
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Sustain. Food Syst. , 22 February 2021
Sec. Crop Biology and Sustainability
Volume 5 - 2021 | https://doi.org/10.3389/fsufs.2021.605195
This article is part of the Research Topic Plant Growth-Promoting Microorganisms for Sustainable Agricultural Production View all 50 articles
Bacillus spp. are well-characterized as efficient bioinoculants for sustainable plant growth promotion and biocontrol of phytopathogens. Members of this spp. exhibit the multifaceted beneficial traits that are involved in plant nutrition and antimicrobial activities against phytopathogens. Keeping in view their diverse potential, this study targeted the detailed characterization of three root-colonizing Bacillus strains namely B. amyloliquefaciens, B. subtilis, and B. tequilensis, characterized based on 16S rRNA sequencing homology. The strains exhibited better plant growth promotion and potent broad-spectrum antifungal activities and exerted 43–86% in-vitro inhibition of growth of eight fungal pathogens. All strains produced indole acetic acid (IAA) in the range of 0.067–0.147 μM and were positive for the production of extracellular enzymes such as cellulase, lipase, and protease. Ultra-performance Liquid Chromatography-Electrospray Ionization-Mass Spectrometry (UPLC-ESI-MS/MS) analysis revealed the production of antifungal metabolites (AFMs) such as surfactins, iturins, fengycins, macrolactins, bacillomycin-D, and catechol-based siderophore bacillibactin which were further confirmed by amplifying the genes involved in the biosynthesis of these antimicrobial lipopeptides. When compared for the amounts of different cyclic-peptides produced by three Bacillus strains, B. amyloliquefaciens SB-1 showed the most noticeable amounts of all the antifungal compounds. Plant experiment results revealed that inoculation with phytohormone producing Bacillus spp. strains demonstrated substantial growth improvement of wheat biomass, number of spikes, and dry weight of shoots and roots. Results of this study indicate the biocontrol and biofertilizer potential of Bacillus spp. for sustainable plant nutrient management, growth promotion, and effective biocontrol of crop plants, particularly cultivated in the South Asian region.
Increasing environmental concerns and food safety issues gave momentum to the use of bio-fertilizers and biological control agents in agriculture sector world-wide. Moreover, to combat the threatening challenges of non-degradable and recalcitrant agrochemicals, and increased pesticide resistance, plant growth promoting bacteria (PGPB), and their bioactive metabolites got global attention (Bhardwaj et al., 2014). PGPB exert multifaceted benefits to the plants and enrich soil nutrients by means of phosphate/potassium/zinc mineralization, release of plant growth promoting regulators and hormones, and nitrogen fixation (Hayat et al., 2010; Saharan and Nehra, 2011). Furthermore, their contribution in plant defense through the production of metabolites make them significant and key players in sustainable agriculture (Sinha et al., 2014).
Currently, main plant growth promoting rhizobacteria (PGPR) include Azotobacter, Azospirillum, and Rhizobium [N2-fixers], Gluconacetobacter, Enterobacter, Azoarcus, Klebsiella, Burkholderia, Pantoea, Stenotrophomonas spp. [mineral mobilizers and plant-hormone regulators], Bacillus and Pseudomonas [PGP and biocontrol agents]. Among these predominant and the most widely studied are Bacillus and Pseudomonas spp. for their characteristic roles in nutrient acquisition and assimilation, secretion and modulation of hormones, plant beneficial secondary metabolites, antifungal metabolites (AFMs), antibiotics, and various extracellular signaling compounds (Ahemad and Kibret, 2014; Agler et al., 2016). Bacillus spp. have been in limelight for their long shelf-life in bio-formulations, effective colonization of plant tissues, and broad-spectrum antifungal abilities (Souza et al., 2014). Almost 4–5% genome of Bacillus spp. is dedicated for the production of structurally diverse antimicrobial compounds that have demonstrated varying antagonism against fungal and bacterial phytopathogens. Most significant among these antimicrobials are cyclic-lipopeptides (CLPs) constituting iturins, fengycins, and surfactins that are pivotal in root colonization by Bacillus spp. (Aira et al., 2010; Carvalhais et al., 2013).
Many research studies have investigated the biofertilizer and biocontrol potential of Bacillus spp., however, less is known about the complete metabolomic profile, account of the plant growth-promoting traits, secondary metabolites and their variants produced by a single strain. For instance, endophytic B. subtilis strain ALB629 was evaluated for its plant growth-promoting traits but its antagonistic secondary metabolites were not identified (Falcäo et al., 2014). Likewise, PGPB Bacillus strains RM-2 and BPR7 were evaluated for in-vitro antagonistic activities, nonetheless, they were not subjected to detailed characterization of their secondary metabolites (Kumar et al., 2012; Minaxi et al., 2012; Shobana et al., 2020).
The following study aimed to identify antifungal Bacillus spp., characterization and relative quantification of their strain-specific antagonistic metabolites, and analysis of their biofertilizer potential. Based on the biocontrol potential, three Bacillus spp. isolates, i.e., SB-1, A-2, A-3, were hypothesized to be active against phytopathogens of important economic crops and subjected to detailed characterization. Antagonistic metabolites of these isolates were comprehensively documented using polyphasic approach with the identification of putative genes involved in the production of these metabolites. Furthermore, bacterial isolates were screened for their plant growth-promoting (PGP) traits and their effects as bioinoculants for wheat were evaluated in-vivo to be used as multifaceted biofertilizer and biopesticides for various crop plants.
Bacteria were isolated from sugarcane, rice (Lahore, Pakistan, 31.5204° N, 74.3587° E), and corn (Kasur, Pakistan, 31.1179° N, 74.4408° E), using standard serial dilution plating (Somasegaran and Hoben, 1985, Supplementary Tables 1–3). Pure bacterial cultures were obtained by streaking single colonies on Luria-Bertani (LB) agar plates incubated at 37°C for 48 h. Antagonistic activity of bacterial isolates was checked against fungal phytopathogens (diseases, affected crops, host plants are described in Supplementary Table 4) including Aspergillus niger (NCBI accession no. MN786323), A. flavus, Fusarium oxysporum (NCBI accession no. MN636869), F. moniliforme (NCBI accession no. MN636870), F. solani, Colletotrichum falcatum, Curvularia sp. (NCBI accession no. MN636871), and Rhizopus sp. (NCBI accession no. MN636450), using plate bioassays. Fungal cultures were obtained from the culture collection of Mycology Laboratory, Forman Christian College (A Chartered University) Lahore, Pakistan and maintained on potato-dextrose agar (PDA) plates at 26–28°C.
Dual culture assay described by Sakthivel and Gnanama-nickam (1986) was used to determine the antagonistic activity of bacterial isolates against fungal phytopathogens. In a complementary antagonistic experiment, agar well-diffusion assay was performed (Magaldi et al., 2004). Results were expressed as percentage inhibition zones by antagonistic bacteria to suppress the growth of fungal mycelia. Each experiment was performed twice in triplicates and percentage inhibition was calculated as
Where
I = % inhibition, C = fungal diameter in control plate, T = fungal diameter in test plate.
Based on antifungal activity, three bacterial isolates [SB-1 (sugarcane), A-2 (rice), and A-3 (corn)] were selected for detailed studies. These isolates were biochemically characterized using QTS-24 bacterial identification kits (DESTO Laboratories, Karachi, Pakistan) using manufacturer's protocol. Bacterial DNA was isolated using GeneJET genomic DNA purification kit (Thermo Fisher Scientific, USA, catalog number: K0721). 16S rRNA gene was amplified from genomic DNA of isolates SB-1, A2, and A3. Primers and PCR conditions are described in Table 1. A reaction mixture of 50 μL was prepared by using Taq buffer 5 μL (10X), MgCl2 2 μL (25 mM), Taq polymerase (5 U) 2 μL, dNTPs 2 μL (2.5 mM), each of forward and reverse primer (20 pmol) 1 μL, dH2O 35 μL, and the template DNA 2 μL (>50 ng/μL). All PCR reagents were purchased from Fermentas (Thermo Fisher Scientific, USA). PCR products were purified using GeneJET PCR purification kit (Thermo Fisher Scientific, USA, catalog number: K0701) and sequenced with both, forward and reverse primers, by Eurofins Genomics, USA. Amplified sequences were compared with NCBI GenBank sequence database, using BLAST search tool and phylogenetic analysis was performed using MEGA5 platform (Tamura et al., 2011). Nucleotide sequences of isolates SB-1, A2, and A3 were aligned with Clustal X 2.1 program. Bootstrap confidence analysis was performed on 1,000 replicates to determine the reliability of the distance tree topologies obtained. The evolutionary distances were computed using the Maximum Composite Likelihood method and are in the units of number of base substitutions per site. All positions containing gaps and missing data were eliminated from the dataset (complete deletion option).
To identify secondary metabolites, bacterial isolates were individually inoculated to 100 mL LB broth and incubated at 37°C for 48 h in shaking incubator at 180 rpm. Bacterial cultures were pelleted by centrifugation (3,900 × g and 4°C for 20 min) and the pH of the supernatants was adjusted to 2.0 with 6N HCl. Secondary metabolites were precipitated by incubating the supernatants overnight at 4°C. The precipitates were recovered by centrifugation (3,900 × g and 4°C for 20 min) and dissolved in 5 mL of methanol: H2O (2:1, v/v). The extracts were re-centrifuged (3,900 × g and 4°C for 20 min), supernatants were dried under vacuum and, re-suspended in 2 mL of methanol, and 10 μL were injected for each LC-MS analysis.
Sample solutions were injected onto an Eclipse Plus C18 RRHD column (2.1 × 100 mm, 1.8 μm; Agilent, Santa Clara, CA, USA) and separated by reversed-phase liquid chromatography using a Waters Acquity UPLC system (Milford, MA, USA) at a flow rate of 0.4 mL/min. The UPLC system was coupled to an Orbitrap Fusion Tribrid mass spectrometer (Thermo Fisher Scientific, San Jose, CA, USA) equipped with an EASY-Max NG electrospray ion source. The mobile phases were water/0.01% formic acid (A) and 100% acetonitrile/0.01% formic acid (B). Secondary metabolites were separated on the column with a 20 min binary solvent elution gradient (0 min, 5% B; 0–15 min, 5–100% B; 15–17 min, 100% B) followed by a 3-min column equilibration at 5% B between injections. The MS parameters were as follows: positive-ion electrospray spray voltage, 3.9 kV; negative-ion electrospray spray voltage, 2.5 kV; capillary temperature, 325°C; S-lens Level, 60%; sheath gas, 50; sweep gas, 1; and auxiliary gas 10. The MS survey scan was carried out over the mass range m/z 100–2,000, with the data recorded in the centroid mode and at a mass resolution of 120 K FWHM (m/z 200), AGC target 4E5, and one microscan with a maximum inject time of 50 ms in the quadruple isolation mode. A lock mass at m/z 391.28426 from di(2-ethylhexyl) phthalate (a ubiquitous plasticizer) was used for real-time internal mass calibration during the LC-MS runs with the Fourier Transform (FT) MS detection. In the LC-MS/MS runs, the top five most intense ions, with their charge state of 1 and ion counts of >5,000 in each survey scan were selected for MS/MS by collision-induced dissociation (CID) in the linear ion trap or by “higher-energy” collisional dissociation (HCD) in the collision cell in the upfront of the C-trap. Other parameters included: activation isolation window, 2 Da; AGC target for MS/MS, 5E4; maximum inject time, 30 ms; activation time, 10 ms; activation Q, 0.25; and normalized collision energy at 35% for the CID operations. Activation isolation window, 2 Da; AGC target for MS/MS, 5E4; maximum inject time, 30 ms; activation time, 10 ms; activation Q, 0.25; and normalized collision energy at 25% were used for the HCD operations. Raw data files were recorded and processed using the XCalibur 4.1.31.9 (Thermo Scientific) software suite. For relative quantification of secondary metabolites produced by Bacillus strains, each sample solution was injected in triplicate and the peak areas were normalized. Bar graphs were plotted with error bars to show the analytical standard deviations.
Detection of the genes encoding surfactin, iturin, bacillomycin, and fengycin was done using gene-specific primers. Primers used in this study and PCR conditions are shown in Table 1. An individual reaction mixture of 50 μL was prepared for all the reactions as described above. PCR products were analyzed on 1% agarose gel. The respective bands were excised and purified using a gel extraction kit. PCR products were sequenced (Eurofins) and analyzed using the NCBI BLAST (blastn) and alignment tools.
Bacterial cultures were grown in 10 mL of LB medium at 180 rpm and 37°C temperature, supplemented with L-tryptophan (100 mg/L). After 7 days of growth, bacterial cells were centrifuged at 3,900 × g for 30 min (Allegra TM X-22R Centrifuge, Beckman Coulter, California, USA) and the pH of the supernatants was adjusted to pH 2.0 with 6 N HCl. The acidified supernatants were extracted twice with an equal volume (10 mL) of ethyl acetate. After centrifugation at 3,900 × g for 30 min, the upper organic layers were collected, pooled, and evaporated to dryness.
The reference standard of indole-3-acetic acid (IAA, C10H9NO2, monoisotopic neutral mass = 175.0633, Sigma-Aldrich, USA) was dissolved in methanol:chloroform:H2O [50:25:25, v/v] at 0.25 mM to form stock standard solution (Supplementary Table 5). Ten-μL aliquots of each standard and sample were injected into the column for the LC-FTMS (Fourier Transform Mass Spectrometry) runs, as described above. The concentrations in the samples were determined from the peak areas of the extracted ion chromatograms of IAA detected at m/z 176.0706 [M+H]+ by interpolation of the linear-regression calibration curve for this compound (Supplementary Figure 1).
Lipase production was checked on 1% Tween-20 LB agar plates according to the method of Sierra (1957). Pikovskaya agar medium was used for the detection of phosphatase enzyme (Pikovskaya, 1948). Protease production was checked using skim milk agar plates (Alnahdi, 2012). Extracellular cellulase production was checked on 1% (Carboxymethyl Cellulose) CMC-LB agar plates (Kasana et al., 2008). In-vitro zinc solubilization was assessed using the method of Sharma et al. (2012). Siderophore production was detected as per O-CAS method (Louden et al., 2011). Qualitative determination of hydrocyanic acid (HCN) production carried out as per alkaline-picrate method (Millar and Higgins, 1970).
In-vivo effect of Bacillus spp. as bioinoculants was evaluated using pot experiments in climate control room. Soil used for this experiment was taken (0–50 cm) from wheat field, demonstrated the following physico-chemical properties: temperature (°C) = 21.7, pH = 7.65, moisture (%) = 23, texture = silty loam, electrical conductivity (EC1 : 1 dS/m) = 1.15, organic matter (OM g/kg) = 29.2, and potential acidity (H+ Al mg/kg) = 47.3. Soil was sieved and sterilized in drying oven at 90°C overnight, to eliminate native microbial communities and analyzed post-sterilization to detetermine any microbial population. Pots were filled with 300 g soil (per pot) and 25 mL of half-strength Hoagland's solution per pot was added (Hoagland and Arnon, 1970). For seed sterilization, 100 seeds of wheat variety, i.e., Faisalabad-2008 were soaked in 100 mL of 0.1 N bleach (sodium hypochlorite; NaClO) solution for 15 min, and given four successive washes of 10 min each with sterile dH2O (Chen et al., 2018). Seeds were transferred to 1% water-agar plates and incubated at 22–24°C. Three days old germinated seedlings were transferred in pots (1 seed/pot) and inoculated with 1 mL of individual bacterial culture containing 1 × 107 cells/mL in 10 replicates for each treatment. Plants were kept in a climate control room at relative humidity of 60% with a 12 h photoperiod (200 μM·m −2·s−1 at pot heights with fluorescent lights, 15/20°C). The experiment was set up in a completely randomized block design (CRBD). The temperature in climate room was maintained at 20 ± 2°C; with light source of 6,000 ± 500 FLUX and light period of 10 ± 1 h. To provide moisture, plants were watered at alternate days using autoclaved distilled water and second dose of half strength Hoagland's solution was applied after 15 days. Plants were harvested after 50 days, roots were thoroughly washed and the root and shoot lengths of individual plants of each treatment were noted. Roots and shoots were detached to determine fresh weights of roots and shoots following drying in an oven for 72 h at 70°C to record dry weights of shoots and roots.
All the experments were prformed in triplicate and the average of each data was taken. Data was statistically analyzed using the Statistical Package for the Social Sciences (SPSS) software (IBM Statistics 23.0) at α = 0.05.
Three isolates; SB-1 (sugarcane stem), A-2 (rice rhizosphere), and A-3 (corn rhizosphere) exhibited broad-spectrum in-vitro antifungal activities against all the pathogens tested (Figure 1A). SB-1 showed ~86% inhibition of F. oxysporum, whereas, Bacillus strains A-2 and A-3 exhibited ~64 and ~50% inhibition of F. oxysporum, respectively. Figure 1B highlights the variable growth inhibition of fungal pathogens by the bacterial isolates. Strain SB-1 also demonstrated >70% inhibition of mycelial growth of F. moniliforme, F. solani, A. flavus, A. Niger, and Curvularia sp. Whereas, 60~65% suppression of these pathogens was shown by strains A-2 and A-3. Minimum inhibition (~43%) was noted for the sugarcane red-rot pathogen C. falcatum by the strain A-3, however, SB-1 exhibited ~66% suppression of this pathogen. Rhizopus sp. was moderately inhibited by all three strains tested (52~65%).
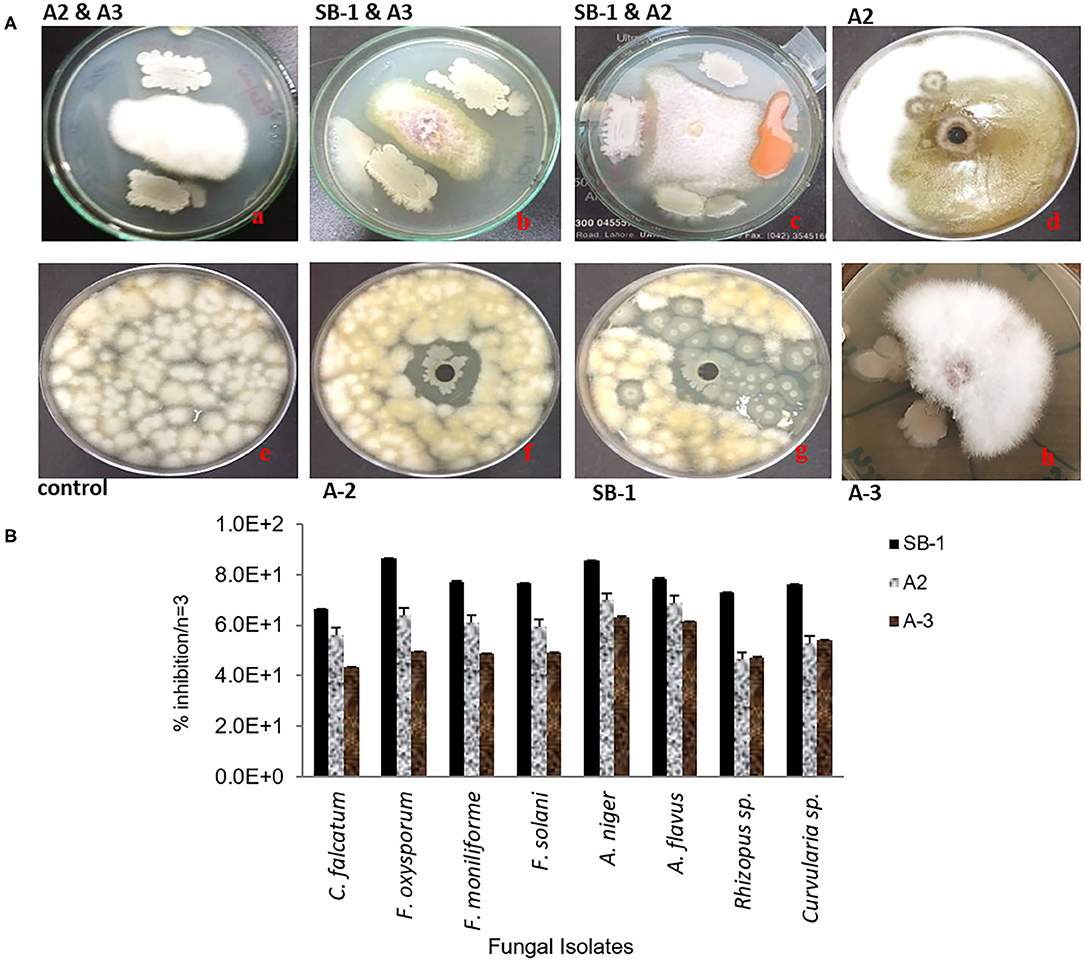
Figure 1. Antifungal dual plate-assay of Bacillus spp. strains with plant-pathogenic fungi; F. oxysporum, F. moniliforme, and F. solani. (A) percentage inhibition of fungal plant pathogens by Bacillus spp. with agar well-diffusion assay (B).
Bacterial isolates were biochemically characterized using QTS-24 bacterial identification kits (Supplementary Table 6). For 16S rRNA gene, sequence data were searched through NCBI BLAST. Results confirmed the strain SB-1 as B. amyloliquefaciens, and A-2 as B. subtilis as they showed 100% homology with 16S rRNA sequences of these strains reported in database. Whereas, strain A-3 exhibited 100%, homology with B. tequilensis strain (KU179328) and 99.79% homology with B. subtilis (KU179327). It was confirmed as B. tequilensis due to 99.86 and 99.79% homologies with other reported 16S rRNA sequences of B. tequilensis in database (Supplementary Table 7). Phylogenetic tree based on 16S rRNA gene sequences showed the homology of these strains with closely related strains (Supplementary Figure 2). Sequence lengths, accession numbers, and percent homology of each strain are summarized in Supplementary Table 3. There were a total of 1,388 positions in the final dataset of SB-1, 1,387 of A-2 and, 1,362 of A-3. Sequences were deposited in the GenBank database and accession numbers were obtained (SB-1; MF171193, A-2; MF574398, A-3; MF574399).
Secondary metabolites produced by Bacillus strains were first detected by LC/ESI-FTMS in the full-mass scan mode (Figures 3A,B). The first set of peaks observed belong to the surfactin cyclic-lipopeptide (CLPs) family, where strong signals corresponding m/z [M+H]+ were recorded for surfactin C12-C17, i.e., C12 m/z [M+H]+ = 994.644, C13 m/z [M+H]+ = 1008.661, C14 m/z [M+H]+ = 1022.676, C15 m/z [M+H]+ = 1036.692, C16 m/z [M+H]+ = 1050.708, C17 m/z [M+H]+ = 1086.704, produced by all Bacillus strains (Figure 2, Supplementary Figures 3, 5A–K, Supplementary Table 8). Weak signals for sodiated ions [M+Na]+ and deprotonated molecules [M-H]− of surfactin variants were also recorded. Similarly, peaks for iturin homologs were apparent in both positive and negative ion-modes (Supplementary Figure 3). ESI-MS showed strong signals for iturin D m/z [M+H]+ = 1043.553, iturin C m/z [M+H]+ = 1044.536, and iturin A1 m/z [M+H]+ = 1029.538 from B. amyloliquefaciens strain SB-1 (Figure 2, Supplementary Figures 6A–8B, Supplementary Table 8). The molecular weights of these iturin homologs and their variants were in agreement with iturin structures confimring the acyl chains of C14, C15, C16, and C17. Other than B. amyloliquefaciens strain SB-1, B. subtilis A-2, and B. tequilensis A-3 also demonstrated strong signals for iturin D. However, only weak [M+H]+ signals for iturin A1 and iturin C were recorded for these two strains whereas no [M-H]− peaks for iturin A1 and iturin C were observed in the extracts of strains A-2 and A-3. Next family of CLPs observed was of fengycin where strong signals corresponding to fengycin m/z [M+H]+ = 1463.804 and fengycin A m/z [M+H]+ = 1477.820 were observed in all three bacterial extracts (Figure 2, Supplementary Figures 9A–10B, Supplementary Table 8). Moreover, equally strong sodiated ions [M+Na]+ i.e., fengycin m/z [M+Na]+ = 1485.62 and fengycin A m/z [M+Na]+ = 1499.65 were also noted for this class.
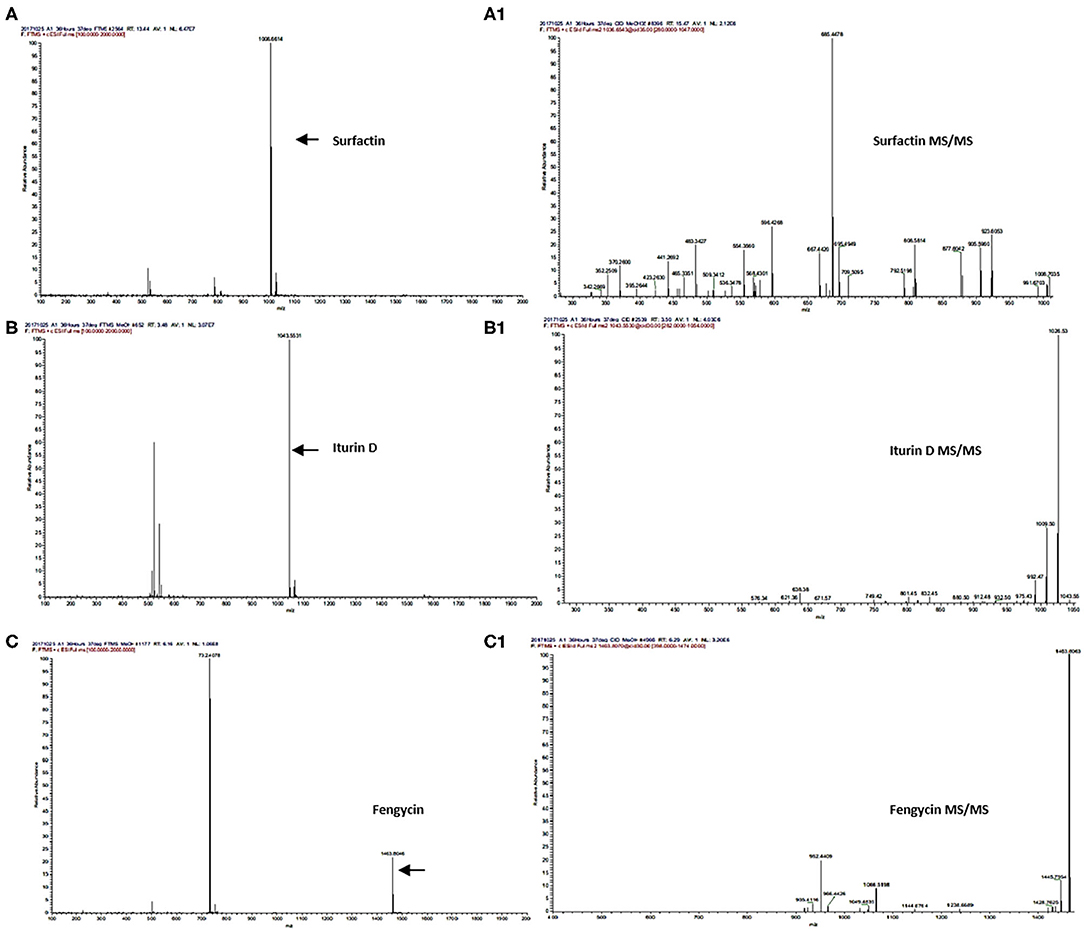
Figure 2. UPLC-MS/MS chromatograms of surfactin (A,A1), iturin (B,B1), and fengycin (C,C1), molecular ion-species detected in Bacillus spp. in positive ion-mode.
Next strong signals observed in sugarcane endophyte B. amyloliquefaciens strain SB-1 corresponded to iturin-like antifungal polypeptide bacillomycin D m/z [M+H]+ = 989.494. Its sodiated ions were not observed, however, weak signals of deprotonated ions were recorded (Supplementary Figure 11). Production of bacillibactin, a catechol-based siderophore, was observed in all three Bacillus strains. Peaks corresponding to m/z [M+H]+ = 883.262 at retention time of 1.52 were detected in bacterial extracts giving the exact fragmentation pattern of daughter ions, confirming the bacillibactin production (Figure 2, Supplementary Figures 12A,B, Supplementary Table 8). Next two molecular ions detected belonged to bacillaene m/z [M+H]+ = 581.358, and bacilysocin m/z [M+H]+ = 471.271, both of which are polyene and phospholipid antibiotics (Figure 2, Supplementary Figures 13A–14B, Supplementary Table 8). Strong signals for bacilysocin were detected in all three strains however, weak signals for bacillaene were observed in strains A-2 and A-3. Besides these, signals for lactone ring harboring macrolide compounds i.e., macrolactin A m/z [M+H]+ = 403.247, macrolactin E m/z [M+H]+ = 401.232, and macrolactin U m/z [M+H]+ = 481.331 were also noted and literature demonstrates their characteristic production from Bacillus spp. (Figure 2, Supplementary Figures 15A–F, Supplementary Table 8). The MS2 fragmentation showed the exact pattern of daugter ions as reported in literature and identified molecules were structurally confirmed by LC-MS/MS. The MS/MS spectra obtained from ion fragmentation in HCD and CID mode were searched against the available MS/MS databases including CFM-ID (Allen et al., 2014), METLIN (Smith et al., 2005), and GNPS (Wang et al., 2016). The MS/MS spectra were also compared with previously published spectra in the literature, especially for those compounds still not entered into any database. Chemical formulas, monoisotopic neutral masses, observed m/z values, and retention times of these compounds have been described in Table 2.
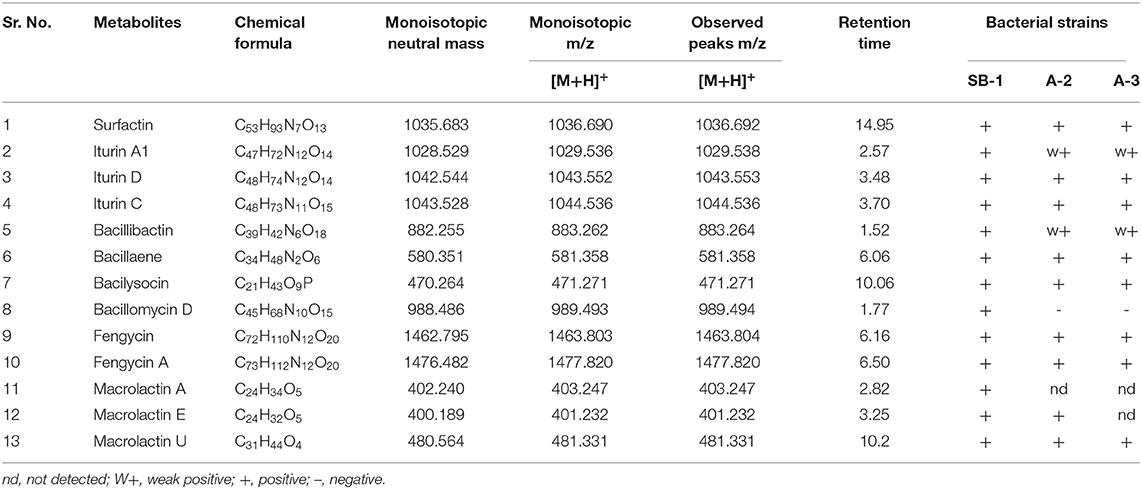
Table 2. Chemical formulas, monoisotopic neutral mass, m/z and observed peaks of detected metabolites.
All the extracts were analyzed in triplicates following the normalization of peak areas. Average of peak area values were plotted to visualize the comparative amounts of secondary metabolites in three different Bacillus strains. Surfactin homologs were abundantly produced by all Bacillus strains and amongst them, surfactin C13 m/z [M+H]+ = 1008.661, and C15 m/z [M+H]+ = 1036.692 were observed in high amounts followed by surfactin C14 m/z [M+H]+ = 1022.676 (Figures 3C–E). Minimum values were observed for surfactin C17 m/z [M+H]+ = 1086.704 which contributed 0.2–1% of the total surfactins produced by three strains. Likewise, maximum production of iturin variants was recorded for the strain B. amyloliquefaciens SB-1. The Highest amounts were noted for iturin C followed by iturin D in all three Bacillus strains, however, very minute amount of iturin A1 was detected for B. subtilis A-2 and B. tequilensis A-3 (Figure 3F).
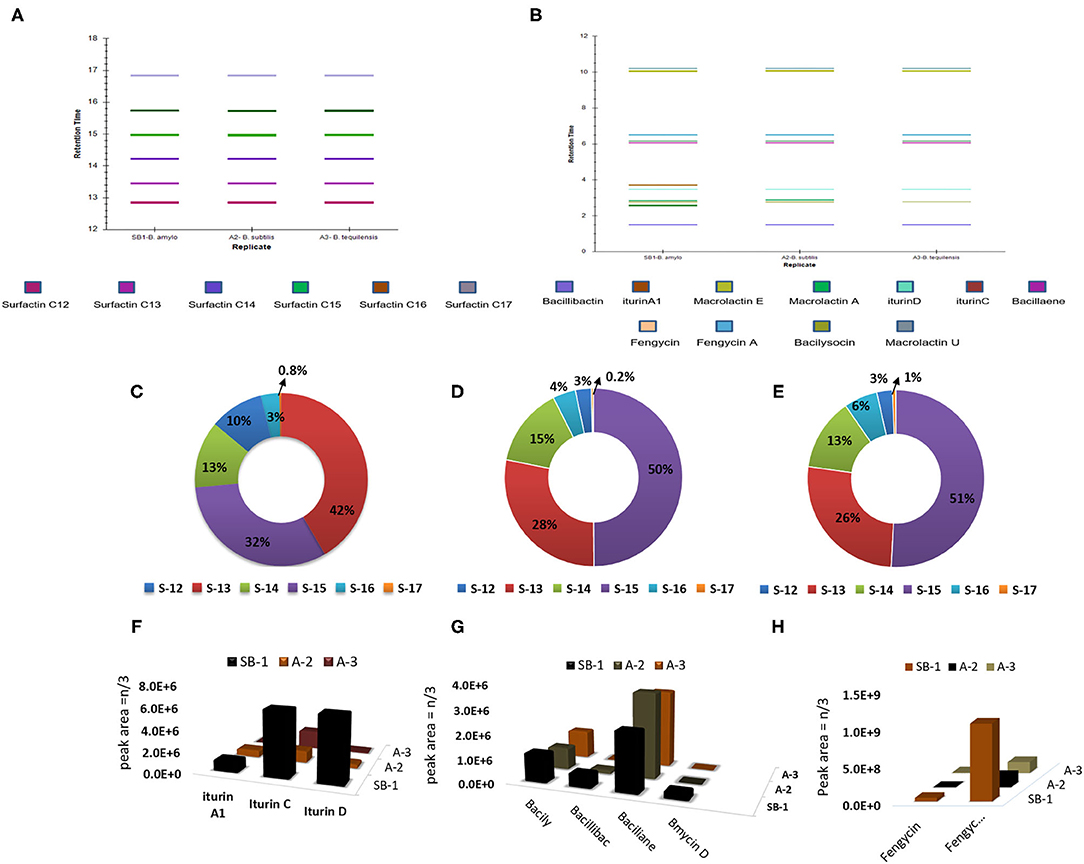
Figure 3. Retention times of bioactive metabolites produced by Bacillus strains SB-1, A2, and A3 (A,B), percentage of various surfactin homologs in B. amyloliquefaciens SB-1 (C), B. subtilis A-2, (D), and B. tequilensis A-3, (E), and amount of various antimicrobials detected in Bacillus spp. strains (F–H).
Amongst other metabolites, all three Bacillus strains produced large amounts of polyene antibiotic bacillaene. Its maximum amount was observed in B. subtilis followed by B. tequilensis A-3. Moreover, all three strains produced phospholipid-based antibiotic bacilysocin and its production was prominently noted in strains B. amyloliquefaciens SB-1 and B. tequilensis A-3. Although the production of catechol-based siderophores was noted in all Bacillus strains, however, its maximum amount was seen in B. amyloliquefaciens SB-1. Production of Iturin-like polypeptide bacillomycin D was nevertheless, variable. Its production was seen only in sugarcane endophyte B. amyloliquefaciens SB-1 and no peak was recorded with the same mass in other two strains (Figure 3G). Bacillus spp. were also compared for the production of cyclic-lipopeptide fengycins and demonstrated high values for fengycin A production. Maximum amounts of fengycin and fengycin A were seen by the strain SB-1 B. amyloliquefaciens followed by strains A-3 and A-2 (Figure 3H).
Gene sfp has been reported as the marker for identification of surfactin production by Bacillus spp. PCR amplification of sfp gene (675 bp) was shown by all three strains. Similarly, ituD gene amplification (1,200 bp) was also noted in all three Bacillus spp. which is the characteristic biomarker for iturin A production and encodes a putative malonyl coenzymeA transacylase. Amplification of 300 bp fragment with fengycin primers confirmed that all three Bacillus strains harbor the gene for production of fengycin lipopeptide. However, bacillomycin D showed the amplification of 375 bp only in B. amyloliquefaciens SB-1 (Figures 4A–D). Moreover, peaks corresponding to bacillomycin D m/z [M+H]+ = 989.494 were only seen in SB-1 confirming that strain A-2 and A-3 do not produce this iturin-like polypeptide. Sequencing of PCR products were searched through BLAST search and confimred homologies with respective gene sequences reported in database.
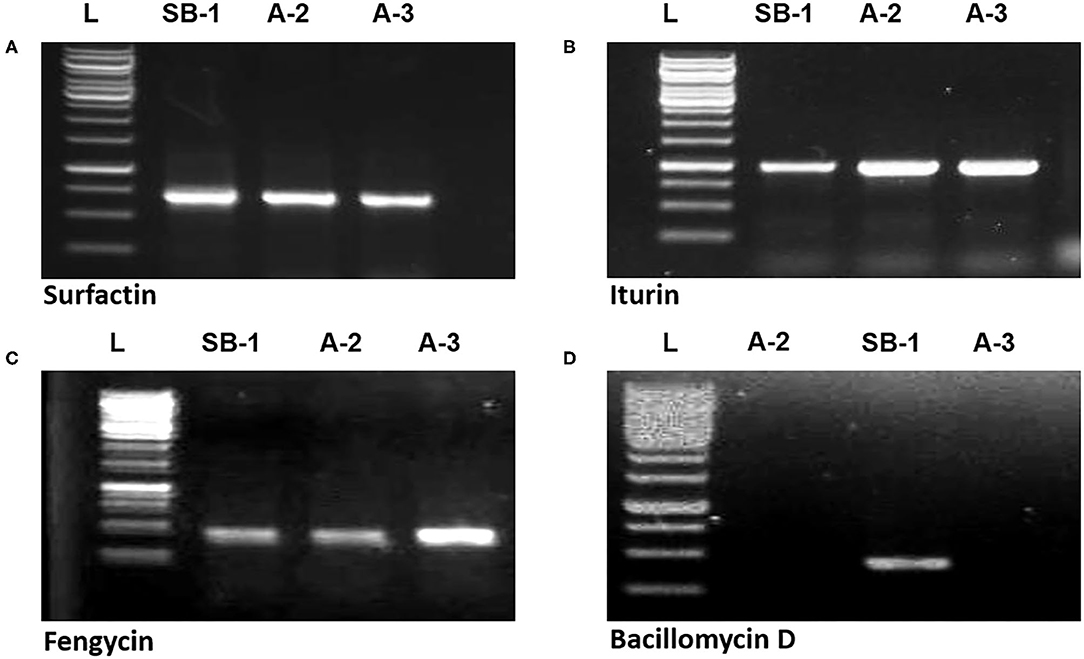
Figure 4. Amplification of sfp (A), ituD (B), fen (C), and bmy (D) genes of Bacillus spp. strains, L represents 1 kb DNA ladder.
All three Bacillus strains produced plant-growth promoting auxin, indole-3-acetic acid. Highest amount of IAA, i.e., 0.147 μM was produced by B. amyloliquefaciens SB-1 followed by B. tequilensis A-3 which produced 0.132 μM. Least amounts of IAA were noted by B. subtilis strain A-2 which produced 0.067 μM of IAA. Furthermore, production of extracellular hydrolytic enzymes including protease, lipase, and cellulase was noted by all three Bacillus spp. in plate bioassays. None of the strains was positive for volatile HCN production, whereas, all three strains produced catechol-type siderophores in plate bioassays. When checked for insoluble mineral solubilization, only B. amyloliquefaciens SB-1 solubilized tris-minimal agar medium supplemented with ZnCO3 (2.1 ± 0.18 SI) and Pikovskya's agar, i.e., 23.2 ± 0.21 μg/mL (Supplementary Table 9).
Significant effects of Bacillus spp. bioinoculants were noted on the growth of wheat plants. Results manifested that bacterial inoculation stimulated plant growth as compared to the un-inoculated controls. Plants inoculated with B. amyloliquefaciens SB-1 demonstrated maximum shoot lengths followed by B. tequilensis A-3, and B. subtilis A-2, respectively. Maximum root lengths were however noted for B. subtilis A-2 inoculated plants followed by B. tequilensis A-3 (Figures 5A,B). When compared for dry weights of shoot and root, most significant difference was recorded for the B. amyloliquefaciens SB-1 inoculated plants followed by B. subtilis A-2. Likewise, B. amyloliquefaciens SB-1 substantially increased the overall plant biomass (~42%) as compared to uninoculated controls. B. subtilis A-2 inoculated plants were next in the que that showed considerable increase in overall plant biomass (~31%). Although all the inoculated plants demonstrated more spikes in comparison to uninoculated ones, however, B. amyloliquefaciens SB-1 inoculated plants were notably significant (Figures 5C,D). Overall results suggested B. amyloliquefaciens SB-1 as the most successful bioinoculum for the wheat.
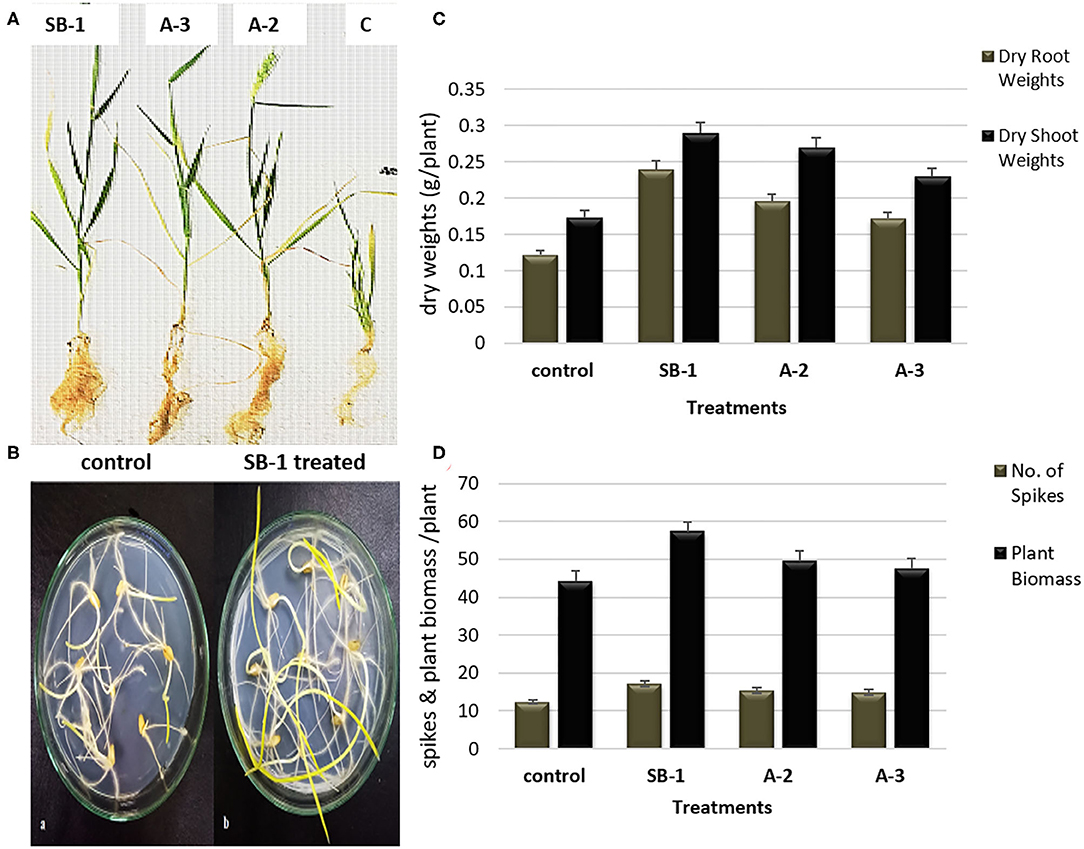
Figure 5. Effect of Bacillus spp. inoculation on shoots and roots growth (A), comparison of un-inoculated control and B. amyloliquefaciens SB-1 treated seedlings in 1% water-agar plates (B), effect of Bacillus spp. inoculation on dry weight of shoot and root (C), and plant biomass, and number of spikes of wheat plants (D).
Bacillus spp. are omnipresent in nature and constitute a major microbial community of rhizomicrobiome. Root-colonizing Bacillus spp. have been excessively characterized for their range of agricultural, environmental, and industrial applications and are regarded as eco-friendly substitutes of chemical fertilizers (Cardinale et al., 2015). Besides possessing diverse plant growth-stimulating factors, Bacillus spp. are also bestowed with the immense capacity of producing antimicrobial compounds active against broad range of plant-pathogens. Among these antimicrobials, the most significant are cyclic-lipopeptides which make them versatile tool for phytopathogenic biocontrol (Santoyo et al., 2012; Smith et al., 2017).
This study describes the isolation and polyphasic characterization of three antagonistic Bacillus spp. from different plant sources. Based on 16S rRNA sequencing, these strains were characterized as B. amyloliquefaciens SB-1, B. subtilis A-2, and B. tequilensis A-3. Strain SB-1 showed 100% homology with reported database strains of B. amyloliquefaciens. Likewise, rice rhizosphere isolate B. subtilis A-2 demonstrated 100% homology with B. subtilis KX692273, and 99.93% homology with other reported B. subtilis strains accession nos.; KU551205, AB735984, and AY775778. Corn isolate A-3 however, showed 100% homology with one B. tequilensis strain (accession no. MF417796) and exhibited 99.79% similarity with two other reported strains of B. tequilensis (accession no. KU179328, KU179329). Additionally, this strain showed 99.86% homology with B. subtilis strain (accession no. KU179327). Close homologies with two different species can be further resolved with additional biomarkers including DNA-DNA based hybridization. These three Bacillus strains demonstrated broad-spectrum in-vitro antifungal activities against eight phyto-pathogens including F. oxysporum, F. solani, F. moniliforme, C. falcatum, A. niger, A. flavus, Curvularia sp. and Rhizopus sp. Among these, maximum antagonism was observed by B. amyloliquefaciens SB-1 showing >85% inhibition of F. oxysporum and ~60–70% suppression of other fungal pathogens tested. Previous in-vitro studies have shown the ability of Bacillus spp. in inhibiting the growth of several fungal pathogens. For instance, B. subtilis was evaluated for the biocontrol of C. gloeosporioides OGC causing anthracnose disease of chili (Luna-Bulbarela et al., 2018). Similarly, B. subtilis strain BPR7 isolated from common bean rhizosphere demonstrated antagonsim against several plant-pathogenic fungi including M. phaseolina, F. oxysporum, F. solani, S. sclerotiorum, and Colletotrichum sp. (Kumar et al., 2012). Likewise, B. amyloliquefaciens CNU114001 showed effective biocontrol of sclerotinia rot of cucumber fruit (Ji et al., 2013). However, biocontrol potential of B. tequilensis have been explored in recent years and one of the latest research studies evaluated the efficacy of B. tequilensis strain MML2476 in biocontrol of rhizome-rot disease of turmeric (Chenniappan et al., 2019).
The underlying mechanism of biocontrol of plant diseases (caused by aerial and soil-borne fungi), by Bacillus spp. is usually attributed to the production of various antibiotics by this genus. Suppression of plant-pathogenic fungi by B. amyloliquefaciens SB-1, B. subtilis A-2, and B. tequilensis A-3 suggested their secondary metabolites as potential biocontrol agents. When subjected to detailed ESI-LC-MS/MS analysis, all three Bacillus spp. strains showed the production of three main classes of cyclic-peptides (CLPs) including surfactins, iturins, and fengycins. These cyclic-lipopetide bioactive metabolites have been well-evaluated for their biocontrol and pharmaceutical applications. Iturins constitute the group of iturin A~E, bacillomycins, and mycosubtilins which induct ion-conducting pores in the membranes of growing pathogens and damage mycelia (Zhang et al., 2013). Surfactins however, act by membrane disruption and solubilization and considered as the strong bio-surfactants. Many studies illustrate the synergistic effects of surfactins on broadening the biological properties of iturins and where two of them act in a coordinated manner (Fan et al., 2017). Similarly, fengycins have been demonstrated as key factors in antagonsim of several Bacillus spp. which together with surfactins can elicit induced systemic resistance (ISR) in plants (Fira et al., 2018). Such as, these CLPs were characterized as the major functional bioactive metabolites of B. amyloliquefaciens C06 toward suppression of M. fructicola (Liu et al., 2011). Likewise, CLPs from B. amyloliquefaciens DH-4 were identified as the main antagonizing agents against citrus green mold P. digitatum (Chen et al., 2018). UV-MALDI-TOF-MS analysis of bioactive compounds of B. subtilis subsp. subtilis PGPMori7 revealed iturins, fengycins, and surfactins in triggering the immunity and microbial competition against M. phaseolina (Torres et al., 2016). Recently, genomic insights into the endophytic B. tequilensis 7PJ-16 strain displayed its biocontrol potential against mulberry fruit sclerotiniose (Xu et al., 2019).
The strains B. amyloliquefaciens SB-1, B. subtilis A-2, and B. tequilensis A-3 used in this study, were able to produce various homologs of surfactins, iturins, and fengycin compounds. Well-ionized peaks for surfactin C12-C17, m/z 994.644–1086.704, iturin A1, D, and C, and fengycin were noted in positive ion mode in all strains. Maximum amounts of these CLPs were however noted for the strain B. amyloliquefaciens SB-1 as compared to other two Bacillus strains. These homologs were further confirmed through ESI-MS/MS and results were in accordance with previously published mass-spectrometry data for these compounds. Furthermore, the fragmentation patterns of daughter ions were also confirmed by latest databases CFM-ID and GNPS (Smith et al., 2005; Allen et al., 2014). Moreover, amplification of biosynthetic genes responsible for the synthesis of surfactins, iturins and fengycins confirmed the production of these metabolites by the strains under investigation. All strains showed 675 bp amplification for sfp gene coding for surfactin, 1.2 kb fragment for iturin D, and 300 bp fragment for fenD. Following amplification, PCR products were sequenced and compared with reported sequences in database.
Surfactins from B. subtilis has been evaluated for its role in root-colonization and protection against plant-pathogen namely P. syringae in Arabidopsis thaliana (Jourdan et al., 2009) and are known for their potential role in initiation of systemic resistance and stimulation of plant-defense (Bais et al., 2004; Hsieh et al., 2004). Likewise, iturin cyclic-peptides are strong antifungal CLPs produced by many of Bacillus strains and act by forming small pores in the membranes of the encountering phytopathogens (Zohora et al., 2016; Zhao et al., 2018). Iturins from B. amyloliquefaciens and B. subtilis were evaluated for effectively managing anthracnose disease of chili caused by C. gloeosporioides OGC1 (Ashwini and Srividya, 2014). Similarly, fengycin family homologs have been characterized for suppressing the growth of filamentous fungi by inducing reactive oxygen species (ROS) production and chromatin condensation (Zhang and Sun, 2018). Moreover, Romero et al. (2007) investigated the contribution of iturins and fengycins from four B. subtilis strains UMAF6614, UMAF6616, UMAF6639, and UMAF8561 in the suppression of powdery mildew of cucurbits caused by Podosphaera fusca.
Furthermore, iturin-like antifungal peptide, bacillomycin D was only detected in B. amyloliquefaciens SB-1 and its PCR analysis also revealed the 375 bp product coding for bacilliomycin gene. The result was in accordance with LC-MS/MS results which showed the characteristic peak corresponding to bacillomycin m/z [M+H] = 989.494 in the extracts of strain B. amyloliquefaciens SB-1 only. Previous research studies manifest the production of bacillomycin D from B. amyloliquefaciens and its contribution in inhibiting spore germination. For instance, B. amyloliquefaciens strain 83 was extensively investigated for its potential to produce certain bacillomycin D homologs effectively inhibiting fungal spores and mycelia (Luna-Bulbarela et al., 2018). Detailed ESI-LC-MS/MS analysis of Bacillus strains also showed the production of a phospholipid- antibiotic, bacilysocin by all strains. Bacilysocin was characteristically reported from B. subtilis 168 and was highlighted for its potential competition against other microorganisms during spore germination (Tamehiro et al., 2002). However, genomic insights into the operational group B. amyloliquefaciens illustrate the presence of bacilysocin, iron-siderophore bacillibactin, and bacillaene polyene in several strains of B. amyloliquefaciens, B. velezensis, and B. siamensis, characteritically produced by strains SB-1, A-2, and A-3 in this study (Fan et al., 2017).
Macrolactins produced by Bacillus spp. have been demonstrated for their efficient antibacterial properties and were recently found in reshaping the soil bacterial communities (Yuan et al., 2016). Production of macrolides A, E, and U was also noticed by the strains used in this study. B. amyloliquefaciens SB-1 showed the presence of all three macrolactin homologs whereas, B. subtilis A-2 showed the presence of two macrolactins; E and U. B. tequilensis A-3, however, showed the production of only macrolactin U which was further confirmed for its daughter ion fragmentation by MS/MS.
In addition to be efficient biocontrol agents, these strains also manifested to be strong biofertilizer candidates. When screening for plant growth-promoting traits, all strains produced IAA (0.067–0.147 μM), lipase, cellulase, protease, and catechole-based siderophore in-vitro. Several studies have shown the production these hydrolytic enzymes and siderophores by Bacillus spp. (Sayyed et al., 2004; Shaikh and Sayyed, 2014; Jadhav and Sayyed, 2016; Shaikh et al., 2018). Zinc and phosphorous solubilization was however exclusive to B. amyloliquefaciens SB-1 which solubilized 23.2 ± 0.21 μg/mL of insoluble phosphorus and zinc carbonate (SI = 2.1 ± 0.18).
To be used as biofertilizers, the in-vivo impact of Bacillus spp. inoculation was also investigated in this study on the growth of wheat plants. Substantial increase in the overall wheat biomass, dry roots and shoots weight, and number of spikes was noted for the plants inoculated with Bacillus bio-inoculants as compared to un-inoculated controls. The most significant results were shown by B. amyloliquefaciens SB-1 as bioinoculant over un-inoculated control plants which caused ~42% increase in the total biomass.
Conclusively, detailed physiological, genetic and metabolomic analyses of three Bacillus spp. strains expolored their ability as biocontrol and biofertilizer agents. Comprehensive analysis of the secondary metabolites of these strains provided new insights about their adaptation to diverse environmental niches and their strain-specific metabolites. Among these, sugarcane endophyte B. amyloliquefaciens SB-1 showed ideal plant growth-promoting and biocontrol abilities and demonstrated all the attributes to be employed as user-friendly, single-strain inoculum. Presumbly, its ability to solubilize phosphorous and zinc can convert more elements into the bioavailable forms to stimulate plant growth and yield. Furthermore, in future, different consortia and single-strains bioinoculum of these plant growth stimualting Bacillus spp. can be used for other crops for the competitive suppression of phytopathogens and as biofertilizer agents.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
IS performed experiments and wrote the manuscript. JH and CB conceptualized the idea. SH helped to prepare illustrations. KM provided workspace. SM edited the manuscript. All authors contributed to the article and approved the submitted version.
This work received financial support from Higher Education Commission (HEC) of Pakistan (Grant No. 1-8/HEC/HRD/2017/7332). The funding was awarded to IS for Ph.D. research work. The metabolomics research at the UVic-Genome BC Proteomics Center was supported by funding to The Metabolomics Innovation Center (TMIC) through the Genome Innovations Network (GIN) from Genome Canada, Genome Alberta, and Genome British Columbia for operations (205MET and 7203) and for technology development (215MET and MC3T) in metabolomics.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors would like to thank Dr. Carol E. Parker for assistance with English language editing.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fsufs.2021.605195/full#supplementary-material
Agler, M. T., Ruhe, J., Kroll, S., Morhenn, C., Kim, S. T., and Weigel, D. (2016). Microbial hub taxa link host and abiotic factors to plant microbiome variation. PLoS Biol. 14:e1002352. doi: 10.1371/journal.pbio.1002352
Ahemad, M., and Kibret, M. (2014). Mechanisms and applications of plant growth promoting rhizobacteria: current perspective. J. King Saud Univ. Sci. 26, 1–20. doi: 10.1016/j.jksus.2013.05.001
Aira, M., Gómez-Brandón, M., Lazcano, C., Baath, E., and Domínguez, J. (2010). Plant genotype strongly modifies the structure and growth of maize rhizosphere microbial communities. Soil Biol. Biochem. 42, 2276–2281 doi: 10.1016/j.soilbio.2010.08.029
Allen, F., Pon, A., Wilson, M., Greiner, R., and Wishart, D. (2014). CFM-ID: a web server for annotation, spectrum prediction and metabolite identification from tandem mass spectra. Nucleic Acid Res. 42, W94–W99. doi: 10.1093/nar/gku436
Alnahdi, H. S. (2012). Isolation and screening of extracellular proteases produced by new isolated Bacillus sp. J. Appl. Pharm. Sci. 2, 71–74. doi: 10.7324/JAPS.2012.2915
Ashwini, N., and Srividya, S. (2014). Potentiality of Bacillus subtilis as biocontrol agent for management of anthracnose disease of chilli caused by Colletotrichum gloeosporioides OGC1. 3 Biotech. 4, 127–136. doi: 10.1007/s13205-013-0134-4
Bais, H. P., Park, S. W., Weir, T. L., Callaway, R. M., and Vivanco, J. M. (2004). How plants communicate using the underground information superhighway. Trends Plant Sci. 9, 26–32. doi: 10.1016/j.tplants.2003.11.008
Bhardwaj, D., Ansari, M. W., Sahoo, R. K., and Tuteja, N. (2014). Biofertilizers function as key player in sustainable agriculture by improving soil fertility, plant tolerance and crop productivity. Microb. Cell Fact. 13:66. doi: 10.1186/1475-2859-13-66
Cardinale, M., Ratering, S., Suarez, C., Zapata, M. A. M., Geissler-Plaum, R., and Schnell, S. (2015). Paradox of plant growth promotion potential of rhizobacteria and their actual promotion effect on growth of barley (Hordeumvulgare L.) under salt stress. Microbiol. Res. 181, 22–32. doi: 10.1016/j.micres.2015.08.002
Carvalhais, L. C., Dennis, P. G., Fan, B., Fedoseyenko, D., Kierul, K., Becker, A., et al. (2013). Linking plant nutritional status to plant-microbe interactions. PLoS ONE 8:e68555. doi: 10.1371/journal.pone.0068555
Chen, K., Tian, Z., Luo, Y., Cheng, Y., and Long, C. A. (2018). Antagonistic activity and the mechanism of Bacillus amyloliquefaciens DH-4 against citrus green mold. Phytopathol 108, 1253–1262. doi: 10.1094/PHYTO-01-17-0032-R
Chenniappan, C., Narayanasamy, M., Daniel, G. M., Ramaraj, G. B., Ponnusamy, P., Sekar, J., et al. (2019). Biocontrol efficiency of native plant growth promoting rhizobacteria against rhizome rot disease of turmeric. Biol. Control 129, 55–64 doi: 10.1016/j.biocontrol.2018.07.002
Falcäo, L., Silva-Werneck, J., Vilarinho, B., da Silva, J., Pomella, A., and Marcellino, L. (2014). Antimicrobial and plant growth-promoting properties of the cacao endophyte Bacillus subtilis ALB629. J. Appl. Microbiol. 116, 1584–1592. doi: 10.1111/jam.12485
Fan, B., Blom, J., Hans-Peter, K., and Rainer, B. (2017). Bacillus amyloliquefaciens, Bacillus velezensis, and Bacillus siamensis form an “Operational Group B. amyloliquefaciens” within the B. subtilis species complex. Front. Microbiol. 8:22. doi: 10.3389/fmicb.2017.00022
Fira, D., Dimkić, I., Berić, T., Lozo, J., and Stanković, S. (2018). Biological control of plant pathogens by Bacillus species. J. Biotechnol. 285, 44–55. doi: 10.1016/j.jbiotec.2018.07.044
Hayat, R., Ali, S., Amara, U., Khalid, R., and Ahmed, I. (2010). Soil beneficial bacteria and their role in plant growth promotion: a review. Ann. Microbiol. 60, 579–598. doi: 10.1007/s13213-010-0117-1
Hoagland, D. R., and Arnon, D. I. (1970). The Water-Culture Method for Growing Plants without Soil. Berkley, CA: California Agricultural Experimental Station Publications and College of Agriculture, University of California.
Hsieh, F. C., Li, M. C., and Lin, T. C. (2004). Rapid detection and characterization of surfactin-producing Bacillus subtilis and closely related species based on PCR. Curr. Microbiol. 49, 186–191. doi: 10.1007/s00284-004-4314-7
Jadhav, H. P., and Sayyed, R. Z. (2016). Hydrolytic enzymes of rhizospheric microbes in crop protection. MOJ Cell Sci. Rep. 3, 135–136. doi: 10.15406/mojcsr.2016.03.00070
Ji, S. H., Paul, N. C., Deng, J. X., Kim, Y. S., Yun, B. S., and Yu, S. H. (2013). Biocontrol activity of Bacillus amyloliquefaciens CNU114001 against fungal plant diseases. Mycobiology 41, 234–242. doi: 10.5941/MYCO.2013.41.4.234
Jourdan, E., Henry, G., Duby, F., Dommes, J., Barthélemy, J. P., Thonart, P., et al. (2009). Insights into the defense-related events occurring in plant cells following perception of surfactin-type lipopeptide from Bacillus subtilis. Mol. Plant Microbe Interact. 22, 456–468. doi: 10.1094/MPMI-22-4-0456
Kasana, R. C., Salwan, R., Dhar, H., Dutt, S., and Gulatti, A. (2008). A rapid and easy method for the detection of microbial cellulases on agar plates using gram's iodine. Curr. Microbiol. 57, 503–507. doi: 10.1007/s00284-008-9276-8
Kefi, A., Ben, S. I., Karkouch, I., Rihouey, C., Azaeiz, S., Bejaoui, M., et al. (2015). Characterization of endophytic Bacillus strains from tomato plants (Lycopersicon esculentum) displaying antifungal activity against Botrytis cinerea Pers. W. J. Microbiol. Biotechnol. 31, 1967–1976. doi: 10.1007/s11274-015-1943-x
Kim, P. I., Ryu, J., Kim, Y. H., and Chi, Y. T. (2010). Production of biosurfactant lipopeptides iturin A, fengycin and surfactin A from Bacillus subtilis CMB32 for control of Colletotrichum gloeosporioides. J. Microbiol. Biotechnol. 20, 138–145. doi: 10.4014/jmb.0905.05007
Kumar, P., Dubey, R. C., and Maheshwari, D. K. (2012). Bacillus strains isolated from rhizosphere showed plant growth promoting and antagonistic activity against phytopathogens. Microbiol. Res. 167, 493–499. doi: 10.1016/j.micres.2012.05.002
Liu, J., Zhou, T., He, D., Li, X. Z., Wu, H., Liu, W., et al. (2011). Functions of lipopeptides bacillomycin D and fengycin in antagonism of Bacillus amyloliquefaciens C06 towards Monilinia fructicola. J. Mol. Microbiol. Biotechnol. 20, 43–52. doi: 10.1159/000323501
Louden, B. C., Haarmann, D., and Lynne, A. M. (2011). Use of blue agar CAS assay for siderophore detection. JMBE 12, 51–53. doi: 10.1128/jmbe.v12i1.249
Luna-Bulbarela, G., Tinoco-Valencia, R., Corzo, G., Kazuma, K., Konno, K., Galindo, E., et al. (2018). Effects of bacillomycin D homologues produced by Bacillus amyloliquefaciens 83 on growth and viability of Colletotrichum gloeosporioides at different physiological stages. Biol. Control 127, 145–154. doi: 10.1016/j.biocontrol.2018.08.004
Magaldi, S., Mata-Essayag, S., and Hartungde, C. (2004). Well diffusion for antifungal susceptibility testing. Int. J. Infect. Dis. 8, 39–45. doi: 10.1016/j.ijid.2003.03.002
Millar, R., and Higgins, V. J. (1970). Association of cyanide with infection of bird‘s foot trefoil by Stemphylium loti. Phytopathology 60, 104–110. doi: 10.1094/Phyto-60-104
Minaxi, L. N., Yadav, R. C., and Saxena, J. (2012). Characterization of multifaceted Bacillus sp. RM-2 for its use as plant growth promoting bioinoculant for crops grown in semi arid deserts. Appl. Soil Ecol. 59, 124–135. doi: 10.1016/j.apsoil.2011.08.001
Normand, P. (1995). Utilisation des séquences 16S pour le positionnement phylétique d'un organisme inconnu. Oceanis 21, 31–56.
Pikovskaya, R. (1948). Mobilization of phosphorus in soil in connection with vital activity of some microbial species. Mikrobiologiya 17:e370.
Romero, D., de-Vicente, A., Rakotoaly, R. H., Dufour, S. E., Veening, J. W., Arrebola, E., et al. (2007). The iturin and fengycin families of lipopeptides are key factors in antagonism of Bacillus subtilis towards Podosphaera fusca. Mol. Plant Microbe Interact. 20, 430–440. doi: 10.1094/MPMI-20-4-0430
Saharan, B., and Nehra, V. (2011). Plant growth promoting rhizobacteria: a critical review. Life Sci. Med. Res. 21.
Sakthivel, N., and Gnanama-nickam, S. (1986). Toxicity of Pseudomonas fluorescens towards rice sheath -rot pathogen Acrocylindrium oryzae. Curr. Sci. 5, 106–107.
Santoyo, G., Orozco-Mosqueda, M. D., and Govindappa, M. (2012). Mechanisms of biocontrol and plant growth-promoting activity in soil bacterial species of Bacillus and Pseudomonas: a review. Biocont. Sci. Technol. 22, 855–872. doi: 10.1080/09583157.2012.694413
Sayyed, A. H., Omar, D., and Wright, D. J. (2004). Genetics of spinosad resistance in a multi-resistant field-selected population of Plutella xylostella. Pesticide Sci. 60, 827–832.
Shaikh, S. S., and Sayyed, R. (2014). “Role of plant growth-promoting rhizobacteria and their formulation in biocontrol of plant diseases,” in Plant Microbes Symbiosis: Applied Facets ed. N. K. Arora (New Delhi: Springer), 337–351. doi: 10.1007/978-81-322-2068-8_18
Shaikh, S., Wani, S., and Sayyed, R. (2018). Impact of interactions between rhizosphere and rhizobacteria. J. Bacteriol. Mycol. 5:1058.
Sharma, S. K., Sharma, M. P., Ramesh, A., and Joshi, O. P. (2012). Characterization of zinc solubilizing Bacillus isolates and their potential to influence zinc assimilation in soybean seeds. J. Microbiol. Biotechnol. 22, 352–359. doi: 10.4014/jmb.1106.05063
Shobana, N., Thangappan, S., and Uthandi, S. (2020). Plant growth-promoting Bacillus sp. cahoots moisture stress alleviation in rice genotypes by triggering antioxidant defense system. Microbiol. Res. 2:126518. doi: 10.1016/j.micres.2020.126518
Sierra, G. (1957). A simple method for the detection of lipolytic activity of micro-648 organisms and some observations on the influence of the contact between cells and fatty 649 acid substrates. Antonie Leeuwenhoek 23, 15–22. doi: 10.1007/BF02545855
Sinha, R. K., Valani, D., Chauhan, K., and Agarwal, S. (2014). Embarking on a second green revolution for sustainable agriculture by vermi-culture biotechnology using earthworms: reviving the dreams of Sir Charles Darwin. Int. J. Agric. Health Saf. 1, 50–64. doi: 10.5897/JABSD.9000017
Smith, C. A., O'Maille, G., Want, E. J., Qin, C., Trauger, S. A., Brandon, T. R., et al. (2005). METLIN: a metabolite mass spectral database. Ther. Drug Monit. 27, 747–751. doi: 10.1097/01.ftd.0000179845.53213.39
Smith, D. L., Gravel, V., and Yergeau, E. (2017). Editorial: signaling in the phytomicrobiome. Front. Plant Sci. 8:611. doi: 10.3389/fpls.2017.00611
Somasegaran, P., and Hoben, H. J. (1985). Methods in Legume-Rhizobium Technology, Paia. Maui: University of Hawaii NifTAL Project and MIRCEN, Department of Agronomy and Soil Science, Hawaii Institute of Tropical Agriculture and Human Resources, College of Tropical Agriculture and Human Resources. Hawaii: Springer. 367.
Souza, R., Meyer, J., Schoenfeld, R., Costa, P. B., and Passaglia, L. M. P. (2014). Characterization of plant growth-promoting bacteria associated with rice cropped in iron-stressed soils. Ann. Microbiol. 65, 951–964. doi: 10.1007/s13213-014-0939-3
Tamehiro, N., Okamoto-Hosoya, Y., Okamoto, S., Ubukata, M., Hamada, M., Naganawa, H., et al. (2002). Bacilysocin, a novel phospholipid antibiotic produced by Bacillus subtilis 168. Antimicrob. Agents Chemother. 46, 315–320. doi: 10.1128/AAC.46.2.315-320.2002
Tamura, K., Dudley, J., Nei, M., and Kumar, S. (2011). MEGA 5: molecular evolutionary genetics analysis using maximum likelihood evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739. doi: 10.1093/molbev/msr121
Torres, M. J., Pérez Brandan, C., Petroselli, G., Erra-Balsells, R., and Audisio, M. C. (2016). Antagonistic effects of Bacillus Subtilis subsp. subtilis and B. amyloliquefaciens against Macrophomina phaseolina: SEM study of fungal changes and UV-MALDI-TOF MS analysis of their bioactive compounds. Microbiol. Res. 182, 31–39. doi: 10.1016/j.micres.2015.09.005
Wang, M., Carver, J. J., Phelan, V. V., Sanchez, L. M., Garg, N., Peng, Y., et al. (2016). Sharing and community curation of mass spectrometry data with global natural products social molecular networking. Nat. Biotechnol. 34, 828–837. doi: 10.1038/nbt.3597
Xu, W., Ren, H., Ou, T., Lei, T., Wei, J. H., Huang, C. S., et al. (2019). Genomic and functional characterization of the endophytic Bacillus subtilis 7PJ-16 strain, a potential biocontrol agent of mulberry fruit sclerotiniose. Microb. Ecol. 77, 651–663. doi: 10.1007/s00248-018-1247-4
Yuan, J., Zhao, M., Li, R., Huang, Q., Rensing, C., Raza, W., et al. (2016). Antibacterial compounds-macrolactin alters the soil bacterial community and abundance of the gene encoding PKS. Front. Microbiol. 7:1904. doi: 10.3389/fmicb.2016.01904
Zhang, L., and Sun, C. (2018). Fengycins, cyclic lipopeptides from marine Bacillus subtilis strains, kill the plant-pathogenic fungus Magnaporthe grisea by inducing reactive oxygen species production and chromatin condensation. Appl. Environ. Microbiol. 84, e00445–e00418. doi: 10.1128/AEM.00445-18
Zhang, X., Li, B., Wang, Y., Guo, Q., Lu, X., Li, S., et al. (2013). Lipopeptides, a novel protein, and volatile compounds contribute to the antifungal activity of the biocontrol agent Bacillus atrophaeus CAB-1. Appl. Microbiol. Biotechnol. 97, 9525–9534. doi: 10.1007/s00253-013-5198-x
Zhao, H., Xu, X., Lei, S., Shao, D., Jiang, C., Shi, J., et al. (2018). Iturin A-like lipopeptides from Bacillus subtilis trigger apoptosis, paraptosis, and autophagy in Caco-2 cells. J. Cell Physiol. 234, 6414–6427. doi: 10.1002/jcp.27377
Keywords: biopriming, Bacillus amyloliquefaciens, cyclic-lipopeptides, mass spectrometry, siderophores, biofertilizer
Citation: Shahid I, Han J, Hanooq S, Malik KA, Borchers CH and Mehnaz S (2021) Profiling of Metabolites of Bacillus spp. and Their Application in Sustainable Plant Growth Promotion and Biocontrol. Front. Sustain. Food Syst. 5:605195. doi: 10.3389/fsufs.2021.605195
Received: 11 September 2020; Accepted: 25 January 2021;
Published: 22 February 2021.
Edited by:
Duraisamy Saravanakumar, The University of the West Indies St. Augustine, Trinidad and TobagoReviewed by:
M Loganathan, Indian Council of Agricultural Research (ICAR), IndiaCopyright © 2021 Shahid, Han, Hanooq, Malik, Borchers and Mehnaz. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Samina Mehnaz, c2FtaW5hbWVobmF6QGZjY29sbGVnZS5lZHUucGs=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.