- 1Department of Biochemistry, Punjab Agricultural University, Ludhiana, India
- 2Biochemistry Section, ICAR-Indian Institute of Maize Research, Ludhiana, India
- 3ICAR Stands for Indian Council of Agricultural Research (ICAR)-National Bureau of Plant Genetic Resources, New Delhi, India
The development of quality protein maize (QPM) was considered a significant leap toward improvement in the nutritional status of rural masses in developing countries. The nutritional quality of QPM is attributed to the higher concentration of essential amino acids, particularly lysine and tryptophan, in its kernel endosperm. However, the similarity in the grains of QPM and normal maize necessitates the development of a standard protocol to assess the protein quality of maize. The present study aimed at improving the protocol of protein quality assessment in QPM. For this purpose, endosperm defatting and protein estimation procedures were restandardized and optimized with respect to the protocol duration and its amenability for high-throughput analysis. Unlike normal maize, QPM and opaque-2 mutants were completely defatted within a 48 h period. It was observed that the tryptophan content, calculated at each defatting interval, increased in the samples defatted for a longer duration. No significant differences were observed in the tryptophan content analyzed in the samples defatted for 48 and 72 h. Moreover, the endosperm protein estimated by using the Bradford method with certain modifications strongly correlated with the micro-Kjeldahl method (r = 0.9). Relative to the micro-Kjeldahl method, the Bradford method was found to be precise, rapid, and hazard-free. The present findings enable a testing protocol of reduced time duration that can be used in resource-poor settings for the determination of a protein quality assay in QPM. Overall, the present study effectively helped in reducing the defatting time by 24 h and protein estimation by 3 h as compared to the already established International Maize and Wheat Improvement Center protocol. This is expected to enable the aggregation of high-protein-quality maize to facilitate its commercialization.
Introduction
Maize is a globally important crop and the third most important cereal in India, in terms of production (FAOSTAT, 2020). It played a major role in alleviating protein-energy malnutrition, micronutrient deficiencies, and vision impairment worldwide. Generally, plant-based foods are economically and environmentally suitable for human protein nutrition (Langyan et al., 2022). Due to its use in the staple diet and processed food, it is a good candidate to manage the health of vulnerable populations. A number of technologies, including molecular breeding (Olsen and Phillips, 2009, Prasanna et al., 2020), genetic engineering (Pérez-Massot et al., 2013; Guo et al., 2018), and gene editing, have the potential to deliver nutritious maize (Ku and Ha, 2020; Singh et al., 2020). Quality protein maize (QPM) is a nutritionally improved commodity and is reported to be equivalent to the milk protein casein in terms of nutritional composition (Arsenault and Brown, 2017). In the presence of a specific opaque-2 gene, the protein profile of QPM is improved. Thus, QPM has about twice the level of lysine and tryptophan (4.15 and 1%) than normal maize (2 and 0.5%) and also increased levels of histidine, arginine, aspartic acid, and glycine but a reduced level of leucine (Wu et al., 2010; Onireti and Ikujenlola, 2020). The enhanced levels of lysine and tryptophan in QPM, which are otherwise limiting in normal maize, make them a good source of biologically usable protein along with several nutritional benefits. QPM is thus a valuable resource for philanthropic efforts toward the nutritional upliftment of vulnerable populations, as various studies on children and adults in a controlled manner have established its nutritional superiority. These studies demonstrated that children fed with QPM had fewer sick days, were more probable to escape death due to diarrhea, and had reduced stunting and better growth-enhancing capabilities (Nedi et al., 2016). Furthermore, these studies also revealed that as animal feed, QPM is more economical and productive than purchased rations (Nurit et al., 2009).
QPM is a product of conventional breeding. The QPM breeding program involves the introduction of three distinct genetic systems, viz., (i) the homozygous recessive and mutated allele of the opaque-2 gene that downregulates the expression of 22-kDa α-zein and 15-kDa β-zein genes, leading to a decrease in nutritionally poor zein protein, (ii) endosperm hardness modifier genes to eliminate the pleotropic effects like a soft and chalky kernel appearance associated with opaque-2 mutants, and (iii) amino acid modifier genes (Arruda et al., 2000; Gavazzi et al., 2007; Tripathy et al., 2017). The opaque-2 mutation has been extensively studied at the biochemical, genetic, molecular and phenotypic levels. Due to the unavailability of the rapid protein assay, the nutritional potential of QPM could not be harnessed as the farmers were unable to fetch any remunerative prices for their QPM produce as it becomes difficult to distinguish QPM from normal maize at grain procurement centers. The similarity in the kernel texture between QPM and normal maize, therefore, necessitates the development of a rapid assay methodology to assess the protein quality of QPM. As per the standard protocol devised by the International Maize and Wheat Improvement Center (CIMMYT), the concentration of tryptophan in the kernel endosperm of QPM should be more than 0.6% of endosperm protein. The method involves the defatting of the maize endosperm for 72 h, followed by protein hydrolysis for 16 h using papain. The tryptophan is then measured spectrophotometrically and expressed as a quality index by calculating the ratio of tryptophan in the endosperm protein. Although the method is precise and accurate, it is lengthy and time consuming. Presently, researchers are seeking a concise and precise method for the protein quality assay. The objective of the present study was, therefore, to develop a rapid laboratory assay to expedite the tryptophan estimation process by optimizing the defatting process and protein estimation in maize.
Materials and Methods
Chemicals and Reagents
For the defatting of samples, petroleum ether (40–60°C) was procured from Sigma-Aldrich (3050 Spruce street, St. Louis, MO 63103 USA 314-771-5765 SIGMA ALDRICH CHEMIE GmbH, Riedstr, 2D-89555 Steinhelm 497329970). For the analysis of tryptophan content, the glacial acetic acid, ferric chloride, papain, DL-tryptophan, and sulfuric acid were obtained from Merck (Merck Lifescience Private Limited, Godrej one, 8th floor, Pirojshanagar, Eastern Express Highway, Vikhroli (East), Mumbai 400079). For the extraction of the maize protein, sodium acetate was procured from Fischer Scientific (Thermo Fischer Scientific India Pvt. Ltd. 403-404, B wing, Delhi, Hiranandani, Business park Powai, Mumbai-400076); ethanol and the Bradford reagent were procured from Himedia (HiMedia Laboratories Pvt. Ltd. 23, Vodhani Ind. Est., LBS Marg, Mumbai-400086, India).
Experimental Material
The genetically pure seeds of a diverse panel of inbreds consisting of 25 lines of normal maize and 13 lines each of opaque-2 maize and QPM were used for the present study. Each plot consisted of 5 rows of 3 m with a row-to-row distance of 60 cm and plant-to-plant distance of 20 cm, resulting in 75 plants of each genotype. The experimental lines varied in kernel color, ranging from deep yellow to deep orange. The complete set of experimental material was grown in a single block at the experimental fields of Indian Council of Agricultural Research (ICAR)-Indian Institute of Maize Research, Ludhiana, India. All the individual entries were pollinated separately. Proper care was taken to maintain the genetic purity of each genotype.
Sample Collection and Screening
The samples from individual entries were collected from four different ears. An equal number of kernels were pooled and treated as one sample in order to reduce the effect of biological variation between ears on gene expression. A minimum of three technical replicates were used for each experiment. Further samples were screened using a light box and assigned a modification score ranging from 1 to 5 as per the percent opaqueness observed in the kernel (Sethi et al., 2020). It is a preliminary measure to segregate opaque-2, QPM, and normal lines on the basis of their endosperm textures. Following screening, the endosperms were extracted from the maize kernels and dried in a hot air oven; the dried samples were ground using a Cyclotec sample mill.
Oil Estimation
The oil content was estimated as per the Ankom method with certain modifications (Ankom, 2005). For this purpose, 2 g of milled maize flour was encapsulated in a filter bag and completely demoisturized prior to defatting. To optimize the defatting time, the dried filter bags containing samples were defatted for different time durations, viz., 6, 12, 24, 48, and 72 h using a non-polar solvent such as petroleum ether (40–60°C).
Tryptophan Estimation
The tryptophan content was estimated in the samples defatted for different time intervals (Hernandez and Bates, 1969). For this purpose, 100 mg of defatted sample was digested using papain. After incubation (65°C for 16 h), the supernatant was treated with reagent C. The colored complex was measured at 545 nm in a UV-Vis spectrophotometer.
Protein Estimation
Protein was estimated using both micro-Kjeldahl and Bradford methods. The micro-Kjeldahl method measures the crude protein content (AOAC, 1975). The protein estimated by the Bradford method (Bradford, 1976; Affolter et al., 2021) involves protein extraction via sonication for 30 min with 0.1 N NaOH (pH-12.5), followed by 55% ethanol (Shukla and Cheryan, 2001), and the final extract was estimated as per the Bradford method.
Statistical Analysis
The F test and ANOVA for multivariate data were conducted to check the level of significance for the time course evaluation of % oil removed in the experimental lines. Similarly, the level of significance, i.e., p-value, was calculated for the tryptophan content estimated at different time intervals. Further, the Pearson's correlation coefficient (r) and coefficient of determination (R2) were calculated for the total protein content estimated via the Bradford method and micro-Kjeldahl method.
Results
Optimization of Endosperm Defatting in the Experimental Lines
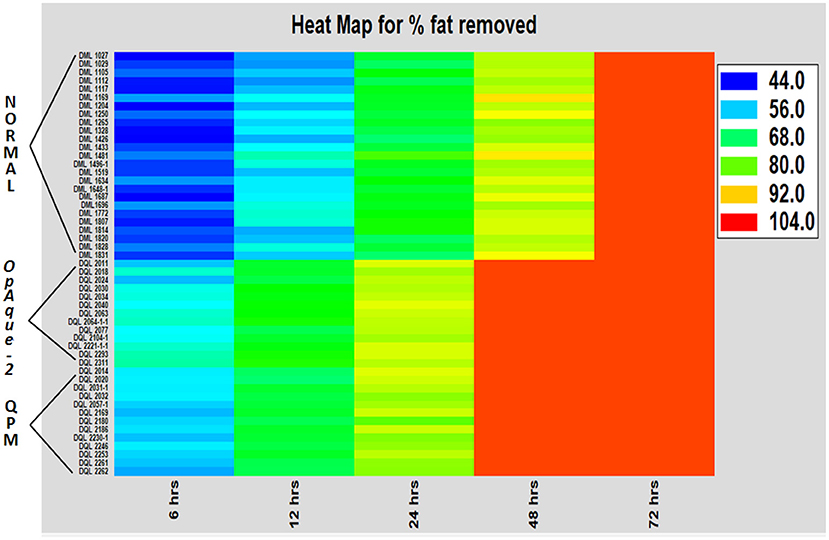
Defatting for different time intervals was done to optimize the oil extraction protocol. The results revealed that the proportion of oil content extracted is directly proportional to the duration of defatting in all the experimental genotypes (Figure 1). As the duration for defatting is increased, the extracted oil content also increased. However, the rate of defatting is maximum in opaque-2, followed by QPM and normal genotypes. The ANOVA for the multivariate data for defatting revealed that a significant difference exists between normal, opaque-2, and QPM with P ≤ 0.005. It is also observed that for complete defatting, normal lines require 72 h, whereas opaque-2 and QPM require only 48 h of extraction. The vitreous and hard kernel texture in normal maize may be primarily responsible for slower defatting, whereas the softness in the opaque-2 kernel assists the fat component to leave the kernel at a faster rate. The delayed extraction of oil in normal maize is also evident from the heat map of defatting (Figure 2).
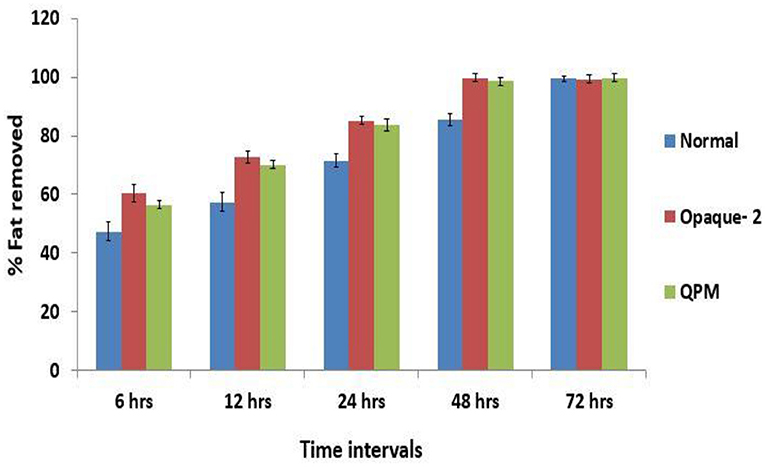
Figure 1. Comparative evaluation of rate of oil extracted (%) at different time intervals in the experimental genotypes. Error bars denote overall ±SD of the genotypes at different time intervals. The difference in the extracted oil (%) within the experimental materials at different defatting time was significant at P ≤ 0.05.
Effect of Defatting on Tryptophan Estimation
A significant difference exists in the tryptophan content estimated in triplicates in all experimental lines, which were defatted for different time intervals (p = 0.0011; Figure 3). The tryptophan content increases as the time duration for defatting increases. The maximum tryptophan content was observed in the samples defatted for 72 h, and the minimum tryptophan content was observed in the samples defatted for 6 h in all 51 lines taken together (Figure 3), indicating that the oil content has a profound effect on the tryptophan estimation process. It was also observed that the tryptophan content is almost similar in non-defatted samples and the samples defatted for 6 h. The comparisons in the tryptophan content among normal, opaque-2, and QPM lines defatted for different time intervals is presented in Figure 3. Although the estimated tryptophan content increases as the time duration of defatting increases, no appreciable differences were observed in the experimental lines defatted for 48 and 72 h. The mean values of tryptophan content at 48 and 72 h was observed to be 0.38 and 0.41%, respectively, for normal, 0.64 and 0.67% for opaque-2, and 0.62 and 0.65% for QPM.
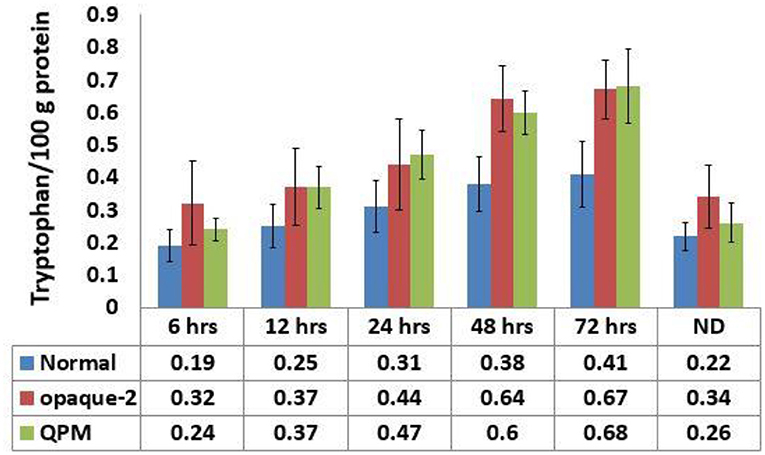
Figure 3. Comparative tryptophan content in the experimental genotypes estimated at different defatting time. Error bars denote overall ±SD of the genotypes at particular time interval. The difference between the tryptophan content (%) among all experimental lines at different defatting time was significant at P ≤ 0.05. ND, non-defatted samples.
Comparison of Total Protein by Micro-Kjeldahl and Bradford Method
The total protein estimation in the kernel endosperm is significant in the analysis of the protein quality index. The protein quality index is the ratio of the concentration of tryptophan to the endosperm protein. The protein content was estimated in triplicates in all 51 experimental lines by micro-Kjeldahl as well as Bradford methods. The comparative means of protein content estimated through Bradford and micro-Kjeldahl methods in 25 normal, 13 opaque-2, and 13 QPM lines is depicted in Figure 4. A significant difference was observed between the protein content estimated by both the methods (p < 0.05). The total protein content determined by the Bradford method was found to be underestimated as the mean values of the protein content in normal, opaque-2, and QPM lines are around 20% less as compared to the samples analyzed through the micro-Kjeldahl method (Figure 4). However, the protein estimations by the modified Bradford method and micro-Kjeldahl method are highly correlated. The linear regression coefficients of 0.89, 0.72, and 0.89 were observed between the protein estimated by the modified Bradford method and micro-Kjeldahl method in normal, opaque-2, and QPM lines, respectively (Figures 5A–C). However, a strong positive correlation (r = 0.9) with coefficient of determination 0.81 was observed between the protein content estimated through micro-Kjeldahl and Bradford methods in all the 51 lines taken together (Figures 5D, 6).
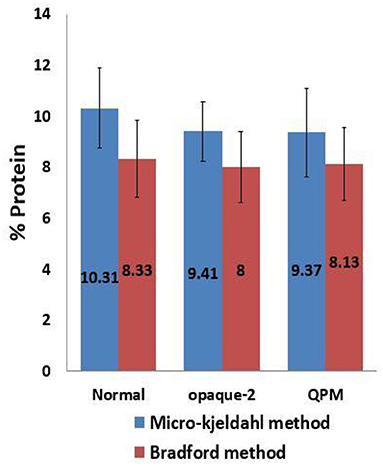
Figure 4. Comparative evaluation of protein content estimated by micro-Kjeldahl and Bradford methods. Error bars denote overall ±SD of the genotypes at particular time interval.
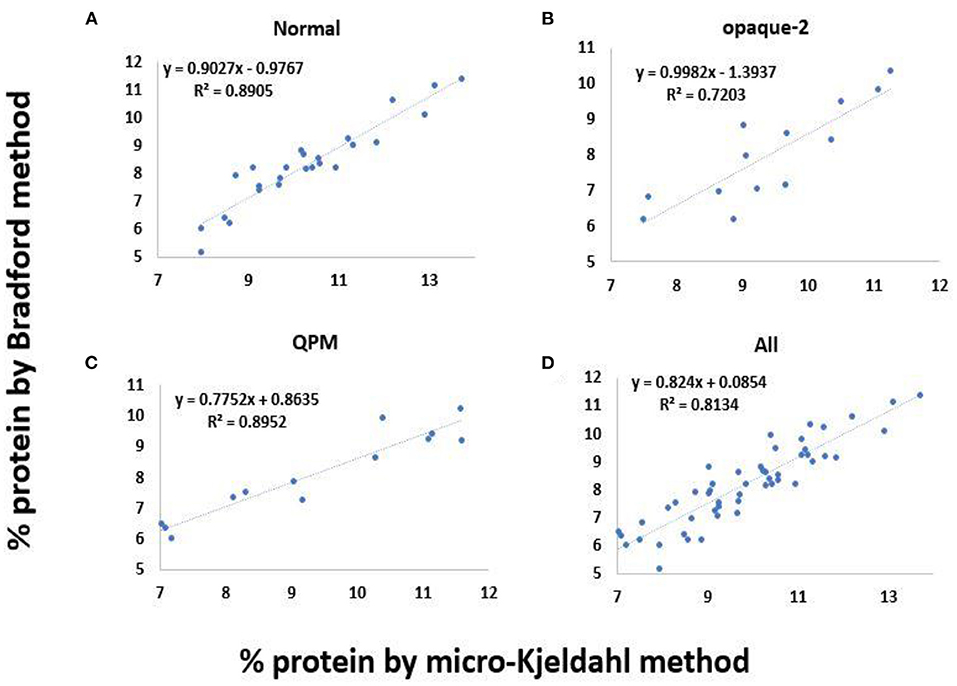
Figure 5. Correlation between protein content estimated by micro-Kjeldahl and Bradford methods. (A) Normal maize genotypes (sample size = 25), (B) opaque-2 maize genotypes (sample size = 13), (C) QPM genotypes (sample size = 13), and (D) Composite of above three samples (sample size = 51).
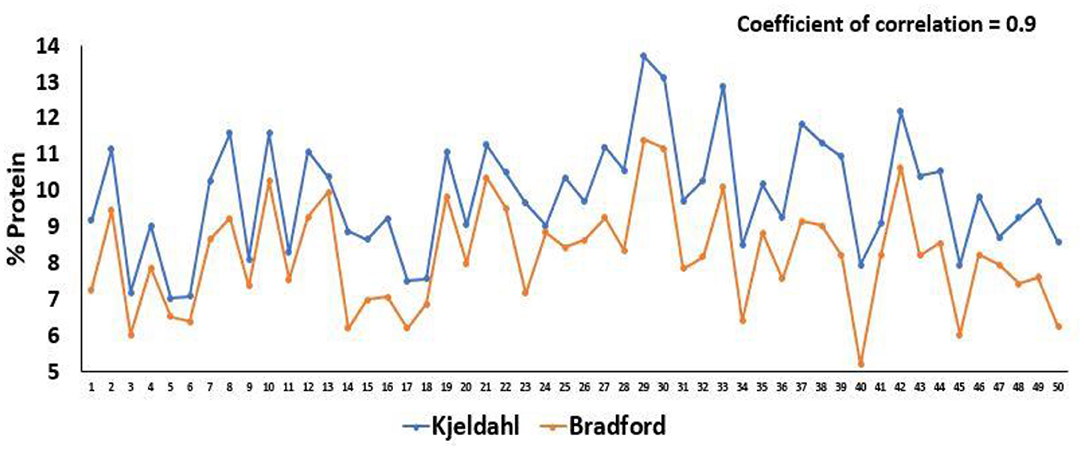
Figure 6. Protein content estimated by micro-Kjeldahl and Bradford methods in 51 experimental lines.
Discussion
Maize kernels usually contain between 2 and 4% of oil, which is primarily located in the germ portion (Chaudhary et al., 2012). It has been shown that maize protein fractions accumulate in the developing kernel differentially (Sethi et al., 2020). With the progressive accumulation of zein/prolamin and prolamin-like protein fractions, the overall kernel hardness increases, but this affects the amount of lysine and tryptophan (Sethi et al., 2021). Oil interferes with the estimation of other components, including tryptophan. The tryptophan estimation of maize requires a papain hydrolysis of the protein to release the amino acids, subsequently specifically estimating the tryptophan content using the acetic acid method (Hernandez and Bates, 1969; Bjarnason and Vasal, 1992). For the proper enzymatic hydrolysis of maize protein using papain, the acetic acid method for tryptophan estimation requires prior defatting of samples. The tryptophan content was analyzed in the samples defatted for different time intervals using the papain hydrolysis method. As mentioned above, the tryptophan content increased as the duration for defatting is increased. It was observed that QPM and opaque-2 samples were completely defatted in 48 h (Figure 1). As per the standard protocol, 72 h of defatting is required for an accurate estimation of tryptophan in maize flour (Hernandez and Bates, 1969; Shetti et al., 2020). A reduction of 24 h for the complete defatting of opaque-2 and QPM and without the use of special equipment would immensely help toward the fast screening of high-protein-quality material for the benefit of farmers as well as seed procurement agencies involved in QPM aggregation on a commercial scale. Unlike opaque-2 and QPM, normal maize has a hard endosperm. The positive correlation between the tryptophan content and kernel opacity has already been established by a previous study, which reported that opaque-2 mutants retained higher tryptophan content as compared to normal lines (Gupta et al., 2013). Opaque-2 mutation is also responsible for the increased concentration of free amino acids as reported by earlier studies (Brochetto-Braga et al., 1992; Wang et al., 2001; Manicacci et al., 2009; Sethi et al., 2021). The softness of the endosperm due to opaque-2 mutation leads to a distorted protein–starch matrix in opaque-2 and QPM germplasm. This results in a chalky and soft kernel texture, which is hardened through the action of endosperm modifiers. Despite this, the differences in the organization of the starch–protein matrix remain to some extent (Holding, 2014). These difference in starch–protein complexation in normal maize vs. the opaque-2 and QPM may be responsible for the enhanced defatting rate of the opaque-2 and QPM as compared to the normal maize. We have demonstrated that a defatting time of 48 h, without the use of specialized equipment, is sufficient to screen for high-protein-quality material in maize (Figure 3).
In order to select high-protein-quality maize, a parameter termed “protein quality index” needs to be analyzed. A protein quality index is analyzed by determining the ratio of tryptophan to protein content in the kernel endosperm. The protein content is usually measured by the micro-Kjeldahl method, whereby the total nitrogen content is measured through digestion, distillation, and titration. This requires expensive instrumentation, skilled workforce, a constant running cost to cover maintenance charges, and certain consumables that are hazardous in nature. Moreover, it is not very amenable for high-throughput analysis. The whole process requires 4–5 h, and a maximum of 16 samples can be analyzed in a day by this protocol. This is in contrast to the spectrophotometric tryptophan estimation described above, which can be done in a high-throughput format. In order to screen for high-protein-quality material, which requires the determination of both tryptophan and protein contents, the necessity of a rapid, inexpensive, and high-throughput method for maize protein estimation was felt. Keeping this consideration in mind, we standardized the Bradford method for the estimation of maize protein. A maize endosperm contains the prolamin zein, which accounts for 60% of the total protein, glutelins (26% of total protein), and albumins and globulins (6% of total protein) (Anderson and Lamsal, 2011). In our study, the extraction procedure mainly focused on the extraction of the major component of maize protein, i.e., glutelins and zeins. To solubilize the glutelins and zeins, a sequential extraction using alkali and alcohol was employed. The extraction protocol involved sonication for 30 min with 0.1 N NaOH (pH 12.5) followed by 55% ethanol (Shukla and Cheryan, 2001), and the pooled extract was used for protein estimation via the Bradford method. This procedure requires only 2 h, and then the protein content is estimated within a few minutes in a large number of samples.
The Coomassie dye-based Bradford protein assay is suitable for the rapid estimation of protein extracts. The protein content determined through the Bradford method was found to be slightly reduced as compared to the protein content estimated through the micro-Kjeldahl method (Figure 4). A correlation of 0.94, 0.84, and 0.94 was observed between the protein content estimated by modified Bradford and micro-Kjeldahl methods in normal, opaque-2, and QPM lines, respectively, with the coefficient of determination as 0.89, 0.72, and 0.89 (Figures 5A–C). An overall significantly positive correlation of 0.9 (R2 = 0.81) was observed for the protein estimations done via the Bradford method and micro-Kjeldahl method in all the 51 lines (Figures 5D, 6). The biggest advantage of the Bradford protein assay is its inexpensive nature and amenability for high-throughput analysis. Once the test has been set up, each sample requires only 5 min of incubation to form a stable blue color required for estimation. This allows several samples to be analyzed in a short span of time, and the retesting of a sample can be quickly performed. Although Nurit et al. (2009) described a rapid tryptophan protocol, they used specialized equipment. Further, as mentioned above, in the absence of a rapid protein estimation method, the protein quality index cannot be determined, which is the international standard for screening high-protein-quality maize.
We propose a workflow for the high-throughput screening of high-protein-quality maize using the standardizations done in the present study (Figure 7). For the screening of high-protein-quality material containing higher tryptophan per unit protein, a defatting of 48 h is sufficient. The defatted sample is then analyzed for the protein content using the modified Bradford method, which does not require expensive equipment and hazardous chemicals. Both the tryptophan and the protein content are used to calculate the protein quality index, which is an important parameter of the protein quality of a particular maize sample. This workflow can be utilized by resource-poor laboratories to enable the aggregation of the high-protein-quality material and extend the benefits of science and technology for the societal benefit.
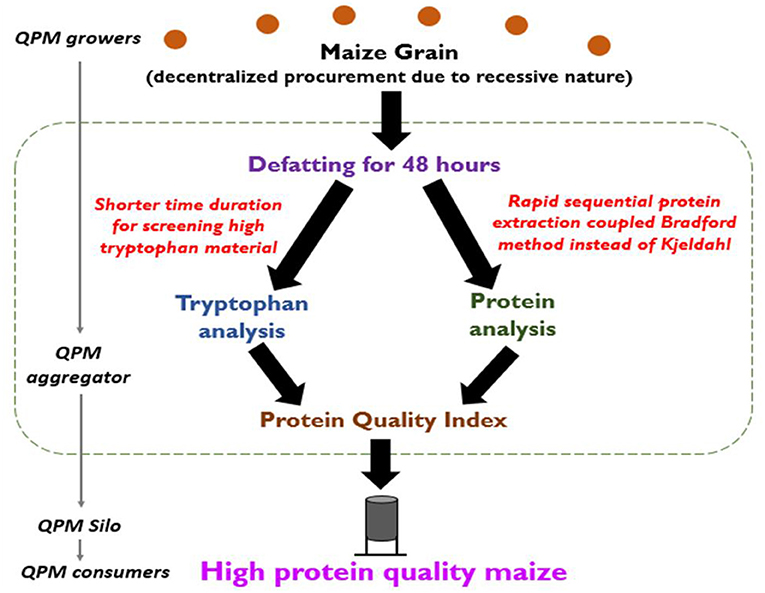
Figure 7. Workflow to screen high-protein-quality maize for aggregation of QPM material. Shorter duration of defatting for tryptophan estimation coupled with the use of defatted maize for protein estimation by the modified Bradford method would enable the resource-poor laboratories to screen QPM and extend the technology for societal benefits.
Conclusion
The present study has helped in optimizing the protein quality assay in maize. It is concluded that the rate of defatting is maximum in opaque-2 followed by QPM and normal maize. QPM and opaque-2 maize defats completely in 48 h of extraction. Since no significant difference is observed in the samples defatted for 48 and 72 h, therefore, 48 h of defatting is optimized for the precise estimation of tryptophan in the maize endosperm. Defatting for 48 h would be sufficient for screening high- protein-quality material (QPM and opaque-2) in the bulk material. Further, the present study optimized the Bradford method for protein estimation in the kernel endosperm of maize, which shows a strong positive correlation (r = 0.90) with the micro-Kjeldahl method with a regression coefficient (R2 = 0.81). Thus, the present study effectively helped in reducing the defatting time by 24 h and protein estimation by 3 h as compared to the already established CIMMYT protocol. In addition, the requirement of sophisticated equipment involving a significant capital cost is avoided by the use of the modified Bradford method. This would enable the agencies with low resource settings to take advantage of the developed protocols for the assessment of protein quality. This is expected to facilitate the aggregation of high-quality material for empowering nutrition with the utilization of QPM material.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author Contributions
CK conducted the experiment, analyzed the data, interpreted the results, and wrote the manuscript. AS envisaged the concept, analyzed the data, interpreted the results, and revised the manuscript. MS conducted the experiments. VD conducted the experiments RP contributed in plant material acquisition of samples at regular intervals. BB and SL revised the manuscript. SR envisaged the concept and revised the manuscript. DC envisaged the concept, analyzed the data, interpreted the results, and wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by ICAR-Indian Institute of Maize Research, Ludhiana. Experimental material was contributed by Punjab Agricultural University, Ludhiana and ICAR-Indian Institute of Maize Research, Ludhiana. AS thanks Science for Equity, Empowerment and Development (SEED), Department of Science and Technology, Government of India for a Research Grant (SP/YO/217/2018).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Affolter, M., Thakkar, S. K., and Garcia-Rodenas, C. L. (2021). Proteins in human milk: an overview. Human Milk. 69–90. doi: 10.1016/B978-0-12-815350-5.00003-6
Anderson, T. J., and Lamsal, B. P. (2011). Zein extraction from corn, corn products, and coproducts and modifications for various applications: a review. Cereal Chem. 88, 159–173. doi: 10.1094/CCHEM-06-10-0091
Ankom (2005). XT10 Extractor. Available online at: www.ankom.com (accessed January 17, 2022).
AOAC (1975). Official Methods of Analysis of the Association of Official Agricultural Chemicals. Tenth Edn. Washington DC: AOAC, 744–745.
Arruda, P., Kemper, E. L., Papes, F., and Leite, S. (2000). Regulation of lysine catabolism in higher plants. T. Plant Sci. 5, 324–330. doi: 10.1016/S1360-1385(00)01688-5
Arsenault, E. J., and Brown, H. K. (2017). Effects of protein or amino-acid supplementation on the physical growth of young children in low-income countries. Nutr. Rev. 75, 699–717. doi: 10.1093/nutrit/nux027
Bjarnason, M., and Vasal, S. K. (1992). Breeding of quality protein maize (QPM). Plant Breed Rev. 9, 181–186. doi: 10.1002/9780470650363.ch7
Bradford, M. M.. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254. doi: 10.1016/0003-2697(76)90527-3
Brochetto-Braga, M. R., Leite, A., and Arruda, P. (1992). Partial purification and characterization of lysine-ketoglutaratereductase in normal and opaque-2 maize endosperm. Plant Physiol. 98, 1139–1147. doi: 10.1104/pp.98.3.1139
Chaudhary, D. P., Sapna, S., and Mandhania Kumar, R. (2012). Interrelationship among nutritional quality parameters of maize (Zea mays) genotypes. Indian J. Agric. Sci. 82, 681–686.
FAOSTAT (2020). Food and Agriculture Data – FAOSTAT. Available online at: https://www.fao.org/faostat/en (accessed January 19, 2022).
Gavazzi, F., Lazzari, B., Ciceri, P., Gianazza, E., and Viotti, A. (2007). Wild-type opaque-2 and defective opaque-2 polypeptides form complexes in maize endosperm cells and bind the opaque-2zein target site. Plant Physiol. 145, 933–945. doi: 10.1104/pp.107.103606
Guo, X., Duan, X., Wu, Y., Cheng, J., Zhang, J., Zhang, H., et al. (2018). Genetic engineering of maize (Zea mays L.) with improved grain nutrients. J. Agric. Food Chem. 66, 1670–1677. doi: 10.1021/acs.jafc.7b05390
Gupta, H. S., Raman, B., Agrawal, P. K., Mahajan, V., Hossain, F., and Thirunavukkarasu, N. (2013). Accelerated development of quality protein maize hybrid through marker-assisted introgression of opaque-2 allele. Plant Breed. 132, 77–82. doi: 10.1111/pbr.12009
Hernandez, H., and Bates, L. S. (1969). “A modified method for rapid tryptophan analysis of maize,” in Res Bull, nineenth Edn, eds H. Horacio Hernandez, and L. S. Bates (Mexico: International Maize and Wheat Improvement Centre), 1–42.
Holding, D. R.. (2014). Recent advances in the study of prolamin storage protein organization and function. Front. Plant. Sci. 5, 276. doi: 10.3389/fpls.2014.00276
Ku, H. K., and Ha, S. H. (2020). Improving nutritional and functional quality by genome editing of crops: status and perspectives. Front. Plant Sci. 11, 1514. doi: 10.3389/fpls.2020.577313
Langyan, S., Yadava, P., Khan, F. N., Dar, Z. A., Singh, R., and Kumar, A. (2022). Sustaining protein nutrition through plant-based foods. Front. Nutr. 8, 772573. doi: 10.3389/fnut.2021.772573
Manicacci, D., Camus-Kulandaivelu, L., Fourmann, M., Arar, C., Barrault, S., Rousselet, A., et al. (2009). Epistatic interactions between Opaque2 transcriptional activator and its target gene CyPPDK1 control kernel trait variation in maize. Plant Physiol. 150, 506–520. doi: 10.1104/pp.108.131888
Nedi, G., Alamerew, S., and Tulu, L. (2016). Review on quality protein maize breeding for Ethiopia. J. Biol. Agric. Healthc. 6, 84–96.
Nurit, E., Tiessen, A., Pixley, K. V., and Rojas, N. P. (2009). Reliable and inexpensive colorimetric method for determining protein-bound tryptophan in maize kernels. J. Agric. Food Chem. 57, 7233–7238. doi: 10.1021/jf901315x
Olsen, M. S., and Phillips, R. L. (2009). Molecular genetic improvement of protein quality in maize. Impact. Agric. Hum. Health Nutr. 2, 60.
Onireti, F. M., and Ikujenlola, A. V. (2020). Nutrients, antinutrients and amino acids profile of malted quality protein maize (Zea Mays L.) based ready-to-eat breakfast cereal fortifiedwith vegetable biomaterials. Croat. J. Food Sci. Technol. 12, 258–267. doi: 10.17508/CJFST.2020.12.2.15
Pérez-Massot, E., Banakar, R., Gómez-Galera, S., Zorrilla-López, U., Sanahuja, G., Arjó, G., et al. (2013). The contribution of transgenic plants to better health through improved nutrition: opportunities and constraints. Genes Nutr. 8, 29–41. doi: 10.1007/s12263-012-0315-5
Prasanna, B. M., Palacios-Rojas, N., Hossain, F., Muthusamy, V., Menkir, A., Dhliwayo, T., et al. (2020). Molecular breeding for nutritionally enriched maize: status and prospects. Front. Genet. 10, 1392. doi: 10.3389/fgene.2019.01392
Sethi, M., Kumar, S., Singh, A., and Chaudhary, D. P. (2020). Temporal profiling of essential amino acids in developing maize kernel of normal, opaque-2 and QPM germplasm. Physiol Mol Biol Plants. 26, 341–351. doi: 10.1007/s12298-019-00724-x
Sethi, M., Singh, A., Kaur, H., Phagna, R. K., Rakshit, S., and Chaudhary, D. P. (2021). Expression profileof protein fractions in the developing kernel of normal, Opaque-2 and quality proteinmaize. Sci. Rep. 11, 1–9. doi: 10.1038/s41598-021-81906-0
Shetti, P., Sagare, D. B., Surender, M., and Reddy, S. S. (2020). Development of lysine and tryptophan rich maize (Zea mays) inbreds employing marker assisted backcross breeding. Plant Gene. 23, 100236. doi: 10.1016/j.plgene.2020.100236
Shukla, R., and Cheryan, M. (2001). Zein: the industrial protein from corn. Ind. Crops Prod. 13, 171–192. doi: 10.1016/S0926-6690(00)00064-9
Singh, A., Karjagi, C., and Rakshit, S. (2020). Minimally altering a critical kinase for low-phytate maize. Sci. Rep. 10, 1–5. doi: 10.1038/s41598-020-63016-5
Tripathy, S. K., Ithape, D. M., Maharana, M., and Prusty, A. M. (2017). Quality protein maize (QPM): genetic basis and breeding perspective. Trop Plant Res. 4, 145–152. doi: 10.22271/tpr.2017.v4.i1.021
Wang, X., Stumpf, D. K., and Larkins, B. (2001). Asparate kinase 2 a candidate gene of a quantitative trait locus influencing free amino acid content in maize endosperm. Plant Physiol. 125, 1778–1787. doi: 10.1104/pp.125.4.1778
Keywords: quality protein maize (QPM), opaque-2, defatting time, tryptophan, Bradford, Kjeldahl
Citation: Kaur C, Singh A, Sethi M, Devi V, Chaudhary DP, Phagna RK, Langyan S, Bhushan B and Rakshit S (2022) Optimization of Protein Quality Assay in Normal, opaque-2, and Quality Protein Maize. Front. Sustain. Food Syst. 6:743019. doi: 10.3389/fsufs.2022.743019
Received: 17 July 2021; Accepted: 18 February 2022;
Published: 04 April 2022.
Edited by:
Uma Tiwari, Technological University Dublin, IrelandReviewed by:
Edmore Gasura, University of Zimbabwe, ZimbabweSofija Bozinovic, Maize Research Institute Zemun Polje, Serbia
Copyright © 2022 Kaur, Singh, Sethi, Devi, Chaudhary, Phagna, Langyan, Bhushan and Rakshit. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dharam Paul Chaudhary, Y2hhdWRoYXJ5ZHBAZ21haWwuY29t
 Charanjeet Kaur
Charanjeet Kaur Alla Singh2
Alla Singh2 Mehak Sethi
Mehak Sethi Dharam Paul Chaudhary
Dharam Paul Chaudhary Ramesh Kumar Phagna
Ramesh Kumar Phagna Sapna Langyan
Sapna Langyan Bharat Bhushan
Bharat Bhushan