
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Sustain. Food Syst., 08 May 2018
Sec. Agro-Food Safety
Volume 2 - 2018 | https://doi.org/10.3389/fsufs.2018.00014
Hepatitis E Virus (HEV) is endemic in areas with poor sanitation and has traditionally been classified as a water-borne virus. Until recently, cases of HEV in industrialized countries were associated with travel to those areas. In the last decade, locally acquired cases of HEV have increased in the European Union, leading to the investigation of potential foodborne transmission of the virus. In the mid-1990's HEV was found to be unique among other water- and food-borne viruses because of the observation of zoonotic transmission of the virus. HEV is endemic on domestic swine farms worldwide and can infect pigs of all ages. Consequently, pork liver and pork liver containing products have been identified as the source of many of the foodborne HEV outbreaks in Europe. Other pork products and game meats have also been implicated in HEV outbreaks. Finally, anecdotal evidence exists for HEV transmission via shellfish and produce. HEV disease presentation is typically a self-limiting acute hepatitis; however, chronic hepatitis and extrahepatic manifestations occur in high-risk populations. Detection and control of HEV remains challenging because an efficient cell culture system has yet to be developed. Thus, detection relies upon molecular and serological methods. No standardized method exists for the detection of HEV in foods and research on the stability of HEV in foods and the environment has been limited. This review summarizes the current knowledge available on foodborne HEV.
The main unifying trait of foodborne viruses is that they are non-enveloped viruses, lacking a lipid envelope. Non-enveloped viruses, in general, are resistant to environmental stress such as heat, extreme pH, desiccation, organic solvents, etc. Typical treatments used to inactivate vegetative foodborne bacterial pathogens or enveloped viruses (such as influenza virus) are not effective in inactivating non-enveloped viruses. This allows non-enveloped viruses to be maintained in foods and the environment for long periods. The stability of non-enveloped viruses also makes them more resistant to common sanitation methods and food processing technologies. Foodborne viruses, primarily noroviruses, are a major cause of foodborne disease in industrialized countries (De Aceituno et al., 2013). However, there are emerging foodborne viruses, such as hepatitis E virus (HEV) and sapoviruses, increasing in prevalence in the European Union (EU) and Asia (Ruggeri et al., 2013). Of the emerging foodborne viruses, HEV is unique because it has known animal reservoirs and is zoonotic (Meng et al., 1997). Norovirus and rotavirus have genotypes or serotypes that can infect animals, but these viruses are not known to be transmitted to humans (i.e., not zoonotic) (Bank-Wolf et al., 2010). A summary of food-borne viruses is found in Table 1.
In 1973, a large-scale epidemic of hepatitis was observed in Kashmir affecting over 200,000 individuals in an area with very poor sanitation (Khuroo, 1980; Khuroo et al., 2016). Epidemics in India continued to be observed over the next 14 years. Patient serum samples collected during this period were tested for antibodies against hepatitis A virus (HAV) and hepatitis B virus (HBV). The serological tests ruled out HAV and HBV as the cause of the epidemics, leading to the identification of a non-A, non-B hepatitis virus (Khuroo, 1980). An outbreak at a Russian military outpost a few years later led to symptoms in patients like those observed in the 1978 Kashmir epidemic. Ingestion of pooled patient fecal samples by human volunteers led to the development of the prior observed non-A, non-B hepatitis and shedding of virus particles in the feces (Balayan et al., 1983). Immune electron microscopy revealed that the virus isolated from the subject was antigenically distinct from both Hepatitis A and Hepatitis C (Balayan et al., 1983). Following this, the physiological traits of the virus were characterized using virus collected from challenge studies conducted in non-human primates (Bradley et al., 1991). In 1991, the full-length HEV genome was sequenced and an enzyme immunoassay was developed for clinical diagnosis (Tam et al., 1991).
HEV is a member of the Hepeviridae family in the genus Orthohepevirus (Meng, 2013; Cossaboom et al., 2016). The genus Orthohepevirus is divided into 4 species, Orthohepevirus A-D, of which Orthohepevirus A includes HEVs infecting humans (Meng, 2013; Cossaboom et al., 2016). Orthohepevirus A contains 8 genotypes, HEV1-8, which are determined by amino acid sequence comparisons of concatenated open reading frames 1 and 2 (ORF1 and ORF2) sequences (Smith et al., 2016; Sridhar et al., 2017). HEV1 and HEV2 are specific to humans and these viruses are often the cause of waterborne HEV outbreaks (Geng and Wang, 2016) (Table 2). HEV3 and HEV4 infect a wide variety of hosts including humans, pigs, wild boar, deer, primates, and rabbits (Cossaboom et al., 2011; Doceul et al., 2016; Smith et al., 2016) (Table 2). HEV3 and HEV4 are zoonotic and are the causative agents of foodborne HEV infections in industrialized countries (Hughes et al., 2010; Meng, 2010; Teo, 2010; Yugo et al., 2014).
An estimated 2.3 billion people, one third of the world's population, have been exposed to HEV (Teshale and Hu, 2011; Pérez-Gracia et al., 2013). HEV is traditionally endemic in developing countries where sanitation is poor, which leads to waterborne outbreaks caused by HEV1 and HEV2 strains (Geng and Wang, 2016). Foodborne HEV outbreaks are most often associated zoonotic HEV3 and HEV4 strains. The transmission mode of HEV is generally the fecal oral route. HEV infections have been associated with direct contact with reservoir animals and consumption of contaminated water and foods (Cossaboom et al., 2016). High-risk foods for HEV contamination include raw or undercooked meat of infected animals, filter feeding bivalve shellfish, and produce (Brassard et al., 2012; Cossaboom et al., 2016; Mansuy et al., 2016; Hazards et al., 2017). In 2004, cases of HEV acquired after blood transfusion were reported in Japan and India (Khuroo et al., 2004; Matsubayashi et al., 2004). Since then, transfusion-associated HEV has been reported worldwide and seems to be highly prevalent among donors (Dreier and Juhl, 2014; Al-Sadeq et al., 2017). Foodborne HEV can also indirectly affect patients receiving blood transfusions. In Japan, a case of transfusion-acquired HEV was linked to a donor who had become infected after consuming pig liver and intestines (Matsubayashi et al., 2008).
HEV infection is typically self-limiting with clinical presentation ranging from asymptomatic infection to acute liver failure (Debing et al., 2016). Pregnant women tend to have symptomatic infections and are at higher risk of acute liver failure and death than other infected individuals (Abravanel et al., 2013). Interestingly, the high morbidity and mortality amongst pregnant women has only been observed following HEV1 and HEV2 infections and not infection with zoonotic HEV strains (Abravanel et al., 2013). HEV was exclusively described as an acute infection until 2008 when significant numbers of chronic HEV infections were reported amongst organ transplant recipients in France (Kamar et al., 2008). Subsequently, chronic HEV infection has been reported in other groups of immunocompromised patients including those with human immunodeficiency virus (HIV) and those with hematological cancers (Péron et al., 2006; Colson et al., 2009; Kamar et al., 2012). To date, chronic HEV infections have only been associated with HEV3 and HEV4 infections (Xin and Xiao, 2016). There is also evidence of neurological manifestations of acute and chronic HEV infection. HEV RNA has been detected in the cerebral spinal fluid of patients with peripheral neuropathy, which resolved upon clearance of the virus (Kamar et al., 2011).
HEV has a single stranded positive sense RNA genome (ss+RNA) that is approximately 7.2 kb in length (Cao and Meng, 2012; Emerson and Purcell, 2013; Debing et al., 2016). The genome is divided into three ORFs, has a 5′ cap structure, and is polyadenylated at the 3′ end (Cao and Meng, 2012). An overview of the HEV genome structure in presented in Figure 1. ORF1 encodes the nonstructural proteins, ORF2 encodes the capsid protein, and ORF3 encodes a small multifunctional phosphoprotein (Cao and Meng, 2012) (Table 3). Proteins translated from ORF2 and ORF3 are encoded by a biscistronic subgenomic mRNA (Graff et al., 2006).
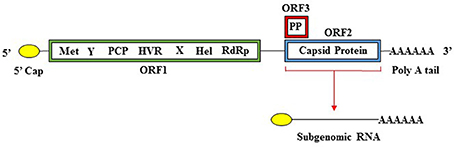
Figure 1. Organization of the HEV genome. The HEV genome has a 5′ cap structure, followed by three open reading frames (ORFs), and is polyadenylated at the 3′ end. ORF1 encodes a polyprotein containing the nonstructural domains (Met, methyltransferase; Y, Y domain; PCP, papain-like cysteine protease; HVR, hypervariable region; X, X domain; Hel, helicase; RdRp, RNA-dependent RNA polymerase. ORF2 encodes the capsid protein and ORF3 encodes a phosphoprotein (PP). ORF1 is translated from full length genomic RNA while ORF2 and ORF3 are translated from a 2kb bicistronic subgenomic RNA.
ORF1 spans the genome from the 5′ non-coding region (NCR) to position 5,082 and encodes a 1,693 amino acid polypeptide. This polypeptide includes domains, which function as the methyltransferase (Met), papain-like cysteine protease (PCP), helicase (Hel), RNA dependent RNA polymerase (RdRp), as well as other domains with unknown functions (Y domain, X domain, and hypervariable region) (Koonin, 1991; Koonin et al., 1992; Agrawal et al., 2001; Magden et al., 2001; Zhang et al., 2001; Emerson et al., 2004; Pudupakam et al., 2009, 2011; Cao and Meng, 2012). ORF2 encodes the 660-amino acid capsid protein, which has a molecular weight of 72 kDa (Cao and Meng, 2012). The HEV virion has T = 3 icosahedral symmetry and contains 180 copies of the capsid protein (Guu et al., 2009; Yamashita et al., 2009; Xing et al., 2010; Cao and Meng, 2012; Emerson and Purcell, 2013). The capsid protein makes up the viral particle structure, interacts with the host cell receptor, and is immunogenic (Li et al., 1997, 2005b; Guu et al., 2009; Yamashita et al., 2009; Cao and Meng, 2012). The HEV capsid structure is more homologous to that of small plant viruses than that of Norovirus or Hepatitis A (Guu et al., 2009).
ORF3 encodes a small, 114 amino acid, cytoskeleton associated phosphoprotein (Zafrullah et al., 1997; Cao and Meng, 2012). This phosophoprotein is theorized to have a regulatory role in assembly of HEV virions, pathogenesis, and release of membrane-associated viral particles (Tyagi et al., 2002; Surjit et al., 2006; Moin et al., 2007; Ding et al., 2017).
HEV replication follows the basic scheme of other ss+RNA viruses. The host translational machinery is exploited to translate the viral non-structural polyprotein encoded by ORF1. Evidence suggests that the ORF1 polyprotein does not undergo proteolytic processing to carry out viral replication functions (Suppiah et al., 2011; Perttilä et al., 2013). The RdRp, translated from the 3′ end of ORF1, synthesizes a complimentary negative-strand RNA (Debing et al., 2016). Genomic RNA is then synthesized from the negative-strand RNA and the two strands are unwound by the HEV helicase (Karpe and Lole, 2010a). In addition, a single bicistronic subgenomic mRNA is synthesized from the complementary negative-strand RNA and is used as a template to translate ORF2 and ORF3 (Zhang et al., 1999; Graff et al., 2006). The formation of the 5′ cap of the genomic RNA begins with the cleavage of 5′-gamma-phosphate by the helicase-associated 5′-triphosphatase (Magden et al., 2001; Karpe and Lole, 2010b). The P110 enzyme facilitates the methylation of guanosine triphosphate to form 7-methylguanosine, which is transferred to the 5′ end of the genome (Magden et al., 2001).
Due to the lack of an efficient cell culture system, little else is known about the replication process (von Nordheim et al., 2016). A RNA element located within ORF2 may also play a role in genomic RNA synthesis but further research is needed to determine the exact function (Emerson et al., 2013). This element likely does not play a role in HEV host adaptation since HEV-1 could replicate in porcine liver cells when only ORF1 was replaced with that of HEV-4 (Chatterjee et al., 2016). The exact location of replication has yet to be determined but evidence suggests that at least part of the process may take place in the endoplasmic reticulum-Golgi intermediate compartment (Perttilä et al., 2013).
Recently, quasi-enveloped HEV particles were observed in blood despite HEV being classified as a non-enveloped virus (Takahashi et al., 2008; Nagashima et al., 2017). The concept of quasi-enveloped viruses was first described for HAV (McKnight et al., 2017). HAV has evolved to be released from cells in exosomes following infection. The HAV particles are covered by a cellular lipid bilayer upon release from the cell and therefore are not recognized by the host immune system (Feng et al., 2013). HAV is shed in the feces in its non-enveloped form but the virus released from the cell appears to be exclusively quasi-enveloped (eHAV) (Feng et al., 2013). HAV found in the feces is derived from eHAV (Hirai-Yuki et al., 2016). Some eHAV is released across the apical membrane, which leads to the biliary tract where bile acids degrade the quasi-envelope. Despite being phylogenetically unrelated, HEV could plausibly utilize this process prior to being shed in the feces. Regardless of the mechanism, quasi-enveloped HEV likely plays a role HEV evasion of the host immune system.
Unlike common foodborne viruses such as HAV and norovirus, HEV3 and HEV4 strains are zoonotic. As previously mentioned, humans, pigs, wild boar, deer, primates, and rabbits are all infected by HEV3 and HEV4 strains (Cossaboom et al., 2011; Smith et al., 2016). The first zoonotic HEV strain belonged to genotype 3 and was a swine strain that was transmitted to humans (Meng et al., 1998; Schlauder et al., 1998). This seminal discovery lead to the subsequent finding that in addition to pigs, wild boar and rabbits are also reservoirs for zoonotic HEV strains (Doceul et al., 2016).
HEV has also been shown to be transmitted between different animal species. Pigs inoculated with a rabbit strain developed viremia and fecal shedding (Cossaboom et al., 2012). Virus shed from rabbit HEV-infected pigs produced viremia and fecal shedding in rabbits, which confirms the infectivity of rabbit HEV in pigs (Cossaboom et al., 2012). Rabbit HEV has also been shown to be infectious in cynomologus macaques, which suggests that this virus may also be infectious to humans (Liu et al., 2013).
HEV is considered endemic on conventional swine farms worldwide. A nationwide survey conducted in France in 2008–2009 found that 31% of pigs tested (1,069/6,565) were seropositive for HEV and 65% of pig farms (137/186) had at least one HEV seropositive pig (Rose et al., 2011). In 2011, 50.21% (714/1,422) of pigs tested in Northern Italy were HEV seropositive and 97.43% (38/39) of farms sampled had least one seropositive pig (Martinelli et al., 2011). Similar farm-level results were obtained in Spain, Norway, and New Zealand (Garkavenko et al., 2001; Seminati et al., 2008; Lange et al., 2016). Seroprevalence levels tend to vary between different types of farms and different age groups. Farms involved in breeding (farrow to finish or farrow to weaning) had higher HEV seroprevalence than fattening farms (Martinelli et al., 2011). Between individual pigs, seroprevalence was highest among sows and lowest among recently weaned pigs (weaners), which indicates that the circulation of HEV is influenced by maternal immunity (Martinelli et al., 2011). Because weaners no longer receive maternal antibodies, they are more vulnerable to HEV infection. Therefore, seroconversion is likely to occur shortly after weaning. While seroprevalence studies can provide information about HEV exposure and predict age of infection, this type of study does not provide information on whether the virus itself is present in the pig.
HEV RNA has been detected in pigs of all ages and the prevalence in pigs close to slaughter age is of particular interest. In a survey of pig farms in Portugal, 32% (16/50) of fattener pigs were shedding the virus in their feces (Berto et al., 2012c). A similar prevalence was found in a study of pigs between 2 and 4 months of age from 6 US states (Huang et al., 2002). A six-country (United Kingdom, Czech Republic, Italy, Portugal, Spain, and the Netherlands) study in Europe found HEV prevalence ranging from 8 to 73% among fattener pigs (Berto et al., 2012a). The wide variation could be attributed to the characteristics of the farms (number of animals, farrow to finish vs. fattening, etc.) and the predominating practices (organic vs. conventional, biosecurity measures, etc.) of the farms in each country (Salines et al., 2017). The sample size may be a major factor as well because the prevalence data for Spain and the Netherlands came from one farm in each country. Such a small sample size can skew the results, thus future studies should include a much larger number of farms. Extensive national surveys of near slaughter age pigs in the United States and Europe should be utilized to elucidate the true prevalence of HEV infection.
Pork liver and pork liver products are the most obvious source of foodborne HEV and have been extensively studied in Europe. In a survey of foods containing raw pork liver sold in France, 68 out of 394 (17.3%) samples tested positive for HEV with prevalence for various products ranging from 3% (dried salted liver) to 30% (figatelli) (Pavio et al., 2014). In a year-long study in the United Kingdom, HEV was detected in 6 of 63 pork sausages (10%) collected from 3 retail outlets with 5 of the 6 positive samples coming from a single batch (Berto et al., 2012b). HEV has also been detected in sausages sold in Spain (Di Bartolo et al., 2012) and Italy (Di Bartolo et al., 2015), in livers and sausages sold in Germany (Wenzel et al., 2011; Szabo et al., 2015), and livers sold in the Netherlands (Bouwknegt et al., 2007). Although fewer studies have been conducted in North America, HEV has been detected in pork liver and pork liver products sold in Canada and the United States (U.S.). Forty-seven percentage of pork pâtés (36/76) and 10.5% of pork livers (2/19) purchased from grocery stores in Ottawa between March 2014 and September 2015 tested positive for HEV (Mykytczuk et al., 2017). In 2007, HEV was detected in 11% (14/127) of pork livers purchased from grocery stores in Virginia and Iowa (Feagins et al., 2007).
Pork liver products are not as commonly consumed in the US as they are in Europe. Therefore, other pork products need to be considered as a source of foodborne HEV. Cossaboom et al. (2016) specifically targeted non-liver pork products and detected HEV in 25% (3/12) of pork chitterling packages purchased in southwestern Virginia grocery stores. Sausages, skeletal muscle, and nervous tissue were also tested but were negative for HEV. This survey, along with the previously mentioned pork liver survey conducted in Virginia and Iowa, had a relatively small sample size and covered a small region of the US. Larger and more expansive surveys are needed to truly understand the prevalence of HEV in pork products.
The number of HEV cases in the EU each year has been rising since 2005 with the largest increase occurring between 2011 and 2015 (Aspinall et al., 2017). 98.5% of cases with known travel history were locally-acquired. Clusters of cases linked to raw or undercooked pork have been reported in France. In 2013, seventeen people contracted HEV after consuming spit-roasted piglet stuffed with raw stuffing made from the liver: three developed hepatitis while the other 14 were asymptomatic (Guillois et al., 2016). Figatelli, a type of raw pork liver sausage, has been linked to two clusters of HEV cases in Marseille (Colson et al., 2012) (Colson et al., 2010) and two cases (1 symptomatic, 1 asymptomatic) in a small French resort town (Renou et al., 2014). In Spain, an outbreak within a family was traced to wild boar meat the family had consumed (Rivero-Juarez et al., 2017). Several outbreaks occurred in Germany but no food source of infection was identified (Hazards et al., 2017). HEV surveillance programs exist in 20 EU countries including France, Spain, and Germany (Aspinall et al., 2017).
According to a 2014 epidemiological survey, 6% of the US population is HEV seropositive (Ditah et al., 2014). Data on outbreaks and the number of cases occurring each year is not readily available due to the lack of a surveillance program. Cases of acute hepatitis not caused by Hepatitis A, B, or C viruses may not be investigated further for a viral causative agent. Although HEV is not considered a reportable disease in the US, cases should be documented so that the disease burden of HEV can be elucidated. HEV should be ruled out in cases of non-A, B, or C hepatitis and in cases of unexplained chronic hepatitis.
HEV 239 (Hecolin; Innovax; Xiamen, China), a recombinant vaccine produced using a genetically modified strain of Escherichia coli, has been available in China since 2012 and is approved for use in people over 16 years old (Park, 2012; Li et al., 2015). No vaccine is available elsewhere. The immunogenic constituent of Hecolin is derived from the ORF2 protein of an HEV1 strain (Li et al., 2005a; Wen et al., 2016). Vaccination with Hecolin confers protection against genotypes 1 and 4 for up to 4.5 years (Zhu et al., 2010; Zhang et al., 2015). However, a recent study showed that sera from people vaccinated with Hecolin produced a stronger antibody response to HEV1 and HEV2 than to HEV3 and HEV4 (Wen et al., 2016). Antibody titers induced by the HEV p179 vaccine, derived from the ORF2 protein of HEV4, were 2-fold higher for HEV3 and HEV4 compared to HEV1 and HEV2 (Wen et al., 2016). HEV p179 was shown to be safe and well tolerated in a phase 1 randomized open-label study (Cao et al., 2017) and could potentially be protective against HEV3. Future vaccine design could include ORF1 proteins as they have been shown to induce T-cell responses in patients with acute, resolved and chronic hepatitis E (Al-Ayoubi et al., 2018).
No vaccine is currently available for animal use. Theoretical models have been used to test the effects of HEV vaccination in pigs. In scenarios where the HEV is not eliminated from the pig population, reduced transmission rates resulted in an increase in the rates of infectious pigs at slaughter age while a shortened infectious period resulted in a decrease (Backer et al., 2012). More research is needed to determine whether vaccination would be an effective strategy to control HEV in the domestic pig population. Future work should take passive immunity, husbandry practices, and other pathogens into consideration (Salines et al., 2017).
Without an efficient cell culture system, study of inactivation methods has been severely limited. Using a swine bioassay, where HEV contaminated pork liver was injected into the ear veins of naïve pigs, Feagins et al. (2008) found that cooking pork liver to an internal temperature of 71°C (161°F) was sufficient to fully inactivate HEV. However, heating to an internal temperature of 56°C (132°F) for 1 h lead to subsequent infection in inoculated pigs. In another study, it was found that HEV RNA persisted in liver suspension stored for 50 days at 4°C and 70 days at 22°C and 37°C, indicating that the virus is highly stable in foods during long-term storage (Schielke et al., 2011). Heating the HEV contaminated liver suspension at 56°C for 30 min only resulted in an approximate 2-log reduction in viral genomic equivalents (Schielke et al., 2011).
Identifying an appropriate surrogate virus for HEV studies will also aide in advancing knowledge of HEV stability and inactivation in foods. Cutthroat trout virus (CTV) is also a member of the Hepeviridae family, has a similar genome organization to HEV, and is quasi-enveloped (von Nordheim et al., 2016). CTV replication in cell culture is robust and is a promising model for study of the molecular biology of HEV (von Nordheim et al., 2016). These features could also be useful for testing the effectiveness of various inactivation strategies.
Advancements are being achieved in adapting HEV to cell culture. Several available immortalized cells lines have been shown to support increases in HEV RNA following infection, however these increases have not been sustained in serial passages (Tanaka et al., 2007; Okamoto, 2013). Cell lines shown to support HEV genome replication included A549 (human lung cell cancer) and PLC/PRF/5 (human hepatocellular carcinoma) (Tanaka et al., 2007; Okamoto, 2013). The swine kidney epithelial cell line, LLC-PK1, was also shown to support the replication of a HEV3 strain (Shukla et al., 2011; Okamoto, 2013). A subsequent study using PLC/PRF/5 cells grown in 3-D culture for HEV infection showed increases in HEV RNA genome, release of viral particles, and subsequent infection in serial passages (Berto et al., 2013). Optimization of these in vitro HEV cultivation techniques will greatly advance our understanding of the replication and pathogenesis of the virus, as well as aid in the development of mitigation strategies for this virus.
Because HEV and other foodborne viruses are difficult to culture, molecular methods are used for detection. Even so, molecular detection in food can be challenging due to low levels of virus in contaminated sample and the presence of inhibitory compounds in the food matrix (De Aceituno et al., 2013). While other foodborne viruses likely are restricted to the food surface, HEV is also present inside the cells of meat thus adequate breakdown of the food matrix is critical for detection (Szabo et al., 2015). Common molecular methods used include RNA detection using PCR based methods and viral antigen detection using ELISA based methods.
Although no standard method currently exists for the detection of HEV RNA in foods, all protocols follow three general steps: (1) virus elution and concentration; (2) extraction of viral RNA; (3) RNA detection (De Aceituno et al., 2013; Martin-Latil et al., 2014; Hazards et al., 2017). A variety of methods have been described but few comparison studies have been done to optimize the elution-concentration and RNA extraction processes for HEV detection. Martin-Latil et al. (2014) found that homogenization in distilled water by stomacher yielded the highest HEV recovery rates from pork liver sausages compared to phosphate buffered saline (PBS) and Tris-HCl, glycine, beef extract (TGBE). Also, purification using the organic solvent choroform:butanol before or after concentration by polyethylene glycol (PEG) did not result in significantly higher recovery rates. When compared with homogenization in TRIzol® reagent, homogenization in distilled water and concentration with PEG resulted in a lower mean HEV recovery rate from pork liver sausages (Szabo et al., 2015). Ultrafiltration was shown to be more efficient in concentrating HEV from pork liver than PEG (Son et al., 2014). Commercial kits are widely used for RNA extraction but have occasionally been shown to be less efficient in extracting viral RNA than protocols developed by individual laboratories (Bouwknegt et al., 2007; Martínez-Martínez et al., 2011). The elution-concentration and extraction method chosen also influence the resulting prevalence data, thus it is important that a standard protocol for HEV elution-concentration and extraction RNA be developed to combat this emerging foodborne virus. Larger studies encompassing multiple laboratories should be conducted to optimize the conditions and develop a validated method for HEV detection in foods.
Reverse transcriptase polymerase chain reaction (RT-PCR) is commonly used to detect viral RNA in food. One complication is developing cross-reactive primer sets that can detect all HEV genotypes. Several groups have developed degenerate primers or genotype specific primers for use in HEV RT-PCR assays. These primer sets typically target the ORF2/3 overlap region as it is conserved between genotypes. However, no standardized reagents are yet available commercially for the detection of HEV RNA. The RT-PCR method itself has the disadvantages of being time consuming and providing only qualitative results. Real-time RT-PCR is faster, provides quantitative results, and has been proven to be more sensitive than RT-PCR for the detection of HEV specifically (Son et al., 2014). However, RT-PCR has not been eliminated completely. DNA fragments generated by real-time RT-PCR are not long enough for sequencing and typing, which are useful for determining genotype and determining relatedness among HEV strains (Hazards et al., 2017).
HEV is an emerging foodborne pathogen, which needs to be researched more extensively. The lack of an efficient cell culture system has hindered study of HEV molecular biology. The receptor used to enter cells has yet to be identified and may be a vital clue to the wide tissue tropism of HEV. Many of the prevalence studies in pigs and pork products have been small-scale, which can skew results. Large scale studies are needed to assess the true prevalence of HEV in pigs and pork products. Lack of standardized detection methods is also an obstacle for HEV research. Methods can vary among individual labs, which can result in wide variation in virus recovery rates. Developing a gold standard method will ensure accuracy and precision, which will enhance the quality of information used to determine prevalence in foodborne HEV worldwide. Overall, more research is needed to understand and combat this emerging foodborne pathogen.
LH was responsible for writing the text of the majority of the manuscript. ED revised the manuscript text, prepared tables and figures.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
This work was supported by the USDA National Institute of Food and Agriculture, Hatch/Multi State project S1056-1012080.
Abravanel, F., Lhomme, S., Dubois, M., Peron, J. M., Alric, L., Kamar, N., et al. (2013). Hepatitis E virus. Méd. Mal. Infect. 43, 263–270. doi: 10.1016/j.medmal.2013.03.005
Agrawal, S., Gupta, D., and Panda, S. K. (2001). The 3′ end of hepatitis E virus (HEV) genome binds specifically to the viral RNA-dependent RNA polymerase (RdRp). Virology 282, 87–101. doi: 10.1006/viro.2000.0819
Al-Ayoubi, J., Behrendt, P., Bremer, B., Suneetha, P. V., Gisa, A., Rinker, F., et al. (2018). Hepatitis E virus ORF 1 induces proliferative and functional T-cell responses in patients with ongoing and resolved hepatitis E. Liver Int. 38, 266–277. doi: 10.1111/liv.13521
Al-Sadeq, D. W., Majdalawieh, A. F., and Nasrallah, G. K. (2017). Seroprevalence and incidence of hepatitis E virus among blood donors: a review. Rev. Med. Virol. 27:e1937. doi: 10.1002/rmv.1937
Aspinall, E. J., Couturier, E., Faber, M., Said, B., Ijaz, S., Tavoschi, L., et al. (2017). Hepatitis E virus infection in Europe: surveillance and descriptive epidemiology of confirmed cases, 2005 to 2015. Eurosurveillance 22:30561. doi: 10.2807/1560-7917.ES.2017.22.26.30561
Backer, J. A., Berto, A., Mccreary, C., Martelli, F., and Van Der Poel, W. H. M. (2012). Transmission dynamics of hepatitis E virus in pigs: estimation from field data and effect of vaccination. Epidemics 4, 86–92. doi: 10.1016/j.epidem.2012.02.002
Balayan, M. S., Andjaparidze, A. G., Savinskaya, S. S., Ketiladze, E. S., Braginsky, D. M., Savinov, A. P., et al. (1983). Evidence for a virus in non-A, non-B hepatitis transmitted via the fecal-oral route. Intervirology 20, 23–31. doi: 10.1159/000149370
Bank-Wolf, B. R., König, M., and Thiel, H.-J. (2010). Zoonotic aspects of infections with noroviruses and sapoviruses. Vet. Microbiol. 140, 204–212. doi: 10.1016/j.vetmic.2009.08.021
Berto, A., Backer, J. A., Mesquita, J. R., Nascimento, M. S., Banks, M., Martelli, F., et al. (2012a). Prevalence and transmission of hepatitis E virus in domestic swine populations in different European countries. BMC Res. Notes 5:190. doi: 10.1186/1756-0500-5-190
Berto, A., Martelli, F., Grierson, S., and Banks, M. (2012b). Hepatitis E virus in pork food chain, United Kingdom, 2009–2010. Emerging Infect. Dis. 18, 1358–1360. doi: 10.3201/eid1808.111647
Berto, A., Mesquita, J. R., Hakze-Van Der Honing, R., Nascimento, M. S. J., and Van Der Poel, W. H. M. (2012c). Detection and characterization of hepatitis E virus in domestic pigs of different ages in Portugal. Zoonoses Public Health 59, 477–481. doi: 10.1111/j.1863-2378.2012.01488.x
Berto, A., Van der Poel, W. H., Hakze-van der Honing, R., Martelli, F., and La Ragione, R. M. (2013). Replication of hepatitis E virus in three-dimensional cell culture. J. Virol. Methods 187, 327–332. doi: 10.1016/j.jviromet.2012.10.017
Bouwknegt, M., Lodder-Verschoor, F., Poel, W. A., Rutjes, S., and De Roda Husman, A. M. (2007). Hepatitis E virus RNA in commercial porcine livers in the Netherlands. J. Food Prot. 70, 2889–2895. doi: 10.4315/0362-028X-70.12.2889
Bradley, D. W., Purdy, M. A., and Reyes, G. R. (1991). Hepatitis E virus genome: molecular features, expression of immunoreactive proteins and sequence divergence. J. Hepatol. 13, S152–S154. doi: 10.1016/0168-8278(91)90049-H
Brassard, J., Gagne, M. J., Genereux, M., and Cote, C. (2012). Detection of human food-borne and zoonotic viruses on irrigated, field-grown strawberries. Appl. Environ. Microbiol. 78, 3763–3766. doi: 10.1128/AEM.00251-12
Cao, D., and Meng, X.-J. (2012). Molecular biology and replication of hepatitis E virus. Emerg. Microbes Infect. 1:e17. doi: 10.1038/emi.2012.7
Cao, Y.-F., Tao, H., Hu, Y.-M., Shi, C.-B., Wu, X., Liang, Q., et al. (2017). A phase 1 randomized open-label clinical study to evaluate the safety and tolerability of a novel recombinant hepatitis E vaccine. Vaccine 35, 5073–5080. doi: 10.1016/j.vaccine.2017.05.072
Chatterjee, S. N., Devhare, P. B., Pingle, S. Y., Paingankar, M. S., Arankalle, V. A., and Lole, K. S. (2016). Hepatitis E virus (HEV)-1 harbouring HEV-4 non-structural protein (ORF1) replicates in transfected porcine kidney cells. J. General Virol. 97, 1829–1840. doi: 10.1099/jgv.0.000478
Colson, P., Romanet, P., Moal, V., Borentain, P., Purgus, R., Benezech, A., et al. (2012). Autochthonous infections with hepatitis E virus genotype 4, France. Emerg. Infect. Dis. J. 18, 1361–1364. doi: 10.3201/eid1808.111827
Colson, P., Borentain, P., Queyriaux, B., Kaba, M., Moal, V., Gallian, P., et al. (2010). Pig liver sausage as a source of hepatitis E virus transmission to humans. J. Infect. Dis. 202, 825–834. doi: 10.1086/655898
Colson, P., Kaba, M., Moreau, J., and Brouqui, P. (2009). Hepatitis E in an HIV-infected patient. J. Clin. Virol. 45, 269–271. doi: 10.1016/j.jcv.2009.06.002
Cossaboom, C. M., Córdoba, L., Dryman, B. A., and Meng, X.-J. (2011). Hepatitis E virus in Rabbits, Virginia, USA. Emerg. Infect. Dis. 17, 2047–2049. doi: 10.3201/eid1711.110428
Cossaboom, C. M., Córdoba, L., Sanford, B. J., Piñeyro, P., Kenney, S. P., Dryman, B. A., et al. (2012). Cross-species infection of pigs with a novel rabbit, but not rat, strain of hepatitis E virus isolated in the United States. J. Gen. Virol. 93, 1687–1695. doi: 10.1099/vir.0.041509-0
Cossaboom, C. M., Heffron, C. L., Cao, D., Yugo, D. M., Houk-Miles, A. E., Lindsay, D. S., et al. (2016). Risk factors and sources of foodborne hepatitis E virus infection in the United States. J. Med. Virol. 88, 1641–1645. doi: 10.1002/jmv.24497
De Aceituno, A. M. F., Rocks, J. J., Jaykus, L. A., and Leon, J. S. (2013). “Foodborne viruses,” in Guide to Foodborne Pathogens, 2nd Edn, eds R. G. Labbé and S. García. (Oxford: John Wiley and Sons, Ltd.), 352–376.
Debing, Y., Moradpour, D., Neyts, J., and Gouttenoire, J. (2016). Update on hepatitis E virology: implications for clinical practice. J. Hepatol. 65, 200–212. doi: 10.1016/j.jhep.2016.02.045
Di Bartolo, I., Diez-Valcarce, M., Vasickova, P., Kralik, P., Hernandez, M., Angeloni, G., et al. (2012). Hepatitis E virus in pork production chain in Czech Republic, Italy, and Spain, 2010. Emerg. Infect. Dis. J. 18, 1282–1289. doi: 10.3201/eid1808.111783
Di Bartolo, I., Angeloni, G., Ponterio, E., Ostanello, F., and Ruggeri, F. M. (2015). Detection of hepatitis E virus in pork liver sausages. Int. J. Food Microbiol. 193, 29–33. doi: 10.1016/j.ijfoodmicro.2014.10.005
Ding, Q., Heller, B., Capuccino, J. M. V., Song, B. K., Nimgaonkar, I., Hrebikova, G., et al. (2017). Hepatitis E virus ORF3 is a functional ion channel required for release of infectious particles. Proc. Natl. Acad. Sci. U.S.A. 114, 1147–1152. doi: 10.1073/pnas.1614955114
Ditah, I., Ditah, F., Devaki, P., Ditah, C., Kamath, P. S., and Charlton, M. (2014). Current epidemiology of hepatitis E virus infection in the United States: low seroprevalence in the national health and nutrition evaluation survey. Hepatology 60, 815–822. doi: 10.1002/hep.27219
Doceul, V., Bagdassarian, E., Demange, A., and Pavio, N. (2016). Zoonotic hepatitis E virus: classification, animal reservoirs and transmission routes. Viruses 8:270. doi: 10.3390/v8100270
Dreier, J., and Juhl, D. (2014). Autochthonous hepatitis E virus infections: a new transfusion-associated risk? Transfus. Med. Hemother. 41, 29–39. doi: 10.1159/000357098
Emerson, S. U., and Purcell, R. H. (2013). “Hepatitis E virus,” in Fields Virology: 6th Edn, eds D. M. Knipe and P. M. Howley (Philadelphia, PA: Wolters Kluwer Health/Lippencott Williams and Wilkins), 2242–2258.
Emerson, S. U., Nguyen, H. T., Torian, U., Mather, K., and Firth, A. E. (2013). An essential RNA element resides in a central region of hepatitis E virus ORF2. J. Gen. Virol. 94, 1468–1476. doi: 10.1099/vir.0.051870-0
Emerson, S. U., Nguyen, H., Graff, J., Stephany, D. A., Brockington, A., and Purcell, R. H. (2004). In vitro replication of hepatitis E virus (HEV) genomes and of an HEV replicon expressing green fluorescent protein. J. Virol. 78, 4838–4846. doi: 10.1128/JVI.78.9.4838-4846.2004
Feagins, A. R., Opriessnig, T., Guenette, D. K., Halbur, P. G., and Meng, X.-J. (2007). Detection and characterization of infectious Hepatitis E virus from commercial pig livers sold in local grocery stores in the USA. J. Gen. Virol. 88, 912–917. doi: 10.1099/vir.0.82613-0
Feagins, A. R., Opriessnig, T., Guenette, D. K., Halbur, P. G., and Meng, X. J. (2008). Inactivation of infectious hepatitis E virus present in commercial pig livers sold in local grocery stores in the United States. Int. J. Food Microbiol. 123, 32–37. doi: 10.1016/j.ijfoodmicro.2007.11.068
Feng, Z., Hensley, L., Mcknight, K. L., Hu, F., Madden, V., Ping, L., et al. (2013). A pathogenic picornavirus acquires an envelope by hijacking cellular membranes. Nature 496, 367–371. doi: 10.1038/nature12029
Garkavenko, O., Obriadina, A., Meng, J., Anderson, D. A., Benard, H. J., Schroeder, B. A., et al. (2001). Detection and characterisation of swine hepatitis E virus in New Zealand. J. Med. Virol. 65, 525–529. doi: 10.1002/jmv.2067
Geng, Y., and Wang, Y. (2016). “Epidemiology of Hepatitis E,” in Hepatitis E Virus, ed Y. Wang (Dordrecht: Springer Netherlands), 39–59.
Graff, J., Torian, U., Nguyen, H., and Emerson, S. U. (2006). A bicistronic subgenomic mRNA encodes both the ORF2 and ORF3 proteins of hepatitis E Virus. J. Virol. 80, 5919–5926. doi: 10.1128/JVI.00046-06
Guillois, Y., Abravanel, F., Miura, T., Pavio, N., Vaillant, V., Lhomme, S., et al. (2016). High proportion of asymptomatic infections in an outbreak of hepatitis E associated with a spit-roasted piglet, France, 2013. Clin. Infect. Dis. 62, 351–357. doi: 10.1093/cid/civ862
Guu, T. S. Y., Liu, Z., Ye, Q., Mata, D. A., Li, K., Yin, C., et al. (2009). Structure of the hepatitis E virus-like particle suggests mechanisms for virus assembly and receptor binding. Proc. Natl. Acad. Sci. U.S.A. 106, 12992–12997. doi: 10.1073/pnas.0904848106
Hazards, E. P. B., Ricci, A., Allende, A., Bolton, D., Chemaly, M., Davies, R., et al. (2017). Public health risks associated with hepatitis E virus (HEV) as a food-borne pathogen. EFSA J. 15:e04886-n/a. doi: 10.2903/j.efsa.2017.4886
Hirai-Yuki, A., Hensley, L., Whitmire, J. K., and Lemon, S. M. (2016). Biliary secretion of quasi-enveloped human hepatitis A virus. mBio 7:e01998–16. doi: 10.1128/mBio.01998-16
Huang, F. F., Haqshenas, G., Guenette, D. K., Halbur, P. G., Schommer, S. K., Pierson, F. W., et al. (2002). Detection by reverse transcription-PCR and genetic characterization of field isolates of swine hepatitis E virus from pigs in different geographic regions of the United States. J. Clin. Microbiol. 40, 1326–1332. doi: 10.1128/JCM.40.4.1326-1332.2002
Hughes, J. M., Wilson, M. E., Teshale, E. H., Hu, D. J., and Holmberg, S. D. (2010). The two faces of hepatitis E virus. Clin. Infect. Dis. 51, 328–334. doi: 10.1086/653943
Kamar, N., Bendall, R. P., Peron, J. M., Cintas, P., Prudhomme, L., Mansuy, J. M., et al. (2011). Hepatitis E virus and neurologic disorders. Emerg. Infect. Dis. 17, 173–179. doi: 10.3201/eid1702.100856
Kamar, N., Bendall, R., Legrand-Abravanel, F., Xia, N.-S., Ijaz, S., Izopet, J., et al. (2012). Hepatitis E. Lancet 379, 2477–2488. doi: 10.1016/S0140-6736(11)61849-7
Kamar, N., Selves, J., Mansuy, J.-M., Ouezzani, L., Péron, J.-M., Guitard, J., et al. (2008). Hepatitis E virus and chronic hepatitis in organ-transplant recipients. N. Engl. J. Med. 358, 811–817. doi: 10.1056/NEJMoa0706992
Karpe, Y. A., and Lole, K. S. (2010a). NTPase and 5′ to 3′ RNA duplex-unwinding activities of the hepatitis E virus helicase domain. J. Virol. 84, 3595–3602. doi: 10.1128/JVI.02130-09
Karpe, Y. A., and Lole, K. S. (2010b). RNA 5′-triphosphatase activity of the hepatitis E virus helicase domain. J. Virol. 84, 9637–9641. doi: 10.1128/JVI.00492-10
Khuroo, M. S. (1980). Study of an epidemic of non-A, non-B hepatitis: possibility of another human hepatitis virus distinct from post-transfusion non-A, non-B type. Am. J. Med. 68, 818–824. doi: 10.1016/0002-9343(80)90200-4
Khuroo, M. S., Kamili, S., and Yattoo Ghulam, N. (2004). Hepatitis E virus infection may be transmitted through blood transfusions in an endemic area. J. Gastroenterol. Hepatol. 19, 778–784. doi: 10.1111/j.1440-1746.2004.03437.x
Khuroo, M. S., Khuroo, M. S., and Khuroo, N. S. (2016). Hepatitis E: discovery, global impact, control and cure. World J. Gastroenterol. 22, 7030–7045. doi: 10.3748/wjg.v22.i31.7030
Koonin, E. V. (1991). The phylogeny of RNA-dependent RNA polymerases of positive-strand RNA viruses. J. Gen. Virol. 72(Pt 9), 2197–2206. doi: 10.1099/0022-1317-72-9-2197
Koonin, E. V., Gorbalenya, A. E., Purdy, M. A., Rozanov, M. N., Reyes, G. R., and Bradley, D. W. (1992). Computer-assisted assignment of functional domains in the nonstructural polyprotein of hepatitis E virus: delineation of an additional group of positive-strand RNA plant and animal viruses. Proc. Natl. Acad. Sci. U.S.A. 89, 8259–8263. doi: 10.1073/pnas.89.17.8259
Lange, H., Øverbø, J., Borgen, K., Dudman, S., Hoddevik, G., Urdahl, A. M., et al. (2016). Hepatitis E in Norway: seroprevalence in humans and swine. Epidemiol. Infect. 145, 181–186. doi: 10.1017/S0950268816002144
Li, S. W., Zhang, J., Li, Y. M., Ou, S. H., Huang, G. Y., He, Z. Q., et al. (2005a). A bacterially expressed particulate hepatitis E vaccine: antigenicity, immunogenicity and protectivity on primates. Vaccine 23, 2893–2901. doi: 10.1016/j.vaccine.2004.11.064
Li, S. W., Zhao, Q., Wu, T., Chen, S., Zhang, J., and Xia, N.-S. (2015). The development of a recombinant hepatitis E vaccine HEV 239. Hum. Vaccin. Immunother. 11, 908–914. doi: 10.1080/21645515.2015.1008870
Li, T.-C., Takeda, N., Miyamura, T., Matsuura, Y., Wang, J. C. Y., Engvall, H., et al. (2005b). Essential elements of the capsid protein for self-assembly into empty virus-like particles of hepatitis E virus. J. Virol. 79, 12999–13006. doi: 10.1128/JVI.79.20.12999-13006.2005
Li, T.-C., Yamakawa, Y., Suzuki, K., Tatsumi, M., Razak, M. A., Uchida, T., et al. (1997). Expression and self-assembly of empty virus-like particles of hepatitis E virus. J. Virol. 71, 7207–7213.
Liu, P., Bu, Q.-N., Wang, L., Han, J., Du, R.-J., Lei, Y.-X., et al. (2013). Transmission of hepatitis E virus from rabbits to Cynomolgus Macaques. Emerg. Infect. Dis. 19, 559–565. doi: 10.3201/eid1904.120827
Magden, J., Takeda, N., Li, T., Auvinen, P., Ahola, T., Miyamura, T., et al. (2001). Virus-specific mRNA capping enzyme encoded by hepatitis E virus. J. Virol. 75, 6249–6255. doi: 10.1128/JVI.75.14.6249-6255.2001
Mansuy, J. M., Gallian, P., Dimeglio, C., Saune, K., Arnaud, C., Pelletier, B., et al. (2016). A nationwide survey of hepatitis E viral infection in French blood donors. Hepatology 63, 1145–1154. doi: 10.1002/hep.28436
Martinelli, N., Luppi, A., Cordioli, P., Lombardi, G., and Lavazza, A. (2011). Prevalence of hepatitis E virus antibodies in pigs in Northern Italy. Infect. Ecol. Epidemiol. 1:7331. doi: 10.3402/iee.v1i0.7331
Martínez-Martínez, M., Diez-Valcarce, M., Cook, N., Hernández, M., and Rodríguez-Lázaro, D. (2011). Evaluation of extraction methods for efficient detection of enteric viruses in pork meat products. Food Anal. Methods 4, 13–22. doi: 10.1007/s12161-010-9165-1
Martin-Latil, S., Hennechart-Collette, C., Guillier, L., and Perelle, S. (2014). Method for HEV detection in raw pig liver products and its implementation for naturally contaminated food. Int. J. Food Microbiol. 176, 1–8. doi: 10.1016/j.ijfoodmicro.2014.01.016
Matsubayashi, K., Kang, J. H., Sakata, H., Takahashi, K., Shindo, M., Kato, M., et al. (2008). A case of transfusion-transmitted hepatitis E caused by blood from a donor infected with hepatitis E virus via zoonotic food-borne route. Transfusion 48, 1368–1375. doi: 10.1111/j.1537-2995.2008.01722.x
Matsubayashi, K., Nagaoka, Y., Sakata, H., Sato, S., Fukai, K., Kato, T., et al. (2004). Transfusion-transmitted hepatitis E caused by apparently indigenous hepatitis E virus strain in Hokkaido, Japan. Transfusion 44, 934–940. doi: 10.1111/j.1537-2995.2004.03300.x
McKnight, K. L., Xie, L., González-López, O., Rivera-Serrano, E. E., Chen, X., and Lemon, S. M. (2017). Protein composition of the hepatitis A virus quasi-envelope. Proc. Natl. Acad. Sci. U.S.A. 114, 6587–6592. doi: 10.1073/pnas.1619519114
Meng, X.-J. (2010). Hepatitis E virus: animal reservoirs and zoonotic risk. Vet. Microbiol. 140, 256–265. doi: 10.1016/j.vetmic.2009.03.017
Meng, X.-J. (2013). Zoonotic and foodborne transmission of hepatitis E virus. Semin. Liver Dis. 33, 41–49. doi: 10.1055/s-0033-1338113
Meng, X.-J., Halbur, P. G., Shapiro, M. S., Govindarajan, S., Bruna, J. D., Mushahwar, I. K., et al. (1998). Genetic and experimental evidence for cross-species infection by swine hepatitis E virus. J. Virol. 72, 9714–9721.
Meng, X.-J., Purcell, R. H., Halbur, P. G., Lehman, J. R., Webb, D. M., Tsareva, T. S., et al. (1997). A novel virus in swine is closely related to the human hepatitis E virus. Proc. Natl. Acad. Sci. U.S.A. 94, 9860–9865. doi: 10.1073/pnas.94.18.9860
Moin, S. M., Panteva, M., and Jameel, S. (2007). The hepatitis E virus Orf3 protein protects cells from mitochondrial depolarization and death. J. Biol. Chem. 282, 21124–21133. doi: 10.1074/jbc.M701696200
Mykytczuk, O., Harlow, J., Bidawid, S., Corneau, N., and Nasheri, N. (2017). Prevalence and molecular characterization of the hepatitis E virus in retail pork products marketed in Canada. Food Environ. Virol. 9, 208–218. doi: 10.1007/s12560-017-9281-9
Nagashima, S., Takahashi, M., Kobayashi, T., Tanggis Nishizawa, T., Nishiyama, T., et al. (2017). Characterization of the quasi-enveloped hepatitis E virus particles released by the cellular exosomal pathway. J. Virol. 91:e00822–17. doi: 10.1128/JVI.00822-17
Okamoto, H. (2013). Culture systems for hepatitis E virus. J. Gastroenterol. 48, 147–158. doi: 10.1007/s00535-012-0682-0
Pavio, N., Merbah, T., and Thébault, A. (2014). Frequent hepatitis E virus contamination in food containing raw pork liver, France. Emerg. Infect. Dis. J. 20, 1925–1927. doi: 10.3201/eid2011.140891
Pérez-Gracia, M. T., Mateos Lindemann, M. L., and Montalvo Villalba, M. C. (2013). Hepatitis E: current status. Rev. Med. Virol. 23, 384–398. doi: 10.1002/rmv.1759
Péron, J.-M., Mansuy, J.-M., Récher, C., Bureau, C., Poirson, H., Alric, L., et al. (2006). Prolonged hepatitis E in an immunocompromised patient. J. Gastroenterol. Hepatol. 21, 1223–1224. doi: 10.1111/j.1440-1746.2006.04209.x
Perttilä, J., Spuul, P., and Ahola, T. (2013). Early secretory pathway localization and lack of processing for hepatitis E virus replication protein pORF1. J. Gen. Virol. 94, 807–816. doi: 10.1099/vir.0.049577-0
Pudupakam, R. S., Huang, Y. W., Opriessnig, T., Halbur, P. G., Pierson, F. W., and Meng, X. J. (2009). Deletions of the hypervariable region (HVR) in open reading frame 1 of hepatitis E virus do not abolish virus infectivity: evidence for attenuation of HVR deletion mutants in vivo. J. Virol. 83, 384–395. doi: 10.1128/JVI.01854-08
Pudupakam, R. S., Kenney, S. P., Córdoba, L., Huang, Y.-W., Dryman, B. A., Leroith, T., et al. (2011). Mutational analysis of the hypervariable region of hepatitis E virus reveals its involvement in the efficiency of viral RNA replication. J. Virol. 85, 10031–10040. doi: 10.1128/JVI.00763-11
Renou, C., Roque-Afonso, A. M., and Pavio, N. (2014). Foodborne transmission of HEPATITIS E virus from raw pork liver sausage, France. Emerg. Infect. Dis. J. 20, 1945–1947. doi: 10.3201/eid2011.140791
Rivero-Juarez, A., Frias, M., Martinez-Peinado, A., Risalde, M. A., Rodriguez-Cano, D., Camacho, A., et al. (2017). Familial hepatitis E outbreak linked to wild boar meat consumption. Zoonoses Public Health 64, 561–565. doi: 10.1111/zph.12343
Rose, N., Lunazzi, A., Dorenlor, V., Merbah, T., Eono, F., Eloit, M., et al. (2011). High prevalence of hepatitis E virus in French domestic pigs. Comp. Immunol. Microbiol. Infect. Dis. 34, 419–427. doi: 10.1016/j.cimid.2011.07.003
Ruggeri, F. M., Di Bartolo, I., Ostanello, F., and Trevisani, M. (2013). “Epidemiology of the Human HEV Infection,” in Hepatitis E Virus: An Emerging Zoonotic and Foodborne Pathogen, eds F. M. Ruggeri, I. Di Bartolo, F. Ostanello, and M. Trevisani. (New York, NY: Springer New York), 11–21.
Salines, M., Andraud, M., and Rose, N. (2017). From the epidemiology of hepatitis E virus (HEV) within the swine reservoir to public health risk mitigation strategies: a comprehensive review. Vet. Res. 48:31. doi: 10.1186/s13567-017-0436-3
Schielke, A., Filter, M., Appel, B., and Johne, R. (2011). Thermal stability of hepatitis E virus assessed by a molecular biological approach. Virol. J. 8:487. doi: 10.1186/1743-422X-8-487
Schlauder, G. G., Dawson, G. J., Erker, J. C., Kwo, P. Y., Knigge, M. F., Smalley, D. L., et al. (1998). The sequence and phylogenetic analysis of a novel hepatitis E virus isolated from a patient with acute hepatitis reported in the United States. J. Gen. Virol. 79(Pt 3), 447–456. doi: 10.1099/0022-1317-79-3-447
Seminati, C., Mateu, E., Peralta, B., De Deus, N., and Martin, M. (2008). Distribution of hepatitis E virus infection and its prevalence in pigs on commercial farms in Spain. Vet. J. 175, 130–132. doi: 10.1016/j.tvjl.2006.11.018
Shukla, P., Nguyen, H. T., Torian, U., Engle, R. E., Faulk, K., Dalton, H. R., et al. (2011). Cross-species infections of cultured cells by hepatitis E virus and discovery of an infectious virus–host recombinant. Proc. Natl. Acad. Sci. U.S.A. 108, 2438–2443. doi: 10.1073/pnas.1018878108
Smith, D. B., Simmonds, P., Izopet, J., Oliveira-Filho, E. F., Ulrich, R., Johne, R., et al. (2016). Proposed reference sequences for Hepatitis E virus subtypes. J. Gen. Virol. 97, 537–542. doi: 10.1099/jgv.0.000393
Son, N. R., Seo, D. J., Lee, M. H., Seo, S., Wang, X., Lee, B.-H., et al. (2014). Optimization of the elution buffer and concentration method for detecting hepatitis E virus in swine liver using a nested reverse transcription-polymerase chain reaction and real-time reverse transcription-polymerase chain reaction. J. Virol. Methods 206, 99–104. doi: 10.1016/j.jviromet.2014.05.026
Sridhar, S., Teng, J., Chiu, T.-H., Lau, S., and Woo, P. (2017). Hepatitis E virus genotypes and evolution: emergence of camel hepatitis E variants. Int. J. Mol. Sci. 18:869. doi: 10.3390/ijms18040869
Suppiah, S., Zhou, Y., and Frey, T. K. (2011). Lack of processing of the expressed ORF1 gene product of hepatitis E virus. Virol. J. 8:245. doi: 10.1186/1743-422X-8-245
Surjit, M., Oberoi, R., Kumar, R., and Lal, S. K. (2006). Enhanced α1 microglobulin secretion from hepatitis E virus ORF3-expressing human hepatoma cells is mediated by the tumor susceptibility gene 101. J. Biol. Chem. 281, 8135–8142. doi: 10.1074/jbc.M509568200
Szabo, K., Trojnar, E., Anheyer-Behmenburg, H., Binder, A., Schotte, U., Ellerbroek, L., et al. (2015). Detection of hepatitis E virus RNA in raw sausages and liver sausages from retail in Germany using an optimized method. Int. J. Food Microbiol. 215, 149–156. doi: 10.1016/j.ijfoodmicro.2015.09.013
Takahashi, M., Yamada, K., Hoshino, Y., Takahashi, H., Ichiyama, K., Tanaka, T., et al. (2008). Monoclonal antibodies raised against the ORF3 protein of hepatitis E virus (HEV) can capture HEV particles in culture supernatant and serum but not those in feces. Arch. Virol. 153, 1703–1713. doi: 10.1007/s00705-008-0179-6
Tam, A. W., Smith, M. M., Guerra, M. E., Huang, C.-C., Bradley, D. W., Fry, K. E., et al. (1991). Hepatitis E virus (HEV): molecular cloning and sequencing of the full-length viral genome. Virology 185, 120–131. doi: 10.1016/0042-6822(91)90760-9
Tanaka, T., Takahashi, M., Kusano, E., and Okamoto, H. (2007). Development and evaluation of an efficient cell-culture system for hepatitis E virus. J. Gen. Virol. 88, 903–911. doi: 10.1099/vir.0.82535-0
Teo, C. G. (2010). Much meat, much malady: changing perceptions of the epidemiology of hepatitis E. Clin. Microbiol. Infect. 16, 24–32. doi: 10.1111/j.1469-0691.2009.03111.x
Teshale, E. H., and Hu, D. J. (2011). Hepatitis E: epidemiology and prevention. World J. Hepatol. 3, 285–291. doi: 10.4254/wjh.v3.i12.285
Tyagi, S., Korkaya, H., Zafrullah, M., Jameel, S., and Lal, S. K. (2002). The phosphorylated form of the ORF3 protein of hepatitis E virus interacts with its non-glycosylated form of the major capsid protein, ORF2. J. Biol. Chem. 277, 22759–22767. doi: 10.1074/jbc.M200185200
von Nordheim, M., Boinay, M., Leisi, R., Kempf, C., and Ros, C. (2016). Cutthroat trout virus—towards a virus model to support hepatitis E research. Viruses 8:289. doi: 10.3390/v8100289
Wen, J., Behloul, N., Dai, X., Dong, C., Liang, J., Zhang, M., et al. (2016). Immunogenicity difference between two hepatitis E vaccines derived from genotype 1 and 4. Antiviral Res. 128, 36–42. doi: 10.1016/j.antiviral.2016.02.002
Wenzel, J. J., Preiß, J., Schemmerer, M., Huber, B., Plentz, A., and Jilg, W. (2011). Detection of hepatitis E virus (HEV) from porcine livers in Southeastern Germany and high sequence homology to human HEV isolates. J. Clin. Virol. 52, 50–54. doi: 10.1016/j.jcv.2011.06.006
Xin, S., and Xiao, L. (2016). “Clinical Manifestations of Hepatitis E,” in Hepatitis E Virus, ed. Y. Wang (Dordrecht: Springer Netherlands), 175–189.
Xing, L., Li, T.-C., Mayazaki, N., Simon, M. N., Wall, J. S., Moore, M., et al. (2010). Structure of Hepatitis E virion-sized particle reveals an RNA-dependent viral assembly pathway. J. Biol. Chem. 285, 33175–33183. doi: 10.1074/jbc.M110.106336
Yamashita, T., Mori, Y., Miyazaki, N., Cheng, R. H., Yoshimura, M., Unno, H., et al. (2009). Biological and immunological characteristics of hepatitis E virus-like particles based on the crystal structure. Proc. Natl. Acad. Sci. U.S.A. 106, 12986–12991. doi: 10.1073/pnas.0903699106
Yugo, D. M., Cossaboom, C. M., and Meng, X.-J. (2014). Naturally occurring animal models of human hepatitis E virus infection. ILAR J. 55, 187–199. doi: 10.1093/ilar/ilu007
Zafrullah, M., Ozdener, M. H., Panda, S. K., and Jameel, S. (1997). The ORF3 protein of hepatitis E virus is a phosphoprotein that associates with the cytoskeleton. J. Virol. 71, 9045–9053.
Zhang, G., Slowinski, V., and White, K. A. (1999). Subgenomic mRNA regulation by a distal RNA element in a (+)-strand RNA virus. RNA 5, 550–561. doi: 10.1017/S1355838299982080
Zhang, J., Zhang, X.-F., Huang, S.-J., Wu, T., Hu, Y.-M., Wang, Z.-Z., et al. (2015). Long-term efficacy of a hepatitis E vaccine. N. Engl. J. Med. 372, 914–922. doi: 10.1056/NEJMoa1406011
Zhang, M., Purcell, R. H., and Emerson, S. U. (2001). Identification of the 5′ terminal sequence of the SAR-55 and MEX-14 strains of hepatitis E virus and confirmation that the genome is capped. J. Med. Virol. 65, 293–295. doi: 10.1002/jmv.2032
Keywords: hepatitis E virus, foodborne virus, foodborne illness, zoonosis, swine
Citation: Harrison L and DiCaprio E (2018) Hepatitis E Virus: An Emerging Foodborne Pathogen. Front. Sustain. Food Syst. 2:14. doi: 10.3389/fsufs.2018.00014
Received: 22 February 2018; Accepted: 17 April 2018;
Published: 08 May 2018.
Edited by:
Joshua B. Gurtler, Agricultural Research Service (USDA), United StatesReviewed by:
Aguang Dai, University of Texas at Austin, United StatesCopyright © 2018 Harrison and DiCaprio. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Erin DiCaprio, ZWxkaWNhcHJpb0B1Y2RhdmlzLmVkdQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.