
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Sustain. Food Syst., 11 November 2021
Sec. Crop Biology and Sustainability
Volume 5 - 2021 | https://doi.org/10.3389/fsufs.2021.761611
This article is part of the Research TopicCoffee: From the Field to the CupView all 14 articles
 Sarada Krishnan1*†
Sarada Krishnan1*† Solene Pruvot-Woehl2
Solene Pruvot-Woehl2 Aaron P. Davis3†
Aaron P. Davis3† Tim Schilling2
Tim Schilling2 Justin Moat3†
Justin Moat3† William Solano4
William Solano4 Amin Al Hakimi5
Amin Al Hakimi5 Christophe Montagnon6†
Christophe Montagnon6†Cultivated Arabica coffee outside Ethiopia is plagued by low genetic diversity, compromising disease resistance, climate resiliency and sensory potential. Access to the wider genetic diversity of this species may circumvent some of these problems. In addition to Ethiopia, South Sudan has been postulated as a center of origin for Arabica coffee, but this has never been genetically confirmed. We used simple sequence repeat (SSR) markers to assess the genetic diversity of wild and cultivated populations of Arabica coffee from the Boma Plateau in South Sudan, against farmed accessions (of wild origin) from Ethiopia, Yemen, and global cultivars. Our results not only validate Boma Plateau as part of the natural distribution and as a center of origin for Arabica coffee but also indicate that wild populations in South Sudan are genetically distinct from Ethiopian Arabica. This newly identified genetic diversity within Arabica could have the potential for crop improvement through selection and use in breeding programs. Observations and analyses show that the extent and health of the wild population of Arabica in South Sudan have declined. Urgent action should be taken to conserve (in situ and ex situ) the unique, remaining genetic diversity of wild Arabica populations in South Sudan.
Coffee is a globally traded agricultural commodity with an estimated export value of US$ 30 billion in 2019 (http://www.worldstopexports.com/coffee-exports-country/), of which Arabica coffee (Coffea arabica) accounts for ~56% of the total production (International Coffee Organization (ICO), 2021). Despite the documented consumption of this coffee species as a beverage since at least the sixth century, and its contribution to the economy of many countries, it is remarkable that there was no factual data confirming native status in Ethiopia until the twentieth century (Meyer, 1965). The first documented cultivation of Arabica is in Yemen from around the fifteenth century, from where two main genetic types (Bourbon and Typica) were disseminated, giving rise to most of the commercial cultivars of Arabica grown worldwide (Anthony et al., 2002; Scalabrin et al., 2020; Krishnan et al., 2021; Montagnon et al., 2021).
The genetic diversity of farmed Arabica coffee is amongst the lowest of cultivated crops (Scalabrin et al., 2020), although the diversity of farmed (and wild) coffee in Ethiopia has still not been fully explored (Davis et al., 2018). Regardless, crossing global cultivars with cultivated Ethiopian accessions of wild origin has proven to bring significant crop improvement as compared to traditional cultivars (Van der Vossen, 2017; Marie et al., 2020). For example, a new genetic group of cultivated Arabica was revealed recently for Yemen (Montagnon et al., 2021). Additional genetic diversity in the species could be an opportunity for further progress. This is important as the Arabica coffee sector faces serious challenges, including climate change (Davis et al., 2012, 2019, 2021; Bunn et al., 2015; Moat et al., 2017, 2019), coffee leaf rust (Hemileia vastatrix) epidemics (Avelino et al., 2015), and the need for new and distinct flavor profiles (Davis et al., 2019; Montagnon et al., 2019).
The first recorded occurrence of C. arabica in the Boma Plateau of South Sudan (Anglo-Egyptian Sudan at that time) was in 1929 by A. Chevalier (Thomas, 1942). Following this, J. G. Myers, ecological advisor to the Sudan Government, visited the plateau in 1938 and confirmed C. arabica growing wild in the forests. Subsequent to this visit he invited A. S. Thomas, an economic botanist with the Ugandan Department of Agriculture, to accompany him on a second visit in December 1941 to collect seed for breeding purposes (Thomas, 1942). In his paper, Thomas described wild Arabica coffee growing abundantly on the slopes of Nelichu in the two localities of Barbuk and Rume (Figure 1). In the Barbuk area, trees growing up to a height of 18 ft. (5.5 m) were documented with much variation in leaves and fruits. Young seedlings were noticed growing near the larger trees as well as beside paths, which were attributed to dropped fruits collected by the local Kichepo tribe. The trees and shrubs listed by Thomas in his account of Barbuk are characteristic of the forests containing wild Arabica coffee in Ethiopia, which are classified as Moist Evergreen Afromontane Forest (MAF), and Transitional Rain Forest (TRF), (Friis et al., 2010). In the Rume area, about 2–3 miles south of Barbuk, coffee was found growing in areas cleared for growing maize (Zea mays)—not deliberately planted, but rather relics of the cleared forests. The coffee trees in Rume were shorter than the trees in Barbuk with green-tipped leaves compared to copper-tipped in Barbuk. During this expedition, seeds were collected and sent to the East African Agricultural Research Station in Amani (Tanzania), Coffee Research Station, Lyamungu (Tanzania), Scott Laboratories, Nairobi (Kenya), and various stations in Uganda, and these accessions are likely to be the progenitors of the cultivars C. arabica ‘Rume Sudan’ and ‘Barbuk Sudan,’ which are in cultivation today. Thomas (1942) noted the bold beans of Barbuk coffee, the relatively low elevation (compared to Barbuk) where the Rume coffee plants coffee were growing, and the lack of noticeable infection by coffee leaf rust (Hemileia vastatrix), which were identified as attributes for expanding the useful traits of cultivated Arabica coffee.
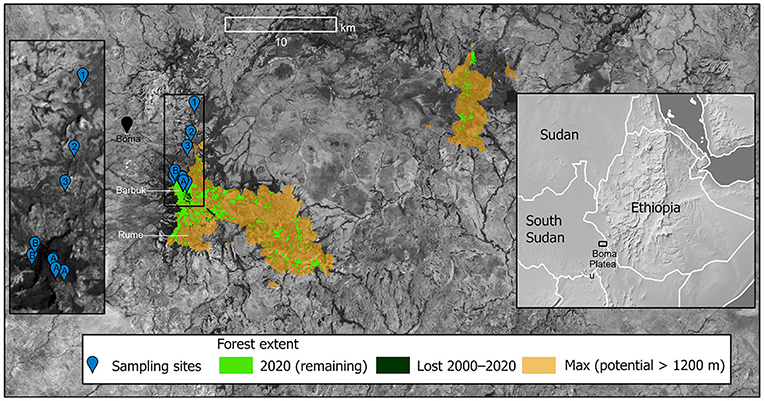
Figure 1. Study area: Boma Plateau of South Sudan. Cultivated collecting sites: 1 = Bayen; 2 = Jonglei; 3 = Kaiwa. Wild collecting area: A = Rumit (Barbuk), on plateau [includes some cultivated plants (see text)]; B = Ngelecho (Barbuk), on escarpment. Inset map right shows regional location of study area; inset map left shows location detail of main sampling sites.
There are only a few accessions available in living coffee research collections claimed to be representative of the original wild Arabica collections from the Boma Plateau (Thomas, 1942; Meyer, 1965), although ‘Rume Sudan’ is cultivated on a few farms as high quality (specialty) coffee, such as in Colombia. In the Centro Agronómico Tropical de Investigación y Enseñanza (CATIE) international coffee collections, two accessions of ‘Rume Sudan’ and one accession of ‘Barbuk Sudan’ are available. Records indicate that the two ‘Rume Sudan’ accessions (T.02724 and T.02744) and the one ‘Barbuk Sudan’ accession (T.02758) were introduced to CATIE in July 1953 from Kenya and Sudan (this southern part is now South Sudan) respectively. All accessions are documented to have been collected by A. S. Thomas from the Boma Plateau, in 1941. Other than this, the details of their collection are non-existent. No further collection or in situ assessment of Arabica from South Sudan has been made since 1941 (Thomas, 1942).
In 2012, as part of a larger team including counterparts from South Sudan, we (SK, APD, TS) undertook fieldwork on the Boma Plateau, which is the only known location of Arabica populations in South Sudan, to collect data of wild and cultivated C. arabica. Based on this survey, the objectives of the present study were: (1) to compare the genetic identity of the C. arabica from the Boma Plateau against available cultivated accessions from Ethiopia (originally of wild origin), landraces from Yemen, and a selection of Arabica cultivars from around the world; and (2) assess the conservation status of C. arabica metapopulation on the Boma Plateau.
Two genetic studies were conducted: (1) study of all the samples from the field survey of C. arabica from the Boma Plateau in South Sudan; and (2) comparison of a subset of South Sudanese samples from Study 1 (South Sudan) with cultivated Ethiopian Arabica (Ethiopian Landraces) and worldwide Arabica cultivar accessions (World Wide Cultivars) from the international coffee collection held at CATIE gene bank, and Yemen landraces from Sana'a University, Yemen.
Fieldwork on the Boma Plateau (Upper Boma, Jonglei State, South Sudan) was conducted 9–12 April 2012. Figure 1 shows the area covered by the expedition, during four days of fieldwork. Collections were made from cultivated and wild plants in local villages (Rumit/Zoch, Kaiwa, Jonglei and Bayen) and two forest locations (Rumit and Ngelecho), respectively. A total of 74 samples were collected and used in this study. Table 1 provides a summary of the cultivated and wild accessions sampled. In addition to the samples collected in South Sudan, two domesticated C. arabica accessions cultivated at Denver Botanic Gardens (DBG) were added to the study as controls. An accession list is provided in Supplementary Table 1. For Study 1, since sampling was from the field collections in South Sudan, we were restricted by the number of trees we were able to access in the forests, leading to unequal sample sizes.
Location coordinates were recorded using a Garmin eTrex Vista NCx and a Holux M-241. Several leaves of each individual plant were collected and placed in plastic bags with silica gel (Chase and Hillis, 1991). Voucher specimens of selected samples were collected in replicates of three, and are housed at the herbaria at the Royal Botanic Gardens, Kew (K) and Missouri Botanical Garden (MO), with the third replicate destined for a suitable location in South Sudan (herbarium not yet established). As collections were being made, individual trees were tagged with numbered aluminum tags nailed to the ground, and with orange tree tape with the corresponding number tied to the truck of the tree. Tagging was done for ease of location and plant identification for subsequent seed collecting expeditions.
For this study, we used 180 samples. Following the identified groupings and nomenclature of Scalabrin et al. (2020), Pruvot-Woehl et al. (2020), and Montagnon et al. (2021), the following categories were considered (Table 2):
- Ethiopian landraces: 116 accessions (all from CATIE)
- Worldwide cultivars: 34 accessions (which included 5 accessions from the South Sudan survey and 29 accessions from CATIE)
- Yemen landraces: 10 accessions
- South Sudan wild populations: 20 accessions
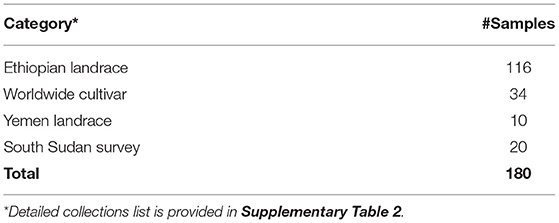
Table 2. Summary of C. arabica samples used in study 2 for comparing South Sudanese samples with worldwide collections.
The accessions from the CATIE collection are predominantly from the germplasm collecting missions undertaken by the Food and Agriculture Organization (FAO, 1968) and the French organization ORSTOM—Office de la Recherche Scientifique et Technique Outre-Mer (now known as Institute de Recherche pour le Développement—IRD), (Charrier, 1978).
This sampling subset was designed to understand the genetic diversity of coffee on the Boma Plateau, which is potentially the last remaining stronghold of Arabica diversity in South Sudan, and how it compares with those in collections worldwide. Hence, the sample sizes are unequal. Based on the genetic assignment from Study 1, the 25 South Sudan subsamples were allocated as 20 to South Sudan Survey and five to Worldwide cultivars (Table 2). The samples used in Study 2 are provided in Table 2; details of these collections are provided in Supplementary Table 2.
Genomic DNA for Study 1 was extracted from 10 mg of silica-dried leaf material using GenCatch™ Plant Genomic DNA Purification kits (Epoch Biolabs) at the Conservation Genetics lab at Denver Botanic Gardens. Slight modifications were made to the extraction protocols: a detailed account of the extraction procedure is described in Krishnan (2011). Extracted DNA was sent to Nevada Genomics, Reno, Nevada, USA, for quantification, optimization, fragment analysis, and scoring using microsatellite markers. Initially, 20 microsatellite markers were selected based on Combes et al. (2000), Rovelli et al. (2000), Poncet et al. (2004), and Cubry et al. (2008). These markers were selected due to their high polymorphism in C. arabica. Of these 20 markers, one did not amplify (M253), and so this marker was dropped from the study. The remaining 19 markers were used (Table 3).
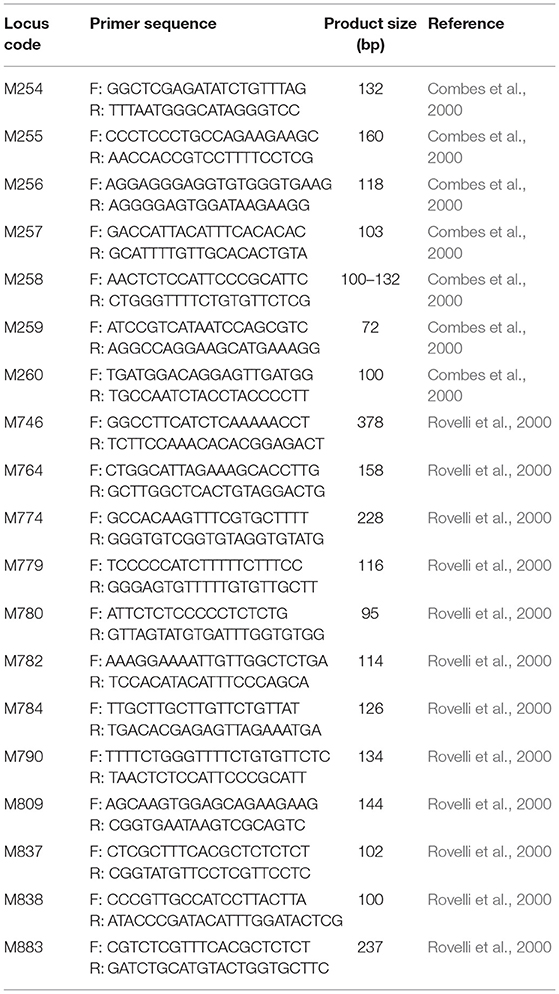
Table 3. List of microsatellite markers used in study 1 with their locus code, primer sequences, and product size.
The DNA was quantified and normalized to 5 ng/μl. PCR amplifications were conducted using an MJ thermocycler. Each 10 μl PCR amplification reaction contained 4 μl of 5 ng/μl genomic DNA, 1 μl Primer Panel mix, and 5 μl Qiagen Multiplex PCR Mix. The 19 microsatellite markers were multiplexed in four panels with a different annealing temperature for each panel. The PCR cycling conditions were as follows: 95°C for 15 min; 95°C for 30 s; specific panel annealing temperature for 45 s; 40 cycles at 72°C for 45 s; 72°C for 30 min. Panel 1 consisted of M780, M790, M259, M254, and M257 with an annealing temperature of 60°C. Panel 2 consisted of M258, M746, M782, M837, and M784 with an annealing temperature of 61°C. Panel 3 consisted of M256, M838, M255, M809, and M883 with an annealing temperature of 63°C. Panel 4 consisted of M260, M779, M774, and M764 with an annealing temperature of 62°C. The samples were run on an Applied Biosystems Prism 3730 DNA Analyzer. The filter set used was G5, which detects the fluorescent dyes 6-FAM, VIC, NED, and PET. The samples were run with the 500 MW size standards labeled with LIZ. The fragment analysis results were scored using GeneMarker 2.2.0 Software by SoftGenetics®.
DNA extraction and SSR marker analysis were performed by the ADNiD laboratory of the Qualtech company in the South of France (http://www.qualtech-groupe.com/en/). Genomic DNA was extracted from ~20 mg of dried tissue using 1 ml of SDS buffer. DNA was then purified with magnetic bead (Agencourt AMPure XP, Beckman Coulter, Brea, California, USA) followed by elution in Tris Edta (TE) buffer. The DNA concentration was estimated with an Enspire spectrofluorimeter (Perkin Elmer) with a bisbenzimide DNA intercalator (Hoechst 33258) and by comparison with known standards of DNA.
Eight SSR primer pairs (Table 4) selected after Combes et al. (2000) and whose wide discrimination power was confirmed by Pruvot-Woehl et al. (2020) were used. This reduced set of markers were demonstrated to be efficient. PCR was performed in a 15 μl final volume comprising 30 ng genomic DNA and 7.5 μl of 2 × PCR buffer (Type-it Microsatellite PCR Kit, Qiagen), 1.0 μM each of forward and reverse primer (10 μM). Amplifications were carried out in thermal cycler (Eppendorf) programmed at 94°C for 5 min for initial denaturation, followed by 94°C for 30 s, annealing temperature depending on the primer used for 30 s and 72°C for 1 min for 35 cycles followed by a final step of extension at 72°C for 5 min. The final holding temperature was 4°C. PCR samples were run on a capillary electrophoresis, ABI 3130XL with an internal standard: GeneScan 500 LIZ size standard (Applied Biosystems). Alleles were scored using GeneMapper v.4.1 software (Applied Biosystems) and then visually inspected.
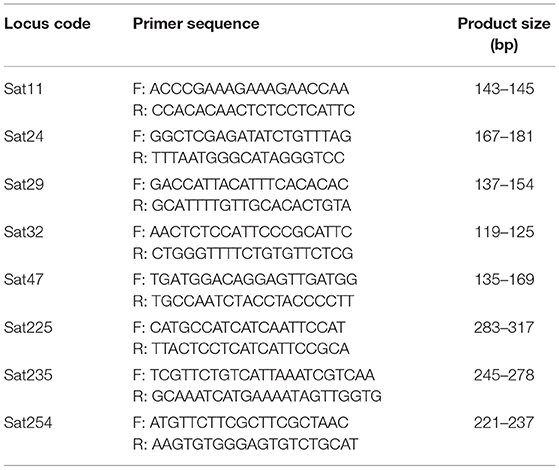
Table 4. List of microsatellite markers used in study 2 with their locus code, primer sequences, and product size.
To identify genetic clusters in populations, DARwin6 software (Perrier and Jacquemoud-Collet, 2006) was used with single data files. The data were entered into a binary matrix as discrete variables, one for presence and zero for absence of the character. The dissimilarity matrix was calculated using Dice Index and was the basis for the execution of the Principal Coordinates Analysis (PCoA). PCoA provides an overall representation of the diversity (Perrier and Jacquemoud-Collet, 2006) and produces graphical representations on Euclidean plans which preserve the distances between units.
All the genotypes were scored for the presence and absence of the SSR bands. The data were entered into a binary matrix as discrete variables, one for presence and zero for absence of the character and this data matrix was subjected to further analysis. Discriminant Analysis of Principal Components (DAPC) was performed to identify and describe clusters of genetically related individuals using the adegenet package version 2.1.3. The optimal number of clusters was determined using the function “find.clusters” which applies successive K-means clustering. The bayesian information criterion (BIC) was visually examined to define the number of clusters. Then, the “dapc” function was applied. The graphs were designed using ggplot2 package Version 3.3.2. PCA-based clustering was also done using the subroutine EIGEN.
The aims of the conservation analyses were to determine the current extent of C. arabica habitat on the Boma Plateau, compared to a potential maximum, and measure recent forest loss (2000–2020). The suitable forest types for C. arabica on the Boma Plateau are the same as that in neighboring Ethiopia, i.e., Moist Afromontane Forest (MAF) and Transitional Rain Forest (TRF), [found at 650–2,600 m (450–3,000 m including extremes)], although coffee is mostly confined to 1,200–2,200 m (Moat et al., 2017). To achieve our objectives for this part of the study we used Google Earth Engine (Gorelick et al., 2017) to query high-resolution elevation data (Jarvis et al., 2008), tree cover for 2000 and global forest change from 2000–2020 (Hansen et al., 2013). For our maximum forest cover we used a conservative minimum elevation threshold of 1,200 m [as per our observations and those of Thomas (1942)], for forest cover at the year 2000, with a tree cover of 50% or more. Forest cover for 2020 was calculated by removing the area of deforestation as identified by the updated dataset from Hansen et al. (2013).
Cluster analysis following PCoA revealed three population clusters (Table 5; Figure 2). The first cluster consisted mainly of plants growing wild in the forests of Ngelecho while the second cluster consisted of plants growing wild in the forests of Rumit. The third cluster consisted of cultivated plants from DBG (control) together with plants cultivated at Bayen village. Two samples from Ngelecho wild population fell into cluster 2 and two samples from the Rumit wild population fell into cluster 1. Plants cultivated in the village of Rumit (identified as two different populations based on distance between populations, Table 1; Figure 2) were split between clusters 1 and 2, with 22 plants falling in cluster 1 (Ngelecho Wild) and 20 plants falling in cluster 2 (Rumit Wild). The one plant cultivated at Kaiwa village clustered with Ngelecho Wild (cluster 1) and the one plant from Jonglei village clustered with Rumit Wild (cluster 2). Cluster 3 identified as plants belonging to cultivated and introduced variants consisted of the three plants collected at Bayen village and the two plants from the collections at Denver Botanic Gardens (DBG). Since the Bayen population was closely related to cultivated plants, for the subsequent study (Study 2), these five plants (Bayen and DBG) were allocated to the Worldwide Cultivar category.
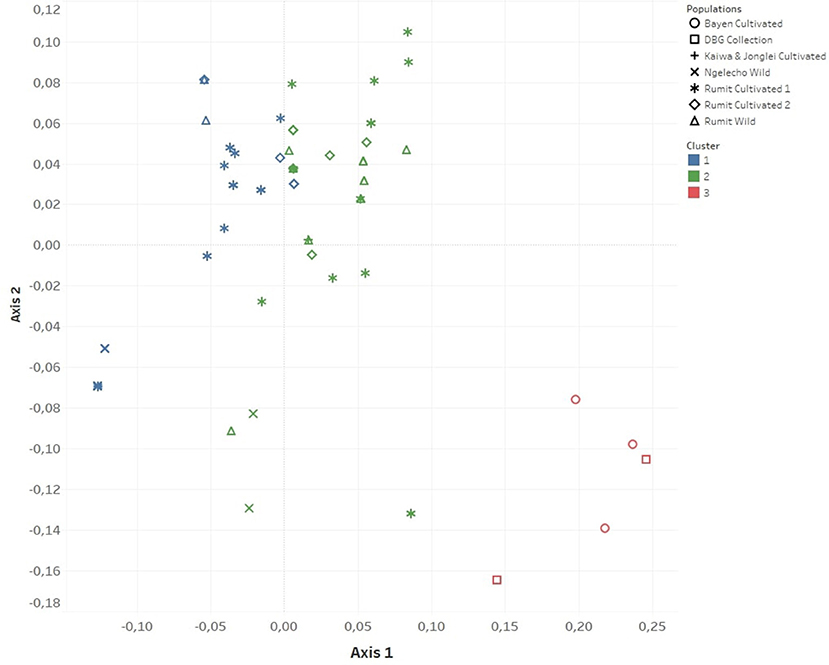
Figure 2. Clustering of South Sudan survey populations (Study 1) on a graph on the first two axes of the Principal Coordinates Analysis (PCoA) based on dissimilarity matrix.
The 180 samples used in this study were partitioned into six clusters after DAPC. One group was only made of South Sudan accessions, which we named “South Sudan.” Three clusters were made of Ethiopian accessions only; we named them EL1, EL2, and EL3. Only Worldwide Cultivar SL-06 was in EL3 cluster. Two other clusters included some Ethiopian accessions, some Yemen Landrace and some Worldwide cultivars. We named these clusters WWC1 and WWC2 (Table 6; Figure 3).
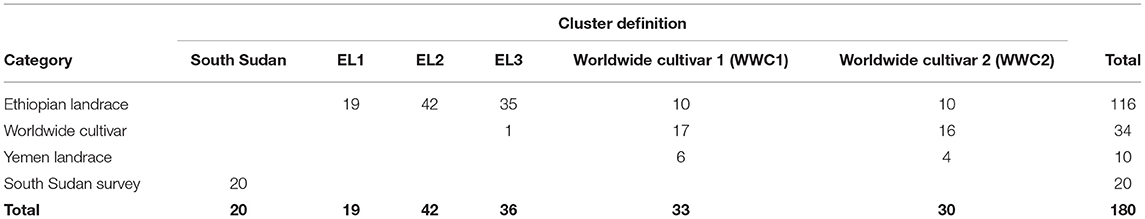
Table 6. Clustering of populations identified as Ethiopian landrace, worldwide cultivar, Yemen landrace and South Sudan survey.
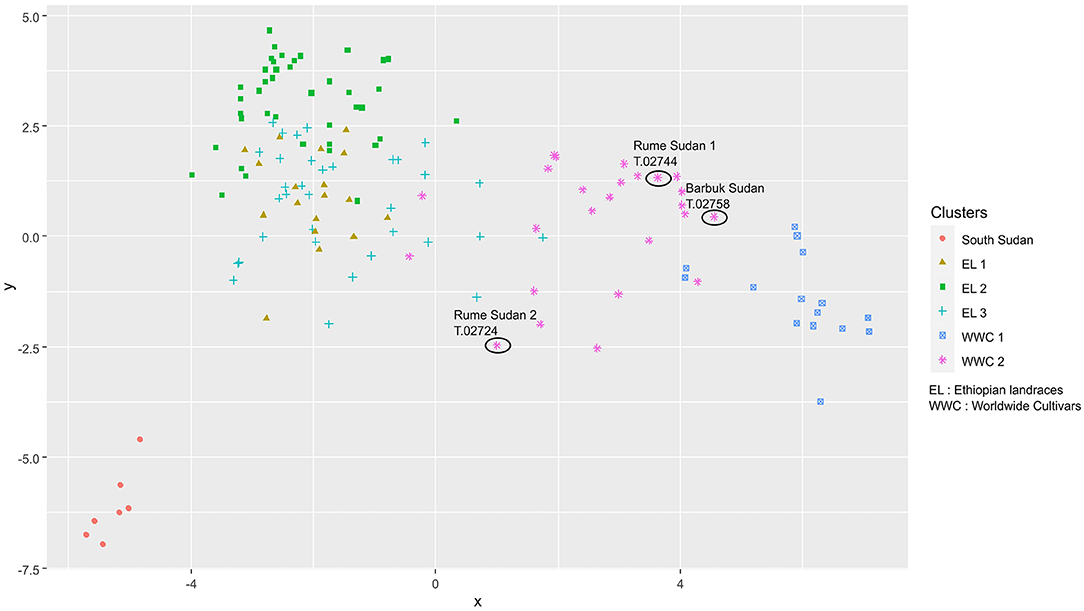
Figure 3. Clustering of genetically related individuals using Discriminant Analysis of Principal Components (DAPC) analysis in Study 2.
The South Sudan group had one unique allele (allele 154 of Sat24). Another allele (allele 127 of Sat 47) had a frequency of 1.00 in the South Sudan group and a frequency of 0.00 in all other groups except WWC2 where the frequency was 0.10. In WWC2, this allele was borne by 3 accessions: one of the “Rume Sudan” accessions (CATIE code: T.02724), SL-14 (T.02747) and SL-17 (T.02745). The two other supposed South Sudanese accessions from CATIE did not bear any of these two alleles and were also part of the WWC2 cluster: “Rume Sudan” (T.02744) and “Barbuk Sudan” (T.02758), (Data not shown), (Figure 3; Supplementary Table 2).
The total suitable forest cover for C. arabica in the Boma Plateau area for 2020 was 15.3 km2, compared to 16.2 km2 in 2000, representing a forest cover loss of 5.8% over 20 years (Figure 1). The maximum potential forest cover suitable for C. arabica (i.e., humid forest above 1,200 m) for the Boma Plateau area was calculated to be 97 km2, which would represent a potential forest loss of 81.7 km2, i.e., a loss of 84.2%. The wild populations of C. arabica on the Boma Plateau are 88 km from the nearest metapopulation in Ethiopia (around Geba); satellite data also show that this forested area is also being rapidly lost (Hansen et al., 2013).
If we use these data for a regional/country-level, by applying International Union for the Conservation of Nature (IUCN) Red List Categories and Criteria (IUCN Standards Petitions Subcommittee, 2017), the Area of Occurrence (AOO) of 15.3 km2 would place it into the Endangered category (EN), approaching the Critically Endangered category (CR). However, under climate change (Davis et al., 2012; Moat et al., 2019) the Boma Plateau area will lose climate suitability for C. arabica by 2010–2039. This would push a regional assessment to CR using a conservative generation length of under 6 years (Moat et al., 2019 recommend a generation length of 21 years for C. arabica).
The unique outcome of this study is the identification of the metapopulation of wild Arabica on the Boma Plateau (South Sudan) as a genetically separate entity, distinct from Ethiopian landraces, global Worldwide Cultivars and Yemen landraces. Thomas (1942) suggested that the C. arabica growing on the Boma Plateau (at Barbuk, Rume and Nelichu) could have been brought for cultivation from Abyssinia (present-day Ethiopia). He reasoned that the Boma Plateau and the adjacent wild coffee forests of the Ethiopian Highlands were separated by low, open woodland (mostly Combretum-Terminalia Woodland and Wooded Grassland (Friis et al., 2010) unfavorable for the growth of C. arabica (Davis et al., 2012). However, the distance between the Boma Plateau and suitable forested areas (even based on current forest cover and climate) is only 80 km, and pre-Anthropocene there would have been numerous suitable forest patch “stepping stones” between Boma and the Ethiopian Highlands. These observations argue for a natural distribution across the Ethiopian Highlands and a small area of eastern South Sudan (including the Boma Plateau). There is certainly some disjunction between the Boma Plateau and Ethiopian populations, but this also is the case with the Ethiopian Highlands (Davis et al., 2018).
The two wild populations collected from Boma Plateau, Ngelecho Wild and Rumit Wild were distinct from each other, forming separate clusters. The plants cultivated in nearby villages were a mix of both populations. Thomas (1942) documented young coffee plants growing beside the paths just inside the edge of the forest and noted that these probably arose from fruits dropped by the Kichepo from harvests made in the forests. This movement of harvested fruits from the forests could account for the presence of both wild populations among the two cultivated populations sampled.
Our current study shows the coffee growing in the Boma Plateau of South Sudan to be genetically distinct from that collected in Ethiopia by the FAO and ORSTOM missions in the 1960s and those in cultivation, and that there is strong support for South Sudan as the center of origin of Arabica. This extension of the genetic diversity for Arabica is important, given that until now Ethiopia was the only established origin of diversity, and that the genetic diversity of cultivated Arabica outside Ethiopia is desperately low (Scalabrin et al., 2020). Other key outcomes are given in the following narrative.
The CATIE collection houses three accessions recorded as being collected in Sudan by Thomas (1942): two ‘Rume Sudan’ (T.02724 and T.02744) and one ‘Barbuk Sudan’ (T.02758) accessions. Genebank records indicate that the ‘Rume Sudan’ and ‘Barbuk Sudan’ accessions were introduced to CATIE in July 1953 from Kenya. The Scott Agricultural Laboratories in Kenya and Lyamungo Research Station in Tanganyika became the main centers for coffee breeding in East Africa (Van der Vossen, 1985), both of which received the seed of ‘Rume Sudan’ and ‘Barbuk Sudan’ from the Thomas expedition in 1941. Passport data indicate the two ‘Rume Sudan’ accessions collected in Rume region of Boma Plateau and the ‘Barbuk Sudan’ collected in the Barbuk region of Boma Plateau in 1941 by A. S. Thomas. There is also documentation of ‘Rume’ and ‘Barbuk Sudan’ accessions in the USDA collections dating to 1953 at Glendale, Maryland and 1957 at Puerto Rico, though these collections do not exist at the current time (M. Winterstein, personal communication). The two ‘Rume Sudan’ and one ‘Barbuk Sudan’ cultivars from the CATIE collection however did not group with the South Sudan samples. These accessions clustered with the Worldwide Cultivar group (Figure 3). However, ‘Rume Sudan’ (T.02724) possesses one of two alleles that are specific to the South Sudan group. Hence, this ‘Rume Sudan’ accession may have originated from a true ‘Rume Sudan’ population but cross-pollinated with other Arabica accessions in germplasm collections where they were held before being introduced to CATIE gene bank. The other two accessions, one ‘Rume Sudan’ and one ‘Barbuk Sudan’ have either been more diluted through cross pollination or perhaps mislabeled in the CATIE collection or previous germplasm collection. There is also the possibility that the ‘Rume Sudan’ and ‘Barbuk Sudan’ currently in cultivation as well as in global gene banks are possibly genetically contaminated from growing in the Scott Agricultural Laboratories research stations with other cultivars, or plants distributed globally from Kenya and Tanzania could have been mislabeled. Hence, future research needs to be undertaken to genotype all Rume and Barbuk Sudan in gene banks and farmers' fields to track the original collections made by Thomas. Two other accessions of the CATIE germplasm collection had one of the two specific alleles to the South Sudan group: SL-14 (T.02747) and SL-17 (T.02745). Interestingly, these two accessions represented one specific mother population (named ‘SL-17’) in the study by Montagnon et al. (2021). The accessions that are part of that mother population do not seem to have followed the Yemeni domestication route. It might well be that this SL-17 mother population is somehow related to South Sudanese accessions.
There were no clear geographical repartition patterns between the three Ethiopian groups (EL1, EL2 and EL3). The Ethiopian accessions used in our study were originally from the FAO (1968) and ORSTOM (Charrier, 1978) coffee surveys, which mostly covered coffee in cultivation and wild coffee forest areas to the west of the Rift Valley, in the South West and Rift coffee zones (Davis et al., 2018). Furthermore, the Ethiopian accessions in our study are part of the core collection established by World Coffee Research (WCR) and CATIE. Scalabrin et al. (2020) included the more than 500 Ethiopian accessions held by CATIE in their study, using SNPs, but only found a weak population structure with a slight West to East geographical cline. Despite this large sample size, no studies have yet comprehensively sampled the entire geographical range of wild and cultivated coffee areas in Ethiopia. Notable exceptions, where either no or very few accessions have been sampled, include the coffee areas west of the Rift Valley, in Amhara, Wellega, northern Illubabor, and Bench Maji; and East of the Rift Valley, in Sidamo, Bale, Central Eastern Highlands, Arsi, West Hararge and East Hararage (Davis et al., 2018). The wild Arabica populations in the forest of western Wellega, northern Illubabor, western Bench Maji and Bale (Davis et al., 2018) will no doubt be found to represent key centers of diversity using molecular data and may help to elucidate the genetic-geographical structure of wild Arabica in Ethiopia. Consistent with other studies (Pruvot-Woehl et al., 2020; Scalabrin et al., 2020; Montagnon et al., 2021), SL-06 was found to be included in an Ethiopian cluster. We therefore suggest to include this accession in the Ethiopian Landrace accessions, instead of in the Worldwide Cultivars. There was no clear interpretation of the two Worldwide Cultivars genetic groups. This is not surprising, however, as only by adding more diverse samples from Yemen were Montagnon et al. (2021) able to give a complete description of the genetic structure of the worldwide Arabica coffee cultivars.
Former studies (Anthony et al., 2002; da Silva et al., 2019; Benti et al., 2020) had proven the efficiency of SSR markers to study the genetic diversity of Arabica. In our study, we confirm that the reduced set of SSR markers established by Pruvot-Woehl et al. (2020) are efficient to describe a new genetic group in Yemen (Montagnon et al., 2021) is again efficient to describe a new genetic group in this species: the South Sudan group. Scalabrin et al. (2020) found the vast majority of the SNPs found were of low-frequency alleles. Recently, apparently more workable SNPs were published (Zhang et al., 2021); testing these SNPs on the South Sudan samples might be a good way to confirm their accuracy.
The three cultivated trees from Bayen village did not cluster with the other two wild South Sudanese clusters. Instead, they clustered with the cultivated trees from the collections at Denver Botanic Gardens, suggesting that these were more closely related to cultivated genotypes. In the second study comparing the South Sudanese survey collections with Ethiopian, Yemeni and Worldwide Cultivars, two of the Bayen trees were identified within the WWC genetic groups. The inference made from this is that the Bayen Cultivated population is of a different origin, and probably representing introduced cultivated material. Discussion with several individuals from the local village in Upper Boma revealed that in the late 1940s, the British had attempted coffee cultivation in this area and may have introduced foreign germplasm at that time.
During the 2012 expedition, even though we did not observe coffee being harvested and used commercially, the observation of cultivated trees in Rumit, Kaiwa and Jonglei villages clearly point to the use of coffee by the villagers. The wild coffee from the forests of Rumit and Ngelecho formed two genetic clusters with limited admixture (Table 5). The cultivated coffee in Rumit village consisted of a mix of Rumit wild and Ngelecho wild genotypes indicating collections made from both forests by the local villagers. The single tree in Kaiwa village had been collected from the Ngelecho forest whereas the single tree in Jonglei village has been collected from the Rumit forest (Supplementary Table 2).
We demonstrate that the humid forests (MAF and TRF; Friis et al., 2010) of the Boma Plateau, where C. arabica exists as a wild plant, have experienced large-scale deforestation. Even in the 1940s it was noted (Thomas, 1942) that the natural forests of Boma Plateau had experienced a long history of human disturbance. Compared to what would be expected prior to human intervention (potential forest cover) we estimate an 81.7 km2 (84.2%) loss of forest cover, and over the last 20 years an 0.9 km2 (5.8%) loss of forest cover. However, during our field survey in 2012 we observed that even in areas with more than 50% forest cover, the understory of the forest is largely unsuitable for the existence of spontaneous wild C. arabica in many places, due to understory clearance, particularly on the forested plateau. At Barbuk (Figure 1) the understory vegetation has been largely removed or degraded, with evidence of widespread human disturbance, including ephemeral habitation. Some of the canopy trees were in poor health, especially the emergent canopy Manilkara butugi (Sapotaceae), with some standing dead trees. At the edges of Barbuk, on the escarpment (western edge of plateau; Figure 1), there was evidence of fire encroachment (i.e. a charcoal layer under the leaf litter), due to the seasonal burning of the surrounding Combretum-Terminalia woodland/grassland. At Barbuk we found very few wild C. arabica trees, which were of limited age classes (only small trees), and there were few or no seedlings. Our observations are thus in stark contrast to those made by Thomas (1942). It is clear from Thomas (1942) that when he visited Barbuk the forest was in good condition, as he states: “At Barbuk there was a large area of closed forest at an altitude of about 4,700 ft. (1,350 m). The forest was dense, with an evergreen canopy of larger trees”… “Lianes were abundant. There was a thick undergrowth of shrubs ….” Thomas (1942) observed wild C. arabica as being “locally frequent,” with the presence of many mature trees, and plentiful seedlings. The difference between the two surveys are no doubt due to the change in forest health at this locality, but also perhaps because nearby villagers collect C. arabica coffee leaves for the production of coffee leaf tea (Campa et al., 2012).
The forested area at Rume (Figure 1) was already largely deforested when visited by Thomas in 1941 (Thomas, 1942). He states: “Until 4 or 5 years ago the valley had been with forest, but since the Italian invasion of Abyssinia much clearance and settlement had taken place. At the time of our visit many trees remained; some had fallen but had not yet rotted; others were still alive”… “After clearing, the land had been planted with maize which, together with considerable amounts of small-leaved tobacco, covered the valley floor.”… “Standing out dark among the ripening maize were scattered bushes of Coffea arabica, either single or in groups. We were told that none of these bushes had been planted and that they were relicts of the original undergrowth of the forest, retained when the other species were cut and burnt.” Thomas reports a point elevation for Rume of 4,100 ft. (1,250 m), which is supported by satellite data (GoogleEarth®), and thus generally of lower elevation than Barbuk (see above). The generally flatter topology or Rume may have either made it easier for clearance or made it more suitable for agriculture, and probably both. When visiting in 2012 there was no remaining forest at Rume, although we did not examine the area for remnant C. arabica trees within either the cultivated areas or small patches of secondary vegetation. Satellite imagery confirms these observations (Figure 1). C. arabica is intolerant of disturbance (Davis et al., 2012; Moat et al., 2019) and after forest removal rapidly declines and disappears.
Overall, observations made in the most suitable areas on the Boma Plateau indicated that the C. arabica populations are in poor health (loss of aged individuals, meager population density, zero or minimal seedling recruitment, low ratio of flower bud development) compared to 70 years ago (Thomas, 1942).
It is unlikely that any C. arabica plants exist now at Rume. If all cultivated accessions of C. arabica ‘Rume’ are compromised, as found in the samples we used in our SSR survey, the genetic diversity from Rume may no longer exist in its original form, if at all. The net result of all anthropogenic influences has been the reduction in the range, density and health of C. arabica populations on the Boma Plateau, which has no doubt resulted in a loss of their genetic diversity.
A country-level conservation assessment for C. arabica in South Sudan, when applying the Red List Categories and Criteria (IUCN Standards Petitions Subcommittee, 2017), returns a threat level of Endangered (EN), with the major threats being deforestation and understory clearance. When factoring in climate change (Moat et al., 2019) the threat level would rise to Critically Endangered (CR). Climate change projection analyses show that climatic suitability for C. arabica on the Boma Plateau (and therefore this species in South Sudan) would cease to exist between 2010 and 2039 (Davis et al., 2012; Moat et al., 2019).
This study reveals that in addition to Ethiopia, the Boma Plateau of South Sudan is a center of origin of C. arabica, and supports the assumption that it is part of the natural range of this species (Thomas, 1942; Davis et al., 2012). During his 1941 expedition, Thomas (1942) observed significant phenotypic variation in the coffee plants as well as a lack of infection by coffee leaf rust (Thomas, 1942). Partial resistance to coffee leaf rust in Boma Arabica has since been documented (Van der Vossen, 1985) and South Sudan Arabica germplasm has been used in breeding programs (Marie et al., 2020). Its sensory qualities are appreciated by the specialty coffee sector. These attributes provide useful resources for breeding and utilization in crop improvement programs for Arabica coffee. Our 2012 expedition was able to verify that Arabica coffee still exists in the forested area of Barbuk, where it was found growing wild by Thomas (1942), but also show that the populations there today are in poor health. Field observation in 2012 and satellite imagery (Figure 1) show that the second main location on Boma Plateau, i.e., Rume, is now in a deforested state, although even in 1941 this area was already mostly deforested (Thomas, 1942). Over the last several decades the impact of human activities, and possibly climate change, has had a major impact on the extent and health of the Boma Plateau populations. Given that the Boma populations represent a distinct genetic entity for Arabica diversity, and that there is still substantial genetic diversity within the Boma metapopulation, it is imperative that all attempts be made to conserve this diversity.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
SK, AD, and TS conducted the expedition to South Sudan. SK conducted the genetic diversity studies of South Sudan samples, following which SP-W and CM compared the genetic diversity of South Sudan samples with accessions from CATIE and Yemen provided by WS and AA respectively. JM and AD conducted the conservation assessment. SK took the lead on writing the manuscript with participation from SP-W, CM, and AD. All authors reviewed the manuscript.
The 2012 South Sudan expedition was funded by USAID under the cooperative Agreement Number AID-668-A-11-00001 with the Norman Borlaug Institute for International Agriculture of the Texas A&M University titled Support to Dr. John Garang Memorial University of Science and Technology: A Consortium for Development.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We also acknowledge the following participants who were part of the 2012 expedition: Emma Bladyka (Sage), Specialty Coffee Association (now with Coffee Quality Institute). Lindsey Bolger, Green Mountain Coffee (now Keurig Dr Pepper, Inc.). Solomon Kuol Deng, State Ministry of Agriculture and Forestry, Jonglei/Bor. Richard Yona, John Garang Memorial University of Science and Technology. Majok Ayuen, John Garang Memorial University of Science and Technology. Thon Nyok Dor, John Garang Memorial University of Science and Technology.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fsufs.2021.761611/full#supplementary-material
Anthony, F., Combes, M. C., Astorga, C., Bertrand, B., Graziosi, G., and Lashermes, P. (2002). The origin of cultivated Coffea arabica L. varieties revealed by AFLP and SSR markers. Theor. Appl. Genet. 104, 894–900. doi: 10.1007/s00122-001-0798-8
Avelino, J., Cristancho, M., Georgiou, S., Imbach, P., Aguilar, L., Bornemann, G., et al. (2015). The coffee rust crises in Colombia and Central America (2008–2013): impacts, plausible causes and proposed solutions. Food Sec. 7, 303–321. doi: 10.1007/s12571-015-0446-9
Benti, T., Gebre, E., Tesfaye, K., Berecha, G., Lashermes, P., Kyallo, M., et al. (2020). Genetic diversity among commercial arabica coffee (Coffea arabica L.) varieties in Ethiopia using simple sequence repeat markers. J. Crop Improv. 35, 1-22. doi: 10.1080/15427528.2020.1803169
Bunn, C., Läderach, P., Ovalle Rivera, O., and Kirschke, D. (2015). A bitter cup: climate change profile of global production of Arabica and Robusta coffee. Clim. Change 129, 89–101. doi: 10.1007/s10584-014-1306-x
Campa, C., Mondolot, L., Rakotondravao, A., Bidel, L. P. R., Gargadennec, A., Couturon, E., et al. (2012). A survey of mangiferin and hydroxycinnamic acid ester accumulation in coffee (Coffea) leaves: biological implications and uses. Ann. Bot. 110, 595–613. doi: 10.1093/aob/mcs119
Charrier, A. (1978). Etude de la structure et de la variabilité génétique des caféiers: Résultats des études et des expérimentations réalisées au Cameroun, en Côte d'Ivoire et à Madagascar sur l'espèce Coffea arabica L. collectée en Ethiopie par une mission Orstom en 1966. Bulletin IFCC n° 14, FRA.
Chase, M. W., and Hillis, H. H. (1991). Silica gel: an ideal material for field preservation of leaf samples for DNA studies. Taxon 40, 215–220. doi: 10.2307/1222975
Combes, M. C., Andrzejewski, S., Anthony, F., Bertrand, B., Rovelli, P., Graziosi, G., et al. (2000). Characterization of microsatellite loci in Coffea arabica and related coffee species. Mol. Ecol. 9, 1178–1180. doi: 10.1046/j.1365-294x.2000.00954-5.x
Cubry, P., Musoli, P., Legnate, H., Pot, D., de Bellis, F., Poncet, V., et al. (2008). Diversity in coffee assessed with SSR markers: structure of the genus Coffea and perspectives for breeding. Genome 51, 50–63. doi: 10.1139/G07-096
da Silva, B. S. R., Sant'Ana, G. C., Chaves, C. L., Androcioli, L. G., Ferreira, R. V., Sera, G. H., et al. (2019). Population structure and genetic relationships between Ethiopian and Brazilian Coffea arabica genotypes revealed by SSR markers. Genetica 147, 205–216 doi: 10.1007/s10709-019-00064-4
Davis, A. P., Chadburn, H., Moat, J., O'Sullivan, R., Hargreaves, S., and Nic Lughadha, E. (2019). High extinction risk for wild coffee species and implications for coffee sector sustainability. Sci. Adv. 5:eaav3473. doi: 10.1126/sciadv.aav3473
Davis, A. P., Gole, T. W., Baena, S., and Moat, J. (2012). The impact of climate change on indigenous Arabica Coffee (Coffea arabica): predicting future trends and identifying priorities. PLoS ONE 7:e47981. doi: 10.1371/journal.pone.0047981
Davis, A. P., Mieulet, D., Moat, J., Sarmu, D., and Haggar, J. (2021). Arabica-like flavour in a heat-tolerant wild coffee species. Nat. Plants 7, 413–418. doi: 10.1038/s41477-021-00891-4
Davis, A. P., Wilkinson, T., Challa, Z. K., Williams, J., Baena, S., Gole, T. W., et al. (2018). Coffee Atlas of Ethiopia. Kew: Royal Botanic Gardens.
Friis, I., Demissew, S., and Breugel, P. V. (2010). Atlas of the potential vegetation of Ethiopia. Biol. Skrif. 58, 1–307. doi: 10.1093/aob/mcq242
Gorelick, N., Hancher, M., Dixon, M., Ilyushchenko, S., Thau, D., and Moore, R. (2017). Google Earth Engine: planetary-scale geospatial analysis for everyone. Remote Sens. Environ. 202, 18–27. doi: 10.1016/j.rse.2017.06.031
Hansen, M. C., Potapov, P. V., Moore, R., Hancher, M., Turubanova, S. A., Tyukavina, A., et al. (2013). High-resolution global maps of 21st-century forest cover change. Science 342, 850–853. doi: 10.1126/science.1244693
International Coffee Organization (ICO) (2021). Coffee Production by Exporting Countries. Available online at: http://www.ico.org/prices/po-production.pdf (accessed January 1, 2021).
IUCN Standards Petitions Subcommittee (2017). Guidelines for Using the IUCN Red List Categories and Criteria. Version 13. Prepared by the Standards and Petitions Subcommittee. Available online at: www.iucnredlist.org/documents/RedListGuidelines.pdf (accessed January 1, 2021).
Jarvis, A., Reuter, H. I., Nelson, A., and Guevara, E. (2008). Hole-Filled SRTM for the Globe Version 4. 2008. Available online at: http://srtm.csi.cgiar.org (accessed June 28, 2021).
Krishnan, S. (2011). Conservation genetics of wild coffee (Coffea spp.) in Madagascar (dissertation). University of Colorado, Boulder, CO, USA.
Krishnan, S., Matsumoto, T., Nagai, C., Falconer, J., Shriner, S., Long, J., et al. (2021). Vulnerability of coffee (Coffea spp.) genetic resources in the United States. Genet. Resour. Crop Evol. 68, 2691-2710. doi: 10.1007/s10722-021-01217-1
Marie, L., Abdallah, C., Campa, C., Courtel, P., Bordeaux, M., Navarini, L., et al. (2020). G × E interactions on yield and quality in Coffea arabica: new F1 hybrids outperform American cultivars. Euphytica. 216:78. doi: 10.1007/s10681-020-02608-8
Meyer, F. G. (1965). Notes on wild Coffea arabica from southwestern Ethiopia, with some historical considerations. Econ. Bot. 19, 136–151. doi: 10.1007/BF02862825
Moat, J., Gole, T. W., and Davis, A. P. (2019). Least concern to endangered: applying climate change projections profoundly influences the extinction risk assessment for wild Arabica coffee. Glob. Chang. Biol. 25, 390–403. doi: 10.1111/gcb.14341
Moat, J., Williams, J., Baena, S., Wilkinson, T., Gole, T. W., Challa, Z. K., et al. (2017). Resilience potential of the Ethiopian coffee sector under climate change. Nat. Plants. 3:17081. doi: 10.1038/nplants.2017.81
Montagnon, C., Mahyoub, A., Solano, W., and Sheibani, F. (2021). Unveiling a unique genetic diversity of cultivated Coffea arabica in its main domestication center: Yemen. Genet. Resour. Crop Evol. 68, 2411–2422. doi: 10.1007/s10722-021-01139-y
Montagnon, C., Marraccini, P., and Bertrand, B. (2019). “Breeding for coffee quality,” in Specialty Coffee-Managing Quality, 2nd Edn, eds T. Oberthur, P. Laderach, H. A. J. Pohlan, J. H. Cock, and P. L. Tan (Innsbruck: Cropster), 109–143.
Perrier, X., and Jacquemoud-Collet, J. P. (2006). DARwin Software. Available online at: https://darwin.cirad.fr/ (accessed January 1, 2021).
Poncet, V., Hamon, P., Minier, J., Carasco, C., Hamon, S., and Noirot, M. (2004). SSR cross-amplification and variation within coffee trees (Coffea spp.). Genome 47, 1071–1081. doi: 10.1139/g04-064
Pruvot-Woehl, S., Krishnan, S., Solano, W., Schilling, T., Toniutti, L., Bertrand, B., et al. (2020). Authentication of Coffea arabica varieties through DNA fingerprinting and its significance for the coffee sector. J. AOAC Int. 103, 325–334. doi: 10.1093/jaocint/qsz003
Rovelli, P., Mettulio, R., Anthony, F., Anzueto, F., Lashermes, P., and Graziosi, G. (2000). “Microsatellites in Coffea arabica L.,” in Coffee Biotechnology and Quality, eds. T. Sera, C. R. Soccol, A. Pandey, and S. Roussos (Dordrecht: Kluwer Academic Publishers), 123–133. doi: 10.1007/978-94-017-1068-8_9
Scalabrin, S., Toniutti, L., Di Gaspero, G., Scaglione, D., Magris, G., Vidotto, M., et al. (2020). A single polyploidization event at the origin of the tetraploid genome of Coffea arabica is responsible for the extremely low genetic variation in wild and cultivated germplasm. Sci. Rep. 10:4642. doi: 10.1038/s41598-020-61216-7
Thomas, A. S. (1942). The wild Arabica coffee on the Boma Plateau, Angle-Egyptian Sudan. Empire J. Exp. Agr. 10, 207–212.
Van der Vossen, H. A. (1985). “Coffee selection and breeding,” in Coffee Botany, Biochemistry and Production of Bean and Beverage, eds. M. N. Clifford, and C. K. Willson (London: Croom Helm), 48–96. doi: 10.1007/978-1-4615-6657-1_3
Van der Vossen, H. A. M. (2017). “Developing varieties of Arabica coffee,” in Achieving Sustainable Cultivation of Coffee, ed P. Lashermes (Cambridge: Burleigh Dodds Science Publishing), 83–113. doi: 10.19103/AS.2017.0022.05
Keywords: Boma Plateau, center of origin, coffee, conservation, genetic diversity, simple sequence repeats (SSR), South Sudan
Citation: Krishnan S, Pruvot-Woehl S, Davis AP, Schilling T, Moat J, Solano W, Al Hakimi A and Montagnon C (2021) Validating South Sudan as a Center of Origin for Coffea arabica: Implications for Conservation and Coffee Crop Improvement. Front. Sustain. Food Syst. 5:761611. doi: 10.3389/fsufs.2021.761611
Received: 20 August 2021; Accepted: 20 October 2021;
Published: 11 November 2021.
Edited by:
Matteo Balderacchi, Independent Researcher, Piacenza, ItalyReviewed by:
Riza Putranto, Indonesian Research Institute for Biotechnology and Bioindustry (IRIBB), IndonesiaCopyright © 2021 Krishnan, Pruvot-Woehl, Davis, Schilling, Moat, Solano, Al Hakimi and Montagnon. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sarada Krishnan, c2FyYWRha3Jpc2huYW5AbXNuLmNvbQ==
†ORCID: Sarada Krishnan orcid.org/0000-0002-2491-4160
Aaron P. Davis orcid.org/0000-0001-9213-4353
Justin Moat orcid.org/0000-0002-5513-3615
Christophe Montagnon orcid.org/0000-0002-6967-9256
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.