
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Sustain. Food Syst., 08 February 2023
Sec. Land, Livelihoods and Food Security
Volume 7 - 2023 | https://doi.org/10.3389/fsufs.2023.1027842
This article is part of the Research TopicEnhancing Food Production System Resilience for Food Security Facing a Changing EnvironmentView all 15 articles
 Sarah Nanyiti1*
Sarah Nanyiti1* Richard Kabaalu1
Richard Kabaalu1 Titus Alicai1
Titus Alicai1 Phillip Abidrabo1
Phillip Abidrabo1 Susan E. Seal2
Susan E. Seal2 Sophie Bouvaine2
Sophie Bouvaine2 Andy M. Bailey3
Andy M. Bailey3 Gary D. Foster3
Gary D. Foster3Cassava is an important staple food in Africa and a major source of carbohydrates for 800 million people globally. However, cassava suffers severe yield losses caused by many factors including pests and diseases. A devastating disease of cassava is cassava brown streak disease (CBSD) caused by the cassava brown streak ipomoviruses (CBSIs) (family Potyviridae), Cassava brown streak virus (CBSV), and Ugandan cassava brown streak virus (UCBSV). Spread of CBSD is mainly through planting infected stem cuttings used for propagation. Transmission of CBSIs by the insect vector (Bemisia tabaci) has been reported. However, experimental transmission efficiencies of CBSIs are usually low. Recent research has showed the occurrence of a DAG motif associated with aphid transmission in other potyviruses, within the coat protein gene of CBSV. Consequently this study aimed to explore the possibility that besides whiteflies, aphids may transmit CBSIs. Cassava plants were assessed during a survey for occurrence of CBSD and aphids as potential alternative CBSIs vectors. We collected aphids from CBSD-symptomatic and symptomless cassava plants within farmers' fields in Uganda during April–July 2020. The aphids were analyzed for the presence of CBSIs by reverse transcriptase-polymerase chain reaction (RT-PCR) and to determine aphid species using mitochondrial cytochrome oxidase (mtCOI) barcoding. Unusual aphid infestation of cassava plants was observed at 35 locations in nine districts across Uganda and on 11 other plant species within or adjacent to cassava fields. This is the first report of aphids infesting cassava in Uganda. Molecular analysis of the aphid confirmed presence of three different aphid species in the surveyed cassava fields, namely, Aphis solanella, Aphis fabae mordvilkoi, and Rhopalosiphum sp. mtCOI nucleotide sequences for the aphids in which CBSIs were detected are deposited with Genbank under accession numbers OP223337-40. Both UCBSV and CBSV were detected by RT-PCR in aphids collected from cassava fields with CBSD-affected plants. The CBSIs were detected in 14 aphid samples collected from 19 CBSD-symptomatic cassava plants. These results suggest the ability of aphids to acquire CBSIs, but transmission experiments are required on their vector potential.
Cassava (Maninot esculenta Crantz, family Euphorbiaceae) is a major staple food and income generation crop for farmers across sub-Saharan Africa. In addition, cassava is a hardy crop that is resilient to drought and performs well on marginal soils where other crops would fail. Cassava has also been prioritized in many African countries as a commodity that can drive industrialization through its use as a raw material in the production of livestock feed, starch, alcohol, pharmaceuticals, biofuels, and bio-polymers. However, the crop is affected by a number of biotic stresses including devastation by pests and diseases. The most significant yield losses are caused by two major viral diseases, namely, cassava mosaic disease (CMD) and cassava brown streak disease (CBSD), reported to cause an estimated annual loss of about US$1 billion (Hillocks et al., 2001; Mohammed et al., 2012). A pandemic of CBSD is currently spreading in eastern, central, and southern Africa, threatening cassava production and the livelihoods of communities that primarily depend on the crop (Alicai et al., 2007; Legg et al., 2011). CBSD-induced necrosis reduces the quantity and quality of storage roots, causing yield losses of up to 70% (Hillocks et al., 2001). A study in Kenya, Tanzania, Malawi, and Uganda estimated losses due to CBSD to be US$750 million annually (Maruthi et al., 2005).
Cassava brown streak disease is caused by two positive-sense single-stranded RNA (+ssRNA) viral species, Cassava brown streak virus (CBSV) and Ugandan cassava brown streak virus (UCBSV), collectively referred to as CBSIs belonging to the genus Ipomovirus in the family Potyviridae (Mbanzibwa et al., 2009a; Tomlinson et al., 2018). The genome of the CBSIs is about 8.9–10.8 kb, which is translated into a polypeptide of 10 proteins (5′-3′), namely, P1, P3, 6KD1, cylindrical inclusion (CI), 6KD2, VPg, nuclear inclusion protein a (NIa), nuclear inclusion protein b (NIb), HAM1h, and coat protein (CP) (Monger et al., 2001; Mbanzibwa et al., 2009b; Winter et al., 2010; Tomlinson et al., 2019). All the proteins serve different roles in different stages in the viral infection cycle, including viral replication, translation, cell-to-cell movement within the host, and transmission between vectors and the host (Llave et al., 2002).
Often, CBSD is unrecognized in the field because of its inconspicuous foliar symptoms compared to those for CMD and cassava green mite (CGM). Symptoms of CBSD on the leaf can take different forms depending on the cassava variety, the age of the plant, and the conditions under which it is growing (Hillocks and Jennings, 2003). The disease gets its name from “brown streak”, a term which refers to the brown lesions often observed on the stem of CBSD-affected plants. Symptoms can appear on either leaves, stems, fruits, storage roots or are exhibited on more than one organ. The necrosis on the storage roots is a symptom of great economic importance (Hillocks et al., 1996).
Currently, CBSIs vector specificity is unclear. This is mainly because most of the vector transmission studies have shown very low transmission efficiencies. For instance, transmission with the whitefly vector (B. tabaci) was only 22% (Maruthi et al., 2005). The CBSIs have been shown to be transmitted by cassava whitefly (B. tabaci) in a semi-persistent manner. However, the observed low transmission efficiencies of CBSIs by whiteflies do not match the rapid CBSD spread observed in the fields, especially under epidemic conditions (Maruthi et al., 2005). This observation suggests possibly another vector involved in CBSI's transmission other than whiteflies, and this needs further investigation. Determining if there are other vectors for CBSIs is important in integrated pest management (IPM) programs required to control the spread of the disease (Dombrovsky et al., 2014).
Interestingly, unusual occurrence of a DAG motif within the coat protein gene of CBSVand implications for its vector transmission was recently reported (Ateka et al., 2017). The highly conserved DAG motif is associated with aphid transmission of potyviruses. Presence of the motif in the CBSV genome denotes aphid transmission possibility. Initially, aphid transmission was not considered possible for CBSIs, as they do not encode HC-Pro proteins, which in other Potyviridae are involved in aphid transmission. The HC-Pro acts as a “bridge” between the aphid stylet and the virus particle. The HC-Pro has an affinity for both the viral coat protein (CP) DAG motif (through its PTK motif) and the receptors in the aphid (through its KITC motif) (Whitfield et al., 2015). This interaction thus acts as a bridge to allow the retention of the virus particles in the aphid stylet. Hence, the presence of the highly conserved DAG and PTK motif in the CBSV CP and the KITC in the CBSV P1 sequences have been highlighted, which raises the possibility for CBSV to be transmitted by aphids in addition to whiteflies (López-Moya et al., 1999).
Arising from these previous findings, it was important to undertake studies that characterize and specify the vector(s) responsible for transmission of CBSIs.
Aphids were initially collected from fields at and near the National Crops Resources Research Institute (NaCRRI), Namulonge, during the months of May to December 2019. For this location, there were few aphids on cassava plants compared to other plant species growing within or near cassava fields.
Ten different districts in Uganda were surveyed during the months of April to July 2020, and aphids collected from cassava fields. During the survey, considerations were made to collect aphids from fields having CBSD and those without any CBSD symptoms. For the fields with CBSD, the incidence of the disease was recorded.
The collection of aphids considered districts that have been reported to have high incidences of cassava brown streak disease (CBSD) and were considered to be representative of CBSD disease situation in Uganda. Some districts, like Bukwo, had previously been reported to have high incidences of aphids in cassava fields and were thus included in this survey.
Thus, in this survey, 10 districts in Uganda were selected. Of these, eight districts were in the Eastern parts of Uganda, including Kamuli, Iganga, Budaka, Bukedea, Bulambuli, Kapchorwa, Kween, and Bukwo. Two districts, were in the Central parts of Uganda including Luwero and Wakiso districts.
Data was recorded in the field using Excel-field datasheets which captured information about the date and time. Details of location were recorded and this included the district, sub-county, village, and farmers' names. Geographical referencing (longitude, latitude, and altitude) of each field was obtained using the global positioning system (GPS) software. Information was recorded on cassava varieties grown, number of cassava fields nearby, field size, age of cassava plants and intercrops.
Surveys were conducted in targeted districts mentioned above, and the sampling domain was a district. In each district, fields were randomly sampled. Fields were sampled along major and accessible roads within the districts. Fields were selected at regular intervals of 7 km between sites or thereafter when a suitable cassava field is found. The age of cassava plants sampled was between 3–10 months old cassava fields, and this information was provided by the farmer. At each site (cassava field), 30 plants were assessed at regular intervals across the two diagonals, and data were collected from any cassava variety that occur along the diagonals. Information on the cassava varieties that occur in the sampled field was also recorded.
Parameters measured in the survey included CBSD severity and incidence and number of aphids on a plant. To determine whether they contain and can possibly transmit CBSIs, aphids isolates were collected and characterized.
CBSD severity was assessed on the stems and leaves of cassava plants. This was assessed for each of the 30 plants using a scale of 1–5, where 1 represents no symptoms and 5 the most severe symptoms that include stem streaking and shoot tip die-back (see scale below) (Ogwok et al., 2010; Ano et al., 2021).
Scale: Symptom description
1. No apparent symptoms of CBSD
2. Mild vein yellowing/chlorotic blotches on some leaves, but no stem lesions
3. Pronounced vein yellowing/chlorotic blotches on most leaves and mild stem lesions
4. Pronounced vein yellowing/chlorotic blotches on most leaves, moderate lesions or streaks on stems, or wilting but no die-back
5. Pronounced vein yellowing/chlorotic blotches on leaves, severe lesions or streaks on stems, defoliation, and die-back.
Adult aphids found on cassava plants were collected using an aspirator from the fields in which they occured. The aphid samples from such fields were separately put in a 1.5-ml sample tube containing 80% ethanol and stored at room temperature until nucleic acid extraction.
For each plant from which aphids were collected, a corresponding cassava leaf was picked, rolled carefully, and put into a 2.0-ml microfuge tube containing 1.0 ml of RNA-later or CTAB solution for preservation. The tubes were labeled appropriately and sealed. The samples were kept at room temperature until RNA extraction.
CTAB was used to extract nucleic acids from cassava leaves (Abarshi et al., 2010). 100 mg of leaf tissue was ground in 1,000 μl of extraction buffer 2% CTAB (cetyltrimethyl ammonium bromide) containing 100 mM Tris–HCl, 20 mM EDTA, 1.4 M NaCl, and 0.2% β-mercaptoethanol (v/v) in a sterile motor and pestle. 600 μl of sap was poured into a sterile tube and vortexed briefly and then incubated in a water bath at 65°C for 10 min. 600 μl of chloroform:isoamyl alcohol (24:1) was added and the emulsion was mixed by inverting the tube. The mixture was centrifuged at maximum speed for 10 min at room temperature. About 450 μl of aqueous layer was removed, and the nucleic acid precipitated with 450 μl ice-cold isopropanol (20°C). The mixture was incubated at −20°C for 30 min. After the incubation, the mixture was centrifuged for 10 min at 6500 rpm to pellet the nucleic acids. The pellet was washed with 70% ethanol before air drying. The nucleic acids pellets were dissolved in 50 μl sterile distilled water. The isolated total nucleic acids samples were then stored at −80°C until they are ready to be used. The concentrations of the extracted nucleic acid were measured using a Nanodrop, and 50 ng of genomic RNA was used for cDNA synthesis using the Revert Aid First Strand cDNA Synthesis Kit (Thermo Scientific™).
The chelex extraction method was used to extract nucleic acid from single aphids (Asghar et al., 2015). The aphids were initially soaked on a paper towel to remove any residual ethanol before using them in the extraction. The aphids were then crushed with the Chelex resin (100 μl resin +10 μl of Proteinase K) in a 1.5-ml Eppendorf tube using a micropestle. The mixture was incubated at 56°C for 20 min, and the Proteinase K heat inactivated by incubating the mixture at 94°C for 5 min. Centrifugation was done at 13,000 rpm for 5 min and 100 μl of the supernatant containing the extracted nucleic acid was taken and stored in an Eppendorf tube. The concentrations of the extracted nucleic acid were measured on a Nanodrop, and the nucleic acid was used for cDNA synthesis with the RevertAid First Strand cDNA Synthesis Kit (Thermo Scientific™).
First-strand cDNA was synthesized using the RevertAid First Strand cDNA Synthesis Kit (Thermo Scientific™) according to the manufacturer's instructions, and oligo (dT)18 was used as the initial primer to synthesize the first strand of cDNA. The synthesized cDNA was then used as a template in a PCR reaction. The reaction set up for cDNA synthesis was as follows. Template RNA (Total RNA) 0.1–5 μg, oligo (dT)18 Primer (10 μM) 1 μl, Nuclease Free Water 11 μl. The samples were then incubated at 65°C for 5 min, centrifuged, and then chilled on ice. To the reaction above, the following were added in order: 5× Reaction Buffer, 4 μl, RiboLock RNase inhibitor 1 μl (20 U/μl), 10 mM dNTP Mix 2 μl; RevertAid RT 2 μl (20 U/μl), Nuclease Free Water 20 μl total volume. The reaction was incubated at 42°C for 60 min and then terminated by incubation at 70°C for 5 min.
The synthesized cDNA was used in RT-PCR reactions which was performed in a 20 μl of final reaction volume that contained 7 μl of sterile distilled water, 10 μl of 2× PCR ready mix containing Taq DNA polymerase (Thermo Scientific™), and 1 μl each of the forward (10 μM) and reverse (10 μM) primers. The appropriate positive and negative controls were included for each of the PCR.
For the detection of CBSIs in the cassava leaves and aphids, a universal diagnostic primer pair CBSDDF2 (5′-GCTMGAAATGCYGGRTAYACAA-3′) and CBSDDR (5′-GGATATGGAGAAAGRKCTCC-3′) for detection of both CBSV and UCBSV in one reaction was used with an expected product size of 344 bp and 438–440 bp, respectively (Mbanzibwa et al., 2011).
For mtCOI barcoding of the aphids, primers HCO2198-puc (5′-TAAACTTCWGGRTGWCCAAARAATC-3′) and HCO-2198 (5′-TAAACTTCAGGGTGACCAAAAAATCA-3′) amplifying the 5′-terminal of the mitochondrial cytochrome C oxidase subunit 1 gene (COI) in insects (Folmer et al., 1994) were used.
PCR was carried out in a PTC-200 Engine thermocycler (Bio-Rad) with the program as an initial denaturation at 95°C for 2 min, followed by 36 cycles of denaturation at 95°C for 30 s, annealing for 30 s (temperature varying with primers) and extension at 72°C for 1 min per kb (depending on the length of the expected PCR product) and a final extension at 72°C for 10 min. PCR products were analyzed by gel electrophoresis in a 1X TAE buffer on a 1.0% agarose gel, stained with ethidium bromide, visualized under UV light, and documented the gel picture.
To determine the aphid species infesting cassava in Uganda, Sanger sequencing was performed on the PCR products from aphids that had been confirmed to contain U/CBSV. The PCR amplicons were sent to the Netherlands. Sequencing was carried out at Macrogen Inc. (Macrogen Europe Laboratory, Amsterdam, The Netherlands). The sequencing results were analyzed by BLAST analysis using the NCBI BLASTn tool.
In a preliminary assessment, aphids were found on cassava, and 11 other plant species near and in cassava fields around Namulonge in central Uganda. Individual aphids collected from the different plant species were analyzed by molecular characterization. It was observed that there were more aphids on weed plants adjacent to cassava fields than on cassava plants at Namulonge. The identities of these plants are indicated in Supplementary Table 1. The aphids present on weed plants are shown in Figure 1.
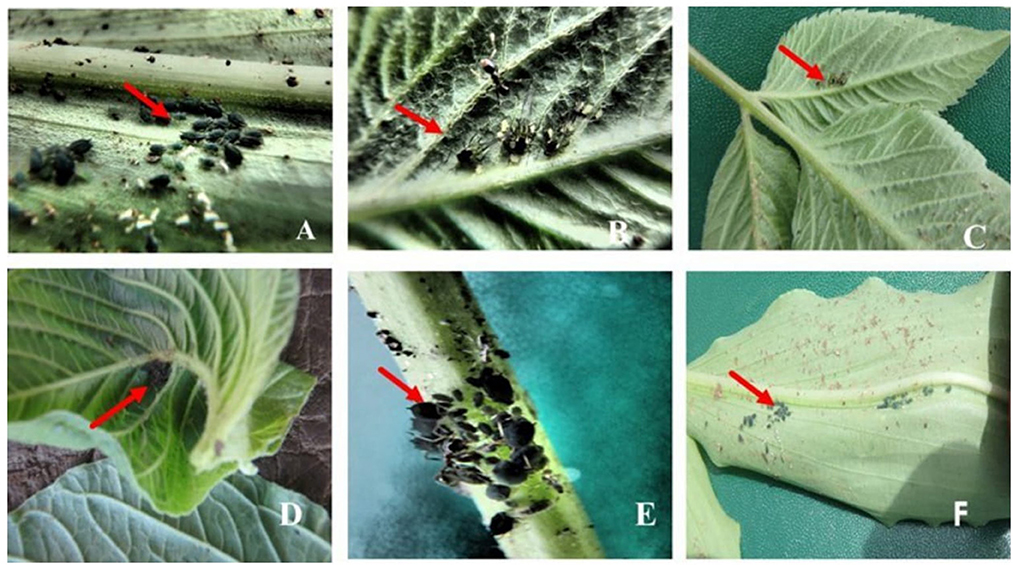
Figure 1. Aphids found on other plant species growing within or near cassava fields. (A) Commelina africana (Nanda—local name) with black wingless aphids. (B) Bidens Pilosa (blackjack) with black winged aphids. (C) Blackjack with black winged aphids. (D) Aspilia Africana (Makayi—local name) with black wingless aphids. (E) Blackjack stem with black wingless aphids. (F) Commelina africana with nymphs.
Of the 10 districts surveyed, up to seven cassava fields were assessed per district, and samples were collected (Supplementary Table 2; Figure 2). In Kween district, there were no cassava fields found as the main crops grown were bananas and maize. Aphids were found on cassava plants at 35 locations in nine districts of Uganda (Supplementary Table 2; Figures 2, 3). For the four districts of Kamuli, Iganga, Budaka, and Bukedea, aphid populations on cassava were low (Figure 2). Districts with the highest prevalence of aphids were Bukwo (aphids found in 80% of visited fields) and Bulambuli (all, 100% of visited cassava fields). Cassava fields surveyed in Bulambuli district had CBSD on most of the sampled cassava plants. Therefore aphids collected from the fields in the Bulambuli district were selected for molecular analysis for the detection of CBSIs. In contrast, no CBSD was observed within the cassava fields in Bukwo, though high aphid populations were recorded. In Luwero district, a field in which CBSD was observed had a few aphids collected from the CBSD symptomatic cassava plants. Most cassava varieties from which aphid samples were collected are known to be CBSD susceptible. In total, 196 aphid samples were collected from farmers' cassava fields, and the same number of cassava leaf samples were collected (Supplementary Table 2). This study provides the first clear record on occurrence of aphids on cassava in Uganda and requires further understanding. Unusual occurrence of heavy aphid infestation on cassava was previously observed in eastern Uganda in Bukwo and Sironko districts, but remained undocumented. Leaves of affected cassava plants appeared distorted and curled inward displaying a typical symptom of feeding damage by aphids. Although such symptoms were not observed during the present survey, aphid populations remained higher in that region compared to others.
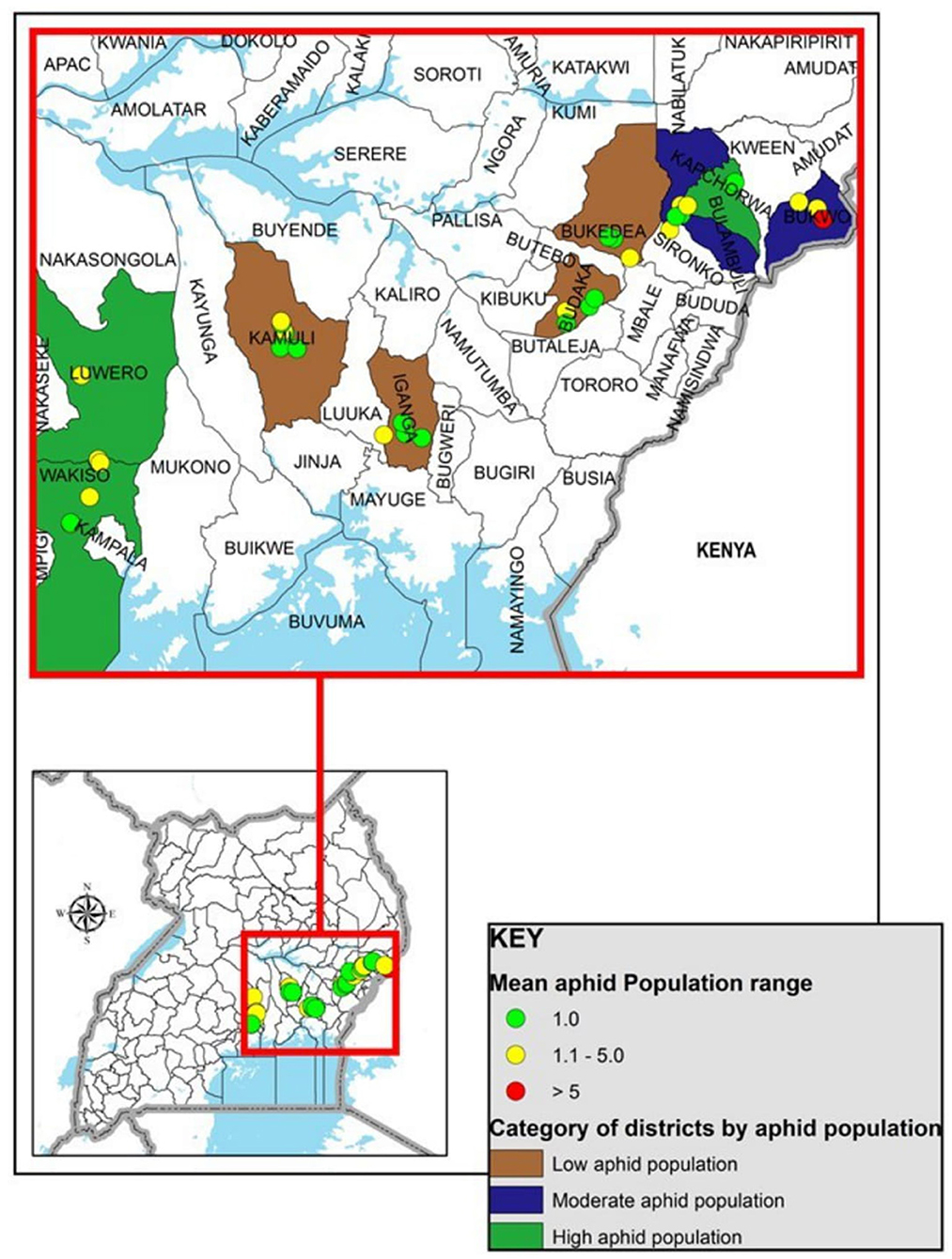
Figure 2. Map of Uganda showing cassava fields surveyed for aphids occurrence and sample collection. Locations where aphids were collected; Green, low aphid population in cassava fields (one aphid per plant); Yellow, moderate aphid population (two to five aphids per plant); Red, high aphid population (more than five aphids per plant).

Figure 3. Images of aphids found on cassava plants in surveyed districts. (A) Aphids on leaves of a CBSD asymptomatic (healthy) cassava plant in Bukwo district. (B) CBSD symptomatic cassava leaves in a field in the Bulambuli district; aphids were collected in this field. (C) Asymptomatic cassava leaves with aphids. (D) Aphids on the lower side of cassava with CBSD symptoms. (E) CBSD symptomatic plant in Budaka district; aphids were collected from this field. (F, G) Aphids on the lower side of cassava leaves. (H) Aphid nymphs on cassava leaf.
Thirty individual aphid samples from different host plant species collected within or near cassava fields at Namulonge and Bulambuli were analyzed by PCR for the mtCOI gene and a 700-bp fragment was amplified (Figure 4). The aphid samples were collected from different plant species, as indicated in Supplementary Table 1. The final results of the mtCOI barcoding allowed definitive identification of aphid species prevalent on cassava that can be used in follow-up studies.
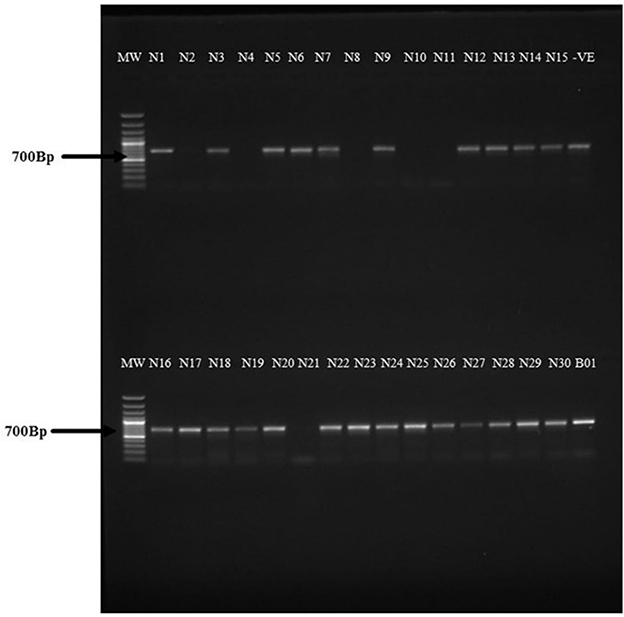
Figure 4. Agarose gel electrophoresis for the PCR products amplifying the mtCOI gene in aphids using the mtCOI primers HCO2198-puc (5′-TAAACTTCWGGRTGWCCAAARAATC-3′) and HCO-2198 (5′-TAAACTTCAGGGTGACCAAAAAATCA-3′). Lane 1: 1 kb MW ladder. Samples N1–N30 from different aphids collected in and around cassava fields at NaCRRI, Namulonge. Samples B01 was an aphid sample collected from a cassava plant in the Bulambuli district (Buginyanya). A 700-bp PCR product was amplified representing the mtCOI gene in the aphids.
Overall, 196 cassava leaf samples were collected corresponding to each plant from which aphids were collected from farmers' fields. Of these, leaf samples from representative plants showing clear CBSD symptoms were selected to confirm the presence of CBSIs in the cassava fields from which aphids were collected. Priority was given to four samples from fields in Luwero (1), Wakiso (1), and Bulambuli (2) districts because these had plants with CBSD symptoms and high aphid populations. RT-PCR products of 340 bp (CBSV) and 440 bp (UCBSV) were obtained (Figure 5). The sample from Luwero had mixed infection (both CBSV and UCBSV), while that from Wakiso tested positive for CBSV and samples from Bulambuli contained UCBSV.
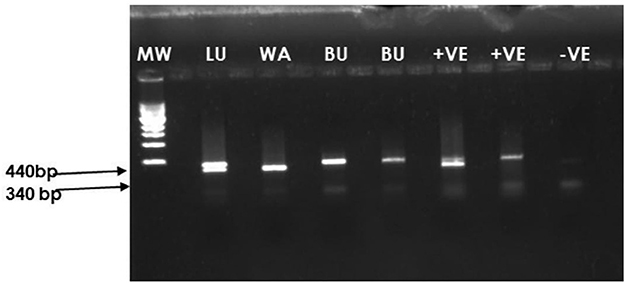
Figure 5. Agarose gel electrophoresis of the PCR products amplified using CBSIs diagnostic primers CBSDDF2 (5′-GCTMGAAATGCYGGRTAYACAA-3′) and CBSDDR (5′-GGATATGGAGAAAGRKCTCC-3′). Lane 1: 1 kb MW ladder. Lane 2: Cassava sample collected from Luwero (LU) district from a field with CBSD and aphids. Lane 3: Cassava sample collected from Wakiso (WA) district from a field with CBSD and aphids. Lanes 4 and 5: Cassava samples collected from the Bulambuli district (BU) from a field with CBSD and aphids. Lanes 6 and 7: Positive control. Lane 8: Negative control.
A sub-sample of 20 aphids was selected from two fields in the Bulambuli district for detection of CBSIs in the aphids. These samples were analyzed by RT-PCR following a protocol for simultaneous detection of CBSV and UCBSV. 14 out of 19 (73.7%) aphid samples were collected from CBSD symptomatic cassava plants tested positive for CBSIs (Figure 6; Supplementary Table 3). One aphid sample taken from a healthy cassava plant tested negative by RT-PCR. All the aphids (14) that tested positive had CBSV, and 8 of those also had UCBSV in addition [double bands for the 440 bp (UCBSV) and 340 bp (CBSV)]. It was also noted that the intensity of the bands was low in the aphids (Figure 6) as compared to the cassava plants (Figure 5), which may be indicative of low virus titers of CBSIs in the aphids. To further confirm species identities of the aphids in which the CBSIs were detected, the same aphid samples were characterized by mtCOI barcoding and respective PCR products sent for Sanger sequencing. The sequence results were analyzed by Blastn in the NCBI database. This confirmed three aphid species as those infecting the cassava namely, Aphis solanella, Aphis fabae mordvilkoi, and Rhopalosiphum sp. Overall Aphis solanella was the most prevalent species for the analyzed samples (Supplementary Table 3). Nucleotide sequences for the aphids were deposited in the GenBank database under accession numbers OP223337, OP223338, OP223339, and OP223340.
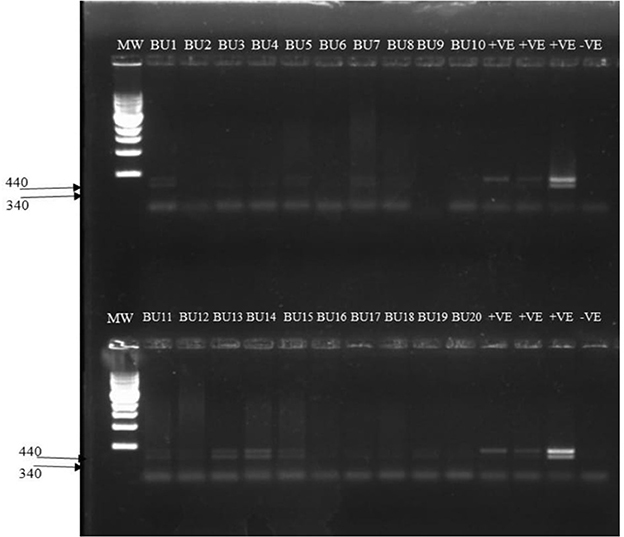
Figure 6. Amplification of CBSV and UCBSV in aphids using diagnostic primers CBSDDF2 (5′-GCTMGAAATGCYGGRTAYACAA-3′) and CBSDDR (5′-GGATATGGAGAAAGRKCTCC-3′). Lane 1: 1 kb MW ladder Lanes BU1-BU20: Aphid samples collected from CBSD infected fields in Bulambuli district. Double bands are indicative of mixed infection of CBSV (340 bp) and UCBSV (440 bp). Lanes labeled +ve are cassava plants from which the aphids were collected. Lane labeled –ve is the negative control.
Aphids are important insect pests of plants and vectors for plant-infecting viruses (Ng and Perry, 2004; Gadhave et al., 2020). To successfully transit a virus, there are a number of different viral-vector interactions that come into play with viruses encoding proteins that play a role in virus transmission. The viral HC-Pro and CP have been reported to be the main proteins involved in aphid transmission (López-Moya et al., 1999; Ala-Poikela et al., 2011; Ateka et al., 2017).
The major reported cassava virus diseases in Africa are cassava mosaic disease (CMD) caused by cassava mosaic begomoviruses (Family Geminiviridae and genus: Begomovirus) and CBSD with whiteflies (B. tabaci) as vector for both (Thresh et al., 1994; Hillocks and Thresh, 2000). However, low transmission efficiency of CBSIs by whiteflies has continued to generate interest in the possibility of other unexplored vectors (Maruthi et al., 2005).
In this study, it was observed that aphids were found in cassava fields infected with CBSIs and on other plant species near or around cassava fields in Uganda. This observation may indicate that, besides spread by planting infected plant materials and whiteflies, aphids may possibly be other unexplored vectors responsible for CBSD spread (Ateka et al., 2017). This observation confirms that aphids can acquire and retain the CBSIs; however, transmission studies are required to confirm if the aphids can transmit the CBSIs (Ng and Perry, 2004; Gadhave et al., 2020). This study follows the notion from an earlier report that a DAG motif present in the CP of CBSV is indicative of aphid transmission because potyviruses require a DAG motif to bind to the aphid stylet in a non-persistent transmission mode (Ateka et al., 2017). To our knowledge, this is the first record for detection of CBSIs in aphids found feeding on CBSD-symptomatic cassava plants.
Aphids were found on a wide host range of weed plants growing near cassava fields. This presents possibility of aphids moving from the weed plants to cassava and vice versa. RT-PCR was performed on aphids and the CBSD-symptomatic cassava leaves using the diagnostic primers reported in Mbanzibwa et al. (2011). Results confirmed that the CBSD-symptomatic cassava leaves and the aphids contained CBSIs. Such CBSIs-infected plants can be considered to be likely sources of the CBSIs that are acquired by the aphids. However, agarose gel electrophoresis of the PCR products for the CBSIs in the aphids showed faint bands compared to the distinct PCR bands observed in the CBSD-symptomatic cassava plant leaves. This may be indicative of low CBSIs titers present in the aphids as compared CBSD-symptomatic cassava plants.
Molecular characterization of the aphids targeting the mtCOI gene of insects using the primers reported by Folmer et al. (1994) gave a 700-bp PCR product and on sequencing, it was confirmed that three different aphid species were present in the surveyed fields in Uganda. The aphids identified included Aphis solanella, Aphis fabae mordvilkoi, and Rhopalosiphum sp with Aphis solanella being the most prevalent species in the samples that were analyzed by molecular characterization (Supplementary Table 3). Previous studies have reported seasonal effect on aphid diversity and this could explain the observations on the diversity of aphids reported in this study (Wamonje et al., 2017). On the contrary, the generalist green peach aphid (Myzus persicae) was not observed to be present in the surveyed cassava fields. The green peach aphid has been reported to be one of the major aphid species involved in the transmission of potyviruses (Gadhave et al., 2020). Most of the sampled aphids were black winged (alate) aphids. The presence of wings on the aphids facilitates their fast movement in the field and hence presents challenges in controlling these aphids using insecticides as they keep moving from one plant to the other. These findings indicate the potential of the aphids to acquire CBSIs, but virus retention, replication, circulation, and transmission remain unknown and require further investigation.
For efficient transmission, plant viruses usually encode proteins that facilitate the virus-vector transmission process. For instance, for potyviruses, the CP and the HC-Pro act as a bridge with the host to aid in viral transmission (Ateka et al., 2017). The detection of both CBSV and UCBSV is interesting because previous reports suggest likely aphid transmission of only CBSV owing to the presence of a DAG (Asp-Ala-Gly) motif in its genome. The DAG motif is implicated in aphid transmission of potyviruses, and this has been reported to be present in the genome of CBSV, whereas that in UCBSV is DEG (Asp-Glu-Gly) (Ateka et al., 2017). It would be rather unusual for very related viruses to be transmitted by such diverse insects, as ideally it is expected that UCBSV and CBSV being related viruses, should be transmitted by the same vector (Whitfield et al., 2015). Although the DAG has been implicated in the aphid transmissibility of viruses, there have been reports were potyviruses with motifs other than the DAG motif are also aphid transmissible (López-Moya et al., 1999).
Interestingly, the DAG motif is also present in the coat protein (CP) of Squash vein yellowing virus (SqVYV) a member of the family Potyviridae and genus Ipomovirus, to which the cassava brown streak ipomoviruses (CBSV and UCBSV) belong. However, aphid transmission studies using Myzus persicae aphids in the transmission of SqVYV demonstrated that the virus was not aphid transmissible (Adkins et al., 2007). The possible explanation for the failed aphid transmission in these experiments was attributed to possibly incorrect sequence context surrounding the DAG motif (López-Moya et al., 1999) or the DAG motif occurring too far away from the N-terminus to be exposed (Shukla and Ward, 1988). With the availability of infectious clones for both UCBSV and CBSV (Duff-Farrier et al., 2018), the DAG and DEG motifs in CBSV and UCBSV infectious clones can be mutated, and aphid transmissibility studies can be performed to confirm if the motifs are necessary for aphid transmission. In conclusion, it is recommended that transmission studies are undertaken to confirm if aphids can transmit CBSIs.
The generated sequence datasets for this study can be found in the GenBank under accession numbers OP223337, OP223338, OP223339, and OP223340.
SN: data collection, analysis, and writing the manuscript. RK: collection of data from the fields. PA: analysis of data. SB, TA, and SS: experimental design and data collection. AB: experimental design. GF: financial support. All authors contributed to the article and approved the submitted version.
This study was funded by CONNECTED-BBSRC Grant No:BB/R005397/1 project in addition to the IsDB-TWAS 2021-Postdoctoral Fellowship.
The funders are acknowledged for the financial support in addition to the cassava farmers in Uganda who allowed the researchers to access and collect data from their cassava fields.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fsufs.2023.1027842/full#supplementary-material
Abarshi, M. M., Mohammed, I. U., Wasswa, P., Hillocks, R. J., Holt, J., Legg, J. P., et al. (2010). Optimization of diagnostic RT-PCR protocols and sampling procedures for the reliable and cost-effective detection of Cassava brown streak virus. J. Virol. 163, 353–359. doi: 10.1016/j.jviromet.2009.10.023
Adkins, S., Webb, S. E., Achor, D., Roberts, P. D., and Baker, C. A. (2007). Identification and characterization of a novel whitefly-transmitted member of the family potyviridae isolated from cucurbits in Florida. Phytopathology 97, 145–154. doi: 10.1094/PHYTO-97-2-0145
Ala-Poikela, M., Goytia, E., Haikonen, T., Rajamäki, M.-L., and Valkonen, J. P. T. (2011). Helper component proteinase of the genus Potyvirus is an interaction partner of translation initiation factors eIF(iso)4E and eIF4E and contains a 4E binding motif. J. Virol. 85, 6784–6794. doi: 10.1128/JVI.00485-11
Alicai, T., Omongo, C. A., Maruthi, M. N., Hillocks, R. J., Baguma, Y., Kawuki, R., et al. (2007). Re-emergence of cassava brown streak disease in Uganda. Plant Dis. 91, 24–29. doi: 10.1094/PD-91-0024
Ano, C. U., Ochwo-Ssemakula, M., Ibanda, A., Ozimati, A., Gibson, P., Onyeka, J., et al. (2021). Cassava brown streak disease response and association with agronomic traits in elite Nigerian cassava cultivars. Front. Plant Sci. 12, 1–15. doi: 10.3389/fpls.2021.720532
Asghar, U., Malik, M. F., Anwar, F., Javed, A., and Raza, A. (2015). DNA extraction from insects by using different techniques: a review. Adv. Entomol. 3, 132–138. doi: 10.4236/ae.2015.34016
Ateka, E., Alicai, T., Ndunguru, J., Tairo, F., Sseruwagi, P., Kiarie, S., et al. (2017). Unusual occurrence of a DAG motif in the Ipomovirus Cassava brown streak virus and implications for its vector transmission. PLoS ONE 12:e0187883. doi: 10.1371/journal.pone.0187883
Dombrovsky, A., Reingold, V., and Antignus, Y. (2014). Ipomovirus–an atypical genus in the family Potyviridae transmitted by whiteflies. Pest Manag. Sci. 70, 1553–1567. doi: 10.1002/ps.3735
Duff-Farrier, C. R. A., Mbanzibwa, D. R., Nanyiti, S., Bunawan, H., Pablo-Rodriguez, J. L., Tomlinson, K. R., et al. (2018). Strategies for the construction of cassava brown streak disease viral infectious clones. Mol. Biotechnol. 61, 93–101. doi: 10.1007/s12033-018-0139-7
Folmer, O., Black, M., Hoeh, W., Lutz, R., and Vrijenhoek, R. (1994). DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol. Marine Biol. Biotechnol. 3, 294–299.
Gadhave, K. R., Gautam, S., Rasmussen, D. A., and Srinivasan, R. (2020). Aphid transmission of potyvirus : the largest plant-infecting RNA virus genus. Viruses 12, 773. doi: 10.3390/v12070773
Hillocks, R., and Jennings, D. (2003). Cassava brown streak disease: a review of present knowledge and research needs. Int. J. Pest Manag. 49, 225–234. doi: 10.1080/0967087031000101061
Hillocks, R. J., Raya, M., and Thresh, J. M. (1996). The association between root necrosis and above-ground symptoms of brown streak virus infection of cassava in southern Tanzania. Int. J. Manag. 42, 285–289. doi: 10.1080/09670879609372008
Hillocks, R. J., Raya, M. D., Mtunda, K., and Kiozia, H. (2001). Effects of brown streak virus disease on yield and quality of cassava in Tanzania. J. Phytopathol. 149, 389–394. doi: 10.1046/j.1439-0434.2001.00641.x
Hillocks, R. J., and Thresh, J. M. (2000). Cassava mosaic and cassava brown streak virus diseases in Africa. a comparative guide to symptoms and aetiologies. Roots 7, 1–8.
Legg, J. P., Jeremiah, S. C., Obiero, H. M., Maruthi, M. N., Ndyetabula, I., Okao-Okuja, G., et al. (2011). Comparing the regional epidemiology of cassava mosaic and cassava brown streak virus pandemics in Africa. Virus Res. 159, 161–170. doi: 10.1016/j.virusres.2011.04.018
Llave, C., Martínez, B., Díaz-Ruíz, J. R., and López-Abella, D. (2002). Amino acid substitutions within the Cys-rich domain of the tobacco etch potyvirus HC-Pro result in loss of transmissibility by aphids. Arch. Virol. 147, 2365–2375. doi: 10.1007/s00705-002-0884-5
López-Moya, J. J., Wang, R. Y., and Pirone, T. P. (1999). Context of the coat protein DAG motif affects potyvirus transmissibility by aphids. J. Gen. Virol. 80, 3281–3288. doi: 10.1099/0022-1317-80-12-3281
Maruthi, M. N., Jeremiah, S. C., Mohammed, I. U., and Legg, J. P. (2005). Transmission of Cassava brown streak virus by Bemisia tabaci (Gennadius). J. Phytopathol. 153, 307–312. doi: 10.1111/j.1439-0434.2005.00974.x
Mbanzibwa, D. R., Tian, Y., Mukasa, S. B., and Valkonen, J. P. T. (2009a). Cassava brown streak virus (Potyviridae) encodes a putative Maf/HAM1 pyrophosphatase implicated in reduction of mutations and a P1 proteinase that suppresses RNA silencing but contains no HC-Pro. J. Virol. 83, 6934–6940. doi: 10.1128/JVI.00537-09
Mbanzibwa, D. R., Tian, Y. P., Tugume, A. K., Mukasa, S. B., Tairo, F., Kyamanywa, S., et al. (2009b). Genetically distinct strains of Cassava brown streak virus in the Lake Victoria basin and the Indian Ocean coastal area of East Africa. Arch. Virol. 154, 353–359. doi: 10.1007/s00705-008-0301-9
Mbanzibwa, D. R., Tian, Y. P., Tugume, A. K., Mukasa, S. B., Tairo, F., Kyamanywa, S., et al. (2011). Simultaneous virus-specific detection of the two cassava brown streak-associated viruses by RT-PCR reveals wide distribution in East Africa, mixed infections, and infections in Manihot glaziovii. J. Virol. Methods 171, 394–400. doi: 10.1016/j.jviromet.2010.09.024
Mohammed, I. U., Abarshi, M. M., Muli, B., Hillocks, R. J., and Maruthi, M. N. (2012). The symptom and genetic diversity of cassava brown streak viruses infecting cassava in East Africa. Adv. Virol. 2012, 1–10. doi: 10.1155/2012/795697
Monger, W. A., Seal, S., Cotton, S., and Foster, G. D. (2001). Identification of different isolates of Cassava brown streak virus and development of a diagnostic test. Plant Pathol. 50, 768–775. doi: 10.1046/j.1365-3059.2001.00647.x
Ng, J. C. K., and Perry, K. L. (2004). Transmission of plant viruses by aphid vectors. Mol. Plant Pathol. 5, 505–511. doi: 10.1111/j.1364-3703.2004.00240.x
Ogwok, E., Patil, B. L., Alicai, T., and Fauquet, C. M. (2010). Transmission studies with Cassava brown streak Uganda virus (Potyviridae: Ipomovirus) and its interaction with abiotic and biotic factors in Nicotiana benthamiana. J. Virol. Methods 169, 296–304. doi: 10.1016/j.jviromet.2010.07.030
Shukla, D. D., and Ward, C. W. (1988). Amino acid sequence homology of coat proteins as a basis for identification and classification of the potyvirus group. J. Gen. Virol. 69, 2703–2710. doi: 10.1099/0022-1317-69-11-2703
Thresh, J. M., Fargette, D., and Otim-Nape, G. (1994). The virus and virus diseases of cassava in Africa. African Crop Science Journal. 2, 459–478.
Tomlinson, K. R., Bailey, A. M., Alicai, T., Seal, S., and Foster, G. D. (2018). Cassava brown streak disease: historical timeline, current knowledge and future prospects. Mol. Plant Pathol. 19, 1282–1294. doi: 10.1111/mpp.12613
Tomlinson, K. R., Pablo-Rodriguez, J. L., Bunawan, H., Nanyiti, S., Green, P., Miller, J., et al. (2019). Cassava brown streak virus Ham1 protein hydrolyses mutagenic nucleotides and is a necrosis determinant. Mol. Plant Pathol. 20, 1080–1092. doi: 10.1111/mpp.12813
Wamonje, F. O., Michuki, G. N., Braidwood, L. A., Njuguna, J. N., Mutuku, J. M., Djikeng, A., et al. (2017). Viral metagenomics of aphids present in bean and maize plots on mixed-use farms in Kenya reveals the presence of three dicistroviruses including a novel Big Sioux River virus-like dicistrovirus. Virol. J. 14, 1–13. doi: 10.1186/s12985-017-0854-x
Whitfield, A. E., Falk, B. W., and Rotenberg, D. (2015). Insect vector-mediated transmission of plant viruses. Virology 479–480, 278–289. doi: 10.1016/j.virol.2015.03.026
Keywords: cassava, disease, aphids, cassava brown streak ipomoviruses, cassava brown streak disease, food security
Citation: Nanyiti S, Kabaalu R, Alicai T, Abidrabo P, Seal SE, Bouvaine S, Bailey AM and Foster GD (2023) Detection of cassava brown streak ipomoviruses in aphids collected from cassava plants. Front. Sustain. Food Syst. 7:1027842. doi: 10.3389/fsufs.2023.1027842
Received: 25 August 2022; Accepted: 04 January 2023;
Published: 08 February 2023.
Edited by:
Ping Zhu, Royal Observatory of Belgium, BelgiumReviewed by:
Samar Sheat, German Collection of Microorganisms and Cell Cultures GmbH (DSMZ), GermanyCopyright © 2023 Nanyiti, Kabaalu, Alicai, Abidrabo, Seal, Bouvaine, Bailey and Foster. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sarah Nanyiti,  c2FyYWgubmFueWl0aS4yMDEyQG15LmJyaXN0b2wuYWMudWs=
c2FyYWgubmFueWl0aS4yMDEyQG15LmJyaXN0b2wuYWMudWs=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.