- 1Department of Clinical Analysis, Federal University of Paraná (UFPR), Curitiba, Brazil
- 2Laboratory of Nitrogen Fixation, Biochemistry and Molecular Biology Department, Federal University of Paraná (UFPR), Curitiba, Brazil
Plant growth stimulation by microorganisms that interact in a mutually beneficial manner remains poorly understood. Understanding the nature of plant-bacteria interactions may open new routes for plant productivity enhancement, especially cereal crops consumed by humans. Proteomic and metabolomic analyses are particularly useful for elucidating these mechanisms. A complete depiction of these mechanisms will prompt researchers to develop more efficient plant-bacteria associations. The success of microorganisms as biofertilizers may replace the current massive use of chemical fertilizers, mitigating many environmental and economic issues. In this review, we discuss the recent advances and current state of the art in proteomics and metabolomics studies involving grass-bacteria associations. We also discuss essential subjects involved in the bacterial plant-growth promotion, such, nitrogen fixation, plant stress, defense responses, and siderophore production.
Introduction
World food demand is increasing due to population growth and dietary changes. From an estimated 7.7 billion people worldwide in 2019, was projected that the global population could grow to around 8.5 billion in 2030, 9.7 billion in 2050, and 10.9 billion in 2100 (United Nations, New York, 2019). Consequently, by 2050, annual world demand for maize (Zea mays L.), rice (Oryza sativa L.), and wheat (Triticum aestivum L.) is expected to reach 3.3 billion tons or 800 million tons more than 2014's combined harvest (Reeves et al., 2016). Cereals provide 42.5% of human dietary calories consumption (Reeves et al., 2016). To meet this crop production demand in the next decades, more nitrogen, phosphorus, and potassium, are expected to be applied to fields. Cereal agriculture, such as wheat, rice and maize, account for ~60% of the global fertilizer used, and they are expected to account for half of the fertilizer consumed in 2050 (Alexandratos and Bruinsma, 2012).
Applying fertilizers to the soil is accompanied by negative environmental impacts. The first problem is a loss of substantial levels of fertilizer, especially nitrogen fertilizer, through natural erosion or leaching soil, which may result in aquatic habitats eutrophication (Carpenter et al., 1998). Also, nitrogen oxide gases, such as the nitrous oxide (N2O), generated by the microbial transformation in the soil, contribute to the greenhouse effect (Hall et al., 1996). Industrial fertilizer production per se is an additional contributor to the “green problem.” Nitrogen fertilizers are produced in large-scale through the Haber–Bosch process, in which atmospheric N2 reacts with H2 or methane at very high pressure (200 atm) and temperature (450°C) over an iron catalyst yielding ammonia or urea. The electricity or natural gas consumed to generate ammonia by this process composes 90% of the total cost (Erisman et al., 2008). Furthermore, other fertilizers, such as phosphorus and potassium, are refined from rocks or sediment; however, the global reserves are susceptible to depletion (Galloway and Cowling, 2002; Erisman et al., 2008; Curatti and Rubio, 2014). As an alternative, sustainable agricultural policies have contributed alleviating the impact on the environment (Tilman et al., 2002).
Potential sustainable agriculture includes microbial inoculants or biofertilizers that aid in environmentally sustainable crop production (Johansson et al., 2004; Singh et al., 2011; Bashan et al., 2014; Bhardwaj et al., 2014; Backer et al., 2018). Plant growth-promoting rhizobacteria (PGPR) are those that beneficially interact with plants and promote their growth (Lugtenberg and Kamilova, 2009; Babalola, 2010; Dutta and Podile, 2010; Bhattacharyya and Jha, 2012; Fibach-Paldi et al., 2012; Vacheron et al., 2013; Nadeem et al., 2014). These bacteria can be found in the rhizosphere at the root surface or inside the plant. The bacteria that colonize inside of plants are commonly referred to as endophytic. Several species from the genera Pseudomonas, Azospirillum, Herbaspirillum, Azotobacter, Klebsiella, Burkholderia, Bacillus, Serratia, Rhizobium, and Bradyrhizobium, among others, can promote plant growth (Johansson et al., 2004; Singh et al., 2011; Vacheron et al., 2013; Nadeem et al., 2014; Fukami et al., 2018). Poaceae family, also called Gramineae or true grasses, are commonly associated with cyanobacteria, Azospirillum, Azotobacter, Gluconacetobacter, Azoarcus, Herbaspirillum, and Burkholderia, in associative or endophytic forms (Pérez-Montaño et al., 2014).
There was an increase of ~23% in total biomass and 65% in grain yield after inoculation of rice with PGPR (Tariq et al., 2007). Rice independently inoculated with Azospirillum brasilense R1, A. lipoferum RSWT1 and Pseudomonas sp. Ky1 exhibited increased grain weight. Significant increases in the total biomass of rice and sugarcane plants were also observed in plants inoculated with Herbaspirillum seropedicae (Boddey et al., 1995; Baldani et al., 2000; James, 2000; Gyaneshwar et al., 2002). Wheat and maize inoculated with a mix of A. brasilense Ab-V5 and Ab-V6 exhibited a 31 and 27% increased yield, respectively (Hungria et al., 2010). In a Cerrado Oxisol with a low capacity to supply N, the same mix increased maize grain yield in more than 30% (Martins et al., 2018). H. seropedicae and H. rubrisubalbicans are components of the commercial inoculant for sugarcane currently recommended by the Brazilian Agricultural Research Company (EMBRAPA) (Oliveira et al., 2006, 2009).
Interestingly, another Brazilian study showed that inoculation of sugar cane varieties grown in two different soils with a consortium of diazotrophic bacteria did not show significant differences concerning N fertilization, but concluded that the growth promotion was due to the positive environmental impact of biofertilizer (Schultz et al., 2017).
Despite the widespread use of biofertilizers in cereal crops, the molecular mechanisms are not fully understood. Omics techniques such as transcriptomics, metabolomics, and proteomics have considerable potential to discover novel mechanisms on plant-bacteria interaction due to the capacity to analyse pools of molecules fast and simultaneously. For instance, proteomics is the appropriated technique to explore differential levels of protein expression and modification before and after plant inoculation with bacteria. In this review, we provide state of the art on beneficial grass-bacteria association studies through proteomic and metabolomic analysis and discuss how these data can pave the way to isolate or develop better biofertilizer strains.
Proteomic Strategies for Studying the Grass-PGPR Association
Proteomics analysis is a technique with great potential for globally understanding processes involving simultaneous modulation of several proteins expression. This analysis can be applied to a range of systems from simpler ones such as cultured cells to more complex systems, such as a population of organisms in a particular niche (metaproteomics) or an association between different organisms (e.g., plant-microbe association). Proteomics relies on three pillars, as follows: (i) extraction of proteins, (ii) fragmentation of proteins in peptides and identification of peptides by mass spectrometry, and (iii) quantification of protein expression and post-translational modifications.
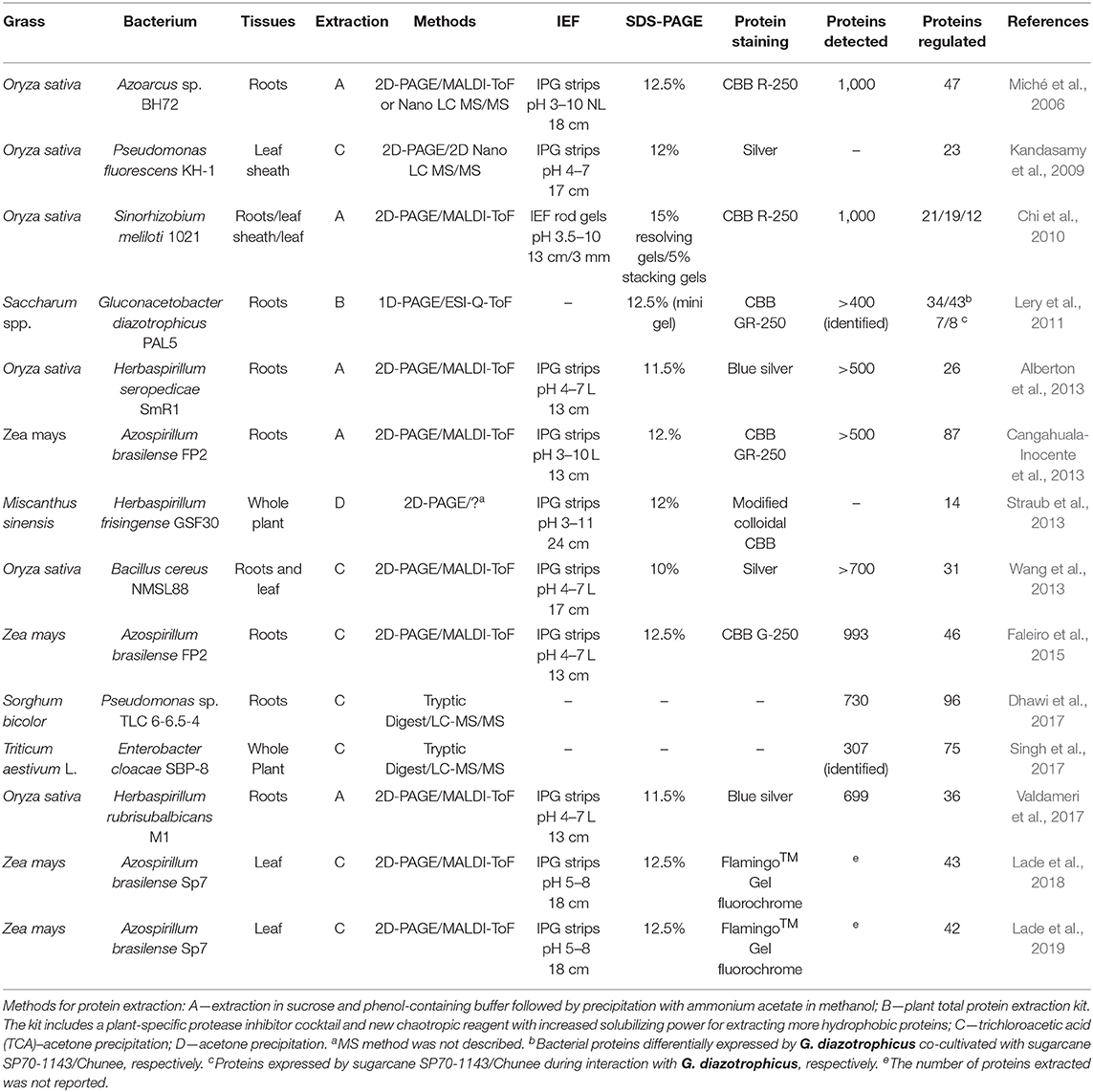
For instance, proteomics has been used to study the Rhizobia and legumes interaction (Cheng et al., 2010; Khatabi et al., 2019), the effects of abiotic stress in plants (Ghosh and Xu, 2014; Prinsi et al., 2018), and the pathogenicity of fungi and certain bacteria (Quirino et al., 2010). However, to our knowledge, only 14 proteomics studies have focused on beneficial grass-bacteria interactions (Table 1); most of the studies were performed using rice, likely because its genome sequence is known (Goff et al., 2002; Yu et al., 2002).
Genome sequencing of the Oryza sativa ssp. japonica cultivar Nipponbare (IRGSP, 2005) was completed, and its genome colinearity with other grass species was identified (Keller and Feuillet, 2000; Bennetzen and Ma, 2003); therefore, rice occupies a prominent position among grasses as an experimental model. In addition, rice is economically significant due to its high global consumption. Thus, several proteomic analyses on rice have focused on abiotic and biotic stresses (Kim et al., 2014), which facilitate a better understanding of different molecular mechanisms triggered by stress factors.
Maize and wheat genomes were also sequenced. The Maize GDB (Maize Genetics Executive Committee) is a global repository of information on genetics, genomics, and breeding research, being a crucial tool to advance the understanding of beneficial plant-maize (Schnable et al., 2009; Portwood et al., 2019). In 2018, the International Wheat Genome Sequencing Consortium (IWGSC) published a detailed analysis of the whole reference sequence of the bread wheat (Triticum aestivum) genome (Alaux et al., 2018; IWGSC, 2018). The maize and wheat genomes are key tools for advances in proteomic studies on bacteria-grass associations. The genomes for the PGPR models used in the seven studies were fully sequenced, which is extremely useful for large protein identification projects by peptide mass fingerprint (PMF).
Most studies employed 2D electrophoresis for protein separation (Table 2). According to Fey and Larsen, “the 2D gel electrophoresis is the technology that everyone loves to hate” (Fey and Larsen, 2001). The main reason that 2D gel electrophoresis remains the preferred method for protein separation is its ability to separate many proteins and isoforms from complex samples with high resolution and high reproducibility, and it facilitates both label-free quantitative analyses and post-translational modifications (Cheng et al., 2010). Furthermore, the known technical limitations, as the analysis of the highly acidic and alkaline proteins, low-abundance and hydrophobic proteins are not obstacles, but challenges that have led to improvements in analyses for these types of proteins (Görg et al., 2009).
For protein identification, nine out of 14 studies used MALDI-TOF MS (Matrix-assisted laser desorption/ionization-time of flight mass spectrometry) (Table 2). The other four studies applied LC/MS (liquid chromatography/mass spectrometry) methods. Accordingly, 2-D electrophoresis/MALDI-TOF is the proteomic strategy most employed for studying plant-PGPR interactions (Afroz et al., 2013). Surprisingly, no study so far has reported a proteomic shotgun analysis to evaluate a grass-PGPR association. A shotgun proteomics approach refers to the identification of proteins from complex mixtures of peptides generated by protein digestion with proteases employing a combination of high-performance liquid chromatography combined with mass spectrometry (Figure 1). This approach typically identifies many more proteins compared with 2D electrophoresis, is fast and adaptable to analyse several samples simultaneously. However, the high complexity of proteins coming from the host and bacteria might be challenging to separate adequately in the chromatographic step. In any case, the establishment of shotgun methods for the analysis of grass-bacteria interaction may yield important advances to the field.
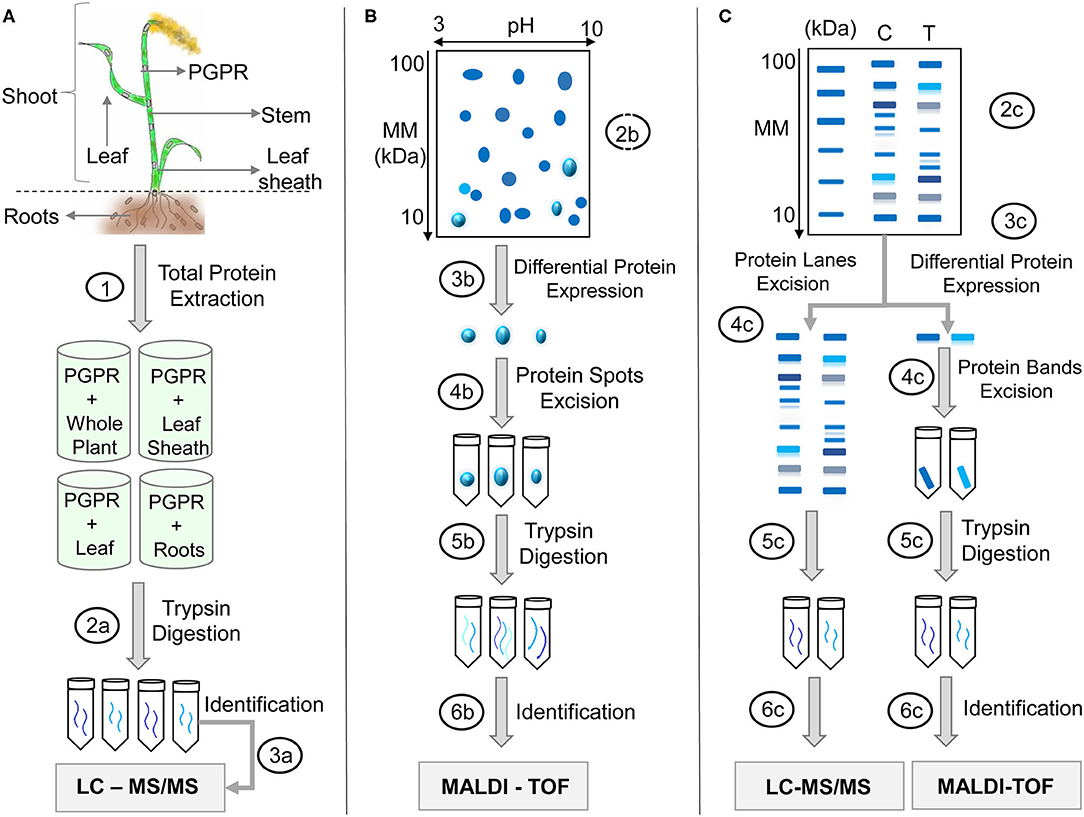
Figure 1. Proteomics applied to PGPR—grasses interaction studies. (A) After colonization by PGPR, whole plant or parts of the plant (shoots, roots, leaf) is submitted to protein extraction (1). The extracted proteins can be: (2a) proteolyzed by trypsin and the peptides separated by (3a) liquid chromatography and its mass/charge analyzed by spectrometer mass (LC-MS/MS) with subsequent identification of the proteins by specific software. The peptides from different samples can be labeled with peptides or other molecules of known mass to perform simultaneous analysis and quantification. If the samples are analyzed individually, the quantification can be done by peptide counts known as label-free quantification. (B) Proteins are firstly separated by bidimensional electrophoresis (2D-SDS-PAGE) (2b), analyzed to differential expression (3b), the differentially expressed protein spots are removed (4b), the protein spots are digested with trypsin (5b), and the peptides analyzed in a MALDI-TOF spectrometer with subsequent identification of the proteins by computational analysis (6b). (C) The protein extract can be separated by unidimensional electrophoresis (SDS-PAGE) (2c), the entire lanes can be sliced in several pieces (4c), proteins are trypsinized (5c), and peptides analyzed by LC-MS (6c). On the other hand, differentially expressed proteins can be identified and extracted form the gel (4c), proteins are trypsinized (5c), and peptides analyzed by MALDI-ToF MS (6c).
Of the studies in Table 2, the Saccharum spp. and Gluconacetobacter diazotrophicus PAL5 proteome study was the only study performed by combining 15N/14N metabolic protein labeling, one-dimensional gel electrophoresis (1D), and LC-electrospray ionization quantitative time-of-flight (ESI-Q-TOF) (Lery et al., 2011). The authors indicated that 15N/14N protein labeling was used for the first time during co-culture of a bacterium and its plant host. About 542 proteins were identified, and 78 proteins presented differential expression levels (Table 2), indicating this is a powerful strategy for such studies.
Regarding the protein extraction protocol employed (Table 2), one study used a commercial extraction kit (Lery et al., 2011), while the other studies used protocols based on phenol/sucrose buffer extraction followed by TCA/acetone precipitation (Isaacson et al., 2006). A version of the integrated protein extraction protocol for crop proteomics analyses was proposed by Wu et al. (2014). The protocol combines three steps: (1) TCA/acetone precipitation to remove non-protein compounds, (2) SDS extraction, which resolubilizes the proteins precipitated by TCA/acetone, and (3) phenol extraction that promotes the denaturation and solubilization of proteins into the organic phase separating them from water-soluble compounds. The use of this protocol may mitigate problems during isoelectric focusing and electrophoresis, which are commonly associated with non-protein compounds released from plant tissues during extraction. Considering that the protein extraction protocols are laborious and time-consuming, a modified TCA/acetone precipitation was established for plant proteomics. This protocol can be performed in 45 min, avoiding chemical protein modification and degradation as occur with the classical TCA/acetone or acetone method (Niu et al., 2018, 2019).
In most studies in Table 1, only proteins from plants were identified. For the Saccharum spp. and G.diazotrophicus interaction proteome (Lery et al., 2011), two sugarcane genotypes (SP70-1143 and Chunee) were inoculated. The G. diazotrophicus co-cultivated with SP70-1143 and Chunee genotypes resulted in 34, and 43 bacterial proteins differently expressed, respectively. The up-regulated proteins were involved in metabolism and signaling pathways. Proteins from Herbaspirillum seropedicae SmR1 colonizing rice roots were also identified, highlighting proteins involved in nitrogen fixation and assimilation (Alberton et al., 2013). Among 87 differentially expressed proteins of maize roots (DKB240 variety) inoculated with A. brasilense FP2, four spots were identified as bacterial proteins: malate dehydrogenase (2 spots), H+-transporting F-type ATPase beta (1 spot) and major outer membrane protein OmaA precursor (1 spot). The latter acts as adhesin involved in root adsorption and cell aggregation (Cangahuala-Inocente et al., 2013). Michè and collaborators detected potential bacterial proteins in the Azoarcus sp. and rice proteome study, but these proteins were not identified because the bacterial genome had not been completed (Miché et al., 2006). For the grass proteins, most of the studies identified defense proteins. In the proteomic study of sorghum colonized by Pseudomonas sp. TLC 6-6.5, 54 bacterial proteins were detected (Dhawi et al., 2017).
Hormonal Modulation in Plant-PGPR Interactions; What Does Proteomics Tell Us?
The phytohormones produced by PGPR are probably responsible for increasing plant growth and yield (Drogue et al., 2012). In such complex networks, several compounds, such as auxins, gibberellins, ethylene, jasmonates, cytokinins, abscisic acid, brassinosteroids and salicylic acid, regulate root hair proliferation and elongation. Phytohormones also coordinate plant responses to environmental factors, leading to nutrient uptake enhancement from the soil and activate defense mechanisms against pathogens (Dodd et al., 2010; Dicke and van Loon, 2014). Reviews on phytohormone biosynthesis, regulation, and function are (Mano and Nemoto, 2012; Duca et al., 2014; Tsukanova et al., 2017; Lymperopoulos et al., 2018). Therefore, we briefly describe the existing models for certain phytohormones that explain the participation of these molecules in PGPR-plant interactions.
Certain PGPR stimulate root proliferation through phytostimulant auxin biosynthesis; indole-3-acetic acid (IAA) is the most well-known auxin and enhancing mineral uptake from the soil (Masciarelli et al., 2013) and root exudation, which stimulates root colonization (Spaepen and Vanderleyden, 2011). However, higher concentrations of bacterial IAA can also inhibit root growth (Dobbelaere et al., 1999). Bacterial and plant IAA biosynthesis can be tryptophan dependent or independent (Duca et al., 2014), and although some intermediates differ, most bacterial pathways are similar to the pathways described in plants (Woodward, 2005; Spaepen et al., 2007; Karnwal, 2009).
High concentrations of auxin promote plant growth through ethylene biosynthesis activation (Hardoim et al., 2008). The gaseous phytohormone ethylene regulates physiological processes throughout the plant life cycle. However, during biotic and abiotic stress conditions, production of this hormone is exacerbated, which leads to deleterious consequences in plants (Bleecker and Kende, 2000). Ethylene is synthesized from S-adenosyl-L-methionine (AdoMet) in two steps. (i) 1-Amino-cyclopropane-1-carboxylic acid (ACC) synthase (ACS) catalyzes the conversion of S-adenosyl-methionine (AdoMet) into ACC and the by-product 5′-methylthioadenosine (MTA). (ii) ACC oxidase (ACO) catalyzes the conversion of ACC and O2 into ethylene, CO2 and cyanide (Rzewuski and Sauter, 2008). The 5′-methylthioadenosine (MTA) is recycled back into methionine through a salvage pathway. Glick and colleagues proposed a modulation mechanism for plant ethylene biosynthesis by PGPR (Glick et al., 1998). Briefly, plant-exuded tryptophan is taken up by bacteria for the IAA synthesis. IAA secreted by bacteria and also synthesized by plants can stimulate the activity of ACC synthase in plants, converting Adomet into ACC, which can be taken up by the bacteria and hydrolyzed by ACC deaminase into ammonia and α-ketobutyrate. Through IAA bacterial activation, the plant synthesizes more ACC, and the bacteria utilize it as a nitrogen and carbon source; consequently, the lower levels of ethylene favor plant development (Figure 2).
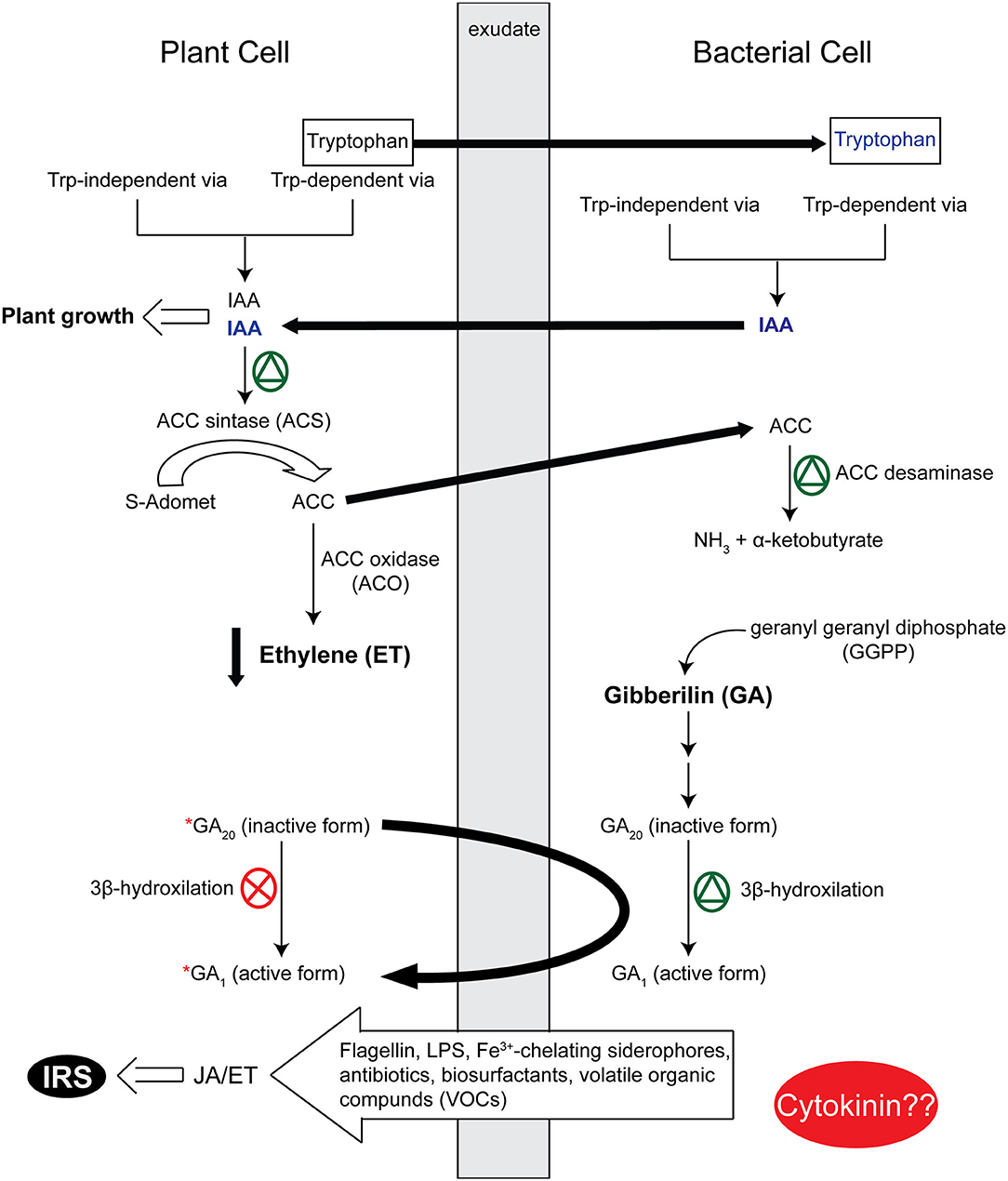
Figure 2. A schematic representation of phytohormone modulation by plant-bacteria interactions. Indol-3-acetic acid (IAA) produced by bacteria and plants through tryptophan-dependent or/and tryptophan-independent pathways activates ACC synthase in plants. ACC synthase stimulates ACC production; ACC is hydrolyzed by bacterial ACC deaminase in NH3 and α-ketobutyrate, decreasing ethylene levels in plants (Glick et al., 1998; Glick, 2012). After inoculation with the Azospirillum lipoferum strain USA5b in mutant rice plants that present genetic lesions in the 3β-hydroxylation step in gibberillin synthesis, the active form of gibberillin (GA1) was detected, which indicates active gibberillin synthesis in bacteria (Cassán, F., et al., 2001; Cassán et al., 2001). The response to jasmonic acid (JA), its derivatives and ethylene is necessary for induced systemic resistance (IRS), and different bacterial determinants are responsible for initiating priming in these hormones. Little information is available on bacterial cytokinin role in plant-bacteria interactions.  activation;
activation;  inhibition.
inhibition.
Gibberellins (GAs) are also involved in PGPR-plant interaction. Present in plant, fungi, and bacteria (MacMillan, 2001), ~136 GAs have been identified (see http://www.phytohormones.info/gibberellins.htm). However, little is known on how gibberellins participate in this interaction (Rademacher, 1994; Bottini et al., 2004). Studies on Azospirillum sp. in gibberellin-deficient mutants of rice demonstrated that Azospirillum could convert the inactive form of gibberellins in plants (GA20) into the active form (GA1) through 3β-hydroxylation, which, consequently, promotes plant growth (Cassán, F., et al., 2001; Cassán et al., 2001) (Figure 2). The inoculation of seed-borne endophytic B. amyloliquefaciens RWL-1 in rice plants promoted a significant increase in the content of GA, which led to faster growth of shoot and roots of inoculated rice plants (Shahzad et al., 2016).
Other phytohormones produced by PGPR, such as with cytokinin, aid in plant growth promotion, but their underlying mechanisms remain unclear. Lettuce plants (Lactuca sativa L., cv Lolla Rossa) inoculated with Bacillus sp. strain IB-22 had larger shoots than control plants, but the root length decreased, and consequently, the root to shoot ratio (Arkhipova et al., 2007). According to Vacheron and colleagues, the contribution of cytokinin by PGPR to root system architecture modifications remains speculative (Vacheron et al., 2013) (Figure 2).
Jasmonic acid (JA) and the derivative jasmonates are hormones important to the plant immune system acting in the induced systemic resistance (IRS), which reduces pathogen invasion and colonization (Pozo et al., 2008) (Figure 2). In rice, ISR mediated by Pseudomonas fluorescens WCS374 against Magnaporthe oryzae depends on jasmonate and the ethylene-regulated signaling pathway (De Vleesschauwer et al., 2008).
Few proteins related to phytohormone biosynthesis and signaling pathways have been identified through proteomics analyses on PGPR-grass interactions. Proteomics analyses of the interaction between Sinorhizobium meliloti 1021 and rice demonstrated that one auxin-induced protein was up-regulated in leaves and that IAA-amino acid hydrolase is down-regulated in roots; thus, IAA biosynthesis may be induced by rhizobia through a tryptophan-independent pathway (Chi et al., 2010). Although it was not a study using grass as host, proteomic analysis of Pseudomonas putida UW4 exposed to canola exudates showed the expression of an ACC deaminase 1.64-fold higher compared with the control condition (without exudates). In order to confirm the ACC deaminase activity of P. putida UW4, ACC, and IAA concentrations were measured on the root exudates of canola treated with the wild-type bacteria and their ACC deaminase negative mutants. Lower ACC and higher IAA concentrations were detected in the exudates of the canola root treated with the wild P. putida UW4, confirming the action of ACC bacterial deaminase on the ACC levels of the plant (Cheng et al., 2009).
G. diazotrophicus co-cultivated in the presence of sugarcane plantlets induced the expression of the GreA transcription factor. According to the authors, greA is close to the ilvA, which encodes a threonine dehydratase, indicating that greA and ilvA might be co-regulated. IlvA is an enzyme of the isoleucine biosynthesis pathway. Isoleucine is conjugated with JA to affect plant development and interactions with microorganisms (dos Santos et al., 2010). The rice root proteome upon treatment with JA and the Azoarcus sp. strain BH72 exhibited induction of a putative ACC oxidase only in the JA-treatment condition, indicating that the ethylene and JA pathways interact with each other (Miché et al., 2006). Although most responses mediated by ethylene and JA are independent, specific pathways, such as induction of the plant defensin gene, PDF1.2, require both hormones (Penninckx et al., 1998; Pieterse et al., 2000; Wang et al., 2002).
Xyloglucan endotransglucosylase (XTH) was 2.8-fold more expressed in rice leaves colonized by Bacillus cereus NMSL88 (Wang et al., 2002) and was also up-regulated in wheat colonized by Enterobacter cloacae SBP-8 (Singh et al., 2017). This enzyme is involved in xyloglucan metabolism in the primary cell wall through xyloglucan endo-cleavage and transferring the new reducing ends to other polymeric or oligomeric xyloglucan molecules, which are important to plant growth and development (Fry et al., 1992; Cosgrove, 2005). In the rice leaf sheath, the XTH gene was up-regulated by GA3, one of the active forms of gibberellin (Jan et al., 2004).
Colonization of the grass Miscanthus sinensis by Herbaspirillum frisingense over 3 h up-regulated the genes BGAF-1, BGAF-2, and dirigent protein gene (DIR) in roots and shoots involved in jasmonate biosynthesis and signaling (Straub et al., 2013). However, after 3 weeks of colonization, these genes were repressed, especially in the roots. BGAF is a β-glucosidase-aggregating factor that interacts with maize β-glucosidase, forming large insoluble complexes (Blanchard et al., 2001; Kittur et al., 2007). This protein is induced by JA and contains two domains: a jacalin-related lectin and a disease-response or dirigent protein (Blanchard et al., 2001; Kittur et al., 2007); one of its key functions in plants is the defense response against insects and pathogens (Williams et al., 2002).
H. frisingense induced low expression of the auxin response factors ARF1, ARF6, IAA18, and IAA20 in the Miscanthus sinensis shoots and roots. At the same time, ethylene modulation was demonstrated by up-regulation of the ethylene receptors (ETR1, SCER1a, and SCER1b) and downregulation of the ethylene response factor (SCERF1-like) in shoots and roots, which suggests ACC deaminase activity from H. frisingense. These results were obtained by transcriptomic and quantitative real-time PCR (qRT-PCR) analyses. The proteins identified by differential proteomics did not match those found by RNA transcripts quantification (Straub et al., 2013).
Conversely, proteome analysis of rice roots colonized by H. seropedicae SmR1 showed up-regulation of proteins involved in methionine recycling and the 1-aminocyclopropane-1-carboxylic acid (ACC) synthase which synthesizes ACC, the precursor for ethylene biosynthesis. In addition, the ethylene levels were lower in plants inoculated with H. seropedicae. Although the ACC deaminase and IAA pathway enzymes were not identified, this result suggests that H. seropedicae modulates the ethylene levels in rice roots, decreasing the stress response promoted by this hormone (Alberton et al., 2013).
In summary, PGPR can affect plant growth through phytohormones. However, few pathways related to this interaction were elucidated, which indicates that more studies are necessary to demonstrate how the phytohormones produced by PGPR alter plant development. Although few proteins related to phytohormone biosynthesis and degradation have been identified in the different proteomes, we can infer from proteomics data that differential expression of the defense and metabolism proteins is likely due to the complex phytohormone network.
Proteome and Biological Nitrogen Fixation in Grass
Nitrogen is an essential component for all living organisms, and its availability is one of the major factors that limit plant growth. The most abundant nitrogen source is dinitrogen gas (N2), which composes 78% of the atmosphere; however, this form is effectively inert. Only certain bacteria and archaea can catalyze biological nitrogen fixation and convert N2 into a plant-assimilable form (Dixon and Kahn, 2004). These microorganisms are diazotrophs and encode nitrogenase, which is the metalloenzyme complex that reduces N2 to NH3 through the following chemical reaction (Burgess and Lowe, 1996):
Nitrogenase is composed of two components that use the electron transfer cascade to reduce its substrate. The dinitrogenase reductase (NifH or Fe protein) transfers electrons in an ATP-dependent manner to the second nitrogenase component, the dinitrogenase (NifDK or MoFe protein), which contains the active site FeMo-cofactor (Seefeldt et al., 2009). In addition to the nitrogenase structural components encoded by nifHDK, diazotrophs have three essential FeMo-cofactor biosynthetic components encoded by nifENB (dos Santos et al., 2012) and a variable set of genes to optimize nitrogenase biosynthesis, activity and regulation. Essentially, biological nitrogen fixation is a natural fertilization process, and its optimization plays a significant role in sustainable agriculture, reducing the economic and environmental impacts of nitrogen inputs for increasing crop productivity (Beatty and Good, 2011; Oldroyd and Dixon, 2014).
The first evidence of nitrogen fixation with plant-associated bacteria was demonstrated through the symbiotic interaction between rhizobia and legume nodules (Smith et al., 2004). Since then, the interaction between legumes and bacteria that participate in nitrogen-fixing symbiosis has been extensively studied (Cheng et al., 2010). Briefly, a series of proteomic studies showed that the key transcription factors responsible for differentiating nitrogen-fixing nodules, then proteins involved in nitrogen metabolism, mainly NifHDK and regulatory PII proteins, are expressed to initiate nitrogen fixation a few days after inoculation (for more details, please refer to Salavati et al., 2013 and Muneer et al., 2012). Recent studies show that the most important crops worldwide, such as rice, wheat, and maize, interact with nitrogen-fixing bacteria, and in addition to the nitrogen supplied by the soil, biological nitrogen fixation significantly contributes to nitrogen accumulation in plants (James, 2000). Although rhizobia-legume symbiosis is the most important and likely the most efficient nitrogen-fixing system, nitrogen fixation in crops consumed by humans has received particular attention in the past few decades (Franche et al., 2009; Beatty and Good, 2011).
Nitrogen biologically fixed by endophytic/endosymbiotic diazotrophs can be transferred to grass, which is inferred mainly by plant dry weight, nitrogen content, and nitrogenase gene expression of their hosts (Egener et al., 1999; Roncato-Maccari et al., 2003; Camilios-Neto et al., 2014). However, few proteomics studies have identified bacterial proteins related to nitrogen fixation or nitrogen metabolism during grass colonization. Only two proteomics studies have identified proteins involved in nitrogen fixation and ammonium assimilation, NifH and glutamine synthetase (GS), respectively, which are up-regulated in rice roots colonized by the H. seropedicae SmR1 (Alberton et al., 2013) or the H. rubrisubalbicans M1Sm300 (Valdameri et al., 2017). These findings suggest that the cells are under nitrogen-fixing conditions after 7 days of inoculation. G. diazotrophicus GS was also up-regulated upon interaction with rooted sugarcane plantlets after 1 day of inoculation (Lery et al., 2011). Differently, this GS expression seems unrelated to fixed nitrogen availability because it was observed with low and high nitrogen input. In contrast, it could be involved in osmoregulatory responses and plant development (Sleator, 2001; Miflin and Habash, 2002).
Interaction with nitrogen-fixing bacteria also affects the regulation of host proteins involved in nitrogen metabolism. G. diazotrophicus inoculation of sugarcane roots led to host's glutamate-ammonia lyase up-regulation, which suggests that nitrogen metabolism was stimulated (Lery et al., 2011). The assimilatory ammonium enzymes, cytosolic GS, and glutamate dehydrogenase (GDH), were down-regulated upon S. meliloti 1021 inoculation of rice seedlings, leaf sheaths, and roots, respectively (Chi et al., 2010). GS catalyzes ATP-dependent conversion of glutamate to glutamine using ammonium as a substrate, which is followed by a condensation reaction involving 2-oxoglutarate (2-OG) and glutamine to produce glutamate, which is catalyzed by glutamate synthase (GOGAT). However, in an alternative pathway, GDH catalyzes the reversible deamination of glutamate into 2-OG. Together, the down-regulation of GS and GDH suggests that glutamate accumulates, and the lower 2-OG levels support down-regulated aspartate aminotransferase. To maintain glutamate homeostasis, this precursor might be used for proline biosynthesis in agreement with the up-regulation of a putative peptidyl-prolyl isomerase under the same conditions (Wang et al., 2007; Broyart et al., 2010). GS plays a key role in controlling plant productivity, and researchers have suggested using GS as a molecular marker for nitrogen utilization and uptake efficiency (Swarbreck et al., 2011). Thus, addressing the regulatory mechanism for GS protein content and activity regulation, isoforms, and localization through proteomics approaches could lead to strategies for higher nitrogen efficiency, allowing for less use of nitrogen fertilizers without lowering crop yield.
Other proteins involved in nitrogen metabolism have not been identified through proteomics analyses of PGPR-grass interactions, likely because low-abundance bacterial proteins are challenging to detect in complex protein mixtures. In addition, Fe-S metallo-centers in nitrogenase can be irreversibly damaged by O2, which leads to protein degradation and prevents identification through proteomics after a lengthy sample preparation procedure (Hartmann and Burris, 1987; Fisher and Newton, 2005).
Stress and Defense Responses in Grass-PGPR Interactions as Seen by a Proteomics Approach
Plant defense mechanisms induced by pathogens in non-compatible interactions has been intensely studied (Senthil-Kumar and Mysore, 2013; Kannojia et al., 2019). However, PGPR has been described to elicit or activate defense mechanisms in host plants, which suggests that even beneficial interactions can trigger plant-defense cell signaling (Goh et al., 2013). Proteomics approaches can be used to understand the molecular basis underlying the modulation of plant proteins in the presence of PGPR, which requires additional studies. Here, we selected stress and defense-related proteins identified in more than one single, comparative proteomic analysis of grass-PGPR to address common pathways.
Stress Response
Peroxidases (POX) are widely recognized as important antioxidant proteins involved in cellular protection against reactive oxygen species (ROS) (Sharma et al., 2012). Two POX isoforms were detected up-regulated in two Oryza sativa cultivars, IR36 and IR42. The colonization study showed that IR36 was successfully and IR42 poorly colonized by the endophyte Azoarcus sp., respectively, (Miché et al., 2006). Interestingly, POX isoforms were not regulated in both cultivars upon inoculation with Azoarcus sp., but when the two cultivars were treated with jasmonic acid, the two POX were up-regulated. Furthermore, POX was also up-regulated in rice roots colonized by B. cereus or S. meliloti 1021 (Chi et al., 2010; Wang et al., 2013). In addition to POX, catalase is a key antioxidant enzyme and was up-regulated in rice leaves and sheaths inoculated with S. meliloti 1021 (Chi et al., 2010).
Interestingly, rice root colonization by H. seropedicae down-regulated two putative L-ascorbate peroxidases (APX), which is related to a lower antioxidant defense (Alberton et al., 2013). In contrast, proteomic analysis of G. diazotrophicus-sugarcane interaction showed an opposite response, wherein APX was exclusively expressed by sugarcane SP70-1143 during its interaction with G. diazotrophicus (Lery et al., 2011). Furthermore, the differential proteomics analysis in M. sinensis seedlings grown in the presence or absence of H. frisingense revealed that APX1 was up-regulated and, in stark contrast, APX2 was down-regulated in inoculated plants (Straub et al., 2013). Given the contradictory data obtained using proteomic approaches, the direct effect of APX regulation in grass inoculated with PGPR must be further investigated. It is likely interplay of different isoforms, cultivar-dependent response and time-dependent expression of APX.
Treatment of rice (cv. IR36) seedlings with JA or inoculation with Azoarcus sp. showed two isoforms of the rice salt-stress induced protein (SalT) up-regulated (Miché et al., 2006). Besides, SalT was also up-regulated in rice roots inoculated with H. seropedicae (Alberton et al., 2013). Up-regulated proteins involved in plant defense, including the salt stress root protein RS1, were also observed in the interaction between Bacillus cereus and rice (cv. Kyou818) (Wang et al., 2013). Together, these data suggest that SalT is up-regulated in rice inoculated with different PGPR. Based on these proteomics studies, SalT may be suggested as a molecular marker for beneficial grass-bacteria interactions. In addition, a BlastP analysis using the Oryza sativa Indica cultivar-group SalT (Uniprot ML OsI_001780) as query against the protein database showed that SalT homologs are widely distributed among grasses, including rice, sorghum, wheat, maize, sugarcane, Setaria grass, Tausch's goatgrass, and Ravenna grass, with at least a 38% identity and an expected value (E-value) of 1 × 10−23.
Wheat colonized by Enterobacter cloacae SBP-8 and submitted to salt stress (200 mM NaCl) enhanced the “calcium channel protein” which is part of the ROS-responsive Ca2+ channel, mediating the salinity -induced Ca2+ influx in the leaves (Singh et al., 2017). Ca2+ is a plant second messenger released under salt stress (Choi et al., 2014). For a review of the beneficial effect of PGPR on the development of plants under salt stress, refer to Bhat et al. (2020).
Defense Response
PGPR can benefit plant development by activating defense mechanisms against attackers. It is already known that PGPR can elicit induced systemic resistance (ISR) against infection by pathogens (fungi, viruses, nematodes, and bacteria) (Jetiyanon and Kloepper, 2002; Walters et al., 2013). The ISR elicited by PGPR is mainly dependent on jasmonic acid-ethylene (JA-ET) signaling (Pieterse et al., 2014; Kannojia et al., 2019).
Azoarcus inoculation promoted weak induction of two different PR proteins in Oryza sativa cv. IR36, PR-10b, and RSOsPR10 (Miché et al., 2006). One putative exoglucanase precursor, classified as an important PR protein, was induced in rice seedlings inoculated with S. meliloti 1021 (Chi et al., 2010). Azoarcus also induced overexpression of a PR protein (Prb1) in cv. IR42 (Miché et al., 2006).
Probenazole-induced protein (PBZ1) is a PR protein with an unknown biological function, although its ribonuclease activity has been described (Bantignies et al., 2000). The PBZ1 expression levels were not regulated upon inoculation of rice with Azoarcus sp., but JA induced PBZ1 up-regulation (Miché et al., 2006). Interestingly, rice inoculated with H. seropedicae exhibited lower PBZ1 mRNA expression levels in roots (Brusamarello-Santos et al., 2012, 2019). These results suggest that specific rice defense targets, such as PBZ1, present opposing behaviors depending on the bacteria and treatment used.
This opposite expression pattern of PBZ1 in rice induced by Azoarcus sp. and H. seropedicae was also observed for glutathione S-transferase (GST) because it was not regulated and up-regulated by rice inoculation, respectively (Miché et al., 2006; Alberton et al., 2013). Similarly, up-regulated GST was observed in rice plants upon JA treatment or inoculation with Bacillus cereus and P. fluorescens (Miché et al., 2006; Kandasamy et al., 2009; Wang et al., 2013). GST was also up-regulated in maize leaves colonized by Azospirillum brasilense Sp7 for 16 days (Lade et al., 2018). The receptor-like protein kinase (RLK) DUF26 is involved in both symbiosis and pathogen defense and was up-regulated in rice seedlings inoculated with Bacillus cereus (Wang et al., 2013). In addition, a putative receptor-like kinase was up-regulated in rice roots following either Azoarcus inoculation or JA treatment (Wang et al., 2013).
S-adenosylmethionine synthetase (SAMS, also known as methionine adenosyltransferase) is involved in methionine recycling and under stress is related to alleviating biotic and abiotic induced-damage; SAMS was up-regulated in rice leaf sheaths inoculated with S. meliloti 1021 (Chi et al., 2010). Two spots corresponding to SAMS and SMAS2 were up-regulated in rice roots inoculated with H. seropedicae (Spaepen et al., 2007). Interestingly, SAMS was up-regulated in wheat grown under salt stress. The inoculation with Enterobacter cloacae SBP-8 reduced SAMS expression under the same salt stress condition (Singh et al., 2017). Proteins involved in methionine recycling, including methylthioribose kinase and acireductone dioxygenase 1, were also induced by H. seropedicae inoculation. Under the same conditions but analyzed using a different approach, the mRNA that codes enolase, which is also involved in methionine recycling, was up-regulated in rice upon H. seropedicae colonization (Alberton et al., 2013). Accordingly, the spots that corresponded to enolase were up-regulated in rice inoculated with S. meliloti 1021 (Chi et al., 2010). These data suggest that some beneficial bacteria might induce methionine recycling, which is involved in ethylene hormone biosynthesis in the plant host.
Two proteomic studies on the interaction between Azospirillum brasilense Sp7 and maize pointed out the maize beta-glucosidase 2 up-regulation (Lade et al., 2018, 2019). Beta-glucosidase 2 catalyzes the cleavage hydroxamic acid glucosides, specifically the glucoside of 4-hydroxy-7-methoxy-l,4-benzoxazin-3-one (DIMBOA), a secondary metabolite plant against pathogens (Babcock and Esen, 1994; Frey et al., 1997). After 16 days of inoculation with A. brasilense Sp7, beta-glucosidase 2 from maize leaf was 2-fold more expressed than non-inoculated maize plants (Lade et al., 2018). In another study, when the maize plants were co-treated with A. brasilense Sp7 and the maize dwarf mosaic virus (MDMV), after 16 days, the Sp7 promoted up-regulation around 1.8 of the beta-glucosidase 2 (Lade et al., 2019). Lade et al. (2018, 2019) concluded that Azospirillum brasilense Sp7 activates the systemic resistance (IRS). Tobacco plants expressing β-glucosidase from Trichoderma reesei showed increased internode length, height, biomass, leaf area, and trichome density than control plants and all these developmental parameters are regulated by gibberellins (Jin et al., 2011). Therefore, beta-glucosidase possibly acted on inactive glucosyl-gibberellin conjugates, releasing the active form of this hormone, as well as of the hormones zeatin and indole-3-acetic acid. In addition, plants expressing beta-glucosidase also showed resistance against whitefly (Bemisia tabaci) and aphid (Myzus persicae) attacks, due to protection provided by the sugar esters exudated from globular trichomes of the leaves (Jin et al., 2011).
Siderophore Production by PGPR
Rice inoculation with H. seropedicae showed that plant proteins involved in methionine recycling and mugineic acid biosynthesis were up-regulated using a proteomics approach and RT-qPCR, respectively. One of the up-regulated genes that code for key enzymes in the phytosiderophore synthesis pathway was nicotianamine synthase 1 (NAS1); the mRNA level was ~100-fold higher in inoculated plants (Alberton et al., 2013). The NAS1 protein was also up-regulated in rice seedlings upon inoculation with Bacillus cereus, as demonstrated by a comparative proteomics analysis (Wang et al., 2013). Siderophore produced by PGPR may improve host plant iron absorption using an Fe-siderophore complex (Dimkpa et al., 2009). Given that H. seropedicae and B. cereus increased the NAS1 expression levels, we hypothesized that inoculated plants might naturally accumulate iron increasing the nutritional value, which is a strategy recently introduced in transgenic rice overexpressing the NAS1 gene (Dimkpa et al., 2009).
To address this hypothesis, we measured by RT-qPCR the mRNA levels for two rice ferritin isoforms, which is one of the most important proteins involved in iron storage, to determine whether the rice ferritin levels increased in rice roots upon inoculation with H. seropedicae. Surprisingly, the mRNA expression levels for both rice ferritin genes (Fer552 and Fer768) were repressed by H. seropedicae inoculation (Figure 3). This finding is inconsistent with our first assumption and leads to two assumptions for further assessment: (i) ferritins might be not involved in iron storage under this condition and (ii) the bacteria reduced the iron availability, which induced the NAS1 mRNA levels to improve iron uptake by the plant. Brusamarello-Santos et al. (2019) detected a bacterioferritin gene induced (2.2-fold) in the study of the interaction between H. seropedicae SmR1 and roots rice. However, other genes related to siderophore biosynthesis were not identified.
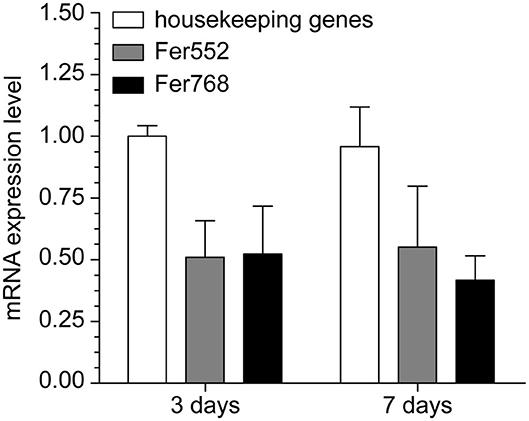
Figure 3. Rice gene expression levels determined using reverse-transcription quantitative-PCR. The levels of mRNA that code two ferritins exhibited differential expression in rice roots inoculated with Herbaspirillum seropedicae. Fer552 corresponds to similar to ferritin 1, chloroplast precursor (EC 1.16.3.1) (ZmFer1), RAPDB Os11t0106700-01, primers forward 5′ CCGCCGCCGCCGCGCCTACC 3′ and reverse 5′ CCTTCCCTTTCCCGGCGGCC. Fer768 corresponds to similar to ferritin 1, chloroplast precursor (EC 1.16.3.1) (ZmFer1), RAPDB Os12t0106000-01, primers forward 5′ ATTCCTTGAGGAGCAGGTTG 3′ and reverse 5′ GCTTCTTCCTCAAGCAGCTTC 3′. The expression levels were normalized using three housekeeping genes (actin 1, tubulin beta-2 chain, and protein kinase). Three independent RNA samples from inoculated and uninoculated rice roots were used, and each sample was analyzed in triplicate.
The Metabolomic View of the Interaction Between PGPR and Grasses
While proteomics identifies and quantifies proteins and their modifications, metabolomics identifies and quantifies metabolites concentration variations. Metabolites are small molecules (<1,500 kDa) and the end products of gene expression. Therefore, the metabolomic picture of a cell or tissue can be directly correlated to the phenotype (Tugizimana et al., 2018). Both proteomics and metabolomics employ the previous extraction with solvents to obtain the analytes, which will be identified and quantified by mass spectrometers. Metabolomic studies also can use Nuclear Magnetic Resonance (NMR) spectroscopy, as in the study of the interaction between plant growth-promoting bacteria and mycorrhiza with Setaria italica, a livestock grass and a potential source of carbon for biofuel production as well (Dhawi et al., 2018). In this study, the endomycorrhizal consortium alone, composed by Glomus intraradices, G. mosseae, G. aggregatum, and G. etunicatum, or combined with the PGPR Pseudomonas TLC6-6-5.4, showed a positive effect on biomass, plant height and chlorophyll content compared to non-inoculated plants. The same was determined for plants inoculated with the ectomycorrhizal consortium, composed by Rhizopogon villosullus, R. luteolus, R. amylogpogon, R. fulvigleba, Pisolithus tinctorius, Scleroderma cepa, and S. citrinum. In inoculated plants, the fructose and glucose concentrations were decreased, having a negative or no correlation with the S. italica biomass amount, while gallate, gluconate, and malate increased positively correlated with plant height and chlorophyll amount (Dhawi et al., 2018).
Pseudomonas TLC6-6-5.4 and the endomycorrhizal blend were inoculated in maize seedlings grown in stamp sands. After 62 days of the seeds germination, the metabolites of the plants were extracted and analyzed by GC-MS (gas chromatography-mass spectrometry), showing upregulation of glyoxylate and dicarboxylate metabolism. In addition, PGPR alone or associated with endomycorrhizal mix increases the concentrations of mannitol, palmitic acid, lysine, stearic acid, and sucrose. In the plants inoculated only with the endomycorrhizal consortium, sucrose concentration was significantly decreased (Dhawi et al., 2015). Regardless of the metabolic variations, the maize biomass was increased in the group inoculated with the combination of endomycorrhiza and PGPR (Dhawi et al., 2015).
Metabolomic analyses of plant-bacteria association have been useful to study the mechanisms by which some bacteria elicit plant resistance against fungi. For instance, the inoculation of rice with Rhizobium leguminosarum bv phaseoli RRE6 and R. leguminosarum bv trifolii ANU 843 increased the concentrations of phenolic acids, such as cinnamic, ferulic, gallic, and tannic acids, increasing the resistance against the pathogen Rhizoctonia solani (Mishra et al., 2006). Noteworthily, the R. leguminosarum RRE6 was the most efficient strain eliciting resistance and phenolic compounds synthesis.
The plant inoculation with PGPR can also activate antimicrobial production. Maize inoculated with Azospirillum lipoferum CRT1, and Azospirillum brasilense strains CFN-535 and UAP-154 were transferred to non-sterile soil under greenhouse conditions. After 10 days of inoculation, metabolomic analyses showed that the concentrations of benzoxazinoids were increased (Walker et al., 2011). Benzoxazinoids are secondary metabolites of plant defense and allelopathy. Maize inoculated with Azospirillum lipoferum CRT1 and transferred to the field showed higher benzoxazinoids amounts than non-inoculated control (Walker et al., 2012). These findings show that metabolomics is a useful tool to screen large libraries of plants and microbes associations, searching for up-regulation of relevant compounds for plant resistance against pathogens and abiotic stresses. Indeed, such kind of technology will be critical to find better and competitive biofertilizers.
Metabolomics has been showing to be a useful tool to compare strains of different genetic background and deepen our understanding of plant-bacteria association. The metabolic profiles of maize roots inoculated with nitrogen-fixing bacteria (Herbaspirillum seropedicae SmR1 and Azospirillum brasilense FP2) presented more alterations than those inoculated with non-nitrogen-fixing mutants (Herbaspirillum seropedicae SmR54 and Azospirillum brasilense FP10). In the roots of the maize variety FV252, the concentrations of mannitol, trehalose, isocitrate, aminoadipate, malonate, gluconate, cysteine, threonate, and trans-aconitate were increased for the group inoculated with nitrogen-fixing strains of H. seropedicae or A. brasilense. For instance, the concentration of mannitol in roots inoculated with H. seropedicae SmR1 was 50-fold higher in comparison to roots inoculated with H. seropedicae SmR54. Similarly, a 33-fold increasing in mannitol concentration was measured in roots inoculated with Azospirillum brasilense FP2. Interestingly, the metabolic changes promoted by the diazotrophic strains in FV252 were significantly reduced in roots of the FV2 variety, indicating that early events of interaction and plant recognition are essential for metabolic adaptability (Brusamarello-Santos et al., 2017).
The metabolic response of different cultivars of rice (Cigalon and Nipponbare) was tracked after inoculation with the epiphyte Azospirillum lipoferum 4B or the endophyte Azospirillum sp. B510. Both strains colonize the rhizoplane of both rice varieties, while only Azospirillum sp. B510 was found within the roots of Cigalon and Nipponbare. Although A. lipoferum 4B colonizes only the root surface, it stimulated accumulation of tryptophan in roots of both cultivars. Tryptophan is the precursor of auxin synthesis like indole acetic acid (IAA) in bacteria and plants. On the other hand, A. lipoferum 4B induced modifications in the secondary metabolism only in Cigalon roots, while B510 promoted secondary metabolic variations on shoots and roots of both cultivars (Chamam et al., 2013). The different combinations of the interactions of the two rice cultivars and two bacteria revealed that the endophytic strain B510 seemed to have little effect on root morphology of both cultivars compared to the non-endophytic strain 4B. However, in Nipponbare, its original host cultivar, the B510 induced the highest increase of shoot and root biomass. Interestingly, 4B induced variations in flavonoids concentrations in both cultivars and to hydroxycinnamic acid-based compounds in Cigalon, particularly p-coumaric acid and feruloyl quinic acid. p-Coumaric and ferulic acids are among the main phenolic compounds of rice (Dixon et al., 2002; Chung et al., 2018). Phenolic compounds are important for resistance against pathogens and allelopathy in rice (Chung et al., 2018). For instance, feruloyl quinic acid confers plant resistance against Burkholderia andropogonis (Mareya et al., 2019). Some C-glycosylated flavones, like luteolin-6-C-arabinoside-8-C-glucoside, isoorientin-200-glucoside, apigenin-6-C-arabinoside-8-C-glucoside, 6-C-diglucose coumaroyl isoscoparin, and isoscoparin-200-glucoside had their concentrations varying in the shoots of colonized Cigalon and Nipponbare. Azospirillum sp. B510 increased the concentration of luteolin-6-C-arabinoside-8-C-glucoside in shoots of Nipponbare, as well as the concentration of apigenin-6-C-arabinoside-8-C-glucoside in shoots of Cigalon. As these flavonoids are described having antioxidant and antimicrobial activity, the B510 strain might induce systemic resistance in rice (Yasuda et al., 2009; Compant et al., 2010).
Except for the metabolomic analysis performed with HPLC in the work of Mishra et al. (2006) and with NMR by Dhawi et al. (2018), the other metabolomic studies employed LC-MS or GC-MS. Agtuca et al. (2020) studied the metabolome of Setaria viridis roots colonized by Herbaspirillum seropedicae SmR1 (Fix+) and the SmR54 mutant (Fix−) using laser-ablation electrospray ionization (LAESI) (Agtuca et al., 2020). LAESI employs a mid-infrared to ablate material from a sample surface, and then, the ablated material is ionized by an electrospray source (Etalo et al., 2018). The LAESI-MS allowed the detection of several metabolites from different pathways. Metabolites from the purine, zeatin (cytokinin), and riboflavin biosynthesis pathways were significantly more abundant in roots inoculated with SmR1 and SmR54 than when uninoculated. However, the diazotrophic strain SmR1 enriched a large number of different types of metabolites in the Setaria roots (Agtuca et al., 2020). Roots inoculated with SmR54 or uninoculated have a decreasing in the concentration of metabolites involved in nitrogen, starch, and sucrose metabolisms, possibly due to N limitation. Interestingly, indole-alkaloid biosynthesis metabolites were more abundant in the roots colonized by SmR54, perhaps reflecting a plant defense response (Agtuca et al., 2020).
Concluding Remarks
Only few proteomic and metabolomic studies have addressed the PGPR interactions with grass so far. However, the inverse is true when the two organisms are evaluated separately. Thus, why are there few proteomic and metabolic studies on PGPR-grass interactions? One limitation in PGPR and grass proteomics studies is the lack of genomic information for plant models, especially for grass. However, high throughput genome sequencing platforms have significantly increased the genomic databank and, consequently, led to more proteomics studies. For the metabolomics studies, likely the low concentration of some metabolites might be challenging to measure confidently with the current techniques. Besides, the extraction of metabolites is usually lengthy and do not guarantee that the quenching of metabolites during the process is sufficient to avoid misinterpretation of the data. Indeed, improved techniques based on LAESI or MALDI-Imaging that can extract and measure the metabolites in situ with high sensitivity will be a breakthrough in the area.
Another important point is the “transcriptomic supremacy.” The recent trend focuses on transcriptomic analyses to obtain global data on plant-bacteria interactions, including the grass-bacteria interactions, for which the number of studies is remarkably more significant. High throughput gene expression analysis, such as cDNA microarrays and transcriptome analyses through next-generation sequencing (RNA-seq), has aided in elucidating plant responses to bacteria colonization. However, proteomics has certain advantages over transcriptome analyses because it analyses protein levels, avoiding misinterpretation from post-transcriptional and translational regulation when only mRNA levels are considered. Therefore, proteomics analyses are still essential for understanding the complex molecular mechanisms involved in plant-bacteria associations, specifically, the beneficial PGPR and grass interactions. Of course, as the concentrations of the metabolites into the cell are dependent on the transcriptomic and proteomic status, metabolomics analysis turns indispensable to have a complete understanding of the grasses-microbe interactions.
Another question is whether the PGPR and grass interactions are an uninteresting topic for proteomics and metabolomics research. The answer seems to be no. The beneficial PGPR and grass interactions involve numerous pathways, which are a scientific topic that must be solved using—omics techniques. In addition, the considerable agriculture interest in grass farming and replacing chemical fertilizer for more sustainable forms of fertilization suggest that employing proteomics/metabolomics to study PGPR-grass interactions will be a fruitful subject for omics research in the next years.
Author Contributions
All authors reviewed the literature and contribute for the manuscript writing and revision.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank the Brazilian Research Council (CNPq), CAPES, Fundação Araucária and Institute of Science and Technology of Biological Nitrogen Fixation (CNPq/INCT-FBN) for financial support.
References
Afroz, A., Zahur, M., Zeeshan, N., and Komatsu, S. (2013). Plant-bacterium interactions analyzed by proteomics. Front. Plant Sci. 4:21. doi: 10.3389/fpls.2013.00021
Agtuca, B. J., Stopka, S. A., Tuleski, T. R., Do Amaral, F. P., Evans, S., Liu, Y., et al. (2020). In-situ metabolomic analysis of Setaria viridis roots colonized by beneficial endophytic bacteria. Mol. Plant Microbe Interact. 33, 272–283. doi: 10.1094/MPMI-06-19-0174-R
Alaux, M., Rogers, J., Letellier, T., Flores, R., Alfama, F., Pommier, C., et al. (2018). Linking the international wheat genome sequencing consortium bread wheat reference genome sequence to wheat genetic and phenomic data. Genome Biol. 19:111. doi: 10.1186/s13059-018-1491-4
Alberton, D., Müller-Santos, M., Brusamarello-Santos, L. C. C., Valdameri, G., Cordeiro, F. A., Yates, M. G., et al. (2013). Comparative proteomics analysis of the rice roots colonized by Herbaspirillum seropedicae strain SmR1 reveals induction of the methionine recycling in the plant host. J. Proteome Res. 12, 4757–4768. doi: 10.1021/pr400425f
Alexandratos, N., and Bruinsma, J. (2012). World Agriculture Towards 2030/2050. The 2012 Revision. 12, 1–146.
Arkhipova, T. N., Prinsen, E., Veselov, S. U., Martinenko, E. V., Melentiev, A. I., and Kudoyarova, G. R. (2007). Cytokinin producing bacteria enhance plant growth in drying soil. Plant Soil. 292, 305–315. doi: 10.1007/s11104-007-9233-5
Babalola, O. O. (2010). Beneficial bacteria of agricultural importance. Biotechnol. Lett. 32, 1559–1570. doi: 10.1007/s10529-010-0347-0
Babcock, G. D., and Esen, A. (1994). Substrate specificity of maize β-glucosidase. Plant Sci.101, 31–39. doi: 10.1016/0168-9452(94)90162-7
Backer, R., Rokem, J. S., Ilangumaran, G., Lamont, J., Praslickova, D., Ricci, E., et al. (2018). Plant growth-promoting rhizobacteria: context, mechanisms of action, and roadmap to commercialization of biostimulants for sustainable agriculture. Front. Plant Sci. 9:1473. doi: 10.3389/fpls.2018.01473
Baldani, V. L. D., Baldani, J. I., and Döbereiner, J. (2000). Inoculation of rice plants with the endophytic diazotrophs Herbaspirillum seropedicae and Burkholderia spp. Biol. Fertil. Soils. 30, 485–491. doi: 10.1007/s003740050027
Bantignies, B., Séguin, J., Muzac, I., Dédaldéchamp, F., Gulick, P., and Ibrahim, R. (2000). Direct evidence for ribonucleolytic activity of a PR-10-like protein from white lupin roots. Plant Mol. Biol. 42, 871–881. doi: 10.1023/A:1006475303115
Bashan, Y., de-Bashan, L. E., Prabhu, S. R., and Hernandez, J. P. (2014). Advances in plant growth-promoting bacterial inoculant technology: Formulations and practical perspectives (1998-2013). Plant Soil. 378, 1–33. doi: 10.1007/s11104-013-1956-x
Beatty, P. H., and Good, A. G. (2011). Plant science. Future prospects for cereals that fix nitrogen. Science 333, 416–417. doi: 10.1126/science.1209467
Bennetzen, J. L., and Ma, J. (2003). The genetic colinearity of rice and other cereals on the basis of genomic sequence analysis. Curr. Opin. Plant Biol. 6, 128–133. doi: 10.1016/S1369-5266(03)00015-3
Bhardwaj, D., Ansari, M. W., Sahoo, R. K., and Tuteja, N. (2014). Biofertilizers function as key player in sustainable agriculture by improving soil fertility, plant tolerance and crop productivity. Microb. Cell Fact. 13:66. doi: 10.1186/1475-2859-13-66
Bhat, M. A., Kumar, V., Bhat, M. A., Wani, I. A., Dar, F. L., Farooq, I., et al. (2020). Mechanistic insights of the interaction of plant growth-promoting rhizobacteria (PGPR) with plant roots toward enhancing plant productivity by alleviating salinity stress. Front. Microbiol. 11, 1–20. doi: 10.3389/fmicb.2020.01952
Bhattacharyya, P. N., and Jha, D. K. (2012). Plant growth-promoting rhizobacteria (PGPR): emergence in agriculture. World J. Microbiol. Biotechnol. 28, 1327–1350. doi: 10.1007/s11274-011-0979-9
Blanchard, D. J., Cicek, M., Chen, J., and Esen, A. (2001). Identification of β-glucosidase aggregating factor (BGAF) and mapping of BGAF binding regions on maize β-glucosidase. J. Biol. Chem. 276, 11895–11901. doi: 10.1074/jbc.M008872200
Bleecker, A. B., and Kende, H. (2000). Ethylene: a gaseous signal molecule in plants. Annu. Rev. Cell Dev. Biol. 16, 1–18. doi: 10.1146/annurev.cellbio.16.1.1
Boddey, R. M., de Oliveira, O. C., Urquiaga, S., Reis, V. M., de Olivares, F. L., Baldani, V. L. D., et al. (1995). Biological nitrogen fixation associated with sugar cane and rice: contributions and prospects for improvement. Plant Soil. 174, 195–209. doi: 10.1007/BF00032247
Bottini, R., Cassán, F., and Piccoli, P. (2004). Gibberellin production by bacteria and its involvement in plant growth promotion and yield increase. Appl. Microbiol. Biotechnol. 65, 497–503. doi: 10.1007/s00253-004-1696-1
Broyart, C., Fontaine, J.-X., Molinié, R., Cailleu, D., Tercé-Laforgue, T., Dubois, F., et al. (2010). Metabolic profiling of maize mutants deficient for two glutamine synthetase isoenzymes using 1 H-NMR-based metabolomics. Phytochem. Anal. 21, 102–109. doi: 10.1002/pca.1177
Brusamarello-Santos, L. C., Gilard, F., Brulé, L., Quilleré, I., Gourion, B., Ratet, P., et al. (2017). Metabolic profiling of two maize (Zea mays L.) inbred lines inoculated with the nitrogen fixing plant-interacting bacteria Herbaspirillum seropedicae and Azospirillum brasilense. PLoS ONE 12:e0174576. doi: 10.1371/journal.pone.0174576
Brusamarello-Santos, L. C. C., Alberton, D., Valdameri, G., Camilios-Neto, D., Covre, R., Lopes, K., et al. (2019). Modulation of defence and iron homeostasis genes in rice roots by the diazotrophic endophyte Herbaspirillum seropedicae. Sci. Rep. 9:10573. doi: 10.1038/s41598-019-45866-w
Brusamarello-Santos, L. C. C., Pacheco, F., Aljanabi, S. M. M., Monteiro, R. A., Cruz, L. M., Baura, V. A., et al. (2012). Differential gene expression of rice roots inoculated with the diazotroph Herbaspirillum seropedicae. Plant Soil 356, 113–125. doi: 10.1007/s11104-011-1044-z
Burgess, B. K., and Lowe, D. J. (1996). Mechanism of molybdenum nitrogenase. Chem. Rev. 96, 2983–3012. doi: 10.1021/cr950055x
Camilios-Neto, D., Bonato, P., Wassem, R., Tadra-Sfeir, M. Z., Brusamarello-Santos, L. C. C., Valdameri, G., et al. (2014). Dual RNA-seq transcriptional analysis of wheat roots colonized by Azospirillum brasilense reveals up-regulation of nutrient acquisition and cell cycle genes. BMC Genomics 15:378. doi: 10.1186/1471-2164-15-378
Cangahuala-Inocente, G. C., Plucani do Amaral, F., Faleiro, A. C., Huergo, L. F., and Maisonnave Arisi, A. C. (2013). Identification of six differentially accumulated proteins of Zea mays seedlings (DKB240 variety) inoculated with Azospirillum brasilense strain FP2. Eur. J. Soil Biol. 58, 45–50. doi: 10.1016/j.ejsobi.2013.06.002
Carpenter, S. R., Caraco, N. F., Correll, D. L., Howarth, R. W., Sharpley, A. N., and Smith, V. H. (1998). Nonpoint pollution of surface waters with phosphorus and nitrogen. Ecol. Appl. 8, 559–568. doi: 10.1890/1051-076119980080559:NPOSWW2.0.CO;2
Cassán, F., Bottini, R., Schneider, G., and Piccoli, P. (2001). Azospirillum brasilense and Azospirillum lipoferum hydrolyze conjugates of Ga20 and metabolize the resultant aglycones to Ga1 in seedlings of rice dwarf mutants. Plant Physiol. 125, 2053–2058. doi: 10.1104/pp.125.4.2053
Cassán, F. D., Lucangeli, C. D., Bottini, R., and Piccoli, P. N. (2001). Azospirillum spp. metabolize [17,17-2H2] gibberellin A20 to [17,17-2H2] gibberellin A1 in vivo in dy rice mutant seedlings. Plant Cell Physiol. 42, 763–767. doi: 10.1093/pcp/pce099
Chamam, A., Sanguin, H., Bellvert, F., Meiffren, G., Comte, G., Wisniewski-Dyé, F., et al. (2013). Plant secondary metabolite profiling evidences strain-dependent effect in the Azospirillum-Oryza sativa association. Phytochemistry 87, 65–77. doi: 10.1016/j.phytochem.2012.11.009
Cheng, Z., Duan, J., Hao, Y., McConkey, B. J., and Glick, B. R. (2009). Identification of bacterial proteins mediating the interactions between Pseudomonas putida UW4 and Brassica napus (Canola). Mol. Plant. Microbe. Interact. 22, 686–694. doi: 10.1094/MPMI-22-6-0686
Cheng, Z., McConkey, B. J., and Glick, B. R. (2010). Proteomic studies of plant-bacterial interactions. Soil Biol. Biochem. 42, 1673–1684. doi: 10.1016/j.soilbio.2010.05.033
Chi, F., Yang, P., Han, F., Jing, Y., and Shen, S. (2010). Proteomic analysis of rice seedlings infected by Sinorhizobium meliloti 1021. Proteomics 10, 1861–1874. doi: 10.1002/pmic.200900694
Choi, W. G., Toyota, M., Kim, S. H., Hilleary, R., and Gilroy, S. (2014). Salt stress-induced Ca2+ waves are associated with rapid, long-distance root-to-shoot signaling in plants. Proc. Natl. Acad. Sci. U.S.A. 111, 6497–6502. doi: 10.1073/pnas.1319955111
Chung, I. M., Park, S. K., Thiruvengadam, M., Lee, J. H., Kim, S. H., and Rajakumar, G. (2018). Review of the biotechnological applications of rice allelopathy in agricultural production. Weed Biol. Manag. 18, 63–74. doi: 10.1111/wbm.12145
Compant, S., Clément, C., and Sessitsch, A. (2010). Plant growth-promoting bacteria in the rhizo- and endosphere of plants: Their role, colonization, mechanisms involved and prospects for utilization. Soil Biol. Biochem. 42, 669–678. doi: 10.1016/j.soilbio.2009.11.024
Cosgrove, D. J. (2005). Growth of the plant cell wall. Nat. Rev. Mol. Cell Biol. 6, 850–861. doi: 10.1038/nrm1746
Curatti, L., and Rubio, L. M. (2014). Challenges to develop nitrogen-fixing cereals by direct nif-gene transfer. Plant Sci. 225, 130–137. doi: 10.1016/j.plantsci.2014.06.003
De Vleesschauwer, D., Djavaheri, M., Bakker, P. A. H. M., and Höfte, M. (2008). Pseudomonas fluorescens WCS374r-induced systemic resistance in rice against Magnaporthe oryzae is based on pseudobactin-mediated priming for a salicylic acid-repressible multifaceted defense response. Plant Physiol. 148, 1996–2012. doi: 10.1104/pp.108.127878
Dhawi, F., Datta, R., and Ramakrishna, W. (2015). Mycorrhiza and PGPB modulate maize biomass, nutrient uptake and metabolic pathways in maize grown in mining-impacted soil. Plant Physiol. Biochem. 97, 390–399. doi: 10.1016/j.plaphy.2015.10.028
Dhawi, F., Datta, R., and Ramakrishna, W. (2017). Proteomics provides insights into biological pathways altered by plant growth promoting bacteria and arbuscular mycorrhiza in sorghum grown in marginal soil. Biochim. Biophys. Acta Proteins Proteomics 1865, 243–251. doi: 10.1016/j.bbapap.2016.11.015
Dhawi, F., Datta, R., and Ramakrishna, W. (2018). Metabolomics, biomass and lignocellulosic total sugars analysis in foxtail millet (Setaria italica) inoculated with different combinations of plant growth promoting bacteria and mycorrhiza. Commun. Plant Sci. 8, 8–14. doi: 10.26814/cps2018002
Dicke, M., and van Loon, J. J. A. (2014). Chemical ecology of phytohormones: how plants integrate responses to complex and dynamic environments. J. Chem. Ecol. 40, 653–656. doi: 10.1007/s10886-014-0479-0
Dimkpa, C., Weinand, T., and Asch, F. (2009). Plant-rhizobacteria interactions alleviate abiotic stress conditions. Plant Cell Environ. 32, 1682–1694. doi: 10.1111/j.1365-3040.2009.02028.x
Dixon, R., and Kahn, D. (2004). Genetic regulation of biological nitrogen fixation. Nat. Rev. Microbiol. 2, 621–631. doi: 10.1038/nrmicro954
Dixon, R. A., Achnine, L., Kota, P., Liu, C.-J., Reddy, M. S. S., and Wang, L. (2002). The phenylpropanoid pathway and plant defence-a genomics perspective. Mol. Plant Pathol. 3, 371–390. doi: 10.1046/j.1364-3703.2002.00131.x
Dobbelaere, S., Croonenborghs, A., Thys, A., Vande Broek, A., and Vanderleyden, J. (1999). Phytostimulatory effect of Azospirillum brasilense wild type and mutant strains altered in IAA production on wheat. Plant Soil 212, 53–162. doi: 10.1023/A:1004658000815
Dodd, I. C., Zinovkina, N. Y., Safronova, V. I., and Belimov, A. A. (2010). Rhizobacterial mediation of plant hormone status. Ann. Appl. Biol. 157, 361–379. doi: 10.1111/j.1744-7348.2010.00439.x
dos Santos, M. F., Muniz de Pádua, V. L., de Matos Nogueira, E., Hemerly, A. S., and Domont, G. B. (2010). Proteome of Gluconacetobacter diazotrophicus co-cultivated with sugarcane plantlets. J. Proteomics 73, 917–931. doi: 10.1016/j.jprot.2009.12.005
dos Santos, P. C., Fang, Z., Mason, S. W., Setubal, J. C., and Dixon, R. (2012). Distribution of nitrogen fixation and nitrogenase-like sequences amongst microbial genomes. BMC Genomics 13:162. doi: 10.1186/1471-2164-13-162
Drogue, B., Doré, H., Borland, S., Wisniewski-Dyé, F., and Prigent-Combaret, C. (2012). Which specificity in cooperation between phytostimulating rhizobacteria and plants? Res. Microbiol. 163, 500–510. doi: 10.1016/j.resmic.2012.08.006
Duca, D., Lorv, J., Patten, C. L., Rose, D., and Glick, B. R. (2014). Indole-3-acetic acid in plant–microbe interactions. Antonie Van Leeuwenhoek 106, 85–125. doi: 10.1007/s10482-013-0095-y
Dutta, S., and Podile, A. R. (2010). Plant growth promoting rhizobacteria (PGPR): the bugs to debug the root zone. Crit. Rev. Microbiol. 36, 232–244. doi: 10.3109/10408411003766806
Egener, T., Hurek, T., and Reinhold-Hurek, B. (1999). Endophytic expression of nif genes of Azoarcus sp. strain BH72 in rice roots. Mol. Plant Microbe Interact. 12, 813–819. doi: 10.1094/MPMI.1999.12.9.813
Erisman, J. W., Sutton, M. A., Galloway, J., Klimont, Z., and Winiwarter, W. (2008). How a century of ammonia synthesis changed the world. Nat. Geosci. 1, 636–639. doi: 10.1038/ngeo325
Etalo, D. W., Díez-Simón, C., de Vos, R. C. H., and Hall, R. D. (2018). Laser ablation electrospray ionization-mass spectrometry imaging (LAESI-MS) for spatially resolved plant metabolomics. Methods Mol. Biol. 1778, 253–267. doi: 10.1007/978-1-4939-7819-9_18
Faleiro, A. C., Neto, P. A. V., de Souza, T. V., Santos, M., and Arisi, A. C. M. (2015). Microscopic and proteomic analysis of Zea mays roots (P30F53 variety) inoculated with Azospirillum brasilense strain FP2. J. Crop Sci. Biotech. 18, 63–71. doi: 10.1007/s12892-014-0061-x
Fey, S. J., and Larsen, P. M. (2001). 2D or not 2D. Curr. Opin. Chem. Biol. 5, 26–33. doi: 10.1016/S1367-5931(00)00167-8
Fibach-Paldi, S., Burdman, S., and Okon, Y. (2012). Key physiological properties contributing to rhizosphere adaptation and plant growth promotion abilities of Azospirillum brasilense. FEMS Microbiol. Lett. 326, 99–108. doi: 10.1111/j.1574-6968.2011.02407.x
Fisher, K., and Newton, W. E. (2005). Nitrogenase proteins from Gluconacetobacter diazotrophicus, a sugarcane-colonizing bacterium. Biochim. Biophys. Acta Proteins Proteomics 1750, 154–165. doi: 10.1016/j.bbapap.2005.04.010
Franche, C., Lindström, K., and Elmerich, C. (2009). Nitrogen-fixing bacteria associated with leguminous and non-leguminous plants. Plant Soil 321, 35–59. doi: 10.1007/s11104-008-9833-8
Frey, M., Chomet, P., Glawischnig, E., Stettner, C., Grün, S., Winklmair, A., et al. (1997). Analysis of a chemical plant defense mechanism in grasses. Science 277, 696–699. doi: 10.1126/science.277.5326.696
Fry, S. C., Smith, R. C., Renwick, K. F., Martin, D. J., Hodge, S. K., and Matthews, K. J. (1992). Xyloglucan endotransglycosylase, a new wall-loosening enzyme activity from plants. Biochem. J. 282, 821–828. doi: 10.1042/bj2820821
Fukami, J., Cerezini, P., and Hungria, M. (2018). Azospirillum: benefits that go far beyond biological nitrogen fixation. AMB Express 8:73. doi: 10.1186/s13568-018-0608-1
Galloway, J. N., and Cowling, E. B. (2002). Reactive nitrogen and the world: 200 years of change. AMBIO. 31, 64–71. doi: 10.1579/0044-7447-31.2.64
Ghosh, D., and Xu, J. (2014). Abiotic stress responses in plant roots: a proteomics perspective. Front. Plant Sci. 5:6. doi: 10.3389/fpls.2014.00006
Glick, B. R. (2012). Plant growth-promoting bacteria: mechanisms and applications. Scientifica 2012:963401. doi: 10.6064/2012/963401
Glick, B. R., Penrose, D. M., and Li, J. (1998). A model for the lowering of plant ethylene concentrations by plant growth-promoting bacteria. J. Theor. Biol. 190, 63–68. doi: 10.1006/jtbi.1997.0532
Goff, S. A., Ricke, D., Lan, T. H., Presting, G., Wang, R., Dunn, M., et al. (2002). A draft sequence of the rice genome (Oryza sativa L. ssp. japonica). Science 296, 92–100. doi: 10.1126/science.1068275
Goh, C.-H., Veliz Vallejos, D. F., Nicotra, A. B., and Mathesius, U. (2013). The impact of beneficial plant-associated microbes on plant phenotypic plasticity. J. Chem. Ecol. 39, 826–839. doi: 10.1007/s10886-013-0326-8
Görg, A., Drews, O., Lück, C., Weiland, F., and Weiss, W. (2009). 2-DE with IPGs. Electrophoresis 30, S122–S132. doi: 10.1002/elps.200900051
Gyaneshwar, P., James, E. K., Reddy, P. M., and Ladha, J. K. (2002). Herbaspirillum colonization increases growth and nitrogen accumulation in aluminium-tolerant rice varieties. New Phytol. 154, 131–145. doi: 10.1046/j.1469-8137.2002.00371.x
Hall, S. J., Matson, P. A., and Roth, P. M. (1996). NOx emissions from soil: implications for air quality modeling in agricultural regions. Annu. Rev. Energy Environ. 21, 311–346. doi: 10.1146/annurev.energy.21.1.311
Hardoim, P. R., van Overbeek, L. S., and van Elsas, J. D. (2008). Properties of bacterial endophytes and their proposed role in plant growth. Trends Microbiol. 16, 463–471. doi: 10.1016/j.tim.2008.07.008
Hartmann, A., and Burris, R. H. (1987). Regulation of nitrogenase activity by oxygen in Azospirillum brasilense and Azospirillum lipoferum. J. Bacteriol. 169, 944–948. doi: 10.1128/JB.169.3.944-948.1987
Hungria, M., Campo, R. J., Souza, E. M., and Pedrosa, F. O. (2010). Inoculation with selected strains of Azospirillum brasilense and A. lipoferum improves yields of maize and wheat in Brazil. Plant Soil 331, 413–425. doi: 10.1007/s11104-009-0262-0
IRGSP (International Rice Genome Sequencing Project). (2005). The map-based sequence of the rice genome. Nature 436, 793–800. doi: 10.1038/nature03895
Isaacson, T., Damasceno, C. M. B., Saravanan, R. S., He, Y., Catalá, C., Saladié, M., et al. (2006). Sample extraction techniques for enhanced proteomic analysis of plant tissues. Nat. Protoc. 1, 769–774. doi: 10.1038/nprot.2006.102
IWGSC (2018). International wheat genome sequencing consortium(IWGSC). Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 361:eaar7191. doi: 10.1126/science.aar7191
James, E. K. (2000). Nitrogen fixation in endophytic and associative symbiosis. Field Crop. Res. 65, 197–209. doi: 10.1016/S0378-4290(99)00087-8
Jan, A., Yang, G., Nakamura, H., Ichikawa, H., Kitano, H., Matsuoka, M., et al. (2004). Characterization of a xyloglucan endotransglucosylase gene that is up-regulated by gibberellin in rice. Plant Physiol. 136, 3670–3681. doi: 10.1104/pp.104.052274
Jetiyanon, K., and Kloepper, J. W. (2002). Mixtures of plant growth-promoting rhizobacteria for induction of systemic resistance against multiple plant diseases. Biol. Control. 24, 285–291. doi: 10.1016/S1049-9644(02)00022-1
Jin, S., Kanagaraj, A., Verma, D., Lange, T., and Daniell, H. (2011). Release of hormones from conjugates: chloroplast expression of β-glucosidase results in elevated phytohormone levels associated with significant increase in biomass and protection from aphids or whiteflies conferred by sucrose esters. Plant Physiol. 155, 222–235. doi: 10.1104/pp.110.160754
Johansson, J. F., Paul, L. R., and Finlay, R. D. (2004). Microbial interactions in the mycorrhizosphere and their significance for sustainable agriculture. FEMS Microbiol. Ecol. 48, 1–13. doi: 10.1016/j.femsec.2003.11.012
Kandasamy, S., Loganathan, K., Muthuraj, R., Duraisamy, S., Seetharaman, S., Thiruvengadam, R., et al. (2009). Understanding the molecular basis of plant growth promotional effect of Pseudomonas fluorescens on rice through protein profiling. Proteome Sci. 7:47. doi: 10.1186/1477-5956-7-47
Kannojia, P., Choudhary, K. K., Srivastava, A. K., and Singh, A. K. (2019). “Chapter Four - PGPR bioelicitors: induced systemic resistance (ISR) and proteomic perspective on biocontrol,” in PGPR Amelioration in Sustainable Agriculture. Food Security and Environmental Management, 67–84.
Karnwal, A. (2009). Production of indole acetic acid by fluorescent Pseudomonas in the presence of L-tryptophan and rice root exudates. J. Plant Pathol. 91, 61–63.
Keller, B., and Feuillet, C. (2000). Colinearity and gene density in grass genomes. Trends Plant Sci. 5, 246–251. doi: 10.1016/S1360-1385(00)01629-0
Khatabi, B., Gharechahi, J., Ghaffari, M. R., Liu, D., Haynes, P. A., McKay, M. J., et al. (2019). Plant–microbe symbiosis: what has proteomics taught us? Proteomics 19:1800105. doi: 10.1002/pmic.201800105
Kim, S. T., Kim, S. G., Agrawal, G. K., Kikuchi, S., and Rakwal, R. (2014). Rice proteomics: a model system for crop improvement and food security. Proteomics 14, 593–610. doi: 10.1002/pmic.201300388
Kittur, F. S., Lalgondar, M., Yu, H. Y., Bevan, D. R., and Esen, A. (2007). Maize β-glucosidase-aggregating factor is a polyspecific jacalin-related chimeric lectin, and its lectin domain is responsible for β-glucosidase aggregation. J. Biol. Chem. 282, 7299–7311. doi: 10.1074/jbc.M607417200
Lade, S. B., Román, C., Cueto-Ginzo, A. I., Serrano, L., Sin, E., Achón, M. A., et al. (2018). Host-specific proteomic and growth analysis of maize and tomato seedlings inoculated with Azospirillum brasilense Sp7. Plant Physiol. Biochem. 129, 381–393. doi: 10.1016/j.plaphy.2018.06.024
Lade, S. B., Román, C., del Cueto-Ginzo, A. I., Serrano, L., Sin, E., Achón, M. A., et al. (2019). Differential proteomics analysis reveals that Azospirillium brasilense (Sp7) promotes virus tolerance in maize and tomato seedlings. Eur. J. Plant Pathol. 155, 1241–1263. doi: 10.1007/s10658-019-01852-6
Lery, L. M. S., Hemerly, A. S., Nogueira, E. M., von Krüger, W. M. A., and Bisch, P. M. (2011). Quantitative proteomic analysis of the interaction between the endophytic plant-growth-promoting bacterium Gluconacetobacter diazotrophicus and sugarcane. Mol. Plant Microbe Interact. 24, 562–576. doi: 10.1094/MPMI-08-10-0178
Lugtenberg, B., and Kamilova, F. (2009). Plant-growth-promoting rhizobacteria. Annu. Rev. Microbiol. 63, 541–556. doi: 10.1146/annurev.micro.62.081307.162918
Lymperopoulos, P., Msanne, J., and Rabara, R. (2018). Phytochrome and phytohormones: working in tandem for plant growth and development. Front. Plant Sci. 9:1037. doi: 10.3389/fpls.2018.01037
MacMillan, J. (2001). Occurrence of gibberellins in vascular plants, fungi, and bacteria. J. Plant Growth Regul. 20, 387–442. doi: 10.1007/s003440010038
Mano, Y., and Nemoto, K. (2012). The pathway of auxin biosynthesis in plants. J. Exp. Bot. 63, 2853–2872. doi: 10.1093/jxb/ers091
Mareya, C. R., Tugizimana, F., Piater, L. A., Madala, N. E., Steenkamp, P. A., and Dubery, I. A. (2019). Untargeted metabolomics reveal defensome-related metabolic reprogramming in Sorghum bicolor against infection by Burkholderia andropogonis. Metabolites 9, 1–8. doi: 10.3390/metabo9010008
Martins, M. R., Jantalia, C. P., Reis, V. M., Döwich, I., Polidoro, J. C., Alves, B. J. R., et al. (2018). Impact of plant growth-promoting bacteria on grain yield, protein content, and urea-15 N recovery by maize in a cerrado oxisol. Plant Soil 422, 239–250. doi: 10.1007/s11104-017-3193-1
Masciarelli, O., Urbani, L., Reinoso, H., and Luna, V. (2013). Alternative mechanism for the evaluation of indole-3-acetic acid (IAA) production by Azospirillum brasilense strains and its effects on the germination and growth of maize seedlings. J. Microbiol. 51, 590–597. doi: 10.1007/s12275-013-3136-3
Miché, L., Battistoni, F., Gemmer, S., Belghazi, M., and Reinhold-Hurek, B. (2006). Upregulation of jasmonate-inducible defense proteins and differential colonization of roots of Oryza sativa cultivars with the endophyte Azoarcus sp. Mol. Plant Microbe Interact. 19, 502–511. doi: 10.1094/MPMI-19-0502
Miflin, B. J., and Habash, D. Z. (2002). The role of glutamine synthetase and glutamate dehydrogenase in nitrogen assimilation and possibilities for improvement in the nitrogen utilization of crops. J. Exp. Bot. 53, 979–987. doi: 10.1093/jexbot/53.370.979
Mishra, R. P. N., Singh, R. K., Jaiswal, H. K., Kumar, V., and Maurya, S. (2006). Rhizobium-mediated induction of phenolics and plant growth promotion in rice (Oryza sativa L.). Curr. Microbiol. 52, 383–389. doi: 10.1007/s00284-005-0296-3
Muneer, S., Ahmad, J., Bashir, H., and Qureshi, M. I. (2012). Proteomics of nitrogen fixing nodules under various environmental stresses. Plant Omics. 5, 167–176.
Nadeem, S. M., Ahmad, M., Zahir, Z. A., Javaid, A., and Ashraf, M. (2014). The role of mycorrhizae and plant growth promoting rhizobacteria (PGPR) in improving crop productivity under stressful environments. Biotechnol. Adv. 32, 429–448. doi: 10.1016/j.biotechadv.2013.12.005
Niu, L., Zhang, H., Wu, Z., Wang, Y., Liu, H., Wu, X., et al. (2018). Modified TCA/acetone precipitation of plant proteins for proteomic analysis. PLoS ONE 13:e0202238. doi: 10.1371/journal.pone.0202238
Niu, L., Zhang, H., Wu, Z., Wang, Y., Liu, H., Wu, X., et al. (2019). Correction: modified TCA/acetone precipitation of plant proteins for proteomic analysis. PLoS ONE 14:e0211612. doi: 10.1371/journal.pone.0211612
Oldroyd, G. E. D., and Dixon, R. (2014). Biotechnological solutions to the nitrogen problem. Curr. Opin. Biotechnol. 26, 19–24. doi: 10.1016/j.copbio.2013.08.006
Oliveira, A. L. M., de Canuto, E. L., Urquiaga, S., Reis, V. M., and Baldani, J. I. (2006). Yield of micropropagated sugarcane varieties in different soil types following inoculation with diazotrophic bacteria. Plant Soil 284, 23–32. doi: 10.1007/s11104-006-0025-0
Oliveira, A. L. M., Stoffels, M., Schmid, M., Reis, V. M., Baldani, J. I., and Hartmann, A. (2009). Colonization of sugarcane plantlets by mixed inoculations with diazotrophic bacteria. Eur. J. Soil Biol. 45, 106–113. doi: 10.1016/j.ejsobi.2008.09.004
Penninckx, I. A. M. A., Thomma, B. P. H. J., Buchala, A., Métraux, J.-P., and Broekaert, W. F. (1998). Concomitant activation of jasmonate and ethylene response pathways is required for induction of a plant defensin gene in Arabidopsis. Plant Cell 10, 2103–2113. doi: 10.1105/tpc.10.12.2103
Pérez-Montaño, F., Alías-Villegas, C., Bellogín, R. A., Del Cerro, P., Espuny, M. R., Jiménez-Guerrero, I., et al. (2014). Plant growth promotion in cereal and leguminous agricultural important plants: from microorganism capacities to crop production. Microbiol. Res. 169, 325–336. doi: 10.1016/j.micres.2013.09.011
Pieterse, C. M. J., Van Pelt, J. A., Ton, J., Parchmann, S., Mueller, M. J., Buchala, A. J., et al. (2000). Rhizobacteria-mediated induced systemic resistance (ISR) in Arabidopsis requires sensitivity to jasmonate and ethylene but is not accompanied by an increase in their production. Physiol. Mol. Plant Pathol. 57, 123–134. doi: 10.1006/pmpp.2000.0291
Pieterse, C. M. J., Zamioudis, C., Berendsen, R. L., Weller, D. M., Van Wees, S. C. M., and Bakker, P. A. H. M. (2014). Induced systemic resistance by beneficial microbes. Annu. Rev. Phytopathol. 52, 347–375. doi: 10.1146/annurev-phyto-082712-102340
Portwood, J. L., Woodhouse, M. R., Cannon, E. K., Gardiner, J. M., Harper, L. C., Schaeffer, M. L., et al. (2019). MaizeGDB 2018: the maize multi-genome genetics and genomics database. Nucl. Acids Res. 47, D1146–D1154. doi: 10.1093/nar/gky1046
Pozo, M. J., Van Der Ent, S., Van Loon, L. C., and Pieterse, C. M. J. (2008). Transcription factor MYC2 is involved in priming for enhanced defense during rhizobacteria-induced systemic resistance in Arabidopsis thaliana. New Phytol. 180, 511–523. doi: 10.1111/j.1469-8137.2008.02578.x
Prinsi, B., Negri, A. S., Failla, O., Scienza, A., and Espen, L. (2018). Root proteomic and metabolic analyses reveal specific responses to drought stress in differently tolerant grapevine rootstocks. BMC Plant Biol. 18:126. doi: 10.1186/s12870-018-1343-0
Quirino, B. F., Candido, E. S., Campos, P. F., Franco, O. L., and Krüger, R. H. (2010). Proteomic approaches to study plant–pathogen interactions. Phytochemistry 71, 351–362. doi: 10.1016/j.phytochem.2009.11.005
Rademacher, W. (1994). Gibberellin formation in microorganisms. Plant Growth Regul. 15, 303–314. doi: 10.1007/BF00029903
Reeves, T. G., Thomas, G., and Ramsay, G. (2016). Save and Grow in Practice: Maize, Rice Wheat. A Guide to Sustainable Cereal Production. Roma: Food and Agriculture Organization of the United Nations (FAO).
Roncato-Maccari, L. D. B., Ramos, H. J. O., Pedrosa, F. O., Alquini, Y., Chubatsu, L. S., Yates, M. G., et al. (2003). Endophytic Herbaspirillum seropedicae expresses nif genes in gramineous plants. FEMS Microbiol. Ecol. 45, 39–47. doi: 10.1016/S0168-6496(03)00108-9
Rzewuski, G., and Sauter, M. (2008). Ethylene biosynthesis and signaling in rice. Plant Sci. 175, 32–42. doi: 10.1016/j.plantsci.2008.01.012
Salavati, A., Shafeinia, A., Klubicova, K., Bushehri, A. A. S., and Komatsu, S. (2013). Proteomic insights into intra- and intercellular plant-bacteria symbiotic association during root nodule formation. Front. Plant Sci. 4:28. doi: 10.3389/fpls.2013.00028
Schnable, P. S., Ware, D., Fulton, R. S., Stein, J. C., Wei, F., Pasternak, S., et al. (2009). The B73 maize genome: complexity, diversity, and dynamics. Science 326, 1122–1115. doi: 10.1126/science.1178534
Schultz, N., Pereira, W., de Albuquerque Silva, P., Baldani, J. I., Boddey, R. M., Alves, B. J. R., et al. (2017). Yield of sugarcane varieties and their sugar quality grown in different soil types and inoculated with a diazotrophic bacteria consortium. Plant Prod. Sci. 20, 366–374. doi: 10.1080/1343943X.2017.1374869
Seefeldt, L. C., Hoffman, B. M., and Dean, D. R. (2009). Mechanism of mo-dependent nitrogenase. Annu. Rev. Biochem. 78, 701–722. doi: 10.1146/annurev.biochem.78.070907.103812
Senthil-Kumar, M., and Mysore, K. S. (2013). Nonhost resistance against bacterial pathogens: retrospectives and prospects. Annu. Rev. Phytopathol. 51, 407–427. doi: 10.1146/annurev-phyto-082712-102319
Shahzad, R., Waqas, M., Khan, A. L., Asaf, S., Khan, M. A., Kang, S.-M., et al. (2016). Seed-borne endophytic Bacillus amyloliquefaciens RWL-1 produces gibberellins and regulates endogenous phytohormones of Oryza sativa. Plant Physiol. Biochem. 106, 236–243. doi: 10.1016/j.plaphy.2016.05.006
Sharma, P., Jha, A. B., Dubey, R. S., and Pessarakli, M. (2012). Reactive oxygen species, oxidative damage, and antioxidative defense mechanism in plants under stressful conditions. J. Bot. 2012:217037. doi: 10.1155/2012/217037
Singh, J. S., Pandey, V. C., and Singh, D. P. (2011). Efficient soil microorganisms: a new dimension for sustainable agriculture and environmental development. Agric. Ecosyst. Environ. 140, 339–353. doi: 10.1016/j.agee.2011.01.017
Singh, R. P., Runthala, A., Khan, S., and Jha, P. N. (2017). Quantitative proteomics analysis reveals the tolerance of wheat to salt stress in response to Enterobacter cloacae SBP-8. PLoS ONE 12:e0183513. doi: 10.1371/journal.pone.0183513
Sleator, R. (2001). Bacterial osmoadaptation: the role of osmolytes in bacterial stress and virulence. FEMS Microbiol. Rev. 26, 49–51. doi: 10.1111/j.1574-6976.2002.tb00598.x
Smith, B. E., Richards, R. L., and Newton, W. E. (2004). Catalysts for Nitrogen Fixation: Nitrogenases, Relevant Chemical Models and Commercial Processes. Boston, MA: Kluwer Academic Publishers.
Spaepen, S., and Vanderleyden, J. (2011). Auxin and plant-microbe interactions. Cold Spring Harb. Perspect. Biol. 3:a001438. doi: 10.1101/cshperspect.a001438
Spaepen, S., Vanderleyden, J., and Remans, R. (2007). Indole-3-acetic acid in microbial and microorganism-plant signaling. FEMS Microbiol. Rev. 31, 425–448. doi: 10.1111/j.1574-6976.2007.00072.x
Straub, D., Yang, H., Liu, Y., Tsap, T., and Ludewig, U. (2013). Root ethylene signalling is involved in Miscanthus sinensis growth promotion by the bacterial endophyte Herbaspirillum frisingense GSF30 T. J. Exp. Bot. 64, 4603–15. doi: 10.1093/jxb/ert276
Swarbreck, S. M., Defoin-Platel, M., Hindle, M., Saqi, M., and Habash, D. Z. (2011). New perspectives on glutamine synthetase in grasses. J. Exp. Bot. 62, 1511–1522. doi: 10.1093/jxb/erq356
Tariq, M., Hameed, S., Malik, K. A., and Hafeez, F. Y. (2007). Plant root associated bacteria for zinc mobilization in rice. Pakistan J. Bot. 39, 245–253.
Tilman, D., Cassman, K. G., Matson, P. A., Naylor, R., and Polasky, S. (2002). Agricultural sustainability and intensive production practices. Nature 418, 671–677. doi: 10.1038/nature01014
Tsukanova, K. A., Chebotar, V., Meyer, J. J. M., and Bibikova, T. N. (2017). Effect of plant growth-promoting Rhizobacteria on plant hormone homeostasis. South Afr. J. Bot. 113, 91–102. doi: 10.1016/j.sajb.2017.07.007
Tugizimana, F., Mhlongo, M. I., Piater, L. A., and Dubery, I. A. (2018). Metabolomics in plant priming research: the way forward? Int. J. Mol. Sci. 19:1759. doi: 10.3390/ijms19061759
United Nations, New York (2019). World Population Prospects 2019: Highlights. Department of Economic and Social Affairs, Population Division (ST/ESA/SER.A/423).
Vacheron, J., Desbrosses, G., Bouffaud, M.-L., Touraine, B., Moënne-Loccoz, Y., Muller, D., et al. (2013). Plant growth-promoting rhizobacteria and root system functioning. Front. Plant Sci. 4:356. doi: 10.3389/fpls.2013.00356
Valdameri, G., Alberton, D., Moure, V. R., Kokot, T. B., Kukolj, C., Brusamarello-Santos, L. C. C., et al. (2017). Herbaspirillum rubrisubalbicans, a mild pathogen impairs growth of rice by augmenting ethylene levels. Plant Mol. Biol. 94, 625–640. doi: 10.1007/s11103-017-0629-1
Walker, V., Bertrand, C., Bellvert, F., Moënne-Loccoz, Y., Bally, R., and Comte, G. (2011). Host plant secondary metabolite profiling shows a complex, strain-dependent response of maize to plant growth-promoting rhizobacteria of the genus Azospirillum. New Phytol. 189, 494–506. doi: 10.1111/j.1469-8137.2010.03484.x
Walker, V., Couillerot, O., Von Felten, A., Bellvert, F., Jansa, J., Maurhofer, M., et al. (2012). Variation of secondary metabolite levels in maize seedling roots induced by inoculation with Azospirillum, Pseudomonas and Glomus consortium under field conditions. Plant Soil 356, 151–163. doi: 10.1007/s11104-011-0960-2
Walters, D. R., Ratsep, J., and Havis, N. D. (2013). Controlling crop diseases using induced resistance: challenges for the future. J. Exp. Bot. 64, 1263–80. doi: 10.1093/jxb/ert026
Wang, K. L. C., Li, H., and Ecker, J. R. (2002). Ethylene biosynthesis and signaling networks. Plant Cell. 14, S131–151. doi: 10.1105/tpc.001768
Wang, W., Chen, L. N., Wu, H., Zang, H., Gao, S., Yang, Y., et al. (2013). Comparative proteomic analysis of rice seedlings in response to inoculation with Bacillus cereus. Lett. Appl. Microbiol. 56, 208–15. doi: 10.1111/lam.12035
Wang, Z. Q., Yuan, Y. Z., Ou, J. Q., Lin, Q. H., and Zhang, C. F. (2007). Glutamine synthetase and glutamate dehydrogenase contribute differentially to proline accumulation in leaves of wheat (Triticum aestivum) seedlings exposed to different salinity. J. Plant Physiol. 164, 695–701. doi: 10.1016/j.jplph.2006.05.001
Williams, C. E., Collier, C. C., Nemacheck, J. A., Liang, C., and Cambron, S. E. (2002). A lectin-like wheat gene responds systemically to attempted feeding by avirulent first-instar hessian fly larvae. J. Chem. Ecol. 28, 1411–28. doi: 10.1023/A:1016200619766
Woodward, A. W. (2005). Auxin: regulation, action, and interaction. Ann. Bot. 95, 707–735. doi: 10.1093/aob/mci083
Wu, X., Xiong, E., Wang, W., Scali, M., and Cresti, M. (2014). Universal sample preparation method integrating trichloroacetic acid/acetone precipitation with phenol extraction for crop proteomic analysis. Nat. Protoc. 9, 362–374. doi: 10.1038/nprot.2014.022
Yasuda, M., Isawa, T., Shinozaki, S., Minamisawa, K., and Nakashita, H. (2009). Effects of Colonization of a Bacterial Endophyte, Azospirillum sp. B510, on Disease Resistance in Rice. Biosci. Biotechnol. Biochem. 73, 2595–2599. doi: 10.1271/bbb.90402
Keywords: plant-bacteria association, Plant growth-promoting rhizobacteria (PGPR), grass, nitrogen fixation, proteomics, metabolomics
Citation: Alberton D, Valdameri G, Moure VR, Monteiro RA, Pedrosa FO, Müller-Santos M and de Souza EM (2020) What Did We Learn From Plant Growth-Promoting Rhizobacteria (PGPR)-Grass Associations Studies Through Proteomic and Metabolomic Approaches? Front. Sustain. Food Syst. 4:607343. doi: 10.3389/fsufs.2020.607343
Received: 17 September 2020; Accepted: 03 December 2020;
Published: 23 December 2020.
Edited by:
Everlon Cid Rigobelo, Universidade Estadual Paulista, BrazilReviewed by:
Naeem Khan, University of Florida, United StatesAjar Nath Yadav, Eternal University, India
Copyright © 2020 Alberton, Valdameri, Moure, Monteiro, Pedrosa, Müller-Santos and de Souza. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Emanuel Maltempi de Souza, c291emFlbUB1ZnByLmJy
 Dayane Alberton
Dayane Alberton Glaucio Valdameri1
Glaucio Valdameri1 Vivian Rotuno Moure
Vivian Rotuno Moure Marcelo Müller-Santos
Marcelo Müller-Santos Emanuel Maltempi de Souza
Emanuel Maltempi de Souza