- 1Key Laboratory of Agricultural Microbiology, College of Agriculture, Guizhou University, Guiyang, China
- 2State Key Laboratory Breeding Base of Green Pesticide and Agricultural Bioengineering, Ministry of Education, Guizhou University, Guiyang, China
- 3Key Laboratory of Green Pesticide and Agricultural Bioengineering, Ministry of Education, Guizhou University, Guiyang, China
The southern rice black-streaked dwarf virus (SRBSDV) is a severe threat to the yield and quality of rice products worldwide. Traditional detection methods for diagnosing SRBSDV infection show several false positives and thus provide inaccurate findings. However, Western blotting (WB) can precisely solve this problem. In this study, P6—a viral RNA-silencing suppressor—was expressed and purified in vitro. Two polyclonal P6 antibodies were obtained and quantified by enzyme-linked immunosorbent assay and WB. Subsequently, WB was performed using the P6 antibodies to identify SRBSDV antigens derived from the suspected rice samples collected from nine districts in Guizhou, China. The assay results showed that Libo, Pingtang, Huishui, Dushan, and Anshun districts had experienced an SRBSDV outbreak. The virus content in the sampled rice tissues was quantified by WB. Our results revealed that SRBSDV mainly accumulated in rice stems rather than rice leaves. Thus, the findings of our study show that the SRBSDV P6 antibody can be used in WB for detecting and monitoring SRBSDV infection in infected rice plants.
Introduction
Southern rice black-streaked dwarf virus (SRBSDV) is a severe threat to both the yield and quality of rice products worldwide (Wang et al., 2019). In 2010, ~2.97 million acres of agricultural land in China was affected by SRBSDV infection (Wu et al., 2017).
Similar to rice black-streaked dwarf virus (RBSDV), SRBSDV is an icosahedral virus with a diameter of 70–75 nm. The SRBSDV genome comprises ten segments (S1–S10) in the decreasing order of molecular weight. Ten SRBSDV segments encode five putative structural proteins (P2, P3, P4, P8, and P10) and eight putative nonstructural proteins (P1, P5-1, P5-2, P6, P7-1, P7-2, P9-1, and P9-2) (Mao et al., 2013; Wang et al., 2013; Li et al., 2017; Yu et al., 2017). Among these proteins, P6 is a viral RNA-silencing suppressor that may affect its interaction with other viral proteins such as P7-1 and P9-1 (Wang et al., 2011; Jia et al., 2012; Wu et al., 2013). Thus, P7-1 and P9-1 were used as targets for detecting SRBSDV infection (Wang et al., 2015; Ran et al., 2018).
Several methods have been used to detect SRBSDV infection such as polymerase chain reaction (PCR) with SRBSDV using S7-1, S9-1, and S10 genes (Wang et al., 2015); immunoassays with SRBSDV P9-1 (Wang et al., 2015); dot enzyme-linked immunosorbent assay (dot-ELISA) with SRBSDV P7-1, P9-1, and P10 (Chen et al., 2012; Wang et al., 2012; Uehara-Ichiki et al., 2013); yeast two-hybrid screening assay with P9-1 (Wang et al., 2015); and proteomics with SRBSDV proteins (Wang et al., 2017; Yang et al., 2017; Yu et al., 2017). Dot-ELISA is a simple and rapid method, but it shows an increased number of false positives. However, Western blotting (WB) can precisely solve this problem. In Spodoptera frugiperda cells induced by SRBSDV infection, P6 is recruited in the whole viroplasm matrix by directly interacting with P9-1 and P5 (Li et al., 2013, 2015a). To date, no study has reported the detection of SRBSDV with P6 using WB.
In the present study, P6 was expressed and regarded as a potential target of antiviral compound because of the importance of P6 in the whole viroplasm matrix (Wang et al., 2015). In our research, P6 was initially expressed and purified in vitro. Two polyclonal P6 antibodies were obtained and measured using ELISA. Subsequently, a WB approach was developed for identifying SRBSDV antigens derived from SRBSDV-infected rice samples collected from nine districts in Guizhou, China. The primary aim of the present study was to use WB to diagnose suspected SRBSDV infection in rice plants. To the best of our knowledge, this is the first study to diagnose SRBSDV infection in rice using WB with P6.
Materials and Methods
Plant Materials
Suspected SRBSDV-infected rice samples with typical dwarf symptoms were collected from Tianzhu, Meitan, Dejiang, Puan, Libo, Pingtang, Huishui, Dushan, and Anshun districts in Guizhou, China (Figure 1). The samples were kept in liquid nitrogen and subsequently stored at −80°C.
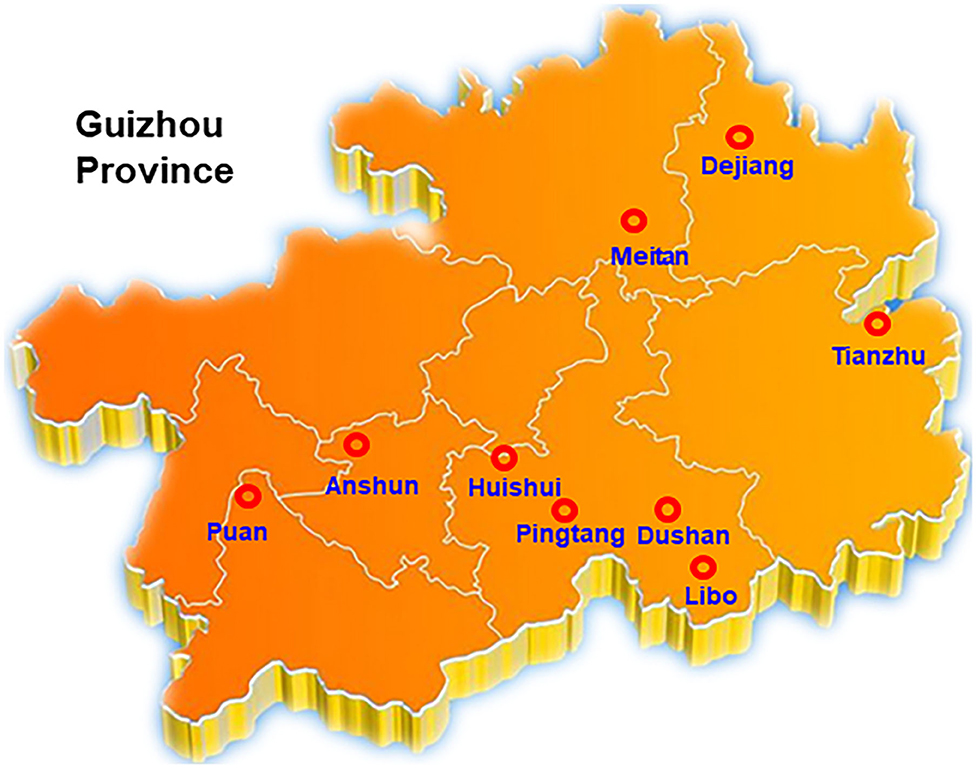
Figure 1. Investigation and collection of SRBSDV in Guizhou Province. Rice samples from Tianzhu, Meitan, Dejiang, Puan, Libo, Pingtang, Huishui, Dushan, and Anshun 9 districts of Guizhou were selected in this assay.
Gene Cloning and Plasmid Construction
SRBSDV RNA was extracted from SRBSDV-inoculated rice leaves using the TRIzol reagent method (TaKaRa, Japan), and the cDNA of P6 was obtained through cDNA synthesis. P6 gene was amplified using forward and reverse primers (5′-CGGGATCCATGTCTACCAACCTCACGAACATA-3′) and (5′-GCTCTAGATTACTCTGAAATAAGTTGCCACAA-3′), with a BamH I and Xho I at both ends, respectively. The target gene and the pCold-SUMO vector (Novagen, USA) were digested, purified through electrophoresis, and ligated with T4 ligase. The ligation products were transformed into DH5α competent cells. Colonies were identified by PCR. Positive clones were sequenced and identified by restriction enzyme digestion.
P6 Protein Expression and Purification
The recombinant SRBSDV P6 plasmid was transformed into the Escherichia coli strain BL21 (DE3). The cells were harvested at 4 h after inoculation and after isopropyl-β-d-thiogalactopyranoside (IPTG) induction at 25°C. Subsequently, the cells were resuspended in a lysis buffer (50 mM Tris pH 7.5, 100 mM NaCl, 1 mM EDTA, 1% Triton X-100, 1 mg/mL lysozyme, and bacterial protease inhibitor cocktail) and sonicated for 2 min on ice. The lysates were incubated on ice for 30 min.
Protein purification was performed according to the protocols reported previously (Ran et al., 2018). The extracted cell lysates were incubated with equilibrated agarose beads (GE Healthcare, IL, USA) for at least 2 h at 4°C. Centrifugation was performed at 2,000 × g for 2 min to remove the supernatant. Thereafter, the beads were washed 5 times with a wash buffer [1 × phosphate-buffered saline (PBS), 0.5% Triton X-100, and 1 mM EDTA]. The purified protein was eluted in a freshly prepared elution buffer (10 mM reduced glutathione, and 50 mM Tris pH 8.8).
Rabbit Immunization
Immunization and blood sample collection were performed at a biotechnology company (Youke, China). In this assay, two rabbits were injected with the suspended antigen in Freund's Adjuvant solution (Sigma, USA). On day 1, each rabbit was subcutaneously injected with 2 mL antigen solution (1 mL antigen with 1 mL Freund's Complete Adjuvant solution). The rabbits were administered three booster doses on days 15, 29, and 43 after immunization with 2 mL antigen solution (1 mL antigen with 1 mL Freund's Incomplete Adjuvant solution) as per the protocol in the first immunization. Pre- and post-immunization blood samples were collected, and 55 mL of each post-immunization blood sample was purified using protein A (Millipore, MA, USA).
Antibody Titer Detection
The titers of the purified antibody were tested by ELISA. Antigens were diluted to 1 μg/mL with citrate-buffered saline buffer (15 mM Na2CO3, 35 mM NaHCO3, pH 9.6) and blocked with 5% non-fat milk in PBS. Incubation was performed with the first and second antibodies. Then, the results of ELISA were detected using a spectrophotometer at A450 nm after the addition of the 3,3′,5,5′-tetramethylbenzidine reaction buffer (CWBio, China). For the titer test, antigens were diluted in a series ratio as follows: 1:1250, 1:2500, 1:5000, 1:10000, 1:20000, 1:40000, 1:80000, 1:160000, 1:320000, 1:640000, and 1:1280000. A healthy rabbit was used as the control.
Protein Extraction and Western Blotting Analysis
Rice leaves and stems were collected and ground in liquid nitrogen and were then resuspended in a protein lysis buffer (50 mM Tris PH 7.5, 6 M urea, 150 mM NaCl, 0.1% NP40, and 1 mM PMSF). The lysates were vortexed for 30 s and placed on ice for 30 min. Following lysis, the protein mixture was centrifuged at 20,000 × g for 20 min at 4°C to eliminate insoluble materials. The extracted protein was boiled with the sodium dodecyl sulfate (SDS)-loading buffer (250 mM Tris PH 6.8, 10% SDS, 0.5% bromophenol blue, and 50% glycerol) for 5 min and cooled on ice.
WB was conducted via the following steps. The extracted protein was loaded and run on SDS-polyacrylamide gel electrophoresis (SDS-PAGE) gel for protein electrophoresis. The protein in the gel was transferred onto a PVDF membrane for antibody detection (the PVDF membrane was soaked for 30 s in methanol before use). The PVDF membrane was blocked for 2 h at room temperature in 5% non-fat milk in PBS Tween-20 (PBST: PBS with 0.5% Tween-20). The first and second antibodies were incubated for 2 and 1 h, respectively, at room temperature in 5% non-fat milk in PBST. Signals were detected by adding the horseradish peroxidase substrate (Millipore, cat. WBKLS0100, USA).
Results
SRBSDV and Symptoms in Rice
Our group collected 50 rice samples with suspected SRBSDV infection from nine districts in Guizhou, China, from June to September 2017 and 2018. All the samples were collected from Tianzhu, Meitan, Dejiang, Puan, Libo, Pingtang, Huishui, Dushan, and Anshun districts in Guizhou, China (Figure 1). Most of the samples had at least one of the typical symptoms of SRBSDV infection such as serious dwarfing, wrinkled leaves, and tumor-like protrusion symptoms on their stems (Figure 2).
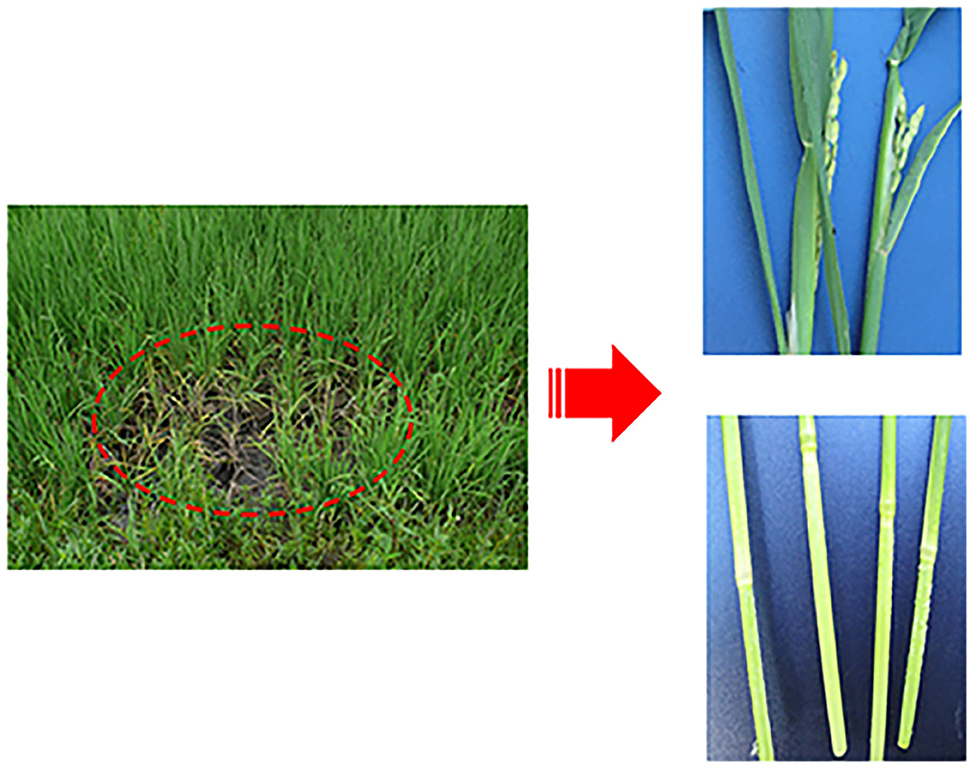
Figure 2. Symptoms of rice infected with SRBSDV. Dashed oval represents rice infected by SRBSDV in the field. The right figures are enlarged symptoms of infected rice leaves (Upper) and stems (Lower) by SRBSDV.
P6 Cloning, Expression and Purification
The results of 1% agarose gel electrophoresis showed that 2,382 bp of the P6 fragment was cut in pCold-SUMO-P6 by BamH I and Xho I, and the target gene segment was matched with the reported length of the P6 gene (Supplementary Table 1). P6 was overexpressed at 101 kDa (with a SUMO tag about 11 kDa) when the final concentration was increased to 0.7 mM IPTG and when the solution was left for 16 h at 25°C. These His-tagged fusion proteins were purified in an Ni-NTA column (GE Healthcare). More than 90% of P6 was eluted from the beads with 100–300 mM imidazole in 50 mM Tris-HCl and 100 mM NaCl buffer at pH 7.5, as revealed by the results of 8% SDS-PAGE analysis (Supplementary Table 2). These purified P6 at 101 kDa were then concentrated to ~1 mg/mL in 20 mM Tris-HCl and 100 mM NaCl at pH 7.5 for further analysis.
Rabbit Immunization and Antibody Induction
To detect SRBSDV, we produced SRBSDV antibodies in rabbits (rabbits 1 and 2). Two rabbits were injected with the purified P6. At 40 days after immunization, rabbit serum was collected and purified to obtain the P6 antibody. To verify the capability of this antibody, the purified P6 and rice leaves infected by SRBSDV were used as antigens. Our results showed that the P6 antibody displays specificity in detecting SRBSDV using WB (~101 kDa) (Figure 3A). Subsequently, the titer of the P6 antibody was tested by ELISA. As shown in Figure 4, the titers of the P6 antibody from rabbit one and rabbit two were about 1:80000 and 1:160000, respectively. Thus, the antibody generated from rabbit two was used for further assays. The antibody from rabbit two was further confirmed by WB (Figure 3B). The P6 antibody could detect SRBSDV, although the antigen was diluted 320,000 times (Figure 3B).
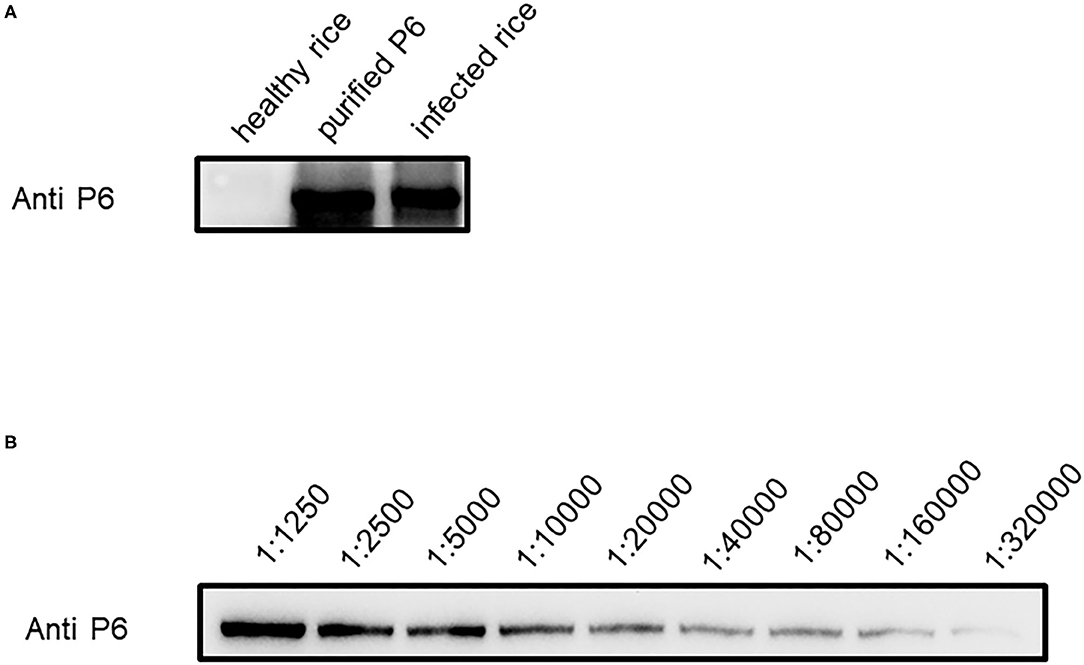
Figure 3. Efficiency of P6 antibody in detecting SRBSDV. (A) SRBSDV detection of P6 antibody in healthy rice, protein antigen, and infected rice. (B) SRBSDV detection sensitivity of P6 antibody. Antigen diluted in series times such as 1:1250, 1:2500, 1:5000, 1:10000, 1:20000, 1:40000, 1:80000, and 1:160000 was used to test for P6 antibody detection limitation.
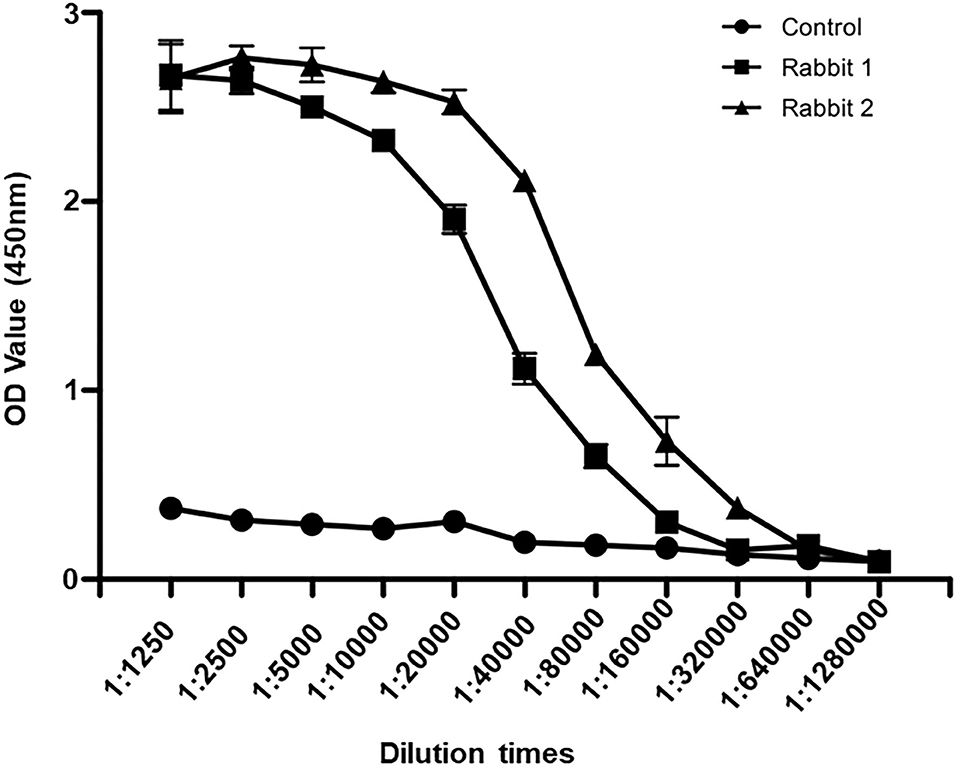
Figure 4. Antibody titer detection by ELISA. Sera from rabbit one, rabbit two, and healthy rabbit were diluted to 1:1250, 1:2500, 1:5000, 1:10000, 1:20000, 1:40000, 1:80000, 1:160000, 1:320000, 1:640000, and 1:1280000. The values of OD450 were recorded.
Detection of SRBSDV in Guizhou Province
To shed light on the dynamics of the occurrence of SRBSDV in Guizhou, China, we collected rice samples showing SRBSDV infection symptoms (Figure 2) in rice-growing areas such as Dejiang, Meitan, Tianzhu, Anshun, Puan, Huishui, Pingtang, Dushan, and Libo (Figure 1). Samples from three different locations of the same district were harvested and stocked in liquid nitrogen immediately. Their proteins were extracted and detected using the P6 antibody for identifying SRBSDV. As shown in Figure 5, the rice samples from Anshun, Huishui, Pingtang, Dushan, and Libo, which are the southern districts, showed the P6 band, indicating that the rice plants in these areas were infected by SRBSDV. However, we did not detect the P6 band in the samples from the northern and western districts such as Dejiang, Meitan, Tianzhu, and Puan, thereby indicating that no SRBSDV existed in these districts. These findings implied that SRBSDV in Guizhou may have spread from the south to north of China.
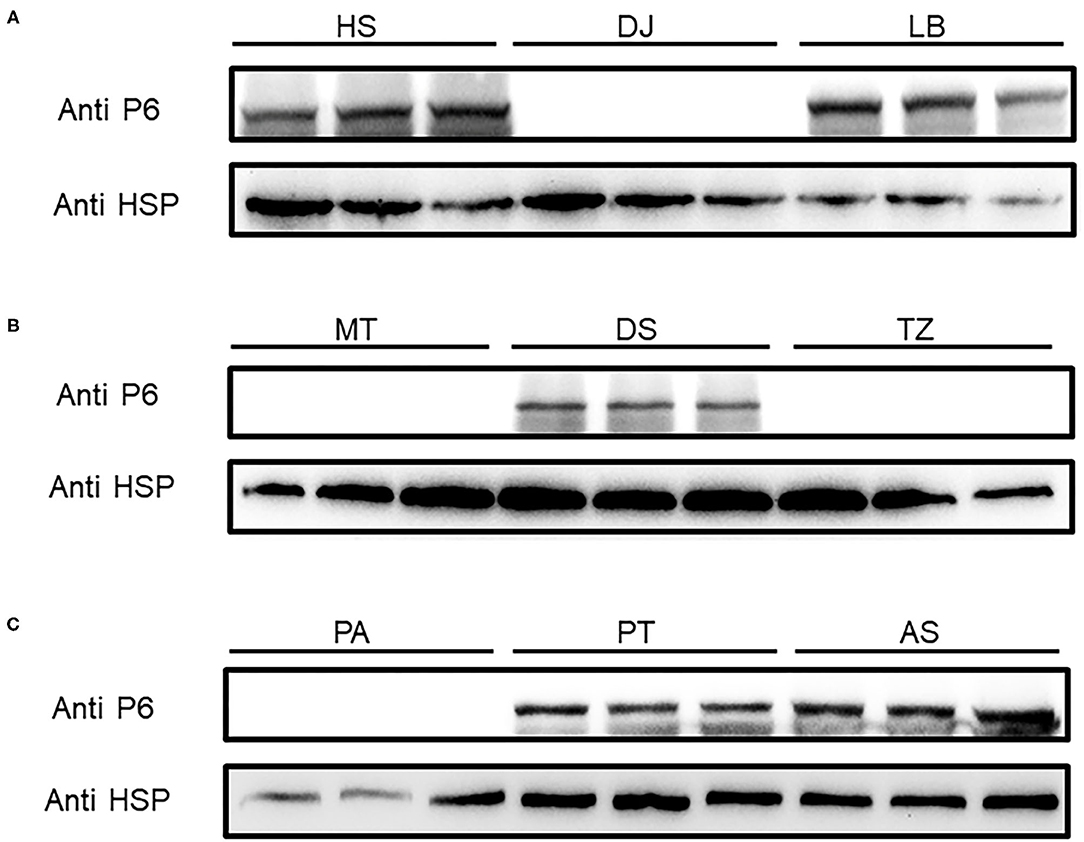
Figure 5. SRBSDV detection in nine districts of Guizhou province. Rice samples from Huishui (HS), Dejiang (DJ), Libo (LB) (A), Meitan (MT), Dushan (DS), Tianzhu (TZ) (B), and Puan (PA), Pingtang (PT), and Anshun (AS) (C) were investigated by Western blot. P6 antibody was used to detect SRBSDV, and rice protein HSP90 was used as the internal control.
Quantitative Comparison of SRBSDV Content in Rice Tissues
The virus content could vary across the plant tissues (Rettcher et al., 2015; Morán et al., 2018). To determine the SRBSDV content in rice leaves and stems that showed different symptoms when infected with SRBSDV (as illustrated in Figure 2), we quantified the SRBSDV content in rice leaves and stems using the P6 antibody. SRBSDV was detected in the rice leaves and stems from Huishui, Pingtang, and Libo districts. As shown in Figure 6, the SRBSDV content in rice stems was much higher than that in rice leaves. These findings indicate that SRBSDV is mainly accumulated in rice stems and then transmitted to rice leaves.
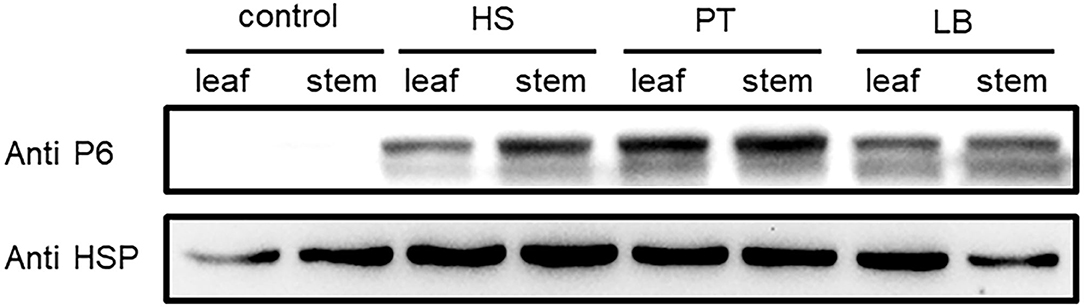
Figure 6. Comparison of SRBSDV content in rice tissues. Rice leaves and stems from Huishui (HS), Pingtang (PT), and Libo (LB) were used to evaluate SRBSDV content. Healthy rice was used as a negative control, and rice internal control HSP90 was used as the internal control.
Discussion
Rice plants infected by SRBSDV have caused a significant economic loss mainly in China and other Asian countries in recent years (Alonso et al., 2019). Moreover, rice plants infected by SRBSDV show a long latent period and are difficult to detect at an early stage, but they result in serious destruction at the later stage. Therefore, a reliable early monitoring and detection method will be useful for managing this infection and evaluating its risk (Zhang et al., 2008; Zhou et al., 2008; Hoang et al., 2011). To date, several methods have been used for detecting plant viruses, such as reverse transcription (RT) loop-mediated isothermal amplification, RT–PCR, and RT–qPCR; however, these methods provide false positive results, thereby causing inaccurate determination (López et al., 2006; Londoño et al., 2016; Malandraki et al., 2017; Treder et al., 2018). WB is a stable, reliable, and highly sensitive method that has been widely used for determining protein interaction and identification, quantifying protein expression, and detecting pathogen and virus movement (Samuilova et al., 2013; Huo et al., 2018; Wu et al., 2018). In our study, we established a WB assay to detect and quantify SRBSDV in infected rice plants. Using P6—a non-structural protein in SRBSDV—as the target, we could effectively detect SRBSDV in a rice paddy field and quantify the concentration of SRBSDV in different rice tissues, indicating that this assay is a salutary method for SRBSDV monitoring and forecasting as well as controlling and managing SRBSDV infection in the early stage.
Monoclonal antibodies have been produced and applied for detecting several plant virus infections (Tian et al., 2014; Li et al., 2015b; Zhang et al., 2018), but compared with polyclonal antibodies, monoclonal antibodies are much more expensive; they have a longer production period and are at more risk for failure. In this study, we aimed to detect SRBSDV infection in rice plants using polyclonal antibodies. First, the solubility of P6 was determined and then P6 was purified in E. coli. Second, two polyclonal P6 antibodies were obtained and quantified using WB. Then, a WB assay was developed for identifying SRBSDV infection in Guizhou, China. Finally, our results had shown for the first time that WB could be used to identify SRBSDV infected using the SRBSDV P6 antigen.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
XX and XL: conceptualization and funding acquisition. JJ and ZQ: methodology. JJ: validation and data curation. MH: formal analysis. MC: investigation. XL: resources, writing—review and editing, supervision, and project administration. XX: writing—original draft preparation. All authors contributed to the article and approved the submitted version.
Funding
This research was funded by the National Natural Science Foundation of China (Nos. 31801691, 31960546, and 32060614), the Science and Technology Foundation of Guizhou Province ([2019]2408), the Advanced Programs of Guizhou Province for the Returned Overseas Scholars ([2018]02), the Ph.D. Funding of Guizhou University, (No. 201754) and the Project of Guizhou Province [No. (2018)5781], Program of Introducing Talents of Discipline to Universities of China (111 Program, D20023).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fsufs.2021.637382/full#supplementary-material
References
Alonso, P., Gladieux, P., Moubset, O., Shih, P., Mournet, P., Frouin, J., et al. (2019). Emergence of southern rice black-streaked dwarf virus in the centuries-old Chinese Yuanyang agrosyst of rice landraces. Viruses 11:985. doi: 10.3390/v11110985
Chen, Z., Liu, J., Zeng, M., Wang, Z., Yu, D., Yin, C., et al. (2012). Dot immunobinding assay method with chlorophyll removal for the detection of southern rice black-streaked dwarf virus. Molecules 17, 6886–6900. doi: 10.3390/molecules17066886
Hoang, A. T., Zhang, H. M., Yang, J., Chen, J. P., Hébrard, E., Zhou, G. H., et al. (2011). Identification, characterization, and distribution of southern rice black-streaked dwarf virus in Vietnam. Plant Dis. 95, 1063–1069. doi: 10.1094/PDIS-07-10-0535
Huo, Y., Yu, Y., Chen, L., Li, Q., Zhang, M., Song, Z., et al. (2018). Insect tissue-specific vitellogenin facilitates transmission of plant virus. PLoS Pathog. 14:e1006909. doi: 10.1371/journal.ppat.1006909
Jia, D., Chen, H., Zheng, A., Chen, Q., Liu, Q., Xie, L., et al. (2012). Development of an insect vector cell culture and RNA interference system to investigate the functional role of fijivirus replication protein. J. Virol. 86, 5800–5807. doi: 10.1128/JVI.07121-11
Li, J., Xue, J., Zhang, H., Yang, J., Lv, M., Xie, L., et al. (2013). Interactions between the P6 and P5-1 proteins of southern rice black-streaked dwarf fijivirus in yeast and plant cells. Arch. Virol. 158, 1649–1659. doi: 10.1007/s00705-013-1660-4
Li, J., Xue, J., Zhang, H., Yang, J., Xie, L., and Chen, J. (2015a). Characterization of homologous and heterologous interactions between viroplasm proteins P6 and P9-1 of the fijivirus southern rice black-streaked dwarf virus. Arch. Virol. 160, 453–457. doi: 10.1007/s00705-014-2268-z
Li, N., Chen, Z., Liu, Y., Liu, Y., Zhou, X., and Wu, J. (2015b). Development of monoclonal antibodies and serological assays specific for Barley yellow dwarf virus GAV strain. Virol. J. 12:136. doi: 10.1186/s12985-015-0367-4
Li, S., Zhang, T., Zhu, Y., and Zhou, G. (2017). Co-infection of two reoviruses increases both viruses accumulation in rice by up-regulating of viroplasm components and movement proteins bilaterally and RNA silencing suppressor unilaterally. Virol. J. 14:150. doi: 10.1186/s12985-017-0819-0
Londoño, M. A., Harmon, C. L., and Polston, J. E. (2016). Evaluation of recombinase polymerase amplification for detection of begomoviruses by plant diagnostic clinics. Virol. J. 13:48. doi: 10.1186/s12985-016-0504-8
López, R., Asensio, C., Guzman, M. M., and Boonham, N. (2006). Development of real-time and conventional RT-PCR assays for the detection of potato yellow vein virus (PYVV). J. Virol. Methods. 136, 24–29. doi: 10.1016/j.jviromet.2006.03.026
Malandraki, I., Beris, D., Isaioglou, I., Olmos, A., Varveri, C., and Vassilakos, N. (2017). Simultaneous detection of three pome fruit tree viruses by one-step multiplex quantitative RT-PCR. PLoS ONE 12:e0180877. doi: 10.1371/journal.pone.0180877
Mao, Q., Zheng, S., Han, Q., Chen, H., Ma, Y., Jia, D., et al. (2013). New Model for the Genesis and maturation of viroplasms induced by fijiviruses in insect vector cells. J. Virol. 87, 6819–6828. doi: 10.1128/JVI.00409-13
Morán, F., Olmos, A., Lotos, L., Predajna, L., Katis, N., Glasa, M., et al. (2018). A novel specific duplex real-time RT-PCR method for absolute quantitation of Grapevine Pinot gris virus in plant material and single mites. PLoS ONE 13:e0197237. doi: 10.1371/journal.pone.0197237
Ran, L., Ding, Y., Luo, L., Gan, X., Li, X., Chen, Y., et al. (2018). Binding constants of Southern rice black-streaked dwarf virus coat protein with ferulic acid derivatives. Data Brief. 17, 321–324. doi: 10.1016/j.dib.2018.01.031
Rettcher, S., Jungk, F., Kühn, C., Krause, H. J., Nölke, G., Commandeur, U., et al. (2015). Simple and portable magnetic immunoassay for rapid detection and sensitive quantification of plant viruses. Appl. Environ. Microbiol. 81, 3039–3048. doi: 10.1128/AEM.03667-14
Samuilova, O., Santala, J., and Valkonen, J. P. (2013). Tyrosine phosphorylation of the triple gene block protein 3 regulates cell-to-cell movement and protein interactions of potato mop-top virus. J. Virol. 87, 4313–4321. doi: 10.1128/JVI.03388-12
Tian, Y. P., Hepojoki, J., Ranki, H., Lankinen, H., and Valkonen, J. P. (2014). Analysis of potato virus Y coat protein epitopes recognized by three commercial monoclonal antibodies. PLoS ONE 9:e115766. doi: 10.1371/journal.pone.0115766
Treder, K., Chołuj, J., Zacharzewska, B., Babujee, L., Mielczarek, M., Burzyński, A., and Rakotondrafara, A. M. (2018). Optimization of a magnetic capture RT-LAMP assay for fast and real-time detection of potato virus Y and differentiation of N and O serotypes. Arch. Virol. 163, 447–458. doi: 10.1007/s00705-017-3635-3
Uehara-Ichiki, T., Shiba, T., Matsukura, K., Ueno, T., Hirae, M., and Sasaya, T. (2013). Detection and diagnosis of rice-infecting viruses. Front. Microbiol. 4:289. doi: 10.3389/fmicb.2013.00289
Wang, D., Xie, X., Gao, D., Chen, K., Chen, Z., Jin, L., et al. (2019). Dufulin intervenes the viroplasmic proteins as the mechanism of action against southern rice black-streaked dwarf virus. J. Agric. Food Chem. 67, 11380–11387. doi: 10.1021/acs.jafc.9b05793
Wang, Q., Tao, T., Han, Y., Chen, X., Fan, Z., Li, D., et al. (2013). Nonstructural protein P7-2 encoded by rice black-streaked dwarf virus interacts with SKP1, a core subunit of SCF ubiquitin ligase. Virol. J. 10:325. doi: 10.1186/1743-422X-10-325
Wang, Q., Tao, T., Zhang, Y., Wu, W., Li, D., Yu, J., et al. (2011). Rice black-streaked dwarf virus P6 self-interacts to form punctate, viroplasm-like structures in the cytoplasm and recruits viroplasm-associated protein P9-1. Virol. J. 8:24. doi: 10.1186/1743-422X-8-24
Wang, Z., Li, X., Wang, W., Zhang, W., Yu, L., Hu, D., et al. (2015). Interaction research on the antiviral molecule dufulin targeting on southern rice black streaked dwarf virus p9-1 nonstructural protein. Viruses 7, 1454–1473. doi: 10.3390/v7031454
Wang, Z., Yu, D., Li, X., Zeng, M., Chen, Z., Bi, L., et al. (2012). The development and application of a Dot-ELISA assay for diagnosis of southern rice black-streaked dwarf disease in the field. Viruses 4, 167–183. doi: 10.3390/v4010167
Wang, Z., Yu, L., Jin, L., Wang, W., Zhao, Q., Ran, L., et al. (2017). Evaluation of rice resistance to southern rice black-streaked dwarf virus and rice ragged stunt virus through combined field tests, quantitative real-time PCR, and proteome analysis. Viruses 9:37. doi: 10.3390/v9020037
Wu, G., Cui, X., Chen, H., Renaud, J. B., Yu, K., Chen, X., et al. (2018). Dynamin-like proteins of endocytosis in plants are coopted by potyviruses to enhance virus infection. J. Virol. 92, e01320–e01318. doi: 10.1128/JVI.01320-18
Wu, J., Li, J., Mao, X., Wang, W., Cheng, Z., Zhou, Y., et al. (2013). Viroplasm protein P9-1 of rice black-streaked dwarf virus preferentially binds to single-stranded RNA in its octamer form, and the central interior structure formed by this octamer constitutes the major RNA binding site. J. Virol. 87, 12885–12899. doi: 10.1128/JVI.02264-13
Wu, Y., Zhang, G., Chen, X., Li, X., Xiong, K., Cao, S., et al. (2017). The influence of Sogatella furcifera (Hemiptera: Delphacidae) migratory events on the southern rice black-streaked dwarf virus epidemics. J. Econ. Entomol. 110, 854–864. doi: 10.1093/jee/tox062
Yang, A., Yu, L., Chen, Z., Zhang, S., Shi, J., Zhao, X., et al. (2017). Label-free quantitative proteomic analysis of chitosan oligosaccharide-treated rice infected with southern rice black-streaked dwarf virus. Viruses 9:115. doi: 10.3390/v9050115
Yu, L., Shi, J., Cao, L., Zhang, G., Wang, W., Hu, D., et al. (2017). A novel method for transmitting southern rice black-streaked dwarf virus to rice without insect vector. Virol. J. 14:155. doi: 10.1186/s12985-017-0815-4
Zhang, H. M., Yang, J., Chen, J. P., and Adams, M. J. (2008). A black-streaked dwarf disease on rice in China is caused by a novel fijivirus. Arch. Virol. 153, 1893–1898. doi: 10.1007/s00705-008-0209-4
Zhang, M., Chen, R., Zhou, X., and Wu, J. (2018). Monoclonal antibody-based serological detection methods for wheat dwarf virus. Virol. Sin. 33, 173–180. doi: 10.1007/s12250-018-0024-3
Keywords: SRBSDV, P6, antibody production, virus detection, western blot
Citation: Xie X, Jiang J, Huang M, Chen M, Qu Z and Li X (2021) Detection of Southern Rice Black-Streaked Dwarf Virus Using Western Blotting With P6. Front. Sustain. Food Syst. 5:637382. doi: 10.3389/fsufs.2021.637382
Received: 03 December 2020; Accepted: 07 January 2021;
Published: 28 January 2021.
Edited by:
Wei Li, Chinese Academy of Agricultural Sciences, ChinaReviewed by:
Xiaoting Zhang, Henan Agricultural University, ChinaXiaofeng Su, Biotechnology Research Institute (CAAS), China
Copyright © 2021 Xie, Jiang, Huang, Chen, Qu and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiangyang Li, eHlsaTFAZ3p1LmVkdS5jbg==
†These authors have contributed equally to this work
 Xin Xie
Xin Xie Junmei Jiang1†
Junmei Jiang1†