- 1Department of Food Science, New York State Agricultural Experiment Station, Cornell University, Geneva, NY, United States
- 2Department of Food Science, Cornell University, Ithaca, NY, United States
- 3Department of Neurobiology and Behavior, Cornell University, Ithaca, NY, United States
The antimicrobial nature of honey and its related apiological origins typically focus on basic chemical analysis without attempting to understand the diversity of the microbial component. The antibacterial activity, chemical characterization, and diversity of bacteria isolated from Apis mellifera honey stomachs and hive honey collected throughout the honey production season are presented. After screening >2,000 isolates, 50 isolates were selected and characterized by 16S rRNA gene homology, Gram stain, catalase and protease tests, as well as for antibacterial activity against select indicators. Antibacterial-producing isolates were predominantly from the Pseudomonas, Paenibacillus, Lonsdalea, Serratia, and Bacillus genera. Isolates collected from honey stomachs in April displayed the highest level of activity (27%). While April isolates did not demonstrate activity against the Gram-negative bacteria tested. Whereas 59% of July isolates, 33% of September isolates, and 100% of the honey isolates did. The predominant honey stomach isolates were Pseudomonas spp. (April), Paenibacillus polymyxa (July, Sept.), and Lonsdalea iberica (Sept.). Chemical characterizations of the antimicrobial compounds show most to be antibiotic in nature with the minority being potential bacteriocins. This study offers the first glimpse into the variability and diversity of the bacteria/host interactions found within the honey stomach of the domestic honey bee while revealing a novel source of potentially beneficial antimicrobial compounds.
1. Introduction
In addition to being a food source for bees and humans alike, honey is a complex concentrated mixture of carbohydrates, enzymes, processed plant nectar, bacterial endospores, as well as a multitude of other materials, all unique to the particular type of honey being produced. The specific types of nectars gathered by the worker bees greatly influence the final product giving rise to a variety of flavors and physical/chemical/medicinal properties. The process is contingent on the function of a structure known as the honey stomach, a secondary stomach-like pouch used to enzymatically break down plant nectars, found within the domestic honey bee, Apis mellifera. These various nectars are processed in the honey stomach of the worker bees before being regurgitated into the honeycombs of the hive. The initial water content of most honey ranges from 60 to 75% and is reduced, primarily through evaporation, to levels below 20% during honey ripening (Crane, 1975).
Different aspects of the antimicrobial nature of honey have been previously studied to varying degrees. Its antimicrobial properties have been recognized for thousands of years (Stomfay-Stitz, 1960; Crane, 1975), and it has been used to treat a variety of ailments and infections (Stomfay-Stitz, 1960; Zumla and Lulat, 1989), including some that are unresponsive to conventional treatments (Efem, 1988; Cooper et al., 2002). However, honey contains inconsistent antimicrobial activity, varying between and even within the same floral sources or bee species (Molan, 1995; Mundo et al., 2004).
Most research has attributed honey's efficacy to its acidity, osmolarity, enzymatic generation of hydrogen peroxide from glucose oxidase, viscosity, aromatic acids, and other compounds of unknown origin (Molan, 1992; Weston et al., 1999). It was recently reported that methylglyoxal, a metabolic side product of glycolysis, was identified as being the primary chemical responsible for the antibacterial nature of Manuka honey, a type of honey that is produced from processing only the nectar of the Manuka tree (Leptospermum scoparium) (Mavric et al., 2008). While this and other studies attempt to identify the various chemical constituents of the honey itself, the antimicrobial activity associated with the microbiological composition of the honey formation process has been studied extensively. For example, lactic acid bacteria (LAB) have been isolated from the honey stomach of honey bees and different types of raw honey (Olofsson and Vásquez, 2008; Endo and Salminen, 2013; Iorizzo et al., 2022). Many of these LAB isolates, including Lactobacillus and Bifidobacterium, possess antimicrobial abilities and protect honey bees against pathogens (Meulen et al., 2006; Olofsson and Vásquez, 2008; Olofsson et al., 2016).
As a means to protect their food source, many documented systems of antimicrobial compounds have been found in the collected nectar used to make honey (Gilliam et al., 1983). While some of these protective chemicals are thought to be transferred to the honey during processing and ripening, the honey itself has still been identified as an additional source of microbiota due to the introduction of contaminating microbes originating from pollen foraging and feeding. Furthermore, as young honey bee larvae, pupae, and emerging worker honey bees are initially free of internal microbiota, the bacterial diversity seen in their alimentary tracts has been attributed to similar pollen contamination brought into the colony during foraging (Gilliam et al., 1983). More than 6,000 microbial strains have been associated with honey bees and their food, predominantly Gram-variable bacteria, Bacillus spp., yeasts, and molds (Gilliam and Prest, 1987). It is postulated that the honey stomachs themselves may serve as a source of antimicrobial activity directly associated with the microbiological component housed within (Jeyaprakash et al., 2003). The gastrointestinal tract of honey bees is dominated by bacteria in the phyla Proteobacteria, Actinobacteria, Bacteroidetes, and Firmicutes (Kwong et al., 2014; Kwong and Moran, 2016; Iorizzo et al., 2022). Most likely, these bacteria compete for carbohydrates until the lack of available water induces sporulation. As a result, while honey contains few vegetative microbes, it may often contain high levels of bacterial spores (Snowdon and Cliver, 1996).
Secondary metabolites such as antibiotics and bacteriocins secreted from honey bee microbiota have been extensively investigated as a source of antibacterial activity (Pajor et al., 2018; Zendo et al., 2020; Xiong et al., 2022). Bacteriocins are antimicrobial proteinaceous substances secreted by select bacteria that are active against microorganisms closely related to the producer strain (Klaenhammer, 1988). Bacilli have been isolated from bee intestines, queen bee organs, brood comb, and hive floor sources (Jeyaprakash et al., 2003; Piccini et al., 2004). Among the isolates of prominent importance from these sources are Bacillus coagulans (Le Marrec et al., 2000), Bacillus cereus (Oscariz et al., 1999), and Bacillus subtilis (Teo and Tan, 2005), all known to be bacteriocin producers. Paenibacillus (formerly Bacillus) polymyxa (Svetoch et al., 2005) and Paenibacillus kobensis (Martin et al., 2003) were similarly identified, and both produce antibiotics that are active against a wide variety of bacteria (Pajor et al., 2020). Lactic acid bacteria isolated from honeycombs were able to produce bacteriocin-like substances and inhibit spoilage microorganisms and foodborne pathogens (Voidarou et al., 2020).
The purpose of this research was to further characterize the antimicrobial aspects of this system by focusing on the rich microbiota environment residing within the honey processing center of the worker bee, namely, the honey stomach. We began with the initial goal of isolating, characterizing, and identifying bacterial species collected from honey bee honey stomachs. While this offered a wealth of information into the ecological diversity of this rarely studied micro-environment, its primary result was to assay each isolate's antimicrobial activity against a number of indicator organisms in vitro. Isolates showing antibacterial activity were characterized based on their respective antibacterial spectra, and the diversity and distribution of isolates were correlated to the early, middle, and late periods of the honey production season.
2. Materials and methods
2.1. Sample collection of honey stomachs and isolation of bacterial biota
The honey stomachs were excised from domestic honey bees kept in Ithaca, NY. Forager bees were captured as they returned to the hive and subsequently chilled for 20 min at −10°C. A total of 10 intact honey stomachs (pooled weight of 0.10 g) were removed from the bees using sterile dissecting forceps and pooled in a sterile 1 ml vial containing 150 μl of 100% glycerol at −10°C. One milliliter of sterile deionized water was added to the stomachs that were then macerated by repeated pipetting using sterile plugged wide bore tips; 100 μl aliquots of the solution were then spread plated on each of ten tryptic soy agar (TSA) (3% tryptic soy broth [TSB] and 1.5% agar) plates and incubated at 37°C for 24 h. Individual bacterial isolates were selected based on colony morphology and transferred to fresh TSA plates, similarly incubated, and then preserved at 4°C for further study. Honey stomach samples were taken three times (April, July, and September 2005) to capture differences in microbiota at the early, middle, and late periods of the honey production season. In April, the primary nectar source was maple trees (Acer spp.); in July, it was white clover (Trifolium repens); and in September, it was goldenrod (Solidago spp.). Proper aseptic techniques were used in all experiments.
2.2. Indicator microorganisms and culture conditions
Eight indicator microorganisms were used for screening the honey stomach isolates for antibacterial activity: Staphylococcus aureus ATCC 9144, S. aureus ATCC 25923, Listeria monocytogenes F2-586-1053, Paenibacillus larvae ATCC 25747, P. larvae ATCC 9545, Bacillus subtilis ATCC 6633, B. cereus ATCC 11778, and B. cereus ATCC F4552 (Pajor et al., 2018). These bacterial indicators include both honey bee and human opportunistic pathogens that are either Gram-positive or Gram-negative. Bacterial cultures were previously purchased or isolated in the lab, and all cultures were kept frozen at −80°C at Cornell University (Ithaca, NY). Bacterial cultures were grown aerobically in TSB (3%) while shaking at 250 rpm, except for P. larvae, which was grown in brain-heart infusion (BHI) broth (3.7%) supplemented with thiamin hydrochloride (1 μg/ml) (Lee et al., 2009b). B. cereus and B. subtilis were grown at 30°C; all others were grown at 37°C. Additional assays were performed to determine the antibacterial activity of select isolates against the Gram-negative bacteria (also grown at 37°C in TSB) Escherichia coli ATCC 25922, E. coli O157:H7 ATCC 933, and Salmonella enterica serovar Rubislaw (Mazzotta, 2001; Lee et al., 2009b).
2.3. Antimicrobial spectrum of bacterial isolates from honey stomachs
Deferred inhibition assays were used to determine the antimicrobial spectrum of each honey stomach isolate (Ahn and Stiles, 1990). Briefly, individual isolated colonies gathered from the contents of the pooled honey stomachs were spotted in groups of 10 on TSA plates and grown for 18–24 h at 37°C. Each of these spotted isolate plates was then overlaid with 7 ml molten soft TSA or BHI soft agar (45°C, 0.75% agar) inoculated with 50 μl of an overnight liquid incubation of one of the nine indicator test strains. Each spotted isolate plate was assayed against each of the indicator strains. The plates were then incubated at 30°C or 37°C for 18–24 h, after which the absence or presence of inhibition zones was noted, and the relative sizes of the zones were recorded. In the same fashion, isolates were additionally tested for antibacterial activity against the Gram-negative bacteria E. coli and Salmonella. Isolates demonstrating visible antibacterial activities (now known as producer strains) as determined by the presence of an inhibition zone around the isolate were selected for identification. Producer strains were characterized by Gram staining and catalase production following standard protocol and identified using 16S rRNA gene sequencing (Edwards et al., 1989; Kelley and Post, 1989).
2.4. Catalase and protease sensitivity testing
A volume of 5 μl of catalase (10 mg/ml), α-chymotrypsin (25 mg/ml), proteinase K (20 mg/ml), and pronase E (10 mg/ml) (Sigma-Aldrich; St. Louis, MO) was spotted adjacent to an isolated colony and allowed to dry. Plates were overlaid as described in the previous section and incubated overnight. The absence or presence of inhibition zones and/or half-moon halos indicating sensitivity was recorded (Ahn and Stiles, 1990; Lee et al., 2008; Chiang et al., 2012; Xiong et al., 2022).
2.5. Sensitivity to honey
Honey itself employs a variety of bacterial inhibition strategies. High levels of sugar, hydrogen peroxide formation, and proteinaceous compounds all contribute to the honey's antibacterial functions. Bacterial isolates from the honey stomach and hive honey should possess some level of resistance against honey. To characterize this honey resistance property, bacterial isolates and indicator species were tested for sensitivity to honey in a well-diffusion assay, using a 50% manuka honey solution (Holder and Boyce, 1994; Magaldi et al., 2004). Briefly, a core plug of agar was removed from the TSA plates, filled with 100 μl of a 50% manuka honey solution, and allowed to diffuse into the agar. The plate was then overlaid with 7 ml of soft (0.75%) TSA containing 50 μl liquid culture of either an isolate or indicator strain and subsequently incubated overnight at 37°C. Raw manuka honey (honey derived mainly from the plant nectar of the manuka tree, Leptospermum scoparium) was used for this assay due to its broad range of antimicrobial properties (Molan, 1995). Diluted honey solutions (i.e., 50%) have been shown to produce hydrogen peroxide as an antimicrobial strategy (White et al., 1963). Isolates demonstrating sensitivity to 50% manuka honey were similarly tested for sensitivity to a 50% synthetic honey solution (3.85 g fructose, 3.10 g glucose, 1.34 g sucrose, and 11.71 g water) in order to assess the sugar's role in observed sensitivity.
2.6. 16S rRNA gene sequencing
Chromosomal extraction was performed for each bacterial isolate followed by an in vitro amplification via the polymerase chain reaction (PCR) in which a pair of primers (5′-AGAGTTTGATCCTGGCTCAG-3′; 5′-AAGGAGGTGAGCCAGCCGCA-3′) was designed to amplify the 16S rRNA gene sequences (Stratagene Robocycler Gradient 40; La Jolla, CA, USA) (Edwards et al., 1989; Sha et al., 2017, 2018). The PCR products were purified from agarose using an Eppendorf Perfectprep® Gel Cleanup Kit (Eppendorf; Westbury, NY, USA) and sequenced using an ABI Prism 373 DNA sequencer (Applied Biosystems; Foster City, CA, USA). The resulting sequences were analyzed by homology comparison using the National Center for Biotechnology Information (NCBI) nucleotide Basic Local Alignment Search Tool (BLAST) database (http://www.ncbi.nlm.nih.gov/BLAST/Blast.cgi). Sequence data collected in this study were deposited under the NCBI GenBank submission SUB11959034.
2.7. Isolation and characterization of bacterial isolates directly from honey
Honey from the same colony in which the forager bees were collected was sampled in August for its microbial content. The honey was diluted to a 50% final concentration with sterile water, and 100 μl of the solution was then spread plated on each of the 10 TSA plates and incubated at 37°C for 24 h. The honey was not heated prior to plating in order to assay for vegetative and spore-forming bacteria contained in the honey. Honey isolates were similarly identified and characterized by the tests described previously. The honey itself also underwent testing to determine antibacterial activity and sensitivity to catalase and proteases using a well-diffusion assay (Mundo et al., 2004).
2.8. Statistics
Chi-square analyses were performed for all comparisons involving two data sets (Fleiss, 1981). The correlations between antibacterial activity and the seasonal origin of the isolates were tested using a generalized linear model with a quasi-binomial likelihood and logit link. All statistical analyses were performed using Minitab software (Minitab version 10.51Xtra; State College, PA).
3. Results
3.1. Antimicrobial activities of isolates
A total of 2,154 isolates were recovered from the bee honey stomachs: 654 in April (A), and 750 each in July (J) and September (S). In addition, 15 isolates were recovered from the honey itself (TH) collected in August. Table 1 indicates the percentage of isolates from each batch showing antibacterial activity against the various indicator organisms. Global chi-square analysis for each indicator strain shows strong statistically significant differences in the activity of the isolates between seasons (p < 0.001 for all comparisons). Chi-square comparisons between groups further illustrate these seasonal variations. For all indicator strains except P. larvae 25747, bacterial isolates collected in April exhibited the highest percentage of antibacterial activity when compared against those taken in July or September (B. subtilis 6633, p < 0.05; all others, p < 0.001). S. aureus 9144, S. aureus 25923, B. subtilis 6633, and B. cereus 11778 showed further statistically significant differences when comparing July and September isolates (S. aureus 9144, p < 0.05; all others, p < 0.001).
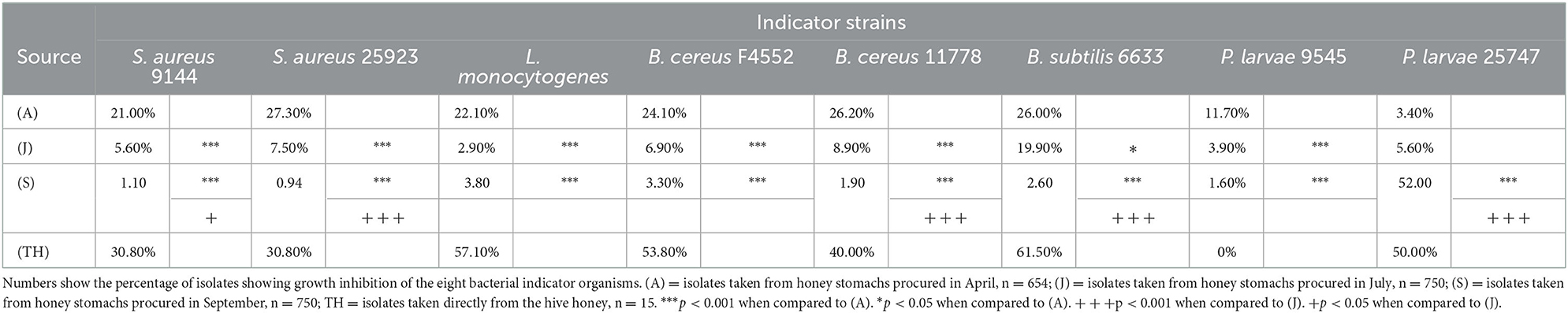
Table 1. Differential antibacterial activity of bee honey stomach and honey isolates against given indicator organisms arranged by sampling month.
For all indicator strains except P. larvae 25747, the seasonal variations of the antibacterial activity against the selected indicators show a statistically similar correlation between season and activity. From April through July and in September, the antibacterial activity against these indicator strains decreased. The trend was reversed for P. larvae 25747 (p < 0.001).
3.2. Select isolate characterization
A total of 50 isolates showing the highest antibacterial activity and broadest spectrum of activity were selected for identification (Table 2) and further characterization of the effectiveness of their antibacterial activity against the indicator strains (Table 3).
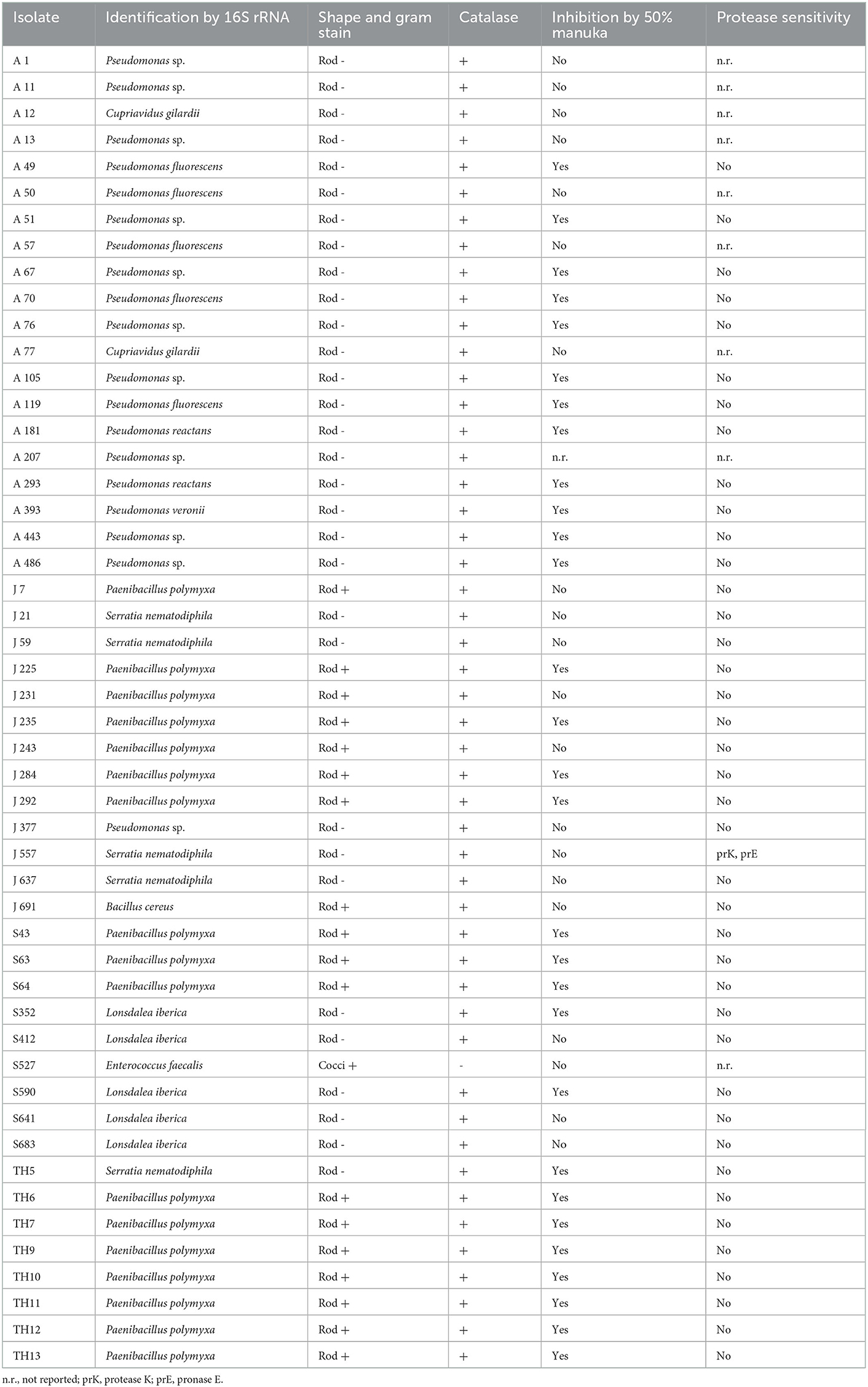
Table 2. Characteristics of select honey stomach isolates collected in April (A), June (J), and September (S), and directly from August hive honey (TH).
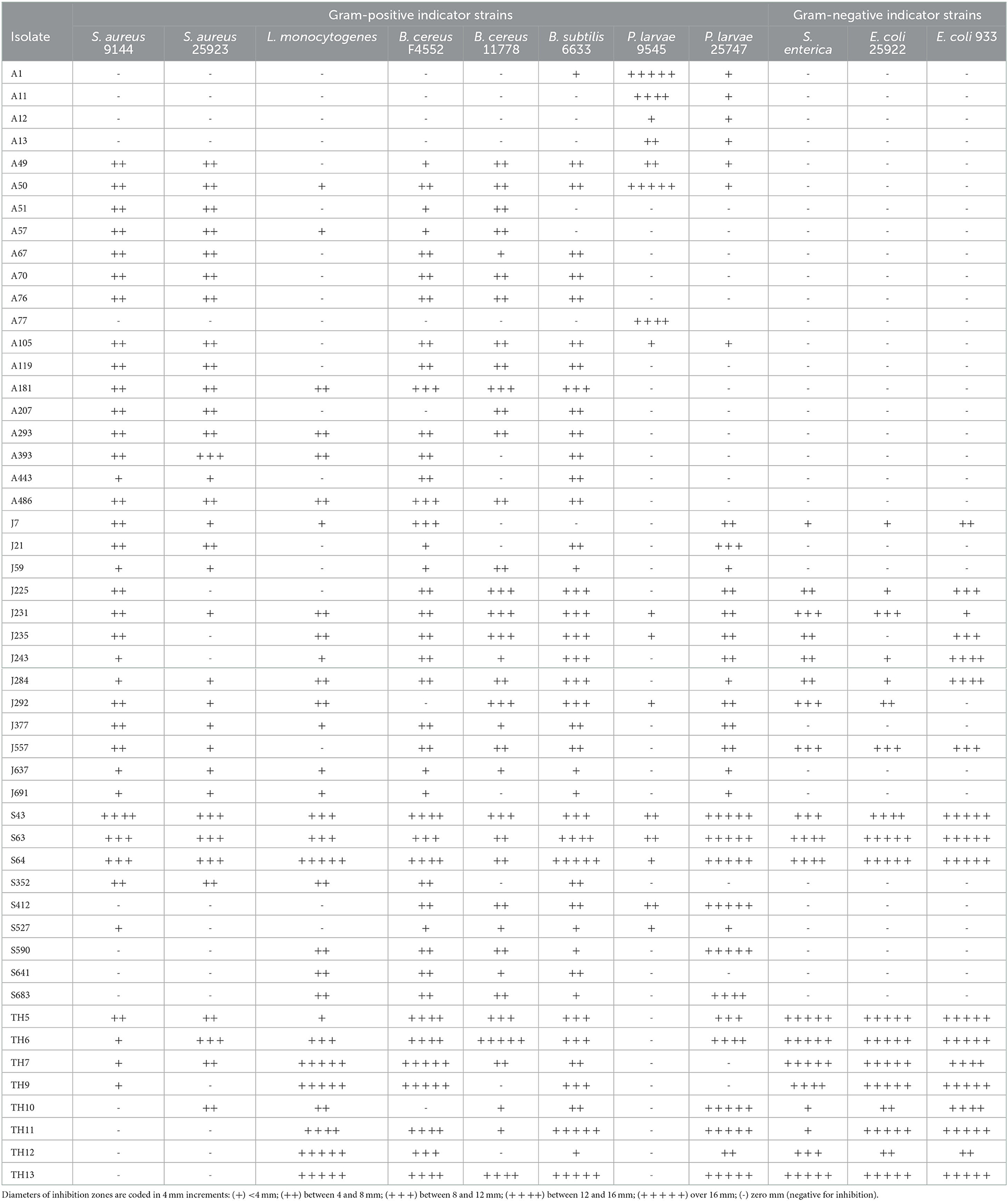
Table 3. The spectrum of antibacterial activity of isolates from April (A), June (J), and September (S) honey stomachs and August hive honey (TH).
All but one of the studied isolates produced catalase (Table 2) as shown by the evolution of bubbles when hydrogen peroxide was added to the cultures. The catalase-negative isolate S527 was later identified as Enterococcus faecalis. The vast majority of honey stomach isolates did not demonstrate sensitivity to catalase or any of the proteases tested (Table 2). Only J557 exhibited sensitivity to proteinase K and pronase E. Approximately one-third of the honey stomach isolates and all of the honey isolates did not grow in the presence of a 50% manuka honey solution (Table 2). Most of these isolates were not inhibited by a 50% artificial honey solution but some had exhibited reduced sensitivity to the artificial solution relative to the honey indicating that sugar may have contributed to but did not account fully for the inhibition. Catalase spotting confirmed that the antibacterial activity from the manuka honey was not due to hydrogen peroxide generation.
NCBI nucleotide BLAST search results of the 16S rRNA gene sequences of the honey stomach and honey isolates revealed homologies to a variety of bacteria (Table 2). Isolates were identified as belonging to the following genera: Pseudomonas, Paenibacillus, Bacillus, Lonsdalea, Serratia, and Enterococcus.
The antibacterial activities of the selected isolates against the indicator strains are given in Table 3. As expected, the targets of antibacterial activity varied between producer organisms, but differences were also evident within a genus or even within a species.
3.3. Seasonal variation of selected bacterial isolates
Three factors illustrating the diversity of bacterial isolates procured from honey stomachs in the three time periods under consideration are summarized in Figure 1. We chose the following factors to best illustrate select seasonal differences in collected isolates: (1) % of Gram-positive bacteria, (2) % antagonistic against E. coli or S. enterica (and hence most likely an antibiotic producer), and (3) % of Pseudomonad spp. All three factors varied greatly throughout the honey-producing season.
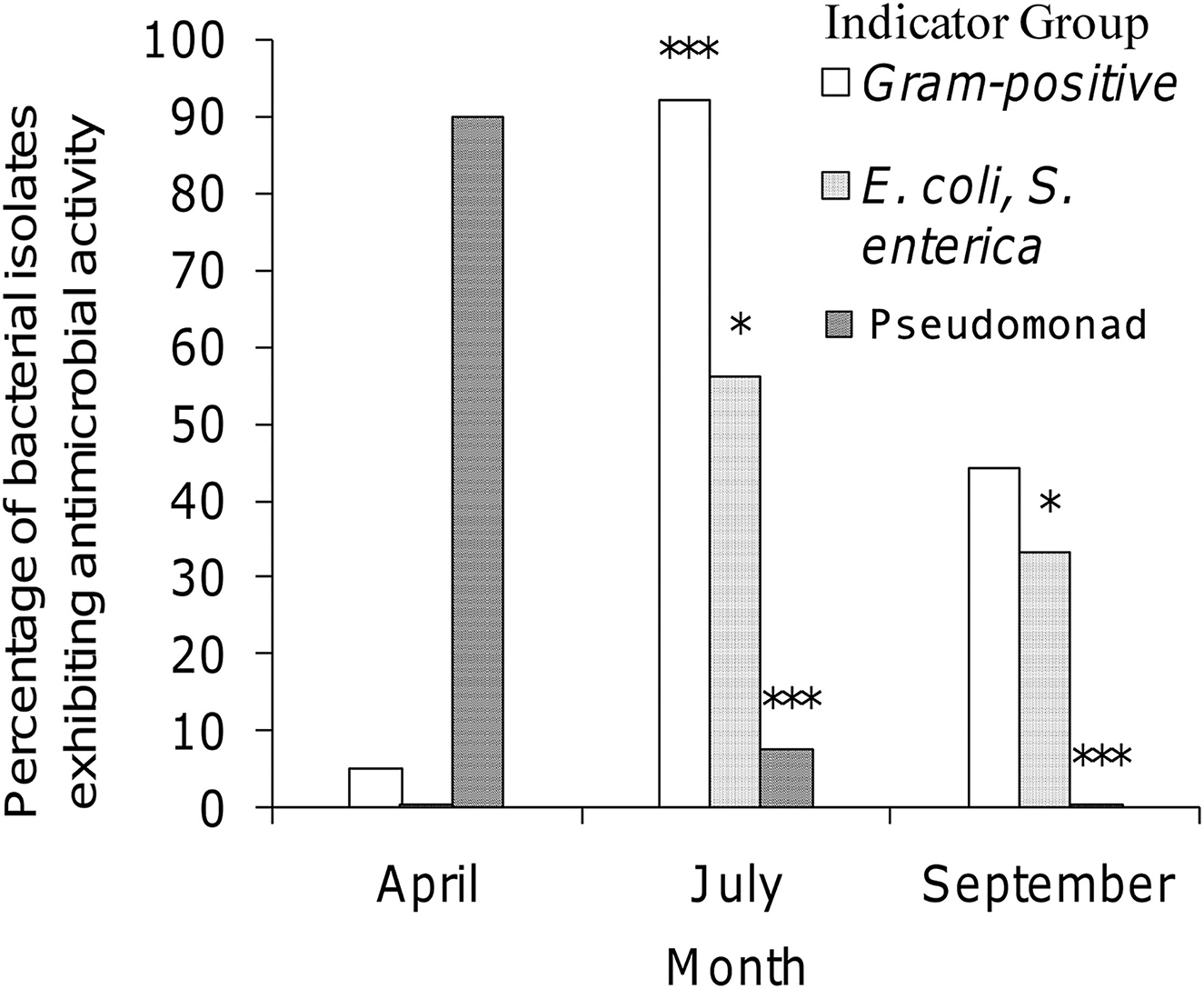
Figure 1. Seasonal variation of selected isolates procured from honey stomachs. Open bar = percentage of bacterial isolates being Gram-positive; dotted bar = percentage of bacterial isolates being antagonistic against E. coli or S. enterica; striped bar = percentage of bacterial isolates being Pseudomonad sp. ***p < 0.001 when compared to April (A). *p < 0.05 when compared to April (A).
July showed a statistically significant higher amount of Gram-positive isolates when compared to April (p < 0.001). All other differences were not significant. April showed a statistically significant lower amount of isolates exhibiting antagonistic activity against E. coli and S. enterica indicators when compared to either July or September (p < 0.05). All other differences were not significant. April showed a statistically significant higher percentage of Pseudomonad isolates when compared to either July or September (p < 0.001). The differences between July and September were not found to be significant.
4. Discussion
Many of the seasonal variations observed in this study can be linked back to the source, variety, and content of the material the worker bees collected during the time periods analyzed. The honey stomachs excised from the bees in April contained water and nectar from maple trees, pussy willows, and various ornamental flowers. However, they contained very little nectar since the foraging season had only just begun. In contrast, the honey stomachs excised in July were full of nectar from a wide variety of wildflowers, especially white clover. By September, most flowers were no longer in bloom; therefore, less nectar was available for the bees to collect. As a result of these differences, the gross number of isolates recovered from the honey stomachs was lowest in April. This allowed for every isolate collected during this time period to be tested for antimicrobial production and bacterial identification. For subsequent sampling sessions, the number of isolates dramatically increased, and isolate sampling was capped at 750 colonies.
The seasonal availability of nectar and its sources may have an influence on the variations in bacterial genera seen from honey stomach isolates (Figure 1). Pseudomonads can survive in cold temperatures and, as such, were the most abundant bacteria demonstrating antibacterial activity from the April sampling session. In July, the dynamics of nectar foraging changed dramatically. The bees are out of the hive for longer periods. They not only have a larger supply of nectar from individual plants but also have a larger variety of nectar sources. Subsequently, in July, they are exposed to a more diverse range of microorganisms found in the plant/nectar environment. It is therefore unsurprising that the colony morphology of bacteria isolated from the July honey stomachs was the most diverse when bees had so numerous different sources of nectar. By September, flowers were scarce and plants were decaying. The bees must then shift their foraging efforts to what little nutrient and nectar sources are available. Therefore, at this point in the sampling season, Lonsdalea spp. (previously identified as Erwinia spp.), a bacterium typically associated with fruit and/or plant rot, began to make a showing in our collections (Brady et al., 2012; Li et al., 2017).
Most of the active isolates were Gram-negative in April, but Gram-positive in July and equally distributed in September. Additionally, while no April isolate exhibited antagonistic activity against Salmonella or E. coli, half of the July isolates and one-third of the September isolates did (Figure 1). This shows direct evidence that the shift in forager honey bee microbiota over time is clearly affected by the bees' environment. Furthermore, our findings support previous research indicating that while Gram-negative rods may continually be present at low levels, the more typically Gram-positive bacilli are the predominant microbiota of honey stomachs (El-Leithy and El-Sibaei, 1973). Indeed, Bacillus spp. and Paenibacillus spp. were highly represented among samples isolated from the honey stomachs for all three sampling periods. Owing to the antibacterial nature of the collected bacilli, these ever-present indigenous honey bee microbiota may provide yet another novel source of antibacterial activity.
The honey itself contained few microorganisms in comparison to other honey, but more than half of these isolates exhibited a high degree of antibacterial activity against most of the indicator organisms (Lee et al., 2008). The honey isolates were in all likelihood spore-forming bacteria since the honey environment is too harsh to allow for the survival of vegetative bacterial cells. All but one of the Paenibacillus isolates collected from the honey (and honey stomachs) produced antibacterial compounds effective against Gram-negative bacteria and were insensitive to three common proteases. Most likely these antibacterial compounds from the honey isolates are antibiotics rather than bacteriocins as they demonstrate a broad spectrum of activity and are active against Gram-negative bacteria. Bacteriocins typically exhibit a narrow spectrum of antibacterial activity against closely related species and are not active against Gram-negative E. coli or Salmonella. A bacterial isolate from honey was reported as producing a unique bacteriocin as well as a novel class of antifungal compound (Lee et al., 2009a; Sit et al., 2011; Manns et al., 2012). The genetic and chemical characteristics of the antibacterial peptide were unique in that the bacteriocin structural gene was present as tandem repeats in a single transcript, allowing for higher expression levels of the antimicrobial peptide. In addition, the tertiary structure of the antibacterial peptide revealed a hairpin structure with alpha helix twists with sulfur cross-linkages evenly spaced across the folded peptide.
While bacteriocins are ribosomally synthesized peptides and proteins that may be further modified posttranslationally, antibiotics are constructed non-ribosomally, instead relying on the very specific enzymatic assembly line organized solely for their production. The chemical and functional diversity of antibiotics, most of which have established medical applications, runs the gamut from β-lactam-built cephalosporins and penicillins to protease-resistant polypeptide-based polymyxins. Polymyxin B, for example, is used in topical antibacterial ointments to prevent infection. P. polymyxa produces the antibiotic Polymyxin B; P. kobensis produces Polymyxin M (Martin et al., 2003). Indeed, from our collection, isolate J7 was shown to be a P. polymyxa that produces antibacterial compounds that are insensitive to proteases. Additional studies of the antibacterial compound produced by this isolate found that the characteristics of the compound were consistent with previously published identifiers for polymyxins (results not shown).
From the information presented in this study, we can not only offer definitive evidence that the honey stomach microbial content imparted to the product honey can be quite beneficial from an antimicrobial standpoint but also show that during a single foraging period, the bacterial population dynamically changes throughout the honey producing season (Table 2), thereby imparting a differential antimicrobial activity spectrum (Table 3). Many of the bacteria associated with honey bees were shown to produce antagonistic compounds effective against a number of indicator species in a seasonally mediated manner. The number and genera of isolates found as well as their antibacterial activity against indicator species varied considerably during the honey production season. Interestingly, while April showed the lowest number of possible isolates and the least variable of identified bacterial species, isolates from this time point showed the highest antibacterial activity against the indicator species tested. However, all time points, including samples taken from the hive honey in August, showed considerable antibacterial activity as well. It is important to note that selected bacterial isolates from our study cannot represent the honey stomach or honey microbiota, since our selection criteria were based on colony morphology and our culturing methods were limited. Many probiotic microorganisms, like lactic acid bacteria, were not isolated by our plating methods (Ghatani et al., 2022). Next-generation sequencing (NGS) technologies should be used in future studies to obtain a comprehensive overview of the honey stomach and honey microbiota.
In conclusion, our study demonstrated the seasonal variation of antimicrobial activities of honey stomach isolates. Taxonomy classification, biochemical testing, and antimicrobial characterization were performed for select bacterial isolates. Due to the sensitivity or lack of sensitivity to various proteases, the majority of the antibacterial compounds employed by the isolates tested are hypothesized to be antibiotics with a minority possibly being bacteriocin in nature. With a novel source such as this, the ensuing step is to consider the chemical diversity and biological importance of the individual compounds produced by these isolates. With the rise in antibiotic-resistant microorganisms, we must continually turn to both nature and our own ingenuity for new sources of antimicrobial compounds. In attempting to stay one step ahead of microbiological impairments, nature has always provided both the problem and solution to our concerns. From this study, we see yet again how an unexploited and overlooked microcosm can provide new insights that showcase the diversity of nature as well as offer potentially new raw materials for both therapeutics and food protectants.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
Author contributions
MM contributed to data collection, data analysis, and writing–original draft. All authors interpreted the results, contributed to the study, and approved the submitted version.
Funding
This research was funded by the New York State Agricultural Experiment Station and the National Honey Board (Longmont, CO, USA).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Ahn, C., and Stiles, M. E. (1990). Antibacterial activity of lactic acid bacteria isolated from vacuum-packaged meats. J. Appl. Bacteriol. 69, 302–310. doi: 10.1111/j.1365-2672.1990.tb01520.x
Brady, C. L., Cleenwerck, I., Denman, S., Venter, S. N., Rodríguez-Palenzuela, P., Coutinho, T. A., et al. (2012). Proposal to reclassify Brenneria quercina (Hildebrand and Schroth 1967) Hauben et al. 1999 into a new genus, Lonsdalea gen. nov., as Lonsdalea quercina comb. nov., descriptions of Lonsdalea quercina subsp. quercina comb. nov., Lonsdalea quercina subsp. iberica subsp. nov. and Lonsdalea quercina subsp. britannica subsp. nov., emendation of the description of the genus Brenneria, reclassification of Dickeya dieffenbachiae as Dickeya dadantii subsp. dieffenbachiae comb. nov., and emendation of the description of Dickeya dadantii. Int. J. Syst. Evol. Microbiol. 62, 1592–1602. doi: 10.1099/ijs.0.035055-0
Chiang, I. Y., Worobo, R. W., Churey, J. J., and Henick-Kling, T. (2012). Growth inhibition of foodborne pathogens by Oenococcus oeni. J. Food Sci. 77, M15–19. doi: 10.1111/j.1750-3841.2011.02446.x
Cooper, R. A., Molan, P. C., and Harding, K. G. (2002). The sensitivity to honey of Gram-positive cocci of clinical significance isolated from wounds. J. Appl. Microbiol. 93, 857–863. doi: 10.1046/j.1365-2672.2002.01761.x
Edwards, U., Rogall, T., Blocker, H., Emde, M., and Bottger, E. C. (1989). Isolation and direct complete nucleotide determination of entire genes. Characterization of a gene coding for 16S ribosomal RNA. Nucleic Acids Res. 17, 7843–7853. doi: 10.1093/nar/17.19.7843
Efem, S. E. (1988). Clinical observations on the wound healing properties of honey. Br. J. Surg. 75, 679–681. doi: 10.1002/bjs.1800750718
El-Leithy, M., and El-Sibaei, K. (1973). External and internal microflora of the honey bees (Apis mellifera L.). J. Microbiol. United Arab Republic. 2013, 79–87.
Endo, A., and Salminen, S. (2013). Honeybees and beehives are rich sources for fructophilic lactic acid bacteria. Syst. Appl. Microbiol. 36, 444–448. doi: 10.1016/j.syapm.2013.06.002
Ghatani, K., Thapa, S., Sha, S. P., Sarkar, S., Modak, D., and Bhattacharjee, S. (2022). Revealing probiotic potential of enterococcus strains isolated from traditionally fermented chhurpi and healthy human gut. Front. Microbiol. 13, 909987. doi: 10.3389/fmicb.2022.909987
Gilliam, M., Moffett, J. O., and Kauffeld, N. (1983). Examination of floral nectar of citrus, cotton, and Arizona desert plants for microbes. Apidologie. 14, 299–302. doi: 10.1051/apido:19830403
Gilliam, M., and Prest, D. B. (1987). Microbiology of feces of the larval honey bee, Apis mellifera. J. Invertebr. Pathol. 49, 70–75. doi: 10.1016/0022-2011(87)90127-3
Holder, I. A., and Boyce, S. T. (1994). Agar well diffusion assay testing of bacterial susceptibility to various antimicrobials in concentrations non-toxic for human cells in culture. Burns 20, 426–429. doi: 10.1016/0305-4179(94)90035-3
Iorizzo, M., Letizia, F., Ganassi, S., Testa, B., Petrarca, S., Albanese, G., et al. (2022). Functional properties and antimicrobial activity from lactic acid bacteria as resources to improve the health and welfare of honey bees. Insects 13, 308. doi: 10.3390/insects13030308
Jeyaprakash, A., Hoy, M. A., and Allsopp, M. H. (2003). Bacterial diversity in worker adults of Apis mellifera capensis and Apis mellifera scutellata (Insecta: Hymenoptera) assessed using 16S rRNA sequences. J. Invertebr. Pathol. 84, 96–103. doi: 10.1016/j.jip.2003.08.007
Kelley, S. G., and Post, F. J. (1989). Basic Microbiology Techniques. Belmont, CA: Star Publishing Company.
Klaenhammer, T. R. (1988). Bacteriocins of lactic acid bacteria. Biochimie 70, 337–349. doi: 10.1016/0300-9084(88)90206-4
Kwong, W. K., Engel, P., Koch, H., and Moran, N. A. (2014). Genomics and host specialization of honey bee and bumble bee gut symbionts. Proc. Nat. Acad. Sci. 111, 11509–11514. doi: 10.1073/pnas.1405838111
Kwong, W. K., and Moran, N. A. (2016). Gut microbial communities of social bees. Nat. Rev. Microbiol. 14, 374–384. doi: 10.1038/nrmicro.2016.43
Le Marrec, C., Hyronimus, B., Bressollier, P., Verneuil, B., and Urdaci, M. C. (2000). Biochemical and genetic characterization of coagulin, a new antilisterial bacteriocin in the pediocin family of bacteriocins, produced by Bacillus coagulans I(4). Appl. Environ. Microbiol. 66, 5213–5220. doi: 10.1128/AEM.66.12.5213-5220.2000
Lee, H., Churey, J. J., and Worobo, R. W. (2008). Antimicrobial activity of bacterial isolates from different floral sources of honey. Int. J. Food Microbiol. 126, 240–244. doi: 10.1016/j.ijfoodmicro.2008.04.030
Lee, H., Churey, J. J., and Worobo, R. W. (2009a). Biosynthesis and transcriptional analysis of thurincin H, a tandem repeated bacteriocin genetic locus, produced by Bacillus thuringiensis SF361. FEMS Microbiol. Lett. 299, 205–213. doi: 10.1111/j.1574-6968.2009.01749.x
Lee, H., Churey, J. J., and Worobo, R. W. (2009b). Isolation and characterization of a protective bacterial culture isolated from honey active against American Foulbrood disease. FEMS Microbiol. Lett. 296, 39–44. doi: 10.1111/j.1574-6968.2009.01615.x
Li, Y., Xue, H., Guo, L. M., Koltay, A., Palacio-Bielsa, A., Chang, J., et al. (2017). Elevation of three subspecies of Lonsdalea quercina to species level: Lonsdalea britannica sp. nov., Lonsdalea iberica sp. nov. and Lonsdalea populi sp. nov. Int. J. Syst. Evol. Microbiol. 67, 4680–4684. doi: 10.1099/ijsem.0.002353
Magaldi, S., Mata-Essayag, S., Hartung de Capriles, C., Perez, C., Colella, M. T., Olaizola, C., et al. (2004). Well diffusion for antifungal susceptibility testing. Int. J. Infect. Dis. 8, 39–45. doi: 10.1016/j.ijid.2003.03.002
Manns, D. C., Churey, J. J., and Worobo, R. W. (2012). Functional assignment of YvgO, a novel set of purified and chemically characterized proteinaceous antifungal variants produced by Bacillus thuringiensis SF361. Appl. Environ. Microbiol. 78, 2543–2552. doi: 10.1128/AEM.07727-11
Martin, N. I., Hu, H., Moake, M. M., Churey, J. J., Whittal, R., Worobo, R. W., et al. (2003). Isolation, structural characterization, and properties of mattacin (polymyxin M), a cyclic peptide antibiotic produced by Paenibacillus kobensis M. J. Biol. Chem. 278, 13124–13132. doi: 10.1074/jbc.M212364200
Mavric, E., Wittmann, S., Barth, G., and Henle, T. (2008). Identification and quantification of methylglyoxal as the dominant antibacterial constituent of Manuka (Leptospermum scoparium) honeys from New Zealand. Mol. Nutr. Food Res. 52, 483–489. doi: 10.1002/mnfr.200700282
Mazzotta, A. S. (2001). Thermal inactivation of stationary-phase and acid-adapted Escherichia coli O157:H7, Salmonella, and Listeria monocytogenes in fruit juices. J. Food Prot. 64, 315–320. doi: 10.4315/0362-028X-64.3.315
Meulen, R. V. d, Adriany, T., Verbrugghe, K., and Vuyst, L. D. (2006). Kinetic Analysis of Bifidobacterial Metabolism Reveals a Minor Role for Succinic Acid in the Regeneration of NAD+ through Its Growth-Associated Production. Appl. Environ. Microbiol. 72, 5204–5210. doi: 10.1128/AEM.00146-06
Molan, P. C. (1992). The antibacterial activity of honey: 1. The nature of the antibacterial activity. Bee world 73, 5–28. doi: 10.1080/0005772X.1992.11099109
Mundo, M. A., Padilla-Zakour, O. I., and Worobo, R. W. (2004). Growth inhibition of foodborne pathogens and food spoilage organisms by select raw honeys. Int. J. Food Microbiol. 97, 1–8. doi: 10.1016/j.ijfoodmicro.2004.03.025
Olofsson, T. C., Butler, È., Markowicz, P., Lindholm, C., Larsson, L., and Vásquez, A. (2016). Lactic acid bacterial symbionts in honeybees – an unknown key to honey's antimicrobial and therapeutic activities. Int. Wound J. 13, 668–679. doi: 10.1111/iwj.12345
Olofsson, T. C., and Vásquez, A. (2008). Detection and identification of a novel lactic acid bacterial flora within the honey stomach of the honeybee Apis mellifera. Curr. Microbiol. 57, 356–363. doi: 10.1007/s00284-008-9202-0
Oscariz, J. C., Lasa, I., and Pisabarro, A. G. (1999). Detection and characterization of cerein 7, a new bacteriocin produced by Bacillus cereus with a broad spectrum of activity. FEMS Microbiol. Lett. 178, 337–341. doi: 10.1111/j.1574-6968.1999.tb08696.x
Pajor, M., Worobo, R. W., Milewski, S., and Szweda, P. (2018). The antimicrobial potential of bacteria isolated from honey samples produced in the apiaries located in pomeranian voivodeship in Northern Poland. Int. J. Environ. Res. Public Health 15, 2002. doi: 10.3390/ijerph15092002
Pajor, M., Xiong, Z. R., Worobo, R. W., and Szweda, P. (2020). Paenibacillus alvei MP1 as a producer of the proteinaceous compound with activity against important human pathogens, including staphylococcus aureus and listeria monocytogenes. Pathogens 9, 319. doi: 10.3390/pathogens9050319
Piccini, C., Antúnez, K., and Zunino, P. (2004). An approach to the characterization of the honey bee hive bacterial flora. J. Apic. Res. 43, 101–104. doi: 10.1080/00218839.2004.11101118
Sha, S. P., Jani, K., Sharma, A., Anupma, A., Pradhan, P., Shouche, Y., et al. (2017). Analysis of bacterial and fungal communities in Marcha and Thiat, traditionally prepared amylolytic starters of India. Sci. Rep. 7, 10967. doi: 10.1038/s41598-017-11609-y
Sha, S. P., Suryavanshi, M. V., Jani, K., Sharma, A., Shouche, Y., and Tamang, J. P. (2018). Diversity of yeasts and molds by culture-dependent and culture-independent methods for mycobiome surveillance of traditionally prepared dried starters for the production of Indian alcoholic beverages. Front. Microbiol. 9, 2237. doi: 10.3389/fmicb.2018.02237
Sit, C. S., van Belkum, M. J., McKay, R. T., Worobo, R. W., and Vederas, J. C. (2011). The 3D solution structure of thurincin H, a bacteriocin with four sulfur to α-carbon crosslinks. Angew. Chem. Int. Ed Engl. 50, 8718–8721. doi: 10.1002/anie.201102527
Snowdon, J. A., and Cliver, D. O. (1996). Microorganisms in honey. Int. J. Food Microbiol. 31, 1–26. doi: 10.1016/0168-1605(96)00970-1
Svetoch, E. A., Stern, N. J., Eruslanov, B. V., Kovalev, Y. N., Volodina, L. I., Perelygin, V. V., et al. (2005). Isolation of Bacillus circulans and Paenibacillus polymyxa strains inhibitory to Campylobacter jejuni and characterization of associated bacteriocins. J. Food Prot. 68, 11–17. doi: 10.4315/0362-028X-68.1.11
Teo, A. Y., and Tan, H. M. (2005). Inhibition of Clostridium perfringens by a novel strain of Bacillus subtilis isolated from the gastrointestinal tracts of healthy chickens. Appl. Environ. Microbiol. 71, 4185–4190. doi: 10.1128/AEM.71.8.4185-4190.2005
Voidarou, C., Alexopoulos, A., Tsinas, A., Rozos, G., Tzora, A., Skoufos, I., et al. (2020). Effectiveness of bacteriocin-producing lactic acid bacteria and bifidobacterium isolated from honeycombs against spoilage microorganisms and pathogens isolated from fruits and vegetables. Appl. Sci. 10, 7309. doi: 10.3390/app10207309
Weston, R. J., Mitchell, K. R., and Allen, K. L. (1999). Antibacterial phenolic components of New Zealand manuka honey. Food Chem. 64, 295–301. doi: 10.1016/S0308-8146(98)00100-9
White, J. W., Subers, M. H., and Schepartz, A. I. (1963). The identification of inhibine, the antibacterial factor in honey, as hydrogen peroxide and its origin in a honey glucose-oxidase system. Biochim. Biophys. Acta 73, 57–70. doi: 10.1016/0926-6569(63)90108-1
Xiong, Z. R., Cobo, M., Whittal, R. M., Snyder, A. B., and Worobo, R. W. (2022). Purification and characterization of antifungal lipopeptide produced by Bacillus velezensis isolated from raw honey. PLoS ONE 17, e0266470. doi: 10.1371/journal.pone.0266470
Zendo, T., Ohashi, C., Maeno, S., Piao, X., Salminen, S., Sonomoto, K., et al. (2020). Kunkecin A, a new nisin variant bacteriocin produced by the fructophilic lactic acid bacterium, apilactobacillus kunkeei FF30-6 isolated from honey bees. Front. Microbiol. 11:571903. doi: 10.3389/fmicb.2020.571903
Keywords: honey stomach, bee microbiota, antibacterial assay, seasonality, chemical characterization
Citation: Mundo MA, Xiong ZR, Galasong Y, Manns DC, Seeley TD, Vegdahl AC and Worobo RW (2023) Diversity, antimicrobial production, and seasonal variation of honey bee microbiota isolated from the honey stomachs of the domestic honey bee, Apis mellifera. Front. Sustain. Food Syst. 6:931363. doi: 10.3389/fsufs.2022.931363
Received: 28 April 2022; Accepted: 28 December 2022;
Published: 16 January 2023.
Edited by:
Dimitris Mossialos, University of Thessaly, GreeceReviewed by:
Elizabeth Harry, University of Technology Sydney, AustraliaShankar Prasad Sha, Kurseong College, India
Copyright © 2023 Mundo, Xiong, Galasong, Manns, Seeley, Vegdahl and Worobo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zirui Ray Xiong,  eng5N0Bjb3JuZWxsLmVkdQ==
eng5N0Bjb3JuZWxsLmVkdQ==
 Melissa A. Mundo1
Melissa A. Mundo1 Zirui Ray Xiong
Zirui Ray Xiong Yupawadee Galasong
Yupawadee Galasong Thomas D. Seeley
Thomas D. Seeley Ann Charles Vegdahl
Ann Charles Vegdahl Randy W. Worobo
Randy W. Worobo