- 1Department of Botany and Plant Sciences, University of California, Riverside, Riverside, CA, United States
- 2California Agriculture and Food Enterprise (CAFÉ), University of California, Riverside, Riverside, CA, United States
- 3Department of Microbiology and Plant Pathology, University of California, Riverside, Riverside, CA, United States
- 4Department of Chemical and Environmental Engineering, University of California, Riverside, Riverside, CA, United States
- 5Department of Environmental Sciences, University of California, Riverside, Riverside, CA, United States
- 6River Road Research, Buffalo, NY, United States
The global production of food waste is a far-reaching problem with sizable financial, ethical, social, and environmental costs. Over 66 million tons of food waste is produced annually in the United States alone. This waste can be converted into valuable digestate by-products that promote a circular economy within agri-food systems. The present work investigated the use of two liquid digestates of microaerobic fermentation from mixed food waste and beer mash, respectively, as biostimulants for non-bearing citrus plants (nursery stock) grown in a zero-runoff greenhouse system with recirculating irrigation. The digestates' impact on the structure and diversity of the microbiota was determined on the irrigation water, soil, leaves, roots, and rhizosphere of citrus plants. A combination of culture-dependent (selective media) and culture-independent approaches (Next-Generation Sequencing) was used to assess the composition of the microbial communities and to single out the presence of foodborne pathogens. Our results suggest that the use of digestates is safe (i.e., no human or plant pathogens were present in the digestates or enriched in the plant production system following amendments). Digestates application to the irrigation water reduced the bacterial diversity within 24–48 h and selectively and significantly stimulated beneficial resident host-associated microorganisms (Pseudomonas putida) by two to three orders of magnitude. Carbon dynamics were analyzed in the nutrient solutions by measuring dissolved organic carbon and characterizing carbon species through gas chromatography-electron ionization-mass spectrometry. Our results indicate that dissolved organic carbon in the recirculating irrigation water spikes after each digestate amendment and it is quickly metabolized by bacteria, plateauing 24 h after application. Soil carbon, nitrogen, and nutrient dynamics were also analyzed, and results suggest that digestates increased the concentration of some plant nutrients in soils without causing a surge of potentially toxic elements. This study represents a proof-of-concept for the safe re-use of organic wastes, from farming and consumers, in agriculture. Implementing this type of integrated plant production system could reduce the environmental impact of food waste and benefit the public by improving soil health, reducing agricultural footprint, and increasing crop fitness by deploying a method based on a circular economy and sustainable food production approaches.
Introduction
The current use of natural resources, accelerated by anthropogenic pressures at many levels, threatens to exceed Earth's carrying capacity [World Health Organization Regional Office for Europe (WHO), 2018]. The way we produce, choose, consume, and waste food, poses a serious threat to our planet. As much as one-third of all food produced is either lost before it reaches a store or wasted before it reaches consumers' plates [Food and Agriculture Organization of the United Nations (FAO), 2013]. Food waste has deleterious effects on the environment, our society, and the economy. In the United States (US) alone, 40–50 percent (%) of all food is thrown away, with an estimated annual cost of about US $750 billion every year (FAO, 2019). Most of this waste is not recycled but instead ends up in landfills, taking up to 21% of America's landfill volume and contributing 9% to the total US greenhouse gas emissions [United States Environmental Protection Agency (EPA), 2017; Friedlingstein et al., 2019]. With agriculture utilizing 70% of the water throughout the world (FAO, 2017), food waste also represents a significant loss of freshwater resources. Nevertheless, issues associated with food waste expand beyond its environmental impact: with millions of low-income consumers struggling to gain access to nutrient-rich foods on a daily basis including fresh fruits, vegetables, and meat, while many smallholder farmers are living on the margins of food insecurity; hence better understanding of how to minimize food waste would have a significant societal impact as well. Improving the efficiency of the food supply chain can lower production costs to farmers, while also lowering the cost of food to the consumer and thus increase access to both (FAO, 2015; Horton et al., 2019; Wright et al., 2019).
In recent decades, the need to transition from a linear economy based on the “take-make-consume-dispose” model toward a circular economy has received increased attention worldwide (Ellen MacArthur Foundation, 2013; Wright et al., 2019; Gao et al., 2020). In the circular economy concept, the value of resources is maintained in the system as long as possible, and products are re-claimed to extract as much value as possible before safely returning them to the biosphere. Food waste is rich in energy, water, and nutrients. With the physical, chemical and biological degradation of agricultural soils intensifying, and the demand of mineral fertilizers and pesticides continuously rising, nutrients from organic wastes have potential to be recycled into high-value agricultural products and used as biostimulants (e.g., Plant Probiotic Microorganisms, bioprotectants, biocontrollers, and biofertilizers) (Xu and Geelen, 2018). This approach requires transforming organic wastes through treatments and their reuse in agriculture. While a variety of methods and food-waste management practices exist (e.g., aerated composting) the development of new processes for nutrient-reuse was identified as one of the main challenges in waste management within a circular economy (Mahanty et al., 2017; Ioannidou et al., 2020). Most of the current composting processes are time-, space-, and energy-consuming, and have sizable greenhouse gas (GHG) footprints, and poor carbon sequestration efficiency (Green and Popa, 2011). In recent years, anaerobic digestion for treatments of organic materials from agricultural and food waste have become more common [Monlau et al., 2015; USDA's Agricultural Marketing Services (AMS), 2017]. Controlled bioprocess producing valuable liquid or solid digestates bioproducts have also been developed (Cerda et al., 2019; Elsharkawy et al., 2019; Giwa et al., 2019). Within this concept, the Bokashi fermentation became increasingly popular worldwide, especially for smallholder farmers across Asia and Latin America (Xu et al., 2001; Gómez-Velasco et al., 2014). Unlike compost, Bokashi is mainly driven by microbial anaerobic to microaerophilic fermentation toward acidic conditions. Bokashi's positive impact on crop productivity and the environment has been shown (Möller and Müller, 2012) however, little is known on the effect of Bokashi liquid digestates on soil and plant microbiota and soil carbon dynamics (Christel, 2017). Efficient and cost-effective reuse of waste by-products depends on their relative agronomic efficiency, their stability, pollution risks and safety but more importantly on how they impact the soil and plant microbiome and their intricate network (Pandey et al., 2016; Bernal, 2017; Vanotti et al., 2019).
In recent years, sustainable methods have been developed for increasing crop production while simultaneously reducing chemical inputs such as synthetic chemical fertilizers and pesticides (Lampridi et al., 2019). Harnessing resident soil and plant-associated microbes will be instrumental to make agriculture production systems more sustainable in the long-term (Lugtenberg and Kamilova, 2009; Mendes et al., 2011; Berendsen et al., 2012; Turner et al., 2013; Schlaeppi and Bulgarelli, 2015; Ciancio et al., 2016; Lareen et al., 2016; Berg et al., 2017; Riera et al., 2017; Amaya-Gómez et al., 2020). Plant growth-promoting microbes (PGPM) (i.e., bacteria and fungi), have been widely reported for their beneficial roles in protecting plants from biotic and abiotic stresses and improving crop productivity (Ruzzi and Aroca, 2015; Abhilash et al., 2016). However, many issues need to be addressed before PGPMs can be used on large-scale to support sustainable food production. One of the main challenges is the inconsistent performance of bio-inoculants under field conditions (Timmusk et al., 2017) primarily due to their inability to maintain critical population thresholds (Ruzzi and Aroca, 2015; Berg et al., 2017) and to offset the antagonism from the native microbiome (Acea et al., 1988; van Veen et al., 1997). It has become evident that PGPMs will not be effective for an extended time unless the environment is modified to make it more conducive for their growth and survival (Pagliaccia et al., 2007, 2008; Zhou et al., 2020). On the other hand, it might not be necessary to introduce foreign microorganisms (i.e., inoculant) since soils already hold a vast reservoir of microbes, and plants can enrich their own root-associated microbiome which can function as biocontol agents and PGPMs (Pagliaccia et al., 2007; Lebeis, 2015; Reinhold-Hurek et al., 2015; Lareen et al., 2016; Timmusk et al., 2017; Zhang et al., 2017; Köhl et al., 2019; Raymaekers et al., 2020).
In California, the citrus industry is valued at $3.4 billion with a total economic impact of over $7 billion (Babcock, 2018). California's citriculture is facing historical drought and water management challenges [University of California Agriculture and Natural Resources (UCANR), 2015; Environmental Defense Fund (EDF), 2016] as well as pest (e.g., Asian citrus psyllid, Diaphorina citri Kuwayama) and disease (Huanglongbing, HLB) threats. As result, programs such as the “Citrus Pest and Disease Prevention Program” and the “Citrus Nursery Stock Pest Cleanliness Program” have transformed pest management efforts and citrus nursery production in the state [da Graça et al., 2016; Babcock, 2018; Dai et al., 2019; McRoberts et al., 2019; California Department of Food and Agriculture (CDFA), 2020]. This demanding and dynamic agricultural environment has created a unique opportunity to adopt alternative citrus fruit and tree production systems.
To develop a sustainable next-generation citriculture, it is imperative to capitalize on soil carbon dynamics, soil and plant microbiota, and microbial traits beneficial to citrus, the environment, and the economy. In this context, the development of novel practices that deploy targeted manipulation of microorganisms to stimulate soil life and harness internal ecosystem processes offers a promising approach to enhance agricultural sustainability (Philippot et al., 2013; Bender et al., 2016) and adopt circular economy-based solutions. As demonstrated in other cases, controlled management of liquid digestates through drip irrigation systems in open field or protected citrus orchards could be implemented to enhance native soil beneficial microbial communities. Furthermore, improvement of indoor citrus nursery production could come from adopting zero run-off irrigation systems where 100% of irrigation/fertigation water is recycled and reused however, these are understudied areas (Bailey and Lazarovits, 2003; Bonilla et al., 2012; Cao et al., 2016; Ciancio et al., 2016; Barzee et al., 2019).
Recent studies have focused on understanding the role of the citrus microbiome and its impact on plant health and productivity (Trivedi et al., 2010, 2012; Xu et al., 2018). It has been suggested that members of the citrus microbiome influence the interaction between Candidatus Liberibacter asiaticus (the bacterium associated with HLB) and citrus (Blaustein et al., 2017; Wang et al., 2017; Blacutt et al., 2020). Several studies have investigated why some citrus trees identified in severely HLB-diseased citrus groves (with the same genotype as symptomatic trees), evaded the HLB disease. Preliminary published research suggests that the HLB-escaped trees differ from the diseased ones because of their associated native microbial community composition (Sagaram et al., 2009). The microbial community of escaped plants seems to be enriched in beneficial traits as compared to those of symptomatic trees (Trivedi et al., 2011, 2012; Riera et al., 2017).
Taken together these elements led to the theory that native soil and plant root-associated microorganisms, endowed with plant growth promoting capability and digestate-metabolizing abilities, can be used to recycle food waste while supporting sustainable agroecosystems with reduced synthetic chemical input. We formulated the following hypothesis for our study: (1) digestate products are suitable for delivery to citrus through drip irrigation lines; (2) digestates are safe to use (i.e., no plant or human pathogens); and (3) digestates will selectively increase certain bacterial groups (such as Pseudomonads) that are known to improve plant health. In this study, we tested two types of digestates, one derived from mixed post-consumer food waste (FW) and one from beer mash (BM).
To unravel the potential use of digestates in zero-runoff indoor citrus plant production, we investigated the effects of FW and BM liquid digestates products as biostimulants. Their impact on the structure and diversity of the microbiota in the recirculating nutrient solution and leaves, roots, and rhizosphere of citrus plants, was studied using a combination of culture-dependent (selective media substrates) and culture-independent approaches (next-generation sequencing). The taxonomic composition of the microbial communities and the bacteria isolates enriched by FW and BM applications, were identified, and their beneficial traits investigated. Simultaneously, potting soil and nutrient solution chemical analyses were performed to determine whether FW and BM amendments significantly changed soil carbon and nitrogen content over the course of the treatments. Using gas chromatography-electron ionization-mass spectrometry (GC/EI-MS) we demonstrated the dependence of microbial growth on available dissolved organic carbon in the nutrient solution and identified overall variation in dissolved organic carbon speciation.
Materials and Methods
Materials, Analyses, and Processing
Two digestate products, one obtained from mixed food waste (FW) and one from beer mash (BM), were used as amendments (applied through drip irrigation) to the recirculating nutrient solution (Figure 1). The two microaerobic fermentation digestates, provided by River Road Research, were produced under a controlled anaerobic Bokashi-like fermentation technology in controlled conditions (Popa et al., 2017). Digestates were stored at 4°C in anaerobic conditions in collapsible plastic containers. We selected these two digestates to test because they are commonly available wastes in southern California urban surroundings; however, the spent grain in the beer mash represents a uniform feedstock while food waste composition is more variable and could contain undesirable elements such as sodium from processed food. Food Waste Digestate samples were sent via next day delivery to A&L Western Agricultural Labs, Modesto CA, an agricultural and environmental testing laboratory, for fertilizer and metal analyses. Fertilizer analyses were performed using several different methods (Please see Supplementary Table 3). Metal analyses were performed using an EPA Method SW846-6010 except for mercury for which the method used was EPA Method SW846-7471A. Fertilizer and metal analysis results are reported in Table 1 and Supplementary Table 3. Dissolved organic carbon species in the two digestates were analyzed by GC/EI-MS at UCR, and results are reported in Supplementary Table 1.
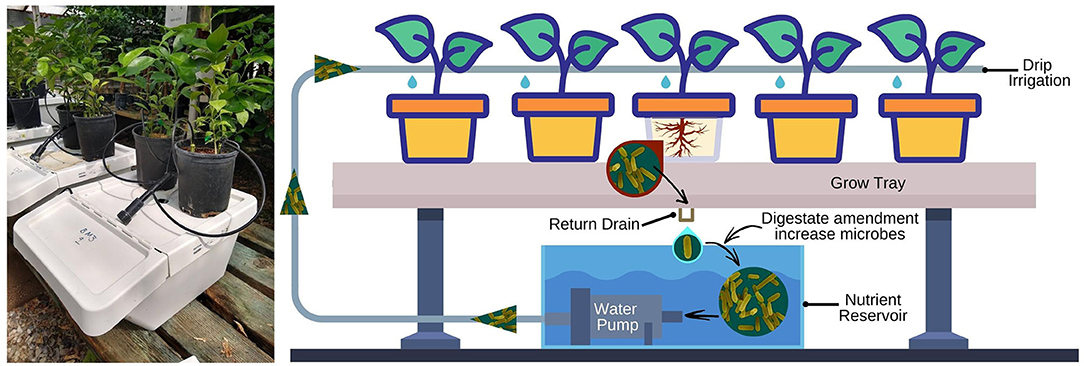
Figure 1. On the left, citrus seedlings in drip-irrigated hydroponic units in a temperature-controlled greenhouse. On the right, simplified layout of a recirculating hydroponic system. FW and BM digestates, at 4 ml/L of nutrient solution, were amended to the reservoir at 7 days interval.
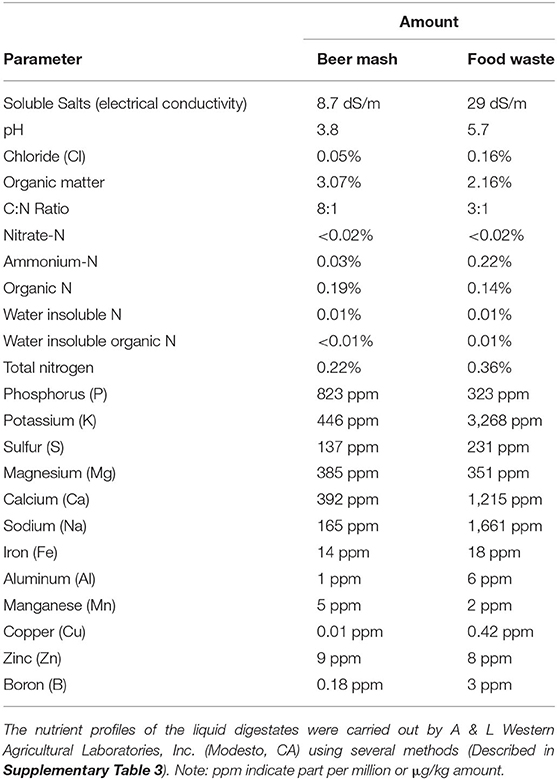
Table 1. Nutrient analysis of Beer Mash and Food digestates used as amendment for all greenhouse experiments.
A&L Western Agricultural labs tested the two digestates for fecal coliforms and Salmonella presence. Further analysis using culture-dependent (King's B, Nutrient Agar, and Tryptic Soy Agar selective media substrates) and culture-independent approaches (next-generation sequencing) were carried out at UCR to assess the presence of pathogens and any active bacteria in the digestates. Upon arrival, each digestate was plated out on King's B, Nutrient Agar, and Tryptic Soy Agar selective media substrates to identify any possible alive culturable bacteria residual. Both stocks were also collected onto sterile nitrocellulose membranes of two different sizes, 0.22 and 8 μm (Millipore, nitrocellulose). The filter-concentrated bacterial cells were then processed as described below to extract DNA and carry out Illumina sequencing of 16S rRNA gene amplicons. Laboratory tests for salmonella (A&L Labs) were negative, while fecal coliform counts were less than the reporting limit of 3.0 MPN/g. Our lab culturing on the digestates were negative. NGS results are discussed and reported in Supplementary Figure 2.
Zero-Runoff Cultural Systems
Experiments were conducted in temperature-controlled greenhouses (20–32°C) using 9 separate recirculating zero-runoff units with top drip irrigated systems. Pineapple sweet orange seedlings (Citrus sinensis L. Osbeck) grown in 1-gallon plastic pots containing a potting soil mix (made of 33% coarse silica sand, 33% redwood shavings and 33% fine coir) were used as a host plant to assess the change of citrus, soil and nutrient solution microbiota after waste by-products amendments through the drip-irrigated system. Each zero-runoff system (located on a bench ca. 40 centimeters (cm) above ground level) consisted of an upper plastic trough, for plant growth, and a lower reservoir where the nutrient solution is kept and recycled (Figure 1).
Each trough held 2 pots with 3 citrus seedlings each. The nutrient solution was pumped from the reservoir to the trough and distributed to each plant via drip tubing with 7.5 liters per hour (L/h) emitters. Standard oxygenation of the nutrient solution was accomplished by air pumps bubbling air into the nutrient solution 20 min every 2 h throughout the day. The excess nutrient solution from each pot drained by gravity back to the reservoir and was recirculated. Plants were irrigated for 30 minutes (min) twice a day between 8:00 A.M. until 8:00 P.M. The solution pH was adjusted manually to 5.8 ± 0.4 with H3PO4 and KOH, every week. The electrical conductivity (EC 1.6 ± 0.2 mS/cm2) and the nutrient solution volume in reservoirs (24 L) were monitored once a week and levels maintained as required. The final element concentrations in the nutrient solution were as follows: Total nitrogen, N 250 (Ammoniacal nitrogen, NH4 86.9; Nitrate Nitrogen, NO3 150; Urea Nitrogen 13); Phosphate, P2O5 59.5; potassium oxide, K20 238.1; boron, B 0.31; copper, Cu 0.31; iron, Fe 1.25; manganese, Mn 0.63; molybdenum, Mo 0.15; and zinc, Zn 0.63 μg/ml.
Citrus seedlings were obtained from the Citrus Clonal Protection Program (CCPP) at the University of California, Riverside. One-year-old seedlings were placed into the zero-runoff systems described above. Plants were kept for 4 days on the recirculating zero-runoff systems before the first treatment to allow the bacteria from the plant root and rhizosphere to be dispersed into the nutrient solution reservoirs. There were three experiments in which the reservoirs were amended with different treatments. Each experiment consisted of three treatments, 1) FW by-product, 2) BM by-product, and non-amended control nutrient solution, with 3 replications per treatment and 18 total plants (6 plants for each replication) per treatment. FW and BM digestates, at 4 ml/L of nutrient solution, were first applied 5 days after the start of the experiment and reapplied at 7 days interval for 63 days for a total of 9 treatments applications. In all greenhouse experiments, the by-products were amended to the nutrient solution 30 min before the start of the irrigation. Between experiments, each zero-runoff system was sterilized with 10% bleach and rinse with water.
Culture Dependent Characterization of Bacterial Populations in Nutrient Solutions
Ten-ml samples of the nutrient solution were collected at different intervals at 0, 24, and 48 hours (h) after the waste by-products amendment from the reservoir of each treatment. After serial 10-fold dilutions, aliquots were plated in quadruplicates, manually, onto 10-cm diameter Petri dishes containing King's B medium (KB) (Sigma-Aldrich, USA) for enumeration of fluorescent pseudomonads, and Tryptic Soy Agar (TSA) (Sigma-Aldrich, USA) for enumeration of aerobic heterotrophic bacteria. Colonies were counted after 24 h incubation at 28°C. Fluorescent pseudomonads were counted under UV light. Enumerations were reported as colony forming units (CFUs).
Bacterial Collection and Identification in Nutrient Solutions and Bacterial Beneficial Traits
To investigate the effect of the digestates amendments on the selection of specific culturable bacteria in the recirculating nutrient solution, 231 single bacterial colonies were isolated from the nutrient solutions from the greenhouse experiments. Forty-two colonies of representative predominant bacteria were then selected for DNA extraction and sequencing to provide genetic identification. Thirty-four isolates were collected from FW and BM treated and nine from non-treated nutrient solutions. For the selection, Petri dishes containing 50–100 single bacterial colonies were chosen, and single colonies were randomly picked from a different sector of each plate. Each isolated colony was purified by repeated streaking onto King B agar. Purified isolates were stored at room temperature on KB agar plates or maintained for long-term storage at −70°C in sterile water supplemented with 15% glycerol. Single colonies were picked, mixed, and vortexed in 100 μl sterile distilled deionized water, out of which 1.25 μl of the mixture was used as DNA template in PCR, amplifying subunits of ribosomal RNA genes or 16S rDNA (dos Santos et al., 2019). Forward 27F (5′-AGAGTTTGATCMTGGCTCAG-3′) and reverse 1492R (5′- TACGGYTACCTTGTTACGACTT-3′) primers were used for 16S rDNA PCR amplification (Suzuki and Giovannoni, 1996). The PCR mixture consisted of 1 × PCR buffer, MgCl2 (2.5 mM), 250 μM each dNTP, 0.4 μM each primer, and 2.5 U of DNA polymerase (JumpstartTM Taq DNA polymerase, Sigma-Aldrich, USA). The thermal cycling conditions used were as follows: 94°C for 5 min; 35 cycles of 94°C for 20 s, 48°C for 20s, and 72°C for 1 min; then 72°C for 10 min. The PCR products were purified using a QIAquick PCR purification kit (Qiagen, Chatsworth, CA, USA) and then separated in 2.0% agarose gel electrophoresis. Resulting DNA bands were ~1,400 bp large. The purified PCR products were then sequenced using the chain termination method (Sanger sequencing). Nucleotide sequences of 43 PCR amplicons were edited and assembled utilizing the Vector NTI software package (Thermo Fisher Scientific, USA). The consensus sequences of individual contig (5 total; GenBank accession numbers MW144948-MW144952) with different lengths were subjected to a remote BLAST search (megablast) in the NCBI nr/nt database using an Entrez query with default setting. The top species IDs with their accession number, % coverage and identity are reported in Table 2.
To acquire preliminary information on potential beneficial traits possessed by the most abundant bacteria isolated from the treated nutrient solution (listed in Table 2) the Pathosystems Resource Integration Center (PATRIC) database was used to search within the 16S rDNA gene sequences. Briefly, consensus sequences obtained from the 43 16S rDNA-sequences described above were used to carry out de novo functional annotation analysis with PATRIC and obtain reference genomes that best match the 16S region. The reference whole genomes were then analyzed with the PATRIC Protein family sorter to obtain the heatmap of relevant genes. Results are reported in Supplementary Figure 1. The beneficial traits investigated here were the one typically reported to be involved in phosphate solubilization, siderophore production, and iron acquisition, VOC production, osmoprotection, and osmotic tolerance, phytohormone production, antagonism, and nutrient competition (Riera et al., 2017).
Because members of the genus Pseudomonas dominated the bacterial community at 24 and 48 h, but the ITs analysis reported the most abundant Pseudomonads as “Unknown,” we further investigated these Amplicon sequence variants (ASV)s by performing a BLAST search against the nucleotide database using default parameters. The results are reported in Table 3.
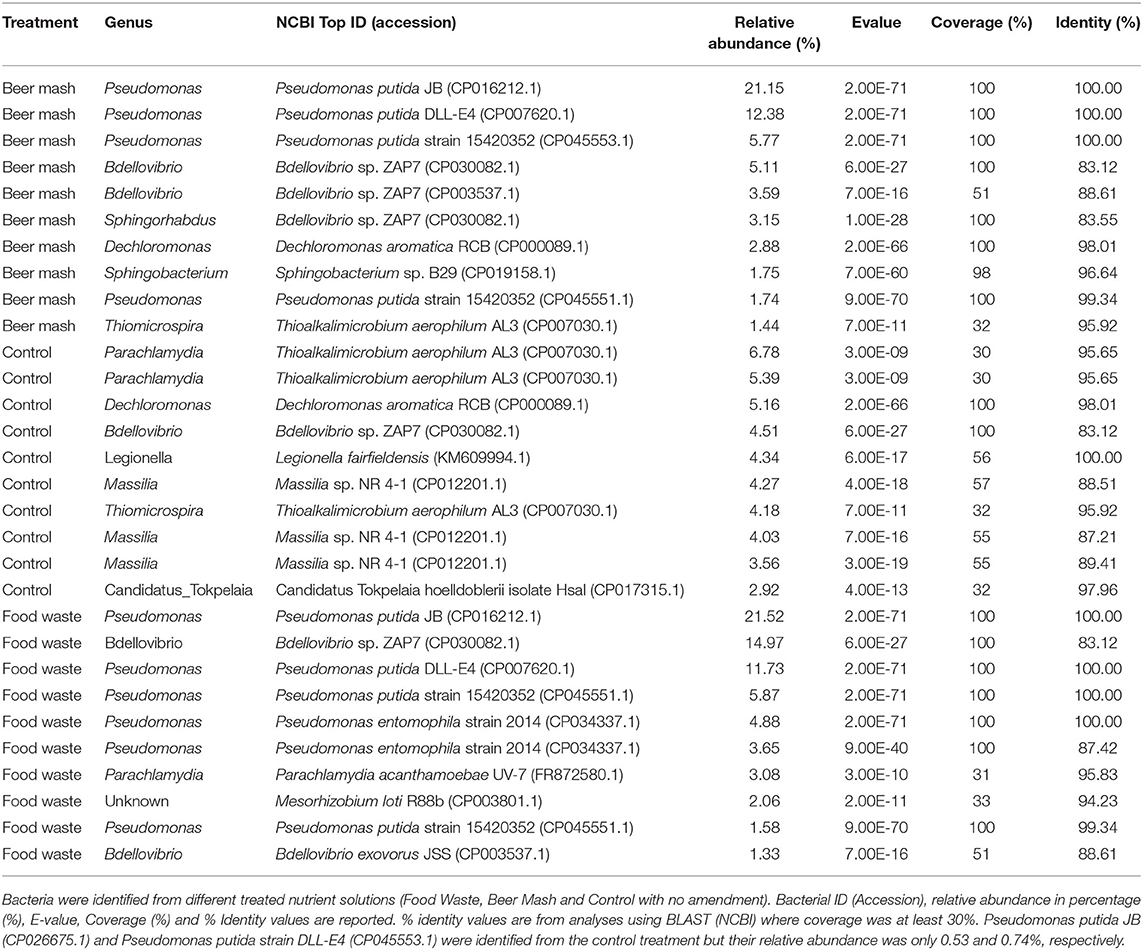
Table 3. Top 10 most abundant bacterial species identified in the nutrient solution by NGS of ITS rRNA encoding region.
Nutrient Solution Samples Collection for Culture-Independent Characterization of Microbial Communities
Following the preliminary results from the bacterial culturing on KB and TSA media, samples of the nutrient solution were collected at different times (0, 24, and 48 h after treatments) for 4 weeks (on 1st, 2nd, 8th, and 9th week) to extract bacterial DNA and provide next-generation sequencing (NGS) datasets of the bacteria associated with recirculating nutrient solution. One liter of nutrient solution from the reservoirs from each treatment was collected onto sterile nitrocellulose membranes of two different sizes, 0.22 and 8 μm (Millipore, nitrocellulose), using a sterile vacuum aspirator filtering apparatus. The filter-concentrated bacterial cells were resuspended in 10–30 ml buffer 50 mM Tris–HCl pH 9, 20 mM EDTA, 400 mM NaCl, 0.75 M sucrose, and stored until extraction procedure in −80°C freezer. Before extraction, the filters were sonicated in their original resuspension buffers by placing the 50 ml falcon tubes in cold water for 40 min. Before sonication, ice water in the Branson CPX952518R sonicator (Branson Ultrasonics™, Danbury, CT, USA) was degassed for 5 min. During sonication, the frequency was set on 40 kHz. Genomic DNA was extracted from 200 μl of bacterial suspension using the PowerSoil® DNA Extraction Kit (MO BIO, Carlsbad, CA, USA). The purity and the concentration of the DNA template were assessed with an Infinite M1000 Pro (Tecan, Männedorf, Switzerland). The final DNA was eluted in 100 μl of DNA Elution Buffer. The DNA was aliquoted into three 1.5 ml Eppendorf DNA LoBind Tubes with 32 μl in each tube and dried using a SpeedVac concentrator (Thermo Fisher Scientific). The dried DNA was stored at −20°C until further use for bacterial Illumina library construction.
Citrus Leaves, Roots, and Soil Samples Collection for Culture-Independent Characterization of the Microbial Community
Two weeks after the last digestates amendment (when the total bacterial population in all nutrient solutions reservoirs had reached 104-105 CFU/ml), plants were taken out from the zero-runoff system and leaves, roots, and soil samples taken. At least 10 mature and fully expanded leaves were randomly collected from each pot (containing 3 plants of Citrus sinensis each) and pooled into 50 ml falcon conical tubes. Feeder roots were sampled from two sides of the tree after the root system was taken out from the plastic pot and topsoil was removed. Feeder roots were pulled apart, shaken to remove soil, and placed into 50 ml conical tubes. Between each sampled tree, the clippers and forceps were cleaned by spraying a 20% household bleach solution on them, and then wiping them clean with paper towels. Researchers also changed their gloves between each potted tree that was sampled. Thirty grams (g) of potting soil samples taken ~10 cm from the base of the trunk, was randomly collected from each pot, and pooled into 50 ml falcon tubes (Corning). All samples were immediately placed in a cooler with ice and then stored at 4°C. All tissue samples were prepared and lyophilized within 24 h from sample collection. Samples were not surface-sterilized; thus, our microbial NGS datasets and analyses included both epiphytes and endophytes. Leaf samples were cut into 1 to 2 cm long pieces and ~6 g of leaves were placed into 50 ml conical tubes and stored at −80°C; root samples were rinsed with autoclaved purified water (Barnstead Mega-Pure System MP-6a, Thermo Fisher Scientific, Waltham, MA, USA) and ~5 g of rinsed root tissue was placed into 50 ml conical tubes and stored at −80°C. Tissue samples were then lyophilized with a bench-top freeze dryer (Labconco FreeZone 4.5L, Kansas City, MO, USA) for 16 to 20 h. The lyophilized tissues were stored at −80°C.
DNA from freeze-dried leaves and roots were obtained as described previously by Ginnan et al. (2018). The purity and the concentration of the DNA template were assessed as described above. The DNA was aliquoted into three 1.5 ml Eppendorf DNA LoBind Tubes with 32 μl in each tube and dried using a SpeedVac concentrator (Thermo Fisher Scientific). The dried DNA was stored at −20°C until further use for bacterial Illumina library construction. DNA from 100 mg of freeze-dried soil was extracted using the MoBio PowerSoil® DNA kit (MoBio Laboratories Inc., CA, USA) and eluted as described above.
Library Construction, Sequence Processing, and Bioinformatics Workflow
Illumina bacterial ITS rRNA gene libraries were constructed as follows. PCRs were performed in an MJ Research PTC-200 thermal cycler (Bio-Rad Inc., Hercules, CA, USA) as 25 μl reactions containing: 50 mM Tris (pH 8.3), 500 μg/ml bovine serum albumin (BSA), 2.5 mM MgCl2, 250 μM of each deoxynucleotide triphosphate (dNTP), 400 nM of the forward PCR primer, 200 nM of each reverse PCR primer, 2.5 μl of DNA template, and 0.625 units JumpStart Taq DNA polymerase (Sigma-Aldrich, St. Louis, MO, USA). PCR primers ITS-1507F (GGTGAAGTCGTAACAAGGTA) and ITS-23SR (GGGTTBCCCCATTCRG) were used to target a small portion of the small subunit, the large subunit, and the hypervariable ITS region, with the reverse primers including a 12 bp barcode (Hunt et al., 2006; Frank et al., 2008). Thermal cycling parameters were 94°C for 5 min; 35 cycles of 94°C for 20 s, 56°C for 20 s, and 72°C for 40 s, and followed by 72°C for 5 min. PCR products were purified using the Qiagen QIAquickPCR Purification Kit (Qiagen, Valencia, CA, USA).
DNA sequencing was performed using an Illumina MiSeq (Illumina, Inc, San Diego, CA, USA). Sequences were demultiplexed using Qiime 1.9.1 (Caporaso et al., 2012). Further quality control of the reads was conducted through the DADA2 v 1.14.0 pipeline (Callahan et al., 2016) including removal of PhiX, quality control (maxEE=1, min length=135bp, maxN=0), dereplicating, and creation of the Amplicon sequence variants (ASV) table. In the dataset, there were 11,119 ASVs in the first experiment and 12,861 ASVs in the second experiment initially identified. After removing sequences which did not belong to the kingdom bacteria or had an unknown phylum identification and did not occur in the dataset at least 3 times (singletons and doubletons) 8,855 and 10,264 ASVs remained. Samples that did not have at least 1,000 sequences were removed leaving a total of 77 and 63 samples, respectively, in the two experiments. Taxonomy identification was assigned using custom scripts and BLAST. Phyloseq v 1.30.3 (McMurdie and Holmes, 2013) was used to assist in the graphical representation of the data. For taxonomy barplots, data were aggregated to the genus level and transformed to relative abundance. Taxa that did not occur > 1% relative abundance were removed. Shannon's index was used to assess alpha diversity. Statistical differences for the alpha diversity and the taxonomic data were assessed with Kruskal–Wallis and pairwise Wilcoxon tests. Beta diversity was assessed using Bray-Curtis dissimilarity, which was depicted using Principal Coordinates Analysis (PCoA), and statistically assessed by performing Adonis tests [999 permutations]. The bacterial sequences have been deposited in the National Center for Biotechnology Information (NCBI)'s Sequence Read Archive (SRA) under bioproject PRJNA670208.
Dissolved Organic Carbon (DOC) Analysis of the Nutrient Solution and Amendments
Samples of nutrient solution were taken from all 9 replicates- 25 ml total per zero-runoff system, as well as samples of fresh nutrient solution before it went into the system, and samples of the original liquid amendment for each treatment - FW and BM. Samples were taken before the amendment, after amendment, and after 24, 48, and 72 h on weeks 1 and 4. Immediately after sampling, all samples were stored on ice and filtered using 0.22 μm syringe filters to filter out microorganisms and root tips, leaving only dissolved organic carbon compounds. 3 ml samples were analyzed by a DOC analyzer.
Gas Chromatography/Electron Ionization-Mass Spectrometry (GC/EI-MS) Analysis
Nutrient solution and amendments were analyzed by GC/EI-MS to characterize dissolved organic carbon species. A protocol similar to Chen et al. (2019) was followed with some modifications. 3 ml of filtered samples were dried using N2 gas evaporation and then trimethylsilylated with 100 μL of N,O-bis(trimethylsilyl)trifluoroacetamide (BSTFA; 99 μL) with trimethylchlorosilane (TMCS; 1 μL) (≥99%, SUPELCO) and 50 μL anhydrous pyridine (≥99%, EMD Millipore) at 70°C for 1 h. A blank (control) made of 50 μL of anhydrous pyridine and 100 μL of BSTFA was used. This derivatization procedure converts hydroxyl functional groups into volatile trimethylsilyl (TMS) derivatives that can be analyzed with GC/EI-MS (Agilent 6890N GC coupled with 5975 MSD). An Agilent 7683B series autosampler was used to inject 5 μL of each derivatized sample in splitless mode. A J&W Scientific DB-5 column (30 m × 0.25 mm i.d., 0.25 μm film) was used to separate the TMS derivatives. The carrier gas used was helium at a flow rate of 1 ml/min. The GC ran for an overall time of 65.17 min at the following temperature program: initial temperature held at 60°C for 1 min, temperature ramp of 3°C min−1 up to 200°C, temperature held at 200°C for 2 min, temperature ramp of 20°C min−1 up to 310°C, and finally temperature held at 310°C for 10 min.; a solvent delay time of 10.5 min was programmed. The GC inlet temperature was set at 250°C and detector at 280°C. The MS scan was performed in the m/z range of 50-550. GC/EI-MS data, including peak ID, peak height and area, and likely compound or molecule match, was processed using Agilent GC/MSD ChemStation software. We then used the O/C vs. H/C ratios of elemental formulas to get an overview of the compound categories within each amendment and generated van Krevelen diagrams that allow for classifying compounds general compound classes such as lipids, proteins, amino sugars, carbohydrates, lignin, tannins, condensed aromatics, and unsaturated hydrocarbons (Minor et al., 2014; Rivas-Ubach et al., 2018). Dissolved organic carbon species characterized in Beer Mash and Food digestates GC/EI-MS analyses are reported in Supplementary Material.
Carbon, Nitrogen, and C:N Ratio Changes in the Potting Soil
Five grams of soil were sampled on Week 1 and Week 10, in composite from each of the 3 replicates for Control, Beer Mash, and Food Waste, then sieved (2 μm), dried, and ground for analysis. Three mg samples of each of the replicates from each of the treatments were prepared (9 samples per week, 18 samples total) for analysis of total C, total N, and C:N using the Costech ECS 4010-Delta V Plus Isotope Ratio Mass Spectrometer. These standards were used: Acet, Glycine, Peach, EM Soil, USGS64, and USGS66, and samples were combusted completely in EA at 88% content CO2 and 1020C combustion T set.
Elemental Composition Changes in the Potting Soil
On week 1 and week 10, the first and last weeks of the experiment, composite samples were taken from each of the 3 replicates for each treatment (Control, Beer Mash and Food Waste), homogenized, sieved (2 μm) and dried for analysis by XRF (X-ray fluorescence) to determine the elemental composition of the soils and whether it changed over time with treatment.
Statistical Analyses and Diversity Measurements
Data from the Dissolved Organic Carbon (DOC) changes in the nutrient solution at different time points (0, 0.1, 24, and 48 h) after digestates amendment and data from carbon, nitrogen, and C:N ratio changes in the potting soil were analyzed by one-way analysis of variance (ANOVA) followed by the all Pairwise Multiple Comparison Procedures (Holm-Sidak method), at P = 0.05 and at P = 0.01, respectively. Data from each elemental composition changes (μg/g) between week 1 and 10 in soil samples from different treatments were analyzed by ANOVA tests followed by multiple comparisons vs. control group procedures (Holm-Sidak method), at P = 0.01. Cumulative data from NGS analysis on relative abundance of fluorescent pseudomonads in the nutrient solution samples taken from 4 weeks out of 9 weeks experiments were analyzed by means of Kruskal–Wallis test at p < 0.05. Data were analyzed using SigmaPlot version 14.0, from Systat Software, Inc., San Jose, CA, USA. Measurements included 3 technical replicates. All experiments were repeated at least 3 times, and results from a single representative experiment were chosen for presentation. The bioinformatics and statistical procedures for all Illumina MiSeq data are described within the “library construction, sequence processing, and bioinformatics workflow” section above.
Results
Effects of Mixed Food Waste and Beer Mash Digestates on Culturable Bacterial Populations in the Nutrient Solution
The culturable bacterial populations were monitored in the nutrient solution before and after the amendment. Before the addition of FW and BM, the recirculating nutrient solution used to irrigate citrus plants harbored ca. 5 log CFU/ml of aerobic heterotrophic bacteria (AHB; Figure 2A) and ca. 4 log CFU/ml level of fluorescent pseudomonads (Figure 2B). However, within 24 h after the addition of FW and BM, the average population of fluorescent pseudomonads in amended treatments (ca. 6-7 log CFU/ml) were two to three orders of magnitude greater than that in non-amended treatments (ca. 3 logs CFU/ml; Figure 2B). The population surge occurred after each consecutive addition (9 weeks) of FW and BM to the nutrient solution. Within 24 to 48 h after the addition of FW and BM, the average fluorescent Pseudomonads population accounted for ca. 97.51% (ranging from 13.74 to 88.13%) and 44.84% (ranging from 2.35 to 94.57%) of the total AHB population, respectively, in the FW and BM -amended solutions but only ca. 39.73% (ranging from 1.34 to 86.63%) in the non-amended nutrient solution.
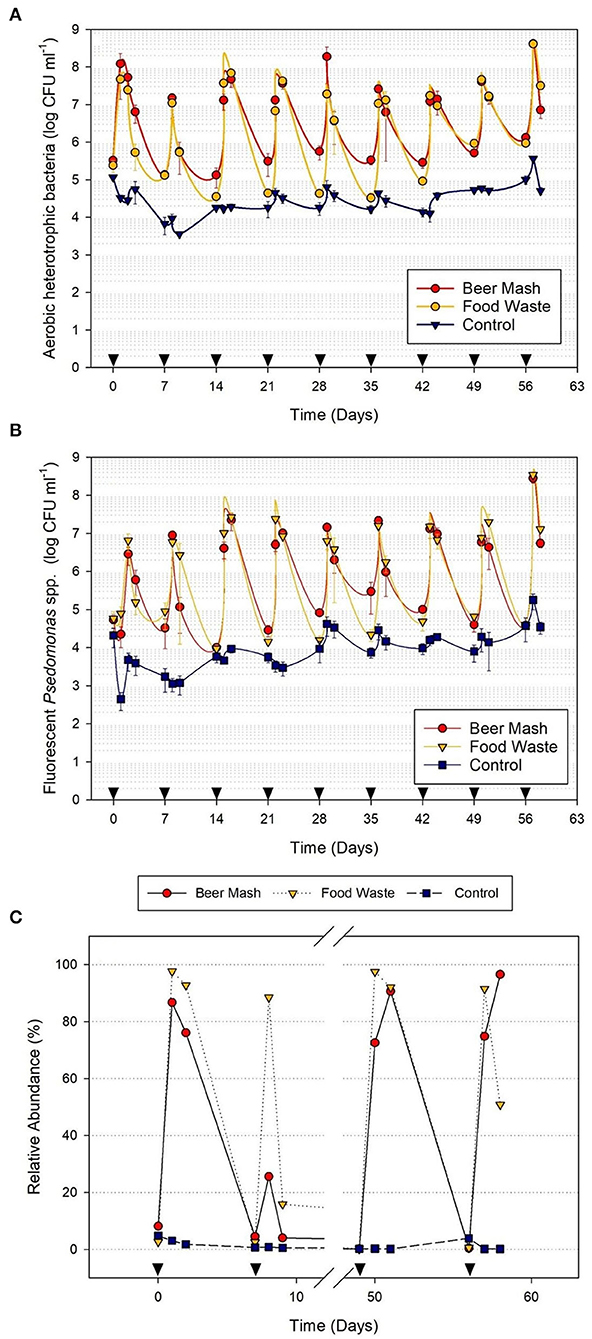
Figure 2. Population dynamics of (A) aerobic heterotrophic bacteria and (B) fluorescent pseudomonads in the nutrient solution as detected by selective media in a greenhouse experiment with citrus as the host plant. Treatments consisted of nutrient solution amended and not amended (control) with beer mash BM and food waste FW digestate every week for 9 weeks. (C) Relative abundance of fluorescent pseudomonads as detected by NGS analysis of samples taken from 4 weeks out of 9 weeks experiments. Cumulative data from NGS were analyzed by Kruskall–Wallis tests to verify statistical differences among treatment groups (See Supplementary Figure 3). Head arrows indicate the timing of FW and BM applications (4 ml/L). Error bars represent the SE of the mean (n = 4) where variation was great enough to be presented.
Taxonomy-Based Bacterial Diversity in the Nutrient Solution and Bacterial Identification
The microbiome in the nutrient solution used in a zero-runoff production system was characterized from samples collected from FW and BM-treated and untreated nutrient solutions from 2 experiments. Our results from the culture-independent NGS method confirmed the results obtained by the culture-dependent methods reported above. The bacterial composition of the nutrient solutions (Figure 3A, FW and BM-treated and untreated) was distributed amongst 11 defined phyla with Proteobacteria being the most abundant. Barplots depict the top 15 defined genera. For data visualization, all other genera were grouped into “other” and colored gray (Figure 3A). Overall, the relative abundance of the bacterial communities in the 24 h and 48 h untreated control samples was not significantly different compared with the 0 h samples. The largest number of bacterial sequences identified in the untreated nutrient solutions belonged to the genera Parachiamydia (19.4%), Massillia (17.3%), and Thiomicrospira (9.58%), with no significant differences in their relative abundance between 0, 24, and 48 h. Figure 3A shows that while the genera Bdellovibrio, Parachiamydia, Massillia, and Thiomicrospira were the most dominant in all 3 treatments (control, FW and BM) at time 0 h, their relative abundance decreased to undetectable levels through 24 and 48 h following the amendment of the nutrient solution with FW and BM, with the exception of Bdellovibrio, which increased again to 22.9 and 10.2%, respectively, in both FW and BM at 48 h. At the same time, the largest number of bacterial sequences identified in the FW and BM treated nutrient solutions at 24 and 48 h belonged to the genus Pseudomonas. In control samples and at time 0 h, Pseudomonas never exceeded more than 4% of the total relative abundance. In BM samples at 24 and 48 h, Pseudomonas represented 66.83 and 64.91%, respectively. While in FW, Pseudomonas represented 93.79 and 62.89% of the relative abundances for 24 and 48 h, respectively (Figure 3A). Principal Coordinates Analysis (PCoA) of Bray-Curtis distances revealed that nutrient solution samples formed two distinct clusters according to sample time collection and amendment, with samples collected at time 24 and 48 h following FW and BM amendments forming one cluster and samples collected at time 0 h with all treatments in the other (P < 0.01 [Adonis]) (Figure 3B).
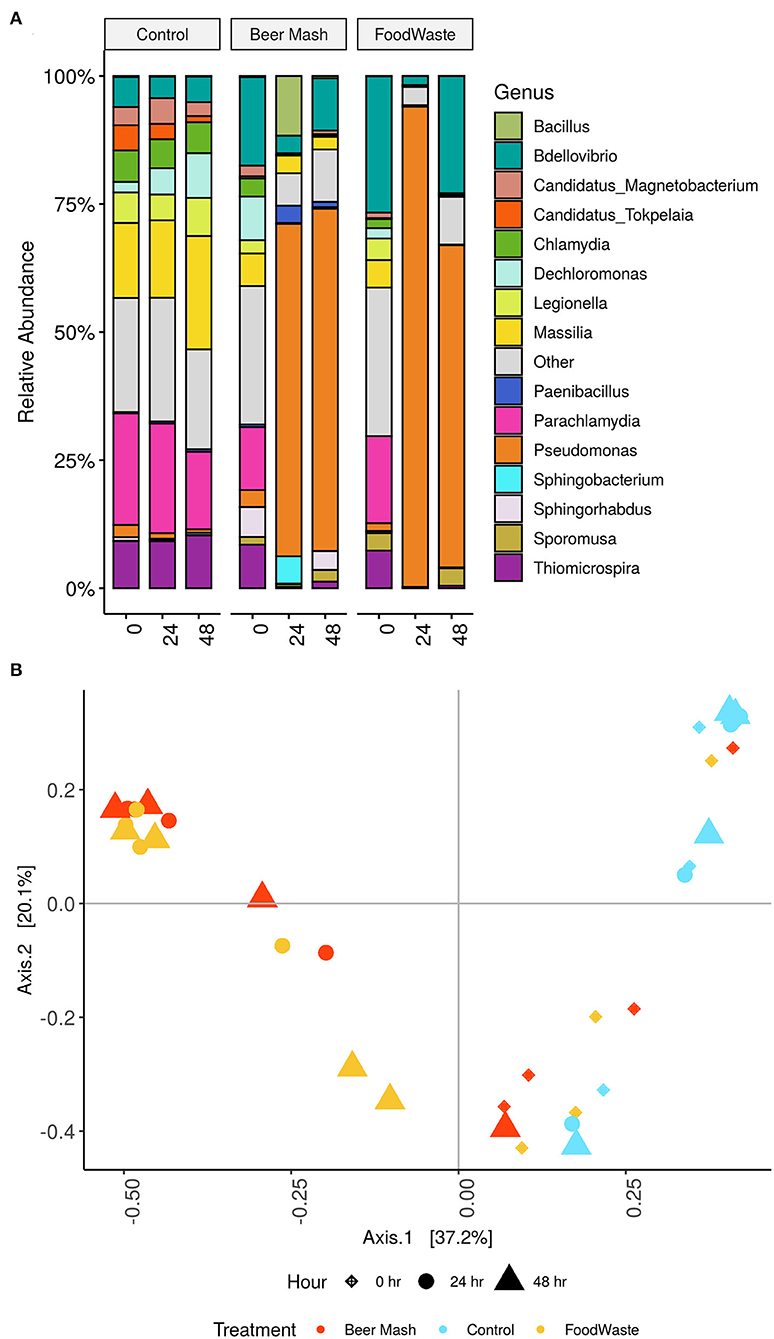
Figure 3. (A) Stacked column bar graph representing the relative abundance and distribution of the 15 most abundant genera across the three nutrient solutions treated with FW and BM or not treated (control) and across time at 0, 24, and 48 h. (B) Clustering of samples, following amendment of nutrient solution with FW and BM or not amended (control), by PCoA based on Bray-Curtis similarity distance. The first two principal coordinates are plotted on the x- and y-axes, respectively representing 57.3% of the total variation). Nutrient solution collections sampled at three different time points are represented by shape, with diamond (♦) for 0 h, circle (•) for 24 h and triangle (▴) for 48 h. Amendments are indicated by color, with light blue color for the not amended (control), red for BM amended and yellow for FW amended nutrient solutions.
Because members of the genus Pseudomonas dominated the bacterial community at 24 and 48 h, we analyzed their relative abundance within this genus and the taxa within the top 10 most abundant bacterial species (Figure 2C and Table 3). Cumulative results from the NGS analysis of samples taken from 4 weeks out of 9 weeks experiments, confirmed similar significant increases in the resident fluorescent pseudomonads population, compared to the non-amended control treatment after the addition of FW and BM to the recirculating nutrient solution. In BM samples at 24 and 48 h, Pseudomonas represented 86.69, 25.58, 72.56, 74.83, and 76.07, 4.03, 90.64, and 96.58% in week 1, 2, 8, and 9, respectively. While in FW, Pseudomonas represented 97.64, 88.47, 97.49, 91.55, and 92.77, 15.83, 92.10, and 50.84% of the relative abundances for 24 and 48 h in week 1, 2, 8, and 9, respectively (Figure 2C and Supplementary Figure 3). At the highest resolution, two strains closely related to Pseudomonas putida (strain JB, DLL-E4 and 15420352) were found to be the most abundant taxa, with a relative abundance of around 39% in both treatments, within the pseudomonas species identified among 103 ASVs present in both BM and FW treated nutrient solutions. A Bdellovibrio sp. was the 2nd highest abundant taxa with a relative abundance of around 14% in both treatments (Table 3). Bacterial species identified by 16S rRNA gene Sanger sequencing and a culturing approach confirmed that Pseudomonas sp. (strain HAL1, Contig 19) and Pseudomonas moorei (strain 28, Contig 14) as the most abundant isolates in both BM and FW treated nutrient solutions (Table 2). When the taxonomic affiliation of the same 2 contigs (#14 and 19) was investigated, and the referenced whole genomes were analyzed against the PATRIC database, the closest strain to both contigs in both BM and FW treated nutrient solutions was again Pseudomonas putida (strain JB), which shared 99.0 and 100% gene sequence similarity, respectively (Supplementary Table 2).
Effect of Waste By-products on Citrus Leaves, Roots, Rhizosphere Soil Bacterial Populations
Samples of citrus leaves, roots, and soil around the roots were taken on week 11 from all 9 replicate zero-runoff production systems. Samples were taken 2 weeks after the last amendment of FW and BM into the system. The bacterial composition on citrus leaves, roots, and soil at the end of the greenhouse experiment was distributed amongst 18 defined phyla. For graphical representations, the top 15 genera from each tissue type are highlighted and the remaining genera are grouped in “other” and colored in gray (Figure 4). Overall, the relative abundance of the bacterial taxa in the untreated control samples were similar to the FW and BM-treated samples. The largest number of bacterial sequences identified in the leaf samples were Bacillus, comprising 17.2, 16.4, and 9.4% of the relative abundance of Control, BM, and FW samples, respectively. Gemmatimonas (16.6%), Burkholderia (0.09%), and Paraburkholderia (7.68%) were the most abundant genera in the untreated root samples. Gemmatimonas abundance was enhanced in both BM (30.7%) and FW (26.0%) treated root samples. In root samples, the genus Thauera was found in much higher abundance in FW (14%) samples than BM or control (<1%). Pseudomonas was found in control (5.34%), BM (5.23%), and FW (4.28%) root samples but not in an appreciable amount in soil (<1%). Gemmatimonas abundance was also enhanced in both BM and FW treated soil samples, accounting for 44.9 and 29.6%, respectively, compared to the control (19.6%). In addition to Gemmatimonas, soil samples treated with BM (15.9%) also had a higher abundance of Brevundimonas compared to the FW (2.5%) and control (0.1%) samples.
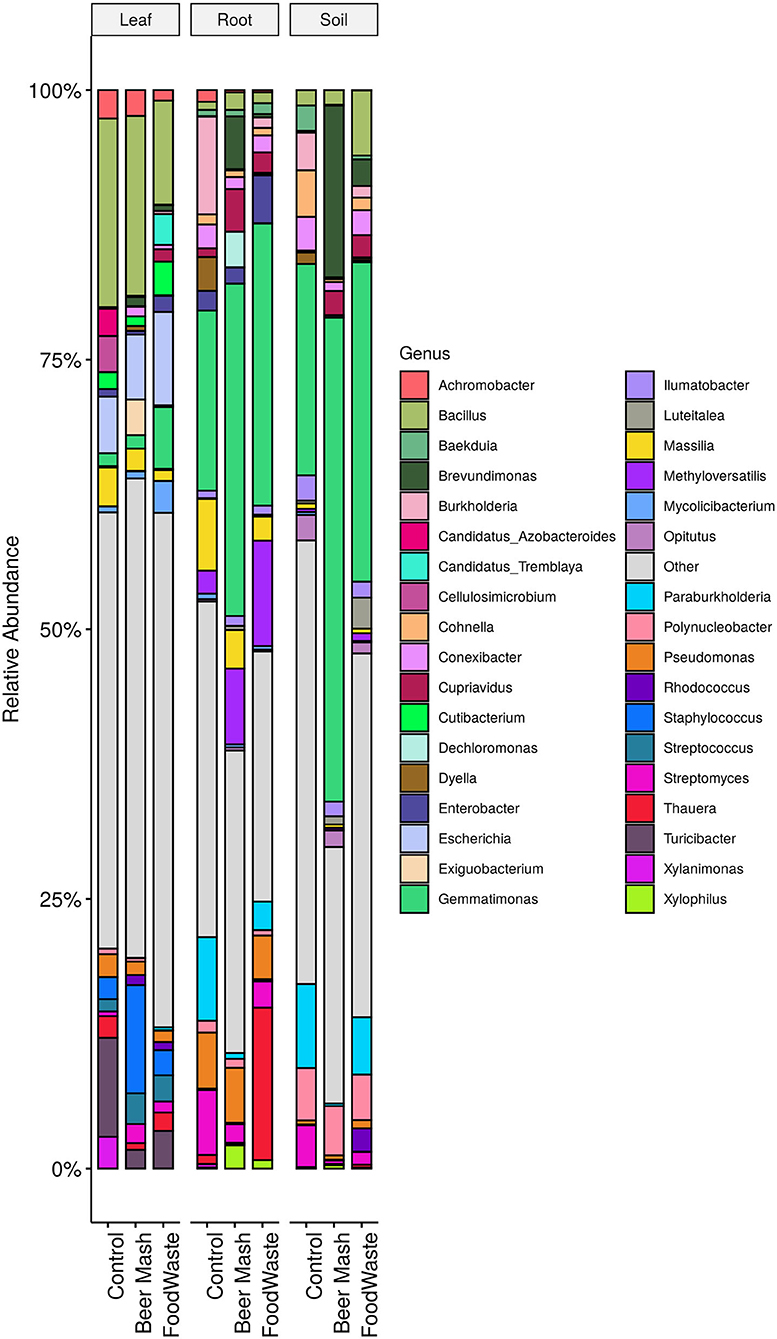
Figure 4. Stacked column bar graph representing the relative abundance and distribution, at the end of the greenhouse experiment, of the 15 most abundant genera from each tissue type, leaf, root and potting soil, treated with food waste and beer mash digestate or not treated (control). Samples were taken 2 weeks after the last FW and BM digestate application into the system.
The samples' alpha diversity was analyzed using Shannon's diversity index (Figures 5A,B). ASV diversity was highly dependent on plant compartment (P < 0.01 [Kruskal-Wallis]) with leaf having a significantly lower Shannon index than root and soil (P < 0.05 [pairwise Wilcox]). High diversity values for soil (4.15 ± 0.198) and root (3.82 ± 0.0977) and consistently decreased diversity estimates in the leaf samples (3.04 ± 0.120) were observed. Shannon indices of the leaf control samples (2.94 ± 0.219) were comparable with those of the leaves of FW-treated and BM-treated samples (2.95 ± 0.277; 3.20 ± 0.105), with no significant difference among the treatments. While at the soil and root level there was a clear separation between FW-treated (4.44 ± 0.197; 3.59 ± 0.144) and BM-treated (3.38 ± 0.390; 3.71 ± 0.150) and untreated samples (4.77 ± 0.145; 4.26 ± 0.163) with the soil control having the highest diversity and being significantly higher than the BM soil (P < 0.05 [pairwise Wilcox]). Control roots had a significantly higher Shannon index than the FW-treated roots (P < 0.05 [pairwise Wilcox]).
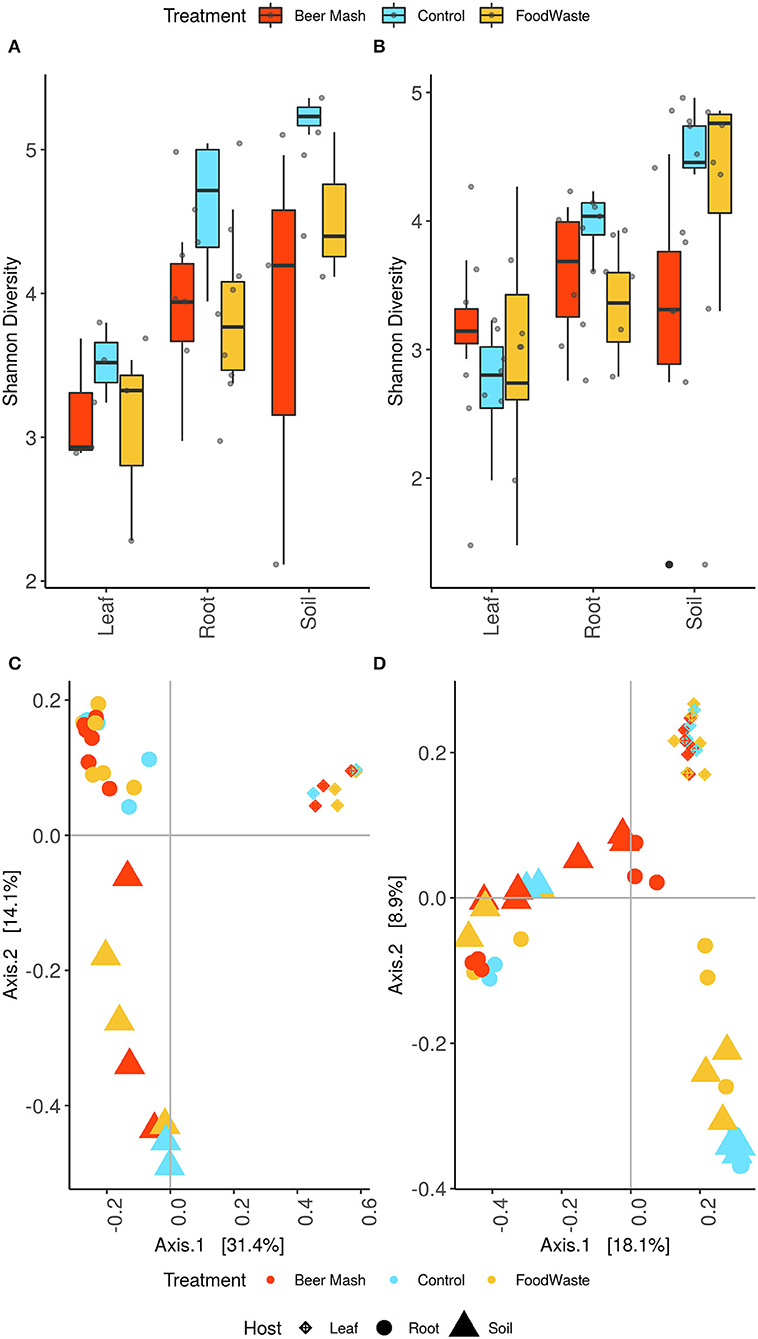
Figure 5. Alpha diversity estimates of the bacterial communities in leaves, root and potting soil samples, for experiment 1 (A) and 2 (B), were estimated using the Shannon diversity index. Box plots display the first (25%) and third (75%) quartiles, the median and the maximum and minimum observed values within each data set. Alpha diversity estimates represent biological replicates for the soil (N = 8; 17) root (N = 16; 17) and leaf samples (N = 8; 17). Data were analyzed by means of Kruskall–Wallis and pairwise Wilcoxon tests were run to verify statistical differences among groups. The overall plant compartment and wastes digestates treatment effects F (DFn, DFd) and P-value are displayed at the top of each graph. Significant differences (P < 0.05) across plant compartments are indicated with lowercase letters. Bray-Curtis beta diversity for experiment 1 (C) and experiment 2 (D). Points represent individual sample communities and are colored by treatment and shapes represent bio-compartment.
Beta diversity analyses revealed that leaves, root and rhizosphere soil samples formed three distinct clusters that were significantly different (P < 0.01 [Adonis]) (Figures 5C,D). Both FW and BM-treated samples separated along the axis 1 with the soil samples separating on the axis 2 as well (P < 0.05 [Adonis]).
Carbon Dynamics in the Nutrient Solution
In the closed zero-runoff systems used in these experiments, there are only 4 main sources of organic carbon compounds in the recirculating nutrient solution: incoming water, plant roots, microbes, and organic growing medium (Waechter-Kristensen et al., 1999). Temporal trends in dissolved organic carbon concentrations are mirrored by changes in bacterial populations where DOC concentrations spike immediately after amendment followed by rapid consumption by microbes within the solution. After 24 h, DOC concentrations return to pre-treatment levels for both FW and BM treatments, demonstrating rapid and complete consumption of added organic carbon. DOC concentrations remained stable throughout the experiment in controls without amendments. These results demonstrate that dissolved organic carbon availability dictates bacterial population growth in the presence of both amendments (Figure 6).
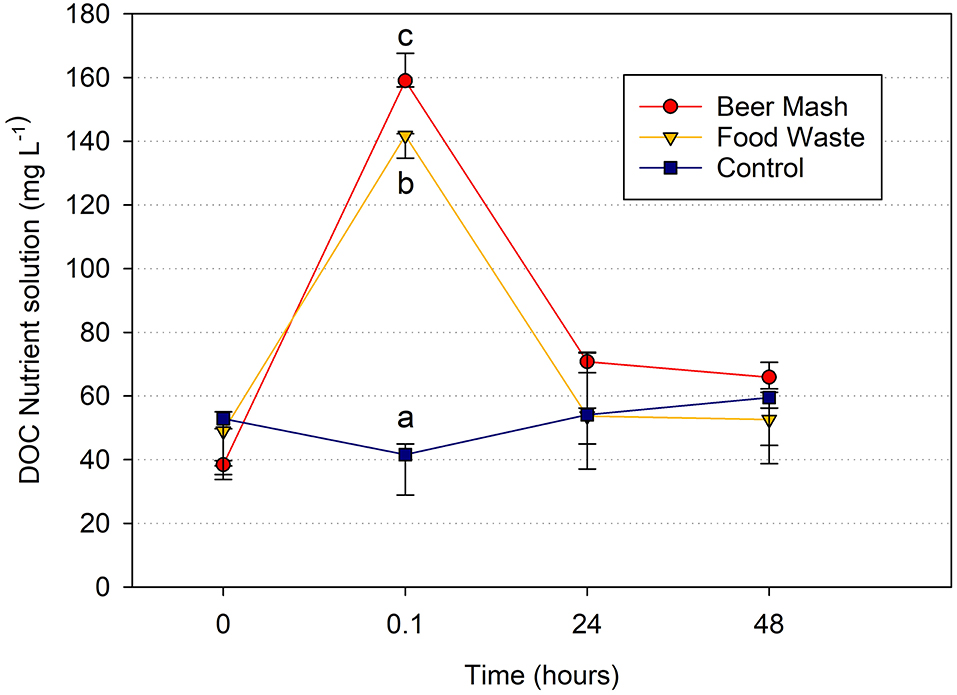
Figure 6. Dissolved Organic Carbon (DOC) changes in the nutrient solution. Average DOC concentration vs. time detected by DOC analyzer in a greenhouse experiment with citrus as the host plant at week 4. We chose to report week 4 because it is representative and because it was a middle point in the experiment. Treatments consisted of nutrient solution receiving no digestate (Control) or amended with beer mash BM or food waste FW digestate at time zero. Time points (0, 0.1, 24, and 48 h) with different letters are significantly different at P = 0.01 according to the all pairwise multiple comparison procedures (Holm-Sidak method). Error bars represent the SE of the mean (n = 3) where variation was great enough to be presented.
In addition to total concentration of DOC having a strong influence on microbial population dynamics within the nutrient solution, the speciation of available organic carbon compounds provided by the two amendments may provide insight on selectivity of microbes for specific carbon sources. A subset of organic carbon compounds within the nutrient solution was characterized using GC/EI-MS after derivatization, where compounds can then be categorized into general classes (i.e., lipids, condensed hydrocarbons, proteins, lignin, amino sugars, and carbohydrates) based on their O/C and H/C composition (Figure 7). These compounds may have originated from the formation of microbial metabolites, from the potting soil, or the amendment.
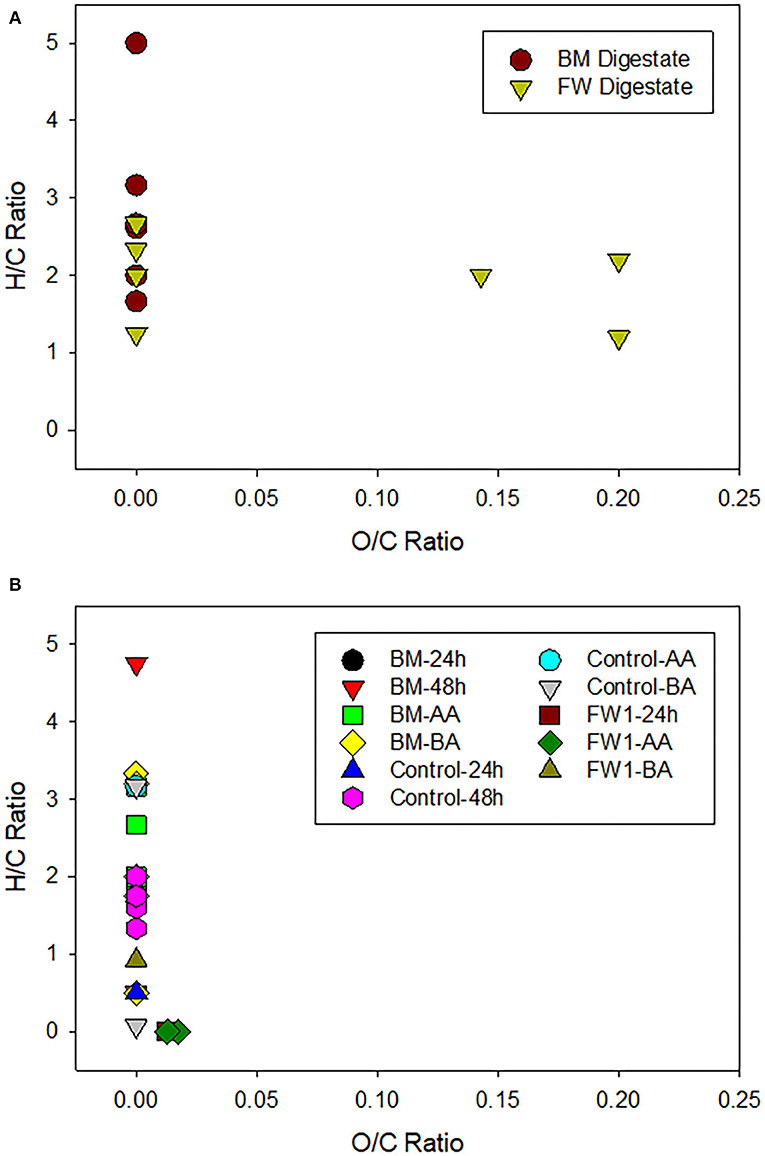
Figure 7. Gas-chromatography/electron ionization mass-spectrometry (GC/EI-MS) analysis of (A) beer mash (BM) and food waste (FW) digestates amendments and (B) nutrient solutions sampled from controls (Not treated), BM, and FW treatments. Samples were taken before and after application (-BA and -AA), 24 and 48 h after digestate application. O/C vs. H/C ratio of elemental formulas was plotted to provide an overview of the compound categories within each amendment. Data were assigned nominally into categories based on a van Krevelen empirically derived metabolite map depicting chemical compound classes and their relative boundaries as assigned in Figure 5 in Kim et al. (2003) study. FW-AA, FW-BA, and FW2 samples had values below detection-limit and are not reported here.
The Van Krevelan diagram (Minor et al., 2014; Rivas-Ubach et al., 2018) of beer mash and food waste digestates prior to addition to nutrient solution shows that organic compounds are dominated by condensed hydrocarbons and lipids, with lignin detected only in the FW digestate. The starting materials (digestates) were electron-rich (i.e., low O:C) capable of fueling aerobic microbial respiration. Analysis of treatments and controls showed the presence of condensed hydrocarbons in both FW and BM treatments and controls at various time points (Figure 7). Carbohydrates were only detected in controls, which is unexpected given that both amendments are expected to contain carbohydrate compounds (Popa et al., 2017). Lipids were detected in nutrient solution after BM amendment and were detected up to 48 h after amendment addition. Though lipids were also detected in nutrient solution after FW treatment, they were not detected after 24 h. Overall, organic compound composition generally became dominated by more oxidized compounds (increase in O:C ratio) and more saturated state (i.e., increase in H:C ratio over time).
Nutrient Dynamic in the Potting Soil
Nutrient dynamics were monitored in the potting soil overtime after amendment. First, the soil solid phase C and N content were measured by flash combustion (Figure 8). Both Food Waste and Beer Mash treatments significantly increased soil C and N concentrations as compared to the control (no treatment), with respective p-values of 0.026 (for N) and 0.005 (for C) through analysis of variance and pairwise comparisons between treatments. Beer Mash amendment increased average N by 166% and average C by 164% while the Food Waste amendment increased average N concentrations by 152% and C by 259% (Figures 8A,B). Overall, the treatments BM and FW did not vary significantly from one another with regards to C and N contributions. Average C:N ratios of all soils were near or within the range of 25–35, which is considered optimal for microbial use (Bernal et al., 2017). Soil amendments with C:N ratio outside of this range require supplementation to prevent N loss (Ibid) (Figure 8C).
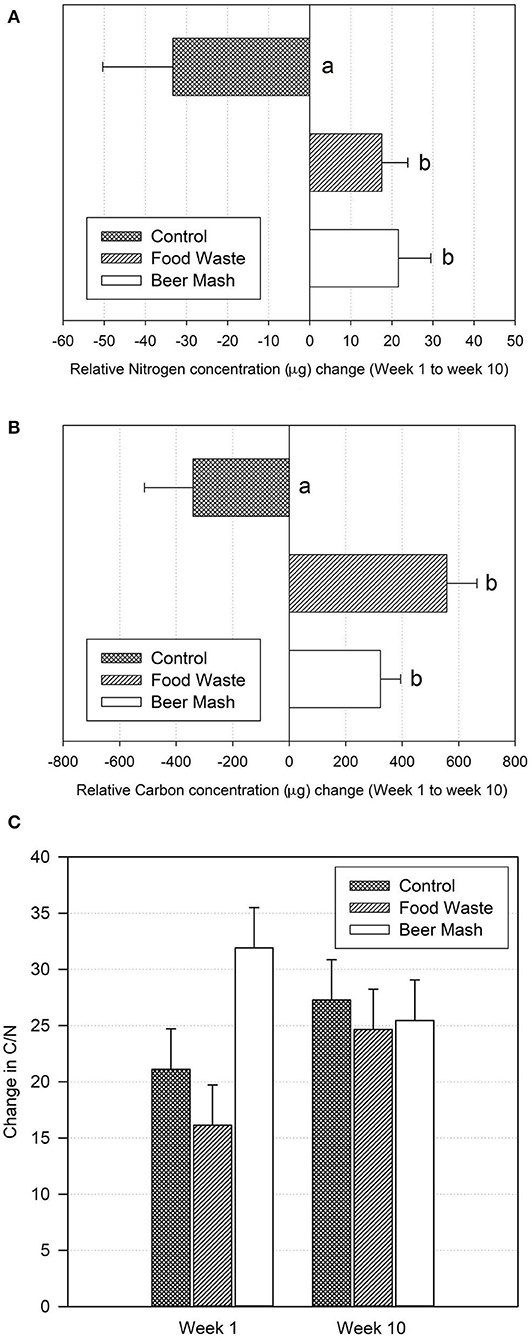
Figure 8. (A) Total N, and (B) total C concentrations and (C) C:N ratio changes between week one and 10 in potting soil samples from different treatments as measured by elemental analysis. Treatments consisted of zero runoff plant production systems set up in a greenhouse where nutrient solution was amended and not amended (control) with beer mash BM and food waste FW digestate at 7 days interval for a total of 9 treatments applications. For (A,B) values are the means of three replicates. Bars with different letters are significantly different at P = 0.05 according to the all pairwise multiple comparison procedures (Holm-Sidak method). Error bars represent the SE of the mean (n = 3) where variation was great enough to be presented. For (C) data were analyzed by two-way analysis of variance. No statistically significant difference was found within treatments (P = 0.109), within weeks (P = 0.369) and no interaction between treatments and weeks (P = 0.123).
Figure 9 shows the changes in elemental composition of soil solid phase collected at Week 1 and 10. We categorized elements into two groups: plant nutrients (Mo, Mg, K, P, Se, Ca, Cl, Fe, Mn, S, and Zn) and potentially toxic elements (Na, Al, Co, Cu, Hg, Pb, V, Cr, As, Cd). While there were changes in the elemental composition with BM and FW treatment, the change in concentrations were not statistically significant for most of the elements analyzed. There were some outliers, notably magnesium (Mg), an essential plant nutrient, which increased in concentration significantly by 1,650% through treatment with FW and 3,242% through treatment with BM (p = 0.003). Phosphorus, Ca, and Co also increased significantly, but only with BM treatment (p = 0.008, 0.004, and 0.028, respectively), while K concentrations decreased significantly (p = 0.042). Overall, amending soils with BM lead to the highest increase in the majority of plant nutrient element concentrations.
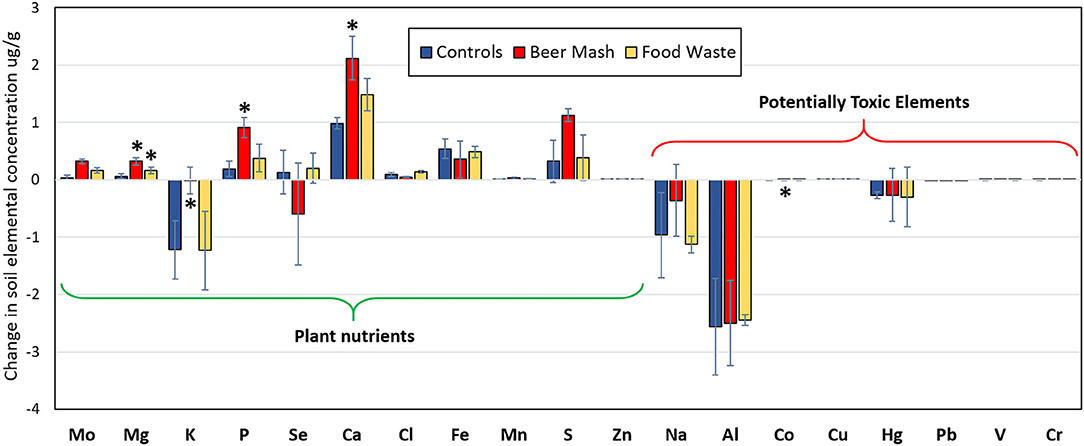
Figure 9. Elemental composition changes (μg/g) between week 1 and 10 in potting soil samples from different treatments as detected by X-ray fluorescence spectrometer. Treatment consisted of nutrient solution not amended (Control) in blue, or amended with Beer mash (BM) in red, food waste (FW) in yellow. Plant nutrient elements tested included: molybdenum (Mo), magnesium (Mg), potassium (K), phosphorus (P), selenium (Se), calcium (Ca), chlorine (Cl), iron (Fe), manganese (Mn), sulfur (S), zinc (Zn); Potentially toxic elements tested included: sodium (Na), aluminum (Al), cobalt (Co), copper (Cu), mercury (Hg), lead (Pb), vanadium (V), and chromium (Cr). Arsenic (As) and cadmium (Cd) concentrations were considered too low to include. Error bars represent standard deviations of the mean (n = 3) where variation was great enough to be presented. For each element, significant differences observed by ANOVA tests (at P = 0.05) followed by multiple comparisons vs. control group procedures (Holm-Sidak method) are indicated with asterisks *.
Because some elements are toxic to plants and humans and thus undesirable in agricultural amendments, we then analyzed potentially toxic elements (PTEs) separately. Although there was a change in soil concentrations of the measured PTEs over time, there was no statistically significant increase or decrease from Beer Mash and Food Waste treatment as compared to untreated soils for most PTEs, except for Co concentration which decreased significantly (p = 0.028) with BM treatment (Figure 9).
Discussion
The present study demonstrated the effect of digestates, derived from FW and BM, on the structure and diversity of bacterial populations in the microbiome of the recirculating nutrient solution, potting soil, roots, and leaves of non-bearing citrus plants. Data obtained using different approaches (culture-dependent and -independent) were combined to gather insights into the distribution of specific taxa and on the ability of strains belonging to these taxa to promote PGPR beneficial traits. Additional analyses of carbon and nutrients dynamics provided preliminary understandings of how changes in the microbial composition, resulting from digestate application, influenced ecosystem functions and below-ground processes such as carbon and nutrient cycling. The implications of such effects and primarily the enhancement of native microorganisms in intensive indoor and potentially field production systems, as well as the safety of digestates, are also discussed here.
Our results from culture-dependent analyses indicated that, for nine consecutive weeks, both types of digestates selectively and significantly enhanced the native population of aerobic heterotrophic bacteria and fluorescent pseudomonads in the recirculating nutrient solution used to fertigate indoor-grown citrus plants (Figures 2A,B). These results agree with earlier findings on the effect of exotic substrates in selecting and stimulating a specific microbial taxon (i.e., Pseudomonas spp.) in the irrigation water that was used in non-woody plants grown in indoor closed production systems (Pagliaccia et al., 2007, 2008). Other studies have also shown that nutritional amendments such as salicylate, ammonium sulfate, methionine, and olive oil, when added to the soil or recirculating nutrient solutions, enhanced the efficacy of several bacterial and fungal biological control agents (Colbert et al., 1993a,b; Yamada and Ogiso, 1997; Ji and Wilson, 2003; Luziatelli et al., 2019).
Soils and plant systems represent biologically diverse habitats. Their biota supports many ecosystem functions and they respond to natural perturbations and human interventions (Bender et al., 2016). Our results support the Bender et al. (2016) concept of soil ecological engineering for agricultural sustainability, where targeted manipulation of soil organisms stimulates soil life and harnesses internally regulated ecosystem processes (Philippot et al., 2013; Bender et al., 2016). Targeting and enhancing beneficial native rhizosphere microbial communities combined with currently available state-of-the-art farming technologies (e.g., closed production systems and precision agriculture with cloud-based applications for optimized plant growth parameters) could help maintain yields while reducing the dependence on external inputs (e.g., chemical fertilizers and pesticides) in agrosystems (Delgado et al., 2019).
Using a variety of experimental approaches, including culture-independent analyses, we showed that the relative abundances of Pseudomonas spp. and to a lesser extent Bdellovibrio spp., increased in the irrigation water treated with both digestates, while the bacterial diversity decreased within 24–48 h compared to the non-treated systems. The low microbial diversity and high Pseudomonas spp. abundance in the nutrient solution persisted for 48 h, whereas no significant difference in the abundance of the Pseudomonads was observed 7 days after treatment (Figures 2C, 3A, and Supplementary Figure 3). These results and the ecological diversity measures (Figure 3B) suggest that, once the nutrient sources are depleted, the bacterial community structures reset to a more diverse composition. In our study, Bdellovibrio was the only genus that survived overtime in the nutrient solution (e.g., longer than 72 h) and it took over when Pseudomonas spp. abundance decreased (Figure 3A). This is not surprising since Bdellovibrio has been shown to be an effective predator of Gram-negative bacteria; it also has biological control properties in environmental as well as medical microbiological settings (Harini et al., 2013) however, further studies are needed to evaluate its potential as PGPMs for agricultural use. Wu et al. (2018) and Wright et al. (2019) also observed a similar effect on the reduction of the bacterial biodiversity, studying the dynamics of the bacterial community using dissolved organic matter (DOM) extracted from sediments adjacent to groundwater and chitin, respectively. In both studies, bacteria able to use DOM and chitin as nutrient sources were able to proliferate and became dominant. In agreement with these studies, we also identified bacteria belonging to Pseudomonas spp. that were enhanced by the organic components of the digestates.
The carbon dynamics assays supported the findings of the microbial analyses. DOC in the recirculating nutrient solution followed a similar trend as the bacterial populations, spiking after each digestate amendment, then plateauing again 24 h post-treatment. These results suggest that the bacteria used organic carbon from FW and BM for metabolic processes and replication. In the recirculating nutrient solution of the closed zero-runoff systems, there are four primary sources of organic carbon: influent water, plant roots, microbes, and organic growing medium. The plateau in DOC concentration in the digestate treated nutrient solutions (Figure 6) likely indicates that depletion of labile carbon in the nutrient solution induced a change in microbial activity from growth to maintenance.
Unlike the DOC data, changes in carbon speciation over time did not show a temporal trend, though there was a slight trend toward more saturated compounds dominating over time, and the amendments starting out highly reduced (electron-rich) (Figure 7). Organic compounds exhibited increasing O:C over time likely due to microbial oxidation during aerobic respiration and some contribution from abiotic oxidation from the nutrient solution being open to the air. A previous study showed a gradual increase toward carbonyl/carboxyl structures attributed to metabolites such as nucleobases, lipids, and amino acids/peptides/protein products of microbial metabolisms (Wu et al., 2018). The authors compared van Krevelen diagrams of detected compounds with and without microbes and found a clear disappearance of protein-like compounds with elemental composition of carbon, hydrogen, oxygen, and nitrogen (CHON), indicating utilization or degradation of these compounds by microbes over the experimental period.
Many studies have demonstrated that soil microbiota drive a wide range of biogeochemical processes, with many species mediating organic matter transformation, which in turn shape microbial community composition over time (Jouraiphy et al., 2005; Kramer and Gleixner, 2008; Gougoulias et al., 2014; Boye et al., 2017). Shifts in microbial community composition have been significantly correlated to changes in the relative proportions of tannin-, protein-, condensed aromatic-, lipid-, and lignin-like compounds in other studies, where relatively labile tannin and protein compounds are correlated with the microbial community in early stages and microbial community composition changes over time to be more correlated with less labile lipid and lignin-like compounds (Wu et al., 2018). Gilmour et al. (2003) suggested that stable microbial decomposition of the organic matter by anaerobic digestion and composting produces a biosolid organic matter residue that is relatively recalcitrant. This supports the results in our study, where the relative proportions of lignin-like and CHO compounds continuously increased during incubation (Figure 7), likely because of their relative resistance to biodegradation, contribution of microbial metabolites, and/or oxidation of certain DOM moieties (Fuchs et al., 2011).
Both our previous and current work suggest that, after amendments, microorganisms preferably utilize labile C in DOC (i.e., aromatic compounds), as indicated by the rapid increase in cell counts (Pagliaccia et al., 2007, 2008). In Wu et al. (2018) study, microbial community structure analysis showed a rapid enrichment of Pseudomonas spp. in early stages. Pseudomonas spp. was often reported to be the dominant genus in microcosms amended with labile C, such as acetate (Cui et al., 2011) or glucose (Ghosh and Leff, 2013). Also, many Pseudomonas species are known to be efficient competitors for resources through secretion of antibiotics (Hibbing et al., 2010) and siderophore-based metal chelators (Stubbendieck and Straight, 2016). It is therefore not surprising that Pseudomonas became the most abundant species in the nutrient solution in the early incubation stage when labile C was still available. In our study, conventional identification (culturing plus Sanger sequencing) and Illumina MiSeq sequencing showed that both BM and FW applications changed the structure and composition of the Pseudomonas community in the nutrient solution.
Culture-dependent analysis identified Pseudomonas sp. strain HAL1 as the most abundant bacteria in the nutrient solution samples treated with either BM or FW after 24 and 48 h. P. moorei was also enriched after digestates amendment (Table 2). Culture-independent analysis identified Pseudomonas putida (strain JB, DLL-E4, and 15420352) as the most abundant bacteria in both BM and FW treated nutrient solutions while no Pseudomonas spp. were found within the most abundant bacteria in the control nutrient solutions (Table 3). These results are in agreement with current literature on phytoremediation principles, which indicates that bacteria with PGPR traits can facilitate the removal of inorganic contaminants stimulating either plant growth or phytoremediation activity (Glick, 2010; Mahanty et al., 2017; Luziatelli et al., 2019). This principle could be used to ensure the selection of a pseudomonas population from the rhizosphere and the nutrient solution, for beneficial purposes such as biocontrol, biofertilization, phytostimulation, and disease management in both indoor nursery and citrus field production.
Plant Growth-Promoting Microbes have a direct effect on phytopathogens through the production of metabolites with antibiotic activity, or by competition for space and nutrients; and an indirect beneficial effect on the plant by enhancing defense responses or promoting plant growth (Mendes et al., 2011; Ma et al., 2019). Our preliminary bioinformatics analysis (PATRIC) indicated that all the bacteria species enriched by BM and FW amendments possess genes involved in the production of siderophores and iron acquisition (Supplementary Figure 1A, e.g., Ferric Siderophore Transport system- ABC efflux system and TonB family proteins). Some of these bacteria also possess genes putatively involved in direct antagonism, phosphate solubilization, and nutrient facilitation (Supplementary Figure 1B, e.g., Acetolactate synthase, Transcriptional regulator PadR). Their enrichment in the recirculating nutrient solutions could be very valuable for the plant since as shown in this work, the fluorescent Pseudomonas spp. reached a consistent population level in the nutrient solution of 107-108 CFU/ml after each digestate application. This level of bacterial cell concentration is well-known to induce the activation of quorum sensing regulatory systems (Hibbing et al., 2010). The regulation of genes through quorum sensing controls essential biological functions in many bacteria, including several regulatory mechanisms for the production of beneficial secondary metabolites in Pseudomonas spp. (i.e., antibiotics and siderophores) (Sahu et al., 2018). This finding is particularly promising for the development of new approaches for integrated plant production and disease management practices, especially under zero runoff indoor production systems, where the beneficial bacterial metabolites produced in the nutrient solution would reach the plant roots at every irrigation cycle (i.e., once to several times a day). This approach could also be applied under field conditions where targeted management of soil community composition could be promoted around the plant root system with the application of digestates through drip fertigation systems. Barzee et al. (2019) investigated the feasibility of drip fertigation for the application of digestate biofertilizers (rich in N, P, K as well as Cu, Ni, Mg, and Zn) to tomato crops in farm-scale cultivation experiments. Their results demonstrated that digestate biofertilizers could also be effectively delivered to the tomato plants through a subsurface drip irrigation system, with increased crop yields.
Digestate materials could have potential agronomic value because they contain high amounts of nutrients and organic matter (Möller and Müller, 2012), as well as volatile organic acids and complex organics from undigested particles (Franke-Whittle et al., 2014). However, the literature is divergent about the effect of digestates on soil quality, and the fate of the digestates' C and N in soils is still an understudied topic (Möller, 2015). We carried out a preliminary analysis to elucidate the agronomic value of BM and FW and their potential impact on long-term sustainability in terms of soil fertility and environmental impact. From a review of the available literature on this subject, Möller (2015) stated that due to the lower portion of easily degradable compounds in digestate, the organic fractions in the digestate can contribute to soil organic matter (SOM) stabilization (i.e., increase stable soil organic carbon), which can lead to changes in biological, chemical and physical soil characteristics. In our study, both BM and FW significantly increased N and C in the potting soil around the roots (Figures 8A,B). Regarding the C/N ratio, a range of 25 to 30 is considered optimal for microbial activity, balanced composting, and reduced nitrogen loss (Bernal et al., 2017). In our case, no statistically significant difference was found in the C/N ratio of samples analyzed between treatments and between the first and last time points; however, the C/N ratio did increase from 16 to 25 in soils treated with food waste digestates. These results suggest that in the short term the high N content in the BM and FW digestates can deliver immediate nutrients to citrus trees in the field, while in the long term, the high C content could provide a slower release of nutrients (i.e., through PGPR mediated processes) and help reduce nutrient leaching. Furthermore, the bokashi fermentation used to produce both BM and FW digestates significantly reduces gaseous emission with a GHG footprint of < 0.22 MtCO2 equivalent (million metric tons) and 1% of CO2 released during fermentation (Alattar et al., 2012) compared to standard aerobic composting which has a GHG footprint of ca. 1.2 MtCO2 equivalent (million metric tons) and ca. 50% carbon released as CO2. Overall, our results are in agreement with Möller (2015) and support the hypothesis that although the digestate's potential as fertilizers is of minor relevance, their indirect effect on the microbial community, the stabilization of the organic matter, and indirect reduction of gaseous emissions have the greatest relevance. Appropriate application rate and time need to be investigated further to optimize the long-term effects of the digestates and to stabilize the positive microbial activities (i.e., keep the bacteria population-level above 105 CFU/ml). Further studies are also needed to investigate which types of feedstock would produce the most valuable digestates in terms of agronomic value.
Based on previous findings (Pagliaccia et al., 2007), and the recurring re-inoculation of the rhizosphere through the recirculating irrigation cycle, we would have expected the Pseudomonas populations enriched in the nutrient solution to colonize the citrus roots system. To our surprise, the bacterial analyses of citrus leaves, roots, and potting soil produced different patterns at the end of the greenhouse experiment. NGS sequencing showed that, compared with non-treated roots, roots treated with BM and FW had higher relative abundances of Gemmatimonas spp. or Brevundimonas, yet no significant difference in the abundance of Pseudomonas spp. was observed. Additionally, we found greater variation in beta-diversity for both root and potting soil microbiomes, while all leaf samples were very similar, but with the lowest diversity and evenness estimates within all treatments (Figure 5). This contrasting pattern was surprising but could reflect divergent microbial responses in different environments (nutrient solution vs. soil) but also within different plant compartments (root vs. leaves) and plant species. In their extensive review, Philippot et al. (2013) discussed how soil properties but also plant species influence microbial community composition and structure in the rhizosphere. Further work with other citrus species and varieties as well as other crops will be essential to decipher the mechanisms underlying host plant effects on the root microbiome, as well as any fitness benefits to the plant caused by the increased Pseudomonas abundance in the irrigation water. On the other hand, our results raise the intriguing hypothesis that the citrus genotype might have mediated changes in the host-microbiome under our abiotic induced conditions (i.e., digestates application). In our study, the citrus cultivar used (“Pineapple sweet orange,” Citrus sinensis (L.) Osbeck) seemed to have lost the capability to recruit the Pseudomonas onto the root system (i.e., Pseudomonads high population of 6-7 log CFU/ml from irrigation water did not colonize root). The “Pineapple sweet orange” has been continuously propagated and cultivated in commercial USA settings since the 1800's (Hodgson, 1967). It has been shown that, due to cultivation practices and intense artificial selection for yield and other desirable agronomic characteristics, quite a few plant cultivars have lost, to some extent, their ability to recruit and associate with beneficial soil biota (Sawers et al., 2008; Porter and Sachs, 2020). Mueller and Sachs (2015), discussed the native microbiome and anticipated that host-mediated microbiome engineering would be more efficient using wild hosts rather than hosts that have experienced domestication, or adaptation to microbially depauperated experimental environments.
In our study, we also tested changes in toxic vs. non-toxic micronutrients in the potting soil following BM and FW application, and found negligible signs of accumulation for both, except for Mg, which increased significantly with both digestates compared to the non-treated system. P and Ca also accumulated, but only with BM treatment while K decreased (Figure 9). Some of the results were unexpected since the FW digestate amendment contained a higher amount of K and Ca (Table 1). Although the FW digestate contained a much higher amount of Na (Table 1), which is expected due to the higher content of Na typically present as a food preservative in food waste (Popa et al., 2017), our analysis showed a decrease in this element, which is a very desirable trait in agriculture and especially in zero-run off plant production systems. These results agree with Barzee et al. (2019), who showed that digestates do not cause a higher risk for salt accumulation. However, as in Barzee's study, long-term trials with drip irrigation systems need to be carried out to understand the impact of repeated digestates applications on the soil chemical properties in both greenhouse and in the field and, most importantly, their effect on the rhizospheric soil and citrus plant biota, especially the roots.
The validation of the digestates' agronomic values also demand that they do not contain or enhance human pathogens and therefore pose a threat to workers and consumers' health. Our results suggest that, although DNA of bacterial groups belonging to the genus Enterococcus, known to be human pathogens, were detected by NGS analysis in the digestates stocks (Supplementary Figure 2), such bacteria were never cultured or detected from the digestates stocks or ever identified by NGS analysis in any bio compartment tested (i.e., soil, root, leaves) in the plant production system after digestates application (Table 2, Figures 3A, 4). Testing for fecal coliform and Salmonella (A & L Western Agricultural Laboratories) confirmed the absence of known human pathogens (data not shown). Therefore, we concluded that under our experimental conditions, the use of Bokashi fermentation digestates is safe. However, additional physical or chemical processes might be necessary to treat digestates to ensure their safe use in diverse non-experimental agricultural systems. Additional investigations are also necessary to assess cause and effect, such as Koch's postulates experimentation, to further define Pseudomonas spp.'s roles in controlling plant diseases in planta. Regarding the potential use of these bioproducts to increase or accelerate citrus growth, the overall results indicate that their application could be beneficial for the citrus industry, inside and outside California, but could also be translated to other crops. However, because citrus plants are slow-growing woody plants, the current tests did not provide any evident biostimulant effect. Longer-term trials with additional citrus varieties will be required to answer this question. Alternatively, further tests with faster-growing plants could be used to evaluate the effect of the digestates on plant growth rate, fitness, etc. A fascinating question to answer is whether, within the same plant species, different crop varieties (new commercial variety vs. a wild relative) will be colonized differently by the digestate-enhanced Pseudomonas spp.
Based on our results, digestates derived from agriculture and food waste can be safely used at very low concentrations as biostimulants and offer a promising solution to enhance the native ecosystem while reducing environmental impact. Further research is needed to determine if such applications would have any impact on plant performance (e.g., yield increase or disease resistance) and to identify the underlying mechanisms of such effects.
Conclusions
More sustainable agri-food systems can be achieved by adopting biotechnological processes that allow nutrient recycling, and the implementation of climate-smart agricultural (CSA) approaches. This study addresses the pressing need to have easily deployable mitigation strategies that integrate the management of soil carbon dynamics, favor beneficial microbiota, and create alternative disposal for agricultural and food waste.
We showed that there is no need to introduce PGPMs into a cropping system (rhizosphere and nutrient solution). Developing novel agricultural practices that deploy targeted manipulation of soil organisms to stimulate soil life and harness internally regulated ecosystem processes offer a more sustainable approach to enhance crop production. Farmers can benefit from the resident microbiota because the plant rhizosphere already contains prospective members that can function as plant growth-promoting (PGP) and as plant disease controlling (PDC) agents when enhanced by digestate by-products obtained from agricultural and food wastes. Digestates can be used as biostimulants to generate a positive shift in the native microbial communities with a parallel increase in beneficial microbiota (i.e., fluorescent Pseudomonads) and their related bioprocesses. This could offer a very promising alternative to synthetic chemicals to stimulate plant growth traits and increase plant defense responses (priming) to pathogens.
Combined with their widespread sourcing from agricultural and industrial processes, waste digestates could help reduce dependency on synthetic chemicals and foster the establishment of eco-friendly, circular economies for agricultural production. More research is needed to elucidate the best feedstock to use for digestate production and the many indirect effects digestates have on plant pathogens, crop yield, N emissions in indoor and in the field production, and the possible long-term effects on the microbiome and soil fertility.
Data Availability Statement
The original contributions generated for the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Author Contributions
DP wrote the manuscript, was primarily responsible for conceptualization and experimental design, contributed to data collection, analysis, and all manuscript drafts. SB and XC were responsible for greenhouse operation, carried out the data collection and analysis for all the microbial work, and assisted with manuscript revisions and tables creation. DS contributed to data collection and analysis for the carbon dynamic work, wrote with SY some manuscript sections for the carbon dynamic work, performed manuscript revisions, and assisted to create some figures and tables. ED performed the sequence processing, bioinformatics, created the figures on the NGS data, and contributed to the manuscript revisions. AD contributed extensively to the manuscript revisions, figures, and tables creation. SY was responsible for the conceptualization and experimental design and collection for the carbon dynamic work, revised, and corrected the manuscript. BP assisted SB and XC for the NGS Library construction. JB, PR, BP, NE, PR, RP, and GV assisted with the experimental design, revised and corrected the manuscript. JB, SY, and GV provided greenhouse and laboratory resources for this study. All authors contributed to the article and approved the submitted version.
Funding
This research was funded in part by the USDA-NIFA, Hatch (project 1020106), the National Clean Plant Network (NCPN) (AP18PPQS&T00C107 and AP19PPQS&T00C148), and the Citrus Research Board (CRB) project Citrus Clonal Protection Program (CCPP) (6100) awarded to GV. Additional support was provided by the USDA-NIFA, Hatch (project 81361) awarded to NE, the Department of Botany and Plant Sciences, and the College of Natural and Agricultural Sciences (CNAS) under the California Agriculture and Food Enterprise (CAFÉ) program at UC Riverside.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Tyler Dang and Greg Greer for their valuable assistance with research protocols and materials.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fsufs.2020.593568/full#supplementary-material
References
Abhilash, P. C., Dubey, R., Tripathi, V., Gupta, V. K., and Singh, H. B. (2016). Plant growth-promoting microorganisms for environmental sustainability. Trends Biotechnol. 34, 847–850. doi: 10.1016/j.tibtech.2016.05.005
Acea, M. J., Moore, C. R., and Alexander, M. (1988). Survival and growth of bacteria introduced into soil. Soil. Biol. Biochem. 20, 509–515. doi: 10.1016/0038-0717(88)90066-1
Alattar, M. A., Green, T. R., Henry, J., Gulca, V., Tizazu, M., Bergstrom, R., et al. (2012). Effect of microaerobic fermentation in preprocessing fibrous lignocellulosic materials. Appl. Biochem. Biotechnol. 167, 909–917. doi: 10.1007/s12010-012-9717-5
Amaya-Gómez, C. V., Porcel, M., Mesa-Garriga, L., and Gómez-Álvarez, M. I. (2020). A framework for the selection of plant growth-promoting rhizobacteria based on bacterial competence mechanisms. Appl. Environ. Microbiol. 86:e00760. doi: 10.1128/AEM.00760-20
Babcock, B. (2018). Economic impact of California's citrus industry. Citrograph 9, 36–39. Available online at: https://www.citrusresearch.org/uncategorized/california-citrus-economic-impact-final-report-citrus-research-board-quantifies-industry-importance/ (accessed November 25, 2020).
Bailey, K. L., and Lazarovits, G. (2003). Suppressing soil-borne diseases with residue management and organic amendments. Soil Tillage Res. 72, 169–180. doi: 10.1016/S0167-1987(03)00086-2
Barzee, T. J., Edalati, A., El-Mashad, H., Wang, D., Scow, K., and Zhang, R. (2019). Digestate biofertilizers support similar or higher tomato yields and quality than mineral fertilizer in a subsurface drip fertigation system. Front. Sustain. Food Syst. 3:58. doi: 10.3389/fsufs.2019.00058
Bender, S. F., Wagg, C., and van der Heijden, M. G. A. (2016). An underground revolution: biodiversity and soil ecological engineering for agricultural sustainability. Trends Ecol. Evol. 31, 440–452. doi: 10.1016/j.tree.2016.02.016
Berendsen, R. L., Pieterse, C. M., and Bakker, P. A. (2012). The rhizosphere microbiome and plant health. Trends Plant Sci. 17, 478–486. doi: 10.1016/j.tplants.2012.04.001
Berg, G., Köberl, M., Rybakova, D., Müller, H., Grosch, R., and Smalla, K. (2017). Plant microbial diversity is suggested as the key to future biocontrol and health trends. FEMS Microbiol. Ecol. 93:fix050. doi: 10.1093/femsec/fix050
Bernal, M. P. (2017). Grand challenges in waste management in agroecosystems. Front. Sustain. Food Syst. 1:1. doi: 10.3389/fsufs.2017.00001
Bernal, M. P., Sommer, S. G., Chadwick, D., Qing, C., Guoxue, L., and Michel, F. C. (2017). “Chapter three - current approaches and future trends in compost quality criteria for agronomic, environmental, and human health benefits” in Advances in Agronomy, vol 144, ed D. L. Sparks (Amsterdam, Netherlands: Elsevier Inc), 143–233. doi: 10.1016/bs.agron.2017.03.002
Blacutt, A., Ginnan, N., Dang, T., Bodaghi, S., Vidalakis, G., Ruegger, P., et al. (2020). An in vitro pipeline for screening and selection of citrus-associated microbiota with potential anti-“Candidatus Liberibacter asiaticus” properties. Appl. Environ. Microbiol. 86:e02883–19. doi: 10.1128/AEM.02883-19
Blaustein, R. A., Lorca, G. L., Meyer, J. L., Gonzalez, C. F., and Teplitski, M. (2017). Defining the core citrus leaf- and root-associated microbiota: factors associated with community structure and implications for managing huanglongbing (Citrus Greening) disease. Appl. Environ. Microbiol. 83, e00210–e00217. doi: 10.1128/AEM.00210-17
Bonilla, N., Gutiérrez-Barranquero, J. A., de Vicente, A., and Cazorla, F. M. (2012). Enhancing soil quality and plant health through suppressive organic amendments. Diversity 4, 475–491. doi: 10.3390/d4040475
Boye, K., Noël, V., Tfaily, M., Bone, S. E., Williams, K. H., Bargar, J. R., et al. (2017). Thermodynamically controlled preservation of organic carbon in floodplains. Nature Geosci. 10, 415–419. doi: 10.1038/ngeo2940
California Department of Food Agriculture (CDFA), (2020). Asian Citrus Psyllid (ACP) / Huanglongbing. Available online at: https://www.cdfa.ca.gov/plant/acp/ (accessed May 16, 2020).
Callahan, B. J., McMurdie, P. J., Rosen, M. J., Han, A. W., Johnson, A. J., and Holmes, S. P. (2016). DADA2: high-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583. doi: 10.1038/nmeth.3869
Cao, Y., Wang, J., Wu, H., Yan, S., Guo, D., Wang, G., et al. (2016). Soil chemical and microbial responses to biogas slurry amendment and its effect on Fusarium wilt suppression. Appl. Soil Ecol. 107, 116–123. doi: 10.1016/j.apsoil.2016.05.010
Caporaso, J. G., Lauber, C. L., Walters, W. A., Berg-Lyons, D., Huntley, J., Fierer, N., et al. (2012). Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 6, 1621–1624. doi: 10.1038/ismej.2012.8
Cerda, A., Artola, A., Barrena, R., Font, X., Gea, T., and Sánchez, A. (2019). Innovative production of bioproducts from organic waste through solid-state fermentation. Front. Sustain. Food Syst. 3:63. doi: 10.3389/fsufs.2019.00063
Chen, J. Y., Jiang, H., Chen, S. J., Cullen, C., Sabbir Ahmed, C. M., and Lin, Y. H. (2019). Characterization of electrophilicity and oxidative potential of atmospheric carbonyls. Environ. Sci. Process Impacts 21, 856–866. doi: 10.1039/C9EM00033J
Christel, D. M. (2017). The Use of Bokashi As A Soil Fertility Amendment in Organic Spinach Cultivation. [Dissertation]. Burlington, VT: University of Vermont. Available online at: https://scholarworks.uvm.edu/graddis/678/
Ciancio, A., Pieterse, C. M. J., and Mercado-Blanco, J. (2016). Harnessing useful rhizosphere microorganisms for pathogen and pest biocontrol. Front. Microbiol. 7:1620. doi: 10.3389/fmicb.2016.01620
Colbert, S. F., Isakeit, T., Ferri, M., Weinhold, A. R., Hendson, M., and Schroth, M. N. (1993a). Use of an exotic carbon source to selectively increase metabolic activity and growth of Pseudomonas putida in soil. Appl. Environ. Microbiol. 59, 2056–2063. doi: 10.1128/AEM.59.7.2056-2063.1993
Colbert, S. F., Schroth, M. N., Weinhold, A. R., and Hendson, M. (1993b). Enhancement of population densities of Pseudomonas putida PpG7 in agricultural ecosystems by selective feeding with the carbon source salicylate. Appl. Environ. Microbiol. 59, 2064–2070. doi: 10.1128/AEM.59.7.2064-2070.1993
Cui, X., Cai, J., Zhang, Y., Li, R., and Feng, T. (2011). Kinetics of transesterification of methyl acetate and N-butanol catalyzed by ionic liquid. Ind. Eng. Chem. Res. 50, 11521–11527. doi: 10.1021/ie2000715
da Graça, J. V., Douhan, G. W., Halbert, S. E., Keremane, M. L., Lee, R. F., Vidalakis, G., et al. (2016). Huanglongbing: an overview of a complex pathosystem ravaging the world's citrus. J. Integr. Plant Biol. 58, 373–387. doi: 10.1111/jipb.12437
Dai, Z., Wu, F., Zheng, Z., Yokomi, R., Kumagai, L., Cai, W., et al. (2019). Prophage diversity of ‘Candidatus Liberibacter asiaticus’ strains in California. Phytopathology 109, 551–559. doi: 10.1094/PHYTO-06-18-0185-R
Delgado, J. A., Short, N. M., Roberts, D. P., and vandenberg, B. (2019). Big data analysis for sustainable agriculture on a geospatial cloud framework. Front. Sustain. Food Syst. 3:54. doi: 10.3389/fsufs.2019.00054
dos Santos, H. R. M., Argolo, C. S., Argôlo-Filho, R. C., and Lopes Loguercio, L. (2019). A 16S rDNA PCR-based theoretical to actual delta approach on culturable mock communities revealed severe losses of diversity information. BMC Microbiol. 19:74. doi: 10.1186/s12866-019-1446-2
Ellen MacArthur Foundation (2013). Towards the Circular Economy. Opportunities for the Consumer Goods Sector. Available online at: https://www.mckinsey.com/~/media/mckinsey/dotcom/client_service/sustainability/pdfs/towards_the_circular_economy.ashx (accessed May 15, 2020).
Elsharkawy, K., Elsamadony, M., and Afify, H. (2019). Comparative analysis of common full scale reactors for dry anaerobic digestion process. E3S Web Conf. 83:01011. doi: 10.1051/e3sconf/20198301011
Environmental Defense Fund (EDF), (2016). How “Fertigation” is Helping this Citrus Grower Beat the Drought. Available online at: http://blogs.edf.org/growingreturns/2016/01/05/how-fertigation-is-helping-this-citrus-grower-beat-the-drought-2/ (accessed May 16, 2020).
FAO. (2015). The State of Food Insecurity in the World. Available online at: http://www.fao.org/3/a-i4646e.pdf (accessed November 25, 2020)
FAO. (2017). Water for Sustainable Food and Agriculture. A report produced for the G20 Presidency of Germany. Rome. ISBN 978-92-5-109977-3. Available online at: http://www.fao.org/3/a-i7959e.pdf (accessed November 25, 2020).
FAO. (2019). The State of Food and Agriculture 2019. Moving Forward on Food Loss and Waste Reduction. The State of the World series. Rome. Licence: CC BY-NC-SA 3.0 IGO. Available online at: http://www.fao.org/3/ca6030en/ca6030en.pdf (accessed November 25, 2020)
Food Agriculture Organization of the United Nations (FAO), (2013). Food Wastage Footprint. Impacts on Natural resources. Summary Report. ISBN 978-92-5-107752-8. Available online at: http://www.fao.org/3/i3347e/i3347e.pdf (accessed November 25, 2020).
Frank, J. A., Reich, C. I., Sharma, S., Weisbaum, J. S., Wilson, B. A., and Olsen, G. J. (2008). Critical evaluation of two primers commonly used for amplification of bacterial 16S rRNA genes. Appl. Environ. Microbiol. 74, 2461–2470. doi: 10.1128/AEM.02272-07
Franke-Whittle, I. H., Walter, A., Ebner, C., and Insam, H. (2014). Investigation into the effect of high concentrations of volatile fatty acids in anaerobic digestion on methanogenic communities. Waste Manag. 34, 2080–2089. doi: 10.1016/j.wasman.2014.07.020
Friedlingstein, P., Jones, M. W., O'Sullivan, M., Andrew, R. M., Hauck, J., Peters, G. P., et al. (2019). Global carbon budget 2019. Earth Syst. Sci. Data 11, 1783–1838. doi: 10.5194/essd-11-1783-2019
Fuchs, G., Boll, M., and Heider, J. (2011). Microbial degradation of aromatic compounds - from one strategy to four. Nat. Rev. Microbiol. 9, 803–816. doi: 10.1038/nrmicro2652
Gao, S., Song, W., and Guo, M. (2020). The integral role of bioproducts in the growing bioeconomy. Ind. Biotechnol. 16, 13–25. doi: 10.1089/ind.2019.0033
Ghosh, S., and Leff, L. G. (2013). Impacts of labile organic carbon concentration on organic and inorganic nitrogen utilization by a stream biofilm bacterial community. Appl. Environ. Microbiol. 79, 7130–7141. doi: 10.1128/AEM.01694-13
Gilmour, J. T., Cogger, C. G., Jacobs, L. W., Evanylo, G. K., and Sullivan, D. M. (2003). Decomposition and plant-available nitrogen in biosolids. J. Environ. Qual. 32, 1498–1507. doi: 10.2134/jeq2003.1498
Ginnan, N. A., Dang, T., Bodaghi, S., Ruegger, P. M., Peacock, B. B., McCollum, G., et al. (2018). Bacterial and fungal next generation sequencing datasets and metadata from citrus infected with ‘Candidatus Liberibacter asiaticus.’ Phytobiomes 2, 64–70. doi: 10.1094/PBIOMES-08-17-0032-A
Giwa, A. S., Xu, H., Chang, F., Zhang, X., Ali, N., Yuan, J., et al. (2019). Pyrolysis coupled anaerobic digestion process for food waste and recalcitrant residues: fundamentals, challenges, and considerations. Energy Sci. Eng. 7, 2250–2264. doi: 10.1002/ese3.503
Glick, B. R. (2010). Using soil bacteria to facilitate phytoremediation. Biotech. Adv. 28, 367–374. doi: 10.1016/j.biotechadv.2010.02.001
Gómez-Velasco, D. A., Álvarez-Solís, J. D., Ruiz-Valdiviezo, V. M., Abud-Archila, M., Montes-Molina, J. A., Dendooven, L., et al. (2014). Enzymatic activities in soil cultivated with coffee (Coffea arabica L. cv. “Bourbon”) and amended with organic material. Commun. Soil Sci. Plan. 45, 2529–2538. doi: 10.1080/00103624.2014.932375
Gougoulias, C., Clark, J. M., and Shaw, L. J. (2014). The role of soil microbes in the global carbon cycle: tracking the below-ground microbial processing of plant-derived carbon for manipulating carbon dynamics in agricultural systems. J. Sci. Food Agric. 94, 2362–2371. doi: 10.1002/jsfa.6577
Green, T. R., and Popa, R. (2011). Turnover of carbohydrate-rich vegetal matter during microaerobic composting and after amendment in soil. Appl. Biochem. Biotechnol. 165, 270–278. doi: 10.1007/s12010-011-9249-4
Harini, K., Ajila, V., and Hegde, S. (2013). Bdellovibrio bacteriovorus : a future antimicrobial agent? J. Indian Soc. Periodontol. 17, 823–825. doi: 10.4103/0972-124X.124534
Hibbing, M. E., Fuqua, C., Parsek, M. R., and Peterson, S. B. (2010). Bacterial competition: surviving and thriving in the microbial jungle. Nat. Rev. Microbiol. 8, 15–25. doi: 10.1038/nrmicro2259
Hodgson, R. W. (1967). “Chapter 4: Horticultural Varieties of Citrus” in The Citrus Industry, (Vol. 1), eds W. Reuther, H. J. Webber, and L. D. Batchelor (Berkeley: University of California), 431–459.
Horton, P., Bruce, R., Reynolds, C., and Milligan, G. (2019). Food chain inefficiency (FCI): accounting conversion efficiencies across entire food supply chains to re-define food loss and waste. Front. Sustain. Food Syst. 3:79. doi: 10.3389/fsufs.2019.00079
Hunt, D. E., Klepac-Ceraj, V., Acinas, S. G., Gautier, C., Bertilsson, S., and Polz, M. F. (2006). Evaluation of 23S rRNA PCR primers for use in phylogenetic studies of bacterial diversity. Appl. Environ. Microbiol. 72, 2221–2225. doi: 10.1128/AEM.72.3.2221-2225.2006
Ioannidou, S. M., Pateraki, C., Ladakis, D., Papapostolou, H., Tsakona, M., Vlysidis, A., et al. (2020). Sustainable production of bio-based chemicals and polymers via integrated biomass refining and bioprocessing in a circular bioeconomy context. Bioresour. Technol. 307:123093. doi: 10.1016/j.biortech.2020.123093
Ji, P., and Wilson, M. (2003). Enhancement of population size of a biological control agent and efficacy in control of bacterial speck of tomato through salicylate and ammonium sulfate amendments. Appl. Environ. Microbiol. 69, 1290–1294. doi: 10.1128/AEM.69.2.1290-1294.2003
Jouraiphy, A., Amir, S., El Gharous, M., Revel, J. C., and Hafidi, M. (2005). Chemical and spectroscopic analysis of organic matter transformation during composting of sewage sludge and green plant waste. Int. Biodeter. Biodegradation 56, 101–108. doi: 10.1016/j.ibiod.2005.06.002
Kim, S., Kramer, R. W., and Hatcher, P. G. (2003). Graphical method for analysis of ultrahigh-resolution broadband mass spectra of natural organic matter, the Van Krevelen diagram. Anal. Chem. 75, 5336–5344. doi: 10.1021/ac034415p
Köhl, J., Kolnaar, R., and Ravensberg, W. J. (2019). Mode of action of microbial biological control agents against plant diseases: relevance beyond efficacy. Front. Plant Sci. 10:845. doi: 10.3389/fpls.2019.00845
Kramer, C., and Gleixner, G. (2008). Soil organic matter in soil depth profiles: distinct carbon preferences of microbial groups during carbon transformation. Soil Biol. Biochem. 40, 425–433. doi: 10.1016/j.soilbio.2007.09.016
Lampridi, M. G., Sørensen, C. G., and Bochtis, D. (2019). Agricultural sustainability: a review of concepts and methods. Sustainability 11:5120. doi: 10.3390/su11185120
Lareen, A., Burton, F., and Schafer, P. (2016). Plant root-microbe communication in shaping root microbiomes. Plant Mol. Biol. 90, 575–587. doi: 10.1007/s11103-015-0417-8
Lebeis, S. L. (2015). Greater than the sum of their parts: characterizing plant microbiomes at the community-level. Curr. Opin. Plant Biol. 24, 82–86. doi: 10.1016/j.pbi.2015.02.004
Lugtenberg, B., and Kamilova, F. (2009). Plant-growth-promoting rhizobacteria. Annu. Rev. Microbiol. 63, 541–556. doi: 10.1146/annurev.micro.62.081307.162918
Luziatelli, F., Ficca, A. G., Colla, G., Baldassarre, S. E., and Ruzzi, M. (2019). Foliar application of vegetal-derived bioactive compounds stimulates the growth of beneficial bacteria and enhances microbiome biodiversity in lettuce. Front. Plant Sci. 10:60. doi: 10.3389/fpls.2019.00060
Ma, Y., Vosátka, M., and Freitas, H. (2019). Editorial: beneficial microbes alleviate climatic stresses in plants. Front. Plant Sci. 10:595. doi: 10.3389/fpls.2019.00595
Mahanty, T., Bhattacharjee, S., Goswami, M., Bhattacharyya, P., Das, B., Ghosh, A., et al. (2017). Biofertilizers: a potential approach for sustainable agriculture development. Environ. Sci. Pollut. Res. Int. 24, 3315–3335. doi: 10.1007/s11356-016-8104-0
McMurdie, P. J., and Holmes, S. (2013). Phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 8:e61217. doi: 10.1371/journal.pone.0061217
McRoberts, N., Figuera, S. G., Olkowski, S., McGuire, B., Luo, W., Posny, D., et al. (2019). Using models to provide rapid programme support for California's efforts to suppress Huanglongbing disease of citrus. Philos. Trans. R. Soc. B 374:20180281. doi: 10.1098/rstb.2018.0281
Mendes, R., Kruijt, M., de Bruijn, I., Dekkers, E., van der Voort, M., Schneider, J. H. M., et al. (2011). Deciphering the rhizosphere microbiome for disease-suppressive bacteria. Science 332, 1097–1100. doi: 10.1126/science.1203980
Minor, E. C., Swenson, M. M., Mattson, B. M., and Oyler, A. R. (2014). Structural characterization of dissolved organic matter: a review of current techniques for isolation and analysis. Environ. Sci. Processes Impacts 16, 2064–2079. doi: 10.1039/C4EM00062E
Möller, K. (2015). Effects of anaerobic digestion on soil carbon and nitrogen turnover, N emissions, and soil biological activity. A review. Agron. Sustain. Dev. 35, 1021–1041. doi: 10.1007/s13593-015-0284-3
Möller, K., and Müller, T. (2012). Effects of anaerobic digestion on digestate nutrient availability and crop growth: a review. Eng. Life Sci. 12, 242–257. doi: 10.1002/elsc.201100085
Monlau, F., Sambusiti, C., Ficara, E., Aboulkas, A., Barakata, A., and Carrere, H. (2015). New opportunities for agricultural digestate valorization: current situation and perspectives. Energy Environ. Sci. 8:2600. doi: 10.1039/C5EE01633A
Mueller, U. G., and Sachs, J. L. (2015). Engineering microbiomes to improve plant and animal health. Trends Microbiol. 23, 606–617. doi: 10.1016/j.tim.2015.07.009
Pagliaccia, D., Ferrin, D., and Stanghellini, M. E. (2007). Chemo-biological suppression of root-infecting zoosporic pathogens in recirculating hydroponic systems. Plant Soil 299, 163–179. doi: 10.1007/s11104-007-9373-7
Pagliaccia, D., Merhaut, D., Colao, M. C., Ruzzi, M., Saccardo, F., and Stanghellini, M. E. (2008). Selective enhancement of the fluorescent pseudomonad population after amending the recirculating nutrient solution of hydroponically grown plants with a nitrogen stabilizer. Microb. Ecol. 56, 538–554. doi: 10.1007/s00248-008-9373-z
Pandey, P., Lejeune, M., Biswas, S., Morash, D., Weimer, B., and Young, G. (2016). A new method for converting food waste into pathogen free soil amendment for enhancing agricultural sustainability. J. Clean. Prod. 112, 205–213. doi: 10.1016/j.jclepro.2015.09.045
Philippot, L., Raaijmakers, J., Lemanceau, P., and van der Putten, W. H. (2013). Going back to the roots: the microbial ecology of the rhizosphere. Nat. Rev. Microbiol. 11, 789–799. doi: 10.1038/nrmicro3109
Popa, R., Nealson, K. H., and Schechter, M. (2017). Method for Converting Food Waste and Other Biological Waste into Invertebrate Feed. US Patent No 10588331. Tonawanda, NY: U.S. Patent and Trademark Office.
Porter, S. S., and Sachs, J. L. (2020). Agriculture and the disruption of plant–microbial symbiosis. Evol. 35, 426 Trends Ecol.−439. doi: 10.1016/j.tree.2020.01.006
Raymaekers, K., Ponet, L., Holtappels, D., Berckmans, B., and Cammue, B. P. A. (2020). Screening for novel biocontrol agents applicable in plant disease management – a review. Biol. Control 144:104240. doi: 10.1016/j.biocontrol.2020.104240
Reinhold-Hurek, B., Bünger, W., Burbano, C. S., Sabale, M., and Hurek, T. (2015). Roots shaping their microbiome: global hotspots for microbial activity. Annu. Rev. Phytopathol. 53, 403–424. doi: 10.1146/annurev-phyto-082712-102342
Riera, N., Handique, U., Zhang, Y., Dewdney, M. M., and Wang, N. (2017). Characterization of antimicrobial-producing beneficial bacteria isolated from huanglongbing escape citrus trees. Front. Microbiol. 8:2415. doi: 10.3389/fmicb.2017.02415
Rivas-Ubach, A., Liu, Y., Bianchi, T. S., Tolić, N., Jansson, C., and Paša-Toli,ć, L. (2018). Moving beyond the van Krevelen diagram: a new stoichiometric approach for compound classification in organisms. Anal. Chem. 90, 6152–6160. doi: 10.1021/acs.analchem.8b00529
Ruzzi, M., and Aroca, R. (2015). Plant growth-promoting rhizobacteria act as biostimulants in horticulture. Sci. Hortic. 196, 124–134. doi: 10.1016/j.scienta.2015.08.042
Sagaram, U. S., Deangelis, K. M., Trivedi, P., Andersen, G. L., Lu, S. E., and Wang, N. (2009). Bacterial diversity analysis of huanglongbing pathogen-infected citrus, using phylochip arrays and 16S rRNA gene clone library sequencing. App. Environ. Microbiol. 75, 1566–1574. doi: 10.1128/AEM.02404-08
Sahu, B., Singh, J., Shankar, G., and Pradhan, A. (2018). Pseudomonas fluorescens PGPR bacteria as well as biocontrol agent: a review. Int. J. Chem. Stud. 6, 1–7. Available online at: https://www.chemijournal.com/archives/2018/vol6issue2/PartA/6-1-326-637.pdf (accessed November 25, 2020).
Sawers, R. J., Gutjahr, C., and Paszkowski, U. (2008). Cereal mycorrhiza: an ancient symbiosis in modern agriculture. Trends Plant Sci. 13, 93–97. doi: 10.1016/j.tplants.2007.11.006
Schlaeppi, K., and Bulgarelli, D. (2015). The plant microbiome at work. Mol. Plant Microbe Interact. 28, 212–217. doi: 10.1094/MPMI-10-14-0334-FI
Stubbendieck, R. M., and Straight, P. D. (2016). Multifaceted interfaces of bacterial competition. J. Bacteriol. 198, 2145–2155. doi: 10.1128/JB.00275-16
Suzuki, M. T., and Giovannoni, S. J. (1996). Bias caused by template annealing in the amplification of mixtures of 16S rRNA genes by PCR. Appl. Environ. Microbiol. 62, 625–630. doi: 10.1128/AEM.62.2.625-630.1996
Timmusk, S., Behers, L., Muthoni, J., Muraya, A., and Aronsson, A. C. (2017). Perspectives and challenges of microbial application for crop improvement. Front. Plant Sci. 8:49. doi: 10.3389/fpls.2017.00049
Trivedi, P., Duan, Y., and Wang, N. (2010). Huanglongbing, a systemic disease, restructures the bacterial community associated with citrus roots. Appl. Environ. Microbiol. 76, 3427–3436. doi: 10.1128/AEM.02901-09
Trivedi, P., He, Z., van Nostrand, J. D., Albrigo, G., Zhou, J., and Wang, N. (2012). Huanglongbing alters the structure and functional diversity of microbial communities associated with citrus rhizosphere. ISME J. 6, 363–383. doi: 10.1038/ismej.2011.100
Trivedi, P., Spann, T., and Wang, N. (2011). Isolation and characterization of beneficial bacteria associated with citrus roots in Florida. Microb. Ecol. 62, 324–336. doi: 10.1007/s00248-011-9822-y
Turner, T. R., James, E. K., and Poole, P. S. (2013). The plant microbiome. Genome Biol. 14:209. doi: 10.1186/gb-2013-14-6-209
United States Environmental Protection Agency (EPA), (2017). United States 2030 Food Loss and Waste Reduction Goal. Available online at: https://www.epa.gov/sustainable-management-food/united-states-2030-food-loss-and-waste-reduction-goal (accessed January 19, 2017).
University of California Agriculture and Natural Resources (UCANR), (2015). Drought Tip. Irrigating Citrus with Limited Water. ANR Pub. 8549, 1–6. Available online at: https://anrcatalog.ucanr.edu/pdf/8549.pdf (accessed November 25, 2020).
USDA's Agricultural Marketing Services (AMS), (2017). Anaerobic Digestate. Technical Evaluation Report. Available online at: https://www.ams.usda.gov/sites/default/files/media/AnaerobicDigestateTR5152017.pdf (accessed May 15, 2017).
van Veen, J. A., van Overbeek, L. S., and van Elsas, J. D. (1997). Fate and activity of microorganisms introduced into soil. Microbiol. Mol. Biol. Rev. 61, 121–135. doi: 10.1128/0.61.2.121-135.1997
Vanotti, M. B., García-González, M. C., Molinuevo-Salces, B., and Riaño, B. (2019). Editorial: new processes for nutrient recovery from wastes. Front. Sustain. Food Syst. 3:81. doi: 10.3389/fsufs.2019.00081
Waechter-Kristensen, B., Caspersen, S., Adalsteinsson, S., Sundin, P., and Jensén, P. (1999). Organic compounds and microorganisms in closed, hydroponic culture: occurrence and effects on plant growth and mineral nutrition. Acta Hortic. 481:20. doi: 10.17660/ActaHortic.1999.481.20
Wang, N., Stelinski, L. L., Pelz-Stelinski, K. S., Graham, J. H., and Zhang, Y. (2017). Tale of the huanglongbing disease pyramid in the context of the citrus microbiome. Phytopathology 107, 380–387. doi: 10.1094/PHYTO-12-16-0426-RVW
World Health Organization Regional Office for Europe (WHO), (2018). Circular Economy and Health. Opportunities and Risks. Available online at: http://www.euro.who.int/__data/assets/pdf_file/0004/374917/Circular-Economy_EN_WHO_web_august-2018.pdf?ua=1 (accessed November 25, 2020).
Wright, C. Y., Godfrey, L., Armiento, G., Haywood, L. K., Inglesi-Lotz, R., Lyne, K., et al. (2019). Circular economy and environmental health in low- and middle-income countries. Global Health 15:65. doi: 10.1186/s12992-019-0501-y
Wu, X., Wu, L., Liu, Y., Zhang, P., Li, Q., Zhou, J., et al. (2018). Microbial interactions with dissolved organic matter drive carbon dynamics and community succession. Front. Microbiol. 9:1234. doi: 10.3389/fmicb.2018.01234
Xu, H. L., Wang, R., and Mridha, M. A. U. (2001). Effects of organic fertilizers and a microbial inoculant on leaf photosynthesis and fruit yield and quality of tomato plants. J. Crop Prod. 3, 173–182. doi: 10.1300/J144v03n01_15
Xu, J., Zhang, Y., Zhang, P., Trivedi, P., Riera, N., Wang, Y., et al. (2018). The structure and function of the global citrus rhizosphere microbiome. Nat. Commun. 9:4894. doi: 10.1038/s41467-018-07343-2
Xu, L., and Geelen, D. (2018). Developing biostimulants from agro-food and industrial by-products. Front. Plant Sci. 9:1567. doi: 10.3389/fpls.2018.01567
Yamada, M., and Ogiso, M. (1997). Control of soil-borne diseases using antagonistic microorganisms. IV. Study on the available substrates for antagonistic bacterial strains to control Fusarium wilt of tomatoes. Res. Bull. Aichi-Ken Agric. Res. Cent. 29, 141–144.
Zhang, Y., Xu, J., Riera, N., Jin, J., Li, J., and Wang, N. (2017). Huanglongbing impairs the rhizosphere-to-rhizoplane enrichment process of the citrus root-associated microbiome. Microbiome 5:97. doi: 10.1186/s40168-017-0304-4
Keywords: circular economy, PGPM microorganisms, biostimulants, microbiome, plant health, food waste, anaerobic digestate, indoor agriculture
Citation: Pagliaccia D, Bodaghi S, Chen X, Stevenson D, Deyett E, De Francesco A, Borneman J, Ruegger P, Peacock B, Ellstrand N, Rolshausen PE, Popa R, Ying S and Vidalakis G (2020) Two Food Waste By-Products Selectively Stimulate Beneficial Resident Citrus Host-Associated Microbes in a Zero-Runoff Indoor Plant Production System. Front. Sustain. Food Syst. 4:593568. doi: 10.3389/fsufs.2020.593568
Received: 11 August 2020; Accepted: 17 November 2020;
Published: 14 December 2020.
Edited by:
Caitlin C. Rering, Agricultural and Veterinary Entomology, United StatesReviewed by:
Arthur Rudolph, Agricultural and Veterinary Entomology, United StatesMukesh Kumar Awasthi, Northwest A&F University, China
Copyright © 2020 Pagliaccia, Bodaghi, Chen, Stevenson, Deyett, De Francesco, Borneman, Ruegger, Peacock, Ellstrand, Rolshausen, Popa, Ying and Vidalakis. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Deborah Pagliaccia, ZGVib3JhaHBAdWNyLmVkdQ==
†These authors have contributed equally to this work and share first authorship
‡These authors have contributed equally to this work and share last authorship
 Deborah Pagliaccia
Deborah Pagliaccia Sohrab Bodaghi
Sohrab Bodaghi Xingyu Chen
Xingyu Chen Danielle Stevenson
Danielle Stevenson Elizabeth Deyett
Elizabeth Deyett Agustina De Francesco
Agustina De Francesco James Borneman
James Borneman Paul Ruegger3
Paul Ruegger3 Norman Ellstrand
Norman Ellstrand Philippe Eric Rolshausen
Philippe Eric Rolshausen Samantha Ying
Samantha Ying