE profile in the ladder. Supplementary Figure S1 shows an example of the DGGE profile. In samples C, the band corresponding to Lcb. casei was never detected. The strain was found in all samples A and its relative band was visible up to a dilution of 10−6. Lcb. casei N87 adapted under respiratory conditions was found at each time of sampling and the band was detected up to a dilution of 10−6 until day 5, 10−4 on day 7, and 10−3 from day 15 until the end of the seasoning. Lcb. casei strains inoculated in fermented sausages have been investigated by several authors using molecular techniques (RAPD-PCR or DGGE); their findings showed the potential of this species to survive until the end of ripening (Rubio et al., 2014; Sidira et al., 2014; Bagdatli and Kundakci, 2016). However, in other studies, Lcb. casei and Lcb. paracasei strains have been found only until 40 days of ripening (Coppola et al., 2000; Andrighetto et al., 2001; Pisacane et al., 2015). These results demonstrated that the starter used was able to survive in the environment and compete with the natural microbiota, independently from the adaptation method.
3.4. Evaluation of the physico-chemical parameters
In the Mediterranean area, salami are dry or semi-dry, with ripening period higher or lower than 4 weeks and Aw lesser than 0.90 or between 0.90 and 0.95, respectively (Lücke, 2000). The final pH can be about 5 or comprised between 5.3 and 6.3 (Aquilanti et al., 2016). The results of this study were similar to those ranges (Figure 3). The pH value of C and R samples were in accordance with several authors, who investigated fermented sausages production in this part of Italy (Friuli-Venezia Giulia) (Comi et al., 2005; Urso et al., 2006; Cocolin et al., 2009). In contrast, the pH of samples A was lower, similar to the pHs of fermented sausages characterized in other studies (with or without the addition of starter cultures; Coppola et al., 2000; Aquilanti et al., 2007; Tabanelli et al., 2012; Wójciak et al., 2012; Ranucci et al., 2013; Trzaskowska et al., 2014) (Figure 3A). It has been noticed that the activation of respiratory metabolism in LAB decreases lactic acid production, which may have led to a lower acidification (Duwat et al., 2001), although in accordance with the control (C). The measure of pH is an indirect measure of lactic acid production, since acidification in sausages mainly depends on the production of lactic acid. In the different assays an increment of lactic acid production, and consequently a decrease of pH of about 1 unit can be observed in fermentation A. This difference could have a direct impact also on the organoleptic perception of the product. Viceversa, fermentation R didn't show significant differences with the control. Aw values during and at the end of ripening did not show significant differences (p > 0.05) (Figure 3B). The averages of the weight losses were 36.45% ± 2.45, 35.48% ±1.14, and 40.26 ±1.79 for samples C, A, and R, respectively (Figure 4). This weight loss was similar to data reported by other studies (Coffey et al., 1998; Moretti et al., 2004; Tabanelli et al., 2012; Bagdatli and Kundakci, 2016), also confirming that the ripening period was optimal to guarantee the safety of the products. In fact, a weight loss of 30% is desired for the product to enter an Aw range that does not allow the growth of Listeria monocytogenes. In this study, different color parameters were considered. No appreciable differences were found among the treatments. Examined samples showed stable red index (a*) and chroma rate, whereas the hue angle (red-purplish color) and the lightness (L*) decreased over time (Figure 5). The weight loss influences the L* value, as was found by Bagdatli and Kundakci (2016). Conversely, in other studies, the L* value decreased during ripening (Olivares et al., 2010; Ercoskun and Özkal, 2011). The stability of a* during ripening suggested that the formation of nitrosylmiyoglobin was not affected by microbial and enzymatic degradation. Moreover, no significant differences were observed between assay C, containing sodium nitrate, and theses A and R, without the use of E252.
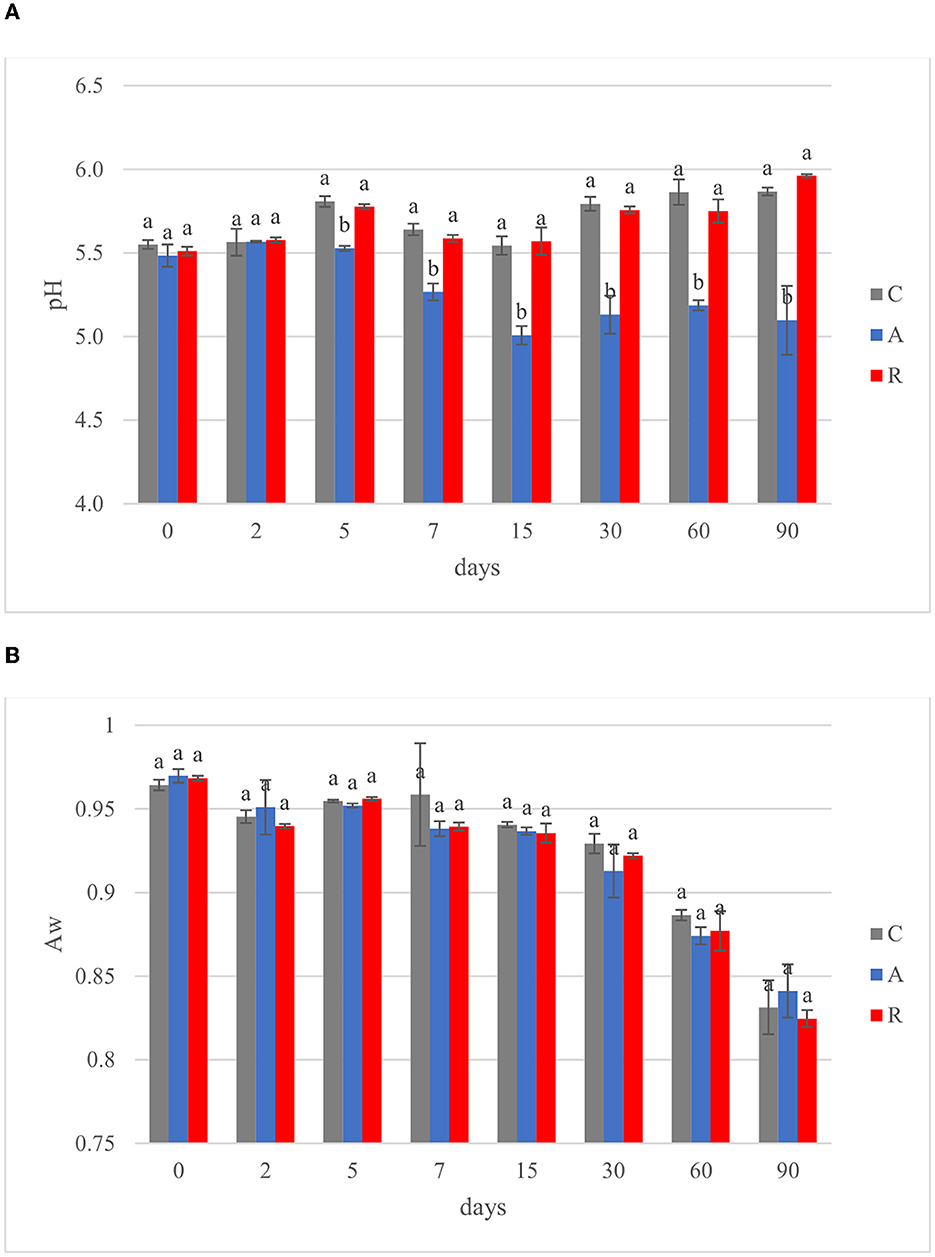
Figure 3. Evolution of (A) pH and (B) water activity (Aw) during ripening of: non-inoculated sausages, C; sausages inoculated with Lcb. casei N87 cultivated under anaerobiosis, A; sausages inoculated with Lcb. casei N87 cultivated under respiratory conditions, R. Error bars refer to the standard deviations obtained from triplicate sample analysis. Different letters (a, b) indicate significant differences among different batches at the same fermentation time (p < 0.05).
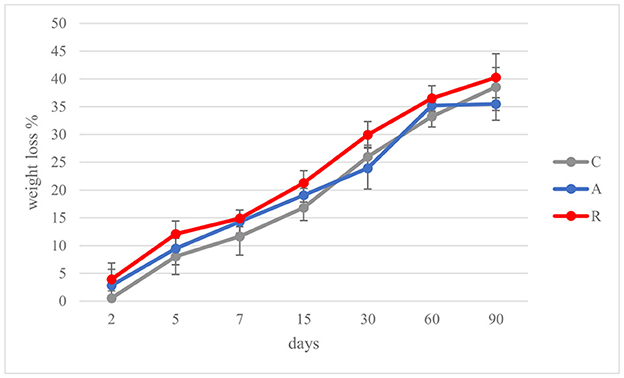
Figure 4. Weight loss (%) during ripening of: non-inoculated sausages, C; sausages inoculated with Lcb. casei N87 cultivated under anaerobiosis, A; sausages inoculated with Lcb. casei N87 cultivated under respiratory conditions, R. Error bars refer to the standard deviations obtained from triplicate sample analysis.
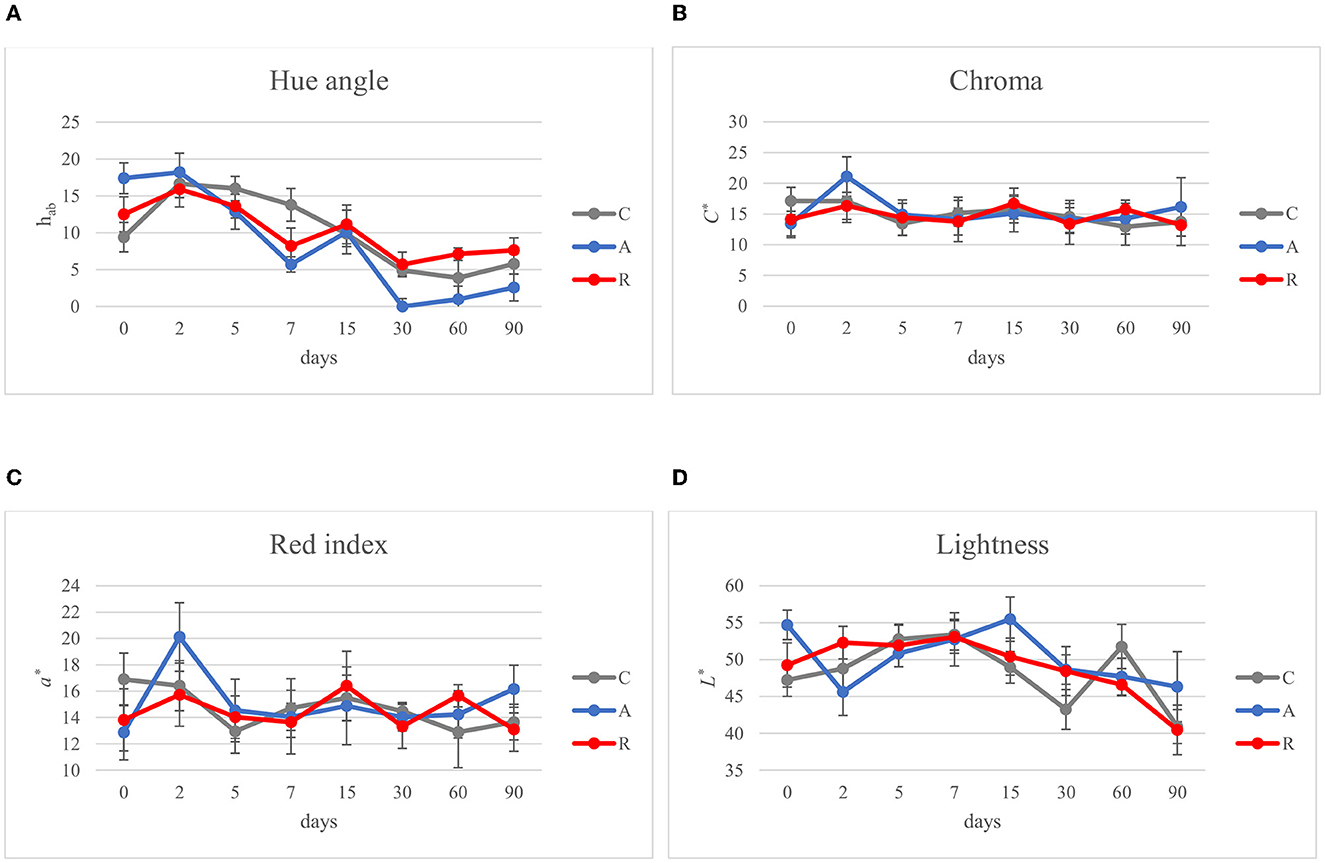
Figure 5. Evolution of color parameters during ripening of: non-inoculated sausages, C; sausages inoculated with Lcb. casei N87 cultivated under anaerobiosis A; sausages inoculated with Lcb. casei N87 cultivated under respiratory conditions R. (A) Hue angle, hab; (B) Chroma, C*; (C) Red/green value, a*; (D) Lightness, L*. Error bars refer to the standard deviations obtained from triplicate sample analysis.
3.5. Antioxidant abilities and volatiles profiles
The activation of respirative metabolism in LAB leads to changes in metabolic pathways and therefore it may affect the flavor development of fermented products (Pedersen et al., 2012; Zotta et al., 2017). In addition, the capability of this strain to scavenge free radicals and reduce lipid and protein oxidation when it is cultivated under respiratory conditions has been confirmed in several in vitro and in vivo studies, in which, the same strain cultivated under anaerobic conditions had lower antioxidant capabilities (Ianniello et al., 2015, 2016; Reale et al., 2016b; Ricciardi et al., 2019). Volatile compounds of raw material and final products were reported in Table 1. Terpenes and terpenoids detected and deriving from pepper and garlic were not considered (Meynier et al., 1999; Sidira et al., 2015; Montanari et al., 2016). Forty-eight compounds were identified and were comprised of 13 aldehydes, 3 ketones, 9 esters, 8 acids, 11 alcohols, and 3 hydrocarbons. Alcohols were the predominant group of volatile compounds, represented by ethanol. The other main components were (E)-2-hexen-1-ol, characterizing only the raw material, 1-hexanol and phenylethyl alcohol, found in all the samples and 1-octen-3-ol, detected only at end of ripening. Alcohol biosynthesis in fermented meat is the result of many metabolic pathways such as lipid oxidation, amino acid metabolism, and methyl ketone reduction (Wójciak and Dolatowski, 2012; Sidira et al., 2015). Ethanol is a product of carbohydrate metabolism, however, the addition of wine during manufacturing can have an impact on the amount detected. Among the identified compounds, 1-octen 3-ol is produced by bacterial metabolism (lipid β-oxidation) (Marco and Navarro, 2006), in fact, it was not detected in the raw material, and imparts mushroom flavor (Tabanelli et al., 2012). As found by Gardini et al. (2013) phenylethyl alcohol decreased during ripening. Aldehydes were one of the predominant groups of volatile compounds. They are produced mainly during lipid autoxidation and hexanal, octanal, and non-anal are commonly used as a marker of the oxidation process (Procida et al., 1999; Marco and Navarro, 2006; Bianchi et al., 2007; Tabanelli et al., 2013; Sidira et al., 2015). In the samples analyzed, hexanal, which has a characteristic odor of green leaves (Meynier et al., 1999; Di Cagno et al., 2008), was the most abundant compound of aldehydes but no noticeable differences among the samples at the end of ripening were observed. Among the identified compounds, benzene-acetaldehyde is derived from phenylalanine metabolism and imparts a floral aroma (Tabanelli et al., 2012). Using aldehydes (hexanal, octanal, and non-anal) as a marker for oxidation, the control and R samples were less oxidized compared to A samples. Similar findings have been observed in the studies conducted by Reale et al. (2016b) in which Cheddar-type cheese was inoculated with Lcb. casei N87 grown under anaerobiosis and respiration conditions. These authors observed lower lipid oxidation in the samples inoculated with the culture grown under respiratory conditions. As it is well known, one of the effects of nitrate is delaying oxidative rancidity in several meat products. Therefore, its reduction or elimination in fermented sausage production may increase the oxidative degradation of lipids and protein during ripening, leading to a decrease in the overall quality of the product. The results obtained in the present study are very promising since the fermented sausages produced using Lcb. casei N87 adapted under respiratory conditions had a similar oxidative state of those produced with the use of nitrate at the maximum permitted level. Other quantitatively relevant compounds were hydrocarbons, they were found also by other authors (Meynier et al., 1999; Tabanelli et al., 2012) but they did not influence the odor profile due to their high odor thresholds (Marco and Navarro, 2006). Nine esters were found in this study, they are often present in fermented sausages, having low sensory threshold values, and providing fruity notes to the product (Demeyer et al., 2000; Marco and Navarro, 2006). They could be formed by alcohol oxidation or esterification of acids and alcohol (Meynier et al., 1999; Bianchi et al., 2007). Esters were represented mainly by ethyl esters, which were detected only at the end of ripening. Their formation during ripening was attributed to LAB and Micrococcaceae (Benito et al., 2004). Among the different ethyl esters detected, ethyl acetate and ethyl ester-2-hydroxy-propanoic acid were present in higher concentrations in the A samples compared to the others. The less abundant groups of volatile compounds were acids and ketones. Acetic acid is a product derived from carbohydrates fermentation (Sidira et al., 2015). The higher content of ethyl acetate detected in the A samples may be the result of the esterification of acetic acid with ethanol, as it is known that esters can be formed by the esterification of alcohols and carboxylic acids (Ferrocino et al., 2018). Samples A presented the higher content of acetic acid, vice versa the lower concentration of this compound was detected in R samples. These results are in contrast with those of other authors, who observed a high concentration of acetic acid in the sourdough produced using respirative culture of Lcb. casei N87. However, it must be considered that the source and concentration of sugars in sourdough and fermented meat are completely different, as well as the environmental conditions and therefore the behavior of the starter culture could be differently affected. The ketones group is composed of acetone, 2,3-octanedione, and 2-nonanone, all found in samples at the end of ripening. Diketones (2,3-octanedione) were produced during Maillard reaction (Bianchi et al., 2007) and are characterized by a butter-creamy flavor. Acetone was the most abundant and it is produced during fermentation, although the involved pathway is not completely clear (Procida et al., 1999; Tabanelli et al., 2012). 2-non-anone was detected, in low concentration, only in the control samples at the end of ripening and is produced by bacterial metabolism through lipid β-oxidation (Marco and Navarro, 2006). To better demonstrate the correlation between the inoculum and the aroma profile and the concentration of volatile compounds detected, two principal component analyses (PCA) were performed, using the concentration of the volatiles reported in Table 1. Figure 6A shows the distance among control and inoculated samples, while Figure 6B reports the coordinates of the variable factor. Meat mixture (CT0) is positioned in the quadrant characterized by 4 alcohol [phenyethyl alcohol, (Z)-3-hexen-1-ol, 1-hexanol and (E)-2-hexen-1-ol), 2 acids (2-hexanoic acid and acetic acid] and 2 aldehydes [non-anal and (E,E)-2,4-decadienal]. After 90 days of ripening, the control is in another quadrant, divided by raw materials and inoculated samples. Five molecules characterize CT90: 1 ketone (2-non-anone), 1 aldehyde (benzenacetaldehyde), and 2 alcohols (3-methyl-2-butanol and 2,3-butanediol). However, alcohols were the main representative compounds in these samples. As mentioned above, the addition of wine in the mixture affected the presence of alcohols in samples. AT90 and RT90 clustered together and were characterized by 3 aldehydes [2,4-nonadienal, (E)-2-nonenal, and octanal] and 3 esters (ethyl ester-2-hydroxy-propanoic acid, ethyl ester dodecanoic acid and ethyl ester hexadecanoic acid). All the other compounds are located among samples at the end of ripening. The fermented sausages inoculated with the starter culture Lcb. casei N87 differed from the spontaneously fermented sausages, indicating the ability of the starter used to produce a distinctive volatile profile. An additional PCA analysis was performed, considering the total amount of each chemical class. The differentiation among meat mixture and samples at the end of casing, as can be seen in Figure 7, is still clear and characterized by the alcohols present in the CT0 samples. In particular, samples A at the end of ripening differed from the other samples for high levels of total aldehydes and esters. A similar amount of the latter compounds was also found in samples R, whereas the control presented the lower amount. Esters, especially ethyl esters, have been correlated with sausage flavor and therefore are essential to obtain the proper fermented sausage aroma (Flores et al., 2004).
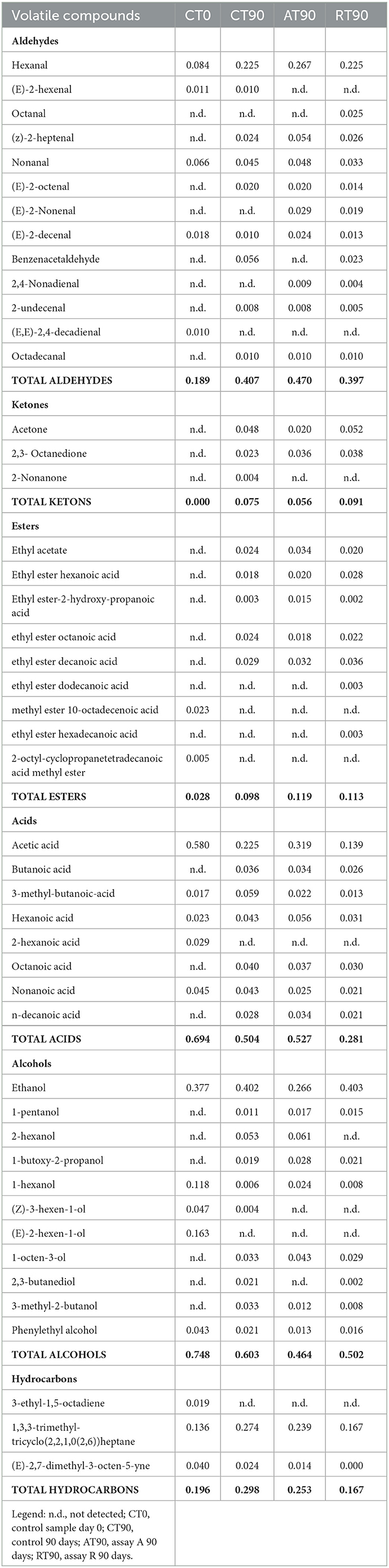
Table 1. Volatile compounds (mg/kg) identified at the end of ripening (90 days) in the different assay.
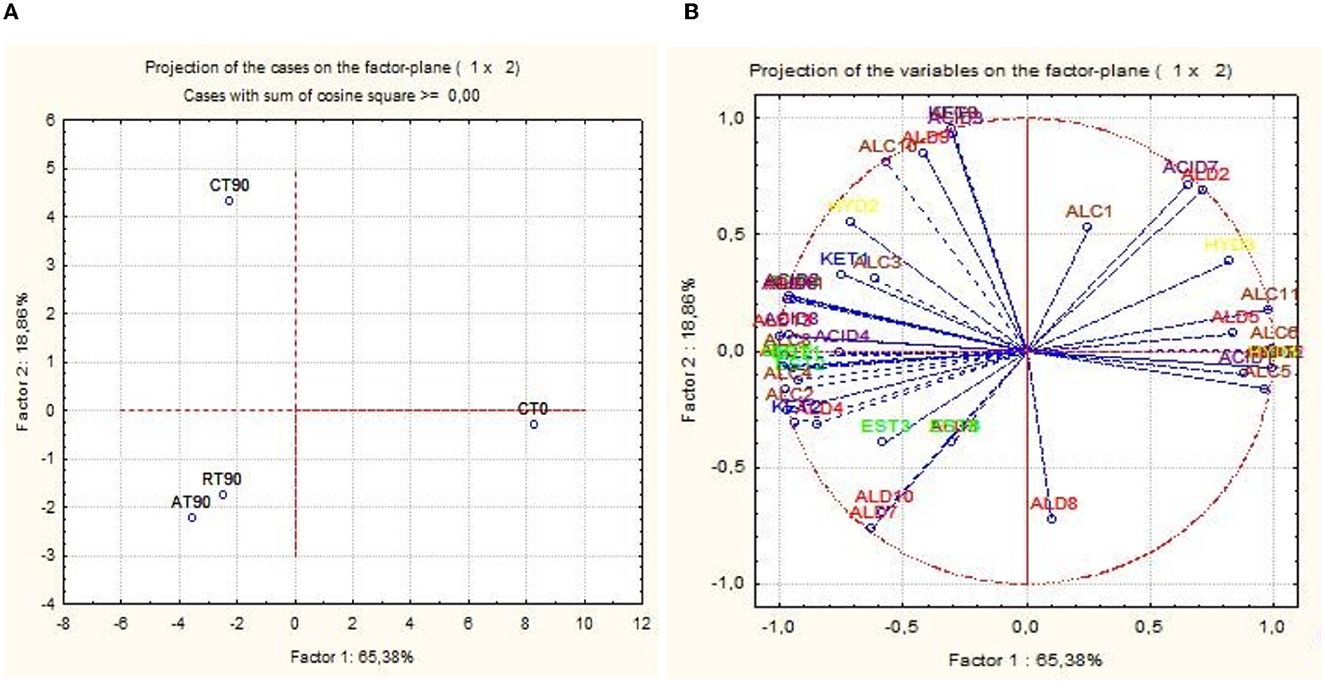
Figure 6. Principal component analysis (PCA) scores (A) and loadings (B) plots of fermented sausages (CT0, meat mixture; CT90, control non-inoculated at 90 days; AT90, sausages inoculated with Lcb. casei N87 cultivated under anaerobiosis at 90 days; RT90, sausages inoculated with Lcb. casei N87 cultivated under respiratory conditions at 90 days) and volatile compounds. ALD1, Hexanal; ALD2, (E)-2-hexenal; ALD3, Octanal; ALD4, (z)-2-heptenal; ALD5, Nonanal; ALD6, (E)-2-octenal; ALD7, (E)-2-Nonenal; ALD8, (E)-2-decenal; ALD9, Benzenacetaldehyde; ALD10, 2,4-Nonadienal; ALD11, 2-undecenal; ALD12, (E,E)- 2,4-decadienal; ALD13, Octadecanal; KET1, Acetone; KET2, 2,3- Octanedione; KET3, 2-Nonanone; EST1, Ethyl acetate; EST2, Ethyl ester hexanoic acid; EST3, Ethyl ester-2-hydroxy-propanoic acid; EST4, ethyl ester octanoic acid; EST5, ethyl ester decanoic acid; EST6, ethyl ester dodecanoic acid; EST7, methyl ester 10-octadecenoic acid; EST8, ethyl ester hexadecanoic acid; EST9, 2-octyl-cyclopropanetetradecanoic acid methyl ester; ACID1, Acetic acid; ACID2, Butanoic acid; ACID3, 3-methyl-butanoic-acid; ACID4, Hexanoic acid; ACID5, 2-hexanoic acid; ACID6, Octanoic acid; ACID7, Nonanoic acid; ACID8, n-decanoic acid; ALC1, Ethanol; ALC2, 1-pentanol; ALC3, 2-hexanol; ALC4, 1-butoxy-2-propanol; ALC5, 1-hexanol; ALC6, (Z)-3-hexen-1-ol; ALC7, (E)-2-hexen-1-ol; ALC8, 1-octen-3-ol; ALC9, 2,3-butanediol; ALC10, 3-methyl-2-butanol; ALC11, Phenylethyl alcohol; HYD1, 3-ethyl-1,5-octadiene; HYD2, 1,3,3-trimethyl-tricyclo(2,2,1,0(2,6))heptane; HYD3, (E)-2,7-dimethyl-3-octen-5-yne.
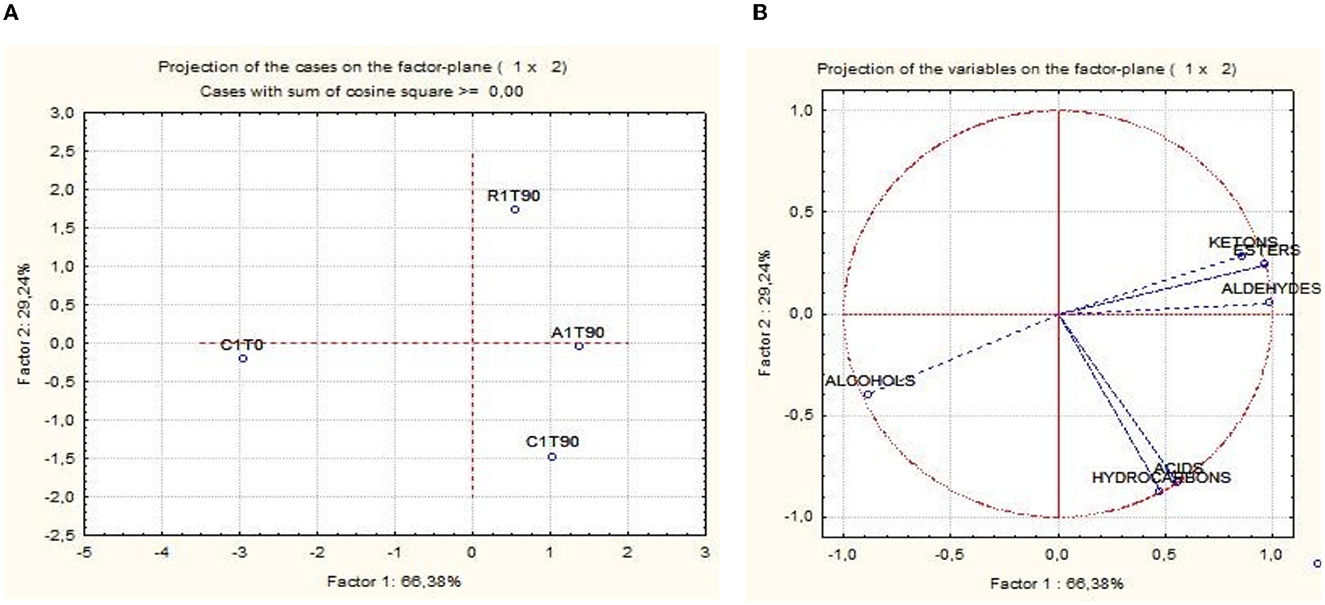
Figure 7. Principal component analysis (PCA) scores (A) and loadings (B) plots of fermented sausages (CT0, meat mixture; CT90, control non-inoculated at 90 days; AT90, sausages inoculated with Lcb. casei N87 cultivated under anaerobiosis at 90 days; RT90, sausages inoculated with Lcb. casei N87 cultivated under respiratory conditions at 90 days) and groups of volatile compounds.
3.6. Proteolysis assessment
During meat fermentation, the main enzymes involved in the initial breakdown of sarcoplasmic and myofibrillar proteins are muscle proteinases (Fadda et al., 2010). Nevertheless, LAB also take part in this process, their proteolytic activity system, consisting of various intracellular peptidases and cell-wall-bound proteinases, is complex (Savijoki et al., 2006). In dry fermented sausages, lactobacilli's enzymes seem to be responsible for the production of amino acid and small peptides, which are precursors of several flavor molecules (Fadda et al., 1998; Sanz et al., 1999a,b). The SDS-PAGE, performed during the fermentation, showed changes in the proteolytic profile. The presence of bands higher than 140 kDa was detected in the meat mixture, while lower bands were found during the overall ripening period. Bands lower than 18 kDa were discovered in C and A samples, except for 15 and 13 kDa bands that were detected only in samples A. In the heatmap (Figure 8) is possible to observe 7 main clusters, bands molecular weight are grouped in 4 main clusters (A, B, C and D) whereas the samples are grouped in 3 main clusters (I, II, and III). The samples belonging to the same assay were clusterized together. Within cluster III, is possible to see two other classifications, samples from 0 to 7 days and 15 to 90 days of ripening. Samples C and R had a similar proteolytic profile. Samples A, probably due to the effects of lower pH on the proteolytic activity (Candogan et al., 2009), differs from the other, also for the presence of bands with a low molecular weight. Similar results were found in cluster I, except for samples at 2 and 15 days of casing that clustered in a different way. Clusters concerning bands were 4, including heterogeneous molecular weight bands. After 15 days, the band at 146 kDa disappeared in all the samples, several authors have reported a lower presence of myosin heavy chain concentration during fermented sausage ripening (Fadda et al., 2001; Casaburi et al., 2008; Spaziani et al., 2009). Cathepsin D, aspartic and cysteine proteinases, originating from meat were considered the more involved enzymes in the production of small fragments of myofibrillar proteins (122, 38, 29, 13 kDa) (Sanz et al., 1999b; Demeyer et al., 2000). In agreement with the study of Fadda et al. (2010), the 45 kDa band (presumably actin) was detected in all the samples until the end of ripening. Bands with a molecular weight lower than 24 kDa were also found and they probably correspond to myosin light chain (Hughes et al., 2002; Fadda et al., 2010). At the end of ripening, the 17 kDa band (myoglobin), with the exclusion of C samples, was completely degraded. Other bands were detected indiscriminately during the 90 days. The proteolytic activity of LAB on meat proteins had been described by several authors through in vitro and in vivo studies (Fadda et al., 1998, 1999, 2001, 2002, 2010; Hierro et al., 1999; Sanz et al., 1999a,b; Demeyer et al., 2000; Hughes et al., 2002; Spaziani et al., 2009; Aro et al., 2010). These studies suggested that the microbial enzymes are more involved in the latter stages of ripening, whereas the initial breakdown of the muscle proteins is due to the activity of meat proteinases. Mold and catalase-positive and negative-cocci could be also implicated in protein degradation (Lücke, 1997; Rouhi et al., 2013). Acids, alcohols, and aldehydes, developed from the degradation of several amino acids (methionine, leucine, valine, phenylalanine), are known to have a very low threshold. Amino acids conversion in volatile compounds is of considerable importance for inducing characteristic flavor in salami or fermented sausage (Rouhi et al., 2013).
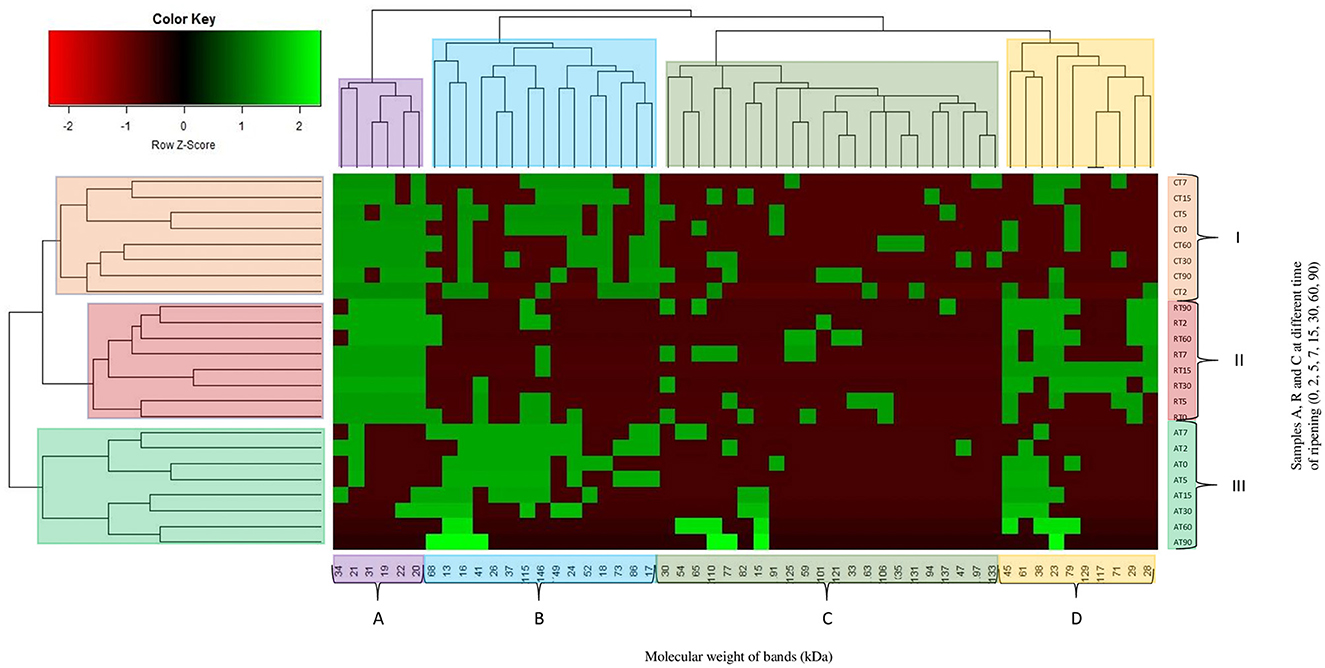
Figure 8. Hierarchical clustering analysis and heatmap visualization of SDS fingerprint (bands and their molecular weight [kDa]) and samples (non-inoculated sausages, C; sausages inoculated with Lcb. casei N87 cultivated under anaerobiosis A; sausages inoculated with Lcb. casei N87 cultivated under respiratory conditions R) during ripening (T0, T2, T5, T7, T15, T30, T60, and T90). Colors are used to identify the different clusters.
4. Conclusions
To date, concerning the sausages production, no valuable alternatives to nitrate have been found. However, the application of different factors, known as “hurdles technology”, may help to substitute the overall effects of nitrate during the manufacturing process. In this study, the starter culture Lcb. casei N87, cultivate under different conditions, was used for sausages production with the aim to assess the possibility to use it for color stabilization and the reduction of lipid oxidation, and therefore, reduce the use of nitrite. It has been seen that the physiological and technological characteristics of lactic acid bacteria can be related to the type of metabolism and growth conditions. In vitro and in vivo experiments have highlighted that the activation of respiratory metabolism in Lcb. casei N87 can lead to changes in the active enzymatic profile of the strains that can impact the organoleptic and nutritional properties of food products. In the specific context the capability of the enzymatic profile of Lcb. casei N87 to be able to protect the product from oxidation when activated under respiratory conditions was evaluated. Lcb. casei N87 was able to effectively carried out sausages fermentation, regardless the type of adaptation conditions of the culture. The strain was able to survive and growth in this type of food matrix, conferring a characteristic volatile profile, which was quite different from the control. Regarding physico-chemical parameters, Aw did not show differences among samples, whereas the pH was lower in A (5.2) samples and the weight loss in R samples (2.5% less than the others). The color parameters of the samples inoculated with the starter cultures did not show noticeable differences with respect to the control added with nitrate, confirming a protection activity performed by Lcb. casei N87. In addition, the fermented sausages produced using the starter culture adapted under respiratory conditions had a similar oxidative state of those produced using the maximum permitted level of nitrate. Conversely, sample A resulted to be more oxidized, confirming the importance of the type of adaptation during the starter culture production steps. The proteolysis that occurred during ripening indicates a differentiation of the A samples from the others. Nonetheless, the volatile profiles of the inoculated fermented sausages were similar. This study demonstrated the possibility to eliminate the use of nitrate in fermented sausage production using Lcb. casei N87 starter culture adapted under respiratory conditions without affecting the color parameters and the amount of aldehydes related to oxidative processes, improving the nutritional properties (reduction of aldehydes and elimination of nitrates) of fermented sausages. However, the bioprotective activity of this starter vs. Cl. botulinum was not evaluated. For this reason, a complete elimination of nitrate from fermented meat must be associated with the use of alternatives to fulfill the safety of the product (such as a NaCl concentration ≥ 2.7% in the recipe or others). As long as some alternative in the protection from Cl. botulinum is not used, a reasonably feasible way is the reduction of the nitrate/nitrite concentration used in conjunction with Lcb. casei N87 starter culture inoculation. Lastly, the use of Lcb. casei N87 can increase the added value of the product and its beneficial properties, due to their probiotic characteristics.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author contributions
LC and MP performed the experiments and wrote a first draft of the manuscript and performed the data analysis. MP and LI wrote the final draft of the manuscript. LI and GC revised the manuscript, edited the language, and PhD student supervision. MP, LI, and GC interpreted the results. LI conceptualization, experimental design, and funding acquisition. All authors have read and approved the submitted version of the manuscript.
Funding
This work was funded by Ministero dell' Istruzione, dell' Università e della Ricerca, Rome, Italy, FIRB n. RBFR107VML.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fsufs.2023.1044357/full#supplementary-material
Supplementary Figure S1. Example of DGGE profile obtained by the DNA extracted from patinas of MRS petri dishes at different dilutions (-2, 10−2;−3, 10−3;−4, 10−4;−5, 10−5;−6, 10−6) of the different samples (non-inoculated sausages, C; sausages inoculated with Lcb. casei N87 cultivated under anaerobiosis A; sausages inoculated with Lcb. casei N87 cultivated under respiratory conditions R) at different time of ripening (T0, day 0; T2, day 2; T5, day 5). Arrows indicates Lcb. casei N87 amplicons in identification ladder (MIX)”.
Supplementary Table S1. Results of microbiological analysis carried out on the fermented sausages (non-inoculated, C; inoculated with Lcb. casei N87 cultivated under respiratory conditions, A; inoculated with Lcb. casei N87 cultivated under anaerobiosis, R) during ripening (Log CFU/g).
References
Altschul, S. F., Madden, T. L., Shaffer, A. A., Zhang, J., Zhang, Z., Miller, W., et al. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucl. Acids Res. 25, 3389–3402. doi: 10.1093/nar/25.17.3389
Ampe, F., Ben Omar, N., and Guyot, J. P. (1999). Culture-independent quantification of physiologically active microbial groups in fermented foods using rRNA-targeted oligonucleotide probes: application to pozol, a Mexican lactic acid fermented maize dough. Appl. Environ. Microbiol. 87, 131–140. doi: 10.1046/j.1365-2672.1999.00803.x
Andrighetto, C., Zampese, L., and Lombardi, A. (2001). RAPD-PCR characterization of lactobacilli isolated from artisanal meat plants and traditional fermented sausages of Veneto region (Italy). Lett. Appl. Microbiol. 33, 26–30. doi: 10.1046/j.1472-765X.2001.00939.x
Aquilanti, L., Santarelli, S., Silvestri, G., Osimani, A., Petruzzelli, A., and Clementi, F. (2007). The microbial ecology of a typical Italian salami during its natural fermentation. Int. J. Food Microbiol. 120, 136–145. doi: 10.1016/j.ijfoodmicro.2007.06.010
Aquilanti, L., Garofalo, C., Osimani, A., and Clementi, F. (2016). Ecology of lactic acid bacteria and coagulase negative cocci in fermented dry sausages manufactured in Italy and other Mediterranean countries: an overview. Int. Food Res. J. 23, 429–445.
Aro, J. M., Nyam-Osor, P., Tsuji, K., Shimada, K., ichiro, Fukushima, M., and Sekikawa, M. (2010). The effect of starter cultures on proteolytic changes and amino acid content in fermented sausages. Food Chem. 119, 279–285. doi: 10.1016/j.foodchem.2009.06.025
Bagdatli, A., and Kundakci, A. (2016). Optimization of compositional and structural properties in probiotic sausage production. J. Food Sci. Technol. 53, 1679–1689. doi: 10.1007/s13197-015-2098-3
Benito, M. J., Rodriguez, M., Martin, A., Aranda, E., and Córdoba, J. J. (2004). Effect of the fungal protease EPg222 on the sensory characteristics of dry fermented sausage “salchichón” ripened with commercial starter cultures. Meat Sci. 67, 497–505. doi: 10.1016/j.meatsci.2003.11.023
Berardi, G., Albenzio, M., Marino, R., D'Amore, T., Di Taranto, A., Vita, V., et al. (2021). Different use of nitrite and nitrate in meats: a survey on typical and commercial Italian products as a contribution to risk assessment. LWT 150, 112004. doi: 10.1016/j.lwt.2021.112004
Bianchi, F., Cantoni, C., Careri, M., Chiesa, L., Musci, M., and Pinna, A. (2007). Characterization of the aromatic profile for the authentication and differentiation of typical Italian dry-sausages. Talanta 72, 1552–1563. doi: 10.1016/j.talanta.2007.02.019
Candogan, K., Wardlaw, F. B., and Acton, J. C. (2009). Effect of starter culture on proteolytic changes during processing of fermented beef sausages. Food Chem. 116, 731–737. doi: 10.1016/j.foodchem.2009.03.065
Casaburi, A., Di Monaco, R., Cavella, S., Toldrá, F., Ercolini, D., and Villani, F. (2008). Proteolytic and lipolytic starter cultures and their effect on traditional fermented sausages ripening and sensory traits. Food Microbiol. 25, 335–347. doi: 10.1016/j.fm.2007.10.006
Cenci-Goga, B. T., Ranucci, D., Miraglia, D., and Cioffi, A. (2008). Use of starter cultures of dairy origin in the production of Salame nostrano, an Italian dry-cured sausage. Meat Sci. 78, 381–390. doi: 10.1016/j.meatsci.2007.07.001
Cocolin, L., Dolci, P., Rantsiou, K., Urso, R., Cantoni, C., and Comi, G. (2009). Lactic acid bacteria ecology of three traditional fermented sausages produced in the North of Italy as determined by molecular methods. Meat Sci. 82, 125–132. doi: 10.1016/j.meatsci.2009.01.004
Cocolin, L., Urso, R., Rantsiou, K., Cantoni, C., and Comi, G. (2006). Dynamics and characterization of yeasts during natural fermentation of Italian sausages. FEMS Yeast Res. 6, 692–701. doi: 10.1111/j.1567-1364.2006.00050.x
Coffey, A., Ryan, M., Ross, R. P., Hill, C., Arendt, E., and Schwarz, G. (1998). Use of a broad-host-range bacteriocin-producing Lactococcus lactis transconjugant as an alternative starter for salami manufacture. Int. J. Food Microbiol. 43, 231–235. doi: 10.1016/S0168-1605(98)00115-9
Comi, G., Andyanto, D., Manzano, M., and Iacumin, L. (2016). Lactococcus lactis and Lactobacillus sakei as bio-protective culture to eliminate Leuconostoc mesenteroides spoilage and improve the shelf life and sensorial characteristics of commercial cooked bacon. Food Microbiol. 58, 16–22. doi: 10.1016/j.fm.2016.03.001
Comi, G., Muzzin, A., Corazzin, M., and Iacumin, L. (2020). Lactic acid bacteria: Variability due to different pork breeds, breeding systems and fermented sausage production technology. Foods 9, 338. doi: 10.3390/foods9030338
Comi, G., Urso, R., Iacumin, L., Rantsiou, K., Cattaneo, P., Cantoni, C., et al. (2005). Characterisation of naturally fermented sausages produced in the North East of Italy. Meat Sci. 69, 381–392. doi: 10.1016/j.meatsci.2004.08.007
Coppola, S., Mauriello, G., Aponte, M., Moschetti, G., and Villani, F. (2000). Microbial succession during ripening of Naples-type salami, a southern Italian fermented sausage. Meat Sci. 56, 321–329. doi: 10.1016/S0309-1740(00)00046-2
Còrdoba, J. J., Antequera, T., Ventanas, J., López-Bote, C., García, C., and Asensio, M. A. (1994). Hydrolysis and loss of extractability of proteins during ripening of iberian ham. Meat Sci. 37, 217–227. doi: 10.1016/0309-1740(94)90082-5
Demeyer, D., Raemaekers, M., Rizzo, A., Holck, A., Smedt, A., Ten Brink, B., et al. (2000). Control of bioflavour and safety in fermented sausages: first results of a European project. Food Res. Int. 33, 171–180. doi: 10.1016/S0963-9969(00)00031-4
Di Cagno, R., Chaves Lòpez, C., Tofalo, R., Gallo, G., De Angelis, M., Paparella, A., et al. (2008). Comparison of the compositional, microbiological, biochemical and volatile profile characteristics of three Italian PDO fermented sausages. Meat Sci. 79, 224–235. doi: 10.1016/j.meatsci.2007.09.006
Duwat, P., Sourice, S., Cesselin, B., Lamberet, G., Vido, K., Gaudu, P., et al. (2001). Respiration capacity of the fermenting bacterium Lactococcus lactis and its positive effects on growth and survival. J. Bacteriol. 183, 4509–4516. doi: 10.1128/JB.183.15.4509-4516.2001
Ercolini, D., Moschetti, G., Blaiotta, G., and Coppola, S. (2001). The potential of a polyphasic PCR-DGGE Approach in evaluating microbial diversity of natural whey cultures for water-buffalo mozzarella cheese production: bias of culture-dependent and culture-independent analyses. Syst. Appl. Microbiol. 24, 610–617. doi: 10.1078/0723-2020-00076
Ercoskun, H., and Özkal, S. G. (2011). Kinetics of traditional Turkish sausage quality aspects during fermentation. Food Contr. 22, 165–172. doi: 10.1016/j.foodcont.2010.06.015
Fadda, S., López, C., and Vignolo, G. (2010). Role of lactic acid bacteria during meat conditioning and fermentation: Peptides generated as sensorial and hygienic biomarkers. Meat Sci. 86, 66–79. doi: 10.1016/j.meatsci.2010.04.023
Fadda, S., Olivier, G., and Vignolo, G. (2002). Protein degradation by Lactobacillus plantarum and Lactobacillus casei in a sausage model system. Food Microbiol. Saf. 67, 1179–1183. doi: 10.1111/j.1365-2621.2002.tb09473.x
Fadda, S., Sanz, Y., Vignolo, G., Aristoy, M., Oliver, G., and Toldr,á, F. (1999). Characterization of muscle sarcoplasmic and myofibrillar protein hydrolysis caused by Lactobacillus plantarum. Appl. Environ. Microbiol. 65, 3540–3546. doi: 10.1128/AEM.65.8.3540-3546.1999
Fadda, S., Vignolo, G., Aristoy, M. C., Oliver, G., and Toldr,á, F. (2001). Effect of curing conditions and Lactobacillus casei CRL705 on the hydrolysis of meat proteins. J. Appl. Microbiol. 91, 478–487. doi: 10.1046/j.1365-2672.2001.01408.x
Fadda, S., Vignolo, G., Holgado, A. P. R., and Oliver, G. (1998). Proteolytic activity of Lactobacillus strains isolated from dry- fermented sausages on muscle sarcoplasmic proteins. Meat Sci. 49, 11–18. doi: 10.1016/S0309-1740(97)00097-1
Ferrocino, I., Bellio, A., Giordano, M., Macori, G., Romano, A., Rantsiou, K., et al. (2018). Shotgun metagenomics and volatilome profile of the microbiota of fermented sausages. Appl. Environ. Microbiol. 84, e02120–e02117. doi: 10.1128/AEM.02120-17
Ferysiuk, K., and Wójciak, K. M. (2020). Reduction of nitrite in meat products through the application of various plant-based ingredients. Antiox. 9, 711. doi: 10.3390/antiox9080711
Flores, M., Durá, M. A., Marco, A., and Toldrá, F. (2004). Effect of Debaryomyces spp. on aroma formation and sensory quality of dry-fermented sausages. Meat Sci. 68, 439–446. doi: 10.1016/j.meatsci.2003.04.001
Francesca, N., Sannino, C., Moschetti, G., and Settanni, L. (2013). Microbial characterisation of fermented meat products from the Sicilian swine breed “suino Nero Dei Nebrodi.” Ann. Microbiol. 63, 53–62. doi: 10.1007/s13213-012-0444-5
Gao, X., Kong, J., Zhu, H., Mao, B., Cui, S., and Zhao, J. (2022). Lactobacillus, Bifidobacterium and Lactococcus response to environmental stress: mechanisms and application of cross-protection to improve resistance against freeze-drying. J. Appl. Microbiol. 132, 802–821. doi: 10.1111/jam.15251
Gardini, F., Suzzi, G., Lombardi, A., Galgano, F., Crudele, M. A., Andrighetto, C., et al. (2001). A survey of yeasts in traditional sausages of southern Italy. FEMS Yeast Res. 1, 161–167. doi: 10.1111/j.1567-1364.2001.tb00027.x
Gardini, F., Tabanelli, G., Lanciotti, R., Montanari, C., Luppi, M., Coloretti, F., et al. (2013). Biogenic amine content and aromatic profile of Salama da sugo, a typical cooked fermented sausage produced in Emilia Romagna Region (Italy). Food Contr. 32, 638–643. doi: 10.1016/j.foodcont.2013.01.039
Hierro, E., de la Hoz, L., and Ordóñez, J. A. (1999). Contribution of the microbial and meat endogenous enzymes to the free amino acid and amine contents of dry fermented sausages. J. Agricult. Food Chem. 47, 1156–1161. doi: 10.1021/jf980834p
Honikel, K. O. (2008). The use and control of nitrate and nitrite for the processing of meat products. Meat Sci. 78, 68–76. doi: 10.1016/j.meatsci.2007.05.030
Huang, C. H., Chang, M. T., Huang, M. C., and Lee, F. L. (2011). Application of the SNaPshot minisequencing assay to species identification in the Lactobacillus casei group. Molecul. Cell. Prob. 25, 153–157. doi: 10.1016/j.mcp.2011.03.002
Hughes, M. C., Kerry, J. P., Arendt, E. K., Kenneally, P. M., McSweeney, P. L. H., and O'Neill, E. E. (2002). Characterization of proteolysis during the ripening of semi-dry fermented sausages. Meat Sci. 62, 205–216. doi: 10.1016/S0309-1740(01)00248-0
Iacumin, L., Cappellari, G., Pellegrini, M., Basso, M., and Comi, G. (2021). Analysis of the bioprotective potential of different lactic acid bacteria against Listeria monocytogenes in cold-smoked sea bass, a new product packaged under vacuum and stored at 6±2° C. Front. Microbiol. 12, 796655. doi: 10.3389/fmicb.2021.796655
Iacumin, L., Cattaneo, P., Zuccolo, C., Galanetto, S., Acquafredda, A., and Comi, G. (2019). Natural levels of nitrites and nitrates in San Daniele dry cured ham PDO, and in meat, salt and sugna used for its production. Food Contr. 100, 257–261. doi: 10.1016/j.foodcont.2019.01.016
Iacumin, L., Comi, G., Cantoni, C., and Cocolin, L. (2006). Ecology and dynamics of coagulase-negative cocci isolated from naturally fermented Italian sausages. Syst. Appl. Microbiol. 29, 480–486. doi: 10.1016/j.syapm.2005.11.006
Iacumin, L., Jayasinghe, A. S., Pellegrini, M., and Comi, G. (2022). Evaluation of different techniques, including modified atmosphere, under vacuum packaging, washing, and Latilactobacillus sakei as a Bioprotective Agent, to increase the shelf-life of fresh gutted sea bass (Dicentrarchus labrax) and sea bream (Sparus aurata) stored at 6±2° C. Biology 11, 217. doi: 10.3390/biology11020217
Ianniello, R. G., Ricciardi, A., Parente, E., Tramutola, A., Reale, A., and Zotta, T. (2015). Aeration and supplementation with heme and menaquinone affect survival to stresses and antioxidant capability of Lactobacillus casei strains. LWT 60, 817–824. doi: 10.1016/j.lwt.2014.10.020
Ianniello, R. G., Zotta, T., Matera, A., Genovese, F., Parente, E., and Ricciardi, A. (2016). Investigation of factors affecting aerobic and respiratory growth in the oxygen-tolerant strain Lactobacillus casei N87. PloS ONE 11, e0164065. doi: 10.1371/journal.pone.0164065
Lazar, I., and Lazar, I. (2010). Gel Analyzer 2010a: Freeware 1D gel electrophoresis image analysis software. Version 19.1. Available online at: www.gelanalyzer.com (accessed June 25, 2020).
Lechardeur, D., Cesselin, B., Fernandez, A., Lamberet, G., Garrigues, C., Pedersen, M., et al. (2011). Using heme as an energy boost for lactic acid bacteria. Curr. Op. Biotechnol. 22, 143–149. doi: 10.1016/j.copbio.2010.12.001
Lücke, F. K. (1997). Microbiology of Fermented Foods. Microbiology of Fermented Foods. Boston, MA: Springer.
Lücke, F. K. (2000). Utilization of microbes to process and preserve meat. Meat Sci. 56, 105–115. doi: 10.1016/S0309-1740(00)00029-2
Majou, D., and Christieans, S. (2018). Mechanisms of the bactericidal effects of nitrate and nitrite in cured meats. Meat Sci. 145, 273–284. doi: 10.1016/j.meatsci.2018.06.013
Marco, A., and Navarro, L. (2006). The influence of nitrite and nitrate on microbial, chemical and sensory parameters of slow dry fermented sausage. Meat Sci. 73, 660–673. doi: 10.1016/j.meatsci.2006.03.011
Martínez, L., Bastida, P., Castillo, J., Ros, G., and Nieto, G. (2019). Green alternatives to synthetic antioxidants, antimicrobials, nitrates, and nitrites in clean label Spanish chorizo. Antiox. 8, 184. doi: 10.3390/antiox8060184
Meynier, A., Novelli, E., Chizzolini, R., Zanardi, E., and Gandemer, G. (1999). Volatile compounds of commercial Milano salami. Meat Sci. 51, 175–183. doi: 10.1016/S0309-1740(98)00122-3
Montanari, C., Bargossi, E., Gardini, A., Lanciotti, R., Magnani, R., Gardini, F., et al. (2016). Correlation between volatile profiles of Italian fermented sausages and their size and starter culture. Food Chem. 192, 736–744. doi: 10.1016/j.foodchem.2015.07.062
Moretti, V. M., Madonia, G., Diaferia, C., Mentasti, T., Paleari, M. A., Panseri, S., et al. (2004). Chemical and microbiological parameters and sensory attributes of a typical Sicilian salami ripened in different conditions. Meat Sci. 6, 845–854. doi: 10.1016/j.meatsci.2003.08.006
Morot-Bizot, S. C., Leroy, S., and Talon, R. (2006). Staphylococcal community of a small unit manufacturing traditional dry fermented sausages. Int. J. Food Microbiol. 108, 210–217. doi: 10.1016/j.ijfoodmicro.2005.12.006
Muyzer, G., Waal, E. C. D. E., and Uitierlinden, A. G. (1993). Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59, 695–700. doi: 10.1128/aem.59.3.695-700.1993
Olivares, A., Navarro, J. L., Salvador, A., and Flores, M. (2010). Sensory acceptability of slow fermented sausages based on fat content and ripening time. Meat Sci. 86, 251–257. doi: 10.1016/j.meatsci.2010.04.005
Pedersen, M. B., Gaudu, P., Lechardeur, D., Petit, M. A., and Gruss, A. (2012). Aerobic respiration metabolism in lactic acid bacteria and uses in biotechnology. Annual Rev. Food Sci. Technol. 3, 37–58. doi: 10.1146/annurev-food-022811-101255
Pisacane, V., Callegari, M. L., Puglisi, E., Dallolio, G., and Rebecchi, A. (2015). Microbial analyses of traditional Italian salami reveal microorganisms transfer from the natural casing to the meat matrix. Int. J. Food Microbiol. 207, 57–65. doi: 10.1016/j.ijfoodmicro.2015.04.029
Polka, J., Rebecchi, A., Pisacane, V., Morelli, L., and Puglisi, E. (2015). Bacterial diversity in typical Italian salami at different ripening stages as revealed by high-throughput sequencing of 16S rRNA amplicons. Food Microbiol. 46, 342–356. doi: 10.1016/j.fm.2014.08.023
Procida, G., Conte, L. S., Fiorasi, S., Comi, G., and Favretto, L. G. (1999). Study on volatile components in salami by reverse carrier gas headspace gas chromatography-mass spectrometry. J. Chromato. A 830, 175–182. doi: 10.1016/S0021-9673(98)00792-4
Rantsiou, K., Urso, R., Iacumin, L., Cantoni, C., Cattaneo, P., Comi, G., et al. (2005). Culture-dependent and-independent methods to investigate the microbial ecology of Italian fermented sausages. Appl. Environ. Microbiol. 71, 1977–1986. doi: 10.1128/AEM.71.4.1977-1986.2005
Ranucci, D., Branciari, R., Acuti, G., Della Casa, G., Trabalza-Marinucci, M., and Miraglia, D. (2013). Quality traits of Ciauscolo salami from meat of pigs fed rosemary extract enriched diet. Int. J. Food Saf. 2, 16. doi: 10.4081/ijfs.2013.e16
Reale, A., Di Renzo, T., Zotta, T., Preziuso, M., Boscaino, F., Ianniello, R., et al. (2016a). Effect of respirative cultures of Lactobacillus casei on model sourdough fermentation. LWT 73, 622–629. doi: 10.1016/j.lwt.2016.06.065
Reale, A., Ianniello, R. G., Ciocia, F., Di Renzo, T., Boscaino, F., Ricciardi, A., et al. (2016b). Effect of respirative and catalase-positive Lactobacillus casei adjuncts on the production and quality of Cheddar-type cheese. Int. Dairy J. 63, 78–87. doi: 10.1016/j.idairyj.2016.08.005
Ricciardi, A., Zotta, T., Ianniello, R. G., Boscaino, F., Matera, A., and Parente, E. (2019). Effect of respiratory growth on the metabolite production and stress robustness of Lactobacillus casei N87 cultivated in cheese whey permeate medium. Front. Microbiol. 10, 851. doi: 10.3389/fmicb.2019.00851
Rouhi, M., Sohrabvandi, S., and Mortazavian, M. (2013). Probiotic fermented sausage: viability of probiotic microorganisms and sensory characteristics. Crit. Rev. Food Sci. Nutri. 53, 331–348. doi: 10.1080/10408398.2010.531407
Rubio, R., Jofr,é, A., Aymerich, T., Guàrdia, M. D., and Garriga, M. (2014). Nutritionally enhanced fermented sausages as a vehicle for potential probiotic lactobacilli delivery. Meat Sci. 96, 937–942. doi: 10.1016/j.meatsci.2013.09.008
Salueña, B. H., Gamasa, C. S., Rubial, J. M. D., and Odriozola, C. A. (2019). CIELAB color paths during meat shelf life. Meat Sci. 157, 107889. doi: 10.1016/j.meatsci.2019.107889
Sanz, Y., Fadda, S., Vignolo, G., Aristoy, M. C., Oliver, G., and Toldr,á, F. (1999a). Hydrolysis of pork muscle sarcoplasmic proteins by Lactobacillus curvatus and Lactobacillus sake. Int. J. Food Microbiol. 53, 115–125. doi: 10.1016/S0168-1605(99)00134-8
Sanz, Y., Fadda, S., Vignolo, G., Aristoy, M. C., Oliver, G., and Toldr,á, F. (1999b). Hydrolytic action of Lactobacillus casei CRL 705 on pork muscle sarcoplasmic and myofibrillar proteins. J. Agricult. Food Chem. 47, 3441–3448. doi: 10.1021/jf981255n
Savijoki, K., Ingmer, H., and Varmanen, P. (2006). Proteolytic systems of lactic acid bacteria. Appl. Microbiol. Biotechnol. 71, 394–406. doi: 10.1007/s00253-006-0427-1
Serrazanetti, D. I., Ndagijimana, M., Sado-Kamdem, S. L., Corsetti, A., Vogel, R. F., Ehrmann, M., et al. (2011). Acid stress-mediated metabolic shift in Lactobacillus sanfranciscensis LSCE1. Appl. Environ. Microbiol. 77, 2656–2666. doi: 10.1128/AEM.01826-10
Sidira, M., Galanis, A., Nikolaou, A., Kanellaki, M., and Kourkoutas, Y. (2014). Evaluation of Lactobacillus casei ATCC 393 protective effect against spoilage of probiotic dry-fermented sausages. Food Contr. 42, 315–320. doi: 10.1016/j.foodcont.2014.02.024
Sidira, M., Kandylis, P., Kanellaki, M., and Kourkoutas, Y. (2015). Effect of immobilized Lactobacillus casei on volatile compounds of heat treated probiotic dry-fermented sausages. Food Chem. 178, 201–207. doi: 10.1016/j.foodchem.2015.01.068
Silvestri, G., Santarelli, S., Aquilanti, L., Beccaceci, A., Osimani, A., Tonucci, F., et al. (2007). Investigation of the microbial ecology of Ciauscolo, a traditional Italian salami, by culture-dependent techniques and PCR-DGGE. Meat Sci. 77, 413–423. doi: 10.1016/j.meatsci.2007.04.015
Sindelar, J. J., and Milkowski, A. L. (2012). Human safety controversies surrounding nitrate and nitrite in the diet. Nitric Oxide 26, 259–266. doi: 10.1016/j.niox.2012.03.011
Spaziani, M., Torre, M., and Del Stecchini, M. L. (2009). Changes of physicochemical, microbiological, and textural properties during ripening of Italian low-acid sausages. Proteolysis, sensory and volatile profiles. Meat Sci. 81, 77–85. doi: 10.1016/j.meatsci.2008.06.017
Tabanelli, G., Coloretti, F., Chiavari, C., Grazia, L., Lanciotti, R., and Gardini, F. (2012). Effects of starter cultures and fermentation climate on the properties of two types of typical Italian dry fermented sausages produced under industrial conditions. Food Contr. 26, 416–426. doi: 10.1016/j.foodcont.2012.01.049
Tabanelli, G., Montanari, C., Grazia, L., Lanciotti, R., and Gardini, F. (2013). Effects of aw at packaging time and atmosphere composition on aroma profile, biogenic amine content and microbiological features of dry fermented sausages. Meat Sci. 94, 177–186. doi: 10.1016/j.meatsci.2013.01.018
Trzaskowska, M., Kołozyn-Krajewska, D., Wójciak, K., and Dolatowski, Z. (2014). Microbiological quality of raw-fermented sausages with Lactobacillus casei LOCK 0900 probiotic strain. Food Contr. 35, 184–191. doi: 10.1016/j.foodcont.2013.07.002
Ursachi, C. Ş., Perţa-Crişan, S., and Munteanu, F. D. (2020). Strategies to improve meat products' quality. Foods 9, 1883. doi: 10.3390/foods9121883
Urso, R., Comi, G., and Cocolin, L. (2006). Ecology of lactic acid bacteria in Italian fermented sausages: isolation, identification and molecular characterization. Syst. Appl. Microbiol. 29, 671–680. doi: 10.1016/j.syapm.2006.01.012
Wójciak, K. M., Dolatowski, Z. J., Kolozyn-Krajewska, D., and Trzaskowska, M. (2012). The Effect of the Lactobacillus casei Lock 0900 probiotic strain on the quality of dry-fermented sausage during chilling storage. J. Food Qual. 35, 353–365. doi: 10.1111/j.1745-4557.2012.00458.x
Wójciak, K. M., and Dolatowski, Z. J. (2012). Oxidative stability of fermented meat products. Acta Sci. Pol. Technol. Al. 11, 99–109.
Zotta, T., Parente, E., and Ricciardi, A. (2017). Aerobic metabolism in the genus Lactobacillus: impact on stress response and potential applications in the food industry. J. Appl. Microbiol. 122, 857–869. doi: 10.1111/jam.13399
Keywords: fermented sausages, bioprotection, Lacticaseibacillus casei, lactic acid bacteria, respiration, aerobic vs. anaerobic growth, antioxidants, probiotic
Citation: Camprini L, Pellegrini M, Comi G and Iacumin L (2023) Effects of anaerobic and respiratory adaptation of Lacticaseibacillus casei N87 on fermented sausages production. Front. Sustain. Food Syst. 7:1044357. doi: 10.3389/fsufs.2023.1044357
Received: 14 September 2022; Accepted: 12 January 2023;
Published: 09 February 2023.
Edited by:
Veronica Ortiz Alvarenga, Federal University of Minas Gerais, BrazilReviewed by:
Larissa Margalho, State University of Campinas, BrazilFred Breidt, Agricultural Research Service (USDA), United States
Copyright © 2023 Camprini, Pellegrini, Comi and Iacumin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lucilla Iacumin,  bHVjaWxsYS5pYWN1bWluQHVuaXVkLml0
bHVjaWxsYS5pYWN1bWluQHVuaXVkLml0
†These authors have contributed equally to this work and share first authorship
 Lucia Camprini
Lucia Camprini Michela Pellegrini
Michela Pellegrini Giuseppe Comi
Giuseppe Comi Lucilla Iacumin
Lucilla Iacumin