- 1Department of Microbiology, Cornell University, Ithaca, NY, United States
- 2Department of Food Science, Cornell University, Ithaca, NY, United States
Introduction
Classification of bacterial pathogens into species has traditionally been an essential prerequisite for many aspects of infection control and food safety. Consequently, control measures, regulations, industry specifications, and other guidance documents for foodborne pathogens typically specify a pathogen species (or genus) as target. For example, current Codex Alimentarius guidelines recommend absence of Salmonella in two 375 g samples for infant formula (FAO, 2020). For other ready-to-eat (RTE) foods, absence in smaller samples size (e.g., 25 g) is typically expected. However, there is increasing evidence for Salmonella, as well as other foodborne pathogens, that subgroups, serovars, and/or clonal groups within a given species may differ considerably in their ability to cause human disease. Importantly, there is emerging evidence that use of “sub-species” (or “sub-genus”) classification for defining food safety hazards may allow for more effective management of foodborne pathogens. The genus Salmonella, a pathogen where all members of the genus are considered a food safety hazard, represents one key foodborne pathogen where there is increasing evidence that subtypes differ considerably in their likelihood of causing human disease and where science-based subtype-specific and risk-based approaches of control may have a positive public health impact. There is clear evidence that certain Salmonella subtypes (e.g., certain subtypes within serovars Kentucky and Cerro) show reduced human virulence, while being frequently isolated from food animal species (specifically poultry and cattle) (Rodriguez-Rivera et al., 2014; Haley et al., 2016). Additionally, under the current approach of considering all Salmonella equally likely to cause human disease, limited progress has been made worldwide to reduce foodborne salmonellosis cases (Havelaar et al., 2015; Tack et al., 2020). In some situations, there are counterproductive incentives to prioritize control of highly frequent Salmonella strains with limited human public health relevance (e.g., through poultry vaccines targeting S. Kentucky) over control of less frequent Salmonella strains commonly linked to human salmonellosis cases. We outline a new paradigm for incorporating strain-specific assessment of human public health significance into regulations and control strategies, which we propose should be used to incentivize control of Salmonella subtypes with the greatest public health impact, with a focus on pre-harvest control in meat and poultry. This paradigm could subsequently be applied to other foodborne pathogens, such as Listeria monocytogenes.
Salmonella is Not a “One Size Fits All” Pathogen
The genus Salmonella consists of two species, Salmonella enterica and Salmonella bongori; S. enterica can be further divided into six recognized subspecies, the most clinically relevant of which is S. enterica subsp. enterica (Worley et al., 2018), a pathogen of warm-blooded mammals including humans. Within S. enterica subsp. enterica (hereafter referred to as “S.”), there are >1,500 serovars (Issenhuth-Jeanjean et al., 2014), which differ in their ability to cause human disease (Brenner et al., 2000). Typhoidal (i.e., S. Typhi) and paratyphoidal (e.g., S. Paratyphi A) serovars cause a systemic illness characterized by fever, headache, malaise, abdominal pain, and rash, while non-typhoidal serovars (e.g., S. Typhimurium) typically cause a self-limiting gastroenteritis. Additionally, S. Typhi and S. Paratyphi A are considered to be human-restricted (i.e., they only cause disease in and are spread by humans) serovars and are typically transmitted through food and/or water contaminated with human feces due to poor sanitation. Non-typhoidal serovars on the other hand are transmitted through both direct animal contact and food (as well as fomites) with contamination often linked to animal hosts and foods derived from warm-blooded animals (e.g., raw meat and poultry, raw eggs, raw milk and dairy products products) (Gal-Mor et al., 2014). However, a variety of foods have been sources of non-typhoidal Salmonella cases and outbreaks, including fruits and vegetables as well as dry products (e.g., spices, chocolate) and non-typhoidal Salmonella have been shown to also survive for extended times in extra-host environments such as soil and even processing plant environments (Waldner et al., 2012; Jechalke et al., 2019). Because typhoidal and paratyphoidal serovars typically cause systemic disease, they have traditionally been considered a higher public health risk than non-typhoidal serovars. Non-typhoidal serovars, despite differing in their ability to cause disease in humans, are considered an equal public health risk. With this current approach, efforts to decrease Salmonella human clinical cases in the United States have essentially failed (Tack et al., 2020). While Healthy People (2020) targeted a reduction in Salmonella incidence from 15.0 cases to 11.4 cases per 100,000 people by 2020, Salmonella incidence reached 17.1 cases per 100,000 people in 2019 (Tack et al., 2020). Similarly, at a global level, Salmonella continues to be the foodborne pathogen with the greatest public health impact based on disability-adjusted life years (Havelaar et al., 2015).
While regulations in many countries consider all non-typhoidal Salmonella (NTS) serovars a hazard and equal public health risk, there is strong scientific evidence that NTS serovars and clonal groups differ in the public health risk they represent, as supported by (i) presence or absence of virulence genes that encode fully functional virulence factors; (ii) phenotypic data (e.g., in tissue culture or animal models); and (iii) epidemiological evidence (e.g., under-representation of certain serovars or clonal groups among human clinical cases relative to their presence in food, raw materials, or animals). Salmonella serovars or clonal groups may also show strong geographical associations which may need to be taken into account when assessing epidemiological evidence for hypo- or hypervirulence. Instances where all three lines of evidence suggest reduced virulence in specific serovars or clonal groups represent a particularly strong case for classifying a group as a reduced public health risk. However, even two lines of evidence [e.g., (i) and (iii)] may be sufficient in some cases to identify groups that differ in the public health risk they represent. This general approach is essentially what has been used to identify Escherichia coli serovars and groups (e.g., Enterohemorrhagic E. coli [EHEC]) that represent a particular public health risk. Importantly, as detailed below for NTS, sufficient data appear to be already available for some NTS to allow for science-based classification of virulence groups.
It is well-established that Salmonella serovars and clonal groups within a serovar can differ greatly in the virulence factors they encode. One important example being differences in gene content on Salmonella Pathogenicity Islands (SPIs), which are large islets of genes that contribute to a particular virulence phenotype (Marcus et al., 2000). Currently, there are 24 recognized SPIs (Cheng et al., 2019); data on presence/absence of these SPIs and genes in these SPIs, as well as data on the functional conservation of genes in these SPIs (e.g., absence of premature stop codons), can be used to assess the virulence potential and public health risks associated with Salmonella serovars or clonal groups. For example, SPI-7, which encodes the Vi polysaccharide (Helena, 2008), is found in multiple serovars associated with invasive human disease, including serovars Typhi (Faucher et al., 2006; Lee et al., 2006), Paratyphi C (Daniels et al., 1989), and Dublin (Pickard et al., 2003). On the other hand, S. Cerro, a serovar rarely associated with human disease, is missing several genes found in SPIs 10, 12, and 13 (Rodriguez-Rivera et al., 2014), and contains premature stop codons in genes involved in pathogenesis, phsA and sopA (Rodriguez-Rivera et al., 2014; Kovac et al., 2017b). Existing evidence thus suggests that genomic information can help to assess both (i) the likelihood that a given Salmonella serovar will cause human disease and (ii) the likelihood of causing more severe or mild human disease.
Additionally, epidemiological evidence suggests that some serovars show reduced likelihood of causing human disease. This type of evidence is particularly strong for meat and poultry associated Salmonella, where large datasets on serovars present in raw meat and poultry are available. For example, in the US, S. Cerro represents the most common serovar isolated from cattle without clinical signs of salmonellosis (National Salmonella Surveillance, 2018b), but this serovar only accounted for 38 culture-confirmed cases of human salmonellosis in the US in 2016 (National Salmonella Surveillance, 2018a). Similarly, S. Kentucky was the most frequently isolated serovar isolated from US broiler chickens between 1998 and 2014 (USDA FSIS, 2014), but this serovar is also isolated from very few cases of human clinical illnesses in the US (e.g., 63 in 2016) (National Salmonella Surveillance, 2018b). Hence, regulations and control efforts that consider all NTS serovars and clonal groups an equal public health risk may negatively impact the ability to effectively control serovars that represent the largest public health burden. For example, a framework where all NTS are considered an equal public health risk would encourage vaccination of live animals (e.g., poultry) against those serovars most commonly found in a given population, even if those serovars are characterized by mutations that confer reduced human virulence and are thus unlikely to cause human disease. This is a particular concern as vaccination of live animals against a large number of Salmonella serovars is not possible and as most polyvalent animal Salmonella vaccines typically target at most three serovars (Berghaus et al., 2011).
Current Regulatory and Industry Approaches to Control Salmonella in Foods
Salmonella is a pathogen of concern in a wide range of products, from infant formula, to Ready-To-Eat (RTE) foods (e.g., spices, chocolate) to produce, raw meat and poultry. Regulatory approaches to control this pathogen reflect the diversity of products where Salmonella is a concern. While absence of Salmonella in specified sample sizes (e.g., one 25 g sample) is typically the limit for RTE foods, many countries do not have clear regulatory limits for Salmonella in raw meat and poultry. For example, the US has a Salmonella verification program, which was created in 1996 through the Pathogen Reduction, Hazard Analysis and Critical Control Point (PR/HACCP) Systems Final Rule (USDA FSIS, 2019). The purpose of this program is to verify that establishments that process raw meat and poultry have implemented proper process control strategies to prevent Salmonella contamination. The current system specifies a maximum acceptable number of % positive samples within a moving 52-week window with a different threshold for each USDA FSIS-regulated product. While the USDA FSIS performance standards target all Salmonella spp. and serovars, regulatory programs targeting specific Salmonella serovars also exist. For example, the US National Poultry Improvement Plan includes a focus on control of two specific Salmonella serovars: S. Enteritidis and S. Gallinarum biovars Pullorum and Gallinarum (Shivaprasad, 2000; USDA APHIS, 2020); the focus on S. Enteritidis specifically addresses the serovar that causes the largest number of poultry and egg associated salmonellosis cases in the US. A similar focus on control of S. Enteritidis (and other serovars of particular human health concern) is embedded in regulations in other countries, such as European Commission Regulations (EC) No. 646/2007 (12 June 2007) and 584/2008 (20 June 2008), which target reduction of S. Enteritidis and S. Typhimurium prevalence in broilers and turkeys, respectively.
Reflecting the diversity of foods associated with human salmonellosis cases, control strategies for different products and commodities also vary. For RTE products, effective control can be achieved through implementation of HACCP or FSMA-compliant food safety plans with appropriate preventive controls. In RTE products, detection of Salmonella at low levels (1 Salmonella cell in 25 or 375 g) would indicate a failure of the HACCP or food safety plan; hence regulatory standards that target detection of Salmonella spp. are warranted for RTE products. However, control of Salmonella in raw meat and poultry, is considerably more challenging due to the lack of a kill step (a processing step that substantially reduces target pathogen levels [e.g., by 5 log]) and involves a combination of strategies, including some that target all Salmonella (and in most cases a larger group, such as all Enterobacteriaceae) as well as some that by design or necessity can only target specific Salmonella serovars or clonal groups. For example, vaccination of poultry against Salmonella, while a highly effective control strategy, can typically only target a few Salmonella serovars. Supporting the efficacy of vaccinations, Dórea et al. showed that compared to a farm without a vaccination strategy, a farm vaccinating breeder chickens against serovars Typhimurium, Berta, and Kentucky displayed significantly lower prevalence of Salmonella in the ceca and reproductive tracts of hens and their progeny, as well as significantly lower prevalence of Salmonella in environmental samples (Dórea et al., 2010). This study however also highlights one of the challenges of using vaccines to control Salmonella in meat and poultry; if regulations and performance standards target all Salmonella, then the vaccine used would typically be selected to target the most common serovars present in a given animal species or in a given operation and not necessarily the serovars representing the largest public health risk. Consistent with this, the Dórea et al. study, which was conducted in the US, included serovar Kentucky as one of the three vaccine strains, most likely since this serovar is common in poultry; however, as detailed above, in the US, the most prevalent S. Kentucky sequence type, ST152, is rarely associated with human cases (Haley et al., 2016) and inclusion of another serovar would likely have a larger public health benefit.
The poultry industry currently relies on two types of vaccines to vaccinate flocks against Salmonella: autogenous and live attenuated. Autogenous vaccines are derived from Salmonella isolated from infected birds and are used in the flock the Salmonella was isolated from. These vaccines must be redesigned every 2 years and administration of these vaccines is often labor-intensive as birds must be given them multiple times. Thus, autogenous vaccines are typically used for a short amount of time and only in cases in which no other alternatives exist. Live attenuated vaccines only protect against certain serovars, but are administered through less labor-intensive methods, including drinking water or sprays. However, they require approval by the USDA, which can often take up to 7 years (Eskin, 2020). With limited options available, farmers must focus vaccination efforts on vaccine options that are available to them or those serovars most prevalent in their flocks, often resulting in vaccination against serovars with limited public health relevance. To maximize the benefits of public health risk-based classification of Salmonella, vaccination options thus need to expand to facilitate vaccination against serovars with high public health relevance. This illustrates why a regulatory approach and framework that incentivizes control of Salmonella serovars that represent a higher public health risk would likely have significant public health benefits, particularly with regard to human salmonellosis cases attributed to raw meat and poultry. In addition to facilitating vaccine deployment that emphasizes positive public health outcomes, a public health and serovar focused approach to control Salmonella in raw meat and poultry could also facilitate and accelerate innovation with regard to development of effective serovar-specific control strategies that can be applied at the farm and raw product level (e.g., phage-based control strategies).
A Framework for Serovar and Clonal Group Specific and Risk-Based Salmonella Control Strategies in Raw Meat and Poultry
As detailed above, there is clear evidence that new approaches are needed to improve our ability to control Salmonella transmission through foods. Improved control of Salmonella in raw meat and poultry in particular would likely have a significant public health impact, as these products are a frequent source of human Salmonella infections (IFSAC, 2020). However, with the frequent presence of Salmonella in poultry and meat animals, risk-based approaches that target prevention and control will be essential to make public health gains. Possible risk-based strategies would be to focus control efforts on animal populations (e.g., herds, flocks) that have a high Salmonella prevalence and that contribute high Salmonella loads and/or to focus control efforts on the specific animals shedding high levels of Salmonella (so called “supershedders”); a focus on supershedders can however be difficult as these are rare and hence, difficult to identify. Another opportunity to shift toward a risk-based approach to Salmonella control is to develop and implement a framework that facilitates interventions focusing on Salmonella serovars that represent the largest public health risk and a reduced focus on serovars that represent a low public health risk. Importantly, a strategy that considers all Salmonella an equal public health risk (which, with few exceptions, is the de facto status quo in many countries around the world) leads to the unintended consequence that resources and efforts are focused on control of the most prevalent serovars and clonal groups. This is a particular concern for meat and poultry, where, at least in some countries and as detailed above, some of the most frequently isolated Salmonella (e.g., serovars Cerro and Kentucky ST152 in the US) show clear evidence for reduced human virulence. Hence, we propose that a shift toward public health prioritization of meat and poultry associated Salmonella with an initial focus on strategies that define virulence attenuated Salmonella and reduce unintentional incentives to focus on intervention efforts (e.g., vaccines) that target these Salmonella groups will improve our ability to reduce the public health burden of human salmonellosis.
A key challenge is how to develop an effective and science-based approach to classify Salmonella serovars based on their human public health risk. Current approaches for public health classification of foodborne pathogens typically take a binary approach and classify bacterial groups (genera, species, or serovars) as a public health hazard (or not). An alternative approach, which we propose for Salmonella control in raw meat and poultry, would be to develop and implement a quantitative and risk-based approach to public health classification. This could be particularly valuable to allow for modification of current performance standards (such as the USDA FSIS performance standard detailed above), where a numerical “virulence modifier” could be used to adjust the contributions of samples positive for virulence attenuated Salmonella (Figure 1A). Defining science-based numerical modifiers (or “adjustment factors”) will not be simple and likely introduces additional challenges. We propose that a stepwise approach to defining adjustment factors for different Salmonella serovars or clonal groups, focusing first on common serovars where there is scientific support for reduced ability to cause human disease (e.g., serovar Cerro, serovar Kentucky ST152) (Haley et al., 2016; National Salmonella Surveillance, 2018b), represents the most likely path to success. While one can envision numerous possible strategies to define adjustment factors, we will briefly outline two possible approaches. As detailed above, initial high confidence identification and characterization of virulence attenuated Salmonella serovars or clonal groups is essential; ideally this classification should be based on a convergence of (i) presence/absence of well-characterized virulence-associated genes, (ii) phenotypic evidence from cell culture and/or animal models, and (iii) epidemiological evidence. Synthesis of these data will reduce the impact of potential biases (e.g., sampling or sequencing biases) that may result in falsely predicting virulence attenuation of some serovars or clonal groups.
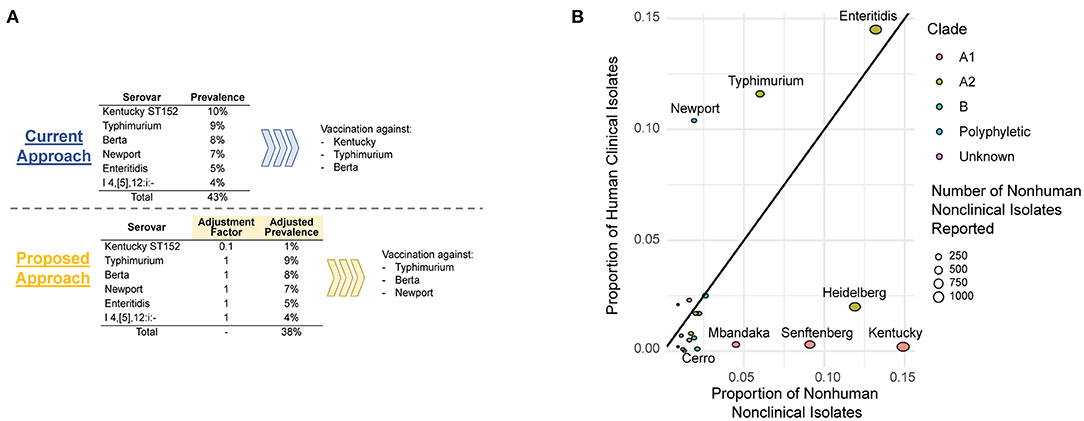
Figure 1. (A) A hypothetical example of how adjustment factors could be used to inform intervention strategies to decrease prevalence of NTS serovars of public health significance in chicken flocks. (B) Over- and underrepresentation of human and animal isolates based on data from Centers for Disease Control and Prevention's 2012 National Salmonella Surveillance Annual Report (National Salmonella Surveillance, 2014). Bubbles are colored based on the phylogenetic clade of the serovar as determined previously (Worley et al., 2018); serovars that are known to be polyphyletic are shown in blue.
One approach that can be used to define an adjustment factor would be based on the relative prevalence of a given serovar or clonal group among the source population (e.g., poultry) and human clinical cases; a similar approach has been used to classify L. monocytogenes clonal complexes into hypo- and hypervirulent clones (Maury et al., 2016). Conceptually, this approach quantifies over- and underrepresentation of pathogen groups (e.g., serovars) among human cases relative to exposure; for example, a factor of 0.1 would indicate that a given group is 10 times less likely to cause human disease relative to the “average” Salmonella. Consequently, this number could be directly used as an adjustment factor (Figure 1A). Possible challenges and drawbacks of this approach include the importance of representative sampling of different sources (to allow for a reliable estimate of this factor) as well as other possible biases in the data used to calculate this factor. A more involved, but likely more robust approach to define an adjustment factor, would be to define the probability of different Salmonella to cause human disease, for example using the r value defined for a given serovar or clonal group. The r value represents the probability of a single pathogen cell causing illness (Chen et al., 2006); the log10 r value for the genus Salmonella has been estimated to be around 2.6 (i.e., 400 cells) (FAO/WHO, 2002). A possible approach for calculating r values for different strains or subgroups of a foodborne pathogen has previously been reported for L. monocytogenes (Chen et al., 2011); mean log10 r values were estimated to be 8.1 and 10.7 for L. monocytogenes subtypes with genes encoding a full-length and a truncated version of the key virulence gene inlA, respectively. A similar approach could be used to initially define r values for all Salmonella and for putative virulence attenuated serovars or clonal groups, which could subsequently be used to calculate an adjustment factor. For example, if the r value for all Salmonella was 400, while the r value for S. Cerro was estimated to be 4,000, then an adjustment factor of 0.1 may be appropriate for use with S. Cerro.
Importantly, a risk-based system that accounts for differences in the ability of Salmonella serovars or clonal groups to cause human disease, requires effective surveillance and monitoring strategies that can identify changes in virulence and emergence of clonal groups with unique hypo- or hypervirulence phenotypes. The widespread use of whole genome sequencing (WGS) for foodborne disease surveillance in an increasing number of countries represents an effective tool for “virulence surveillance,” which is essential for reliable and safe implementation of a risk-based approach to Salmonella control as described above. The NCBI Pathogen Detection browser (ncbi.nlm.nih.gov/pathogen) integrates WGS information for human clinical, animal, food, and environmental isolates and as of March 24, 2021, includes WGS data for >350,000 Salmonella isolates, which are classified into single nucleotide polymorphism (SNP) clusters; an isolate is grouped into a given SNP cluster if it differs by <50 SNPs from at least one isolate already in the cluster. NCBI Pathogen Detection thus provides an essential infrastructure for implementation of modern, risk-based precision food safety approaches. Importantly, this dataset can be used to define both potential hypo- and hypervirulent serovars and clonal groups, by identifying groups with over and under-representation of human isolates (Figure 1B). These WGS data can also be used to identify genomic features and mutations that are likely to be associated with attenuated human virulence. With this strategy, representative and reliable WGS data for human, food, and animal isolates are essential. Currently, CDC, USDA FSIS, and FDA all perform WGS on Salmonella isolates obtained through their respective surveillance systems. For example, USDA FSIS uses WGS to characterize Salmonella isolates from product samples, National Antimicrobial Resistance Monitoring System (NARMS) cecal samples, and USDA Agriculture Marketing Service samples (USDA FSIS, 2020). CDC and state health department partners perform WGS on all human Salmonella isolates obtained in the US. Continued routine collection of WGS data is essential to ensure that the system proposed here works effectively and appropriately. In addition, increased funding and continued efforts to standardize WGS approaches across regulatory and public health agencies will be extremely valuable for this type of approach. On the other hand, it is important to note that the food industry is still in early phases of implementing the use of WGS data. Currently, the food industry uses WGS data for root cause analyses in the case of pathogen contamination events and for identification of virulence factors or antimicrobial resistance genes in a pathogen, but it is rarely used for routine surveillance (Jagadeesan et al., 2019). As such, industry WGS data is currently limited and may be potentially biased (e.g., WGS may be performed predominantly for strains that cause clinical disease).
Virulence and public health risk associated with a given clonal group (e.g., a SNP cluster) and a serovar is not static; use of NCBI Pathogen Detection (or a similar dataset or repository) to monitor for evidence of changes in virulence potential will be essential. Examples of an indication for increased virulence potential of a clonal group previously considered to represent a reduced public health would be if (i) isolates of a given clonal group newly added to a database are found to contain mutations or mobile genetic elements (e.g., plasmids and phages) that are predicted to restore virulence (Miller et al., 2020; Tassinari et al., 2020) or if (ii) addition of human clinical isolates significantly increases the number of human isolates in this clonal group (indicating an increased frequency of human cases by a clonal group previously assumed to be “hypo-virulent”). Additionally, monitoring WGS and other data for indications of an increased prevalence of a given serovar or clonal group can provide important data on the changing ecology and transmission of Salmonella. For example, a campaign to eliminate S. Gallinarum in chickens during the early 20th century resulted in other serovars, like S. Enteritidis, exploiting a newly unoccupied niche and causing it to become a major cause of human salmonellosis (Callaway et al., 2008). As such, increased monitoring of WGS data will also allow for faster detection of similar phenomena and thus, faster response to prevent a similar outcome. Finally, virulence capacity can also vary over time or based on geographical location with specific hypo- or hypervirulent Salmonella serovars or clonal groups being associated with a specific region. Increased monitoring of WGS data will be useful in quickly identifying so-called “geographic hotspots” where hypo- or hypervirulent clonal groups or serovars have increased in prevalence. For example, S. Kentucky appears to include clonal groups that differ in the ability to cause human disease with some regions characterized by circulation of fully virulent strains (e.g., ST198 in Europe), while other regions may be characterized by circulation of strains that show reduced likelihood of causing human disease (e.g., ST152 in the US) (Haley et al., 2016). Importantly, these examples illustrate that implementation of food safety systems that consider Salmonella virulence differences require a sophisticated food safety and public heath infrastructure.
Conclusions
While hazard-based approaches (e.g., HACCP) have been credited to have played an important role in reducing the public health burden associated with bacterial foodborne illnesses, there is increasing evidence that continuous food safety improvements will require improved implementation of risk-based and precision food safety approaches (Kovac et al., 2017a). While risk-based food safety systems can consider a number of different factors that affect risk (e.g., pathogen loads in food, ability of a food to support pathogen growth, host susceptibility), differences in the ability of a given pathogen group (e.g., species, genus) to cause human disease are rarely explicitly considered in regulations, performance standards, or food safety strategies. With the emergence of new tools and data (e.g., large WGS datasets for foodborne pathogen isolates from diverse sources), it is now feasible to develop food safety strategies that consider differences in ability to cause human disease, among members of a given species or genus that currently may be considered a “carte blanche” pathogen. As detailed here, the genus Salmonella represents a pathogen group where stepwise implementation of a new approach that considers the public health risk associated with different serovars and clonal groups may provide for a considerably improved ability to prevent human Salmonella infections linked to raw meat and poultry, by focusing directed interventions (e.g., vaccination) on those Salmonella most likely to cause human infections.
Author Contributions
This opinion piece was conceptualized by RC, RO, and MW. AC and MW wrote, reviewed, and edited the original and final drafts. RO and RC reviewed and edited the final drafts. Funding was secured by MW. All authors contributed to the article and approved the submitted version.
Funding
This work was funded by Pew Charitable Trusts contract number 34063. RC was supported by USDA 2020-67034-31905.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Pew Charitable Trusts for their support in the publication of this opinion piece.
References
Berghaus, R. D., Thayer, S. G., Maurer, J. J., and Hofacre, C. L. (2011). Effect of vaccinating breeder chickens with a killed Salmonella vaccine on Salmonella prevalences and loads in breeder and broiler chicken flocks. J. Food Protect. 74, 727–734. doi: 10.4315/0362-028X.JFP-10-542
Brenner, F. W., Villar, R. G., Angulo, F. J., Tauxe, R., and Swaminathan, B. (2000). Salmonella nomenclature. J. Clin. Microbiol. 38, 2465–2467. doi: 10.1128/JCM.38.7.2465-2467.2000
Callaway, T. R., Edrington, T. S., Anderson, R. C., Byrd, J. A., and Nisbet, D. J. (2008). Gastrointestinal microbial ecology and the safety of our food supply as related to Salmonella. J. Anim. Sci. 86, E163–172. doi: 10.2527/jas.2007-0457
Chen, Y., Ross, W. H., Gray, M. J., Wiedmann, M., Whiting, R. C., and Scott, V. N. (2006). Attributing risk to Listeria monocytogenes subgroups: dose response in relation to genetic lineages. J. Food Protect. 69, 335–344. doi: 10.4315/0362-028X-69.2.335
Chen, Y., Ross, W. H., Whiting, R. C., Van Stelten, A., Nightingale, K. K., Wiedmann, M., et al. (2011). Variation in Listeria monocytogenes dose responses in relation to subtypes encoding a full-length or truncated internalin A. Appl. Environ. Microbiol. 77, 1171–1180. doi: 10.1128/AEM.01564-10
Cheng, R. A., Eade, C. R., and Wiedmann, M. (2019). Embracing diversity: differences in virulence mechanisms, disease severity, and host adaptations contribute to the success of nontyphoidal Salmonella as a foodborne pathogen. Front. Microbiol. 10, 1368–1368. doi: 10.3389/fmicb.2019.01368
Daniels, E. M., Schneerson, R., Egan, W. M., Szu, S. C., and Robbins, J. B. (1989). Characterization of the Salmonella paratyphi C Vi polysaccharide. Infect. Immun. 57, 3159–3164. doi: 10.1128/IAI.57.10.3159-3164.1989
Dórea, F. C., Cole, D. J., Hofacre, C., Zamperini, K., Mathis, D., Doyle, M. P., et al. (2010). Effect of Salmonella vaccination of breeder chickens on contamination of broiler chicken carcasses in integrated poultry operations. Appl. Environ. Microbiol. 76, 7820–7825. doi: 10.1128/AEM.01320-10
Eskin, S. (2020). Vaccines for Poultry Are Crucial for Preventing Salmonella Contamination [Online]. pewtrusts.org. Available online at: https://www.pewtrusts.org/en/research-and-analysis/articles/2020/09/24/vaccines-for-poultry-are-crucial-for-preventing-salmonella-contamination (accessed March 22, 2021).
FAO (2020). Codex Alimentarius: International Food Standards [Online]. Available online at: https://fao.org (accessed January 10, 2021).
FAO/WHO (2002). Risk Assessments of Salmonella in Eggs and Broiler Chickens. Food and Agricultural Organization of the United Nations. World Health Organization.
Faucher, S. P., Porwollik, S., Dozois, C. M., Mcclelland, M., and Daigle, F. (2006). Transcriptome of Salmonella enterica serovar Typhi within macrophages revealed through the selective capture of transcribed sequences. Proc. Natl. Acad. Sci. U. S. A. 103, 1906–1911. doi: 10.1073/pnas.0509183103
Gal-Mor, O., Boyle, E. C., and Grassl, G. A. (2014). Same species, different diseases: how and why typhoidal and non-typhoidal Salmonella enterica serovars differ. Front. Microbiol. 5:391. doi: 10.3389/fmicb.2014.00391
Haley, B. J., Kim, S. W., Pettengill, J., Luo, Y., Karns, J. S., Van Kessel, J., et al. (2016). Genomic and evolutionary analysis of two Salmonella enterica serovar Kentucky sequence types isolated from bovine and poultry sources in North America. PLoS ONE 11:e0161225. doi: 10.1371/journal.pone.0161225
Havelaar, A. H., Kirk, M. D., Torgerson, P. R., Gibb, H. J., Hald, T., Lake, R. J., et al. (2015). World Health Organization global estimates and regional comparisons of the burden of foodborne disease in 2010. PLoS Med 12:e1001923. doi: 10.1371/journal.pmed.1001923
Healthy People (2020). Healthy People 2020 Objectives and Topics: Food Safety [Online]. healthypeople.gov: Office of Disease Prevention and Health Promotion (accessed February 02, 2021).
Helena, M. B. S.-S. (2008). SPI-7: Salmonella's Vi-encoding pathogenicity island. J. Infect. Dev. Countr. 2, 267–271. doi: 10.3855/jidc.220
IFSAC (2020). Foodborne Illness Source Attribution Estimates for 2018 for Salmonella, Escherichia coli O157, Listeria monocytogenes, and Campylobacter Using Multi-Year Outbreak Surveillance Data, United States. T.I.F.S.A. Collaboration (cdc.gov).
Issenhuth-Jeanjean, S., Roggentin, P., Mikoleit, M., Guibourdenche, M., De Pinna, E., Nair, S., et al. (2014). Supplement 2008-2010 (no. 48) to the White-Kauffmann-Le Minor scheme. Res. Microbiol. 165, 526–530. doi: 10.1016/j.resmic.2014.07.004
Jagadeesan, B., Gerner-Smidt, P., Allard, M. W., Leuillet, S., Winkler, A., Xiao, Y., et al. (2019). The use of next generation sequencing for improving food safety: translation into practice. Food Microbiol. 79, 96–115. doi: 10.1016/j.fm.2018.11.005
Jechalke, S., Schierstaedt, J., Becker, M., Flemer, B., Grosch, R., Smalla, K., et al. (2019). Salmonella establishment in agricultural soil and colonization of crop plants depend on soil type and plant species. Front. Microbiol. 10:967. doi: 10.3389/fmicb.2019.00967
Kovac, J., Bakker, H. D., Carroll, L. M., and Wiedmann, M. (2017a). Precision food safety: a systems approach to food safety facilitated by genomics tools. TRAC - Trend Anal. Chem. 96, 52–61. doi: 10.1016/j.trac.2017.06.001
Kovac, J., Cummings, K. J., Rodriguez-Rivera, L. D., Carroll, L. M., Thachil, A., and Wiedmann, M. (2017b). Temporal genomic phylogeny reconstruction indicates a geospatial transmission path of Salmonella Cerro in the United States and a clade-specific loss of hydrogen sulfide production. Front. Microbiol. 8:737. doi: 10.3389/fmicb.2017.00737
Lee, F. K. M., Morris, C., and Hackett, J. (2006). The Salmonella enterica serovar Typhi Vi capsule and self-association pili share controls on expression. FEMS Microbiol. Lett. 261, 41–46. doi: 10.1111/j.1574-6968.2006.00338.x
Marcus, S. L., Brumell, J. H., Pfeifer, C. G., and Finlay, B. B. (2000). Salmonella pathogenicity islands: big virulence in small packages. Microbes Infect. 2, 145–156. doi: 10.1016/S1286-4579(00)00273-2
Maury, M. M., Tsai, Y. H., Charlier, C., Touchon, M., Chenal-Francisque, V., Leclercq, A., et al. (2016). Uncovering Listeria monocytogenes hypervirulence by harnessing its biodiversity. Nat. Genet. 48, 308–313. doi: 10.1038/ng.3501
Miller, E. A., Elnekave, E., Flores-Figueroa, C., Johnson, A., Kearney, A., Munoz-Aguayo, J., et al. (2020). Emergence of a novel Salmonella enterica serotype Reading clonal group is linked to its expansion in commercial turkey production, resulting in unanticipated human illness in North America. mSphere 5:e00056-20. doi: 10.1128/mSphere.00056-20
National Salmonella Surveillance CDC. (2014). National Salmonella Surveillance Annual Report, 2012. National Salmonella Surveillance, Centers for Disease Control and Prevention (cdc.gov).
National Salmonella Surveillance CDC. (2018a). National Salmonella Surveillance 2016 Supplement: Animals Isolate Report. USDA National Veterinary Services Laboratory (cdc.gov).
National Salmonella Surveillance CDC. (2018b). Salmonella Annual Summary, 2016. National Salmonella Surveillance (cdc.gov: Centers for Disease Control and Prevention).
Pickard, D., Wain, J., Baker, S., Line, A., Chohan, S., Fookes, M., et al. (2003). Composition, acquisition, and distribution of the Vi exopolysaccharide-encoding Salmonella enterica pathogenicity island SPI-7. J. Bacteriol. 185, 5055–5065. doi: 10.1128/JB.185.17.5055-5065.2003
Rodriguez-Rivera, L. D., Moreno Switt, A. I., Degoricija, L., Fang, R., Cummings, C. A., Furtado, M. R., et al. (2014). Genomic characterization of Salmonella Cerro ST367, an emerging Salmonella subtype in cattle in the United States. BMC Genom. 15:427. doi: 10.1186/1471-2164-15-427
Shivaprasad, H. L. (2000). Fowl typhoid and pullorum disease. Rev. Sci. Tech. 19, 405–424. doi: 10.20506/rst.19.2.1222
Tack, D. M., Ray, L., Griffin, P. M., Cieslak, P. R., Dunn, J., Rissman, T., et al. (2020). Preliminary incidence and trends of infections with pathogens transmitted commonly through food - Foodborne Disease Active Surveillance Network, 10 U.S. Sites, 2016-2019. Morbid. Mortal. W. 69, 509–514. doi: 10.15585/mmwr.mm6917a1
Tassinari, E., Bawn, M., Thilliez, G., Charity, O., Acton, L., Kirkwood, M., et al. (2020). Whole-genome epidemiology links phage-mediated acquisition of a virulence gene to the clonal expansion of a pandemic Salmonella enterica serovar Typhimurium clone. Microb. Genom. 6:456. doi: 10.1099/mgen.0.000456
USDA APHIS (2020). National Poultry Improvement Plan (NPIP) [Online]. aphis.usda.gov (accessed January 09, 2021).
USDA FSIS (2014). Serotypes Profile of Salmonella Isolates from Meat and Poultry Products January 1998 through December 2014. fsis.usda.gov: USDA FSIS. (accessed March 31, 2021).
USDA FSIS (2019). Pathogen Reduction - Salmonella and Campylobacter Performance Standards Verification Testing [Online]. fsis.usda.gov: USDA (accessed March 31, 2021).
USDA FSIS (2020). Roadmap to Reducing Salmonella. USDA FSIS (fsis.usda.gov).
Waldner, L. L., Mackenzie, K. D., Köster, W., and White, A. P. (2012). From exit to entry: long-term survival and transmission of Salmonella. Pathogens 1, 128–155. doi: 10.3390/pathogens1020128
Keywords: Salmonella, risk assessment, performance standards, food safety, whole genome sequencing
Citation: Cohn AR, Cheng RA, Orsi RH and Wiedmann M (2021) Moving Past Species Classifications for Risk-Based Approaches to Food Safety: Salmonella as a Case Study. Front. Sustain. Food Syst. 5:652132. doi: 10.3389/fsufs.2021.652132
Received: 11 January 2021; Accepted: 13 April 2021;
Published: 13 May 2021.
Edited by:
Teresa M. Bergholz, Michigan State University, United StatesReviewed by:
Young Min Kwon, University of Arkansas, United StatesSi Hong Park, Oregon State University, United States
Bradley L. Bearson, United States Department of Agriculture (USDA), United States
Copyright © 2021 Cohn, Cheng, Orsi and Wiedmann. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Martin Wiedmann, bWFydGluLndpZWRtYW5uQGNvcm5lbGwuZWR1
 Alexa R. Cohn
Alexa R. Cohn Rachel A. Cheng
Rachel A. Cheng Renato H. Orsi
Renato H. Orsi Martin Wiedmann
Martin Wiedmann