Explore article hub
- 1Mathematical Modelling of Infectious Diseases Unit, Institut Pasteur, Université Paris Cité, Centre National dete de Recherche (CNRS) UMR2000, Paris, France
- 2Division of Cell Matrix Biology and Regenerative Medicine, University of Manchester, Manchester, United Kingdom
- 3Division of Neuroscience, Istituto di Ricovero e Cura a Carattere Scientifico (IRCCS) Ospedale San Raffaele, Milan, Italy
- 4Muscle Research Unit, Charité Medical Faculty and Max Delbrück Center, Berlin, Germany
- 5Metabolism and Nutrition Research Group, Louvain Drug Research Institute, Université Catholique de Louvain, Brussels, Belgium
- 6Systems Immunology Department, Weisman Institute of Science, Rehovot, Israel
- 7Microbiome and Cancer Division, German Cancer Research Center (DKFZ), Heidelberg, Germany
- 8Collège de France, Paris, France
- 9Paediatric Immuno-Hematology and Rheumatology Unit, Necker Enfants Malades University Hospital, Assistance Publique-Hôpitaux de Paris, Paris, France
- 10Paris-Cité University, Imagine Institute, Paris, France
- 11Paris-Cité University, Inserm UMR 1163, Paris, France
- 12Académie des Sciences, Paris, France
- 13Center for Alternatives to Animal Testing (CAAT), Johns Hopkins University, Bloomberg School of Public Health and Whiting School of Engineering, Baltimore, MD, United States
- 14Center for Alternatives to Animal Testing (CAAT) Europe, University of Konstanz, Konstanz, Germany
- 15The European Institute for Innovation Through Health Data, Ghent, Belgium
- 16Department of Internal Medicine and Radboud Center for Infectious Diseases, Radboud University Nijmegen Medical Centre, Nijmegen, Netherlands
- 17Department for Immunology and Metabolism, Life and Medical Sciences Institute (LIMES), University of Bonn, Bonn, Germany
- 18Laboratory of Virology and Chemotherapy, Department of Microbiology and Immunology, Rega Institute for Medical Research, Katholieke Universiteit (KU) Leuven, Leuven, Belgium
- 19The VirusBank Platform, Leuven, Belgium
- 20Global Virus Network (GVN), Baltimore, MD, United States
- 21Monoclonal Antibody Discovery (MAD) Lab, Fondazione Toscana Life Sciences, Siena, Italy
- 22Department of Biotechnology, Chemistry and Pharmacy, University of Siena, Siena, Italy
- 23Fondazione Biotecnopolo di Siena, Siena, Italy
- 24Department of Life Sciences, Imperial College London, London, United Kingdom
- 25Vaccine R&D, Coalition for Epidemic Preparedness Innovations (CEPI), London, United Kingdom
- 26Medable, Inc, Palo Alto, CA, United States
- 27Michael G. DeGroote (MG) Institute for Infectious Disease Research, Department of Biochemistry and Biomedical Sciences, McMaster University, Hamilton, ON, Canada
- 28Unité de Pathogénie Microbienne Moléculaire, Institut Pasteur, Paris, France
- 29Institute for Interdisciplinary Innovation in Healthcare, Université Libre de Bruxelles, Brussels, Belgium
The COVID-19 pandemic accelerated research and innovation across numerous fields of medicine. It emphasized how disease concepts must reflect dynamic and heterogeneous interrelationships between physical characteristics, genetics, co-morbidities, environmental exposures, and socioeconomic determinants of health throughout life. This article explores how scientists and other stakeholders must collaborate in novel, interdisciplinary ways at these new frontiers of medicine, focusing on communicable diseases, precision/personalized medicine, systems medicine, and data science. The pandemic highlighted the critical protective role of vaccines against current and emerging threats. Radical efficiency gains in vaccine development (through mRNA technologies, public and private investment, and regulatory measures) must be leveraged in the future together with continued innovation in the area of monoclonal antibodies, novel antimicrobials, and multisectoral, international action against communicable diseases. Inter-individual heterogeneity in the pathophysiology of COVID-19 prompted the development of targeted therapeutics. Beyond COVID-19, medicine will become increasingly personalized via advanced omics-based technologies and systems biology—for example targeting the role of the gut microbiome and specific mechanisms underlying immunoinflammatory diseases and genetic conditions. Modeling proved critical to strengthening risk assessment and supporting COVID-19 decision-making. Advanced computational analytics and artificial intelligence (AI) may help integrate epidemic modeling, clinical features, genomics, immune factors, microbiome data, and other anthropometric measures into a “systems medicine” approach. The pandemic also accelerated digital medicine, giving telehealth and digital therapeutics critical roles in health system resilience and patient care. New research methods employed during COVID-19, including decentralized trials, could benefit evidence generation and decision-making more widely. In conclusion, the future of medicine will be shaped by interdisciplinary multistakeholder collaborations that address complex molecular, clinical, and social interrelationships, fostering precision medicine while improving public health. Open science, innovative partnerships, and patient-centricity will be key to success.
Key points
- The number of medical and health science articles published during the COVID-19 pandemic years 2020–2022 was 9% higher than trend-based predictions, reflecting accelerated research and technological progress across many areas of biomedical science.
- Innovations unleashed by the pandemic for combatting communicable diseases—including in vaccine, monoclonal antibody, and antimicrobial development, in mathematical modeling, and in multisectoral, international collaboration—must be leveraged to further improve patient care and public health.
- The pandemic underlined the importance of advancing personalized/precision medicine, taking individual, environmental, and social determinants of health into account; advanced omics-based technologies enabled by systems biology, advanced computational analytics, artificial intelligence (AI), and new clinical trial designs, offer unprecedented opportunities.
- The pandemic accelerated digital medicine: telehealth and digital therapeutics are driving improvements both in clinical care and health systems resilience.
- Achieving this future of medicine requires new forms interdisciplinarity that integrate knowledge and skills across biomedical science, healthcare, public health, regulation, policy, economics, social science, and more—supported by radical reform of research funding and policies to ensure equitable access to medical innovation across populations and countries.
Introduction
The COVID-19 pandemic has emphasized the importance of rethinking our concepts of disease based on dynamic and heterogeneous interrelationships: patients suffering from SARS-CoV-2 infection exhibit a high degree of heterogeneity in susceptibility to infection, disease manifestations, and outcomes. This has been determined with respect to age, sex, race, underlying genetic variation, differential immune responses, and preexisting co-morbidities, which are in turn subject to environmental and socioeconomic determinants (1). These complex connections led some to label COVID-19 a “syndemic”, prompting calls for integrated healthcare and public health responses that tackle these factors in concert with research into the mechanistic pathways underlying the interrelationships (2–4).
The pandemic triggered a substantial increase in research across many fields of biomedical science. Overall, there was an increase of 9% in the number of articles published in the fields of medical and health sciences in the period 2020–2022 as compared with the volume that would have been expected, and approximately 7% of all articles published over this period concerned or included reference to SARS-CoV-2/COVID-19 (Figure 1). The lessons of COVID-19 extend far beyond infectious disease science. They underline the need for a comprehensive new vision of health as a complex equilibrium that balances numerous bodily systems and their interactions with the totality of health-related exposures during life. This is known as the “exposome” (5). Indeed, the pandemic revealed new frontiers in medicine, where scientists and other stakeholders must collaborate in novel, interdisciplinary ways to unravel and address the complex interrelationships linking communicable and non-communicable diseases—studying the molecular mechanisms of disease in the context of environmental and social determinants of health.
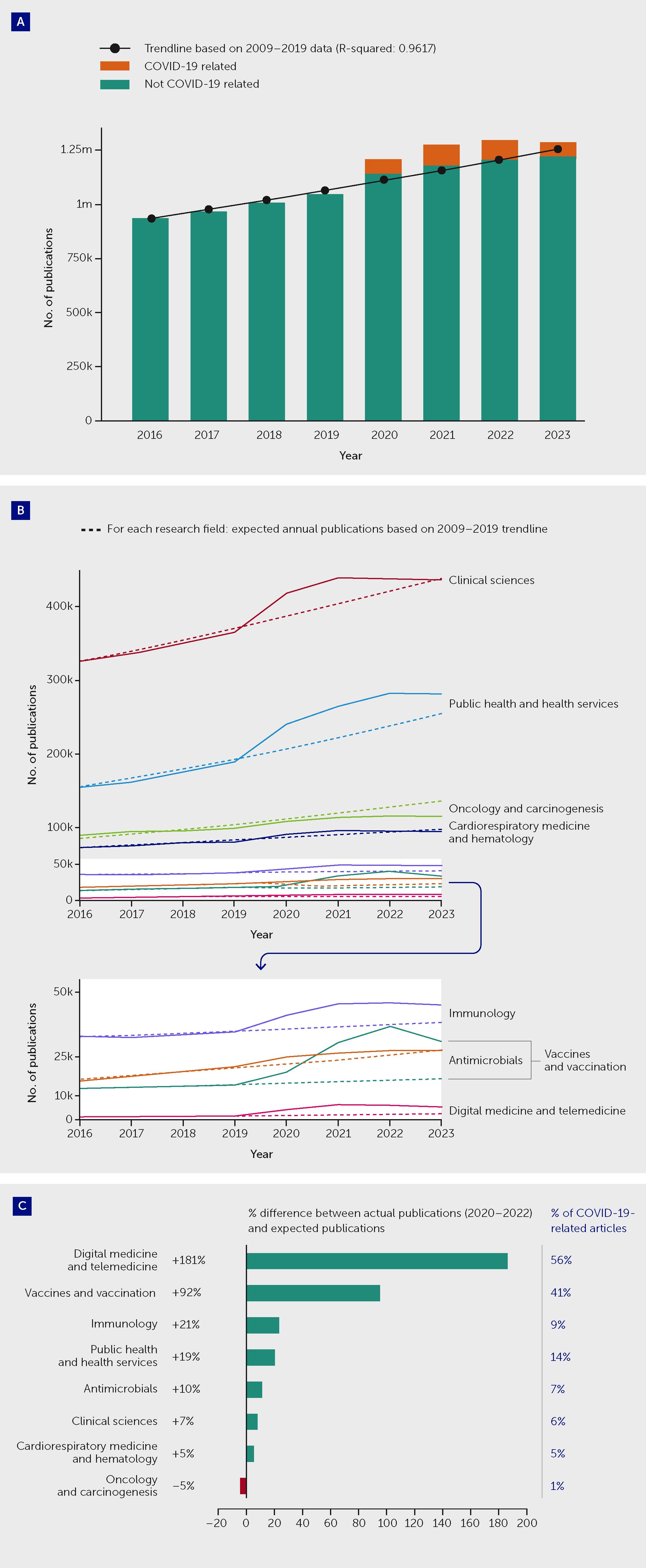
Figure 1 The COVID-19 pandemic triggered an increase in medical and health sciences publications overall but with variations between fields. (A) Actual and expected publications across all medical and health sciences, and proportion of all articles that were COVID-19–related, by year between 2016 and 2023. Overall, there was an increase of approximately 9% in the number of articles published during the “COVID-19 years,” during the period 2020–2022, as compared with the volume that would have been expected based on 2009–2019 trends. Approximately 7% of all articles published in 2020–2022 concerned or included reference to SARS-CoV-2/COVID-19. After the increase in medical and health sciences publications between 2020 and 2022, 2023 saw a decrease in publication volume. (B) Actual and expected publications in selected research fields by year between 2016 and 2023. (C) Difference between actual and expected publication numbers between 2020 and 2023 in selected research fields (expressed as a percentage of expected publications) and the proportion of all actual articles that were COVID-19-related. The 2008 Australian and New Zealand Standard Research Classification (ANZSRC) was applied to identify publications in the field of “Medical and Health Sciences” within Dimensions (data until 1 January 2024), using machine learning to sort publications into Fields of Research. Expected publication numbers for 2020–2022 were calculated based on an exponential trendline fitted on the actual 2009–2019 annual publication numbers in relevant fields. An estimated total publication volume for 2023 was used to correct for the indexation lag of publications. See Appendix for further methodological details.
The authors of this article come from different disciplines and backgrounds, but we hold in common our views on the medical and scientific challenges and opportunities that we now face. We aim to provide an interdisciplinary perspective on the future of medicine in the following domains, based on insights gleaned during the COVID-19 pandemic:
● Innovative strategies to combat communicable diseases;
● Precision/personalized medicine to better address individual patient needs;
● Systems medicine and data science, including modeling, artificial intelligence (AI), digital medicine, and novel trial designs;
● Public health science, aiming to better inform health policies and address health inequalities.
Pandemic responses and preparedness will not be addressed directly, as these have been the subject of several reports, including the report of the Independent Panel for Pandemic Preparedness and Response (6). Similarly, while we touch on aspects of global health and gaps to be filled in low- and middle-income countries, these important questions are dealt with in a companion article and elsewhere (6–8).
Innovative strategies to combat communicable diseases
The Spanish flu claimed between 50 and 100 million lives at the start of the 20th century, and it is estimated that the COVID-19 pandemic has already caused approximately 20 million deaths worldwide (9). Vaccination led to the first major successes in the fight against infectious threats following the pioneering experiments of Edward Jenner at the end of the 18th century and Louis Pasteur in the late 19th century. More than two centuries later, vaccines are still the key to limiting the public health and societal disasters inflicted by infectious organisms, including SARS-CoV-2. Vaccines undoubtedly represent one of the greatest medical achievements of modern civilization, and we can anticipate that they will remain at the core of future strategies to combat communicable diseases.
While the development of vaccines allowed us to effectively prevent infectious diseases, it was not until the discovery of antibiotics at the beginning of the 20th century that infections could be efficiently treated at scale. The spectacular success of antibiotics in addressing bacterial infections led many people to forget for a time that populations remain vulnerable to other types of infections, namely those caused by viruses and parasites. The AIDS epidemic, in the early 1980s, reawakened this consciousness and resulted in an unprecedented mobilization of resources that eventually led to a new generation of powerful antiretroviral drugs. In addition to controlling HIV infection, these drugs served as a model for new treatments for hepatitis C—another devastating viral infection. The resurgence of tuberculosis concomitant with the spread of AIDS, along with the heavy malaria burden in low- and middle-income countries, helped to put the fight against neglected diseases back on the international agenda. These efforts must still be scaled up to address not only future pandemics but also the great threat posed by antimicrobial resistance (AMR).
While the innovative vaccines and therapies discussed below are essential for fighting the COVID-19 pandemic, a broader, global approach must be developed to prevent future pandemics and mitigate their impact. The factors contributing to the transmission of microbes from animals to humans must be addressed, as must the impact of travel, population growth, pollution, and global warming (10). The concept of “One Health” underlies this holistic approach (11). As discussed throughout this article, the unprecedented efforts needed to keep the world’s population safe from future infectious threats will require mobilization of competencies across many disciplines, and collaboration with multiple stakeholders from both the public and private sectors.
Vaccines of the future
Thanks to new technologies and innovative approaches to vaccine development, COVID-19 vaccines were developed at an unprecedented pace. These vaccines quickly demonstrated their value in preventing infections, decreasing hospitalizations, and reducing mortality—preventing up to 20 million deaths within the first year of their use (12) and saving the global economy several trillion dollars. The field of vaccinology was transformed by the technologies and methodologies employed to accelerate vaccine development during the pandemic, and, as a result, we expect that an increasing number of diseases will soon be conquered by vaccination.
The COVID-19 pandemic radically changed the vaccine development process. Before the pandemic, the time from the discovery phase until proof of concept in the laboratory generally spanned more than 10 years (Figure 2). The discovery phase was followed by the early-development phase, which included scaling up production; characterization of the antigens; formulation, toxicity and immunogenicity testing in animal models; and production under Good Manufacturing Practices conditions. Following regulatory authorization, vaccines then entered phase I clinical trials, followed by phase II clinical trials to achieve proof of concept with respect to safety and efficacy in the clinic. Only at that point would the industry invest in the expensive late-development phase, which includes phase III clinical trials and the construction of dedicated manufacturing facilities. Overall investment was approximately US$1 billion per vaccine, of which 70% was spent in the late-development phase. During the COVID-19 pandemic, new technologies—including synthetic biology, mRNA constructs, and viral vectors—dramatically accelerated the process, with phase I clinical trials beginning only 2 months after vaccine development began.
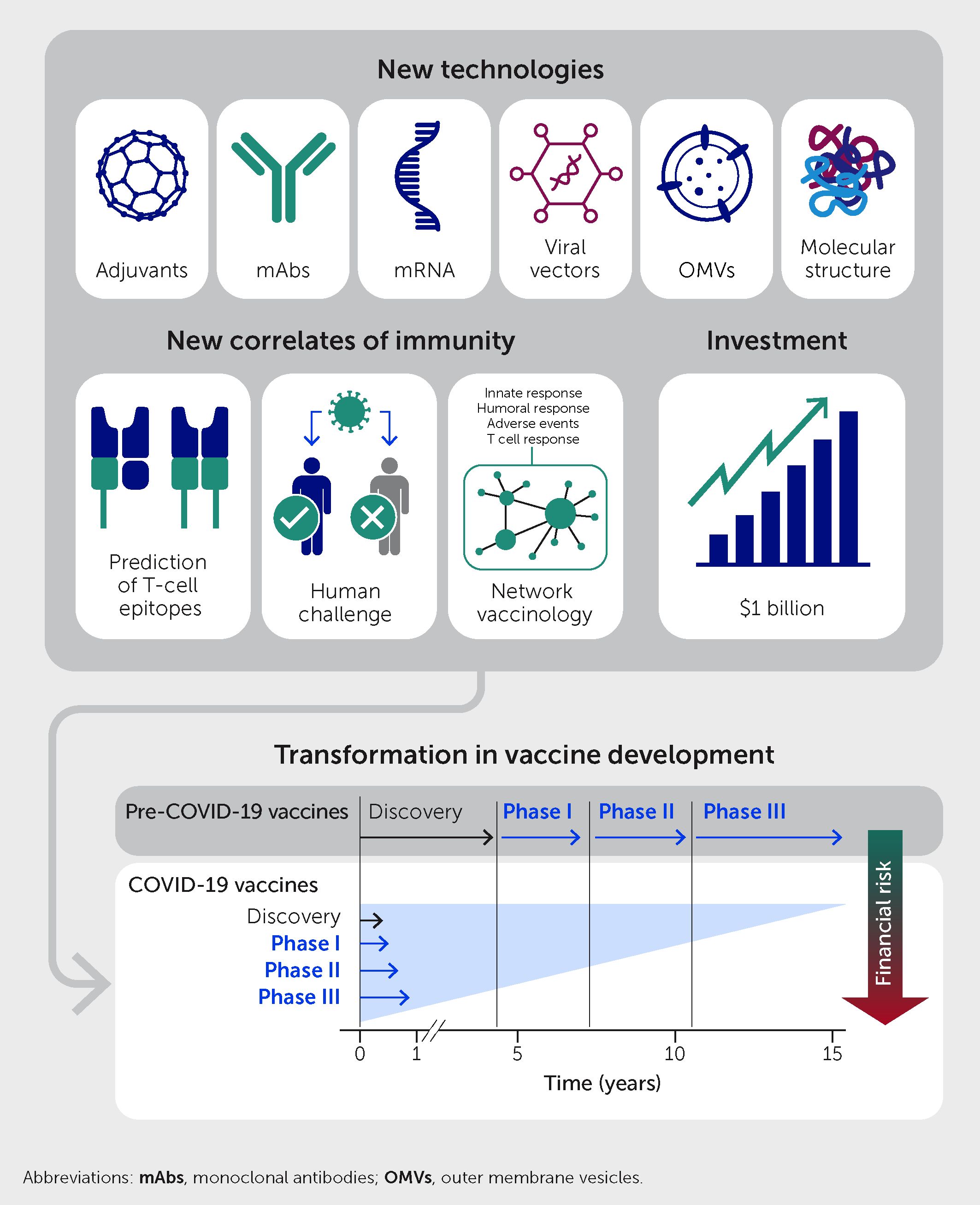
Figure 2 The COVID-19 pandemic radically changed the vaccine development process. Pre-pandemic, the development process entailed sequential phases of discovery, early-development, and phase I–III clinical trials over a period of 15 years or so. For COVID-19 vaccines, new technologies, new correlates of immunity, and early public investment allowed all steps to establish safety and efficacy to be completed in parallel within 10 months and with less financial risk to developers.
Two seminal discoveries over the last decade enabled the development of mRNA vaccines (13). The first was the demonstration that modified nucleosides enhance the stability of mRNA and reduce its inflammatory potential (14, 15); the second was the discovery that encapsulation of mRNA in lipid nanoparticles could both facilitate the delivery of mRNA in vivo and serve as an adjuvant (16, 17). In parallel, the public sector spent more than US$15 billion to encourage companies to invest in manufacturing facilities prior to having a vaccine in hand and to perform phase I, II, and III clinical trials in parallel rather than sequentially. This public investment removed the financial risk from the private sector and allowed COVID-19 vaccines to be developed in just 10 months without skipping any of the steps necessary to establish safety and efficacy (Figure 2). In conclusion, the COVID-19 pandemic accelerated the availability of novel technologies, validated the incredible contribution of vaccines to society and the global economy, and emphasized the critical role of the public sector in creating the necessary conditions for rapid vaccine development. In the process, the financial, regulatory, and scientific approaches to vaccine development were irrevocably altered.
Major investments will continue to improve future COVID-19 vaccines, with the aim of blocking virus transmission and preventing immune evasion (18). Furthermore, lessons learned during the creation of COVID-19 vaccines will undoubtedly facilitate the development of new strategies to fight not only emerging infections but also other health challenges such as AMR, chronic infectious diseases, and cancer (19–21).
Development of vaccines against antibiotic-resistant bacteria such as Escherichia coli, Staphylococcus aureus, Clostridium difficile, Klebsiella pneumoniae, Pseudomonas aeruginosa, Neisseria gonorrhoeae, Salmonella typhi, Shigella, Acinetobacter baumannii, Enterococcus faecium, and Campylobacter is possible and can help to mitigate the risk of AMR. In addition, the recent success of vaccines against the varicella zoster virus (22) shows that vaccines can now be developed against microorganisms that are normally controlled by the immune system but which can cause chronic infections when the immune system is weakened by concomitant infections, immunosuppressive pharmacological treatments, or aging. These include cytomegalovirus, herpes simplex virus (types 1, 2, 6, and 7), Epstein-Barr virus, and bacteria such as Mycobacterium tuberculosis. Some other diseases entirely defeat the immune system, and these include certain cancers and chronic infections caused by HIV, human papillomavirus, and hepatitis B and C viruses. We still lack scientific solutions for such diseases, but the progress achieved in vaccinology during the last few years suggests that vaccine-based solutions could be identified soon.
In conclusion, vaccines represent a sustainable method for improving human health and well-being because they prevent and control new infectious diseases, limit pandemics, mitigate AMR, and address critical public health issues in both high- and low-income countries.
Toward a new generation of antibacterial agents to tackle antimicrobial resistance
Bacterial infections remain a major cause of disease and death globally. In 2019, 33 bacterial genera are estimated to have caused 7.7 million deaths across 11 infectious syndromes—this being equivalent to 13.6% of all global deaths, making bacterial infections the second leading cause of death globally (23). Globally, bacterial AMR is estimated to have directly caused 1.27 million deaths in 2019 based on a counterfactual of drug-susceptible infection (23). The COVID-19 pandemic has significantly increased AMR among bacteria. In the early stages of the pandemic, biocides and antibiotics were used widely in clinical treatment to address concerns of secondary bacterial or fungal infections in COVID-19 patients. Furthermore, most critically ill patients admitted to crowded intensive care facilities received prophylactic or therapeutic antibiotics. According to the United States Centers for Disease Control and Prevention (CDC), carbapenem-resistant Acinetobacter infections increased by 78% from 2019 to 2020 in United States hospitals, while carbapenem-resistant Enterobacterales producing extended-spectrum β-lactamases (ESBL) rose by more than 30%, and antifungal-resistant Candida auris infections increased by 60% over the same period (24). The result is an increased need for new antimicrobials, yet the past two decades have witnessed a decrease in their discovery and development while AMR has become a global health crisis threatening the medical advances made over the past century (23).
The challenges are twofold: economic and scientific. Traditional return on investment, based on drug cost and volume of sales, is no longer valid for antimicrobials because they are relatively inexpensive and their use is increasingly rationed by healthcare providers (25). In response to this dilemma, several countries are developing policies and economic models that incentivize companies to bring needed antimicrobials to patients (26). However, elimination of these economic barriers still requires new drug candidates, and the scientific challenges are significant. A principal roadblock is the lack of novel chemicals with suitable properties to provide a basis for development. Historically, many antibiotics were developed from natural products isolated from microbes, but this source was largely abandoned in the 1980s owing to the frequent re-isolation of known compounds. Fully synthetic approaches, which dominate drug development in other fields, have proven unsuitable for developing leads for new antibiotics (27).
Fortunately, these difficulties are being met with creativity and innovation in fundamental antimicrobial research. Efforts to codify the unique physical-chemical properties of antibiotic compounds offer an opportunity to build tailored libraries for antibiotic discovery (27, 28). Furthermore, the development of genome-based efforts to “mine” for new natural products, paired with synthetic biology tools to optimize these compounds, are opening promising routes to new antibiotics (29, 30). Another method involves pairing existing antibiotics with resistance inhibitors. This approach continues to be highly successful in maintaining the efficacy of β-lactam antibiotics such as carbapenems and cephalosporins, and several new combination drugs have been approved for clinical use over the last decade (31).
As antibiotic efficacy has waned, orthogonal strategies have gained prominence. The use of phages, for example, is gaining traction as a strategy to address particularly intransigent bacterial infections (32). This highly personalized medical approach is increasingly saving lives and offers promise for the most difficult-to-treat antibiotic-resistant infections. Many of these innovations occur in academic labs or small biotechnology companies. With renewed efforts to incentivize antimicrobial discovery and development, there is good reason to believe that we will be able to address the challenge of AMR.
Toward a new generation of antiviral drugs
During the pandemic, remdesivir, molnupiravir, and the combination of nirmaltrelvir and ritonavir (Paxlovid®) rapidly emerged as antiviral drugs active against SARS-CoV-2 (33). These anti-SARS-Cov-2 agents complete the armamentarium of small molecule antiviral drugs available for the treatment and sometimes prophylaxis of infections with herpes viruses, HIV, hepatitis B and C viruses, and influenza viruses. Yet, there are no antiviral drugs available for the treatment of several viral infections, including emerging and/or neglected viruses such as flaviviruses (e.g., dengue), paramyxoviruses (e.g., Nipah), togaviruses (e.g., Chikungunya), arenaviruses (e.g., Lassa), filoviruses (e.g., Ebola and Marburg), bunyaviruses (e.g., Rift Valley fever and Crimean Congo hemorrhagic fever), enteroviruses (e.g., EV68 and EV71), and several others.
A key takeaway from COVID-19 is that we have the tools to develop new efficient antiviral therapies. Relevant targets include RNA-dependent RNA polymerase, viral proteases, and RNA capping machinery (34). Among the innovative approaches currently used to develop new antiviral therapeutics, the PROteolysis TArgeting Chimeras (PROTAC) technology—aimed at degradation of pathogenic proteins by hijacking of the ubiquitin-proteasome-system—is a promising strategy despite some limitations (35). As discussed below, discovery of new antivirals will also benefit from innovative tools based on AI and machine learning approaches.
The revival of passive immunotherapy
Passive immunization entails the administration of antibodies to individuals to control infectious diseases. Unlike traditional serum polyclonal antibodies, monoclonal antibodies (mAbs) are much more specific in their targeting activity, with a high degree of consistency among manufactured lots. The safety profile of mAbs is also better, particularly for the new generation of fully human mAbs.
Two mAbs against the SARS-CoV-2 virus were approved in the United States in November 2020 after their combined use was shown to reduce the risk of severe COVID-19 in highly vulnerable patients (36). Later, another combination of mAbs was successfully developed for pre-exposure prevention in immunosuppressed individuals not protected by vaccines (37). This product ensures several months of protection thanks to a genetically engineered Fc region that prolongs its half-life (38). However, the therapeutic efficiency of most anti-SARS-CoV-2 mAbs has been hampered by the emergence of viral variants that escape their neutralizing activity (39). Treatment with mAbs might actually favor the emergence of escape variants in immunosuppressed individuals (40). For these reasons, major research efforts are focusing on the development of long-acting, broadly reactive mAbs with optimal binding and neutralizing properties (38, 41). Such antibodies will be essential to protect against COVID-19 in patients who cannot benefit from vaccines.
Insights gained during the research and development of anti-COVID-19 mAbs will greatly aid the ongoing development of novel mAbs against several other infectious diseases, Currently, the most widely used mAbs in the clinic are palivizumab and nirsevimab for the prophylaxis of respiratory syncytial virus in high-risk infants (42, 43). mAbs were also approved against C. difficile (44) and Ebola virus (45). New targets under development include Zika virus and HIV infections (46), malaria (47), and antimicrobial-resistant bacterial infections (48).
Statistical and mathematical modeling of epidemic data
The COVID-19 pandemic highlighted the major role of statistical and mathematical modeling to strengthen epidemic risk assessment and support decision-making during epidemics. It also highlighted important challenges that remain when performing such risk assessments in situations characterized by limited data and volatile epidemic trajectories driven by multiple factors (including interventions, behaviors, population immunity, variants, and vaccines).
First, these methods proved essential to estimate key characteristics of the emerging pathogen, such as its transmissibility and severity. Estimating these parameters can be challenging because the epidemic process is imperfectly observed. For example, even though chains of transmission were rarely available, the transmission potential of the new virus was quickly derived from the analysis of epidemic growth (49, 50). Early on during viral emergence, severity estimation may be biased upward if asymptomatic and mild infections go undetected. However, the joint analysis of multiple types of data with dedicated modeling techniques (50–52) ensured effective estimation of severity later confirmed by serosurveys (53). Modeling also helped monitor key quantities, such as the proportion infected, which was essential to capture population immunity and determine the likelihood of future waves. The analysis of viral sequences with phylodynamic techniques provided unique insights into the dynamics of spread (54). Early estimates of severity might be the most important quantity to anticipate the potential impact of a starting pandemic and determine the intensity of the response. Even though these estimates proved correct for SARS-CoV-2, estimation of this quantity remains highly uncertain early on because it is difficult to identify mild or asymptomatic infections in the absence of important testing capacity and serology. It is essential to develop study designs and modeling techniques that make the most of available resources to improve the early estimation of severity.
Early in the pandemic, in the absence of treatments and vaccines, the only way to mitigate pandemic impact was through non-pharmaceutical measures. A broad range of measures, including school and place closures, curfews, and lockdowns were considered, and modeling played a key role in evaluating their impact even though disentangling the effects of individual measures proved difficult given their concomitant implementation (51, 55, 56) The impact of these interventions depends on population adherence. This behavioral response could change over time (for example due to pandemic fatigue) and was often difficult to anticipate, complicating epidemic forecasting. We also often lacked a description of contact patterns that was granular enough to ascertain the impact of specific measures, such as bar closures. Modeling also helped design and evaluate new approaches to SARS-CoV-2 control, such as digital contact tracing (55, 57) or contact bubbles (56). While some argued that non-pharmaceutical measures should be restricted to frail individuals, the detailed analysis of porous transmission dynamics between age groups through mathematical modeling helped demonstrate that such approaches would not avoid saturation of hospitals and that an effort from all was necessary (58).
When vaccines became available, modeling was used to determine optimal vaccine use depending on vaccine characteristics, first with respect to the question of prioritization in a context of limited resources (59, 60) and later to address more specific issues such as the management of adverse events (61) and booster doses (62). As SARS-CoV-2 immunity has progressively become more complex, with individuals being exposed to multiple variants and vaccines, the modeling of vaccine impact has become harder as well.
Epidemic forecasting has been another important area of research with the aim to support healthcare planning in a context of major stress on healthcare systems. Such forecasting has proved difficult as the dynamics of COVID-19 cases changed rapidly with the implementation and relaxation of control measures, behavioral changes, the emergence of new variants, and mass vaccination. As a result, the forecasting horizon in a country such as France was limited to a couple of weeks (63). Models were also used to build scenarios over longer time periods, with the aim to show how the pandemic might progress depending on a specific set of assumptions (e.g., regarding the intensity of control measures, vaccine coverage, and seasonality) (64). A communication challenge for modeling teams was to explain the distinction between forecasts and modeling scenarios.
Prior to the pandemic, only a few countries, such as the United Kingdom, regularly used mathematical modeling to support decision-making during epidemics. In many countries, the COVID-19 pandemic has cemented modeling as an important tool to inform decision-makers during epidemics. The way such scientific input is provided varies by country. In the United Kingdom for example, the Scientific Pandemic Influenza Group on Modelling, Operational sub-group (SPI-M-O), a subcommittee attached to the Scientific Advisory Group for Emergencies (SAGE), comprised many modeling teams and met regularly to provide a consensus of evidence (65), whereas the process was often more informal in other countries. The pandemic has also strengthened interesting collaborative projects, such as the United States and European modeling hubs, in which multiple teams aim to address the same question. The comparison of the results of the different teams helps assess the robustness of findings.
The COVID-19 pandemic has helped strengthen analytical capacity for epidemic data globally. A key goal for future epidemics is to ensure that this capacity can be maintained as modelers move back to their “peacetime” research. It is also important to develop models that can better capture and anticipate behavior changes during epidemics. This requires strong collaboration between modelers and scientists from the behavioral and social sciences (66). As data that can inform epidemic dynamics become more complex and diverse (encompassing syndromic and virological surveillance; epidemiological studies; phylogenies reconstructed from the analysis of viral sequences; data on human mobility, contacts and behaviors; and experimental data), it is essential to develop methods that can jointly integrate these data in a coherent analytical framework. The development of an interdisciplinary perspective on epidemic dynamics will be key to improving our understanding of infectious disease spread and control.
Multistakeholder public-private collaborative initiatives
From the University of Oxford’s influential pairing with pharmaceutical company AstraZeneca to the leading collaboration between Pfizer and BioNTech, COVID-19 research and development partnerships revolutionized our response to COVID-19 (67, 68) and perhaps future pandemics. The COVID-19 crisis triggered a wave of international and intersectoral scientific collaborations between diverse sets of partners and sectors, thanks to unparalleled digital connectivity. Indeed, nearly one-third of SARS-CoV-2 vaccine candidates were developed through partnerships (68).
Such collaborative efforts enabled research to benefit from a broad and unprecedented pool of expertise, experience, and data, which will inspire interdisciplinary research collaborations for years to come. Lessons learned from the creation of these initiatives will be critical to advance the next generation of COVID-19 vaccine candidates and, in the longer term, to tackle challenges posed by neglected diseases and future pandemics. These lessons include the need for rapid data sharing, significant up-front funding, further global diversification of research and development and manufacturing efforts, and agreements with pharmaceutical entities that enable equitable access to vaccines in advance of clinical testing. A white paper published by the Coalition for Epidemic Preparedness Innovations (CEPI), in collaboration with the Africa Centres for Disease Control and Prevention and the International Federation of Pharmaceutical Manufacturers and Associations, details these lessons (67).
CEPI’s multisectoral approach is accelerating vaccine research and development not only for COVID-19 but also for Lassa fever, Nipah, Middle East respiratory syndrome (MERS), and other emerging infectious diseases. The European and Developing Countries Clinical Trials Partnership (EDCTP) is another public–public partnership between countries in Europe and sub-Saharan Africa, supported by the European Union. Like CEPI, it addresses major diseases endemic in African countries by developing clinical research capacities in those countries.
Indeed, public–private partnerships (PPPs) proved to be useful in accelerating the development of drugs and vaccines in several domains for which the pharmaceutical industry does not anticipate sufficient return on investment. The experiences of the Innovative Medicines Initiative, which gave rise to the Innovative Health Initiative (IHI), and of the Critical Path Institute in the United States demonstrate that PPPs allow us to combine the strength of the industry with the creativity present in academia for the benefit of patients with a variety of disorders suffering from insufficient investments, such as dementia, autism, and AMR (69). Projects focusing on pre-competitive research allowed unprecedented collaborations even between companies which are competing on the market (70). The search for novel biomarkers of efficacy and safety, and the establishment of platforms for early drug screening are typical examples of such pre-competitive research. Although PPPs have proven their usefulness, their modus operandi must be further improved by reducing administrative/bureaucratic burdens, avoiding sterile competition, and fostering more collaboration between partners.
Toward a global and inclusive approach to public health
The COVID-19 pandemic highlighted the key role public health science has to play during epidemics to ascertain and monitor the epidemiological situation, identify risk groups, and design and evaluate control measures. This clearly requires a better integration of public health science in health sciences. The introduction of innovative study designs can provide key complements to traditional surveillance systems. For example, in a context where many infections were not detected by surveillance, the Real-time Assessment of Community Transmission (REACT) study in the United Kingdom (71) measured viral circulation by randomly testing about 100,000 persons per month for 2 years. The study proved extremely informative for SARS-CoV-2 surveillance and research, and it is hoped that the design will be redeployed in other settings as well. It seems essential that countries develop and maintain solid capacity and infrastructures for public health during inter-pandemic periods to be able to respond more robustly to future pandemics.
Most importantly, the pandemic underlined the need for new approaches to address health inequalities. The COVID-19 pandemic was marked by the striking over-representation of socially disadvantaged populations in terms of disease burden. Economic, geographic, and ethnic variables doubled—or even tripled—death rates (72), warranting specific explanations and responses. People without housing, people who are incarcerated, and undocumented migrants were heavily impacted (8). Lessons must be learned from these disparities, including longer-term disparities in mortality due to economic disadvantage, as has been observed following previous economic crises (73, 74).
The lack of global solidarity, particularly in the distribution of vaccines to low-and middle-income countries was obvious, especially with regard to the African continent. Undoubtedly, it is of utmost importance to better integrate the specific needs of low- and middle-income countries in the strategies developed as part of the preparedness and response to pandemics and other health emergencies. This key lesson from COVID-19 is addressed more fully elsewhere (6–8).
The COVID-19 pandemic also demonstrated the critical importance of public understanding and support in the fight against infectious diseases, with wider implications for the future of medicine. Public support is dependent on the conveyance of the proper information at the proper time to the proper audience. Indeed, health literacy appears to be key to translating scientific advances into efficient preventive and therapeutic strategies. COVID-19 has revealed, irrespective of the level of economic development, an unexpected degree of scientific and sanitary “illiteracy” in the populations and in the media. As a result, the equal weight given to science-based information and irrational statements created a cognitive dissonance, opening avenues to conspiracy theories and fake news. It is time to restore the place of scientific and health education at all levels, starting from primary school levels.
Precision medicine for COVID-19 and beyond
Precision medicine is commonly viewed as a response to the phenotypic heterogeneity of diseases. It is about deciphering the molecular mechanisms underlying the specific pathological process within individuals to provide the “right treatment at the right time to the right patient”. From a clinical standpoint, it is important to consider precision medicine from the perspective of personalized medicine, which considers the individual characteristics of the patient. As stated by William Osler (1849–1919) more than 100 years ago, “The good physician treats the disease; the great physician treats the patient who has the disease".
Precision medicine’s proof of principle has come from its application in the treatment of cancer based on the identification of driver mutations or by gene correction for inherited diseases. Does the concept of precision medicine apply to an infectious disease affecting hundreds of millions of people and which kills millions of them? The new reality of COVID-19 showed us that this is the case. Indeed, as El-Sadr et al. recently discussed, now is the time to move away from population-wide universal recommendations toward a more tailored approach that considers the characteristics of people and the pathogen (75). In this section we explore some lessons learned from COVID-19 for the future of precision/personalized medicine.
Precision medicine during the COVID-19 pandemic
Understanding at the molecular level of mechanisms underlying host/virus interactions has been key to addressing the pathogenesis of COVID-19 disease, its treatment, and its prevention.
The phenotypic heterogeneity of COVID-19 was striking from the onset of the pandemic: it was immediately apparent that the same virus could be deadly in some individuals and yet go unnoticed in others. Although genetic factors controlling the type 1 interferon pathways as well as autoantibodies to type I interferon were found to explain some cases of severe disease in young and healthy individuals (76, 77), it rapidly became clear that old age was a major risk factor alongside obesity, type 2 diabetes, and other co-morbidities (78). These observations led to the development of targeted therapies for vulnerable individuals, including nirmatrelvir/ritonavir and anti-SARS-CoV-2 mAbs as mentioned above, and subsequently to the investigation of additional therapeutics (79).
Post-acute COVID-19 sequelae are drawing increasing attention because of their high incidence and public health impact. Although they are regrouped under the same denomination of long COVID, post-COVID-19 damages are very diverse and probably involve several mechanisms (80). Because of its huge phenotypic heterogeneity, long COVID is an area for which a precision medicine approach is much needed. In this vein, a recent study suggests that immune profiling combined with machine learning might be helpful by defining subgroups of patients with long COVID (81).
Precision medicine beyond COVID-19
Toward new classifications of patients with chronic diseases
While oncology is currently the main field where precision medicine is applied in practice, several patients with other non-communicable disorders will increasingly benefit from similar approaches, especially patients with immunoinflammatory disorders.
Dysregulated immune responses are critically involved in several chronic inflammatory disorders such as rheumatic diseases, inflammatory bowel and skin diseases, and multiple sclerosis. Sustained efforts have been made to develop immunotherapeutic approaches aimed at rebalancing immune responses and, thus, improving outcomes for patients. Notable successes have been attained, especially through the development of anti-cytokine therapies (82, 83). However, these successes were partial, with patient responses ranging from 25 to 60%. The main reason for this limited success is the heterogeneity of these disorders, which should be better qualified as syndromes with different possible causes involving the genome, the epigenome, and signaling pathways. This should lead to a new taxonomy of immune-inflammatory diseases that are presently often named according to clinical observations made several decades ago.
Immunotherapies will fulfill their promise only if biomarker-driven personalized immunotherapy becomes the standard of care. The emergence of cutting-edge omics technologies (from genomics to transcriptomics, proteomics, and metabolomics) has led to the discovery of novel pathways and biomarkers of disease, description of immune endotypes of various diseases, and ‘theranostics’ approaches to combine advanced diagnostics and therapies (84). A new drive to use advanced omics-based technologies combined with sophisticated systems biology integration to reclassify patients in a novel manner will transform our understanding of disease pathophysiology, as well as the way we will apply personalized immunotherapy. As discussed in the next section, this approach will be based on new knowledge in systems medicine and will take advantage of AI tools.
Among other diseases which might benefit from a new patients’ classification, one can mention type 2 diabetes and Alzheimer’s disease. In type 2 diabetes, patients clustered according to different variables associated with different outcomes might benefit from tailored management (85). In Alzheimer’s disease, it will be important to identify the subgroups of patients for whom beneficial or detrimental effects of novel therapies can be anticipated. It is already apparent that patients homozygous for the APOE4 gene should be managed with caution (86).
Targeting the microbiome
The COVID-19 pandemic introduced new challenges related to the complex interactions between commensal and pathogenic microbes, with direct and indirect ramifications on human health (87). On the one hand, a variety of COVID-related factors may contribute to compositional and functional alterations in the gut microbiome (dysbiosis). These changes include the presence of SARS-CoV-2 virus in the gastrointestinal tract, the systemic inflammation accompanying SARS-CoV-2 infection, effects of natural and vaccination-induced antiviral immune reactivity on the microbiome, and the effects of antibiotics, which are frequently administered to patients with severe COVID-19. On the other hand, there is evidence that the severity of COVID-19 is linked to alterations in the composition and function of the gut microbiota—alterations that persist even after viral clearance and symptomatic recovery (88). Indeed, a prospective study of patients with a spectrum of COVID-19 severity revealed that the gut microbiota composition at admission was associated with the development of long COVID-19 syndrome, and the microbiota composition was found to impact immune responses during COVID-19 and following vaccination (89).
These findings are not surprising given that, from birth to death, the gut microbial ecosystem plays major roles in host metabolic, immune, and neuroendocrine functions. The adult gut microbiome appears as a “signature” unique to everyone, and although it is considered quite resilient, the gut microbiome is constantly modulated by environmental factors, including nutrition and xenobiotic exposure (e.g., drugs, pollutants, and food additives) (90). Dysbiosis has been associated with several conditions, including diabetes and obesity (91–93), and gut microbiome characteristics can explain therapeutic responses in several settings, including cancer. This reveals the importance of the gut microbiome to personalized medicine (94). Several pharmacological approaches targeting the microbiome are under development, including precision probiotics optimized to colonize the gastrointestinal tract and commensal bacteria engineered to secrete health-promoting bioactive metabolites. One might therefore anticipate that the gut microbiome will continue to elicit major interest as a therapeutic target for an increased number of communicable and non-communicable diseases. Recent research even suggests that the gut microbiome represents an important pathway linking environmental exposures—in turn related to social, political, and economic forces—to health inequities (95).
The exposome concept
The clustering of severe COVID-19 cases in certain Italian regions revealed the impact of air pollution on the outcome of COVID-19 (96). Indeed, current efforts for improved responses and preparedness to new pandemics should include consideration of the environmental exposures on the outcome of infectious diseases, both in the short and long term.
Environmental insults occurring throughout the life course will have to be integrated into precision medicine approaches for non-infectious diseases as well. Taken together, they represent the exposome. It includes chemical factors of natural and anthropic origin, physical factors (such as temperature, noise, and ionizing and non-ionizing radiation), as well as psychosocial and behavioral factors (Figure 3) (5).
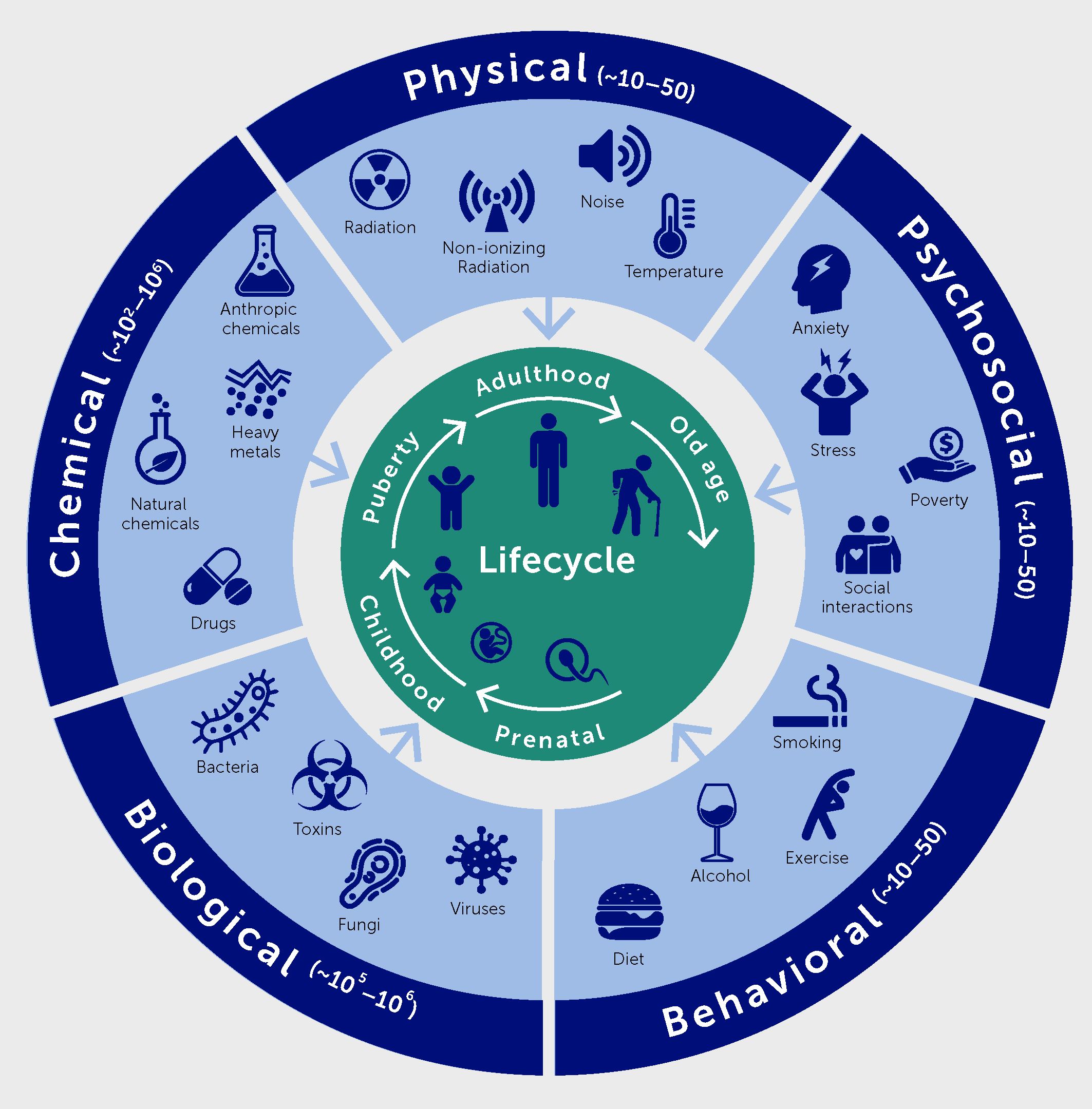
Figure 3 The exposome comprises five domains, namely physical, chemical, biological, psychosocial, and behavioral factors.
Research on the impact of the exposome on health include many aspects: i) the description of the different factors contributing to the exposome, their spatial and temporal variations, as well as their variations according to socioeconomic categories; ii) the identification of the impact of environmental factors on all components of health; iii) the identification of synergy or antagonism between environmental factors; iv) the understanding of the biological mechanisms underlying these effects; and v) the quantification of the corresponding health and societal burdens, in terms of attributable deaths, disease cases, disability-adjusted life-years lost, and possibly cost.
Regenerative medicine and genome-based therapies
The COVID-19 pandemic occurred while the field of regenerative medicine was in a renaissance after long years when hurdles to safe and efficacious treatments appeared difficult to overcome. A growing list of previously incurable diseases, mainly but not exclusively of a genetic nature, can indeed be treated with cell- and gene-therapy products that have the potential to cure patients. Success stories in this field include therapies for congenital immune deficiencies, lysosomal storage diseases, spinal muscular atrophy, diseases of the retina, unilateral burns of the cornea, epidermolysis bullosa, hemophilias, and beta thalassemia (97). Most successful treatments are based upon either ex vivo gene therapy using hematopoietic and epithelial stem cells and retro- and lentivectors (98, 99) or in vivo gene therapy using adeno-associated virus-based vectors (AAV). Despite successes, caution is still warranted when considering these ultimate forms of precision medicine. For example, direct in vivo delivery of adeno-associated vectors has shown excellent results in eye diseases (100) and spinal muscular atrophy (101), but severe adverse events have been observed in several cases of muscular dystrophy (102).
Additional major problems remain to be addressed before regenerative medicine can become mainstream. For in vivo gene therapy, immunogenicity, duration of therapeutic effect—potentially no longer a problem with genome editing (103)—and production costs greatly limit its widespread application. For ex vivo gene therapy, success has been limited to tissues in which ablation of diseased cells is possible (e.g., epithelia and bone marrow), subsequently allowing high engraftment of genetically corrected cells—something that is impossible for the brain, heart, and other critical organs (104). In the treatment of lysosomal storage diseases, hematopoietic stem cells (HSC) are being genetically engineered to overexpress specific lysosomal enzymes to correct deficiencies (105). In vivo genome editing based on second generation CRISPR-Cas technology (i.e., base or prime editing) shows interesting promise of application to the treatment of numerous diseases. So far, ex vivo knockdown of genetic elements in hematopoietic stem cells by CRISPR-Cas9 successfully revigorated fetal hemoglobin expression by red blood cells in patients with sickle cell disease—a remarkable achievement (106).
Wide access to new therapeutic solutions for regenerating diseased tissues and organs is hampered by economic hurdles that must still be addressed. Even when successful cell/gene therapy products receive marketing authorization, extremely high costs prevent access to many patients who should benefit from it. Furthermore, these products may be abruptly withdrawn from the market—owing either to barriers to their adoption by national health systems or insurance companies, or to abrupt changes in corporate strategy—depriving patients of life-saving therapies (107). Solving these issues is crucial for the continuous progress of regenerative medicine as a field (108, 109).
Creation of human leukocyte antigen (HLA)-depleted, immune-privileged lines from embryonic or induced pluripotent stem cells (110) may lead to banks of universal donor cells, moving beyond personalized medicine and dramatically cutting costs and prices while enhancing safety. With respect to regulation and economics, early engagement of relevant stakeholders during product development and transparent negotiation of prices may prevent reimbursement problems. Furthermore, if an authorized, efficacious product is abandoned by the manufacturer, government-supported non-profit facilities should step in to take over production.
So far, only patients from rich countries might have the opportunity to get access to regenerative medicine and genome-based therapies. To prevent further deepening of health inequalities across populations and across countries, it is of utmost importance to design new international programs to ensure that whoever they are and wherever they live, patients can benefit from those disruptive therapies derived from recent scientific advances.
Leveraging systems medicine: artificial intelligence, digital medicine, and novel clinical trial designs
The COVID-19 pandemic emphasized how public health decisions depend on numbers: disease incidences, positivity rates, hospitalization rates, and mortalities became daily fodder not only for decision-makers and public health managers but for a broad general audience. It also highlighted the need for sound analytical methods to interpret a range of complex data, both to support policymaking in a context of high uncertainty and to gain critical scientific insights.
The pandemic also provided a unique opportunity to utilize our increasing ability to gather “big data” from individuals and factor it into precision medicine. Advanced computational analytics and AI can facilitate improved apprehension of personalized reactiveness to environmental insults such as COVID-19 infection. Integrating clinical features, genomics, immune factors, microbiome data, and a variety of other anthropometric measures into a “systems medicine” approach may enable existing medical interventions to be repurposed in a personalized fashion (111). As the field of systems medicine develops, issues related to reproducibility, data availability, and ethical concerns will need to be resolved (112). It is also essential that racial and ethnic differences are fully considered in the collection and analysis of data. Challenges notwithstanding, systems medicine may enable us to better adapt currently available and new diagnostics and treatments to the individual while minimizing failures and adverse effects.
Artificial intelligence during and after the COVID-19 pandemic
AI has become a core technology of the fourth industrial revolution, if not representing the fifth industrial revolution on its own, with enormous uptake across science and modern life. The World Health Organization, the Organisation for Economic Co-operation and Development, and others quickly highlighted the potential of AI in managing COVID-19, the first pandemic in a hyperconnected world, and in improving future preparedness and resilience (113).
By early November 2020, about 9 months into the pandemic, only around 300 of >55,000 scientific articles on COVID-19 listed in PubMed included the terms AI or machine learning. By January 2021, the number of COVID-19 articles mentioning AI had quadrupled while total COVID-19 articles rose by only 60% (T. Hartung, unpublished observations). As the pandemic progressed and more data became available, AI started to make contributions. A systematic review of studies of AI applications in COVID-19 included 78 studies, some with impressive predictivities: 46 studies used AI to support diagnosis, 14 to evaluate prognosis, nine to predict the epidemiology of the pandemic, eight to explore potential drugs, and one to predict vaccine targets (114). The delayed application of AI early in the pandemic reflected the need for sufficient data collection to allow these techniques to be applied (115). Moreover, “big data” means more than many data; it is defined by volume, variety, and velocity, which represent three of the five “Vs” (the others being veracity and value). While volume and velocity were present early in the pandemic, the variety of interconnected data available was limited. Most data lacked metadata—the connections between datapoints and information describing how they were generated. For example, early in the pandemic the generation of many false positives and some false negatives by diagnostics of varying quality (116) made accurate monitoring of infection rates impossible.
AI revolutionized drug and vaccine design in the fight against COVID-19 by facilitating the rapid identification of potential drug candidates, enabling the discovery of new drugs through computational methods. In the future, artificial neural networks will certainly represent important tools in the identification of novel antivirals and other drug candidates (117). Most importantly, AI can be instrumental for drug repurposing, which consists of the use of existing drugs in novel clinical indications. As the safety profile of these drugs is already established, their development and regulatory approval are greatly accelerated. This was the case during the COVID-19 pandemic for baricitinib, an antirheumatic drug identified through AI to also target the SARS-CoV-2 virus (118). The field of drug safety itself will also be increasingly impacted by AI, as conventional toxicology studies in animals will progressively be replaced by in silico tools (119, 120).
AI also serves science through the mining of scientific publications. This advance is particularly relevant to COVID-19 because the pandemic triggered broad open access to scientific information on this topic. For example, the COVID-19 Open Research Dataset Challenge (CORD-19) has amassed over 1 million COVID-19-related articles (400,000 full-text) for researchers to use in AI techniques to generate new insights (121). This database was used, for example, to identify the role of blood glucose levels in the severity of COVID-19 (122), a feat of analysis that could never be achieved by human beings (123). While open access is, unfortunately, still the exception and not the rule, this project showcases its potential. However, we are still far away from machine-readable publishing, standardized data and metadata sharing, and other standards that would boost the natural language processing of articles to take full advantage of AI in handling vast quantities of scientific information. There is no reason to stop at scientific articles—databases and the gray literature of the internet could similarly contribute to the minable knowledge base.
Despite its promise, AI also raises concerns about fairness, reliability, accountability, privacy, transparency, and safety. COVID-19 is the perfect illustration of these issues, as the dominance of certain countries in terms of AI might have deepened differences in pandemic responses. Flaws in the data used for AI will translate into biases in results—to find an answer, that answer must be in the data. A major problem related to AI is its “black box” nature, meaning its methods are not readily open to scrutiny. Explainable, or “white box”, AI is still in its infancy. For decisions in which human life is at stake, the limitations of AI may be difficult to accept, but the choice not to leverage this powerful technology would cost lives as well. To take full advantage of AI, it is essential to improve AI literacy among those using its results as well as the embedded ethics and transparency among data curators and miners.
Digital medicine
Digital medicine is a potent area of growth and innovation, with the potential to strengthen the resilience of the healthcare system and reduce healthcare costs, while concurrently empowering patients to manage long-term health conditions and improve health outcomes (124) (see https://www.dimesociety.org).
The overwhelming impact of COVID-19 during the height of the pandemic has emphasized the importance of increasing healthcare system resilience. To avoid a repeat scenario in the face of future pandemics, we must reduce our absolute reliance on face-to-face contact with patients when managing their health trajectories (125). During the pandemic period, telemedicine has been increasingly adopted across the world to compensate for the severe reduction in face-to-face contact (126).
Although digital medicine spans a diversity of products and services, the greatest opportunity for impact may lie with digital therapeutics (see https://dtxalliance.org). Digital therapeutics are digital tools, often apps but sometimes coupled with portable or wearable sensors, that provide patients with real-time feedback on the status of a disease (such as continuous blood glucose monitoring), coupled with smart guidance on what to do (what dose of insulin to take and when, for example, or possibly even automating insulin delivery). Digital therapeutics can also detect deteriorations in various parameters and alert patients or health professionals to these changes. For example, home monitoring of heart failure has been shown to improve cardiac function over conventional clinic visits, reduce hospitalizations, and improve quality of life (127, 128). Such tools are becoming more reliable, convenient, and inexpensive. The adoption of digital therapeutics across multiple disease areas increased by 38-fold in the United States during the COVID-19 pandemic (Figure 4) (129), and a substantial increase in digital health research was evident (Figure 1). The global digital health market is projected to grow by a compound annual growth rate of around 28% over the coming decade (130). However, it remains challenging for health systems to adopt digital therapeutics. Several countries have introduced formal assessment frameworks and policies that allow digital health tools to be “prescribed” or for patients to be directly reimbursed for using them, such as the Digitale Gesundheitsanwendungen (DiGA) model in Germany (131). Indeed, a recent study comparing the digitalization of healthcare systems revealed important inequalities among European countries (132). This digital gap should be addressed as a priority.
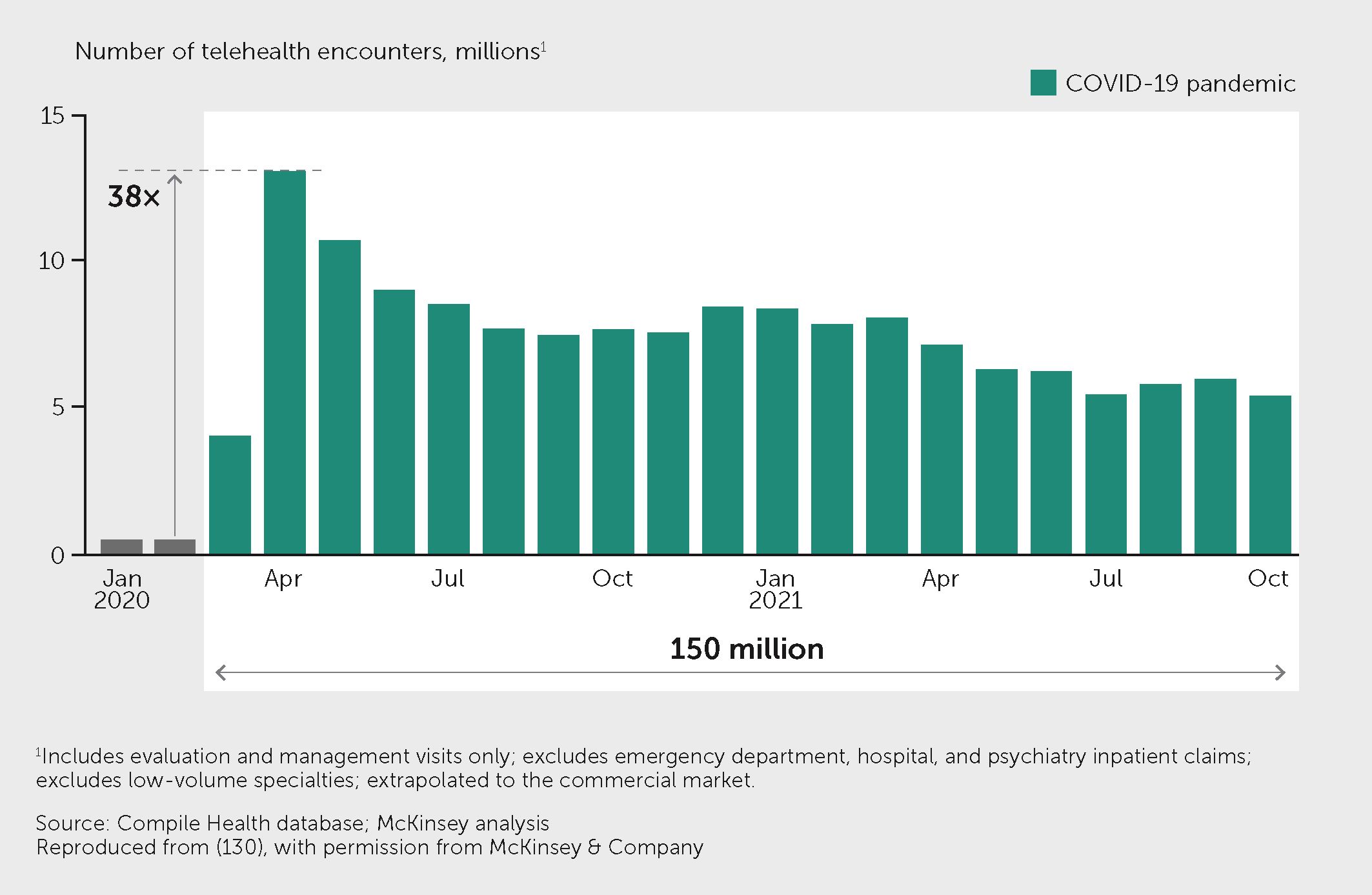
Figure 4 Telehealth use increased rapidly during the pandemic. Figure shows the telehealth encounters in the United States from February 2020 to October 2021, at which point the level of telehealth visits remained more than 1,300% higher than before the pandemic. Exhibit from (129): “The next frontier of care delivery in healthcare”, March 2022, McKinsey & Company, www.mckinsey.com. Copyright (c) 2024 McKinsey & Company. All rights reserved. Reprinted by permission.
More must be done to help digital therapeutics developers generate the effectivity and economic evidence required by these frameworks (133) and to support the health-system transformations needed to transition to digital therapeutics, which include modernizing care pathways, educating patients about how to use them and become more autonomous, and educating health professionals about how to be care facilitators as well as care providers. There is an understandable professional concern about the increasing workload that could result from digital therapeutics, and we should look to AI to help triage and filter incoming data streams and determine the necessity for professional intervention. Trustworthy health record data is the critical fuel for well-functioning digital medicine, so health systems must increase their investment in capturing and communicating high-quality and interoperable health data (134, 135).
The use of telehealth, digital therapeutics, and other forms of remote-care services are vital components of any strategy to increase the resilience of health systems and society. Not only will such methods reduce the dependence on face-to-face contacts in the case of future pandemics, but they will also reduce healthcare delivery costs, improve health outcomes, and enhance both the convenience and empowerment of the patient experience.
Novel clinical trial designs
The randomized controlled trial remains the gold standard for generating evidence to support clinical decision-making by regulators, clinicians, and patients. Data from such trials have advanced the understanding of the benefits and risks associated with the use of medical products. In recent decades, however, clinical trials have become prohibitively expensive, slow to finish, and are often under-enrolled—presenting physicians and patients with major gaps in knowledge about the appropriateness of treatments. The emergence of the COVID-19 pandemic highlighted these issues, both in terms of the pandemic’s effect on new and ongoing clinical trials and on the implications of the current trial infrastructure for the development of COVID-19 treatments.
Traditional “brick-and-mortar” clinical trials were difficult to initiate or to continue during the early stages of the pandemic. From February to May 2020, the monthly activation of United States-based clinical studies made up only 57% of the expected estimate, while the initiation of non-United States-based studies decreased by 27% (136). From June 2020 onward, numbers of phase II and III interventional trials in the United States returned to the levels seen in 2019, while in Europe, although the initial decrease was less pronounced, trial numbers did not rebound to 2019 levels until February 2021 (137). Disruptions in multiple aspects of clinical trials—from shifts in focus and resources to COVID-19, to decreased enrollment and adaptation of protocols to reflect social distancing measures—slowed the progress of clinical research and drug development, ultimately negatively impacting patient care.
Early in the pandemic, many clinical COVID-19 treatment trials began swiftly and were widespread (138). Yet, analysis showed that <5% of clinical trial arms would be capable of delivering actionable results in terms of regulatory decision-making regarding the safety and efficacy COVID-19 treatments owing to low randomization rates and underpowered outcome data. This tells us that, in the face of a pandemic or other public health crises, our current trial infrastructure is insufficient.
In response to the shortcomings in the clinical trial infrastructure uncovered by the COVID-19 pandemic, newer clinical trial designs were more widely adopted, including the use of master protocols. Master protocols use a single trial design infrastructure to efficiently evaluate multiple treatments concurrently in a disease (139). While most master protocols have been used to study cancer treatments, they have also been used to evaluate the efficacy and safety of COVID-19 treatments (140). The RECOVERY study was one of the first master protocols to unequivocally demonstrate a 30% reduction in death when dexamethasone was administered in hospitalized patients with severe respiratory symptoms (141). Following that success, efficacy and safety determinations were made for nine additional medical products. Both the REMAP-CAP trial and the I-SPY COVID trial also made helpful treatment discoveries quickly (142, 143). In March 2022, the United States Food and Drug Administration (FDA) published a guidance document for the industry containing advice for the use of master protocols to study drugs or biologics for cancer treatment (144). Wider use of master protocols could serve to increase the overall efficiency of clinical trials, expediting the development of treatments not only for cancer and COVID-19 but also for emerging infectious diseases and future public health crises.
Another modification increasingly adopted since the start of the COVID-19 pandemic involves decentralized clinical trials, which broaden methods of participation and decrease or eliminate the need for patients to travel to site-based trials. During the pandemic, a shift to incorporating digital technologies, including smartphones and wearable health devices, has allowed critical research to continue while giving study participants more choices and the ability to participate while adhering to social-distancing practices. In addition to being more patient-centric, decentralized trials may serve to increase the resiliency of the clinical trials system in the face of future pandemics or other public health emergencies.
In summary, the COVID-19 pandemic has clearly demonstrated that changes must be made to the structure and procedures of clinical trials to improve both the efficiency of trials and their resiliency in the face of public health crises. Advances in trial design, including master protocols and decentralized trials, should continue to be adopted across the clinical trials landscape. Such advances could boost the production of evidence required for critical health-related decision making, which will ultimately improve, prolong, or even save the lives of untold numbers of patients.
Promoting an interdisciplinary future for medicine
As underlined throughout this article, the COVID-19 pandemic highlighted the importance of novel approaches to manage global health crises, integrating knowledge and skills in various fields including virology, immunology, neurosciences, clinical medicine, digital healthcare, and public health. Besides science and healthcare, policy, economics, and social and behavioral sciences are also essential in addressing the complex challenges posed by pandemics. Integration means more than juxtaposing expertise from different fields. It is about developing a joint interdisciplinary approach to solve complex issues that cannot be successfully addressed within a single discipline. Scientists engaged in these interdisciplinarity endeavors must accept having to work outside their comfort zones to reach a common objective. Interdisciplinarity underlies the concept of convergence developed in a seminal report of the Massachusetts Institute of Technology (MIT); this defined convergence science as “an approach to problem solving that integrates expertise from life sciences with physical, mathematical, and computational sciences, as well as engineering, to form comprehensive frameworks that merge areas of knowledge from multiple fields to address specific challenges” (145). Convergence builds on fundamental progress made within individual disciplines and cuts across disciplinary boundaries in these fields (146, 147). Indeed, it goes beyond collaboration: as further explained in the MIT report, “Convergence is the integration of historically distinct disciplines and technologies into a unified whole that creates fundamentally new opportunities for life science and medical practice.” Unfortunately, few initiatives were launched to foster interdisciplinarity and Convergence since the publication of the MIT report in 2016. The turmoil elicited by the COVID-19 pandemic offers the impetus and opportunity to implement the major changes that are needed to translate these concepts into novel research and innovation approaches that are implemented widely in practice (Figure 5). Since the ultimate goal of health research is to translate scientific advances into products, services, and tools for the benefit of citizens, interdisciplinarity should extend across the different healthcare stakeholders, including the regulatory agencies in charge of the approval of novel pharmaceuticals. Indeed, regulatory science is interdisciplinary by nature.
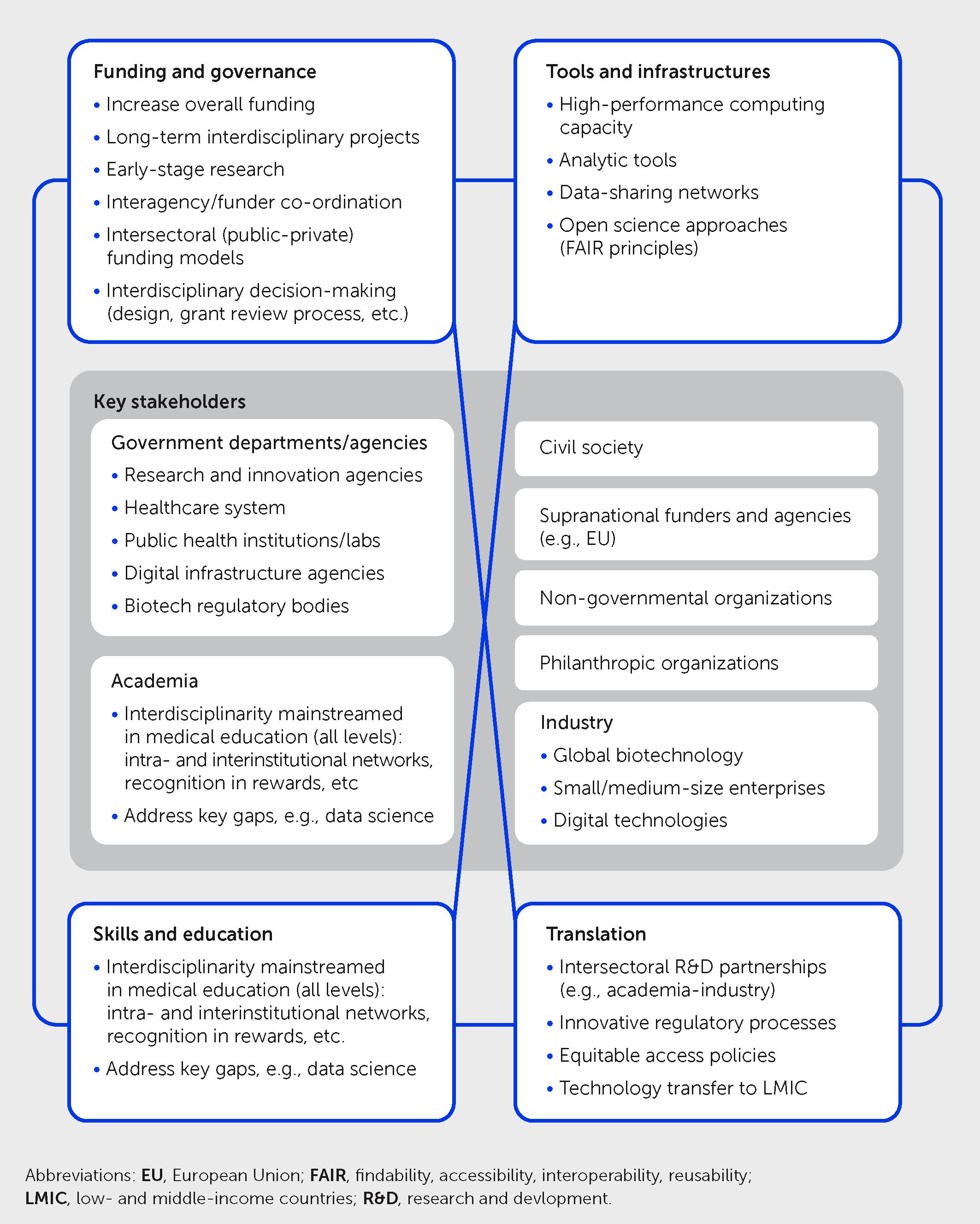
Figure 5 Priorities for promoting interdisciplinarity in future health sciences research. Schematic illustrates four key priority areas for multistakeholder action to foster greater interdisciplinarity to explore new frontiers of medicine and address future health challenges, including pandemics—namely in governance and funding, tools and infrastructures, skills and education, and translation. This framework takes into account previous convergence science recommendations by the Massachusetts Institute of Technology (145) and the United States National Academies (148) together with COVID-19 learnings and other developments.
All these actions require radical changes in the governance and funding of health research. As of today, research is still very much organized in silos, and projects of interdisciplinary nature are often considered to be not focused enough to get significant support (149). Approaches to promote interdisciplinary research in the future should include greater interagency/inter-funder co-ordination, interdisciplinarity participation in the processes of research programming and decision-making, and novel inter-sectoral collaborations—learning from those developed during COVID-19. Likewise, scientists with interdisciplinary skills often do not get the credit and the promotion that they deserve. There is a clear need to engage in a profound reform of governmental research agencies, especially in Europe, which lags behind the United States both in terms of global funding and allocation of resources dedicated to health research.
Finally, and crucially, lessons must be learned from the pandemic about how (interdisciplinary) science can best inform policy. A recent reevaluation (66) of the evidence relating to an earlier set of social and behavioral science insights that influenced COVID-19 policies (150) underscored the need for approaches that ensure better communication and collaboration between decision-makers and professionals and all relevant academia to ensure that insights generated by research are translated into evidence-based practice guidance and policies.
Conclusion
The future of medicine will be shaped by the convergence of biomedical sciences with several other disciplines. Interdisciplinary approaches will be essential to foster both precision medicine tailored to the individual characteristics of each patient and public health for the benefit of large populations. This new vision should be implemented in low- and middle-income countries as well as in rich countries to make medical science of the future a science for all.
Statements
Author contributions
SC: Writing – original draft, Writing – review & editing. GC: Writing – original draft, Writing – review & editing. ND: Writing – original draft, Writing – review & editing. EE: Writing – original draft, Writing – review & editing. DF: Writing – original draft, Writing – review & editing. AF: Writing – original draft, Writing – review & editing. TH: Writing – original draft, Writing – review & editing. DK: Writing – original draft, Writing – review & editing. MN: Writing – original draft, Writing – review & editing. JN: Writing – original draft, Writing – review & editing. RR: Writing – original draft, Writing – review & editing. MP: Writing – original draft, Writing – review & editing. MS: Writing – original draft, Writing – review & editing. PT: Writing – original draft, Writing – review & editing. GW: Writing – original draft, Writing – review & editing. PS: Writing – original draft, Writing – review & editing, Conceptualization, Project administration, Supervision, Visualization. MG: Writing – original draft, Writing – review & editing, Conceptualization, Project administration, Supervision, Visualization.
Data availability statement
The original contributions presented in the study are included in the article/supplementary materials, further inquiries can be directed to the corresponding author/s.
Funding
MN received funding from the European Research Council (ERC-Adv-833247). GW received funding from the Canadian Institutes for Health Research. The funders were not involved in the study design, collection, analysis, interpretation of data, the writing of this article, or the decision to submit it for publication.
Conflict of interest
PT is employed by Medable, Inc. EE is a scientific cofounder of DayTwo and BiomX and an advisor to Purposebio, Igen, Aposense, and Zoe in topics unrelated to this work. JN is the co-founder of AstriVax www.astrivax.com, a vaccine technology platform. MS declares shares in Sanofi S.A. The companies mentioned above were not involved in the study design, collection, analysis, interpretation of data, the writing of this article, or the decision to submit it for publication. The Frontiers Business Intelligence team assisted in the creation of Figure 1 in this article by selecting, grouping, filtering and analyzing data generated from Dimensions AI. Licensing restrictions apply. The filter configuration used by the Frontiers Business Intelligence team is available in the figure caption and the Appendix section below.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors MG, GW, MP, RR, MN, DK, TH, ND declared that they were an editorial board member of Frontiers at the time of submission. This had no impact on the peer review process and the final decision.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Pereira NL, Ahmad F, Byku M, Cummins NW, Morris AA, Owens A, et al. COVID-19: Understanding inter-individual variability and implications for precision medicine. Mayo Clin Proc (2021) 96:446–63. doi: 10.1016/J.MAYOCP.2020.11.024
2. Bambra C, Riordan R, Ford J, Matthews F. The COVID-19 pandemic and health inequalities. J Epidemiol Community Health (1978) (2020) 74:964–68. doi: 10.1136/JECH-2020–214401
3. Courtin E, Vineis P. COVID-19 as a syndemic. Front Public Health (2021) 9:763830. doi: 10.3389/FPUBH.2021.763830
4. Mendenhall E, Kohrt BA, Logie CH, Tsai AC. Syndemics and clinical science. Nat Med (2022) 28:1359–62. doi: 10.1038/S41591–022-01888-Y
5. Wild CP. Complementing the genome with an “exposome”: The outstanding challenge of environmental exposure measurement in molecular epidemiology. Cancer Epidemiol Biomarkers Prev (2005) 14:1847–50. doi: 10.1158/1055–9965.EPI-05–0456
6. The independent Panel for Pandemic Preparedness and Response. COVID-19: Make it the last pandemic (2021). Available at: https://theindependentpanel.org/covid-19-make-it-the-last-pandemic_final/.
7. Adeyi O, Yadav P, Kazatchkine M. Frontiers of medicine unveiled: equitable access is an imperative. Front Sci (2024) 2:1422583. doi: 10.3389/fsci.2024.1422583
8. Sachs JD, Karim SSA, Aknin L, Allen J, Brosbøl K, Colombo F, et al. The Lancet Commission on lessons for the future from the COVID-19 pandemic. Lancet (2022) 400:1224–80. doi: 10.1016/S0140–6736(22)01585–9
9. Wang H, Paulson KR, Pease SA, Watson S, Comfort H, Zheng P, et al. Estimating excess mortality due to the COVID-19 pandemic: A systematic analysis of COVID-19-related mortality, 2020–21. Lancet (2022) 399:1513–36. doi: 10.1016/S0140–6736(21)02796–3
10. Baker RE, Mahmud AS, Miller IF, Rajeev M, Rasambainarivo F, Rice BL, et al. Infectious disease in an era of global change. Nat Rev Microbiol (2022) 20:193–205. doi: 10.1038/S41579–021-00639-Z
11. Sinclair JR. Importance of a One Health approach in advancing global health security and the sustainable development goals. Rev Sci Tech (2019) 38:145–54. doi: 10.20506/RST.38.1.2949
12. Watson OJ, Barnsley G, Toor J, Hogan AB, Winskill P, Ghani AC. Global impact of the first year of COVID-19 vaccination: A mathematical modelling study. Lancet Infect Dis (2022) 22:1293–302. doi: 10.1016/S1473–3099(22)00320–6
13. Casadevall A. The mRNA vaccine revolution is the dividend from decades of basic science research. J Clin Invest (2021) 131(19):e153721. doi: 10.1172/JCI153721
14. Karikó K, Buckstein M, Ni H, Weissman D. Suppression of RNA recognition by toll-like receptors: The impact of nucleoside modification and the evolutionary origin of RNA. Immunity (2005) 23:165–75. doi: 10.1016/j.immuni.2005.06.008
15. Karikó K, Muramatsu H, Welsh FA, Ludwig J, Kato H, Akira S, et al. Incorporation of pseudouridine into mRNA yields superior nonimmunogenic vector with increased translational capacity and biological stability. Mol Ther (2008) 16:1833–40. doi: 10.1038/mt.2008.200
16. Geall AJ, Verma A, Otten GR, Shaw CA, Hekele A, Banerjee K, et al. Nonviral delivery of self-amplifying RNA vaccines. Proc Natl Acad Sci USA (2012) 109:14604–9. doi: 10.1073/PNAS.1209367109/-/DCSUPPLEMENTAL
17. Alameh MG, Tombácz I, Bettini E, Lederer K, Sittplangkoon C, Wilmore JR, et al. Lipid nanoparticles enhance the efficacy of mRNA and protein subunit vaccines by inducing robust T follicular helper cell and humoral responses. Immunity (2021) 54:2877–2892.e7. doi: 10.1016/j.immuni.2021.11.001
18. Altmann DM, Boyton RJ. COVID-19 vaccination: The road ahead. Science (2022) 375:1127–32. doi: 10.1126/science.abn1755
19. Rappuoli R, de Gregorio E, Del Giudice G, Phogat S, Pecetta S, Pizza M, et al. Vaccinology in the post-COVID-19 era. Proc Natl Acad Sci USA (2021) 118(3):e2020368118. doi: 10.1073/PNAS.2020368118
20. Pardi N, Hogan MJ, Porter FW, Weissman D. mRNA vaccines — A new era in vaccinology. Nat Rev Drug Discovery (2018) 17:261–79. doi: 10.1038/nrd.2017.243
21. Deng Z, Tian Y, Song J, An G, Yang P. mRNA vaccines: The dawn of a New Era of cancer immunotherapy. Front Immunol (2022) 13:887125/BIBTEX. doi: 10.3389/FIMMU.2022.887125/BIBTEX
22. Winston DJ, Mullane KM, Cornely OA, Boeckh MJ, Brown JW, Pergam SA, et al. Inactivated Varicella zoster vaccine in autologous haemopoietic stem-cell transplant recipients: An international, multicentre, randomised, double-blind, placebo-controlled trial. Lancet (2018) 391:2116–27. doi: 10.1016/S0140–6736(18)30631–7
23. Murray CJ, Ikuta KS, Sharara F, Swetschinski L, Robles Aguilar G, Gray A, et al. Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. Lancet (2022) 399(10325):629–55. doi: 10.1016/S0140–6736(21)02724–0
24. Centers for Disease Control and Prevention. COVID-19: US impact on antimicrobial resistance, special report 2022. Atlanta, GA: US Department of Health and Human Services, CDC (2022). Available at: https://www.cdc.gov/drugresistance/covid19.html.
25. Årdal C, Balasegaram M, Laxminarayan R, McAdams D, Outterson K, Rex JH, et al. Antibiotic development — Economic, regulatory and societal challenges. Nat Rev Microbiol (2020) 18:267–74. doi: 10.1038/s41579–019-0293–3
26. Outterson K. Estimating the appropriate size of global pull incentives for antibacterial medicines. Health Aff (Millwood) (2021) 40:1758–65. doi: 10.1377/HLTHAFF.2021.00688
27. O’Shea R, Moser HE. Physicochemical properties of antibacterial compounds: Implications for drug discovery. J Med Chem (2008) 51:2871–78. doi: 10.1021/jm700967e
28. Richter MF, Drown BS, Riley AP, Garcia A, Shirai T, Svec RL, et al. Predictive compound accumulation rules yield a broad-spectrum antibiotic. Nature (2017) 545:299–304. doi: 10.1038/NATURE22308
29. Hemmerling F, Piel J. Strategies to access biosynthetic novelty in bacterial genomes for drug discovery. Nat Rev Drug Discov (2022) 21:359–78. doi: 10.1038/s41573–022-00414–6
30. Privalsky TM, Soohoo AM, Wang J, Walsh CT, Wright GD, Gordon EM, et al. Prospects for antibacterial discovery and development. J Am Chem Soc (2021) 143:21127–42. doi: 10.1021/JACS.1C10200
31. Bush K. Game changers: New β-lactamase inhibitor combinations targeting antibiotic resistance in Gram-negative bacteria. ACS Infect Dis (2018) 4:84–7. doi: 10.1021/ACSINFECDIS.7B00243
32. Broncano-Lavado A, Santamaría-Corral G, Esteban J, García-Quintanilla M. Advances in bacteriophage therapy against relevant multidrug-resistant pathogens. Antibiotics (Basel) (2021) 10(6):672. doi: 10.3390/ANTIBIOTICS10060672
33. Hammond J, Leister-Tebbe H, Gardner A, Abreu P, Bao W, Wisemandle W, et al. Oral nirmatrelvir for high-risk, nonhospitalized adults with Covid-19. N Engl J Med (2022) 386:1397–408. doi: 10.1056/NEJMoa2118542
34. Geraghty RJ, Aliota MT, Bonnac LF. Broad-spectrum antiviral strategies and nucleoside analogues. Viruses (2021) 13(4):667. doi: 10.3390/V13040667
35. Liang J, Wu Y, Lan K, Dong C, Wu S, Li S, et al. Antiviral PROTACs: Opportunity borne with challenge. Cell Insight (2023) 2:100092. doi: 10.1016/J.CELLIN.2023.100092
36. Weinreich DM, Sivapalasingam S, Norton T, Ali S, Gao H, Bhore R, et al. REGN-COV2, a neutralizing antibody cocktail, in outpatients with Covid-19. N Engl J Med (2021) 384:238–51. doi: 10.1056/NEJMoa2035002
37. Levin MJ, Ustianowski A, De Wit S, Launay O, Avila M, Templeton A, et al. Intramuscular AZD7442 (Tixagevimab–Cilgavimab) for prevention of covid-19. N Engl J Med (2022) 386:2188–200. doi: 10.1056/NEJMoa2116620
38. Crowe JE. Human antibodies for viral infections. Annu Rev Immunol (2022) 40:349–86. doi: 10.1146/annurev-immunol-042718-041309
39. Casadevall A, Focosi D. SARS-CoV-2 variants resistant to monoclonal antibodies in immunocompromised patients is a public health concern. J Clin Invest (2023) 133(6):e168603. doi: 10.1172/JCI168603
40. Sabin AP, Richmond CS, Kenny PA. Emergence and onward transmission of a SARS-CoV-2 E484K variant among household contacts of a bamlanivimab-treated patient. Diagn Microbiol Infect Dis (2022) 103:115656. doi: 10.1016/J.DIAGMICROBIO.2022.115656
41. Dacon C, Tucker C, Peng L, Lee CD, Lin TH, Yuan M, et al. Broadly neutralizing antibodies target the coronavirus fusion peptide. Science (2022) 377:728–35. doi: 10.1126/SCIENCE.ABQ3773
42. Sandritter T. Palivizumab for respiratory syncytial virus prophylaxis. J Pediatr Health Care (1999) 13:191–5. doi: 10.1016/S0891–5245(99)90039–1
43. Hammitt LL, Dagan R, Yuan Y, Baca Cots M, Bosheva M, Madhi SA, et al. Nirsevimab for prevention of RSV in healthy late-preterm and term infants. N Engl J Med (2022) 386:837–46. doi: 10.1056/NEJMoa2110275
44. Navalkele BD, Chopra T. Bezlotoxumab: An emerging monoclonal antibody therapy for prevention of recurrent Clostridium difficile infection. Biologics (2018) 12:11–21. doi: 10.2147/BTT.S127099
45. Tshiani Mbaya O, Mukumbayi P, Mulangu S. Review: Insights on current FDA-approved monoclonal antibodies against Ebola virus infection. Front Immunol (2021) 12:721328. doi: 10.3389/FIMMU.2021.721328
46. Abernathy ME, Dam KA, Esswein SR, Jette CA, Bjorkman PJ. How antibodies recognize pathogenic viruses: Structural correlates of antibody neutralization of HIV-1, SARS-CoV-2, and Zika. Viruses (2021) 13(10):2106. doi: 10.3390/V13102106
47. Kayentao K, Ongoiba A, Preston AC, Healy SA, Doumbo S, Doumtabe D, et al. Safety and efficacy of a monoclonal antibody against malaria in Mali. N Engl J Med (2022) 387:1833–42. doi: 10.1056/NEJMoa2206966
48. Troisi M, Marini E, Abbiento V, Stazzoni S, Andreano E, Rappuoli R. A new dawn for monoclonal antibodies against antimicrobial resistant bacteria. Front Microbiol (2022) 13:1080059. doi: 10.3389/fmicb.2022.1080059
49. Wu JT, Leung K, Leung GM. Nowcasting and forecasting the potential domestic and international spread of the 2019-nCoV outbreak originating in Wuhan, China: A modelling study. Lancet (2020) 395:689–97. doi: 10.1016/S0140–6736(20)30260–9
50. Salje H, Tran Kiem CT, Lefrancq N, Courtejoie N, Bosetti P, Paireau J, et al. Estimating the burden of SARS-CoV-2 in France. Science (2020) 369:208–11. doi: 10.1126/SCIENCE.ABC3517
51. Verity R, Okell LC, Dorigatti I, Winskill P, Whittaker C, Imai N, et al. Estimates of the severity of coronavirus disease 2019: A model-based analysis. Lancet Infect Dis (2020) 20:669–77. doi: 10.1016/S1473–3099(20)30243–7
52. Russell TW, Hellewell J, Jarvis CI, van Zandvoort K, Abbott S, Ratnayake R, et al. Estimating the infection and case fatality ratio for coronavirus disease (COVID-19) using age-adjusted data from the outbreak on the Diamond Princess cruise ship, February 2020. Euro Surveill (2020) 25(12):2000256. doi: 10.2807/1560–7917.ES.2020.25.12.2000256
53. O’Driscoll M, Ribeiro dos Santos G, Wang L, Cummings DAT, Azman AS, Paireau J, et al. Age-specific mortality and immunity patterns of SARS-CoV-2. Nature (2021) 590:140–5. doi: 10.1038/S41586–020-2918–0
54. Lemey P, Ruktanonchai N, Hong SL, Colizza V, Poletto C, Van den Broeck F, et al. Untangling introductions and persistence in COVID-19 resurgence in Europe. Nature (2021) 595:713–7. doi: 10.1038/S41586–021-03754–2
55. Wymant C, Ferretti L, Tsallis D, Charalambides M, Abeler-Dörner L, Bonsall D, et al. The epidemiological impact of the NHS COVID-19 app. Nature (2021) 594:40812. doi: 10.1038/S41586–021-03606-Z
56. Willem L, Abrams S, Libin PJK, Coletti P, Kuylen E, Petrof O, et al. The impact of contact tracing and household bubbles on deconfinement strategies for COVID-19. Nat Commun (2021) 12:1524. doi: 10.1038/S41467–021-21747–7
57. Ferretti L, Wymant C, Kendall M, Zhao L, Nurtay A, Abeler-Dörner L, et al. Quantifying SARS-CoV-2 transmission suggests epidemic control with digital contact tracing. Science (2020) 368(6491):eabb6936. doi: 10.1126/SCIENCE.ABB6936
58. Tran Kiem C, Bosetti P, Paireau J, Crépey P, Salje H, Lefrancq N, et al. SARS-CoV-2 transmission across age groups in France and implications for control. Nat Commun (2021) 12:6895. doi: 10.1038/S41467–021-27163–1
59. Bubar KM, Reinholt K, Kissler SM, Lipsitch M, Cobey S, Grad YH, et al. Model-informed COVID-19 vaccine prioritization strategies by age and serostatus. Science (2021) 371:916–21. doi: 10.1126/SCIENCE.ABE6959
60. Tran Kiem C, Massonnaud CR, Levy-Bruhl D, Poletto C, Colizza V, Bosetti P, et al. A modelling study investigating short and medium-term challenges for COVID-19 vaccination: From prioritisation to the relaxation of measures. EClinicalmedicine (2021) 38:101001. doi: 10.1016/J.ECLINM.2021.101001
61. Tran Kiem C, Crépey P, Bosetti P, Levy Bruhl D, Yazdanpanah Y, Salje H, et al. Lockdown as a last resort option in case of COVID-19 epidemic rebound: A modelling study. Euro Surveill (2021) 26(22):2001536. doi: 10.2807/1560–7917.ES.2021.26.22.2001536
62. Bosetti P, Tran Kiem CT, Andronico A, Paireau J, Levy-Bruhl D, Alter L, et al. Impact of booster vaccination on the control of COVID-19 Delta wave in the context of waning immunity: Application to France in the winter 2021/22. Euro Surveill (2022) 27(1):2101125. doi: 10.2807/1560–7917.ES.2022.27.1.2101125
63. Paireau J, Andronico A, Hozé N, Layan M, Crépey P, Roumagnac A, et al. An ensemble model based on early predictors to forecast COVID-19 health care demand in France. Proc Natl Acad Sci USA (2022) 119:e2103302119. doi: 10.1073/PNAS.2103302119/-/DCSUPPLEMENTAL
64. Sonabend R, Whittles LK, Imai N, Perez-Guzman PN, Knock ES, Rawson T, et al. Non-pharmaceutical interventions, vaccination, and the SARS-CoV-2 delta variant in England: A mathematical modelling study. Lancet (2021) 398:1825–35. doi: 10.1016/S0140–6736(21)02276–5
65. Medley GF. A consensus of evidence: The role of SPI-M-O in the UK COVID-19 response. Adv Biol Regul (2022) 86:100918. doi: 10.1016/j.jbior.2022.100918
66. Ruggeri K, Stock F, Haslam SA, Capraro V, Boggio P, Ellemers N, et al. A synthesis of evidence for policy from behavioural science during COVID-19. Nature (2024) 625:134–47. doi: 10.1038/s41586–023-06840–9
67. COVAX: key learnings for future pandemic preparedness and response (2022). Available at: https://cepi.net/wp-content/uploads/2022/09/COVAX_Key-Learnings-for-Future-PPR_Sept-2022_FINAL_Updated-artwork.pdf.
68. Druedahl LC, Minssen T, Price WN. Collaboration in times of crisis: A study on COVID-19 vaccine R&D partnerships. Vaccine (2021) 39:6291–5. doi: 10.1016/J.VACCINE.2021.08.101
69. Goldman M. The Innovative Medicines Initiative: A European response to the innovation challenge. Clin Pharmacol Ther (2012) 91:418–25. doi: 10.1038/clpt.2011.321
70. Goldman M. Reflections on the innovative medicines initiative. Nat Rev Drug Discov (2011) 10:321–2. doi: 10.1038/nrd3434
71. Riley S, Ainslie KEC, Eales O, Walters CE, Wang H, Atchison C, et al. Resurgence of SARS-CoV-2: Detection by community viral surveillance. Science (2021) 372:990–5. doi: 10.1126/SCIENCE.ABF0874
72. Acosta AM, Garg S, Pham H, Whitaker M, Anglin O, O’halloran A, et al. Racial and ethnic disparities in rates of COVID-19–associated hospitalization, Intensive Care Unit admission, and in-hospital death in the United States from March 2020 to February 2021. JAMA Netw Open (2021) 4:e2130479. doi: 10.1001/JAMANETWORKOPEN.2021.30479
73. Aldridge RW, Story A, Hwang SW, Nordentoft M, Luchenski SA, Hartwell G, et al. Morbidity and mortality in homeless individuals, prisoners, sex workers, and individuals with substance use disorders in high-income countries: A systematic review and meta-analysis. Lancet (2017) 6736:1–10. doi: 10.1016/S0140–6736(17)31869-X
74. Benach J, Padilla-Pozo Á, Martínez-Herrera E, Molina-Betancur JC, Gutiérrez M, Pericàs JM, et al. What do we know about the impact of economic recessions on mortality inequalities? A critical review. Soc Sci Med (2022) 296:114733. doi: 10.1016/J.SOCSCIMED.2022.114733
75. El-Sadr WM, Vasan A, El-Mohandes A. Facing the new Covid-19 reality. N Engl J Med (2023) 388:385–7. doi: 10.1056/NEJMp2213920
76. Zhang Q, Bastard P, Liu Z, Le Pen J, Moncada-Velez M, Chen J, et al. Inborn errors of type I IFN immunity in patients with life-threatening COVID-19. Science (2020) 370(6515):eabd4570. doi: 10.1126/SCIENCE.ABD4570
77. Bastard P, Rosen LB, Zhang Q, Michailidis E, Hoffmann HH, Zhang Y, et al. Autoantibodies against type I IFNs in patients with life-threatening COVID-19. Science (2020) 370(6515):eabd4585. doi: 10.1126/SCIENCE.ABD4585
78. Clark A, Jit M, Warren-Gash C, Guthrie B, Wang HHX, Mercer SW, et al. Global, regional, and national estimates of the population at increased risk of severe COVID-19 due to underlying health conditions in 2020: A modelling study. Lancet Glob Health (2020) 8:e1003–17. doi: 10.1016/S2214–109X(20)30264–3
79. Li G, Hilgenfeld R, Whitley R, De Clercq E. Therapeutic strategies for COVID-19: Progress and lessons learned. Nat Rev Drug Discov (2023) 22:449–75. doi: 10.1038/S41573–023-00672-Y
80. Davis HE, McCorkell L, Vogel JM, Topol EJ. Long COVID: Major findings, mechanisms and recommendations. Nat Rev Microbiol (2023) 21:133–46. doi: 10.1038/s41579–022-00846–2
81. Klein J, Wood J, Jaycox JR, Dhodapkar RM, Lu P, Gehlhausen JR, et al. Distinguishing features of long COVID identified through immune profiling. Nature (2023) 623:139–48. doi: 10.1038/s41586–023-06651-y
82. Atzeni F, Sarzi-Puttini P. Anti-cytokine antibodies for rheumatic diseases. Curr Opin Investig Drugs (2009) 10:1204–11.
83. Gubernatorova EO, Namakanova OA, Gorshkova EA, Medvedovskaya AD, Nedospasov SA, Drutskaya MS. Novel anti-cytokine strategies for prevention and treatment of respiratory allergic diseases. Front Immunol (2021) 12:601842. doi: 10.3389/FIMMU.2021.601842
84. Rajewsky N, Almouzni G, Gorski SA, Aerts S, Amit I, Bertero MG, et al. LifeTime and improving European healthcare through cell-based interceptive medicine. Nature (2020) 587:377–86. doi: 10.1038/S41586–020-2715–9
85. Ahlqvist E, Storm P, Käräjämäki A, Martinell M, Dorkhan M, Carlsson A, et al. Novel subgroups of adult-onset diabetes and their association with outcomes: A data-driven cluster analysis of six variables. Lancet Diabetes Endocrinol (2018) 8587:1–9. doi: 10.1016/S2213–8587(18)30051–2
86. Loomis SJ, Miller R, Castrillo-Viguera C, Umans K, Cheng W, O’Gorman J, et al. Genome-wide association studies of ARIA from the aducanumab Phase 3 ENGAGE and EMERGE studies. Neurology (2024) 102:e207919. doi: 10.1212/WNL.0000000000207919
87. Finlay BB, Amato KR, Azad M, Blaser MJ, Bosch TCG, Chu H, et al. The hygiene hypothesis, the COVID pandemic, and consequences for the human microbiome. Proc Natl Acad Sci USA (2021) 118:e2010217118. doi: 10.1073/pnas.2010217118
88. Sarkar A, Harty S, Moeller AH, Klein SL, Erdman SE, Friston KJ, et al. The gut microbiome as a biomarker of differential susceptibility to SARS-CoV-2. Trends Mol Med (2021) 27:1115–34. doi: 10.1016/J.MOLMED.2021.09.009
89. Yeoh YK, Zuo T, Lui GCY, Zhang F, Liu Q, Li AYL, et al. Gut microbiota composition reflects disease severity and dysfunctional immune responses in patients with COVID-19. Gut (2021) 70:698–706. doi: 10.1136/GUTJNL-2020–323020
90. Ahn J, Hayes RB. Environmental influences on the human microbiome and implications for noncommunicable disease. Annu Rev Public Health (2021) 42:277–92. doi: 10.1146/ANNUREV-PUBLHEALTH-012420–105020
91. Wang J, Qin J, Li Y, Cai Z, Li S, Zhu J, et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature (2012) 490:55–60:7418. doi: 10.1038/nature11450
92. Guinane CM, Cotter PD. Role of the gut microbiota in health and chronic gastrointestinal disease: Understanding a hidden metabolic organ. Ther Adv Gastroenterol (2013) 6:295–308. doi: 10.1177/1756283X13482996
93. Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology: Human gut microbes associated with obesity. Nature (2006) 444:1022–23. doi: 10.1038/4441022a
94. Rodriguez J, Hiel S, Neyrinck AM, Le Roy T, Pötgens SA, Leyrolle Q, et al. Discovery of the gut microbial signature driving the efficacy of prebiotic intervention in obese patients. Gut (2020) 69:1975–87. doi: 10.1136/GUTJNL-2019–319726
95. Amato KR, Arrieta MC, Azad MB, Bailey MT, Broussard JL, Bruggeling CE, et al. The human gut microbiome and health inequities. Proc Natl Acad Sci USA (2021) 118(25):e2017947118. doi: 10.1073/PNAS.2017947118
96. Perone G. Assessing the impact of long-term exposure to nine outdoor air pollutants on COVID-19 spatial spread and related mortality in 107 Italian provinces. Sci Rep (2022) 12:13317. doi: 10.1038/s41598–022-17215-x
97. Anguela XM, High KA. Entering the modern era of gene therapy. Annu Rev Med (2019) 70:273–88. doi: 10.1146/ANNUREV-MED-012017–043332
98. De Luca M, Aiuti A, Cossu G, Parmar M, Pellegrini G, Robey PG. Advances in stem cell research and therapeutic development. Nat Cell Biol (2019) 21:801–11. doi: 10.1038/S41556–019-0344-Z
99. Tucci F, Galimberti S, Naldini L, Valsecchi MG, Aiuti A. A systematic review and meta-analysis of gene therapy with hematopoietic stem and progenitor cells for monogenic disorders. Nat Commun (2022) 13:1315. doi: 10.1038/S41467–022-28762–2
100. Moore NA, Morral N, Ciulla TA, Bracha P. Gene therapy for inherited retinal and optic nerve degenerations. Expert Opin Biol Ther (2018) 18:37–49. doi: 10.1080/14712598.2018.1389886
101. Mercuri E. Spinal muscular atrophy: From rags to riches. Neuromuscul Disord (2021) 31:998–1003. doi: 10.1016/J.NMD.2021.08.009
102. Duan D. Lethal immunotoxicity in high-dose systemic AAV therapy. Mol Ther (2023) 31:3123–6. doi: 10.1016/J.YMTHE.2023.10.015
103. Stevanovic M, Piotter E, McClements ME, MacLaren RE. CRISPR systems suitable for single AAV vector delivery. Curr Gene Ther (2022) 22:1–14. doi: 10.2174/1566523221666211006120355
104. Cossu G, Birchall M, Brown T, De Coppi P, Culme-Seymour E, Gibbon S, et al. Lancet Commission: Stem cells and regenerative medicine. Lancet (2018) 391:883–910. doi: 10.1016/S0140–6736(17)31366–1
105. Penati R, Fumagalli F, Calbi V, Bernardo ME, Aiuti A. Gene therapy for lysosomal storage disorders: Recent advances for metachromatic leukodystrophy and mucopolysaccaridosis I. J Inherit Metab Dis (2017) 40:543–54. doi: 10.1007/S10545–017-0052–4
106. Frangoul H, Altshuler D, Cappellini MD, Chen YS, Domm J, Eustace BK, et al. CRISPR-Cas9 gene editing for sickle cell disease and β-thalassemia. N Engl J Med (2021) 384:252–60. doi: 10.1056/NEJMoa2031054
107. Driscoll D, Farnia S, Kefalas P, Maziarz RT. Concise review: The high cost of high tech medicine: Planning ahead for market access. Stem Cells Transl Med (2017) 6:1723–9. doi: 10.1002/SCTM.16–0487
108. Aiuti A, Pasinelli F, Naldini L. Ensuring a future for gene therapy for rare diseases. Nat Med (2022) 28:1985–8. doi: 10.1038/S41591–022-01934–9
109. De Luca M, Cossu G. Cost and availability of novel cell and gene therapies: Can we avoid a catastrophic second valley of death?: Can we avoid a catastrophic second valley of death? EMBO Rep (2023) 24:e56661. doi: 10.15252/EMBR.202256661
110. Thongsin N, Wattanapanitch M. Generation of B2M bi-allelic knockout human induced pluripotent stem cells (MUSIi001-A-1) using a CRISPR/Cas9 system. Stem Cell Res (2021) 56:102551. doi: 10.1016/J.SCR.2021.102551
111. Adlung L, Cohen Y, Mor U, Elinav E. Machine learning in clinical decision making. Med (2021) 2:642–65. doi: 10.1016/J.MEDJ.2021.04.006
112. Federici S, Suez J, Elinav E. Our microbiome: On the challenges, promises, and hype. Results Probl Cell Differ (2020) 69:539–57. doi: 10.1007/978–3-030–51849-3_20
113. Piccialli F, di Cola VS, Giampaolo F, Cuomo S. The role of artificial intelligence in fighting the COVID-19 pandemic. Inf Syst Front (2021) 23:1467–97. doi: 10.1007/S10796–021-10131-X
114. Wang L, Zhang Y, Wang D, Tong X, Liu T, Zhang S, et al. Artificial intelligence for COVID-19: A systematic review. Front Med (Lausanne) (2021) 8:704256. doi: 10.3389/fmed.2021.704256
115. Hartung T. Making big sense from big data. Front Big Data (2018) 1:5. doi: 10.3389/FDATA.2018.00005
116. Ioannidis JPA. Over- and under-estimation of COVID-19 deaths. Eur J Epidemiol (2021) 36:581–8. doi: 10.1007/S10654–021-00787–9
117. Timmons PB, Hewage CM. ENNAVIA is a novel method which employs neural networks for antiviral and anti-coronavirus activity prediction for therapeutic peptides. Brief Bioinform (2021) 22(6):bbab258. doi: 10.1093/BIB/BBAB258
118. Mehta P, Titanji BK. Baricitinib in COVID-19: A coming-of-age from artificial intelligence to reducing mortality. Lancet (2022) 400:338–9. doi: 10.1016/S0140–6736(22)01295–8
119. Hartung T. ToxAIcology - The evolving role of artificial intelligence in advancing toxicology and modernizing regulatory science. ALTEX Altern Anim Exp (2023) 40:559–70. doi: 10.14573/ALTEX.2309191
120. Hartung T. Artificial intelligence as the new frontier in chemical risk assessment. Front Artif Intell (2023) 6:1269932. doi: 10.3389/frai.2023.1269932
121. Beranová L, Joachimiak MP, Kliegr T, Rabby G, Sklenák V. Why was this cited? Explainable machine learning applied to COVID-19 research literature. Scientometrics (2022) 127:2313–49. doi: 10.1007/s11192–022-04314–9
122. Logette E, Lorin C, Favreau C, Oshurko E, Coggan JS, Casalegno F, et al. A machine-generated view of the role of blood glucose levels in the severity of COVID-19. Front Public Health (2021) 9:695139. doi: 10.3389/FPUBH.2021.695139
123. Hartung T. Evidence integration in the era of information flooding—The Advent of the comprehensive review. Front Public Health (2021) 9:763828. doi: 10.3389/fpubh.2021.763828
124. Stern AD, Bronneke J, Debatin JF, Hagen J, Matthies H, Patel S. Advancing digital health applications: Priorities for innovation in real-world evidence generation. Lancet Digit Health (2022) 4:E200–6. doi: 10.1016/S2589–7500(21)00292–2
125. Horgan D, Hackett J, Westphalen CB, Kalra D, Richer E, Romao M, et al. Digitalisation and COVID-19: the perfect storm. BioMed Hub (2020) 5:1341–63. doi: 10.1159/000511232
126. Nittari G, Savva D, Tomassoni D, Tayebati SK, Amenta F. Telemedicine in the COVID-19 era: A narrative review based on current evidence. Int J Environ Res Public Health (2022) 19:5101. doi: 10.3390/IJERPH19095101
127. Ware P, Ross HJ, Cafazzo JA, Boodoo C, Munnery M, Seto E. Outcomes of a heart failure telemonitoring program implemented as the standard of care in an outpatient heart function clinic: Pretest-posttest pragmatic study. J Med Internet Res (2020) 22:e16538. doi: 10.2196/16538
128. Koehler F, Koehler K, Prescher S, Kirwan BA, Wegscheider K, Vettorazzi E, et al. Mortality and morbidity 1 year after stopping a remote patient management intervention: Extended follow-up results from the telemedical interventional management in patients with heart failure II (TIM-HF2) randomised trial. Lancet Digit Health (2020) 2:e16–24. doi: 10.1016/S2589–7500(19)30195–5
129. Singhal S, Vinjamoori N, Radha M. The next frontiers of healthcare practice. McKinsey & Company (2022). Available at: https://www.mckinsey.com/~/media/mckinsey/industries/healthcare%20systems%20and%20services/our%20insights/the%20next%20frontier%20in%20health%20care/the-next-frontier-of-healthcare-delivery.pdf.
130. Global Industry Analysts, Inc. Digital therapeutics: Global strategic business report (2023). Available at: https://www.researchandmarkets.com/reports/4804793/digital-therapeutics-global-market-trajectory.
131. Lauer W, Löbker W, Höfgen B. Digital health applications (DiGA): Assessment of reimbursability by means of the “DiGA Fast Track” procedure at the Federal Institute for Drugs and Medical Devices (BfArM). Bundesgesundheitsblatt Gesundheitsforschung Gesundheitsschutz (2021) 64:1232–40. doi: 10.1007/s00103–021-03409–7
132. Majcherek D, Hegerty SW, Kowalski AM, Lewandowska MS, Dikova D. Opportunities for healthcare digitalization in Europe: Comparative analysis of inequalities in access to medical services. Health Policy (2024) 139:104950. doi: 10.1016/J.HEALTHPOL.2023.104950
133. Kalra D. Scaling up the Big Health Data ecosystem: Engaging all stakeholders!. Journal of the International Society for Telemedicine and EHealth (2020) 8(e16):1–5. doi: 10.29086/JISfTeH.8.e16
134. Lewy H, Barkan R, Sela T. Personalized health systems—Past, present, and future of research development and implementation in real-life environment. Front Med (Lausanne) (2019) 6:149/BIBTEX. doi: 10.3389/FMED.2019.00149/BIBTEX
135. Ahlqvist J, Kalliola M. How can digital therapeutics help Europe? SITRA (2021). Available at: https://www.sitra.fi/en/publications/how-can-digital-therapeutics-help-europe.
136. Unger JM, Xiao H. The COVID-19 pandemic and new clinical trial activations. Trials (2021) 22:1–5. doi: 10.1186/S13063–021-05219–3/TABLES/1
137. Lasch F, Psarelli EE, Herold R, Mattsson A, Guizzaro L, Pétavy F, et al. The impact of COVID‐19 on the initiation of clinical trials in Europe and the United States. Clin Pharmacol Ther (2022) 111:1093–02. doi: 10.1002/CPT.2534
138. Bugin K, Woodcock J. Trends in COVID-19 therapeutic clinical trials. Nat Rev Drug Discov (2021) 20:254–5. doi: 10.1038/D41573–021-00037–3
139. Woodcock J, LaVange LM. Master protocols to study multiple therapies, multiple diseases, or both. N Engl J Med (2017) 377:62–70. doi: 10.1056/NEJMra1510062
140. LaVange L, Adam SJ, Currier JS, Higgs ES, Reineck LA, Hughes EA, et al. Accelerating COVID-19 therapeutic interventions and vaccines (ACTIV): Designing master protocols for evaluation of candidate COVID-19 therapeutics. Ann Intern Med (2021) 174:1293–300. doi: 10.7326/M21–1269
141. RECOVERY Collaborative Group, Horby P, Lim WS, Emberson JR, Mafham M, Bell JL, et al. Dexamethasone in hospitalized patients with Covid-19. N Engl J Med (2021) 384:693–704. doi: 10.1056/NEJMoa2021436
142. Angus DC, Berry S, Lewis RJ, Al-Beidh F, Arabi Y, van Bentum-Puijk W, et al. The remap-cap (Randomized embedded multifactorial adaptive platform for community-acquired pneumonia) Study rationale and design. Ann Am Thorac Soc (2020) 17:879–91. doi: 10.1513/AnnalsATS.202003–192SD
143. Files DC, Aggarwal N, Albertson T, Auld S, Beitler JR, Berger P, et al. Report of the first seven agents in the I-spy COVID trial: A phase 2, open label, adaptive platform randomised controlled trial. EClinicalmedicine (2023) 58:101889. doi: 10.1016/j.eclinm.2023.101889
144. US Food and Drug Administration. Master protocols: Efficient clinical trial design strategies to expedite development of oncology drugs and biologics guidance for industry. Silver Spring, MD: FDA (2022). Available at: https://www.fda.gov/media/120721/download.
145. Sharp P, Hockfield S, Jacks T. Massachusetts Institute of Technology. Convergence: The future of health. Cambridge, MA: MIT (2016). Available at: https://www.convergencerevolution.net/2016-report.
146. Sharp P, Jacks T, Hockfield S. Capitalizing on convergence for health care. Science (2016) 352:1522–3. doi: 10.1126/science.aag2350
147. Dzau VJ, Ogilvie JL, McGinnis M. Improving health through convergence science: Reimagining our approach to solving the world’s biggest challenges. PNAS Nexus (2022) 1:pgac007. doi: 10.1093/PNASNEXUS/PGAC007
148. National Research Council. Convergence: facilitating transdisciplinary integration of life sciences, physical sciences, engineering, and beyond. Washington, DC: National Academies Press (2014). doi: 10.17226/18722
149. Bromham L, Dinnage R, Hua X. Interdisciplinary research has consistently lower funding success. Nature (2016) 534:684–7. doi: 10.1038/NATURE18315
150. Bavel JJV, Baicker K, Boggio PS, Capraro V, Cichocka A, Cikara M, et al. Using social and behavioural science to support COVID-19 pandemic response. Nat Hum Behav (2020) 4:460–71. doi: 10.1038/s41562–020-0884-z
Appendix
Additional methodological details for Figure 1 are as follows.
The 2008 Australian and New Zealand Standard Research Classification (ANZSRC) system was applied to identify all actual publications in relevant years in the field of “Medical and Health Sciences” within Dimensions, using machine learning to sort publications into Fields of Research. Expected publication numbers for 2020–2022 were calculated based on an exponential trendline fitted on the actual 2009–2019 annual publication numbers in relevant fields. All fields included in the analysis showed a reliable trend 2009–2019 (R-squared > 0.8). In the line charts, an estimated total publication volume for 2023 was used to correct for the indexation lag of publications. For each field, we looked at the difference in publication numbers between incomplete and complete data of the previous year and applied this difference as a correction factor to the 2023 publication volumes retrieved in the January 2024 data.
For the fields “Clinical sciences”, “Public health and health services,” “Oncology and carcinogenesis”, “Cardiorespiratory medicine and hematology”, and “Immunology”, field segmentation was based on subcategories of the ANZSRC 2008 Medical and Health Sciences Fields of Research. Publications were assigned to fields based on fractional counting. The segmentation of other fields was approximated by the identification of relevant terms in the abstracts of articles within the Medical and Health Sciences Field of Research. Terms used were as follows: for “Antimicrobials”, a combination of “antibiotic” or “antimicrobial” or “antiviral” or “antibacterial” or “antifungal” or “antiparasitic” and “treatment” or “therapy” or “use” or “drug”; “Vaccines and vaccination”, “vaccine” or “vaccination”; and “Digital medicine and telemedicine”, “digital medicine” or “digital health” or “telemedicine” or “telehealth”.
The proportion of articles that were COVID-related was calculated based on the appearance of SARS-CoV-2/COVID-19 terms within abstracts. Terms used were as follows: “coronavirus disease 2019” or “COVID” or “COVID-19” or “COVID-19 disease” or “SARS-COV-2” or “SARS-COV2” or “SARS-CoV-2/COVID” or “COVID-19 pandemic” or “COVID19” or “COVID-19 virus” or “coronavirus SARS-CoV2” or “coronavirus SARS-CoV-2”. Only articles without missing data on the terms present in their abstracts were used in the calculation determining the share of COVID-related articles. An average of 1% of articles per field had no terms data and were discounted from the calculation.
Analyses were performed by Frontiers using Dimensions data (until 1 January 2024).
Keywords: COVID-19, precision medicine, public health, digital health, clinical trial, pandemic, infectious diseases, systems medicine
Citation: Cauchemez S, Cossu G, Delzenne N, Elinav E, Fassin D, Fischer A, Hartung T, Kalra D, Netea M, Neyts J, Rappuoli R, Pizza M, Saville M, Tenaerts P, Wright G, Sansonetti P and Goldman M. Standing the test of COVID-19: charting the new frontiers of medicine. Front Sci (2024) 2:1236919. doi: 10.3389/fsci.2024.1236919
Received: 08 June 2023; Accepted: 30 April 2024;
Published: 23 May 2024.
Edited by:
Bruno Villoutreix, Hôpital Robert Debré, FranceReviewed by:
Michel Kazatchkine, Geneva Graduate Institute, SwitzerlandJohan Frostegard, Karolinska Institutet (KI), Sweden
Copyright © 2024 Cauchemez, Cossu, Delzenne, Elinav, Fassin, Fischer, Hartung, Kalra, Netea, Neyts, Rappuoli, Pizza, Saville, Tenaerts, Wright, Sansonetti and Goldman. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Michel Goldman, bWdvbGRtYW5AaTNoZWFsdGguZXU=
†MG and PS acted as lead authors; contributions among other authors were equal
 Simon Cauchemez
Simon Cauchemez Giulio Cossu
Giulio Cossu Nathalie Delzenne
Nathalie Delzenne Eran Elinav
Eran Elinav Didier Fassin
Didier Fassin Alain Fischer
Alain Fischer Thomas Hartung
Thomas Hartung Dipak Kalra
Dipak Kalra Mihai Netea
Mihai Netea Johan Neyts
Johan Neyts Rino Rappuoli
Rino Rappuoli Mariagrazia Pizza
Mariagrazia Pizza Melanie Saville
Melanie Saville Pamela Tenaerts
Pamela Tenaerts Gerry Wright
Gerry Wright Philippe Sansonetti
Philippe Sansonetti Michel Goldman
Michel Goldman