- 1Key Laboratory of Plant Functional Genomics of the Ministry of Education, Jiangsu Key Laboratory of Crop Genomics and Molecular Breeding, Agricultural College of Yangzhou University, Yangzhou, China
- 2Key Laboratory of Crop Genetics and Physiology of Jiangsu Province, Co-innovation Center for Modern Production Technology of Grain Crops of Jiangsu Province, Yangzhou University, Yangzhou, China
- 3Joint International Research Laboratory of Agriculture and Agri-Product Safety, The Ministry of Education of China, Institutes of Agricultural Science and Technology Development, Yangzhou University, Yangzhou, China
Lesion-mimic mutants are useful materials to dissect mechanisms controlling programmed cell death (PCD) and defense response in plants. Although dozens of lesion-mimic mutant genes have been identified in plants, the molecular mechanisms underlying PCD and defense response remain to be extensively elucidated. Here, we identified a rice lesion mimic mutant, named lesion mimic 42 (lm42), from an ethylmethylsulfone (EMS)-induced mutant population. The lm42 mutant displayed flame-red spots on the leaves and sheaths at the 3-leaf developmental stage and exhibited impaired photosynthetic capacity with decreased chlorophyll content and decomposed chloroplast thylakoids. The lesion development of lm42 was light- and temperature-dependent. We identified a single base mutation (T38A), changing a Leu to Gln, in the first exon of LOC_Os12g16720 (LM42), which encodes a tryptamine 5-hydroxylase, by map-based cloning. We carried out transgenic complementation to confirm that this mutation caused the lm42 phenotype. We further knocked out the LM42 gene by CRISPR/Cas9 to recreate the lm42 phenotype. LM42 is highly expressed in leaves, leaf sheaths and roots. Loss-of-function of LM42 activated expression of ROS-generating genes and inhibited expression of ROS-scavenging genes, leading to ROS accumulation and eventually cell death. Furthermore, its disruption induced expression of defense-response genes and enhanced host resistance to both fungal pathogen Magnaporthe oryzae and bacterial pathogen Xanthomonas oryzae pv. oryzae. Our transcriptomic data suggested that the way lm42 led to lesion-mimic was probably by affecting ribosome development. Overall, our results demonstrate that tryptamine 5-hydroxylase-coding gene LM42 is required for suppression of cell death and uncontrolled activation of defense responses in rice.
Introduction
Plants have developed sophisticated mechanisms to prevent pathogen attack, including hypersensitive response (HR) which triggers rapid programmed cell death (PCD) at infection sites to inhibit further pathogen invasion or spread to adjacent cells (Heath, 2000; Williams and Dickman, 2008). Typical physiological events associated with HR include bursts of reactive oxygen species (ROS), followed by accumulation of antibacterial compounds, expression of pathogenesis-related (PR) genes, and cell wall strengthening through callose deposition or lignin reinforcement (Shirsekar et al., 2014). Although HR plays a critical role in plant immunity, its molecular mechanisms have not been fully clarified.
Lesion-mimic mutants (LMMs) or spotted-leaf (spl) mutants that spontaneously produce HR-like cell death lesions in the absence of biotic stresses or mechanical damage have been reported in a range of plant species (Zhu et al., 2020; Yang et al., 2021). It has been reported that cell death in LMMs mainly results from increased accumulation of ROS including hydrogen peroxide (H2O2), superoxide anion (O2−), and singlet oxygen (Qiao et al., 2010; Huang et al., 2016; Chen et al., 2018), which are widely known to kill cells when excessively accumulated. In many cases, LMMs constitutively activate immune responses without pathogen infection, suggesting that most of LMM genes are negative regulators of PCD and defense response (Zhu et al., 2020). Therefore, LMMs are considered ideal materials for deciphering the mechanisms underlying PCD and defense response, providing a useful tool to the development of broad-spectrum disease resistance in plants (Wang et al., 2015).
The first rice LMM named sekiguchi lesion (sl) was discovered and confirmed to be controlled by a single recessive gene in 1970 (Kiyosawa, 1970). Since then, various LMMs have been identified based on different lesion phenotypes, including spotted leaf (spl), cell death and resistance (cdr), brown leaf spot (bl), blast lesion mimic mutants (blm), zebra necrosis (zn), probenazol (pbz) and yellow leaf spot (ysl) (Qian et al., 2021). To date, at least 41 genes have been cloned from LMMs in rice that encode a broad range of proteins, such as E3 ubiquitin ligase, protein kinase, chlorophyll synthesis enzyme, ATPase protein, transcription factors, fatty acids/lipids biosynthesis eEF1A-like protein and so on (Zeng et al., 2004; Vega-Sánchez et al., 2008; Sakuraba et al., 2013; Fekih et al., 2015; Matsui et al., 2015; Liu et al., 2017; Hu et al., 2021; Ma et al., 2021; Qiu et al., 2021). Thus, the molecular mechanisms triggering LMMs phenotypes and associated defense responses are not uniform, indicating that the complex network regulating PCD and defense responses needs more investigation.
Here, we identified a stably inherited LMM named lesion mimic 42 (lm42) generated by ethylmethylsulfone (EMS) mutagenesis. At the 3-leaf stage, lm42 showed flame-red pots on leaves and leaf sheaths and significantly lower quality on main yield-related agronomic traits than wild-type (WT). Through map-based cloning, we identified a single base substitution (T38A) in the first exon of LOC_Os12g16720. We further confirmed this single base substitution was the cause of the lesion mimic phenotype of lm42 by transgenic complementation and CRISPR/Cas9-mediated knockout of LM42. Loss-of-function of LM42 led to cell death, ROS accumulation and enhanced resistance to blast and bacterial blight diseases. RNA-seq data showed that the ribosome abnormality in lm42 that likely led to its lesion mimic phenotype. Our study provides in-depth insights to the molecular mechanism that regulates PCD and disease resistance in rice.
Materials and Methods
Plant Materials
The rice (Oryza sativa L) lm42 mutant was obtained from japonica rice Taigeng 394 (T394) mutagenized with EMS treatment. For morphological and genetic analysis, all plants were grown under natural conditions with temperature between 28 and 37°C in Yangzhou City, Jiangsu Province.
Determination of Chlorophyll and Carotenoid Contents
The leaves of WT and lm42 at tillering stage were harvested to determine chlorophyll and carotenoid contents. Weigh fresh chopped leaves with 0.1 g quartz sand (veins removed), calcium carbonate and 95% ethanol were grinded in a mortar. Add 95% ethanol and continue grinding until the tissues turn white. Let stand at room temperature for 5 min. Filter with a filter paper and funnel, use 95% ethanol to 25 mL, shake well. The chloroplast pigment extract was poured into a colorimetric dish. Using 95% ethanol as blank, the absorbance was measured by spectrophotometer at wavelengths of 470, 646, and 663 nm. Formula for calculating chlorophyll and carotenoid contents: Chla = (12.21* D663-2.81*D646) *0.025/w Chlb = (20.13*D646-5.03* D663) *0.025/w; Chl (a+b) = (17.32*D646-7.18*D663) *0.025/w; Car = (1000*D470-3.27* Chla-104* Chlb)/229*0.025/w.
DNA Extraction and Molecular Marker Development
Genomic DNA was extracted from frozen young leaves using the cetyltrimethylammonium bromide (CTAB) method. Insertion and deletion (InDel) markers were developed and listed in Supplementary Tables 3, 4.
Histochemical Analysis
Leaves of lm42 and WT were used in histochemical assays at different leaf stages. A lactic acid-phenol-trypan blue solution was used to evaluate cell death, and 3,3'-diaminobenzidine (DAB) solution and tetranitro blue tetrazolium chloride (NBT) solution was used to evaluate H2O2 and O2− accumulation, respectively. Staining was conducted as previously described (Wang et al., 2017).
Physiological and Biochemical Analysis
POD and CAT enzyme activities and H2O2 and O2− contents at different leaf stages of WT and mutant were measured using kits purchased from Solarbio, Beijing, following manufacturer's instructions.
Temperature and Light Treatment Experiment
For temperature treatment, WT and lm42 plants growing to the 2-leaf stage were treated at 25, 30, and 35°C separately. At the booting stage, the number of leaves with lesion mimics was continuously identified and recorded, with three repetitions. For light treatment, leaves of WT and mutant at the tillering stage were shaded with 2 cm aluminum foil for 10 days, observed and photos taken.
RNA Extract and RT-qPCR Analysis
RNA samples were extracted using a Trizol reagent (Vazyme). Reverse transcription and fluorescence quantitative PCR analysis were performed using a RT-qPCR system from the company. Primers for RT-qPCR analysis are listed in Supplementary Table 5.
Map-Based Cloning, Complementation, and Knock-Out Tests
For genetic analysis, the mapping population was generated from a cross between the lm42 mutant and the indica cultivar 9311. Individuals with lesion-visible leaves from the F2 generation were selected for initial mapping. For fine mapping, we used SSR markers and designed Indel markers in the localized regions (Supplementary Tables 2, 3). For complementation of the lm42 mutant, the promoter and coding region of the LM42 gene were inserted into the pCAMBIA2300 vector, and the resulting plasmid (pCAMBIA-2300-LM42) was introduced into the lm42 mutant. To knock out the LM42 gene, an 18-bp gene-specific guide RNA sequence targeting LM42 was cloned into the binary vector containing a Cas9 expression cassette using the Vazyme Enzyme mix. The primers are listed in Supplementary Table 6.
Inoculation of Rice Blast and Bacterial Blight
Isolates of 2 Magnaporthe oryzae pathotypes (2018-263 and XZ602-3), virulent to WT rice, were used to infect lm42 plants. Inoculum preparation and seedling inoculation with the isolates followed a previously described procedure (Li et al., 2008). The disease index was calculated as described previously (Wang et al., 2013). Xanthomonas oryzae pv. oryzae (Xoo) isolate PX099A, virulent to WT rice, was used to evaluate resistance to bacterial blight in lm42 plants as previously described (Chen et al., 2018). Lesion lengths were measured two weeks after inoculation.
RNA-Seq and Functional Classification of Differentially Expressed Genes
RNA-seq was used to compare transcriptomic differences between WT and lm42 at the 3-4 leaf stage when the mutant showed visible lesions. The screening criteria for differentially expressed genes were “FC=2” and “p-value < 0.05.” Gene Ontology (GO) analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) were performed using the DAVID Resources 6.7 (http://david.abcc.ncifcrf.gov/) and KEGG analysis website (http://www.kegg.jp/), respectively (Wei et al., 2009; Minoru et al., 2014).
Results
Phenotypic Characterization of the lm42 Mutant
We identified a stable rice mutant, lm42, from an EMS mutagenized population of japonica rice Taigeng 394 (T394). The lm42 mutant carries random flame-red spots that appear on leaves and leaf sheaths starting at the 3-leaf stage (Figures 1a,b). At the tillering stage, the distribution and size of spots on lm42 leaves appears random and follows no particular orders (Figure 1c). Lower leaves and leaf sheaths dry out significantly earlier in lm42 than in WT (Figure 1d). The lm42 mutation also affects agronomic traits, including plant height, effective panicle number, panicle length, grain number per panicle, seed setting rate and 1,000-grain weight, significantly decreasing their yield-related performance in field tests (Figure 1d, Supplementary Table 1).
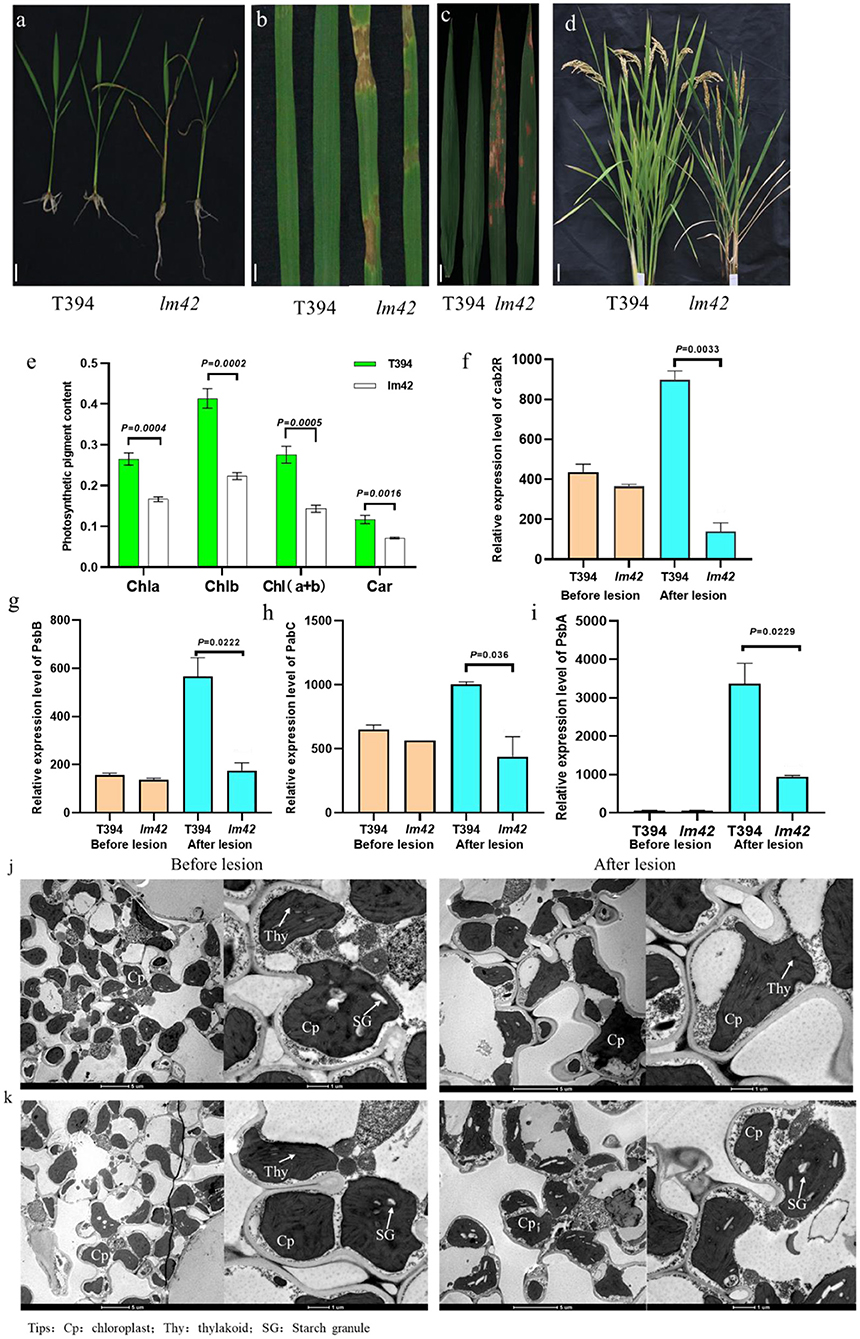
Figure 1. Phenotypic characterization of lm42. (a,b) The lm42 lesion mimic phenotype at seedling stage. (c) Location and size of lesions on lm42 leaves at tillering stage. (d) Plant stature and leaf senescence of lm42 at maturity stage, scale bars, 1 cm. (e) Contents of photosynthetic pigments at tillering stage. (f–i) RNA levels of photosynthesis-related genes determined by RT-qPCR. (j,k) Observation of chloroplasts by TEM in WT (j) and lm42 (k), scale bars, 5 and 1 μm.
To explore whether the photosynthetic capacity of lm42 was affected, we examined chlorophyll and carotenoid contents, and found that chlorophyll a, chlorophyll b and carotenoid contents in lm42 were significantly lower than in WT (Figure 1e). In addition, RT-qPCR showed that the expression levels of four photosynthesis-related genes cab2R, PsbB, Pabc, and PsbA were significantly lower in lm42 than in WT (Figures 1f–i). Examination with transmission electron microscopy (TEM) showed that the number of thylakoid and thylakoid grana decreased, demonstrating the chloroplast thylakoids decomposed after the appearance of lesions in lm42 leaves at the tillering stage (Figures 1j,k). These results suggest that the reduction on photosynthetic ability in lm42 is likely due to the abnormal chloroplasts with decreased chlorophyll and carotenoid contents.
Lesion Occurrence in lm42 Depends on Light and Temperature
The lesion initiation of many LMMs depends on light or temperature (Huang et al., 2010). In order to determine whether the lesion phenotype of lm42 was affected by light and temperature, we first treated lm42 and WT at 2-3 leaf stage (before appearance of lesions) at 25, 30, and 35°C separately (Figure 2a). We found that although all three temperatures caused the lesion phenotype at the 3rd day, high temperature produced significantly more leaves with lesions than low temperature (Figures 2b–e). Shading treatment was then performed on lm42 leaves by covering lm42 leaves with a piece of 2 cm aluminum foil for 7 days. The results showed that lesions did not appear in the shaded leaf areas, while lesions emerged in the unshaded leaf areas in lm42 (Figure 2f). These data indicate that the lesion formation in lm42 was regulated by light and temperature.
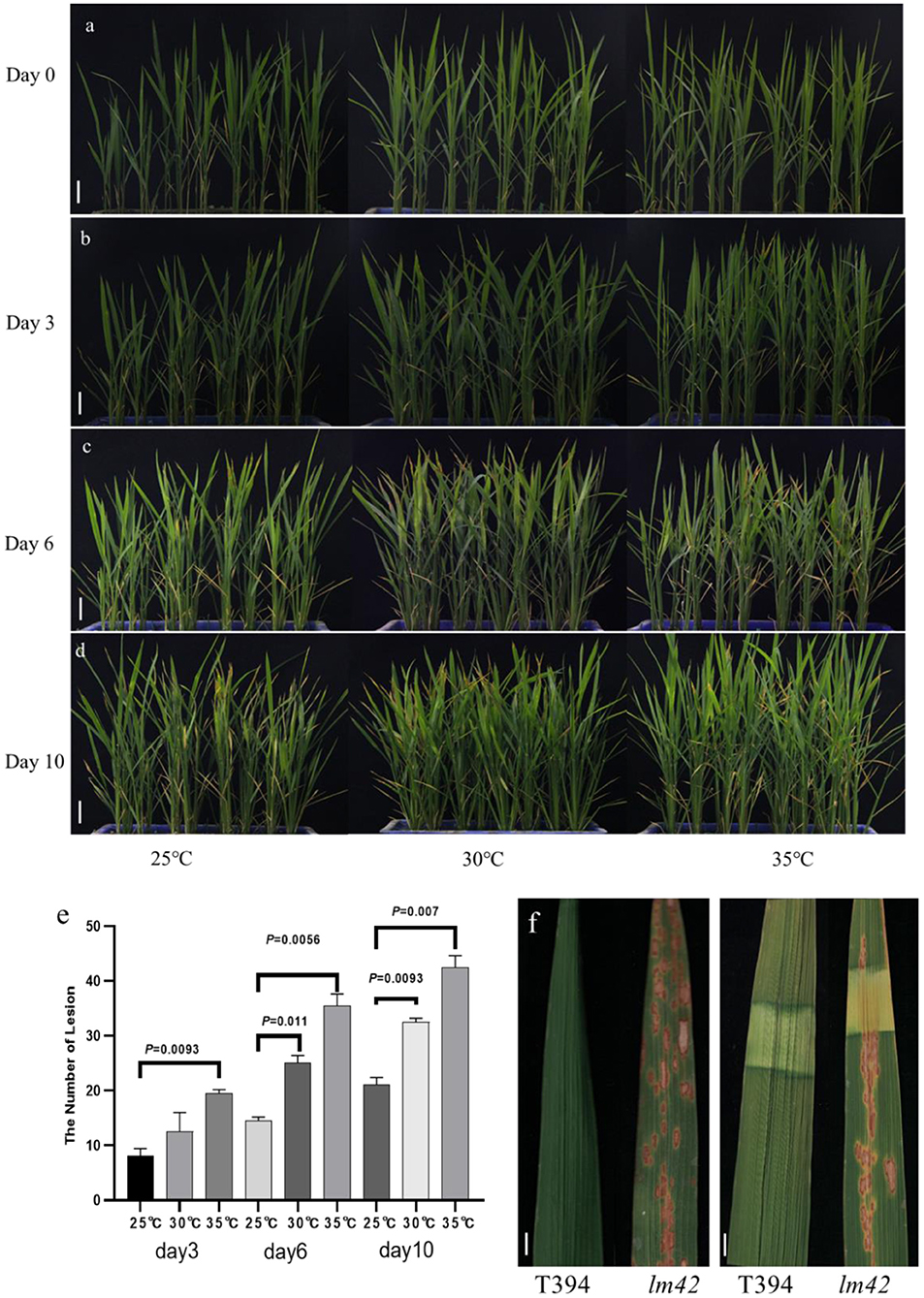
Figure 2. Light and temperature influence lesion formation in lm42. (a–d) The phenotypes of WT and lm42 were observed at 0 (a), 3 (b), 6 (c), and 10 (d) days at different temperatures. (e) Number of lesion mimic leaves in lm42 at 3, 6, and 10 days at different temperatures. (f) Effects of shading treatment on lesion formation, scale bars, 1 cm.
Map-Based Cloning Identified a Single Base Substitution in lm42
To assess the genetic nature of the lm42 lesion phenotype, we carried out a cross between lm42 and WT, and found that the hybrid F1 plants all exhibited WT phenotype during the whole growth period, suggesting a recessive nature for the phenotype. In the self-pollinated progeny (F2 population) of F1 plants, we observed a segregation of WT and lm42 phenotypes in a 3:1 ratio (Supplementary Table 2). These data indicate that the lm42 phenotype is conferred by a single recessive gene.
To map the LM42 locus, we developed an F2 population from the cross of lm42 and indica cv. 9311. We first genotyped 15 F2 individuals with WT phenotype and another 15 with mutant phenotype using 280 SSR markers randomly distributed on the rice genome, yielding a preliminary map region containing LM42 located between markers RM27808 and RM1337 on chromosome 12 (Figure 3A, Supplementary Table 3). We further developed 6 polymorphic Indel markers to fine-map the LM42 gene with a total of 745 F2 plants with mutant phenotype, which allowed us to finally narrowed down the LM42 gene to a 30 kb region between markers Indel13 and Indel38 (Figure 3B, Supplementary Table 4). According to the rice Genome Interpretation database (https://www.gramene.org/), seven open reading frames (ORFs) were annotated in this region (Figure 3C). Comparison of our sequencing data revealed that the fourth ORF (LOC_Os12g16720) carried a single-base substitution (T38A) in lm42 (Supplementary Table 5), leading to a single amino acid change from leucine (Leu) to glutamine (Gln). The other six ORFs showed no differences between lm42 and WT (Figure 3D). We subsequently developed a dCAPs marker for this T/A SNP: the PCR product from lm42 is digested by the Alu I enzyme into two fragments (126 and 39 bp), whereas the WT PCR product is not cut. A screen of the F2 population with this dCAPs marker confirmed the association of this SNP with the phenotype (Supplementary Figure 1). Therefore, LOC_Os12g16720, encoding a tryptamine 5-hydroxylase/cytochrome P450 monooxygenase, is the candidate gene for LM42.
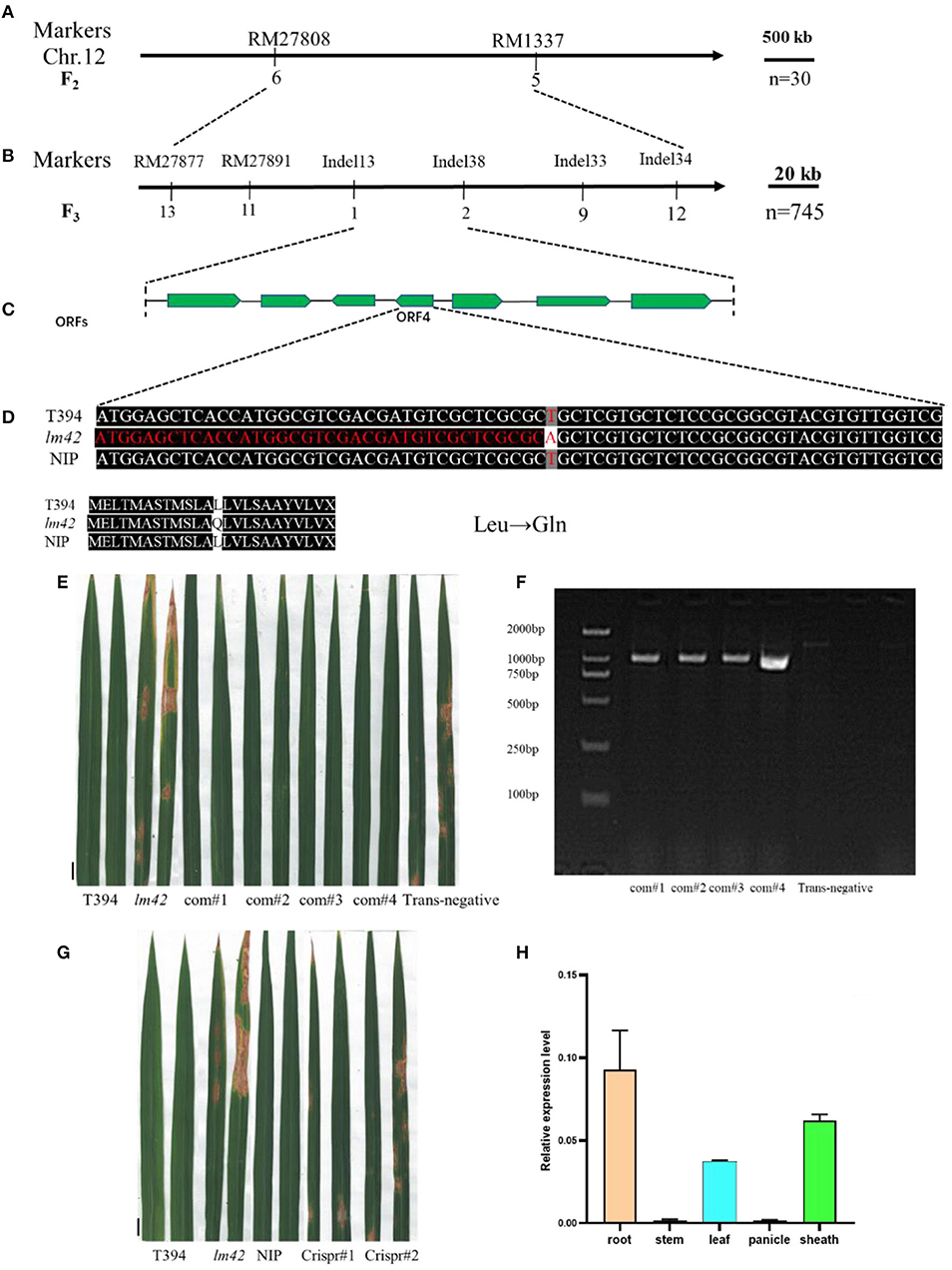
Figure 3. Map-based identification of the LM42 gene. (A) LM42 was initially mapped on chromosome 12 between markers RM27808 and RM1337. (B) The LM42 locus was fine-mapped to a 30-kb genomic region between markers Indel13 and Indel38; the number below a primer represents the number of recombinants. (C) Seven putative ORFs are located in the ~30-kb identified region. (D) Sequence comparison at the mutation site for wild type and the lm42 mutant; a T-to-A point mutation is present, resulting in a substitution of Leu by Gln. (E) Lesion mimic phenotype of LOC_Os12g16720 knock-out lines. (F) Lesion mimic phenotype of LOC_Os12g16720 complemented lines, scale bars, 1 cm. (G) PCR products of LOC_Os12g16720 for positive complementation lines, scale bars, 1 cm. (H) LM42 RNA expression in various tissues.
Transgenic Complementation and CRISPR/Cas9-Mediated Knockout Validate the LM42 Gene
In order to verify the candidate gene, we generated a construct (pCAMBIA-2300-LM42) containing the upstream 2.5 kb promoter and the full-length coding region genomic DNA of LM42 from WT for complementation. We introduced the LM42 gene into the lm42 mutant through Agrobacterium-mediated transformation, and obtained 4 T0 transgenic plants confirmed by PCR genotyping. We found that all four independent transformants showed WT phenotype exhibiting no lesion mimics across the whole life cycle, whereas negative controls regenerated from the same tissue culture containing no transgene exhibited the mutant phenotype (Figures 3E,F). Also, the overall agronomic traits of the positive transformants were similar with that of WT (Supplementary Figure 2A, Supplementary Table 8). This result demonstrated that the WT candidate gene can rescue the lesion phenotype of the lm42 mutant.
Furthermore, we generated two LM42 knockout plants in the japonica variety Nipponbare (NIP) background using the CRISPR/Cas9 technology. After sequencing candidate transgenic lines, we obtained two LM42 knockout (ko) T0 plants: one contained a heterozygous two-base insertion and the other a one-base insertion, both causing loss-of-function due to frameshifts (Supplementary Figure 2B). For the two heterozygous LM42-ko T0 plants, we observed no lesion mimics throughout the whole growth stage, whereas we found that some T1 plants of each line showed clear lesion mimic phenotype, like the lm42 mutant (Figure 3G). What's more, the major agronomic phenotype of LM42-ko lines were similar with that of lm42 mutant (Supplementary Figure 2C, Supplementary Table 9). This result indicates that knockouts of LOC_Os12g16720 cause the lm42 lesion mimic phenotype. Together, these results demonstrate that the lm42 lesion mimic phenotype is caused by mutations that block the function of LOC_Os12g16720.
ROS Accumulation and Cell Death Occur in the lm42 Mutant
To assess the expression pattern of LM42, we ran RT-qPCR to monitor LM42 transcript levels and found that LM42 was mainly expressed in roots, leaves and leaf sheaths (Figure 3H). To detect whether cell death occurred in lm42, we firstly carried out trypan blue staining (Qiao et al., 2010) and observed more and darker blue spots on lm42 than on WT leaves (Figure 4A), indicating that severe cell death occurred in lm42. We next assessed expression levels of metacaspase family genes that are usually associated with PCD (Fagundes et al., 2015) and found changes in their expression levels: OSMC1-OSMC4 were down-regulated whereas OSMC5-OSMC7 were up-regulated in lm42 (Supplementary Figure 3A).
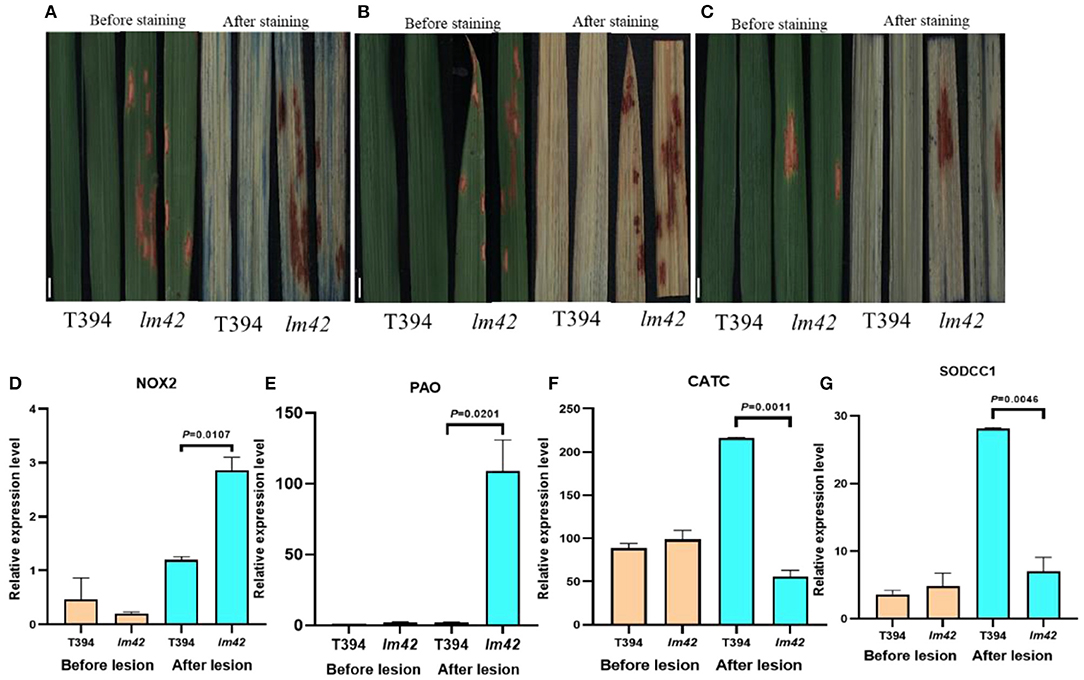
Figure 4. Cell death and ROS accumulation occur in lm42. (A) Comparison of trypan blue staining between WT and lm42 leaves at tillering stage, scale bars, 1 cm. (B) Comparison of DAB staining between WT and lm42 leaves at tillering stage, scale bars, 1 cm. (C) Comparison of NBT staining between WT and lm42 leaves at tillering stage, scale bars, 1 cm. (D,E) Expression levels of genes encoding ROS-generating enzymes NOX2 and PAO. (F,G) Expression levels of genes encoding ROS-scavenging enzymes CATC and SODCC1.
ROS are major signal molecules in plant PCD and may lead to cellular damage (Khanna-Chopra, 2012). To determine ROS levels in lm42, we firstly carried out 3,3'-diaminobenzidine (DAB) staining, an indicator of H2O2 accumulation, and found more brown precipitates around lesion sites in lm42 leaves than in WT leaves (Figure 4B). In agreement with DAB staining, H2O2 contents were significantly higher in lm42 than in WT when quantified (Supplementary Figure 3B). In addition, we found that the content of CAT in mutant was higher than that in WT, indicating that the increased content of CAT was not enough to remove excessive ROS in mutant (Supplementary Figure 3C). We next assessed O2− contents by staining leaves with tetranitro blue tetrazolium chloride (NBT), an indicator of O2− accumulation. We observed much more blue spots on lm42 than on WT leaves (Figure 4C), indicating higher O2− contents in lm42 (Supplementary Figures 3D,E). We further found that genes encoding ROS-scavenging enzymes (CATC and SODCC1) were significantly down-regulated whereas ROS-generating enzymes (NOX2 and PAO) were upregulated in lm42 compared to WT (Figures 4D–G). Together, these results indicate excess accumulation of ROS in lm42, which likely causes cell death in lm42.
lm42 Confers Enhanced Resistance to Bacterial Blight and Blast Diseases
The lesion mimic phenotype of lm42 resembles the HR that occurs in plants following infection by many pathogens. Many LMMs show enhanced resistance to fungal and bacterial pathogens (Zhu et al., 2020). Thus, we tested whether lm42 also gained disease resistance. We inoculated lm42 and WT plants with two common rice fungal and bacterial pathogens: two M. oryzae isolates and a Xoo pathotype that are virulent on WT plants. Compared with WT, we found that lm42 developed significantly shorter lesion lengths than WT for all isolates of the two pathogens, demonstrating that the lm42 mutant significantly enhanced resistance to the two common rice bacterial and fungal pathogens (Figures 5A–D).
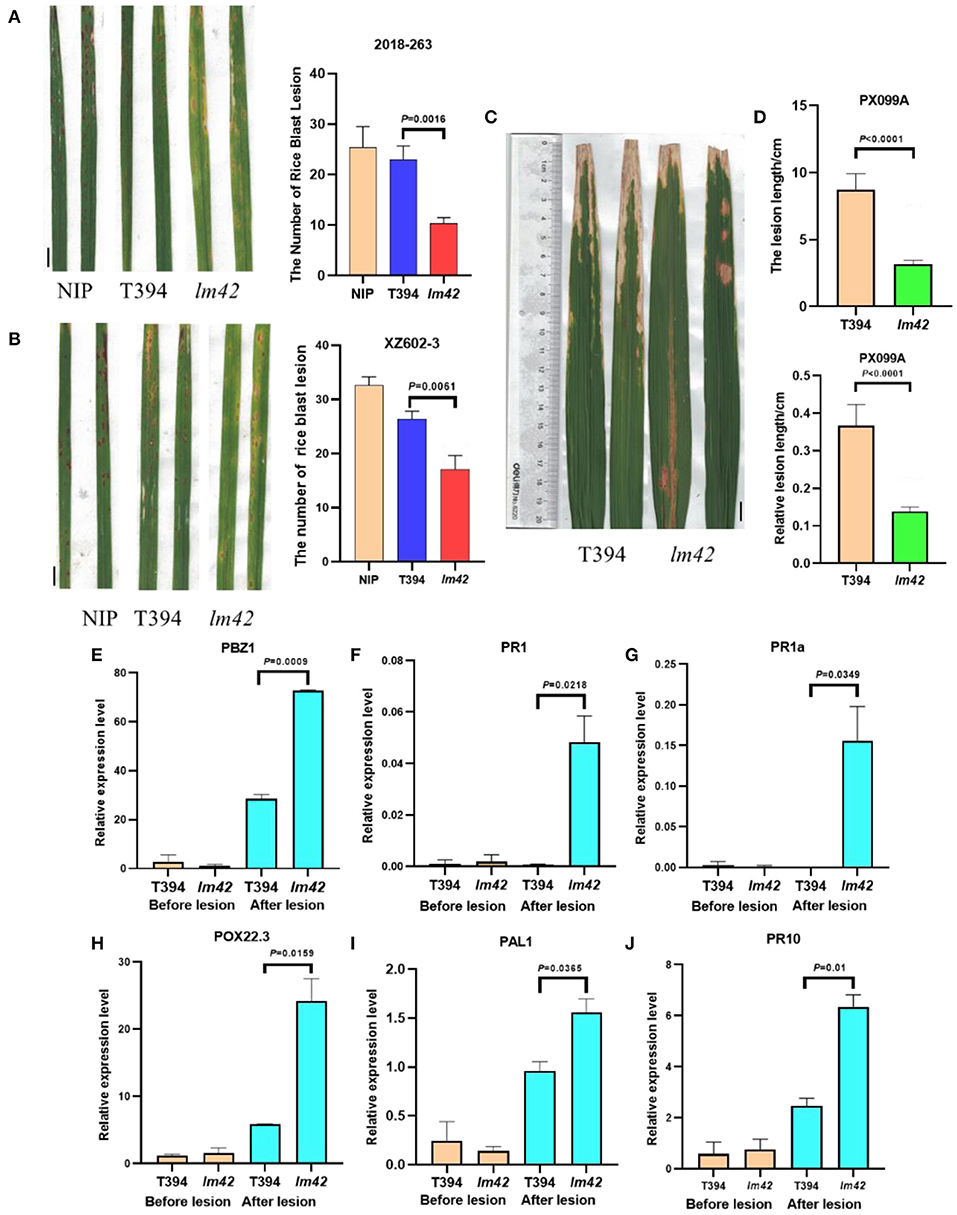
Figure 5. Defense responses are activated in lm42. (A,B) Lesions on WT and lm42 leaves developed after infection with rice blast races 2018-263 (A) and XZ602-3 (B), scale bars, 1 cm. (C,D) Lesion lengths on WT and lm42 leaves developed 14 days post inoculation with bacterial blight race PX099A, scale bars, 1 cm. (E–J) Expression levels of defense-related genes before (orange bars) and after (cyan bars) the appearance of lesions in lm42.
To determine whether defense response genes were induced in lm42, we assessed the expression levels of defense-related genes PBZ1, PR1, PR1a, POX22.3, PAL1, and PR10 (Hou et al., 2012) in lm42 and WT plants at 2-leaf stage (before appearance of visible lesions) and 3-4 leaf stage (clearly visible lesions) by RT-qPCR. At 2-leaf stage, we found that these genes showed similar expression levels in WT and lm42. In contrast, at 3-4 leaf stage, their expression levels were significantly elevated in lm42 than in WT (Figures 5E–J). These results suggest that LM42/LOC_Os12g16720 functions to suppress the defense response and disease resistance.
LM42 Affects Multiple Biological Pathways
To probe the molecular basis underlying the cell death and defense activation in lm42, we performed RNA-seq to assess transcriptomes of lm42 and WT at the 3-4 leaf stage when mutant plants showed visible lesions. Based on our RNA-seq data, we identified 6,409 differentially expressed genes (DEGs) comparing lm42 to WT, including 3,789 up-regulated and 2,620 down-regulated genes (Figures 6A,B). GO functional enrichment including cellular components, biological processes and molecular functions was performed on these 6,409 DEGs (Figure 6B). For cellular components, ribosome was the most prevalent class including 164 DEGs, followed by proteasome complex, photosynthesis synthesis and mitochondrial inner membrane. For biological processes, translation was the most prevalent class including 144 DEGs, followed by glycolysis and photosynthesis. For molecular function, structural constituent of ribosome and iron ion binding were the two most prevalent classes each including nearly 150 DEGs, followed by oxidoreductase activity of degrading ROS, metal ion binding, transferase activity, magnesium ion binding and pyridoxal phosphate binding.
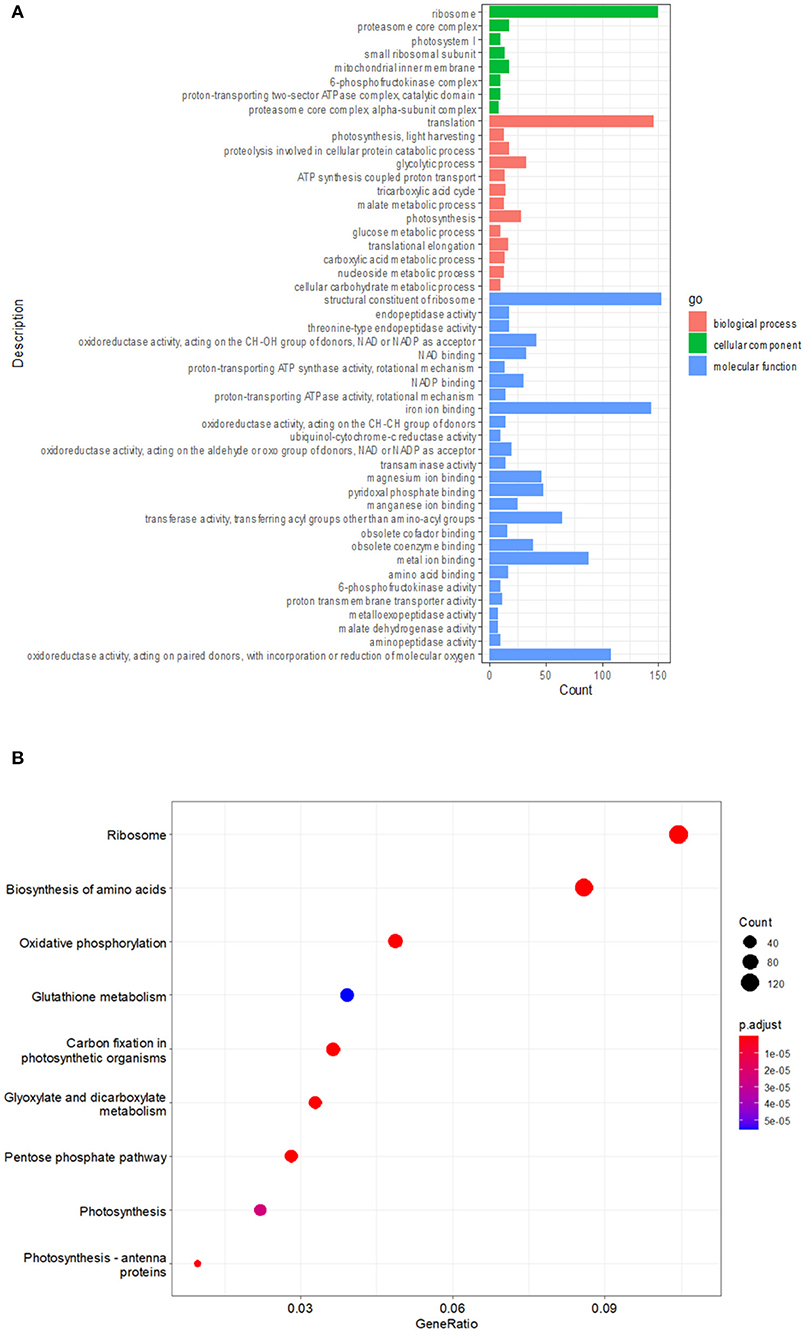
Figure 6. Multiple biological pathways affected in lm42 as revealed by RNA-seq. (A) GO enrichment analysis of differentially expressed genes before and after lesion appearance. (B) KEGG analysis of differentially expressed genes before and after lesion appearance.
To further explore the biological pathways in which LM42 may be involved, we performed KEGG enrichment analysis for the DEGs, and found that 9 pathways were significantly (P < 0.01) enriched (Figure 6B). The highly enriched pathways are mainly related to protein synthesis (including ribosome and biosynthesis of amino acids), redox (including oxidative phosphorylation and glutathione metabolism), carbon metabolism (including carbon fixation in photosynthetic organisms, glyoxylate and dicarboxylate metabolism and pentose phosphate pathway), and photosynthesis. Together, these results suggest that LM42 likely regulates cell death and plant defense through affecting these functional pathways.
Discussion
LMMs are often used as a tool to study plant PCD and defense response because they autonomously form necrotic lesions resembling activation of the immune response in the absence of pathogen attack (Sun et al., 2011). In this study, a mutant allele of the previously characterized gene SL was identified (Fujiwara et al., 2010). We named the mutant lm42 and cloned the LM42 gene via a map-based cloning strategy. SL encodes a cytochrome P450 monooxygenase, which has tryptamine 5-hydroxylase activity and can catalyze the conversion of tryptamine to 5-hydroxytryptamine. However, previous report showed no detailed analysis on how this gene caused lesion mimic symptoms (Fujiwara et al., 2010). Through various staining analyses and physiological and biochemical tests, we confirmed that irreversible ROS bursts and PCD occurred in mutant leaves after the appearance of lesions. In the same leaf, ROS accumulation and PCD only occurred at the lesion sites, but not at areas without lesions, indicating that the occurrence of lesions in lm42 is tightly associated with ROS bursts (Figures 4B,C). Recently, Cui et al. (2020) identified another mutant allele of the SL gene in the ell1 mutant and also detected excessive accumulation of ROS, which is consistent with our results. However, the mechanism underlying how lm42/ell1/sl mutations cause excessive accumulation of ROS and PCD remains unclear. Therefore, we performed the transcriptomic sequencing for WT and lm42 at the 3-4 leaf stage when lesions are clearly visible (Figure 6). Combined with the functional analysis results of DEGs, we hypothesize that the lm42 mutation may lead to ribosome abnormality which ultimately triggers PCD. This hypothesis is based on two aspects: First, LM42/ELL1/SL is located in the endoplasmic reticulum (ER) rather than in chloroplasts (Cui et al., 2020). Ribosomes are attached to the membranes of the ER. Second, our GO and KEGG functional enrichment analyses of DEGs showed that DEGs involved in ribosome function or assembly were most prevalent (Figures 6C,D). Ribosomes are the key organelle for the biosynthesis and transport of all proteins. If damaged, it will in particular affect the biosynthesis and transport of many ion binding proteins, which are necessary for the maintenance of oxidoreductase activities associated with ROS-scavenging and photosynthesis (Suo et al., 2020). As a result, ROS were excessively accumulated, and toxic substances produced in the process of photosynthesis could not be timely degraded either, which ultimately cause damages in chloroplasts leading to cell death.
Mutations in many LMM genes lead to significantly enhanced disease resistance and their molecular mechanisms regulating defense response are very diverse and complicated (Zhu et al., 2020; Qian et al., 2021). Previous research found that LM42/ELL1/SL was induced by the chitin elicitor and by infection with Magnaporthe oryzae. Also, application of 5-hydroxytryptamine induced expression of defense genes and cell death in rice (Fujiwara et al., 2010; Tian et al., 2020). In this study, we further confirmed that LM42 negatively regulates defense-response gene expression and resistance to both fungal pathogen M. oryzae and bacterial pathogen Xoo (Figures 5A–D). However, to date, the molecular mechanism of LM42/ELL1/SL-mediated immune response is not all clear. It was reported that there is mutual antagonistic regulation between 5-hydroxytryptamine biosynthesis and salicylic acid (SA) biosynthesis. Their biosynthesis pathways share the common source substance—chorismic acid, and they actively inhibit the activity of biosynthesis genes of the other pathway (Lu et al., 2018). SA is a well-known defense signal against biotrophic pathogens (Zavaliev et al., 2020). Thus, we reason that the molecular mechanism underlying the lm42/ell1/sl-mediated immune response is likely related to a release of the suppression of SA biosynthesis and signaling when the 5-hydroxytryptamine biosynthesis pathway is blocked due to the lm42 mutation. Whether this is true or not requires further investigation.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
WS, ZF, KH, WC, ML, and RJ conceived and designed the experiments. WS, ZF, ML, and RJ performed the experiments and analyzed the data. ZC was responsible for material plant and field management. WS and ZF wrote the manuscript. SZ revised the manuscript. All authors read and approved the manuscript.
Funding
This study was supported by the Natural Science Foundation of China (31872013 and 32000362) and of Jiangsu Province (BK20200930), a Project Funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD), and the Scientific Research Innovation Practice Project for postgraduate students of Jiangsu (KYCX21_3248).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fsufs.2022.857760/full#supplementary-material
References
Chen, Z., Chen, T., Sathe, A., He, Y., Zhang, X., and Wu, J. (2018). Identification of a novel semi-dominant spotted-leaf mutant with enhanced resistance to Xanthomonas oryzae pv. oryzae in rice. Int. J. Mol. Sci. 19:3766. doi: 10.3390/ijms19123766
Cui, Y., Peng, Y., Zhang, Q., Xia, S., Ruan, B., Xu, Q., et al. (2020). Disruption of Early Lesion Leaf 1, encoding a cytochrome P450 monooxygenase, induces ROS accumulation and cell death in rice. Plant J. 105, 942–956. doi: 10.1111/tpj.15079
Fagundes, D., Bohn, B., Cabreira, C., Leipelt, F., Dias, N., Bodanese-Zanettini, M., et al. (2015). Caspases in plants: metacaspase gene family in plant stress responses. Funct. Integr. Genom. 15, 639–649. doi: 10.1007/s10142-015-0459-7
Fekih, R., Tamiru, M., Kanzaki, H., Kanzaki, H., Abe, A., Yoshida, K., et al. (2015). The rice (Oryza sativa L.) lesion mimic resembling, which encodes an AAA-type ATPase, is implicated in defense response. Mol. Genet. Genom. 29, 611–622. doi: 10.1007/s00438-014-0944-z
Fujiwara, T., Maisonneuve, S., Isshiki, M., Mizutani, M., Chen, L., Wong, H., et al. (2010). Sekiguchi lesion gene encodes a cytochrome P450 monooxygenase that catalyzes conversion of tryptamine to serotonin in rice. J. Biol. Chem. 285, 11308–11313. doi: 10.1074/jbc.M109.091371
Heath, M.. (2000). Hypersensitive response-related death. Plant Mol. Biol. 44, 77–90. doi: 10.1023/A:1026592509060
Hou, M., Xu, W., Bai, H., Liu, Y., Li, L., Liu, L., et al. (2012). Characteristic expression of rice pathogenesis-related proteins in rice leaves during interactions with Xanthomonas oryzae pv. oryzae. Plant Cell Rep. 31, 895–904. doi: 10.1007/s00299-011-1210-z
Hu, H., Ren, D., Hu, J., Jiang, H., Chen, P., Zeng, D., et al. (2021). White and Lesion-mimic leaf1, encoding a lumazine synthase, affects ROS balance and chloroplast development in rice. Plant J. 108, 1690–1703. doi: 10.1111/tpj.15537
Huang, Q., Shi, Y., Zhang, X., Song, L., Feng, B., Wang, H., et al. (2016). Single base substitution in OsCDC48 is responsible for premature senescence and death phenotype in rice. J. Integr. Plant Biol. 58, 12–28. doi: 10.1111/jipb.12372
Huang, Q., Yang, Y., Shi, Y., Chen, J., and Wu, J. (2010). Spotted-leaf mutants of rice (Oryza sativa). Rice Sci. 17, 247–256. doi: 10.1016/S1672-6308(09)60024-X
Khanna-Chopra, R.. (2012). Leaf senescence and abiotic stresses share reactive oxygen species-mediated chloroplast degradation. Protoplasma 249, 469–481. doi: 10.1007/s00709-011-0308-z
Kiyosawa, S.. (1970). Inheritance of a particular sensitivity of the rice variety, Sekiguchi-Asahi, to pathogens and chemicals, and linkage relationship with blast resistance. Nogyo Gijutsu Kenkyusho Hokoku 21, 61–71.
Li, W., Lei, C., Cheng, Z., Jia, Y., Huang, D., Wang, J., et al. (2008). Identification of SSR markers for a broad-spectrum blast resistance gene Pi20(t) for marker-assisted breeding. Mol. Breed. 22, 141–149. doi: 10.1007/s11032-008-9163-9
Liu, Q., Ning, Y., Zhang, Y., Yu, N., Zhao, C., Zhan, X., et al. (2017). OsCUL3a negatively regulates cell death and immunity by degrading OsNPR1 in rice. Plant Cell 292, 345–359. doi: 10.1105/tpc.16.00650
Lu, H., Luo, T., Fu, H., Wang, L., Tan, Y., Huang, J., et al. (2018). Resistance of rice to insect pests mediated by suppression of serotonin biosynthesis. Nat. Plants 4, 338–344. doi: 10.1038/s41477-018-0152-7
Ma, H., Li, J., Ma, L., Wang, P., Xue, Y., Yin, P., et al. (2021). Pathogen-inducible OsMPKK10.2-OsMPK6 cascade phosphorylates the Raf-like kinase OsEDR1 and inhibits its scaffold function to promote rice disease resistance. Mol. Plant 14, 620–632. doi: 10.1016/j.molp.2021.01.008
Matsui, H., Takahashi, A., and Hirochika, H. (2015). Rice immune regulator, OsPti1a, is specifically phosphorylated at the plasma membrane. Plant Signal. Behav. 10:e991569. doi: 10.4161/15592324.2014.991569
Minoru, K., Susumu, G., Yoko, S., Kawashima, M., Furumichi, M., and Tanabe, M. (2014). Data, information, knowledge and principle: back to metabolism in KEGG. Nucl. Acids Res. 42, 199–205. doi: 10.1093/nar/gkt1076
Qian, J., Liu, F., Qu, C., and Yue, W. (2021). Research progress on cloning and mechanism of rice lesion mimic mutation gene. Mol. Plant Breed. 19, 3274–3280. doi: 10.5376/rgg.2020.11.0002
Qiao, Y., Jiang, W., Lee, J., Park, B., Choi, M., Piao, R., et al. (2010). SPL28 encodes a clathrinassociated adaptor protein complex 1, medium subunit micro 1 (AP1M1) and is responsible for spotted leaf and early senescence in rice (Oryza sativa). N. Phytologist 18, 258–274. doi: 10.1111/j.1469-8137.2009.03047.x
Qiu, T., Zhao, X., Feng, H., Qi, L., Yang, J., Penget, Y., et al. (2021). OsNBL3, a mitochondrion-localized pentatricopeptide repeat protein, is involved in splicing nad5 intron 4 and its disruption causes lesion mimic phenotype with enhanced resistance to biotic and abiotic stresses. Plant Biotechnol. J. 19, 2277–2290. doi: 10.1111/pbi.13659
Sakuraba, Y., Rahman, M., Cho, S., Kim, Y., Koh, H., Yoo, S., et al. (2013). The rice faded green leaf locus encodes protochlorophyllide oxidoreductase B and is essential for chlorophyll synthesis under high light conditions. Plant J. 74, 122–133. doi: 10.1111/tpj.12110
Shirsekar, G., Vega-Sanchez, M., Bordeos, A., Baraoidan, M., Swisshelm, A., Fan, J., et al. (2014). Identification and characterization of suppressor mutants of spl11-mediated cell death in rice. Mol. Plant Microbe Interact. 27, 528–536. doi: 10.1094/MPMI-08-13-0259-R
Sun, C., Liu, L., Tang, J., Lin, A., Zhang, F., Fang, J., et al. (2011). RLIN1, encoding a putative coproporphyrinogen III oxidase, is involved in lesion initiation in rice. J. Genet. Genom. 38, 29–37. doi: 10.1016/j.jcg.2010.12.001
Suo, J., Zhang, H., Zhao, Q., Zhang, N., Zhang, Y., Li, Y., et al. (2020). Na2CO3-responsive photosynthetic and ROS scavenging mechanisms in chloroplasts of alkaligrass revealed by phosphoproteomics. Genom. Proteom. Bioinformat. 18, 271–288. doi: 10.1016/j.gpb.2018.10.011
Tian, D., Fang, F., Niu, Y., Lin, Y., Chen, Z., Li, G., et al. (2020). Loss function of SL (sekiguchi lesion) in the rice cultivar Minghui 86 leads to enhanced resistance to (hemi)biotrophic pathogens. BMC Plant Biol. 20:507. doi: 10.1186/s12870-020-02724-6
Vega-Sánchez, M., Zeng, L., Chen, S., Leung, H., and Wang, G. (2008). SPIN1, a komology domain protein negatively regulated and ubiquitinated by the E3 ubiquitin ligase SPL11, is involved in flowering time control in rice. Plant Cell 20, 1456–1469. doi: 10.1105/tpc.108.058610
Wang, S., Lei, C., Wang, J., Ma, J., Tang, S., Wang, C., et al. (2017). SPL33, encoding an eEF1A-like protein, negatively regulates cell death and defense responses in rice. J. Exp. Bot. 68, 899–913. doi: 10.1093/jxb/erx001
Wang, Y., Liu, S., Mao, X., Zhang, Z., Jiang, H., Chai, R., et al. (2013). Identification and characterization of rhizosphere fungal strain MF-91 antagonistic to rice blast and sheath blight pathogens. J. Appl. Microbiol. 114, 1480–1490. doi: 10.1111/jam.12153
Wang, Z., Wang, Y., Hong, X., Hu, D., Liu, C., Yang, J., et al. (2015). Functional inactivation of UDP-N-acetylglucosamine pyrophosphorylase 1 (UAP1) induces early leaf senescence and defence responses in rice. J. Exp. Bot. 66, 973–987. doi: 10.1093/jxb/eru456
Wei, H., Sherman, B., and Lempicki, R. (2009). Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucl. Acids Res. 37, 1–13. doi: 10.1093/nar/gkn923
Williams, B., and Dickman, M. (2008). Plant programmed cell death: can't live with it; can't live without it. Mol. Plant Pathol. 9, 534–544. doi: 10.1111/j.1364-3703.2008.00473.x
Yang, Y., Lin, Q., Chen, X., Liang, W., Fu, Y., Xu, Z., et al. (2021). Characterization and proteomic analysis of novel rice lesion mimic mutant with enhanced disease resistance. Rice Sci. 28, 466–487. doi: 10.1016/j.rsci.2021.07.007
Zavaliev, R., Mohan, R., Chen, T., and Dong, X. (2020). Formation of NPR1 condensates promotes cell survival during the plant immune response. Cell 182, 1093–1108. doi: 10.1016/j.cell.2020.07.016
Zeng, L., Qu, S., Bordeos, A., Yang, C., Baraoidan, M., Yan, H., et al. (2004). Spotted leaf 11, a negative regulator of plant cell death and defense, encodes a U-box/armadillo repeat protein endowed with E3 ubiquitin ligase activity. Plant Cell 16, 2795–2808. doi: 10.1105/tpc.104.025171
Keywords: Oryza sativa, lesion mimic mutant, defense responses, map-based cloning, programmed cell death, RNA-seq
Citation: Shen W, Feng Z, Hu K, Cao W, Li M, Ju R, Zhang Y, Chen Z and Zuo S (2022) Tryptamine 5-Hydroxylase Is Required for Suppression of Cell Death and Uncontrolled Defense Activation in Rice. Front. Sustain. Food Syst. 6:857760. doi: 10.3389/fsufs.2022.857760
Received: 19 January 2022; Accepted: 07 February 2022;
Published: 08 March 2022.
Edited by:
Wei Li, Hunan Agricultural University, ChinaReviewed by:
Jiangbo Fan, Shanghai Jiao Tong University, ChinaXuli Wang, Chinese Academy of Agricultural Sciences (CAAS), China
Mawsheng Chern, University of California, Davis, United States
Copyright © 2022 Shen, Feng, Hu, Cao, Li, Ju, Zhang, Chen and Zuo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shimin Zuo, c216dW9AeXp1LmVkdS5jbg==
†These authors have contributed equally to this work
 Wangxin Shen
Wangxin Shen Zhiming Feng1,2†
Zhiming Feng1,2† Keming Hu
Keming Hu Wenlei Cao
Wenlei Cao Shimin Zuo
Shimin Zuo