- 1State Key Laboratory of Rice Biology, Key Laboratory for Zhejiang Super Rice Research, China National Rice Research Institute, Fuyang, China
- 2Northern Center of China National Rice Research Institute, China National Rice Research Institute, Fuyang, China
- 3Zhejiang Guodao High-Tech Seed Industry Co., Ltd., Fuyang, China
Lesion mimic mutants (LMMs) exhibit spots on leaves without fungal infection pressure. The spots confer variable resistance to pathogens in different LMM, making them useful research materials. It is unclear how the rice immune system responds to infection with the fungal pathogen Magnaporthe oryzae (M. oryzae). Here, we identified a rice LMM, lm8015-3, which shows reduced resistance to M. oryzae. We used Quantitative Real-Time PCR (qRT-PCR) to observe the immune system response to M. oryzae–induced lm8015-3. Lm8015-3, obtained from an ethyl methane sulfonate (EMS)–induced Zhonghui8015 (ZH8015) library, showed orange-yellow spots starting in the seedling stage and accumulated more H2O2, resulting in severe degradation of the chloroplast. With map-based cloning, the target gene was located on chromosome 12. Once inoculated with M. oryzae, the expression level of pathogen-related genes of lm8015-3 was downregulated between 48 and 72 h. In addition, more germinating spores appeared in lm8015-3. Therefore, we conclude that M. oryzae weakening the immune system of lm8015-3 from 48 to 72 h makes lm8015-3 more susceptible to M. oryzae. These results suggested that understanding how LMMs defend against M. oryzae infection will contribute to improving rice breeding.
Introduction
Rice is the staple food for the majority of people in China. Improving its grain production has thus been a major research focus for many years. However, many factors reduce rice yield production, especially rice blast fungus, M. oryzae. M. oryzae is considered the most threatening pathogen to rice production, partly because it develops into spots to spread and infect more plants.
Plants have developed a strict defense response to restrict pathogens from spreading. The pathogen-associated molecular pattern (PAMP)–triggered immunity and effector-triggered immunity are two barriers in the plant defense system. The conserved PAMPs or specific effectors of microbes are essential for the plant to identify the pathogen. In the interaction between plants and pathogens (Jones and Dangl, 2006; Bent and Mackey, 2007), many molecular reactions are activated to assist the immune system, such as the production of reactive oxygen species (ROS), the secondary messengers of plant–pathogen interactions (Cheval et al., 2013; Hetmann and Kowalczyk, 2018). Hypersensitive response (HR), a symptom of programmed cell death, finally occurs within the infected area to avoid infection (Chen et al., 2012).
Lesion mimic mutants (LMMs) display spontaneous spots on leaves similar to the HR and usually exhibit differential resistance to pathogens without external stresses (Yoshimura et al., 1997). The first LMM, sl, was found in 1965 (Fujiwara et al., 2010), and since then, researchers have identified more rice mutants to explain the function of immune system genes in responding to pathogens. Most LMMs have enhanced resistance to Pyricularia oryzae or Xanthomonas oryzae pv. oryzae except for spl3 and spl6 (Kang et al., 2007; Zhang et al., 2011), which means that the immune mechanism of LMMs still needs to be substantiated.
Results
Phenotypic Characterization of lm8015-3
The rice LMM lm8015-3 was obtained from the ethyl methane sulfonate (EMS)–induced Zhonghui8015 (ZH8015) mutant library. The lower leaves first exhibited small orange-yellow spots in the seedling stage. The spots gradually spread throughout the leaves from the early tillering stage to the active tillering stage. During the harvest period, the initial spots fused into a few big spots, resulting in the death of leaves (Figures 1A–F). In addition, the plant height, number of productive tillers per plant, and the lengths of flag leaves of lm8015-3 were significantly smaller than in ZH8015 (Figures 1G–I). These results revealed that the occurrence of leaf spots affected the growth of lm8015-3.
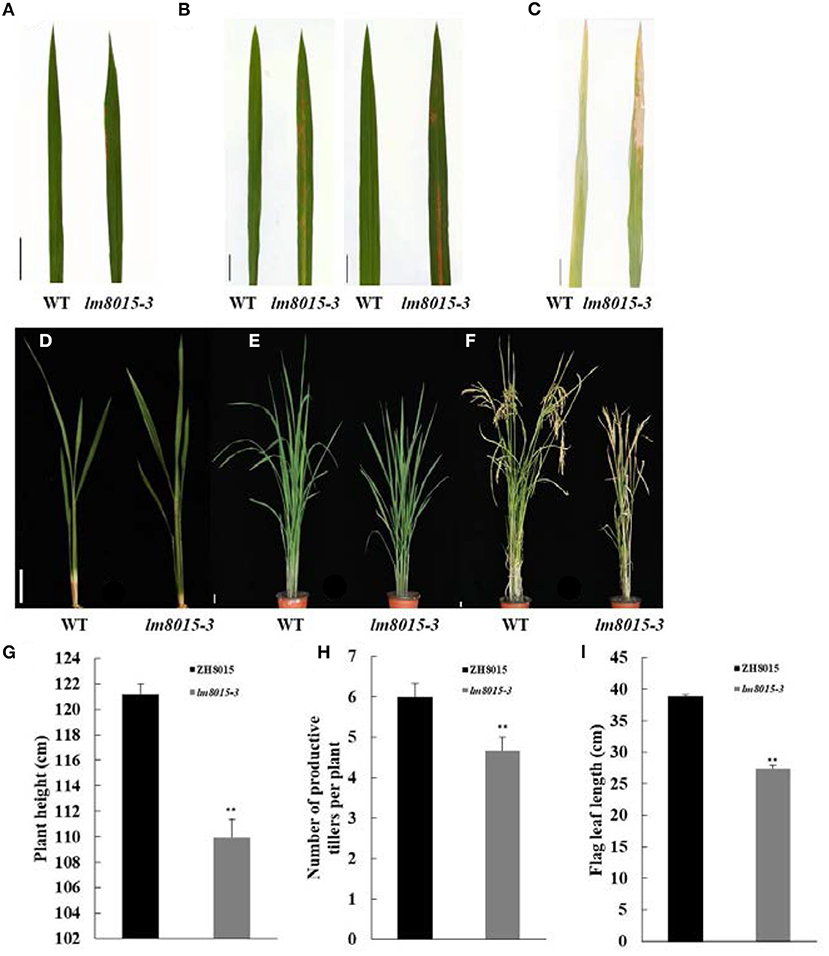
Figure 1. Phenotypic characterization of the lm8015-3 mutant. (A–C) Leaf phenotypes of ZH8015 and lm8015-3 at the seedling stage, tillering stage, and mature stage. (D–F) Plant architecture of ZH8015 and lm8015-3 at the seedling stage, tillering stage, and mature stage. Scale bar = 2 cm. (G–I) Bar graphs of plant height, number of productive tillers per plant, and flag leaf lengths of ZH8015 and lm8015-3 measured at the mature stage. **Significant at P < 0.01 (t-test); Error bars represent SD (n = 3).
Light-Dependent Formation and Chloroplast Degradation of lm8015-3
Light influences spot formation of LMMs. Therefore, we covered part of the leaf tip of lm8015-3 with foil to avoid exposure to natural light when the spots did not appear. One week later, we found spots on the lm8015-3 leaves exposed to sunlight, whereas no spots formed in the leaves without exposure (Figures 2A,B), indicating that spot formation of lm8015-3 was induced by natural light. Photosynthetic pigments are essential to plant growth, so we measured their contents. Compared with ZH8015, the contents of chlorophyll (Chl) and carotenoid (Car) of lm8015-3 were significantly decreased at the tillering stage (Figure 2C), suggesting a deficiency in the photosynthetic pigments. By transmission electron microscopy (TEM), the chloroplast structure of leaves of lm8015-3 was disrupted, whereas it was normal in ZH8015 (Figures 2D,E).
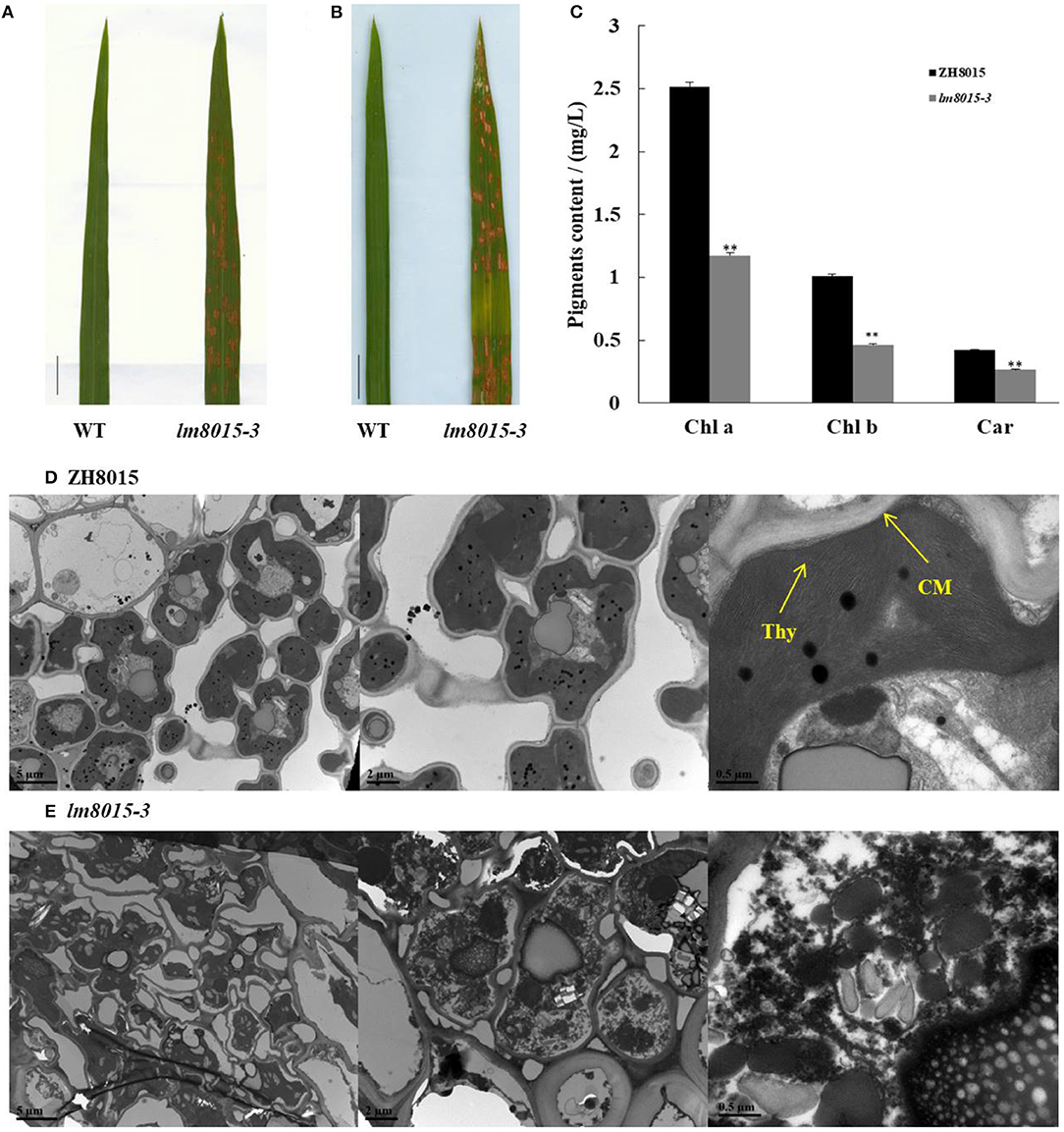
Figure 2. Identification of leaves of lm8015-3. (A,B) Shading experiment of the second top leaves of ZH8015 and lm8015-3 at the tillering stage. The leaves were exposed to light, and the leaves in (B) were covered with foil for 7 days; scale bar = 2 cm. (C) Determination of pigment contents of ZH8015 and lm8015-3 at the tillering stage. **Significant at P < 0.01 (t-test); Error bars represent SD (n = 3). (D,E) TEM observation of the chloroplasts in ZH8015 (D) and lm8015-3 (E) at the tillering stage. Thy, thylakoid lamellae; CM, chloroplast membrane.
ROS Accumulation and Cell Death in lm8015-3
LMMs always correspond with abnormal antioxidation metabolism, such as the accumulation of H2O2 or O2−. We observed H2O2 accumulation and apoptosis of lm8015-3 with diaminobenzidine (DAB) and Evans Blue (EB) staining. Compared with ZH8015, more H2O2 and dead cells accumulated in the leaves of lm8015-3 (Figures 3A,B).
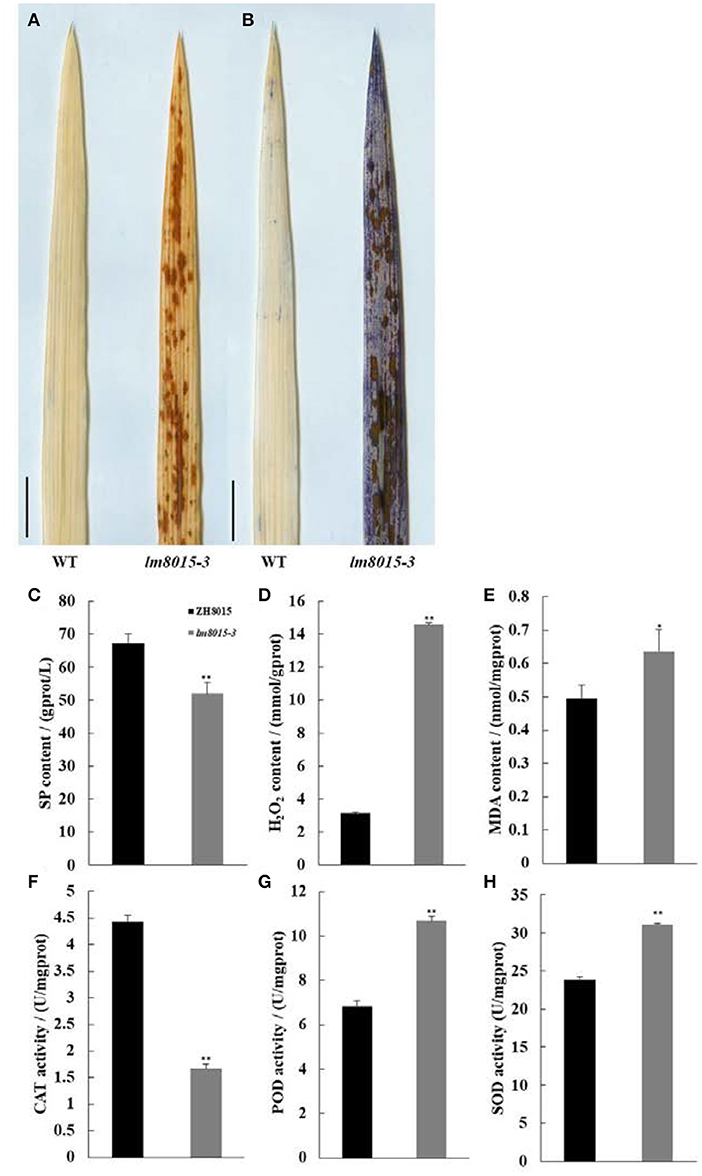
Figure 3. Staining and physiological indices of ZH8015 and lm8015-3. (A,B) DAB staining and EB staining of the leaves of the wild type (ZH8015) and lm8015-3 at the tillering stage. Scale bar = 2 cm. (C–H) Measurements of SP content, H2O2 content, MDA content, CAT activity, POD activity, and SOD activity of ZH8015 and lm8015-3 at the tillering stage. *Significant at P < 0.05 (t-test); **Significant at P < 0.01 (t-test); Error bars represent SD (n = 3).
Furthermore, we detected data of changes in physiological indices. The contents of soluble protein (SP) and malondialdehyde (MDA) are two key indexes that describe the extent of lipid peroxidation damage to plants. Compared with ZH8015, the SP content of lm8015-3 decreased significantly, whereas the MDA and H2O2 contents increased significantly (Figures 3C–E). These results mean that the excessive H2O2 accumulation seriously damaged the lipid peroxidation in lm8015-3 leaves. Superoxide dismutase (SOD) converts O2− into H2O2, and catalase (CAT) and peroxidase (POD) immediately convert H2O2 into water. However, CAT activity decreased significantly in the leaves of lm8015-3, opposite to the changes in SOD and POD (Figures 3F–H). The abnormal enzyme activity and metabolic product content of lm8015-3 indicated that SOD accumulation in lm8015-3 destroyed its metabolic system.
Gene Mapping of lm8015-3
To identify the candidate gene related to spot formation of lm8015-3, we crossed 02428 and lm8015-3 to generate F1 individuals. All F1 individuals showed a normal phenotype. The F2 generation displayed a 3:1 ratio between typical plants and spotted plants, indicating that a single nuclear recessive gene controlled the spotted phenotype of lm8015-3. Then, we used 96 F2 individuals with spots to generate primers over the 12 rice chromosomes (from our laboratory), and found the target gene on chromosome 12 (Figure 4A). We designed a series of primers to shorten the linkage interval, including C35-16, W35-6, W35-7, and W35-1 (Figure 4B). Finally, lm8015-3 was localized to a 544-kb interval between C35-16 and W35-6. The sequencing results showed a single base substitution (T to A) in the first exon of LOC_Os12g16720, which resulted in the substitution of aspartic acid to valine (Figure 4C). To verify the mutation, we constructed knockout plants under the Nipponbare background. The knockout plants showed the same orange spots on leaves similar to lm8015-3 (Figure 4D), as a result of the deletion or insertion of a single base in LOC_Os12g16720.
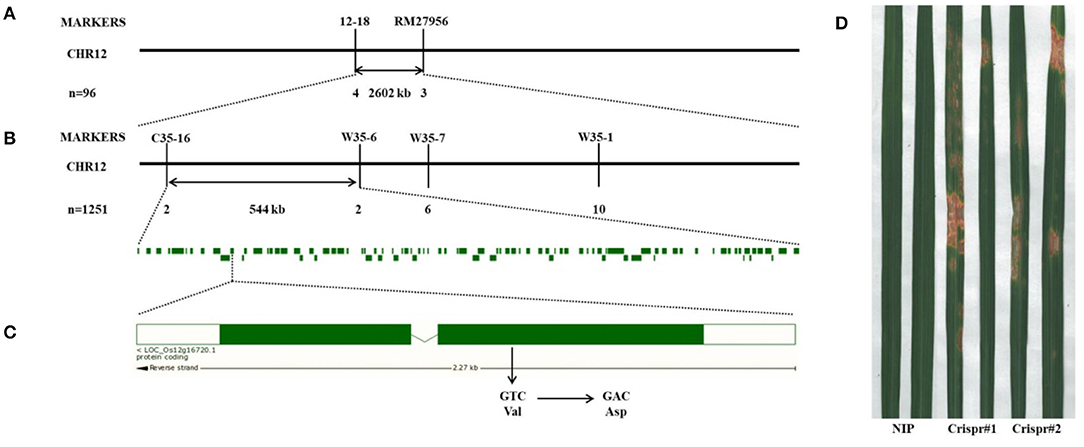
Figure 4. Map-based cloning of lm8015-3. (A) LM8015-3 was first mapped on chromosome 12 between markers 12–18 and RM27956 with 96 F2 individuals. (B) LM8015-3 was finally mapped between markers C35-16 and W35-6 with 1,251 F2 individuals. (C) A single base substitution (T to A) occurred in the first exon of LOC_Os12g16720 resulting in the mutation of Val to Asp. (D) The knockout plant of LOC_Os12g16720 in Nipponbare showed the same phenotype with lm8015-3.
Reduced Resistance of lm8015-3 to M. oryzae
In LMMs, changes in the expression levels of some defense gene always accompany the appearance of spots. We examined the expression levels of defense genes between the leaves with spots and the leaves with no spots. PR10 and WRKY45 were highly expressed in the leaves without spots and downregulated in leaves of lm8015-3 with spots. The expression level of PR1b decreased in the leaves without spots, opposite to other genes, whereas it also displayed the same decreased tendency in general (Figures 5A,B). These results indicated that the appearance of spots in lm8015-3 leads to downregulated expression of defense genes.
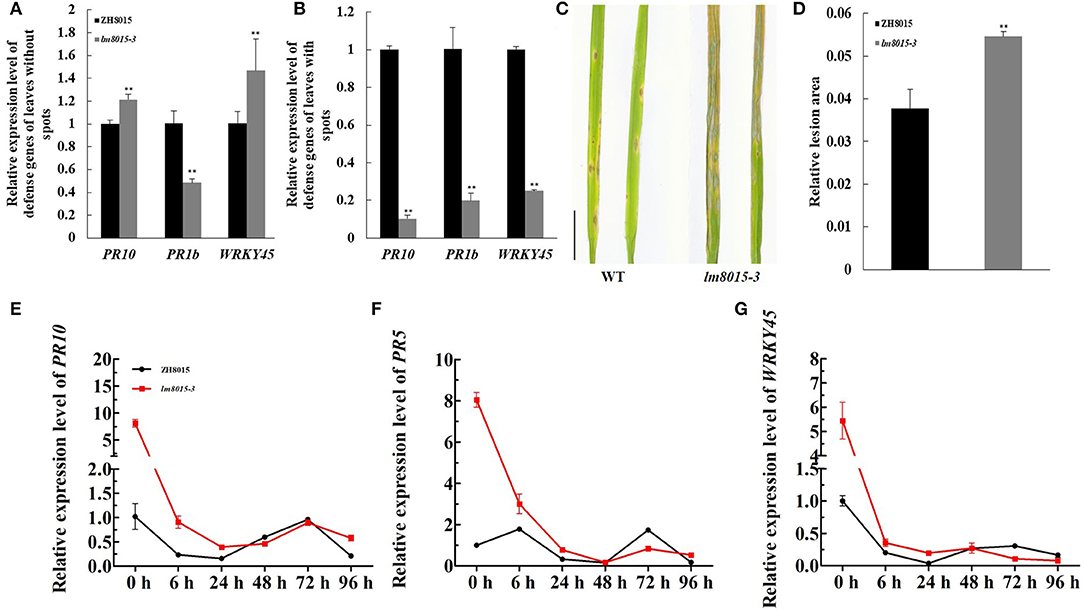
Figure 5. M. oryzae resistance of ZH8015 and lm8015-3. (A,B) Relative expression analysis of defense genes of the leaves with and without spots. The leaves without spots were younger leaves of lm8015-3 with no lesions at the tillering stage, whereas the leaves with spots were older leaves of lm8015-3 with lesions at the tillering stage. (C,D) Spray inoculation and statistics of relative lesion area of the 3-week-old wild type (ZH8015) and lm8015-3. (E–G) Analysis of relative expression levels of PR10, PR5, and WRKY45 of the 3-week-old ZH8015 and lm8015-3 after inoculation. **Significant at P < 0.01 (t-test); Error bars represent SD (n = 3).
LMMs have differential resistance to pathogens. Using the spray method to induce M. oryzae infection, 3-week-old lm8015-3 exhibited a more significant lesion area than ZH8015 (Figures 5C,D), meaning that formation of spots reduced the resistance of lm8015-3 to M. oryzae. The lesions of lm8015-3 appeared at 72 h after inoculation (HAI), earlier than in ZH8015. It seemed that 72 h is needed for M. oryzae to infect lm8015-3. After inoculation, we used the leaves of lm8015-3 and ZH8015, ranging from 0 to 96 HAI, for qRT-PCR of defense genes (PR10/PR5/WRKY45). The expression levels of all three genes decreased over time (Figures 5E–G), suggesting that the infection of M. oryzae suppressed the expression of defense genes. Expression levels of defense genes in lm8015-3 decreased before 24 h and gradually approached ZH8015 within 48 HAI, indicating that the early defense reaction of lm8015-3 was more sensitive than ZH8015. Expression levels of defense genes in lm8015-3 were significantly lower than those in ZH8015 from 48 to 72 HAI, resulting in the successful infection of lm8015-3 with M. oryzae. The expression level decreased from 72 to 96 HAI, which explained the delayed defense reaction of ZH8015. Therefore, lm8015-3 was susceptible to M. oryzae.
As mentioned before, 72 h is required for M. oryzae to infect lm8015-3. We used leaf-sheath inoculation to determine the M. oryzae growth conditions in ZH8015 and lm8015-3 at 72 h. The number of spores in lm8015-3 was higher than in ZH8015 (Figure 6A) under the identified growth conditions. Most of the spores were germinating, and there were more germinating conidia in the leaves of lm8015-3 (Figures 6B,C). However, the numbers of uninfected conidia and the successfully infected conidia were lower in lm8015-3 than in ZH8015.
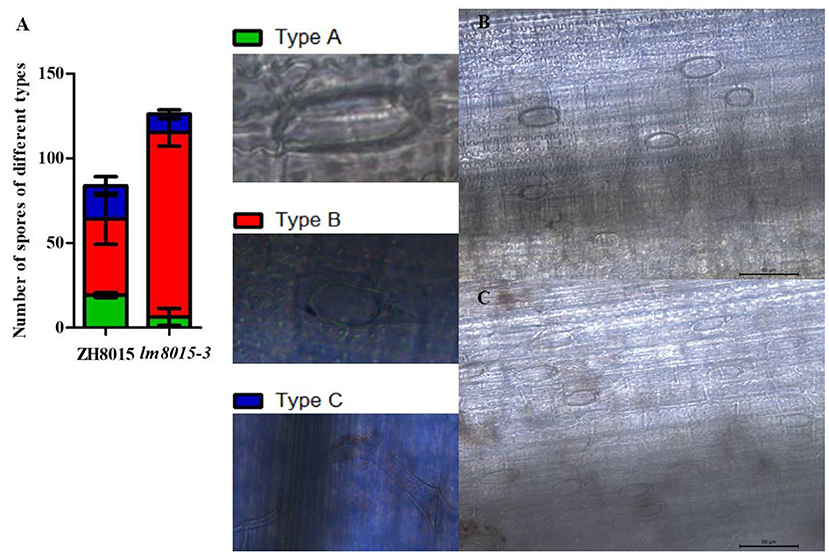
Figure 6. Statistics of different spore types of ZH8015 and lm8015-3 at 72 h after inoculation. (A) Graph of different types of spores of 3-week-old ZH8015 and lm8015-3 at 72 h after inoculation; Error bars represent SD (n = 3). (B,C) The 72-h–inoculated leaf sheath of 3-week-old ZH8015 and lm8015-3. Type A, uninfected conidium; Type B, germinating conidium; Type C, successfully infected conidium.
Discussion
Previous studies have demonstrated that sl encodes a cytochrome P450 monooxygenase. Cytochrome P450 monooxygenase is a large family of self-oxidizing ferrous hemeprotein enzymes, named for their specific absorption peaks at 450 nm. They are involved in the metabolism of endogenous and exogenous substances, including drugs and environmental compounds. Fujiwara et al. (2010) found that sl possesses tryptamine 5-hydroxylase activity, catalyzing tryptamine conversion to serotonin. However, the serotonin and salicylic acid (SA) pathways are antagonistic and trace back to chorismate (Maeda and Dudareva, 2012). As Lu et al. (2018) found, insect damage suppresses serotonin while inducing SA biosynthesis. Moreover, serotonin has high antioxidant activity, which could function as an oxygen radical scavenger to restrict the M. oryzae-induced lesion area from expanding (Hayashi et al., 2016).
Researchers previously identified more than five mutants related to LOC_Os12g16720; however, they exhibited differential resistance to M. oryzae. For example, sl showed reduced resistance, whereas sl-MH did not (Tian et al., 2020). We speculate that there are two main reasons for this phenomenon. One is the background differences between the wild type used, which might influence the genome sequence. The other more important reason is the mutation sites, which change the expression activity of LOC_Os12g16720. As mentioned before, 72 h is required for M. oryzae to infect lm8015-3. From 48 to 72 HAI, the pathogen-related genes of lm8015-3 were downregulated compared to ZH8015, a symbol of an ineffective immune system response. Os4CL5 and PAL genes are essential for lignin biosynthesis (Gui et al., 2011; Zhou et al., 2018), and OsbHLH6 encodes a factor to regulate the signal antagonism of SA and JA (Meng et al., 2020). Os4CL5 and PAL7 of lm8015-3 were downregulated, whereas OsbHLH6 had the opposite pattern (Supplementary Figure S1). The biosynthesis pathways of SA, serotonin, and melatonin were identified to be involved by Lu et al. (2018), so we speculated that the SA pathway of chorismate was damaged, which means that the serotonin pathway is active. The increased level of serotonin might be the real reason for the reduced resistance of lm8015-3. However, the mechanism of how M. oryzae induces serotonin accumulation in lm8015-3 is still unsubstantiated.
Conclusion
Pathogen resistance is an essential issue for rice breeding, as it can severely affect crop yield. This study identified the LMM lm8015-3, which exhibited reduced resistance to M. oryzae. The expression levels of most defense genes in lm8015-3 were downregulated when spots appeared. The defense reaction of lm8015-3 thus appears to be sensitive to M. oryzae. After the inoculation with M. oryzae, lm8015-3 was easier to infect than ZH8015. Furthermore, the infected lesions on lm8015-3 appeared 2 days earlier than ZH8015 because of the disordered immune system from 48 to 72 HAI in lm8015-3.
Methods
Plant Materials and Blast Isolates
ZhongHui8015, an indica rice restorer line, was bred at the China National Rice Research Institute (CNRRI), Hangzhou, Zhejiang, China. The LMM lm8015-3 was isolated from the ZH8015 EMS-induced library. The F2 population derived from 02428/lm8015-3 was used for genetic analysis and fine mapping. We measured the agronomic characteristics of lm8015-3 and ZH8015 at the harvest period with three biological replicates, including plant height, numbers of productive tillers per plant, and flag leaf length. All materials were grown in the FuYang experimental base of CNRRI. The M. oryzae strain 14-1 was virulent to ZH8015.
Shading Experiment
At the tillering stage, the second top leaves without lesions of lm8015-3 and ZH8015 were shaded with 4-cm-wide foil for 7 days, and then, the foil was removed and the leaves were photographed.
Measurements of Photosynthetic Pigment Contents
As described before (Arnon, 1949; Schippers et al., 2015), the second top leaves of plants with lesions at the tillering stage were used to measure the contents of photosynthetic pigments. Each experiment was performed with three biological replicates. We used the means of three replicates for the t-test.
Chloroplast Structure Observation
For the TEM observation of chloroplast structure, the leaves of lm8015-3 and ZH8015 at the tillering stage were first soaked in 2.5% glutaraldehyde for 24 h. Then, we sent samples to Zhejiang University for subsequent treatments.
Histochemical Analysis
The leaves of lm8015-3 and ZH8015 at the tillering stage were stained with DAB and EB to observe the H2O2 accumulation and cell death as described previously (Thordal-Christensen et al., 1997; Li et al., 2004).
Physiological Indexes
Following the instructions of kits from the Nanjing JianCheng Bioengineering Institute, we ground the leaves of lm8015-3 and ZH8015 at the tillering stage into homogenates to test the content of SP, MDA, H2O2, and the activities of SOD, POD, and CAT. Three replications were used for each group. The means of three replicates were used for the t-test.
Gene Mapping
As described previously (Rogers and Bendich, 1985), DNA of the mapping group was extracted using Cetrimonium Bromide (CTAB). Genome sequences of rice cultivars were downloaded from http://ensembl.gramene.org/ and http://rice.genomics.org.cn/, and differences between the genome sequences of the japonica cultivar Nipponbare and the indica cultivar 9311 were used to design primers for map-based cloning of lm8015-3. With the databases GRAMENE (http://ensembl.gramene.org/) and MSU (http://rice.uga.edu/), we designed primers for genes and analyzed the sequence differences. The PCR followed the instructions of Vazyme (P222-01). To construct knockout plants of LOC_Os12g16720, we used pCAMBIA1305.1 vector to delete or insert a single base in the exon. The primers for gene mapping and confirmation are listed in Supplementary Table S1.
qRT-PCR Analysis
Following kit instructions for Invitrogen TRIzol (Thermo Fisher), all RNA samples were extracted. To determine whether lesion occurrence changed the expression levels of defense genes, we extracted RNA of the leaves with or without lesions of lm8015-3 at the tillering stage. In the qRT-PCR of defense genes after inoculation, the 3-week-old leaves RNA of lm8015-3 and ZH8015 at 0, 6, 24, 48, 72, and 96 HAI were extracted. With SYBR qPCR Master Mix of Vazyme (Q511-02), three replicates were used for each group. The means of three replicates were used for the t-test. The primers for qRT-PCR are listed in Supplementary Table S1.
Blast Inoculation
M. oryzae strain 14-1, which was virulent to ZH8015, was cultured on oatmeal agar medium for 2 weeks under light for sporulation. We sprayed the 3-week-old leaves of lm8015-3 and ZH8015 with a conidial suspension of 2 × 105 spores/ml in the growth chamber. We measured the relative infected-lesion area of lm8015-3 and ZH8015 by relative pixels with Photoshop. The detached sheath of 3-week-old lm8015-3 and ZH8015 leaves was injected with a conidial suspension of 1 × 106 spores/ml in the growth chamber by leaf sheath inoculation (Li et al., 2011). After 72 h, the sample was placed under a microscope and observed with a 40× objective. The number of each spore type, observed in 10 random fields of view, was used to calculate one biological replicate. Three biological replicates were used for each group. The means of three replications were used for the t-test.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
XZ and QL contributed to conception and design of the study. CW finished all the experiments and wrote the manuscript. BW, LC, YXZ, YG, YC, and YZ contributed to the completion of the study. All authors contributed to manuscript revision, read, and approved the submitted version.
Funding
This study was supported by grants from the National Natural Science Foundation of China (31871236 and 31801726) and the Agricultural Science and Technology Innovation Program of the Chinese Academy of the Agricultural Sciences (CAAS-ASTIP-2013-CNRRI).
Conflict of Interest
XZ was employed by the Zhejiang Guodao High-Tech Seed Industry Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fsufs.2022.809008/full#supplementary-material
References
Arnon, D. I. (1949). Copper enzymes in isolated chloroplasts. Polyphenoloxidase in beta vulgaris. Plant Physiol. 24, 1–15. doi: 10.1104/pp.24.1.1
Bent, A. F., and Mackey, D. (2007). Elicitors, effectors, and R genes: the new paradigm and a lifetime supply of questions. Annu. Rev. Phytopathol. 45, 399–436. doi: 10.1146/annurev.phyto.45.062806.094427
Chen, M., Zeng, H., Qiu, D., Guo, L., Yang, X., Shi, H., Zhou, T., and Zhao, J. (2012). Purification and characterization of a novel hypersensitive response-inducing elicitor from Magnaporthe oryzae that triggers defense response in rice. PLoS ONE. 7, e37654. doi: 10.1371/journal.pone.0037654
Cheval, C., Aldon, D., Galaud, J. P., and Ranty, B. (2013). Calcium/calmodulin-mediated regulation of plant immunity. Biochim. Biophys. Acta. 1833, 1766–1771. doi: 10.1016/j.bbamcr.2013.01.031
Fujiwara, T., Maisonneuve, S., Isshiki, M., Mizutani, M., Chen, L., Wong, H. L., et al. (2010). Sekiguchi lesion gene encodes a cytochrome P450 monooxygenase that catalyzes conversion of tryptamine to serotonin in rice. J. Biol. Chem. 285, 11308–11313. doi: 10.1074/jbc.M109.091371
Gui, J., Shen, J., and Li, L. (2011). Functional characterization of evolutionarily divergent 4-coumarate:coenzyme a ligases in rice. Plant Physiol. 157, 574–586. doi: 10.1104/pp.111.178301
Hayashi, K., Fujita, Y., Ashizawa, T., Suzuki, F., Nagamura, Y., and Hayano-Saito, Y. (2016). Serotonin attenuates biotic stress and leads to lesion browning caused by a hypersensitive response to Magnaporthe oryzae penetration in rice. Plant J. 85, 46–56. doi: 10.1111/tpj.13083
Hetmann, A., and Kowalczyk, S. (2018). Membrane receptors recognizing MAMP/PAMP and DAMP molecules that activate first line of defence in plant immune system. Postepy Biochem. 64, 29–45. doi: 10.18388/pb.2018_103
Jones, J. D., and Dangl, J. L. (2006). The plant immune system. Nature. 444, 323–329. doi: 10.1038/nature05286
Kang, S. G., Matin, M. N., Bae, H., and Natarajan, S. (2007). Proteome analysis and characterization of phenotypes of lesion mimic mutant spotted leaf 6 in rice. Proteomics. 7, 2447–2458. doi: 10.1002/pmic.200600961
Li, R., Lqn, S. Y., and Xu, Z. X. (2004). Studies on the programmed cell death in rice during starchy endosperm development. Agric. Sci. China. 3, 663–670.
Li, W., Zhong, S., Li, G., Li, Q., Mao, B., Deng, Y., et al. (2011). Rice RING protein OsBBI1 with E3 ligase activity confers broad-spectrum resistance against Magnaporthe oryzae by modifying the cell wall defence. Cell Res. 21, 835–848. doi: 10.1038/cr.2011.4
Lu, H. P., Luo, T., Fu, H. W., Wang, L., Tan, Y. Y., Huang, J. Z., et al. (2018). Resistance of rice to insect pests mediated by suppression of serotonin biosynthesis. Nat. Plants. 4, 338–344. doi: 10.1038/s41477-018-0152-7
Maeda, H., and Dudareva, N. (2012). The shikimate pathway and aromatic amino acid biosynthesis in plants. Annu. Rev. Plant Biol. 63, 73–105. doi: 10.1146/annurev-arplant-042811-105439
Meng, F., Yang, C., Cao, J., Chen, H., Pang, J., Zhao, Q., et al. (2020). A bHLH transcription activator regulates defense signaling by nucleo-cytosolic trafficking in rice. J. Integr. Plant Biol. 62, 1552–1573. doi: 10.1111/jipb.12922
Rogers, S. O., and Bendich, A. J. (1985). Extraction of DNA from milligram amounts of fresh, herbarium and mummified plant tissues. Plant Mol. Biol. 5, 69–76. doi: 10.1007/BF00020088
Schippers, J. H., Schmidt, R., Wagstaff, C., and Jing, H. C. (2015). Living to die and dying to live: the survival strategy behind leaf senescence. Plant Physiol. 169, 914–930. doi: 10.1104/pp.15.00498
Thordal-Christensen, H., Zhang, Z. G., Wei, Y. D., and Collinge, D. B. (1997). Subcellular localization of H2O2 in plants. H2O2 accumulation in papillae and hypersensitive response during the barley-powdery mildew interaction. Plant J. 11, 1187–1194. doi: 10.1046/j.1365-313X.1997.11061187.x
Tian, D., Yang, F., Niu, Y., Lin, Y., Chen, Z., Li, G., et al. (2020). Loss function of SL (sekiguchi lesion) in the rice cultivar Minghui 86 leads to enhanced resistance to (hemi) biotrophic pathogens. BMC Plant Biol. 20, 507. doi: 10.1186/s12870-020-02724-6
Yoshimura, A., Ideta, O., and Iwata, N. (1997). Linkage map of phenotype and RFLP markers in rice. Plant Mol. Biol. 35, 49–60. doi: 10.1023/A:1005764026871
Zhang, H., Liu, K., Zhang, X., Tang, W., Wang, J., Guo, M., et al. (2011). Two phosphodiesterase genes, PDEL and PDEH, regulate development and pathogenicity by modulating intracellular cyclic AMP levels in Magnaporthe oryzae. PLos ONE. 6, e17241. doi: 10.1371/journal.pone.0017241
Keywords: rice, LMM, ZH8015, lm8015-3, defense
Citation: Wang C, Wang B, Cao L, Zhang Yx, Gao Y, Cao Y, Zhang Y, Liu Q and Zhang X (2022) Identification and Gene Mapping of the Lesion Mimic Mutant lm8015-3. Front. Sustain. Food Syst. 6:809008. doi: 10.3389/fsufs.2022.809008
Received: 04 November 2021; Accepted: 03 February 2022;
Published: 25 March 2022.
Edited by:
Wei Li, Hunan Agricultural University, ChinaReviewed by:
Yuese Ning, Institute of Plant Protection (CAAS), ChinaShimin Zuo, Yangzhou University, China
Zhiqiang Li, Institute of Plant Protection (CAAS), China
Copyright © 2022 Wang, Wang, Cao, Zhang, Gao, Cao, Zhang, Liu and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qunen Liu, bGl1cXVuZW4yMDJAMTYzLmNvbQ==; Xiaohui Zhang, emhhbmd4aWFvaHVpMDJAY2Fhcy5jbg==
 Chen Wang
Chen Wang Beifang Wang
Beifang Wang Liyong Cao1,2
Liyong Cao1,2 Yingxin Zhang
Yingxin Zhang Yue Zhang
Yue Zhang Qunen Liu
Qunen Liu