- 1Key Laboratory of South Subtropical Fruit Biology and Genetic Resource Utilization, Guangdong Provincial Key Laboratory of Tropical and Subtropical Fruit Tree Research, Institute of Fruit Tree Research, Guangdong Academy of Agricultural Sciences, Ministry of Agriculture and Rural Affairs, Guangzhou, China
- 2College of Life Science, Langfang Normal University, Langfang, Hebei, China
- 3Key Laboratory of Horticultural Plant Biology, Ministry of Education, College of Horticulture and Forestry, Huazhong Agricultural University, Wuhan, China
- 4Genomics Applications Platform, National Agricultural Research Organisation (NARO), Entebbe, Uganda
Introduction: Reconstructive breeding based on autotetraploids to generate triploid varieties is a promising breeding strategy in banana (Musa spp.). Therefore understanding the molecular mechanisms underlying the phenotypic differences between the original diploid and its autopolyploid derivatives is of significant importance in such breeding programs of banana.
Methods: In this study, a number of non-chimeric autotetraploid plants, confirmed by flow cytometry and chromosome counting were obtained using colchicine treatment of ‘Pisang Berlin' (AA Group), a diploid banana cultivar highly resistant to Fusarium wilt Tropical Race 4 (Foc TR4) and widely cultivated in Asia.
Results and discussion: The autotetraploids showed significant increase in plant height, pseudostem diameter, root length, leaf thickness, leaf area, and leaf chlorophyll content. Transcriptomic analysis indicated that differentially expressed genes were mainly enriched in plant hormone signal transduction, mitogen-activated protein kinase (MAPK) signaling pathway, and carbon fixation in photosynthetic organelles. The genes related to the metabolism, transport or signaling of auxin, abscisic acid (ABA), cytokinin (CTK) and gibberellin (GA), as well as the genes encoding essential enzymes in photosynthetic CO2 fixation were differentially expressed in leaves of autotetraploids and most of them were up-regulated. Metabolomic analysis revealed that the differentially accumulated metabolites were mainly involved in plant hormone signal transduction, porphyrin and chlorophyll metabolism, indole alkaloid biosynthesis, and carbon fixation in photosynthetic organelles. The results therefore, demonstrate that the hormones IAA, ABA, and photosynthetic regulation may play a vital role in the observed enhancement in the autotetraploids. These could be used as molecular and biochemical markers to facilitate the generation of triploid progenies as suitable new varieties for cultivation.
Introduction
Banana (Musa spp.) is one of the major cash crops in the tropics and subtropics, as well as one of the major food crops in most developing countries (Heslop-Harrison and Schwarzacher, 2007). Banana production can be affected by a number of pests and diseases as well as climatic change (Dita et al., 2011). Development of resistant varieties is considered the most sustainable solution to banana productivity challenges. However, traditional banana breeding is very complicated due to most cultivated bananas being triploid (2n = 3x = 33) and very low fertility or complete sterility. The lack of pest and disease resistance genes has also been noted in most edible varieties (Poerba et al., 2019). Several of the desired characteristics are however, available in the diploid germplasm (AA and AB groups), including high hand number, parthenocarpy, slim fruit fingers, dwarf plant height, pest and disease resistance but with low yields, and generally non-parthenogenesis (Ortiz and Vuylsteke, 1995; Silva et al., 2001; Ssebuliba et al., 2006).
Polyploidization is a breeding technique that offers an opportunity to create novel genetically improved polyploid germplasm with increased interspecific fertility that can further be used to generate triploid varieties through conventional crossing programs (Sattler et al., 2016). Polyploid induction is thus put forward as an important step in banana breeding. Triploid bananas can be generated by crossing between diploid and autotetraploid plants and this has become one of the most important strategies in crossbreeding of bananas and has led to the successful commercialization of banana varieties in the Cavendish subgroup (Jenny et al., 2002; Perrier et al., 2019).
Autotetraploid plants can be obtained by crossing triploids with diploids, chromosome doubling of diploids, somatic hybridization, or genetic engineerin (Do Amaral et al., 2015; Borges et al., 2016; De Carvalho Santos et al., 2019). Induction of chromosome doubling using colchicine is widely used in polyploid mutagenesis of various plants. To date, polyploidy has been successfully induced in the crops like apple (Zhang F. et al., 2015), citrus (Pablo et al., 2011; Wu et al., 2012), and pear (Kadota and Niimi, 2002; Sun et al., 2009). However, rather than the desired euploids, the aneuploids and even hyperpolyploidy may occur in the chemical mutagenesis of chromosome doubling. Polyploids generally showed organ enlargement than the diploids in kiwifruits (Wu et al., 2012) and rice (Zhang J. et al., 2015). Allario et al. (2011) found that the autotetraploid leaves, roots, stems, and other organs of lemon became larger. Polyploids also tended to be better adapted to the environment. Polyploids of Hordeum vulgare, Citrus limonia as well as Manihot esculenta showed enhanced drought resistance compared to the corresponding diploid parents (Chen and Tang, 1945; Nassar, 2006; Allario et al., 2013). Apple autotetraploids showed higher drought and salt resistance (Xue et al., 2015; Zhang F. et al., 2015). Moreover, it was found that polyploidy can accumulate more quality related metabolites, like sugars, amino acids, and organic acids (Cohen et al., 2013). However, the growth of some autopolyploids was weaker than those of the diploids, exhibiting the characteristics of delayed development and dwarfing. For example, the autotetraploids of Arabidopsis spp. were weaker than the diploids (Fort et al., 2015), and the autotetraploid orange and apple plants were reported to be dwarfing (Syvertsen et al., 2000; Allario et al., 2011).
Efforts have been made to generate desired banana genotypes through polyploidy induction (Poerba et al., 2019). Banana plants generated by chromosome doubling mainly showed increased pseudostem size, stomatal length, fruit, and bunch weight (Hamill et al., 1992). The autotetraploid plants obtained from the wild diploids were taller and more robust, but with droopy leaves, fewer suckers, more sparse roots, changed fruit size and shape, and doubled anthocyanin content in the leaves (Vakili, 1967). Chromosome doubling of SH-3362 (AA) resulted in stronger autotetraploid plants with thicker pseudostems, thicker roots, and wider leaves (Hamill et al., 1992). In genotypes ‘Kluai Sa' (AA) and ‘Kluai Leb Mu Nang' (AA), leaf and fruit shape changed in autotetraploid plants relative to their diploid counterparts under the same growth conditions (Kanchanapoom and Koarapatchaikul, 2012). Chromosome doubling of ‘Pisang Lilin' (AA) showed increased pseudostem diameter and plant height. Leaf number at flowering and harvest stages as well as bunch and fruit weights significantly increased in the autotetraploids (Do Amaral et al., 2015). However, apart from the phenotypic changes, little is known on molecular mechanisms underlying the phenotypic difference between diploid and its autopolyploid counterparts.
The diploid banana variety Pisang Berlin (AA) is highly resistant to Fusarium wilt Tropical Race 4 (Foc TR4), and is widely grown in southern China and other Southeast Asian countries (Hapsari and Lestari, 2016; Zuo et al., 2018). It has superior fruit quality, but the yields are low due to its small hands and fruits size. Therefore in our breeding program, we attempted to increase fruit size through polyploidization and therefore improve the yields of the diploids with good fruit quality characteristics. The present study focused on the growth of the autotetraploid seedlings of Pisang Berlin and explored the molecular mechanisms underlying phenotypic variation caused by ploidy changes, which would allow better understanding and utilizing the autotetraploids in the banana breeding program.
Materials and methods
Plant materials
The diploid banana variety Pisang Berlin (Musa acuminata, AA genome; ITC0611, doi: 10.18730/9K96E) was used in this study. The meristematic shoot tips of this variety were obtained from the China's National Banana Collection at Guangzhou, and propagated in the solid Murashige and Skoog (MS) medium under sterile conditions at 25°C (Swamy et al., 1983). The pH of the medium was adjusted between 5.7 and 5.8.
Chromosome doubling
In vitro induction of autotetraploids on shoot tips of Pisang Berlin (treated by colchicine) was carried out according to the method described by Bakry et al. (2007). Thereafter, the shoot tips were grown in vitro in the liquid MS medium (125 mL) consisting of adenine sulfate (651.5 μM) and 6-benzylaminopurine (22.2 μM, BAP). After that, they were kept at 27°C on a gyratory shaker at 80 rpm in liquid MS medium for 7 days under 16-h/8-h light/dark cycle. Selected shoot tips were treated with 1.25 mmol/L colchicine added to the 125 ml of liquid MS medium. The treated shoot tips were cultured in darkness for 48 h at 80 rpm. Thereafter, the shoots were rinsed in sterile water for 48 h at 80 rpm, followed by transfer into liquid MS medium for additional 7 days culture. Finally, the shoots were transferred into the solid MS medium for further sub-culture.
Rooting culture and plant regeneration
After 30-days of post-initial sub-culture, the surviving shoot tips were further sub-cultured twice or more at every 35–40 days, followed by transfer into the rooting medium (MS that contained 30 g/L sucrose and 7 g/L agar solidification). Thereafter, all shoots were maintained in the growing stage at 25 ± 2°C and 16-h/8-h light-dark cycle conditions. After 30-40 days, the rooted plantlets were transferred into liquid Hoagland nutrient solutions and retained for further 30 days (Hoagland and Arnon, 1950).
FCM-based ploidy degree determination
In order to determine the ploidy level, FCM was used using the method proposed by Roux et al. (2004). Meanwhile, sample preparation was conducted following the descriptions of Poerba et al. (2019). Leaf tissues (20–30 mg) were sampled using a sharp scalpel blade and placed in glass petri dishes.. 1 mL LB01 buffer (363 mg Tris, 148.9 mg Na2EDTA, 20.2 mg spermine, 1.193 g KCl, 233.8 mg NaCl, 200 ul Triton X-100, 220 ul mercaptoethanol 200 ml deionized water with a pH of 7.5) (DoleŽel et al., 1989) was added to the plate. The nuclear DNA was stained with 2 μg/ml PI solution. Thereafter, the nuclear suspension was filtered using the 50-μm nylon mesh, followed by maintenance on ice till further experiments. FCM was performed to measure DNA concentrations (C units) within the samples, with 1C indicating the DNA concentration in the haploid chromosome set (n). Following FCM, the fluorescence intensity distribution (relative DNA concentration) was generally determined in arbitrary units (channel numbers). In this study, one Musa acuminata ssp. malaccensis (2n = 22) sample was utilized as the diploid reference (Poerba et al., 2019). The flow cytometer was adjusted to ensure that the peak represented the G1 nuclei appearing on channel 100. Later, additional samples were normalized to locations relative to the G1 peak.
Detection of chromosome number in the root tips
Young root tips of plants were selected and sliced by wall-removing low permeability and flame drying (Chen et al., 1979). Enzymolysis was carried out in a mixed enzyme solution (3% cellulase + 1% macerozyme) for 1.5 h. The smear was fixed after cleaning with water, dried by alcohol lamp flame, and then examined after staining with 5% Giemsa dye. A preliminary microscopic examination was performed under a 10-fold Olympus microscope. The splits were photographed under a 40x lens.
Phenotype analyses of diploid and autotetraploid
Diploid (control) and autotetraploid plantlets were grown in liquid Hoagland nutrient solutions for 30 days at 25 ± 2°C and 16-h/8-h light-dark cycle conditions, photographed, and the phenotypes were recorded as described by An et al. (2009). Plant height, length, and width of leaves were measured by measuring tape, stem diameter and leaf thickness were measured by an electronic micrometer, and chlorophyll value was measured by a portable chlorophyll meter. The leaf area was calculated as described by Liu et al. (2013).
cDNA preparation and illumina sequencing
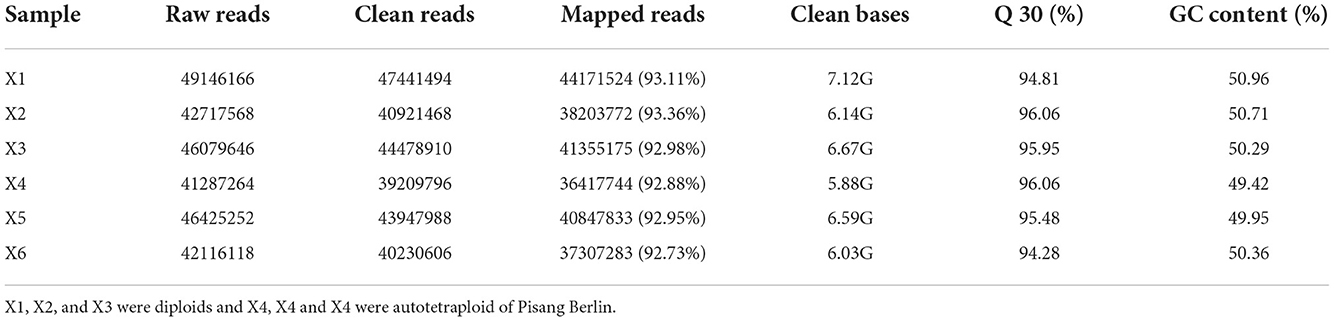
Total RNA was isolated from the cultured leaves of diploids (X1, X2, and X3) and autotetraploids (X4, X5, and X6) of banana plants after 30 days with EASY spin Plus Plant RNA Kit (AidLab, Beijing). RNA concentration was detected by Nanodrop. Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA) was employed for detecting RIN and 28S/18S values. Then, RNA integrity and quality were measured by 1% AGE. Additionally, 18 cDNA libraries were prepared in line with specific protocols using NEB Next Ultra RNA Library Prep Kit. This was followed by the sequencing of cDNA libraries by Novogene (Beijing, China), adopting Illumina HiSeq TM 2000 platform. The quality of raw reads was checked using FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/) before data analysis. The impure reads were filtered, whereas the high-quality ones were acquired using Trinity by adopting default parameters, which were later utilized for constructing the distinct consensus sequences (Grabherr et al., 2011). Filtered reads were compared with the Musa acuminata subsp. malaccensis (ID: ensemblplants_musa_acuminata_subsp_malaccensis_asm31385v1_gca_000313855_1) using TopHat (v2.0.10) (Trapnell et al., 2012).
Gene analysis and functional annotation
Using the RPKM (Reads Per kb per Million reads) approach, unigene expression was estimated by cuffdiff (Trapnell et al., 2012). DEGseq v1.14.0 software was used for identifying differentially expressed genes (DEGs). All genes were assigned with corresponding p-values, and DEGs were selected upon the thresholds of |log2|≥1 and p ≤ 0.05. Later, acquired unigenes were mapped against Nr (Non-redundant Protein Sequence Database in GenBank), Swiss-Prot (Protein Sequence Database), GO (gene ontology; Conesa et al., 2005), COG (Cluster of Orthologous Groups of proteins; Tatusov et al., 2003), and KEGG (Kyoto Encyclopedia of Genes and Genomes; Kanehisa et al., 2006) databases.
Metabolic profiling
The leaf tissue samples of diploid and autotetraploid of banana plants were freeze dried in a lyophilizer (Scientz-100F) under vacuum, followed by grinding into fine powders. Later, 80% methanol (500 μL), added with 0.1% formic acid, to dissolve the powders (100 mg). After overnight preservation at 4 °C the samples were vortexed six times to enhance extraction efficiency. Later, the samples were centrifuged for 10 min at 15,000 g and the supernatants were then filtered using a 0.22 μm microporous filter, followed by processing for Ultra Performance Liquid Chromatography (UPLC)-mass spectrometry (MS/MS) (Want et al., 2010; Dunn et al., 2011). To characterize the secondary metabolites, around 0.1 g lyophilized leaf sample powders were subjected to methanol (80%) extraction. Then, the QTOF 6520 mass spectrometer (Agilent Technologies, Palo Alto, CA, USA) was employed for profiling secondary metabolites by adopting the 1200 series Rapid Resolution HPLC system. Typical fragment ions were compared to the reference for identifying secondary metabolites as well as additional amino acids (Wang et al., 2016). Metabolites that shared close fragment ions were deemed as identical compounds. Statistical analysis of secondary metabolite data was performed using Analyst 1.6.1 software (AB SCIEX, Ontario, Canada). Variable importance in projection (VIP) values were determined through partial least squares discriminant analysis. The differentially changed metabolites (DCMs) were selected based on the thresholds of VIP > 1, p < 0.05, and FC > 1.5 or <0.667.
Reverse transcription of and qRT-PCR
M-MLV reverse transcriptase was adopted for synthesizing 2 μg RNA in line with Evo M-MLV RT Kit instructions (Accurate biotechnology, China) and further diluted to 1:6 for subsequent experiments. Ten genes were validated by qRT-PCR with thermal cycler apparatus (BIO-RAD iQ3 7700, Applied Biosystems). The 20 mm3 qRT-PCR reaction system contained 2 × SYBR Green Pro Taq HS Premix (10 mm3), ROX Reference Dye (0.4 mm3), respective primers (10 μmol × dm−3, 0.4 mm3 each), RNase Free ddH2O (7.8 mm3) and cDNA (1 mm3). The thermal profile was comprised of two segments: 30-S at 95°C; 5-S denaturation at 95 °C, and 30-S annealing at 60 °C for altogether 40 cycles. Every assay was repeated thrice. Primer Express 2.0 Software (PE Applied Biosystems, USA) was applied in primer designing with default parameters. Supplementary Table S1 displayed the sequences of all the primers. Excel software and 2−ΔΔCt (Livak and Schmittgen, 2001) were used for data analysis with MaCAC-Q as the reference.
Statistical analysis
ANOVA was adopted using the SPSS 19.0 statistics software for comparing phenotypic data of different samples. The Fisher's least significance difference (LSD) test was applied for multiple comparisons of variables at 0.05 (p<0.05).
Results
Autotetraploids generated by colchicine treatment
We carried out the chromosome doubling by treating shoot tips of Pisang Berlin (Musa spp. AA genome) with colchicine. Using flow cytometry, we checked the ploidy levels of the shoot tips in each sub-culturing step (without damaging their meristematic cells) in order to dissociate chimeras by repeated vegetative propagation. After three successive sub cultures, autotetraploid shoots were confirmed and then transferred into the rooting culture medium. Accurate chromosome counts in root tips were carried out to verify ploidy levels of the regenerated autotetraploid plantlets (Figure 1). Compared to the diploid plants, autotetraploid plants exhibited a better growth performance, exhibiting significant increase in pseudostem diameter, plant height, maximum root length, leaf thickness, leaf area, and chlorophyll content (Figure 2).
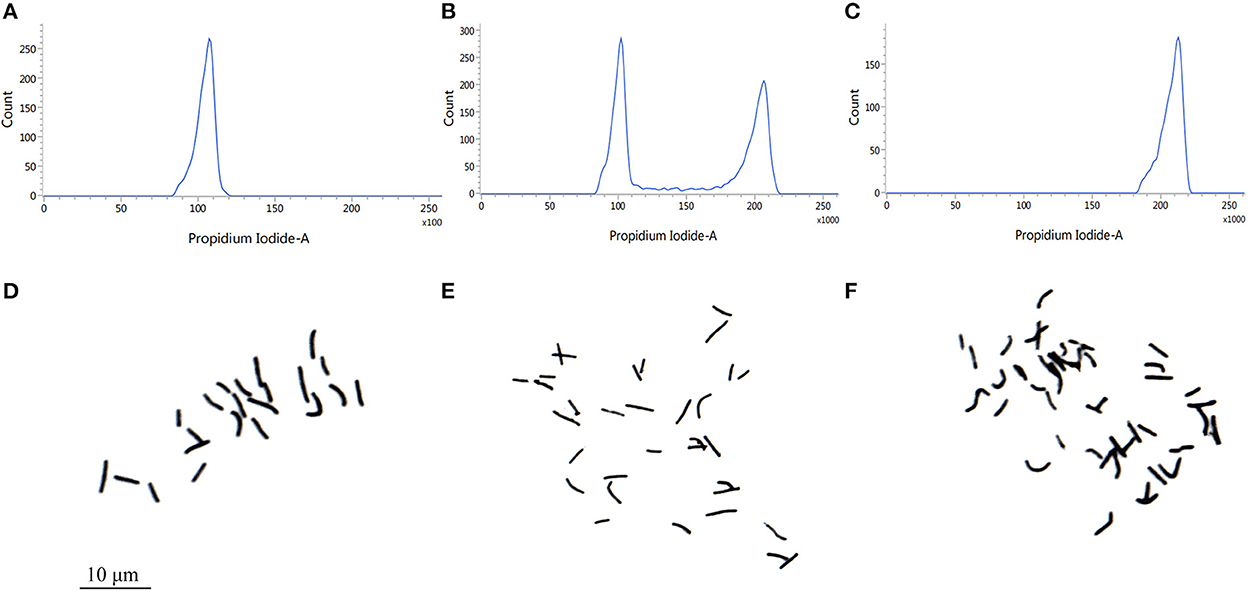
Figure 1. Ploidy analysis of of Pisang Berlin (AA). (A) DNA content of diploids (the main peak at channel 100). (B) DNA content of triploids (the main peak at channel 100–200). (C) DNA content of autotetraploid (the main peak at channel 200). (D) Chromosome number of diploids (2n = 2X = 22). (E) Chromosome number of triploids (2n = 3X = 33). (F) Chromosome number of autotetraploids (2n = 4X = 44).
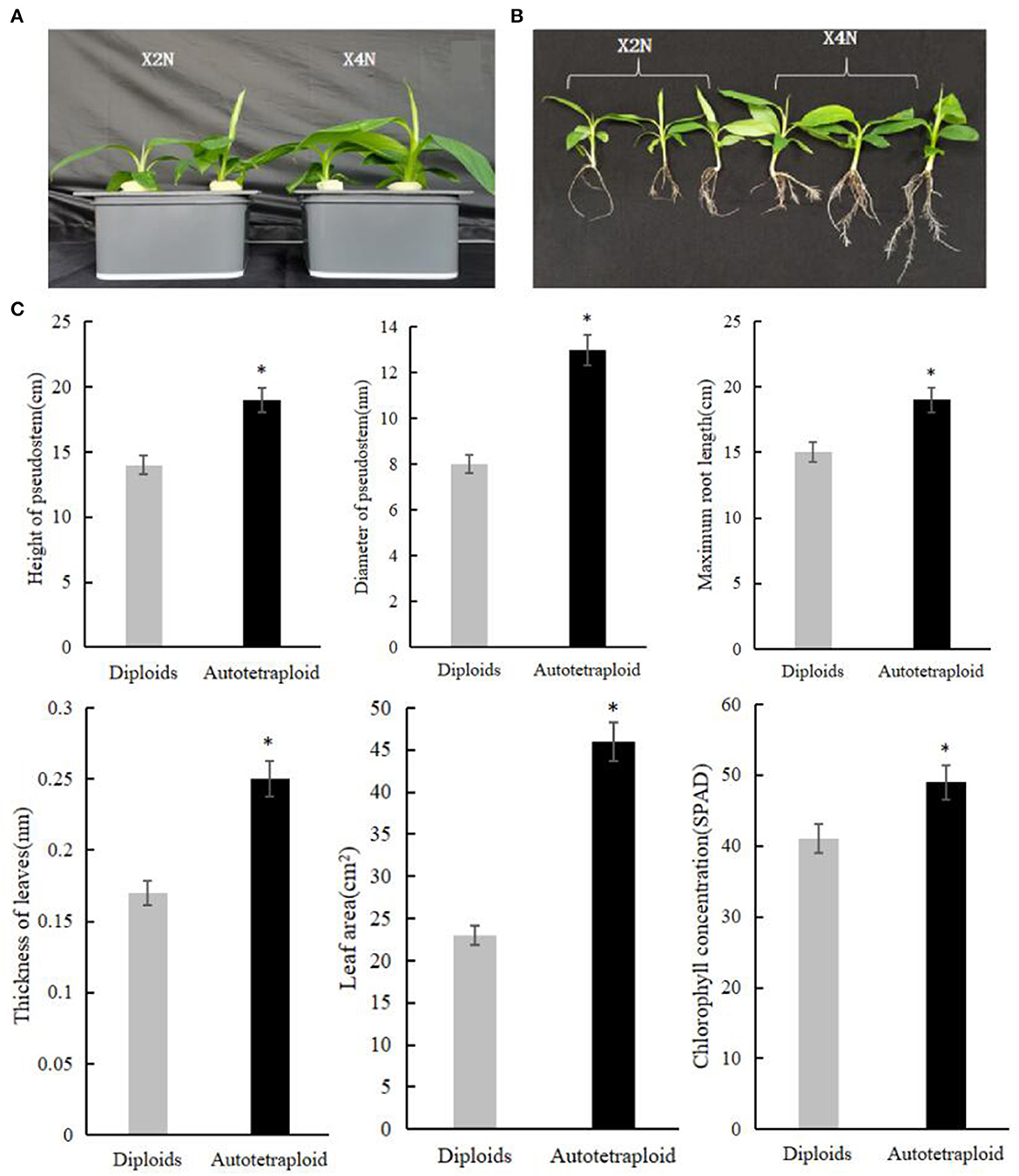
Figure 2. Phenotypic characterization of autotetraploid and diploid Pisang Berlin seedlings grown under the same conditions. (A, B) Phenotypic comparison of autotetraploid and diploid Pisang Berlin seedlings. (C) The pseudostem diameter, plant height, maximum root length, leaf thickness, leaf area, and chlorophyll content of autotetraploid and diploid Pisang Berlin seedlings. X2N represented diploids of Pisang Berlin. X4N represented autotetraploids of Pisang Berlin. * in the figure represent significant differences at P < 0.05 level.
Transcriptomic analysis and differentially expressed genes
Transcriptomic analysis was carried out on the diploid and autotetraploid plants to explore the genes responsible for the differential growth. The overall raw/clean reads within every sample ranged between 41,287,264 and 49,146,166. The sequence reads were aligned into the reference genome of banana, and results indicated that > 92% were map-able. This yielded altogether of 38.43 Gb of clean data. The > 94% Q30 and 50% GC concentrations suggested a high-quality transcriptomic results for the subsequent analyses (Table 1). The PCA analysis classified overall variation in two major components as 51.68 and 17.65%. These observations were validated through PCA, which suggested that diploids tissues fall away from autotetraploid tissues (Figure 3). These samples were also subject to hierarchical clustering on the basis of RPKM values. This approach classified six samples into two main groups: one (X2N) containing X1, X2, and X3, while the other (X4N) comprised X4, X5, and X6 (Figure 3).
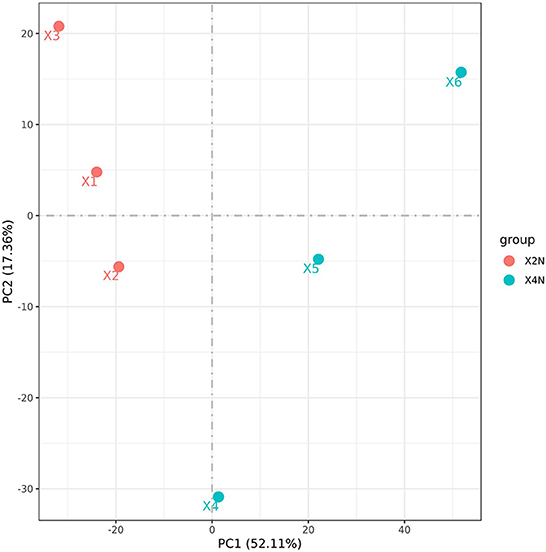
Figure 3. Principle component analysis (PCA) of diploids and autotetraploids of Pisang Berlin samples. X2N represented the diploids of Pisang Berlin (X1, X2, and X3 represented sample replicates). X4N represented the autotetraploids of Pisang Berlin (X4, X5, and X6 represented sample replicates).
There were 25334 DEGs in X4N vs. X2N. GO-function classification statistics were further performed on DEGs in autotetraploids and diploids after doubling. The DEGs were mainly found enriched in the plastid and chloroplast parts in the cellular composition of Pisang Berlin (X4N.vs.X2N) (Figure 4A). KEGG enrichment further indicated DEGs were mainly enriched in plant hormone signal transduction, MAPK signaling pathway, and carbon fixation in photosynthetic organisms (Figure 4B). The change in polyploid phenotype was tightly associated with plant endogenous hormones (Dai et al., 2015). Therefore, we focused on the DEGs related to plant hormone between autotetraploids and diploids because we found that the growth of autotetraploids at the seedling stage had a better plant performance compared with those of diploids (Figure 2).
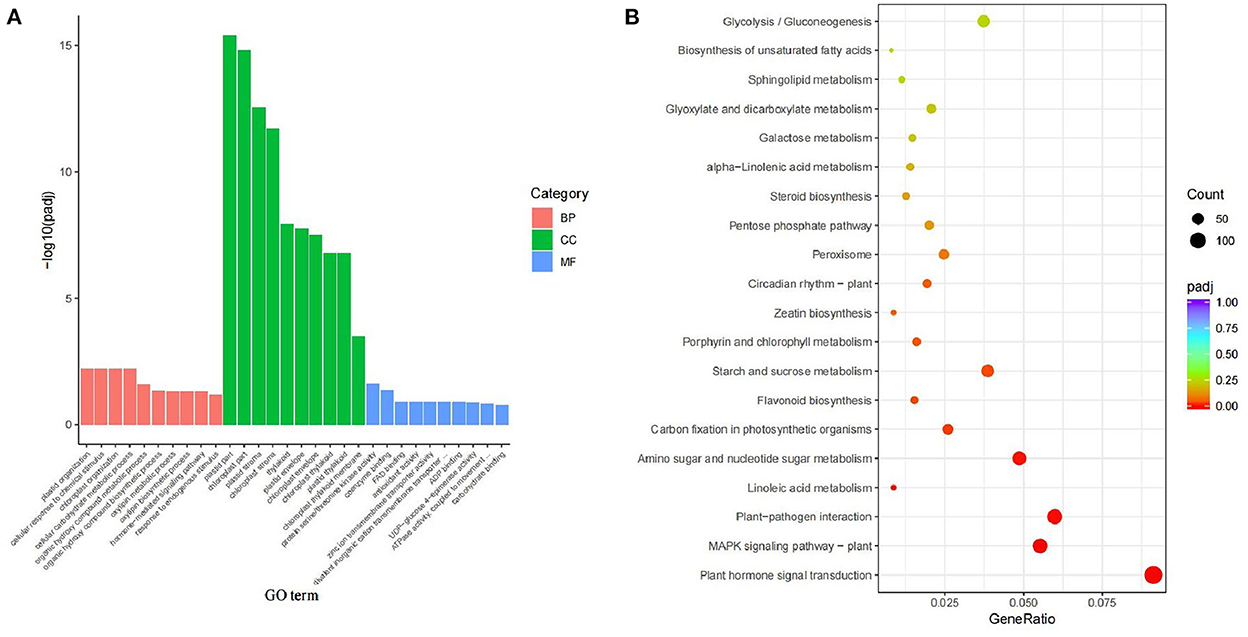
Figure 4. GO enrichment and KEGG enrichment of differentially expressed genes in diploids and autotetraploids of Pisang Berlin. (A) GO enrichment and different colors represented different functional categories, including BP biological processes, CC cellular components, and MF molecular functions. (B) KEGG enrichment and gene ratio represented the ration of the number of differentially expressed genes to the total gene number in the specific pathway, green indicated a low Q-value, while red indicated a high Q-value.
There were eight auxin-related genes within the autotetraploids of Pisang Berlin, including four with up-regulation and four with down-regulation. The up-regulated DEGs included three Ma5NG4 genes (GSMUA_Achr2G22430_001, GSMUA_Achr4G12460_001, and GSMUA_Achr4G04580_001 encoding auxin-induced 5NG4), which were auxin-induced protein with transport function and played a role in signaling of auxin (Kaur et al., 2017); MaIAA30 (GSMUA_Achr4G22520_001) encoded an auxin-responsive protein (Sato and Yamamoto, 2008). The down-regulated DEGs included MaARF18 gene (GSMUA_AchrUn_randomG11300_001 encoding auxin-response factor 18) which inhibited activity on downstream auxin genes (Chen et al., 2021); MaARF7 gene (GSMUA_Achr6G29070_001 encoding auxin-response factor 7) negatively regulated auxin (Jong et al., 2009); Two MaPIN genes (GSMUA_Achr8G23120_001 and GSMUA_Achr10G07210_001) were auxin efflux carrier component and inhibited cell growth (Xu et al., 2005) (Figure 5; Supplementary Table S2).
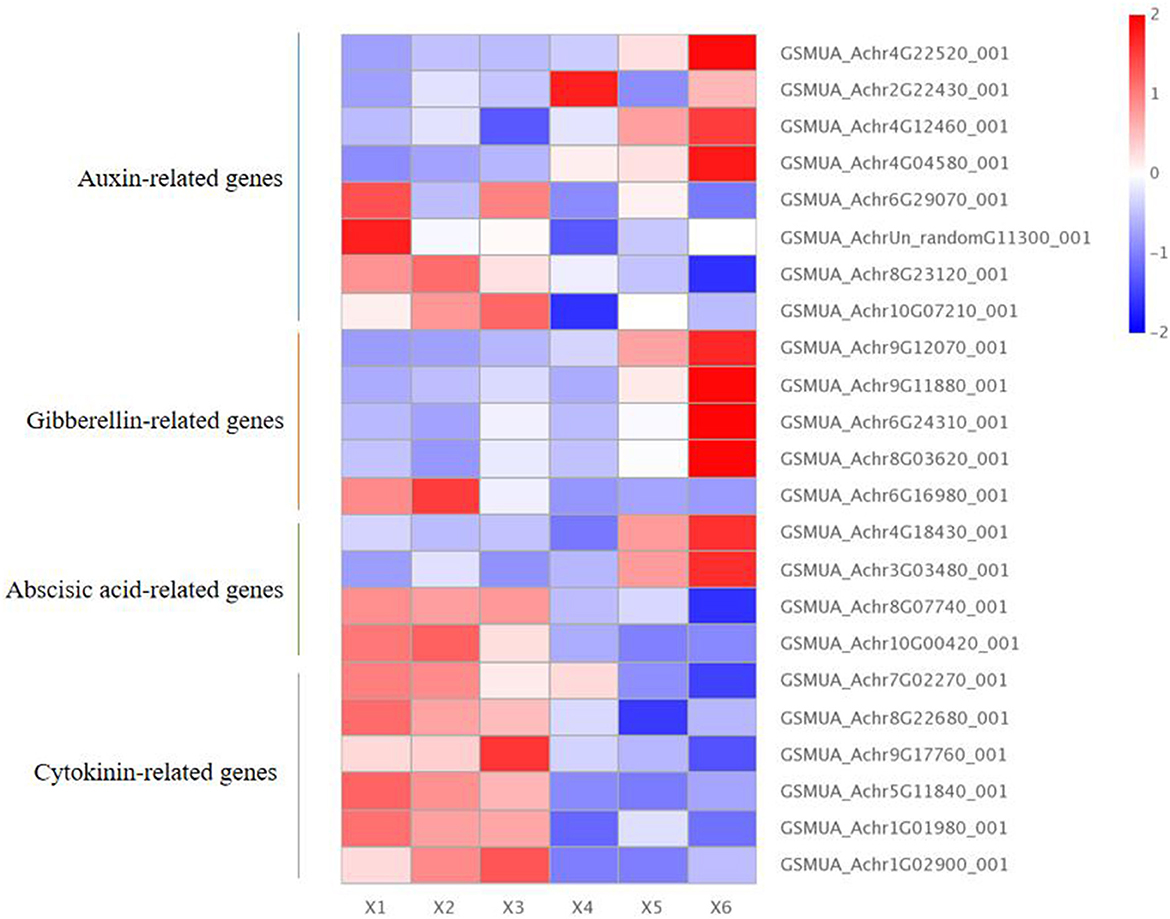
Figure 5. Endogenous hormone differentially expressed genes in diploid and autotetraploid of Pisang Berlin. X1, X2, and X3 represented the diploid banana plants, and X4, X5, and X6 represented the autotetraploid banana plants.
Furthermore, we screened five gibberellin (GA)-related DEGs, including four DEGs up-regulated and one down-regulated in the autotetraploids. The four up-regulated DEGs included three MaCIGR1 genes and one MaGA2ox8 gene. The MaCIGR1 genes (GSMUA_Achr9G12070_001, GSMUA_Achr6G24310_001, and GSMUA_Achr8G03620_001), are belonged to the GRAS family and encode chitin-inducible gibberellin-responsive protein 1, which could play an importance role in regulating plant height (Kovi et al., 2011). The MaGA2ox8 gene (GSMUA_Achr9G11880_001) encodes gibberellin 2-beta-dioxygenase 8 and may regulate GA levels (Zhai et al., 2019). The down-regulated DEGs included MaGID1 (GSMUA_Achr6G16980_001 encoding gibberellin receptor GID1) occluded gibberellin in a deep binding pocket covered by its N-terminal helical switch region (Murase et al., 2008) (Figure 5; Supplementary Table S2).
Moreover, there were four abscisic acid (ABA)-related DEGs within the autotetraploids, including two that were up-regulated and two that were down-regulated. The two up-regulated DEGs included one ABA 8'-hydroxylase 1 gene (GSMUA_Achr4G18430_001) promoting the content of ABA (Kitahata et al., 2005), and one MaPYL8 gene (GSMUA_Achr3G03480_001 encoding abscisic acid receptor PYL8) positively regulating ABA signaling during germination and abiotic stress responses (Saavedra et al., 2010). The two down-regulated DEGs were MaABI5 genes (GSMUA_Achr8G07740_001 and GSMUA_Achr10G00420_001 encoding abscisic acid-insensitive 5) which would negatively regulate ABA signaling (Brocard, 2002; Figure 5; Supplementary Table S2).
It was noted that six down-regulated DEGs related with cytokinin, and the six MaCKX genes (GSMUA_Achr7G02270_001,GSMUA_Achr8G22680_001,GSMUA_Achr9G17760_001,GSMUA_Achr5G11840_001, GSMUA_Achr1G01980_001, GSMUA_Achr1G02900_001 encoding cytokinin dehydrogenase) inactivated cytokinin (Chen et al., 2009; Figure 5; Supplementary Table S2).
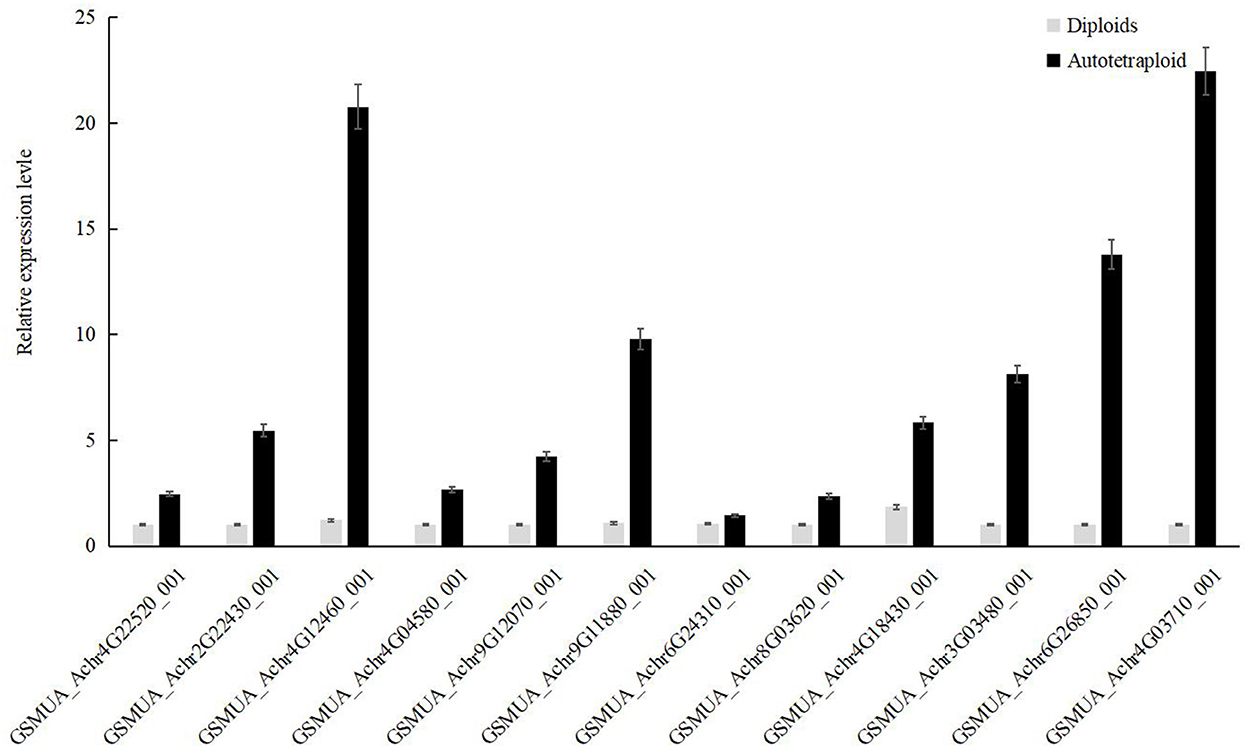
Our results showed that there were seven differentially expressed MaRubisco genes encoding Ribulose 1,5-bisphosphate carboxylase, which were down-regulated in autotetraploids of Pisang Berlin. There is one MaPEPC (GSMUA_Achr6G26850_001 encoding phosphoenolpyruvate carboxylase), which was up-regulated in the autotetraploids of Pisang Berlin. PEPC catalyzed the irreversible carboxylation of phosphoenolpyruvate (PEP) to form oxaloacetate which played a key role in photosynthesis (Yasushi et al., 2003). There were three MaRPI DEGs encoding ribose-5-phosphate isomerase DEGs, including one with up-regulation (GSMUA_Achr4G03710_001) and the other two with down-regulation in the autotetraploids. There were five MaMDH DEGs encoding malate dehydrogenase and three MaFBP DEGs encoding fructose-1,6-bisphosphatase down-regulated in the autotetraploids of Pisang Berlin (Figure 6; Supplementary Table S3). The twelve DEGs that were up-regulated in the autotetraploids were further screened for qRT-PCR analysis, and the results were consistent with the transcriptome results (Figure 7).
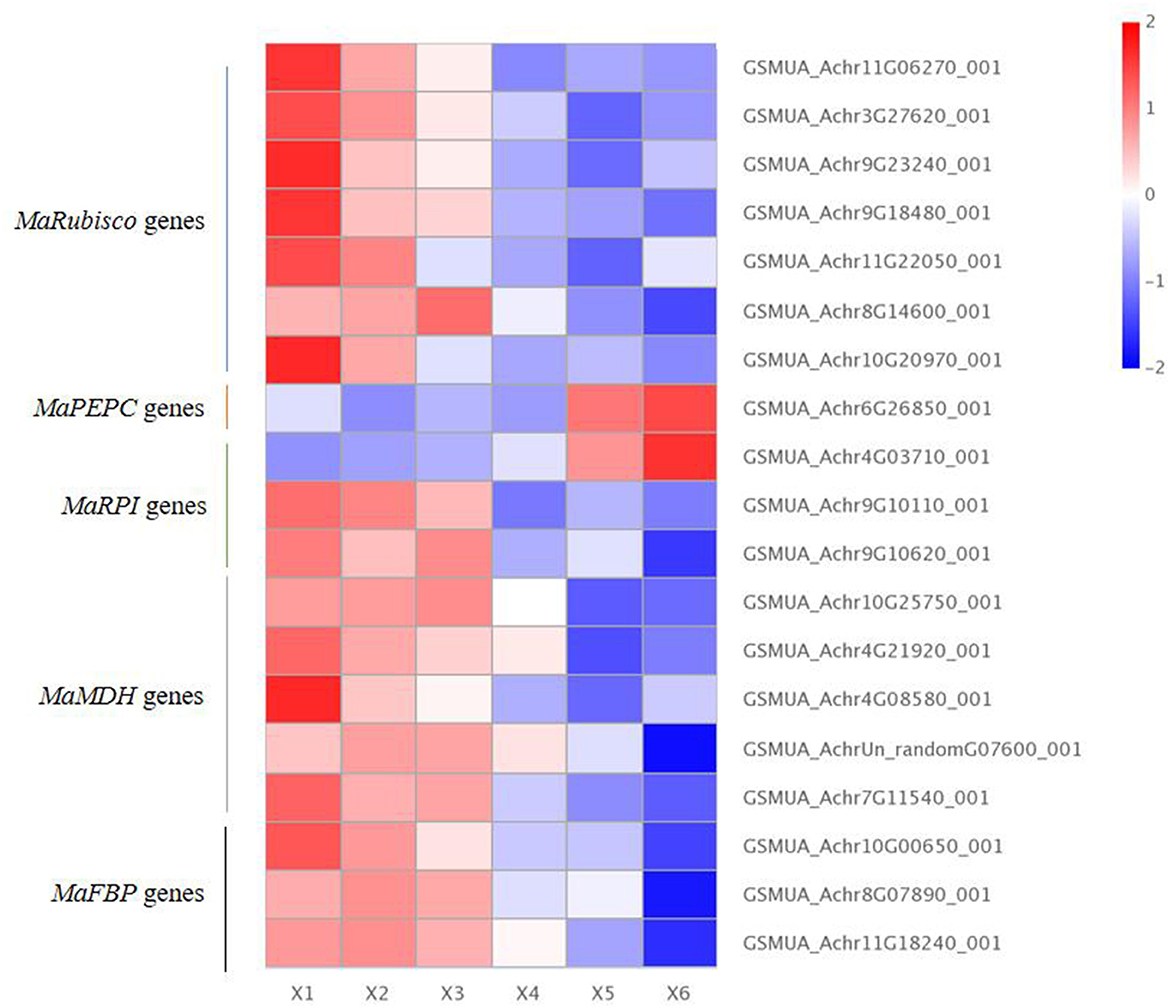
Figure 6. Photosynthesis differentially expressed genes in diploids and autotetraploids of Pisang Berlin. X1, X2, and X3 represented the diploid banana plants, and X4, X5, and X6 represented the autotetraploid banana plants.
Metabolome analysis and differential metabolites
PCA analysis classified overall variation as PC1 and PC2, contributing 16.92/18.17%, respectively (Figure 8). The correlations between samples with the same ploidy were high, indicating good repeatability of samples, as well as stability and reliability of the experimental data. The results of metabolome analysis revealed that metabolites of autotetraploid seedlings were significantly different from those of diploid banana seedlings (Figure 8).
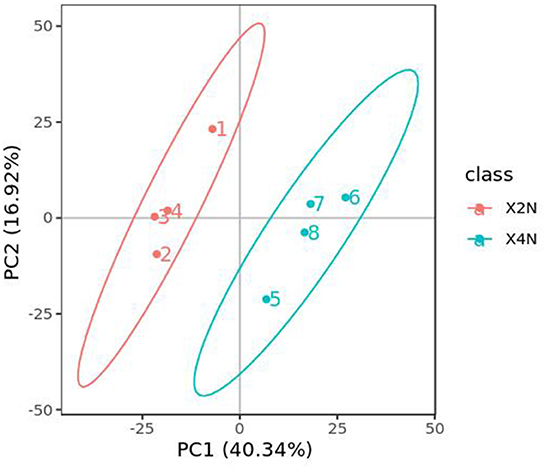
Figure 8. Principal component analysis (PCA) analysis of diploids and autotetraploids of Pisang Berlin samples.The ordinate represented the clustering of samples and the abscissa represented the clustering of metabolites. The diploids and autotetraploids of Pisang Berlin were represented as following, X2N represented diploids of Pisang Berlin (1, 2, 3, and 4 represented sample replicates), X4N represented autotetraploid of Pisang Berlin (5, 6, 7, and eight represented sample replicates).
Our results indicated that there were 276 differential metabolites in the autotetraploids and diploids of Pisang Berlin. We found 107 differential metabolites that were up-regulated in the autotetraploids. Furthermore, the KEGG pathway enrichment analysis showed that the significantly enriched pathways were biosynthesis of amino acids, porphyrin and chlorophyll metabolism, indole alkaloid biosynthesis, and carbon fixation in photosynthetic organisms (Figure 9; Supplementary Table S4).
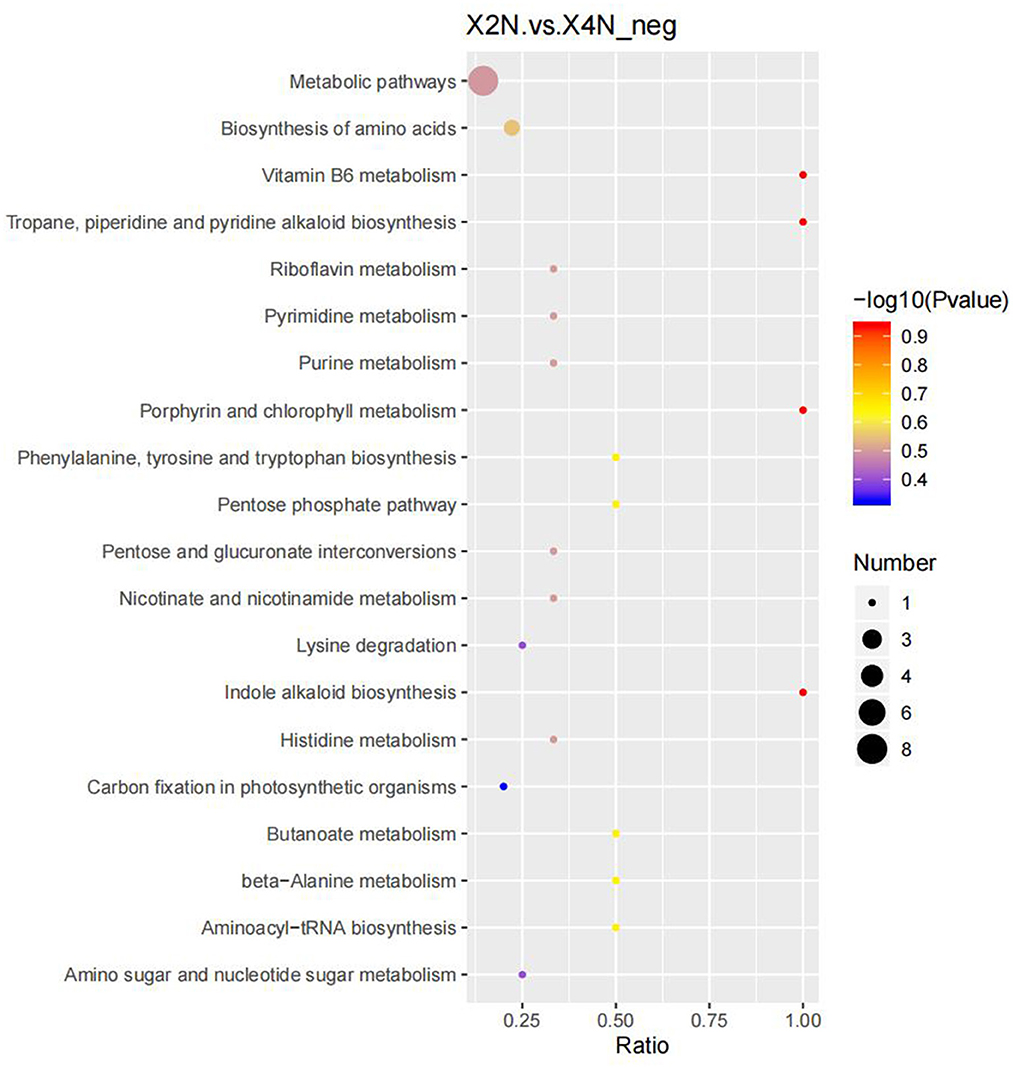
Figure 9. Scatter plot of 20 KEGG pathways between diploids and autotetraploids of Pisang Berlin of the differentially accumulated metabolites enrichment analysis. The degree of enrichment is shown by ratio, P-value, and the number of metabolites enriched in each pathway. X2N represented diploids of Pisang Berlin; X4N represented autotetraploid of Pisang Berlin.
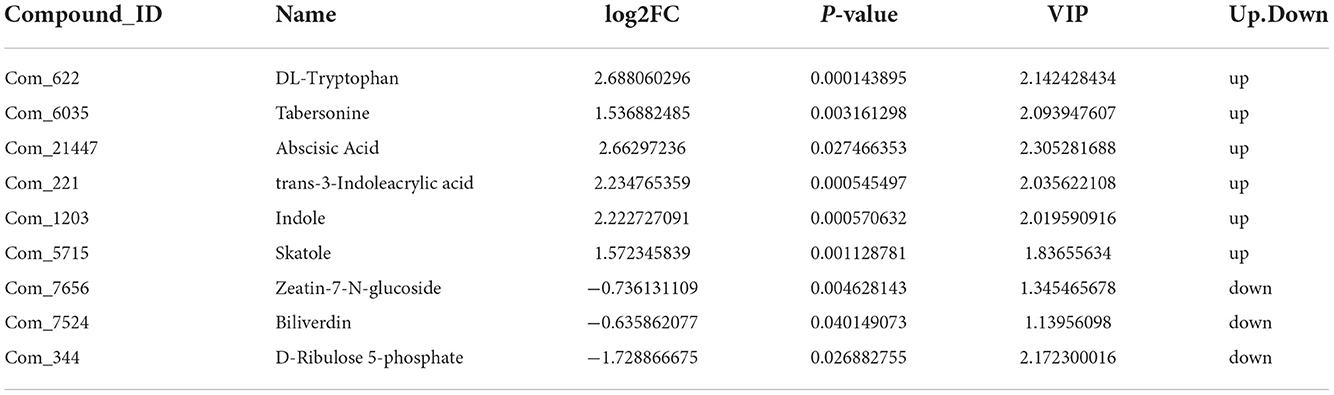
We further analyzed metabolites associated with hormones and photosynthesis. We found that DL-tryptophan, tabersonine, trans-3-Indoleacrylic acid, Indole, and skatole related to auxin, as well as Abscisic acid, present at high levels in the autotetraploids. In contrast, zeatin-7-N-glucoside related to cytokinin, and biliverdin, D-ribulose 5-phosphate related to photosynthesis present at high levels in diploids of Pisang Berlin (Table 2).
Combined analysis of the metabolome and transcriptome revealed that, DEGs and differential metabolites in diploids and autotetraploids of Pisang Berlin, were found to be enriched in 30 metabolic pathways. DEGs and DCMs were mostly related to phenylpropanoid and amino acid biosynthesis, as well as plant hormone signaling in Pisang Berlin (Figure 10).
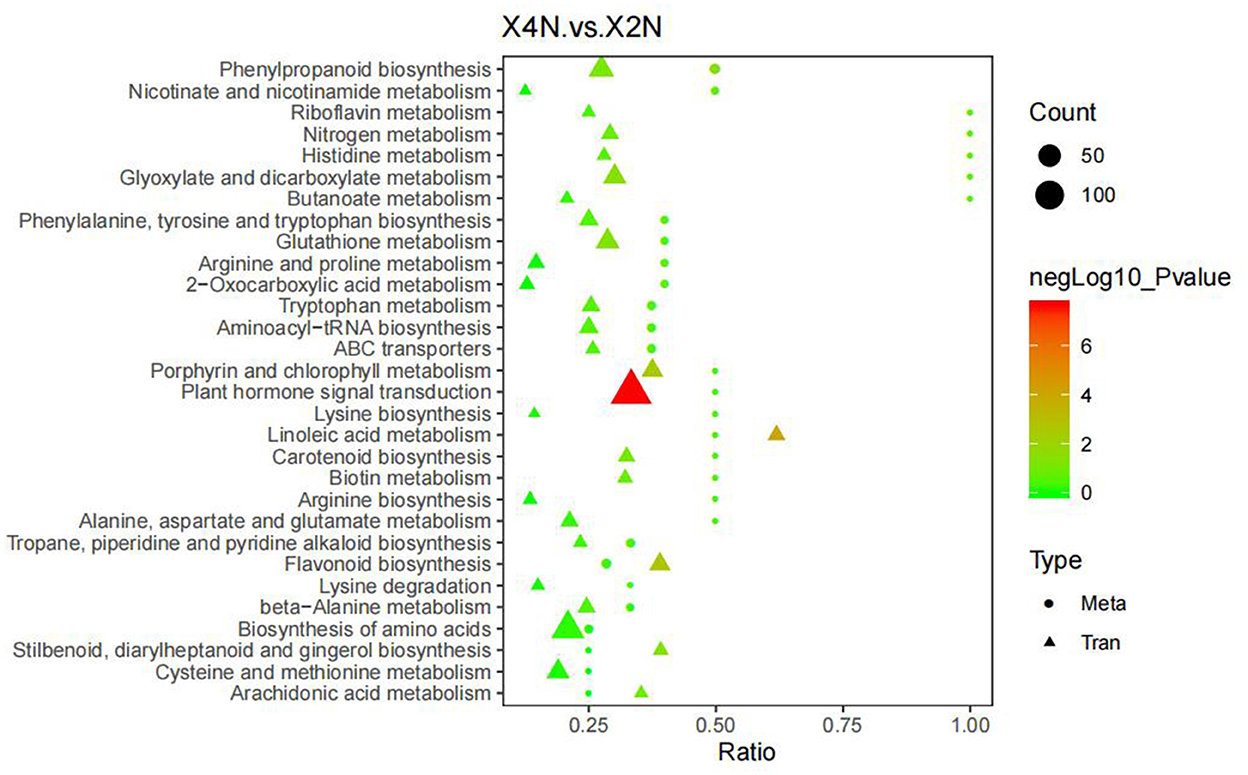
Figure 10. Association analysis of differential expression genes and differential metabolites. The abscissa represented the ratio of the metabolites or genes enriched in this pathway to the number of metabolites or genes annotated in this pathway, the ordinate represented the KEGG pathway enriched by metabolome and transcriptome. Count represented the number of metabolites or genes enriched in the pathway. X2N, diploids of Pisang Berlin; X4N, autotetraploid of Pisang Berlin.
Discussion
Banana breeding for triploids is believed to be the optimum for cultivation in the banana industry, since the triploids exhibit better agronomic characteristics, good fruit quality and infertile or (seedless; Bakry et al., 2007). The reconstructive breeding to generate banana (Musa spp.) triploid varieties is a promising breeding strategy based on artificial autotetraploids as a breeding step. Induction of autotetraploid plants during mitosis with spindle fiber-inhibiting chemicals (such as colchicines and oryzalin) is a feasible approach to develop autotetraploids, but chimeras may occur during the polyploidy induction by colchicine, while some plants of ploidy variation could return to their original ploidy (De Carvalho Santos et al., 2019). In this study, regenerated plants of autotetraploids were obtained by colchicine induction (Figure 1). The mixoploidy phenomenon observed after the first and second generation of propagation was detected by the FCM approach (Pio et al., 2014), and this phenomenon, known as “retromutation” usually occurs in polyploid cells (Bakry et al., 2007). To avoid this phenomenon, we used the strategy of repeated vegetative propagation to dissociate chimeras (Roux et al., 2001), and obtained the non-chimeral autotetraploids (Figure 1).
Polyploids that were generated by chromosome doubling could have some superior agronomic features, including increased fruit size, disease tolerance, and seedless (Sanford, 1983; Predieri, 2001; Kanchanapoom and Koarapatchaikul, 2012). In this study, autotetraploids were obtained from treatment with colchicine on meristematic shoot tips of diploids Pisang Berlin (AA). The autotetraploid plantlets exhibited better plant performance, displaying significantly increases in height and diameter of pseudostem, root length, thickness and area of leaves, as well as leaf chlorophyll content, compared to the diploid control (Figure 2). This effect seems to be associated with the gene dosage effect as would be expected. Previous research based on colchicine derived polyploidy also obtained similar results (Vakili, 1967; Hamill et al., 1992). Polyploidy has the general effect of increasing gene expression levels, which would result in enhancing plant resistance and improving adaptation to adverse environmental conditions (Sattler et al., 2016).
Endogenous plant hormones are synthesized and transported to various parts that regulate growth and development by forming different concentration gradients and ratios (Arnao and Hernández-Ruiz, 2018). Polyploidy plants usually show organ hypertrophy and longer growth periods, mainly due to the alterations of endogenous hormone levels. Previous reports showed that the DEGs related to cell development and carbohydrate metabolism, showed up-regulation within poplar polyploids (Cheng et al., 2015). In autotetraploid plants of Atractylodes lancea, their increasing stress resistance and phenotypic changes in leaves, may be related to the DEGs including DnaK/Hsp70, ACO, ETR/ERS, FAD2, AOS, LOX, PHS1/PAS2 and YWHAE (Ul Haq et al., 2019). Another study found that in autotetraploids of birch (Betula platyphylla) and mulberry (Morus alba), the phenotypic changes would be attributed to the differentially expressed genes involved in signal transduction of plant hormones (including cytokinin, gibberellins, ethylene, and auxin; Mu et al., 2012; Dai et al., 2015).
The metabolic changes caused by polyploidy usually endue plants with stronger environmental adaptability. For example in willow plants, it was observed that the endogenous hormones (auxin and gibberellin) accumulated more in autotetraploids' root tips (Dudits et al., 2016), in comparison to the diploids. In our study, we analyzed genes associated with endogenous hormones. In autotetraploids of Pisang Berlin (AA), the genes Ma5NG4 and MaIAA30 were up-regulated, whereas the genes MaARF18, MaARF7 and MaPIN were down-regulated, which may result in higher auxin content and afterwards lead to growth promotion (Figures 2, 5, Supplementary Table S2). The gibberellin-related genes, MaCIGR1 and MaGA2ox8 were up-regulated, MaGID1 was down-regulated, indicating that these genes might play an importance role in increasing the plant height of autotetraploids (Supplementary Table S2).
Abscisic acid (ABA) has the effect on the slow growth when in vitro preservation of Mahesak (Tectona grandis) (Tongsad et al., 2020). In our study, the up-regulation of ABA 8'-hydroxylase 1 and MaPYL8, as well as down-regulation of MaABI5, may lead to the high levels of abscisic acid found in the autotetraploids (Figure 5, Supplementary Table S2). Interestingly, we recently observed the slow plant growth occurred in autotetraploid plants of Pisang Berlin in the field, which also could be due to the inhibition effect caused by ABA (Humplík et al., 2017). Furthermore, we found that six genes of MaCKX cytokinins in the autotetraploids were down-regulated, and zeatin-7-N-glucoside related to cytokinin present at low levels (Figure 5, Supplementary Table S2). These evidences confirmed that MaCKX genes might play an important role in inactivation of cytokinins.
The photosynthetic process is considered as a series of enzymatic reactions, which involves genes related to Ribulose 1,5-bisphosphate carboxylase (Rubisco), malate dehydrogenase (MDH), ribose-5-phosphate isomerase (RPI), phosphoenolpyruvate carboxylase (PEP), fructose-1,6-bisphosphatase (FBP) (Whitney and Andrews, 2001; Liang et al., 2011; Wang et al., 2011; Ciou et al., 2015). Rubisco is responsible for the first step of the dark reaction in photosynthesis, but its catalytic efficiency is very low (Sharwood, 2017; Galmés et al., 2019). From our work, we identified the down-regulated Rubisco DEGs, as well as the up-regulated DEGs of PEP and RPI (Figure 6, Supplementary Table S3), which would possibly have critical positive effects on photosynthesis of the autotetraploids. Dai et al. (2015) also found that, the larger plants of autotetraploid mulberry trees were associated with the differentially expressed genes involved in photosynthesis biosynthesis.
Conclusion
Through in vitro chromosome doubling, we obtained the non-chimeral autotetraploids of M. acuminata (AA) “Pisang Berlin”, a highly resistant banana diploid variety to Foc TR4. The autotetraploids showed significant increases in plant height, pseudostem diameter, main root length, and leaf thickness, area, and chlorophyll content. The evidences from transcriptome and metabolome profiling demonstrated that the hormones IAA, ABA, and photosynthetic regulation may play a vital role in the plant growth of autotetraploids in bananas. However, we contend that further investigations are needed to analyze the autotetraploids cultivated and hybridized with diploid elite cultivars for producing superior triploid hybrids that might have better values in sustainable breeding programs.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found here: https://www.ncbi.nlm.nih.gov/bioproject/PRJNA900716.
Author contributions
OS, GY, NZ, LL, and LZ provided substantial contributions to the conception and design of the work. NZ, LL, and LZ performed the experiments, analyzed and interpreted the data, and wrote the manuscript. OS, WH, QY, FB, and GD revised the manuscript. OS, NZ, and AK reviewed and finalized manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was jointly funded by National Key R&D Program of China (2019YFD1000203 and 2019YFD1000900), the earmarked fund for CARS (CARS-31-01), GDAAS (202102TD, R2020PY-JX002, and BZ202012), Laboratory of Lingnan Modern Agriculture Project (NT2021004), and Maoming Branch Grant (2021TDQD003).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fsufs.2022.1070108/full#supplementary-material
References
Allario, T., Brumos, J., Colmenero-Flores, J. M., Iglesias, D. J., Pina, J. A., Navarro, L., et al. (2013). Tetraploid Rangpur lime rootstock increases drought tolerance via enhanced constitutive root abscisic acid production. Plant Cell Environ. 36, 856–868. doi: 10.1111/pce.12021
Allario, T., Brumos, J., Colmenero-Flores, J. M., Tadeo, F., Froelicher, Y., Talon, M., et al. (2011). Large changes in anatomy and physiology between diploid Rangpur lime (Citrus limonia) and its autotetraploid are not associated with large changes in leaf gene expression. J. Exp. Bot. 62, 2507–2519. doi: 10.1093/jxb/erq467
An, J. J., Li, M. F., Huang, M. J., and Li, S. P. (2009). Regression model building on leaf area of banana. Guangxi Agr. Sci. 40, 724–727
Arnao, M. B., and Hernández-Ruiz, J. (2018). Melatonin and its relationship to plant hormones. Ann. Bot. 121, 195–207. doi: 10.1093/aob/mcx114
Bakry, F., De La Reberdiere, N. P., Pichot, S., and Jenny, C. (2007). In liquid medium colchicine treatment induces non chimerical doubled-diploids in a wide range of mono-and interspecific diploid banana clones. Fruits 62, 3–12. doi: 10.1051/fruits:2006043
Borges, V. P., Marques, T. D. S., Reis, A. S. D., Oliveira, N. H. C., Jesus, J. A., Silveira, D. G., et al. (2016). Desenvolvimento in vitro de bananeira'Ouro'após poliploidização com antimitóticos. Pesquisa Agro. Bra. 51, 1789–1793. doi: 10.1590/s0100-204x2016001000011
Brocard, I. M. (2002). Regulation and role of the Arabidopsis abscisic acid-insensitive 5 gene in abscisic acid, sugar, and stress response. Plant Physiol. 129, 1533–1543. doi: 10.1104/pp.005793
Chen, J. W., Li, Y., Li, Y., Li, Y. Q., Wang, Y., Jiang, C. Y., et al. (2021). auxin response factor 18–histone deacetylase 6 module regulates floral organ identity in rose (Rosa hybrida). Plant Physiol. 186, 1074–1087. doi: 10.1093/plphys/kiab130
Chen, L., Zhao, J., Song, J., and Jameson, P. E. (2009). Cytokinin dehydrogenase: a genetic target for yield improvement in wheat. Plant Biotechnol. J. 18, 614–630. doi: 10.1111/pbi.13305
Chen, R. Y., Song, W. Q., and Li, X. L. (1979). A new method for preparing mitotic chromosomes from plant. J. Integr. Plant Biol. 21, 297–298.
Chen, S. L., and Tang, P. (1945). Studies on colchicine-induced autotetraploid barley. III. Physiological studies. Am. J. Bot. 32, 177–179. doi: 10.1002/j.1537-2197.1945.tb05104.x
Cheng, S., Huang, Z., Li, Y., Liao, T., Suo, Y. J., Zhang, P. D., et al. (2015). Differential transcriptome analysis between Populus and its synthesized allotriploids driven by second-division restitution. J. Integr. Plant. Biol. 57, 1031–1045. doi: 10.1111/jipb.12328
Ciou, S. C., Chou, Y. T., Liu, Y. L., Lu, J. W., Huang, S. F., Chou, Y. T., et al. (2015). Ribose-5-phosphate isomerase A regulates hepatocarcinogenesis via PP2A and ERK signaling. Int. J. Cancer 137, 104–115. doi: 10.1002/ijc.29361
Cohen, H., Fait, A., and Tel-Zur, N. (2013). Morphological, cytological and metabolic consequences of autopolyploidization in Hylocereus (Cactaceae) species. BMC Plant Biol. 13, 1–14. doi: 10.1186/1471-2229-13-173
Conesa, A., Götz, S., García-Gómez, J. M., Terol, J., Talón, M., and Robles, M. (2005). Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21, 3674–3676. doi: 10.1093/bioinformatics/bti610
Dai, F., Wang, Z., Luo, G., and Tang, C. (2015). Phenotypic and transcriptomic analyses of autotetraploid and diploid mulberry (Morus alba L.). Int. J. Mol. Sci. 16, 22938–22956. doi: 10.3390/ijms160922938
De Carvalho Santos, T. T., De Oliveira Amorim, V. B., Dos Santos-Serejo, J. A., Da Silva Ledo, C. A., Haddad, F., Ferreira, C. F., et al. (2019). Genetic variability among autotetraploid populations of banana plants derived from wild diploids through chromosome doubling using SSR and molecular markers based on retrotransposons. Mol. Breed 39, 1–13. doi: 10.1007/s11032-019-0996-1
Dita, M., Teixeira, L., Li, C., Zheng, S., O'Neill, W., and Daniels, J. (2011). Phenotyping Musa spp. for host reaction to Fusarium oxysporum f. sp. cubense, under greenhouse and field conditions In editor M Dita, Practical Guidelines for Early Screening and Field Evaluation of Banana against Fusarium Wilt, Pseudocercospora Leaf Spots and Drought (Montpellier France: Bioversity International), 5–18.
Do Amaral, C. M., De Almeida Dos Santos-Serejo, J., De Oliveira E Silva, S., Da Silva Ledo, C. A., and Amorim, E. P. (2015). Agronomic characterization of autotetraploid banana plants derived from ‘Pisang Lilin'(AA) obtained through chromosome doubling. Euphytica. 202, 435–443. doi: 10.1007/s10681-014-1320-0
DoleŽel, J., Binarová, P., and Lcretti, S. (1989). Analysis of nuclear DNA content in plant cells by flow cytometry. Biol. Plant 31, 113–120. doi: 10.1007/BF02907241
Dudits, D., Török, K., Cseri, A., Paul, K., Nagy, A. V., Nagy, B., et al. (2016). Response of organ structure and physiology to autotetraploidization in early development of energy willow Salix viminalis. Plant Physiol. 170, 1504–1523. doi: 10.1104/pp.15.01679
Dunn, W. B., Broadhurst, D., Begley, P., Zelena, E., Francis-McIntyre, S., Anderson, N., et al. (2011). Procedures for large-scale metabolic profiling of serum and plasma using gas chromatography and liquid chromatography coupled to mass spectrometry. Nat. Protocols 6, 1060–1083. doi: 10.1038/nprot.2011.335
Fort, A., Ryder, P., McKeown, P. C., Wijnen, C., Aartset, M. G., Sulpice, R., et al. (2015). Disaggregating polyploidy, parental genome dosage and hybridity contributions to heterosis in Arabidopsis thaliana. New Phytol. 209, 590–599. doi: 10.1111/nph.13650
Galmés, J., Capó-Bauçà, S., Niinemets, Ü., and Iñiguez, C. (2019). Potential improvement of photosynthetic CO2 assimilation in crops by exploiting the natural variation in the temperature response of Rubisco catalytic traits. Curr. Opin. Plant Biol. 49, 60–67. doi: 10.1016/j.pbi.2019.05.002
Grabherr, M. G., Haas, B. J., Yassour, M., Thompson, D. A., and Amit, I. (2011). Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 29, 644–652. doi: 10.1038/nbt.1883
Hamill, S., Smith, M., and Dodd, W. (1992). In vitro induction of banana autotetraploids by colchicine treatment of micropropagated diploids. Austral. J. Bot. 40, 887–896. doi: 10.1071/BT9920887
Hapsari, L., and Lestari, D. A. (2016). Fruit characteristic and nutrient values of four Indonesian banana cultivars (Musa spp.) at different genomic groups. J. Agri. Sci. 38, 303–311. doi: 10.17503/agrivita.v38i3.696
Heslop-Harrison, J. S., and Schwarzacher, T. (2007). Domestication, genomics and the future for banana. Ann. Bot. 100, 1073–1084. doi: 10.1093/aob/mcm191
Hoagland, D. R., and Arnon, D. I. (1950). The Water-Culture Method for Growing Plants Without Soil. California: Agricultural Experiment Station, 1–32.
Humplík, J. F., Bergougnoux, V., and Van, V. E. (2017). To stimulate or inhibit? That is the question for the function of abscisic acid. Trends Plant Sci. 22, 830–841. doi: 10.1016/j.tplants.2017.07.009
Jenny, C., Tomekpe, K., Bakry, F., and Escalant, J. (2002). “Conventional breeding of bananas” in Mycosphaerella Leaf Spot Diseases of Bananas: Present Status and Outlook, ed L. Jacome (Rome: International Plant Genetic Resources Institute, Italy), 199.
Jong, M. D., Wolters-Arts, M., Feron, R., Mariani, C., and Vriezen, W. H. (2009). The Solanum lycopersicum auxin response factor 7 (SlARF7) regulates auxin signaling during tomato fruit set and development. Plant J. 57, 160–170. doi: 10.1111/j.1365-313X.2008.03671.x
Kadota, M., and Niimi, Y. (2002). In vitro induction of tetraploid plants from a diploid Japanese pear cultivar (Pyrus pyrifolia N. cv. Hosui). Plant Cell Rep. 21, 282–286. doi: 10.1007/s00299-002-0509-1
Kanchanapoom, K., and Koarapatchaikul, K. (2012). In vitro induction of tetraploid plants from callus cultures of diploid bananas (Musa acuminata, AA group) ‘Kluai Leb Mu Nang'and ‘Kluai Sa'. Euphytica 183, 111–117. doi: 10.1007/s10681-011-0516-9
Kanehisa, M., Goto, S., Hattori, M., Aoki-Kinoshita, K. F., Itoh, M., Kawashima, S., et al. (2006). From genomics to chemical genomics: new developments in KEGG. Nucl Acids Res. 34, D354–D357. doi: 10.1093/nar/gkj102
Kaur, S., Zhang, X., Mohan, A., Vikram, P., Singh, S., Zhang, Z. W., et al. (2017). Genome-wide association study reveals novel genes associated with culm cellulose content in bread wheat (Triticum aestivum, L.). Front. Plant Sci. 8, 1913. doi: 10.3389/fpls.2017.01913
Kitahata, N., Saito, S., Miyazawa, Y., Umezawa, T., Shimada, Y., Min, Y. K., et al. (2005). Chemical regulation of abscisic acid catabolism in plants by cytochrome P450 inhibitors. Bioorgan. Med. Chem. 13, 4491–4498. doi: 10.1016/j.bmc.2005.04.036
Kovi, M. R., Zhang, Y., Yu, S., Yang, G. Y., Yan, W. H., and Xing, Y. Z. (2011). Candidacy of a chitin-inducible gibberellin-responsive gene for a major locus affecting plant height in rice that is closely linked to Green Revolution gene sd1. Theor. Appl. Genet. 123, 705–714. doi: 10.1007/s00122-011-1620-x
Liang, W., Ouyang, S., Shaw, N., Joachimiak, A., Zhang, R., and Liu, Z. J. (2011). Conversion of D-ribulose 5-phosphate to D-xylulose 5-phosphate: new insights from structural and biochemical studies on human RPE. FASEB J. 25, 497–504. doi: 10.1096/fj.10-171207
Liu, Y. X., Zhou, Z. X., Tang, F. L., He, Y. D., Cao, H. X., Wang, B. Z., et al. (2013). Quantitative analysis of relationship between brazilian banana's leaf Area and its morphological characteristics. Chin. J. Tropic Crops 34, 1641–1645. doi: 10.3969/j.issn.1000-2561.2013.09.003
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Mu, H. Z., Liu, Z. J., Lin, L., Li, H. Y., Jiang, J., and Liu, G. F. (2012). Transcriptomic analysis of phenotypic changes in birch (Betula platyphylla) autotetraploids. Int. J. Mol. Sci. 13, 13012–13029. doi: 10.3390/ijms131013012
Murase, K., Hlirano, Y., Sun, T. P., and Hakoshima, T. (2008). Gibberellin-induced DELLA recognition by the gibberellin receptor GID1. Nature 456, 459–463. doi: 10.1038/nature07519
Nassar, N. M. (2006). Chromosome doubling induces apomixis in a cassava × Manihot anomala hybrid. Hereditas 143, 246–248. doi: 10.1111/j.2006.0018-0661.01957.x
Ortiz, R., and Vuylsteke, D. (1995). Effect of the parthenocarpy gene P1 and ploidy on fruit and bunch traits of plantain-banana hybrids. Heredity 75, 460–465. doi: 10.1038/hdy.1995.162
Pablo, A., Yann, F., Sergio, S., Manuel, A., María, H., Jos,é, J., et al. (2011). Tetraploidization events by chromosome doubling of nucellar cells are frequent in apomictic citrus and are dependent on genotype and environment. Ann. Bot. 108, 37–50. doi: 10.1093/aob/mcr099
Perrier, X., Jenny, C., Bakry, F., Karamura, D., Kitavi, M., Dubois, C., et al. (2019). East African diploid and triploid bananas: a genetic complex transported from South-East Asia. Ann. Bot. 123, 19–36. doi: 10.1093/aob/mcy156
Pio, L. S., Pasqual, M. E, Silva, S. D. O., Rocha, H. S., Magalhaes, H. M., et al. (2014). Inducing and identifying artificially-induced polyploidy in bananas. African J. Biotechnol. 13, 3748–3758. doi: 10.5897/AJB2014.14009
Poerba, Y., Martanti, D., and Handayani, T. (2019). Morphology and reproductive function of induced autotetraploid banana by chromosome doubling. SABRAO J Breed Genet. 51, 175–190.
Predieri, S. (2001). Mutation induction and tissue culture in improving fruits. Plant Cell Tissue Organ. Cult. 64, 185–210. doi: 10.1023/A:1010623203554
Roux, N., Dolezel, J., Swennen, R., and Zapata-Arias, F. J. (2001). Effectiveness of three micropropagation techniques to dissociate cytochimeras in Musa spp. Plant Cell Tissue Organ. Cult. 66, 189–197. doi: 10.1023/A:1010624005192
Roux, N., Strosse, H., Toloza, A., and Swennen, R. (2004). Detecting Ploidy Level Instability of Banana Embryogenic Cell Suspension Cultures by Flow Cytometry, in: Banana Improvement: Cellular Molecular Biology and Induced Mutations. Enfield: Science Publishers Inc.
Saavedra, X., Modrego, A., Rodríguez, D., González-García, M. P., Sanz, L., and Lorenzo, G. N. (2010). The nuclear interactor PYL8/RCAR3 of fagus sylvatica FsPP2C1 Is a positive regulator of abscisic acid signaling in seeds and stress. Plant Physiol. 152, 133–150. doi: 10.1104/pp.109.146381
Sanford, J. (1983). Ploidy Manipulations in Methods in Fruit Breeding. West Lafayette, IN: Purdue University Press, 100–123.
Sato, A., and Yamamoto, K. T. (2008). What's the physiological role of domain II-less Aux/IAA proteins? Plant Signal. Behav. 3, 496–497. doi: 10.4161/psb.3.7.5994
Sattler, M. C., Carvalho, C. R., and Clarindo, W. R. (2016). The polyploidy and its key role in plant breeding. Planta 243, 281–296. doi: 10.1007/s00425-015-2450-x
Sharwood, R. E. (2017). Engineering chloroplasts to improve Rubisco catalysis: prospects for translating improvements into food and fiber crops. New Phytol. 213, 494–510. doi: 10.1111/nph.14351
Silva, S. D. O., Junior, M. T. S., Alves, É. J., Silveira, J. R. S., and Lima, M. B. (2001). Banana breeding program at Embrapa. Crop Breed Appl Biotechnol. 1, 399–436. doi: 10.13082/1984-7033.v01n04a09
Ssebuliba, R., Magambo, M., Talengera, D., Makumbi, D., Tenkouano, A., Rubaihayo, P., et al. (2006). Biological factors affecting seed production in East African Highland bananas. J. Crop Improv. 16, 67–79. doi: 10.1300/J411v16n01_05
Sun, Q., Sun, H., Li, L., and Bell, R. L. (2009). In vitro colchicine-induced polyploid plantlet production and regeneration from leaf explants of the diploid pear (Pyrus communis L.) cultivar,‘Fertility'. J. Horticult Sci. Biotechnol. 84, 548–552. doi: 10.1080/14620316.2009.11512564
Swamy, R. D., Rao, N. S., and Chacko, E. K. (1983). Tissue-culture propagation of banana. Sci Horticult. 18, 247–252. doi: 10.1016/0304-4238(83)90028-6
Syvertsen, J., Lee, L., and Grosser, J. (2000). Limitations on growth and net gas exchange of diploid and tetraploid Citrus rootstock cultivars grown at elevated CO2. J Am Soc Horticult Sci. 125:228–234. doi: 10.21273/JASHS.125.2.228
Tatusov, R.L., Fedorova, N.D., Jackson, J.D., Jacobs, A., Kiryutin, B., Koonin, E., et al. (2003). The COG database: an updated version includes eukaryotes. BMC Bioinform. 4, 41. doi: 10.1186/1471-2105-4-41
Tongsad, P., Laipasu, P., Chareonsap, P. P., and Poeaim, A. (2020). The effects of abscisic acid and sorbitol on the slow growth in vitro preservation of Mahesak (Tectona grandis L.). Int J. Agri. Technol. 16, 1277–1286.
Trapnell, C., Roberts, A., Goff, L., Pertea, G., Kim, D., Kelley, D. R., et al. (2012). Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protocols 7, 562–578. doi: 10.1038/nprot.2012.016
Ul Haq, S., Khan, A., Ali, M., Khattak, A. M., Gai, W. X., Zhang, H. X., et al. (2019). Heat shock proteins: dynamic biomolecules to counter plant biotic and abiotic stresses. Int. J. Mol. Sci. 20, E5321. doi: 10.3390/ijms20215321
Vakili, N. G. (1967). The experimental formation of polyploidy and its effect in the genus Musa. Am. J. Bot. 54, 24–36. doi: 10.1002/j.1537-2197.1967.tb06889.x
Wang, S., Tu, H., Wan, J., Chen, W., Liu, X., Luo, J., et al. (2016). Spatio-temporal distribution and natural variation of metabolites in citrus fruits. Food Chem. 199, 8–17. doi: 10.1016/j.foodchem.2015.11.113
Wang, Y. Q., He, F., Tan, X. F., Su, J. K., Xing, W. Y., and Bin-Linga, A. (2011). EST-based analysis of gene expression of photosynthesis in Ginkgo biloba mature leaves. J. Central South Univ. Forest Technol. 31, 54–59. doi: 10.14067/j.cnki.1673-923x.2011.03.024
Want, E. J., Wilson, I. D., Gika, H., Theodoridis, G., Plumb, R. S., Shockcor, J., et al. (2010). Global metabolic profiling procedures for urine using UPLC–MS. Nat. Protocols 5, 1005–1018. doi: 10.1038/nprot.2010.50
Whitney, S. M., and Andrews, T. J. (2001). The gene for the ribulose-1, 5-bisphosphate carboxylase/oxygenase (Rubisco) small subunit relocated to the plastid genome of tobacco directs the synthesis of small subunits that assemble into Rubisco. Plant Cell. 13, 193–205. doi: 10.1105/tpc.13.1.193
Wu, R. M., Walton, E. F., Richardson, A. C., Wood, M., Hellens, R. P., and Varkonyi-Gasic, E. (2012). Conservation and divergence of four kiwifruit SVP-like MADS-box genes suggest distinct roles in kiwifruit bud dormancy and flowering. J. Exp. Bot. 63, 797–807. doi: 10.1093/jxb/err304
Xu, M., Zhu, L., Shou, H., and Wu, P. (2005). A PIN1 family gene, OsPIN1, involved in auxin-dependent adventitious root emergence and tillering in rice. Plant Cell Physiol. 46, 1674–1681. doi: 10.1093/pcp/pci183
Xue, H., Zhang, F., Zhang, Z. H., Fu, J. F., Wang, F., Zhang, B., et al. (2015). Differences in salt tolerance between diploid and autotetraploid apple seedlings exposed to salt stress. Sci. Horticult. 190, 24–30. doi: 10.1016/j.scienta.2015.04.009
Yasushi, K., Hiroyoshi, M., and Katsura, I. (2003). Phosphoenolpyruvate carboxylase: three-dimensional structure and molecular mechanisms. Arch. Biochem. Biophys. 414, 170–179. doi: 10.1016/S0003-9861(03)00170-X
Zhai, R., Wang, Z. G., Yang, C. Q., Wang, K. L., Espley, R., Liu, J. L., et al. (2019). PbGA2ox8 induces vascular-related anthocyanin accumulation and contributes to red stripe formation on pear fruit. Hortic. Res. 6, 137. doi: 10.1038/s41438-019-0220-9
Zhang, F., Xue, H., Lu, X., Zhang, B., Wang, F., Ma, Y., et al. (2015). Autotetraploidization enhances drought stress tolerance in two apple cultivars. Trees 29, 1773–1780. doi: 10.1007/s00468-015-1258-4
Zhang, J., Liu, Y., Xia, E. H., Yao, Q. Y., Liu, X. D., and Gao, L. Z. (2015). Autotetraploid rice methylome analysis reveals methylation variation of transposable elements and their effects on gene expression. Proc. Natl. Acad. Sci. 112, E7022–E7029. 10.1073/pnas.1515170112 doi: 10.1073/pnas.1515170112
Keywords: autotetraploid, diploid, hormonal regulation, transcriptome, metabolome, colchicine, banana, Musa spp.
Citation: Zhan N, Li L, Zhang L, He W, Yang Q, Bi F, Deng G, Kiggundu A, Yi G and Sheng O (2023) Transcriptome and metabolome profiling provide insights into hormone-mediated enhanced growth in autotetraploid seedlings of banana (Musa spp.). Front. Sustain. Food Syst. 6:1070108. doi: 10.3389/fsufs.2022.1070108
Received: 14 October 2022; Accepted: 23 November 2022;
Published: 04 January 2023.
Edited by:
Mohamed Ait El Mokhtar, Université Hassan II Mohammedia, MoroccoReviewed by:
Jingyang Li, Chinese Academy of Tropical Agricultural Sciences, ChinaNoor Saidi, Putra Malaysia University, Malaysia
Copyright © 2023 Zhan, Li, Zhang, He, Yang, Bi, Deng, Kiggundu, Yi and Sheng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ou Sheng,  c2hlbmdvdTZAMTI2LmNvbQ==
c2hlbmdvdTZAMTI2LmNvbQ==
†These authors have contributed equally to this work
 Ni Zhan
Ni Zhan Li Li1,3†
Li Li1,3† Qiaosong Yang
Qiaosong Yang Fangcheng Bi
Fangcheng Bi Ganjun Yi
Ganjun Yi Ou Sheng
Ou Sheng