
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Food. Sci. Technol. , 19 July 2023
Sec. Food Safety and Quality Control
Volume 3 - 2023 | https://doi.org/10.3389/frfst.2023.1168896
 Gianluigi Ferri1*
Gianluigi Ferri1* Alessio Buonavoglia2
Alessio Buonavoglia2 Muhammad Farooq3
Muhammad Farooq3 Anna Rita Festino1
Anna Rita Festino1 Fausto Ruffini4
Fausto Ruffini4 Domenico Paludi1
Domenico Paludi1 Cristina E. Di Francesco3
Cristina E. Di Francesco3 Alberto Vergara1†
Alberto Vergara1† Camilla Smoglica3†
Camilla Smoglica3†In this study a multidisciplinary approach was applied in order to determine the diffusion of resistant bacteria and selected antibiotic resistance genes in antibiotic-free and conventional broiler farms. Litter samples coming from the two farming types and surface sponges obtained from carcasses at slaughterhouse level were screened by end-point PCR targeting specific resistance for tetracycline, ampicillin, sulfonamide, aminoglycoside, carbapenem, nitrofurantoin, vancomycin, quinupristin-dalfopristin, lincomycin, linezolid, chloramphenicol molecules. Microbiological investigations were conducted from the carcasses to determine phenotypical and genetic resistance patterns from pathogenic and commensal Gram-negative and Gram-positive strains. At farm level, catA1, sul2, blaTEM and aadA2 genes were amplified in all samples, while from carcasses the most representative genes were sul2, blaTEM, along with the vatD, relative to quinupristin-dalfopristin resistance. Gram-negative isolates included Aeromonas, Salmonella, Proteus spp. And Escherichia coli, while the Gram-positive were represented by Enterococcus strains. Phenotypical and genetic analysis revealed multidrug resistance patterns in Salmonella, E. coli and Serratia isolates, followed by the Enterococcus species. The comparison between antibiotic-free and conventional farming systems showed some difference regarding the distribution of resistance genes at farm level but no significance was obtained comparing the phenotypical resistance profiles of bacterial strains from both groups of samples, suggesting a poor influence of farming model on the diffusion of antibiotic resistance in poultry meat production chain.
The antimicrobial resistance (AMR) is a consolidated reality in many animals’ productive systems and represents a crucial One Health issue which involves human, animal, and environmental medicine. It is also strictly related to inappropriate antibiotic administrations in the zootechnic sector producing selective pressures for certain resistant bacterial strains (Hernando-Amado et al., 2019).
This condition poses the basis for a critical situation regarding drugs’ administration and their management. Consequently, it imposes further responsibilities to the pharma surveillance institutions with special attention focusing on the necessity to preserve any efficacious molecules for human patients, especially due to the spreading of multidrug (MDR) or pan-resistant nosocomial microorganisms (World Health Organization, 2022). For this reason, the European Food Safety Authority (EFSA) decided to include any antibiotic classes (as carbapenems, vancomycin, nitrofurans) in the so-called CIA (Critical Important Antimicrobials) list, including molecules which usage is indicated to the human medicine (EFSA Panel on Animal Health and Welfare et al., 2021). However, antibiotics result necessary, especially in the intensive breeding systems due to the high animal density. Indeed, in these realities, any environmental bio-chemical parameters (i.e., aw, pH, and macromolecular substrates, etc.) provide ideal conditions for pathogens and commensal multiplication in receptive hosts (EFSA Panel on Biological Hazards et al., 2021).
The AMR and antibiotic residues (usually directly related to inappropriate and massive usage) have consequently repercussions on the animal origin food matrices, representing potential risks for the final consumers’ health (Feng et al., 2016).
For these reasons, the European Legislator has introduced different Regulations: starting from the EU Reg. No. 37/2010, in which specific acceptable cut-offs were clearly defined, till the EU Reg. No. 625/2017, where for the first time, the fundamental role covered by the “environmental protection” was introduced (EFSA Panel on Animal Health and Welfare et al., 2021). High animal densities and the last-mentioned environmental aspects enforce possible horizontal transmissions of oligonucleotide resistance elements leading to the spreading of AMR in human microbiota known as antibiotic resistance genes (ARGs) drivers (EFSA Panel on Biological Hazards et al., 2021).
From the zootechnic perspective, poultry productions systems have revolutionized their approach regarding breeding phases resulting more efficient than mammals’ ones. Every year, poultry meat productive volumes increase due to the low carbon foot-print emission, if compared to the other animal species (Barthelmie, 2022).
The growing attention focused on sustainability has induced the main broiler producers to provide the so-called “Sustainability Report”, as indicated by the International Standard ISO 14001/2018, in order to certify their production systems as low environmental impact industries (EFSA Panel on Biological Hazards et al., 2021).
Indeed, the avian sector has further advantages, i.e., short productions’ times from the hatching to the carcasses, and consequently low economic losses that characterizes its trades, high nutritive value of poultry meat and fewer religious or social taboos on its consumption (Food and Agriculture Organization of the United Nations, 2013).
Nowadays, two kinds of poultry husbandries have been identified: the “old-system” or conventional ones C), in which antibiotics are administrated, for therapeutic purposes only, and on the other hand, the antibiotic-free (AF) ones (EFSA Panel on Animal Health and Welfare et al., 2021).The AF model avoids the antibiotic usage through the improvement of preventive measures, i.e., biosecurity, productive flows, vaccination programs, animal welfare, in accordance with the “Animal Health Law” EU Reg. No. 2016/429.
Published data regarding the effects of AF production system on the reduction of AMR spreading are still incomplete and controversial. Previous works aimed to compare the resistance patterns of selected bacterial species, as commensal Escherichia coli, Campylobacter and Salmonella isolates (Bailey et al., 2019; Bailey et al., 2020; Pesciaroli et al., 2020; Musa et al., 2021; Racewicz et al., 2022), while preliminary results were provided by the direct detection of ARGs (De Cesare et al., 2022; Farooq et al., 2022).
The aim of this research paper was to compare the distribution of antibiotic resistant bacteria and the relative genetic determinants (ARGs) between C and AF poultry meat productive chain.
From the farms to the slaughterhouse, applying a multidisciplinary approach coupling the direct amplification of resistance genes from litter and carcasses with the microbiological analysis of the isolates recovered from carcasses.
Sampling activities involved broiler antibiotic-free (AF) and conventional C) flocks selected based on farming size (18,000–28,000 animals) and geographical location (Central Italy). In AF flocks no antibiotic treatment was applied for the entire cycle of production, while in the conventional farming systems antimicrobial molecules (amoxicillin) were included for therapeutic purposes when necessary. Environmental temperature and humidity were set at 18°C–19°C and 60% of relative humidity. All flocks belonged to the same poultry industry characterized by an integrated supply chain, from hatchery to the slaughtering, including feed manufacturing process (Figure 1).
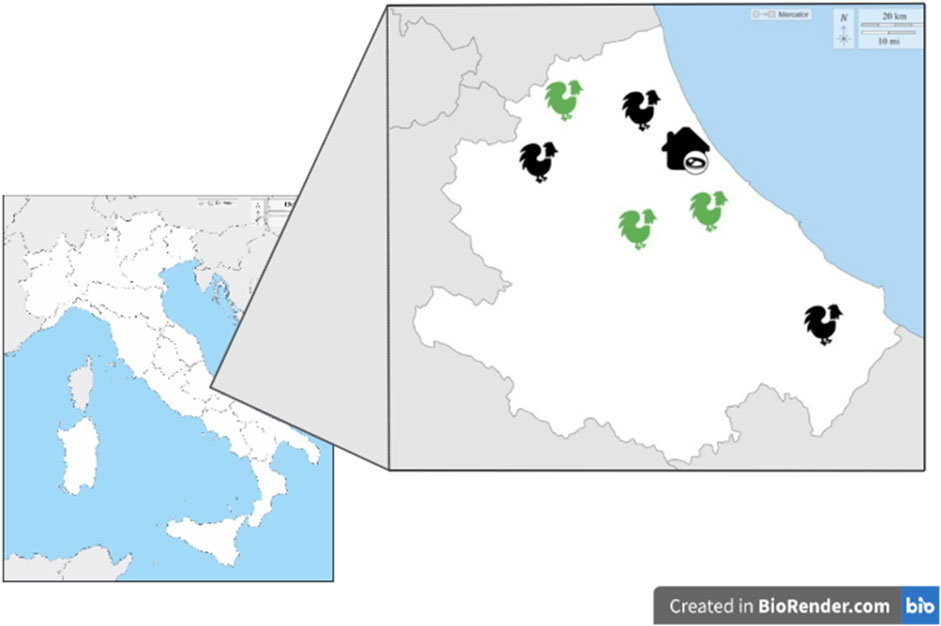
FIGURE 1. AF and C farms and slaughterhouse distributions in Central Italy. Green: antibiotic-free farm; Black: conventional farm.
For each flock sampling were performed at farming step recovering litter pools and, finally, at slaughtering step collecting surface sponges from carcasses. In order to make the fecal samples as homogeneous as possible, several pools of litter at different points in the shed (at the center and four corners) were collected and the sampling were performed twice, at 4–11 and 27–34 days of animals age for a total of 6 pooled environmental feces (3 from AF and 3 from C flocks). The carcasses were sampled at the end of slaughtering line in three different phases where a total of 60 samples were collected each time; 30 samples were taken from carcasses belonging to the AF farm and 30 samples were taken from carcasses of C farm, respectively. For each sampling activity, a systematic method was applied, selecting the carcasses at equal intervals (at the beginning, at the midway and at the ending) of the batch associated to each farm of origin. The whole chicken carcass was sampled by swabbing with the commercially available sponge (Polywipes® RPM Technology, LLC, Reno, NV, United States) (Capita et al., 2004), in agreement with the ISO 17604:2015. Each sponge was hydrated with 5 mL of Buffered Peptone Water (BPW; LIOFILCHEM® s.r.l., Roseto Degli Abruzzi, Italy) in a sterile plastic container (Polywipes). Five sponges, coming from the same farm, were grouped (25 mL), placed in sterile stomacher bags (BagMixer®, Interscience, Puycapel, Cantal, France) and diluted with 225 mL aliquots of PBS (1:10 dilution) broth, to obtain a pooled sample, for a total of 36 pooled samples (n. 18 from AF farms and n. 18 from C farms) (Table 1).
The litter samples and pooled carcasses were investigated by means of a culture-independent approach, directly performing a molecular screening of ARGs. Total DNA was obtained from all samples using the Maxwell® 11 Instrument (Promega, Italy) and Maxwell kit® 11 Tissue DNA Purification (Promega, Italy), following the manufacturer’s instructions. At the end of the extraction protocol, the quality and quantity of DNA were checked by using the spectrophotometer and fluorometer Denovix DS-11FX (Wilmington, United States) and 100 of high-quality bacterial DNA was recovered from each sample and stored to −20°C until the PCR testing.
For molecular screening, single or multiplex end-point PCR protocols were applied, combining the different primers pairs as reported in the Supplementary Table S1, for the simultaneous detection of ARGs specific for the commonly used in veterinary poultry practice molecules (tetracycline, lincomycin, aminoglycosides, quinolones; nitrofurans; sulfonamides; beta-lactams) and critically important antimicrobials for human medicine (chloramphenicol, vancomycin and carbapenems) (WHO, 2019), in accordance with the European Legislation. The reaction volumes ranged from 25 to 50 μL, the thermocycler settings were adapted, especially for the annealing time-temperature ratio, according to the different primers’ references. The PCR products were visualized by agarose gel electrophoresis. Appropriate ATCC bacterial strains were included as positive controls following the respective references, while sterile distilled water was used as negative one.
All pooled samples obtained from the carcasses were stomached for 60 s and, consequently, incubated at 37°C for 24 h.
After incubation, the aliquots were directly plated onto specific cultured media for each microorganism, conducing to a qualitative investigation especially focusing on pathogen species (in accordance with specific UNI EN ISO), as reported in Supplementary Table S2.
The bacterial identification was firstly performed through the VITEK® 2 system (bioMérieux, France). Gram-positive and negative strains were screened with specific cards VITEK® ID-GP and VITEK® ID-GN, respectively. For Salmonella serovar typing qualitative PCR assays were performed as previously described (Park et al., 2009).
For each bacterial strain, antibiotic susceptibility was performed by using VITEK® 2 system as automated instrument for finding phenotypic antibiotic resistance which is also able to elaborate the Minimum Inhibitory Concentration (MIC) value. Bacterial suspensions and their relative turbidities were measured and adjusted by using a densitometer (DENSICHEK, bioMerieux, Marcy-l’Etoile, France) to the values 0.5–0.63 McFarland standard. One hundred and 45 µl for Gram-positive microorganisms and 280 µL for Gram-negative ones respectively, were added to 3 mL of VITEK 0.45% saline solution (bioMérieux, 2013). The VITEK2 system tested automatically cards and bacterial suspensions and after incubation period (ranged between 10 and 12 h) produced susceptibility results.
In the present study, VITEK® 2 AST cards (bioMérieux, France) were employed. For Gram-positive cocci AST-P658 and AST-P659were used, including the following antibiotics: amoxicillin/clavulanic acid, ampicillin, ampicillin/sulbactam, imipenem, gentamicin, kanamycin, streptomycin, ciprofloxacin, levofloxacin, quinupristin/dalfopristin, linezolid, daptomycin, teicoplanin, vancomycin, tigecycline, nitrofurantoin, trimethoprim, trimethoprim/sulfamethoxazole, cefoxitin, benzylpenicillin, oxacillin, ceftaroline, clindamycin, erythromycin, tetracycline, fusidic acid, mupirocin, rifampicin. Gram-negative were tested by means of the card AST-N379 that includes: ESBL, amoxicillin/clavulanic acid, piperacillin/tazobactam, cefotaxime, ceftazidime, cefepime, ertapenem, imipenem, meropenem, amikacin, gentamycin, ciprofloxacin, fosfomycin, nitrofurantoin, trimethoprim/sulfamethoxazole.
The MIC value was evaluated using the Clinical and Laboratory Standards Institute (CLSI) break-points (CLSI, 2017).
Based on the phenotypic resistance results, all resistant bacteria were screened to detect the presence of specific resistance genes through single or multiple PCR assays and using the same primers used for litter samples ARGs biomolecular screenings (Supplementary Table S1).
PCR reactions were performed in a total volume of 25 μL by using the Green Master Mix (Promega®, Italy). Thermocycler settings were adapted, especially for the annealing time-temperature ratio, according to the different primers’ references. The PCR products were visualized by agarose gel electrophoresis.
Descriptive and statistical analysis was performed using the software STATA (StataCorp, 2019). The t-test was applied to analyze the ARGs frequency in litter and carcasses samples between AF and C farming types, while the chi-square (χ2) test was used to compare the phenotypical resistance profiles in bacterial strains recovered from AF and C carcasses. A p-value < 0.05 was applied as threshold of significance.
The details of ARGs results were reported in Table 2. Considering the litter samples, in conventional farms catA1 (specific for chloramphenicol resistance), aadA2 (for aminoglycosides resistance), sul2 (sulfonamides resistance) and blaTEM (for ampicillin resistance) resulted to be present in all samples, followed by tetracyclines genes amplified in at least one farm, except for tetB(P) and tetC.

TABLE 2. Results of ARGs screening performed from litter and carcasses pooled samples. For carcasses the number of positive samples were reported in brackets.
The lincomycin ARGs (lnuA and lnuB) were recovered from C1 and C3 farms, while no positivity was observed for vancomycin, carbapenems, quinolones, nitrofurantoin, linezolid and quinupristin/dalfopristin ARGs along with the aadB and aac3IV) aminoglycosides genes.
The occurrence of tet genes resulted higher in the AF farms where all target fragments were amplified.
The genes aadA2,sul1, sul2,blaTEM, and msrC were detected in all AF farms, followed by catA1, recovered from two AF farms. Finally, the aminoglycosides aac3IV) and quinupristin/dalfopristin vatE resistance genes were present in AF1 sample. No amplification was obtained for remaining vancomycin, carbapenems, quinolones, nitrofurantoin, linezolid genes.
The ARGs screening carried out on the carcass samples confirmed the negativity for vancomycin, carbapenems, quinolones, nitrofurantoin and linezolid genes.
Considering the tetracycline ARGs, the tetM and tetL were the only amplified genes in carcasses from C farms; similar occurrence was observed in carcasses from AF farms for tetM and tetB.
The most representative genes were sul2 and blaTEM, both amplified in all investigated conventional and antibiotic-free carcasses, followed by quinupristin/dalfopristin vatD resistance gene amplified in a total of 12 samples coming from C1, C2 and AF2 farms.
Among ARGs for aminoglycosides, the aadB was amplified in both groups (3 from AF and 5 from C farms), while the aadA2 gene was present only in two samples from C1 farm. No other positivity was observed for the remaining ARGs target.
Ninety-three strains were isolated from poultry carcasses, as reported in Table 3. The Gram-negative strains (59/93) resulted mainly distributed among the Aeromonas, Salmonella and Proteus genera. The serovar typing PCR for Salmonella revealed n. Five Typhi (n. Three from AF carcasses and n. Two from C farms), n. Five Thyphimurium (n. Three from C farms and n. Two from AF farms) and n. Three Enteritidis (n. One from C and 2 from AF farms) strains. All Gram-positive microorganisms (34/93) belonged to the genus Enterococcus and the most abundant species resulted Enterococcus faecalis (18/34; 52.9%).
In Tables 4 and 5 the results regarding bacterial susceptibilities were reported.
Concerning Gram-negative, 5 Aeromonas strains resulted resistant to at least one molecule among meropenem, piperacillin-tazobactam and trimethoprim-sulfamethoxazole with 2 isolates presenting a resistance to all above-mentioned antibiotics. Two Salmonella isolates (S. Typhi and S. Typhimurium) were MDR strains (resistant to cephalosporines, nitrofuransand sulfonamides). Additional MDR patterns were observed for two additional strains belonging to E. coli species and Serratia genus. Among enterococci, the higher number of resistant strains were observed in E. faecalis species for tetracyclines and streptomycin molecules (10/18 and 7/18 isolates, respectively), followed by E. gallinarumfor quinupristin-dalfopristin (4/4 isolates) and by E. avium and E. casseliflavus for tetracyclines (4/5 isolates and 3/3 isolates, respectively).
All Aeromonas strains did not present any ARGs related to the phenotypic resistance profiles. Similar results were observed in two Salmonella Enteritidis E. coli and Serratia fonticola isolates along with one strain of Raoultella ornithinolytica, Cronobacter sakazaki and Klebsiella pneumoniae. No evidence of nitrofurantoin ARGs were amplified starting from E. coli strains. The highest number of ARGs were observed from Salmonella and E. coli strains coming from the same farming type AF, Among the quinopristin-dalfopristin resistant enterococci, one strain of E. gallinarum and two strains of E. casseliflavus did not harbored the relative ARGs. The vancomycin phenotypic resistance observed in E. faecalis was related to the vanC1 gene presence. Similarly, the linezolid resistance with the cfr gene was observed in the same strain (Tables 6 and 7).
A significant difference was observed in litter (p = 0.02) and carcasses (p = 0.04) specimens for ARGs frequency detected in AF and C farms. On the contrary, no significance was obtained comparing the phenotypical resistance profiles observed in bacterial strains from AF and C farms.
In this study a multidisciplinary approach, based on culture-dependent and culture-independent investigations, was applied involving an integrated food supply chain, from the farms to slaughterhouse, in order to evaluate the potential drivers of antimicrobial resistant bacteria and their genetic determinants.
At the farm level the biomolecular screening by means of endpoint PCR test allowed to confirm the role of broiler litter as environmental source of ARGs. In this respect a wide panel of genetic targets was selected including not only the most administrated molecules in veterinary medicine but considering also the so-called CIAs restricted to human use (WHO, 2019), to obtain a more comprehensive overview of the AMR in the poultry industry. In addition, two different types of farming systems were considered, including conventional and antibiotic-free.
The ARGs screening revealed that sul, blaTEM and tet genes were the most representative ARGs, in agreement with previous investigations carried out in Italian poultry farming (Di Francesco et al., 2021; Salerno et al., 2022), suggesting the environmental spreading of bacteria potentially resistant to sulfonamide, beta-lactam and tetracycline molecules. These antibiotics are widely used in veterinary practice for enteric and respiratory infections treatment of poultry (Agyare et al., 2018), and they represent the most abundant molecules sold in European Union in the years 2019–2020, as reported by the European Surveillance of Veterinary Antimicrobial Consumption (ESVAC) (EFSA Panel on Animal Health and Welfare et al., 2021).
Similarly, a recent study highlighted a major abundance of sul1 and tetA genes in antibiotic-free layer hen and broiler manure, which correspond to the major classes of antibiotics used in animal industry, in comparison with other food producing animals, such as swine and beef cattle (Rothrock et al., 2021). A similar distribution was observed by Cadena et al. (2018) analyzing the soil from organic farms, suggesting a diffuse environmental spreading of these ARGs.
In our study, the antibiotic-free farms showed a major heterogeneity of ARGs in litter, more specifically referring to the tet genes. The statistical analysis supported this evidence, highlighting a significant difference between the two farming systems. Accordingly, a recent study demonstrated a relative abundance of tetA in environmental samples collected from a broiler farm where the antibiotics have never been used (Salerno et al., 2022). However, previous investigations reported opposite results, highlighting that no statistically significant differences in antibiotic resistance were found comparing enterococci obtained from conventional and antibiotic-free broiler fecal samples (Semedo-Lemsaddek et al., 2021). Furthermore, the conventional broiler flocks appeared to carry more frequently resistant E. coli in comparison with antibiotic-free and organic types (Pesciaroli et al., 2020; Musa et al., 2021). Probably, other resistance determinants, apart from the farming production conditions, can be involved in the environmental persistence of ARGs at flock level, as past antibiotic treatments, able to exert a long-term selective pressure (Khine et al., 2022) or different sources of genetic elements, as water or wild birds (EFSA Panel on Animal Health and Welfare et al., 2021; Storey et al., 2022).
The blaTEM gene has been selected as representative of genes responsible for beta-lactamase production, widely harbored by Enterobacteriaceae. Indeed, all farms belonging to the both conventional and antibiotic-free systems presented this target gene in litter samples confirming the results of previous epidemiological surveys carried in Europe (Cantón et al., 2008; Uyanik et al., 2021).
Noteworthy, the catA1 gene, specific for chloramphenicol resistance, resulted also widely diffused despite its administration is not allowed since 1994 (CVMP, 1994). Chloramphenicol resistant bacteria continue to be recovered from avian samples in different countries worldwide (El-Sharkawy et al., 2017; Yaqoob et al., 2018), while in Italy this pattern seems to be not relevant in broiler farms (Busani et al., 2004; Camarda et al., 2013). Probably, the detection of catA1 gene in the studied farms could be a consequence of its localization on the mobile elements carrying other ARGs, i.e., for aminoglycosides class (Lei et al., 2020). To support this consideration, the aadA2 was detected in all farms. Furthermore, the resistance pattern against chloramphenicol has been described because of repeated antibiotic treatments with amoxicillin in broilers (Jiménez-Belenguer et al., 2016), although the catA1 gene was also recovered in two AF flocks. Therefore, all ARGs investigated in our study were located in mobile genetic elements as transposons or plasmids, leading to an increased spreading of resistance patterns in different bacteria species, including pathogens responsible of infections in animals and humans (EFSA Panel on Animal Health and Welfare et al., 2021).
The ARGs screening of carcasses samples confirmed a wide distribution of sul and blaTEM genes, while the occurrence of the remaining genes fragments appeared to be of minor importance. However, the carcasses from two conventional and one antibiotic-free flocks resulted positive for the quinupristin/dalfopristin resistance gene vatD, that has been not detected from respective litter samples. This gene was identified in several enterococci isolates resulted resistant to quinupristin/dalfopristin and coming from human, farm animals and meat products (Donabedian et al., 2006; Jackson et al., 2007; Hwang et al., 2010), suggesting multiple sources of contamination, potentially able to reach the carcasses at slaughterhouse level. Probably, the PCR test failed to detect the vatD gene from litter samples, but additional cross-contaminations, due to the workers or processing surfaces, should be considered, as suggested by other authors (Musa et al., 2021; Salerno et al., 2022). The Good Hygiene and Good Manufacturing Practices resulted correctly applied by the food operators in sampled slaughterhouse, however some critical points (i.e., evisceration process) could lead to carcasses contamination (EU Reg. 852 del 2004).
The ARGs distribution resulted lower in antibiotic-free carcasses as demonstrated by the statistical analysis. This trend is in accordance with previous investigations that revealed a statistically significant lower occurrence of resistance genes in commensal E. coli isolated from broiler carcasses produced without the use of antibiotics (Gambi et al., 2022), and a minor occurrence of resistant Campylobacter strains in antibiotic-free chicken products (Price et al., 2005). However, similarly to that reported for litter specimens, the results are still controversial based on the different classes of antibiotics, as recently highlighted in nalidixic acid-resistant E. coli isolates, significantly higher in antibiotic-free than conventional system, and in the MDR E. coli strains, equally distributed in both types of meat production (Musa et al., 2021). Similar conclusions were achieved by previous studies focusing on Salmonella, E. coli and Campylobacter resistant strains recovered from fresh retail chicken (Millman et al., 2013; Mollenkopf et al., 2014).
In line with these data, the distribution of total bacteria strains in the carcasses observed in present study did not appear to be affected by the farming type, except for few Gram-negative isolates (Cronobacter, Citrobacter, and Raoultella genera), only recovered from C carcasses, and for Enterobacter spp. And E. durans from AF carcasses.
Considering the phenotypic resistance patterns, the nitrofurantoin, cefotaxime and trimethoprim/sulfamethoxazole resistance profiles were the most diffused resistances observed in Gram-negative strains. The published researches, mainly focused on Salmonella spp., E. coli and Campylobacter spp. Isolates, showed similar resistance profiles (Nguyen et al., 2021; Sarowska et al., 2022; Ščerbová et al., 2022). In addition, two investigated Salmonella strains showed MDR patterns, harboring the correspondent target genes sul and nfs, arising a potential risk for transmission to consumers of these pathogens (Parvin et al., 2020).
Other potential pathogens discovered in this study are E. coli strains, one of which resulted multidrug resistant. However, additional investigations aimed to identify any virulence factors produced by these bacteria should be performed, to assess their effective role as foodborne pathogens (Hamilton et al., 2018; Jang et al., 2020; Pakbin et al., 2021; Wareth and Neubauer, 2021). Noteworthy, a Klebsiella pneumonia strain, resistant to nitrofurans, was recovered from AF carcasses. Klebsiella pneumonia is recognized as responsible of respiratory, urinary and bloodstream infections in humans and it belongs to the ESKAPE group, including antibiotic resistant pathogens (World Health Organization, 2017).
In enterococci the resistance for tetracycline, and streptomycin were mostly detected, while other studies, involving enterococci isolates recovered from broilers, revealed the resistance also for vancomycin, gentamicin and quinupristin-dalfopristin in Portugal (Novais et al., 2005), Turkey (Ünal et al., 2020) and, more recently, in Bangladesh (Roy et al., 2022) and Russia (Makarov et al., 2022). This evidence represents an important public health concern, considering that resistant enterococci of animal origin could be drivers of resistance patterns for human strains responsible of various nosocomial infections (Hammerum et al., 2010).
The molecular investigations of ARGs in resistant strains returned some divergent results, as mainly remarked from Aeromonas isolates, two Salmonella enteritidis and 1 E. coli, which were phenotypically resistant to beta-lactams including carbapenems, nitrofurantoin and sulfonamides, but negative for the genetic determinants. Basing on nitrofurantoin phenotypic resistant results, the respective nfs genes lacked in E. coli, Klebsiella and Cronobacter spp. Probably, the detection limit of PCR applied in this study was not efficient to amplify the before-mentioned ARGs, or the phenotypically resistance was related to other genetic elements not included in this study (Jian et al., 2021; Liao et al., 2021).
Comparing the profiles between AF and C bacterial strains, no significant difference was observed as highlighted by statistical analysis. As already mentioned, studies published to date provided inconclusive results regarding the effect of antibiotic-free or organic farming models on the mitigation of antibiotic resistance diffusion (Price et al., 2005; Pesciaroli et al., 2020; Musa et al., 2021; Gambi et al., 2022). In contrast with our results, the farming system without the antibiotic use seems to support a lower prevalence of resistant Salmonella (Bailey et al., 2020). However, same authors, in a previous study, were not able to demonstrate a protective role of AF farming type for the reduction of resistant Campylobacter in chicken (Bailey et al., 2019).
Apparently, the AF farming model investigated in this study did not appear to be related to a major risk to transfer MDR bacteria, including pathogenic species, to the final consumers. However, it is not possible to exclude that other factors can influence the occurrence of resistant strains in meat production systems. More in detail, a potential cross-contamination along the slaughter lines can occur, whereas a distinction between AF and C animals is generally not applied, or other environmental sources (food operator’s hands, surfaces, and devices) cannot be ruled out. This hypothesis may be considered in accordance with the isolation of Salmonella Typhi serovar from both production lines. The serovar Typhi is generally recognized as human host-adapted pathogen responsible for systemic febrile illness transmitted through the oro-fecal route. This finding was justified by the food operator contamination instead of animal source (Kidgell et al., 2002).
In this respect, further studies are mandatory to better clarify the effective role of different variables in chicken food chain production in the diffusion of MDR bacterial strains.
In this study a biomolecular screening for selected ARGs, firstly carried out at farm level and then at the slaughterhouse, was complemented with the phenotypic AMR analyses of pathogenic and commensal bacteria isolated from broiler carcasses. The performed biomolecular screenings provided us a wide understanding about the distribution of antibiotic resistance determinants sampling from the beginning to the end of the chicken meat production chain. The most representative resistance patterns were relative to the most used antimicrobials in veterinary medicine but different CIA (carbapenems, third generation cephalosporins and vancomycin) were also observed, arising a critical issue for human health. The investigated farming systems did not reveal a significant difference in AMR diffusion between AF and C samples. Alternative farming systems continue to be an expanding meat production source, able to satisfy the consumers demand for sustainable, healthy and more respectful of animal welfare products (EFSA Panel on Biological Hazards et al., 2021). However, additional studies are mandatory to better clarify the effective influence of different environmental and management factors on the AMR persistence.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
CD, AV: conceptualization, GF, CD, MF: writing. GF, MF, AF, CS, FR, AB, AV, CD: review and editing. GF, MF, CS, RF, AV, CD: methodology and data curation. GF, MF, AF, CS, CD: analysis. GF, CS, CD: writing—original draft preparation. AV, CD: supervision, project administration, and funding acquisition. All authors contributed to the article and approved the submitted version.
The present study had been founded by the University of Teramo, Italy (Founds for Basic Research Activities—FARBID “Antimicrobial resistance genes in poultry industry”).
Author FR was employed by Gesco Cons, Coop Arl.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/frfst.2023.1168896/full#supplementary-material
Agyare, C., Boamah, V. E., and Osei, C. N. Z. (2018). “Antibiotic use in poultry production and its effects on bacterial resistance,” in Antimicrobial resistance - a global threat. Editor Y. Kumar (London: IntechOpen), 34–40.
Bailey, M. A., Taylor, R. M., Brar, J. S., Corkran, S. C., Velásquez, C., Novoa Rama, E., et al. (2019). Prevalence and antimicrobial resistance of Campylobacter from antibiotic-free broilers during organic and conventional processing. Poult. Sci. 98, 1447–1454. doi:10.3382/ps/pey486
Bailey, M., Taylor, R., Brar, J., Corkran, S., Velásquez, C., Novoa-Rama, E., et al. (2020). Prevalence and antimicrobial resistance of Salmonella from antibiotic-free broilers during organic and conventional processing. J. Food Prot. 83, 491–496. doi:10.4315/0362-028X.JFP-19-269
Barthelmie, R. J. (2022). Impact of dietary meat and animal products on GHG footprints: The UK and the US. Climate 10, 43. doi:10.3390/cli10030043
Busani, L., Del Grosso, M., Paladini, C., Graziani, C., Pantosti, A., Biavasco, F., et al. (2004). Antimicrobial susceptibility of vancomy-cin-susceptible and -resistant enterococci isolated in Italy from raw meat products, farm animals, and human infections. Int. J. Food Microbiol. 97, 17–22. doi:10.1016/j.ijfoodmicro.2004.04.008
Cadena, M., Durso, L. M., Miller, D. N., Waldrip, H. M., Castleberry, B. L., Drijber, R. A., et al. (2018). Tetracycline and sulfonamide antibiotic resistance genes in soils from Nebraska organic farming operations. Front. Microbiol. 9, 1283. doi:10.3389/fmicb.2018.01283
Camarda, A., Pugliese, N., Pupillo, A., Oliva, M., Circella, E., Dionisi, A. M., et al. (2013). Resistance genes, phage types and pulsed field gel electrophoresis pulsotypes in Salmonella entericastrains from laying hen farms in southern Italy. Int. J. Environ. Res. Public Health. 10, 3347–3362. doi:10.3390/ijerph10083347
Cantón, R., Novais, A., Valverde, A., Machado, E., Peixe, L., Baquero, F., et al. (2008). Prevalence and spread of extended-spectrum be-ta-lactamase-producing Enterobacteriaceae in Europe. Clin. Microbiol. Infect. 1, 144–153. doi:10.1111/j.1469-0691.2007.01850.x
Capita, R., Prieto, M., and Alonso-Calleja, C. (2004). Sampling methods for microbiological analysis of red meat and poultry carcasses. J. Food Protec 67 (6), 1303–1308. doi:10.4315/0362-028x-67.6.1303
Clinical and Laboratory Standards Institute (CLSI) (2017). M100: Performance Standards for antimicrobial susceptibility testing. 27th ed. Wayne, Pennsylvania: Clinical and Laboratory Standards Institute. CLSI supplement M100 (ISBN 1-56238-804-5 [Print]; ISBN 1-56238-805-3 [Electronic]) 950 West Valley Road, Suite 2500.
CVMP (Committee for Veterinary Medicinal Products) (1994). Chloramphenicol. Summary report. Available at: http://www.ema.europa.eu/docs/en_GB/document_library/Maximum _Residue_Limits_-_Report/2009/11/WC500012060.pdf (Accessed July 5, 2022).
De Cesare, A., Oliveri, C., Lucchi, A., Savini, F., Manfreda, G., and Sala, C. (2022). Pilot study on poultry meat from antibiotic free and conventional farms: Can metagenomics detect any difference? Foods 11, 249. doi:10.3390/foods11030249
Di Francesco, C. E., Smoglica, C., Profeta, F., Farooq, M., Di Giannatale, E., Toscani, T., et al. (2021). Research Note: Detection of antibiotic-resistance genes in commercial poultry and Turkey flocks from Italy. Poult. Sci. 100, 101084. doi:10.1016/j.psj.2021.101084
Donabedian, S. M., Perri, M. B., Vager, D., Hershberger, E., Malani, P., Simjee, S., et al. (2006). Quinupristin-dalfopristin resistance in Enterococcus faecium isolates from humans, farm animals, and grocery store meat in the United States. J. Clin. Microbiol. 44, 3361–3365. doi:10.1128/JCM.02412-05
El-Sharkawy, H., Tahoun, A., El-Gohary, A. E. A., El-Abasy, M., El-Khayat, F., Gillespie, T., et al. (2017). Epidemiological, molecular characterization and antibiotic resistance of Salmonella enterica serovars isolated from chicken farms in Egypt. Gut Pathog. 9, 8. doi:10.1186/s13099-017-0157-1
EFSA Panel on Biological Hazards (BIOHAZ), Koutsoumanis, K., Allende, A., Álvarez-Ordóñez, A., Bolton, D., Bover-Cid, S., Chemaly, M., et al. (2021). Role played by the environment in the emergence and spread of antimicrobial resistance (AMR) through the food chain. EFSA J. 19, e06651. doi:10.2903/j.efsa.2021.6651
EFSA Panel on Animal Health and Welfare (AHAW), Nielsen, S. S., Bicout, D. J., Calistri, P., Canali, E., Drewe, J. A., Garin-Bastuji, B., et al. (2021). Assessment of animal diseases caused by bacteria resistant to antimicrobials: Poultry. EFSA J. 19, e07114. doi:10.2903/j.efsa.2021.7114
Farooq, M., Smoglica, C., Ruffini, F., Soldati, L., Marsilio, F., and Di Francesco, C. E. (2022). Antibiotic resistance genes occurrence in conventional and antibiotic-free poultry farming, Italy. Animals 12, 2310. doi:10.3390/ani12182310
Feng, M. X., Wang, G. N., Yang, K., Liu, H. Z., and Wang, J. P. (2016). Molecularly imprinted polymer-high performance liquid chromatography for the determination of tetracycline drugs in animal derived foods. Food control. 69, 171–176. doi:10.1016/j.foodcont.2016.04.050
Food and Agriculture Organization of the United Nations (FAO) (2013). Poultry development review. Available at: https://www.fao.org/3/i3531e/i3531e.pdf (Accessed July 10, 2022).
Gambi, L., Crippa, C., Lucchi, A., De Cesare, A., Parisi, A., Manfreda, G., et al. (2022). The resistome of commensal Escherichia coli isolated from broiler carcasses "produced without the use of antibiotics. Poult. Sci. 101, 101770. doi:10.1016/j.psj.2022.101770
Hamilton, A. L., Kamm, M. A., Ng, S. C., and Morrison, M. (2018). Proteus spp. as putative gastrointestinal pathogens. Clin. Microbiol. Rev. 31, e00085-17. doi:10.1128/CMR.00085-17
Hammerum, A. M., Lester, C. H., and Heuer, O. E. (2010). Antimicrobial-resistant enterococci in animals and meat: A human health hazard? Foodborne Pathog. Dis. 7, 1137–1146. doi:10.1089/fpd.2010.0552
Hernando-Amado, S., Coque, T. M., Baquero, F., and Martínez, J. L. (2019). Defining and combating antibiotic resistance from one health and global health perspectives. Nat. Microbiol. 4, 1432–1442. doi:10.1038/s41564-019-0503-9
Hwang, I. Y., Ku, H. O., Lim, S. K., Lee, K. J., Park, C. K., Jung, G. S., et al. (2010). Distribution of streptogramin resistance genes and genetic relatedness among quinupristin/dalfopristin-resistant Enterococcus faecium recovered from pigs and chickens in Korea. Res. Vet. Sci. 89, 1–4. doi:10.1016/j.rvsc.2010.01.011
Jackson, C. R., Fedorka-Cray, P. J., Barrett, J. B., Hiott, L. M., and Woodley, T. A. (2007). Prevalence of streptogramin resistance in enterococci from animals: Identification of vatD from animal sources in the USA. Int. J. Antimicrob. Agents. 30, 60–66. doi:10.1016/j.ijantimicag.2007.03.010
Jang, H., Gopinath, G. R., Eshwar, A., Srikumar, S., Nguyen, S., Gangiredla, J., et al. (2020). The secretion of toxins and other exoproteins of cronobacter: Role in virulence, adaption, and persistence. Microorganisms 8, 229. doi:10.3390/microorganisms8020229
Jian, Z., Zeng, L., Xu, T., Sun, S., Yan, S., Yang, L., et al. (2021). Antibiotic resistance genes in bacteria: Occurrence, spread, and control. J. Basic Microbiol. 61, 1049–1070. doi:10.1002/jobm.202100201
Jiménez-Belenguer, A., Doménech, E., Villagrá, A., Fenollar, A., and Ferrús, M. A. (2016). Antimicrobial resistance of Escherichia coli isolated in newly-hatched chickens and effect of amoxicillin treatment during their growth. Avian Pathol. 45, 501–507. doi:10.1080/03079457.2016.1168515
Khine, N. O., Lugsomya, K., Niyomtham, W., Pongpan, T., Hampson, D. J., and Prapasarakul, N. (2022). Longitudinal monitoring reveals persistence of colistin-resistant Escherichia coli on a pig farm following cessation of colistin use. Front. Vet. Sci. 9, 845746. doi:10.3389/fvets.2022.845746
Kidgell, C., Reichard, U., Wain, J., Linz, B., Torpdahl, M., Dougan, G., et al. (2002). Salmonella typhi, the causative agent of typhoid fever, is approximately 50,000 years old. Infect. Genet. Evol. 2 (1), 39–45. doi:10.1016/s1567-1348(02)00089-8
Lei, C. W., Yao, T. G., Yan, J., Li, B. Y., Wang, X. C., Zhang, Y., et al. (2020). Identification of Proteus genomic island 2 variants in two clonal Proteus mirabilis isolates with coexistence of a novel genomic resistance island PmGRI1. J. Antimicrob. Chemother. 75, 2503–2507. doi:10.1093/jac/dkaa215
Liao, C. Y., Balasubramanian, B., Peng, J. J., Tao, S. R., Liu, W. C., and Ma, Y. (2021). Antimicrobial resistance of Escherichia coli from aquaculture farms and their environment in zhanjiang, China. Front. Vet. Sci. 8, 806653. doi:10.3389/fvets.2021.806653
Makarov, D. A., Ivanova, O. E., Pomazkova, A. V., Egoreva, M. A., Prasolova, O. V., Lenev, S. V., et al. (2022). Antimicrobial resistance of commensal Enterococcus faecalis and Enterococcus faecium from food-producing animals in Russia. Vet. World 15, 611–621. doi:10.14202/vetworld.2022.611-621
Millman, J. M., Waits, K., Grande, H., Marks, A. R., Marks, J. C., Price, L. B., et al. (2013). Prevalence of antibiotic-resistant E. coli in retail chicken: Comparing conventional, organic, kosher, and raised without antibiotics. F1000Res 2, 155. doi:10.12688/f1000research.2-155.v2
Mollenkopf, D. F., Cenera, J. K., Bryant, E. M., King, C. A., Kashoma, I., Anand, K., et al. (2014). Organic or antibiotic-free labeling does not impact the recovery of enteric pathogens and antimicrobial-resistant Escherichia coli from fresh retail chicken. Foodborne Pathog. Dis. 11, 920–929. doi:10.1089/fpd.2014.1808
Musa, L., Proietti, P. C., Marenzoni, M. L., Stefanetti, V., Kika, T. S., Blasi, F., et al. (2021). Susceptibility of commensal E. coli isolated from conventional, antibiotic-free, and organic meat chickens on farms and at slaughter toward antimicrobials with public health relevance. Antibiotics 10, 1321. doi:10.3390/antibiotics10111321
Nguyen, T. K., Nguyen, L. T., Chau, T. T. H., Nguyen, T. T., Tran, B. N., Taniguchi, T., et al. (2021). Prevalence and antibiotic resistance of Salmonella isolated from poultry and its environment in the Mekong Delta, Vietnam. Vet. World 14, 3216–3223. doi:10.14202/vetworld.2021.3216-3223
Novais, C., Coque, T. M., Costa, M. J., Sousa, J. C., Baquero, F., and Peixe, L. V. (2005). High occurrence and persistence of antibiotic-resistant enterococci in poultry food samples in Portugal. J. Antimicrob. Chemother. 56, 1139–1143. doi:10.1093/jac/dki360
Pakbin, B., Brück, W. M., and Rossen, J. W. A. (2021). Virulence factors of enteric pathogenic Escherichia coli: A review. Int. J. Mol. Sci. 22, 9922. doi:10.3390/ijms22189922
Park, S. H., Kim, H. J., Cho, W. H., Kim, J. H., Oh, M. H., Kim, S. H., et al. (2009). Identification of Salmonella enterica subspecies I, Salmonella enterica serovars Typhimurium, Enteritidis and Typhi using multiplex PCR. FEMS Microbiol. Lett. 301, 137–146. doi:10.1111/j.1574-6968.2009.01809.x
Parvin, M. S., Hasan, M. M., Ali, M. Y., Chowdhury, E. H., Rahman, M. T., Islam, M. T., et al. (2020). Prevalence and Multidrug Resistance Pattern of Salmonella Carrying Extended-Spectrum β-Lactamase in Frozen Chicken Meat in Bangladesh. J. Food Prot. 83 (12), 2107–2121. doi:10.4315/JFP-20-172
Pesciaroli, M., Magistrali, C. F., Filippini, G., Epifanio, E. M., Lovito, C., Marchi, L., et al. (2020). Antibiotic-resistant commensal Escherichia coli are less frequently isolated from poultry raised using non-conventional management systems than from conventional broiler. Int. J. Food Microbiol. 314, 108391. doi:10.1016/j.ijfoodmicro.2019.108391
Price, L. B., Johnson, E., Vailes, R., and Silbergeld, E. (2005). Fluoroquinolone-resistant Campylobacter isolates from conventional and antibiotic-free chicken products. Environ. Health Perspect. 113, 557–560. doi:10.1289/ehp.7647
Racewicz, P., Majewski, M., Biesiada, H., Nowaczewski, S., Wilczyński, J., Wystalska, D., et al. (2022). Prevalence and characterisation of antimicrobial resistance genes and class 1 and 2 integrons in multiresistant Escherichia coli isolated from poultry production. Sci. Rep. 12 (1), 6062. doi:10.1038/s41598-022-09996-y
Rothrock, M. J., Min, B. R., Castleberry, L., Waldrip, H., Parker, D., Brauer, D., et al. (2021). Antibiotic resistance, antimicrobial residues, and bacterial community diversity in pasture-raised poultry, swine, and beef cattle manures. J. Anim. Sci. 99, skab144. doi:10.1093/jas/skab144
Roy, K., Islam, M. S., Paul, A., Ievy, S., Talukder, M., Sobur, M. A., et al. (2022). Molecular detection and antibio-typing of multi-drug resistant Enterococcus faecium from healthy broiler chickens in Bangladesh. Vet. Med. Sci. 8, 200–210. doi:10.1002/vms3.669
Salerno, B., Furlan, M., Sabatino, R., Di Cesare, A., Leati, M., Volanti, M., et al. (2022). Antibiotic resistance genes load in an antibiotic free organic broiler farm. Poult. Sci. 101, 101675. doi:10.1016/j.psj.2021.101675
Sarowska, J., Olszak, T., Jama-Kmiecik, A., Frej-Madrzak, M., Futoma-Koloch, B., Gawel, A., et al. (2022). Comparative characteristics and pathogenic potential of Escherichia coli isolates originating from poultry farms, retail meat, and human urinary tract infection. Life (Basel). 12, 845. doi:10.3390/life12060845
Ščerbová, J., Lauková, A., Losasso, C., and Barco, L. (2022). Antimicrobial susceptibility to natural substances of Campylobacter jejuni and Campylobacter coli isolated from Italian poultry. Foodborne Pathog. Dis. 19, 266–271. doi:10.1089/fpd.2021.0085
Semedo-Lemsaddek, T., Bettencourt Cota, J., Ribeiro, T., Pimentel, A., Tavares, L., Bernando, F., et al. (2021). Resistance and virulence distribution in enterococci isolated from broilers reared in two farming systems. Ir. Vet. J. 74, 22. doi:10.1186/s13620-021-00201-6
Storey, N., Cawthraw, S., Turner, O., Rambaldi, M., Lemma, F., Horton, R., et al. (2022). Use of genomics to explore AMR persistence in an outdoor pig farm with low antimicrobial usage. Microb. Genom 8, 000782. doi:10.1099/mgen.0.000782
Ünal, N., Bal, E., Karagöz, A., Altun, B., and Koçak, N. (2020). Detection of vancomycin-resistant enterococci in samples from broiler flocks and houses in Turkey. Acta Vet. hung. 68, 117–122. doi:10.1556/004.2020.00024
Uyanik, T., Gülel, G. T., and Alişarli, M. (2021). Characterization of extended-spectrum beta-lactamase-producing Enterobacterales from organic and conventional chicken meats. Lett. Appl. Microbiol. 72, 783–790. doi:10.1111/lam.13472
Wareth, G., and Neubauer, H. (2021). The animal-foods-environment interface of Klebsiella pneumoniae in Germany: An observational study on pathogenicity, resistance development and the current situation. Vet. Res. 52, 16. doi:10.1186/s13567-020-00875-w
WHO (2019). Critically important antimicrobials for human medicine, 6th revision. Geneva: World Health Organization. Licence: CC BY-NC-SA 3.0 IGO.
World Health Organization (2022). Comprehensive review of the WHO global action plan on antimicrobial resistance: Management response – march 2022. Available at: https://www.who.int/publications/m/item/comprehensive-review-of-the-who-global-action-plan-on-antimicrobial-resistance-management-response---march-2022 (Accessed October 12, 2022).
World Health Organization (2017). Global priority list of antibiotic-resistant bacteria to guide research, discovery, and development of new antibiotics. New York, United States: WHO Press, 1–7.
Keywords: broiler, litter, carcasses, antibiotic resistance genes, phenotypic resistance, antibiotic-free, poultry meat, one-health
Citation: Ferri G, Buonavoglia A, Farooq M, Festino AR, Ruffini F, Paludi D, Di Francesco CE, Vergara A and Smoglica C (2023) Antibiotic resistance in Italian poultry meat production chain: a one-health perspective comparing antibiotic free and conventional systems from the farming to the slaughterhouse. Front. Food. Sci. Technol. 3:1168896. doi: 10.3389/frfst.2023.1168896
Received: 18 February 2023; Accepted: 11 July 2023;
Published: 19 July 2023.
Edited by:
Fernando Perez Rodriguez, University of Cordoba, SpainReviewed by:
Ali Akbar, University of Balochistan, PakistanCopyright © 2023 Ferri, Buonavoglia, Farooq, Festino, Ruffini, Paludi, Di Francesco, Vergara and Smoglica. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gianluigi Ferri, Z2ZlcnJpQHVuaXRlLml0
†These authors share last authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.