
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Sustain. Food Syst. , 24 November 2022
Sec. Agroecology and Ecosystem Services
Volume 6 - 2022 | https://doi.org/10.3389/fsufs.2022.1004373
Diet is a major factor in influencing the growth performance and the microbial community of lambs. This study aimed to investigate how diverse diets influence their growth performance and rumen microbiota. Ninety male lambs were randomly allocated into three groups in a completely randomized design with equal lambs: non-pelleted native grass hay (HA) as the control diet and pelleted native grass hay (GP) and pelleted native grass hay with concentrate (GPC) as experimental diets. The rumen fluid samples of the lambs in the HA, GP, and GPC groups were used to study rumen microbiota diversity through 16S rDNA high-throughput sequencing. In the present study, the final body weight, dry matter intake, and average daily gain differed significantly (p < 0.05) among the HA, GP, and GPC groups. Compared to the HA group, higher final body weight, dry matter intake, and average daily gain were found in the GP group. Similarly, better animal performance was observed in the GPC group than in the GP group. The principal coordinates analysis displayed that the composition of the rumen microbiota in the three groups was distinctly separated from each other. Bacteroidetes and Firmicutes were the dominant members of the community in the HA and GP groups, while Bacteroidetes, Firmicutes, and Proteobacteria became the predominant members in the GPC group. The comparison among these groups showed significant (p < 0.05) differences in Rikenellaceae_RC9_gut_group, Prevotella_1, Ruminococcaceae_NK4A214_group, and Succiniclasticum. These results suggest that the GP and GPC diets are more beneficial for growth performance than the HA diet and also indicate that the rumen microbiota varied in response to different feed types. In conclusion, these results could provide strategies to influence rumen microbiota for better growth and a healthier ecosystem on the Mongolian Plateau and lay the theoretical groundwork for feeding the pelleted native grass diet.
The Ujimqin sheep, inhabiting the Mongolian Plateau in China, have been selected by the unique environment and natural conditions for thousands of years. The animal is the critical variety of coarse wool sheep in China. Many anatomical and physiological traits enable Ujimqin lambs to produce sufficient wool and meat for consumers in harsh living conditions (Zeng et al., 2017).
Similar to other ruminants, the lambs' rumen microbiota comprises bacteria, protozoa, archaea, and fungi (Guo et al., 2020; Mizrahi et al., 2021), and the microbes provide protein, vitamins, and short-chain organic acids for the animal by an efficient bioconversion of carbohydrates, lignocellulose, and others (Xing et al., 2020; Bhujbal et al., 2022). Among this microbiome, bacteria are essential to the viability of the ruminant and make the highest contribution to digestion (Kim et al., 2015). Previously published studies revealed that animal performance and the health, compositions, and metabolism of rumen microbiota are strongly influenced by animal age, feed type, and feeding system (Zened et al., 2013; Trabi et al., 2019).
Traditionally, the Ujimqin lambs are stronger and fatter when grazing in summer and autumn on native grassland and thinner and weaker (even leading to death) when fed in winter and spring with hay as their only diet (Du et al., 2020). Pellets, total mixed rations, and supplements with concentrate diets have become common by increasing the dry matter intake (DMI) and the average daily gain (ADG) in sheep production systems. However, the total mixed ratio is usually compromised by the selection of animals (Sarker et al., 2019), which results in higher labor costs (Trabi et al., 2019). Consequently, pellets or supplements with concentrate diets have been widely applied because these feeding systems could reduce feed waste and make management easy (Masoudzadeh et al., 2020). Previous reports found that the pellet diets could reduce the bacterial richness in the rumen (McClements and Xiao, 2012), while others found that the pelleted-hay diets had a greater increase in bacterial richness (Ishaq et al., 2019).
Additionally, some studies indicated that high-concentrate diets could affect the composition of the rumen microbiota (Plaizier et al., 2012), and high-forage diets are beneficial for some microorganisms, such as those in the genus Firmicutes (Khafipour et al., 2009). Previous reports also found that the growth performance, carcass traits, and meat quality of lambs were significantly influenced by the native grass diet on the Mongolian Plateau (Du et al., 2019, 2022; Bu et al., 2021). However, the related research about the effects of non-pelleted native grass without or with concentrate or pellets on the ruminal microbiota is still limited.
Therefore, in the current study, we hypothesized that non-pelleted native grass hay without or with concentrate or pellet diets could directly affect the rumen microbiota and growth performance in lamb. This study aimed to reveal lamb's ruminal microbiota and growth performance in response to non-pelleted native grass hay without or with concentrate or pellet diets.
The animal study was approved by the College of Grassland, Resources, and Environment, Inner Mongolian Agricultural University, China. All experiments were performed following the Regulations for the Administration of Affairs Concerning Experimental Animals.
The feeding trial was carried out at the Lvye Grass-based Livestock Husbandry Development Co., Ltd., Xilin Hot, China, from August to November 2020. The experiment lasted 75 days, with a 15-day adaptation period for diet and environment and a 60-day data for sample collection. We selected the hay from the same grassland in the typical steppe to minimize the effects of the materials. Stipa gigantea L., and Leymus chinensis (Trin.) Tzvel. are the dominant species in this grassland, also including Lespedeza davurica (Laxm.) Schindl, Allium mongolicum Regel, Thalictrum petaloideum Linn., Bupleurum chinensis DC., Serratula centauroides Linn., Caragana microphylla Lam, and others. A total of 90 six-month-old male Ujimqin lambs with body weights (BW) of 26.83 ± 0.26 kg were selected for this experiment. All lambs were randomly assigned to 18 units (five lambs per unit) based on similar BW before morning feeding in a completely randomized design. Then, the 18 units were randomly assigned to the three treatments, with six units per treatment. Three diets were used in the experiment (Table 1). The lambs received non-pelleted native grass hay (HA) as the control diet and pelleted native grass hay (GP), and pelleted native grass hay with concentrate (GPC) as the experimental diets. Pellets were produced from native grass hay with a length of 2–3 cm through pellet machines (H.S. 508 Pellets Mill; Liyang Weifang Equipment Co Ltd., Liyang, China). The concentrate was purchased from a local company (Ruiyuan Agriculture and Animal Husbandry Co. Ltd., Xilinhot, China). The concentrate consisted of maize, soybean meal, distillers dried grains with solubles, and wheat bran in 25, 8, 4, and 3%, respectively. The feed was offered at 110% of their expected intake to evaluate voluntary intake. The diets, feeding, management, and analysis of chemical compositions followed the previously published study (Bu et al., 2021).
The amounts of feed offered and the residue were recorded daily in the experiment to calculate the voluntary feed intake and were expressed on a dry matter (DM) basis. All lambs were weighed in the morning (06:00–07:00 h) before the morning feeding with an empty stomach and without fasting throughout the experimental period. At intervals of 7 days, the initial BW and the final BW were also recorded. The growth performance was analyzed based on the difference between the final BW and the initial BW (Du et al., 2022). At the end of the experiment, all of the lambs were transferred and slaughtered at a commercial slaughterhouse (Xilin Hot, China). Rumen samples were collected from all lambs, and the rumen content sample of each lamb in the same pen was first homogenized, and then, the same volume of rumen contents was mixed to reduce localized effects and strained through four layers of cheesecloth. At last, ~40 ml of rumen fluid was collected in the sterilized polypropylene centrifuge tube. Therefore, a total of 18 rumen fluid samples were immediately frozen in liquid nitrogen and then stored at −80°C for further analysis (Wang et al., 2019).
The E.Z.N.A. ®Stool DNAKit (D4015, Omega, Inc., USA) was used to extract the DNA of rumen samples following the manufacturer's instructions. The NanoDrop 2000 UV-vis Spectrophotometer (Thermo Scientific, Wilmington, USA) was used to determine the concentration and purity of the extracted DNA, and 1% agarose gel electrophoresis was used to assess the extracted DNA quality (Bai et al., 2020).
The polymerase chain reaction (PCR) amplification and bioinformatics analysis were performed at LC-Bio Technology Co., Ltd. (Hang Zhou, China). The variable regions V3–V4 of the bacterial16S rDNA gene were amplified with slightly modified versions of primers 341F (5′-CCTACGGGNGGCWGCAG-3′) and 805R (5′-GACTACHVGGGTATCTAATCC-3′) (Logue et al., 2016). The 16S amplification was conducted following the description of the previous report (Tanja and Salzberg, 2011). Sequences have been deposited into the Sequence Read Archive (SRA) with the BioProject accession number PRJNA825331.
Paired-end reads were assigned to the samples based on their unique barcode and truncated by cutting off the barcode and primer sequence and merged with FLASH (v1.2.8) (Tanja and Salzberg, 2011). Operational taxonomic units (OTUs) with a 97% similarity cutoff (Liu et al., 2019) were clustered using UPARSE (version 7.1, http://drive5.com/uparse/), and the high-quality sequences with ≥97% similarity were assigned to the same operational taxonomic units (OTUs) (Rognes et al., 2016). The OTUs were classified using the SILVA database (https://www.arbsilva.de/) with a confidence threshold of 70%. The false discovery rate (FDR)-adjusted Kruskal–Wallis multiple comparisons (p < 0.05) were used to detect the bacterial community structure and analyze at the phylum and genus levels (Omontese et al., 2022). A permutational multivariate analysis of variance (PERMANOVA) test was used to test the significant difference (Chambers and Hastie, 1992) in R (version 2.5.4). Principal coordinates analysis (PCoA) was conducted on the OTU level by R (version 3.3.1) software based on weighted UniFrac distances, and the ellipses represent the confidence in the fitting. The common and unique OTUs were used to construct the Venn diagram by R (version 1.6.2). The alpha diversity for these samples used the Chao1 value and the Shannon index to evaluate richness and diversity (Muwonge et al., 2021), and Good's coverage was calculated through QIIME software. The main differentially abundant genera were analyzed by the linear discrimination analysis coupled with the effect size (LEFSe) method (Segata et al., 2011). The PICRUSt was applied to predict metabolic genes based on the 16S rRNA data. The Kyoto Encyclopedia of Genes and Genomes (KEGG) database was also used to assign the genes to the metabolic pathways (Su et al., 2021). The heatmap package of R (R Core Team, 2014) was applied to generate heat maps of genera and Level 3-predicted microbial gene functions. Bar plots were generated in GraphPad Prism 7 (San Diego, CA, United States).
The ADI and DMI data were analyzed using SAS software (version 9.0) (SAS Inst., Inc., Cary, NC) as a completely randomized design per unit used as experimental units. Data were checked for normality using the PROC UNIVARIATE procedure of SAS (SAS Inst., Inc., Cary, NC). The ADI and DMI data were calculated from measurements collected throughout the study and analyzed as repeated measures using the PROC MIXED procedure of SAS (SAS Inst., Inc., Cary, NC). Significant differences among treatments were analyzed using a one-way analysis of variance according to the statistical model, Y = μ + α + ε, where Y = observation, μ = overall mean, α = diet effect, and ε = error, and Duncan's tests, with a p < 0.05 as statistical significance using SAS ver. 9.0.
The chemical composition of the diet used in this study is shown in Table 1. The DM contents in the HA, GP, and GPC groups were 93.90, 91.30, and 92.94%, respectively. The organic matter and the ether extract in the three groups were similar, while the crude protein and metabolizable energy in the GPC group were higher than those in the HA and GP groups, and the acid detergent fiber and neutral detergent fiber contents were lower than those in the HA and GP groups. The contents and composition of the fatty acids in the diet were various.
The results of the intake and growth performance of lambs fed different diets are presented in Table 2. The influence of feed types on the growth performance of lambs was studied, and it revealed that the feed types could significantly affect the DMI and ADG. As expected, no significant (p > 0.05) difference was found among the HA, GP, and GPC groups on the initial BW. The final BW, DMI, and ADG differed significantly (p < 0.05) in the HA, GP, and GPC groups. Compared to the final BW (30.00 and 31.75 kg), DMI (1.53 and 1.58 kg), and ADG (52.08 and 81.25 g/d) found for the HA and GP groups, the final BW (34.13 kg), DMI (1.67 kg), and ADG (122.92 g/d) were highest in the GPC group.
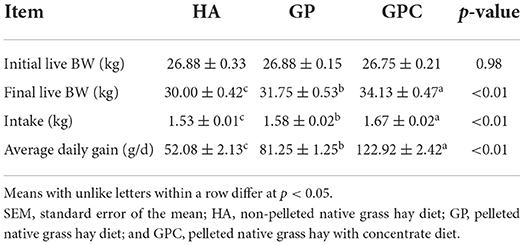
Table 2. Intake and growth performance of lambs fed non-pelleted native grass hay without or with concentrate or pellets.
A total of 18 rumen fluid samples from the lambs in the HA, GP, and GPC groups were used to evaluate the rumen microbiota diversity through 16S rDNA sequencing. The present study analyzed the differences in ruminal bacterial diversity among the HA, GP, and GPC groups, and the alpha and beta diversity were also calculated. A total of 1,520,890 raw reads were obtained for 18 rumen samples, with an average of 84,494 sequences for each rumen sample, and overall, 1,347,069 valid reads were obtained for 18 rumen samples, with an average of 74,837 sequences for each rumen sample and the sequencing depth at 20,000 (Table 1). According to the sampling depth at which rarefaction curves tend to plateau, all samples were rarefied to OTU numbers 1,338 (HA group), 1,134 (GP group), and 1,020 (GPC group) with ≥97% similarity identified (Table 3), and no statistically significant (p > 0.05) differences were observed on OTU number among the three groups. Similarly, the Good's coverage index was higher than 99% for all rumen samples, indicating that the sequencing depth was adequate to represent each ruminal bacterial community. No significant (p > 0.05) differences were observed among the three groups. However, significant (p < 0.05) differences were observed in Shannon indices, which indicate higher degree of species richness in the HA group and lower degree of species richness in the GP and GPC groups (Table 3).
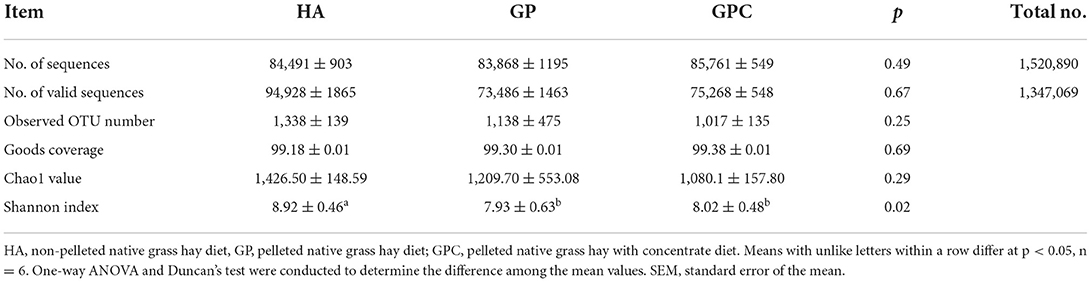
Table 3. Diversity indices of ruminal microbiome of lambs fed non-pelleted native grass hay without or with concentrate or pellets.
The effect of feed types on the OTUs analysis is demonstrated in Figure 1. The Venn diagram showed the common and unique OTUs in the three groups. In addition, the principal coordinate analysis (PCoA) profile displayed that the structure of the rumen microbiota in groups was distinctly separated from each other (Figure 2). The samples in the HA, GP, and GPC groups were clustered together, and no interaction was found between the three groups.
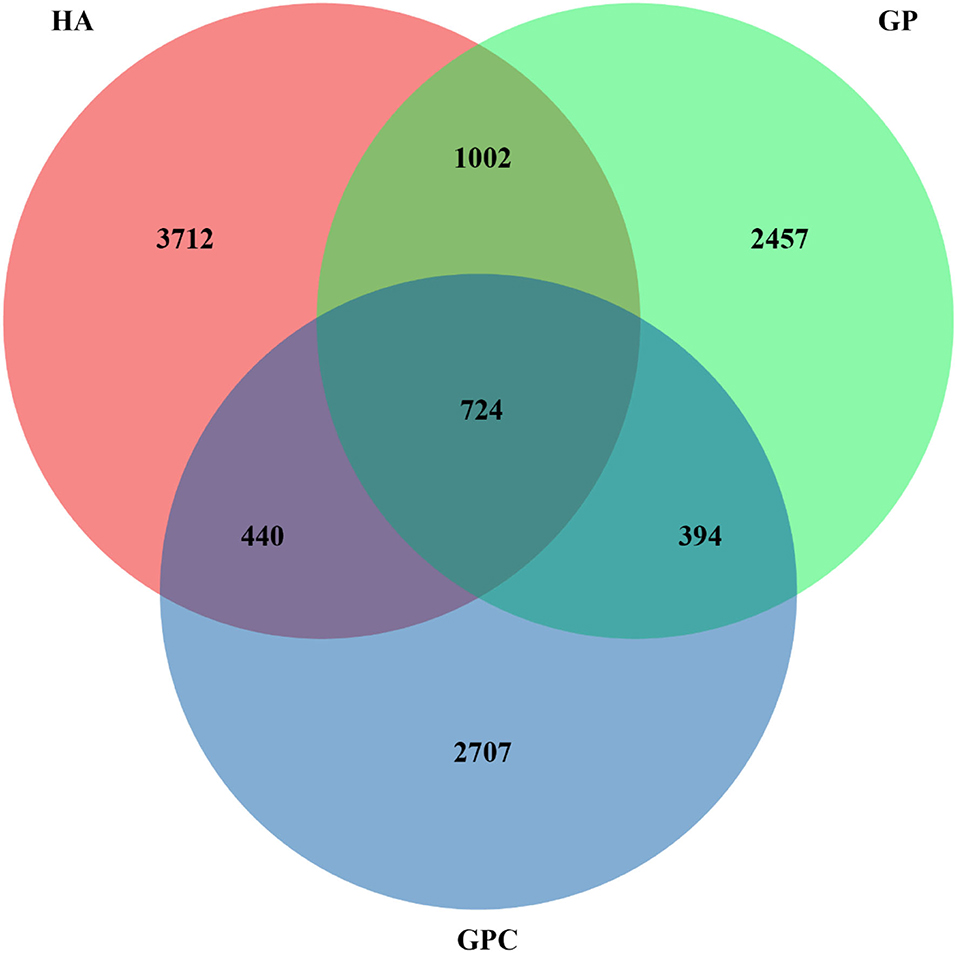
Figure 1. Venn diagram representing the common and unique OTUs found at each diet (n = 6). HA, non-pelleted native grass hay diet, GP, pelleted native grass hay diet; and GPC, pelleted native grass hay with concentrate diet.
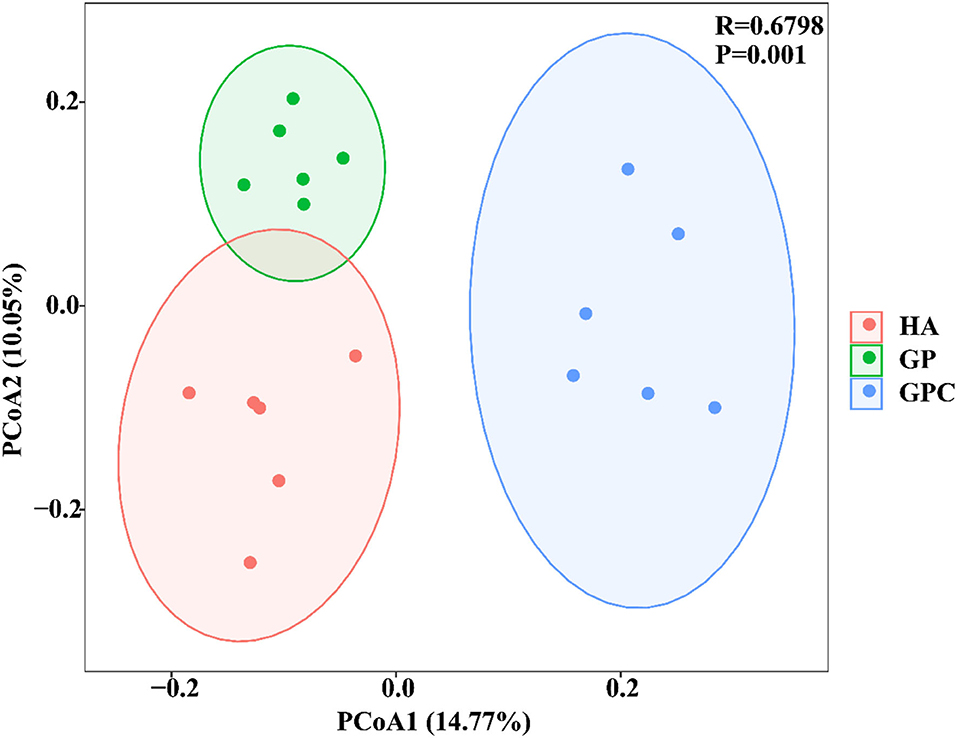
Figure 2. Principal coordinate analysis (PCoA) of rumen microbial communities based on unweighted UniFrac distance (n = 6). HA, non-pelleted native grass hay diet, GP, pelleted native grass hay diet; and GPC, pelleted native grass hay with concentrate diet.
The influence of feed types on the relative abundance of taxa at the phylum and genus levels is shown in Figure 3. Taxonomic analysis of the reads classified them into 425 genera belonging to 26 phyla. Seven phyla were referred to as the detected bacterial phyla at the phylum level. At the phylum level, Bacteroidetes and Firmicutes were the predominant phyla in the HA and GP groups, while Bacteroidetes, Firmicutes, and Proteobacteria were the predominant phyla in the GPC group (Figure 3A). There were significant (p < 0.05) differences in the abundance of Firmicutes and Proteobacteria among the three groups (Figure 3B).
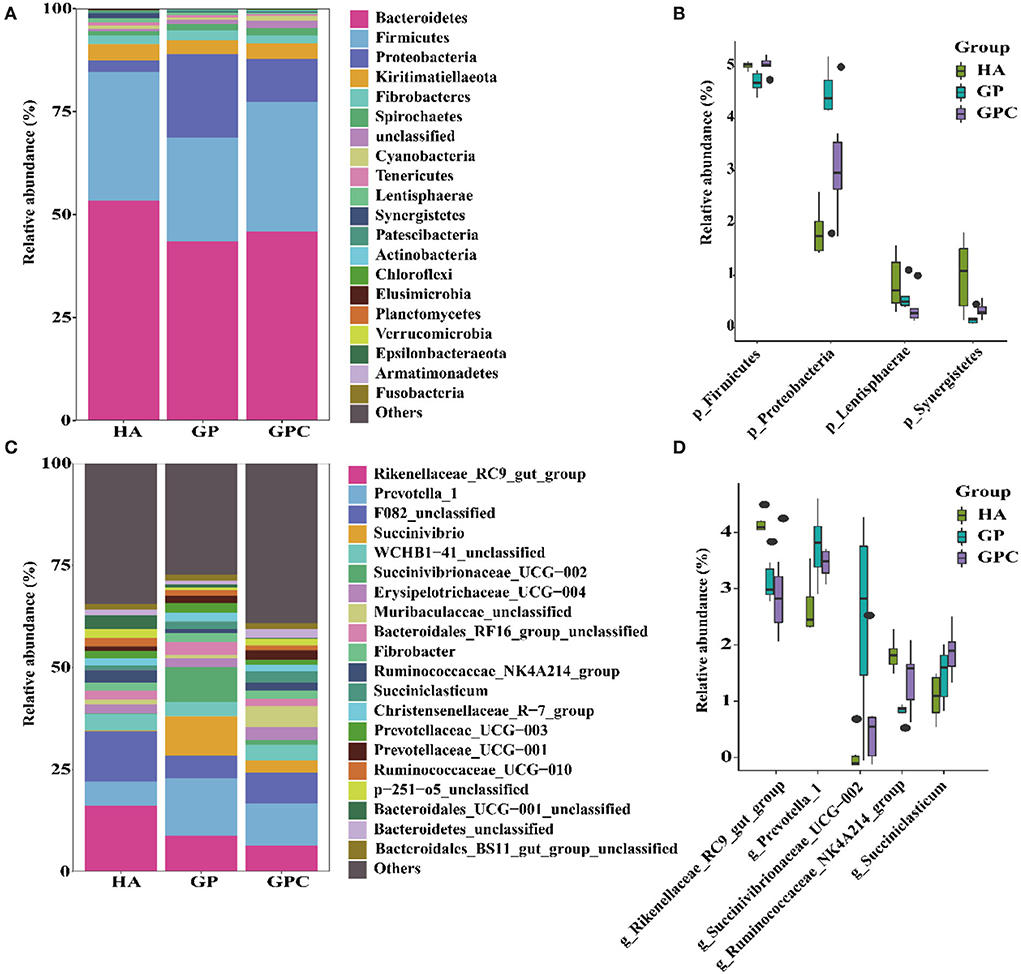
Figure 3. The relative abundance (%) of bacterial phyla (1% at least in one group) of ruminal microbiome of lambs fed non-pelleted native grass hay without or with concentrate or pellets (n = 6). (A) Phylum level. (B) Extended error bar plot showing the bacteria at the phylum level that had significant differences among the HA, GP, and GPC groups. (C) Genus level. (D) Extended error bar plot showing the bacteria at the genus level that had significant differences among the HA, GP, and GPC groups. HA, non-pelleted native grass hay diet, GP, pelleted native grass hay diet; GPC, pelleted native grass hay with concentrate diet.
At the genus level, Rikenellaceae_RC9_gut_group, Prevotella_1, F082_unclassified, and WCHB1-41_unclassified were the most abundant in the HA group, Rikenellaceae_RC9_gut_group, Prevotella_1, F082_unclassified, Succinivibrio, and Succinivibrionaceae_UCG-002 were the primary genera in the GP group; Rikenellaceae_RC9_gut_group, Prevotella_1, F082_unclassified, and Muribaculaceae_unclassified were the predominant genera in the GPC group (Figure 3C). Significant (p < 0.05) differences were observed in Rikenellaceae_RC9_gut_group, Prevotella_1, Ruminococcaceae_NK4A214_group, and Succiniclasticum among the three groups (Figure 3D).
The results of the linear discriminant analysis (LDA) effect size (LEFSe) showed significantly different taxonomies among the rumen microbiota of lambs fed the three diets (Figure 4). The genera Rikenellaceae_RC9_gut_group and Ruminococcaceae were enriched in the HA group, and the microbiota of the GP group was enriched with Succinivibrio, Succinivibrionaceae_UCG-002, and Prevotella_1. In contrast, the microbiota of the GPC group was enriched with Ruminobacter and Muribaculaceae_unclassified.
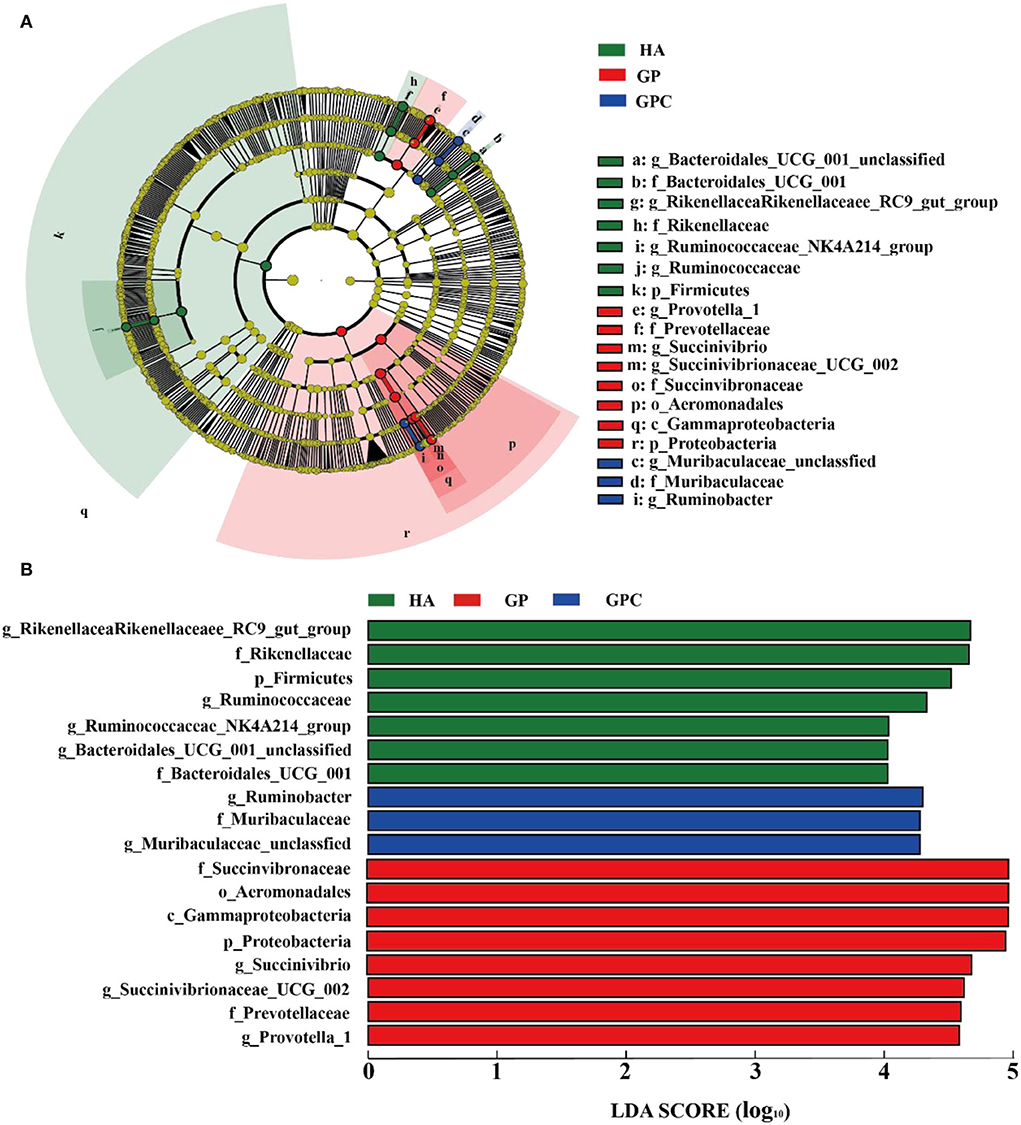
Figure 4. Linear discrimination analysis (LDA) coupled with effect size (LEFSe) analysis of the rumen microbial community of lamb in the HA, GP and GPC groups (n = 6). (A) Cladogram showing microbial species with significant differences among the three treatments. Red, green, and blue represent different groups. Species classification at the phylum, class, order, family, and genus level are displayed from inner to outer layers. The red, green, and blue nodes represent microbial species in the phylogenetic tree that play important roles in the HA, GP, and GPC groups, respectively. Yellow nodes represent no significant difference between species. (B) Significantly different species with an LDA score greater than the estimated value (default score = 4). The length of the histogram represents the LDA score of different species in the three groups. HA, non-pelleted native grass hay diet, GP, pelleted native grass hay diet; and GPC, pelleted native grass hay with concentrated diets.
The predicted functional genes of the ruminal bacterial community were predicted using the PICRUSTS-dependent Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway database. The main predicted functional genes at level 1 in the three groups were assigned to metabolism (55.72–56.61%), genetic information processing (26.59–26.79%), and environmental information processing (11.05–11.60%), respectively (Figure 5A), which indicated different pathways (Figure 5B). Notably, amino acid metabolism, carbohydrate metabolism, energy metabolism, and nucleotide metabolism were higher than 5% of the enriched pathways of the lambs fed in the three diets. Furthermore, the sequences related to amino acid metabolism, energy metabolism, and metabolism of terpenoids and polyketides were significantly (p < 0.05) enriched in the HA group, while folding, sorting, degradation, and cellular processes and signaling were statistically (p < 0.05) enriched in the GP group. Additionally, the HA and GP groups markedly enriched genetic information processing (p < 0.05). At level 3 of the microbial genes that predicted the functions of bacteria, some differences were observed in the three groups (Figure 5C). The abundance of a majority of the genes related to amino acid metabolism (amino acid-related enzymes, alanine, aspartate, and glutamate metabolism, arginine, and proline metabolism, phenylalanine, tyrosine, and tryptophan biosynthesis), glycolysis/gluconeogenesis, pyruvate metabolism, and amino sugar and nucleotide sugar metabolism) increased dramatically (p < 0.05) in the HA group. Similarly, the genes associated with translation, transcription, replication and repair, folding, sorting, and degradation, such as ribosome biogenesis, transcription machinery, chaperones, and folding catalysts, were markedly (p < 0.05) enriched in the GP group. Interestingly, the abundance of most genes assigned to amino acid metabolism, nucleotide metabolism, and energy metabolism was decreased in the GPC group.
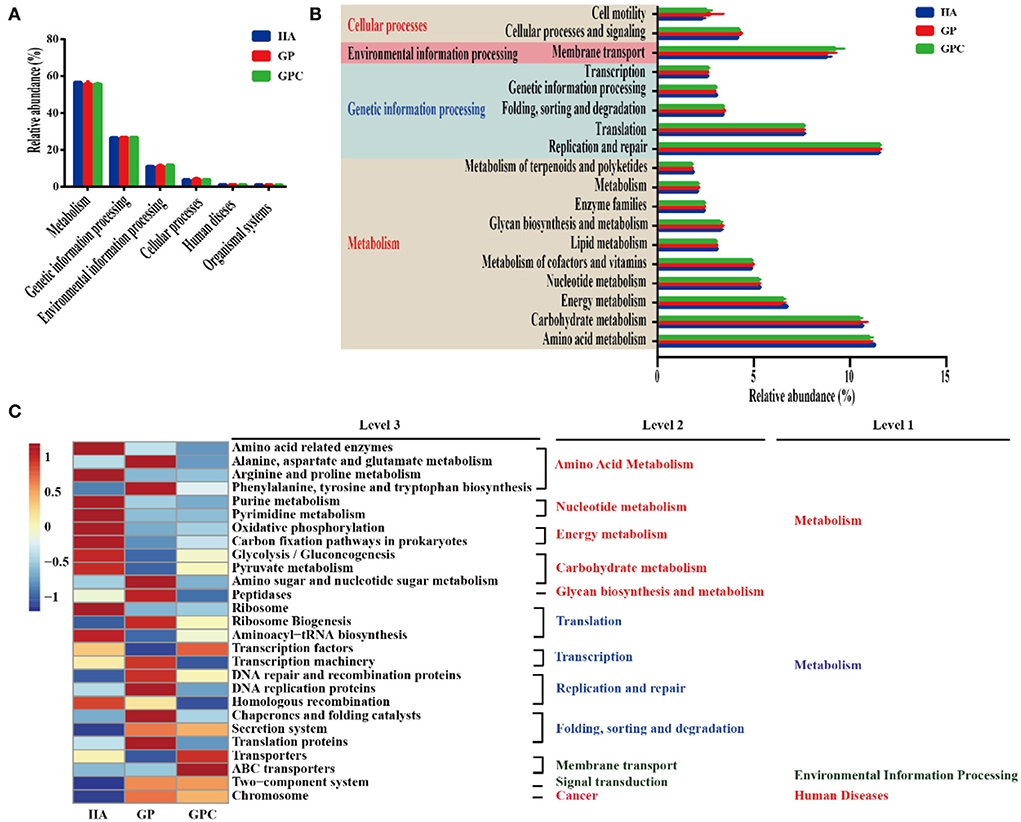
Figure 5. Dynamics of rumen bacterial predicted functional profiles fed with different diets analyzed using PICRUSt (n = 6). (A) Level 1 metabolic pathways. (B) Level 2 Kyoto Encyclopedia of Genes and Genomes (KEGG) ortholog functional predictions of the relative abundances of the top 20 metabolic functions. (C) Level 3 KEGG ortholog functional predictions of the relative abundances of the top 30 metabolic functions. HA, non-pelleted native grass hay diet, GP, pelleted native grass hay diet; and GPC, pelleted native grass hay with concentrate diet.
In the current study, as compared to the HA group, the final BW and ADG were statistically increased in the GP group, which is consistent with the studies of Du et al. (2019) and Trabi et al. (2019), who found that the grinding and pelleting processes could directly reduce the feed particle size and increase the intake. The physical characteristic (freshness, flavor, color, texture) and form difference between grass and pellets may be the reason for the enhanced intake (Prachumchai et al., 2021, 2022). Therefore, higher final BW and ADG were observed in the GP group. Furthermore, the ADG increased in the GPC group compared to the GP group, which could be explained by the higher DMI and energy intakes in the GPC group compared to the lambs fed with the GP diet (Trabi et al., 2019).
Diets have dominant effects on the compositions of rumen microbiota (Zhou et al., 2010; Chen et al., 2011). In the present study, compared to the HA group, the number of OTUs, species richness estimators (Chao1 value), and the Shannon index decreased in the GP and GPC groups, which is in agreement with various studies in the rumen with high concentrate or pelleted diets (Mao et al., 2013; Huo et al., 2014; Trabi et al., 2019). These results indicated that the diversity and richness of the bacterial community can be influenced by feed types. Previous reports also found that a decreased microbial community was associated with a decrease in forage percentages and pellets (Hook et al., 2011; Henderson et al., 2015), which could be explained by the increased DMI and higher energy intake that directly influence the bacterial growth rate (Dagaew et al., 2020; Prachumchai et al., 2022). To address the effects of diets on beta diversity, the ruminal bacterial community was characterized based on unweighted UniFrac distance across all ruminal samples. The PCoA diagram was used to display the changes in the bacterial community among the three groups. Macroscopically, the different diets led to a distinguishable separation in the bacterial community among three groups, which is consistent with the previous reports, in that a noticeable separation of the microbial structure was observed among pelleted, concentrate, and forage diets (Zhang et al., 2017; Trabi et al., 2019; Lin L. et al., 2021), and various diets drove the diverse pH environment, which may be the main reason (Logue et al., 2016).
Changes in diversity were tightly correlated to phylogenetic modifications (Amin et al., 2021). The phyla Firmicutes and Bacteroidetes were detected as predominant phyla in this study, following the previously published studies that also identified phyla Firmicutes and Bacteroidetes as the dominant phyla in the rumen microbiome of sheep, which include cellulose degraders, such as Fibrobacter succinogenes, Ruminococcus flavefaciens, and Ruminococcus albus (Henderson et al., 2015; Zhang et al., 2019; Mizrahi et al., 2021). In contrast, there were some differences in these phyla among the three groups. The predominant bacterial phylum, Bacteroidetes, which is associated with the degradation of carbohydrates and proteins, was observed in all groups, and the highest relative abundance of this phylum was detected in the HA group, which is in agreement with the study of Lopes et al. (2015), who found that Bacteroidetes is the dominant phylum in the sheep rumen microbiome. The pelleting process or highly concentrated diets could be the main reason leading to the decreased relative abundance of Bacteroidetes (Trabi et al., 2019; Chao et al., 2020). Firmicutes play a critical role in the degradation of fiber and cellulose and are associated with the composition of polysaccharides and the utilization of energy (Singh et al., 2012; Crisol-Martínez et al., 2017). The abundance of Firmicutes decreased in the GP groups, following previous reports that indicate that feeding pellets could make the abundance of Firmicutes drop (Trabi et al., 2019).
The higher abundance of Firmicutes was detected in the GPC group, which is similar with the previous report who found the abundance increased with the increasing concentrate proportions (Lourenco et al., 2020). Interestingly, it is known that Proteobacteria are a biomarker for inflammatory progression and enhance the progression of the disease (Larsen et al., 2015; Shin et al., 2015). The higher relative abundance of Proteobacteria was found in the GP and GPC groups compared to the HA group, which could be attributed to the HA group increasing the ruminal fermentation rate, decreasing the ruminal pH, and inhibiting the growth of Proteobacteria (Larsen et al., 2015; Shin et al., 2015). Furthermore, hydrogenation also influenced fatty acid compositions by the bacterial structure, and the rumen microbiota was diverse (Harfoot, 1978; Martin et al., 2016).
The present study also analyzed the changes in the taxa at the genus level. At the genus level, the most abundant bacteria were Prevotella_1, Rikenellaceae_RC9_gut_group, F082_unclassfied, and Succinivibrio, which are highly efficient in hemicellulose and structural carbohydrate degradation (Zened et al., 2013; Morais and Mizrahi, 2019; Li et al., 2021; Mizrahi et al., 2021). At the genus level, Rikenellaceae_RC9_gu_group was the most abundant bacterial group. It was significantly decreased in the GP and GPC groups, following the study of Ebeid et al., which found a high level of Rikenellaceae_RC9_gut_group in lambs in the forage feeding systems (Ebeid et al., 2020). Wang et al. (2016) reported that the differences in ruminal microflora probably came from the physical effects. Members of the Prevotella genera have been particularly studied as dominant species in the colonization and degradation of the hemicellulose portion of the fiber because of a broader fiber-related enzymatic repertoire that could enable them to digest the available fiber (Jami et al., 2013; Piao et al., 2014; Huws et al., 2016). The genera Prevotella_1 was the most abundant bacterial genus in the rumen of lambs, which was reported by Vibart et al. (2019) and Zhang et al. (2019). The abundance of Prevotella_1 increased in the GP and GPC groups, which could attributed to its sensitivity to the forage percentages (Li et al., 2015; Solden et al., 2016; Bainbridge et al., 2018), as well as the insufficient fiber contents that did not effectively ensure proper rumen fermentation (Kleefisch et al., 2017). Previously published research found that C16:0 and C18:0 could influence the genera Prevotella and Butyrivibrio (Maczulak et al., 1981). In the present study, the C16:0 and C18:0 contents could increase the abundance of the genus Prevotella (Maczulak et al., 1981), and the higher C16:0 and C18:0 contents were also found in the GP group compared to that in the HA group. However, compared to the HA group, the markedly higher intake of the GPC group increased the ingestion of C16:0 and C18:0 contents. Therefore, the abundance of Prevotella_1 increased in the GP and GPC groups. The Bacteroidetes unclassified family F082 was also reduced in the GP and GPC groups compared to the HA group. These results indicate that significant ecological diversity exists in the host and feed types within this genus and uncultured family. However, the ruminal role of the Bacteroidetes unclassified family F082 is still unclear (Chiariotti et al., 2020) and will be researched in further study. Besides, the authors found that the abundance of Succinivibrio, a short-chain fatty acid-producing bacteria (Louis and Flint, 2009; Ticinesi et al., 2017), was significantly increased in the GP group, which could be explained by the starch granules used during pellet processing, which made it stay longer in the rumen (Lin T. et al., 2021). The genus Succiniclasticum is the main participant in producing propionate, the critical precursor of glucose in ruminants (Van Gylswyk, 1995; Liu et al., 2019). In this study, Succiniclasticum_UCG-002 was significantly increased in the GP group, which is consistent with previous reports that Succiniclasticum increased in pellet diets. It contributes to the pelleting process (Trabi et al., 2019). Indeed, the different substrates of the diet affect the growth of rumen microbiota (Maslowski and Mackay, 2011; Li et al., 2012; Kasparovska et al., 2016). Previous reports found that Prevotella_1, Rikenellaceae_RC9_gut_group, F082_unclassfied, and Succinivibrio were the genera of the core-microbiome (Du et al., 2022; Zhou et al., 2022). In the present study, these genera were influenced by the physical forms and chemical compositions, whereas the stability of the most abundant bacteria can reflect the presence of the genus-core microbiome (Henderson et al., 2015; Lin T. et al., 2021). The ruminal microbial ecosystem could provide ~70% of the energy that ruminants need, and the rumen microbiome has been linked to host feed efficiency and animal performance (Anderson et al., 2017; Bickhart and Weimer, 2018; Rosenberg and Zilber-Rosenberg, 2018). However, prior research found no significant positive or negative correlations between the dominant genera and animal performance in lambs (Du et al., 2022).
The results of KEGG analysis at gene levels 1 through 3 were consistent. Compared to the GP and GPC groups, the abundance of amino acid and carbohydrate metabolism genes was decreased, which could contribute to cellulose metabolism, and hemicellulose could provide many compounds beneficial for bacterial growth (Toledo et al., 2017; Su et al., 2021). Amino acid metabolism also provides energy and carbon sources for bacteria (Sanchez et al., 2017). These results revealed that the degradation of large carbohydrates and proteins resulted in increased levels of amino acids, which the microbiota could utilize in the HA group. The higher abundance of fiber in the HA group may be the main reason. The gene abundance of folding, sorting, and degradation, and cellular processes and signaling increased in the GP group, which follows the prior research that concentrates on the cellular process and genetic information processing in the high-forage diets (Lin L. et al., 2021). These predicted metabolic functions were related to compound production and cellular processes, suggesting enzyme synthesis. More activity associated with cell metabolism and genetic information processing in the GP group could contribute to the higher DMI, and high-forage diets are mainly devoted to maintaining basic survival capabilities for growth (Lamendella et al., 2011; Lin L. et al., 2021). The abundance associated with amino acid metabolism, nucleotide metabolism, and energy metabolism was detected in the GPC group compared to those in the HA and GP groups. The higher final BW weight and the lower growth rate might be the main reasons (Flint et al., 2012; Furman et al., 2020). The bacteria were correlated with different kinds of metabolic pathways, revealing that the multiple metabolic pathways were active in responding to different feed types.
This study explored the effects of feed types on growth performance and the rumen bacterial microbiota of lambs. It can be concluded that, compared to the HA group, the GP and GPC diets could increase the DMI and ADG of lambs, and the GPC groups performed best among the three groups and altered the composition and function of the rumen microbiota. One of the limitations of the current study was the homogenizing of rumen samples for analysis and reflecting the microbial community of lambs. Regardless, the findings of this study could provide a direct perspective and fundamental knowledge of the rumen microbiota in response to feeding types. In a further study, the relationship between rumen microbiota and function with feed type and meat quality should be explored to identify whether the rumen microbiota can contribute to host growth and meat quality.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/, PRJNA825331.
The animal study was approved by the College of Grassland, Resources and Environment, Inner Mongolian Agricultural University, China. All experiments were performed following the Regulations for the Administration of Affairs Concerning Experimental Animals.
Conceptualization, methodology, data curation, writing—original draft preparation, and writing—review and editing: SD. Investigation and resources: ZB. Writing—original draft preparation: SY. Methodology: JB. Project administration and funding acquisition: YJ. All authors have read and agreed to the published version of the manuscript.
This work was funded by the Key Program of the National Dairy Innovation Center, China (2021-National Dairy Innovation Center-1), the National Natural Science Fund (32260346), and Project of Education Department of Inner Mongolia Autonomous Region (NJZY21238).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Amin, N., Schwarzkopf, S., Kinoshita, A., Trscher-Muotter, J., Danicke, S., Camarinha-Silva, A., et al. (2021). Evolution of rumen and oral microbiota in calves is influenced by age and time of weaning. Anim. Microb. 3, 1–15. doi: 10.1186/s42523-021-00095-3
Anderson, C. L., Sullivan, M. B., and Fernando, S. C. (2017). Dietary energy drives the dynamic response of bovine rumen viral communities. Microbiome 5, 155. doi: 10.1186/s40168-017-0374-3
Bai, J., Xu, D., Xie, D., Wang, M., Li, Z., Guo, X., et al. (2020). Effects of antibacterial peptide-producing Bacillus subtilis and Lactobacillus buchneri on fermentation, aerobic stability, and microbial community of alfalfa silage. Bioresour. Technol. 315, 123881. doi: 10.1016/j.biortech.2020.123881
Bainbridge, M. L., Saldinger, L. K., Barlow, J. W., Alvez, J. P., Roman, J., Kraft, J., et al. (2018). Alteration of rumen bacteria and protozoa through grazing regime as a tool to enhance the bioactive fatty acid content of bovine milk. Front. Microbiol. 9, 904–916. doi: 10.3389/fmicb.2018.00904
Bhujbal, S. K., Ghosh, P., Vijay, V. K., Rathour, R., Kumar, M., Singh, L., et al. (2022). Biotechnological potential of rumen microbiota for sustainable bioconversion of lignocellulosic waste to biofuels and value-added products. Sci. Total Environ. 814, 152773. doi: 10.1016/j.scitotenv.2021.152773
Bickhart, D. M., and Weimer, P. J. (2018). Symposium review: host-rumen microbe interactions may be leveraged to improve the productivity of dairy cows. J. Dairy Sci. 101, 7680–7689. doi: 10.3168/jds.2017-13328
Bu, Z. K., Ge, G. T., Jia, Y. S., and Du, S. (2021). Effect of hay with or without concentrate or pellets on growth performance and meat quality of Ujimqin lambs on the Inner Mongolian Plateau. Anim. Sci. J. 92, e13553. doi: 10.1111/asj.13553
Chambers, J. M., and Hastie, T. J. (1992). Statistical Models. Grove, CA: Wadsworth and Brooks/Cole.
Chao, R., Xia, C., Pei, C., Huo, W., Liu, Q., Zhang, C., et al. (2020). Comparison of the microbial communities of alpacas and sheep fed diets with three different ratios of corn stalk to concentrate. J. Anim. Physiol. Anim. Nutr. 1, 26–34. doi: 10.1111/jpn.13442
Chen, Y., Penner, G. B., Li, M., Oba, M., and Guan, L. L. (2011). Changes in bacterial diversity associated with epithelial tissue in the beef cow rumen during the transition to a high-grain diet. Appl. Environ. Microbiol. 77, 5770–5781. doi: 10.1128/AEM.00375-11
Chiariotti, A., Edwards, J. E., Hermes, G. D. A., Catillo, G., Zilio, D. M., Giovanni, S. D., et al. (2020). Increasing the sustainability of maize grain production by using Arbuscular Mycorrhizal fungi does not affect the rumen of dairy cattle (Bos taurus) and buffalo (Bubalus bubalis). Front. Vet. Sci. 7, 556764. doi: 10.3389/fvets.2020.556764
Crisol-Martínez, E., Stanley, D., Geier, M. S., Hughes, R. J., and Moore, R. J. (2017). Understanding the mechanisms of zinc bacitracin and avilamycin on animal production: linking gut microbiota and growth performance in chickens. Appl. Microbiol. Biot. 101, 4547–4559. doi: 10.1007/s00253-017-8193-9
Dagaew, G., Cherdthong, A., Wanapat, M., and Chanjula, P. (2020). In vitro rumen gas production kinetics, hydrocyanic acid concentration and fermentation characteristics of fresh cassava root and feed block sulfur concentration. Anim. Prod. Sci. 60, 659–664. doi: 10.1071/AN18784
Du, S., You, S. H., Bao, J., Ge, G. T., Jia, Y. S., Cai, Y. M., et al. (2019). Evaluation of the growth performance and meat quality of Mongolian lamb fed grass, hay or pellets of Inner Mongolian native grass. Small Ruminant. Res. 181, 34–38. doi: 10.1016/j.smallrumres.2019.10.008
Du, S., You, S. H., Bao, J., Ge, G. T., Jia, Y. S., Cai, Y. M., et al. (2020). Growth performance, carcass characteristics, and meat quality of Mongolian lambs fed native grass or hay with or without concentrate on the Inner Mongolian Plateau. Can. J. Anim. Sci. 100, 470–478. doi: 10.1139/cjas-2019-0126
Du, S., You, S. H., Sun, L., Jia, Y. S., and Zhou, Y. L. (2022). Effects of replacing alfalfa hay with native grass hay in pelleted total mixed ration on physicochemical parameters, fatty acid profile and rumen microbiota in lamb. Front. Microbiol. 13, 861025. doi: 10.3389/fmicb.2022.861025
Ebeid, H. M., Hassan, F., Li, M. W., Peng, L. J., Peng, K. P., Liang, X., et al. (2020). Camelina sativa L. Oil mitigates enteric in vitro methane production, modulates ruminal fermentation, and ruminal bacterial diversity in Buffaloes. Front. Vet. Sci. 7, 550. doi: 10.3389/fvets.2020.00550
Flint, H. J., Scott, K. P., Duncan, S. H., Louis, P., and Forano, E. (2012). Microbial degradation of complex carbohydrates in the gut. Gut Microbes. 3, 289–306. doi: 10.4161/gmic.19897
Furman, O., Shenhav, L., Sasson, G., Kokou, F., Honig, H., Jacoby, S., et al. (2020). Stochasticity constrained by deterministic effects of diet and age drive rumen microbiome assembly dynamics. Nat. Commun. 11, 1904. doi: 10.1038/s41467-020-15652-8
Guo, W., Zhou, M., Ma, T., Bi, S., Wang, W., Zhang, Y., et al. (2020). Survey of rumen microbiota of domestic grazing yak during different growth stages revealed novel maturation patterns of four key microbial groups and their dynamic interactions. Anim. Microb. 2, 23. doi: 10.1186/s42523-020-00042-8
Harfoot, C. G. (1978). Lipid metabolism in the rumen. Prog. Lipid. Res. 17, 2154. doi: 10.1016/0079-6832(78)90004-6
Henderson, G., Cox, F., Ganesh, S., Jonker, A., Young, W., Global, R. C. C., et al. (2015). Rumen microbial community composition varies with diet and host, but a core microbiome is found across a wide geographical range. Sci. Rep. 5, 14567–14581. doi: 10.1038/srep14567
Hook, S. E., Steele, M. A., Northwood, K. S., Dijkstra, J., France, J., Wright, A. D., et al. (2011). Impact of subacute ruminal acidosis (SARA) adaptation and recovery on the density and diversity of bacteria in the rumen of dairy cows. FEMS Microbiol. Ecol. 78, 275–284. doi: 10.1111/j.1574-6941.2011.01154.x
Huo, W., Zhu, W., and Mao, S. (2014). Impact of subacute ruminal acidosis on the diversity of liquid and solid-associated bacteria in the rumen of goats. World. J. Microbiol. Biotechnol. 30, 669–680. doi: 10.1007/s11274-013-1489-8
Huws, S. A., Edwards, J. E., Creevey, C. J., Stevens, P. R., Lin, W. C., Girdwood, S. E., et al. (2016). Temporal dynamics of the metabolically active rumen bacteria colonizing fresh perennial ryegrass. FEMS Microbiol. Ecol. 92, 137. doi: 10.1093/femsec/fiv137
Ishaq, S. L., Lachman, M. M., Wenner, B. A., Baeza, A., Butler, M., Gates, E., et al. (2019). Pelleted-hay alfalfa feed increases sheep wether weight gain and rumen bacterial richness over loose-hay alfalfa feed. PLoS ONE. 14, e0215797. doi: 10.1371/journal.pone.0215797
Jami, E., Israel, A., Kotser, A., and Mizrahi, I. (2013). Exploring the bovine rumen bacterial community from birth to adulthood. ISME. J. 7, 1069–1079. doi: 10.1038/ismej.2013.2
Kasparovska, J., Pecinkova, M., Dadakova, K., Krizova, L., Hadrova, S., Lexa, M., et al. (2016). Effects of isoflavone-enriched feed on the rumen microbiota in dairy cows. PLoS ONE. 11, e0154642. doi: 10.1371/journal.pone.0154642
Khafipour, E., Li, S. C., Plaizier, J. C., and Krause, D. O. (2009). Rumen microbiome composition determined using two nutritional models of subacute ruminal acidosis. Appl. Environ. Microb. 75, 7115–7124. doi: 10.1128/AEM.00739-09
Kim, M., Morrison, M., and Yu, Z. (2015). Status of the phylogenetic diversity census of ruminal microbiomes. FEMS Microbiol. Ecol. 76, 49–63. doi: 10.1111/j.1574-6941.2010.01029.x
Kleefisch, M. T., Zebeli, Q., Humer, E., KrÖger, I., Etrl, P., and Klevenhusen, F. (2017). Effects of the replacement of concentrate and fibre-rich hay by high-quality hay on chewing, rumination and nutrient digestibility in non-lactating Holstein cows. Arch. Anim. Nutr. 71, 21–36. doi: 10.1080/1745039X.2016.1253227
Lamendella, R., Domingo, J. W., Ghosh, S., Martinson, J., and Oerther, D. B. (2011). Comparative fecal metagenomics unveils unique functional capacity of the swine gut. BMC Microbiol. 11, 103. doi: 10.1186/1471-2180-11-103
Larsen, J. M., Musavian, H. S., Butt, T. M., Ingvorsen, C., Thysen, A. H., Brix, S., et al. (2015). Chronic obstructive pulmonary disease and asthma-associated Proteobacteria, but not commensal Prevotella spp, promote Toll-like receptor 2-independent lung inflammation and pathology. Immunology 144, 333. doi: 10.1111/imm.12376
Li, R. W., Connor, E. E., Li, C. J., Baldwin, R. L., and Sparks, M. E. (2012). Characterization of the rumen microbiota of pre-ruminant calves using metagenomic tools. Environ. Microbiol. 14, 129–139. doi: 10.1111/j.1462-2920.2011.02543.x
Li, S., Dong, L., Lian, W. H., Lin, Z. L., Lu, C. Y., Xu, L., et al. (2021). Exploring untapped potential of Streptomyces spp. in Gurbantunggut Desert by use of highly selective culture strategy. Sci. Total. Environ. 790, 148235. doi: 10.1016/j.scitotenv.2021.148235
Li, Z. P., Wright, A. D. G., Liu, H. L., Bao, K., Zhang, T., Wang, K. Y., et al. (2015). Bacterial community composition and fermentation patterns in the rumen of sika deer (Cervus nippon) fed three different diets. Microb. Ecol. 2, 307–318. doi: 10.1007/s00248-014-0497-z
Lin, L., Trabi, E. B., Xie, F., and Mao, S. (2021). Comparison of the fermentation and bacterial community in the colon of Hu sheep fed a low-grain, non-pelleted, or pelleted high-grain diet. Appl. Microbiol. Biotechnol. 105, 2071–2080. doi: 10.1007/s00253-021-11158-5
Lin, T., Wu, P., Lin, Y., and Huang, S. (2021). Gut dysbiosis and mortality in hemodialysis patients. NPJ Biofilms. Microbiomes. 7, 20. doi: 10.1038/s41522-021-00191-x
Liu, C., Wu, H., Liu, S. J., Chai, S. T., Meng, Q. X., Zhou, Z. M., et al. (2019). Dynamic alterations in yak rumen bacteria community and metabolome characteristics in response to feed type. Front. Microbiol. 10, 1116. doi: 10.3389/fmicb.2019.01116
Logue, J. B., Stedmon, C. A., Kellerman, A. M., Nielsen, N. J., Andersson, A. F., Laudon, H., et al. (2016). Experimental insights into the importance of aquatic bacterial community composition to the degradation of dissolved organic matter. ISME J. 10, 533–545. doi: 10.1038/ismej.2015.131
Lopes, L. D., Lima, A. O. D. S., Taketani, R. G., Darias, P., Silva, L. R. F. D., Romagnoli, E. M., et al. (2015). Exploring the sheep rumen microbiome for carbohydrate-active enzymes. Anton. Leeuw. 108, 15–30. doi: 10.1007/s10482-015-0459-6
Louis, P., and Flint, H. J. (2009). Diversity, metabolism and microbial ecology of butyrate-producing bacteria from the human large intestine. FEMS Microbiol. Lett. 294, 1–8. doi: 10.1111/j.1574-6968.2009.01514.x
Lourenco, J. M., Kieran, T. J., Seidel, D. S., Glenn, T. C., Silveira, M. F. D., Callaway, T. R., et al. (2020). Comparison of the ruminal and fecal microbiotas in beef calves supplemented or not with concentrate. PLoS ONE. 15, e0231533. doi: 10.1371/journal.pone.0231533
Maczulak, E., Dehority, B. A., and Palmquist, D. L. (1981). Effects of long-chain fatty acids on growth of rumen bacteria. Appl. Environl. Microb. 42, 856–862. doi: 10.1128/aem.42.5.856-862.1981
Mao, Y. S., Zhang, R. Y., Wang, D. S., and Zhu, W. Y. (2013). Impact of subacute ruminal acidosis (SARA) adaption on rumen microbiota in dairy cattle using pyrosequencing. Anaerobe 24, 12–19. doi: 10.1016/j.anaerobe.2013.08.003
Martin, C., Ferlay, A., Mosoni, P., Rochette, Y., Chilliard, Y., Doreau, M., et al. (2016). Increasing linseed supply in dairy cow diets based on hay or corn silage: effect on enteric methane emission, rumen microbial fermentation, and digestion. J. Dairy. Sci. 99, 3445–3456. doi: 10.3168/jds.2015-10110
Maslowski, K. M., and Mackay, C. R. (2011). Diet, gut microbiota and immuneresponses. Nat. Immunol. 12, 5–9. doi: 10.1038/ni0111-5
Masoudzadeh, S. H., Mohammadabadi, M., Khezri, A., Stavetska, R. V., Oleshko, V. P., Babenko, O. I., et al. (2020). Effects of diets with different levels of fennel (Foeniculum vulgare) seed powder on DLK1 gene expression in brain, adipose tissue, femur muscle and rumen of Kermani lambs. Small Ruminant Res. 193, e106276. doi: 10.1016/j.smallrumres.2020.106276
McClements, D. J., and Xiao, H. (2012). Potential biological fate of ingested nanoemulsions: influence of particle characteristics. Food Funct. 3, 202–220. doi: 10.1039/C1FO10193E
Mizrahi, I., Wallace, R. J., and Moras, S. (2021). The rumen microbiome: balancing food security and environmental impacts. Nat. Rev. Microbiol. 19, 553–566. doi: 10.1038/s41579-021-00543-6
Morais, S., and Mizrahi, I. (2019). Islands in the stream: from individual to communal fiber degradation in the rumen ecosystem. FEMS Rev. Microbiol. 43, 362–379. doi: 10.1093/femsre/fuz007
Muwonge, A., Karuppannan, A. K., and Opriessnig, T. (2021). Probiotics mediated gut microbiota diversity shifts are associated with reduction in histopathology and shedding of Lawsonia intracellularis. Anim. Microb. 3, 22. doi: 10.1186/s42523-021-00084-6
Omontese, B. O., Sharma, A. K., Davison, S., Jacobson, E., DiConstanzo, A., Webb, M. J., et al. (2022). Microbiome network traits in the rumen predict average daily gain in beef cattle under different backgrounding systems. Anim. Microb. 4, 25. doi: 10.1186/s42523-022-00175-y
Piao, H., Lachman, M., Malfatti, S., Sczyrba, A., Knierim, B., Auer, M., et al. (2014). Temporal dynamics of fibrolytic and methanogenicrumen microorganisms during in situ incubation of switchgrass determined by 16S rRNA gene profiling. Front. Microbiol. 5, 307. doi: 10.3389/fmicb.2014.00307
Plaizier, J. C., Khafipour, E., Li, S., Gozho, G. N., and Krause, D. O. (2012). Subacute ruminal acidosis (SARA), endotoxins and health consequences. Anim. Feed Sci. Technol. 172, 9–21. doi: 10.1016/j.anifeedsci.2011.12.004
Prachumchai, R., Cherdthong, A., and Wanapat, M. (2021). A screening of cyanide-utilizing bacteria from rumen and in vitro evaluation of fresh cassava root utilization with pellet containing high sulfur diet. Vet. Sci. 8, 10. doi: 10.3390/vetsci8010010
Prachumchai, R., Cherdthong, A., Wanapat, M., So, S., and Polyorach, S. (2022). Fresh cassava root replacing cassava chip could enhance milk production of lactating dairy cows fed diets based on high sulfur-containing pellet. Sci. Rep. 12, 3809. doi: 10.1038/s41598-022-07825-w
R Core Team. (2014). R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing.
Rognes, T., Flouri, T., Nichols, B., Quince, C., and Mah,é, F. (2016). Vsearch: a versatile open source tool for metagenomics. Peer J. 4, e2584. doi: 10.7717/peerj.2584
Rosenberg, E., and Zilber-Rosenberg, I. (2018). The hologenome concept of evolution after 10 years. Microbiome 6, 78. doi: 10.1186/s40168-018-0457-9
Sanchez, O. J., Ospina, D. A., and Montoya, S. (2017). Compost supplementation with nutrients and microorganisms in composting process. Waste Manag. 69, 136–153. doi: 10.1016/j.wasman.2017.08.012
Sarker, N. R., Yeasmin, D., Habib, M. A., and Tabassum, F. (2019). Feeding effect of total mixed ration on milk yield, nutrient intake, digestibility and rumen environment in red chittagong cows. Asian J. Medical and Biological Res. 5, 71–77.
Segata, N., Izard, J., Waldron, L., Gevers, D., Miropolsky, L., Garrett, W., et al. (2011). Metagenomic biomarker discovery and explanation. Genome Boil. 12, R60. doi: 10.1186/gb-2011-12-6-r60
Shin, N. R., Whon, T. W., and Bae, J. W. (2015). Proteobacteria: microbial signature of dysbiosis in gut microbiota. Trends. Biotechnol. 33, 496–503. doi: 10.1016/j.tibtech.2015.06.011
Singh, K. M., Shah, T., Deshpande, S., Jakhesara, S. J., Koringa, P. G., Rank, D. N., et al. (2012). High through put 16S rRNA gene-based pyrosequencing analysis of the fecal microbiota of high FCR and low FCR broiler growers. Mol. Biol. Rep. 39, 10595–10602. doi: 10.1007/s11033-012-1947-7
Solden, L. M., Hoyt, D. W., Collins, W. B., Plank, J. E., Daly, R. A., Hildebrand, E., et al. (2016). New roles in hemicellulosic sugar fermentation for the uncultivated Bacteroidetes family BS11. ISME J. 11, 691–703. doi: 10.1038/ismej.2016.150
Su, W., Jiang, Z., Hao, L., Li, W., Gong, T., Zhang, Y., et al. (2021). Variations of soybean meal and corn mixed substrates in physicochemical characteristics and microbiota during two-stage solid-state fermentation. Front. Microbiol. 12, 688839. doi: 10.3389/fmicb.2021.688839
Tanja, M., and Salzberg, S. L. (2011). FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27, 2957–2963. doi: 10.1093/bioinformatics/btr507
Ticinesi, A., Lauretani, F., Milani, C., Nouvenne, A., Meschi, T., Rio, D. D., et al. (2017). Aging gut microbiota at the cross-road between nutrition, physical frailty, and sarcopenia: is there a gut-muscle axis? Nutrients 30, 1303. doi: 10.3390/nu9121303
Toledo, M., Gutiérrez, M. C., Siles, J. A., García-Olmo, J., and Martín, M. A. (2017). Chemometric analysis and NIR spectroscopy to evaluate odorous impact during the composting of different raw materials. J. Clean. Prod. 7, 154–162. doi: 10.1016/j.jclepro.2017.08.163
Trabi, E. B., Seddik, H. E., Xie, F., Lin, L. M., and Mao, S. Y. (2019). Comparison of the rumen bacterial community, rumen fermentation and growth performance of fattening lambs fed low grain, pelleted or non-pelleted high grain total mixed ration. Anim. Feed Sci. Technol. 3, 1–12. doi: 10.1016/j.anifeedsci.2019.05.001
Van Gylswyk, N. O. (1995). Succiniclasticum ruminis gen. nov, sp. nov, aruminal bacterium converting succinate to propionate as the sole energy yielding mechanism. Int. J. Syst. Bacteriol. 45, 297–300. doi: 10.1099/00207713-45-2-297
Vibart, R. E., Ganesh, S., Kirk, M. R., Kittelmann, S., Leahy, S. C., Janssen, P. H., et al. (2019). Temporal fermentation and microbial community dynamics in rumens of sheep grazing a ryegrass-based pasture offered either in the morning or in the afternoon. Animal 2, 2251. doi: 10.1017/S1751731119000168
Wang, B., Ma, M. P., Diao, Q. Y., and Tu, Y. (2019). Saponin-Induced shifts in the rumen microbiome and metabolome of young cattle. Front. Microbiol. 10, 356. doi: 10.3389/fmicb.2019.00356
Wang, L., Xu, Q., Kong, F., Yang, Y., Wu, D., Mishra, S., et al. (2016). Exploring the goat rumen microbiome from seven days to two years. PLoS ONE. 11, e0154354. doi: 10.1371/journal.pone.0154354
Xing, B. S., Cao, S. F., Han, Y. L., Wang, X. C. C., Wen, J. W., Zhang, K. D., et al. (2020). A comparative study of artificial cow and sheep rumen fermentation of corn straw and food waste: Batch and continuous operation. Sci. Total Environ. 745, 140731. doi: 10.1016/j.scitotenv.2020.140731
Zened, A., Combes, S., Cauquil, L., Mariette, J., Klopp, C., Bouchez, O., et al. (2013). Microbial ecology of the rumen evaluated by 454 GS FLX pyrosequencing is affected by starch and oil supplementation of diets. FEMS Microbiol. Ecol. 83, 504–514. doi: 10.1111/1574-6941.12011
Zeng, Y., Zeng, D., Ni, X., Zhu, H., Jian, P., Zhou, Y., et al. (2017). Microbial community compositions in the gastrointestinal tract of Chinese Mongolian sheep using Illumina MiSeq sequencing revealed high microbial diversity. AMB Exp. 7, 75. doi: 10.1186/s13568-017-0378-1
Zhang, J., Shi, H. T., Wang, Y. J., Li, S. L., Cao, Z. J., Ji, S. K., et al. (2017). Effect of dietary forage to concentrate ratios on dynamic profile changes and interactions of ruminal microbiota and metabolites in Holstein heifers. Front. Microbiol. 8, 2206. doi: 10.3389/fmicb.2017.02206
Zhang, Q., Li, C., Niu, X., Zhang, Z., and Li, F. (2019). The effects of milk replacer allowance and weaning age on the performance, nutrients digestibility, and ruminal microbiota communities of lambs. Anim. Feed. Sci. Technol. 257, 114263. doi: 10.1016/j.anifeedsci.2019.114263
Zhou, M., Hernandez-Sanabria, E., and Guan, L. L. (2010). Characterization of variation in rumen methanogenic communities under different dietary and host feed efficiency conditions, as determined by PCR-denaturing gradient gel electrophoresis analysis. Appl. Environ. Microbiol. 76, 3776–3786. doi: 10.1128/AEM.00010-10
Zhou, Y. L., Sun, L., Cheng, Q. M., Li, Y. C., Chen, J. X., Zhao, B., et al. (2022). Effect of pelleted alfalfa or native grass total mixed ration on the rumen bacterial community and growth performance of lambs on the Mongolian Plateau. Small Ruminant Res. 207, 106610. doi: 10.1016/j.smallrumres.2021.106610
Keywords: lamb, native grass diet, concentrate, rumen microbiota, growth performance
Citation: Du S, Bu Z, You S, Bao J and Jia Y (2022) Diversity of growth performance and rumen microbiota vary with feed types. Front. Sustain. Food Syst. 6:1004373. doi: 10.3389/fsufs.2022.1004373
Received: 27 July 2022; Accepted: 28 October 2022;
Published: 24 November 2022.
Edited by:
Yu Liu, Northwest A&F University, ChinaReviewed by:
Blaz Stres, University of Ljubljana, SloveniaCopyright © 2022 Du, Bu, You, Bao and Jia. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shuai Du, ZHVzaHVhaV9ubUBzaW5hLmNvbQ==; Yushan Jia, anlzX25tQHNpbmEuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.