
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Sustain. Food Syst. , 14 October 2022
Sec. Crop Biology and Sustainability
Volume 6 - 2022 | https://doi.org/10.3389/fsufs.2022.1036111
This article is part of the Research Topic Soil Microbiome Metabolomics: A Way Forward to Sustainable Intensification View all 8 articles
 Fahim Bashir1,2
Fahim Bashir1,2 Arif Bashir3
Arif Bashir3 Vishnu D. Rajput4
Vishnu D. Rajput4 Noureddine Bouaïcha5
Noureddine Bouaïcha5 Khalid M. Fazili6
Khalid M. Fazili6 Samrat Adhikari7
Samrat Adhikari7 Yogesh Negi7
Yogesh Negi7 Tatiana Minkina4
Tatiana Minkina4 Waleed Hassan Almalki8
Waleed Hassan Almalki8 Bashir Ahmad Ganai1,2*
Bashir Ahmad Ganai1,2*Cyanobacterial harmful algal blooms (CHABs) are increasing at an alarming rate in different water bodies worldwide. In India, CHAB events in water bodies such as Dal Lake have been sporadically reported with no study done to characterize the cyanobacterial species and their associated toxins. We hypothesized that this Lake is contaminated with toxic cyanobacterial species with the possibility of the presence of cyanotoxin biosynthetic genes. We, therefore, used some of the molecular tools such as 16S ribosomal DNA, PCR, and phylogenetic analysis to explore cyanobacterial species and their associated toxins. A 3-year (2018–2020) survey was conducted at three different sampling sites of Dal Lake namely, Grand Palace Gath (S1), Nigeen basin (S2), and Gagribal basin (S3). Two strains of Dolichospermum sp. AE01 and AE02 (S3 and S1 site) and one strain of Microcystis sp. AE03 (S2 site) was isolated, cultured, and characterized phylogenetically by 16S ribosomal DNA sequencing. The presence of cyanotoxin genes from the isolates was evaluated by PCR of microcystins (mcyB), anatoxins (anaC), and cylindrospermopsins (pks) biosynthesis genes. Results revealed the presence of both mcyB and pks gene in Microcystis sp. AE03, and only anaC gene in Dolichospermum sp. AE02 strain. However, Dolichospermum sp. AE01 strain was not found to harbor any such genes. Our findings, for the first time, reported the coexistence of pks and mcyB in a Microcystis AE03 strain. This study has opened a new door to further characterize the unexplored cyanobacterial species, their associated cyanotoxin biosynthetic genes, and the intervention of high-end proteomic techniques to characterize the cyanotoxins.
Cyanobacterial harmful algal blooms (CHABs) and their associated toxins are becoming more common in aquatic systems. Scientists predict that harmful blooms in nutrient-rich lakes will increase in frequency, intensity, and duration as global warming and atmospheric carbon dioxide concentrations rise. As a result of eutrophication, anthropogenic pressures, and global warming, toxic blooms have become a worldwide problem, including in India as well (Massey et al., 2020; Mohan et al., 2020; Chorus et al., 2021; Khan et al., 2021). These toxic blooms can adversely affect the quality of freshwater resources through the production of potent toxins known as cyanotoxins (Svirčev et al., 2019; Mehinto et al., 2021). Different cyanotoxins and their variants have been reported in aquatic ecosystems that includes: cylindrospermopsins (CYNs), hepatotoxins: microcystins (MCs), ad neurotoxins: anatoxins (ATXs) (Huang and Zimba, 2019). Exposure to these cyanotoxins can lead to illness and even death in humans, animals, and other eukaryotic organisms (Svirčev et al., 2019; Huang et al., 2020; Mehinto et al., 2021).
MCs are extensively studied cyanotoxin that is produced by an array of cyanobacteria, including Dolichospermum, Microcystis, Planktothrix, Oscillatoria, and Anabaenopsis (Bernard et al., 2017; Chapman and Foss, 2019). So far, more than 300 variants of it have been characterized (Bouaïcha et al., 2019; Jones et al., 2021). They are hepatotoxins with the potential affinity toward serine/threonine phosphatases, thereby dephosphorylating different proteins in many important biochemical pathways. Inhibition of many important biochemical pathways is the main mechanism leading to liver damage and side effects on other organs as well. It also acts as a tumorogenic agent and can induce oxidative stress in cells. CYN is a cyclic guanidine alkaloid. The species producing this toxin belong to certain cyanobacterial genera including Raphidiopsis, Aphanizomenon, Dolichospermum, and Lyngbya (Chapman and Foss, 2019). It primarily shows its toxicity on liver, kidney, heart, spleen, etc. It also acts as a genotoxic agent. Among the different organs of the body, kidney has been found most sensitive to its toxicity. Another alkaloid, namely ATX-a is the most frequently reported of the known ATXs with neurotoxic activity. ATX-a binds to nicotinic acetylcholine receptors present on pheripheral nerve cells that can lead to respiratory arrest (Colas et al., 2021). More than 41 ATX-a producing species of freshwater cyanobacteria have been identified to date including Dolichospermum, Planktothrix, Oscillatoria, Microcystis, Aphanizomenon, Cylindrospermum, and Phormidium (Chapman and Foss, 2019; Christensen and Khan, 2020).
Algal bloom events in water bodies are less common in India compared to other parts of the world (Svirčev et al., 2019). Indian coastlines are suffering from climate change and other anthropogenic influences, and as a result, episodes of CHABs are predicted to significantly rise. This will have long-term negative effects on many of the fragile coastal biotopes and the related blue economy (Mishra et al., 2020; Maniyar et al., 2022). These CHABs can also cause skin lesions, gastric disorders, mass fish mortalities, and even death (Padmakumar et al., 2012). Recently, M. aerugonisa blooms were reported in two freshwater ponds (9°580 6.700N; 76°160 56.800E, and 9°570 3400N; 76°170 3200E) at Ernakulam District of Kerala (India) during early summer (February) and pre-summer monsoon (May) (Mohan et al., 2020). Additionally, it was reported that the Ganga River (26°27.539N; 80°22.859E, 25°25.695N; 81°52.767E, and 25°18.373N; 83°00.585E) has the highest concentration of cyanobacteria-producing MCs in May, and the lowest concentration in August (Dixit et al., 2017).
Among cyanobacterial genera-producing cyanotoxins, the genus Microcystis was observed as the dominant genus in two freshwater ponds, Lakshmikund and Sankuldhara (Varanasi, Uttar-Pradesh, India) (25°29′69′N; 82°99'33” E and 25°30′94″N, 82°99'93” E), constituting ~97 and 67% of the MC population of cyanobacteria, respectively (Singh et al., 2015). The Dal Lake (Kashmir, India) is presently under great anthropogenic pressures such as the discharge of synthetic fertilizers, diesel leakage from dredger machines, nearby sanitary influx, plastic debris, untreated sewage water, and water runoff from agricultural practices (Qadir and Singh, 2018; Ahmad et al., 2020). Excess phosphorus (P) and nitrogen (N) influx into freshwater ecosystems has been linked in several studies to the global spread of toxic cyanobacterial blooms (Griffith and Gobler, 2020). In freshwater ecosystems, P is typically regarded as the limiting nutrient for cyanobacterial growth. As a result, cyanobacterial blooms are frequently associated with high quantities of this nutrient (Xu et al., 2015; Gobler et al., 2016; Harke et al., 2016).
The overall P concentration in Dal Lake has gradually increased from 0.1–0.4 mg/l in 1997 to 6 mg/l in 2017, which is conducive to cyanobacterial growth, particularly bloom-forming cyanobacteria (Kumar et al., 2022). This has led to a significant reduction of the surface of Dal Lake from 31 to 24 km2 between 1859 and 2013, respectively; and a sharp decrease in water quality, decreasing fish stocks, and negative impacts on the lake's recreational activities via cyanobacterial blooms (Rashid et al., 2017). In Dal Lake, many cyanobacterial blooms have been recorded during the last three decades. Red cyanobacterial scum was observed on the surface of Dal Lake in August 1991 (Shafiq-Ur-Rehman, 1998). In April 1998, a bloom of the genus Cladophora was reported in the Pokhribal area of Nigeen basin (Western basin of Dal Lake) that stench like untreated sewage and choked the waterways near the Amir Khan Canal exit point (Kundangar, 1999). Many reports of CHABs in various Indian water bodies and reservoirs in 2019 (hottest-year recorded) were reported (Narayana et al., 2020; Inaotombi and Sarma, 2021). Apart from the coastal waters on India's East and West coasts, Ukai Dam in Gujarat, Chilika Lagoon in Odisha, Gandhisagar and Bargi Dams in Madhya Pradesh, Stanley Reservoir in Tamil Nadu, and Dal Lake in Kashmir, India are just a few examples of affected Indian inland water bodies (Mishra et al., 2020; Ray et al., 2021). Due to the increased frequency, duration, and intensity of CHABs in Dal Lake and its socio-economic importance in the region for tourism, fisheries, agriculture, food, water supply, and recreational opportunities to the local population, it highly advocates for a broad spectrum molecular biology-based studies to identify different cyanobacterial species, characterizing their toxigenic potential, and taking immediate preventive steps to mitigate the cyanobacterial growth.
In the present study, a 3-year (2018–2020) survey was conducted at three sampling sites in Dal Lake, Kashmir. The dominant cyanobacteria morphospecies were isolated and cultured in sterile conditions and their identification was subsequently confirmed by following a phylogenetic analysis approach. The cultured strains were further screened for the cyanotoxin biosynthetic gene clusters (MCs, CYNs, and ATXs) by PCR amplification that highlighted the co-occurrence of two major toxins (cylindrospermopsin and microcystin) producing strains in Dal Lake, Srinagar, Kashmir.
Dal Lake (34.0625°N, 74.5010°E) is a Himalayan urban lake located in Srinagar, Kashmir. It is India's northernmost union territory with an elevation of 1,586 meters above sea level. The total water surface area of the lake is ~24 km2, and its maximum depth is about 6 m. In this study, a 3-year (2018–2020) survey was conducted at the three sampling sites of Dal Lake (Figure 1) namely, S1: Grand Palace Gath (34.1000°N, 74.8766°E), S2: Nigeen basin (34.1182°N, 74.8317°E), and S3: Gagribal basin (34.0824°N, 74.8493°E).
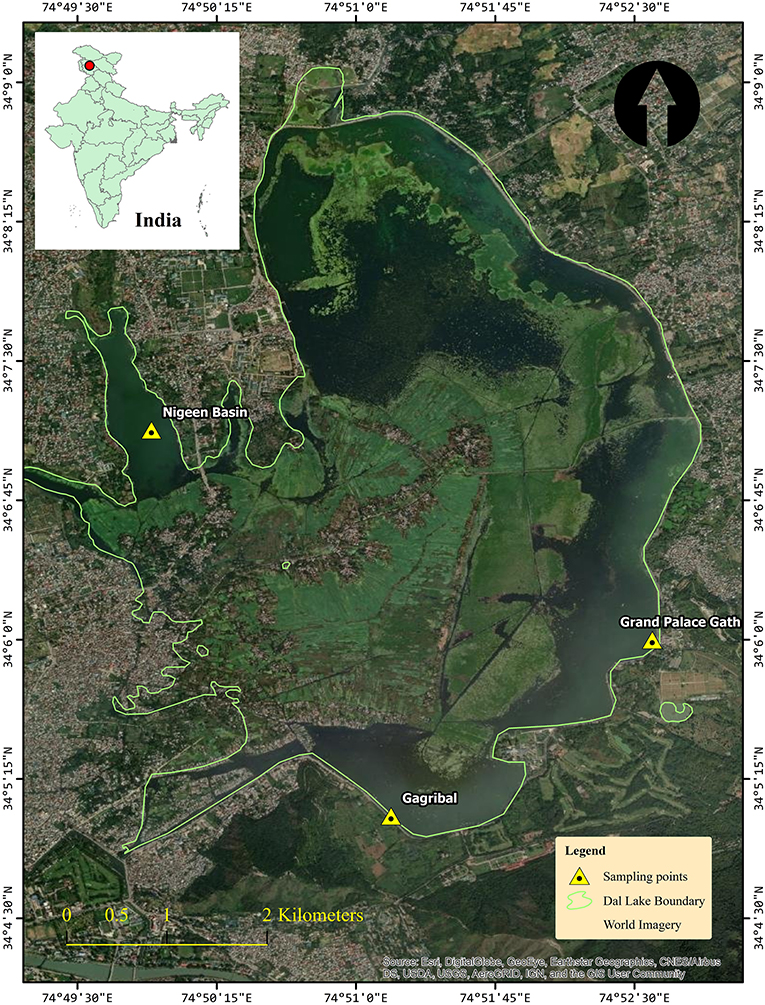
Figure 1. Study Map and locations of the Dal Lake showing the three sampling sites, Grand Palace Gath (Site 1), Nigeen basin (Site 2), and Gagribal basin (Site 3).
Phytoplankton samples were harvested during the blooms period in October 2018 at the Nigeen basin (S2), in August 2019 at the Grand Palace Gath (S1), and in September 2020 at the Gagribal basin (S3). The bloom samples (~0.5 L of water) were collected from the three different sampling locations and were immediately stored in a refrigerator (4°C) before being transported to the laboratory for further identification, isolation, and culture of dominant cyanobacterial morphospecies and analysis of cyanotoxins.
For the morphological identification of the dominant forming-bloom cyanobacteria morphospecies at each sampling site, freshly cyanobacterial bloom samples were observed using a phase-contrast microscope, attached with a charged coupled device camera, and examined under 400X magnification. Different cyanobacteria morphospecies were identified using several morphological taxonomic keys, including colony/filament type, cell size, and shape, whether or not having specialized cells (Komárek, 2016, 2020), and other web databases and websites such as CyanoDB (Hauer and Komárek, 2020), AlgaeBase (Guiry and Guiry, 2020), and Cyanosite (https://www-cyanosite.bio.purdue.edu) online database tools were used for the taxonomic classification and nomenclature of toxigenic cyanobacteria. After a preliminary microscopic examination of the different bloom samples, we switched over to evaluate the dominant morphospecies. An aliquot from each fresh cyanobacteria bloom sample was diluted in a sterilized BG-11 liquid medium. An aliquot of each sample was spread over the 1.5% agarose gel Petri dish prepared with the sterile BG-11 solid medium.
Petri dishes were covered with Parafilm and incubated in an air-conditioned chamber at 22 ± 2°C with a light intensity of 55 μmol of photons m−2 s−1 coupled with the white fluorescent tube lights (12 h:12 h light/dark cycle). After incubation for 2 weeks, each strain was selected from a contaminant-free individual colony and diluted in a sterile 5 mL BG-11 liquid medium. An aliquot was observed using an inverted microscope to identify the cyanobacterial species and the remaining solution was again spread over the freshly prepared Petri dishes comprising of 1.5% agarose gel and incubated as described previously. The procedure was repeated three to four times until a pure culture of each strain was achieved. Each isolated strain was subsequently cultured in batch culture in a sterile 500 mL flasks containing 200 mL of a sterile BG-11 liquid medium at 23°C with an incident light intensity of 55 μmol of photons m−2 s−1 provided by white fluorescent tube lights (12 h:12 h light/dark cycle). Cell biomass samples were obtained after 3 weeks of cultivation by centrifuging at 12,000 rpm for 15 min. Samples were subsequently lyophilized for further molecular analysis.
Exponentially grown cyanobacterial strains (Dolichospermum sp. strain AE01, AE02, and Microcystis sp. strain AE03) were harvested, and genomic DNA was extracted using the Wizard® Genomic DNA Purification Kit (Promega Corporation, USA), by strictly following the manufacturer's protocol. BioRad's Nanodrop® was used to check the quality and quantity of the extracted genomic DNA from the three cyanobacterial strains. The integrity of DNA was confirmed by running it in 1% agarose gel in 1X TAE running buffer. BIO-RAD T100TM Thermal Cycler (Applied Biosystems, Singapore) was used to amplify the 16S ribosomal DNA (16S rDNA) with a specific primer set (Cya106F/23S30R) (Table 1). For PCR amplification, the GoTaq® Flexi DNA Polymerase (Promega Corporation, USA) reagents were used in a final volume of 40 μL. Each PCR reaction was prepared with 2 μL of 25 mM MgCl2 solution, 4 μL of 5X colorless GoTaq® Flexi Buffer, 1 μL of 10 mM of deoxynucleotide triphosphate mix, 1 μL of forward and reverse primer, 1 μL of DNA templates, and 0.2 μL of GoTaq® Flexi DNA Polymerase (5 U/μL). Genomic DNA concentrations were found in the range of 80–193 ng/μL. For 16S rDNA amplification, the PCR reaction conditions were set at an initial denaturation for 3 min at 95°C, followed by 30 cycles of amplification: 30 s at 94°C, 45 s at 55°C, 2 min at 72°C, and a final extension for 4 min at 72°C.
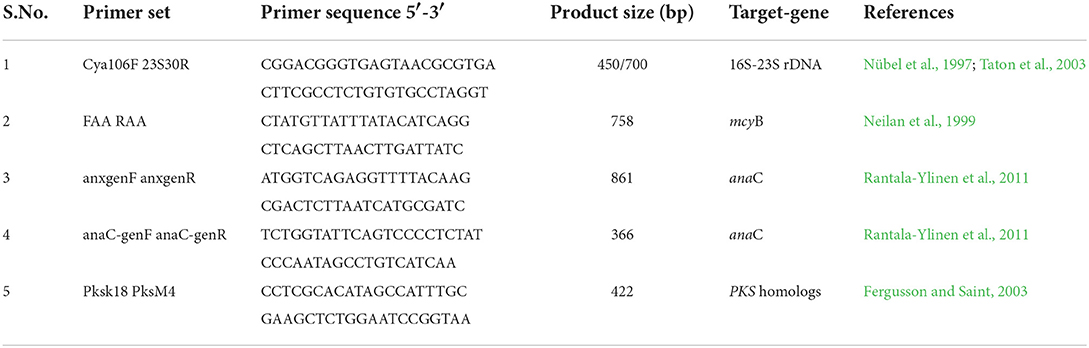
Table 1. Primer sets used in conventional PCR for the phylogenic characterization of cyanobacteria and detection of cyanotoxin biosynthesis gene clusters.
Amplified PCR products were purified by gel extraction kit (Promega, USA). The purified PCR products were sent to Applied Biosystems, Bangaluru, India for Sanger DNA sequencing (Barcode Biosciences, Bangalore, India). Each purified PCR product was directly sequenced using Cya106F/23S30R for 16S rDNA gene cluster sequencing. The sequences obtained were checked for low quality and edited using the Sequence scanner tool. The sequences obtained were finally submitted to National Center for Biotechnology Information (NCBI) and gene accession numbers were obtained. Sequences were then compared using the mega blast search program (NCBI) for 16S rDNA. The neighbor-joining (NJ) algorithm was used to create a phylogenetic tree for the 16S rDNA gene sequences with n = 500 bootstrap replicates. Mega-X software and the Jukes-Cantor model were used in NJ analysis to evaluate the evolutionary substitution model (Kumar et al., 2018).
Cyanotoxin biosynthetic gene cluster of the three isolated cyanobacterial strains Dolichospermum sp. strain AE01, AE02, and Microcystis sp. AE03 was analyzed through PCR amplification of mcyB, anaC, and pks gene cluster, which are indicative of the presence of microcystins, anatoxins, and cylindropsermopsins biosynthesis genes clusters, respectively (Neilan et al., 1999; Fergusson and Saint, 2003; Rantala-Ylinen et al., 2011). The primers listed in Table 1 were used to amplify these cyanotoxin biosynthetic gene clusters. The amplification of mcyB gene was carried out by using gene-specific primer sets (FAA/RAA) (Neilan et al., 1999). The thermal cycler was set at an initial denaturation at 94°C for 3 min, followed by 30 cycles of amplification: 30 s at 95°C, 30 s at 43°C, 1 min at 72°C, and final elongation for 7 min at 72°C. For pks amplification, a conventional PCR was developed by using the primer sets (pksK18/pksM4) (Fergusson and Saint, 2003). The PCR conditions were set at an initial denaturation for 5 min at 94°C, followed by 30 cycles of amplification: 30 s at 94°C, 30 s at 55°C, 1 min at 72°C, and final elongation for 5 min at 72°C. For anaC gene detection, nested PCR was performed with the primer pairs anaC-genF/anaC-genR, and anxgenF/anxgenR (Rantala-Ylinen et al., 2011). The first PCR was performed with the anxgen primers (anxgenF/anxgenR) and the thermal cycler program was set at 94°C for 5 min, followed by 30 cycles of amplification: 30 s at 95°C, 30 s at 52°C, 30 s at 72°C, and final elongation for 7 min at 72°C. Furthermore, to improve the amplification signal of the anaC gene, the second PCR with purified PCR products from the first PCR was tested with the anaC-gen primers (anaC-genF/anaC-genR), and the thermal cycler was set at an initial denaturation for 5 min at 94°C, followed by 30 cycles of amplification: 30 s at 94°C, 30 s at 58°C, 30 s at 72°C, and final elongation for 7 min at 72°C. For DNA sequencing, all PCR products were purified using the QIAquick® Gel Extraction Kit (Hilden, Germany), and the sequences were compared and analyzed using the nucleotide basic local alignment search tool (BLAST).
Preliminary microscopic examinations of the three bloom samples collected in October 2018, August 2019, and September 2020, respectively, from Dal Lake showed the prevalence of three dominant cyanobacteria morphospecies (Figures 2d–f). The dominant morphospecies observed in the samples collected from Grand Palace Gath (S1) and Gagribal basin (S3) were found to be filamentous along with the presence of specialized heterocyte cells.
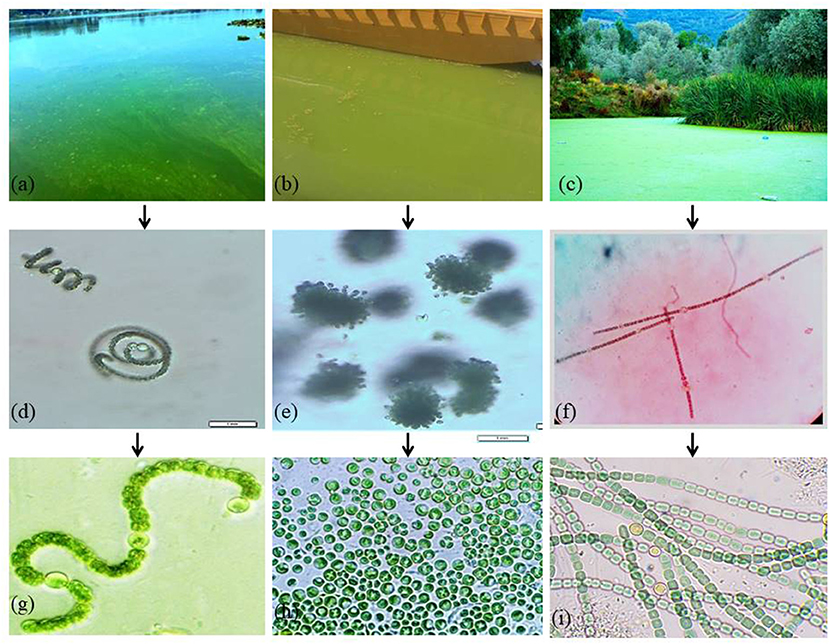
Figure 2. Cyanobacterial blooms at Dal Lake: (a) Grand Palace Gath (Site 1) (b) Nigeen basin (Site 2) and (c) Gagribal basin (Site 3). (d–f) Microscopic examination of dominant cyanobacterial species in natural bloom samples. Photos of isolated cyanobacterial strains: (g) Dolichospermum sp. strain AE02 (h) Microcystis sp. strain AE03, and (i) Dolichospermum sp. strain AE01.
Thus, representing typical characteristic features exhibited by members of the family Nostocaceae (Figures 2d,f). However, both the filamentous morphospecies differed markedly in the overall similitude of the filaments, owing to their multicellular nature that arises as a result of the linking pattern of individual cells imparting the filament its inimitable structure. Filaments of the morphospecies observed in the S1 (Figure 2d) were found to be coiled into a spiral architecture; individual vegetative cells appeared globose in shape with prominently differentiated heterocyte cells. However, the individual cells in the morphospecies observed in the S3 (Figure 2f) were cylindrically shaped vegetative cells with ovoid heterocyte linked in a way that manifests into more ordered, straight and unbranched filaments, thereby facilitating a marked contrast between the two morphospecies. The dominant morphospecies collected from the Nigeen basin (S2) showed spherical cells grouped in colony-forming behavior which are remarkable characteristics of the genus Microcystis (Figure 2e). These results indicate the morphologically different cyanobacterial species species existing at each site. Furthermore, molecular identification of the three isolates was confirmed using the 16S rDNA gene cluster as a molecular marker. PCR amplification of 16S rDNA sequencing, and BLAST analysis revealed that each isolate has the highest similarity with several strains of particular cyanobacteria strains. The nucleotide sequences of the three amplified products were submitted to the NCBI GenBank (Supplementary Table S1). Isolate 1, collected from Grand Palace Gath (S1) showed the highest similarity with several strains of Dolichospermum, and hence the isolate was designated as Dolichospermum sp. strain AE02 (Accession No. MW828342). Isolate 2 purified from Nigeen basin (S2) showed the highest homology with Microcystis and hence typed as Microcystis sp. strain AE03 (Genbank Accession No. MW856452), and Isolate 3 from Gagribal basin (S3) as Dolichospermum sp. strain AE01 (Accession No. MW807366) (Figures 2g–i).
The nucleotide sequences obtained with the best BLAST hit results against the querying 16S rDNA sequence were retrieved from the NCBI database and were subjected to phylogenetic analysis using the MEGA-X software. The Jukes-Cantor model was chosen to be the most accurate evolutionary model for carrying out such an analysis.
Phylogenetic tree construction using the 16S rDNA sequence of Dolichospermum sp. strain AE02 (Accession No. MW828342 and 505 bp) revealed its close similarity with the various strains of Dolichospermum, in particular, with the D. flos-aquae strain OF8 (Accession No. JQ894511 and 597 bp) and thus clustered together into a single clade (Figure 3A). The strain “OF8” was isolated from crude oil/hydrocarbon contaminated soil at Noonmati Refinery, Guwahati, India suggesting its potential role in bioremediation of polluted sediments (Akoijam et al., 2015). Similarly, AE02 strain, an isolate from a bloom water sample, concurrently polluted with chemical fertilizers discharged into the Dal Lake, Kashmir, India shows its adaptability to survive in such a hostile environment. Thus, both strains display their shared ability to cope and thrive in contaminated habitats. The targeted 16S rDNA gene cluster of Microcystis sp. strain AE03 (Accession No. MW856452) yielded a PCR amplicon of size 591 bp length.
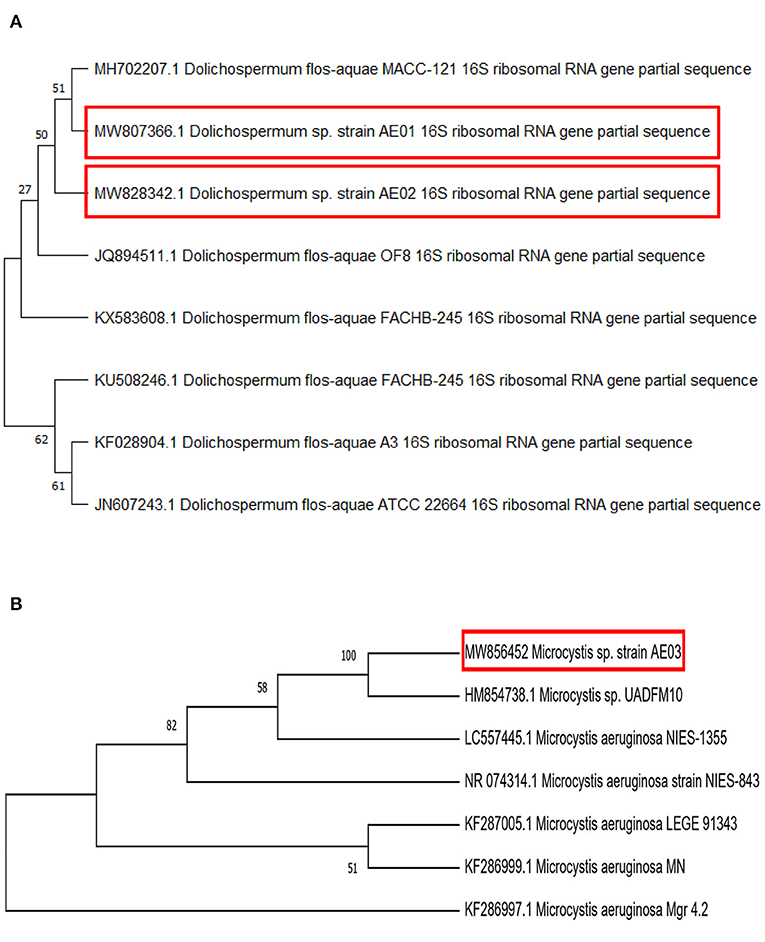
Figure 3. A Cladogram, using the Neighbor-Joining method of selected 16S rDNA nucleotide sequences of (A) Dolichospermum sp. strain AE02, and AE01 (B) Microcystis sp. strain AE03. Bootstrap replicates 500, constructed with the MEGA X program. The evolutionary distances were computed using the Jukes-Cantor method.
This strain AE03 showed its similarity principally with the Microcystis sp. strain UADFM10 (Accession No. HM854738 and 710 bp) and other members of the genus Microcystis from the phylogenetic relationship analysis (Figure 3B). The strain “UADFM10” is an isolate from Vela Lake (Western Central Portugal) (De Figueiredo et al., 2009). Both strains are lake-dwelling and possess toxic potential. The 16S rDNA nucleotide sequence of the third isolate, Dolichospermum sp. strain AE01 (Accession No. MW807366 and 591 bp), was phylogenetically analyzed and found to have the highest level of identity with Dolichospermum flos-aquae MACC-121 (Accession No. MH702207 and 1,311 bp) (Makra et al., 2019) (Figure 3A). Thus, a total of three cyanobacterial strains were isolated, identified, and characterized from the surface bloom water samples from Dal Lake (India).
Microcystin-, cylindrospermopsin-, and anatoxin-producing potential of the three isolated cyanobacteria strains were analyzed by performing PCR. Specific primers targeting cyanotoxin genes biosynthesis were used. A total of four different primer sets (Table 1) were selected, which included 4 primer sets anxgenF/anxgenR and anaC-genF/anaC-genR for anaC gene (anatoxin-a synthetase); 2 primer sets FAA/RAA for mcyB gene (microcystins synthetase) and 2 primer sets pksK18/pksM4 for pks gene (cylindrospermopsin synthetase). For prominent, specific, and reproducible amplification of target genes, PCR reactions were repeated thrice. The PCR results indicated that out of four primer sets used, two of the primer pairs; FAA/RAA and pksK18/pksM4 showed successful amplified products of the expected size range (758 and 422 bp, respectively) for Microcystis sp. strain AE03, confirming the presence of mcyB and pks genes in its genome (Figures 4B,C). However, no amplification was observed for anaC gene using the extracted genomic DNA of Microcystis sp. strain AE03 as a template. For Dolichospermum sp. strain AE02, only the primer pair, anaC-genF/anaC-genR and anxgenF/anxgenR confirmed the presence of the targeted anaC gene with an expected product size of 366 bp (Figure 4A), while no amplifications were observed for mcyB and pks genes (Figures 4B,C). Unlike the other two isolates, Dolichospermum sp. strain AE01 showed no amplification for any of the targeted toxin genes (Figures 4A–C). Thus, in our present study, PCR-based screening using cyanotoxin-specific primers indicated the potential toxigenic nature of Microcystis sp. strain AE03 and Dolichospermum sp. strain AE02 for harboring cyanotoxin-producing gene(s).
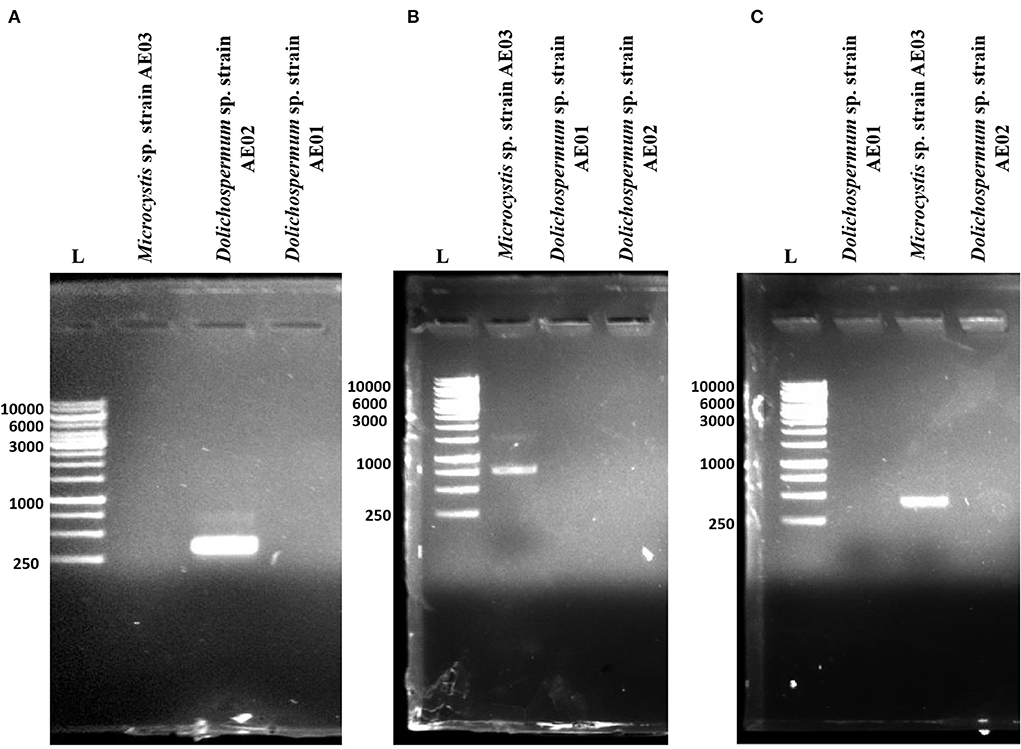
Figure 4. PCR amplification of targeted toxin genes responsible for (A) anatoxin (anaC), (B) microcystin (mcyB), and (C) cylindrospermopsin (polyketide synthetase, pks) production in the isolated cyanobacterial strains from Dal Lake, Kashmir. L: 1kb DNA Ladder.
The nucleotide sequences of the three amplified products were submitted to GenBank under accession numbers MW874625 for mcyB gene and MW874626 for pks gene present in Microcystis sp. strain AE03, and MW874627 for anaC gene present in Dolichospermum sp. strain AE02 (Supplementary Table S1). Identification of mcyB gene in the Microcystis sp. strain AE03 was based on comparison with mcyB gene of Microcystis aeruginosa strain CHAB437 (Accession No. KJ818140) with the percentage identity of 99.58% while the sequence of the pks gene shows 94.48% identity with pks clone from a toxic environmental cyanobacterial bloom sample (Accession No. EF157681). The sequence of the anaC gene in the isolated Dolichospermum sp. strain AE02 showed 86.85% identity with the sequence of the anaC gene of the Oscillatoria sp. strain PCC 10601 (Accession No. JF803652).
This study was aimed to set up a simple and reliable method to easily and simultaneously detect cyanotoxin-producing strains in natural bloom samples of Dal Lake. We observed that dense cyanobacterial blooms occur in Dal Lake annually, more specifically, we observed these blooms in October 2018, August 2019, and September 2020 (Figures 2a–c). Three dominant cyanobacterial strains Dolichospermum sp. strain AE01 (Accession No. MW807366), Dolichospermum sp. strain AE02 (Accession No. MW828342), and Microcystis sp. strain AE03 (Accession No. MW856452) were identified during these three bloom periods. The 16S rDNA sequence analysis of these three strains revealed a high degree of similarity (>99% sequence identity) among them and the reference strain Dolichospermum flos-aquae MACC-121 (Accession No. MH702207 and 1,311 bp), Dolichospermum flos-aquae strain OF8 (Accession No. JQ894511), and Microcystis sp. strain UADFM10 (Accession No. HM854738), respectively (Figure 3).
Interestingly, these three dominant strains did not co-exist during the same bloom episode and at the same site in Dal Lake. The Microcystis sp. AE03 strain was dominant in the Nigeen basin (S2) during October 2018, Dolichospermum sp. AE02 strain was dominant in the Grand Palace Gath (S1) during August 2019, and Dolichospermum sp. AE01 strain was dominant in the Gagribal basin (S3) during September 2020. Therefore, one would suggest that the emergence of different cyanobacterial strains could depend on abiotic factors such as nutrient concentrations in the water bodies that favor their growth, distribution, and potential for toxigenicity. A country-wide cyanobacterial bloom evaluation for Indian inland waters was recently carried out in 2018, highlighting the geographical distribution of blooms across the country. The findings revealed that blooms are dominant during the post-monsoon season (September–October) when nutrient concentrations in the water bodies are at their peak, and begin to decline as winter approaches (November–December) (Maniyar et al., 2022). Recently, (Kaloo and Amin, 2020) reported that the major cyanobacterial genera present in the Dal Lake are Microcystis, Merismopedia, Anabaena, Nodularia, Glalotrichia, Oscillatoria, Anacystis, Gomphosphaeria, Nostoc, Chroococcus, Arthrospira, and Aphonacapsa. Among the cyanobacterial genera-producing cyanotoxins, Microcystis is often observed as the dominant one in various surface waters across India (Kundangar, 1999; Singh et al., 2015; Mohan et al., 2020; Gurao and Sangolkar, 2022; Kesari et al., 2022). Microcystis was the most frequently encountered genus in Central America, tropical Africa, and Asia (Svirčev et al., 2019; Kaloudis et al., 2022; Van Hassel et al., 2022).
The isolated strains from Dal Lake have cyanotoxin biosynthetic gene clusters that have been linked to geographical distribution and toxicity worldwide (Diez-Quijada et al., 2022; Svirčev et al., 2022; Zhang et al., 2022). Microcystis spp. are also known as a microcystins-producing species, and Dolichospermum spp. are known as anatoxins-, cylindrospermopsins-, microcystins-, and saxitoxins-producing species (Huisman et al., 2018). The PCR amplification of the N-methyl transferase (NMT) domain of the microcystin synthetase gene mcyB, anatoxin-a synthetase gene anaC, and cylindrospermopsin synthetase gene pks gene (Figure 4) showed positive PCR amplification for both mcyB and pks genes for the Microcystis sp. AE03 strain (Figures 4B,C). The PCR amplification was positive only for anaC gene harboring in the genome of filamentous species Dolichospermum sp. AE02 strain (Figure 4A). These results indicate the toxic potential of these two isolated cyanobacterial species. However, for the other filamentous strain Dolichospermum sp. AE01 strain none of the three genes were detected (Figures 4A–C). Therefore, for the first time we detected cylindrospermopsin synthetase gene pks co-existing with the microcystin synthetase gene mcyB in an isolated Microcystis strain. The identification of this strain capable of producing two cyanotoxins shows the evolutionary changes taking place in cyanobacteria. This change could be speculated as an adaptive process to sustain their living in many water bodies across the globe. The water samples representative of the surface blooms collected from Grand Palace Gath (S1) and Nigeen basin (S2) serve as suitable microhabitats for the growth of Dolichospermum sp. strain AE02 and Microcystis sp. strain AE03 (toxin-producing cyanobacteria identified), respectively as evident from PCR based screening and thus, possibly causing CHABs. Yet, the water sample from the Gagribal basin (S3) did not comprised of any dominantly occurring toxicogenic cyanobacterium despite being collected from the bloom site. This probably indicates that some other uncultured species responsible for the observed blooms could not sustain themselves in the BG-11-defined medium. To date, ATX-a, MC, and CYN was reported in several countries worldwide in both lakes and rivers (Ballot et al., 2020; Bauer et al., 2020; Jones et al., 2021; Kaloudis et al., 2022; Van Hassel et al., 2022; You et al., 2022).
In India, the presence of MC has already been reported in several lakes (Srivastava et al., 2016; Dixit et al., 2017; Mohan et al., 2020; Kesari et al., 2022). However, to our undertsanding, this is the first time, we detected anatoxin-a synthetase gene (anaC) and cylindrospermopsin synthetase gene (pks) in Dal Lake, Kashmir (India).
Microcystins (MCs) are among the most diverse cyanotoxins found in water bodies around the world. The “mcyB” gene is part of the MC biosynthetic gene cluster encoding a “peptide synthetase” involved in MC biosynthesis. The peptide synthetase contains two modules with each module performing several processing functions such as, adenylation, thiolation, and condensation (Neilan et al., 1999; Tillett et al., 2000). The encoded products are low molecular weight cyclic heptapeptides known for their toxic effects on liver and are potent inhibitors of eukaryotic protein phosphatases 1 and 2A. These toxins are frequently responsible for the deaths of wild animals and livestock worldwide. The cytotoxic guanidine alkaloid, CYN is hepatotoxic, neurotoxic, carcinogenic, and general cytotoxic in nature. This can be attributed with its ability to inhibit glutathione and protein synthesis as well as cytochrome P450 inhibition (Runnegar et al., 1994, 1995, 2002; Froscio et al., 2003). Subsequently, several studies have shown that this toxin can also be produced by other different species belonging to the genera Umezakia, Aphanizomenon, Dolichospermum, Lyngbya, and Oscillotoria; excluding the genus Microcystis (Adamski et al., 2020). According to several investigations, a single cyanobacterial strain can produce multiple variants of the same class of cyanotoxins, such as Microcystis CAWBG11 that has at least 27 identified microcystin variants (Puddick et al., 2014), Raphidiopsis raciborskii with 3 cylindrospermopsin congeners (Wimmer et al., 2014), and a Raphidiopsis mediterranea strain LBRI48 producing simultaneously anatoxin-a, homoanatoxin-a and 4-hydroxyhomoanatoxin-a (Namikoshi et al., 2003). We have also identified Dolichospermum strain AE02 strain is capable of synthesizing anatoxin (anaC). It act as a powerful agonist of the acetylcholine receptor leading to sustained excitation at the neuromuscular junction, leading to depolarization of the cells, opening of the voltage-sensitive Ca2+ and Na+ channels that contributes to muscle paralysis and death by asphyxiation (Méjean et al., 2009).
This study revealed the presence of toxigenic CHABs in Dal Lake, Kashmir along with their potential cyanotoxin-producing genes (ana, pks, and mcy) in their genome. The present study reports the co-occurrence of the microcystin synthetase (mcyB), and cylindrospermopsin synthetase (pks) genes in a single strain of Microcystis isolated from this lake. The co-occurrence of more than one cyanotoxin at the sampling sites used for agriculture irrigation, recreation, and drinking water highlights its detrimental effect on aquatic life and human health as well. Keeping in view the present conditions of Dal Lake, further investigations are required with a special emphasis on the mitigation of CHABs through abiotic control mechanism.
For the first time, we took an initiative to study the cyanobacterial strains in Dal Lake. We were able to explore some of the dominant cyanobacteria species. In addition, we also explored their toxic nature by screening the cyanobacterial biosynthetic gene clusters. However, the work can be extended by considering the other area of the Lake to explore the other toxic cyanobacterial species. The monoculture cyanobacterium could be studied in vivo more extensively by studying the effect of various abiotic factors like P and N, change in temperature and light intensity, etc. on the expression of cyanotoxin biosynthetic gene clusters at transcription (RNA next-generation sequencing/transcriptome) and translation level (Liquid chromatography-mass spectrometry). The concentration of these secreted toxins could be quantified by using the techniques like enzyme-linked immunosorbent assay. Different abiotic factors are responsible for the emergence of harmful cyanobacterial blooms, such as nutrient influx, wind speed, sediment deposition, slow water flow, increased salinity, fluctuation in temperature, and global warming. These abiotic factors can be controlled to a minimum level to eradicate or reduce the bloom incidence. However, the methods applied for bloom suppression should be biologically sustainable without affecting the aquatic environment. Multiple strategies have been proposed such as chemical, physical, biological, and other approaches for alleviating the harmful cyanobacterial bloom incidences. The implementation of these strategies could possibly help us to reduce the cyanobacterial burden in Dal Lake.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
FB: investigation and article writing. NB: draft editing and logical inputs. VDR, TM, and WHA: technical inputs. KMF: logical inputs and lab accessibility. AB: idea and draft editing. BAG: concept designing and logical inputs. YN and SA: valuable feedback for providing some logical inputs for the isolation of cyanobacterial species. All the authors contributed to the article and approved the submitted version.
All reagents were bought personally and some were kindly provided by the Department of Environmental Science, University of Kashmir. Instrumentation access was kindly provided by Dr. Fazili's UPR Signaling Laboratory, Department of Biotechnology, University of Kashmir. WHA acknowledges the Deanship of Scientific Research at Umm Al Qura University, Makkah, Saudi Arabia, through Grant Code (Project Code: 22UQU4310387DSR28). VDR and TM acknowledge the support by the Laboratory Soil Health of the Southern Federal University with the financial support of the Ministry of Science and Higher Education of the Russian Federation, Agreement No. 075-15-2022-1122.
The authors would like to thank the Deanship of Scientific Research at Umm Al-Qura University for supporting this work by Grant code (22UQU4310387DSR28).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fsufs.2022.1036111/full#supplementary-material
Adamski, M., Wołowski, K., Kaminski, A., and Hindáková, A. (2020). Cyanotoxin cylindrospermopsin producers and the catalytic decomposition process: a review. Harmful Algae 98, 101894. doi: 10.1016/j.hal.2020.101894
Ahmad, T., Gupta, G., Sharma, A., Kaur, B., Alsahli, A. A., and Ahmad, P. (2020). Multivariate statistical approach to study spatiotemporal variations in water quality of a himalayan urban fresh water lake. Water 12, 2365.
Akoijam, C., Langpoklakpam, J. S., Chettri, B., and Singh, A. K. (2015). Cyanobacterial diversity in hydrocarbon-polluted sediments and their possible role in bioremediation. Intll. Biodeterior. Biodegr. 103, 97–104. doi: 10.1016/j.ibiod.2015.03.035
Ballot, A., Swe, T., Mjelde, M., Cerasino, L., Hostyeva, V., and Miles, C. O. (2020). Cylindrospermopsin-and deoxycylindrospermopsin-producing Raphidiopsis raciborskii and microcystin-producing Microcystis spp. in Meiktila Lake, Myanmar. Toxins 12, 232.
Bauer, F., Fastner, J., Bartha-Dima, B., Breuer, W., Falkenau, A., Mayer, C., et al. (2020). Mass occurrence of anatoxin-a-and dihydroanatoxin-a-producing Tychonema sp. in mesotrophic reservoir Mandichosee (River Lech, Germany) as a cause of neurotoxicosis in dogs. Toxins 12, 726.
Bernard, C., Ballot, A., Thomazeau, S., Maloufi, S., Furey, A., Mankiewicz-Boczek, J., et al. (2017). “Cyanobacteria associated with the production of cyanotoxins,” in Handbook on Cyanobacterial Monitoring and Cyanotoxin Analysis, eds J. Meriluoto, L. Spoof, G. A. Codd (Wiley), 501–525. Available online at: http://hdl.handle.net/10449/36453
Bouaïcha, N., Miles, C. O., Beach, D. G., Labidi, Z., Djabri, A., Benayache, N. Y., et al. (2019). Structural diversity, characterization and toxicology of microcystins. Toxins 11, 714. doi: 10.3390/toxins11120714
Chapman, A., and Foss, A. (2019). GreenWater Laboratories Potentially Toxigenic (PTOX) Cyanobacteria List. Florida: Greenwater Laboratories. doi: 10.13140/RG.2.2.32214.50243/1
Chorus, I., Fastner, J., and Welker, M. (2021). Cyanobacteria and cyanotoxins in a changing environment: concepts, controversies, challenges. Water 13, 2463. doi: 10.3390/w13182463
Christensen, V. G., and Khan, E. (2020). Freshwater neurotoxins and concerns for human, animal, and ecosystem health: a review of anatoxin-a and saxitoxin. Sci. Total Environ. 736, 139515. doi: 10.1016/j.scitotenv.2020.139515
Colas, S., Marie, B., Lance, E., Quiblier, C., Tricoire-Leignel, H., and Mattei, C. (2021). Anatoxin-a: overview on a harmful cyanobacterial neurotoxin from the environmental scale to the molecular target. Environ. Res. 193, 110590. doi: 10.1016/j.envres.2020.110590
De Figueiredo, D. R., Alves, A., Pereira, M. J., and Correia, A. (2009). Molecular characterization of bloom-forming aphanizomenon strains isolated from Vela Lake (Western Central Portugal). J. Plankton. Res. 32, 239–252. doi: 10.1093/plankt/fbp111
Diez-Quijada, L., Puerto, M., Gutiérrez-Praena, D., Turkina, M. V., Campos, A., Vasconcelos, V., et al. (2022). In vitro toxicity evaluation of cyanotoxins cylindrospermopsin and microcystin-LR on human kidney HEK293 cells. Toxins 14, 429. doi: 10.3390/toxins14070429
Dixit, R. B., Patel, A. K., Toppo, K., and Nayaka, S. (2017). Emergence of toxic cyanobacterial species in the Ganga River, India, due to excessive nutrient loading. Ecol. Indic. 72, 420–427. doi: 10.1016/j.ecolind.2016.08.038
Fergusson, K. M., and Saint, C. P. (2003). Multiplex PCR assay for Cylindrospermopsis raciborskii and cylindrospermopsin-producing cyanobacteria. Environ Toxicol: An International Journal 18, 120–125. doi: 10.1002/tox.10108
Froscio, S. M., Humpage, A. R., Burcham, P. C., and Falconer, I. R. (2003). Cylindrospermopsin-induced protein synthesis inhibition and its dissociation from acute toxicity in mouse hepatocytes. Environ. Toxicol. Int. J. 18, 243–251. doi: 10.1002/tox.10121
Gobler, C. J., Burkholder, J. M., Davis, T. W., Harke, M. J., Johengen, T., Stow, C. A., et al. (2016). The dual role of nitrogen supply in controlling the growth and toxicity of cyanobacterial blooms. Harmful. Algae 54, 87–97. doi: 10.1016/j.hal.2016.01.010
Griffith, A. W., and Gobler, C. J. (2020). Harmful algal blooms: a climate change co-stressor in marine and freshwater ecosystems. Harmful. Algae 91, 101590. doi: 10.1016/j.hal.2019.03.008
Guiry, M. D., Guiry, G. M., and 2020 (2020). AlgaeBase. World-Wide Electronic Publication. Galway: National University of Ireland. Available online at: http://www.algaebase.org/ (accessed June 5, 2020).
Gurao, S. N., and Sangolkar, L. N. (2022). Seasonal variation of microcystis and microcystin in lakes at Nagpur (MS), India. Int. J. Algae 24, 129–144. doi: 10.1615/InterJAlgae.v24.i2.30
Harke, M. J., Davis, T. W., Watson, S. B., and Gobler, C. J. (2016). Nutrient-controlled niche differentiation of western Lake Erie cyanobacterial populations revealed via metatranscriptomic surveys. Environ. Sci. Technol. 50, 604–615. doi: 10.1021/acs.est.5b03931
Hauer, T., and Komárek, J. (2020). CyanoDB 2.0-On-line Database of Cyanobacterial Genera. World-wide Electronic Publication, University of South Bohemia and Institution of Botany, Academy of Sciences of the Czech Republic. Available online at: http://www.cyanodb.cz/ (accessed March 24, 2020).
Huang, I.-S., Pietrasiak, N., Gobler, C. J., Johansen, J. R., Burkholder, J. M., D'antonio, S., et al. (2020). Diversity of bioactive compound content across 71 genera of marine, freshwater, and terrestrial cyanobacteria. Harmful. Algae 109, 102116. doi: 10.1016/j.hal.2021.102116
Huang, I.-S., and Zimba, P. V. (2019). Cyanobacterial bioactive metabolites—a review of their chemistry and biology. Harmful. Algae 86, 139–209. doi: 10.1016/j.hal.2019.05.001
Huisman, J., Codd, G. A., Paerl, H. W., Ibelings, B. W., Verspagen, J. M., and Visser, P. M. (2018). Cyanobacterial blooms. Nat. Rev. Microbiol. 16, 471–483. doi: 10.1038/s41579-018-0040-1
Inaotombi, S., and Sarma, D. (2021). Factors influencing distribution patterns of cyanobacteria in an upland lake of the Kumaun Himalayas, India. Arch. Environ. Occup. Health 76, 123–133. doi: 10.1080/19338244.2020.1760190
Jones, M. R., Pinto, E., Torres, M. A., Dörr, F., Mazur-Marzec, H., Szubert, K., et al. (2021). CyanoMetDB, a comprehensive public database of secondary metabolites from cyanobacteria. Water Res. 196, 8. doi: 10.1016/j.watres.2021.117017
Kaloo, Z. A., and Amin, S. (2020). Algal diversity in Jammu and Kashmir state, Biodiversity of the Himalaya: Jammu and Kashmir State. Springer, 18 pp. 285–317. doi: 10.1007/978-981-32-9174-4_13
Kaloudis, T., Hiskia, A., and Triantis, T. M. (2022). Cyanotoxins in bloom: ever-increasing occurrence and global distribution of freshwater cyanotoxins from Planktic and Benthic cyanobacteria. Toxins 14, 264. doi: 10.3390/toxins14040264
Kesari, V., Kumar, S., Yadav, I., Chatterjee, A., Rai, S., and Pandey, S. (2022). Ganga river water quality assessment using combined approaches: physico-chemical parameters and cyanobacterial toxicity detection with special reference to microcystins and molecular characterization of microcystin synthetase (mcy) genes carrying cyanobacteria. Environ. Sci. Pollut. Res. 29, 13122–13140. doi: 10.1007/s11356-021-16589-1
Khan, R. M., Salehi, B., Mahdianpari, M., Mohammadimanesh, F., Mountrakis, G., and Quackenbush, L. J. (2021). A meta-analysis on harmful algal bloom (HAB) detection and monitoring: a remote sensing perspective. Remote Sens. 13, 4347. doi: 10.3390/rs13214347
Komárek, J. (2016). A polyphasic approach for the taxonomy of cyanobacteria: principles and applications. Eur. J. Phycol. 51, 346–353. doi: 10.1080/09670262.2016.1163738
Komárek, J. (2020). Quo vadis, taxonomy of cyanobacteria 2019. Fottea 20, 104–110. doi: 10.5507/fot.2019.020
Kumar, R., Parvaze, S., Huda, M. B., and Allaie, S. P. (2022). The changing water quality of lakes—a case study of Dal Lake, Kashmir Valley. Environ. Monit. Assess 194, 228. doi: 10.1007/s10661-022-09869-x
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Kundangar, M. (1999). “Is water quality at pokhribal-nigeen fit for potable purposes,” in 5th. Nat. Seminar on Fish and Their Env. (abst). Kashmir: PG Deptt. of zoology Univ. of Kashmir.
Makra, N., Gell, G., Juhász, A., Soós, V., Kiss, T., Molnár, Z., et al. (2019). Molecular taxonomic evaluation of Anabaena and Nostoc strains from the Mosonmagyaróvár algal culture collection. S. Afr. J. Bot. 124, 80–86. doi: 10.1016/j.sajb.2019.03.008
Maniyar, C. B., Kumar, A., and Mishra, D. R. (2022). Continuous and synoptic assessment of Indian inland waters for harmful algae blooms. Harmful. Algae 111, 102160. doi: 10.1016/j.hal.2021.102160
Massey, I. Y., Al Osman, M., and Yang, F. (2020). An overview on cyanobacterial blooms and toxins production: their occurrence and influencing factors. Toxin. Rev. 41, 326–346. doi: 10.1080/15569543.2020.1843060
Mehinto, A. C., Smith, J., Wenger, E., Stanton, B., Linville, R., Brooks, B. W., et al. (2021). Synthesis of ecotoxicological studies on cyanotoxins in freshwater habitats–evaluating the basis for developing thresholds protective of aquatic life in the United States. Sci. Total Environ. 795, 148864. doi: 10.1016/j.scitotenv.2021.148864
Méjean, A., Mann, S., Maldiney, T., Vassiliadis, G., Lequin, O., and Ploux, O. (2009). Evidence that biosynthesis of the neurotoxic alkaloids anatoxin-a and homoanatoxin-a in the cyanobacterium Oscillatoria PCC 6506 occurs on a modular polyketide synthase initiated by L-proline. J. Am. Chem. Soc. 131, 7512–7513. doi: 10.1021/ja9024353
Mishra, D. R., Kumar, A., Muduli, P. R., Equeenuddin, S. M., Rastogi, G., Acharyya, T., et al. (2020). Decline in phytoplankton biomass along indian coastal waters due to COVID-19 lockdown. Remote Sen. 12, 2584. doi: 10.3390/rs12162584
Mohan, R., Sathish, T., and Padmakumar, K. B. (2020). Occurrence of potentially toxic cyanobacteria Microcystis aeruginosa in aquatic ecosystems of central Kerala (south India). Ann. Limnologie-Int. J. Limnol EDP Sci. 18, 15. doi: 10.1051/limn/2020015
Namikoshi, M., Murakami, T., Watanabe, M. F., Oda, T., Yamada, J., Tsujimura, S., et al. (2003). Simultaneous production of homoanatoxin-a, anatoxin-a, and a new non-toxic 4-hydroxyhomoanatoxin-a by the cyanobacterium Raphidiopsis mediterranea Skuja. Toxicon 42, 533–538. doi: 10.1016/S0041-0101(03)00233-2
Narayana, S., Tapase, S., Thamke, V., Kodam, K., and Mohanraju, R. (2020). Primary screening for the toxicity of marine cyanobacteria Lyngbya bouillonii (Cyanophyceae: Oscillatoriales) recorded for the first time from Indian Ocean. Reg. Stud. Mar. Sci. 40, 101510. doi: 10.1016/j.rsma.2020.101510
Neilan, B. A., Dittmann, E., Rouhiainen, L., Bass, R. A., Schaub, V., Sivonen, K., et al. (1999). Nonribosomal peptide synthesis and toxigenicity of cyanobacteria. J. Bacteriol. 181, 4089–4097. doi: 10.1128/JB.181.13.4089-4097.1999
Nübel, U., Garcia-Pichel, F., and Muyzer, G. (1997). PCR primers to amplify 16S rRNA genes from cyanobacteria. Appl. Environ. Microbiol. 63, 3327–3332. doi: 10.1128/aem.63.8.3327-3332.1997
Padmakumar, K., Menon, N., and Sanjeevan, V. (2012). Is occurrence of harmful algal blooms in the exclusive economic zone of India on the rise? Int. J. Oceanogr. 2012, 263946. doi: 10.1155/2012/263946
Puddick, J., Prinsep, M. R., Wood, S. A., Kaufononga, S. A., Cary, S. C., and Hamilton, D. P. (2014). High levels of structural diversity observed in microcystins from Microcystis CAWBG11 and characterization of six new microcystin congeners. Mar. Drugs 12, 5372–5395. doi: 10.3390/md12115372
Qadir, J., and Singh, P. (2018). Land use/cover mapping and assessing the impact of solid waste on water quality of Dal Lake catchment using remote sensing and GIS (Srinagar, India). SN Appl. Sci. 1, 25. doi: 10.1007/s42452-018-0027-6
Rantala-Ylinen, A., Känä, S., Wang, H., Rouhiainen, L., Wahlsten, M., Rizzi, E., et al. (2011). Anatoxin-a synthetase gene cluster of the cyanobacterium Anabaena sp. strain 37 and molecular methods to detect potential producers. Appl. Environ. Microbiol. 77, 7271–7278. doi: 10.1128/AEM.06022-11
Rashid, I., Romshoo, S. A., Amin, M., Khanday, S. A., and Chauhan, P. (2017). Linking human-biophysical interactions with the trophic status of Dal Lake, Kashmir Himalaya, India. Limnologica 62, 84–96. doi: 10.1016/j.limno.2016.11.008
Ray, J. G., Santhakumaran, P., and Kookal, S. (2021). Phytoplankton communities of eutrophic freshwater bodies (Kerala, India) in relation to the physicochemical water quality parameters. Environ. Dev. Sustain. 23, 259–290. doi: 10.1007/s10668-019-00579-y
Runnegar, M. T., Kong, S.-M., Zhong, Y.-Z., Ge, J.-L., and Lu, S. C. (1994). The role of glutathione in the toxicity of a novel cyanobacterial alkaloid cylindrospermopsin in cultured rat hepatocytes. Biochem. Biophys. Res. Commun. 201, 235–241. doi: 10.1006/bbrc.1994.1694
Runnegar, M. T., Kong, S.-M., Zhong, Y.-Z., and Lu, S. C. (1995). Inhibition of reduced glutathione synthesis by cyanobacterial alkaloid cylindrospermopsin in cultured rat hepatocytes. Biochem. Pharmacol. 49, 219–225. doi: 10.1016/S0006-2952(94)00466-8
Runnegar, M. T., Xie, C., Snider, B. B., Wallace, G. A., Weinreb, S. M., and Kuhlenkamp, J. (2002). In vitro hepatotoxicity of the cyanobacterial alkaloid cylindrospermopsin and related synthetic analogues. Toxicol. Sci. 67, 81–87. doi: 10.1093/toxsci/67.1.81
Shafiq-Ur-Rehman (1998). A red bloom of Euglena shafiqii, a new species, in Dal Lake, Srinagar, Kashmir. Water Air Soil Pollut. 108, 69–82. doi: 10.1023/A:1005037531147
Singh, S., Rai, P. K., Chau, R., Ravi, A. K., Neilan, B. A., and Asthana, R. K. (2015). Temporal variations in microcystin-producing cells and microcystin concentrations in two fresh water ponds. Water Res. 69, 131–142. doi: 10.1016/j.watres.2014.11.015
Srivastava, A., Ko, S. R., Ahn, C. Y., Oh, H. M., Ravi, A. K., and Asthana, R. K. (2016). Microcystin biosynthesis and mcyA expression in geographically distinct microcystis strains under different nitrogen, phosphorus, and boron regimes. Biomed Res Int. 5985987, 10.
Svirčev, Z., Chen, L., Sántha, K., Drobac Backović, D., Šušak, S., Vulin, A., et al. (2022). A review and assessment of cyanobacterial toxins as cardiovascular health hazards. Arch. Toxicol. 23, 1–35. doi: 10.1007/s00204-022-03354-7
Svirčev, Z., Lalić, D., Savić, G. B., Tokodi, N., Backović, D. D., Chen, L., et al. (2019). Global geographical and historical overview of cyanotoxin distribution and cyanobacterial poisonings. Arch. Toxicol. 93, 2429–2481. doi: 10.1007/s00204-019-02524-4
Taton, A., Grubisic, S., Brambilla, E., De Wit, R., and Wilmotte, A. (2003). Cyanobacterial diversity in natural and artificial microbial mats of Lake Fryxell (McMurdo Dry Valleys, Antarctica): a morphological and molecular approach. Appl. Environ. Microbiol. 69, 5157–5169. doi: 10.1128/AEM.69.9.5157-5169.2003
Tillett, D., Dittmann, E., Erhard, M., Von Döhren, H., Börner, T., and Neilan, B. A. (2000). Structural organization of microcystin biosynthesis in Microcystis aeruginosa PCC7806: an integrated peptide–polyketide synthetase system. Chem. Biol. 7, 753–764. doi: 10.1016/S1074-5521(00)00021-1
Van Hassel, W. H. R., Andjelkovic, M., Durieu, B., Marroquin, V. A., Masquelier, J., Huybrechts, B., et al. (2022). A summer of cyanobacterial blooms in belgian waterbodies: microcystin quantification and molecular characterizations. Toxins 14, 61. doi: 10.3390/toxins14010061
Wimmer, K. M., Strangman, W. K., and Wright, J. L. (2014). 7-Deoxy-desulfo-cylindrospermopsin and 7-deoxy-desulfo-12-acetylcylindrospermopsin: two new cylindrospermopsin analogs isolated from a Thai strain of Cylindrospermopsis raciborskii. Harmful. Algae 37, 203–206. doi: 10.1016/j.hal.2014.06.006
Xu, H., Paerl, H., Qin, B., Zhu, G., Hall, N., and Wu, Y. (2015). Determining critical nutrient thresholds needed to control harmful cyanobacterial blooms in eutrophic Lake Taihu, China. Environ. Sci. Technol. 49, 1051–1059. doi: 10.1021/es503744q
You, L., Tong, X., Te, S. H., Tran, N. H., bte Sukarji, N. H., He, Y., et al. (2022). Multi-class secondary metabolites in cyanobacterial blooms from a tropical water body: Distribution patterns and real-time prediction. Water Res. 212, 118129.
Keywords: Dal Lake, Dolichospermum sp. strain AE01, Dolichospermum sp. strain AE02, Microcystis sp. strain AE03, anatoxin-a, microcystin, cylindrospermopsin
Citation: Bashir F, Bashir A, Rajput VD, Bouaïcha N, Fazili KM, Adhikari S, Negi Y, Minkina T, Almalki WH and Ganai BA (2022) Microcystis sp. AE03 strain in Dal Lake harbors cylindrospermopsin and microcystin synthetase gene cluster. Front. Sustain. Food Syst. 6:1036111. doi: 10.3389/fsufs.2022.1036111
Received: 03 September 2022; Accepted: 26 September 2022;
Published: 14 October 2022.
Edited by:
Riyaz Zafar Sayyed, PSGVP Mandal's ASC College, IndiaReviewed by:
Shalini Rai, Society for Higher Education & Practical Applications, IndiaCopyright © 2022 Bashir, Bashir, Rajput, Bouaïcha, Fazili, Adhikari, Negi, Minkina, Almalki and Ganai. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bashir Ahmad Ganai, YmJjZ2FuYWlAZ21haWwuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.