- 1Hunan Provincial Key Laboratory for Biology and Control of Plant Diseases and Insect Pests, Hunan Agricultural University, Changsha, China
- 2College of Plant Protection, Hunan Agricultural University, Changsha, China
GRAS transcription factors are widely present in the plant kingdom and play important roles in regulating multiple plant physiological processes. Brachypodium distachyon is a model for grasses for researching plant-pathogen interactions. However, little is known about the BdGRAS family genes involved in plant response to biotic stress. In this study, we identified 63 genes of the GRAS family in B. distachyon. The phylogenetic analysis showed that BdGRAS genes were divided into ten subfamilies and unevenly distributed on five chromosomes. qRT-PCR results showed that the BdGRAS family genes were involved in the growth and development of B. distachyon. Moreover, the expression of the HAM subfamily genes of BdGRAS changed during the interaction between B. distachyon and Magnaporthe oryzae. Interestingly, BdGRAS31 in the HAM subfamily was regulated by miR171 after inoculation with M. oryzae. These results provide insight into the potential functions of the BdGRAS family in disease resistance.
Introduction
The GRAS protein family is an important transcription factor family named after the function of three members: Gibberellic acid insensitive (GAI), Repressor of GAI (RGA), and Scarecrow (SCR; Chen et al., 2019). Genome-wide analysis of the family showed that GRAS genes are also widely distributed in the plant kingdom (Tian et al., 2004). GRAS proteins typically consist of 400–700 amino acids. The sequences at the C-terminus are highly conserved and are divided into five conserved motifs: LHRI, VHIID, LHRII, PFYRE, and SAW (Pysh et al., 1999; Tian et al., 2004). VHIID, PFYRE, and SAW are important for maintaining the structural stability and function of GRAS proteins (Itoh et al., 2002; Smit et al., 2005; Hofmann, 2016; Li et al., 2016); the VHIID sequence can interact with other proteins (Gao et al., 2004). LHRI and VHIID have abundant leucine repeats, which affect protein dimerization. The N-terminal sequence is highly variable and can be used as bait in molecular research activities (Tian et al., 2004). Some GRAS proteins contain the DELLA domain, which plays important roles in gibberellin signaling (Silverstone et al., 1998).
The GRAS family is widely involved in regulating plant growth and development, including gibberellin signaling (Peng et al., 1997; Ikeda et al., 2001; Hirsch and Oldroyd, 2009), formation of axillary meristems (Greb et al., 2003; Li et al., 2006), root radial patterning (Helariutta et al., 2000), photosensitive signal transduction (Bolle et al., 2000), and male gametogenesis (Morohashi et al., 2003). For example, two GRAS proteins are required for root rot formation in legumes (Kaló et al., 2005; Heckmann et al., 2006). Moreover, the GRAS family has been shown to participate in the hormone signaling pathway that regulates plant growth and development (Davière and Achard, 2016; Van De Velde et al., 2017). DELLA proteins also participate in the signaling pathways of auxin, brassinosteroids (BRs), abscisic acid (ABA), and ethylene (Chen et al., 2019). AtSCL3 (an Arabidopsis thaliana GRAS gene) mediates the gibberellic acid pathway by attenuating the DELLA repressors during root development (Heo et al., 2011). In addition, GRAS genes participate in the regulation of jasmonic acid (JA) signaling by interacting with the JAZ1 protein (repressor of JA signal pathway) (De Lucas et al., 2008; Feng et al., 2008; Hou et al., 2010).
GRAS genes also participate in the process of stress resistance. For example, the expression of GRAS6, GRAS37, GRAS50, GRAS68, and GRAS69 was significantly induced in sweet potato under salt stress treatment (Chen et al., 2019). OsGRAS23-overexpressing plants demonstrated less H2O2 accumulation under antioxidant stress and enhanced drought resistance (Xu et al., 2015). Moreover, GRAS genes are involved in the interaction between plants and fungi. The GRAS transcription factor CIGR2 activates OsHsf23 to cause hypersensitive cell death and inhibits excessive cell death in the incompatible interaction between rice (Oryza sativa) and Magnaporthe oryzae (Tanabe et al., 2016).
Brachypodium distachyon is a new model of gramineous plants and has a short growth cycle, small plant size, small genome, and easy growth. B. distachyon can be infected by M. oryzae, which causes rice blast, a devastating disease of rice. In the process of M. oryzae infecting B. distachyon, conidia first germinated and formed appressoriaand invaded epidermal cells. Invasive hyphae were found in the cells, and the hyphae continued to develop and spread to adjacent cells, causing cell death (Routledge et al., 2004). The infection process and symptoms developed on B. distachyon were very similar to those in rice, including the degree of occurrence, time of emergence, size of the lesions, and growth rate (Routledge et al., 2004).
In this study, we performed chromosomal mapping, constructed a phylogenetic tree, and performed gene structure analysis of the BdGRAS family. Furthermore, we analyzed the expression levels of BdGRAS family of B. distachyon after inoculation with M.oryzae. Our results will provide evidence for further study of GRAS gene function in disease resistance.
Materials and Methods
Experimental Materials and Treatments
B. distachyon (Bd21-3 genotype) seeds were grown in the soil (nutrient soil and stone 1:1) in a greenhouse under a 16 h light/8 h dark photoperiod, at a constant temperature of 23°C and humidity of 60%. Two-month-old B. distachyon seedlings were used in the experiment (Wang et al., 2015). Four different B. distachyon tissues (roots, stems, leaves, and seeds) were used for total RNA extraction.
The seedlings were treated with jasmonic acid(JA100 μmol/mL), salicylic acid (SA, 100 μmol/mL), indoleacetic acid (IAA, 100 μmol/mL), and abscisic acid (ABA, 100 μmol/mL). Fifteen seedlings were used for each treatment. Three leaves from the treated samples were harvested at 0 and 1 h post-treatment (hpt).
The rice blast fungus (M. oryzae) strain RO1-1 was grown on an oat medium at 28°C in the light in an incubator. After 20 days, the spores were washed with sterile water and filtered through gauze. The seedlings were challenged with M. oryzae spore (1 × 105 spores mL−1) by spraying (Wei et al., 2013). Inoculated leaves were harvested at 0, 24, and 48 h post inoculation (hpi).
Identification of BdGRAS Genes and Phylogenetic Analysis
All B. distachyon GRAS genes were derived from the Plant Transcription Factor Database (http://planttfdb.gao-lab.org/). The sequences of the GRAS genes of rice, wheat and A. thaliana for analysis of the phylogeny of the BdGRAS family were also obtained from the Plant Transcription Factor Database (Supplementary Table 1).
The protein sequences of the BdGRAS, OsGRAS, and AtGRAS family were analyzed to generate a phylogenetic tree using MEGA7.0 (Kumar et al., 2016). ClustalW was used for sequence alignment to infer the evolutionary history of multiple sequences. The neighbor-joining method was used to set the parameters to the P-distance model, and 1,000 bootstrap replicates were used to generate phylogenetic trees (Wu et al., 2021). We performed miRNA target analysis using the webtool psRNATarget (http://plantgrn.noble.org/psRNATarget/).
Analysis of Conserved Domains and Motifs for the Chromosomal Location of BdGRAS Protein
We used NCBI to search for each gene annotation in the BdGRAS family to confirm the specific location of each gene on the chromosome. The genetic map of the BdGRAS family was visualized using MapChart software. We used Multiple Em for Motif Elicitation Version 5.1.1 (MEME) (http://meme-suite.org/tools/meme) to analyze BdGRAS family motifs. For the analysis of the conserved motifs of BdGRAS family genes, we first used the NCBI Web CD-search Tool (https://www.ncbi.nlm.nih.gov/Structure/bwrpsb/bwrpsb.cgi) to predict the conserved domains of genes and then used TBtools to realize visualization. We also performed cis-acting regulatory element analysis, found the 2000 bp DNA promoter sequence upstream of the start site of each gene, and used PlangtCare (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/).
RT-PCR and qRT-PCR Analysis
Total RNA (roots, stems, leaves and seeds, and leaves treated by hormone and rice blast) was extracted using TRIzol Reagent (Ambion, Waltham, USA). cDNA synthesis was performed using TranScript One-Step gDNA Removal and cDNA Synthesis SuperMix (TransGen Biotech, Beijing, China) for RT-PCR (reverse transcription-polymerase chain reaction). The specific primers for BdGRAS family genes were designed using Primer5.0 (Singh et al., 1998) (Supplementary Table 2). The BdUBC gene (Bradi4g00660) was used as the internal reference. For PCR amplification, 2× EasyTaq PCR SuperMix (+dye) (TransGen Biotech, Beijing, China) was used. The reaction procedure was as follows: 94°C for 3 min, 34 cycles of 15 s at 94°C, 30 s at 56°C, and 72°C for 1 min (Chen et al., 2015).
First-strand cDNA was synthesized using the RevertAid First Stand cDNA Synthesis Kit (Thermo Fisher Scientific, Waltham, USA) for qRT-PCR. The qRT-PCR reaction sample adopts 20 μl systems, of which 10 μM forward and reverse primers are 0.4 μl each, TransStart Tip Green qPCR SuperMix 10 μl, template 2 μl, 7.2 μl ddH2O. qRT-PCR conditions were as follows: 94°C for 1 min, 30 cycles (94°C for 20 s, 56°C for 15 s, and 1 min at 72°C), and 72°C for 5 min. qRT-PCR was performed using the CFX 96 qPCR instrument (BioRad, California, USA) (Tong et al., 2020). The BdUBC gene was used as the internal reference. Similar results were obtained from three biological replicates.
Transcriptional Activation Assay
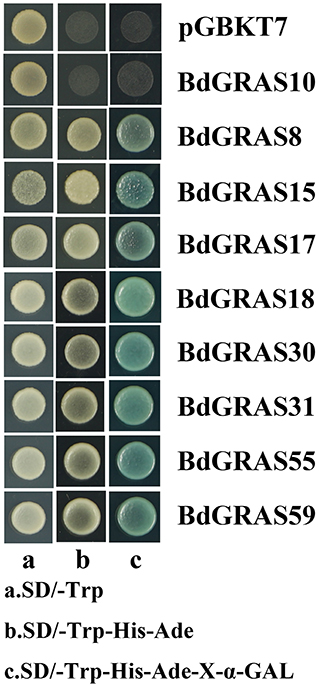
The GRAS genes were constructed on the pGBKT7 vector and then transformed into AH109 yeast competent cells. Transformed cells were streaked onto SD/-Trp and SD/-Trp-His-Ade medium. The growth status of AH109 transformed cells was evaluated after incubation at 30°C for 2 days. Transformed cells (OD600 = 1.0) were tested on the selection medium (SD/-Trp and SD/-Trp-His-Ade medium). The α-galactosidase activity assay was performed using X-α-Gal as a substrate (Wang et al., 2018a).
Results
Phylogenetic Evolution Analysis and Chromosome Location of GRAS Family
Through in silico prediction, 63 candidate GRAS genes were identified in B. distachyon. The proteins of the BdGRAS family ranged from 185 to 806 amino acids with molecular weights ranging from 30 to 89 kDa. The isoelectric point of BdGRAS was between 4.6 and 9.8, with an average of 6.7 (Supplementary Table 3). A phylogenetic tree was constructed using 60 GRAS genes from B. distachyon, 50 GRAS genes from O. sativa, and 33 GRAS genes from A. thaliana. Atypical GRAS family genes, including, 3 BdGRAS genes, 10 OsGRAS genes, and 1 AtGRAS gene, were also observed and were not included in the phylogenetic tree. The phylogenetic analysis showed they were divided into ten subfamilies, namely, DELLA, SCL3, SCL4/7, LAS, SCR, HAM, PAT1, SHR, DLT, and LISCL. Among these, the LISCL subfamily contained the greatest number of BdGRAS genes (11 members), the DLT subfamily contained only three genes. Moreover, 16 BdGRAS family genes were not classified into any subfamilies (Figure 1A). According to the clade support values and the classification of orthologs in wheat and B. distachyon. They can also be divided into 10 subfamilies using the phylogenetic analysis (Supplementary Figure 1).These BdGRAS genes were unevenly distributed on five chromosomes. Chr4 contained the greatest number of BdGRAS genes (n = 28), followed by Chr1 (n = 18). Furthermore, nine genes were distributed on Chr2, and five genes were distributed on Chr3. Only three genes were distributed on Chr5 (Figure 1B).
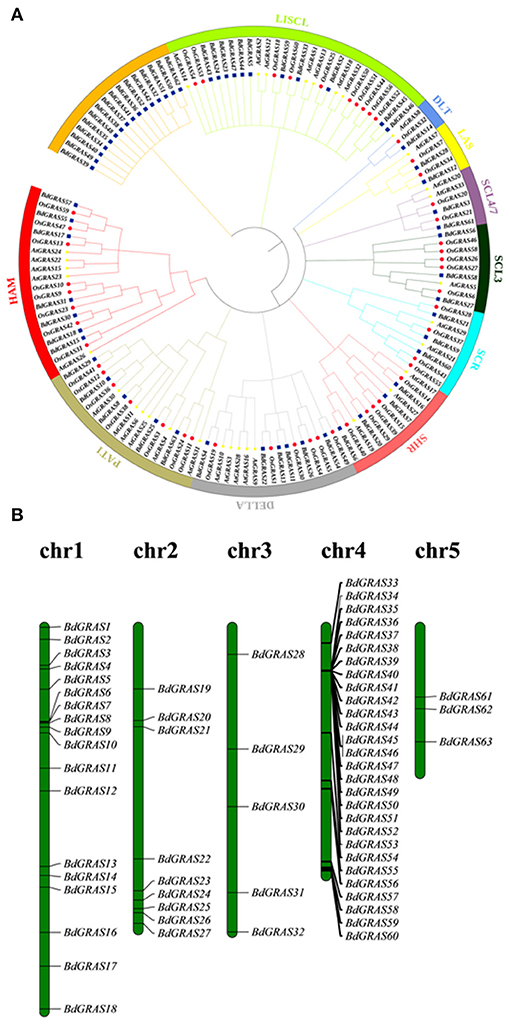
Figure 1. Phylogenetic analysis of GRAS proteins in Arabidopsis thaliana, Oryza sativa and Brachypodium distachyon and chromosomal mapping of GRAS gene in Brachypodium distachyon. (A) The phylogenetic tree is divided into 10 different subfamilies, and the different subfamilies are shown with branches and outer ring of different colors. The blue solid square represents B. distachyon GRAS protein, the red solid circle represents O. sativa protein, and the yellow five-pointed star represents Arabidopsis protein. (B) Chromosomal locations of GRAS genes in B. distachyon.
B. distachyon GRAS Family Collinearity Analysis and Structural Domain Analysis
Protein domain analysis showed that most members contained typical GRAS regions (Supplementary Figure 2). In each subfamily, the amino acid sequences of BdGRAS proteins showed high homology. All BdGRAS proteins have five conserved motifs in the C-terminus, namely, LHRI, VHIID, LHRII, PFYRE, and SAW; meanwhile, the N-terminus of BdGRAS proteins was highly variable (Figure 2B). To compare the genetic relationship of BdGRAS in B. distachyon, we used TBtools to identify paralogous genes in the B. distachyon genome. Five pairs of homologous genes were identified in BdGRAS, indicating that B. distachyon has experienced genome replication events during evolution (Figure 2A). Through the analysis of the transcriptional regulation mechanism of GRAS gene, it is found that BdGRAS gene mainly involves 15 biological pathways, most of which are involved in stress response, such as low temperature and light signal. It has been detected that BdGRAS gene is related to hormones, such as salicylic acid and jasmonic acid (Supplementary Figure 3).
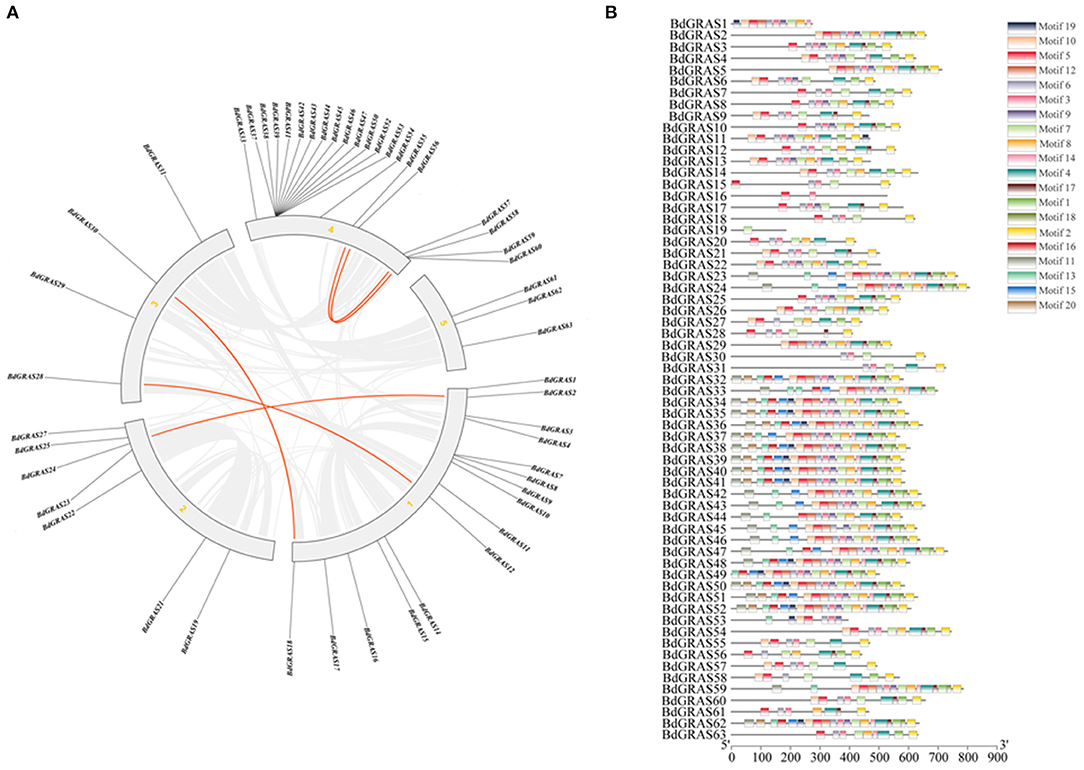
Figure 2. BdGRAS gene replication and conserved domain analysis. (A) Connecting lines indicate duplicated genes where in red lines represent GRAS genes while gray lines signify collinear blocks in whole genome. Composition of the GRAS protein motif of B. distachyon. (B) Identify the BdGRAS protein through the MEME website, and allow up to 20 motifs. Others are the default settings, and 20 different motifs are represented by different colored rectangles.
Expression Profiles of the BdGRAS Family Genes in Different Tissues and Hormone Treatments
We tested the expression levels of BdGRAS family genes in different tissues of B. distachyon, including the roots, stems, leaves, and seeds, by RT-PCR. It was found that most of the genes were identified in a wide range of tissues, such as BdGRAS33, BdGRAS57, and BdGRAS58 (Figure 3). Moreover, some genes were highly expressed in some tissues. For instance, the expression of BdGRAS34 to BdGRAS40 was high in the roots and seeds. In contrast, the expression of BdGRAS31 was low in the seeds.
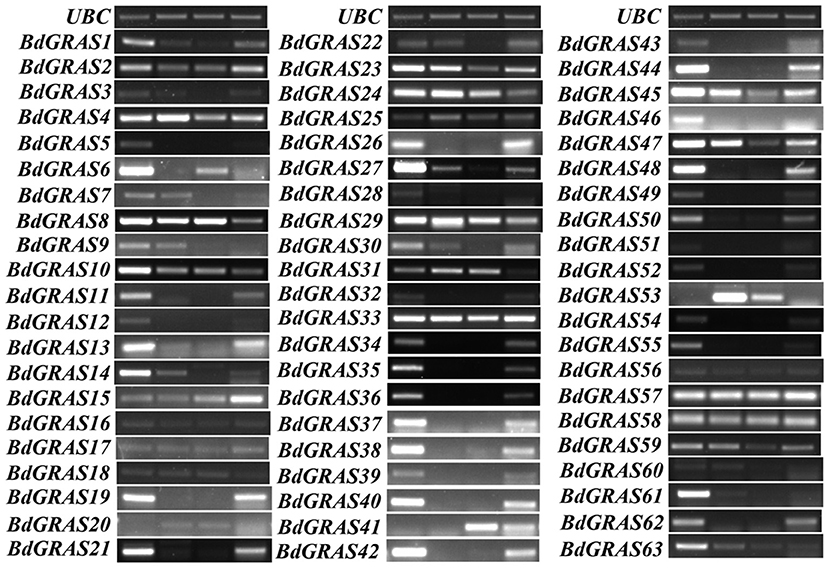
Figure 3. The GRAS genes expression pattern in different tissues of Brachypodium distachyon. The expression level of the BdGRAS gene in the root, stem, leaf, and seeds of B. distachyon, using UBC as a control.
We also investigated the expression levels of BdGRAS genes under four different hormone treatments by qRT-RCR (SA, JA, IAA, and ABA). The expression levels of BdGRAS48, BdGRAS49, BdGRAS51, BdGRAS52, and BdGRAS55 significantly increased after SA hormone treatment at 1 hpt; BdGRAS2, BdGRAS8, BdGRAS10, BdGRAS23, BdGRAS47, and BdGRAS59 were significantly induced after JA hormone treatment. Moreover, BdGRAS47 expression levels significantly increased after ABA hormone treatment. After IAA treatment, the expression levels of most GRAS genes significantly reduced. The expression of BdGRAS34, BdGRAS36, BdGRAS37, BdGRAS38, BdGRAS39, BdGRAS40, BdGRAS41, BdGRAS42, and BdGRAS45 was inhibited under all four different hormone treatments (Figure 4). These results indicated that the GRAS family genes participate in different hormone signaling pathways.
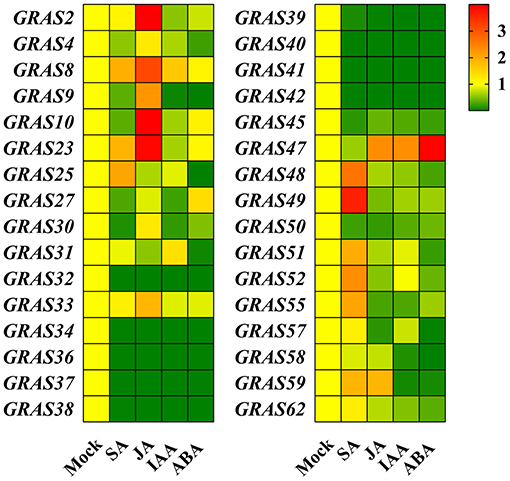
Figure 4. BdGRAS genes transcription level expression analysis under different hormone treatments. The changes of BdGRAS genes expression levels in B. distachyon under salicylic acid (SA), jasmonic acid (JA), indoleacetic acid (IAA), and abscisic acid (ABA) treatments at 1 hpi. Green and red represent relatively low and high expression levels.
Expression Levels of BdGRAS Family Genes in B. distachyon and M. oryzae Interaction
To investigate the roles of BdGRAS genes in response to biotic stress, we analyzed the expression levels of BdGRAS genes in B. distachyon inoculated with M. oryzae RO1-1 at 0, 24, and 48 hpi. The results showed that the expression of BdGRAS4, BdGRAS8, BdGRAS10, BdGRAS30, and BdGRAS31 gradually decreased after inoculation with M. oryzae. Moreover, expression levels of BdGRAS2, BdGRAS9, BdGRAS18, BdGRAS23, BdGRAS33, BdGRAS34, BdGRAS47, BdGRAS48, BdGRAS49, BdGRAS51, BdGRAS52, BdGRAS57, and BdGRAS58 significantly increased at 24 hpi (Figure 5). These results indicate that the BdGRAS family participates in the interaction between B. distachyon and M. oryzae, and plays a role in the resistance of B. distachyon to rice blast.
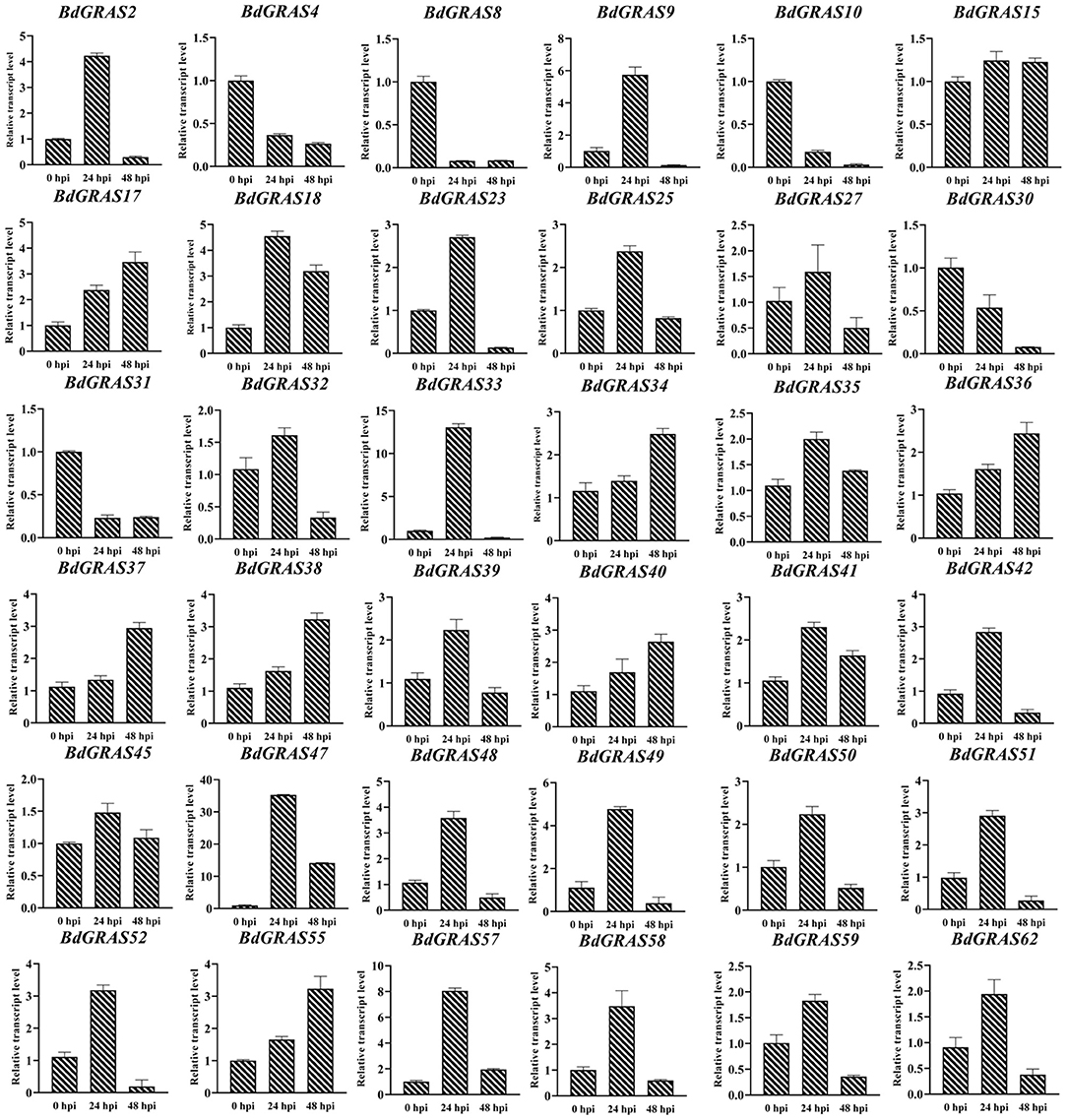
Figure 5. qRT-PCR analysis of the expression pattern of BdGRAS gene after inoculation with Magnaporthe oryzae. The Y-axis is the relative expression of BdGRAS gene, and the X-axis is different time points after inoculation with M. oryzae, including 0, 24, and 48 hpi. UBC is used as an internal control (hour post inoculation, hpi).
BdGRAS31 Is the Target of miR171 in B. distachyon
Some members of the GRAS gene family are potential regulatory targets of miRNAs. We predicted that 23 BdGRAS genes were targeted by six miRNA families (Supplementary Table 4). Furthermore, we identified that BdGRAS15, BdGRAS18, and BdGRAS31 were candidate targets of Bd-miR171 by degradome deep sequencing (Figure 6A). We used qRT-PCR to detect the expression levels of the three candidate target genes and Bd-miR171 after inoculation with M. oryzae (Figure 6B). The expression of Bd-miR171 gradually increased within 48 hpi with RO1-1, while that of BdGRAS31 gradually decreased within 48 hpi. The expression level of BdGRAS15 did not change significantly, and the expression of BdGRAS18 increased at 24 hpi and began to decrease at 48 hpi (Supplementary Figure 4). Therefore, we inferred that BdGRAS31 is regulated by Bd-miR171 in the interactions between B. distachyon and M. oryzae.
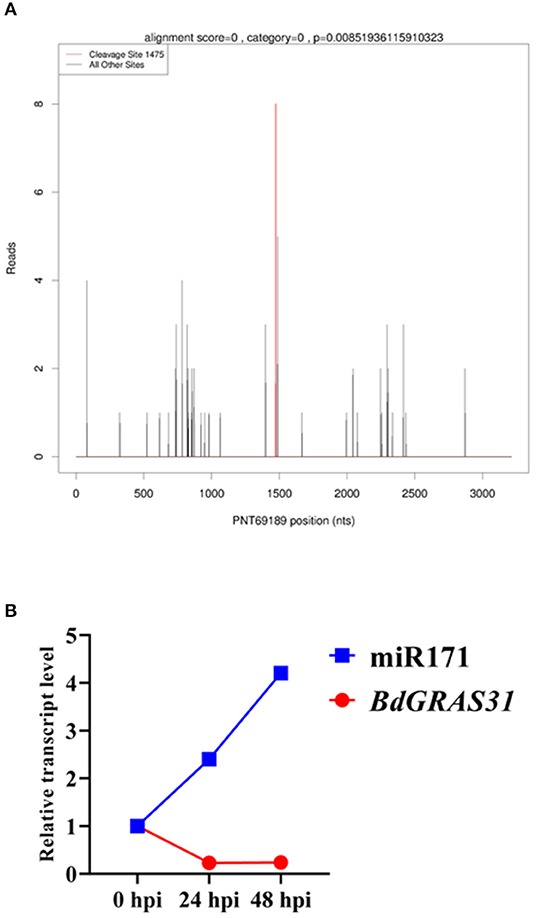
Figure 6. BdGRAS31 is the target gene of miR171. (A) Identification of BdGRAS31 was the target of Bd-miR171 by degradome sequencing. (B) The expression of miR171 and BdGRAS31 in Brachypodium distachyon after inoculation with M. oryzae.
Verification of Transcriptional Activity of BdGRAS Genes
To detect whether the BdGRAS family has transcriptional activity, we selected nine genes-BdGRAS8, BdGRAS10, BdGRAS515, BdGRAS17, BdGRAS18, BdGRAS30, BdGRA31, BdGRA55, and BdGRA59 for testing transcriptional activation. It was found that eight transformants harboring BdGRAS genes could grow on SD/-Trp and SD/-Trp-His-Ade and turned blue on SD-Trp-His-Ade-X-α-GAL, indicating that BdGRAS8, BdGRAS15, BdGRAS17, BdGRAS18, BdGRAS30, BdGRA31, BdGRA55, and BdGRA59 show transcriptional activity (Figure 7). However, BdGRAS10 did not show transcriptional activity.
Discussion
The GRAS family is an important plant-specific transcription factor family divided into multiple subfamilies (Bolle, 2004). Recent studies have divided the GRAS family into 10–17 subfamilies in multiple plants (Bolle, 2016; Wang et al., 2018b,c). It is reported there were 48 BdGRAS genes, which were divided into ten known subfamilies (Niu et al., 2019). In this study, 63 BdGRAS family genes were identified by domain analysis. The phylogenetic tree showed that BdGRAS family genes were divided into ten subfamilies according to their genetic relationship. Moreover, most B. distachyon GRAS genes are divided into the same clade as that of Arabidopsis or rice (Niu et al., 2019). For example, the LISCL subfamily contains 11 BdGRAS genes. The conserved function of putative orthologs in each subfamily was indicated by the functional characterization of GRAS proteins. Therefore, we inferred that BdGRAS genes are also involved in multiple plant physiological processes by different regulation.
Each subfamily of GRAS genes has specific functions in plant development (Chen et al., 2019). For example, in Lilium longiflorum, the LISCL gene is involved in regulating microsporogenesis (Morohashi et al., 2003). The LISCL subfamily members were identified in a wide range of tissues in Brassica napus (Guo et al., 2019). The HAM subfamily is considered to play an important role in the growth and development of root tips, stem tips, and shoot apex meristem (Stuurman et al., 2002). In this study, we identified some LISCL genes, such as BdGRAS2, BdGRAS23, BdGRAS45, and BdGRAS59, in multiple tissues. Moreover, we found that members of the HAM subfamily showed differences in tissue expression. BdGRAS55 is only highly expressed in roots. Therefore, the results suggested that BdGRAS genes play an important role in controlling the growth and development of B. distachyon.
Multiple plant physiological processes are regulated by a diverse group of hormones (Huang et al., 2015). IAA is involved in almost all aspects of plant growth and development. JA and SA play important roles in biotic stresses. The HcSCL13 gene of Halostachys Caspica was overexpressed in Arabidopsis, under the treatment of hormone ABA, the HcSCL13 gene was induced rapidly, reached the highest peak at 1 h, and then decreased to the control level (Zhang et al., 2020). In the potato, the expression levels of StGRAS34, StGRAS36, and StGRAS46 are increased under the treatment of IAA, ABA, and GA3 hormones (Wang et al., 2019). In this study, we observed that BdGRAS47 expression increased significantly under ABA and IAA treatment; this gene may play important roles in plant development. Moreover, the expression of BdGRAS2, BdGRAS8, BdGRAS10, BdGRAS23, and BdGRAS47 increased significantly under JA treatment. The expression of BdGRAS48, BdGRAS49, BdGRAS51, BdGRAS52, and BdGRAS55 significantly increased after SA treatment. These results suggest that BdGRAS genes may be involved in the plant response to biotic stress.
GRAS genes play important roles in the plant response to pathogens. For example, VviRGA5 is upregulated in grape berries at the initial stage of fungal infection (Agudelo-Romero et al., 2014). SlGRAS1 is involved in the plant response to biotic stress (Mayrose et al., 2006). CIGR2 negatively regulated resistance to M. oryzae in rice (Tanabe et al., 2016). In this study, the expression levels of 28 BdGRAS genes changed after inoculation with rice blast fungus. We found that the expression levels of most members of the HAM subfamily, including BdGRAS17, BdGRAS18, BdGRAS55, and BdGRAS57, increased significantly after inoculation with M. oryzae, while the expression levels of BdGRAS30 and BdGRAS31 significantly decreased. In B. napus, HAM subfamilies comprise two clades and perform different functions (Guo et al., 2019). We inferred that BdGRAS genes of HAM subfamilies may play multiply function in B. distachyon and M. oryzae interaction. Moreover, HAM genes are candidate targets of miR171 (Fan et al., 2015). We found 4 HAM subfamilies genes as candidate targets of Bd-miR171, and confirmed that BdGRAS31 could be regulated by degradome deep sequencing and qRT-PCR. We also identified the transcriptional activity of the members of GRAS genes. Most BdGRAS genes showed transcriptional activity, except BdGRAS10. These results suggested that BdGRAS regulated target genes through multiple approaches.
In this study, we found that the HAM subfamily of BdGRAS genes may play essential roles in disease resistance. These results provide a basis for further research on the BdGRAS family and also provide a reference for the study of disease resistance and stress resistance of grasses.
Data Availability Statement
The data presented in the study are deposited in the Plant Transcription Factor Database (http://planttfdb.gao-lab.org/index.php) repository, accession number Bradi1g00219.1.p-Bradi5g19190.1.p; LOC_Os01g45860-LOC_Os12g38490; AT1G0 7520-AT5G67411; TRAES3BF021600230CFD_t1-Traes_7DL_44FCA17C4.1, see Supplementary Table 1 for details.
Author Contributions
ZT and NS are the experimental designers and executors of the experimental research, completing data analysis, and writing the first draft of the paper. WP, YY, TQ, and CH participated in the experimental design and analysis of the experimental results. LD and BW are the creators and leaders of the project, directing experimental design, data analysis, thesis writing, and revision.
Funding
This study was supported by grants from the National Natural Science Foundation of China (31801721; 31672017), National Key Research and Development Project (2016YFD0200800; 2016YFD0300707), the Natural Science Foundation of Hunan Province, China (2020JJ5240), the Scientific Research Fund of Hunan Provincial Education Department (19B247), Commonweal Plan Project of Ningbo City (202002N3004), and the Youth Fund Project of Hunan Agricultural University (19QN31).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fsufs.2021.675177/full#supplementary-material
Supplementary Figure 1. Phylogenetic analysis of GRAS proteins in Triticum aestivum and Brachypodium distachyon.
Supplementary Figure 2. The composition of the GRAS protein domain of Brachypodium distachyon. Green represents the GRAS domain, yellow represents the GRAS superfamily domain, pink represents the DELLA domain, and blue represents the PA28_alpha superfamily domain.
Supplementary Figure 3. Brachypodium distachyon GRAS cis-acting regulatory element analysis.
Supplementary Figure 4. The expression level of candidate targets of Bd-miR171.
Supplementary Table 1. GRAS family genes in wheat, rice and Arabidopsis thaliana.
Supplementary Table 2. Brachypodium distachyon GRAS family genes specific primers for qRT-PCR and RT-PCR analysis.
Supplementary Table 3. Parameters of Brachypodium distachyon GRAS family genes.
Supplementary Table 4. Prediction of target RNA of GRAS family in Brachypodium distachyon.
References
Agudelo-Romero, P., Ali, K., Choi, Y. H., Sousa, L., Verpoorte, R., Tiburcio, A. F., et al. (2014). Perturbation of polyamine catabolism affects grape ripening of Vitis vinifera cv. Trincadeira. Plant Physiol. Biochem. 74, 141–155. doi: 10.1016/j.plaphy.2013.11.002
Bolle, C. (2016). Structure and evolution of plant GRAS family proteins. Plant Transcription Factors 153–161. doi: 10.1016/B978-0-12-800854-6.00010-5
Bolle, C. (2004). The role of GRAS proteins in plant signal transduction and development. Planta 218, 683–692. doi: 10.1007/s00425-004-1203-z
Bolle, C., Koncz, C., and Chua, N. H. (2000). PAT1, a new member of the GRAS family, is involved in phytochrome A signal transduction. Genes Dev. 14, 1269–1278. doi: 10.1101/gad.14.10.1269
Chen, H., Li, H., Lu, X., Chen, L., Liu, J., and Wu, H. (2019). Identification and expression analysis of GRAS transcription factors to elucidate candidate genes related to stolons, fruit ripening and abiotic stresses in woodland strawberry (Fragaria vesca). Int. J. Mol. Sci. 20:4593. doi: 10.3390/ijms20184593
Chen, Y. Q., Tai, S. S., Wang, D. W., Ding, A. M., Sun, T. T., Wang, W. F., et al. (2015). Homology-based analysis of the GRAS gene family in tobacco. Genet. Mol. Res. 14, 15188–15200. doi: 10.4238/2015.November.25.7
Davière, J. M., and Achard, P. (2016). A pivotal role of DELLAs in regulating multiple hormone signals. Mol. Plant 9, 10–20. doi: 10.1016/j.molp.2015.09.011
De Lucas, M., Daviere, J. M., Rodríguez-Falcón, M., Pontin, M., Iglesias-Pedraz, J. M., Lorrain, S., et al. (2008). A molecular framework for light and gibberellin control of cell elongation. Nature 451, 480–484. doi: 10.1038/nature06520
Fan, T., Li, X., Yang, W., Xia, K., Ouyang, J., and Zhang, M. (2015). Rice osa-miR171c mediates phase change from vegetative to reproductive development and shoot apical meristem maintenance by repressing four OsHAM transcription factors. PLoS ONE 10:e0125833. doi: 10.1371/journal.pone.0125833
Feng, S., Martinez, C., Gusmaroli, G., Wang, Y., Zhou, J., Wang, F., et al. (2008). Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature 451, 475–479. doi: 10.1038/nature06448
Gao, M. J., Parkin, I., Lydiate, D., and Hannoufa, A. (2004). An auxin-responsive SCARECROW-like transcriptional activator interacts with histone deacetylase. Plant Mol. Biol. 55, 417–431. doi: 10.1007/s11103-004-0892-9
Greb, T., Clarenz, O., Schäfer, E., Müller, D., Herrero, R., Schmitz, G., et al. (2003). Molecular analysis of the LATERAL SUPPRESSOR gene in Arabidopsis reveals a conserved control mechanism for axillary meristem formation. Genes Dev. 17, 1175–1187. doi: 10.1101/gad.260703
Guo, P., Wen, J., Yang, J., Ke, Y., Wang, M., Liu, M., et al. (2019). Genome-wide survey and expression analyses of the GRAS gene family in Brassica napus reveals their roles in root development and stress response. Planta 250, 1051–1072. doi: 10.1007/s00425-019-03199-y
Heckmann, A. B., Lombardo, F., Miwa, H., Perry, J. A., Bunnewell, S., Parniske, M., et al. (2006). Lotus japonicus nodulation requires two GRAS domain regulators, one of which is functionally conserved in a non-legume. Plant Physiol. 142, 1739–1750. doi: 10.1104/pp.106.089508
Helariutta, Y., Fukaki, H., Wysocka-Diller, J., Nakajima, K., Jung, J., Sena, G., et al. (2000). The SHORT-ROOT gene controls radial patterning of the Arabidopsis root through radial signaling. Cell 101, 555–567. doi: 10.1016/S0092-8674(00)80865-X
Heo, J. O., Chang, K. S., Kim, I. A., Lee, M. H., Lee, S. A., Song, S. K., et al. (2011). Funneling of gibberellin signaling by the GRAS transcription regulator scarecrow-like 3 in the Arabidopsis root. Proc. Natl. Acad. Sci. U.S.A. 108, 2166–2171. doi: 10.1073/pnas.1012215108
Hirsch, S., and Oldroyd, G. E. (2009). GRAS-domain transcription factors that regulate plant development. Plant Signal. Behav. 4, 698–700. doi: 10.4161/psb.4.8.9176
Hofmann, N. R. (2016). A structure for plant-specific transcription factors. Plant Cell 28, 993–994. doi: 10.1105/tpc.16.00309
Hou, X., Lee, L. Y. C., Xia, K., Yan, Y., and Yu, H. (2010). DELLAs modulate jasmonate signaling via competitive binding to JAZs. Dev. Cell 19, 884–894. doi: 10.1016/j.devcel.2010.10.024
Huang, W., Xian, Z., Kang, X., Tang, N., and Li, Z. (2015). Genome-wide identification, phylogeny and expression analysis of GRAS gene family in tomato. BMC Plant Biol. 15, 1–18. doi: 10.1186/s12870-015-0590-6
Ikeda, A., Ueguchi-Tanaka, M., Sonoda, Y., Kitano, H., Koshioka, M., Futsuhara, Y., et al. (2001). slender rice, a constitutive gibberellin response mutant, is caused by a null mutation of the SLR1 gene, an ortholog of the height-regulating gene GAI/RGA/RHT/D8. Plant Cell 13, 999–1010. doi: 10.1105/tpc.13.5.999
Itoh, H., Ueguchi-Tanaka, M., Sato, Y., Ashikari, M., and Matsuoka, M. (2002). The gibberellin signaling pathway is regulated by the appearance and disappearance of SLENDER RICE1 in nuclei. Plant Cell 14, 57–70. doi: 10.1105/tpc.010319
Kaló, P., Gleason, C., Edwards, A., Marsh, J., Mitra, R. M., Hirsch, S., et al. (2005). Nodulation signaling in legumes requires NSP2, a member of the GRAS family of transcriptional regulators. Science 308, 1786–1789. doi: 10.1126/science.1110951
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Li, S., Zhao, Y., Zhao, Z., Wu, X., Sun, L., Liu, Q., et al. (2016). Crystal structure of the GRAS domain of SCARECROW-LIKE7 in Oryza sativa. Plant Cell 28, 1025–1034. doi: 10.1105/tpc.16.00018
Li, X., Duan, X., Jiang, H., Sun, Y., Tang, Y., Yuan, Z., et al. (2006). Genome-wide analysis of basic/helix-loop-helix transcription factor family in rice and Arabidopsis. Plant Physiol. 141, 1167–1184. doi: 10.1104/pp.106.080580
Mayrose, M., Ekengren, S. K., Melech-Bonfil, S. H. I. R. I., Martin, G. B., and Sessa, G. (2006). A novel link between tomato GRAS genes, plant disease resistance and mechanical stress response. Mol. Plant Pathol. 7, 593–604. doi: 10.1111/j.1364-3703.2006.00364.x
Morohashi, K., Minami, M., Takase, H., Hotta, Y., and Hiratsuka, K. (2003). Isolation and characterization of a novel GRAS gene that regulates meiosis-associated gene expression. J. Biol. Chem. 278, 20865–20873. doi: 10.1074/jbc.M301712200
Niu, X., Chen, S., Li, J., Liu, Y., Ji, W., and Li, H. (2019). Genome-wide identification of GRAS genes in Brachypodium distachyon and functional characterization of BdSLR1 and BdSLRL1. BMC Genomics 20, 1–18. doi: 10.1186/s12864-019-5985-6
Peng, J., Carol, P., Richards, D. E., King, K. E., Cowling, R. J., Murphy, G. P., et al. (1997). The Arabidopsis GAI gene defines a signaling pathway that negatively regulates gibberellin responses. Genes Dev. 11, 3194–3205. doi: 10.1101/gad.11.23.3194
Pysh, L. D., Wysocka-Diller, J. W., Camilleri, C., Bouchez, D., and Benfey, P. N. (1999). The GRAS gene family in Arabidopsis: sequence characterization and basic expression analysis of the SCARECROW-LIKE genes. Plant J. 18, 111–119. doi: 10.1046/j.1365-313X.1999.00431.x
Routledge, A. P., Shelley, G., Smith, J. V., Talbot, N. J., Draper, J., and Mur, L. A. (2004). Magnaporthe grisea interactions with the model grass Brachypodium distachyon closely resemble those with rice (Oryza sativa). Mol. Plant Pathol. 5, 253–265. doi: 10.1111/j.1364-3703.2004.00224.x
Silverstone, A. L., Ciampaglio, C. N., and Sun, T. P. (1998). The Arabidopsis RGA gene encodes a transcriptional regulator repressing the gibberellin signal transduction pathway. Plant Cell 10, 155–169. doi: 10.1105/tpc.10.2.155
Singh, V. K., Mangalam, A. K., Dwivedi, S., and Naik, S. (1998). Primer premier: program for design of degenerate primers from a protein sequence. Biotechniques 24, 318–319. doi: 10.2144/98242pf02
Smit, P., Raedts, J., Portyanko, V., Debellé, F., Gough, C., Bisseling, T., et al. (2005). NSP1 of the GRAS protein family is essential for rhizobial Nod factor-induced transcription. Science 308, 1789–1791. doi: 10.1126/science.1111025
Stuurman, J., Jäggi, F., and Kuhlemeier, C. (2002). Shoot meristem maintenance is controlled by a GRAS-gene mediated signal from differentiating cells. Genes Dev. 16, 2213–2218. doi: 10.1101/gad.230702
Tanabe, S., Onodera, H., Hara, N., Ishii-Minami, N., Day, B., Fujisawa, Y., et al. (2016). The elicitor-responsive gene for a GRAS family protein, CIGR2, suppresses cell death in rice inoculated with rice blast fungus via activation of a heat shock transcription factor, OsHsf23. Biosci. Biotechnol. Biochem. 80, 145–151. doi: 10.1080/09168451.2015.1075866
Tian, C., Wan, P., Sun, S., Li, J., and Chen, M. (2004). Genome-wide analysis of the GRAS gene family in rice and Arabidopsis. Plant Mol. Biol. 54, 519–532. doi: 10.1023/B:PLAN.0000038256.89809.57
Tong, J., Walk, T. C., Han, P., Chen, L., Shen, X., Li, Y., et al. (2020). Genome-wide identification and analysis of high-affinity nitrate transporter 2 (NRT2) family genes in rapeseed (Brassica napus L.) and their responses to various stresses. BMC Plant Biol. 20, 1–16. doi: 10.1186/s12870-020-02648-1
Van De Velde, K., Ruelens, P., Geuten, K., Rohde, A., and Van Der Straeten, D. (2017). Exploiting DELLA signaling in cereals. Trends Plant Sci. 22, 880–893. doi: 10.1016/j.tplants.2017.07.010
Wang, B., Wei, J., Song, N., Wang, N., Zhao, J., and Kang, Z. (2018a). A novel wheat NAC transcription factor, TaNAC30, negatively regulates resistance of wheat to stripe rust. J. Integrative Plant Biol. 60, 432–443. doi: 10.1111/jipb.12627
Wang, L., Hu, W., Sun, J., Liang, X., Yang, X., Wei, S., et al. (2015). Genome-wide analysis of SnRK gene family in Brachypodium distachyon and functional characterization of BdSnRK2. 9. Plant Sci. 237, 33–45. doi: 10.1016/j.plantsci.2015.05.008
Wang, M., Xu, Z., Ding, A., and Kong, Y. (2018b). Genome-wide identification and expression profiling analysis of the xyloglucan endotransglucosylase/hydrolase gene family in tobacco (Nicotiana tabacum L.). Genes 9:273. doi: 10.3390/genes9060273
Wang, S., Zhang, N., Zhu, X., Yang, J., Li, S., Che, Y., et al. (2019). Identification and expression analysis of StGRAS gene family in potato (Solanum tuberosum L.). Comput. Biol. Chem. 80, 195–205. doi: 10.1016/j.compbiolchem.2019.03.020
Wang, Y. X., Liu, Z. W., Wu, Z. J., Li, H., Wang, W. L., Cui, X., et al. (2018c). Genome-wide identification and expression analysis of GRAS family transcription factors in tea plant (Camellia sinensis). Sci. Rep. 8, 1–19. doi: 10.1038/s41598-018-22275-z
Wei, T., Ou, B., Li, J., Zhao, Y., Guo, D., Zhu, Y., et al. (2013). Transcriptional profiling of rice early response to Magnaporthe oryzae identified OsWRKYs as important regulators in rice blast resistance. PLoS ONE 8:e59720. doi: 10.1371/journal.pone.0059720
Wu, J., Liu, H., Lu, S., Hua, J., and Zou, B. (2021). Identification and expression analysis of chloroplast ribonucleoproteins (cpRNPs) in Arabidopsis and rice. Genome 99, 1–10. doi: 10.1139/gen-2020-0007
Xu, K., Chen, S., Li, T., Ma, X., Liang, X., Ding, X., et al. (2015). OsGRAS23, a rice GRAS transcription factor gene, is involved in drought stress response through regulating expression of stress-responsive genes. BMC Plant Biol. 15, 1–13. doi: 10.1186/s12870-015-0532-3
Keywords: GRAS family, transcription factor, Brachypodium distachyon, Magnaporthe oryzae, HAM subfamily
Citation: Tang Z, Song N, Peng W, Yang Y, Qiu T, Huang C, Dai L and Wang B (2021) Genome Identification and Expression Analysis of GRAS Family Related to Development, Hormone and Pathogen Stress in Brachypodium distachyon. Front. Sustain. Food Syst. 5:675177. doi: 10.3389/fsufs.2021.675177
Received: 02 March 2021; Accepted: 12 April 2021;
Published: 14 May 2021.
Edited by:
Wen Xie, Chinese Academy of Agricultural Sciences, ChinaReviewed by:
Yong Liu, Hunan Academy of Agricultural Sciences (CAAS), ChinaSuprasanna Penna, Bhabha Atomic Research Centre (BARC), India
Copyright © 2021 Tang, Song, Peng, Yang, Qiu, Huang, Dai and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bing Wang, emh1ZnVAaHVuYXUuZWR1LmNu; Liangying Dai, ZGFpbHlAaHVuYXUubmV0
†These authors have contributed equally to this work
 Zejun Tang
Zejun Tang Na Song1,2†
Na Song1,2† Weiye Peng
Weiye Peng Bing Wang
Bing Wang