
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Sustain. Food Syst. , 23 March 2021
Sec. Crop Biology and Sustainability
Volume 5 - 2021 | https://doi.org/10.3389/fsufs.2021.643225
This article is part of the Research Topic Plant Growth-Promoting Microorganisms for Sustainable Agricultural Production View all 50 articles
The One Strain Many Compounds approach (OSMAC) is a powerful and comprehensive method that enables the chemo-diversity evaluation of microorganisms. This is achieved by variations of physicochemical cultivation parameters and by providing biotic and abiotic triggers to mimic microorganisms' natural environment in the lab. This approach can reactivate the silent biosynthetic routes of specific metabolites typically not biosynthesized under standard laboratory conditions. In the present study, we combined the OSMAC approach with static headspace solid-phase microextraction-gas chromatography-mass spectrometry (SPME-GC-MS), high-performance liquid chromatography-high-resolution tandem mass spectrometry (HPLC-HRMSn), and matrix-assisted laser desorption/ionization high-resolution mass spectrometry imaging (MALDI-HRMSI) to evaluate the chemoecological significance of an apple root-associated endophytic Streptomyces pulveraceus strain ES16. We employed the OSMAC approach by cultivating the endophyte in six different media conditions and performed temporal studies over 14 days. Analysis of the volatilome revealed that only under stressful conditions associated with sporulation, endophytic S. pulveraceus ES16 produces geosmin, a volatile semiochemical known to attract the soil arthropods Collembola (springtails) specifically. Subsequently, targeted metabolic profiling revealed polycyclic tetramate macrolactams (PTMs) production by the endophyte under stress, which are bioactive against various pathogens. Additionally, the endophyte produced the iron-chelating siderophore, mirubactin, under the same conditions. The structures of the compounds were evaluated using HRMSn and by comparison with literature data. Finally, MALDI-HRMSI revealed the produced compounds' spatial-temporal distribution over 14 days. The compounds were profusely secreted into the medium after production. Our results indicate that endophytic S. pulveraceus ES16 can release the signal molecule geosmin, chemical defense compounds such as the PTMs, as well as the siderophore mirubactin into the host plant apoplast or the soil for ecologically meaningful purposes, which are discussed.
Plant-associated mutualistic microorganisms such as endophytic microorganisms, commonly known as endophytes, which colonize plants' internal tissues, frequently contribute to host metabolic function and protect plants against pests and diseases by producing biocontrol traits such as bioactive secondary metabolites (Carrión et al., 2019; White et al., 2019; Morales-Cedeño et al., 2020; Newman and Cragg, 2020). For example, endophytes are known to produce natural products that possess a plethora of biological activities ranging fromantibacterial, antifungal, cytotoxic, anticancer, insecticidal to communication molecules (Kusari et al., 2017; Ancheeva et al., 2020; Joo et al., 2020). These biocontrol traits might be amenable to utilization by genetic, epigenetic, and metabolic fine-tuning of endophytes that can then be used as biologics (or biological control agents) to protect host plants in the field conditions without the negative environmental impacts of conventional pesticides. However, to date, biologics or endophytic microbial biocontrol agents have mostly failed to work in field conditions. This is frequently attributed to insufficient expression of biocontrol traits in planta or the introduced organism's failure to be competitive in the host environment. Whole-genome sequencing data shows that only a minor portion of endophytes' biosynthetic potential is expressed under standard in vitro culture conditions (Khare et al., 2018). Current evidence indicates that the synthesis of these “cryptic” compounds is induced in planta in the presence of other interacting micro- and macro-organisms through transcriptional and epigenetic regulatory mechanisms (Mhlongo et al., 2018; Cheng et al., 2019; Jones et al., 2019; Babalola et al., 2020). Chemical interactions among endophytes or between endophytes and interacting partners such as associated endophytes, host plant, invading pathogens, insects, and soil arthropods lead to sustained production of physiologically relevant metabolites, and interspecies crosstalk is often necessary for triggering the production of natural products that are not produced without a suitable trigger (Mhlongo et al., 2018; Cheng et al., 2019; Jones et al., 2019; Babalola et al., 2020). Only suitable interactions lead to the continuous and sustained production of one or more metabolites in an ecological niche.
Hence, it is necessary to understand the chemical-ecological interactions of endophytes on a case-by-case basis to fully exploit the potential of natural product biosynthesis and sustainable production for agriculture and industrial production of desired pharmaceutically relevant compounds, as well as to elucidate the intricate mesh of endophytic microbial chemoecological networks with coexisting organisms. A comprehensive and robust methodological approach employed to achieve this is called the One Strain Many Compounds (OSMAC) approach (Bode et al., 2002; Maghembe et al., 2020). The OSMAC approach enables establishing a comprehensive metabolic profile for each microorganism, revealing the compounds produced by the organism in its natural ecological niche (Romano et al., 2018; Pan et al., 2019; Maghembe et al., 2020). In vitro, culture-based OSMAC approach includes modulation of the physiochemical culture parameters, the addition of ecological niche-mimetic biotic and abiotic stress factors and triggers, and bioprocess optimization in a bid to selectively express cryptic gene clusters that are only expressed under particular conditions in the original natural habitat of the microorganism under study (Pan et al., 2019; Maghembe et al., 2020).
Numerous studies have substantiated the biosynthetic capacity of Streptomyces species, many of which exhibit an endophytic lifestyle, synthesize various bioactive metabolites, and communication molecules that are not only important from a biochemical and molecular standpoint but also from an ecological perspective (van der Meij et al., 2017; Spasic et al., 2018; Law et al., 2020; Quinn et al., 2020). Over the past few decades, Streptomyces have received a significant amount of attention due to their vast production of secondary metabolites and their contribution to modern medicine, agriculture, and veterinary practice, among others. These species also play a significant role in ecology by processing and breaking down enzymes, as well as plants and fungi cell walls. For example, Streptomyces reticuli can degrade complex carbohydrates and cellulose (Wibberg et al., 2016), whereas Streptomyces sp. TH-11 has shown chitinolytic activities (Hoang et al., 2010). Bafilomycins produced by an endophytic Streptomyces species demonstrate antifungal, antitumor, and ionophore properties (Yu et al., 2011). In another study, endophytic Streptomyces spp. have successfully reduced the disease severity of Fusarium spp. on wheat (Colombo et al., 2019). Apart from inhibiting phytopathogens, they also promote plant growth through phytohormones and facilitate the uptake of nutrients by producing compounds such as siderophores (Terra et al., 2020).
The replant disease of apple orchards is a major recurring problem in different regions of the world, the etiology of which is agreed to be a combination of various biotic factors, including multiple pathogenic fungal complexes and is influenced by abiotic factors (Mazzola and Manici, 2012; Winkelmann et al., 2019). Apple Replant Disease (ARD) typically presents a stressful environment for apple plants, endophytes, and associated microbiome (Mazzola and Manici, 2012; Yim et al., 2015; Radl et al., 2019; Winkelmann et al., 2019). The plant roots show signs of browning and blackening, tip necrosis, and reduced root hairs to exemplify this (Grunewaldt-Stöcker et al., 2019). Above the soil, stunted growth and reduced plant biomass are observed, as shown in several studies (Yim et al., 2013, 2015; Weiß et al., 2017). There is a decrease in the fruit yield, and the fruit size and taste are negatively influenced. Building upon an earlier study (Nicola et al., 2018), we recently found an increased abundance of actinobacteria (particularly Streptomyces species) in roots grown in ARD soils compared to healthy roots grown in non-ARD soils using molecular barcoding (Mahnkopp-Dirks et al., 2020). Interestingly, we found that the increased abundance of Streptomyces in apple roots grown in ARD soils negatively correlated with increased shoot length and fresh mass. Furthermore, gene sequencing revealed that Streptomyces taxa are selectively enriched in the rhizosphere in ARD soils in split-root experiments (Balbín-Suárez et al., 2020). However, whether these observations are causal or correlated remains unclear. Further, the involvement and the precise role of yet-unexplored “ecological links” in the form of interacting micro- or macro-organisms such as associated endophytes, rhizosphere microbes, or soil arthropods, among others, is not fully known. Notably, the chemical basis of these observations, particularly the role of natural products such as signal or communication molecules and compounds with niche-relevant biological activities that mediate the underlying chemical ecology, has not been studied.
Therefore, we investigated a cultivable endophytic Streptomyces pulveraceus strain ES16 that we isolated from apple plant roots (Malus domestica) within the Central Experiment 1 setup (CE1; Ellerhoop, Spring 2018) of the BMBF BonaRes program ORDIAmur (Mahnkopp et al., 2018). In particular, we wanted to answer the following three questions about the endophytic S. pulveraceus strain ES16:
1. What are the secreted volatile semiochemicals or signal molecules?
2. What are the bioactive, chemical defense compounds produced?
3. What are the spatial-temporal dynamics of the produced compounds?
To answer the above questions, we employed an OSMAC approach by cultivating endophytic S. pulveraceus ES16 in six different media compositions ranging from nutrient-rich to minimal conditions, including media to stress the organism under submerged conditions (broth) as well as on agar (Figure 1). Under the influence of different OSMAC conditions, we investigated the volatilome produced by the strain by static headspace solid-phase microextraction-gas chromatography-mass spectrometry (SPME-GC-MS) focusing on volatile semiochemicals or signal compounds. After that, we established the selectively expressed metabolic profiles under the different OSMAC conditions, typically focusing on bioactive compounds such as antimicrobial, antiprotozoal, and cytotoxic activities using high-performance liquid chromatography-high resolution tandem mass spectrometry (HPLC-HRMSn). Finally, we employed matrix-assisted laser desorption/ionization high-resolution mass spectrometry imaging (MALDI-HRMSI) to visualize the production, distribution, and release of target compounds in high spatial resolution and temporally for 14 days. We discuss the results obtained concerning the possible chemoecological significance of apple root-associated endophytic S. pulveraceus strain ES16.
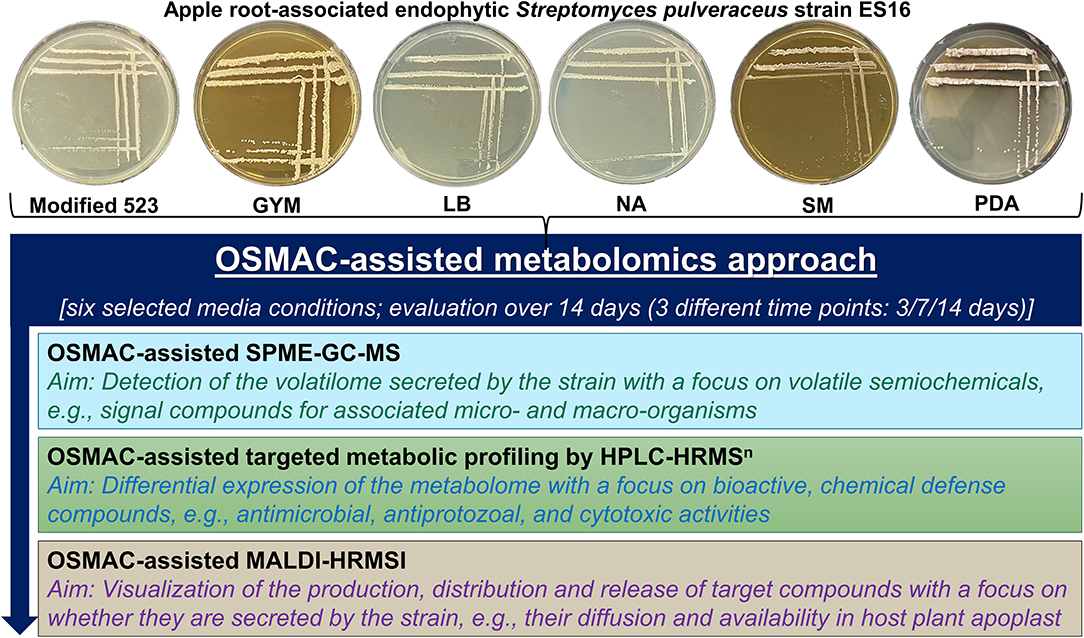
Figure 1. Schematic representation of the overview of workflow employed in the present study. GYM, glucose yeast malt medium; LB, lysogeny medium; NA, nutrient agar; SM, streptomyces medium; PDA, potato dextrose agar.
Endophytic Streptomyces pulveraceus strain ES16 (“Ellerhoop Spring Isolate No. 16”) was isolated from surface-disinfected apple roots (Malus domestica) grown in grass control (ARD-unaffected) soil within the Central Experiment setup 1 (CE1; Ellerhoop, Spring 2018) of the BMBF BonaRes ORDIAmur program (Mahnkopp et al., 2018). For isolation, 1 cm pieces of surface-disinfected fine roots (Ø < 2 mm) were placed in Petri dishes containing 523 medium (Viss et al., 1991) and stored for ~7 days at room temperature. After this, emerging colonies were picked and incubated for 1–7 days in liquid 523 medium until growth was visible. 1 mL of suspension was used for DNA extraction based on Quambusch et al. (2014). The 16S rRNA gene was amplified using the primers 27f (AGAGTTTGATCCTGGCTCAG) and 1492r (GGYTACCTTGTTACGACTT) (Weisburg et al., 1991). 16S rDNA fragments were sequenced with the Sanger method (Sanger et al., 1977) by Microsynth Seqlab (Göttingen, Germany), the obtained sequence (Supplementary Table 1) was blasted (Blastn) (Zhang et al., 2000) against the NCBI database (https://www.ncbi.nlm.nih.gov/), and submitted (accession no. MW580619).
We optimized our OSMAC approach established for endophytic bacteria (Eckelmann et al., 2018) to cultivate endophytic Streptomyces pulveraceus strain ES16. Thus, the endophyte was cultivated in six different attuned media conditions, ranging from nutrient-rich to minimal conditions, for selecting different phenotypes (e.g., sporulation under stress) and metabolic expression patterns for this particular strain, both under submerged conditions (broth) as well as on agar. These included the enriched media viz. modified 523 medium and streptomyces medium (SM), the moderately nutrient-rich lysogeny medium (LB) and basal nutrient medium (agar, NA; broth, NB), as well as glucose yeast malt medium (GYM) and potato dextrose medium (agar, PDA; broth, PDB) that served as minimal media for this strain by selectively limiting phosphate and/or nitrogen, thereby inducing moderate to high-stress and sporulation. The components of the media and related references are detailed in Supplementary Table 2. The cultures were incubated at 28°C (Memmert Incubator, Schwabach, Germany), and the bacterial growth, morphology, and OSMAC-relevant phenotypic characteristics were monitored and documented regularly over 14 days. For submerged cultivation and fermentation, 500 mL broth of each of the six media was prepared in 1,000 mL Erlenmeyer flasks and autoclaved at 121°C for 15 min (Autoclave VB-55, Systec, Wettenberg, Germany). One loop of the bacteria was added to the broth under sterile conditions, and the flask was sealed. The inoculated broths were placed in a shaker incubator (Lab-Rotation Incubator Multitron 2, INFORS HT, Einsbach, Germany) and shaken at 125 rpm at 28°C for 3, 7, and 14 days, respectively. As negative controls, 100 mL of all six sterile, uninoculated media broths were incubated simultaneously. All setups and experiments were performed in biological triplicates.
The 3-, 7- and 14-day old bacterial agar plates and negative control plates were crushed to small pieces using a spatula. Each plate was covered with 20 mL HPLC-grade MeOH (J. T. Baker, Deventer, The Netherlands) and was mixed well. The mixture was transferred to a beaker and was extracted in an ultrasonic bath (Sonorex Longlife, Bandelin, Berlin, Germany). After 15 min, the mixture was filtered. This step was repeated thrice (i.e., extraction with 3 × 20 mL fresh MeOH). The filtrate was concentrated to dryness under reduced pressure in a rotary evaporator (Laborota 4001 Efficient, Heidolph, Schwabach, Germany) and resuspended in 4 mL HPLC-grade MeOH (J. T. Baker, Deventer, The Netherlands) for further analyses.
On the 3rd, 7th, and 14th day of cultivation and under sterile conditions, 100 mL of each inoculated broth was transferred to a 250 mL Erlenmeyer flask. Each flask was placed in an ice bath and ultrasonicated thrice for 15 min in the Sonoplus ultrasonic device (Bandelin, Berlin, Germany) equipped with ultrasonic lance UW 3200 (Bandelin, Berlin, Germany). The flasks' content was transferred to Eppendorf centrifuge tubes and was centrifuged at 10,000 rpm for 10 min to separate the biomass (Centrifuge Allegra™ IR, Beckman Coulter GmbH, Krefeld, Germany). The supernatant was decantated, frozen, and was freeze-dried overnight in a Vaco 5 freeze dryer (ZIRBUS technology GmbH, Bad Grund, Germany). The residue was extracted thrice with 20 mL HPLC-grade MeOH (J. T. Baker, Deventer, The Netherlands) in an ultrasonic bath (Sonorex Longlife, Bandelin, Berlin, Germany) for 15 min, and filtered. The filtrate was concentrated to dryness under reduced pressure in a rotary evaporator (Laborota 4001 Efficient, Heidolph, Schwabach, Germany), and was resuspended in 4 mL HPLC-grade MeOH (J. T. Baker, Deventer, The Netherlands). The uninoculated negative control blanks were extracted following the same procedure.
Static headspace SPME-GC-MS was performed according to the published procedure (Yalage Don et al., 2020), suitably modified. Agar plates seeded with S. pulveraceus ES16 were incubated for 5 days at 28°C, and ca. one gram (small pieces) of each bacterial agar plate was cut under sterile conditions and transferred to a headspace vial. The vial was then incubated for another 2 days so that the vial atmosphere was enriched with the volatilome secreted by the bacterium cultivated in a specific agar medium. After that, the vial septum was punctured with a solid-phase microextraction (SPME) needle coating, and the fiber (PDME, 75 μm) was exposed to absorb the bacterial volatilome. After 15 min, the fiber was retracted, and the SPME was inserted directly into the GC-MS, where the bacterial volatilome was thermally desorbed. The analysis of volatile organic compounds (VOCs) was performed with a Trace GC Ultra system (Thermo Fisher Scientific, Waltham, USA), coupled to an ISQ mass spectrometer (Thermo Fisher Scientific, Waltham, USA). The separation of the compounds was achieved via an Optima 5 MS (30 m, 0.25 mm ID, 1 μm, Macherey-Nagel, Düren, Germany) GC column. The initial oven temperature was set to 60°C for the first 2 min and was ramped to 175°C at a rate of 20°C/min. The temperature was again increased to 260°C with the second ramp of 4°C/min and held for 2 min resulting in a total run time of 30 min. The mass spectrometer equipped with an electron impact source (IE) was operated at 70 eV with an ion source temperature of 200°C and a scan range of m/z 50–550.
100 μL of each extract was transferred to an HPLC vial and was concentrated to dryness in a concentrator (Savant SDP1010 SpeedVac, Thermo Fisher Scientific, Waltham, USA). 100 μL of a 2:1 H2O and MeOH mixture was added to the vial, and the residue was reconstituted by vortexing (Vortex mixer VF2, IKA-Werke GmbH, Staufen, Germany). The sample was centrifuged at 6,600 rpm for 5 min (Mini Centrifuge, MCF-2360, LMS CO., LTD., Tokyo, Japan), and the supernatant was transferred to an inlet and measured. The measurements were either carried out with an Agilent 1200 system HPLC (Waldbronn, Germany) coupled to an LTQ-Orbitrap XL mass spectrometer (Thermo Fisher Scientific, Waltham, USA) or a Nexera X2 HPLC (Shimadzu Scientific Instruments, Maryland, USA) coupled with an LTQ-Orbitrap mass spectrometer (Thermo Fisher Scientific, Waltham, USA). A Nucleoshell C18 reverse-phase column (2.7 μm, 150 × 4.6 mm, Macherey-Nagel, Düren, Germany) was used for chromatographic separation at 30°C with H2O (+ 0.1% HCOOH) (A) and MeOH (+ 0.1% HCOOH) (B) gradient (flow rate 300 μL min−1). The gradient program was as follows: 95% A isocratic for 2 min, linear gradient to 100% B over 26 min, 100% B isocratic for 6 min, the system returned within 0.5 min to initial conditions of 95% A and was equilibrated for 2.5 min. The LTQ-Orbitrap XL and the LTQ-Orbitrap were equipped with a HESI ion source with 5 kV voltage at 350°C. The ion source was operated with He as collision gas, and N2 as sheath- (40 arbitrary units), and auxiliary gas (8 arbitrary units). The spectrometers were operated in positive modes with a mass range of m/z 100–1,600 with a nominal mass resolving power of 60,000 at m/z 400 with a scan rate of 1 Hz, with the internal lock masses N-butylbenzenesulfonamide ([M+H]+ m/z 214.0896) and dibutyl phthalate ([M+H]+ m/z 279.1591). The analyses were performed using Xcalibur software v. 2.2 SP1.48 (Thermo Scientific, Bremen, Germany). The acquired masses were sorted by intensity (I > 1.00E3). A maximum mass tolerance of 2 ppm was accepted. For structure elucidation, tandem HRMS experiments were performed with the LTQ-Orbitrap mass spectrometer with collision-induced dissociation (CID) energies of 15, 25, and 35 eV. Background subtraction was performed as required using Xcalibur software v. 2.2 SP1.48 (Thermo Scientific, Bremen, Germany). Scifinder, Knapsack, and PubChem were used as reference databases.
MALDI-HRMSI experiments were performed using our previously established procedure (Eckelmann et al., 2018), suitably modified. Areas of interest of the respective agar plates with the seeded or cultivated microorganisms were cut to square pieces with a razor blade, transferred, fixed onto a 4 × 1 cm microscopical glass slide, and dried for 24 h. A photographic image was taken for each sample using a specialized digital microscope (VHX-5000, Keyence Deutschland GmbH, Neu-Isenburg, Germany) to evaluate the measured area and record the optical image. A SMALDI Prep spray device (TransMIT GmbH, Giessen, Germany) was utilized for matrix spraying. The samples were sprayed uniformly with matrix MBT (2-mercaptobenzothiazole; 5 mg/mL) prepared in H2O:acetone 1:4 (v/v). The spray application was performed using the Matrix Sprayer Control [v. 1.9.2890] (TransMIT GmbH, Giessen, Germany) employing the following parameters: matrix flow 16 μL/min; gas flow 3 L/min; 27 min under the Area Mode. The measurements were carried out at a resolution of 30 μm with an AP-SMALDI5 ionization source (TransMIT GmbH, Giessen, Germany) coupled to a Thermo Q-Exactive mass spectrometer (Thermo Scientific Inc., Bremen, Germany). Data processing and mapping of mass pixels corresponding to the target compounds were done using the software package Mirion V3.36.4.13 (TransMIT GmbH, Giessen, Germany). Ion images were generated with a bin width of ±5.0 ppm. Mass pixels are shown as false colors.
Endophytic S. pulveraceus ES16 colonies' growth on modified 523 agar started within the first 24 h of incubation. After 3 days, the colonies were visible to the naked eye and continued to increase in size until the 7th day of incubation. However, the colonies' growth stopped after the 1st week, as the colonies did not continue to grow in diameter until 14 days. The size of each colony did not exceed 1 mm and therefore could be characterized as punctiform. On this medium, S. pulveraceus ES16 formed circular colonies with a minor convex elevation. They showed a smooth texture with an opaque light beige/white color during the first few incubation days. The color altered slightly after a week to a light pink color. There were no visible changes in the medium's color, i.e., no visible microbial exudates were observed. No sporulation was observed (Figure 2A). This bacterium demonstrated a soft colony consistency when touched with the loop on the 3rd day of incubation, which gradually hardened over the 14-day experimental period.
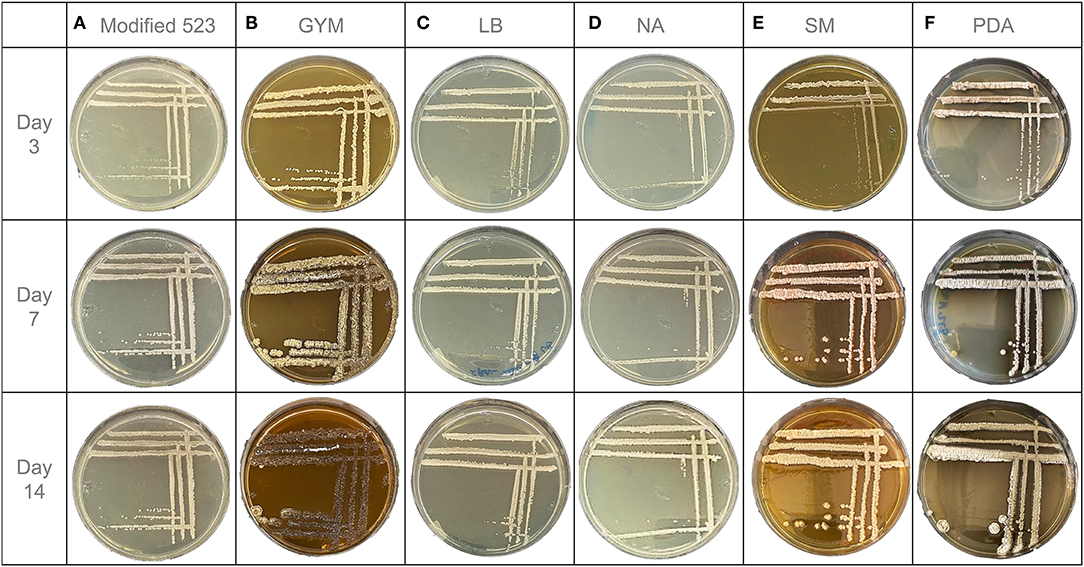
Figure 2. OSMAC-assisted phenotypic characteristics of endophytic S. pulveraceus ES16 grown on six different agar media for 14 days. (A) Modified 523 medium. (B) Glucose Yeast Malt medium (GYM). (C) Lysogeny medium (LB). (D) Nutrient agar (NA). (E) Streptomyces medium (SM). (F) Potato dextrose agar (PDA).
However, the endophyte demonstrated markedly different morphology on GYM agar than that on modified 523 agar (Figure 2B). The colonies started forming after just 12 h of incubation and were completely visible at 48 h. The size of the colonies had a relatively considerable increase with time. On day 3, a single colony's diameter was measured around 1 mm, and it increased to 4 mm and 6 mm on days 7 and 14, respectively. Apart from growth in size, a drastic change in the bacterium's color and shape was observed with incubation time. The initial colony shape of S. pulveraceus ES16 on GYM agar was an irregular convex form with a wrinkled surface. The colonies were firstly small and had a light beige color. On day 7, it was noticeable that some parts of the colonies had started to sporulate while other parts had a darker brown color and no longer had an elevated convex colony character. After 14 days of incubation, the bacterium's color altered almost entirely to dark brown, which indicated that the bacteria could be in the stationary or decline phase. On day 7, a potting soil odor was detected as the plate was opened. The change in the medium color around the colonies to a darker shade over time was indicative of visual exudates secreted by the endophyte. On day 3, the bacterial colony consistency was soft and smooth, whereas it was brittle on day 7. On day 14, the bacterium had developed a hard and dry texture.
The growth of S. pulveraceus ES16 on LB agar was typically similar to that on modified 523 agar (Figure 2C). The colonies' formation started 24–36 h after the incubation, and after 3 days, the colonies were visible to the naked eye. The single colonies showed a minimal increase in size from day 3 to day 7, and the overall diameter of a single colony did not exceed 1 mm. The endophytic colonies were light beige/white with a smooth texture. The colonies were circular and raised. The color of the bacteria remained the same for the first 7 days. After 14 days of incubation, the color altered to light yellow. There were no visible changes in the agar's texture and shade, and no sporulation was observed. When touched with the inoculation loop, S. pulveraceus ES16 showed a viscid consistency throughout the 14 days of observation.
In the case of NA, the colonies of S. pulveraceus ES16 were not visible on agar until 48 h after incubation. Like that of modified 523 and LB agar, the colonies were relatively small (1 mm) and fully grown by day 7. They maintained the same light beige/white color throughout the 14-day incubation time. The colonies had a circular shape with a smooth texture. No alterations in the color of the medium, and no sporulation were observed (Figure 2D).
The colonies of S. pulveraceus were visible within the first 6 h after cultivation on SM agar. On day 3 of incubation, tiny punctiform colonies (<1 mm) could be observed (Figure 2E). From day 3 till day 7, a drastic increase in colony size was observed. The colony color was initially opaque white/light beige, similar to the other tested media. On day 7, the observed color had transformed into pink/beige. The endophyte formed irregular curled colonies with a pulvinate elevation and demonstrated a rough, brittle surface. No sporulation was observed.
Entirely different morphology of S. pulveraceus ES16 was observed on PDA (Figure 2F). The first colonies appeared after 24 h. The colony size increased with time while remaining ca. 1 mm on day 3, 3 mm on day 7, and 5 mm on day 14. At 48 h of incubation, the colonies had a similar morphology to that of modified 523 agar, NA, and LB agar with light beige color and circular shapes. However, by day 3, the bacterium began to sporulate vigorously, revealing a high-stress environment. The colony color changed rapidly to light gray and darkened after 7 and 14 days, respectively. By day 7, extensive sporulation of the colonies was noted. Despite sporulation, the colonies remained circular and convex in shape. Strikingly, from around day 7 of the incubation, the bacterial plate emitted a potting soil scent, which intensified by day 14. Another significant observation on the PDA plates was the pigmentation of the medium. Already by day 3, the color of the medium changed from yellow to dark brown, which intensified during the 14 days of incubation time, revealing secretion of visible exudates. S. pulveraceus ES16 had a texture similar to that of a fungal mycelium on PDA. Once in contact with the loop, it was viscid and soft.
SPME-GC-MS was employed to evaluate the production of volatile organic compounds (VOCs) (Tholl et al., 2006; Iijima, 2014; Yalage Don et al., 2020) by endophytic S. pulveraceus ES16 in different media conditions. Notably, the strong earthy odor released by the endophyte, when growing on PDA plates, coupled with extensive sporulation, provided a scientific handle to focus on stress-induced compounds and semiochemicals (signal molecules). Extensive analyses of the samples compared to the NIST EI-MS spectral library revealed the endophytic production of the volatile sesquiterpene, geosmin (trans-1,10-dimethyl-trans-9-decalol; compound 1; Figure 3) specifically on PDA but not in other media conditions. The fragmentation pattern of the geosmin (Figure 4) matched with the reference NIST EI-MS spectral library, confirming its identity. Geosmin (1) production was only observed when the endophyte was subjected to stressful conditions, concomitant with extensive sporulation and a strong earthy smell. We also searched for other known signal molecules that might be produced along with geosmin, including 2-methylisoborneol (2-MIB), germacerene D, germacradienol (Becher et al., 2020), among others. However, these compounds were not detected (< LOD) while analyzing the expressed volatilome of endophytic S. pulveraceus ES16.
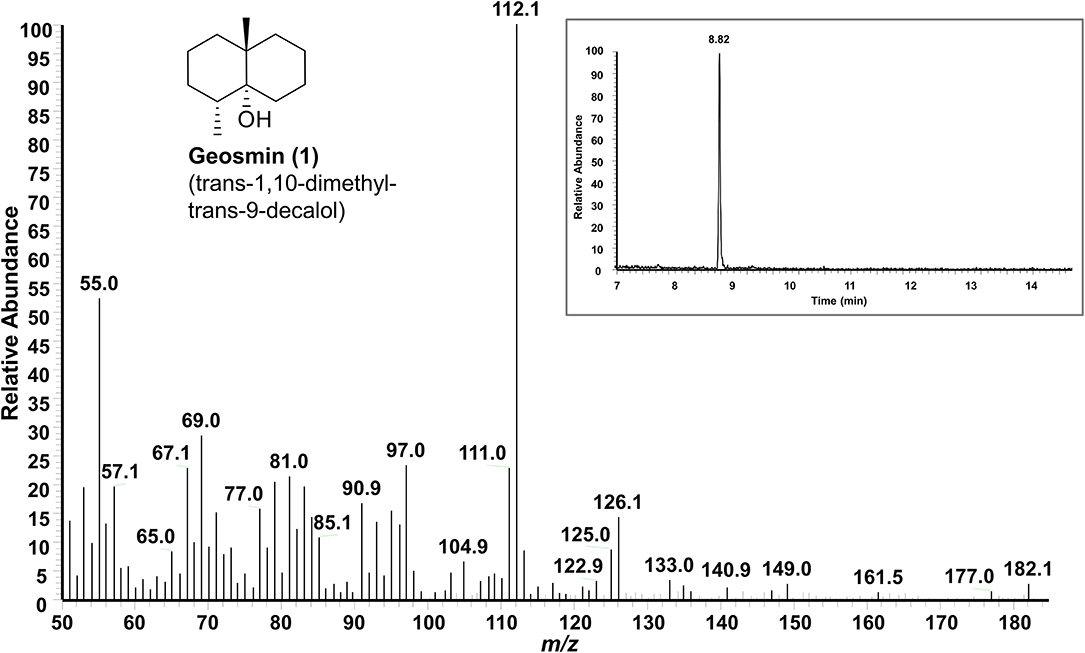
Figure 4. GC-MS spectrum of the volatile semiochemical geosmin (1) produced by endophytic S. pulveraceus ES16 on PDA under stress and during sporulation. Insert shows the GC chromatogram (RT 8.82 min) of geosmin (1).
Guided by the OSMAC approach, the chemo-diversity of endophytic S. pulveraceus ES16 cultivated in different media conditions was investigated using HPLC-HRMS. The extracts of plates and broths were analyzed in parallel to examine the compounds produced by the bacterium both under submerged cultivation conditions and agar (solid media). Moreover, HPLC-HRMS based temporal analyses were performed after cultivation for 3-, 7- and 14 days, respectively, to unravel the production pattern of the compounds and set up the basis of further MALDI-HRMSI experiments. Furthermore, a bioactivity-guided selection of the produced compounds was carried out to target the “hits,” focusing on antibacterial, antifungal, antiprotozoal, and relevant plant beneficial compounds, and the targeted metabolic profile of the endophyte was established. Finally, the target compounds were identified by extensive HRMSn experiments and by comparison with the literature. Endophytic S. pulveraceus ES16 demonstrated the production of two classes of compounds with relevant bioactivities: polycyclic tetramate macrolactams (PTMs) and siderophores.
While examining the secondary metabolites of endophytic S. pulveraceus ES16 using the OSMAC approach, a family of polycyclic tetramate macrolactams (PTMs) was detected (Figure 3, compounds 2–7). Interestingly, the PTMs were only produced in two specific media conditions that induced stress in the organism, exemplified by sporulation and visible exudation into the media, namely GYM agar, and PDA. The production of these compounds (2–7) was first detected by the 7th day of incubation, which increased till day-14 of observation. Overall, six different compounds/masses belonging to this family were detected. Apart from m/z 525.2601 (2), which was only detected on GYM agar, all five other compounds (3–7) were produced in both media conditions.
Although various PTMs are known in the literature, their analysis was restricted to NMR techniques such as NOESY and COSY. However, as expected, in our study, endophytic S. pulveraceus ES16 produced the PTMs (2–7) in low, physiologically-relevant amounts, which did not allow the isolation and purification of PTMs (2–7) using preparative HPLC. Therefore, the structures of compounds 2–7 were dereplicated using HRMSn. Due to similarities in the structures of the compounds, we performed HRMS2 and HRMS3 to evaluate their structures based on typical fragmentation patterns and by comparison with the literature (HRMS2 could not be done for compound 2 due to low production). The HRMSn spectra of compounds 2–7 are presented in Supplementary Figures 1–10. Yu et al. (2017) and Qi et al. (2018) have reported the MSn fragmentation of some PTMs, which enabled us to compare the fragmentation patterns of compounds 2–7 and assign their plausible structures.
In addition to the bioactive PTMs (2–7), endophytic S. pulveraceus ES16 also produces an unusual siderophore called mirubactin (compound 8, Figure 3). This siderophore contains an O-acyl hydroxamic acid ester group with a high affinity to the valuable ferric ions in iron-depleted environments. It also consists of one D-arginine and one d-hfOrn unit in addition to two DHB units. Our OSMAC approach revealed that this compound's production critically depends on the medium on which the bacterium is cultivated. Mirubactin (8) was detected only on GYM agar and on PDA ([M+H]+, m/z 605.2192, ± 2 ppm, RT 15.02 min). Interestingly, the endophyte produces this siderophore 10-fold more pronounced on PDA than on GYM agar (PDAintensity 4.2E5; GYMintensity 1.9E4) concomitant with the strain's higher stress and extensive sporulation on PDA.
HRMSn experiments confirmed the structure of mirubactin (8) (Figure 5). The HRMS2 measurement yielded the primary fragment ion with an m/z of 469.2042 after one DHB unit's cleavage. Consecutive HRMS3 of the fragment m/z 469.2042 yielded in three additional fragments: m/z 333.1877 is the result of the loss of the two dehydrated DHB groups from each side, while m/z 293.1242 arises from the cleavage of one DHB moiety together with the fhOrn; the fragment m/z 311.1349 follows the same fragmentation pattern as m/z 293.1242 after a rearrangement (Giessen et al., 2012). Strikingly, the mirubactin-iron complex (C26H30FeN6O11, [M+H]+, m/z 658.1316, ± 2 ppm) was also produced by the endophyte on both GYM agar and PDA.
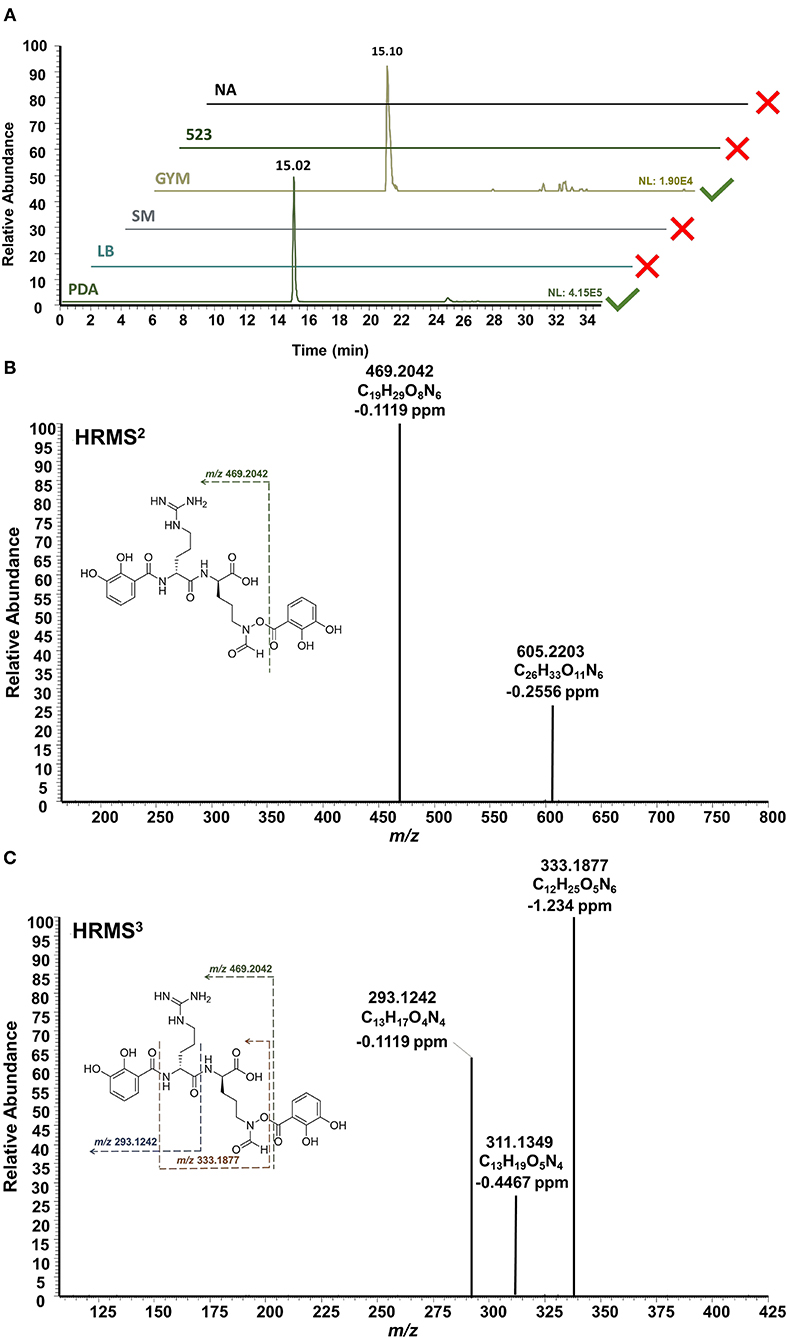
Figure 5. HPLC-HRMSn analysis of mirubactin (8). (A) Extracted ion chromatograms ([M+H]+, m/z 605.2192, ±2 ppm] shows the selective production of mirubactin (8) only on GYM agar and on PDA. (B) HRMS2 of mirubactin (8). (C) HRMS3 of mirubactin (8). The proposed mass spectral fragmentation pathway is annotated on the compound structure.
In order to visualize the production, distribution, and secretion of the above compounds produced by endophytic S. pulveraceus ES16 in high spatial resolution, we performed MALDI-HRMSI to monitor the target compounds over 14 days. Figure 6 depicts the ion intensity images of the PTMs 3–7 on day 14 ([M+H]+ and [M+K]+ adducts). Compound 2 was below the limit of detection (< LOD) and could not be mapped. MALDI-HRMSI revealed that compounds 5–7 were secreted more profusely compared to the other PTMs. These results are consistent with the data obtained from inoculated agar extractions and HPLC-HRMS analyses. The spatial-temporal distribution of the produced PTMs showed that the compounds were readily secreted into the medium after production. The catecholate siderophore, mirubactin (8), produced on PDA and GYM agar, was also subjected to visualization by MALDI-HRMSI (Supplementary Figure 11). Similar to the PTMs, the endophyte also secretes mirubactin (8) into the media over 14 days.
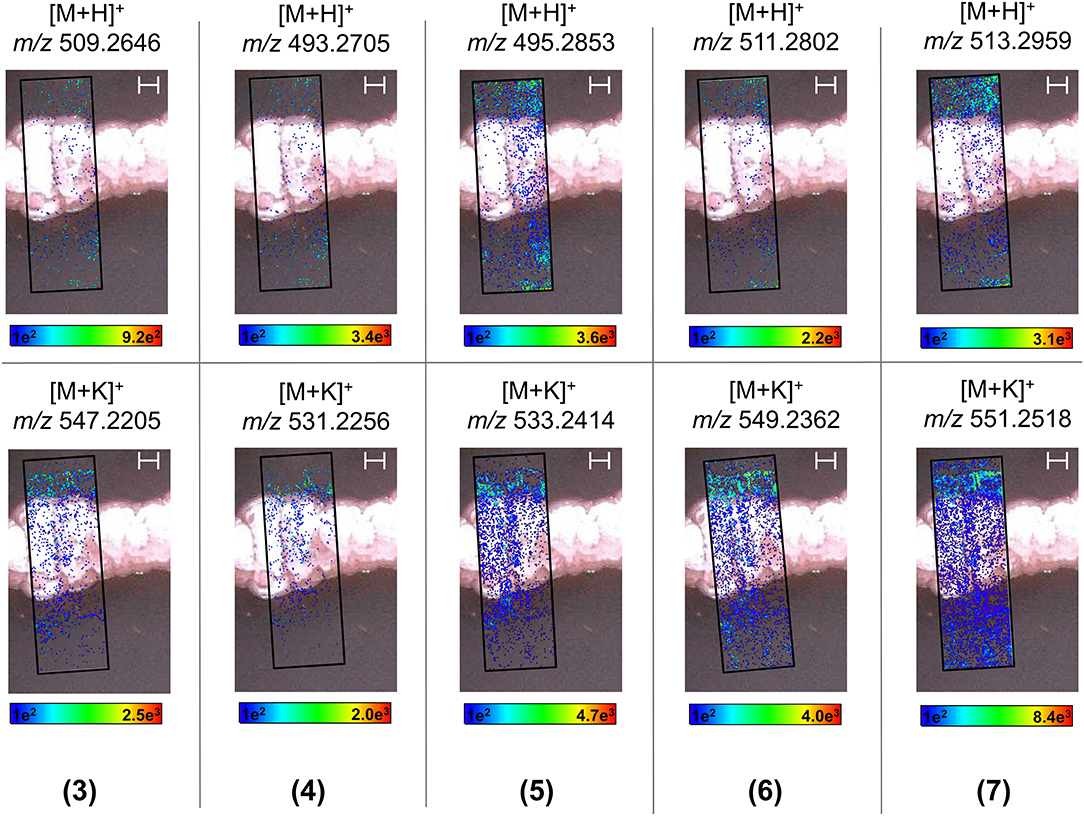
Figure 6. MALDI-HRMSI of PTMs (compounds 3–7) produced by endophytic S. pulveraceus ES16. Selected ion images depict the localization and distribution of the PTMs ([M+H]+ and [M+K]+; ±5 ppm; spatial resolution = 30 μm; matrix used = MBT) after cultivation on PDA for 14 days. The assigned scale bar represents 1,000 μm. The black insert depicts the scanned area in each case.
The stressful and dysbiotic environment characteristically associated with ARD, not only for apple plants but also for associated microbiota, is well-established (Mazzola and Manici, 2012; Yim et al., 2013, 2015; Weiß et al., 2017; Grunewaldt-Stöcker et al., 2019; Radl et al., 2019; Winkelmann et al., 2019; Balbín-Suárez et al., 2020). For example, an increased abundance of environmental stress sensing genes, particularly in the rhizosphere microbiome of ARD soils compared to ARD-unaffected soils, was recently unraveled by a metagenomics approach (Radl et al., 2019). Against this background, in the present study, we aimed to study apple root-associated endophytic S. pulveraceus ES16 by employing a chemical OSMAC approach (Romano et al., 2018; Pan et al., 2019; Maghembe et al., 2020) to induce high stress and sporulation in the organism. Earlier studies have substantiated that Streptomyces species are stressed and forced to sporulate by regulation of the pleiotropic transcriptional factors such as GlnR, MtrA, and PhoP, which can be achieved by manipulating the nature of carbon and nitrogen sources, their quantity (specifically the carbon/nitrogen ratio as well as their depletion), and phosphate availability in the medium (Karandikar et al., 1996; Martín and Liras, 2020). In particular, by limiting the nitrogen availability, Streptomyces species can be starved of essential amino acids leading to a stringent response that restricts transcription of numerous genes involved in vegetative growth (i.e., induces sporulation) and stimulates the expression of stress response genes (Bibb, 2005; Tiffert et al., 2008, 2011; Dalebroux and Swanson, 2012; Martín and Liras, 2020). Additionally, phosphate starvation influences the two-component phosphate control of the metabolism system, PhoR-PhoP (Hutchings et al., 2004; Martín et al., 2012), slows down primary metabolism and triggers the production of secondary metabolites (Sola-Landa et al., 2003, 2005; Martínez-Castro et al., 2018). Therefore, we successfully employed PDA limited in both nitrogen and phosphate (Supplementary Table 2) as a high stress-inducing and sporulating medium for endophytic S. pulveraceus ES16. Earlier studies have also demonstrated how PDA can induce sporulation and production of antimicrobial compounds by different marine Streptomyces species (Vijayakumar et al., 2012; English et al., 2017). Furthermore, we used the GYM medium following the recommendation of Shirling and Gottlieb (1966) in the International Streptomyces Project (ISP), which is limited in phosphate but not in nitrogen and has been used earlier to induce sporulation in some Streptomyces strains (Arasu et al., 2009). Endophytic S. pulveraceus ES16 sporulated mildly on GYM agar. We also used a modified 523 medium that does not limit nitrogen or phosphate and provides the disaccharide saccharose as the carbon source. Along with the basal medium NA that does not induce sporulation and nutrient-rich LB and SM media (favorable growing conditions), this medium is used for optimal cultivation of Streptomyces species (Atlas, 2010; Vijayakumar et al., 2012; English et al., 2017). Taken together, we evaluated the phenotypic differentiation of endophytic S. pulveraceus ES16 under stress compared to unstressed as well as optimal growth conditions, which laid a scientific handle to study the concomitant effects on its secreted volatilome and expressed metabolome.
Several reports have documented the production of geosmin (1) by a plethora of microorganisms inhabiting a variety of ecological niches, including filamentous fungi, actinomycetes, cyanobacteria, and proteobacteria, to name a few (Yamada et al., 2015; Vurukonda et al., 2018; Churro et al., 2020; Melo et al., 2020). Becher et al. (2020) recently unraveled this compound's central role in Streptomyces species' lifecycle since it plays a specific role as the volatile signal molecule attracting the soil arthropods Collembola (springtails) to the producer strains. Several studies have demonstrated that Collembola illustrates biocontrol potential by feeding on pathogenic fungi. Curl (1979) examined the biocontrol potential of Collembolan species for the first time by observing their interactions with pathogenic fungi. He suggested that the Collembola species Proisotoma minuta and Thalassaphorura encarpata decreased pathogenic fungal growth rate such as Rhizoctonia solani and Fusarium oxysporum, which are associated with root disease. Shiraishi et al. (2003) showed the suppression of the damping-off disease in cabbage and Chinese cabbage by the Collembola Folsomia hidakana. Recently, Innocenti and Sabatini (2018) reported that Collembola prefers feeding on pathogenic fungi such as Fusarium culmorum rather than biocontrol fungi such as Trichoderma harzianum. In the present study, the phenotypic observations of endophytic S. pulveraceus ES16 under specific OSMAC parameters revealed an earthy soil odor release during this bacterium's prolific sporulation on PDA plates that induced stress to the organism. One important reason why we employed the six different defined media conditions to cultivate the organism was to study its secreted volatilome and metabolome expressed under the influence of stressful media conditions compared to nutrient-rich and optimum conditions (Romano et al., 2018; Pan et al., 2019; Maghembe et al., 2020). OSMAC-guided SPME-GC-MS analyses further revealed that the organism produces the volatile signal compound geosmin (compound 1, Figure 3; γεω-“geo”: earth, o- “osmi”: smell) only under stressful conditions and during sporulation, and it is responsible for this scent. The endophyte did not emit this signal compound in the other tested media conditions. As expected, endophytic S. pulveraceus ES16 mildly sporulated on GYM agar. However, we could not detect the compound's production, possibly because its production is only associated with high stress and extensive sporulation or produced in meager amounts (< LOD). Moreover, the volatile terpenoid 2-methylisoborneol (2-MIB), which is also responsible for the earthy smell similar to geosmin (1) and found to be secreted by some Streptomyces species (Rabe et al., 2013; Yamada et al., 2015), was also not detected (< LOD) on PDA or GYM agar.
PTMs are natural products containing one tetramic acid and a polycyclic system fused to a macrolactam. Their polycyclic system is categorized into three main groups: the PTMs containing a 5/5, those with a 5/6/5, or PTMs with a 5/5/6 ring system in their structure (Luo et al., 2013; Liu et al., 2019). PTMs are well-known for possessing various biological activities such as antibacterial, antifungal, antiprotozoal, and antiviral (Luo et al., 2013; Liu et al., 2019; Jiang et al., 2020). For example, capsimycin B was discovered from Streptomyces sp. Tü 6239 and showed antibiotic activities against Gram-positive bacteria (Bertasso et al., 2003). In another study, Lysobacter enzymogenes and Streptomyces spp. both produced dihydromaltophilin (also known as HSAF) (Li et al., 2008). HSAF is an antifungal agent capable of harming fungal biosynthetic pathways and is therefore suitable against phytopathogens. Recently, marine sponge-associated Streptomyces zhaozhouensis strain MCCB267 was shown to produce clifednamide A that demonstrated remarkable antiproliferative activities by inducing apoptosis in lung carcinoma cells (Dhaneesha et al., 2019). A detailed examination of endophytic S. pulveraceus ES16 metabolome guided by the OSMAC approach revealed the production of several bioactive PTMs (compounds 2–7, Figure 3), a series of antibiotic agents. MALDI-HRMSI experiments further revealed that the produced PTMs are part of the endophytic S. pulveraceus ES16 strain's secretome and are released into the media. Accordingly, the compounds can be considered to be released by the endophyte into the host plant, leading to an impact on the plant itself. Further experiments are required to elucidate the positive (e.g., warding off pathogens or associated endophytes) or negative (e.g., phytotoxic) effects of the endophytic PTMs (2–7) on its host plant.
Siderophores play an essential role in host-pathogen interactions (Kramer et al., 2020). For instance, van Loon et al. (2008) showed that the production of siderophores by endophytes contributes to protecting the host plant through induced systemic resistance (ISR), a mechanism in the plant activated by an infection. Furthermore, siderophores are considered potential biocontrol agents; endophytic siderophore production reduces the amount of ferric ions available for uptake by pathogens, contributing to the plant defense (Ghosh et al., 2020; Kramer et al., 2020). Endophytic S. pulveraceus ES16 can produce the rare iron-chelating siderophore mirubactin (compound 8, Figure 3), which it also secretes out as revealed by MALDI-HRMSI. The isolation, structure elucidation, and biosynthesis of this compound have been reported by Giessen et al. (2012). This siderophore's production has only been reported from Actinosynnema mirum (Giessen et al., 2012), and gene sequencing has been employed to predict the possibility of its production by Streptomyces species (Jackson et al., 2018; Almeida et al., 2019). To the best of our knowledge, we report for the first time the in vitro production of mirubactin (8) by a Streptomyces species using targeted metabolic profiling.
In the present study, we employed a targeted metabolomics approach on endophytic S. pulveraceus ES16 comprising of three different methodologies, namely SPME-GC-MS to evaluate the secreted volatilome, HPLC-HRMSn to evaluate the expressed metabolome, and MALDI-HRMSI to visualize the spatial dynamics of target compounds, all guided by the OSMAC approach. We used the cultivable apple root-associated endophytic S. pulveraceus strain ES16 as a representative working model to investigate its chemoecological relevance.
Firstly, the results of our volatilome analyses led us to propose that S. pulveraceus ES16 succumbs to high stress during the onset of ARD (concomitant to its host plant), starts sporulating, and synthesizes and secretes geosmin (1). The precise function of this semiochemical inside apple plants, whether roots further secrete it into the soil along with plant exudates, and its function in ARD soil remains to be tested in future experiments. Given the recently-established ecological function of geosmin (1) for Streptomyces (Becher et al., 2020), it can be hypothesized that if the compound is released into the soil, it can signal selected soil arthropod species. This further opens up a conceivable chemoecological “link” between the endophyte, its host plant, and associated organisms in soil that can receive and respond to the endophytic signaling cues. Secondly, it is plausible that production and secretion of the bioactive PTMs (2–7) by endophytic S. pulveraceus ES16 can provide a competitive advantage to the bacterium in the host plant apoplast because it can ward off a plethora of pathogens or associated endophytes, thereby enabling better colonization and increased abundance of the endophyte in the host. However, some of the endophytic PTMs possess cytotoxic activities in addition to having antimicrobial efficacies (Luo et al., 2013; Liu et al., 2019; Jiang et al., 2020). Whether these compounds also negatively affect the host plant by exerting phytotoxic effects should be studied in the future. Thirdly, mirubactin (8) was also secreted by the endophyte, as evidenced by the MALDI-HRMSI, similar to the PTMs (2–7). Therefore, it can be anticipated that this siderophore is also released into the plant apoplast by endophytic S. pulveraceus ES16 and can add to the PTMs-mediated “warfare” against various microorganisms residing in the immediate environment by chelating ferric ions available for uptake by associated or invading microorganisms (Ghosh et al., 2020; Kramer et al., 2020).
Future studies, for instance, by combining phytotoxicity assays, plant inoculation experiments, and MALDI-HRMSI, can precisely unravel the production, release, and effect of the compounds in planta. Noteworthy, apple roots in ARD soil are often highly disturbed so that endophytic S. pulveraceus ES16, as well as the chemicals produced by the organism, can also be assumed to enter the rhizosphere soil, whose precise function in the soil remains to be explored. Furthermore, our results provide a basis for employing a similar approach to other Streptomyces strains and species or other apple root-associated organisms to comprehend the chemical basis of their ecological functions and interactions. It would also be useful to investigate the phenotypic and metabolic effect of ARD biomarkers such as phytoalexins (Reim et al., 2020; Rohr et al., 2020) and apple root exudates or compounds therein (e.g., phlorizin) on apple root-associated microorganisms and vice versa. Further investigations on the etiology of ARD as well as designing effective and sustainable biocontrol remedies must take into account both the molecular and the chemical networks between associated ecological partners in order to gain a holistic view.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
RA performed the OSMAC experiments, phenotypic characterizations, SPME-GC-MS, HPLC-HRMSn, MALDI-HRMSI, and analyzed and interpreted the data. SZ enabled realization and optimization of mass spectrometry and MALDI-HRMSI experiments, and interpreted the data. FM-D undertook the central experiments, isolated, identified, and established the axenic culture of the Streptomyces strain. TW conceptualized the central experimental designs, supervised FM-D, and coordinated the BonaRes ORDIAmur project consortia. SK conceived and led this research, designed the experiments, supervised RA, interpreted the data, and wrote the manuscript. RA and SK prepared the figures and tables. All co-authors contributed to the manuscript and reviewed and approved the final version.
The German Federal Ministry of Education and Research (BMBF) funded this work through the project ORDIAmur (Overcoming Replant Disease by Applying an Integrated Approach) within the framework of the BonaRes program (FKZ 031B0512E to SK and FKZ 031B0512A to TW).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank the Faculty of Chemistry and Chemical Biology (CCB) of Technische Universität Dortmund for providing laboratory space and research amenities, and Till Bebenroth, Gabriele Hardes, and Eva Wieczorek for technical assistance.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fsufs.2021.643225/full#supplementary-material
ARD, Apple Replant Disease; CID, Collision Induced Dissociation; COSY, Correlation Spectroscopy; ESI, Electrospray Ionization; DHB, 2,5-Dihydroxybenzoic Acid; EI, Electron Ionization; fhOrn, δ-N-formyl-δ-N-hydroxyornithine; GC, Gas Chromatography; GYM, Glucose Yeast Malt; HESI, Heated Electrospray Ionization; HPLC, High-Performance Liquid Chromatography; HR, High-Resolution; HRMS, High-Resolution Mass Spectrometry; HSAF, Heat Stable Antifungal Factor; MALDI, Matrix-Assisted Laser Desorption/Ionization; min, Minute; LB, Lysogeny Broth; LC, Liquid Chromatography; LOD, Limit of Detection; LTQ, Linear Trap Quadrupole; MeOH, Methanol; MS, Mass Spectrometry; MSn, Tandem Mass Spectrometry, m/z, Mass-to-Charge Ratio; NA, Nutrient Agar; NCBI, National Center for Biotechnology Information; NIST, Natural Institute of Standards and Technology; NMR, Nuclear Magnetic Resonance; NOESY, Nuclear Overhauser Effect Spectroscopy; OSMAC, One Strain Many Compounds; PDA, Potato Dextrose Agar; ppm, Parts Per Million; PTM, Polycyclic Tetramate Macrolactam; Rpm, Rotations Per Minute; RT, Retention Time; SM, Streptomyces Medium; SPME, Solid Phase Microextraction; TIC, Total Ion Current; UV, Ultra-Violet; VOC, Volatile Organic Compound.
Almeida, E. L., Carrillo Rincón, A. F., Jackson, S. A., and Dobson, A. D. W. (2019). Comparative genomics of marine sponge-derived Streptomyces spp. isolates SM17 and SM18 with their closest terrestrial relatives provides novel insights into environmental niche adaptations and secondary metabolite biosynthesis potential. Front. Microbiol. 10:1713. doi: 10.3389/fmicb.2019.02213
Ancheeva, E., Daletos, G., and Proksch, P. (2020). Bioactive secondary metabolites from endophytic fungi. Curr. Med. Chem. 27, 1836–1854. doi: 10.2174/0929867326666190916144709
Arasu, M. V., Duraipandiyan, V., Agastian, P., and Ignacimuthu, S. (2009). In vitro antimicrobial activity of Streptomyces spp. ERI-3 isolated from Western Ghats rock soil (India). J. Mycol. Méd. 19, 22–28. doi: 10.1016/j.mycmed.2008.12.002
Atlas, R. M. (2010). Handbook of Microbiological Media. 4th Edn. Boca Raton, FL: CRC Press. doi: 10.1201/EBK1439804063
Babalola, O. O., Fadiji, A. E., Enagbonma, B. J., Alori, E. T., Ayilara, M. S., and Ayangbenro, A. S. (2020). The nexus between plant and plant microbiome: revelation of the networking strategies. Front. Microbiol. 11:548037. doi: 10.3389/fmicb.2020.548037
Balbín-Suárez, A., Lucas, M., Vetterlein, D., Sørensen, S. J., Winkelmann, T., Smalla, K., et al. (2020). Exploring microbial determinants of apple replant disease (ARD): a microhabitat approach under split-root design. FEMS Microbiol. Ecol. 96:fiaa211. doi: 10.1093/femsec/fiaa211
Becher, P. G., Verschut, V., Bibb, M. J., Bush, M. J., Molnár, B. P., Barane, E., et al. (2020). Developmentally regulated volatiles geosmin and 2-methylisoborneol attract a soil arthropod to Streptomyces bacteria promoting spore dispersal. Nat. Microbiol. 5, 821–829. doi: 10.1038/s41564-020-0697-x
Bertasso, M., Holzenkämpfer, M., Zeeck, A., Stackebrandt, E., Beil, W., and Fiedler, H. P. (2003). Ripromycin and other polycyclic macrolactams from Streptomyces sp. Tü 6239: taxonomy, fermentation, isolation and biological properties. J. Antibiot. 56, 364–371. doi: 10.7164/antibiotics.56.364
Bibb, M. J. (2005). Regulation of secondary metabolism in streptomycetes. Curr. Opin. Microbiol. 8, 208–215. doi: 10.1016/j.mib.2005.02.016
Bode, H. B., Bethe, B., Höfs, R., and Zeeck, A. (2002). Big effects from small changes: possible ways to explore nature's chemical diversity. Chembiochem 3, 619–627. doi: 10.1002/1439-7633(20020703)3:7<619::AID-CBIC619>3.0.CO;2-9
Carrión, V. J., Perez-Jaramillo, J., Cordovez, V., Tracanna, V., de Hollander, M., Ruiz-Buck, D., et al. (2019). Pathogen-induced activation of disease-suppressive functions in the endophytic root microbiome. Science 366, 606–612. doi: 10.1126/science.aaw9285
Cheng, Y. T., Zhang, L., and He, S. Y. (2019). Plant-microbe interactions facing environmental challenge. Cell Host Microbe 26, 183–192. doi: 10.1016/j.chom.2019.07.009
Churro, C., Semedo-Aguiar, A. P., Silva, A. D., Pereira-Leal, J. B., and Leite, R. B. (2020). A novel cyanobacterial geosmin producer, revising GeoA distribution and dispersion patterns in bacteria. Sci. Rep. 10:8679. doi: 10.1038/s41598-020-64774-y
Colombo, E. M., Kunova, A., Pizzatti, C., Saracchi, M., Cortesi, P., and Pasquali, M. (2019). Selection of an endophytic Streptomyces sp. strain DEF09 from wheat roots as a biocontrol agent against Fusarium graminearum. Front. Microbiol. 10:2356. doi: 10.3389/fmicb.2019.02356
Curl, E. A. (1979). “Effects of myeophagous Collembola on Rhizoctonia solani and cotton seedling disease,” in Soil-borne Plant Pathogens, eds. B. Schippers, and W. Garns (London: Academic Press), 253–269.
Dalebroux, Z. D., and Swanson, M. S. (2012). ppGpp: magic beyond RNA polymerase. Nat. Rev. Microbiol. 10, 203–212. doi: 10.1038/nrmicro2720
Dhaneesha, M., Hasin, O., Sivakumar, K. C., Ravinesh, R., Naman, C. B., Carmeli, S., et al. (2019). DNA binding and molecular dynamic studies of polycyclic tetramate macrolactams (PTM) with potential anticancer activity isolated from a sponge-associated Streptomyces zhaozhouensis subsp. mycale subsp. nov. Mar. Biotechnol. 21, 124–137. doi: 10.1007/s10126-018-9866-9
Eckelmann, D., Spiteller, M., and Kusari, S. (2018). Spatial-temporal profiling of prodiginines and serratamolides produced by endophytic Serratia marcescens harbored in Maytenus serrata. Sci. Rep. 8:5283. doi: 10.1038/s41598-018-23538-5
English, A. L., Boufridi, A., Quinn, R. J., and Kurtböke, D. I. (2017). Evaluation of fermentation conditions triggering increased antibacterial activity from a near-shore marine intertidal environment-associated Streptomyces species. Synth. Syst. Biotechnol. 2, 28–38. doi: 10.1016/j.synbio.2016.09.005
Ghosh, S. K., Bera, T., and Chakrabarty, A. M. (2020). Microbial siderophore – a boon to agricultural sciences. Biol. Cont. 144:104214. doi: 10.1016/j.biocontrol.2020.104214
Giessen, T. W., Franke, K. B., Knappe, T. A., Kraas, F. I., Bosello, M., Xie, X., et al. (2012). Isolation, structure elucidation, and biosynthesis of an unusual hydroxamic acid ester-containing siderophore from Actinosynnema mirum. J. Nat. Prod. 75, 905–914. doi: 10.1021/np300046k
Grunewaldt-Stöcker, G., Mahnkopp, F., Popp, C., Maiss, E., and Winkelmann, T. (2019). Diagnosis of apple replant disease (ARD): microscopic evidence of early symptoms in fine roots of different apple rootstock genotypes, Sci. Hortic. 243, 583–594. doi: 10.1016/j.scienta.2018.09.014
Hoang, K. C., Lai, T. H., Lin, C. S., Chen, Y. T., and Liau, C. Y. (2010). The chitinolytic activities of Streptomyces sp. TH-11. Int. J. Mol. Sci. 12, 56–65. doi: 10.3390/ijms12010056
Hutchings, M. I., Hoskisson, P. A., Chandra, G., and Buttner, M. J. (2004). Sensing and responding to diverse extracellular signals? Analysis of the sensor kinases and response regulators of Streptomyces coelicolor A3(2). Microbiology 150, 2795–2806. doi: 10.1099/mic.0.27181-0
Iijima, Y. (2014). Recent advances in the application of metabolomics to studies of biogenic volatile organic compounds (BVOC) produced by plant. Metabolites 4, 699–721. doi: 10.3390/metabo4030699
Innocenti, G., and Sabatini, M. A. (2018). Collembola and plant pathogenic, antagonistic and arbuscular mycorrhizal fungi: a review. Bull. Insectol. 71, 71–76.
Jackson, S. A., Crossman, L., Almeida, E. L., Margassery, L. M., Kennedy, J., and Dobson, A. D. W. (2018). Diverse and abundant secondary metabolism biosynthetic gene clusters in the genomes of marine sponge derived Streptomyces spp. isolates. Mar. Drugs 16:67. doi: 10.3390/md16020067
Jiang, M., Chen, S., Li, J., and Liu, L. (2020). The biological and chemical diversity of tetramic acid compounds from marine-derived microorganisms. Mar. Drugs 18:114. doi: 10.3390/md18020114
Jones, P., Garcia, B. J., Furches, A., Tuskan, G. A., and Jacobson, D. (2019). Plant host-associated mechanisms for microbial selection. Front. Plant Sci. 10:862. doi: 10.3389/fpls.2019.00862
Joo, H. S., Deyrup, S. T., and Shim, S. H. (2020). Endophyte-produced antimicrobials: a review of potential lead compounds with a focus on quorum-sensing disruptors. Phytochem. Rev. doi: 10.1007/s11101-020-09711-7 [Epub ahead of print].
Karandikar, A., Sharples, G. P., and Hobbs, G. (1996). Influence of medium composition on sporulation by Streptomyces coelicolor A3(2) grown on defined solid media. Biotechnol. Tech. 10, 79–82. doi: 10.1007/BF00765186
Khare, E., Mishra, J., and Arora, N. K. (2018). Multifaceted Interactions between endophytes and plant: developments and prospects. Front. Microbiol. 9:2732. doi: 10.3389/fmicb.2018.02732
Kramer, J., Özkaya, Ö., and Kümmerli, R. (2020). Bacterial siderophores in community and host interactions. Nat. Rev. Microbiol. 18, 152–163. doi: 10.1038/s41579-019-0284-4
Kusari, S., Kusari, P., and Spiteller, M. (2017). “Novel insights in plant-endophyte interactions,” in Chemical Biology of Natural Products, eds D. J. Newman, G. M. Cragg, and P. G. Grothaus (Boca Raton, FL: CRC Press), 205–231. doi: 10.1201/9781315117089-7
Law, J. W., Law, L. N., Letchumanan, V., Tan, L. T., Wong, S. H., Chan, K. G., et al. (2020). Anticancer drug discovery from microbial sources: the unique mangrove streptomycetes. Molecules 25:5365. doi: 10.3390/molecules25225365
Li, S., Jochum, C. C., Yu, F., Zaleta-Rivera, K., Du, L., Harris, S. D., et al. (2008). An antibiotic complex from Lysobacter enzymogenes strain C3: antimicrobial activity and role in plant disease control. Phytopathology 98, 695–701. doi: 10.1094/PHYTO-98-6-0695
Liu, W., Zhang, W., Jin, H., Zhang, Q., Chen, Y., Jiang, X., et al. (2019). Genome mining of marine-derived Streptomyces sp. SCSIO 40010 leads to cytotoxic new polycyclic tetramate macrolactams. Mar. Drugs 17:663. doi: 10.3390/md17120663
Luo, Y., Huang, H., Liang, J., Wang, M., Lu, L., Shao, Z., et al. (2013). Activation and characterization of a cryptic polycyclic tetramate macrolactam biosynthetic gene cluster. Nat. Commun. 4:2894. doi: 10.1038/ncomms3894
Maghembe, R., Damian, D., Makaranga, A., Nyandoro, S. S., Lyantagaye, S. L., Kusari, S., et al. (2020). Omics for bioprospecting and drug discovery from bacteria and microalgae. Antibiotics 9:229. doi: 10.3390/antibiotics9050229
Mahnkopp, F., Simon, M., Lehndorff, E., Pätzold, S., Wrede, A., and Winkelmann, T. (2018). Induction and diagnosis of apple replant disease (ARD): a matter of heterogeneous soil properties? Sci. Hortic. 241, 167–177. doi: 10.1016/j.scienta.2018.06.076
Mahnkopp-Dirks, F., Radl, V., Kublik, S., Gschwendtner, S., Schloter, M., and Winkelmann, T. (2020). Molecular barcoding reveals the genus Streptomyces as associated root endophytes of apple (Malus domestica) plants grown in soils affected by apple replant disease. Phytobiomes J. doi: 10.1094/PBIOMES-07-20-0053-R [Epub ahead of print].
Martín, J. F., and Liras, P. (2020). The balance metabolism safety net: integration of stress signals by interacting transcriptional factors in Streptomyces and related actinobacteria. Front. Microbiol. 10:3120. doi: 10.3389/fmicb.2019.03120
Martín, J. F., Sola-Landa, A., and Rodríguez-García, A. (2012). “Two-component systems in Streptomyces,” in Two-component Systems in Bacteria, eds R. Gross, and D. Beier (Würzburg: Caister Academic), 315–332.
Martínez-Castro, M., Barreiro, C., and Martín, J. F. (2018). Analysis and validation of the pho regulon in the tacrolimus-producer strain Streptomyces tsukubaensis: differences with the model organism Streptomyces coelicolor. Appl. Microbiol. Biotechnol. 102, 7029–7045. doi: 10.1007/s00253-018-9140-0
Mazzola, M., and Manici, L. M. (2012). Apple replant disease: role of microbial ecology in cause and control. Annu. Rev. Phytopathol. 50, 45–65. doi: 10.1146/annurev-phyto-081211-173005
Melo, N., Wolff, G. H., Costa-da-Silva, A. L., Arribas, R., Triana, M. F., Gugger, M., et al. (2020). Geosmin attracts Aedes aegypti mosquitoes to oviposition sites. Curr. Biol. 30, 127–134. doi: 10.1016/j.cub.2019.11.002
Mhlongo, M. I., Piater, L. A., Madala, N. E., Labuschagne, N., and Dubery, I. A. (2018). The chemistry of plant-microbe interactions in the rhizosphere and the potential for metabolomics to reveal signaling related to defense priming and induced systemic resistance. Front. Plant Sci. 9:112. doi: 10.3389/fpls.2018.00112
Morales-Cedeño, L. R., Orozco-Mosqueda, M. D. C., Loeza-Lara, P. D., Parra-Cota, F. I., de Los Santos-Villalobos, S., and Santoyo, G. (2020). Plant growth-promoting bacterial endophytes as biocontrol agents of pre- and post-harvest diseases: fundamentals, methods of application and future perspectives. Microbiol. Res. 242:126612. doi: 10.1016/j.micres.2020.126612
Newman, D. J., and Cragg, G. M. (2020). Plant endophytes and epiphytes: burgeoning sources of known and “unknown” cytotoxic and antibiotic agents? Planta Med. 86, 891–905. doi: 10.1055/a-1095-1111
Nicola, L., Insam, H., Pertot, I., and Stres, B. (2018). Reanalysis of microbiomes in soils affected by apple replant disease (ARD): old foes and novel suspects lead to the proposal of extended model of disease development. Agric. Ecosyst. Environ. Appl. Soil Ecol. 129, 24–33. doi: 10.1016/j.apsoil.2018.04.010
Pan, R., Bai, X., Chen, J., Zhang, H., and Wang, H. (2019). Exploring structural diversity of microbe secondary metabolites using OSMAC strategy: a literature review. Front. Microbiol. 10:294. doi: 10.3389/fmicb.2019.00294
Qi, Y., Ding, E., and Blodgett, J. A. V. (2018). Native and engineered clifednamide biosynthesis in multiple Streptomyces spp. ACS Synth. Biol. 7, 357–362. doi: 10.1021/acssynbio.7b00349
Quambusch, M., Pirttilä, A. M., Tejesvi, M. V., Winkelmann, T., and Bartsch, M. (2014). Endophytic bacteria in plant tissue culture: differences between easy- and difficult-to-propagate Prunus avium genotypes. Tree Physiol. 34, 524–533. doi: 10.1093/treephys/tpu027
Quinn, G. A., Banat, A. M., Abdelhameed, A. M., and Banat, I. M. (2020). Streptomyces from traditional medicine: sources of new innovations in antibiotic discovery. J. Med. Microbiol. 69, 1040–1048. doi: 10.1099/jmm.0.001232
Rabe, P., Citron, C. A., and Dickschat, J. S. (2013). Volatile terpenes from actinomycetes: a biosynthetic study correlating chemical analyses to genome data. Chembiochem 14, 2345–2354. doi: 10.1002/cbic.201300329
Radl, V., Winkler, J. B., Kublik, S., Yang, L., Winkelmann, T., Vestergaard, G., et al. (2019). Reduced microbial potential for the degradation of phenolic compounds in the rhizosphere of apple plantlets grown in soils affected by replant disease. Environ. Microbiome 14:8. doi: 10.1186/s40793-019-0351-5
Reim, S., Rohr, A. D., Winkelmann, T., Weiß, S., Liu, B., Beerhues, L., et al. (2020). Genes involved in stress response and especially in phytoalexin biosynthesis are upregulated in four Malus genotypes in response to apple replant disease. Front. Plant Sci. 10:1724. doi: 10.3389/fpls.2019.01724
Rohr, A. D., Schimmel, J., Liu, B., Beerhues, L., Guggenberger, G., and Winkelmann, T. (2020). Identification and validation of early genetic biomarkers for apple replant disease. PLoS ONE 15:e0238876. doi: 10.1371/journal.pone.0238876
Romano, S., Jackson, S. A., Patry, S., and Dobson, A. D. W. (2018). Extending the “One Strain Many Compounds” (OSMAC) principle to marine microorganisms. Mar. Drugs 16:244. doi: 10.3390/md16070244
Sanger, F., Nicklen, S., and Coulson, A. R. (1977). DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. U. S. A. 74, 5463–5437. doi: 10.1073/pnas.74.12.5463
Shiraishi, H., Enami, Y., and Okano, S. (2003). Folsomia hidakana (Collembola) prevents damping-off disease in cabbage and Chinese cabbage by Rhizoctonia solani. Pedobiologia 47, 33–38. doi: 10.1078/0031-4056-00167
Shirling, J. L., and Gottlieb, D. (1966). Methods for characterization of Streptomyces species. Int. J. Syst. Bacteriol. 16, 313–340. doi: 10.1099/00207713-16-3-313
Sola-Landa, A., Moura, R. S., and Martín, J. F. (2003). The two component PhoR-PhoP system controls both primary metabolism and secondary metabolite biosynthesis in Streptomyces lividans. Proc. Natl. Acad. Sci. U. S. A. 100, 6133–6138. doi: 10.1073/pnas.0931429100
Sola-Landa, A., Rodríguez-García, A., Franco-Domínguez, E., and Martín, J. F. (2005). Binding of PhoP to promoters of phosphate regulated genes in Streptomyces coelicolor: identification of PHO boxes. Mol. Microbiol. 56, 1373–1385. doi: 10.1111/j.1365-2958.2005.04631.x
Spasic, J., Mandic, M., Djokic, L., and Nikodinovic-Runic, J. (2018). Streptomyces spp. in the biocatalysis toolbox. Appl. Microbiol. Biotechnol. 102, 3513–3536. doi: 10.1007/s00253-018-8884-x
Terra, L., Dyson, P., Ratcliffe, N., Castro, H. C., and Vicente, A. C. P. (2020). Biotechnological potential of Streptomyces siderophores as new antibiotics. Curr. Med. Chem. doi: 10.2174/0929867327666200510235512 [Epub ahead of print].
Tholl, D., Boland, W., Hansel, A., Loreto, F., Röse, U. S., and Schnitzler, J. P. (2006). Practical approaches to plant volatile analysis. Plant J. 45, 540–560. doi: 10.1111/j.1365-313X.2005.02612.x
Tiffert, Y., Franz-Wachtel, M., Fladerer, C., Nordheim, A., Reuther, J., Wohlleben, W., et al. (2011). Proteomic analysis of the GlnR-mediated response to nitrogen limitation in Streptomyces coelicolor M145. Appl. Microbiol. Biotechnol. 89, 1149–1159. doi: 10.1007/s00253-011-3086-9
Tiffert, Y., Supra, P., Wurm, R., Wohlleben, W., Wagner, R., and Reuther, J. (2008). The Streptomyces coelicolor GlnR regulon: identification of new GlnR targets and evidence for a central role of GlnR in nitrogen metabolism in actinomycetes. Mol. Microbiol. 67, 861–880. doi: 10.1111/j.1365-2958.2007.06092.x
van der Meij, A., Worsley, S. F., Hutchings, M. I., and van Wezel, G. P. (2017). Chemical ecology of antibiotic production by actinomycetes. FEMS Microbiol. Rev. 41, 392–416. doi: 10.1093/femsre/fux005
van Loon, L. C., Bakker, P. A., van der Heijdt, W. H., Wendehenne, D., and Pugin, A. (2008). Early responses of tobacco suspension cells to rhizobacterial elicitors of induced systemic resistance. Mol. Plant Microbe Interact. 21, 1609–1621. doi: 10.1094/MPMI-21-12-1609
Vijayakumar, R., Panneerselvam, K., Muthukumar, C., Thajuddin, N., Panneerselvam, A., and Saravanamuthu, R. (2012). Optimization of antimicrobial production by a marine actinomycete Streptomyces afghaniensis VPTS3-1 isolated from Palk Strait, East Coast of India. Indian J. Microbiol. 52, 230–239. doi: 10.1007/s12088-011-0138-x
Viss, P., Brooks, E., and Driver, J. (1991). A simplified method for the control of bacterial contamination in woody plant tissue culture. In Vitro Cell. Dev. Biol. 27:42. doi: 10.1007/BF02632060
Vurukonda, S. S. K. P., Giovanardi, D., and Stefani, E. (2018). Plant growth promoting and biocontrol activity of Streptomyces spp. as endophytes. Int. J. Mol. Sci. 19:952. doi: 10.3390/ijms19040952
Weisburg, W. G., Barns, S. M., Pelletier, D. A., and Lane, D. J. (1991). 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173, 697–703. doi: 10.1128/JB.173.2.697-703.1991
Weiß, S., Liu, B., Reckwell, D., Beerhues, L., and Winkelmann, T. (2017). Impaired defense reactions in apple replant disease-affected roots of Malus domestica 'M26'. Tree Physiol. 37, 1672–1685. doi: 10.1093/treephys/tpx108
White, J. F., Kingsley, K. L., Zhang, Q., Verma, R., Obi, N., Dvinskikh, S., et al. (2019). Review: Endophytic microbes and their potential applications in crop management. Pest Manag. Sci. 75, 2558–2565. doi: 10.1002/ps.5527
Wibberg, D., Al-Dilaimi, A., Busche, T., Wedderhoff, I., Schrempf, H., Kalinowski, J., et al. (2016). Complete genome sequence of Streptomyces reticuli, an efficient degrader of crystalline cellulose. J. Biotechnol. 222, 13–14. doi: 10.1016/j.jbiotec.2016.02.002
Winkelmann, T., Smalla, K., Amelung, W., Baab, G., Grunewaldt-Stöcker, G., Kanfra, X., et al. (2019). Apple replant disease: causes and mitigation strategies. Curr. Issues Mol. Biol. 30, 89–106. doi: 10.21775/cimb.030.089
Yalage Don, S. M., Schmidtke, L. M., Gambetta, J. M., and Steel, C. C. (2020). Aureobasidium pullulans volatilome identified by a novel, quantitative approach employing SPME-GC-MS, suppressed Botrytis cinerea and Alternaria alternata in vitro. Sci. Rep. 10:4498. doi: 10.1038/s41598-020-61471-8
Yamada, Y., Kuzuyama, T., Komatsu, M., Shin-Ya, K., Omura, S., Cane, D. E., et al. (2015). Terpene synthases are widely distributed in bacteria. Proc. Natl. Acad. Sci. U. S. A. 112, 857–862. doi: 10.1073/pnas.1422108112
Yim, B., Smalla, K., and Winkelmann, T. (2013). Evaluation of apple replant problems based on different soil disinfection treatments-links to soil microbial community structure? Plant Soil 366, 617–631. doi: 10.1007/s11104-012-1454-6
Yim, B., Winkelmann, T., Ding, G. C., and Smalla, K. (2015). Different bacterial communities in heat and gamma irradiation treated replant disease soils revealed by 16S rRNA gene analysis - contribution to improved aboveground apple plant growth? Front. Microbiol. 6:1224. doi: 10.3389/fmicb.2015.01224
Yu, H. L., Jiang, S. H., Bu, X. L., Wang, J. H., Weng, J. Y., Yang, X. M., et al. (2017). Structural diversity of anti-pancreatic cancer capsimycins identified in mangrove-derived Streptomyces xiamenensis 318 and post-modification via a novel cytochrome P450 monooxygenase. Sci. Rep. 7:40689. doi: 10.1038/srep40689
Yu, Z., Zhao, L. X., Jiang, C. L., Duan, Y., Wong, L., Carver, K. C., et al. (2011). Bafilomycins produced by an endophytic actinomycete Streptomyces sp. YIM56209. J. Antibiot. 64, 159–162. doi: 10.1038/ja.2010.147
Keywords: apple replant disease, geosmin, HPLC-HRMS, MALDI-HRMSI, mirubactin, OSMAC, SPME-GC-MS, Streptomyces pulveraceus
Citation: Armin R, Zühlke S, Mahnkopp-Dirks F, Winkelmann T and Kusari S (2021) Evaluation of Apple Root-Associated Endophytic Streptomyces pulveraceus Strain ES16 by an OSMAC-Assisted Metabolomics Approach. Front. Sustain. Food Syst. 5:643225. doi: 10.3389/fsufs.2021.643225
Received: 17 December 2020; Accepted: 15 February 2021;
Published: 23 March 2021.
Edited by:
Duraisamy Saravanakumar, The University of the West Indies St. Augustine, Trinidad and TobagoReviewed by:
Amarendra Narayan Misra, Central University of Jharkhand, IndiaCopyright © 2021 Armin, Zühlke, Mahnkopp-Dirks, Winkelmann and Kusari. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Souvik Kusari, c291dmlrLmt1c2FyaUB0dS1kb3J0bXVuZC5kZQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.