
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Plant Sci. , 28 February 2025
Sec. Plant Abiotic Stress
Volume 16 - 2025 | https://doi.org/10.3389/fpls.2025.1527952
Soil salinity is a significant environmental challenge that threatens plant growth and development, adversely affecting global food crop production. This underscores the critical need to elucidate the molecular mechanisms underlying plant salt tolerance, which has profound implications for agricultural advancement. Recent progress in plant salt tolerance has greatly improved our understanding of the molecular mechanisms of plant responses to salt stress and precision design breeding as an effective strategy for developing new salt-tolerant crop varieties. This review focuses on the model plant species Arabidopsis thaliana and important crops, namely, wheat (Triticum aestivum), maize (Zea mays), and rice (Oryza sativa). It summarizes current knowledge on plant salt tolerance, emphasizing key aspects such as the perception and response to salt stress, Na+ transport, Na+ compartmentalization and clearance, changes in reactive oxygen species induced by salt stress, and regulation of plant stem cell development under salt stress conditions. The review might provide new and valuable information for understanding the molecular mechanisms of plant response and adaptation to salt stress.
Plants, as sessile organisms, face numerous abiotic stresses throughout their life cycle, including drought, salinity, temperature fluctuations, heavy metal ion exposure, ultraviolet radiation, and other physical disturbances (Sun H. et al., 2020; Markham and Greenham, 2021; Zhang et al., 2022a; Kopecka et al., 2023). These stresses not only influence the geographical distribution of plants but also significantly affect their growth, development, and agricultural productivity (Villalobos-Lopez et al., 2022; Zhang et al., 2023a).
Among these challenges, salt stress is particularly critical, with soil salinization posing a major environmental threat to global crop sustainability. During crop domestication, their tolerance to abiotic stress has typically diminished. Soil salinity currently impacts approximately 7% of the world’s land area, covering approximately 950 million hectares (Munns and Tester, 2008; Yang and Guo, 2018a). Of the 230 million hectares of irrigated land globally, 20% to 30% are affected by various degrees of salinization, a figure that continues to grow due to land clearance and agricultural irrigation (Ismail and Horie, 2017; Liang et al., 2024). Projections indicate that global climate change will increase the frequency, duration, and severity of extreme weather events such as droughts and heat waves (Pareek et al., 2020a, b). Prolonged droughts and high temperatures have driven the expansion of irrigation systems worldwide, further exacerbating land salinization. Currently, 52% of the global population, across 13 countries, faces severe salinization issues (Liu M. et al., 2020). As freshwater resources dwindle, agriculture in the 21st century is increasingly contending with saline conditions (Rawat et al., 2022).
Despite the challenges posed by environmental factors like soil salinization, the global population is projected to reach nearly 10 billion by 2050 (Bailey-Serres et al., 2019; Hickey et al., 2019), driving a 60% increase in food demand. At the same time, urbanization is rapidly reducing the available arable land. In combination with drastic environmental changes, key food crops, such as wheat, rice, and maize, are particularly sensitive to salinity. Therefore, crop breeders must focus on developing high-yielding, salt-tolerant varieties to improve productivity on salt-affected land and expand cultivable areas. This strategy is essential to address climate change and ensure global food security. In this review, we focus on summarizing our current understanding of plant salt tolerance mechanisms. We discuss plant perception and response to salt stress; Na+ transport, compartmentalization, and clearance; reactive oxygen species (ROS) changes induced by salt stress; and regulation of plant stem cell development under saline conditions (Figure 1). Finally, we explore future challenges and opportunities in understanding crop salt tolerance mechanisms and breeding new, high-yielding, and salt-tolerant varieties.
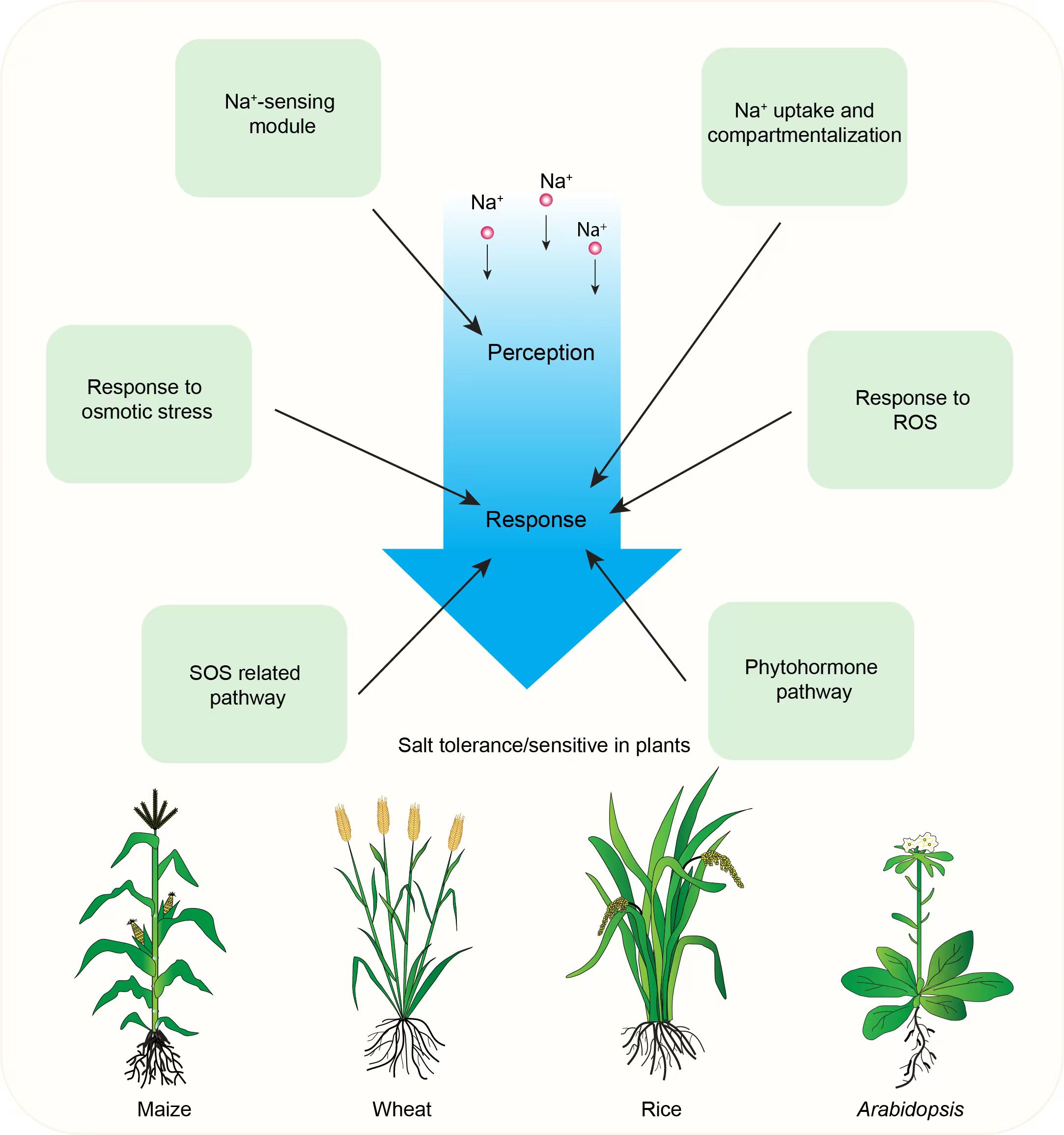
Figure 1. Illustration of plant salt stress responses and mechanisms. Green boxes represent key aspects of plant responses to salt stress, including the processes of salt ion perception and response. The salt stress perception process involves a well-characterized Na+-sensing module. Key mechanisms depicted include the SOS pathway, Na+ uptake and compartmentalization, osmotic stress response, reactive oxygen species response, and phytohormone signaling pathways.
Under high-salt stress conditions, plants have evolved a range of complex physiological responses to adapt and resist these adverse environments. This suggests that plant roots possess Na+ receptors, enabling salt-avoidance growth strategies (Sun et al., 2008; Galvan-Ampudia et al., 2013; Wang et al., 2022b). When exposed to high salinity, plants must contend not only with osmotic stress but also with the ionic stress caused by Na+ ions. Therefore, Na+ uptake can potentially contribute to ionic stress. These ionic stresses are often accompanied by tissue-specific Ca²+ oscillations and amplitude changes within the Na+ signaling pathway (Knight et al., 1997; Ma et al., 2019; Steinhorst et al., 2022). The Monocation-Induced [Ca2+]i Increases1 (MOCA1) gene encodes a glucuronyl transferase responsible for adding a negatively charged glucuronic acid (GlcA) group to inositol phosphorylated ceramide (IPC), thereby generating glycosyl inositol phosphorylated ceramide (GIPC) sphingolipids. These GIPC sphingolipids bind Na+ cations, causing cell membrane depolarization and salt-dependent intracellular Ca²+ oscillations. However, the specific Ca²+ channels involved in this reaction remain unclear (Cao et al., 2019; Jiang et al., 2019).
Studies show that MOCA1-dependent GIPC functions as a sensor for environmental Na+ fluctuations, involving Ca²+ transporters. In Arabidopsis, two highly relevant Ca²+ permeable transporters, ANNEXIN1 (AtANN1) and AtANN4, are crucial in responding to salt-induced Ca²+ signals (Laohavisit et al., 2013; Ma et al., 2019). AtANN1 is essential for promoting salt-activated Ca²+ influx in the plasma membranes of root epidermal cells, while AtANN4 plays a pivotal role in increasing salt-induced [Ca²+]cyt and activating the Salt Overly Sensitive (SOS) pathway (Ma et al., 2019). AtANN4 is regulated by SOS2 protein phosphorylation, and in the presence of its interacting protein SCaBP8 (also known as CBL10), it modulates salt-induced Ca²+ signaling through a phosphorylation-dependent negative feedback loop (Ma et al., 2019).
Salt stress not only disrupts the plasma membrane but also negatively affects multiple organelles within plant cells. Each organelle must be capable of sensing and responding to salt stress signals (Zhao et al., 2020). In Arabidopsis, the LRX-RALF-FER module—including Cell Wall Leucine-Rich Repeat Extensins 3/4/5 (LRX3/4/5), Rapid Alkalinization Factor 22/23 (RALF22/23), and Receptor-Like Kinase FERONIA (FER)—plays a key role in sensing salt stress and regulating salt tolerance (Zhu, 2016; Byrt et al., 2018; Feng et al., 2018; Zhao et al., 2018, 2020). This module senses changes in the cell wall during salt stress, such as the displacement of pectin-cross-linked Ca²+ and the accumulation of reactive oxygen species (ROS), triggering signal transduction pathways that maintain cell wall integrity (CWI) and prevent cell damage (Feng et al., 2018; Zhao et al., 2020).
LRX3/4/5 proteins, located in the cell wall, regulate CWI by interacting with RALF22/23 peptides, preventing their binding to FER under normal conditions. However, under salt stress, the mature RALF22/23 proteins are released and interact with FER, inducing its internalization via an endosomal pathway (Zhao et al., 2018). Mutants such as lrx 345 and fer-4, as well as plants overexpressing RALF22/23, exhibit slow growth and heightened sensitivity to salt stress (Zhao et al., 2018, 2021). Therefore, the LRX3/4/5–RALF22/23–FER module plays a critical role in regulating plant growth, maintaining CWI, and responding to salt stress.
The Arabidopsis TZF1 protein, along with the rice proteins OsBAG6 and OsCaM1-1, plays essential roles in plant perception and response to salt stress (He et al., 2024; Wang et al., 2024b). The Arabidopsis tandem CCCH zinc finger (TZF) protein, TZF1, binds to and promotes the degradation of autoinhibited Ca2+-ATPase 11 (ACA11) mRNA, enhancing salt tolerance. ACA11 encodes a tonoplast-localized calcium pump responsible for exporting calcium, thereby modulating key signal transduction pathways critical for salt tolerance (He et al., 2024). In rice, the mitochondrial-localized chaperone regulator OsBAG6, a member of the B-cell lymphoma 2 (Bcl-2)-associated athanogene family, acts as a novel negative regulator of salt-alkali stress tolerance. Loss-of-function mutations in osbag6 reduce sensitivity to salt-alkali stress. Under non-stress conditions, OsBAG6 binds to the calcium sensor OsCaM1-1, but as Ca²+ levels rise, OsBAG6 releases OsCaM1-1, allowing the Ca²+-saturated OsCaM1-1 to regulate downstream stress-responsive genes as part of the salt-alkali stress response (Wang et al., 2024b). Additionally, functional variants in ZmCBL8 (Calcineurin B-like protein), a component of the Salt Overly Sensitive pathway, have been found to confer salt tolerance in maize (Wang et al., 2024d). In addition, recent studies suggest that cellulose and β-1,4-galactan have biological functions in regulating plant salt tolerance (Zhang et al., 2016b; Yan J. et al., 2021; Yan J. et al., 2023). Although considerable progress has been made in identifying genes and signaling pathways related to plant salt perception, the precise mechanisms through which plants sense salt ions, including the intracellular Na+ receptors, remain largely unresolved.
Plants respond to high-salt environments by altering their osmotic potential. Over time, they have evolved a range of sensing and response mechanisms that regulate osmotic stress signals induced by salt stress (Christmann et al., 2013; Shabala et al., 2016; Gong et al., 2020; Wang et al., 2022b; Zhang et al., 2023a). Among these mechanisms, Ca²+ functions as a crucial secondary messenger in sensing and signaling under various abiotic stresses (Gong et al., 2020; Dong et al., 2022; Zhang et al., 2022a). In plants, the Reduced Hyperosmolality-Induced [Ca2+]i Increase1 (OSCA1) gene encodes a hyperosmolarity-gated calcium channel that acts as an osmotic stress sensor. This channel detects rapid increases in cytosolic Ca²+ ([Ca²+]cyt) levels triggered by osmotic stress (Knight et al., 1997; Yuan et al., 2014). In osca1 mutants, Ca²+ influx under high osmolarity is impaired, leading to reduced leaf transpiration and hindered root growth (Jojoa-Cruz et al., 2018; Liu et al., 2018; Maity et al., 2019). Similarly, in Arabidopsis thaliana, the MSCS-LIKE 8 (MSL8) gene encodes a membrane tension-gated ion channel that responds to osmotic stress by upregulating expression during low osmotic conditions. This process facilitates ion efflux, helping protect cells from hypoosmotic stress (Hamilton et al., 2015). These genes are central to osmotic stress sensing in plants.
Recent research has revealed that Ca²+-responsive proteins such as BONZAI (Chen et al., 2020a), plastid K+ exchange antiporters (Stephan et al., 2016), small-conductance mechanosensitive ion channel-like channels (Hamilton et al., 2015), and MID1-COMPLEMENTING ACTIVITY (MCA) channels (Kurusu et al., 2012a, b) activate downstream pathways in response to osmotic stress-induced changes in cell turgor pressure. Furthermore, SEUSS, a transcriptional co-regulator of AGAMOUS, plays a critical role in osmotic stress responses. Under hyperosmotic stress, SEUSS forms liquid-like nuclear condensates, which are associated with osmotic stress tolerance and gene expression regulation. The absence of SEUSS significantly impairs the expression of osmotic stress tolerance genes (Wang et al., 2022a).
Abscisic acid (ABA), a key plant hormone, regulates various physiological processes that allow plants to survive under adverse environmental conditions. The role of ABA in plant adaptation to drought, cold, and salinity stress is well established (Zhu, 2016). Salt-induced osmotic stress triggers the expression of ABA biosynthesis genes, including NINECIS-EPOXYCAROTENOID DIOXYGENASEs (NCEDs), ABA DEFICIENT (ABA), and ABA Aldehyde Oxidase (AAO3), promoting ABA accumulation and enhancing environmental adaptability (van Zelm et al., 2020; Du et al., 2023). Protein phosphorylation is key to ABA signal transduction under stress conditions. Conserved in A. thaliana, rice, and maize, clade A-type 2C protein phosphatases (PP2Cs) and subclass III SNF1-related protein kinase 2s (SnRK2s) are central to this process (Geiger et al., 2009; Sirichandra et al., 2009; Klingler et al., 2010; Sun et al., 2016; Min et al., 2019; Wu et al., 2019; Wang et al., 2020c). Salt-induced ABA accumulation is detected by the Pyrabactin Resistance 1/PYR1-Like/Regulatory Component of ABA Receptors (PYR/PYL/RCAR) family, which inhibits PP2Cs, leading to the release of SnRK2s. These SnRK2s phosphorylate downstream anion efflux channels and transcription factors, regulating stomatal closure and gene expression (Negi et al., 2008; Chen et al., 2020b; Takahashi et al., 2020). This phosphorylation activates ABA-responsive element (ABRE)/ABRE-binding factor transcription factors, which promote the expression of stress-responsive genes like β-AMYLASE1 and α-AMYLASE, contribute to osmolyte accumulation, and improve water and nutrient uptake (Thalmann et al., 2016).
ABA is synthesized in various plant tissues, including vascular tissues and guard cells, and is transported to long distances to ensure proper signal transduction (Christmann et al., 2005; Li et al., 2018). For instance, under dehydration stress, the CLAVATA3/Embryo-Surrounding Region-Related 25 (CLE25) peptide is induced in the roots and transported to the leaves via the vascular system, where it triggers ABA biosynthesis and accumulation, promoting stomatal closure (Takahashi et al., 2018). CLE25 loss-of-function mutants exhibit heightened sensitivity to salt stress (Takahashi et al., 2018).
In addition to PP2Cs and SnRK2s, mitogen-activated protein kinase (MAPK) cascades, composed of MAP kinase kinase kinases (MKKKs/MAP3Ks/MEKKs), MAP kinase kinases (MKKs/MAP2Ks/MEKs), and MAP kinases (MAPKs/MPKs), are pivotal in plant responses to osmotic stress (Gasulla et al., 2016). These MAPK cascades exhibit functional diversity, allowing plants to adapt to various environmental stimuli and internal demands (Zhang et al., 2022a). In Arabidopsis, the MKK4-MPK3 and MKKK20-MPK6 modules are vital in osmotic stress responses, and loss-of-function mutations in MKKs improve tolerance to dehydration and salinity (Kim et al., 2011, 2012). The bread wheat TMPK3 (wheat Mitogen-Activated Protein Kinase) plays a vital role in plant tolerance to salt and osmotic stresses. TMPK3 can autophosphorylate in vitro and is phosphorylated by the constitutively active Arabidopsis kinase AtMKK2. Additionally, TMPK3 phosphorylates its substrate, MAP kinase phosphatase 1. Notably, TMPK3 can compensate for the salt sensitivity in the Arabidopsis mpk3-1 loss-of-function mutant, and its overexpression significantly enhances plant tolerance to salt and osmotic stresses beyond the levels observed in wild-type plants (Ghorbel et al., 2023). Similarly, Raf-like protein kinases (RAFs), which act as MKKKs, are essential for ABA-triggered SnRK2 activation under salt-induced osmotic stress (Lin et al., 2021).
Accumulation of metabolic substances like proline, hydroxyproline, glycine betaine, sugars, and polyamines helps stabilize osmotic pressure changes under salt stress. These osmolytes mitigate water loss and improve cell turgor (Pommerrenig et al., 2007; Henry et al., 2015; Mansour and Ali, 2017). Salt-induced proline accumulation maintains H3K4me3 levels at the Δ1-PYRROLINE-5-CARBOXYLATE SYNTHETASE 1 locus (Feng et al., 2016). A metabolomics analysis of 266 maize inbred lines identified 37 metabolic biomarkers (METOs) associated with salt-induced osmotic stress (SIOS). Research further revealed that ZmCS3 (citrate synthase), ZmUGT (glucosyltransferase), and ZmCYP709B2 (cytochrome P450) contribute to METO-SIOS tolerance (Liang et al., 2021).
Multiple studies suggest that salt stress impacts plants through osmotic stress, to which they have developed various adaptive mechanisms. These include Ca²+ signaling, ABA pathways, protein phosphorylation, kinase cascades, and osmolyte accumulation.
Plants exposed to high-salt environments face two primary types of stress that are exposed to high-salt environments: osmotic stress, caused by changes in osmotic potential that reduce water absorption, and ionic toxicity, due to the accumulation of toxic ions such as Na+. While osmotic stress has been extensively reviewed, the study of Na+ transport and accumulation is equally critical for understanding salt stress in plants. As sessile organisms, plants rely primarily on their roots to absorb water, nutrients, and ions from their surroundings. In high-salt environments, Na+, the most abundant soluble cation, enters plants through various ion channels and transporters in the roots, including calcium-permeable non-selective cation channels (NSCCs) and high-affinity K+ transporters (HKTs) (Tester and Davenport, 2003; Mian et al., 2011) (Figure 2).
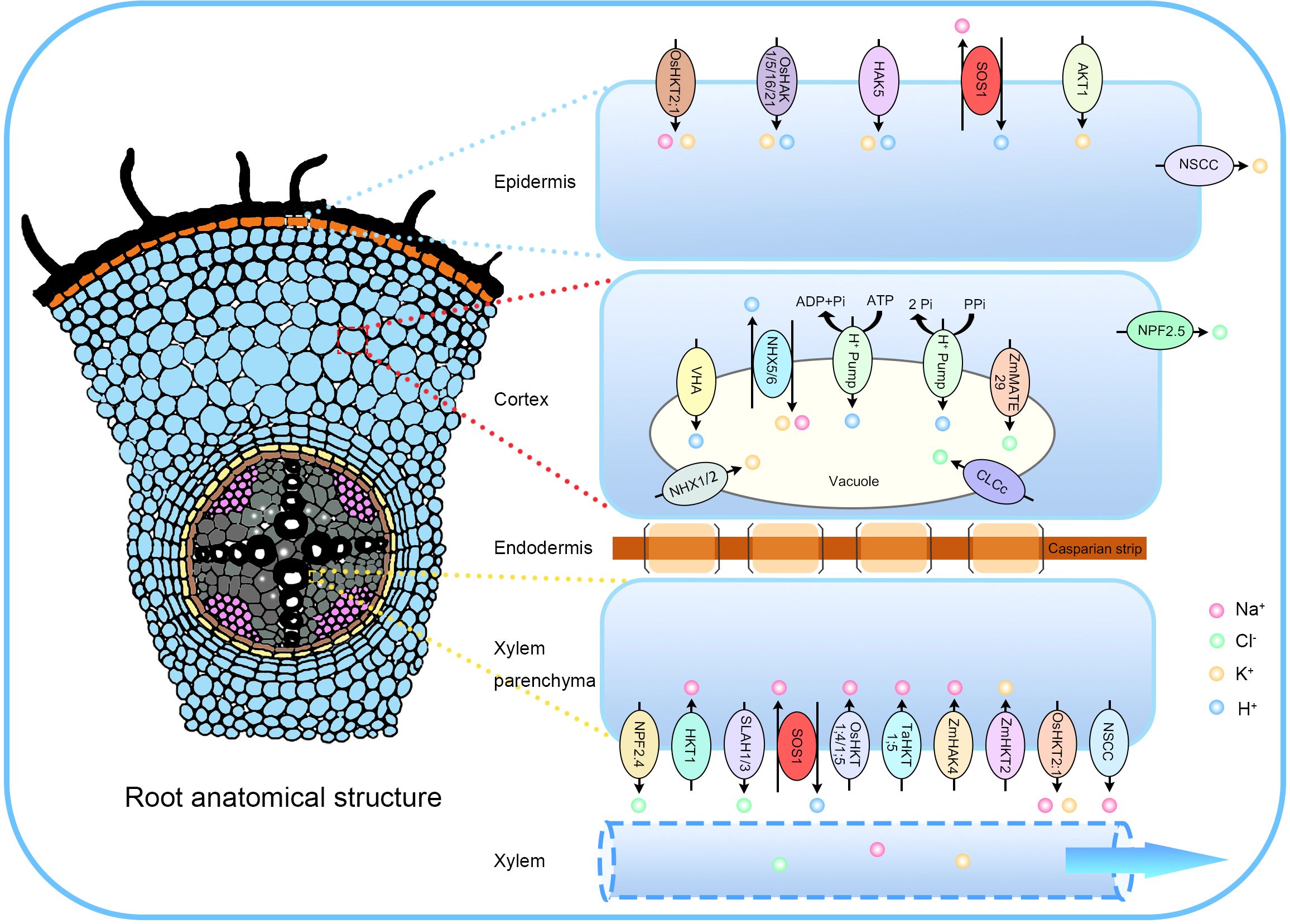
Figure 2. The translocation and compartmentalization of Na+, K+, and Cl− facilitated by transporters. Salt ions move symplastically across the epidermis, cortex, and endodermis, entering the xylem via xylem parenchyma cells. Na+ enters root cells through the plasma membrane (PM) via Ca²+-permeable non-selective cation channels (NSCCs) and the high-affinity K+ transporter OsHKT2;1 in rice. The SOS1 protein mediates Na+ extrusion, while K+-permeable NSCCs contribute to salt-induced K+ loss. To maintain K+ homeostasis, the influx of K+ is facilitated by PM inward K+ channels, including HAK5 and AKT1. Within cortical cells, NPF2.5 removes Cl− from the roots, while ZmMATE29 sequesters Cl− into vacuoles. Na+ sequestration into root vacuoles is mediated by the antiporter NHX, while Cl− sequestration occurs through the chloride channel CLCc, which operates with energy from H+-ATPase or H+-pyrophosphatase (H+-PPase) pumps. The Casparian strip prevents apoplastic entry of Na+ and Cl− into the stele. In xylem parenchyma cells, passive Na+ loading occurs via NSCCs and OsHKT2;1, while active Na+ loading is mediated by SOS1. Transporters such as HKT1, OsHKT1;4, OsHKT1;5, TaHKT1;5, and ZmHAK4 are involved in the retrieval of Na+ from the root xylem to distribute Na+ between the root and shoot. OsHKT2;1 is responsible for loading K+ into the xylem, while ZmHKT2 removes K+ from the xylem. Finally, NPF2.4, SLAH1, and SLAH3 are key facilitators of Cl− transport into the root xylem.
Transpiration leads to significant water loss, which in turn increases the concentration of ions like Na+ within plant tissues. To combat this, plants have developed sophisticated mechanisms to exclude excess salts from their cells (Munns et al., 2020). Studies have shown that plants primarily rely on membrane transporters responsible for Na+ uptake, export, and compartmentalization to limit Na+ accumulation, thereby mitigating salt stress. These mechanisms are vital for maintaining intracellular ion homeostasis and preserving normal physiological functions in salt-stressed plants (Gong et al., 2020; Liang et al., 2024). In recent years, an increasing number of studies have reported that membrane transporters and membrane proteins play a crucial role in the salt tolerance of plants (Isayenkov and Maathuis, 2019; Banik and Dutta, 2023; Dutta, 2023; Mishra et al., 2024). While these processes have been extensively studied in A. thaliana, they have also been observed in a variety of crop species, highlighting their broader relevance.
Roots serve as the primary organs for Na+ absorption in plants. The ion traverses the root epidermis, cortex, and endodermis before entering the xylem, where it is transported to the stem tissue. Excessive accumulation of Na+ in the stem can lead to toxic effects, prompting plants to evolve various mechanisms to limit Na+ entry into the root xylem and facilitate its recovery, thereby reducing transport to the stem.
HKT-type transporter proteins play a pivotal role in the retrieval of Na+ from the root xylem, significantly contributing to plant salt tolerance (Gong et al., 2020). Members of the HKT1 subfamily are known to reclaim Na+ from the root xylem, thereby minimizing its transport from roots to stems. This includes AtHKT1 in Arabidopsis; OsHKT1;1, OsHKT1;4, and OsHKT1;5 in rice; TaHKT1;4 and TaHKT1;5 in wheat; and ZmHKT1;1 and ZmHKT1;2 in maize (Ren et al., 2005; Huang et al., 2006; Munns et al., 2012; Byrt et al., 2014; Campbell et al., 2017; Oda et al., 2018; Zhang et al., 2018, 2023b; Wang et al., 2024c). In Arabidopsis, the athkt1 mutants exhibit excessive Na+ accumulation and hypersensitivity to Na+ in the stem (Berthomieu et al., 2003). AtHKT1 functions in root parenchyma cells to retrieve Na+ from the root xylem, thereby mitigating Na+ accumulation in the stem. Specifically, overexpression of AtHKT1 in the root stele enhances salt tolerance in Arabidopsis (Moller et al., 2009). Under salt stress, PP2C49, a type G PP2C, inhibits Na+ permeability of AtHKT1 (Chu et al., 2021).
In rice, OsHKT1;5 is crucial for removing Na+ from the root xylem, thus conferring salt tolerance to the plant (Ren et al., 2005; Kobayashi et al., 2017). Salt stress enhances the interaction between OsDNAJ15 and OsBAG4 in the nucleus, promoting the formation of transcriptional complexes that activate OsHKT1;5 (Liu et al., 2023). Additionally, the loss of function of OsWRKY53 facilitates Na+ homeostasis mediated by OsMKK10.2 and OsHKT1;5 (Yu J. et al., 2023). Literature also suggests that epigenetic modifications are involved in gene expression mediated by AtHKT1 and OsHKT1;5 under salt stress (Baek et al., 2011; Wang et al., 2020a; Liu et al., 2022a, b). Mutations in OsHKT1;1 can affect Na+ accumulation in rice roots and alter salt tolerance (Campbell et al., 2017).
In wheat, TaHKT1;4 and TaHKT1;5 are major genes that reduce Na+ transport and accumulation, enhancing salt tolerance. TaHKT1;4 sequesters Na+ in the leaf sheath, minimizing its transport and accumulation in leaf blades (Huang et al., 2006; James et al., 2006). Conversely, TaHKT1;5 functions similarly to AtHKT1, facilitating the retrieval of Na+ from the root xylem (Byrt et al., 2007). The SPL6-HKT1;5 module offers a target for the molecular breeding of salt-tolerant crops (Wang et al., 2024c). Notably, TaHKT1;5 enables durum wheat to increase grain yield by 25% in saline soils, significantly enhancing its economic value for developing salt-tolerant, high-yielding crop varieties (Munns et al., 2012).
In maize, the functions of ZmHKT1;1 and ZmHKT1;2 are closely linked to salt tolerance. Mutations in ZmHKT1;1 result in increased Na+ content in the leaves and a salt-sensitive phenotype (Zhang et al., 2018). ZmHKT1;2 is also associated with Na+ content in the shoots of maize seedlings under salt stress (Zhang et al., 2019, 2023b). Modern research indicates that salt tolerance in maize is a complex trait governed by multiple genes that influence Na+ transport and distribution within the plant.
The HKT1 subfamily of transporters primarily mediates sodium ion transport. In contrast, the HKT2 subfamily, exclusive to monocotyledonous plants, uniquely transports both sodium and potassium ions (Hamamoto et al., 2015; Ali et al., 2019). However, HKT2 transporters can also mediate Na+ uptake. For instance, ZmNC2/ZmHAK4 functions as a Na+-selective transporter located in the xylem parenchyma cells of the root stele, reducing Na+ accumulation in the stem by facilitating Na+ removal from the root xylem (Zhang et al., 2019). In rice, OsHKT2;1, localized to the plasma membrane, mediates Na+ uptake but is inhibited under salt stress (Horie et al., 2007). OsHAK12 promotes Na+ exclusion from stem tissues via a mechanism akin to ZmHAK4 (Figure 2) (Zhang et al., 2021).
Salt stress also promotes the development of the Casparian strip (CS), which forms a barrier that prevents salt ions from entering the root stele through the apoplastic pathway (Chen et al., 2011; Wang et al., 2022c). In summary, by minimizing the transport of Na+ from roots to shoots and enhancing its retrieval from the root xylem, plants can effectively mitigate the toxic effects of salt stress.
The discovery of SOS genes has provided a foundational understanding of plant salt tolerance. Over the past few decades, scientists have increasingly recognized the central role of the SOS signaling pathway in mediating Na+ extrusion from plant roots (Shi et al., 2002; Yang and Guo, 2018b). Through mutant screening experiments, researchers have identified various salt-sensitive mutants (sos mutants), cloned SOS genes, and thoroughly characterized their functions. It is now widely accepted that the SOS signaling pathway comprises three key proteins: SOS1, SOS2, and SOS3 (Zhu, 2001).
The SOS1 gene is highly conserved across sequenced genomes and encodes a plasma membrane Na+/H+ antiporter that utilizes the H+ gradient to drive Na+ extrusion, thereby lowering cytoplasmic Na+ concentrations (Figure 2) (Shi et al., 2000, 2002, 2003; Steinhorst et al., 2022; Zhang et al., 2023c, d). In A. thaliana, SOS1 uniquely possesses a long cytoplasmic C-terminal region exceeding 700 amino acid residues (Shi et al., 2000). This C-terminal region contains a self-inhibitory domain that interacts with upstream sequences harboring putative cyclic nucleotide monophosphate (cNMP) binding motifs, inhibiting SOS1 transport activity under normal conditions. These motifs are essential for SOS1 function, and mutations in these motifs lead to its inactivation (Shi et al., 2000). This suggests that SOS1 may be regulated by cyclic nucleotides, signaling molecules that mediate plant environmental adaptation and play roles in responses to salt and osmotic stress (Shabala et al., 2015; Swiezawska et al., 2018). The C-terminal region also interacts with numerous regulatory proteins and undergoes phosphorylation, modulating SOS1 antiporter activity (Fliegel, 2019). Under high-salt conditions, serine residues within this self-inhibitory domain are phosphorylated by SOS2, activating SOS1 to transport Na+ out of the cell and prevent salt toxicity (Quintero et al., 2011). The SOS2 gene encodes a member of the SNF1-related kinase 3 (SnRK3s), also known as CBL-interacting protein kinase 24 (CIPK24), which comprises 25 members in A. thaliana. Under high-salt conditions, the autoinhibition of SOS2 is relieved, enabling it to activate SOS1 through phosphorylation (Liu et al., 2000; Guo et al., 2001; Quintero et al., 2002). Meanwhile, SOS3 (also known as CBL4) encodes a Ca²+-binding protein with three EF-hand motifs that senses the elevation of Ca²+ levels triggered by salt stress. Its family includes 10 members (Liu and Zhu, 1998; Ishitani et al., 2000; Kim et al., 2007; Quan et al., 2007; Steinhorst et al., 2022; Yang and Guo, 2018a; Zhu, 2016).
The SOS3–SOS2–SOS1 module plays a critical role in plant salt tolerance. Mutants lacking any of these three genes exhibit heightened sensitivity to NaCl, underscoring their essential function in conferring salt tolerance. In this signaling pathway, SOS3 detects the salt-induced rise in Ca²+ levels and binds to the autoinhibitory domain of SOS2, activating it. The N-terminus of SOS3 is myristoylated, allowing it to bind to the plasma membrane and recruit the SOS3–SOS2 complex, where SOS2 phosphorylates SOS1. This phosphorylation enhances the Na+/H+ antiporter activity of SOS1, promoting Na+ extrusion and lowering intracellular Na+ concentrations. The Na+ extrusion mediated by the SOS3–SOS2–SOS1 module is a critical mechanism for maintaining ion homeostasis and preventing salt toxicity under high-salt conditions (Halfter et al., 2000; Qiu et al., 2002; Kim et al., 2007; Quan et al., 2007; Quintero et al., 2011; Steinhorst et al., 2022; Zhang et al., 2023c, d).
The CBL–CIPK signaling pathway plays a pivotal role in plant responses to various abiotic stresses (Weinl and Kudla, 2009; Kudla et al., 2018). Under salt stress, SOS2 specifically interacts with and phosphorylates SCaBP8 (also known as CBL10), promoting the recruitment of SOS2 to the plasma membrane, where SOS2 then phosphorylates SOS1, enhancing its Na+ extrusion transporter activity (Quan et al., 2007; Lin et al., 2009). Additionally, the SOS2–SCaBP8 complex phosphorylates and inhibits the putative Ca²+-permeable transporter AtANN4, forming a negative feedback loop to fine-tune Ca²+ signaling in response to salt stress (Ma et al., 2019). Research indicates that the SOS2–SOS3 complex primarily functions in the roots, while the SOS2–SCaBP8 complex acts predominantly in the shoots. These complexes confer salt tolerance by enhancing SOS1 transport activity (Quan et al., 2007; Lin et al., 2009; Yang and Guo, 2018a). Furthermore, the cbl8 mutant exhibits hypersensitivity to severe salt stress, highlighting the importance of CBL8 in salt stress responses (Steinhorst et al., 2022). CBL5, an ortholog of CBL4 and CBL10 in Arabidopsis, interacts with and recruits CIPK8 and CIPK24 to the plasma membrane. This interaction is essential during seed germination, where CBL5 helps protect seeds and germinating seedlings from salt stress via the CBL5-CIPK8/CIPK24-SOS1 pathway (Xie et al., 2024). Another member of the CIPK family, CIPK8, forms a complex with SCaBP8/CBL10 and SOS1, promoting salt tolerance (Yin et al., 2020).
Numerous regulatory factors fine-tune the SOS signaling pathway, enhancing plant responses to salt stress (Ali et al., 2023). In parallel with the regulation at the post-translational level, several protein complexes have been identified recently that regulate the SOS pathway core components transcriptionally. Among them, the histone linker protein HIS1-3 (negatively) and the transcription factor WRKY1 (positively) are two proteins that regulate all three core elements of the SOS pathway. HIS1-3 and WRKY1 bind to the same loci on the chromatin of the SOS pathway genes and regulate their transcription (Wu et al., 2022). Short Root in Salt Medium1(RSA1)-RSA1 Interacting Transcription Factor1(RITF1) is another complex that positively regulates the SOS1 transcript through a nuclear calcium signaling pathway (Shi et al., 2003; Guan et al., 2013). MPK4-MYB42 is a module that activates SOS2 transcription in a UBC1/UBC2-dependent manner (Sun Y. et al., 2020; Zarreen et al., 2022). The transcription factor plant AT-rich sequence and zinc-binding protein 2 (PLATZ2) suppresses SOS3/SCaBP8 transcription, thereby negatively regulating salt stress tolerance (Liu S. et al., 2020). Under normal conditions, ABI2 (Ohta et al., 2003), GIGANTEA (GI) (Kim et al., 2013), 14-3-3 proteins, and SOS2-like protein kinase 5 (PKS5) (Zhou et al., 2014; Yang et al., 2019c) interact with SOS2 to inhibit its kinase activity. The interaction between SOS2 and ABI2 suggests a potential link between the salt stress response and ABA signaling pathways. Under high-salt conditions, GI is degraded, releasing its inhibition of SOS2, which subsequently activates SOS2 to promote salt tolerance (Kim et al., 2013). PKS5-mediated phosphorylation of SOS2 enhances its interaction with 14-3-3 proteins, but during salt stress, elevated Ca²+ levels promote 14-3-3 binding to PKS5, inhibiting PKS5 kinase activity and weakening the interaction between 14-3-3 and SOS2, thereby activating SOS2 (Yang et al., 2019c). GRIK1 phosphorylates and promotes SOS2 activity (Barajas-Lopez et al., 2018), while VPS23A enhances the SOS2–SOS3 interaction, promoting SOS2 localization to the plasma membrane (PM) (Lou et al., 2020). The SOS signaling pathway has been shown to connect salt stress with plant hormone signaling pathways (Yu et al., 2020). SOS2 enhances salt tolerance by inhibiting the kinase activity of CONSTITUTIVE TRIPLE RESPONSE1 (CTR1) through phosphorylation at serine 87 (S87), thereby activating the ethylene signaling response (Li et al., 2024). In addition, AFP2, a plant-specific ABI5-binding protein, serves as a negative regulator in the ABA signaling pathway and plays a key role in salt tolerance during seed germination. AFP2 mutations increase sensitivity to salt stress. SOS2 physically interacts with and stabilizes AFP2, promoting the degradation of ABI5, a transcription factor that negatively regulates seed germination under salt stress. These findings suggest a potential link between salt stress and the ABA signaling pathway, opening new possibilities for enhancing plant resilience to environmental challenges (Wang et al., 2024a). Under salt stress, SCaBP8 inhibits the activity and PM localization of two type-D protein phosphatases (PP2C D6/D7), thereby activating SOS1 (Fu et al., 2023). PAMP-induced secreted peptide 3 (PIP3) binds and activates receptor-like kinase 7 (RLK7), which in turn activates MPK3 and MPK6, enhancing SOS1 activity and promoting Na+ homeostasis via ethylene/ROS signaling (Yu et al., 2010; Li et al., 2014; Zhou et al., 2022a). Salt-induced cytosolic Ca²+ increases facilitate the activation of phospholipase D (PLD) at the PM, generating the lipid messenger phosphatidic acid (PA) (Wang et al., 2006; Do et al., 2019). PA accumulation promotes the localization and enhances the kinase activity of SOS2 and MPK6 at the PM, facilitating SOS1-mediated Na+ efflux (Yu et al., 2010; Li et al., 2023a).
Beyond Arabidopsis, the SOS pathway is conserved in rice (Martinez-Atienza et al., 2007; El Mahi et al., 2019), maize (Zhang et al., 2016a; Zhou et al., 2022b; Li et al., 2023a, b), wheat (Feki et al., 2011; Yang et al., 2024), soybean (Zhang et al., 2022b), and tomato (Wang et al., 2021; Hong et al., 2023). In summary, the SOS pathway and its regulatory factors play crucial roles in promoting Na+ efflux from the roots into the soil solution and enhancing salt tolerance in shoot tissues (Quan et al., 2007; Liang et al., 2024).
Researchers have long recognized that Na+ compartmentalization serves as an effective mechanism for plants to mitigate Na+ toxicity under salt stress conditions. Excess Na+ absorbed by plant roots can be sequestered into vacuoles or transported to the aerial parts of the plant. The central vacuole is particularly well-suited for Na+ sequestration, as it reduces cytoplasmic Na+ accumulation. This sequestration offers several advantages: it decreases Na+ concentration in the cytoplasm, preventing transport from underground parts to aerial tissues, thereby protecting plants from salt-induced toxicity. Furthermore, Na+ accumulation in vacuoles can act as an osmoregulatory substance, reducing cell water potential and promoting water uptake under salt stress (Yang and Guo, 2018a). While the size of plant cells is finite, the process of sequestering Na+ into vacuoles is an effective cellular strategy throughout the plant’s life cycle, safeguarding against Na+ toxicity caused by salt stress.
The sequestration of Na+ in vacuoles is mediated by vacuolar Na+/H+ exchangers (NHXs), which rely on the H+ gradient established by vacuolar H+-ATPases (V-ATPase, VHA) and H+-pyrophosphatases (V-PPase, VP) (Drozdowicz and Rea, 2001; Brini and Masmoudi, 2012). The NHX gene family is conserved across various plant species, including Arabidopsis, tomato, and several crops (Apse et al., 1999; Zhang and Blumwald, 2001; Zhang et al., 2001; Zhang and Shi, 2013). In Arabidopsis, the NHX family comprises six members, and overexpression of the AtNHX1 gene has been shown to enhance salt tolerance in multiple plant species (Zhang and Shi, 2013). Studies indicate that the C-terminal region of AtNHX1 interacts with calmodulin (CaM) within the vacuole, inhibiting the transport activity of AtNHX1 in a Ca²+-dependent manner (Yamaguchi et al., 2003, 2005). Gain-of-function mutations in AtNHX1 can suppress salt hypersensitivity in sos1 mutants by limiting Na+ accumulation in the cytoplasm and its transport to the shoots (Pabuayon et al., 2021). Additionally, AtNHX1 and AtNHX2 are instrumental in K+ accumulation in vacuoles and the regulation of vacuolar pH (Bassil et al., 2011b; Barragan et al., 2012). AtNHX5 and AtNHX6, localized in the Golgi, trans-Golgi vesicles, and prevacuolar compartments, are critical for maintaining the pH of these compartments (Bassil et al., 2011a; Reguera et al., 2015). The nhx5 nhx6 double mutants exhibit salt hypersensitivity, likely due to the mislocalization of vacuolar transporters essential for Na+ sequestration (Bassil et al., 2011b).
In addition to NHX proteins, the vacuolar membrane-localized SCaBP8 also plays a pivotal role in Na+ sequestration by interacting with SOS2 (Kim et al., 2007; Yang et al., 2019b). Moreover, salt-induced endocytosis contributes to Na+ accumulation in vacuoles. For example, overexpression of AtRab7, a gene involved in vesicle trafficking regulation, enhances endocytosis in protoplasts, roots, and leaves, promoting Na+ accumulation in vacuoles (Mazel et al., 2004; Hamaji et al., 2009; Golani et al., 2013). In summary, NHX-type Na+/H+ antiporters are essential for mediating Na+ sequestration in plant cells, thereby promoting salt tolerance, growth, and development (Figure 2) (Zhang and Shi, 2013). However, the membrane transport mechanisms involved in intracellular Na+ sequestration remain poorly understood and warrant further investigation (Liang et al., 2024).
In addition to Na+, maintaining K+ homeostasis under high salinity conditions is equally crucial for plant growth and development. Achieving K+ homeostasis in saline environments necessitates facilitating K+ influx while inhibiting K+ efflux (Yang and Guo, 2018a, b). K+ influx primarily relies on transporters such as HAKs and AKT (Gierth et al., 2005). In Arabidopsis, the HAK5 protein promotes high-affinity K+ uptake induced by K+ deprivation (Ragel et al., 2015). Similarly, in rice, OsHAK1, OsHAK5, OsHAK16, and OsHAK21 function analogously to Arabidopsis HAK5, enhancing K+ uptake under salt stress (Song et al., 2021). The AKT1 transporter is responsible for primary K+ uptake in Arabidopsis roots (Lagarde et al., 1996). Under salt stress, SOS2 promotes the phosphorylation of SCaBP8, alleviating its inhibitory effect on AKT1 and enhancing K+ uptake in the roots (Li et al., 2023). Conversely, the ZmHKT2 protein in maize negatively regulates K+ accumulation in the stems, which reduces the salt tolerance of maize (Cao et al., 2019). TaHKT9-B is a K+-preferring HKT transporter. The tae-miR390/TaTAS3/TaARF4/TaHKT9-B module has been identified as a crucial regulatory pathway in wheat under salt stress, offering valuable genetic resources for breeders aiming to enhance wheat salt tolerance (Du et al., 2024). These findings underscore the significance of maintaining K+ homeostasis for plant salt tolerance, emphasizing the need to study K+ transport mechanisms to improve crop salt tolerance.
Excessive Cl− accumulation under salt stress can limit NO3− uptake, transport, and assimilation (Li et al., 2017), potentially leading to Cl− toxicity (Ren et al., 2021). Consequently, Cl− uptake and transport processes are intricately linked to plant salt tolerance. Reported transporters include the Arabidopsis nitrate transporter/peptide transporters (AtNPF2.4 and AtNPF2.5), slow-type anion channel associated 1 homologs (AtSLAH1 and AtSLAH3), cation/Cl− cotransporters (AtCLCc and AtCLCg), aluminum-activated malate transporter 9 (ALMT9), and the rice MATE (multidrug and toxic compound extrusion family) transporter BIG RICE GRAIN 1 (BIRG1). Additionally, ZmMATE29, type-A response regulator (ZmRR1), and histidine phosphotransfer protein 2 (ZmHP2) in maize maintain Cl− homeostasis by facilitating Cl− retrieval from the root xylem, vacuolar Cl− accumulation, or Cl− efflux from the roots, thereby regulating plant salt tolerance (Figure 2) (Colmenero-Flores et al., 2007; Negi et al., 2008; Jossier et al., 2010; Teakle and Tyerman, 2010; De Angeli et al., 2013; Baetz et al., 2016; Cubero-Font et al., 2016; Li et al., 2016a, b; Henderson et al., 2018; Ren et al., 2021; Yin et al., 2023).
Throughout their life cycle, plants are constantly subjected to both biotic and abiotic stresses, which often lead to the accumulation of ROS and trigger stress responses, with salt stress being a significant factor. The generation of ROS in plants primarily involves four forms: hydroxyl radicals, hydrogen peroxide (H2O2), superoxide anions, and singlet oxygen (Yang et al., 2023; Jiang et al., 2024). Salt stress induces the transcription of genes encoding Respiratory Burst Oxidase Homologs D (RBOHD) and RBOHF, which subsequently catalyze the production of H2O2 (Miller et al., 2010). This process also triggers changes in Ca²+ signaling, mediating the overall response of plants to local salt stress through the RBOHD–RBOHF–H2O2–Ca²+ coupled signaling network (Evans et al., 2016). Excessive ROS accumulation can damage cells, leading to lipid peroxidation of cell membranes, DNA damage, protein denaturation, carbohydrate oxidation, pigment decomposition, and impaired enzyme activity (Noctor and Foyer, 1998).
To cope with elevated ROS levels induced by stress, plants synthesize antioxidant enzymes and non-enzymatic antioxidants to maintain ROS homeostasis. Antioxidant enzymes include superoxide dismutase (SOD), catalase (CAT), ascorbic acid peroxidase (APX), glutathione peroxidase (GPX), glutathione reductase (GR), dehydroascorbate reductase (DHAR), monodehydroascorbate reductase (MDHAR), and glutathione S-transferase (GST) (Das and Roychoudhury, 2014). Non-enzymatic antioxidants comprise glutathione, ascorbic acid, flavonoids, carotenoids, phenolic compounds, and tocopherols (Yang and Guo, 2018a). In high-salt environments, maize enhances salt tolerance by inducing ROS accumulation and activating the synthesis of various antioxidant enzymes through moderate expression of the male sterility gene Open Reading Frame 355 (ZmORF355) (Xiao S. et al., 2023). Salt stress-induced ROS accumulation inhibits the accumulation of maize microRNA ZmmiR169q, promoting the transcription of the antioxidant enzyme gene peroxidase 1 (ZmPER1), which mediates ROS elimination and protects against salt stress (Xing et al., 2022). Stress-induced ROS also inhibits the accumulation of maize miR408, increasing the transcription levels of its target genes LACCASE 9 (ZmLAC9) and ZmLAC18, thereby promoting cell wall development and salt tolerance by regulating the polymerization of lignin monomers (Qin et al., 2023). ZmIAA9, a member of the maize Aux/IAA gene family, acts as a positive regulator of salt tolerance in maize, accompanied by increased ROS detoxification and elevated expression of ROS-scavenging genes. The transcription factor ZmbHLH32, part of the bHLH family, directly binds to the promoter region of ZmIAA9, activating its expression. The ZmbHLH32-ZmIAA9-ZmARF1 module is therefore crucial in regulating salt tolerance in maize (Yan Z. et al., 2023). Overexpression of rice SALT TOLERANCE RECEPTOR-LIKE CYTOPLASMIC KINASE 1 (OsSTRK1) leads to phosphorylation and activation of CATALASE C (CatC), resulting in higher CAT activity, reduced H2O2 accumulation, and enhanced salt tolerance compared to controls (Zhou et al., 2018). Transgenic rice lines expressing MIM396 and OE-GRF6 show reduced H2O2 levels and increased activities of ROS-scavenging enzymes (CAT, SOD, and POD), contributing to significant enhancement of salt tolerance mediated by the miR396b/GRF6 module (Yuan et al., 2024). The rice OsTET5 (tetraspanins) protein regulates reactive oxygen species homeostasis by modulating the expression and activity of antioxidant pathway enzyme genes, as well as the accumulation of proline. This regulation contributes to enhancing the salt tolerance of rice (Mani et al., 2024). OsNF-YC5 encodes a putative subunit of the NF-Y transcription factor in rice. The osnf-yc5 mutant exhibits reduced levels of H2O2 and malondialdehyde (MDA), as well as increased CAT activity under salt stress. Furthermore, in the mutant lines, both ABA-dependent marker genes (OsABI2 and OsLEA3) and ABA-independent marker genes (OsDREB1A, OsDREB1B, and OsDREB2A) are upregulated in response to salt stress. These results suggest that knocking out OsNF-YC5 enhances rice salt tolerance by boosting CAT enzyme activity and modulating gene expression in both ABA-dependent and ABA-independent pathways (Yan et al., 2024). Furthermore, ABA demonstrates a notable role in managing salt stress-induced oxidative stress in salt-sensitive rice cultivars. Under salt stress conditions, ABA-treated Swarna sub1 (a salt-sensitive rice variety) exhibits increased relative water content, an elevated K+/Na+ ratio, and enhanced cell membrane stability. Additionally, ABA treatment reduces cell wall peroxidation, leading to the enlargement of the endodermal lumen and a decrease in malondialdehyde content. In summary, rice varieties with higher accumulation of ABA can improve their salt tolerance (Deng et al., 2023; Das et al., 2024). The mutation in the gene encoding a cytochrome b561 domain-containing protein (OsCYBDOMG1) results in decreased ascorbic acid (AsA) content and AsA/DHA (dehydroascorbate) ratio, leading to increased H2O2 accumulation and reduced salt stress tolerance (Deng et al., 2023). Salt stress induces the expression of GDP-mannose pyrophosphorylase (vitamin C1, VTC1), promoting ascorbic acid (AsA) synthesis and salt tolerance in A. thaliana (Zhang et al., 2012). RADICAL-INDUCED CELL DEATH 1 (RCD1) is identified as an essential player in salt stress response. Arabidopsis NAC domain-containing protein 17 (ANAC017) functions downstream of RCD1 in the salt stress response and plays a negative role by impairing SOD enzyme activity. RCD1 promotes salt stress response and maintains ROS homeostasis by inhibiting the activity of ANAC017 (Tao et al., 2023). The wheat WRKY transcription factor TaWRKY17 enhances salt stress tolerance by regulating ABA/ROS-related and stress-responsive genes, as well as by increasing antioxidative stress capabilities. Overexpression of TaWRKY17 significantly improves the plant’s tolerance to salt stress. Under salt stress conditions, compared to the wild type (WT), transgenic wheat plants overexpressing TaWRKY17 exhibit increased enzyme activities of SOD, POD, and CAT, while the accumulation of H2O2 is reduced. Furthermore, the transgenic wheat plants show regulated expression of ABA/ROS-related and stress-responsive genes, leading to enhanced tolerance to salt stress (Yu Y. et al., 2023). Under salt stress conditions, the levels of SOD and proline in wheat overexpressing TaGB1-B (G-Protein β-Subunit Gene) were higher than those in the control, while the concentration of MDA was lower. This indicates that TaGB1-B enhances the salt tolerance of wheat by scavenging ROS (Xiong et al., 2023). The overexpression of the wheat 2-Cys peroxiredoxin gene TaBAS1 enhances tolerance to oxidative stress by promoting the activity of ROS-scavenging enzymes and reducing the accumulation of ROS under salt stress, thereby enhancing salt tolerance at both the germination and seedling stages of wheat (Xiao G. et al., 2023). Furthermore, under salt stress conditions, the ectopic expression of wheat BR synthesis gene TaDWF4 and BR signaling gene TaBAK1 can enhance plant salt tolerance by balancing the levels of ROS in the roots (Hou et al., 2024). Through these enzymatic and non-enzymatic reactions, plants maintain ROS stability to cope with various abiotic stresses, including salt stress (Zhang et al., 2014, 2017; Wang et al., 2020d). Additionally, the salt-induced ABA signaling pathway regulates the production and distribution of ROS within plants, mitigating damage caused by stress-induced ROS accumulation and enhancing plant salt tolerance (Zhang et al., 2009; He et al., 2022).
Pluripotent stem cells play a pivotal role throughout the entire life cycle of biological development, governing the continuous regeneration of tissues and organs. This sustained regenerative capacity enables many plants to survive for hundreds or even thousands of years (Sang et al., 2018; McKim, 2019; Hata and Kyozuka, 2021; Umeda et al., 2021). The localization of stem cells within specific microenvironments is crucial for cellular differentiation, allowing these initially undifferentiated cells to retain robust self-renewal capabilities (Aichinger et al., 2012; Wang et al., 2020b; Nicolas and Laufs, 2022). In recent years, stem cell research has primarily focused on elucidating the processes of stem cell initiation, maintenance, and signal transduction (Sablowski, 2011; Janocha and Lohmann, 2018; Motte et al., 2019; Fujinami et al., 2020; Xue et al., 2020). As sessile organisms, plants constantly face challenges from abiotic stresses throughout their life cycle, including drought, salinity, temperature fluctuations, heavy metal ion exposure, and ultraviolet radiation (Sun H. et al., 2020; Markham and Greenham, 2021; Zhang et al., 2022a; Kopecka et al., 2023).
Elevated soil salinity affects plant development, inevitably leading to reduced crop yields. The SOS signaling pathway serves as a conserved and vital regulatory mechanism for excluding Na+ and mitigating its toxic long-distance transport (Shi et al., 2002; El Mahi et al., 2019). This pathway involves Ca²+ sensors, kinases, and Na+/H+ exchanger modules. Salt stress induces the expression of GSO1 in the endodermis and meristems (Chen et al., 2023; Jiang et al., 2024). The GSO1–SOS2–SOS1 module functions as a dedicated intracellular Na+ detoxification channel, enabling roots to continue growing in high-salt environments (Chen et al., 2023). The plant transcription factors PLETHORA1/2 (PLT1/2), featuring the AP2 domain, play a pivotal role in regulating responses to salt stress. Under salt stress conditions, the SOS2 protein phosphorylates PLT1/2, stabilizing them and thereby preserving the activity of meristematic tissues, which facilitates the recovery of root growth once the salt stress abates (Hao et al., 2023).
Simultaneously, the resilience and adaptability of apical meristem stem cells to salt stress are closely tied to redox reactions, ROS, nitric oxide (NO), microRNAs, and plant hormones such as auxin and cytokinins (Yang and Lee, 2023; Yu et al., 2020). Plant hormones are crucial for regulating responses to salt stress, which typically impedes plant growth. Research has shown that stress hormones, including ABA, SA, JA, and ethylene, as well as growth hormones like auxin, cytokinins (CKs), gibberellins (GAs), and brassinosteroids (BRs), play vital roles in mediating salt stress signaling while balancing growth and stress responses. Some hormones positively influence salt tolerance, while others can have inhibitory effects (Yu et al., 2020; Jiang et al., 2024). Salt stress has been associated with altered redox status in Arabidopsis root meristems, affecting root growth (Jiang et al., 2016). Under optimal conditions, the region with the lowest redox potential in the quiescent center (QC) coincides with the area of maximum auxin accumulation (Jiang et al., 2016; Smolko et al., 2021). However, under salt stress, auxin signaling in this region is notably reduced (Smolko et al., 2021). Auxin is known to support developmental plasticity under abiotic stresses, including salinity, and promotes ROS production via NADPH oxidase activation (Bartoli et al., 2013; Korver et al., 2018). These findings suggest a link between redox balance and auxin regulation under salt stress, collectively influencing adaptive root growth. Salt stress also induces miR393 expression, which suppresses auxin receptor proteins, such as TRANSPORT INHIBITOR RESISTANT1 (TIR1) and AUXIN SIGNALING F-BOX (AFB), at the post-transcriptional level (Iglesias et al., 2014). In mir393ab and tir1 afb2 mutants, the inhibitory effect of salt on root growth is less pronounced compared to WT plants (Iglesias et al., 2010, 2014). This suggests a complex network of salt-responsive miRNAs, redox status, and auxin dynamics in the root meristem, which is crucial for root plasticity under salt stress. NO also plays a significant role in plant growth and development, particularly in the root system (Ma et al., 2020). Salt stress-induced NO signaling reduces cell division and promotes cell differentiation in the root meristem (Fernandez-Marcos et al., 2011). Moreover, using NO biosynthesis inhibitors can mitigate the salt-induced inhibition of root meristem growth (Liu et al., 2015). The protein PIN1, which regulates auxin distribution, modulates Rho-of-plant 2 (ROP2) GTPase-mediated endocytic recycling in the root meristem, essential for NO-mediated root growth inhibition (Kenesi et al., 2023). Additionally, salt stress decreases the expression of miR165 and miR166, leading to an upregulation of PHABULOSA (PHB) and increased production of cytokinins, which are associated with pre-differentiation signaling in root meristems (Figure 3) (Scintu et al., 2023).
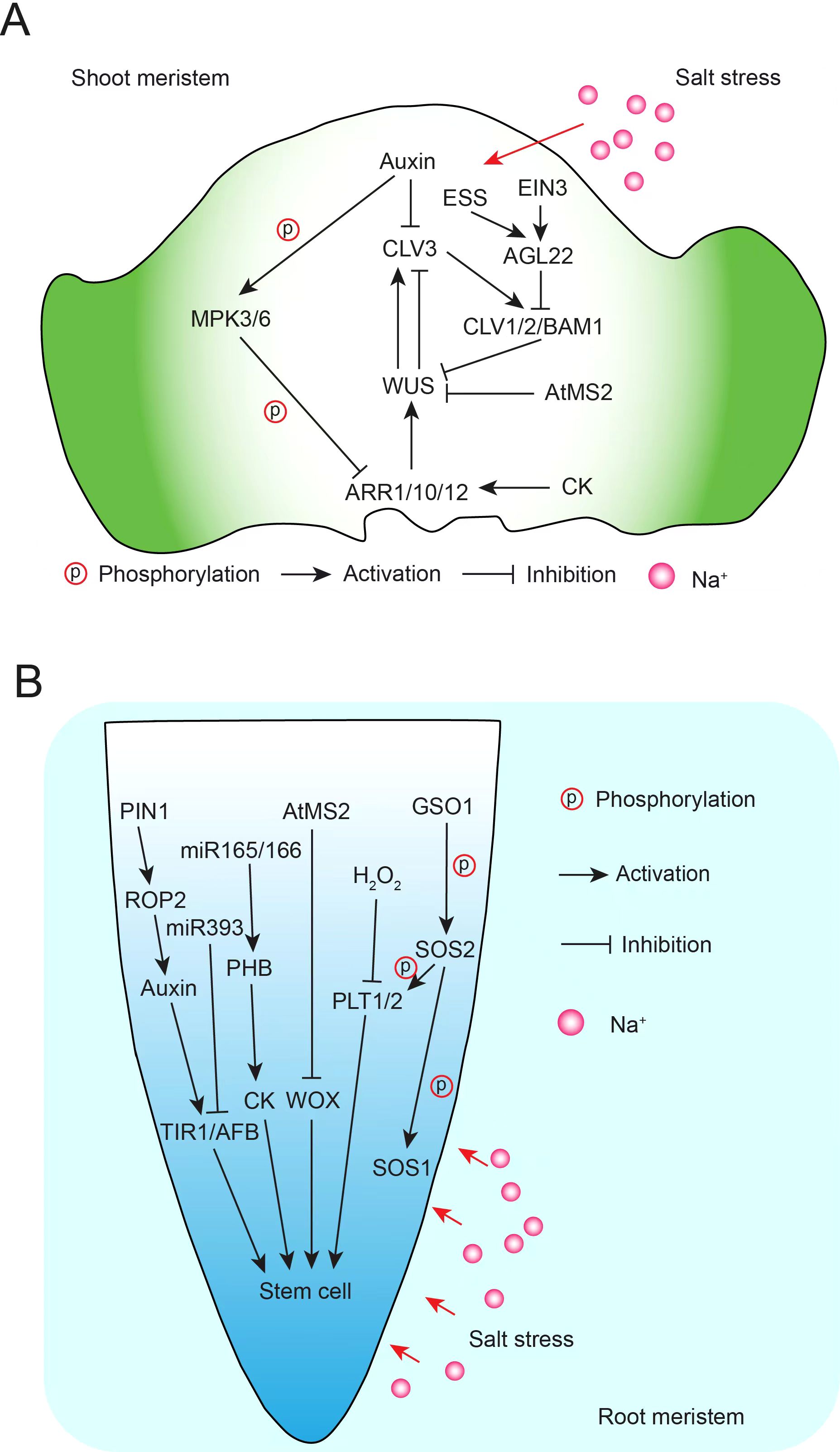
Figure 3. A schematic diagram of stem cell development and salt stress. (A) Signaling pathways of shoot meristem stem cell development under salt stress. (B) Signaling pathways of root meristem stem cell development under salt stress.
Stem cell homeostasis in the shoot apical meristem (SAM) is primarily governed by a feedback loop between CLAVATA3 (CLV3) and WUSCHEL (WUS) (Kitagawa and Jackson, 2019). Interestingly, CLV3 loss-of-function mutants, which enhance stem cell signaling in the SAM, exhibit a salt-tolerant phenotype in overall stem growth compared to WT plants under salt stress (Jun et al., 2019). Additionally, double mutants with loss-of-function in CLV1 and BAM1, receptors for the CLV3 peptide in the SAM, show higher survival rates under salt stress (Jun et al., 2019; Lee et al., 2019). These findings suggest that CLV3p-CLV1/BAM1 signaling may contribute to stress tolerance, including salinity. The SAM’s activity, indicated by cell division in clv3-2 mutants, appears essential for this salt tolerance effect. Moreover, MPK3 and MPK6 are implicated in the salt stress response. Under salt stress, these kinases phosphorylate and degrade Arabidopsis Response Regulators ARR1, ARR10, and ARR12, enhancing stress tolerance (Yan Z. et al., 2021). ARR proteins are central to CK signaling, integral to plant development and stem cell maintenance (Hwang et al., 2012). CK signaling has been shown to inhibit growth adaptation under high-salt conditions (Nishiyama et al., 2011). In addition, the MKK7–MPK6 module modulates SAM growth, as constitutive MKK7 expression leads to SAM defects (Doczi et al., 2019), underscoring the probable importance of MAPK signaling in salt-induced stress adaptation in the SAM. Proline, an osmolyte and ROS scavenger, accumulates in many plant species under salt-induced osmotic stress (Verslues and Sharma, 2010; Meena et al., 2019). Proline-mediated regulation bridges salt stress and SAM plasticity, aiding SAM growth adaptation (Mattioli et al., 2008; Szekely et al., 2008; Mattioli et al., 2009, 2022). Similar to root plasticity, the redox balance maintained by ROS is crucial for regulating SAM development under salt stress by managing the balance between stem cell proliferation and differentiation (Lee, 2018). Endogenous stress-related signals (ESS), including stress hormones, regulate stem cell maintenance in the SAM under natural growth conditions. Ethylene signaling, mediated by the transcription factor EIN3, activates AGAMOUS-LIKE 22 (AGL22), which represses CLV1 and CLV2 and is a key regulator in stress-responsive gene networks (Gregis et al., 2013; Bechtold et al., 2016). AGL22 thus functions as a signaling hub for the SAM’s developmental plasticity in response to ESS and external stress like salinity (Zeng et al., 2021). Recent studies have also highlighted the role of the prion-like domain (PrD) in the SAM regulator SHOOT MERISTEMLESS (STM), which forms nuclear condensates under salt stress, enhancing plant salt tolerance (Cao et al., 2023). A combined metabolomic and transcriptomic analysis of CK signaling-deficient Arabidopsis mutants (ahp2,3,5 and arr1,10,12) under salt stress demonstrated that CK signaling reprograms gene-metabolic networks linked to salinity responses (Abdelrahman et al., 2021). Furthermore, Arabidopsis methionine synthase 2 (AtMS2) inhibits stem cell maintenance under salt stress. While primarily cytoplasmic under normal conditions, AtMS2 accumulates in the nucleus under salt stress, where it interacts with WUS/WOX proteins to repress WUS/WOX expression, thus limiting stem cell maintenance. Mutations in AtMS2 result in increased salt tolerance, indicating that AtMS2 acts as a negative regulator of stem cell maintenance under stress (Qiu et al., 2024). Collectively, these findings suggest intricate interactions between salt stress signaling and the regulatory pathways governing meristem stem cell homeostasis (Figure 3) (Yang and Lee, 2023).
In recent years, global climate change has intensified, leading to more frequent and prolonged environmental stress events that pose significant challenges to plant growth, development, and crop yield. As sessile organisms, plants have evolved intricate systems to withstand abiotic stresses. This article reviews the findings of researchers on salt stress in plants such as A. thaliana and important crops including wheat, maize, and rice. It summarizes key issues such as how plants perceive salt (Na+) stress; the mechanisms of response; Na+ transport, compartmentalization, and clearance; and changes in ROS induced by salt stress. Furthermore, researchers have explored the regulation of plant stem cell development by salt stress and the interplay between plant hormones and salt stress regulation. Meanwhile, we also summarize some genes and protein factors involved in the above biological processes (Supplementary Table S1). Additionally, there has been a growing focus on the advantages of salt stress proteins and rhizosphere microbes in enhancing plant salt tolerance and increasing crop yields under high-salinity conditions (Athar et al., 2022; Liu et al., 2022c).
However, many questions and challenges persist regarding how plants, especially crops, cope with salt stress and how we can apply our understanding of plant salt tolerance to boost food crop yields in saline–alkali soils while developing new salt-tolerant varieties (Liang et al., 2024). Currently, advancements in single-cell and spatial transcriptomics, as well as high-throughput phenomics platforms, have facilitated a comprehensive understanding of plant responses to salt stress at the molecular, cellular, and subcellular levels. Quantitative trait locus (QTL) mapping and genome-wide association studies (GWAS) have gradually identified crucial abiotic stress response regulators and natural allelic variations in crop species. Integrated analyses of transcriptomics, spatial transcriptomics, proteomics, metabolomics, and phenomics provide effective and rapid tools for mining crop salt tolerance genes and screening for high-quality salt-tolerant lines. The development of various gene-editing technologies will expedite the utilization of salt tolerance genes identified through forward or reverse genetics in breeding new salt-tolerant varieties. Combining big data-based artificial intelligence (AI) with foundational knowledge of plant salt tolerance will enable the simulation and prediction of crop responses to salt stress, facilitating the molecular design of salt-tolerant crops and the breeding of new varieties that are both salt-tolerant and high-yielding.
Given the global importance of wheat, maize, and rice for food security, conducting salt tolerance research on these crops and breeding salt-tolerant, high-yield varieties are invaluable for optimizing the utilization of saline–alkali soils and ensuring a stable food supply. With continuous advancements in scientific and technological methods, the mechanisms underlying crop salt tolerance are gradually being unraveled. These studies are expected to contribute to the cultivation of more salt-tolerant crop varieties and promote the development of high and stable crop yields in high-salinity environments.
JZ: Writing – original draft. CY: Writing – review & editing. QZ: Writing – review & editing. ZQ: Writing – review & editing. XZ: Writing – review & editing. YH: Writing – review & editing. HZ: Writing – original draft.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by grants from the Taishan Scholar Foundation of Shandong Province (tsqnz20231242) and the Key R&D Program of Shandong Province, China (ZR202211070163).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
The author(s) declare that no Generative AI was used in the creation of this manuscript.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2025.1527952/full#supplementary-material
Abdelrahman, M., Nishiyama, R., Tran, C. D., Kusano, M., Nakabayashi, R., Okazaki, Y., et al. (2021). Defective cytokinin signaling reprograms lipid and flavonoid gene-to-metabolite networks to mitigate high salinity in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 118, e2105021118. doi: 10.1073/pnas.2105021118
Aichinger, E., Kornet, N., Friedrich, T., Laux, T. (2012). Plant stem cell niches. Annu. Rev. Plant Biol. 63, 615–636. doi: 10.1146/annurev-arplant-042811-105555
Ali, A., Maggio, A., Bressan, R. A., Yun, D. J. (2019). Role and functional differences of HKT1-Type transporters in plants under salt stress. Int. J. Mol. Sci. 20, 1059. doi: 10.3390/ijms20051059
Ali, A., Petrov, V., Yun, D. J., Gechev, T. (2023). Revisiting plant salt tolerance: novel components of the SOS pathway. Trends Plant Sci. 28, 1060–1069. doi: 10.1016/j.tplants.2023.04.003
Apse, M. P., Aharon, G. S., Snedden, W. A., Blumwald, E. (1999). Salt tolerance conferred by overexpression of a vacuolar Na+/H+ antiport in Arabidopsis. Science 285, 1256–1258. doi: 10.1126/science.285.5431.1256
Athar, H. U., Zulfiqar, F., Moosa, A., Ashraf, M., Zafar, Z. U., Zhang, L., et al. (2022). Salt stress proteins in plants: An overview. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.999058
Baek, D., Jiang, J., Chung, J. S., Wang, B., Chen, J., Xin, Z., et al. (2011). Regulated AtHKT1 gene expression by a distal enhancer element and DNA methylation in the promoter plays an important role in salt tolerance. Plant Cell Physiol. 52, 149–161. doi: 10.1093/pcp/pcq182
Baetz, U., Eisenach, C., Tohge, T., Martinoia, E., De Angeli, A. (2016). Vacuolar chloride fluxes impact ion content and distribution during early salinity stress. Plant Physiol. 172, 1167–1181. doi: 10.1104/pp.16.00183
Bailey-Serres, J., Parker, J. E., Ainsworth, E. A., Oldroyd, G. E. D., Schroeder, J. I. (2019). Genetic strategies for improving crop yields. Nature 575, 109–118. doi: 10.1038/s41586-019-1679-0
Banik, S., Dutta, D. (2023). Membrane proteins in plant salinity stress perception, sensing, and response. J. Membr Biol. 256, 109–124. doi: 10.1007/s00232-023-00279-9
Barajas-Lopez, J. D., Moreno, J. R., Gamez-Arjona, F. M., Pardo, J. M., Punkkinen, M., Zhu, J. K., et al. (2018). Upstream kinases of plant SnRKs are involved in salt stress tolerance. Plant J. 93, 107–118. doi: 10.1111/tpj.13761
Barragan, V., Leidi, E. O., Andres, Z., Rubio, L., De Luca, A., Fernandez, J. A., et al. (2012). Ion exchangers NHX1 and NHX2 mediate active potassium uptake into vacuoles to regulate cell turgor and stomatal function in Arabidopsis. Plant Cell 24, 1127–1142. doi: 10.1105/tpc.111.095273
Bartoli, C. G., Casalongué, C. A., Simontacchi, M., Marquez-Garcia, B., Foyer, C. H. (2013). Interactions between hormone and redox signalling pathways in the control of growth and cross tolerance to stress. Environ. Exp. Bot. 94, 73–88. doi: 10.1016/j.envexpbot.2012.05.003
Bassil, E., Ohto, M. A., Esumi, T., Tajima, H., Zhu, Z., Cagnac, O., et al. (2011a). The Arabidopsis intracellular Na+/H+ antiporters NHX5 and NHX6 are endosome associated and necessary for plant growth and development. Plant Cell 23, 224–239. doi: 10.1105/tpc.110.079426
Bassil, E., Tajima, H., Liang, Y. C., Ohto, M. A., Ushijima, K., Nakano, R., et al. (2011b). The Arabidopsis Na+/H+ antiporters NHX1 and NHX2 control vacuolar pH and K+ homeostasis to regulate growth, flower development, and reproduction. Plant Cell 23, 3482–3497. doi: 10.1105/tpc.111.089581
Bechtold, U., Penfold, C. A., Jenkins, D. J., Legaie, R., Moore, J. D., Lawson, T., et al. (2016). Time-series transcriptomics reveals that AGAMOUS-LIKE22 affects primary metabolism and developmental processes in drought-stressed Arabidopsis. Plant Cell 28, 345–366. doi: 10.1105/tpc.15.00910
Berthomieu, P., Conejero, G., Nublat, A., Brackenbury, W. J., Lambert, C., Savio, C., et al. (2003). Functional analysis of AtHKT1 in Arabidopsis shows that Na+ recirculation by the phloem is crucial for salt tolerance. EMBO J. 22, 2004–2014. doi: 10.1093/emboj/cdg207
Brini, F., Masmoudi, K. (2012). Ion transporters and abiotic stress tolerance in plants. ISRN Mol. Biol. 2012, 927436. doi: 10.5402/2012/927436
Byrt, C. S., Munns, R., Burton, R. A., Gilliham, M., Wege, S. (2018). Root cell wall solutions for crop plants in saline soils. Plant Sci. 269, 47–55. doi: 10.1016/j.plantsci.2017.12.012
Byrt, C. S., Platten, J. D., Spielmeyer, W., James, R. A., Lagudah, E. S., Dennis, E. S., et al. (2007). HKT1;5-like cation transporters linked to Na+ exclusion loci in wheat, Nax2 and Kna1. Plant Physiol. 143, 1918–1928. doi: 10.1104/pp.106.093476
Byrt, C. S., Xu, B., Krishnan, M., Lightfoot, D. J., Athman, A., Jacobs, A. K., et al. (2014). The Na+ transporter, TaHKT1;5-D, limits shoot Na+ accumulation in bread wheat. Plant J. 80, 516–526. doi: 10.1111/tpj.12651
Campbell, M. T., Bandillo, N., Al Shiblawi, F. R. A., Sharma, S., Liu, K., Du, Q., et al. (2017). Allelic variants of OsHKT1;1 underlie the divergence between indica and japonica subspecies of rice (Oryza sativa) for root sodium content. PloS Genet. 13, e1006823. doi: 10.1371/journal.pgen.1006823
Cao, X., Du, Q., Guo, Y., Wang, Y., Jiao, Y. (2023). Condensation of STM is critical for shoot meristem maintenance and salt tolerance in Arabidopsis. Mol. Plant 16, 1445–1459. doi: 10.1016/j.molp.2023.09.005
Cao, Y., Liang, X., Yin, P., Zhang, M., Jiang, C. (2019). A domestication-associated reduction in K+ -preferring HKT transporter activity underlies maize shoot K+ accumulation and salt tolerance. New Phytol. 222, 301–317. doi: 10.1111/nph.15605
Chen, C., He, G., Li, J., Perez-Hormaeche, J., Becker, T., Luo, M., et al. (2023). A salt stress-activated GSO1-SOS2-SOS1 module protects the Arabidopsis root stem cell niche by enhancing sodium ion extrusion. EMBO J. 42, e113004. doi: 10.15252/embj.2022113004
Chen, K., Gao, J., Sun, S., Zhang, Z., Yu, B., Li, J., et al. (2020a). BONZAI proteins control global osmotic stress responses in plants. Curr. Biol. 30, 4815–4825.e4814. doi: 10.1016/j.cub.2020.09.016
Chen, K., Li, G. J., Bressan, R. A., Song, C. P., Zhu, J. K., Zhao, Y. (2020b). Abscisic acid dynamics, signaling, and functions in plants. J. Integr. Plant Biol. 62, 25–54. doi: 10.1111/jipb.12899
Chen, T., Cai, X., Wu, X., Karahara, I., Schreiber, L., Lin, J. (2011). Casparian strip development and its potential function in salt tolerance. Plant Signal Behav. 6, 1499–1502. doi: 10.4161/psb.6.10.17054
Christmann, A., Grill, E., Huang, J. (2013). Hydraulic signals in long-distance signaling. Curr. Opin. Plant Biol. 16, 293–300. doi: 10.1016/j.pbi.2013.02.011
Christmann, A., Hoffmann, T., Teplova, I., Grill, E., Muller, A. (2005). Generation of active pools of abscisic acid revealed by in vivo imaging of water-stressed Arabidopsis. Plant Physiol. 137, 209–219. doi: 10.1104/pp.104.053082
Chu, M., Chen, P., Meng, S., Xu, P., Lan, W. (2021). The Arabidopsis phosphatase PP2C49 negatively regulates salt tolerance through inhibition of AtHKT1;1. J. Integr. Plant Biol. 63, 528–542. doi: 10.1111/jipb.13008
Colmenero-Flores, J. M., Martinez, G., Gamba, G., Vazquez, N., Iglesias, D. J., Brumos, J., et al. (2007). Identification and functional characterization of cation-chloride cotransporters in plants. Plant J. 50, 278–292. doi: 10.1111/j.1365-313X.2007.03048.x
Cubero-Font, P., Maierhofer, T., Jaslan, J., Rosales, M. A., Espartero, J., Diaz-Rueda, P., et al. (2016). Silent S-type anion channel subunit SLAH1 gates SLAH3 open for chloride root-to-shoot translocation. Curr. Biol. 26, 2213–2220. doi: 10.1016/j.cub.2016.06.045
Das, A., Pal, S., Chakraborty, N., Hasanuzzaman, M., Adak, M. K. (2024). Regulation of reactive oxygen species metabolism and oxidative stress signaling by abscisic acid pretreatment in rice (Oryza sativa L.) seedlings through sub1A QTL under salinity. Plant Stress 11, 100422. doi: 10.1016/j.stress.2024.100422
Das, K., Roychoudhury, A. (2014). Reactive oxygen species (ROS) and response of antioxidants as ROS-scavengers during environmental stress in plants. Front. Environ. Sci. 2. doi: 10.3389/fenvs.2014.00053
De Angeli, A., Zhang, J., Meyer, S., Martinoia, E. (2013). AtALMT9 is a malate-activated vacuolar chloride channel required for stomatal opening in Arabidopsis. Nat. Commun. 4, 1804. doi: 10.1038/ncomms2815
Deng, P., Cao, C., Shi, X., Jiang, Q., Ge, J., Shen, L., et al. (2023). OsCYBDOMG1, a cytochrome b561 domain-containing protein, regulates salt tolerance and grain yield in rice. Theor. Appl. Genet. 136, 76. doi: 10.1007/s00122-023-04302-4
Do, T. D., Vuong, T. D., Dunn, D., Clubb, M., Valliyodan, B., Patil, G., et al. (2019). Identification of new loci for salt tolerance in soybean by high-resolution genome-wide association mapping. BMC Genomics 20, 318. doi: 10.1186/s12864-019-5662-9
Doczi, R., Hatzimasoura, E., Farahi Bilooei, S., Ahmad, Z., Ditengou, F. A., Lopez-Juez, E., et al. (2019). The MKK7-MPK6 MAP kinase module is a regulator of meristem quiescence or active growth in Arabidopsis. Front. Plant Sci. 10. doi: 10.3389/fpls.2019.00202
Dong, Q., Wallrad, L., Almutairi, B. O., Kudla, J. (2022). Ca2+ signaling in plant responses to abiotic stresses. J. Integr. Plant Biol. 64, 287–300. doi: 10.1111/jipb.13228
Drozdowicz, Y. M., Rea, P. A. (2001). Vacuolar H+ pyrophosphatases: from the evolutionary backwaters into the mainstream. Trends Plant Sci. 6, 206–211. doi: 10.1016/s1360-1385(01)01923-9
Du, L., Ding, L., Huang, X., Tang, D., Chen, B., Tian, H., et al. (2024). Natural variation in a K(+) -preferring HKT transporter contributes to wheat shoot K(+) accumulation and salt tolerance. Plant Cell Environ. 47, 540–556. doi: 10.1111/pce.14746
Du, F., Wang, Y., Wang, J., Li, Y., Zhang, Y., Zhao, X., et al. (2023). The basic helix-loop-helix transcription factor gene, OsbHLH38, plays a key role in controlling rice salt tolerance. J. Integr. Plant Biol. 65, 1859–1873. doi: 10.1111/jipb.13489
Dutta, D. (2023). Interplay between membrane proteins and membrane protein-lipid pertaining to plant salinity stress. Cell Biochem. Funct. 41, 399–412. doi: 10.1002/cbf.3798
El Mahi, H., Perez-Hormaeche, J., De Luca, A., Villalta, I., Espartero, J., Gamez-Arjona, F., et al. (2019). A critical role of sodium flux via the plasma membrane Na+/H+ exchanger SOS1 in the salt tolerance of rice. Plant Physiol. 180, 1046–1065. doi: 10.1104/pp.19.00324
Evans, M. J., Choi, W. G., Gilroy, S., Morris, R. J. (2016). A ROS-assisted calcium wave dependent on the AtRBOHD NADPH oxidase and TPC1 cation channel propagates the systemic response to salt stress. Plant Physiol. 171, 1771–1784. doi: 10.1104/pp.16.00215
Feki, K., Quintero, F. J., Pardo, J. M., Masmoudi, K. (2011). Regulation of durum wheat Na+/H+ exchanger TdSOS1 by phosphorylation. Plant Mol. Biol. 76, 545–556. doi: 10.1007/s11103-011-9787-8
Feng, W., Kita, D., Peaucelle, A., Cartwright, H. N., Doan, V., Duan, Q., et al. (2018). The FERONIA receptor kinase maintains cell-wall integrity during salt stress through Ca2+ signaling. Curr. Biol. 28, 666–675.e665. doi: 10.1016/j.cub.2018.01.023
Feng, X. J., Li, J. R., Qi, S. L., Lin, Q. F., Jin, J. B., Hua, X. J. (2016). Light affects salt stress-induced transcriptional memory of P5CS1 in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 113, E8335–E8343. doi: 10.1073/pnas.1610670114
Fernandez-Marcos, M., Sanz, L., Lewis, D. R., Muday, G. K., Lorenzo, O. (2011). Nitric oxide causes root apical meristem defects and growth inhibition while reducing PIN-FORMED 1 (PIN1)-dependent acropetal auxin transport. Proc. Natl. Acad. Sci. U.S.A. 108, 18506–18511. doi: 10.1073/pnas.1108644108
Fliegel, L. (2019). Structural and functional changes in the Na+/H+ exchanger isoform 1, induced by erk1/2 phosphorylation. Int. J. Mol. Sci. 20, 2378. doi: 10.3390/ijms20102378
Fu, H., Yu, X., Jiang, Y., Wang, Y., Yang, Y., Chen, S., et al. (2023). SALT OVERLY SENSITIVE 1 is inhibited by clade D Protein phosphatase 2C D6 and D7 in Arabidopsis thaliana. Plant Cell 35, 279–297. doi: 10.1093/plcell/koac283
Fujinami, R., Yamada, T., Imaichi, R. (2020). Root apical meristem diversity and the origin of roots: insights from extant lycophytes. J. Plant Res. 133, 291–296. doi: 10.1007/s10265-020-01167-2
Galvan-Ampudia, C. S., Julkowska, M. M., Darwish, E., Gandullo, J., Korver, R. A., Brunoud, G., et al. (2013). Halotropism is a response of plant roots to avoid a saline environment. Curr. Biol. 23, 2044–2050. doi: 10.1016/j.cub.2013.08.042
Gasulla, F., Barreno, E., Parages, M. L., Camara, J., Jimenez, C., Dormann, P., et al. (2016). The role of phospholipase D and MAPK signaling cascades in the adaption of lichen microalgae to desiccation: changes in membrane lipids and phosphoproteome. Plant Cell Physiol. 57, 1908–1920. doi: 10.1093/pcp/pcw111
Geiger, D., Scherzer, S., Mumm, P., Stange, A., Marten, I., Bauer, H., et al. (2009). Activity of guard cell anion channel SLAC1 is controlled by drought-stress signaling kinase-phosphatase pair. Proc. Natl. Acad. Sci. U.S.A. 106, 21425–21430. doi: 10.1073/pnas.0912021106
Ghorbel, M., Zaidi, I., Ebel, C., Brini, F., Hanin, M. (2023). The wheat Mitogen Activated Protein Kinase TMPK3 plays a positive role in salt and osmotic stress response. Acta Physiologiae Plantarum 45. doi: 10.1007/s11738-023-03548-1
Gierth, M., Maser, P., Schroeder, J. I. (2005). The potassium transporter AtHAK5 functions in K+ deprivation-induced high-affinity K+ uptake and AKT1 K+ channel contribution to K+ uptake kinetics in Arabidopsis roots. Plant Physiol. 137, 1105–1114. doi: 10.1104/pp.104.057216
Golani, Y., Kaye, Y., Gilhar, O., Ercetin, M., Gillaspy, G., Levine, A. (2013). Inositol polyphosphate phosphatidylinositol 5-phosphatase9 (At5ptase9) controls plant salt tolerance by regulating endocytosis. Mol. Plant 6, 1781–1794. doi: 10.1093/mp/sst072
Gong, Z., Xiong, L., Shi, H., Yang, S., Herrera-Estrella, L. R., Xu, G., et al. (2020). Plant abiotic stress response and nutrient use efficiency. Sci. China Life Sci. 63, 635–674. doi: 10.1007/s11427-020-1683-x
Gregis, V., Andres, F., Sessa, A., Guerra, R. F., Simonini, S., Mateos, J. L., et al. (2013). Identification of pathways directly regulated by SHORT VEGETATIVE PHASE during vegetative and reproductive development in Arabidopsis. Genome Biol. 14, R56. doi: 10.1186/gb-2013-14-6-r56
Guan, Q., Wu, J., Yue, X., Zhang, Y., Zhu, J. (2013). A nuclear calcium-sensing pathway is critical for gene regulation and salt stress tolerance in Arabidopsis. PloS Genet. 9, e1003755. doi: 10.1371/journal.pgen.1003755
Guo, Y., Halfter, U., Ishitani, M., Zhu, J. K. (2001). Molecular characterization of functional domains in the protein kinase SOS2 that is required for plant salt tolerance. Plant Cell 13, 1383–1400. doi: 10.1105/tpc.13.6.1383
Halfter, U., Ishitani, M., Zhu, J. K. (2000). The Arabidopsis SOS2 protein kinase physically interacts with and is activated by the calcium-binding protein SOS3. Proc. Natl. Acad. Sci. U.S.A. 97, 3735–3740. doi: 10.1073/pnas.97.7.3735
Hamaji, K., Nagira, M., Yoshida, K., Ohnishi, M., Oda, Y., Uemura, T., et al. (2009). Dynamic aspects of ion accumulation by vesicle traffic under salt stress in Arabidopsis. Plant Cell Physiol. 50, 2023–2033. doi: 10.1093/pcp/pcp143
Hamamoto, S., Horie, T., Hauser, F., Deinlein, U., Schroeder, J. I., Uozumi, N. (2015). HKT transporters mediate salt stress resistance in plants: from structure and function to the field. Curr. Opin. Biotechnol. 32, 113–120. doi: 10.1016/j.copbio.2014.11.025
Hamilton, E. S., Jensen, G. S., Maksaev, G., Katims, A., Sherp, A. M., Haswell, E. S. (2015). Mechanosensitive channel MSL8 regulates osmotic forces during pollen hydration and germination. Science 350, 438–441. doi: 10.1126/science.aac6014
Hao, R., Zhou, W., Li, J., Luo, M., Scheres, B., Guo, Y. (2023). On salt stress, PLETHORA signaling maintains root meristems. Dev. Cell 58, 1657–1669.e1655. doi: 10.1016/j.devcel.2023.06.012
Hata, Y., Kyozuka, J. (2021). Fundamental mechanisms of the stem cell regulation in land plants: lesson from shoot apical cells in bryophytes. Plant Mol. Biol. 107, 213–225. doi: 10.1007/s11103-021-01126-y
He, Q. Y., Jin, J. F., Lou, H. Q., Dang, F. F., Xu, J. M., Zheng, S. J., et al. (2022). Abscisic acid-dependent PMT1 expression regulates salt tolerance by alleviating abscisic acid-mediated reactive oxygen species production in Arabidopsis. J. Integr. Plant Biol. 64, 1803–1820. doi: 10.1111/jipb.13326
He, S. L., Li, B., Zahurancik, W. J., Arthur, H. C., Sidharthan, V., Gopalan, V., et al. (2024). Overexpression of stress granule protein TZF1 enhances salt stress tolerance by targeting ACA11 mRNA for degradation in Arabidopsis. Front. Plant Sci. 15. doi: 10.3389/fpls.2024.1375478
Henderson, S. W., Wege, S., Gilliham, M. (2018). Plant Cation-Chloride Cotransporters (CCC): evolutionary origins and functional insights. Int. J. Mol. Sci. 19, 492. doi: 10.3390/ijms19020492
Henry, C., Bledsoe, S. W., Griffiths, C. A., Kollman, A., Paul, M. J., Sakr, S., et al. (2015). Differential role for trehalose metabolism in salt-stressed maize. Plant Physiol. 169, 1072–1089. doi: 10.1104/pp.15.00729
Hickey, L. T., H., A. N., Robinson, H., Jackson, S. A., Leal-Bertioli, S. C. M., Tester, M., et al. (2019). Breeding crops to feed 10 billion. Nat. Biotechnol. 37, 744–754. doi: 10.1038/s41587-019-0152-9
Hong, Y., Guan, X., Wang, X., Kong, D., Yu, S., Wang, Z., et al. (2023). Natural variation in SlSOS2 promoter hinders salt resistance during tomato domestication. Hortic. Res. 10, uhac244. doi: 10.1093/hr/uhac244
Horie, T., Costa, A., Kim, T. H., Han, M. J., Horie, R., Leung, H. Y., et al. (2007). Rice OsHKT2;1 transporter mediates large Na+ influx component into K+-starved roots for growth. EMBO J. 26, 3003–3014. doi: 10.1038/sj.emboj.7601732
Hou, L., Liu, Z., Zhang, D., Liu, S., Chen, Z., Wu, Q., et al. (2024). BR regulates wheat root salt tolerance by maintaining ROS homeostasis. Planta 260, 5. doi: 10.1007/s00425-024-04429-8
Huang, S., Spielmeyer, W., Lagudah, E. S., James, R. A., Platten, J. D., Dennis, E. S., et al. (2006). A sodium transporter (HKT7) is a candidate for Nax1, a gene for salt tolerance in durum wheat. Plant Physiol. 142, 1718–1727. doi: 10.1104/pp.106.088864
Hwang, I., Sheen, J., Muller, B. (2012). Cytokinin signaling networks. Annu. Rev. Plant Biol. 63, 353–380. doi: 10.1146/annurev-arplant-042811-105503
Iglesias, M. J., Terrile, M. C., Bartoli, C. G., D’Ippolito, S., Casalongue, C. A. (2010). Auxin signaling participates in the adaptative response against oxidative stress and salinity by interacting with redox metabolism in Arabidopsis. Plant Mol. Biol. 74, 215–222. doi: 10.1007/s11103-010-9667-7
Iglesias, M. J., Terrile, M. C., Windels, D., Lombardo, M. C., Bartoli, C. G., Vazquez, F., et al. (2014). MiR393 regulation of auxin signaling and redox-related components during acclimation to salinity in Arabidopsis. PLoS One 9, e107678. doi: 10.1371/journal.pone.0107678
Isayenkov, S. V., Maathuis, F. J. M. (2019). Plant salinity stress: many unanswered questions remain. Front. Plant Sci. 10. doi: 10.3389/fpls.2019.00080
Ishitani, M., Liu, J., Halfter, U., Kim, C. S., Shi, W., Zhu, J. K. (2000). SOS3 function in plant salt tolerance requires N-myristoylation and calcium binding. Plant Cell 12, 1667–1678. doi: 10.1105/tpc.12.9.1667
Ismail, A. M., Horie, T. (2017). Genomics, physiology, and molecular breeding approaches for improving salt tolerance. Annu. Rev. Plant Biol. 68, 405–434. doi: 10.1146/annurev-arplant-042916-040936
James, R. A., Davenport, R. J., Munns, R. (2006). Physiological characterization of two genes for Na+ exclusion in durum wheat, Nax1 and Nax2. Plant Physiol. 142, 1537–1547. doi: 10.1104/pp.106.086538
Janocha, D., Lohmann, J. U. (2018). From signals to stem cells and back again. Curr. Opin. Plant Biol. 45, 136–142. doi: 10.1016/j.pbi.2018.06.005
Jiang, S., Lan, Z., Zhang, Y., Kang, X., Zhao, L., Wu, X., et al. (2024). Mechanisms by which exogenous substances enhance plant salt tolerance through the modulation of ion membrane transport and reactive oxygen species metabolism. Antioxidants (Basel) 13, 1050. doi: 10.3390/antiox13091050
Jiang, K., Moe-Lange, J., Hennet, L., Feldman, L. J. (2016). Salt stress affects the redox status of Arabidopsis root meristems. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.00081
Jiang, Z., Zhou, X., Tao, M., Yuan, F., Liu, L., Wu, F., et al. (2019). Plant cell-surface GIPC sphingolipids sense salt to trigger Ca2+ influx. Nature 572, 341–346. doi: 10.1038/s41586-019-1449-z
Jojoa-Cruz, S., Saotome, K., Murthy, S. E., Tsui, C. C. A., Sansom, M. S., Patapoutian, A., et al. (2018). Cryo-EM structure of the mechanically activated ion channel OSCA1.2. Elife 7, e41845. doi: 10.7554/eLife.41845
Jossier, M., Kroniewicz, L., Dalmas, F., Le Thiec, D., Ephritikhine, G., Thomine, S., et al. (2010). The Arabidopsis vacuolar anion transporter, AtCLCc, is involved in the regulation of stomatal movements and contributes to salt tolerance. Plant J. 64, 563–576. doi: 10.1111/j.1365-313X.2010.04352.x
Jun, Y. S., Cha, O.-K., Kim, J. H., Lee, H. (2019). Shoot meristem activity is involved in salt tolerance on Arabidopsis shoot growth. J. Plant Biol. 62, 410–418. doi: 10.1007/s12374-019-0348-z
Kenesi, E., Kolbert, Z., Kaszler, N., Klement, E., Menesi, D., Molnar, A., et al. (2023). The ROP2 GTPase participates in Nitric Oxide (NO)-induced root shortening in Arabidopsis. Plants (Basel) 12, 750. doi: 10.3390/plants12040750
Kim, W. Y., Ali, Z., Park, H. J., Park, S. J., Cha, J. Y., Perez-Hormaeche, J., et al. (2013). Release of SOS2 kinase from sequestration with GIGANTEA determines salt tolerance in Arabidopsis. Nat. Commun. 4, 1352. doi: 10.1038/ncomms2357
Kim, B. G., Waadt, R., Cheong, Y. H., Pandey, G. K., Dominguez-Solis, J. R., Schultke, S., et al. (2007). The calcium sensor CBL10 mediates salt tolerance by regulating ion homeostasis in Arabidopsis. Plant J. 52, 473–484. doi: 10.1111/j.1365-313X.2007.03249.x
Kim, S. H., Woo, D. H., Kim, J. M., Lee, S. Y., Chung, W. S., Moon, Y. H. (2011). Arabidopsis MKK4 mediates osmotic-stress response via its regulation of MPK3 activity. Biochem. Biophys. Res. Commun. 412, 150–154. doi: 10.1016/j.bbrc.2011.07.064
Kim, J. M., Woo, D. H., Kim, S. H., Lee, S. Y., Park, H. Y., Seok, H. Y., et al. (2012). Arabidopsis MKKK20 is involved in osmotic stress response via regulation of MPK6 activity. Plant Cell Rep. 31, 217–224. doi: 10.1007/s00299-011-1157-0
Kitagawa, M., Jackson, D. (2019). Control of meristem size. Annu. Rev. Plant Biol. 70, 269–291. doi: 10.1146/annurev-arplant-042817-040549
Klingler, J. P., Batelli, G., Zhu, J. K. (2010). ABA receptors: the START of a new paradigm in phytohormone signalling. J. Exp. Bot. 61, 3199–3210. doi: 10.1093/jxb/erq151
Knight, H., Trewavas, A. J., Knight, M. R. (1997). Calcium signalling in Arabidopsis thaliana responding to drought and salinity. Plant J. 12, 1067–1078. doi: 10.1046/j.1365-313x.1997.12051067.x
Kobayashi, N. I., Yamaji, N., Yamamoto, H., Okubo, K., Ueno, H., Costa, A., et al. (2017). OsHKT1;5 mediates Na+ exclusion in the vasculature to protect leaf blades and reproductive tissues from salt toxicity in rice. Plant J. 91, 657–670. doi: 10.1111/tpj.13595
Kopecka, R., Kameniarova, M., Cerny, M., Brzobohaty, B., Novak, J. (2023). Abiotic stress in crop production. Int. J. Mol. Sci. 24, 6603. doi: 10.3390/ijms24076603
Korver, R. A., Koevoets, I. T., Testerink, C. (2018). Out of shape during stress: A key role for auxin. Trends Plant Sci. 23, 783–793. doi: 10.1016/j.tplants.2018.05.011
Kudla, J., Becker, D., Grill, E., Hedrich, R., Hippler, M., Kummer, U., et al. (2018). Advances and current challenges in calcium signaling. New Phytol. 218, 414–431. doi: 10.1111/nph.14966
Kurusu, T., Nishikawa, D., Yamazaki, Y., Gotoh, M., Nakano, M., Hamada, H., et al. (2012a). Plasma membrane protein OsMCA1 is involved in regulation of hypo-osmotic shock-induced Ca2+ influx and modulates generation of reactive oxygen species in cultured rice cells. BMC Plant Biol. 12, 11. doi: 10.1186/1471-2229-12-11
Kurusu, T., Yamanaka, T., Nakano, M., Takiguchi, A., Ogasawara, Y., Hayashi, T., et al. (2012b). Involvement of the putative Ca2+-permeable mechanosensitive channels, NtMCA1 and NtMCA2, in Ca2+ uptake, Ca2+-dependent cell proliferation and mechanical stress-induced gene expression in tobacco (Nicotiana tabacum) BY-2 cells. J. Plant Res. 125, 555–568. doi: 10.1007/s10265-011-0462-6
Lagarde, D., Basset, M., Lepetit, M., Conejero, G., Gaymard, F., Astruc, S., et al. (1996). Tissue-specific expression of Arabidopsis AKT1 gene is consistent with a role in K+ nutrition. Plant J. 9, 195–203. doi: 10.1046/j.1365-313x.1996.09020195.x
Laohavisit, A., Richards, S. L., Shabala, L., Chen, C., Colaco, R. D., Swarbreck, S. M., et al. (2013). Salinity-induced calcium signaling and root adaptation in Arabidopsis require the calcium regulatory protein annexin1. Plant Physiol. 163, 253–262. doi: 10.1104/pp.113.217810
Lee, H. (2018). Stem cell maintenance and abiotic stress response in shoot apical meristem for developmental plasticity. J. Plant Biol. 61, 358–365. doi: 10.1007/s12374-018-0301-6
Lee, H., Jun, Y. S., Cha, O. K., Sheen, J. (2019). Mitogen-activated protein kinases MPK3 and MPK6 are required for stem cell maintenance in the Arabidopsis shoot apical meristem. Plant Cell Rep. 38, 311–319. doi: 10.1007/s00299-018-2367-5
Li, B., Byrt, C., Qiu, J., Baumann, U., Hrmova, M., Evrard, A., et al. (2016a). Identification of a stelar-localized transport protein that facilitates root-to-shoot transfer of chloride in Arabidopsis. Plant Physiol. 170, 1014–1029. doi: 10.1104/pp.15.01163
Li, W., de Ollas, C., Dodd, I. C. (2018). Long-distance ABA transport can mediate distal tissue responses by affecting local ABA concentrations. J. Integr. Plant Biol. 60, 16–33. doi: 10.1111/jipb.12605
Li, B., Qiu, J., Jayakannan, M., Xu, B., Li, Y., Mayo, G. M., et al. (2016b). AtNPF2.5 modulates chloride Cl- efflux from roots of Arabidopsis thaliana. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.02013
Li, J., Shen, L., Han, X., He, G., Fan, W., Li, Y., et al. (2023a). Phosphatidic acid-regulated SOS2 controls sodium and potassium homeostasis in Arabidopsis under salt stress. EMBO J. 42, e112401. doi: 10.15252/embj.2022112401
Li, B., Tester, M., Gilliham, M. (2017). Chloride on the move. Trends Plant Sci. 22, 236–248. doi: 10.1016/j.tplants.2016.12.004
Li, C. H., Wang, G., Zhao, J. L., Zhang, L. Q., Ai, L. F., Han, Y. F., et al. (2014). The Receptor-like kinase SIT1 mediates salt sensitivity by activating MAPK3/6 and regulating ethylene homeostasis in rice. Plant Cell 26, 2538–2553. doi: 10.1105/tpc.114.125187
Li, J., Zhou, X., Wang, Y., Song, S., Ma, L., He, Q., et al. (2023b). Inhibition of the maize salt overly sensitive pathway by ZmSK3 and ZmSK4. J. Genet. Genomics 50, 960–970. doi: 10.1016/j.jgg.2023.04.010
Liang, X., Li, J., Yang, Y., Jiang, C., Guo, Y. (2024). Designing salt stress-resilient crops: Current progress and future challenges. J. Integr. Plant Biol. 66, 303–329. doi: 10.1111/jipb.13599
Li, Q., Fu, H., Yu, X., Wen, X., Guo, H., Guo, Y., et al (2024). The SALT OVERLY SENSITIVE 2–CONSTITUTIVE TRIPLE RESPONSE1 module coordinates plant growth and salt tolerance in Arabidopsis. J. Exp. Bot. 75, 391–404.
Liang, X., Liu, S., Wang, T., Li, F., Cheng, J., Lai, J., et al. (2021). Metabolomics-driven gene mining and genetic improvement of tolerance to salt-induced osmotic stress in maize. New Phytol. 230, 2355–2370. doi: 10.1111/nph.17323
Lin, Z., Li, Y., Wang, Y., Liu, X., Ma, L., Zhang, Z., et al. (2021). Initiation and amplification of SnRK2 activation in abscisic acid signaling. Nat. Commun. 12, 2456. doi: 10.1038/s41467-021-22812-x
Lin, H., Yang, Y., Quan, R., Mendoza, I., Wu, Y., Du, W., et al. (2009). Phosphorylation of SOS3-LIKE CALCIUM BINDING PROTEIN8 by SOS2 protein kinase stabilizes their protein complex and regulates salt tolerance in Arabidopsis. Plant Cell 21, 1607–1619. doi: 10.1105/tpc.109.066217
Liu, J., Ishitani, M., Halfter, U., Kim, C. S., Zhu, J. K. (2000). The Arabidopsis thaliana SOS2 gene encodes a protein kinase that is required for salt tolerance. Proc. Natl. Acad. Sci. U.S.A. 97, 3730–3734. doi: 10.1073/pnas.97.7.3730
Liu, G., Jiang, W., Tian, L., Fu, Y., Tan, L., Zhu, Z., et al. (2022a). Polyamine oxidase 3 is involved in salt tolerance at the germination stage in rice. J. Genet. Genomics 49, 458–468. doi: 10.1016/j.jgg.2022.01.007
Liu, W., Li, R. J., Han, T. T., Cai, W., Fu, Z. W., Lu, Y. T. (2015). Salt stress reduces root meristem size by nitric oxide-mediated modulation of auxin accumulation and signaling in Arabidopsis. Plant Physiol. 168, 343–356. doi: 10.1104/pp.15.00030
Liu, Y., Li, M., Yu, J., Ma, A., Wang, J., Yun, D. J., et al. (2023). Plasma membrane-localized Hsp40/DNAJ chaperone protein facilitates OsSUVH7-OsBAG4-OsMYB106 transcriptional complex formation for OsHKT1;5 activation. J. Integr. Plant Biol. 65, 265–279. doi: 10.1111/jipb.13403
Liu, M., Pan, T., Allakhverdiev, S. I., Yu, M., Shabala, S. (2020). Crop halophytism: an environmentally sustainable solution for global food security. Trends Plant Sci. 25, 630–634. doi: 10.1016/j.tplants.2020.04.008
Liu, L., Song, W., Huang, S., Jiang, K., Moriwaki, Y., Wang, Y., et al. (2022b). Extracellular pH sensing by plant cell-surface peptide-receptor complexes. Cell 185, 3341–3355.e3313. doi: 10.1016/j.cell.2022.07.012
Liu, X., Wang, J., Sun, L. (2018). Structure of the hyperosmolality-gated calcium-permeable channel OSCA1.2. Nat. Commun. 9, 5060. doi: 10.1038/s41467-018-07564-5
Liu, Y., Xun, W., Chen, L., Xu, Z., Zhang, N., Feng, H., et al. (2022c). Rhizosphere microbes enhance plant salt tolerance: Toward crop production in saline soil. Comput. Struct. Biotechnol. J. 20, 6543–6551. doi: 10.1016/j.csbj.2022.11.046
Liu, S., Yang, R., Liu, M., Zhang, S., Yan, K., Yang, G., et al. (2020). PLATZ2 negatively regulates salt tolerance in Arabidopsis seedlings by directly suppressing the expression of the CBL4/SOS3 and CBL10/SCaBP8 genes. J. Exp. Bot. 71, 5589–5602. doi: 10.1093/jxb/eraa259
Liu, J., Zhu, J. K. (1998). A calcium sensor homolog required for plant salt tolerance. Science 280, 1943–1945. doi: 10.1126/science.280.5371.1943
Lou, L., Yu, F., Tian, M., Liu, G., Wu, Y., Wu, Y., et al. (2020). ESCRT-I component VPS23A sustains salt tolerance by strengthening the SOS module in Arabidopsis. Mol. Plant 13, 1134–1148. doi: 10.1016/j.molp.2020.05.010
Ma, M., Wendehenne, D., Philippot, L., Hansch, R., Flemetakis, E., Hu, B., et al. (2020). Physiological significance of pedospheric nitric oxide for root growth, development and organismic interactions. Plant Cell Environ. 43, 2336–2354. doi: 10.1111/pce.13850
Ma, L., Ye, J., Yang, Y., Lin, H., Yue, L., Luo, J., et al. (2019). The SOS2-SCaBP8 complex generates and fine-tunes an AtANN4-dependent calcium signature under salt stress. Dev. Cell 48, 697–709.e695. doi: 10.1016/j.devcel.2019.02.010
Maity, K., Heumann, J. M., McGrath, A. P., Kopcho, N. J., Hsu, P. K., Lee, C. W., et al. (2019). Cryo-EM structure of OSCA1.2 from Oryza sativa elucidates the mechanical basis of potential membrane hyperosmolality gating. Proc. Natl. Acad. Sci. U.S.A. 116, 14309–14318. doi: 10.1073/pnas.1900774116
Mani, B., Kaur, I., Dhingra, Y., Saxena, V., Krishna, G. K., Kumar, R., et al. (2024). Tetraspanin 5 orchestrates resilience to salt stress through the regulation of ion and reactive oxygen species homeostasis in rice. Plant Biotechnol. J. 23, 51–71. doi: 10.1111/pbi.14476
Mansour, M. M. F., Ali, E. F. (2017). Evaluation of proline functions in saline conditions. Phytochemistry 140, 52–68. doi: 10.1016/j.phytochem.2017.04.016
Markham, K. K., Greenham, K. (2021). Abiotic stress through time. New Phytol. 231, 40–46. doi: 10.1111/nph.17367
Martinez-Atienza, J., Jiang, X., Garciadeblas, B., Mendoza, I., Zhu, J. K., Pardo, J. M., et al. (2007). Conservation of the salt overly sensitive pathway in rice. Plant Physiol. 143, 1001–1012. doi: 10.1104/pp.106.092635
Mattioli, R., Falasca, G., Sabatini, S., Altamura, M. M., Costantino, P., Trovato, M. (2009). The proline biosynthetic genes P5CS1 and P5CS2 play overlapping roles in Arabidopsis flower transition but not in embryo development. Physiol. Plant 137, 72–85. doi: 10.1111/j.1399-3054.2009.01261.x
Mattioli, R., Francioso, A., Trovato, M. (2022). Proline affects flowering time in Arabidopsis by modulating FLC expression: A clue of epigenetic regulation? Plants (Basel) 11, 2348. doi: 10.3390/plants11182348
Mattioli, R., Marchese, D., D’Angeli, S., Altamura, M. M., Costantino, P., Trovato, M. (2008). Modulation of intracellular proline levels affects flowering time and inflorescence architecture in Arabidopsis. Plant Mol. Biol. 66, 277–288. doi: 10.1007/s11103-007-9269-1
Mazel, A., Leshem, Y., Tiwari, B. S., Levine, A. (2004). Induction of salt and osmotic stress tolerance by overexpression of an intracellular vesicle trafficking protein AtRab7 (AtRabG3e). Plant Physiol. 134, 118–128. doi: 10.1104/pp.103.025379
McKim, S. M. (2019). How plants grow up. J. Integr. Plant Biol. 61, 257–277. doi: 10.1111/jipb.12786
Meena, M., Divyanshu, K., Kumar, S., Swapnil, P., Zehra, A., Shukla, V., et al. (2019). Regulation of L-proline biosynthesis, signal transduction, transport, accumulation and its vital role in plants during variable environmental conditions. Heliyon 5, e02952. doi: 10.1016/j.heliyon.2019.e02952
Mian, A., Oomen, R. J., Isayenkov, S., Sentenac, H., Maathuis, F. J., Very, A. A. (2011). Over-expression of an Na+-and K+-permeable HKT transporter in barley improves salt tolerance. Plant J. 68, 468–479. doi: 10.1111/j.1365-313X.2011.04701.x
Miller, G., Suzuki, N., Ciftci-Yilmaz, S., Mittler, R. (2010). Reactive oxygen species homeostasis and signalling during drought and salinity stresses. Plant Cell Environ. 33, 453–467. doi: 10.1111/j.1365-3040.2009.02041.x
Min, M. K., Choi, E. H., Kim, J. A., Yoon, I. S., Han, S., Lee, Y., et al. (2019). Two clade A phosphatase 2Cs expressed in guard cells physically interact with abscisic acid signaling components to induce stomatal closure in rice. Rice (N Y) 12, 37. doi: 10.1186/s12284-019-0297-7
Mishra, G., Mohapatra, S. K., Rout, G. R. (2024). Plant membrane transporters function under abiotic stresses: a review. Planta 260, 125. doi: 10.1007/s00425-024-04548-2
Moller, I. S., Gilliham, M., Jha, D., Mayo, G. M., Roy, S. J., Coates, J. C., et al. (2009). Shoot Na+ exclusion and increased salinity tolerance engineered by cell type-specific alteration of Na+ transport in Arabidopsis. Plant Cell 21, 2163–2178. doi: 10.1105/tpc.108.064568
Motte, H., Vanneste, S., Beeckman, T. (2019). Molecular and environmental regulation of root development. Annu. Rev. Plant Biol. 70, 465–488. doi: 10.1146/annurev-arplant-050718-100423
Munns, R., James, R. A., Xu, B., Athman, A., Conn, S. J., Jordans, C., et al. (2012). Wheat grain yield on saline soils is improved by an ancestral Na+ transporter gene. Nat. Biotechnol. 30, 360–364. doi: 10.1038/nbt.2120
Munns, R., Passioura, J. B., Colmer, T. D., Byrt, C. S. (2020). Osmotic adjustment and energy limitations to plant growth in saline soil. New Phytol. 225, 1091–1096. doi: 10.1111/nph.15862
Munns, R., Tester, M. (2008). Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 59, 651–681. doi: 10.1146/annurev.arplant.59.032607.092911
Negi, J., Matsuda, O., Nagasawa, T., Oba, Y., Takahashi, H., Kawai-Yamada, M., et al. (2008). CO2 regulator SLAC1 and its homologues are essential for anion homeostasis in plant cells. Nature 452, 483–486. doi: 10.1038/nature06720
Nicolas, A., Laufs, P. (2022). Meristem initiation and de novo stem cell formation. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.891228
Nishiyama, R., Watanabe, Y., Fujita, Y., Le, D. T., Kojima, M., Werner, T., et al. (2011). Analysis of cytokinin mutants and regulation of cytokinin metabolic genes reveals important regulatory roles of cytokinins in drought, salt and abscisic acid responses, and abscisic acid biosynthesis. Plant Cell 23, 2169–2183. doi: 10.1105/tpc.111.087395
Noctor, G., Foyer, C. H. (1998). ASCORBATE AND GLUTATHIONE: Keeping active oxygen under control. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49, 249–279. doi: 10.1146/annurev.arplant.49.1.249
Oda, Y., Kobayashi, N. I., Tanoi, K., Ma, J. F., Itou, Y., Katsuhara, M., et al. (2018). T-DNA tagging-Based gain-of-function of OsHKT1;4 reinforces Na+ exclusion from leaves and stems but triggers Na+ toxicity in roots of rice under salt stress. Int. J. Mol. Sci. 19. doi: 10.3390/ijms19010235
Ohta, M., Guo, Y., Halfter, U., Zhu, J. K. (2003). A novel domain in the protein kinase SOS2 mediates interaction with the protein phosphatase 2C ABI2. Proc. Natl. Acad. Sci. U.S.A. 100, 11771–11776. doi: 10.1073/pnas.2034853100
Pabuayon, I. C. M., Jiang, J., Qian, H., Chung, J. S., Shi, H. (2021). Gain-of-function mutations of AtNHX1 suppress sos1 salt sensitivity and improve salt tolerance in Arabidopsis. Stress Biol. 1, 14. doi: 10.1007/s44154-021-00014-1
Pareek, A., Dhankher, O. P., Foyer, C. H. (2020a). Mitigating the impact of climate change on plant productivity and ecosystem sustainability. J. Exp. Bot. 71, 451–456. doi: 10.1093/jxb/erz518
Pareek, A., Joshi, R., Gupta, K. J., Singla-Pareek, S. L., Foyer, C. (2020b). Sensing and signalling in plant stress responses: ensuring sustainable food security in an era of climate change. New Phytol. 228, 823–827. doi: 10.1111/nph.16893
Pommerrenig, B., Papini-Terzi, F. S., Sauer, N. (2007). Differential regulation of sorbitol and sucrose loading into the phloem of Plantago major in response to salt stress. Plant Physiol. 144, 1029–1038. doi: 10.1104/pp.106.089151
Qin, R., Hu, Y., Chen, H., Du, Q., Yang, J., Li, W. X. (2023). MicroRNA408 negatively regulates salt tolerance by affecting secondary cell wall development in maize. Plant Physiol. 192, 1569–1583. doi: 10.1093/plphys/kiad135
Qiu, J., Chen, M., Lu, F., Chen, X., Cai, Z., Huang, T. (2024). Methionine Synthase 2 represses stem cell maintenance of Arabidopsis thaliana in response to salt stress. Plants (Basel) 13, 2224. doi: 10.3390/plants13162224
Qiu, Q. S., Guo, Y., Dietrich, M. A., Schumaker, K. S., Zhu, J. K. (2002). Regulation of SOS1, a plasma membrane Na+/H+ exchanger in Arabidopsis thaliana, by SOS2 and SOS3. Proc. Natl. Acad. Sci. U.S.A. 99, 8436–8441. doi: 10.1073/pnas.122224699
Quan, R., Lin, H., Mendoza, I., Zhang, Y., Cao, W., Yang, Y., et al. (2007). SCABP8/CBL10, a putative calcium sensor, interacts with the protein kinase SOS2 to protect Arabidopsis shoots from salt stress. Plant Cell 19, 1415–1431. doi: 10.1105/tpc.106.042291
Quintero, F. J., Martinez-Atienza, J., Villalta, I., Jiang, X., Kim, W. Y., Ali, Z., et al. (2011). Activation of the plasma membrane Na+/H+ antiporter Salt-Overly-Sensitive 1 (SOS1) by phosphorylation of an auto-inhibitory C-terminal domain. Proc. Natl. Acad. Sci. U.S.A. 108, 2611–2616. doi: 10.1073/pnas.1018921108
Quintero, F. J., Ohta, M., Shi, H., Zhu, J. K., Pardo, J. M. (2002). Reconstitution in yeast of the Arabidopsis SOS signaling pathway for Na+ homeostasis. Proc. Natl. Acad. Sci. U.S.A. 99, 9061–9066. doi: 10.1073/pnas.132092099
Ragel, P., Rodenas, R., Garcia-Martin, E., Andres, Z., Villalta, I., Nieves-Cordones, M., et al. (2015). The CBL-interacting protein kinase CIPK23 regulates HAK5-mediated high-affinity K+ uptake in Arabidopsis roots. Plant Physiol. 169, 2863–2873. doi: 10.1104/pp.15.01401
Rawat, N., Wungrampha, S., Singla-Pareek, S. L., Yu, M., Shabala, S., Pareek, A. (2022). Rewilding staple crops for the lost halophytism: Toward sustainability and profitability of agricultural production systems. Mol. Plant 15, 45–64. doi: 10.1016/j.molp.2021.12.003
Reguera, M., Bassil, E., Tajima, H., Wimmer, M., Chanoca, A., Otegui, M. S., et al. (2015). pH regulation by NHX-type antiporters is required for receptor-mediated protein trafficking to the vacuole in Arabidopsis. Plant Cell 27, 1200–1217. doi: 10.1105/tpc.114.135699
Ren, Z., Bai, F., Xu, J., Wang, L., Wang, X., Zhang, Q., et al. (2021). A chloride efflux transporter, BIG RICE GRAIN 1, is involved in mediating grain size and salt tolerance in rice. J. Integr. Plant Biol. 63, 2150–2163. doi: 10.1111/jipb.13178
Ren, Z. H., Gao, J. P., Li, L. G., Cai, X. L., Huang, W., Chao, D. Y., et al. (2005). A rice quantitative trait locus for salt tolerance encodes a sodium transporter. Nat. Genet. 37, 1141–1146. doi: 10.1038/ng1643
Sablowski, R. (2011). Plant stem cell niches: from signalling to execution. Curr. Opin. Plant Biol. 14, 4–9. doi: 10.1016/j.pbi.2010.08.001
Sang, Y. L., Cheng, Z. J., Zhang, X. S. (2018). iPSCs: A comparison between animals and plants. Trends Plant Sci. 23, 660–666. doi: 10.1016/j.tplants.2018.05.008
Scintu, D., Scacchi, E., Cazzaniga, F., Vinciarelli, F., De Vivo, M., Shtin, M., et al. (2023). microRNA165 and 166 modulate response of the Arabidopsis root apical meristem to salt stress. Commun. Biol. 6, 834. doi: 10.1038/s42003-023-05201-6
Shabala, S., Wu, H., Bose, J. (2015). Salt stress sensing and early signalling events in plant roots: Current knowledge and hypothesis. Plant Sci. 241, 109–119. doi: 10.1016/j.plantsci.2015.10.003
Shabala, L., Zhang, J., Pottosin, I., Bose, J., Zhu, M., Fuglsang, A. T., et al. (2016). Cell-type-specific H+-ATPase activity in root tissues enables K+ retention and mediates acclimation of Barley (Hordeum vulgare) to salinity stress. Plant Physiol. 172, 2445–2458. doi: 10.1104/pp.16.01347
Shi, H., Ishitani, M., Kim, C., Zhu, J. K. (2000). The Arabidopsis thaliana salt tolerance gene SOS1 encodes a putative Na+/H+ antiporter. Proc. Natl. Acad. Sci. U.S.A. 97, 6896–6901. doi: 10.1073/pnas.120170197
Shi, H., Lee, B. H., Wu, S. J., Zhu, J. K. (2003). Overexpression of a plasma membrane Na+/H+ antiporter gene improves salt tolerance in Arabidopsis thaliana. Nat. Biotechnol. 21, 81–85. doi: 10.1038/nbt766
Shi, H., Quintero, F. J., Pardo, J. M., Zhu, J. K. (2002). The putative plasma membrane Na+/H+ antiporter SOS1 controls long-distance Na+ transport in plants. Plant Cell 14, 465–477. doi: 10.1105/tpc.010371
Sirichandra, C., Gu, D., Hu, H. C., Davanture, M., Lee, S., Djaoui, M., et al. (2009). Phosphorylation of the Arabidopsis AtrbohF NADPH oxidase by OST1 protein kinase. FEBS Lett. 583, 2982–2986. doi: 10.1016/j.febslet.2009.08.033
Smolko, A., Bauer, N., Pavlovic, I., Pencik, A., Novak, O., Salopek-Sondi, B. (2021). Altered root growth, auxin metabolism and distribution in Arabidopsis thaliana exposed to salt and osmotic stress. Int. J. Mol. Sci. 22, 7793. doi: 10.3390/ijms22157993
Song, T., Shi, Y., Shen, L., Cao, C., Shen, Y., Jing, W., et al. (2021). An endoplasmic reticulum-localized cytochrome b(5) regulates high-affinity K+ transport in response to salt stress in rice. Proc. Natl. Acad. Sci. U.S.A. 118, e2114347118. doi: 10.1073/pnas.2114347118
Steinhorst, L., He, G., Moore, L. K., Schultke, S., Schmitz-Thom, I., Cao, Y., et al. (2022). A Ca2+-sensor switch for tolerance to elevated salt stress in Arabidopsis. Dev. Cell 57, 2081–2094.e2087. doi: 10.1016/j.devcel.2022.08.001
Stephan, A. B., Kunz, H. H., Yang, E., Schroeder, J. I. (2016). Rapid hyperosmotic-induced Ca2+ responses in Arabidopsis thaliana exhibit sensory potentiation and involvement of plastidial KEA transporters. Proc. Natl. Acad. Sci. U.S.A. 113, E5242–E5249. doi: 10.1073/pnas.1519555113
Sun, S. J., Qi, G. N., Gao, Q. F., Wang, H. Q., Yao, F. Y., Hussain, J., et al. (2016). Protein kinase OsSAPK8 functions as an essential activator of S-type anion channel OsSLAC1, which is nitrate-selective in rice. Planta 243, 489–500. doi: 10.1007/s00425-015-2418-x
Sun, H., Sun, X., Wang, H., Ma, X. (2020). Advances in salt tolerance molecular mechanism in tobacco plants. Hereditas 157, 5. doi: 10.1186/s41065-020-00118-0
Sun, F., Zhang, W., Hu, H., Li, B., Wang, Y., Zhao, Y., et al. (2008). Salt modulates gravity signaling pathway to regulate growth direction of primary roots in Arabidopsis. Plant Physiol. 146, 178–188. doi: 10.1104/pp.107.109413
Sun, Y., Zhao, J., Li, X., Li, Y. (2020). E2 conjugases UBC1 and UBC2 regulate MYB42-mediated SOS pathway in response to salt stress in Arabidopsis. New Phytol. 227, 455–472. doi: 10.1111/nph.16538
Swiezawska, B., Duszyn, M., Jaworski, K., Szmidt-Jaworska, A. (2018). Downstream targets of cyclic nucleotides in plants. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.01428
Szekely, G., Abraham, E., Cseplo, A., Rigo, G., Zsigmond, L., Csiszar, J., et al. (2008). Duplicated P5CS genes of Arabidopsis play distinct roles in stress regulation and developmental control of proline biosynthesis. Plant J. 53, 11–28. doi: 10.1111/j.1365-313X.2007.03318.x
Takahashi, F., Suzuki, T., Osakabe, Y., Betsuyaku, S., Kondo, Y., Dohmae, N., et al. (2018). A small peptide modulates stomatal control via abscisic acid in long-distance signalling. Nature 556, 235–238. doi: 10.1038/s41586-018-0009-2
Takahashi, Y., Zhang, J., Hsu, P. K., Ceciliato, P. H. O., Zhang, L., Dubeaux, G., et al. (2020). MAP3 Kinase-dependent SnRK2-kinase activation is required for abscisic acid signal transduction and rapid osmotic stress response. Nat. Commun. 11, 12. doi: 10.1038/s41467-019-13875-y
Tao, J., Wu, F., Wen, H., Liu, X., Luo, W., Gao, L., et al. (2023). RCD1 promotes salt stress tolerance in Arabidopsis by repressing ANAC017 activity. Int. J. Mol. Sci. 24, 9793. doi: 10.3390/ijms24129793
Teakle, N. L., Tyerman, S. D. (2010). Mechanisms of Cl- transport contributing to salt tolerance. Plant Cell Environ. 33, 566–589. doi: 10.1111/j.1365-3040.2009.02060.x
Tester, M., Davenport, R. (2003). Na+ tolerance and Na+ transport in higher plants. Ann. Bot. 91, 503–527. doi: 10.1093/aob/mcg058
Thalmann, M., Pazmino, D., Seung, D., Horrer, D., Nigro, A., Meier, T., et al. (2016). Regulation of leaf starch degradation by Abscisic Acid is important for osmotic stress tolerance in plants. Plant Cell 28, 1860–1878. doi: 10.1105/tpc.16.00143
Umeda, M., Ikeuchi, M., Ishikawa, M., Ito, T., Nishihama, R., Kyozuka, J., et al. (2021). Plant stem cell research is uncovering the secrets of longevity and persistent growth. Plant J. 106, 326–335. doi: 10.1111/tpj.15184
van Zelm, E., Zhang, Y., Testerink, C. (2020). Salt tolerance mechanisms of plants. Annu. Rev. Plant Biol. 71, 403–433. doi: 10.1146/annurev-arplant-050718-100005
Verslues, P. E., Sharma, S. (2010). Proline metabolism and its implications for plant-environment interaction. Arabidopsis Book 8, e0140. doi: 10.1199/tab.0140
Villalobos-Lopez, M. A., Arroyo-Becerra, A., Quintero-Jimenez, A., Iturriaga, G. (2022). Biotechnological advances to improve abiotic stress tolerance in crops. Int. J. Mol. Sci. 23, 12053. doi: 10.3390/ijms231912053
Wang, J., Ao, M., Ma, A., Yu, J., Guo, P., Huang, S., et al. (2024b). A mitochondrial localized chaperone regulator OsBAG6 functions in saline-alkaline stress tolerance in rice. Rice (N Y) 17, 10. doi: 10.1186/s12284-024-00686-z
Wang, Y., Cao, Y., Liang, X., Zhuang, J., Wang, X., Qin, F., et al. (2022c). A dirigent family protein confers variation of Casparian strip thickness and salt tolerance in maize. Nat. Commun. 13, 2222. doi: 10.1038/s41467-022-29809-0
Wang, R., Chen, P., Han, M., Wang, W., Hu, X., He, R., et al. (2024d). Calcineurin B-like protein ZmCBL8-1 promotes salt stress resistance in Arabidopsis. Planta 259, 49. doi: 10.1007/s00425-024-04330-4
Wang, M., Cheng, J., Wu, J., Chen, J., Liu, D., Wang, C., et al. (2024c). Variation in TaSPL6-D confers salinity tolerance in bread wheat by activating TaHKT1;5-D while preserving yield-related traits. Nat. Genet. 56, 1257–1269. doi: 10.1038/s41588-024-01762-2
Wang, X., Devaiah, S. P., Zhang, W., Welti, R. (2006). Signaling functions of phosphatidic acid. Prog. Lipid Res. 45, 250–278. doi: 10.1016/j.plipres.2006.01.005
Wang, C. F., Han, G. L., Yang, Z. R., Li, Y. X., Wang, B. S. (2022b). Plant salinity sensors: current understanding and future directions. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.859224
Wang, Z., Hong, Y., Li, Y., Shi, H., Yao, J., Liu, X., et al. (2021). Natural variations in SlSOS1 contribute to the loss of salt tolerance during tomato domestication. Plant Biotechnol. J. 19, 20–22. doi: 10.1111/pbi.13443
Wang, P., Hsu, C. C., Du, Y., Zhu, P., Zhao, C., Fu, X., et al. (2020c). Mapping proteome-wide targets of protein kinases in plant stress responses. Proc. Natl. Acad. Sci. U.S.A. 117, 3270–3280. doi: 10.1073/pnas.1919901117
Wang, J., Nan, N., Li, N., Liu, Y., Wang, T. J., Hwang, I., et al. (2020a). A DNA methylation reader-chaperone regulator-transcription factor complex activates OsHKT1;5 expression during salinity stress. Plant Cell 32, 3535–3558. doi: 10.1105/tpc.20.00301
Wang, J., Su, Y., Kong, X., Ding, Z., Zhang, X. S. (2020b). Initiation and maintenance of plant stem cells in root and shoot apical meristems. aBIOTECH 1, 194–204. doi: 10.1007/s42994-020-00020-3
Wang, W., Wang, W., Wu, Y., Li, Q., Zhang, G., Shi, R., et al. (2020d). The involvement of wheat U-box E3 ubiquitin ligase TaPUB1 in salt stress tolerance. J. Integr. Plant Biol. 62, 631–651. doi: 10.1111/jipb.12842
Wang, C., Wen, J., Liu, Y., Yu, B., Yang, S. (2024a). SOS2-AFP2 module regulates seed germination by inducing ABI5 degradation in response to salt stress in Arabidopsis. Biochem. Biophys. Res. Commun. 723, 150190. doi: 10.1016/j.bbrc.2024.150190
Wang, B., Zhang, H., Huai, J., Peng, F., Wu, J., Lin, R., et al. (2022a). Condensation of SEUSS promotes hyperosmotic stress tolerance in Arabidopsis. Nat. Chem. Biol. 18, 1361–1369. doi: 10.1038/s41589-022-01196-z
Weinl, S., Kudla, J. (2009). The CBL-CIPK Ca2+-decoding signaling network: function and perspectives. New Phytol. 184, 517–528. doi: 10.1111/j.1469-8137.2009.02938.x
Wu, Q., Wang, M., Shen, J., Chen, D., Zheng, Y., Zhang, W. (2019). ZmOST1 mediates abscisic acid regulation of guard cell ion channels and drought stress responses. J. Integr. Plant Biol. 61, 478–491. doi: 10.1111/jipb.12714
Wu, X., Xu, J., Meng, X., Fang, X., Xia, M., Zhang, J., et al. (2022). Linker histone variant HIS1-3 and WRKY1 oppositely regulate salt stress tolerance in Arabidopsis. Plant Physiol. 189, 1833–1847. doi: 10.1093/plphys/kiac174
Xiao, S., Song, W., Xing, J., Su, A., Zhao, Y., Li, C., et al. (2023). ORF355 confers enhanced salinity stress adaptability to S-type cytoplasmic male sterility maize by modulating the mitochondrial metabolic homeostasis. J. Integr. Plant Biol. 65, 656–673. doi: 10.1111/jipb.13382
Xiao, G., Zhao, M., Liu, Q., Zhou, J., Cheng, Z., Wang, Q., et al. (2023). TaBAS1 encoding a typical 2-Cys peroxiredoxin enhances salt tolerance in wheat. Front. Plant Sci. 14. doi: 10.3389/fpls.2023.1152375
Xie, Q., Yin, X., Wang, Y., Qi, Y., Pan, C., Sulaymanov, S., et al. (2024). The signalling pathways, calcineurin B-like protein 5 (CBL5)-CBL-interacting protein kinase 8 (CIPK8)/CIPK24-salt overly sensitive 1 (SOS1), transduce salt signals in seed germination in Arabidopsis. Plant Cell Environ. 47, 1486–1502. doi: 10.1111/pce.14820
Xing, L., Zhu, M., Luan, M., Zhang, M., Jin, L., Liu, Y., et al. (2022). miR169q and NUCLEAR FACTOR YA8 enhance salt tolerance by activating PEROXIDASE1 expression in response to ROS. Plant Physiol. 188, 608–623. doi: 10.1093/plphys/kiab498
Xiong, X. X., Liu, Y., Zhang, L. L., Li, X. J., Zhao, Y., Zheng, Y., et al. (2023). G-protein beta-subunit gene TaGB1-B enhances drought and salt resistance in wheat. Int. J. Mol. Sci. 24, 7337. doi: 10.3390/ijms24087337
Xue, Z., Liu, L., Zhang, C. (2020). Regulation of shoot apical meristem and axillary meristem development in plants. Int. J. Mol. Sci. 21, 2917. doi: 10.3390/ijms21082917
Yamaguchi, T., Aharon, G. S., Sottosanto, J. B., Blumwald, E. (2005). Vacuolar Na+/H+ antiporter cation selectivity is regulated by calmodulin from within the vacuole in a Ca2+- and pH-dependent manner. Proc. Natl. Acad. Sci. U.S.A. 102, 16107–16112. doi: 10.1073/pnas.0504437102
Yamaguchi, T., Apse, M. P., Shi, H., Blumwald, E. (2003). Topological analysis of a plant vacuolar Na+/H+ antiporter reveals a luminal C terminus that regulates antiporter cation selectivity. Proc. Natl. Acad. Sci. U.S.A. 100, 12510–12515. doi: 10.1073/pnas.2034966100
Yan, X., Han, M., Li, S., Liang, Z., Ouyang, J., Wang, X., et al. (2024). A member of NF-Y family, OsNF-YC5 negatively regulates salt tolerance in rice. Gene 892, 147869. doi: 10.1016/j.gene.2023.147869
Yan, Z., Li, K., Li, Y., Wang, W., Leng, B., Yao, G., et al. (2023). The ZmbHLH32-ZmIAA9-ZmARF1 module regulates salt tolerance in maize. Int. J. Biol. Macromol 253, 126978. doi: 10.1016/j.ijbiomac.2023.126978
Yan, J., Liu, Y., Yan, J., Liu, Z., Lou, H., Wu, J. (2023). The salt-activated CBF1/CBF2/CBF3-GALS1 module fine-tunes galactan-induced salt hypersensitivity in Arabidopsis. J. Integr. Plant Biol. 65, 1904–1917. doi: 10.1111/jipb.13501
Yan, J., Liu, Y., Yang, L., He, H., Huang, Y., Fang, L., et al. (2021). Cell wall beta-1,4-galactan regulated by the BPC1/BPC2-GALS1 module aggravates salt sensitivity in Arabidopsis thaliana. Mol. Plant 14, 411–425. doi: 10.1016/j.molp.2020.11.023
Yan, Z., Wang, J., Wang, F., Xie, C., Lv, B., Yu, Z., et al. (2021). MPK3/6-induced degradation of ARR1/10/12 promotes salt tolerance in Arabidopsis. EMBO Rep. 22, e52457. doi: 10.15252/embr.202152457
Yang, Z., Cao, Y., Shi, Y., Qin, F., Jiang, C., Yang, S. (2023). Genetic and molecular exploration of maize environmental stress resilience: Toward sustainable agriculture. Mol. Plant 16, 1496–1517. doi: 10.1016/j.molp.2023.07.005
Yang, Y., Guo, Y. (2018a). Elucidating the molecular mechanisms mediating plant salt-stress responses. New Phytol. 217, 523–539. doi: 10.1111/nph.14920
Yang, Y., Guo, Y. (2018b). Unraveling salt stress signaling in plants. J. Integr. Plant Biol. 60, 796–804. doi: 10.1111/jipb.12689
Yang, S., Lee, H. (2023). Salinity-triggered responses in plant apical meristems for developmental plasticity. Int. J. Mol. Sci. 24, 6647. doi: 10.3390/ijms24076647
Yang, Z., Wang, C., Xue, Y., Liu, X., Chen, S., Song, C., et al. (2019c). Calcium-activated 14-3-3 proteins as a molecular switch in salt stress tolerance. Nat. Commun. 10, 1199. doi: 10.1038/s41467-019-09181-2
Yang, Z., Yang, R., Bai, W., Chen, W., Kong, X., Zhou, Y., et al. (2024). Q negatively regulates wheat salt tolerance through directly repressing the expression of TaSOS1 and reactive oxygen species scavenging genes. Plant J. 119, 478–489. doi: 10.1111/tpj.16777
Yang, Y., Zhang, C., Tang, R. J., Xu, H. X., Lan, W. Z., Zhao, F., et al. (2019b). Calcineurin B-Like proteins CBL4 and CBL10 mediate two independent salt tolerance pathways in Arabidopsis. Int. J. Mol. Sci. 20, 2421. doi: 10.3390/ijms20102421
Yin, P., Liang, X., Zhao, H., Xu, Z., Chen, L., Yang, X., et al. (2023). Cytokinin signaling promotes salt tolerance by modulating shoot chloride exclusion in maize. Mol. Plant 16, 1031–1047. doi: 10.1016/j.molp.2023.04.011
Yin, X., Xia, Y., Xie, Q., Cao, Y., Wang, Z., Hao, G., et al. (2020). The protein kinase complex CBL10-CIPK8-SOS1 functions in Arabidopsis to regulate salt tolerance. J. Exp. Bot. 71, 1801–1814. doi: 10.1093/jxb/erz549
Yu, Z., Duan, X., Luo, L., Dai, S., Ding, Z., Xia, G. (2020). How plant hormones mediate salt stress responses. Trends Plant Sci. 25, 1117–1130. doi: 10.1016/j.tplants.2020.06.008
Yu, L., Nie, J., Cao, C., Jin, Y., Yan, M., Wang, F., et al. (2010). Phosphatidic acid mediates salt stress response by regulation of MPK6 in Arabidopsis thaliana. New Phytol. 188, 762–773. doi: 10.1111/j.1469-8137.2010.03422.x
Yu, Y., Wu, Y., He, L. (2023). A wheat WRKY transcription factor TaWRKY17 enhances tolerance to salt stress in transgenic Arabidopsis and wheat plant. Plant Mol. Biol. 113, 171–191. doi: 10.1007/s11103-023-01381-1
Yu, J., Zhu, C., Xuan, W., An, H., Tian, Y., Wang, B., et al. (2023). Genome-wide association studies identify OsWRKY53 as a key regulator of salt tolerance in rice. Nat. Commun. 14, 3550. doi: 10.1038/s41467-023-39167-0
Yuan, H., Cheng, M., Wang, R., Wang, Z., Fan, F., Wang, W., et al. (2024). miR396b/GRF6 module contributes to salt tolerance in rice. Plant Biotechnol. J. 22, 2079–2092. doi: 10.1111/pbi.14326
Yuan, F., Yang, H., Xue, Y., Kong, D., Ye, R., Li, C., et al. (2014). OSCA1 mediates osmotic-stress-evoked Ca2+ increases vital for osmosensing in Arabidopsis. Nature 514, 367–371. doi: 10.1038/nature13593
Zarreen, F., Karim, M. J., Chakraborty, S. (2022). The diverse roles of histone 2B monoubiquitination in the life of plants. J. Exp. Bot. 73, 3854–3865. doi: 10.1093/jxb/erac120
Zeng, J., Li, X., Ge, Q., Dong, Z., Luo, L., Tian, Z., et al. (2021). Endogenous stress-related signal directs shoot stem cell fate in Arabidopsis thaliana. Nat. Plants 7, 1276–1287. doi: 10.1038/s41477-021-00985-z
Zhang, H. X., Blumwald, E. (2001). Transgenic salt-tolerant tomato plants accumulate salt in foliage but not in fruit. Nat. Biotechnol. 19, 765–768. doi: 10.1038/90824
Zhang, M., Cao, Y., Wang, Z., Wang, Z. Q., Shi, J., Liang, X., et al. (2018). A retrotransposon in an HKT1 family sodium transporter causes variation of leaf Na+ exclusion and salt tolerance in maize. New Phytol. 217, 1161–1176. doi: 10.1111/nph.14882
Zhang, M., Cao, J., Zhang, T., Xu, T., Yang, L., Li, X., et al. (2022b). A putative plasma membrane Na+/H+ antiporter GmSOS1 is critical for salt stress tolerance in glycine max. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.870695
Zhang, H. X., Hodson, J. N., Williams, J. P., Blumwald, E. (2001). Engineering salt-tolerant Brassica plants: characterization of yield and seed oil quality in transgenic plants with increased vacuolar sodium accumulation. Proc. Natl. Acad. Sci. U.S.A. 98, 12832–12836. doi: 10.1073/pnas.231476498
Zhang, D., Jiang, S., Pan, J., Kong, X., Zhou, Y., Liu, Y., et al. (2014). The overexpression of a maize mitogen-activated protein kinase gene (ZmMPK5) confers salt stress tolerance and induces defence responses in tobacco. Plant Biol. (Stuttg) 16, 558–570. doi: 10.1111/plb.12084
Zhang, F., Li, L., Jiao, Z., Chen, Y., Liu, H., Chen, X., et al. (2016a). Characterization of the calcineurin B-Like (CBL) gene family in maize and functional analysis of ZmCBL9 under abscisic acid and abiotic stress treatments. Plant Sci. 253, 118–129. doi: 10.1016/j.plantsci.2016.09.011
Zhang, M., Li, Y., Liang, X., Lu, M., Lai, J., Song, W., et al. (2023b). A teosinte-derived allele of an HKT1 family sodium transporter improves salt tolerance in maize. Plant Biotechnol. J. 21, 97–108. doi: 10.1111/pbi.13927
Zhang, M., Liang, X., Wang, L., Cao, Y., Song, W., Shi, J., et al. (2019). A HAK family Na+ transporter confers natural variation of salt tolerance in maize. Nat. Plants 5, 1297–1308. doi: 10.1038/s41477-019-0565-y
Zhang, H., Mu, Y., Zhang, H., Yu, C. (2023a). Maintenance of stem cell activity in plant development and stress responses. Front. Plant Sci. 14. doi: 10.3389/fpls.2023.1302046
Zhang, J. L., Shi, H. (2013). Physiological and molecular mechanisms of plant salt tolerance. Photosynth Res. 115, 1–22. doi: 10.1007/s11120-013-9813-6
Zhang, S. S., Sun, L., Dong, X., Lu, S. J., Tian, W., Liu, J. X. (2016b). Cellulose synthesis genes CESA6 and CSI1 are important for salt stress tolerance in Arabidopsis. J. Integr. Plant Biol. 58, 623–626. doi: 10.1111/jipb.12442
Zhang, L., Sun, X., Li, Y., Luo, X., Song, S., Chen, Y., et al. (2021). Rice Na+-permeable transporter OsHAK12 mediates shoots Na+ exclusion in response to salt stress. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.771746
Zhang, Y., Tan, J., Guo, Z., Lu, S., He, S., Shu, W., et al. (2009). Increased abscisic acid levels in transgenic tobacco over-expressing 9 cis-epoxycarotenoid dioxygenase influence H2O2 and NO production and antioxidant defences. Plant Cell Environ. 32, 509–519. doi: 10.1111/j.1365-3040.2009.01945.x
Zhang, X. Y., Tang, L. H., Nie, J. W., Zhang, C. R., Han, X., Li, Q. Y., et al. (2023c). Structure and activation mechanism of the rice Salt Overly Sensitive 1 (SOS1) Na+/H+ antiporter. Nat. Plants 9, 1924–1936. doi: 10.1038/s41477-023-01551-5
Zhang, Z., Wang, J., Zhang, R., Huang, R. (2012). The ethylene response factor AtERF98 enhances tolerance to salt through the transcriptional activation of ascorbic acid synthesis in Arabidopsis. Plant J. 71, 273–287. doi: 10.1111/j.1365-313X.2012.04996.x
Zhang, G., Zhang, M., Zhao, Z., Ren, Y., Li, Q., Wang, W. (2017). Wheat TaPUB1 modulates plant drought stress resistance by improving antioxidant capability. Sci. Rep. 7, 7549. doi: 10.1038/s41598-017-08181-w
Zhang, Y., Zhou, J., Ni, X., Wang, Q., Jia, Y., Xu, X., et al. (2023d). Structural basis for the activity regulation of Salt Overly Sensitive 1 in Arabidopsis salt tolerance. Nat. Plants 9, 1915–1923. doi: 10.1038/s41477-023-01550-6
Zhang, H., Zhu, J., Gong, Z., Zhu, J. K. (2022a). Abiotic stress responses in plants. Nat. Rev. Genet. 23, 104–119. doi: 10.1038/s41576-021-00413-0
Zhao, C., Jiang, W., Zayed, O., Liu, X., Tang, K., Nie, W., et al. (2021). The LRXs-RALFs-FER module controls plant growth and salt stress responses by modulating multiple plant hormones. Natl. Sci. Rev. 8, nwaa149. doi: 10.1093/nsr/nwaa149
Zhao, C., Zayed, O., Yu, Z., Jiang, W., Zhu, P., Hsu, C. C., et al. (2018). Leucine-rich repeat extensin proteins regulate plant salt tolerance in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 115, 13123–13128. doi: 10.1073/pnas.1816991115
Zhao, C., Zhang, H., Song, C., Zhu, J. K., Shabala, S. (2020). Mechanisms of plant responses and adaptation to soil salinity. Innovation (Camb) 1, 100017. doi: 10.1016/j.xinn.2020.100017
Zhou, X., Li, J., Wang, Y., Liang, X., Zhang, M., Lu, M., et al. (2022b). The classical SOS pathway confers natural variation of salt tolerance in maize. New Phytol. 236, 479–494. doi: 10.1111/nph.18278
Zhou, H., Lin, H., Chen, S., Becker, K., Yang, Y., Zhao, J., et al. (2014). Inhibition of the Arabidopsis salt overly sensitive pathway by 14-3-3 proteins. Plant Cell 26, 1166–1182. doi: 10.1105/tpc.113.117069
Zhou, Y. B., Liu, C., Tang, D. Y., Yan, L., Wang, D., Yang, Y. Z., et al. (2018). The receptor-like cytoplasmic kinase STRK1 phosphorylates and activates CatC, thereby regulating H2O2 homeostasis and improving salt tolerance in rice. Plant Cell 30, 1100–1118. doi: 10.1105/tpc.17.01000
Zhou, H., Xiao, F., Zheng, Y., Liu, G., Zhuang, Y., Wang, Z., et al. (2022a). PAMP-INDUCED SECRETED PEPTIDE 3 modulates salt tolerance through RECEPTOR-LIKE KINASE 7 in plants. Plant Cell 34, 927–944. doi: 10.1093/plcell/koab292
Zhu, J. K. (2001). Cell signaling under salt, water and cold stresses. Curr. Opin. Plant Biol. 4, 401–406. doi: 10.1016/s1369-5266(00)00192-8
Keywords: salt stress, molecular design breeding, crop production, Na+, molecular mechanisms
Citation: Zhang H, Yu C, Zhang Q, Qiu Z, Zhang X, Hou Y and Zang J (2025) Salinity survival: molecular mechanisms and adaptive strategies in plants. Front. Plant Sci. 16:1527952. doi: 10.3389/fpls.2025.1527952
Received: 14 November 2024; Accepted: 28 January 2025;
Published: 28 February 2025.
Edited by:
Marko S. Sabovljevic, University of Belgrade, SerbiaReviewed by:
Malay Kumar Adak, University of Kalyani, IndiaCopyright © 2025 Zhang, Yu, Zhang, Qiu, Zhang, Hou and Zang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jie Zang, jie.zang@pku-iaas.edu.cn; Yifeng Hou, yifeng.hou@pku-iaas.edu.cn
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.