
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Sustain. Food Syst. , 06 October 2020
Sec. Agroecology and Ecosystem Services
Volume 4 - 2020 | https://doi.org/10.3389/fsufs.2020.547751
This article is part of the Research Topic Microbiome Research in Agroecosystems View all 8 articles
This review emphasizes the potential and use of microbes in sustainable insect pest management. We first review the diverse insect traits shaped by insect-microbe associations that span nutrition, immunity, ecological interactions with natural enemy, insecticide resistance, and behavior. This is followed by discussing different microbiome manipulation approaches to alter pest traits, describing some of the opportunities and obstacles for each approach. We then highlight microbiomes as untapped chemical inventories to discover novel biopesticides, including plant-incorporated protectants and semiochemicals. The last topic covered is the use of beneficial microbes to improve mass-reared insects' performance for autocidal programs, including sterile insect technique and incompatible insect technique, in which we identify topics where data are limited or inconclusive, for future research.
According to the Entomological Society of America's (ESA) recent position statement, invasive insects incur control costs of over $2.5 billion and cause economic damages to crops, lawns, forests, and pastures totaling $18 billion per year (The Not-So-Hidden Dangers of Invasive Species., 2018). The threat of invasive crop pests to food security continues to be driven by the complex dynamics of human movements, global trade activities, climate change, and changing agricultural practices. Some of these factors also facilitate the dramatic expansion of urban pests and disease vector insects. Chemical insecticides have been central to insect pest control. However, a growing demand for reduced agricultural chemical use due to environmental and human health concerns in addition to pesticide resistance issues are fueling interests in innovative approaches to manage insect pests. For decades, the role of microbes in pest management has been largely confined to using entomopathogens, with only a handful of microbial species being developed into bioinsecticides. The paradigm is shifting owing to the advent of high-throughput sequencing, functional omics, and gene editing technologies, which significantly accelerate microbial discovery, plus a better understanding of microbial functions in complex communities across biological systems. There is also overwhelming evidence that symbiotic microbes play pivotal roles in shaping various insect traits. The collection of microbes associated with a given environment (both biotic and abiotic) and their collective genetic materials is termed the microbiome. Research in the microbiomes of insects, plants, and natural resources could be leveraged to develop novel management tools for insect pests. Figure 1 provides a graphical summary of the potential microbiome-based insect management strategies discussed in this review.
Insects are associated with diverse microbial communities and in many cases, these associations are crucial for insect survival and development. Symbiotic microbes in the gut, hemolymph, as well as in specialized cells, carry an arsenal of enzymes that provide specialized services to the insect hosts (Blow and Douglas, 2019). Supplies of essential nutrients (particularly amino acids and B vitamins) by endosymbionts have been well documented in a number of crop pests, particularly plant sap-sucking Hemipteran insects such as aphids, whiteflies, and psyllids (Douglas, 1998; Thao et al., 2000; Thao and Baumann, 2004; Luan et al., 2015), and in human disease vectors and urban pests such as tsetse flies in the genus Glossina and the common bed bug (Cimex lectularius). Some symbionts can degrade complex polysaccharides or recycle nitrogen for insects, such as the termites (Raychoudhury et al., 2013; Brune, 2014) and cockroaches (Berlanga et al., 2016). The production of antimicrobials by symbionts aids the immune system to fight against invading pathogens, as was shown in the beewolf digger wasps (Kroiss et al., 2010) and cotton leafworm (Shao et al., 2017).
Beside nutritional and immune services, symbionts can shape the ecological interactions between insects and their natural enemies. For instance, the secondary symbiont of aphids Hamiltonella defensa increased the chance of host survival from parasitoid wasp attacks by disrupting wasp embryogenesis, mediated by its bacteriophage-encoded toxins (Oliver et al., 2003, 2005; Moran et al., 2005; Vorburger et al., 2009; Schmid et al., 2012; Brandt et al., 2017). H. defensa was also shown to attenuate volatile release in aphid-infested plants, thus reducing parasitic wasp recruitment (Frago et al., 2012, 2017). Similarly, symbiont manipulation of plant physiology that facilitates insect colonization was observed in whiteflies and the Colorado potato beetle (Chung et al., 2013; Su et al., 2015). Modification of body color by facultative symbionts may determine aphid susceptibility to predation or parasitism (Xu et al., 2009). In particular, Rickettisella infection in the pea aphid Acyrthosiphon pisum increased the synthesis of blue-green polycyclic quinone pigments, turning the host from red to green. This symbiont-dependent color variation is believed to affect the aphid's relative risks between predation and parasitism, as their predators such as the ladybird beetles preferentially prey on the red morphs, while parasitoids preferentially attack the green morphs (Libbrecht et al., 2007; Tsuchida et al., 2010, 2014).
Symbionts have also been shown to influence insecticide resistance in various insect pests. In the bean bug (Riptortus pedestris) and the oriental fruit fly (Bactrocera dorsalis), resistance to organophosphorous pesticides has been attributed to direct detoxification by their symbionts (Kikuchi et al., 2012; Cheng et al., 2017). The insecticidal activity of Bacillus thuringiensis (Bt) in the gypsy moth (Lymantria dispar) larvae was shown to be dependent on the presence of symbiotic midgut bacteria (Broderick et al., 2006). Microbiome surveys from field-collected samples indicated that Bt resistance in cotton bollworm (Helicoverpa armigera) was associated with distinct microbiome compositions (Paramasiva et al., 2014; Visweshwar et al., 2015). Xia et al. observed varied levels of susceptibility to chlorpyrifos in the diamondback moth (Plutella xylostella) treated with antibiotics and then recolonized with different gut-associated bacteria (Xia et al., 2013, 2018). The contribution of gut bacteria to insecticide resistance was also reported in mosquitoes that showed reduced mortality to insecticides when fed with different gut commensal bacteria (Barnard et al., 2019).
Accumulating evidences have suggested that microbes play a more prominent role in shaping insect behavior than previously thought (McFall-Ngai et al., 2013). Insect behaviors that have been shown to be affected by microbes include long-term dispersal, oviposition, mating, host searching, and kin recognition (Ezenwa et al., 2012). Studies also suggest that microbiomes can influence host behavior via production of metabolites acting on host neuro-endocrine circuits (Adamo, 2013; Hemarajata and Versalovic, 2013; Wang and Kasper, 2014), a phenomenon termed the “gut-brain axis.” There is a plethora of research on the gut-brain axis, a majority of which has been focused on mammalian systems. However, this subject is still in infancy. One example is a recent study suggesting that the microbiome of Drosophila melanogaster influences host olfactory-guided foraging preferences toward foods of different microbial content (Wong et al., 2017). Similar microbiome-priming effects on fly behaviors were found in subsequent studies by Farine et al. (2017) and Qiao et al. (2019).
Advances in high throughput sequencing and functional genomics have enabled scientists to accomplish two milestones in insect microbiome research: 1. Explore previously uncharacterized microbiomes in a greater variety of insects, leading to a better understanding of host and environmental factors that shape insect microbiome diversity and composition. Some examples include microbial communities associated with Drosophilid and Tephritid fruit flies (Behar et al., 2008b; Wong et al., 2011, 2013; Ras et al., 2017), ants (Ramalho et al., 2017; Shin et al., 2017), bees (Engel et al., 2016), mosquitoes (Minard et al., 2013; Novakova et al., 2017), ticks (Narasimhan and Fikrig, 2015), beetles (Hulcr et al., 2012), and midges (Halpern and Senderovich, 2015), among others. 2. Attribute microbiome functions to specific microbial taxa or consortia. While the diversity and stability of microbiomes vary by insects, microbial influence on insect invasive traits is widely supported. For example, the invasiveness of the sweet potato whitefly (Bemisia tabaci) was promoted by a recent sweep of a Rickettsia sp. into the pest population, from 1% infected in 2000 to 97% by 2006) which led to faster development, higher survival rate to adulthood, and higher fecundity of the host (Himler et al., 2011). Studies on the powerful lab model Drosophila melanogaster showed that its microbiome, consisting of 5–20 microbial species, accelerates larval development, affects host foraging preference and reproduction (Ridley et al., 2012, 2013; Wong et al., 2014, 2017; Morimoto et al., 2017), which are all important parameters for invasiveness. Its close relative, Drosophila suzukii, or spotted wing drosophila (SWD), a significant pest of small fruits, relies on the microbiome to develop in ripening fruits (Bing et al., 2018). Among the highly destructive crop pests in the family Tephritidae, microbial symbiosis was first described in the olive fruit fly Bactrocera oleae (Petri, 1909, 1910). Unlike other fruit feeding Bactrocera species, B. oleae is associated with an obligate bacterial symbiont (Candidatus Erwinia dacicola) maintained in larval midgut caeca. The symbiont is currently uncultured but studies have shown that it facilitates fly development and reproduction by providing essential amino acids and metabolizing urea from various sources such as bird droppings, making nitrogen available to adult flies (Capuzzo et al., 2005; Ben-Yosef et al., 2014). It also helps the larvae develop in unripe olives by counteracting a plant defensive metabolite, oleuropein (Capuzzo et al., 2005; Estes et al., 2009). Interestingly, the symbiont has been found to be absent in domesticated B. oleae reared on artificial media containing antibiotics, demonstrating the impact of rearing on symbiont selection (Estes et al., 2012; Sacchetti et al., 2019). Other bacterial species associated with the guts of B. oleae include Pantoea sp. and Burkholderia sp., although their nutritional role is unclear (Ras et al., 2017). Microbiome-dependent larval development has also been seen in the medfly (Ceratitis capitata) and apple maggot fly (Rhagoletis pomonella). In medfly, microbial nutrient provisioning involves diazotrophs that express nitrogen reductase gene (nifH) in the gut (Behar et al., 2005). These medfly-associated bacteria include the genera Citrobacter, Klebsiella, Pectobacteria, Enterobacter, and Pantoea (Behar et al., 2008b). The community has been shown to support fly development, reproduction, and extend longevity when administered as probiotics (Behar et al., 2008b), and increase male copulatory success (Ben-Yosef et al., 2008). In addition to the dominant Enterobacteriaceae, a minor but stable community associated with medfly gut includes Pseudomonas spp. (Behar et al., 2008b). Using a metabarcoding approach, differences in microbial community was observed at different instar stages of medfly (Malacrinò et al., 2018). Burkholderia was found to be dominant in early instars and adults and may play a role in nitrogen fixation, as suggested in Tetraponera ants (Borm et al., 2002). Similarly, bacteria belonging to the genera Sphingomonas, Pseudomonas and an unidentified bacterium belonging to family Methylobacteraceae were found more abundant in late instars of medfly, whereas Leuconostoc, Weissella, Acetobacter, Gluconobacter and an unidentified bacterium belonging to family Xanthomonadaceae were more abundant at pupal stage (Malacrinò et al., 2018). In addition, variable microbial community was observed in medfly fed on different host plants. For example, medfly larvae feeding on Ficus carica (common fig) fruits were found to harbor Acinetobacter and Gluconobacter, while Acetobacter and Leuconostoc were found to be more abundant when fed on Prunus persica (peaches) (Malacrinò et al., 2018). Acinetobacter and Gluconostoc have been suggested to play a role in detoxifying phenolic glycosides (Mason et al., 2014). Similarly, apple maggot flies are associated with a diverse range of Enterobacteria in their gut, such as Pantoea, Klebsiella, and Enterobacter (Behar et al., 2009). The bacteria are deposited into the fruit during oviposition, thereby providing essential nutrients and proteins required for larval development (Lauzon, 2003; Behar et al., 2008a). In some Tephritids of the sub-families Dacinae and Trypetinae, symbiotic bacteria support larval development by metabolizing sugars, increasing the level of organic nitrogen, and synthesizing vitamins (Lauzon, 2003; Behar et al., 2009; Hamden et al., 2013). However, their functions in adult flies are unclear (Ben-Yosef et al., 2014).
The contribution of microbiomes to diverse insect invasiveness-related traits represents a rich array of resources that could be targeted for pest management. A simple strategy is to eliminate or disrupt insect symbiosis using biochemicals (Baumann, 2005). For example, ingestion of antibiotics such as tetracycline and penicillin has been shown to render tsetse flies sterile by affecting the obligate mutualist Wigglesworthia, impede the development of immature ticks and diminish reproduction of adult ticks by reducing their symbiont load (Zhong et al., 2007). The use of antimicrobial peptides (AMPs) has also been explored to manipulate insect symbionts, though more commonly used to control human or plant pathogens vectored by insects (Carter et al., 2013). AMPs (including diverse amphiphilic and cationic oligopeptides) are an integral part of insect innate immunity. They confer protection against a variety of microbes, including bacteria, fungi and viruses. Cecropin was the first AMP isolated from the pupae of Hylaophora cecropia (Hultmark et al., 1980; Steiner et al., 1981) and since then, about 150 AMPs have been purified from insects (Yi et al., 2014). On the basis of amino acid sequences and structures they have been classified into four groups: the α-helical peptides (e.g., moricin and cecropin), cysteine-rich peptides (e.g., defensin and drosomycin), proline-rich peptides (e.g., apidaecin, drosocin and lebocin), and glycine-rich proteins (e.g., attacin and gloverin). Some of the AMPs (e.g., defensins, cecropins, attacins, and proline-rich peptides) are present across insect orders, whereas others (e.g., moricin and gloverin) are restricted only to certain orders (e.g. Lepidoptera). The antimicrobial activity of AMPs is attributed to their positively charged surface which enables them to bind negatively charged microbial surface via charge-charge interactions, thereby interfering with the integrity of bacterial cell wall (Wu et al., 2018). AMPs have been engineered in plants and some insects to confer resistance to bacterial, fungal, and some eukaryotic parasites. For example, insect defensins (gallerimycin from Galleria mellonella) and cecropin (sarcotoxin-IA from Sarcophaga peregrina) have been shown to confer resistance to pathogenic fungi in transgenic tobacco (Mitsuhara et al., 2000). Similarly, transgenic expression of cecropins has also been shown in conferring resistance to fungal and bacterial pathogens in rice and tomato plants (Sharma et al., 2000; Jan et al., 2010). Cecropins also show activity against protozoan parasites, such as Plasmodium and Trypanosoma (Rodriguez et al., 1995; Fieck et al., 2010). Transgenic expression of cecropin in Anopheles gambiae, a vector for human parasite has been shown to reduce the number of Plasmodium berghei oocysts by 60% (Kim et al., 2004). In addition, transgenic co-expression of defensin-A and cecropin-A in Aedes aegypti has been shown to cooperatively block the transmission of Plasmodium parasite (Kokoza et al., 2010).
A key limitation of using antibiotics or AMPs is the possibility of non-target effects, which can lead to disruption of the native microbiome in non-target insects. Other limitations of using AMPs include low bioavailability, instability, and emergence of antimicrobial resistance (Shen et al., 2018). Nonetheless, AMPs with enhanced specificities can be generated by methods such as fusion with antibody fragments with affinity toward specific antigens (Peschen et al., 2004), protein engineering and synthetic biology techniques (e.g., substitution of amino acids, chemical modifications) (Cao et al., 2018). Improved delivery of AMPs to different living systems using nanotechnology is also being actively researched (Biswaro et al., 2018).
Introducing a foreign microbe to insects or replacing a symbiont with another microbe is also a viable strategy for manipulating insect traits. Experimental replacement of specific cultured and uncultured insect symbionts has been demonstrated in stink bugs and aphids. Hosokawa et al. (2007) showed that the swapping of the Ishikawaella symbionts between stink bug Megacopta punctatissima, a regular pest of soybean and other legumes, and a closely related non-pest species, Megacopta cribraria, resulted in poor M. punctatissima egg hatching on the plants. Experimental replacement of the primary symbiont Buchnera with a different genotype by microinjection in the pea aphid has also been shown to alter pest thermal tolerance (Moran and Yun, 2015).
Multiple protocols have been established to generate gnotobiotic insects (i.e., insects with experimentally controlled microbiomes) in the lab, such as Drosophila (Ridley et al., 2013), mosquitoes (Coon et al., 2014), and honey bees (Kešnerová et al., 2017). The protocol often involved cleansing the insect eggs to remove maternally deposited microbes on the surface, or treating larval or adult insects with antibiotics, followed by feeding on food seeded with cultured microbes or microbe-laden materials (e.g., fecal transplantation). While the gnotobiotic approach has been instrumental in the discoveries of microbiome impact on insect traits such as development, physiology, behavior, and insecticide resistance (Jaenike et al., 2010; Ridley et al., 2012; Coon et al., 2014; Wong et al., 2017; Xia et al., 2018), its use in pest management remains largely conceptual. A major exception is the rapidly growing interest in utilizing Wolbachia to control mosquitoes and mosquito-borne infectious agents. Wolbachia is a widely distributed, vertically transmitted endosymbiont in arthropods, estimated to infect over 60% of all insects (Hilgenboecker et al., 2008). The bacterium is known primarily for its ability to manipulate host reproduction through induction of cytoplasmic incompatibility (CI), feminization, parthenogenesis, and male killing (Stouthamer et al., 1999). By distorting sex ratios toward females, Wolbachia-infected females have a reproductive advantage over uninfected females, facilitating their propagation in a population. The incompatible insect technique (IIT) employs Wolbachia-induced CI as a strategy to control mosquitoes and other insect pests (Werren, 1997; Stouthamer et al., 1999). CI results in embryonic lethality (Bourtzis et al., 1998) and can be induced unidirectionally in crosses between Wolbachia-infected males and uninfected females, or bidirectionally in crosses between infected individuals bearing different strains of Wolbachia (Saridaki and Bourtzis, 2010). Wolbachia-induced sterility does not compromise the male mating competitiveness and their survival, as observed in Aedes aegypti (Segoli et al., 2014). In IIT, Wolbachia-infected males are repeatedly released to compete with the wild populations (Zabalou et al., 2004, 2009; Bourtzis, 2008; Berasategui et al., 2016). Extensive research has been carried out to use IIT against several insect pests including Rhagoletis cerasi, Ceratitits capitata, the tsetse fly, and disease vectors including Culex pipiens, Aedes albopictus and Culex quinquefasciatus (Zabalou et al., 2004, 2009; Alam et al., 2011; Bourtzis et al., 2014; Zhang et al., 2015; Atyame et al., 2016). Wolbachia can be transinfected into a novel host which are not naturally infected by Wolbachia strains that can induce CI (Zabalou et al., 2004, 2008; O'Connor et al., 2012; Zhang et al., 2015). For example, Wolbachia strain wSuz naturally infects D. suzukii but it does not induce CI (Ometto et al., 2013; Siozios et al., 2013; Hamm et al., 2014; Cattel et al., 2016). Two CI-inducing Wolbachia strains (wHa and wTei) from other Drosophila species have been identified and successfully introduced to D. suzukii, setting up the stage for IIT for this pest (Cattel et al., 2018; Nikolouli et al., 2018). However, the stability of the association, consequences of Wolbachia transinfection on host fitness including mating competitiveness need to be critically assessed before field applications can be exercised (O'Connor et al., 2012). In addition, the genotype of IIT insects should be considered since host genotype plays an important role in Wolbachia density and phenotypic expression of infection in hosts, including CI (Mouton et al., 2007).
Recently, significant progress has been made in identifying the molecular factors for Wolbachia-mediated CI. LePage et al. (2017) identified two differentially transcribed wMel genes, cifA and cifB (encoded by WO prophage), that functionally reiterated CI when expressed dually in uninfected males. Both genes are incapable of inducing CI independently. Further studies demonstrated that transgenic expression of cifA gene independently rescues CI and abolishes the embryonic lethality caused by wMel Wolbachia in Drosophila (Shropshire et al., 2018). In another study, Beckmann et al. (2017) demonstrated that interaction of Wolbachia deubiquitylating enzymes (DUB) cidA and cidB induces CI in transgenic Drosophila.
In addition to CI, another important trait of Wolbachia for mosquito control concerns its pathogen-blocking ability. Aedes aegypti, the vector for many clinically important arboviruses, do not generally bear Wolbachia in nature, but when transinfected with Wolbachia derived from Drosophila or other mosquitoes (e.g., Aedes albopictus and Culex quinquefasciatus), display drastically reduced competence for dengue, chikungunya, yellow fever, and Zika viruses (Bian et al., 2010; Van den Hurk et al., 2012; Aliota et al., 2016; Carrington et al., 2018), as well as Plasmodium and filarial nematodes (Kambris et al., 2009; Moreira et al., 2009). The exact mechanism of Wolbachia-mediated pathogen blocking is under investigation. Several hypotheses have been proposed, including priming of the immune system, changes in cholesterol and lipid droplets production and trafficking (Geoghegan et al., 2017), and (viral) RNA degradation (Thomas et al., 2018). In 2011, Ae. aegypti carrying the wMel strain were released into the wild near Cairns, Australia, marking the first trial of microbiome manipulation of a wild insect population for the purpose of reducing vector competence (Hoffmann et al., 2011). A follow-up investigation 2 years later suggested that the Wolbachia infection has stably established in the mosquito population (Hoffmann et al., 2014). More importantly, the release effectively stopped dengue transmission in Cairns and surrounding locations in northern Queensland, Australia. As of late 2019, mean Wolbachia frequencies in the original Cairns populations remains around 95%, with a 96% reduction in dengue cases. Subsequent releases have established Wolbachia throughout northern Queensland, as well as Yogyakarta, Indonesia, and Kuala Lumpur, Malaysia (Nazni et al., 2019; Ryan et al., 2020; Tantowijoyo et al., 2020).
A related strategy gaining traction in recent years is to genetically modify microbes to express desired effects in insects, known as paratransgenesis (Aksoy et al., 2008; Coutinho-Abreu et al., 2010; Caragata and Walker, 2012). Instead of transforming the insects (i.e., transgenesis), paratransgenesis bypasses the disadvantages of fitness cost associated with introducing a transgene into the insects and transgene instability in insect genomes. This approach is particularly suited for microbes that can be cultured, transformed, and readily reintroduced into the insect hosts. Paratransgenesis was proposed back in the early 90's, but most of the research progress has been focusing on human disease vectors and a few Hemipteran crop pests. Beard et al. (1992, 1993, 2000) demonstrated that the gut symbiont Rhodococcus rhodnii of the triatomine bug (Rhodnius prolixus) can be genetically modified to express effector molecules (cecropin A and related pore-forming molecules) against the protozoan Trypanosoma cruzi, the causative agent of Chagas disease. The symbiont can be introduced to insect progeny by inoculating eggshells or food with feces seeded with the engineered symbiont. Durvasula et al. (1999) also transformed the symbiont with an anti-trypanosome single-chain antibody and showed significant reduction in parasite load. The promising results from laboratory studies led to subsequent field trials in testing the transmission efficiency of engineered R. rhodnii to the triatomine bug using simulated triatomine-fecal materials called CRUZIGARD which consisted of an inert guar gum matrix dyed with India ink (Hurwitz et al., 2011a,b). More recently, a study has integrated paratransgenesis with RNA interference (RNAi) technology to control Rhodnius prolixus. Oral administration of an Escherichia coli strain HT115 or R. rhodnii engineered to express dsRNA targeting the antioxidant genes-heme-binding protein (RHBP) and catalase (CAT) genes in R. prolixus was shown to trigger systemic RNAi to silence these genes, resulting in poor development of nymphs and reduced fecundity of females (Taracena et al., 2015). Similar paratransgenic approaches have been tested on tsetse flies, using engineered symbiont Sodalis glossinidius that expressed antigen binding molecules targeting Trypanosoma brucei, the causative agent of sleeping sickness. Sodalis is found in the hemolymph, midgut, and milk gland and can be transmitted vertically through the milk glands (De Vooght et al., 2012, 2014). In mosquitoes, several paratransgenic strategies have been explored to prevent the transmission of malaria-causing Plasmodium parasites, using bacteria and fungi isolated from the mosquito midguts and ovaries. The Gram-negative Asaia bogorensis was selected for paratransgenesis against Plasmodium berghei since it was shown to persist in the midguts of mosquitoes and can spread quickly within a population, both vertically and horizontally (Bisi and Lampe, 2011; Dinparast Djadid et al., 2011). Genetically modified Asaia strains were constructed by fusing the siderophore receptor gene with anti-plasmodial effector genes. These genes included the scorpine antimicrobial peptide and a synthetic anti-Pbs21 scFv-Shiva1 immunotoxin composed of a single chain antibody (scFv) against P. berghei ookinete surface protein 21-Shiva1 fusion protein. Parasite development was significantly inhibited after Anopheles stephensi mosquitoes were fed with the transformed Asaia and challenged with P. berghei infected blood (Bongio and Lampe, 2015). Previously, a common symbiotic bacterium of mosquitoes, Pantoea agglomerans, was engineered to express anti-Plasmodium effector proteins using Type I hemolysin secretion system derived from E. coli. These engineered P. agglomerans strains were found to inhibit the development of P. falciparum and P. berghei in the midgut of Anopheles mosquitoes (Wang et al., 2012). An entomopathogenic fungus, Metarhizium anisopliae, has also been manipulated to secrete the antimicrobial scorpine and anti-plasmodial peptide SM1 capable of inhibiting the development of Plasmodium parasite (Fang et al., 2011). A study by Wang et al. (2017) discovered that gut and ovaries of A. stephensi are colonized by a novel Serratia sp. AS1. The AS1 strain was both sexually and vertically transmitted, persisting for at least three generations. Mosquitoes infected with an engineered Serratia AS1 containing five different anti-plasmodium effector molecules (Shiva1, a cecropin-like synthetic antimicrobial peptide; MP2, midgut peptide 2; EPIP, enolase-plasminogen interaction peptide (lysine-rich enolase peptide); scorpine, scorpion Pandinus imperator venom antimicrobial peptide; and mPLA2, inactive bee venom phospholipase A2) displayed a reduction in the oocyte load by 93% (Wang et al., 2017).
The potential of paratransgenesis in crop protection against insect pests or insect-vectored pathogens is demonstrated in the Glassy-winged sharpshooter (GWSS), Homalodisca coagulata. GWSS is a vector of Xylella fastidiosa, the bacterial pathogen that causes Pierce's disease in grapes by producing exopolysaccharides, which helps them colonize the xylem of its host plant, blocking the xylem flow (Hopkins and Purcell, 2002; Hackett, 2003; Bextine et al., 2004; Killiny et al., 2013). Among the different bacterial species isolated from GWSS, Alcaligenes xylosoxidans var. denitrificans (Axd), was chosen as a candidate bacterium for genetic manipulation. It has been consistently found in the xylem of host plants; the same niche occupied by the pathogen. Genetically modified Axd containing a DsRed fluorescent protein gene was successful delivered from injected stems to GWSS. It was found to colonize the insect foregut, suggesting a paratransgenic approach to clear X. fastidiosa from GWSS is feasible (Bextine et al., 2004). However, the use of transformed Axd in plants has potential drawbacks since this genus of bacteria has been reported as nosocomial human pathogen implicated in causing lung infection in cystic fibrosis patients (Krzewinski et al., 2001; Saiman et al., 2001). To ease the safety concern, an endophytic bacteria of grapes Pantoea agglomerans E325 (an EPA-approved agent for managing fire blight in pears and apples) was genetically engineered to express anti-Xylella effector proteins melittin and a scorpine-like AMP, and successfully colonized in the foregut of GWSS through an artificial feeding system (AFS) (Arora et al., 2015). Furthermore, targeted delivery of recombinant Pantoea agglomerans E325 to the gut of GWSS using microencapsulation strategy has been established to control Pierce's disease under simulated field conditions. Microencapsulation strategy may be useful for field application as it could prevent the environmental spread of foreign genetic material (Arora et al., 2015).
More recently, Leonard et al. (2020) developed a paratransgenic approach in honeybees and showed that it improved bee survival against viral infection and varroa mites in the lab. The authors engineered a symbiotic gut bacterium Snodgrassella alvi, expressing dsRNA specifically targeted to interfere with bee, viral or mite genes.
Successful application of gnotobiotic or paratransgenic approaches to control insect pests largely depends on the choice of microbes, the genetic design, and implementation of the treated insects. The microbes should ideally be specific to target insects or harmless to non-target hosts, another obstacle concerns the stability of the association. Persistent association ensures microbial-mediated impacts on host insects will be lasting, but some microbes may be “lost” from the insects due to environmental selective pressure or antagonistic effects with other microbes. Despite these caveats, it is anticipated that research and development on gnotobiotic and paratransgenic insects will continue to grow.
A key challenge to crop protection concerns the emergence of insecticide resistance, and for many insect pests there are no alternative control methods. Development of new synthetic pesticides has become increasingly costly and challenging, partly due to the difficulty of converting a lead compound into a product that can pass through strict environmental and safety regulations. A recent report estimated that in the US, the development of a synthetic pesticide now costs over $300 million and takes nearly 12 years (McDougall, 2016). The need to overcome resistance problems and to promote sustainable agriculture necessitates the discovery of new insecticidal agents.
In recent years, the push to develop novel insecticides with minimal environmental impact has led to a resurgence of interest in biopesticides (i.e., pesticides based on living organisms or their natural products, including their genes and metabolites). Currently, the global market for biopesticides is valued at $3 billion and is expected to grow by 15% in the next 4 years, outpacing the market growth of synthetics by 10-fold (Damalas and Koutroubas, 2018). Biopesticides fall into one of the three main categories: biochemical pesticides, which are naturally occurring chemicals that can alter pest behavior or physiology; microbial pesticides, which use pathogenic or toxic microbes as the active ingredient, and plant-incorporated protectants (PIPs), transgenic plants that produce pesticidal compounds (USEPAO, 2015).
Historically, microbial insecticides have taken the form of living microbes or spores applied directly to the field. Among the most common microbial insecticides are the entomopathogenic fungi, or EPFs, which are used in half of all classical biological control programs (Hajek and Delalibera, 2010). Several attributes of EPFs make them ideal candidates for biological control agents: many can be mass cultured in vitro; fungal spores have a long shelf life compared to other biological control agents; and they are often capable of persisting in the host population without repeated introductions (Hajek and Delalibera, 2010). The majority of economically significant species are soil inhabiting Hypocreales, including Metarhizium anisopliae, Beauveria bassiana, and Entomophaga maimaiga. Entomopathogenic nematodes (EPNs) of the families Steinernematidae and Heterorhabditidae, and their respective bacterial symbionts Xenorhabdus and Photorhabdus, are also important biological control agents of insects. EPNs have a broad host range, kill rapidly, and actively search for hosts to infect. However, they have a short shelf-life and are sensitive to environmental conditions such as low moisture levels and UV exposure (Lacey and Georgis, 2012).
Entomopathogens as biological control agents have several advantages over applying conventional insecticides: they are often host-specific, reducing impact on non-target species, and usually harmless to humans; they can be used in organic farming and Integrated Pest Management Programs; and are less susceptible to the pest resistance issues of conventional products. However, they can be difficult and expensive to mass-produce, with many requiring a host to complete their life cycle; they are also sensitive to environmental conditions such as sun exposure and humidity; and tend to have a short shelf life. A list of entomopathogens (bacteria, fungi, protozoa, and viruses) is listed in Table 1.
Most of the bacterial insecticidal products are derived from a small number of species, more significantly Bacillus thuringenesis (Bt), a Gram-positive, spore-forming species common in soils. Bt was first discovered in 1901, as the causal agent of sotto disease in silkworms, Bombyx mori (Ibrahim et al., 2010). However, the mechanism of Bt-induced insect mortality was not understood until the 1950's, when it was discovered the Cry proteins of certain strains are toxic to insects (Schnepf et al., 1998; Bravo et al., 2007).
Bt remained a niche product used primarily in forestry and organic farming until 1995, when transgenic crops expressing Cry proteins became commercially available. Development of Bt transgenic crops completely transformed Bt application: the toxin becomes constitutively expressed, covers all tissues, and non-target effects were minimized, as only insects that consume the plants are affected (Sanchis, 2011). Today, Bt varieties of maize, cotton, soy, and eggplant are widely grown, and new Bt varieties are constantly being developed (ISAAA's GM Approval Database, 2019). In the United States, more than 80% of maize and cotton acreage is planted with Bt varieties (Wechsler, 2018). Furthermore, over 300 distinct forms of the cry gene have been identified so far, with specificity against Lepidopteran, Dipteran, Coleopteran, or nematode hosts (Ibrahim et al., 2010). However, in recent years, the emergence of Bt resistance threatens the efficacy of Bt crops, underlining the importance of insect resistance management (IRM) practice and the need to search for new microbial insecticides.
Spinosad is another successful commercial insecticide derived from microbial natural products (NPs). It is the result of a natural product discovery program, conducted in the mid-1980's by Dow Agrosciences. The program involved exposing larval mosquitoes and Lepidoptera to fermentation broths derived from soil samples. One particular soil sample collected at a Virgin Islands rum distillery showed insecticidal activity toward both taxa, which was determined to be caused by secondary metabolites produced by an unknown actinomycete, described later as Saccharopolyspora spinosa (Mertz and Yao, 1990). These metabolites, the spinosyns, were found to be broad-spectrum insecticides with low toxicity to vertebrates and a novel mode of action targeting nicotinic acetylcholine and γ-aminobutyric acid (GABA) receptors. Spinosad, a formulation combining spinosyn A and spinosyn D in an approximately 5:1 ratio, first received regulatory approval in South Korea and the United States in the late 1900's, and was approved in the US for over 150 conventional and organic crops by 2004 (Racke, 2006).
This relatively low number of microbial insecticides is likely non-representative of the true diversity of microbial insecticidal products. Screens of microbial NPs for insecticidal properties are rare in the literature; the majority of NP discovery efforts focus on potential pharmaceuticals. Additionally, in the past 30 years, identifying novel NPs using traditional, culture-dependent screenings has become increasingly difficult, due to the high rate of rediscovery of known compounds (Katz et al., 2016). However, considering the percentage of successfully cultured microbial taxa is frequently estimated to be 1% or less, it is likely that many bioactive microbial NPs have yet to be discovered (Hofer, 2018).
The development of culture-independent, high-throughput metagenomics techniques has begun a renaissance in the field of NP discovery. Microbial toxins could be “mined” by extracting DNA from environmental samples, cloning into plasmids and expressing in recombinant expression systems. However, this method assumes the expression system possesses the molecular machinery necessary for heterologous expression and folding of the desired gene products, including regulatory elements (such as promoters and ribosomal binding sites), chemical substrates and biosynthetic precursors, an assumption which does not always hold. For example, in an experiment by Gabor et al. (2004), shotgun cloning of environmental microbial DNA recovered only ~40% of enzyme activity in the E. coli vector. This issue can be mitigated by conducting functional screenings in a broad range of expression systems, typically through the use of a specially constructed shuttle vector (Katz et al., 2016).
Insects rely on chemical communication to adjust their behaviors in response to the environment. These behaviors include foraging, mating, hazard avoidance, kin recognition, and social interactions. Chemicals that convey signals between organisms leading to changes in the behavior of recipient organisms are referred to as semiochemicals. A sustainable approach to pest management is to harness these semiochemicals to manipulate insect behavior. Insect pheromones (defined as semiochemicals that mediate interspecific interactions) constitute a large market share of attractants for plant and urban pests. Pheromone-baited traps are used in both monitoring and control of a variety of agricultural, forestry, and urban pests, such as the cotton boll weevil Anthonomus grandis, the gypsy moth Lymantria dispar, and the house fly Musca domestica (Witzgall et al., 2010). Field application of synthetic sex pheromones has also been shown to effectively control multiple Lepidopteran pests, including codling moth Cydia pomonella, pink bollworm Pectinophora gossypiella, and diamondback moth, by inhibiting their ability to find mates, a process known as mating disruption (Cardé and Minks, 1995).
Many commercially used insect semiochemicals are not insect pheromones, but rather, plant or microbial volatile organic compounds (VOCs) (Davis et al., 2013). VOCs are a large class of low-molecular weight compounds signified by high volatility and low vapor pressure. VOCs can be produced through different metabolic pathways, such as fermentation, amino acid catabolism, sulfur reduction, and terpenoid biosynthesis (Lemfack et al., 2017; Choudoir et al., 2019). Many VOCs are effective semiochemicals, requiring only small quantities to elicit responses from insects, which tend to have highly sensitive chemosensory (especially olfactory) systems. VOCs permit ranged communication by traveling through the air and serve as important means of environmental perception.
Recently, there is a growing interest in the discovery and implementation of microbial VOCs (mVOCs) as pest management tools. Multiple factors have contributed to this trend. First, according to mVOC 2.0, an online database of over 2,000 microbial volatiles from nearly 1,000 microbial species (Lemfack et al., 2017), <10% of known mVOCs are studied or assigned with functions. Given that an estimated 1018 microbial species are expected to exist on earth, many mVOCs have yet to be identified, Second, there is an increasing demand for natural products in agricultural pest management, as such products are generally better perceived by the public than synthetic chemicals in terms of safety and environmental sustainability.
Microbes have existed for over three billion years and through the processes of evolution, their metabolism is extremely diverse and versatile. The metabolic capabilities of microbiomes often far exceed than those of their eukaryotic hosts. Additionally, microbial metabolism can be augmented by genetic engineering using recombinant and synthetic techniques. This implies that microbial production of desired metabolites (including mVOCs) can be readily scaled up to meet the commercial needs. Advances in “-omics” techniques and computational biology to study the microbial metabolism can also help to accelerate the discoveries of mVOCs in a number of ways. Notably, high-throughput sequencing is illuminating the biosynthetic potential of microbiomes that extend beyond from cultured microbes, to uncultured ones found in plants, soils, and other natural resources. Computational tools are also becoming available to assemble complex metagenomes and metatranscriptomes, and to predict biosynthetic gene clusters from mining big data sets of DNA/RNA sequences (Medema et al., 2011; Weber et al., 2015). Although the vast majority of microbes on earth are yet to be cultured such that they are not readily amenable to conventional culture-dependent approaches, biologically-active natural products can be obtained from uncultured microbes by cloning environmental DNA into plasmid vectors and expressing in recombinant systems, such as E. coli, yeast, or baculovirus (Rappé and Giovannoni, 2003; Hofer, 2018). This approach has been actively pursued in screening for new antibiotics and therapeutic drugs in recent years, and in principle can be applied to the discoveries of microbial-based compounds for agricultural uses, including novel semiochemicals as well as biopesticides (Clardy et al., 2006).
Besides acting on insects directly, microbes can influence insect behavioral interactions with plants by altering plant VOC emission profiles. Ballhorn et al. (2013) showed that lima bean plants colonized with Rhizobia emitted a different VOC profile from the non-colonized plant in response to stimulation by jasmonic acid (JA), an important plant hormone involved in anti-pest defense, growth, and development. The distinct VOC profile from colonized plants corresponds to significantly greater Epilachna varivestis (Mexican bean beetle) repellency and was attributed to increased indole secretion (Ballhorn et al., 2013). Additionally, several symbiotic fungi, including endophytic fungi and arbuscular mycorrhizas, were shown to modify plant volatile emissions and consequently plant susceptibility to various insect pests (Fontana et al., 2009). These results hint at indirect beneficial effects of symbiotic microbes in plant defense that can be developed into probiotics for plants. On the other hand, some pathogenic microbes are known to manipulate plant volatile emissions to attract pests and potential vectors. Examples include the bacterium Candidatus Liberibacter asiasticus that infects citrus plants and causes Huanglongbing (HLB), also known as citrus greening disease. Candidatus L. asiaticus infection of citrus plants increased methyl salicylate levels, which was believed to promote attraction of its vector insect, the Asian citrus psyllid (Diaphorina citri) (Li et al., 2017). A number of plant viruses such as the Cucumber mosaic virus and Barley yellow dwarf luteovirus were also found to modify plant volatile emission to attract its vector insects (aphids) to improve their transmission (Sharifi et al., 2018).
Applications of microbial-based attractants or repellents in pest management have been backed by laboratory studies and field trials. For example, the invasive Drosophila suzukii is a major pest of small fruits and distinct from other non-pest Drosophila species in that it attacks intact, ripe, and ripening fruits (Keesey et al., 2015). Research has shown that Drosophila species are strongly attracted to fermentation products (Keesey et al., 2015), and by adding fermenting sugar-yeast mixture to a commonly used non-microbial trap bait (apple cider vinegar + 10% ethanol), D. suzukii capture rate increased by up to 15-fold in both laboratory and field conditions (Lasa et al., 2017). Huang et al. (2017) also showed combination baits of yeast-wine-vinegar-sugar mixtures are superior attractants compared to wine-vinegar alone. Ishii et al. (2015) genetically modified the acetic acid bacterium (AAB) Komagataeibacter europaeus with increased acetoin (a key ingredient in vinegar) production and was able to trap significantly more Drosophila using the mutant strain compared to other strains loaded on sticky traps. Similarly, microbial semiochemicals have been shown to attract tephritid fruit flies. For instance, a set of 21 compounds including alcohols, ketones, pyrazines, phenols and acids were identified from fermented broth of Klebsiella pneumoniae that attracted the Mexican fruit fly (Anastrepha ludens) (Lee et al., 1995). The Caribbean fruit fly (Anastrepha suspensa) females were shown to be attracted to VOCs 3-methyl-1-butanol and ammonia emitted from Enterobacter agglomerans, a bacterium isolated from larvae infested fruits and fly adults (Epsky et al., 1998). Some mVOCs, especially those produced by fungi, have potential to be developed into insect repellents. Geosmin was first described to be emitted by mold, sensed and avoided by Drosophila melanogaster (Stensmyr et al., 2012). It was later shown to induce aversive behaviors in SWD and may serve as an oviposition deterrent on crops (Stensmyr et al., 2012; Wallingford et al., 2016). The establishment of certain fungal species (Phoma spp., Fusarium spp., or Rhizopus spp.) on chicken feces also significantly reduced oviposition by the housefly, Musca domestica, and the effect was associated with emission of dimethyl trisulfide and 2-phenylethanol (Lam et al., 2010). Plant-associated symbionts could be another source of VOCs. For instance, Muscodor vitigenus, an endophyte of the tropical liana Paullina paullinoides, produces naphthalene and is strongly repelled by insects (Daisy et al., 2002). Microbial catalysis of repellent chemicals has been demonstrated in fungi including Penicillium, Aspergillus, and Fusarium that can convert JA derivatives dihydrojasmone and cis-jasmone to bioactive compounds such as (+)-(R)-4-hydroxydihydrojasmone and (–)-4-hydroxyjasmone (Gliszczyńska et al., 2015). These compounds were then found to repel aphids in a food-choice behavioral assay. In a study by Skrobiszewski et al. (2018) (±)-β-aryl-γ-ethylidene-γ-lactones (compounds which have previously been determined to have phagodetterent, antifungal, antibacterial, and anticancer activity) were enantioselectively hydrolyzed by a strain of Aspergillus ochraceus to form (–)-(S)-γ-ethylidene-γ-lactones and (+)-(R)-γ-ketoacids (Skrobiszewski et al., 2018). The phagodeterrent/repellent properties of these derived chemicals were then demonstrated against the lesser mealworm Alphitobius diaperinus. Many entomopathogenic bacteria, such as Bacillus thuringiensis (Bt), Pseudomonas entomophila, and nematode-associated species in the genus of Xenorhabdus and Photorhabdus have also been shown to have insect repellent properties (Bode, 2009; Kajla et al., 2019). For example, a recent study showed that a cocktail of bioactive secondary metabolites (primarily fabclavines) isolated from Xenorhabdus budapestensis exhibited greater repellency against Aedes aegypti than DEET and required a smaller concentration to achieve the same repellent effect (Kajla et al., 2019). More examples of microbial semiochemicals and their interaction with insects has been described by Leroy et al. (2011).
By identifying the different pathways from large-scale omics data, scientists will be able to make accurate inferences of microbial regulatory networks in mVOC production. Another emerging field is to dissect synergistic multi-species interactions (i.e., consortia of microbes) in metabolite production (Schulz-Bohm et al., 2015; Kai et al., 2018). Together, these studies hold promise that microbial-based insect attractants or repellents produce more diverse, effective, and versatile products for agricultural applications. One of the applications is to incorporate microbial based semiochemicals into push-pull strategies (Figure 2). In a typical push-pull system, a target pest is repelled or deterred away (“push”) from a protected source (a valuable crop or farm animal) using stimuli that make it unattractive. In parallel, the pest is diverted to attractive stimuli (“pull”) placed outside of the protected source. Stimuli commonly in use include different crop plants (intercrops or trap crops), visual signs, pheromones, synthetic volatiles, phagostimulants/repellents, and antifeedants (Cook et al., 2006). Microbial products can replace or be used in conjunction to these stimuli to improve the efficacy of future push-pull strategies.
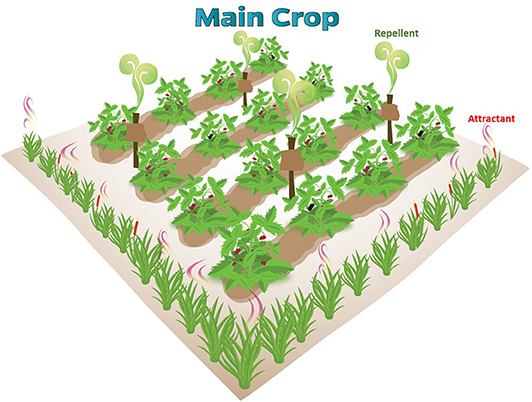
Figure 2. A generic pull-push system for agricultural pest management. Microbial-based semiochemicals can replace or synergize non-microbial attractants and repellents in the pull-push system.
Advances in nanotechnology are promoting effective and sustainable use of biopesticides in field applications. For instance, the potency of entomopathogenic bacteria, including Bt and Photorhabdus luminescens, increased significantly when they were applied to insect pests in the form of nanoparticles (Kim and Je, 2012; Kulkarni et al., 2017). Murthy et al. (2014) demonstrated that larvae subjected to Bt nanoparticles resulted in a faster and higher mortality as compared to unhomogenized Bt powder, owing to increased solubility of the Cry toxin in the alkaline midguts. Additionally, the development of nanoformulation delivery systems, such as nanoemulsion, nanocapsule, and nanosuspension, enhances the durability and stability of biopesticides under variable environmental conditions including exposure to UV light and humidity (Li et al., 2015; Sabbour and Singer, 2016; Zhang et al., 2016; Damalas and Koutroubas, 2018; Vassilev et al., 2020). These delivery systems are ecofriendly since they are often made of biodegradable natural polymers.
An emerging theme led from the research on nanoformulation is smart nanopesticides, in which the active ingredients are encapsulated in stimuli-responsive carriers such that their release can be controlled by stimuli such as light, temperature, humidity, or pH (Khot et al., 2012; Camara et al., 2019; Kumar et al., 2019). The stimuli-responsive delivery approach enables more precise spatial delivery while reduces the dosage and frequency of pesticide applications, therefore minimizing the impacts on the environment. In case of VOCs, nanoformulations have the potential to fine-tune the thermal stability of the compounds, leading to optimal release and longevity of the semiochemicals. Comprehensive reviews on different types of nanopesticides and stimuli-responsive delivery systems have been published by Huang et al. (2018), Camara et al. (2019).
Currently, the use of nanotechnology in agrochemical applications is still at an early stage. However, commercial products in this niche are beginning to be introduced to the market. An example is Seltima®, a fungicide launched by BASF in 2016 to protect rice crops. Seltima uses humidity responsive polymers to encapsulate pyraclostrobin, a fungicide chemical that is highly toxic to aquatic organisms. This encapsulation is resistant to water, thus allowing controlled release of the fungicide to the rice leaf surface and minimizing contamination of the surrounding aquatic ecosystems.
Based on the expectation that new microbes and microbiome-derived chemicals are being discovered exponentially, there will be unforeseen obstacles in the handling, storage, and delivery of these products for agricultural uses. Integration of microbial products with nanotechnology offers versatile means to improve their efficacy and stability, while controlling their environmental dispersal. Together, the combined efforts of microbiome mining and nanotechnology offers great potential in generating transformative bioinsecticide applications in near future.
Probiotics strategies to promote populations of beneficial insects (e.g., pollinators, natural enemies of pests) has been described by Engel and Moran (2013). Here, we focus on microbial strategies to promote insect rearing for autocidal programs, such as, the incompatible insect technique (IIT, described earlier in the section “Manipulation of insect-associated microbiomes for pest management”) and the sterile insect technique (SIT).
SIT is an environmentally-friendly insect pest control method involving mass-rearing of the target pests, where sterile male insects are generated by exposure to non-lethal-doses of ionizing radiation (Dyck et al., 2006; Nikolouli et al., 2018). Large numbers of fully sterile males are then released to mate with wild females, resulting in suppression or eradication of the pest population (Knipling, 1979; Dyck et al., 2006). A derivative of the SIT termed inherited sterility (IS) has also been developed for insect pests, mainly lepidopterans, that require high radiation dose to achieve complete sterility. In IS, partially sterile males are generated by an exposure to sub-sterilizing doses of radiation and then mated with wild females, resulting in reduced egg viability and highly sterile offspring that are predominately male (Vreysen et al., 2010). This approach avoids deleterious effects triggered by high-dose radiation on the insects. Studies have shown that males with inherited sterility suppress wild populations more effectively than fully sterile males for several lepidopteran (moth) species.
Over the past decades, SIT has been used successfully to control several high-profile pests, including the primary screwworm (Cochliomyia hominivorax), medfly (Ceratitis capitata), the Mexican fruit fly (Anastrepha ludens), various Lepidoptera (moths), and tsetse flies (Pereira et al., 2013; Calla et al., 2014; Vreysen et al., 2014; Barnes et al., 2015; Lees et al., 2015; Zhang et al., 2015; Bourtzis et al., 2016; Munhenga et al., 2016). However, a major setback for pest control through autocidal programs is the cost, because large numbers of treated insects are required to be released to compete with wild males and sometimes repeated releases are necessary. For SIT, a ratio up to 100 sterile insects for each wild insect is required. Less data is available for IIT, but a study on Wolbachia-infected mosquitoes showed that a release at 10:1 ratio of sterile to wild males was sufficient to suppress local mosquito population (Harris et al., 2012). Additionally, irradiation and handling in mass rearing facilities compromise the survival and performance of insects, which also contributes to the high cost of autocidal programs. Empirical evidence suggested that reduced performance, such as competence in attracting and mating with wild females, of mass-reared and irradiated males could be linked to disruption of their gut microbiomes. Agricultural pests and disease vectors reared under laboratory conditions tend to have fewer gut microbial taxa than their wild counterparts (Xiang et al., 2006; Rani et al., 2009; Rinke et al., 2011; Wang et al., 2011; Morrow et al., 2015; Liu et al., 2016; Waltmann et al., 2019; Raza et al., 2020). Such reduction in species richness or diversity can be due to limited microbial exposure, as insects are believed to acquire their gut microbes from their natural habitats and food sources but these microbes are mostly absent from the artificial environment and diets (Drew and Yuval, 2000; Wong et al., 2017; Stockton et al., 2019). Additionally, artificial rearing exerts distinct selection pressure on the insect gut microbiome composition. For instance, insect diets in the laboratory are commonly added with antimicrobials as preservatives. These antimicrobials could wipe out most of the microbes naturally associated with the insects, as shown in the B. oleae example discussed earlier. Considering the microbiome as an ecosystem, losses in species richness or diversity may lead to poor host outcomes, especially if the microbes provide specialized services to the host.
While a considerable body of literature exists on the impact of artificial rearing on the microbiomes of different insects, very little is known about how irradiation alters the insect microbiome. Most of the research has been conducted on tephritid fruit flies because several significant pests belonging to this group have been the foci of SIT applications. In medfly, newly eclosed irradiated males had lower levels of several dominant gut bacteria (especially Klebsiella sp.) but higher level of Pseudomonas sp. that was not presented in wild flies (Ami et al., 2010) However, some of these bacteria appeared to recover in abundance after a few days post irradiation. Similarly, in the oriental fruit fly, irradiation treatment selectively increased the abundance of Lactococcus and Orbus but diminished Lactobacillus, members of the Orbacecae family, and Morganella (Stathopoulou et al., 2019). More research is needed to define how these transient changes in the gut microbiome influence fly recovery from irradiation, and their long-term effects on fly fitness.
Based on the assumption that reversing the microbiome changes associated with artificial rearing or irradiation treatment can improve the fitness of the insects, scientists have been investigating the effects of dietary supplementation with bacteria on the performance of irradiated tephritid fruit flies. A summary of the current findings is shown in Table 2, showing that probiotic effects were reported in many, but not all studies. The discrepancies of results among these studies highlight that different parameters likely affect the outcome of probiotic treatment on insects, such as host age, genotype, microbial strain, diet and rearing condition. It is also worth noting that studies done so far have only tested single bacterium or cocktails of a few bacteria as supplement. Other approaches to manipulate the microbiomes of artificially reared or irradiated insects remain to be tested. One approach would be microbiome transplantation, either by rearing the treated insects on diet seeded with feces from freshly collected wild insects, or co-housing treated insects with wild insects, although this may be challenging to scale up for mass rearing. Another option would be to modify the diet recipe to stimulate the growth of beneficial microbes in the insect gut (“prebiotics”), which includes optimizing the nutrient composition and possibly incorporating natural food substances into their diets.
Clearly, more work is still needed to define how probiotics improve the fitness of artificially reared, sterile insects, and to identify treatment parameters that affect insect outcomes in response to the different microbial intervention approaches mentioned above. From a basic research angle, insects are excellent models to tease apart the complexities of microbial interactions and mechanisms, since their microbiomes are generally less diverse than in the mammalian systems, where most probiotics research has been conducted.
The ongoing advances and reduced cost of high throughput sequencing and omics technologies have enabled scientists to conduct in-depth analyses of microbiomes in various agroecosystems, including insects, plants, and other natural resources. It signifies an exciting era to discover new microbes or microbiome functions with potentials to be applied for insect pest management. Progresses in cutting-edge gene editing, microbial engineering, and nanotechnologies allow scientists to continue to refine procedures to extract bioactive products from uncultured microbes and apply them into pest management. These approaches will prove to be highly valuable for agricultural innovations.
AW conceived of the article and generated Figure 1. AW, MQ, SS, KG, and JH conducted the literature search, writing, and editing of the manuscript. MQ, SS, and KG generated Table 1. MQ generated Table 2. All authors have approved the final version of the manuscript.
This research was supported by the BARD US-Israel Agricultural Research and Development Fund US-5179-19 (AW).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors would like to thank Dr. Dorota Porazinska, Adam Robert, and Jesse Blair for their critical comments and editing of the manuscript. We also thank Ms. Susan Rodriguez for drawing the push-pull system figure.
Adamo, S. A. (2013). Parasites: evolution's neurobiologists. J. Exp. Biol. 216, 3–10. doi: 10.1242/jeb.073601
Aksoy, S., Weiss, B., and Attardo, G. (2008). “Paratransgenesis applied for control of tsetse transmitted sleeping sickness,” in Transgenesis and the Management of Vector-Borne Disease. Advances in Experimental Medicine and Biology, Vol. 627, eds S. Aksoy (New York, NY: Springer), 35–48. doi: 10.1007/978-0-387-78225-6_3
Alam, U., Medlock, J., Brelsfoard, C., Pais, R., Lohs, C., Balmand, S., et al. (2011). Wolbachia symbiont infections induce strong cytoplasmic incompatibility in the tsetse fly glossina morsitans. PLoS Pathog. 7:e1002415. doi: 10.1371/journal.ppat.1002415
Aliota, M. T., Peinado, S. A., Velez, I. D., and Osorio, J. (2016). The wMel strain of Wolbachia reduces transmission of zika virus by Aedes aegypti. Sci. Rep. 6:28792. doi: 10.1038/srep28792
Ami, E. B., Yuval, B., and Jurkevitch, E. (2010). Manipulation of the microbiota of mass-reared mediterranean fruit flies ceratitis capitata (diptera: tephritidae) improves sterile male sexual performance. ISME J. 4, 28–37. doi: 10.1038/ismej.2009.82
Arora, A. K., Forshaw, A., Miller, T. A., and Durvasula, R. (2015). A delivery system for field application of paratransgenic control. BMC Biotechnol. 15:59. doi: 10.1186/s12896-015-0175-3
Atyame, C. M., Labbé, P., Lebon, C., Weill, M., Moretti, R., Marini, F., et al. (2016). Comparison of irradiation and wolbachia based approaches for sterile-male strategies targeting aedes albopictus. PLoS ONE. 11:e0146834. doi: 10.1371/journal.pone.0146834
Augustinos, A. A., Kyritsis, G. A., Papadopoulos, N. T., Abd-Alla, A. M., Caceres, C., and Bourtzis, K. (2015). Exploitation of the medfly gut microbiota for the enhancement of sterile insect technique: use of Enterobacter sp. in larval diet-based probiotic applications. PLoS ONE. 10:e0136459. doi: 10.1371/journal.pone.0136459
Aw, K. M. S., and Hue, S. M. (2017). Mode of infection of Metarhizium spp. fungus and their potential as biological control agents. J. Fungi. 3:30. doi: 10.3390/jof3020030
Ballhorn, D. J., Kautz, S., and Schädler, M. (2013). Induced plant defense via volatile production is dependent on rhizobial symbiosis. Oecologia 172, 833–846. doi: 10.1007/s00442-012-2539-x
Barnard, K., Jeanrenaud, A. C. S. N., Brooke, B. D., and Oliver, S. (2019). The contribution of gut bacteria to insecticide resistance and the life histories of the major malaria vector Anopheles arabiensis (Diptera: Culicidae). Sci. Rep. 9:9117. doi: 10.1038/s41598-019-45499-z
Barnes, B., Hofmeyr, H., Groenewald, S., Conlong, D., and Wohlfarter, M. (2015). The sterile insect technique in agricultural crops in south Africa: a metamorphosis…. but will it fly? Afr. Entomol. 23, 1–18. doi: 10.4001/003.023.0103
Baumann, P. (2005). Biology bacteriocyte-associated endosymbionts of plant sap-sucking insects. Annu. Rev. Microbiol. 59, 155–189. doi: 10.1146/annurev.micro.59.030804.121041
Beard, C., O'Neill, S. L., Tesh, R., Richards, F., and Aksoy, S. (1993). Modification of arthropod vector competence via symbiotic bacteria. Parasitol Today. 9, 179–183. doi: 10.1016/0169-4758(93)90142-3
Beard, C. B., Durvasula, R. V., and Richards, F. F. (2000). “Bacterial symbiont transformation in Chagas disease vectors,” in Insect Transgenesis: Methods and Applications, eds A. M. Handler and A. A. James (Boca Raton: CRC Press), 289–303. doi: 10.1201/9781420039399.ch16
Beard, C. B., Mason, P. W., Aksoy, S., Tesh, R. B., and Ritchards, F. F. (1992). Transformation of an insect symbiont and expression of a foreign gene in the chagas' disease vector rhodnius prolixus. Am. J. Trop. Med. Hyg. 46, 195–200. doi: 10.4269/ajtmh.1992.46.195
Beckmann, J. F., Ronau, J. A., and Hochstrasser, M. (2017). A wolbachia deubiquitylating enzyme induces cytoplasmic incompatibility. Nat Microbiol. 2:17007. doi: 10.1038/nmicrobiol.2017.7
Behar, A., Ben-Yosef, M., Lauzon, C. R., Yuval, B., and Jurkevich, E. (2009). Structure and function of the bacterial community associated with the mediterranean fruit fly. Insect Symbiosis. 3, 251–271. doi: 10.1201/9781420064117.ch11
Behar, A., Jurkevitch, E., and Yuval, B. (2008a). Bringing back the fruit into fruit fly–bacteria interactions. Mol. Ecol. 17, 1375–1386. doi: 10.1111/j.1365-294X.2008.03674.x
Behar, A., Yuval, B., and Jurkevitch, E. (2005). Enterobacteria-mediated nitrogen fixation in natural populations of the fruit fly ceratitis capitata. Mol. Ecol. 14, 2637–2643. doi: 10.1111/j.1365-294X.2005.02615.x
Behar, A., Yuval, B., and Jurkevitch, E. (2008b). Gut bacterial communities in the mediterranean fruit fly (Ceratitis capitata) and their impact on host longevity. J. Insect Physiol. 54, 1377–1383. doi: 10.1016/j.jinsphys.2008.07.011
Ben-Yosef, M., Jurkevitch, E., and Yuval, B. (2008). Effect of bacteria on nutritional status and reproductive success of the mediterranean fruit fly Ceratitis capitata. Physiol. Entomol. 33, 145–154. doi: 10.1111/j.1365-3032.2008.00617.x
Ben-Yosef, M., Pasternak, Z., Jurkevitch, E., and Yuval, B. (2014). Symbiotic bacteria enable olive flies (Bactrocera oleae) to exploit intractable sources of nitrogen. J. Evol. Biol. 27, 2695–2705. doi: 10.1111/jeb.12527
Berasategui, A., Shukla, S., Salem, H., and Kaltenpoth, M. (2016). Potential applications of insect symbionts in biotechnology. Appl. Microbiol. Biotechnol. 100, 1567–1577. doi: 10.1007/s00253-015-7186-9
Berlanga, M., Llorens, C., Comas, J., and Guerrero, R. (2016). Gut bacterial community of the Xylophagous cockroaches Cryptocercus punctulatus and Parasphaeria boleiriana. PLoS ONE. 11:e0152400. doi: 10.1371/journal.pone.0152400
Bextine, B. R., Harshman, D., Johnson, M. C., and Miller, T. A. (2004). Impact of pymetrozine on glassy-winged sharpshooter feeding behavior and rate of Xylella fastidiosa transmission. J. Insect. Sci. 4:34. doi: 10.1673/031.004.3401
Bian, G., Xu, Y., Lu, P., Xie, Y., and Xi, Z. (2010). The endosymbiotic bacterium wolbachia induces resistance to dengue virus in aedes aegypti. PLoS Pathog. 6:e1000833. doi: 10.1371/journal.ppat.1000833
Bing, X., Gerlach, J., Loeb, G., and Buchon, N. (2018). Nutrient-dependent impact of microbes on drosophila suzukii development. MBio. 9, e02199–17. doi: 10.1128/mBio.02199-17
Bisi, D. C., and Lampe, D. J. (2011). Secretion of anti-plasmodium effector proteins from a natural pantoea agglomerans isolate by using PelB and HlyA secretion signals. Appl. Environ. Microbiol. 77, 4669–4675. doi: 10.1128/AEM.00514-11
Biswaro, L. S., da Costa Sousa, M. G., Rezende, T. M. B., Dias, S. C., Franco, O., et al. (2018). Antimicrobial peptides and nanotechnology, recent advances and challenges. Front. Microbiol. 9:855. doi: 10.3389/fmicb.2018.00855
Blow, F., and Douglas, A. E. (2019). The hemolymph microbiome of insects. J. Insect. Physiol. 115, 33–39. doi: 10.1016/j.jinsphys.2019.04.002
Bode, H. B. (2009). Entomopathogenic bacteria as a source of secondary metabolites. Curr. Opin. Chem. Biol. 13, 224–230. doi: 10.1016/j.cbpa.2009.02.037
Bongio, N. J., and Lampe, D. J. (2015). Inhibition of plasmodium berghei development in mosquitoes by effector proteins secreted from Asaia sp. Bacteria using a novel native secretion signal. PLoS ONE. 10:e0143541. doi: 10.1371/journal.pone.0143541
Borm, S., van Buschinger, A., Boomsma, J. J., and Billen, J. (2002). Tetraponera ants have gut symbionts related to nitrogen–fixing root–nodule bacteria. Proc. R Soc. Lond. B Biol. Sci. 269, 2023–2027. doi: 10.1098/rspb.2002.2101
Bourtzis, K. (2008). “Wolbachia- based technologies for insect pest population control,” in Transgenesis and the Management of Vector-Borne Disease, Advances in Experimental Medicine and Biology, ed. S. Aksoy (New York, NY: Springer), 104–113. doi: 10.1007/978-0-387-78225-6_9
Bourtzis, K., Dobson, S. L., Braig, H. R., and O'Neill, S. L. (1998). Rescuing wolbachia have been overlooked. Nature 391, 852–853. doi: 10.1038/36017
Bourtzis, K., Dobson, S. L., Xi, Z., Rasgon, J. L., Calvitti, M., Moreira, L. A., et al. (2014). Harnessing mosquito-wolbachia symbiosis for vector and disease control. Acta Trop. 132, S150–163. doi: 10.1016/j.actatropica.2013.11.004
Bourtzis, K., Lees, R. S., Hendrichs, J., and Vreysen, M. J. B. (2016). More than one rabbit out of the hat: Radiation, transgenic and symbiont-based approaches for sustainable management of mosquito and tsetse fly populations. Acta Trop. 157, 115–130. doi: 10.1016/j.actatropica.2016.01.009
Brandt, J. W., Chevignon, G., Oliver, K. M., and Strand, M. R. (2017). Culture of an aphid heritable symbiont demonstrates its direct role in defence against parasitoids. Proc. R Soc. B Biol. Sci. 284:20171925. doi: 10.1098/rspb.2017.1925
Bravo, A., Gill, S. S., and Soberón, M. (2007). Mode of action of bacillus thuringiensis cry and cyt toxins and their potential for insect control. Toxicon 49, 423–435. doi: 10.1016/j.toxicon.2006.11.022
Broderick, N. A., Raffa, K. F., and Handelsman, J. (2006). Midgut bacteria required for bacillus thuringiensis insecticidal activity. Proc. Natl. Acad. Sci. U. S. A. 103, 15196–15199. doi: 10.1073/pnas.0604865103
Brune, A. (2014). Symbiotic digestion of lignocellulose in termite guts. Nat. Rev. Microbiol. 12, 168–180. doi: 10.1038/nrmicro3182
Cai, Z., Yao, Z., Li, Y., Xi, Z., Bourtzis, K., Zhao, Z., et al. (2018). Intestinal probiotics restore the ecological fitness decline of Bactrocera dorsalis by irradiation. Evol. Appl. 11, 1946–1963. doi: 10.1111/eva.12698
Calla, B., Hall, B., Hou, S., and Geib, S. M. (2014). A genomic perspective to assessing quality of mass-reared SIT flies used in mediterranean fruit fly (Ceratitis capitata) eradication in California. BMC Genomics. 15:98. doi: 10.1186/1471-2164-15-98
Camara, M. C., Campos, E. V. R., Monteiro, R. A., do Espirito Santo Pereira, A., de Freitas Proenca, P. L., and Fraceto, L. F. (2019). Development of stimuli-responsive nano-based pesticides: emerging opportunities for agriculture. J. Nanobiotechnol. 17:100. doi: 10.1186/s12951-019-0533-8
Cao, J., de la Fuente-Nunez, C., Ou, R. W., Torres, M. D. T., Pande, S. G., Sinskey, A. J., et al. (2018). Yeast-based synthetic biology platform for antimicrobial peptide production. ACS Synth. Biol. 7, 896–902. doi: 10.1021/acssynbio.7b00396
Capuzzo, C., Firrao, G., Mazzon, L., Squartini, A., and Girolami, V. (2005). ‘Candidatus erwinia dacicola’, a coevolved symbiotic bacterium of the olive fly bactrocera oleae (Gmelin). Int. J. Syst. Evol. Microbiol. 55, 1641–1647. doi: 10.1099/ijs.0.63653-0
Caragata, E. P., and Walker, T. (2012). Using bacteria to treat diseases. Expert Opin. Biol. Ther. 12, 701–712. doi: 10.1517/14712598.2012.677429
Cardé, R. T., and Minks, A. K. (1995). Control of moth pests by mating disruption: successes and constraints. Annu. Rev. Entomol. 40, 559–585. doi: 10.1146/annurev.en.40.010195.003015
Carrington, L. B., Tran, B. C. N., Le, N. T. H., Luong, T. T. H., Nguyen, T. T., Nguyen, P. T., et al. (2018). Field- and clinically derived estimates of wolbachia-mediated blocking of dengue virus transmission potential in Aedes aegypti mosquitoes. Proc. Natl. Acad. Sci. U. S. A. 115, 361–366. doi: 10.1073/pnas.1715788115
Carter, V., Underhill, A., Baber, I., Sylla, L., Baby, M., Larget-Thiery, I., et al. (2013). Killer bee molecules: antimicrobial peptides as effector molecules to target sporogonic stages of plasmodium. PLoS Pathog. 9:e1003790. doi: 10.1371/journal.ppat.1003790
Castro, M. E., Souza, M. L., Araújo, S., and Bilimoria, S. (1997). Replication of Anticarsia gemmatalis nuclear polyhedrosis virus in four lepidopteran cell lines. J. Invertebr. Pathol. 69, 40–45. doi: 10.1006/jipa.1996.4624
Cattel, J., Kaur, R., Gibert, P., Martinez, J., Fraimout, A., Jiggins, F., et al. (2016). Wolbachia in European populations of the invasive pest drosophila suzukii: regional variation in infection frequencies. PLoS ONE. 11:e0147766. doi: 10.1371/journal.pone.0147766
Cattel, J., Nikolouli, K., Andrieux, T., Martinez, J., Jiggins, F., Charlat, S., et al. (2018). Back and forth wolbachia transfers reveal efficient strains to control spotted wing drosophila populations. J. Appl. Ecol. 55, 2408–2418. doi: 10.1111/1365-2664.13101
Cheng, D., Guo, Z., Riegler, M., Xi, Z., Liang, G., and Xu, Y. (2017). Gut symbiont enhances insecticide resistance in a significant pest, the oriental fruit fly Bactrocera dorsalis (Hendel). Microbiome. 5:13. doi: 10.1186/s40168-017-0236-z
Chiu, E., Coulibaly, F., and Metcalf, P. (2012). Insect virus polyhedra, infectious protein crystals that contain virus particles. Curr. Opin. Struct. Biol. 22, 234–240. doi: 10.1016/j.sbi.2012.02.003
Choudoir, M., Rossabi, S., Gebert, M., Helmig, D., and Fierer, N. (2019). A phylogenetic and functional perspective on volatile organic compound production by actinobacteria. MSystems. 4, e00295–18. doi: 10.1128/mSystems.00295-18
Chung, S. H., Rosa, C., Scully, E. D., Peiffer, M., Tooker, J. F., Hoover, K., et al. (2013). Herbivore exploits orally secreted bacteria to suppress plant defenses. Proc. Natl. Acad. Sci. U. S. A. 110, 15728–15733. doi: 10.1073/pnas.1308867110
Clardy, J., Fischbach, M. A., and Walsh, C. T. (2006). New antibiotics from bacterial natural products. Nat. Biotechnol. 24, 1541–1550. doi: 10.1038/nbt1266
Cook, S. M., Khan, Z. R., and Pickett, J. A. (2006). The use of push-pull strategies in integrated pest management. Annu. Rev. Entomol. 52, 375–400. doi: 10.1146/annurev.ento.52.110405.091407
Coon, K. L., Vogel, K. J., Brown, M. R., and Strand, M. (2014). Mosquitoes rely on their gut microbiota for development. Mol. Ecol. 23, 2727–2739. doi: 10.1111/mec.12771
Coutinho-Abreu, I. V., Zhu, K. Y., and Ramalho-Ortigao, M. (2010). Transgenesis and paratransgenesis to control insect-borne diseases: current status and future challenges. Parasitol. Int. 59, 1–8. doi: 10.1016/j.parint.2009.10.002
Daisy, B. H., Strobel, G. A., Castillo, U., Ezra, D., Sears, J., Weaver, D. K., et al. (2002). Naphthalene, an insect repellent, is produced by muscodor vitigenus, a novel endophytic fungus. Microbiology 148, 3737–3741. doi: 10.1099/00221287-148-11-3737
Damalas, C. A., and Koutroubas, S. D. (2018). Current status and recent developments in biopesticide use. Agriculture 8:13. doi: 10.3390/agriculture8010013
Davis, T. S., Crippen, T. L., Hofstetter, R. W., and Tomberlin, J. (2013). Microbial volatile emissions as insect semiochemicals. J. Chem. Ecol. 39, 840–859. doi: 10.1007/s10886-013-0306-z
De Vooght, L., Caljon, G., De Ridder, K., and Van Den Abbeele, J. (2014). Delivery of a functional anti-trypanosome nanobody in different tsetse fly tissues via a bacterial symbiont, Sodalis glossinidius. Microb. Cell Fact. 13:156. doi: 10.1186/s12934-014-0156-6
De Vooght, L., Caljon, G., Stijlemans, B., De Baetselier, P., Coosemans, M., and Van Den Abbeele, J. (2012). Expression and extracellular release of a functional anti-trypanosome nanobody® in Sodalis glossinidius, a bacterial symbiont of the tsetse fly. Microb. Cell Fact. 11:23. doi: 10.1186/1475-2859-11-23
Dinparast Djadid, N., Jazayeri, H., Raz, A., Favia, G., Ricci, I., and Zakeri, S. (2011). Identification of the midgut microbiota of an. stephensi and an. maculipennis for their application as a paratransgenic tool against malaria. PLoS ONE. 6:e0028484. doi: 10.1371/journal.pone.0028484
Douglas, A. E. (1998). Nutritional interactions in insect-microbial symbioses: aphids and their symbiotic bacteria buchnera. Annu. Rev. Entomol. 43, 17–37. doi: 10.1146/annurev.ento.43.1.17
Drew, R. A., and Yuval, B. (2000). “The evolution of fruit fly feeding behavior,” in Fruit Flies (Tephritidae) Phylogeny and Evolution of Behavior, eds eds M. Aluja and A. L. Norrbom (Boca Raton: CRC Press), 749–768. doi: 10.1201/9781420074468.ch27
Durvasula, R. V., Kroger, A., Goodwin, M., Panackal, A., Kruglov, O., Taneja, J., et al. (1999). Strategy for introduction of foreign genes into field populations of chagas disease vectors. Ann. Entomol. Soc. Am. 92, 937–943. doi: 10.1093/aesa/92.6.937
Dyck, V. A., Hendrichs, J., and Robinson, A. S. (Eds.) (2006). Sterile Insect Technique: Principles and Practice in Area-Wide Integrated Pest Management. (Dordrecht: Springer), 39–68. doi: 10.1007/1-4020-4051-2
Engel, P., Kwong, W. K., McFrederick, Q., Anderson, K. E., Barribeau, S. M., Chandler, J. A., et al. (2016). The bee microbiome: impact on bee health and model for evolution and ecology of host-microbe interactions. mBio 7:e02164–15. doi: 10.1128/mBio.02164-15
Engel, P., and Moran, N. A. (2013). The gut microbiota of insects – diversity in structure and function. FEMS Microbiol. Rev. 37, 699–735. doi: 10.1111/1574-6976.12025
Epsky, N. D., Heath, R. R., Dueben, B. D., Lauzon, C. R., Proveaux, A. T., and MacCollom, G. B. (1998). Attraction of 3-methyl-1-butanol and ammonia identified from Enterobacter agglomerans to Anastrepha suspensa. J. Chem. Ecol. 24, 1867–1880. doi: 10.1023/A:1022363718193
Estes, A. M., Hearn, D. J., Bronstein, J. L., and Pierson, E. A. (2009). The olive fly endosymbiont, “Candidatus erwinia dacicola,” switches from an intracellular existence to an extracellular existence during host insect development. Appl. Environ. Microbiol. 75, 7097–7106. doi: 10.1128/AEM.00778-09
Estes, A. M., Hearn, D. J., Burrack, H. J., Rempoulakis, P., and Pierson, E. A. (2012). Prevalence of Candidatus erwinia dacicola in wild and laboratory olive fruit fly populations and across developmental stages. Env. Entom. 41, 265–274. doi: 10.1603/EN11245
Ezenwa, V. O., Gerardo, N. M., Inouye, D. W., Medina, M., and Xavier, J. B. (2012). Animal behavior and the microbiome. Science 338, 198–199. doi: 10.1126/science.1227412
Fang, W., Vega-Rodríguez, J., Ghosh, A. K., Jacobs-Lorena, M., Kang, A., and St. Leger, R. J. (2011). Development of transgenic fungi that kill human malaria parasites in mosquitoes. Science 331, 1074–1077. doi: 10.1126/science.1199115
Farine, J.-P., Habbachi, W., Cortot, J., Roche, S., and Ferveur, J.-F. (2017). Maternally-transmitted microbiota affects odor emission and preference in drosophila larva. Sci. Rep. 7:6062. doi: 10.1038/s41598-017-04922-z
Fieck, A., Hurwitz, I., Kang, A. S., and Durvasula, R. (2010). Trypanosoma cruzi: synergistic cytotoxicity of multiple amphipathic anti-microbial peptides to T. cruzi and potential bacterial hosts. Exp. Parasitol. 125, 342–347. doi: 10.1016/j.exppara.2010.02.016
Fontana, A., Reichelt, M., Hempel, S., Gershenzon, J., and Unsicker, S. B. (2009). The effects of arbuscular mycorrhizal fungi on direct and indirect defense metabolites of plantago lanceolata L. J. Chem. Ecol. 35, 833–843. doi: 10.1007/s10886-009-9654-0
Frago, E., Dicke, M., and Godfray, H. C. J. (2012). Insect symbionts as hidden players in insect–plant interactions. Trends Ecol. Evol. 27, 705–711. doi: 10.1016/j.tree.2012.08.013
Frago, E., Mala, M., Weldegergis, B. T., Yang, C., McLean, A., Godfray, H. C. J., et al. (2017). Symbionts protect aphids from parasitic wasps by attenuating herbivore-induced plant volatiles. Nat. Commun. 8:1860. doi: 10.1038/s41467-017-01935-0
Gabor, E. M., Alkema, W. B. L., and Janssen, D. B. (2004). Quantifying the accessibility of the metagenome by random expression cloning techniques. Environ. Microbiol. 6, 879–886. doi: 10.1111/j.1462-2920.2004.00640.x
Gassman, A. J., and Clifton, E. H. (2017). “Current and potential applications of biopesticides to manage insect pests of maize,” in Microbial Control of Insect and Mite Pests: From Theory to Practice, ed L. A. Lacey (Amsterdam: Academic Press), 173–184.
Gavriel, S., Jurkevitch, E., Gazit, Y., and Yuval, B. (2011). Bacterially enriched diet improves sexual performance of sterile male mediterranean fruit flies: bacteria and medfly sexual performance. J. Appl. Entomol. 135, 564–573. doi: 10.1111/j.1439-0418.2010.01605.x
Geoghegan, V., Stainton, K., Rainey, S. M., Ant, T. H., Dowle, A. A., Larson, T., et al. (2017). Perturbed cholesterol and vesicular trafficking associated with dengue blocking in wolbachia-infected aedes aegypti cells. Nat. Commun. 8:526. doi: 10.1038/s41467-017-00610-8
Gliszczyńska, A., Gladkowski, W., Dancewicz, K., and Gabryś, B. (2015). Enantioselective microbial hydroxylation as a useful tool in the production of jasmonate derivatives with aphid deterrent activity. Curr. Microbiol. 71, 83–94. doi: 10.1007/s00284-015-0831-9
Hajek, A. E., and Delalibera, I. (2010). Fungal pathogens as classical biological control agents against arthropods. BioControl 55, 147–158. doi: 10.1007/s10526-009-9253-6
Halpern, M., and Senderovich, Y. (2015). Chironomid microbiome. Microb. Ecol. 70, 1–8. doi: 10.1007/s00248-014-0536-9
Hamden, H., Guerfali, M. M., Fadhl, S., Saidi, M., and Chevrier, C. (2013). Fitness improvement of mass-reared sterile males of ceratitis capitata (vienna 8 strain) (Diptera: Tephritidae) after gut enrichment with probiotics. J. Econ. Entomol. 106, 641–647. doi: 10.1603/EC12362
Hamm, C. A., Begun, D. J., Vo, A., Smith, C. C. R., Saelao, P., Shaver, A. O., et al. (2014). Wolbachia do not live by reproductive manipulation alone: infection polymorphism in Drosophila suzukii and D. subpulchrella. Mol. Ecol. 23, 4871–4885. doi: 10.1111/mec.12901
Harris, A. F., McKemey, A. R., Nimmo, D., Curtis, Z., Black, I., Morgan, S. A., et al. (2012). Successful suppression of a field mosquito population by sustained release of engineered male mosquitoes. Nat. Biotechnol. 30, 828–830. doi: 10.1038/nbt.2350
Hemarajata, P., and Versalovic, J. (2013). Effects of probiotics on gut microbiota: mechanisms of intestinal immunomodulation and neuromodulation. Therap. Adv. Gastroenterol. 6, 39–51. doi: 10.1177/1756283X12459294
Hilgenboecker, K., Hammerstein, P., Schlattmann, P., Telschow, A., and Werren, J. H. (2008). How many species are infected with wolbachia?—a statistical analysis of current data. FEMS Microbiol Lett. 281, 215–220. doi: 10.1111/j.1574-6968.2008.01110.x
Himler, A. G., Adachi-Hagimori, T., Bergen, J. E., Kozuch, A., Kelly, S. E., Tabashnik, B. E., et al. (2011). Rapid spread of a bacterial symbiont in an invasive whitefly is driven by fitness benefits and female bias. Science 332, 254–256. doi: 10.1126/science.1199410
Hofer, U. (2018). The majority is uncultured. Nat. Rev. Microbiol. 16, 716–717. doi: 10.1038/s41579-018-0097-x
Hoffmann, A. A., Iturbe-Ormaetxe, I., Callahan, A. G., Phillips, B. L., Billington, K., Axford, J. K., et al. (2014). Stability of the Wmel wolbachia infection following invasion into Aedes aegypti populations. PLoS Negl. Trop. Dis. 8:e3115. doi: 10.1371/journal.pntd.0003115
Hoffmann, A. A., Montgomery, B. L., Popovici, J., Iturbe-Ormaetxe, I., Johnson, P. H., Muzzi, F., et al. (2011). Successful establishment of wolbachia in aedes populations to suppress dengue transmission. Nature 476, 454–457. doi: 10.1038/nature10356
Hopkins, D., and Purcell, A. (2002). Xylella fastidiosa : cause of pierce's disease of grapevine and other emergent diseases. Plant Dis. Plant Dis. 86, 1056–1066. doi: 10.1094/PDIS.2002.86.10.1056
Hosokawa, T., Kikuchi, Y., Shimada, M., and Fukatsu, T. (2007). Obligate symbiont involved in pest status of host insect. Proc. Biol. Sci. 274, 1979–1984. doi: 10.1098/rspb.2007.0620
Huang, B., Chen, F., Shen, Y., Qian, K., Wang, Y., Sun, C., et al. (2018). Advances in targeted pesticides with environmentally responsive controlled release by nanotechnology. Nanomaterials 8:102. doi: 10.3390/nano8020102
Huang, J., Gut, L., and Grieshop, M. (2017). Evaluation of food-based attractants for Drosophila suzukii (Diptera: Drosophilidae). Environ. Entomol. 46, 878–884. doi: 10.1093/ee/nvx097
Hulcr, J., Rountree, N. R., Diamond, S. E., Stelinski, L. L., Fierer, N., and Dunn, R. R. (2012). Mycangia of ambrosia beetles host communities of bacteria. Microb. Ecol. 64, 784–793. doi: 10.1007/s00248-012-0055-5
Hultmark, D., Steiner, H., Rasmuson, T., and Boman, H. G. (1980). Insect immunity. Purification and properties of three inducible bactericidal proteins from hemolymph of immunized pupae of hyalophora cecropia. Eur. J. Biochem. 106, 7–16. doi: 10.1111/j.1432-1033.1980.tb05991.x
Hurst, M. R., Glare, T. R., Jackson, T. A., and Ronson, C. W. (2000). Plasmid-located pathogenicity determinants of Serratia entomophila, the causal agent of amber disease of grass grub, show similarity to the insecticidal toxins of Photorhabdus luminescens. J. Bacteriol. 182, 5127–5138. doi: 10.1128/jb.182.18.5127-5138.2000
Hurwitz, I., Fieck, A., Read, A., Hillesland, H., Klein, N., Kang, A., et al. (2011a). Paratransgenic control of vector borne diseases. Int. J. Biol. Sci. 7:1334. doi: 10.7150/ijbs.7.1334
Hurwitz, I., Hillesland, H., Fieck, A., Das, P., and Durvasula, R. (2011b). The paratransgenic sand fly: a platform for control of Leishmania transmission. Parasit. Vectors. 4:82. doi: 10.1186/1756-3305-4-82
Ibrahim, M. A., Griko, N., Junker, M., and Bulla, L. A. (2010). Bacillus thuringiensis. Bioeng. Bugs. 1, 31–50. doi: 10.4161/bbug.1.1.10519
ISAAA's GM Approval Database (2019). International Service for the Aquisition of Agri-Biotech Applications. ISAAA's GM Approval Database. Available online at: http://www.isaaa.org/gmapprovaldatabase/default.asp (accessed January, 2020).
Ishii, Y., Akasaka, N., Goda, I., Sakoda, H., and Fujiwara, S. (2015). Effective trapping of fruit flies with cultures of metabolically modified acetic acid bacteria. Appl. Environ. Microbiol. 81, 2265–2273. doi: 10.1128/AEM.03678-14
Jaenike, J., Unckless, R., Cockburn, S. N., Boelio, L. M., and Perlman, S. J. (2010). Adaptation via symbiosis: recent spread of a drosophila defensive symbiont. Science 329, 212–215. doi: 10.1126/science.1188235
Jan, P.-S., Huang, H.-Y., and Chen, H.-M. (2010). Expression of a Synthesized gene encoding cationic peptide cecropin B in transgenic tomato plants protects against bacterial diseases. Appl. Environ. Microbiol. 76, 769–775. doi: 10.1128/AEM.00698-09
Ji, D., Yi, Y., Kang, G. H., Choi, Y. H., Kim, P., Baek, N. I., et al. (2004). Identification of an antibacterial compound, benzylideneacetone, from Xenorhabdus nematophila against major plant-pathogenic bacteria. FEMS Microbiol. Lett. 239, 241–248. doi: 10.1016/j.femsle.2004.08.041
Kai, M., Effmert, U., Lemfack, M. C., and Piechulla, B. (2018). Interspecific formation of the antimicrobial volatile schleiferon. Sci. Rep. 8:16852. doi: 10.1038/s41598-018-35341-3
Kajla, M. K., Barrett-Wilt, G. A., and Paskewitz, S. M. (2019). Bacteria: a novel source for potent mosquito feeding-deterrents. Sci. Adv. 5:eaau6141. doi: 10.1126/sciadv.aau6141
Kambris, Z., Cook, P. E., Phuc, H. K., and Sinkins, S. P. (2009). Immune activation by life-shortening wolbachia and reduced filarial competence in mosquitoes. Science 326, 134–136. doi: 10.1126/science.1177531
Katz, M., Hover, B. M., and Brady, S. F. (2016). Culture-independent discovery of natural products from soil metagenomes. J. Ind. Microbiol. Biotechnol. 43, 129–141. doi: 10.1007/s10295-015-1706-6
Keesey, I. W., Knaden, M., and Hansson, B.S. (2015). Olfactory specialization in Drosophila suzukii supports an ecological shift in host preference from rotten to fresh fruit. J. Chem. Ecol. 41, 121–128. doi: 10.1007/s10886-015-0544-3
Kešnerová, L., Mars, R. A. T., Ellegaard, K. M., Troilo, M., Sauer, U., and Engel, P. (2017). Disentangling metabolic functions of bacteria in the honey bee gut. PLoS Biol. 15:e2003467. doi: 10.1371/journal.pbio.2003467
Khot, L. R., Sankaran, S., Maja, J. M., Ehsani, R., and Schuster, E. W. (2012). Applications of nanomaterials in agricultural production and crop protection: a review. Crop Protect. 35, 64–70. doi: 10.1016/j.cropro.2012.01.007
Kikuchi, Y., Hayatsu, M., Hosokawa, T., Nagayama, A., Tago, K., and Fukatsu, T. (2012). Symbiont-mediated insecticide resistance. Proc. Natl. Acad. Sci. U. S. A. 109, 8618–8622. doi: 10.1073/pnas.1200231109
Killiny, N., Martinez, R. H., Dumenyo, C. K., Cooksey, D. A., and Almeida, R. P. P. (2013). The exopolysaccharide of Xylella fastidiosa is essential for biofilm formation, plant virulence, and vector transmission. Mol. Plant Microbe Interact. 26, 10444–53. doi: 10.1094/MPMI-09-12-0211-R
Kim, J. S., and Je, Y. H. (2012). Milling effect on the control efficacy of spray-dried Bacillus thuringiensis technical powder against diamondback moths. Pest Manag. Sci. 68, 321–323. doi: 10.1002/ps.2330
Kim, W., Koo, H., Richman, A. M., Seeley, D., Vizioli, J., Klocko, A. D., et al. (2004). Ectopic expression of a cecropin transgene in the human malaria vector mosquito anopheles gambiae (Diptera: Culicidae): effects on susceptibility to plasmodium. J. Med. Entomol. 41, 447–455. doi: 10.1603/0022-2585-41.3.447
Kokoza, V., Ahmed, A., Woon Shin, S., Okafor, N., Zou, Z., and Raikhel, A. S. (2010). Blocking of Plasmodium transmission by cooperative action of cecropin A and defensin A in transgenic Aedes aegypti mosquitoes. Proc. Natl. Acad. Sci. U. S. A. 107, 8111–8116. doi: 10.1073/pnas.1003056107
Kroiss, J., Kaltenpoth, M., Schneider, B., Schwinger, M. G., Hertweck, C., Maddula, R. K., et al. (2010). Symbiotic streptomycetes provide antibiotic combination prophylaxis for wasp offspring. Nat. Chem. Biol. 6, 261–263. doi: 10.1038/nchembio.331
Krzewinski, J. W., Nguyen, C. D., Foster, J. M., and Burns, J. L. (2001). Use of random amplified polymorphic DNA PCR to examine epidemiology of Stenotrophomonas maltophilia and Achromobacter (alcaligenes) xylosoxidans from patients with cystic fibrosis. J. Clin. Microbiol. 39, 3597–3602. doi: 10.1128/JCM.39.10.3597-3602.2001
Kulkarni, R. A., Prabhuraj, A., Ashoka, J., Hanchinal, S. G., and Hiregoudar, S. (2017). Generation and evaluation of nanoparticles of supernatant of photorhabdus luminescens (thomas and poinar) against mite and aphid pests of cotton for enhanced. Curr. Sci. 112:2312. doi: 10.18520/cs/v112/i11/2312-2316
Kumar, S., Nehra, M., Dilbaghi, N., Marrazza, G., Hassan, A. A., and Kim, K. H. (2019). Nano-based smart pesticide formulations: emerging opportunities for agriculture. J. Controlled Release. 294, 131–153. doi: 10.1016/j.jconrel.2018.12.012
Lacey, L. A., Frutos, R., Kaya, H. K., and Vail, P. (2015) Insect pathogens as biological control agents: back to the future. J. Invertebr. Pathol. 132, 1–41. doi: 10.1006/bcon.2001.0938
Lacey, L. A., and Georgis, R. (2012). Entomopathogenic nematodes for control of insect pests above and below ground with comments on commercial production. J. Nematol. 44, 218–225.
Lam, K., Tsang, M., Labrie, A., Gries, R., and Gries, G. (2010). Semiochemical-mediated oviposition avoidance by female house flies, musca domestica, on animal feces colonized with harmful fungi. J. Chem. Ecol. 36, 141–147. doi: 10.1007/s10886-010-9741-2
Lasa, R., Tadeo, E., Toledo-Hérnandez, R. A., Carmona, L., Lima, I., and Williams, T. (2017). Improved capture of drosophila suzukii by a trap baited with two attractants in the same device. PLoS ONE. 12:e0188350. doi: 10.1371/journal.pone.0188350
Lauzon, C. (2003). “Symbiotic relationships of tephritids,” Insect Symbiosis, Vol. 2, eds. K. Bourtzis and T. Miller (Boca Raton: CRC) 115–129. doi: 10.1201/9780203009918.ch8
Lee, C.-J., DeMilo, A. B., Moreno, D. S., and Martinez, A. J (1995). Analysis of the volatile components of a bacterial fermentation attractive to the mexican fruit fly (Anastrepha ludens). J. Agric. Food Chem. 43, 1348–1351. doi: 10.1021/jf00053a041
Lees, R. S., Gilles, J. R., Hendrichs, J., Vreysen, M. J., and Bourtzis, K. (2015). Back to the future: the sterile insect technique against mosquito disease vectors. Curr. Opin. Insect Sci. 10, 156–162. doi: 10.1016/j.cois.2015.05.011
Lemfack, M. C., Gohlke, B.-O., Toguem, S. M. T., Preissner, S., Piechulla, B., and Preissner, R. (2017). mVOC 2.0: a database of microbial volatiles. Nucleic Acids Res. 46, D1261–D1265. doi: 10.1093/nar/gkx1016
Leonard, S. P., Powell, J. E., Perutka, J., Geng, P., Heckmann, L. C., Horak, R. D., et al. (2020). Engineered symbionts activate honey bee immunity and limit pathogens. Science 367, 573–576. doi: 10.1126/science.aax9039
LePage, D. P., Metcalf, J. A., Bordenstein, S. R., On, J., Perlmutter, J. I., Shropshire, J. D., et al. (2017). Prophage WO genes recapitulate and enhance wolbachia-induced cytoplasmic incompatibility. Nature 543, 243–247. doi: 10.1038/nature21391
Leroy, P. D., Sabri, A., Verheggen, F. J., Francis, F., Thonart, P., and Haubruge, E. (2011). The semiochemically mediated interactions between bacteria and insects. Chemoecology 21, 113–122. doi: 10.1007/s00049-011-0074-6
Li, J., Pang, Z., Trivedi, P., Zhou, X., Ying, X., Jia, H., et al. (2017). ‘Candidatus liberibacter asiaticus’ encodes a functional salicylic acid (SA) hydroxylase that degrades SA to suppress plant defenses. Mol. Plant Microbe Interact. 30, 620–630. doi: 10.1094/MPMI-12-16-0257-R
Li, L., Chen, J., Yang, Z., Li, D., Wu, W., Rao, W., et al. (2015). The effect of nano-Mg (OH)2 on insecticidal activity and UV resistance of bacillus thuringiensis protein. J. Agric. Biotechnol. 23, 1452–1457.
Libbrecht, R., Gwynn, D. M., and Fellowes, M. D. E. (2007). Aphidius ervi preferentially attacks the green morph of the pea aphid, acyrthosiphon pisum. J. Insect Behav. 20, 25–32. doi: 10.1007/s10905-006-9055-y
Liu, L. J., Martinez-Sañudo, I., Mazzon, L., Prabhakar, C. S., Girolami, V., Deng, Y. L., et al. (2016). Bacterial communities associated with invasive populations of Bactrocera dorsalis (Diptera: Tephritidae) in China. Bull. Entomol. Res. 106, 718–728. doi: 10.1017/S0007485316000390
Luan, J. B., Chen, W., Hasegawa, D. K., Simmons, A. M., Wintermantel, W. M., Ling, K. S., et al. (2015). Metabolic coevolution in the bacterial symbiosis of whiteflies and related plant sap-feeding insects. Genome Biol. Evol. 7, 2635–2647. doi: 10.1093/gbe/evv170
Malacrinò, A., Campolo, O., Medina, R. F., and Palmeri, V. (2018). Instar- and host-associated differentiation of bacterial communities in the mediterranean fruit fly Ceratitis capitata. PLoS ONE. 13:e0194131. doi: 10.1371/journal.pone.0194131
Mason, C. J., Couture, J. J., and Raffa, K. F. (2014). Plant-associated bacteria degrade defense chemicals and reduce their adverse effects on an insect defoliator. Oecologia 175, 901–910. doi: 10.1007/s00442-014-2950-6
McDougall, P. (2016). The Cost of New Agrochemical Product Discovery, Development and Registration in (1995). (2000). 2005–8 and 2010–2014. R&D expenditure in 2014 and expectations for (2019). A Consultancy Study for CropLife International, CropLife America and the European Crop Protection Association. Consultancy Study for CropLife International, CropLife America and the European Crop Protection Association.
McFall-Ngai, M., Hadfield, M. G., Bosch, T. C., Carey, H. V., Domazet-Lošo, T., Douglas, A. E., et al. (2013). Animals in a bacterial world, a new imperative for the life sciences. Proc. Natl. Acad. Sci. U. S. A. 110, 3229–3236. doi: 10.1073/pnas.1218525110
Medema, M. H., Blin, K., Cimermancic, P., de Jager, V., Zakrzewski, P., Fischbach, M., et al. (2011). antiSMASH: rapid identification, annotation and analysis of secondary metabolite biosynthesis gene clusters in bacterial and fungal genome sequences. Nucleic Acids Res. 39, W339–W346. doi: 10.1093/nar/gkr466
Mertz, F. P., and Yao, R. C. (1990). Saccharopolyspora spinosa sp. nov. isolated from soil collected in a sugar mill rum still. Int. J. Syst. Evol. Microbiol. 40, 34–39. doi: 10.1099/00207713-40-1-34
Minard, G., Mavingui, P., and Moro, C. V. (2013). Diversity and function of bacterial microbiota in the mosquito holobiont. Parasit. Vectors. 6:146. doi: 10.1186/1756-3305-6-146
Mitsuhara, I., Matsufuru, H., Ohshima, M., Kaku, H., Nakajima, Y., Murai, N., et al. (2000). Induced expression of sarcotoxin IA enhanced host resistance against both bacterial and fungal pathogens in transgenic tobacco. MPMI. 13, 860–868. doi: 10.1094/MPMI.2000.13.8.860
Moran, N. A., Russell, J. A., Koga, R., and Fukatsu, T. (2005). Evolutionary relationships of three new species of enterobacteriaceae living as symbionts of aphids and other insects. Appl. Environ. Microbiol. 71, 3302–3310. doi: 10.1128/AEM.71.6.3302-3310.2005
Moran, N. A., and Yun, Y. (2015). Experimental replacement of an obligate insect symbiont. Proc. Natl. Acad. Sci. U. S. A. 112, 2093–2096. doi: 10.1073/pnas.1420037112
Moreira, L. A., Iturbe-Ormaetxe, I., Jeffery, J. A., Lu, G., Pyke, A. T., Hedges, L. M., et al. (2009). A wolbachia symbiont in Aedes aegypti limits infection with dengue, chikungunya, and plasmodium. Cell 139, 1268–1278. doi: 10.1016/j.cell.2009.11.042
Morimoto, J., Simpson, S. J., and Ponton, F. (2017). Direct and trans-generational effects of male and female gut microbiota in drosophila melanogaster. Biol. Lett. 13:20160966. doi: 10.1098/rsbl.2016.0966
Morrow, J. L., Frommer, M., Shearman, D. C., and Riegler, M. (2015). The microbiome of field-caught and laboratory-adapted Australian tephritid fruit fly species with different host plant use and specialisation. Microb. Ecol. 70, 498–508. doi: 10.1007/s00248-015-0571-1
Mouton, L., Henri, H., Charif, D., Boulétreau, M., and Vavre, F. (2007). Interaction between host genotype and environmental conditions affects bacterial density in wolbachia symbiosis. Biol. Lett. 3, 210–213. doi: 10.1098/rsbl.2006.0590
Munhenga, G., Brooke, B. D., Gilles, J. R., Slabbert, K., Kemp, A., Dandalo, L. C., et al. (2016). Mating competitiveness of sterile genetic sexing strain males (GAMA) under laboratory and semi-field conditions: steps towards the use of the sterile insect technique to control the major malaria vector anopheles arabiensis in South Africa. Parasit Vectors. 9:122. doi: 10.1186/s13071-016-1385-9
Murthy, K. S., Vineela, V., and Vimala Devi, P. S. (2014). Generation of nanoparticles from technical powder of the insecticidal bacterium Bacillus thuringiensis var. kurstaki for improving efficacy. Int. J. Biomed. Nanosci. Nanotechnol. 3, 236–250. doi: 10.1504/IJBNN.2014.065470
Narasimhan, S., and Fikrig, E. (2015). Tick microbiome: the force within. Trends Parasitol. 31, 315–323. doi: 10.1016/j.pt.2015.03.010
Nazni, W. A., Hoffmann, A. A., NoorAfizah, A., Cheong, Y. L., Mancini, M. V., Golding, N., et al. (2019). Establishment of wolbachia strain wAlbB in malaysian populations of Aedes aegypti for dengue control. Curr. Biol. 29, 4241–4248.e5. doi: 10.1016/j.cub.2019.11.007
Nikolouli, K., Colinet, H., Renault, D., Enriquez, T., Mouton, L., Gibert, P., et al. (2018). Sterile insect technique and wolbachia symbiosis as potential tools for the control of the invasive species Drosophila suzukii. J. Pest. Sci. 91, 489–503. doi: 10.1007/s10340-017-0944-y
Niyazi, N., Lauzon, C. R., and Shelly, T. E. (2004). Effect of probiotic adult diets on fitness components of sterile male mediterranean fruit flies (Diptera: Tephritidae) under laboratory and field cage conditions. J. Econ. Entomol. 97, 1570–1580. doi: 10.1603/0022-0493-97.5.1570
Novakova, E., Woodhams, D. C., Rodríguez-Ruano, S. M., Brucker, R. M., Leff, J. W., Maharaj, A., et al. (2017). Mosquito microbiome dynamics, a background for prevalence and seasonality of west nile virus. Front. Microbiol. 8:526. doi: 10.3389/fmicb.2017.00526
O'Connor, L., Plichart, C., Sang, A. C., Brelsfoard, C. L., Bossin, H. C., et al. (2012). Open release of male mosquitoes infected with a wolbachia biopesticide: field performance and infection containment. PLoS Negl. Trop. Dis. 6:e1797. doi: 10.1371/journal.pntd.0001797
Oliver, K. M., Moran, N. A., and Hunter, M. S. (2005). Variation in resistance to parasitism in aphids is due to symbionts not host genotype. Proc. Natl. Acad. Sci. U. S. A. 102, 12795–12800. doi: 10.1073/pnas.0506131102
Oliver, K. M., Russell, J. A., Moran, N. A., and Hunter, M. S. (2003). Facultative bacterial symbionts in aphids confer resistance to parasitic wasps. Proc. Natl. Acad. Sci. U. S. A. 100, 1803–1807. doi: 10.1073/pnas.0335320100
Ometto, L., Cestaro, A., Ramasamy, S., Grassi, A., Revadi, S., Siozios, S., et al. (2013). Linking genomics and ecology to investigate the complex evolution of an invasive drosophila pest. Genome Biol. Evol. 5, 745–757. doi: 10.1093/gbe/evt034
Paramasiva, I., Sharma, H. C., and Krishnayya, P. V. (2014). Antibiotics influence the toxicity of the delta endotoxins of Bacillus thuringiensis towards the cotton bollworm, helicoverpa armigera. BMC Microbiol. 14:200. doi: 10.1186/1471-2180-14-200
Pereira, R., Yuval, B., Liedo, P., Teal, P. E. A., Shelly, T., Mcinnis, D. O., et al. (2013). Improving sterile male performance in support of programmes integrating the sterile insect technique against fruit flies: improving sterile male performance in fruit fly SIT. J. Appl. Entomol. 137, 178–190. doi: 10.1111/j.1439-0418.2011.01664.x
Peschen, D., Li, H.-P., Fischer, R., Kreuzaler, F., and Liao, Y.-C. (2004). Fusion proteins comprising a fusarium -specific antibody linked to antifungal peptides protect plants against a fungal pathogen. Nat. Biotechnol. 22, 732–738. doi: 10.1038/nbt970
Petri, L. (1909). Ricerche sopra i batteri intestinali della mosca olearia. Tipografia nazionale di G. Bertero e c.
Petri, L. (1910). Untersuchung uber die darmbakterien der olivenfliege. zentralblatt bakteriol. parasitenkd. Infekt. Hyg. 26, 357–367.
Podgwaite, J. D., Rush, P., Hall, D., and Gerald, S. W. (1984). Efficacy of the Neodiprion sertifer (Hymenoptera: Diprionidae) nucleopolyhedrosis virus (Baculovirus) product, Neochek-S. J. Econ. Entomol. 77, 525–528. doi: 10.1093/jee/77.2.525
Qiao, H., Keesey, I. W., Hansson, B. S., and Knaden, M. (2019). Gut microbiota affects development and olfactory behavior in drosophila melanogaster. J. Exp. Biol. 222:jeb192500. doi: 10.1242/jeb.192500
Racke, K. D. (2006). “A reduced risk insecticide for organic agriculture: Spinosad case study," in Crop Protection Products for Organic Agriculture: Environmental, Health and Efficacy Assessment, ACS Symposium Series, Vol. 947, eds A. S. Felsot and K. D. Racke (Washington, DC: American Chemical Society), 92–108. doi: 10.1021/bk-2007-0947.ch007
Ramalho, M., Bueno, O., and Saux Moreau, C. (2017). Species-specific signatures of the microbiome from camponotus and colobopsis ants across developmental stages. PLoS ONE. 12:e0187461. doi: 10.1371/journal.pone.0187461
Rani, A., Sharma, A., Rajagopal, R., Adak, T., and Bhatnagar, R. K. (2009). Bacterial diversity analysis of larvae and adult midgut microflora using culture-dependent and culture-independent methods in lab-reared and field-collected anopheles stephensi-an Asian malarial vector. BMC Microbiol. 9:96. doi: 10.1186/1471-2180-9-96
Rappé, M. S., and Giovannoni, S. J. (2003). The uncultured microbial majority. Annu. Rev. Microbiol. 57, 369–394. doi: 10.1146/annurev.micro.57.030502.090759
Ras, E., Beukeboom, L. W., Cáceres, C., and Bourtzis, K. (2017). Review of the role of gut microbiota in mass rearing of the olive fruit fly, bactrocera oleae, and its parasitoids. Entomol. Exp. Appl. 164, 237–256. doi: 10.1111/eea.12609
Raychoudhury, R., Sen, R., Cai, Y., Sun, Y., Lietze, V.-U., Boucias, D., et al. (2013). Comparative metatranscriptomic signatures of wood and paper feeding in the gut of the termite reticulitermes flavipes (Isoptera: Rhinotermitidae). Insect Mol. Biol. 22, 155–171. doi: 10.1111/imb.12011
Raza, M. F., Yao, Z., Bai, S., Cai, Z., and Zhang, H. (2020). Tephritidae fruit fly gut microbiome diversity, function and potential for applications. Bull. Entomol. Res. 110, 423–437. doi: 10.1017/S0007485319000853
Ridley, E. V., Wong, A. C.-N., Westmiller, S., and Douglas, A. E. (2012). Impact of the resident microbiota on the nutritional phenotype of Drosophila melanogaster. PLoS ONE. 7:e36765. doi: 10.1371/journal.pone.0036765
Ridley, E. V., Wong, A. C. N., and Douglas, A. E. (2013). Microbe-dependent and nonspecific effects of procedures to eliminate the resident microbiota from Drosophila melanogaster. Appl. Environ. Microbiol. 79, 3209–3214. doi: 10.1128/AEM.00206-13
Rinke, R., Costa, A. S., Fonseca, F. P., Almeida, L. C., Delalibera Júnior, I., and Henrique-Silva, F. (2011). Microbial diversity in the larval gut of field and laboratory populations of the sugarcane weevil sphenophorus levis (Coleoptera, Curculionidae). Genet. Mol. Res. 10, 2679–91. doi: 10.4238/2011.November.1.1
Rippere, K. E., Tran, M. T., Yousten, A. A., Hilu, K. H., and Klein, M. G. (1998). Bacillus popilliae and Bacillus lentimorbus, bacteria causing milky disease in Japanese beetles and related scarab larvae. Int. J. Syst. Bacteriol. 48, 395–402. doi: 10.1099/00207713-48-2-395
Rodriguez, M. D., Zamudio, F., Torres, J. A., Gonzalezceron, L., Possani, L., Rodriguez, M. H., et al. (1995). Effect of a cecropin-like synthetic peptide (shiva-3) on the sporogonic development of Plasmodium berghei. Exp. Parasitol. 80, 596–604. doi: 10.1006/expr.1995.1075
Ryan, P. A., Turley, A. P., Wilson, G., Hurst, T. P., Retzki, K., Brown-Kenyon, J., et al. (2020). Establishment of wMel wolbachia in aedes aegypti mosquitoes and reduction of local dengue transmission in cairns and surrounding locations in northern Queensland, Australia. Gates Open Res. 3:1547. doi: 10.12688/gatesopenres.13061.2
Sabbour, M. M., and Singer, S. M. (2016). Observations of the effect of two isolated nano Bacillus thuringiensis on Tuta absoluta infestation under laboratory and field condition. Res. J. Pharm. Biol. Chem. Sci. 7, 1891–1897.
Sacchetti, P., Ghiardi, B., Granchietti, A., Stefanini, F. M., and Belcari, A. (2014). Development of probiotic diets for the olive fly: evaluation of their effects on fly longevity and fecundity: effects of probiotic diets on the olive fly. Ann. Appl. Biol. 164, 138–150. doi: 10.1111/aab.12088
Sacchetti, P., Pastorelli, R., Bigiotti, G., Guidi, R., Ruschioni, S., Viti, C., et al. (2019). Olive fruit fly rearing procedures affect the vertical transmission of the bacterial symbiont candidatus Erwinia dacicola. BMC Biotechnol. 19:91. doi: 10.1186/s12896-019-0582-y
Saiman, L., Chen, Y., Tabibi, S., Gabriel, P. S., Zhou, J., Liu, Z., et al. (2001). Identification and antimicrobial susceptibility of Alcaligenes xylosoxidans isolated from patients with cystic fibrosis. J. Clin. Microbiol. 39, 3942–3945. doi: 10.1128/JCM.39.11.3942-3945.2001
Sanchis, V. (2011). From microbial sprays to insect-resistant transgenic plants: history of the biospesticide Bacillus thuringiensis. A review. Agron. Sustain. Dev. 31, 217–231. doi: 10.1051/agro/2010027
Saridaki, A., and Bourtzis, K. (2010). Wolbachia: more than just a bug in insects genitals. Curr. Opin. Microbiol. 13, 67–72. doi: 10.1016/j.mib.2009.11.005
Sauer, A. J., Schulze-Bopp, S., Fritsch, E., Undorf-Spahn, K., and Jehle, J. A. (2017). A third type of resistance to Cydia pomonella granulovirus in codling moths shows a mixed Z-linked and autosomal inheritance pattern. Appl. Environ. Microbiol. 83, e01036–e01017. doi: 10.1128/AEM.01036-17
Schmid, M., Sieber, R., Zimmermann, Y.-S., and Vorburger, C. (2012). Development, specificity and sublethal effects of symbiont-conferred resistance to parasitoids in aphids. Funct. Ecol. 26, 207–215. doi: 10.1111/j.1365-2435.2011.01904.x
Schnepf, E., Crickmore, N., Van Rie, J., Lereclus, D., Baum, J., Feitelson, J., et al. (1998). Bacillus thuringiensis and its pesticidal crystal proteins. Microbiol. Mol. Biol. Rev. 62, 775–806. doi: 10.1128/MMBR.62.3.775-806.1998
Schulz-Bohm, K., Zweers, H., de Boer, W., and Garbeva, P. (2015). A fragrant neighborhood: volatile mediated bacterial interactions in soil. Front. Microbiol. 6:1212. doi: 10.3389/fmicb.2015.01212
Segoli, M., Hoffmann, A. A., Lloyd, J., Omodei, G. J., and Ritchie, S. A. (2014). The effect of virus-blocking wolbachia on male competitiveness of the dengue vector mosquito, Aedes aegypti. PLoS Negl. Trop. Dis. 8:e3294. doi: 10.1371/journal.pntd.0003294
Shao, Y., Chen, B., Sun, C., Ishida, K., Hertweck, C., and Boland, W. (2017). Symbiont-derived antimicrobials contribute to the control of the lepidopteran gut microbiota. Cell. Chem. Biol. 24, 66–75. doi: 10.1016/j.chembiol.2016.11.015
Sharifi, R., Lee, S.-M., and Ryu, C.-M. (2018). Microbe-induced plant volatiles. N. Phytol. 220, 684–691. doi: 10.1111/nph.14955
Sharma, A., Sharma, R., Imamura, M., Yamakawa, M., and Machii, H. (2000). Transgenic expression of cecropin B, an antibacterial peptide from Bombyx mori, confers enhanced resistance to bacterial leaf blight in rice. FEBS Lett. 484, 7–11. doi: 10.1016/S0014-5793(00)02106-2
Shen, W., He, P., Xiao, C., and Chen, X. (2018). From antimicrobial peptides to antimicrobial poly(α-amino acid)s. Adv. Healthc. Mater. 7:e1800354. doi: 10.1002/adhm.201800354
Shin, B., Park, S. H., Kim, B.-Y., Jo, S.-I., Lee, S. K., Shin, J., et al. (2017). Deinococcucins A–D, aminoglycolipids from Deinococcus sp., a gut bacterium of the carpenter ant camponotus japonicus. J. Nat. Prod. 80, 2910–2916. doi: 10.1021/acs.jnatprod.7b00426
Shropshire, J. D., On, J., Layton, E. M., Zhou, H., and Bordenstein, S. R. (2018). One prophage WO gene rescues cytoplasmic incompatibility in Drosophila melanogaster. Proc. Natl. Acad. Sci. U. S. A. 115, 4987–4991. doi: 10.1073/pnas.1800650115
Shuttleworth, L. A., Khan, M. A. M., Osborne, T., Collins, D., Srivastava, M., and Reynolds, O. L. (2019). A walk on the wild side: gut bacteria fed to mass-reared larvae of Queensland fruit fly [Bactrocera tryoni (froggatt)] influence development. BMC Biotechnol. 19:95. doi: 10.1186/s12896-019-0579-6
Sinha, K.K., Choudhary, A. K., and Kumari, P. (2016). “Entomopathogenic fungi,” in Ecofriendly Pest Management for Food Security (Amsterdam: Elsevier Inc.), 475–505.
Siozios, S., Cestaro, A., Kaur, R., Pertot, I., Rota-Stabelli, O., and Anfora, G. (2013). Draft genome sequence of the wolbachia endosymbiont of Drosophila suzukii. Genome Announce. 1:e00032–13. doi: 10.1128/genomeA.00032-13
Skrobiszewski, A., Gladkowski, W., Mazur, M., Szczepanik, M., Maciejewska, G., and Wawrzeńczyk, C. (2018). Microbial hydrolysis of racemic β-Aryl-γ-ethylidene-γ-lactones and antifeedant activity of the products against alphitobius diaperinus panzer. Molecules 23:1516. doi: 10.3390/molecules23071516
Solter, L. F., Becnel, J. J., and Oi, D. H. (2012). “Chapter 7: microsporidian entomopathogens,” in Insect Pathology, 2nd edn, eds F. E. Vega, and H. K. Kaya (San Diego, CA: Elsevier), 1–45.
Stathopoulou, P., Asimakis, E. D., Khan, M., Caceres, C., Bourtzis, K., and Tsiamis, E. (2019). Irradiation effect on the structure of bacterial communities associated with the oriental fruit fly, bactrocera dorsalis. Entomol. Exp. Appl. 167, 209–219. doi: 10.1111/eea.12770
Steiner, H., Hultmark, D., Engström, A., Bennich, H., and Boman, H. G. (1981). Sequence and specificity of two antibacterial proteins involved in insect immunity. Nature 292, 246–248. doi: 10.1038/292246a0
Stensmyr, M. C., Dweck, H. K. M., Farhan, A., Ibba, I., Strutz, A., Mukunda, L., et al. (2012). A conserved dedicated olfactory circuit for detecting harmful microbes in Drosophila. Cell 151, 1345–1357. doi: 10.1016/j.cell.2012.09.046
Stilwell, M. D., Cao, M., Goodrich-Blair, H., and Weibel, D. B. (2018). Studying the symbiotic bacterium Xenorhabdus nematophila in individual, living Steinernema carpocapsae nematodes using microfluidic systems. mSphere 3:e00530-17. doi: 10.1128/mSphere.00530-17
Stockton, D. G., Brown, R., and Loeb, G. M. (2019). Not berry hungry? Discovering the hidden food sources of a small fruit specialist, Drosophila suzukii. Ecol. Entomol. 44, 810–822. doi: 10.1111/een.12766
Stouthamer, R., Breeuwer, J. A., and Hurst, G. D. (1999). Wolbachia pipientis: microbial manipulator of arthropod reproduction. Annu. Rev. Microbiol. 53, 71–102. doi: 10.1146/annurev.micro.53.1.71
Su, Q., Oliver, K. M., Xie, W., Wu, Q., Wang, S., and Zhang, Y. (2015). The whitefly-associated facultative symbiont Hamiltonella defensa suppresses induced plant defences in tomato. Funct. Ecol. 29, 1007–1018. doi: 10.1111/1365-2435.12405
Suh, C. P., and Axtell, R. C. (1999). Lagenidium giganteum Zoospores: effects of concentration, movement, light, and temperature on infection of mosquito larvae. Biol. Control 15, 33–38.
Tantowijoyo, W., Andari, B., Arguni, E., Budiwati, N., Nurhayati, I., Fitriana, I., et al. (2020). Stable establishment of wMel wolbachia in Aedes aegypti populations in yogyakarta, Indonesia. PLoS Negl. Trop. Dis. 14:e0008157. doi: 10.1371/journal.pntd.0008157
Taracena, M. L., Oliveira, P. L., Almendares, O., Umaña, C., Lowenberger, C., Dotson, E. M., et al. (2015). Genetically modifying the insect gut microbiota to control chagas disease vectors through systemic RNAi. PLoS Negl. Trop Dis. 9:e0003358. doi: 10.1371/journal.pntd.0003358
Thao, M. L., and Baumann, P. (2004). Evolutionary relationships of primary prokaryotic endosymbionts of whiteflies and their hosts. Appl. Environ. Microbiol. 70, 3401–3406. doi: 10.1128/AEM.70.6.3401-3406.2004
Thao, M. L., Moran, N. A., Abbot, P., Brennan, E. B., Burckhardt, D. H., and Baumann, P. (2000). Cospeciation of psyllids and their primary prokaryotic endosymbionts. Appl. Environ. Microbiol. 66, 2898–2905. doi: 10.1128/AEM.66.7.2898-2905.2000
The Not-So-Hidden Dangers of Invasive Species. (2018). The Not-So-Hidden Dangers of Invasive Species. Ann. Entomol. Soc. Am. 111, 79–80. doi: 10.1093/aesa/sax069
Thomas, S., Verma, J., Woolfit, M., and O'Neill, S. L. (2018). Wolbachia-mediated virus blocking in mosquito cells is dependent on XRN1-mediated viral RNA degradation and influenced by viral replication rate. PLoS Pathog. 14:e1006879. doi: 10.1371/journal.ppat.1006879
Tsuchida, T., Koga, R., Fujiwara, A., and Fukatsu, T. (2014). Phenotypic effect of “Candidatus Rickettsiella viridis,” a facultative symbiont of the pea aphid (Acyrthosiphon pisum), and its interaction with a coexisting symbiont. Appl. Environ. Microbiol. 80, 525–533. doi: 10.1128/AEM.03049-13
Tsuchida, T., Koga, R., Horikawa, M., Tsunoda, T., Maoka, T., Matsumoto, S., et al. (2010). Symbiotic bacterium modifies aphid body color. Science 330, 1102–1104. doi: 10.1126/science.1195463
USEPAO (2015). What are Biopesticides? US EPA. Available online at: https://www.epa.gov/pesticides/biopesticides#what (accessed February, 2020).
Van den Hurk, A. F., Hall-Mendelin, S., Pyke, A., T. Frentiu, F. D., McElroy, K., Day, A., et al. (2012). Impact of wolbachia on infection with chikungunya and yellow fever viruses in the mosquito vector aedes aegypti. PLoS Negl. Trop. Dis. 6:e1892. doi: 10.1371/journal.pntd.0001892
Vassilev, N., Vassileva, M., Martos, V., del Moral, L. F. G., Kowalska, J., Tylkowski, B., et al. (2020). Formulation of microbial inoculants by encapsulation in natural polysaccharides: focus on beneficial properties of carrier additives and derivatives. Front. Plant Sci. 11:270. doi: 10.3389/fpls.2020.00270
Visweshwar, R., Sharma, H. C., Akbar, S. M. D., and Sreeramulu, K. (2015). Elimination of gut microbes with antibiotics confers resistance to bacillus thuringiensis toxin proteins in helicoverpa armigera (Hubner). Appl. Biochem. Biotechnol. 177, 1621–1637. doi: 10.1007/s12010-015-1841-6
Vorburger, C., Sandrock, C., Gouskov, A., Castañeda, L. E., and Ferrari, J. (2009). Genotypic variation and the role of defensive endosymbionts in an all-parthenogenetic host–parasitoid interaction. Evol. Int. J. Organ. Evol. 63, 1439–1450. doi: 10.1111/j.1558-5646.2009.00660.x
Vreysen, M. J., Saleh, K., Mramba, F., Parker, A., Feldmann, U., Dyck, V., et al. (2014). Sterile insects to enhance agricultural development: the case of sustainable tsetse eradication on Unguja Island, Zanzibar, using an area-wide integrated pest management approach. PLoS Negl. Trop. Dis. 8:e2857. doi: 10.1371/journal.pntd.0002857
Vreysen, M. J. B., Carpenter, J. E., and Marec, F. (2010). Improvement of the sterile insect technique for codling moth cydia pomonella (linnaeus)(lepidoptera tortricidae) to facilitate expansion of field application. J. Appl. Entomol. 134, 165–181. doi: 10.1111/j.1439-0418.2009.01430.x
Wallingford, A. K., Hesler, S. P., Cha, D. H., and Loeb, G. (2016). Behavioral response of spotted-wing drosophila, Drosophila suzukii matsumura, to aversive odors and a potential oviposition deterrent in the field. Pest Manag. Sci. 72, 701–706. doi: 10.1002/ps.4040
Waltmann, A., Willcox, A. C., Balasubramanian, S., Mayori, K. B., Guerrero, S. M., Sanchez, R. S. S., et al. (2019). Hindgut microbiota in laboratory-reared and wild triatoma infestans. PLoS Negl. Trop. Dis. 13:e0007383. doi: 10.1371/journal.pntd.0007383
Wang, H., Jin, L., and Zhang, H. (2011). Comparison of the diversity of the bacterial communities in the intestinal tract of adult Bactrocera dorsalis from three different populations. J. Appl. Microbiol. 110, 1390–1401. doi: 10.1111/j.1365-2672.2011.05001.x
Wang, S., Dos-Santos, A. L. A., Huang, W., Liu, K. C., Oshaghi, M. A., Wei, G., et al. (2017). Driving mosquito refractoriness to Plasmodium falciparum with engineered symbiotic bacteria. Science 357, 1399–1402. doi: 10.1126/science.aan5478
Wang, S., Ghosh, A. K., Bongio, N., Stebbings, K. A., Lampe, D. J., and Jacobs-Lorena, M. (2012). Fighting malaria with engineered symbiotic bacteria from vector mosquitoes. Proc. Natl. Acad. Sci. USA. 109, 12734–12739. doi: 10.1073/pnas.1204158109
Wang, Y., and Kasper, L. H. (2014). The role of microbiome in central nervous system disorders. Brain Behav. Immun. 38, 1–12. doi: 10.1016/j.bbi.2013.12.015
Weber, T., Blin, K., Duddela, S., Krug, D., Kim, H. U., Bruccoleri, R., et al. (2015). antiSMASH 3.0—a comprehensive resource for the genome mining of biosynthetic gene clusters. Nucleic Acids Res. 43, W237–W243. doi: 10.1093/nar/gkv437
Wechsler, S. (2018). USDA ERS - Recent Trends in GE Adoption. USDA. Available online at: https://www.ers.usda.gov/data-products/adoption-of-genetically-engineered-crops-in-the-us/recent-trends-in-ge-adoption.aspx (accessed July, 2020).
Werren, J. H. (1997). Biology of wolbachia. Annu. Rev. Entomol. 42, 587–609. doi: 10.1146/annurev.ento.42.1.587
Witzgall, P., Kirsch, P., and Cork, A. (2010). Sex pheromones and their impact on pest management. J. Chem. Ecol. 36, 80–100. doi: 10.1007/s10886-009-9737-y
Wong, A. C.-N., Chaston, J. M., and Douglas, A. E. (2013). The inconstant gut microbiota of drosophila species revealed by 16S rRNA gene analysis. ISME J. 7, 1922–1932. doi: 10.1038/ismej.2013.86
Wong, A. C.-N., Dobson, A. J., and Douglas, A. E. (2014). Gut microbiota dictates the metabolic response of drosophila to diet. J. Exp. Biol. 217, 1894–1901. doi: 10.1242/jeb.101725
Wong, A. C.-N., Wang, Q.-P., Morimoto, J., Senior, A. M., Lihoreau, M., Neely, G. G., et al. (2017). Gut microbiota modifies olfactory-guided microbial preferences and foraging decisions in Drosophila. Curr. Biol. 27, 2397–2404.e4. doi: 10.1016/j.cub.2017.07.022
Wong, C. N. A., Ng, P., and Douglas, A. E. (2011). Low-diversity bacterial community in the gut of the fruitfly Drosophila melanogaster. Environ. Microbiol. 13, 1889–1900. doi: 10.1111/j.1462-2920.2011.02511.x
Wright, D. E., Zydenbos, S. E, Wessman, P., O'Callaghan, M., Townsend, R. J., Jackson, T. A., et al. (2017). Surface coating aids survival of Serratia entomophila (Enterobacteriaceae) in granules for surface application. Biocontr. Sci. Technol. 27, 1383–1399. doi: 10.1080/09583157.2017.1402861
Wu, Q., Patočka, J., and Kuča, K. (2018). Insect antimicrobial peptides, a mini review. Toxins. 10:461. doi: 10.3390/toxins10110461
Xia, X., Sun, B., Gurr, G. M., Vasseur, L., Xue, M., and You, M (2018). Gut microbiota mediate insecticide resistance in the diamondback moth, Plutella xylostella (L.). Front. Microbiol. 9:25. doi: 10.3389/fmicb.2018.00025
Xia, X., Zheng, D., Zhong, H., Qin, B., Gurr, G. M., Vasseur, L., et al. (2013). DNA sequencing reveals the midgut microbiota of diamondback moth, Plutella xylostella (L.) and a possible relationship with insecticide resistance. PLoS ONE. 8:e68852. doi: 10.1371/journal.pone.0068852
Xiang, H., Wei, G.-F., Jia, S., Huang, J., Miao, X.-X., Zhou, Z., et al. (2006). Microbial communities in the larval midgut of laboratory and field populations of cotton bollworm (Helicoverpa armigera). Can. J. Microbiol. 52, 1085–1092. doi: 10.1139/w06-064
Xu, H., Zheng, X., Liu, S., Ye, G., and Lu, Z. (2009). The role of endosymbionts in insect host resistance against adverse factors. Chin. Bull. Entomol. 46, 350–354.
Yao, M., Zhang, H., Cai, P., Gu, X., Wang, D., and Ji, Q. (2017). Enhanced fitness of a bactrocera cucurbitae genetic sexing strain based on the addition of gut-isolated probiotics (enterobacter spec.) to the larval diet. Entomol. Exp. Appl. 162, 197–203. doi: 10.1111/eea.12529
Yi, H.-Y., Chowdhury, M., Huang, Y.-D., and Yu, X.-Q. (2014). Insect antimicrobial peptides and their applications. Appl. Microbiol. Biotechnol. 98, 5807–5822. doi: 10.1007/s00253-014-5792-6
Zabalou, S., Apostolaki, A., Livadaras, I., Franz, G., Robinson, A. S., Savakis, C., et al. (2009). Incompatible insect technique: incompatible males from a Ceratitis capitata genetic sexing strain. Entomol. Exp. Appl. 132, 232–240. doi: 10.1111/j.1570-7458.2009.00886.x
Zabalou, S., Apostolaki, A., Pattas, S., Veneti, Z., Paraskevopoulos, C., Livadaras, I., et al. (2008). Multiple rescue factors within a wolbachia strain. Genetics 178, 2145–2160. doi: 10.1534/genetics.107.086488
Zabalou, S., Riegler, M., Theodorakopoulou, M., Stauffer, C., Savakis, C., and Bourtzis, K. (2004). Wolbachia-induced cytoplasmic incompatibility as a means for insect pest population control. Proc. Natl. Acad. Sci. U. S. A. 101, 15042–15045. doi: 10.1073/pnas.0403853101
Zhang, D., Zheng, X., Xi, Z., Bourtzis, K., and Gilles, J. R. (2015). Combining the sterile insect technique with the incompatible insect technique: i-impact of wolbachia infection on the fitness of triple- and double-infected strains of Aedes albopictus. PLoS ONE. 10:e0121126. doi: 10.1371/journal.pone.0121126
Zhang, L., Zhang, X., Zhang, Y., Wu, S., Gelbič, I., Xu, L., et al. (2016). A new formulation of Bacillus thuringiensis: UV protection and sustained release mosquito larvae studies. Sci. Rep. 6:39425. doi: 10.1038/srep39425
Keywords: invasive species, agroecosystems, omics, pest management, microbiome
Citation: Qadri M, Short S, Gast K, Hernandez J and Wong AC-N (2020) Microbiome Innovation in Agriculture: Development of Microbial Based Tools for Insect Pest Management. Front. Sustain. Food Syst. 4:547751. doi: 10.3389/fsufs.2020.547751
Received: 31 March 2020; Accepted: 25 August 2020;
Published: 06 October 2020.
Edited by:
Johann G. Zaller, University of Natural Resources and Life Sciences Vienna, AustriaReviewed by:
Ram Prasad, Mahatma Gandhi Central University, Motihari, IndiaCopyright © 2020 Qadri, Short, Gast, Hernandez and Wong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Adam Chun-Nin Wong, YWRhbWNud29uZ0B1ZmwuZWR1
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.