
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Sustain. Food Syst. , 08 April 2022
Sec. Sustainable Food Processing
Volume 6 - 2022 | https://doi.org/10.3389/fsufs.2022.869430
Naturally occurring autochthonous microbes associated with ethnic beer starters are diverse and important as they play different functional roles in beer fermentations. The study on culturable microbes from the ethnic rice beer starter “emao” of the Bodo community of Assam is limited. Here we isolated and identified the culturable fungal diversity associated with emao and screened them for beer-producing capability from glucose and starch substrates. Based on morphology and molecular characterization, the species identified were Candida glabrata (Cgla_RF2), Cyberlindnera fabianii (Cfab_RF37), Hyphopichia burtonii (Hbur_RF19), Mucor circinelloides (Mcir_RF48), Mucor indicus (Mind_RF25), Penicillium citrinum (Pcit_RF32), Rhodosporidiobolus ruineniae (Rrui_RF4 & Rrui_RF43), Saccharomyces cerevisiae (Scer_RF6), Saccharomycopsis fibuligera (Sfib_RF11), and Wickerhamomyces anomalus (Wano_RF3) among which the relative abundance (RA) of W. anomalus was the highest (24%) followed by C. glabrata and H. burtonii (16% in each). Five (Hbur_RF19, Sfib_RF11, Mind_RF25, Mcir_RF48, and Pcit_RF32) of eleven isolates showed amylase positive in the starch medium. Scer_RF6 showed the highest ethanol tolerance (14% v/v) followed by Hbur_RF19 (12% v/v), Cgla_RF2 (11% v/v) and Wano_RF3 (11% v/v). The amylase-positive strains produced beer-containing ethanol in the range of 3.17–7.3 (% v/v) from rice substrate. Although the rice beer produced by amylase-positive strains showed negligible pH difference, other parameters like ethanol, ascorbic acid, total phenol, and antioxidant properties were varied from beer to beer. Antibacterial activities shown by Mcir_RF48 and Pcit_RF32 against the test bacteria were higher with a 23–35 mm zone of inhibition than the other isolates. The present findings reveal the presence of fungi with antibacterial, amylolytic, ethanol fermenting, and antioxidant producing capacity in emao which could the source for future bioprospection.
Asian ethnic starters used in beer fermentation generally comprise several microbes, mostly molds, yeasts, and bacteria (Zheng et al., 2012; Bora et al., 2016; Hui et al., 2017; Sha et al., 2018). As the cultural practice of starter culture preparation and the beer fermentation processes differ from community to community, the beer quality also varies accordingly in terms of ethanol content, taste, flavor, and other beer attributes (Missbach et al., 2017). In addition to the associated microbes, the selection of substrates and herbs for brewing might also contribute to the beer quality. Earlier culture-dependent and culture-independent studies revealed the association of a large number of microbial species to the ethnic beer starters (Chen et al., 2014; Sha et al., 2018; Anupama and Tamang, 2020; Narzary et al., 2021; Piraine et al., 2021). Although the metagenomic approach can give us better information on microbial diversity in an environmental sample, the isolation of microbial pure cultures and their activity study is still a gold standard in microbiology and helps us know the specific roles played by a microbial species in a growing condition.
Microorganisms are physiologically diverse, and differences are also seen at the strain level in many cases. Both amylolytic and ethanol-producing activities are important in beer production from starch that is often done by different groups of microbes. However, reports of microbes having both amylolytic and ethanol-producing capacity are not rare in the traditional beer starters of the Asian region (Carroll et al., 2017). Ethanol tolerance capacity of microbes is an important factor in beer fermentation which highly differs from microbes to microbes or strain to strain. As the ethanol concentration gradually increases after a certain period of beer fermentation, ethanol-sensitive microbes generally die and only the ethanol tolerant microbes thrive at the later stages of beer fermentations (Hao et al., 2021). High ethanol tolerant microbes, such as wild Saccharomyces cerevisiae lack amylolytic activity (Kregiel et al., 2017). Therefore, such microbes are always dependent on amylolytic microbes for their growth, development, and ethanol production while in a starch medium.
Some fungi associated with ethnic foods were found antagonistic to pathogenic bacteria (Osamwonyi and Wakil, 2012). Therefore, the presence of such antibacterial fungi is desirable in fermented foods or liquors so far food safety is a concern. Traditionally fermented beers are prepared in semi-aseptic conditions, and therefore, chances of harmful bacterial contamination are always there in such conditions. But, the presence of antibacterial fungi in traditional beer starters generally gives us safe beer products to drink. Besides, the presence of some nutritional (amino acids and vitamins) and functional (antioxidants) attributes are reported from some ethnic rice beers which are often contributed by the associated microbes in beer fermentations (Vanderhaegen et al., 2003; Budroni et al., 2017; Callejo et al., 2017). Some microbes are also found to enhance flavor in beer (Budroni et al., 2017).
The ethnic rice beer “jou” (also called “zu” or “zumai”) of the Bodo tribe of the North East India is highly refreshing and generally harmless in regular low dose consumption (Das et al., 2012). The starter culture emao is used traditionally for jou production by the Bodo community and the process of emao and jou preparations are already available somewhere else (Basumatary and Gogoi, 2014; Narzary et al., 2021). Some non-Saccharomyces yeasts were reported from emao but their ethanol-producing capacity is missing (Buragohain et al., 2013; Parashar et al., 2017). Recently, microbial diversity, community structure, carbohydrate-active enzymes, and biofuel-producing enzymes have been reported by Narzary et al. (2021) in a metagenomics study on emao. Much of the microbial roles and their potentialities are yet to be explored from emao to know who does what in beer fermentation at the species or strain level. In this study, we isolated the pure cultures of fungi associated with an emao sample for screening their amylolytic activity and beer-producing attributes.
The traditionally prepared “emao” sample was collected in a sterile container from a local brewer of Rajapanichanda, Rani, Kamrup, Assam, and processed immediately for culturing fungi as described below.
The dried starter culture was crushed into powder with the help of a sterile spatula, and 10 g of it was homogenized in 90 mL of 0.1% phosphate-buffered saline (PBS) for 2–5 min from which serial dilutions (10−1–10−7) were made. Three diluents, i.e., 10−3, 10−5, and 10−7 were used for inoculation in the Sabouraud dextrose agar (SDA) medium. In each 50 mL capacity Petri plate containing 30 mL of the agar medium, 300 μl of a diluent was spread with the help of a glass spreader. SDA medium was supplemented with tetracycline (25 μg/mL working concentration) to stop the bacterial growth. The plates were then incubated at 25 ± 1°C for 3–4 days till the appearance of the colonies. The colony-forming unit (CFU) was counted from three replica plates inoculated with a suitable dilution and the average CFU was calculated from the replicates. Distinct non-overlapping colonies were sub-cultured in fresh agar medium to get the pure cultures. The pure cultures were stored at 4°C in agar slants for routine work, and at −80°C in broth medium amended with 20% glycerol for long time preservation.
The colony and cellular morphology of all the pure culture isolates were studied to group the isolates according to their morphological similarities. Each pure culture was grown in a PDA medium incubating at 25 ± 1°C for 72 h for studying its morphological characteristics. Colony morphology was studied with the help of a stereo-zoom microscope (LM52, Lawrence and Mayo). The thallus structure was stained with lactophenol blue and observed under a trinocular compound microscope (Evos FL Cell Imaging System, Invitrogen). Both morphological and microscopic characters were compared with the characters of species described in Kurtzman and Fell (1998) for identification.
The representative fungal isolates were considered for genomic DNA isolation following the method of Cenis (1992). The nuclear DNA internal transcribed spacer (nrDNA ITS) regions were amplified in a thermal cycler (Mastercycler GSX1, Eppendorf, Germany) using the universal primer pairs: ITS1 (5′-TCCGTAGGTGAACCTGCGG-3′) and ITS4 (5′-TCCTCCGCTTATTGATATGC-3′). The polymerase chain reaction (PCR) contained 1 × PCR master-mix (Bangalore Genei), 0.4 μM each of forward and reverse primers and nuclease-free water in a 25 μl reaction volume. The PCR program was a pre-denaturation at 94°C for 3 min, 35 cycles of denaturation at 94°C for 1 min, annealing at 50°C for 30 s, extension at 72°C for 1 min, followed by a final extension at 72°C for 5 min and preservation at 4°C. The PCR products were separated in 1% agarose gel containing ethidium bromide (0.2 μg/mL) and visualized in a UV transilluminator (UVITEC, Cambridge). The amplified DNA bands were excised and extracted using QIAquick Gel Extraction Kit (QIAGEN) and were sequenced by outsourcing. The DNA sequences were deposited in the NCBI database.
The representative fungal isolates were tested for three different enzyme activities viz. amylase, cellulase, and protease in agar plates following the well-diffusion method. The amylase production and activity were tested in a starch agar medium by following the method of Stolp and Gadkeri (1981). In the case of yeasts, each pure culture isolate was inoculated at three equally distant points in an agar plate, whereas in the case of molds three separate plates were considered and inoculated with inoculum at the center of the plate to avoid the overlapping of colonies due to their vigorous growth. All the inoculated culture plates were incubated at 27 ± 1°C for 72 h and thereafter treated with Lugol's iodine solution for 15 min. Excess iodine was drained off and the culture plates were observed for the formation of a halo zone around the colonies. A colorless halo zone formation was considered amylase positive result, while blue or purple coloration was a negative result.
The cellulase activity of the isolates was tested using a CMC-Bushnell Haas Agar medium by following the method of Singh et al. (2013). The pure culture isolates were inoculated and incubated as described above for the amylase activity test. After 72 h of incubation, 0.3% of Congo red solution was poured over the culture media and kept for 20 min. Thereafter, the excess stain was drained off and the plates were washed with 1 M NaCl solution. The formation of a halo zone around the colonies indicates the cellulase positive result, while its absence indicates the negative result.
The protease enzyme activity was screened in Skim Milk Agar (SM) plates following the method of Atlas (2006). The inoculation and incubation procedure was the same as the amylase test described above. The formation of the halo zone around the colonies indicates protease positive, while the absence of the halo zone indicates the negative result.
The semi-quantitative enzymatic index was calculated for amylase, cellulase, and protease from the following equation (Coronado-Ruiz et al., 2018):
For determining the optimum growth temperature, the representative fungal isolates were inoculated in PDA plates and incubated at five different temperatures, i.e., 25, 30, 35, 40, and 45°C for 3 days. The experiments were set in triplicates for each isolate along with one negative control. The growth performance of the isolates was measured by the colony diameter size. The optimum temperature for growth was determined based on the maximum diameter of the colonies for each isolate.
To determine the optimum growth pH, each representative isolate was inoculated in three separate test tubes containing potato dextrose broth (PDB) medium adjusted to a particular pH. The pH considered were 1.5, 2, 2.5, 3, 3.5,.………..up to 10 for the experiment. A volume of 400 mL PDB was prepared and the pH was adjusted by adding either NaOH (for basic pH) or citric acid (for acidic pH). Experiments were set in 10 mL volume in triplicates for each isolate and three negative controls for each pH without adding inoculum. Actively growing pure culture isolates in PDA plates were used for inoculation in PDB tubes with the help of a sterile inoculating needle. The inoculated tubes were screw-capped and incubated at 30°C for 3 days in an orbital shaker. The tubes were centrifuged at 7,000 rpm for 15 min to collect the biomass and the supernatant was decanted off. The biomass was dried overnight in a hot air oven at 50 ± 2°C. The difference in weight of the isolates showing maximum biomass weight among the different pH was considered to have optimum growth at that pH.
The PDB medium containing different percentages of ethanol ranging from 0 to 16% was prepared for the ethanol tolerance assay of the representative isolates. The setting of experiments, inoculation, incubation, and biomass estimation of the fungal isolates was done as described above for the determination of optimum pH for growth.
The agar well diffusion method as described by Padhi et al. (2016) was followed for this experiment. To extract the crude fungal metabolites, each fungal isolate was grown in a 250 mL capacity Erlenmeyer flask containing 100 mL of sterile Potato dextrose broth (PDB) and incubated at 30°C for 7 days by shaking at 120 rpm in an incubator. The fungal culture was filtered through a Whatman filter paper 1 to remove the mycelial mass. The filtrate was taken in a separating funnel and extracted with an equal volume of ethyl acetate (99.8%) by shaking for 20 min. Two distinct phases appeared when the separating funnel was standstill for a few minutes. The lower aqueous phase containing debris was discarded and the upper phase containing ethyl acetate was collected and transferred to a heating rotary evaporator. The solution was heated at constant temperature (40°C) and pressure (239 mbar) to evaporate the solvent and the leftover dry mass was thereafter dissolved in 1 mL of 10% dimethyl sulfoxide (DMSO) and stored at 4°C for further use.
Antibacterial activity was performed against three bacterial test organisms, viz. Escherichia coli (MTCC 433), Pseudomonas aeruginosa (MTCC 2582), and Staphylococcus aureus (MTCC 9886) were procured from the Institute of Microbial Technology (IMTECH), Chandigarh, India. The test organisms were grown separately in 10 mL of nutrient broth at 35 ± 2°C for 24 h and thereafter 0.2 mL of each bacterial suspension was evenly spread over a fresh nutrient agar plate with the help of a sterile glass spreader. Three agar wells were made at equal distances in each bacterial agar plate with the help of a sterile cork-borer (8 mm in diameter). Each well was then filled with 0.1 mL of crude metabolite extracts of the fungal isolates and the plates were incubated at 35 ± 2°C for another 24 h. The antibacterial activity against the test organisms was determined based on the halo zone formations around the agar wells. DMSO (10%) was used as a negative control along with the antibacterial assay for each fungal extract.
The ethanol-producing capability of the representative fungal isolates from the glucose solution (containing 2% glucose, 1% peptone, 1% yeast extract) and starch solution (containing 2% soluble starch, 1% peptone, 1% yeast extract) was tried for all the representative isolates one by one in a liquid state fermentation. Each isolate was grown in 10 mL of sterile PDB for 12 h at 30 ± 1°C before using 100 μl of it as an inoculum for ethanol production test in 150 mL of glucose or starch solution. Each isolate was set in triplicates along with one negative control for both glucose and starch solution. The inoculated glucose and starch solutions with fungal isolates were incubated at 30 ± 1°C for 7 days; thereafter the fermented solutions were processed immediately for ethanol estimation.
The amylolytic positive fungal strains which produced ethanol from soluble starch were considered for further beer production from rice substrates. The whole experiment was performed in two steps: inoculum preparation and beer fermentation. Each representative isolate was inoculated in a 250 mL Erlenmeyer flask containing 100 mL of sterile PDB and incubated at 30 ± 1°C for 7 days to get a sufficient amount of fungal biomass for the inoculum preparation. After 1 week, the biomass was harvested by centrifugation at 7,000 rpm for 15 min in a 50 mL capacity centrifuge tube. The biomass was washed twice with sterile distilled water by centrifugation. Finally, 2 g (fresh weight) of pure culture pellet was re-suspended in 30 mL of distilled water and homogenized properly before inoculating it to the rice substrates. For beer fermentation, 180 g of dehusked glutinous rice (Oryza sativa var. glutinosa) was washed and baked in a 500 mL capacity borosilicate glass bottle (Borosil) by autoclaving at 15 lbs pressure for 15 min. The experiment was set in triplicates for each isolate. The autoclaved bottles with rice substrates were brought inside a laminar airflow hood and allowed to cool up to ~48°C. The inoculum prepared as above was inoculated to the rice substrate and mixed properly with the help of a sterile glass rod. Thereafter, the fermentation bottles were loosely capped and incubated at 30 ± 1°C for 30 days. The same procedure was followed for all the isolates, except for the isolate Mind_RF25 which took 6 months incubation period to give the beer product. The fermentation progress was monitored every two alternate days.
Rice beers produced by the amylolytic fungal isolates were recovered after 30 days of fermentation, except for Mind_RF25 which was recovered after 180 days due to its slow-growing. All the beer samples were analyzed for the following parameters.
The pH of all the rice beer samples was taken in a calibrated digital pH meter (MK-VI, Systronics).
The concentration of ethanol produced by different fungal strains was determined by following a method described by Pai et al. (2015) with minor modifications as described below. Acid dichromate solution was prepared by dissolving 0.75 g of potassium dichromate in 70 mL concentrated sulphuric acid and adjusting the final volume up to 250 mL with distilled water. Beer samples were diluted at 1:20 with distilled water and 1 mL of each was taken in a sample holder and placed suspended above 10 mL of acid dichromate solution inside a conical flask. The whole system was sealed using a rubber stopper and incubated at 35 ± 2°C overnight. Thereafter, 100 mL distilled water and 1 mL of 20% (w/v) potassium iodide solution was added to the acid dichromate and was titrated with 0.03 mol/L sodium thiosulphate solution using 1% starch solution (w/v) as an indicator until the blue color disappeared. Ethanol concentration was determined using the formula:
where, Vb – the volume of thiosulphate consumed for blank, Vs – the volume of thiosulphate consumed for the sample.
Ethanol concentration was expressed in v/v, conversion made by dividing the concentration in w/v with ethanol's specific gravity at 25°C.
The total phenol estimation of the rice beer samples was determined following the method of Singleton et al. (1999). A volume of 0.02 mL beer sample was diluted with 1.58 mL of double-distilled water, and a volume of 0.1 mL of Folin-Ciocalteu reagent was added to it and mixed properly and kept standing still for 5 min. Thereafter, 0.3 mL of a saturated sodium carbonate solution was added to the mixture and incubated at room temperature for 2 h and the absorbance was taken at 765 nm. Double distilled water was used as a blank, whereas gallic acid of different concentrations was used as standard.
Ascorbate content of beer samples was estimated following the redox titration method as described by Costa et al. (2012). A volume of 25 mL beer sample was subjected to an iodometric titration with 1% iodine solution using 1% starch solution as an indicator. The endpoint was observed by the formation of a blue-black color in the solution which persisted for 10–15 s. Ascorbic acid of different concentrations (0.2–1 mg/mL) was used as standard.
Three types of antioxidant properties viz. DPPH radical scavenging activity (SADPPH), total reducing power (TRP), and total antioxidant activity (TAA) were tested for the beer samples.
The DPPH free radical scavenging activity of beer samples was determined by a method described by Chen et al. (2010). A 5 mL of 0.1 mmol/L DPPH (1,1-diphenyl-2-picrylhydrazyl) prepared in methanol was mixed with a 5 mL aqueous solution containing 150 μL of a beer sample. The sample was incubated at 30°C for 30 min in the dark and the absorbance was measured at 517 nm wavelength. An ascorbic acid solution (0.2 mg/mL) was used as a standard. SADPPH was expressed in percentage as below:
The reducing power of the beer samples was determined using a method as described by Bhalodia et al. (2013). A 1 mL volume of 10 times diluted beer sample was mixed subsequently with 2.5 mL of phosphate buffer (0.2 M, pH 6.6) and 2.5 mL of potassium ferricyanide (1% w/v). The mixture was incubated in a pre-set water bath at 50°C for 20 min. Thereafter, it was cooled to room temperature, 2.5 mL of trichloroacetic acid (10% w/v) was added, and then the mixture was centrifuged at 3,000 rpm for 10 min. The upper phase of the solution was extracted and 2.5 mL of it was mixed with 2.5 mL of distilled water and 0.5 mL of ferric chloride (0.1%) solution. The absorbance of the mixture was taken at 700 nm. Ascorbic acid was used as standard. The higher the absorbance, the higher is the reducing potential of a sample.
TAA was determined following the phosphomolybdate assay as described by Prieto et al. (1999) with some minor modifications. A 0.3 mL of beer sample was mixed with 3 mL of phosphomolybdate reagent containing 0.6 M sulphuric acid, 20 mM sodium phosphate, and 4 mM ammonium molybdate in a test tube and incubated at 90°C for 90 min. After incubation, the mixture was cooled to room temperature and the absorbance was taken at 765 nm wavelength in a spectrophotometer. Ascorbic acid (0.2 mg/mL) was used as a standard. A standard curve was prepared using different concentrations of ascorbic acid in the range of 20 mg/L to 500 mg/L. The antioxidant activity of beer samples was expressed in μg ascorbic acid equivalent (AAE)/mL.
The average CFU calculated for three agar plates inoculated with 10−5 diluents was 13.53 × 107 per gram emao sample. A total of 50 fungal isolates were randomly recovered from the mixed culture plates that were sub-cultured to get the pure cultures. The pure culture isolates were segregated into 11 distinct groups based on their morphology (Table 1, Supplementary Data S1). One representative isolate from each group was considered for nrDNA ITS sequencing and DNA sequence homology search in the NCBI database. The representative isolates were identified as Candida glabrata (Cgla_RF2), Cyberlindnera fabianii (Cfab_RF37), Hyphopichia burtonii (Hbur_RF19), Mucor circinelloides (Mcir_RF48), Mucor indicus (Mind_RF25), Penicillium citrinum (Pcit_RF32), Rhodosporidiobolus ruineniae (Rrui_RF4 & Rrui_RF43), Saccharomyces cerevisiae (Scer_RF6), Saccharomycopsis fibuligera (Sfib_RF11), and Wickerhamomyces anomalus (Wano_RF3). In a relative abundance (RA) analysis of a total of 50 isolates, W. anomalus was found dominant with 24% RA followed by C. glabrata and H. burtonii with 16% RA each; whereas M. circinelloides, M. indicus, and P. citrinum were the least with only 2% RA (Figure 1).
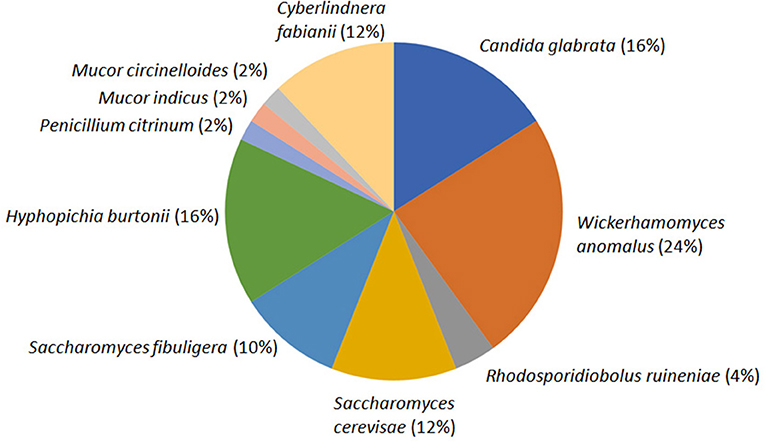
Figure 1. Species diversity and relative abundance of culturable fungi as recorded in the beer starter “emao.” Values within parentheses indicate the respective relative abundance of the species.
All the yeast isolates (Cgla_RF2, Hbur_RF19, Rrui_RF4, Rrui_RF43, Scer_RF6, Sfib_RF11, and Wano_RF3), except Cfab_RF37, were showed optimum growth at 30°C, whereas all three mold isolates (Mind_RF25, Mcir_RF48, and Pcit_RF32) showed optimum growth at 35°C like Cfab_RF37 (Table 2, Supplementary Data S2). The isolates Rrui_RF4 and Rrui_RF43 did not grow beyond 30°C, whereas the rest of the yeast isolates did not grow beyond 35°C, in contrast to the molds that also grew at 45°C.
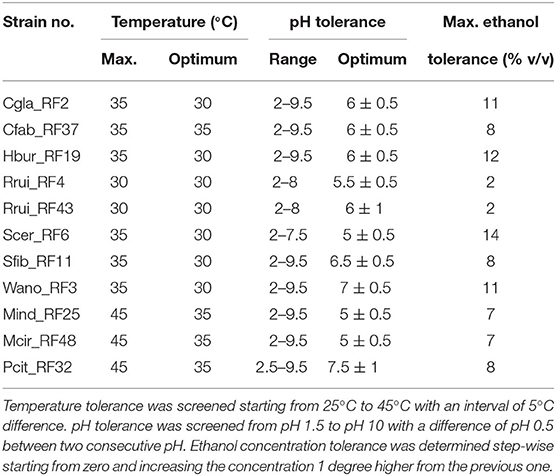
Table 2. Range of pH, maximum temperature, and maximum ethanol tolerance to the fungal isolates obtained from emao.
The optimum growth pH and the pH tolerance range varied among the isolates. The growth was found optimum at pH 5 ± 0.5 for Scer_RF6, Mind_RF25, and Mcir_RF48; at pH 5.5 ± 0.5 for Rrui_RF4; at pH 6 ± 0.5 for Cgla_RF2, Cfab_RF37, and Hbur_RF19; at pH 6.5 ± 0.5 for Sfib_RF11; at pH 7 ± 0.5 for Wano_RF3; and at pH7.5 for Pcit_RF32 (Table 2, Supplementary Data S3). The pH tolerance range also varied among the isolates, i.e., pH 2–7.5 in the case of Scer_RF6, pH 2–8 in the case of Rrui_RF4, and Rrui_RF43, pH 2.5–9.5, and pH 2–9.5 in the rest of the isolates. None of the isolates grew at pH 1.5 and 10.
In the ethanol tolerance assay, Scer_RF6 was found tolerant to as high as 14% (v/v) ethanol, followed by Hbur_RF19 (12%), Cgla_RF2 (11%), and Wano_RF3 (11%) in decreasing order (Table 2, Supplementary Data S4). Rrui_RF4 and Rrui_RF43 showed the least tolerance to ethanol, i.e., only up to 2% ethanol.
Of the 11 representative fungal isolates considered for amylase, protease, and cellulase activity test, five isolates (Hbur_RF19, Sfib_RF11, Mind_RF25, Mcir_RF48, and Pcit_RF32) exhibited amylase positive, only three isolates (Hbur_RF19, Sfib_RF11, and Pcit_RF32) exhibited protease positive, and only one isolate (Pcit_RF32) exhibited cellulase positive (Table 3). The amylase activity exhibited by Sfib_RF11 was the highest with an EI of 2.9 ± 0.02 among the amylase-positive isolates.
Nine of eleven isolates produced ethanol from glucose substrates with varied concentrations (Table 4). The isolates Rrui_RF4 and Rrui_RF43 did not produce ethanol from glucose substrates. The ethanol produced by Scer_RF6 was the highest (11.45 ± 0.11% v/v) followed by Hbur_RF19 (9.43 ± 007%) and Mcir_RF48 (8.05 ± 0.07%), respectively, after 1 week of liquid-state fermentation. Five of nine isolates (Hbur_RF19, Sfib_RF11, Mind_RF25, Mcir_RF48, and Pcit_RF32) produced ethanol from soluble starch in liquid state fermentation. The concentration of ethanol was observed slightly lower in the beer produced from starch than in the beer produced from glucose for all the ethanol-producing isolates from both the substrates (Table 4).
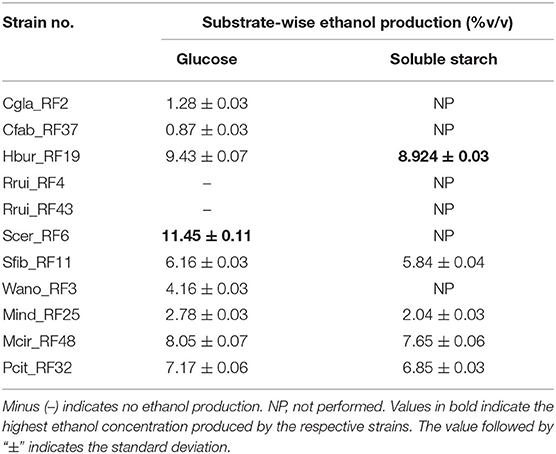
Table 4. Ethanol production assay with the fungal isolates using glucose and soluble starch as substrates.
The crude metabolic extract of all the representative fungal isolates showed antibacterial activity against E. coli (MTCC 433) and S. aureus (MTCC 9886) with inhibition zones in the range of 11–32 mm and 9–35 mm, respectively, in agar well diffusion plates (Figure 2). On the other hand, only eight fungal isolates (Cgla_RF2, Hbur_RF19, Rrui_RF4, Rrui_RF43, Sfib_RF11, Mind_RF25, Mcir_RF48, and Pcit_RF32) exhibited antibacterial activity against P. aeruginosa (MTCC 2582). The antibacterial activities exhibited by Mcir_RF48, Pcit_RF32, and Hbur_RF19 to all the test organisms were noteworthy among the fungal isolates considered in this study. The three isolates (Cfab_RF37, Scer_RF6, and Wano_RF3) that showed no inhibition to P. aeruginosa (MTCC 2582) also showed weak inhibition zone to the other test organisms, E. coli (MTCC 433) and S. aureus (MTCC 9886) (Figure 2).
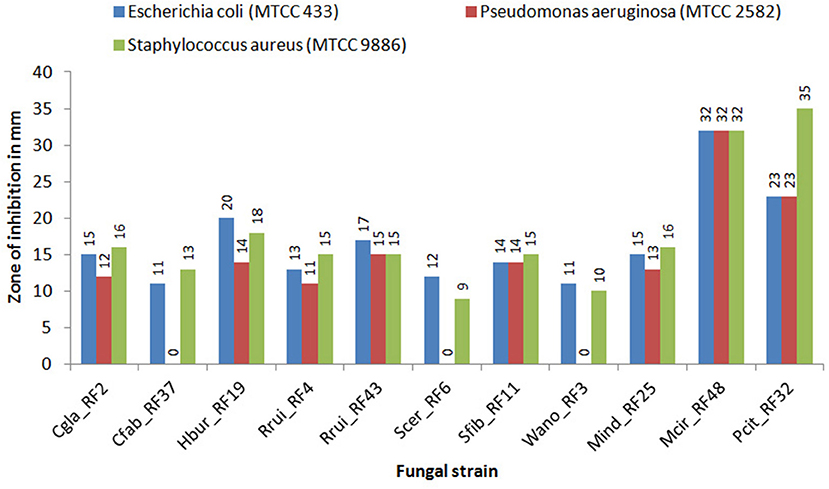
Figure 2. Antibacterial activity of the fungal isolates against test bacteria. Experiments were set in triplicate for each fungus-bacterium pair. The average zone of inhibitions was having a standard error of <1 mm in each case, this is not shown here.
The pH of the rice beers was almost the same, i.e., pH 3.54 ± 0.2 in all the samples. After 30 days of rice beer fermentation, the ethanol concentration in the beer produced by Hbur_RF19 was the highest (7.3 ± 0.09% v/v) followed by Mcir_RF48, Pcit_RF32, and Sfib_RF11 (in decreasing order) (Table 4). The isolate Mind_RF25 did not produce rice beer after 30 days of the fermentation period, but after 6 months of the fermentation period, the ethanol concentration was 3.17 ± 0.06% (v/v) which was the least among the isolates. The fungal isolates produced beer containing ascorbic acids, total phenol, SADPPH, TRP, and TAC in the range of 15.26–31.1 mg/100 mL, 186.66–241.33 mg/L, 56.85–82.84%, 0.173–0.396 absorbance at A700, and 0.174–0.364 g AAE/L, respectively (Table 5). The ascorbic acid, total phenol, and antioxidant properties of the beer produced by Hbur_RF19 were higher, except for SADPPH than the beers produced by the other four isolates. The SADPPH of the beer produced by Mcir_RF48 was the highest (82.84 ± 0.27%) followed by the beers produced by Pcit_RF32, Mind_RF25, Sfib_RF11, and Hbur_RF19 in decreasing order.
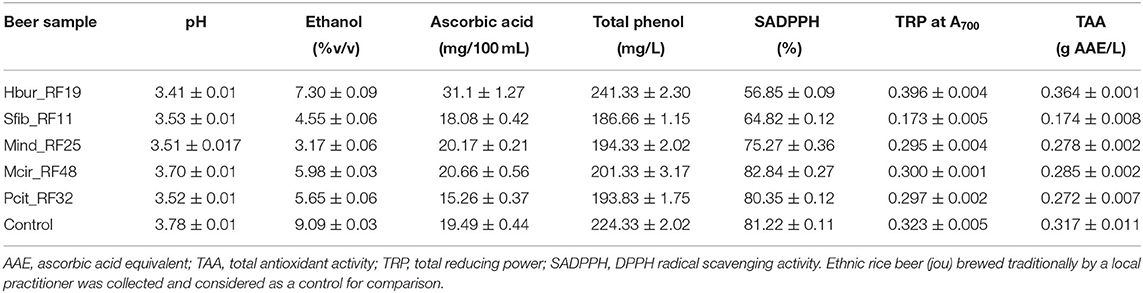
Table 5. Biochemical parameters of rice beer produced by the amylolytic fungal strains after 30 days of the fermentation period, except for Mind_RF25 that produced beer after 6 months of the fermentation period.
In a Pearson correlation analysis, ethanol content was found positively correlated to ascorbic acid, total phenol, total reducing power (TRP), and total antioxidant activity (TAA) of the beers (Table 6). Of the three antioxidant properties, TRP and TAA showed a positive correlation to each other, whereas SADPPH was found negatively correlated to TRP and TAA. Ascorbic acid and total phenol showed a positive correlation to each other as well as to TRP and TAA, but a negative correlation to SADPPH of the beer. The pH was found positively correlated only to SADPPH.
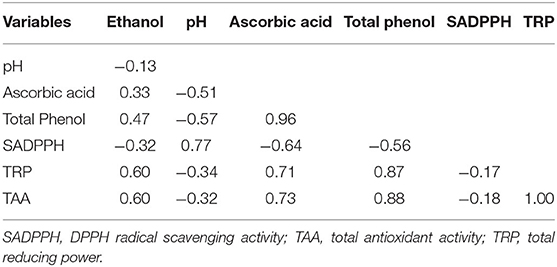
Table 6. Pearson correlation of the biochemical parameters of the rice beers produced by five amylolytic fungal strains individually.
In a PCA biplot (Figure 3) of biochemical parameters of beer samples that produced by the fungal trains, two strains (Mcir_RF48 and Pcit_RF32) with higher SADPPH producing in rice beer appeared together in a quarter, whereas Hbur_RF19 producing higher ascorbic acid, total phenol, TRP, and TAA in beer appeared separately in another quarter of the biplot. Similarly, Sfib_RF11 and Mind_RF25 appeared separately in another quarter of the biplot. The positively correlated biochemical parameters of the beer (ascorbic acid, ethanol, total phenol, TAA, and TRP appeared together in the F1 coordinate contributing 65.83% variability, whereas the negatively correlated parameters (pH and SADPPH) appeared in the F2 coordinate contributing 20.70% variability in the biplot. In one-way ANOVA, the p-value was >0.05, thus accepting the null hypothesis that the beers produced by all the five fungal isolates had no significant difference from each other (Supplementary Data S5).
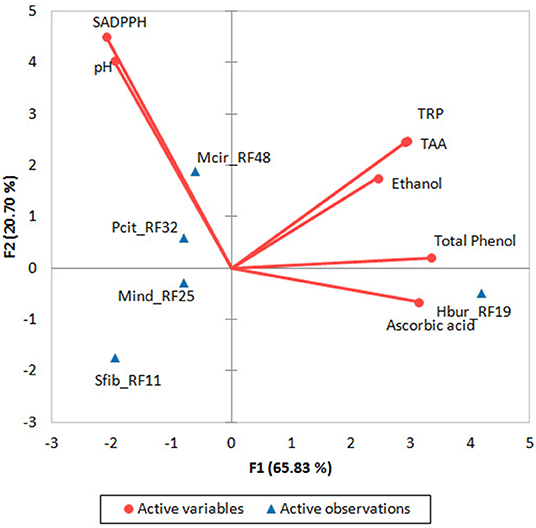
Figure 3. PCA biplot for the rice beer-producing fungal isolates (Hbur_RF19, Sfib_RF11, Mind_RF25, Mcir_RF48, and Pcit_RF32) and their beer properties. SADPPH, DPPH radical scavenging activity; total antioxidant activity; TRP, total reducing power.
Molds, yeasts, and bacteria are the groups of microorganisms mostly reported from the traditional beer starters of the Asian region. But the microbial species reported in different beer starters are often found different from one another which might be due to differences in cultural practices and ingredients used. Besides, strain level difference within a species is a common phenomenon that encounters even in the same starter.
The fungal species C. glabrata, C. fabianii, H. burtonii, M. circinelloides, M. indicus, P. citrinum, S. cerevisiae, S. fibuligera, and W. anomalus recorded in emao were also reported earlier from other traditional beer starters viz. apop, balma, dawdim, hamei, humao, khekhrii, marcha, thempra, and thiat from India (Teramoto et al., 2002; Buragohain et al., 2013; Bharadwaj et al., 2015; Parashar et al., 2017; Sha et al., 2018; Anupama and Tamang, 2020); banh-men from Vietnam (Dung et al., 2007); daku from China (Shi et al., 2009; Zheng et al., 2012); loogpang from Thailand (Taechavasonyoo et al., 2013), and nuruk from Korea (Bal et al., 2014). However, R. ruineniae reported from emao in this study was not reported earlier. Both the strains of R. ruineniae were found non-amylolytic, non-proteolytic, non-cellulolytic, and non-ethanol producing with a low ethanol tolerance, i.e., up to 2% (v/v) only in this study (Tables 3, 4). Thus, their function in beer fermentation is unclear; rather they can be the opportunistic microbe that came by chance from the herbs used in the beer starters. It was earlier reported as an endophyte from several plant species (Santiago et al., 2017).
In ethnic beer fermentations, dozens of naturally occurring microorganisms with unknown species information generally take part in beer fermentation in contrast to the use of only the known and well-characterized microbes in beer industries. As the metabolic potentials of microbes vary greatly from species to species or strain to strain, it is necessary to characterize and understand each microbial species/strain for its industrial uses. Low-ethanol beer fortified with health-beneficial components is in demand nowadays since the beer with high ethanol content and also less or without nutritional components is injurious to health, and can cause irreparable harm to the regular consumers. The shooting taste of most of the ethnic beers is due to their moderate to high ethanol content along with high organic acids. The flavor and aroma of ethnic beer vary due to the use of different substrates, herbal ingredients, and different cultural practices.
The naturally associated microorganisms that are either maintained through the traditional practices or originated from the ingredients used might play major roles in beer fermentations as they can perform some specific activities like the production of ethanol, enzymes, organic acids, antimicrobials, vitamins, amino acids, flavor compounds, etc. However, strains vary greatly in their activity and performance as evident from several studies including the present one. For example, the amylase activity index of different fungal strains was in the range of 1.21–2.09, whereas for protease it was in the range of 1.2–1.6 among the isolates in this study (Table 3). Accordingly, the ethanol concentrations of the beer produced by the pure culture of amylolytic fungi were in the range of 3.17–7.3% (v/v) in this study. It is noteworthy to mention that the ethanol concentration of rice beer “jou” fermented with the traditional starter “emao” of the Bodo community usually contains about 12–19% (v/v) ethanol (Das et al., 2019). Earlier findings of lower ethanol production (Hashem et al., 2021) and lower ethanol tolerance (Yamaoka et al., 2014; Garcia et al., 2021) of some yeast while growing individually corroborate our present findings of the low ethanol tolerance and low ethanol production of the fungal isolates that were grown separately on rice substrates (Tables 2, 5). Besides, the presence of a good amount of ascorbic acid, total phenols, and antioxidant activities in the beer produced by the fungal strains are important from the nutritional point of view. Polyphenols contribute to antioxidant properties, health-promoting qualities, stability, and foam retention in beer (Callemien and Collin, 2009). The presence of ascorbic acid in beer increases the preservation or shelf-life of beer or wine by preventing the oxidation of beer (Jeney-Nagymate and Fodor, 2007). Therefore, immense scope lies for new product development, improvement, or value addition to the existing products through the selection and harnessing of such potential microbes having specific functions not only in beer industries but also in other food and nutraceutical industries.
S. cerevisiae is well-known for producing high ethanol concentrations and is, therefore, industrially exploited for bio-ethanol production. Wild S. cerevisiae recovered from fermented beverages can produce more than 10% (v/v) ethanol from simple carbohydrates but not directly from starch (Tesfaw et al., 2021) which is also evident from this study. But, those wild strains of S. cerevisiae can tolerate and produce higher concentrations of ethanol if they grow in association with the lactic acid bacteria (Senne de Oliveira Lino et al., 2021). Both high ethanol producing and low ethanol-producing microorganisms are reported from several ethnic beer starters. Low ethanol-producing microbes are gaining importance for low-ethanol-containing beers, in contrast to high-ethanol producing microbes that are in demand for bio-ethanol production in industries.
The ethanol produced by fungi is of economic importance to the people. In our study, we reported the ethanol-producing capacity of the amylolytic fungi and non-amylolytic fungi in the soluble starch, glucose, and rice substrate. Hbur_RF19 showed highest ethanol production (7–8% v/v) in the soluble starch (Table 4) and rice substrates (Table 5) among the amylolytic fungi followed by Mcir_RF48 (5–7% v/v), Pcit_RF32 (5–6% v/v), and Sfib_RF11 (4–5% v/v) whereas Scer_RF6 showed the highest ethanol content of 11.45% (v/v) in glucose among the non-amylolytic fungi followed by Wano_RF3 (4.16%), Cgla_RF2 (1.28%), and Cfab_RF37 (0.87%). Tsuyoshi et al. (2005) reported the presence of S. fibuligera and H. burtonii in traditional starter culture “Marcha” that has no ethanol-producing capacity. Satari et al. (2015) reported the production of ethanol (0.4 g/L) from pretreated lignocellulose by Mucor indicus. The ethanol production by Candida glabrata, Cyberlindnera fabianii, Saccharomyces cerevisiae, and Wickerhamomyces anomalus from different carbohydrate sources was reported earlier (Van Rijswijck et al., 2017; Chang et al., 2018; Atitallah et al., 2020; Vincent et al., 2021).
Antioxidants such as phenols and ascorbic acids play an important role in health-promoting factors such as scavenging free radicals, anti-inflammatory and also to the regulation of pathogens and stability of beer. In our study, the total polyphenol in the beer samples ranged from 186 to 241 mg/L, which was higher if compared to 47–60 mg/L polyphenol of Cambodian rice wine (Chay et al., 2020), 3.36–9.63 mg/L polyphenol of Brazilian beer (Moura-Nunes et al., 2016), 140–157 mg/L polyphenol of Xaj (Handique et al., 2020), and 138–148 mg/L polyphenol of Apong (Handique et al., 2020). Ambra et al. (2021) reported that the content and concentration of polyphenols depend on the use of a variety of substrates (hops or barley) for beer production. The ascorbic acid content in beer was in the range of 15.26–31.1 mg/100 mL in the present study, which is comparable to the ascorbic acid content of grape wine (15–16 mg/100 mL) and orange juice (39–41 mg/100 mL) (Pisoschi et al., 2011).
We found that all the fungal species considered in the present study were tolerant to ethanol in the range of 7–14% (v/v) ethanol, except R. ruineniae. But such ethanol tolerance did not support that all of them are good ethanol producers because the concentrations of ethanol produced by Cgla_RF2, Cfab_RF37, and Mind_RF25 were <3% (v/v). Crude metabolite extracts of low ethanol producing strains including Wano_RF3 showed lesser inhibition than the crude extracts of higher ethanol producing strains to the test organisms. In contrast, the non-ethanol producing strains Rrui_RF4 and Rrui_RF43 exhibited antibacterial activity as good as the ethanol-producing strains (Figure 2). Such fungi having antibacterial activity against pathogenic bacteria could be useful for maintaining the quality of beer starters, and thus their presence could be the quality indication in beer fermentations.
In the present study, only a subsection of yeast and mold isolates produced ethanol from rice substrates. Six yeast isolates produced ethanol from glucose, whereas only two of them produced ethanol from starch (Table 4). In contrast, all the three mold isolates (Mind_RF25, Mcir_RF48, and Pcit_RF32) produced ethanol from both glucose and starch. This is the first time to document Penicillium citrinum (Pcit_RF32) as an ethanol producing from glucose and starch substrates. Six of 11 fungal isolates were found non-amylolytic, thus are likely to depend on the amylolytic microbes when starch is used as the sole carbon source in beer fermentations. Such microbial interaction, cross-feeding, and succession are the common phenomena that occur in all-natural ecosystems including ethnic beer fermentations. Therefore, the understanding of the functional roles of each microbial species is important for process manipulation and product improvements in fermentation biology.
Certain physiological conditions of ethanol production such as Crabtree and Pasteur effects (Moulin et al., 1984; Panikov, 2011; Pfeiffer and Morley, 2014; Hagman and Piškur, 2015) are well-studied and documented in yeasts. However, such physiological study of ethanol production by filamentous fungi is limited although there are several reports of ethanol production by filamentous fungi from different substrates. For example, Mucor circinelloides is a Crabtree-positive organism and can produce ethanol in both aerobic and anaerobic conditions (Lubbehusen et al., 2004). Accordingly, M. indicus and Rhizopus oryzae can produced ethanol anaerobically from enzyme hydrolysate of pure cellulose and acid pre-treated hydrolysate of rice straw (Abedinifar et al., 2009). M. indicus (Sharifia et al., 2008), Mucor hiemalis (Esmaeili and Keikhosro, 2018), Aspergillus flavus, A. niger, A. oryzae, and A. terreus (Liaud et al., 2014) were reported to produce ethanol from glucose. Other molds such as Amylomyces rouxii, R. oryzae, R. oligosporus, R. stolonifer was reported to produce ethanol from cooked rice (Dung et al., 2006; Song et al., 2018). Fusarium oxysporum (Ruiz et al., 2007), F. verticilloides (De Almeida et al., 2013) and F. oxysporum f. sp. lycopersici (Ueng and Gong, 1982) were also reported to produce ethanol from glucose, sucrose and xylose. Therefore, detail physiological study is required to understand the mechanism of ethanol production in filamentous fungi which would help us better understand the precise mechanism of traditional beer/wine fermentations.
During beer fermentation, the growth of many pathogenic and unwanted organisms could be arrested by several factors like ethanol, pH, lack of oxygen, low nutrient availability, and antimicrobial effects (Menz et al., 2011). However, some of the undesirable and pathogenic bacteria can survive for a prolonged period at a different range of temperatures and pH (Kim et al., 2014). The presence of such unwanted microbes in beer is a concern because such microbes can spoil the beer products mostly with undesirable flavor (Vriesekoop et al., 2013).
Fungi are reported to produce various secondary metabolites or bioactive compounds which inhibit the growth of other microorganisms. In beverage fermentations, the antagonistic effect of fungi against other microbes is due to the production of antimicrobial compounds, competition for nutrients, ethanol stress, or changes in pH (Marquina et al., 2002; Gil-Rodriguez and Garcia-Gutierrez, 2020). In the antibacterial activity assay of the fungal isolates against the three bacterial test organisms (E. coli MTCC 433, P. aeruginosa MTCC 2582, and S. aureus MTCC 9886), MCir_RF48 showed the highest inhibition against the Gram-negative bacteria whereas Pcir_RF32 showed the highest inhibition against the Gram-positive bacteria in the present study. Other than molds, the yeast Hbur_RF19 showed high inhibition against E. coli and S. aureus whereas Rrui_RF43 showed high inhibition against P. aeruginosa. Fakruddin et al. (2017) reported the antibacterial activity of S. cerevisiae against E. coli (8.7 mm), S. aureus (4.9 mm), and P. aeruginosa (9.1 mm). In contrast, S. cerevisiae showed antibacterial activity against only two organisms E. coli (12 mm) and S. aureus (9 mm) in the present study. Takahashi et al. (2008) reported Penicillium citrinum from soil having no antibacterial activity against the three test organisms, but we found it showing antibacterial activity which could be due to the strain differentiation. Mohammad et al. (2020) also reported the antibacterial activity of Mucor circinelloides against E. coli (13 mm) and S. aureus (15 mm). However, reports on the antibacterial activity of yeasts are meager. The antibacterial activity of C. glabrata, C. fabianii, H. burtonii, M. indicus, M. circinelloides, S. fibuligera, and R. ruineniae against E. coli, P. aeruginosa, and S. aureus have been reported for the first time in our study. Some bacteria are also reported to grow as contaminants during fermentation which can affect the growth of beneficial yeasts and ethanol production (Thomas et al., 2001; Skinner and Leathers, 2004). Watanabe et al. (2008) reported the fermentation of ethanol with the co-culture of microbes with antimicrobial activity that inhibits the contaminants without affecting the production of ethanol. Therefore, the knowledge of the specific role of each microbial species is of paramount significance for microbial resource management and utilization in fermentation biology.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
DN conceptualized the research project. NB did the experimental works. DN and NB jointly analyzed the data and wrote the manuscript. All authors contributed to the article and approved the submitted version.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The authors acknowledge the Department of Botany, Gauhati University for providing necessary facilities. DN is thankful to the DST-SERB, Govt. of India, for research funding (SB/EMEQ-443/2014). NB is thankful to the UGC, Govt. of India for the research fellowship. The authors thank Ms. Deepa Kachari who provided the emao samples and the relevant information related to their brewing tradition.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fsufs.2022.869430/full#supplementary-material
Abedinifar, S., Karimi, K., Khanahmadi, M., and Taherzadeh, M. J. (2009). Ethanol production by Mucor indicus and Rhizopus oryzae from straw by separate hydrolysis and fermentation. Biomass Bioenergy 33, 828–833. doi: 10.1016/j.biombioe.2009.01.003
Ambra, R., Pastore, G., and Lucchetti, S. (2021). The role of bioactive phenolic compounds on the impact of beer on health. Molecules 26, 486. doi: 10.3390/molecules26020486
Anupama, A., and Tamang, J. P. (2020). Diversity of filamentous fungi isolated from some amylase and alcohol producing starters of India. Front. Microbiol. 11, 1–16. doi: 10.3389/fmicb.2020.00905
Atitallah, I. B., Ntaikou, I., Antonopoulou, G., Alexandropoulou, M., Brysch-Herzberg, M., Nasri, M., et al. (2020). Evaluation of the non-conventional yeast strain Wickerhamomyces anomalus (Pichia anomala) X19 for enhanced bioethanol production using date palm sap as renewable feedstock. Renew. Energy 154, 71–81. doi: 10.1016/j.renene.2020.03.010
Atlas, R. (2006). Handbook of Microbiological Media for the Examination of Food, 2nd Edn. Boca Raton, FL: CRC Press; Taylor and Francis Group.
Bal, J., Yun, S. H., Song, H. Y., Yeo, S. H., Kim, J. H., Kim, J. M., et al. (2014). Microflora dynamics analysis of Korean traditional wheat-based nuruk. J. Microbiol. 52, 1025–1029. doi: 10.1007/s12275-014-4620-0
Basumatary, M., and Gogoi, M. (2014). A traditional alcoholic beverage Jou: prepared by Bodo community of Assam, India. Int. J. Multidiscip. Res. Dev. 1, 307–313. Available online at: http://www.allsubjectjournal.com/vol1/issue7/PartH/pdf/36.2.pdf
Bhalodia, N. R., Nariya, P. B., Acharya, R. N., and Shukla, V. J. (2013). In vitro antioxidant activity of hydro alcoholic extract from the fruit pulp of Cassia fistula Linn. Ayu 34, 209–214. doi: 10.4103/0974-8520.119684
Bharadwaj, K. N., Jain, K. K., Kumar, S., and Kuhad, R. C. (2015). Microbiological analysis of traditional alcoholic beverage (Chhang) and its starter (Balma) prepared by the Bhotia tribe of Uttrakhand, India. Indian J. Microbiol. 56, 28–34. doi: 10.1007/s12088-015-0560-6
Bora, S. S., Keot, J., Das, S., Sharma, K., and Borooah, M. (2016). Metagenomic analysis of microbial communities associated with a traditional rice wine starter culture (Xaj-pitha) of Assam, India. 3 Biotech. 6, 1–13. doi: 10.1007/s13205-016-0471-1
Budroni, M., Zara, G., Ciani, M., and Comitini, F. (2017). Saccharomyces and Non-Saccharomyces Starter Yeasts. London: Brewing Technology, Makoto Kanauchi, IntechOpen. doi: 10.5772/intechopen.68792
Buragohain, A. K., Tanti, B., Sharma, H. K., Barman, P., and Das, K. (2013). Characterization of yeast starter cultures used in household alcoholic beverage preparation by a few ethnic communities of North East India. Ann. Microbiol. 63, 863–869. doi: 10.1007/s13213-012-0537-1
Callejo, M. J., Gonzalez, C., and Morata, A. (2017). Use of Non-Saccharomyces Yeasts in Bottle Fermentation of Aged Beers. London: Brewing Technology, Makoto Kanauchi, IntechOpen. doi: 10.5772/intechopen.68793
Callemien, D., and Collin, S. (2009). Structure, organoleptic properties, quantification methods and stability of phenolic compounds in beer-A review. Food Rev. Int. 26, 1–84. doi: 10.1080/87559120903157954
Carroll, E., Trinh, T. N., Son, H., Lee, Y. W., and Seo, J. A. (2017). Comparative analysis of fungal diversity and enzyme activity in Nuruk, a Korean fermenting starter, for acquiring useful fungi. J. Microbiol. 55, 357–365. doi: 10.1007/s12275-017-7114-z
Cenis, J. L. (1992). Rapid extraction of fungal DNA for PCR amplification. Nucleic Acids Res. 20, 2380. doi: 10.1093/nar/20.9.2380
Chang, Y. H., Chang, K. S., Chen, C. Y., Hsu, C. L., Chang, T. C., and Jang, H. D. (2018). Enhancement of the efficiency of bioethanol production by Saccharomyces cerevisiae via gradually batch-wise and fed-batch increasing the glucose concentration. Fermentation 4, 45. doi: 10.3390/fermentation4020045
Chay, C., Dizon, E. I., Hurtada, W. A., Elegado, F. B., Norng, C., and Raymundo, L. C. (2020). Total phenolic content and antioxidant activity of rice wine from waxy pigmented and non-pigmented rice varieties produced by traditional and multi-parallel fermentation. Food Res. 4, 199–206. doi: 10.26656/fr.2017.4(1).036
Chen, B., Wu, Q., and Xu, Y. (2014). Filamentous fungal diversity and community structure associated with the solid-state fermentation of Chinese Maotai-flavor liquor. Int. J. Food Microbiol. 179, 80–84. doi: 10.1016/j.ijfoodmicro.2014.03.011
Chen, L. S., Ma, Y., Chen, L. J., Zhao, C. H., Maubois, J. L., Jiang, T. M., et al. (2010). Antioxidant activity of two yeasts and their attenuation effect on 4-nitroquinoline 1-oxide induced in vitro lipid peroxidation. Int. J. Food Sci. Technol. 45, 555–561. doi: 10.1111/j.1365-2621.2009.02165.x
Coronado-Ruiz, C., Avendano, R., Escudero-Leyva, E., Conejo-Barboza, G., Chaverri, P., and Chavarria, M. (2018). Two new cellulolytic fungal speies isolated from a 19th-century art collection. Sci. Rep. 8, 7492. doi: 10.1038/s41598-018-24934-7
Costa, A. S. G., Nunes, M. A., Almeida, I. M. C., Carvalho, M. R., Barroso, M. F., Alves, R. C., et al. (2012). Teas, dietary supplements and fruit juices: a comparative study regarding antioxidant activity and bioactive compounds. LWT Food Sci. Technol. 49, 324–328. doi: 10.1016/j.lwt.2012.02.030
Das, A. J., Deka, S. C., and Miyaji, T. (2012). Methodology of rice beer preparation and various plant materials used in starter culture preparations by some tribal communities of North-East India: a survey. Int. Food Res. J. 19, 101–107. Available online at: http://www.ifrj.upm.edu.my/19%20(01)%202011/(14)IFRJ-2011-137%20Deka.pdf
Das, S., Deb, D., Adak, A., and Kham, M. R. (2019). Exploring the microbiota and metabolites of traditional rice beer varities of Assam and their functionalities. 3 Biotech 9, 174. doi: 10.1007/s13205-019-1702-z
De Almeida, M. N., Guimaraes, V. M., Falkoski, D. L., Visser, E. M., Siqueira, G. A., Milagres, A. M. F., et al. (2013). Direct ethanol production from glucose, xylose and sugarcane bagasse by the corn endophytic fungi Fusarium verticilloides and Acremonium zeae. J. Biotechnol. 168, 71–77. doi: 10.1016/j.jbiotec.2013.07.032
Dung, N. T. P., Rombouts, F. M., and Nout, M. J. R. (2006). Functionality of selected strains of moulds and yeasts from Vietnamese rice wine starters. Food Microbiol. 23, 331–340. doi: 10.1016/j.fm.2005.05.002
Dung, N. T. P., Rombouts, F. M., and Nout, M. J. R. (2007). Characteristics of some traditional Vietnamese starch-based rice wine fermentation starters (men). LWT Food Sci. Technol. 40, 130–135. doi: 10.1016/j.lwt.2005.08.004
Esmaeili, H., and Keikhosro, K. (2018). Optimization of fermentation conditions for efficient ethanol production by Mucor hiemalis. Turk. J. Biochem. 43, 587–594. doi: 10.1515/tjb-2017-0290
Fakruddin, M., Hossain, M. N., and Ahmed, M. M. (2017). Antimicrobial and antioxidant activities of Saccharomyces cerevisiae IFST062013, a potential probiotic. BMC Complement Altern. Med. 17, 1–11. doi: 10.1186/s12906-017-1591-9
Garcia, M., Crespo, J., Cabellos, J. M., and Arroyo, T. (2021). Growth of non-saccharomyces native strains under different fermentative stress conditions. Fermentation 7, 124. doi: 10.3390/fermentation7030124
Gil-Rodriguez, A. M., and Garcia-Gutierrez, E. (2020). Antimicrobial Mechanisms and Applications of Yeasts. Cambridge, MA: Academic Press.
Hagman, A., and Piškur, J. (2015). A study on the fundamental mechanism and the evolutionary driving forces behind aerobic fermentation in yeast. PLoS ONE 10, e0116942. doi: 10.1371/journal.pone.0116942
Handique, P., Deka, A. K., and Deka, D. C. (2020). Antioxidant properties and phenolic contents of traditional rice-based alcoholic beverages of Assam, India. Natl. Acad. Sci. Lett. doi: 10.1007/s40009-020-00903-5
Hao, F., Tan, Y., Lv, X., Chen, L., Yang, F., Wang, H., et al. (2021). Microbial community succession and its environment driving factors during initial fermentation of Maotai-flavor Baijiu. Front. Microbiol. 12, 669201. doi: 10.3389/fmicb.2021.669201
Hashem, M., Alamri, S. A., Asseri, T. A. Y., Mostafa, Y. S., Lyberatos, G., and Ntaikou, I. (2021). On the optimization of fermentation conditions for enhanced bioethanol yields from starchy biowaste via yeast co-cultures. Sustainability 13, 1890. doi: 10.3390/su13041890
Hui, W., Hou, Q., Cao, C., Xu, H., Zhen, Y., Kwok, L. Y., et al. (2017). Identification of microbial profile of Koji using single molecule, real-time sequencing technology. J. Food Sci. 82, 1193–1199. doi: 10.1111/1750-3841.13699
Jeney-Nagymate, E., and Fodor, P. (2007). Examination of the effect of vitamin E and C addition on the Beer's lag time parameter. J. Inst. Brew. 113, 28–33. doi: 10.1002/j.2050-0416.2007.tb00252.x
Kim, S. A., Kim, N. H., Lee, S. H., Hwang, I. G., and Rhee, M. S. (2014). Survival of foodborne pathogenic bacteria (Bacillus cereus, Escherichia coliO157:H7, Salmonella enterica, Serovar typhimurium, Staphylococcus aureus and Listeria monocytogenes) and Bacillus cereus spores in fermented alcoholic beverages (Beer and refined rice wine). J. Food Prot. 77, 419–426. doi: 10.4315/0362-028X.JFP-13-234
Kregiel, D., Pawlikowska, E., and Antolak, H. (2017). Non-conventional Yeasts in Fermentation Processes: Potentialities and Limitations. London: Old Yeasts- New Questions, Candida Lucas ad Celia Pai, IntechOpen. doi: 10.5772/intechopen.70404
Kurtzman, C. P., and Fell, J. W. (1998). The Yeasts, a Taxonomic Study. 4th Edn. Amsterdam: Elsevier.
Liaud, N., Ginies, C., Navarro, D., Fabre, N., Crapart, S., Herpoel-Gimbert, I., et al. (2014). Exploring fungal biodiversity: organic acid production by 66 strains of filamentous fungi. Fungal Biol. Biotechnol. 1, 1. doi: 10.1186/s40694-014-0001-z
Lubbehusen, T. L., Nielsen, J., and Mclntyre, M. (2004). Aerobic and anaerobic ethanol production by Mucor circinelloides during submerged growth. Appl. Microbiol. Biotehnol. 63, 543–548. doi: 10.1007/s00253-003-1394-4
Marquina, D., Santos, A., and Peinado, J. M. (2002). Biology of killer yeasts. Int. Microbiol. 5, 65–71. doi: 10.1007/s10123-002-0066-z
Menz, G., Aldred, P., and Vriesekoop, F. (2011). Growth and survival of foodborne pathogens in beer. J. Food Prot. 74, 1670–1675. doi: 10.4315/0362-028X.JFP-10-546
Missbach, B., Majchrzak, D., Sulzner, R., Wansink, B., Reichel, M., and Koenig, J. (2017). Exploring the flavor life cycle of beers with varying alcohol content. Food Sci. Nutr. 5, 889–895. doi: 10.1002/fsn3.472
Mohammad, H., El-Shanawany, A. R., Shah, A. M., Nazir, Y., Naz, T., Ullah, S., et al. (2020). Comparative analysis of different isolated oleaginous Mucoromycota fungi for their γ-linolenic acid and carotenoid production. Biomed. Res. Int. 2020, 1–13. doi: 10.1155/2020/3621543
Moulin, G., Boze, H., and Galzy, P. (1984). Inhibition of alcoholic fermentation. Biotechnol. Genetic Eng. Rev. 2, 365–382. doi: 10.1080/02648725.1984.10647805
Moura-Nunes, N., Brito, T. C., da-Fonseca, N. D., de Agular, P. F., Monteiro, M., Perrone, D., et al. (2016). Phenolic compounds of Brazilian beers from different types and styles and application of chemometrics for modelling antioxidant capacity. Food Chem. 199, 105–113. doi: 10.1016/j.foodchem.2015.11.133
Narzary, D., Boro, N., Borah, A., Okubo, T., and Takami, H. (2021). Community structure and metabolic potentials of the traditional rice beer starter ‘Emao'. Sci. Rep. 11, 14628. doi: 10.1038/s41598-021-94059-x
Osamwonyi, U. O., and Wakil, S. M. (2012). Isolation of fungal species from fermenting pearl millet gruel and determination of their antagonistic activities against indicator bacterial species. Niger. Food J. 30, 35–42. doi: 10.1016/S0189-7241(15)30011-4
Padhi, S., Das, D., Panja, S., and Tayung, K. (2016). Molecular characterization and antimicrobial activity of an endolichenic fungus, Aspergillus sp. isolated from Parmelia caperata of Similipal Biosphere Reserve, India. Interdiscip. Sci. Comput. Life Sci. 9, 237–246. doi: 10.1007/s12539-016-0146-y
Pai, T. V., Sawant, S. Y., Ghatak, A. A., Chaturvedi, P. A., Gupte, A. M., and Desai, N. S. (2015). Characterization of Indian beers: chemical composition and antioxidant potential. J. Food Sci. Technol. 52, 1414–1423. doi: 10.1007/s13197-013-1152-2
Panikov, N. S. (2011). “Microbial growth dynamics,” in Comprehensive Biotechnology, Vol. 1, Scientific Fundamentals of Biotechnology, 2nd Edition, ed M. Moo-Young (Barcelona: Elsevier), 257–283. doi: 10.1016/B978-0-08-088504-9.00032-5
Parashar, D. P., Sharma, H. K., and Kotoky, J. (2017). Exploring the genealogy and phenomic divergences of indigenous domesticated yeasts cultivated by six ethnic communities of Assam, India. J. Biol. Sci. 17, 91–105. doi: 10.3923/jbs.2017.91.105
Pfeiffer, T., and Morley, A. (2014). An evolutionary perspective on the Crabtree effect. Front. Mol. Biosci. 1, 1–6. doi: 10.3389/fmolb.2014.00017
Piraine, R. E. A., Leite, F. P. L., and Bochman, M. L. (2021). Mixed-culture metagenomics of the microbes making sour beer. Fermentation 7, 174. doi: 10.3390/fermentation7030174
Pisoschi, A. M., Pop, A., Negulescu, G. P., and Pisoschi, A. (2011). Determination of ascorbic acid content of some fruit juices and wine by voltammetry performed at Pt and carbon paste electrodes. Molecules 16, 1349–1365. doi: 10.3390/molecules16021349
Prieto, P., Pineda, M., and Aguilar, M. (1999). Spectrophotometric quantification of antioxidant capacity through the formation of a phosphomolybdenum complex: specific application to the determination of vitamin E. Anal. Biochem. 269, 337–341. doi: 10.1006/abio.1999.4019
Ruiz, E., Romero, I., Moya, M., Sanchez, S., Bravo, V., and Castro, E. (2007). Sugar fermentation by Fusarium oxysporum to produce ethanol. World J. Microbiol. Biotechnol. 23, 259–267. doi: 10.1007/s11274-006-9222-5
Santiago, I. F., Rosa, C. A., and Rosa, L. H. (2017). Endophytic symbiont yeasts associated with the Antarctic angiosperms Deschampsia antartica and Colobanthus quitensis. Polar Biol. 40, 177–183. doi: 10.1007/s00300-016-1940-z
Satari, B., Karimi, K., and Zamani, A. (2015). Oil, chitosan and ethanol production by dimorphic fungus Mucor indicus from different lignocelluloses. J. Chem. Technol. Biotechnol. 91, 1835–1843. doi: 10.1002/jctb.4776
Senne de Oliveira Lino, F., Bajic, D., Vila, J. C. C., Sanchez, A., and Sommer, M. O. A. (2021). Complex yeast-bacteria interactions affect the yield of industrial ethanol fermentation. Nat. Commun. 12, 1498. doi: 10.1038/s41467-021-21844-7
Sha, S. P., Suryavanshi, M. V., Jani, K., Sharma, A., Souche, Y., and Tamang, J. P. (2018). Diversity of yeasts and molds by culture dependent and culture-independent methods for microbiome surveillance of traditionally prepared dried starters for the production of Indian alcoholic beverages. Front. Microbiol. 9, 1–15. doi: 10.3389/fmicb.2018.02237
Sharifia, M., Karimi, K., and Taherzadeh, M. J. (2008). Production of ethanol by filamentous and yeast-like forms of Mucor indicus from fructose, glucose, sucrose and molasses. J. Ind. Microbiol. Biotechnol. 35, 1253–1259. doi: 10.1007/s10295-008-0422-x
Shi, J. H., Xiao, Y. P., Li, X. R., Ma, E. B., Du, X. W., and Quan, Z. X. (2009). Analysis of microbial consortia in the starter of Fen Liquor. Lett. Appl. Microbiol. 48, 478–485. doi: 10.1111/j.1472-765X.2009.02554.x
Singh, S., Moholkar, V. S., and Goyal, A. (2013). Isolation, identification and characterization of a cellulolytic Bacillus amyloliquefaciens Strain SS35 from rhinocerous dung. ISRN Microbiol. 2013, 728134. doi: 10.1155/2013/728134
Singleton, V. L., Orthofer, R., and Lamuela-Raventos, R. M. (1999). Analysis of total phenols and other oxidation substrates and antioxidants by means of folin-ciocalteu reagent. Methods Enzymol. 299, 152–178. doi: 10.1016/s0076-6879(99)99017-1
Skinner, K. A., and Leathers, T. D. (2004). Bacterial contaminants in fuel ethanol production. J. Ind. Microbiol. Biotechnol. 31, 401–408. doi: 10.1007/s10295-004-0159-0
Song, J. H., Kim, J. H., Ahn, B. H., and Lee, J. S. (2018). Screening of functional Rhizopus stolonifer for alcohol fermentation and production of high quality Korean traditional rice wine. Mycobiology 38, 122–127. doi: 10.4489/MYCO.2010.38.2.122
Stolp, H., and Gadkeri, D. (1981). “Non-pathogenic members of genus pseudomonas,” in The Prokaryotes, eds M. P. Starr, H. Stolp, H. G. Truper, A. Balows, and H. G. Schlegel (Berlin: Springer), 719–741.
Taechavasonyoo, A., Thaniyavarn, J., and Yompakdee, C. (2013). Identification of the moulds and yeasts characteristic of a superior Loogpang, starter of the Thai rice-based alcoholic beverage Sato. As. J. Food. Ag Ind. 6, 24–38. Available online at: https://www.ajofai.info/Abstract/Identification%20of%20the%20moulds%20and%20yeasts%20characteristic%20of%20a%20superior%20loogpang,%20starter%20of%20thai%20rice-based%20alcoholic%20beverage%20sato.pdf
Takahashi, J. A., Monteiro de Castro, M. C., Souza, G. G., Lucas, E. M. F., Bracarense, A. A. P., Abreu, L. M., et al. (2008). Isolation and screening of fungal species isolated from Brazilian cerrado soil for antibacterial activity against Escherichia coli, Staphylococcus aureus, Salmonella typhimurium, Streptococcus pyogenes and Listeria monocytogenes. J. Mycol. Med. 18, 198–204. doi: 10.1016/j.mycmed.2008.08.001
Teramoto, Y., Yoshida, S., and Ueda, S. (2002). Characteristics of a rice beer (Zutho) and a yeast isolated from the fermented product in Nagaland, India. World J. Microb. Biot. 18, 813–816. doi: 10.1023/A:1021293804327
Tesfaw, A., Oner, E. T., and Assefa, F. (2021). Optimization of ethanol production using newly isolated ethanologenic yeasts. Biochem. Biophys. Rep. 25, 100886. doi: 10.1016/j.bbrep.2020.100886
Thomas, K. C., Hynes, S. H., and Ingledew, W. M. (2001). Effect of lactobacilli on yeast growth, viability and batch and semi-continuous alcoholic fermentation of corn mash. J. Appl. Microbiol. 90, 819–828. doi: 10.1046/j.1365-2672.2001.01311.x
Tsuyoshi, N., Fudou, R., Yamanaka, S., Kozaki, M., Tamang, N., Thapa, S., et al. (2005). Identification of yeast strains isolated from marcha in Sikkim, a microbial starter for amylolytic fermentation. Int. J. Food Microbiol. 99, 135–146. doi: 10.1016/j.ijfoodmicro.2004.08.011
Ueng, P. P., and Gong, C. (1982). Ethanol production from pentoses and sugar-cane bagasse hemicellulose hydrolysate by Mucor and Fusarium species. Enzyme Microb. Technol. 4, 169–171. doi: 10.1016/0141-0229(82)90111-9
Van Rijswijck, I. M. H., Wolkers-Rooijackers, J. C. M., Abee, T., and Smid, E. J. (2017). Perofrmance of non-conventional yeasts in co-culture with brewers' yeast for steering ethanol and aroma production. Microb. Biotechnol. 10, 1591–1602. doi: 10.1111/1751-7915.12717
Vanderhaegen, B., Neven, H., Coghe, S., Verstrepen, K. J., Derdelinckx, G., and Verachtert, H. (2003). Bioflavoring and beer refermentation. Appl. Microbiol. Biotechnol. 62, 140–150. doi: 10.1007/s00253-003-1340-5
Vincent, M., Johnny, Q., Adeni, D. S. A., and Suhaili, N. (2021). Potential of Candida glabrata from ragi as a bioethanol producer using selected carbohydrate substrates. Nusantara Biosci. 13, 1–10. doi: 10.13057/nusbiosci/n130101
Vriesekoop, F., Krahl, M., Hucker, B., and Menz, G. (2013). Bacteria in brewing: the good, the bad and the ugly. J. Inst. Brew. 118, 335–345. doi: 10.1002/jib.49
Watanabe, I., Nakamura, T., and Shima, J. (2008). A strategy to prevent the occurrence of Lactobacillus strains using lactate tolerant yeast Candida glabrata in bioethanol production. J. Ind. Microbiol. Biotechnol. 35, 1117–1122. doi: 10.1007/s10295-008-0390-1
Yamaoka, C., Kurita, O., and Kubo, T. (2014). Improved ethanol tolerance of Saccharomyces cerevisiae in mixed cultures with Kluyveromyces lactis on high-sugar fermentation. Microbiol. Res. 169, 907–914. doi: 10.1016/j.micres.2014.04.007
Keywords: emao, ethnic starter, amylolytic fungi, ethanol-producing, antioxidant, antimicrobial
Citation: Boro N and Narzary D (2022) Amylolytic Fungi in the Ethnic Beer Starter “emao” and Their Beer-Producing Attributes. Front. Sustain. Food Syst. 6:869430. doi: 10.3389/fsufs.2022.869430
Received: 04 February 2022; Accepted: 14 March 2022;
Published: 08 April 2022.
Edited by:
Ricardo N. Pereira, University of Minho, PortugalReviewed by:
Nicolás Oscar Soto-Cruz, TecNM-Instituto Tecnológico de Durango, MexicoCopyright © 2022 Boro and Narzary. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Diganta Narzary, ZF9uYXJ6YXJ5QGdhdWhhdGkuYWMuaW4=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.