
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Sustain. Food Syst. , 01 February 2021
Sec. Crop Biology and Sustainability
Volume 4 - 2020 | https://doi.org/10.3389/fsufs.2020.606618
This article is part of the Research Topic Plant Growth-Promoting Microorganisms for Sustainable Agricultural Production View all 50 articles
Organic farming systems are gaining popularity as agronomically and environmentally sound soil management strategies with potential to enhance soil microbial diversity and fertility, environmental quality and sustainable crop production. This work aimed at understanding the effect of organic and conventional farming on the diversity of soybean nodulating bradyrhizobia species. Field trapping of indigenous soybean Bradyrhizobium was done by planting promiscuous soybeans varieties SB16 and SC squire as well as non-promiscuous Gazelle in three organic and three conventional farms in Tharaka-Nithi County of Kenya. After 45 days of growth, 108 nodule isolates were obtained from the soybean nodules and placed into 13 groups based on their morphological characteristics. Genetic diversity was done by polymerase chain reaction (PCR) targeting 16S rDNA gene using universal primers P5-R and P3-F and sequencing was carried out using the same primer. High morphological and genetic diversity of the nodule isolates was observed in organic farms as opposed to conventional farms. There was little or no genetic differentiation between the nodule isolates from the different farms with the highest molecular variation (91.12%) being partitioned within populations as opposed to among populations (8.88%). All the isolates were identified as bradyrhizobia with close evolutionary ties with Bradyrhizobium japonicum and Bradyrhizobium yuanminense. Organic farming systems favor the proliferation of bradyrhizobia species and therefore a suitable environmentally friendly alternative for enhancing soybean production.
Human population has risen steadily over time, soils lost fertility and arable land are scarce leading to increased pressure on agricultural systems to provide food for the ever-increasing population. This has led to drastic land-use changes that involve intensive agricultural practices that has affected biodiversity driven ecosystem processes on the local and global scale (Foley et al., 2005). Most African countries have agriculture as a main component of their economies and organic agriculture plays an important role for not only assuring sustainable food security but also essential for environmental management (Grandi, 2008; Mutoni et al., 2017). Organic farming enhances the population and diversity of beneficial indigenous bradyrhizobia in the soil which in turn boosts soybean production through biological nitrogen fixation (Wongphatcharachai et al., 2015). Therefore, it is crucial to evaluate the effects of organic and conventional farming systems on soil microbial diversity since farming systems may either increase or suppress the diversity and activity of soil microorganisms.
Conventional farming system of soybean involves the use of inorganic fertilizers, herbicides and pesticides and other farm inputs to optimize production. Most of these inputs contribute significantly to environmental degradation (Abou-Shanab et al., 2017) and are not sustainable in smallholder farming systems. Soil nutrient management strategies can enhance soybean production without detrimental effects to the environment (Hellal and Abdelhamid, 2013). Soil microorganisms that inhabit the plant rhizosphere are important as pest biocontrol agents, enhancement of nutrient utilization and promotion of plant growth (Igiehon et al., 2019). Conventional farm management practices have caused soil erosion, increased salinization, pollution due to agrochemicals, degrading effect on the environment, danger to food security; quality and safety, reduced biodiversity and lack of sustainability in agriculture. The adverse effects of prolonged use of inorganic agrochemicals in conventional farming system have led to increased demands to make agricultural systems more sustainable by reforming agricultural practices. These concerns and negative impacts posed by conventional farming gave birth to organic farming.
Organic farming is a system which avoids the use of synthetic inputs but embrace biological system of nutrient mobilization and plant protection. Organic farming system is characterized by strict limitation of chemical fertilizers, herbicides and pesticides, where soil management is through addition of organic materials and other strategies such as crop rotation (IFOAM, 2006). This mode of farming relies more on agro-ecological services than on the use of expensive external inputs. Therefore, compared to conventional system, organic farming is sustainable, cost effective, environmentally friendly, and enhances soil fertility and biodiversity.
Beneficial soil microorganisms are critical in organic farming systems due to their potential to promote soil fertility and maximize on nutrient cycles. Symbiotic bradyrhizobia-legume interaction that results to biological nitrogen fixation (BNF) can be a suitable alternative for improvement of soybean production (Thuita et al., 2012). Inoculation of soybean with effective indigenous or exotic nitrogen fixing bacteria has been touted as the most environmentally friendly way of improving its production (Ulzen et al., 2016; Ndusha et al., 2019). However, the inoculant isolates should be selected based on their effectiveness in fixing nitrogen and their competitiveness. Introduction of exotic inoculant strains may experience indigenous isolates competition in the soil that limits their nitrogen fixing ability leading to little or no realization of their potential (Heerwaarden et al., 2018). Other than nitrogen fixation, some bradyrhizobia species produce indole-3-acetic acid (IAA) which enhances the survival of soybean crops (Igiehon et al., 2019). Soybean varieties have been reported to express host specificity and therefore, fail to form symbiotic interactions with indigenous bradyrhizobia when introduced in farms for the first time (Thuita et al., 2012). Tropical Glycine cross (TGx) genotypes of soybean that were developed by International Institute of Tropical Agriculture (IITA) are promiscuous and capable of effective symbiotic interaction with indigenous bradyrhizobia communities in African soils (Chibeba et al., 2017). Soils with no previous history of soybean cultivation may not have suitable population of bradyrhizobia for appropriate nodulation of the crop, that poses another challenge to the reliance on indigenous bradyrhizobia isolates for soybean production enhancement (Abbasi et al., 2008).
Farming practices including tillage, use of organic and inorganic fertilizers and cropping patterns adjust the soil physicochemical parameters, which in turn influence the diversity and number of soil microorganisms (Schmidt et al., 2019). Various studies have demonstrated that the diversity of bradyrhizobia in agroecosystems is as a result of interaction between the bradyrhizobia and legume species, other bradyrhizobia as well as biotic and abiotic factors in that ecosystem (Yan et al., 2014). The diversity of soybean nodulating bradyrhizobia in agroecosystems is not only related to the crop genotype and geographical location of the site but also the soil conditions including pH, available phosphorus and organic matter (Yan et al., 2014).
Farmers in Sub-Sahara Africa mainly cultivate traditional crops as the main bulk of their agricultural production. However, non-traditional crops like soybean can be a worthy investment for enhanced agricultural productivity (Chianu et al., 2009). Soybean is one of the most traded legumes in the world accounting for over 84.5% of the traded grain legumes (Varshney et al., 2019). It is a highly traded crop due to its importance, as a major source of vegetable oil and a rich protein source for human food and animal feeds (Abou-Shanab et al., 2017). Kenya requires 120,000 metric tons of soybean grain per year. However, it produces only 2007 metric tons covering the deficit through importation (Murage et al., 2019). Between the years 2013 to 2016, Sub Saharan African countries imported 6.8 million tons of soybean grain of which Kenya imported 18% of this volume. This signifies continually rising demand for soybean in Africa (Muriithi et al., 2016).
As a result of climate change, global crop production is estimated to decline owing to stressful biotic and abiotic factors (Raza et al., 2019). Soybean is a hardy crop that can easily be cultivated in soils varying greatly in texture, fertility and in fluctuating climatic conditions (Cely et al., 2016). Soybean yield losses due to soil deficiencies have been reported. However, they differ depending on the nutrients; with soil N deficiency contributing 10 percent of the yield losses (Hellal and Abdelhamid, 2013).
Soybean is an exotic crop that originated in China and introduced in Kenya in the early 1960s (Abate et al., 2008). In Kenya, the legume is cultivated as a food diet and for income by poor rural smallholder farmers (Ndungu et al., 2018). However, the growth of soybean in Eastern Kenya by smallholder farmers has not been sustainable in the past one decade (Njeru et al., 2013). Several factors such as nutrient deficiency in soil (Caliskan et al., 2008), inappropriate use of fertilizers (Brahim et al., 2017), and poor soil management practices (Samson et al., 2019) pose a threat to farmers in their cultivation efforts. Although field studies show that organic farming and inoculation tend to increase soybean production; the range of morphological and genetic diversity of indigenous bradyrhizobia that nodulate soybean under the two farming systems in Eastern Kenya has not been tested fully. Moreover, little attention has been focused on the possibility of raising the inoculum potential of indigenous bradyrhizobia by appropriate agricultural management practices; a strategy that would be fundamental in crop production. Knowledge of how indigenous bradyrhizobia strains are affected by agronomic practices is a crucial step toward optimizing utilization of bradyrhizobia in sustainable crop production. Additionally, a better understanding of the diversity of bradyrhizobia strains, their interactions in different agronomic practices and role is important in the development of sustainable agriculture, increased food production, enhanced soil fertility and biodiversity.
In this research, we tested the hypotheses that bradyrhizobia strains that nodulate soybeans under organic and conventional farming are morphologically and genetically diverse. The objectives of this study were to (i) assess the morphological characteristics of soybean nodulating indigenous bradyrhizobia isolated from nodules under organic and conventional farming in Eastern Kenya and (ii) evaluate the genetic diversity of indigenous bradyrhizobia nodulating soybean.
Field experiments were conducted in six family farming systems in Tharaka-Nithi County, Kenya at 0°19′49.7″S and 37°38′52.9″E, three strictly organic and conventional, respectively. All the organic farms had 3 years after conversion. The organic farmers are trained and certified. The area is in the Upper midland 2 and 3 (UM2 and UM3) agroecological zones and has an altitude of ~1,373 m above sea level. It experiences bimodal rains, which range from 1,200 to 1,400 mm and a mean temperature of 20°C annually. The long rains are from March to June, and the short rains are from October to December. The soils are Humic Nitisols, commonly called the red Kikuyu loams; deep, well weathered, typically acidic, and free draining with a friable clay texture and moderate to high fertility (Jaetzold et al., 2006).
Soils samples were collected in September 2015 from the six selected organic and conventionally managed farms. The upper 20 cm of soil was sampled from six different points along, diagonally and across every selected farm prior to the onset of short rain. Samples from each farm were mixed thoroughly to make a homogenous composite sample. The soils were air-dried, ground and passed through a 2-mm sieve prior to analysis. Soil pH was determined using a pH meter in a prepared soil-water suspension ratio of 1:2.5. The soil organic carbon was determined by Walkley-Black combustion method (Ashworth et al., 2014) while percentage nitrogen was determined following the Kjeldahl method (Hanon K9840 Kjeldahl apparatus) (Vauclare et al., 2013). The available phosphorus (P) and potassium (K) were determined according to Mehlich-3 (M-3) procedures (Furseth et al., 2012).
The farms were plowed and prepared for planting in October 2015 prior to the onset of short rains. Field trapping of indigenous bradyrhizobia was carried out in all the six farms using soybeans obtained from Kenya Seed Company Limited, Nairobi. Three soybean varieties; two promiscuous (SC squire and SB 16) and one non-promiscuous (Gazelle) were planted in the identified organic farms designated as HO, JO and GO as well as in conventionally managed farms designated as HC, JC and GC during October to December short rain season. The experiment was set up in a split plot design with farm management as the main factor and soybean variety as the sub-factor and in triplicate, giving a total of 18 treatments. None of the farms had previous history of bradyrhizobia inoculation or soybean cultivation. Organic farms had no recent history of herbicide, pesticide and inorganic fertilizer application while conventional farms had a long history of herbicide, pesticide and inorganic fertilizer application. Forty-five (45) days after crop emergence, three healthy soybean plants were randomly selected and harvested from each of the treatment for nodulation and shoot dry weight analysis. A total of 54 soybean plants were sampled and cut at the cotyledonary node separating shoots from the roots. The roots were carefully washed with distilled water and nodules from each sampled plant were detached, collected, nodule number per plant determined and packed in sterile sample vials containing cotton wool and silica gel to prevent desiccation. The nodules and shoots were transported to the Microbiology laboratory at Kenyatta University where they were air-dried and dry weight assessed. Nodules were stored at 4°C for bradyrhizobia isolation. At physiological maturity (after 125 days), three plants per plot were randomly selected, manually harvested, pods detached and threshed. The recovered seeds were dried to a constant dry weight and weight recorded in g plant−1.
In the laboratory, undamaged nodules showing red or pink coloration were selected from the preserved nodules. The air-dried intact nodules were placed in sterile distilled water and allowed to imbibe water for 1 h. The nodules were then rinsed with sterile distilled water and immersed in ethanol (95% v/v) for 30 s to reduce the surface tension and remove air bubbles from the tissues. The nodules were then sterilized using 3.8% sodium hypochlorite solution for 4 min and finally rinsed with six changes of sterile double-distilled water (Thuita et al., 2012). After rinsing, 10 nodules from each of the treatment samples were picked and crushed with a sterile glass rod in a drop of sterile distilled water using a blunt-tipped pair of flame-sterilized forceps. A loopful of the nodule suspension was aseptically streaked onto a Petri dish plate containing Yeast Extract Mannitol Agar (YEMA) media supplemented with 25 μg/ml Congo red and incubated at 28°C in the dark. Daily observations on colony emergence were made after incubation for 5 days and well-isolated colonies were streaked on YEMA plates containing Congo red. The single colonies were identified and purified by sub-culturing on YEMA media supplemented with Congo red (Mungai and Karubiu, 2011).
After purification, the cultural and biochemical characteristics of the nodule isolates were established through presumptive tests. The indigenous bradyrhizobia isolates were authenticated, confirmed as nodule-forming bacterial strains and grouped based on morphological characteristics (Odee et al., 1997). The nodule isolates were also evaluated for their ability to alkalize or acidify the media by growing them on YEMA media containing 25 mg/l of bromothymol blue. The Gram reaction of each isolate was determined using standard Gram staining procedures. Bradyrhizobia do not absorb or they absorb little Congo red in the dark. This characteristic was used to identify possible bradyrhizobia colonies after culturing the isolates in YEMA media supplemented with 0.25 g/ml Congo red stain and incubating in the dark (Bala et al., 2002; Chianu et al., 2009). After confirming the purity of each single type of colony, the isolates were maintained as cultures and stored on YEMA slants in McCartney screwed cap bottles at 4°C.
DNA extraction was done from randomly selected representative nodule isolates in each group using a modified CTAB genomic DNA extraction procedure. Three-day old pure bacteria isolate plate culture colonies were put into 1.5 ml microcentrifuge tubes containing 600 μl of normal saline. The mixture was then vortexed at maximum speed for 1 min followed by centrifugation at 13,000 rpm for 5 min. The liquid phase was then decanted off and the process repeated several times to get rid of all the extracellular polysaccharides. The cell pellets were resuspended through vortexing in 600 μl of CTAB genomic lysis buffer.
The incubation of the samples was carried out in a water bath at 65°C for 1 h. The samples were then centrifuged at 13,000 rpm for 10 min and the liquid phase pipetted into sterilized 1.5 ml micro centrifuge tubes. Six hundred microliters of chilled absolute alcohol were then added and the samples incubated at −20°C in a freezer. This was followed by centrifugation of the samples at 13,000 rpm for 5 min and the liquid phase discarded leaving behind DNA pellets. To clean the DNA pellets, 600 μl of chilled 70% alcohol was added followed by centrifugation of the samples at 13,000 rpm for 3 min. The alcohol was then decanted off gently. After air-drying, the DNA pellets were dissolved in 60 μl of elution buffer (TE). Storage of the DNA was done at −20°C.
DNA yield was quantified employing a NanoDrop® ND-1000 spectrophotometer (Inqaba Biotech, South Africa) using the convention that one absorbance unit at 260 nm equals 50 μg/ml DNA. The purity of the DNA was judged on the basis of A260:A280 ratio and its integrity and overall quality judged by inspection of the genomic DNA band on the agarose gel. On running the agarose gel stained with SYBR green, DNA was resolved in 1% agarose gel and 80 V for 30 min followed by observation of the DNA in a gel documentation system.
Polymerase chain reaction (PCR) was done on the genomic DNA after dilution and standardizing concentration to 50 ng/ul. The PCR machine used was 9800 Fast Thermal Cycler from Applied Biosystems. PCR targeting the 16S rDNA was done using universal primers P3 (forward, >5′-ATTAGATACCCTGGTAGTCC-3′) and P5 (reverse, >3′-GGTTACCTTGTTACGACTTC-5′). The PCR reaction mix contained 12.5 μl one Taq 2X mastermix with standard buffer from Biolabs, 1.25 μl of 10 μM primer P3F, 1.25 μl of 10 μM primer P5R, 9.0 μl sterile nuclease free PCR water, and 1.0 μl of sample DNA. After optimization, the PCR conditions were as follows: an initial DNA denaturation at 94°C for 3 min then denaturation at 94°C for 45 s, annealing at 53°C for 45 s, and extension at 72°C for 2 min (35 cycles). The final extension was carried out at 72°C for 5 min. Amplified DNA was held at 4°C.
The PCR products stained with SYBR-Green were separated by gel electrophoresis in 1.5% agarose in 0.5X TAE buffer at 100 V for 1 h. A 100 bp DNA ladder (thermoscientific) was used to estimate the molecular sizes of the bands. Gel visualization was done using a UV trans-illuminator and photographed using a digital camera. The PCR products were purified using QIAquick® PCR purification Kit following the manufacturer's instructions and run again on an agarose gel to confirm the presence of the product. After purification of the PCR products, Sanger sequencing targeting the 16S rDNA was done using primers P3 (forward) and P5 (reverse) in MacroGen Inc (Netherlands).
Qualitative data on the isolate characteristics were coded into numeric characters and then used to cluster the nodule isolates. The cluster analysis based on morphology was done using the neighbor-joining method and Euclidean similarity index using PAST software version 3 (Hammer et al., 2001). Principal component analysis displaying the relationship between the isolates was also done using PAST version 3 and DARwin version 6.0.17 software. Nodule isolate diversity indices were determined using PAST software. Dendrogram on morphological diversity was drawn using PAST software version 3. Redundancy analysis (RDA) was done using Canoco software version 5 to determine the relationship between soil characteristics and nodule isolate diversity.
Sequence editing was done using Bioedit software. Consensus sequences were created using DNA Baser software followed by comparison with the sequences in the NCBI database using Basic Local Alignment Search Tool (BLAST). The consensus sequences and aligned matching sequences from the NCBI database were then aligned using Muscle in Mega X software, MEGA X (Kumar et al., 2018). Aligned sequences were then used to construct a neighbor-joining phylogenetic tree employing the Maximum Composite Likelihood method (Tamura et al., 2004). Genetic diversity and conversion of sequence data to haplotypes was done using DNA SP6 software. The haplotype data was then used to determine the isolates' genetic differentiation and molecular variance using Arlequin 3.5.2.2 software.
Generally, the physicochemical properties of the soil varied depending on farm management practice. Interestingly, the organic farm soils had higher nutrient and chemical content compared to conventional farm soils. The soil tested characteristically acidic with conventional farm soils recording lower pH (4.57) than the organic farm soils (5.70). On average, the organic soil contained 2.82% organic carbon (OC), 0.27% available nitrogen (N), 27.73 ppm available phosphorus (P), 1.00 cmol kg−1 potassium (K) and 4.86% soil organic matter (SOM) which was higher than in conventional soil that had 2.54% OC, 0.22% N, 25.50 ppm P, 0.96 cmol kg−1 K and 4.38% SOM (Table 1).
Farm management practices had influence of soil acidity, nutrient levels and various soybean growth parameters. Organic farm management practice significantly enhanced soybean nodulation, growth and yield production. Soybean grown in less acidic organic farm soils recorded significantly higher nodule number (p < 0.0001), nodule dry weight (p < 0.0001), shoot dry weight (p < 0.0001) and seed dry weight (p = 0.001) compared to these in slightly more acidic conventional farms. No significance difference was noted among the individual farms in terms of nodulation. However, the three organic farms (GO, HO, JO,) recorded higher nodule number and nodule dry weight compared to the three conventional farms (GC, HC, JC). A significant variation was noted in shoot dry weight (p = 0.015) and seed dry weight (p = 0.037) among the individual organic and conventional farms (Table 2).
In this study, 108 pure isolates were obtained from the root nodules of soybean grown in the field trapping experiment and placed into 13 groups based on their morphological characteristics. A majority of the isolates (constituting 62%) had colonies visible after 5 days and characterized by alkalizing YEMA media supplemented with Bromothymol blue (BTB) which turned color from deep green to blue. Minority of the isolates (accounting for 38%) turned BTB medium from deep green to yellow due to production of acid substances in the medium which is a typical characteristic of rhizobia that show visible growth after only 3 days (Table 3).
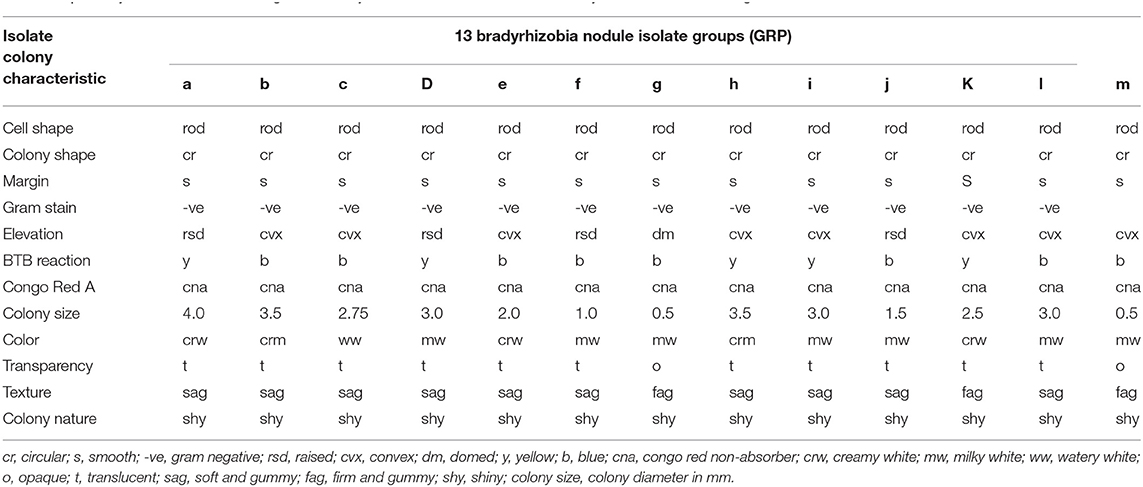
Table 3. Colony characteristics of indigenous bradyrhizobia isolates obtained from soybean nodules from organic and conventional farms.
In addition, all the isolates absorbed little or no Congo red in the dark. During Gram staining reaction, the isolates obtained had color change that signified they were Gram negative. The isolates were rod shaped with varied colony diameter, i.e. 15.4% formed small colonies (≤0.5 mm), while 30.8% had diameters ≥2.5 mm. The isolates with larger colonies with a diameter ranging between 2 and 4 mm and constituted 53.8% of the isolates showed production of copious extracellular polysaccharides on the growth medium (Table 3). The colony size/diameter was correlated with growth rate. The rhizobial that had visible colonies after 3 days produced large colony sizes, while those that had colonies visible after 5 days tended to produce small or very small sized colonies.
The colonies were shiny, with circular shape while the elevation was convex, domed, or raised. All colonies had entire margins and colony appearance was either opaque or translucent and the color was white, creamy, milky, and watery. The colony texture was either gummy, soft gummy and firm gummy with rod shaped cells (Table 3).
The morphological features of the isolates had notable variations and the 108 isolates obtained were placed into 13 groups (GRP) designated as GRPa to GRPm based on their morphological characteristics. The most abundant isolates were in group GRPh accounting for 42.05% of the total isolates while group GRPb followed with 33.28%. Groups GRPl, GRPg, and GRPm had the rarest isolates accounting for 1.07, 0.74, and 0.38%, respectively (Table 4). Organic farms had higher representation in the 13 groups compared to conventional farms. Among the organic farms, farm GO had the highest representation at 85% while farm GC scored the highest (69%) among the conventional farms as derived from Table 4.
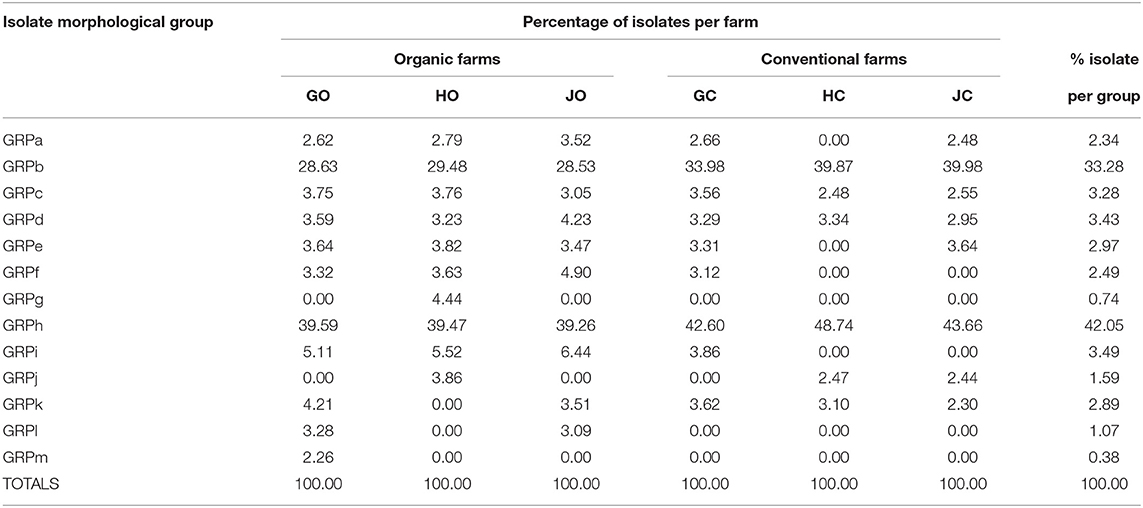
Table 4. Percentage abundance of bradyrhizobia isolates obtained from soybean nodules grown in organic and conventional farms.
A neighbor-joining tree using Euclidian similarity index and principal component analysis using the same similarity index partitioned the isolates into five morphological groups. In the neighbor-joining tree there were five clusters (A, B, C, D, and E). Isolate groups GRPe, GRPk and GRPa partitioned in cluster A. Groups GRPb and GRPh partitioned in cluster B. Group GRPc was independent in cluster C. Groups GRPg, GRPm, GRPf, and GRPj partitioned in cluster D while groups GRPi, GRPl, and GRPd were in cluster E (Figure 1A). The clustering of the groups in the neighbor-joining tree was similar to the clustering of the isolate groups using principal component analysis (Figure 1B). The isolates were partitioned in the PCA and appeared in the four quadrats with the same grouping pattern as the neighbor-joining tree (Figure 1B).
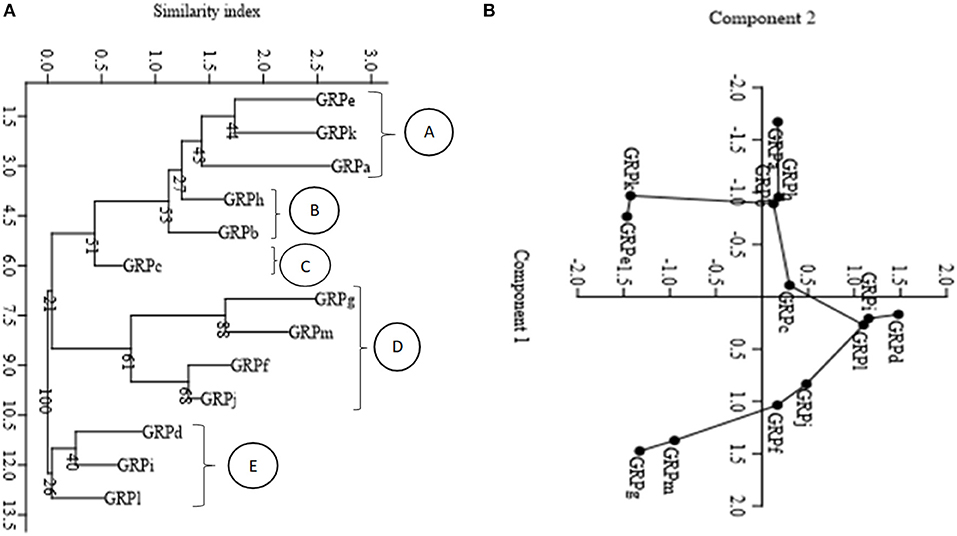
Figure 1. Distance dendrogram and principal component analysis diagram displaying morphological distribution of the nodule isolates. (A) Neighbor joining dendrogram based on Euclidian similarity index; (B) Principal component analysis (PCA) (Variance and Eigen values in the first two components; 1st component variance = 59.32%, Eigen value = 3.59 and 2nd component variance = 21.21%, Eigen value 1.28). The numbers displayed at the nodes of figure (A) (dendrogram) indicate the bootstrap support values in percentage after 1,000 resampling.
The number of isolate taxa varied according to the farm management practices. The three organic farms GO, HO, and JO had the highest number of isolate taxa between 10 and 11 while the conventional farm HC had the lowest number of taxa (6). According to Shannon (H) diversity index all the organic farms had the highest diversity of bradyrhizobia isolates while conventional farm bradyrhizobia isolates had the lowest diversity. Based on Shannon diversity index (H), farm GO had the highest diversity index (1.78) while farm HC had the lowest diversity index of 1.121 (Table 5). The use of Simpson 1-D diversity index revealed that organic farms GO and JO had the highest diversity of bradyrhizobia isolates while conventional farms bradyrhizobia isolates population had the lowest. The organic farm JO had the highest diversity of rhizobia isolates at 0.75 while conventional farms HC had the lowest bradyrhizobia diversity at 0.60 based on Simpson diversity index. The rhizobia diversity evenness also varied according to the farm management practices. The lowest rhizobia diversity evenness was observed in conventional farm JC having a low of 0.4658 while organic farm JO had the highest isolate distribution evenness at 0.5749 (Table 5). The conventional farms (GC, HC, and JC) had the highest species dominance D while the organic farms (GO, HO, and JO) had the lowest species dominance. The highest dominance was observed in conventional farm HC with 0.3998 while organic farm JO had the lowest Dominance-D at 0.249 (Table 5).
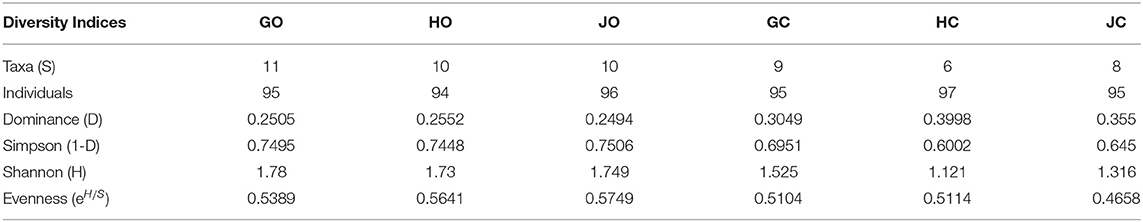
Table 5. Diversity indices of bradyrhizobia isolates obtained from soybeans nodules grown in organic and conventional farms.
A neighbor-joining cluster diagram based on morphological diversity of the nodule isolates partitioned the farms into two main clusters (A and B). Organic farms GO, HO, and JO clustered together in cluster A while conventional farms HC, JC, and GC clustered together in cluster B (Figure 2).
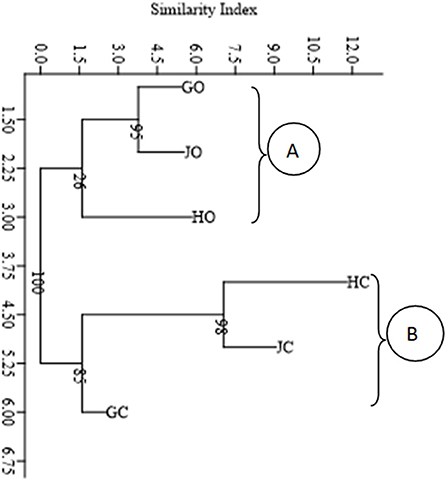
Figure 2. Cluster diagram showing morphological distribution of the nodule isolates based on Neighbor joining method and Euclidian similarity index. The numbers shown at the nodes of the dendrogram indicate the percentage bootstrap support value after 1,000 resampling.
Based on redundancy analysis (RDA) with soil characteristics as the explanatory variable, the soil characteristics accounted for 98.7% of the variation. The variation as explained in the first four axes was 96.96% (Axis 1), 98.13% (Axis 2), 98.72% (Axis 3), and 98.75% (Axis 4). The Eigen values for the first four axes were 0.9696 (Axis 1), 0.0117 (Axis 2), 0.0059 (Axis 3), and 0.0002 (Axis 4) with the first axis having the highest Eigen values. The soil chemical parameters, P, K, organic carbon, soil organic matter, and pH correlated positively with the number of taxa per farm, the Shannon diversity index and Evenness of isolate distribution in the farms. Species dominance had negative correlation with all the measured soil chemical parameters (Figure 3).
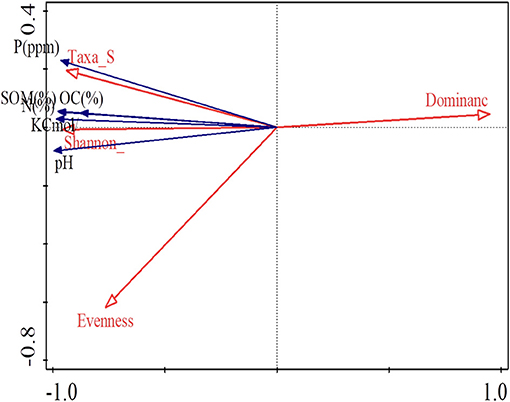
Figure 3. Redundancy analysis showing the relationship between the soil characteristics and nodule isolate diversity.
Using the procedures described in DNA extraction, good concentration, and quality of DNA was realized for all the 38 representative samples. The PCR process targeting 16S rDNA of the bradyrhizobia isolates were amplified by using primer P3 and P5; and yielded a single DNA band of ~1,500 bp.
Haplotype diversity was high in all populations. Population in farms GC, GO, HO and JO had high haplotype diversity (Hd) of 1 except population in farm JC (0.985). Nucleotide diversity, pi was lowest (0.711) in farm HO population and highest (0.748) in farm JO population (Table 6).
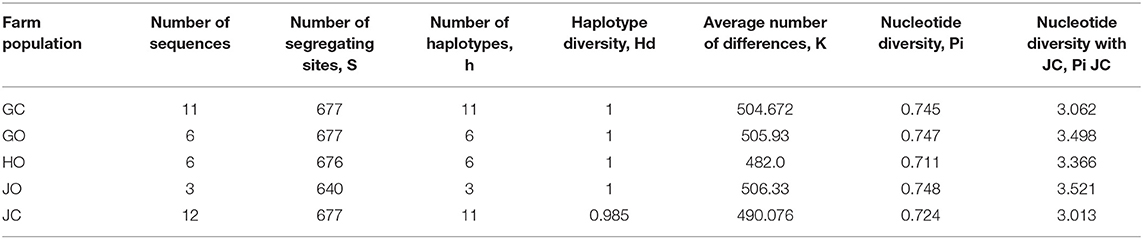
Table 6. Genetic diversity of bradyrhizobia isolates obtained from trapping of soybean in organic and conventional farms.
The nucleotide diversity with Jukes and Cantor correction, Pi (JC) was also highest among farm HO isolates and lowest in farm JC. The analysis showed that the isolates were genetically diverse with bradyrhizobia population from JC farm having the lowest genetic diversity estimate compared to the other farms which recorded the highest diversity (Table 6).
Based on analysis of molecular variance, there was significant variation (p ≤ 0.05) between isolates from the various farms. The highest variation was within populations (91.12%) while the lowest variation was among populations at 8.88% (Table 7).
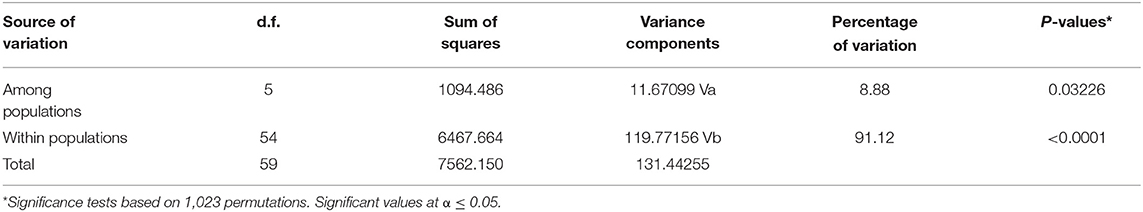
Table 7. Analysis of molecular variance (AMOVA) of nodule isolates obtained from organic and conventional farms.
Pairwise differentiation analysis demonstrated that there was no significant differentiation (p > 0.05) between most of the populations. Pairwise FST values for populations in farm GO and GC, JO and GC, JO and GO, and JO and JC were all negative (zero). However, a significant differentiation (p = 0.018) was observed between populations in farms JC and HO with FST value of 0.041 (Table 8).
Phylogenetic analysis based on the sequence data clustered the isolates into three clusters (A, B, and C) (Figure 4). Most of the isolates clustered with Bradyrhizobium sp. Strain UFLA05-136 in cluster C. Cluster C isolates related closely with cluster A isolates that also had above 98% match with Bradyrhizobium sp. Strain UFLA05-136 after BLAST analysis. Cluster B had the least number of isolates and two sub-clusters. In the major sub-cluster in cluster B, supported by 100% bootstrap value, the isolates related closely with isolates Bradyrhizobium sp. CCBAU 51511 (KF114656.1:780-1457) and uncultured Bradyrhizobiaceae bacterium clone SIF008 (JF733143.1:431-1043) from the NCBI database. In the minor sub-cluster in cluster B, supported by 95% bootstrap value the isolates clustered with Bradyrhizobium sp. strain R-46313 (FR753119.1:737-1104), Bradyrhizobium yuanmingense (LC460889.1:726-1013), and Bradyrhizobium japonicum PRY65 (AF239848.2:713-1394).
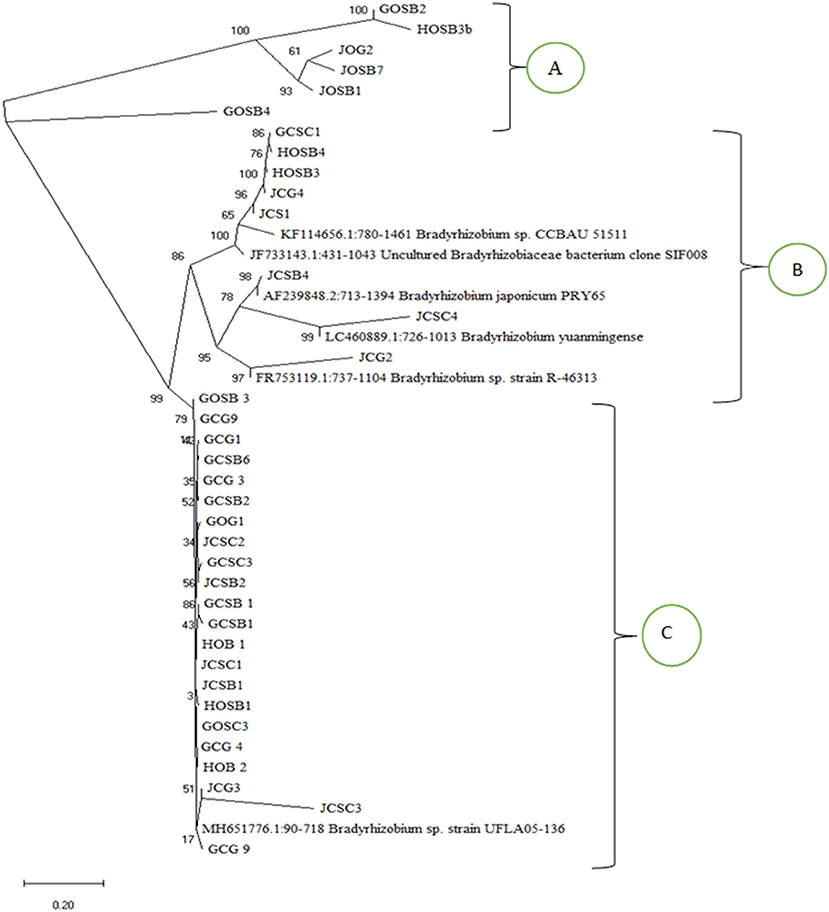
Figure 4. Phylogenetic tree based on the partial 16S rDNA sequences using the Neighbor joining method (Saitou and Nei, 1987) showing the phylogenetic relationship of the nodule isolates and matching isolates from the NCBI database. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1,000 replicates) are shown next to the branches.
The conservation of fertile agricultural soils is important for sustainable food production systems and soil nutrient contents can be maintained at an optimum level over time if good farm management practices are adhered to by farmers. The soil properties varied from farm to farm and largely depended on farm management system in practice. The conventional farm soils typically recorded higher acidity and lower fertility levels compared to organic farms; which could be due to extensive application of inorganic fertilizers and other agrochemicals, heavy leaching and runoff. Conventional farming renders the soil more acidic and less fertile due to leaching or salinization (Aktar et al., 2009). The low level of soil fertility in conventional farms as compared to organic farms; could be attributed to acidity, runoff and leaching; following the use of inorganic chemical fertilizers (Oberson et al., 2013; Hunt et al., 2019). The relatively higher acidity of the conventional soil, which is typical of highly weathered East African soils (Okalebo et al., 2002), debilitates soil nitrogen cycling and phosphorous solubility and availability. The results revealed that conventional farming systems rendered the soil less fertile due to increased soil acidity, loss of beneficial soil microorganisms and increased nutrient leaching. The continuous application of chemical fertilizers as practiced in conventional farming promotes soil acidification, leaching and mineralization of important ions like phosphorus, nitrogen and potassium, which play a vital role in plant growth (Wyngaard et al., 2016).
The less acidic soil in organic farms may be due to application of organic fertilizers and amendments that neutralize the low soil acidity and maintain beneficial soil microorganisms that maintain soil nutrient recycling. The application of animals' manure and compost promote the bioactivity and diversity of soil microbes (Hassink, 1995) that promote nutrient cycling rate and hence enhancing soil properties and plant productivity (Gajda et al., 2000). Organic farming system that involves the application of organic fertilizer, obliterate the use of inorganic chemical fertilizers and other agrochemicals that leads to loss of biodiversity, decline in soil fertility, pollute the environment, costly and unaffordable to most resource constrained smallholder farmers. Therefore, organic farming promotes diversity of bradyrhizobia species compared to conventional system; diverse species that could be responsible for enhanced nitrogen fixation and nutrient solubility that improve levels of soil nutrient content.
Soil properties varied from farm to farm and this explains the observed differences in various soybean growth parameters such as nodulation, plant biomass and seed yields. Generally, organic farming platform allows good nutrient management practices that are affordable, favor the soil properties, legume growth and sustainable yield production. The organic farms had better soybean performance compared with the conventional farms, which appears to result from the favorable soil conditions for biological nitrogen fixation (Pandey et al., 2017; Schneider et al., 2017). The high acidity of conventionally managed farm soil could have contributed to lower nodulation in the respective farms compared to organic farms with higher soil pH and high nodule number. Conventional farm soils are highly acidic and this hinders the establishment of effective nodulation (Kawaka et al., 2014; Koskey et al., 2017). Soil pH below 5.5 does not favor soybean production due to suppression of nodule formation and symbiotic nitrogen fixation and limits the availability of essential elements like nitrogen and phosphorous (Mairura et al., 2008). The higher seed dry weight revealed among soybean grown in organic farms in the current study resonates with Enrico et al. (2018) who documented increased soybean biomass and seed yield after intensive organic farm management practices.
Non-application of chemical fertilizers in organic farming is a smart strategy of producing organic products that are perceived to be healthier, safer and contain lower levels of agrochemical residues. The growing and unmet consumer demand for healthier organic food is high in markets worldwide and has fueled unprecedented interest in organic farming. Organic farm management practices positively influence soil fertility and biodiversity (Tuomisto et al., 2012), reduces erosion (Seitz et al., 2019) and negative environmental impacts and healthwise, organic foods contain lower levels of agrochemical residues (Smith-Spangler et al., 2012). Therefore, organic farming is gaining popularity and significance due to its beneficial role in producing organic food with lower amounts of chemical residues, enhanced soil fertility, maintenance of environmental quality and increased biodiversity. Profitable organic farm management system is increasing in popularity due to high consumer demand for healthier food, soil fertility improvement, the need for environmental conservation and restoration of soil biodiversity (Bett and Ayieko, 2017). Additionally, organic farming involves the application of locally available, affordable and environmentally friendly organic farm yard manure; that reduce the cost of production and maintain environmental quality; hence promote sustainability in smallholder farming systems.
The morphological diversity of the indigenous bradyrhizobia isolates nodulating soybeans in organic and conventional farms in Eastern Kenya was confirmed by calculating the diversity indices. The morphological diversity estimates calculated showed differences in the nodule isolate diversity in the farms investigated. The nodule isolate diversity indices were higher among organic farming systems as compared to conventional farming systems. According to Liao et al. (2018), different farm management practices affect the structure of the soil which in turn influences the microbial community structure in that agro-ecosystem. Moreover, organic farming systems have been reported to enhance soil organic carbon providing much needed energy for microorganisms and hence enhancing their diversity and abundance unlike conventional farming systems (Liao et al., 2018).
According to Shannon diversity (H), Simpson 1-D and other diversity estimates, organic farms had the highest diversity of bradyrhizobia isolates. Among the different farms, organic farms GO and JO had the highest rhizobia diversity while conventional farm HC and JC had the lowest diversity. The organic farms with the greatest bradyrhizobia diversity indices also had the highest levels of isolate distribution evenness. Similar findings where observed by César et al. (2015) when working on rhizobia isolates from stem and root nodules of Discolobium and Aeschynomene. The bradyrhizobia species dominance values were lower in farms with high rhizobia diversity in the present study. This is in agreement with research by Valley et al. (2009) who reported lower dominance values in areas with higher cowpea rhizobia diversity in Fransisco River Valley.
Cluster dendrogram and principal component analysis of the isolates based on their diversity indices clustered organic farms GO, HO and JO isolates together and the conventional farms GC, HC and JC with the lowest diversity of bradyrhizobia isolates in an independent group. This variation could be due to different farm management practices and the characteristics of the farms soil such as pH and nutrient content. The bradyrhizobia diversity variation based on farm management practices has also been documented in previous study findings. Working in both organic and conventional farming systems, Liao et al. (2018) noted similar clustering of microbial diversity based on the farming system. According to Yan et al. (2014), rhizobia diversity results due to different land use histories, different fertilizer supplements, planting of legume or non-legume plants and rotation or monoculture of the host plant. Mason et al. (2018), while working on effect of soil characteristics and flooding on bradyrhizobia diversity in the Philippines also emphasized on the ability of soil management to influence bradyrhizobia diversity and species dominance. Chemical soil fertilization also decreases rhizobia diversity which explains the lower diversity of rhizobia in conventional farming systems (Yan et al., 2014).
Redundancy analysis showed a positive correlation between bradyrhizobia Shannon diversity index and soil properties like soil organic matter, Nitrogen, Potassium, Phosphorus, and pH. This is in agreement with the findings by other researchers who have reported positive correlation between rhizobia diversity and soil characteristics (Yan et al., 2014; Li et al., 2016). Yan et al. (2014) also reported positive correlation between soybean bradyrhizobia diversity and soil organic carbon, available potassium and phosphates. The positive correlation between soil organic matter and bradyrhizobia diversity could be due to enhanced soil organic matter, which is a key source of energy for microorganisms and usually a major indicator of the quality of soil (Li et al., 2016). Soil pH has also been reported to be an important component in the management of soybean bradyrhizobia diversity and distribution in the soil (Mason et al., 2018). Alterations in soil pH are believed to influence the bioavailability of soil carbon and nitrogen which are important nutrients for proliferation of microorganisms (Sharma et al., 2013; Han et al., 2019). The positive correlation between the bradyrhizobia diversity and soil nitrogen is contrary to findings by Lin et al. (2019), however it suggests low nitrogen content in the soils hence the need for nitrogen fixing microorganisms. The biological nitrogen fixation is a high energy demanding process that utilizes large amounts of adenosine triphosphate (ATP). This possibly explains the positive correlation between phosphorus and the trapped bradyrhizobia diversity (Han et al., 2019).
The representative of the 108 bradyrhizobia isolates obtained from farm trapping experiments were evaluated for their genetic diversity using the 16S rDNA. The amplification of the target 16S rDNA gene resulted to a single band of 1.5 kb in size. Previous results reported that the band size obtained correspond to the expected size due to the use of primers specific to rhizobia (Zhang et al., 2014; Ondieki et al., 2017). The molecular analysis findings demonstrated a high variation in the 38 indigenous representatives isolates which is an indication that the soils from the trapping farms harbor populations of highly diverse bradyrhizobia species. Similar results were reported by Atieno et al. (2012) and Shiro et al. (2013) from other parts of the world.
Haplotype diversity and nucleotide diversity are common measures of genetic diversity of living organisms in a population. There was a high haplotype and nucleotide diversity of bradyrhizobia isolates in all the populations which is in accordance with that of previous research studies. High genetic diversity of rhizobia isolates from Lower Eastern Kenya was also reported by Ondieki et al. (2017) among nodule isolates from cowpea plant hosts. Previous study by Koskey et al. (2018) reported a high genetic diversity of rhizobia isolates from nodules of mid altitude climbing beans planted in Embu and Tharaka-Nithi regions of Eastern Kenya. High genetic diversity of soybean nodulating bradyrhizobia in Kenya was also reported by Wasike et al. (2009) in different soils. The isolates from different farms showed high nucleotide and haplotype diversity. This agrees with findings by Naamala et al. (2016) on high diversity of soybean nodulating isolates in South Africa based on 16S rDNA analysis. The marginally lower haplotype diversity in JC populations as compared to the other populations could be due the cultivation history in the farm or even the soil characteristics as influenced by different farm management practices at the study farm. This is in line with findings by Appunu et al. (2008) and Stefan et al. (2015) who reported variations in soybean bradyrhizobia haplotype diversity with even higher diversity in areas that had no history of soybean cultivation in various agro ecological zones. The geographical locality also had an effect on all the biodiversity measures including the nucleotide and haplotype diversity that is concordant with findings by Wasike et al. (2009) who reported a similar trend in diversity of soybean nodulating bradyrhizobia from two localities in Kenya.
The analyses of molecular variance (AMOVA) based on amplified 16S rDNA showed that most of the genetic variation of native bradyrhizobia isolates was within the bradyrhizobia populations and not among the populations. Similar findings were reported by Elboutahiri et al. (2015) who observed a larger proportion of significant genetic variation distributed within regions (89%) than among regions (11%) in Sinorhizobium meliloti and Sinorhizobium medicae obtained from drought and salt affected regions of Morocco. Risal et al. (2010) and Vauclare et al. (2013) recorded high variation of rhizobia isolates within populations and a small variation across different regions.
The AMOVA analysis results demonstrated that there is low level of differentiation and gene flow between the various farms trapping isolates obtained from different regions. Similar observations, however on climbing beans nodulating rhizobia from Embu and Tharaka-Nithi Counties of Kenya were also reported by Koskey et al. (2018). Wu et al. (2011) also reported similar findings on low variation of rhizobia isolates between populations; which increases the probability of the crop host to get a wide variety of the same indigenous nitrogen fixers in any of the farms. Stefan et al. (2015) recorded high variation of clover nodulating rhizobia isolates within populations and a small variation across different ecosystems in Northern Romania. The low differentiation between the populations signifies high gene flow between the populations (Omondi et al., 2010). The low genetic variation across regions also supports the position that geographical distance between populations of particular organisms does not always translate to increased variation and there is high possibility of limited barriers to gene flow between the populations for many years (Degani et al., 2013).
A comparison of the analysis of molecular variance and the pairwise difference analysis of the current study showed no significant differentiation between most of the populations. This meant that most of the bradyrhizobia isolates were not secluded based on farm or even distance from where they were isolated (Jiang et al., 2019). In this study, the highest significant differentiation (p = 0.018) was observed between populations JC and HO with FST value of 0.041. The study results are in line with finding by Koskey et al. (2018) who reported some differentiation of climbing bean nodulating rhizobia isolates from different sites in Embu and Tharaka-Nithi Counties. Wu et al. (2011) also reported differentiation of soybean nodulating and non-nodulating bacteria from different ecological regions in China. Pairwise FST values for populations GO and GC, JO and GC, JO and GO, and JO and JC were all negative (zero); meaning that there was no genetic differentiation between the populations. The results are in agreement with previous study by Appunu et al. (2008) who reported no genetic differentiation between soybean nodulating bradyrhizobia from different ecoclimatic regions in India. According to Krause and Whitaker (2015), most microorganisms obey the clonal ecotype model of natural variation in their genomes due the high possibility of little or no genetic recombination within populations as a result of clonal reproduction in microorganisms. This explains the low gene flow that was observed between the populations.
Phylogenetic analysis based on the sequence data separated the isolates into three clusters. All the isolates from both organic and inorganic farming systems in the three clusters were identified as bradyrhizobia spp. This supports findings by Yan et al. (2014) who reported bradyrhizobia species to be the common micro symbionts of soybean. A majority of the isolates clustered with Bradyrhizobium japonicum and Bradyrhizobium yuanminense. These results are supported by finding by Wasike et al. (2009) that isolated the two isolates from nodules of promiscuous soybean grown in Mitunguu in Lower Eastern Kenya and therefore the isolates are resident in Kenyan soils. Bradyrhizobium japonicum has also been identified as symbiont of soybean in other countries in Africa including South Africa, Ethiopia and Zibambwe (Jaiswal et al., 2019). A big number of the isolates had a similar evolutionary trend with Bradyrhizobium sp. Strain UFLA05-136 that is a bradyrhizobia that has not yet been classified and not yet reported in African soils as a common soybean micro symbiont, but was first isolated from Arachis pintoi in Brazil.
According to this study, the farm management system influences bradyrhizobia species diversity and abundance; with organic farming systems enhancing the diversity of bradyrhizobia as compared to conventional farming systems. Eastern Kenya soils have indigenous bradyrhizobia species that can form symbiotic interaction with soybean in both organic and conventional farming systems. There was also no genetic differentiation between the nodule isolates between the organic and conventional farms which signifies the presence of similar bradyrhizobia species across the region. Organic farming system favors the proliferation of bradyrhizobia species and is therefore, a suitable environmentally friendly alternative for enhancing soybean production.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
NG, EN, JM, and RC conceived and designed the research and data collection tools, and participated in drafting the manuscript. NG collected the data and participated in data analyses. EN, JM, and RC supervised the project and data evaluation. All authors read the manuscript and approved the submission.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We express our sincere gratitude to National Research Fund (NRF) for supporting this study and laboratory staff from microbiology and tissue culture, Kenyatta University for their participation, cooperation, time, and efforts during the collection of molecular data and analysis.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fsufs.2020.606618/full#supplementary-material
Abate, T., Shiferaw, B., Gebeyehu, S., and Eshet, T. N. (2008). Variation in the pattern of Nitrogen accumulation and distribution ion legumes. Crop. Sci. 37, 1682–1886. doi: 10.2135/cropsci1997.0011183X003700050011x
Abbasi, M. K., Majeed, A., Sadiq, A., and Khan, S. R. (2008). Application of Bradyrhizobium japonicum and phosphorus fertilization improved growth, yield and nodulation of soybean in the sub-humid hilly region of Azad Jammu and Kashmir, Pakistan. Plant Prod. Sci. 11, 368–376. doi: 10.1626/pps.11.368
Abou-Shanab, R. A. I., Wongphatcharachai, M., Sheaffer, C. C., Orf, J. C., and Sadowsky, M. J. (2017). Competition between introduced Bradyrhizobium japonicum strains and indigenous bradyrhizobia in Minnesota organic farming systems. Symbiosis 73, 155–163. doi: 10.1007/s13199-017-0505-4
Aktar, W., Sengupta, D., and Chowdhury, A. (2009). Impact of pesticides use in agriculture: their benefits and hazards. Interdiscip. Toxicol. 2, 1–12. doi: 10.2478/v10102-009-0001-7
Appunu, C., N'Zoue, A., and Laguerre, G. (2008). Genetic diversity of native bradyrhizobia isolated from soybeans (Glycine max L.) in different agricultural-ecological-climatic regions of India. Appl. Environ. Microbiol. 74, 5991–5996. doi: 10.1128/AEM.01320-08
Ashworth, A. J., Allen, F. L., Wight, J. P., Saxton, A. M., Tyler, D. D., and Sams, C. E. (2014). Soil organic carbon sequestration rates under crop sequence diversity, bio-covers, and no-tillage. Soil Sci. Soc. Am. J. 78:1726. doi: 10.2136/sssaj2013.09.0422
Atieno, M., Herrmann, L., Okalebo, R., and Lesueur, D. (2012). Efficiency of different formulations of Bradyrhizobium japonicum and effect of co-inoculation of Bacillus subtilis with two different strains of Bradyrhizobium japonicum. World J. Microbiol. Biotechnol. 28, 2541–2550. doi: 10.1007/s11274-012-1062-x
Bala, A., Murphy, P., and Giller, K. E. (2002). Occurrence and genetic diversity of rhizobia nodulating Sesbastian in African soils. Soil Biol. Biochem. 34, 28–30. doi: 10.1016/S0038-0717(02)00163-3
Bett, E. K., and Ayieko, D. M. (2017). Economic potential for conversion to organic farming: a net present value analysis in the East Mau Catchment, Nakuru, Kenya. Environ. Develop. Sustain. 19, 1307–1325. doi: 10.1007/s10668-016-9800-0
Brahim, S., Niess, A., Pflipsen, M., Neuhoff, D., and Scherer, H. (2017). Effect of combined fertilization with rock phosphate and elemental sulphur on yield and nutrient uptake of soybean. Plant Soil Environ. 63, 89–95. doi: 10.17221/22/2017-PSE
Caliskan, S., Ozkaya, I., Caliskan, M. E., and Arslan, M. (2008). The effects of nitrogen and iron fertilization on growth, yield and fertilizer use efficiency of soybean in a Mediterranean-type soil. Field Crops Res. 108, 126–132. doi: 10.1016/j.fcr.2008.04.005
Cely, M. V. T., de Oliveira, A. G., de Freitas, V. F., de Luca, M. B., Barazetti, A. R., dos Santos, I. M. O., et al. (2016). Inoculant of arbuscular mycorrhizal fungi (Rhizophagus clarus) increase yield of soybean and cotton under field conditions. Front. Microbiol. 7:720. doi: 10.3389/fmicb.2016.00720
César, J., Martins, R., Dolores, A., Freitas, S., de Simões, R., and Menezes, C. (2015). Nitrogen symbiotically fixed by cowpea and gliricidia in traditional and agroforestry systems under semiarid conditions. Pesq. Agropec. Bras. 1, 178–184. doi: 10.1590/S0100-204X2015000200010
Chianu, J. N., Nkonya, E. M., Mairura, F. S., Chianu, J. N., and Akinnifesi, F. K. (2009). Biological nitrogen fixation and socioeconomic factors for legume production in sub-Saharan Africa. Sustain. Agric. 2, 309–329. doi: 10.1007/978-94-007-0394-0_17
Chibeba, A. M., Kyei-Boahen, S., Guimarães, M. de F., Nogueira, M. A., and Hungria, M. (2017). Isolation, characterization and selection of indigenous Bradyrhizobium strains with outstanding symbiotic performance to increase soybean yields in Mozambique. Agric. Ecosyst. Environ. 246, 291–305. doi: 10.1016/j.agee.2017.06.017
Degani, G., Goldberg, T., Gasith, A., Elron, E., and Nevo, E. (2013). DNA variations of the green toad Pseudepidalea viridis (syn. Bufo viridis) from various habitats. Zool. Stud. 52:1. doi: 10.1186/1810-522X-52-18
Elboutahiri, N., Thami-Alami, I., and Udupa, S. M. (2015). Phenotypic and genetic diversity in Sinorhizobium meliloti and S. medicae from drought and salt affected regions of Morocco. BMC Microbiol. 10:15. doi: 10.1186/1471-2180-10-15
Enrico, J. M., García, F., Stewart, M., Francisco, E., Balboa, G., Ciampitti, I., et al. (2018). Integrating crop and fertilization management strategies for soybean in the Central Pampas. Better Crops Plant Food 102, 6–10. doi: 10.24047/BC10226
Foley, J. A., Defries, R., Asner, G. P., Barford, C., Bonan, G., Carpenter, S. R., et al. (2005). Global consequences of land use. Science 309, 570–574. doi: 10.1126/science.1111772
Furseth, B. J., Conley, S. P., and Ané, J. M. (2012). Soybean response to soil rhizobia and seed-applied rhizobia inoculants in Wisconsin. Crop Sci. 52, 339–344. doi: 10.2135/cropsci2011.01.0041
Gajda, A., Martyniuk, S., Stachyra, A., Wróblewska, B., and Zieba, S. (2000). Relations between microbiological and biochemical properties of soil under different agrotechnical conditions and its productivity. Polish J. Soil Sci. 33, 55–60. doi: 10.17951/pjss/2019.52.2.259
Grandi, C. (2008). Organic agriculture enhances agrobiodiversity. Biodiversity 9, 33–35. doi: 10.1080/14888386.2008.9712878
Hammer, O., Harper, D. A. T., and Ryan, P. D. (2001). PAST: Paleontological statistics software package for education and data analysis. Palaeontol. Electron. 4:9. doi: 10.12691/aees-3-6-5
Han, L. L., Wang, Q., Shen, J. P., Di, H. J., Wang, J. T., Wei, W. X., et al. (2019). Multiple factors drive the abundance and diversity of the diazotrophic community in typical farmland soils of China. FEMS Microbiol. Ecol. 95, 1–10. doi: 10.1093/femsec/fiz113
Hassink, J. (1995). Density fractions of soil macroorganic matter and microbial biomass as predictors of C and N mineralization. Soil Biol. Biochem. 27, 1099–1108. doi: 10.1016/0038-0717(95)00027-C
Heerwaarden, J., Van Baijukya, F., Kyei-boahen, S., Adjei-nsiah, S., Ebanyat, P., Kamai, N., et al. (2018). Soyabean response to rhizobium inoculation across sub-Saharan Africa : patterns of variation and the role of promiscuity. Agric. Ecosyst. Environ. 261, 211–218. doi: 10.1016/j.agee.2017.08.016
Hellal, F. A., and Abdelhamid, M. T. (2013). Prácticas de gestión de nutrientes para mejoramiento en la producción de soja (Glycine max L.). Acta Biol. Colomb. 18, 3–14.
Hunt, N. D., Hill, J. D., and Liebman, M. (2019). Cropping system diversity effects on nutrient discharge, soil erosion, and agronomic performance. Environ. Sci. Technol. 53, 1344–1352. doi: 10.1021/acs.est.8b02193
IFOAM (2006). The IFOAM Basic Standards for Organic Production and Processing. Version 2005. Bonn: IFOAM Publications.
Igiehon, N. O., Babalola, O. O., and Aremu, B. R. (2019). Genomic insights into plant growth promoting rhizobia capable of enhancing soybean germination under drought stress. BMC Microbiol. 19:159. doi: 10.1186/s12866-019-1536-1
Jaetzold, R., Schimdt, H., Hornetz, B., and Shisanya, C. (2006). Farm Management Handbook of Kenya. Natural Conditions and Farm Information Vol II/C. Nairobi: Ministry of Agriculture.
Jaiswal, S. K., Dakora, F. D., and Ramírez-bahena, M. H. (2019). Widespread distribution of highly adapted Bradyrhizobium species nodulating diverse legumes in Africa. Front. Microbiol. 10:310. doi: 10.3389/fmicb.2019.00310
Jiang, S., Luo, M. X., Gao, R. H., Zhang, W., Yang, Y. Z., Li, Y. J., et al. (2019). Isolation-by-environment as a driver of genetic differentiation among populations of the only broad-leaved evergreen shrub Ammopiptanthus mongolicus in Asian temperate deserts. Sci. Rep. 9, 1–14. doi: 10.1038/s41598-019-48472-y
Kawaka, F., Mathews, M., Dida, M. M., Opala, P. A., Ombori, O., Maingi, J., et al. (2014). Symbiotic efficiency of native rhizobia nodulating common bean (Phaseolus vulgaris L.) in soils of Western Kenya. Int. Scholar. Res. Not. 2014:258497. doi: 10.1155/2014/258497
Koskey, G., Mburu, S. W., Kimiti, J. M., Ombori, O., Maingi, J. M., and Njeru, E. M. (2018). Genetic characterization and diversity of Rhizobium isolated from root nodules of mid-altitude climbing bean (Phaseolus vulgaris L.) varieties. Front. Microbiol. 9:968. doi: 10.3389/fmicb.2018.00968
Koskey, G., Mburu, S. W., Njeru, E. M., Kimiti, J. M., Ombori, O., and Maingi, J. M. (2017). Potential of native rhizobia in enhancing nitrogen fixation and yields of climbing beans (Phaseolus vulgaris L.) in contrasting environments of Eastern Kenya. Front. Plant Sci. 8:443. doi: 10.3389/fpls.2017.00443
Krause, D. J., and Whitaker, R. J. (2015). Inferring speciation processes from patterns of natural variation in microbial genomes. Syst. Biol. 64, 926–935. doi: 10.1093/sysbio/syv050
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Li, Y. C., Li, Z., Li, Z. W., Jiang, Y. H., Weng, B. Q., and Lin, W. X. (2016). Variations of rhizosphere bacterial communities in tea (Camellia sinensis L.) continuous cropping soil by high-throughput pyrosequencing approach. J. Appl. Microbiol. 121, 787–799. doi: 10.1111/jam.13225
Liao, J., Liang, Y., and Huang, D. (2018). Organic farming improves soil microbial abundance and diversity under greenhouse condition: a case study in Shanghai (Eastern China). Sustainability (Switzerland) 10:3825. doi: 10.3390/su10103825
Lin, Y., Te Lin, Y. F., Tsai, I. J., Chang, E. H., Jien, S. H., Lin, Y. J., et al. (2019). Structure and diversity of soil bacterial communities in offshore islands. Sci. Rep. 9, 1–9. doi: 10.1038/s41598-019-41170-9
Mairura, F. S., Mugendi, D. N., Mwanje, J. I., Ramisch, J. J., Mbugua, P. K., and Chianu, J. N. (2008). Scientific evaluation of smallholder land use knowledge in Central Kenya. Land Degr. Dev. 19, 77–90. doi: 10.1002/ldr.815
Mason, M. L. T., Tabing, B. L. C., Yamamoto, A., and Saeki, Y. (2018). Influence of flooding and soil properties on the genetic diversity and distribution of indigenous soybean-nodulating bradyrhizobia in the Philippines. Heliyon 4:e00921. doi: 10.1016/j.heliyon.2018.e00921
Mungai, N. W., and Karubiu, N. M. (2011). Effectiveness of rhizobia isolates from Njoro soils (Kenya) and commercial inoculants in nodulation of common beans (phaseolus vulgaris). J. Agric. Sci. Technol. 12, 47–60.
Murage, F. M., Mugwe, J. N., Ngetich, K. F., Mucheru-Muna, M. M., and Mugendi, D. N. (2019). Adoption of soybean by smallholder farmers in the Central Highlands of Kenya. Afr. J. Agric. Econ. Rural Dev. 7, 1–12. Available online at: www.internationalscholarsjournals.org
Muriithi, H. M., Beed, F., Tukamuhabwa, P., Thomma, B. P. H. J., and Joosten, M. H. A. J. (2016). Soybean production in eastern and southern Africa and the the threat of yield loss due to soybean rust caused by Phakopsorapachyrhizi. Plant Pathol. 65, 176–188. doi: 10.1111/ppa.12457
Mutoni, C. K., Nyongesa, D., Esilaba, A. O., and Mabele, R. B. (2017). The economics and gender factor in soya bean production and profitability in Kenya: a case of smallholder farms in Western Kenya. Int. J. Agric. Resour. Gov. Ecol. 13, 211–240. doi: 10.1504/IJARGE.2017.10008042
Naamala, J., Sanjay, K. J., and Dakora, F. D. (2016). Microsymbiont diversity and phylogeny of native Bradyrhizobia associated with soybean (Glycine Max L. Merr.) nodulation in South African soils. Syst. Appl. Microbiol. 39, 336–344. doi: 10.1016/j.syapm.2016.05.009
Ndungu, S. M., Messmer, M. M., Ziegler, D., Gamper, H. A., Mészáros, É., Thuita, M., et al. (2018). Cowpea (Vigna unguiculata L. Walp) hosts several widespread bradyrhizobial root nodule symbionts across contrasting agro-ecological production areas in Kenya. Agric. Ecosyst. Environ. 261, 161–171. doi: 10.1016/j.agee.2017.12.014
Ndusha, B. N., Onwonga, R. N., Nabahungu, L. S., Mushagalusa, N. G., Matendo, R. E., and Keya, S. O. (2019). Indigenous rhizobia strains: the silver bullet for enhanced biological nitrogen fixation and soybean (Glycine max (L.) Merr.) yield under different soil conditions in South Kivu province, Democratic Republic of Congo. Afr. J. Agric. Res. 14, 2038–2047. doi: 10.5897/AJAR2019.14457
Njeru, E. M., Maingi, J. M., Cheruiyot, R., and Mburugu, G. N. (2013). Managing soybean for enhanced food production and soil bio-fertility in smallholder systems through maximized fertilizer use efficiency. Int. J. Agric. Forestry 3, 191–197. doi: 10.5923/j.ijaf.20130305.01
Oberson, A., Frossard, E., Bühlmann, C., Mayer, J., Mäder, P., and Lüscher, A. (2013). Nitrogen fixation and transfer in grass-clover leys under organic and conventional cropping systems. Plant Soil 371, 237–255. doi: 10.1007/s11104-013-1666-4
Odee, D. W., Sutherland, J. M., Makatiani, E. T., McInroy, S. G., and Sprent, J. I. (1997). Phenotypic characteristics and composition of rhizobia associated with woody legumes growing in diverse Kenyan conditions. Plant Soil 188, 65–75. doi: 10.1023/A:1004204413140
Okalebo, R. J., Gathua, K. W., and Woomer, P. L. (2002). Laboratory Methods of Soil and Plant Analysis: A Working Manual. 2nd Edn. Nairobi: TSBF-CIAT and Sacred Africa.
Omondi, S. F., Kireger, E., Danga suk, O. G., Chikamai, B., Odee, D. W., Cavers, S., et al. (2010). Genetic diversity and population structure of Acacia senegal (L) Willd. in Kenya. Trop. Plant Biol. 3, 59–70. doi: 10.1007/s12042-009-9037-2
Ondieki, D. K., Nyaboga, E. N., Wagacha, J. M., and Mwaura, F. B. (2017). Morphological and genetic diversity of rhizobia nodulating cowpea (Vigna unguiculata L.) from agricultural soils of Lower Eastern Kenya. Int. J. Microbiol. 2017:8684921. doi: 10.1155/2017/8684921
Pandey, A., Li, F., Askegaard, M., and Olesen, J. E. (2017). Biological nitrogen fixation in three long-term organic and conventional arable crop rotation experiments in Denmark. Eur. J. Agron. 90, 87–95. doi: 10.1016/j.eja.2017.07.009
Raza, A., Razzaq, A., Mehmood, S. S., Zou, X., Zhang, X., and Lv, Y. (2019). Impact of climate change on crops adaptation and strategies to tackle its outcome : a review. Plants (Basel, Switzerland) 8:34. doi: 10.3390/plants8020034
Risal, C. P., Yokoyama, T., Ohkama-Ohtsu, N., Djedidi, S., and Sekimoto, H. (2010). Genetic diversity of native soybean bradyrhizobia from different topographical regions along the southern slopes of the Himalayan Mountains in Nepal. Syst. Appl. Microbiol. 33, 416–425. doi: 10.1016/j.syapm.2010.06.008
Saitou, N., and Nei, M. (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425.
Samson, M. E., Menasseri-Aubry, S., Chantigny, M. H., Angers, D. A., Royer, I., and Vanasse, A. (2019). Crop response to soil management practices is driven by interactions among practices, crop species and soil type. Field Crops Res. 243:107623. doi: 10.1016/j.fcr.2019.107623
Schmidt, J. E., Kent, A. D., Brisson, V. L., and Gaudin, A. C. M. (2019). Agricultural management and plant selection interactively affect rhizosphere microbial community structure and nitrogen cycling. Microbiome 7, 1–18. doi: 10.1186/s40168-019-0756-9
Schneider, K. D., Lynch, D. H., Bünemann, E. K., and Voroney, R. P. (2017). Vegetative composition, arbuscular mycorrhizal fungi root colonization, and biological nitrogen fixation distinguish organic and conventional perennial forage systems. Agron. J. 109:1697. doi: 10.2134/agronj2016.12.0700
Seitz, S., Goebes, P., Puerta, V. L., Pereira, E. I. P., Wittwer, R., Six, J., et al. (2019). Conservation tillage and organic farming reduce soil erosion. Agron. Sustain. Dev. 39, 1–10. doi: 10.1007/s13593-018-0545-z
Sharma, S. R., Rao, N. K., Gokhale, T. S., and Ismail, S. (2013). Isolation and characterization of salt-tolerant rhizobia native to the desert soils of United Arab Emirates. Emir. J. Food Agric. 25, 102–108. doi: 10.9755/ejfa.v25i2.7590
Shiro, S., Matsuura, S., Saiki, R., Sigua, G. C., Yamamoto, A., and Umehara. (2013). Genetic diversity and geographical distribution of indigenous soybean-nodulating bradyrhizobia in the United States. Appl. Environ. Microbiol. 79, 3610–3618. doi: 10.1128/AEM.00236-13
Smith-Spangler, C., Brandeau, M. L., Hunter, G. E., Bavinger, J. C., Pearson, M., Eschbach, P. J., et al. (2012). Are organic foods safer or healthier than conventional alternatives? Ann. Intern. Med. 157, 348–366. doi: 10.7326/0003-4819-157-5-201209040-00007
Stefan, A., Rosu, C. M., Stedel, C., Gorgan, L. D., and Efrose, R. C. (2015). RAPD-inferred genetic variability of some indigenous rhizobium leguminosarum isolates from red clover (Trifolium pratense L.) nodules. Acta Biol. Hung. 66, 316–325. doi: 10.1556/018.66.2015.3.7
Tamura, K., Nei, M., and Kumar, S. (2004). Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc. Natl. Acad. Sci. U. S. A. 101, 11030–11035. doi: 10.1073/pnas.0404206101
Thuita, M., Pypers, P., Herrmann, L., Okalebo, R. J., Othieno, C., Muema, E., et al. (2012). Commercial rhizobial inoculants significantly enhance growth and nitrogen fixation of a promiscuous soybean variety in Kenyan soils. Biol. Fertil. Soils 48, 87–96. doi: 10.1007/s00374-011-0611-z
Tuomisto, H. L., Hodge, I. D., Riordan, P., and Macdonald, D. W. (2012). Does organic farming reduce environmental impacts? - a meta-analysis of European research. J. Environ. Manage. 112, 309–320. doi: 10.1016/j.jenvman.2012.08.018
Ulzen, J., Abaidoo, R. C., Mensah, N. E., Masso, C., and AbdelGadir, A. A. H. (2016). Bradyrhizobium inoculants enhance grain yields of soybean and cowpea in Northern Ghana. Front. Plant Sci. 7:1770. doi: 10.3389/fpls.2016.01770
Valley, F. R., Leite, J., Seido, S. L., Passos, S. R., Ribeiro, G., Rumjanek, N. G., et al. (2009). Biodiversity of rhizobia associated with cowpea cultivars in soils of the lower half of the são. R. Bras. Ci. Solo 1, 1215–1226. doi: 10.1590/S0100-06832009000500015
Varshney, R. K., Thudi, M., Roorkiwal, M., He, W., Upadhyaya, H. D., Yang, W., et al. (2019). Resequencing of 429 chickpea accessions from 45 countries provides insights into genome diversity, domestication and agronomic traits. Nat. Genet. 51, 857–864. doi: 10.1038/s41588-019-0401-3
Vauclare, P., Bligny, R., Gout, E., and Widmer, F. (2013). An overview of the metabolic differences between Bradyrhizobium japonicum 110 bacteria and differentiated bacteroids from soybean (Glycine max) root nodules: an in vitro 13C- and 31P-nuclear magnetic resonance spectroscopy study. FEMS Microbiol. Lett. 343, 49–56. doi: 10.1111/1574-6968.12124
Wasike, V. W., Lesueur, D., Wachira, F. N., Mungai, N. W., Mumera, L. M., Sanginga, N., et al. (2009). Genetic diversity of indigenous Bradyrhizobium nodulating promiscuous soybean [Glycine max (L) Merr.] varieties in Kenya: impact of phosphorus and lime fertilization in two contrasting sites. Plant Soil 322, 151–163. doi: 10.1007/s11104-009-9902-7
Wongphatcharachai, M., Staley, C., Wang, P., Moncada, K. M., Sheaffer, C. C., and Sadowsky, M. J. (2015). Predominant populations of indigenous soybean-nodulating Bradyrhizobium japonicum strains obtained from organic farming systems in Minnesota. J. Appl. Microbiol. 118, 1152–1164. doi: 10.1111/jam.12771
Wu, L. J., Wang, H. Q., Wang, E. T., Chen, W. X., and Tian, C. F. (2011). Genetic diversity of nodulating and non-nodulating rhizobia associated with wild soybean (Glycine soja Sieb. and Zucc.) in different ecoregions of China. FEMS Microbiol. Ecol. 76, 439–450. doi: 10.1111/j.1574-6941.2011.01064.x
Wyngaard, N., Franklin, D. H., Habteselassie, M. Y., Mundepi, A., and Cabrera, M. L. (2016). Legacy effect of fertilization and tillage systems on nitrogen mineralization and microbial communities. Soil Sci. Soc. Am. J. 80:1262. doi: 10.2136/sssaj2016.03.0070
Yan, J., Han, Z., Ji, J., Li, Y., Wang, T., Xie, H., et al. (2014). Abundance and diversity of soybean-nodulating rhizobia in black soil are impacted by land use and crop management. Appl. Environ. Microbiol. 80, 5394–5402. doi: 10.1128/AEM.01135-14
Zhang, X. X., Guo, H. J., Wang, R., Sui, X. H., Zhang, Y. M., and Wang. (2014). Genetic divergence of Bradyrhizobium strains nodulating soybeans as revealed by multilocus sequence analysis of genes inside and outside the symbiosis island. Appl. Environ. Microbiol. 80, 3181–3190. doi: 10.1128/AEM.00044-14
Keywords: soybeans, bradyrhizobia, genetic diversity, organic and conventional farming, Kenya
Citation: Gitonga NM, Njeru EM, Cheruiyot R and Maingi JM (2021) Genetic and Morphological Diversity of Indigenous Bradyrhizobium Nodulating Soybean in Organic and Conventional Family Farming Systems. Front. Sustain. Food Syst. 4:606618. doi: 10.3389/fsufs.2020.606618
Received: 15 September 2020; Accepted: 31 December 2020;
Published: 01 February 2021.
Edited by:
Saveetha Kandasamy, A&L Canada Laboratories, CanadaReviewed by:
Mark Paul Running, University of Louisville, United StatesCopyright © 2021 Gitonga, Njeru, Cheruiyot and Maingi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ezekiel Mugendi Njeru, bmplcnVlemVrQGdtYWlsLmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.